Abstract
Histopathology has historically been the critical technique for the diagnosis and treatment of human disease. Today, genomics, transcriptomics, and proteomics from specific cells, rather than bulk tissue, have become key to understanding underlying disease mechanisms and rendering useful diagnostic information. Extraction of desired analytes, i.e. nucleic acids or proteins, from easily accessible formalin-fixed paraffin-embedded (FFPE) tissues allows for clinically-relevant activities, such as sequencing biomarker mutations or typing amyloidogenic proteins. Genetic profiling has become routine for cancers as varied as non-small cell lung cancer and prostatic carcinoma. The five main tissue dissection techniques that have been developed thus far include: bulk scraping, manual macrodissection (MMa), manual microdissection (MMi), laser-capture microdissection (LCM), and expression microdissection (xMD). In this review, we discuss the importance of tissue dissection in clinical practice and research, the basic methods, applications, as well as some advantages and disadvantages for each modality.
Keywords: tissue dissection, microdissection, nucleic acid isolation
Introduction
In this era of personalized medicine, histological patterns are augmented by the identification of genetic mutations and other pathological features essential to diagnose human disease, to offer prognostic information, and to determine optimal treatment strategies. As a result, biomolecular testing (i.e. genomics, transcriptomics, and proteomics) from surgical tissue specimens is now all but required for individualized care [1–3]. This evolution resulted from the combined understanding of the role of genetics in medicine and the advances in extracting analytes (i.e. nucleic acids and proteins) from standard histologic specimens.
Histopathology has been central for diagnostics, prognostics, and theragnostics for over a century. Pathologists evaluate slides derived from fresh-frozen or formalin-fixed paraffin-embedded (FFPE) tissues using histological or immunohistochemical (IHC) stains to offer specific diagnoses [4–5]. FFPE tissue blocks are easy to store, maintain, and section, making them a desirable, long-term substrate for biomolecular testing [6–7]. Although formalin fixation is known to cause nucleic acid degradation, newer extraction methods are able to mitigate this complication [8–10]. Formalin fixation has also been shown to cause protein crosslinking and modifications, most of which can be reversed using heat-induced antigen retrieval [11–12]. Other variables, such as sample age, sectioning, and storage conditions, can also impact the quality and quantity of extracted analytes. Further research into how preanalytical factors of FFPE tissues influence biomolecular extraction is needed, but that is outside of the scope of this review [13–14]. Still, the majority of current studies indicate little problem with obtaining sufficient materials for downstream applications [6; 15–17].
Post-histopathological review, the best described utility of biomolecular diagnostics with FFPE tissues is biomarker mutation detection. Identification of mutations in oncogenes and tumor suppressors, such as Erb-B2 receptor tyrosine kinase 2 (ERBB2); KRAS proto-oncogene, GTPase (KRAS); B-Raf proto-oncogene, serine/threonine kinase (BRAF); epidermal growth factor receptor (EGFR); and tumor protein p53 (TP53), can be used to inform diagnostics, prognostics, and theragnostics. As will be discussed, several of these mutations can be targeted by monoclonal antibodies or small-molecule drugs [7; 18–19]. To acquire this molecular information, different approaches to enrich for specific analytes by dissecting FFPE tissues have been developed, each optimized for certain types of testing.
The general purpose of tissue dissection is to remove any unnecessary portions of a tissue section, and then to extract DNA, RNA, or proteins from the desired areas, enriching for the molecule(s) of interest [20]. Next-generation DNA sequencing of specific biomarker panels is now well-established to identify clinically actionable mutations across a range of malignancies [3; 7; 21]. RNA and protein profiling are still being established, but qRT-PCR, microarrays, Nanostring RNA hybridization arrays, RNA-seq, reverse-phase protein array analysis, mass spectrometry, and enzyme-linked immunosorbent assays are among those that are being implemented [21–24]. There are five main types of tissue dissection that have been described, including bulk scraping, manual macrodissection (MMa), manual microdissection (MMi), laser-capture microdissection (LCM), and expression microdissection (xMD) [20; 25–28]. Microdissection techniques have become more popular over the past couple decades due to improved accuracy and precision, as well as the advent of single-cell technology [20; 29].
Tissue sections are typically heterogeneous composites of diverse cell types, which can lead to masking of the desired analytes [20]. For example, the tumor microenvironment contains a variety of immune cells, fibroblasts, and stem cells, which can result in a very low proportion of neoplastic cells to non-neoplastic cells [30–31]. In the absence of enrichment, the percentage of malignant cells relative to benign cells may be so low that mutations of interest fall below the limit of detection (LOD) for reporting variants using current methodologies and quality control metrics [7]. Modern deep sequencing systems have a purported assay-specific minimum LOD of around 5%, which would require tumor cellularity of at least 10%, assuming allelic heterozygosity of the biomarker [32–33]. Recent studies, however, have demonstrated an accurate minimum LOD of 3% for single nucleotide variants, but this would still require at least 6% tumor cellularity [32; 34]. Therefore, by specifically capturing only the tumor cells, or at least enriching for them, evaluation of their DNA will ideally allow for the successful identification of actionable biomarker mutations.
This review describes the five major tissue dissection methods, examples of their current usage, and proposed future uses, within the clinical biomolecular diagnostic setting. While this review will focus on dissection methods for FFPE tissue, it is important to note that these techniques can also be used on cytological samples, such as fine needle aspirates or cellular smears. Furthermore, flow cytometry is a useful alternative to microdissection techniques [20; 35–36]. Other reviews have evaluated downstream mutation detection methods and we invite the reader to explore this other literature [37–38].
Macrodissection Methods
Tissue macrodissection refers to a dissection that is performed without the use of a microscope or specialized equipment. Macrodissections are typically performed on uniform tissue sections, such as large tumors demarcated without any additional magnification needed.
Bulk Scraping
The first and most simple method of tissue dissection is referred to as bulk scraping. For this procedure, the entire tissue section is removed from a slide using a razorblade or scalpel (shown in Fig. 1a) [26; 39]. Typically, the tissue adheres to the blade by electrostatic forces, but hydrostatic forces may also be incorporated [40]. The tissue scroll is then transferred to a microcentrifuge tube for further processing (shown in Fig. 1b). Some labs generate a tissue scroll, or “curl,” directly from the FFPE block, which is then placed into a microcentrifuge tube, instead of first being attached to a slide.
Fig. 1.

Bulk scraping dissection technique. The full tissue section is first removed from the slide using a razorblade (a). The tissue is then transferred to a microcentrifuge tube for further processing (b).
