Abstract
The plant hormone abscisic acid (ABA) plays a central role in the regulation of seed maturation and dormancy. ABA also restrains germination under abiotic-stress conditions. Here, we show in tomato (Solanum lycopersicum) that the ABA importer ABA-IMPORTING TRANSPORTER 1.1 (AIT1.1/NPF4.6) has a role in radicle emergence under salinity conditions. AIT1.1 expression was upregulated following seed imbibition, and CRISPR/Cas9-derived ait1.1 mutants exhibited faster radicle emergence, increased germination and partial resistance to ABA. AIT1.1 was highly expressed in the endosperm, but not in the embryo, and ait1.1 isolated embryos did not show resistance to ABA. On the other hand, loss of AIT1.1 activity promoted the expression of endosperm-weakening-related genes, and seed-coat scarification eliminated the promoting effect of ait1.1 on radicle emergence. Therefore, we propose that imbibition-induced AIT1.1 expression in the micropylar endosperm mediates ABA-uptake into micropylar cells to restrain endosperm weakening. While salinity conditions strongly inhibited wild-type M82 seed germination, high salinity had a much weaker effect on ait1.1 germination. We suggest that AIT1.1 evolved to inhibit germination under unfavorable conditions, such as salinity. Unlike other ABA mutants, ait1.1 exhibited normal seed longevity, and therefore, the ait1.1 allele may be exploited to improve seed germination in crops.
The tomato ABA-IMPORTING TRANSPORTER 1.1 gene is induced in the endosperm following seed imbibition to restrain endosperm weakening and germination under high salinity conditions.
Introduction
The timing of seed germination is important for seedling establishment (Donohue et al., 2010), and due to its irreversible nature, this process is highly regulated. Germination is regulated by environmental and internal cues including light, temperature, water availability, and hormones. The plant hormones gibberellin (GA) and abscisic acid (ABA) promote and inhibit germination, respectively (Nonogaki, 2014).
ABA accumulates during seed maturation and induces seed dormancy (Finch-Savage and Leubner-Metzger, 2006). ABA-induced seed dormancy is orchestrated by dormancy-related genes such as ABSCISIC ACID INSENSITIVE 3 (ABI3), FUSCA 3 (FUS3), DELAY OF GERMINATION1 (DOG1), and LEAFY COTYLEDON 1 (LEC1) and LEC2 (Holdsworth et al., 2008). The output of these regulatory machinery promotes storage compound accumulation, inhibits viviparous germination, reduces metabolic activity, and leads to the acquisition of desiccation tolerance, enabling embryo survival under long periods of dry environment (Graeber et al., 2012).
During the early stages of seed imbibition, dormancy is released due to ABA catabolism, which precedes the accumulation of GA (Toyomasu et al., 1998; Ogawa et al., 2003; Nambara and Marion-Poll, 2005; Raghavendra et al., 2010). These increase the GA to ABA ratio and promote seed germination (Piskurewicz et al., 2008; Liu and Hou, 2018, Shu et al., 2018). In dicot seeds, the high GA to ABA ratio induces the accumulation of cell-wall-loosening enzymes, leading to endosperm and seed coat (testa) weakening (Muller et al., 2006). GA also promotes radicle elongation, and the growing radicle ruptures the weakened endosperm and testa, leading to radicle emergence and germination (Steinbrecher and Leubner-Metzger, 2018). Unfavorable conditions, such as salinity, inhibiting ABA catabolism, reducing GA to ABA ratio, and inhibiting endosperm weakening, embryo growth, and germination (Shu et al., 2017; Ruggiero et al., 2019; Lai et al., 2020).
ABA activity is regulated at the level of metabolism and signaling, but also by its transport to target tissues and cells (Anfang and Shani, 2021). The earliest characterized ABA transporters were the ATP-BINDING CASSETTE (ABC) transporters ABCG25 (Kuromori et al., 2010) and ABCG40 (Kang et al., 2010). ABCG25 functions as an exporter, and its loss of function resulted in reduced germination on ABA, while the loss of ABCG40, which is an ABA importer, exhibited increased germination in the presence of ABA. The integrated activity of ABCG ABA importers and exporters in the control of seed germination was demonstrated in Arabidopsis (Arabidopsis thaliana) and Medicago (Medicago truncatula) (Kang et al., 2015; Pawela et al., 2019). It was recently shown that the Arabidopsis ABA importers ABCG17 and ABCG18 redundantly mediate stomatal aperture and shoot-to-root ABA translocation to regulate lateral root emergence (Zhang et al., 2021a, 2021b). Another ABA transporter called Detoxification Efflux Carriers (DTX)/Multidrug and Toxic Compound Extrusion (MATE) DTX50 is upregulated by ABA and expressed mainly in guard cells and vascular tissue (Zhang et al., 2014). The dtx50 mutant plants exhibited hypersensitivity to ABA during seed germination and increased tolerance to drought. Altogether these studies suggest an important role for ABA transport in key physiological and developmental processes in plants (Kuromori et al., 2018).
An additional ABA transporter from the NITRATE TRANSPORTER1 (NRT1)/PTR TRANSPORTER FAMILY (NPF) named ABA-IMPORTING TRANSPORTER 1 (AIT1), NRT1.2, or NPF4.6 according to the nomencalture suggested by the community (Léran et al., 2014) 2014 was characterized in Arabidopsis (Kanno et al., 2012). AIT1 is expressed mainly in vascular tissue of the inflorescence stem, but it is also expressed in leaf vascular tissue and guard cells. The ait1 mutant exhibits increased transpiration due to larger stomatal aperture and a partial resistance to ABA (Shimizu et al., 2021). Three AIT1 homologs were found in Arabidopsis; AIT2, which exhibits reduced ABA transport activity compared with AIT1 (Kanno et al., 2012), AIT3, which exhibits rather high ABA transport activity (Kanno et al., 2012), but in contrast to AIT1, it also transports jasmonoyl-isoleucine and GA (Chiba et al., 2015), and AIT4, which has a weak ABA transport activity but transports also GA (Kanno et al., 2012; Chiba et al., 2015). The tomato (Solanum lycopersicum) AIT1 putative ortholog was recently characterized (AIT1.1, Solyc05g006990; Shohat et al., 2020). A CRISPR/Cas9-derived ait1.1 mutant exhibited increased transpiration and reduced stomatal closure in response to ABA.
Tomato seeds exhibit primary non-deep physiological dormancy and can be stored dry for years (Hilhorst and Downie, 1995; Baskin and Baskin, 2004). The tomato embryo is enclosed in a rigid endosperm and testa, which act as a physical barrier to restrict radicle emergence (Yan et al., 2014). During seed imbibition, the endosperm at the radicle tip region (i.e. micropylar endosperm) is weakened due to GA-induced cell-wall-loosening enzyme activity, enabling radicle emergence (Nonogaki et al., 2000). Here, we show that the tomato AIT1.1 is upregulated following seed imbibition. ait1.1 mutant seeds exhibited improved germination under normal and salinity conditions. Our results suggest that AIT1.1 acts in the endosperm following imbibition to mediate ABA-suppression of endosperm weakening and germination under normal and salinity conditions.
