SUMMARY
A central factor in the maintenance of tissue integrity is the response of stem cells to variations in the levels of niche signals. In the gut, intestinal stem cells (ISCs) depend on Wnt ligands for self-renewal and proliferation. Transient increases in Wnt signaling promote regeneration after injury or in inflammatory bowel diseases, whereas constitutive activation of this pathway leads to colorectal cancer. Here, we report that Discs large 1 (Dlg1), while dispensable for polarity and cellular turnover during intestinal homeostasis, is required for ISC survival in the context of increased Wnt signaling. RNA sequencing (RNAseq) and genetic mouse models demonstrated that DLG1 regulates the cellular response to increased canonical Wnt ligands. This occurs via transcriptional regulation of Arhgap31, a GTPase-activating protein that deactivates CDC42, an effector of the non-canonical Wnt pathway. These findings reveal a DLG1-ARHGAP31-CDC42 axis that is essential for the ISC response to increased niche Wnt signaling.
Keywords: stem cells, Wnt, niche, regeneration, Dlg1, Arhgap31, Cdgap, Cdc42, Rac1, organoid, intestine, epithelium, cell death
Graphical Abstract
eTOC Blurb
Throughout an organism’s life, the activity of niche signals fluctuates due to injury, and stem cells need to respond accordingly. Klein and colleagues found that during high Wnt activity observed in intestinal regeneration, niche canonical WNT ligands activate non-canonical Wnt signaling via a DLG1-ARHGAP31-CDC42 axis required for stem cell-driven regeneration.
INTRODUCTION
Epithelial tissues have extraordinary resilience against physical and chemical damage, in part due to rapid regeneration fueled by stem cells1. Stem cells are essential for maintaining tissue function and ensuring return to homeostasis after injury2. The small intestinal epithelium is a multifunctional tissue that performs essential tasks, including nutrient absorption and maintenance of a barrier against harmful pathogens and carcinogens that are ingested. The epithelial lining is made up of repetitive units of villus-crypt structures. At the base of each villus, tubular crypts house proliferative LGR5+ intestinal stem cells (ISCs)3 that continuously replenish the entire human and mouse epithelium within 3 to 5 days. Indeed, the gut lining is the most rapidly cycling epithelium in the mammalian body and thus provides an excellent model to study somatic stem cells4,5. ISCs actively proliferate to self-renew and generate transit-amplifying (TA) progenitor cells, which in turn give rise to differentiated absorptive and secretory cells3,6.
The regenerative potential of stem cells depends on microenvironmental cues from the stem cell niche7. As a result of ever-changing tissue needs, such as growth during development and repair during regeneration, niche signals required for stem cell activity fluctuate throughout life8-10. In the intestine, epithelial and mesenchymal Wnt signals are a central part of the molecular milieu responsible for ISC function6,11-19. During intestinal homeostasis, Wnt signals regulate ISCs to maintain steady-state conditions20-24. After injury, such as by irradiation, inflammation, resection or infection, regeneration of the intestinal epithelium is driven by transient increases in expression of Wnt ligands followed by robust pathway activation that promotes proliferation and stemness17,25-30. However, constitutive activation of Wnt signaling can lead to tumorigenesis31-35, which in the intestine often occurs through mutation of the tumor suppressor Adenomatous Polyposis Coli (APC)20-24,36. Even though the Wnt pathway has been extensively studied in the intestinal epithelium, it remains unclear how stem cells contend with the transient increases in Wnt signaling that occur throughout life.
APC interacts with DLG137-39, a tumor suppressor protein that is part of Scribble polarity complex (together with SCRIBBLE and LGL) and is classically known to regulate cell polarity by directing formation of basolateral membranes37,39-41. Deletion of Scribble polarity proteins leads to severe developmental abnormalities in several organs in invertebrates and vertebrates42-45. Likewise, deletion of intestinal Scribble (Scrib) has a profound effect on apical basal polarity in the adult intestine46. However, deletion of Drosophila midgut Dlg is dispensable for midgut polarity47.
Potential intersections between Wnt/β-catenin signaling and DLG1-mediated polarity have been reported using in vitro approaches. These studies demonstrated the importance of interactions between APC and DLG1 for various cellular functions: polarization of migrating astrocytes48, cell cycle progression in fibroblasts49, and migration of isolated Xenopus epithelial cells50. Deletion of Dlg1 in the intestine decreases the survival of mice harboring tumorigenic Apc mutations, demonstrating the involvement of DLG1 in the Wnt signaling pathway51. However, the molecular and cellular mechanisms underlying the role of DLG1 in Wnt signaling is not clear.
In this study, we investigated the link between DLG1 and canonical Wnt signaling in the intestinal epithelium by conditionally deleting Dlg1 using intestinal VilCreER and ISC-specific Lgr5CreER drivers. During homeostasis, intestinal cell turnover and apical-basal polarization were not affected by the absence of Dlg1, consistent with data from the Drosophila midgut47. However, we found that Dlg1 is required for the proper intestinal response to increased canonical Wnt levels. Dlg1 regulates the expression of Arhgap31, a GTPase-activating protein that regulates the activity of the small GTPase CDC42, which in turn is an effector of the non-canonical Wnt planar cell polarity pathway52,53. These findings shed light on a previously underappreciated crosstalk between canonical Wnt ligands and the non-canonical Wnt pathway which ISCs rely on during WNT-dependent regeneration.
RESULTS
Dlg1 is dispensable for mammalian intestinal polarity maintenance and epithelial integrity during homeostasis
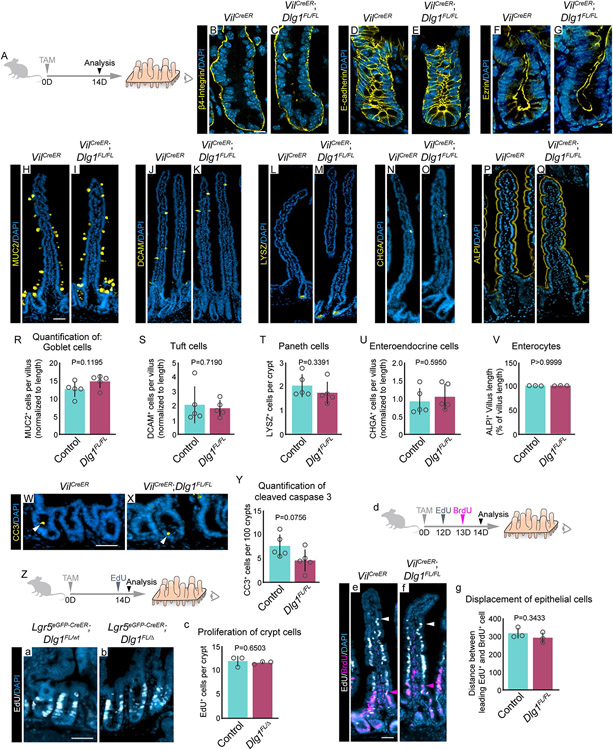
To investigate the role of Dlg1 in the mouse adult intestine, we conditionally deleted Dlg1 throughout the intestinal epithelium using VilCreERT2;Dlg1FL/FL (Dlg1FL/FL) mice54,55 (Figures S1A-C”). In addition, to specifically delete Dlg1 in ISCs, we used Lgr5eGFp-CreERT2;Dlg1FL/Δ (Dlg1FL/Δ) mice3,56, in which mosaic recombination takes place in LGR5+ ISCs (Figures S1D-E”). Both genetic models generate DLG1− cells. In control mice, Dlg1 mRNA was expressed and the protein was basolaterally localized in intestinal epithelial cells (Figures S1A, B-B”, and D-D”). After a single tamoxifen induction, Dlg1 expression was lost at 7 days (Figure S1A); DLG1 protein was completely absent from the epithelium in Dlg1FL/FL mice (Figure S1C-C”) and diminished in Dlg1FL/Δ mice (Figure S1E-E”). To assess the requirement of DLG1 for the maintenance of intestinal epithelial cell polarity, we analyzed membrane distribution of key proteins involved in cell adhesion that mediate cell polarity, including basally located hemidesmosomes by staining β4-Integrin (Figures 1A-C), basolateral adherens junctions by staining E-cadherin (Figures 1D-E), and apically localized Ezrin (Figures 1F-G)57-62. Surprisingly, conditional deletion of Dlg1 in vilCreERT2 mice did not affect cellular distribution of β4-Integrin, E-cadherin, or Ezrin, whereas in all other tissues that have been examined, diminished Dlg1 expression resulted in impaired cell polarity43,56,63-66. Furthermore, conditional deletion of Scrib in the intestinal epithelium leads to disrupted crypt apicobasal polarity and hyperproliferation of crypt cells46. Thus, we next sought to explore whether other crypt functions were disrupted by loss of Dlg1 in the intestine. Although loss of Dlg1 results in cell fate abnormalities in other tissues43,56,66, we found that DLG1 is not required for differentiation into goblet cells, tuft cells, Paneth cells, enteroendocrine cells, and enterocytes (Figures 1H-V). In addition, we observed no significant difference in the frequency of crypt cell death between controls and DLG1− intestines. (Figures 1W-Y). Similarly, the number of crypt cells that actively incorporated EdU over the course of 2 hours was not significantly different between controls and Dlg1FL/Δ mice (Figures 1Z-c). Finally, we analyzed the distance that cells moved toward the villus tips over 24 hours by injecting control and Dlg1FL/FL mice with EdU and BrdU at 48 and 24 hours, respectively, before tissue analysis (Figure 1d). Measuring the distance between the leading EdU+ and BrdU+ cells revealed a similar rate of cell displacement in control and DLG1− intestines (Figures 1e-g). Thus, in contrast to previous observations showing a requirement for DLG1 in mammalian tissue polarity, loss of Dlg1 expression during homeostasis does not affect polarity, integrity, or the ability of the intestinal epithelium to renew.
Figure 1. Polarity and integrity of the intestinal epithelium do not depend on the canonical polarity factor DLG1.
(A) Mice were injected with one dose of tamoxifen (TAM) and the small intestinal epithelium was analyzed 14 days later. (B-Q) Immunofluorescence images of mAbs for adhesion molecules and differentiation markers. Scale bar = 10 μm (B-G) and 50 μm (H-Q). (R-V) Quantification of intestinal cell populations. N = 5 x 30-40 villi, or 10 crypts per condition, mean ± SD, unpaired t test with Welch’s correction. (W and X) Immunofluorescence images of anti-CC3. Scale bar = 20 μm. (Y) Quantification of cleaved caspase-3 in crypt region. N = 5 x 10 crypts per condition, mean ± SD, unpaired t test with Welch’s correction. (Z) Experimental schematic for analyzing incorporation of EdU by crypt cells. (a and b) Immunofluorescence images of EdU. Scale bar = 20 μm. (c) Quantification of EdU incorporation by crypt cells. N = 3 x 50 crypts per condition, mean ± SD, unpaired t test with Welch’s correction. (d) Experimental schematic for analyzing the migration of crypt cells toward the villus tip. (e and f) Immunofluorescence images of EdU and anti-BrdU. Arrowheads indicate the leading EdU+ cell (white) and BrdU+ cell (magenta). Scale bar = 50 μm. (g) Quantification of epithelial cell migration towards villus tip by measuring the distance between leading EdU+ and BrdU+ cells. N = 3 x 50 villi per condition, mean ± SD, unpaired t test with Welch’s correction. Nuclei counterstained with DAPI.
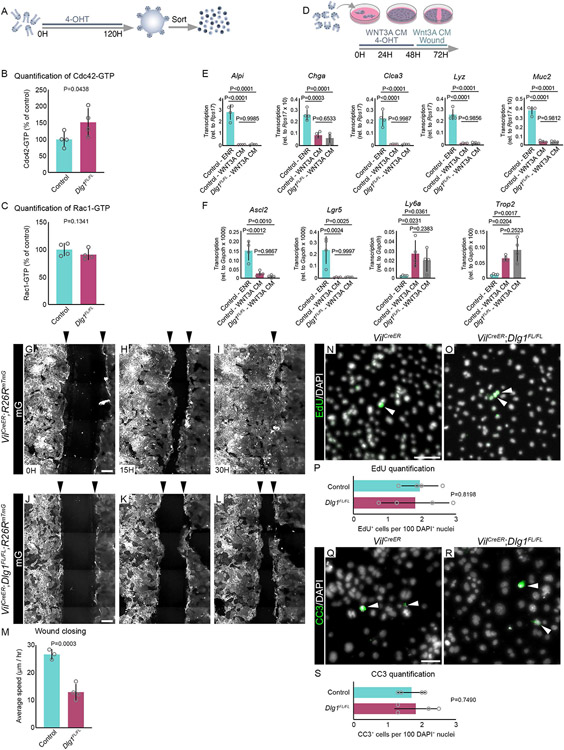
Wnt signaling activation leads to increased cell death in crypts lacking Dlg1
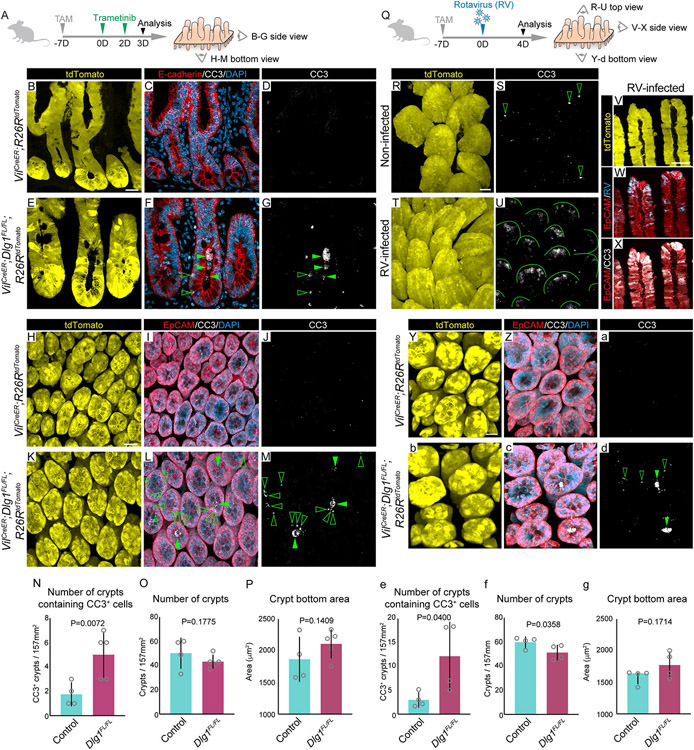
Previous reports showed that loss-of-function mutations in the Dlg family of genes contribute to tumorigenesis and inflammatory bowel disease, and these diseases also involve upregulated Wnt signaling51,67-73. Therefore, we asked whether DLG1 regulates the response of the intestinal epithelium to elevated Wnt signaling by treating VilCreERT2;Dlg1FL/FL;R26RtdTomato (Dlg1FL/FL;tdTomato) mice with intraperitoneal injections of trametinib, a MEK1/2 inhibitor that activates canonical Wnt signaling in the intestine74. Mice received two doses of trametinib 7 days after tamoxifen induction, and intestines were isolated 3 days after initial trametinib treatment (Figure 2A). Following this paradigm, we confirmed the activation of canonical Wnt signaling downstream target genes in crypts (Figure S2A). Whole mount and tissue section analysis of small intestines demonstrated robust Cre recombination in the epithelium, assessed by tdTomato expression (Figures 2B, E, H, K). The overall intestinal morphology was preserved in trametinib-treated control and DLG1− mice, but we observed accumulation of apoptotic cells in the luminal space of DLG1− crypts, as indicated by labeling with cleaved caspase-3 (CC3) compared to controls (Figures 2B-N). In images of whole mounted tissue, we found that dying cells were concentrated also in the epithelium of DLG1− crypts (Figures 2G and M; Videos 1A-B). However, the number of crypts remained the same as in control intestines (Figure 2O), and control and DLG1− crypts showed a similar crypt bottom area (Figures 2P, S2B-C).
Figure 2. Wnt signaling activation in vivo by trametinib or rotavirus leads to increased cell death in crypts lacking DLG1.
