Abstract
Biphasic production of reactive oxygen species (ROS) has been observed in plants treated with avirulent bacterial strains. The first transient peak corresponds to pattern-triggered immunity (PTI)-ROS, whereas the second long-lasting peak corresponds to effector-triggered immunity (ETI)-ROS. PTI-ROS are produced in the apoplast by plasma membrane-localized NADPH oxidases, and the recognition of an avirulent effector increases the PTI-ROS regulatory module, leading to ETI-ROS accumulation in the apoplast. However, how apoplastic ETI-ROS signaling is relayed to the cytosol is still unknown. Here, we found that in the absence of cytosolic ascorbate peroxidase 1 (APX1), the second phase of ETI-ROS accumulation was undetectable in Arabidopsis (Arabidopsis thaliana) using luminol-based assays. In addition to being a scavenger of cytosolic H2O2, we discovered that APX1 served as a catalyst in this chemiluminescence ROS assay by employing luminol as an electron donor. A horseradish peroxidase (HRP)-mimicking APX1 mutation (APX1W41F) further enhanced its catalytic activity toward luminol, whereas an HRP-dead APX1 mutation (APX1R38H) reduced its luminol oxidation activity. The cytosolic localization of APX1 implies that ETI-ROS might accumulate in the cytosol. When ROS were detected using a fluorescent dye, green fluorescence was observed in the cytosol 6 h after infiltration with an avirulent bacterial strain. Collectively, these results indicate that ETI-ROS eventually accumulate in the cytosol, and cytosolic APX1 catalyzes luminol oxidation and allows monitoring of the kinetics of ETI-ROS in the cytosol. Our study provides important insights into the spatial dynamics of ROS accumulation in plant immunity.
Ascorbate peroxidase 1 catalyzes luminol-based chemiluminescence assays and allows monitoring of effector-triggered reactive oxygen species that accumulate in the cytosol.
Introduction
Plants have evolved a sophisticated innate immune system as a protection mechanism against pathogen attacks. When pathogens reach the physical barriers on the plant surface, immunity-related responses are initiated by the cell surface receptor-mediated recognition of pathogen-associated molecular patterns (PAMPs), thereby activating PAMP-triggered immunity (PTI) (Boller and Felix, 2009; Zipfel, 2014; Bigeard et al., 2015). Through long-term interactions with their hosts, pathogens have evolved specific effector proteins. These are in turn recognized by intracellular nucleotide-binding, leucine-rich repeat receptors in plants, activating effector-triggered immunity (ETI) (Jones and Dangl, 2006; Wang et al., 2019; Ma et al., 2020). Although PTI and ETI are initiated by 2 classes of receptors, they both use similar signals, including the influx of calcium, production of reactive oxygen species (ROS), and activation of mitogen-activated protein kinases (MAPKs) (Qi et al., 2011; Yuan et al., 2021b). Notably, PTI and ETI signals are mutually potentiated to activate strong immune responses to arrest pathogens (Ngou et al., 2021; Yuan et al., 2021a).
ROS are not only the by-products of normal metabolism during plant growth and development but also act as signaling molecules in response to many environmental stresses (Mittler et al., 2011; Suzuki et al., 2012; Baxter et al., 2014; Camejo et al., 2016; Mittler, 2017; Qi et al., 2017). Previous studies have reported biphasic ROS production in plants inoculated with avirulent bacterial strains; however, the kinetics of the 2 ROS bursts are very different (Doke, 1983; Baker and Orlandi, 1995; Lamb and Dixon, 1997; Mur et al., 2009). The first ROS peak within 1 h corresponds to PTI-ROS, whereas the second instance of ROS accumulation after approximately 8–24 h corresponds to ETI-ROS (Doke, 1983; Baker and Orlandi, 1995; Lamb and Dixon, 1997). Unlike most typical PAMPs, lipopolysaccharides (LPS), abundant in the outer cell envelope of gram-negative bacteria, trigger a biphasic ROS burst (Shang-Guan et al., 2018; Li et al., 2019, 2021; Wu et al., 2022). The transient first-phase ROS peak at approximately 0.5 h is similar to the PTI-ROS peak but much weaker than that induced by flg22 (a 22 amino acid peptide derived from bacterial flagellin). The strong second-phase ROS burst peaks at approximately 8 h and continues for several hours, a profile that bears an intense similarity to previous observations during bacteria-triggered ETI (Levine et al., 1994; Chandra et al., 1996; Shang-Guan et al., 2018; Li et al., 2021; Wu et al., 2022). Similar patterns of biphasic ROS production have been reported in plants subjected to free medium-chain 3-hydroxy fatty acids (e.g. 3-hydroxydecanoic acid, 3-OH-C10:0), identified as a contaminant in LPS preparations; however, the second peak occurs earlier and is lower than that induced by LPS (Kutschera et al., 2019; Li et al., 2021). Abiotic stresses, such as ozone and mild salt concentrations, can also trigger a biphasic ROS burst in Arabidopsis (Arabidopsis thaliana), in which ROS production reaches the first and second peaks at approximately 1 h and 6–18 h after treatment, respectively. In these biphasic ROS production patterns, the first peak is usually transient and rapid, whereas the second peak is slow and prolonged (Joo et al., 2005; Xie et al., 2011). In Arabidopsis, ROS production during an immune response is mainly mediated by the respiratory burst oxidase homolog D (RBOHD), a plasma membrane nicotinamide adenine dinucleotide phosphate (NADPH) oxidase (Torres et al., 2002). During PTI-ROS, RBOHD is activated by phosphorylation and the binding of Ca2+ and phosphatidic acid. This leads to the reduction of oxygen to superoxide, which is rapidly dismutated into hydrogen peroxide (H2O2) by superoxide dismutase (SOD) in the apoplast (Brandes et al., 2014; Kadota et al., 2014, 2015; Li et al., 2014, 2021). The ectopic expression of an avirulent bacterial effector enhanced the activities of key regulatory components of PTI-ROS and thus induced a long-lasting ETI-ROS burst in the apoplast (Yuan et al., 2021a).
Prolonged accumulation of H2O2 is toxic to cells. To avoid the toxic effects, all organisms contain a sophisticated system to detoxify oxidative stress, including enzymes such as ascorbate peroxidases (APX), catalases (CAT), and glutathione peroxidases (GPX), as well as a nonenzyme ascorbate-glutathione system (Apel and Hirt, 2004; Gest et al., 2013; Shigeoka and Maruta, 2014). APXs are mostly found in photosynthetic organisms, and different cellular compartments in plants contain their own APX isoforms, including cytosolic APX (cAPX), thylakoid membrane-bound APX (tAPX) in chloroplasts, stromal APX (sAPX), and microbody membrane-bound APX (mAPX) (Shigeoka et al., 2002). The different APX isoforms respond differently to metabolic and environmental signals (Caverzan et al., 2012). APX has a high specificity toward ascorbic acid as an electron donor and reduces H2O2 to water with the concomitant generation of monodehydroascorbate, a univalent oxidant of ascorbate that spontaneously disproportionates to ascorbic acid and dehydroascorbate (Smirnoff, 2000; Smirnoff and Wheeler, 2000). Cytosolic APX1 has been characterized as a central peroxidase that protects the chloroplast from oxidative stress (Davletova et al., 2005; Koussevitzky et al., 2008; Pandey et al., 2017). Under normal growth conditions, apx1 mutants show retarded growth and reduced photosynthetic activity. Under high light, drought, and cold conditions, the leaves of apx1 mutant plants show increased H2O2 accumulation, leading to reduced tolerance to these stresses (Davletova et al., 2005; Koussevitzky et al., 2008).
The study of ROS production is challenging because ROS are highly reactive and extremely unstable molecules. Therefore, the detection of ROS relies on measuring the end products formed when ROS react with specific probes (Zulfugarov et al., 2011; Griendling et al., 2016). Several colorimetric, fluorescent, and chemiluminescent probes have been developed to detect ROS production after treatment with immune elicitors or pathogens (Zulfugarov et al., 2011). 3,3-diaminobenzidine (DAB), which reacts with H2O2 to form a reddish-brown polymer, is a colorimetric probe typically used to detect H2O2, especially after infiltration with avirulent bacterial strains (Großkinsky et al., 2012; Kadota et al., 2019). Membrane permeable 2,7-dichlorodihydrofluorescein diacetate (H2DCFDA) and membrane impermeable Amplex Ultra Red (AUR) are 2 fluorescent probes that are widely used in combination with confocal microscopy to visualize the spatial distribution of ROS (Ashtamker et al., 2007; Tian et al., 2016). Compared with colorimetric and fluorescent probes, chemiluminescent probes can interact with ROS and emit photons that can be detected by a luminometer or a sensitive CCD camera system (Khan et al., 2014; Yang et al., 2020). Most available luminescence detection systems are accompanied by a computerized system that allows continuous monitoring of the lights emitted in response to various stimuli (Khan et al., 2014). Although the chemiluminescence method is not suitable for absolute quantitation of ROS production, it provides several advantages, such as the monitoring of ROS kinetics, imaging ROS production in vivo, and screening genetic mutants on a large scale (Dikalov et al., 2007; Khan et al., 2014; Smith and Heese, 2014; Griendling et al., 2016).
The most commonly used chemiluminescent probe is luminol (5-amino-2,3-dihydrophthalazine-1,4-dione), which emits light at a maximum wavelength of 425 nm when oxidized (Khan et al., 2014). Exogenous addition of peroxidases such as the frequently used and commercially available horseradish peroxidase (HRP) could enhance luminol oxidation (Coyle et al., 1986; Thorpe and Kricka, 1986). Luminol/HRP-dependent chemiluminescence assays have been widely used to study ROS kinetics in leaf disks after immune elicitor treatment (Smith and Heese, 2014; Sang and Macho, 2017). Although luminol is a small molecule that can cross plasma membranes, HRP cannot penetrate cell membranes; therefore, the luminol/HRP-based assay is typically considered to measure extracellular “oxidative bursts.” Recently, this approach has also been used to measure the biphasic ROS burst (Shang-Guan et al., 2018; Ngou et al., 2021; Yuan et al., 2021a).
In this study, we monitored ROS production using a luminol-based chemiluminescence assay and found that Arabidopsis apx1 mutants showed reduced long-lasting ROS accumulation in response to LPS, 3-OH-C10:0, and Pseudomonas syringae pv. tomato (Pst) DC3000 (avrRpt2). However, apx1 mutants displayed increased ROS production when it was measured using DAB staining. Remarkably, we found that APX1 acts as a catalyst in the luminol-H2O2 reaction, and the W41F mutation in APX1 that mimics the critical residue of HRP enhanced its catalytic activity toward luminol. As APX1 is a cytosolic peroxidase, these results imply that sustained ETI-ROS might accumulate in the cytosol. Indeed, we did observe cytosolic accumulation of ETI-ROS in the cytosol when ROS were measured using DCF staining. Taken together, the in vivo catalytic activity of APX1 in the luminol-based chemiluminescence system reveals that ETI-ROS triggered by avirulent bacterial strains eventually accumulate in the cytosol.
