Abstract
Next-generation sequencing has revolutionized the diagnostic process, making broadscale testing affordable and applicable to almost all specialties; however, there remain several challenges in its widespread implementation. Barriers such as lack of infrastructure or expertise within local health systems and complex result interpretation or counseling make it harder for frontline clinicians to incorporate genomic testing in their existing workflow. The general population is more informed and interested in pursuing genetic testing, and this has been coupled with the increasing accessibility of direct-to-consumer testing. As a result of these changes, primary care physicians and nongenetics specialty providers find themselves seeing patients for whom genetic testing would be beneficial but managing genetic test results that are out of their scope of practice. In this report, we present a practical and centralized approach to providing genomic services through an independent, enterprise-wide clinical service model. We present 4 years of clinical experience, with >3400 referrals, toward designing and implementing the clinical service, maximizing resources, identifying barriers, and improving patient care. We provide a framework that can be implemented at other institutions to support and integrate genomic services across the enterprise.
Next-generation sequencing has revolutionized the diagnostic process in clinical genetics. It has helped clinicians rapidly identify new syndromes, better manage diseases, and reduce diagnostic odysseys.1 As genetic and genomic testing has become more comprehensive, it has also become more challenging to identify the most appropriate test, obtain insurance authorization, interpret test results that require a high level of expertise, and return results to families in a meaningful way. Paradoxically, despite these complexities, the advent of clinical genomic testing has resulted in a broader applicability of genetic testing to patient populations not traditionally serviced by genetic specialists. In response to these challenges, several academic medical centers and health systems have instituted programs offering personalized genomic medicine services.2 The need for responsible genetic test ordering, insurance navigation, genetic counseling, consenting, and return of results across all clinical practices and divisions at the Children’s Hospital of Philadelphia (CHOP) led to the establishment of the Roberts Individualized Medical Genetics Center (RIMGC).
Exome sequencing (ES) was first implemented as a research tool to identify an underlying molecular cause for various clinical presentations, with the first causal pathogenic variant reported in 2009,3 and rapidly transitioned to the clinical setting in 2010. In 2013, Yang et al4 reported the results of ES in 250 probands, and with additional reports, it is estimated that ES has a diagnostic rate of ~25% to 30%.5 Despite the proven diagnostic benefits of applying ES in the clinical setting, there remain several challenges in its widespread implementation.
The first challenge is the gap in knowledge and access to genetics specialists. It is estimated that there are ~75 000 genetic tests on the market, and at least 10 new tests are added every day.6 On the other hand, it is estimated that there is 1 full-time clinical geneticist and 10 certified genetic counselors per 600 000 people in the United States7 (National Society of Genetic Counselors). With the widespread uptake of complex genomic testing in patient populations not traditionally seen in clinical genetics practices, the onus of ordering ES tests often falls on clinicians who do not have genetic training or expertise. Many nongenetics providers lack the time during routinely scheduled clinic appointments to provide adequate pre- and posttest genetic counseling.8,9 Additionally, in a recent survey of 488 primary care physicians on their views of genetic testing, only 25% felt prepared to order a genetic test and 14% felt confident interpreting the test results.10 There is also a large gap in understanding the significance of each variant detected by ES and what is considered medically actionable. Other challenges include lack of appreciation that genomics will actually improve clinical care,2 limited return on institutional investment, ownership of patient care, integration of actionable findings into the electronic health record (EHR), communication of genomic findings, management of secondary findings, follow-up for the patients and other affected family members, and insurance reimbursement.
The RIMGC, a joint initiative between the departments of pediatrics and pathology at CHOP, was established in 2014 to provide a centralized resource to bridge these needs across all clinical divisions and practices. For referring clinicians, the RIMGC facilitates selection and review of the most appropriate genetic testing strategy on the basis of a complete review of the patient’s symptoms, findings, family history, and physical examination. It also assists with patient education, consent, educational materials, insurance authorizations, and clinical interpretation of results. The RIMGC works closely with the division of genomic diagnostics (DGD) at CHOP and participates in several steps of the clinical ES pipeline including (1) accurate phenotype capture during clinical evaluation, including generation of human phenotype ontology (HPO) terms for each case; (2) a clinical correlation step; and (3) ES test result medical interpretation as part of the final clinical report.
