Abstract
Ferns are one of the most significant plant groupings that comprise a substantial proportion of the plant flora due to the fact of their great diversity, especially in tropical areas. The biodiversity of fungi associated with ferns and fern-like hosts has also received little attention in studies. Plant samples were collected from diseased and dead plants of ten fern or fern-like species from Chiang Rai in northern Thailand. Forty-one isolates were selected from the obtained isolates for molecular and morphological analysis, with a focus on pathogenic fungal genera and consideration of the diversity in host and geographical location. Twenty-six species belonging to seven genera (Colletotrichum, Curvularia, Diaporthe, Fusarium, Lasiodiplodia, Neopestalotiopsis, and Pestalotiopsis) in six families were identified. Thirty new hosts, eight new geographical hosts, and one new species, Colletotrichum polypodialium, are described. Nepestalotiopsis phangngaensis, N. pandancola, Diaporthe tectonendophytica, D. chiangraiensis, and D. delonicis were isolated for the first time from leaf spots. Additionally, new reservoirs and geographical locations for species previously isolated from leaf spots or whose pathogenicity was established were found. However, more studies are necessary to prove the pathogenicity of the fungi isolated from the leaf spots and to identify the fungi associated with other species of ferns.
Keywords: Colletotrichum, Curvularia, Diaporthe, Fusarium, Lasiodiplodia, pestalotiod fungi
1. Introduction
Ferns are considered an important component of plant diversity in Thailand, which consists of approximately 5–7% of the total flora in this country, and 670 taxa have been estimated [1]. Ferns also comprise a large proportion of the world’s plant flora (9000–12,000 species) [2,3]. They are used as ethnomedicine, pests controllers, food, and ornamental plants [4]. Additionally, they play a crucial role in ecology and interact with a variety of organisms, including insects and saprobic, endophytic, pathogenic, and mycorrhizal fungi [5].
Endophytes related to ferns have not been extensively studied. An investigation of the endophytes of seven fern species in Costa Rica revealed that more than 95% of the fungi belonging to Ascomycota, with Dothideomycetes, Eurotiomycetes, and Sordariomycetes dominating [6], with a prevalence of Xylariales [6]. Pathogenic Ascomycota and Basidiomycota have been isolated from ferns worldwide [7]. Inocyclus angularis was reported to cause tar spots on Pleopeltis astrolepis in Brazil [8] and Milesina dryopteridis to cause rust on Rumohra adiantiformis and Pteris fauriei in Japan [9]. Pestalotiopsis maculans causes leaf spots on Lygodium venustum in Argentina [10] and Colletotrichum gloeosporioides on Lygodium microphyllum and L. japonicum in the USA [11]. Additionally, C. acutatum causes anthracnosis in leather ferns in the USA [12] and Fusarium thapsinum brown rot in Azolla microphylla in India [13].
In contrast to endophytes and pathogens, saprobes have been frequently identified in ferns [5], accounting for 21 ascomycetes on the fronds and rachises of various ferns in Mexico [14]. More recently, new species have been introduced from this plant group: Venustosynnema reniformisporum from dead leaves of Selaginella moellendorffii [15], Pestalotiopsis magna from dead leaves of Pteridium sp. [16], and Monilochaetes pteridophytophila from the dead stalk of Alsophila costularis [17].
Given that Thailand is a center of fern diversity and few studies have investigated their diversity, it is assumed that this plant category is colonized by a wide range of fungal species. The current study aimed to identify saprobes and leaf spot fungi associated with ferns and fern-like species using molecular and morphological methods.
2. Results
This study identified 26 fungal species from fern and fern-like hosts in Chiang Rai, northern Thailand. The phylogenetic results and species descriptions for all species are provided below.
Dothideomycetes O.E. Erikss. & Winka;
Botryosphaeriales C.L. Schoch, Crous & Shoemaker, Mycologia 98 (6): 1050 (2007).
This order includes six families: Botryosphaeriaceae, Melanopsaceae, Phyllostictaceae, Planistromellaceae, and Saccharataceae [18].
Botryosphaeriaceae Theiss. & Syd (as “Botryosphaeriacae”), Annls mycol. 16(1/2): 16 (1918).
Members of this family are pathogens, endophytes, or saprobes on a wide range of hosts. They can also function as opportunists and primary plant pathogens [19,20]. Twenty-two genera are accepted in this family [18].
Lasiodiplodia Ellis & Everh., Botanical Gazette Crawfordsville, 21: 92 (1896).
Based on molecular data, 35 species have been identified for this genus [18]. In the present study, two isolates obtained from ferns were identified as L. thailandica based on morphological and phylogenetic analyses of ITS, tef1, and tub2 loci.
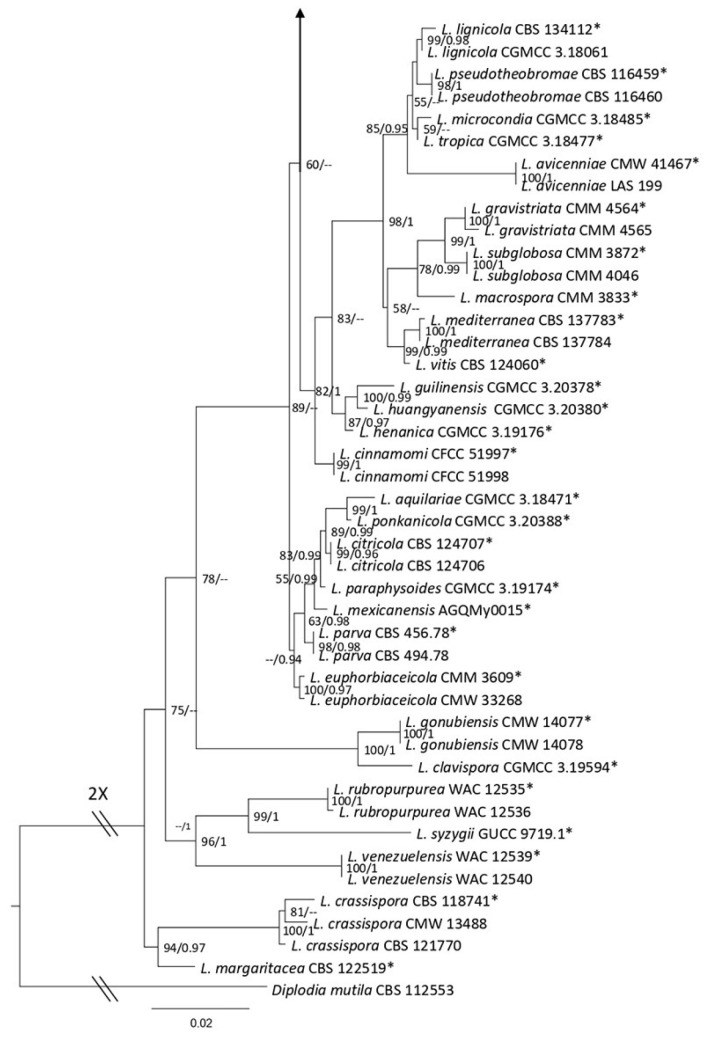
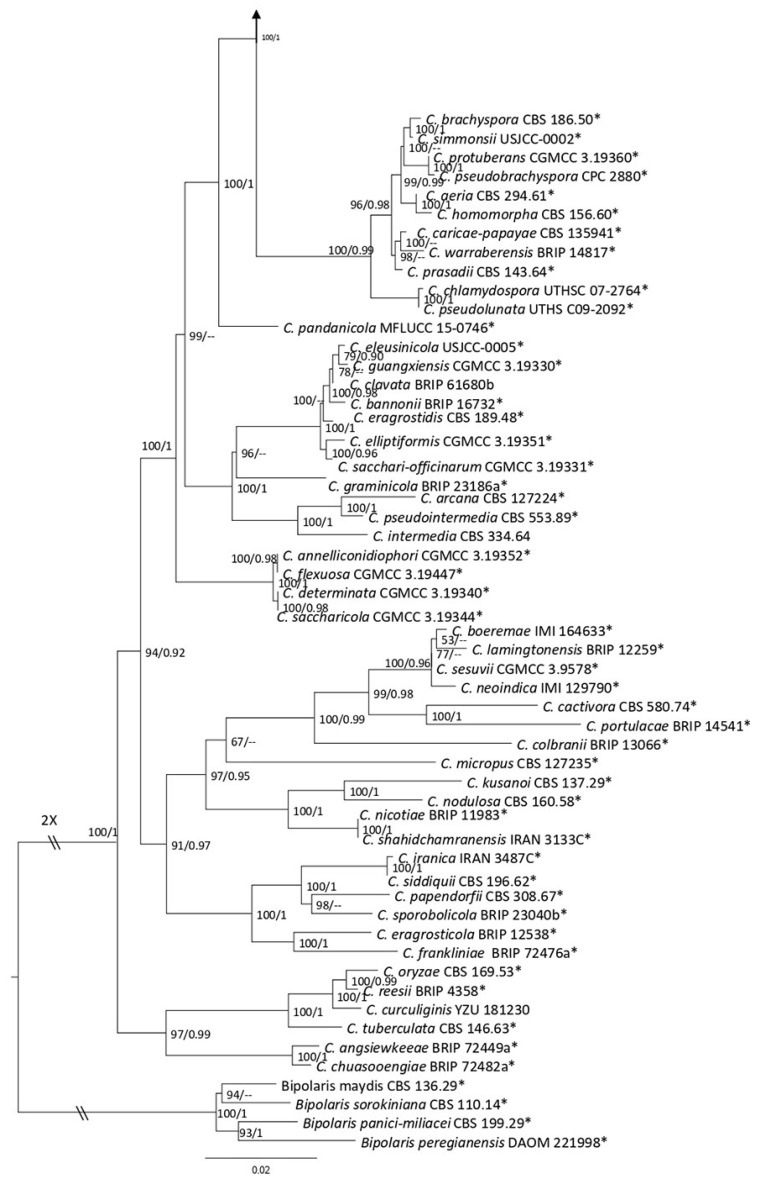
Figure 1.
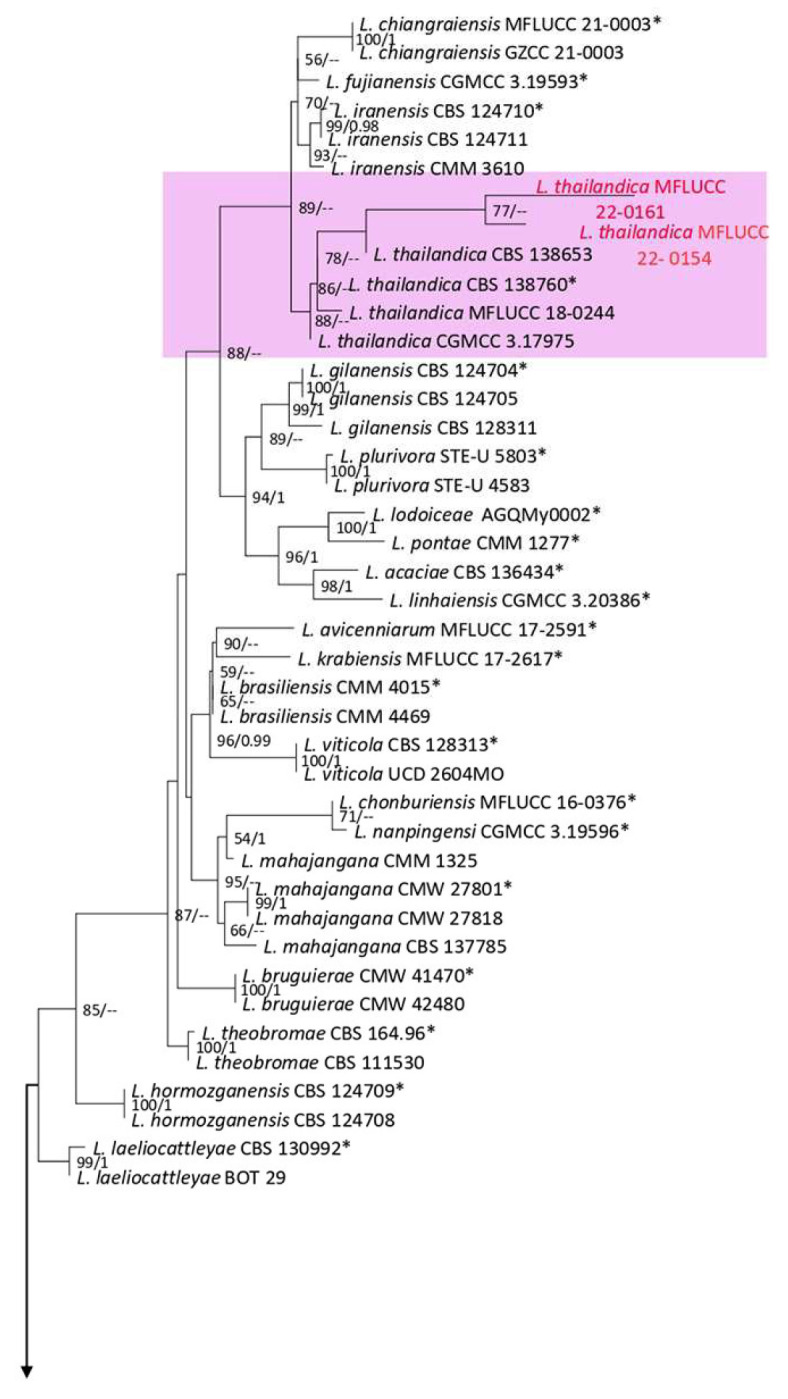
Maximum likelihood (ML) phylogenetic tree of Lasiodiplodia spp. obtained from the combined ITS, tef1, and tub2 sequence data. The ultrafast maximum likelihood bootstrap support (%) ≥ 50 and BYPP ≥ 0.95 are shown at the nodes. The ex-type strains are marked with an asterisk. The tree is rooted in Diplodia mutila (CBS 112553).
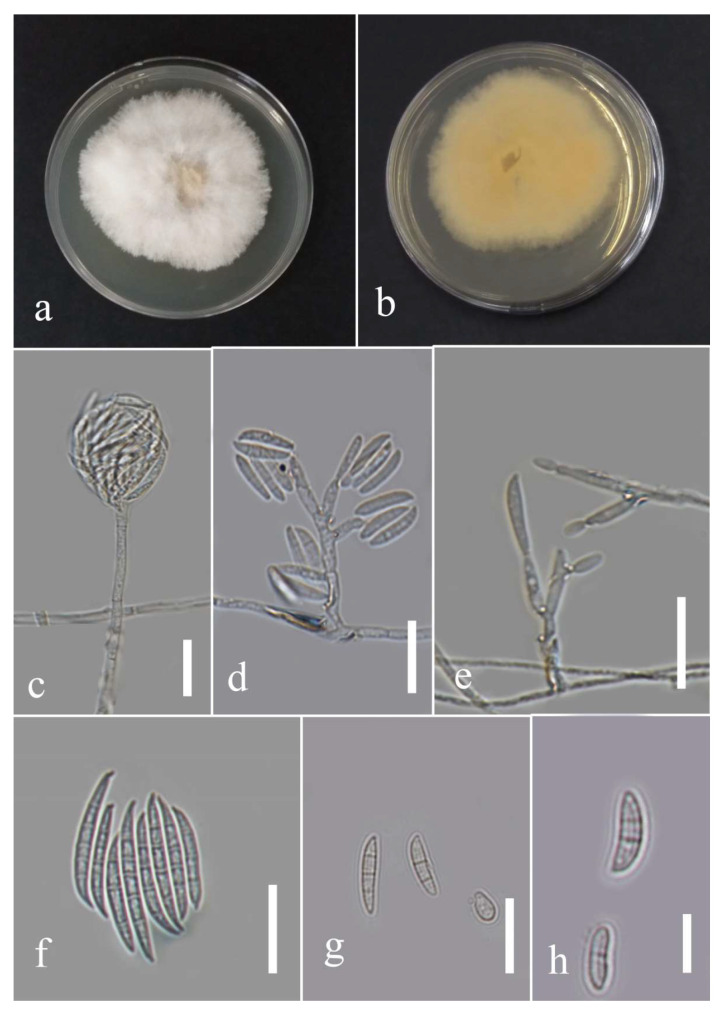
Lasiodiplodia thailandica Trakun., L. Lombard & Crous, 2014.
Facesoffungi number: FoF 09333; Figure 2.
Associated with Asplenium nidus leaf spot. Sexual morph: not observed. Asexual morph: Conidiomata pycnidium, a few, scattered, singular, unilocular, superficial, and semi-immersed, globes, black with dark-brown outer layer and hyaline inner layer with hyphae tips around the outer wall of the pycnidium, brown, septate and rounded tips. Conidiophores reduced to conidiogenous cells. Conidiogenous cells cylindrical, hyaline, holoblastic, and thin walled. Paraphyses originated from the inner cells of pycnidium, cylindrical, hyaline, 1–3 septate, the basal cell swollen, thin walled. Conidia elliptic with granular content, hyaline at the immature stage, pale brown at maturity, aseptate, thick walled (1.1–1.7 µm), 15–26 × 9–16 µm (mean = 21.0 × 12.9, n = 30).
Culture characteristics: Colonies filled a 90 mm Petri dish at seven days on PDA at 28 °C; fluffy, circular, and dull surface; medium density, and without pigmentation in the medium and fruiting body. The upper view was white, and the reverse primrose.
Material examined: Thailand, Chiang Rai Province, Muang District, Huai Sak, on leaf spots of Asplenium nidus (Aspleniaceae); 17 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0207) and living culture MFLUCC 22-0161; Thailand, Chiang Rai Province, Muang District, Thasud, on dead leaves of Nephrolepis cordifolia (Nephrolepidaceae); 3 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0208) and living culture MFLUCC 22-0154.
Notes: Isolated from blight spots on Asplenium nidus. The isolates obtained in this study (MFLUCC 22-0161 and MFLUCC 22-0154) were clustered with L. thailandica with 78% ML bootstrap support (Figure 1). A comparison of the ex-type strain L. thailandica (CBS 138760) with strains MFLUCC 22-0161 and MFLUCC 22-0154 revealed identical sequences in ITS (for both isolates) and tef1 (only for MFLUCC 22-0154). The sequences of tub2 for the type strain and tef1 for strain MFLUCC 22-0161 are not available. This species was first reported from Mangifera indica and Phyllanthus acidus in Thailand [21], and here it was isolated from Asplenium nidus and Nephrolepis cordifolia. Hence, we provide new host records for this species.
Figure 2.
Morphology of Lasiodiplodia thailandica (MFLUCC 22-0161): (a) blight spots on Asplenium nidus; (b) upper reverse view of the colony after seven days of growth on PDA at 28 °C; (c) conidiomata; (d) conidiogenous cells; (e–h) conidia. Scale bars (d) = 20 µm and (e–h) = 10 µm.
Pleosporales Luttr. ex M.E. Barr, Prodromus to class Loculoascomycetes: 67 (1987).
Pleosporales is the largest order of Dothideomycetes, which covers a quarter of the fungi in this class and, currently, it comprises 92 families (OUTLINE OF FUNGI 2023). According to these authors, Pleosporales species are reported as pathogens, saprobes, endophytes, epiphytes, pathogen-like organisms on fungi or insects, and lichenized fungi.
Pleosporaceae Nitschke, Verh. naturh. Ver. preuss. Rheinl. 26: 74 (1869).
Based on molecular and morphological data, 23 genera are accepted in Pleosporaceae, including human pathogens and plant saprobes [22].
Curvularia Boedijn, Bull. Jard. bot. Buitenz, 3 Sér. 13(1): 123 (1933).
Curvularia includes more than 170 epithets (Index Fungorum 2022). In this study, one isolate obtained from ferns was identified as C. lunata based on phylogenetic analyses of the ITS, gapdh, and tef1 sequences and morphological data.
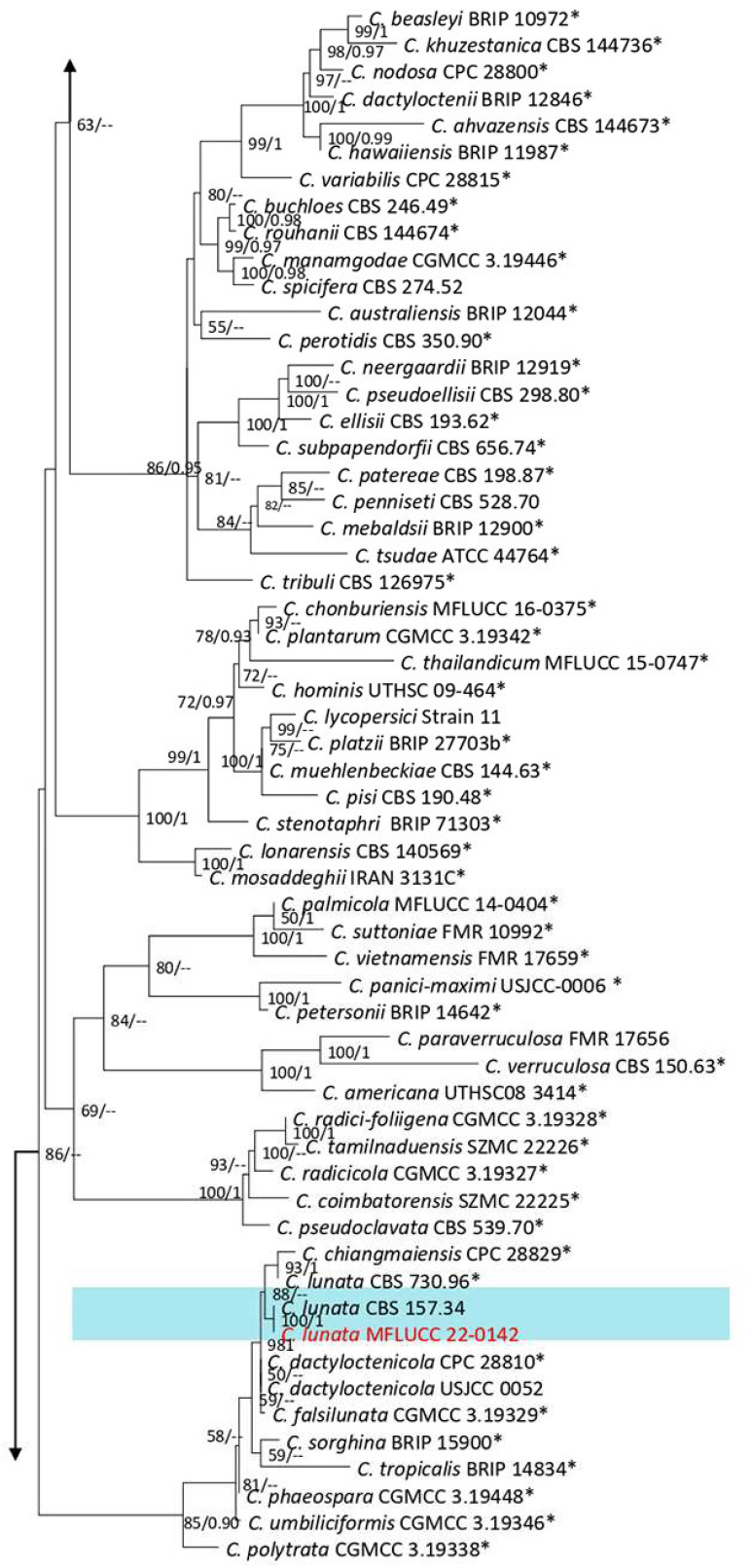
Figure 3.
Maximum likelihood (ML) phylogenetic tree of Curvularia spp. Obtained from combined ITS, gapdh, and tef1 sequence data. The ultrafast maximum likelihood bootstrap support (%) ≥ 50 and BYPP ≥ 0.90 are shown at the nodes. The ex-type strains are marked with an asterisk. The tree is rooted in Bipolaris panici-miliacei (CBS 199.29), B. peregianensis (DAOM 221998), B. sorokiniana (CBS 110.14), and B. maydis (CBS 136.29).
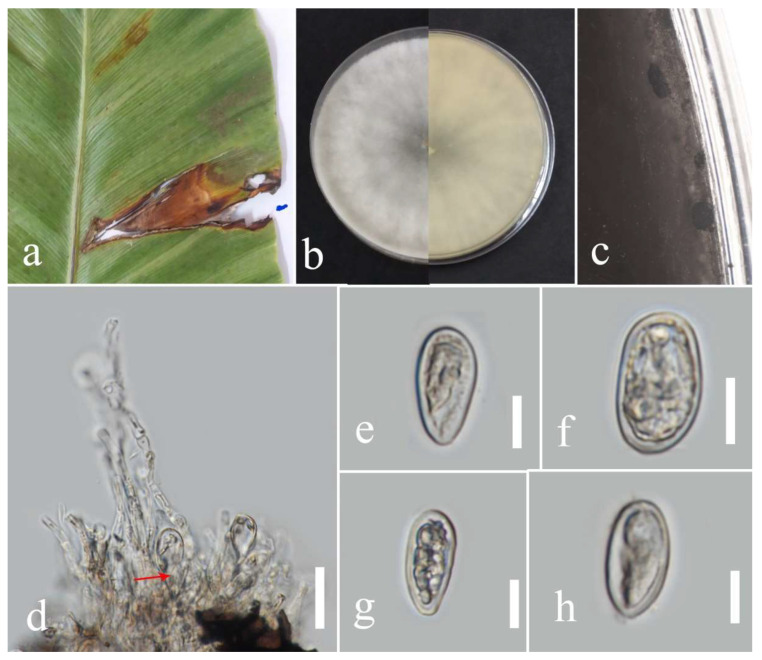
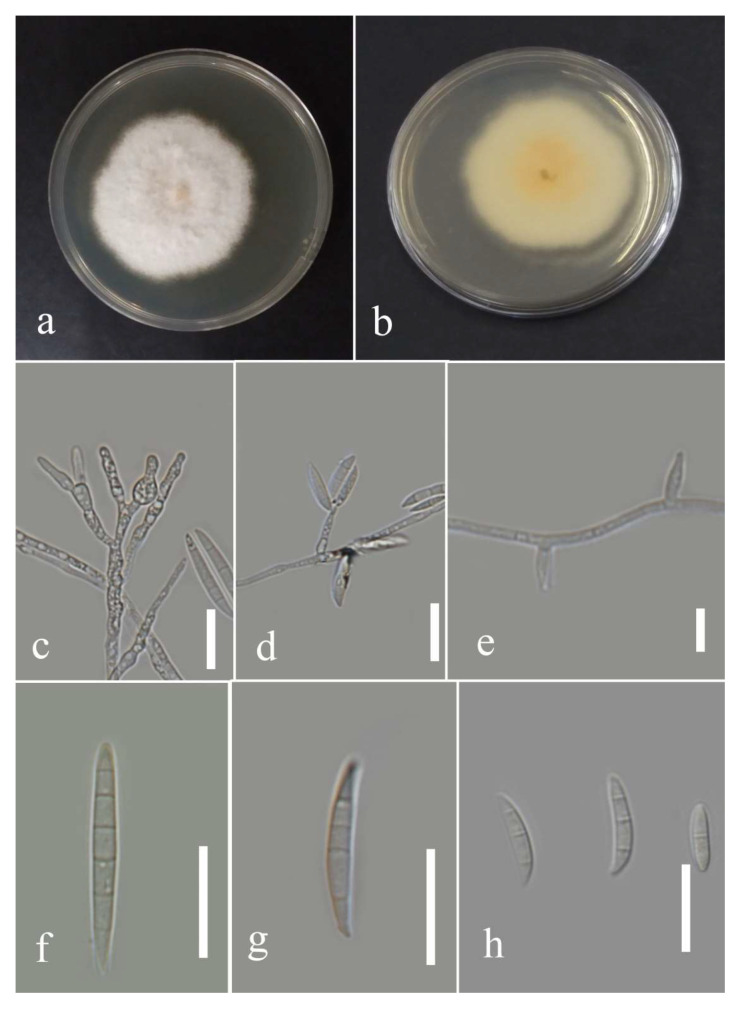
Curvularia lunata (Wakker) Boedijn, 1933; Figure 4.
