Abstract
Purpose
To develop models based on radiomics and genomics for predicting the histopathologic nuclear grade with localized clear cell renal cell carcinoma (ccRCC) and to assess whether macro-radiomics models can predict the microscopic pathological changes.
Method
In this multi-institutional retrospective study, a computerized tomography (CT) radiomic model for nuclear grade prediction was developed. Utilizing a genomics analysis cohort, nuclear grade-associated gene modules were identified, and a gene model was constructed based on top 30 hub mRNA to predict the nuclear grade. Using a radiogenomic development cohort, biological pathways were enriched by hub genes and a radiogenomic map was created.
Results
The four-features-based SVM model predicted nuclear grade with an area under the curve (AUC) score of 0.94 in validation sets, while a five-gene-based model predicted nuclear grade with an AUC of 0.73 in the genomics analysis cohort. A total of five gene modules were identified to be associated with the nuclear grade. Radiomic features were only associated with 271 out of 603 genes in five gene modules and eight top 30 hub genes. Differences existed in the enrichment pathway between associated and un-associated with radiomic features, which were associated with two genes of five-gene signatures in the mRNA model.
Conclusion
The CT radiomics models exhibited higher predictive performance than mRNA models. The association between radiomic features and mRNA related to nuclear grade is not universal.
Abbreviations: ccRCC, Clear cell renal cell carcinoma; WHO/ISUP, World Health Organization and International Society of Urological Pathology; FDR, False discovery rate; GLRLM, Gray level run length matrix; GLSZM, Gray level size matrix; NGTDM, Neighborhood gray tone difference matrix; KEGG, KOBAS-Kyoto Encyclopedia of Genes and Genomes; TCGA, The cancer genome atlas; TCIA, The cancer imaging archive; WGCNA, Weighted gene co-expression network; PPI, Protein-Protein Interaction Networks
Keywords: Kidney, Radiomics, Neoplasms-Primary, Computer Applications, Radiogenomics, Pathological nuclear grade
Highlights
-
•
A radiomics model and genomics model was developed for predicting the histopathologic nuclear grade with localized clear cell renal cell carcinoma (ccRCC).
-
•
The CT radiomics models exhibited higher predictive performance than mRNA models.
-
•
The association between radiomics features and mRNA related to nuclear grade is not universal.
1. Introduction
Radiogenomics focus on discovering correlations between radiomic features and genomics [1]. Based on the principle that medical images are the outcome of complex interactions at the genetic and molecular levels [2], radiogenomic studies have revealed associations between imaging features and genetic information of tumours in glioblastoma, gastric, ovarian and other cancers [3], [4], [5]. Nuclear grade of ccRCC is considered to affect prognosis and surgical benefits, and its preoperative evaluation can provide effective guidelines for clinical treatments [6]. Therefore, predicting ccRCC nuclear grade is important in clinical practice. Studies evaluating the ccRCC nuclear grade prediction revealed that performance of a CT-radiomics-based model was higher than that of a The Cancer Genomics Atlas (TCGA) transcriptomics-based model, as indicated by the AUCs in the validation set which were 0.90 and 0.82, respectively [7], [8]. Radiogenomic studies for prognosis and prediction of ccRCC suggested the existence of correlations between radiomic features and prognosis-related pathways such as cell cycle and microvascular infiltration [9], [10], [11]. The answer to predicting nuclear grade of ccRCC may lie in deciphering the association between nuclear grade-related radiomic features and genomics. However, the underlying mechanisms governing this association have yet to be elucidated. Under this light, the present study aimed to a) predict the nuclear grade of ccRCC by investigating the correlation between CT radiomic features and mRNA data, b) decipher the underlying genomic mechanisms of the tested radiomic models, and c) provide insights in the interpretation of relevant radiomic features.
2. Materials and methods
2.1. Study design
The design of the study included three main stages: First, we extracted the suitable CT radiomic features for building a model. Second, we identified gene modules related to nuclear grade and hub genes for building the genomics model. Finally, we mapped the radiomic features with gene modules, hub genes and gene signatures (Fig. 1).
Fig. 1.
Overview of the study cohort and design.
2.2. Patient information sources
In this multi-institutional retrospective study, a total of 645 images of patients were retrieved from the kits21 public imaging dataset, The Cancer Imaging Archive (TCIA), The Cancer Genomics Atlas (TCGA), and the radiology department of local hospital from January 2015 to December 2021. The acquisition of images and genomic data from public database sources such as TCIA and TCGA and, the approval of this study by the ethics committee of the local institution, waived the requirement to obtain informed patient consent. The study included all patients with pathologically confirmed ccRCC that underwent a preoperative enhanced CT scan, except those with multiple regional tumours. Patients were divided into four subgroups. Subgroup 1 comprised of 103 patients with preoperative CT images (corticomedullary) collected from the kits21 dataset and used to develop the radiomics model. Subgroup 2 included 23 patients with images from local hospitals, used to construct a validation set with subgroup 4 for radiomics analysis. Subgroup 3 included 519 patients with mRNA sequencing data obtained from TCGA, used as a genomics analysis cohort for gene module identification and development of gene models. Subgroup 4 has served as a subset of subgroup 3, consisting of 34 patients with both preoperative CT corticomedullary images and mRNA sequencing data. Subgroup 4 was used in combination with subgroup 2 as a radiomics validation set and for analysing the potential association between radiomic features and mRNA expression.
2.3. Evaluation of pathological nuclear grade
Nuclear grade information from the local hospital originated from the pathology department of local hospital and evaluated by two independent pathologists according to the WHO/ISUP grade system, while all kidney biopsy specimens were re-examined independently by a pathologist with 7 years of genitourinary pathology experience. Grades between 1 and 2 of the WHO grade system were considered as low and grades between 3 and 4 were considered as high.
2.4. CT images
All CT examinations at the local hospital were conducted using spiral scanners to acquire images of corticomedullary phase on the CT system. All patients were given 400 mL of water orally 25 min before the examination and injected with a dose of 1.5 mL/kg of a non-ionic contrast agent. After 25–30 s, diaphragm to kidney images of the corticomedullary phase were obtained. The image parameters were as follows: (1) manufacturer: Siemens; (2) gantry rotation time 0.5 s; (3) tube voltage: 120 kV; (4) detector collimation: 64 × 0.6 mm. all images were reconstructed as a 5 mm thick reconstructed section.
