Abstract
Increasing planting density is one of the most effective ways to improve crop yield. However, one major factor that limits crop planting density is the weakened immunity of plants to pathogens and insects caused by dim light (DL) under shade conditions. The molecular mechanism underlying how DL compromises plant immunity remains unclear. Here, we report that DL reduces rice (Oryza sativa) resistance against brown planthopper (BPH; Nilaparvata lugens) by elevating ethylene (ET) biosynthesis and signaling in a Phytochrome B (OsPHYB)-dependent manner. The DL-reduced BPH resistance is relieved in osphyB mutants, but aggravated in OsPHYB overexpressing plants. Further, we found that DL reduces the nuclear accumulation of OsphyB, thus alleviating Phytochrome Interacting Factor Like14 (OsPIL14) degradation, consequently leading to the up-regulation of 1-Aminocyclopropane-1-Carboxylate Oxidase1 (OsACO1) and an increase in ET levels. In addition, we found that nuclear OsphyB stabilizes Ethylene Insensitive Like2 (OsEIL2) by competitively interacting with EIN3 Binding F-Box Protein (OsEBF1) to enhance ET signaling in rice, which contrasts with previous findings that phyB blocks ET signaling by facilitating Ethylene Insensitive3 (EIN3) degradation in other plant species. Thus, enhanced ET biosynthesis and signaling reduces BPH resistance under DL conditions. Our findings provide insights into the molecular mechanism of the light-regulated ET pathway and host-insect interactions and potential strategies for sustainable insect management.
Introduction
Rice (Oryza sativa) is a staple food crop for over half of the world's population. The brown planthopper (BPH; Nilaparvata lugens) is the most destructive insect pest of rice, causing an annual direct loss of several billion dollars (Catindig, 2009; FAO, 2020). In addition to the direct feeding damage caused by piercing and sucking sap from the vascular tissues of plant, BPH also transmits two virus diseases, namely the rice grassy stunt and the rugged stunt viruses, further aggravating the damage (Cheng et al., 2013). Pesticides are the currently common strategy for controlling BPH, which are costly and detrimental to the environment. Breeding resistant rice cultivars is believed to be the most cost-effective and environmentally friendly strategy for BPH management (Liu et al., 2015).
Over the past few decades, increasing planting density has been one of the most effective ways to improve crop yields per unit land area. However, a key factor that limits high-density planting is the shade avoidance syndrome (SAS) in plants, which is triggered by a drop in the ratio of red (R) to far-red (FR) light (R:FR) or quantity of light irradiation under shade conditions (Xie et al., 2017; Wei et al., 2018). Typical shade avoidance syndrome includes increased stem elongation, elevated leaf angle (to the horizontal), reduced branching, early flowering (Franklin, 2008; Casal, 2012; Xie et al., 2017) and decreased immunity to microbial pathogens and pests (Augspurger and Kelly, 1984; Bell et al., 2006; Roberts and Paul, 2006; Ballare, 2014).
The red/far-red light photoreceptors, phytochromes (phys), are key regulators of SAS. Among the phytochromes, phyB plays a major role in suppressing SAS. PHYB loss-of-function mutations in Arabidopsis (Arabidopsis thaliana), wheat (Triticum aestivum), or maize (Zea mays) commonly display constitutive SAS (Neff et al., 2000; Kebrom et al., 2006; Kippes et al., 2020). In addition, it has been shown that under shade conditions, the activity of phyB is suppressed due to conversion of its active, far-red light absorbing form (Pfr) into the inactive, red light-absorbing form (Pr), which abrogates the inhibitory effect of phyB-Pfr on the activity of phytochrome-interacting factors (PIFs) (Quail, 2002). In turn, PIFs can induce the expression of auxin biosynthetic or response genes to promote stem elongation and plant height (Fernández-Milmanda and Ballaré, 2021; Xi et al., 2021). However, unlike other plant species, the internode elongation is suppressed in osphyB mutants of rice (Takano et al., 2009), and osphyB mutants display decreased plant height phenotype (Sun et al., 2017a). These studies indicate that phyB in rice plays a distinguished role in SAS compared with its role in other plant species, but the underlying mechanism remains poorly understood.
Most studies on SAS have been focused on plant growth and development while little is known about how SAS weakens the defense response to pests and pathogens in plants. Several recent studies showed that phyB plays a positive role in light-modulated resistance to pathogens and insects in some dicot plant species (Campos et al., 2016; Fernández-Milmanda et al., 2020). For example, it was reported that in Arabidopsis thaliana, increased accumulation of PIFs under low R:FR conditions facilitates the expression of ST2a, which encodes a sulfotransferase, thus reducing the precursor pool of active forms of jasmonates (JA) (Fernández-Milmanda et al., 2020). In addition, the stability of the JA signaling repressor Jasmonate ZIM-domain10 (JAZ10) is enhanced under low R:FR conditions, resulting in weakened defense against the necrotrophic pathogen Botrytis cinerea and chewing insect Spodoptera littoralis (Cerrudo et al., 2012; Ballare, 2014; Fernández-Milmanda et al., 2020). Despite the molecular mechanism governing light-modulated biotic resistance has been relatively well elucidated in the model dicot plant Arabidopsis thaliana, little is known regarding the molecular mechanisms in monocot crops.
Ethylene (ET) as a gas phytohormone plays a critical role in regulating various aspects of plant growth and defense response (Abeles, 1992; Chen et al., 2005b). Previous studies demonstrate that ET should play diverse roles in plant immunity. Increasing ET level or enhancing ET signal can substantially increase the resistance against pathogens and chewing herbivores in various plant species (Marta and Antonio, 2004; Onkokesung et al., 2010). However, reducing the ET level by down-regulating the expression of OsACS2 (1-Aminocyclopropane-1-Carboxylate Synthase2) substantially increased the resistance against BPH in rice (Lu et al., 2014). OsERF3 (Ethylene responsive factor 3) silenced plants displayed higher resistance against BPH, while OsERF3 overexpressing plants were more susceptible to BPH (Lu et al., 2011). Moreover, a recent study reported that OsEIL1 (EIN3-like gene 1, a key positive regulator in ET pathway) play negative role in BPH resistance, while OsEBF1 (EIN3 Binding F-Box Protein, which facilitates the degradation of EIN3) positively regulated the BPH resistance (Ma et al., 2019). These studies indicate that ET should negatively regulate the resistance against BPH in rice.
It has been demonstrated that during seedling photomorphogenesis, phyB acts as a negative regulator of ethylene biosynthesis and signaling in Arabidopsis thaliana. On one hand, light-activated phyB inhibits the function of PIF proteins, thereby reducing ET biosynthesis by repressing the expression of ET biosynthesis-related genes ACSs (Khanna et al., 2007; Song et al., 2018). On the other hand, phyB promotes the interaction between EIN3 (Ethylene Insensitive 3) and EBF1/2, thereby stimulating the degradation of EIN3 to turn off ET signaling (Shi et al., 2016). Thus, the inhibition of ET signaling mediated by phyB promotes cotyledon opening and expansion, enabling the seedlings to adopt a photomorphogenic mode of growth as soon as they emerge from the soil. However, it is unclear whether the molecular mechanism of light regulating ET pathway is conserved between monocots and dicots, and whether the crosstalk between phyB-mediated light signaling and ET signaling is implicated in plant immunity.
Here, our studies revealed a distinct function of phyB in Arabidopsis and rice. Unlike in Arabidopsis, we found that OsphyB negatively regulates insect resistance in rice under dim light (DL) conditions. Mechanistically, the nuclear OsphyB interacts with OsPIL14 (Phytochrome-Interacting Factor-Like14), resulting in the degradation of OsPIL14. Compared with normal light (NL) conditions, DL reduces the accumulation of OsphyB in the nucleus, resulting in the accumulation of OsPIL14 to up-regulate the expression of OsACO1 (1-Aminocyclopropane-1-Carboxylate Oxidase1) and increase ET biosynthesis. This in turn up-regulates the expression of OsEIL2. In contrast to the negative role of phyB in regulating ET signaling in Arabidopsis thaliana, our further studies showed that OsphyB remaining in the nucleus stabilizes ET signaling pathway by competitively interacting with OsEBF1 to block the degradation of OsEIL2. In addition, we found that increasing ET level or enhancing ET signaling can substantially reduce the resistance against BPH. However, ET and DL-reduced BPH resistance is relieved in osphyB mutants and OsEIL2 RNAi plants. Together, we discovered a role of OsphyB in regulating ET pathway and suggest that OsphyB negatively regulates insect resistance through increasing the accumulation of ET and stabilizing ET signaling under DL conditions in rice.
Results
Dim light reduces BPH resistance in an OsPHYB-dependent manner
To investigate whether light intensity affects the resistance to BPH, we assessed the BPH resistance in two japonica rice cultivars Nipponbare (NIP) and Zhonghua 11 (ZH11) under strong light (SL, 650 μmol m−2 s−1), normal light (NL, 200 μmol m−2 s−1), and dim light (DL, 50 μmol m−2 s−1) conditions (Liu et al., 2019), respectively. Two-week-old seedlings growing under NL condition were pretreated with SL, NL, or DL for 24 h, then infested with BPH and continuously exposed to SL, NL or DL treatment, respectively. The results showed that BPH resistance was gradually reduced as the light intensity decreased both in NIP and ZH11 (Supplemental Figure 1, A–F). To determine whether the effect of light on the BPH resistance varies among rice cultivars, we also included two indica rice cultivars, 93–11 (a BPH susceptible cultivar) and IR64 (a BPH-resistant cultivar, carrying the BPH resistance gene Bph1), to evaluate the BPH resistance under different light conditions. Compared with NL conditions, the BPH resistance in both 93–11 and IR64 was significantly reduced under DL, but enhanced under SL (Supplemental Figure 1, G–L). These results suggest that light intensity positively regulates BPH resistance in a wide range of rice cultivars.