Results
Arabidopsis apx1 mutants show abolished long-lasting ROS production after LPS treatment
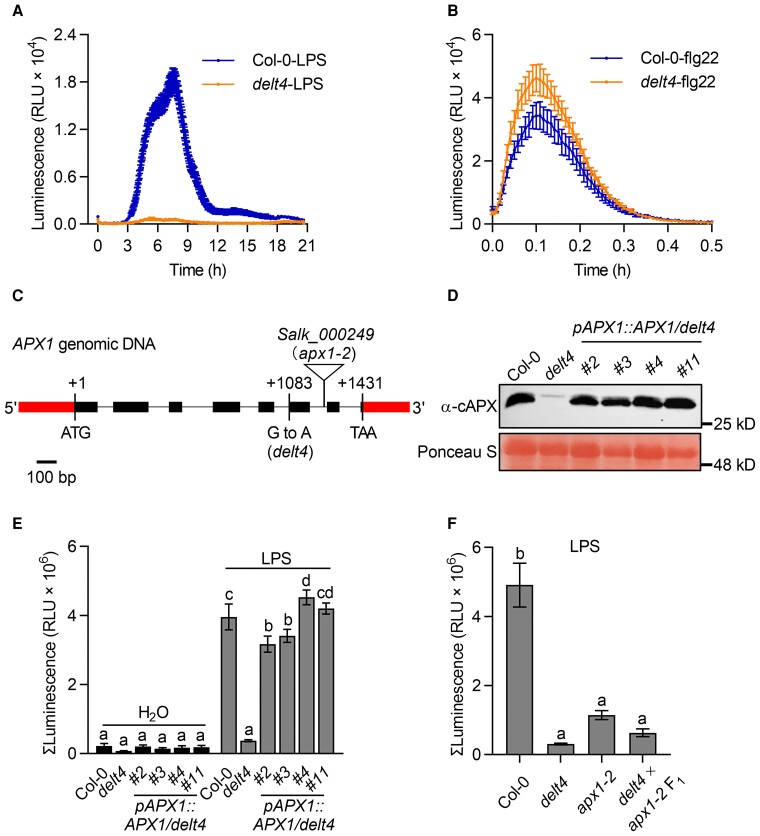
The long-lasting ROS bursts triggered by LPS, 3-OH-C10:0, and avirulent bacteria shared similar kinetics (Supplemental Figure 1). To investigate the molecular mechanism underlying the regulation of the long-lasting ROS burst, we first isolated mutants defective in LPS-triggered ROS production (delt) from ethylmethanesulfonate (EMS)-mutagenized Arabidopsis plants. The delt4 mutants showed almost no long-lasting ROS production after LPS treatment (Figure 1A), whereas the transient ROS burst triggered by flg22 was slightly higher in delt4 mutants than that in the wild type (Figure 1B). ROS production was monitored using a luminol-based chemiluminescence assay, and we found that the addition of HRP did not significantly promote chemiluminescent signals when flg22- and LPS-induced ROS levels were high enough to oxidize luminol to produce chemiluminescence (Supplemental Figure 2). Therefore, we did not add HRP in the following ROS measurement unless otherwise mentioned. The delt4 mutant was backcrossed to the wild type, and the LPS-triggered ROS burst was evaluated in the resulting F1 and F2 populations. All tested F1 plants (n = 12) showed a level of ROS similar to that of the wild type after LPS treatment. Among the 365 tested F2 plants, 70 showed no second ROS burst after LPS treatment, whereas the others were like the wild type (with ROS: without ROS = 295:70, χ2 = 6.6, P > 0.01), indicating that the DELT4 mutation is recessive in a single nuclear gene. The original delt4 mutants were backcrossed with the wild type twice, and the homozygous progenies of F3 or subsequent generations were used in all the experiments described below.
Figure 1.
The apx1 mutants show abolished long-lasting ROS signals after LPS treatment. A, The delt4 mutants showed abolished long-lasting ROS signals after treatment with 50 μg mL−1 LPS. The dynamics of ROS signals were monitored for 21 h using a chemiluminescence assay with luminol as a substrate. Data are presented as the mean ± SE (n = 16). B, The delt4 mutants showed increased ROS signals after treatment with flg22 (100 nM). The dynamics of the ROS signals were monitored for 30 min using a chemiluminescence assay with luminol as a substrate. Data are shown as the mean ± SE (n = 6). C, Schematic representation of the APX1 gene. Mutations in the delt4 and T-DNA insertion sites in the apx1-2 are indicated. The start codon (ATG) and stop codon (TAA) are indicated. D, APX1 protein levels in 4 independent pAPX1::APX1/delt4 transgenic lines. The genomic DNA of APX1 was expressed under the control of the native APX1 promoter in the delt4 mutants background (referred to as pAPX1::APX1/delt4). Total proteins were extracted from 8-day-old seedlings and detected by immunoblot analysis using an α-cAPX antibody, and Ponceau S staining of the membrane served as a loading control. E, The APX1 transgene complemented the ROS signals in delt4 mutants. The bar graph indicates the total integrated photon counts within 1–21 h after 50 μg mL−1 LPS treatment. Data are presented as the mean ± SE (n = 8). Different letters above the bars indicate significant differences between the genotypes (P ≤ 0.05, one-way ANOVA). F, delt4 is allelic to apx1-2. F1 plants were generated by crossing delt4 to apx1-2. The experimental conditions were similar to those used in (E). Data are presented as the mean ± SE (n = 8–12). Different letters above the bars indicate significant differences between the genotypes (P ≤ 0.05, one-way ANOVA). All experiments were repeated 3 times with similar results.
To genetically map DELT4, we generated an F2 population derived from the cross between delt4 mutants and the Landsberg erecta (Ler) ecotype. We roughly mapped DELT4 to Chromosome 1 at a position between 2 molecular markers—F10K1 and F22O13—that are 0.57 Mb apart (Supplemental Figure 3A). We also identified single nucleotide polymorphisms (SNPs) in delt4 mutants compared with the wild-type gene using bulked-segregant analysis. The F2 progenies generated from the backcross between delt4 and Col-0 were divided into 2 pools according to the mutant or wild-type ROS levels. Genomic DNA from 25 individuals was pooled and subjected to whole-genome sequencing (Supplemental Figure 3B). After filtering for detected SNPs, 2 nonsynonymous SNPs were found in the linkage region of the At1g07890 and At1g07700 genes (Supplemental Figure 3C). To identify which mutation caused the delt4 phenotype, complementation tests were performed by transferring wild-type At1g07890 and At1g07700 to the delt4 mutants. Accordingly, At1g07890 (encoding the APX1 protein) complemented the ROS levels in delt4 mutants (Figure 1, C–E), but At1g07700 did not (Supplemental Figure 3D). The delt4 mutant harbors a G-to-A mutation at nucleotide 1083 of APX1, which is the splicing site of the fifth intron (Figure 1C). To complement the delt4 mutants, the genomic DNA of APX1 was expressed under the control of the APX1 native promoter in the delt4 mutants background (referred to as pAPX1::APX1/delt4). In 4 independent transgenic lines, the LPS-induced ROS levels were rescued in delt4 (Figure 1, D and E). Moreover, similar to apx1-2 T-DNA insertion mutants, delt4 showed significantly low levels of APX1 transcripts and no detectable APX1 proteins (Supplemental Figure 4). Allelic tests conducted by crossing delt4 to apx1-2 indicated that they were allelic to each other (Figure 1F). Together, these results indicate that the reduced ROS levels in delt4 after LPS treatment are caused by the mutation in APX1.
APX1 is required for the production of long-lasting ROS triggered by 3-OH-C10:0 and Pst (avrRpt2)
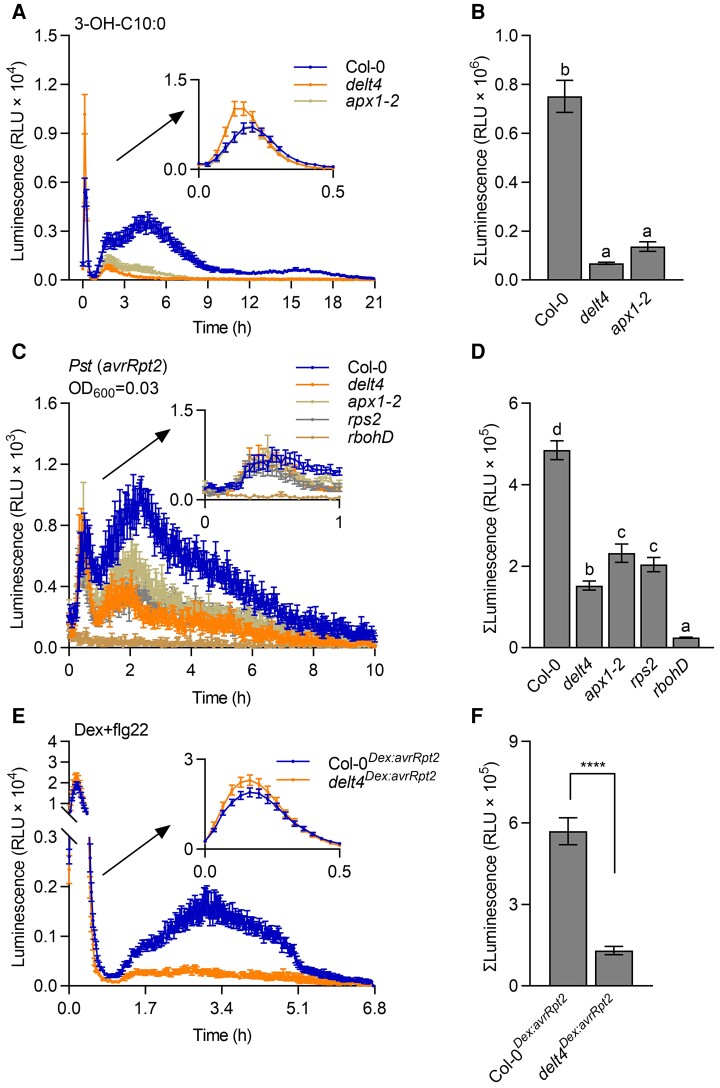
LPS preparations may be contaminated with 3-OH-C10:0, which also induces similar biphasic ROS production (Kutschera et al., 2019). Although the first transient burst triggered by 3-OH-C10:0 is stronger than that induced by LPS, the second sustained ROS peak occurs earlier at approximately 3–6 h but is much weaker than that induced by LPS (Supplemental Figure 1). Therefore, we also examined ROS production in delt4 and apx1-2 plants after treatment with 3-OH-C10:0. Similar to the response to LPS, samples from delt4 and apx1-2 plants showed reduced long-lasting ROS signals, but also showed an enhanced transient early ROS burst (Figure 2, A and B). Furthermore, we also examined whether APX1 is required for effector-triggered long-lasting ROS. Avirulent Pst (avrRpt2) bacteria activate resistance to P. syringae 2 (RPS2)-dependent ETI responses in Arabidopsis. We monitored the ROS production in leaf disks after incubation with avirulent Pst (avrRpt2) bacteria using a luminol-based chemiluminescence assay. The pattern of biphasic ROS production triggered by avirulent Pst (avrRpt2) bacteria is similar to that induced by LPS/3-OH-C10:0. The first ROS peak occurring within approximately 0.5 h corresponds to PTI-ROS, whereas the second kinetically slower ETI-ROS peak at 3–6 h after treatment lasts for several hours (Supplemental Figure 1). We found that the ETI-ROS signals were significantly reduced in delt4 and apx1-2 mutants compared with those in the wild type (Figure 2, C and D, Supplemental Figure 5, A–D). The rbohD and rps2 mutants were used as negative controls for this experiment. High concentrations of bacteria (OD600 = 0.3 or 0.03) induced a strong early ROS burst, whereas the early transient ROS peak in delt4 and apx1-2 mutants was like that in the wild type (Figure 2C, Supplemental Figure 5A). When low concentrations of bacteria (OD600 = 0.003) were used, the early transient ROS peak was nearly undetectable while the long-lasting ROS peak was slower than that triggered by high bacterial concentrations (Figure 2C, Supplemental Figure 5, A and C). However, regardless of the bacterial concentration, the prolonged ETI-ROS signals were significantly reduced in apx1 mutants (Figure 2D, Supplemental Figure 5, B and D). The addition of HRP slightly enhanced the luminescent signals in both wild type and mutants, but the decrease of luminescent signals in apx1 mutants was similar to that without HRP (Supplemental Figure 6), suggesting that exogenous application of HRP cannot rescue the reduced ETI-ROS signals in apx1 mutants, as monitored by a luminol-based chemiluminescence assay.
Figure 2.
APX1 is required for the long-lasting luminol-based light signals triggered by 3-OH-C10:0 and an avirulent effector AvrRpt2. A, The delt4 and apx1-2 mutants showed reduced long-lasting ROS signals after treatment with 3-OH-C10:0 (5 μM). ROS signals were monitored for 21 h using a chemiluminescence assay with luminol as a substrate. Data are shown as the mean ± SE (n = 16). B, Total ROS signals within 1–21 h from (A). Data are shown as the mean ± SE (n = 16). Different letters above the bars indicate significant differences (P ≤ 0.05, one-way ANOVA). C, The delt4 and apx1-2 mutants showed reduced long-lasting ROS signals after treatment with Pseudomonas syringae pv. tomato (Pst) DC3000 (avrRpt2). Signals were monitored in the leaf disks for 10 h using a luminol-based chemiluminescent assay after treatment with bacteria (OD600 = 0.03). The first peak within 1 h is magnified in the upper right corner of the figure. The rps2 and rbohD mutants were used as negative controls. Data are shown as the mean ± SE (n = 9). D, Total ROS signals within 1–10 h from (C). Data are shown as the mean ± SE (n = 9). Different letters above the bars indicate significant differences between different genotypes (P ≤ 0.05, one-way ANOVA). E, The delt4Dex:avrRpt2 leaves showed reduced long-lasting ROS signals after co-treatment with dexamethasone (dex, 10 μM) and flg22 (200 nM). Signals were monitored using a luminol-based chemiluminescent assay in the leaf disks of Col-0Dex:avrRpt2 and delt4Dex:avrRpt2 transgenic plants. Data are shown as the mean ± SE (n = 18). F, Total ROS signals after 1 h from (E). Data are shown as the mean ± SE (n = 18, ****P ≤ 0.0001, t test).