2.5. Radiomic analysis
2.5.1. Tumour segmentation
Image voxels were resampled to 1 × 1 mm2, and 25 Gray levels were used to discretize their intensities. Pre-testing of 60 randomly selected patients showed no significant difference in predictive performance between ROI (region of interest) and VOI (volume of interest) (Supplement Fig. 1). Hence, segmentation was performed in the largest diameter cross-section along the tumour border by a radiologist with 8 years of experience using the ITK-snap software. Additionally, 30 patients were randomly selected for repeat segmentation by another senior radiologist to select for robust radiomic features.
2.5.2. Feature selection and radiomic model construction
Features of the segmented 2D tumour were extracted from the original images of cases using python package named Pyradiomics, following an intraclass correlation coefficient (ICC) test, the radiomic features with ICC less than 0.80 were excluded. The univariate Wilcoxon rank sum test was used to screen for radiomic features significantly associated with nuclear grade. Only radiomic features with ICC equal or more than 0.8 and univariate Wilcox-rank sum test p values less than 0.01 qualified for further analysis. Consequently, the maximum Relevance and Minimum Redundancy (mRMR) and Boruta's algorithms were used to select the features with a mean absolute correlation greater than 0.9. The selected radiomic features were used to construct radiomic models by using six classified algorithmic methods, which included Logistic regression (LR), Naive Bayes (NB), Decision Tree (DT), Random Forest (RF), Support vector machine (SVM), and XGBoost (XGB). Prediction performance was estimated by the AUC of the receiver operator characteristic curve (ROC). The optimal cut-off value was obtained from the Jorden index of the ROC curve in the training set. The accuracy (ACC), sensitivity (Se), specificity (Sp), negative prediction rate (NPV), and positive prediction rate (PPV) of the model were also calculated. The DeLong test was used for comparison between different AUCs.
2.6. Nuclear grade-related gene module identification
2.6.1. Differential gene expression analysis for nuclear grade
The mRNA data of the genomics analysis cohort were processed using the "limma" R package to identify differentially expressed genes, with a differential expression threshold calculated according to the following formula: . Significance of differential genes was evaluated using a t-test, and differential expressed genes were considered the ones having false discovery rates (FDRs) less than 0.05 and fold changes (FCs) greater than the threshold, the differentially expressed genes (DEGs) was subsequently used in the WGCNA algorithm for further analysis.
2.6.2. Construction of weighted gene co-expression networks
Based on the genomics analysis cohort (subgroup 3), a scale-free network of DEGs was constructed using the WGCNA algorithm to identify gene modules related to nuclear grades. A soft threshold was calculated based on the corresponding scale independence (R²) and average connectivity. An adjacency matrix was constructed based on the soft threshold and the Pearson correlation coefficient between pairs of genes, which was then transformed into a topological overlap matrix (TOM) and the dissimilarity (1-TOM) was calculated. According to the similarity matrix, similar genes were grouped into one gene co-expression modules by hierarchical clustering, setting the number of genes within a module to be no less than 30 and the height threshold for gene module merging to 0.25. The association between gene modules and nuclear grade was visualized as a heat map, and the respective significance was expressed as gene significance (GS). Associations with a false discovery rate (FDR) corrected p value less than 0.05 were considered significant.
2.7. Construction of radiogenomic maps
Radiogenomic association maps were created based on the radiogenomic development and genomic analysis cohorts including the gene modules related to nuclear grade, using the Spearman correlation with a significance p value of 0.05.
2.7.1. Screening of nuclear grade-related hub genes
Based on genomics analysis cohort, Protein-Protein Interaction Networks (PPI) were constructed by submitting the WGCNA results into the STRING database and setting the minimum interaction scores to 0.4. Then the results were imported into the Cytoscape software (v3.8.2), and the Degree Centrality of genes was calculated using the Degree algorithm. Top 30 genes were regarded as hub genes.
2.8. Identification of biological pathways for hub genes
The biological pathways of hub genes were identified using the R package “Cluster Profiler” for KOBAS Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, based on the radiogenomic development cohort. A false positive rate (FDR) p-value less than 0.05 was considered as significant.
2.9. Multigene model for predicting nuclear grade
Based on the genomics cohort analysis, the genetic sample was randomly divided into training and validation sets in a 7:3 ratio. The training and validation sets were balanced using the Synthetic Minority Over-Sampling Technique (SMOTE), and the dataset was normalized using the minmax method. The Pearson correlation coefficient (PCC) was calculated for each gene pair, and any one of the genes in pairs with PCC> 0.99 was randomly removed. Gene selection was performed using recursive feature elimination (RFE), before the construction of RF, SVM, DT, and LR models. All the above processes were performed by running the FAE software (V0.5.3) on python (3.7.6) (https://github.com/salan668/FAE).
3. Statistical analysis of clinical data
Data was analysed using R 4.1.2. Nuclear grade, sex and age were analysed using Chi-square or Wilcoxon test. Statistical differences were significant at a p value less than 0.05.
4. Results
4.1. Clinical information of patients
The general clinical information of the study cohort is shown in Table 1. With the exception of the nuclear grade of ccRCC, no significant differences were found between age and gender.
Table 1.
Demographic and clinical information of the study cohorts.
