Abstract
Infectious diseases emerge unprecedentedly, posing serious challenges to public health and the global economy. Virulence factors (VFs) enable pathogens to adhere, reproduce and cause damage to host cells, and antibiotic resistance genes (ARGs) allow pathogens to evade otherwise curable treatments. Simultaneous identification of VFs and ARGs can save pathogen surveillance time, especially in situ epidemic pathogen detection. However, most tools can only predict either VFs or ARGs. Few tools that predict VFs and ARGs simultaneously usually have high false-negative rates, are sensitive to the cutoff thresholds and can only identify conserved genes. For better simultaneous prediction of VFs and ARGs, we propose a hybrid deep ensemble learning approach called HyperVR. By considering both best hit scores and statistical gene sequence patterns, HyperVR combines classical machine learning and deep learning to simultaneously and accurately predict VFs, ARGs and negative genes (neither VFs nor ARGs). For the prediction of individual VFs and ARGs, in silico spike-in experiment (the VFs and ARGs in real metagenomic data), and pseudo-VFs and -ARGs (gene fragments), HyperVR outperforms the current state-of-the-art prediction tools. HyperVR uses only gene sequence information without strict cutoff thresholds, hence making prediction straightforward and reliable.
INTRODUCTION
Microbiome is critical to the inner ecosystem of hosts, e.g. humans, animals and plants, as well as to maintain the external environment (1–4). In particular, pathogenic microorganisms cause diseases and even threaten the life of the host by carrying virulence factors (VFs) and/or antibiotic resistance genes (ARGs) (5,6). Accurate and timely identification of VFs and ARGs can effectively guide medical treatments, decrease host morbidity and mortality, and reduce the economic losses in husbandry, aquaculture, etc.
The VFs in pathogenic microorganisms can induce the pathogenicity, with the ability to assist the microorganisms in colonizing their hosts at the cellular level (7). The success of pathogenic microorganisms leans on their power to utilize VFs to cause infection, survive in the hostile host environment and cause the disease. The assorted expressions, organizations and combinations of VFs are answerable for different clinical symptoms of pathogenic infection (8). VFs can be classified into various categories such as adhesion, colonization, exotoxin, endotoxin, iron transport, etc. The different VFs work in concert to enable the pathogenic microorganisms, such as bacteria and fungi, to successfully adhere, reproduce and cause damage to the host cells (9).
The ARGs are barriers to the treatment of pathogenic infections, exacerbating the pathogenic ability of microorganisms (10). Antibiotics have proven effective in treating a variety of microbial infections, especially bacterial infections over the years (11). However, the treatment for bacterial infections is increasingly limited worldwide as bacterial pathogens become increasingly resistant to antibiotics. The previous effective treatment options even do not exist for some patients, such as those caused by multidrug-resistant (MDR) bacteria, which are now unresponsive to conventional first-line treatments (12). Some examples of these MDR bacteria are vancomycin-resistant enterococci, which are global nosocomial pathogens with important clinical implications (13); methicillin-resistant Staphylococcus aureus, which is the leading cause of hospital- and community-acquired infections, leading to severe mortality and morbidity (14); and colistin–carbapenem-resistant Escherichia coli, which develops resistance to the last-resort drug by acquiring ARGs blaNDM-1 and mcr-1 (15).
Despite the different evolutionary pathways, VFs and ARGs have common features that are necessary for pathogenic bacteria to adapt to and survive in a competitive microbial environment. Specifically, both VFs and ARGs are frequently transferred between bacteria through horizontal gene transfer and both utilize similar systems (i.e. two-component systems, efflux pumps, cell wall alterations and porins) to activate or repress the expression of various genes (16). Pathogens can use VFs to cause diseases in their hosts, while they can colonize an environment with selective antibiotic pressure through the acquisition or presence of ARGs (17–19). Therefore, to understand the causal relationship among microbiome composition, function and disease, both VFs and ARGs must be identified.
Recently, the use of bioinformatic tools to predict VFs/ARGs is gaining momentum particularly with the development of high-throughput sequencing technology. Most of these bioinformatic tools fall into two categories: the ‘best hit’ method-based tools and the computational method-based tools. Specifically, the ‘best hit’ methods are currently the primary means to identify VFs or ARGs by comparing gene sequences with existing online databases using programs such as BLAST (20), DIAMOND (21) or Bowtie (22). Basically, these methods align predicted open reading frames from assembled contigs or raw reads to known gene databases and then use an alignment length requirement or sequence similarity cutoffs to predict or assign classes of genes. For example, Underwood et al. (23) proposed the Virulence Searcher tool, which used FingerPRINTScan (24) to compare protein sequences against a curated set of VF sequence motifs. Lakin et al. (25) proposed the AMRPlusPlus tool, which directly aligns short reads to a custom reference database using BWA (26) to predict the presence of ARGs. On the other hand, the computational methods utilizing machine learning or deep learning can potentially learn the statistical patterns of VFs or ARGs, and be able to predict novel ones. For example, Garg and Gupta (27) proposed a bacterial virulent protein prediction method (VirulentPred) based on bilayer cascade support vector machine. Arango-Argoty et al. (28) developed the deep learning models (DeepARG) to offer a more accurate tool for antimicrobial resistance annotation.
Simultaneous prediction of VFs and ARGs can save pathogen surveillance time, especially in situ detection of epidemic pathogens. However, the bioinformatic tools for the identification of ARGs or VFs usually focus on predicting ARGs or VFs independently. To the best of our knowledge, only two tools, including VRprofile (29) and PathoFact (30), are currently available to simultaneously identify/predict VFs and ARGs utilizing the ‘best hit’ approach. In particular, VRprofile enables the identification of VFs and/or ARGs and their transfer-associated gene clusters by performing homology searches on the genome sequences of pathogenic bacteria. PathoFact combined different existing modules and databases to build a pipeline for the identification of VFs, toxin genes and ARGs.
It is noted that the ‘best hit’ approaches for simultaneously predicting ARGs and VFs can only identify conserved VFs/ARGs, but fail to identify novel VFs/ARGs that are evolutionary distant from known VFs/ARGs. Also, these approaches typically have low false-positive rates (31), i.e. few non-VFs and non-ARGs are predicted to be VFs and ARGs, but high false-negative rates (32,33), i.e. a large number of actual VFs and ARGs are predicted to be non-VFs and non-ARGs. In addition, these approaches are very sensitive to the cutoff threshold: when the cutoff value is high, the predicted results have high precision but low recall; when the cutoff value is low, the predicted results have high recall but low precision, making ‘best hit’ approaches impossible for novice bioinformaticians to decide on an appropriate cutoff threshold.
To address the limitations of current best hit approaches used in simultaneous identification of VFs and ARGs, we proposed a hybrid computational deep ensemble learning approach called HyperVR by potentially learning the traditional best hit scores and statistical patterns of VFs and ARGs at the same time. Specifically, HyperVR integrates multiple key genetic features, including bit score-based similarity feature, physicochemical property-based features, evolutionary information-based features and one-hot encoding feature, and then combines classical ensemble learning methods and deep learning for training and prediction (see Figure 1). HyperVR was validated using the 5-fold cross-validation method, resulting in good performance for predicting ARGs, VFs and negative genes (neither VFs nor ARGs) simultaneously and individually. HyperVR was also validated using novel VFs and ARGs, in silico spike-in experiment (the VFs and ARGs in real metagenomic data), and pseudo-VFs and -ARGs (gene fragments). In all of the above experiments, HyperVR showed better predictive capability, outperforming the current state-of-the-art tools in terms of precision and recall. Finally, three specific pathogen strain datasets were selected to test the ability of HyperVR to simultaneously identify ARGs and VFs in real pathogenic bacteria, and HyperVR accurately annotated a large number of VFs and ARGs.
Figure 1.
Schematic illustration of HyperVR for simultaneous prediction of ARGs and VFs. (A) The overview of HyperVR. The first step was to collect and organize the dataset, including removing characters other than the 20 natural amino acid abbreviations from the sequences (remove X) and using CD-HIT to remove identical or duplicate sequences from the three types of samples to ensure the accuracy of the model prediction results. The second step is to predict whether the gene is an ARG, including feature extraction (generation of bit score-based similarity feature using the DIAMOND program and evolutionary information-based features using the BLAST program) and stacking model training and prediction (classical ensemble learning and deep learning); if the judgment result is no, go to the third step; otherwise, output the prediction score of the gene belonging to ARGs. The third step is then to predict whether the gene is a VF, again including feature extraction (generation of sequence-based features, one-hot encoding feature and evolutionary information-based features using the BLAST program) and stacking model training and prediction (classical ensemble learning, boosting learning and deep learning); if the judgment result is no, output the prediction score of the gene belonging to negative samples (NSs); otherwise, output the prediction score of the gene belonging to VFs. (B) The detailed procedures for data processing. Yellow represents the ARG dataset, blue represents the VF dataset and orange represents the NS dataset; 2000 ARGs, VFs and NSs in the UniProt database were used for model training and validation to address the data imbalance; 209 ARGs, VFs and NSs in the UniProt database were used for the independent test dataset. (C) The detailed flowchart of HyperVR. The top half of the flowchart can be used individually to predict ARGs (HyperVR-ARGs) and the bottom half of the flowchart can be used individually to predict VFs (HyperVR-VFs).