Associated with Pteris grandifolia leaf blight. Sexual morph: not observed. Asexual morph: hyphae pale to medium brown; septate, 1.9–3.9 µm wide. Conidiophores straight to flexible, medium brown, septate, unbranched, geniculate toward the tip, 27–270 µm long. Conidiogenous cell sympodial, brown, terminal or intercalary, polytretic with dark scars. Conidia cylindrical to elliptic, pale to medium brown, 3-distoseptate, rounded at tips, the third cell from base swollen on one side in some spores, straight on one side, and convex on the opposite side, swollen cell larger with a similar color to other cells, 14–23 µm (mean = 17.96, n = 30) × 5–11 µm (mean = 7.8, n = 30).
Culture characteristics: Colonies reached 17–27 mm after seven days of growth on PDA at 28 °C; felted, irregular shape, dull surface, lobate edge, well-defined margin, and medium density with pigmentation in media. Upper view smoke gray and reverse rust to chestnut.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, leaf spot on Pteris grandifolia (Pteridaceae), 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0209) and living culture MFLUCC 22-0142.
Notes: Isolated from blight spot on Pteris grandifolia leaves. The isolate obtained (MFLUCC 22-0142) clustered with C. lunata, with 100% ML bootstrap support and 1.0 BYPP (Figure 3). A pairwise comparison of the neotype strain (CBS 730.96) and the strain MFLUCC 22-0142 revealed a 0.82% nucleotide differences in gapdh (4 nucleotides) and 0.11% in tef1, while the ITS sequences were identical. This species was first reported from a lung biopsy in the USA and redescribed using a neotype [23]. It was found on Brassica rapa, Morus sp., Oryza sativa, Panicum sp., and Zea mays in Thailand [23,24,25,26]. Here, we provide a new host record for C. lunata. In the phylogenetic tree, C. lunata and C. chiangmaiensis clustered in the same clade with 90% ML bootstrap support, suggesting they may belong to the same species. Pairwise comparison of the DNA sequence data of the type strain of C. lunata (CBS 730.96) and C. chiangmaiensis (CPC 28829) revealed only 0.27% nucleotide differences in ITS (one nucleotide) and 0.34% in tef1 (three nucleotides), while the sequences of gapdh were identical.
Figure 4.
Morphology of Curvularia lunata (MFLUCC 22-0142): (a) blight on Pteris grandifolia; (b,c) upper and reverse views of the culture after seven days on PDA at 28 °C; (d–f) conidiophores and sympodial conidiogenous cells; (g–j) conidia. Scale bars (d) = 50 µm, (e–f) = 25 µm, and (g–j) = 10 µm.
Sordariomycetes O.E. Erikss. & Winka.
Amphisphaeriales D. Hawksw. & O.E. Erikss., Systema Ascomycetum 5: 177 (1986).
This order comprises 88 genera and 17 families: Amphisphaeriaceae, Apiosporaceae, Beltraniaceae, Castanediellaceae, Clypeophysalosporaceae, Cylindriaceae, Hyponectriaceae, Iodosphaeriaceae, Melogrammataceae, Oxydothidaceae, Phlogicylindriaceae, Pseudomassariaceae, Pseudosporidesmiaceae, Pseudotruncatellaceae, Sporocadaceae, Vialaeaceae, and Xyladictyochaetaceae. The taxonomic treatment of this order is based on Hyde et al. [27].
Sporocadaceae Corda, Icones fungorum hucusque cognitorum 5: 34 (1842).
This family included 23 genera, including endophytes, saprobes, plant pathogens, and parasites of animals and humans. Members of Sporocadaceae are characterized by acervuli, septate, hyaline, and pale to dark brown conidia. Additionally, they are distinguished by the sequence data of ITS, LSU, and rpb2 [27].
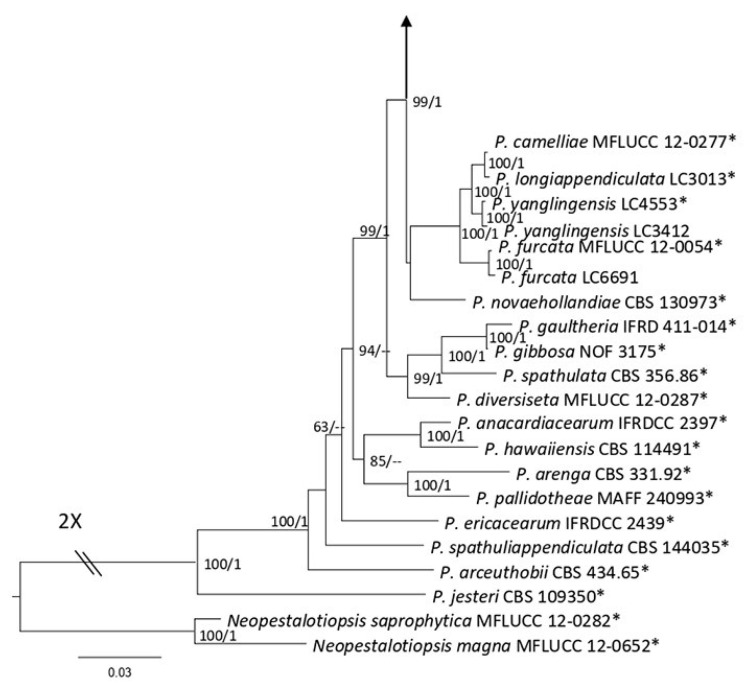
Neopestalotiopsis Maharachch., K.D. Hyde & Crous, Studies in Mycology 79: 135.
Neopestalotiopsis species are found as saprobes or plant pathogens. Due to the versicolorous median cells of the conidia, this genus was recognized from Pesatalotiopsis [28]. This study reports seven Neopestalotiopsis species associated with ferns based on morphomolecular justification: N. guajavicola, N. hydeana, N. musae, N. pandanicola, N. phangngaensis, N. psidii, and N. saprophytica.
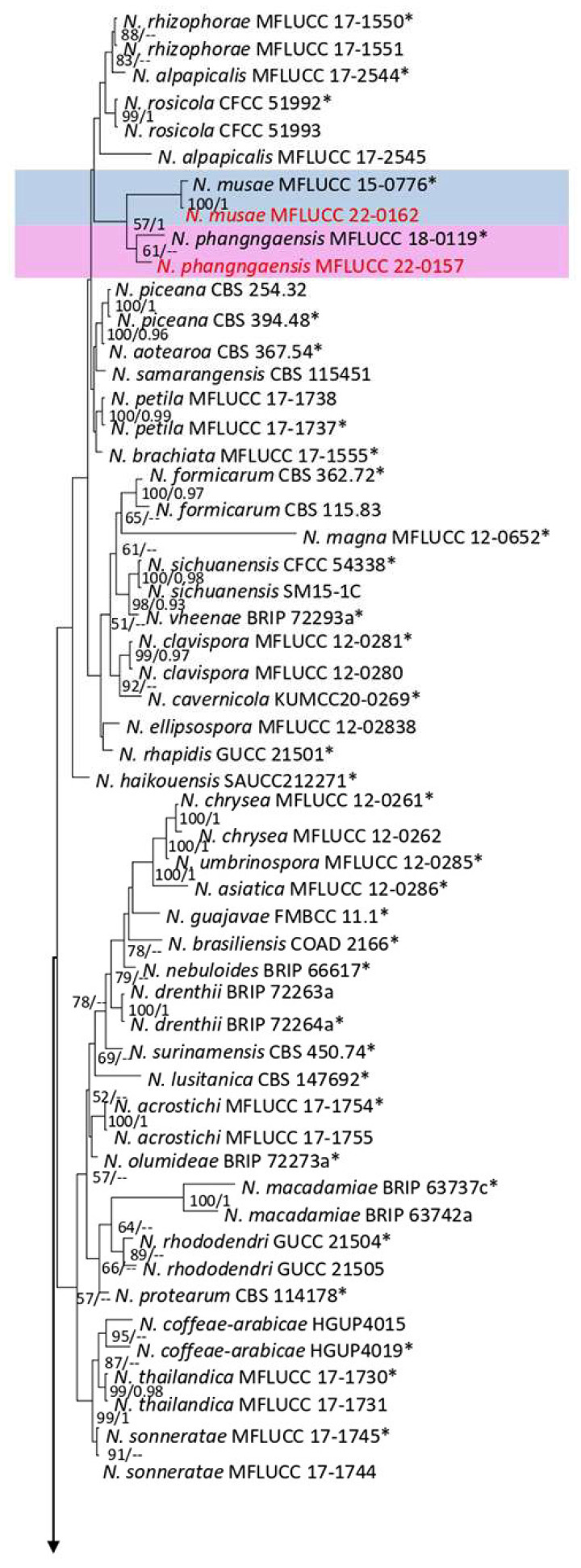
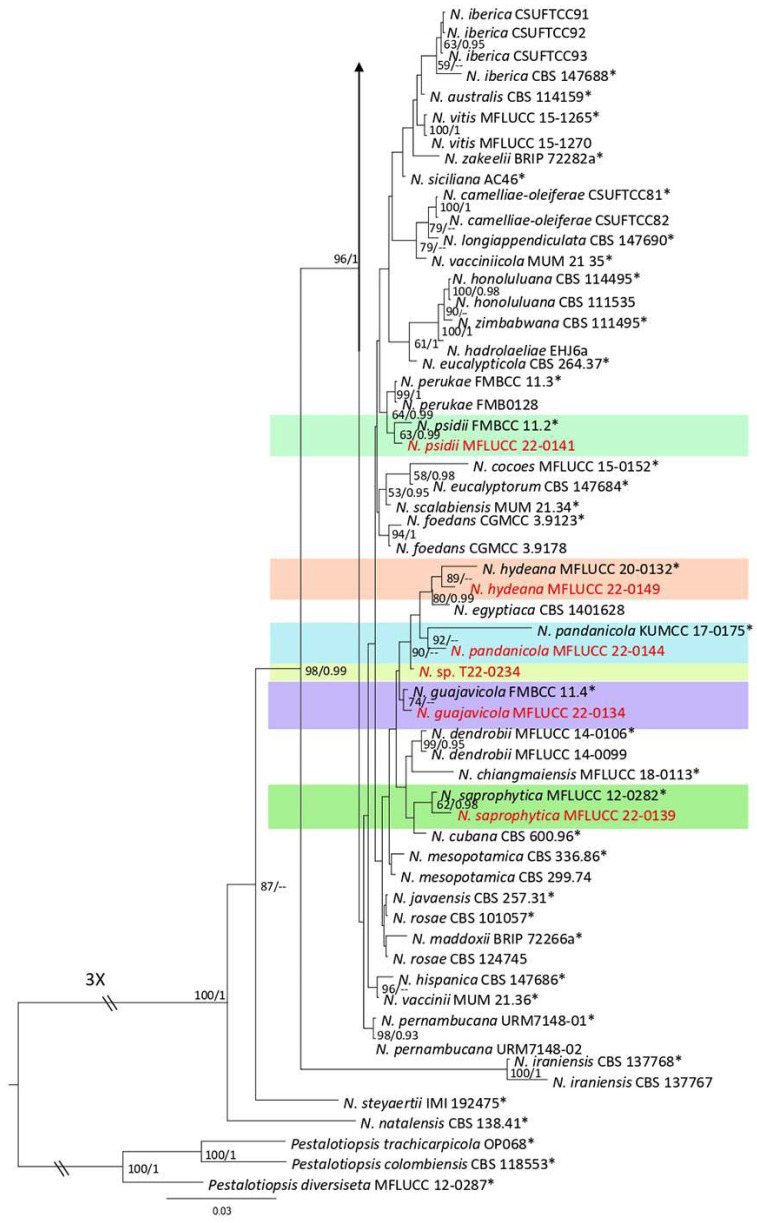
Figure 5.
Bayesian phylogenetic tree of Neopestalotiopsis spp. Obtained from the combined ITS, tef1, and tub2 sequence data. The ultrafast maximum likelihood bootstrap support (%) ≥ 50 and BYPP ≥ 0.95 are shown at the nodes. The ex-type strains are marked with an asterisk. The tree is rooted in Pestalotiopsis trachicarpicola (OP068), P. colombiensis (CBS 118553), and P. diversiseta (MFLUCC 12-0287).
Neopestalotiopsis guajavicola I.U. Haq, Ijaz & N.A. Khan, 2021.
Facesoffungi number: FoF 13402; Figure 6.
Associated with Nephrolepis sp. leaf spot. Sexual morph: not observed. Asexual morph: conidiomata ample, aggregated and scattered, immersed and semi-immersed, exuding black conidial mass. Conidiophores reduced to conidiogenous cells. Conidiogenous cells subcylindrical to ampliform, hyaline to pale brown. Conidia fusiform, straight or curved, 4 septate, 17–22 × 4–8 µm (mean = 20.1 × 6.5, n = 30). Basal cell conoid with truncate base, hyaline, 2–4 µm long; median cells, versicolored, darker spectate than other cells, 12–16 × 5–8 µm (mean = 14.3 × 6.4, n = 30) (second cell from the base pale brown, 3.5–5.9 µm long; third cell from the base medium brown, 3–7 µm long; forth cell from the base pale to medium brown, 3–6 µm long); apical cell conic, hyaline, 2–5 µm with 2–3 tubular apical appendage (often three) filiform, hyaline, unbranched, and 11–21 µm long (mean = 16.1, n = 30); basal appendage filiform, hyaline, unbranched, solitary, and 1–7 µm long.
Culture characteristics: The colonies reached 72–80 mm after seven days of growth on PDA at 28 °C; cottony, irregular shape, dull surface, undulated edge, fluffy margin, medium density, and without pigmentation in the medium and conidial mass. Upper view is white, reverse pale luteous in the center, and primrose in other areas.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, on leaf spot of Nephrolepis sp. (Nephrolepidaceae), 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0210) and living culture MFLUCC 22-0134.
Notes: Isolated from a blight on leaves of Nephrolepis sp. The obtained isolate (MFLUCC 22-0134) clustered with N. guajavicola with a 74% ML bootstrap (Figure 5). Comparison of the DNA sequences of N. guajavicola strains (ex-type FMBCC 11.4 and MFLUCC 22-0134) revealed 0.59% nucleotide differences in tef1 (1 gap) and 0.24% in tub2 (one nucleotide) genes, while the sequences of ITS were identical. This species was first reported from a guava tree in Pakistan [29] and, here, it was isolated from Nephrolepis sp. In Thailand. Herein, we provided a new host and a geographical record for N. guajavicola.
Figure 6.
Morphology of Neopestalotiopsis guajavicola (MFLUCC 22-0134): (a) blight on Nephrolepis sp.; (b,c) upper and reverse views of the culture after seven days of growth on PDA at 28 °C; (d) conidiomata; (e) conidiogenous cells; (f,g) conidia. Scale bars = 10 µm.
Neopestalotiopsis hydeana Huanraluek & Jayaward., 2021.
Facesoffungi number: FoF 09459; Figure 7.
Associated with leaf spot of Cyclosorus sp. Sexual morph: not observed. Asexual morph: conidiomata not observed. Conidia were found rarely, fusiform, straight, 4 septate, 19–27 × 4–7 µm (mean = 23.3 × 6.2, n =20). Basal cell conoid with a truncated base, hyaline, 3.9–5.4 µm; median cell versicolored, darker septate than other cells, 9–17 × 4–9 µm (mean = 15.1 × 6.0, n = 20) (second cell from the base pale brown, 4–7 µm long; third cell medium brown, 4–6 µm long; forth cell medium brown, 3–6 µm long); apical cell conoid, hyaline, 1–5 µm long with 2–3 apical appendages tubular, filiform, hyaline, unbranched, and 4–13 µm long (mean = 9.1, n = 20); basal appendage filiform, hyaline, unbranched, single, and 2–6 µm long.
Culture characteristics: The colonies reached 65–67 mm after seven days of growth on PDA at 28 °C; cottony, entire edge, fluffy margin, medium density, and without pigmentation in the medium and fruiting body. Upper view white and the reverse primrose.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, on leaf spot of Cyclosorus sp. (Thelypteridaceae), 17 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0211) and living culture MFLUCC 22-0149.
Notes: Isolated from a necrotic spot with a dark-brown margin. The obtained isolate in this study (MFLUCC 22-0149) clustered with N. hydeana by 89% ML bootstrap support (Figure 5). Comparison of the DNA sequences of N. hydeana strains (MFLUCC 20-0132 and ex-type strain T22-0333) showed 0.83% nucleotide differences in ITS (three nucleotides and one gap), 0.91% in tef1 (four nucleotides), and 0.26% in tub2 (one nucleotide). This species was isolated from fruit rot in Annona squamosal and Garcinia mangostana and leaf spots on Alpinia malaccensis and Garcinia mangostana in Thailand [30] and, here, it was isolated from Cyclosorus sp. Hence, we provide a new host record for N. hydeana.
Figure 7.
Morphology of Neopestalotiopsis hydeana (MFLUCC 22-0149): (a) necrotic spot in Cyclosorus sp.; (b,c) upper and reverse views of the colony after seven days of growth on PDA at 28 °C; (d) immature conidium; (e) mature conidium. Scale bars = 10 µm.
Neopestalotiopsis musae Norphanph., T.C. Wen & K.D. Hyde, 2016; Figure 8.
Associated with Asplenium nidus leaf spot. Sexual morph: not observed. Asexual morph: conidiomata ample, scattered, and aggregated, immersed and semi-immersed, with black conidial mass. Conidiophores reduced to conidiogenous cells. Conidiogenous cells subcylindrical to ampulliform and hyaline to medium brown. Conidia fusiform, straight to slightly curved, 4 septate, 19–26 × 5–9 µm (mean = 22.7 × 6.9, n = 30). Basal cell conical, hyaline to pale brown, and 3–6 µm; three median cells doliiform, versicolored, and darker septate than the other cells, 12–17 × 5–9 µm (mean = 14.55 × 6.75, n = 30) (second cell from the base pale brown, 3–7 µm long; third cell pale to medium brown, 3–7 µm long; forth cell pale to medium brown, 3–6 µm long); apical cell conical, hyaline, 3–6 µm long with 2–3 apical appendages (often, two appendages), filiform, hyaline, unbranched, and 13–28 µm long (mean = 22.5, n = 30); basal appendage filiform, hyaline, unbranched, singular, and 3–7 µm long.
Culture characteristics: Colonies reached 31–33 mm after seven days of growth on PDA at 28 °C; felted to cottony, dull surface, entire edge, regular and fluffy margin, medium density, and without pigmentation in the medium and fruiting body. Upper view white and the reverse primrose to straw.
Material examined: Thailand, Chiang Rai Province, Muang District, Huai Sak, on leaf spot of Asplenium nidus, 17 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0212) and living culture MFLUCC 22-0162.
Notes: Isolated from blights on leaves of Asplenium nidus. The isolate obtained in this study (MFLUCC 22-0162) grouped with N. musae in the same clade by 100% ML bootstrap support and 1.0 BYPP (Figure 5). Comparison of the DNA sequences of N. musae strains (MFLUCC 15-0776 and ex-type strainT22-0164) showed 0.21% nucleotide differences in the ITS (1 gap), 0.49% in tub2 (1 nucleotide and 1 gap) genomic regions, while the tef1 sequences were identical. This species was first reported from Musa sp. in Thailand [31] and, here, it was isolated from Asplenium nidus, providing a new host record.
Figure 8.
Morphology of Neopestalotiopsis musae based on the isolate MFLUCC 22-0162: (a) leaf spot in Asplenium nidus; (b,c) upper and reverse views of the colony after seven days of growth on PDA at 28 °C; (d) conidiomata; (e,f) conidiogenous cells; (g–i) conidia. Scale bars = 10 µm.
Neopestalotiopsis pandanicola Tibpromma & K.D. Hyde, 2018.
Facesoffungi number: FoF 13403; Figure 9.
Associated with leaf spot. Sexual morph: Not observed. Asexual morph: Conidiomata ample, scattered, and aggregated, immersed and semi-immersed with black conidial mass. Conidiophores reduced to conidiogenous cells. Conidiogenous cells subcylidrical to ampulliform, hyaline. Conidia fusiform, straight to slightly curved, 4-septate, 16–25 µm × 4–7 µm (mean = 21.1 × 5.5, n= 30). Basal cell conical, hyaline, 3–7 µm long; three median cells doliform, concoloured, pale to medium brown, darker septate than the other cells, and 10–16 × 4–7 µm (mean = 13.6 × 5.4, n = 30) (the second cell from the base 4–6 µm long; the third cell 3–6 µm long; and the fourth cell 3–7 µm); apical cell conical to cylindrical, hyaline, 3–7 µm long, with 2–3 apical appendages, filiform, hyaline, unbranched, and 7–30 µm long (mean = 17.6, n = 30); basal appendage filiform, hyaline, unbranched, single, and 3–7 µm long.
Culture characteristics: Colonies reached 16–21 mm after seven days of growth on PDA at 28 °C; felted, dull surface, irregular shape, crenate edge, and wrinkled aspect, dense density, and without pigmentation in the medium and fruiting body. Upper view olivaceous buff in center and white in other areas, and reverse pale luteous with ochreous parts.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, on leaf spot of Cyclosorus sp., 4 December 2021, Elaheh Seifollahi, (MFLU 22-0213), living culture MFLUCC 22-0144.
Notes: Isolated from a necrotic spot with a brown margin on Cyclosorus sp. The isolate obtained in this study (MFLUCC 22-0144) clustered with N. pandanicola in the same clade by 92% ML bootstrap support (Figure 5). Comparison of the DNA sequences of the ex-type strain of N. pandanicola (KUMCC 17-0175) with strain MFLUCC 22-0144 revealed 4.29% nucleotide differences in tef1 (11 nucleotides and eight gaps) and 0.26% in tub2 (1 nucleotide) genes. The ITS sequence for the type strain was not available. This species was introduced from Pandanus sp. in China [32] and, here, it was isolated from Cyclosorus sp. in Thailand, providing a new host and geographical records.
Figure 9.
Morphology of Neopestalotiopsis pandanicola based on the isolate MFLUCC 22-0144: (a) leaf spot in Cyclosorus sp.; (b,c) upper and reverse views of the colony after seven days of growth on PDA at 28 °C; (d) conidiomata; (e) conidiogenous cell; (f–h) conidia. Scale bars = 10 µm.
Neopestalotiopsis phangngaensis Tibpromma & K.D. Hyde, 2018.
Facesoffungi number: FoF 04527; Figure 10.
Associated with Cyclosorus sp. leaf spot. Sexual morph: not observed. Asexual morph: conidiomata rarely formed, scattered, semi-immersed, with black conidial mass. Conidiophores reduced to conidiogenous cells. Conidiogenous cells subcylindrical to ampulliform, hyaline, entroblastic, and thin walled. Conidia fusiform, straight or slightly curved, 4 septate, and 20–26 × 4–8 µm (mean = 22.6 × 6.0, n = 30); basal cell conical, hyaline to pale brown, 3–6 µm long; three median cells doliiform, versicolored with darker septate than the rest of the cells, and 12–17 µm × 4–8 (mean = 14.7 × 6.1, n = 30) (second cell from the base pale brown, 3–6 µm long; third cell medium brown, 4–7 µm long; forth cell pale to medium brown, 2–7 µm long), apical cell conical, hyaline, 3–6 µm long with 2–3 apical appendages, filiform, hyaline, unbranched, and 8–26 µm long; basal appendage filiform, hyaline, unbranched, singular, and 4–8 µm long.
Culture characteristics: Colonies reached 75–90 mm after seven days of growth on PDA at 28 °C; fluffy to cottony, irregular shape, dull surface, undulate edge, fluffy margin, wrinkled folded aspect in some areas, medium density and without pigmentation in medium and conidial mass. Upper view white and the reverse primrose.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, on leaf spot of Cyclosorus sp., 17 December 2021, Elaheh Seifollah, dried culture (MFLU 22-0214) and living culture (MFLUCC 22-0157).
Notes: Isolated from brown leaf spots on Cyclosorus sp. The isolate obtained in this study (MFLUCC 22-0157) grouped with N. phangngaensis in the same cluster by 61% ML bootstrap support (Figure 5). In comparing the DNA sequences of the ex-type strain (MFLCC 18-0119) with MFLUCC 22-0157, the tef1 for the type strain was removed in analyses because of ambiguities in the sequence. Comparison of the DNA sequences showed 0.22% nucleotide differences in ITS (one gap) and 0.51% in tub2 (two nucleotides) genomic regions. N. phangngaensis was first isolated from dead leaves of Pandanus sp. in Thailand [32] and, here, it was recorded on Cyclosorus sp., providing a new host occurrence.
Figure 10.
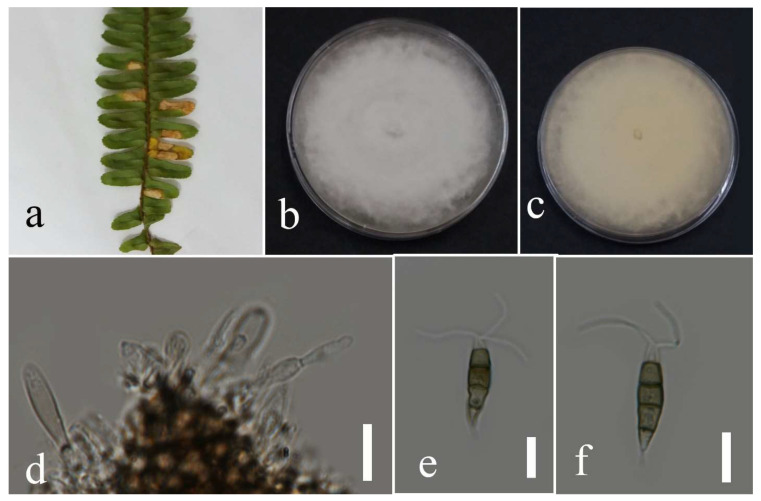
Morphology of Neopestalotiopsis phangngaensis (MFLUCC 22-0157): (a) brown leaf spot in Cyclosorus sp.; (b,c) upper and reverse views of the colony after seven days of growth on PDA at 28 °C; (d) conidiogenous cells; (e,f) conidia. Scale bars = 10 µm.
