Abstract
The ability to generate haematopoietic stem cells in vitro would have immeasurable impact upon many areas of clinical practice, including trauma, cancer, and congenital disease. In this protocol, we describe a stepwise approach to convert adult murine endothelial cells (ECs) to haematopoietic stem cells (HSCs), termed reprogrammed endothelial cells into haematopoietic stem and progenitor cells (rEC-HSPCs). The conversion is achieved without cells transitioning through a pluripotent state and comprises three phases: induction, specification, and expansion. Adult ECs are first isolated from Runx1-IRES-GFP; Rosa26-rtTa mice and maintained in culture under EC growth factor stimulation and Tgfβ inhibition. In the first (induction) phase of conversion (days 0–8), four transcription factors—FosB, Gfi1, Runx1, and Spi1 (FGRS)—are expressed transiently, which results in endogenous Runx1 expression. During the second (specification) phase (days 8–20), endogenous Runx1+ FGRS-transduced ECs commit to a haematopoietic fate and no longer require exogenous FGRS expression. Finally, the vascular niche drives robust proliferation of rEC-HSPCs during the expansion phase (days 20–28). The resulting converted cells possess a transcriptomic signature and long-term self-renewal capacity indistinguishable from those of adult HSCs. In this protocol we also describe functional in vitro and in vivo assays that can be used to demonstrate that rEC-HSPCs are competent for clonal engraftment and possess multi-lineage reconstitution potential, including antigen-dependent adaptive immune function. This approach thus provides a tractable strategy to interrogate the generation of engraftable haematopoietic cells, advancing the mechanistic understanding of haematopoietic development and HSC self-renewal.
Keywords: reprogramming, direct conversion, endothelial cells, haematopoietic stem cells, endothelial-to-haematopoietic transition, EHT, transcription factor overexpression, vascular niche, angiocrine signaling, homing and engraftment, bone marrow reconstitution, bone marrow engraftment
EDITORIAL SUMMARY
Adult endothelial cells from mice are reprogrammed to haematopoietic stem cells without cells transitioning through a pluripotent state. The protocol also describes in vitro and in vivo assays to confirm the resulting cells function as expected.
INTRODUCTION
The ability to generate cells of the haematopoietic lineage in vitro would have myriad research and clinical applications. In the short term, this technology would open the door to disease modelling in humanized mice and facilitate the development of therapeutic strategies to counter genetic blood disorders. Ultimately, the de novo generation and scalability of haematopoietic cells, multipotent or terminally differentiated, are tied to the promise of autologous bone marrow transplants for cancer therapy and blood transfusions in the event of trauma12.
All haematopoietic stem cells (HSCs) arise from a population of endothelial cells in the aorta-gonad-mesonephros (AGM) of the mammalian embryo. More specifically, VE-Cadherin+CD45− cells lining the dorsal aspect of the mouse AGM have been shown to transition into VE-Cadherin−CD45+ cells that are able to both reconstitute the myeloablated adult bone marrow and differentiate into all blood cell lineages1. This process, termed endothelial-to-haematopoietic transition (EHT), is only known to occur in utero. However, adult endothelium, a somatic, terminally differentiated tissue type with no in vivo or spontaneous haemogenic capacity reported to date, has been found to maintain and expand immature haematopoietic cells, or haematopoietic stem and progenitor cells (HSPCs), both in vivo and in vitro2–5. These synergistic interactions between endothelial and haematopoietic cells vary across organs and ages. For example, the expansion and maintenance potential of ECs derived from the bone marrow, a primary haematopoietic organ, is higher than that of ECs derived from other vascular beds, such as the lungs’, or even that of their stromal neighbors in the bone marrow6. Also, co-culture of ECs and HSPCs derived from younger donors yield superior engraftment and repopulation potentials than those of older donors as measured by limiting dilution transplantations. Even co-infusion of bone marrow ECs along with HSPCs confers a radioprotective effect post-myeloablation7.
The relationship between endothelial and haematopoietic cells, however, is more than angiocrine signaling, it goes back to a common cellular identity in development. Parting from this understanding, we developed a platform to convert, or reprogram, adult endothelial cells into haematopoietic cells. The haematopoietic cells thus generated are phenotypically and functionally indistinguishable from naïve HSCs, i.e., the resulting reprogrammed cells (rEC-HSPCs) are able to engraft and repopulate the bone marrow as well as differentiate into all blood cell lineages in vivo8,9. In this protocol we provide detailed instructions to produce haematopoietic cells from adult murine endothelial cells and to confirm their functionality.
Development of the protocol
We compared the transcriptomic profiles of human umbilical vein endothelial cells (HUVECs) and cord blood-derived HSPCs and identified over 20 transcription factors that were differentially expressed. We overexpressed these transcription factors in various combinations and consequently found that expression of four—FOSB, GFI1, RUNX1, and SPI1 (FGRS)—was sufficient to convert endothelial cells to a haematopoietic state. Concurrently, from the HSPC expansion technology also established in our lab, we had learned that haematopoietic cell maintenance in vitro is aided by xenobiotic-free culture conditions as well as angiocrine signaling, that is, growth factor and cytokine signaling provided by endothelial cells in co-culture. We found that overexpression of the E4ORF1 protein from adenovirus 5 enabled ECs alone to grow in serum- and xenobiotic-free conditions. ECs with this ability are thus able to provide the requisite angiocrine signaling to facilitate HSPC expansion and maintenance4,10. The vascular niche we established consists of HUVECs transduced with a lentiviral E4ORF1 vector (E4ECs), and only FGRS-transduced cells in co-culture with E4ECs give rise to rEC-HSPCs.
We first reprogrammed adult human endothelium using lentiviral vectors encoding FGRS under constitutive promoters. To demonstrate that the resulting rEC-HSPCs functioned as anticipated, we transplanted them into immuno-compromised mice9. These rEC-HSPCs maintained exogenous expression of the four transcription factors in vivo post-transplantation. Constitutive expression of Spi1 has been shown to hinder lymphoid differentiation of rEC-HSPCs in vivo by blocking T lymphopoiesis at the pro-T cell stage11. In addition, mice of the NSG strain cannot educate native B or T cells to maturity. Our system of constitutive exogenous FGRS expression was therefore unable to confer immune-compromised recipient animals with the ability to generate an adaptive immune response. We addressed these drawbacks by adapting our platform to the mouse system and using inducible lentiviral vectors to overexpress FGRS only temporarily. Adult murine endothelium yielded rEC-HSPCs that were transplantable into immuno-competent, syngeneic mice and cloning FGRS under a doxycycline-inducible promoter enabled exogenous expression to be turned off after in vitro reprogramming. Recipient mice had the ability to generate T cell subsets with adaptive immune function, as measured by sequencing of TCR diversity, chicken ovalbumin vaccination, interferon-γ upregulation, and CD8+ cytotoxicity assays8,9.
In summary, the major requirements of our reprogramming approach are concurrent exogenous expression of transcription factors and vascular niche induction. The former is a requirement of most reprogramming technologies; the latter results from our previous understanding of endothelial-haematopoietic interactions in vitro. Both requirements take advantage of the vestigial endothelial identity present in all haematopoietic cells.
Comparison with other methods
Other approaches to produce HSCs involve HSC-directed differentiation (using cytokines or overexpression of transcription factors) from induced or native pluripotent cells, or conversion from non-endothelial somatic cell types such as fibroblasts or B cell progenitors9,13–16. Directed differentiation of putative haematopoietic stem and progenitor cells from pluripotent cell sources (PCS) is probably the most common avenue for research in the field. In one attempt, directed differentiation of PCS into putative HSPCs recapitulated the different waves of haematopoiesis (primitive followed by definitive) in the dish; however, the resulting in vitro-derived populations lacked robust engraftment and lymphoid differentiation potential14,17,18.
Most recently, an approach published by Sugimura et al. relied on overexpression of seven transcription factors—two of which (RUNX1 and SPI1) were common to our cocktail of four, and another (ERG) is constitutively expressed by endothelial cells19—to generate HSPCs from human PCS. The resulting cells were detectable in NSG mice assayed 14 weeks following primary and secondary transplantations and produced a modest adaptive immune response at eight weeks20. Notably, these PCS-derived HSPCs were delivered via intra-femoral injection, circumventing the process of homing and engraftment that bona fide HSPCs are capable of undergoing by way of intravenous injection post-myeloablation. Thus it was not demonstrated that these HSPC could home or engraft. Also, the genetic and epigenetic stability of the differentiated PCS-derived HSPCs has not yet been reported beyond 14 weeks following transplantation.
The inability to generate fully functional HSCs from PCS alone could be due to the lack of appropriate environmental cues to support haematopoietic commitment. Prior to the publication of Sugimura et al.20, Riddell et al. converted murine lymphoid cells into engraftable HSCs (iHSCs) by overexpressing eight transcription factors (TFs) and immediately transplanting the cells, allowing them to convert in vivo20,21. This approach may have been successful due to the iHSCs utlilising the niche of the recipient to support fate reversal. By the same means, in the system described by Sugimura et al., PCS-derived cells were driven forward, though also to a multipotent haematopoietic fate, when placed in the bone marrow niche shortly after transduction. While exposure to the normal niche signals endowed the resulting HSPCs with self-renewal and lymphoid differentiation capacity, neither of these experimental strategies is suitable for mechanistic studies of endothelial-to-haematopoietic transition.
In contrast, our method combines the exogenous expression of transcription factors with the reestablishment of physiological microenvironmental cues in vitro to convert organotypic murine ECs into functional HSCs. Our method recapitulates EHT in vivo8 to a degree that the resulting cells are able to reconstitute the myeloablated bone marrow indistinguishably from naïve controls. This approach has uncovered the ability of niche-derived signals to choreograph EHT in addition to bringing us closer to therapeutic autologous HSC transplantation.
Experimental design
FGRS virus production and transduction.
We cloned open reading frames (Fosb: NM_008036.2, Gfi1: NM_010278.1, Runx1: BC069929.1, Sfpi1: BC003815.1) into doxycycline-inducible pLVx TET-3G lentiviral plasmids obtained from GeneCopoeia (www.genecopoeia.com/). We produce lentivirus as described in Ginsberg et al.22 with the modifications described in Reagent Setup.
Murine EC maintenance.
Naïve murine ECs are prone to losing endothelial identity, i.e., VE-Cadherin/CD31 expression, if cultured carelessly. We exhort first-time researchers to (i) limit first-time isolations to the vascular beds of the lungs and/or liver, as these two organs yield the more durable endothelial populations in vitro as well as the least haematopoietic contamination; (ii) maintain the oxygen concentration within the tissue culture incubator at 5% for the duration of the conversion experiment; (iii) always supplement murine EC medium with 5 μM SB431542 to block competition from non-endothelial cells and prevent endothelial-to-mesenchymal transition; (iv) always expand the cells 1:2, as sparse culture hinders their growth; and (v) perform FACS prior to any costly or long-term assay to ensure the bulk identity of the cells of interest has remained at least 90% endothelial, i.e., VE-Cadherin+CD31+CD45−. All of these recommendations and precautions can be found within the various steps of the Procedure section. We include 2 methods to isolate murine ECs in the procedure, one using direct FACS of primary cells and another using magnetic bead binding. However, if magnetic bead binding is used at least one round of cell sorting via FACS will also be required to rid the cultures of non-endothelial cells.
HUVEC isolation and E4EC generation.