| train | test | p value | |
|---|---|---|---|
| N = 103 | N = 57 | ||
| Age | 61.000 [51.000;69.000] | 60.000 [50.000;75.500] | 0.056 |
| Sex | 0.052 | ||
| female | 32 (31.068%) | 14 (24.561%) | |
| male | 71 (68.932%) | 43 (75.439%) | |
| Pathological-nuclear-grading | 0.001 | ||
| low | 59 (57.282%) | 31 (54.386%) | |
| high | 44 (42.718%) | 26 (45.614%) |
4.2. Radiomic feature selection and model construction
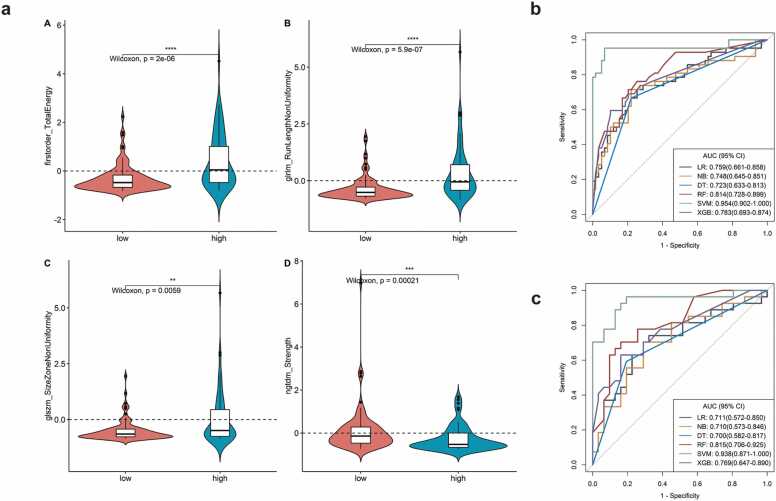
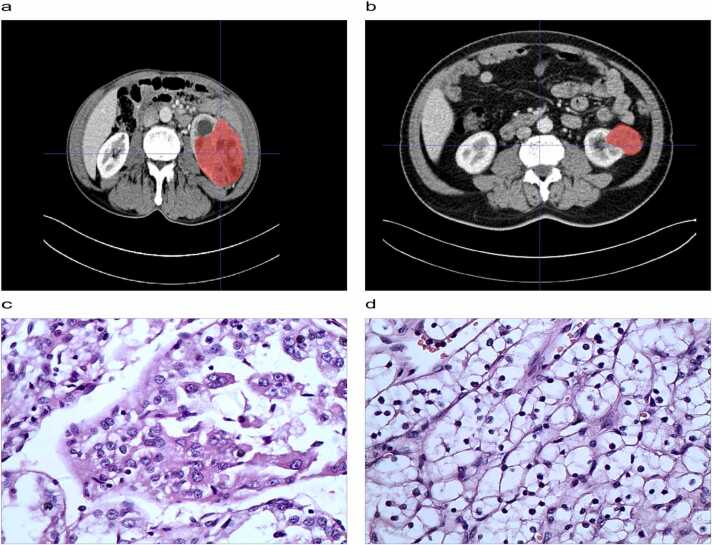
A total of 107 radiomic features were extracted, including 14 morphological feature, 18 first-order features and 75 texture features. The result of ICC test revealed that 102 radiomic features was identified. After univariate Wilcox-rank sum test analysis, 11 radiomic features was screened. Finally, the four radiomic features were selected by mRMR and Boruta's algorithms (Fig. 2a). The values of neighbourhood Gray-tone difference matrix (NGTDM)-strength were reduced, while values of the other three radiomic features increased in the higher grades. The prediction performance of the six classifiers is shown in Fig. 2b and c. The SVM classification method achieved AUC and ACC values of 0.94 and 0.85 in the validation set, reaching the best performance among the six methods in both the training and validation sets with a cut-off value of 0.37 (Table 2). The results of the DeLong test between the SVM classifier and other classifiers also demonstrate that the prediction effectiveness of SVM was higher than the other five classes of classifiers (Table 3a and b). We showed corticomedullary images and histopathological images in a patient with high-grade ccRCC and a patient with low-grade ccRCC in the cohort (Fig. 3a-d).
Fig. 2.
Selection of radiomic features and the development of radiomics model. a: Values of four selected radiomic features of high and low nuclear grades, b: ROC curve of six classified algorithms in the training set, c: The ROC curve of six classified algorithms in the validation set.
Table 2.
Prediction performance of radiomics models constructed by six classifiers.
| method | feature_numbers | train_auc | train_acc | train_sen | train_spe | train_ppv | train_npv | test_auc | test_acc | test_sen | test_spe | test_ppv | test_npv | cutoff |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LR | 4 | 0.759(0.661–0.858) | 0.733(0.635–0.816) | 0.714(0.499–0.833) | 0.746(0.424–0.865) | 0.667(0.583–0.700) | 0.786(0.676–0.810) | 0.711(0.572–0.850) | 0.707(0.573–0.819) | 0.704(0.370–0.889) | 0.710(0.290–0.903) | 0.679(0.526–0.727) | 0.733(0.529–0.778) | 0.36104249901878 |
| NB | 4 | 0.748(0.645–0.851) | 0.752(0.657–0.833) | 0.714(0.404–0.857) | 0.780(0.423–0.864) | 0.698(0.566–0.735) | 0.793(0.675–0.810) | 0.710(0.573–0.846) | 0.672(0.537–0.790) | 0.519(0.222–0.816) | 0.806(0.612–0.936) | 0.700(0.500–0.786) | 0.658(0.593–0.691) | 0.101875041416575 |
| DT | 4 | 0.723(0.633–0.813) | 0.733(0.635–0.816) | 0.667(0.371–0.799) | 0.780(0.508–0.872) | 0.683(0.545–0.721) | 0.767(0.682–0.786) | 0.700(0.582–0.817) | 0.707(0.573–0.819) | 0.593(0.333–0.751) | 0.806(0.506–0.923) | 0.727(0.600–0.772) | 0.694(0.588–0.722) | 0.479414682539682 |
| RF | 4 | 0.814(0.728–0.899) | 0.762(0.667–0.841) | 0.714(0.452–0.858) | 0.797(0.542–0.898) | 0.714(0.613–0.750) | 0.797(0.727–0.815) | 0.815(0.706–0.925) | 0.759(0.628–0.861) | 0.667(0.283–0.889) | 0.839(0.548–0.968) | 0.783(0.605–0.828) | 0.743(0.654–0.769) | 0.43 |
| SVM | 4 | 0.954(0.902–1.000) | 0.941(0.875–0.978) | 0.952(0.714–1.000) | 0.932(0.153–0.983) | 0.909(0.882–0.913) | 0.965(0.818–0.967) | 0.938(0.871–1.000) | 0.845(0.726–0.927) | 0.963(0.852–1.000) | 0.742(0.097–0.935) | 0.765(0.742–0.771) | 0.958(0.750–0.967) | 0.370222946320497 |
| XGB | 4 | 0.783(0.693–0.874) | 0.772(0.678–0.850) | 0.595(0.333–0.715) | 0.898(0.729–0.966) | 0.806(0.700–0.833) | 0.757(0.717–0.770) | 0.769(0.647–0.890) | 0.707(0.573–0.819) | 0.444(0.222–0.741) | 0.935(0.710–1.000) | 0.857(0.750–0.909) | 0.659(0.595–0.674) | 0.603537037968635 |
Table 3a.