This technique is by far the fastest and least expensive method for analyte extraction. Additionally, this method has been shown to yield the greatest amount of molecular material [41–42]. Nevertheless, for most clinical profiling, bulk scraping has insufficient precision, and thus it is not ideal if a particular cell type or analyte is desired for enrichment [20; 42–43]. Until recently, this technique had frequently been used to obtain DNA for biomarker mutation testing, but it has become less popular due to the rise of microdissection and single-cell analysis [20; 29]. As noted previously, most tissue sections are heterogeneous/admixed and collecting all of the cells present can mask the desired product.
One common clinical application for bulk scraping is the evaluation of DNA, RNA, or protein quality prior to performing a microdissection method [44]. Since microdissection itself can influence analyte quality, it is important to understand the starting quality before attempting further analysis. The most basic way to measure DNA or RNA quality isolated from bulk scraped material is through spectrophotometry, i.e. the 260/280 nm and 260/230 nm absorbance ratios. However, these ratios may be affected by contaminants, such as phenol, so more specific methods may be required. Newer methods based on target-specific fluorescent dyes (e.g. the Invitrogen Qubit) have now become routine along with spectrophotometry [45]. Capillary electrophoresis systems, such as the Agilent TapeStation and the Agilent Bioanalyzer, can accurately and efficiently calculate nucleic acid quantity, degree of fragmentation, and quality indices (DIN or RIN, respectively) [46–49]. qPCR can be used to measure DNA quality by taking the ratio of two amplicons, one smaller and one larger, generated from a reference gene [46]. RNA quality can be measured using qRT-PCR to take the 5’/3’ ratio of amplicons generated from a reference gene, or by calculating the 28S/18S rRNA ratio [50]. Protein quality can be measured by spectrometric and electrophoretic techniques [51].
Manual Macrodissection (MMa)
The other large-scale method for tissue dissection is MMa. In contrast to bulk scraping, only an ROI is collected, not the entire tissue section [25; 52–55]. For this method, a pathologist first examines an H&E-stained slide, circling part of the tissue with a marker to note a general area of enrichment for the desired cell type [53–55]. This is frequently performed during the normal handling of the case in anticipation of biomolecular testing. This annotated slide is then aligned with an unstained slide, and the ROI is removed from the unstained slide with a razorblade or scalpel (shown in Fig. 2a) [25; 52–55]. Afterwards, the dissected tissue is transferred to a microcentrifuge tube for further processing (shown in Fig. 2b).
Fig. 2.
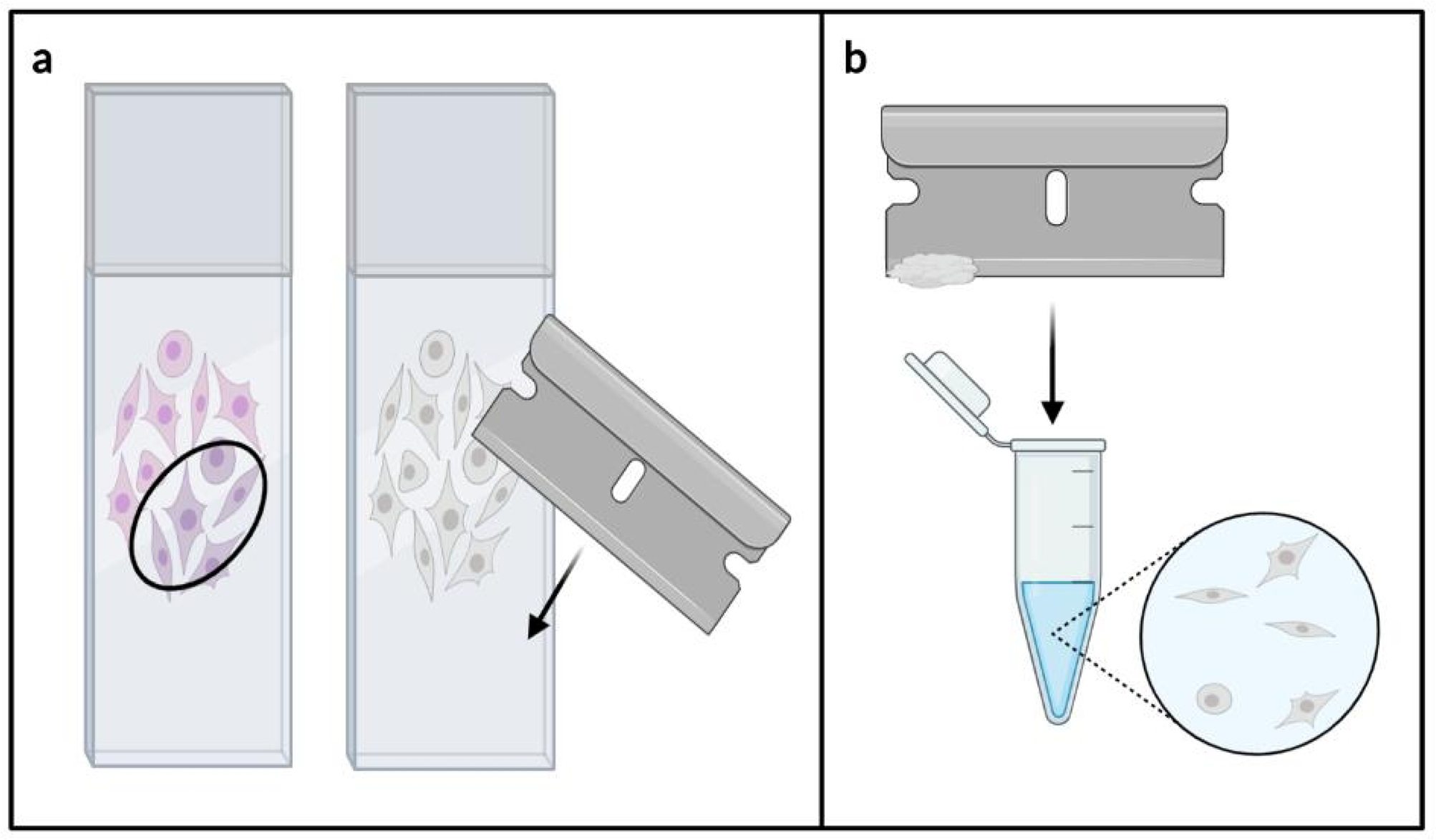
Manual macrodissection dissection technique. An annotated H&E stained slide is aligned to an unstained slide, and the region of interest is removed with a razorblade (a). The tissue is then transferred to a microcentrifuge tube for further processing (b).