We also crossed delt4 mutants with a transgenic line carrying dexamethasone (Dex)-induced expression of the bacterial effector gene, avrRpt2 (referred to as Col-0Dex:avrRpt2). Consistent with previous results (Yuan et al., 2021a), without the activation of PTI, Dex only induced a weak ETI-ROS burst (Supplemental Figure 5, E and F). Co-treatment with flg22 and Dex induced a strong ETI-ROS signal in Col-0Dex:avrRpt2, whereas delt4Dex:avrRpt2 showed almost no long-lasting signals (Figure 2, E and F). Collectively, these results suggest that apx1 mutants show reduced long-lasting ROS signals triggered by 3-OH-C10:0 and an avirulent effector AvrRpt2, as monitored by a luminol-based chemiluminescence assay.
Loss of APX1 does not substantially enhance the expression of other H2O2-scavenging enzymes
As APX1 is an H2O2-scavenging enzyme, it is intriguing to observe a reduced long-lasting ROS burst in apx1 mutants. However, the absence of APX1 has been reported to elevate other H2O2-scavenging enzymes, such as CATs or GPXs (Jiang et al., 2016). Therefore, we compared the expression of transcripts encoding other cytosolic APX, CAT, and GPX enzymes involved in stress responses between the wild type and apx1 mutants (Mittler, 2002). Our results indicated that these genes were not substantially altered in apx1 mutants after LPS treatment (Supplemental Figure 7). Furthermore, apx2, cat2, and cat3 mutants did not show altered long-lasting ROS production in response to LPS (Supplemental Figure 8). These results suggest that the reduced long-lasting ROS in apx1 mutants is not due to elevated H2O2-scavenging enzymes. In addition, cytosolic APX1 is a central component that regulates redox homeostasis by protecting chloroplasts under abiotic stresses (Davletova et al., 2005). Therefore, we collected 4-week-old mature leaves 4 h after LPS treatment and compared the chloroplast structure between the wild type and delt4 mutants under transmission electron microscopy. We found that the number and size of chloroplasts, plastoglobules, and thylakoids in delt4 mutants were not considerable different from those in the wild type (Supplemental Figure 9A). In a previous study, we had found that the addition of LPS impaired chloroplast function in young seedlings, as evidenced by the release of green fluorescent protein (GFP)-tagged small subunit of ribulose-1.5-bisphosphate carboxylase (SSU-GFP) from the chloroplasts (Shang-Guan et al., 2018). Here, as ROS production was measured in mature plants, we re-evaluated the SSU-GFP release using 4-week-old leaves. We did not observe a substantial release of SSU-GFP from chloroplasts 4 h after LPS treatment, likely because of the stronger stress tolerance in mature leaves. Compared with the wild type, delt4 mutants showed no obvious release of SSU-GFP from the chloroplasts, suggesting that delt4 mutants did not exhibit considerable chloroplast dysfunction after LPS treatment (Supplemental Figure 9B). Taken together, these results suggest that the loss of APX1 function does not enhance the expression of other H2O2-scavenging enzymes or cause the dysfunction of chloroplasts.
Loss of APX1 does not affect the abundance of RBOHD
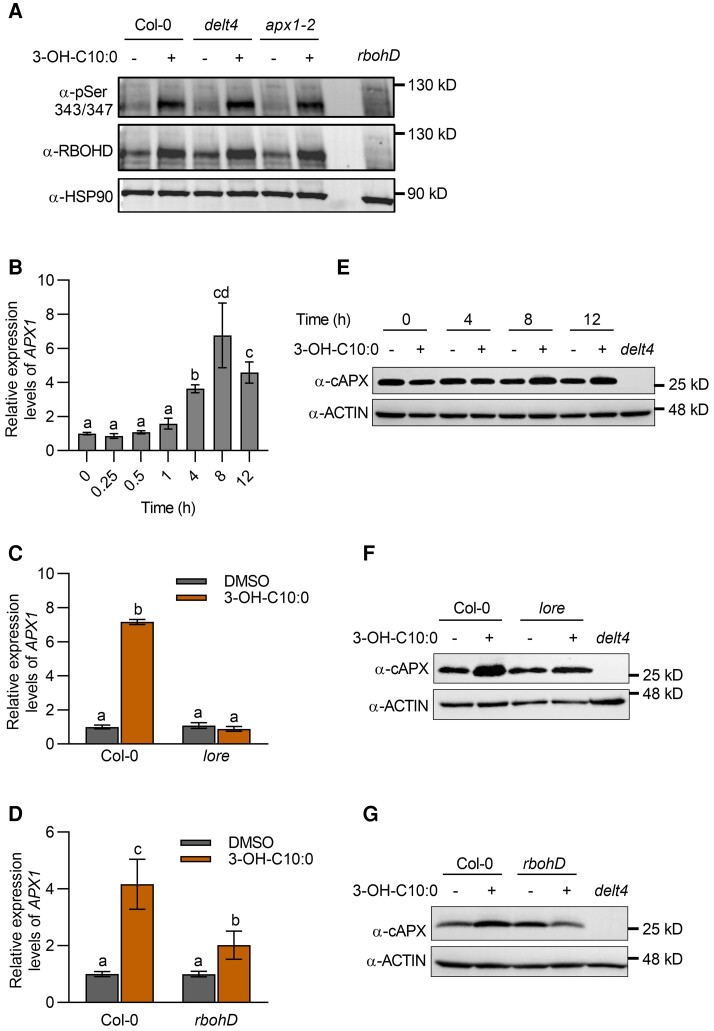
ROS production in plant immunity is mainly mediated by RBOHD; therefore, we compared the RBOHD levels between the wild type and apx1 mutants 3 h after 3-OH-C10:0 treatment. We found that RBOHD accumulation and the phosphorylation of S343/S347 (2 main residues phosphorylated in immunity) in the delt4 and apx1-2 mutants were not substantially different from those in the wild type (Figure 3A). APX1 transcripts were significantly up-regulated at 4, 8, and 12 h after 3-OH-C10:0 treatment (Figure 3B). In Arabidopsis, 3-OH-C10:0 is sensed by LIPOOLIGOSACCHARIDE-SPECIFIC REDUCED ELICITATION (LORE), a lectin receptor-like kinase. Treatment with 3-OH-C10:0 did not induce APX1 up-regulation in lore mutants, suggesting that 3-OH-C10:0-triggered APX1 up-regulation depends on its receptor (Figure 3C). We also examined whether RBOHD is required for 3-OH-C10:0-triggered up-regulation of APX1 and found that the induction of APX1 transcripts was significantly reduced in rbohD mutants when compared to that in the wild type (Figure 3D). Furthermore, we measured the APX1 protein levels by immunoblotting with an α-cAPX antibody after 3-OH-C10:0 treatment. We found that the APX1 proteins were accumulated 8 and 12 h after treatment (Figure 3E), and the accumulation was reduced in lore and rbohD mutants (Figure 3, F and G). Notably, the second ROS peak (3–6 h) in response to 3-OH-C10:0 treatment preceded APX1 up-regulation, suggesting that rather than being a prerequisite for ROS production, APX1 up-regulation may be a downstream response to ROS accumulation. Collectively, these results suggest that the reduced long-lasting chemiluminescent signals in apx1 mutants are not due to the impairment of RBOHD-mediated ROS production.
Figure 3.
Loss of APX1 does not affect the abundance of RBOHD. A, RBOHD accumulation and phosphorylation after treatment with 3-OH-C10:0 (5 μM). Proteins were detected by immunoblot analysis using α-RBOHD and α-pSer343/347 antibodies, with α-HSP90 used as a loading control. B, APX1 transcript levels at different time points. Total RNA was prepared at the indicated time points after treatment with 3-OH-C10:0 (5 μM). APX1 transcript levels were determined using RT-qPCR. Data are shown as the mean ± SE (n = 3–4). Different letters above the bars indicate significant differences (P ≤ 0.05, one-way ANOVA). C, LORE was required for 3-OH-C10:0-triggered up-regulation of APX1 transcripts. Total RNA was prepared from wild type and lore mutants 8 h after treatment with 3-OH-C10:0 (5 μM). Detection of APX1 transcript levels was similar to that used in (B). Data are shown as the mean ± SE (n = 3–4). Different letters above the bars indicate significant differences (P ≤ 0.05, one-way ANOVA). D, RBOHD contributed to 3-OH-C10:0-triggered up-regulation of APX1 transcripts. The experimental conditions were similar to those used in (C). Data are shown as the mean ± SE (n = 3–5). Different letters above the bars indicate significant differences (P ≤ 0.05, one-way ANOVA). E, APX1 protein abundance at different time points. Total proteins were extracted at the indicated time points after treatment with 3-OH-C10:0 (5 μM). APX1 protein abundance was detected by immunoblot analysis using an α-cAPX antibody, with α-ACTIN used as a loading control. F, LORE was required for 3-OH-C10:0-triggered APX1 protein accumulation. Total proteins were extracted from wild type and lore mutants 8 h after treatment with 3-OH-C10:0 (5 μM). Detection of APX1 protein abundance was similar to that used in (E). G, RBOHD contributed to 3-OH-C10:0-triggered APX1 protein accumulation. The experimental conditions were similar to those used in (F). Experiments for immunoblot analysis in this figure were repeated at least twice with similar trends.
The apx1 mutants show increased ROS accumulation, as detected by DAB staining
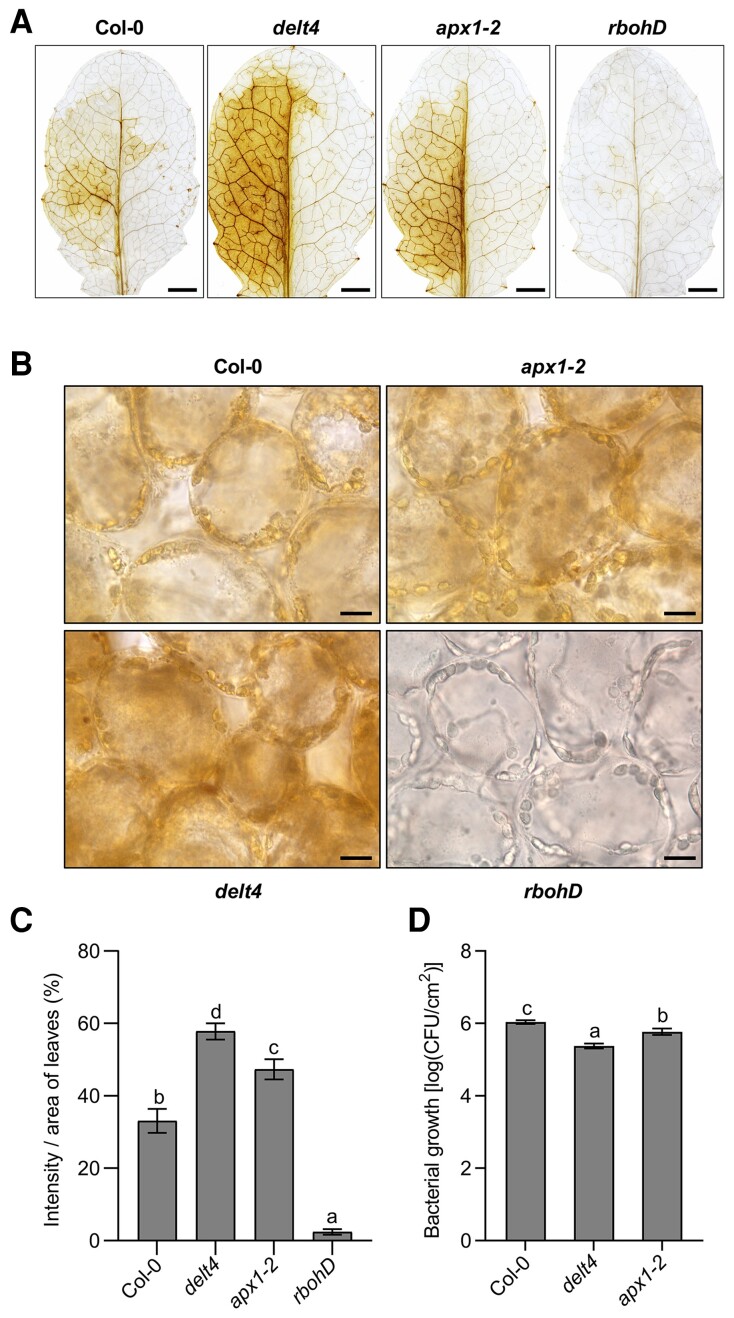
Avirulent bacteria-induced ROS accumulation could be visualized in leaves by DAB staining, and the presence of H2O2 is indicated by a dark-brown precipitate. To relate with the timing of the kinetics of the luminol-emitted light signals, we performed DAB staining 5 h after infiltration with avirulent Pst (avrRpt2) bacteria. Unfortunately, we did not observe a clear brown color 5 h after bacterial infiltration at an OD600 of 0.003. However, a clear brown color was observed 10 h after infiltration with avirulent Pst (avrRpt2) bacteria, whereas no visible brownness was observed after the control treatment (Figure 4A). The sensitivity of DAB staining is probably weaker than luminol; therefore, we could only detect the accumulated ROS at the end point of ROS production using DAB staining. We then quantified the levels of the brown precipitate using the ImageJ software and found that the delt4 and apx1-2 mutants showed color intensity higher than the wild type (Figure 4C). The rbohD mutants were used as a negative control to confirm that this brown color was derived from RBOHD-mediated ROS production. The brown precipitates were microscopically observed in the cytosol and associated with the chloroplasts, which were more pronounced in the apx1 mutants than the wild type (Figure 4B). Consistent with the function of increased ROS levels, apx1 mutants conferred enhanced resistance to Pst (avrRpt2) compared with the wild type (Figure 4D). Taken together, these results suggest that apx1 mutants showed increased ROS accumulation when detected using DAB staining.