DeLong test among radiomics models of training set.
| name | p value |
|---|---|
| LR-NB | 0.519020801 |
| LR-DT | 0.228144417 |
| LR-RF | 0.111431884 |
| LR-SVM | 0.0000743 |
| LR-XGB | 0.272050649 |
| NB-DT | 0.393096654 |
| NB-RF | 0.061762627 |
| NB-SVM | 0.0000552 |
| NB-XGB | 0.116238452 |
| DT-RF | 0.00091159 |
| DT-SVM | 0.000000251 |
| DT-XGB | 0.013907988 |
| RF-SVM | 0.000426971 |
| RF-XGB | 0.214224916 |
| SVM-XGB | 0.0000602 |
Table 3b.
DeLong test among radiomics models of validation set.
| name | p value |
|---|---|
| LR-NB | 0.97683832 |
| LR-DT | 0.810259242 |
| LR-RF | 0.044645072 |
| LR-SVM | 0.000698819 |
| LR-XGB | 0.13932625 |
| NB-DT | 0.862149834 |
| NB-RF | 0.046600886 |
| NB-SVM | 0.00037982 |
| NB-XGB | 0.194359811 |
| DT-RF | 0.003248959 |
| DT-SVM | 0.0000378 |
| DT-XGB | 0.148733046 |
| RF-SVM | 0.016589642 |
| RF-XGB | 0.299101041 |
| SVM-XGB | 0.001676355 |
Fig. 3.
Two examples of radiomic features nuclear grade of ccRCC. a: corticomedullary images of high nuclear grade of ccRCC, b: corticomedullary images of low nuclear grade of ccRCC, c: histopathological images of high nuclear grade of ccRCC, d: histopathological images of low nuclear grade of ccRCC.
4.3. Radiogenomic analysis and hub-related pathway identification
4.3.1. Differential genes associated with the nuclear grade
Based on genomics analysis cohort, mRNAs differential analysis was performed using a threshold value of 0.579. The differential expression analysis revealed 887 gene, including 451 down-regulated and 436 up-regulated genes.
4.3.2. Identification of gene modules associated with nuclear grade
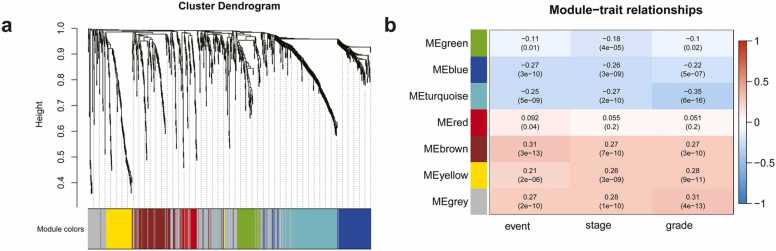
Based on genomics analysis cohort, we selected β = 10 as a suitable soft threshold to construct the scale-free network, and 887 differential genes were divided into five gene modules (Fig. 4a). Five gene modules (brown, yellow, turquoise, blue, and green modules) containing 603 genes were associated with nuclear grade (p < 0.05) (Fig. 4b), which were associated with radiomic features in the radiogenomic development cohort.
Fig. 4.
Results of the WGCNA analysis of genomics analysis cohort. a: Genes with the same characteristics were grouped into a gene module, and each module was assigned a colour, b: The five-gene module that is shown to be related with the nuclear grade.
4.3.3. Construction of radiogenomic maps, hub gene selection and pathway identification
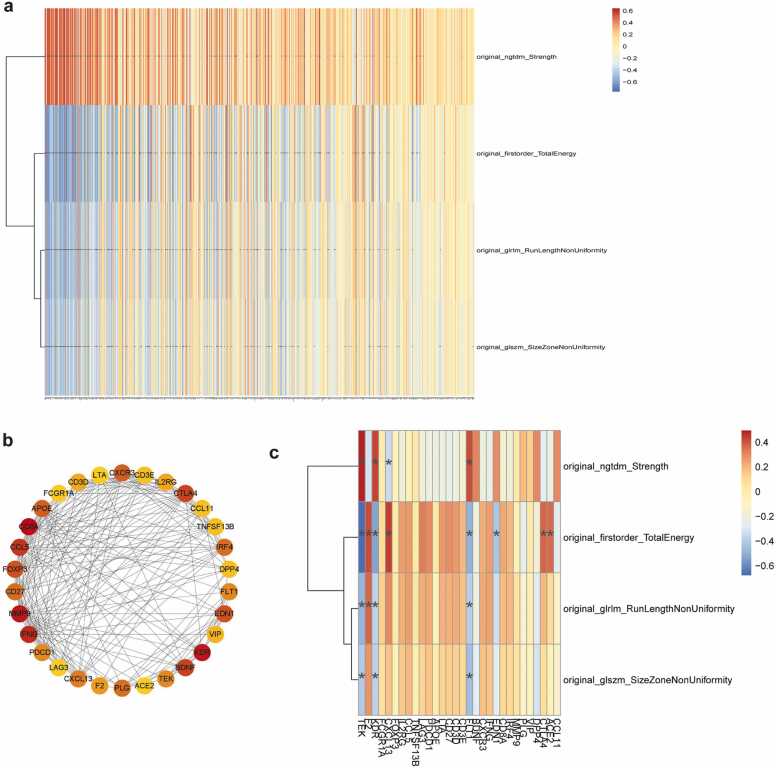
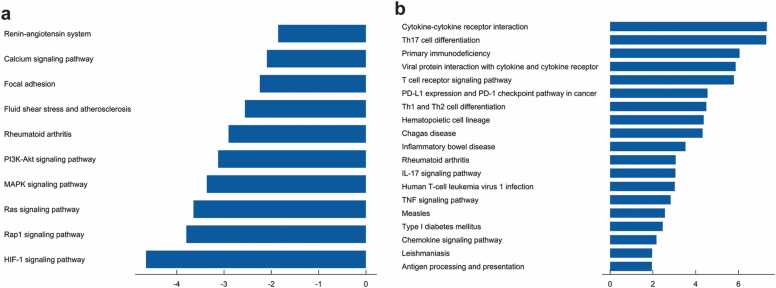
The radiogenomic development cohort revealed that 271 genes were associated with radiomic features while the remaining 332 genes were not associated with radiomic features, as shown in the respective radiogenomic map (Fig. 5a). The results of the hub genes are shown in Fig. 5b. The radiogenomic map revealed that 8 hub genes were associated with radiomic features, while 22 were not associated with the radiomic features (Fig. 5c). The radiomic features-related 8 hub genes were enriched in 10 KEGG pathways, including PI3K-Akt pathway, HIF-1 signalling pathway, and MAPK signalling pathway (Fig. 6a). The remaining 22 hub genes were enriched in 19 KEGG pathways, including IL-17, Th17 cell differentiation and PD-L1 immune checkpoint signalling pathway, and other tumour-related signalling pathways (Fig. 6b).