Results
AIT1.1 is upregulated during tomato seed imbibition to restrain germination
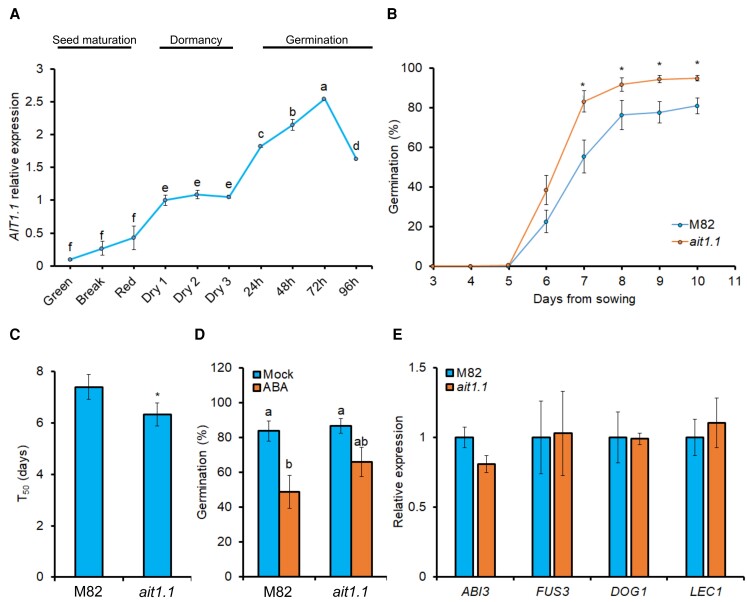
ABA accumulates during seed maturation and promotes seed dormancy, whereas following imbibition, it is catabolized to enable germination (Braybrook and Harada, 2008). To study the possible role of the ABA transporter AIT1.1 in seed maturation, dormancy and/or germination, we first examined its expression in wild-type (WT) M82 seeds during different developmental and physiological stages. We collected seeds from green, color break and red ripped tomato fruits, dry seeds 1 day, 2 weeks, and 4 weeks after extraction from red ripen fruits and at different times following seed imbibition (0–96 h). AIT1.1 expression was low during seed maturation and slightly upregulated in dry seeds (Figure 1A). It was strongly upregulated following imbibition (up to 72 h), and after radicle emergence (96 h), its expression was downregulated. These results imply that AIT1.1 has a role following seed imbibition, perhaps to restrain germination. To verify that this expression dynamic is reliable, we used additional reference housekeeping genes acting in tomato seeds (Dekkers et al., 2012) and all showed similar results (Supplemental Figure 1A).
Figure 1.
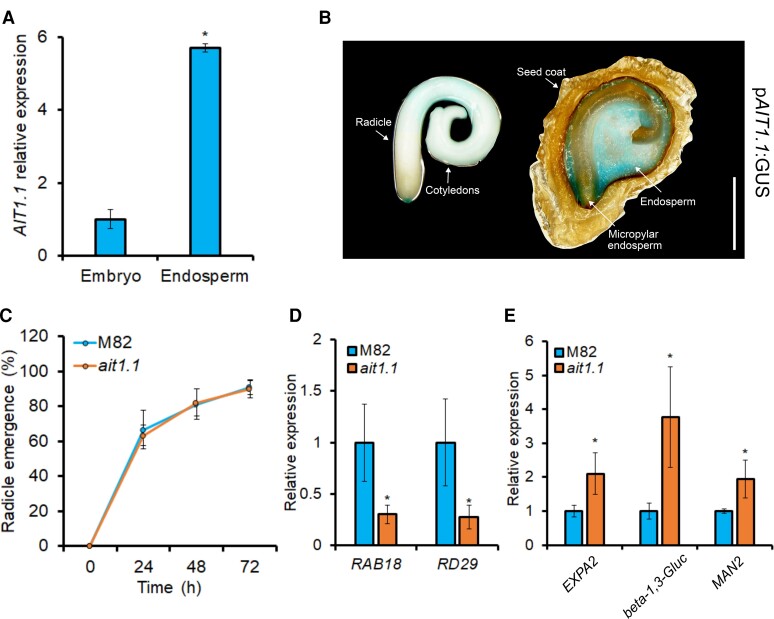
AIT1.1 is upregulated following seed imbibition and restrains germination. (A) Relative expression of AIT1.1 in seeds taken from green, color break and red ripped tomato fruits, as well as, from fresh dry seeds for 1 day (dry 1), 2 weeks (dry 2), or 4 weeks (dry 3) after extraction from red ripen fruits, and imbibed seeds (0–96 h imbibition after 4 weeks of dry storage). (B) Germination of M82 and ait1.1 seeds in soil. (C) Germination rate (T50) of M82 and ait1.1. (D) Percentages of germinated seeds after 10 days in petri dishes containing MS medium with (or without) 1 µM abscisic acid (ABA). (E) Relative expression of central regulators of seed dormancy genes ABI3, FUS3, DOG1, and LEC1 in M82 and ait1.1 dry seeds stored for 1 month. Values in A and E are means of 4 replicates, each contains 10 seeds ± Se. Values in B–D are means of four independent replicates each contained 100 (B, C) or 50 (D) seeds ± Se. Small letters in A and stars in B and C represent significant differences between respective treatments by Student's t test (P < 0.05). Small letters in D represent significant differences between respective treatments by Tukey–Kramer HSD test (P < 0.05).
We next examined whether the loss of AIT1.1 activity in ait1.1 CRISPR/Cas9-derived mutant (Shohat et al., 2020) affects germination. Ten days after sowing in the soil, the mutant seeds reached 95% germination, whereas M82 seeds reached only 81% (Figure 1B). Increased germination was also found in another independent allele, ait1.1#2 (Supplemental Figures 2A and 3A). Germination rate can be determined by the time (days) it takes to reach 50% germination (T50, Coolbear et al., 1984). T50 of ait1.1 was 6.3, whereas that of M82 was 7.4 (Figure 1C). In a germination assay on MS media containing ABA, seeds of the two independent alleles of ait1.1 exhibited partial resistance to the hormone and germinated at higher percentage than M82 (Figure 1D, Supplemental Figure 3B).
We tested whether ait1.1 also affects dormancy by analyzing the transcriptional profile of the central regulators of seed dormancy ABI3, FUS3, DOG1, and LEC1. Loss of AIT1.1 activity had no effect on the expression of these genes (Figure 1E, Supplemental Figure 1, C–E). It was shown before that reduced ABA accumulation or activity decreases seed desiccation tolerance and, therefore, longevity (Ooms et al., 1993). We tested the germination of M82 and ait1.1 seeds that were stored for 1 or 18 months. The loss of AIT1.1 did not affect the longevity of the seeds, and they exhibited normal germination after the long storage, suggesting that AIT1.1 has no role in seed desiccation tolerance (Supplemental Figure 4). On the other hand, seeds of the ABA-deficient sitiens mutant (sit, Groot and Karssen, 1992) exhibited reduced germination after long storage (15 months, Supplemental Figure 5). Taken together, the results suggest that AIT1.1 does not affect dormancy or desiccation tolerance but probably postimbibition-related processes.