Figure 4.
The apx1 mutants show increased effector-triggered accumulation of ROS, as detected by 3,3′-DAB. A, Images of representative leaves after DAB staining. The seventh to ninth leaves of 4-week-old Col-0, delt4, apx1-2, and rbohD plants were infiltrated with Pseudomonas syringae pv. tomato (Pst) DC3000 (avrRpt2) at an OD600 of 0.003, with MgCl2 as the control. Bacterial suspensions were infiltrated into the left side of the leaf and the mock control (MgCl2) was into the right side. The leaves were collected 10 h after infiltration and stained with DAB solution. ROS production was visualized as dark-brown precipitates in the detached leaves. Scale bars, 100 μm. B, The subcellular distribution of effector-triggered ROS accumulation. Images were taken from the left half of DAB-stained leaves in (A) under microscopy. Scale bars, 10 μm. C, The intensity of brownness from DAB-stained leaves in (A). DAB staining per unit area of leaves was quantified using the ImageJ software. Data are shown as the mean ± SE (n = 10). Different letters above the bars indicate significant differences (P ≤ 0.05, one-way ANOVA). The experiment was repeated 3 times with similar results. D, apx1 mutants showed enhanced resistance against Pst (avrRpt2). Leaves derived from 5-week-old plants with the indicated genotypes were inoculated with Pst (avrRpt2) at an OD600 of 0.0001. The bacterial population in the leaf was determined 3 dpi (day postinoculation). Data are presented as the mean ± SE (n = 9). Different letters above the bars indicate significant differences between different genotypes (P ≤ 0.05, one-way ANOVA).
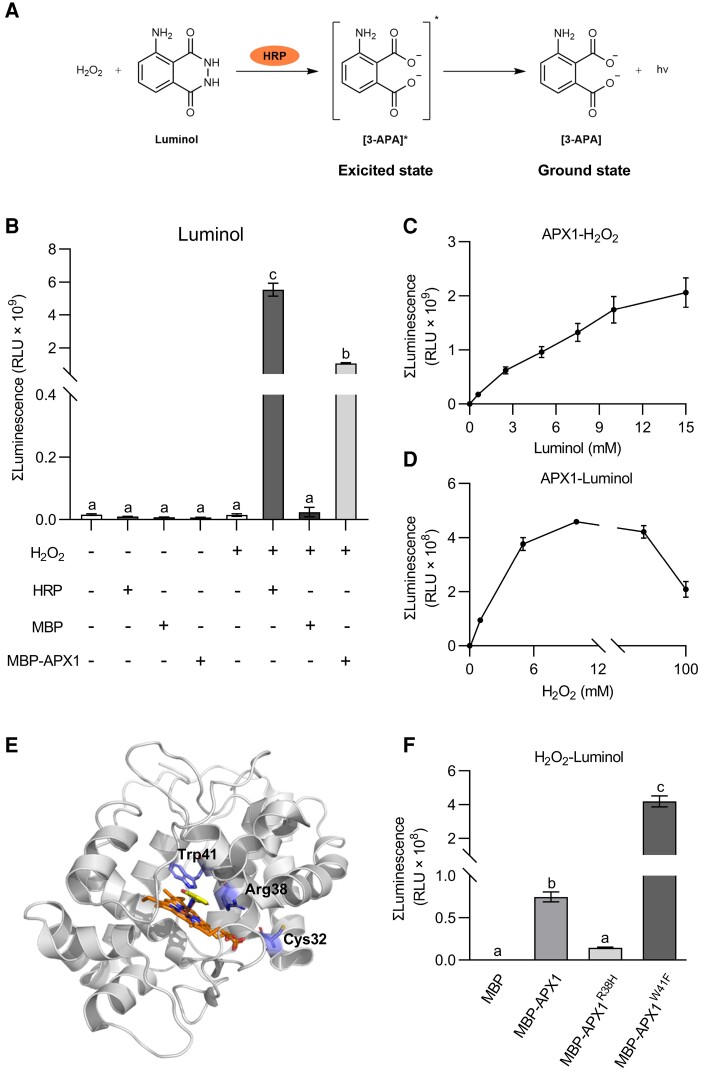
APX1 catalyzes luminol oxidation in vitro
The contrasting results obtained between the luminol-based chemiluminescence assay and the DAB staining assay led us to hypothesize that APX1 might be a catalyst for the luminol-H2O2 reaction. Luminol reacts with H2O2 in the presence of HRP, helping to form 3-aminophthalate (3-APA*), and light is emitted during the decay of the excited-state 3-APA* (Figure 5, A and B). To analyze the catalytic activity of APX1 in luminol-H2O2 reaction, APX1 proteins were fused to maltose-binding protein (MBP), and the recombinant proteins were purified from Escherichia coli. Compared with MBP alone, MBP-APX1 recombinant proteins significantly enhanced the light emission of the luminol-H2O2 reaction (Figure 5B). We then evaluated the enzyme kinetics of APX1 as a catalyst for the luminol-H2O2 reaction. Accordingly, in this study, the catalytic activity of APX1 corresponded with the concentrations of luminol (<15 mM) and H2O2 (<10 mM) (Figure 5, C and D). High concentrations of H2O2 (>10 mM) may inhibit APX1 activity (Figure 5D). Taken together, these results suggest that APX1 can catalyze the oxidation of luminol by H2O2 in vitro.
Figure 5.
APX1 catalyzes the luminol-H2O2 reaction in vitro. A, HRP catalyzes the luminol-H2O2 reaction. In the presence of HRP, luminol is oxidized by H2O2 to create an excited-state product, 3-aminophthalate (3-APA*), whose decay to the ground state results in photon emission. B, APX1 catalyzed the chemiluminescence reaction of luminol. MBP-tagged APX1 recombinant proteins were purified from E. coli. Luminol (300 μM) and MBP, MBP-APX1, or HRP (0.04 mg mL−1) were added first, and light signals were recorded immediately after the addition of H2O2 (10 mM). Values represent the total photon counts within 10 min. Data are shown as the mean ± SE (n = 4). Different letters above the bars indicate significant differences (P ≤ 0.05, one-way ANOVA). C, APX1 activity was corresponded with luminol concentrations below 15 mM. Data are shown as the mean ± SE (n = 5). D, APX1 activity corresponded with H2O2 concentrations below 10 mM. Data are represented as the mean ± SE (n = 6). E, Homology modeling of the structure of APX1 using the 3D coordination of the soybean cAPX1 (PDB-1V0H) as a template. The heme cofactor and substrate salicylhydroxamic acid locate in the middle. The 3 important residues, Cys32, Arg38 and Trp41, are marked F, Catalytic abilities of APX1 variants, APX1R38H and APX1W41F. Luminol (300 μM) and MBP, MBP-APX1, MBP-APX1R38H, or MBP-APX1W41F (0.09 mg mL−1) were added first, and light signals were recorded immediately after the addition of H2O2 (1 mM). Values represent the total photon counts within 10 min. Data are shown as the mean ± SE (n = 8). Different letters above the bars indicate significant differences (P ≤ 0.05, one-way ANOVA). All experiments were repeated 3 times with similar results.
The catalytic mechanism of HRP has been extensively studied using crystallography and site-directed mutagenesis (Savenkova et al., 1998; Martell et al., 2012). Arg38, Phe41, and His42 are critical catalytic residues of HRP that may be involved in heme binding and coordination, and mutations of these residues have a severe impact on peroxidase activity. Compared to HRP, APX1 contains conserved Arg38 and His42 residues, but not Phe41 residue (known as Trp41 in APX1, Figure 5E, Supplemental Figure 10). Therefore, we mutated the Trp41 residue of APX1 to Phe to mimic HRP (referred to as APX1W41F). As expected, compared with the wild-type APX1, APX1W41F showed increased catalytic activity in the luminol-H2O2 reaction (Figure 5F). However, the R38H mutation of APX1 reduced the light emission of the luminol-H2O2 reaction (Figure 5F). Overall, these results suggest that APX1 acts as a peroxidase similar to HRP to catalyze the luminol-H2O2 reaction in vitro.
APX1 catalyzes luminol oxidation in vivo
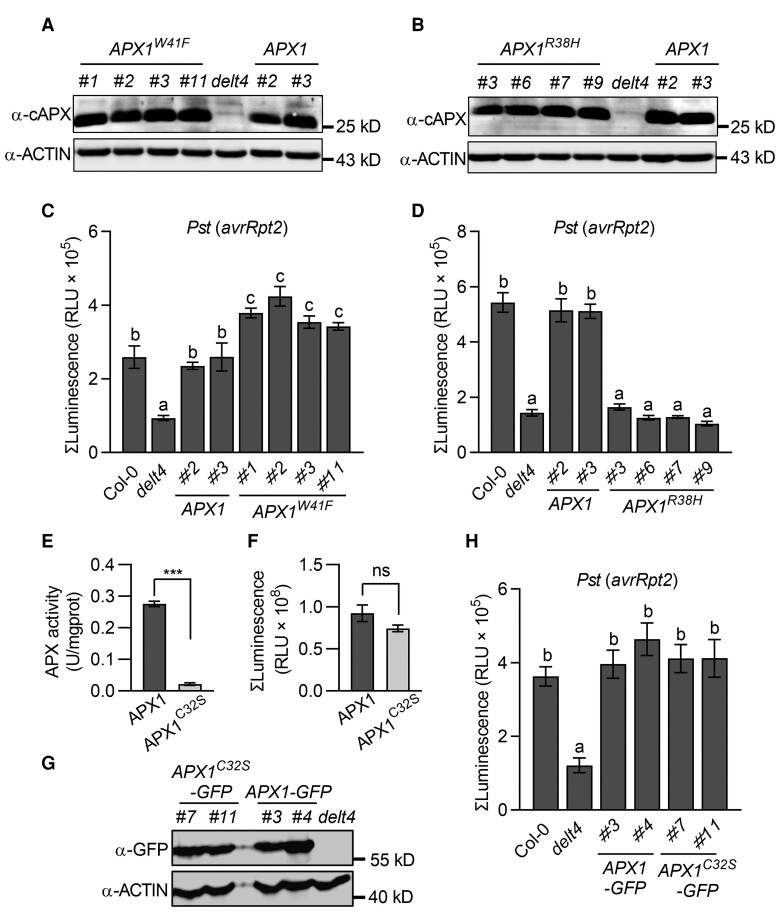
To evaluate whether APX1 acts as a catalyst of luminol in vivo, we transformed APX1W41Fand APX1R38H back to delt4 mutants under the control of the native promoter (Figure 6, A and B). Compared with pAPX1::APX1/delt4 plants, 4 independent transgenic lines of pAPX1::APX1W41F/delt4 displayed significantly increased light intensity in response to Pst (avrRpt2) when using luminol as a substrate, whereas transgene APX1R38H failed to complement the light intensity of delt4 (Figure 6, C and 6D). Similar results were observed after LPS treatment (Supplemental Figure 11, A and B). Collectively, these results further demonstrate that APX1 can catalyze the oxidation of luminol in vivo.
Figure 6.