Neopestalotiopsis psidii I.U. Haq, Ijaz & N.A. Khan, 2021.
Facesoffungi number: FoF 13404; Figure 11.
Associated with Nephrolepis cordifolia leaf spot. Sexual morph: not observed. Asexual morph: conidiomata ample, scattered, or aggregated, immersed and semi-immersed, with black conidial mass. Conidiophores reduced to conidiogenous cells. Conidiogenous cells cylindrical to ampulliform, hyaline, entroblastic, and thin walled. Conidia fusiform, straight to slightly curved, 4-septate, 17–30 µm × 4–8 µm (mean = 22.2 × 6.1, n = 30); basal cell conical, hyaline, thin walled, 2–5 µm long; three median cells doliiform, versicolored with darker septate than the other cells, 12–19 µm × 5–8 (mean = 14.6 × 6.5, n = 30) (second cell from the base pale brown, 3–7 µm long; third cell medium brown, 2–6 µm long; forth cell pale to medium brown, 3–6 µm long); apical cell conical, hyaline, with thin wall, 2–5 µm long with 2–3 apical appendages filiform, hyaline, unbranched, 10–19 µm (mean = 13.99, n = 30); basal appendage filiform, hyaline, unbranched, singular, 1–5 µm long.
Culture characteristics: Colonies filled 90 mm Petri dish after seven days of growth on PDA at 28 °C, cottony, dull surface, irregular margin, undulate edge, medium density, and without pigmentation in medium and conidial mass. Upper view white, and reverse primrose.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, on leaf spot of Nephrolepis cordifolia, 4 December 2021, Elaheh Seifollah, dried culture (MFLU 22-0215), living culture MFLUCC 22-0141.
Notes: Isolated from a necrotic leaf spot with a brown margin on Nephrolepis cordifolia. The isolate obtained in this study (MFLUCC 22-0141) clustered with N. psidii in the same clade by 63% ML bootstrap support and 0.99 BYPP (Figure 5). Comparison of the DNA sequences of N. psidii strains (FMBCC 11.2 and ex-type strain T22-0247) revealed 2.13% nucleotide differences in tef1 (seven nucleotides and one gap) gene, while the sequences of ITS and tub2 were identical. This species was first reported from guava in Pakistan [29] and, here, it was isolated from Nephrolepis cordifolia in Thailand as a new host and geographical record.
Figure 11.
Morphology of Neopestalotiopsis psidii based on isolate MFLUCC 22-0141: (a) leaf spot in Nephrolepis cordifolia; (b,c) upper and reverse views of the colony after seven days of growth on PDA at 28 °C; (d) conidiogenous cells; (e,f) conidia. Scale bars = 10 µm.
Neopestalotiopsis saprophytica Maharachch., K.D. Hyde & Crous, 2014.
Facesoffungi number: FoF 13405; Figure 12.
Associated with leaf spot. Sexual morph: not observed. Asexual morph: conidiomata ample, scattered, and aggregated, semi-immersed with black conidial mass. Conidiophores reduced to conidiogenous cells. Conidiogenous cells lageniform, cylindrical to subcylindrical, and hyaline. Conidia fusiform, straight to slightly curved, 4 septate, 17–29 × 5–8 µm (mean = 21.9 × 6.8, n = 30); basal cell conical, hyaline to pale brown, thin walled, 3–6 µm long; three median cells doliiform, versicolored, and darker septate than the other cells, and 11–18 × 5–8 µm (mean = 14.5 × 6.5, n = 30) (second cell from the base pale brown to olivaceous, 3–6 µm long; third cell from base medium to dark brown, 2–6 µm long; forth cell medium to dark brown, 3–6 µm long); apical cell conical, hyaline, thin walled, 3–6 µm long with 2–3 apical appendages (mostly 3), tubular, hyaline, unbranched, and 13–26 µm (mean = 19.4, n = 30) long; basal appendage filiform, hyaline, unbranched, singular, and 2–10 µm long.
Culture characteristics: Colonies reached 72–80 mm after seven days of growth on PDA at 28 °C; cottony irregular shape, dull surface, undulate edge, medium density, and without pigmentation in the medium and conidial mass. Upper view white and the reverse primrose.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, on leaf spot of Cyclosorus sp., 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0216) and living culture MFLUCC 22-0139.
Notes: Isolated from a brown leaf spot on Cyclosorus sp. The isolate obtained in this study (MFLUCC 22-0139) clustered with N. saprophytica with 62% ML bootstrap support and 0.98 BYPP (Figure 5). Comparison of the DNA sequences of N. saprophytica strains (ex-type strain MFLUCC 12-0282 and MFLUCC 22-0139) revealed 1.03% nucleotide differences in ITS (five nucleotides) and 0.90% in tef1 (four gaps), while the tub2 sequences were identical. The mentioned species was first reported from Litsea rotundifolia and Magnolia sp. in China [28] and, here, it was isolated from Cyclosorus sp. in Thailand as the new host and geographical records.
Figure 12.
Morphology of Neopestalotiopsis saprophytica (MFLUCC 22-0139): (a) brown leaf spot in Cyclosorus sp.; (b,c) upper and reverse views of the colony after seven days of growth on PDA at 28 °C; (d) coidiomata; (e,f) conidiogenous cells; (g–i) conidia. Scale bars = 10 µm.
Pestalotiopsis Steyaert, Bulletin du Jardin Botanique de l’État à Bruxelles, 19 (3): 300 (1949).
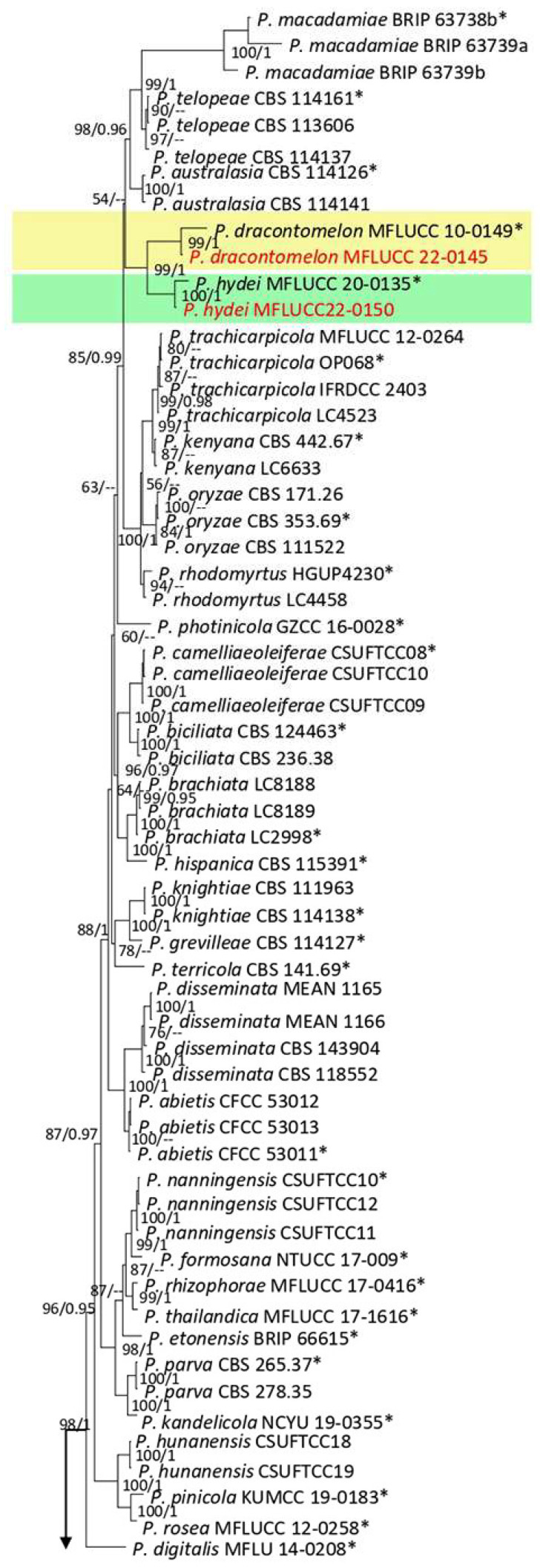
The species of this genus occur as endophytes, saprobes, and opportunistic pathogens on various plants. Pestalotiopsis is distinguished from Nepestalotiopsis by concolorous median cells of the conidia [28]. In the current study, P. hydei and P. dracontomelon were obtained from leaf spots on ferns by morphological characters and phylogeny of the ITS, tub2, and tef1 gene regions.
Figure 13.
Bayesian phylogenetic tree of Pestalotiopsis spp. obtained from the combined ITS, tef1, and tub2 sequence data. The ultrafast maximum likelihood bootstrap support (%) ≤ 50 and BYPP ≥ 0.95 are shown at the nodes. The ex-type strains are marked with an asterisk. The tree is rooted in Neopestalotiopsis saprophytica (MFLUCC 12-0282) and N. magna (MFLUCC 12-0652).
Pestalotiopsis dracontomelon Maharachch. & K.D. Hyde, 2015.
Facesoffungi number: FOF00457; Figure 14.
Associated with Nephrolepis cordifolia leaf spot. Sexual morph: not observed. Asexual morph: conidiomata a few, scattered, semi-immersed to immersed, with black conidial mass. Conidiophores reduced to conidiogenous cells. Conidiogenous cells cylindrical, hyaline to pale brown, entroblastic. Conidia fusiform, straight to slightly curved, 4 septate, 16–22 × 4–7 µm (mean = 19.8 × 6.0, n = 30); basal cell conical, hyaline, thin walled, and 3–5 µm long; three median cells doliiform, concoloured, olivaceous, darker septate than the other cells, and 10–15 µ × 4–8 µm (mean = 12.9 × 6.0, n = 30) (second cell from the base 3–5 µm long; third cell 3–6 µm long; and forth cell 2–6 µm long); apical cell conical, hyaline, thin walled, and 2–5 µm long with 2–3 apical appendages filiform, hyaline, unbranched, and 7–20 µm long (mean = 13.7, n = 30); basal appendage tubular, hyaline, unbranched, singular, and 1–8 µm long.
Culture characteristics: Colonies reached 32–36 mm after seven days of growth on PDA at 28 °C; felted, regular and entire edge, dull surface and well-defined margin, medium density and without pigmentation in medium and conidial mass. Upper view white, and the reverse greenish black, buff, and primrose circles from center to margin.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, on leaf spot of Nephrolepis cordifolia, 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0217) and living culture MFLUCC 22-0145.
Notes: Isolated from a blight on leaves’margin of Nephrolepis cordifolia. The isolate obtained in this study (MFLUCC 22-0145) clustered with P. dracontomelon in the same clade by 99% ML bootstrap support and 1.0 BYPP (Figure 13). Comparison of the DNA sequences of P. dracontomelon strains (ex-type strain MFLUCC 10-149 with MFLUCC 22-0145) showed 1.02% nucleotide differences in ITS (five nucleotides) and 0.23% in tef1 (one gap). Sequence data for tub2 were unavailable for the type strain (MFLUCC 10-149). This species was introduced from Dracontomelon dao in Thailand [33]. Here, we provide a new host record for P. dracontomelon.
Figure 14.
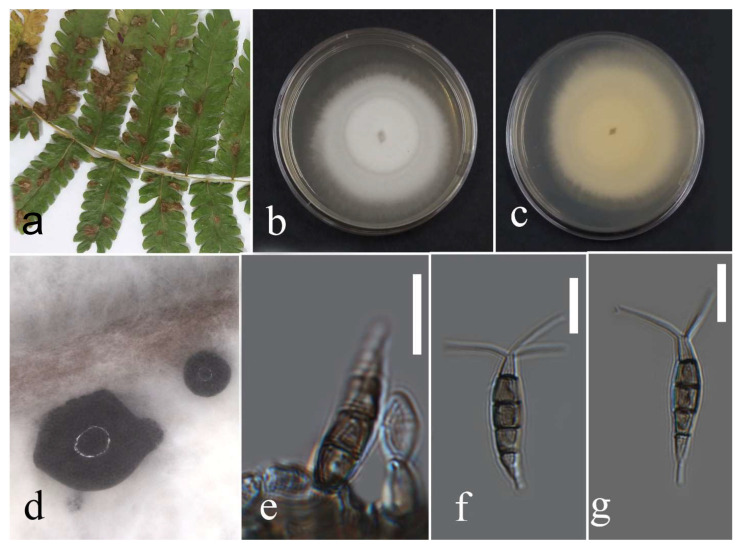
Morphology of Pestalotiopsis dracontomelon (MFLUCC 22-0145): (a) blight in Nephrolepis cordifolia; (b,c) upper and reverse views of the colony after seven days of growth on PDA at 28 °C; (d) conidiomata; (e,f) conidiogenous cells; (g–i) conidia. Scale bars = 10 µm.
Pestalotiopsis hydei Huanraluek & Jayaward., 2021.
Facesoffungi number: FoF 09460; Figure 15.
Associated with Cyclosorus sp. leaf spot. Sexual morph: not observed. Asexual morph: conidiomata a few, scattered, immersed, or semi-immersed, with black conidial mass. Conidiphores reduced to conidiogenous cells. Conidiogenous cells lageniform to cylindrical, hyaline, and entroblastic. Conidia fusiform, straight to slightly curved, 4 septate, and 18–28 × 4–7 µm (mean = 23.6 × 5.7, n = 30); basal cell conical, hyaline, thin walled, and 3–6 µm long; three median cell doliiform, concolourous, olivaceous, darker septate than the other cells, and 13–18 × 4–7 µm (mean = 15.6 × 5.6, n = 30) (second cell from the base 3–7 µm long; third cell 3–7 µm long; and forth cell 4–7 µm long); apical cell conical, hyaline, thin walled, 3–6 µm long with 2–3 apical appendages tubular, hyaline, unbranched, and 5–13 (mean = 9.4, n = 30); basal appendage filiform, hyaline, unbranched, singular, and 2–8 µm.
Culture characteristics: Colonies reached 73–82 mm after seven days of growth on PDA at 28 °C; felted, circular shape, dull surface, entire edge, fluffy margin, medium dense density, and without pigmentation in medium and conidial mass. Upper view white and the reverse primrose.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, on leaf spot of Cyclosorus sp., 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0218) and living culture (MFLUCC 22-0150).
Notes: Th strain MFLUCC 22-0150 was isolated from necrotic brown leaf spots on Cyclosorus sp. The isolate obtained in this study (MFLUCC 22-0150) clustered with P. hydei in the same cluster by 100% ML bootstrap support and 1.0 BYPP (Figure 13). Comparison of the DNA sequence data of P. hydei strains (ex-type strain MFLUCC 20-0135 and MFLUCC 22-0150) revealed 0.62% nucleotide differences in ITS (three nucleotides), while the sequences of tef1 and tub2 were identical. This species was first reported from Litsea petiolata in Thailand [30] and, here, it was isolated from Cyclosorus sp., providing a new host record for this species.
Figure 15.
Morphology of Pestalotiopsis hydei based on isolate MFLUCC 22-0150: (a) leaf spot in Cyclosorus sp.; (b,c) upper and reverse views of the colony after seven days of growth on PDA at 28 °C; (d) conidiomata; (e) conidiogenous cell; (f,g) conidia. Scale bars = 10 µm.
Diaporthales Nannf., Nova Acta Regiae Societatis Scientiarum Upsaliensis. 8 (2): 53 (1932).
Diaporthales comprises 30 families and 181 genera and is characterized by solitary or aggregated perithecia, sometimes with long papilla, unitunicate asci, with a conspicuous refractive ring. The asexual morph is mostly coelomycetes and rarely hyphomycetes [27]. Species of this order can be plant and animal pathogens and are found in the soil and as saprobes or endophytes [34].
Diaporthaceae Höhn. ex Wehm., American Journal of Botany. 13: 638 (1926).
This family comprised endophytes, pathogens, and saprobes on submerged plants or terrestrials [27]. Diaporthaceae is divided into 15 genera: Apioporthella, Apiosphaeria, Chaetoconis, Chiangraiomyces, Diaporthe, Hyaliappendispora, Leucodiaporthe, Massariothea, Mazzantia, Ophiodiaporthe, Paradiaporthe, Phaeocytostroma, Phaeodiaporthe, Pustulomyces, and Stenocarpella [27].
Diaporthe Nitschke, Pyrenomycetes Germanici. 2: 240 (1870).
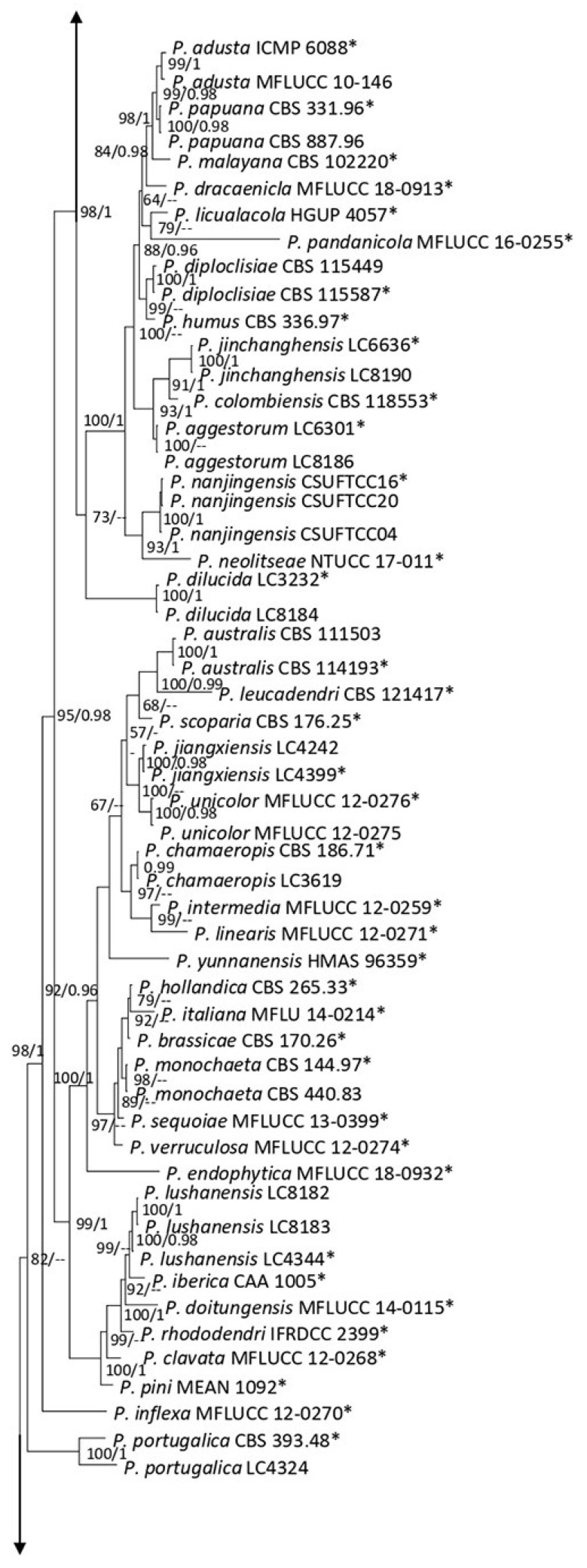
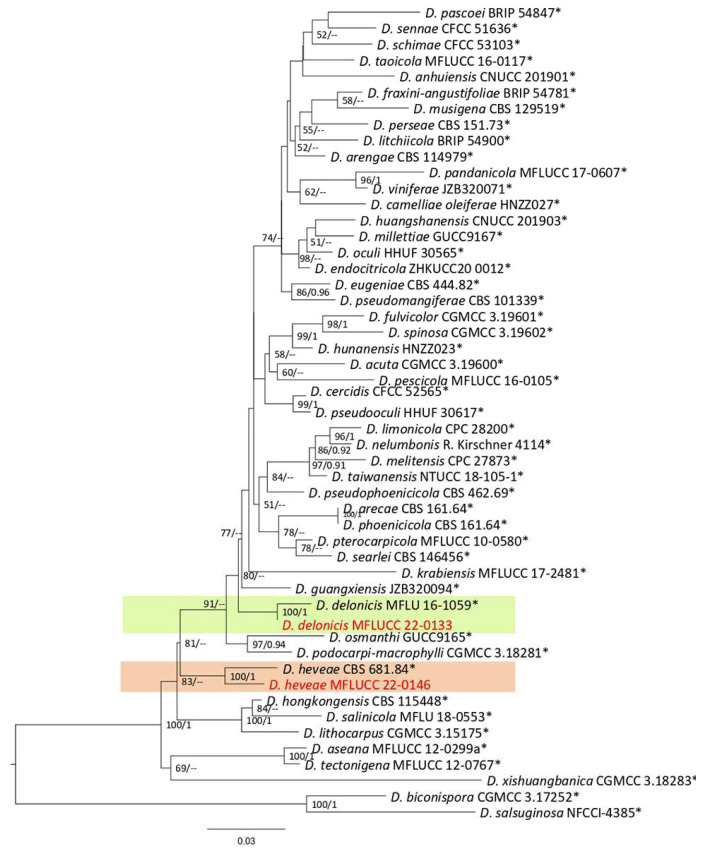
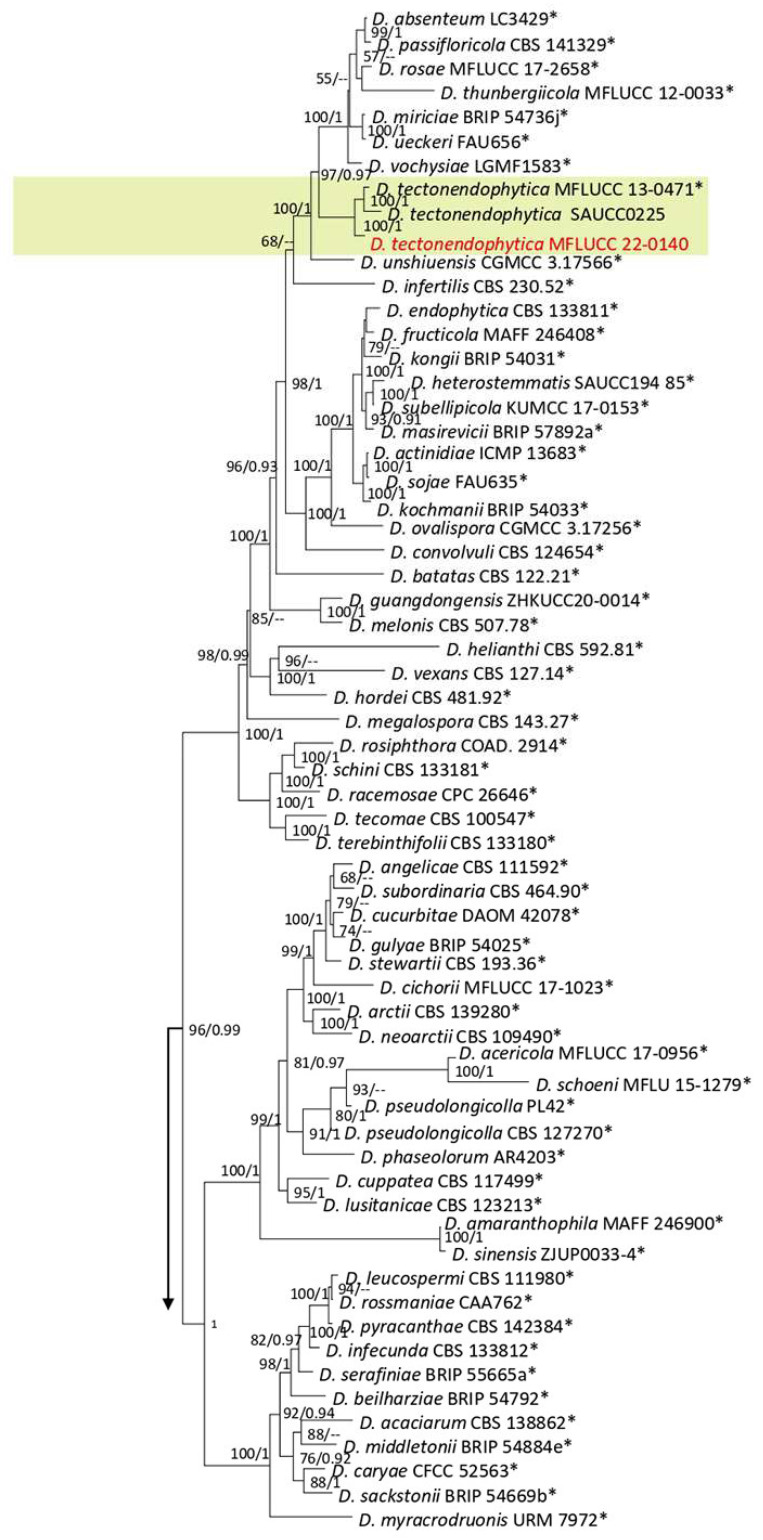
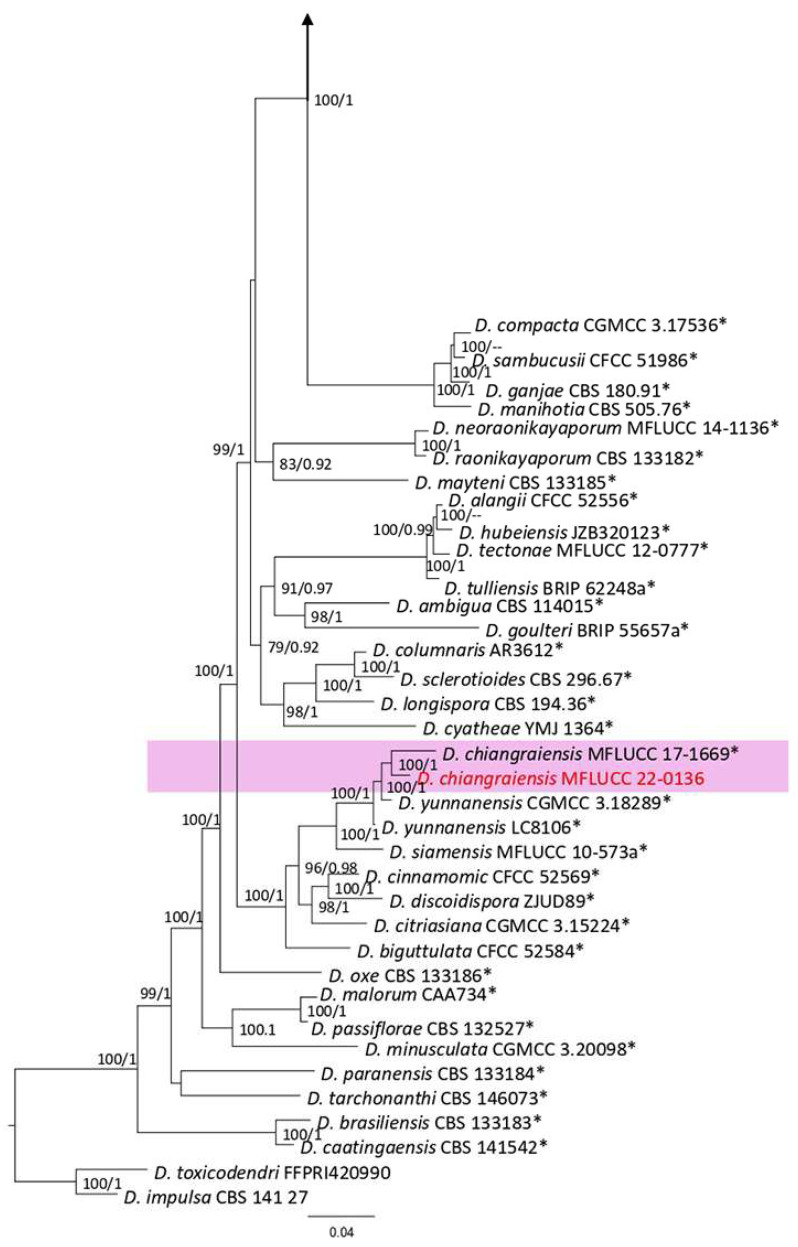
Diaporthe comprises 13 species complexes and nine singleton species based on phylogenetic analyses of ITS, tef1, tub2, cal, and his3 [35]. Diaporthe species can live on various hosts as saprobes, endophytes, or pathogens [35]. The present study identified four species (D. chiangraiensis, D. delonicis, D. heveae, and D. tectonendophytica) from the D. arecae and D. sojae complexes on ferns, based on the morphology and phylogeny of the ITS, tef1, tub2, cal, and his3 sequence data.