We routinely isolate human umbilical vein endothelial cells (HUVECs) following the method outlined in Rafii et al.23,24. Commercially available HUVEC lines may be used instead, but cell sorting may be necessary to ensure a pure endothelial identity of the starting cell population. We generate HUVEC-E4ORF1 (E4ECs) as described in Seandel et al.25 but with the following modifications. The E4ORF1 gene from adenovirus serotype 5 was cloned into the pCCL-PGK lentivirus vector. We routinely produce viral particles using the same method as that used to generate FGRS lentiviruses (see Reagent Setup). Consistently, 14 ml lentivirus supernatant produced by transfection of one 100-mm plates 293T cells for each of the 4 transcription factors (Fosb, Gfi1, Spi1, and Runx1) is sufficient for several reprogramming experiments, as E4ECs can be expanded and frozen down repeatedly. We usually follow the Lenti-X Packaging Single Shot system for convenience when making large batches of various vectors at the same time, but E4ORF1 lentivirus can also generated reliably by the calcium precipitation method, cotransfecting 15 μg of lentiviral vector, 3 μg of pENV/VSV-G, 5 μg of pRRE, and 2.5 μg of pRSV-REV in 293T Lenti-X cells (passage 8–10; subconfluent). Supernatants are collected only once, 48 h after transfection, then filtered, concentrated, and stored as already described. To assess the quality of virus produced, we recommend performing ELISA against p24 to estimate the concentration of viral capsids and qRT-PCR to detect lentiviral genome copies. We perform transduction in a ~ 60% confluent well of a six-well plate of HUVECs using a total volume of 1 ml human EC medium, 10,000 pico -gram concentrated E4ORF1 lentivirus, and 5 μg polybrene. Cells and lentivirus are cultured undisturbed for 72 h and then selected in serum-free medium for ~ 10 days. Confluent populations of E4ECs are contact inhibited, non-transformed, and able to propagate as homogenous monolayers, providing an ideal instructive niche for reprogramming and sustaining FGRS-transduced endothelial cells into rEC-HSCs. E4ECs can be cultured until their Hayflick limit, generally 20–25 passages from the initial harvest. E4ECs are not commercially available but can be provided through an MTA with our lab (Rafii lab). The E4ORF1 adenoviral cassette can be obtained from Addgene (Plasmid no. 38063).
Animals:
Animals are required in the protocol for EC isolation as well as transplantation purposes. We used Runx1-IRES-GFP (Runx1tm4Dow) mice, obtained from James Downing at St. Jude Hospital. These are knock-in reporter mice with an IRES-GFP-pA-neo cassette inserted downstream of the coding region of the Runx1 gene. They are useful to ascertain the progress of conversion as the cells adopt a haematopoietic (Runx1+) identity and shed their endothelial (Runx1−) one (see Fig. 3c). Runx1-IRES-GFP mice were crossed with Rosa26-rtTa mice (B6.Cg-Gt(ROSA)26Sortm1(rtTA*M2)Jae/J, The Jackson Laboratory, strain cat. no. 006965) to produce Runx1-IRES-GFP; Rosa26-rtTa mice (referred to as Runx1-rtTA). The Runx1-IRES-GFP reporter mouse is used to facilitate the enrichment of likely reprogramming events for single-cell conversion (see Box 3) with two goals in mind: (i) quantifying the number of rEC-HSPCs generated and (ii) assessing the systemic fate of different clones in vivo. The ubiquitous rtTA transgenic mouse, on the other hand, offers a way to ensure the presence of the transactivator protein in all of the cells harvested, reducing the likelihood of successful reprogramming events to the stoichiometry of only the four transcription factors at play: Fosb, Gfi1, Runx1, and Spi1. Homozygosity of either allele is not required for its intended purpose to be measurable: all our experiments have made use of Runx1-rtTA mice with heterozygous Runx1-IRES-GFP and rtTa alleles, respectively. Neither strain is required for successful reprogramming. For rEC-HSPC transplantation, we use CD45.1+ congenic recipients (B6.SJL-Ptprca Pepcb/BoyJ, Jackson Laboratory, strain cat. no. 002014) and CD45.2+ (C57BL/6J, Jackson Laboratory, strain cat. no. 000664) control donors.
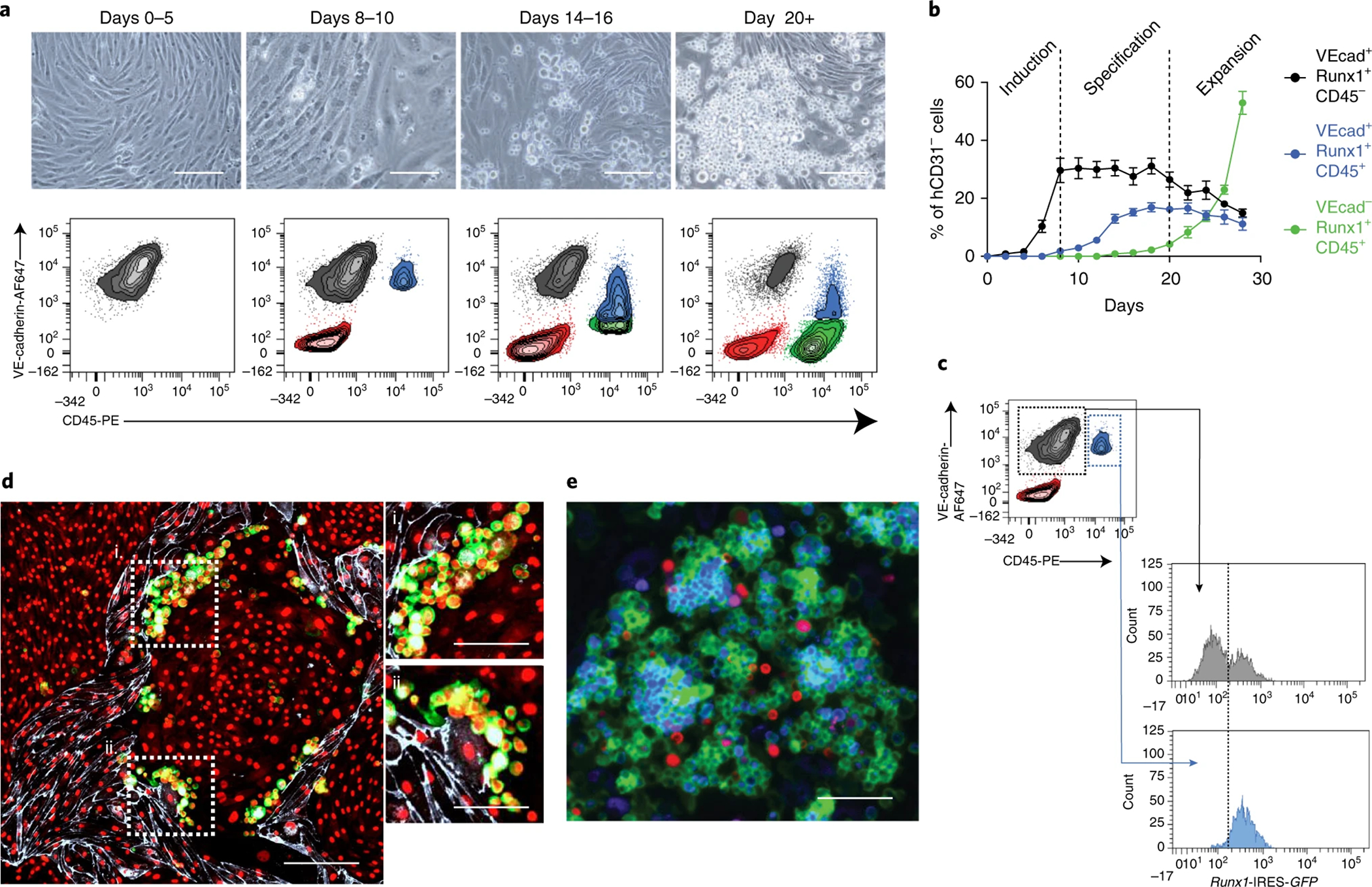
Figure 3. Conditional expression of FGRS in adult murine ECs generates haematopoietic cells.
a, Time-course depicting the emergence of CD45+ cells in the vicinity of vascular niche ECs (HUVEC-E4ORF1, or E4ECs). Representative phase contrast microscopy (top panels; original magnification, ×10; scale bars, 100 μm) and representative flow cytometry plot of anti-mouse VE-Cadherin-AF647 vs. anti-mouse CD45-PE after events are discriminated by size, granularity and viability (see Fig. 2a) and E4ECs are excluded by negative anti-human CD31-BV421 gating (bottom panels). The VE-Cadherin−CD45− population represents cells that have lost endothelial identity in the reprogramming process (red); the VE-Cadherin+CD45− population represents non-reprogrammed endothelial cells (gray); the VE-Cadherin+CD45+ population represents cells undergoing reprogramming (blue); and the VE-Cadherin−CD45+ population represents fully reprogrammed rEC-HSPCs, amenable to downstream assaying (green). b, Quantification of gray, blue, and green populations every two days over the course of reprogramming. Stage of reprogramming is indicated in black within dotted lines. Data represent percent mean ± s.e.m. (n = 5). c, Representative flow cytometry showing in vitro tracking of endothelial-to-haematopoietic transition using cells isolated from a Runx1-IRES-GFP reporter mouse. Data shown correspond to day 8 of reprogramming. The VE-Cadherin+CD45− (gray) population has partially induced endogenous Runx1 expression, while the VE-Cadherin+CD45+ (blue) population has inducing endogenous Runx1 to greater extent. Both maintain VE-Cadherin expression, but the blue population is said to be farther along the reprogramming process. d, Representative fluorescence microscopy showing the emergence of CD45+ cells in the vicinity of E4ECs. Cells were stained on day 10 of reprogramming using DAPI (red), anti-mouse CD45-PE (green), and anti-human CD31-BV421 (white; original magnification, ×10; scale bars, 100 μm). e, Representative fluorescence microscopy showing the emergence of LKS cells in the vicinity of E4ECs. Cells were stained on day 23 of reprogramming using anti-mouse CD45-PE (green), anti-mouse Lin-BV421 (blue), anti-mouse cKit-APC, and anti-mouse Sca1-APC (both red; original magnification, ×10; scale bars, 100 μm). Flow cytometry data was obtained was analyzed using a BD FACSAria II and BD FACSDiva software. All imaging was performed using a Zeiss 710 META confocal microscope and ZEN software.
BOX 3.
Single-cell conversion of Runx1+ ECs to rEC-HSC · TIMING ~ 8 weeks
On 7 day of conversion, i.e., after 7 days of incubating the reprogramming co-cultures in conversion medium (step 30), prepare 15 gelatin-coated 96 well plates and seed 2,500 E4ECs in each well.
-
On day 8, sort single cells from the co-cultures as set up on step 29 for Runx1-GFP expression by flow cytometry. We use the single-cell purity mask of a BD FACSAria II.
Critical point: Seed single Runx1+ ECs, defined as VE-Cadherin+CD31+Runx1+CD45−, into individual wells of the 15 96-well plates that were seeded with 2,500 E4ECs in step 1.
Quickly centrifuge the plates at 100g for 5 min and place in a humidified incubator at 37 °C, 5% CO2, and 5% O2.
Feed cells with 20 μl conversion medium and 1 μg ml−1 doxycycline as specified on step 32 of the main protocol until haematopoietic colonies appear (See Fig. 3d; Extended Data Fig. 4 and 5 from Lis et al.)
Colony Formation Assay.
In parallel with investigating the function of our derived cells by means of transplantation, we also perform Colony-Forming Unit (CFU) assays in vitro. These are well-established, pseudo-functional assays that allow the researcher to test the ability of haematopoietic stem and progenitor cells to form colonies that are associated with different stages of differentiation. Albeit entirely qualitative, CFU assays are useful in the context of reprogramming because only haematopoietic cells give rise to colonies under these conditions and native haematopoietic cells are readily available positive controls. We perform CFU assays using cytokine-rich methylcellulose and following the manufacturer’s instructions, adding doxycycline to the CFU mixture during setup for some of the assays. We score haematopoietic colonies with a dissection microscope after 4 and 10 days to evaluate the various CFU phenotypes.
MATERIALS
REAGENTS
Dulbecco’s Phosphate-Buffered Saline (PBS; Corning, cat. no. 21-031-CU)
EDTA, Biotechnology Grade (VWR, cat. no. E522)
Bovine Serum Albumin (BSA; Sigma, cat. no. A7906)
-
293T Lenti-X cell line (Clontech, cat. no. 632180)
! CAUTION: The cell lines used in your research should be regularly checked to ensure they are authentic and are not infected with mycoplasma.
