Abstract
Introduction
We evaluated tissue tumor mutational burden (tTMB) and mutations in STK11, KEAP1, and KRAS as biomarkers for outcomes with pembrolizumab plus platinum-based chemotherapy (pembrolizumab-combination) for NSCLC among patients in the phase 3 KEYNOTE-189 (ClinicalTrials.gov, NCT02578680; nonsquamous) and KEYNOTE-407 (ClinicalTrials.gov, NCT02775435; squamous) trials.
Methods
This retrospective exploratory analysis evaluated prevalence of high tTMB and STK11, KEAP1, and KRAS mutations in patients enrolled in KEYNOTE-189 and KEYNOTE-407 and the relationship between these potential biomarkers and clinical outcomes. tTMB and STK11, KEAP1, and KRAS mutation status was assessed using whole-exome sequencing in patients with available tumor and matched normal DNA. The clinical utility of tTMB was assessed using a prespecified cutpoint of 175 mutations/exome.
Results
Among patients with evaluable data from whole-exome sequencing for evaluation of tTMB (KEYNOTE-189, n = 293; KEYNOTE-407, n = 312) and matched normal DNA, no association was found between continuous tTMB score and overall survival (OS) or progression-free survival for pembrolizumab-combination (Wald test, one-sided p > 0.05) or placebo-combination (Wald test, two-sided p > 0.05) in patients with squamous or nonsquamous histology. Pembrolizumab-combination improved outcomes for patients with tTMB greater than or equal to 175 compared with tTMB less than 175 mutations/exome in KEYNOTE-189 (OS, hazard ratio = 0.64 [95% confidence interval (CI): 0.38‒1.07] and 0.64 [95% CI: 0.42‒0.97], respectively) and KEYNOTE-407 (OS, hazard ratio = 0.74 [95% CI: 0.50‒1.08 and 0.86 [95% CI: 0.57‒1.28], respectively) versus placebo-combination. Treatment outcomes were similar regardless of KEAP1, STK11, or KRAS mutation status.
Conclusions
These findings support pembrolizumab-combination as first-line treatment in patients with metastatic NSCLC and do not suggest the utility of tTMB, STK11, KEAP1, or KRAS mutation status as a biomarker for this regimen.
Keywords: Tissue tumor mutational burden, Single-gene genetic alterations, Pembrolizumab, Metastatic non‒small-cell lung cancer, Biomarker
Introduction
Pembrolizumab plus platinum-based chemotherapy (pembrolizumab-combination) is a standard-of-care first-line treatment for patients with metastatic NSCLC irrespective of tumor programmed death-ligand 1 (PD-L1) expression.1,2 The role of pembrolizumab-combination in treatment is supported by two placebo-controlled randomized phase 3 studies that reported improved overall survival (OS) and progression-free survival (PFS) among patients with previously untreated metastatic nonsquamous NSCLC without sensitizing EGFR mutation or ALK alteration (KEYNOTE-189; NCT02578680) or metastatic squamous NSCLC (KEYNOTE-407; NCT02775435) irrespective of PD-L1 expression levels.3,4
Tumor mutational burden (TMB), defined as the number of somatic mutations in the tumor genome, is of interest as a biomarker for immune checkpoint inhibitors.5, 6, 7 There is discordant evidence for TMB as a biomarker for treatment outcomes with first-line immunotherapy versus immunotherapy plus chemotherapy in advanced NSCLC.8 Using whole-exome sequencing (WES) of patients with NSCLC who received pembrolizumab monotherapy, tissue TMB (tTMB) was found to be associated with OS and PFS benefit.9 Findings from other studies of immunotherapy with anti‒PD-(L)1 agents alone or in combination with anti‒CTLA-4 therapy suggest that tTMB may have clinical utility as a biomarker for treatment outcomes.5,10, 11, 12, 13, 14
Mutations in driver genes, including in STK11 (also known as LKB1), KEAP1, and KRAS, occur in a meaningful proportion of patients with advanced or metastatic NSCLC and are of interest as potential biomarkers for outcomes with anti‒PD-(L)1 therapy.15, 16, 17 Mutations in STK11 and KRAS are more common in patients with nonsquamous histology (including adenocarcinoma) than those with squamous histology.18 KRAS G12C is the most frequently occurring KRAS mutation in NSCLC, comprising approximately 35% of identified KRAS mutations.18,19 Mutations in KRAS are a common oncogenic driver in nonsquamous NSCLC,18 and some studies have suggested that KRAS mutations may be associated with improved outcomes with anti‒PD-(L)1 therapy plus chemotherapy.17 STK11 and KEAP1 mutations have been associated with poor outcomes in nonsquamous NSCLC, may occur concurrently with KRAS mutations, and have been associated with a potential lack of benefit with anti‒PD-(L)1 therapy plus chemotherapy.15, 16, 17,20,21
To investigate the prevalence and potential clinical utility of tTMB and STK11, KEAP1, and KRAS mutations as biomarkers of outcomes, we conducted separate exploratory analyses of the KEYNOTE-189 and KEYNOTE-407 trials in patients with metastatic NSCLC who received pembrolizumab or placebo plus platinum-based chemotherapy.
Materials and Methods
Study Design and Patients
The KEYNOTE-189 (ClinicalTrials.gov, NCT02578680)3 and KEYNOTE-407 (ClinicalTrials.gov, NCT02775435)4 trials were randomized, double-blind, placebo-controlled phase 3 trials enrolling patients with previously untreated metastatic NSCLC regardless of tumor PD-L1 expression. Patients had nonsquamous NSCLC without sensitizing EGFR or ALK alterations in KEYNOTE-189 and squamous NSCLC in KEYNOTE-407. The study protocols and all amendments were approved by the appropriate ethics committee at each study site. Patients provided written informed consent before participation.
Treatment
In KEYNOTE-189, patients were randomized 2:1 to receive four 3-week cycles of intravenous pembrolizumab 200 mg or placebo, plus pemetrexed 500 mg/m2 and either cisplatin (75 mg/m2) or carboplatin (area under the concentration‒time curve = 5 mg/mL/min) followed by pembrolizumab or placebo once every 3 weeks for an additional 31 cycles (35 cycles in total) and indefinite pemetrexed maintenance therapy.
In KEYNOTE-407, patients were randomized 1:1 to four 3-week cycles of intravenous pembrolizumab 200 mg or placebo plus carboplatin (area under the concentration‒time curve = 6 mg/mL/min) and either paclitaxel (200 mg/m2) or nab-paclitaxel (100 mg/m2) followed by pembrolizumab or placebo once every 3 weeks for an additional 31 cycles (35 cycles in total).
