Abstract
Objective:
Prior studies identified SNPs associated with physical activity (PA) level in a natural environment and intervention study: rs978656-DNAPTP6, rs10887741-PAPSS2, rs7279064-C18orf2, and rs6265-BDNF. Using the 4 SNPs’ polygenic score (PGS), we examined whether PGS moderate a lifestyle intervention’s effect on changes in PA level and cardiorespiratory fitness (CRF).
Methods:
This is a secondary analysis of Look AHEAD, a multi-center randomized controlled trial designed to test the health benefits of a lifestyle intervention among 2,675 participants with overweight/obesity and type 2 diabetes (ages 45–76). Using linear mixed effects models, level of PA (Paffenbarger PA questionnaire) and treadmill-assessed CRF were each regressed on 4 SNPs’ PGS, study time (baseline, year1, and year4), intervention arm, and interactions between the three. Models adjusted for age, sex, body mass index, ancestry principal components (population stratification), and study sites, with Bonferroni corrections for multiple testing (alpha<0.005). Effect modification by age was examined.
Results:
PGS was not predictive of change in CRF or PA level in response to intervention. In analyses without PGS×intervention×time, the relationships between PGS and PA phenotypes were modified by age (p-interaction=0.048 for CRF and 0.058 for PA), such that one unit increase in PGS was associated with 24 kcal·week−1 more in moderate intensity PA and 0.004 MET higher CRF only among older groups (age>55 year for CRF, >60 year for PA level).
Conclusion:
The effects of the intervention on PA and CRF were not moderated by the 4 SNPs. Future studies with extended SNP list should confirm the findings on effect modification by age.
Keywords: physical activity, intervention, genomics, prediction behavioral medicine
INTRODUCTION
Over the past seventy years, biomedical and epidemiological research has shown that regular physical activity (PA) is critical for physical and mental health (1–3). However, only half (51.6%) of US adults meet the national guidelines of expending 500–1000 metabolic equivalent (MET) minutes (~1000kacl) per week through PA (4). In particular, a sharp decline in PA is observed after age 45–50 (5–7). Thus, enhancing PA rate among middle-aged adults has high public health relevance.
It is widely accepted that low rates of PA stem primarily from environmental factors, such as cars and elevators that have allowed people to move less (8, 9) but individuals respond differently to exogenous stimuli (e.g., obesogenic environment) due to an endogenous factors, such as age and sex (8). Similarly, a substantial proportion of variance in PA behavior is accounted for by genomic variability. A large body of behavioral genetics research indicates that genomic factors can predict a substantial proportion of variance in PA (30–78%), exercise participation (48–71%), and sports participation (35–83%), as evidenced by twin studies (10–15). Given the nontrivial contribution of genomic factors in predicting variance in PA (~30%), it deserves to be considered in more detail.
Human genetics research identifying the specific genetic variants, or single nucleotide polymorphisms (SNPs), associated with PA has been stagnant in the past decade since De Moor, Liu (16) identified 37 SNPs clustered near 3 genes (DNAPTP6, PAPSS2, and C18orf2), chiefly due to lack of studies with sufficiently large sample sizes. In recent years, however, the field has observed an emergence of genome-wide association studies (GWAS) from large cohorts. These include, but are not limited to, the UK Biobank (17, 18), Japan Multi-Institutional Collaborative Cohort (19), as well as aggregation of Framingham Heart Study, Women’s Health Initiative and Jackson Heart Study (20). More GWAS are expected to come through Precision Medicine Initiative cohorts (e.g., “All of Us” and “Million Veterans Program”). To this effect, it is reasonable to be optimistic that researchers may soon identify which SNPs are associated with PA in the natural environment.
While naturally occurring PA level at a single time point is the most widely used PA phenotype in large GWAS, these phenotypes are considered relatively shallow as they yield insufficient knowledge regarding how we can use genomic data to change PA behavior, and ultimately the public’s health. A study using a deeper phenotype, such as changes in PA level or PA-related health outcomes in response to an intervention, can provide more useful information for increasing PA rate. For example, in a randomized controlled PA promotion trial, a SNP near Brain derived neurotropic factor gene (BDNF), moderated the efficacy of a PA promotion intervention on PA level (21), such that the A allele carriers engaged in more PA than the GG homozygotes randomized to the PA promotion arm, but not in the control group. This study highlights the possibility of using genomic data for personalized interventions for PA interventions. However, the major weakness of this intervention research is the small sample size (n<300), and the candidate gene approach, which has shown a low replication rate (22).
Taken together, investigating the genetic influence on changes in PA level and PA-related health outcomes using a large sample size with GWAS-identified SNPs adds significant evidence to the existing body of literature on the genetic basis of PA.
Thus, the objective of the present study is to examine whether genetic variations moderate a lifestyle intervention’s effect on changes in PA level and cardiorespiratory fitness (CRF) using a large data set (n=2,675). To quantify genetic variation, we computed a polygenic score (PGS) by adding the number of effect alleles that predispose individuals to be more physically active from four SNPs1—three from the GWAS of PA (i.e., G of rs978656, T of rs10887741, G of rs7279064) and one from the intervention study (i.e., A of rs6265)—with weights equal to the published per-allele effects (Table S1, Supplemental Digital Content). We used data from the Action for Health in Diabetes (Look AHEAD) trial, a multi-center randomized controlled trial designed to test the health benefits of a lifestyle intervention among participants with overweight/obesity and type 2 diabetes (ages 45–76). The central hypothesis is that those who are genetically predisposed to high PA (based on PGS of the 4 SNPs) may respond better to a PA promotion intervention than those who are prone to low PA.
METHODS
Study Cohort
We used data from Look AHEAD participants. The primary objective of the Look AHEAD trial was to assess the long-term effects (up to 11.5 years) of an intensive lifestyle intervention (ILI) in individuals with overweight and obesity as well as type 2 diabetes. The baseline characteristics of the Look AHEAD participants as well as the design and methods have been reported elsewhere (23). Briefly, Look AHEAD, which started in June 2001, was a randomized controlled trial investigating the efficacy of an ILI on cardiovascular outcomes compared to diabetes support and education (DSE). Both ILI and DSE groups received one session of education on cardiovascular risk factors and diabetes. The Look AHEAD trial was approved by local Institutional Review Boards, including genetic analyses. The current data analysis was approved by the Miriam Hospital Institutional Review Board.
