Abstract
Plants accumulate several metabolites in response to drought stress, including branched-chain amino acids (BCAAs). However, the roles of BCAAs in plant drought responses and the underlying molecular mechanisms for BCAA accumulation remain elusive. Here, we demonstrate that rice (Oryza sativa) DROUGHT-INDUCED BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE (OsDIAT) mediates the accumulation of BCAAs in rice in response to drought stress. An in vitro enzyme activity assay indicated that OsDIAT is a branched-chain amino acid aminotransferase, and subcellular localization analysis revealed that OsDIAT localizes to the cytoplasm. The expression of OsDIAT was induced in plants upon exposure to abiotic stress. OsDIAT-overexpressing (OsDIATOX) plants were more tolerant to drought stress, whereas osdiat plants were more susceptible to drought stress compared with nontransgenic (NT) plants. Amino acid analysis revealed that BCAA levels were higher in OsDIATOX but lower in osdiat compared with in NT plants. Finally, the exogenous application of BCAAs improved plant tolerance to osmotic stress compared with that in control plants. Collectively, these findings suggest that OsDIAT mediates drought tolerance by promoting the accumulation of BCAAs.
Branched-chain amino acid aminotransferase mediates the accumulation of branched-chain amino acids in response to drought stress, which confers drought tolerance in rice.
Introduction
Drought is a major abiotic stress that adversely affects rice (Oryza sativa) yield due to drought-induced phenomena such as reduced spikelet number and low grain-filling rates (Jin et al., 2013). Recent global climate change has been increasing the frequency and severity of drought in important cultivation areas. These changes have motivated efforts to improve crop productivity by manipulating drought tolerance mechanisms.
To cope with drought stress, plants have evolved protective mechanisms that allow them to acclimate to drought stress naturally (Valliyodan and Nguyen, 2006; Harb et al., 2010; Todaka et al., 2015). One such mechanism that is widely conserved in plant species is the accumulation of small organic molecules—amino acids, betaines, sugars, organic acids, and other osmolytes—induced by drought stress (Delauney and Verma, 1993; Urano et al., 2009; Joshi et al., 2010; Bowne et al., 2012). Metabolic acclimation via the accumulation of compatible osmolytes has long been considered a general protective strategy of plants under drought stress (Fiehn et al., 2000; Yancey, 2005; Chen et al., 2007; Urano et al., 2009; Huang and Jander, 2017; Ullah et al., 2017). Compatible osmolytes enhance tolerance to drought conditions by adjusting the osmotic balance, protecting protein structure, and maintaining membrane integrity (Yancey et al., 1982; Hare et al., 1998; Wani et al., 2013; Khan et al., 2015). Therefore, determining the molecular mechanism and engineering of compatible osmolyte biosynthesis pathways have become a focus of study as a way to improve drought tolerance in plants (Chen et al., 2007; Wani et al., 2013; Khan et al., 2015; Todaka et al., 2015).
There is extensive evidence of amino acids and their metabolites providing stress tolerance by functioning as compatible osmolytes (Campalans et al., 1999; Ashraf and Foolad, 2007; Joshi et al., 2010; Abid et al., 2018). For instance, the accumulation of proline has been observed not only in plants but also in eubacteria, protozoa, and algae (Maggio et al., 2002; Liang et al., 2013) and is positively associated with stress tolerance (Delauney and Verma, 1993; Lehmann et al., 2010). On the other hand, proline metabolism plays an important role in regulating cell redox status (NADP/NADPH ratio) to enhance plant adaptation and tolerance to drought stress (Sharma et al., 2011; Zheng et al., 2021). Moreover, proline is not the only amino acid that accumulates in plants in response to abiotic stress. Three essential amino acids, isoleucine, leucine, and valine, which are collectively referred to as branched-chain amino acids (BCAAs) due to their short branched hydrophobic side chains, also accumulate copiously in plants in response to drought stress (Urano et al., 2009; Joshi et al., 2010; Bowne et al., 2012). Like proline, BCAAs might help enhance plant drought tolerance by acting as compatible osmolytes or alternative energy sources (Taylor et al., 2004; Joshi et al., 2010; Huang and Jander, 2017; Fabregas and Fernie, 2019). However, it is still unclear how BCAA accumulation is regulated by drought stress and how it confers drought tolerance in plants.
The biosynthesis of BCAAs mainly occurs in plastids, and many enzymes involved in BCAA biosynthesis contain plastid-localizing signal peptides (Diebold et al., 2002; Graham and Eastmond, 2002; Maloney et al., 2010; Xing and Last, 2017). BCAA biosynthesis is unique in that four identical enzymes are involved in reactions with different substrates to synthesize the corresponding BCAAs. Acetohydroxy acid synthase (EC 4.1.3.18) is responsible for the first step of BCAA biosynthesis by catalyzing the condensation of two pyruvates, as well as pyruvate and 2-ketobutyrate. Ketol-acid reductoisomerase (EC 1.1.1.86) and dihydroxy-acid dehydratase (EC 4.2.1.9) sequentially catalyze the next two steps to produce branched-chain 2-oxo acids. The final transamination step in BCAA biosynthesis is catalyzed by branched-chain amino acid aminotransferase (BCAT, EC 2.6.1.42) (Binder, 2010; Pratelli and Pilot, 2014).
Unlike BCAA biosynthesis, BCAA degradation occurs in the mitochondria or peroxisomes. Besides the final step in BCAA biosynthesis, BCAT is also involved in the first step in BCAA degradation. The branched-chain α-keto acids are then further degraded by branched-chain keto acid dehydrogenase (BCKD, EC 1.2.4.4) and isovaleryl-CoA dehydrogenase (EC 1.3.99.10). Allosteric inhibition of these committed enzymes tightly regulates BCAA homeostasis by altering the levels of their end-products at the protein level. However, the transcriptional regulation of genes involved in BCAA metabolism is also thought to play a critical role in plants, especially under stress conditions (Urano et al., 2009; Binder, 2010; Chen et al., 2010; Joshi et al., 2010). BCAT is thought to be a key regulator of BCAA accumulation in plants under stress conditions due to its stress-inducible expression patterns and association with BCAA levels (Urano et al., 2009; Joshi et al., 2010). For instance, Arabidopsis (Arabidopsis thaliana) BCAT2 was identified as the gene responsible for abscisic acid (ABA)-dependent accumulation of BCAA (Urano et al., 2009). In addition, the expression of Arabidopsis BCAT1 and BCAT2 is induced under salt stress (Joshi et al., 2010). Like BCAT, BCAA catabolic enzyme genes BCKD and IVD are also upregulated at the transcriptional level under stress conditions (Peng et al., 2015). The activation of BCAA degradation is beneficial for plant survival under stress conditions because BCAA catabolites can also be used as an alternative energy source.
In this study, we investigated the molecular functions of rice DROUGHT-INDUCED BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE (OsDIAT), a rice BCAT belonging to the aminotransferase IV family, in BCAA biosynthesis and drought tolerance. Overexpression of OsDIAT resulted in the increased accumulation of BCAAs under both normal and drought conditions. OsDIAT-overexpressing (OsDIATOX) plants showed higher survival rates and grain yield under drought conditions compared with the wild type. By contrast, osdiat plants accumulated fewer BCAAs and showed reduced tolerance to drought stress. Finally, the exogenous application of BCAAs enhanced plant tolerance to polyethylene glycol (PEG)-driven osmotic stress. These results suggest that OsDIAT regulates the drought-inducible accumulation of BCAAs, which in turn enhances drought tolerance in plants.
