Abstract
Biomedical ontologies provide formalized information and knowledge in the biomedical domain. Over the years, biomedical ontologies have played an important role in facilitating biomedical research and applications. Common quality issues of biomedical ontologies include inconsistent naming of concepts, redundant concepts, redundant relations, incomplete/incorrect concept definitions, and incomplete/incorrect class hierarchies. In this work, we focus on addressing the incompleteness of the class hierarchy in SNOMED CT. We develop a substring replacement approach, leveraging concepts’ lexical features and existing IS-A relations to identify potential missing IS-A relations in SNOMED CT. To evaluate the effectiveness of our approach, we performed both automated and manual validation. For the automated evaluation, we leverage relations from external terminologies in the Unified Medical Language System (UMLS) to validate the identified missing IS-A relations. For the manual validation, a randomly selected 100 samples from the results are reviewed by a domain expert. Applying our approach to the March 2022 release of SNOMED CT US Edition, we identified 3,228 potential missing IS-A relations, among which 63 were validated through the UMLS. The evaluation by the domain expert revealed that 89 out of 100 (a precision of 89%) missing IS-A relations are valid cases, showing the effectiveness of this substring replacement approach to facilitate the quality assurance of IS-A relations in SNOMED CT.
Keywords: Ontologies and Terminologies, Ontology Quality Assurance, SNOMED CT, UMLS
I. Introduction
Over the past few decades, an enormous amount of unstructured text has been generated clinically and in research in biomedicine, such as medical reports, physician notes, and scientific papers. Such continuously growing textual content needs to be organized, curated, and managed so as to obtain effective use for clinical and research purposes [1]. This presents unique challenges due to data heterogeneity, ambiguity, complexity, and size.
Biomedical ontologies address such challenges by serving as conceptual frameworks that model concepts in the biomedical domains in a manner that is understandable to both humans and machines [2]. Over the years, biomedical ontologies have played a vital role in facilitating biomedical research and applications. They can facilitate data sharing, data integration, information retrieval, natural language processing, and decision support [3]. For example, biomedical ontologies provide standards for encoding diagnoses and problem lists in electronic health records (EHRs) [4] as well as physician billing and insurance claims [5]. Moreover, biomedical ontologies are used in systems biology and systems medicine to generate knowledge-based representations of simulation models [2].
Biomedical ontologies are rapidly evolving as knowledge in the biomedical domain is constantly growing [6]. Although curators of biomedical ontologies seek ways to assure they are accurate and comprehensive as possible, quality issues inevitably exist [7], which may lead to ambiguity, complexity, and inaccuracy in down-stream ontology-based biomedical applications. There are various types of common quality issues (e.g., inconsistent naming of concepts, concept redundancy, relation redundancy, incomplete/incorrect concept definitions, incomplete/incorrect class hierarchies) [8]. Proper quality assurance (QA) techniques need to be developed to address such issues. QA focuses on identifying modeling errors and inconsistencies in ontologies to improve their quality [7]. In this work, we focus on the issue of incompleteness in the class hierarchy of SNOMED CT. Hierarchical or IS-A relations form the backbone structure of SNOMED CT and missing IS-A relations may have a significant impact on downstream tasks (such as patient cohort identification). We propose a substring replacement approach, leveraging concepts’ lexical features and existing IS-A relations to identify potential missing IS-A relations in SNOMED CT.
The remainder of this paper is arranged as follows. Section II presents some background information on SNOMED CT and UMLS, as well as related work on identifying missing relations in biomedical ontologies. In Section III, we introduce our substring replacement approach for identifying missing IS-A relations in SNOMED CT. Section IV reports the results we obtained. The contributions and limitations of our work, as well as future work are discussed in Section V. Section VI concludes this paper.
II. Background
A. SNOMED CT
SNOMED CT is a comprehensive, multilingual clinical health terminology that supports the development of high-quality electronic health records [9]. It provides a common terminology that supports effective communication between different specialties and sites of care. SNOMED CT is essential for indexing, storing, retrieving, and aggregating clinical data [10]. The United States (US) Edition of SNOMED CT is the official source for use in US healthcare systems. It includes content from both the US Extension and the International releases of SNOMED CT [11].
The logical model shown in Fig. 1 demonstrates the core components of SNOMED CT: concepts, descriptions, and relationships [12]. A concept represents a certain clinical meaning with a unique numerical identifier [13]. Descriptions are human readable terms that are used to refer to these concepts. Two types of description are used to represent every concept: fully specified name (FSN) and synonym. Each concept has a FSN that represents the meaning of the concept in a unique and unambiguous way [14]. FSNs contain a semantic tag (parenthesized towards the end of the FSN) indicating the domain of the concept. A synonym is an acceptable way to express the meaning of a concept in a certain language or dialect [15]. The synonym that is considered as the most clinically suitable way to express a concept in a clinical record is marked as “preferred term” [16]. Relationships reflect associations between concepts and are used to logically define the meaning of a concept in such a way that can be processed by a computer. There are two types of relationships available within SNOMED CT [12]: IS-A relationship and attribute relationship (e.g., Finding site, Associated morphology). Each concept has at least one IS-A relationship and can have as many attribute relationships as needed. SNOMED CT releases both stated and inferred logical definitions for all concepts. Stated logical definitions include only assertions made by SNOMED CT authors. Inferred logical definitions are logically derived by applying a description logic classifier on the stated logical definitions [17]. We use inferred logical definitions of concepts in this work.
Fig. 1.