MMa is perhaps the most popular dissection technique used in clinical oncology for biomolecular testing today, as most resected tumors have reasonably distinct boundaries and a high percentage of tumor cells to non-tumor cells [53–55]. This technique allows one to avoid areas of low yield (e.g. necrosis), high admixture with non-neoplastic elements (e.g. inflammation), and adjacent normal areas or zones with low frequency of neoplastic cells (e.g. fibrosis). Like bulk scraping, this method is fast, inexpensive, and generates a large amount of molecular material [53–55]. Nonetheless, this method lacks the precision necessary for cancers with low neoplastic cell content [53–55]. Furthermore, a pathologist or other trained histologist is needed to examine the tissue prior to dissection, which can increase cost, delay turnaround time, and increases the risk of subjectivity.
As noted, MMa has been, and continues to be, an important tool in clinical oncology, especially in extracting DNA for targeted biomarker sequencing. For example, MMa has been employed to examine the mutational status of BRAF in patients with metastatic melanoma [56]. The Cobas 4800 BRAF V600 Mutation Test was used on these samples, and reported cases specifically with the V600E variant (a newer version of the test also identifies V600D, V600E2, and V600K). A cohort of 45 patients in the study were then treated with either dabrafenib or vemurafenib, both of which target nonsynonymous mutations at the BRAF V600 codon, based on their mutational status.
Another study used MMa coupled with the SNaPshot Assay to examine the mutational status of KRAS in patients with pancreatic ductal adenocarcinoma [57]. KRAS point mutations were found in 92% of tested tumor samples (cohort = 46 patients). However, in this population treated with a combination of selumetinib plus erlotinib, both EGFR signaling inhibitors, this combination therapy showed limited success.
Finally, a study utilized MMa to survey the expression of KIAA1549-BRAF fusions in pilocytic astrocytoma [58]. Unlike the two previous studies, this study extracted RNA and used qRT-PCR to detect gene fusions. Thirty-two patients were enrolled, with 75% exhibiting a KIAA1549-BRAF fusion. Testing for BRAF fusions is valuable for glioma diagnostics, and the specific gene fusions are correlated with tumor location.
These studies highlight the importance of MMa in clinical diagnostics and theragnostics, albeit with the inherent limitations of not having cell-level resolution.
Microdissection Methods
In contrast to macrodissection, tissue microdissection refers to a dissection that is performed using a microscope and/or specialized equipment. Microdissections are typically performed on highly heterogeneous/admixed tissues as well as small, defined tissue regions/functional units, such as glomeruli.
Manual Microdissection (MMi)
The need for microdissection techniques due to tissue heterogeneity was advanced in the 1970s by Lowry and Passonneau [59]. MMi is similar to MMa, with the major difference being the use of a microscope. Either a compound microscope or a stereomicroscope may be employed, but a stereomicroscope has the added benefits of more space for hand movement and a non-inverted field of view [40]. For this technique, the previously identified ROI(s) is/are removed by gentle scraping with a needle, scalpel, or pipette tip (shown in Fig. 3a) [40; 59]. Then, the dissected tissue is transferred to a microcentrifuge tube for further processing (shown in Fig. 3b).
Fig. 3.

Manual microdissection dissection technique. Regions of interest are removed by hand with a needle under a microscope (a). The tissue is then transferred to a microcentrifuge tube for further processing (b).
This technique is much more precise than the two previously discussed methods as microscopic architectural features can be dissected, although cell-level resolution is still not achieved. As a result of this precision, tumor DNA in highly heterogeneous samples can be enriched for well beyond the limits of macrodissection techniques. It also remains relatively inexpensive as no specialized equipment is required. Still, this technique can lead to a loss of tissue integrity, making it difficult to verify accuracy [20]. To mitigate these concerns, some researchers have started using automated cutting/milling instruments with digital guidance, such as the AVENIO Millisect System, at an increased cost [54–55]. However, these methods are generally time intensive, require considerable patience, manual dexterity, and pathology expertise [20; 40; 54–55].
Due to cost restraints, MMi is the most popular method for microdissection in clinical practice today. However, it is still not nearly as common as MMa, as it is a more complex and time-consuming procedure. Thus, MMi is typically reserved for cases with low neoplastic cellularity. For example, one study used MMa for lung adenocarcinoma samples with >50% tumor cellularity, and used MMi for the remaining low-density samples [60]. Of the 493 patient samples that were included in this study, 474 had sufficient tumor cellularity for EGFR sequencing to compare sequencing results from the Cobas 4800 EGFR Mutation Test to traditional Sanger sequencing. The authors found that both methods should be used together for clinical sequencing to ensure the greatest depth of coverage.
In a breast cancer study, MMa was used for samples with >20% tumor cellularity, and MMi was used for the remaining samples [61]. A cohort of 415 patient samples were used for this study, with 281 mutations detected in the 46 genes tested by the Ion AmpliSeq Cancer Panel. Two of the most commonly mutated genes were phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) and AKT serine/threonine kinase 1 (AKT1), which can be targeted by current or future therapeutic agents.
Finally, a variant of MMi, the Pinpoint Slide DNA Isolation System (Zymo Research), was used to study oncogenic mutations in melanoma samples [62]. This approach is based on a proprietary solution that adheres, and essentially embeds, the ROI(s), allowing for the whole region to be removed at once. Of the 502 patient samples that were studied, 474 had great enough tumor cellularity for an oncogene sequencing panel, which assessed mutations in BRAF, NRAS proto-oncogene, GTPase (NRAS), KRAS, HRAS proto-oncogene, GTPase (HRAS), PIK3CA, and KIT proto-oncogene, receptor tyrosine kinase (KIT), finding variable rates of mutation in each gene. Mutation testing across these multiple genes has important implications for prognostics and theragnostics.
Laser-Capture Microdissection (LCM)
LCM is a popular form of tissue dissection in basic research. As the name implies, an infrared or ultraviolet laser is employed to remove the ROI(s) from a tissue section under microscopic guidance (shown in Fig. 4).
Fig. 4.