Assessments
tTMB and select single-gene mutations (STK11, KEAP1, and KRAS) were assessed centrally by WES of tumor tissue and matched normal DNA as previously described.22 tTMB was assessed using a prespecified cutpoint of 175 mutations/exome (mut/exome) to define subgroups with high tTMB (≥175 mut/exome; tTMB-high) versus low tTMB (<175 mut/exome; tTMB-low). This cutpoint was derived using GEP and WES TMB data from a training set of patients with multiple tumor types across the pembrolizumab clinical program, in which 175 mut/exome yielded the most statistically significant difference in the distribution of a gene expression profile comprising 18 genes.6,23, 24, 25 This cutpoint most closely approximates the 10 mutations per megabase used by the updated pipeline FoundationOne F1Dx_v3.2 assay (FoundationMedicine, Cambridge, MA).26, 27, 28 Full methodology for WES analysis is included in Supplementary Methods.
End Points
The clinical objectives of KEYNOTE-189 and KEYNOTE-407 have been reported previously.3,4 The objectives of these analyses were to evaluate the prevalence of high tTMB and STK11, KEAP1, and KRAS mutations in patients enrolled in KEYNOTE-189 and KEYNOTE-407 and to evaluate the relationship between these potential biomarkers and clinical outcomes (OS, PFS, and objective response rate [ORR]) in patients treated with pembrolizumab-combination and placebo-combination. Additional objectives were to investigate the relationship between tTMB and tumor PD-L1 expression, the association between tTMB and treatment efficacy, and the clinical utility of tTMB as a predictor of efficacy. Exploratory biomarker analyses were prespecified in the study protocol for each study. The statistical analysis plan was prespecified before merging clinical and biomarker data.
Statistical Analysis
Efficacy was assessed in the biomarker-evaluable populations, which comprised randomized patients who had evaluable samples for WES and received one or more doses of study treatment. The association between tTMB, assessed as a continuous log10-transformed variable, and treatment efficacy were evaluated separately for each trial, with the significance level set at 0.05 and no multiplicity adjustment. Wald tests on the tTMB regression coefficients were used to calculate one-sided p values for pembrolizumab, under the hypothesis that higher tTMB positively associates with improved outcomes. Two-sided p values were calculated for chemotherapy because there was no a priori hypothesis regarding the direction of the association. Descriptive analyses were performed to assess the association between STK11, KEAP1, and KRAS status and clinical outcomes (OS, PFS, and ORR). The prespecified statistical analysis plan is described in Supplementary Methods.
Results
Patients
In KEYNOTE-189, 293 of 616 (47.6%) randomized patients had evaluable WES data and were included in the tTMB-evaluable population (pembrolizumab, n = 207; control, n = 86), and 289 (46.9%) had matched normal DNA and were included in the single-gene mutation-evaluable population (STK11, KEAP1, and KRAS). The data cutoff for all analyses from KEYNOTE-189 was September 21, 2018 (Supplementary Fig. 1A). In KEYNOTE-407, 312 of 559 (55.8%) randomized patients with evaluable WES data were included in the tTMB-evaluable population (pembrolizumab, n = 143; control, n = 169), and 285 (46.9%) were included in the single-gene mutation-evaluable population (KEAP1; STK11 and KRAS were not evaluated in patients with squamous NSCLC owing to the low prevalence of these mutations in squamous NSCLC). The data cutoff date for all analyses from KEYNOTE-407 was May 9, 2019 (Supplementary Fig. 1B). Demographics and baseline clinical characteristics are described in Table 1.
Table 1.
Demographics and Baseline Clinical Characteristics in the tTMB-Evaluable Populations in Each Study
| Characteristics | KEYNOTE-189 |
KEYNOTE-407 |
||||
|---|---|---|---|---|---|---|
| tTMB-Evaluable Population (n = 293) | Single-Gene Mutation-Evaluable Population (n = 289) | Total Population (n = 616) | tTMB-Evaluable Population (n = 312) | Single-Gene Mutation-Evaluable Populationa (n = 285) | Total Population (n = 559) | |
| Median age, y (IQR) | 64 (56–69) | 63 (56–69) | 64 (57–69) | 66 (60–71) | 66 (60–71) | 65 (60–71) |
| Male | 166 (56.7) | 162 (56.1) | 363 (58.9) | 252 (80.8) | 230 (80.7) | 455 (81.4) |
| ECOG performance status 1 | 164 (55.9) | 162 (56.1) | 346 (56.2) | 215 (68.9) | 194 (68.1) | 396 (70.8) |
| Former or current smoker | 260 (88.7) | 256 (88.6) | 543 (88.1) | 291 (93.3) | 265 (93.0) | 518 (92.7) |
| PD-L1 TPS | ||||||
| <1% | 99 (33.8) | 98 (33.9) | 190 (30.8) | 111 (35.6) | 100 (35.1) | 194 (34.7) |
| 1%–49% | 910 (31.1) | 90 (31.1) | 186 (30.2) | 117 (37.5) | 111 (38.9) | 207 (37.0) |
| ≥50% | 98 (33.4) | 96 (33.2) | 202 (32.8) | 83 (26.6) | 74 (26.0) | 146 (26.1) |
| Could not be evaluatedb | 5 (1.7) | 5 (1.7) | 38 (6.2) | 0 (0.0) | 0 (0.0) | 12 (2.1) |
ECOG, Eastern Cooperative Oncology Group; IQR, interquartile range; PD-L1 TPS, programmed death-ligand 1 tumor proportion score; tTMB, tissue tumor mutational burden; WES, whole-exome sequencing.
Note: Data are presented as n (%) unless otherwise noted.
KRAS and STK11 mutation data were excluded for KEYNOTE-407 because these mutations are rare in squamous NSCLC, and the number of patients with these mutations who also had evaluable WES data from both tumor and normal DNA was small.
Specimens had an inadequate number of tumor cells or no tumor cells.
Clinical Outcomes in the tTMB-Evaluable Population and Association of tTMB With Efficacy
In each study, clinical outcomes (i.e., OS, PFS, and ORR) in the tTMB-evaluable groups for pembrolizumab-combination versus placebo-combination were similar to those in the intent-to-treat population (Supplementary Table 1). tTMB and PD-L1 tumor proportion score (TPS) were not strongly associated with one another in either treatment arm in either study (Supplementary Fig. 2A and B). For assessment of the association of tTMB with efficacy, on the basis of the area under the receiver operating characteristic curve for ORR, higher tTMB assessed as a continuous variable was not associated with ORR in either treatment arm (Fig. 1A and C). No association was found between tTMB (assessed as a continuous variable) and ORR (in logistic regression analyses) or OS and PFS (in Cox proportional hazard regression analyses) in either treatment arm in either study (Wald test one-sided, p > 0.05 for the pembrolizumab-combination arm and two-sided p > 0.05 for the placebo-combination arm in each study; Fig. 1B and D).