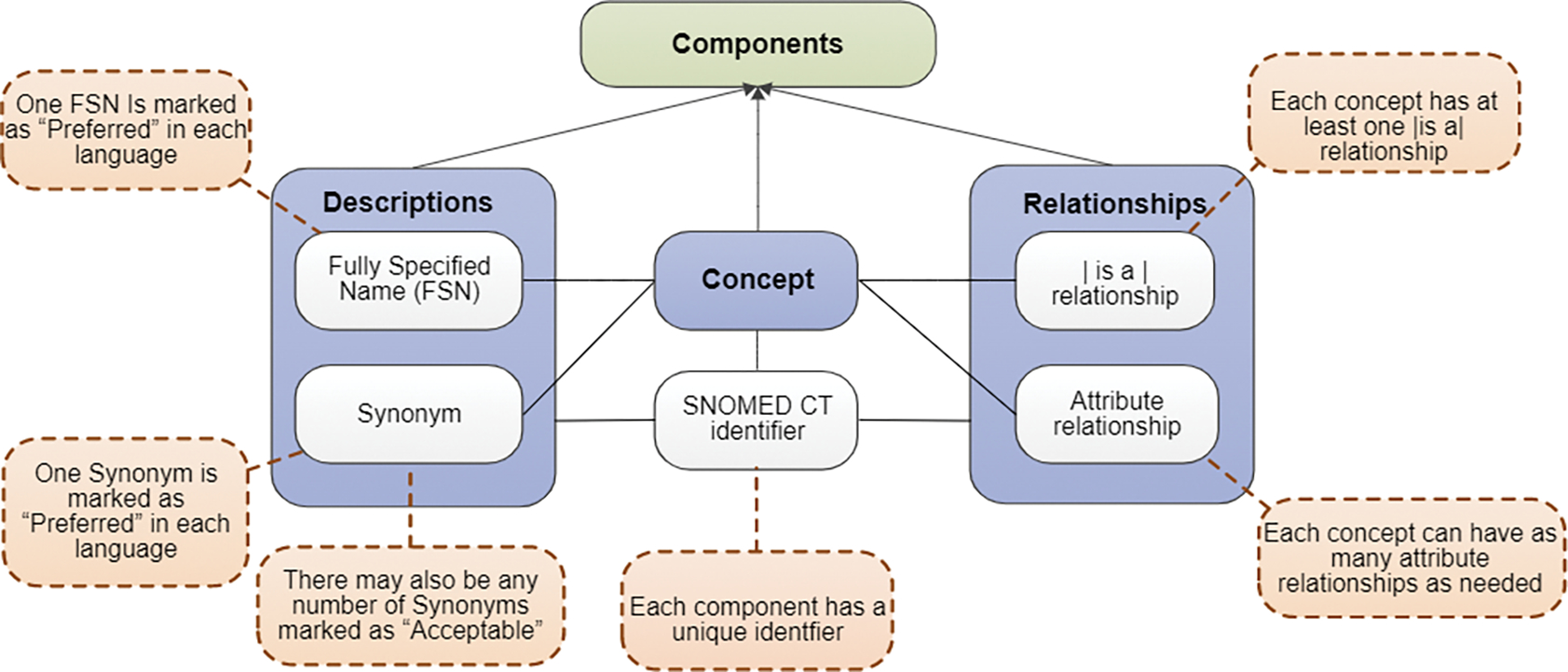
SNOMED CT logical model. This figure is adopted from SNOMED CT Starter Guide [12].
B. Unified Medical Language System
The UMLS incorporates a multitude of different vocabularies such as National Cancer Institute (NCI) thesaurus, Gene Ontology (GO), RxNorm, Logical Observation Identifiers Names and Codes (LOINC) and comprises millions of biomedical concepts [18]. Concept names from different source vocabularies, which are known as atoms, form the basic building block of the UMLS. Each atom is assigned an Atom Unique Identifies (AUI) in the UMLS. The UMLS concepts are formed by aggregating and linking concept names (atoms) from different source vocabularies that convey the same meaning [19]. All of the atoms associated with a concept are synonyms. Each UMLS concept is assigned a Concept Unique Identifier (CUI) and is aggregated from at least one atom [18]. For example, UMLS atoms “Disorder of cornea” with AUI “A6924805” from SNOMED CT, “Corneal Disorder” with AUI “A7591856” from NCIt, and “Corneal Disease” with AUI “A0042855” from MeSH are 3 example atoms that are aggregated under the UMLS concept “Corneal Diseases” with CUI “C0010034”.
C. Related work on identifying missing relations
A number of approaches have been investigated to identify missing relations including IS-A in different biomedical ontologies. Recent approaches include structural approaches [20], lexical approaches [21]–[26], structural-lexical approaches [27], [28], and deep learning approaches [29], [30].
For instance, Zheng et al. investigated abstraction networks to identify missing lateral relationships among top-level concepts of a biomedical ontology [20]. Abstraction networks summarizes the hierarchy of an ontology. An anomalous feature of the abstraction network was utilized to guide the search for missing relations. Expert review was performed on the concepts that were deemed to have a higher likelihood of having missing relations.
Bodenreider proposed a lexical approach where lexical features in concept names are leveraged to identify missing IS-A relations in SNOMED CT [25]. This approach relies on the lexical features to construct logical definitions of concepts. Description logic reasoning was performed on the resulting logical definitions to generate a new IS-A hierarchy, which was compared with the original hierarchy of SNOMED CT. The difference between the original and new hierarchy was leveraged to identify missing IS-A relations.
In [26], Zheng et al. proposed a lexical, transformation-based method to identify missing IS-A relations in biomedical ontologies in the UMLS. For each concept, its base and secondary noun chunks were identified and replaced with more general terms to generate potential supertypes of the concept. If an IS-A relation did not already exist between the concept and the generated potential supertype, a potential missing IS-A relation was identified.
In [27], Cui et al. introduced a hybrid, structural-lexical method for scalable and systematic discovery of missing hierarchical relations in SNOMED CT. Four lexical patterns in non-lattice subgraphs were investigated, where non-lattice subgraphs are graph fragments in an ontology violating the lattice property, a desirable hierarchical property for a well-constructed ontology. Three of the lexical patterns uncovered missing IS-A relation, while one lexical pattern revealed missing concepts.
Liu et. al. presented a deep learning-based approach using a Convolutional Neural Network (CNN) to discover missing IS-A relations for Neoplasm concepts in the NCI thesaurus [29]. They constructed training data from the NCI thesaurus by considering concept-pairs having IS-A relations as positive samples and uncle-nephew-pairs as negative samples. For each concept, they created a textual document by leveraging the concept’s identifier, name, and names of its ancestors and children. By using a Doc2vec model, embeddings for concept documents were obtained. These were fed into the CNN model, which was trained to predict IS-A relations.