Lenti-X Packaging Single Shot (Clontech, cat. no. 631275)
Lenti-X Concentrator (Clontech, cat. no. 631232)
RBC lysis buffer (Biolegend cat. no. 420301)
KCl (Sigma-Aldrich cat. no. P9333)
NaCl (Thermo, cat. no. AM9760G)
HEPES (Invitrogen, cat. no. 15630080)
CaCl2 (Sigma-Aldrich cat. no. C1016)
MgCl2 (Sigma-Aldrich cat. no. M8266)
Collagenase A (Roche cat. no. 11088793001)
Dispase II (Roche cat. no. 04942078001)
DNase I (Roche cat. no. 10104159001)
HBSS (Corning cat. no. 21-020-CV)
SB431542 (R&D, cat. no. 1614)
Dimethyl sulfoxide (DMSO; Sigma, cat. no. D2650)
Doxycycline (Stemgent, cat. no. 04-0016)
Fibronectin (Sigma, cat. no. F1141)
DMEM/Ham’s F-12 (Sigma, cat. no. D6421)
M199 (Sigma, cat. no. M4530)
X-Vivo serum-free medium (Lonza, cat. no. 04448Q)
StemSpan SFEM (STEMCELL Technologies, cat. no. 09650)
Fetal Bovine Serum (FBS; Corning, cat. no. 35-016-CV)
KnockOut Serum Replacement (Invitrogen, cat. no. 10828028)
Heparin (Sigma, cat. no. H3393)
Endothelial mitogen (Alfa Aeser cat. no. J65416),
GlutaMAX (Gibco, cat. no. 35050-061)
MEM Nonessential Amino Acids (Corning, cat. no. 25-025-CI)
Recombinant human FGF-basic (bFGF; Peprotech, cat. no. 100-18B)
Recombinant murine SCF (SCF; Peprotech, cat. no. 250-03).
Cytokine-rich methylcellulose (StemCell Technologies, cat. no. 03444)
RNeasy Micro Kit (Qiagen, cat. no. 74106)
SuperScript III reverse transcriptase (Invitrogen, cat. no. 18080093)
SYBR GreenER qPCR SuperMix Universal (SYBR green; Life Technologies, cat. no. 11762500)
MicroAmp Fast Optical 96-Well Reaction Plate with Barcode (qPCR plate(s); Life Technologies, cat. no. 4346906)
MicroAmp Optical Adhesive Film (adhesive film; Applied Biosystems, cat. no. 4311971)
IsoThesia Solution (isoflurane; Henry Schein cat. no. 1169567761)
Accutase (Thermo, cat. no. A1110501)
Trypsin (Corning, cat. no. 25-052-CI)
-
Human umbilical vein endothelial cells. We routinely isolate human umbilical vein endothelial cells (HUVECs) following the method outlined in Rafii et al.23,24 and culture in hEC medium in a humidified incubator at 37 °C, 5% CO2, and 5% O2. However, HUVECs are also available commercially (for example, Lonza, cat. no. C2519AS; ATCC, cat. no. CRL-1730)
! CAUTION: The cell lines used in your research should be regularly checked to ensure they are authentic and are not infected with mycoplasma.
E4ORF1 adenoviral cassette (Addgene, Plasmid cat. no. 38063)
Polybrene (Sigma, cat. no. TR-1003)
DAPI (Biolegend, cat. no. 422801)
Antibodies as listed in Table 1
Mice for EC isolation. We have used Runx1-IRES-GFP (Runx1tm4Dow) mice (obtained from James Downing at St. Jude Hospital) crossed with Rosa26-rtTa mice (B6.Cg-Gt(ROSA)26Sortm1(rtTA*M2)Jae/J, The Jackson Laboratory, strain cat. no. 006965) to produce Runx1-IRES-GFP; Rosa26-rtTa mice (referred to as Runx1-rtTA). Alternatively, the rtTA transgenic mouse can be used without crossing, but any mouse model could in theory be used for reprogramming.
Mice for rEC-HSPC transplantation (experimental and control recipients). We use CD45.1+ congenic recipients (B6.SJL-Ptprca Pepcb/BoyJ, Jackson Laboratory, strain cat. no. 002014).
-
Mice for rEC-HSPC transplantation (control donors). We use CD45.2+ congenic recipients (C57BL/6J, Jackson Laboratory, strain cat. no. 000664) control donors.
! CAUTION: All animal experiments were performed under the approval of Weill Cornell Medicine Institutional Animal Care and Use Committee.
! CAUTION: Experiments using rodents must conform to Institutional and National regulations
Table 1.
FACS antibodies.
| Antigen | Reactivity | Clone | Manufacturer | Concentration (in μg million cells−1) |
|---|---|---|---|---|
| VE-Cadherin | mouse | BV13 | Biolegend | 0.2 |
| CD45 | mouse | 30-F11 | Biolegend | 0.2 |
| c-Kit | mouse | 2B8 | Biolegend | 0.2 |
| Sca1 | mouse | D7 | Biolegend | 0.2 |
| Lineage | mouse | 145-2c11, RB6-8C5, RA3-6B2, Ter-119, M1/70 | Biolegend | 0.2 |
| Ter119 | mouse | Ter11 | Biolegend | 0.2 |
| CD31 | mouse | 390 | Biolegend | 0.2 |
| CD31 | human | WM59 | Biolegend | 0.2 |
| CD45.2 | mouse | 104 | Biolegend | 0.2 |
| CD45.1 | mouse | A20 | Biolegend | 0.2 |
| CD3 | mouse | 17A2 | Biolegend | 0.2 |
| CD4 | mouse | RM4-5 | Biolegend | 0.2 |
| CD8 | mouse | YTS156.7.7 | Biolegend | 0.2 |
| B220 | mouse | RA3-6B2 | Biolegend | 0.2 |
| Gr1 | mouse | RB6-8C5 | Biolegend | 0.2 |
| CD11b | mouse | M1/70 | Biolegend | 0.2 |
| CD150 | mouse | TC15-12F12.2 | Biolegend | 0.2 |
| CD48 | mouse | HM48-1 | Biolegend | 0.2 |
| CD16/32 | mouse | 2.4G2 | Biolegend | 0.2 |
EQUIPMENT
Biosafety Level II laboratory
Laminar flow hood (NuAire Biosafety Level 2 cat. no. NU-425-600)
Forma Series II Water Jacket CO2 Incubator (Thermo, cat. no. 3130)
Tissue culture centrifuge (Thermo, Sorvall RG3 cat. no. 750003433)
Refrigerated microcentrifuge (Eppendorf, cat. no. 5415R)
Dissection microscope (Olympus, ZSX16)
Sterile pipettes, 10 ml (Denville cat. no. P7128)
Sterile micropipettes (Denville); 1000 μl (cat. no. P1126), 200 μl (cat. no. P1122), 20 μl (cat. no. P1121), and 10μl (cat. no. P1096-FR)
Micropipettors (Gilson Pipetman cat. no. F167700)
Tissue culture plates (Falcon); 12-well (cat. no. 353224) and six-well (cat. no. 353043)
Conical tubes (Falcon); 15 ml (cat. no. 352097) and 50 ml (cat. no. 352098)
Round-bottom polystyrene tube (Falcon, cat. no. 352058)
Sterile microcentrifuge tubes, 1.5 ml (Denville, cat. no. C2172); 2 ml (USA Scientific, cat. no. 1620-2700)
Vacuum Filter/Storage Bottle System, 500 ml, 0.22 μm (filter bottle; Corning, cat. no. 430769)
Syringe Filters with Acrylic Housing 0.22 μm; 0.45 μm (PES filter; VWR, cat. no. 28145-501, 28145-505, respectively)
Syringe with Luer-Lok® Tip, 1 ml; 3 ml; 30 ml; 60 ml (syringe; VWR, cat. no. BD309628, BD309657, BD309832, BD309653, respectively)
Cell strainers, 40 μm (Falcon, cat. no. 352340)
Mini LabRoller rotator (rotator; Sigma, cat. no. Z674583)
Isoflurane Vaporizer and Anesthesia Chamber (Summit Anesthesia Solutions, cat. no. 2022043)
MaxQ™ 4000 Benchtop Orbital Shaker (Thermo, cat. no. SHKE4000-7)
Flow cytometer equipped with multiple lasers (BD model FACSAria II)
Confocal microscope with multiple lasers (Zeiss 710 META).
NanoDrop spectrophotometer (VWR, cat. no. 15170-263)
C1000 Touch™ Thermal Cycler (thermocycler; Bio-Rad, cat. no. 1851148)
ViiA 7 Real-Time PCR system (Applied Biosystems, cat. no. 4453552)
Cesium-137 or X-Ray irradiator (usually provided by a core facility)
REAGENT SETUP
Lentivirus storage buffer.
Combine 50 mM Tris (pH 7.8), 1 mM EDTA, and 130 mM NaCl in 50 ml dH2O. Filter through a 0.22-μm PES filter attached to a 60-ml syringe and store at 4 °C for up to one year.
Collagenase/Dispase/DNAse digestion solution.
Prepare digestion buffer by combining 5 mM KCl, 10 mM HEPES, 2 mM CaCl2, and 1.3 mM MgCl2 in PBS (pH 7.4). In a flask with a stir bar, reconstitute the following enzymes to the indicated concentrations in digestion buffer: Collagenase A: 20 mg ml−1; Dispase II: 8 units ml−1; and DNase I: 400 μg ml−1. This mix yields an 8× digestion solution. Dispense 0.5 ml aliquots into tubes and store at −20 °C for up to 6 months. To digest ~ 350 mg mouse tissue, which is approximately the weight of the lungs of an adult male mouse, add 3.5 ml HBSS to a 0.5-ml aliquot of 8× digestion solution to produce working concentrations of collagenase A (2.5 mg ml−1), dispase II (1 unit ml−1), and DNase (50 μg ml−1).
SB431542.
Add 470 μl DMSO to 10 mg SB431542 and filter through a 0.22-μm PES filter attached to a 1-ml syringe to generate a 50-mM stock solution of SB431542 (10,000×). Dispense 50-μl aliquots stock solution into sterile microcentrifuge tubes and store at −80 °C for up to 1 year.
Doxycycline.
Dissolve 10 mg doxycycline into 5 ml dH2O to prepare a 2-mg ml−1 stock solution of doxycycline (2,000×). Dispense 0.5-ml aliquots into 1.5 ml micro-centrifuge tubes to avoid repeated freeze-thaw cycles and store at −20 °C for up to 6 months. When treating cells with doxycycline, thaw one aliquot, or the amount to be used for the duration of the experiment, and store it at 4 °C for 1 month. Doxycycline is light-sensitive and should be stored accordingly.
Fibronectin-coated plates.
Coat all plates for cell culture with fibronectin solution (1 mg ml−1 fibronectin in PBS) before plating cells. Add enough fibronectin solution to each plate to cover its surface and place at room temperature (20–25 °C) in a tissue culture hood for at least 30 min. Aspirate fibronectin solution and allow remaining moisture to evaporate. Plates can be used right away or stored at 4 °C for 3–4 weeks.
Suspension buffer.
Combine PBS, 0.5% BSA, and 2 mM EDTA. Filter through a 0.45-μm filter bottle and store at 4 °C for 1 month.
Murine EC (mEC) medium.
Combine DMEM/Ham’s F-12 supplemented with 20% heat-inactivated FBS, 100 μg ml−1 heparin, 50 μg ml−1 endothelial mitogen, 20 mM HEPES, 1× GlutaMAX, and 1× MEM Nonessential Amino Acids. Filter through a 0.45-μm filter bottle. Add 5 μM SB431542 to filtered medium and store at 4 °C for 2 weeks.
Human EC (hEC) medium.
Combine M199, 10% FBS, 100 μg ml−1 heparin, 50 μg ml−1 endothelial mitogen, 20 mM HEPES, and 1× GlutaMAX. Filter through a 0.45-μm filter bottle. Store at 4 °C for 2 weeks.
Serum-free EC medium.
Combine X-Vivo serum-free medium, 100 μg ml−1 heparin, 20 mM HEPES, and 1× GlutaMAX. Filter through a 0.45-μm filter bottle. Store at 4 °C for 1 month.
Conversion medium.
Combine StemSpan SFEM, 10% KnockOut Serum Replacement, 100 μg ml−1 heparin, 20 mM HEPES, and 1× GlutaMAX. Filter through a 0.45-μm filter bottle and freeze in 50-ml aliquots. Store at −20 °C for up to 6 months. Thaw at room temperature or 4 °C in the dark. Add 10 ng ml−1 bFGF and 50 ng ml−1 SCF to thawed aliquots, which can be stored at 4° C for one week.
Lentivirus preparation.