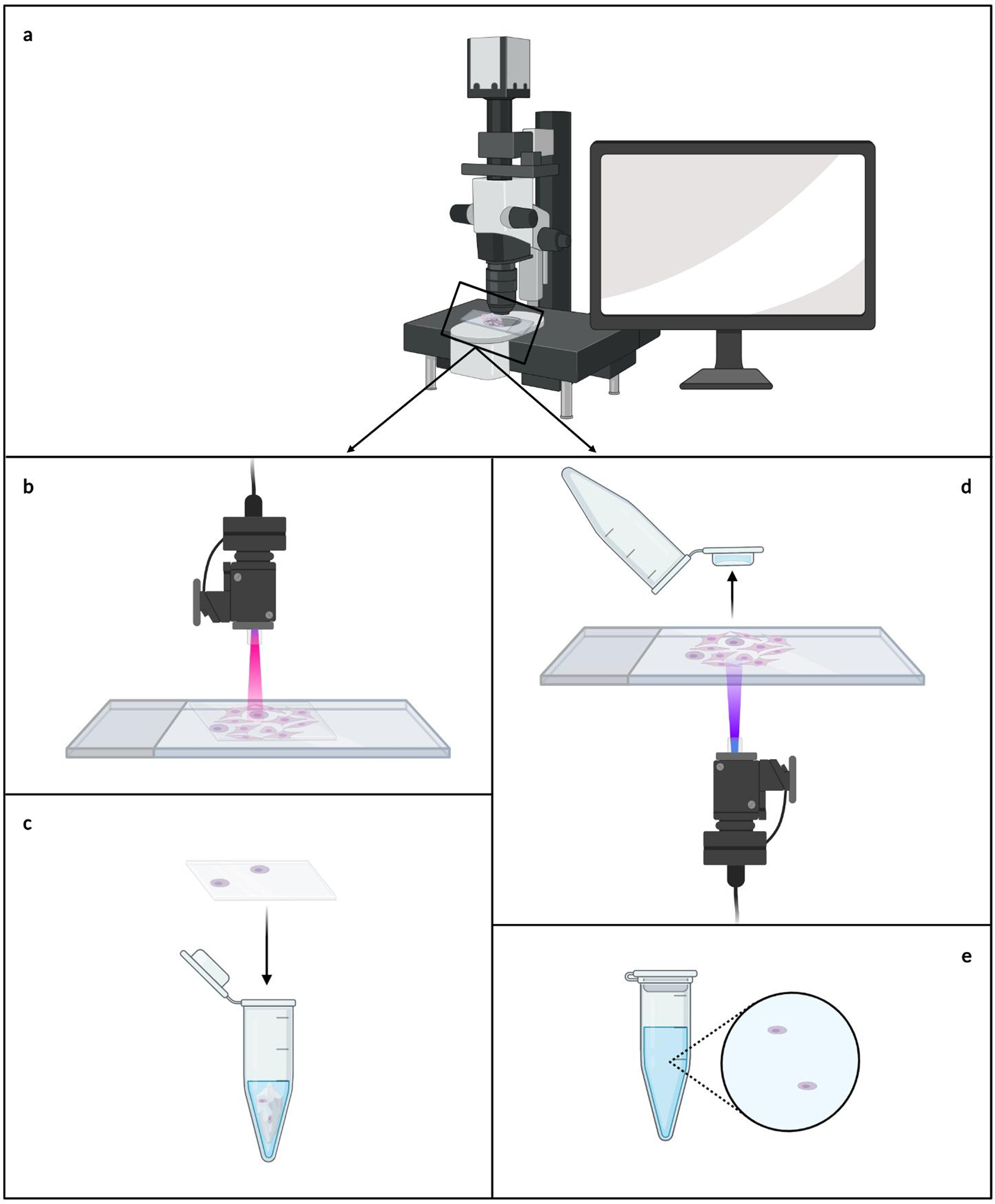
Laser-capture microdissection dissection techniques. The regions of interest are determined microscopically with the aid of a computer and a digital camera (a). For infrared laser-capture microdissection, the cells of interest are focally adhered to a membrane, and the membrane is transferred to a microcentrifuge tube for further processing (b-c). For ultraviolet laser-capture microdissection, the cells of interest are cut from the tissue and propelled into a microcentrifuge cap, where they are then ready for further processing (d-e).
IR-LCM was first developed in 1996 by Emmert-Buck and colleagues [63]. For this technique, a tissue section is covered with ethylene-vinyl acetate (EVA) film directly or used with a specialized EVA coated microcentrifuge cap (e.g. CapSure LCM Caps, Thermo Fisher Scientific). The ROI(s) is first selected and marked in real time using image processing software with a digital camera mounted on a microscope that is part of the LCM apparatus (shown in Fig. 4a) [20; 40; 63–64]. The ROI(s) is/are then focally adhered to the film by short, localized pulses of the laser that melts the EVA and binds it to the subjacent tissues/cells within the ROI(s) (shown in Fig. 4b) [20; 40; 63–64]. The adherence of the cells to the EVA film surpasses their adhesion to the slide, allowing for the cells of interest to be removed along with the film, while leaving the remaining tissue intact [63–64]. Lastly, the film is transferred to a microcentrifuge tube for further processing (shown in Fig. 4c). This system is commercially available as the Arcturus XT LCM (ThermoFisher).
A variation on LCM was developed by Schütze and Lahr in 1998 wherein they introduced UV-LCM [63]. Similar to IR-LCM, the ROI(s) is selected and marked in real time using a digital camera attached to the LCM system (shown in Fig. 4a). Next, a laser is used to cut around the ROI(s), removing it from the surrounding tissue [40; 64–65]. The laser power is then increased, catapulting the cells into a collection chamber, typically a microcentrifuge cap filled with a few microliters of lysis buffer (shown in Fig. 4d) [20; 64–65]. The microcentrifuge tube can then be closed, and the cells are ready for further processing (shown in Fig. 4e). This system is commercially available as the PALM Microbeam (Zeiss). Although specialized slides are available for both types of LCM, standard glass slides can be used effectively [64].
LCM systems today are primarily either semi- or fully-automated, and have combinations of the IR and UV systems, increasing precision and limiting human error [64; 66]. Although it is considerably faster than manual microdissection, LCM remains a slow and low-throughput procedure, especially in the case of IR-LCM [64; 66]. In addition, a pathologist or other trained histologist is needed to examine the tissue and mark ROI(s) in real time, which adds to time-consumption and subjectivity [64; 66]. There is active research using machine learning to identify ROI(s) in order to mitigate the amount of time required and to limit subjectivity. One example is Spatially Invariant Vector Quantization (SIVQ), which along with other algorithms, coupled to LCM, showing promise for increasing accuracy and throughput [67–68]. Still, LCM involves specialized instrumentation and specific consumables, making the cost prohibitive for most clinical uses [64; 66].
Likely due to cost and time concerns, LCM has not been well integrated into the clinical setting like the other methods that have been discussed thus far. One area that LCM has been employed in, though, is amyloid protein typing via mass spectrometry. Amyloidosis is a rare condition in which misfolded proteins aggregate at various sites throughout the body, ultimately leading to organ dysfunction or failure [69–70]. There are over 25 known proteins that can cause amyloidosis, with some of the most common being the immunoglobulin light chains, transthyretin, serum amyloid A protein, and the alpha chain of fibrinogen. Similar to cancer biomarkers, correctly identifying the amyloidogenic protein is crucial for prognosis and treatment. Several institutions, including the Mayo Clinic, currently employ LCM to enrich for amyloidogenic proteins before further analysis with mass spectrometry on the polypeptides [69–70].
