Abstract
Grass inflorescences support floral structures that each bear a single grain, where variation in branch architecture directly impacts yield. The maize (Zea mays) RAMOSA1 (ZmRA1) transcription factor acts as a key regulator of inflorescence development by imposing branch meristem determinacy. Here, we show RA1 transcripts accumulate in boundary domains adjacent to spikelet meristems in sorghum (Sorghum bicolor, Sb) and green millet (Setaria viridis, Sv) inflorescences similar as in the developing maize tassel and ear. To evaluate the functional conservation of syntenic RA1 orthologs and promoter cis sequences in maize, sorghum, and setaria, we utilized interspecies gene transfer and assayed genetic complementation in a common inbred background by quantifying recovery of normal branching in highly ramified ra1-R mutants. A ZmRA1 transgene that includes endogenous upstream and downstream flanking sequences recovered normal tassel and ear branching in ra1-R. Interspecies expression of two transgene variants of the SbRA1 locus, modeled as the entire endogenous tandem duplication or just the nonframeshifted downstream copy, complemented ra1-R branching defects and induced unusual fasciation and branch patterns. The SvRA1 locus lacks conserved, upstream noncoding cis sequences found in maize and sorghum; interspecies expression of a SvRA1 transgene did not or only partially recovered normal inflorescence forms. Driving expression of the SvRA1 coding region by the ZmRA1 upstream region, however, recovered normal inflorescence morphology in ra1-R. These data leveraging interspecies gene transfer suggest that cis-encoded temporal regulation of RA1 expression is a key factor in modulating branch meristem determinacy that ultimately impacts grass inflorescence architecture.
Gene transfer between species suggests that cis-encoded temporal regulation of RAMOSA1 expression helps modulate the activity of branch meristems to impact grass inflorescence architecture.
Introduction
Understanding the genetic basis of morphological diversity between and within species is a key objective in biology (Carroll, 2008). Grass (Poaceae) inflorescences display tremendous intra- and interspecific variation (Kellogg, 2015) and are an effective model for studying genetic mechanisms that underly evolutionary change in morphology. Inflorescence diversity is well-documented in the cereal crops rice (Oryza spp.) (Yamaki et al., 2010; Crowell et al., 2016), millet (Setaria spp.) (Doust and Kellogg, 2002; Doust et al., 2005; Huang and Feldman, 2017), sorghum (Sorghum spp.) (Harlan and de Wet, 1972; Brown et al., 2006; Zhou et al., 2019; Li et al., 2020), and maize (Zea mays spp.) (Upadyayula et al., 2006a; Upadyayula et al., 2006b; Brown et al., 2011; Wu et al., 2016; Xu et al., 2017). As inflorescences in the Poaceae ultimately support reproduction and the floral structures that bear a single grain, variation in inflorescence morphology directly impacts yield in cereal crops and weedy grass species. Despite such agronomical and ecological importance, the genes that underlie diverse inflorescence forms in the grasses have not been fully elucidated, and tests of functional conservation of syntenic orthologous genes are limited.
Mature inflorescence traits are patterned early in development through variation in size, identity, and the timing and duration of maturation schedules of active pluripotent stem cell tissues called meristems. These variations impact the number, arrangement, and elaboration of lateral organs that arise from meristems (Doust and Kellogg, 2002; Vollbrecht et al., 2005; Prusinkiewicz et al., 2007; Whipple et al., 2010; Kellogg et al., 2013; Lemmon et al., 2016; Zhu et al., 2018; Leiboff and Hake, 2019). A general framework for the ontogeny of grass inflorescences (Kellogg et al., 2013) follows: When internal and external cues signal the reproductive transition, inflorescence development ensues as a vegetative shoot apical meristem, which elaborates leaf primordia at its flanks, converts to a reproductive inflorescence meristem (IM) that elaborates lateral meristems at its flanks. The IM is indeterminate, i.e. capable of producing an unspecified number of lateral primordia, and the lateral meristems can be either relatively indeterminate in which case they may also initiate additional lateral meristems, or relatively determinate (producing a specified number of lateral primordia). Indeterminate grass IMs are called branch meristems (BMs) and show diverse indeterminacy across and even within grass species, while all grass inflorescences ultimately produce determinate meristems called spikelet meristems (SMs). Thus, in the general framework, IMs initiate BMs, and both IMs and BMs initiate SMs at their flanks. An SM gives rise to two glumes (bract) primordia, followed by one or multiple florets which altogether comprise the spikelet, the central unit of a grass inflorescence (Clifford, 1987). SMs in some grass species are more determinate (e.g. members of the subfamily Aristidoideae, and members of the subtribe Poinae, i.e. Agrostopoa, Apera, Cinna, Limnas, and Phleum) in that they terminate by converting to a floral meristem that is consumed in the production of floral organs, whereas in other species SMs are somewhat indeterminate (e.g. members of the subtribe Tripogoninae, i.e. Eragrostiella, Tripogon, and members of Arundinoideae, i.e. Alloeochaete, Phragmites) and produce multiple floral meristems, and therefore multiple florets, before terminating (Kellogg, 2015). Diverse morphological complexity among grass inflorescences arises though variation in type, activity, and determinacy of IMs, BMs, and SMs.
The family Poaceae consists of over 11,500 species (Kellogg, 2015) distributed about equally among two major lineages known as the PACMAD and BOP clades. In the PACMAD clade, the largest subfamily Panicoideae has over 3,300 species that include global staple cereal crops maize (Zea mays ssp. mays), sorghum (Sorghum bicolor [L.] Moench), and foxtail millet (Setaria italica) (Kellogg, 2015). Maize and sorghum are among the ∼1,200 species in tribe Andropogoneae; Setaria is in the tribe Paniceae (Kellogg, 2015). Unlike most of the Panicoideae, where spikelets are unpaired, the Andropogoneae are distinguished by producing their spikelets in pairs; specialized, determinate BMs called spikelet pair meristems (SPMs) each produce two SMs. Thus, spikelet pairs (SPs) and long branches (LBs), which commonly coexist in the same inflorescence, are branches that differ by length (short versus long, respectively) and meristem determinacy at origin (SPMs versus BMs, respectively). By contrast, within the tribe, Paniceae or the “bristle clade” are a few hundred grass species, including the foxtail millet progenitor Setaria viridis where adjacent meristems differentiate into either single spikelets or sterile branches called bristles (Doust and Kellogg, 2002, Hodge and Doust, 2017). Developmental and morphological studies in Setaria lend support to the ontogenetic pairing of a single spikelet with a bristle, but spikelets are not paired (Doust and Kellogg, 2002).
Maize and sorghum are estimated to have diverged from a common ancestor approximately 12 million years ago (MYA) (Swigonová et al., 2004); Setaria diverged from maize and sorghum approximately 26–27 MYA (Bennetzen et al., 2012; Zhang et al., 2012). Sorghum and Setaria genomes show extensive synteny (Bennetzen et al., 2012; Zhang et al., 2012). Likewise, approximately 60% of annotated genes are syntenically conserved between maize and sorghum, and this gene set accounts for 90% of all genes characterized by forward genetics in maize (Schnable and Freeling, 2011; Schnable, 2015). Regulation and expression of syntenic orthologous genes are often conserved in related species, suggesting a retention of ancestral function (Dewey, 2011; Davidson et al., 2012). However, to date, functional conservation between syntenic orthologs in related grass species remains widely untested.
The maize RAMOSA1 (ZmRA1) locus is a key regulator of tassel and ear development and morphology (Vollbrecht et al., 2005). ZmRA1 was a target of selection during maize domestication (Sigmon and Vollbrecht, 2010), co-localizes with nucleotide polymorphisms for inflorescence branching traits in genome-wide association studies of diverse maize breeding lines (Brown et al., 2011; Wu et al., 2016; Xu et al., 2017) and is a candidate quantitative trait locus for tassel branch number in the Mexican highland maize landrace Palomero Toluqueño (Perez-Limón et al., 2021). Strong maize ra1 mutants were recognized over a century ago as resembling inflorescences of other grasses (Collins, 1917), and more recently, comparisons to the complexly branched sorghum panicle have been drawn at developmental and molecular levels (Vollbrecht et al., 2005; Leiboff and Hake, 2019). Whereas normal inflorescence branching in maize produces only SPs or LBs bearing SPs, mutations in ZmRA1 relax the determinacy normally imposed on SPMs such that SPs are replaced by LBs bearing several unpaired, single spikelets (spikelet multimers), or by LBs bearing a mix of single and/or paired spikelets (Vollbrecht et al., 2005). The graded, multiple orders of inflorescence branching in ra1 mutants reveal a general determinacy function of ZmRA1 in addition to or that includes a specific role for ZmRA1 activity in producing the canonical SP. RA1 encodes a C2H2 zinc-finger transcription factor with EAR repression motifs (Vollbrecht et al., 2005). Mutations in the maize C2H2 zinc-finger domain or C-terminal EAR motif result in severe ra1 mutants that display highly ramified tassels and ears (Vollbrecht et al., 2005; Gallavotti et al., 2010). One mechanism by which RA1 imposes SPM determinacy in maize is through interactions with RA1 ENHANCER LOCUS2 (REL2), orthologous to the co-repressor TOPLESS (Gallavotti et al., 2010). ZmRA1 transcripts and ZmRA1 protein accumulate in a boundary domain between the inflorescence or branch axis and the determinate meristems it regulates (Vollbrecht et al., 2005; Eveland et al., 2014). The noncell-autonomous nature of ZmRA1 suggests that it regulates a trafficable signal for meristem determinacy, or its gene product is capable of trafficking to the adjacent meristem (Vollbrecht et al., 2005). Genetic and molecular data support that RA1 expression in maize impacts branch complexity through regulating SPM determinacy (Vollbrecht et al., 2005). Variation in timing of RA1 expression, presumably imposed by variation in promoter cis sequences, in Miscanthus (Vollbrecht et al., 2005), sorghum (Vollbrecht et al., 2005; Leiboff and Hake, 2019), and S. viridis (Zhu et al., 2018) corresponds with degree of branch activity and distinct inflorescence morphologies. Thus, heterochronic RA1 expression and regulation of RA1 activity are hypothesized to impact inflorescence branching directly by modulating meristem determinacy. To date, ra1 mutants have not been reported outside of maize, leaving open the question of RA1 function in the Panicoideae with respect to evolutionarily and agronomically important characters such as meristem determinacy, branch length, and pairing of spikelets.
Here, we report on genetic tests for the functional conservation of syntenic orthologous RA1 genes in maize, sorghum, and setaria. We show that RA1 expression marks boundary domains adjacent to meristems in sorghum and setaria inflorescences in concordance with RA1 transcript accumulation in maize. We generated RA1 transgenes from maize (Zm), sorghum (Sb), and setaria (Sv) loci and utilized the strong maize ra1-R mutant to investigate the impact of expressing ZmRA1, SbRA1, and SvRA1 transgenes on the regulation of branching in maize tassels and ears. Expression as a transgene of ZmRA1 including flanking upstream and downstream sequences recovered normal inflorescence morphologies in ra1-R mutants. Interspecies expression of two transgene variants of the SbRA1 locus, one modeled as the entire endogenous tandem duplication and the other as only the nonframeshifted downstream gene copy, yielded a range of ra1-R inflorescence architectures, showing partial recovery with or without unusual branch patterns and fasciation. We found that interspecies expression of a SvRA1 transgene, which lacks cis-promoter sequences conserved in maize, sorghum and other Andropogoneae species, either not at all or only partially recovered normal inflorescence forms in ra1-R mutants, whereas fusing the SvRA1 coding region to the ZmRA1 upstream region recovered normal inflorescence morphology in ra1-R mutants. Our functional tests of RA1 sufficiency indicate that heterochronic modulation of meristem determinacy that results from cis-regulatory differences impacts ear and tassel morphology, and is an important factor in shaping inflorescence diversity throughout the grasses.