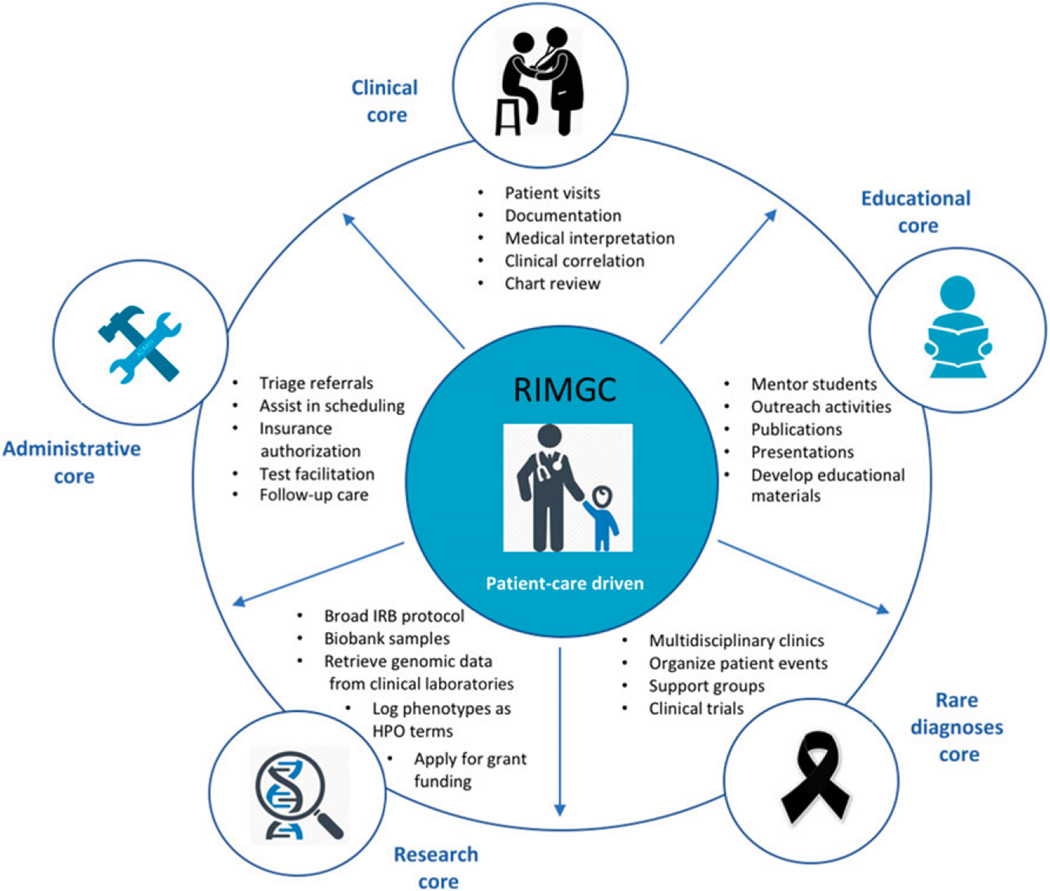
As the RIMGC has grown in response to the clinical and research demands, specific cores have been established to consolidate and streamline operations including clinical, educational, research, rare diagnoses, and administrative cores (Fig 1). Each core leads focused efforts in areas detailed in Fig 1 to better serve our patient population, clinicians, and investigators. The clinical core is focused on optimizing clinical workflows, standardizing visit templates, and navigating insurance authorizations. The educational core developed online educational modules for patients and families and maintains the content, organizes annual lectureships for primary care and pediatric specialty physicians, compiles and publishes annual reports, and coordinates clinical rotations for rotating trainees (medical students, genetic counseling students, residents, postdoctoral trainees, etc). The research core developed and implemented a broad institutional review board–approved research protocol and coordinated participation in various clinical research projects. The rare diagnoses core is focused on running diagnosis-specific multispecialty clinics, supporting rare diagnoses family support group and foundation meetings, and developing clinical care and management plans for rare diagnoses. The administrative core provides logistics and administrative support for all RIMGC activities.
FIGURE 1.
Overview of the RIMGC infrastructure. Descriptions of each core and organization are shown.
In this report, we summarize the use of genomic diagnostics across diverse pediatric specialties for 4 years, with 3483 referrals to the RIMGC, toward establishing optimal approaches to implementing genomic diagnostics into the clinical workflow, maximizing resource use, reducing the diagnostic odyssey for families, and improving counseling and management.
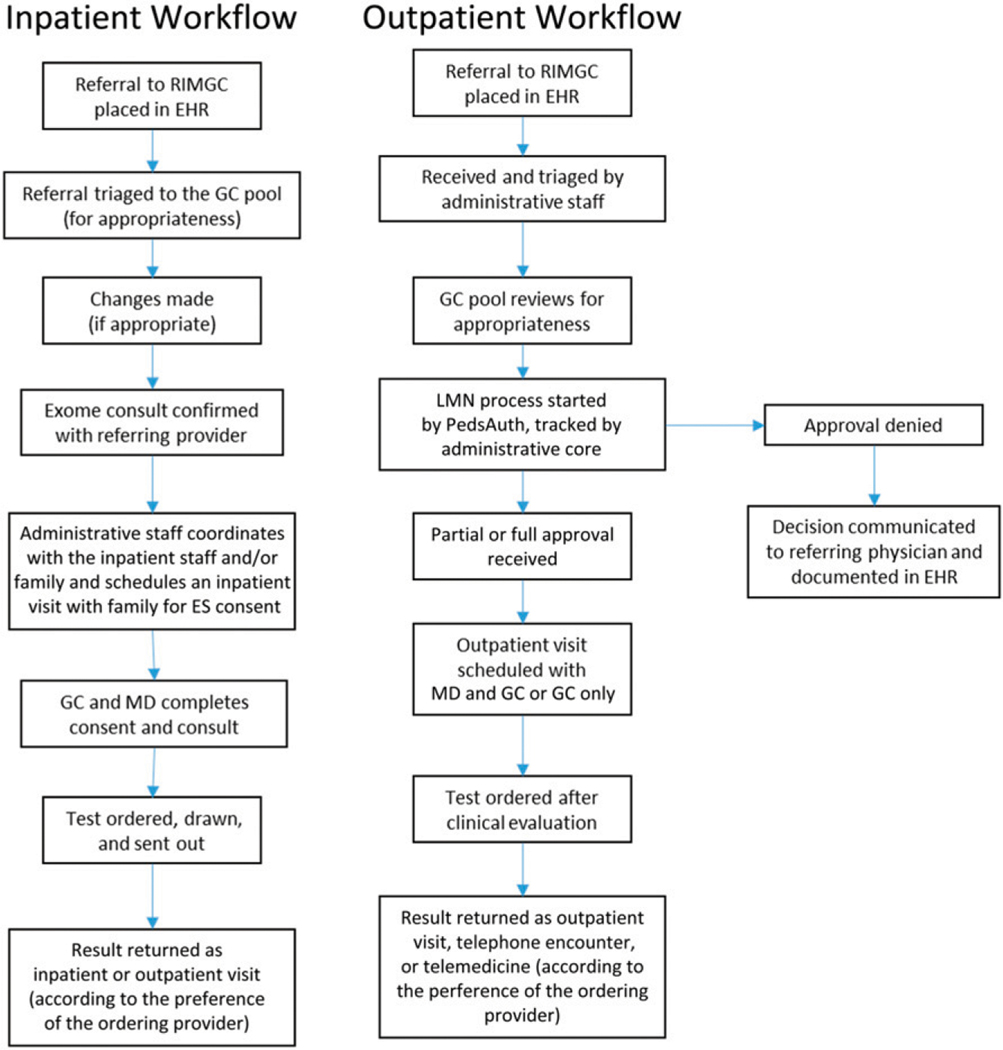
To assess the use and effectiveness of the RIMGC, the program built a comprehensive custom database and established metrics that were tracked for all patient referrals for quality improvement purposes (July 1, 2014–June 30, 2018). The data were stored in a custom FileMaker database housed in CHOP-managed servers, which was designed to capture all elements of clinical and research involvement. Every RIMGC clinician, administrator, and coordinator updates the database after every interaction with a provider or patient. Each case was tracked starting from the patient referral through return of results and follow-up care with data on referring provider specialty, requested tests, performed tests, reasons for not completing recommended testing, insurance authorization and/or approval steps and outcomes, test results, patient preferences for result options (eg, including secondary findings or not), and billed and unbilled encounters. For in-house ES cases we recorded the following metrics: number of chart reviews, number of HPO terms generated, and clinical correlation of filtered variants. Data were also collected for research activities that included enrollment data, sample collection, HPO terms, and retrieval of binary alignment map files from clinically run tests. The database was regularly maintained by an administrator, who would troubleshoot errors and update fields to capture different nuances of the clinical workflows. The RIMGC database was also correlated with data extracted from the EHR for a subset of the metrics analyzed below. The details of the clinical workflow for inpatient and outpatient referrals have been outlined in Fig 2.