APX1 catalyzes the luminol-H2O2 reaction in vivo. A, APX1 protein levels in the 4 independent pAPX1::APX1W41F/delt4 transgenic lines. Total proteins were extracted from 4-week-old leaves and detected by immunoblot analysis using an α-cAPX antibody, with α-ACTIN used as a loading control. B, APX1 protein levels in the 4 independent pAPX1::APX1R38H/delt4 transgenic lines. The experimental conditions were similar to those used in (A). C, Transformation of APX1W41F rescued the luminol-emitted long-lasting light signals of delt4 mutants. Light signals were measured using a luminol-based assay after treatment with Pseudomonas syringae pv. tomato (Pst) DC3000 (avrRpt2). The bar graph indicates the total integrated photon counts within 1–10 h after incubation with avirulent bacteria (OD600 = 0.03). D, Transformation APX1R38H failed to complement the luminol-emitted long-lasting light signals of delt4 mutants in response to Pst (avrRpt2). The experimental conditions were similar to those used in (C). E, Analysis of the enzymatic activity of MBP-APX1 and MBP-APX1C32S recombinant proteins using ascorbate as a substrate. Data are shown as the mean ± SE (n = 3, ***P ≤ 0.001, t test). F, Analysis of the enzymatic activity of MBP-APX1 and MBP-APX1C32S recombinant proteins using luminol as a substrate. Values represent the total photon counts within 10 min. Data are shown as the mean ± SE (n = 7). Ns stands for no significant differences (t test). G, APX1 protein levels in the 2 independent pAPX1::APX1C32S-GFP/delt4 transgenic lines. Total proteins were extracted from 4-week-old leaves and detected by immunoblot analysis using an α-GFP antibody, with α-ACTIN used as a loading control. H, Transformation of APX1C32S complemented the luminol-emitted long-lasting light signals of delt4 mutants in response to Pst (avrRpt2). The experimental conditions were similar to those used in (C). Data are presented as the mean ± SE (n = 7 in C; 10 in D, 8 in H). Different letters above the bars indicate significant differences between different genotypes (P ≤ 0.05, one-way ANOVA).
To evaluate whether the APX activity of APX1 contributes to its catalytic activity in the luminol-H2O2 reaction, we mutated the Cys32 residue, which is the target for S-nitrosylation and S-sulfhydration and is important for the APX activity (Aroca et al., 2015; Yang et al., 2015). Consistent with previous studies, APX1C32S abolished peroxidase activity using ascorbate as a substrate in vitro (Figure 6E). However, compared with the purified MBP-APX1, MBP-APX1C32S exhibited similar catalytic activity in the in vitro luminol-H2O2 system (Figure 6F). Consistently, the transgene APX1C32S complemented the long-lasting luminol-emitted light signals of delt4 mutants in response to Pst (avrRpt2) and LPS (Figure 6, G and H, Supplemental Figure 11C). Therefore, these results suggest that APX1 acts as a peroxidase to catalyze the luminol-ROS reaction in vivo, which might be unrelated to its APX activity.
ETI-ROS were observed in the cytosol at later time points, as detected by H2DCFDA staining
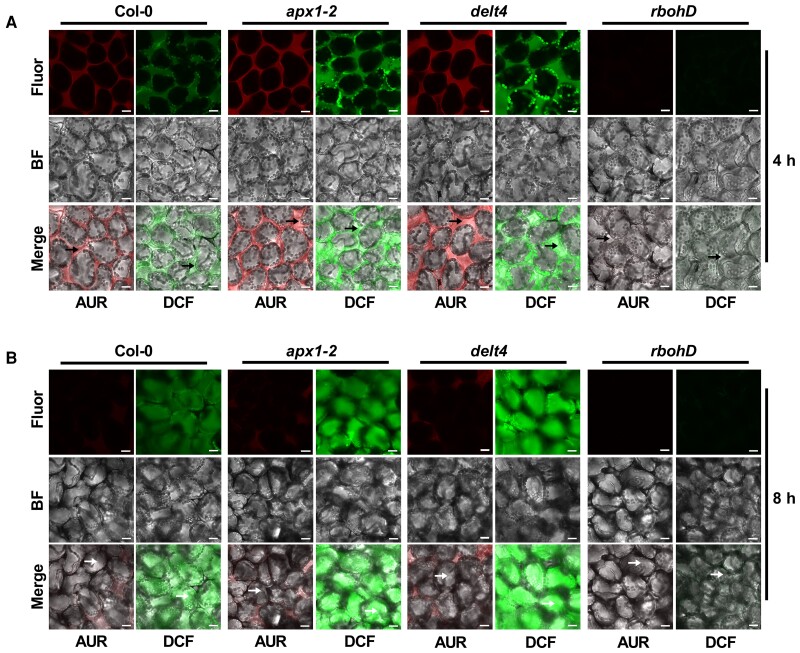
ETI-ROS are produced by RBOHD in the apoplast (Yuan et al., 2021a), but APX1 is known in the cytosol (Panchuk et al., 2002), thus raising a possibility that effectors might also trigger the accumulation of ROS in the cytosol where APX1 catalyzes the luminol oxidation. To test this possibility, we performed H2DCFDA and AUR staining to measure the temporal and spatial accumulation of ROS in leaves after infiltration with Pst (avrRpt2) bacteria. H2DCFDA is a cell-permeable probe that can visualize the apoplastic and cytosolic ROS, while AUR is impermeable to membranes and thus can only probe apoplastic ROS (Ashtamker et al., 2007; Tian et al., 2016). Red and green fluorescent signals were observed in the apoplastic spaces of wild type leaves 4 h after infiltration, as detected by the staining with AUR and H2DCFDA, respectively (Figure 7A, Supplemental Figure 12). Interestingly, green fluorescent signals in the cytosol began to appear at 6 h and became obvious 8 h after infiltration of Pst (avrRpt2), whereas fluorescent signals in apoplasts became much weaker at later time points (Figure 7B, Supplemental Figure 12). Furthermore, compared with the wild type, the apx1 mutants showed stronger apoplastic and intracellular fluorescence signals (Figure 7, Supplemental Figures 12 and 13). Likewise, green fluorescent signals were also observed in the cytosol 4 h after LPS treatment and were largely associated with chloroplasts (Supplemental Figure 14). Collectively, our findings support the notion that ETI-ROS are eventually accumulated in the cytosol.
Figure 7.
Fluorescence imaging of ROS accumulation in response to avirulent bacteria. ROS accumulation was detected by AUR or H2DCFDA staining. The abaxial side of 4-week-old leaves from Col-0, apx1-2, delt4, and rbohD plants was infiltrated with Pst (avrRpt2) at an OD600 of 0.03. Fluorescence was observed at 4 (A) and 8 h (B) after bacterial infiltration. Before microscopic observation, leaves were infiltrated with a solution of AUR or H2DCFDA and incubated for 15–30 min. At least 6 leaves were observed for each treatment, and representative images were shown. Black arrowheads indicate the apoplast space, while white arrowheads indicate the cytoplasm. Scale bars, 15 μm. Fluor, fluorescence; BF, bright field. The experiment was repeated 3 times with similar results.
Discussion
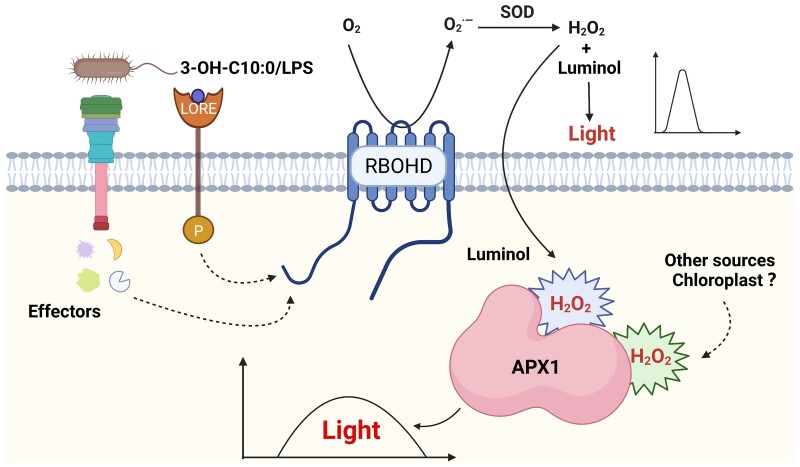
It has been known for a long time that avirulent bacteria trigger a biphasic ROS burst; however, the molecular mechanism underlying the regulation of ETI-ROS and the spatial distribution of ETI-ROS are still unclear. In this study, we found that ETI-ROS were produced in the apoplast at early time points but later accumulated in the cytosol, as detected by H2DCFDA staining. APX1, a cytosolic protein that normally functions as a ROS scavenger, acts as a catalyst of the luminol-ROS reaction in vitro and in vivo to allow the monitoring of long-lasting ROS accumulation in the cytosol using a luminol-based chemiluminescence assay (Figure 8). Our study provides important insights into the spatial dynamics of ROS accumulation in plant immunity and reveals an unexpected role of APX1 protein in catalyzing luminol-ROS reaction, raising cautions in the detection and interpretation of ROS production widely studied in diverse biological processes of plants.
Figure 8.
A schematic model illustrating APX1-mediated monitoring of the kinetics of long-lasting production of ROS in the cytosol. LPS, 3-OH-C10:0, or bacterial effectors trigger a long-lasting ROS burst in the cytosol. When ROS production is monitored by a luminol-based chemiluminescence assay, cytosolic APX1 serves as a catalyst for luminol oxidation. The model was created with BioRender.com.
APXs are characterized by their high specificity to ascorbate, which provides an electron source for detoxifying H2O2 in the cytosol (Raven, 2003). However, previous studies suggest that APXs may also play other roles besides scavenging toxic H2O2. The APXs in Brachypodium distachyon and Arabidopsis can serve as coumarate 3-hydroxylases (C3H) involved in the early steps of lignin biosynthesis that catalyzes the 3-hydroxylation of 4-coumarate to caffeate (Barros et al., 2019). Rice (Oryza sativa) APX2 and Arabidopsis APX1 proteins have been found to function as chaperone molecules (Hong et al., 2018; Kaur et al., 2021). Moreover, APX1 might show peroxidase activity toward other aromatic electron donors, which is unrelated to its physiological functions; for example, an engineered ascorbate peroxidase (APEX2) has been successfully used for electron microscopic imaging in mammalian cells (Lam et al., 2015; Martell et al., 2017). This technique was developed based on the function of HRP in catalyzing DAB and Amplex UltraRed as substrates. However, HRP requires posttranslational modification and is not active in the cytosol, thus limiting its use in vivo (Porstmann et al., 1985). Unlike HRP, APX1 is naturally active in the cytosol; therefore, it was chosen as an HRP mimic for labeling cytosolic proteins in mammalian cells (Lam et al., 2015; Martell et al., 2017). However, in this study, the in vivo APX1 may not be required for DAB staining in Arabidopsis when measuring H2O2 production. The apx1 mutants did not show a decrease in brown precipitates after infiltration with Pst (avrRpt2) strain, suggesting that Arabidopsis may contain other peroxidases that show higher catalytic activity toward DAB than APX1 does. Instead, in this study, APX1 showed catalytic activity toward luminol and that APX1W41F, the most HRP-like mutant that can facilitate the incorporation of aromatic residues into the active site, displayed higher catalytic activity toward the luminol substrate than that displayed by wild-type APX1. Likewise, APEX2, which contains a W41F mutation in APX1, has been developed because of its high activity toward the DAB substrate in mammalian cells (Lam et al., 2015; Martell et al., 2017). Therefore, we believe that the catalytic activity of APX1 toward luminol provides important information that may help to engineer a cytosolic peroxidase that allows real-time monitoring of intracellular ROS production without interfering with the biological ROS levels in mammalian cells.
Many methods have been developed for ROS detection, including chemiluminescent probes, colorimetric dyes, fluorescent dyes, CeCl3 staining, and genetically encoded biosensors. Each has advantages and potential limitations (Dikalov et al., 2007; Griendling et al., 2016; Mamone et al., 2016; Zhang et al., 2018). Among these methods, fluorescent dyes, CeCl3 staining, and genetically encoded biosensors can measure subcellular ROS production. However, the limitations include nonspecific fluorescence, labor-intensive protocols, or the requirement for transgenic plants (Zulfugarov et al., 2011). Colorimetric dyes are relatively easier to use and do not require other instruments but show high levels of nonspecific staining (as they react with any redox molecules). The luminol-based chemiluminescence assay for detecting biological ROS production has several advantages. The biggest advantage of this assay is the real-time detection of ROS kinetics, as the emission of light can be monitored every second by a photon detector or a CCD camera (Khan et al., 2014; Yang et al., 2020). In addition, luminol-based chemiluminescence assays have higher sensitivity compared to other probes and are less expensive than fluorescent probes and CeCl3 (Kim et al., 2019). This method is also suitable for large-scale genetic mutant screens because it can be set up in a 96-well plate (Smith and Heese, 2014). Luminol-based chemiluminescence assays have been successfully used to isolate mutants defective in flg22-induced ROS bursts and to identify several important signaling components in response to PAMPs (Boutrot et al., 2010; Macho et al., 2012; Li et al., 2019, 2021; Wu et al., 2022). In this study, we found that APX1 acts as an intracellular peroxidase that catalyzes the oxidation of luminol and allows the use of a luminol-based chemiluminescence assay to monitor intracellular ROS production. However, more attention should be paid when this method is used. Because APX1 is an H2O2-scavenging enzyme, a lack of APX1 would yield higher H2O2 levels in the cytosol. If APX1 is impaired during a luminol-based chemiluminescence assay to monitor intracellular ROS production, researchers may draw wrong conclusions regarding the actual intracellular ROS levels. To avoid this, it is advisable to use other methods to confirm the results of experiments using this method.