Results and discussion
Inflorescence architectures and RA1 alleles in PACMAD and Panicoid grasses
Mature maize inflorescences are spatially and morphologically distinct and produce dimorphic, unisexual florets: a terminal tassel bearing staminate florets and a lateral ear with pistillate florets (Figure 1, A and B). Mutations in the ZmRA1 gene, typified by the strong ra1-R allele (Vollbrecht et al., 2005), result in multiple orders of branching in the tassel and the ear (Figure 1, C and D) that resemble complexly branched inflorescences of other grasses, such as terminal panicles of sorghum (Figure 1E) and setaria (Figure 1F) which have unimorphic, bisexual florets. The conspicuous diversity of mature inflorescence morphologies in maize, sorghum, and setaria, largely attributed to variation in degree of branching, manifests early in development (Supplemental Figure 1). Maize, sorghum, and setaria belong to the subfamily Panicoideae, and within this large clade of grasses, maize and sorghum are members of tribe Andropogoneae, whereas setaria is a member of tribe Paniceae (Kellogg, 2015). Maize and sorghum inflorescences produce a multitude of spikelets in pairs as is characteristic of related species in the Andropogoneae, whereas the setaria inflorescence is dense with single spikelets that each develops in close association with a bristle (Doust and Kellogg, 2002; Kellogg, 2015).
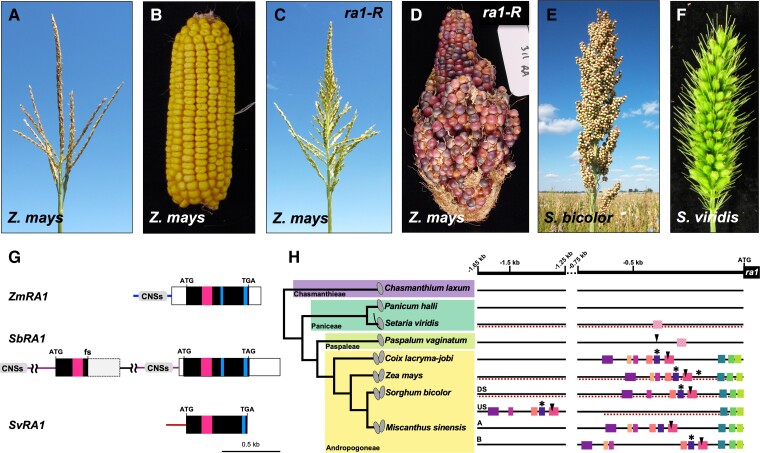
Figure 1.
Architecture of maize, maize ra1-R mutant, sorghum, and S. viridis inflorescences and genomic relationship of RA1. Normal inbred B73 maize (A) tassel and (B) ear. Maize ra1-R mutant (C) tassel and (D) ear. (E), Sorghum bicolor panicle. (F), Setaria viridis panicle. (A–F), Inflorescences not to scale. (G), Annotated gene structure for RA1 homologs. Tandem duplication of SbRA1 locus is shown with indicated frameshift mutation (fs) in upstream copy of SbRA1. Predicted promoter regions are indicated by color lines. CNSs box, conserved noncoding cis sequences (see 1H). Open box, UTR sequences. Magenta box, encoded C2H2 zinc finger domain. Blue box, encoded EAR motif. (H) Conserved noncoding cis sequences in the RA1 promoters of Panicoid grasses. Among species in the tribe Andropogoneae, the promoter regions of RA1 display different motifs conserved in sequence and arrangement (correspondingly colored boxes are conserved; Supplemental Figure 3C) compared with other tribes in the Panicoideae family. Upstream (US) and downstream (DS) tandem duplicate SbRA1 copies and duplicate MsRA1 copies A and B are indicated. Dashed lines underscore promoter regions incorporated into transgene cassettes. Some conserved sequences contained binding motifs for well-known transcriptional regulators, such as LEAFY and Clade A ARFs (Supplemental Figure 3B). Solid squares, P-values ≤ 1−20; cross-hatched squares, P-values ≤ 1−5; arrowhead—LEAFY-binding motifs; asterisks—Clade A Auxin Response Factor (ARF)-binding motifs. Character state of spikelets (paired, single, or with a bristle) is indicated on the phylogeny.
Comparative genomic data indicate the RA1 locus is specific to the PACMAD clade, whose largest subfamilies are the Panicoideae and Chloridoideae, where the intronless structure and unique QGLGGH motif within the C2H2 zinc finger present in maize (Vollbrecht et al., 2005) appear conserved. For example, a syntenic copy of RA1 is absent from the genomes of BOP clade members rice (Oryza sativa), Brachypodium distachyon, and wheat (Triticum aestivum) (Supplemental Figure 2A) (Vollbrecht et al., 2005; Sigmon, 2010), but is present in the genome assemblies of Chloridoideae species teff (Eragrostis tef) and Oropetium thomaeum (Schnable, 2019), and of finger millet (Eleusine coracana). Within the Panicoideae RA1 resides as a single-copy gene in maize and setaria and as a single-locus tandem duplication in sorghum (comprised of SbRA1 upstream [SbRA1US] and SbRA1 downstream [SbRA1DS] copies); however, a frameshift mutation in SbRA1US introduces a stop codon after the C2H2 zinc finger domain, rendering it presumably nonfunctional (Figure 1G) (Vollbrecht et al., 2005; Sigmon, 2010). Previously published RT-PCR and transcript profiling data indicate that SbRA1US is not expressed in inflorescences of the sorghum reference line BTx623, while SbRA1DS is (Vollbrecht et al., 2005; Wang et al., 2018; Leiboff and Hake, 2019). Broad sampling of diverse cultivated and wild sorghums found that, in all cultivated accessions, (1) SbRA1US contains the same frameshift and that the SbRA1DS open reading frame (ORF) encodes a predicted full-length RA1 protein; (2) the SbRA1 tandem duplication likely originated relatively recently with the Sorghum genus and may not be present in other grass species (Sigmon, 2010). Two RA1 loci are present in miscanthus (Figure 1H), but these are segmental duplicates in this paleotetraploid species (Sigmon, 2010; Mitros et al., 2020). The encoded SbRA1DS protein of cultivated sorghums, hereafter referred to as SbRA1, is ∼69% identical to the ZmRA1 protein and ∼56% identical to the SvRA1 protein. ZmRA1 and SvRA1 proteins are ∼65% identical. ZmRA1, SbRA1, and SvRA1 proteins share a highly conserved C2H2 zinc-finger domain and a conserved C-terminal EAR motif (Figure 1, G; Supplemental Figure 2B). Biochemical experiments have demonstrated the C2H2 zinc-finger domain binds DNA (Dathan et al., 2002), and the EAR motif acts as a potent transcriptional repressor (Hiratsu et al., 2004; Tiwari et al., 2004). The motifs and their positioning are highly conserved between ZmRA1, SbRA1, and SvRA1 proteins. The C2H2 zinc-finger domain between ZmRA1 and SbRA1 differs by one conservative amino acid variant (I67V, position relative to ZmRA1) that is identical (V) between SbRA1 and SvRA1. Relative to ZmRA1 and SbRA1, the SvRA1 zinc-finger domain differs at three positions, none of them among invariant core C2H2 residues (Vollbrecht et al., 2005). The C-terminal EAR motif is conserved between ZmRA1 and SbRA1 and varies by one residue (Q169E) in SvRA1. A second EAR motif adjacent to the C2H2 zinc-finger domain (Sigmon 2010, Gallavotti et al., 2010) is highly conserved between ZmRA1 and SbRA1 but absent from SvRA1 (Figure 1, G; Supplemental Figure 2B). Physical interaction between ZmRA1 and REL2 involves both EAR motifs (Gallavotti et al., 2010); however, functional sufficiency of the maize C-terminal EAR motif has not been demonstrated.
By mining 2 kb of the RA1 promoter region from eight Panicoideae taxa across the Chasmanthieae, Paniceae, Paspaleae, and Andropogoneae tribes, we identified several blocks of highly conserved, noncoding cis sequence restricted to the Andropogoneae, where spikelets are paired (Figure 1, H; Supplemental Figure 3). These conserved cis sequences located in the promoter region of ZmRA1 and SbRA1DS (Sigmon, 2010), were absent from the ∼0.7 kb promoter region included in our SbRA1US transgene construct and were largely absent or not well conserved outside the Andropogoneae, including in SvRA1 (Figure 1, H; Supplemental Figure 3, A and B). Within the four Andropogoneae tribe taxa, where there are six promoter regions due to gene duplications, the conserved noncoding cis sequences harbored 48 putative transcription factor binding sites present among at least five of six sequences queried (Supplemental Dataset 1). In maize, some of the conserved cis sequence overlaps with accessible chromatin profiled from developing ears but not accessible in leaves (Supplemental Figure 3C; Ricci et al., 2019). Indeed, coinciding with the region of accessible chromatin, we found that DNA affinity purification sequencing of maize AUXIN RESPONSE FACTOR (ARF) transcription factors identified binding peaks (Supplemental Figure 3C; Galli et al., 2018) centered on a putative ARF binding motif, providing a possible additional link between auxin signaling and response and branch development (Gallavotti et al., 2008; Eveland et al., 2014). Also, within the region of accessible chromatin and within a conserved noncoding cis sequence, we identified a putative LEAFY (LFY) transcription factor binding motif (Winter et al., 2011) in all six Andropogoneae sequences queried (Figure 1, H; Supplemental Figure 3, B and C and Dataset 1). LFY is bifunctional as an activator and repressor in Arabidopsis (Arabidopsis thaliana) (William et al., 2004; Winter et al., 2011). Within the Andropogoneae, the protein-coding regions of the LFY-like genes are highly conserved suggesting purifying selection and constraint on amino acid sequence (Bomblies and Doebley, 2005). Interestingly, in maize, transcripts of the LFY homologs Zea FLORICAULA/LEAFY1 (ZFL1) and ZFL2 (Bomblies et al., 2003) accumulate in SPMs in a pattern that would likely border ZmRA1 transcript accumulation (Vollbrecht et al., 2005). Tassel branch number is decreased in zfl1; zfl2 double mutants, and positively correlates with ZFL2 copy number (Bomblies et al., 2003; Bomblies and Doebley, 2006). These ZFL data are consistent with negative regulation of ZmRA1 activity by ZFL gene activity, making it tempting to speculate that ZFL could repress ZmRA1 where their expression domains are adjacent in boundary cells at the margin of SPMs.
RA1 marks boundary domains adjacent to meristems in sorghum and setaria panicles
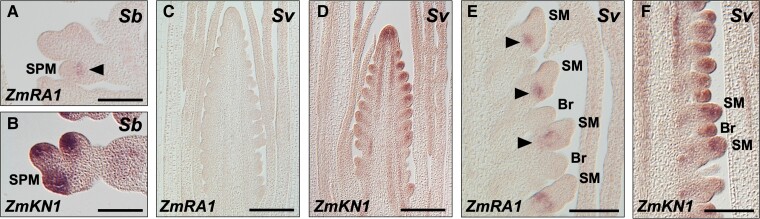
To determine the accumulation of RA1 transcripts in sorghum and setaria inflorescences, we performed RNA in situ hybridization with an antisense probe for ZmRA1, along with the meristem marker gene KNOTTED1 (KN1; Jackson et al., 1994). In sorghum, RA1 transcripts accumulated in a boundary domain directly adjacent to the SPM, as marked by the accumulation of KN1 transcripts (Figure 2, A, B; Supplemental Figure 4, A and B). RA1 transcripts were not detected in early-staged setaria inflorescences initiating BMs, as shown by the accumulation of KN1 (Figure 2, C and D), consistent with transcriptomic profiling of setaria inflorescence development (Zhu et al., 2018). In later-staged setaria inflorescences marked by SMs and bristles, we detected RA1 transcripts in accordance with transcriptomic data (Zhu et al., 2018), which showed boundary domain accumulation adjacent to the SM (Figure 2, E, F; Supplemental Figure 4, C–F). We consistently did not detect RA1 transcript accumulation in or adjacent to bristles, further distinguishing them from the spikelets they are paired with. In maize, RA1 transcripts accumulate between recently-initiated SPMs and the inflorescence or branch axis (Vollbrecht et al., 2005). These results demonstrate (1) a conserved spatial pattern of RA1 transcript accumulation that marks boundary domains adjacent to spikelet-associated short BMs in sorghum, setaria, and maize inflorescences, whether SMs (setaria) or SPMs (maize and sorghum), (2) a conserved lack of expression associated with BMs and LBs and other branch types (i.e. the bristle in setaria), and (3) distinct temporal patterns consistent with discrete branching ontogenies.