Figure 16.
The Diaporthe arecae complex maximum likelihood phylogenetic tree obtained from the combined ITS, tef1, tub2, cal, and his3 sequence data. The ultrafast maximum likelihood bootstrap support (%) ≥ 50 and BYPP ≥ 0.90 are shown at the nodes. The ex-type strains are marked with an asterisk. The tree is rooted in D. biconispora (CGMCC 3.17252) and D. salsuginosa (NFCCI-4385).
Diaporthe delonicis R.H. Perera, E.B.G. Jones & K.D. Hyde, 2020.
Facesoffungi number: FoF 07754; Figure 17.
Associated with Pteris grandifolia leaf spot. Sexual morph: not observed. Asexual morph: conidiomata pycnidia, aggregated, semi-immersed, globes to nearly globes, black, with cream conidial mass. Conidiphores subcylindrical, hyaline, and 14–23 × 1–2 µm. Alfa conidia absent. Beta conidia filiform, slightly curved at one end, hyaline, aseptate, rounded tips, and 13–25 µm (mean = 18.7, n = 30) × 1–2 µm (mean = 1.5, n = 30).
Culture characteristics: Colonies filled a 90 mm Petri dish after seven days of growth on PDA at 28 °C; fluffy, circular shape, dull surface, undulate edge and fluffy margin, medium sparse density, and without pigmentation in the medium and fruiting body. Upper view with circles of white and olivaceous buff and the reverse primrose with greenish olivaceous areas.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, leaf spot on Pteris grandifolia, 3 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0219) and living culture (MFLUCC 22-0133).
Notes: Isolated from dark-brown spots on Pteris grandifolia leaves. The isolate obtained in this study (MFLUCC 22-0133) clustered with D. delonicis by 100% ML bootstrap support and 0.98 BYPP (Figure 16). The sequence data of cal and tef1 are not available for the type strain of D. delonicis. Comparing sequences of the ex-type strain and our strain (MFLUCC 22-0133) revealed 1.32% nucleotide differences in ITS (six nucleotides) and 0.66% in tub2 (three nucleotides). This species was first reported from dried seed pods of Delonix regia in Thailand [36]. It was isolated from Pteris grandifolia, providing a new host record for D. delonicis.
Figure 17.
Morphology of Diaporthe delonicis (MFLUCC 22-0133): (a) leaf brown spots in Pteris grandifolia; (b,c) upper and reverse views of the colony after seven days of growth on PDA at 28 °C; (d) pycnidia and conidial mass; (e) conidiophores; (f,g) beta conidia. Scale bars = 10 µm.
Diaporthe heveae
Facesoffungi number: FoF 13406; Figure 18.
Associated with Nephrolepis cordifolia leaf spot. Sexual morph: not observed. Asexual morph: conidiomata pycnidia, aggregated, semi-immersed, subglobes, black with cream conidial mass. Conidiophores cylindrical, hyaline, septate, branched and unbranched, rounded at the tip and wider at the base, and 15–38 × 1–2 µm. Alfa and gamma conidia absent. Beta conidia filiform, curved at one tip, hyaline, aseptate, rounded at tips, and 14–30 µm (mean = 25.4, n = 30) × 1–2 µm (mean = 1.4, n = 30).
Culture characteristics: Colonies reached 33–43 mm after seven days of growth on PDA at 28 °C; felted, dull surface, circular shape, entire edge, fluffy margin, puckered aspect, medium density without pigmentation in the medium and fruiting body. Upper view white with olivaceous buff areas and the reverse primrose and buff areas.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, leaf spot on Nephrolepis cordifolia, 3 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0220), living culture (MFLUCC 22-0146).
Notes: Isolated from a blight on the leaf margin of Nephrolepis cordifolia. The isolate obtained in this study (MFLUCC 22-0146) was clustered with D. heveae by 100% ML bootstrap support and 1.0 BYPP (Figure 16). The sequence of his3 is not available for the strain MFLUCC 22-0146 in this study, while the reference sequences of D. heveae had this gene region. Comparison of the sequence data of the reference strain for D. heveae with our strain (MFLUCC 22-0146) showed 2.87% nucleotide differences in cal (7 nucleotides and four gaps), 1.10% in ITS (5 nucleotides), 3.22% in tef1 (10 nucleotides), and 3.36% in tub2 (15 nucleotides). This species was first reported from Hevea brasiliensis as a die-back agent of the seedlings of the mentioned host in Thailand, Brazil, China, Indonesia, Malaysia, and Sri Lanka [37]. Here, it is first reported in Nephrolepis cordifolia.
Figure 18.
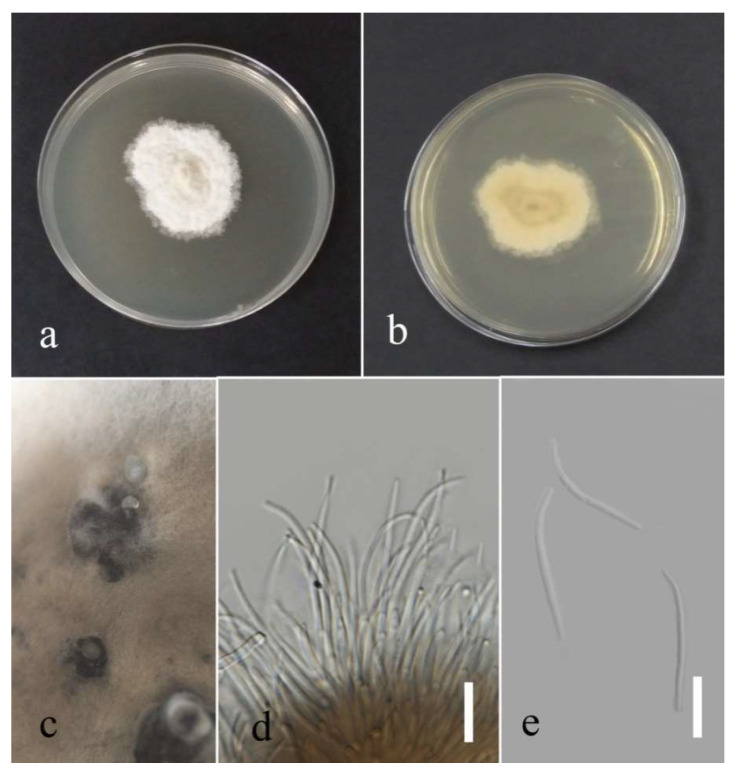
Morphology of Diaporthe heveae based on isolate MFLUCC 22-0146: (a,b) upper and reverse views of the colony after seven days of growth on PDA at 28 °C; (c) pycnidia and conidial mass; (d) conidiophores; (e) beta conidia. Scale bars (d,e) = 10 µm.
Figure 19.
The Diaporthe sojae complex. Bayesian phylogenetic tree obtained from the combined ITS, tef1, tub2, cal, and his3 sequence data. The tree was constructed using MrBayes 3.2.7a on CIPRES Science Gateway. The ultrafast maximum likelihood bootstrap support (%) ≥ 50 and BYPP ≥ 0.90 are shown at the nodes. The ex-type strains are marked with an asterisk. The tree is rooted in D. toxicodendri (FFPRI420990) and D. impulse (CBS 141.27).
Diaporthe chiangraiensis (Senan & Hyde) Norph., 2022.
Facesoffungi number: FoF 13407.
Assocciated with Asplenium nidus leaf spot. Sexual morph: not observed. Asexual morph: conidiomata, pycnidium, immersed and semi-immersed, a few, aggregated and black. Sporulation, not observed (for the conidial morphology, see the description of Chiangraiomyces bauhiniae [38]).
Culture characteristics: Colonies reached 75–80 mm after seven days of growth on PDA at 28 °C; felted, dull surface, circular shape, undulate edge, fluffy margin, and medium sparse density without pigmentation in the medium and fruiting body. Upper view with circles of white and primrose and the reverse with circles of straw and olivaceous buff.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, leaf spot on Asplenium nidus, 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0221) and living culture (MFLUCC 22-0136).
Notes: Isolated from a small blight on leaves of Asplenium nidus. The isolate obtained in this study (MFLUCC 22-0136) grouped with D. chiangraiensis with 100% ML bootstrap support and 1.0 BYPP (Figure 19). Sequence data of cal and tub2 were unavailable for the ex-type strain of D. chiangraiensis. Comparing sequences of D. chiangraiensis strains (MFLUCC 22-0136 and ex-type strain MFLUCC 17-1669) revealed DNA sequence differences (including gaps) in 2% ITS (nine nucleotides) and 4.73% in tef1 (nine nucleotides and six gaps) genomic regions. This species was first reported from dead twigs of Bauhinia sp. (Fabaceae) in Thailand [38], and it was synonymized as D. chiangraiensis based on the phylogenetic analysis [35]. Here, we provide a new host record for D. chiangraiensis.
Diaporthe tectonendophytica Doilom, Dissan. & K.D. Hyde, 2016; Figure 20.
Associated with Nephrolepis cordifolia leaf spot. Sexual morph: not observed. Asexual morph: conidiomata pycnidia, aggregated, semi-immersed, pyriform or subglobes with longish neck, black with hyaline conidial mass exuding from central ostioles after one month on PDA containing a tooth pick under florescent light at 28 °C. Conidiophores cylindrical, straight or slightly curved, hyaline, septate, unbranched, rounded at the tip and wider at the base, and 11–19 × 1–3 µm. Alfa conidia absent. Beta conidia filiform, curved at one end, hyaline, aseptate, rounded at tips, and 11–24 µm (mean = 18.93, n = 30) × 1–2 µm (mean = 1.4, n = 30).
Culture characteristics: Colonies filled a 90 mm Petri dish after seven days growth on PDA at 28 °C; fluffy to felted, dull surface, and fluffy margin, medium sparse density without pigmentation in medium and fruiting body. Upper view white and primrose circles and the reverse view, honey in the center and primrose in other areas with honey spots. Gamma conidia, absent.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, leaf spot on Nephrolepis cordifolia, 3 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0222) living culture (MFLUCC 22-0140).
Notes: Isolated from a blight on Nephrolepis cordifolia leaves. The isolate obtained in this study (MFLUCC 22-0140) was grouped with D. tectonendophytica by 100% ML bootstrap support and 1.0 BYPP (Figure 19). Comparison of the DNA sequence data of D. tectonendophytica strains (MFLUCC 22-0140 and ex-type strain MFLUCC 13-0471) showed 1.46% nucleotide differences in cal (seven nucleotides), 0.67% in ITS (three nucleotides), 1.02% in tef1 (two nucleotides and one gap), and 0.98% in tub2 (two nucleotides and two gaps) genomic regions. This species was first introduced from Tectona grandis in Thailand [39]. Here, we provide a new host record for D. tectonendophytica.
Figure 20.
Morphology of Diaporthe tectonendophytica (MFLUCC 22-0140): (a,b) upper and reverse views of the colony after seven days of growth on PDA at 28 °C; (c) pycnidia on media; (d) pycnidium; (e) peridium; (f) conidiophores; (g) beta conidia. Scale bars (d) = 20 µm and (f,g) = 10 µm.
Glomerellales Chadef. ex Réblová, W. Gams & Seifert, Studies in Mycology, 68: 170 (2011).
The taxonomic structure of this order was clarified, and its name was published validly by Réblová et al. [40]. This order comprises five families: Australiascaceae, Glomerellaceae, Malaysiascaceae, Plectosphaerellaceae, and Reticulascaceae [27]. The members of this order are characterized by apostromatal and endostromatal ascomata and hyaline aseptate ascospores, which can be endophytes or plant parasites [40].
Glomerellaceae Locq., Mycologie générale et structurale: 175 (1984).
This is a monotypic family with Colletotrichum asexual morph and Glomerella sexual morph. Members of this family are plant endophytes, pathogens, and saprobes [27].
Colletotrichum Corda, Deutschlands Flora, Abt. III. Die Pilze Deutschlands. 3 (12): 41; (1831).
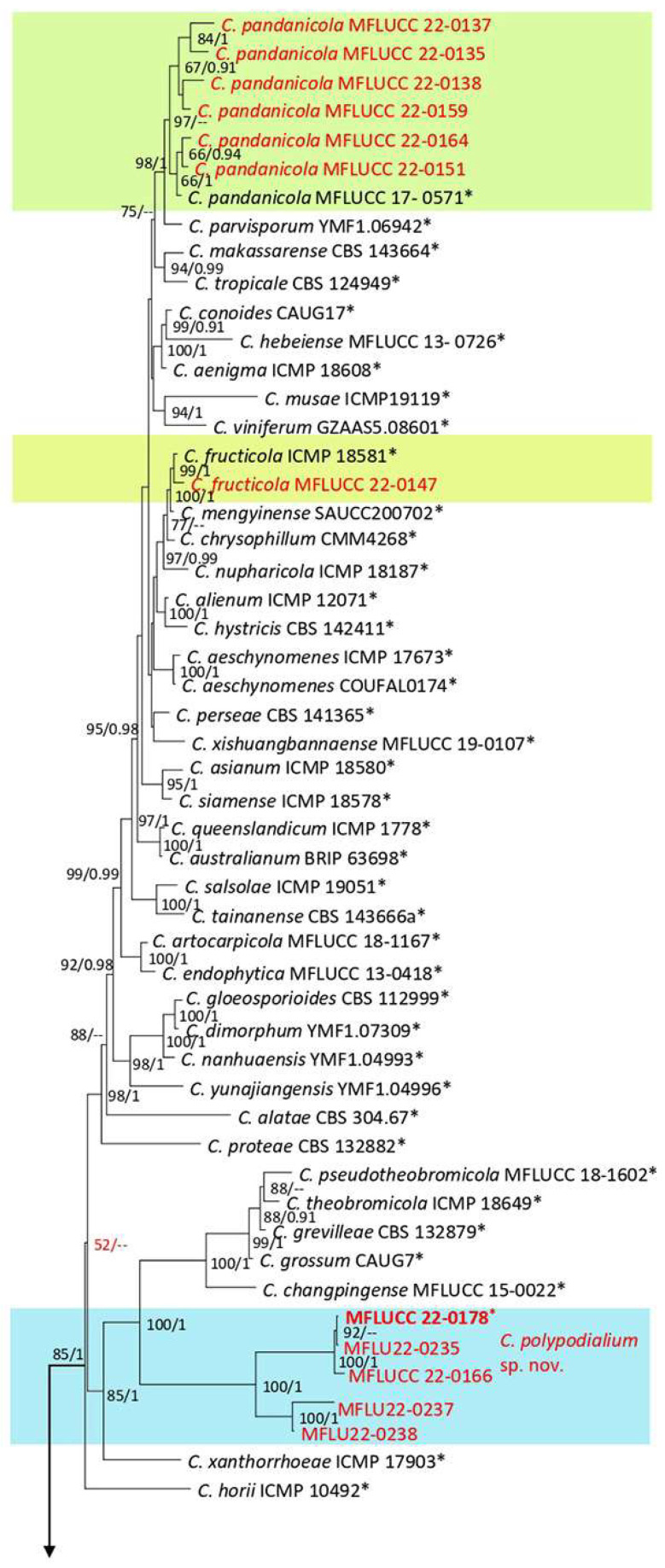
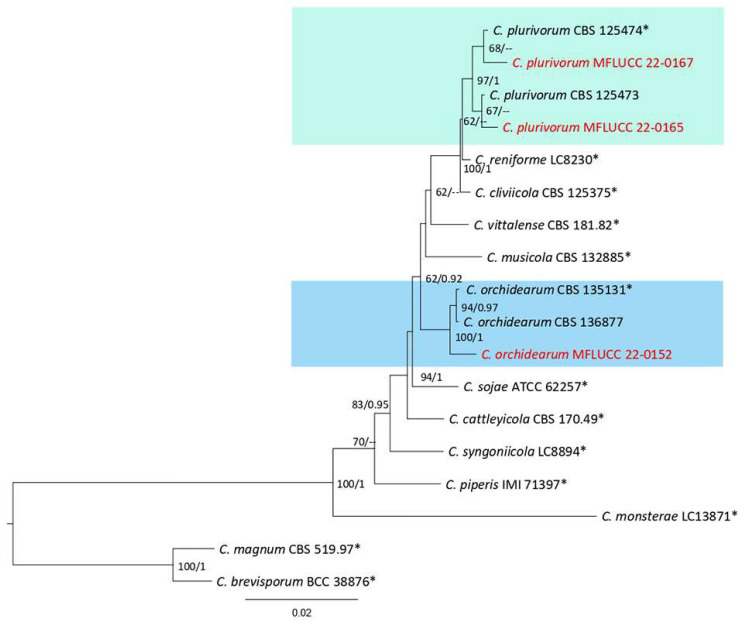
This genus comprises endophytes, saprobes, plant, insect, and human pathogens. Species of Colletotrichum are distributed in 16 species complex and 15 singleton species based on morphological and phylogenetic data [41]. In this study, seven species (Colletotrichum polypodialium, C. fructicola, C. gigasporum, C. orchidearum, C. pandanicola, C. plurivorum, and C. truncatum) belonging to C. gloesporioides, C. gigasporum, C. orchidearum, and C. truncatum complexes reported on ferns and fern-like hosts based on the morphology and combined phylogeny of the ITS, tub2, act, gapdh, and chs-1 sequence data.
Figure 21.
The Colletotrichum gloesporioides complex. The Bayesian phylogenetic tree obtained from the combined act, chs-1, gapdh, ITS, and tub2 sequence data. The ultrafast maximum likelihood bootstrap support (%) ≥ 50 and BYPP ≥ 0.90 are shown at the nodes. The ex-type strains are marked with an asterisk. The tree is rooted in C. braseiliense (CBS 128501) and C. boninense (CBS 123755).
Colletotrichum fructicola Prihast., L. Cai & K.D. Hyde, 2009.
Facesoffungi number: FoF 06767; Figure 22.
Associated with Nephrolepis cordifolia leaf spot. Sexual morph: not observed. Asexual morph: on PDA Conidiomata acervulus, semi-immersed, with orange conidial mass. Setae absent. Conidiophores reduced to conidiogenous cells. Conidiogenous cells cylindrical, hyaline, and entroblastic. Conidia cylindrical, hyaline, rounded apices, and 11–15 µm (mean = 13.2, n = 30) × 4–6 (mean = 5.3, n = 30). Appressoria clavate, ovoid and slightly irregular or regular shape, brown to dark-brown, and s6–14 µm (mean = 9. 5, n = 20) × 4–11 µm (mean = 7.4, n = 20).
Culture characteristics: Colony reached 59–68 mm after seven days of growth on PDA at 28 °C; cottony, circular shape, dull surface, entire edge, well-defined margin, and medium density, without conidial mass and pigmentation. Upper view pale olivaceous gray in the center, smoke gray in the middle, and white margin, and the reverse with circles of dull green, greenish gray, and primrose margin.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, leaf spot on Nephrolepis cordifolia, 3 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0223) and living culture (MFLUCC 22-0147).
Notes: Isolated from a blight on the leaf margin of Nephrolepis cordifolia. The isolate obtained in this study (MFLUCC 22-0147) clustered with C. fructicola by 99% ML bootstrap support and 1.00 BYPP (Figure 21). The DNA sequence data of C. fructicola strains (MFLUCC 22-0147 and ex-type strain ICMP 18581) differed 1.05% in act (one nucleotide and one gap) and 0.41% in tub2 (two nucleotides), while the sequences of chs-1, gapdh, and ITS were identical. This species was first reported from Coffea arabica in Thailand [42]. It was also isolated from Capsicum annuum, Carica papaya, Cymbopogon citratus, Dendrobium sp., Dimocarpus longan, Freycinetia sp., Freycinetia sp., Pandanus sp., and Pennisetum purpureum in Thailand [43]. Here, we provide a new host record for C. fructicola.
Figure 22.
Morphology of Colletotrichum fructicola (MFLUCC 22-0147) obtained from Nephrolepis cordifolia: (a) blight on the leaf margin in the host; (b,c) upper and reverse views of the colony after seven days of growth on PDA; (d) conidiogenous cell; (e) appressorium; (f,g) conidia. Scale bars = 10 µm.
Colletotrichum pandanicola Tibpromma & K.D. Hyde.
Facesoffungi number: FoF 04534; Figure 23.
Associated with Nephrolepis cordifolia leaf spot. Sexual morph: not observed on PDA. Asexual morph: conidiomata acervulus, semi-immersed, scattered with orange conidial mass. Setae formed on PDA after two months, brown, 3–4 cell, and 255–358 × 10–18 µm. Conidiohpores reduced to conidiogenous cells. Conidiogenous cells cylindrical or clavate, hyaline, enteroblastic, and 4–9 µm (mean = 6. 8, n = 20) × 2–5 µm (mean = 3.4, n = 20). Conidia cylindrical, hyaline with rounded apices and 10–15 µm (mean = 12.6, n = 30) × 2–6 µm (mean = 4.1, n = 30). Appressoria formed in slide culture on SNA medium after one week with diverse shapes, lobated, knobbed brown to dark-brown, and 6–14 µm (mean = 8.9, n = 20) × 2–8 µm (mean = 5.7, n = 20).
Culture characteristics: Colonies reached 56–71 mm diameter after seven days of growth on PDA at 28 °C; fluffy, dull surface, entire edge, fluffy margin, and medium density, without pigmentation in media and conidial mass. Upper view smoke gray in the center and white in other parts and the reverse primrose.
Material examined: Thailand, Chiang Rai Province, Muang District, Huai Sak, leaf spot on Nephrolepis cordifolia (Nephrolepidaceae), 17 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0224) and living (MFLUCC 22-0137); ibid., Thasud, leaf spot on Nephrolepis cordifolia, 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0227) and living culture (MFLUCC 22-0159); ibid., leaf spot on Pteris ensiformis (Pteridaceae), 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0225) and living culture (MFLUCC 22-0135); ibid., leaf spot on Cyclosorus sp., 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0226) and living culture (MFLUCC 22-0138); ibid., leaf spot on Cyclosorus sp. (Thelypteridaceae), 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0228) and living culture (MFLUCC 22-0164); ibid., leaf spot on Cyclosorus sp., 17 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0229) and living culture (MFLUCC 22-0151).
Notes: Isolated from a dark necrotic margin (Cyclosorus sp.), brown spot (Pteris ensiformis), and necrotic lesions (Nephrolepis cordifolia). In this study, the isolates obtained from fern species (MFLUCC 22-0137, MFLUCC 22-0135, MFLUCC 22-0138, MFLUCC 22-0159, T22-0231, and MFLUCC 22-0151) clustered in the same clade with C. pandanicola, with 97–98% ML bootstrap support (Figure 21). Pairwise nucleotide comparison of the ex-type strain (MFLUCC 17-0571) with strains in this study are shown in Table 1. This species was first reported from Pandanus sp. in Thailand as an endophytic fungus [44]. This is the first report of C. pandanicola from Nephrolepis cordifolia, Pteris ensiformis, and Cyclosorus.
Table 1.
Pairwise differences of the DNA sequences data of the ex-type strain with strains obtained in this study.
| Isolate Name | act | chs -1 | gapdh | ITS | tub2 |
|---|---|---|---|---|---|
| MFLUCC 22-0137 | 3.62% (7 N *, 1 G *) | 1.98% (2 N, 3 G) | 0% | 1.18% (6 N) | 0% |
| MFLUCC 22-0135 | 1.44% (3 N) | 1.61% (4 N) | 0% | 1.57% (7 N, 1 G) | 0% |
| MFLUCC 22-0138 | 1.75% (2 N, 2 G) | 1.20% (3 N) | 0.48% (1 N) | 1.13% (6 N) | 0% |
| MFLUCC 22-0159 | 1.75% (4 N) | 0.40% (1 N) | 0.48% (1 N) | 0.19% (1 G) | 0% |
| MFLUCC 22-0164 | 0.44% (1 G) | 0.80% (1 N, 1 G) | 0% | 0.40% (1 N, 1 G) | 0.65% (3 G) |
| MFLUCC 22-0151 | 0% | 1.21% (3 N) | 0% | 0.40% (1 N, 1 G) | 0% |
* N = nucleotide; G = gap.
Figure 23.
Morphology of Colletotrichum pandanicola (MFLUCC 22-0137) obtained from Nephrolepis cordifolia: (a,b) upper and reverse views of the colony after seven days of growth on PDA at 28 °C; (c) setae; (d,e) conidiogenous cells; (f,g) appresoria; (h–j) conidia. Scales bars = 10 µm.
