Abstract
Numerous genes for monogenic kidney diseases with classical patterns of inheritance as well as for complex kidney diseases that manifest in combination with environmental factors have been discovered. Genetic findings are increasingly used to inform clinical management of nephropathies, and have led to improved diagnostics, disease surveillance, choice of therapy, and family counseling. All of these rely on accurate interpretation of genetic data, which can be outpaced by current rates of data collection. In March of 2021, KDIGO (Kidney Diseases: Improving Global Outcomes) held a Controversies Conference on Genetics in Chronic Kidney Disease (CKD) to review the current state of understanding of monogenic and complex (polygenic) kidney diseases, processes for applying genetic findings in clinical medicine, and using genomics for defining and stratifying CKD. Given the important contribution of genetic variants to CKD, practitioners with CKD patients are advised to “think genetic,” which specifically involves obtaining a family history, detailed information on age of CKD onset, clinical examination for extra-renal symptoms, and considering genetic testing. To improve implementation of genetics in nephrology, meeting participants advise developing an advanced training or subspecialty track for nephrologists, guidelines for testing and treatment, and education of patients, students, and practitioners. Key areas of future research, including clinical interpretation of genome variation, electronic phenotyping, global representation, kidney-specific molecular data, polygenic scores, translational epidemiology, and open data resources, were also identified.
Keywords: genetic kidney disease, monogenic, polygenic, genome-wide association studies, single nucleotide polymorphism
INTRODUCTION
Chronic kidney disease (CKD) affects approximately 10% of the global adult population.1 Multiple genetic and environmental risk factors contribute to kidney diseases, making it difficult to identify the underlying pathophysiologic mechanisms. However, the advent of high-throughput genotyping and massively parallel sequencing combined with the availability of large datasets of genomic and health information have led to rapid advances in our understanding of the genetic basis of kidney function and disease.
To date, more than 600 genes have been implicated in monogenic kidney diseases,2 and known single-gene disorders account for up to 50% of non-diabetic CKD in pediatric cohorts and 30% in adult cohorts.3-10 In addition, genetic variation plays an important role for kidney function in the normal range,11-16 and common genetic variants account for approximately 20% of the estimated genetic heritability of estimated glomerular filtration rate (eGFR).13 Common genetic variants have also been shown to contribute to disorders such as IgA nephropathy (IgAN),17, 18 membranous nephropathy,19, 20 or nephrotic syndrome.21-23 Hence the pathogenesis model for many kidney diseases has expanded to include multiple genetic and environmental factors that together contribute to the pathology, commonly referred to as “complex disease.”
Genetic findings are increasingly used to inform clinical management of many nephropathies, enabling more precise diagnostics, targeted disease surveillance, and better-informed choices of therapy and family counseling.24 Clinical management relies on accurate interpretation of genomic data, a labor intensive process that can be outpaced by speed of discovery.25 To realize the promises of genomic medicine for kidney disease, many technical, logistical, ethical, and scientific questions must be addressed 24 In March of 2021, KDIGO (Kidney Diseases: Improving Global Outcomes) held a Controversies Conferences on Genetic in CKD to review the current state of understanding of monogenic and complex kidney diseases, processes for applying genetic findings in clinical medicine, and use of genomics for defining and stratifying CKD. Participants identified areas of consensus, gaps in knowledge, and priorities for research (Table 1).
Table 1.
Summary Points From the Genetics in CKD Controversies Conference
| Consensus |
|
| Ongoing controversies |
Definitions/terminology
|
| Processes for improving data capture and analysis |
|
| Priorities for Implementation |
|
CKD, chronic kidney disease; VUS, variants of uncertain significance
DEFINITIONS AND EPIDEMIOLOGY OF GENETIC KIDNEY DISEASES
Familial aggregation and substantial heritability of CKD is well described across the world. Recent large-scale analyses of electronic medical records estimated observational heritability of CKD to be in the range of 25-44%, with higher estimates for patients of African ancestry.26 These estimates are generally consistent with traditional family-based heritability studies of CKD and glomerular filtration rate.27-29 Relatively high heritability of CKD is likely attributable to both monogenic causes as well as complex or polygenic factors.
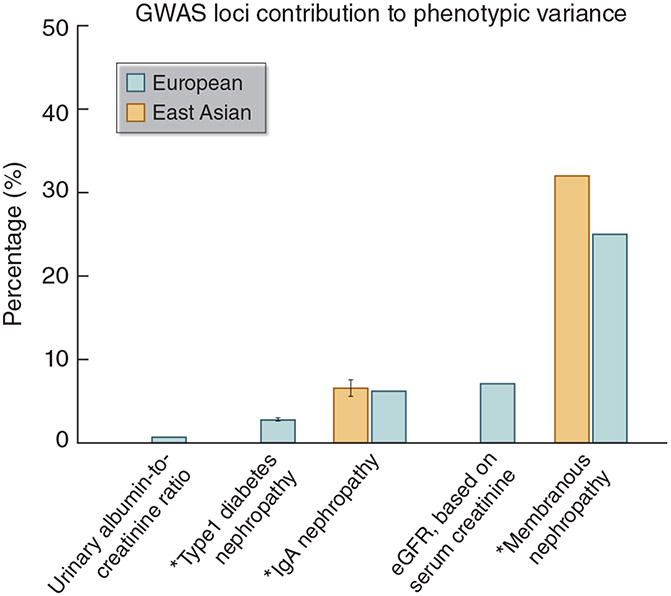
Monogenic (also termed “Mendelian”) CKD generally refers to diseases caused by rare, pathogenic variants in a single gene (Table 2); there is a strong genotype-to-phenotype relationship, and environmental factors have limited influence. Oligogenic disorders are determined by rare variants in a few genes. Complex or polygenic diseases lack simple patterns of inheritance (e.g., dominant, recessive, or sex-linked) and instead are influenced by the aggregate effect of many common genetic variants in multiple genomic regions as well as environmental factors.30 Such aggregate effects of common variants (or single nucleotide polymorphisms, SNPs) can be quantified by SNP-based heritability, which has been estimated for various types of kidney disorders to range from 14% for renal cancer among individuals of European ancestry to 43% for membranous nephropathy among individuals of East Asian ancestry. The proportions of variance explained by known loci of these diseases are smaller, ranging from <1% for urinary albumin-to-creatinine ratio to 32% for membranous nephropathy among individuals of East Asian ancestry (Figure 1).13, 14, 17, 19, 31 However, common genetic factors may also influence the age of onset, severity, rate of progression, and associated extra-renal complications of monogenic diseases, which often have variable expression.32, 33 In addition to CKD attributed to specific etiologies, genetic studies also use phenotypic readouts such as measures of kidney function or damage (e.g., eGFR, albuminuria), kidney histology classification, or molecular injury markers to define CKD (Table 3).34, 35
Table 2.
Characteristics of Monogenic Versus Complex Genetic Diseases
| Monogenic (Mendelian) | Polygenic (Complex) | |
|---|---|---|
| Allele/variant frequency | Rare | Can be common |
| Effect size of major driving gene | Large | Small |
| Penetrance | High | Low |
| Role of environment | Limited | Strong |
| Inheritance model | Mendelian | None apparent |
Figure 1. Common Variant Contributions to Kidney Diseases and Traits.13, 14, 17, 19, 31.
*For binary outcomes, the proportions of phenotypic variance explained by loci from genome-wide association studies (GWAS) were estimated from Nagelkerke’s or McKelvey & Zavoina pseudo R2.
SNP, single nucleotide polymorphism.
Table 3.
Disease Definitions for Genetic Studies Based on Kidney Function, Kidney Histology, or Molecular Markers
| Advantages | Disadvantages | |
|---|---|---|
| Kidney function markers (e.g., eGFR, albuminuria) | ||
|
|
|
| Kidney histology | ||
|
|
|
| Non-traditional molecular markers (e.g., markers quantified with high-throughput omics technologies) | ||
|
|
|
FSGS, focal segmental glomerulosclerosis.
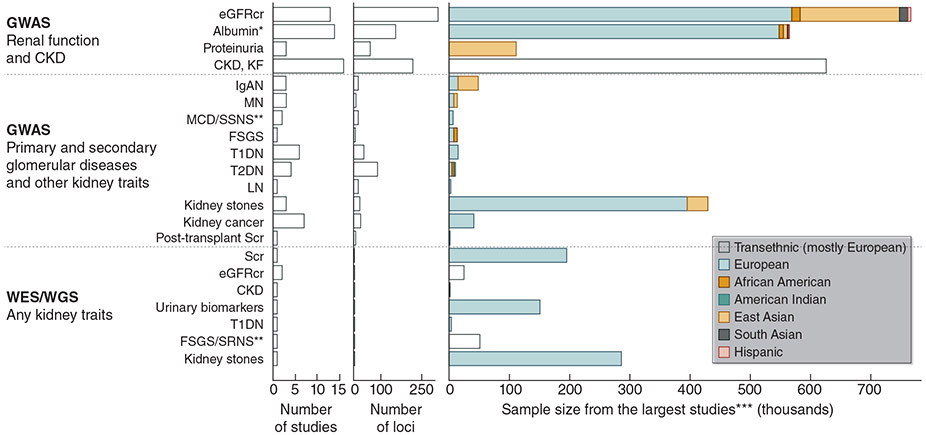
Monogenic variants account for approximately 30-50% of cases of CKD stages G3b-G5 in children3-5, 36, 37 and 10-30% in adults.3-10 Diagnostic yields differ between 12-65% among studies, with selection bias likely contributing to the variability. However, prevalence estimations for genetic diseases are likely to change over time as genetics-first approaches to diagnosis become more common (where sequence data is obtained first, followed by characterization of associated phenotypes).38 Many common variants associated with specific kidney function measures or complex kidney diseases have been identified through genome-wide association studies (GWAS) and exome or genome sequencing studies of large population samples—usually of European or East Asian ancestry (Figure 2).13, 14, 17, 19, 31, 39-41 The largest number of loci, genomic regions containing associated SNPs, were discovered for the continuous kidney function measure eGFR with studies based on data from >1 million individuals reporting more than 250 such loci.12-14, 17, 19, 22, 23, 31, 40, 42-66
Figure 2. GWAS, Exome, or Genome Sequencing Studies. 13, 14, 17, 19, 31, 39-41.