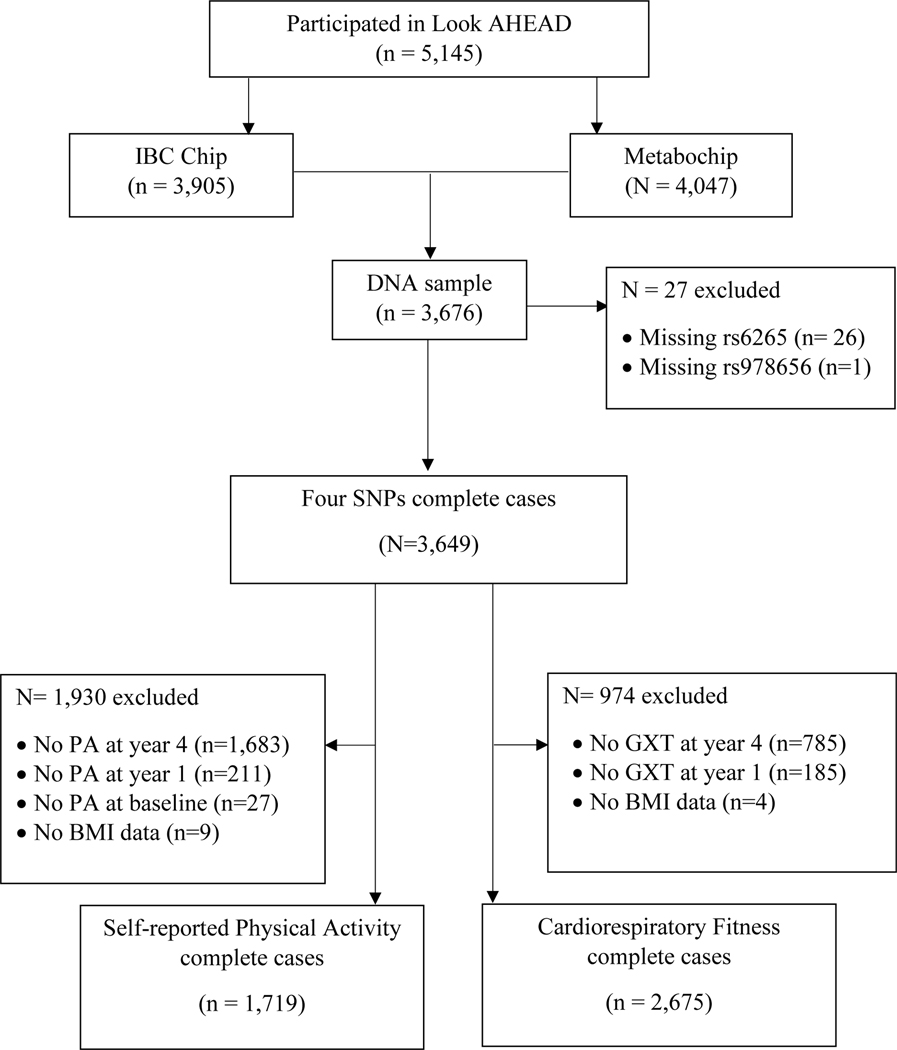
The flow diagram illustrating the study sample is presented in Figure 1. From 2001 to 2004, the Look AHEAD trial enrolled 5,145 ethnically diverse subjects with overweight and obesity as well as type 2 diabetes between the age of 45–76 years from 16 centers (24). Of these, 1,038 did not provide genetic consent to be included in a genetic ancillary study, including all participants from three Southwest American Indian sites, 10 withdrew consent for genotyping and 60 were identified to have no or a low concentration of DNA. This left 4,037 individuals, of which, 3,905 provided genetic samples that were characterized on the Illumina CARe iSelect (IBC) that passed genotyping quality control procedures, and 4,047 did so for the Metabochip. Together, 3,676 subjects contributed DNA samples on both the IBC and Metabochip, and 3,649 participants were successfully genotyped for all the four SNPs that were examined in the present study. Out of a total of 17 study sites, the Look AHEAD team collected self-reported PA data in eight study sites at baseline, year 1, and year 4. Using data from these 3,649 participants, two data sets were created for analysis: 1) the sample with complete data for both self-reported PA at baseline, year 1, year 4 and covariates (n=1,719), and 2) the sample with complete data for CRF at baseline, year 1, year 4, and covariates (n=2,675).
Figure 1.
Flow diagram of subject inclusion and exclusion
Main outcome measures
Self-reported PA was assessed using the Paffenbarger PA Questionnaire (25), which estimates weekly energy expenditure (kcal·week−1) from moderate intensity PA (e.g., climbing stairs, walking, and other fitness, sport, and recreational activities).
CRF was operationalized as the estimated metabolic equivalent (MET) level based on the treadmill workload (i.e., speed and grade) achieved at the point of termination of the graded exercise treadmill test. At baseline, a maximal graded exercise test was used and at years 1 and 4 a submaximal graded exercise test (i.e., 80%) was used. Additionally, at baseline, data were recorded on the MET value at 80% of maximum heart rate (=220-age), to correspond with data collected at follow-up time points. Participants were instructed not to exercise on their own before the graded exercise test and not to drink alcohol 24 hours prior to the testing.
Weight and height were measured by study staff and BMI calculated, and blood samples were collected for DNA extraction.
Genotyping
The IBC chip is a gene-centric 50,000-SNP array designed to assess potentially relevant loci across a range of cardiovascular, metabolic, and inflammatory syndromes (26). The Metabochip is a custom genotyping array that provides accurate genotyping of nearly 200,000 SNPs chosen based on GWAS meta-analyses of cardiovascular disease and diabetes risk traits (27). IBC chip genotyping was carried out at the Children’s Hospital of Philadelphia, and Cardio-Metabo chip genotyping was carried out at Center for Inherited Disease Research at Johns Hopkins University. On both platforms, the Look AHEAD study team excluded participants with failed genotyping, sex inconsistency, or familial relatedness (kinship coefficient > 0.025). SNP assays failing on an individual DNA sample indicates a poor-quality DNA sample. Thus, SNPs with a genotyping call rate less than 95% in any ethnic group (i.e., samples with low genotype frequency) were also excluded. After quality control procedures, the mean genotyping success rate per SNP was greater than 99.9%. Consistent with prior Look AHEAD publications (28) and data from other studies (27), SNPs not directly represented on either the IBC chip or the Cardio-Metabo chip were replaced by proxies (r2 ≥ 0.90) using phased genotype data from the 1000 Genomes Project and the SNP Annotation and Proxy Search tool (29) (see Table S1, Supplemental Digital Content).