Results
OsDIAT is a drought-induced gene encoding a cytoplasmic BCAT
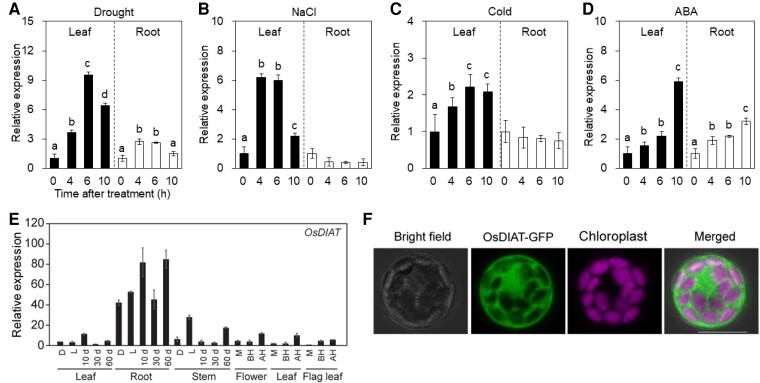
BCAAs are synthesized via multiple enzymatic reactions. BCATs are strong candidate enzymes that are likely responsible for increasing BCAA levels under abiotic stress conditions (Matsui et al., 2008; Urano et al., 2009). Phylogenetic analysis grouped putative rice BCAT genes into two distinct subgroups (Supplemental Figure 1). OsDIAT (Os05g0244700) belongs to a subgroup with two additional putative BCAT genes (Os01g0238500 and Os02g0273100), while five other genes form another subgroup with Arabidopsis plastidial BCATs (Binder, 2010). We examined the expression patterns of these putative BCAT genes under drought conditions based on previously reported RNA-sequencing data (Chung et al., 2016). Among the 20 genes, eight genes showed more than two-fold expression changes under drought conditions. The expression change of OsDIAT was most prominent among eight genes; therefore, we focused on the OsDIAT instead of other genes (Supplemental Table 1). OsDIAT expression was induced 1 day after drought treatment and further induced 2–3 days after drought treatment. To verify the induction of OsDIAT expression under drought and other abiotic stress conditions, we performed reverse transcription quantitative PCR (RT-qPCR) analysis using total RNA from the leaves and roots of plants exposed to drought, high salinity, and low-temperature conditions (Figures 1A–C). OsDIAT expression was significantly induced in both leaves and roots under drought conditions, with stronger induction in leaves (Figure 1A). The abiotic stress conditions of high salinity and low-temperature treatment also induced OsDIAT expression in leaf tissues (Figure 1, B and C). Therefore, OsDIAT expression is predominantly induced in leaves by abiotic stress.
Figure 1.
Expression patterns of OsDIAT in response to abiotic stress. (A–D) Relative expression patterns of OsDIAT in response to four different abiotic stresses and ABA treatment. Two-week-old seedlings were exposed to air-drying (drought) (A), 400 mM NaCl (high salinity) (B), 4°C (low temperature) (C), and 100 μM abscisic acid (ABA) (D). Leaves and roots of the plants were harvested at the indicated time points after treatment. (A–D) Different letters indicate significant differences between NT and transgenic plants at P < 0.05 (ANOVA followed by Tukey's honestly significant difference) (E) RT-qPCR analysis of OsDIAT expression in rice tissues at different developmental stages. D, day; D, dark; L, light; M, meiosis; BH, before heading; AH, after heading. (A–E) Rice UBIQUITIN1 (OsUBI1) was used as an internal control for normalization, and data represent mean values ± standard deviation (SD) of three independent samples (n = 5). (F) Subcellular localization of OsDIAT in rice protoplasts. Protoplasts were transiently transfected with OsDIAT-GFP expression constructs. Fluorescence was observed in protoplasts 12 h after transfection under a confocal microscope. Scale bar, 10 μm.
ABA is required for BCAA accumulation under drought conditions (Urano et al., 2009; Huang and Jander, 2017). To investigate the potential role of ABA in regulating OsDIAT expression, we analyzed the promoter region of OsDIAT in silico and identified two ABA-responsive elements within the 2-kb promoter region (Supplemental Figure 2A). To examine the expression of OsDIAT in response to ABA treatment, we performed RT-qPCR analysis (Figure 1D; Supplemental Figure 2). As a positive control, we used rice DEHYDRATION-INDUCIBLE PROTEIN1 (OsDIP1), whose expression increases in response to ABA (Supplemental Figure 2B). OsDIAT expression showed dosage- and time-dependent induction in response to ABA treatment in both leaves and roots, as did OsDIP1 (Figure 1D; Supplemental Figure 2C). We investigated the developmental regulation of OsDIAT expression by performing spatiotemporal expression profiling, finding that OsDIAT was more highly expressed in roots than in other tissues (Figure 1E).
Since the functions of BCATs depend on their subcellular localization in plants (Diebold et al., 2002), we examined the subcellular localization of OsDIAT. OsDIAT was predicted to be a cytoplasmic BCAT by in silico analysis (Target P-2.0; https://services.healthtech.dtu.dk/service.php? TargetP-2.0). To confirm the subcellular localization of OsDIAT, we generated a construct expressing the coding sequence of OsDIAT translationally fused with green fluorescent protein (GFP) under the control of the 35S promoter (35S::OsDIAT-GFP). We transiently expressed this plasmid in rice. The GFP signal was detected in the cytosol but not in the mitochondria, a site of BCAA degradation (Binder, 2010) (Figure 1F). In order to investigate the kinetic properties of OsDIAT, the protein was expressed in Escherichia coli and purified. Enzyme assays were performed with the recombinant OsDIAT in both the forward (synthesis) and reverse (degradation) directions. Table 1 shows the kinetic values with all six branched-chain substrates. OsDIAT exhibits higher efficiency in the forward direction than the reverse, having higher affinity for the BCKAs than BCAAs. Taken together, these results suggest that OsDIAT is a drought-induced cytosolic BCAT that converts α-keto acids to BCAAs.
Table 1.
Kinetic parameters of OsDIAT. Activities of purified recombinant OsDIAT protein on all BCAA and BCKA substrates. Km is presented as average ± Se. Kinetic data were obtained using GraphPad Prism 8 software
| Substrate | K m | V max | K cat | K cat /Km |
|---|---|---|---|---|
| mM | nkat mg−1 | s−1 | µM−1 s−1 | |
| KIC | 0.37 ± 0.07 | 1.2 | 33.7 | 0.091 |
| KMV | 0.44 ± 0.08 | 1.4 | 37.4 | 0.085 |
| KIV | 0.64 ± 0.06 | 1.7 | 47.4 | 0.074 |
| Leu | 4.89 ± 0.53 | 4.1 | 112.5 | 0.023 |
| Ile | 5.14 ± 0.45 | 6.5 | 179.9 | 0.035 |
| Val | 5.42 ± 0.72 | 3.7 | 103.0 | 0.019 |
OsDIAT mediates drought tolerance
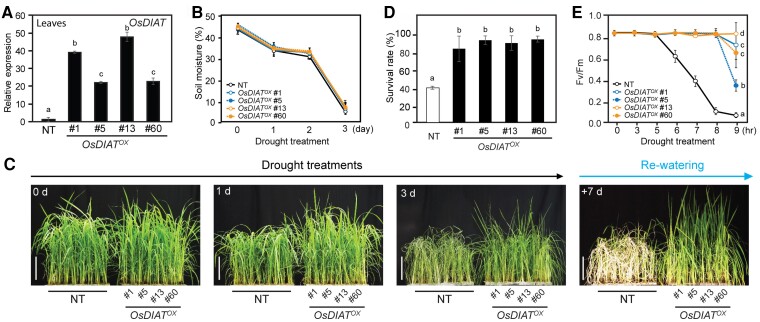
To investigate the biological functions of OsDIAT in rice under drought conditions, we generated OsDIAT-overexpressing and osdiat mutant plants. Among the 70 independent OsDIAT-overexpressing transgenic lines produced, we selected plants that did not show stunting to eliminate the effects of somaclonal variations. Based on the expression levels of OsDIAT in transgenic plants, we chose four independent single-copy homozygous transgenic lines (OsDIATOX #1, #5, #13, and #60) for further analysis (Figure 2A). To examine the performance of plants under drought conditions at the vegetative stage, we grew the selected OsDIATOX and nontransgenic (NT, Dongjin) plants in a greenhouse for 4 weeks and exposed them to drought stress by withholding water (Figure 2, B–E). Soil moisture content, an indicator of drought treatment, showed a similar rate of decrease across the plants, confirming that the stress treatment was uniformly applied (Figure 2B). Drought-induced symptoms, such as wilting, leaf rolling, and chlorosis, appeared earlier in NT plants compared with in OsDIATOX plants during drought treatment (Figure 2C). The OsDIATOX plants also showed better recovery rates compared with that of NT plants after being relieved from drought stress by re-watering (Figure 2, C and D). The OsDIATOX plants showed an 82% to 93% survival rate at 7 days after re-watering, whereas NT plants only showed a 40% survival rate (Figure 2D). To further verify the performance of the plants under drought conditions, the Fv/Fm values, an indicator of the photochemical efficiency of photosystem II, were measured in plants exposed to drought stress. The Fv/Fm values in NT plants started to decrease rapidly at 5 days after drought treatment, whereas values in OsDIATOX plants showed a decrease at 9 days after treatment (Figure 2E). Although OsDIAT expression level is different among transgenic OsDIATOX lines, all the transgenic lines showed similar drought-tolerance phenotypes (Figure 2C–E), suggesting that expression level of #5 and #60 lines were already high enough for the performance, and expression variation among the transgenic lines did not substantially affect the performance.
Figure 2.