In summary, the novelty of HyperVR mainly includes the following aspects: (i) in application: to the best of our knowledge, HyperVR is the first computational tool to simultaneously predict VFs and ARGs in microbial data; (ii) in theory: HyperVR is the first to integrate multiple key genetic features, including bit score-based similarity feature, physicochemical property-based features, evolutionary information-based features and one-hot encoding feature, and then combines classical ensemble learning methods and deep learning to predict VFs and ARGs; and (iii) in performance: HyperVR addresses the limits of traditional alignment-based methods and is more effective and robust than the current state-of-the-art tools. In general, researchers can filter ARGs and VFs in real pathogenic strains based on HyperVR’s prediction scores, which narrows down the scope to conduct biological experiments and greatly saves their time and effort.
MATERIALS AND METHODS
Data collection and annotation
Antibiotic resistance genes
The original ARGs in this work were first derived from DeepARG-DB (28), including three major databases: UniProt (34), CARD (35) and ARDB (36). For the UniProt dataset, all genes containing the antibiotic resistance keyword (KW-0046) or a metadata description were selected and further annotated through a manual inspection and text mining, and sequences annotated as conferring resistance by single-nucleotide polymorphisms were removed. During the construction of the dataset, we followed the pre-established experimental approaches described in (28,37) by using the CD-HIT (38) to cluster all 100% identical or duplicate ARG and VF sequences, and then keeping one of the fully identical or duplicate genes in the final database. The non-identical or non-duplicate sequences would not cause bias or label leakage in our model due to the fact that genes of the same type are inherently similar, some with high similarity and some with slightly lower similarity, and this can be considered as similarity characteristics between genes. Highly similar genes are not equivalent to duplicate genes, and the model must also be able to make accurate judgments about these genes. Finally, 14 933 ARGs were retained for downstream analysis, including 10 602 UniProt, 2203 CARD and 2128 ARDB.
Virulence factors
The original VFs in this work were collected from four public databases: PATRIC (39), Victors (40), VFDB (8) and UniProt (41). Specifically, we first downloaded 1293, 4964, and 28 913 VFs from PATRIC, Victors and VFDB, respectively. For the UniProt dataset, 4085 genes that contained the virulence factor keyword (KW-0843) were selected. Second, all sequences from the four datasets (PATRIC + Victors + VFDB + UniProt) were clustered using CD-HIT similar to the ARG operation by setting the identity parameter to 100%. Finally, 33 154 VFs were retained for downstream analysis, including 3887 PATRIC, 615 Victors, 26 443 VFDB and 2209 UniProt.
Negative samples
For NSs, i.e. genes that were neither ARGs nor VFs, we first randomly collected 20 000 genes from UniProt (41) (3.5% of the original UniProt genes). Second, all the ARGs and VFs mentioned earlier and genes containing the virulence or antibiotic keywords (Supplementary Table S1) were removed. Third, the remaining genes and all the ARGs and VFs mentioned earlier were clustered using CD-HIT to ensure maximum cleanliness of the NSs. Finally, 4880 genes were obtained as NSs.
HyperVR-DB
The resulting database, HyperVR-DB, comprises 52 967 genes, including 14 933 ARGs, 33 154 VFs and 4880 NSs. To make reasonable use of the bit score-based similarity feature, we divided the ARG and VF dataset into two major parts: 10 602 ARGs and 2209 VFs in the UniProt database collected manually by us as much as possible were used for model training and validation; ARGs and VFs in other public datasets were used to represent known ARGs and VFs. Finally, to address the data imbalance problem and avoid the prediction bias of the model, we selected an equal number of 2209 ARGs and VFs in the UniProt. On this basis, 2000 ARGs, VFs and NSs in the UniProt database were used for model training and validation; 209 ARGs, VFs and NSs in the UniProt database were used for the independent test dataset; genes in the remaining database were used to represent known ARGs and VFs (see Figure 1B).
Feature extraction
To obtain a superior predictive power for ARGs and VFs, HyperVR considers a variety of gene-related features, including the following five categories: bit score-based similarity feature, sequence information-based features, physicochemical property-based features, evolutionary information-based features and one-hot encoding feature. Detailed feature descriptions are presented in the following subsections.
Bit score-based similarity feature
The bit score-based similarity feature (28) consists of the bit scores between full gene length sequences and known ARGs and VFs, which considers the similarity distribution of sequences in the ARG and VF databases, not just the best hits. The bit score is used as a similarity metric because it considers the identity extent between sequences and, unlike the e-value, it is independent of the size of the database (42). In this work, we chose the DIAMOND program, which is faster than BLAST, to align the gene sequences in the training dataset with the remaining known 12 724 (14 933 − 2209) ARGs and 30 945 (33 154 − 2209) VFs used for comparison in HyperVR-DB under the more sensitive parameter. It should be noted that the training dataset has been de-duplicated using CD-HIT program with the dataset used for comparison to avoid the possibility of label leakage (refer to the ‘Data collection and annotation’ section). Then, the bit scores are normalized to the [0, 1] interval to represent the similarity of the sequences in terms of distance. Finally, the bit score-based similarity feature of each gene sequence in the training dataset is transformed into a fixed 12 724 + 30 945 = 43 669-dimensional feature vector, where each dimension is the bit score output by DIAMOND program between full gene length sequences and each available ARG and VF in the comparison dataset. The feature vector contains information about the length of the query sequence, and the similarity features differ for query sequences of different lengths. Subsequently, the deep learning model could discriminates relevant features without the need of human intervention and was trained by taking into account the identity distance distribution of a sequence to all known ARGs and VFs.
Sequence-based features
Amino acid composition
The amino acid composition (AAC) feature (43) indicates the frequencies of 20 natural amino acids (i.e. ‘ACDEFGHIKLMNPQRSTVWY’) in a protein or peptide sequence and can be calculated as follows:
 |
(1) |
where N(a) denotes the number of a given amino acid a, N denotes the sequence length of the protein or peptide, and f(a) denotes the final generated 20-dimensional feature vector.
Dipeptide composition
The dipeptide composition (DPC) feature (44) indicates the frequencies of dipeptide in a protein or peptide sequence and can be calculated as follows:
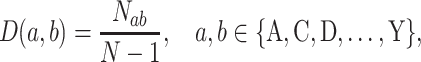 |
(2) |
where Nab denotes the number of a given dipeptide ab, N denotes the sequence length of the protein or peptide, and D(a, b) denotes the final generated 20 × 20 = 400-dimensional feature vector.
Dipeptide deviation from expected mean
The dipeptide deviation from expected mean (DDE) feature (44) is a combination of three features: theoretical mean (TM), DPC and theoretical variance (TV). Specifically, the TM feature is calculated as follows:
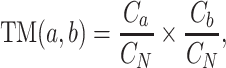 |
(3) |
where Ca and Cb, respectively, denote the codon numbers encoding amino acids a and b, and CN equals 61, denoting the total number of possible codons without including the three stop codons. The calculation of DPC feature refers to the previous description. The TV feature is calculated as follows:
 |
(4) |
Finally, DDE(a, b) is calculated as follows:
 |
(5) |
Physicochemical property-based features
Pseudo-amino acid composition
The pseudo-amino acid composition (PAAC) feature (45,46) contains two aspects. First, the original side chain masses, hydrophobicity and hydrophilicity of the 20 natural amino acids are defined as Mo(i), 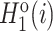 and
and 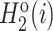 for
for  , respectively. Second, they are normalized as follows:
, respectively. Second, they are normalized as follows:
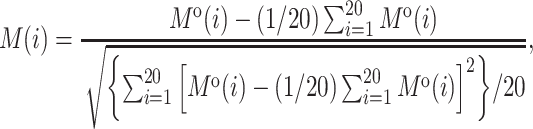 |
(6) |
where 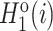 and
and 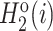 are normalized in the same way. Third, the correlation between Ri and Rj that possess a set of n amino acid properties is defined as follows:
are normalized in the same way. Third, the correlation between Ri and Rj that possess a set of n amino acid properties is defined as follows:
 |
(7) |
where Hk(Ri) denotes the kth amino acid property of Ri. Fourth, a set of sequence order-correlated factors is defined as follows:
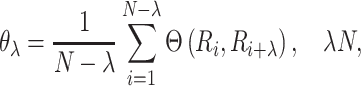 |
(8) |
where λ denotes a positive integer and N is the integer used to define the maximum value of λ. Finally, the PAAC feature for a protein sequence is defined as follows:
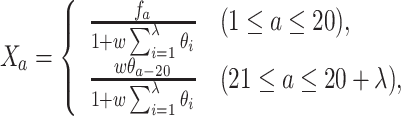 |
(9) |
where fa denotes the normalized frequency of occurrence of amino acid a in the protein sequence and 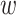 denotes the weighting factor for the sequence-order effect.
denotes the weighting factor for the sequence-order effect.