III. Method
In this work, we use the March 2022 release of SNOMED CT US Edition. The crux of our approach is as follows. Given a concept, if a parent of the concept appears as a substring in the concept’s own name, then we replace the substring with the parent’s ancestors to generate new concept names. If a newly generated concept already exists in SNOMED CT and is not already linked with the original concept through IS-A relations, then we suggest a potential missing IS-A relation between the original concept and the new concept. Our approach contains five major steps: (1) preprocessing concept names; (2) identifying replacement candidates; (3) suggesting potential missing IS-A relations; (4) removing redundant suggestions; and (5) validating suggested missing IS-A relations.
A. Preprocessing concept names
Each concept’s FSN is preprocessed by performing lexical normalization and single-word synonym replacement as follows.
1). Lexical normalization:
For each concept, we first convert its FSN to lowercase and remove punctuation (such as !, ”, #, $, %) and extra white-spaces. Next, we remove the stop words (such as “of”, “and”, “the”) from the FSN. For this, we use the open-source python library Natural Language Toolkit (NLTK) [31], [32]. Finally, the FSN is lemmatized using the WordNet lemmatizer in NLTK.
For example, the result of lexical normalization for the FSN “Open fracture of facial bones (disorder)” is “open fracture facial bone (disorder)”. Note that the stop word “of” has been removed and the word “bones” has been lemmatized to “bone”.
2). Single-word synonym replacement:
For each word in a concept’s FSN, if a synonym can be found in the SNOMED CT, then we replace the word with its preferred synonym. Synonyms for words in SNOMED CT can be obtained as follows. First, we extract all the SNOMED CT concepts with one-word FSNs (not considering the semantic tag). Then, among the synonyms of such a concept, we identify the one marked as “preferred term”, which we consider as the preferred synonym for that particular word. Therefore, in all circumstances where we encounter other single-word synonyms of this concept, then we can replace them with the preferred synonym [33].
For instance, the concept “Contusion (disorder)” has a preferred term “Contusion” and a synonym “Bruise”. Therefore, “Contusion” is considered as the preferred synonym for “Bruise”. Hence, whenever the FSN of any concept contains the word “Bruise”, we replace it with the preferred word “Contusion”. For instance, the result of single-word synonym replacement for the FSN “Bruise of toe (disorder)” is “Contusion of toe (disorder)”.
B. Identifying replacement candidates
Given a concept C, we represent the preprocessed FSN of the concept as a sequence
where w1 to wn are the words in the FSN and sc is the semantic tag of the concept. If there exists a concept P that is a parent of C and F(P) = [wi, wi+1, …, wj, sp] where wi, wi+1, …, wj are consecutive sequence of words in F(C), then we consider the concept P as a replacement candidate for C. In other words, if a parent concept’s FSN without its semantic tag appears as a substring in a concept’s FSN, then the parent is considered as a replacement candidate for the said concept.
For instance, Fig. 2 shows the inferred logical definitions for the concept “Open fracture of lateral malleolus (disorder)”. This concept has two parents: “Fracture of lateral malleolus (disorder)” and “Open fracture of distal fibula (disorder)”. Out of these two, the concept “Fracture of lateral malleolus” appears as a substring of “Open fracture of lateral malleolus (disorder)” and hence, is considered as a replacement candidate.
Fig. 2.
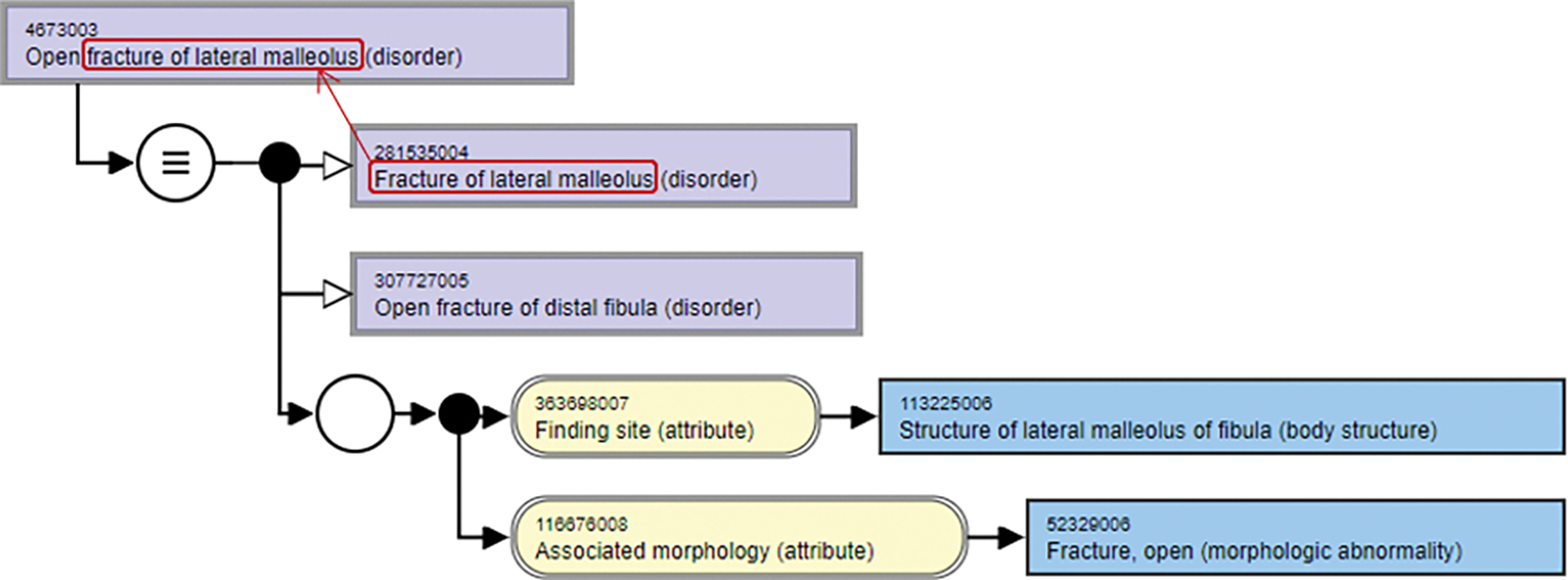
Identifying the replacement candidates for concept “Open fracture of lateral malleolus (disorder)”.