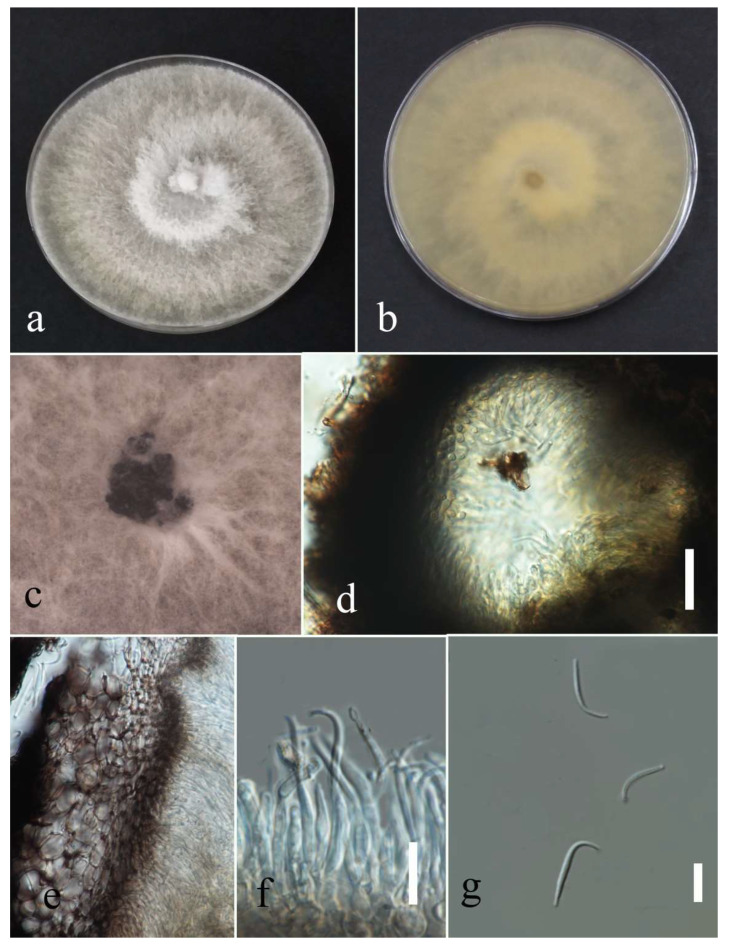
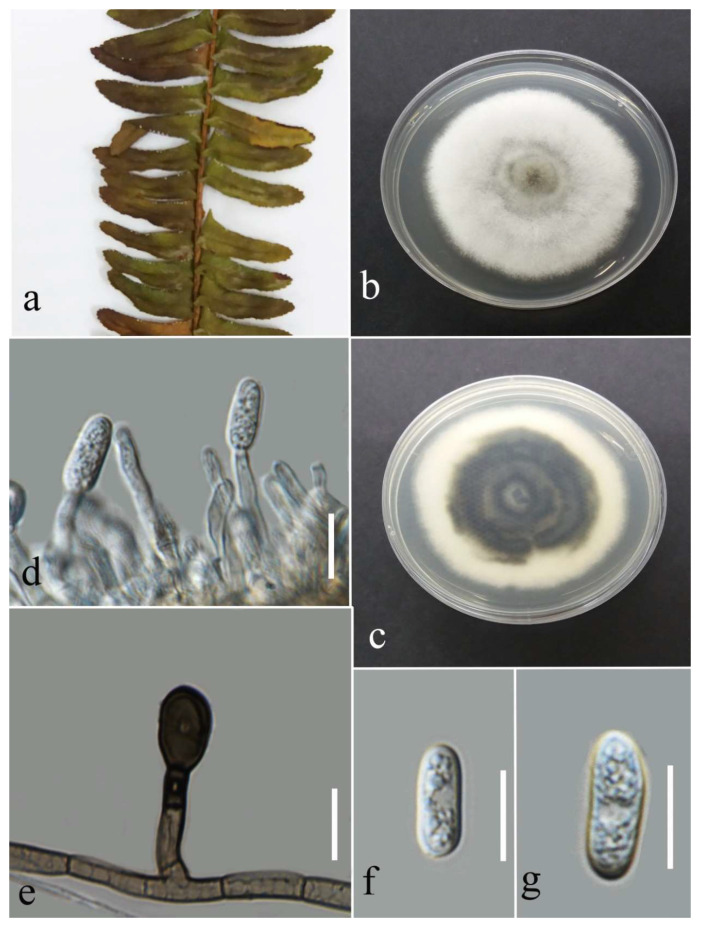
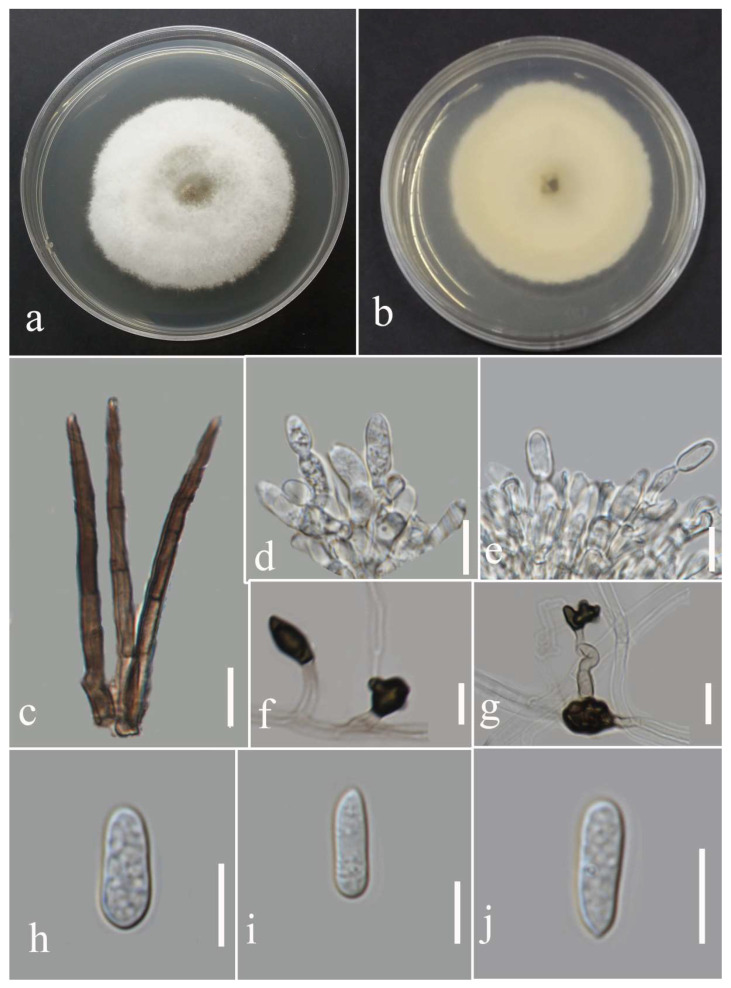
Colletotrichum polypodialium Seifollahi, Jayaward., K.D. Hyde.
Index Fungorum number: IF 559325; Facesoffungi number: FoF 13408; Figure 24.
Etymology: The species epithet refers to the plant order (Polypodiales) from which it was isolated.
Saprobe and associated with Nephrolepis sp. leaf spots and other leaves necrotic symptoms. Sexual morph: ascomata, perithecia aggregated, semi-immersed, ostiolate, pyriform, medium to dark brown, 360–600 µm high, and 125–240 µm wide. Asci cylindrical to clavate, hyaline, unitunicate, eight ascospores, uniseriate arrangement, truncate at tips, and 67–99 × 7–11 µm. Paraphyses septate. Ascospores fusiform, hyaline, aseptate, rounded at tips, and 13–18 µm (mean = 15.0, n = 30) × 3–7 µm (mean = 5.4, n = 30). Asexual morph: conidiomata acervulus with orange conidial mass. Setae absent. Conidiophores reduced to conidiogenous cells. Conidiogenous cells cylindrical, hyaline to pale brown, and 7–13 × 2–5 µm. Conidia cylindrical, hyaline, aseptate, round at tips, and 11–16 µm (mean = 13.6, n = 30) × 3–8 µm (mean = 4.8, n = 30). Appressoria lobate, pale to medium brown, irregular outline, singular, and 5–13 µm (mean = 8.8, n = 20) × 5–15 (mean = 9.3, n = 20).
Culture characteristics: Colonies reached 78–82 mm after seven days of growth on PDA at 28 °C; fluffy, circular, dull surface, entire to lobate edge, fluffy margin, and medium density without conidial mass and pigmentation. Upper view white and the reverse straw.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, leaf spot on Nephrolepis sp., 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0234, holotype); ex-type living culture (MFLUCC 22-0178) ibid., dried culture (MFLU 22-0235); ibid., leaf spot on Phymatosorus sp. (Polypodiaceae), 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0236); living culture (MFLUCC 22-0166); ibid., dried culture (MFLU 22-0238); ibid., dead leaves of Phymatosorus sp., 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0237).
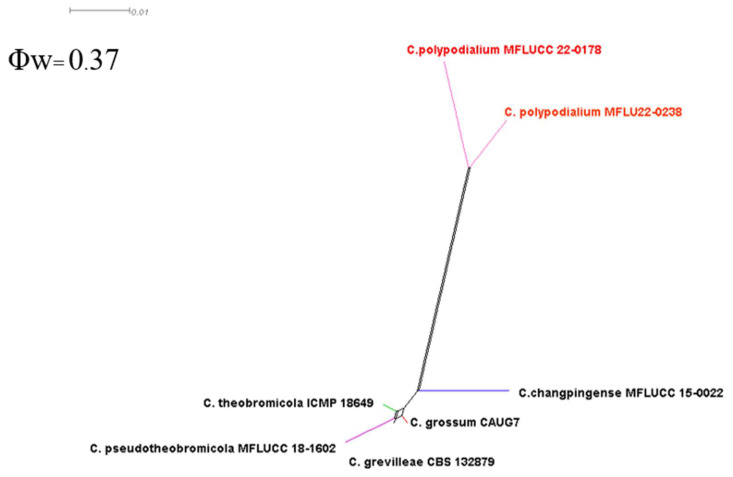
Notes: Associated with leaf spots, dark necrotic margin or anthracnosis lesion on Nephrelepis sp., and brown spots and dark-brown necrotic margin lesion on Phymatosorus sp. Perithecium was formed in the isolates MFLU 22-0235, MFLUCC 22-0178, and MFLUCC 22-0166 on PDA after two months of disruption with toothpicks, while it was not observed in the isolates MFLU 22-0238 and MFLU 22-0237. After six months, we could not maintain a living culture for the isolates MFLU 22-0235, MFLU 22-0237, and MFLU 22-0238 on PDA, 15% glycerol and distilled water at 16, −21, and 25 °C, respectively. Our isolates (MFLU 22-0235, MFLU 22-0234, MFLU 22-0236, MFLU 22-0238, MFLUCC 22-0166, and MFLU 22-0237) clustered in a separate group with 100% ML ultrafast bootstrap supports and 1.00 BYPP (Figure 21), closely related to C. pseudotheobromicola, C. theobromicola, C. grevilleae, C. grossum, and C. changpingense (Figure 21). Pairwise comparison of the ex-type strain sequences of C. polypodialium and C. changpingense revealed 6.88% nucleotide differences in act (15 nucleotides), 7.79% in chs-1 (18 nucleotides), 17.22% in gapdh (35 nucleotides and one gap), 1.78% in ITS (8 nucleotides and one gap), and 8.11% in tub2 (36 nucleotides and one gap). The PHI test did not show significant recombination between the C. polypodialium and C. changpingense isolates (Φw = 0.37) (Figure 25). Based on the recommendations of Chethana et al. [45] and Jayawardena et al. [46,47], we introduce C. polypodialium to accommodate the newly obtained isolates.
Figure 24.
Morphology of Colletotrichum polypodialium (ex-type: MFLUCC 22-0178 for asexual morph and culture; MFLU 22-0235 for sexual morph): (a,b) symptoms on Nephrolepis sp. and Phymatosorus sp.; (c) upper and reverse views of culture after seven days of growth on PDA at 28 °C; (d) perithecium; (e) peridium; (f) paraphyses; (g) immature asci; (h) mature ascus; (i) conidiogenous cells; (j) ascospore; (k–m) conidia; (n) appressoria. Scale bars (d) = 100 µm, (f) = 20 µm, and (g–n) = 10 µm.
Figure 25.
Pairwise homoplasy index (PHI) test of closely related species using LogDet transformation. The PHI test results (Φw) < 0.05 indicate significant recombination within the dataset.
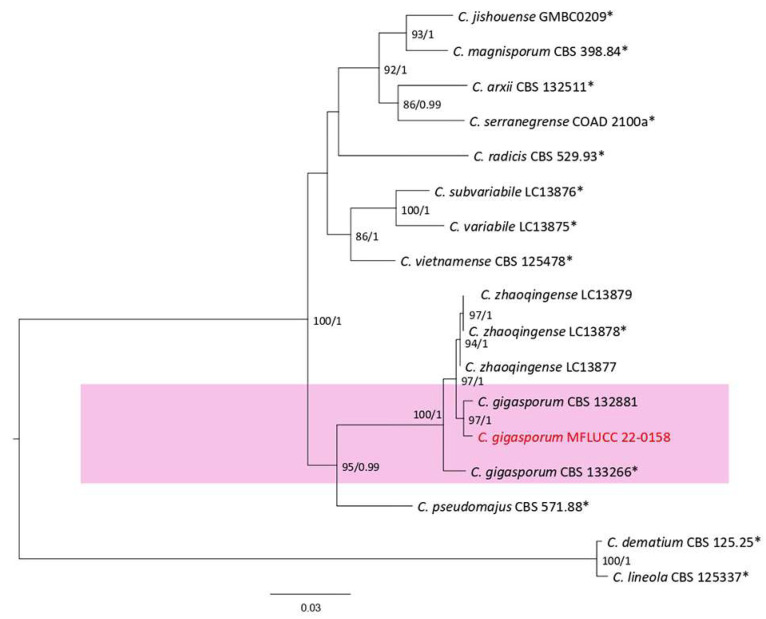
Figure 26.
The Colletotrichum gigasporum complex maximum likelihood phylogenetic tree obtained from the combined act, chs-1, gapdh, ITS, and tub2 sequence data. The ultrafast maximum likelihood bootstrap support (%) ≥ 50 and BYPP ≥ 0.90 are shown at the nodes. Th ex-type strains are marked with an asterisk. The tree is rooted in C. dematium (CBS 125.25) and C. lineola (CBS 125337).
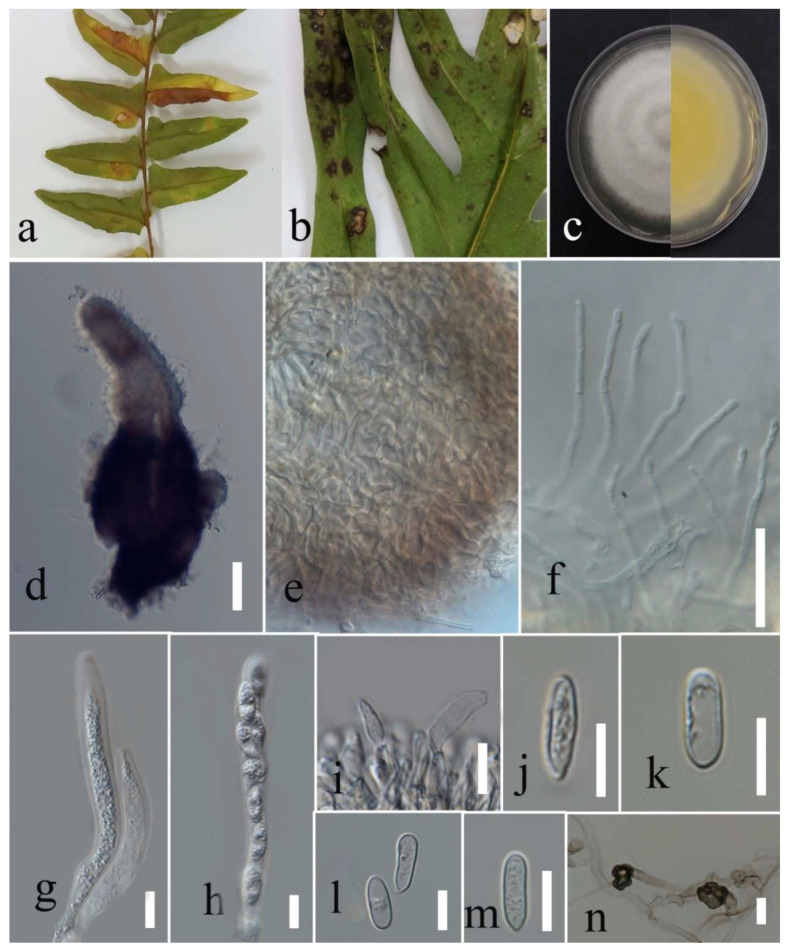
Colletotrichum gigasporum Rakotonir. & Munaut, 2013; Figure 27.
Associated with Nephrolepis cordifolia leaf blight. Sexual morph formed on PDA containing tooth pick after two months. Ascomata, perithecia solitary or aggregated, obpyriform to subglobose, and covered with mycelia. Asci cylindrical to clavate, hyaline, unitunicate, eight ascospores, truncate at apex, 85–124 × 16–22 µm. Ascospores falcate, hyaline, aseptate, rounded tips, and 34–60 µm (mean = 49.1, n = 30) × 4–10 µm (mean = 7.5, n = 30).
Asexual morph: Conidiomata condiophores and conidiogenous cells not observed. Conidia cylindrical, hyaline, aseptate, and rounded at both tips, and 21–32 µm (mean = 26.6, n = 30) × 6–10 µm (mean = 7.55, n = 30). Appresorria clavate, pale to medium brown, regular or irregular outline with a germ pore, and 17–20 × 8–12 µm.
Culture characteristics: Colonies reached 47–52 mm after seven days of growth on PDA at 28 °C; cottony, circular shape, dull surface, entire edge, and well-defined margin with medium density and without pigmentation in media and conidial mass. Upper view white and the reverse dull green in the center and primrose in other parts.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, leaf spot on Nephrolepis cordifolia, 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0239) and living culture (MFLUCC 22-0158).
Notes: Strain MFLUCC 22-0158 was isolated from a necrotic spot with a brown margin on Nephrolepis cordifolia leaves. The isolate obtained in this study (MFLUCC 22-0158) clustered with C. gigasporum in the same clade by 97% ML bootstrap support and 1.0 BYPP (Figure 26). The DNA sequence data of C. gigasporum strains (MFLUCC 22-0158 and CBS 133266) differed 2.13% in ITS (seven nucleotides), 1.94% in chs-1 (one gap and four nucleotides), 2.83% in gapdh (seven nucleotides), and 1.44% in tub2 (one gap and six nucleotides). The sequence of act for the type strain is not available. Colletotrichum gigasporum was first introduced from Centella asiatica, Stylosanthes guianensis, and Coffea arabica from Madagascar, Mexico, and Colombia [48]. It was also reported on Acacia auriculiformis, Alocasia sp. and Hibiscus rosa-sinensis in Thailand [43,47,49]. Here, we first report it from N. cordifolia.
Figure 27.
Morphology of Colletotrichum gigasporum (MFLUCC 22-0158): (a) leaf blight on Nephrolepis cordifolia; (b,c) upper and reverse views of the culture after seven days of growth on PDA at 28 °C; (d) immature ascus; (e) mature ascus; (f) appressorium; (g) ascospore; (h,i) Conidia. Scale bars (d,e) = 20 µm and (f–i) = 10 µm.
Figure 28.
The Colletotrichum orchidearum complex. Bayesian phylogenetic tree obtained from combined act, chs-1, gapdh, ITS, and tub2 sequence data. The tree was constructed using MrBayes 3.2.7a on CIPRES Science Gateway. The ultrafast maximum likelihood bootstrap support (%) ≥ 50 and BYPP ≥ 0.90 are shown at the nodes. The ex-type strains are marked with an asterisk. The tree is rooted in C. magnum (CBS 519.97) and C. boninense (BCC 38876).
Colletotrichum orchidearum Allesch., Rabenhorst’s Kryptogamen-Flora, Pilze, 1902.
Facesoffungi number: FoF 13409; Figure 29.
Associated with Cyclosorus sp. necrotic lesion. Sexual morph: not observed. Asexual morph: conidiomata acervulus. Setae present, medium brown to dark-brown, 3–4-septate, and tip acute to round. Conidiophores reduced to conidiogenous cells. Conidiogenous cells cylindrical to doliiform, hyaline to pale brown, and 2–11 × 3–5 µm. Conidia cylindrical, hyaline, aseptate, rounded apices, and 10–18 (mean = 14.3, n = 30) × 3–7 µm (mean = 5.4, n = 30). Appressoria absent.
Culture characteristics: Colonies reached 65–72 mm after seven days of growth on PDA at 28 °C; fluffy, circular, entire edge, fluffy margin, dull surface, and medium density, without pigmentation in media and conidial mass. Upper view pale olivaceous with white margin and the reverse gray olivaceous to dull green with primrose margin.
Material examined: Thailand, Chiang Rai Province, Muang District, Huai sak, leaf spot on Cyclosorus sp., 17 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0240) and living culture (MFLUCC 22-0152).
Notes: The isolate obtained in this study (MFLUCC 22-0152) clustered with C. orchidearum in the same clade by 100% ML bootstrap support and 1.0 BYPP (Figure 28). The DNA sequence data of C. orchidearum strains (ex-type strain CBS 135131 and T22-0385) showed 1.17% nucleotide differences in chs-1 (three nucleotides), 0.48% in gapdh (one gap), and 1.03% in tub2 (three nucleotides and two gaps), while the sequences of ITS and act were identical. Colletotrichum orchidearum was revised using obtained isolates from Eria javanica, Epipremnum aureum, Dendrobium nobile, and Hymenocallis sp. in Germany, Iran, Netherlands, and Thailand [50]. Here, we provided a new host record for this species.
Figure 29.
Morphology of Colletotrichum orchidearum (MFLUCC 22-0152): (a) necrotic lesion on Cyclosorus sp.; (b–c) upper and reverse views of the colony after seven days of growth on PDA at 28 °C; (d) conidiogenous cells; (e) satae; (f–h) conidia. Scale bars (e) = 20 µm and (d,f–h) = 10 µm.
Colletotrichum plurivorum Damm, Alizadeh & Toy. Sato, 2018; Figure 30.
Associated with Cyclosorus sp. leaf necrotic lesions. Sexual morph: formed on PDA containing tooth pick at room temperature after two months, ascomata, perithecia aggregated, semi-immersed, globes, and covered with sparse white aerial mycelia. Asci cylindrical to clavate, hyaline, unitunicate, eight ascospores, and 67–86 × 10–12 µm. Ascospores allantoid to fusiform, initially hyaline, aseptate, rounded tips, and 12–31 µm (mean = 18.2, n = 30) × 4–8 µm (mean = 5.5, n = 30).
Asexual morph: conidiomata acervulus. Setae absent. conidiophores reduced to conidiogenous cells. Conidiogenous cells cylindrical and hyaline. Conidia cylindrical, hyaline, aseptate with rounded apices, and 10–21 µm (mean = 16.3, n = 30) × 3–6 µ (mean = 4.7, n = 30). Appressoria diverse shape, bullet-shaped to lobate, brown, regular or irregular margin, and 7–13 µm (mean = 9.5, n = 20) × 4–8 (mean = 6.2, n = 20).
Culture characteristics: Colonies filled the 90 mm Petri dish after seven days of growth on PDA at 28 °C; fluffy to cotton, circular shape, dull surface, entire edge, with fluffy margin and medium density, without pigmentation in media and conidial mass. Upper view circles of smoke gray and white and the reverse circles of greenish olivaceous and primrose with greenish olivaceous dots.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, leaf necrotic lesions on Cyclosorus sp., 17 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0242) and living culture (MFLUCC 22-0165); ibid., leaf spot on Cyclosorus sp., 17 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0241) and living culture (MFLUCC 22-0167).
Notes: Strains MFLUCC 22-0165 and MFLUCC 22-0167 were isolated from leaf necrotic lesions with a dark-brown margin and anthracnose, respectively, on Cyclosorus species. Conidiomata, formed in isolate MFLUCC 22-0165, while it was not observed for isolate MFLUCC 22-0167. Additionally, conidiophores and conidiogenous cells were not observed in isolate MFLUCC 22-0167. The isolates obtained in this study clustered with C. plurivorum in the same clade by 97% ML bootstrap support and 1.0 BYPP (Figure 28). Pairwise comparison sequences of the ex-type strain and strain MFLUCC 22-0167 revealed 0.39% nucleotide differences in chs-1 (one nucleotide), 0.98% in gapdh (one nucleotide and one gap), 0.39% in ITS (two gaps), and 0.62% in tub2 (two nucleotides and one gap). The pairwise comparison results for strain MFLUCC 22-0165 showed 0.86% nucleotide differences in act (two nucleotides), 0.39% in chs-1 (one nucleotide), 0.97% in gapdh (one nucleotide and one gap), 0.19% in ITS (one nucleotide), and 1.11% in tub2 (four nucleotides and two gaps). Colletotrichum plurivorum was introduced causing anthracnose from Phaseolus lunatus, Gossypium sp., Spathiphyllum wallisii, Phaseolus vulgaris, and Coffea sp. in Benin, Brazil, Iran, and Vietnam, [50]. Later, it was reported on Capsicum annuum and Capsicum sp. in Thailand [51]. This study introduces Cyclosorus sp. as a new host of C. plurivorum.
Figure 30.
Morphology of Colletotrichum plurivorum based (MFLUCC 22-0167): (a) symptoms Cyclosorus sp.; (b,c) upper and reverse views of the colony after seven days of growth on PDA at 28 °C; (d) ascomata on PDA; (e) asci; (f) ascospore; (g) conidia; (h–i) appressoria. Scales bars (e) = 20 µm (f–i) = 10 µm.
Figure 31.
The Colletotrichum truncatum complex maximum likelihood phylogenetic tree obtained from combined act, chs-1, gapdh, ITS, and tub2 sequence data. The ultrafast maximum likelihood bootstrap support (%) ≥ 50 and BYPP ≥ 0.90 are shown at the nodes. The ex-type strains are marked with an asterisk. The tree is rooted in C. boninense (CBS 123755) and C. lineola (CBS 128501).