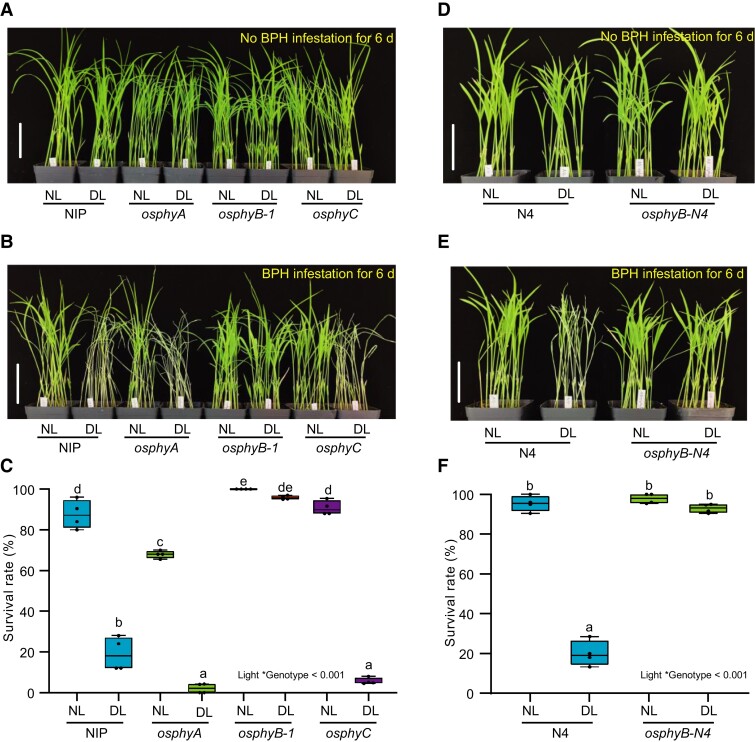
We next investigated whether phytochromes affect the light-regulated BPH resistance. We assessed the BPH resistance in three distinct phytochrome mutants (osphyA, osphyB-1, and osphyC) under NL and DL conditions. Under NL conditions, the osphyA mutant displayed increased susceptibility to BPH, but the osphyB-1 and osphyC mutants showed similar resistance to the wild type (WT) control (Figure 1, A–C). Like the WT, the BPH resistance of osphyA and osphyC were significantly reduced by DL, but osphyB-1 displayed comparable resistance levels under both NL and DL conditions (Figure 1, A–C). Further, we used two additional lines of OsPHYB allelic mutants (osphyB-2 and osphyB-N4) to verify these results (Supplemental Figure 2). Consistently, both osphyB-2 and osphyB-N4 showed similar BPH resistance under NL and DL conditions (Supplemental Figure 3; Figure 1, D–F). These results suggest that OsPHYB, rather than OsPHYA or OsPHYC, is involved in DL-induced reduction of BPH resistance in rice.
Figure 1.
Dim light reduces BPH resistance through OsPHYB. A, Two-week-old seedlings of wild type Nipponbare (NIP), osphyA, osphyB-1, and osphyC mutants grown under Normal light conditions (NL) and pretreated with NL and dim light (DL) for 24 h, then continuously exposed to NL and DL conditions without BPH infestation for 6 d. Bar, 7 cm. B and C, Representative images (B) and seedling survival rate (C) of NIP, osphyA, osphyB-1, and osphyC at 6 d post infestation (dpi) with BPH under NL and DL treatments. Bar, 7 cm. D, Two-week-old seedlings of wild type Ningjing 4 (N4) and OsPHYB mutant (osphyB-N4) grown under NL conditions and pretreated with NL and DL for 24 h, then continuously exposed to NL and DL conditions without BPH infestation for 6 d. Bar, 7 cm. E and F, Representative images (E) and seedling survival rate (F) of N4 and osphyB-N4 at 6 dpi with BPH under NL and DL treatments. Bar, 7 cm. In C and F, the mean separation tests are shown with box plots. The center lines of the box plots indicate the median, the bounds of the box show the 25th and the 75th percentiles, and the whiskers indicate 1.5 × IQR. n ≥ 3 independent experiments, with each pool including 25 individual plants. Different letters designate significantly different means by 2-way ANOVA + Tukey's post hoc test (P < 0.05). Interaction P value is shown in the inset.
To further confirm that the DL-triggered attenuated BPH resistance depends on OsPHYB, we next generated two independent transgenic rice overexpressing OsPHYB (OsPHYB -OX#2 and OsPHYB -OX#3) (Supplemental Figure 4). Compared with the WT, these two OsPHYB overexpressing transgenic lines were both more susceptible to BPH under DL conditions (Supplemental Figure 4). Together, these results demonstrate that OsPHYB plays a crucial role in light-modulated BPH resistance.
OsphyB represses the expression of OsACO1 but activates the downstream signaling pathway of OsEIL2
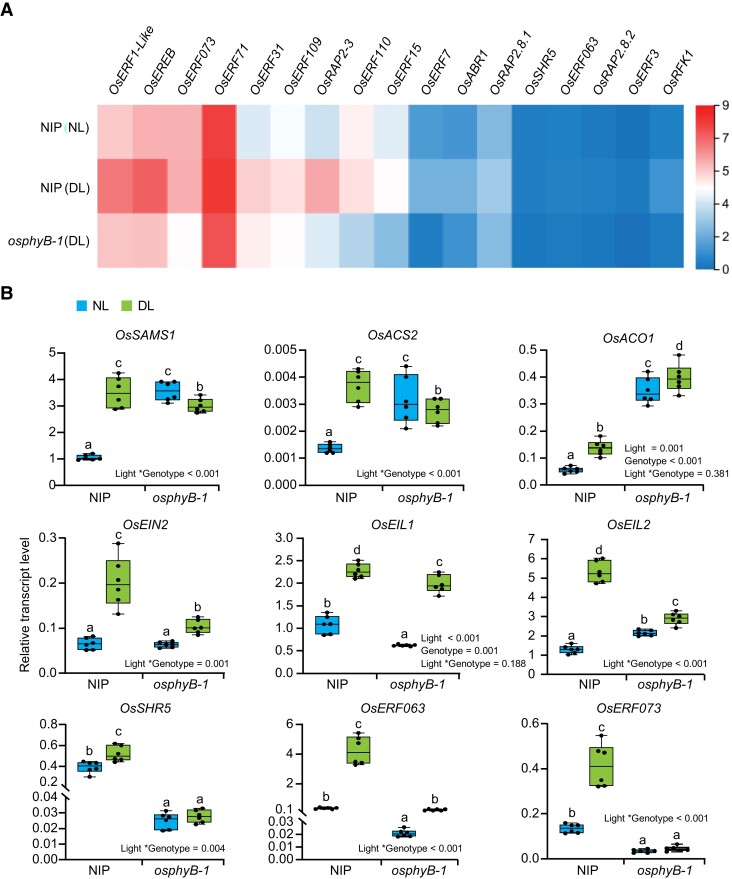
To further investigate the molecular mechanisms underlying OsPHYB negatively regulating BPH resistance under DL condition, we performed an RNA-Sequencing (RNA-seq) assay using two-week-old seedlings of osphyB-1 mutant and WT plants pretreated under NL and DL conditions for 4 h. Approximately 1,186 genes were significantly up-regulated and 969 genes were significantly down-regulated in WT grown in DL when compared with those grown in NL (Supplemental Datasets 1 and 2). KEGG analysis showed that the pathway with the largest number of genes up-regulated by DL was hormone signal transduction, with an overrepresentation of ET and auxin signal pathways (Supplemental Figure 5 and Supplemental Datasets 1 and 2). Among these hormones signaling-related genes, both ET biosynthesis and signaling-related genes were significantly up-regulated by DL in WT (Figure 2A; Supplemental Figure 5; Supplemental Dataset 3). Moreover, the up-regulation was also confirmed by reverse transcription quantitative PCR (RT-qPCR) assay (Figure 2B). In the osphyB-1 mutant, the expression of OsACO1 was significantly up-regulated compared with WT under both NL and DL conditions, but the expression of other ET biosynthesis-related genes (OsSAMS1 (S-Adenosyl-L-Methionine Synthetase) and OsACS2) and ET signaling pathway genes (OsEIN2, OsEIL1, and OsEIL2) were not significantly up-regulated in the osphyB-1 mutant under both NL and DL conditions (Figure 2B; Supplemental Dataset 3).
Figure 2.
Dim light up-regulates the expression of the ethylene pathway-related genes. A, Heatmap showing the expression of downstream target genes of OsEIL2 in WT (NIP) under NL (top), NIP under DL (middle) and osphyB-1 under DL (bottom). The color scale represents log2 fold change. B, RT-qPCR assay showing the transcript levels of ET pathway-related genes in NIP and osphyB-1 under NL and DL. The mean separation tests are shown with box plots. The center lines of the box plots indicate the median, the bounds of the box show the 25th and the 75th percentiles, and the whiskers indicate 1.5 × IQR. n ≥ 3 independent experiments. Different letters designate significantly different means by 2-way ANOVA + Tukey's post hoc test (P < 0.05). Interaction P value is shown in the inset.
Among the six EIL homologs in rice, OsEIL1 and OsEIL2 exhibit the highest similarity to the Arabidopsis EIN3 protein, and have been reported to regulate root and coleoptile, respectively (Ma et al., 2010; Yang et al., 2015a, 2015b). Notably, we found that the expression of several OsEIL2 controlled genes such as OsSHR5, OsERF063 and OsERF073 (Yang et al., 2015b), was up-regulated in WT under DL conditions, but was significantly reduced in the osphyB-1 mutant under both NL and DL conditions (Figure 2B; Supplemental Datasets 1 and 2). In addition, the expression of these genes was also decreased in the osphyB-2 and osphyB-N4 mutants under both NL and DL conditions (Supplemental Figure 6A), but significantly increased in the OsPHYB-OX#2 and OsPHYB-OX#3 transgenic plants (Supplemental Figure 6B). Also notably, the expression of EIL2-regulated genes was significantly induced by BPH infestation in WT, but the induction was repressed in the osphyB mutants (Supplemental Figure 6C), suggesting that OsPHYB positively regulates BPH-induced ET signaling process. These observations show that OsphyB likely plays an opposite role in regulating ET biosynthesis and signaling, repressing the expression of ET biosynthetic gene OsACO1 while promoting the expression of OsEIL2 target genes.