SNP selection and Polygenic Score
The four SNPs examined in the proposed research are derived from two prior studies. The first SNP, rs6265, is identified from a randomized controlled PA promotion trial (21), in which an interaction between the rs6265 genotype and the efficacy of a PA promotion intervention was observed. Rest of the three SNPs are from the only previous GWAS that we were aware of when the present team obtained data from Look AHEAD (16). Specifically, De Moor, Liu (16) identified 37 SNPs clustered in three genomic regions (i.e., 2q33.1, 10q23.2, and 18p11.32), and we selected one SNP from each region: rs978656 near DNAPTP6, rs10887741 near PAPSS2, rs7279064 near C18orf2. Because two of the GWAS-identified SNPs were not in Cardio-Metabo chip or IBC chip, we examined proxy SNPs (see Table S1, Supplemental Digital Content), which were identified using the SNP Annotation and Proxy Search tool (29).
To examine cumulative effects of the 4 SNPs, we computed a PGS by adding the number of effect alleles that predispose individuals to be more physically active from four SNPs (G of rs978656, T of rs10887741, G of rs7279064, and A of rs6265) with weights equal to the published per-allele effects (see Table S1). To calculate per-allele-effects, odds ratios reported in the GWAS were used (i.e., > 4 metabolic equivalents-hours per week), in which a binary PA variable was used as the dependent variable. The genotype from each of the four SNPs were coded as 0, 1, or 2 in accordance with the number of effect alleles. Then, each SNP was weighted by its effect size based on the assumption that all SNPs have independent effects—and an additive effect of each allele within each SNP—on PA phenotypes. The highest possible value of the PGS is 10.8 (i.e., having two effect alleles for all four SNPs), while the lowest score is zero.
Statistical Analysis
To control for admixed study population, all IBC SNPs were examined by principal component analysis using the EIGENSTRAT algorithm (30) as implemented in Golden Helix version 7.1 (Bozeman, MT, USA). Principal component analysis results indicated that the majority of the variance among the multi-racial Look AHEAD cohort was accounted for by the first two principal components, which agreed with self-reported race/ethnicity in distinguishing Caucasians from African-Americans, and Hispanics from these other 2 groups (31).
Using a series of longitudinal linear-mixed models, we assessed the effect of PGS on PA phenotypes by treatment arm over time. Baseline PA phenotypes as well as treatment response (i.e., change in PA phenotypes from pre- to post-intervention) can be influenced by the SNPs. Therefore, baseline was modeled as the first time point in longitudinal analyses (32). In the model, we adjusted for baseline PA, time, treatment group, and genetic variation, as well as all one, two and three-way interactions between treatment, time and genetic variation. A Bonferroni correction was used to account for multiple comparisons of 10 tests, i.e., 5 (PGS + 4 SNPs) × 2 phenotypes. Thus, p-values below 0.005 (=0.05/10 tests) were considered statistically significant. Finally, Paffenbarger PA variables included some zero values (n=499), and since the linear-mixed models cannot include zero values as outcome variables, these zero were changed to one. We then performed sensitivity analyses excluding the 499 participants who reported zero for Paffenbarger PA.
Power analysis was computed a priori using G*Power (33). With 1,719 participants and 6 predictors (PGS, age, sex, BMI, study site, and population stratification) in the self-reported PA data set, there was 80% power to detect r2 of 0.00511 or 95% power to detect r2 of 0.007955. With 2,675 participants and 6 predictors (PGS, age, sex, BMI, study site, and population stratification) in the CRF data set, there was 80% power to detect r2 of 0.008 or 95% power to detect r2 of 0.0122.
We conducted several secondary analyses. A wealth of research on PA determinants has consistently shown that age is a strong determinant of PA (8, 34). Thus, to test potential effect modification by age in the association between PGS and PA phenotypes, we included PGS × age in the linear mixed models. We also examined each SNP individually × time × intervention, controlling for all covariates. All four SNPs were in separate chromosomes. Hardy Weinberg Equilibrium was determined using χ2 (Rodriguez, Gaunt, & Day, 2009). For analyses for individual SNPs (but not PGS), when minor allele frequency was below 20%, the minor allele homozygotes were combined with the heterozygotes type, resulting binary variables: dominant in effect allele A (e.g., AA/AB vs. BB) or recessive in effect allele A (e.g., AA vs. AB/BB). Except rs10887741, minor allele frequencies of all three SNPs (rs6265, rs7279064, and rs978656) were below 20%. Thus, these three SNPS were examined as binary variables: rs6265 (GG vs AA/GA), rs7279064 (TT vs GG/TG), and rs978656 (AA/GA vs GG). All analyses were adjusted for age, sex, BMI, study site, and population stratification. All analyses were performed in R statistical software (35). Longitudinal mixed effect models were performed using nlme package (36).
RESULTS
Descriptive Statistics
Participant characteristics are summarized in Table 1a and 2b. Individuals were evenly distributed between the intervention arms, with respect to age, sex, and ethnicity. Generally, compared to those who provide their self-reported PA data, those who did not were more frequently Hispanic (10.3% vs 3.8%; p<0.001), had lower BMI (35.95 ± 5.84 kg.m−2 vs. 36.47 ± 6.13 kg.m−2; p=0.01), and higher PGS (5.45 ± 1.64 vs. 5.30 ± 1.63; p=0.005). Compared to those who provided CRF, those who did not, had higher BMI (37.18 ± 6.43 kg.m−2 vs. 35.84 ± 5.77 kg.m−2; p<0.001), were older (59.98 ± 7.23 years vs. 58.81 ± 6.67; p<0.001), and were less frequently assigned to the treatment arm (47.1% vs 51.1%; p=0.037). The slightly higher PGS in ILI group came out statistically significant in Table 1b (n=2,675) but not Table 1a (n=1,719), which is likely due to the larger sample size. Genotype distribution of all studied SNPs in this population confirmed the HWE assumption (p>0.001, see Table S1, Supplemental Digital Content). Minor allele frequencies of all four SNPs are summarized in Table S1.
Table 1a.