Figure 2.
RNA in situ hybridization in sorghum and S. viridis inflorescences. Antisense RNA probes to (A, C, E) ZmRA1 or (B, D, F) ZmKN1 were hybridized to longitudinal sections of developing inflorescences from (A, B) sorghum, Sb or (C–F) S. viridis, Sv. Arrowheads denote RA1 transcript accumulation in boundary domains. SPM, spikelet pair meristem; SM, spikelet meristem; Br, bristle. Scale bars, 100 µm.
Expression of a ZmRA1 transgene largely recovers normal inflorescence architectures in ra1-R mutants
To study the function of promoter cis and coding sequence diversity of RA1 loci in shaping the inflorescences of maize, sorghum, and setaria, we generated a suite of transgenic experiments using interspecies gene transfer (Nikolov and Tsiantis, 2015). Maize, sorghum, and setaria RA1 genes and one chimeric maize-setaria RA1 gene were introduced into maize and backcrossed into the B73 inbred genetic background containing the ra1-R mutant allele. During backcrosses, the events were scored for evidence of a heritable, single-locus, herbicide-resistance phenotype as an indicator of stable expression of the 35S::BAR component of the transgene cassette. In total, 17 independent transgenic events satisfied these genetic segregation criteria and these were also scored qualitatively for their capacity to complement the ra1-R mutant phenotype; from among them, we selected nine events for detailed analysis (Supplemental Tables 1 and 2 and Methods).
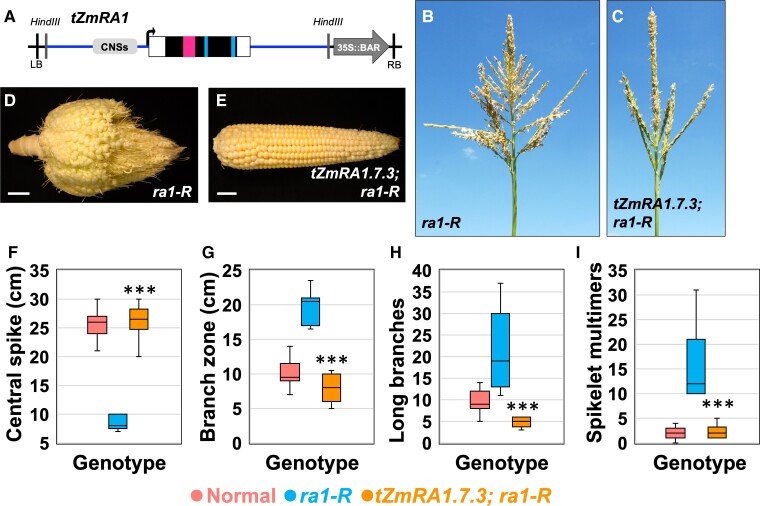
To examine maize RA1 gene function, we first asked if normal tassel and ear morphologies could be recovered in severe ra1-R mutants expressing a reintroduced ZmRA1 genomic fragment containing 2.95 kb of the promoter region including the conserved cis sequences as well as 2.35 kb of sequence downstream of the CDS. We refer to this transgenic cassette as “tZmRA1’ (Figure 3A; Supplemental Table 1). Five independent, stable, single-locus transgene events were generated for tZmRA1. Four of them showed similar effects on the ra1-R phenotype and minimal pleiotropy, while the fifth was markedly pleiotropic (Supplemental Table 1), conferring a dwarfed plant stature and severely reduced tassels and ears. We studied the effects of tZmRA1 in a single, nonpleiotropic insertion event, i.e. tZmRA1.7.3 (nomenclature construct.callus.plant, used throughout for all constructs; see Methods) (Figure 3; Supplemental Table 2). Gross tassel and ear morphology of ra1-R mutants expressing tZmRA1 appeared normal relative to nontransgenic ra1-R siblings (cf. Figure 3, B–E toFigure 1, A and B; Supplemental Figure 6A). Notably, ra1-R ears expressing tZmRA1 were fully unbranched, and kernels were in straight parallel rows along the ear axis; in contrast, kernel rowing was crooked in highly ramified ra1-R ears (Figure 3, D and E) (Vollbrecht et al., 2005).
Figure 3.
Expression of the ZmRA1 locus as a transgene in the ra1-R mutant background. (A), tZmRA1 cassette for expression of ZmRA1 containing 2.9 kb of upstream sequence including conserved noncoding cis regions. Cassette not to scale; see Supplemental Table 1 for details. Open box, UTR sequences. Magenta box, encoded C2H2 zinc finger domain. Blue box, encoded EAR motif. Vertical black lines, left and right borders (LB, RB); Vertical gray lines, HindIII restriction enzyme sites. (B), ra1-R tassel. (C), ra1-R tassel expressing tZmRA1.7.3. (D), ra1-R ear. (E), ra1-R ear expressing tZmRA1.7.3. Scale bars, 2 cm. (F), Central spike length. (G), Branch zone length. (H), Number of long branches. (I), Number of spikelet multimers. For all box and whisker plots, the bottom and top boxes represent the first and third quartile, respectively, the middle line is the median, and the whiskers represent the minimum and maximum values, outlier data points are displayed as individual dots. Two-tailed Student's t test for transgene versus ra1-R ***P < 0.001; normal, n = 20; ra1-R, n = 10; tZmRA1.7.3, n = 8.
We quantified degree of branching, including branch type, lengths, and spikelet pair density (SPD) (Supplemental Figure 5) among inflorescences of segregating normal, ra1-R mutants expressing tZmRA1 and nontransgenic ra1-R siblings to evaluate the degree of normal phenotype recovery. Along the primary axis of the tassel, normal maize produces LBs at the base with an immediate shift to short branches of SPs on the central spike (CS) (Figure 1A). ra1-R mutants produce LBs at the tassel base, then a variable number of transformed, mixed-fate branches bearing both SPs and single spikelets, followed by transformed branches (spikelet multimers) with multiple, single spikelets and finally an abbreviated CS predominantly of short branches of SPs (Figure 3B) (Vollbrecht et al., 2005). The length of the CS between normal and ra1-R expressing tZmRA1 were nearly equivalent (mean difference +0.95 cm); CS was significantly longer in ra1-R with the transgene compared with nontransgenic ra1-R siblings (mean difference −17.68 cm) (Figure 3F). The length of the long branch zone (LBZ) was slightly shorter in ra1-R expressing tZmRA1 relative to normal (mean difference −1.68 cm), whereas LBZ was significantly shorter in transgene-positive ra1-R compared with nontransgenic ra1-R siblings (mean difference −11.84 cm) (Figure 3G). Normal tassels produced on average 4.9 more LBs compared with ra1-R tassels expressing tZmRA1, whereas nontransgenic ra1-R siblings produced on average 17.4 more LBs than ra1-R expressing tZmRA1 (Figure 3H). We observed a negligible difference in spikelet multimers (referred to as “multimers” throughout) between normal and ra1-R transgene-expressing tassels, but nontransgenic ra1-R siblings produced on average 14 more multimers than ra1-R expressing tZmRA1 (Figure 3I). SPD taken from a circumference of 1 cm at the CS midpoint was lower in ra1-R transgene-positive plants compared with both normal and nontransgenic ra1-R siblings (−3.2 and −2.67 SPs, respectively) (Supplemental Figure 6B). The three most-basal tassel LBs were longer in ra1-R expressing tZmRA1 compared with both normal and nontransgenic ra1-R siblings (Supplemental Figure 6C). Collectively, these results indicate that the ZmRA1 transgene is sufficient to recover normal inflorescence architectures in the ra1-R mutant background.
Interspecies expression of a tandem duplicated SbRA1 modeled transgene produces novel ra1-R inflorescence architectures
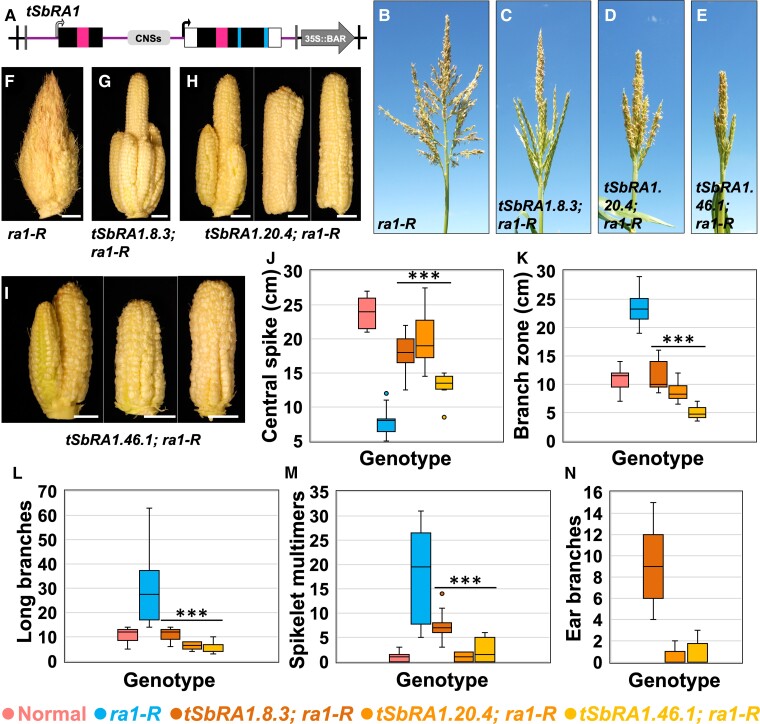
We next asked if interspecies expression of the canonical tandem duplicated SbRA1 locus could recover normal tassel and ear morphologies in ra1-R mutants. We modeled the tandem duplicated SbRA1 transgenic cassette as a 6 kb genomic DNA fragment that includes ∼0.7 kb promoter region of SbRA1US, the SbRA1US paralogous coding region followed by the contiguous 2.03 kb (including the conserved cis sequences) between the SbRA1US paralogous stop codon and the beginning of the SbRA1DS predicted ORF, the predicted ORF, and 2.17 kb downstream of the SbRA1DS stop codon. We refer to this construct as “tSbRA1” (Figure 4A; Supplemental Table 1). Three independent, stable, single-locus transgene events were generated for tSbRA1 and backcrossed into the B73 background; we studied its effects on the ra1-R mutant in all three (Supplemental Table 2).
Figure 4.
Interspecies expression of the tandem duplicated SbRA1 modeled transgene in the ra1-R mutant background. (A), tSbRA1 cassette for interspecies expression of the tandem duplicated SbRA1 locus. Cassette not to scale; see Supplemental Table 1 for details. Open box, UTR sequences. Magenta box, encoded C2H2 zinc finger domain. Blue box, encoded EAR motif. Vertical black lines, left and right borders (LB, RB); Vertical gray lines, HindIII restriction enzyme sites. (B), ra1-R tassel. (C-E), ra1-R tassels expressing (C) tSbRA1.8.3, (D) tSbRA1.20.4, and (E) tSbRA1.46.1 transgenes. (F), ra1-R ear. (G-I), ra1-R ears expressing (G) tSbRA1.8.3, (H) tSbRA1.20.4 and (I) tSbRA1.46.1 transgenes. Scale bars, 2 cm. (J), Central spike length. (K), Branch zone length. (L), Number of long branches. (M), Number of spikelet multimers. (N), Number of ear branches. For all box and whisker plots, the bottom and top boxes represent the first and third quartile, respectively, the middle line is the median, and the whiskers represent the minimum and maximum values, outlier data points are displayed as individual dots. Two-tailed Student's t test for transgene versus ra1-R ***P < 0.001; ra1-R, n = 18; tSbRA1.8.3, n = 11; tSbRA1.20.4, n = 12; tSbRA1.46.1, n = 11.