Generate and titer the FosB, Gfi1, Runx1, Spi1 lentiviral particles as described in Ginsberg et al22, with the following modifications. Briefly, produce viral particles in 293T Lenti-X cells (passage 8–10; subconfluent) using the Lenti-X Packaging Single Shot system and following the manufacturer’s instructions. Harvest supernatant only once, as second harvests often yield considerably less lentivirus, filter through a 0.45-μm PES filter attached to a 30-ml syringe, and concentrate using Lenti-X Concentrator, also following the manufacturer’s instructions. Resuspend the concentrate from one 100-mm plate of 293T cells in 100 μl lentivirus storage buffer and store at −80 °C for up to 1 year. Usually, 28 ml lentivirus supernatant produced by transfection of two 100-mm plates 293T cells for each of the 4 transcription factors (Fosb, Gfi1, Spi1, and Runx1) is sufficient for one reprogramming experiment. To assess the quality of virus produced, we recommend performing ELISA against p24 to estimate the concentration of viral capsids and qRT-PCR to detect lentiviral genome copies. We perform transduction in a ~ 60% confluent well of a six-well plate of murine ECs using a total volume of 1 ml murine EC medium, 10,000 pico-gram each concentrated Fosb, Gfi1, Spi1, and Runx1 lentiviruses, and 5 μg polybrene. Culture cells and lentiviruses undisturbed for 72 h. Although alternative methods may be used to generate lentivirus, we cannot guarantee similar efficacy when using the reprogramming platform described herein.
! CAUTION: Lentivirus production should be conducted in a tissue culture hood, and waste products should be disposed of properly.
! CAUTION: Contact your local biosafety officer before working with lentiviral vectors and follow all biosafety level 2 regulations, such as using personal protection equipment and properly disposing of waste. Safety regulations are available on the website of the US National Institutes of Health (https://osp.od.nih.gov/biotechnology/biosafety-guidance/).
PROCEDURE
! CAUTION: All steps in which cells are manipulated should be conducted in a tissue culture hood, and waste products should be disposed of properly.
Adult murine endothelial cell isolation by FACS · TIMING ~ 3 weeks
Critical point: If you wish to replace FACS isolation with isolation using magnetic beads, follow the procedure in Box 1 in place of steps 9–15.
BOX 1.
Adult murine endothelial cell isolation by magnetic beads · TIMING ~ 3 weeks
If access to cell sorting facilities is limited or, from experience, more than one FACS-enabled purification step is often needed to definitively separate haematopoietic and other contaminating cell populations from endothelial cells, the initial isolation can be performed using magnetic beads. To do this, perform steps 1–8 of the main procedure after step 10 of the following procedure.
Additional materials required:
Dynamag-2 Magnet (magnetic particle concentrator; Thermo, cat. no. 12321D)
Dynabeads CD31 Endothelial Cell (Dynabeads; Thermo, cat. no. 11155D)
Anti-mouse CD31 antibody (Biolegend, cat. no. 102502; clone MEC13.3)
-
Dilute 25 μl Dynabeads for every 350 mg murine tissue with 250 μl suspension buffer in a sterile 2 ml microcentrifuge tube.
Critical point: To ensure optimal bead washing and conjugation, one tube should be used to accommodate no more than 100 μl beads and 1 ml suspension buffer, i.e., four whole ~ 350 mg lung preparations, or the equivalent weight of another organ.
Gently pipette up and down to dissociate beads.
Capture beads using a magnetic particle concentrator and aspirate supernatant.
Add 1 ml suspension buffer to each tube (the volume of suspension buffer to add is independent of the number of preparations in a single tube).
Gently pipette up and down to dissociate beads and then capture beads using a magnetic particle concentrator and aspirate supernatant.
Repeat washing steps (steps iv and v) once and aspirate supernatant.
Resuspend beads in 250 μl suspension buffer and add 5 μg anti-CD31 antibody for each 350 mg preparation. Adjust quantities for more/different preparations proportionally.
Conjugate antibody to beads at 4 °C for 45 min on a rotator.
Capture conjugated beads using a magnetic particle concentrator and wash 3× in 1 ml suspension buffer regardless of the number of preparations in a single tube.
-
Resuspend beads in 250 μl of suspension buffer for one 350 mg preparation and store at 4 °C until use.
Pause point: Conjugated beads can be stored at 4 °C for up to a week.
Follow steps 1–8 from the main procedure.
-
Add cell suspension to 2 ml microcentrifuge tube containing the same volume conjugated beads. For one 350 mg preparation, this should yield 0.5 ml conjugated beads and cells.
Critical point: We recommend using one 2 ml microcentrifuge tube for each 350 mg preparation for this and subsequent steps so as to maximize bead-antibody-cell conjugation.
Incubate bead-cell suspension at 4 °C for 45 min on a rotator.
-
Capture bead-cell conjugates with a magnetic particle concentrator for 1 min, turning upside down several times. Remove supernatant using a P1000 micropipette.
Critical point: Do not aspirate discard liquid to avoid losing loosely conjugated cells.
-
Wash beads 3× by adding 1 ml suspension buffer to each sample, removing the magnet from the magnetic particle concentrator, inverting 5–6 times or until the beads diffuse, and placing the magnet back on the concentrator. Remove supernatant using a P1000 micropipette.
Critical point: Do not aspirate discard liquid to avoid losing loosely conjugated cells.
Wash beads 1× with 1 ml ice-cold PBS to remove EDTA following the method described in step 15.
Resuspend the beads in mEC medium and culture one 350 mg preparation onto a single well of a fibronectin-coated six-well plate in a humidified incubator at 37 °C, 5% CO2, and 5% O2 for 48 h.
Without removing any medium, add fresh mEC medium onto plated cells and continue culture for another 48 h.
Aspirate medium, wash 1× with PBS, and place fresh mEC onto cells. Replace medium every 48 hours until cells are ~ 90% confluent.
-
Harvest cells along with remaining beads using 0.5 ml Accutase for every well of a six-well plate in use. The cells will detach after ~ 1 min at 37 °C. Centrifuge cell suspension(s) at 500g for 5 min and resuspend pellet(s) in 0.5 ml trypsin for no more than 30 s. Capture unconjugated beads using a magnetic particle concentrator and transfer the supernatant to a 15 ml tube containing 5 ml mEC medium, then centrifuge at 500g for 5 min.
Critical point: When murine ECs are bead-isolated, trypsin can help purify cells by dissociating bead-cell conjugates. To increase viability and the retention of endothelial identity, do not overexpose the cells to trypsin. In every other instance, murine ECs can be passaged using Accutase followed by a single 5-min centrifugation at 500g.
Remove supernatant and plate cells 1:2 in mEC medium onto fibronectin-coated plates. Incubate in a humidified incubator at 37 °C, 5% CO2, and 5% O2 until further use (see step 19 of main protocol).
-
1
Anesthetize mice with a mixture of O2 and isoflurane within an anesthesia chamber connected to an isoflurane vaporizer. Turn the vaporizer knob to 3 and set the flow rate to 3 L min−1. Wait 2–3 min until mice are completely anesthetized.
! CAUTION: The method you use to anesthetize mice should be conducted following a protocol approved by your Institutional Animal Care and Use Committees protocol.
-
2
Inject 25 μg anti-VE-Cadherin-AF647 antibody resuspended in 50 μl PBS retro-orbitally into 8-to-10-week-old Runx1-rtTA mice. Return mice to their cages for 8 min.
-
Critical point: VE-cadherin becomes internalized during tissue digestion. While ECs could be stained ex vivo, we have observed much greater yields when VE-cadherin is intra-vitally labelled.
! CAUTION: This step should be conducted following a protocol approved by your Institutional Animal Care and Use Committees protocol. Contact your animal care facility to receive the appropriate training to perform this step.
-
-
3
Euthanize mice using CO2, excise the organs for harvest, and place them on ice-cold PBS.
-
Critical point: We have successfully converted ECs from the lungs, liver, and brain following this method. See Fig. 2e for an estimatated number of cells that can be isolated from various organs.
! CAUTION: This step should be conducted following a protocol approved by your Institutional Animal Care and Use Committees.
-
-
4
Wash the organs twice in ice-cold PBS to get rid of excess blood.
-
5
Mince organ(s) of interest to a slurry using a double-edged safety razor blade and incubate with 1× Dispase/Collagenase/DNAse mix in HBSS at 37 °C on an orbital shaker (250 rpm) for 20–30 min to digest tissue.
Critical point: The lungs of an adult male C57BL/6 mouse weigh ~ 350 mg. We routinely digest both lungs, or an equivalent weight of the organ of interest, in 0.5 ml 8× Dispase/Collagenase/DNAse and 3.5 ml HBSS. These amounts should yield a viable population of sorted cells or bead-isolated ECs (see Box 1).
-
6
Add 10 ml suspension buffer to tissue digest, pipette up and down to homogenize mixture, and filter through a 40-μm strainer.
-
7
Spin down at 500g for 5 min, wash with 10 ml suspension buffer, and spin down again at 500g for 5 min.
-
8
Resuspend cells in 250 μl suspension buffer and transfer to a sterile 1.5 ml microcentrifuge tube.
-
9
Block single cell suspension by adding anti-mouse CD16/32 antibody at the concentration specified on Table 1 for 5 min on ice.
-
10
Stain single cells with anti-mouse CD31-PE/Cy7, anti-mouse Ter119-BV421 and anti-mouse CD45-PE antibodies at the concentrations specified on Table 1 for 30 min on ice.
-
11
Wash with 1 ml suspension buffer and centrifuge at 500g for 5 min.
-
12
Resuspend cells in 500 μl suspension buffer 300 nM DAPI.
-
13
Sort 250,000 to 300,000 adult murine ECs per well of a fibronectin-coated 12-well plate into a 1.5 ml microcentrifuge tube containing 250 μl mEC medium. Adult murine ECs can be sorted with a BD FACSAria II, at 25 psi, and using a 100 μm nozzle. The FACS setup and gating strategy for EC isolations is depicted on Fig. 2a–d.
Critical point: flow rate on the BD FACSAria II should not exceed 5.0 to maximize adult endothelial cell viability.
-
14
Following cell sorting, centrifuge plates at 100g for 5 min, carefully aspirate supernatant and add 1 ml fresh mEC medium per well.
Critical point: sorted cells may be placed on ice momentarily, but we recommend plating them as soon as possible to improve viability. Sorted endothelial cells should not spend more than 45 min in suspension. For this reason, we also recommend sorting no more than 350 mg digested tissue at a time (see step 5).
-
15
Culture purified adult murine EC cultures on fibronectin-coated plates in mEC medium in a humidified incubator at 37 °C, 5% CO2, and 5% O2 until further use (see step 19).
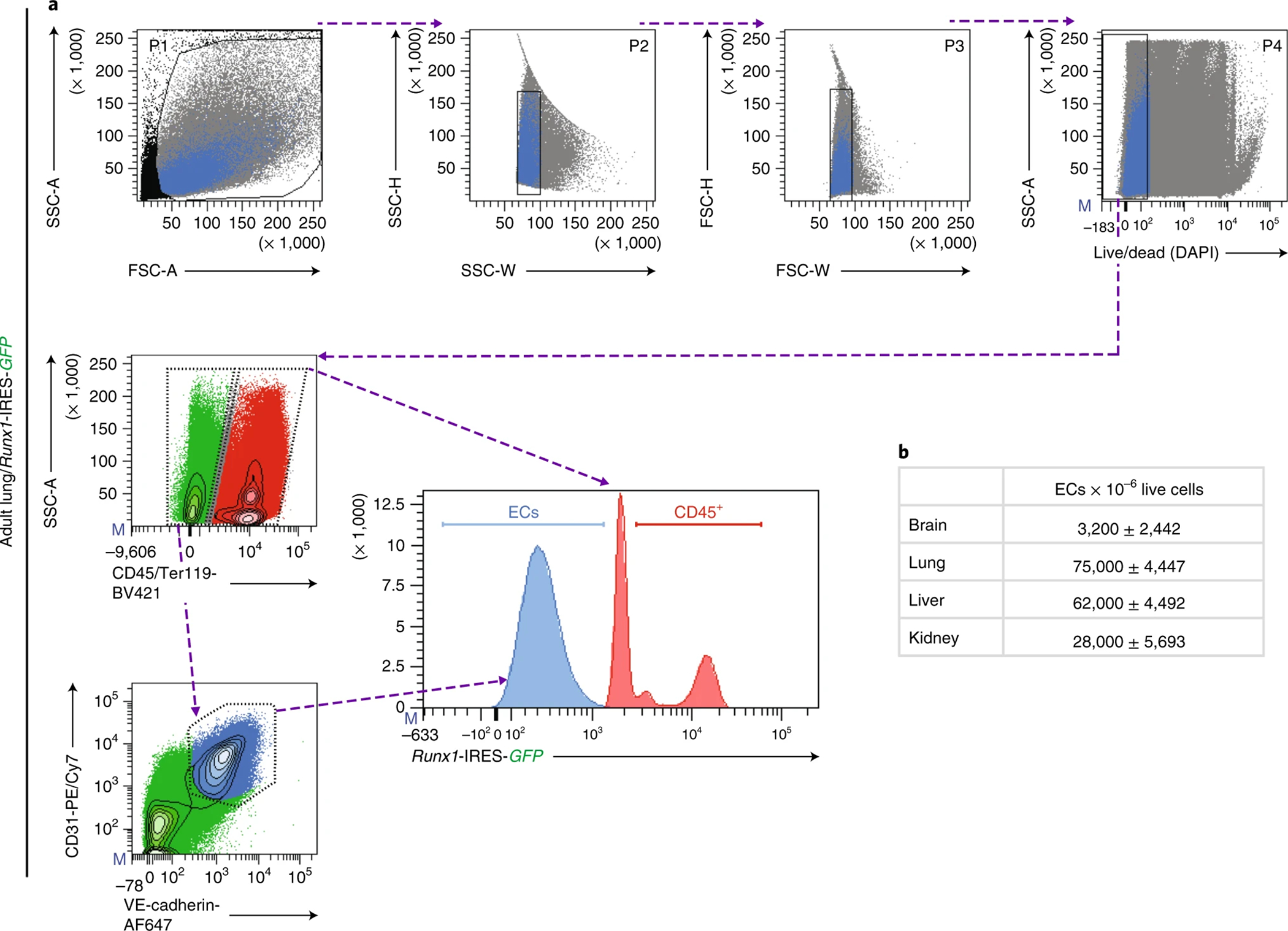
Figure 2. Isolation of murine endothelial cells by flow cytometry.