*The largest study focused on urinary albumin-to-creatinine ratio. Several included serum albumin studies.
**Pediatric population.
***For case-control studies, the total sample sizes were plotted.
CKD, chronic kidney disease; eGFR, estimated glomerular filtration rate; FSGS, focal segmental glomerulosclerosis; GWAS, genome-wide association studies; IgAN, IgA nephropathy; KF, kidney failure; LN, lupus nephritis; MCD, minimal change disease; MN, membranous nephropathy; Scr, serum creatinine; SRNS and SSNS, steroid-resistant and steroid-sensitive nephrotic syndrome; T1DM and T2DM, types 1 and 2 diabetes mellitus; WES, whole exome sequencing; WGS, whole genome sequencing.
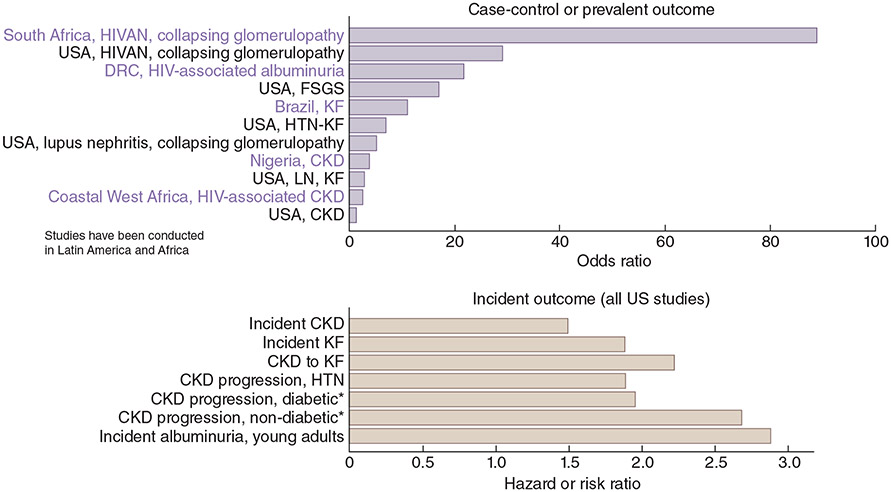
Although distinguishing monogenic versus polygenic diseases provides a useful practical framework, genetic risk variants for kidney diseases occur on a spectrum from rare variants with large effects to common variants with small effects, and many diseases do not fit neatly into either category. For example, APOL1 (apolipoprotein L1)-associated kidney risk variants are common among some populations of African ancestry and impart a relatively high risk under a recessive mode of inheritance, but these variants are not considered monogenic. The magnitude of the risk associated with APOL1 variants varies significantly for different forms of nephropathy. For example, black South Africans with untreated HIV and two APOL1 risk alleles have been reported to have a more than 80-fold increased risk of developing HIV-associated nephropathy, but the magnitude of the risk conferred by the same risk alleles ranged between 1.2 and 2 for CKD or non-diabetic kidney failure (Figure 3).67-83 Similarly, the combination of two common variants in the HLA-DR and PLA2R1 loci imparts a high risk of the complex disease membranous nephropathy, defying the common variant/small effect paradigm.84, 85
Figure 3. Associations of APOL1 High Risk Genotype.67-83.
APOL1 high-risk genotype: G1G1, G2G2, or G1G2
Studies were ordered by PMID, a proxy for publication date.
*Compared with white patients
APOL1, apolipoprotein L1; CKD, chronic kidney disease; DRC, Democratic Republic of Congo; FSGS, focal segmental glomerulosclerosis; HIVAN, HIV-associated nephropathy; HTN, hypertension; KF, kidney failure; LN, lupus nephritis; PMID, PubMed identifier.
CONSIDERATIONS FOR GENETIC TESTING
A positive family history, early age of onset, and presence of extra-renal symptoms are associated with a higher probability of monogenic disease. In addition, the clinical diagnosis is highly predictive of diagnostic yield and will also guide the choice of genetic tests, motivating a thorough clinical workup prior to genetic testing. For example, glomerular and tubulointerstitial disorders are associated with a higher diagnostic yield than diabetic kidney disease. In general, because of the genetic heterogeneity of most forms of nephropathy, genetic testing with phenotype-driven gene panels, or exome or genome sequencing is more efficient than sequential single-gene analyses.
Genetic testing is usually performed subsequent to a clinical work up, but there may be some situations when early genetic testing can be advantageous. For example, prospective kidney donors related to a recipient with a known genetic condition should be tested early during the donor evaluation process. Other situations where early genetic testing may be considered are listed in Table 4. In healthy children or adults, there are currently no data supporting predictive or presymptomatic genetic testing even if there is a family history. Nevertheless, once a pathogenic variant is identified in a proband, cascade testing of family members and genetic counseling in mutation carriers is the standard practice in clinical genetics.
Table 4.
Potential Indications for Genetic Testing for Monogenic Forms of CKD
|
ADPKD, autosomal dominant polycystic kidney disease; aHUS, atypical hemolytic uremic syndrome; CKD, chronic kidney disease; TMA, thrombotic microangiopathy
Most countries do not have guidelines regarding which nephrology patients should be referred to genetic testing and counseling. Nephrology communities would therefore benefit from developing guidelines based on best evidence and practices in clinical genetics. Overall, guidance should take into account the potential benefit of a genetic diagnosis for the specific patient and their family (e.g., treatment changes, family planning, ending a diagnostic odyssey) and balance the risk of false positive results that could engender unnecessary clinical workup for the patient and their families. A position paper by the ERA-EDTA Working Group for Inherited Kidney Diseases (WGIKD) and the Molecular Diagnostics Taskforce of the European Rare Kidney Disease Reference Network (ERKNet) has been recently issued to delineate indications for genetic testing in chronic kidney diseases.86
Defining Actionable Genes in Kidney Diseases
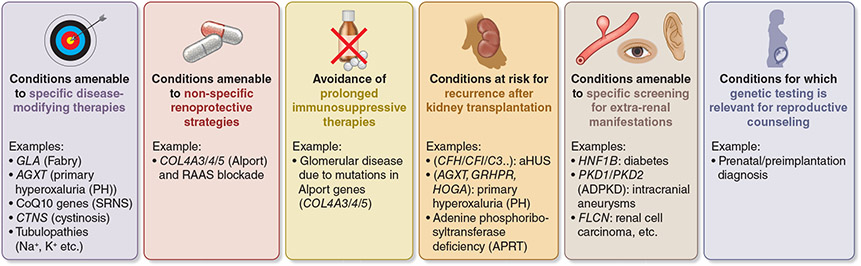
Actionable genes in kidney diseases refer to genes that, when significantly altered, confer a high risk of serious disease that could be prevented or mitigated if the risk were known.87 A set of 73 actionable genes have been proposed by the American College of Medical Genetics and Genomics (ACMG), many of which are associated with phenotypes relevant to nephrology (PALB2, GLA, HNF1A, MEN1, MAX, RET, SDHAF2, SDHB, SDHC, SDHD, VHL, TMEM127, TSC1, TSC2, WT1). While these genes were selected based on the possibility of preventing overall morbidity and/or mortality, one can conceive additional, kidney-specific actionable genes, nominated based on availability of interventions that could prevent renal morbidity (Figure 4). Examples include early initiation of general renoprotective therapies (e.g. reninangiotensin blockade for carriers of pathogenic variants in type IV collagen genes); initiation of targeted therapies (e.g. enzyme therapy for Fabry disease or CoQ10 supplementation for nephrotic syndrome due to CoQ10 deficiency); avoidance of treatment that would be futile and perhaps even deleterious (e.g., prolonged immunosuppressive therapies for genetic podocytopathies); or surveillance for recurrence of disease after kidney transplantation (e.g. atypical hemolytic uremic syndrome/thrombotic microangiopathy [aHUS/TMA], primary hyperoxaluria). ClinGen, an international initiative to define robust disease-gene associations and curate pathogenic variants,87 now has a kidney expert work group that is developing a stable list of nephropathy-associated genes and variants. It is expected that this group would also provide guidance for actionability for kidney genes and nominate them for the ACMG list. Awareness of the ClinGen Initiative should be promoted in the kidney community, along with messaging regarding the importance of variant submission to public databases such as ClinVar and the value of creating interdisciplinary expert boards to discuss controversial variants of uncertain significance (VUS) and discussing the most complex cases. Additional efforts to harmonize gene and gene panel curation such as the Genomics England panel app (https://panelapp.genomicsengland.co.uk) are listed in Supplementary Table S1.
Figure 4. Actionable Genes in Kidney Diseases.
Actionability refers to the potential for genetic test results to lead to specific clinical actions for prevention or treatment of a condition, supported by recommendations based on evidence. aHUS, atypical hemolytic uremic syndrome; SRNS, steroid-resistant nephrotic syndrome; RAAS, renin-angiotensin-aldosterone system.
In addition to rare pathogenic variants, common genetic variants or polygenic scores may become appropriate for clinical reporting if they are shown to alter patient management, indicate need for surveillance for progression or associated comorbidities, or inform familial screening.88 In complex diseases, the current best candidates for reporting include APOL1 risk alleles,89, 90 genetic risk score for membranous nephropathy based on PLA2R1, NFKB1, IRF4, and HLA risk alleles,19 extremes of a polygenic risk score for eGFR,91 and pharmacogenetic variants that are informative about risk of adverse events, pharmacokinetics, and pharmacodynamics for specific drugs, some of which may be especially relevant to CKD patients (for example, azathioprine, tacrolimus, warfarin, clopidogrel, simvastatin, voriconazole, allopurinol). However, we currently lack evidence for actionability for polygenic scores, i.e. evidence that reporting can improve clinical outcomes.
APOL1 presents a special case in clinical nephrology because biallelic inheritance of two common variants in this gene, present at high frequency in some populations of African ancestry, increases risk for several kidney disorders.89, 90 Potential benefits for APOL1 screening include improved risk stratification and opportunities for education. However, only a minority of patients with APOL1 risk genotypes develop nephropathy, and currently no data support early intervention in asymptomatic individuals to reduce future risk of disease. Potential drawbacks to screening include potential for anxiety, stigma, or apathy and the lack of evidence-based interventions.92, 93 Combined, these drawbacks could lead to misunderstanding among patients, mistrust of the medical system, and perceived or real racial bias given that APOL1 risk variants are predominantly found in those with African ancestry. On the other hand, the failure to offer a test that could be most informative in a specific ancestry group could also be perceived as bias. For transplant patients, APOL1 screening could prevent harm to living donors and meet recipient right to know, but screening could also reduce rates of living donation, waste deceased donor kidneys, and exacerbate shortage of organs. The APOLLO study, which is in progress and expected to end in 2023, is prospectively evaluating the impact of APOL1 risk alleles on donor and recipient outcomes.94 Moreover, the initiation of genotype driven clinical trials may change the approach to diagnostic testing for APOL1 and other genetic disorders. These considerations emphasize the importance of further research into the usefulness of APOL1 testing.