Figure 1.
Association of tTMB with efficacy outcomes in (A) and (B) KEYNOTE-189 and (C) and (D) KEYNOTE-407. In panels A and C, the graph illustrates the area under the ROC curve for ORR. Panels B and D provide p values for OS, PFS, and ORR in each respective study from logistic regression analysis. ap values were calculated using the Wald test and are one-sided for pembrolizumab-combination (a priori hypothesis that tTMB was positively associated with improved outcomes for pembrolizumab-combination) and two-sided for placebo-combination (no a priori hypothesis regarding the direction of the association between tTMB and outcomes) with significance level set at 0.05 and no multiplicity adjustment. tTMB was graphed on a log10 scale for the ROC curve. AUC, area under the curve; CI, confidence interval; ORR, objective response rate; OS, overall survival; PD-L1, programmed death-ligand 1; PFS, progression-free survival; r, correlation coefficient; ROC, receiver operating characteristics; tTMB, tissue tumor mutational burden; TPS, tumor proportion score.
Clinical Outcomes in Patients With tTMB Greater Than or Equal to 175 Mutations/Exome and tTMB Less Than 175 Mutations/Exome
In KEYNOTE-189, 134 patients had tTMB greater than or equal to 175 mut/exome (pembrolizumab-combination, n = 100; placebo-combination, n = 34) and 159 had tTMB less than 175 mut/exome (pembrolizumab-combination, n = 107; placebo-combination, n = 52). Hazard ratios (HRs) (95% confidence interval [CI]) for OS favored the pembrolizumab-combination group in patients with tTMB greater than or equal to 175 mut/exome (0.64, 0.38‒1.07) and in patients with tTMB less than 175 mut/exome (0.64, 0.42‒0.97) (Fig. 2A). HRs (95% CI) for PFS favored the pembrolizumab-combination group in patients with tTMB greater than or equal to 175 mut/exome (0.32, 0.21‒0.51) and in patients with tTMB less than 175 mut/exome (0.51, 0.35‒0.74) (Fig. 2B). In the tTMB greater than or equal to 175 mut/exome group, the ORR (95% CI) was 50.0% (39.8%‒60.2%) with pembrolizumab-combination versus 11.8% (3.3%‒27.5%) with placebo-combination. For patients with tTMB less than 175 mut/exome, the ORR (95% CI) was 40.2% (30.8%‒50.1%) versus 19.2% (9.6%‒32.5%), respectively (Supplementary Fig. 3A).
Figure 2.
Clinical utility of tTMB for OS and PFS in each study at cutpoints of greater than or equal to 175 mut/exome and less than 175 mut/exome. Kaplan-Meier estimates of OS (A) and PFS (B) in KEYNOTE-189. Kaplan-Meier estimates of OS (C) and PFS (D) in KEYNOTE-407. Chemo, chemotherapy; CI, confidence interval; HR, hazard ratio; mut, mutation; NR, not reached; OS, overall survival; PFS, progression-free survival; tTMB, tissue tumor mutational burden.
In KEYNOTE-407, 162 patients had tTMB greater than or equal to 175 mut/exome (pembrolizumab-combination, n = 73; placebo-combination, n = 89) and 150 patients had tTMB less than 175 mut/exome (pembrolizumab-combination, n = 70; placebo-combination, n = 80). The HRs for OS favored pembrolizumab-combination in the tTMB greater than or equal to 175 mut/exome group (0.74; 95% CI 0.50‒1.08) and less than 175 mut/exome group (0.86; 95% CI: 0.57‒1.28) (Fig. 2C). PFS was improved with pembrolizumab-combination among patients with tTMB greater than or equal to 175 mut/exome (HR = 0.57; 95% CI: 0.41‒0.81) and less than 175 mut/exome (HR = 0.68; 95% CI: 0.48‒0.96) (Fig. 2D). In the tTMB greater than or equal to 175 mut/exome group, ORR (95% CI) was 58.9% (46.8%‒70.3%) with pembrolizumab-combination versus 44.9% (34.4%‒55.9%) with placebo-combination. For patients with tTMB less than 175 mut/exome, ORR (95% CI) was 64.3% (51.9%‒75.4%) versus 38.8% (28.1%‒50.3%), respectively (Supplementary Fig. 3B).
Clinical Outcomes in Patients With Versus Without Single-Gene Mutations
STK11
Of the 289 evaluable patients in KEYNOTE-189, 54 (18.7%) had STK11 mutations. In KEYNOTE-407, 8 of 285 (2.8%) evaluable patients had STK11 mutations. Because STK11 mutations occurred infrequently in KEYNOTE-407, associations between STK11 status and PD-L1, tTMB, or outcomes were not evaluated.
In KEYNOTE-189, the median (interquartile range [IQR]) PD-L1 TPS tended to be numerically lower (0% [0‒16] versus 15% [0‒75]) and the median (IQR) TMB scores (209 [132‒265] versus 146 [89‒264] mut/exome) tended to be numerically higher among patients with versus without an STK11 mutation (Supplementary Fig. 4A). The prevalence of STK11 mutations by PD-L1 (TPS) and tTMB score (mut/exome) in the STK11-evaluable population is illustrated in Supplementary Figure 4B.
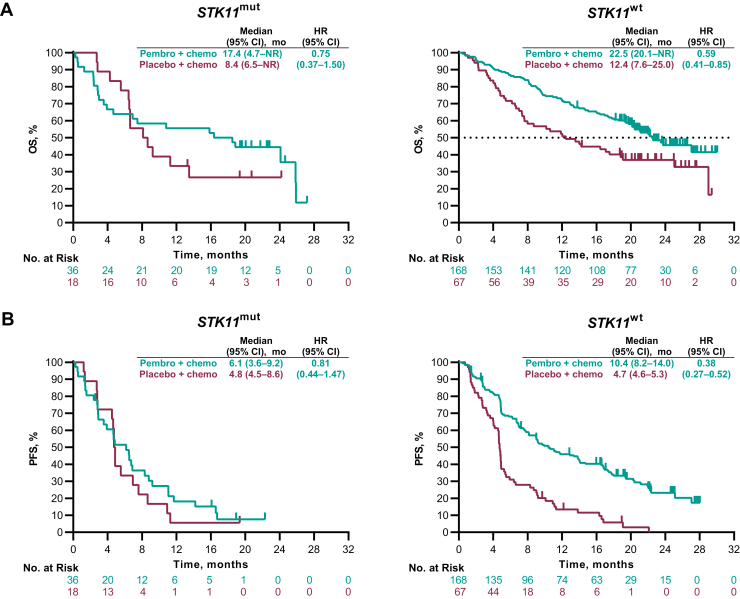
In KEYNOTE-189, the HRs (95% CI) for OS among patients with an STK11 mutation were 0.75 (0.37‒1.50) and 0.59 (0.41‒0.85) with wild-type STK11 (Fig. 3A). The HRs (95% CI) for PFS were 0.81 (0.44‒1.47) in patients with an STK11 mutation and 0.38 (0.27‒0.52) with wild-type STK11 (Fig. 3B). The ORRs (95% CI) for pembrolizumab-combination versus placebo-combination were 30.6% (16.4%‒48.1%) versus 16.7% (3.6%‒41.4%), respectively, in the STK11 mutation group and 48.8% (41.0%‒56.6%) versus 16.4% (8.5%‒27.5%), respectively, in the STK11 wild-type group (Supplementary Fig. 5).