OsDIAT overexpression enhances drought resistance. A, Relative expression levels of OsDIAT in the leaves of nontransgenic (NT) and four independent T3 homozygous lines of GOS2::OsDIAT (OsDIATOX) plants. Total RNA extracted from 2-week-old seedlings was used for RT-qPCR analysis (n > 5, biological repeat >3). OsUBI1 was used as an internal control for normalization. B, Measurement of soil moisture contents (n > 30, biological repeat >3). (A, B) Data represent mean values ± standard deviation (SD). Different letters indicate significant differences between NT and transgenic plants at P < 0.05 (ANOVA followed by Tukey's honestly significant difference). C, Phenotypes of OsDIATOX transgenic and NT plants during drought treatment. Two-month-old plants from four independent T3 homozygous OsDIATOX lines and NT plants were exposed to drought stress for 3 days and then re-watered. Numbers in the images indicate the duration of drought treatment and re-watering. Scale bars, 20 cm. D, Survival rates of OsDIATOX transgenic and NT plants after re-watering (n > 30, biological repeat >3). Chlorophyll fluorescence (Fv/Fm) values of OsDIATOX transgenic and NT plants under drought conditions. Five-week-old plants from four independent T3 homozygous lines of OsDIATOX and NT plants were exposed to drought stress for 9 days. Chlorophyll fluorescence was measured in the dark at the indicated time points using a Handy-PEA fluorometer. (n > 10, biological repeat >3) (D–E) Data represent mean values ± standard deviation (SD). Different letters indicate significant differences between NT and transgenic plants at P < 0.05 (ANOVA followed by Tukey's honestly significant difference).
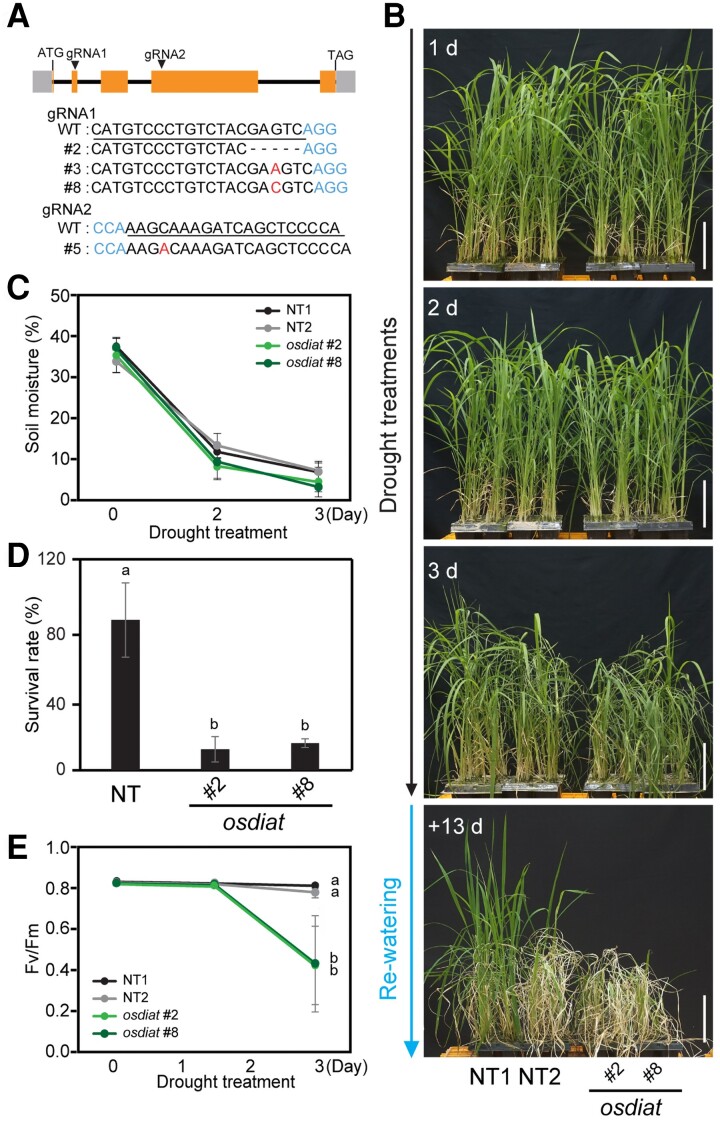
We also investigated the drought responses of osdiat plants. We generated four independent osdiat mutants via CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 (CRISPR associated protein 9)-mediated gene editing with two independent single-guide RNAs (Figure 3A; Supplemental Figure 3). Each mutant showed different patterns of mutation, deletion, and insertion in the OsDIAT locus. RT-qPCR results showed that osdiat mutants still produce the transcripts (Supplemental Figure 3B); however, sequencing results demonstrated that they are abnormal transcripts (Supplemental Figure 3C). The osdiat plants were more susceptible to a time course of drought treatment than NT plants (Figure 3B), while soil moisture contents were reduced to similar levels for both osdiat and NT plants (Figure 3C). The osdiat plants showed lower survival rates following drought treatment than NT control plants (Figure 3D). The Fv/Fm values of osdiat plants started to decrease more rapidly (at 2 days after drought treatment) than those of NT plants (Figure 3E). These results suggest that OsDIAT is required for drought responses in rice and that overexpressing OsDIAT enhances drought tolerance in plants at the vegetative stage.
Figure 3.
Drought responses of osdiat plants. A, Mutation patterns of osdiat plants. Blue, protospacer adjacent motif (PAM) sequence; red, nucleotide insertion; underline, sgRNA sequence; hyphen, nucleotide deletion. B, The drought responses of osdiat and NT plants during drought treatment. One-month-old plants of two independent osdiat lines (#2 and #8) and NT plants were exposed to drought stress for 3 days and then re-watered. Numbers in the images indicate the duration of drought treatment and re-watering. Scale bars, 10 cm. C, Measurement of soil moisture contents. D, Survival rates of osdiat and NT plants after re-watering. € Chlorophyll fluorescence (Fv/Fm) values of osdiat transgenic (#2 and #8) and NT control plants under drought conditions. Four-week-old plants were exposed to drought stress for 3 days. Chlorophyll fluorescence was measured in the dark at the indicated time points using a Handy-PEA fluorometer. (C–E) Data represent mean values ± standard deviation (SD). (n > 30, biological repeat >3) Different letters indicate significant differences between NT and transgenic plants at P < 0.05 (ANOVA followed by Tukey's honestly significant difference).
Overexpression of OsDIAT increases grain yield under field drought conditions
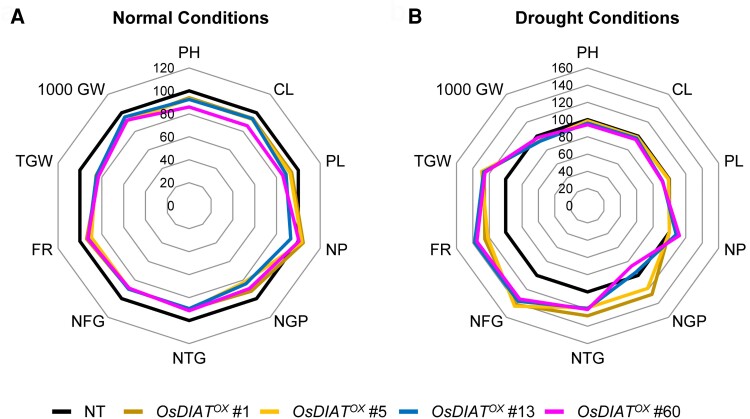
Yield components, including grain-filling rate, number, and weight, are essential criteria used to evaluate plant drought tolerance at the reproductive stage (Atkinson and Urwin, 2012; Jin et al., 2013; Sehgal et al., 2018). To test the drought tolerance of OsDIATOX transgenic plants against drought at the reproductive stage of growth, we evaluated the yield parameters of OsDIATOX plants under both normal and drought conditions in the field. We planted four independent T5 homozygous OsDIATOX lines and NT plants in a paddy field and scored yield parameters in 30 plants per independent line with two replicates (Figure 4, A and B; Supplemental Table 2). OsDIATOX plants exposed to drought stress conditions at the reproductive stage showed dramatically higher total grain weight, grain-filling rate, and the number of filled grains compared with NT plants (Figure 4B; Supplemental Table 2). Specifically, the total grain weight of OsDIATOX plants was 25%–29% higher than that of NT plants. Similarly, the grain-filling rate and the number of filled grains were significantly higher in OsDIATOX plants versus NT plants (21%–37% higher and 34%–44% higher, respectively). On the other hand, OsDIATOX plants did not exhibited significant difference in yield parameters compared with NT plants under the normal growth condition although there were some variations among transgenic lines (Figure 4A; Supplemental Table 2). The significant increases in yield components of OsDIATOX plants under drought conditions suggest that overexpression of OsDIAT enhanced drought tolerance and relieved the damage by drought stress during the reproduction stage.
Figure 4.
Agronomic traits of OsDIATOX transgenic plants under field conditions. Four independent T5 OsDIATOX and nontransgenic (NT) plants were grown in the field under normal and drought conditions. Agronomic traits were analyzed from 10 plants per line. Data represent the percentage of the mean value with NT plants assigned a reference value of 100%. PH, plant height; CL, culm length; PL, panicle length; NP, number of panicles per hill; NGP, number of grains per panicle; NTG, number of total grains per hill; NFG, number of filled grains per hill; FR, filling rate; TGW, total grain weight; 1,000 GW, thousand-grain weight.