REPORTING AND TERMINOLOGY STANDARDS
Differences in how diagnostic laboratories evaluate and report variants is a significant challenge in molecular diagnosis, and there is agreement that standardization of evaluation and reporting among different laboratories and countries is a key priority. The determination for pathogenicity is a semi-quantitative process that takes into account variant allele frequency, predicted impact on protein function, and prior reports of occurrence with disease. The ACMG and the Association for Molecular Pathology (AMP) published standards and guidelines for the interpretation of sequence variants.95 These guidelines are periodically reviewed and refined by the ClinGen Initiative to reduce discrepancies in variant interpretation between laboratories and clinicians.
The ACMG criteria classify variants into 1 of 5 tiers, with tiers 4 and 5 (i.e. likely pathogenic and pathogenic) classified as diagnostic variants.95 All variant classes can later be upgraded or downgraded based on novel information or interpretation, perhaps necessitating periodic review of clinical genetic reports. However, the abundance of class 3 VUS has created a particular challenge and urgency for improving evaluation and reporting. The definition and relevance of VUS may be unclear to physicians or patients, causing incorrect assignment of diagnoses and/or psychological distress to patients and families. This situation necessitates proper communication with the patients to inform and educate them about the possibility of VUS, in which case familial segregation analysis might be recommended. Additionally, VUS should be reported only after interdisciplinary contact between the clinician and geneticist.96 Future reinterpretation of variants can be facilitated by diagnostic reports that provide detailed description of ACMG classification criteria that were applied at the time of reporting. Although there are currently no existing guidelines, incidental carrier status for autosomal recessive inheritance is not routinely reported in standard diagnostic reports. Guidelines for systematic reporting of these variants should be developed. Heterozygosity associated with a mild phenotype is increasingly recognized in human genetics, for example for COL4A3/COL4A4 variants.97
Unified Disease Terminology
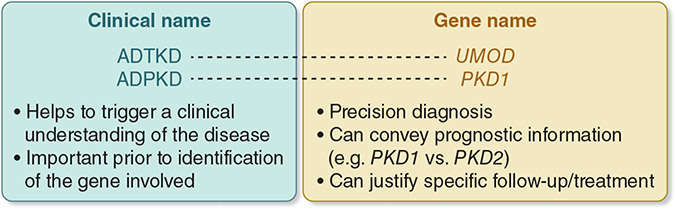
There was consensus that establishing a unified disease terminology that takes into account genetic disease nomenclature is an important goal for the community. In support of unified, precise disease terminology, a suggested approach is two-part (“dyadic”) naming comprising both the clinical condition and gene name (Figure 5), although there is some controversy around this approach.98, 99 An important example is adoption of two-part naming in autosomal dominant tubulointerstitial kidney disease (ADTKD), in which ADTKD is followed by reference to the underlying genetic defect, such as ADTKD-UMOD and ADTKD-MUC1.100 Two-part names provide flexibility, in that some users (patients/clinicians) can use the first part (ADTKD) while others (patients/clinicians/researchers) can use the whole name (ADTKD-UMOD). When clinical presentation is unspecific, or very heterogeneous, use of gene name followed by “kidney disease” (e.g., PAX2-kidney disease) is encouraged. Potential limits to this approach include the possibility of classifying a patient with a benign prognosis as having a potentially progressive disorder, as well as the challenge of adding a second or gene name to conditions already described in International Classification of Disease codes. To that end, participants of this KDIGO controversies conference did not reach consensus regarding renaming traditional disease terms, such as Alport Syndrome.
Figure 5. Unified Disease Terminology.
Two-part (“dyadic”) naming comprises both the clinical condition and gene name. An example would be autosomal dominant tubulointerstitial kidney disease (ADTKD), in which ADTKD is followed by reference to the underlying genetic defect, such as ADTKD-UMOD. ADTKD, autosomal dominant tubulointerstitial kidney disease; PKD, polycystic kidney disease.
GENOMIC DISCOVERY AND IMPLICATIONS FOR CHRONIC KIDNEY DISEASES
As demonstrated by the first GWAS for eGFR, common genetic variants that are associated with complex kidney traits usually have small effects and therefore require very large sample sizes for discovery.101 Accordingly, there has been limited success in identifying common kidney disease susceptibility variants in individual observational studies of adult 102-105 or pediatric 106-108 CKD. Conference participants therefore recognized the importance of collaborative consortia, such as CKDGen,11, 109 CHARGE,66, 110 iGeneTRAiN,111 or COGENT Kidney,112, 113 that aggregate and harmonize genetic and phenotypic data across multiple studies for combined genetic discovery. In addition to enlarging sample size and providing a platform for replication studies, expanding consortia to international sites can enable studies of more ancestrally and geographically diverse populations. For more specific but less frequent primary kidney disorders such as IgAN, membranous nephropathy, or steroid-sensitive nephrotic syndrome, aggregating multiple international case-control cohorts is even more important to assure adequate power. Additionally, more diverse ancestral composition of analyzed cohorts facilitates fine-mapping of GWAS loci, enables discovery of ancestry-specific effects, and assures broader generalizability of genetic findings.
The identification of causal genes and variants underlying GWAS associations and defining their pleiotropic effects are recognized as important challenges in the field. Examples such as UMOD the locus with the strongest common variant association with CKD,66 support the existence of a spectrum of risk variants from monogenic to complex. There are currently no examples for successful translation of insights from GWAS in CKD to new therapies, but the discovery of the MYH9 locus,114, 115 followed by the identification of APOL1 as the causal gene,69 refinement of nephrotoxic mechanisms of APOL1 risk variants,89 and an ongoing phase IIa study of a small molecule APOL1 inhibitors (ClinicalTrials.gov identifier NCT04340362) represent promising steps to that end.
Conference participants recognized the emerging importance of electronic health record (EHR)-based genetic research for linking genetic information with a wide range of laboratory parameters and medical conditions. EHR-linkage is possible in various settings, ranging from existing biobanks in research settings, hospitals, or healthcare systems to entire countries such as Iceland, Estonia, or Finland. Examples of EHR-linked biobanks, institutions, health care systems, or country-wide efforts are UK Biobank,116 MVP,117 HUNT,118 deCODE,119 FinnGen,60 Biobank Japan,120 BioVU,121 MGI,122, eMERGE,123 and All of Us.124 The development of standardized, scalable, and portable computable phenotypes is time consuming and represents many challenges,125 but it can empower future genetic studies by automated identification of kidney disease patients in large EHR databases.26, 126 Notably, it is just as important (and often harder) to accurately define those without a disease versus those with the disease to serve as healthy controls in genetic studies. We envision that computable phenotyping can be used to find patients with or without CKD, hypertension, kidney stones, or glomerular disease, as well as patients who have received a kidney biopsy or kidney transplant. In nephrology, computable phenotyping is underway,26, 127-129 with CKD phenotyping perhaps best positioned for widespread implementation given the availability of new algorithms based on ICD codes and laboratory values routinely measured in clinical practice.26, 126
In addition to genomic discovery, EHR-linked genetic research may allow for recontacting of patients with a specific genotype for detailed clinical and molecular studies. Linking EHR and genetic data can also be used to investigate pleiotropic associations of genetic variants originally discovered for a specific condition (e.g. APOL1 or UMOD) with additional traits captured in medical records using phenome-wide association approaches.14, 26, 42, 130 Such studies can be further complemented with Mendelian randomization methods to clarify associations between genetic variants, biomarkers, and phenotypes.131
Despite the large size of consortia and EHR-linked studies, certain groups of patients are still underrepresented in genetic research. For instance, the paucity of pediatric patients with genetic information has limited longitudinal phenotype analyses from childhood to adulthood and the ability to identify genetic drivers of kidney diseases or traits of childhood. There is also an urgent need to expand ancestral diversity of participants in genetic studies, specifically aiming to increase the representation of non-European populations.132 Additional challenges include harmonizing data for rare kidney conditions that necessitate aggregating cases from across several biobanks and EHRs; identifying ancestry-matched controls for case-control analyses; handling of missing data; and harmonizing genotypes in the presence of different types of available genetic data.133, 134
Partnerships between academic labs and industry allow efficient exchange of ideas and resources to promote investigation of disease mechanisms, biomarkers, and therapeutic targets. Such partnerships can enable academic labs, biobanks, and institutions and health care systems to conduct large-scale multi-omic studies that would not be feasible with only support from internal funds or extramural grants and facilitate follow-up studies to “functionalize” key genes or genetic variants. Successful partnerships must achieve a balance between a companies’ incentive to invest and the academic freedom in research and publishing. Key principles and processes, such as intellectual property, publications, and data sharing and access, also must be aligned. These partnerships have been particularly valuable for generating functional genomic data from primary kidney tissues and allow for rapid implementation of new methods.135-138 Generation of additional such data from primary kidney tissue and cell types should continue to be a research priority, because the kidney is underrepresented in many existing public databases, including ENCODE,139 Roadmap Epigenomics,140 and GTEx Projects.141 The Kidney Precision Medicine Project (KPMP)142 and similar new initiatives aim to address some of these important gaps by generating and harmonizing new multi-dimensional molecular data for human kidney tissue in health and disease.