FIGURE 2.
Clinical workflow for inpatient and outpatient referrals for RIMGC. ENT, ear, nose, and throat; IRB, institutional review board
REFERRAL CHARACTERISTICS
In the 4-year period between fiscal year (FY) 2015 and FY 2018 (July 1, 2014–June 30, 2018), the RIMGC received 3483 referrals (412 in FY 2015, 795 in FY 2016, 952 in FY 2017, and 1324 in FY 2018) from 30 unique pediatric divisions and/or clinical programs. The top 10 referral sources by FY are detailed in Table 1: ear, nose, and throat (828); general genetics (834); neurology (311); endocrinology (213); ophthalmology (153); rheumatology (126); metabolism (131); immunology (123); gastroenterology (113); dermatology (64); and cardiology (38). Some services that traditionally refer to genetics (such as cardiology) are underrepresented in these data because they have their own genetic support and specialty clinics. The “all others” category was made of referrals from a number of other divisions and services in the hospital, with the top 5 of these referrals coming from child development, audiology, hematology, neonatology, and primary care.
TABLE1.
Breakdown of the 3483 Referrals by Source and FY
| Referral Source | FY 2015 | FY 2016 | FY 2017 | FY 2018 | Total |
|---|---|---|---|---|---|
| ENT | 123 | 178 | 252 | 275 | 828 |
| General genetics | 122 | 202 | 230 | 280 | 834 |
| Neurology | 58 | 81 | 64 | 108 | 311 |
| Endocrine | 6 | 67 | 66 | 74 | 213 |
| Ophthalmology | 11 | 54 | 48 | 40 | 153 |
| Rheumatology | 25 | 31 | 30 | 40 | 126 |
| Gastroenterology | 23 | 33 | 34 | 23 | 113 |
| Immunology | 25 | 36 | 29 | 33 | 123 |
| Metabolism | 3 | 32 | 52 | 44 | 131 |
| Dermatology | 0 | 18 | 11 | 35 | 64 |
| Cardiology | 6 | 4 | 20 | 8 | 38 |
| All others | 10 | 59 | 116 | 364 | 549 |
| Total | 412 | 795 | 952 | 1324 | 3483 |
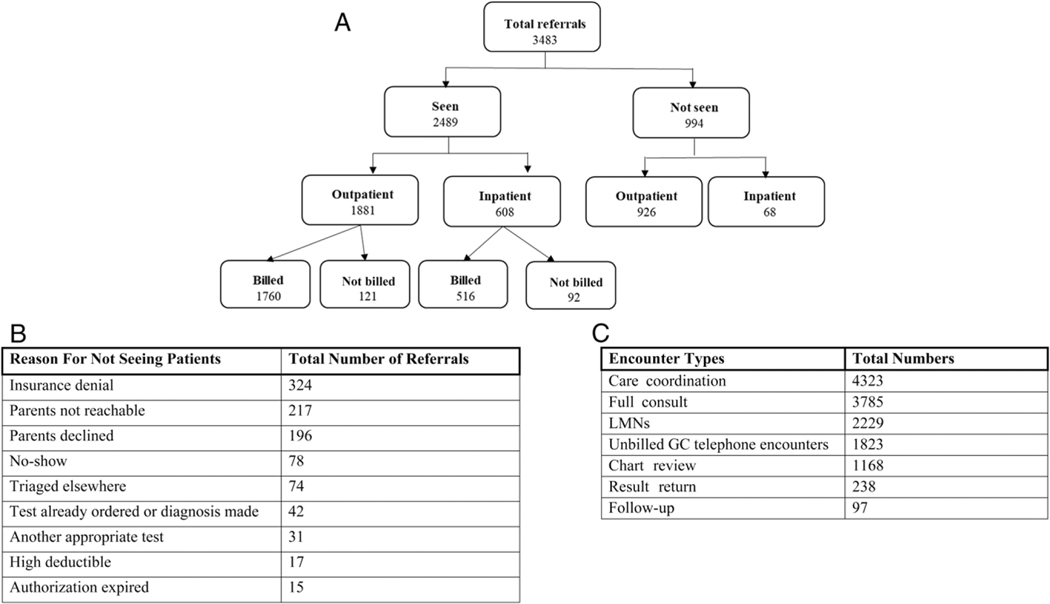
Of the 3483 referrals (Fig 3A), RIMGC staffed 2489 patient encounters. Reasons for 994 referrals (28.5%) not being seen included (Fig 3B) insurance denials (324), parents not reachable (217), parent or patient declined visits (196), triaged elsewhere (74), already ordered or another diagnosis made (42), another appropriate test ordered (31), authorization expired (15), high deductible (17), and not showing for a scheduled appointment (78). Of the 2489 patient encounters, 1881 were outpatient visits (75.57%) and 608 were inpatient consults (24.42%). Of the 1881 outpatient encounters, 1760 were billed (93.56%) and 121 were not billed (6.43%). Of the 608 inpatient encounters, 516 were billed (84.86%) and 92 were not billed (15.13%). The general makeup of the inpatient and outpatient encounters after reviewing data from the CHOP EHR over this time period was (Fig 3C) care coordination (4323), new patient consult (2789), letter of medical necessity (LMN) (2229), unbilled telephone encounters (1823), chart reviews and clinical correlations for DGD (1168), and established patient encounters (335) (result return [238] and follow-up visits [97]). Several inpatient and outpatient encounters were also not billed for the following reasons: (1) follow-up genetic counseling provided by a genetic counselor without a physician geneticist present, (2) test facilitation for family members of the proband, (3) return of positive test results with the parents without the proband present, (4) patient encounters on same day as visits with other genetics specialists, and (5) errors in medical documentation meeting internal billing standards.