C. Suggesting potential missing IS-A relations
After identifying the replacement candidates as mentioned above, we replace them as follows to generate new concept names. Given a concept C with F(C) = [w1, w2, …, wi, wi+1, …, wj, …, wn, sc] and a replacement candidate P with F(P) = [wi, wi+1, …, wj, sp]. For each ancestor A of P (i.e., P IS-A A) with F(A) = [ak, ak+1, …, al, sa], if A and C share the same semantic tag (i.e., sa = sc), then we replace the substring wi, wi+1, …, wj in F(C) with ak, ak+1, …, al (i.e., A’s FSN excluding the semantic tag) to generate a new concept name: [w1, w2, …, ak, ak+1, …, al, …, wn, sc].
For each newly generated concept name, if the following two conditions are met:
there is an existing concept B in SNOMED CT whose FSN is the same as the newly generated concept name [w1, w2, …, ak, ak+1, …, al, …, wn, sc], and
B is not already an ancestor of C,
then we suggest a potential missing IS-A relation between C and B (i.e., C IS-A B).
For example, in Fig. 2, the replacement candidate “Fracture of lateral malleolus (disorder)” of the concept “Open fracture of lateral malleolus (disorder)” has an ancestor “Fracture of ankle (disorder)”. After replacement by this ancestor, a new concept name “Open fracture of ankle (disorder)” is generated, which is the FSN of an existing concept (with the SNOMED CT identifier 48187004) in SNOMED CT. Since “Open fracture of ankle (disorder)” is not an ancestor of “Open fracture of lateral malleolus (disorder)” in the current release of SNOMED CT, we suggest a potential missing IS-A relation: “Open fracture of lateral malleolus (disorder)” IS-A “Open fracture of ankle (disorder)”.
D. Redundant missing IS-A removal
If a suggested potential missing IS-A relation can be inferred by combining the rest of the suggested potential missing IS-A relations and the existing IS-A relations in SNOMED CT, it is considered to be redundant and removed from the final result. This is because the other suggested potential missing IS-A relations and the existing IS-A relations indirectly suggest this particular potential missing IS-A relation.
E. Validating suggested missing IS-A relations
We validate the suggested potential missing IS-A relations in two ways: (1) automated validation leveraging the UMLS, and (2) manual validation by a domain expert (author JS).
1). Automated validation by the UMLS:
For validation purposes, we used the English concepts in the 2022-AA-full version of the UMLS, which contains more than 4 million UMLS concepts and over 16 million terms aggregated from 222 source vocabularies [34].
We first preprocess the UMLS atoms by performing lexical normalization and single-word synonym replacement similar to how FSNs were preprocessed earlier. Then we try to match FSNs of both the concepts in the potential missing IS-A relations to UMLS atoms belonging to external terminologies (i.e., source vocabularies in the UMLS other than SNOMED CT). Note that we do not consider the semantic tag here. If matching atoms are found, we further check if there exists a direct or indirect IS-A relation between them. If so, then the potential missing IS-A relation is considered to be validated.
2). Manual validation by the domain expert:
For expert evaluation, we randomly select a collection of samples from the suggested potential missing relations for manual review by the domain expert to assess their validity.
IV. Results
There existed a total of 361,780 concepts in the March 2022 release of the SNOMED CT US Edition. Out of these, we identified 80,516 concepts with replacement candidates. After replacing with the ancestors of the replacement candidates, we generated 1,192,065 new concept names. Among these, 43,922 were the FSNs of existing concepts in SNOMED CT. After examining their relations with the original concepts, we identified 3,966 potential missing IS-A relations in total. It was seen that 738 of these were redundant and hence were removed. This finally resulted in 3,228 potential missing IS-A relations.
Table I shows a breakdown of the 3,228 potential missing IS-A by the 19 subhierarchies of SNOMED CT. For example, “Clinical finding (finding)” was the subhierarchy where we identified the most number of potential missing IS-A relations. It should also be noted that this is the largest subhierarchy of SNOMED CT with 120,609 concepts.
TABLE I.
Breakdown of the number of concepts, identified potential missing IS-A relations, and validated missing IS-A relations by the 19 subhierarchies of SNOMED CT.
| Top-level domain | # of concepts | # of potential missing relations | # Validated through UMLS | # Validated through expert |
|---|---|---|---|---|
| Clinical finding (finding) | 120,609 | 1,661 | 51 | 51 |
| Procedure (procedure) | 59,489 | 554 | 2 | 13 |
| Body structure (body structure) | 40,275 | 396 | 9 | 9 |
| Organism (organism) | 33,158 | 20 | 0 | 0 |
| Substance (substance) | 27,332 | 260 | 1 | 4 |
| Pharmaceutical / biologic product (product) | 24,076 | 0 | 0 | 0 |
| Physical object (physical object) | 13,736 | 110 | 0 | 3 |
| Qualifier value (qualifier value) | 11,423 | 33 | 0 | 2 |
| Observable entity (observable entity) | 10,109 | 88 | 0 | 4 |
| Situation with explicit context (situation) | 5,340 | 4 | 0 | 0 |
| Social context (social concept) | 4,478 | 33 | 0 | 0 |
| Event (event) | 3,265 | 21 | 0 | 1 |
| SNOMED CT Model Component (metadata) | 1,849 | 2 | 0 | 0 |
| Environment or geographical location (environment / location) | 1,832 | 16 | 0 | 0 |
| Specimen (specimen)) | 1,784 | 27 | 0 | 2 |
| Staging and scales (staging scale) | 1,685 | 0 | 0 | 0 |
| Special concept (special concept) | 636 | 0 | 0 | 0 |
| Record artifact (record artifact) | 528 | 3 | 0 | 0 |
| Physical force (physical force) | 171 | 0 | 0 | 0 |
| Total | 361,775 | 3,228 | 63 | 89 |
A. Automated validation by the UMLS
Among the 3,228 potential missing IS-A relations identified in this work, there were 817 of them where both the concepts were matched to UMLS atoms belonging to external terminologies. From those, 63 were found to be connected by IS-A relations in the respective source vocabularies. Hence, this automated validation method confirmed 63 cases as valid missing IS-A relations. Table I also shows the breakdown of these 63 cases by subhierarchy. For instance, in the “Clinical finding (finding)” subhierarchy, 51 suggested missing IS-A relations were validated through UMLS.