Population Characteristics in Look AHEAD Genetic Sub-cohort for those completed Self-reported Physical Activity Questionnaire (n=1,719)
| Characteristic | Total (n=1,719) |
DSE (n=845) |
ILI (n=874) |
|---|---|---|---|
| Women (%) | 968 (56.3) | 482 (57.0) | 486 (55.6) |
| Ethnicity (%) | |||
| African American/Black | 314 (18.3) | 156 (18.5) | 158 (18.1) |
| American Indian/Alaskan native | 9 (0.5) | 3 (0.4) | 6 (0.7) |
| Asian/Pacific Islander | 6 (0.3) | 2 (0.2) | 4 (0.5) |
| White | 1,288 (74.9) | 621 (73.5) | 667 (76.3) |
| Other | 102 (6.0) | 63 (7.5) | 40 (4.4) |
| Hispanic (%) | 65 (3.8) | 42 (5.1) | 22 (2.5) |
| rs6265 near BDNF (%) | |||
| GG | 1,219 (70.9) | 593 (70.2) | 626 (71.6) |
| GA | 451 (26.2) | 232 (27.5) | 219 (25.1) |
| AA | 49 (2.9) | 20 (2.4) | 29 (3.3) |
| rs7279064 near C18orf2 (%) | |||
| TT | 853 (49.6) | 436 (51.6) | 417 (47.7) |
| TG | 695 (40.4) | 332 (39.3) | 363 (41.5) |
| GG | 171 (9.9) | 77 (9.1) | 94 (10.8) |
| rs978656 near DNAPTP6 (%) | |||
| AA | 58 (3.4) | 29 (3.4) | 29 (3.3) |
| GA | 452 (26.3) | 223 (26.4) | 229 (26.2) |
| GG | 1,209 (70.3) | 593 (70.2) | 616 (70.5) |
| rs10887741 near PAPSS2 (%) | |||
| CC | 237 (13.8) | 120 (14.2) | 117 (13.4) |
| TC | 760 (44.2) | 374 (44.3) | 386 (44.2) |
| TT | 722 (42.0) | 351 (41.5) | 371 (42.4) |
| Age (year) | 59.18 ± 6.85 | 59.14 ± 6.82 | 59.22 ± 6.87 |
| Body mass index (kg·m−2) | |||
| Baseline | 36.47 ± 6.13 | 36.46 ± 6.01 | 36.47 ± 6.25 |
| Year 1 | 34.64 ± 6.32 | 36.18 ± 6.10 | 33.15 ± 6.17 a |
| Year 4 | 35.31 ± 6.43 | 36.04 ± 6.37 | 34.80 ± 6.42 a |
| Self-reported Physical Activity (kcal·week−1) | |||
| Baseline | 867.42 ± 1169.982 | 859.77 ± 1244.72 | 874.81 ± 1093.19 |
| Year 1 | 1,388.70 ± 1573.15 | 986.99 ± 1419.66 | 1,777.09 ± 1616.19 a |
| Year 4 | 1,098.71 ± 1346.63 | 933.53 ± 1198.05 | 1,240.71 ± 1461.02 a |
| Polygenic Score of Physical Activity | 5.30 ± 1.63 | 5.25 ± 1.16 | 5.34 ± 1.65 |
Abbreviations: BMI, body mass index; DSE, diabetes support and education; ILI, intensive lifestyle intervention; PGS, polygenic score; DNAPTP6, DNA polymerase trans activated protein 6 gene; PAPSS2, 3’-phosphoadenosine 5’-phosphosulfate synthase 2 gene; C18orf2, chromosome 18 open reading frame 2 gene; and BDNF, Brain Derived Neurotropic Factor gene
p<0.001 (DSE vs ILI)
Table 2b.
Linear Mixed Effect Models: Cardiorespiratory Fitness regressed on Genetic Score and Individual Single Nucleotide Polymorphism
| ß | 95% CI | p-value | ||
|---|---|---|---|---|
| Total sample | ||||
| Model with no interaction term | PGS | 0.016 | −0.013 ~ 0.044 | 0.28 |
| Model with 2-way interaction term | PGS*intervention | −0.030 | −0.087 ~ 0.027 | 0.31 |
| Model with 3-way interaction term | PGS*intervention*year1 | −0.023 | −0.080 ~ 0.035 | 0.44 |
| PGS*intervention*year4 | 0.021 | −0.042 ~ 0.084 | 0.50 | |
|
| ||||
| Effect modification: age | ||||
| Model with 2-way interaction term | PGS*age | 0.004 | 0.00 ~ 0.008 | 0.05 |
| Model with 3-way interaction term | PGS*intervention*age | 0.002 | −0.006 ~ 0.010 | 0.63 |
| Model with 4-way interaction term | PGS*intervention*year1*age | 0.004 | −0.004 ~ 0.012 | 0.35 |
| PGS*intervention*year4*age | −0.003 | −0.013 ~ 0.005 | 0.48 | |
PGS, polygenic score; 1 unit increase in PGS roughly equates to one additional effect allele for higher physical activity. All models were controlled for age, sex, baseline body mass index, principal component 1 and principal component 2, and study sites as well as all subcomponents of the interaction terms (e.g., PGS*age included PGS and age separately).
Table 1b.