FIGURE 3.
Patient referral and encounter details. A, Breakdown of referrals seen and billed. B, Reasons for not seeing referrals to RIMGC. C, Details of encounter types.
The top 5 HPO terms from referrals were “bilateral sensorineural hearing loss,” “global developmental delay,”“short stature,” “failure to thrive,” and microcephaly.”
TEST AUTHORIZATION EFFORTS
When a referral to the RIMGC is received, each referral is triaged by an administrative coordinator to a genetic counselor for determination of appropriateness of the requested test. Once a determination of appropriateness is established, insurance preauthorization is initiated and an LMN is drafted on the basis of EHR review, which is required for most insurance plans. In the past 4 years, 2229 LMNs have been written. Some letters were written for >1 test. An LMN was needed for 97% of ES referrals (approval rate: 73.9%), 87% of gene panel referrals (approval rate: 77%), 78% of single-gene test referrals (approval rate: 87.9%), 77% of the single-nucleotide polymorphism (SNP) array referrals (approval rate: 86%), 85% of the familial variant testing referrals (approval rate: 93%), and 92% of the karyotype referrals (approval rate: 97%) (Table 2). We were most successful in obtaining approvals for karyotypes and least successful for ES (Table 3). Genetic testing was denied in 26.5% of referrals (exome >gene panels>SNP array>single gene testing) .
TABLE 2.
LMN Determinability (Details of the LMNs Required by Test Type)
| Test Type | LMN Needed, n (%) |
|---|---|
| Exome | 1299 (97.8) |
| Panels | 1217 (86.8) |
| Single gene | 364 (77.7) |
| SNP array | 550 (77.9) |
| Familial variant | 72 (85.7) |
| Karyotype | 38 (92.7) |
| Total | 3540 (87.8) |
TABLE 3.
Outcome After LMN Submission (Details of the LMN Percentage Approval by Test Type)
| Test Type | Testing Approved, n (%) |
|---|---|
| Exome | 961 (73.9) |
| Panels | 940 (77.2) |
| Single gene | 320 (87.9) |
| SNP array | 473 (86.0) |
| Familial variant | 67 (93.0) |
| Karyotype | 37 (97.4) |
| Total | 2688 (78.4) |
In FY 2018, a peer-to-peer call with the medical geneticist or certified genetic counselor was also warranted in 30 cases. On average, the staff spends 45 to 65 minutes per referral from referral receipt to scheduling an appointment (10 minutes for referral triage, 10–30 minutes for LMN writing based on the complexity of referral, 10 minutes for care coordination, and 15 minutes for scheduling and preparing for appointments).
TESTS ORDERED BY RIMGC
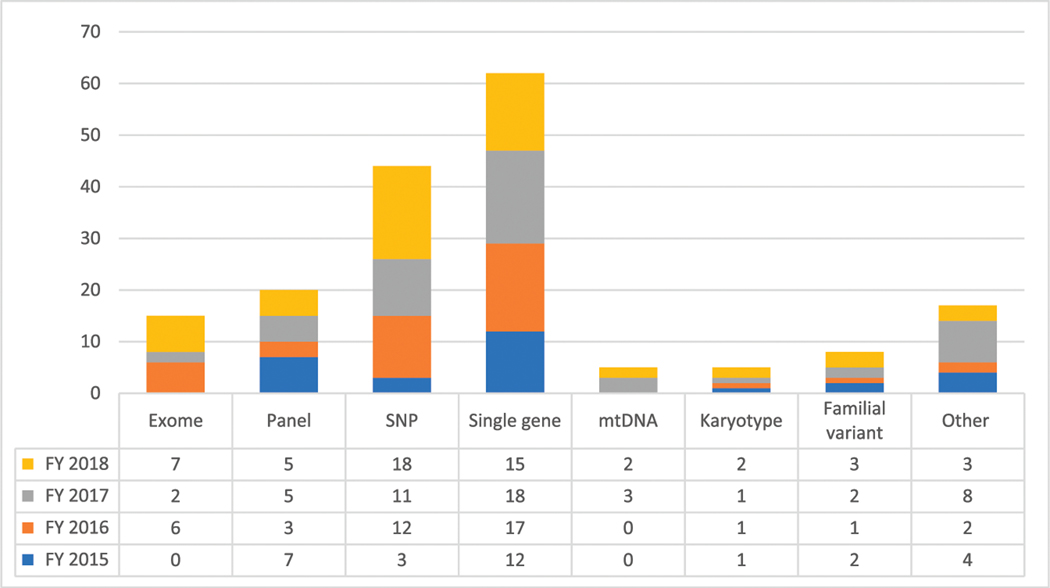
After receiving test authorization, the patient is scheduled for a clinic visit. The RIMGC clinicians ordered a total of 2518 tests from FY 2015 to FY 2018. Some patients had >1 test ordered. Testing included 1080 ES tests (42.89%), 92 rapid ES tests (3.65%), 296 SNP arrays (11.75%), 564 gene panel tests (22.39%), 257 single-gene tests (10.20%), 144 mitochondrial DNA (mtDNA) analysis tests (5.71%), 15 karyotypes (0.59%), and 70 familial variant tests (2.77%) (Table 4). In addition to the facilitation of test requests from physicians across the CHOP enterprise, there were 176 instances where the RIMGC clinicians changed the test ordered or added additional testing on the basis of the chart review or examination of the referred patient (Fig 4). A positive diagnosis result was made in 29 cases (16%) where RIMGC clinicians modified the test order. In some cases, the originally ordered test (eg, ES) would have identified the molecular diagnosis, but a targeted test was chosen on the basis of the clinical presentation (Supplemental Information). In other cases, an addition of copy number variant testing or triplet repeat testing, not readily detectable on most ES platforms, led to the molecular diagnosis.