Polygenic Scores
Polygenic scores (PGS) are based on the results of GWAS and aggregate the effects of trait- or disease-associated variants across the genome. PGS capture a greater proportion of genetic variance compared to individual SNPs and may potentially be useful to risk-stratify populations, enhance screening, and ultimately inform diagnosis, prognosis, and/or treatment. PGS have been shown to modify the penetrance of monogenic variants for hypercholesterolemia, hereditary breast and colon cancer, and obesity,32, 33 although this effect has not yet been examined for kidney diseases. PGS for kidney disease can be constructed using a smaller set of genome-wide significant SNPs only, such as a 147-SNP score for eGFR (odds ratio of ~2 for individuals in the highest 10% of the score)13 or a 5-SNP score for membranous nephropathy (odds ratio of >20 for those in the highest 10% of the score),19 or by using genome-wide scores with hundreds of thousands of variants, such as the UK Biobank score for CKD91 Currently, most scores are derived from European populations and do not include rare or population-specific variation, potentially creating a new health disparity between individuals from European descent and others.132 Since scores are constructed from GWAS for complex traits and diseases, they may reflect heterogeneous mechanisms and therefore not necessarily point to targeted interventions.
Conference participants agreed that before applying PGS in clinical nephrology, more research is needed to derive the most accurate and cosmopolitan scores for kidney disease. Also necessary are proof of clinical utility in surveillance, diagnosis, prognosis, or treatment of kidney disease; a better understanding of dependence on the clinical context, including disease stage, ancestry, sex or demographics;143 and cost-effectiveness and added value beyond standard clinical risk factors. PGS computation would need to be robust, open-source, and able to be incorporated into points of care. Quality standards for PGS have recently been defined by ClinGen,144-146 providing a framework for evaluating clinical translation and utility.
ACHIEVING IMPLEMENTATION IN CLINICAL MEDICINE
Clinical Knowledge
Often, insufficient experience and knowledge is a major barrier for implementing genetic evaluation in nephrology practice. To ensure equitable access to genetic testing, all nephrologists should have a sufficient knowledge base for discerning which patients would benefit from genetic testing and, at minimum, be able to collect personal and family histories. While it would be best for all nephrologists to also be able to recommend screening for at-risk family members if applicable; conceptually understand types of genetic tests, including their risks and benefits; and remain aware of local regulations around genetic testing, nephrologists lacking experience in these domains should collaborate with a clinical geneticist and/or a genetic counselor. In addition, reporting of positive genetic results to patients necessitates individual and family counseling and referrals. Hence, a multidisciplinary approach is key for successfully implementing genetics in the clinic.
Participants recognized workforce education as a critical need. Genetics is currently not part of the nephrology fellowship curriculum in the Unites States,147 and indeed, fellows report lacking competency in genetic renal disease.148 Similarly, in Australia, less than half of nephrologists feel confident in using results of genomic testing in clinical practice.149 There are no genetics core competency guidelines for nephrologists, nor guidelines for evaluating competencies for clinical genetic consent and return of results. Based on published data and information,150 a compiled list of core competencies expected from nephrologists at different levels of expertise can be found in Supplementary Table S2. These gaps can be remedied by including more robust genetics curricula in medical school, residency, and fellowship training. Education for current practicing nephrologists can be achieved via workshops at national and international societies, continuing medical education, review papers in nephrology journals, and introduction of clinical genetic questions to re-licensing tests.151 One can also envision an advanced training or subspecialty track in genetic nephrology, similar to transplant, oncology, or glomerular diseases sub-specialization. Supplemental Table S1 provides an overview of clinical genetics web resources to aid nephrologists.
Clinical Practice
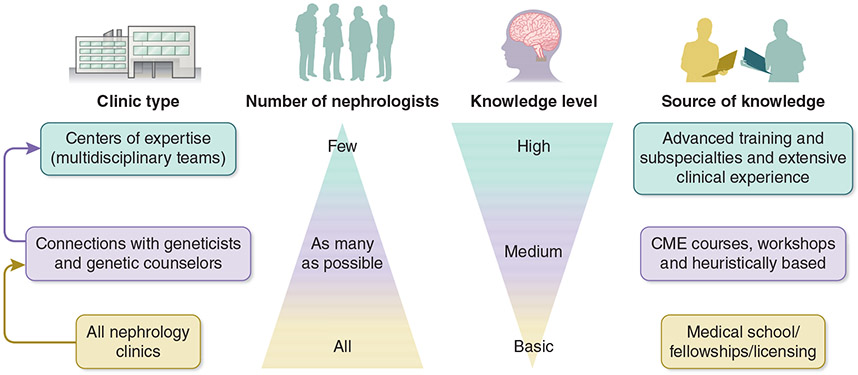
Centers of expertise are sites where patients can receive comprehensive, coordinated care from a multidisciplinary team that includes a relatively small number of nephrologists with a high skill set for genetic diagnosis (Figure 6). These centers also play an important role in training and research. Centers of expertise, or reference, are concentrated in Europe, with ERKNet constituting a consortium of more than 30 centers in 12 countries, supported by the European Union. In most regions of the globe, including the United States, there are no centralized accreditation mechanisms for developing centers of expertise or reference. The establishment of such centers can facilitate standardized variant interpretation, identify “actionable” genes associated with kidney diseases, train the future generation of physicians with dual expertise in genetics and nephrology, develop guidelines for referral and testing of patients with kidney diseases, disseminate implementation knowledge, and develop collaborative research projects and clinical trials for rare disorders.
Figure 6. Proposed Organization for Implementing Genetics in Nephrology.
Within a health system, multiple center types, provider specialties, and education strategies are needed for best implementing genetics in nephrology. A three-tiered organization model includes 1) a basic, common level of knowledge in genetics among all nephrologists, 2) clinical connections between nephrologists and geneticists and genetic counselors, and 3) centers of expertise where nephrologists with genetic expertise collaborate with geneticists and genetic counselors. CME, continuing medical education.
Cost and Access
Often, genetic testing is not affordable for either patients or healthcare systems. In regions where there is cost coverage or reimbursement, access can still be unequal since genetic testing is based on clinical presentation, and obtaining coverage is often easier in children than in adults. Many countries do have genetic protection acts, laws, or regulations to ensure equitable access to genetic testing without fear of discrimination. However, legislation alone is not always sufficient for allaying patient concerns about the potential for prejudice.
Logistically, remote sample collection and telemedicine have potential for increasing access to genetic counseling. However, adequate physical evaluation and identification of extra-renal manifestations can be more complicated or impossible with telemedicine. In addition, although the SARS-CoV-2 pandemic has accelerated the deployment of telemedicine across many health systems, not all patients and physicians are comfortable with remote, video-based communications.
For most genetic conditions, we lack large-scale cost-effectiveness analyses to demonstrate the benefits of genetic testing. Recent data suggest that genetic testing has a high diagnostic yield in patients with CKD of unknown etiology and may reduce costly diagnostic workups, hopefully increasing the coverage of genetic testing for those patients.3, 8 It is also important to demonstrate the clinical value of genetic testing beyond diagnosis, such as impact on long-term outcomes and health economics. A comparison of the cost-effectiveness of genetic testing in nephrology across different healthcare coverage systems could provide key insights and an evidence base for expanding testing.
Patient Voice
Patient engagement is vital for successful treatment and advances in research. To advocate for their own genetic testing, patients need to have an awareness of and education regarding genetics and kidney disease and the relative benefits and risks of genetic testing.152 The complex ethical, psychosocial and familial implications for genetic testing, including presymptomatic testing, can make decision-making challenging and require an understanding of patient values, goals, and priorities.153 To engage and activate patients and patient communities, educational content needs to be accessible and sensitive to patients in terms of culture, language, and literacy as well as be shared across multiple platforms.154
The topics of race and ancestry have been widely debated in genetics as well as nephrology.155-158 In specific terms, race is a social, categorical construct, whereas ancestry is based on inherited genetic variants without categorization. In principle, genetics research is agnostic to race,157 and identifying disease causing variants could obviate reliance on race or ancestry as a proxy for probability of carrying a risk allele.132, 158
Within nephrology, patient reported outcome measures (PROMs) can provide doctors, investigators, and policymakers with important insights into patient symptoms and experiences that cannot be identified through laboratory or imaging studies alone.159 Research communities that engage with patients and include the patient voice can better advocate for more research and development in rare kidney diseases.
Research in Implementation
Evidence-based frameworks for evaluating quality of care in genetic testing have been put forth by ACMG,160 ERKNet, and others.161, 162 These cover different methods for evaluating analytic and clinical validity as well as clinical utility of genetic tests. Nephrology outcomes used in clinical trials have included those that are disease-specific or represent more general longer-term outcomes, such as kidney failure, cardiovascular death, or mortality, which require large datasets. Yet this space is evolving, as demonstrated by development of novel trial designs using Bayesian methodology, inclusion of patient-reported outcomes, and additional economic evaluation of genetic risk. Steps for expanding measures to best inform value-based implementation and quality assurance of clinical genetics in nephrology are listed in Table 5. This is a large and critical space underpinning clinical translation and mainstreaming, with much research and work anticipated in the coming years.
Table 5.
Recommended Practices for Value-Based Measures of Implementation and Quality Assurance of Clinical Genetics in Nephrology
| Measure nephrologist adoption of genetic testing and appropriate referral to genetic testing |
| Measure nephrologist utilization of genetic results (to determine if appropriate changes in diagnosis and care have occurred) |
Define disease-specific outcomes that can be measured
|
| Define and measure potential harmful impacts of genetic testing (e.g., wrongful impact on change of treatment) |
| Define audits/assessments for centers that offer genetic testing in nephrology as quality assurance activity |
| Potentially apply USPSTF and EGAPP methods to analyze the implementation of genetic testing for kidney diseases |
USPSTF, United States Preventive Services Task Force; EGAPP, Evaluation of Genomic Applications in Practice and Prevention.
CONCLUSIONS
This KDIGO Controversies Conference on Genetics in Chronic Kidney Disease discussed many technical, logistical, ethical, and/or research questions related to the definition and epidemiology of monogenic and complex kidney diseases, applications of genetic findings in clinical medicine, and utilization of genomics for defining and stratifying CKD. Identified areas of consensus and future research priorities provide a roadmap towards realizing the promises of genomic medicine for nephrology.
The conference agenda, discussion questions, and plenary session presentations are available on the KDIGO website: https://kdigo.org/conferences/genetics-in-ckd/.
Supplementary Material
Table S1. Online Clinical Genetics Resources to Aid Nephrologists
Table S2. Recommended Competencies in Genetic Nephrology
ACKNOWLEDGMENT
This conference was sponsored by KDIGO and supported in part by unrestricted educational grants from American Kidney Fund, AstraZeneca, Chinook Therapeutics, Natera, Otsuka, Reata Pharmaceuticals, and Sanofi.