Overall, tassels of ra1-R mutants that expressed tSbRA1 were much less branched and ranged from normal-appearing overall (events tSbRA1.8.3 and tSbRA1.20.4) to compact (event tSbRA1.46.1) relative to highly branched nontransgenic ra1-R siblings (Figure 4, B–E; Supplemental Figure 7A). Similarly, tSbRA1-expressing ra1-R ears displayed a range in gross phenotype (Figure 4, F–I, N), but were overall much less branched than ra1-R sibling ears. For event tSbRA1.8.3, ear branching was reminiscent of weak ra1 mutant alleles (Figure 4G) (Vollbrecht et al., 2005; Gallavotti et al., 2010). Ears from event tSbRA1.20.4 and tSbRA1.46.1 were occasionally fasciated and branched, and frequently had crooked kernel rows (Figure 4, H, I, and N). Ears from event tSbRA1.46.1 were consistently short and compact (Figure 4I).
To understand the impact of tSbRA1 on ra1-R inflorescences, we quantified branch phenotypes for the three events. When compared with nontransgenic ra1-R siblings, mean CS lengths were significantly longer (range of differences from +5.61 to +12.15 cm), and mean LBZ lengths were significantly shorter in ra1-R carrying the tSbRA1 transgene (range of differences from −12.13 to −18.36 cm) (Figure 4, J and K). Nontransgenic ra1-R siblings produced on average 29.11 LBs and 18 multimers, which was significantly more compared with the mean range of 5.17–10.73 LBs and 2.33–7.09 multimers in tSbRA1-expressing ra1-R siblings (Figure 4, L and M). SPD had a mean range of differences from −0.25 to +5.5 SPs between ra1-R expressing the tSbRA1 transgene and nontransgenic ra1-R siblings (Supplemental Figure 7B). The three most basal LBs were significantly shorter in ra1-R tassels that expressed the tSbRA1 transgene compared with nontransgenic ra1-R siblings (Supplemental Figure 7C).
LBs are completely suppressed in normal ears (Figure 1B), whereas LBs are de-repressed by mutations in ZmRA1 (Figure 1D) (Vollbrecht et al., 2005). For example, ears of strong ra1 mutant alleles, such as ra1-RSd, produce over 200 branches (Weeks, 2013). Ra1-R ears expressing the tSbRA1 transgene were dramatically less branched compared with highly branched ears of nontransgenic ra1-R siblings (Figure 4, N, F, G, and H). Event tSbRA1.8.3 had a mean ear branch number of 9.3, similar to previously reported mean ear branch totals for weak alleles, ra1-63.3359 (11.2 branches) or ra1-RS (12.1 branches) (Weeks, 2013). Events tSbRA1.20.4 and tSbRA1.46.1 had a mean of <1 branch (Figure 4N). Transcripts of the tSbRA1 transgene accumulated in developing tassels beyond the stages when the endogenous ZmRA1 transcript accumulation are highest (Supplemental Figure 8A), supporting heterochronic expression of the transgene in the tassel.
Taken together, expression of the tSbRA1 transgene reduced the order of branching in ra1-R mutant inflorescences, but curiously also produced novel ra1-R phenotypes that included compact tassels and ears, and ear fasciation (Figure 4, D, E, H, and I). Pleiotropic fasciation and stubbiness in the main axis suggest effects on the main IM, where ra1 expression was not detected in normal maize or sorghum. Strong, likely null, maize ra1 alleles have genetic lesions in the C2H2 zinc finger domain (Vollbrecht et al., 2005), a putative DNA-binding domain (Dathan et al., 2002). Indeed, ZmRA1 is suggested to bind and modulate the expression of hundreds of genes during tassel and ear development, which includes the putative direct targeting and repression of COMPACT PLANT2 (CT2; Bommert et al., 2013; Eveland et al., 2014). Loss-of-function ct2 mutants have compact inflorescences and fasciated ears (Bommert et al., 2013), similar to what was observed to be conditioned by the tSbRA1 transgene (Figure 4, B–I). To explain the novel ra1-R phenotypes, we hypothesize that the tSbRA1 transgene may function ectopically and affect expression of target genes like CT2 outside of the spatiotemporally normal expression domain for RA1. Misregulation of RA1 could occur if the upstream copy competes with the downstream copy for binding of regulatory factors, or if the gene duplication itself alters regulation, for example, by changing the distance between cis-regulatory elements or by creating novel ones. Another potential mechanism for the novel phenotypes could be at the level of the gene product. For example, given that the truncated upstream RA1 copy encodes a C2H2 zinc finger domain (Figures 1G and4A), expression from both copies could lead to binding interference between SbRAUS (truncated) and SbRADS (complete) proteins, where SbRADS is required at sufficient levels to impose meristem determinacy. Similar interference mechanisms for dominant negative alleles have been reported to influence flowering in Arabidopsis (Ahn et al., 2006) and sunflower (Helianthus annuus) (Blackman et al., 2010). Although SbRAUS expression is barely detectable in sorghum inflorescences (Vollbrecht et al., 2005), we did not assay its expression in the transgenic lines.
Interspecies expression of the downstream SbRA1 modeled transgene partially recovers normal inflorescence architectures in ra1-R mutants
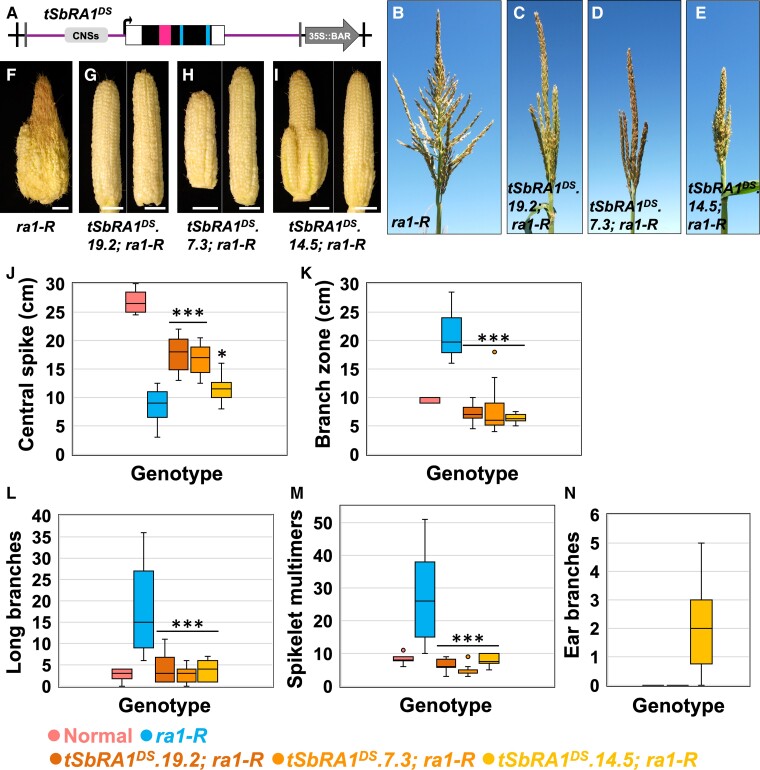
Because tSbRA1 conditioned novel phenotypic changes in addition to complementation, we asked if normal tassel and ear morphologies in ra1-R mutants could be recovered by interspecies expression of only the downstream SbRA1 locus, which does not contain frameshifts or apparent deleterious mutations. The downstream SbRA1 transgenic cassette SbRA1DS was modeled to include its predicted ORF and 1.68 kb upstream, including the conserved cis sequences plus 2.17 kb downstream of the stop codon, and we refer to this construct as “tSbRA1DS’ (Figure 5A; Supplemental Table 1). Three independent, stable, single-locus transgene events were generated for tSbRA1DS and backcrossed to the ra1-R mutant in B73, and we studied its effects in all three (Supplemental Table 2).
Figure 5.
Interspecies expression of the downstream SbRA1 modeled transgene in the ra1-R mutant background. (A), tSbRA1DS cassette for interspecies expression of the downstream SbRA1 locus. Cassette not to scale; see Supplemental Table 1 for details. Open box, UTR sequences. Magenta box, encoded C2H2 zinc finger domain. Blue box, encoded EAR motif. Vertical black lines, left and right borders (LB, RB); Vertical gray lines, HindIII restriction enzyme sites. (B), ra1-R tassel. (C–E), ra1-R tassels expressing (C) tSbRA1DS.19.2, (D) tSbRA1DS.7.3, and (E) tSbRA1DS.14.5 transgenes. (F), ra1-R ear. (G-I), ra1-R ears expressing (G) tSbRA1DS.19.2, (H) tSbRA1DS.7.3, and (I) tSbRA1DS.14.5 transgenes. Scale bars, 2 cm. (J), Central spike length. (K), Branch zone length. (L), Number of long branches. (M), Number of spikelet multimers. (N), Number of ear branches. For all box and whisker plots, the bottom and top boxes represent the first and third quartile, respectively, the middle line is the median, and the whiskers represent the minimum and maximum values, outlier data points are displayed as individual dots. Two-tailed Student's t test for transgene versus ra1-R ***P < 0.001, *P < 0.05; ra1-R, n = 15; tSbRA1DS.19.2, n = 10; tSbRA1DS.7.3, n = 12; tSbRA1DS.14.5, n = 10.
Overall, tassels from ra1-R mutants that expressed tSbRA1DS were less branched and ranged from normal (events tSbRA1DS.19.2 and tSbRA1DS.7.3) to moderately compact (event tSbRA1DS.14.5) architectures relative to highly ramified architecture of nontransgenic ra1-R siblings (Figure 5, B–E; Supplemental Figure 9A). Similarly, ra1-R ears expressing the tSbRA1DS transgene displayed a range in gross phenotype (Figure 5, F–I, N). Events tSbRA1DS.19.2 and tSbRA1DS.7.3 produced unbranched ears with straight rows of kernels along the ear axis (Figure 5, G and H), whereas event tSbRA1DS.14.5 showed ear branching reminiscent of weak ra1 mutant alleles (Figure 5I) (Vollbrecht et al., 2005; Gallavotti et al., 2010).
To characterize the impact of tSbRA1DS on ra1-R inflorescences in detail, we quantified tassel branch phenotypes for the three events. When compared with nontransgenic ra1-R siblings, mean CS lengths were significantly longer (range of differences from +2.75 to +8.95 cm), and mean LBZ lengths were significantly shorter in ra1-R that carried the tSbRA1DS transgene (range of differences from −12.71 to −14.15 cm) (Figure 5, J and K). Nontransgenic ra1-R siblings produced on average 26.8 LBs and 17 multimers, which was significantly more compared with the mean range of 4.58 to 8 LBs and 2.83 to 4.2 multimers in ra1-R expressing the tSbRA1DS transgene (Figure 5, L and M). SPD had a mean range of differences from +1.53 to +7.33 SPs between ra1-R with the tSbRA1DS transgene and transgene-free ra1-R siblings (Supplemental Figure 9B). The three most basal LBs were significantly shorter in ra1-R tassels with the tSbRA1DS transgene compared with nontransgenic ra1-R siblings (Supplemental Figure 9C). Interspecies expression of tSbRA1DS was sufficient to impose SPM determinacy in ra1-R ears for events tSbRA1DS.19.2 and tSbRA1DS.7.3, where branch suppression was fully penetrant. Event tSbRA1DS.14.5 had on average two branches (Figure 5N), which was substantially less than average ear branch number for weak ra1 alleles (7.1 branches; Weeks, 2013). Transcripts of the tSbRA1DS transgene accumulated in developing tassels beyond the stages when the endogenous ZmRA1 transcript accumulation are highest (Supplemental Figure 8B), supporting heterochronic expression of the transgene in the tassel.
Collectively, interspecies expression of tSbRA1DS restored more normal ear inflorescences with less branching and straighter rows, and less pleiotropy with respect to fasciation and shortened axes, relative to the tSbRA1 cassette. Furthermore, both the tSbRA1 and tSbRA1DS constructs substantially remediated ra1-R tassel branching. Given that the tSbRA1DS transgene eliminates the SbRA1US locus present in the tSbRA1 construct, these results suggest functional cis-regulatory element(s) that reside in the 1.68 kb sequence promoter region of the SbRA1DS locus are affected by their proximity to SbRA1US in the tandem duplication, especially in the maize ear. Our data on ra1-R mutants expressing either tSbRA1 or tSbRA1DS cassettes are consistent with a hypothesis raised previously (Vollbrecht et al., 2005): variation in inflorescence architecture, and thus degrees of determinacy, is attributed to the developmental timing of RA1 expression and its activity, as reflected in the range of branch types observed among maize mutant alleles, transgene versions, or genetic diversity of RA1 in maize and other grasses. Furthermore, these results suggest that the developmental context of RA1 activity in the tassel and ear is crucial in regulating determinacy (cf. ear and tassel phenotypes in Figures 4 and 5). Indeed, quantification of ear and tassel branch number in the F1 hybrid generation of B73×Mo17 introgressions homozygous for the weak allele ra1-63.3359 showed additive effects on ear branching and over-dominance effects on tassel branching (Weeks, 2013).