ETI-ROS were also found to be accumulated in the cytosol by CeCl3 staining (Hamdoun et al., 2013). We monitored the temporal and spatial accumulation of ROS by H2DCFDA staining after infiltration with an avirulent bacterial strain and found that ROS were produced in the apoplast, but accumulated in the cytosol at later time points. The accumulation of ETI-ROS in the cytosol could be partially due to the translocation of H2O2 from the apoplast into the cytosol. It has been suggested that biomembrane channels (aquaporins) could mediate the transmembrane diffusion of H2O2 during immune signaling (Bienert and Chaumont, 2014; Tian et al., 2016; Rodrigues et al., 2017). However, we could not rule out the possibility that RBOHD-mediated early ROS signaling may be conveyed to the intracellular environment, which is a prerequisite for inducing other ROS sources, such as chloroplasts, mitochondria, and peroxisomes, which then release ROS into the cytosol (Asada, 2006; de Torres Zabala et al., 2015; Mignolet-Spruyt et al., 2016; Sun et al., 2018). Moreover, apoplastic ROS might induce a secondary signal, such as calcium influx into the cells, and thus induce a second long-lasting ROS burst (Ogasawara et al., 2008; Kimura et al., 2012; Gilroy et al., 2016). In addition, RBOHD is prone to be endocytosed and the endocytosed RBOHD may still function in producing superoxide in the cytosol. Indeed, this mechanism has been found in mammals. For example, the NADPH oxidase NOX1 is endocytosed to generate ROS within smooth muscle cells in response to TNF-α (Miller et al., 2010). Recently, in Arabidopsis, endocytosis of RBOHD was also found to be necessary for salt-induced ROS production in roots (Lee et al., 2022), and salt stress could trigger intracellular ROS accumulation (Lin et al., 2006; Xie et al., 2011; Ben Rejeb et al., 2015; Petrov et al., 2015; Mao et al., 2018; Jin et al., 2021), suggesting that RBOHD might be endocytosed to produce ROS in the cytosol after salt treatment. Nevertheless, how the ETI-ROS were accumulated into the cytosol require further investigation.
In conclusion, ETI-ROS were accumulated in the cytosol, and APX1 catalyzes the luminol-ROS reaction, allowing the measurement of the kinetics of intracellular ROS production using a luminol-based approach.
Materials and methods
Plant materials and growth conditions
Seeds of the Arabidopsis (Arabidopsis thaliana) rbohD (CS9555), lore (Sail_857_E06), cat2-2 (Salk_057998), cat3 (Salk_092911), and rps2 (101C) mutants were obtained from the Arabidopsis Biological Resource Center (Ohio State University, USA). Seeds of the Arabidopsis apx1-2 (Salk_000249), apx2-1 (Salk_091880), and apx2-2 (Salk_057686) mutants were obtained from the Non-Profit Arabidopsis Share Center (https://www.arashare.cn/). The Arabidopsis rps2Dex:avrRpt2 transgenic plants were kindly provided by Prof. Jun Liu at the China Agricultural University. The Arabidopsis Col-0Dex:avrRpt2 and delt4Dex:avrRpt2 transgenic plants were obtained from F2 segregating progenies by crossing rps2Dex:avrRpt2 plants with Col-0 and delt4 plants, respectively. For young seedling assays, the seeds were sterilized with 10% (v/v) sodium hypochlorite and grown on half-strength Murashige-Skoog agar plates in a growth chamber (model A1000AR, Conviron, Manitoba, Canada) under a 16 h photoperiod, 75% humidity, and 22°C. For mature plant assays, 10-day-old seedlings were transferred to pots containing a peat-based compost (Sun Gro Horticulture, Agawam, MA, USA) and grown under a 14-h photoperiod for ROS assays or a 8-h photoperiod for bacterial growth assays.
Map-based cloning of the DELT4 gene
The delt4 mutant in the Col-0 background was isolated, as previously described (Li et al., 2021). To generate a mapping population, the delt4 mutant was crossed with the Landsberg erecta (Ler) ecotype, and individuals with reduced ROS production after LPS treatment were isolated from the F2 generation. Genomic DNA was extracted from each individual for linkage analysis. The PCR markers were developed using the Arabidopsis Mapping Platform (https://www.arabidopsis.org/browse/Cereon/help.jsp), and the primers used are listed in Supplemental Table 1.
Next-generation sequencing from bulked-segregant analysis
The delt4 mutant was backcrossed to Col-0, and the generated F1 progeny was self-pollinated to produce F2 seeds. Individuals with or without ROS production after LPS treatment were pooled separately and subjected to whole-genome sequencing using the Illumina X-ten system (Gene Denovo Biotechnology, Guangzhou, China). Illumina short reads generated from the bulked DNA samples were cleaned and filtered according to their Phred quality score and aligned against the public wild-type reference genome using the Burrows-Wheeler Aligner software (Li and Durbin, 2009). Alignment files were converted to sequence alignment map/binary alignment map files using SAMtools (Li et al., 2009). A sliding window analysis was applied to the frequency distribution of SNPs (SNP-index) in the population of bulked individuals. The Δ(SNP-index) values were calculated by subtracting the SNP-index values between the 2 bulk pools.
Plasmid construction and generation of transgenic plants
All the primers used for gene cloning in this study are listed in Supplemental Table 1. The coding or genomic sequences of the indicated genes were amplified and cloned into a pDONR-Zeo plasmid via BP cloning (Thermo Fisher Scientific, Waltham, MA, USA). The inserts were verified by sequencing and cloned into the destination plasmids via LR cloning (Thermo Fisher Scientific). PCR-based site-directed mutagenesis was used to construct plasmids with point mutations in a gene of interest. All constructs used for the generation of transgenic plants were verified by sequencing and are listed in Supplemental Table 2. Each resulting construct was introduced into Agrobacterium tumefaciens strain GV3101, and the Agrobacterium-mediated floral dip method was used to generate transgenic Arabidopsis plants.
RNA isolation and reverse transcription quantitative PCR (RT-qPCR)
Total RNA was extracted from 8-day-old seedlings using an Easy Plant RNA kit (Easy-Do, Hangzhou, China) according to the manufacturer's instructions. First-strand complementary DNA (cDNA) was synthesized from 1 μg of isolated RNA using HiScript II reverse transcriptase (Vazyme Biotech, Nanjing, China). RT-qPCR reactions were performed using an SYBR Green Master Mix (Vazyme Biotech). The relative levels of gene expression were calculated using the 2−ΔΔCt method with ELONGATION FACTOR1α (EF1α) as an internal control. All primers used for the RT-qPCR are listed in Supplemental Table 1.
Expression and purification of recombinant proteins
The recombinant proteins were expressed and purified as previously described (Li et al., 2021). APX1 cDNA was cloned into pMAL-c2x-GW (gateway) vectors, and the resulting plasmid was transformed into E. coli BL21. All constructs used for prokaryotic expression were verified by sequencing and are listed in Supplemental Table 2. The bacterial cultures were grown in liquid Luria-Bertani medium at 37°C for approximately 3 h. When the optical density at 600 nm (OD600) reached approximately 0.8–1.0, 0.2 mM of isopropyl-β-D-thiogalactoside was added to induce the expression of MBP-tagged APX1. The cultures were collected 6 h after induction, and MBP-tagged recombinant proteins were purified using amylose resin (New England Biolabs, Ipswich, MA, USA) following the manufacturer's instructions.
Protein extraction and immunoblot analysis
Detection of RBOHD using immunoblot analysis was performed as previously described (Li et al., 2021). To detect APX1, total proteins were extracted with a buffer containing 50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 1% Triton X-100 (v/v), and 1× protease inhibitor cocktail (Merck KGaA, Darmstadt, Germany). The extracted proteins were separated by 12% SDS-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Merck KGaA). Proteins were detected with an α-cAPX antibody (Agrisera, Vännäs, Sweden) and an α-ACTIN antibody (ABclonal, Wuhan, China).
ROS detection by luminol-based chemiluminescent assay
ROS detection by luminol-based chemiluminescence assay was performed as previously described, with some modifications (Liang et al., 2013). Leaf disks (0.2 cm2) were obtained from the seventh to ninth leaves of 4-week-old plants and incubated overnight with water in a 96-well plate. The following day, 600 μM luminol (Merck KGaA) was added along with the indicated elicitors, including LPS (L9143, Merck KGaA), flg22 (GenScript, Nanjing, China), 3-OH-C10:0 (Bide Pharmatech Ltd., Shanghai, China), and dexamethasone (MedChemExpress, Monmouth Junction, NJ, USA). The chemiluminescent signal was immediately recorded using a Photek camera (HRPCS5, Photek, East Sussex, UK). When bacteria were used for the luminol-based assay, the bacterial cells were freshly diluted to the desired concentration as previously described (Smith and Heese, 2014). HRP (Merck KGaA) was exogenously applied for the luminol-based assay at 20 mg L−1 (w/v).
ROS detection by histochemical DAB staining
DAB staining was performed as described previously, with some modifications (Shang-Guan et al., 2018). The seventh to ninth leaves of 4-week-old plants were infiltrated with Pst (avrRpt2) at an OD600 of 0.003. The leaves were detached 10 h post infiltration, vacuum-infiltrated with DAB staining solution (1 mg mL−1, pH 6.0), and incubated at 24°C room temperature for 2 h. Chlorophyll was removed by incubation with glacial acetic acid/glycerol/ethanol (1:1:3), and the dark-brown colored leaves (visualizing the generation of H2O2) were observed under a light microscope (Nikon, Japan). The intensity of DAB staining per unit area of leaves was quantified using the ImageJ software (https://imagej.nih.gov/ij/).
ROS detection by fluorescent staining
AUR and H2DCFDA staining were performed as described previously, with some modifications (Tian et al., 2016; Yuan et al., 2021a). The seventh to ninth leaves of 4-week-old plants were infiltrated with Pst (avrRpt2) (OD600 = 0.03) or LPS (100 μg mL−1). For bacterial infiltration, leaves were directly infiltrated with a solution of 10 μM AUR (Merck KGaA) or 10 μM H2DCFDA (MedChemExpress) and incubated for 10–30 min; for LPS treatment, leaves were detached and incubated with an H2DCFDA solution (10 μM) for 30 min. Images were captured under an Olympus FV3000 confocal laser scanning microscope with the following setting of laser/detection wavelength: oxidized H2DCFDA (488/500–540), oxidized AUR (561/565–620), and chlorophyll autofluorescence (640/650–750). The laser transmissivity was set as 0.2% in all fluorescence detection.
Bacterial growth assay
For disease assay, Pst (avrRpt2) was cultured in a selective King's B (KB) medium overnight at 28°C to an OD600 of 0.8–1.0. Bacteria were collected by centrifugation and resuspended with 10 mM MgCl2 and the concentration was adjusted to an OD600 of 0.0001. Using a needleless syringe, the resuspended bacterial solution was infiltrated into the abaxial surfaces of 5-week-old Arabidopsis leaves. After being air-dried in the plant growth room, the inoculated plants were covered with a transparent plastic dome for at least 6 h to maintain a high humidity for disease to develop. For quantification at 3 dpi, 3 leaf discs from 3 different leaves were collected with an 8-mm-diameter cork borer (with disc area of 0.502 cm2) as 1 biological repeat, and 9 repeats were taken for each genotype. Samples were ground and diluted in 10 mM MgCl2, and the extraction solutions were then plated on selective KB medium plates (100 μL per plate). The number of colonies was counted 36 h after incubation at 28°C, and bacterial growth was represented as CFU/cm2 of leaf tissue.
Measurement of APX activity
APX activity was measured as previously described with some modifications (Nakano and Asada, 1981). Briefly, the reaction mixture contained 25 mM PBS (pH = 7.0) with 0.1 mM EDTA, 0.5 mM ascorbate, 2 mM H2O2, and 100 μg purified proteins (MBP-APX1 and MBP-APX1C32S). After the initiation of the reaction by adding H2O2. the samples were immediately analyzed with Tecan Spark Multimode Microplate Reader by measuring the absorbance at A290 every 5 s for a 2-min recording time. Protein concentration was determined with Coomassie Brilliant Blue G-250 according to the method of Bradford using bovine serum albumin as the standard.