Abbreviations:
- ACMG
American College of Medical Genetics and Genomics
- ADPKD
autosomal dominant polycystic kidney disease
- ADTKD
autosomal dominant tubulointerstitial kidney disease
- aHUS
atypical hemolytic uremic syndrome
- APOL1
apolipoprotein L1
- CKD
chronic kidney disease
- eGFR
estimated glomerular filtration rate
- ERKNet
European Reference Network for Rare Kidney Diseases
- FSGS
focal segmental glomerulosclerosis
- GWAS
genome-wide association studies
- IgAN
IgA nephropathy
- KDIGO
Kidney Disease: Improving Global Outcomes
- PROM
patient-reported outcome measure
- SNP
single nucleotide polymorphism
- TMA
thrombotic microangiopathy
- VUS
variants of uncertain significance
- WGIKD
ERA-EDTA Working Group for Inherited Kidney Diseases
APPENDIX
Listing of KDIGO Conference Participants
Anna Köttgen1*, Emilie Cornec-Le Gall2**, Jan Halbritter3**, Krzysztof Kiryluk4**, Andrew J. Mallett5**, Rulan S. Parekh6**, Hila Milo Rasouly4**, Matthew G. Sampson7**, Adrienne Tin8**, Corinne Antignac9, Elisabet Ars10, Carsten Bergmann11,12, Anthony J. Bleyer13, Detlef Bockenhauer14, Olivier Devuyst15, Jose C. Florez16, Kevin J. Fowler17, Nora Franceschini18, Masafumi Fukagawa19, Daniel P. Gale20, Rasheed A. Gbadegesin21, David B. Goldstein22, Morgan E. Grams23, Anna Greka24, Oliver Gross25, Lisa M. Guay-Woodford26, Peter C. Harris27, Julia Hoefele28, Adriana M. Hung29, Nine V.A.M. Knoers30, Jeffrey B. Kopp31, Matthias Kretzler32, Matthew B. Lanktree33, Beata S. Lipska-Ziętkiewicz34, Kathleen Nicholls35, Kandai Nozu36, Akinlolu Ojo37, Afshin Parsa38, Cristian Pattaro39, York Pei40, Martin R. Pollak41, Eugene P. Rhee42, Simone Sanna-Cherchi43, Judy Savige44, John A. Sayer45, Francesco Scolari46, John R. Sedor47, Xueling Sim48, Stefan Somlo49, Katalin Susztak50, Bamidele O. Tayo51, Roser Torra52, Albertien M. van Eerde53, André Weinstock54, Cheryl A. Winkler55, Matthias Wuttke56, Hong Zhang57, Jennifer M. King58, Michael Cheung59, Michel Jadoul60, Wolfgang C. Winkelmayer61, and Ali G. Gharavi4*
*Conference Co-Chairs: Anna Köttgen, Ali G. Gharavi
**Steering Committee Members: Emilie Cornec-Le Gall, Jan Halbritter, Krzysztof Kiryluk, Andrew J. Mallett, Rulan S. Parekh, Hila Milo Rasouly, Matthew G. Sampson, Adrienne Tin. All Steering Committee Members contributed equally.
The conference planning and the drafting and critical revision of this manuscript were performed by the Steering Committee Members, the Conference Co-Chairs, and Jennifer King, with important intellectual content contributions provided by the remaining authors.
Affiliation information:
1Institute of Genetic Epidemiology, Faculty of Medicine and Medical Center - University of Freiburg, Freiburg, Germany
2Univ. Brest, INSERM, UMR 1078, GGB, CHU Brest, F-29200 Brest, France
3Division of Nephrology, Department of Internal Medicine, University Hospital Leipzig, Leipzig, Germany; Department of Nephrology and Medical Intensive Care, Charité - Universitätsmedizin Berlin
4Division of Nephrology and Center for Precision Medicine and Genomics, Department of Medicine, Columbia University Irving Medical Center, New York, New York, USA
5Institute for Molecular Bioscience (IMB), The University of Queensland, Brisbane, Queensland, Australia; Department of Nephrology, Townsville University Hospital; College of Medicine, James Cook University, Townsville, Queensland, Australia; KidGen Collaborative, Australian Genomics Health Alliance, Melbourne, Victoria, Australia
6Department of Medicine and Paediatrics, University of Toronto; Division of Nephrology, Women’s College Hospital, The Hospital for Sick Children; Dalla Lana School of Public Health, and Health Policy, Management and Evaluation, University of Toronto, Toronto, Ontario, Canada
7Division of Nephrology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts; Broad Institute, Cambridge, Massachusetts, USA
8Division of Nephrology, University of Mississippi Medical Center, Jackson, Mississippi, USA
9Laboratory of Hereditary Kidney Disease, Imagine Institute, INSERM U1163, Université de Paris, Paris, France. Department of Genetics, Necker Hospital, APHP, Paris, France
10Molecular Biology Laboratory, Fundació Puigvert, Instituto de Investigaciones Biomédicas Sant Pau (IIB Sant Pau), Universitat Autònoma de Barcelona, Barcelona, Catalonia, Spain
11Medizinische Genetik Mainz, Limbach Genetics, Mainz, Germany
12Department of Nephrology, Faculty of Medicine and Medical Center - University of Freiburg, Freiburg, Germany
13Section on Nephrology, Wake Forest School of Medicine, Winston-Salem, North Carolina, USA
14Renal Unit, Great Ormond Street Hospital for Children NHS Foundation Trust, London, UK; Department of Renal Medicine, University College London, London, UK
15Division of Nephrology, Cliniques Universitaires Saint-Luc, Brussels, Belgium; Institut de Recherche Expérimentale et Clinique, UCLouvain, Brussels, Belgium; Department of Physiology, Mechanisms of Inherited Kidney Disorders Group, University of Zurich, Zurich, Switzerland
16Programs in Metabolism and Medical & Population Genetics, Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA; Diabetes Unit and Center for Genomic Medicine, Massachusetts General Hospital, Boston, Massachusetts, USA; Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA
17The Voice of the Patient, Inc, Elmhurst, Illinois, USA
18Department of Epidemiology, Gillings School of Global Public Health, University of North Carolina, Chapel Hill, North Carolina, USA
19Division of Nephrology, Endocrinology and Metabolism, Tokai University School of Medicine, Isehara, Japan
20Department of Renal Medicine, University College London, London, UK; Rare Renal Disease Registry, UK Renal Registry, Bristol, UK
21Department of Pediatrics, Division of Nephrology, Duke University Medical Center, Durham, North Carolina, USA
22Institute for Genomic Medicine, Columbia University, New York, New York, USA; Department of Genetics and Development, Columbia University, New York, New York, USA
23Department of Nephrology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA
24Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, Massachusetts, USA; Broad Institute of MIT and Harvard, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA
25Clinic for Nephrology and Rheumatology, University Medical Center Göttingen, 37075 Göttingen, Germany
26Center for Translational Science, Children's National Health System, Washington, District of Columbia, USA
27Division of Nephrology and Hypertension, Mayo Clinic, Rochester, Minnesota, USA
28Institute of Human Genetics, Klinikum rechts der Isar, Technical University of Munich, School of Medicine, Munich, Germany
29VA Tennessee Valley Healthcare System, Division of Nephrology and Hypertension, Department of Medicine, Vanderbilt Center for Kidney Disease, Vanderbilt Precision Nephrology Program, Vanderbilt University Medical Center, Nashville, Tennessee, USA
30Department of Genetics, University Medical Center Groningen, Groningen, The Netherlands
31Kidney Disease Section, National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), NIH, Bethesda, Maryland, USA
32Division of Nephrology, Department of Internal Medicine, Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan, USA
33Division of Nephrology, St. Joseph’s Healthcare Hamilton and Department of Medicine, McMaster University, Hamilton, Ontario, Canada
34Rare Diseases Centre and Clinical Genetics Unit, Department of Biology and Medical Genetics, Medical University of Gdansk, Gdansk, Poland
35Department of Nephrology, Royal Melbourne Hospital, University of Melbourne, Parkville, Victoria, Australia
36Department of Pediatrics, Kobe University Graduate School of Medicine, Kobe 650-0017, Japan
37University of Kansas School of Medicine, Kansas City, Kansas, USA
38Division of Kidney, Urologic and Hematologic Diseases, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland, USA; Department of Medicine, University of Maryland School of Medicine, Baltimore, Maryland, USA
39Eurac Research, Institute for Biomedicine (affiliated with the University of Lübeck), Bolzano, Italy
40Division of Nephrology, University Health Network and University of Toronto, Toronto, ON, Canada
41Division of Nephrology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA
42Division of Nephrology, Department of Medicine, Massachusetts General Hospital, Boston, Massachusetts, USA
43Division of Nephrology, Department of Medicine, Columbia University, New York, New York, USA
44Department of Medicine, Melbourne and Northern Health, The University of Melbourne, Parkville, Victoria, 3050, Australia
45Translational and Clinical Research Institute, Faculty of Medical Sciences, Newcastle University, Central Parkway, Newcastle upon Tyne, UK; The Newcastle upon Tyne NHS Hospitals Foundation Trust, Newcastle upon Tyne, UK; NIHR Newcastle Biomedical Research Centre, Newcastle upon Tyne, UK
46Division and Chair of Nephrology, ASST-Spedali Civili and University of Brescia, Brescia, Italy
47Lerner Research and Glickman Urology and Kidney Institutes, Cleveland Clinic, Cleveland, Ohio, USA; Department of Physiology and Biophysics, Case Western Reserve University, Cleveland, Ohio, USA
48Saw Swee Hock School of Public Health, National University of Singapore and National University Health System, Singapore, Singapore
49Department of Internal Medicine, Yale University, New Haven, Connecticut, USA; Department of Genetics, Yale University, New Haven, Connecticut, USA
50Department of Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA; Department of Genetics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA
51Department of Public Health Sciences, Loyola University Chicago, Maywood, Illinois, USA
52Inherited Kidney Disorders, Nephrology Department, Fundacio Puigvert, IIB Sant Pau, Universitat Autonoma de Barcelona, Barcelona, Spain
53Department of Genetics, University Medical Center Utrecht, Utrecht, The Netherlands
54Alport Syndrome Foundation, Phoenix, Arizona, USA
55Basic Science Program, Frederick National Laboratory and Basic Research Laboratory, National Cancer Institute, Frederick, Maryland, USA
56Institute of Genetic Epidemiology, Dep. of Biometry, Epidemiology, and Medical Bioinformatics, Faculty of Medicine and Medical Center - University of Freiburg, Freiburg, Germany
57Renal Division, Peking University First Hospital; Peking University, Institute of Nephrology, Key Laboratory of Renal Disease, Ministry of Health of China, Beijing, China
58August Editorial, Durham, North Carolina, USA
59KDIGO, Brussels, Belgium
60Cliniques Universitaires Saint Luc, Université Catholique de Louvain, Brussels, Belgium
61Selzman Institute for Kidney Health, Section of Nephrology, Department of Medicine, Baylor College of Medicine, Houston, Texas, USA
Footnotes
DISCLOSURES