Interspecies expression of SvRA1 only recovers near normal inflorescence branching in ra1-R mutants when chimeric with the ZmRA1 promoter region
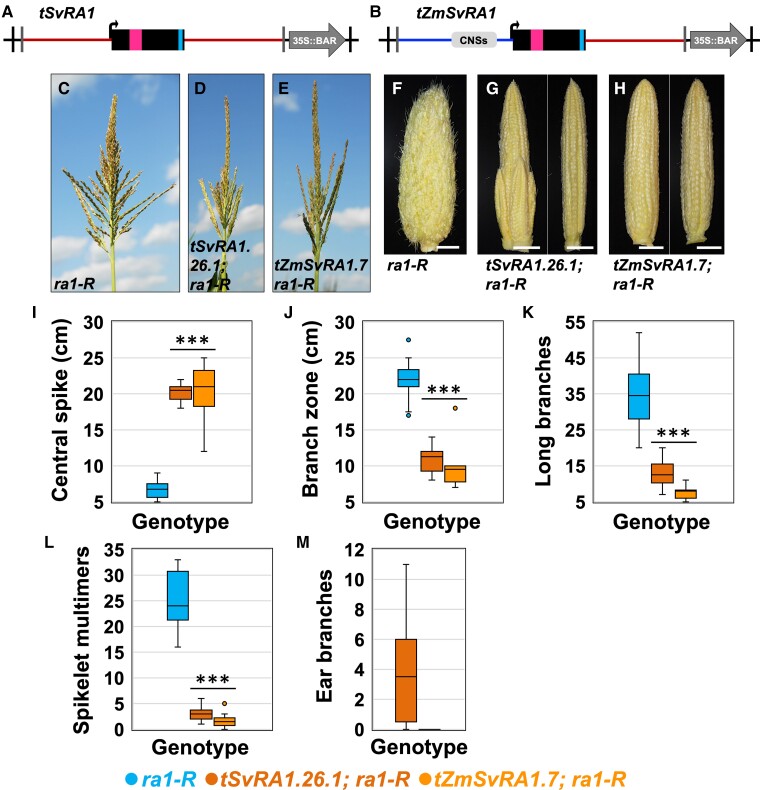
Given the complex genetic nature of the SbRA1 locus, we sought to explore the impact of the single-copy SvRA1 on inflorescence morphology. We were also interested in testing the impact of the cis sequences found in promoter regions of ZmRA1 and SbRA1DS and conserved among Andropogoneae grasses, as well as sufficiency of the single EAR motif in SvRA1. We, therefore, compared and contrasted interspecies expression of the SvRA1 coding region with its endogenous promoter region that largely lacks the conserved cis sequences with expression of the SvRA1 gene body in cis with the maize promoter region (pZmRA1). We modeled the SvRA1 transgene cassette to include 1.53 kb of the predicted SvRA1 promoter region, the coding region and 1.97 kb downstream of the stop codon and we refer to it as “tSvRA1” hereafter (Figure 6A; Supplemental Table 1). Additionally, we generated a chimeric gene cassette termed pZmRA1::SvRA1 where 2.95 kb of ZmRA1 promoter region and five-prime untranslated region was fused upstream of the SvRA1 coding sequence and 1.97 kb of downstream SvRA1 sequence and we refer to the construct as “tZmSvRA1’ hereafter (Figure 6B; Supplemental Table 1). Four independent, stably herbicide-resistant and single-locus transgene events were identified for tSvRA1 during backcrossing to the B73 tester line (Supplemental Table 2). Of those events, three were unique among all stable, herbicide-resistant transgenics we propagated in this study, across all five constructs, in that they showed no notable effect on the strong ra1-R mutant phenotype or any other plant phenotypes examined. Thus, quantitative phenotyping was not performed for these three events, which strongly suggests the SvRA1 transgene has little or no functional activity in maize. The fourth event for tSvRA1 showed some reduction of vegetative shoot stature and effects on inflorescence branching and was therefore examined for ear and tassel phenotype, although we consider it an outlier or unusual event among the four tSvRA1 transgenic lines. One stable, single-locus transgene event was generated for tZmSvRA1 and backcrossed to B73 and it affected inflorescences but was nonpleiotropic for vegetative plant characteristics. Thus, we studied the effects of tSvRA1 and tZmSvRA1 in single-locus events backcrossed in the ra1-R mutant background (Supplemental Table 2).
Figure 6.
Interspecies expression of an outlier SvRA1 event or of chimeric ZmSvRA1 as a transgene in the ra1-R mutant background. Three of four SvRA1 events failed to complement the ra1-R mutant phenotype; see text and Supplemental Table 1 for details. The weakly complementing event tSvRA1.26.1 is thus considered an outlier, but its phenotype is analyzed here for comparison and to demonstrate functional potential of SvRA1 in maize. (A), tSvRA1 cassette for interspecies expression of the SvRA1 locus. Cassette not to scale; see Supplemental Table 1 for details. Magenta box, encoded C2H2 zinc finger domain. Blue box, encoded EAR motif. Vertical black lines, left and right borders (LB, RB); vertical gray lines, HindIII restriction enzyme sites. (B), tZmSvRA1 cassette for expression of the SvRA1 coding region fused to the 2.9 kb Zm upstream region including conserved noncoding cis sequences. Cassette not to scale; see Supplemental Table 1 for details. Open box, UTR sequences. Magenta box, encoded C2H2 zinc finger domain. Blue box, encoded EAR motif. Vertical black lines, left and right borders (LB, RB); vertical gray lines, HindIII restriction enzyme sites. (C), ra1-R tassel. (D, E) ra1-R tassels expressing (D) the outlier event tSvRA1.26.1 and (E) tZmSvRA1.7 transgenes. (F), ra1-R ear. (G, H) ra1-R ears expressing (G) the outlier event tSvRA1.26.1 and (H) tZmSvRA1.7 transgenes. Scale bars, 2 cm. (I), Central spike length. (J), Branch zone length. (K), Number of long branches. (L), Number of spikelet multimers. (M), Number of ear branches. For all box and whisker plots, the bottom and top boxes represent the first and third quartile, respectively, the middle line is the median, and the whiskers represent the minimum and maximum values, outlier data points are displayed as individual dots. Two-tailed Student's t test for transgene versus ra1-R ***P < 0.001; ra1-R, n = 15; tSvRA1.26.1, n = 12; tZmSvRA1.7, n = 9.
Overall, tassels from ra1-R mutants that expressed the unusual tSvRA1 event or expressed the tZmSvRA1 transgene were less branched and had normal architectures relative to the highly branched architecture of nontransgenic ra1-R siblings (Figure 6, C–E; Supplemental Figure 10A). Similarly, ra1-R ears expressing tSvRA1 displayed a range in gross phenotype from unbranched ears with straight rows of kernels along the ear axis and no branches to those with crooked rows and a low degree of branching (Figure 6, F, G, and M). In contrast, ra1-R ears expressing tZmSvRA1 were fully unbranched with kernels in straight parallel rows along the ear axis (Figure 6, H and M).
To understand the impact of the unusual tSvRA1 event or of tZmSvRA1 on ra1-R tassels and ears, we quantified branch phenotypes. When compared with nontransgenic ra1-R siblings, mean CS lengths were significantly longer (difference +13.67 cm for both tSvRA1 and tZmSvRA1), and mean LBZ lengths were significantly shorter in ra1-R tassels expressing either tSvRA1 or tZmSvRA1 transgenes (difference −11 cm for tSvRA1 and −12.1 cm for tZmSvRA1) (Figure 6, I and J). Relative to normal tassels, mean CS lengths were shorter (difference −5.0 cm for both tSvRA1 and tZmSvRA1), and mean LBZ lengths were marginally longer in ra1-R tassels with either tSvRA1 or tZmSvRA1 (difference +1.27 cm for tSvRA1 and +0.17 cm for tZmSvRA1) (cf. Figure 3, F and G to Figure 6, I and J). Nontransgenic ra1-R sibling tassels produced on average 35.1 LBs and 25.1 multimers, which were significantly more compared with averages of 12.7 LBs and 3.1 multimers for tSvRA1, and 7.5 LBs and 1.7 multimers for tZmSvRA1 expressing ra1-R siblings (Figure 6, K and L). Compared with a mean of 9.6 LBs and 2 multimers for normal tassels, tSvRA1 expressing ra1-R tassels produced on average 3.1 more LBs and 0.9 more multimers, whereas tZmSvRA1 expressing ra1-R tassels had 2.1 fewer LBs and 0.3 fewer multimers (cf. Figures 3, H and I to 6, K and L). For SPD, ra1-R tassels with tSvRA1 had on average 3.2 more SPs along the CS compared with nontransgenic ra1-R siblings, and similarly, ra1-R tassels with tZmSvRA1 had 2.2 more SPs (Supplemental Figure 10B). Relative to SPD for normal tassels, ra1-R expressing tSvRA1 had on average 0.7 fewer SPs and ra1-R expressing tZmSvRA1 had 1.7 fewer SPs along the CS (cf. Supplemental Figures 6B to 10B). The three most basal LBs were consistently shorter in ra1-R tassels that carried the tSvRA1 transgene compared with nontransgenic ra1-R siblings; LBs were of similar length between ra1-R expressing the tZmSvRA1 transgene and nontransgenic ra1-R siblings (Supplemental Figure 10C). Compared with normal tassels, the three most basal LBs of ra1-R tassels expressing either tSvRA1 or tZmSvRA1 were shorter (cf. Supplemental Figures 6C to 10C).
Establishment of SPM determinacy during ear development differed conspicuously between ra1-R expressing the tZmSvRA1 transgene and expressing the unusual tSvRA1 event. ra1-R with the tSvRA1 transgene produced an average of four branches, whereas ra1-R ears carrying the tZmSvRA1 transgene were unbranched (Figure 6M). Overall, the tZmSvRA1 transgene behaved most similarly to the tZmRA1 endogenous maize construct.
Collectively, the transgene constructs containing SvRA1 conferred degrees of complementation from non to partial to nearly complete, all without inducing the novel inflorescence phenotypes of sorghum transgenes. Whereas in most SvRA1 (tSvRA1) lines the intact SvRA1 gene did not complement the ra1-R mutant phenotype, we saw some effects in one line. Similarly, the tZmRA1 and tSvRA1events were not all identical in their phenotypic effects, as is not unusual among transgene events integrated into different chromosomal regions. We speculate that the unusual SvRA1 (tSvRA1) event may be integrated in a genomic context that results in effectively ectopic expression, and therefore suggesting a lack of appropriate cis-regulatory components in the SvRA1 promoter region while revealing some functional potential of the SvRA1 gene product. It is also possible that such regulatory components do exist in setaria but are located distantly and not included in the tSvRA1 construct; however, the relatively compact nature of the setaria genome and the fact that similarly sized maize and sorghum fragments did complement may argue against that hypothesis.