Figure 3.
Kaplan-Meier estimates of OS and PFS by STK11 status in the single-gene mutation-evaluable population in KEYNOTE-189. (A) OS and (B) PFS. Chemo, chemotherapy; CI, confidence interval; HR, hazard ratio; mut, mutation; NR, not reached; OS, overall survival; mut, mutation; Pembro, pembrolizumab; PFS, progression-free survival; wt, wild-type.
KEAP1
Of the 289 patients in KEYNOTE-189 with evaluable WES data from matched tumor and normal DNA, 68 (23.5%) had KEAP1 mutations. In KEYNOTE-407, 285 patients had evaluable WES data from matched tumor and normal DNA, and 36 (12.6%) had KEAP1 mutations.
Among patients in KEYNOTE-189 with KEAP1 mutations, the median ([IQR]) PD-L1 TPS was numerically lower (1% [0‒13] versus 20% [0‒75]), and the median (IQR) tTMB score was numerically higher versus wild-type KEAP1 (173 [124‒267] versus 147 [89‒263] mut/exome) (Supplementary Fig. 6A). The prevalence of KEAP1 mutations by PD-L1 (TPS) and tTMB score (mut/exome) in the KEAP1-evaluable population is illustrated in Supplementary Figure 6B. Among patients in KEYNOTE-407 with KEAP1 mutations, the median (IQR) PD-L1 TPS (11% [1‒57]) and median (IQR) tTMB scores (205 [140‒296]) were numerically higher versus patients with wild-type KEAP1 (Supplementary Fig. 6C). No association between PD-L1 (TPS) and tTMB score (mut/exome) in the KEAP1-evaluable population was observed (Supplementary Fig. 6D).
In KEYNOTE-189, pembrolizumab-combination was associated with improved OS and PFS compared with placebo-combination, regardless of KEAP1 mutation status (KEAP1 mutation HR [95% CI] for OS, 0.81 [0.44‒1.49]; KEAP1 wild-type HR [95% CI] for OS, 0.57 [0.39‒0.84]) (Fig. 4A). The HRs (95% CI) for PFS were 0.65 (0.38‒1.12) in patients with KEAP1 mutations and 0.38 (0.28‒0.53) with KEAP1 wild-type (Fig. 4C). The ORR (95% CI) for pembrolizumab-combination versus placebo-combination was 35.6% (21.9%‒51.2%) versus 17.4% (5.0%‒38.8%), respectively, in patients with KEAP1 mutations and 48.4% (40.4%‒56.5%) versus 16.1% (8.0%‒27.7%), respectively, in patients with wild-type KEAP1 (Supplementary Fig. 7A).
Figure 4.
Kaplan-Meier estimates of OS and PFS by KEAP1 status in the single-gene mutation-evaluable populations. OS in (A) KEYNOTE-189 and (B) KEYNOTE-407. PFS in (C) KEYNOTE-189 and (D) KEYNOTE-407. Chemo, chemotherapy; CI, confidence interval; HR, hazard ratio; mut, mutation; NR, not reached; OS, overall survival; Pembro, pembrolizumab; PFS, progression-free survival; wt, wild-type.
In KEYNOTE-407, the HRs (95% CI) for OS were 1.08 (0.48‒2.41) in patients with KEAP1 mutations and 0.75 (0.55‒1.02) for wild-type KEAP1 (Fig. 4B). The HRs (95% CI) for PFS were 0.40 (0.19‒0.86) in patients with KEAP1 mutations and 0.63 (0.48‒0.83) in patients with wild-type KEAP1 (Fig. 4D). The ORRs (95% CI) for pembrolizumab-combination versus placebo-combination were 66.7% (34.9%‒90.1%) versus 54.2% (32.8%‒74.5%), respectively, in patients with KEAP1 mutations and 61.7% (52.4%‒70.4%) versus 41.9% (33.2%‒50.9%), respectively, in patients with wild-type KEAP1 (Supplementary Fig. 7B).
KRAS
Of the 289 evaluable patients in KEYNOTE-189, 89 (32.2%) had KRAS mutations, of which 37 (12.8%) were KRAS G12C mutations. In KEYNOTE-407, 14 out of 285 (4.9%) patients had KRAS mutations; none were KRAS G12C. Because KRAS occurred infrequently in KEYNOTE-407 (squamous NSCLC), associations between KRAS status and PD-L1, tTMB, or outcomes were not evaluated.
In KEYNOTE-189, the median (IQR) PD-L1 TPS (30% [1%‒71%] versus 5% [0%‒60%]) and median (IQR) TMB scores (204 [137‒276] versus 141 [85‒252] mut/exome) tended to be higher in patients with versus without KRAS mutations (Supplementary Fig. 8A). Joint association between PD-L1 (TPS) and tTMB score (mut/exome) for KRAS-mutant and KRAS wild-type patients is illustrated in Supplementary Figure 8B.