Fig. 5.
Analysis of the radiogenomic development cohort. a: The radiogenomic map constructed by four radiomic features and related genes, b: Top 30 hub genes by the Degree algorithm, c: The radiogenomic map constructed by four radiomic features and 30 hub genes.
Fig. 6.
The KEGG results for hub genes distinguished by association. a: The pathway underlying radiomic feature-correlated genes, b: The pathway underlying radiomics feature non-correlated genes.
4.4. Gene signature for nuclear grade prediction
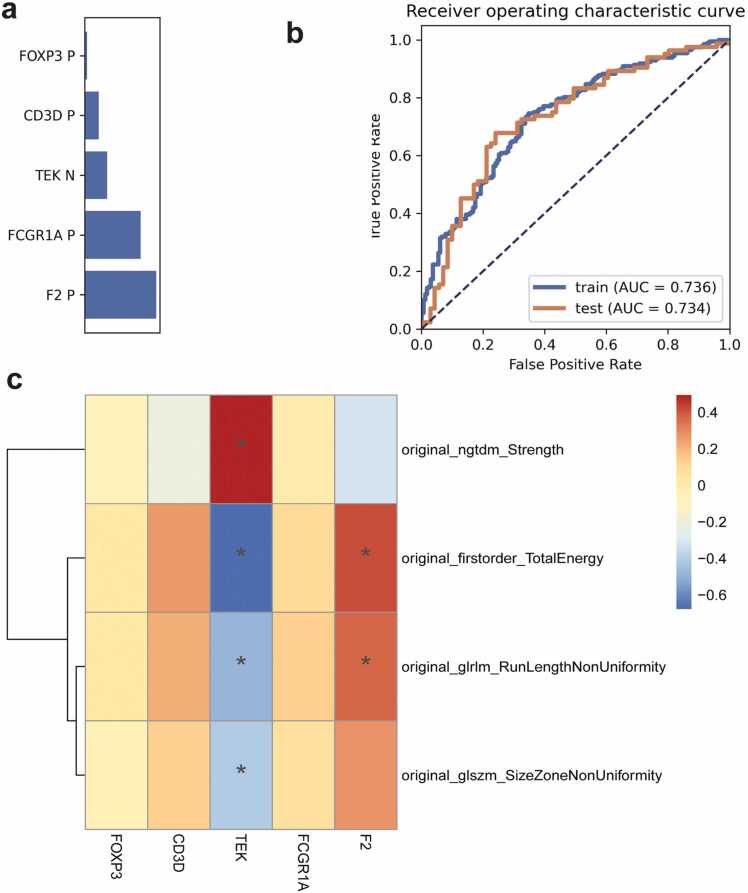
Five hub genes were selected and used to develop a LR model with the highest AUC value of 0.72 in the validation set (Fig. 7a). The prediction performance and result of DeLong test among four algorithms is shown in Table 4a, Table 4b, Table 5a, Table 5b, respectively. The results of the analysis between radiomic features and gene signatures showed that F2, TEK was associated with the four radiomic features (Fig. 7b).
Fig. 7.
Prediction performance of the multigene model. a: The contribution of gene signatures in the final multigene model, b: ROC curve of the multigene model in training and validation sets, c: The radiogenomic map constructed by combining genes of the multigene model and radiomic features.
Table 4a.
Prediction performance of the genomics models constructed by four classifiers of training set.
| feature-selection of training set | Number | PosNum | NegNum | AUC | 95% CIs | Std | Acc | Youden Index | Sen | Spe | PPV | NPV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MinMax_PCC_RFE_5_LR | 364 | 197 | 167 | 0.7363 | [0.6882–0.7895] | 0.0262 | 0.7033 | 0.4541 | 0.7462 | 0.6527 | 0.7171 | 0.6855 |
| MinMax_PCC_RFE_9_SVM | 364 | 197 | 167 | 0.7251 | [0.6726–0.7781] | 0.0265 | 0.6868 | 0.4691 | 0.7716 | 0.5868 | 0.6878 | 0.6853 |
| MinMax_PCC_RFE_7_RF | 364 | 197 | 167 | 1 | [1.0000–1.0000] | 0 | 1 | 0.6 | 1 | 1 | 1 | 1 |
| MinMax_PCC_RFE_8_DT | 364 | 197 | 167 | 1 | [1.0000–1.0000] | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
Table 4b.
Prediction performance of the genomics models constructed by four classifiers of validation set.
| feature-selection of training set | Number | PosNum | NegNum | AUC | 95% CIs | Std | Acc | Youden Index | Sen | Spe | PPV | NPV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MinMax_PCC_RFE_5_LR | 155 | 84 | 71 | 0.7341 | [0.6496–0.8155] | 0.0419 | 0.7161 | 0.4904 | 0.6786 | 0.7606 | 0.7703 | 0.6667 |
| MinMax_PCC_RFE_9_SVM | 155 | 84 | 71 | 0.7202 | [0.6353–0.8042] | 0.0417 | 0.6774 | 0.562 | 0.5595 | 0.8169 | 0.7833 | 0.6105 |
| MinMax_PCC_RFE_7_RF | 155 | 84 | 71 | 0.7076 | [0.6278–0.7898] | 0.0404 | 0.6581 | 0.550 | 0.5595 | 0.7746 | 0.746 | 0.5978 |
| MinMax_PCC_RFE_8_DT | 155 | 84 | 71 | 0.5804 | [0.5067–0.6600] | 00.0394 | 0.5806 | 1 | 0.583 | 0.575 | 0.6203 | 0.5395 |
Table 5a.
Results of the DeLong test between genomics models of training set.
| Classifier name | p value |
|---|---|
| LR-RF | < 0.0001 |
| LR-SVM | 0.402 |
| LR-DT | < 0.0001 |
| RF-SVM | < 0.0001 |
| RF-DT | 1 |
| SVM-DT | < 0.0001 |
Table 5b.