Quasi-sequence order
The quasi-sequence order (QSO) feature utilizes two specific distance matrices to describe the occurrence probability of amino acids in a protein sequence, including the Schneider–Wrede physicochemical distance matrix (47) and the chemical distance matrix (48). Specifically, the dth rank sequence-order-coupling number is first defined as follows:
 |
(10) |
where N denotes the protein or peptide sequence length, di,i+d denotes the element in row i and column i + d of the distance matrix and nlag denotes the maximum lag value. Then, the first 20 QSO features can be defined as follows:
 |
(11) |
Furthermore, the other 30 QSO features are defined as follows:
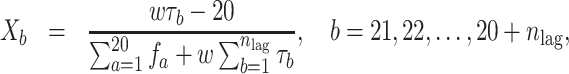 |
(12) |
where fa denotes the normalized frequency of occurrence of amino acid a in the protein sequence and 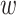 denotes the weighting factor.
denotes the weighting factor.
Evolutionary information-based features
The position-specific scoring matrix (PSSM) consists of a set of probability scores for each amino acid (or gap) at each position in the alignment table and is used to estimate the evolutionary conservation of genes. The basic idea of PSSM is to match query sequences in a database to sequences in an alignment table, giving higher weights to conserved positions than to variable positions. In recent years, the PSSM profiles have been successfully used in various fields, including identifying functional residues, binding residues and proteins of different fold types, etc. In this work, we generated the original PSSM profiles by utilizing the PSI-BLAST program (version blast-2.12.0) (20) to iteratively (three times) search distantly related homologous sequence of proteins against the database UniRef50 (49) with a specified e-value score (0.001).
PSSM composition
The PSSM composition feature (50) eliminates the variability introduced by protein sequence length by summing and averaging all rows of the original PSSM profile for each naturally occurring amino acid type, and is defined as follows:
 |
(13) |
subject to
 |
(14) |
where Ri denotes the ith row of the PSSM composite feature matrix, rk denotes the kth row of the normalized PSSM profile, pk denotes the kth amino acid in the protein sequence and ai denotes the ith amino acid of the 20 standard amino acids.
RPM-PSSM
The RPM-PSSM feature (51) transforms the original PSSM by filtering the negative values to 0 and leaving the positive values unchanged. The idea of the method is derived from the residue probing method, where each amino acid corresponding to a specific column in the PSSM is considered as a probe. Ultimately, the original PSSM is transformed into a 400-dimensional feature vector, and is defined as follows:
 |
(15) |
subject to
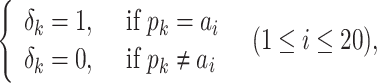 |
(16) |
where Mi denotes the ith row of the RPM-PSSM feature matrix, mk denotes the kth row of the original PSSM profile, pk denotes the kth amino acid in the protein sequence and ai denotes the ith amino acid of the 20 standard amino acids.
AADP-PSSM
The AADP-PSSM (52) feature extends the traditional AAC and DPC concepts to PSSM. First, the AAC-PSSM is transformed into a fixed-length 20-dimensional feature vector by averaging the columns of the original PSSM profile, defined as follows:
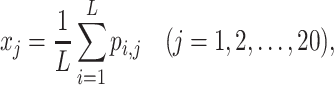 |
(17) |
where xj denotes the jth row of the AAC-PSSM feature matrix, representing the average proportion of amino acid mutations in the evolutionary process, and pi,j denotes the entity in row i and column j of the original PSSM profile. Second, the DPC-PSSM is transformed into a fixed-length 400-dimensional feature vector to avoid information loss due to X in the protein, defined as follows:
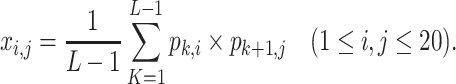 |
(18) |
Finally, the AADP-PSSM was transformed into a fixed-length 20 + 400 = 420-dimensional feature vector by combining these two components.
One-hot encoding feature
The one-hot encoding (53) feature conveniently converts protein sequences into numerical vectors as inputs for deep learning, where each of the 20 natural amino acids is converted into a 20-dimensional feature vector, with the amino acid locations set to 1 and the remaining locations set to 0 in alphabetical order. In addition, the gene sequence lengths were intercepted uniformly at 2000, as the distribution of gene sequence lengths in our dataset mostly falls within this range. Finally, each gene sequence is transformed into a 20 × 2000 = 40 000-dimensional feature vector.
Hybrid model training and stacking
To obtain superior predictive performance for ARGs and VFs, HyperVR ensembles the power of classical machine learning methods and deep learning in a stacking strategy (see Figure 1C). Stacking is an ensemble learning technique that has been proven to have better prediction results in several fields and is first recommended in many high-level competitions (54,55). It integrates multiple base-level classification or regression models through a single meta-classifier or meta-regressor. The base-level model does the training using the entire training dataset, and the meta-model uses the output of the base-level models as features for training. To address the overfitting phenomenon in the final prediction, we further utilized the 5-fold cross-validation method to, respectively, train the base-level models. The detailed training process is shown in Supplementary Figure S1.
In particular, classical machine learning methods in HyperVR are mainly chosen to produce better predictive performance with ensemble learning methods, including Random Forest (56), Extra Trees classifier (57), Xgboost (58), GradientBoosting (59) and Adaboost (60). The stacking algorithm in HyperVR is represented by the pseudocode shown in Algorithm 1.

In detail, the sklearn package in Python and the corresponding default parameters are adopted for Random Forest, Extra Trees classifier, Adaboost and GradientBoosting in our work. Furthermore, the stand-alone Xgboost package in Python is adopted, and the maximum number of estimators to terminate the boost is set to 500, the booster is selected to GBTree and the other parameters are set to their default values. In this work, deep learning is used to automatically target the training of two classes of the bit score-based similarity and one-hot encoding features that do not require prior knowledge. Specifically, we construct the deep learning framework containing six hidden layers, where the number of neurons is, respectively, 212, 210, 28, 26, 24 and 22. Furthermore, a dropout layer follows each hidden layer to avoid overfitting, and the dropout rate is set to 0.05. The model ends with the sigmoid layer, which is used to output the final predicted scores. In addition, the framework is written by the TensorFlow program and compiled with the following parameters, where the optimizer is the stochastic gradient descent method with a learning rate of 0.05, the loss function is BinaryCrossentropy, the evaluation metric is BinaryAccuracy, the training epoch is 500 and early stopping mechanism is added.
Evaluation criteria
In this work, HyperVR was evaluated using the common multilabel standard performance metric, including precision, recall, F1 score and their micro-averages. The specific formulas are defined as follows:
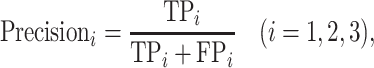 |
(19) |
 |
(20) |
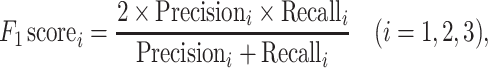 |
(21) |
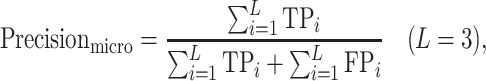 |
(22) |
 |
(23) |
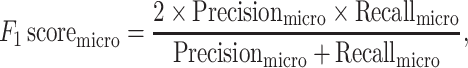 |
(24) |
where TP (true positive), FP (false positive), TN (true negative) and FN (false negative), respectively, denote the number of positive samples correctly labeled, negative samples incorrectly labeled, negative samples correctly labeled and positive samples incorrectly labeled. The i = 1, 2, 3, respectively, represents the ARG, VF and NS categories, and L = 3 is the total number of categories.