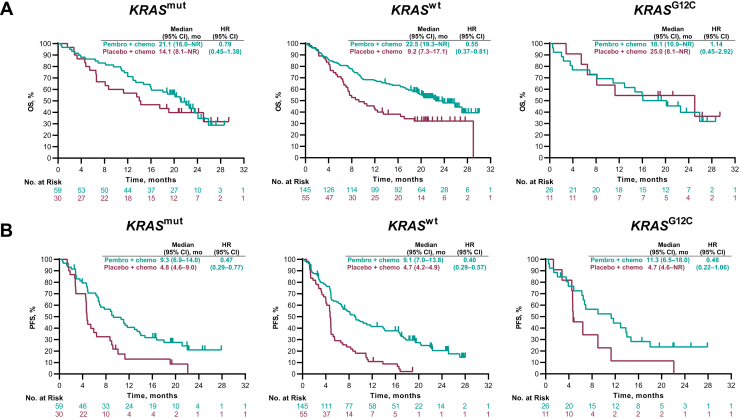
The HRs (95% CI) for OS were 0.79 (0.45‒1.38) for any KRAS mutation and 0.55 (0.37‒0.81) for KRAS wild-type (Fig. 5A). For PFS, the HRs (95% CI) were 0.47 (0.29‒0.77) for any KRAS mutation and 0.40 (0.29‒0.57) for KRAS wild-type (Fig. 5B). The ORR (95% CI) for pembrolizumab-combination versus placebo-combination was 40.7% (28.1%‒54.3%) versus 26.7% (12.3%‒45.9%) for any KRAS mutation and 47.6% (39.2%‒56.0%) versus 10.9% (4.1%‒22.3%) for wild-type KRAS (Supplementary Fig. 9).
Figure 5.
Kaplan-Meier estimates of OS and PFS by KRAS status in the single-gene mutation-evaluable population in KEYNOTE-189. (A) OS and (B) PFS. Chemo, chemotherapy; CI, confidence interval; HR, hazard ratio; mut, mutation; NR, not reached; OS, overall survival; Pembro, pembrolizumab; PFS, progression-free survival; wt, wild-type.
For the subgroup of patients with KRAS G12C mutation (pembrolizumab-combination, n = 26; placebo-combination, n = 11), the HRs for patients who received pembrolizumab-combination or placebo-combination were 1.14 (0.45‒2.92) and 0.48 (0.22‒1.06) for the OS and PFS, respectively (Fig. 5A and B). The corresponding ORRs were 50.0% (29.9%‒70.1%) and 18.2% (2.3%‒51.8%), respectively (Supplementary Fig. 9).
Discussion
Among patients with advanced NSCLC in the KEYNOTE-189 (nonsquamous) and KEYNOTE-407 (squamous) studies, first-line treatment with platinum-based chemotherapy with or without pembrolizumab revealed no association between tTMB, KEAP1 mutation (nonsquamous or squamous) or STK11, or KRAS mutation (nonsquamous) and treatment outcomes. There was no significant association between tTMB for either treatment arm and NSCLC histology. Furthermore, there was no strong correlation between tTMB and PD-L1 TPS in either treatment arm in either study. Pembrolizumab-combination revealed improved clinical benefit versus placebo-combination irrespective of mutations in STK11, KEAP1, and KRAS. These findings do not support the clinical utility of tTMB as a biomarker for pembrolizumab plus platinum-based chemotherapy for metastatic squamous or nonsquamous NSCLC.
The prevalence of tTMB, STK11, KEAP1, and KRAS mutations were generally consistent with that previously reported.7,17,29 The predictive value of tTMB as a biomarker for outcomes with anti‒PD-(L)1 therapy may vary when administered as monotherapy or in combination with chemotherapy. In an exploratory analysis of biomarker-evaluable data from the phase 3 KEYNOTE-042 trial of pembrolizumab monotherapy in patients with PD-L1 TPS greater than or equal to 1% advanced NSCLC that used a similar analytical approach, higher tTMB levels were associated with improved outcomes with pembrolizumab but not with chemotherapy. Moreover, patients with tTMB greater than or equal to 175 mut/exome had improved OS and PFS compared with chemotherapy, whereas those with tTMB less than 175 mut/exome did not.30 In other studies of anti‒PD-(L)1 therapies in NSCLC, a relationship between tissue or plasma TMB and clinical outcomes has been reported for studies of both monotherapies, including pembrolizumab,9 nivolumab,14 and atezolizumab,12 and immunotherapy combination therapies, such as nivolumab plus ipilimumab11,13,31 and durvalumab plus tremelimumab.32 A review of multiple studies of anti‒PD-(L)1 therapy given as single agents across various solid tumor types, including nonsquamous and squamous NSCLC, revealed a significant correlation between increasing TMB and increasing ORR (p < 0.001).33 This finding of an association between tTMB and outcomes with pembrolizumab monotherapy but not with pembrolizumab-combination represents a parallel to the utility of PD-L1 as a biomarker in first-line NSCLC: PD-L1 provides a biomarker for response with pembrolizumab monotherapy,34, 35, 36 but its predictive value is diminished among patients receiving pembrolizumab-combination.3,4 For patients with PD-L1‒negative disease (who are not eligible for pembrolizumab monotherapy), pembrolizumab plus chemotherapy remains an appropriate treatment option irrespective of tTMB.
We also investigated relationships between mutations in STK11, KEAP1, and KRAS and clinical outcomes in KEYNOTE-189 and KEYNOTE-407, each of which has been suggested to be potentially associated with outcomes among patients receiving anti‒PD-(L)1 therapy.17 Our results indicate that OS benefit persisted among patients who received pembrolizumab-combination regardless of STK11 or KEAP1 mutation status. There was no difference in PFS; however, given the relatively small number of patients, there is low precision for estimating the HRs for OS and PFS, as reflected in the very wide confidence intervals. Pembrolizumab-combination was generally associated with improved clinical outcomes compared with placebo-combination regardless of STK11, KEAP1, or KRAS mutation status; nevertheless, the magnitude of benefit in some groups remains uncertain. In KEYNOTE-407, the HR for OS was 0.96 versus 0.76 among patients with KEAP1 mutations versus wild-type KEAP1. However, given the small number of patients with a mutation, there is insufficient evidence to support the hypothesis of no benefit for pembrolizumab-combination in patients with squamous NSCLC with KEAP1 mutations or vice versa. KEYNOTE-189 did not provide evidence of an association between KRAS mutation status and outcomes with pembrolizumab-combination. Among patients with KRAS G12C mutation, the HR (95% CI) for OS was 1.14 (0.45‒2.92), although the sample size was too small to make definitive conclusions. The improvement in OS, PFS, and ORR with pembrolizumab-combination versus placebo-combination was observed irrespective of KRAS mutation status. These findings are consistent with other studies of the associations between these mutations and response and resistance to anti‒PD-(L)1 therapies, with KRAS mutations generally associated with improved outcomes and STK11 and KEAP1 mutations being associated with poorer outcomes compared with the corresponding wild-types.9,17,37, 38, 39, 40 In contrast with studies that have suggested STK11 and KEAP1 mutations confer resistance to anti‒PD-(L)1 therapies, patients with these mutations were found to have improved outcomes with pembrolizumab monotherapy versus chemotherapy in patients with advanced NSCLC in the KEYNOTE-042 study.41
These analyses were exploratory with few patients in some groups. Biomarker analyses were prespecified in the study protocol for both KEYNOTE-407 (squamous) and KEYNOTE-189 (nonsquamous) and the analysis plan was prespecified before the clinical and biomarker data were merged. Furthermore, our analysis only included patients with WES-evaluable samples, resulting in small sizes for certain groups. Notably, improvements in clinical outcomes observed with pembrolizumab-combination versus placebo-combination in the biomarker-evaluable populations were similar to the total populations of each study. As discussed, there is discordant evidence for TMB as a biomarker for treatment outcomes with first-line immunotherapy versus chemotherapy compared with immunotherapy plus chemotherapy versus chemotherapy in advanced NSCLC.8 Although WES is considered the gold-standard measurement of TMB, this technique is time-consuming, costly, and laborious.5 In addition, although there can be variations across cancer types,42 TMB 175 mut/exome assessed by WES has been shown to be well aligned with the FoundationOne CDx (Foundation Medicine, Cambridge, MA) TMB cutpoint of 10 mutations per megabase that is known to enrich for response across multiple solid tumor types, including NSCLC.28
In conclusion, the results of this exploratory analysis suggest that tTMB and STK11, KEAP1, and KRAS mutation status have limited clinical utility as biomarkers for patients treated with first-line pembrolizumab plus platinum-based chemotherapy in metastatic nonsquamous and squamous NSCLC. Our findings support the use of pembrolizumab plus platinum-based chemotherapy as a standard first-line combination therapy for patients with metastatic nonsquamous NSCLC, regardless of tTMB or STK11, KEAP1, or KRAS mutation status.