Results of the DeLong test between genomics models of validation set.
| Classifier name | p value |
|---|---|
| LR-RF | 0.399 |
| LR-SVM | 0.514 |
| LR-DT | 0.0003 |
| RF-SVM | 0.717 |
| RF-DT | 0.003 |
| SVM-DT | 0.002 |
5. Discussion
In conclusion, a radiomics model used to predict the ccRCC nuclear grade was developed, and radiomic features were correlated with genes in order to classify the nuclear grade and explore the driving pathway of the radiomic features. In comparison with this study, most previous radiogenomic studies identified gene modules associated with radiomic features, but not gene modules that were not associated with radiomic features but which may also predict the clinical outcome. In order to avoid selection bias of gene modules, we identified gene modules based on clinical outcomes, including gene modules un-associated with the radiomic features. Furthermore, previous studies have not evaluated the predictive performance of gene modules, whereas in our study we constructed a hub gene model for predicting nuclear grade.
CT radiomic features able to identify the nuclear grade of ccRCC in the validation cohort with an AUC of 0.81 have been previously reported [12]. The respective CT based SVM model performed well in the validation cohort (AUC=0.82) [13]. These results are consistent with our study, where the constructed SVM model had a comparable performance in the validation set (AUC=0.94). In addition, we constructed a hub mRNA model for predicting the nuclear grade. Biological processes always involve interactions among genes expressed by interaction networks which are controlled by driving hub genes [14], [15]. Exploring the predictive performance of hub genes can help understand the association between radiomic features and genes. Our hub mRNA genes achieved a comparable AUC (0.73) in the validation set, which is lower than that of similar previous studies (AUC=0.82) [8]. The different predictive performance between the two models suggests that single-dimension genes such as mRNA genes may not reflect the entire variation observed in nuclear grades. mRNA is also regulated by other factors such as lncRNA, miRNA, and methylation modifications [16], [17], [18]. This is evident by the fact that combined models of mRNA and lncRNA achieved higher predictive efficacy compared with the two individual models [19]. Furthermore, a multi-omics model significantly outperformed the single-omics model [20]. These findings may explain the reason why the gene model we developed had a lower predictive performance. In a similar way, the differences in predictive performance between the radiomics model and the mRNA model utilized in our study suggest that radiomics may be influenced by and reflect multi-dimensional biological information. Previous studies have indicated that CT radiomic features can predict variation of different dimensions such as individual key genes and methylation phenotypes [21], [22], [23], [24]. These results are also consistent with the theory that radiomic features can reflect changes in different dimensions, containing information not found in the genomic features [25]. By connecting different dimensions of biological information, the values of radiomic features may serve as indicators of the product of multi-omics molecule interaction through activation cascade.
One of the limitations in deploying radiomics analysis in clinical practice is the lack of interpretability. Previous studies have shown the ability of the four imaging features to reflect tumour progression. Firstorder_TotalEnergy has been reported to predict the aggressiveness of papillary thyroid cancer and the T-stage of colorectal cancer [26], [27]. Both glrlm_RunLengthNonUniformity and glszm_SizeZoneNonUniformity can distinguish large nodular medullary thyroid carcinoma from papillary thyroid carcinoma. In addition, glszm_SizeZoneNonUniformity can predict the non-perfusion volume ratio (NPVR) after high-intensity focused ultrasound ablation of uterine fibroids, which could reflect the prognosis after treatment [28], [29], [30]. NGTDM_strength was found to be an independent preoperative predictor of T-stage in rectal cancer [31]. The rising popularity of radiogenomics suggests that radiomic features are increasingly relied on to elucidate biological mechanisms related to genes and pathways. A total of 13 MRI radiomic features were associated with immune regulation, cell function, tumour proliferation and treatment response in glioblastoma [32]. CT radiomics have been shown to be predictive of postoperative metastasis of ccRCC, which was correlated with ECM-receptor interactions, focal adhesion and PI3-Akt pathways [33]. Our KEGG results are in agreement with prior research revealing the critical role of these pathways in relation to the nuclear grade. Other ccRCC-related pathways include the MAPK pathway, which has been associated with tumour aggressiveness [34] and the HIF-1 pathway demonstrated to be associated with nuclear grade [35]. Furthermore, the Ras signalling pathway, which is related to the HIF pathway, has been reported to be involved in the regulation of ccRCC progression and drug resistance [36]. CT radiomic features correlate with genes associated with clinical outcomes, such as postoperative distant metastasis, overall survival, and immune subtypes of ccRCC [10], [33], [37]. Radiomic features-associated genes was enriched in critical pathways for ccRCC development, suggesting that four radiomic features was related to the MAPK, HIF-1 and Ras signalling pathways.
However, previous radiogenomic studies have not yet investigated the generality of this association. Interestingly, in the present study we found that hub genes which were not associated with radiomic features exhibited comparable ccRCC nuclear grade predictive performance. Our findings indicate that non-associated genes may also constitute an added value to the radiomic model. A glioma research study comparing a radiomic model separately with a combined genetic, clinical and radiomics model revealed that the C-index improved from 0.75 to 0.81 [38]. A deep learning survival prediction model combining MRI radiomic features with gene expression data, indicated that the added genetic markers contributed to improving the predictive performance of the radiomics model [39]. These studies indicate that genes have an added value for radiomics and deep learning models. Genomics could complement genomic-related information which cannot be reflected by radiomic features. Radiomics may contain biological information beyond a single dimension. Considering the possible complementary relationship between radiomics and genomics, combined models of radiogenomics may help comprehensively decipher tumour-related biological information.
However novel, our study has several limitations. First, the radiogenomic development cohort had a small sample size. Nevertheless, we have used multi-institutional data for radiomics analysis and utilized WGCNA results from genomics analysis for the radiogenomic development cohort ensuring the robustness of gene modules and radiomic features in distinguishing nuclear grades. Second, although we detected genes which were not associated with the radiomic features, we did not estimate the added value of those genes in the prediction performance of our model. The above shortcomings will be addressed in a follow-up study, where more samples of both imaging, gene expression as well as multi-omics data will be processed in order to enhance the validity and interpretability of radiomics. Finally, the corticomedullary phase used in this study may be difficult to obtain highly reproducible radiomic features, although we validated its stable predictive efficacy in the test set, and in the future study we will use the arterial phase to validate the efficacy of the classification of the four radiomic features.
In conclusion, the major findings of this study were the following: a) Both radiomic model constructed based on CT cortex medullary phase and the mRNA model from the TCGA database can predict nuclear grade, and b) The predictive radiomic features were associated with only a part of the hub genes related to nuclear grade. In conclusion, this study revealed differences in the performance between radiomic and genomic mRNA models for predicting nuclear grade of ccRCC. A non-universal association between the radiomic features and genetic data at the mRNA level was found, which could provide useful insights for future radiogenomic studies.