RESULTS AND DISCUSSION
Implementation details
The classic machine learning model and deep learning network of HyperVR were, respectively, implemented on Python 3.8 with Scikit-learn 1.0.1 and Keras 2.8.0 with Tensorflow-gpu 2.8.0 backend. In addition, we utilized Keras multi-GPU processing to increase the training speed significantly. The experiments were performed on a Linux system server with 16x Intel® Xeon® Bronze 3106 CPUs (1.70 GHz) featuring 128 CPU cores in total, 4x NVIDIA V100 GPUs and 250 GB of RAM.
HyperVR can accurately predict ARGs, VFs and NSs simultaneously
To accurately evaluate the performance of HyperVR in predicting ARGs, VFs and NSs simultaneously, the 5-fold cross-validation method was employed in this section, resulting in an average accuracy of 91.94%. We implemented the rigorous procedure in the cross-validation step to enable an unbiased evaluation of the effectiveness of HyperVR. After training HyperVR with 80% of the selected raw data from HyperVR-DB, the remaining 20% held-out data were utilized to evaluate its generalization capabilities, and so on for five trials repeated (Figure 2A).
Figure 2.
The prediction results of HyperVR to simultaneously predict ARGs, VFs and NSs under 5-fold cross-validation. (A) The dataset used by HyperVR under 5-fold cross-validation. (B) The confusion matrix generated by HyperVR under 5-fold cross-validation. (C) The histogram with error bar of evaluation metrics under 5-fold cross-validation.
We recorded detailed classification reports, including accuracy, precision, recall, F1 score and micro-average metrics (Supplementary Table S2), and plotted confusion matrices (Supplementary Figure S2) for each fold. The mean and standard deviation of all metrics derived from all five cross-validation experiments were calculated and reported in Table 1. Figure 2B and C, respectively, visualizes the final confusion matrix generated in the experiment and the error bar histograms of three evaluation indicators. All the results reported above demonstrate that HyperVR can accurately classify ARGs, VFs and NSs at the same time, especially with the excellent performance of 99.85% precision and 99% recall for ARGs, and 89.76% accuracy and 86.45% recall for VFs. Furthermore, the standard deviation of 0.65% for the micro-average results under five independent experiments further demonstrates the stability of HyperVR.
Table 1.
The detailed results of HyperVR to simultaneously predict ARGs, VFs and NSs under 5-fold cross-validation
| Precision | Recall | F 1 score | |
|---|---|---|---|
| ARGs | 0.9985a ± 0.0022b | 0.9900 ± 0.0039 | 0.9942 ± 0.0018 |
| VFs | 0.8976 ± 0.0171 | 0.8645 ± 0.0211 | 0.8805 ± 0.0113 |
| NSs | 0.8653 ± 0.0149 | 0.9040 ± 0.0155 | 0.8841 ± 0.0085 |
| Accuracy | 0.9194 ± 0.0065 | ||
| Macro-average | 0.9204 ± 0.0063 | 0.9194 ± 0.0065 | 0.9196 ± 0.0065 |
a The mean of all metrics under 5-fold cross-validation.
b The standard deviation of all metrics under 5-fold cross-validation.
Two-step strategy for more accurate simultaneous predictions
We consider two main advantages to use the two-step strategy. First, the two-step strategy allows for greater model flexibility, allowing users to run different parts of the model separately for predicting specific VFs or ARGs individually. Second, the two-step strategy allows the model to, respectively, select different features as well as classification methods for VFs and ARGs, thus annotating them more accurately. To demonstrate the advantages of the two-step strategy, we take an example of bit score-based similarity feature and then use deep learning method for multiclass classification. The bit score-based similarity feature consists of the bit scores between full gene length sequences and known ARGs and VFs, which considers the similarity distribution of sequences in the ARG and VF databases, not just the best hits. The process of obtaining this feature is described in detail in the ‘Feature extraction’ section. After that, a deep learning multiclass model was trained by taking into account the 4/5 ARGs, VFs and NSs in the UniProt. Finally, the output layer of the deep neural network consists of three units that correspond to the three categories and uses a softMax activation function that predicts the probability of the remaining 1/5 input sequence against three categories. Table 2 shows the prediction results of the deep learning multiclass model with the bit score-based similarity feature. From the results, it can be seen that the bit score-based similarity feature is significant for the classification of ARGs when the deep neural network converges. However, the feature is poorly effective in discriminating between VFs and NSs, posing difficulties for their classification. The two-step strategy can solve the problem well by selecting different features and classification methods for different categories of classification, so as to achieve better classification results in our manuscript.
Table 2.
The prediction results of the deep learning multiclass model with the bit score-based similarity feature
| Precision | Recall | F 1 score | |
|---|---|---|---|
| ARGs | 0.9963 | 0.9905 | 0.9927 |
| VFs | 0.8255 | 0.4797 | 0.6032 |
| NSs | 0.6335 | 0.9032 | 0.7479 |
| Accuracy | 0.7865 | ||
| Macro-average | 0.8190 | 0.7845 | 0.7789 |
Comparison experiment between HyperVR and the published individual tools
Furthermore, we have further compared HyperVR used by the two-step strategy for ARGs (HyperVR-ARGs) and VFs (HyperVR-VFs) with the published individual tools, say, HMD-ARG (37) for ARGs and VFanalyzer (8) for VFs. The same novel dataset as in the ‘Validation of HyperVR through novel ARGs, VFs and NSs’ section was used, and we first annotated the novel ARGs in the dataset using HMD-ARG (http://www.cbrc.kaust.edu.sa/HMDARG/), followed by the VFs using VFanalyzer (http://www.mgc.ac.cn/cgi-bin/VFs/v5/main.cgi?func=VFanalyzer). It should be noted that for VFanalyzer, we conducted 32 sets of experiments corresponding to a total of the 32 genera of the genome in the method, so as to ensure the accuracy of VFanalyzer results as much as possible. Figure 3 shows the comparison results of the individual tools with HyperVR, including the final confusion matrix, precision, recall and F1 score. From the figure, we can see that the prediction results of HyperVR for novel ARGs are better than the latest independent computational tool HMD-ARG, probably due to the fact that HMD-ARG only considers the one-hot feature of the gene sequence, and the prediction of novel VFs is much better than that of the independent alignment tool VFanalyzer, which once again confirms the drawback of high false-negative rate of the current ‘best hit’ tools.
Figure 3.
The comparison results between HyperVR and the published individual tools.
HyperVR-ARGs can flexibly and accurately predict ARGs individually
To further evaluate the performance of HyperVR in predicting ARGs individually (shorthand HyperVR-ARGs), the same 5-fold cross-validation method was employed in this section, resulting in an excellent average cross-validation accuracy of 99.85%. The detailed cross-validation results for each part in HyperVR-ARGs, including accuracy (Acc.), sensitivity (Sen.), specificity (Spec.), precision (Prec.), Matthews correlation coefficient (MCC) and the area under the receiver operating characteristic (ROC) curve (AUC), were recorded in Supplementary Table S3. The final results of HyperVR-ARGs under 5-fold cross-validation are shown in Table 3. On the other hand, we plotted the ROC curve (Figure 4A) and the precision–recall (PR) curve (Figure 4B) to visualize the performance of HyperVR-ARGs. Figure 4C visualized the training process of the DNN for the bit score-based similarity feature. Figure 4D further visualized the prediction results of each part in HyperVR-ARGs under 5-fold cross-validation through the error bar histogram. The detailed confusion matrices of HyperVR-ARGs under 5-fold cross-validation are shown in Supplementary Figure S3.
Table 3.
The 5-fold cross-validation results of HyperVR-ARGs to predict ARGs individually
| Fold | Acc. | Sen. | Spec. | Prec. | MCC | AUC |
|---|---|---|---|---|---|---|
| 1st | 0.9967 | 0.9950 | 0.9975 | 0.9950 | 0.9925 | 0.9985 |
| 2nd | 0.9958 | 0.9925 | 0.9975 | 0.9950 | 0.9906 | 0.9984 |
| 3rd | 0.9958 | 0.9875 | 1.0000 | 1.0000 | 0.9906 | 0.9984 |
| 4th | 0.9967 | 0.9950 | 0.9975 | 0.9950 | 0.9925 | 0.9985 |
| 5th | 0.9942 | 0.9825 | 1.0000 | 1.0000 | 0.9869 | 0.9984 |
| Average ± SD | 0.9958 ± 0.0010 | 0.9905 ± 0.0054 | 0.9985 ± 0.0014 | 0.9970 ± 0.0027 | 0.9906 ± 0.0023 | 0.9984 ± 0.0001 |
Figure 4.