Several authors of this paper are members of the European Reference Network for rare Kidney Diseases (ERKNet)- project ID No 739532. AMvE receives support from the Dutch Kidney Foundation (#18OKG19).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Global Burden of Disease Chronic Kidney Disease Collaboration. Global, regional, and national burden of chronic kidney disease, 1990-2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2020; 395: 709–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rasouly HM, Groopman EE, Heyman-Kantor R, et al. The burden of candidate pathogenic variants for kidney and genitourinary disorders emerging from exome sequencing. Ann Intern Med 2019; 170: 11–21. [DOI] [PubMed] [Google Scholar]
- 3.Groopman EE, Marasa M, Cameron-Christie S, et al. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med 2019; 380: 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Domingo-Gallego A, Pybus M, Bullich G, et al. Clinical utility of genetic testing in early-onset kidney disease: seven genes are the main players. Nephrol Dial Transplant 2021. [DOI] [PubMed] [Google Scholar]
- 5.Bullich G, Domingo-Gallego A, Vargas I, et al. A kidney-disease gene panel allows a comprehensive genetic diagnosis of cystic and glomerular inherited kidney diseases. Kidney Int 2018; 94: 363–371. [DOI] [PubMed] [Google Scholar]
- 6.Mallett A, Patel C, Salisbury A, et al. The prevalence and epidemiology of genetic renal disease amongst adults with chronic kidney disease in Australia. Orphanet J Rare Dis 2014; 9: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snoek R, van Jaarsveld RH, Nguyen TQ, et al. Genetics-first approach improves diagnostics of ESKD patients younger than 50 years. Nephrol Dial Transplant 2020. [DOI] [PubMed] [Google Scholar]
- 8.Ottlewski I, Munch J, Wagner T, et al. Value of renal gene panel diagnostics in adults waiting for kidney transplantation due to undetermined end-stage renal disease. Kidney Int 2019; 96: 222–230. [DOI] [PubMed] [Google Scholar]
- 9.Connaughton DM, Kennedy C, Shril S, et al. Monogenic causes of chronic kidney disease in adults. Kidney Int 2019; 95: 914–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snoek R, van Setten J, Keating BJ, et al. NPHP1 (Nephrocystin-1) gene deletions cause adult-onset ESRD. J Am Soc Nephrol 2018; 29: 1772–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kottgen A, Pattaro C, Boger CA, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet 2010; 42: 376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanai M, Akiyama M, Takahashi A, et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat Genet 2018; 50: 390–400. [DOI] [PubMed] [Google Scholar]
- 13.Wuttke M, Li Y, Li M, et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet 2019; 51: 957–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teumer A, Li Y, Ghasemi S, et al. Genome-wide association meta-analyses and fine-mapping elucidate pathways influencing albuminuria. Nat Commun 2019; 10: 4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okada Y, Terao C, Ikari K, et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat Genet 2012; 44: 511–516. [DOI] [PubMed] [Google Scholar]
- 16.Morris AP, Le TH, Wu H, et al. Trans-ethnic kidney function association study reveals putative causal genes and effects on kidney-specific disease aetiologies. Nat Commun 2019; 10: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiryluk K, Li Y, Scolari F, et al. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet 2014; 46: 1187–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gharavi AG, Kiryluk K, Choi M, et al. Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet 2011; 43: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie J, Liu L, Mladkova N, et al. The genetic architecture of membranous nephropathy and its potential to improve non-invasive diagnosis. Nat Commun 2020; 11: 1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanescu HC, Arcos-Burgos M, Medlar A, et al. Risk HLA-DQA1 and PLA(2)R1 alleles in idiopathic membranous nephropathy. N Engl J Med 2011; 364: 616–626. [DOI] [PubMed] [Google Scholar]
- 21.Gbadegesin RA, Adeyemo A, Webb NJ, et al. HLA-DQA1 and PLCG2 are candidate risk loci for childhood-onset steroid-sensitive nephrotic syndrome. J Am Soc Nephrol 2015; 26: 1701–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dufek S, Cheshire C, Levine AP, et al. Genetic identification of two novel loci associated with steroid-sensitive nephrotic syndrome. J Am Soc Nephrol 2019; 30: 1375–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia X, Yamamura T, Gbadegesin R, et al. Common risk variants in NPHS1 and TNFSF15 are associated with childhood steroid-sensitive nephrotic syndrome. Kidney Int 2020; 98: 1308–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groopman EE, Rasouly HM, Gharavi AG. Genomic medicine for kidney disease. Nat Rev Nephrol 2018; 14: 83–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gale DP, Mallett A, Patel C, et al. Diagnoses of uncertain significance: Kidney genetics in the 21st century. Nat Rev Nephrol 2020. [DOI] [PubMed] [Google Scholar]
- 26.Shang N, Khan A, Polubriaginof F, et al. Medical records-based chronic kidney disease phenotype for clinical care and "big data" observational and genetic studies. NPJ Digit Med 2021; 4: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fox CS, Yang Q, Cupples LA, et al. Genomewide linkage analysis to serum creatinine, GFR, and creatinine clearance in a community-based population: the Framingham Heart Study. J Am Soc Nephrol 2004; 15: 2457–2461. [DOI] [PubMed] [Google Scholar]
- 28.Mottl AK, Vupputuri S, Cole SA, et al. Linkage analysis of glomerular filtration rate in American Indians. Kidney Int 2008; 74: 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langefeld CD, Beck SR, Bowden DW, et al. Heritability of GFR and albuminuria in Caucasians with type 2 diabetes mellitus. Am J Kidney Dis 2004; 43: 796–800. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh S, Collins FS. The geneticist's approach to complex disease. Annu Rev Med 1996; 47: 333–353. [DOI] [PubMed] [Google Scholar]
- 31.Salem RM, Todd JN, Sandholm N, et al. Genome-wide association study of diabetic kidney disease highlights biology involved in glomerular basement membrane collagen. J Am Soc Nephrol 2019; 30: 2000–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akbari P, Gilani A, Sosina O, et al. Sequencing of 640,000 exomes identifies GPR75 variants associated with protection from obesity. Science 2021; 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fahed AC, Wang M, Homburger JR, et al. Polygenic background modifies penetrance of monogenic variants for tier 1 genomic conditions. Nat Commun 2020; 11: 3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levey AS, de Jong PE, Coresh J, et al. The definition, classification, and prognosis of chronic kidney disease: a KDIGO Controversies Conference report. Kidney Int 2011; 80: 17–28. [DOI] [PubMed] [Google Scholar]
- 35.KDIGO 2012 Clinical Practice Guideline for the Evaluation and Management of Chronic Kidney Disease. Chapter 2: General principles in the management of glomerular disease. Kidney Int Suppl (2011) 2012; 2: 156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mann N, Braun DA, Amann K, et al. Whole-exome sequencing enables a precision medicine approach for kidney transplant recipients. J Am Soc Nephrol 2019; 30: 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Ven AT, Connaughton DM, Ityel H, et al. Whole-exome sequencing identifies causative mutations in families with congenital anomalies of the kidney and urinary tract. J Am Soc Nephrol 2018; 29: 2348–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stessman HA, Bernier R, Eichler EE. A genotype-first approach to defining the subtypes of a complex disease. Cell 2014; 156: 872–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandholm N, Van Zuydam N, Ahlqvist E, et al. The genetic landscape of renal complications in Type 1 diabetes. J Am Soc Nephrol 2017; 28: 557–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verbitsky M, Krithivasan P, Batourina E, et al. Copy number variant analysis and genome-wide association study identify loci with large effect for vesicoureteral reflux. J Am Soc Nephrol 2021; 32: 805–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scelo G, Purdue MP, Brown KM, et al. Genome-wide association study identifies multiple risk loci for renal cell carcinoma. Nat Commun 2017; 8: 15724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hellwege JN, Velez Edwards DR, Giri A, et al. Mapping eGFR loci to the renal transcriptome and phenome in the VA Million Veteran Program. Nat Commun 2019; 10: 3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qian H, Kowalski MH, Kramer HJ, et al. Genome-wide association of kidney traits in Hispanics/Latinos using dense imputed whole-genome sequencing data: The Hispanic Community Health Study/Study of Latinos. Circ Genom Precis Med 2020; 13: e002891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okuda H, Okamoto K, Abe M, et al. Genome-wide association study identifies new loci for albuminuria in the Japanese population. Clin Exp Nephrol 2020; 24: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorski M, Tin A, Garnaas M, et al. Genome-wide association study of kidney function decline in individuals of European descent. Kidney Int 2015; 87: 1017–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yun S, Han M, Kim HJ, et al. Genetic risk score raises the risk of incidence of chronic kidney disease in Korean general population-based cohort. Clin Exp Nephrol 2019; 23: 995–1003. [DOI] [PubMed] [Google Scholar]
- 47.Langefeld CD, Comeau ME, Ng MCY, et al. Genome-wide association studies suggest that APOL1-environment interactions more likely trigger kidney disease in African Americans with nondiabetic nephropathy than strong APOL1-second gene interactions. Kidney Int 2018; 94: 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parsa A, Kanetsky PA, Xiao R, et al. Genome-wide association of CKD progression: The Chronic Renal Insufficiency Cohort Study. J Am Soc Nephrol 2017; 28: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li M, Foo JN, Wang JQ, et al. Identification of new susceptibility loci for IgA nephropathy in Han Chinese. Nat Commun 2015; 6: 7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Genovese G, Tonna SJ, Knob AU, et al. A risk allele for focal segmental glomerulosclerosis in African Americans is located within a region containing APOL1 and MYH9. Kidney Int 2010; 78: 698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iyengar SK, Sedor JR, Freedman BI, et al. Genome-wide association and trans-ethnic meta-analysis for advanced diabetic kidney disease: Family Investigation of Nephropathy and Diabetes (FIND). PLoS Genet 2015; 11: e1005352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taira M, Imamura M, Takahashi A, et al. A variant within the FTO confers susceptibility to diabetic nephropathy in Japanese patients with type 2 diabetes. PLoS One 2018; 13: e0208654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guan M, Keaton JM, Dimitrov L, et al. Genome-wide association study identifies novel loci for type 2 diabetes-attributed end-stage kidney disease in African Americans. Hum Genomics 2019; 13: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chung SA, Brown