Fund information
This study was supported by the entrepreneurship project of Gansu University of Chinese Medicine (grant number. LCCX2021006).
CRediT authorship contribution statement
Xuan-ming He: Investigation, Formal analysis, Data curation, Writing – original draft, Visualization, Software. Jian-xin Zhao: Resources, Data curation. Di-liang He: Resources, Data curation. Jia-liang Ren: Formal analysis, Visualization, Software. Lian-ping Zhao: Project administration. Gang Huang: Conceptualization, Methodology, Validation, Writing – review & editing, Supervision, Funding acquisition.
Acknowledgements
Thanks to contribution of TCGA database for their platforms and datasets. And thanks to the researchers and study participants for their contributions and finance support of grant from innovation and entrepreneurship project of Gansu University of Chinese medicine. We would like to thank Editage (www.editage.cn) for English language editing.
Consent for publication
The authors affirm that human research participants provided informed consent for publication of all the images.
Author contributions
All authors contributed to the study conception and design, GH and XH designed the experimental flow. JZ and DH collected and downloaded the data, XH and RJ analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval
The studies involving human participants were reviewed and approved by The Ethics Committee of Gansu Provincial Hospital. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Footnotes
Grant number: GSWSKY2020-15 and grant number. LCCX2021006.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.ejro.2023.100476.
Appendix A. Supplementary material
Supplemental Figure. 1 Performance Comparison between ROI and VOI model. a: Predictive performance of the 2D-ROI model on the dataset adjusted using the SMOTE algorithm. b: Predictive performance of the 3D-VOI model on the dataset adjusted using the SMOTE algorithm. Supplementary material
.
References
- 1.Kuo M.D., Jamshidi N. Behind the numbers: decoding molecular phenotypes with radiogenomics--guiding principles and technical considerations. Radiology. 2014;270:320–325. doi: 10.1148/radiol.13132195. [DOI] [PubMed] [Google Scholar]
- 2.Pinker K., Chin J., Melsaether A.N., Morris E.A., Moy L. Precision medicine and radiogenomics in breast cancer: new approaches toward diagnosis and treatment. Radiology. 2018;287:732–747. doi: 10.1148/radiol.2018172171. [DOI] [PubMed] [Google Scholar]
- 3.Beig N., Bera K., Prasanna P., Antunes J., Correa R., Singh S., et al. Radiogenomic-based survival risk stratification of tumor habitat on Gd-T1w MRI Is associated with biological processes in glioblastoma. Clin. Cancer Res. 2020;26:1866–1876. doi: 10.1158/1078-0432.Ccr-19-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu H., Wang Y., Liu Y., Lin D., Zhang C., Zhao Y., et al. Contrast-enhanced computed tomography-based radiogenomics analysis for predicting prognosis in gastric cancer. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.882786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vargas H.A., Huang E.P., Lakhman Y., Ippolito J.E., Bhosale P., Mellnick V., et al. Radiogenomics of high-grade serous ovarian cancer: multireader multi-institutional study from the cancer genome atlas ovarian cancer imaging research group. Radiology. 2017;285:482–492. doi: 10.1148/radiol.2017161870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J., Cao N., Li S., Wang Y. Identification of a risk stratification model to predict overall survival and surgical benefit in clear cell renal cell carcinoma with distant metastasis. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.630842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shu J., Wen D., Xi Y., Xia Y., Cai Z., Xu W., et al. Clear cell renal cell carcinoma: Machine learning-based computed tomography radiomics analysis for the prediction of WHO/ISUP grade. Eur. J. Radio. 2019;121 doi: 10.1016/j.ejrad.2019.108738. [DOI] [PubMed] [Google Scholar]
- 8.Wan F., Zhu Y., Han C., Xu Q., Wu J., Dai B., et al. Identification and validation of an eight-gene expression signature for predicting high Fuhrman grade renal cell carcinoma. Int J. Cancer. 2017;140:1199–1208. doi: 10.1002/ijc.30535. [DOI] [PubMed] [Google Scholar]
- 9.Jamshidi N., Jonasch E., Zapala M., Korn R.L., Aganovic L., Zhao H., et al. The radiogenomic risk score: construction of a prognostic quantitative, noninvasive image-based molecular assay for renal cell carcinoma. Radiology. 2015;277:114–123. doi: 10.1148/radiol.2015150800. [DOI] [PubMed] [Google Scholar]
- 10.Gao J., Ye F., Han F., Jiang H., Zhang J. A radiogenomics biomarker based on immunological heterogeneity for non-invasive prognosis of renal clear cell carcinoma. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.956679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng H., Chen L., Wang M., Luo Y., Huang Y., Ma X. Integrative radiogenomics analysis for predicting molecular features and survival in clear cell renal cell carcinoma. Aging (Albany NY) 2021;13:9960–9975. doi: 10.18632/aging.202752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang R., Hu Z., Shen X., Wang Q., Zhang L., Wang M., et al. Computed tomography-based radiomics model for predicting the WHO/ISUP grade of clear cell renal cell carcinoma preoperatively: a multicenter study. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.543854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xv Y., Lv F., Guo H., Zhou X., Tan H., Xiao M., et al. Machine learning-based CT radiomics approach for predicting WHO/ISUP nuclear grade of clear cell renal cell carcinoma: an exploratory and comparative study. Insights Imaging. 2021;12:170. doi: 10.1186/s13244-021-01107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogelstein B., Lane D., Levine A.J. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 15.Fu M., Hu Y., Lan T., Guan K.L., Luo T., Luo M. The Hippo signalling pathway and its implications in human health and diseases. Signal Transduct. Target Ther. 2022;7:376. doi: 10.1038/s41392-022-01191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y., Feng Y.C., Gan Y., Teng L., Wang L., La T., et al. LncRNA MILIP links YBX1 to translational activation of Snai1 and promotes metastasis in clear cell renal cell carcinoma. J. Exp. Clin. Cancer Res. 2022;41:260. doi: 10.1186/s13046-022-02452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Y.F., Chou J.L., Chang J.S., Chiu I.J., Chiu H.W., Lin Y.F. Dysregulation of the miR-25-IMPA2 axis promotes metastatic progression in clear cell renal cell carcinoma. EBioMedicine. 