ET negatively regulates the resistance to BPH in rice
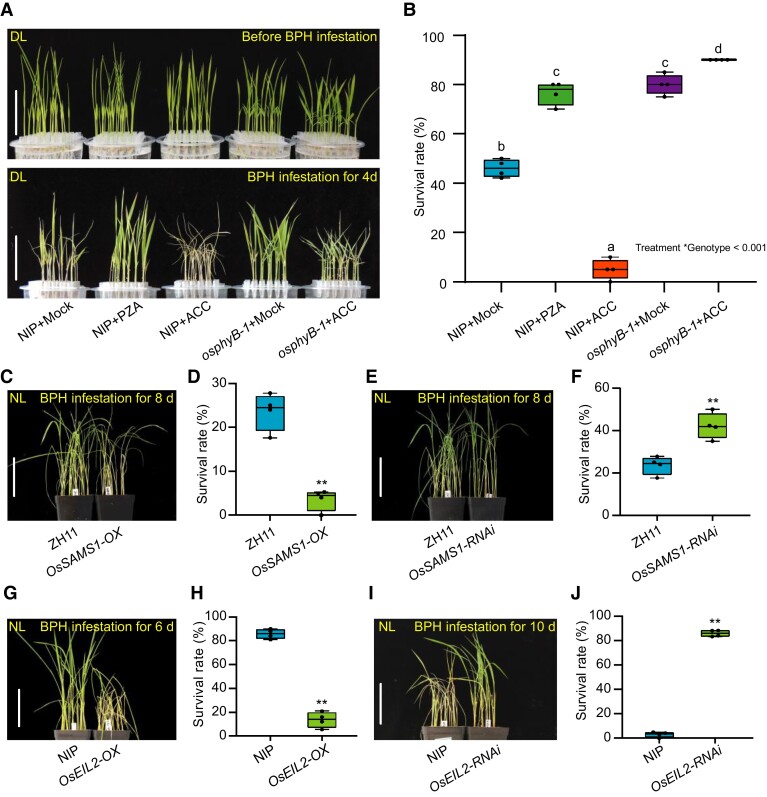
To determine whether the ET pathway participates in BPH resistance, we pretreated rice seedlings with exogenous ET biosynthesis precursor ACC and ET biosynthesis inhibitor pyrazinamide (PZA) (Sun et al., 2017b) under DL. The results showed that exogenous ACC treatment significantly reduced BPH resistance in the NIP seedlings. In contrast, PZA treatment significantly enhanced the resistance compared with the mock treatment (Figure 3, A and B). To verify these observations, we evaluated BPH resistance in the OsSAMS1 overexpressing (OsSAMS1-OX) or knockdown (OsSAMS1-RNAi) plants under NL, which has increased or reduced levels of endogenous ET, respectively (Zhao et al., 2017). As shown in Figure 3, C–F, the OsSAMS1-OX plants were more susceptible to BPH compared with WT, while the OsSAMS1-RNAi plants displayed higher BPH resistance. These results indicate that ET negatively affects the BPH resistance in rice.
Figure 3.
ET negatively regulates BPH resistance in rice. A, Seedlings of WT (NIP) or osphyB-1 were incubated in Hoagland solution under NL conditions for one week, and were pretreated with Hoagland solution (mock), 50 μM ACC or 50 μM pyrazinamide (PZA) for 2 d (upper), and then infested with BPH for 4 d (bottom) under DL. Bar, 5 cm. B, Seedling survival rate of one-week old NIP and osphyB-1 seedlings in Hoagland solution pre-incubated with mock, 50 µM PZA or 50 µM ACC for 2 d and infested with BPH for 4 d. The mean separation tests are shown with box plots. The center lines of the box plots indicate the median, the bounds of the box show the 25th and the 75th percentiles, and the whiskers indicate 1.5 × IQR. n ≥ 3 independent experiments, with each pooled with 20 individual plants. Different letters designate significantly different means by 2-way ANOVA + Tukey's post hoc test (P < 0.05). Interaction P value is shown in the inset. C and D, Representative images (C) and seedling survival rate (D) of WT (ZH11) and OsSAMS1 overexpressing transgenic plants (OsSAMS1-OX) at 8 dpi with BPH under NL. Bar, 7 cm. E and F, Representative images (E) and seedling survival rate (F) of ZH11 and OsSAMS1-RNAi at 8 dpi with BPH under NL. Bar, 7 cm. G and H, Representative images (G) and seedling survival rate (H) of WT (NIP) and OsEIL2-OX at 6 dpi with BPH infestation under NL. Bar, 7 cm. I and J, Representative images (I) and seedling survival rate (J) of WT (NIP) and OsEIL2-RNAi at 10 dpi with BPH infestation under NL. Bar, 7 cm. In D, F, H, and J, the center lines of the box plots indicate the median, the bounds of the box show the 25th and the 75th percentiles, and the whiskers indicate 1.5 × IQR. (n ≥ 3 independent experiments, with each pooled with 25 individual plants), ** P < 0.01 by Student's t-test.
OsEIL2 is the key positive regulator of ET pathway (Yang et al., 2015a; Yang et al., 2015b), the expression of OsEIL2 was significantly induced by ET (Supplemental Figure 7). To further investigate the role of ET signaling in BPH resistance, we evaluated BPH resistance in the OsEIL2 overexpressing (OsEIL2-OX) or knockdown (OsEIL2-RNAi) plants under NL. Compared with the control plants, the OsEIL2-OX plants was more susceptibility to BPH, while the OsEIL2-RNAi plants was more resistant (Figure 3, G–J). Constitutive Triple Response2 (OsCTR2) and OsEIN2 play a positive and negative role in ET signaling in rice, respectively (Hoon et al., 2004; Wang et al., 2013). Thus, we also evaluated BPH resistance in the OsCTR2 mutants (osctr2) and OsEIN2 overexpressing (OsEIN2-OX#2 and #3) plants under NL. As expected, both the osctr2 and OsEIN2-OX plants displayed increased susceptibility to BPH (Supplemental Figure 8). Collectively, these results validate our hypothesis that the ET pathway plays a negative role in BPH resistance.
OsphyB interacts with OsPIL14 and promotes its degradation
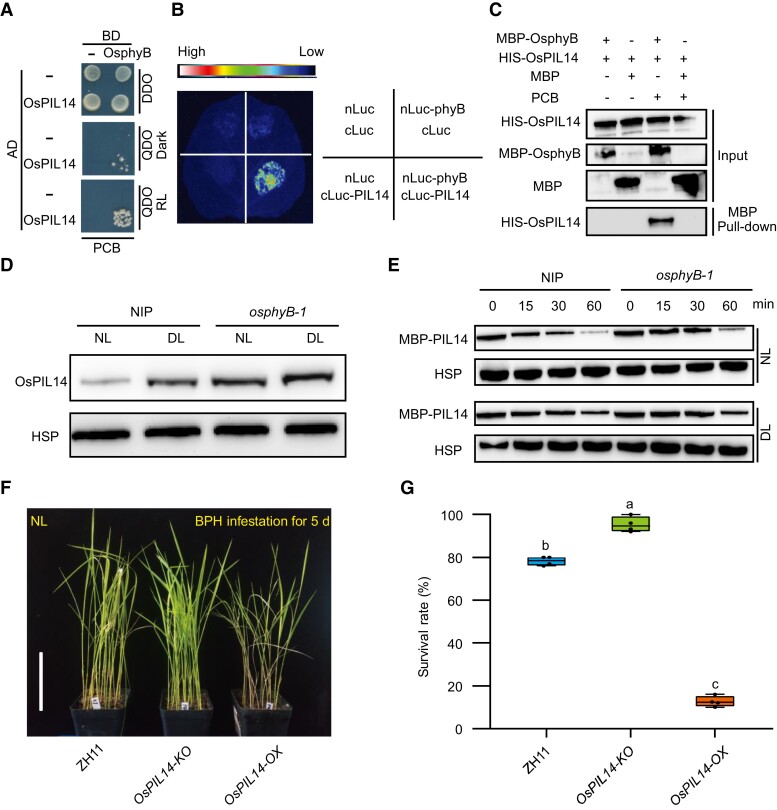
To elucidate the molecular mechanisms of OsphyB regulating the ET pathway, we performed a yeast two-hybrid (Y2H) assay using OsphyB as a bait, and identified OsPIL14 as an interacting partner of OsphyB (Figure 4A). The interaction was further verified using an in vivo split-luciferase (LUC) complementation assay in Nicotiana benthamianan and in vitro pull-down assay (Figure 4, B and C). RT-qPCR assay showed that DL promoted the expression of OsPIL14 in WT plants (Supplemental Figure 9). Consistently, we detected higher accumulation of OsPIL14 protein under DL conditions than under NL conditions (Figure 4D). Intriguingly, though DL-induced OsPIL14 expression was inhibited in the osphyB-1 mutants (Supplemental Figure 9), higher OsPIL14 protein accumulation was detected in the osphyB-1 mutant than in WT under both NL and DL conditions (Figure 4D). Next, we analyzed the effect of OsphyB on the stability of OsPIL14 using a cell-free protein degradation assay. The OsPIL14-MBP recombinant protein was incubated with total cell extracts from WT and osphyB-1 mutant seedlings, respectively. The degradation rate of OsPIL14 was slower when incubated with the cell extracts from osphyB-1 mutant plants than incubated with cell extract from WT (Figure 4E). These results suggest that OsphyB interacts with and facilitates the degradation of OsPIL14.
Figure 4.
OsphyB interacts with and promotes the degradation of OsPIL14. A, Y2H assay shows that OsphyB interacts with OsPIL14 in a red light-dependent manner. DDO, SD-Leu/-Trp. QDO, SD-Leu/-Trp/-His/-Ade. BD, binding domain. Ad, active domain. PCB, phycocyanobilin. B, Split-LUC complementation assay showing the interaction between OsphyB and OsPIL14 in N. benthamiana leaves. cLUC, C terminus of LUC; nLUC, N terminus of LUC. C, In vitro pull-down assay shows that MBP-OsphyB interacts with HIS-OsPIL14. MBP-OsphyB was detected using anti-MBP antibody; HIS-OsPIL14 was detected using anti-HIS antibody. D, OsPIL14 protein levels in two-weeks old seedlings of NIP and osphyB-1 cultivated under NL, which were treated with NL and DL for 4 h before protein extraction. OsHSP indicates loading control. E, Cell-free degradation assay of MBP-OsPIL14 incubated with extracts from two-weeks old seedlings of NIP and osphyB-1 rice plants treated with NL and DL for 4 h. OsHSP was used as a loading control. The experiment was repeated three times. F and G, Representative images (F) and seedling survival rate (G) of WT (ZH11), OsPIL14-KO, and OsPIL14-OX at 5 dpi with BPH under NL. Bar, 7 cm. The center lines of the box plots indicate the median, the bounds of the box show the 25th and the 75th percentiles, and the whiskers indicate 1.5 ×IQR. n ≥ 3 independent experiments, with each pooled with 25 individual plants. Different letters at the top of each box plot indicate a significant difference at P < 0.05, by one-way ANOVA test.