BCAA levels are significantly elevated in OsDIATOX leaves
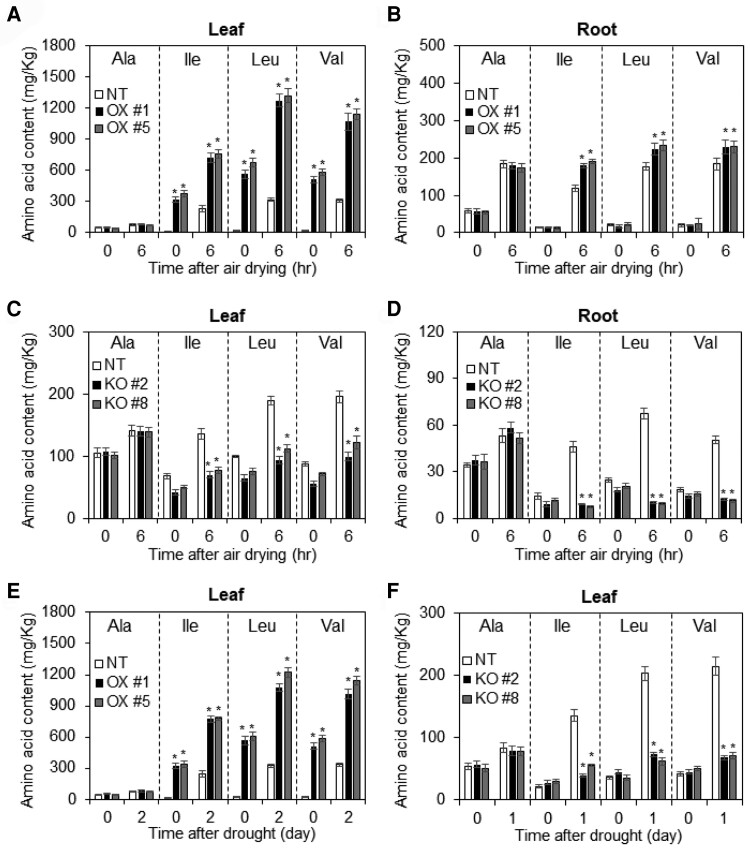
BCAA accumulates when plants are exposed to drought stress (Huang and Jander, 2017), but whether BCAA accumulation directly contributes to drought tolerance had not previously been tested. To examine the effects of BCAA contents on drought tolerance, we measured the BCAA levels of NT plants under both normal and drought conditions (Figure 5). Under normal conditions, BCAA levels in the leaf and root tissues of NT plants were comparable. However, BCAA levels in both tissues were significantly higher under drought stress conditions (Figure 5A–F). These results confirm the finding that plants accumulate BCAAs when exposed to drought stress. We then analyzed BCAA levels in OsDIATOX and osdiat plants to determine whether OsDIAT overexpression or mutation alters BCAA accumulation in plants (Figures 5A–F). BCAA levels were significantly higher in leaf tissues of OsDIATOX versus NT plants under normal growth conditions and further increased in OsDIATOX plants when both groups were exposed to drought stress (Figure 5, A and E). By contrast, the BCAA levels in root tissues of OsDIATOX plants remained similar to those of NT plants under normal growth conditions, although they were higher in OsDIATOX than in NT roots under drought-stress conditions (Figure 5B). Interestingly, the BCAA levels in leaf and root tissues of osdiat plants were similar to those of NT plants under normal growth conditions (Figure 5, C and D) but were significantly lower than those of NT plants under drought conditions (Figure 5, C, D and F). These results suggest that drought induces BCAA accumulation in plants and that OsDIAT mediates this process.
Figure 5.
Branched-chain amino acid contents of OsDIATOX and osdiat plants. Three-week-old OsDIATOX, osdiat, and nontransgenic (NT) plants were exposed to (A–D) dehydration (air-drying) and (E–F) drought (soil) stress. BCAA contents in (A) leaves and (B) roots of OsDIATOX and NT plants after air-drying treatment. BCAA contents in (C) leaves and (D) roots in osdiat and NT plants after air-drying treatment. BCAA contents in leaves OsDIATOX € and osdiat plants (F) after drought treatment (6-week-old soil-grown plants). Data represent mean values ± SD (n > 5, biological repeat >3). Significant differences from the NT control are indicated by asterisks (ANOVA followed by Tukey's honestly significant difference, *P < 0.05).
Exogenous BCAAs enhance osmotic stress tolerance
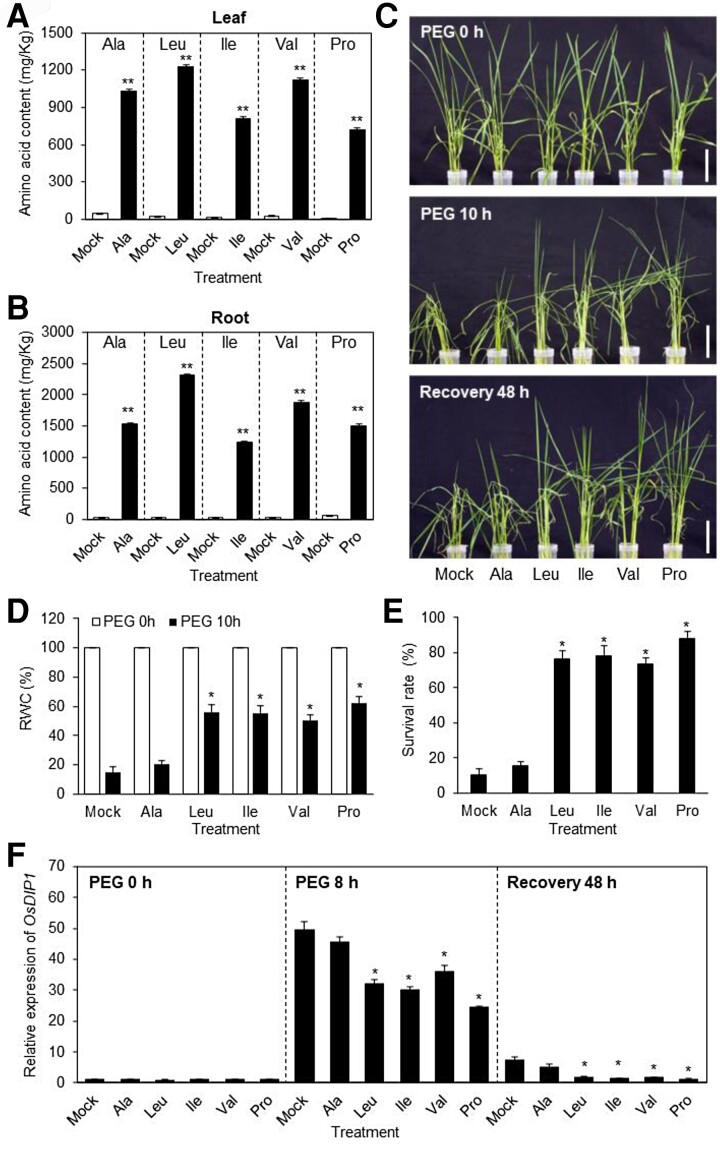
To investigate the effects of BCAAs on osmotic stress tolerance, we examined the response of plants treated with exogenous BCAAs under osmotic stress conditions driven by PEG treatment (Figure 6). To increase internal BCAA levels, roots were pretreated with alanine (negative control), leucine, isoleucine, valine, or proline (positive control). We analyzed BCAA levels in plants pretreated with exogenous BCAAs using HPLC. The exogenous application of BCAAs resulted in increased levels of the corresponding BCAAs in both leaf and root tissues (Figure 6, A and B). To investigate the enhanced drought tolerance of plants induced by exogenous application of BCAAs, we transferred plants pretreated with BCAA to PEG solution and monitored osmotic stress-induced symptoms. During PEG treatment, osmotic stress-related symptoms such as leaf-rolling and wilting appeared earlier in mock-treated plants and plants pretreated with alanine compared with plants pretreated with BCAA or proline (Figure 6C). The relative water content (RWC) of plants pretreated with BCAA or proline was significantly higher than that of mock-treated plants or plants pretreated with alanine (Figure 6D). Pretreatment with BCAA also improved the recovery of plants after they were relieved from osmotic stress via transfer to normal growth medium (Figure 6E). Finally, we monitored the degree of dehydration stress caused by PEG treatment by analyzing the expression of OsDIP1. OsDIP1 expression was induced in plants pretreated with BCAAs during PEG treatments, but the levels of OsDIP1 induction in plants pretreated with BCAA or proline were lower than those in mock-treated plants or plants pretreated with alanine (Figure 6F). Moreover, the expression levels of OsDIP1 were lower in plants pretreated with BCAA than mock control plants after recovery (Figure 6F). These results suggest that the accumulation of BCAAs is sufficient to induce osmotic stress tolerance in plants.