Representative flow cytometry and gating strategy to isolate adult murine lung endothelial cells (ECs). a, First, all events are discriminated by size, granularity, and viability (top panels). The first gate (P1) is drawn on a plot of SSC-A vs. FSC-A (far left), excluding a narrow strip of debris and enucleated cells, such as erythrocytes and platelets. P1 is then visualized on consecutive plots of SSC-H vs. SSC-W and FSC-H vs. FSC-W (or vice versa) to discern single cells from two or more cells in close proximity to each other (center), yielding P2 and P3, respectively. Next, DAPI staining allows live-dead cell discrimination (far right). The resulting P4 gate should consist of single, live cells. b, Negative gating of anti-mouse CD45-PE and anti-mouse Ter119-BV421 is used to exclude miscellaneous haematopoietic and erythroid cells (red), respectively. c, Single, live, non-haematopoietic cells (green) are visualized on a plot of anti-mouse VE-Cadherin-AF647 vs. anti-mouse CD31-PE/Cy7. Adult murine ECs (blue) are defined and sorted as DAPI−CD45−Ter119−VE-Cadherin+CD31+. d, Cells isolated from a Runx1-IRES-GFP reporter can be plotted on a histogram to ensure that the EC population to be sorted (blue) and the excluded miscellaneous haematopoietic and erythroid cells (red) do not overlap (center panel). e, Absolute numbers of ECs (DAPICD45−Ter119−VE-Cadherin+CD31+) for every million live cells (P4) isolated from indicated organ. Data represent mean ± s.d. (n = 4). Flow cytometry data was obtained was analyzed using a BD FACSAria II and BD FACSDiva software.
Isolation of human umbilical vein endothelial cells (HUVECs) and generation of E4ORF1-HUVECs (E4ECs) · TIMING ~ 3 weeks
! CAUTION: Contact your local biosafety officer before working with lentiviral vectors and follow all biosafety level 2 regulations, such as using personal protection equipment and properly disposing of waste. Safety regulations are available on the website of the US National Institutes of Health (https://osp.od.nih.gov/biotechnology/biosafety-guidance/).
-
16
Transduce ~ 60% confluent HUVECs with E4ORF1 lentivirus (prepared as described in Reagent Setup) at 10,000 pico-gram × 10−5 cells (~ 1 well of a six-well plate) in the presence of 5 μg polybrene. Culture in hEC medium undisturbed in hEC medium in a humidified incubator at 37 °C, 5% CO2, and 5% O2 for 72 h.
-
17
Change media to serum-free EC medium for ~ 10 days, or until a control well of untransduced HUVECs dies off.
Critical point: This will select for a population of cells able to survive under serum- and xenobiotic-free conditions, i.e. E4ORF1-HUVECs (E4ECs). E4ECs will remain viable and provide the angiocrine factors that are essential for rEC-HSPC generation for the length of the experiment (days 0–28).
-
18
Continue to culture E4ECs in hEC medium until co-culture setup (step 29) in a humidified incubator at 37 °C, 5% CO2, and 5% O2. In the meantime, proceed with next step.
In vitro bulk conversion of adult murine ECs into haematopoietic stem and progenitor cells (rEC-HSPCs) · TIMING ~ 5 weeks
-
19
Seed adult murine Runx1-rtTA ECs from step 15 (or step 21 from Box 1) at a concentration of 250,000 to 300,000 cells in 1 well of a fibronectin-coated 12-well plate. Maintain cell cultures in a humidified incubator at 37 °C, 5% CO2, and 5% O2.
-
20
Refresh mEC media every 48 h.
-
21
Grow ECs to ~ 90% confluency in mEC medium and expand 1:2 until they cover 6 wells of a six-well plate. To harvest cells, use 0.25 ml Accutase for every well of a 12-well plate in use. The cells will detach after ~ 1 min at 37 °C. Centrifuge cell suspension at 500g for 5 min, resuspend pellet in mEC medium, and plate cells 1:2 onto a fibronectin-coated plate. Maintain cell cultures in a humidified incubator at 37 °C, 5% CO2, and 5% O2.
-
22
Repeat steps 8–15 to sort-purify Runx1-rtTA EC cultures by flow cytometry once a full six-well plate is ~ 90% confluent, which is usually ~ 15 days after the initial isolation. To harvest cells, use 0.5 ml Accutase for every well of a 6-well plate in use. The cells will detach after ~ 1 min at 37 °C. Centrifuge cell suspension at 500g for 5 min.
Critical point: Perform this additional sorting step to exclude any non-endothelial contaminating cells from the culture prior to transduction with lentiviral FGRS. If cells were isolated using the method in Box 1, then this sort will also serve as confirmation of their endothelial phenotype, i.e., VE-Cadherin+CD31+CD45−.
-
23
Grow purified ECs to ~ 90% confluency in mEC medium and expand 1:2 until they cover at least a full six-well plate at ~ 60% confluency (see step 21 for how to passage the cells).
-
24
Transduce 60% confluent Runx1-rtTA ECs with FosB, Gfi1, Runx1, and Spi1 lentivirus at 10,000 pico-gram × 10−6 cells (~ 1 well of a six-well plate) in the presence of 5 μg polybrene. Culture in mEC medium undisturbed for 72 h.
-
25
Induce FGRS expression by adding 1 μg ml−1 doxycycline to the mEC medium. Add a further 1 μg ml−1 doxycycline after 24 h and incubate cells for a further 24 h.
-
26
Check the phenotype of the transduced ECs for CD31, VE-Cadherin, and CD45 one more time by assaying one well of cells by flow cytometry (repeat step 22 without sorting). If VE-Cadherin+CD31+CD45− Runx1-rtTA cells represent less than 90% of the culture, restart the isolation process from step 1.
Critical point: It is not uncommon for primary murine ECs to lose VE-Cadherin and CD31 expression after repeated passaging and purification, so it is vital to confirm that the cells about to undergo reprogramming remain VE-Cadherin+CD31+CD45− until they are co-cultured with E4ECs. A small VE-Cadherin−CD31−CD45− subpopulation of undefined stromal cells can outcompete ECs in serum-free conditions, and VE-Cadherin−CD31−CD45+ cells would represent a contaminating haematopoietic population.
-
27
Extract RNA from one well of FGRS-transduced Runx1-rtTA ECs 48 h after doxycycline induction to confirm successful lentiviral transduction by performing a qRT-PCR experiment as described in Box 2.
-
28
Harvest E4ECs from step 18 using 0.5 ml Accutase for every well of a six-well plate in use. The cells will detach after ~ 1 min at 37 °C. Centrifuge cell suspension at 500g for 5 min, resuspend pellet in hEC medium, and plate at 250,000 cells per six-well plate. Grow to ~ 40% confluency overnight at in a humidified incubator at 37 °C, 5% CO2, and 5% O2.
-
29
Harvest FGRS-transduced, doxycycline-induced Runx1-rtTA ECs from step 25 (after performing steps 26 and 27) using 0.5 ml Accutase for every well of a six-well plate in use. The cells will detach after ~ 1 min at 37 °C. Centrifuge cell suspension at 500g for 5 min, resuspend pellet in mEC medium with 1 μg ml−1 doxycycline, and plate cells 1:6 onto 40% confluent E4ECs from step 28 (e.g., one well of a six-well plate Runx1-rtTA ECs onto six wells of a six-well plate 40% confluent E4ECs).
-
30
When the co-culture reaches confluency, replace mEC media with conversion media.
Critical point: This marks day 0 of conversion.
-
31
Continue to add 1 μg ml−1 doxycycline every 24 h for 28 days.
-
32
Feed cells every 48 h by adding 1 ml conversion medium to each well of a six-well plate containing the reprogramming co-cultures.
Critical point: Converted haematopoietic cells grow in suspension. To avoid the loss of converting or converted cells whilst feeding them, do not aspirate media. Instead, collect old media and centrifuge it at 300g for 5 min at 4°C, making sure to place 0.5 ml fresh medium in each well of the reprogramming co-cultures to prevent drying. Aspirate centrifuged media and resuspend pellet in 0.5 fresh conversion medium for each well. Feed cells with this suspension.
-
33
If using ECs derived from a haematopoietic reporter mouse, such as the Runx1-IRES-GFP (Runx1tm4Dow) mouse, follow the procedure given in Box 3 to carry out single-cell conversion into rEC-HSPCs.
BOX 2.
RNA Isolation and qRT-PCR · TIMING ~ 2 hours
We perform qRT-PCR to measure FGRS overexpression in murine ECs prior to reprogramming.
Extract RNA from 1 well of a six-well plate at least 48 h after addition of doxycycline (vector; step 24 from main procedure) using an RNeasy Micro Kit and following the manufacturer’s instructions. As controls, extract total RNA from a sample that has not been exposed to doxycycline (vector control) as well as from a sample of cells that have not been transduced with FGRS lentiviruses but have been exposed to doxycycline for the same amount of time as the experimental group (doxycycline control).
-
Measure total RNA concentration using a spectrophotometer.
Critical point: This method usually yields 200–300 ng μl−1 total RNA with a ratio of absorbance at 260 nm and 280 nm ~ 2.0. If RNA concentration or quality are less than 20 ng μl−1 or 1.8, respectively, repeat step 1.
Critical point: Maintain all RNA and DNA material on ice throughout the qRT-PCR assay.
- Convert 500 ng total RNA to cDNA in 16 μl dH2O added to 4 μl SuperScript III reverse transcriptase using the thermocycler program below.
Cycle number Annealing Synthesis Denaturation 1 5 min at 25 °C 2 30 min at 42 °C 3 5 min at 85°C Dilute the resulting cDNA by adding 100 μl dH2O, yielding enough material for an extra round of qPCR.
-
Prepare four master mixes (for detection of each transcription factor: Fosb, Gfi1, Runx1, and Spi1) by combining the components listed below.
- Critical point: Calculate master mix volumes taking into consideration number of samples as well as technical replicates. To perform qRT-PCR on three samples (vector, vector control, and doxycycline control) in triplicate, increase the volumes below by a factor of 9.
2× SYBR green 10 μl fwd + rev primer working stock, 2.5 μM each 4 μl dH2O 1 μl Critical point: HPLC-purified oligonucleotide primer sequences (Integrated DNA Technologies) are provided in Table 2. Reconstitute primers in dH2O to prepare 100 μM stock solutions and store indefinitely at −20 °C. Working stocks consist of 12.5 μl forward primer and 12.5 μl reverse primer in 475 μl dH2O, which yields 500 μl 2.5 μM forward and reverse primer mix.
Lay out an experimental scheme to prevent errors when dispensing reagents. For three samples (vector, vector control, and doxycycline control) in triplicate, assign 9 wells of a qPCR plate for each of the four master mixes (Fosb, Gfi1, Runx1, and Spi1). As such, 36 wells are required for this qRT-PCR experiment.