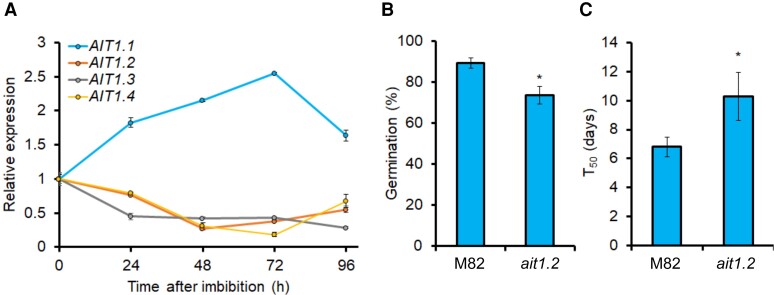
Tomato has three uncharacterized AIT1.1 paralogs (Shohat et al., 2020). Since the sequences of all three proteins show higher similarity to the Arabidopsis AIT1 compared with AIT2/AIT3/AIT4, we named them AIT1.2 (Solyc05g007000), AIT1.3 (Solyc04g005790), and AIT1.4 (Solyc03g113250) (Shohat et al., 2020). We examined their expression pattern following imbibition to uncover possible functional redundancy. While AIT1.1 was upregulated following imbibition, AIT1.2, AIT1.3, and AIT1.4 were downregulated (Figure 2A), suggesting that AIT1.1 has a unique role in tomato seeds. The expression of AIT1.2/3/4 was not affected in the ait1.1 mutant seeds (Supplemental Figure 6). Since AIT1.2 is the closest homolog of AIT1.1 (86% identity of the amino acid sequences), we generated CRISPR/Cas9-derived ait1.2 mutant. The ait1.2 mutant gene had 5 bp deletion, causing a frame shift and a premature stop codon (Supplemental Figure 2B). Homozygous mutant plants did not show any clear phenotypic alteration. We performed germination assay and found that ait1.2 seeds exhibit reduced germination compared with M82 (Figure 2, B and C). It should be noted that these results are based on the analysis of a single allele and, therefore, should be taken with caution.
Figure 2.
Expression dynamics of the tomato AIT1 genes following seed imbibition. A, Relative expression of the tomato AIT1 genes in seeds following imbibition (0–96 h imbibition following 4 weeks of dry storage). Values are means of four replicates each containing 10 seeds ± Se. (B, C) Germination (B) and germination rate (C) of M82 and ait1.2 seeds in soil after 10 days. Values are means of four replicates (plates) each contained 70 seeds ± Se. Stars in B and C represent significant differences between respective treatments by Student's t test (P < 0.05).
AIT1.1 acts in the endosperm to mediate seed germination
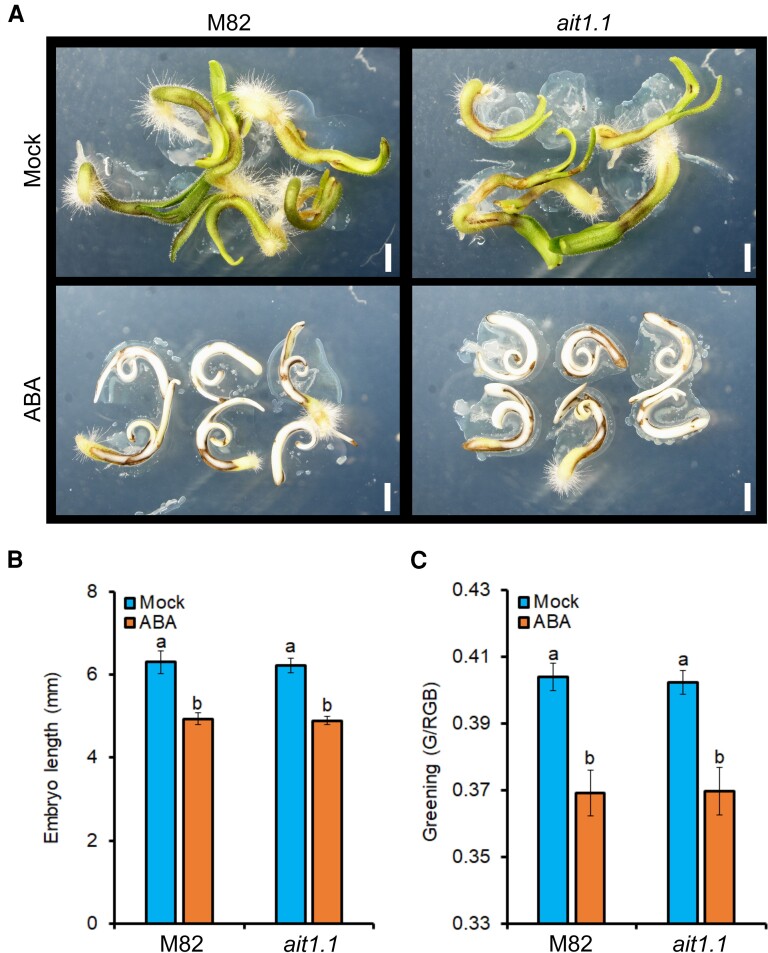
Tomato seed germination is controlled by two parallel processes: embryo growth and endosperm weakening (Steinbrecher and Leubner-Metzger, 2017). We first tested whether AIT1.1 acts in the embryo to mediate seed germination. To this aim, we imbibed M82 and ait1.1 seeds for several hours, then rescued the embryos from the seeds and grew them for 72 h on MS medium with or without ABA (Figure 3A). We analyzed two ABA-inhibited developmental processes: embryo growth and greening. We did not find differences in growth and greening between M82 and ait1.1 embryos on MS with or without ABA (Figure 3, B and C), suggesting that AIT1.1 does not affect germination via its activity in the embryo.
Figure 3.
AIT1.1 did not affect embryo growth. A, Representative isolated embryos grown for 72 h on petri dishes containing MS medium with or without 10 µM abscisic acid (ABA). B, Length of M82 and ait1.1 isolated embryos grown for 72 h on Petri dishes containing MS medium with (or without) 10 µM ABA. C, Greening of M82 and ait1.1 isolated embryos grown for 72 h on Petri dishes containing MS medium with (or without) 10 µM ABA. Values in B and C are means of 10 embyos ± Se. Small letters in B and C represent significant differences between respective treatments by Tukey–Kramer HSD test (P < 0.05). Scale bar in A = 1 mm.