(A) Experimental schematic for analyzing the epithelial response to trametinib driven increased Wnt signaling. Mice were injected with one dose of TAM on day 7 followed by two doses of trametinib on days 0 and 2 prior to sacrifice on day 3. (B-G) Side view of crypts. (H-M) Bottom view of crypts. (B, E, H, K) Images of endogenous tdTomato. (C, F, I, L) Immunofluorescence images of anti-E-cadherin or anti-EpCAM with anti-CC3. (D, G, J, M) Immunofluorescence images of anti-CC3 with highlighted CC3+ dying cells in the lumen (closed arrowhead) and in the epithelium (open arrowhead). Scale bar for side view images = 20 μm, for bottom view images = 30 μm. (N) Quantification of crypts containing CC3+ cells per 157 mm2 field of view. (O) Quantification of the number of crypts per 157 mm2 field of view. (P) Quantification of crypt bottom area. N = 4-5 x 10 fields of view per condition, mean ± SD, unpaired t test with Welch’s correction. (Q) Experimental schematic for analyzing the epithelial response to rotavirus (RV) infection17. Mice were injected with one dose of TAM on day 7 followed by rotavirus infection on day 0 prior to sacrifice on day 4. (R-U) Top view of villi. (V-X) Side view of villi. (Y-d) Bottom view of crypts. (R, T, V, Y, b) Images of endogenous tdTomato. (S and U) Immunofluorescence images of anti-CC3 with highlighted CC3+ dying cells in villi tips of control mice (open arrowheads) and RV-infected mice (lines). (W and X) Immunofluorescence images of anti-Epcam with anti-RV or anti-CC3. (Z, c) Immunofluorescence images of anti-EpCAM and anti-CC3. (a and d) Immunofluorescence images of anti-CC3 with highlighted CC3+ dying cells in the lumen (closed arrowhead) and in the epithelium (open arrowhead). Scale bar for top and side view images = 50 μm, for bottom view images = 20 μm. (e) Quantification of crypts containing CC3+ cells per 157 mm2 field of view. (f) Quantification of the number of crypts per 157 mm2 field of view. (g) Quantification of crypt bottom area. N = 4 x 6 fields of view per condition. e-f: mean ± SD, unpaired t test with Welch’s correction. g: median ± interquartile range, unpaired Mann-Whitney test. Nuclei counterstained with DAPI.
To further test for the requirement of DLG1 in ISCs when exposed to Wnt signaling upregulation, we next used the enteric pathogen, rotavirus (RV). RV infects and induces cell death of differentiated villus tip cells, while ISCs remain intact17,75-77. RV-induced damage promotes canonical Wnt signaling activation in crypts leading to ISC proliferation and differentiation17. Mice received a single RV inoculation via oral gavage 7 days after tamoxifen induction, and intestines were isolated 4 days after infection at the peak of viral shedding (Figure 2Q)17. Fecal pellets were collected for ELISA to confirm rotavirus viral load on day 4 post-infection (Figure S2D). We collected whole mount tissue to assess RV-induced damage. We observed robust CC3 staining extending the length of villus tips in infected intestines (Figures 2R-U). As expected, infected cells were specifically localized to the tip of the villi and colocalized with the staining of CC3 (Figures 2V-X). Similar to trametinib, RV-induced Wnt signaling activation promoted the accumulation of dying cells in DLG1− crypts (Figures 2Y-e; Videos 2A-B). Unlike trametinib treatment, RV infection led to a significant decrease in the number of crypts of DLG1− intestines compared to controls (Figure 2f). In both trametinib and RV, the remaining crypts in DLG1− intestines had a similar crypt bottom area as controls (Figure 2g). Taken together, these results show that high levels of Wnt signaling trigger increased cell death in epithelial crypts lacking DLG1.
Elevated levels of canonical Wnt ligands promote rapid loss of DLG1− organoids
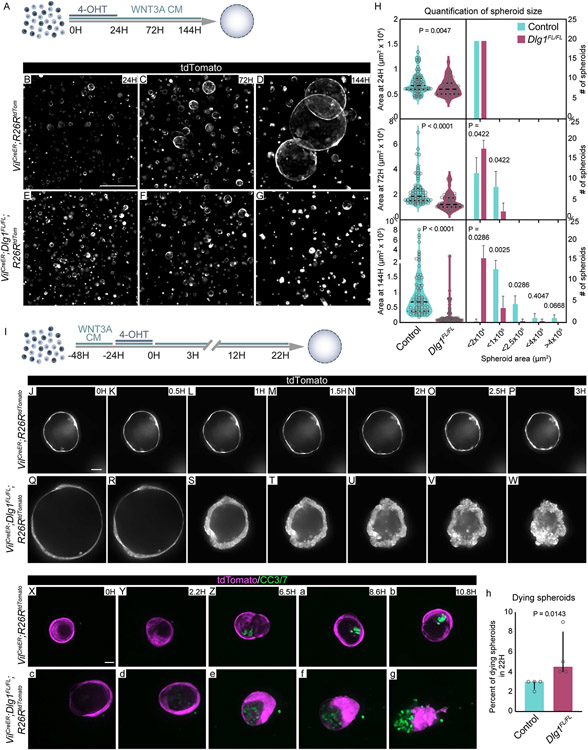
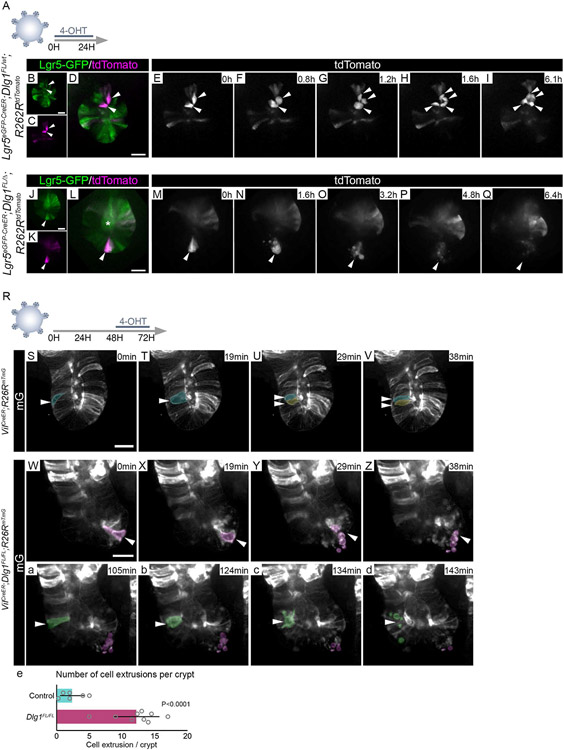
Intestinal organoids enable the functional dissection of how specific niche signals affect the epithelium. To determine if the increase in cell death in vivo was linked to the response of ISCs to Wnt signaling, we generated intestinal organoids from control and Dlg1FL/FL;tdTomato intestines and exposed them to exogenous WNT3A conditioned medium (WNT3A CM) (Figures 3A) or recombinant WNT3A (Figures S3I). Exogenous Wnt ligands in organoids mimic tissue regeneration, which is reflected by increased expression of fetal ISC markers like Lgr4, Birc5, Ly6a and Cnx43 (Figures S3A-D), and a sharp decrease of adult ISC markers Lgr5 and Olfm4 (Figures S3E-F)78-81. Loss of expression of Lgr5 and Olfm4 was accompanied by decreased expression of their regulator Ascl2 (Figure S3G). Additionally, after exogenous Wnt exposure, organoids morphologically change from budding crypts to spheroids that are composed of hyperproliferative ISCs11,79,82,83. Organoids were passaged by cell dissociation, then treated with 4-hydroxy-tamoxifen (4-OHT) to genetically ablate Dlg1 (Figure S3H), and concurrently exposed to 50% WNT3A CM (Figure 3A). Between 24 and 144 hours, control cells formed spheroids and continued growing, whereas the majority of DLG1− cells did not give rise to spheroids, and the few spheroids that formed remained significantly smaller than controls (Figures 3B-H). We observed the same behavior in DLG1− crypts when they were treated with 200 ng/ml of recombinant WNT3A (Figures S3I), demonstrating that treatment with WNT3A alone is sufficient to reveal this distinction between the wildtype and DLG1− organoids. (Figures S3J-P). Next, we analyzed the effect of increasing levels of Wnt CM, ranging from 0.5 to 20% on wildtype or DLG1− organoids. All tested concentrations supported spheroid growth of control organoids with no apparent difference between the size of the spheroids in 0.5% to 20% of WNT3A CM, suggesting that even low amounts of Wnt are sufficient to drive the regenerative response (Figures S4A-D, I-M). In contrast, the vast majority of DLG1− organoids could not grow as spheroids at any WNT3A CM concentration (Figures S4E-H, I-M), confirming that Dlg1 is critical for proper ISC response during increased levels of Wnt signaling.
Figure 3. Genetic deletion of Dlg1 leads to ISCs death in high WNT3A conditions.
(A) Experimental schematic for analyzing the ISC response to increased WNT3A levels. Organoids were dissociated into single cells that were treated with ENR medium containing 4-hydroxy-tamoxifen (4-OHT) and WNT3A conditioned medium (WNT3A CM). (B-G) Images of endogenous tdTomato expressing spheroids at (B and E) 24 hours, (C and F) 72 hours, and (D and G) 144 hours after plating. Scale bar = 500 μm. (H) Quantification of spheroid size by binning them according to the area at 24, 72, and 144 hours after plating. N = 4 organoid lines per condition. Area at all timepoints: median ± interquartile range, unpaired Mann-Whitney test. # spheroids at 72H: mean ± SD, unpaired t test with Welch’s correction. # spheroids at 144H: median ± interquartile range, unpaired Mann-Whitney test. (I) Experimental schematic for analyzing DLG1 requirement in control or DLG1− organoids exposed to increased WNT3A levels. Single cells were exposed to WNT3A CM for 24 hours to let them grow into spheroids. Then, spheroids were treated with 4-OHT for 24 hours and imaged for 3 to 22 hours. (J-W) Frames from time-lapse imaging of spheroids expressing tdTomato report. Scale bar = 50 μm. (X-g) Frames from time-lapse imaging of spheroids expressing tdTomato report and incubated with CC3/7 dye prior to and during live imaging. Scale bar = 20 μm. (h) Quantification of the number of dying spheroids. N = 4 organoid lines per condition, median ± interquartile range, unpaired Mann-Whitney test.
In addition to epithelial-derived WNT3A, ISCs are continuously exposed to mesenchymal-derived canonical and non-canonical Wnt ligands. We asked whether the impaired response of DLG1− ISCs is attributed solely to WNT3A or whether this phenotype can be observed with other Wnt ligands. Crypts from control and Dlg1FL/FL;tdTomato intestines were grown in conditioned medium of niche PDGFRαlo cells (PDGFRαlo CM), or 500 ng/ml of non-canonical recombinant WNT5A (Figures S4N). PDGFRαlo CM contains canonical WNT2B and WNT9A, and similarly to WNT3A CM promotes the rapid transformation of organoids into spheroids84. At 72 hours the majority of control crypts grew as spheroids (Figures S4O-P, S), while DLG1− crypts responded poorly, and only a fraction of seeded crypts initiated spheroid growth (Figures S4Q-S). Similar to WNT3A treatment, DLG1− spheroids were significantly smaller compared to controls (Figure S4T). However, in contrast to treatment with WNT3A or canonical ligands derived from PDGFRαlo cells (Figures S3P and S4T), we observed no significant difference in the number or size of spheroids that were formed between DLG1− and control crypts exposed to non-canonical WNT5A (Figure S4U-Z). In agreement with previous reports, WNT5A promoted short-term in vitro survival but not rapid growth as observed with canonical Wnts, and organoids eventually died after 5 days in culture (data not shown)15,85. Overall, these data indicate that DLG1 is required for the normal response of ISCs to high levels of multiple Wnt ligands, but the capacity to respond to non-canonical WNT5A is DLG1 independent.
Next, we explored if endogenous levels of WNT3A coming from Paneth cells are sufficient to phenocopy the loss of organoids observed in increased levels of WNT3A. Therefore, we established organoids from control Lgr5eGFP-CreERT2;Dlg1FL/WT;R26RtdTomato (Dlg1FL/WT;tdTomato) and mutant Lgr5eGFP-CreERT2;Dlg1FL/Δ;R26RtdTomato (Dlg1FL/Δ;tdTomato) mice that had been induced with a single dose of tamoxifen 14 days prior to organoid establishment (Figure S5A). The mosaic nature of Lgr5eGFP-CreERT2 3 results in the presence of both tdTomato−/DLG1+ and tdTomato+/DLG1− organoids within the same well. This enabled comparison of the fraction of live cells between tdTomato−/DLG1+ and tdTomato+/DLG1− organoids by flow cytometry (Figure S5B). In both Dlg1FL/WT;tdTomato and Dlg1FL/Δ;tdTomato organoids, the tdTomato+ cells persisted over 47 days (Figure S5C). Even though we observed a slow decline in numbers of tdTomato+/DLG1− cells, the decrease was negligible compared to the inability to grow observed in high Wnt. qPCR analysis of Dlg1FL/Δ;tdTomato organoids revealed that loss of Dlg1 expression did not affect the expression of major epithelial differentiation markers between tdTomato−/DLG1+ and tdTomato+/DLG1− organoids (Figure S5D-F). In summary, loss of DLG1− organoids positively correlates with the levels of WNT3A that they are exposed to and is not a consequence of impaired cell differentiation.
High Wnt conditions promote ISCs death in the absence of Dlg1
Based on our finding that single cells lacking Dlg1 fail to form spheroids when exposed to exogenous Wnt, we asked whether single cells expressing Dlg1 grow into spheroids and collapse only when they lose DLG1. To test this hypothesis, control and non-induced mutant single cells were grown in high Wnt conditions for 24 hours. Once spheroids formed, Dlg1 ablation was induced by a 24 hour pulse of 4-OHT, followed by a 3 or 22 hour imaging (Figure 3I). Both control and non-induced mutant single cells were able to grow into spheroids when exposed to WNT3A CM (Figures 3J and Q). However, DLG1− spheroids transitioned from thin-walled spheroids to progressively smaller, amorphous, and thick-walled structures, whereas the morphology of control spheroids was unperturbed (Figures 3J-W; Videos 3A-B). As DLG1− spheroids ultimately collapsed, cells that remained within the epithelium became more cuboidal than slender, and cells that had left the epithelium rounded up and accumulated on the basal and apical sides of the epithelium. Together, these events caused the visual effect of epithelial “wall” thickening.
Finally, we asked if the DLG1− spheroid growth phenotype was due to impaired proliferation and/or increased cell death. EdU incorporation demonstrated that DLG1− spheroids had no apparent proliferative defect (Figures S5G-N). To test for cell death, we live-imaged the spheroids for up to 12 hours in the presence of cleaved caspase-3/7 dye (CC3/7). Control organoids accumulated only small amounts of fluorescence coming from the CC3/7 reporter dye in the spheroid lumen over 12 hours (Figures 3X-b, and h; Video 4A). In contrast, DLG1− spheroids accumulated increasing amounts of CC3/7+ cells and eventually collapsed (Figures 3c-h; Video 4B). Together, the in vivo and in vitro data indicate that, instead of initiating a regenerative response following increased Wnt signaling, ISCs lacking DLG1 undergo cell death.
Transcription of CDC42 GTPase-activating protein (Arhgap31) is reduced following Dlg1 loss in ISCs
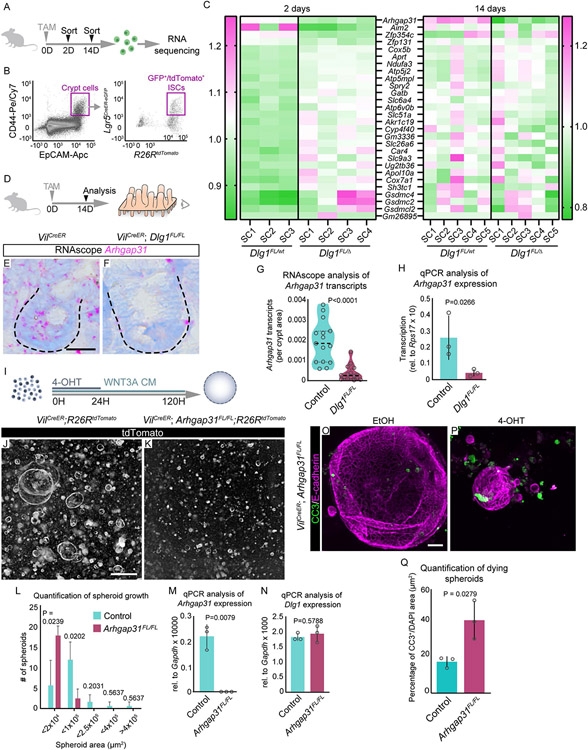
To determine the molecular mechanism responsible for increased epithelial apoptosis in response to WNT3A, we analyzed the transcriptional profile of the DLG1− ISCs. To delete Dlg1 in ISCs, Dlg1FL/WT;tdTomato and Dlg1FL/Δ;tdTomato mice were given 1 dose of tamoxifen. Recombined ISCs lacking Dlg1 were isolated by FACS (DAPI−/EpCAM+/CD44+/Lgr5-eGFP+/tdTomato+) and their actively transcribed genes were assessed by RNA sequencing. To identify changes intrinsic to ISCs as well as changes influenced by the DLG1− microenvironment, we analyzed ISCs at 2 and 14 days after induction (Gene Expression Omnibus GSE198573) (Figures 4A and B). Surprisingly, the only gene that was differentially expressed at both 2 and 14 days was Arhgap31, a CDC42 GTPase-activating protein (Figures 4C). We identified another 27 genes with differential expression in Dlg1FL/Δ ISCs at the 2 day time point, but none of them were significantly up- or down-regulated at 14 days (Figure 4C). At 14 days after tamoxifen induced Dlg1 loss (Figure 4D), the downregulation of Arhgap31 occurred throughout the the entire epithelium, as indicated by mRNA quantification using RNAscope and by qPCR of sorted intestinal epithelial cells (DAPI−/EpCAM+) (Figure 4E-H).