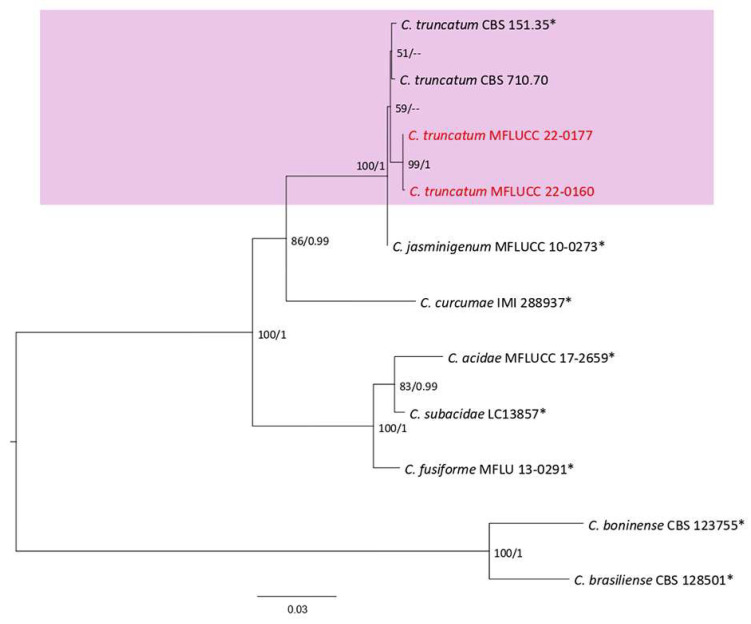
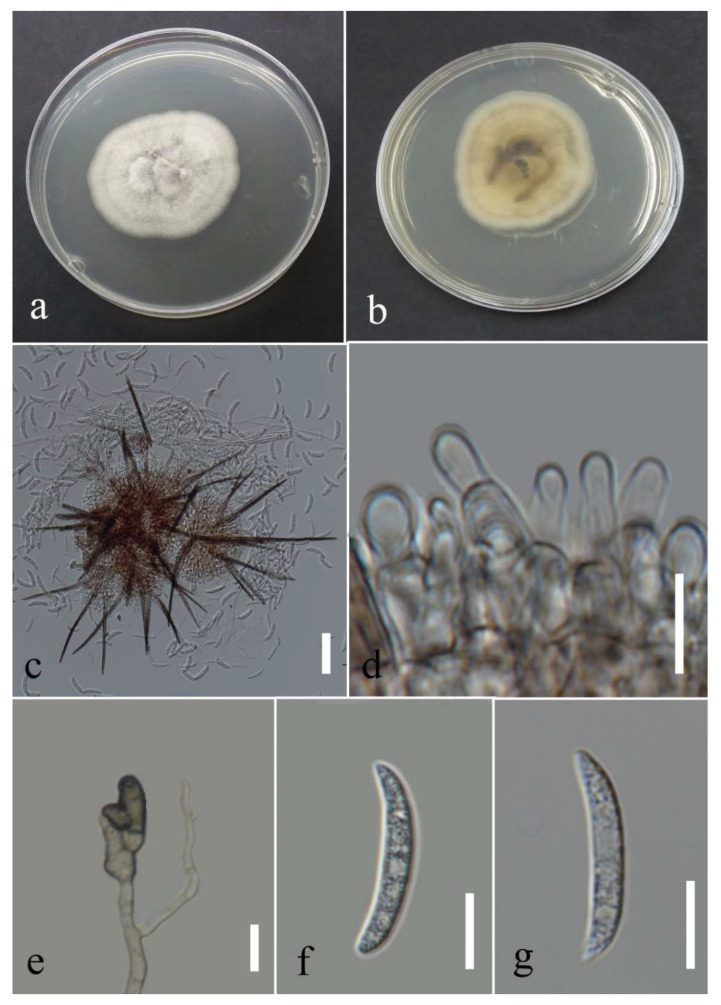
Colletotrichum truncatum (Schwein.) Andrus & W.D. Moore, 1935; Figure 32.
Associated with Cyclosorus sp. leaf spot. Sexual morph: not observed. Asexual morph: conidiomata acervulus. Setae medium to dark brown, 3 septate, round to slightly acute tip, and conical to cylindrical base. Conidiophores reduced to conidiogenous cells. Conidiogenous cells cylindrical, hyaline to pale brown, entroblastic, and 4–10 × 3–5 µm. Conidia falcate, hyaline, aseptate, and 16–27 µm (mean = 22.1, n = 30) × 2–5 µm (mean = 4.1, n = 30). Appressoria diverse shaped, lobed or bullet-shaped, pale to medium brown, entire edge to lobed, in a group or solitary, and 8–16 × 4–8 µm.
Culture characteristics: Colonies reached 40–47 mm after seven days of growth on PDA at 28 °C; fluffy, circular, dull surface, entire edge, well-defined margin, and medium density, without pigmentation in media and conidial mass. Upper view white with smoke gray circles and spots and the reverse olivaceous in the center with gray olivaceous and primrose circles.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, leaf spot on Cyclosorus sp., 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0243), living culture MFLUCC 22-0177; Thailand, Thasud, Muang District, Chiang Rai Province, leaf spot on Cyclosorus sp., 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0244) and living culture (MFLUCC 22-0160).
Notes: Strains MFLUCC 22-0177 and MFLUCC 22-0160 were isolated from small brown spots and necrotic spots with brown margins, respectively, on the leaves of two Cylosorus sp. The isolates from this study (MFLUCC 22-0177 and MFLUCC 22-0160) clustered with C. truncatum in the same clade by 59% ML bootstrap support (Figure 31). A pairwise comparison sequence data of the ex-type strain and strain MFLUCC 22-0177 revealed 1.76% nucleotide differences in act (two nucleotides and two gaps), 1.72% in chs-1 (four nucleotides), 0.89% in gapdh (two nucleotides), and 0.19% in ITS (one gap), while the tub2 sequences were identical. The pairwise comparison results for strain MFLUCC 22-0160 showed 1.76% nucleotide differences in act (two nucleotides and two gaps), 1.72% in chs-1 (four nucleotides), 1.34% in gapdh (three nucleotides), and 0.19% in ITS (one gap), while the tub2 sequences were identical. Colletotrichum truncatum was revised using the obtained isolate from Phaseolus sp., Phaseolus lunatus, Capsicum frutescens, and Arachis hypogaea in the USA and India [52]. It was obtained from Capsicum annuum, C. frutescens, Capsicum sp., Glycine max, Gossypium sp., Hymenocallis sp., Manihot esculenta, Solanum melongena, Stylosanthes hamate, and Vigna sesquipedalis in Thailand [43]. Here, it was isolated from Cyclosorus sp., providing a new host of C. truncatum.
Figure 32.
Morphology of Colletotrichum truncatum (MFLUCC 22-0177) obtained from Cyclosorus sp.: (a,b) upper and reverse views of the colony after seven days growth on PDA at 28 °C; (c) acervulus and setae; (d) conidiogenous cells; (e) appressoria; (f,g) fulcate conidia. Scale bars (c) = 50 µm and (d–g) = 10 µm.
Hypocreales Lindau, Die Natürlichen Pflanzenfamilien nebst ihren Gattungen und wichtigeren Arten 1. (1): 343 (1897).
Based on molecular data, this order comprised 14 families: Bionectriaceae, Calcarisporiaceae, Clavicipitaceae, Cocoonihabitaceae, Cordycipitaceae, Flammocladiellaceae, Hypocreaceae, Myrotheciomycetaceae, Nectriaceae, Niessliaceae, Ophiocordycipitaceae, Sarocladiaceae, Stachybotryaceae, and Tilachlidiaceae [27].
Nectriaceae Tul. & C. Tul., Selecta Fungorum Carpologia: Nectriei-Phacidiei-Pezizei. 3: 3 (1865).
This family, comprising 64 genera, are saprobes, pathogens, and endophytes on human, plant, and insect substrates in aquatic and terrestrial habitats [27]. They also cause important plant diseases and are significant from this point of view [27]. The asexual morph of them is mostly hyphomycetous and, rarely, coelomycetous [27].
Fusarium Link, Magazin der Gesellschaft Naturforschenden Freunde Berlin. 3 (1): 10 (1809).
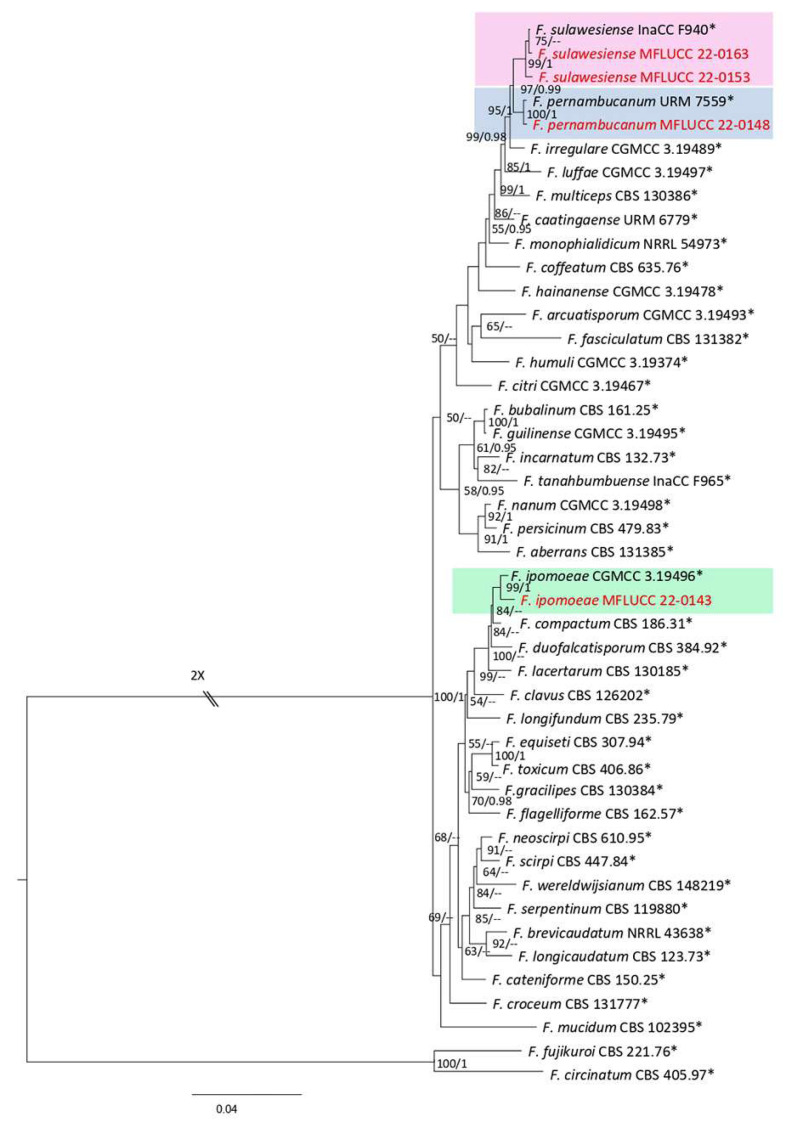
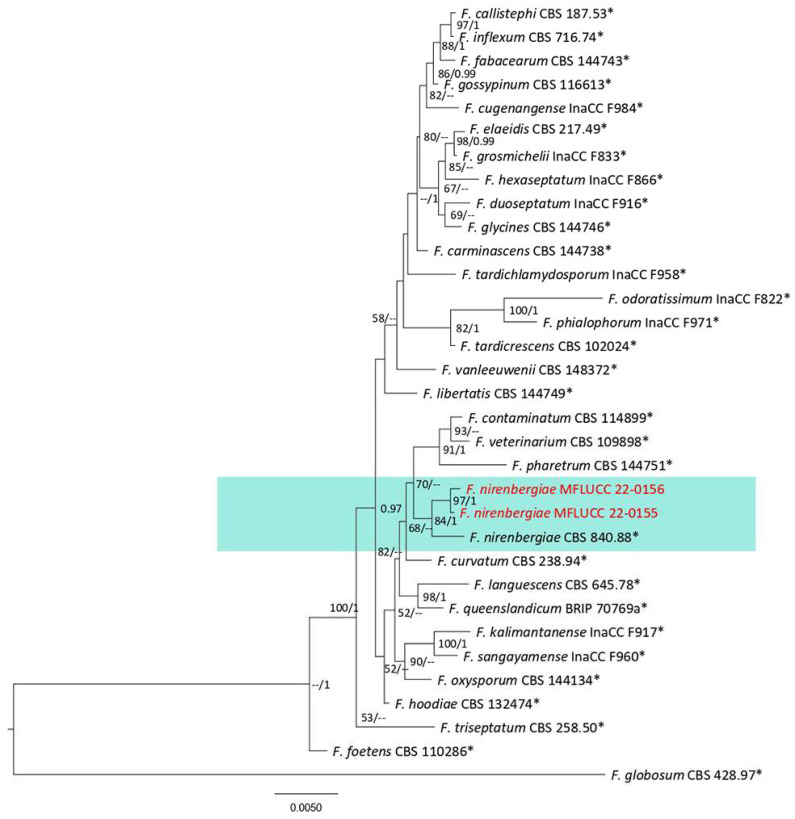
This genus is characterized by thin- or thick-walled macroconidia with various basal or apical cell shapes, production of trichothecene mycotoxin, and Giberrella sexual morph [53]. Species of this genus were identified by morphological characters and phylogenetic data of the tef1, rpb1, and rpb2 gene regions [53]. This study reports four species (Fusarium ipomoeae, F. nirenbergiae, F. pernambucanum, and F. sulawesiense) of the F. incarnatum-equiseti and F. oxysporum complexes obtained from fern and fern-like hosts using morphological data and multilocus phylogeny.
Figure 33.
Fusarium incarnatum complex. Bayesian phylogenetic tree obtained from the combined tef1, rpb1, and rpb2 sequence data. The ultrafast maximum likelihood bootstrap support (%) ≥ 50 and BYPP ≥ 0.95 are shown at the nodes. The ex-type strains are marked with an asterisk. The tree is rooted in F. fujikuroi (CBS 221.76) and F. circinatum (CBS 405.97).
Fusarium ipomoeae M.M. Wang, Qian Chen & L. Cai, 2019.
Facesoffungi number: FoF 13410; Figure 34.
Associated with Pteris grandifolia leaf blight. Arial conidiophores hyaline, branched or unbranched, diverse length, bearing lateral or terminal monophialides and polyphialides, and 15–41 µ × 3–6. Sometimes conidiophores reduced to conidiogenous cells on hyphae. Conidiogenous cells monophialidic, hyaline, and 7–9 × 4–5 µm. Chlamydospore formed on SNA, globes, hyaline, solitary, intercalary, and 10–13 µm diameter. Mesoconidia diverse shape, fusiform or clavate, straight to curved, hyaline, 1–7 cells, aseptate or 1–6 septate, one septate conidia, and 11–24 × 2–5 µm (mean = 16.9 × 3.5, n = 30). Three septate conidia 17–32 × 3–5 µm (mean = 24.0 × 4.0, n = 30). Microconidia not observed. Sporodochium on PDA and SNA not observed.
Culture characteristics: Colonies reached 38–39 mm after seven days of growth on PDA at 28 °C; fluffy, dull surface, entire edge, fluffy margin, medium density, and without pigmentation in the medium. Upper view was white and the reverse primrose.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, leaf spot on Pteris grandifolia, 3 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0245) and living culture (MFLUCC 22-0143).
Notes: Strain MFLUCC 22-0143 was isolated from a leaf blight of Pteris grandifolia. The isolate obtained in this study (MFLUCC 22-0143) clustered with F. ipomoeae in the same clade by 99% ML bootstrap supports and 1.0 BYPP (Figure 33). The comparison DNA sequences of F. ipomoeae strains (ex-type strain GCMCC 3.19496 and MFLUCC 22-0143) showed 0.32% nucleotide differences in tef1 (two nucleotides) and 1.94% in rpb1 (nine nucleotides), while no difference was found for the sequences of rpb2. This species was introduced from Solanum lycopersicum, Hibiscus syriacus, Lagenaria siceraria, Oryza sativa, Rhododendron pulchrum, Vinca major, submerged wood, bamboo, and Capsicum sp. [54]. Here, we provide a new host and geographical records for F. ipomoeae.
Figure 34.
Morphology of Fusarium ipomoeae based on the isolate MFLUCC 22-0143: (a) leaf blight on Pteris grandifolia; (b,c) upper and reverse views of the colony after seven days of growth on PDA at 28 °C; (d) conidiogenous cells; (e) phialides; (f,g) mesoconidia. Scale bars (d–f) = 20 µm and (g) = 10 µm.
Fusarium pernambucanum A.C.S. Santos, C.S. Lima, P.V. Tiago & N.T. Oliveira, 2019.
Facesoffungi number: FoF 13411; Figure 35.
Associated with a dry stem of Asparagus setaceous. Sexual morph: not observed. Aerial conidiophores hyaline, tall or branched sympodially, diverse length, bearing mono and polyphialides, and reduced to conidiogenous cells or not. Monophialides subulate to subcylindrical, hyaline, terminal, or intercalary, and 4–30 µm × 1–4 µ. Polyphialides 7–20 µm × 2–4 µm. Chlamydospores rarely found on PDA, hyaline, intercalary, and solitary. Conidia fusiform, straight to slightly curved, hyaline, 0–6 septate. Four septate conidia 24–33 × 2–5 µm (mean = 29.8 × 3.4, n = 30). One septate conidia 7–16 × 2–5 µm (mean = 11.6 × 3.6, n =30). Microconidia formed in false head on SNA.
Culture characteristics: Colonies reached 47–68 mm after seven days of growth on PDA at 28 °C; fluffy, circular shape, dull surface, entire edge, fluffy margin, medium density, and without pigmentation in the medium. Upper view is salmon in the center, white in other areas, and the reverse in salmon and primrose areas.
Material examined: Thailand, Chiang Rai Province, Muang District, Huai Sak, on a dry stem of Asparagus setaceous (Asparagaceae), 17 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0246) and living culture (MFLUCC 22-0148).
Notes: Strain MFLUCC 22-0148 was isolated from dry stems of Asparagus setaceous. MFLUCC 22-0148 clustered with F. pernambucanum in the same clade with 100% ML bootstrap supports and 1.0 BYPP (Figure 33). Comparison of the sequences of the ex-type strain (URM 7559) of this species with strain MFLUCC 22-0148 showed 0.64% nucleotide differences in tef1 (four nucleotides), while no differences were found in the rpb1 and rpb2 genes. Fusarium pernambucanum was introduced from Aleurocanthus woglumi and Dactylopius opuntiae in Brazil [55]. Here, we provide a new host and geographical records for F. pernambucanum.
Figure 35.
Morphology of Fusarium pernambucanum (MFLUCC 22-0148): (a,b) upper and reverse views of the culture after seven days of growth on PDA at 28 °C; (c) false head; (d,e) phialides; (f–h) conidia. Scale bars (c–g) = 20 µm and (h) = 10 µm.
Fusarium sulawesiense Maryani, Sand.-Den., L. Lombard, Kema & Crous, 2019.
Facesoffungi number: FoF 13412; Figure 36.
Saprobe and associated with Asplenium nidus leaf spot. Sexual morph: not observed. Arial conidiophores hyaline, branched vertically or irregularly, diverse length, bearing monophialides or polyphialides subulate to subcylindrical, terminal, or lateral. Monophialides 4–35 × 2–4 µm. Polyphialides 12–34 × 3–8 µm. Chlamydospores absent. Arial conidia fusiform, straight to slightly curved (falcate), hyaline, and 0–7 septate. One septate conidia 9–21 × 2–5 µm (mean = 13.6 × 3.6, n = 30). Five septate conidia and 30–43 × 3–5 µm (mean = 35.1 × 4.1, n = 30).
Culture characteristics: Colonies reached 57–62 mm after seven days of growth on PDA at 28 °C; fluffy, circular shape, dull surface, entire edge, fluffy margin, medium density, and without pigmentation in the medium. Upper view is white to light salmon and the reverse with salmon and straw circles.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, from leaf spot of Asplenium nidus, 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0247), living culture MFLUCC 22-0163; ibid., on a dead leaf of Asplenium nidus, 4 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0248) and living culture (MFLUCC 22-0153).
Notes: Strain MFLUCC 22-0163 was obtained from a necrotic spot with a brown margin leaf spot on Asplenium nidus. The isolates obtained in this study (MFLUCC 22-0163 and MFLUCC 22-0153) clustered with F. sulawesiense with 99% ML bootstrap support and 1.0 BYPP (Figure 33). The sequence of the rpb1 gene is not available for the type strain of this species. A comparison of the ex-type strains of F. sulawesiense (InaCC F490) with strains MFLUCC 22-0163 and MFLUCC 22-0153 revealed 0.35% nucleotide differences in tef1 (two nucleotides between the ex-type and MFLUCC 22-0153), while the tef1 sequences between the ex-type and MFLUCC 22-0163 and rpb2 sequences (in all comparisons) were identical. This species was first introduced from Musa acuminate in Indonesia [56]. Here, we provide a new host and geographical records for F. sulawesiense.
Figure 36.
Morphology of Fusarium sulawesiense (MFLUCC 22-0163): (a,b) upper and reverse views of the colony after seven days of growth on PDA at 28 °C; (c–e) phialides; (f–h) conidia. Scale bars (c,d,f–h) = 20 µm and (e) = 10 µm.
Figure 37.
Fusarium oxysporum complex. Bayesian phylogenetic tree obtained from the combined of the tef1, rpb1, and rpb2 sequence data. The ultrafast maximum likelihood bootstrap support (%) ≥ 50 and BYPP ≥ 0.95 are shown at the nodes. The ex-type strains are marked with an asterisk. The tree was rooted with F. globosum (CBS 428.97).
Fusarium nirenbergiae L. Lombard & Crous, 2018.
Facesoffungi number: FoF 13413; Figure 38.
Associated with leaf blight of Nephrolepis cordifolia and dried branches of Asparagus setaceous. Sexual morph: not observed. Aerial conidiophores hyaline, branched sparingly or unbranched, diverse length, bearing monophialides subulate to subcylindrical, hyaline, terminal or intercalary, and 2–19 × 2–3 µm. Chalmydospores ample on PDA, globose to subglobose, hyaline, intercalary or terminally, and 6.0–11.4 µm diameter. Conidia in false head on SNA, ellipsoidal to fusiform, straight or curved, hyaline, aseptate, and 4–11 × 1–4 µm (mean = 6.6 × 2.7, n = 30).
Culture characteristics: Colonies reached 57–64 mm after seven days of growth on PDA at 28 °C; fluffy, circular, dull surface, entire edge, fluffy margin, medium density, and without pigmentation in the medium. Upper view white to very light rosy vinaceous and the reverse circles of primrose and buff.
Material examined: Thailand, Chiang Rai Province, Muang District, Thasud, on leaf spot of Nephrolepis cordifolia, 3 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0249) and living culture (MFLUCC 22-0156); ibid., Huai Sak, on diseased dry leaves of Asparagus setaceous (Asparagaceae), 17 December 2021, Elaheh Seifollahi, dried culture (MFLU 22-0250) and living culture (MFLUCC 22-0155).
Notes: Strain MFLUCC 22-0156 was isolated from leaf blight on Nephrolepis cordifolia, while strain MFLUCC 22-0155 was isolated from the dried disease branch of Asparagus setaceous. The isolates obtained in this study (MFLUCC 22-0155 and MFLUCC 22-0156) grouped in the same clade with F. nirenbergiae by 84% ML bootstrap support and 1.0 BYPP (Figure 37). The sequence of the rpb1 gene is not available for the ex-type strain of this species (CBS 840.88). Comparison of the sequences of the ex-type strain with strain MFLUCC 22-0155 revealed 0.58% nucleotide differences in rpb2 (five nucleotides) and 0.16% in tef1 (one nucleotide). The pairwise comparison results between the ex-type and strain MFLUCC 22-0156 were 0.57% nucleotide differences in rpb2 (five nucleotides) and 0.33% in tef1 (two nucleotides). This species was first reported from Dianthus caryophyllus, Passiflora edulis, Bouvardia longiflora, Solanum lycopersicum, Agathosma betulina, amputated human toe, Solanum tuberosum, Chrysanthemum sp., tulip roots, human leg ulcer, Secale cereal, and Musa sp. [57]. Here, we provide a new host and geographical record for F. nirenbergiae.
Figure 38.
Morphology of Fusarium nirenbergiae (MFLUCC 22-0155): (a) symptoms on Asparagus setaceous; (b,c) upper and reverse views of the colony after seven days of growth on PDA at 28 °C; (d) phialides; (e) false head; (f) conidia; (g) chlamydospores. Scale bars (d,e) = 20 µm and (f,g) = 10 µm.
3. Discussion
Ferns and fern-like species comprise a significant portion of the world’s flora and are crucial for economic and ecological reasons [2,4,58]. In this study, we collected fungi associated with ferns and fern-like species in Chaing Rai, Thailand, including ornamental (Asplenium nidus, Asparagus setaceus, Nephrolepis cordifolia, Nephrolepis spp., Phymatosorous sp., Pteris ensiformis, and P. grandifolia) and wild (Cyclosorus spp.). We introduce one new species and report 30 new hosts of 25 fungal species.
Colletotrichum fructicola, C. polypodialium, C. gigasporum, C. orchidearum, and C. truncatum species were isolated from the anthracnose symptoms (Figure 21, Figure 22, Figure 24, Figure 26, Figure 27, Figure 28, Figure 29, Figure 30 and Figure 31). Considering the isolation of these species from anthracnose spots on the other hosts [42,50,52,59] and the proof of pathogenicity of some species by other studies [60], our results indicate that ferns can be important hosts for Colletotrichum species, contributing to their evolution and fitness. This study obtained multiple associations from anthracnose of three hosts: Nephrolepis cordifolia (C. fructicola, C. pandanicola, and C. gigasporum); Cyclosorus spp. (C. pandanicola, C. plurivorum and C. truncatum, C. Orchidearum, and C. truncatum). Therefore, further studies are necessary to identify the dominant species on these hosts to ease controlling the fungus. Moreover, we demonstrated that C. polypodialium is able to complete its life cycle as a saprophyte or parasite on Phymatosorus sp., characterizing an important characteristic of the biology of this in ferns.
We isolated and identified C. lunata from leaf spots on Pteris grandifolia (Figure 3 and Figure 4) in Thailand. Curvularia lunata was first introduced as a saprobe on decaying leaves of sugarcane [23], while it was also introduced as the leaf spot agent in many hosts, such as corn [61], mulberry [25], and olive [62]. Even though we did not do pathogenicity proofs, based on the mentioned studies, the occurrence of C. lunata in P. grandifolia characterizes a reservoir for this pathogen and should be noted in the quarantine program and for managing the disease. In addition, the phylogenetic analysis of this article suggested synonymizing some species in the C. gigasporum complex and Curvularia genus. It is recommended that C. lunata and C. chiangmiensis be labeled the same species, because they cannot be distinguished by phylogenetic analysis (Figure 3), as also shown by Garcia-Aroca et al. [61]. It also appears that C. gigsporum and C. zhaoqingense (Figure 26) are essentially one species, which is supported by the nonseparation of the different isolates of these two species in the phylogenetic tree constructed for the C. gigasporum complex.
We isolated D. heveae and D. tectonendophytica associated with leaf spots on N. cordifolia (Figure 16 and Figure 19), and D. delonicis and D. chiangriensis were associated with leaf spots on Pteris grandifolia and Asplenium nidus, respectively (Figure 16, Figure 17 and Figure 19). Among the Diaporthe species that have been reported with different lifestyles, D. delonicis and D. chiangriensis have been reported as saprobe on dried seed pods of Delonix regia and dead twigs of Bauhinia sp., respectively [36,38], while D. tectonendophytica was introduced as the endophyte [39]. However, all obtained Diaporthe species in this study were isolated from leaf spots. Therefore, this indicates a lifestyle switch from endophytes to pathogens or saprophytes [63], representing an increase in their fitness.