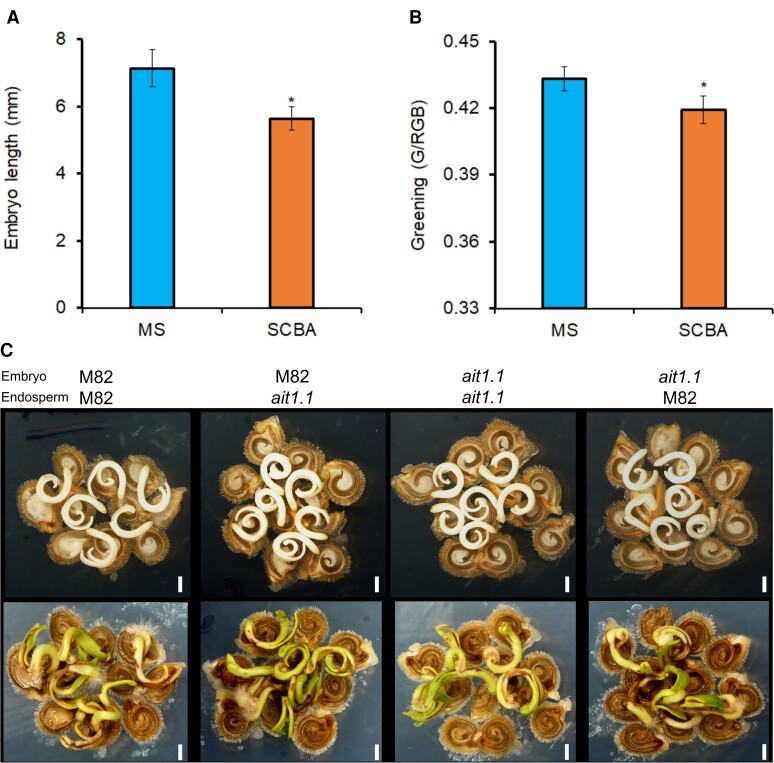
The Arabidopsis ABCG ABA transporters controls ABA transport from the endosperm to the embryo to regulate embryo growth and germination (Kang et al., 2015). To test whether AIT1.1 has a similar role in tomato, we have used the Seed Coat Bedding Assay (SCBA, Lee et al., 2010) that tests the inhibition of embryo growth by endosperm-synthesized ABA. We found that isolated M82 embryos taken from imbibed seeds, developed faster on MS medium compared with embryos that were placed on a layer of embryo-less seed coats and endosperms (Figure 4, A and B). However, we did not find differences in embryo growth and greening between M82 and ait1.1 in reciprocal SCBA assays (Figure 4C, Supplemental Figure 7), suggesting that the increased germination in ait1.1 is probably not a result of inhibition of ABA transport from the endosperm to the embryo.
Figure 4.
AIT1.1 did not affect endosperm inhibition of embryo growth. A, Length of M82 isolated embryos grown for 72 h on Murashige and Skoog (MS) medium or on a layer of endosperm containing seed coat (seed coat bedding assay, SCBA). B, Greening of M82 isolated embryos grown for 72 h on MS medium or on a layer of endosperm containing seed coat (SCBA). C, Representative M82 and ait1.1 reciprocal SCBA assays grown for 72 h on Petri dishes containing MS medium. Values in A and B are means of five biological replicates ±Se. Stars in A and B represent significant differences between respective treatments by Student's t test (P < 0.05). Scale bar = 1 mm.
AIT1.1 expression was significantly higher in the endosperm compared with the embryo (Figure 5A, Supplemental Figure 1B). Moreover, GUS activity in transgenic pAIT1.1:GUS (Shohat et al., 2020) imbibed seeds was substantially higher in the endosperm compared with the embryo (Figure 5B). We, therefore, tested whether the increased germination of ait1.1 seeds is a result of reduced endosperm weakening. We first scarified the seed at the micropylar endosperm region to eliminate the inhibitory effect of the endosperm and the testa on radicle emergence (Liptay and Schopfer, 1983). Following scarification, we did not find differences between M82 and ait1.1 in the time of radicle emergence or in postemergence primary root growth with or without ABA (Figure 5C, Supplemental Figure 8). We then analyzed the expression of the ABA-responsive genes RAB18 and RD29 (Nir et al., 2017) in the micropylar endosperms of M82 and ait1.1, and both genes exhibited lower expression in the mutant, suggesting reduced ABA activity in this site (Figure 5D). We also analyzed the expression of endosperm-weakening-related genes EXPANSIN2 (EXPA2), beta-1,3-Glucanase, and ENDO-BETA-MANNASE 2 (MAN2) following imbibition (Leubner-Metzger et al., 1995; Martınez-Andujar et al., 2012; Graeber et al., 2014). These genes exhibited higher expression in ait1.1 endosperm, suggesting higher endosperm weakening activity in the mutant (Figure 5E).
Figure 5.
AIT1.1 acts in the endosperm to restrain radicle emergence. (A) Relative expression of AIT1.1 in isolated embryos or endosperms after 48 h of imbibition. (B) Representative pAIT1.1:GUS transgenic embryo and embryo-less endosperm, showing GUS staining after 48 h of imbibition. The images were digitally extracted for comparison. (C) Percentages of radicle emergence in M82 and ait1.1 seeds following scarification. (D) Relative expression of ABA-responsive genes RAB18 and RD29 in micropylar endosperms of M82 and ait1.1 48 h after the beginning of imbibition. E, Relative expression of endosperm weakening marker genes EXPA2, beta-1,3-Glucanase and MAN2 in endosperms of M82 and ait1.1 after 48 h of imbibition. Values in A, D, and E are means of four replicates each containing 10 seeds ±Se. Values in C are means of three replicates each containing 30 seeds ±Se. Stars in A, D, and E represent significant differences between respective treatments by Student's t test (P < 0.05). Scale bar in B = 1 mm.