Table II lists 10 examples of valid missing IS-A relations that was confirmed through the UMLS. For instance, our approach suggested the missing IS-A relation: “Open fracture of lateral malleolus (disorder)” IS-A “Open fracture of ankle (disorder)”. The concept “Open fracture of lateral malleolus (disorder)” was mapped to atom “open fracture of lateral malleolus” with AUI “A13568594” in the UMLS that is a term from MEDCIN, and “Open fracture of ankle (disorder)” was mapped to atom “open fracture of ankle” with AUI “A14065675” which is also from MEDCIN. UMLS records an IS-A relation between these atoms: “A13568594” IS-A “A14065675”. This confirms that the missing IS-A relation: “Open fracture of lateral malleolus (disorder)” IS-A “Open fracture of ankle (disorder)” is indeed valid. Note that a potential missing IS-A relation may be validated through multiple source vocabularies in the UMLS.
TABLE II.
Ten missing IS-A relations validated by external terminologies in the UMLS.
| Descendent concept | Ancestor concept | External terminologies |
|---|---|---|
| Open fracture of lateral malleolus (disorder) | Open fracture of ankle (disorder) | MEDCIN |
| Small cell osteosarcoma (morphologic abnormality) | Small cell sarcoma (morphologic abnormality) | NCI |
| Congenital heart block (disorder) | Congenital heart disease (disorder) | MEDCIN |
| Radical retropubic prostatectomy (procedure) | Radical prostatectomy (procedure) | NCI |
| Autoimmune lymphoproliferative syndrome (disorder) | Autoimmune disease (disorder) | MSH, MDR |
| Bilious vomiting of newborn (disorder) | Vomiting in newborn (disorder) | ICD10CM |
| Human immunoglobulin G (substance) | Human immunoglobulin (substance) | MED-RT |
| Allergic conjunctivitis of right eye (disorder) | Allergic conjunctivitis (disorder) | MEDCIN |
| Laceration of heart without hemopericardium (disorder) | Injury of heart without hemopericardium (disorder) | ICD10CM |
| Sweat gland adenoma (morphologic abnormality) | Sweat gland tumor (morphologic abnormality) | NCI |
The suggested missing IS-A relations may not always be direct relations. Therefore, for each validated missing IS-A relation, we further investigated how far the matched atoms are in the source vocabulary. The number of IS-A relation hops among the two atoms in the source vocabulary hierarchy is considered as the distance between the concept-pair. For instance, if the atoms are directly connected by an IS-A relation, then the distance is 1; while if they are grandchild and grandparent, then the distance is 2. Note that whenever there are multiple IS-A paths, we considered the path with the minimum distance.
Fig. 3 demonstrates the distribution of the distances between matched atoms for concept-pairs with a missing IS-A relation. As can be seen from the figure, 50 out of 63 were direct IS-A relations (i.e., distance = 1).
Fig. 3.
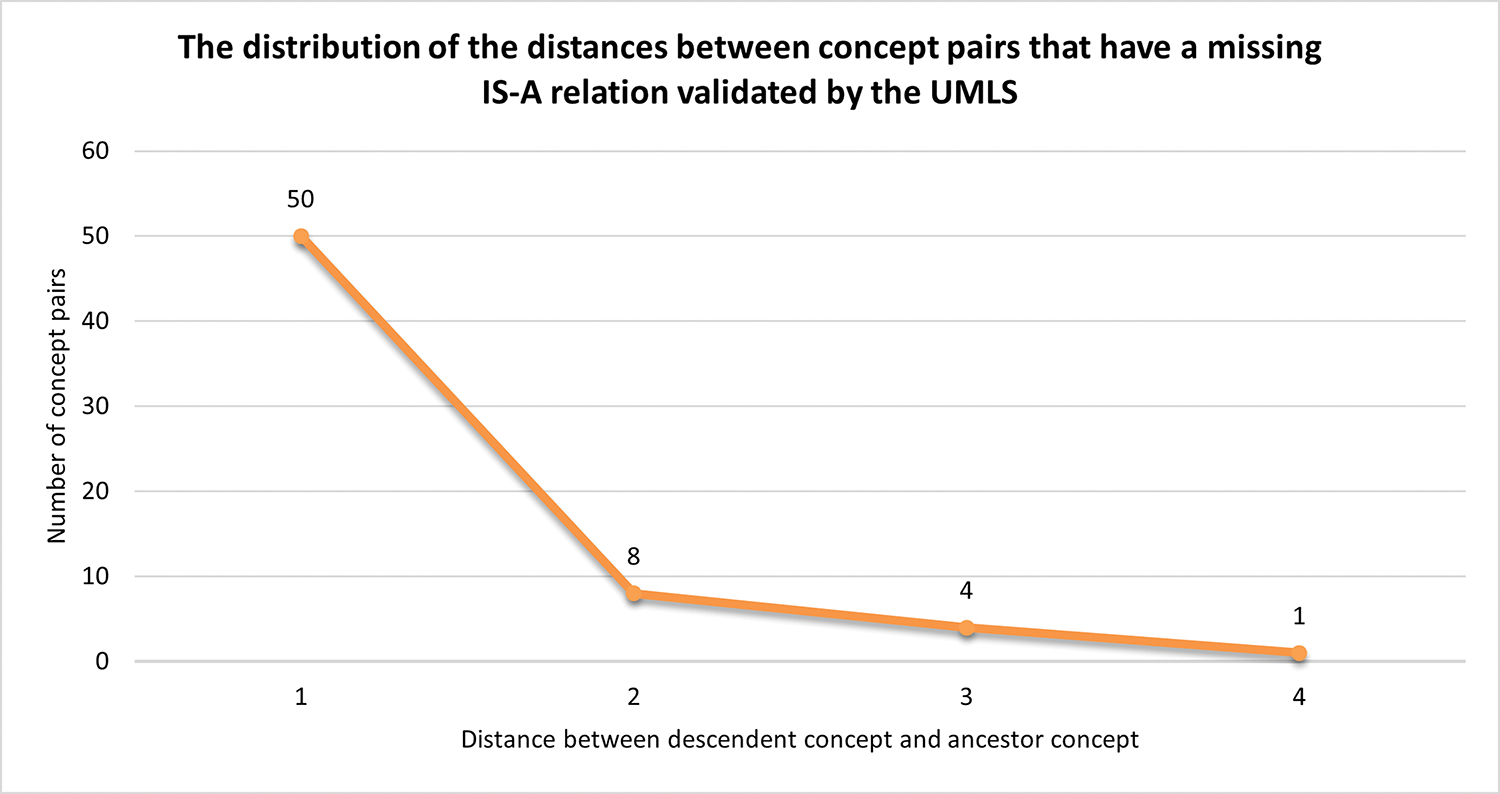
The distribution of the distances between matched UMLS atoms for concept-pairs with a missing IS-A relation.