CRediT Authorship Contribution Statement
Marina C. Garassino, Shirish Gadgeel, Silvia Novello, Balazs Halmos, Enriqueta Felip, Giovanna Speranza, Rina Hui, Edward B. Garon, Hidehito Horinouchi, Shunichi Sugawara, Delvys Rodriguez-Abreu, Martin Reck, Razvan Cristescu, Deepti Aurora-Garg, Andrey Loboda, Jared Lunceford, Julie Kobie, Mark Ayers, Bilal Piperdi, M. Catherine Pietanza, Luis Paz-Ares: Writing - review & editing.
Julie Kobie, Razvan Cristescu: Data curation, Formal analysis.
Marina C. Garassino, Shirish Gadgeel, Silvia Novello, Balazs Halmos, Enriqueta Felip, Giovanna Speranza, Rina Hui, Edward B. Garon, Hidehito Horinouchi, Shunichi Sugawara, Delvys Rodriguez-Abreu, Martin Reck, Luis Paz-Ares: Supervision, Investigation.
Razvan Cristescu, Deepti Aurora-Garg, Andrey Loboda, Jared Lunceford, Julie Kobie, Mark Ayers, Bilal Piperdi, M. Catherine Pietanza: Methodology.
Data-Sharing Statement
Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA (Merck Sharp & Dohme) is committed to providing qualified scientific researchers access to anonymized data and clinical study reports from the company’s clinical trials for the purpose of conducting legitimate scientific research. Merck Sharp & Dohme is also obligated to protect the rights and privacy of trial participants and, as such, has a procedure in place for evaluating and fulfilling requests for sharing company clinical trial data with qualified external scientific researchers. The Merck Sharp & Dohme data-sharing website (available at: http://engagezone.msd.com/ds_documentation.php) outlines the process and requirements for submitting a data request. Applications will be promptly assessed for completeness and policy compliance. Feasible requests will be reviewed by a committee of Merck Sharp & Dohme subject matter experts to assess the scientific validity of the request and the qualifications of the requestors. In line with data privacy legislation, submitters of approved requests must enter into a standard data-sharing agreement with Merck Sharp & Dohme before data access is granted. Data will be made available for request after product approval in the United States and European Union or after product development is discontinued. There are circumstances that may prevent Merck Sharp & Dohme from sharing requested data, including country or region-specific regulations. If the request is declined, it will be communicated to the investigator. Access to genetic or exploratory biomarker data requires a detailed, hypothesis-driven statistical analysis plan that is collaboratively developed by the requestor and Merck Sharp & Dohme subject matter experts; after approval of the statistical analysis plan and execution of a data-sharing agreement, Merck Sharp & Dohme will either perform the proposed analyses and share the results with the requestor or will construct biomarker covariates and add them to a file with clinical data that is uploaded to an analysis portal so that the requestor can perform the proposed analyses.
Acknowledgments
All authors’ institutions received research funding from Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, for the conduct of this study. The authors thank the patients and their families and caregivers for participating in these trials; all the investigators and site personnel; Eli Lilly and company (Indianapolis, IN) for providing pemetrexed; and the Merck & Co., Inc., Rahway, NJ employees who supported the studies and the tissue tumor mutational burden analysis. Medical writing and editorial assistance was provided by Christabel Wilson, MSc, of ICON plc (Blue Bell, PA). This assistance was funded by Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ.