AIT1.1 restrained germination under salinity conditions
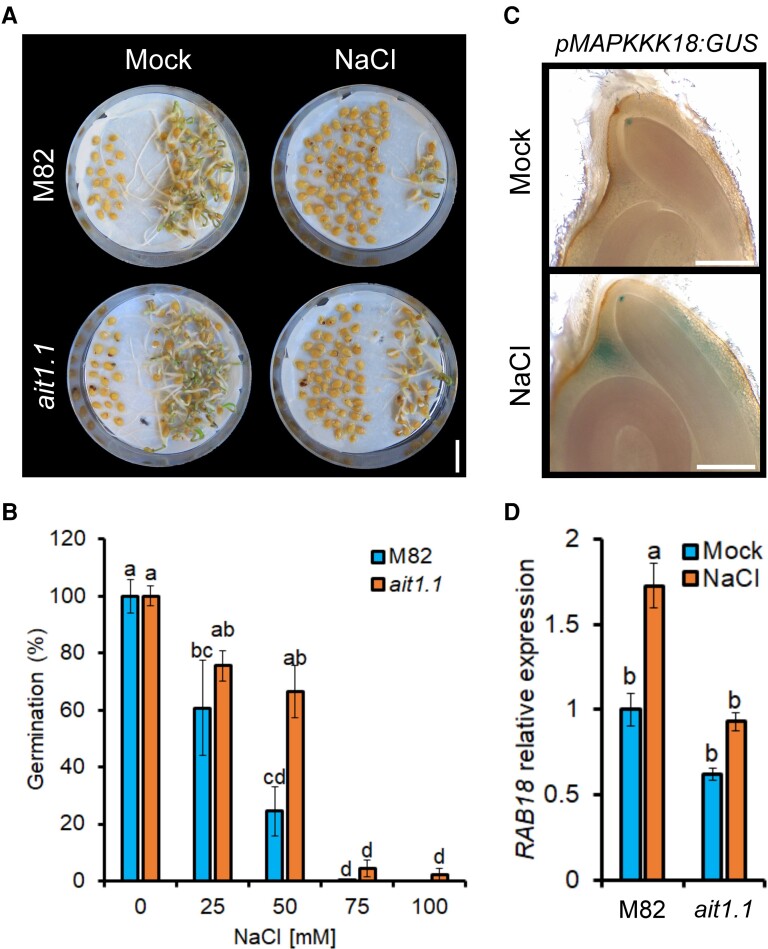
Salinity conditions suppress germination via the inhibition of ABA catabolism (Kong et al., 2017; Xia et al., 2019). We tested if AIT1.1 has a role in the inhibition of germination imposed by salinity. M82 and ait1.1 seeds were germinated on 0, 25, 50, 75, and 100 mM NaCl. In the presence of low NaCl concentration (25 mM), ait1.1 seeds were germinated slightly better than M82 seeds. However, in the presence of higher NaCl concentration (50 mM), M82 seeds exhibited only 24% germination, whereas ait1.1 exhibited 66% (Figure 6, A and B). Similar result was found for the second allele ait1.1 #2 (Supplemental Figure 9). We noticed that the germination of ait1.1 seeds is more uniform than M82. To evaluate uniformity, we analyzed the degree of variability in germination under salinity conditions by using the parameter of coefficient of variation (CV − Sd/mean, Illouz-Eliaz et al., 2020). The variability in M82 seed germination was higher under control and salinity conditions (Supplemental Figure 10), suggesting that inhibition of germination by AIT1.1 affect the uniformity of germination. We further tested the effect of salinity on ABA responses in germinating seeds. To this end, we have used the ABA transcriptional reporter pMAPKKK18:GUS (Okamoto et al., 2013; Shohat et al., 2020). NaCl treatment promoted GUS activity in the micropylar region following imbibition (Figure 6C), indicating that salinity induces ABA activity in germinating seeds. To test whether the increased germination of ait1.1 under salinity conditions is a result of reduced ABA activity, we analyzed the expression of the ABA-inducible gene RAB18 in salt-treated micropylar endosperms of M82 and ait1.1. The results show significantly lower expression of RAB18 in the mutant (Figure 6D), implying that the hyposensitivity of ait1.1 to salt stress is caused by reduced ABA activity in the micropylar endosperm. We did not find any effect of salt or ABA on AIT1.1 expression in imbibed seeds (Supplemental Figure 11), suggesting that although AIT1.1 plays a role in the inhibition of germination under salt stress, it is not regulated by salinity or ABA at the transcriptional level.
Figure 6.
Loss of AIT1.1 promoted germination under salinity conditions. (A) Representative M82 and ait1.1 seeds placed for 5 days on Petri dishes with or without 50 mM NaCl. The images were digitally extracted for comparison. Scale bar = 1 cm. (B) Percentages of germinated seeds after 7 days on Petri dishes containing different concentrations of NaCl. (C) GUS activity in pMAPKKK18:GUS transgenic seeds 48 h after the beginning of imbibition with (or without) 100 mM NaCl. Scale bar = 1 mm. (D) Relative expression of the abscisic acid (ABA)-responsive gene RAB18 in micropylar endosperms of M82 and ait1.1 48 h after the beginning of imbibition with (or without) 75 mM NaCl. Values in B are germination relative to 0 mM NaCl of the same line. Values in B are means of four replicates each containing 80 seeds ± Se. Values in D are means of four replicates each containing 10 seeds ± Se. Small letters in B and D represent significant differences between respective treatments by Tukey–Kramer HSD test (P < 0.05).
Discussion
ABA levels decrease in seeds following imbibition to enable germination (Nambara and Marion-Poll, 2005). However, under salinity-stress conditions, ABA levels remain high and inhibit germination (Kong et al., 2017). All these are eliminated in ABA-deficient mutants that are able to germinate on salt-containing media (Groot and Karssen, 1992; Léon-Kloosterziel et al., 1996). Here, we show in tomato that the ABA importer AIT1.1 acts in the endosperm to mediate the inhibitory effect of ABA on germination under salinity conditions.
Although ABA is the central regulator of seed maturation, dormancy, and desiccation tolerance, the expression of AIT1.1 was relatively low during these stages and was substantially upregulated following imbibition, when ABA level drops (Braybrook and Harada, 2008). Loss of AIT1.1 did not affect the expression of the major dormancy-related genes but reduced ABA activity and increased the expression of genes related to endosperm weakening. These suggest that AIT1.1 promotes ABA inhibition of seed germination but has no role in ABA-mediated seed maturation or dormancy. In line with this, the loss of AIT1.1 had no effect on seed longevity and therefore, it probably has no role in seed desiccation tolerance.
Tomato has three AIT1.1 putative paralogs, but their ABA transport activity has not been characterized. While AIT1.1 was upregulated following imbibition, AIT1.2, AIT1.3, and AIT1.4 were downregulated. These opposite expression dynamics of AIT1 genes imply that they might have roles in different processes, e.g. AIT1.1 in germination and AIT1.2, AIT1.3, and AIT1.4 in maturation, dormancy, or desiccation tolerance. AIT1.1 and AIT1.2 may have an opposite role in germination; while the loss of AIT1.1 improved germination, the loss of AIT1.2 inhibited germination, suggesting that AIT1.1 inhibits, and AIT1.2 promotes germination. However, since only one ait1.2 allele was analyzed, these are preliminary results and more alleles should be analyzed to confirm our suggestion.
Seed germination includes two major paralleled processes: embryo growth and endosperm weakening to allow radicle emergence. Both processes are inhibited by ABA (Belin et al., 2009; Lee et al., 2012). We found that the loss of AIT1.1 had no effect on embryo growth or on radicle emergence following the removal of the micropylar barrier (scarification). Kang et al (2015) suggest that ABA is transported from the endosperm to the embryo to inhibit embryo growth. Our SCBA assays do not support such a role for AIT1.1 in this process. AIT1.1 was predominantly expressed in the endosperm and the expression of endosperm weakening marker genes, such as EXPA2, beta-1,3-Glucanase and MAN2 was upregulated in the ait1.1 endosperm, which also exhibited reduced ABA activity. Taken together, all these results suggest that AIT1.1 mediates the inhibitory effect of ABA on endosperm weakening, but not on embryo growth. We propose that AIT1.1 is upregulated during imbibition to restrain endosperm-weakening and radicle emergence, by promoting ABA-uptake into the micropylar endosperm cells. AIT1.1 exhibited the highest expression just before radicle emergence which in our experiments occurred 60–70 h after the beginning of imbibition. After the emergence of the radicle, AIT1.1 was downregulated and had no role in postemergence primary root growth.