The 5-fold cross-validation performance for HyperVR-ARGs to predict ARGs individually. (A) The ROC curves and interpolated AUC of HyperVR-ARGs under 5-fold cross-validation. (B) The PR curves and interpolated area under PR curve (AUPR) of HyperVR-ARGs under 5-fold cross-validation. (C) The training process of DNN for the bit score-based similarity feature in HyperVR-ARGs. (D) The prediction results of each part in HyperVR-ARGs under 5-fold cross-validation.
The predictive power of HyperVR-ARGs comes from two contributions, including traditional machine learning methods (Extra Trees classifier) utilizing gene evolutionary information in the form of PSSM and DNN utilizing bit score-based similarity features of full gene sequences. From the results in this section, we can see that the DNN can quickly reach high accuracy (∼50 epochs) and converge quickly with the early stopping mechanism, which tells us that using the combination of DNN and bit score-based similarity feature is fast and effective for identifying ARGs. Second, the comparison results in Figure 4D show that excellent performance in identifying ARGs can also be achieved using traditional machine learning methods and the corresponding PSSM features, especially the combination of Extra Trees and RPM-PSSM. Finally, HyperVR-ARGs can achieve better predictive performance by combining the capabilities of different combinations. In summary, HyperVR-ARGs can flexibly and accurately predict ARGs individually.
HyperVR-VFs can flexibly and accurately predict VFs individually
To further estimate the predictive ability of HyperVR in predicting VFs individually (shorthand HyperVR-VFs), the 5-fold cross-validation in this section was performed again, resulting in a good average cross-validation accuracy of 91.83%. The same evaluation strategy as in the ‘HyperVR-ARGs can flexibly and accurately predict ARGs individually’ section was adopted. The detailed cross-validation results for each part in HyperVR-VFs are shown in Supplementary Table S4. The final stacking results of HyperVR-VFs under 5-fold cross-validation are shown in Table 4. We also calculated the AUC of the ROC curves and PR curves of HyperVR-VFs under 5-fold cross-validation, as shown in Figure 5A and B. Figure 5C illustrates the training process of the DNN for the one-hot encoding feature. Figure 5D visualizes the predicted AUC results of each part in HyperVR-VFs under 5-fold cross-validation through the error bar histogram. The detailed confusion matrices of HyperVR-VFs under 5-fold cross-validation are shown in Supplementary Figure S4.
Table 4.
The 5-fold cross-validation results of HyperVR-VFs to predict VFs individually
| Fold | Acc. | Sen. | Spec. | Prec. | MCC | AUC |
|---|---|---|---|---|---|---|
| 1st | 0.9250 | 0.8750 | 0.9500 | 0.8974 | 0.8303 | 0.9767 |
| 2nd | 0.9200 | 0.8425 | 0.9587 | 0.9108 | 0.8179 | 0.9758 |
| 3rd | 0.9167 | 0.8625 | 0.9437 | 0.8846 | 0.8115 | 0.9717 |
| 4th | 0.9133 | 0.8200 | 0.9600 | 0.9111 | 0.8024 | 0.9695 |
| 5th | 0.9167 | 0.8525 | 0.9487 | 0.8927 | 0.8108 | 0.9671 |
| Average ± SD | 0.9183 ± 0.0044 | 0.8505 ± 0.0208 | 0.9522 ± 0.0069 | 0.8993 ± 0.0115 | 0.8145 ± 0.0103 | 0.9722 ± 0.0040 |
Figure 5.
The 5-fold cross-validation performance for HyperVR-VFs to predict VFs individually. (A) The ROC curves and interpolated AUC of HyperVR-VFs under 5-fold cross-validation. (B) The PR curves and interpolated AUPR of HyperVR-VFs under 5-fold cross-validation. (C) The training process of DNN for the one-hot encoding feature in HyperVR-VFs. (D) The predicted AUC results of each part in HyperVR-VFs under 5-fold cross-validation.
The predictive power of HyperVR-VFs comes from more contributions, including traditional ensemble learning methods utilizing sequence-based features, physicochemical property-based features, and gene evolutionary information in the form of the PSSM and DNN utilizing the one-hot encode features. From the results in this section, we can see that the DNN reached best performance and converge on the validation dataset after ∼210 epochs with the early stopping mechanism. The evolutionary information of genes contributes more than sequence-based features, physicochemical property-based features and one-hot encode features for the prediction of VFs. Last but not least, the stacking of these multiple combinations can yield better prediction performance than any one of them. It can be concluded that HyperVR-VFs have a strong and stable ability to predict VFs individually.
Validation of HyperVR through novel ARGs, VFs and NSs
To test the predictive ability of HyperVR on novel ARGs, VFs and NSs, an independent dataset in HyperVR-DB was obtained consisting of 209 ARGs, 209 VFs and 209 NSs. Notably, both the novel genes (test dataset) and the training dataset are derived from the Swiss-Prot dataset in UniProt, a hand-checked, non-redundant, protein dataset with detailed annotation information. Moreover, we also introduced all the current state-of-the-art prediction tools that can be used to predict ARGs and VFs simultaneously: VRprofile (29), PathoFact (30) and the traditional ‘best hit’ approaches as baseline comparison methods. Since both VRprofile and ‘best hit’ methods require a choice of thresholds, we choose three different thresholds for the experiments, respectively, and compare their experimental results with those of HyperVR. Specifically, we chose Ha-value parameters of 0.21, 0.64 and 0.81 for VRprofile. The integrated web interface (https://bioinfo-mml.sjtu.edu.cn/VRprofile/) and the subject dataset (MobilomeDB) such as those listed on the VRprofile website were utilized for consistency and convenience. For the best hit approach, we utilized the DIAMOND program and chose the identity parameters of 0.21 (Diamond-21%), 0.64 (Diamond-64%) and 0.81 (Diamond-81%) for consistency. The known ARGs and VFs (genes in the ARDB, CARD, PATRIC, Victors and VFDB database) in our HyperVR-DB dataset were used as the subject database, which are same as those used for HyperVR and independent from the novel ARGs, VFs and NSs in UniProt.
Table 5 shows the detailed prediction results of HyperVR and three different baseline methods under different parameters for these novel ARGs, VFs and NSs. It should be noted that we find that VRprofile obtains the same prediction results for three different Ha-value parameters, so we only list one of the results. For those that PathoFact cannot determine whether they are VFs or not (the prediction result is unclassified), we treat them as NS categories. Figure 6 visualized the confusion matrices and histograms of results for different baseline methods. Thus, we can draw the following tentative conclusions from the foregoing results. VRprofile and PathoFact have good precision and recall for identifying novel ARGs, but very poor identification of VFs. The best hit approach using the DIAMOND program is sensitive to the identity cutoffs. When the identity cutoff is relatively small, the best hit approach is relatively good at identifying ARGs but still poor at identifying VFs. By increasing the identity cutoff, the identification precision of ARGs and VFs will be improved, but at the same time the recall will be greatly reduced. Overall, the best hit approach does not yield an overall superior prediction for the novel ARGs and VFs. In contrast, HyperVR achieved excellent overall performance in predicting both novel ARGs and VFs simultaneously without parameter selection. This again proves that HyperVR can be used as an effective and simple tool for the identification of ARGs and VFs.
Table 5.
The prediction results of HyperVR and baseline methods for novel ARGs, VFs and NSs
| Precision | Recall | F 1 score | Precision | Recall | F 1 score | ||||
|---|---|---|---|---|---|---|---|---|---|
| ARGs | 0.9801 | 0.9426 | 0.9610 | ARGs | 0.9533 | 0.9761 | 0.9645 | ||
| VFs | 0.5490 | 0.1340 | 0.2154 | VFs | 0.5189 | 0.4593 | 0.4873 | ||
| VRprofilea | NSs | 0.5227 | 0.9378 | 0.6712 | Diamond-21%b | NSs | 0.5175 | 0.5646 | 0.5400 |
| Accuracy | 0.6715 | Accuracy | 0.6667 | ||||||
| Macro-average | 0.6839 | 0.6715 | 0.6159 | Macro-average | 0.6632 | 0.6667 | 0.6640 | ||
| Precision | Recall | F 1 score | Precision | Recall | F 1 score | ||||
| ARGs | 1.0000 | 0.7129 | 0.8324 | ARGs | 1.0000 | 0.4976 | 0.6645 | ||
| VFs | 0.7805 | 0.1531 | 0.2560 | VFs | 0.9375 | 0.1435 | 0.2490 | ||
| Diamond-64%c | NSs | 0.4577 | 0.9569 | 0.6192 | Diamond-81%d | NSs | 0.4216 | 0.9904 | 0.5914 |
| Accuracy | 0.6077 | Accuracy | 0.5439 | ||||||
| Macro-average | 0.7461 | 0.6077 | 0.5692 | Macro-average | 0.7864 | 0.5439 | 0.5016 | ||
| Precision | Recall | F 1 score | Precision | Recall | F 1 score | ||||
| ARGs | 0.9924 | 0.6220 | 0.7647 | ARGs | 0.9952 | 1.0000 | 0.9976 | ||
| VFs | 0.7319 | 0.4833 | 0.5821 | VFs | 0.9351 | 0.8378 | 0.8782 | ||
| PathoFacte | NSs | 0.5084 | 0.8708 | 0.6420 | HyperVR | NSs | 0.8448 | 0.9378 | 0.8889 |
| Accuracy | 0.6586 | Accuracy | 0.9219 | ||||||
| Macro-average | 0.7442 | 0.6587 | 0.6629 | Macro-average | 0.9251 | 0.9219 | 0.9216 |
a Three Ha-value parameters of 0.21, 0.64 and 0.81 for VRprofile get the same results.
b The DIAMOND program with the identity threshold of 0.21 (Diamond-21%).
c The DIAMOND program with the identity threshold of 0.64 (Diamond-64%).
d The DIAMOND program with the identity threshold of 0.81 (Diamond-81%).
e For those that PathoFact cannot determine whether they are VFs or not (the prediction result is unclassified), we treat them as NS categories.