Footnotes
Disclosure: Dr. Garassino received grants and personal fees during the conduct of this study from Merck Sharp & Dohme; grants and personal fees for clinical trials from AstraZeneca, Novartis, Bristol Myers Squibb, Roche, Pfizer, Celgene, Bayer, and Merck Sharp & Dohme; grants from Tiziana Life Sciences, Clovis, Merck Serono, GlaxoSmithKline, and Spectrum Pharmaceuticals; and personal fees from Eli Lilly, Boehringer Ingelheim, Otsuka Pharmaceutical Co., Ltd., Incyte, Inivata, Takeda, and Sanofi-Aventis. Dr. Gadgeel received personal fees from Merck, AstraZeneca, Genentech/Roche, Takeda/Ariad, Novocure, Bristol Myers Squibb, AbbVie, Xcovery, Janssen, Pfizer, Jazz Pharmaceuticals, Blueprint, and Eli Lilly. Prof. Novello reports as speakers bureau/advisor for AstraZeneca, Boehringer Ingelheim, Celgene, AbbVie, Bristol Myers Squibb, Eli Lilly, Pfizer, Takeda, Roche, and Merck Sharp & Dohme. Dr. Halmos received research funding from Merck, Bristol Myers Squibb, Boehringer Ingelheim, Eli Lilly, Novartis, GlaxoSmithKline, Pfizer, AbbVie, AstraZeneca, Mirati, Takeda, Guardant Health, Blueprint, Elevation Oncology, TPT, Amgen, and Advaxis; and served as a health caonsultant for Merck, Novartis, Boehringer Ingelheim, AstraZeneca, Novartis, Pfizer, Guardant Health, Spectrum, Bristol Myers Squibb, and Genentech. Dr. Felip received personal fees as an advisor, consultant, and/or speaker from AbbVie, AstraZeneca, Blueprint Medicines, Boehringer Ingelheim, Bristol Myers Squibb, Celgene, Eli Lilly, Guardant Health, Janssen, Medscape, Merck KGaA, Merck Sharp & Dohme, Novartis, Pfizer, Roche, Takeda, Touchtime, BerGenBio, and Samsung; and is an Independent Member of the Board for Grífols. Prof. Hui received personal fees as an advisor and/or speaker from AstraZeneca, Bristol Myers Squibb, Eli Lilly, Merck, Merck Sharp & Dohme, Novartis, Oncosec, Pfizer, Roche, and Seagen. Dr. Garon received grants and research support to the institution during the conduct of this study from Merck; has received grants from AstraZeneca, Bristol Myers Squibb, Eli Lilly, Genentech, Merck, Novartis, Dynavax, Mirati Therapeutics, and Iovance Biotherapeutics; and payment for participation in advisory boards/steering committees from Dracen Pharmaceuticals, EMD Serono, and Novartis. Dr. Horinouchi reports receiving grants and personal fees from Bristol Myers Squibb, Merck Sharp & Dohme, Chugai, Taiho, Eli Lilly, Ono, and AstraZeneca; and grants from Astellas, Merck Serono, and Genomic Health. Dr. Sugawara received honoraria from AstraZeneca, Bristol Myers Squibb, Chugai Pharmaceuticals, Kyowa Kirin, Eli Lilly, Merck Sharp & Dohme K.K, Nippon Boehringer Ingelheim, Novartis, Ono Pharmaceuticals, Pfizer, Taiho Pharmaceuticals, and Yakult Honsha. Dr. Rodriguez-Abreu received personal fees/honoraria for consultancy and lectures from Roche, AstraZeneca, Bristol Myers Squibb, Merck Sharp & Dohme, Eli Lilly, Pfizer, and Novartis; travel expenses from Roche, Bristol Myers Squibb, Merck Sharp & Dohme, and Novartis; and grant support for institutional studies from Bristol Myers Squibb. Dr. Reck received personal fees from Amgen, AstraZeneca, Bristol Myers Squibb, Boehringer Ingelheim, Eli Lilly, Merck, Mirati, Merck Sharp & Dohme LLC, Novartis, Roche, and Pfizer. Drs. Aurora-Garg, Loboda, and Lunceford, and Mr. Ayers are employees of Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, New Jersey. Drs Cristescu, Kobie, and Pietanza are employees of Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, New Jersey and are stockholders of the company. Dr. Piperdi is a former employee of Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, New Jersey and is a stockholder of the Company. Dr. Paz-Ares reports receiving speaker fees from AZ, Beigene, BMS, Daichii, Eli Lilly, Medscape, Merck Sharp & Dohme Corp, PER, Pharmamar, Roche; has participated in advisory boards for Altum sequencing, Amgen, AstraZeneca, Bayer, Beigene, BMS, Daichii, GSK, Janssen, Eli Lilly, Medscape, Merck Serono, Mirati, Merck Sharp & Dohme Corp, Novartis, PER, Pfizer, Pharmamar, Roche, Sanofi, Takeda; and has received research support from Amgen, BMS, Daiichi Sankyo, Janssen-cilag international NV, Eli Lilly, Merck Sharp & Dohme Corp, Novartis, Pfizer, Pharmamar, Roche, Sanofi, Small Lung Cancer Group, and Takeda. Dr. Speranza declares no conflict of interest.
A portion of these results was previously presented at the 2020 American Association for Cancer Research Annual Meeting, April 27–28 and June 22–24, Virtual Meeting; the 2019 European Society for Medical Oncology Annual Meeting, September 27–October 1, 2019, Barcelona, Spain; the 2019 European Society for Medical Oncology Immuno-Oncology Congress 2019, December 11–14, 2019, Geneva, Switzerland; and the 2019 World Conference on Lung Cancer, September 7–10, 2019, Barcelona, Spain.
Cite this article as: Garassino MC, Gadgeel S, Novello S, et al. Associations of tissue tumor mutational burden and mutational status with clinical outcomes with pembrolizumab plus chemotherapy versus chemotherapy for metastatic NSCLC. JTO Clin Res Rep. 2023;4:100431.
Note: To access the supplementary material accompanying this article, visit the online version of the JTO Clinical and Research Reports at www.jtocrr.org and at https://doi.org/10.1016/j.jtocrr.2022.100431.
Supplementary Data
References
- 1.National Comprehensive Cancer Network NCCN clinical practice guidelines in oncology [NCCN Guidelines]: non-small-cell lung cancer, version 3.2019. https://www.nccn.org/patients/guidelines/cancers.aspx#nsclc
- 2.Planchard D., Popat S., Kerr K., et al. Metastatic non-small cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2019;30:863–870. doi: 10.1093/annonc/mdy474. [DOI] [PubMed] [Google Scholar]
- 3.Gandhi L., Rodriguez-Abreu D., Gadgeel S., et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med. 2018;378:2078–2092. doi: 10.1056/NEJMoa1801005. [DOI] [PubMed] [Google Scholar]
- 4.Paz-Ares L., Luft A., Vicente D., et al. Pembrolizumab plus chemotherapy for squamous non-small-cell lung cancer. N Engl J Med. 2018;379:2040–2051. doi: 10.1056/NEJMoa1810865. [DOI] [PubMed] [Google Scholar]
- 5.Fancello L., Gandini S., Pelicci P.G., Mazzarella L. Tumor mutational burden quantification from targeted gene panels: major advancements and challenges. J Immunother Cancer. 2019;7:183. doi: 10.1186/s40425-019-0647-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stenzinger A., Allen J.D., Maas J., et al. Tumor mutational burden standardization initiatives: recommendations for consistent tumor mutational burden assessment in clinical samples to guide immunotherapy treatment decisions. Genes Chromosomes Cancer. 2019;58:578–588. doi: 10.1002/gcc.22733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yarchoan M., Albacker L.A., Hopkins A.C., et al. PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight. 2019;4 doi: 10.1172/jci.insight.126908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galvano A., Gristina V., Malapelle U., et al. The prognostic impact of tumor mutational burden (TMB) in the first-line management of advanced non-oncogene addicted non-small-cell lung cancer (NSCLC): a systematic review and meta-analysis of randomized controlled trials. ESMO Open. 2021;6 doi: 10.1016/j.esmoop.2021.100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizvi N.A., Hellmann M.D., Snyder A., et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berland L., Heeke S., Humbert O., et al. Current views on tumor mutational burden in patients with non-small cell lung cancer treated by immune checkpoint inhibitors. J Thorac Dis. 2019;11(suppl 1):S71–S80. doi: 10.21037/jtd.2018.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hellmann M.D., Ciuleanu T.E., Pluzanski A., et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378:2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kowanetz M., Zou W., Shames D., et al. Tumor mutation burden (TMB) is associated with improved efficacy of atezolizumab in 1l and 2l+ NSCLC patients. J Thorac Oncol. 2017;12(suppl):S321–S322. [Google Scholar]