In the encoded polypeptides, ZmRA1 and SvRA1 C2H2 zinc-finger domains vary by three amino acid residues, and the C-terminal EAR motif in SvRA1 differs by one residue. However, a conserved EAR motif adjacent to the C2H2 zinc-finger domain in ZmRA1 is absent in SvRA1 (Figure 1, G; Supplemental Figure 2B). In maize, RA1 physically interacts with REL2 via EAR motifs in a large transcriptional repressor complex to impose SPM determinacy (Gallavotti et al., 2010; Liu et al., 2019). Functional importance of the EAR motif adjacent to the C2H2 zinc-finger domain has not been tested genetically. Our data from the tZmSvRA1 chimeric gene cassette suggest the C2H2 -proximal EAR motif, which is by definition dispensable for RA1 function in setaria, is likewise to a large degree nonessential in maize. Whereas complementation was only partial for the unusual tSvRA1 event, it was more complete for the chimeric tZmSvRA1 construct. The promoter region swap data clearly indicate that cis-encoded regulation of RA1 expression is a key functional component in promoting SPM determinacy, especially during ear development. In an evolutionary context, it is interesting to note that while spikelets are normally unpaired in setaria and the SvRA1 gene is insufficient to complement the maize ra1-R mutant with its many unpaired spikelets, under the proper expression conditions, the SvRA1 gene product does confer sufficient determinacy activity to restore SPs to ra1-R maize. These results suggest that within the Panicoideae subfamily of the PACMAD grasses, RA1 has an evolutionarily conserved determinacy function that contributes to specifying short BMs: SMs in setaria and SPMs in maize and sorghum. Our data are all consistent with a hypothesis wherein within the paired-spikelet Andropogoneae tribe, RA1 has adopted a key role in producing the SP by imposing determinacy in the proper developmental context rather than by specifying any strict SPM identity. It would be interesting to test whether the RA1 genes from other Panicoid species as well as from Chloridoid subfamily and/or other PACMAD grasses show similar functions.
The developmental context in which genes and networks operate within meristems and flanking organ boundary domains is critical in determining inflorescence form. Elegant genetic studies on the spatiotemporal regulation and function of transcription factors have shed important light on the mechanisms governing inflorescence branching patterns. Genetic variation in distal regulatory elements (Clark et al., 2006; Studer et al., 2011), proximal or intronic cis-regulatory elements (Arnaud et al., 2011; Wills et al., 2013; Kusters et al., 2015), coding sequences that alter protein function (Wang et al., 2005; Whipple et al., 2010), protein–protein interactions (Bartlett et al., 2016; Abraham-Juarez et al., 2020) or protein–DNA interactions (Maizel et al., 2005; Sayou et al., 2014) are critical drivers of inflorescence branching. Our data leveraging interspecies gene transfer and chimeric transgene expression suggest that cis-encoded regulation of RA1 expression is a key factor in modulating meristem determinacy that ultimately impacts grass inflorescence architecture. With the ability to map hundreds of regulatory regions and transcription factor binding sites across diverse plant genomes (Lu et al., 2019; Galli et al., 2020), it will be important to understand the regulatory context of the conserved cis sequences that reside in RA1 promoters.
Branch determinacy in the grasses is controlled by gene networks that function in boundary domains adjacent to the meristem they positionally regulate. Since their discovery, such “signaling centers” have emerged as a major theme in regulating meristem determinacy, not meristem identity, and are key drivers of complex branching patterns seen in grass inflorescences (Whipple, 2017; Kellogg 2022). Maize RAMOSA genes—RA1 (Vollbrecht et al., 2005), RA2 that encodes a LATERAL ORGAN BOUNDARY domain transcription factor (Bortiri et al., 2006), and the TREHALOSE PHOSPHATE PHOSPHATASE-encoding RA3 (Satoh-Nagasawa et al., 2006)—constitute a “signaling center” as these genes are co-expressed in overlapping boundary domains (Vollbrecht and Schmidt, 2009) and likely regulate a mobile signal that promotes determinacy of adjacent BMs. Similarly, BM determinacy is controlled by the GATA domain zinc-finger and SQUAMOSA PROMOTER BINDING PROTEIN transcription factors encoded by TASSEL SHEATH1 (TSH1) and TSH4 (Whipple et al., 2010; Chuck et al., 2010), and SM identity and determinacy are regulated by boundary expression of BRANCHED SILKLESS1 and INDETERMINANT SPIKELET1 that encode APETALA2 domain transcription factors (Chuck et al., 1998; 2002). BMs, SPMs, and SMs are not meristem types found in eudicot inflorescences, where variation and complexity are largely governed by shifts in meristem identity (Prusinkiewicz et al., 2007; Lemmon et al., 2016). Given that RA1 transcripts accumulate in meristem boundary regions during the development of sorghum and setaria inflorescences, it will be interesting to test the functional consequences of mutating RA1 in these grasses. Meristem identity genes in eudicots are expressed in meristems; genes that regulate inflorescence variation and complexity in the grasses are expressed in adjacent boundary domains to regulate meristem determinacy. Our work on the expression and functional conservation of syntenic RA1 orthologs provides comparative insight into the genetic basis of grass inflorescence diversity, and opens the door for future reverse engineering of grass inflorescence evolution for crop improvement.
Materials and methods
Genetic stocks
This study utilized the maize (Zea mays) ra1-R allele (Vollbrecht et al., 2005) backcrossed seven generations to the B73 background to generate the “recurrent B73 parent” either ra1-R homozygotes or ra1-R/ra1-B73 heterozygotes were used in crossing schemes.
Generation of RA1 transgenes
The 35SBAR fragment from pTF101.1 was modified by PCR to introduce a HindIII site at the 3’ end of the terminator. This allowed a 2.0 kb HindIII restriction fragment containing 35SBAR-terminator to be isolated, treated with DNA polymerase I (Klenow) and dNTPs to generate blunt ends, and ligated into the SmaI site of pSB11 (Komari et al., 2006), creating a vector called pSB11_BAR. This vector, which contains the 35SBAR gene adjacent to and transcribed towards the T-DNA left border, was the precursor to all of the complementation vectors containing the genomic regions described below.
For construct tZmRA1, ZmRA1, and flanking regulatory regions were PCR amplified from Z. mays B73 genomic DNA and ligated with pSB11_BAR at HindIII. To distinguish the tZmRA1_RA1 allele from endogenous allele in subsequent generations after plant transformation, we introduced an AccI restriction site in the RA1 coding DNA sequence. This synonymous SNP (B73_v5 7: 114959005 C > T) is a natural, low-frequency variant found in the maize inbred P39 haplotype (Vollbrecht et al., 2005). For the sorghum (Sorghum bicolor) construct tSbRA1DS, a 6.0 kb XbaI fragment obtained by screening a BTx623-derived BAC library with a ZmRA1 probe was cloned into pBluescript II KS (Agilent) and the HindIII site in the polylinker was used for ligation into pSB11_BAR. Sorghum construct tSbRA1 was generated from tSbRA1DS following introduction of a HindIII site at the 3’ end of the upstream SbRA1 frameshift copy (2: 58699332; Supplemental Table 1), thereby removing a 1.6 kb fragment containing the upstream copy. For the green millet (Setaria viridis) construct tSvRA1, in-fusion cloning methods (Clontech/Takara) were employed to PCR-amplify and clone from S. viridis A10 genomic DNA a 4 kb fragment containing the SvRA1 transcribed region and regulatory sequences into pSB11-BAR as a HindIII-BamHI insertion. The maize/setaria chimeric construct tZmSvRA1 was generated as a translational fusion at the start codon by replacing the Setaria promoter-containing fragment in tSvRA1 with the 2.9 kb maize fragment. The reference genome coordinates of the RA1 genes and regulatory regions are listed in Supplemental Table 1, and all primers used for vector construction are listed in Supplemental Table 3.
Constructs except for tZmSvRA1 were recombined into the pSB1 superbinary vector in Agrobacterium tumefaciens LBA4404 via triparental mating (Komari et al., 2006). These strains were used for Agrobacterium-mediated maize transformation of Hi-II embryos by the Iowa State University Plant Transformation Facility. Transgenic maize plants containing the tZmSvRA1 cassette were generated in Erik Vollbrecht's lab at Iowa State University using particle bombardment of immature Hi-II embryos with the SB11_BAR-derived vector directly (Frame et al., 2000).
Tests for recovery in ra1-R
Transgenic plants derived from a single callus event in the Hi-II transformation system are not necessarily identical; therefore, we implemented a nomenclature that denotes construct, callus number, and plant number (e.g. tZmRA1.7.3). T0 transgenic plants were crossed three times (construct tZmRA1) or four times (constructs tSbRA1, tSbRA1DS, tSvRA1, and tZmSvRA1) to the recurrent B73 parent line before phenotyping. During the introgression generations, plants were treated with a 2.5% v/v Liberty solution applied to a single leaf to assay for 35SBAR gene-mediated resistance to Liberty herbicide (source, BASF). We also used transgene-specific genotype analyses to track integration events and determine transgene locus number by segregation analysis. DNA was made from leaf punches as previously described (Strable et al., 2017) and PCR-based genotype assays were performed using standard conditions with the primers described (Supplemental Table 3). To genotype alleles at the endogenous ZmRA1 locus in the presence of all but the tZmRA1 transgene, a CAPS assay was utilized to detect an SNP within with the ra1-R allele which results in the introduction of an AccI restriction site. The 765 bp amplicon generated by primers RA8 and RA11 is digested by AccI in ra1-R to generate two fragments, 334 and 431 bp. The tZmRA1 transgene contains the same AccI SNP as ra1-R. Thus, in crosses with the tZmRA1 transgene, an additional MscI dCAPS assay that detects the lesion in the ra1-R mutant allele was employed to distinguish the tZmRA1-derived amplicons (i.e. without MscI site to yield 190 bp) from the ra1-R derived amplicons (with the MscI site to yield 155 and 35 bp following digestion).
Transgene events that segregated as single-locus integrations and showed a stable herbicide-resistance phenotype were selected for qualitative or quantitative phenotyping analysis. To produce the segregating populations used for phenotyping tZmRA1, tSbRA1, and tSbRA1DS, plants heterozygous ra1-R/+ and hemizygous for the transgene of interest were crossed as females by ra1-R/ra1-R pollen of the recurrent B73 parent. To produce the tSvRA1 and tZmSvRA1 material for phenotyping, we crossed females homozygous ra1-R/ra1-R and hemizygous for the transgene of interest by ra1-R/ra1-R pollen.
Phenotypic analysis
All maize plant phenotyping was performed on field-grown plants in the summers of 2014 (constructs tZmRA1, tSbRA1, and tSbRA1DS) and 2018 (constructs tSvRA1 and tZmSvRA1), at the same location on the Woodruff Farm in Ames, Iowa. Tassel phenotype characters are summarized in Supplemental Figure 5 and described here. LBZ was measured from the basal-most to the apical-most LBs. CS length was taken from the apical-most long branch to the tip of the tassel and comprised SPs. A long branch was defined as the typical basal LBs in maize, i.e. bearing only SPs, or as bearing a mix of SPs and single spikelets. Spikelet multimers were any branches bearing three or more single spikelets. SPD was taken from a 1 cm band in circumference at the CS midpoint.
RNA in situ hybridization and expression analysis
Field-grown S. bicolor and growth chamber-grown S. viridis panicles were fixed overnight at 4°C in FAA. Samples were dehydrated through a graded ethanol series (50%, 70, 85, 95, and 100) each 1 h, with three changes in 100% ethanol. Samples were then passed through a graded Histo-Clear (National Diagnostics) series (3:1, 1:1, and 1:3 ethanol: Histo-Clear) with three changes in 100% Histo-Clear; all changes were 1 h each at room temperature. Samples were then embedded in Paraplast®Plus (McCormick Scientific), sectioned, and hybridized as described previously (Strable and Vollbrecht, 2019). Hybridizations were performed using antisense digoxygenin-labeled RNA probes to ZmRA1 (Supplemental Table 3) and ZmKN1 (Jackson et al., 1994).
Field-grown, developmentally staged maize tassels were dissected away from leaf primordia and placed individually in 100 µl Trizol (Thermo-Fisher) and stored at −80°C in a 1.5 ml Eppendorf tube until processing. To process, 400 µl Trizol was added, and tassel tissue was thawed and ground in the presence of Trizol using a plastic drill mount pestle. Total RNA was extracted as per the Trizol manufacturer and treated with RQ1 DNase (Promega) following the protocol outlined by the manufacturer, and converted to cDNA using RNA to cDNA EcoDry™ Premix (Double Primed) reagents (Takara Bio, USA). The cDNA was diluted 1:1 with water, and 1.0 µl was used for PCR. PCR followed standard conditions using GoTaq®Green Master Mix (Promega corp.), Ta = 58°C, 1 min. extension at 72°C for 33 cycles. Primers are listed in Supplemental Table 3.