To test whether OsPIL14 participates in BPH resistance, we evaluated BPH resistance of OsPIL14 knockout plants (OsPIL14-KO) and seedlings overexpressing OsPIL14 (OsPIL14-OX) under NL (Figure 4, F and G). As expected, the BPH resistance of OsPIL14-KO and OsPIL14-OX plants was enhanced and weakened, respectively. The results demonstrate that OsPIL14 plays a negative role in light-regulated BPH resistance.
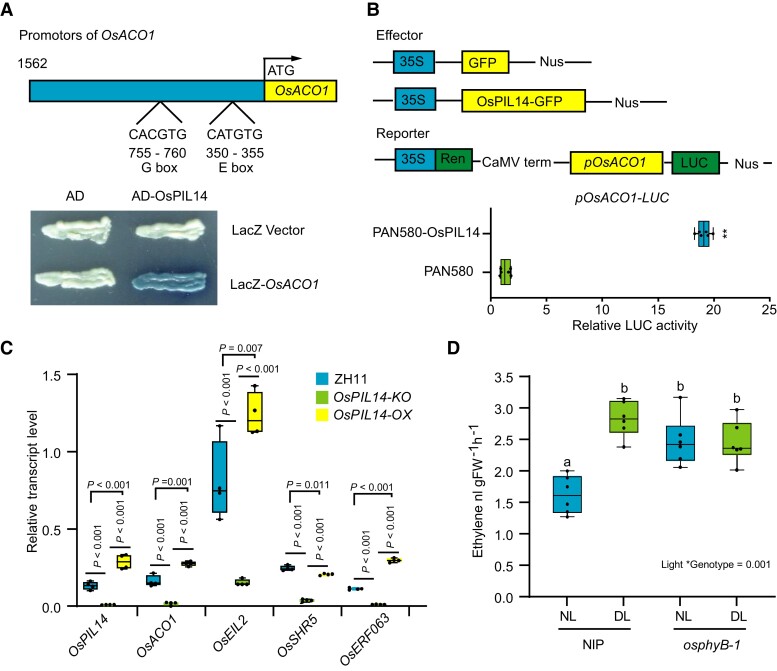
OsPIL14 binds to the promoter of OsACO1 and activates its transcription
Given that both OsACO1 transcription and the OsPIL14 protein level were substantially increased in the osphyB-1 mutant (Figures 2B and 4D), we tested whether OsPIL14 directly activates the transcription of OsACO1. OsPIL14-like proteins acts as transcription factors that prefer to bind to G-box (CACGTG) or E-box (CANNTG) cis-elements (Cordeiro et al., 2016). Thus, we analyzed the promoters of OsACO1, and identified a G-box and an E-box (Figure 5A). We then verified that OsPIL14 bound to the promoter of OsACO1 using a yeast one-hybrid (Y1H) assay (Figure 5A). The LUC/REN ratio was significantly increased when OsPIL14 and OsACO1 promoter-driven LUC were co-introduced compared with the control treatments (Figure 5B), suggesting that OsPIL14 directly binds to the OsACO1 promoter and activates its transcription. To verify this conclusion, we tested the expression of OsACO1 and ET signaling-related genes in the OsPIL14-KO and OsPIL14-OX plants. As expected, the results showed that the expression of OsACO1 and ET signaling-related genes was significantly decreased in the OsPIL14-KO plants, but increased in the OsPIL14-OX plants (Figure 5C). Combining with the results that OsphyB facilitates the degradation of OsPIL14 (Figure 4, D and E) and the expression of OsACO1 were significantly increased in the osphyB mutants (Figure 2B), we demonstrated that OsphyB should repress the expression of OsACO1 by facilitating the degradation of OsPIL14.
Figure 5.
OsPIL14 binds to the promoter and activates transcription of OsACO1. A, YIH assay shows that OsPIL14 binds to the promoters of OsACO1. B, Relative LUC activities in protoplasts transfected with OsPILl4 were compared with that transfected with the empty vector (PAN580). The center lines of the box plots indicate the median, the bounds of the box show the 25th and the 75th percentiles, and the whiskers indicate 1.5 × IQR. n ≥ 3 independent experiments, ** P < 0.01 by Student's t-test. C, RT-qPCR assay showing the expression levels of OsPIL14, OsACO1 and ET-responsive genes in ZH11, OsPIL14-KO and OsPIL14-OX. n ≥ 3 independent experiments, P values were derived by one-way ANOVA test. D, The levels of ET in WT (NIP) and osphyB-1 under NL and DL conditions, respectively. n ≥ 3 independent experiments, Different letters designate significantly different means by 2-way ANOVA + Tukey's post hoc test (P < 0.05). Interaction P value is shown in the inset.
Because light induces phyB translocation into the nucleus (Kircher, 1999; Quail, 2002; Chen et al., 2005a), we hypothesized that the increase in the expression of OsACO1 and the ET content is likely due to less accumulation of OsphyB in the nucleus under DL conditions. Thus, we tested the protein level of OsphyB under NL and DL conditions, and indeed, we detected less OsphyB accumulation in the nucleus under DL conditions than under NL conditions (Supplemental Figure 10). These results suggest that DL conditions reduce OsphyB nuclear accumulation, which stabilizes OsPIL14 in the nucleus, and thus enhancing expression of OsACO1 to promote ET biosynthesis. Consistent with this viewpoint, we found that ET content was significantly increased in the osphyB mutants (Figure 5D).
OsphyB stabilizes OsEIL2 by competitively interacting with OsEBF1
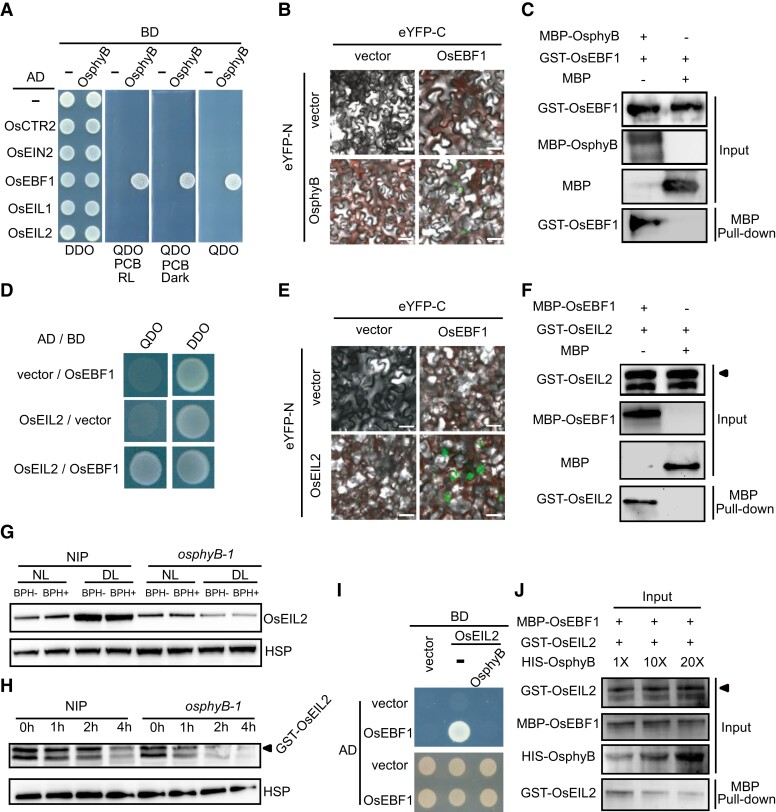
Our Y2H hunting also identified OsEBF1 as an OsphyB interactive partner (Figure 6A). This interaction was also confirmed by bimolecular fluorescence complementation (BiFC) and an in vitro pull-down assay (Figure 6, B and C). Further, we compared the protein levels of OsEBF1 between WT and the osphyB-1 mutant, but found no substantial difference (Supplemental Figure 11A), indicating that OsphyB may not affect the stability of OsEBF1.
Figure 6.
OsphyB stabilizes OsEIL2 by competitively interacting with OsEBF1. A, Y2H assay shows that OsphyB interacts with OsEBF1 independent of light. DDO, SD-Leu/-Trp. QDO, SD-Leu/-Trp/-His/-Ade. BD, binding domain. Ad, active domain. PCB, phycocyanobilin. B, BiFC assay shows that OsphyB interacts with OsEBF1 in Nicotiana benthamiana. eYFPN and eYFPC stand for the N terminus and C terminus of yellow fluorescent protein (YFP), respectively. Bars, 20 µm. C, In vitro pull-down assay shows that MBP-OsphyB interacts with GST-OsEBF1. MBP-OsphyB was detected using anti-MBP antibody; GST-OsEBF1 was detected using anti-GST antibody. D, Y2H assay shows that OsEIL2 interacts with OsEBF1. DO, SD-Leu/-Trp. QDO, SD-Leu/-Trp/-His/-Ade. BD, binding domain. Ad, active domain. E, BiFC assay shows that OsEIL2 interacts with OsEBF1 in Nicotiana benthamiana. Bars, 20 µm. F, In vitro pull-down assay shows that MBP-EBF1 interacts with GST-OsEIL2. G, OsEIL2 protein levels in two-weeks old seedlings of NIP and osphyB-1 with BPH infestation (BPH+) or no BPH infestation (BPH-) under NL and DL for 24 h. OsEIL2 was detected using anti-OsEIL2 antibody. OsHSP indicates loading control. H, Cell-free degradation assay of GST-OsEIL2 incubated with extracts from NIP and osphyB-1 rice plants. OsHSP was used as a loading control. The experiment was repeated three times. I and J, Yeast three-hybrid assay and in vitro pull-down assay show that OsphyB inhibits the interaction between OsEBF1 and OsEIL2. 1×, 10× and 20× indicates the fold of HIS-OsphyB protein.
It was reported that phyB directly interacts with both EIN3 and EBF1, promoting the degradation of EIN3 in Arabidopsis thaliana (Shi et al., 2016). We thus tested whether OsphyB and OsEBF1 interact with OsEIL2, and found that OsEBF1, but not OsphyB, interacted with OsEIL2 (Figure 6, A and D). The interaction between OsEBF1 and OsEIL2 was also verified by BiFC and pull-down assays (Figure 6, E and F). Next, we determined the protein level of OsEIL2 in WT and the osphyB-1 mutant under NL and DL conditions with or without BPH infestation. Consistent with the reduced expression of OsEIL2 downstream genes in the osphyB-1 mutant (Figure 2B), less OsEIL2 accumulation was detected in the osphyB-1 mutant than WT either under NL and DL conditions, and was not affected by BPH infestation (Figure 6G). A cell-free degradation assay also showed that OsphyB has a stabilizing effect on OsEIL2 (Figure 6H). Further, we found higher OsEIL2 accumulation in the OsPHYB-OX plants under DL conditions, compared with the WT (Supplemental Figure 11B). Thus, our results indicate that unlike the role of phyB in promoting the degradation of EIN3 in Arabidopsis thaliana, OsphyB stabilizes OsEIL2 by competitively interacting with OsEBF1 in rice, as verified by a yeast three-hybrid (Y3H) (Figure 6I) and an in vitro pull-down assay (Figure 6J).