Figure 6.
The confusion matrices and histograms representing results for HyperVR and baseline methods to simultaneously predict novel ARGs, VFs and NSs. (A) The confusion matrix of VRprofile for simultaneously predicting novel ARGs, VFs and NSs. (B) The confusion matrix of best hit approach by using DIAMOND program with 21% identity for simultaneously predicting novel ARGs, VFs and NSs. (C) The confusion matrix of best hit approach by using DIAMOND program with 64% identity for simultaneously predicting novel ARGs, VFs and NSs. (D) The confusion matrix of best hit approach by using DIAMOND program with 81% identity for simultaneously predicting novel ARGs, VFs and NSs. (E) The confusion matrix of PathoFact for simultaneously predicting novel ARGs, VFs and NSs. (F) The confusion matrix of HyperVR for simultaneously predicting novel ARGs, VFs and NSs. (G) The histograms representing results for HyperVR and baseline methods to simultaneously predict novel ARGs, VFs and NSs.
Validation of HyperVR through an in silico spike-in experiment
For microbial datasets from real-world samples, ARGs and VFs may represent only a small fraction of the total number of genes. It is essential to evaluate the performance of HyperVR in cases where non-target genes dominate. To assess the ability of HyperVR for predicting a small number of ARGs and VFs among the majority of NSs, we constructed a negative microbial dataset mimicking a spike-in metagenomic experiment. In this section, we are more concerned with the recall score of the method; i.e. all positive data should be identified as much as possible, even if this leads to some false samples being predicted and subsequent biological experiments can help to eliminate. Specifically, we first randomly selected 10 ARGs and 10 VFs from the independent dataset. Next, we reconstructed 10 000 NSs using the NS construction method (refer to the ‘Data collection and annotation’ section). The final spike-in dataset containing 10 020 genes was ensured to have no overlap with the training dataset, and the percentage of positive samples was only (20/10 020)% ≈ 0.19%. In this section, we have chosen the same baseline methods as the ‘Validation of HyperVR through novel ARGs, VFs and NSs’ section for comparison. Table 6 shows the prediction results of HyperVR and baseline methods for positive samples in the spike-in dataset. From the table, we see that VRprofile and PathoFact have a good identification effect for a small number of ARGs among the majority of NSs, but the identification effect for VFs is extremely poor. The best hit approach using the DIAMOND program decreases in identification accuracy as the identity cutoff increases, and the best result is achieved when the identity is 21%, which is better than the VRprofile method. HyperVR obtained the best prediction results among the three methods, with all 10 ARGs predicted correctly and only 1 of 10 VFs predicted incorrectly to NS. This section demonstrates that HyperVR can well predict simultaneously small amounts of ARGs and VFs that exist in a large number of NSs, which is applicable to the fact that ARGs and VFs may represent only a small fraction of the total number of genes in the real world.
Table 6.
The prediction results of HyperVR and baseline methods through an in silico spike-in experiment
| Predicted class | |||||||
|---|---|---|---|---|---|---|---|
| VRprofilea | Diamond-21%b | Diamond-64%c | Diamond-81%d | PathoFacte | HyperVR | ||
| E3XRD1 | VFs | ARGs | ARGs | ARGs | ARGs | ARGs | |
| T2F8F6 | ARGs | VFs | NSs | NSs | NSs | ARGs | |
| A0A141R9W9 | ARGs | ARGs | ARGs | ARGs | NSs | ARGs | |
| A0A141RNL3 | ARGs | ARGs | ARGs | ARGs | ARGs | ARGs | |
| ARGs | A0A084TE22 | VFs | ARGs | ARGs | NSs | ARGs | ARGs |
| A0A0B4ZUG5 | ARGs | ARGs | ARGs | ARGs | ARGs | ARGs | |
| A0A127T2F4 | ARGs | ARGs | ARGs | ARGs | ARGs | ARGs | |
| G0A279 | ARGs | ARGs | ARGs | NSs | ARGs | ARGs | |
| A0A143GH84 | ARGs | ARGs | ARGs | ARGs | ARGs | ARGs | |
| G8AP61 | ARGs | ARGs | NSs | NSs | ARGs | ARGs | |
| Recall | 80% | 90% | 80% | 60% | 80% | 100% | |
| sp|Q4WFS2 | NSs | VFs | NSs | NSs | NSs | VFs | |
| sp|P0DJH1 | NSs | VFs | NSs | NSs | NSs | NSs | |
| sp|A9N230 | NSs | VFs | NSs | NSs | NSs | VFs | |
| sp|B6A877 | NSs | NSs | NSs | NSs | VFs | VFs | |
| VFs | sp|P9WIZ7 | NSs | VFs | NSs | NSs | NSs | VFs |
| sp|Q9I739 | NSs | NSs | NSs | NSs | VFs | VFs | |
| sp|Q8E372 | NSs | NSs | NSs | NSs | NSs | VFs | |
| sp|Q6G2B4 | VFs | VFs | VFs | VFs | VFs | VFs | |
| sp|P0A3W9 | NSs | VFs | VFs | NSs | VFs | VFs | |
| sp|P0C536 | NSs | VFs | VFs | VFs | VFs | VFs | |
| Recall | 10% | 70% | 30% | 20% | 50% | 90% | |
a Three Ha-value parameters of 0.21, 0.64 and 0.81 for VRprofile get the same results.
b The DIAMOND program with the identity threshold of 0.21 (Diamond-21%).
c The DIAMOND program with the identity threshold of 0.64 (Diamond-64%).
d The DIAMOND program with the identity threshold of 0.81 (Diamond-81%).
e For those that PathoFact cannot determine whether they are VFs or not (the prediction result is unclassified), we treat them as NS categories.
Validation of HyperVR through pathogen cases
To verify HyperVR in ARG and VF detection in real pathogenic bacteria, we chose four representative bacterial datasets, including Mycobacterium tuberculosis (strain ATCC 25618/H37Rv, 3993 protein genes) (61), Bacillus anthracis (strain Ames Ancestor, 5493 protein genes) (62), S. aureus (strain NCTC 8325/PS 47, 2889 protein genes) (63) and Klebsiella pneumoniae (strain ATCC 700721/MGH 78578, 5126 protein genes) (64). All four kinds of bacteria are important pathogens worth much attention nowadays. Specifically, M. tuberculosis is, to this day, according to the World Health Organization, the leading killer of adults, with ∼2 million deaths annually worldwide (61). Also, the drug resistance in M. tuberculosis is a major concern in the bacterial infection (61). Bacillus anthracis is an endospore-forming bacterium that causes inhalational anthrax (62). Staphylococcus aureus and K. pneumoniae are the important antibiotic-resistant strains of the ESKAPE (the short form of the most common conditional pathogenic bacteria in hospital infections, namely Enterococcus faecium, S. aureus, K. pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species) group (64).