Although the loss of AIT1.1 promoted germination under normal conditions, its effect was more prominent under salt stress. It is possible that the main role of AIT1.1 in seeds is to transport ABA following imbibition under unfavorable conditions, to inhibit germination. Indeed, the loss of AIT1.1 reduced ABA activity in the micropylar endosperm under salinity conditions. Although ABA and salinity did not affect the expression of AIT1.1 in seeds, it is possible that high ABA levels under salinity conditions posttranslationally regulate AIT1.1 activity to increase ABA transport into endosperm cells. It was recently shown that ABA activates a kinase that phosphorylates the Arabidopsis AIT1 to enhance ABA transport and responses (Zhang et al., 2021a, 2021b). We propose that imbibition under salinity conditions increases ABA levels and, therefore, the contribution of AIT1.1 to the inhibition of germination is more prominent under stress.
Plants have adopted mechanisms to reduce germination under unfavorable conditions. It was demonstrated before that reduced germination-uniformity under abiotic stress conditions promotes the survival of a population (Gioria et al., 2018; Vishal and Kumar, 2018). However, this evolutionary advantage is an undesired trait for agricultural needs (Finch-Savage and Bassel, 2016; Fabrissin et al., 2021). We showed here that the loss of AIT1.1 activity reduced germination variability under salt stress. Since, the ait1.1 mutant exhibited increased and uniform germination but had no effect on seed longevity and in field test exhibited normal fruit quality, size and total yield (Supplemental Figure 12), the ait1.1 allele may have the potential to improve germination in commercial tomato varieties.
Materials and methods
Plant materials, growth conditions, and hormone treatments
Tomato (S. lycopersicum L.) plants in M82 background (sp/sp, Pnueli et al., 1998) were used throughout this study. ait1.1 (Shohat et al., 2020), ait1.2, sit (Nir et al., 2017) and the transgenic lines pAIT1.1:GUS and pMAPKKK18:GUS (Shohat et al., 2020) were backcrossed to or generated in M82 background. Plants were grown in a growth room set to a photoperiod of 12/12 h night/day, light intensity of 150 μmol m−2 s−1°C and 25°C and irrigated to saturation. The seeds were harvested from ripped fruits and treated with 1% sodium hypochlorite followed by 1% (v/v) Na3PO4 12H2O, and incubated with 10% sucrose (w/v) over-night in 37°C. Seeds were stored dry at room temperature. (±)-ABA is dissolved in dimethyl sulfoxide (DMSO, Sigma-Aldrich, St. Louis, USA) and NaCl is dissolved in double-distilled water.
CRISPR/Cas9 mutagenesis, tomato transformation, and selection of mutant alleles
Four single-guide RNAs (sgRNAs, Supplemental Table 1) were designed to target AIT1.1 and AIT1.2, using the CRISPR-P tool (http://cbi.hzau.edu.cn/crispr). Vectors were assembled using the Golden Gate cloning system, as described by Weber et al. (2011). Final binary vectors pAGM4723 was introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. The constructs were transferred into M82 cotyledons using transformation and regeneration methods described by McCormick (1991). Kanamycin-resistant T0 plants were grown and independent transgenic lines were selected and self-pollinated to generate homozygous transgenic lines. The genomic DNA of each plant was extracted and genotyped by PCR for the presence of the Cas9 construct. The CRISPR/Cas9-positive lines were further genotyped for mutations using a forward primer to the upstream sequence of the sgRNA1 target and a reverse primer to the downstream of the sgRNA2 target sequence. AIT1.1 and AIT1.2 were sequenced in all mutant lines. Homozygous lines were identified and selected for further analysis. The Cas9 construct was segregated out by crosses to M82.
Germination assays
Seeds were germinated in soil or petri dishes contained Murashige and Skoog (MS) medium including vitamins (N0224, Duchefa, Haarlem, NL) or wet Whatman filter paper (GE Healthcare, Amersham, UK) in a growth room set to a photoperiod of 12/12-h night/day, with a light intensity of 150 µmol m−2 s−1, and at a temperature of 25°C. In soil, germination was scored when the hypocotyl emerged from the soil, and in petri dishes when the radicle pierced the seed coat.
RNA extraction and cDNA synthesis
Total RNA extracted by RNeasy Plant Mini Kit (Qiagen). For synthesis of cDNA, SuperScript II reverse transcriptase (18064014; Invitrogen, Waltham, MA, USA) and up to 3 mg of total RNA were used, according to the manufacturer's instructions.
RT-qPCR analysis
RT-qPCR analysis was performed using an Absolute Blue qPCR SYBR Green ROX Mix (AB-4162/B) kit (Thermo Fisher Scientific, Waltham, MA, USA). Reactions were performed using a Rotor-Gene 6000 cycler (Corbett Research, Sydney, Australia). A standard curve was obtained using dilutions of the cDNA sample. The expression was quantified using Corbett Research Rotor-Gene software. Three independent technical repeats were performed for each sample. Relative expression was calculated by dividing the expression level of the examined gene to ACTIN, TIP41 or PP2CA1. Gene to ACTIN/TIP41/PP2CA1 ratio was then averaged. All primers sequences are presented in Supplemental Table 2. The values of mock and/or M82 WT treatments were set to 1.
Embryo and radicle length measurements
Embryos and radicles images were captured using a Nikon SMZ1270 stereo microscope equipped with a Nikon DS-Ri2 camera and NIS-ELEMENT software (Nikon Instruments, Melville, NY, USA). Images were than analyzed to determine length using ImageJ software segmented line tool (http://rsb.info.nih.gov/ij/).
Embryo greening analysis
Embryos greening was analyzed using ImageJ software histogram feature. Greening was determined by calculating the green (G) pixels intensity relative to Red + Green + Blue (RGB) intensity. The minimum greening value which calculated from white screen was 0.34, and the maximum value which calculated from 4 days old embryo cotyledons was 0.54.
Embryos isolation and seed coat bedding assay (SCBA)
To isolate embryos, tomato seeds were imbibed for several hours in water, and then the seeds were dissected into embryos and endosperms by using a surgical blade and very fine tweezers under a binocular microscope (Olympus, Waltham, MA, USA). The SCBAs were performed according to Lee et al. (2010). Briefly, isolated embryos were placed on a layer of embryo-less endosperms that laid on a MS medium. Embryos growth and greening were analyzed after 72 h as described above (see Material and methods).
GUS staining
Histochemical detection of GUS activity was performed using 5-bromo-4- chloro-3-indolyl-b-D-glucuronide as described in Donnelly et al. (1999). Samples were photographed under a Nikon SMZ1270 stereo microscope equipped with a Nikon DS-Ri2 camera and NIS-ELEMENT software.
Statistical analyses
All experiments were repeated at least three times, each with three or more biological replicates and analyzed using JMP software (SAS Institute, Cary, NC, USA). Means comparison was conducted using ANOVA with post hoc Tukey–Kramer HSD (for multiple comparisons) and Student's t tests (for one comparison) (P < 0.05).
Accession numbers
Sequence data from this article can be found in the Sol Genomics Network (https://solgenomics.net/) under the following accession numbers: ACTIN, Solyc11g005330; TIP41, Solyc10g049850; PP2AC1, Solyc05g006590; AIT1.1, Solyc05g006990; AIT1.2, Solyc05g007000; AIT1.3, Solyc04g005790; AIT1.4, Solyc03g113250; ABI3, Solyc06g083600; FUS3, Solyc02g094460; DOG1, Solyc03g006120; LEC1, Solyc04g015060; beta-1-3-Glucanase, Solyc01g060010; EXPA2, Solyc09g018020; MAN2, Solyc06g064520; RAB18 Solyc02g084850; RD29, Solyc03g025810.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Using different housekeeping genes (ACTIN, TIP41, and PP2C1A) gave similar results in gene expression analyses in tomato seeds.