To further investigate whether OsphyB suppresses BPH resistance by stabilizing OsEIL2 under DL conditions, we test the influence of exogenous ET and DL treatment on the BPH resistance of OsEIL2 RNAi plants. As expected, ET and DL-reduced BPH resistance were significantly relieved in OsEIL2 RNAi plants compared with WT (Supplemental Figure 12). These results demonstrate ET and DL-reduced BPH resistance should depend on OsEIL2. Further, we tested the sensitivity of the osphyB-1 mutant to ET. Consistent with the findings that OsphyB stabilizes OsEIL2, application of ACC substantially reduced BPH resistance in the WT plants, but not in the osphyB-1 mutant (Figure 3B). In addition, high concentrations of ACC promoted the senescence of the WT seedlings, but had little effect on osphyB-1 (Supplemental Figure 13). These results suggest that the osphyB mutant has reduced sensitivity to ET compared with WT. Thus, we conclude that OsphyB promote the accumulation of OsEIL2 by competitively interacting with OsEBF1 to suppress BPH resistance in rice under DL conditions.
Discussion
Light is one of the most essential environmental factors affecting plant defense against pathogens and herbivores. The resistance to pathogens and herbivores in plants is generally reduced under shade conditions (Ballare, 2014; Pierik and Ballare, 2021), but the underlying mechanism is still not clear. Here, we found that DL-reduced resistance against BPH is dependent on OsphyB in rice. Mechanistically, we show that on one hand, more OsPIL14 proteins are accumulated due to reduced accumulation of biologically active OsphyB in the nucleus under DL conditions, thereby resulting in the up-regulation of OsACO1 and increase of ET content (Figures 4 and 5). This in turn up-regulates the expression of OsEIL2 and promotes ET signal (Figures 2B and 5C). Moreover, OsphyB remaining in the nucleus also interacts with OsEBF1 to block OsEBF1-mediated degradation of OsEIL2, thus stabilizing the ET signaling pathway (Figure 6). Further, we also found that increasing ET biosynthesis and enhancing ET signal substantially reduces the rice resistance to BPH. Thus, our study unravels the molecular mechanism underlying the decreased resistance to BPH caused by DL in rice (Supplemental Figure 14).
Previous studies showed that phyB positively regulates the resistance to pathogens and insects in Arabidopsis thaliana (Campos et al., 2016; Fernández-Milmanda et al., 2020). Surprisingly, our results revealed a negative role of OsphyB in regulating BPH resistance in rice under DL conditions. We collected several lines of evidence to support our claim: 1. BPH resistance of WT were substantially reduced by DL, but DL-triggered reduction in BPH resistance was relieved in the osphyB mutants (osphyB-1, osphyB-2, and osphyB-N4); 2. Compared with the WT, two independent OsPHYB overexpressing transgenic lines were more susceptible to BPH under DL conditions. Notably, the contrasting effects of phyB on insect resistance in Arabidopsis and rice do share similarities to morphological changes of their phyB mutants. Previous studies reported that the Arabidopsis phyB mutants display a constitutive SAS phenotype including elongated internodes phenotype (so do the soybean, maize, and wheat phyB mutants) (Wu et al., 2011; Wies et al., 2019; Kippes et al., 2020). In contrast, the rice phyB mutants show a decreased plant height phenotype (Sun et al., 2017a). In addition to biotic stress, phyB were also reported to positively regulate the tolerance to salinity, drought, and low temperature stresses in Arabidopsis thaliana and tomato (Kim et al., 2002; GonzÁLez et al., 2012; Xu et al., 2019). However, several studies have shown that phyB negatively regulates the tolerance to such stresses in rice (He et al., 2016; Yoo et al., 2017; Kwon et al., 2018). These results indicate that phyB likely plays diverse roles in SAS and stress resistance among different plant species.
Our results also reveal distinct molecular mechanisms regulating the cross talk of phytochrome-mediated light signaling pathway with ET signaling pathway between Arabidopsis and rice. First, it was reported that in Arabidopsis thaliana, light-activated phyB directly interacts with both EIN3 and EBF1/2 to accelerate the degradation of EIN3 and block the ET signaling pathway (Shi et al., 2016). Here we found that OsphyB stabilizes the ET signal pathway by specifically interacting with OsEBF1, but not OsEIL2, to competitively block the interaction of OsEBF1 with OsEIL2 in rice. Consistent with this finding, we also found that the osphyB mutants are more insensitive to exogenous ACC treatment (Supplemental Fig. 13). Secondly, it was reported that in Arabidopsis thaliana, light-activated phyB promotes the degradation of PIFs to repress the expression of ACSs and ET biosynthesis (Khanna et al., 2007; Song et al., 2018), but here we found that the expression of OsACSs was not substantially changed in WT and osphyB. However, OsACO1 expression was substantially increased in the osphyB mutant. Our finding is consistent with previous studies showing that OsACO1 transcription was up-regulated in the osphyB and osphyA osphyB osphyC seedlings (Iwamoto et al., 2011; Iwamoto and Takano, 2011). Moreover, we showed that OsPHYB represses the expression of OsACO1 by regulating the stability of OsPIL14. Together, our findings unveil a mechanism of OsphyB-regulated ET pathway in rice. It will be an interesting avenue to investigate whether the opposite regulatory mechanisms of phyB on ET signaling uncovered in this study broadly affect other developmental processes or responses to other biotic and abiotic responses in Arabidopsis thaliana and rice.
In summary, our results demonstrate that light integrates ET pathway through OsphyB to regulate the BPH resistance in rice (Supplemental Figure 14). Under NL condition, light induces OsphyB to accumulate in nucleus to reduce ET biosynthesis by promoting the degradation of OsPIL14, then down-regulates OsEIL2 to suppress ET signal. Thus, light suppress ET pathway to enhance BPH resistance in rice. Under DL conditions, reduced amount of OsphyB in nucleus results in the accumulation of OsPIL14 protein. OsPIL14 activates the transcription of OsACO1 to increase ET biosynthesis, which up-regulate the expression of OsEIL2. Besides, the OsphyB remaining in the nucleus blocks the degradation of OsEIL2 by competitively binding to OsEBF1. Finally, DL-induced accumulation of OsEIL2 enhances the ET signal and decreases the BPH resistance level in rice. These findings will advance our understanding of the molecular mechanisms underlying light regulating plant–insect interaction. Manipulating OsphyB and/or ET pathways and components may provide new technologies to achieve sustainable insect pest management in crop production.
Materials and methods
Plant materials and growth conditions
Rice (Oryza sativa) mutant or transgenic lines used in this work include osphyA, osphyB-1, osphyB-2, osphyB-N4, osphyC, OsPHYB-OX, OsSAMS1-OX, OsSAMS1-RNAi, osctr2, OsEIN2-OX, OsEIL2-OX, OsEIL2-RNAi, OsPIL14-KO, and OsPIL14-OX. osphyB-1, osphyB-2, osphyA, and osphyC mutants are in the background of japonica cultivar Nipponbare (NIP) and were previously described (Takano et al., 2009). osphyB mutant osphyB-N4 in the background of japonica cultivar Ningjing 4, and OsPHYB overexpressing transgenic plants in the background of japonica cultivar NIP (OsPHYB-OX#2 and OsPHYB-OX#3) were generated in this study. OsSAMS1-OX and OsSAMS1-RNAi are in the background of japonica cultivar Zhonghua 11 (ZH11) (Zhao et al., 2017). osctr2 is an OsCTR2 mutant in the background of japonica cultivar Dongjin (DJ) (Wang et al., 2013). OsEIN2-OX#2, OsEIN2-OX#3, OsEIL2-OX, and OsEIL2-RNAi are in the background of japonica cultivar NIP (Yang et al., 2015b). OsPIL14-KO and OsPIL14-OX transgenic plants are in the background of ZH11 (Mo, et al, 2020). All plants were cultivated under long-day (LD) (14 h: 10 h, light: dark at 28 °C) photoperiods in climate chambers at 60% humidity. Three light intensity conditions were provided by controlling the number of fluorescent white-light tubes, which included DL (dim light, 50 μmol m−2 s−1), NL (normal light, 200 μmol m−2 s−1), and SL (strong light, 650 μmol m−2 s−1) (Liu et al., 2019), respectively.
Maintenance of the brown planthopper (Nilaparvata lugens)
A colony of the BPH was collected from rice fields in Nanjing, and maintained on the susceptible cultivar Taichung Native 1 (TN1) under greenhouse conditions at Nanjing Agricultural University.
Evaluation of rice survival rate in response to BPH
To evaluate the influence of light intensity on BPH resistance, seedlings of wild type, mutants and transgenic lines were first grown for two weeks under NL conditions, pretreated with NL or DL for 24 h, and then infested with BPH and placed in NL or DL conditions, respectively.
BPH resistance was scored according to the standard evaluation systems of International Rice Research Institution (IRRI) (Heinrichs et al., 1985). A bulk seedling test was conducted to evaluate the response of plants to BPH. Seeds were pre-germinated to ensure that the seedlings were at the same growth stage for infestation. Approximately 25 seedlings were grown in 10-cm diameter plastic pots. When the seedlings were at the second-leaf stage, which were thinned to about 20–25 plants per pot and then infested with 2nd- to 3rd-instar BPH nymphs at a density of eight insects per seedling. When the survival rate of the susceptible control plants had reached 10%, the survival rates of the other cultivars and lines were recorded. At least three experiments were performed and each pool including about 20–25 individual plants for each cultivar/transgenic line.
Generation of transgenic plants
The OsPHYB coding region was amplified with gene-specific primers of pCUBi1390-OsPHYB-F/R and cloned into Kpn I and BamH I digested binary vector pCUBi1390 (OsPHYB is driven by the ZmUBI promoter). The recombinant plasmids were transformed into japonica rice cultivar NIP calli by Agrobacterium-mediated method. Hygromycin-resistant calli were grown in artificial incubator to produce transgenic plants. T0 plants were grown in padding field of Nanjing Agricultural University and positive plants were selected and confirmed by PCR and sequencing. The positive plant T2 seedlings were used for BPH resistance evaluation. Primers and restriction sites for vector construction are listed in Supplemental Table 1.