- 13.Ready N., Hellmann M.D., Awad M.M., et al. First-line nivolumab plus ipilimumab in advanced non-small-cell lung cancer (CheckMate 568): outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. J Clin Oncol. 2019;37:992–1000. doi: 10.1200/JCO.18.01042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carbone D.P., Reck M., Paz-Ares L., et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med. 2017;376:2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell J.D., Alexandrov A., Kim J., et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skoulidis F., Heymach J.V. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer. 2019;19:495–509. doi: 10.1038/s41568-019-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.La Fleur L., Falk-Sorqvist E., Smeds P., et al. Mutation patterns in a population-based non-small cell lung cancer cohort and prognostic impact of concomitant mutations in KRAS and TP53 or STK11. Lung Cancer. 2019;130:50–58. doi: 10.1016/j.lungcan.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Aredo J.V., Padda S.K., Kunder C.A., et al. Impact of KRAS mutation subtype and concurrent pathogenic mutations on non-small cell lung cancer outcomes. Lung Cancer. 2019;133:144–150. doi: 10.1016/j.lungcan.2019.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Best S.A., De Souza D.P., Kersbergen A., et al. Synergy between the KEAP1/NRF2 and PI3K pathways drives non-small-cell lung cancer with an altered immune microenvironment. Cell Metab. 2018;27:935–943.e4. doi: 10.1016/j.cmet.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Kadara H., Choi M., Zhang J., et al. Whole-exome sequencing and immune profiling of early-stage lung adenocarcinoma with fully annotated clinical follow-up. Ann Oncol. 2017;28:75–82. doi: 10.1093/annonc/mdw436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cristescu R., Mogg R., Ayers M., et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362:eaar3593. doi: 10.1126/science.aar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merino D.M., McShane L.M., Fabrizio D., et al. Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: phase I of the Friends of Cancer Research TMB Harmonization Project. J Immunother Cancer. 2020;8 doi: 10.1136/jitc-2019-000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayers M., Lunceford J., Nebozhyn M., et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panda A., Betigeri A., Subramanian K., et al. Identifying a clinically applicable mutational burden threshold as a potential biomarker of response to immune checkpoint therapy in solid tumors. JCO Precis Oncol. 2017;2017 doi: 10.1200/PO.17.00146. PO.17.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marabelle A., Fakih M., Lopez J., et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020;21:1353–1365. doi: 10.1016/S1470-2045(20)30445-9. [DOI] [PubMed] [Google Scholar]
- 27.Aurora-Garg D, Albright A, Qiu P, et al. Large-scale evaluation of concordance of genomic scores in whole exome sequencing and Foundation Medicine comprehensive genomic platform across cancer types. Paper presented at: Society for Immunotherapy of Cancer (SITC); November 6–10, 2019. National Harbor, MD.
- 28.Cristescu R., Aurora-Garg D., Albright A., et al. Tumor mutational burden predicts the efficacy of pembrolizumab monotherapy: a pan-tumor retrospective analysis of participants with advanced solid tumors. J Immunother Cancer. 2022;10 doi: 10.1136/jitc-2021-003091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papillon-Cavanagh S., Doshi P., Dobrin R., Szustakowski J., Walsh A.M. STK11 and KEAP1 mutations as prognostic biomarkers in an observational real-world lung adenocarcinoma cohort. ESMO Open. 2020;5 doi: 10.1136/esmoopen-2020-000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herbst R.S., Lopes G., Kowalski D.M., et al. Association between tissue TMB (tTMB) and clinical outcomes with pembrolizumab monotherapy (pembro) in PD-L1-positive advanced NSCLC in the KEYNOTE-010 and −042 trials. Ann Oncol. 2019;30:v916–v917. [Google Scholar]
- 31.Hellmann M.D., Nathanson T., Rizvi H., et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell. 2018;33:843–852. doi: 10.1016/j.ccell.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rizvi N.A., Cho B.C., Reinmuth N., et al. Durvalumab with or without tremelimumab vs standard chemotherapy in first-line treatment of metastatic non-small cell lung cancer: the MYSTIC Phase 3 randomized clinical trial. JAMA Oncol. 2020;6:661–674. doi: 10.1001/jamaoncol.2020.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yarchoan M., Hopkins A., Jaffee E.M. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reck M., Rodriguez-Abreu D., Robinson A.G., et al. Pembrolizumab versus chemotherapy for PD-L1-positive non–small-cell lung cancer. N Engl J Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 35.Reck M., Rodriguez-Abreu D., Robinson A.G., et al. Updated analysis of KEYNOTE-024: pembrolizumab versus platinum-based chemotherapy for advanced non-small-cell lung cancer with PD-L1 tumor proportion score of 50% or greater. J Clin Oncol. 2019;37:537–546. doi: 10.1200/JCO.18.00149. [DOI] [PubMed] [Google Scholar]
- 36.Mok T.S.K., Wu Y.L., Kudaba I., et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. 2019;393:1819–1830. doi: 10.1016/S0140-6736(18)32409-7. [DOI] [PubMed] [Google Scholar]
- 37.Aggarwal C., Thompson J.C., Chien A.L., et al. Baseline plasma tumor mutation burden predicts response to pembrolizumab-based therapy in patients with metastatic non-small cell lung cancer. Clin Cancer Res. 2020;26:2354–2361. doi: 10.1158/1078-0432.CCR-19-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skoulidis F. Inactivating STK11/LKB1 genomic alterations are a major driver of primary resistance to PD-1 axis blockade in non-squamous non-small cell lung cancer. Cancer Res. 2019;79 Abstract nr SY42-02. [Google Scholar]
- 39.Rizvi H., Sanchez-Vega F., La K., et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol. 2018;36:633–641. doi: 10.1200/JCO.2017.75.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miao D., Margolis C.A., Vokes N.I., et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet. 2018;50:1271–1281. doi: 10.1038/s41588-018-0200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho B.C., Lopes G., Kowalski D.M., et al. Relationship between STK11 and KEAP1 mutational status and efficacy in KEYNOTE-042: pembrolizumab monotherapy versus platinum-based chemotherapy as first-line therapy for PD-L1-positive advanced NSCLC. Cancer Res. 2020;80:CT084. [Google Scholar]
- 42.Samstein R.M., Lee C.H., Shoushtari A.N., et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.