Figure 6.
BCAA accumulation confers osmotic stress tolerance in rice. Three-week-old nontransgenic (NT) plants were incubated in a solution containing 10 mM amino acids for 24 h before being transferred to 25% PEG solution for osmotic stress treatment. Amino acid contents in leaf (A) and root (B) tissue of plants pretreated with amino acids. Plants were harvested 24 h after incubation in solution containing 10 mM alanine (Ala), leucine (Leu), isoleucine (Ile), valine (Val), and proline (Pro). (A, B) Data represent mean values ± SD (n > 10, biological repeat >3). Significant differences from the mock-treated control are indicated by asterisks (Student's t test, *P < 0.05, **P < 0.01). C, Phenotypes of NT plants pretreated with BCAAs during PEG treatment. Numbers in the images indicate the duration of PEG treatment and recovery. Scale bars, 5 cm. D, Relative water content (RWC) in the leaf tissues of plants pretreated with different amino acids after PEG treatment. E, Survival rates of plants pretreated with different amino acids after recovery. F, OsDIP1 expression was monitored during PEG treatment and recovery by RT-qPCR. OsUBI1 expression was used as an internal control for normalization. (D–F) Data represent mean values ± SD (n > 10, biological repeat >3). Significant differences from the mock-treated control are indicated by asterisks (ANOVA followed by Tukey's honestly significant difference, *P < 0.05).
Discussion
Due to the importance of BCAAs as essential amino acids and potential protective metabolites, increasing BCAA levels in plants is of great interest for crop engineering. Despite their biological and industrial importance, engineering plants with increased BCAA levels is difficult due to complex feedback regulation in BCAA biosynthesis and degradation pathways (Chen et al., 2010; Gu et al., 2010; Peng et al., 2015; Xing and Last, 2017). Therefore, identifying specific targets that can increase plant BCAA contents will facilitate the development of crops with higher nutritional value and enhanced stress tolerance. BCAT is a strong candidate enzyme responsible for the control of BCAA accumulation in plants. In tomato (Solanum lycopersicum), the two major quantitative trait loci for elevated BCAA levels co-localize with SIBCAT1 and SIBCAT4 (Maloney et al., 2010). Similarly, AtBCAT2 is a causative gene of natural variation in BCAA contents in Arabidopsis (Angelovici et al., 2013). However, whether BCAT is responsible for drought tolerance in plants had not yet been investigated. In this study, we demonstrated that OsDIAT mediates the accumulation of BCAAs under drought stress conditions. Through RNA sequencing and gene expression analysis, we determined that OsDIAT was significantly upregulated under drought conditions (Figure 1; Supplemental Table 1). We also found that OsDIAT expression is controlled by ABA (Figure 1D; Supplemental 2), which is in agreement with the finding that ABA is required for drought-induced accumulation of BCAAs in plants (Urano et al., 2009; Huang and Jander, 2017). These results suggest that OsDIAT alters the accumulation of BCAAs in response to drought stress in an ABA-dependent manner.
Subcellular localization analysis in protoplasts revealed that OsDIAT localizes to the cytosol (Figure 1F). The functions of BCAT enzymes in BCAA metabolism are closely related to their subcellular localizations (Diebold et al., 2002; Binder, 2010; Maloney et al., 2010). For example, plastid-localized BCATs are involved in the biosynthesis of BCAAs, while BCATs localized to mitochondria are thought to function in the degradation of BCAAs. Several BCATs are also localized to the cytoplasm in Arabidopsis (AtBCAT4 and AtBCAT6) and tomato (SIBCAT5) (Diebold et al., 2002; Maloney et al., 2010). Although the role of cytosolic BCATs remains unclear, in vitro enzyme assays suggested that they function in BCAA biosynthesis (Diebold et al., 2002; Maloney et al., 2010). Cytosolic BCATs are also thought to regulate specific developmental processes or plant responses to stress conditions (Binder, 2010; Maloney et al., 2010). The current data support the idea that a cytosolic BCAT might regulate BCAA biosynthesis under stress conditions.
The potential involvement of BCAT in a drought tolerance pathway has been proposed in Arabidopsis. Arabidopsis NAM, ATAF, and CUC family 16 (AtNAC016) negatively regulate drought tolerance by downregulating the ABA-dependent transcription factor gene ABA-RESPONSIVE ELEMENT BINDING PROTEIN1 (Sakuraba et al., 2015). Moreover, BCAT1 expression is negatively regulated by AtNAC016. Finally, BCAT3 is involved in ETHYLENE RESPONSIVE FACTOR 19-dependent drought tolerance mechanisms (Winter et al., 2007).
Similar to what has been seen in other plants species (Ranieri et al., 1989; Urano et al., 2009; Bowne et al., 2012), BCAAs accumulated in both the leaves and roots of rice under drought conditions (Figure 5). These findings suggest that BCAA accumulation is a conserved strategy that helps plants cope with drought stress. Although the detailed mechanisms are not yet fully understood, BCAAs are thought to induce drought tolerance in plants by acting as compatible osmolytes or as a source of alternative energy under stress conditions (Taylor et al., 2004; Joshi et al., 2010). Therefore, the increased drought tolerance of OsDIATOX plants could be explained by the enhanced accumulation of BCAAs (Figure 5). Indeed, the transcriptional regulation of OsDIAT is essential for BCAA accumulation under drought conditions.
Treatment with peptides or amino acids can help protect plants against abiotic stresses such as drought (Colla et al., 2017; Haghighi et al., 2020). We determined that the exogenous application of individual BCAAs ameliorated the adverse effects of PEG-driven osmotic stress. Interestingly, their impact on osmotic stress tolerance was similar to that of proline, the best-characterized metabolite for plant drought tolerance (Figure 6). By contrast, alanine treatment did not significantly affect the osmotic stress tolerance of plants. These results suggest that not all amino acids confer osmotic stress tolerance in plants. It has been proposed that proline plays other roles beyond simply functioning as an osmoprotectant. Proline metabolism is thought to modulate the redox balance and energy status of the cell and contribute to stress tolerance by maintaining the NADPH/NADP+ balance and antioxidant levels (Sharma et al., 2011; Zheng et al., 2021). A recent study showed that the enzyme activity of BCAT, which harbors a redox-active motif (CXXC), is modulated by peroxide-mediated redox status, indicating that BCAA metabolism is also closely associated with cellular redox balance (Conway, 2021). These findings suggest that BCAA metabolism shares a common mechanism with proline metabolism in drought tolerance.
Most of the yield parameters, including total grain number and weight, of OsDIATOX were significantly higher than those of NT plants after drought treatment at the early reproductive stage (Figure 4B; Supplemental Table 2). It has been documented that drought affects rice reproduction primarily by reducing male fertility and grain filling, which causes severe losses of grain yield (Jin et al., 2013; Sehgal et al., 2018). We did not observe a significant difference in vegetative and reproductive growth between OsDIATOX and NT under normal conditions. However, a dramatic difference in filling rate was observed between OsDIATOX and NT plants, while 1,000 seed weight was slightly reduced in NT compared with in OsDIATOX after drought treatment. It suggested that yield improvement in OsDIATOX plants was mainly due to the difference of filling rate (Figure 4B; Supplemental Table 2) and overexpressing OsDIAT greatly relieves the damage by drought stress during fertilization. On the other hand, it appears that overexpressing OsDIAT has a partly negative effect on the agronomic traits, especially the number of total grains per hill, under normal field conditions although the difference in yield parameters between NT and OsDIATOX plants was marginal (Figure 4A; Supplemental Table 2). Constitutive overexpression of OsDIAT might have affected the reproductive growth of the plants by inducing the production of particular amino acids (Lehmann et al., 2010; Stuttmann et al., 2011). For future development, tissue (especially leaf) specific promoter, instead of whole-body constitutive promoter, could improve the yield variations under normal conditions, maintaining the drought tolerance.
In conclusion, we provided direct genetic evidence that BCAAs confer drought tolerance in plants. We also identified a valuable candidate gene for engineering crops with high BCAA contents and enhanced drought tolerance.
Materials and methods
Plant growth conditions and ABA treatment
Rice (Oryza sativa “Dongjin”) seeds were sown on Murashige and Skoog (MS) solid medium and incubated in the dark for 4 days at 28°C. The seedlings were transferred to a growth chamber with a 16-h-light/8-h-dark cycle, the light intensity of 200 μmol m−2 s−1, and relative humidity of 70%. One-week-old seedlings were transferred to hydroponic solution (Yoshida et al., 1976) and grown for two more weeks for gene expression analysis. To examine the ABA concentration-dependent response of OsDIAT, eight seedlings were transferred to 50-ml tubes containing 0, 1, 10, or 100 μM ABA solution (Sigma, USA). Seedlings were harvested 6 h after ABA treatment for RNA extraction. To examine the sensitivity of OsDIAT to ABA treatment, 2-week-old rice seedlings were transferred to 50-ml tubes containing 100 μM ABA. The seedlings were harvested at the indicated time points (0, 2, 4, and 6 h) for RNA extraction.