RNA-Sequencing assay
The two weeks old seedling of NIP and osphyB-1 pretreated with NL and DL for 4 h, respectively. Total RNA was extracted using RNAiso Plus (TaKaRa). Total RNA is processed by mRNA enrichment method or rRNA removal method. Enrich the mRNA with polyA tail using magnetic beads with OligodT. Add appropriate amount of interruption reagent to the obtained mRNA to fragment it under high temperature conditions, use the interrupted mRNA as a template to synthesize the first-strand cDNA, and then configure the two-strand synthesis reaction system to synthesize the second-strand cDNA, and use the kit to purify the recovered End repair, add a base “A” to the 3'end of the cDNA and connect the linker, then select the size of the fragment, and finally perform PCR amplification; the quality of the constructed library is checked and sequenced after passing. High-throughput sequencing was conducted on the BGISEQ-500 platform (BGI, Shenzhen) with 150 bp paired-end reads.
The sequencing data were filtered with SOAPnuke (v1.5.2) by (1) Removing reads containing sequencing adapter; (2) Removing reads whose low-quality base ratio (base quality less than or equal to 5) is more than 20%; (3) Removing reads whose unknownbase (‘N” base) ratio is more than 5%, afterwards clean reads were obtained and stored in FASTQ format. The clean reads were mapped to the reference genome using HISAT2 (v2.0.4). After that, Ericscript (v0.5.5) and rMATS (V3.2.5) were used to fusion genes and differential splicing genes (DSGs), respectively. Bowtie2 (v2.2.5) was applied to align the clean reads to the gene set, a database for this organism built by BGI (Beijing Genomic Institute in ShenZhen), for which known and newly coding transcripts were included, then expression level of gene was calculated by RSEM (v1.2.12). The heatmap was drawn by pheatmap (v1.0.8) according to the gene expression in different samples. Essentially, differential expression analysis was performed using the DESeq2(v1.4.5) with Q value ≤ 0.05. To take insight to the change of phenotype, GO (http://www.geneontology.org/) and KEGG (https://www.kegg.jp/) enrichment analysis of annotated different expression gene was performed by Phyper (https://en.wikipedia.org/wiki/Hypergeometric_distribution) based on Hypergeometric test. The significant levels of terms and pathways were corrected by Q value with a rigorous threshold (Q value ≤ 0.05) by Bonferroni.
Hormone treatments
To evaluate the effects of ET on the BPH resistance of rice, the rice seeds were surface sterilized and germinated in water for 2 d, placed on the petri dishes (10 × 10 cm) with two layers of gauze, incubated in Hoagland solution (pH 6) for eight or two weeks under NL, and treated Hoagland solution containing 50 μM ACC or 50 μM pyrazinamide (PZA) for 2 d. The control group was Hoagland solution only. Then, the seedlings were used for BPH resistance evaluation.
For the ethylene-response assay, seeds were soaked in water at 37 °C for 2 d, and the germinated seeds were placed on stainless nets with a water level below the seeds. High ACC concentration (200 μM) was added into the Hoagland solution. The seedlings were grown at 28 °C in the NL for 5 d. The senescence phenotype of rice was monitored.
RNA extraction and Rt-qPCR
Total RNA was extracted with RNA prep pure Plant kit (TIANGEN Biotech, Beijing, China). RNA was reversed transcribed with PrimeScript Reverse Transcriptase kit (Takara, Otsu, Japan). RT-qPCR reactions were carried out on an ABI PRISM 7,500 device using a SYBR Premix Ex Taq RT–PCR Kit (Takara). Relative transcript levels were calculated using the 2−ΔΔCT method as previously described (Livak and Schmittgen, 2001). The rice Ubiquitin gene (Os03g0234350) was used as an internal control. The primers for RT-qPCR analysis are listed in Supplemental Table 1.
OsPIL14, OsEIL2, and OsphyB protein levels analysis
Samples were ground into fine powder in liquid nitrogen, and then suspended in 2 × volumes of protein extraction buffer (100 mM Tris–HCl pH 8.0, 10 mM MgCl2, 18% (w/v) sucrose, 40 mM β-mercaptoethanol and 1 × Protease inhibitor cocktail) and incubated at 4 °C for 30 min with rotation. After centrifuging at 12,000 g and 4 °C for 10 min, the supernatant was boiled in 5 × SDS sample buffer (250 mM Tris–HCl, PH = 6.8, 10% (w/v) SDS, 0.5% (w/v) bromophenol blue, 50% (v/v) glycerin, 5% (v/v) β-mercaptoethanol). Protein samples were separated on 10% SDS-PAGE gel and then transferred to the NC membrane, detected with antibodies against OsPIL14, OsEIL2, and OsphyB. OsPIL14, OsEIL2, and OsphyB polyclonal antibodies were created by Abclonal (Wuhan Abclonal, WG-03044). Anti-OsHSP antibody (Beijing Protein Innovation, AbM51099–31-PU) was used as an internal reference.
Cell-free degradation
Rice leaves were harvested and ground into fine powder in liquid nitrogen. Total proteins were subsequently extracted with a degradation buffer containing 25 mM Tris–HCl, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 5 mM DTT, as previously described (Wang et al., 2009). The extract was centrifuged at 12,000 rpm for 10 min. The supernatant was collected and adjusted to equal concentration with the degradation buffer. For OsPIL14-MBP, OsEBF1-MBP, and OsEIL2-GST protein degradation, about 10 µg of purified proteins were separately mixed with 40 μl rice total-protein extract at 28 °C. The reactions were incubated at 28 °C for 1 h, then 5 × SDS sample buffer (250 mM Tris–HCl, PH = 6.8, 10% (w/v) SDS, 0.5% (w/v) bromophenol blue, 50% (v/v) glycerin, 5% (v/v) β-mercaptoethanol) was added to stop the reactions. OsPIL14-MBP and OsEBF1-MBP proteins were detected by western blots with the HRP-conjugated Anti-MBP antibody at 1:10,000 dilution. EIL2-GST proteins were detected by western blots with the HRP-conjugated Anti-GST monoclonal antibody at 1:5,000 dilution. Primers and restriction sites for vector construction are listed in Supplemental Table 1.
Nuclear and cytoplasmic protein isolation and immunoblot analysis
Ten-day-old NIP seedlings were placed under NL and DL conditions for 3 h, respectively. The proteins of nucleus and cytoplasm were extracted using Plant Nuclear and Cytoplasmic Protein Extraction Kit (Bestbio, Shanghai, China). Both nuclear and cytoplasmic proteins were quantified using Bradford assay, and 40 µg of the proteins was subjected to SDS-PAGE. The immunoblot experiment was performed using the antibodies (1:2,000 dilution, Wuhan Abclonal, WG-03044) raised against OsphyB, β-Actin (1:5,000 dilution; EarthOx, San Francisco, CA) and Histone 3 (1:5,000 dilution; EarthOx) as the primary antibodies, and the horseradish peroxidase conjugated goat anti-rabbit/mouse IgG (1:3,000 dilution; Sungene Biotechnology, Tian Jin, China) as the secondary antibody.
Yeast one-hybrid assay
For Y1H assay, about 1.5 kb promoter region of OsACO1 was amplified and cloned into the Y1H vector pLacZi (Lin et al., 2007). Full-length of OsPIL14 (NCBI accession No. LOC4342383) was amplified and inserted into the pGADT7 (prey) vector (Clontech, United States) to produce the pGADT7-OsPIL14 construct. The bait plasmids were linearized and transformed into the yeast strain Y1HGold. Positive yeast cells were then transformed with pGADT7-OsPIL14 plasmid. The DNA–protein interaction was determined based on the growth ability of the co-transformants on SD/-Leu medium with Aureobasidin A (AbA). Primers and restriction sites for vector construction are listed in Supplemental Table 1.
LUC assays
For dual-LUC analysis, the promoter of OsACO1 was amplified and cloned into the pGreenII 0800-LUC vector (Zhou et al., 2013). The CDS of OsPIL14 was amplified and cloned into the PAN580 vectors to generate the effector plasmid. The plasmids were transiently expressed in rice protoplasts and the LUC activity was measured using the Dual-Luciferase Reporter Assay System (Promega, E2920). Primers and restriction sites for vector construction are listed in Supplemental Table 1.
Y2H assay
The CDSs of OsPHYB, OsPIL14, OsEBF1, and OsEIL2 were amplified from cDNA of NIP using primers listed in Supplemental Table 1. After sequencing confirmation, OsEBF1 and OsPHYB were cloned into the “BD” vector pGBKT7 (Takara). OsPHYB, OsPIL14, OsCTR2, OsEIN2, OsEBF1, OsEIL1, and OsEIL2 were cloned into the “Ad” vector pGADT7 (Takara). Both the BD and Ad constructs were co-transformed into the yeast (Saccharomyces cerevisiae) strain AH109 (Takara). The resultant yeast strains with OsphyB were suspended in SD-Leu/-Trp liquid culture medium with 20 µM phycocyanobilin (Frontier Scientific, Logan, UT, USA) and incubated for 12 h at 30 °C in darkness. After phycocyanobilin combined with apophytochromes, all of the resultant yeast stains were grown on DDO (SD-Leu/-Trp) plates for 3 d at 30 °C under light. Interactions between the baits and preys were examined on selective media QDO (SD-Leu/-Trp/-His/-Ade). Plates were incubated for 4 d at 30 °C. Primers and restriction sites for vector construction are listed in Supplemental Table 1.
Bimolecular Fluorescence Complementation (BiFC) assay
For BiFC assay, the CDSs of OsPHYB and OsEIL2 were amplified and cloned into the p2YN vector to produce OsPHYB-p2YN and OsEIL2-p2YN, respectively. CDS of OsEBF1 was amplified and inserted into the p2YC vector to produce OsEBF1-p2YC. For transient expression, the recombinant BiFC vectors were then co-transformed together with the virus silencing suppressor Tombusvirus P19 into five-weeks-old Nicotiana benthamiana leaves by A. tumefaciens-mediated transformation as described previously (Waadt and Kudla, 2008). For light treatment, after N. benthamiana leaves were infiltrated, N. benthamiana plants were subsequently incubated under LD conditions for 2 d, then in white light for additional 2 h. eYFP (YFP, yellow fluorescent protein) fluorescent signal of the infiltrated leaves were monitored sequentially using a laser confocal scanning microscope. Fluorescence was excited with the X-Cite Series 120 fluorescence lamp (EXFO) and images were collected at 500–550 nm (eGFP fluorescence) and 670–750 nm (Chl autofluorescence). Primers and restriction sites for vector construction are listed in Supplemental Table 1.