Figure 4. Arhgap31 expression is regulated by DLG1 in ISCs.
(A) Experimental schematic for identifying differentially expressed genes in DLG1− ISCs. Mice were injected with 1 dose of TAM, and 2 or 14 days later ISCs were isolated by FACS-sorting, and mRNA was sequenced. (B) Flow cytometry plot showing gating strategy for sorting specifically ISCs. (C) Heat map showing relative expression of genes at 2 and 14 days in control and DLG1− ISCs. Since Arhgap31 was the only differentially expressed gene at 14 days, to generate the 14 day heat map we used the remaining 27 genes that were identified to be differentially expressed at 2 days. Gene expression in DLG1− samples are displayed as a fraction of expression in DLG+ normalized across all samples. SC: stem cells isolated from individual mice. (D) Experimental schematic for analyzing Arhgap31 transcripts in intestinal crypts. Mice were injected with one dose of TAM two weeks prior to sacrifice. (E and F) RNAscope images of Arhgap31 expression in (E) DLG1+ and (F) DLG1− crypts. Scale bar = 25 μm. (G) Quantification of Arhgap31 transcripts in crypt regions normalized to crypt areas. N = 3 x 5 crypts per condition, median ± interquartile range, unpaired Mann-Whitney test. (H) Transcription levels of Arhgap31 in intestinal epithelial cells analyzed by qPCR. N = 3 mice per condition, mean ± SD, unpaired t test with Welch’s correction. (I) Experimental schematic for analyzing the ISC response to increased WNT3A levels in ARHGAP31− spheroids. (J and K) Images of endogenous tdTomato expressing (J) ARHGAP31+ and (K) ARHGAP31− spheroids grown in the presence of WNT3A CM for 120 hours. Scale bar = 500 μm. (L) Quantification of spheroid by binning them according to size at 120 hours. N = 3-4 organoid lines, per condition, mean ± SD, unpaired t test with Welch’s correction. (M and N) Transcription levels of Arhgap31 and Dlg1 analyzed by qPCR on FACS-sorted live intestinal organoids (DAPI−). N = 3, per condition, mean ± SD, unpaired t test with Welch’s correction. (O and P) Immunofluorescence images of anti-CC3 and anti-E-cadherin stained spheroids treated with (O) ethanol (EtOH) or (P) 4-OHT. Scale bar = 50 μm. (Q) Quantification of the number of dying spheroids. N = 3 organoid lines per condition, mean ± SD, unpaired t test with Welch’s correction.
Next, we asked if the phenotype observed in DLG1− organoids following exposure to exogenous WNT3A was driven by the transcriptional downregulation of Arhgap31 in DLG1− ISCs. We induced recombination of VilCreERT2;Arhgap31FL/FL;R26RtdTomato (Arhgap31FL/FL;tdTomato) organoids and treated them with WNT3A CM for 6 days (Figure 4I). Similar to DLG1− cells, ARHGAP31− cells responded poorly to exogenous WNT3A and failed to form viable spheroids (Figures 4J-L). Importantly, deletion of Arhgap31 did not affect the expression of Dlg1 (Figures 4M-N), suggesting that DLG1 acts upstream of ARHGAP31. Additionally, we found that under high Wnt conditions, ARHGAP31− spheroids underwent cell death, phenocopying DLG1− spheroids (Figures 4O-Q).
To analyze the dynamics of ARHGAP31− organoids in media containing only Paneth cell-derived WNT3A, we established organoids from control and Arhgap31FL/FL;tdTomato, and treated them with ethanol (EtOH) or 4-OHT for 48 hours. Following passaging, tdTomato−/ARHGAP31+ and tdTomato+/ARHGAP31− crypts were mixed together, and the ratio of tdTomato− and tdTomato+ cells was quantified by flow cytometry for 5 passages (Figures S6A). As with tdTomato+/DLG1− organoids (Figure S3Q), tdTomato+/ARHGAP31− organoids (Figure S6B) persisted over 40 days in culture (Figure S6C). Taken together, these data show that loss of Dlg1 reduces Arhgap31 expression and that DLG1 acts upstream of ARHGAP31, which is important when levels of Wnt signaling are increased.
Reduced ARHGAP31 expression leads to activation of CDC42, which negatively impacts ISC migration and cell division
ARHGAP31 deactivates a downstream activator of the Wnt-PCP pathway, CDC42, by promoting hydrolysis of GTP86,87. Since we found that Arhgap31 expression was reduced in DLG1− intestinal epithelium, we hypothesized that CDC42 activity would be elevated in DLG1− ISCs. To test this hypothesis, we established and treated control and Dlg1FL/FL organoids with 4-OHT and measured the levels of active CDC42 in sorted cells (Figure 5A). In order to obtain a sufficient number of DLG1− cells for quantification of CDC42-GTP, we used organoids instead of spheroids, because DLG1− spheroids have a rapid decline in cell numbers caused by cell death. Using the G-LISA assay to measure the active GTP-bound form of CDC42 in FACS-sorted DLG1+ and DLG1− cells, we found that the intracellular level of active GTP-CDC42 was significantly increased in DLG1− organoids compared to DLG1+ organoids (Figure 5B). Interestingly, the intracellular level of active RAC1, another target of ARHGAP3186, was not altered in DLG1− cells compared to control organoids (Figure 5C).
Figure 5. Loss of Dlg1 results in increased levels of activated CDC42 followed by impaired cell migration.
(A) Experimental schematic for analyzing the active forms of CDC42-GTP and RAC1-GTP in organoids. Organoids were mechanically disrupted into crypts that were treated with ENR medium containing 4-OHT. After 5 days, organoids were then enzymatically dissociated into live single cells (DAPI−) that were FACS-sorted and processed for analyzing CDC42-GTP and RAC1-GTP levels by G-LISA. (B) Quantification of CDC42-GTP levels in DLG1+ and DLG1− organoids. N = 4 organoid lines per condition, mean ± SD, unpaired t test with Welch’s correction. (C) Quantification of RAC1-GTP levels in DLG1+ and DLG1− organoids. N = 4 organoid lines per condition, mean ± SD, unpaired t test with Welch’s correction. (D) Experimental schematic for analyzing the migration capacity of 2D organoid monolayers assessed by scratch wound assay. Organoids were mechanically disrupted into crypts that were plated on Matrigel coated 24-well plates and treated with ENR medium containing 4-OHT and WNT3A CM, and grown for 48 hours. After reaching confluency, the center of the well was scratched with a P200 tip and organoids were imaged for 30 hours to record the speed of migration in the direction of the scratch. (E and F) Transcription levels of (E) differentiated cell markers and (F) Wnt target genes analyzed by qPCR from FACS-sorted live (DAPI−) 2D monolayers established from control and DLG1− mice. N = 4 organoid lines per condition, mean ± SD, one-way ANOVA with Tukey’s multiple comparisons test. (G-L) Frames from time-lapse imaging of 2D organoid monolayers expressing mG. Arrowheads above the images indicate the leading edges of the migrating 2D organoids. Scale bar = 200 μm. (M) Quantification of average speed at which organoids migrated in the direction of the scratch/wound. N = 3-4 organoid lines per condition, mean ± SD, unpaired t test with Welch’s correction. (N and O) Immunofluorescence images of EdU stained 2D organoid monolayers at 12 hours post-scratch/wound. Scale bar = 50 μm. (P) Quantification of EdU. N = 4 organoid lines per condition, mean ± SD, unpaired t test with Welch’s correction. (Q and R) Immunofluorescence images of anti-CC3 stained 2D organoid monolayers at 12 hours post-scratch/wound. Scale bar = 50 μm. (S) Quantification of CC3. N = 4 organoid lines per condition, mean ± SD, unpaired t test with Welch’s correction. Nuclei counterstained with DAPI.
ARHGAP31, through its regulation of CDC42 activity, is involved in cellular motility, migration, and proliferation88. In particular, high levels of ARHGAP31 are oncogenic, promoting cell migration and invasion89. To examine if loss of DLG1 with subsequent reduction of ARHGAP31 and increased active CDC42 affects ISC cellular behaviors such as migration, we evaluated ISC motility and migration using a scratch wound assay. To simplify the 3D architecture of spheroids and facilitate the analysis of ISC migration, we cultured VilCreERT2;R26RmTmG or VilCreERT2;Dlg1FL/FL; R26RmTmG (Dlg1FL/FL;mTmG) intestinal monolayers90 in the presence of WNT3A CM and 4-OHT (Figure 5D). WNT3A treatment of 2D monolayers results in the depletion of differentiated cells as quantified by qPCR (Figure 5E). As with spheroids, 2D monolayers expressed a genetic profile associated with regeneration and fetal ISCs (Figure 5F). After the cells reached confluence, we scratched the ISC monolayers to create a wound spanning the entire culture plate and measured the time it took to close the wound. mG+/DLG1+ ISC monolayers closed the wound at 30 hours after scratching (Figures 5G-I; Video 5A), whereas mG+/DLG1− ISC monolayers failed to close the wound (Figures 5J-L; Video 5B). On average, control mG+/DLG1+ ISC monolayers moved at 27 μm/hr, and mutant mG+/DLG1− ISC monolayers were significantly slower, at 13 μm/hr (Figure 5M). To determine if the difference in migration was caused by different rates of proliferation or cell death in the confluent 2D cultures, we quantified EdU+ and CC3+ cell number in control and DLG1− cultures 12 hours after the scratch. No difference was observed in the number of proliferating and dying cells between control and DLG1− cultures (Figures 5N-S), supporting the idea that DLG1− ISCs failed to close the wound due to impaired migration.
CDC42 is a well-known regulator of various cellular functions, including motility, cytoskeletal rearrangement, and proliferation91,92. In the intestine, deletion of CDC42 causes stem and TA cell hyperproliferation and crypt hyperplasia93. We performed live imaging of DLG1− organoid crypts to test if active CDC42 impacts cell division. Using Lgr5eGFP-CreERT2 to delete Dlg1 in ISCs after 24 hours of induction (Figure 6A), we found that cell divisions in control ISCs progressed to completion (Figures 6B-I; Videos 6A and B). However, LGR5+ ISCs lacking DLG1 rounded up, and their cell membrane blebbed (Figure 6J-Q; Videos 6C and D). To better characterize the cellular behavior in DLG1− crypts, we used the mTmG membrane reporter, which allows precise tracking of individual cells. Prior to recombination, Dlg1FL/FL;mTmG cells were able to divide akin to their control counterparts (Videos 7A and B). After a 24 hour pulse of 4-OHT resulting in mosaic deletion of Dlg1 and mG reporter expression, we followed individual cell divisions (Figure 6R). During crypt cell division, mitotic cells round up, progressively moving towards the apical surface of the epithelium where cytokinesis takes place (Figures 6S-U; Video 7C). As mitosis ends and interphase begins, daughter cells elongate, occupying the entire apical-basal axis of the epithelium94 (Figure 6V; Video 7C). In contrast to cell division in recombined control mG+/DLG1+ cells, recombined mutant mG+/DLG1− dividing cells rounded up but failed to complete mitosis, resulting in blebbing, death, and extrusion (Figures 6W-e; Video 7D). Together, these findings indicate that loss of Dlg1 leads to elevated CDC42-GTP in ISCs, impairing cell motility and cell division.
Figure 6. ISCs fail to divide and undergo cell death in the absence of Dlg1.
(A) Experimental schematic for analyzing the cell division of ISCs. Organoids grown in ENR medium for 48 hours were treated with 1 μM 4-OHT for 24 hours prior to live imaging. (B-Q) Frames from time-lapse imaging of organoid crypts expressing tdTomato and Lgr5-GFP. (B-I) Recombined tdTomato+/Lgr5-GFP+ ISCs dividing in DLG1+ organoids. (J-Q) Recombined tdTomato+/Lgr5-GFP+ ISCs failing to divide in DLG1− organoids. Arrowheads follow the fate of recombined ISCs. Asterisk indicates background signal in crypt lumen. Scale bars = 20 μm. (R) Experimental schematic for analyzing the cell division of organoid crypt cells. Organoids grown in ENR medium for 48 hours were treated with 0.25 μM 4-OHT for 24 hours prior live imaging. (S-d) Frames from time-lapse imaging of organoid crypts mosaically expressing mG. Actively dividing cells highlighted in pseudo-colors and indicated by arrowheads. Scale bars = 20 μm. (S, W, a) Dividing cell (arrowhead) is still attached to the basal side and aligned with other cells in the crypt. (T, X, b) Dividing cell (arrowhead) is rounded up and detached from the basal side of the crypt, undergoing cell division. (U, V) Newly divided daughter cells (arrowheads) in DLG1+ crypt. (Y, Z, c, d) Cell blebbing and fragmented cell debris in DLG1− crypt with no daughter cells post-mitosis. (e) Quantification of cell extrusion events per crypt. N = 2-3 organoid lines per condition, mean ± SD, unpaired t test with Welch’s correction.
DISCUSSION
The small intestine has the fastest cell turnover rate of all mammalian epithelia5; as such, its renewal relies heavily on niche signals that ensure proper stem cell function. The niche, or cellular microenvironment, provides factors that guide stem cell behavior, including proliferation, differentiation, and polarity95. Throughout the life cycle of an organism, niche signals change in activity levels due to external stimuli or diseases, and stem cells need to respond accordingly to fulfill tissue renewal, but how stem cells interpret fluctuating niche signals is not clear. Here, we have elucidated cellular and molecular mechanisms that ISCs use to respond to increased levels of canonical Wnt ligands, which are crucial niche signals for ISCs96,97.
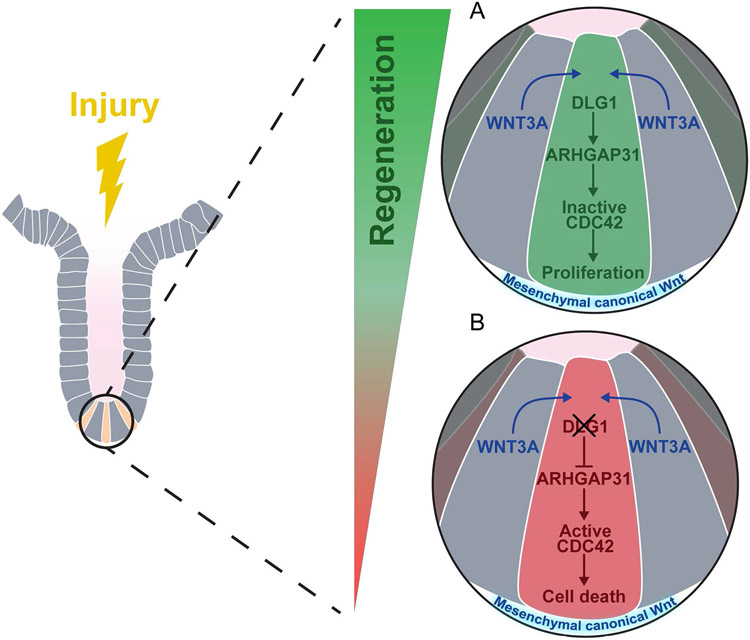
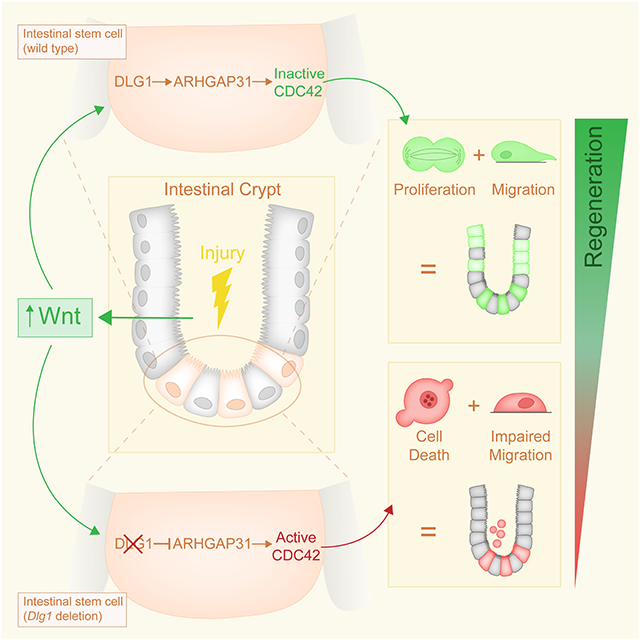
ISCs depend on canonical Wnt signaling for homeostatic self-renewal19. During intestinal injury, increased canonical Wnt signaling is involved in epithelial regeneration 17,25-30, whereas aberrant activation of the pathway causes colorectal cancer 31. We found that canonical WNT3A participates in the non-canonical PCP pathway via a DLG1-ARHGAP31-CDC42 molecular axis. DLG1 in ISCs is required for a proper response to high WNT3A during regeneration by regulating Arhgap31 transcription. Increased ARHGAP31 deactivates CDC42, allowing ISC proliferation required for regeneration (Figure 7A). In the absence of DLG1, regenerative levels of WNT3A become detrimental. Reduced ARHGAP31 causes accumulation of CDC42-GTP, culminating in failed cell division and death (Figure 7B). Furthermore, we confirmed that other canonical Wnt ligands, but not the non-canonical WNT5A ligand, mimic the response of WNT3A.