TABLE 4.
Types of Tests Ordered by RIMGC Clinicians by FY
| Test Type | FY 2015 | FY 2016 | FY 2017 | FY 2018 |
|---|---|---|---|---|
| Exome | 154 | 195 | 307 | 412 |
| Panel | 109 | 104 | 127 | 220 |
| SNP array | 84 | 80 | 63 | 69 |
| Single gene | 32 | 63 | 72 | 83 |
| mtDNA | 30 | 30 | 40 | 46 |
| Rapid exome | 8 | 45 | 19 | 22 |
| Familial variant | 12 | 13 | 17 | 27 |
| Karyotype | 4 | 2 | 4 | 3 |
FIGURE 4.
Details of tests added or changed by the RIMGC clinician based on the clinical review or consultation (value added).
ES TEST FACILITATION AT CHOP
CHOP’s DGD launched its medical exome test in 2014. The RIMGC serves as an extension of the DGD’s clinical arm and works synergistically to interpret ES data. A protocol was developed that included the RIMGC in 2 steps of the exome analysis process. First was a chart review step for all patients undergoing ES through DGD, including patients not seen by the RIMGC clinicians. This step included summarizing clinical and physical findings, as well as generation of HPO terms. Second was a clinical correlation step, which entailed a review and classification of genes to provide clinical insight on whether a specific gene flagged with a sequence change could be related to a patient’s phenotype. It included annotating reasons for the clinical calls (associated, possibly associated, not associated, candidate gene) for each gene. This clinical correlation step was performed by at least 1 clinical geneticist and 1 genetic counselor certified under a Clinical Laboratory Improvement Amendments–approved protocol. The RIMGC staff recognized that there are limitations to the breadth of knowledge required to appropriately interpret and correlate ES variants. In collaboration with the department of pediatrics, 31 “genetic champions” were selected from all divisions and departments at CHOP. The genetic champions are pediatric specialists with a particular interest and expertise in the genetic etiologies of the diagnoses seen in their specialties. They are consulted on cases specific to their specialty. The genetic champions serve vital roles in spearheading collaborative genomic research efforts between their divisions and the RIMGC as well as in identifying needs for novel genetic test expansion and development in collaboration with the DGD.
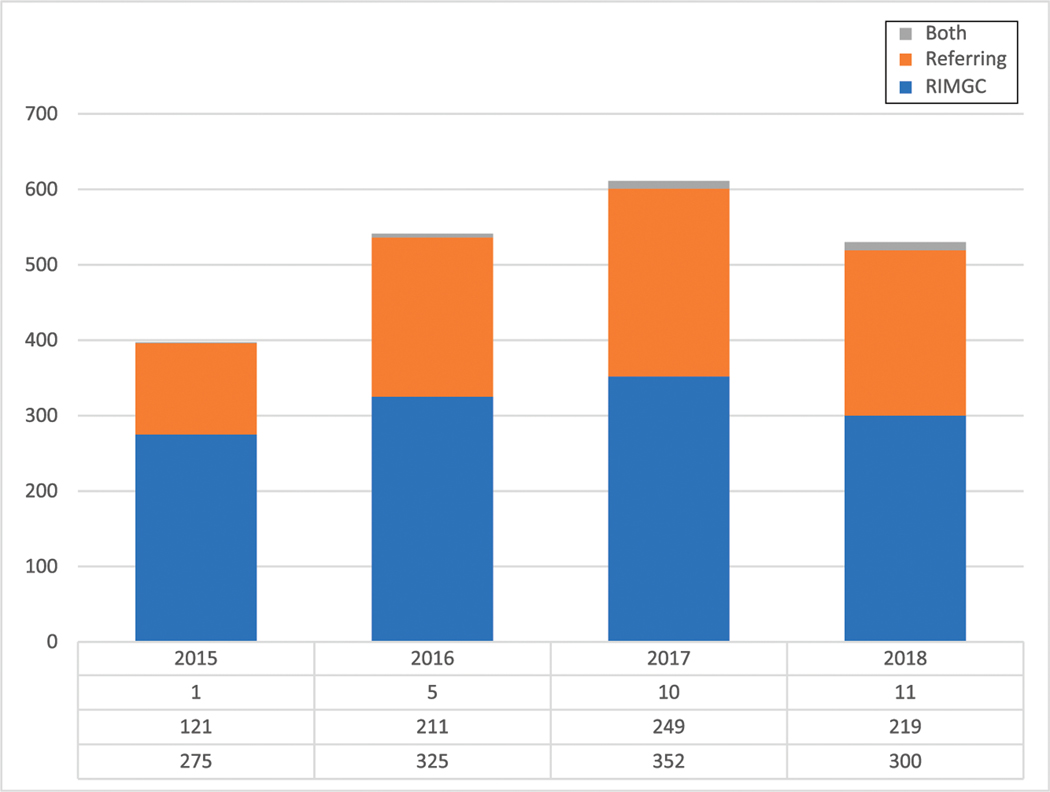
The diagnostic rate of ES facilitated by the RIMGC (n = 1172) is 26% for a positive diagnosis result, 37% for an uncertain diagnosis, 28% for negative result, 6.5% for a possible diagnosis in a candidate gene, and 1.2% for a dual diagnosis (Supplemental Information). A positive American College of Medical Genetics (ACMG) secondary finding was reported in 2.8% of the cases (Supplemental Information). After receiving the ES results from the diagnostic laboratory, they are shared with the referring physician. The RIMGC clinicians were available to assist in result return depending on the comfort level of the referring physician with the genomic findings. Of 2079 RIMGC result returns for all test results (Fig 5), 61% of the test results ordered by RIMGC were returned to the patient by an RIMGC clinician (25% positive results, 36% negative results, and 37% uncertain results), 39% by the referring physician (31% positive results, 36% negative results, and 32% uncertain results), and 1% by both. Of the 39% of results returned by the referring physician, 64% were returned by genetic specialists and 36% were by other specialists. This underscores a preference for a geneticist or genetic counselor to consent and return complex genomic results to families.