LUC complementation assay
The LUC complementation assay was conducted as previously described (Chen et al., 2008). The CDS of OsPHYB was amplified and cloned into the nLuc vector to produce OsPHYB-nLuc. CDS of OsPIL14 were amplified and inserted into the cLuc vector to produce OsPIL14-cLuc. Agrobacteria cells harboring the nLUC and cLUC constructs were co-infiltrated into N. benthamiana leaves. Leaves were harvested 2 d later, 525 incubated with 1 mM luciferin, and the luminescence activity was recorded by the NightSHADE LB 985 (Berthold). Primers and restriction sites for vector construction are listed in Supplemental Table 1.
In vitro pull-down assays
The CDSs of OsPHYB and OsEBF1 were amplified and cloned into the pMal-c2 × vector to generate the OsPHYB-pMal-c2 × and OsEBF1-pMal-c2 × construct, respectively. The CDSs of OsPIL14 was amplified and cloned into the PET-30a to generate the OsPIL14− PET-30a construct, respectively. In addition, CDSs of OsEBF1 and OsEIL2 were amplified and cloned into pGEX-4T-1 to generate OsEBF1-pGEX− 4T-1 and OsEIL2-pGEX-4T-1, respectively. Expression of Maltose-Binding Protein (MBP), MBP-OsphyB, MBP-OsEBF1, HIS-OsPIL14, GST-OsEBF1, and GST-OsEIL2 in BL21 Rosetta cells was induced with 0.5 mM IPTG at16 °C for 16 h. Total proteins were subsequently extracted in a binding buffer (20 mM Tris–HCl, pH 8.0, 200 mM NaCl, and 0.5% Nonidet P40). The total-protein concentration was determined using the Bio-Rad protein assay (Bio-Rad). MBP-OsphyB fusion proteins were incubated with 20 µM phycocyanobilin for 2 h.
For in vitro pull-down, MBP-OsphyB and MBP-OsEBF1 recombination proteins (1 µg) were immobilized to Amylose Resin (New England Biolabs, Ipswich, MA, USA). Lysate of E. coli cells expressing HIS-OsPIL14, GST-OsEBF1, and GST-OsEIL2 (containing 2 mg total proteins) were incubated with Amylose Resin immobilized with MBP fusion proteins at 4 °C for 3 h. The resin was washed five times with the binding buffer. After washing, 50 µl of 1×SDS-PAGE sample buffer was added, then the mixture was denatured. The sample was loaded on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, and the GST and HIS fusion proteins were detected using the HRP-conjugated anti-GST and HIS antibody (1: 10,000, cat. no. #E8038S; New England Biolabs), respectively. The MBP fusion proteins were detected using anti-MBP antibody (1: 5,000, cat. no. A00865–100; GenScript, Nanjing, China) and anti-Mouse IgG antibody (1: 5,000, cat. no. CW0110; Kang Wei Biotech, Beijing, China). Signals of the GST, HIS, and MBP fusion proteins were visualized with enhanced chemiluminescence reagent (Tanon, Shanghai, China). Primers and restriction sites for vector construction are listed in Supplemental Table 1.
Y3H assay
The CDSs of OsPHYB and OsEIL2 were amplified with primers listed in Supplemental Table 1 and cloned into the pBridge vector to produce OsEIL2-pBridge and OsEIL2-OsPHYB-pBridge, respectively. pBridge and pGADT7-OsEBF1 constructs were co-transformed into the S. cerevisiae strain AH109. Then all of the resultant yeast stains were grown on DDO (SD-Leu/-Trp) plates for 3 d at 30 °C under light. Interactions between the baits and preys were examined on selective media QDO (SD-Leu/-Trp/-3His/-Ade). Plates were incubated for 4 d at 30 °C. Primers and restriction sites for vector construction are listed in Supplemental Table 1.
Measurement of ET emission
Two-week-old rice seedlings were pretreated for 24 h under NL and DL conditions, respectively. Then the fourth leaves of rice seedling were detached from the base of the leaf blade. Twenty leaves were put into 40-mL glass vials with 1 mL of water, sealed with a gas-proof septum, and left in a growth cabinet at 24 °C for 3 h under light. One milliliter of gas was withdrawn from the airspace of each tube using a gas-tight syringe (Hamilton) and injected into a gas chromatograph (Shimadzu GC-14B) equipped with an aluminum column (Shumpak-A; Shimazu) and a flame-ionization detector for ET determination.
Accession numbers
Sequence from this study can be downloaded from the rice genome annotation project (http://rice.plantbiology.msu.edu/) with the following accession numbers: OsPHYB, Os03g0309200; OsPIL14, Os07g0143200; OsEBF1, Os06g0605900; OsEIL2, Os07g0685700; OsEIN2, Os07g0155600; OsSAMS1, Os05g0135700; OsACS2, Os04g0578000; OsACO1, Os09g0451000; OsEIL1, Os03g0324300; OsSHR5, Os08g0203400; OsERF063, Os09g0287000; OsERF073, Os09g0286600; OsUBQ, Os03g0234350.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Light intensity positively affects BPH resistance.
Supplemental Figure S2. The mutational sites of the osphyB mutants.
Supplemental Figure S3. BPH resistance in the osphyB-2 mutant under different light intensities.
Supplemental Figure S4. BPH resistance in NIP and OsPHYB-OX lines under DL condition.
Supplemental Figure S5. DL up-regulates the expression of genes involved in hormone signaling.
Supplemental Figure S6. Transcript analysis of genes transcriptionally regulated by OsEIL2.
Supplemental Figure S7. The influence of ET on the transcript level of OsEIL2 and protein accumulation.
Supplemental Figure S8. ET negatively regulates the resistance against BPH in rice.
Supplemental Figure S9. Transcript analysis of OsPIL14 in NIP and osphyB-1 under NL and DL conditions.
Supplemental Figure S10. DL reduces the accumulation of OsphyB in the nucleus.
Supplemental Figure S11. The effect of OsphyB on the stability of OsEBF1 and the accumulation of OsEIL2.
Supplemental Figure S12. ET and DL-reduced BPH resistance dependent upon OsEIL2.
Supplemental Figure S13. The osphyB mutant is less sensitive to exogenous ET.
Supplemental Figure S14. A working model of light integrated ET pathway regulating BPH resistance in rice.
Supplemental Table S1. Primers used in this study.
Supplemental Dataset S1. Supplemental Figure S5 data: 1186 up-regulated genes in DN vs LN.
Supplemental Dataset S2. Supplemental Figure S5 data: 868 down-regulated genes in DN vs LN.
Supplemental Dataset S3. Supplemental Figure S5 data: 44 hormone signal transduction genes in Fig S5A.
Supplementary Material
Acknowledgments
We owe special thanks to Professor Yi Li (State Key Laboratory of Protein and Plant Gene Research, College of Life Sciences, Peking University, Beijing) for providing OsSAMS1-OX and OsSAMS1-RNAi, to Professor Chikuang Wen (National Key Laboratory of Plant Molecular Genetics and National Center for Plant Gene Research, Shanghai) for providing osctr2, to Professor Jinsong Zhang (State Key Laboratory of Plant Genomics, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing) for providing OsEIL2-OX and OsEIL2-RNAi, to Professor Rongcheng Lin (Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing) for providing OsPIL14-KO and OsPIL14-OE, to Professor Dongqing Xu (State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University) for proofreading the manuscript. We also acknowledge support from the Jiangsu plant gene editing Engineering Research Center and the Southern Japonica Rice Research and Development Co., Ltd.
Contributor Information
Jie Huang, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Province and Ministry Co-sponsored Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Ze-Yu Qiu, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Province and Ministry Co-sponsored Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Jun He, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Province and Ministry Co-sponsored Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Hao-Sen Xu, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Province and Ministry Co-sponsored Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Kan Wang, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Province and Ministry Co-sponsored Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Hua-Ying Du, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Province and Ministry Co-sponsored Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Dong Gao, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Province and Ministry Co-sponsored Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Wei-Ning Zhao, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Province and Ministry Co-sponsored Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Quan-Guang Sun, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Province and Ministry Co-sponsored Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Yong-Sheng Wang, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Province and Ministry Co-sponsored Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Pei-Zheng Wen, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Province and Ministry Co-sponsored Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Qi Li, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Province and Ministry Co-sponsored Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Xiao-Ou Dong, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Province and Ministry Co-sponsored Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Xian-Zhi Xie, Shandong Rice Research Institute, Shandong Academy of Agricultural Sciences, Jinan 250100, China.
Ling Jiang, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Province and Ministry Co-sponsored Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Hai-Yang Wang, National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing 100081, China.
Yu-Qiang Liu, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Province and Ministry Co-sponsored Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Jian-Min Wan, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Province and Ministry Co-sponsored Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China; National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing 100081, China.
Funding
This work was funded by the National Natural Science Foundation of China (32088102, 32072030), Major Project of Natural Science Foundation of Jiangsu Province (BK20212010), Jiangsu Provincial Key Research Program (BE2019380), The “JBGS” Project of Seed Industry Revitalization in Jiangsu Province (JBGS202108), Hainan Yazhou Bay Seed Lab (B21HJ1004), and the Fundamental Research Funds for the Central Universities (JCQY201902).