Figure 7. Model of ISC response to canonical WNT ligands via DLG1-ARHGAP31-CDC42 molecular axis.
Intestinal epithelial regeneration relies on the ISC response to increased levels of Wnt signaling. (A) DLG1 is dispensable during homeostasis when levels of Wnt signaling are low. When levels of canonical Wnt are increased in the crypt, DLG1 mediates the cellular response by regulating the expression of GTPase-activating protein Arhgap31, a negative regulator of CDC42 activity, and low levels of active CDC42 promote crypt regeneration114. (B) Deletion of DLG1 in ISCs and crypt cells results in decreased expression of Arhgap31. Due to low levels of ARHGAP31, cells accumulate active CDC42, which impairs the capacity of ISCs to undergo cell division following increased Wnt signaling and ultimately leads to cell death.
The WNT1 family of ligands, including WNT3A, are components of the canonical Wnt/β-catenin signaling pathway that are ubiquitous promoters of cell proliferation98. The non-canonical Wnt/PCP signaling pathway is activated by the WNT5A family of ligands that control polarization of tissues, cell motility, mitosis, and cytoskeletal organization in both healthy and cancerous tissues99-102. Crosstalk between canonical and non-canonical Wnt pathways can regulate various cytoskeletal-dependent cellular behaviors in development and homeostasis, such as migration and adhesion103-107. In particular, WNT3A can activate non-canonical signaling in osteoblasts, chondrocytes, mesenchymal stem cells, and colon cancers108-111. Our findings demonstrate an intersection between the canonical WNT3A ligand and non-canonical Wnt/PCP output and suggest a previously unrecognized mechanism that underlies ISC-driven intestinal regeneration, and we propose that ISCs, once exposed to upregulated WNT3A, require DLG1 for replication. In the context of Dlg1 loss, Wnt/β-catenin target genes in ISCs are transcriptionally unchanged compared to ISCs in control intestines that are exposed to normal Wnt levels. Additionally, in DLG1− ISCs, the only differentially expressed gene at both 2 and 14 days was Arhgap31, which regulates the non-canonical Wnt PCP pathway. Together, these findings raise the intriguing possibility that WNT3A can simultaneously activate canonical and non-canonical Wnt pathways in ISCs.
Prior studies in organisms ranging from Drosophila to mammals have shown the essential role of DLG1 as a member of the Scribble protein complex in maintaining apicobasal polarity of epithelial cells112. However, we found that, after conditional deletion of Dlg1 in the homeostatic adult small intestine, crypts exhibited normal polarity protein localization and cell proliferation. Furthermore, the distance of cell displacement, used as a measure of the speed by which older cells are pushed along the crypt-villus axis by new daughter cells, is unchanged in DLG1− crypts. From these results, we conclude that cell polarity and polarity-dependent phenotypes like cell division are not affected by the loss of Dlg1 under homeostatic Wnt signaling regimes. This may be due to redundancy between Dlg vertebrate paralogs that is sufficient for the maintenance of adult intestinal epithelium cell polarity in mammals or because polarity is controlled by other factors such as the integrin adhesion complex. Our data complement a study in Drosophila showing that, unlike other fly epithelia, midgut cell polarity is controlled by factors other than the classical Scribble polarity complex 47. In addition, a prior report demonstrated that Dlg1 deletion at embryonic day 9 did not affect polarity in adult mouse intestinal epithelium in vivo and in vitro51. However, our results contrast with the latter report regarding delayed cell displacement from crypts. These conflicting results may be due to the constitutive genetic system used in the previous report versus our conditional genetic system.
Our data support a model in which ISC proliferation and motility during regeneration is under the control of CDC42 inactivation by ARHGAP31 (Figure 7). Consistent with this model, ARHGAP31 has been identified as a GAP for CDC42, and CDC42 is a critical regulator cell motility86,88. Furthermore, depletion of ARHGAP31 in prostate cancer cells significantly reduced their cytoskeletal-dependent phenotypes, like migration and invasion89. This molecular model may be a conserved mechanism of non-canonical Wnt directed cell migration, as it has been shown that non-canonical Wnt signaling promotes ISCs movement to sites of injury in the Drosophila intestine113. Aging ISCs have elevated CDC42 activity and reduced regenerative capacity114. Experimentally, an increase of activated CDC42 accelerated ISC aging, while decrease of CDC42 activity enhanced the regenerative capacity of aged ISCs114. This raises the intriguing possibility that the molecular mechanism that we describe during regeneration is impaired in aging ISCs.
A key finding from our live imaging is that Dlg1− ISCs undergo cell death while they try to divide. We observed that mutant cells die at the apical surface of the epithelium, suggesting that failed apical mitosis triggers cell extrusion and death. In C. elegans embryos, S phase arrest during cell cycle ends in cell elimination via extrusion115, suggesting that cell death promoted by cell division stress is an evolutionarily conserved mechanism. Nonetheless, it is unclear if the observed cell division failure is a direct consequence of loss of DLG1 at the midbody during cytokinesis116, or a CDC42-dependent cytoskeletal rearrangement117-119, or both. A parallel molecular axis in the Wnt PCP pathway that phenocopies our observations in cell replication, survival, and motility is RhoA-ROCK. Genetic deletion of RhoA results in cytokinesis failure, leading to cell cycle arrest culminating in cell death. Furthermore, RhoA deletion impairs cellular chemotaxis120. It remains to be explored whether DLG1 can function via the RhoA-ROCK axis and whether decreased motility and increased cell death resulting from loss of Dlg1 contribute to impaired regeneration by independent mechanisms.
In summary, our results demonstrate that ISCs respond to fluctuating signaling pathways and reveal a link between canonical Wnt ligands and a non-canonical Wnt response, providing further insights into the niche-stem cell interaction.
LIMITATIONS OF THE STUDY
Our experiments focused on ISCs, but it is possible that the WNT3A-DLG1-ARHGAP31-CDC42 molecular axis described here is not exclusive to ISCs. For example, this mechanism may also control the more rapidly cycling TA cells, which are exposed to Wnt signals coming from Paneth cells and underlying mesenchyme.
In addition, the scant number of reliable readouts of active Cdc42 and of genetic tools for manipulating its inactive and active states in mice makes it difficult to conduct a deeper dissection of intestinal Cdc42 in the non-canonical Wnt pathway. Thus, it is of particular interest and importance to generate a more robust toolkit for the study of Cdc42, including conditional alleles of the active form of Cdc42, to further explore its role in homeostasis, regeneration, and disease.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents may be directed to, and will be fulfilled by, the lead contact, Ophir Klein (ophir.klein@ucsf.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Bulk RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-CD45, clone 30-F11 | Biolegend | Cat#: 103133 RRID: AB_10899570 |
| Rat monoclonal anti-EpCAM, clone G8.8 | Biolegend | Cat#: 118211 RRID: AB_1134104 |
| Rat monoclonal anti-CD44, clone IM7 | Biolegend | Cat#: 103029 RRID: AB_830786 |
| Mouse anti-Dlg1, Clone 12/Dlg | BD Biosciences | Cat#: 610874 RRID: AB_398192 |
| Mouse monoclonal anti-BrdU | Novus Biologicals | Cat#: NB500-439 RRID:AB_10000514 |
| Rabbit monoclonal anti-E-cadherin | Cell Signaling Technology | Cat#: 3195S RRID:AB_2291471 |
| Rat monoclonal anti-β4-integrin | Abcam | Cat#: ab25254 RRID:AB_2129042 |
| Mouse monoclonal anti-Ezrin | Invitrogen | Cat#: MA5-13862 |
| Rabbit monoclonal anti-RFP | Rockland | Cat#: 200-301-379 RRID:AB_2611063 |
| Rabbit polyclonal anti-cleaved caspase-3 | Cell Signaling | Cat#: 9661 RRID:AB_2341188 |
| Rabbit polyclonal anti-Mucin2 | Novus Biologicals | Cat#: NBP1- 31231 RRID: AB_10003763 |
| Rabbit polyclonal anti-Chromogranin A | Abcam | Cat#: ab45179 RRID:AB_300798 |
| Rabbit polyclonal anti-DCAMKL1 | Abcam | Cat#: ab31704 RRID:AB_873537 |
| Rabbit polyclonal anti-Lysozyme | Dako | Cat#: A0099 RRID:AB_2341230 |
| Bacterial and virus strains | ||
| Rotavirus strain ECWT (P[17], G3) | Mary Estes | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| EdU: 5-ethynyl-2'-deoxyuridine | ThermoFisher | Cat#: C10637 |
| BrdU: 5-Bromo-2'-deoxyuridine | Sigma-Aldrich | Cat#: B5002 |
| Trametinib | ApexBio | Cat#: A3018 |
| Tamoxifen | Sigma-Aldrich | Cat#: T5648 |
| 4-hydroxytamoxifen | Sigma-Aldrich | Cat#: H7904 |
| TryplE | Gibco | Cat#: 12604039 |
| Matrigel | Corning | Cat#: 356231 |
| WNT3A | PeproTech | Cat#: 315-20-10ug |
| WNT5A | R&D Systems | Cat#: 645-WN |
| Nicotine amide | Sigma-Aldrich | Cat#: 72340 |
| Collagenase/Dispase | Roche | Cat#: 10269638001 |
| 293T-HA-RSPO1-FC cells | Trevigen | Cat#: 3710-001-01 |
| L-WNT3A cells | ATCC | Cat#: CRL-2647 |
| Critical commercial assays | ||
| Alkaline Phosphatase Red Substrate Kit | Vector Laboratories | Cat#: SK-5100 |
| Click-iT™ Plus EdU Cell Proliferation Kit for Imaging | Thermo Fisher | Cat#: C10637 |
| CellEvent™ Caspase-3/7 Green ReadyProbes™ Reagent | Thermo Fisher | Cat#: R37111 |
| RNAscope® 2.5 High Definition (HD) – Red Assay | Advanced Cell Diagnostics | Cat#: 322350 |
| G-LISA activation assay kit for Cdc42 | Cytoskeleton | Cat#: BK127 |
| G-LISA activation assay kit for Rac1 | Cytoskeleton | Cat#: BK128 |
| polyA Dynabeads mRNA direct Kit | Invitrogen | Cat#: 61012 |
| SmartSeq DNA library preparation Kits | Takarabio | Cat#: 634471 |
| NexteraXT DNA library preparation Kits | Illumina | Cat#: FC-131-1024 |
| RNeasy Mini Kit | Qiagen | Cat#: 74104 |
| High-capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat#: 4368814 |
| iTaq Universal SYBR Green Supermix | Bio Rad | Cat#: 1725121 |
| Animal-free blocker | Vector Laboratories | Cat#: SP-5030 |
| ProLong Gold Antifade | Thermo Fisher | Cat#: P36930 |
| Deposited data | ||
| Gene Expression Omnibus | This paper | GSE198573 |
| Experimental models: Organisms/strains | ||
| Mouse: Dlg1min | Todd Nystul | Rivera et al., 2009 |
| Mouse: Arhgap31flox | Nathalie LaMarche Vane | Caron et al., 2016 |
| Mouse: Dlg1flox: B6;129-Dlg1tm1Rlh/J | JAX | Strain #:013097 RRID:IMSR_JAX:013 097 |
| Mouse: Lgr5GFP-CreERT2: B6.129P2-Lgr5tm1(cre/ERT2)Cle/J | JAX | Strain #:008875 RRID:IMSR_JAX:008 875 |
| Mouse: VillinCreERT: B6.Cg-Tg(Vil1cre/ERT2)23Syr/J | JAX | Strain #:020282 RRID:IMSR_JAX:020 282 |
| Mouse: ROSA26tdTomato: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | JAX | Strain #:007914 RRID:IMSR_JAX:007 914 |
| Mouse: ROSA26mTmG: B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | JAX | Strain #:007676 RRID:IMSR_JAX:007 676 |
| Mouse: PdgfrαH2B-eGFPB6.129S4-Pdgfratm11(EGFP)Sor/J | JAX | Strain #:007669 RRID:IMSR_JAX:007 669 |
| Oligonucleotides | ||
| RNAscope® probe Mm-Arhgap31 | Advanced Cell Diagnostics | Cat#: 569971 |
| Oligonucleotides | Table S1 | N/A |
| Software and algorithms | ||
| Fiji v2.1.0/1.53c | Open source | https://imagej.net/software/fiji |
| GraphPad Prism9 | GraphPad Software | https://www.graphpad.com |
| Adobe Illustrator v25.4 | Adobe | https://www.adobe.com |
| Adobe Photoshop | Adobe | https://www.adobe.com |
| QuantStudio Real-time PCR software v1.3 | Applied Biosystems | https://www.thermofisher.com |
| LasX v3.4.2 | Leica | https://www.leicamicrosystems.com |
| ZEN Blue v2.5 | Zeiss | https://www.zeiss.com |
| STAR 2.4.2a | Dobin et al., 2013 | https://code.google.com/archive/p/rna-star/ |
| DESeq2 v1.16.1 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| BD FACS Diva v.8.0.1 | BD Biosciences | https://www.bdbiosciences.com/ |
| FlowJo v10.8 | Tree Star | https://www.flowjo.com |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All experimental procedures involving mice were done in accordance with approved protocols by the Institutional Animal Care and Use Committee (IACUC) and Laboratory Animal Resource Center (LARC) at University of California San Francisco, and the mice were handled in accordance with the principles and procedures of the Guide for the Care and Use of Laboratory Animals under the approved protocol AN180876. All mouse strains were maintained on a predominantly C57BL/6 background. Animals were housed under pathogen-free conditions in 12-hour light/dark cycles at 23 ± 1°C and humidity 55 ± 15%. Food and water were provided ad libitum. Animals were weaned 21 to 28 days after birth and handled and euthanized according to procedures approved by LARC of the University of California San Francisco. Mice were at least 8 weeks old at the time of experiments and cell isolations. Mice of the both sexes were used in all experiments, and they were either randomly assigned to the experimental groups or used with littermate controls. The Dlg1min (i.e. Dlg1δ)66 and Arhgap31flox 52 alleles were previously described. Mice harboring Dlg1flox (Jax: 013097121), Lgr5GFP-CreERT2 (Jax 0088753), VillinCreERT2 (Jax: 02028255), ROSA26tdTomato (Jax 007905122), ROSA26mTmG (Jax: 007676123), and PdgfrαH2B-eGFP (JAX: 007669124) alleles were purchased from The Jackson Laboratory.
METHOD DETAILS
Animal treatments
Mice were given an intraperitoneal injection of 1 mg/25 g body weight of 5-ethynyl-2'-deoxyuridine (EdU; ThermoFisher, C10637) and then analyzed at various timepoints after treatment (exact chase timepoints are provided in the main text). Mice received intraperitoneal injection of 1 mg/25 g body weight of 5-Bromo-2'-deoxyuridine (BrdU, Sigma-Aldrich, B5002-250MG) 24 hours before analysis. Mice received two intraperitoneal injections of trametinib at a dose of 2 mg/kg of body weight and intestinal tissue was analyzed 24 hours after the second administration. Trametinib was dissolved in sterile DMSO at 6.15 mg/ml and diluted in sterile 1:1 mixture PBS:DMSO to final concentration 500 μg/ml prior administration. Mice were given an intraperitoneal injection of 2.5 mg/25 g body weight of tamoxifen (Sigma-Aldrich, T5648-5G) dissolved in corn oil (Sigma-Aldrich, C8267) at concentration 25 mg/ml. Rotavirus strain ECWT (P[17], G3) was prepared and administered by oral gavage as described previously125. All animals were housed in a physically separated BSL-2 animal facility. A 1×105 50% infectious dose (ID50) was used to obtain adequate infection125. To monitor rotavirus infection, fecal samples were collected and fecal ELISA was used as described previously125. Mice were sacrificed 4 days following infection at the peak of viral shedding.