Population Characteristics in Look AHEAD Genetic Sub-cohort for those completed Cardiorespiratory Fitness test (n=2,675)
| Characteristic | Total (n=2,675) |
DSE (n=1,308) |
ILI (n=1,367) |
|---|---|---|---|
| Women (%) | 1480 (55.3) | 709 (54.2) | 771 (56.4) |
| Ethnicity (%) | |||
| African American/Black | 416 (15.6) | 198 (15.1) | 218 (15.9) |
| American Indian/Alaskan native | 10 (0.4) | 3 (0.2) | 7 (0.5) |
| Asian/Pacific Islander | 23 (0.9) | 9 (0.7) | 14 (1.0) |
| White | 1,989 (74.4) | 975 (74.5) | 1,014 (74.2) |
| Other | 237 (8.9) | 132 (9.4) | 114 (8.3) |
| Hispanic (%) | 186 (7.8) | 94 (7.2) | 92 (6.7) |
| rs6265 near BDNF (%) | |||
| GG | 1,889 (70.6) | 928 (70.9) | 961 (70.3) |
| GA | 704 (26.3) | 340 (26.0) | 364 (26.6) |
| AA | 82 (3.1) | 40 (3.1) | 42 (3.1) |
| rs7279064 near C18orf2 (%) | |||
| TT | 1,279 (47.8) | 652 (49.8) | 627 (45.9) |
| TG | 1,097 (41.0) | 522 (39.9) | 575 (42.1) |
| GG | 299 (11.2) | 134 (10.2) | 165 (12.1) |
| rs978656 near DNAPTP6 (%) | |||
| AA | 83 (3.1) | 42 (3.2) | 41 (3.0) |
| GA | 712 (26.6) | 346 (26.5) | 366 (26.8) |
| GG | 1,880 (70.3) | 920 (70.3) | 960 (70.2) |
| rs10887741 near PAPSS2 (%) | |||
| CC | 335 (12.5) | 176 (13.5) | 159 (11.6) |
| TC | 1,118 (44.4) | 577 (44.1) | 611 (44.7) |
| TT | 1,152 (43.1) | 555 (42.4) | 597 (43.7) |
| Age (year) | 58.81 ± 6.67 | 58.73 ± 6.59 | 58.89 ± 6.74 |
| Body mass index (kg·m−2) | |||
| Baseline | 35.84 ± 5.77 | 35.72 ± 5.57 | 35.94 ± 5.96 |
| Year 1 | 34.00 ± 5.96 | 35.42 ± 5.68 | 32.64 ± 5.92 a |
| Year 4 | 34.79 ± 6.10 | 35.32 ± 5.97 | 34.29 ± 6.19 a |
| Cardiorespiratory Fitness (MET) | |||
| Baseline | 5.21 ± 1.53 | 5.20 ± 1.55 | 5.21 ± 1.51 |
| Year 1 | 5.86 ± 1.87 | 5.45 ± 1.66 | 6.25 ± 1.97 a |
| Year 4 | 5.25 ± 1.70 | 5.09 ± 1.59 | 5.40 ± 1.77a |
| Polygenic Score of Physical Activity | 5.38 ± 1.64 | 5.31 ± 1.64 | 5.44 ± 1.65 b |
Abbreviations: BMI, body mass index; DSE, diabetes support and education; ILI, intensive lifestyle intervention; PGS, polygenic score; DNAPTP6, DNA polymerase trans activated protein 6 gene; PAPSS2, 3’-phosphoadenosine 5’-phosphosulfate synthase 2 gene; C18orf2, chromosome 18 open reading frame 2 gene; and BDNF, Brain Derived Neurotropic Factor gene
p<0.001 (DSE vs ILI)
p=0.042 (DSE vs ILI)
Genetic Associations with Self-Reported Physical Activity Questionnaire
PGS was not associated with baseline PA level in multivariate or bivariate analyses. The linear mixed models initially included two and three-way interactions between treatment, time, and PGS (PGS × intervention and PGS × intervention × time). However, the interaction terms were not statistically significant in any model (Table 2a), indicating that, in response to randomization to lifestyle intervention, PGS was not related to rate of change in PA level or PA level across time.
Table 2a.
Linear Mixed Effect Models: Self-reported Physical Activity regressed on Polygenic Score of Physical Activity
| ß | 95% CI | p-value | ||
|---|---|---|---|---|
| Total sample | ||||
| Model with no interaction term | PGS | −4 | −33 ~ 25 | 0.79 |
| Model with 2-way interaction term | PGS*intervention | −31 | −88 ~ 26 | 0.28 |
| Model with 3-way interaction term | PGS*intervention*year1 | −22 | −107 ~ 64 | 0.62 |
| PGS*intervention*year4 | −13 | −103 ~ 77 | 0.77 | |
|
| ||||
| Effect modification: age | ||||
| Model with 2-way interaction term | PGS*age | 4 | 0 ~ 8 | 0.05 |
| Model with 3-way interaction term | PGS*intervention*age | −4 | −12 ~ 5 | 0.39 |
| Model with 4-way interaction term | PGS*intervention*year1*age | 11 | −1 ~ 24 | 0.08 |
| PGS*intervention*year4*age | −2 | −14 ~ 12 | 0.81 | |
PGS, polygenic score; 1 unit increase in PGS roughly equates to one additional effect allele for higher physical activity. All models were controlled for age, sex, baseline body mass index, principal component 1 and principal component 2, and study sites as well as all subcomponents of the interaction terms (e.g., PGS*age included PGS and age separately).
In a model with main effects only, PGS was not associated with PA level across time (p=0.79). Further analysis indicated that age was a nearly significant moderator of the association between PGS and PA level (p-interaction=0.058). Specifically, among those age>60 years of age, a one unit increase in PGS was associated with expending an average of 24 kcal∙week−1 more in moderate intensity PA (p=0.34). Conversely, among those age≤60 years of age, a one unit increase in PGS was associated with an average of 19 kcal∙week−1 less in moderate intensity PA (p=0.28).
Models of self-reported PA indicated that the interaction terms for each individual SNP was not significant (p>0.1) (Table S2a, Supplemental Digital Content). For sensitivity analyses, we performed the same analyses among the subsample of participants who reported at least some PA at baseline, year 1, or year 4. That is, those who reported zero for self-reported PA (n=499) were removed, and the overall pattern remained the same.
Genetic Associations with Cardiorespiratory Fitness
PGS was not associated with baseline CRF in multivariate or bivariate analyses. The linear mixed models initially included two and three-way interactions between treatment, time, and PGS (PGS × intervention and PGS × intervention × time). However, the interaction terms were not statistically significant in any model (Table 2b), indicating that, in response to randomization to lifestyle intervention, PGS was not related to rate of change in CRF level or CRF level across time.
In a model with main effects only, CRF was not associated with PA level across time (p=0.28). However, similar to PA level, age appeared to modify the association between PGS and CRF (p-interaction=0.048). Specifically, among those age>55 years of age, a one unit increase in PGS was associated with 0.004 MET higher CRF (p=0.04). Conversely, among those age≤55 years of age, one unit increase in PGS was associated with 0.03 MET lower CRF (p=0.23).