References
- Abeles F (1992). Ethylene in plant biology. Cell 72: 11–12 [Google Scholar]
- Augspurger C, Kelly C (1984) Pathogen mortality of tropical tree seedlings: experimental studies of the effects of dispersal distance, seedling density, and light conditions. Oecologia 60: 211–217 [DOI] [PubMed] [Google Scholar]
- Ballare C (2014) Light regulation of plant defense. Annu Rev Plant Biol 65: 335–363 [DOI] [PubMed] [Google Scholar]
- Bell T, Freckleton R, Lewis O (2006) Plant pathogens drive density-dependent seedling mortality in a tropical tree. Ecol Lett 9: 569–574 [DOI] [PubMed] [Google Scholar]
- Campos M, Yoshida Y, Major I, de Oliveira Ferreira D, Weraduwage S, Froehlich J, Johnson B, Kramer D, Jander G, Sharkey T, et al. (2016) Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat Commun 7: 12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J (2012) Shade avoidance. Arabidopsis Book 10: e0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catindig J, International Rice Research Institute, Manila, Philippines . (2009). Planthoppers: new treats to the sustainability of intensive rice production systems in Asia, 191–220
- Cerrudo I, Keller M, Cargnel M, Demkura P, de Wit M, Patitucci M, Pierik R, Pieterse C, Ballare C (2012) Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiol 158: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Etheridge N, Schaller G (2005b) Ethylene signal transduction. Ann Bot 95: 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Tao Y, Lim J, Shaw A, Chory J (2005a) Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr Biol 15: 637–642 [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou J (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Zhu L, He G (2013) Towards understanding of molecular interactions between rice and the brown planthopper. Mol Plant 6: 621–634 [DOI] [PubMed] [Google Scholar]
- Cordeiro A, Figueiredo D, Tepperman J, Borba A, Lourenco T, Abreu I, Ouwerkerk P, Quail P, Margarida Oliveira M, Saibo N (2016) Rice phytochrome-interacting factor protein OsPIF14 represses OsDREB1B gene expression through an extended N-box and interacts preferentially with the active form of phytochrome B. Biochim Biophys Acta 1859: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2020). FAOSTAT agriculture data. Available online at: http://www.fao.org/faostat/ar/#home
- Fernández-Milmanda G, Ballaré C (2021) Shade avoidance: expanding the color and hormone palette. Trends Plant Sci 26: 509–523 [DOI] [PubMed] [Google Scholar]
- Fernández-Milmanda G, Crocco C, Reichelt M, Mazza C, Köllner T, Zhang T, Cargnel M, Lichy M, Fiorucci A, Fankhauser C, et al. (2020) A light-dependent molecular link between competition cues and defence responses in plants. Nat Plants 6: 223–230 [DOI] [PubMed] [Google Scholar]
- Franklin K (2008) Shade avoidance. New Phytol 179: 930–944 [DOI] [PubMed] [Google Scholar]
- GonzÁLez C, Ibarra S, Piccoli P, Botto J, Boccalandro H (2012) Phytochrome B increases drought tolerance by enhancing ABA sensitivity in Arabidopsis thaliana. Plant Cell Environ 35: 1958–1968 [DOI] [PubMed] [Google Scholar]
- He Y, Li Y, Cui L, Xie L, Zheng C, Zhou G, Zhou J, Xie X (2016) Phytochrome B negatively affects cold tolerance by regulating OsDREB1 gene expression through phytochrome interacting factor-like protein OsPIL16 in rice. Front Plant Sci 7: 1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs E, Medrano F, Rapusas H (1985) Genetic Evaluation for Insect Resitance in Rice. International Rice Research Institute, Philippines [Google Scholar]
- Hoon J, Han M, Lee S, Seo Y, Kim W, An G (2004) OsEIN2 is a positive component in ethylene signaling in rice. Plant Cell Physiol 45: 281–289 [DOI] [PubMed] [Google Scholar]
- Iwamoto M, Kiyota S, Hanada A, Yamaguchi S, Takano M (2011) The multiple contributions of phytochromes to the control of internode elongation in rice. Plant Physiol 157: 1187–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Takano M (2011) Phytochrome-regulated EBL1 contributes to ACO1 upregulation in rice. Biotechnol Lett 33: 173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom T, Burson B, Finlayson S (2006) Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol 140: 1109–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Marion C, Tsuchisaka A, Theologis A, Schafer E, Quail P (2007) The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19: 3915–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Yun K, Park J, Kim J (2002) Light signalling mediated by phytochrome plays an important role in cold-induced gene expression through the C-repeat/dehydration responsive element (C/DRE) in Arabidopsis thaliana. Plant J 29: 693–704 [DOI] [PubMed] [Google Scholar]
- Kippes N, VanGessel C, Hamilton J, Akpinar A, Budak H, Dubcovsky J, Pearce S (2020) Effect of phyB and phyC loss-of-function mutations on the wheat transcriptome under short and long day photoperiods. BMC Plant Biol 20: 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S (1999) Light quality–dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Song G, Kim S, Han J, Yoo S, An G, Kang K, Paek N (2018) Functional deficiency of phytochrome B improves salt tolerance in rice. Environ Exp Bot 148: 100–108 [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll D, Feschotte C, Wang H (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wu H, Chen H, Liu Y, He J, Kang H, Sun Z, Pan G, Wang Q, Hu J, et al. (2015) A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat Biotechnol 33: 301–305 [DOI] [PubMed] [Google Scholar]
- Liu M, Zhang S, Hu J, Sun W, Padilla J, He Y, Li Y, Yin Z, Liu X, Wang W, et al. (2019) Phosphorylation-guarded light-harvesting complex II contributes to broad-spectrum blast resistance in rice. PNAS 116: 17572–17577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K, Schmittgen T (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lu J, Ju H, Zhou G, Zhu C, Erb M, Wang X, Wang P, Lou Y (2011) An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. Plant J 68: 583–596 [DOI] [PubMed] [Google Scholar]
- Lu J, Li J, Ju H, Liu X, Erb M, Wang X, Lou Y (2014) Contrasting effects of ethylene biosynthesis on induced plant resistance against a chewing and a piercing-sucking herbivore in rice. Mol Plant 7: 1670–1682 [DOI] [PubMed] [Google Scholar]
- Ma B, Chen S, Zhang J (2010) Ethylene signaling in rice. Chinese Science Bulletin 55: 2204–2210 [Google Scholar]
- Ma F, Yang X, Shi Z, Miao X (2019) Novel crosstalk between ethylene- and jasmonic acid-pathway responses to a piercing-sucking insect in rice. New Phytol 225: 474–487 [DOI] [PubMed] [Google Scholar]
- Marta B, Antonio M (2004) Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol Plant Microbe Interact 17: 763–770 [DOI] [PubMed] [Google Scholar]
- Mo W, Tang W, Du Y, Jing Y, Bu Q, Lin R (2020) PHYTOCHROME-INTERACTING FACTOR-LIKE14 and SLENDER RICE1 interaction controls seedling growth under salt stress. Plant Physiol 184: 506–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M, Fankhauser C, Chory J (2000) Light: an indicator of time and place. Genes Dev 14: 257–271 [PubMed] [Google Scholar]
- Onkokesung N, Baldwin I, Galis I (2010) The role of jasmonic acid and ethylene crosstalk in direct defense of Nicotiana attenuata plants against chewing herbivores. Plant Signal Behav 5: 1305–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, Ballare C (2021) Control of plant growth and defense by photoreceptors: from mechanisms to opportunities in agriculture. Mol Plant 14: 61–76 [DOI] [PubMed] [Google Scholar]
- Quail P (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Roberts M, Paul N (2006) Seduced by the dark side: integrating molecular and ecological perspectives on the influence of light on plant defence against pests and pathogens. New Phytol 170: 677–699 [DOI] [PubMed] [Google Scholar]
- Shi H, Shen X, Liu R, Xue C, Wei N, Deng X, Zhong S (2016) The red light receptor phytochrome B directly enhances substrate-E3 ligase interactions to attenuate ethylene responses. Dev Cell 39: 597–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Ando A, Xu D, Fang L, Zhang T, Huq E, Qiao H, Deng X, Chen Z (2018) Diurnal down-regulation of ethylene biosynthesis mediates biomass heterosis. PNAS 115: 5606–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Li Y, He W, Ji C, Xia P, Wang Y, Du S, Li H, Raikhel N, Xiao J, et al. (2017b) Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase. Nat Commun 8: 15758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Xu X, Lu X, Xie L, Bai B, Zheng C, Sun H, He Y, Xie X (2017a) The rice phytochrome genes, PHYA and PHYB, have synergistic effects on anther development and pollen viability. Sci Rep 7: 6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano M, Inagaki N, Xie X, Kiyota S, Baba-Kasai A, Tanabata T, Shinomura T (2009) Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice. PNAS 106(34): 14705–14710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Kudla J (2008) In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). CSH Protoc 2008: pdb prot4995. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhang W, Yin Z, Wen C (2013) Rice CONSTITUTIVE TRIPLE-RESPONSE2 is involved in the ethylene-receptor signalling and regulation of various aspects of rice growth and development. J Exp Bot 64: 4863–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhu D, Huang X, Li S, Gong Y, Yao Q, Fu X, Fan L, Deng X (2009) Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21: 2378–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Zhao Y, Xie Y, Wang H (2018) Exploiting SPL genes to improve maize plant architecture tailored for high-density planting. J Exp Bot 69: 4675–4688 [DOI] [PubMed] [Google Scholar]
- Wies G, Mantese A, Casal J, Maddonni G (2019) Phytochrome B enhances plant growth, biomass and grain yield in field-grown maize. Ann Bot 123: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhang X, Li D, Fu Y (2011) Ectopic expression reveals a conserved PHYB homolog in soybean. PLoS One 6: e27737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Yang Y, Yang J, Zhang X, Pan Y, Guo H (2021) IAA3-mediated Repression of PIF proteins coordinates light and auxin signaling in Arabidopsis. PLoS Genet 17: e1009384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Liu Y, Wang H, Ma X, Wang B, Wu G, Wang H (2017) Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat Commun 8: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Marino G, Klingl A, Enderle B, Monte E, Kurth J, Hiltbrunner A, Leister D, Kleine T (2019) Extrachloroplastic PP7L functions in chloroplast development and abiotic stress tolerance. Plant Physiol 180: 323–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Lu X, Ma B, Chen S, Zhang J (2015a) Ethylene signaling in rice and Arabidopsis: conserved and diverged aspects. Mol Plant 8: 495–505 [DOI] [PubMed] [Google Scholar]
- Yang C, Ma B, He S, Xiong Q, Duan K, Yin C, Chen H, Lu X, Chen S, Zhang J (2015b) MAOHUZI6/ETHYLENE INSENSITIVE3-LIKE1 and ETHYLENE INSENSITIVE3-LIKE2 regulate ethylene response of roots and coleoptiles and negatively effect salt tolerance in rice. Plant Physiol 169: 148–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo Y, Nalini Chandran A, Park J, Gho Y, Lee S, An G, Jung K (2017) OsPhyB-mediating novel regulatory pathway for drought tolerance in rice root identified by a global RNA-seq transcriptome analysis of rice genes in response to water deficiencies. Front Plant Sci 8: 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Hong W, Wu J, Wang Y, Ji S, Zhu S, Wei C, Zhang J, Li Y (2017) A viral protein promotes host SAMS1 activity and ethylene production for the benefit of virus infection. Elife 6: e27529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, et al. (2013) D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504: 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.