FIGURE 5.
Composition of results returned by the RIMGC clinician versus the referring physician versus both.
We also evaluated our distribution of efforts to increase efficiency. A significant percent (48%) of RIMGC clinical effort was related to facilitating genomic testing (care coordination and unbilled telephone encounters), 18% toward insurance authorization (LMNs), 9% toward DGD related efforts, and only 25% toward clinic visits or returning results.
DIAGNOSTIC LABORATORY ANALYTIC SUPPORT ACTIVITIES
The RIMGC performed clinical correlation and chart review steps for 1006 ES cases. Eighty-four were performed in FY 2015, 164 in FY 2016, 330 in FY 2017, and 428 in FY 2018. On average, genetic counselors and medical doctors (MDs) spent 30 minutes per case for clinical correlation, 15 minutes for chart review cases in which the patient had been seen in the RIMGC, and at least 1 hour for chart reviews in which the patient had not been seen in the RIMGC.
FINANCES
Exome consent and test facilitation visits were scheduled with a medical geneticist and genetic counselor, and services were charged as new, established, or follow-up visit. When clinically appropriate, a subset of exome pretest counseling and consent and result return visits (not requiring a physical examination) were seen by a certified genetic counselor under Current Procedural Terminology code 96040. Fifty-one percent of our total operating budget in FY 2018 came from patient revenue, with an average collection rate of 38.17%. Of the remaining 49% of the total operating budget, 24.5% came from the department of pathology, and 24.5% came from the Roberts Endowment at CHOP.
Authors of several studies have described their experience of integrating ES but primarily from the perspective of diagnostic laboratories, focusing on diagnostic rates, case reports, and challenges of managing ES finding.11–14 Several broadscale research initiatives like CSER (Clinical Sequencing Exploratory Research), IGNITE (Implementing Genomics in Practice), and eMERGE (Electronic Medical Records and Genomics consortium)15–17 have addressed the issues of integration of research and clinical interface of genomics. In a review of all studies evaluating the diagnostic yield and utility of ES, Smith et al18 reported 8% to 100% of diagnostic yields (with a median yield of 33%), which is consistent with the yield obtained through the RIMGC exome experience.19 Few reports were focused on approaches to scale and implement an enterprise-wide infrastructure.
The RIMGC model is the first of its kind dedicated to pediatrics and implemented in a children’s hospital; however, there are other individualized medical genetics programs that have reported on their efforts. Mayo Clinic in 2012 instituted the Center for Individualized Medicine15 with a similar concept and presented lessons learned from their first year of service. Hamilton et al20 highlighted barriers in integrating genomic medicine in routine health care systems through structured interviews. Machini et al21 provided perspectives from genetic counselors on the lack of complete understanding of the clinical utility of ES and identified areas that prevented them from offering this technology to patients (mostly in prenatal or cancer settings). Vassy et al22 presented data from a randomized trial in the adult primary care setting on outcomes of adding whole genome sequencing. Although some primary care physicians managed the results appropriately, whole-genome sequencing prompted additional clinical actions without evidence of clinical utility. A survey of 4824 physicians (a subset of which were pediatricians) highlighted gaps in knowledge and awareness of available genetic services around them. Some primary care physicians perceived a potential risk of harm and refrained from referring patients to genetics.23,24 There is growing evidence that genomic medicine should be an integral part of every health care institution, but challenges remain on how to identify and implement these services. A good resource to find nearby genetic expertise is provided by the ACMG Web site at https://www.acmg.net/ACMG/Genetic_Services_Directory_Search.aspx.
In this report, we highlight the RIMGC model, which differs from most standard clinical genetics practices in the centralization of services, integration with the diagnostic laboratory, and having a unified approach for all providers and patient populations across the CHOP enterprise. There are several lessons learned from the 4 years of successful clinical operation.
Building a Community
Our rapid growth across 30 disciplines at CHOP since 2014 (Fig 2) was a result of establishing an easy-to-use model for clinicians across the enterprise paired with extensive outreach and educational programs. We built collaborative relationships with the diagnostic laboratories, incorporated clinical correlation and medical interpretation services into the RIMGC model, which helped us enhance the in-house ES test, gene panels, and other genomic testing strategies. Clinical correlation sessions with genetic champions have been invaluable because they reduce the laboratory’s burden of variant interpretation and increase actionability of the test results. These sessions also serve as a valuable educational exercise for trainees and laboratory personnel. The genetic champions also played a significant role in increasing test uptake and test development because they represented different specialties. Ultimately, successful integration of genetic testing requires a team effort.