Antibodies
For flow cytometry of intestinal epithelium, organoids, and PDGFRαlo cells, the following antibodies were used: rat anti-CD45 (BioLegend, 30-F11; 1:100), rat anti-EpCAM (BioLegend, G8.8; 1:200), rat anti-CD44 (BioLegend, IM7; 1:100). Nuclei were counterstained with DAPI (stock concentration 5 mg/ml, Sigma-Aldrich, D9542; 1:1,000 or 10,000).
For immunofluorescence staining of sections and organoids, the following antibodies were used: mouse anti-Dlg1 (BD Biosciences, 610874; 1:200), anti-BrdU (Novus Biologicals, NB500-439; 1:100), rabbit anti-E-cadherin (Cell Signaling Technology, 3195S; 1:200), rat anti-β4-integrin (Abcam, ab25254; 1:200), mouse anti-Ezrin (Invitrogen, MA5-13862; 1:200), rabbit anti-RFP (Rockland, 200-301-379; 1:100), rabbit anti-cleaved caspase-3 (Cell Signaling, 9661; 1:200), rabbit anti-Mucin2 (Novus Biologicals, NBP1-31231; 1:100), rabbit anti-Chromogranin A (Abcam, ab45179; 1:100), rabbit anti-DCAMKL1 (Abcam, ab31704; 1:100), rabbit anti-Lysozyme (Dako, A0099; 1:500), Alkaline Phosphatase Red Substrate Kit (Vector Laboratories, SK-5100), rabbit anti-Histone H4K16ac (Active Motif, 39068; 1:100). EdU was detected using Click-iT™ Plus EdU Cell Proliferation Kit for Imaging, Alexa Fluor™ 488 dye (ThermoFisher, C10637). Caspase-3/7 was detected using CellEvent™ Caspase-3/7 Green ReadyProbes™ Reagent (ThermoFisher, R37111). Nuclei were counterstained with DAPI (1:10,000).
Tissue preparation for immunofluorescence staining and RNAscope
For immunofluorescence staining, harvested intestinal tissue was perfusion fixed. For perfusion fixation, animals were anesthetized by intraperitoneal injection of 250 mg/kg of body weight avertin (2,2,2-tribromoethanol) and transcardially perfused with 4% paraformaldehyde (PFA) in 100 mM PBS. Dissected tissues were post-fixed in 4% PFA for 4 hours at 4°C and cryoprotected in 30% sucrose in 1× PBS overnight at 4°C. Tissue was embedded in OCT compound (Sakura, 4583), frozen, and stored at −80°C. For paraffin-embedded tissues, we post-fixed tissues with 4% PFA in 100 mM PBS for 24 hours at 4°C, followed by paraffin processing, using a standard protocol. For RNAscope in situ analysis, freshly isolated intestinal tissue was immersed into 4% PFA for 24 hours at room temperature, followed by standard dehydration and paraffin embedding protocol.
Immunofluorescence staining of intestinal tissue
Immunofluorescence was performed on 7 μm cryosections or paraffin sections. Cryosections were washed with PBS, blocked with 0.3% Triton X-100 in PBS supplemented with 5% normal goat serum. Paraffin sections were rehydrated, and antigen retrieval was performed by sub-boiling slides in a pressure cooker for 30 min in a citrate buffer (pH 6.2) containing 10 mM citric acid, 2 mM EDTA and 0.05% Tween-20. For BrdU immunostaining, samples were additionally washed with 2 N HCl for 30 min. Paraffin sections were blocked in 1× animal-free blocker (Vector Laboratories, SP-5030) supplemented with 2.5% heat-inactivated goat serum, 0.02% SDS and 0.1% Triton X-100. All of the antibodies were diluted in the same blocking solution without serum and sections were incubated with primary antibodies at 4°C for 12 to 16 hours. Appropriate secondary antibodies from Thermo Fisher Scientific were used at 1:1,000 dilution. Nuclei were counterstained with DAPI. Cryosections and paraffin sections were coverslipped with ProLong Gold Antifade (Thermo Fisher Scientific, P36930).
Quantification of Arhgap31 transcripts using RNAscope
RNA in situ hybridization for Arhgap31 expression was performed on 7 μm paraffin sections using RNAscope® 2.5 High Definition (HD) – Red Assay (Advanced Cell Diagnostics, 322350). Manufacturer’s protocol was followed with 15 min of target retrieval and 30 min of protease digestion, using the RNAscope® probe Mm-Arhgap31 (Advanced Cell Diagnostics, 569971). Quantification of Arhgap31 mRNA transcripts was performed using the open-source platform Fiji126 and the analysis guidelines from Advanced Cell Diagnostics. Area of single probes was measured and used to determine total probe count within probe clusters. Total probe clusters containing at least 10 probes were quantified and normalized to crypt area.
Organoid culture media
Complete organoid 1× ENR medium was prepared from 1× Advanced DMEM/F12, 10 mM HEPES, 2 mM Glutamax, 0.11 mg/ml Penicillin-Streptomycin antibiotics, 1 mM N-Acetylcysteine, 50 ng/ml hEGF, 100 ng/ml Noggin, 2% B-27 Supplement (Thermo Fisher Scientific, 17504044), 1% N-2 supplement (Thermo Fisher Scientific, 17502048) and 5% R-Spondin1 conditioned medium (RSP01 CM). RSPO1 CM was prepared by harvesting conditioned medium from cultured 293T-HA-RSPO1-Fc cells (Trevigen, 3710-001-01) as described (R&D Systems Protocol; https://resources.rndsystems.com/images/site/dw_r-spondinmediumprotocol_34749-web.pdf?v=1). Aliquots of 10 ml were stored at −80°C and were thawed prior mixing the ENR medium. WNT3A conditioned medium (WNT3A CM) was generated by harvesting conditioned medium from cultured L-WNT3A cells (ATCC CRL-2647) at 1× dilution in Advanced DMEM/F12 (ThermoFisher, #11320-033) and supplemented with 10 mM HEPES (UCSF Media Production Core, #CCFGL002), 0.11 mg/ml Penicillin-Streptomycin antibiotics (UCSF Media Production Core, #CCFGK004), 2 mM Glutamax (ThermoFisher, #35050-079), 1 mM N-Acetylcysteine (Sigma, #SKU-A7250-10G) and heat inactivated 10% FBS. Aliquots of 1 ml were stored at −80°C and were thawed just prior preparing complete organoid media containing WNT3A CM. Conditioned medium from PDGFRαlo cells (PDGFRαlo CM) was prepared by digesting small intestinal tissue from PdgfrαH2B-eGFP mouse for 3 h in 6 ml of serum-free DMEM containing 1% Glutamax, 1% Penicillin-Streptomycin and 2 mg/ml of Collagenase/Dispase (Roche, 10269638001) as described previously127, followed by FACS sorting intestinal PDGFRαlo cells (DAPI−/EpCAM−/CD44−/CD45−/PDGFRαH2B-GFP-lo+) as described previously84 100,000 sorted cells were plated into 1 well of 24-well plate, and grown in 500 μl of 1× Advanced DMEM/F12, 10 mM HEPES, 2 mM Glutamax, 0.11 mg/ml Penicillin-Streptomycin antibiotics, 1 mM N-Acetylcysteine, 20% FBS, 50 ng/ml hEGF, 2% B-27 Supplement, 1% N-2 supplement. Conditioned medium was collected every 2-3 days for up to 2 passages, combined together and aliquots of 0.5 ml were stored at −80°C and were thawed prior use.
Complete organoid media containing WNT3A CM or PDGFRαlo CM was prepared from 2× ENR supplemented with 50% WNT3A CM or PDGFRαlo CM , and 10 mM Nicotine amide (Sigma-Aldrich, 72340). Complete organoid media containing recombinant WNT3A or WNT5A was prepared from 1× ENR supplemented with 200 ng/ml recombinant murine WNT3A (PeproTech, 315-20-10ug) or 500 ng/ml recombinant human/mouse WNT5A (R&D Systems; 645-WN), and 10 mM Nicotine amide.
Establishment of intestinal organoids from primary epithelial tissue
Organoid cultures were established from primary tissue as previously described128,129. Briefly, 8- to 16-week old mice were sacrificed and dissected to harvest the small intestine. Tissue was placed in 15 ml of cold 1× PBS supplemented with 0.11 mg/ml Penicillin-Streptomycin antibiotics, 2 mM DTT, 1 mM EDTA, and 10 μM Y-27632, and incubated on ice for 15 minutes. Intestines were then moved to a tube with 20 ml cold PBS with 2 mM DTT, 3 mM EDTA, 10 μM Y-27632 and incubated for additional 60 minutes followed by vigorous shaking for one minute to release crypts into solution. Crypts were separated from villi material by filtering the solution using 70 μm cell strainers, followed by 2 washes with Advanced DMEM/F12 supplemented with 10 mM HEPES, 0.11 mg/ml Penicillin-Streptomycin antibiotics, and 1 mM N-Acetylcysteine. Finally, crypts were resuspended in Matrigel (Corning, 356231) plated on 24-well culture plates and overlaid with 1× ENR medium to initiate organoid cultures (defining passage P0). Organoids were grown at 37°C in 5% CO2 incubator, growth medium was changed at D5 and organoids were passaged at D7.
Mechanical passaging and enzymatic dissociation of intestinal organoids, and treatment
Mechanical passaging of organoids was done according to the protocols described previously (Sato and Clevers, 2013; Sato et al., 2009). Briefly, at day 7 following plating, Matrigel droplets containing organoids were disrupted with P1000 pipette, transferred to 15 ml conical tube and washed twice with 1× DMEM/F12 medium. Washed crypts were plated at a ratio 1:4 into a new Matrigel droplet and plated into plastic 24-well plates or into 12-well glass bottom plate for live-imaging.
For plating the organoids into 2D monolayer, 24-well plastic plate was first coated with 2.5% Matrigel in 1× DMEM/F12 medium. Enzymatic dissociation followed the same protocol as mechanical passaging. However, washed crypts were treated with TryplE (Gibco, 12604039) for 7 minutes at 37°C, followed by a wash with 1× ENR medium. Enzymatically dissociated organoids were resuspended and plated in the complete organoid media containing WNT3A conditioned medium. To culture 2D monolayers in complete ENR medium, organoids were only mechanically passaged and plated.
To delete Dlg1 and Arhgap31 in vitro, crypts, organoids, or 2D monolayers were incubated in the presence of 1 μM 4-hydroxy-tamoxifen (4-OHT) for 24 or 48 hours, unless the concentration was specified in the Figure legend. Stock solution of 20 mM 4-OHT dissolved in 96% ethanol was diluted to 100 μM prior treatment and was added to culture medium at ratio 1:100. Control in vitro cultures were incubated in the presence of 0.0048% ethanol (EtOH).
Immunofluorescence staining of intestinal organoids
Organoids were plated into 50% Matrigel droplet on glass-bottom 12-well plate and grown in complete organoid 1× ENR medium or complete organoid media containing WNT3A conditioned medium. Organoids were washed with 1× PBS and fixed with 4% PFA in PBS for 45 min at room temperature and blocked with 0.3% Triton X-100 in 1× PBS supplemented with 5% normal goat serum. Primary and secondary antibody staining followed the protocol used for staining intestinal tissue described above.
To quantify EdU incorporation in 2D monolayers, 2D cultures were grown to confluency, scratched, and grown for additional 12 hours, followed by treatment with 10 μM EdU in PBS for 30 minutes and fixation by 4% PFA. EdU was detected using Click-iT™ Plus EdU Cell Proliferation Kit for Imaging, Alexa Fluor™ 488 dye (ThermoFisher, C10637).
Flow cytometry
Freshly dissected small intestine was flushed with cold 1× PBS, opened lengthwise, and incubated at 37°C for 20 min in HBSS (Gibco, 14185-020) buffered with 10 mM HEPES (pH 8), and containing 10 mM DTT and 2% FBS, followed by 10 min wash in buffered HBSS containing 2% FBS. Next, intestine was cut into 0.5 mm pieces and incubated at 37°C for 20 min in buffered HBSS containing to 5 mM EDTA and 2% FBS, followed by vigorous shaking to mechanically release the epithelial cells. The suspension was filtered through a 40 μm cell strainer and cells were pelleted by centrifugation at 350 × g for 5 min. The resulting cell pellet was resuspended in buffered HBSS containing to 5 mM EDTA and 2% FBS and stained with antibodies for 30 min on ice. Prior sorting, DAPI was added, and DAPI and doublet exclusions were used in all cases. Cells were sorted using FACSAria II cell sorter (BD Bioscience) cell sorter and data were analyzed using FlowJo (Tree Star). For qPCR analysis intestinal stem cells or organoids were collected into 350 μL RLT buffer (Qiagen, 79216), for RNA sequencing, intestinal stem cells were collected into 500 μL sterile 1× PBS.
Quantitative PCR
Total RNA from 5,000 – 20,000 sorted intestinal cells or organoids were extracted using RNeasy Mini Kit (Qiagen, 74104) according to the manufacturer’s protocol. cDNA was synthesized with High-capacity cDNA Reverse Transcription Kit (Applied Biosystems, 4368814). qPCR reactions were performed using iTaq Universal SYBR Green Supermix (Bio Rad, 1725121) in 384-well plates on a QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific). Primers were purchased from Integrated DNA Technologies (IDT). Primer sequences and corresponding IDT identifiers are listed in Table S1.
RNA sequencing
In total, 5,000 – 10,000 ISCs were FACS sorted into 500 μl sterile PBS, pelleted and mRNA was isolated using the polyA Dynabeads mRNA direct Kit (Invitrogen, 61012) according to the manufacture’s protocol. cDNA and libraries were made by the UCSF Genomic Core Laboratories using the SmartSeq/NexteraXT DNA library preparation Kits (Takarabio, 634471; Illumina, FC-131-1024). Three to five biological replicates were used for each condition. 50 bp single end sequencing was carried out on an Illumina HiSeq 4000. Sequencing reads were aligned to the Mouse reference genome (GRCm38.87) and the Ensembl gene annotation using STAR 2.4.2a130. Analysis for differential expression across the replicates was performed using DESeq2 v1.16.1131. Gene expression data was filtered based on outlier detection, low counts, and no counts per gene. Genes passing a multiple testing correction with p-value of 0.05 (FDR method) were considered significant.
CDC42 and RAC1 G-LISA activation assay
Intracellular levels of active CDC42 and RAC1 were measured using the G-LISA activation assay kit (Cytoskeleton, BK127 and BK128) according to manufacturer’s guidelines. Briefly, intestinal organoids were FACS sorted into PBS, spun down, lysed in ice-cold lysis buffer provided by in the kit and snap frozen. Protein concentration was quantified, and additional lysis buffer was added to each sample to achieve equal protein concentration. Lysates were immediately used for colorimetric G-LISA assays according to the manufacturer’s protocol.
Image acquisition and analysis
Fluorescence and bright-field images were acquired using a Leica inverted DMi8 microscope or Life Technology EVOS tissue culture microscope. Organoid 3D rendered z-stack images were acquired using a Leica inverted DMi8 microscope. Whole mount intestines were imaged using a Zeiss LSM 900 with Airyscan 2 confocal system. Live imaged organoids and stained organoids were acquired with a Zeiss Cell Observer spinning disc confocal system with incubation chamber. Images were processed with the open-source platform Fiji126. Quantifications of immunofluorescence stainings were performed manually using Fiji on the indicated number of villi, crypts, or fields of view per mice. Crypt bottom areas were segmented using the Weka segmentation tool in Fiji. Finally, the segmented crypt bottom areas were measured using the “Analyze Particles” function in Fiji.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical significance between groups was determined using GraphPad Prism 9 (GraphPad Software, La Jolla, CA). The value N represents the number of animals or independent organoid preparations. Normally distributed data were analyzed using parametric Student’s t-test with Welch’s correction or one-way ANOVA with Tukey’s multiple comparisons test. The non-parametric Mann–Whitney U-test was used if the data did not fit a normal distribution. Significance was taken as P < 0.05 with a confidence interval of 95%. Data are presented as mean ± SD for parametric data or as median ± interquartile range for non-parametric data.