Conservation analysis of promoter cis sequences
For mVISTA analysis, genomic sequences (0.5 kb) upstream of the predicted 5'UTR regions of RA1 in Zea mays, Sorghum bicolor, and Setaria viridis were downloaded from https://ensembl.gramene.organd aligned using mVISTA LAGAN alignment (https://genome.lbl.gov/vista/mvista/submit.shtml). The plots depict 100 bp alignment windows at a similarity threshold 70% shaded in red.
To identify conserved noncoding sequences and binding motifs, the coding sequence of Zea mays RA1 (Zm00001eb312340—B73-REFERENCE-NAM-5.0) was used to find likely orthologs in other Panicoideae grasses. Sequences from Chasmanthium laxum (Chala.06G030500—v1.1), Miscanthus sinensis (Misin03G169300 & MisinT268200—v7.1), Panicum halli (Pahal.2G260300—v3.2), Paspalum vaginatum (Pavag06G030400—v.3.1), Setaria viridis (Sevir.2G209800—v2.1), and Sorghum bicolor (Sobic.002G197700 and Sobic.002G197800—v3.1.1) were identified using the BLAST tool in Phytozome v13. Sequence from Coix lacryma-jobi (Adlay0592-017T1) was selected from its own genome site. (http://phyzen.iptime.org/adlay/index.php). From all accessions, we took 2 kb upstream of the translation initiation site. First, conserved noncoding sequences from RA1 promoter region sequence from Andropogoneae was determined using MEME (Bailey and Elkan, 1994). Then, the resulting motifs were searched in the other Panicoideae nonAndropogoneae grasses using FIMO (Grant et al., 2011). All sequences were compared against the nonredundant (Khan et al., 2018) database from plants while using SEA to observe any possible well-known binding sites present internally (Bailey and Grant, 2021). The position of motifs from JASPAR was compared with the position of conserved noncoding sequences to check for overlap. Motifs from Clade “A” ARFs were searched on the different sequences by using FIMO (Galli et al, 2018). Finally, these binding sites from the SEA analysis were used to search again in the Andropogoneae grasses using FIMO to obtain the relative coordinates in the ZmRA1 promoter region (Grant et al., 2011).
Statistical analysis
Comparative statistical analysis for plant phenotyping was assessed by using pairwise t test and ANOVA test (Supplemental Table 4). In the case of ANOVA test, we first evaluated if the data followed the different assumptions to perform this statistical analysis. If the data followed the assumptions, the test was performed, then a Tukey post hoc test was carried out to achieve multiple pairwise comparisons between groups. A Welch ANOVA test was implemented if the data did not meet the homogeneity of variance assumption; this was followed by a Tukey post hoc test. A Kruskal–Wallis test was performed if the data did not meet the normality assumption, and a Dunn's test was executed to make multiple pairwise comparisons. Statically analysis was carried out using the rstatix package in R.
Accession numbers
ZmRA1, Zm00001eb312340; ZmKN1, Zm00001eb055920; SbRA1DS, Sobic.002G197700; SbRA1US, Sobic.002G197800; SvRA1, Sevir.2G209800; ClRA1, Chala.06G030500; MsRA1, Misin03G169300 and MisinT268200; PhRA1, Pahal.2G260300; PvRA1, Pavag06G030400; Cl-j, Adlay0592-017T1, Eleusine coracana RA1 ELECO.r07.6AG0534810.1
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Developing inflorescences of maize, sorghum and S. viridis.
Supplemental Figure S2. RA1 locus and amino acid sequence in Panicoid grass species.
Supplemental Figure S3. Conserved cis sequences in RA1 promoter regions of Panicoid grasses.
Supplemental Figure S4. RNA in situ hybridization in sorghum and S. viridis inflorescences.
Supplemental Figure S5. Tassel traits quantified in this study.
Supplemental Figure S6. Tassel traits in ra1-R mutants expressing the tZmRA1 transgene cassette.
Supplemental Figure S7. Tassel traits in ra1-R mutants expressing the tSbRA1 transgene cassette.
Supplemental Figure S8. Expression of tSbRA1 and tSbRA1DS transgenes and endogenous ZmRA1 in developing tassels.
Supplemental Figure S9. Tassel traits in ra1-R mutants expressing the tSbRA1DS transgene cassette.
Supplemental Figure S10. Tassel traits in ra1-R mutants expressing either the tSvRA1 or tZmSvRA1 transgene cassette.
Supplemental Table S1. Reference genome coordinates and construct and transgenic event information.
Supplemental Table S2. Transgene genetic segregation data.
Supplemental Table S3. Primers used in this study.
Supplemental Table S4. ANOVA tests for transgene effects
Supplemental Dataset S1 . Transcription factor binding sites in selected Andropogoneae conserved noncoding cis sequences.
Supplementary Material
Acknowledgments
We are grateful to Brandi Sigmon for insightful discussion on RA1 in sorghum and for comments on the manuscript. Additionally, we thank Pete Lelonek for assisting with greenhouse management and plant care. Many thanks to former undergraduate students, especially Emery Peyton, Charlie Beeler, Tryggve Rogers, Raven Saunders-Duckett, Matt Hirsch, Nicole Essner, and Jack Schwickerath for their help with summer genetics nurseries and phenotypic analysis. We appreciate the insightful comments on the manuscript from Jack Satterlee.
Contributor Information
Josh Strable, Department of Genetics, Development and Cell Biology, Iowa State University, Ames, Iowa 50011, USA; Department of Molecular and Structural Biochemistry, North Carolina State University, Raleigh, North Carolina 27695, USA.
Erica Unger-Wallace, Department of Genetics, Development and Cell Biology, Iowa State University, Ames, Iowa 50011, USA.
Alejandro Aragón Raygoza, Department of Molecular and Structural Biochemistry, North Carolina State University, Raleigh, North Carolina 27695, USA.
Sarah Briggs, Department of Genetics, Development and Cell Biology, Iowa State University, Ames, Iowa 50011, USA.
Erik Vollbrecht, Department of Genetics, Development and Cell Biology, Iowa State University, Ames, Iowa 50011, USA.
Funding
This work was supported by the National Science Foundation (IOS number 1238202 to E.V.), as well as USDA Hatch project IOW03649 and the Iowa State University Crop Bioengineering Center. J.S. and A.A.R. are supported by North Carolina State University startup funds and USDA Hatch project 1026392.
References
- Abraham-Juarez MJ, Schrager-Lavelle A, Man J, Whipple C, Handakumbura P, Babbitt C, Bartlett M (2020) Evolutionary variation in MADS-box dimerization affects floral development and protein degredation dynamics. Plant Cell 32(11): 3408–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D (2006) A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J 25(3): 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud N, Lawrenson T, Østergaard L, Sablowski R (2011) The same regulatory point mutation changed seed-dispersal structures in evolution and domestication. Curr Biol 21(14): 1215–1219 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Altman RBrutlag DKarp PLathrop R and Searls D, eds, Proceedings of the second international conference on intelligent systems for molecular biology. AAAI Press, Menlo Park, CA, pp. 28–36 [PubMed] [Google Scholar]
- Bailey TL, Grant CE (2021) SEA: simple enrichment analysis of motifs. bioRxiv 10.1101/2021.08.23.457422 [DOI] [Google Scholar]
- Bartlett M, Thompson B, Brabazon H, Del Gizzi R, Zhang T, Whipple C (2016) Evolutionary dynamics of floral homeotic transcription factor protein-protein interactions. Mol Biol Evol 33(6): 1486–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Schmutz J, Wang H, Percifield R, Hawkins J, Pontraoli AC, Estep M, Feng L, Vaughn JN, Grimwood J, et al. (2012) Reference genome sequence of the model plant Setaria. Nat Biotechnol 30(6): 555–561 [DOI] [PubMed] [Google Scholar]
- Blackman BK, Strasburg JL, Raduski AR, Michaels SD, Riesberg LH (2010) The role of recently derived FT paralogs in sunflower domestication. Curr Biol 20(7): 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Doebley JF (2005) Molecular evolution of FLORICAULA/LEAFY orthologs in the Andropogoneae (Poaceae). Mol Biol Evol 22(4): 1082–1094 [DOI] [PubMed] [Google Scholar]
- Bomblies K, Doebley JF (2006) Pleiotropic effects on the duplicate maize FLORICAULA/LEAFY genes zfl1 and zfl2 on traits under selection during maize domestication. Genetics 172(1): 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Wang RL, Ambrose BA, Schmidt RJ, Meeley RJ, Doebley J (2003) Duplicate FLORICAULA/LEAFY homologs zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Development 130(11): 2385–2395 [DOI] [PubMed] [Google Scholar]
- Bommert P, Je BI, Goldschmidt A, Jackson D (2013) The maize Gα gene COMPACT PLANT2 functions in CLAVATA signaling to control shoot meristem size. Nature 502(7472): 555–558 [DOI] [PubMed] [Google Scholar]
- Bortiri E, Chuck G, Vollbrecht E, Rocheford T, Martienssen R, Hake S (2006) Ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18(3): 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJ, Klein PE, Bortiri E, Acharay CB, Rooney WL, Kresovich S (2006) Inheritance of inflorescence architecture in sorghum. Theor Appl Genet 113(5): 931–942 [DOI] [PubMed] [Google Scholar]
- Brown PJ, Upadyayula N, Mahone GS, Tian F, Bradbury PJ, Myles S, Holland JB, Flint-Garcia S, McMullen MD, Buckler ES, et al. (2011) Distinct genetic architectures for male and female infloresence traits of maize. PLoS Genet 11(11): 1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB (2008) Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134(1): 25–36 [DOI] [PubMed] [Google Scholar]
- Chuck G, Meeley RB, Hake S (1998) The control of maize spikelet meristem fate by the APETALA2-like gene indeterminant spikelet1. Gene Dev 12(8): 1145–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Muszynski M, Kellogg E, Hake S, Schmidt RJ (2002) The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 298(5596): 1238–1241 [DOI] [PubMed] [Google Scholar]
- Chuck G, Whipple C, Jackson D, Hake S (2010) The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development 137(8): 1243–1250 [DOI] [PubMed] [Google Scholar]
- Clark RM, Wagler TN, Quijada P, Doebley J (2006) A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat Genet 38(5): 594–597 [DOI] [PubMed] [Google Scholar]
- Clifford HT (1987) Spikelet and floral morphology. In Soderstrom TR, Hilu KW, Campbell CS and Barkworth ME, eds, Grass systematics and evolution. Smithsonian Institution Press, Washington, DC, pp. 21–30 [Google Scholar]
- Collins G (1917) Hybrids of Zea tunicate and Zea ramosa. Proc Natl Acad Sci USA 3(5): 345–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell S, Korniliev P, Falcão A, Ismail A, Gregorio G, Mezey J, McCouch S (2016) Genome-wide association and high-resolution phenotyping link Oryza sativa panicle traits to numerous trait-specific QTL clusters. Nat Comm 7(1): 10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathan N, Zaccaro L, Esposito S, Isernia C, Omichinski JG, Riccio A, Pedone C, Di Blasio B, Fattorusso R, Pedone PV (2002) The Arabidopsis SUPERMAN protein is able to specifically bind DNA through is single Cys2-His2 zinc finger motif. Nucleic Acids Res 30(22): 4945–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RM, Gowda M, Moghe G, Lin H, Vaillancourt B, Shiu SH, Jiang N, Buell CR (2012) Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution. Plant J 71(3): 492–502 [DOI] [PubMed] [Google Scholar]
- Dewey CN (2011) Positional orthology: putting genomic evolutionary relationships into context. Brief Bioinform 12(5): 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doust AN, Devos KM, Gadberry MD, Gale MD, Kellogg EA (2005) The genetic basis for inflorescence variation between foxtail and green millet (Poaceae). Genetics 169(3): 1659–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doust AN, Kellogg EA (2002) Inflorescence diversification in the panicoid “bristle grass” clade (Paniceae, Poaceae): evidence from molecular phylogenies and developmental morphology. Am J Bot 89(8): 1203–1222 [DOI] [PubMed] [Google Scholar]