Examination of the ultrastructure of leaf chloroplasts by transmission electron microscopy
Small fragments (approximately 3 mm2) were excised from freshly harvested leaves of 4-week-old mature plants with a razor blade, and then immediately transferred to a cold fixation buffer [2.5% glutaraldehyde (v/v) in 0.1 M phosphate buffer, pH 7.0]. After incubation with the fixation buffer for at least 4 h, the samples were washed 3 times for 15 min in phosphate buffer (0.1 M, pH 7.0), postfixed for 1–2 h with 1% osmium tetroxide (w/v) in phosphate buffer, and washed again 3 times with phosphate buffer. The doubled-fixed samples were then dehydrated, infiltrated, embedded, sectioned, and stained for observation as described previously (Bestwick et al., 1995). The sections were examined using a transmission electron microscope (H7650, Hitachi, Tokyo, Japan) at an accelerating voltage of 75 kV.
Confocal microscopy observation of SSU-GFP
Transgenic plants expressing SSU-GFP were used as previously described (Shang-Guan et al., 2018). The GFP signals were detected under a confocal microscope (FV3000, Olympus, Tokyo, Japan) with the same parameters as oxidized H2DCFDA.
Modeling the structure of APX1
Sequences were aligned using the freely available web server, ESPript 3.0 (http://espript.ibcp.fr; Robert and Gouet, 2014). Homology modeling of the structure of the APX1 molecule was performed using the 3D coordination of the soybean (Glycine max) cAPX1 (PDB-1V0H) as the template. The images were generated using PyMol (http://pymol.sourceforge.net).
Statistical analysis
The statistical analyses were performed by 2-tailed Student's t test with Office Excel software for paired comparison or by one-way analysis of variance (ANOVA) with SPSS software for multiple comparisons. Groups or samples with statistically significant differences are marked with asterisk(s) (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001, ****P ≤ 0.0001, Student's t test) or different letters (P ≤ 0.05, one-way ANOVA, lowercase letters). Each experiment was repeated at least 3 times and data were represented as the mean ± SE as indicated.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers_: APX1 (At1g07890), RBOHD (At5g47910), LORE (At1g61380), RPS2 (At4g26090), APX2 (At3g09640), APX6 (At4g32320), CAT1 (At1g20630), CAT2 (At4g35090), CAT3 (At1g20620), GPX1 (At2g25080), GPX2 (At2g31570), GPX3 (At2g43350), GPX7 (At4g31870) and avrRpt2 from P. syringae (Q6LAD6).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . The kinetics of ROS burst triggered by LPS/3-OH-C10:0/Pst (avrRpt2).
Supplemental Figure S2 . HRP is not indispensable when ROS level is high enough to oxidize luminol.
Supplemental Figure S3 . Molecular characterization of the DELT4 gene.
Supplemental Figure S4 . APX1 transcripts and protein abundance in apx1 mutants.
Supplemental Figure S5 . The apx1 mutants show reduced effector-triggered long-lasting luminescent signals.
Supplemental Figure S6 . The exogenous addition of HRP cannot restore the reduced ETI-ROS signals in apx1 mutants.
Supplemental Figure S7 . Expression of genes involved in ROS scavenging.
Supplemental Figure S8 . apx2, cat2, and cat3 mutants are not impaired in LPS-triggered long-lasting ROS accumulation.
Supplemental Figure S9 . Chloroplasts of delt4 mutants are not impaired after LPS treatment.
Supplemental Figure S10 . Sequence alignment of APX1 with horseradish peroxidase (HRP) and soybean peroxidase (SBP).
Supplemental Figure S11 . Effect of APX1 mutation on luminol-based light signals triggered by LPS.
Supplemental Figure S12 . The temporal and spatial ETI-ROS detected by H2DCFDA staining.
Supplemental Figure S13 . Detection of ROS accumulation in leaves infiltrated with MgCl2 by AUR and H2DCFDA staining.
Supplemental Figure S14 . apx1 mutants show increased cytosolic ROS accumulation after LPS treatment, as detected by H2DCFDA staining.
Supplemental Table S1 . Primers used in this study.
Supplemental Table S2 . Constructs used in this study.
Supplementary Material
Acknowledgments
We thank Prof. Jun Liu (China Agricultural University, China) for kindly providing rps2Dex:avrRpt2 seeds, Xiao-xiao Feng (Agricultural Experiment Station of Zhejiang University) for her assistance with plant growth, and Prof. Yanni Yin (Zhejiang University, China) for her discussion of the manuscript.
Contributor Information
Xiufang Hong, State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Institute of Biotechnology, Zhejiang University, Hangzhou 310058, China.
Fan Qi, State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Institute of Biotechnology, Zhejiang University, Hangzhou 310058, China.
Ran Wang, State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Institute of Biotechnology, Zhejiang University, Hangzhou 310058, China.
Zhiyi Jia, State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Institute of Biotechnology, Zhejiang University, Hangzhou 310058, China.
Fucheng Lin, State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Institute of Biotechnology, Zhejiang University, Hangzhou 310058, China.
Minhang Yuan, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.
Xiu-Fang Xin, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.
Yan Liang, State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Institute of Biotechnology, Zhejiang University, Hangzhou 310058, China.
Funding
This work was supported by the Key Research and Development Program of Zhejiang Province (2021C02064-7, 2021C02009, and 2022C02016) and National Natural Science Foundation of China (31622006 and 31970279). Xiu-Fang Xin was supported by the Chinese Academy of Sciences Strategic Priority Research Program (type-B; grant number XDB27040211). Minhang Yuan was supported by National Postdoctoral Program for Innovative Talents (BX2021313) and Shanghai Postdoctoral Excellence Program.
References
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55(1): 373–399 [DOI] [PubMed] [Google Scholar]
- Aroca A, Serna A, Gotor C, Romero LC (2015) S-sulfhydration: a cysteine posttranslational modification in plant systems. Plant Physiol 168(1): 334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K (2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol 141(2): 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashtamker C, Kiss V, Sagi M, Davydov O, Fluhr R (2007) Diverse subcellular locations of cryptogein-induced reactive oxygen species production in tobacco bright yellow-2 cells. Plant Physiol 143(4): 1817–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CJ, Orlandi EW (1995) Active oxygen in plant pathogenesis. Annu Rev Phytopathol 33(1): 299–321 [DOI] [PubMed] [Google Scholar]
- Barros J, Escamilla-Trevino L, Song L, Rao X, Serrani-Yarce JC, Palacios MD, Engle N, Choudhury FK, Tschaplinski TJ, Venables BJ, et al. (2019) 4-Coumarate 3-hydroxylase in the lignin biosynthesis pathway is a cytosolic ascorbate peroxidase. Nat Commun 10(1): 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter A, Mittler R, Suzuki N (2014) ROS as key players in plant stress signalling. J Exp Bot 65(5): 1229–1240 [DOI] [PubMed] [Google Scholar]
- Ben Rejeb K, Benzarti M, Debez A, Bailly C, Savouré A, Abdelly C (2015) NADPH oxidase-dependent H2O2 production is required for salt-induced antioxidant defense in Arabidopsis thaliana. J Plant Physiol 174: 5–15 [DOI] [PubMed] [Google Scholar]
- Bestwick CS, Bennett MH, Mansfield JW (1995) Hrp mutant of Pseudomonas syringae pv. phaseolicola induces cell wall alterations but not membrane damage leading to the hypersensitive reaction in lettuce. Plant Physiol 108(2): 503–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert GP, Chaumont F (2014) Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochim Biophys Acta 1840(5): 1596–1604 [DOI] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H (2015) Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant 8(4): 521–539 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60(1): 379–406 [DOI] [PubMed] [Google Scholar]
- Boutrot F, Segonzac C, Chang KN, Qiao H, Ecker JR, Zipfel C, Rathjen JP (2010) Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc Natl Acad Sci U S A 107(32): 14502–14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes RP, Weissmann N, Schröder K (2014) Nox family NADPH oxidases: molecular mechanisms of activation. Free Radic Biol Med 76: 208–226 [DOI] [PubMed] [Google Scholar]
- Camejo D, Guzmán-Cedeño Á, Moreno A (2016) Reactive oxygen species, essential molecules, during plant-pathogen interactions. Plant Physiol Biochem 103: 10–23 [DOI] [PubMed] [Google Scholar]
- Caverzan A, Passaia G, Rosa SB, Ribeiro CW, Lazzarotto F, Margis-Pinheiro M (2012) Plant responses to stresses: role of ascorbate peroxidase in the antioxidant protection. Genet Mol Biol 35(4 Suppl 1): 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S, Martin GB, Low PS (1996) The Pto kinase mediates a signaling pathway leading to the oxidative burst in tomato. Proc Natl Acad Sci U S A 93(23): 13393–13397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle PM, Thorpe GH, Kricka LJ, Whitehead TP (1986) Enhanced luminescent quantitation of horseradish peroxidase conjugates: application in an enzyme immunoassay for digoxin. Ann Clin Biochem 23(1): 42–46 [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17(1): 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres Zabala M, Littlejohn G, Jayaraman S, Studholme D, Bailey T, Lawson T, Tillich M, Licht D, Bolter B, Delfino L, et al. (2015) Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nat Plants 1(6): 15074. [DOI] [PubMed] [Google Scholar]
- Dikalov S, Griendling KK, Harrison DG (2007) Measurement of reactive oxygen species in cardiovascular studies. Hypertension 49(4): 717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doke N (1983) Involvement of superoxide anion generation in the hypersensitive response of potato-tuber tissues to infection with an incompatible race of Phytophthora infestans and to the hyphal wall components. Physiol Plant Pathol 23(3): 345–357 [Google Scholar]
- Gest N, Gautier H, Stevens R (2013) Ascorbate as seen through plant evolution: the rise of a successful molecule? J Exp Bot 64(1): 33–53 [DOI] [PubMed] [Google Scholar]
- Gilroy S, Białasek M, Suzuki N, Górecka M, Devireddy AR, Karpiński S, Mittler R (2016) ROS, calcium, and electric signals: key mediators of rapid systemic signaling in plants. Plant Physiol 171(3): 1606–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griendling KK, Touyz RM, Zweier JL, Dikalov S, Chilian W, Chen YR, Harrison DG, Bhatnagar A, American Heart Association Council on Basic Cardiovascular S (2016) Measurement of reactive oxygen species, reactive nitrogen species, and redox-dependent signaling in the cardiovascular system: a scientific statement from the American Heart Association. Circ Res 119(5): e39–e75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Großkinsky DK, Koffler BE, Roitsch T, Maier R, Zechmann B (2012) Compartment-specific antioxidative defense in Arabidopsis against virulent and avirulent Pseudomonas syringae. Phytopathology 102(7): 662–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdoun S, Liu Z, Gill M, Yao N, Lu H (2013) Dynamics of defense responses and cell fate change during Arabidopsis-Pseudomonas syringae interactions. PLoS One 8(12): e83219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Tripathi BN, Chung MS, Cho C, Lee S, Kim JH, Bai HW, Bae HJ, Cho JY, Chung BY, et al. (2018) Functional switching of ascorbate peroxidase 2 of rice (OsAPX2) between peroxidase and molecular chaperone. Sci Rep 8(1): 9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Chen Z, Gao Q, Ci L, Cao S, Han Y, Wang W (2016) Loss-of-function mutations in the APX1 gene result in enhanced selenium tolerance in Arabidopsis thaliana. Plant Cell Environ 39(10): 2133–2144 [DOI] [PubMed] [Google Scholar]
- Jin J, Li K, Qin J, Yan L, Wang S, Zhang G, Wang X, Bi Y (2021) The response mechanism to salt stress in Arabidopsis transgenic lines over-expressing of GmG6PD. Plant Physiol Biochem 162: 74–85 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444(7117): 323–329 [DOI] [PubMed] [Google Scholar]
- Joo JH, Wang S, Chen JG, Jones AM, Fedoroff NV (2005) Different signaling and cell death roles of heterotrimeric G protein α and β subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell 17(3): 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y, Liebrand TWH, Goto Y, Sklenar J, Derbyshire P, Menke FLH, Torres MA, Molina A, Zipfel C, Coaker G, et al. (2019) Quantitative phosphoproteomic analysis reveals common regulatory mechanisms between effector- and PAMP-triggered immunity in plants. New Phytol 221(4): 2160–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y, Shirasu K, Zipfel C (2015) Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol 56(8): 1472–1480 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, et al. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell 54(1): 43–55 [DOI] [PubMed] [Google Scholar]
