Abstract
Aurora A kinase (AURKA) is a master cell cycle regulator that is often dysregulated in human cancers. Its overexpression has been associated with genome instability and oncogenic transformation. The protein kinase D (PKD) family is an emerging therapeutic target of cancer. Aberrant PKD activation has been implicated in tumor growth and survival, yet the underlying mechanisms remain to be elucidated. This study identified, for the first time, a functional crosstalk between PKD2 and Aurora A kinase in cancer cells. The data demonstrate that PKD2 is catalytically active during the G2/M phases of the cell cycle, and inactivation or depletion of PKD2 causes delay in mitotic entry due to downregulation of Aurora A, an effect that can be rescued by overexpression of Aurora A. Moreover, PKD2 localizes in the centrosome with Aurora A by binding to γ-tubulin. Knockdown of PKD2 caused defects in centrosome separation, elongated G2 phase, mitotic catastrophe and eventually cell death via apoptosis. Mechanistically, PKD2 interferes with Fbxw7 function to protect Aurora A from ubiquitin- and proteasome-dependent degradation. Taken together, these results identify PKD as a cell cycle checkpoint kinase that positively modulates G2-M transition through Aurora A kinase in mammalian cells.
Keywords: Cell cycle, G2/M transition, Protein Kinase D, Aurora A kinase, Cancer
Introduction
Cell cycle is propelled by well-coordinated complex signaling events by which cells grow and divide. Uncontrolled cell division, whereby cells go through the cycle unchecked, lies at the base of cancer formation and progression. Cell cycle occurs through progression of four distinct stages, namely G0/G1, S, G2 and M, which are monitored by a battery of cyclin-dependent kinases (CDKs) and their partner cyclins (1). Mutations in the signaling pathways, aberrant activation of CDKs, genetic lesions in the genes encoding cell cycle-regulatory proteins result in genome instability, abnormal growth and eventually cancer.
The protein kinase D (PKD) family of serine/threonine kinases the Ca++/Calmodulin-dependent protein kinases (CaMKs) superfamily and consists of three isoforms in mammals, notably, PKD1, PKD2 and PKD3, which are highly conserved throughout evolution (2). Structurally, PKD possesses an N-terminal regulatory domain that contains a tandem cysteine-rich Zn-finger like domains (CRD, C1a and C1b) and a plekstrin homology (PH) domain, and a C-terminal catalytic domain (3, 4). In a canonical pathway, diverse physiological factors, such as GPCR agonists, bioactive peptides (5), lipids (6) and growth factors (7) converge to the activation of PKDs through the generation of diacylglycerol (DAG) by phospholipase C (PLC) and the activation of classical or novel protein kinase C (c/nPKC) (8). Activation of PKD is marked by phosphorylation of two conserved serine residues in the activation loop of PKDs and concomitant autophosphorylation of phospho-PKD1Ser916 or phospho-PKD2Ser876 are widely used as a biomarker of PKD activation (3, 9). PKD regulates a wide range of cellular processes including cell proliferation, migration/invasion, angiogenesis, protein trafficking, and gene expression. Its functional importance has been implicated in major human cancers including carcinoma of breast, skin, pancreas and prostate (8).
Aurora kinases are master regulators of the cell cycle (10–13). Human genome encodes three isoforms of Aurora kinases; namely, Aurora kinases A, B and C (14). Aurora A localizes on centrosomes, spindle poles and spindles during mitosis and regulates centrosome function, spindle assembly and mitotic progression. Expression of Aurora A is cell cycle-regulated, that is, the levels of mRNA and protein are low in G1 and S, increase during G2/M, and reduce during mitotic exit. Aurora A is ubiquitinated by APC/Cdh1 and Fbxw7 ubiquitin ligases and degraded in proteasome-dependent pathways (15, 16). A number of Aurora A substrates have been reported, such as TPX2, CDC25B and p53 (17). Aurora A is frequently overexpressed in cancer. Aberrant expression of Aurora A promotes centrosome amplification and aneuploidy, leading to genome instability and consequently oncogenic transformation (18). Thus, Aurora A represents a well-established therapeutic target for cancer. In this study, we uncovered a novel crosstalk between PKD2 and Aurora A kinase. Our study defined the functional impact of this regulatory axis on G2/M transition and mitosis. We have shown that PKD2 is required for stabilization of Aurora A kinase, and that the abrogation of PKD2 expression or inhibition of its catalytic activity causes proteasome-mediated downregulation of Aurora A kinase displaying delayed G2, mitotic catastrophe and cell death. Moreover, we report that PKD2 localizes on the centrosomes during G2 and that it is critical for centrosome separation in early G2. Taken together, our study provides compelling evidence to support the role of PKD2 in G2/M transition though modulating Aurora A stability, and aberrant PKD2 activity may contribute to oncogenesis.