- Eveland AL, Goldschmidt A, Pautler M, Morohashi K, Liseron-Monfils C, Lewis MW, Kumari S, Hiraga S, Yang F, Unger-Wallace E, et al. (2014) Regulatory modules controlling maize inflorescence architecture. Genome Res 24(3): 431–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame BR, Zhang H, Cocciolone SM, Sidorenko LV, Dietrich CR, Pegg SE, Zhen S, Schnable PS, Wang K (2000) Production of transgenic maize from bombarded type II callus: effect of gold particle size and callus morphology on transformation efficiency. In Vitro Cell Dev Biol-Plant 36(1): 21–29 [Google Scholar]
- Gallavotti A, Long JA, Stanfield S, Yang X, Jackson D, Vollbrecht E, Schmidt RJ (2010) The control of axillary meristem fate in the maize ramosa pathway. Development 137(17): 2849–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Yang Y, Schmidt RJ, Jackson D (2008) The relationship between auxin transport and maize branching. Plant Physiol 147(4): 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli M, Feng F, Gallavotti A (2020) Mapping regulatory determinants in plants. Front Genet 11: 591194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli M, Khakhar A, Lu Z, Chen Z, Sen S, Joshi T, Nemhauser JL, Schmitz RJ, Gallavotti A (2018) The DNA binding landscape of the maize AUXIN RESPONSE FACTOR family. Nat Commun 9(1): 4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics 27(7): 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan JR, de Wet JMJ (1972) A simplified classification of cultivated sorghum. Crop Sci 12(2): 172–176 [Google Scholar]
- Hiratsu K, Mitsuda N, Matsui K, Ohme-Takagi M (2004) Identificaiton of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem Biophys Res Commun 321(1): 172–178 [DOI] [PubMed] [Google Scholar]
- Hodge JG, Doust AN (2017) Morphological development of Setaria viridis from germination to flowering. Plant Genet Genomics 19: 161–175 [Google Scholar]
- Huang P, Feldman M (2017) Genetic diversity and geographic distribution of North American Setaria viridis populations, in genetics and genomics of Setaria. Springer, Cham, Switzerland [Google Scholar]
- Jackson D, Veit B, Hake S (1994) Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120(2): 405–413 [Google Scholar]
- Kellogg EA (2015) Flowering plants, monocots: poaceae, vol. 13. Families and genera of vascular plants. Springer, Cham, Switzerland [Google Scholar]
- Kellogg EA (2022) Genetic control of branching patterns in grass inflorescences. Plant Cell 34(7): 2518–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EA, Camara PEAS, Rudall PJ, Ladd P, Malcomber ST, Whipple CJ, Doust AN (2013) Early inflorescence development in the grasses (Poaceae). Front Plant Sci 4: 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Chèneby J, Kulkarni SR, Tan G, et al. (2018) JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res 46(D1): D260–D266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komari T, Takakura Y, Ueki J, Kato N, Ishida Y, Hiei Y (2006) Binary vectors and super-binary vectors. InWang K, eds, Agrobacterium protocols. Methods in molecular biology, vol 343, Ed 2, Vol 1. Humana Press, Totowa, New Jersey, USA, pp 15–41 [DOI] [PubMed] [Google Scholar]
- Kusters E, Della Pina S, Castel R, Souer E, Koes R (2015) Chances in cis-regulatory elements of key floral regulators are associated with divergence of inflorescence architectures. Development 142(16): 2822–2831 [DOI] [PubMed] [Google Scholar]
- Leiboff S, Hake S (2019) Reconstructing the transcriptional ontogeny of maize and sorghum supports an inverse hourglass model of inflorescence development. Curr Biol 29(20): 3410–3419.e3 [DOI] [PubMed] [Google Scholar]
- Lemmon ZH, Park SJ, Jiang K, Van Eck J, Schatz MC, Lippman ZB (2016) The evolution of inflorescence diversity in the nightshades and heterochrony during meristem maturation. Genome Res 26(12): 1676–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Shao MR, Zeng D, Ju T, Kellogg EA, Topp CN (2020) Comprehensive 3D phenotyping reveals continuous morphological variation across genetically diverse sorghum inflorescences. New Phytol 226(6): 1873–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Galli M, Camehl I, Gallavotti A (2019) RAMOSA1 ENHANCER LOCUS2-mediated transcriptional repression regulates vegetative and reproductive architecture. Plant Physiol 179(1): 348–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Marand AP, Ricci WA, Ethridge CL, Zhang X, Schmitz RJ (2019) The prevalence, evolution and chromatin signatures of plant regulatory elements. Nat Plants 5(12): 1250–1259 [DOI] [PubMed] [Google Scholar]
- Maizel A, Busch MA, Tanahashi T, Perkovic J, Kato M, Hasebe M, Weigel D (2005) The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science 308(5719): 260–263 [DOI] [PubMed] [Google Scholar]
- Mitros T, Session AM, James BT, Wu GA, Belaffif MB, Clark LV, Shu S, Dong H, Barling A, Holmes JR, et al. (2020) Genome biology of the paleotetraploid perennial biomass crop Miscanthus. Nat Commun 11(1): 5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov LA, Tsiantis M (2015) Interspecies gene transfer as a method for understanding the genetic basis for evolutionary change: progress, pitfalls and prospsects. Front Plant Sci 6: 1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Limón S, Li M, Cintora-Martinez C, Aguilar-Rangel MR, Salazar-Vidal MN, González-Segovia E, Blöcher-Juárez K, Guerrero-Zavala A, Barrales-Gamez B, Caracaño-Macias J, et al. (2022) A B73xPalomero Toluqueño mapping population reveals local adaptation in Mexican highland maize. G3 12(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E (2007) Evolution and development of inflorescence architectures. Science 316(5830): 1452–1456 [DOI] [PubMed] [Google Scholar]
- Ricci WA, Lu Z, Ji L, Marand AP, Ethridge CL, Murphy NG, Noshay JM, Galli M, Mejía-Guerra MK, Colomé-Tatché M, et al. (2019) Widespread long-range cis-regulatory elements in the maize genome. Nat Plants 5(12): 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D (2006) A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441(7090): 227–230 [DOI] [PubMed] [Google Scholar]
- Sayou C, Monniaux M, Nanao MH, Moyroud E, Brockington SF, Thévenon E, Chahtane H, Warthmann N, Melkonian M, Zhang Y, et al. (2014) A promiscuous intermediate underlies the evolution of LEAFY DNA binding specificity. Science 343(6171): 645–648 [DOI] [PubMed] [Google Scholar]
- Schnable JC (2015) Genome evolution in maize: from genomes back to genes. Annu Rev Plant Biol 66(1): 329–343 [DOI] [PubMed] [Google Scholar]
- Schnable J (2019). Grass Syntenic Gene List sorghum v3 maize v3/4 with teff and oropetium v2. Figshare. https://figshare.com/articles/Grass_-Syntenic_Gene_List_sor-ghum_v3_maize_v3_4_with_teff_and_oropetium_v2/7926674/1
- Schnable JC, Freeling M (2011) Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLoS ONE 6(3): e17855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigmon BA (2010). ramosa1 in the development and evolution of inflorescence architecture in grasses. PhD dissertation. Iowa State University, Ames, IA [Google Scholar]
- Sigmon B, Vollbrecht E (2010) Evidence of selection at the ramosa1 locus during maize domestication. Mol Ecol 19(7): 1296–1311 [DOI] [PubMed] [Google Scholar]
- Strable J, Vollbrecht E (2019) Maize YABBY genes drooping leaf1 and drooping leaf2 regulate floret development and floral meristem determinacy. Development 146(6): dev171181. [DOI] [PubMed] [Google Scholar]
- Strable J, Wallace JG, Unger-Wallace E, Briggs S, Bradbury P, Buckler ES, Vollbrecht E (2017) Maize YABBY genes drooping leaf1 and drooping leaf2 regulating plant architecture. Plant Cell 29(7): 1622–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A, Zhao Q, Ross-Ibarra J, Doebley J (2011) Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet 43(11): 1160–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigoňová Z, Lai J, Ma J, Ramakrishna W, Llaca V, Bennetzen JL, Messing J (2004) Close split of sorghum and maize genome progenitors. Genome Res 14(10a): 1916–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16(2): 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadyayula N, da Silva HS, Bohn MO, Rocheford TR (2006a) Genetic and QTL analysis of maize tassel and ear inflorescence architecture. Theor App Genet 112(4): 592–606 [DOI] [PubMed] [Google Scholar]
- Upadyayula N, Wassom J, Bohn MO, Rocheford TR (2006b) Quantitative trait loci analysis of phenotypic traits and principle components of maize tassel inflorescence architecture. Theor App Genet 113(8): 1395–1407 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Schmidt RJ (2009) Development of the inflorescences, eds, Handbook of Maize: Its Biology. Springer,New York, NY, USA, pp 13–40 [Google Scholar]
- Vollbrecht E, Springer PS, Goh L, Buckler ES, Martienssen R (2005) Architecture of floral branch systems in maize and related grasses. Nature 436(7054): 1119–1126 [DOI] [PubMed] [Google Scholar]
- Wang H, Nussbaum-Wagler T, Li B, Zhao Q, Vigouroux Y, Faller M, Bomblies K, Lukens L, Doebley JF (2005) The origin of the naked grains of maize. Nature 436(7051): 714–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Regulsky M, Tseng E, Olson A, Goodwin S, McCombie WR, Ware D (2018) A comparative transcriptional landscape of maize and sorghum obtained by single-molecule sequencing. Genome Res 28(6): 921–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks BL (2013) Inflorescence branching in maize: a quantitative genetics approach to identifying key players in the inflorescence development pathway. PhD dissertation. Iowa State University, Ames, IA [Google Scholar]
- Whipple CJ (2017) Grass inflorescence architecture and evolution: the origin of novel signaling centers. New Phytol 216(2): 367–372 [DOI] [PubMed] [Google Scholar]
- Whipple CJ, Hall DH, DeBlasio S, Taguchi-Shiobara F, Schmidt RJ, Jackson DP (2010) A conserved mechanism of bract suppression in the grass family. Plant Cell 22(3): 565–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- William DA, Su Y, Smith MR, Lu M, Baldwin DA, Wagner D (2004) Genomic identification of direct target genes of LEAFY. Proc Natl Acad Sci USA 101(6): 1775–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills DM, Whipple CJ, Takuno S, Kursel LE, Shannon LM, Ross-Ibarra J, Doebley JF (2013) From many, one: genetic control of prolificacy during maize domestication. PLoS Genet 9(6): e1003604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter CM, Austin RS, Blanvillain-Baufumé S, Reback MA, Monniaux M, Wu MF, Sang Y, Yamaguchi A, Yamaguchi N, Parker JE, et al. (2011) LEAFY Target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev Cell 20(4): 430–443 [DOI] [PubMed] [Google Scholar]
- Wu X, Li Y, Shi Y, Song Y, Zhang D, Li C, Buckler ES, Li Y, Zhang Z, Wang T (2016) Joint linkage mapping and GWAS reveal extensive genetic loci that regulate male inflorescence size in maize. Plant Biotechnol J 14(7): 1551–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Wang X, Huang C, Xu D, Li D, Tian J, Chen Q, Wang C, Liang Y, Wu Y, et al. (2017) Complex genetic architecture underlies maize tassel domestication. New Phytol 214(2): 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki S, Miyabayashi T, Eiguchi M, Kitano H, Nonomura KI, Kurata N (2010) Diversity of panicle branching patterns of wild relatives of rice. Breeding Sci 60(5): 586–596 [Google Scholar]
- Zhang G, Liu X, Quan Z, Cheng S, Xu X, Pan S, Xie M, Zeng P, Yue Z, Wang W, et al. (2012) Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat Biotechnol 30(6): 549–554 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Srinivasan S, Mirnezami SV, Kusmec A, Fu Q, Attigala L, Salas Fernandes MG, Ganapathysubramanian B, Schnable PS (2019) Semiautomated feature extraction from RGB images for Sorghum panicle architecture GWAS. Plant Physiol 179(1): 24–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Yang J, Box MS, Kellogg EA, Eveland AL (2018) A dynamic co-expression map of early inflorescence development in Setaria viridis provides a resource for gene discovery and comparative genomics. Front Plant Sci 9: 1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.