2019;45:220–230. doi: 10.1016/j.ebiom.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi Y.C., Chen X.Y., Zhang J., Zhu J.S. Novel insights into the interplay between m(6)A modification and noncoding RNAs in cancer. Mol. Cancer. 2020;19:121. doi: 10.1186/s12943-020-01233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Liu S.B., Shen J., Bai L., Zhang X., Cao J., et al. Development and Validation of Prognostic Model for Lung Adenocarcinoma Patients Based on m6A Methylation Related Transcriptomics. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.895148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D., Li R., Lucas A., Verma S.S., Dudek S.M., Ritchie M.D. Using knowledge-driven genomic interactions for multi-omics data analysis: metadimensional models for predicting clinical outcomes in ovarian carcinoma. J. Am. Med Inf. Assoc. 2017;24:577–587. doi: 10.1093/jamia/ocw165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z.C., Zhai G., Zhang J., Wang Z., Liu G., Wu G.Y., et al. Differentiation of clear cell and non-clear cell renal cell carcinomas by all-relevant radiomics features from multiphase CT: a VHL mutation perspective. Eur. Radio. 2019;29:3996–4007. doi: 10.1007/s00330-018-5872-6. [DOI] [PubMed] [Google Scholar]
- 22.Feng Z., Zhang L., Qi Z., Shen Q., Hu Z., Chen F. Identifying BAP1 mutations in clear-cell renal cell carcinoma by CT radiomics: preliminary findings. Front Oncol. 2020;10:279. doi: 10.3389/fonc.2020.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang C., Cintra M., Brennan K., Zhou M., Colevas A.D., Fischbein N., et al. Development and validation of radiomic signatures of head and neck squamous cell carcinoma molecular features and subtypes. EBioMedicine. 2019;45:70–80. doi: 10.1016/j.ebiom.2019.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun R., Limkin E.J., Vakalopoulou M., Dercle L., Champiat S., Han S.R., et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180–1191. doi: 10.1016/s1470-2045(18)30413-3. [DOI] [PubMed] [Google Scholar]
- 25.Mazurowski M.A. Radiogenomics: what it is and why it is important. J. Am. Coll. Radio. 2015;12:862–866. doi: 10.1016/j.jacr.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 26.Wei R., Zhuang Y., Wang L., Sun X., Dai Z., Ge Y., et al. Histogram-based analysis of diffusion-weighted imaging for predicting aggressiveness in papillary thyroid carcinoma. BMC Med Imaging. 2022;22:188. doi: 10.1186/s12880-022-00920-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dou Y., Tang X., Liu Y., Gong Z. T stage prediction of colorectal tumor based on multiparametric functional images. Transl. Cancer Res. 2020;9:522–528. doi: 10.21037/tcr.2019.11.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao L., Ma B. Radiomics features of different sizes of medullary thyroid carcinoma (MTC) and papillary thyroid carcinoma (PTC) tumors: a comparative study. Clin. Med Insights Oncol. 2022;16 doi: 10.1177/11795549221097675. 11795549221097675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei C., Li N., Shi B., Wang C., Wu Y., Lin T., et al. The predictive value of conventional MRI combined with radiomics in the immediate ablation rate of HIFU treatment for uterine fibroids. Int J. Hyperth. 2022;39:475–484. doi: 10.1080/02656736.2022.2046182. [DOI] [PubMed] [Google Scholar]
- 30.Spies J.B. Sustained relief of leiomyoma symptoms by using focused ultrasound surgery. Obstet. Gynecol. 2007;110:1427–1428. doi: 10.1097/01.AOG.0000295979.81092.e5. author reply 1428-1429. [DOI] [PubMed] [Google Scholar]
- 31.You J., Yin J. Performances of whole tumor texture analysis based on MRI: predicting preoperative T stage of rectal carcinomas. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.678441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Q., Chen Y., Liang C., Zhao Y., Lv X., Zou Y., et al. Biologic pathways underlying prognostic radiomics phenotypes from paired MRI and RNA sequencing in glioblastoma. Radiology. 2021;301:654–663. doi: 10.1148/radiol.2021203281. [DOI] [PubMed] [Google Scholar]
- 33.Zhao Y., Liu G., Sun Q., Zhai G., Wu G., Li Z.C. Validation of CT radiomics for prediction of distant metastasis after surgical resection in patients with clear cell renal cell carcinoma: exploring the underlying signaling pathways. Eur. Radio. 2021;31:5032–5040. doi: 10.1007/s00330-020-07590-2. [DOI] [PubMed] [Google Scholar]
- 34.Zhang G., Yu Z., Fu S., Lv C., Dong Q., Fu C., et al. ERCC6L that is up-regulated in high grade of renal cell carcinoma enhances cell viability in vitro and promotes tumor growth in vivo potentially through modulating MAPK signalling pathway. Cancer Gene Ther. 2019;26:323–333. doi: 10.1038/s41417-018-0064-8. [DOI] [PubMed] [Google Scholar]
- 35.Kroeze S.G., Vermaat J.S., van Brussel A., van Melick H.H., Voest E.E., Jonges T.G., et al. Expression of nuclear FIH independently predicts overall survival of clear cell renal cell carcinoma patients. Eur. J. Cancer. 2010;46:3375–3382. doi: 10.1016/j.ejca.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 36.Green Y.S., Sargis T., Reichert E.C., Rudasi E., Fuja D., Jonasch E., et al. Hypoxia-associated factor (HAF) mediates neurofibromin ubiquitination and degradation leading to ras-ERK pathway activation in hypoxia. Mol. Cancer Res. 2019;17:1220–1232. doi: 10.1158/1541-7786.Mcr-18-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Y., Zeng H., Chen L., Luo Y., Ma X., Zhao Y. Exploration of an integrative prognostic model of radiogenomics features with underlying gene expression patterns in clear cell renal cell carcinoma. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.640881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Stefano A.L., Picca A., Saragoussi E., Bielle F., Ducray F., Villa C., et al. Clinical, molecular, and radiomic profile of gliomas with FGFR3-TACC3 fusions. Neuro Oncol. 2020;22:1614–1624. doi: 10.1093/neuonc/noaa121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Islam M., Wijethilake N., Ren H. Glioblastoma multiforme prognosis: MRI missing modality generation, segmentation and radiogenomic survival prediction. Comput. Med Imaging Graph. 2021;91 doi: 10.1016/j.compmedimag.2021.101906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure. 1 Performance Comparison between ROI and VOI model. a: Predictive performance of the 2D-ROI model on the dataset adjusted using the SMOTE algorithm. b: Predictive performance of the 3D-VOI model on the dataset adjusted using the SMOTE algorithm. Supplementary material