In individual SNP analysis, there was a significant three-way interaction between rs978656, time and intervention (p=0.04). Thus, we stratified the data by the rs978656 SNP to see whether intervention effects differ by the genotype. Among A allele carriers of the rs978656 SNP (i.e., low PA prone group), intervention participants had significantly higher CRF at year 1 (b=1.14) and year 4 (b=.52) compared to participants in the control group. Among GG homozygotes of the rs978656 SNP (high PA prone group), intervention participants also had significantly higher CRF at year 1 (b=0.73) and year 4 (b=.31) compared to control, but with a lower magnitude of difference between the intervention versus control conditions. However, this interaction did not persist following Bonferroni correction (alpha<0.005). Figure S1a shows the CRF trajectory by intervention group among GG homozygotes (high PA prone group), and Figure S1b shows the same graph among the T allele carriers. The interaction terms for rest of the three SNP (SNP × intervention time) was not significant (p>0.1) (Table S2a, Supplemental Digital Content).
DISCUSSION
In a multi-center randomized controlled trial designed to test the health benefits of a lifestyle intervention among 2,675 participants with overweight/obesity and type 2 diabetes (ages 45–76), higher PGS was not related to higher rate of change in PA- or CRF-level, or maintaining higher PA- or CRF-level across time, in response to randomization to lifestyle intervention.
Age emerged as an effect modifier in the secondary analyses in both PA level and CRF. PA tends to decline as age increases (44), with a more progressive decline in PA after age 45–50 (5, 6). This phenomenon has a strong biological basis that extends to nonhumans (45), and empirical evidence suggests that, compared to younger adults, the PA levels of middle-aged adults are more strongly associated with environmental factors such as socioeconomic status (21) and randomization to PA promotion intervention (22). Combined with the present findings, genetic and social predispositions appear to manifest more saliently among those who are older, with the directions consistent with prior research (i.e., lower PA is related to lower socioeconomic status as well as lower PA-prone genetic factors). However, these post-hoc analyses should be considered as preliminary results for hypothesis generation, and future replications studies are needed.
In individual SNP analyses, one SNP near DNAPTP6 gene, rs978656, emerged as a moderator of the lifestyle intervention, such that CRF increase was higher in A-allele-carriers of rs978656 (low PA prone group) vs. GG homozygotes (high PA prone group). The biological functions of the proteins coded by DNAPTP6 gene—also known as spermatogenesis associated serine rich 2-like (SPATS2L)—are largely unknown. However, our finding is comparable with one of the recent GWAS of PA by Hara, Hachiya (19) who conducted two-stage genome-wide association analyses using discovery (n = 13,980) and replication (n = 2,036) samples among Japanese adults. The authors found that rs12612420 in DNAPTP6 (i.e., proxy SNP rs978656; Table S1) is associated with self-reported leisure time PA level. Of note, in present study, the rs978656 in DNAPTP6 was associated with CRF, but not PA level. Triangulating the observations from the present and prior studies, DNAPTP6 SNP may influence capacity to perform PA (e.g., CRF), which may in turn influence behavior (e.g., PA rate). However, these are based on speculation and future research examining the biological function of DNAPTP6 with respect to PA phenotypes is needed. The rest of the 3 SNPs examined (i.e., rs10887741-PAPSS2, rs7279064-C18orf2, and rs6265-BDNF) did not moderate the effect of lifestyle intervention on CRF or PA level. Although the SNPs we examined are selected from the only GWAS of PA that was available at the time of the present study, these SNPs were not robustly replicated in a subsequent well-powered GWAS of PA (e.g., UK biobank). Future studies incorporating the recently identified PA-related SNPs from the larger, more recent GWAS (17–20) warrant future investigation.
There are several limitations to the present study. Many participants were missing CRF data at follow-up. Since the major causes of missing CRF data at follow-up were inability or unwillingness to do the test (37), these data are not missing at random. The sample size is considered small compared to most GWAS in this post-genomic era. However, Look AHEAD is the largest genetic study that examined a deep phenotype, such as change in PA and CRF change in response to a randomization to a lifestyle intervention. Self-reported PA is susceptible to biases (36), is known to be unreliable unless restricted to salient re-callable activities (e.g., gyms visits), and there were no measures of reproducibility and validity regarding the Paffenbarger questionnaire in our sample. However, the Paffenbarger PA questionnaire is a well-validated and reliable method for assessing leisure-time PA. There are several strengths of the present study. This is the largest study to examine genetic predictors of PA level and CRF change over time. Moreover, there are methodological advantages to focusing on PA change in response to an intervention, as it constitutes a more fine-grained phenotype compared to naturally occurring PA behavior, a phenotype that is typically used in the GWAS approach. In addition, variations in environmental factors are significantly reduced when the outcome is PA change in response to lifestyle intervention. This is because the latter outcome controls access to a PA intervention. Finally, given that participants were recruited from 16 centers across the US, the present findings have strong generalizability for middle-aged adults with type 2 diabetes in the US.
We recommend future studies to take a similar approach with more SNPs. The low number of SNPs is determined by the progress of genomic research on PA behavior, which has been stagnant in the past decade. However, in this post-genomic era, genetic epidemiology research is evolving rapidly, with four GWAS being published in 2018 (17–20)—9 years after the first GWAS of PA came out (16) and a year after the present study received genotype data from Look AHEAD team. Advanced technology such as next-generation sequencing and wearable devices (e.g., accelerometer), along with federal enterprise such as NIH’s precision medicine initiative cohort (“All of Us”) and Millions Veteran Program, will likely accelerate the discoveries of SNPs that are associated with PA. With an enhanced number of SNPs, future PA promotion studies may be able to predict how genomic factors influence PA phenotypes (i.e., PA rate and CRF) over the years. To improve upon predicting PA level from increasing PA level, future studies examining deeper PA phenotypes—such as the trajectory of PA in a longitudinal cohort study, changes in PA phenotypes in response to an intervention, intermediate phenotypes (or endophenotypes)—are needed. Taken together, future studies incorporating the recently identified PA-related SNPs from larger GWAS (17–20) using various deeper PA phenotypes warrant future investigation.
CONCLUSION
In summary, an individual difference in PA phenotypes in response to a PA promotion intervention was not observed. The genetic predisposition to PA level of CRF may be moderated by age, which should be confirmed by future studies. Finally, future studies incorporating the recently identified PA-related SNPs from larger GWAS using various deeper PA phenotypes warrant future investigation.