Phylogenetic analysis
Phylogenetic analysis was carried out using the amino acid sequences of 49 putative rice aminotransferase proteins using the annotation from the riceXPro public database (http://ricexpro.dna.affrc.go.jp/). The phylogenetic tree was constructed with CLC workbench software using the deduced protein sequences from genes with the following IDs: Os01g0178000, Os01g0238500, Os01g0290100, Os01g0290600, Os01g0729600, Os01g0736400, Os01g0760600, Os02g0236000, Os02g0252600, Os02g0273100, Os02g0302400, Os02g0302700, Os02g0709200, Os02g0797500, Os02g0806900, Os03g0106400, Os03g0171900, Os03g0195100, Os03g0231600, Os03g025800, Os03g0299900, Os03g0338000, Os04g0405700, Os04g0559400, Os04g0614500, Os05g0129100, Os05g0244700, Os05g0475400, Os05g0558400, Os06g0345200, Os06g0548000, Os07g0106700, Os07g0108300, Os07g0461900, Os07g0617800, Os08g0245400, Os08g0532200, Os08g0532200, Os09g0433900, Os10g0189600, Os10g0390500, Os10g0390600, Os10g0484700, Os10g0549500, Os10g0560900, Os11g0209900, Os11g055200, Os11g0644800, and Os12g0131100 (https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-clc-genomics-workbench/).
Plasmid construction and rice transformation
To generate overexpression plants, the coding sequence of OsDIAT (Os05g0244700) was amplified from rice (O. sativa L. ssp. japonica “Nipponbare”) total RNA using the Reverse Transcription System (Promega, USA) and PrimeSTAR HS DNA polymerase (Takara, Japan). The amplified OsDIAT coding sequence was cloned into rice transformation vector p700 carrying the GOS2 promoter for constitutive overexpression using the Gateway Cloning system (Invitrogen, USA) (Supplemental Table 3 and Jeong et al., 2010). The final construct (GOS2::OsDIAT) was introduced into rice (O. sativa “Dongjin”) by Agrobacterium (LBA4404)-mediated transformation as described previously (Jang et al., 1999). The primers used for plasmid construction are listed in Supplemental Table 3. The copy numbers of OsDIAT in the transgenic plants were determined by TaqMan Q-PCR (Thermo Fisher, USA) using probes specific for the Bar gene (which confers resistance to the herbicide Basta). To analyze the copy number of OsDIAT in the transgenic plants, genomic DNA was extracted from 2-week-old seedlings. Genomic DNA extracted from transgenic plants from a previously confirmed homozygous line with a single insert was used as a control. The single-copy insertion lines were allowed to self-fertilize, and homozygous transgenic lines were selected from the T2 generation on MS medium containing phosphinothricin (Duchefa, Netherlands). Four independent single-copy homozygous plants were selected and propagated in a rice paddy field at Kyungpook National University, Gunwi (128:34E/36:15N), Korea.
To generate osdiat mutant plants, two independent single-guide RNAs (sgRNAs) (5′-CATGTCCCTGTCTACGAGTCAGG-3′, 5′-TGGGGAGCTGATCTTTGCTTTGG-3′) were introduced into the pSB11 vector harboring recombinant codon-optimized Streptococcus pyogenes Cas9 for rice (Chung et al., 2020). The constructs were introduced into plants by Agrobacterium (LBA4404)-mediated transformation (Jang et al., 1999). To analyze the mutation patterns, the genomic regions targeted by the sgRNAs were amplified by PCR using gene-specific primers. The PCR products were purified using a QIAquick PCR purification kit (Qiagen, Germany) and subjected to Sanger sequencing (Macrogen, Korea). Homozygous mutants were identified in which both alleles contained the same nucleotide deletion or insertion. T2 mutants used for the experiments were verified to be homozygous for the mutation by genomic DNA sequencing.
RNA isolation and reverse transcription quantitative PCR
Total RNA was extracted from leaf or root tissues using a Hybrid-R RNA Purification kit (GeneAll Biotechnology, Korea) according to the manufacturer's instructions. To generate first-strand complementary DNA (cDNA), 2 µg of total RNA was reverse-transcribed using RevertAid M-MuLV Reverse Transcriptase (Thermo Scientific, USA). Quantitative PCR analysis was performed using 2 × real-time PCR Smart mix (SolGent, Korea) and EvaGreen (SolGent, Korea) in an Mx3000P real-time PCR system (Stratagene, USA). The PCR conditions were an initial denaturation at 95°C for 15 min, followed by 40 cycles of 95°C for 20 s, 60°C for 20 s, and 72°C for 30 s. Rice UBIQUITIN1 (Os06g0681400, OsUBI1) was used as an internal control for normalization, and rice DIP1 (Os02g0669100) was used as a marker of drought and ABA stress treatments. Three biological replicates (independent samples (n = 5)) were analyzed for the quantitative experiments. The primers used for RT-qPCR are listed in Supplemental Table 3.
Rice protoplast preparation and transient gene expression
Rice (O. sativa “Dongjin”) seedlings were grown in the dark for 10 days and transferred to the light for 8–10 h. Leaf sheaths of 50 rice seedlings were cut into 0.5-mm pieces using a sharp blade on a piece of glass. Rice protoplast preparation and transient gene expression were performed as described previously (Shim et al., 2018). The 35S:OsDIAT-GFP plasmid was transferred into protoplasts using PEG-mediated transformation. Following incubation at 28°C, the protoplasts were harvested by centrifugation at 300 × g for 2 min. The subcellular localization of OsDIAT was observed under a Leica SP8 STED confocal laser-scanning microscope (Leica, Germany) with 488 nm excitation wavelength lasers and 512∼560 nm emission wavelength detection filter.
Enzyme activity assay
To test the enzyme activity of OsDIAT in vitro, the OsDIAT coding sequence was cloned into the pGEX-5X protein expression vector through the EcoRI restriction enzyme site using an Infusion Cloning kit (Takara, Japan). The construct was transformed into Escherichia coli BL21(DE3) pLysS for protein expression. In vitro protein expression was induced by adding 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG, Sigma, USA) to the E. coli culture (OD = 0.6) and incubating for 16 h at 20°C with shaking. Induced protein was captured by glutathione-agarose resin (Thermo, USA). The protein was eluted by adding 10 mM glutathione (Sigma, USA). Eluted protein samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Supplemental Figure 4). The concentration of OsDIAT was measured with a protein assay kit based on Bradford assay (Bio-Rad, USA) using bovine serum albumin as a standard. The BCAT assay was performed as described previously (Prohl et al., 2000; Maloney et al., 2010). One microgram of purified DIAT protein was used in each reaction, which was carried out at 25°C for 5 min. Forward assays were recorded at 340 nm and reverse assays at 440 nm using a spectrophotometer. Reactions lacking substrate were used as a negative control. Kinetic data were obtained using GraphPad Prism 8 software (https://www.graphpad.com/scientific-software/prism/).
Drought stress treatments and evaluation of drought tolerance
OsDIAT transgenic and NT control plants (O. sativa “Dongjin”) were sown on MS solid medium and incubated in a dark growth chamber for 4 days at 28°C. The seedlings were transferred to a growth chamber under a 16-h-light/8-h-dark cycle and grown for one additional day before transplanting to soil. Thirty plants per line were transplanted into 10 pots containing soil (4 × 4 × 6 cm: three plants per pot) in a container (59 × 38.5 × 15 cm) and grown for an additional 4 weeks in a greenhouse (16-h-light/8-h-dark cycle, 28°C, and 50∼60% humidity). Drought stress was imposed by sequentially withholding water for 3 days and re-watering for 7 days. Drought-induced symptoms were monitored by imaging transgenic and NT plants at the indicated time points using a NEX-5N camera (Sony, Japan). The soil moisture contents were measured at the indicated time points using an SM150 Soil Moisture Sensor (Delta-T Devices, United Kingdom). Relative water content was measured as described previously (Sharp et al., 1990).
Transient chlorophyll a fluorescence and the performance index were measured using a Handy-PEA fluorimeter (Hansatech Instruments, United Kingdom) as previously described (Redillas et al., 2011). Two-week-old plants were transplanted to pots containing soil (15 × 15 × 14 cm) and grown for 5 weeks. Chlorophyll a fluorescence was measured from the longest leaves of each plant after 1 h of dark adaptation to ensure sufficient opening of the reaction centers. Measurement was performed at the apex, middle, and base regions of leaves using a Handy-PEA fluorimeter. Thirty measurements per line were averaged using HANDY-PEA software (version 1.31). Fv/Fm values and the performance index were calculated according to the equations of the JIP test (Redillas et al., 2011).