B. Manual validation by the domain expert
We randomly selected 100 samples from our total 3,228 suggested missing IS-A relations for the domain expert’s manual review. The domain expert confirmed that 89 out of 100 potential missing IS-A relations are valid cases (a precision of 89%). The breakdown of these validated cases by the subhierarchies of SNOMED CT can be found in Table I. For instance, in the “Clinical finding (finding)” subhierarchy, 51 were validated. Table III contains 10 examples of missing IS-A relations validated by the domain expert. For instance, the domain expert confirmed “Chronic pyonephrosis (disorder)” IS-A “Chronic pyelonephritis (disorder)” as a valid missing IS-A relation.
TABLE III.
Ten missing IS-A relations validated by the domain expert.
| Descendent concept | Ancestor concept |
|---|---|
| Chronic pyonephrosis (disorder) | Chronic pyelonephritis (disorder) |
| Venography of lower extremity using contrast (procedure) | Angiography using contrast (procedure) |
| Structure of proximal epiphysis of lower limb (body structure) | Structure of long bone of lower limb (body structure) |
| Fibrinoid necrotizing inflammation (morphologic abnormality) | Fibrinoid necrosis (morphologic abnormality) |
| Discontinue - dosing instruction imperative (qualifier value) | Then discontinue - dosing instruction fragment (qualifier value) |
| Therapeutic hand stretching (procedure) | Therapeutic procedure on soft tissue (procedure) |
| Neonatal audiological screening service (qualifier value) | Neonatal service (qualifier value) |
| Muscle biopsy sample (specimen) | Muscle specimen (specimen) |
| Pain of toe of left foot (finding) | Pain in left foot (finding) |
| Quadrantanopia of right eye (finding) | Visual field defect of right eye (finding) |
V. Discussion
In this work, we investigated a simple substring replacement approach to automatically identify potential missing IS-A relations in SNOMED CT. Our approach leveraged lexical features of the FSNs of concepts and existing IS-A relations to suggest potential missing IS-A relations. We validated the potential missing IS-A relations identified in two ways: (1) automatically leveraging UMLS, (2) manually with a domain expert.
The automated validation was only able to validate 1.95% (63/3,228) potential missing IS-A relations that our methood suggested. This was in part due to the inability to find matching atoms in the UMLS. Only 25.31% (817/3,228) of the potential missing IS-A cases could be matched to UMLS atoms. In cases where matching atoms were found, only in 7.71% (63/817) we could find IS-A relations. It can also be seen from Table I that the UMLS-based validation only validated cases belonging to subhierarchies: “Clinical finding (finding)”, “Procedure (procedure)”, “Body structure (body structure)”, and “Substance (substance)”. This is a drawback of the UMLS-based validation as it is unable to cover a wide variety of concepts since it depends on the domains covered by external terminologies in the UMLS. However, the advantage of the automated UMLS-based validation is the fact that it is quick as it requires no manual labor.
On the other hand, the manual validation revealed that a vast majority (89%) of randomly picked samples are in fact valid missing IS-A relations. While the manual validation requires more human inspection, with that we are able to cover more cases. It should be also noted that only through manual validation, we are able to identify false positives (i.e., invalid missing IS-A suggestions made by the method).
The distance plot for the UMLS-based validation in Fig. 3 shows that 79.37% (50/63) validated IS-A relations are direct IS-A relations in the respective terminologies that the concepts are mapped to. While needing confirmation from SNOMED CT curators, there is a likelihood that these cases will be direct IS-A relations in SNOMED CT as well. Direct missing IS-A relations are easier to fix than indirect ones. This is because for indirect missing IS-A relations, intermediate missing IS-A relations may need to be further identified that infer the indirect missing IS-A.
A. False positives
Though the review by the domain expert revealed our approach to be effective in identifying missing IS-A relations, it still disclosed some false positives. Table IV demonstrates 5 invalid missing IS-A relations pointed out by the domain expert. For example, our suggested missing IS-A relation “Lower respiratory tract structure (body structure)” IS-A “Lower body structure (body structure)” is invalid, because the respiratory tract is located in the chest which is an upper body structure.
TABLE IV.
Five examples of invalid missing IS-A relations identified in the manual evaluation by the domain expert.