Supplementary Material
Source of Funding:
Funded by the National Institutes of Health through cooperative agreements with the National Institute of Diabetes and Digestive and Kidney Diseases: DK57136, DK57149, DK56990, DK57177, DK57171, DK57151, DK57182, DK57131, DK57002, DK57078, DK57154, DK57178, DK57219, DK57008, DK57135, and DK56992. Additional funding was provided by the National Heart, Lung, and Blood Institute (F31 HL140817); National Institute of Nursing Research; National Center on Minority Health and Health Disparities; NIH Office of Research on Women’s Health; and the Centers for Disease Control and Prevention. This research was supported in part by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases. The Indian Health Service (I.H.S.) provided personnel, medical oversight, and use of facilities. The opinions expressed in this paper are those of the authors and do not necessarily reflect the views of the I.H.S. or other funding sources. Additional support was received from The Johns Hopkins Medical Institutions Bayview General Clinical Research Center (M01RR02719); the Massachusetts General Hospital Mallinckrodt General Clinical Research Center and the Massachusetts Institute of Technology General Clinical Research Center (M01RR01066); the Harvard Clinical and Translational Science Center (RR025758–04); the University of Colorado Health Sciences Center General Clinical Research Center (M01RR00051) and Clinical Nutrition Research Unit (P30 DK48520); the University of Tennessee at Memphis General Clinical Research Center (M01RR0021140); the University of Pittsburgh General Clinical Research Center (GCRC) (M01RR000056), the Clinical Translational Research Center (CTRC) funded by the Clinical & Translational Science Award (UL1 RR 024153) and NIH grant (DK 046204); the VA Puget Sound Health Care System Medical Research Service, Department of Veterans Affairs; and the Frederic C. Bartter General Clinical Research Center (M01RR01346). The following organizations have committed to make major contributions to Look AHEAD: FedEx Corporation; Health Management Resources; LifeScan, Inc., a Johnson & Johnson Company; OPTIFAST® of Nestle HealthCare Nutrition, Inc.; Hoffmann-La Roche Inc.; Abbott Nutrition; and Slim-Fast Brand of Unilever North America. Some of the information contained herein was derived from data provided by the Bureau of Vital Statistics, New York City Department of Health and Mental Hygiene.
Abbreviations:
- GWAS
genome-wide association study
- SNP
single nucleotide polymorphism
- PA
physical activity
- CRF
cardiorespiratory fitness
- PGS
polygenic score
- Look AHEAD
Action for Health in Diabetes
- DSE
diabetes support and education
- ILI
intensive lifestyle intervention
- BMI
body mass index
Footnotes
After receiving relevant genotype data from Look AHEAD team, four additional GWAS of PA came out from UK Biobank (17, 18), Japan Multi-Institutional Collaborative Cohort (19), as well as aggregation of Framingham Heart Study, Women’s Health Initiative and Jackson Heart Study (20), which were not included in this study.
3.7 References
- 1.Horstmann DM. Acute poliomyelitis: relation of physical activity at the time of onset to the course of the disease. Journal of the American Medical Association. 1950;142(4):236–41. [DOI] [PubMed] [Google Scholar]
- 2.Wennesland R, Brown E, Hopper J Jr, Hodges J Jr, Guttentag O, Scott K, Tucker IN, Bradley B. Red cell, plasma and blood volume in healthy men measured by radiochromium (Cr51) cell tagging and hematocrit: influence of age, somatotype and habits of physical activity on the variance after regression of volumes to height and weight combined. Journal of Clinical Investigation. 1959;38(7):1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garber CE, Blissmer B, Deschenes MR, Franklin BA, Lamonte MJ, Lee IM, Nieman DC, Swain DP. American College of Sports Medicine position stand. Quantity and quality of exercise for developing and maintaining cardiorespiratory, musculoskeletal, and neuromotor fitness in apparently healthy adults: guidance for prescribing exercise. Med Sci Sports Exerc. 2011;43:1334–59. [DOI] [PubMed] [Google Scholar]
- 4.Harris CD, Watson KB, Carlson SA, Fulton JE, Dorn JM, Elam-Evans L. Adult participation in aerobic and muscle-strengthening physical activities--United States, 2011. Morbidity and Mortality Weekly Report. 2013;62(17):326–30. [PMC free article] [PubMed] [Google Scholar]
- 5.Troiano RP, Berrigan D, Dodd KW, Masse LC, Tilert T, McDowell M. Physical activity in the United States measured by accelerometer. Medicine and science in sports and exercise. 2008;40(1):181. [DOI] [PubMed] [Google Scholar]
- 6.Luke A, Dugas LR, Durazo-Arvizu RA, Cao G, Cooper RS. Assessing physical activity and its relationship to cardiovascular risk factors: NHANES 2003–2006. BMC public health. 2011;11(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evenson KR, Wilcox S, Pettinger M, Brunner R, King AC, McTiernan A. Vigorous leisure activity through women’s adult life: the Women’s Health Initiative Observational Cohort Study. American Journal of Epidemiology. 2002;156(10):945–53. [DOI] [PubMed] [Google Scholar]
- 8.Bauman AE, Reis RS, Sallis JF, Wells JC, Loos RJ, Martin BW, and Lancet Physical Activity Series Working Group. Correlates of physical activity: why are some people physically active and others not? Lancet. 2012;380:258–71. [DOI] [PubMed] [Google Scholar]
- 9.Sallis JF, Bowles HR, Bauman A, Ainsworth BE, Bull FC, Craig CL, Sjöström M, De Bourdeaudhuij I, Lefevre J, Matsudo V, Matsudo S. Neighborhood environments and physical activity among adults in 11 countries. American journal of preventive medicine. 2009;36(6):484–90. [DOI] [PubMed] [Google Scholar]
- 10.Fisher A, van Jaarsveld CH, Llewellyn CH, Wardle J. Environmental influences on children’s physical activity: quantitative estimates using a twin design. PloS one. 2010;5:e10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlsson S, Andersson T, Lichtenstein P, Michaelsson K, Ahlbom A. Genetic effects on physical activity: results from the Swedish Twin Registry. MedSciSports Exerc. 2006;38:1396–401. [DOI] [PubMed] [Google Scholar]
- 12.Eriksson M, Rasmussen F, Tynelius P. Genetic factors in physical activity and the equal environment assumption-- the Swedish young male twins study. Behavior genetics. 2006;36:238–47. [DOI] [PubMed] [Google Scholar]
- 13.Joosen AM, Gielen M, Vlietinck R, Westerterp KR. Genetic analysis of physical activity in twins. The American Journal of Clinical Nutrition. 2005;82:1253–9. [DOI] [PubMed] [Google Scholar]