EE, Williams AH, et al. Lupus nephritis susceptibility loci in women with systemic lupus erythematosus. J Am Soc Nephrol 2014; 25: 2859–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Howles SA, Wiberg A, Goldsworthy M, et al. Genetic variants of calcium and vitamin D metabolism in kidney stone disease. Nat Commun 2019; 10: 5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stafford-Smith M, Li YJ, Mathew JP, et al. Genome-wide association study of acute kidney injury after coronary bypass graft surgery identifies susceptibility loci. Kidney Int 2015; 88:823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Purdue MP, Ye Y, Wang Z, et al. A genome-wide association study of renal cell carcinoma among African Americans. Cancer Epidemiol Biomarkers Prev 2014; 23: 209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hernandez-Fuentes MP, Franklin C, Rebollo-Mesa I, et al. Long- and short-term outcomes in renal allografts with deceased donors: A large recipient and donor genome-wide association study. Am J Transplant 2018; 18: 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stapleton CP, Heinzel A, Guan W, et al. The impact of donor and recipient common clinical and genetic variation on estimated glomerular filtration rate in a European renal transplant population. Am J Transplant 2019; 19: 2262–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Locke AE, Steinberg KM, Chiang CWK, et al. Exome sequencing of Finnish isolates enhances rare-variant association power. Nature 2019; 572: 323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nanayakkara S, Senevirathna ST, Parahitiyawa NB, et al. Whole-exome sequencing reveals genetic variants associated with chronic kidney disease characterized by tubulointerstitial damages in North Central Region, Sri Lanka. Environ Health Prev Med 2015; 20: 354–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weng PL, Majmundar AJ, Khan K, et al. De novo TRIM8 variants impair its protein localization to nuclear bodies and cause developmental delay, epilepsy, and focal segmental glomerulosclerosis. Am J Hum Genet 2021; 108: 357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin BM, Grinde KE, Brody JA, et al. Whole genome sequence analyses of eGFR in 23,732 people representing multiple ancestries in the NHLBI trans-omics for precision medicine (TOPMed) consortium. EBioMedicine 2021; 63: 103157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benonisdottir S, Kristjansson RP, Oddsson A, et al. Sequence variants associating with urinary biomarkers. Hum Mol Genet 2019; 28: 1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo J, Rackham OJL, Sandholm N, et al. Whole-genome sequencing of Finnish Type 1 diabetic siblings discordant for kidney disease reveals DNA variants associated with diabetic nephropathy. J Am Soc Nephrol 2020; 31: 309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kottgen A, Glazer NL, Dehghan A, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet 2009; 41: 712–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kasembeli AN, Duarte R, Ramsay M, et al. APOL1 risk variants are strongly associated with HIV-associated nephropathy in black South Africans. J Am Soc Nephrol 2015; 26: 2882–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kopp JB, Nelson GW, Sampath K, et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol 2011; 22: 2129–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Genovese G, Friedman DJ, Ross MD, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 2010; 329: 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Riella C, Siemens TA, Wang M, et al. APOL1-associated kidney disease in Brazil. Kidney Int Rep 2019; 4: 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Freedman BI, Langefeld CD, Andringa KK, et al. End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis Rheumatol 2014; 66: 390–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Larsen CP, Beggs ML, Saeed M, et al. Apolipoprotein L1 risk variants associate with systemic lupus erythematosus-associated collapsing glomerulopathy. J Am Soc Nephrol 2013; 24: 722–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ekulu PM, Nkoy AB, Betukumesu DK, et al. APOL1 risk genotypes are associated with early kidney damage in children in Sub-Saharan Africa. Kidney Int Rep 2019; 4: 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ekrikpo UE, Mnika K, Effa EE, et al. Association of genetic polymorphisms of TGF-beta1, HMOX1, and APOL1 With CKD in Nigerian patients with and without HIV. Am J Kidney Dis 2020; 76: 100–108. [DOI] [PubMed] [Google Scholar]
- 75.Naik RP, Irvin MR, Judd S, et al. Sickle cell trait and the risk of ESRD in Blacks. J Am Soc Nephrol 2017; 28: 2180–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mukamal KJ, Tremaglio J, Friedman DJ, et al. APOL1 genotype, kidney and cardiovascular disease, and death in older adults. Arterioscler Thromb Vasc Biol 2016; 36: 398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tayo BO, Kramer H, Salako BL, et al. Genetic variation in APOL1 and MYH9 genes is associated with chronic kidney disease among Nigerians. Int Urol Nephrol 2013; 45: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Foster MC, Coresh J, Fornage M, et al. APOL1 variants associate with increased risk of CKD among African Americans. J Am Soc Nephrol 2013; 24: 1484–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grams ME, Rebholz CM, Chen Y, et al. Race, APOL1 risk, and eGFR decline in the general population. J Am Soc Nephrol 2016; 27: 2842–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen TK, Coresh J, Daya N, et al. Race, APOL1 risk variants, and clinical outcomes among older adults: The ARIC Study. J Am Geriatr Soc 2021; 69: 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peralta CA, Bibbins-Domingo K, Vittinghoff E, et al. APOL1 genotype and race differences in incident albuminuria and renal function decline. J Am Soc Nephrol 2016; 27: 887–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parsa A, Kao WH, Xie D, et al. APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med 2013; 369: 2183–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen TK, Tin A, Peralta CA, et al. APOL1 risk variants, incident proteinuria, and subsequent eGFR decline in Blacks with hypertension-attributed CKD. Clin J Am Soc Nephrol 2017; 12: 1771–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Friedman DJ, Pollak MR. Genetics of kidney failure and the evolving story of APOL1. J Clin Invest 2011; 121: 3367–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Watanabe A, Guaragna MS, Belangero VMS, et al. APOL1 in an ethnically diverse pediatric population with nephrotic syndrome: Implications in focal segmental glomerulosclerosis and other diagnoses. Pediatr Nephrol 2021; 36: 2327–2336. [DOI] [PubMed] [Google Scholar]
- 86.Knoers N, Antignac C, Bergmann C, et al. Genetic testing in the diagnosis of chronic kidney disease: Recommendations for clinical practice. Nephrol Dial Transplant 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rehm HL, Berg JS, Brooks LD, et al. ClinGen--the clinical genome resource. N Engl J Med 2015; 372: 2235–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Clinical Genome Research. ClinGen Complex Disease PRS Reporting Standards. Available at: https://clinicalgenome.org/docs/clingen-complex-disease-prs-reporting-standards/. Accessed March 25, 2021.
- 89.Friedman DJ, Pollak MR. APOL1 nephropathy: From genetics to clinical applications. Clin J Am Soc Nephrol 2021; 16: 294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Surapaneni AL, Ballew SH, Coresh J, et al. APOL1 risk alleles, cardiac markers, and risk of ESKD in African Americans: The Atherosclerosis Risk in Communities Study. Kidney Med 2020; 2: 502–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sinnott-Armstrong N, Tanigawa Y, Amar D, et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat Genet 2021; 53: 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gordon EJ, Amomicronrtegui D, Blancas I, et al. African American living donors' attitudes about APOL1 genetic testing: A mixed methods study. Am J Kidney Dis 2018; 72: 819–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Freedman BI, Burke W, Divers J, et al. Diagnosis, education, and care of patients with APOL1-associated nephropathy: A Delphi consensus and systematic review. J Am Soc Nephrol 2021; 32: 1765–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fuentes F, Kopp JB. Launching APOLLO: The role of APOL1 genetic variants in live- and deceased-donor kidney transplantation. Kidney Int Rep 2020; 5: 252–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ellard S, Baple EL, Callaway A, et al. Association for Clinical Genome Science Best Practices Guidelines for Variant Classification in Rare Disease 2020. Available at: https://www.acgs.uk.com/media/11631/uk-practice-guidelines-for-variant-classification-v4-01-2020.pdf. Accessed March 18, 2021. [Google Scholar]
- 97.Furlano M, Martinez V, Pybus M, et al. Clinical and genetic features of autosomal dominant Alport Syndrome: A cohort study. Am J Kidney Dis 2021; 78: 560–570 e561. [DOI] [PubMed] [Google Scholar]
- 98.Biesecker LG, Adam MP, Alkuraya FS, et al. A dyadic approach to the delineation of diagnostic entities in clinical genomics. Am J Hum Genet 2021; 108: 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hamosh A, Amberger JS, Bocchini CA, et al. Response to Biesecker et al. Am J Hum Genet 2021; 108: 1807–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eckardt KU, Alper SL, Antignac C, et al. Autosomal dominant tubulointerstitial kidney disease: diagnosis, classification, and management--A KDIGO consensus report. Kidney Int 2015; 88: 676–683. [DOI] [PubMed] [Google Scholar]
- 101.Kottgen A, Kao WH, Hwang SJ, et al. Genome-wide association study for renal traits in the Framingham Heart and Atherosclerosis Risk in Communities Studies. BMC Med Genet 2008; 9: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Feldman HI, Appel LJ, Chertow GM, et al. The Chronic Renal Insufficiency Cohort (CRIC) Study: Design and methods. J Am Soc Nephrol 2003; 14: S148–153. [DOI] [PubMed] [Google Scholar]
- 103.Appel LJ, Middleton J, Miller ER 3rd, et al. The rationale and design of the AASK cohort study. J Am Soc Nephrol 2003; 14: S166–172. [DOI] [PubMed] [Google Scholar]
- 104.Klahr S. The modification of diet in renal disease study. N Engl J Med 1989; 320: 864–866. [DOI] [PubMed] [Google Scholar]
- 105.Eckardt KU, Barthlein B, Baid-Agrawal S, et al. The German Chronic Kidney Disease (GCKD) study: Design and methods. Nephrol Dial Transplant 2012; 27: 1454–1460. [DOI] [PubMed] [Google Scholar]
- 106.Furth SL, Cole SR, Moxey-Mims M, et al. Design and methods of the Chronic Kidney Disease in Children (CKiD) prospective cohort study. Clin J Am Soc Nephrol 2006; 1: 1006–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Querfeld U, Anarat A, Bayazit AK, et al. The Cardiovascular Comorbidity in Children with Chronic Kidney Disease (4C) study: Objectives, design, and methodology. Clin J Am Soc Nephrol 2010; 5: 1642–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wuhl E, Trivelli A, Picca S, et al. Escape Trial Group. Strict blood-pressure control and progression of renal failure in children. N Engl J Med 2009; 361: 1639–1650. [DOI] [PubMed] [Google Scholar]