While LCM remains popular in basic science research, forays into clinical practice, such as digital droplet PCR and RNA-seq for clinically relevant gene expression tests, are on the horizon [71–72].
Expression Microdissection (xMD)
The final and newest form of tissue dissection, xMD, was first introduced by Tangrea et al. in 2004 [28]. For this method, the tissue slide is first stained using IHC for a desired cell type or analyte of interest, thus capitalizing on an inherent cellular characteristic to determine the ROI(s) (shown in Fig. 5a) [28; 73–74]. Next, the tissue is covered with EVA film, and the stained cells are bound to the film using a high-intensity flashbulb or laser (shown in Fig. 5b) [28; 73–74]. Unlike IR-LCM, the entire tissue field is irradiated, allowing the EVA film to selectively melt directly over the pigmented cells. The pigmented cells convert light energy to heat, and the bonding of the EVA film to the cells becomes stronger than that of the cells to the slide, thus causing them to transfer from the slide when the film is removed [28]. The film is then placed in a microcentrifuge tube for further processing (shown in Fig. 5c).
Fig. 5.

Expression microdissection dissection technique. The regions of interest are determined using immunohistochemistry. Primary and secondary antibodies bind to cells that express the product of interest, and a pigment is produced by means of horseradish peroxidase (a). The stained cells are then heat-bound to a membrane by means of a high-intensity flashbulb (b). Finally, the membrane with the stained cells is transferred to a microcentrifuge tube for further processing (c).
While EVA film is required for both IR-LCM and xMD, the EVA film used in xMD does not contain a light-absorbing dye [28]. This is critical as it allows for focal melting of the EVA only to the pigmented cells. The most commonly used commercially available films are CoTran 9715 and CoTran 9702, both of which are produced by 3M [73–74]. However, there is ongoing research into other EVA films to allow for greater customizability and quality control management [unpublished data].
At its inception, xMD was performed using the same laser systems as LCM, but newer methods rely on off-the-shelf light sources [28; 73–75]. These devices are significantly less expensive, along with being easier to source and use. A commonly employed device for xMD is the SensEpil broad-spectrum flashbulb manufactured by Silk’n [73–75].
There are substantial benefits for xMD compared to the other techniques discussed in this review. This method combines speed and precision without requiring specialized training, making it the most high-throughput of all the microdissection techniques. Furthermore, it is a user-independent procedure, eliminating all subjectivity in determining the ROI(s) [28; 73–75]. Nonetheless, there are still two major challenges that need to be overcome with xMD to make it more practical. First, the commercially available EVA films are cross-linked, making a remarkably stable polymer that tightly adheres to the cells. Failure to obtain the desired analyte off these films can further reduce the yield [unpublished data]. The development of uncross-linked membranes by our lab have led to significant increases in DNA and miRNA yields [unpublished data]. Additionally, antibodies and other molecular tags do not currently exist for every analyte of interest, which makes this technique inapplicable in several cases [66; 71].
Since xMD is still in development, there are few published studies that incorporate it outside of optimization protocols. Given the benefits of xMD compared to the other four methods described in this review, this method shows great promise for becoming an asset in both clinical and basic science biomolecular testing.
Conclusion
Since the twentieth century, histopathological review of FFPE tissues has been the dominant diagnostic tool for the pathologist. More recently, evaluating nucleic acids and proteins extracted from these same tissues has become critical in precision medicine. The methods of collecting tissues span the range of resolutions from bulk collections, to large ROIs, to most recently single cells. Each method has its advantages and disadvantages, as summarized in Table 1, as well as current/potential uses in clinical medicine. All of these techniques are coupled to downstream sequencing or proteomic methods for biomolecular profiling. As we move forward and better understand the genetic variations that drive malignancy, it will be important for laboratories to adopt the best dissection methods for obtaining actionable molecular material to ensure optimal individualized patient care.
Table 1.
Comparison of Tissue Dissection Techniques with Clinical Applications.
| Technique | Advantages | Disadvantages |
|---|---|---|
|
| ||
| Bulk Scraping | • Time efficient | • Imprecise |
| • Inexpensive | ||
| • High-throughput | ||
| • High yield | ||
|
| ||
| Manual Macrodissection | • Time efficient | • Imprecise |
| • Inexpensive | • Expertise required (subjective) | |
| • High-throughput | ||
| • High yield | ||
|
| ||
| Manual Microdissection | • Precision | • Time consuming |
| • Inexpensive | • Labor intensive | |
| • Expertise required (subjective) | ||
| • Low-throughput | ||
| • Low yield | ||
|
| ||
| Laser-Capture Microdissection | • Precision | • Time consuming |
| • Expertise required (subjective) | ||
| • Expensive | ||
| • Low-throughput | ||
| • Low yield | ||
|
| ||
| Expression Microdissection | • Precision | • Limited antibody availability |
| • Time efficient | ||
| • Inexpensive | • Low yield | |
| • High-throughput | ||
Acknowledgments
The authors thank Avi Rosenberg for his critical reading of the manuscript. Figures were created by E.M.W. with BioRender.com.
Funding Sources
Elise M. Walsh is supported by the Sol Goldman Pancreas Cancer Fund, the National Institutes of Health/National Cancer Institute NIH/NCI (P50-CA062924), and the National Institute of General Medical Sciences (T32GM007814). Marc K. Halushka is supported by NIH grants R01HL137811, and R01GM130564.
Abbreviations
- AKT1
AKT serine/threonine kinase 1
- BRAF
B-Raf proto-oncogene, serine/threonine kinase
- EGFR
epidermal growth factor receptor
- ERBB2
Erb-B2 receptor tyrosine kinase 2
- EVA
ethylene-vinyl acetate
- FFPE
formalin-fixed paraffin-embedded
- HRAS
HRAS proto-oncogene, GTPase
- IHC
immunohistochemistry
- KIT
KIT proto-oncogene, receptor tyrosine kinase
- KRAS
KRAS proto-oncogene, GTPase
- LCM
laser-capture microdissection
- LOD
limit of detection
- MMa
manual macrodissection
- MMi
manual microdissection
- NRAS
NRAS proto-oncogene, GTPase
- PIK3CA
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha
- ROI
region of interest
- TP53
tumor protein p53
- xMD
expression microdissection
Footnotes
Conflict of Interest Statement
The authors declare that they have no conflicts of interests.