Evaluation of the agronomic traits of rice grown in the field
To evaluate yield components of transgenic and NT plants under field conditions, three independent T4 homozygous GOS2::OsDIAT lines and NT plants were planted in the rice paddy field at Kyungpook National University, Gunwi (36°06′48.0′′N,128°38038.0′′E), Korea. The field experiments were performed for 3 years (2018–2020), and similar patterns were observed between NT and GOS2::OsDIAT plants. The representative data of 2020 are presented in Figure 4 and Supplemental Table 2. The experiments were repeated in multiple locations in our field. Climate data from the field in 2020 is provided (Supplemental Dataset 1). Yield parameters were scored from 10 plants collected from three different plots under normal field conditions. To evaluate the yield components of plants under field drought conditions, plants were grown in semi-field conditions under rain-off shelters. The plants were exposed to intermittent drought twice by withholding water during panicle development. Drought treatment was monitored by measuring soil water content using a soil moisture sensor. After two rounds of drought treatment at the reproductive stage, the plants were re-irrigated until the harvesting stage. Yield components were scored from 30 plants per line under drought field conditions. The plant height (PH) panicle length (PL), and culm length (CL) of individual plants were measured in the field. Then, the panicles of individual plant (hill) were harvested separately. We counted the filled and unfilled grains of individual plants, then measured and calculated the yield parameters, including total grain weight (TGW), filling rate (FR), number of filled grains (NFG), number of total grains (NTG), and number of grains per panicle (NGP)
Amino acid content analysis
To measure amino acid contents, NT (O. sativa “Dongjin”) and T3 homozygous transgenic GOS2::OsDIAT (#5) seeds were sown on MS solid medium and incubated in a dark growth chamber for 4 days at 28°C. The seedlings were transferred to a growth chamber under a 16-h-light/8-h-dark cycle and grown for 2 weeks. The 2-week-old seedlings were transferred to hydroponic solution and incubated for an additional week. The 3-week-old plants were air-dried to stimulate dehydration stress and sampled at the indicated time points after stress treatment. Plants not exposed to stress were used as a control. Amino acid contents were measured by high-performance liquid chromatography (HPLC) using an HPLC Ultimate 3000 equipped with a VDSpher 100 C18-E column (4.6 mm × 150 mm, 3.5 μm/VDS, Optilab, USA) and a 1260 FLD FL detector (Agilent, USA) at the National Instrumentation Center for Environmental Management, College of Agriculture and Life Science in Seoul National University.
Amino acid feeding and PEG treatment
Three-week old plants grown in Yoshida liquid medium were fed with 10 mM L-valine, leucine, or isoleucine (Sigma, USA) by dipping their roots into a solution containing the corresponding BCAAs for 24 h. The plants were then transferred to a 50-ml tube containing 25 ml of 25% (w/v) PEG 8000 (Sigma) solution. Plants pretreated with BCAAs were harvested for amino acid analysis. PEG-induced visual symptoms such as leaf rolling and wilting were monitored by imaging plants at the indicated time points using a NEX-5N camera. Plants were harvested at the indicated time point after PEG treatment for RNA extraction. Total RNA extraction, cDNA synthesis, and RT-qPCR analysis were performed as described in the “RNA isolation and reverse transcription quantitative PCR” section. The DIP1 gene was used as a molecular marker for osmotic stress responses in PEG-treated plants.
Statistical analysis
All data are represented as the mean value ± SD. Each data value was separately compared with the control value to determine significant differences using ANOVA or an LSD test. Data were analyzed using Microsoft Excel or IBM SPSS software.
Accession numbers
Sequence data from this article can be found in the Rice Annotation Project Database (https://rapdb.dna.affrc.go.jp/) under the following accession numbers: OsDIAT (Os05g0244700), OsUBI1 (Os06g0681400), and OsDIP1 (Os02g0669100).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Phylogenetic analysis of rice BCATs.
Supplemental Figure S2 . Expression pattern of OsDIAT in response to ABA treatment.
Supplemental Figure S3 . Analysis of osdiat plants generated by CRISPR/Cas9-mediated gene editing.
Supplemental Figure S4 . In vitro protein expression of GST-OsDIAT for enzyme assay.
Supplemental Table S1. Expression of rice BCAT genes in response to drought stress.
Supplemental Table S2. Agronomic traits of OsDIAT-overexpressing transgenic plants grown in the field.
Supplemental Table S3. Primer used in this study.
Supplemental Dataset S1. Climate information of the field in 2020.
Supplementary Material
Acknowledgments
We thank the Rural Development Administration and Kyungpook National University for providing the rice paddy fields.
Contributor Information
Jae Sung Shim, Graduate School of International Agricultural Technology, Seoul National University, Pyeongchang 25354, Korea; School of Biological Sciences and Technology, Chonnam National University, Gwangju 61186, Korea.
Hye In Jeong, Crop Biotechnology Institute/GreenBio Science and Technology, Seoul National University, Pyeongchang 25354, Korea.
Seung Woon Bang, Graduate School of International Agricultural Technology, Seoul National University, Pyeongchang 25354, Korea.
Se Eun Jung, Crop Biotechnology Institute/GreenBio Science and Technology, Seoul National University, Pyeongchang 25354, Korea.
Goeun Kim, Crop Biotechnology Institute/GreenBio Science and Technology, Seoul National University, Pyeongchang 25354, Korea.
Youn Shic Kim, Graduate School of International Agricultural Technology, Seoul National University, Pyeongchang 25354, Korea.
Mark Christian Felipe R Redillas, Graduate School of International Agricultural Technology, Seoul National University, Pyeongchang 25354, Korea; Department of Biology, De La Salle University, Manila 1004, Philippines.
Se-Jun Oh, LaSemilla Co. Ltd., Pyeongchang 25354, Korea.
Jun Sung Seo, Graduate School of International Agricultural Technology, Seoul National University, Pyeongchang 25354, Korea.
Ju-Kon Kim, Graduate School of International Agricultural Technology, Seoul National University, Pyeongchang 25354, Korea; Crop Biotechnology Institute/GreenBio Science and Technology, Seoul National University, Pyeongchang 25354, Korea; LaSemilla Co. Ltd., Pyeongchang 25354, Korea.
Funding
This work was supported by a grant from the New Breeding Technologies Development Program (Project No. PJ01652501 to J.-K.K.) and Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01597601 to J.-K.K.), Rural Development Administration, Republic of Korea.