Dispense 15 μl each master mix into corresponding well of a qPCR plate.
-
Dispense 5 μl vector, vector control, or doxycycline control cDNA into appropriate wells already containing master mix. As such, 60 μl cDNA are required from each sample for this qRT-PCR experiment.
Criticial point: It is crucial to avoid cross-contamination of samples or master mixes. If cross-contamination occurs, discard affected wells from future calculations. Available wells on the same plate may be assigned to replace discarded wells by repeating steps 5–7 using the necessary volumes.
Cover qPCR plate with a layer of adhesive film and centrifuge plates at 100g for 1 min.
Run experiment in an appropriate real-time PCR reader, such as a ViiA 7, using the cycling conditions recommended by the manufacturer.
-
For each primer pair, subtract mean CT values from vector samples from mean CT values from vector controls or doxycycline controls to obtain ΔCTvc and ΔCTdc, respectively (ΔCTvc ≈ ΔCTdc). Calculate relative transcript abundance as 2−ΔCT.
Criticial point: We recommend moving forward with the reprogramming process using ECs that present 2−ΔCT ≥ 50 around the time of co-culture (step 29 from the main protocol).
End of conversion and rEC-HSPC harvest · TIMING ~ 2 hours
-
34
On approximately day 28 of conversion, harvest co-cultures of rEC-HSPCs and E4ECs by collecting non-adherent cells in a 15- or 50-ml conical tube, then adding 0.5 ml Accutase to each well of a six-well plate in use. The cells will detach after ~ 1 min at 37 °C. Collect and centrifuge cell suspension(s) at 500g for 5 min.
-
35
Discard supernatant and resuspend pellet(s) in 5 ml suspension buffer. Wash twice with suspension buffer, centrifuging at 500g for 5 min at 4°C after each wash.
-
36
Resuspend cells in 100 μl suspension buffer, transfer to a microcentrifuge tube, and block by adding anti-mouse CD16/32 antibody at the concentration specified on Table 1 for 5 min on ice.
-
37
Stain single-cell suspension by adding anti-human CD31-BV421, anti-mouse VE-Cadherin-A647, anti-mouse CD31-PE/Cy7, and anti-mouse CD45-PE at the concentrations specified on Table 1 for 40 min on ice.
-
38
Repeat step 35 using 1 ml suspension buffer.
-
39
Re-suspend cells in suspension buffer containing 300 nM DAPI.
-
40
Set up BD FACSAria II to sort rEC-HSPCs at 45 psi and using the 85 μm nozzle.
Critical point: rEC-HSPCs are defined and sorted as VE-Cadherin−mCD31−Runx1-GFP+CD45−DAPI− (see Fig. 3a).
-
41
Sort 33,000 VE-Cadherin−mCD31−Runx1-GFP+CD45+DAPI− cells using the single cell purity mask and a microcentrifuge collection tube containing 300 μl PBS.
Downstream assays
Follow option A to carry out a colony-forming assay. Follow option B to undertake primary and secondary transplantation of rEC-HSPCs.
Critical point: Transplantations can be performed in addition to or independently of colony-forming cell (CFC) assays of rEC-HSPCs.
A). Preparation of cells for colony-forming cell assay · TIMING ~ 2 weeks
Sort 33,000 VE-Cadherin−mCD31−Runx1-GFP+CD45+DAPI− cells into a microcentrifuge tube containing 300 μl PBS (see steps 34–41).
Place sorted cell suspension in its entirety in a single tube of cytokine-rich methylcellulose and vortex for 15 s. This will yield a ~ 3.3-ml suspension.
-
Using a blunt-end needle attached to a 3 ml syringe, aspirate 2 ml cell suspension in methylcellulose and dispense 1 ml per well onto two wells of a meniscus-free six-well plate, respectively.
Critical point: One rEC-HSPC sample will yield two technical replicates. Assay up to two samples (four wells), leaving at least two wells free, in each plate.
Aliquot ~ 3 ml dH2O on the empty wells to prevent drying of the methylcellulose for the duration of the assay.
Maintain methylcellulose experiment(s) in a humidified incubator at 37 °C, 5% CO2, and 5% O2.
Perform the colony count according to manufacturer’s instructions 14 days after setting up the experiment (see Fig. 1e from Lis et al.)
B). Primary and secondary transplantation of rEC-HSPCs · TIMING ~ 40 weeks
-
i
Irradiate 12-week-old CD45.1+ recipient mice of either sex with a lethal dose of radiation, e.g., 950 cGy, using a Caesium-137 or X-Ray irradiator.
! CAUTION: The method you use to irradiate mice should be conducted following a protocol approved by your Institutional Animal Care and Use Committees protocol.
-
ii
Anesthetize mice 18 h post-irradiation with a mixture of O2 and isoflurane within an anesthesia chamber connected to an isoflurane vaporizer. Turn the vaporizer knob to 3 and set the flow rate to 3 L min−1. Wait 2–3 min until mice are completely anesthetized.
! CAUTION: The method you use to anesthetize mice should be conducted following a protocol approved by your Institutional Animal Care and Use Committees protocol.
-
iii
For each mouse receiving a transplant, sort 800,000 VE-Cadherin−mCD31−Runx1-GFP+CD45+DAPI− cells into PBS (see steps 34–41).
-
iv
Inject every mouse with 800,000 VE-Cadherin−mCD31−Runx1-GFP+CD45+DAPI− sorted cells in no more than 100 μl PBS retro-orbitally.
-
Critical point: As a positive control, isolate whole bone marrow (WBM) from CD45.2+ mice (steps xxvii–xxxv) and co-inject a cohort of mice with 800,000 unfractionated WBM cells concomitantly with the experimental cohort receiving rEC-HSPC transplants.
! CAUTION: This step should be conducted following a protocol approved by your Institutional Animal Care and Use Committees protocol. Contact your animal care facility to receive the appropriate training to perform this step.
-
-
v
Maintain transplanted recipients with Sulfatrim-supplemented feed for the first 2 weeks following transplantation. Mice need no other special husbandry and can be housed next to untreated mice for the length of the experiment.
! CAUTION: This step should be accepted and conducted in accordance to your Institutional Animal Care and Use Committees protocol.
-
vi
Determine multi-lineage engraftment every 4 weeks for 20 weeks by flow cytometry of peripheral blood using DAPI to discriminate dead cells (steps viii–xiv).
-
vii
After 20 weeks, proceed to secondary transplantation, isolating WBM from primary-engrafted mice (steps xxvii–xxxv) and transplanting 800,000 unfractionated WBM cells retro-orbitally into lethally irradiated CD45.1+ recipients (steps i–vi).
! CAUTION: This step should be conducted following a protocol approved by your Institutional Animal Care and Use Committees protocol. Contact your animal care facility to receive the appropriate training to perform this step.
Preparation of peripheral blood for staining · TIMING ~ 1.5 h
-
viii
Prepare 1× RBC lysis solution by diluting 10× RBC lysis buffer in dH2O and warm up to room temperature before use.
Critical point: The pH of the RBC lysis solution should be between 7.1 and 7.4; adjust if necessary.
-
ix
Place ~ 35-μl blood sample in a 5-ml round-bottom polystyrene tube containing 130 μl suspension buffer.
-
x
Add 4 ml 1× RBC lysis solution and incubate on ice for 8 min.
-
xi
Centrifuge for 5 min at 450g at 4 °C.
-
xii
Wash sample twice using 4 ml suspension buffer, centrifuging every time at 450g for 5 min at 4 °C.
-
xiii
Discard the supernatant, resuspend pellet in 100 μl suspension buffer, transfer to a microcentrifuge tube, and block by adding anti-mouse CD16/32 antibody at the concentration specified on Table 1 for 5 min on ice.
-
xivPerform FACS staining using the following antibody panels (clone numbers and concentrations can be found on Table 1; representative gating is available on Fig. 4e):
Myeloid/B lymphoid cocktail T lymphoid cocktail BV421/Pacific Blue B220 CD3 GFP Runx1 reporter Runx1 reporter PE Gr1 CD4 PE-Cy7 CD45.1 CD45.1 APC Ter119 Ter119 AF700 CD11b CD45.2 APC-Cy7 CD45.2 CD8
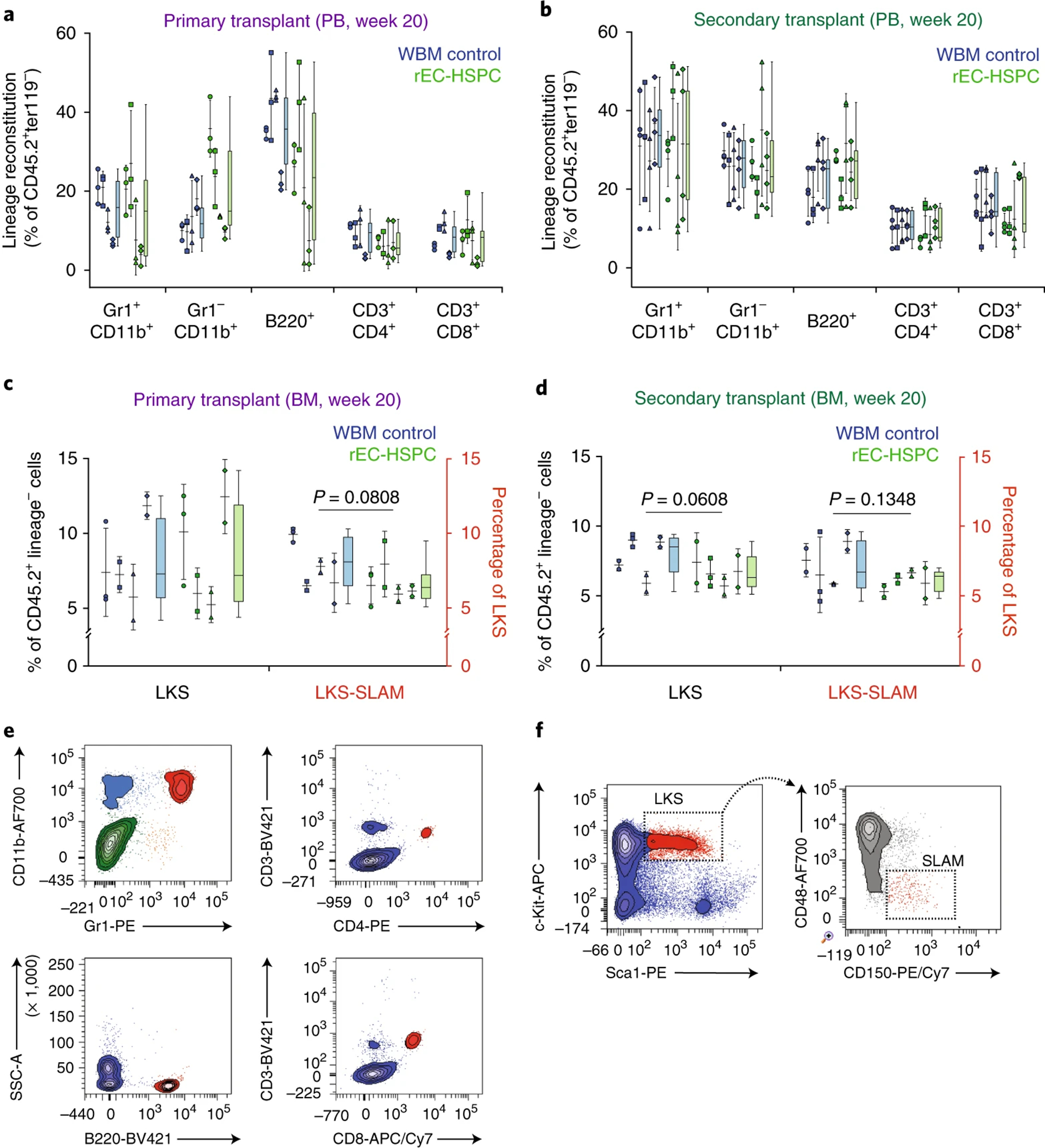
Figure 4. Conditional FGRS expression supports long-term 1° and 2° rEC-HSPC engraftment (adapted from Lis et al.).