Supplementary Material
Video 1A (related to Figure 2). 3D projection of DLG1+ crypt region of VilCreERT2;Dlg1wt/wt;R26RtdTomato mouse treated with trametinib. Color scheme: Yellow – TdTomato; Red – anti-E-cadherin; White – anti-cleaved caspase-3; Blue – DAPI.
Video 1B (related to Figure 2). 3D projection of DLG1− crypt region of VilCreERT2;Dlg1FL/FL;R26RtdTomato mouse treated with trametinib. CC3+ cells are located in both, the epithelium and crypt lumen. Color scheme: Yellow – TdTomato; Red – anti-E-cadherin; White – anti-cleaved caspase-3; Blue – DAPI.
Video 2A (related to Figure 2). 3D projection of DLG1+ crypt region of VilCreERT2;Dlg1wt/wt;R26RtdTomato mouse infected with rotavirus. Color scheme: Yellow – TdTomato; Red – anti-E-cadherin; White – anti-cleaved caspase-3; Blue – DAPI.
Video 2B (related to Figure 2). 3D projection of DLG1− crypt region of VilCreERT2;Dlg1FL/FL;R26RtdTomato mouse infected with rotavirus. CC3+ cells are observed in the epithelium and crypt lumen. Color scheme: Yellow – TdTomato; Red – anti-E-cadherin; White – anti-cleaved caspase-3; Blue – DAPI.
Video 3A (related to Figure 3). Live-imaging of VilCreERT2;Dlg1wt/wt;R26RtdTomato spheroids grown in WNT3A CM for 48 hours followed by a 24 hours pulse of 1 μM 4-OHT. Color scheme: White – tdTomato.
Video 3B (related to Figure 3). Live-imaging of VilCreERT2;Dlg1FL/FL;R26RtdTomato spheroids grown in WNT3A CM for 48 hours followed by a 24 hours pulse of 1 μM 4-OHT. Color scheme: White – tdTomato.
Video 4A (related to Figure 3). Live-imaging of VilCreERT2;Dlg1wt/wt;R26RtdTomato spheroids grown in WNT3A CM for 48 hours followed by a 24 hours pulse of 1 μM 4-OHT and incubated with CC3/7 dye prior and during live-imaging. Color scheme: Magenta – tdTomato; Green – CC3/7 dye.
Video 4B (related to Figure 3). Live-imaging of VilCreERT2;Dlg1FL/FL;R26RtdTomato spheroids grown in WNT3A CM for 48 hours followed by a 24 hours pulse of 1 μM 4-OHT and incubated with CC3/7 dye prior and during live-imaging. Color scheme: Magenta – tdTomato; Green – CC3/7 dye.
Video 5A (related to Figure 5).: Live-imaging of VilCreERT2;Dlg1wt/wt;R26RtdTomato 2D organoids following scratch wound. Color scheme: White – mG.
Video 5B (related to Figure 5).: Live-imaging of VilCreERT2;Dlg1FL/FL;R26RmTmG 2D organoids following scratch wound. Color scheme: White – mG.
Video 6A (related to Figure 6). Live-imaging of Lgr5eGFP-CreERT2;Dlg1wt/FL;R26RtdTomato organoid following 4-OHT treatment. Color scheme: Magenta – tdTomato; Green – GFP. Single tdTomato channel highlighting DLG1+ ISCs is in Video 6B.
Video 6B (related to Figure 6). Live-imaging of Lgr5eGFP-CreERT2;Dlg1wt/FL;R26RtdTomato organoid following 4-OHT treatment. Color scheme: White – tdTomato.
Video 6C (related to Figure 6). Live-imaging of Lgr5eGFP-CreERT2;Dlg1FL/Δ;R26RtdTomato organoid following 4-OHT treatment. Color scheme: Magenta – tdTomato; Green – GFP. Single tdTomato channel highlighting dividing DLG1− ISCs are in Video 6D.
Video 6D (related to Figure 6).: Live-imaging of Lgr5eGFP-CreERT2;Dlg1FL/Δ;R26RtdTomato organoid following 4-OHT treatment. Color scheme: White – tdTomato.
Video 7A (related to Figure 6). Live-imaging of VilCreERT2;Dlg1wt/wt;R26RtdTomato 6RmTmG organoids prior 4-OHT treatment. Dividing cells are highlighted with arrowheads. Color scheme: Magenta – mT.
Video 7B (related to Figure 6).: Live-imaging of VilCreERT2;Dlg1FL/FL;R26RmTmG organoids prior 4-OHT treatment. Dividing cells are highlighted with arrowheads. Color scheme: Magenta – mT.
Video 7C (related to Figure 6). Live-imaging of VilCreERT2;Dlg1wt/wt;R26RtdTomato organoids following 4-OHT treatment. Dividing cells are highlighted with arrowheads. Color scheme: Green – mG.
Video 7D (related to Figure 6). Live-imaging of VilCreERT2;Dlg1FL/FL;R26RmTmG organoids following 4-OHT treatment. Dividing cells are highlighted with arrowheads. Color scheme: Green – mG.
HIGHLIGHTS.
Polarity of the mammalian intestinal epithelium is independent of Dlg1.
Loss of Dlg1 in ISCs results in cell death under high Wnt conditions.
ISCs lacking Dlg1 have impaired cell division and migration.
Canonical WNT ligands activate non-canonical Wnt signaling via DLG1-ARHGAP31-CDC42.
ACKNOWLEDGEMENTS
We thank Brooks Hoehn, Evelyn Sandoval, Asoka Rathnayake, and Sergio Lopez for technical assistance and Drs. Kara L. McKinley, Amnon Sharir, Laura Sanman, and Rachel Zwick for helpful discussions. This work was funded by NIH R35-DE026602, and by U01DK103147 from the Intestinal Stem Cell Consortium, a collaborative research project funded by the NIDDK and the NIAID, to O.D.K. T.N. was supported by NIH R35-GM136348. D. C.-A. was supported by NIH K99-AG071933.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
D.C.-A. is an employee of Genentech, Inc. and shareholder of Roche.
REFERENCES
- 1.Blanpain C, and Fuchs E (2014). Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science 344, 1242281. 10.1126/science.1242281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wells JM, and Watt FM (2018). Diverse mechanisms for endogenous regeneration and repair in mammalian organs. Nature 557, 322–328. 10.1038/S41586-018-0073-7. [DOI] [PubMed] [Google Scholar]
- 3.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, and Clevers H (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 4.Barker N, Bartfeld S, and Clevers H (2010). Tissue-Resident Adult Stem Cell Populations of Rapidly Self-Renewing Organs. Stem Cell 7, 656–670. 10.1016/j.stem.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 5.Sender R, and Milo R (2021). The distribution of cellular turnover in the human body. Nat Med 27, 45–48. 10.1038/s41591-020-01182-9. [DOI] [PubMed] [Google Scholar]
- 6.Sato T, Vries RG, Snipped HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, and Clevers H (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 7.Jones DL, and Wagers AJ (2008). No place like home: anatomy and function of the stem cell niche. Nat Rev Mol Cell Biol 9, 11–21. 10.1038/nrm2319. [DOI] [PubMed] [Google Scholar]
- 8.Scadden DT (2006). The stem-cell niche as an entity of action. Nature 441, 1075–1079. 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 9.Biteau B, Hochmuth CE, and Jasper H (2011). Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell 9, 402–411. 10.1016/j.stem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrison SJ, and Spradling AC (2008). Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132, 598–611. 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stzepourginski I, Nigro G, Jacob JM, Dulauroy S, Sansonetti PJ, Eberl G, and Peduto L (2017). CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc Natl Acad Sci U S A 114, E506–E513. 10.1073/pnas.1620059114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan KS, Janda CY, Chang J, Zheng GXY, Larkin KA, Luca VC, Chia LA, Mah AT, Han A, Terry JM, et al. (2017). Non-equivalence of Wnt and R-spondin ligands during Lgr5(+) intestinal stem-cell self-renewal. Nature 545, 238–242. 10.1038/nature22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, and Clevers H (2011). Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418. 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clevers H, Loh KM, and Nusse R (2014). Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 1248012. 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 15.Valenta T, Degirmenci B, Moor AE, Herr P, Zimmerli D, Moor MB, Hausmann G, Cantu C, Aguet M, and Basler K (2016). Wnt Ligands Secreted by Subepithelial Mesenchymal Cells Are Essential for the Survival of Intestinal Stem Cells and Gut Homeostasis. Cell Rep 15, 911–918. 10.1016/j.celrep.2016.03.088. [DOI] [PubMed] [Google Scholar]
- 16.Shoshkes-Carmel M, Wang YJ, Wangensteen KJ, Tóth B, Kondo A, Massasa EE, Itzkovitz S, and Kaestner KH (2018). Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 557, 242–246. 10.1038/s41586-018-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou WY, Blutt SE, Zeng XL, Chen MS, Lo YH, Castillo-Azofeifa D, Klein OD, Shroyer NF, Donowitz M, and Estes MK (2018). Epithelial WNT Ligands Are Essential Drivers of Intestinal Stem Cell Activation. Cell Rep 22, 1003–1015. 10.1016/j.celrep.2017.12.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregorieff A, Pinto D, Begthel H, Destree O, Kielman M, and Clevers H (2005). Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology 129, 626–638. 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Tikhonova AN, Lasry A, Austin R, and Aifantis I (2020). Cell-by-Cell Deconstruction of Stem Cell Niches. Cell Stem Cell 27, 19–34. 10.1016/j.stem.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fevr T, Robine S, Louvard D, and Huelsken J (2007). Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol Cell Biol 27, 7551–7559. 10.1128/MCB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhnert F, Davis CR, Wang HT, Chu P, Lee M, Yuan J, Nusse R, and Kuo CJ (2004). Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci U S A 101, 266–271. 10.1073/pnas.2536800100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinto D, Gregorieff A, Begthel H, and Clevers H (2003). Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev 17, 1709–1713. 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, and Clevers H (1998). Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet 19, 379–383. 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 24.Hoffman J, Kuhnert F, Davis CR, and Kuo CJ. (2004). Wnts as essential growth factors for the adult small intestine and colon. Cell Cycle. [PubMed] [Google Scholar]
- 25.Davies PS, Dismuke AD, Powell AE, Carroll KH, and Wong MH (2008). Wnt-reporter expression pattern in the mouse intestine during homeostasis. BMC Gastroenterol 8, 57. 10.1186/1471-230X-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moparthi L, and Koch S (2019). Wnt signaling in intestinal inflammation. Differentiation 108, 24–32. 10.1016/j.diff.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Konsavage WM Jr., Jin G, and Yochum GS (2012). The Myc 3' Wnt-responsive element regulates homeostasis and regeneration in the mouse intestinal tract. Mol Cell Biol 32, 3891–3902. 10.1128/MCB.00548-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernal NP, Stehr W, Zhang Y, Profitt S, Erwin CR, and Warner BW (2005). Evidence for active Wnt signaling during postresection intestinal adaptation. J Pediatr Surg 40, 1025–1029; discussion 1029. 10.1016/j.jpedsurg.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 29.Castillo-Azofeifa D, Fazio EN, Nattiv R, Good HJ, Wald T, Pest MA, de Sauvage FJ, Klein OD, and Asfaha S (2019). Atoh1(+) secretory progenitors possess renewal capacity independent of Lgr5(+) cells during colonic regeneration. EMbO J 38. 10.15252/embj.201899984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saha S, Aranda E, Hayakawa Y, Bhanja P, Atay S, Brodin NP, Li J, Asfaha S, Liu L, Tailor Y, et al. (2016). Macrophage-derived extracellular vesicle-packaged WNTs rescue intestinal stem cells and enhance survival after radiation injury. Nat Commun 7, 13096. 10.1038/ncomms13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voloshanenko O, Erdmann G, Dubash TD, Augustin I, Metzig M, Moffa G, Hundsrucker C, Kerr G, Sandmann T, Anchang B, et al. (2013). Wnt secretion is required to maintain high levels of Wnt activity in colon cancer cells. Nat Commun 4, 2610. 10.1038/ncomms3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP, Batlle E, Simon-Assmann P, Clevers H, Nathke IS, et al. (2004). Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev 18, 1385–1390. 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashton GH, Morton JP, Myant K, Phesse TJ, Ridgway RA, Marsh V, Wilkins JA, Athineos D, Muncan V, Kemp R, et al. (2010). Focal adhesion kinase is required for intestinal regeneration and tumorigenesis downstream of Wnt/c-Myc signaling. Dev Cell 19, 259–269. 10.1016/j.devcel.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shibata H, Toyama K, Shioya H, Ito M, Hirota M, Hasegawa S, Matsumoto H, Takano H, Akiyama T, Toyoshima K, et al. (1997). Rapid Colorectal Adenoma Formation Initiated by Conditional Targeting of the Apc Gene. Science 278, 120. 10.1126/science.278.5335.120. [DOI] [PubMed] [Google Scholar]
- 35.Major MB, Camp ND, Berndt JD, Yi X, Goldenberg SJ, Hubbert C, Biechele TL, Gingras A-C, Zheng N, MacCoss MJ, et al. (2007). Wilms Tumor Suppressor WTX Negatively Regulates WNT/ß-Catenin Signaling. Science 316, 1043. 10.1126/science/1141515. [DOI] [PubMed] [Google Scholar]
- 36.Parker TW, and Neufeld KL (2020). APC controls Wnt-induced beta-catenin destruction complex recruitment in human colonocytes. Sci Rep 10, 2957. 10.1038/s41598-020-59899-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsumine A, Ogai A, Senda T, Okumura N, Satoh K, Baeg GH, Kawahara T, Kobayashi S, Okada M, Toyoshima K, and Akiyama T (1996). Binding of APC to the human homolog of the Drosophila discs large tumor suppressor protein. Science 272, 1020–1023. 10.1126/science.272.5264.1020. [DOI] [PubMed] [Google Scholar]
- 38.Yanai H, Satoh K, Matsumine A, and Akiyama T (2000). The colorectal tumour suppressor APC is present in the NMDA-receptor-PSD-95 complex in the brain. Genes Cells 5, 815–822. 10.1046/j.1365-2443.2000.00368.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z, Li H, Chen L, Lu X, Zhang J, Xu P, Lin K, and Wu G (2011). Molecular basis for the recognition of adenomatous polyposis coli by the Discs Large 1 protein. PLoS One 6, e23507. 10.1371/journal.pone.0023507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonello TT, and Peifer M (2019). Scribble: A master scaffold in polarity, adhesion, synaptogenesis, and proliferation. J Cell Biol 218, 742–756. 10.1083/jcb.201810103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearson HB, McGlinn E, Phesse TJ, Schluter H, Srikumar A, Godde NJ, Woelwer CB, Ryan A, Phillips WA, Ernst M, et al. (2015). The polarity protein Scrib mediates epidermal development and exerts a tumor suppressive function during skin carcinogenesis. Mol Cancer 14, 169. 10.1186/s12943-015-0440-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caruana G, and Bernstein A (2001). Craniofacial dysmorphogenesis including cleft palate in mice with an insertional mutation in the discs large gene. Mol Cell Biol 21, 1475–1483. 10.1128/MCB.21.5.1475-1483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naim E, Bernstein A, Bertram JF, and Caruana G (2005). Mutagenesis of the epithelial polarity gene, discs large 1, perturbs nephrogenesis in the developing mouse kidney. Kidney Int 68, 955–965. 10.1111/j.1523-1755.2005.00489.x. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen MM, Nguyen ML, Caruana G, Bernstein A, Lambert PF, and Griep AE (2003). Requirement of PDZ-containing proteins for cell cycle regulation and differentiation in the mouse lens epithelium. Mol Cell Biol 23, 8970–8981. 10.1128/MCB.23.24.8970-8981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iizuka-Kogo A, Senda T, Akiyama T, Shimomura A, Nomura R, Hasegawa Y, Yamamura K, Kogo H, Sawai N, and Matsuzaki T (2015). Requirement of DLG1 for cardiovascular development and tissue elongation during cochlear, enteric, and skeletal development: possible role in convergent extension. PLoS One 10, e0123965. 10.1371/journal.pone.0123965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Z, Zhang F, Davis AK, Xin M, Walz G, Tian W, and Zheng Y (2022). CDC42 controlled apical-basal polarity regulates intestinal stem cell to transit amplifying cell fate transition via YAP-EGF-mTOR signaling. Cell Rep 38, 110009. 10.1016/j.celrep.2021.110009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J, Sayadian AC, Lowe N, Lovegrove HE, and St Johnston D (2018). An alternative mode of epithelial polarity in the Drosophila midgut. PLoS Biol 16, e3000041. 10.1371/journal.pbio.3000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Etienne-Manneville S, Manneville JB, Nicholls S, Ferenczi MA, and Hall A (2005). Cdc42 and Par6-PKCzeta regulate the spatially localized association of Dlg1 and APC to control cell polarization. J Cell Biol 170, 895–901. 10.1083/jcb.200412172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishidate T, Matsumine A, Toyoshima K, and Akiyama T (2000). The APC-hDLG complex negatively regulates cell cycle progression from the G0/G1 to S phase. Oncogene 19, 365–372. 10.1038/sj.onc.1203309. [DOI] [PubMed] [Google Scholar]