| Invalid missing IS-A relation | Replacement candidate’s existing IS-A relation | Domain expert’s comment |
|---|---|---|
| Open repair of strangulated inguinal hernia with prosthesis (procedure) IS-A Open repair of strangulated ventral hernia with prosthesis (procedure) |
Open repair of strangulated inguinal hernia (procedure) IS-A Open repair of strangulated ventral hernia (procedure) |
Inguinal hernia and ventral hernia are located in different anatomical locations. |
| Closed trimalleolar fracture of left ankle (disorder) IS-A Closed bimalleolar fracture of left ankle (disorder) |
Closed trimalleolar fracture (disorder) IS-A Closed bimalleolar fracture (disorder) |
These are distinct clinical entities. |
| Closed fracture finger middle phalanx, head (disorder) IS-A Fracture of bone of head (disorder) |
Closed fracture finger middle phalanx (disorder) IS-A Fracture of bone (disorder) |
The fracture in a finger and fracture of the head (cranium) are completely different. |
| Lower respiratory tract structure (body structure) IS-A Lower body structure (body structure) |
Respiratory tract structure (body structure) IS-A Body structure (body structure) |
The respiratory tract is located in the chest - which is an upper body structure. |
| Visual intelligence quotient (observable entity) IS-A Visual function (observable entity) |
Intelligence quotient (observable entity) IS-A Function (observable entity) |
Visual IQ is a cognitive function. Not visual. |
It should be noted that the domain expert’s comments for some of these cases directly contradict with the existing IS-A relation between the replacement candidate and its ancestor that was leveraged to come up with the missing IS-A suggestion. For example, our suggested missing IS-A relation “Open repair of strangulated inguinal hernia with prosthesis (procedure)” IS-A “Open repair of strangulated ventral hernia with prosthesis (procedure)” is invalid as “inguinal hernia” and “ventral hernia” are located in different anatomical locations. However, this should also apply to the existing IS-A relation between the replacement candidate and its ancestor: “Open repair of strangulated inguinal hernia (procedure)” IS-A “Open repair of strangulated ventral hernia (procedure)”. Such instances illustrate that evaluating the existing IS-A relation between the replacement candidate and its ancestor may have the potential to uncover erroneous existing IS-A relations. The first two examples in Table IV are such cases.
B. Distinction with related work
As mentioned earlier, in [26], Zheng et al. introduced a transformation-based method to identify missing IS-A relations in the source vocabularies in UMLS, where a similar idea of replacement was leveraged to identify missing IS-A relations. The major differences between Zheng et al’s work in [30] and our approach in this paper are: (1) in [30] noun chunks were identified in concept names and replaced by more general terms, while in this work we replace the substring corresponding to the concept’s parent with the parent’s ancestors; and (2) the approach in [30] relies on the IS-A relations in external terminologies in the UMLS to perform replacement, while our approach in this work leverages the IS-A relations within SNOMED CT for replacement. In addition, in this work, we also took semantic tags of concepts into account when suggesting potential missing IS-A relations. Regarding effectiveness of the approaches for identifying missing IS-A relations in SNOMED CT evaluated by domain experts, the approach in [30] achieved a precision of 86.5%, while our approach in this work achieved 89%.
C. Limitations and future work
In this work, we only leveraged the direct IS-A relations (i.e. the parents) of a concept to uncover missing IS-A relations. However, each SNOMED CT concept may also have different attribute relations such as “Finding site(attribute)” and “Associated morphology (attribute)” where the target concepts of those relations may also appear as a substring in the FSN of the concept. Therefore, in the future, we will investigate the possibility of replacing the attribute relation targets with their ancestors to further identify additional cases of missing IS-A relations. In addition, instead of only focusing on parents as replacement candidates, we can consider other indirect ancestors as well. This applies to attribute relations too.
Both the above mentioned improvements would only apply to cases where the substring that we replace appears as a FSN of a concept in SNOMED CT. We expect to investigate on strategies that would identify whether a specific substring is more general than another substring even if both the said substrings do not correspond to FSNs of concepts.
As discussed earlier, during the UMLS-based validation it was found that 817 were mapped to UMLS atoms, out of which only 63 were found to be having IS-A relations. An interesting future work would be to investigate the rest of the 754 cases. Valid missing IS-A relations found among these would not only improve SNOMED CT, but also other external ontologies where the mapped atoms belong to.
As mentioned earlier, some of the false positives identified during the manual validation by the domain expert may lead to the identification of erroneous existing IS-A relations between the replacement candidates and their ancestors. Therefore, in the future we will perform another round of manual review to validate and confirm these cases.
Our method identified a significant number of potential missing IS-A relations in SNOMED CT. We will submit our findings to the SNOMED CT authors to further contribute towards the quality improvement process of SNOMED CT.
VI. Conclusion
In this paper, we developed a substring replacement approach leveraging the lexical features of fully specified names of concepts and existing IS-A relations of concepts to uncover potential missing IS-A relations in SNOMED CT. Applying this approach to the March 2022 release of the SNOMED CT US Edition, a total of 3,228 potential missing relations were suggested, which were validated automatically based on UMLS and manually by a domain expert. The automated evaluation validated 63 missing IS-A relations. The manual domain expert evaluation was performed on a random sample of 100 cases and confirmed 89 valid missing IS-A relations. The results indicate that our substring replacement approach is effective in identifying missing IS-A relations in SNOMED CT.
Acknowledgments
This work was supported by the US National Science Foundation (NSF) under grants 1931134 and 2047001, and National Institutes of Health (NIH) under grants R01LM013335 and R21AG068994. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NSF or NIH.
References
- [1].Jovanović J and Bagheri E, “Semantic annotation in biomedicine: the current landscape,” Journal of biomedical semantics, vol. 8, no. 1, pp. 1–18, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ferreira JD, Teixeira DC, and Pesquita C, “Biomedical ontologies: coverage, access and use,” 2021.
- [3].Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, and Musen MA, “Bioportal: enhanced functionality via new web services from the national center for biomedical ontology to access and use ontologies in software applications,” Nucleic acids research, vol. 39, no. suppl 2, pp. W541–W545, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Agrawal A, He Z, Perl Y, Wei D, Halper M, Elhanan G, and Chen Y, “The readiness of snomed problem list concepts for meaningful use of electronic health records,” Artificial intelligence in medicine, vol. 58, no. 2, pp. 73–80, 2013. [DOI] [PubMed] [Google Scholar]
- [5].Finnegan R, “Icd-9-cm coding for physician billing,” Journal (American Medical Record Association), vol. 60, no. 2, pp. 22–23, 1989. [PubMed] [Google Scholar]
- [6].Duque-Ramos A, Quesada-Martínez M, Iniesta-Moreno M, Fernández-Breis JT, and Stevens R, “Supporting the analysis of ontology evolution processes through the combination of static and dynamic scaling functions in oquare,” Journal of biomedical semantics, vol. 7, no. 1, pp. 1–20, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Amith M, He Z, Bian J, Lossio-Ventura JA, and Tao C, “Assessing the practice of biomedical ontology evaluation: Gaps and opportunities,” Journal of biomedical informatics, vol. 80, pp. 1–13, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhu X, Fan J-W, Baorto DM, Weng C, and Cimino JJ, “A review of auditing methods applied to the content of controlled biomedical terminologies,” Journal of biomedical informatics, vol. 42, no. 3, pp. 413–425, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lee D, de Keizer N, Lau F, and Cornet R, “Literature review of snomed ct use,” Journal of the American Medical Informatics Association, vol. 21, no. e1, pp. e11–e19, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].“Snomed ct managed service - us edition release notes - march 2021,” https://confluence.ihtsdotools.org/display/RMT/SNOMED+CT+Managed+Service+-+US+Edition+Release+Notes+-+March+2021, (Online; accessed June, 2021).