References
- 1.Nair SR. Personalized medicine: Striding from genes to medicines. Perspectives in Clinical Research 2010. Oct;1(4):146–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nony E, Moingeon P. Proteomics in support of immunotherapy: Contribution to model-based precision medicine. Expert Review of Proteomics 2021. Dec:1–10. [DOI] [PubMed] [Google Scholar]
- 3.Hussen BM, Abdullah ST, Salihi A, Sabir DK, Sidiq KR, Rasul MF, et al. The emerging roles of NGS in clinical oncology and personalized medicine. Pathology - Research and Practice 2022. Feb;230:153760. [DOI] [PubMed] [Google Scholar]
- 4.Rosai J, Ackerman LV. The pathology of tumors, part III: Grading, staging & classification. CA: A Cancer Journal for Clinicians 1979. Apr;29(2):66–77. [DOI] [PubMed] [Google Scholar]
- 5.Alturkistani HA, Tashkandi FM, Mohammedsaleh ZM. Histological stains: A literature review and case study. Global Journal of Health Science 2015. Jun;8(3):72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaffney EF, Riegman PH, Grizzle WE, Watson PH. Factors that drive the increasing use of FFPE tissue in basic and translational cancer research. Biotechnic & Histochemistry 2018;93(5):373–386. [DOI] [PubMed] [Google Scholar]
- 7.Fairley JA, Gilmour K, Walsh K. Making the most of pathological specimens: Molecular diagnosis in formalin-fixed, paraffin embedded tissue. Current Drug Targets 2012. Nov;13(12):1475–1487. [DOI] [PubMed] [Google Scholar]
- 8.McDonough SJ, Bhagwate A, Sun Z, Wang C, Zschunke M, Gorman JA, et al. Use of FFPE-derived DNA in next generation sequencing: DNA extraction methods. PloS One 2019;14(4):e0211400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Leng WWJ, Gadellaa-van Hooijdonk CG, Barendregt-Smouter FAS, Koudijs MJ, Nijman I, Hinrichs JWJ, et al. Targeted next generation sequencing as a reliable diagnostic assay for the detection of somatic mutations in tumours using minimal DNA amounts from formalin fixed paraffin embedded material. PloS One 2016;11(2):e0149405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Locy H, Correa RJM, Autaers D, Schiettecatte A, Jonckheere J, Waelput W, et al. Overcoming the challenges of high quality RNA extraction from core needle biopsy. Biomolecules 2021. Apr;11(5):621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gustafsson OJR, Arentz G, Hoffmann P. Proteomic developments in the analysis of formalin-fixed tissue. Biochimica et Biophysica Acta 2015. Jun;1854(6):559–580. [DOI] [PubMed] [Google Scholar]
- 12.Buczak K, Kirkpatrick JM, Truckenmueller F, Santinha D, Ferreira L, Roessler S, et al. Spatially resolved analysis of FFPE tissue proteomes by quantitative mass spectrometry. Nature Protocols 2020. Sep;15(9):2956–2979. [DOI] [PubMed] [Google Scholar]
- 13.Bass BP, Engel KB, Greytak SR, Moore HM. A review of preanalytical factors affecting molecular, protein, and morphological analysis of formalin-fixed, paraffin-embedded (FFPE) tissue: How well do you know your FFPE specimen? Archives of Pathology & Laboratory Medicine 2014. Nov;138(11):1520–1530. [DOI] [PubMed] [Google Scholar]
- 14.Carithers LJ, Agarwal R, Guan P, Odeh H, Sachs MC, Engel KB, et al. The biospecimen preanalytical variables program: A multiassay comparison of effects of delay to fixation and fixation duration on nucleic acid quality. Archives of Pathology & Laboratory Medicine 2019. Sep;143(9):1106–1118. [DOI] [PubMed] [Google Scholar]
- 15.Chionh F, Gebski V, Al-Obaidi SJ, Mooi JK, Bruhn MA, Lee CK, et al. VEGF-A, VEGFR1 and VEGFR2 single nucleotide polymorphisms and outcomes from the AGITG MAX trial of capecitabine, bevacizumab and mitomycin C in metastatic colorectal cancer. Scientific Reports 2022. Jan;12(1):1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang D, Rolfe PA, Foernzler D, O’Rourke D, Zhao S, Scheuenpflug J, et al. Comparison of two Illumina whole transcriptome RNA sequencing library preparation methods using human cancer FFPE specimens. Technology in Cancer Research & Treatment 2022. Dec;21:15330338221076304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossouw SC, Bendou H, Blignaut RJ, Bell L, Rigby J, Christoffels A. Evaluation of protein purification techniques and effects of storage duration on LC-MS/MS analysis of archived FFPE human CRC tissues. Pathology and Oncology Research 2021;27:622855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selleck MJ, Senthil M, Wall NR. Making meaningful clinical use of biomarkers. Biomarker Insights 2017;12:1177271917715236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szeto CW, Kurzrock R, Kato S, Goloubev A, Veerapaneni S, Preble A, et al. Association of differential expression of immunoregulatory molecules and presence of targetable mutations may inform rational design of clinical trials. ESMO Open 2022. Feb;7(1):100396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eltoum IA, Siegal GP, Frost AR. Microdissection of histologic sections: Past, present, and future. Advances in Anatomic Pathology 2002. Sep;9(5):316–322. [DOI] [PubMed] [Google Scholar]
- 21.Malone ER, Oliva M, Sabatini PJB, Stockley TL, Siu LL. Molecular profiling for precision cancer therapies. Genome Medicine 2020. Jan;12(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Yin X, Wang Q, Song X, Xia W, Mao Q, et al. A novel gene expression signature-based on B-cell proportion to predict prognosis of patients with lung adenocarcinoma. BMC Cancer 2021. Oct;21(1):1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chong PY, Iqbal J, Yeong J, Aw TC, Chan KS, Chui P. Immune response in myocardial injury: In situ hybridization and immunohistochemistry techniques for SARS-CoV-2 detection in COVID-19 autopsies. Frontiers in Molecular Biosciences 2021;8:658932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Becker K Lysate preparation for reverse phase protein arrays. Advances in Experimental Medicine and Biology 2019;1188:21–30. [DOI] [PubMed] [Google Scholar]
- 25.Gullo I, Marques A, Pinto R, Cirnes L, Schmitt F. Morphological control for molecular testing: A practical approach. Journal of Clinical Pathology 2021. May;74(5):331–333. [DOI] [PubMed] [Google Scholar]