- 50.Mimori-Kiyosue Y, Matsui C, Sasaki H, and Tsukita S (2007). Adenomatous polyposis coli (APC) protein regulates epithelial cell migration and morphogenesis via PDZ domain-based interactions with plasma membranes. Genes Cells 12, 219–233. 10.1111/j.1365-2443.2007.01045.x. [DOI] [PubMed] [Google Scholar]
- 51.Young MA, May S, Damo A, Yoon YS, Hur MW, Swat W, and Parry L (2019). Epigenetic Regulation of Dlg1, via Kaiso, Alters Mitotic Spindle Polarity and Promotes Intestinal Tumorigenesis. Mol Cancer Res 17, 686–696. 10.1158/1541-7786.MCR-18-0280. [DOI] [PubMed] [Google Scholar]
- 52.Caron C, DeGeer J, Fournier P, Duquette PM, Luangrath V, Ishii H, Karimzadeh F, Lamarche-Vane N, and Royal I (2016). CdGAP/ARHGAP31, a Cdc42/Rac1 GTPase regulator, is critical for vascular development and VEGF-mediated angiogenesis. Sci Rep 6, 27485. 10.1038/srep27485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirjavainen A, Laos M, Anttonen T, and Pirvola U (2015). The Rho GTPase Cdc42 regulates hair cell planar polarity and cellular patterning in the developing cochlea. Biol Open 4, 516–526. 10.1242/bio.20149753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stephenson LM, Sammut B, Graham DB, Chan-Wang J, Brim KL, Huett AS, Miletic AV, Kloeppel T, Landry A, Xavier R, and Swat W (2007). DLGH1 is a negative regulator of T-lymphocyte proliferation. Mol Cell Biol 27, 7574–7581. 10.1128/MCB.00439-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, and Robine S (2004). Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193. 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 56.Mahoney ZX, Sammut B, Xavier RJ, Cunningham J, Go G, Brim KL, Stappenbeck TS, Miner JH, and Swat W (2006). Discs-large homolog 1 regulates smooth muscle orientation in the mouse ureter. Proc Natl Acad Sci U S A 103, 19872–19877. 10.1073/pnas.0609326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gagnoux-Palacios L, Awina H, Audebert S, Rossin A, Mondin M, Borgese F, Planas-Botey C, Mettouchi A, Borg JP, and Hueber AO (2018). Cell polarity and adherens junction formation inhibit epithelial Fas cell death receptor signaling. J Cell Biol 217, 3839–3852. 10.1083/jcb.201805071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Desai RA, Gao L, Raghavan S, Liu WF, and Chen CS (2009). Cell polarity triggered by cell-cell adhesion via E-cadherin. J Cell Sci 122, 905–911. 10.1242/jcs.028183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang X, Dong B, Zhang K, Ji Z, Cheng C, Zhao H, Sheng Y, Li X, Fan L, Xue W, et al. (2018). E-cadherin bridges cell polarity and spindle orientation to ensure prostate epithelial integrity and prevent carcinogenesis in vivo. PLoS Genet 14, e1007609. 10.1371/journal.pgen.1007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weaver VM, Lelievre S, Lakins JN, Chrenek MA, Jones JC, Giancotti F, Werb Z, and Bissell MJ (2002). beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell 2, 205–216. 10.1016/s1535-6108(02)00125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Myllymaki SM, Teravainen TP, and Manninen A (2011). Two distinct integrin-mediated mechanisms contribute to apical lumen formation in epithelial cells. PLoS One 6, e19453. 10.1371/journal.pone.0019453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Casaletto JB, Saotome I, Curto M, and McClatchey AI (2011). Ezrin-mediated apical integrity is required for intestinal homeostasis. Proc Natl Acad Sci U S A 108, 11924–11929. 10.1073/pnas.1103418108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woods DF, Hough C, Peel D, Callaini G, and Bryant PJ (1996). Dlg protein is required for junction structure, cell polarity, and proliferation control in Drosophila epithelia. J Cell Biol 134, 1469–1482. 10.1083/jcb.134.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valdano MB, Cavatorta AL, Morale MG, Marziali F, de Souza Lino V, Steenbergen RDM, Boccardo E, and Gardiol D (2016). Disc large 1 expression is altered by human papillomavirus E6/E7 proteins in organotypic cultures of human keratinocytes. J Gen Virol 97, 453–462. 10.1099/jgv.0.000364. [DOI] [PubMed] [Google Scholar]
- 65.Iizuka-Kogo A, Ishidao T, Akiyama T, and Senda T (2007). Abnormal development of urogenital organs in Dlgh1-deficient mice. Development 134, 1799–1807. 10.1242/dev.02830. [DOI] [PubMed] [Google Scholar]
- 66.Rivera C, Yamben IF, Shatadal S, Waldof M, Robinson ML, and Griep AE (2009). Cell-autonomous requirements for Dlg-1 for lens epithelial cell structure and fiber cell morphogenesis. Dev Dyn 238, 2292–2308. 10.1002/dvdy.22036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Humbert PO, Grzeschik NA, Brumby AM, Galea R, Elsum I, and Richardson HE (2008). Control of tumourigenesis by the Scribble/Dlg/Lgl polarity module. Oncogene 27, 6888–6907. 10.1038/onc.2008.341. [DOI] [PubMed] [Google Scholar]
- 68.Clevers H (2006). Wnt/beta-catenin signaling in development and disease. Cell 127, 469–480. 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 69.Xu S, Zhou F, Tao J, Song L, Ng SC, Wang X, Chen L, Yi F, Ran Z, Zhou R, and Xia B (2014). Exome sequencing identifies DLG1 as a novel gene for potential susceptibility to Crohn's disease in a Chinese family study. PLoS One 9, e99807. 10.1371/journal.pone.0099807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stoll M, Corneliussen B, Costello CM, Waetzig GH, Mellgard B, Koch WA, Rosenstiel P, Albrecht M, Croucher PJ, Seegert D, et al. (2004). Genetic variation in DLG5 is associated with inflammatory bowel disease. Nat Genet 36, 476–480. 10.1038/ng1345. [DOI] [PubMed] [Google Scholar]
- 71.Yoshimi K, Tanaka T, Serikawa T, and Kuramoto T (2013). Tumor suppressor APC protein is essential in mucosal repair from colonic inflammation through angiogenesis. Am J Pathol 182, 1263–1274. 10.1016/j.ajpath.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 72.McGovern DP, Gardet A, Torkvist L, Goyette P, Essers J, Taylor KD, Neale BM, Ong RT, Lagace C, Li C, et al. (2010). Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet 42, 332–337. 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cui H, Shu-Hong Y, and Qiao M (2016). Expression of DLG1 and DLG5 in the Intestinal Epithelium of Patients with CD. Inflamm Bowel Dis 22, E43–E44. 10.1097/MIB.0000000000000948. [DOI] [PubMed] [Google Scholar]
- 74.Zhan T, Ambrosi G, Wandmacher AM, Rauscher B, Betge J, Rindtorff N, Haussler RS, Hinsenkamp I, Bamberg L, Hessling B, et al. (2019). MEK inhibitors activate Wnt signalling and induce stem cell plasticity in colorectal cancer. Nat Commun 10, 2197. 10.1038/s41467-019-09898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Greenberg HB, and Estes MK (2009). Rotaviruses: from pathogenesis to vaccination. Gastroenterology 136, 1939–1951. 10.1053/j.gastro.2009.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramig RF (2004). Pathogenesis of intestinal and systemic rotavirus infection. J Virol 78, 10213–10220. 10.1128/JVI.78.19.10213-10220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Conner ME, and Ramig RF (1997). Viral enteric diseases. In Viral pathogenesis, Nathanson N, ed. (Lippincott-Raven Publishers; ), pp. p. 713–743. [Google Scholar]
- 78.Mustata RC, Vasile G, Fernandez-Vallone V, Strollo S, Lefort A, Libert F, Monteyne D, Perez-Morga D, Vassart G, and Garcia MI (2013). Identification of Lgr5-independent spheroid-generating progenitors of the mouse fetal intestinal epithelium. Cell Rep 5, 421–432. 10.1016/j.celrep.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 79.Nusse YM, Savage AK, Marangoni P, Rosendahl-Huber AKM, Landman TA, de Sauvage FJ, Locksley RM, and Klein OD (2018). Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature 559, 109–113. 10.1038/s41586-018-0257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yui S, Azzolin L, Maimets M, Pedersen MT, Fordham RP, Hansen SL, Larsen HL, Guiu J, Alves MRP, Rundsten CF, et al. (2018). YAP/TAZ-Dependent Reprogramming of Colonic Epithelium Links ECM Remodeling to Tissue Regeneration. Cell Stem Cell 22, 35–49 e37. 10.1016/j.stem.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Merenda A, Fenderico N, and Maurice MM (2020). Wnt Signaling in 3D: Recent Advances in the Applications of Intestinal Organoids. Trends Cell Biol 30, 60–73. 10.1016/j.tcb.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 82.Sprangers J, Zaalberg IC, and Maurice MM (2021). Organoid-based modeling of intestinal development, regeneration, and repair. Cell Death Differ 28, 95–107. 10.1038/S41418-020-00665-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fordham RP, Yui S, Hannan NR, Soendergaard C, Madgwick A, Schweiger PJ, Nielsen OH, Vallier L, Pedersen RA, Nakamura T, et al. (2013). Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell 13, 734–744. 10.1016/j.stem.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McCarthy N, Manieri E, Storm EE, Saadatpour A, Luoma AM, Kapoor VN, Madha S, Gaynor LT, Cox C, Keerthivasan S, et al. (2020). Distinct Mesenchymal Cell Populations Generate the Essential Intestinal BMP Signaling Gradient. Cell Stem Cell 26, 391–402 e395. 10.1016/j.stem.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miyoshi H, Ajima R, Luo CT, Yamaguchi TP, and Stappenbeck TS (2012). Wnt5a Potentiates TGF-β Signaling to Promote Colonic Crypt Regeneration After Tissue Injury. Science 338, 108. 10.1126/science.1223821.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lamarche-Vane N, and Hall A (1998). CdGAP, a novel proline-rich GTPase-activating protein for Cdc42 and Rac. J Biol Chem 273, 29172–29177. 10.1074/jbc.273.44.29172. [DOI] [PubMed] [Google Scholar]
- 87.Tcherkezian J, Triki I, Stenne R, Danek EI, and Lamarche-Vane N (2006). The human orthologue of CdGAP is a phosphoprotein and a GTPase-activating protein for Cdc42 and Rac1 but not RhoA. Biol Cell 98, 445–456. 10.1042/BC20050101. [DOI] [PubMed] [Google Scholar]
- 88.LaLonde DP, Grubinger M, Lamarche-Vane N, and Turner CE (2006). CdGAP associates with actopaxin to regulate integrin-dependent changes in cell morphology and motility. Current biology : CB 16, 1375–1385. 10.1016/j.cub.2006.05.057. [DOI] [PubMed] [Google Scholar]
- 89.Mehra C, Chung JH, He Y, Lara-Marquez M, Goyette MA, Boufaied N, Barres V, Ouellet V, Guerard KP, Delliaux C, et al. (2021). CdGAP promotes prostate cancer metastasis by regulating epithelial-to-mesenchymal transition, cell cycle progression, and apoptosis. Commun Biol 4, 1042. 10.1038/S42003-021-02520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thorne CA, Chen IW, Sanman LE, Cobb MH, Wu LF, and Altschuler SJ (2018). Enteroid Monolayers Reveal an Autonomous WNT and BMP Circuit Controlling Intestinal Epithelial Growth and Organization. Dev Cell 44, 624–633 e624. 10.1016/j.devcel.2018.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lamarche N, Tapon N, Stowers L, Burbelo PD, Aspenstrom P, Bridges T, Chant J, and Hall A (1996). Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell 87, 519–529. 10.1016/s0092-8674(00)81371-9. [DOI] [PubMed] [Google Scholar]
- 92.Ueyama T (2019). Rho-Family Small GTPases: From Highly Polarized Sensory Neurons to Cancer Cells. Cells 8. 10.3390/cells8020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Melendez J, Liu M, Sampson L, Akunuru S, Han X, Vallance J, Witte D, Shroyer N, and Zheng Y (2013). Cdc42 coordinates proliferation, polarity, migration, and differentiation of small intestinal epithelial cells in mice. Gastroenterology 145, 808–819. 10.1053/j.gastro.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McKinley KL, Stuurman N, Royer LA, Schartner C, Castillo-Azofeifa D, Delling M, Klein OD, and Vale RD (2018). Cellular aspect ratio and cell division mechanics underlie the patterning of cell progeny in diverse mammalian epithelia. Elife 7. 10.7554/eLife.36739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lane SW, Williams DA, and Watt FM (2014). Modulating the stem cell niche for tissue regeneration. Nat Biotechnol 32, 795–803. 10.1038/nbt.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gehart H, and Clevers H (2019). Tales from the crypt: new insights into intestinal stem cells. Nat Rev Gastroenterol Hepatol 16, 19–34. 10.1038/s41575-018-0081-y. [DOI] [PubMed] [Google Scholar]
- 97.Tan DW, and Barker N (2014). Intestinal stem cells and their defining niche. Curr Top Dev Biol 107, 77–107. 10.1016/B978-0-12-416022-4.00003-2. [DOI] [PubMed] [Google Scholar]