a, Lineage contribution to Gr1+CD11b+ and Gr1−CD11b+ myeloid cells, B220+ B cells, CD3+CD4+ T cells, and CD3+CD8+ T cells at week 20 post-primary whole bone marrow (WBM) control (blue boxes) or rEC-HSPC (green boxes) transplants in peripheral blood (PB). Data represent mean ± s.d. (n = 4 independent conversion experiments run in technical triplicates for each condition). P-values, two-tailed unpaired t-test. b, Lineage contribution to Gr1+CD11b+ and Gr1−CD11b+ myeloid cells, B220+ B cells, CD3+CD4+ T cells, and CD3+CD8+ T cells at week 20 post-secondary WBM control (blue boxes) or rEC-HSPC (green boxes) transplants in PB. Data represent mean ± s.d. (n = 4 independent conversion experiments run in technical triplicates for each conditions). P-values, two-tailed unpaired t-test. c, Relative representation of LKS and LKS-SLAM cells in bone marrow at week 20 post-primary and d, secondary transplantation of WBM controls (blue boxes) or rEC-HSPC (green boxes). Data represent mean ± s.d. (n = 4 independent conversion experiments run in technical triplicates for each condition). P-values, two-tailed unpaired t-test. e, Representative flow cytometry plots of rEC-HSPC lineage contribution to the peripheral blood after events are discriminated by size, granularity and viability (see Fig. 2a), and CD45.1−CD45.2+ cells are gated by anti-mouse CD45.1-PE/Cy7 and anti-mouse CD45.2-APC/Cy7 (myeloid and B lymphoid panels, top left and bottom left, respectively) or CD45.2-AF700 (T lymphoid panels, right panels) staining. Data shown correspond to one mouse bled on week 20 post-primary transplantation. f, Representative flow cytometry plots of rEC-HSPC LKS-SLAM contribution to the bone marrow after events are discriminated by size, granularity and viability (see Fig. 2a), and CD45.2+Lin− cells are gated by anti-mouse CD45.2-APC/Cy7 and anti-mouse Lin-BV421 staining (left panel) and cKit+Sca1+ cells are gated by anti-mouse cKit-APC and anti-mouse Sca1-PE staining (right panel). Data shown correspond to one mouse femur analyzed on week 20 post-primary transplantation. Flow cytometry data was obtained was analyzed using a BD FACSAria II and BD FACSDiva software. All animal experiments were performed under the approval of Weill Cornell Medicine Institutional Animal Care and Use Committee. Flow cytometry data was obtained was analyzed using a BD FACSAria II and BD FACSDiva software.
Preparation of spleen for staining · TIMING ~ 1.5 h
-
xv
Euthanize end-point transplanted mice (step vii) using CO2 and harvest spleens in ice-cold suspension buffer.
! CAUTION: This step should be accepted and conducted in accordance to your Institutional Animal Care and Use Committees protocol.
-
xvi
Place the spleen on a 40-μm strainer fit atop a 50-ml conical tube.
-
xvii
Push the spleen through the strainer using the rubber end of the plunger of a 1-ml syringe.
-
xviii
Wash the cells off the strainer with ~ 10 ml ice-cold suspension buffer.
-
xix
Centrifuge the cell suspension at 300g for 5 min.
-
xx
Prepare RBC lysis buffer as explained in step viii above.
-
xxi
Aspirate the supernatant from step xix and resuspend pellet in a 50-ml conical tube with 4 ml 1× RBC lysis solution.
-
xxii
Incubate on ice for 8 min.
-
xxiii
Wash cells as described in steps viii and xii above.
-
xxiv
Resuspend twice-washed spleen pellets in 100 μl suspension buffer.
-
xxv
Discard the supernatant, resuspend pellet in 100 μl suspension buffer, transfer to a microcentrifuge tube, and block by adding anti-mouse CD16/32 antibody at the concentration specified on Table 1 for 5 min on ice.
-
xxvi
Perform FACS staining using the following antibody panels (clone numbers and concentrations can be found on Table 1; representative gating is available on Fig. 4f):
- Critical point: Haematopoietic stem cells are defined as Ter119−CD45+Lin−Sca1+cKit+CD48−CD150+.
LKS SLAM-LT HSC BV421/Pacific Blue Lineage cocktail GFP Runx1 reporter PE Sca1 PE-Cy7 CD150 APC c-kit AF700 CD48 APC-Cy7 CD45.2
Preparation of bone marrow for staining · TIMING ~ 1.5 h
-
xxvii
Euthanize end-point transplanted mice (step vii) using CO2 and harvest both femurs in ice-cold PBS.
! CAUTION: This step should be accepted and conducted in accordance to your Institutional Animal Care and Use Committees protocol.
-
xxviii
Excise femurs carefully so as to preserve the bones intact..
! CAUTION: This step should be conducted following a protocol approved by your Institutional Animal Care and Use Committees protocol. Contact your animal care facility to receive the appropriate training to perform this step.
-
xxix
Rid bones of all muscle and connective tissue and place them in a sterile mortar.
-
xxx
Add 3 ml suspension buffer and crush with a pestle for ~ 30 s.
-
xxxi
Using a 1000-μl pipette, transfer as much of the cell suspension as possible to a 40-μm strainer fit atop a 50-ml conical tube, leaving the bones in the mortar.
-
xxxii
Repeat the previous two steps twice.
-
xxxiii
Centrifuge the cell suspension at 300g for 5 min.
-
xxxiv
Discard the supernatant, resuspend pellet in 100 μl suspension buffer, transfer to a microcentrifuge tube, and block by adding anti-mouse CD16/32 antibody at the concentration specified on Table 1 for 5 min on ice.
- xxxv
TROUBLESHOOTING
Troubleshooting guidance is given in Table 4.
Table 4.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 1–5; Box 1 | Little or no murine endothelial cell growth | Poor EC medium quality | Make fresh mEC medium |
| Poor EC sample viability | Proceed with another sample | ||
| Incubation conditions are not correct (5% O2) | Check and calibrate incubators | ||
| 1–5; Box 1 | Fibroblastic outgrowth | Poor EC medium quality | Add 1.5× recommended SB431542 concentration |
| Poor EC isolation quality | Proceed with a new sample | ||
| 16–18 | Little or no HUVEC growth | Poor EC medium quality | Make fresh EC medium (cf. Reagent setup/Cell culture media/Human EC media |
| Poor EC sample viability | Proceed with another sample | ||
| Incubation conditions are not correct (5% O2) | Check and calibrate incubators | ||
| 19–33 | Massive cell death | Lentiviral toxicity | Decrease the viral input |
| rEC-HSC are poorly or not generated | Lentiviral preparation was poor | Remake viruses | |
| Lentiviral protein expression is low | Remake viruses | ||
| Poor conversion medium quality | Make fresh conversion media | ||
| Poor endothelial feeder quality | Isolate new HUVEC feeder layer | ||
| Incubation conditions are off | Check and calibrate the incubators. | ||
| viii–xiv | Blood coagulates | No EDTA in suspension buffer | Add 0.2 mM EDTA to suspension buffer |
| Pellets appear white after RBC lysis | Not enough lysis time | Lyse again (repeat steps ix–xii) | |
| RBC lysis buffer concentration is too low | Remake RBC lysis buffer | ||
| RBC lysis buffer is old or contaminated | Remake RBC lysis buffer from new bottle | ||
| xxvi; xxxv | LKS-SLAM population is under 0.1% live singlets | Poor engraftment | Check the host’s (CD45.1+) haematopoietic compartment for LKS-SLAM |
| Too-low antibody concentration; old, or denatured antibodies | Make sure all antibodies are used in the concentrations suggested on Table 1 | ||
| Too little material processed | Process tibias and hip bones if necessary | ||
| xxxv | Too much debris appears on plot of SCC-A vs. FSC-A | Bone was pulverized | Crush bones without dragging the pestle |
TIMING
Timing is summarized in Fig. 1.
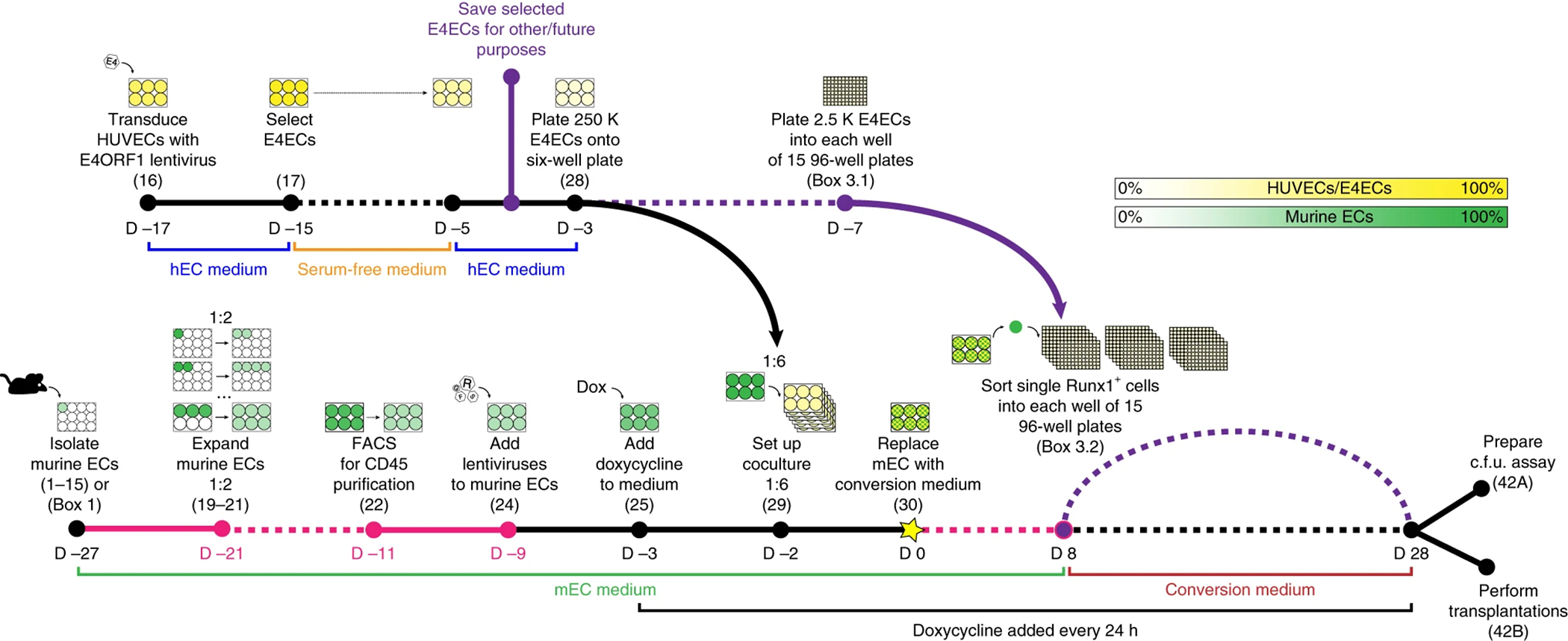
Figure 1. In vitro conversion of adult murine EC into HSPCs: timeline.
Schema for step-wise conversion of adult murine ECs (mECs) into HSPCs. Course of reprogramming is depicted in time from left to right. Key steps are denoted by a filled circle, with a corresponding pictogram, the step number in parentheses, and day (D). D 0, denoted by a star, corresponds to the day when the reprogramming co-culture is placed in cytokine-enriched, serum- and xenobiotic-free medium (see Reagent Setup under Materials). Time points are not spaced to scale, yet dotted lines indicate durations between steps longer than this representation indicates. Fucsia lines indicate steps of variable length. Violet lines indicate optional steps. Human umbilical vein endothelial cells (HUVECs) and E4ORF1-HUVECs (E4ECs) are represented in yellow; mECs are represented in green. Cell confluency in vitro is depicted as a transparency spectrum on the legend at the top right of the figure from 0% (absence of color) to 100% (saturated green/yellow).
Day -30: adult murine Runx1-rtTA ECs isolation (steps 1–15, or Box 1). HUVEC transduction with E4ORF1 gene (step 16) and selection (step 17).
Day -25 to -5: expansion of Runx1-rtTA ECs (steps 19–22) in mEC.
Day -5: transduction of Runx1-rtTA ECs with FGRS lentivirus (step 24).
Day -3: doxycycline addition (1 μg ml−1) to induce FGRS in Runx1-rtTA ECs (step 25).
Day -2: Plating of FGRS-transduced Runx1-rtTA ECs on E4ECs in mEC medium (step 29).
Day 0: switch to conversion media (step 30).