Species of Fusarium were isolated from leaf blight on a dry stem of A. setaceous (F. ipomoeae), dead leaf and leaf spot on A. nidus (F. pernambucanum), leaf spot on N. cordifolia (F. nirenbergiae), and dry leaves of A. setaceous (F. sulawesiense) (Figure 33, Figure 34, Figure 37 and Figure 38). Fusarium sulawesiense and F. ipomoeae are pathogenic on sesame [64] and peanut [65], respectively. Additionally, F. pernambucanum and F. nirenbergiae were obtained from disease symptoms of Musa sp., Passiflora edulis [66], and potato [57]. This article completed our knowledge of the host range of these species by isolating them from symptoms on new hosts. Furthermore, F. nirenbergiae and F. pernambucanum were isolated from the dried disease branches of Asparagus setaceous, indicating that it is essential to identify the dominant species on this host before treating the related disease on this host.
This article reports L. thailandica from a leaf spot on A. nidus and a dead leaf of N. cordifolia (Figure 1 and Figure 2). Lasiodiplodia thailandica was first reported from symptomless twigs of Mangifera indica [21]. It was also isolated from canker on branches of Podocarpus macrophyllus and Albizia chinensis in China [67]. Hence, pathogenicity assays should be carried out to confirm that this species possesses both endophytic and pathogenic phases.
Several species of Neopestalotiopsis (N. guajavicola, N. hydeana, N. musae, N. pandanicola, N. phangngaensis, N. psidii, and N. saprophytica) were also isolated from leaf spots on ferns and fern-like species (Figure 5, Figure 6, Figure 7, Figure 8, Figure 9, Figure 10, Figure 11 and Figure 12). These species have been reported from several hosts worldwide [30,31]. Neopestalotiopsis musae was isolated from a leaf spot on Musa sp. [31], while N. guajavicola and N. psidii were obtained from symptomatic guava tree branches [29]. Additionally, N. hydeana was shown to cause fruit rot in Annona squamosal and Garcinia mangostana and leaf spots on a fern, Alpinia malaccensis, and Garcinia mangostana [30]. However, N. saprophytica was isolated as the saprobe on Litsea rotundifolia and Magnolia [28] and as the pathogen on Elaeis guineensis and Paphiopedilum micranthum [68,69]. Neopestalotiopsis phangngaensis and N. pandanicola were introduced as the saprobes [32]. A novel lifestyle or extending host range was thus discovered for all Neopestalotiopsis species listed in this study, which were all isolated from leaf spots of various ferns. Three species N. phangngaensis, N. pandanicola, and N. saprophytica, were isolated from Cyclosorus spp., a wild fern in Thailand, and should be regarded as the reservoir of these fungi. We also obtained the closely related species, P. dracontomelon and P. hydei, associated with leaf spots on N. cordifolia and Cyclosorus spp., in the present study (Figure 13, Figure 14 and Figure 15). These species were also reported from leaf spots on Dracontomelon dao [33] and Litsea petiolata [30]. The new host revealed the new reservoirs for these fungi, which should be considered in quarantine programs and disease management.
Ninety percent (37 out of 41 strains) of the fungi reported in this article were isolated from leaf spots and blights using the tissue culture method. All strains were collected during the dry season from Mueang district in Chiang Rai Province, Thailand. Among the 41 isolates, 26 species were identified, which shows great diversity. Each isolate of Pestalotiopsis, Neopestalotipsis, and Diaporthe isolated from ferns represented single species, showing 100% biodiversity when compared with the other recorded genera. Only approximately 4% of the fungi identified in this paper represent new species. However, Hyde et al. [43] predicted that 96% of the fungi in northern Thailand are new. Thus, limiting the lifestyle (fungi that cause leaf spot), environmental conditions (e.g., dry or wet season), and geographic area also limits identifying the flora of a given area, such as northern Thailand, which has a high fungal diversity [70,71,72,73,74]. In addition, it was documented that the sampling time on fern species affected the diversity, abundance, and composition of fungal flora [75]. Similar to our findings, only one new species was found in the biodiversity survey of associated fungi with ferns in Taiwan, although it has been estimated that between 800 and 8000 new species can be found in the country [7]. Moreover, only one new saprobe was detected in Alsophila costularis in Thailand [17]. In addition, the fungal biodiversity of two collections of fern in Mexico revealed 14 records among 21 taxa, and no new species was found [14]. However, 15% of species novelty (two new species among 13 strains) and 92% diversity (12 species among 13 strains) were recorded by Kirschner and Liu [15] in Taiwan, and 76% of the novel taxa (15 new species and on new genera among 21 taxa) was detected on amazon ferns [76] and five new Pseudocercospora species were discovered on ferns [77]. Our study showed a snapshot of fungi that can cause spot or blight on ferns and the low or high species novelty on ferns. However, wide collections must be performed in pristine tropical environments to reveal the fungal diversity of ferns and fern-like species.
This study revealed new hosts for 25 fungal species and new lifestyles for some of them (D. delonicis, D. tectonendophytica, D. chiangriensis, N. phangngaensis, and N. pandanicola). Identifying the host range of associated fungi with plants is important from several points of view. To manage plant disease using different strategies, including resistant cultivars, quarantine regulations, crop rotations, eradication of reservoirs, disease forecasting, landscape planning, risk modeling, and knowledge of pathogen host range, is necessary [78]. Furthermore, most pathogens have host specificity, meaning they cannot cause damage to nontarget plants. Therefore, they can be employed as a biocontrol agent alongside herbicides or physical techniques to manage exotic plants in agricultural ecosystems [79].
Furthermore, identifying new endophyte hosts is important because some have both pathogenic and no-pathogenic phases, while others only have nonpathogenic phases [63]. As nonpathogenic endophytes are the cause of resistance to heat, salinity, and heavy metals, identifying endophytes whose advantages have been proven in the past leads to the identification of plants that are resistant to salt and heat stress, as well as the identification of plants that are involved in bioremediation [80,81]. In addition, identifying the pathogenic forms of fungi previously reported as saprophytic, provided that their pathogenicity is proven, is necessary to control plant diseases, because it reveals unrecognized gaps in the disease cycle. No correlation between biodiversity and geographic location was found in the hosts where more than one species of the same genus was isolated from leaf spots. Only a small portion of the fungi associated with ferns in northern Thailand was identified in this work. Given the biodiversity of ferns in tropical regions and the paucity of research in this area, it is advised that additional hosts be collected and surveyed to investigate the diversity of fern-associated fungi.
4. Materials and Methods
4.1. Sampling and Fungal Isolation
Infected leaves and debris of 11 fern and fern-like species were collected from three locations in Chiang Rai Province, Thailand, in December 2021. For the fungal pathogens isolations, they were cut into 4 mm square pieces and sanitized using 1:1 70% alcohol: 1% sodium hypochlorite solution for 1–2 min, followed by a double wash in sterilized distilled water for 1 min [82]. Finally, they were air-dried on sterilized filter paper for 5 min, placed on PDA plates, and incubated in darkness at room temperature. From the debris, cultures were obtained using the imprint or direct culture methods [82]. Pure isolates were obtained using the hyphal tip method.
4.2. Culture Characteristics and Morphological Investigations
Fungal structures were obtained from the sporulating cultures on PDA and SNA, and the morphology of the isolates was investigated. The necessary morphological characters, such as asci, ascospores, conidiomata, conidiophores, phialides, conidiogenous cells, conidia, appressoria, and chlamydospores, were examined. All microscopic characters were observed with a microscope (Nikon Y-TV55, Nikon coopration, Tokyo, Japan), and digital images were captured with a Nikon DS-Ri2 camera. All measurements were made using image framework Version 0.9.7. Images used for photoplates were processed with Adobe Photoshop CS6 v. 13.1.2 (Adobe Systems, USA).
The culture morphology was investigated for all isolates, and the colony character and color were recorded after seven days growth on PDA at 28 ℃. The color was recorded based on Rayner’s color chart [83].
4.3. DNA Extraction and PCR Amplification
The DNA was extracted using the DNA extraction kit (Omega Bio-Tek). Polymerase chain reaction (PCR) amplifications were performed in a total of 25 μL: 1 μL DNA (50-700 ng/μL), 1 μL of each primer, 12.5 μL Go Taq Green Master Mix 2x (PROMEGA), and 9.5 μL molecular grade water [84] and amplified using the primers and conditions listed in Table 2.
Table 2.
PCR conditions and used primers in this study.
| Locus (Primers) | PCR Condition | Genus | Reference |
|---|---|---|---|
| act (ACT-512F/ACT-783R) | 95 °C: 5 min, (95 °C: 30 s, 55 °C: 50 s, 72 °C: 1 min) ×40 cycles, 72 °C: 10 min | Colletotrichum | [85] |
| cal (CL1/CL2A) | 95 °C: 5 min, (95 °C: 30 s, 50.8 °C: 50 s, 72 °C: 1 min) ×40 cycles, 72 °C: 10 min | Diaporthe | [86] |
| chs-1 (CHS79F/CHS345 R) | 94 °C: 5 min, (94 °C: 50 s, 58 °C: 30 s, 72 °C: 1:30 min) ×35 cycles, 72 °C: 10 min | Colletotrichum | [85] |
| gapdh (GDF/GDR) | 95 °C: 5 min, (95 °C: 50 s, 58 °C: 50 s, 72 °C: 1 min) ×40 cycles, 72 °C: 10 min | Colletotrichum | [87] |
| gapdh (gpd1/gpd2) | 94 °C: 5 min, (94 °C: 30 s, 54 °C: 50 s, 72 °C: 1 min) ×35 cycles, 72 °C: 10 min | Curvularia | [88] |
| ITS (ITS4/ITS5) | 95 °C: 5 min, (95 °C: 45 s, 53 °C: 45 s, 72 °C: 2 min) ×40 cycles, 72 °C: 10 min | All genera in this study | [89] |
| rpb1 (Fa/R8) | 94 °C: 5 min, (94 °C: 30 s, 53 °C: 50 s, 72 °C: 1 min) ×35 cycles, 72 °C: 10 min | Fusarium | [90,91] |
| rpb2 (5f2/7cr) | 94 °C: 3 min, (94 °C: 30 s, 58 °C: 1 min, 72 °C: 1:20 min) ×40 cycles, 72 °C: 10 min | Fusarium | [92,93] |
| tef1 (EF-983/EF2218R) | 94 °C: 3 min, (94 °C: 30 s, 58 °C: 1:30 min, 72 °C: 1:20 min) ×40 cycles, 72 °C: 10 min | Curvularia | [94] |
| tef1 (EF-728F/EF2) | 94 °C: 3 min, (94 °C: 30 s, 58 °C: 1:30 min, 72 °C: 1:20 min) ×40 cycles, 72 °C: 10 min | Diaporthe, Lasiodiplodia, Neopestalotiopsis, Pestalotiopsis | [85,95] |
| tef1 (EF1/EF2) | 94 °C: 3 min, (94 °C: 30 s, 58 °C: 1:30 min, 72 °C: 1:20 min) ×40 cycles, 72 °C: 10 min | Fusarium | [95] |
| tub2 (Bt2a/Bt2b) | 94 °C: 3 min, (94 °C: 30 s, 58 °C: 1:30 min, 72 °C: 1:20 min) ×40 cycles, 72 °C: 10 min | Nepestalotiopsis, Pestalotiopsis | [96,97] |
| tub2 (Btub2Fd/Btub4Rd) | 94 °C: 3 min, (94 °C: 30 s, 58 °C: 1:30 min, 72 °C: 1:20 min) ×40 cycles, 72 °C: 10 min | Colletotrichum, Diaporthe, Lasiodiplodia, | [98] |
4.4. Phylogenetic Analysis
The obtained sequences were blasted against the NCBI no-redundant sequence database, and the sequences of the closely related taxa were downloaded from the GenBank database, comprising different datasets (one per genus). The sequences of each dataset were aligned using ClustalW [99] implemented in Geneious Prime® 2019.1.3 (tef1 and tub2 genes for Neopestalotiopsis) or the MAFFT v.7 online server [100] (acceessed on 22 August 2022, https://mafft.cbrc.jp/alignment/server/) for the other cases. Sequences where ambiguities were identified were removed from the alignments. The alignments were automatically treated using TrimAl v 1.2 [101] under the -gapthreshold of 0.8′ (tef1 and tub2 gene regions for Neopestalotiopsis) or the -gappyout option for the other datasets.
Maximum likelihood (ML) inferences were performed using IQ-TREE v. 1.6.12 [102] in the CIBIV web server ([103] http://iqtree.cibiv.univie.ac.at/, acceessed on 25 August 2022), with the automatic substitution model selection [104], 1000 ultrafast bootstraps [105], and 1000 SH-aLRT branch test replicates [106]. Bayesian inference (BI) was performed in the CIPRES Science Gateway portal (https://www.phylo.org/, acceessed on 28 August 2022) [107]. The best evolutionary model for each genomic region was calculated by jModelTest 2 v.2.1.6 on XSEDE [108], and the BI analyses by MrBayes on XSEDE v.3.2.7a [109] with four independent Markov Chain Monte Carlo (MCMC) chains and four runs. The number of the generations and the sample frequency were optimized based on the number of taxa in each genus, and the stop value was set at 0.01. The first 0.25 of the sampled values was discarded as burn-in. The obtained phylogenetic trees were visualized using FigTree v 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/, acceessed on 28 August 2022).
Taxonomic novelties were registered in the Index Fungorum (http://www.indexfungorum.org/names/Names.asp, acceessed on 22 August 2022), and all the obtained sequences of this study were submitted to GenBank (Table 3).
Table 3.
GenBank accession numbers of taxa isolated in the present study.
| Species | ID | Fungarium Code | Host | ITS | tef1 | tub2 | act | cal | chs -1 | gapdh | rpb1 | rpb2 |
| Colletotrichum fructiola | MFLUCC a 22-0147 | MFLU b 22-0223 | Nephrolepis cordifolia | OP802378 | OP801751 | OP801696 | OP801713 | OP801731 | ||||
| C. gigasporum | MFLUCC 22-0158 | MFLU 22-0239 | N. cordifolia | OP802376 | OP801756 | OP801701 | OP801719 | OP801737 | ||||
| C. orchidearum | MFLUCC 22-0152 | MFLU 22-0240 | Cyclosorus sp. | OP802367 | OP801743 | OP801698 | OP801715 | OP801733 | ||||
| C. pandanicola | MFLUCC 22-0137 | MFLU 22-0224 | Nephrolepis cordifolia | OP802362 | OP801740 | OP801686 | OP801703 | OP801721 | ||||
| MFLUCC 22-0135 | MFLU 22-0225 | Pteris ensiformis | OP802374 | OP801748 | OP801693 | OP801710 | OP801728 | |||||
| MFLUCC 22-0138 | MFLU 22-0226 | Cyclosporus sp. | OP802366 | OP801742 | OP801688 | OP801705 | OP801723 | |||||
| MFLUCC 22-0159 | MFLU 22-0227 | N. cordifolia | OP802373 | OP801747 | OP801692 | OP801709 | OP801727 | |||||
| MFLUCC 22-0164 | MFLU 22-0228 | Cyclosporus sp. | OP802369 | OP801744 | OP801689 | OP801706 | OP801724 | |||||
| MFLUCC 22-0151 | MFLU 22-0229 | Cyclosporus sp. | OP802371 | OP801746 | OP801691 | OP801708 | OP801726 | |||||
| C. plurivorum | MFLUCC 22-0167 | MFLU 22-0241 | Cyclosorus sp. | OP802368 | OP801753 | OP801716 | OP801734 | |||||
| MFLUCC 22-0165 | MFLU 22-0242 | Cyclosorus sp. | OP802363 | OP801752 | OP801697 | OP801714 | OP801732 | |||||
| C. polypodialium | MFLUCC 22-0178T | MFLU 22-0234 | Nephrolepis sp. | OP802361 | OP801739 | OP801685 | OP801702 | OP801720 | ||||
| MFLU 22-0235 | Nephrolepis sp. | OP802370 | OP801745 | OP801690 | OP801707 | OP801725 | ||||||
| MFLUCC 22-0166 | MFLU 22-0236 | Phymatosorus sp. | OP802365 | OP801741 | OP801687 | OP801704 | OP801722 | |||||
| Species | ID | Fungarium Code | Host | ITS | tef1 | tub2 | act | cal | chs -1 | gapdh | rpb1 | rpb2 |
| MFLU 22-0237 | Phymatosorus sp. | OP802375 | OP801749 | OP801694 | OP801711 | OP801729 | ||||||
| MFLU 22-0238 | Phymatosorus sp. | OP802377 | OP801750 | OP801695 | OP801712 | OP801730 | ||||||
| C. truncatum | MFLUCC 22-0177 | MFLU 22-0243 | Cyclosorus sp. | OP802364 | OP801754 | OP801699 | OP801717 | OP801735 | ||||
| MFLUCC 22-0160 | MFLU 22-0244 | Cyclosorus sp. | OP802372 | OP801755 | OP801700 | OP801718 | OP801736 | |||||
| Curvularia lunata | MFLUCC 22-0142 |
MFLU 22-0209 | Pteris grandifolia | OP802379 | OP830873 | OP801738 | ||||||
| Diaporthe chiangraiensis | MFLUCC 22-0136 |
MFLU 22-0221 | Asplenium nidus | OP802381 | OP830875 | OP801758 | OP830858 | |||||
| Diaporthe delonicis | MFLUCC 22-0133 |
MFLU 22-0219 | P. grandifolia | OP802382 | OP830876 | OP801759 | OP830859 | |||||
| Diaporthe heveae | MFLUCC 22-0146 |
MFLU 22-0220 | N. cordifolia | OP802380 | OP830874 | OP801757 | OP830857 | |||||
| Diaporthe tectonendophytica | MFLUCC 22-0140 |
MFLU 22-0222 | N. cordifolia | OP802383 | OP830877 | OP801760 | OP830860 | |||||
| F. ipomoeae | MFLUCC 22-0143 |
MFLU 22-0245 | P . grandifolia | OP830879 | OP830862 | OP830868 | ||||||
| F. nirenbergiae | MFLUCC 22-0156 |
MFLU 22-0249 | N. cordifolia | OP830882 | OP830865 | OP830871 | ||||||
| MFLUCC 22-0155 |
MFLU 22-0250 | Asparagus setaceous | OP830883 | OP830866 | OP830872 | |||||||
| Fusarium sulawesiense | MFLUCC 22-0163 | MFLU 22-0247 | A. nidus | OP830878 | OP830861 | OP830867 | ||||||
| MFLUCC a 22-0153 |
MFLU b 22-0248 | A. nidus | OP830881 | OP830864 | OP830870 | |||||||
| F. pernambucanum | MFLUCC 22-0148 |
MFLU 22-0246 | A. setaceous | OP830880 | OP830863 | OP830869 | ||||||
| Lasiodiplodia thailandica | MFLUCC 22-0161 | MFLU 22-0207 | A. nidus | OP802384 | OP801761 | |||||||
| MFLUCC 22-0154 |
MFLU 22-0208 | N . cordifolia | OP802385 | OP830884 | OP801762 | |||||||
| Neopestalotiopsis guajavicola | MFLUCC 22-0134 |
MFLU 22-0210 | Nephrolepis sp. | OP802393 | OP830892 | OP801770 | ||||||
| N. hydeana | MFLUCC 22-0149 | MFLU 22-0211 | Cyclosorus sp. | OP802386 | OP830885 | OP801763 | ||||||
| N. musae | MFLUCC 22-0162 | MFLU 22-0212 | A . nidus | OP802388 | OP830887 | OP801765 | ||||||
| N. pandanicola | MFLUCC 22-0144 |
MFLU 22-0213 | Cyclosorus sp. | OP802391 | OP830890 | OP801768 | ||||||
| N. phangngaensis | MFLUCC 22-0157 | MFLU 22-0214 | Cyclosorus sp. | OP802390 | OP830889 | OP801767 | ||||||
| N . psidii | MFLUCC 22-0141 |
MFLU 22-0215 | N. cordifolia | OP802392 | OP830891 | OP801769 | ||||||
| Species | ID | Fungarium Code | Host | ITS | tef1 | tub2 | act | cal | chs -1 | gapdh | rpb1 | rpb2 |
| N. saprophytica | MFLUCC 22-0139 |
MFLU 22-0216 | Cyclosorus sp. | OP802389 | OP830888 | OP801766 | ||||||
| Neopestalotiopsis sp. | T22-0234 | FT0328 | Cyclosorus sp. | OP802387 | OP830886 | OP801764 | ||||||
| Pestalotiopsis dracontomelon | MFLUCC 22-0145 |
MFLU 22-0217 | N. cordifolia | OP802395 | OP830894 | OP801772 | ||||||
| P. hydei | MFLUCC 22-0150 |
MFLU 22-0218 | Cyclosorus sp. | OP802394 | OP830893 | OP801771 |
New species and ex-type strain are in bold. T = type strain. a Mae Fah Luang University culture collection; b Mae Fah Luang University.
4.5. Pairwise Homoplasy Index Test
To compute the recombination level between the closely related species on the phylogenetic tree, the pairwise homoplasy index (PHI) test [110] was performed by Splits Tree4 v. 4.18.2 [110], using concatenated datasets of the five loci of Colletotrichum species. The relationship between species was observed by the splits tree graph and activation of the splits decomposition and logDet options.
Acknowledgments
S.E. thanks the Center of Excellence in Fungal Research and Mae Fah Luang University (MFU) for the financial support. Ruvishika S. Jayawardena thanks the National Research Council of Thailand (NRCT), grant “Biodiversity, Taxonomy, Phylogeny and Evolution of Colletotrichum in Northern Thailand” (grant no: NRCT-TRG010); Thailand Science Research and Innovation (TSRI) grant “Biodiversity, Taxonomy, Phylogeny and Evolution of Colletotrichum on Avocado, Citrus, Durian and Mango in Northern Thailand (grant no: 652A01003); and Mae Fah Luang University grant “Identification of Fungicolous Fungi in Northern Thailand” (grant no: 651B01010) for funding this research. Kevin D. Hyde thanks the National Research Council of Thailand (NRCT) grant “Total fungal Diversity in a Given Forest Area with Implications towards Species Numbers, Chemical Diversity and Biotechnology” (grant no: N42A650547). The authors thank Mae Fah Luang University grant “Taxonomy, Phylogeny of Micro and Macro Fungi in Mae Fah Luang University Premises” (Grant No: 6431600) and Ministry of Higher Education, Science, Research and Innovation, Thailand, “National Science, Research and Innovation Fund” Reinventing University System Project (652A16049) for supporting the postdoctoral fellowship.