Constantly Adapting to Change
The need to keep pace with rapidly evolving technological advances has led to constant reassessment of the RIMGC organization model. The core team has expanded from 3 full-time employees to 12.4 full-time employees over 4 years to meet the demands of our service. We have also incorporated new areas to increase visibility of our services and revenue (capabilities to bill genetic counselor–only visits, medical interpretations, clinical correlations, implementation of telemedicine). Constant assessment of clinical load, needs, and patient satisfaction was crucial in the design of our business model.
Teamwork and Flexibility
As highlighted in the report, the RIMGC clinicians wear different hats at different times. As “test facilitators,” the majority of our time (48%) was spent on efforts that were unbillable (Fig 3B). RIMGC clinicians completed test authorization or scheduled a visit and retrieved clinic information for 417 (42%) of the referrals that did not result in an actual visit, which equates to ~228 hours of work without reimbursement.
Although most nongenetic providers are aware of the complexity involved in ordering genetic tests, they are not familiar with all of the “behind-the-scenes” effort involved in genomic test ordering and result return. Additionally, insurance providers generally require preauthorization and significant clinical justification to obtain approval for testing. Insurance approval for genetic laboratory tests (not consultations) is the rate-limiting step in providing testing for patients with an LMN required in 87% of the referrals for various genetic tests (Table 3). Data on the number of patients in need of genetic testing who did not receive it because of insurance barriers before the establishment of the RIMGC are not available across the enterprise. Because direct patient care accounted for 28% of staff efforts while the generation of LMNs added an additional 16% effort, there could be an increase in patient volume if insurance barriers were removed. Although care coordination efforts would remain notable for complex testing such as ES, 48% effort spent facilitating genomic testing also included various test coordination efforts in addition to generation of LMNs for each case. With the continued increase in patient referrals each year, we believe that our current model was able to reduce the time and burden of ordering a genetic test for other specialists through centralizing the authorization process. The RIMGC service model, although not fiscally self-sufficient yet, does represent a cost saving to the hospital when compared to other models that require each division and/or clinical center to hire and support dedicated genetic counselors and administrative staff to facilitate genomic testing.
The RIMGC clinicians as “consultants” work with the referring clinicians and identify the most suitable genetic test for the patient. A number of studies have demonstrated the utility of having genetic expertise integrated into a test use strategy to minimize expense and optimize patient satisfaction. Dickerson et al25 demonstrated significant test modification (24%) after review by a panel that included a genetic counselor with an estimated 19% of total cost savings. Similarly, Miller et al26 reported test request changes in 26% of all genetic test requests through ARUP (Associated Regional and University Pathologists) laboratories with a cost saving of ~$48 000 per month to referring institutions when genetic counselors performed preanalytic assessments of complex genetic test orders. The RIMGC was able to bring in our expertise and triage cases that did not require ES or redundant testing and order a different genetic test, thereby reducing health care costs and test burden.
As the RIMGC continues to evolve, certain hurdles remain, such as (1) improving turnaround times and reducing costs of genomic diagnostics, (2) implementation of novel and more comprehensive genomic diagnostic modalities (eg, RNA sequencing), (3) expanding educational services to increase awareness among providers and patient populations to the values and limitations of genetic and genomic diagnostics, (4) making genomic diagnostic test results longitudinal and portable so that patients benefit from the information embedded in these results as new associations are discovered wherever they obtain health care, and (5) integrating clinical testing into collaborative research efforts to help drive discovery.
CONCLUSIONS
As genomics and genomic testing continue to expand and infiltrate all specialties and medical practices, increasing support will be needed to help facilitate the integration of these tests into the care of patients and workflow of clinicians across all specialties. This demand will necessitate a different approach to the provision of genetic and genomic services beyond what traditional genetic divisions and departments are likely able to provide. The experience of the RIMGC as an early provider of centralized genomic services in a large pediatric hospital provides insight into the needs of practitioners across multiple specialties and how these needs can be met and supported.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the patients and their families who participated in clinical care at CHOP. We also thank the other members of the division of human genetics and the DGD at CHOP. Funding for this work was supported by CHOP. The funding body did not participate in the study design, collection, or analysis and interpretation of data, writing of the article, or decision to submit the article for publication.
FUNDING:
Supported by the Children’s Hospital of Philadelphia.
FINANCIAL DISCLOSURE:
The authors have indicated they have no financial relationships relevant to this article to disclose.
ABBREVIATIONS
- ACMG
American College of Medical Genetics
- CHOP
Children’s Hospital of Philadelphia
- DGD
division of genomic diagnostics
- EHR
electronic health record
- ES
exome sequencing
- FY
fiscal year
- HPO
human phenotype ontology
- LMN
letter of medical necessity
- MD
medical doctor
- mtDNA
mitochondrial DNA
- RIMGC
Roberts Individualized Medical Genetics Center
- SNP
single nucleotide polymorphism
Footnotes
Ms Biswas, Ms Medne, and Dr Krantz conceptualized and designed the study and drafted the initial manuscript; Drs Izumi, Deardorff, Skraban, and Pyle and Ms Bedoukian, Ms Tarpinian, Mr Gray, Ms Leonard, Ms Wilkens, and Ms Raible consulted the patients, collected data, entered data in the databases, and contributed to the analysis; Dr Devkota and Ms Berrodin created the database for data collection and summarized and analyzed the data; Ms Weatherly, Ms Fortunato, Ms Montgomery, Ms Williams, Ms McEldrew, and Ms Kaur coordinated and supervised data collection and participated in Roberts Individualized Medical Genetics Center core activities; Dr Spinner participated in data acquisition and critically reviewed the manuscript for important intellectual content; and all authors reviewed and revised the manuscript, approved the final manuscript as submitted, and agree to be accountable for all aspects of the work.