- 26.Kokkat TJ, Patel MS, McGarvey D, LiVolsi VA, Baloch ZW. Archived formalin-fixed paraffin-embedded (FFPE) blocks: A valuable underexploited resource for extraction of DNA, RNA and protein. Biopreservation and Biobanking 2013. Apr;11(2):101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stamis SA, Heath EI, Lucas S, Boerner J, Slusher LB. Alcohol dehydrogenase expression patterns in normal prostate, benign prostatic hyperplasia and prostatic adenocarcinoma in African American and Caucasian men. Prostate 2022. Feb. [DOI] [PubMed] [Google Scholar]
- 28.Tangrea MA, Chuaqui RF, Gillespie JW, Ahram M, Gannot G, Wallis BS, et al. Expression microdissection: Operator-independent retrieval of cells for molecular profiling. Diagnostic Molecular Pathology 2004. Dec;13(4):207–212. [DOI] [PubMed] [Google Scholar]
- 29.Liotta LA, Pappalardo PA, Carpino A, Haymond A, Howard M, Espina V, et al. Laser capture proteomics: Spatial tissue molecular profiling from the bench to personalized medicine. Expert Review of Proteomics 2021. Oct;18(10):845–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandey V, Storz P. Targeting the tumor microenvironment in pancreatic ductal adenocarcinoma. Expert Review of Anticancer Therapy 2019. Jun;19(6):473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell 2011. Mar;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 32.Lih C, Harrington RD, Sims DJ, Harper KN, Bouk CH, Datta V, et al. Analytical validation of the next-generation sequencing assay for a nationwide signal-finding clinical trial: Molecular analysis for therapy choice clinical trial. The Journal of Molecular Diagnostics 2017. Mar;19(2):313–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L, Chen M, Lin J, Chen X, Yu X, Chen Z, et al. Identifying a wide range of actionable variants using capture-based ultra-deep targeted sequencing in treatment-naive patients with primary lung adenocarcinoma. International Journal of Clinical and Experimental Pathology 2020;13(3):525–535. [PMC free article] [PubMed] [Google Scholar]
- 34.Petrackova A, Vasinek M, Sedlarikova L, Dyskova T, Schneiderova P, Novosad T, et al. Standardization of sequencing coverage depth in NGS: Recommendation for detection of clonal and subclonal mutations in cancer diagnostics. Frontiers in Oncology 2019;9:851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bova GS, Eltoum IA, Kiernan JA, Siegal GP, Frost AR, Best CJM, et al. Optimal molecular profiling of tissue and tissue components: Defining the best processing and microdissection methods for biomedical applications. Molecular Biotechnology 2005;29(2):119–152. [DOI] [PubMed] [Google Scholar]
- 36.Roy Chowdhuri S, Hanson J, Cheng J, Rodriguez-Canales J, Fetsch P, Balis U, et al. Semiautomated laser capture microdissection of lung adenocarcinoma cytology samples. Acta Cytologica 2012. Dec;56(6):622–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh RR. Next-generation sequencing in high-sensitive detection of mutations in tumors: Challenges advances and applications. The Journal of Molecular Diagnostics Aug;22(8):994–1007. [DOI] [PubMed] [Google Scholar]
- 38.Tsimberidou AM, Fountzilas E, Nikanjam M, Kurzrock R. Review of precision cancer medicine: Evolution of the treatment paradigm. Cancer Treatment Reviews 2020. Jun;86:102019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dubeau L, Chandler LA, Gralow JR, Nichols PW, Jones PA. Southern blot analysis of DNA extracted from formalin-fixed pathology specimens. Cancer Research 1986. Jun;46(6):2964–2969. [PubMed] [Google Scholar]
- 40.Hunt JL, Finkelstein SD. Microdissection techniques for molecular testing in surgical pathology. Archives of Pathology & Laboratory Medicine 2004. Dec;128(12):1372–1378. [DOI] [PubMed] [Google Scholar]
- 41.Klee EW, Erdogan S, Tillmans L, Kosari F, Sun Z, Wigle DA, et al. Impact of sample acquisition and linear amplification on gene expression profiling of lung adenocarcinoma: Laser capture micro-dissection cell-sampling versus bulk tissue-sampling. BMC Medical Genomics 2009. Mar;2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Marchi T, Braakman RBH, Stingl C, van Duijn MM, Smid M, Foekens JA, et al. The advantage of laser-capture microdissection over whole tissue analysis in proteomic profiling studies. Proteomics 2016. May;16(10):1474–1485. [DOI] [PubMed] [Google Scholar]
- 43.Zhuang Z, Bertheau P, Emmert-Buck MR, Liotta LA, Gnarra J, Linehan WM, et al. A microdissection technique for archival DNA analysis of specific cell populations in lesions < 1 mm in size. The American Journal of Pathology 1995. Mar;146(3):620–625. [PMC free article] [PubMed] [Google Scholar]
- 44.Liu A Laser capture microdissection in the tissue biorepository. Journal of Biomolecular Techniques 2010. Sep;21(3):120–125. [PMC free article] [PubMed] [Google Scholar]
- 45.Masago K, Fujita S, Oya Y, Takahashi Y, Matsushita H, Sasaki E, et al. Comparison between fluorimetry (Qubit) and spectrophotometry (NanoDrop) in the quantification of DNA and RNA extracted from frozen and FFPE tissues from lung cancer patients: A real-world use of genomic tests. Medicina 2021. Dec;57(12):1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dang J, Mendez P, Lee S, Kim JW, Yoon J, Kim TW, et al. Development of a robust DNA quality and quantity assessment qPCR assay for targeted next-generation sequencing library preparation. International Journal of Oncology 2016. Oct;49(4):1755–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kresse SH, Namløs HM, Lorenz S, Berner J, Myklebost O, Bjerkehagen B, et al. Evaluation of commercial DNA and RNA extraction methods for high-throughput sequencing of FFPE samples. PloS One 2018;13(5):e0197456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ziraldo R, Shoura MJ, Fire AZ, Levene SD. Deconvolution of nucleic-acid length distributions: A gel electrophoresis analysis tool and applications. Nucleic Acids Research 2019. Sep;47(16):e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stephenson NL, Hornaday KK, Doktorchik CTA, Lyon AW, Tough SC, Slater DM. Quality assessment of RNA in long-term storage: The All Our Families biorepository. PloS One 2020;15(12):e0242404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vermeulen J, De Preter K, Lefever S, Nuytens J, De Vloed F, Derveaux S, et al. Measurable impact of RNA quality on gene expression results from quantitative PCR. Nucleic Acids Research 2011. May;39(9):e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raynal B, Lenormand P, Baron B, Hoos S, England P. Quality assessment and optimization of purified protein samples: Why and how? Microbial Cell Factories 2014. Dec;13:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goelz SE, Hamilton SR, Vogelstein B. Purification of DNA from formaldehyde fixed and paraffin embedded human tissue. Biochemical and Biophysical Research Communications 1985. Jul;130(1):118–126. [DOI] [PubMed] [Google Scholar]