References
- Abid M, Ali S, Qi LK, Zahoor R, Tian Z, Jiang D, Snider JL, Dai T (2018) Physiological and biochemical changes during drought and recovery periods at tillering and jointing stages in wheat (Triticum aestivum L.). Sci Rep 8(1): 4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Lipka AE, Deason N, Gonzalez-Jorge S, Lin H, Cepela J, Buell R, Gore MA, Dellapenna D (2013) Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. Plant Cell 25(12): 4827–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf M, Foolad MR (2007) Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ Exp Bot 59(2): 206–216 [Google Scholar]
- Atkinson NJ, Urwin PE (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot 63(10): 3523–3543 [DOI] [PubMed] [Google Scholar]
- Binder S (2010) Branched-chain amino acid metabolism in Arabidopsis thaliana. Arabidopsis book 8: e0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowne JB, Erwin TA, Juttner J, Schnurbusch T, Langridge P, Bacic A, Roessner U (2012) Drought responses of leaf tissues from wheat cultivars of differing drought tolerance at the metabolite level. Mol Plant 5(2): 418–429 [DOI] [PubMed] [Google Scholar]
- Campalans A, Messeguer R, Goday A, Pagès M (1999) Plant responses to drought, from ABA signal transduction events to the action of the induced proteins. Plant Physiol Biochem 37(5): 327–340 [Google Scholar]
- Chen H, Saksa K, Zhao F, Qiu J, Xiong L (2010) Genetic analysis of pathway regulation for enhancing branched-chain amino acid biosynthesis in plants. Plant J 63(4): 573–583 [DOI] [PubMed] [Google Scholar]
- Chen Z, Cuin TA, Zhou M, Twomey A, Naidu BP, Shabala S (2007) Compatible solute accumulation and stress-mitigating effects in barley genotypes contrasting in their salt tolerance. J Exp Bot 58(15–16): 4245–4255 [DOI] [PubMed] [Google Scholar]
- Chung PJ, Chung H, Oh N, Choi J, Bang SW, Jung SE, Jung H, Shim JS, Kim JK (2020) Efficiency of recombinant CRISPR/rCas9-mediated miRNA gene editing in rice. Int J Mol Sci 21(24): 9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung PJ, Jung H, Jeong D-H, Ha S-H, Choi YD, Kim J-K (2016) Transcriptome profiling of drought responsive noncoding RNAs and their target genes in rice. BMC Genomics 17(1): 563–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colla G, Hoagland L, Ruzzi M, Cardarelli M, Bonini P, Canaguier R, Rouphael Y (2017) Biostimulant action of protein hydrolysates: unraveling their effects on plant physiology and microbiome. Front Plant Sci 8: 2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway ME (2021) Emerging moonlighting functions of the branched-chain aminotransferase proteins. Antioxid Redox Signal 34(13): 1048–1067 [DOI] [PubMed] [Google Scholar]
- Delauney AJ, Verma DPS (1993) Proline biosynthesis and osmoregulation in plants. Plant J 4(2): 215–223 [Google Scholar]
- Diebold R, Schuster J, Däschner K, Binder S (2002) The branched-chain amino acid transaminase gene family in Arabidopsis encodes plastid and mitochondrial proteins. Plant Physiol 129(2): 540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregas N, Fernie AR (2019) The metabolic response to drought. J Exp Bot 70(4): 1077–1085 [DOI] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L (2000) Metabolite profiling for plant functional genomics. Nat Biotechnol 18(11): 1157–1161 [DOI] [PubMed] [Google Scholar]
- Graham IA, Eastmond PJ (2002) Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog Lipid Res 41(2): 156–181 [DOI] [PubMed] [Google Scholar]
- Gu L, Jones AD, Last RL (2010) Broad connections in the Arabidopsis seed metabolic network revealed by metabolite profiling of an amino acid catabolism mutant. Plant J 61(4): 579–590 [DOI] [PubMed] [Google Scholar]
- Haghighi M, Saadat S, Abbey L (2020) Effect of exogenous amino acids application on growth and nutritional value of cabbage under drought stress. Sci Hortic 272: 109561 [Google Scholar]
- Harb A, Krishnan A, Ambavaram MMR, Pereira A (2010) Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol 154(3): 1254–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare PD, Cress WA, Van Staden J (1998) Dissecting the roles of osmolyte accumulation during stress. Plant Cell Environ 21(6): 535–553 [Google Scholar]
- Huang T, Jander G (2017) Abscisic acid-regulated protein degradation causes osmotic stress-induced accumulation of branched-chain amino acids in Arabidopsis thaliana. Planta 246(4): 737–747 [DOI] [PubMed] [Google Scholar]
- Jang I-C, Nahm BH, Kim J-K (1999) Subcellular targeting of green fluorescent protein to plastids in transgenic rice plants provides a high-level expression system. Mol Breeding 5(5): 453–461 [Google Scholar]
- Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153(1): 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Yang H, Wei Z, Ma H, Ge X (2013) Rice male development under drought stress: phenotypic changes and stage-dependent transcriptomic reprogramming. Mol Plant 6(5): 1630–1645 [DOI] [PubMed] [Google Scholar]
- Joshi V, Joung JG, Fei Z, Jander G (2010) Interdependence of threonine, methionine and isoleucine metabolism in plants: accumulation and transcriptional regulation under abiotic stress. Amino Acids 39(4): 933–947 [DOI] [PubMed] [Google Scholar]
- Khan MS, Ahmad D, Khan MA (2015) Utilization of genes encoding osmoprotectants in transgenic plants for enhanced abiotic stress tolerance. Electron J Biotechnol 18(4): 257–266 [Google Scholar]
- Lehmann S, Funck D, Szabados L, Rentsch D (2010) Proline metabolism and transport in plant development. Amino Acids 39(4): 949–962 [DOI] [PubMed] [Google Scholar]
- Liang X, Zhang L, Natarajan SK, Becker DF (2013) Proline mechanisms of stress survival. Antioxid Redox Signal 19(9): 998–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio A, Miyazaki S, Veronese P, Fujita T, Ibeas JI, Damsz B, Narasimhan ML, Hasegawa PM, Joly RJ, Bressan RA (2002) Does proline accumulation play an active role in stress-induced growth reduction? Plant J 31(6): 699–712 [DOI] [PubMed] [Google Scholar]
- Maloney GS, Kochevenko A, Tieman DM, Tohge T, Krieger U, Zamir D, Taylor MG, Fernie AR, Klee HJ (2010) Characterization of the branched-chain amino acid aminotransferase enzyme family in tomato. Plant Physiol 153(3): 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, Okamoto M, Nambara E, Nakajima M, Kawashima M, et al. (2008) Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 49(8): 1135–1149 [DOI] [PubMed] [Google Scholar]
- Peng C, Uygun S, Shiu SH, Last RL (2015) The impact of the branched-chain ketoacid dehydrogenase complex on amino acid homeostasis in Arabidopsis. Plant Physiol 169(3): 1807–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli R, Pilot G (2014) Regulation of amino acid metabolic enzymes and transporters in plants. J Exp Bot 65(19): 5535–5556 [DOI] [PubMed] [Google Scholar]
- Prohl C, Kispal G, Lill R (2000) Branched-chain-amino-acid transaminases of yeast Saccharomyces cerevisiae. Methods Enzymol 324: 365–375 [DOI] [PubMed] [Google Scholar]
- Ranieri A, Bernardi R, Lanese P, Soldatini GF (1989) Changes in free amino acid content and protein pattern of maize seedlings under water stress. Environ Exp Bot 29(3): 351–357 [Google Scholar]
- Redillas MCFR, Jeong JS, Strasser RJ, Kim YS, Kim J-K (2011) JIP Analysis on rice (Oryza sativa cv Nipponbare) grown under limited nitrogen conditions. J Korean Soc Appl Biol Chem 54(5): 827–832 [Google Scholar]
- Sakuraba Y, Kim YS, Han SH, Lee BD, Paek NC (2015) The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell 27(6): 1771–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Sita K, Siddique KHM, Kumar R, Bhogireddy S, Varshney RK, HanumanthaRao B, Nair RM, Prasad PVV, Nayyar H (2018) Drought or/and heat-stress effects on seed filling in food crops: impacts on functional biochemistry, seed yields, and nutritional quality. Front Plant Sci 9: 1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Villamor JG, Verslues PE (2011) Essential role of tissue-specific proline synthesis and catabolism in growth and redox balance at low water potential. Plant Physiol 157(1): 292–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp RE, Hsiao TC, Silk WK (1990) Growth of the maize primary root at low water potentials: II. Role of growth and deposition of hexose and potassium in osmotic adjustment. Plant Physiol 93(4): 1337–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JS, Oh N, Chung PJ, Kim YS, Choi YD, Kim JK (2018) Overexpression of OsNAC14 improves drought tolerance in rice. Front Plant Sci 9: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuttmann J, Hubberten H-M, Rietz S, Kaur J, Muskett P, Guerois R, Bednarek P, Hoefgen R, Parker JE (2011) Perturbation of Arabidopsis amino acid metabolism causes incompatibility with the adapted biotrophic pathogen Hyaloperonospora arabidopsidis. Plant cell 23(7): 2788–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Day DA, Millar AH (2004) Lipoic acid-dependent oxidative catabolism of α-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiol 134(2): 838–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaka D, Shinozaki K, Yamaguchi-Shinozaki K (2015) Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front Plant Sci 6: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah N, Yüce M, Neslihan Öztürk Gökçe Z, Budak H (2017) Comparative metabolite profiling of drought stress in roots and leaves of seven Triticeae species. BMC Genomics 18(1): 969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano K, Maruyama K, Ogata Y, Morishita Y, Takeda M, Sakurai N, Suzuki H, Saito K, Shibata D, Kobayashi M, et al. (2009) Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J 57(6): 1065–1078 [DOI] [PubMed] [Google Scholar]
- Valliyodan B, Nguyen HT (2006) Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr Opin Plant Biol 9(2): 189–195 [DOI] [PubMed] [Google Scholar]
- Wani SH, Singh NB, Haribhushan A, Mir JI (2013) Compatible solute engineering in plants for abiotic stress tolerance—role of glycine betaine. Curr Genomics 14(3): 157–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2(8): e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing A, Last RL (2017) A regulatory hierarchy of the Arabidopsis branched-chain amino acid metabolic network. Plant Cell 29(6): 1480–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey PH (2005) Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J Exp Biol 208(15): 2819–2830 [DOI] [PubMed] [Google Scholar]
- Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN (1982) Living with water stress: evolution of osmolyte systems. Science 217(4566): 1214–1222 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory Manual for Physiological Studies of Rice. The International Rice Research Institute, Los Banos, Philippines [Google Scholar]
- Zheng Y, Cabassa-Hourton C, Planchais S, Lebreton S, Savoure A (2021) The proline cycle as an eukaryotic redox valve. J Exp Bot 72(20): 6856–6866 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.