- [11].“Snomed ct united states edition,” https://www.nlm.nih.gov/healthit/snomedct/us\_edition.html, (Online; accessed June, 2021).
- [12].“Snomed ct logical model,” https://confluence.ihtsdotools.org/display/DOCSTART/5.+SNOMED+CT+Logical+Model, (Online; accessed June, 2021).
- [13].“Snomed ct identifier,” https://confluence.ihtsdotools.org/display/DOCGLOSS/SNOMED+CT+identifier, (Online; accessed July, 2022).
- [14].“Fully specified name,” https://confluence.ihtsdotools.org/display/DOCEG/Fully+Specified+Name, (Online; accessed June, 2021).
- [15].“Snomed ct synonym,” https://confluence.ihtsdotools.org/display/DOCEG/Synonym, (Online; accessed June, 2021).
- [16].“Preferred term,” https://confluence.ihtsdotools.org/display/DOCEG/Preferred+Term, (Online; accessed August, 2021).
- [17].“Stated and inferred definitions - examples,” https://confluence.ihtsdotools.org/display/DOCRELFMT/D.1+Stated+and+Inferred+Definitions+-+Examples#:~:text=SNOMED%20CT%20concepts%20are%20defined,revised%20by%20SNOMED%20CT%20authors., (Online; accessed August, 2022).
- [18].Bodenreider O, “The unified medical language system (umls): integrating biomedical terminology,” Nucleic acids research, vol. 32, no. suppl_1, pp. D267–D270, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].B. M. N. L. of Medicine (US), “Umls® reference manual,” https://www.ncbi.nlm.nih.gov/books/NBK9684/, 2009, (Online; accessed Aug, 2021).
- [20].Zheng L, Chen Y, Min H, Hildebrand PL, Liu H, Halper M, Geller J, de Coronado S, and Perl Y, “Missing lateral relationships in top-level concepts of an ontology,” BMC Medical Informatics and Decision Making, vol. 20, no. 10, pp. 1–16, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cui L, Bodenreider O, Shi J, and Zhang G-Q, “Auditing snomed ct hierarchical relations based on lexical features of concepts in non-lattice subgraphs,” Journal of biomedical informatics, vol. 78, pp. 177–184, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Abeysinghe R, Zheng F, Hinderer EW, Moseley HN, and Cui L, “A lexical approach to identifying subtype inconsistencies in biomedical terminologies,” in 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). IEEE, 2018, pp. 1982–1989. [Google Scholar]
- [23].Burse R, McArdle G, and Bertolotto M, “Stop-word based contextual auditing to identify inconsistencies in snomed.” in SWH@ ISWC, 2020, pp. 7–18. [Google Scholar]
- [24].Zheng F, Shi J, and Cui L, “A lexical-based approach for exhaustive detection of missing hierarchical is-a relations in snomed ct,” in AMIA Annual Symposium Proceedings, vol. 2020. American Medical Informatics Association, 2020, p. 1392. [PMC free article] [PubMed] [Google Scholar]
- [25].Bodenreider O, “Identifying missing hierarchical relations in snomed ct from logical definitions based on the lexical features of concept names.” ICBO/BioCreative, vol. 2016, 2016. [PMC free article] [PubMed] [Google Scholar]
- [26].Zheng F, Shi J, Yang Y, Zheng WJ, and Cui L, “A transformation-based method for auditing the is-a hierarchy of biomedical terminologies in the unified medical language system,” Journal of the American Medical Informatics Association, vol. 27, no. 10, pp. 1568–1575, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cui L, Zhu W, Tao S, Case JT, Bodenreider O, and Zhang G-Q, “Mining non-lattice subgraphs for detecting missing hierarchical relations and concepts in snomed ct,” Journal of the American Medical Informatics Association, vol. 24, no. 4, pp. 788–798, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Abeysinghe R, Brooks MA, Talbert J, and Licong C, “Quality assurance of nci thesaurus by mining structural-lexical patterns,” in AMIA annual symposium proceedings, vol. 2017. American Medical Informatics Association, 2017, p. 364. [PMC free article] [PubMed] [Google Scholar]
- [29].Liu H, Zheng L, Perl Y, Geller J, and Elhanan G, “Can a convolutional neural network support auditing of nci thesaurus neoplasm concepts?” in ICBO, 2018. [Google Scholar]
- [30].Liu H, Perl Y, and Geller J, “Concept placement using bert trained by transforming and summarizing biomedical ontology structure,” Journal of Biomedical Informatics, vol. 112, p. 103607, 2020. [DOI] [PubMed] [Google Scholar]
- [31].Bird S, “Nltk: the natural language toolkit,” in Proceedings of the COLING/ACL 2006 Interactive Presentation Sessions, 2006, pp. 69–72. [Google Scholar]
- [32].Davydova O, “Text preprocessing in python: Steps, tools, and examples,” https://medium.com/@datamonsters/text-preprocessing-in-python-steps-tools-and-examples-bf025f872908, 2018, (Last accessed Feb 26, 2021). [Google Scholar]
- [33].Zheng F and Cui L, “A lexical-based formal concept analysis method to identify missing concepts in the nci thesaurus,” in 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). IEEE, 2020, pp. 1757–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].“Statistics - 2021aa release,” https://www.nlm.nih.gov/research/umls/knowledge_sources/metathesaurus/release/statistics.html, (Online; accessed Aug, 2021).