- Kaur S, Prakash P, Bak DH, Hong SH, Cho C, Chung MS, Kim JH, Lee S, Bai HW, Lee SY, et al. (2021) Regulation of dual activity of ascorbate peroxidase 1 from Arabidopsis thaliana by conformational changes and posttranslational modifications. Front Plant Sci 12: 678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan P, Idrees D, Moxley MA, Corbett JA, Ahmad F, von Figura G, Sly WS, Waheed A, Hassan MI (2014) Luminol-based chemiluminescent signals: clinical and non-clinical application and future uses. Appl Biochem Biotechnol 173(2): 333–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Jeong K, Murphy JM, Rodriguez YAR, Lim SS (2019) A quantitative method to measure low levels of ROS in nonphagocytic cells by using a chemiluminescent imaging system. Oxid Med Cell Longev 2019: 1754593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Kaya H, Kawarazaki T, Hiraoka G, Senzaki E, Michikawa M, Kuchitsu K (2012) Protein phosphorylation is a prerequisite for the Ca2+-dependent activation of Arabidopsis NADPH oxidases and may function as a trigger for the positive feedback regulation of Ca2+ and reactive oxygen species. Biochim Biophys Acta 1823(2): 398–405 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Suzuki N, Huntington S, Armijo L, Sha W, Cortes D, Shulaev V, Mittler R (2008) Ascorbate peroxidase 1 plays a key role in the response of Arabidopsis thaliana to stress combination. J Biol Chem 283(49): 34197–34203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera A, Dawid C, Gisch N, Schmid C, Raasch L, Gerster T, Schaffer M, Smakowska-Luzan E, Belkhadir Y, Vlot AC, et al. (2019) Bacterial medium-chain 3-hydroxy fatty acid metabolites trigger immunity in Arabidopsis plants. Science 364(6436): 178–181 [DOI] [PubMed] [Google Scholar]
- Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, Ting AY (2015) Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods 12(1): 51–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48(1): 251–275 [DOI] [PubMed] [Google Scholar]
- Lee J, Hanh Nguyen H, Park Y, Lin J, Hwang I (2022) Spatial regulation of RBOHD via AtECA4-mediated recycling and clathrin-mediated endocytosis contributes to ROS accumulation during salt stress response but not flg22-induced immune response. Plant J 109(4): 816–830 [DOI] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79(4): 583–593 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25(14): 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25(16): 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QY, Li P, Myint Phyu Sin Htwe N, Shangguan KK, Liang Y (2019) Antepenultimate residue at the C-terminus of NADPH oxidase RBOHD is critical for its function in the production of reactive oxygen species in Arabidopsis. J Zhejiang Univ Sci B 20(9): 713–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, et al. (2014) The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15(3): 329–338 [DOI] [PubMed] [Google Scholar]
- Li P, Zhao L, Qi F, Htwe N, Li Q, Zhang D, Lin F, Shang-Guan K, Liang Y (2021) The receptor-like cytoplasmic kinase RIPK regulates broad-spectrum ROS signaling in multiple layers of plant immune system. Mol Plant 14(10): 1652–1667 [DOI] [PubMed] [Google Scholar]
- Liang Y, Cao Y, Tanaka K, Thibivilliers S, Wan J, Choi J, Kang C, Qiu J, Stacey G (2013) Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science 341(6152): 1384–1387 [DOI] [PubMed] [Google Scholar]
- Lin J, Wang Y, Wang G (2006) Salt stress-induced programmed cell death in tobacco protoplasts is mediated by reactive oxygen species and mitochondrial permeability transition pore status. J Plant Physiol 163(7): 731–739 [DOI] [PubMed] [Google Scholar]
- Ma S, Lapin D, Liu L, Sun Y, Song W, Zhang X, Logemann E, Yu D, Wang J, Jirschitzka J, et al. (2020) Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 370(6521):eabe3069 [DOI] [PubMed] [Google Scholar]
- Macho AP, Boutrot F, Rathjen JP, Zipfel C (2012) Aspartate oxidase plays an important role in Arabidopsis stomatal immunity. Plant Physiol 159(4): 1845–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamone L, Di Venosa G, Sáenz D, Batlle A, Casas A (2016) Methods for the detection of reactive oxygen species employed in the identification of plant photosensitizers. Methods 109: 73–80 [DOI] [PubMed] [Google Scholar]
- Mao C, Ding J, Zhang B, Xi D, Ming F (2018) OsNAC2 positively affects salt-induced cell death and binds to the OsAP37 and OsCOX11 promoters. Plant J 94(3): 454–468 [DOI] [PubMed] [Google Scholar]
- Martell JD, Deerinck TJ, Lam SS, Ellisman MH, Ting AY (2017) Electron microscopy using the genetically encoded APEX2 tag in cultured mammalian cells. Nat Protoc 12(9): 1792–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY (2012) Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol 30(11): 1143–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignolet-Spruyt L, Xu E, Idänheimo N, Hoeberichts FA, Mühlenbock P, Brosché M, Van Breusegem F, Kangasjärvi J (2016) Spreading the news: subcellular and organellar reactive oxygen species production and signalling. J Exp Bot 67(13): 3831–3844 [DOI] [PubMed] [Google Scholar]
- FJ Miller, Chu X, Stanic B, Tian X, RV Sharma, RL Davisson, FS Lamb (2010) A differential role for endocytosis in receptor-mediated activation of Nox1. Antioxid Redox Signal 12(5): 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7(9): 405–410 [DOI] [PubMed] [Google Scholar]
- Mittler R (2017) ROS are good. Trends Plant Sci 22(1): 11–19 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F (2011) ROS signaling: the new wave? Trends Plant Sci 16(6): 300–309 [DOI] [PubMed] [Google Scholar]
- Mur LA, Lloyd AJ, Cristescu SM, Harren FJ, Hall MA, Smith AR (2009) Biphasic ethylene production during the hypersensitive response in Arabidopsis: a window into defense priming mechanisms? Plant Signal Behav 4(7): 610–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y, Asada K (1981) Hydrogen peroxide is scavenged by ascorbate specific peroxidase in spinach chloroplasts. Plant Cell Physiol 22(5): 867–880 [Google Scholar]
- Ngou BPM, Ahn HK, Ding P, Jones JDG (2021) Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592(7852): 110–115 [DOI] [PubMed] [Google Scholar]
- Ogasawara Y, Kaya H, Hiraoka G, Yumoto F, Kimura S, Kadota Y, Hishinuma H, Senzaki E, Yamagoe S, Nagata K, et al. (2008) Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J Biol Chem 283(14): 8885–8892 [DOI] [PubMed] [Google Scholar]
- Panchuk II, Volkov RA, Schöffl F (2002) Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol 129(2): 838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Fartyal D, Agarwal A, Shukla T, James D, Kaul T, Negi YK, Arora S, Reddy MK (2017) Abiotic stress tolerance in plants: myriad roles of ascorbate peroxidase. Front Plant Sci 8: 581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov V, Hille J, Mueller-Roeber B, Gechev TS (2015) ROS-mediated abiotic stress-induced programmed cell death in plants. Front Plant Sci 6: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann B, Porstmann T, Nugel E, Evers U (1985) Which of the commonly used marker enzymes gives the best results in colorimetric and fluorimetric enzyme immunoassays: horseradish peroxidase, alkaline phosphatase or β-galactosidase? J Immunol Methods 79(1): 27–37 [DOI] [PubMed] [Google Scholar]
- Qi Y, Tsuda K, Glazebrook J, Katagiri F (2011) Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol Plant Pathol 12(7): 702–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Wang J, Gong Z, Zhou JM (2017) Apoplastic ROS signaling in plant immunity. Curr Opin Plant Biol 38: 92–100 [DOI] [PubMed] [Google Scholar]
- Raven EL (2003) Understanding functional diversity and substrate specificity in haem peroxidases: what can we learn from ascorbate peroxidase? Nat Prod Rep 20(4): 367–381 [DOI] [PubMed] [Google Scholar]
- Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42(W1): W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues O, Reshetnyak G, Grondin A, Saijo Y, Leonhardt N, Maurel C, Verdoucq L (2017) Aquaporins facilitate hydrogen peroxide entry into guard cells to mediate ABA- and pathogen-triggered stomatal closure. Proc Natl Acad Sci U S A 114(34): 9200–9205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y, Macho AP (2017) Analysis of PAMP-triggered ROS burst in plant immunity. Methods Mol Biol 1578: 143–153 [DOI] [PubMed] [Google Scholar]
- Savenkova MI, Kuo JM, Ortiz de Montellano PR (1998) Improvement of peroxygenase activity by relocation of a catalytic histidine within the active site of horseradish peroxidase. Biochemistry 37(30): 10828–10836 [DOI] [PubMed] [Google Scholar]
- Shang-Guan K, Wang M, Htwe N, Li P, Li Y, Qi F, Zhang D, Cao M, Kim C, Weng H, et al. (2018) Lipopolysaccharides trigger two successive bursts of reactive oxygen species at distinct cellular locations. Plant Physiol 176(3): 2543–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y, Yoshimura K (2002) Regulation and function of ascorbate peroxidase isoenzymes. J Exp Bot 53(372): 1305–1319 [PubMed] [Google Scholar]
- Shigeoka S, Maruta T (2014) Cellular redox regulation, signaling, and stress response in plants. Biosci Biotechnol Biochem 78(9): 1457–1470 [DOI] [PubMed] [Google Scholar]
- Smirnoff N (2000) Ascorbic acid: metabolism and functions of a multi-facetted molecule. Curr Opin Plant Biol 3(3): 229–235 [PubMed] [Google Scholar]
- Smirnoff N, Wheeler GL (2000) Ascorbic acid in plants: biosynthesis and function. Crit Rev Biochem Mol Biol 35(4): 291–314 [DOI] [PubMed] [Google Scholar]
- Smith JM, Heese A (2014) Rapid bioassay to measure early reactive oxygen species production in Arabidopsis leave tissue in response to living Pseudomonas syringae. Plant Methods 10(1): 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Jiang F, Cen B, Wen J, Zhou Y, Wu Z (2018) Respiratory burst oxidase homologue-dependent H2O2 and chloroplast H2O2 are essential for the maintenance of acquired thermotolerance during recovery after acclimation. Plant Cell Environ 41(10): 2373–2389 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Koussevitzky S, Mittler R, Miller G (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 35(2): 259–270 [DOI] [PubMed] [Google Scholar]
- Thorpe GH, Kricka LJ (1986) Enhanced chemiluminescent reactions catalyzed by horseradish peroxidase. Methods Enzymol 133: 331–353 [DOI] [PubMed] [Google Scholar]
- Tian S, Wang X, Li P, Wang H, Ji H, Xie J, Qiu Q, Shen D, Dong H (2016) Plant aquaporin AtPIP1; 4 links apoplastic H2O2 induction to disease immunity pathways. Plant Physiol 171(3): 1635–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JD (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA 99(1): 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hu M, Wang J, Qi J, Han Z, Wang G, Qi Y, Wang HW, Zhou JM, Chai J (2019) Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364(6435):eaav5870 [DOI] [PubMed] [Google Scholar]
- Wu B, Li P, Hong X, Xu C, Wang R, Liang Y (2022) The receptor-like cytosolic kinase RIPK activates NADP-malic enzyme 2 to generate NADPH for fueling ROS production. Mol Plant 15(5): 887–903 [DOI] [PubMed] [Google Scholar]
- Xie YJ, Xu S, Han B, Wu MZ, Yuan XX, Han Y, Gu Q, Xu DK, Yang Q, Shen WB (2011) Evidence of Arabidopsis salt acclimation induced by up-regulation of HY1 and the regulatory role of RbohD-derived reactive oxygen species synthesis. Plant J 66(2): 280–292 [DOI] [PubMed] [Google Scholar]
- Yang M, Huang J, Fan J, Du J, Pu K, Peng X (2020) Chemiluminescence for bioimaging and therapeutics: recent advances and challenges. Chem Soc Rev 49(19): 6800–6815 [DOI] [PubMed] [Google Scholar]
- Yang H, Mu J, Chen L, Feng J, Hu J, Li L, Zhou JM, Zuo J (2015) S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol 167(4): 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Jiang Z, Bi G, Nomura K, Liu M, Wang Y, Cai B, Zhou JM, He SY, Xin XF (2021a) Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592(7852): 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Ngou BPM, Ding P, Xin XF (2021b) PTI-ETI crosstalk: an integrative view of plant immunity. Curr Opin Plant Biol 62: 102030. [DOI] [PubMed] [Google Scholar]
- Zhang YF, Dai MH, Yuan ZH (2018) Methods for the detection of reactive oxygen species. Anal Methods 10(38): 4625–4638 [Google Scholar]
- Zipfel C (2014) Plant pattern-recognition receptors. Trends Immunol 35(7): 345–351 [DOI] [PubMed] [Google Scholar]
- Zulfugarov IS, Tovuu A, Kim J-H, Lee C-H (2011) Detection of reactive oxygen species in higher plants. J Plant Biol 54(6): 351–357 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.