- 53.Zhong A, Tian Y, Zhang H, Lai M. DNA hydroxymethylation of colorectal primary carcinoma and its association with survival. Journal of Surgical Oncology 2018. Apr;117(5):1029–1037. [DOI] [PubMed] [Google Scholar]
- 54.Geiersbach K, Adey N, Welker N, Elsberry D, Malmberg E, Edwards S, et al. Digitally guided microdissection aids somatic mutation detection in difficult to dissect tumors. Cancer Genetics 2016. Feb;209(1–2):42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qi P, Bai Q, Yao Q, Yang W, Zhou X. Performance of automated dissection on formalin-fixed paraffin-embedded tissue sections for the 21-gene recurrence score assay. Technology in Cancer Research & Treatment 2020. Dec;19:1533033820960760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klein O, Clements A, Menzies AM, O’Toole S, Kefford RF, Long GV. BRAF inhibitor activity in V600R metastatic melanoma. European Journal of Cancer 2013. Mar;49(5):1073–1079. [DOI] [PubMed] [Google Scholar]
- 57.Ko AH, Bekaii-Saab T, Van Ziffle J, Mirzoeva OM, Joseph NM, Talasaz A, et al. A multicenter open-label phase II clinical trial of combined MEK plus EGFR inhibition for chemotherapy-refractory advanced pancreatic adenocarcinoma. Clinical Cancer Research 2016. Jan;22(1):61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Faulkner C, Ellis HP, Shaw A, Penman C, Palmer A, Wragg C, et al. BRAF fusion analysis in pilocytic astrocytomas: KIAA1549-BRAF 15–9 fusions are more frequent in the midline than within the cerebellum. Journal of Neuropathology & Experimental Neurology 2015. Sep;74(9):867–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lowry OH, Passonneau JV. Some recent refinements of quantitative histochemical analysis. Current Problems in Clinical Biochemistry 1971;3:63–84. [PubMed] [Google Scholar]
- 60.Ardakani NM, Giardina T, Grieu-Iacopetta F, Tesfai Y, Carrello A, Taylor J, et al. Detection of epidermal growth factor receptor mutations in lung adenocarcinoma: Comparing Cobas 4800 EGFR assay with Sanger bidirectional sequencing. Clinical Lung Cancer 2016. Sep;17(5):e113–e119. [DOI] [PubMed] [Google Scholar]
- 61.Roy-Chowdhuri S, de Melo Gagliato D, Routbort MJ, Patel KP, Singh RR, Broaddus R, et al. Multigene clinical mutational profiling of breast carcinoma using next-generation sequencing. American Journal of Clinical Pathology 2015. Nov;144(5):713–721. [DOI] [PubMed] [Google Scholar]
- 62.Lokhandwala PM, Tseng L, Rodriguez E, Zheng G, Pallavajjalla A, Gocke CD, et al. Clinical mutational profiling and categorization of BRAF mutations in melanomas using next generation sequencing. BMC Cancer 2019. Jul;19(1):665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, et al. Laser capture microdissection. Science 1996. Nov;274(5289):998–1001. [DOI] [PubMed] [Google Scholar]
- 64.Vandewoestyne M, Goossens K, Burvenich C, Van Soom A, Peelman L, Deforce D. Laser capture microdissection: Should an ultraviolet or infrared laser be used? Analytical Biochemistry 2013. Aug;439(2):88–98. [DOI] [PubMed] [Google Scholar]
- 65.Schütze K, Lahr G. Identification of expressed genes by laser-mediated manipulation of single cells. Nature Biotechnology 1998. Aug;16(8):737–742. [DOI] [PubMed] [Google Scholar]
- 66.Bevilacqua C, Ducos B. Laser microdissection: A powerful tool for genomics at cell level. Molecular Aspects of Medicine 2018. Feb;59:5–27. [DOI] [PubMed] [Google Scholar]
- 67.Hipp JD, Cheng J, Hanson JC, Rosenberg AZ, Emmert-Buck MR, Tangrea MA, et al. SIVQ-LCM protocol for the ArcturusXT instrument. Journal of Visualized Experiments 2014. Jul(89). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hipp JD, Johann DJ, Chen Y, Madabhushi A, Monaco J, Cheng J, et al. Computer-aided laser dissection: A microdissection workflow leveraging image analysis tools. Journal of Pathology Informatics 2018;9:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mollee P, Boros S, Loo D, Ruelcke JE, Lakis VA, Cao KL, et al. Implementation and evaluation of amyloidosis subtyping by laser-capture microdissection and tandem mass spectrometry. Clinical Proteomics 2016;13:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Law S, Gillmore JD. When to suspect and how to approach a diagnosis of amyloidosis. The American Journal of Medicine 2022. Jan:S0002–8. [DOI] [PubMed] [Google Scholar]
- 71.Johann DJ, Laun S, Shin IJ, Weigman R, Stephens O, Roberge A, et al. Microdissection methods utilizing single-cell subtype analysis and the impact on precision medicine. Methods in Molecular Biology 2022;2394:93–107. [DOI] [PubMed] [Google Scholar]
- 72.Hammoudeh SM, Hammoudeh AM, Venkatachalam T, Rawat S, Jayakumar MN, Rahmani M, et al. Enriched transcriptome analysis of laser capture microdissected populations of single cells to investigate intracellular heterogeneity in immunostained FFPE sections. Computational and Structural Biotechnology Journal 2021;19:5198–5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Du Q, Yan W, Burton VH, Hewitt SM, Wang L, Hu N, et al. Validation of esophageal squamous cell carcinoma candidate genes from high-throughput transcriptomic studies. American Journal of Cancer Research 2013;3(4):402–410. [PMC free article] [PubMed] [Google Scholar]
- 74.Rosenberg AZ, Armani MD, Fetsch PA, Xi L, Pham TT, Raffeld M, et al. High-throughput microdissection for next-generation sequencing. PloS One 2016;11(3):e0151775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hanson JC, Tangrea MA, Kim S, Armani MD, Pohida TJ, Bonner RF, et al. Expression microdissection adapted to commercial laser dissection instruments. Nature Protocols 2011. Apr;6(4):457–467. [DOI] [PMC free article] [PubMed] [Google Scholar]