- 109.Kottgen A, Pattaro C. The CKDGen Consortium: Ten years of insights into the genetic basis of kidney function. Kidney Int 2020; 97: 236–242. [DOI] [PubMed] [Google Scholar]
- 110.Psaty BM, O'Donnell CJ, Gudnason V, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet 2009; 2: 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fishman CE, Mohebnasab M, van Setten J, et al. Genome-wide study updates in the International Genetics and Translational Research in Transplantation Network (iGeneTRAiN). Front Genet 2019; 10: 1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Magi R, Horikoshi M, Sofer T, et al. Trans-ethnic meta-regression of genome-wide association studies accounting for ancestry increases power for discovery and improves fine-mapping resolution. Hum Mol Genet 2017; 26: 3639–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Franceschini N, Morris AP. Genetics of kidney traits in worldwide populations: The Continental Origins and Genetic Epidemiology Network (COGENT) Kidney Consortium. Kidney Int 2020; 98: 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kao WH, Klag MJ, Meoni LA, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet 2008; 40: 1185–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kopp JB, Smith MW, Nelson GW, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet 2008; 40: 1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sudlow C, Gallacher J, Allen N, et al. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015; 12: e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gaziano JM, Concato J, Brophy M, et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 2016; 70: 214–223. [DOI] [PubMed] [Google Scholar]
- 118.Krokstad S, Langhammer A, Hveem K, et al. Cohort Profile: The HUNT Study, Norway. Int J Epidemiol 2013; 42: 968–977. [DOI] [PubMed] [Google Scholar]
- 119.Jonsson H, Sulem P, Kehr B, et al. Whole genome characterization of sequence diversity of 15,220 Icelanders. Sci Data 2017; 4: 170115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nagai A, Hirata M, Kamatani Y, et al. Overview of the BioBank Japan Project: Study design and profile. J Epidemiol 2017; 27: S2–S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008; 84: 362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Graham SE, Nielsen JB, Zawistowski M, et al. Sex-specific and pleiotropic effects underlying kidney function identified from GWAS meta-analysis. Nat Commun 2019; 10: 1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.eMERGE Consortium. Lessons learned from the eMERGE Network: balancing genomics in discovery and practice. Human Genetics and Genomics Advances 2021; 2: 100018 https://doi.org/100010.101016/j.xhgg.102020.100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Denny JC, Rutter JL, Goldstein DB, et al. All of Us Research Program Investigators. The "All of Us" Research Program. N Engl J Med 2019; 381: 668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shang N, Liu C, Rasmussen LV, et al. Making work visible for electronic phenotype implementation: Lessons learned from the eMERGE network. J Biomed Inform 2019; 99: 103293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Norton JM, Ali K, Jurkovitz CT, et al. Development and validation of a pragmatic electronic phenotype for CKD. Clin J Am Soc Nephrol 2019; 14: 1306–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Denburg MR, Razzaghi H, Bailey LC, et al. Using electronic health record data to rapidly identify children with glomerular disease for clinical research. J Am Soc Nephrol 2019; 30: 2427–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chan L, Beers K, Yau AA, et al. Natural language processing of electronic health records is superior to billing codes to identify symptom burden in hemodialysis patients. Kidney Int 2020; 97: 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zheng NS, Feng Q, Kerchberger VE, et al. PheMap: A multi-resource knowledge base for high-throughput phenotyping within electronic health records. J Am Med Inform Assoc 2020; 27: 1675–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bajaj A, Ihegword A, Qiu C, et al. Phenome-wide association analysis suggests the APOL1 linked disease spectrum primarily drives kidney-specific pathways. Kidney Int 2020; 97: 1032–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tin A, Kottgen A. Mendelian randomization analysis as a tool to gain insights into causes of diseases: A primer. J Am Soc Nephrol 2021; 32: 2400–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Martin AR, Kanai M, Kamatani Y, et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet 2019; 51: 584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang Z, Hernandez K, Savage J, et al. Uniform genomic data analysis in the NCI Genomic Data Commons. Nat Commun 2021; 12: 1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.eMERGE Consortium. Harmonizing clinical sequencing and interpretation for the eMERGE III Network. Am J Hum Genet 2019; 105: 588–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Eales JM, Jiang X, Xu X, et al. Uncovering genetic mechanisms of hypertension through multi-omic analysis of the kidney. Nat Genet 2021; 53: 630–637. [DOI] [PubMed] [Google Scholar]
- 136.Gillies CE, Putler R, Menon R, et al. An eQTL landscape of kidney tissue in human nephrotic syndrome. Am J Hum Genet 2018; 103: 232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Townsend RR, Guarnieri P, Argyropoulos C, et al. Rationale and design of the Transformative Research in Diabetic Nephropathy (TRIDENT) Study. Kidney Int 2020; 97: 10–13. [DOI] [PubMed] [Google Scholar]
- 138.Abedini A, Zhu YO, Chatterjee S, et al. Urinary single-cell profiling captures the cellular diversity of the kidney. J Am Soc Nephrol 2021; 32: 614–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Consortium EP. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 2004; 306: 636–640. [DOI] [PubMed] [Google Scholar]
- 140.Roadmap Epigenomics C, Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature 2015; 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Consortium GT. Erratum: Genetic effects on gene expression across human tissues. Nature 2018; 553: 530. [DOI] [PubMed] [Google Scholar]
- 142.de Boer IH, Alpers CE, Azeloglu EU, et al. Rationale and design of the Kidney Precision Medicine Project. Kidney Int 2021; 99: 498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mostafavi H, Harpak A, Agarwal I, et al. Variable prediction accuracy of polygenic scores within an ancestry group. Elife 2020; 9: e48376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wand H, Lambert SA, Tamburro C, et al. Improving reporting standards for polygenic scores in risk prediction studies. Nature 2021; 591: 211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Buniello A, MacArthur JAL, Cerezo M, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 2019; 47: D1005–d1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lambert SA, Gil L, Jupp S, et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nat Genet 2021; 53: 420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shaikh A, Patel N, Nair D, et al. Current paradigms and emerging opportunities in nephrology training. Adv Chronic Kidney Dis 2020; 27: 291–296 e291. [DOI] [PubMed] [Google Scholar]
- 148.Berns JS. A survey-based evaluation of self-perceived competency after nephrology fellowship training. Clin J Am Soc Nephrol 2010; 5: 490–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jayasinghe K, Quinlan C, Mallett AJ, et al. Attitudes and practices of Australian nephrologists toward implementation of clinical genomics. Kidney Int Rep 2021; 6: 272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tognetto A, Michelazzo MB, Ricciardi W, et al. Core competencies in genetics for healthcare professionals: Results from a literature review and a Delphi method. BMC Med Educ 2019; 19: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Milo-Rasouly H, Aggarwal V, Bier L, et al. Cases in precision medicine: Genetic testing to predict future risk for disease in a healthy patient. Ann Intern Med 2021; 174: 540–547. [DOI] [PubMed] [Google Scholar]
- 152.Chen J, Fowler KJ, Grams ME. Knowledge is power: Patient education as a tool for patient activation. Am J Kidney Dis 2020; 76: 163–165. [DOI] [PubMed] [Google Scholar]
- 153.Logeman C, Cho Y, Sautenet B, et al. 'A sword of Damocles': Patient and caregiver beliefs, attitudes and perspectives on presymptomatic testing for autosomal dominant polycystic kidney disease: A focus group study. BMJ Open 2020; 10: e038005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Waterman AD, Gleason J, Lerminiaux L, et al. Amplifying the patient voice: Key priorities and opportunities for improved transplant and living donor advocacy and outcomes during COVID-19 and beyond. Curr Transplant Rep 2020: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Vyas DA, Eisenstein LG, Jones DS. Hidden in plain sight - Reconsidering the use of race correction in clinical algorithms. N Engl J Med 2020; 383: 874–882. [DOI] [PubMed] [Google Scholar]
- 156.Borrell LN, Elhawary JR, Fuentes-Afflick E, et al. Race and genetic ancestry in medicine - A time for reckoning with racism. N Engl J Med 2021; 384: 474–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Oni-Orisan A, Mavura Y, Banda Y, et al. Embracing genetic diversity to improve Black health. N Engl J Med 2021; 384: 1163–1167. [DOI] [PubMed] [Google Scholar]
- 158.Race Ethnicity, and Genetics Working Group. The use of racial, ethnic, and ancestral categories in human genetics research. Am J Hum Genet 2005; 77: 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nair D, Wilson FP. Patient-reported outcome measures for adults with kidney disease: Current measures, ongoing initiatives, and future opportunities for incorporation into patient-centered kidney care. Am J Kidney Dis 2019; 74: 791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.ACMG Board of Directors. Clinical utility of genetic and genomic services: A position statement of the American College of Medical Genetics and Genomics. Genet Med 2015; 17: 505–507. [DOI] [PubMed] [Google Scholar]
- 161.Sun F, Bruening W, Erinoff E, et al. Addressing Challenges in Genetic Test Evaluation: Evaluation Frameworks and Assessment of Analytic Validity. Agency for Healthcare Research and Quality. https://effectivehealthcare.ahrq.gov/. 2011. [PubMed] [Google Scholar]
- 162.Giacomini M, Miller F, Browman G. Confronting the "gray zones" of technology assessment: evaluating genetic testing services for public insurance coverage in Canada. Int J Technol Assess Health Care 2003; 19: 301–316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Online Clinical Genetics Resources to Aid Nephrologists
Table S2. Recommended Competencies in Genetic Nephrology