Supplemental Figure S2. Sequence analyses of ait1.1 #2 and ait1.2 CRISPR mutant alleles.
Supplemental Figure S3. Loss of AIT1.1 promoted germination in additional independent allele.
Supplemental Figure S4. Loss of AIT1.1 did not affect seed longevity.
Supplemental Figure S5. ABA deficiency reduced seed longevity.
Supplemental Figure S6. Loss of AIT1.1 did not affect the expression of the other AIT1 genes.
Supplemental Figure S7. AIT1.1 did not affect endosperm inhibition of embryo growth.
Supplemental Figure S8. Loss of AIT1.1 did not affect post-emergence radicle elongation.
Supplemental Figure S9. Increased germination under salinity conditions in additional independent ait1.1 allele.
Supplemental Figure S10. Loss of AIT1.1 increased germination uniformity under normal and stress conditions.
Supplemental Figure S11. Salt or ABA treatments did not affect AIT1.1 expression seeds.
Supplemental Figure S12. Loss of AIT1.1 did not affect fruit yield in the field.
Supplemental Table S1. gRNAs used in this study.
Supplemental Table S2. Primers used in this study.
Supplementary Material
Contributor Information
Hagai Shohat, Institute of Plant Sciences and Genetics in Agriculture, The Hebrew University of Jerusalem, P.O. Box 12, Rehovot 76100, Israel.
Hadar Cheriker, Institute of Plant Sciences and Genetics in Agriculture, The Hebrew University of Jerusalem, P.O. Box 12, Rehovot 76100, Israel.
Amir Cohen, Institute of Plant Sciences and Genetics in Agriculture, The Hebrew University of Jerusalem, P.O. Box 12, Rehovot 76100, Israel.
David Weiss, Institute of Plant Sciences and Genetics in Agriculture, The Hebrew University of Jerusalem, P.O. Box 12, Rehovot 76100, Israel.
Data availability
All data can be found in the manuscript and in the supporting information.
Author contributions
H.S. and D.W. designed the research plan; H.S., H.C., and A,C, performed the research; H.S. and A.C. analyzed data; H.S. and D.W. wrote the paper.
Funding
This research was supported by the Israel Science Foundation to D.W. (617/20).
References
- Anfang M, Shani E (2021) Transport mechanisms of plant hormones. Curr Opin Plant Biol 63: 102055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin JM, Baskin CC (2004) A classification system for seed dormancy. Seed Sci Res 14(1): 1–16 [Google Scholar]
- Belin C, Megies C, Hauserová E, Lopez-Molina L (2009) Abscisic acid represses growth of the Arabidopsis embryonic axis after germination by enhancing auxin signaling. Plant Cell 21(8): 2253–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braybrook SA, Harada JJ (2008) LECs go crazy in embryo development. Trends Plant Sci 13(12): 624–630 [DOI] [PubMed] [Google Scholar]
- Chiba Y, Shimizu T, Miyakawa S, Kanno Y, Koshiba T, Kamiya Y, Seo M (2015) Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. J Plant Res 128(4): 679–686 [DOI] [PubMed] [Google Scholar]
- Coolbear P, Francis A, Grierson D (1984) The effect of low temperature pre-sowing treatment on the germination performance and membrane integrity of artificially aged tomato seeds. J Exp Bot 35(11): 1609–1617 [Google Scholar]
- Dekkers BJW, Willems L, Bassel GW, van Bolderen-Veldkamp RPM, Ligterink W, Hilhorst HWM, Bentsink L (2012) Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol 53(1): 28–37 [DOI] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215(2): 407–419 [DOI] [PubMed] [Google Scholar]
- Donohue K, de Casas RR, Burghardt L, Kovach K, Willis CG (2010) Germination, postgermination adaptation, and species ecological ranges. Ann Rev Ecol Evol Sys 41(1): 293–319 [Google Scholar]
- Fabrissin I, Sano N, Seo M, North HM (2021) Ageing beautifully: can the benefits of seed priming be separated from a reduced lifespan trade-off? J Exp Bot 72(7): 2312–2333 [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Bassel GW (2016) Seed vigour and crop establishment: extending performance beyond adaptation. J Exp Bot 67(3): 567–591 [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G (2006) Seed dormancy and the control of germination. New Phytol 171(3): 501–523 [DOI] [PubMed] [Google Scholar]
- Gioria M, Pysek P, Osborne BA (2018) Timing is everything: does early and late germination favor invasions by herbaceous alien plants? J Plant Ecol 11(1): 4–16 [Google Scholar]
- Graeber K, Linkies A, Steinbrecher T, Mummenhoff K, Tarkowska D, Tureckova V, Ignatz M, Sperber K, Voegele A, de Jong H, et al. (2014) DELAY OF GERMINATION 1 mediates a conserved coat-dormancy mechanism for the temperature- and gibberellin-dependent control of seed germination. Proc Natl Acad Sci USA 111(34): E3571-E3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber K, Nakabayashi K, Miatton E, Leubner-Metzger G, Soppe WJJ (2012) Molecular mechanisms of seed dormancy. Plant Cell Environ 35(10): 1769–17860 [DOI] [PubMed] [Google Scholar]
- Groot SPC, Karssen CM (1992) Dormancy and germination of abscisic acid-deficient tomato seeds. Plant Physiol 99(3): 952–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilhorst HWM, Downie B (1995) Primary dormancy in tomato (Lycopersicon esculentum cv. Moneymaker): studies with the sitiens mutant. J Exp Bot 47(1): 89–97 [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJJ (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179(1): 33–54 [DOI] [PubMed] [Google Scholar]
- Illouz-Eliaz N, Ramon U, Shohat H, Blum S, Livne S, Mendelson D, Weiss D (2020) Multiple gibberellin receptors contribute to phenotypic stability under changing environments. Plant Cell 31(7): 1506–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Hwang JU, Lee M, Kim YY, Assmann S, Martinoia E, Lee Y (2010) PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci USA 107(5): 2355–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Yim S, Choi H, Kim A, Lee KP, Lopez-Molina L, Martinoia E, Lee Y (2015) Abscisic acid transporters cooperate to control seed germination. Nat Commun 6(1): 8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M, Koshiba T, Kamiya Y, Seo M (2012) Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci USA 109(24): 9653–9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Luo Z, Zhang Y, Li W, Dong H (2017) Soaking in H2O2 regulates ABA biosynthesis and GA catabolism in germinating cotton seeds under salt stress. Act Physiol Plant 39(1): 2 [Google Scholar]
- Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, Kamiya A, Moriyama Y, Shinozaki K (2010) ABC Transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci USA 107(5): 2361–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Seo M, Shinozaki K (2018) ABA transport and plant water stress responses. Trends Plant Sci 23(6): 513–522 [DOI] [PubMed] [Google Scholar]