Each of the three strain datasets was, respectively, input to HyperVR as a test dataset for simultaneous identification of ARGs and VFs. We listed the genes that HyperVR considered most likely to be ARGs or VFs (prediction scores >95%). Subsequently, we verified each of these genes in the following two ways. On one hand, the genes were directly queried in existing databases and determined by database annotation. On the other hand, BLAST was used to find similar genes and the type of genes was determined by database annotation of similar genes. The genes predicted by HyperVR in M. tuberculosis, B. anthracis, S. aureus and K. pneumoniae are, respectively, shown in Supplementary Tables S5–S8. It is evident from the results that HyperVR can effectively identify ARGs and VFs in real pathogenic strains, which narrows down the scope for researchers to conduct in vitro experiments and greatly saves them time and effort. Furthermore, it should be noted that, as with all in silico predictions, HyperVR is used to obtain an overview or inference of ARGs and VFs in a pathogenic strain. Some genes are not explicitly annotated as antibiotic resistant or virulence by the database, but it may still be the corresponding gene, e.g. genes annotated as penicillin binding, toxin, etc. Strictly speaking, downstream experiments are required to determined which category the gene truly belongs to.
CONCLUSION
In this study, we proposed a novel hybrid prediction approach called HyperVR for simultaneously predicting VFs and ARGs. HyperVR integrates multiple key genetic features, including bit score-based similarity feature, physicochemical property-based features, evolutionary information-based features and one-hot encoding feature, and then combines the power of classical ensemble learning methods and deep learning. It can accurately predict VFs, ARGs and NSs at the same time, and can be used flexibly and accurately to predict VFs or ARGs individually. HyperVR addresses the drawbacks of traditional ‘best hit’ methods, including high false-negative rates, being sensitive to the cutoff thresholds and only identifying conserved genes. Moreover, HyperVR outperforms the previous tools, in terms of precision and recall on novel VFs and ARGs, in silico spike-in experiment (the VFs and ARGs in real metagenomic data), and pseudo-VFs and -ARGs (gene fragments). To our knowledge, this is the first work to use computational methods including machine learning and deep neural network to predict VFs and ARGs simultaneously, with competitive results compared to all the state-of-the-art VF and/or ARG prediction tools. Overall, HyperVR is an effective, simple prediction tool and requires only gene sequence information without additional expert knowledge input for simultaneously predicting ARGs and VFs. However, HyperVR also has some limitations that we need to further optimize in the future. First, although the prediction accuracy of HyperVR for pseudo-ARGs and -VFs without any false-positive training samples is already better than most of the baseline methods, HyperVR would further try to reduce the false-positive rate. Adding some pseudo-ARGs and -VFs to the NSs during training may improve the prediction accuracy for false-positive samples. Furthermore, we likewise consider adding more authoritative data from the latest literature and wet lab experiments to avoid the overfitting problem caused by excessive false-positive samples. Updating our database and retraining the model will be our next goal. Second, the predictive performance of HyperVR has been demonstrated to be better than most current methods in several ways. However, the computational efficiency of the method for the whole sequence of a pathogenic bacterial strain is also an important factor. Although the prediction time of HyperVR is within the acceptable range, the computational efficiency is really the part that we need to further improve compared to other methods because HyperVR integrates multiple features and uses an ensemble approach for training. Improving the computing power of platform or parallelizing the training of the model will be the ways we further consider.
DATA AVAILABILITY
The data and materials that support the findings in this study are available in GitHub at https://github.com/jiboyalab/HyperVR and Zenodo at https://doi.org/10.5281/zenodo.7558490.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the editor and reviewers for valueble suggestions to improve our manuscript.
Author contributions: All authors were involved in the conceptualization of the HyperVR method. B.J., X.Z. and S.P. conceived and supervised the project. B.J., X.Z. and W.P. designed the study and developed the approach. B.J., X.Z., W.L. and Y.L. implemented and applied the method to microbial data. B.J. and Y.C. collected the data. B.J., X.Z. and S.P. analyzed the results. Y.L., Y.C., X.Z. and S.P. contributed to the review of the manuscript before submission.
Contributor Information
Boya Ji, College of Computer Science and Electronic Engineering, Hunan University, Changsha 410006, People’s Republic of China.
Wending Pi, College of Computer Science and Electronic Engineering, Hunan University, Changsha 410006, People’s Republic of China.
Wenjuan Liu, College of Computer Science and Electronic Engineering, Hunan University, Changsha 410006, People’s Republic of China.
Yannan Liu, Emergency Medicine Clinical Research Center, Beijing Chao-Yang Hospital, Capital Medical University, Beijing 100020, People’s Republic of China.
Yujun Cui, State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing 100071, People’s Republic of China.
Xianglilan Zhang, State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing 100071, People’s Republic of China.
Shaoliang Peng, College of Computer Science and Electronic Engineering, Hunan University, Changsha 410006, People’s Republic of China.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
National Key R&D Program of China [2022YFC3400400]; National Key R&D Program of China [2021YFC2302004, 2019HK125]; NSFC Grants [U19A2067]; National Key R&D Program of China [2022YFC3400404]; Science Foundation for Distinguished Young Scholars of Hunan Province [2020JJ2009]; Science Foundation of Changsha [Z202069420652, kq2004010; JZ20195242029, JH20199142034]; The Funds of State Key Laboratory of Chemo/Biosensing and Chemometrics, the National Supercomputing Center in Changsha (http://nscc.hnu.edu.cn/), and Peng Cheng Lab.
Conflict of interest statement. None declared.
REFERENCES
- 1. Henry L.P., Bruijning M., Forsberg S.K., Ayroles J.F.. The microbiome extends host evolutionary potential. Nat. Commun. 2021; 12:5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stappenbeck T.S., Virgin H.W.. Accounting for reciprocal host–microbiome interactions in experimental science. Nature. 2016; 534:191–199. [DOI] [PubMed] [Google Scholar]
- 3. McFall-Ngai M., Hadfield M.G., Bosch T.C., Carey H.V., Domazet-Lošo T., Douglas A.E., Dubilier N., Eberl G., Fukami T., Gilbert S.F.et al.. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. U.S.A. 2013; 110:3229–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blaser M.J., Falkow S.. What are the consequences of the disappearing human microbiota?. Nat. Rev. Microbiol. 2009; 7:887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bavro V.N., Pietras Z., Furnham N., Pérez-Cano L., Fernández-Recio J., Pei X.Y., Misra R., Luisi B.. Assembly and channel opening in a bacterial drug efflux machine. Mol. Cell. 2008; 30:114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Becker K., Hu Y., Biller-Andorno N.. Infectious diseases—a global challenge. Int. J. Med. Microbiol. 2006; 296:179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sharma A.K., Dhasmana N., Dubey N., Kumar N., Gangwal A., Gupta M., Singh Y.. Bacterial virulence factors: secreted for survival. Ind. J. Microbiol. 2017; 57:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu B., Zheng D., Jin Q., Chen L., Yang J.. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019; 47:D687–D692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Filler S.G., Sheppard D.C.. Fungal invasion of normally non-phagocytic host cells. PLoS Pathog. 2006; 2:e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee E., Li X., Oh J., Kwon N., Kim G., Kim D., Yoon J.. A boronic acid-functionalized phthalocyanine with an aggregation-enhanced photodynamic effect for combating antibiotic-resistant bacteria. Chem. Sci. 2020; 11:5735–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sharma D., Misba L., Khan A.U.. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control. 2019; 8:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gupta A., Landis R.F., Li C.-H., Schnurr M., Das R., Lee Y.-W., Yazdani M., Liu Y., Kozlova A., Rotello V.M.. Engineered polymer nanoparticles with unprecedented antimicrobial efficacy and therapeutic indices against multidrug-resistant bacteria and biofilms. J. Am. Chem. Soc. 2018; 140:12137–12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blaskovich M.A., Hansford K.A., Gong Y., Butler M.S., Muldoon C., Huang J.X., Ramu S., Silva A.B., Cheng M., Kavanagh A.M.et al.. Protein-inspired antibiotics active against vancomycin- and daptomycin-resistant bacteria. Nat. Commun. 2018; 9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reuter S., Ellington M.J., Cartwright E.J., Köser C.U., Török M.E., Gouliouris T., Harris S.R., Brown N.M., Holden M.T., Quail M.et al.. Rapid bacterial whole-genome sequencing to enhance diagnostic and public health microbiology. JAMA Int. Med. 2013; 173:1397–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yao X., Doi Y., Zeng L., Lv L., Liu J.-H.. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect. Dis. 2016; 16:288–289. [DOI] [PubMed] [Google Scholar]