- 98.Nusse R, and Clevers H (2017). Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 169, 985–999. 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 99.Acebron SP, and Niehrs C (2016). beta-Catenin-Independent Roles of Wnt/LRP6 Signaling. Trends Cell Biol 26, 956–967. 10.1016/j.tcb.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 100.Chae WJ, and Bothwell ALM (2018). Canonical and Non-Canonical Wnt Signaling in Immune Cells. Trends in immunology 39, 830–847. 10.1016/j.it.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Humphries AC, and Mlodzik M (2018). From instruction to output: Wnt/PCP signaling in development and cancer. Curr Opin Cell Biol 51, 110–116. 10.1016/j.ceb.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.VanderVorst K, Dreyer CA, Konopelski SE, Lee H, Ho HH, and Carraway KL 3rd (2019). Wnt/PCP Signaling Contribution to Carcinoma Collective Cell Migration and Metastasis. Cancer Res 79, 1719–1729. 10.1158/0008-5472.CAN-18-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pinzon-Daza ML, Salaroglio IC, Kopecka J, Garzon R, Couraud PO, Ghigo D, and Riganti C (2014). The cross-talk between canonical and non-canonical Wnt-dependent pathways regulates P-glycoprotein expression in human blood-brain barrier cells. J Cereb Blood Flow Metab 34, 1258–1269. 10.1038/jcbfm.2014.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Glinka A, Dolde C, Kirsch N, Huang YL, Kazanskaya O, Ingelfinger D, Boutros M, Cruciat CM, and Niehrs C (2011). LGR4 and LGR5 are R-spondin receptors mediating Wnt/beta-catenin and Wnt/PCP signalling. EMBO Rep 12, 1055–1061. 10.1038/embor.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mentink RA, Rella L, Radaszkiewicz TW, Gybel T, Betist MC, Bryja V, and Korswagen HC (2018). The planar cell polarity protein VANG-1/Vangl negatively regulates Wnt/beta-catenin signaling through a Dvl dependent mechanism. PLoS Genet 14, e1007840. 10.1371/journal.pgen.1007840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rella L, Fernandes Povoa EE, Mars J, Ebbing ALP, Schoppink L, Betist MC, and Korswagen HC (2021). A switch from noncanonical to canonical Wnt signaling stops neuroblast migration through a Slt-Robo and RGA-9b/ARHGAP-dependent mechanism. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2013239118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bastias-Candia S, Martinez M, Zolezzi JM, and Inestrosa NC (2019). Wnt Signaling Upregulates Teneurin-3 Expression via Canonical and Non-canonical Wnt Pathway Crosstalk. Front Neurosci 13, 505. 10.3389/fnins.2019.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Qiu W, Chen L, and Kassem M (2011). Activation of non-canonical Wnt/JNK pathway by Wnt3a is associated with differentiation fate determination of human bone marrow stromal (mesenchymal) stem cells. Biochem Biophys Res Commun 413, 98–104. 10.1016/j.bbrc.2011.08.061. [DOI] [PubMed] [Google Scholar]
- 109.Flores-Hernandez E, Velazquez DM, Castaneda-Patlan MC, Fuentes-Garcia G, Fonseca-Camarillo G, Yamamoto-Furusho JK, Romero-Avila MT, Garcia-Sainz JA, and Robles-Flores M (2020). Canonical and non-canonical Wnt signaling are simultaneously activated by Wnts in colon cancer cells. Cell Signal 72, 109636. 10.1016/j.cellsig.2020.109636. [DOI] [PubMed] [Google Scholar]
- 110.Nalesso G, Sherwood J, Bertrand J, Pap T, Ramachandran M, De Bari C, Pitzalis C, and Dell'accio F (2011). WNT-3A modulates articular chondrocyte phenotype by activating both canonical and noncanonical pathways. J Cell Biol 193, 551–564. 10.1083/jcb.201011051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Almeida M, Han L, Bellido T, Manolagas SC, and Kousteni S (2005). Wnt proteins prevent apoptosis of both uncommitted osteoblast progenitors and differentiated osteoblasts by beta-catenin-dependent and -independent signaling cascades involving Src/ERK and phosphatidylinositol 3-kinase/AKT. J Biol Chem 280, 41342–41351. 10.1074/jbc.M502168200. [DOI] [PubMed] [Google Scholar]
- 112.Humbert PO, Dow LE, and Russell SM (2006). The Scribble and Par complexes in polarity and migration: friends or foes? Trends Cell Biol 16, 622–630. 10.1016/j.tcb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 113.Hu DJ, Yun J, Elstrott J, and Jasper H (2021). Non-canonical Wnt signaling promotes directed migration of intestinal stem cells to sites of injury. Nat Commun 12, 7150. 10.1038/s41467-021-27384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nalapareddy K, Hassan A, Sampson LL, Zheng Y, and Geiger H (2021). Suppression of elevated Cdc42 activity promotes the regenerative potential of aged intestinal stem cells. iScience 24, 102362. 10.1016/j.isci.2021.102362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dwivedi VK, Pardo-Pastor C, Droste R, Kong JN, Tucker N, Denning DP, Rosenblatt J, and Horvitz HR (2021). Replication stress promotes cell elimination by extrusion. Nature. 10.1038/s41586-021-03526-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li Y, Junge JA, Arnesano C, Gross GG, Miner JH, Moats R, Roberts RW, Arnold DB, and Fraser SE (2018). Discs large 1 controls daughter-cell polarity after cytokinesis in vertebrate morphogenesis. Proceedings of the National Academy of Sciences 115, E10859–E10868. 10.1073/pnas.1713959115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nobes CD, and Hall A (1995). Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81, 53–62. 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 118.Ridley AJ, and Hall A (1992). The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70, 389–399. 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 119.Symons M (1996). Rho family GTPases: the cytoskeleton and beyond. Trends Biochem Sci 21, 178–181. [PubMed] [Google Scholar]
- 120.Zhou X, Florian MC, Arumugam P, Chen X, Cancelas JA, Lang R, Malik P, Geiger H, and Zheng Y (2013). RhoA GTPase controls cytokinesis and programmed necrosis of hematopoietic progenitors. J Exp Med 210, 2371–2385. 10.1084/jem.20122348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou W, Zhang L, Guoxiang X, Mojsilovic-Petrovic J, Takamaya K, Sattler R, Huganir R, and Kalb R (2008). GluR1 controls dendrite growth through its binding partner, SAP97. J Neurosci 28, 10220–10233. 10.1523/JNEUROSCI.3434-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140. 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 124.Hamilton TG, Klinghoffer RA, Corrin PD, and Soriano P (2003). Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol Cell Biol 23, 4013–4025. 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.O'Neal CM, Crawford SE, Estes MK, and Conner ME (1997). Rotavirus virus-like particles administered mucosally induce protective immunity. J Virol 71, 8707–8717. 10.1128/JVI.71.11.8707-8717.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kabiri Z, Greicius G, Madan B, Biechele S, Zhong Z, Zaribafzadeh H, Edison, Aliyev J, Wu Y, Bunte R, et al. (2014). Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development 141, 2206–2215. 10.1242/dev.104976. [DOI] [PubMed] [Google Scholar]
- 128.Sanman LE, Chen IW, Bieber JM, Steri V, Trentesaux C, Hann B, Klein OD, Wu LF, and Altschuler SJ (2021). Transit-Amplifying Cells Coordinate Changes in Intestinal Epithelial Cell-Type Composition. Dev Cell 56, 356–365 e359. 10.1016/j.devcel.2020.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sanman LE, Chen IW, Bieber JM, Thorne CA, Wu LF, and Altschuler SJ (2020). Generation and Quantitative Imaging of Enteroid Monolayers. Methods Mol Biol 2171, 99–113. 10.1007/978-1-0716-0747-3_6. [DOI] [PubMed] [Google Scholar]
- 130.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video 1A (related to Figure 2). 3D projection of DLG1+ crypt region of VilCreERT2;Dlg1wt/wt;R26RtdTomato mouse treated with trametinib. Color scheme: Yellow – TdTomato; Red – anti-E-cadherin; White – anti-cleaved caspase-3; Blue – DAPI.
Video 1B (related to Figure 2). 3D projection of DLG1− crypt region of VilCreERT2;Dlg1FL/FL;R26RtdTomato mouse treated with trametinib. CC3+ cells are located in both, the epithelium and crypt lumen. Color scheme: Yellow – TdTomato; Red – anti-E-cadherin; White – anti-cleaved caspase-3; Blue – DAPI.
Video 2A (related to Figure 2). 3D projection of DLG1+ crypt region of VilCreERT2;Dlg1wt/wt;R26RtdTomato mouse infected with rotavirus. Color scheme: Yellow – TdTomato; Red – anti-E-cadherin; White – anti-cleaved caspase-3; Blue – DAPI.
Video 2B (related to Figure 2). 3D projection of DLG1− crypt region of VilCreERT2;Dlg1FL/FL;R26RtdTomato mouse infected with rotavirus. CC3+ cells are observed in the epithelium and crypt lumen. Color scheme: Yellow – TdTomato; Red – anti-E-cadherin; White – anti-cleaved caspase-3; Blue – DAPI.
Video 3A (related to Figure 3). Live-imaging of VilCreERT2;Dlg1wt/wt;R26RtdTomato spheroids grown in WNT3A CM for 48 hours followed by a 24 hours pulse of 1 μM 4-OHT. Color scheme: White – tdTomato.
Video 3B (related to Figure 3). Live-imaging of VilCreERT2;Dlg1FL/FL;R26RtdTomato spheroids grown in WNT3A CM for 48 hours followed by a 24 hours pulse of 1 μM 4-OHT. Color scheme: White – tdTomato.
Video 4A (related to Figure 3). Live-imaging of VilCreERT2;Dlg1wt/wt;R26RtdTomato spheroids grown in WNT3A CM for 48 hours followed by a 24 hours pulse of 1 μM 4-OHT and incubated with CC3/7 dye prior and during live-imaging. Color scheme: Magenta – tdTomato; Green – CC3/7 dye.
Video 4B (related to Figure 3). Live-imaging of VilCreERT2;Dlg1FL/FL;R26RtdTomato spheroids grown in WNT3A CM for 48 hours followed by a 24 hours pulse of 1 μM 4-OHT and incubated with CC3/7 dye prior and during live-imaging. Color scheme: Magenta – tdTomato; Green – CC3/7 dye.
Video 5A (related to Figure 5).: Live-imaging of VilCreERT2;Dlg1wt/wt;R26RtdTomato 2D organoids following scratch wound. Color scheme: White – mG.
Video 5B (related to Figure 5).: Live-imaging of VilCreERT2;Dlg1FL/FL;R26RmTmG 2D organoids following scratch wound. Color scheme: White – mG.
Video 6A (related to Figure 6). Live-imaging of Lgr5eGFP-CreERT2;Dlg1wt/FL;R26RtdTomato organoid following 4-OHT treatment. Color scheme: Magenta – tdTomato; Green – GFP. Single tdTomato channel highlighting DLG1+ ISCs is in Video 6B.
Video 6B (related to Figure 6). Live-imaging of Lgr5eGFP-CreERT2;Dlg1wt/FL;R26RtdTomato organoid following 4-OHT treatment. Color scheme: White – tdTomato.
Video 6C (related to Figure 6). Live-imaging of Lgr5eGFP-CreERT2;Dlg1FL/Δ;R26RtdTomato organoid following 4-OHT treatment. Color scheme: Magenta – tdTomato; Green – GFP. Single tdTomato channel highlighting dividing DLG1− ISCs are in Video 6D.
Video 6D (related to Figure 6).: Live-imaging of Lgr5eGFP-CreERT2;Dlg1FL/Δ;R26RtdTomato organoid following 4-OHT treatment. Color scheme: White – tdTomato.
Video 7A (related to Figure 6). Live-imaging of VilCreERT2;Dlg1wt/wt;R26RtdTomato 6RmTmG organoids prior 4-OHT treatment. Dividing cells are highlighted with arrowheads. Color scheme: Magenta – mT.
Video 7B (related to Figure 6).: Live-imaging of VilCreERT2;Dlg1FL/FL;R26RmTmG organoids prior 4-OHT treatment. Dividing cells are highlighted with arrowheads. Color scheme: Magenta – mT.
Video 7C (related to Figure 6). Live-imaging of VilCreERT2;Dlg1wt/wt;R26RtdTomato organoids following 4-OHT treatment. Dividing cells are highlighted with arrowheads. Color scheme: Green – mG.
Video 7D (related to Figure 6). Live-imaging of VilCreERT2;Dlg1FL/FL;R26RmTmG organoids following 4-OHT treatment. Dividing cells are highlighted with arrowheads. Color scheme: Green – mG.
Data Availability Statement
Bulk RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-CD45, clone 30-F11 | Biolegend | Cat#: 103133 RRID: AB_10899570 |
| Rat monoclonal anti-EpCAM, clone G8.8 | Biolegend | Cat#: 118211 RRID: AB_1134104 |
| Rat monoclonal anti-CD44, clone IM7 | Biolegend | Cat#: 103029 RRID: AB_830786 |
| Mouse anti-Dlg1, Clone 12/Dlg | BD Biosciences | Cat#: 610874 RRID: AB_398192 |
| Mouse monoclonal anti-BrdU | Novus Biologicals | Cat#: NB500-439 RRID:AB_10000514 |
| Rabbit monoclonal anti-E-cadherin | Cell Signaling Technology | Cat#: 3195S RRID:AB_2291471 |
| Rat monoclonal anti-β4-integrin | Abcam | Cat#: ab25254 RRID:AB_2129042 |
| Mouse monoclonal anti-Ezrin | Invitrogen | Cat#: MA5-13862 |
| Rabbit monoclonal anti-RFP | Rockland | Cat#: 200-301-379 RRID:AB_2611063 |
| Rabbit polyclonal anti-cleaved caspase-3 | Cell Signaling | Cat#: 9661 RRID:AB_2341188 |
| Rabbit polyclonal anti-Mucin2 | Novus Biologicals | Cat#: NBP1- 31231 RRID: AB_10003763 |
| Rabbit polyclonal anti-Chromogranin A | Abcam | Cat#: ab45179 RRID:AB_300798 |
| Rabbit polyclonal anti-DCAMKL1 | Abcam | Cat#: ab31704 RRID:AB_873537 |
| Rabbit polyclonal anti-Lysozyme | Dako | Cat#: A0099 RRID:AB_2341230 |
| Bacterial and virus strains | ||
| Rotavirus strain ECWT (P[17], G3) | Mary Estes | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| EdU: 5-ethynyl-2'-deoxyuridine | ThermoFisher | Cat#: C10637 |
| BrdU: 5-Bromo-2'-deoxyuridine | Sigma-Aldrich | Cat#: B5002 |
| Trametinib | ApexBio | Cat#: A3018 |
| Tamoxifen | Sigma-Aldrich | Cat#: T5648 |
| 4-hydroxytamoxifen | Sigma-Aldrich | Cat#: H7904 |
| TryplE | Gibco | Cat#: 12604039 |
| Matrigel | Corning | Cat#: 356231 |
| WNT3A | PeproTech | Cat#: 315-20-10ug |
| WNT5A | R&D Systems | Cat#: 645-WN |
| Nicotine amide | Sigma-Aldrich | Cat#: 72340 |
| Collagenase/Dispase | Roche | Cat#: 10269638001 |
| 293T-HA-RSPO1-FC cells | Trevigen | Cat#: 3710-001-01 |
| L-WNT3A cells | ATCC | Cat#: CRL-2647 |
| Critical commercial assays | ||
| Alkaline Phosphatase Red Substrate Kit | Vector Laboratories | Cat#: SK-5100 |
| Click-iT™ Plus EdU Cell Proliferation Kit for Imaging | Thermo Fisher | Cat#: C10637 |
| CellEvent™ Caspase-3/7 Green ReadyProbes™ Reagent | Thermo Fisher | Cat#: R37111 |
| RNAscope® 2.5 High Definition (HD) – Red Assay | Advanced Cell Diagnostics | Cat#: 322350 |
| G-LISA activation assay kit for Cdc42 | Cytoskeleton | Cat#: BK127 |
| G-LISA activation assay kit for Rac1 | Cytoskeleton | Cat#: BK128 |
| polyA Dynabeads mRNA direct Kit | Invitrogen | Cat#: 61012 |
| SmartSeq DNA library preparation Kits | Takarabio | Cat#: 634471 |
| NexteraXT DNA library preparation Kits | Illumina | Cat#: FC-131-1024 |
| RNeasy Mini Kit | Qiagen | Cat#: 74104 |
| High-capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat#: 4368814 |
| iTaq Universal SYBR Green Supermix | Bio Rad | Cat#: 1725121 |
| Animal-free blocker | Vector Laboratories | Cat#: SP-5030 |
| ProLong Gold Antifade | Thermo Fisher | Cat#: P36930 |
| Deposited data | ||
| Gene Expression Omnibus | This paper | GSE198573 |
| Experimental models: Organisms/strains | ||
| Mouse: Dlg1min | Todd Nystul | Rivera et al., 2009 |
| Mouse: Arhgap31flox | Nathalie LaMarche Vane | Caron et al., 2016 |
| Mouse: Dlg1flox: B6;129-Dlg1tm1Rlh/J | JAX | Strain #:013097 RRID:IMSR_JAX:013 097 |
| Mouse: Lgr5GFP-CreERT2: B6.129P2-Lgr5tm1(cre/ERT2)Cle/J | JAX | Strain #:008875 RRID:IMSR_JAX:008 875 |
| Mouse: VillinCreERT: B6.Cg-Tg(Vil1cre/ERT2)23Syr/J | JAX | Strain #:020282 RRID:IMSR_JAX:020 282 |
| Mouse: ROSA26tdTomato: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | JAX | Strain #:007914 RRID:IMSR_JAX:007 914 |
| Mouse: ROSA26mTmG: B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | JAX | Strain #:007676 RRID:IMSR_JAX:007 676 |
| Mouse: PdgfrαH2B-eGFPB6.129S4-Pdgfratm11(EGFP)Sor/J | JAX | Strain #:007669 RRID:IMSR_JAX:007 669 |
| Oligonucleotides | ||
| RNAscope® probe Mm-Arhgap31 | Advanced Cell Diagnostics | Cat#: 569971 |
| Oligonucleotides | Table S1 | N/A |
| Software and algorithms | ||
| Fiji v2.1.0/1.53c | Open source | https://imagej.net/software/fiji |
| GraphPad Prism9 | GraphPad Software | https://www.graphpad.com |
| Adobe Illustrator v25.4 | Adobe | https://www.adobe.com |
| Adobe Photoshop | Adobe | https://www.adobe.com |
| QuantStudio Real-time PCR software v1.3 | Applied Biosystems | https://www.thermofisher.com |
| LasX v3.4.2 | Leica | https://www.leicamicrosystems.com |
| ZEN Blue v2.5 | Zeiss | https://www.zeiss.com |
| STAR 2.4.2a | Dobin et al., 2013 | https://code.google.com/archive/p/rna-star/ |
| DESeq2 v1.16.1 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| BD FACS Diva v.8.0.1 | BD Biosciences | https://www.bdbiosciences.com/ |
| FlowJo v10.8 | Tree Star | https://www.flowjo.com |