POTENTIAL CONFLICT OF INTEREST: The authors have indicated they have no potential conflicts of interest to disclose.
REFERENCES
- 1.Berberich AJ, Ho R, Hegele RA. Whole genome sequencing in the clinic: empowerment or too much information? CMAJ. 2018;190(5): E124–E125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manolio TA, Chisholm RL, Ozenberger B,et al. Implementing genomic medicine in the clinic: the future is here. Genet Med. 2013;15(4):258–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng SB, Bigham AW, Buckingham KJ,et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42(9): 790–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y, Muzny DM, Reid JG, et al. Clinicalwhole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med. 2013;369(16):1502–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee H, Deignan JL, Dorrani N, et al.Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312(18): 1880–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips KA, Deverka PA, Hooker GW,Douglas MP. Genetic test availability and spending: where are we now? Where are we going? Health Aff (Millwood). 2018;37(5):710–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gay EA, Byers PH, Bennett RL, Bird TD,Hisama FM. Trends over 42 years in the Adult Medical Genetics Clinic at the University of Washington. Genet Med. 2019;21(6):1457–1461 [DOI] [PubMed] [Google Scholar]
- 8.Kotzer KE, Riley JD, Conta JH, Anderson CM, Schahl KA, Goodenberger ML. Genetic testing utilization and the role of the laboratory genetic counselor. Clin Chim Acta. 2014;427: 193–195 [DOI] [PubMed] [Google Scholar]
- 9.McCullough LB, Slashinski MJ, McGuire AL, et al. Is whole-exome sequencing an ethically disruptive technology? Perspectives of pediatric oncologists and parents of pediatric patients with solid tumors. Pediatr Blood Cancer. 2016;63(3):511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauser D, Obeng AO, Fei K, Ramos MA,Horowitz CR. Views of primary care providers on testing patients for genetic risks for common chronic diseases. Health Aff (Millwood). 2018; 37(5):793–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valencia CA, Husami A, Holle J, et al.Clinical impact and cost-effectiveness of whole exome sequencing as a diagnostic tool: a pediatric center’s experience. Front Pediatr. 2015;3:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iglesias A, Anyane-Yeboa K, Wynn J, et al.The usefulness of whole-exome sequencing in routine clinical practice. Genet Med. 2014;16(12):922–931 [DOI] [PubMed] [Google Scholar]
- 13.Baldridge D, Heeley J, Vineyard M, et al. The Exome Clinic and the role of medical genetics expertise in the interpretation of exome sequencing results. Genet Med. 2017;19(9): 1040–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowdin SC, Hayeems RZ, Monfared N,Cohn RD, Meyn MS. The SickKids Genome Clinic: developing and evaluating a pediatric model for individualized genomic medicine. Clin Genet. 2016;89(1):10–19 [DOI] [PubMed] [Google Scholar]
- 15.Green RC, Goddard KAB, Jarvik GP, et al. ; CSER Consortium. Clinical Sequencing Exploratory Research consortium: accelerating evidence-based practice of genomic medicine. Am J Hum Genet. 2016;98(6):1051–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weitzel KW, Alexander M, Bernhardt BA,et al. ; IGNITE Network. The IGNITE Network: a model for genomic medicine implementation and research. BMC Med Genomics. 2016;9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottesman O, Kuivaniemi H, Tromp G,et al. ; eMERGE Network. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15(10):761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith HS, Swint JM, Lalani SR, et al.Clinical application of genome and exome sequencing as a diagnostic tool for pediatric patients: a scoping review of the literature. Genet Med. 2019;21(1): 3–16 [DOI] [PubMed] [Google Scholar]
- 19.McEwen JE, Boyer JT, Sun KY, Rothenberg KH, Lockhart NC, Guyer MS. The Ethical, Legal, and Social Implications Program of the National Human Genome Research Institute: reflections on an ongoing experiment. Annu Rev Genomics Hum Genet. 2014;15: 481–505 [DOI] [PubMed] [Google Scholar]
- 20.Hamilton AB, Oishi S, Yano EM, Gammage CE, Marshall NJ, Scheuner MT. Factors influencing organizational adoption and implementation of clinical genetic services. Genet Med. 2014;16(3): 238–245 [DOI] [PubMed] [Google Scholar]
- 21.Machini K, Douglas J, Braxton A, Tsipis J, Kramer K. Genetic counselors’ views and experiences with the clinical integration of genome sequencing. J Genet Couns. 2014;23(4):496–505 [DOI] [PubMed] [Google Scholar]
- 22.Vassy JL, Christensen KD, Schonman EF,et al. ; MedSeq Project. The impact of whole-genome sequencing on the primary care and outcomes of healthy adult patients: a pilot randomized trial. Ann Intern Med. 2017;167(3):159–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayflick SJ, Eiff MP. Role of primary care providers in the delivery of genetics services. Community Genet. 1998;1(1):18–22 [DOI] [PubMed] [Google Scholar]
- 24.Hayflick SJ, Eiff MP, Carpenter L, Steinberger J. Primary care physicians’ utilization and perceptions of genetics services. Genet Med. 1998; 1(1):13–21 [DOI] [PubMed] [Google Scholar]
- 25.Dickerson JA, Cole B, Conta JH, et al.Improving the value of costly genetic reference laboratory testing with active utilization management. Arch Pathol Lab Med. 2014;138(1):110–113 [DOI] [PubMed] [Google Scholar]
- 26.Miller CE, Krautscheid P, Baldwin EE,et al. Genetic counselor review of genetic test orders in a reference laboratory reduces unnecessary testing. Am J Med Genet A. 2014; 164A(5):1094–1101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.