Day 28: transplantation of rEC-HSPCs into lethally irradiated CD45.1 recipients (downstream assay B, steps i–v).
Day 28 + 4, 8, 12, 16 and 20 weeks (primary transplant): determination of multi-lineage engraftment by flow cytometry of peripheral blood (steps viii–xiv).
Day 28 +4, 8, 12, 16 and 20 weeks (secondary transplant): determine multi-lineage engraftment by flow cytometry of peripheral blood (steps viii–xiv).
ANTICIPATED RESULTS
During conversion, cells transition through three sequential phases. During the first phase, the Induction phase (days 0–8), doxycycline addition should up-regulate the expression of conditional FGRS in adult murine VE-Cadherin+Runx1−CD45− Runx1-rtTA ECs co-cultured with E4ECs. Within 8 days, FGRS-transduced Runx1-rtTA ECs (FGRS-ECs) should turn on endogenous Runx1 expression giving rise to haemogenic-like ECs (Fig. 3a and b).
During the second phase, the Specification phase (days 8–20), Runx1+ ECs should commit to a haematopoietic fate and erase their vascular phenotype transitioning from VE-Cadherin+Runx1+CD45− to VE-Cadherin+Runx1+CD45+ to VE-Cadherin−Runx1+CD45+ cells (Fig. 3a–d). This phase is characterized by morphological changes, acquisition of haematopoietic markers, and emergence of committed rEC-HSPCs with colony forming cell (CFC) function independent of FGRS expression. rEC-HSPCs are endowed with multi-lineage progenitor properties, yielding CFC-GEMM, CFC-GM, and BFU-E colonies.
During the final phase, the Expansion phase (days 20–28) the total number of short-term repopulating/radio-protective cells and Lin−c-Kit+Sca1+ (LKS) cells should significantly increase. rEC-LKS cells expand adherent to E4ECs, suggesting angiocrine factors supplied by the latter maintain and expand haematopoietic stem and progenitors cells (Fig. 3e).
To assess HSC function of putative rEC-HSPCs, rEC-HSPCs are transplanted into lethally irradiated congenic recipients. Circulating CD45.2+ haematopoietic cells derived from rEC-HSPCs should be detectable in peripheral blood (PB) 4 to 20 weeks after transplantation. Table 3 indicates the PB contribution of 20 rEC-HSPC transplants as measured every four weeks for 20 weeks (see Extended Data Fig. 2c from Lis et al.) About 70% of the transplanted mice (14 out of 20) show robust (>75%) engraftment of rEC-HSPCs.
Table 3.
Peripheral blood contribution of rEC-HSPCs at indicated time point post-primary transplant (% of host).
| Week 4 | Week 8 | Week 12 | Week 16 | Week 20 | |
|---|---|---|---|---|---|
| Mouse #3 | 93.00% | 93.40% | 89.30% | 92.10% | 91.30% |
| Mouse #1 | 89.30% | 93.30% | 93.30% | 97.30% | 93.40% |
| Mouse #10 | 89.30% | 93.20% | 93.30% | 93.40% | 93.20% |
| Mouse #11 | 87.80% | 92.30% | 93.20% | 93.20% | 92.20% |
| Mouse #12 | 86.40% | 91.30% | 92.70% | 93.20% | 91.20% |
| Mouse #13 | 86.40% | 91.20% | 92.20% | 92.30% | 89.40% |
| Mouse #14 | 86.30% | 90.20% | 92.10% | 91.30% | 89.30% |
| Mouse #15 | 85.70% | 89.30% | 91.60% | 91.20% | 85.90% |
| Mouse #16 | 85.60% | 88.30% | 91.20% | 91.20% | 85.70% |
| Mouse #17 | 85.60% | 87.90% | 89.80% | 91.20% | 85.60% |
| Mouse #18 | 85.40% | 87.80% | 89.60% | 90.20% | 85.40% |
| Mouse #19 | 83.60% | 85.80% | 87.40% | 89.30% | 84.30% |
| Mouse #2 | 83.40% | 84.10% | 85.80% | 87.40% | 82.30% |
| Mouse #20 | 82.60% | 83.60% | 85.30% | 85.70% | 81.30% |
| Mouse #4 | 4.50% | 83.40% | 84.10% | 81.30% | 79.10% |
| Mouse #5 | 2.02% | 4.50% | 3.20% | 2.90% | 1.30% |
| Mouse #6 | 1.53% | 1.23% | 0.60% | 0.30% | 0.30% |
| Mouse #7 | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% |
| Mouse #8 | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% |
| Mouse #9 | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% |
Peripheral blood of recipients transplanted with rEC-HSPCs should present numbers and ratios of donor-derived B cells and circulating T cells equivalent to those seen in control mice transplanted with unfractionated whole bone marrow (WBM; Fig. 4a). The immunophenotyping of bone marrow cells should reveal an indistinguishable frequency of donor-derived, immuno-phenotypically marked HSCs (Ter119−CD45.2+Lin−Sca1+cKit+CD48−CD150+; LKS-SLAM) between mice engrafted by rEC-HSCs and WBM controls (Fig. 4c).
The self-renewal potential of CD45.2+ rEC-HSPCs can be assessed by transplanting unfractionated WBM from primary transplant recipients into lethally irradiated secondary recipients. CD45.2+ myeloid and lymphoid cells should be detectable in the blood of secondary recipients from 4 to 20 weeks post-transplantation (Fig. 4b; Extended Data Fig. 2e from Lis et al). Secondary recipients of primary-transplanted WBM from experimental (rEC-HSPC) and control (WBM) groups should present a comparable number and proportion of LKS-SLAM cells (Fig. 4d).
Table 2.
Oligonucleotide sequences.
| Primer | Sequence | Expected MW (bp) |
|---|---|---|
| Fosb-fwd | agatcgacttcaggcggaaa | 184 |
| Fosb-rev | caaatctctcacctcggcca | |
| Gfi1-fwd | atgtgcggcaagaccttc | 81 |
| Gfi1-rev | acagtcaaagctgcgttcct | |
| Runx1-fwd | gaagtgtaagcccagcacagt | 103 |
| Runx1-rev | ggcgggggattctataattt | |
| Spi1-fwd | ggagaagctgatggcttgg | 75 |
| Spi1-rev | caggcgaatctttttcttgc |
Acknowledgements
We thank D. James for help in providing material and intellectual input. This work was supported by AnsaryStem Cell Institute, Starr Foundation Tri-Institute Stem Cell Initiative – TRI-SCI, grants from the Daedalus Fund for Innovation of Weill Cornell Medicine, the Empire State Stem Cell Board of New York State Department of Health – NYSTEM (C026878, C028117, C029156), a US National Institutes of Health – NIH; the National Heart, Lung, and Blood Institute – NHLBI (R01HL115128, R01HL128158, and R01 HL139056-01A1); the National Institute of Diabetes and Digestive and Kidney Diseases – NIDDK (R01DK095039 and RC2DK114777), and the Qatar National Priorities Research Program – NPRP (8-1898-3-392), and fellowships from AVALON GloboCare Biomedical Research Training Program – AGBRT, and New York Stem Cell Foundation – NYSCF. J.G.B.D. was supported by Ansary Stem Cell Institute, and NIH/NHLBI R01HL139056-01A1. R.L. was supported by Ansary Stem Cell Institute, TRI-SCI, NYSTEM C029156, NIH/NHLBI R01HL139056-01A1, NPRP 8-1898-3-392, and NYSCF Druckenmiller Fellowship D-F56. T.L. was supported by TRI-SCI and AGBRT. S.R. and K.S. were supported by Ansary Stem Cell Institute, TRI-SCI, Daedalus Fund for Innovation, NYSTEM (C026878, C028117, C029156), NIH/NHLBI (R01HL115128, R01HL128158, and R01 HL139056-01A1); NIH/NIDDK (R01DK095039, RC2DK114777), and NPRP 8-1898-3-392.
Footnotes
Competing Interests Statement
S.R. is the founder and a non-paid consultant to Angiocrine Bioscience, New York, New York, USA.
REFERENCES
- 1.de Bruijn MF, Speck NA, Peeters MC & Dzierzak E Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. The EMBO journal 19, 2465–2474, doi: 10.1093/emboj/19.11.2465 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heissig B et al. Recruitment of stem and progenitor cells from the bone marrow niche requires MMP-9 mediated release of kit-ligand. Cell 109, 625–637 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hooper AT et al. Engraftment and reconstitution of hematopoiesis is dependent on VEGFR2-mediated regeneration of sinusoidal endothelial cells. Cell stem cell 4, 263–274, doi: 10.1016/j.stem.2009.01.006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler JM et al. Development of a vascular niche platform for expansion of repopulating human cord blood stem and progenitor cells. Blood 120, 1344–1347, doi: 10.1182/blood-2011-12-398115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler JM et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell stem cell 6, 251–264, doi: 10.1016/j.stem.2010.02.001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poulos MG et al. Vascular Platform to Define Hematopoietic Stem Cell Factors and Enhance Regenerative Hematopoiesis. Stem Cell Reports 5, 881–894, doi: 10.1016/j.stemcr.2015.08.018 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poulos MG et al. Endothelial transplantation rejuvenates aged hematopoietic stem cell function. The Journal of clinical investigation 127, 4163–4178, doi: 10.1172/JCI93940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lis R et al. Conversion of adult endothelium to immunocompetent haematopoietic stem cells. Nature 545, 439–445, doi: 10.1038/nature22326 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandler VM, Lis R & Rafii S Reprogramming human endothelial cells to haematopoietic cells requires vascular induction. Nature 511, 312–318, doi: 10.1038/nature13547 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi H et al. Angiocrine factors from Akt-activated endothelial cells balance self-renewal and differentiation of haematopoietic stem cells. Nature cell biology 12, 1046–1056, doi: 10.1038/ncb2108 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson MK, Weiss AH, Hernandez-Hoyos G, Dionne CJ & Rothenberg EV Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity 16, 285–296 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Wahlster L & Daley GQ Progress towards generation of human haematopoietic stem cells. Nature cell biology 18, 1111–1117, doi: 10.1038/ncb3419 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Batta K, Florkowska M, Kouskoff V & Lacaud G Direct reprogramming of murine fibroblasts to hematopoietic progenitor cells. Cell reports 9, 1871–1884, doi: 10.1016/j.celrep.2014.11.002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doulatov S et al. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell stem cell 13, 459–470, doi: 10.1016/j.stem.2013.09.002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elcheva I et al. Direct induction of haematoendothelial programs in human pluripotent stem cells by transcriptional regulators. Nature communications 5, 4372, doi: 10.1038/ncomms5372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira CF et al. Induction of a hemogenic program in mouse fibroblasts. Cell stem cell 13, 205–218, doi: 10.1016/j.stem.2013.05.024 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sturgeon CM, Ditadi A, Clarke RL & Keller G Defining the path to hematopoietic stem cells. Nature biotechnology 31, 416–418, doi: 10.1038/nbt.2571 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Rafii S et al. Human ESC-derived hemogenic endothelial cells undergo distinct waves of endothelial to hematopoietic transition. Blood 121, 770–780, doi: 10.1182/blood-2012-07-444208 (2013). [DOI] [PubMed] [Google Scholar]
- 19.McLaughlin F et al. Combined genomic and antisense analysis reveals that the transcription factor Erg is implicated in endothelial cell differentiation. Blood 98, 3332–3339 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Sugimura R et al. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545, 432–438, doi: 10.1038/nature22370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riddell J et al. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell 157, 549–564, doi: 10.1016/j.cell.2014.04.006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ginsberg M, Schachterle W, Shido K & Rafii S Direct conversion of human amniotic cells into endothelial cells without transitioning through a pluripotent state. Nat Protoc 10, 1975–1985, doi: 10.1038/nprot.2015.126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rafii S et al. Human bone marrow microvascular endothelial cells support long-term proliferation and differentiation of myeloid and megakaryocytic progenitors. Blood 86, 3353–3363 (1995). [PubMed] [Google Scholar]
- 24.Rafii S et al. Isolation and characterization of human bone marrow microvascular endothelial cells: hematopoietic progenitor cell adhesion. Blood 84, 10–19 (1994). [PubMed] [Google Scholar]
- 25.Seandel M et al. Generation of a functional and durable vascular niche by the adenoviral E4ORF1 gene. Proceedings of the National Academy of Sciences of the United States of America 105, 19288–19293, doi: 10.1073/pnas.0805980105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]