- 16. Burrus V., Waldor M.K.. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 2004; 155:376–386. [DOI] [PubMed] [Google Scholar]
- 17. Cabot G., Zamorano L., Moyà B., Juan C., Navas A., Blázquez J., Oliver A.. Evolution of Pseudomonas aeruginosa antimicrobial resistance and fitness under low and high mutation rates. Antimicrob. Agents Chemother. 2016; 60:1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsai Y.-K., Fung C.-P., Lin J.-C., Chen J.-H., Chang F.-Y., Chen T.-L., Siu L.K.. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 2011; 55:1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barbosa T.M., Levy S.B.. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 2000; 182:3467–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buchfink B., Xie C., Huson D.H.. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015; 12:59–60. [DOI] [PubMed] [Google Scholar]
- 22. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Underwood A.P., Mulder A., Gharbia S., Green J.. Virulence Searcher: a tool for searching raw genome sequences from bacterial genomes for putative virulence factors. Clin. Microbiol. Infect. 2005; 11:770–772. [DOI] [PubMed] [Google Scholar]
- 24. Scordis P., Flower D.R., Attwood T.K.. FingerPRINTScan: intelligent searching of the PRINTS motif database. Bioinformatics. 1999; 15:799–806. [DOI] [PubMed] [Google Scholar]
- 25. Lakin S.M., Dean C., Noyes N.R., Dettenwanger A., Ross A.S., Doster E., Rovira P., Abdo Z., Jones K.L., Ruiz J.et al.. MEGARes: an antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res. 2017; 45:D574–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H., Durbin R.. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garg A., Gupta D.. VirulentPred: a SVM based prediction method for virulent proteins in bacterial pathogens. BMC Bioinformatics. 2008; 9:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arango-Argoty G., Garner E., Pruden A., Heath L.S., Vikesland P., Zhang L.. DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome. 2018; 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li J., Tai C., Deng Z., Zhong W., He Y., Ou H.-Y.. VRprofile: gene-cluster-detection-based profiling of virulence and antibiotic resistance traits encoded within genome sequences of pathogenic bacteria. Brief. Bioinform. 2018; 19:566–574. [DOI] [PubMed] [Google Scholar]
- 30. de Nies L., Lopes S., Busi S.B., Galata V., Heintz-Buschart A., Laczny C.C., May P., Wilmes P.. PathoFact: a pipeline for the prediction of virulence factors and antimicrobial resistance genes in metagenomic data. Microbiome. 2021; 9:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Forsberg K.J., Patel S., Gibson M.K., Lauber C.L., Knight R., Fierer N., Dantas G.. Bacterial phylogeny structures soil resistomes across habitats. Nature. 2014; 509:612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McArthur A.G., Tsang K.K.. Antimicrobial resistance surveillance in the genomic age. Ann. N. Y. Acad. Sci. 2017; 1388:78–91. [DOI] [PubMed] [Google Scholar]
- 33. Yang Y., Jiang X., Chai B., Ma L., Li B., Zhang A., Cole J.R., Tiedje J.M., Zhang T.. ARGs-OAP: online analysis pipeline for antibiotic resistance genes detection from metagenomic data using an integrated structured ARG-database. Bioinformatics. 2016; 32:2346–2351. [DOI] [PubMed] [Google Scholar]
- 34. Apweiler R., Bairoch A., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M.et al.. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2004; 32:D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jia B., Raphenya A.R., Alcock B., Waglechner N., Guo P., Tsang K.K., Lago B.A., Dave B.M., Pereira S., Sharma A.N.et al.. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017; D566–D573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu B., Pop M.. ARDB—antibiotic resistance genes database. Nucleic Acids Res. 2009; 37:D443–D447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Y., Xu Z., Han W., Cao H., Umarov R., Yan A., Fan M., Chen H., Duarte C.M., Li L.et al.. HMD-ARG: hierarchical multi-task deep learning for annotating antibiotic resistance genes. Microbiome. 2021; 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang Y., Niu B., Gao Y., Fu L., Li W.. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010; 26:680–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wattam A.R., Abraham D., Dalay O., Disz T.L., Driscoll T., Gabbard J.L., Gillespie J.J., Gough R., Hix D., Kenyon R.et al.. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014; 42:D581–D591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sayers S., Li L., Ong E., Deng S., Fu G., Lin Y., Yang B., Zhang S., Fa Z., Zhao B.et al.. Victors: a web-based knowledge base of virulence factors in human and animal pathogens. Nucleic Acids Res. 2019; 47:D693–D700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boeckmann B., Bairoch A., Apweiler R., Blatter M.-C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O’Donovan C., Phan I.et al.. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003; 31:365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pearson W.R. An introduction to sequence similarity (‘homology’) searching. Curr. Protoc. Bioinformatics. 2013; Chapter 3:3.1.1–3.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bhasin M., Raghava G.P.. Classification of nuclear receptors based on amino acid composition and dipeptide composition. J. Biol. Chem. 2004; 279:23262–23266. [DOI] [PubMed] [Google Scholar]
- 44. Saravanan V., Gautham N.. Harnessing computational biology for exact linear B-cell epitope prediction: a novel amino acid composition-based feature descriptor. Omics. 2015; 19:648–658. [DOI] [PubMed] [Google Scholar]
- 45. Chou K.-C. Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins. 2001; 43:246–255. [DOI] [PubMed] [Google Scholar]
- 46. Chou K.-C. Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics. 2005; 21:10–19. [DOI] [PubMed] [Google Scholar]
- 47. Schneider G., Wrede P.. The rational design of amino acid sequences by artificial neural networks and simulated molecular evolution: de novo design of an idealized leader peptidase cleavage site. Biophys. J. 1994; 66:335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974; 185:862–864. [DOI] [PubMed] [Google Scholar]
- 49. Suzek B.E., Wang Y., Huang H., McGarvey P.B., Wu C.H., UniProt Consortium. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015; 31:926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zou L., Nan C., Hu F.. Accurate prediction of bacterial type IV secreted effectors using amino acid composition and PSSM profiles. Bioinformatics. 2013; 29:3135–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. cheol Jeong J., Lin X., Chen X.-W.. On position-specific scoring matrix for protein function prediction. IEEE/ACM Trans. Comput. Biol. Bioinformatics. 2010; 8:308–315. [DOI] [PubMed] [Google Scholar]
- 52. Liu T., Zheng X., Wang J.. Prediction of protein structural class for low-similarity sequences using support vector machine and PSI-BLAST profile. Biochimie. 2010; 92:1330–1334. [DOI] [PubMed] [Google Scholar]
- 53. Veltri D., Kamath U., Shehu A.. Deep learning improves antimicrobial peptide recognition. Bioinformatics. 2018; 34:2740–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Charoenkwan P., Chiangjong W., Nantasenamat C., Hasan M.M., Manavalan B., Shoombuatong W.. StackIL6: a stacking ensemble model for improving the prediction of IL-6 inducing peptides. Brief. Bioinform. 2021; 22:bbab172. [DOI] [PubMed] [Google Scholar]
- 55. Ribeiro M.H.D.M., dos Santos Coelho L.. Ensemble approach based on bagging, boosting and stacking for short-term prediction in agribusiness time series. Appl. Soft Comput. 2020; 86:105837. [Google Scholar]
- 56. Breiman L. Random forests. Mach. Learn. 2001; 45:5–32. [Google Scholar]
- 57. Geurts P., Ernst D., Wehenkel L.. Extremely randomized trees. Mach. Learn. 2006; 63:3–42. [Google Scholar]
- 58. Chen T., Guestrin C.. Xgboost: a scalable tree boosting system. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2016; 785–794. [Google Scholar]
- 59. Friedman J.H. Stochastic gradient boosting. Comput. Stat. Data Anal. 2002; 38:367–378. [Google Scholar]
- 60. Freund Y., Schapire R., Abe N.. A short introduction to boosting. J. Jpn. Soc. Artif. Intell. 1999; 14:1612. [Google Scholar]
- 61. Cole S., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S., Eiglmeier K., Gas S., Barry C.R.et al.. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998; 396:190–190. [DOI] [PubMed] [Google Scholar]
- 62. Ravel J., Jiang L., Stanley S.T., Wilson M.R., Decker R.S., Read T.D., Worsham P., Keim P.S., Salzberg S.L., Fraser-Liggett C.M.et al.. The complete genome sequence of Bacillus anthracis Ames ‘Ancestor’. J. Bacteriol. 2009; 191:445–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gillaspy A.F., Worrell V., Orvis J., Roe B.A., Dyer D.W., Iandolo J.J.. Fischetti V.A., Novick R.P., Ferretti J.J., Portnoy D.A., Rood J.I.. The Staphylococcus aureus NCTC 8325 genome. Gram-Positive Pathogens. 2006; Washington, DC: ASM Press; 381–412. [Google Scholar]
- 64. Liu P., Li P., Jiang X., Bi D., Xie Y., Tai C., Deng Z., Rajakumar K., Ou H.-Y.. Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J. Bacteriol. 2012; 194:1841–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and materials that support the findings in this study are available in GitHub at https://github.com/jiboyalab/HyperVR and Zenodo at https://doi.org/10.5281/zenodo.7558490.