Author Contributions
Conceptualization, R.S.J. and K.D.H.; methodology, E.S.; software, E.S.; validation, R.S.J., A.R.G.d.F. and K.D.H.; formal analysis, E.S.; investigation, E.S.; writing—original draft preparation, E.S.; writing—review and editing, R.S.J., A.R.G.d.F. and K.D.H.; visualization, E.S.; supervision, R.S.J. and K.D.H.; funding acquisition, R.S.J. and K.D.H. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by a National Research Council of Thailand (NRCT) grant, “Biodiversity, Taxonomy, Phylogeny and Evolution of Colletotrichum in Northern Thailand” (grant no: NRCT-TRG010); Thailand Science Research and Innovation (TSRI) grant, “Biodiversity, Taxonomy, Phylogeny and Evolution of Colletotrichum on Avocado, Citrus, Durian and Mango in Northern Thailand: (grant no: 652A01003); Mae Fah Luang University grant “Identification of Fungicolous Fungi in Northern Thailand” (grant no: 651B01010); National Research Council of Thailand (NRCT) grant, “Total Fungal diversity in a Given Forest Area with Implications towards Species Numbers, Chemical Diversity and Biotechnology” (grant no. N42A650547); and Mae Fah Luang University grant “Taxonomy, Phylogeny of Micro and Macro Fungi in Mae Fah Luang University Premises” (grant no: 6431600); Ministry of Higher Education, Science, Research and Innovation, Thailand, “National Science, Research and Innovation Fund” Reinventing University System Project (652A16049) for supporting the postdoctoral fellowship.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Middleton D.J. Progress on the Flora of Thailand. Telopea. 2003;10:33–42. doi: 10.7751/telopea20035603. [DOI] [Google Scholar]
- 2.Smith A.R., Pryer K.M., Schuettpelz E., Korall P., Schneider H., Wolf P.G. A classification for extant ferns. Taxon. 2006;55:705–731. doi: 10.2307/25065646. [DOI] [Google Scholar]
- 3.Moran R.C. Diversity, biogeography, and floristics. In: Ranker T.A., Haufler C.H., editors. Biology and Evolution of Ferns and Lycophytes. Cambridge University Press; Cambridge, UK: 2008. pp. 367–394. [DOI] [Google Scholar]
- 4.Mannan M.M., Maridass M., Victor B. A review on the potential uses of ferns. Ethnobot. Leafl. 2008;12:281–285. [Google Scholar]
- 5.Mehltreter K., Walker L.R., Sharpe J.M. Fern Ecology. Cambridge University Press; Cambridge, UK: 2010. pp. 1–429. [DOI] [Google Scholar]
- 6.Del Olmo-Ruiz M., Arnold A.E. Interannual variation and host affiliations of endophytic fungi associated with ferns at La Selva, Costa Rica. Mycologia. 2014;106:8–21. doi: 10.3852/13-098. [DOI] [PubMed] [Google Scholar]
- 7.Kirschner R., Lee P.H., Huang Y.-M. Diversity of fungi on Taiwanese fern plants: Review and new discoveries. Taiwania. 2019;64:163–175. doi: 10.6165/tai.2019.64.163. [DOI] [Google Scholar]
- 8.Guatimosim E., Schwartsburd P.B., Barreto R.W. A new Inocyclus species (Parmulariaceae) on the neotropical fern Pleopeltis astrolepis. IMA Fungus. 2014;5:51–55. doi: 10.5598/imafungus.2014.05.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kakishima M., Ji J.-X., Wang Q., Li Y. First report of rust disease caused by Milesina dryopteridis on two species of ferns, Rumohra adiantiformis and Pteris fauriei, in Japan. Plant Dis. 2016;100:2529. doi: 10.1094/PDIS-05-16-0592-PDN. [DOI] [Google Scholar]
- 10.Bejar J., Luján Luna M., Murace M.A., Nazareno Saparrat M.C. Effects of the fungus Pestalotiopsis maculans (Ascomycota: Amphisphaeriales) on the gametophytic development of the fern Lygodium venustum (Lygodiaceae) Rev. Biol. Trop. 2019;67:1520–1530. doi: 10.15517/rbt.v67i6.35860. [DOI] [Google Scholar]
- 11.Jones K.A., Rayamajhi M.B., Pratt P.D., Van T.K. First report of the pathogenicity of Colletotrichum gloeosporioides on invasive ferns, Lygodium microphyllum and L. japonicum, in Florida. Plant Dis. 2003;87:101. doi: 10.1094/PDIS.2003.87.1.101B. [DOI] [PubMed] [Google Scholar]
- 12.Strandberg J.O. Pathogenicity of the fern anthracnose fungus, Colletotrichum acutatum, on wild and cultivated ferns in Florida. Proc. Fla. State Hortic. Soc. 1999;112:274–276. [Google Scholar]
- 13.Dey S., Hore M., Biswas J., Biswas M., Mandal B.K., Das P., Gupta S. A new record of brown rot disease in water fern Azolla microphylla (Azollaceae): Loss of important bio-resource. Fern Gaz. 2017;20:245–254. [Google Scholar]
- 14.Medel-Ortiz R., Baeza Y., Lorea-Hernández F.G. Pteridicolous Ascomycetes from a cloud forest in eastern Mexico. Mycotaxon. 2020;134:681–705. doi: 10.5248/134.681. [DOI] [Google Scholar]
- 15.Kirschner R., Liu L.-C. Mycosphaerellaceous fungi and new species of Venustosynnema and Zasmidium on ferns and fern allies in Taiwan. Phytotaxa. 2014;176:309–323. doi: 10.11646/phytotaxa.176.1.29. [DOI] [Google Scholar]
- 16.Maharachchikumbura S.S.N., Guo L.-D., Chukeatirote E., Hyde K.D. Improving the backbone tree for the genus Pestalotiopsis; addition of P. steyaertii and P. magna sp. nov. Mycol. Prog. 2014;13:617–624. doi: 10.1007/s11557-013-0944-0. [DOI] [Google Scholar]
- 17.Zhang J., Phookamsak R., Mapook A., Lu Y., Lv M. Monilochaetes pteridophytophila (Australiascaceae, Glomerellales), a new fungus from Tree fern. Biodivers. Data J. 2021;9:e67248. doi: 10.3897/BDJ.9.e67248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hongsanan S., Hyde K.D., Phookamsak R., Wanasinghe D.N., McKenzie E.H.C., Sarma V.V., Lücking R., Boonmee S., Bhat J.D., Liu N.-G., et al. Refined families of Dothideomycetes: Orders and families Incertae sedis in Dothideomycetes. Fungal Divers. 2020;105:17–318. doi: 10.1007/s13225-020-00462-6. [DOI] [Google Scholar]
- 19.Linaldeddu B.T., Deidda A., Scanu B., Franceschini A., Serra S., Berraf-Tebbal A., Zouaoui Boutiti M., Ben Jamâa M.L., Phillips A.J.L. Diversity of Botryosphaeriaceae species associated with grapevine and other woody hosts in Italy, Algeria and Tunisia, with descriptions of Lasiodiplodia exigua and Lasiodiplodia mediterranea sp. nov. Fungal Divers. 2015;71:201–214. doi: 10.1007/s13225-014-0301-x. [DOI] [Google Scholar]
- 20.Chethana K.W.T., Li X., Zhang W., Hyde K.D., Yan J. Trail of Decryption of molecular research on Botryosphaeriaceae in woody plants. Phytopathol. Mediterr. 2016;55:147–171. doi: 10.14601/Phytopathol_Mediterr-16230. [DOI] [Google Scholar]
- 21.Trakunyingcharoen T., Lombard L., Groenewald J.Z., Cheewangkoon R., To-Anun C., Crous P.W. Caulicolous Botryosphaeriales from Thailand. Pers.-Mol. Phylogeny Evol. Fungi. 2015;34:87–99. doi: 10.3767/003158515X685841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hongsanan S., Hyde K.D., Phookamsak R., Wanasinghe D.N., McKenzie E.H.C., Sarma V.V., Boonmee S., Lücking R., Bhat D.J., Liu N.G., et al. Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere. 2020;11:1553–2107. doi: 10.5943/mycosphere/11/1/13. [DOI] [Google Scholar]
- 23.Manamgoda D.S., Cai L., McKenzie E.H.C., Crous P.W., Madrid H., Chukeatirote E., Shivas R.G., Tan Y.P., Hyde K.D. A phylogenetic and taxonomic re-evaluation of the Bipolaris-Cochliobolus-Curvularia complex. Fungal Divers. 2012;56:131–144. doi: 10.1007/s13225-012-0189-2. [DOI] [Google Scholar]
- 24.Wonglom P., Ito S., Sunpapao A. First report of Curvularia lunata causing leaf spot of Brassica rapa subsp. pekinensis in Thailand. New Dis. Rep. 2018;38:588–2044. doi: 10.5197/j.2044-0588.2018.038.015. [DOI] [Google Scholar]
- 25.Bussaban B., Kodchasee P., Apinyanuwat S., Kosawang C., Jonglaekha N. First report of Curvularia lunata causing leaf blight on mulberry (Morus sp.) in Thailand. Plant Dis. 2017;101:1951. doi: 10.1094/PDIS-02-17-0260-PDN. [DOI] [Google Scholar]
- 26.Ariyawansa H.A., Thambugala K.M., Manamgoda D.S., Jayawardena R., Camporesi E., Boonmee S., Wanasinghe D.N., Phookamsak R., Hongsanan S., Singtripop C., et al. Towards a natural classification and backbone tree for Pleosporaceae. Fungal Divers. 2015;71:85–139. doi: 10.1007/s13225-015-0323-z. [DOI] [Google Scholar]
- 27.Hyde K., Norphanphoun C., Maharachchikumbura S., Bhat D., Jones E.B.G., Bundhun D., Chen Y.J., Bao D.F., Boonmee S., Boonmee M.S., et al. Refined families of Sordariomycetes. Mycosphere. 2020;11:305–1059. doi: 10.5943/mycosphere/11/1/7. [DOI] [Google Scholar]
- 28.Maharachchikumbura S.S.N., Hyde K.D., Groenewald J.Z., Xu J., Crous P.W. Pestalotiopsis revisited. Stud. Mycol. 2014;79:121–186. doi: 10.1016/j.simyco.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ul Haq I., Ijaz S., Aslam Khan N. Genealogical concordance of phylogenetic species recognition-based delimitation of Neopestalotiopsis species associated with leaf spots and fruit canker disease affected guava Plants. Pak. J. Agric. Sci. 2021;58:1301–1313. doi: 10.21162/PAKJAS/21.1045. [DOI] [Google Scholar]
- 30.Huanaluek N., Jayawardena R.S., Maharachchikumbura S.S., Harishchandra D.L. Additions to Pestalotioid fungi in Thailand: Neopestalotiopsis hydeana sp. nov. and Pestalotiopsis hydei sp. nov. Phytotaxa. 2021;479:23–43. doi: 10.11646/phytotaxa.479.1.2. [DOI] [Google Scholar]
- 31.Hyde K.D., Hongsanan S., Jeewon R., Bhat D.J., McKenzie E.H.C., Jones E.B.G., Phookamsak R., Ariyawansa H.A., Boonmee S., Zhao Q., et al. Fungal Diversity Notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;80:1–270. doi: 10.1007/s13225-016-0373-x. [DOI] [Google Scholar]
- 32.Tibpromma S., Hyde K.D., McKenzie E.H.C., Bhat D.J., Phillips A.J.L., Wanasinghe D.N., Samarakoon M.C., Jayawardena R.S., Dissanayake A.J., Tennakoon D.S., et al. Fungal Diversity Notes 840–928: Mmicro-fungi associated with Pandanaceae. Fungal Divers. 2018;93:1–160. doi: 10.1007/s13225-018-0408-6. [DOI] [Google Scholar]
- 33.Liu J.K., Hyde K.D., Jones E.B., Ariyawansa H.A., Bhat D.J., Boonmee S., Maharachchikumbura S.S.N., McKenzie E.H.C., Phookamsak R., Phukhamsakda C., et al. Fungal Diversity Notes 1–110: Taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 2015;72:1–197. doi: 10.1007/s13225-015-0324-y. [DOI] [Google Scholar]
- 34.Senanayake I.C., Jeewon R., Chomnunti P., Wanasinghe D.N., Norphanphoun C., Karunarathna A., Pem D., Perera R.H., Camporesi E., McKenzie E.H.C., et al. Taxonomic circumscription of Diaporthales based on multigene phylogeny and morphology. Fungal Divers. 2018;93:241–443. doi: 10.1007/s13225-018-0410-z. [DOI] [Google Scholar]
- 35.Norphanphoun C., Gentekaki E., Hongsana S., Jayawardena R., Senanayake I.C., Manawasinghe I.S., Abeywickrama P.D., Bhunjun C.S., Hyde K.D. Diaporthe: Formalizing the species-group concept. Mycosphere. 2022;13:752–819. doi: 10.5943/mycosphere/13/1/9. [DOI] [Google Scholar]
- 36.Perera R.H., Hyde K.D., Maharachchikumbura S.S.N., Jones E.B.G., McKenzie E.H.C., Stadler M., Lee H.B., Samarakoon M.C., Ekanayaka A.H., Camporesi E., et al. Fungi on wild seeds and fruits. Mycosphere. 2020;11:2108–2480. doi: 10.5943/mycosphere/11/1/14. [DOI] [Google Scholar]
- 37.Gomes R.R., Glienke C., Videira S.I.R., Lombard L., Groenewald J.Z., Crous P.W. Diaporthe: A genus of endophytic, saprobic and plant pathogenic fungi. Pers.-Mol. Phylogeny Evol. Fungi. 2013;31:1–41. doi: 10.3767/003158513X666844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Senanayake I.C., Crous P.W., Groenewald J.Z., Maharachchikumbura S.S.N., Jeewon R., Phillips A.J.L., Bhat J.D., Perera R.H., Li Q.R., Li W.J., et al. Families of Diaporthales based on morphological and phylogenetic evidence. Stud. Mycol. 2017;86:217–296. doi: 10.1016/j.simyco.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doilom M., Dissanayake A.J., Wanasinghe D.N., Boonmee S., Liu J.-K., Bhat D.J., Taylor J.E., Bahkali A., McKenzie E.H.C., Hyde K.D., et al. Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Divers. 2017;82:107–182. doi: 10.1007/s13225-016-0368-7. [DOI] [Google Scholar]
- 40.Réblová M., Gams W., Seifert K.A. Monilochaetes and allied genera of the Glomerellales, and a reconsideration of families in the microascales. Stud. Mycol. 2011;68:163–191. doi: 10.3114/sim.2011.68.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu F., Ma Z.Y., Hou L.W., Diao Y.Z., Wu W.P., Damm U., Song S., Cai L. Updating species diversity of Colletotrichum, with a phylogenomic overview. Stud. Mycol. 2022;101:1–56. doi: 10.3114/sim.2022.101.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prihastuti H., Cai L., Chen H., McKenzie E.H.C., Hyde K.D. Characterization of Colletotrichum species associated with Coffee berries in Northern Thailand. Fungal Divers. 2009;39:89–109. [Google Scholar]
- 43.Hyde K.D., Norphanphoun C., Chen J., Dissanayake A.J., Doilom M., Hongsanan S., Jayawardena R.S., Jeewon R., Perera R.H., Thongbai B., et al. Thailand’s amazing diversity: Up to 96% of fungi in Northern Thailand may be novel. Fungal Divers. 2018;93:215–239. doi: 10.1007/s13225-018-0415-7. [DOI] [Google Scholar]
- 44.Tibpromma S., Hyde K.D., Bhat J.D., Mortimer P.E., Xu J., Promputtha I., Doilom M., Yang J.-B., Tang A.M.C., Karunarathna S.C. Identification of endophytic fungi from leaves of Pandanaceae based on their morphotypes and DNA sequence data from Southern Thailand. MycoKeys. 2018;33:25–67. doi: 10.3897/mycokeys.33.23670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chethana K.W., Manawasinghe I.S., Hurdeal V.G., Bhunjun C.S., Appadoo M.A., Gentekaki E., Raspé O., Promputtha I., Hyde K.D. What are fungal species and how to delineate them? Fungal Divers. 2021;109:1–25. doi: 10.1007/s13225-021-00483-9. [DOI] [Google Scholar]
- 46.Jayawardena R.S., Bhunjun C.S., Hyde K.D., Gentekaki E., Itthayakorn P. Colletotrichum: Lifestyles, biology, morpho-species, species complexes and accepted species. Mycosphere. 2021;12:519–669. doi: 10.5943/mycosphere/12/1/7. [DOI] [Google Scholar]
- 47.Jayawardena R.S., Hyde K.D., de Farias A.R.G., Bhunjun C.S., Ferdinandez H.S., Manamgoda D.S., Udayanga D., Herath I.S., Thambugala K.M., Manawasinghe I.S., et al. What is a species in fungal plant pathogens? Fungal Divers. 2021;109:239–266. doi: 10.1007/s13225-021-00484-8. [DOI] [Google Scholar]
- 48.Rakotoniriana E.F., Scauflaire J., Rabemanantsoa C., Urveg-Ratsimamanga S., Corbisier A.-M., Quetin-Leclercq J., Declerck S., Munaut F. Colletotrichum gigasporum sp. nov., a new species of Colletotrichum producing long straight conidia. Mycol. Prog. 2013;12:403–412. doi: 10.1007/s11557-012-0847-5. [DOI] [Google Scholar]
- 49.Jayawardena R.S., Hyde K.D., Damm U., Cai L., Liu M., Li X.H., Zhang W., Zhao W.S., Yan J.Y. Notes on currently accepted species of Colletotrichum. Mycosphere. 2016;7:1192–1260. doi: 10.5943/mycosphere/si/2c/9. [DOI] [Google Scholar]
- 50.Damm U., Sato T., Alizadeh A., Groenewald J.Z., Crous P.W. The Colletotrichum dracaenophilum, C. magnum and C. orchidearum species complexes. Stud. Mycol. 2018;90:71–118. doi: 10.1016/j.simyco.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Silva D.D., Groenewald J.Z., Crous P.W., Ades P.K., Nasruddin A., Mongkolporn O., Taylor P.W.J. Identification, prevalence and pathogenicity of Colletotrichum species causing anthracnose of Capsicum annuum in Asia. IMA Fungus. 2019;10:8. doi: 10.1186/s43008-019-0001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Damm U., Woudenberg J.H.C., Cannon P.F., Crous P.W. Colletotrichum species with curved conidia from herbaceous hosts. Fungal Divers. 2009;39:45–87. [Google Scholar]
- 53.Crous P.W., Lombard L., Sandoval-Denis M., Seifert K.A., Schroers H.-J., Chaverri P., Gené J., Guarro J., Hirooka Y., Bensch K., et al. Fusarium: More than a node or a foot-shaped basal cell. Stud. Mycol. 2021;98:100116. doi: 10.1016/j.simyco.2021.100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang M.M., Chen Q., Diao Y.Z., Duan W.J., Cai L. Fusarium incarnatum-equiseti complex from China. Pers.-Mol. Phylogeny Evol. Fungi. 2019;43:70–89. doi: 10.3767/persoonia.2019.43.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.da Silva Santos A.C., Trindade J.V.C., Lima C.S., Barbosa R.d.N., da Costa A.F., Tiago P.V., de Oliveira N.T. Morphology, phylogeny, and sexual stage of Fusarium caatingaense and Fusarium pernambucanum, new species of the Fusarium incarnatum-equiseti species complex associated with insects in Brazil. Mycologia. 2019;111:244–259. doi: 10.1080/00275514.2019.1573047. [DOI] [PubMed] [Google Scholar]
- 56.Maryani N., Sandoval-Denis M., Lombard L., Crous P.W., Kema G.H.J. New endemic Fusarium species hitch-hiking with pathogenic Fusarium strains causing Panama disease in small-holder banana plots in Indonesia. Pers.-Mol. Phylogeny Evol. Fungi. 2019;43:48–69. doi: 10.3767/persoonia.2019.43.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lombard L., Sandoval-Denis M., Lamprecht S.C., Crous P.W. Epitypification of Fusarium oxysporum- clearing the taxonomic chaos. Pers.-Mol. Phylogeny Evol. Fungi. 2019;43:1–47. doi: 10.3767/persoonia.2019.43.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mehltreter K. Interactions of ferns with fungi and animals. Fern Ecol. 2010:220–254. doi: 10.1017/CBO9780511844898.008. [DOI] [Google Scholar]
- 59.Mu T., Zhang Z., Liu R., Liu S., Li Z., Zhang X., Xia J. Morphological and molecular phylogenetic analyses reveal three species of Colletotrichum in Shandong province, China. MycoKeys. 2021;85:57–71. doi: 10.3897/mycokeys.85.75944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Costa J.F.O., Kamei S.H., Silva J.R.A., Miranda A.R.G., Netto M.B., da Silva S.J.C., Correia K.C., de Andrade Lima G.S., Assunção I.P. Species diversity of Colletotrichum infecting Annona spp. in Brazil. Eur. J. Plant Pathol. 2019;153:1119–1130. doi: 10.1007/s10658-018-01630-w. [DOI] [Google Scholar]
- 61.Garcia-Aroca T., Doyle V., Singh R., Price T., Collins K. First report of Curvularia leaf spot of corn, caused by Curvularia lunata, in the United States. Plant Health Prog. 2018;19:140–142. doi: 10.1094/PHP-02-18-0008-BR. [DOI] [Google Scholar]
- 62.Mohamed M.D.A., Moharam M.H.A., Ahmed H.A.M. In vitro and in vivo toxicity of nano chitosan against Curvularia lunata, the causal microorganism of fruit rot and blight, a new disease of olive (O. Europaea L.) Eur. J. Plant Pathol. 2021;161:881–894. doi: 10.1007/s10658-021-02371-z. [DOI] [Google Scholar]
- 63.del Carmen Orozco-Mosqueda M., Santoyo G. Plant-microbial endophytes interactions: Scrutinizing their beneficial mechanisms from genomic explorations. Curr. Plant Biol. 2021;25:100189. doi: 10.1016/j.cpb.2020.100189. [DOI] [Google Scholar]
- 64.Kamran M., Ullah I., Ehetisham ul Haq M., Iqbal M., Abbas H., Nazir S., Ali S., Khan Q.A.T., Idrees M. First report of Fusarium sulawesiensis (FIESC-16) associated with stem rot of sesame in Pakistan. Plant Dis. 2020;104:1554. doi: 10.1094/PDIS-11-19-2330-PDN. [DOI] [Google Scholar]
- 65.Xu M., Zhang X., Yu J., Guo Z., Li Y., Wu J., Chi Y. First report of Fusarium ipomoeae causing peanut leaf spot in China. Plant Dis. 2021;105:3754. doi: 10.1094/PDIS-01-21-0226-PDN. [DOI] [PubMed] [Google Scholar]
- 66.Lima E.N., Oster A.H., Bordallo P.N., Araújo A.A.C., Silva D.E.M., Lima C.S. A novel lineage in the Fusarium incarnatum-equiseti species complex is one of the causal agents of Fusarium rot on melon fruits in northeast Brazil. Plant Pathol. 2021;70:133–143. doi: 10.1111/ppa.13271. [DOI] [Google Scholar]
- 67.Dou Z.P., He W., Zhang Y. Does morphology matter in taxonomy of Lasiodiplodia? an answer from Lasiodiplodia hyalina sp. nov. Mycosphere. 2017;8:1014–1027. doi: 10.5943/mycosphere/8/2/5. [DOI] [Google Scholar]
- 68.Ismail S.I., Zulperi D., Norddin S., Ahmad-Hamdani S. First report of Neopestalotiopsis saprophytica causing leaf spot of oil palm (Elaeis guineensis) in Malaysia. Plant Dis. 2017;101:1821. doi: 10.1094/PDIS-02-17-0271-PDN. [DOI] [Google Scholar]
- 69.Qin Q., Lu Z., Lu Z., Ding L., Chi Z., Shan B. First report of leaf spot on Paphiopedilum micranthum caused by Neopestalotiopsis saprophytica in China. Plant Dis. 2020;104:2738. doi: 10.1094/PDIS-02-20-0275-PDN. [DOI] [Google Scholar]
- 70.Boonmee S., Wanasinghe D.N., Calabon M.S., Huanraluek N., Chandrasiri S.K.U., Jones G.E.B., Rossi W., Leonardi M., Singh S.K., Rana S., et al. Fungal Diversity Notes 1387–1511: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2021;111:1–335. doi: 10.1007/s13225-021-00489-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hyde K.D., Tennakoon D.S., Jeewon R., Bhat D.J., Maharachchikumbura S.S.N., Rossi W., Leonardi M., Lee H.B., Mun H.Y., Houbraken J., et al. Fungal Diversity Notes 1036–1150: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2019;96:1–242. doi: 10.1007/s13225-019-00429-2. [DOI] [Google Scholar]
- 72.Phookamsak R., Hyde K.D., Jeewon R., Bhat D.J., Jones E.B., Maharachchikumbura S.S.N., Raspé O., Karunarathna S.C., Wanasinghe D.N., Hongsanan S., et al. Fungal Diversity Notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 2019;95:1–273. doi: 10.1007/s13225-019-00421-w. [DOI] [Google Scholar]
- 73.Hyde K.D., Dong Y., Phookamsak R., Jeewon R., Bhat D.J., Jones E.B., Liu N.-G., Abeywickrama P.D., Mapook A., Wei D., et al. Fungal Diversity Notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020;100:5–277. doi: 10.1007/s13225-020-00439-5. [DOI] [Google Scholar]
- 74.Yuan H.-S., Lu X., Dai Y.-C., Hyde K.D., Kan Y.-H., Kušan I., He S.-H., Liu N.-G., Sarma V.V., Zhao C.-L., et al. Fungal Diversity Notes 1277–1386: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2020;104:1–266. doi: 10.1007/s13225-020-00461-7. [DOI] [Google Scholar]
- 75.Del Olmo-Ruiz M., Arnold A.E. Community structure of fern-affiliated endophytes in three neotropical forests. J. Trop. Ecol. 2017;33:60–73. doi: 10.1017/S0266467416000535. [DOI] [Google Scholar]
- 76.Guatimosim E., Schwartsburd P.B., Barreto R.W., Crous P.W. Novel fungi from an ancient niche: Cercosporoid and related sexual morphs on ferns. Persoonia. 2016;37:106–141. doi: 10.3767/003158516X690934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Braun U., Nakashima C., Crous P.W. Cercosporoid fungi (Mycosphaerellaceae) 1. species on other fungi, Pteridophyta and Gymnospermae. IMA Fungus. 2013;4:265–345. doi: 10.5598/imafungus.2013.04.02.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morris C.E., Moury B. Revisiting the concept of host range of plant pathogens. Annu. Rev. Phytopathol. 2019;57:63–90. doi: 10.1146/annurev-phyto-082718-100034. [DOI] [PubMed] [Google Scholar]
- 79.Barton J. Predictability of pathogen host range in classical biological control of weeds: An update. BioControl. 2012;57:289–305. doi: 10.1007/s10526-011-9401-7. [DOI] [Google Scholar]
- 80.Clay K., Schardl C. Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am. Nat. 2002;160:S99–S127. doi: 10.1086/342161. [DOI] [PubMed] [Google Scholar]
- 81.Rodriguez R.J., Redman R.S., Henson J.M. The role of fungal symbioses in the adaptation of plants to high stress environments. Mitig. Adapt. Strateg. Glob. Chang. 2004;9:261–272. doi: 10.1023/B:MITI.0000029922.31110.97. [DOI] [Google Scholar]
- 82.Senanayake I.C., Rathnayaka A.R., Marasinghe D.S., Calabon M.S., Gentekaki E., Lee H.B., Hurdeal V.G., Pem D., Dissanayake L.S., Wijesinghe S.N., et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere. 2020;11:2678–2754. doi: 10.5943/mycosphere/11/1/20. [DOI] [Google Scholar]
- 83.Rayner R.W. A Mycological Colour Chart. Volume 64. Taylor & Francis, Ltd.; Milton Park, UK: 1970. pp. 230–233. [DOI] [Google Scholar]
- 84.Dissanayake A.J., Bhunjun C.S., Maharachchikumbura S.S.N., Liu J.K. Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere. 2020;11:2652–2676. doi: 10.5943/mycosphere/11/1/18. [DOI] [Google Scholar]
- 85.Carbone I., Kohn L.M. A method for designing primer sets for speciation studies in filamentous Ascomycetes. Mycologia. 1999;91:553–556. doi: 10.1080/00275514.1999.12061051. [DOI] [Google Scholar]
- 86.O’Donnell K., Nirenberg H.I., Aoki T., Cigelnik E. A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience. 2000;41:61–78. doi: 10.1007/BF02464387. [DOI] [Google Scholar]
- 87.Templeton M.D., Rikkerink E.H.A., Solon S.L., Crowhurst R.N. Cloning and molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase encoding gene and cDNA from the plant pathogenic fungus Glomerella cingulata. Gene. 1992;122:225–230. doi: 10.1016/0378-1119(92)90055-T. [DOI] [PubMed] [Google Scholar]
- 88.Berbee M.L., Pirseyedi M., Hubbard S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia. 1999;91:964–977. doi: 10.1080/00275514.1999.12061106. [DOI] [Google Scholar]
- 89.White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. 1990;18:315–322. [Google Scholar]
- 90.Hofstetter V., Miadlikowska J., Kauff F., Lutzoni F. Phylogenetic comparison of protein-coding versus ribosomal RNA coding sequence data: A case study of the Lecanoromycetes (Ascomycota) Mol. Phylogenet. Evol. 2007;44:412–426. doi: 10.1016/j.ympev.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 91.O’Donnell K., Sutton D.A., Rinaldi M.G., Sarver B.A.J., Balajee S.A., Schroers H.-J., Summerbell R.C., Robert V.A.R.G., Crous P.W., Zhang N., et al. Internet-accessible DNA sequence database for identifying Fusaria from human and animal infections. J. Clin. Microbiol. 2010;48:3708–3718. doi: 10.1128/JCM.00989-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reeb V., Lutzoni F., Roux C. Contribution of RPB2 to multilocus phylogenetic studies of the Euascomycetes (Pezizomycotina, fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Mol. Phylogenet. Evol. 2004;32:1036–1060. doi: 10.1016/j.ympev.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 93.Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among Ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 94.Rehner S.A., Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1 sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- 95.O’Donnell K., Kistler H.C., Cigelnik E., Ploetz R.C. Multiple evolutionary origins of the fungus causing panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Appl. Environ. Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.O’Donnell K., Cigelnik E. Two Divergent Intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- 98.Woudenberg J.H.C., Aveskamp M.M., De Gruyter J., Spiers A.G., Crous P.W. Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Pers.-Mol. Phylogeny Evol. Fungi. 2009;22:56–62. doi: 10.3767/003158509X427808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Capella-Gutiérrez S., Silla-Martinez J.M., Gabaldón T. TrimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2014;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Trifinopoulos J., Nguyen L.-T., von Haeseler A., Minh B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2017;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 107.Miller M.A., Pfeiffer W., Schwartz T. The CIPRES Science Gateway: Enabling high-impact science for phylogenetics researchers with limited resources; Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the Extreme to the Campus and beyond; New York, NY, USA. 16–20 July 2012; pp. 1–8. [DOI] [Google Scholar]
- 108.Darriba D., Taboada G.L., Doallo R., Posada D. JModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ronquist F., Teslenko M., Van Der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.