- 14.Aaltonen S, Ortega-Alonso A, Kujala UM, Kaprio J. A longitudinal study on genetic and environmental influences on leisure time physical activity in the Finnish Twin Cohort. Twin research and human genetics : the official journal of the International Society for Twin Studies. 2010;13:475–81. [DOI] [PubMed] [Google Scholar]
- 15.Stubbe JH, Boomsma DI, Vink JM, Cornes BK, Martin NG, Skytthe A, Kyvik KO, Rose RJ, Kujala UM, Kaprio J, Harris JR. Genetic influences on exercise participation in 37.051 twin pairs from seven countries. PloS one. 2006;1(1):e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Moor MH, Liu Y-J, Boomsma DI, Li J, Hamilton JJ, Hottenga J-J, Levy S, Liu XG, Pei YF, Posthuma D, Recker RR. Genome-wide association study of exercise behavior in Dutch and American adults. Medicine and science in sports and exercise. 2009;41(10):1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klimentidis YC, Raichlen DA, Bea J, Garcia DO, Wineinger NE, Mandarino LJ, Alexander GE, Chen Z, Going SB. Genome-wide association study of habitual physical activity in over 377,000 UK Biobank participants identifies multiple variants including CADM2 and APOE. Int J Obes. 2018;42(6):1161–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doherty A, Smith-Byrne K, Ferreira T, Holmes MV, Holmes C, Pulit SL, Lindgren CM. GWAS identifies 14 loci for device-measured physical activity and sleep duration. Nature communications. 2018;9(1):5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hara M, Hachiya T, Sutoh Y, Matsuo K, Nishida Y, Shimanoe C, Tanaka K, Shimizu A, Ohnaka K, Kawaguchi T, Oze I. Genomewide association study of leisure-time exercise behavior in Japanese adults. Medicine and science in sports and exercise. 2018;50(12):2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin X, Chan KK-h, Huang Y-T, Luo X, Liang L, Wilson J, Correa A, Levy D, Liu S. Genetic Determinants for Leisure-Time Physical Activity. Medicine and science in sports and exercise. 2018;50(8):1620–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryan AD, Magnan RE, Hooper AE, Ciccolo JT, Marcus B, Hutchison KE. Colorado stride (COSTRIDE): testing genetic and physiological moderators of response to an intervention to increase physical activity. International Journal of Behavioral Nutrition and Physical Activity. 2013;10(1):139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ioannidis JP, Khoury MJ. Improving validation practices in “omics” research. Science. 2011;334(6060):1230–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bray GA. Baseline characteristics of the randomised cohort from the Look AHEAD (Action for Health in Diabetes) study. Diabetes and Vascular Disease Research. 2006;3(3):202–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan DH, Espeland MA, Foster GD, Haffner SM, Hubbard VS, Johnson KC, Steven EK, William CK, Susan ZY. Look AHEAD (Action for Health in Diabetes): design and methods for a clinical trial of weight loss for the prevention of cardiovascular disease in type 2 diabetes. Controlled clinical trials. 2003;24(5):610–28. [DOI] [PubMed] [Google Scholar]
- 25.Paffenbarger RS Jr, Wing AL, Hyde RT. Physical activity as an index of heart attack risk in college alumni. American Journal of Epidemiology. 1978;108:161–75. [DOI] [PubMed] [Google Scholar]
- 26.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, Chandrupatla HR. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PloS one. 2008;3:e3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, Burtt NP, Fuchsberger C, Li Y, Erdmann J, Frayling TM. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS genetics. 2012;8(8):e1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCaffery JM, Papandonatos GD, Peter I, Huggins GS, Raynor HA, Delahanty LM, Cheskin LJ, Balasubramanyam A, Wagenknecht LE, Wing RR. Obesity susceptibility loci and dietary intake in the Look AHEAD Trial. American Journal of Clinical Nutrition. 2012;95:1477–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’donnell CJ, De Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38(8):904–9. [DOI] [PubMed] [Google Scholar]
- 31.McCaffery J, Papandonatos GD, Huggins GS, Peter I, Kahn SE, Knowler WC, Hudnall GE, Lipkin EW, Kitabchi AE, Wagenknecht LE, Wing RR. FTO predicts weight regain in the Look AHEAD clinical trial. International journal of obesity. 2013;37(12):1545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McArdle P, Whitcomb B. Improper adjustment for baseline in genetic association studies of change in phenotype. Human heredity. 2008;67(3):176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faul F, Erdfelder E, Lang A-G, Buchner AG. Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behavior research methods. 2007;39(2):175–91. [DOI] [PubMed] [Google Scholar]
- 34.Trost SG, Owen N, Bauman AE, Sallis JF, Brown W. Correlates of adults’ participation in physical activity: review and update. Medicine & science in sports & exercise. 2002;34(12):1996–2001. [DOI] [PubMed] [Google Scholar]
- 35.Team RC. R: A Language and environment for statistical computing, R software version 3.3. 1. 2016.
- 36.Pinheiro J, Bates D, DebRoy S, Sarkar D, Heisterkamp S, Van Willigen B, Maintainer R. Package ‘nlme’. Linear and Nonlinear Mixed Effects Models, version. 2017:3–1.
- 37.Jakicic JM, Egan CM, Fabricatore AN, Gaussoin SA, Glasser SP, Hesson LA, Knowler WC, Lang W, Regensteiner JG, Ribisl PM, Ryan DH. Four-year change in cardiorespiratory fitness and influence on glycemic control in adults with type 2 diabetes in a randomized trial: the Look AHEAD Trial. Diabetes care. 2013;36(5):1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulhus DL. Measurement and control of response bias. In: Robinson JP, Shaver PR, Wrightsman LS, editors. Measures of personality and social psychological attitudes. San Diego, CA: Academic Press; 1991. p. 17–59. [Google Scholar]
- 39.Albanes D, Conway JM, Taylor PR, Moe PW, Judd J. Validation and comparison of eight physical activity questionnaires. Epidemiology (Cambridge, Mass). 1990;1:65–71. [DOI] [PubMed] [Google Scholar]
- 40.Cust AE, Smith BJ, Chau J, van der Ploeg HP, Friedenreich CM, Armstrong BK, Bauman A. Validity and repeatability of the EPIC physical activity questionnaire: a validation study using accelerometers as an objective measure. International Journal of Behavioral Nutrition and Physical Activity. 2008;5(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.