- Lai Y, Zhang D, Wang J, Wang J, Ren P, Yao L, Si E, Kong Y, Wang H (2020) Integrative transcriptomic and proteomic analyses of molecular mechanism responding to salt stress during seed germination in Hulless Barley. Int J Mol Sci 21(1): 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Piskurewicz U, Turečková V, Carat S, Chappuis R, Strnad M, Fankhauser C, Lopez-Molina L (2012) Spatially and genetically distinct control of seed germination by phytochromes A and B. Gen Dev 26(17): 1984–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Piskurewicz U, Turečková V, Strnad M, Lopez-Molina L (2010) A seed coat bedding assay shows that RGL2-dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proc Natl Acad Sci USA 107(44): 19108–19113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GS, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10(4): 655–661 [DOI] [PubMed] [Google Scholar]
- Léran S, Varala K, Boyer J-C, Chiurazzi M, Crawford N, Daniel-Vedele F, David L, Dickstein R, Fernandez E, Forde B, et al. (2014) A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trend Plant Sci 19(1): 5–9 [DOI] [PubMed] [Google Scholar]
- Leubner-Metzger G, Frundt C, Vogeli-Lange R, Meins F Jr (1995) Class I beta-1,3-Glucanases in the endosperm of tobacco during germination. Plant physiol 109(3): 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liptay A, Schopfer P (1983) Effect of water stress, seed coat restraint, and abscisic acid upon different germination capabilities of two tomato lines at low temperature. Plant Physiol 73(4): 935–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hou X (2018) Antagonistic regulation of ABA and GA in metabolism and signaling pathways. Front Plant Sci 9: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Andujar C, Pluskota WE, Bassel GW, Asahina M, Pupel P, Nguyen TT, Takeda-Kamiya N, Toubiana D, Bai B, et al. (2012) Mechanisms of hormonal regulation of endosperm cap-specific gene expression in tomato seeds. Plant J 71(4): 575–586 [DOI] [PubMed] [Google Scholar]
- McCormick S (1991) Transformation of tomato with Agrobacterium tumefaciens. Plant Tiss Cult Man B 6: 1–9 [Google Scholar]
- Muller K, Tintelnot S, Leubner-Metzger G (2006) Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of Cress and Arabidopsis thaliana. Plant Cell Physiol 47(7): 864–877 [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Ann Rev Plant Biol 56(1): 165–185 [DOI] [PubMed] [Google Scholar]
- Nir I, Shohat H, Panizel I, Olszewski N, Aharoni A, Weiss D (2017) The tomato DELLA protein PROCERA acts in guard cells to promote stomatal closure. Plant Cell 29(12): 3186–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H (2014) Seed dormancy and germination-emerging mechanisms and new hypotheses. Front Plant Sci 5: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H, Gee OH, Bradford KJ (2000) A germination-specific Endo-b-Mannanase gene is expressed in the micropylar endosperm cap of tomato seeds. Plant Physiol 123(4): 1235–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15(7): 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Peterson FC, Defries A, Park SY, Endo A, Nambara E, Volkman BF, Cutler SR (2013) Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proc Natl Acad Sci USA 110(29): 12132–12137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms JJJ, Leon-Kloosterziel KM, Bartels D, Koornneef M, Karssen CM (1993) Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana. Plant Physiol 102(4): 1185–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawela A, Banasiak J, Biala W, Martinoia E, Jasinski M (2019) MtABCG20 is an ABA exporter influencing root morphology and seed germination of Medicago truncatula. Plant J 98(3): 511–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y, Lopez-Molina L (2008) The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20(10): 2729–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Carmel-Goren L, Hareven D, Gutfinger T, Alvarez J, Ganal M, Zamir D, Lifschitz E (1998) The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125(11): 1979–1989 [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA Perception and signaling. Trends Plant Sci 15(7): 395–401 [DOI] [PubMed] [Google Scholar]
- Ruggiero A, Landi S, Punzo P, Possenti M, Van Oosten MJ, Costa A, Morelli G, Maggio A, Grillo S, Batelli G (2019) Salinity and ABA seed responses in pepper: expression and interaction of ABA core signaling components. Front Plant Sci 10: 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Kanno Y, Suzuki H, Watanabe S, Seo M (2021) Arabidopsis NPF4.6 and NPF5.1 control leaf stomatal aperture by regulating abscisic acid transport. Genes (Basel) 12(6): 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shohat H, Illouz Eliaz N, Kanno Y, Seo M, Weiss D (2020) The tomato DELLA protein PROCERA promotes abscisic acid responses in guard cells by upregulating an abscisic acid transporter. Plant Physiol 184(1): 518–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Qi Y, Chen F, Meng Y, Luo X, Shuai H, Zhou W, Ding J, Du J, Liu J, et al. (2017) Salt stress represses soybean seed germination by negatively regulating GA biosynthesis while positively mediating ABA biosynthesis. Front Plant Sci 8: 1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Zhou W, Chen F, Luo X, Yang W (2018) Abscisic acid and gibberellins antagonistically mediate plant development and abiotic stress responses. Front Plant Sci 9: 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbrecher T, Leubner-Metzger G (2017) The biomechanics of seed germination. J Exp Bot 68(4): 765–783 [DOI] [PubMed] [Google Scholar]
- Steinbrecher T, Leubner-Metzger G (2018) Tissue and cellular mechanics of seeds. Curr Opin Gen Dev 51: 1–10 [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Kawaide H, Mitsuhashi W, Inoue Y, Kamiya Y (1998) Phytochrome regulates gibberellin biosynthesis during germination of photoblastic lettuce seeds. Plant Physiol 118(4): 1517–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishal B, Kumar PP (2018) Regulation of seed germination and abiotic stresses by gibberellins and abscisic acid. Front Plant Sci 9: 838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S (2011) A modular cloning system for standardized assembly of multigene constructs. PLoS One 6(2): e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K, Liu A, Wang Y, Yang W, Jin Y (2019) Mechanism of salt-inhibited early seed germination analysed by transcriptomic sequencing. Seed Sci Res 29(2): 73–84 [Google Scholar]
- Yan D, Duermeyer L, Leoveanu C, Nambara E (2014) The functions of the endosperm during seed germination. Plant Cell Physiol 55(9): 1521–1533 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kilambi HV, Liu J, Bar H, Lazary S, Egbaria A, Ripper D, Charrier L, Belew ZM, Wulff N, et al. (2021b) ABA Homeostasis and long- distance translocation are redundantly regulated by ABCG ABA importers. Sci Adv 7(43): eabf6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yu Z, Xu Y, Yu M, Ren Y, Zhang S, Yang G, Huang J, Yan K, Zheng C, et al. (2021a) Regulation of the stability and ABA import activity of NRT1.2/NPF4.6 by CEPR2-mediated phosphorylation in Arabidopsis. Mol Plant 14(4): 633–646 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhu H, Pan Y, Yu Y, Luan S, Li L (2014) A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol Plant 7(10): 1522–1532 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data can be found in the manuscript and in the supporting information.