Materials and Methods
Cell culture, synchronization, and treatments
Authenticated HeLa, LNCap and PC3 cells were obtained from ATCC more than 6 months ago. After purchase, the cell lines were expanded and frozen according to manufacturer’s instruction after two to three passages. LNCaP was used for no longer than 3 passages and PC3/HeLa cells were used for no longer than 10 passages. All cell cultures were routinely tested for mycoplasma using MycoAlert PLUS Mycoplasma Detection Kit (Lonza). HeLa, LNCaP and PC3 cells were grown in 1X Minimum Essential Medium (MEM) supplemented with Eagle’s salt and L-glutamine (Invitrogen), RPMI-1640 (Invitrogen) or Ham’s F-12 (Thermo Fisher) media respectively, supplemented with 10% FBS and 1X Pen/Strep (Fisher, MT30002CI) and maintained at 370C in a humidified incubator containing 5% CO2. Cells were synchronized at G1-S or the start of M phases by double thymidine or nocodazole, respectively. Briefly, for G1-S synchronization, cells were treated with 2 mM thymidine for 16 hr, washed three times with 1X PBS and refed into fresh culture medium without thymidine for 8 hr. The second block was performed by treating the cells with 2 mM thymidine for another 16 hr. Cell arrest at the start of M phase was performed by treating the cells with nocodazole (100 ng/ml) for 16hr. PKD inhibitors (CRT0066101 and kb-NB142–70) were used at a final concentration of 2 μM for indicated times. Turbofect (Thermo Fisher) and Lipofectamine 3000 (Invitrogen) transfection reagents were used to transfect plasmids and siRNAs respectively, according to the manufacturer’s protocol.
Cell survival assay
Cell survival assay was performed by Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s protocol. Briefly, HeLa and PC3 cells were plated at a density of 4000 cells per well in 96 well plate. After treating the cells with different doses of drugs (CRT0066101 and Kb-NB142–70) for 72 hr, CCK-8 solution was added to cells in each well, followed by incubation for 2 h. Cell proliferation/viability was determined by measuring the OD at 450 nm.
Western blot and densitometry analysis
Cells were lysed in IP lysis buffer (50 mM tris-HCl pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 10% glycerol, 1% Triton X-100, 5 mM EGTA, 1 mM Na3VO4, 10 mM NaF, 1 mM β-glycerophosphate and protease inhibitor cocktail). Cell extracts were resolved by SDS PAGE, transferred to nitrocellulose membrane and probed with following antibodies: PKD2 (CST 8188), phospho PKD2Ser876 (Thermo PA5–64538), Cyclin B1 (CST 12231), phospho CDC25Thr48 (CST 12028), phospho Histone H3Ser10 (CST 3377), Aurora Kinase A (CST 14475), Aurora Kinase B (CST 3094), gamma tubulin (Santa Cruz, sc-17788), PARP (CST 9542), ubiquitin (P4D1, CST 3936), Lamin A (CST 2032), Cdh1 (Santa Cruz, sc-56312), Fbxw7 (Abcam, EPR8069) and GAPDH (Enzo ADI-CSA-335-E). Protein half-life was measured as described previously (44). Briefly, the band intensities were quantified by ImageJ software and the values of target proteins were normalized against the band intensity of GAPDH. The measured protein intensity values were log transformed, fitted to a linear regression plot and decay rate constant (k) was determined. The half-life was calculated from k using the formula .
Immunoprecipitation analysis
For immunoprecipitation assay, whole cell extract was prepared by lysing the cells with IP lysis buffer (50 mM tris-HCl pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 10% glycerol, 1% Triton X-100, 5 mM EGTA, 1 mM Na3VO4, 10 mM NaF, 1 mM β-glycerophosphate and protease inhibitor cocktail). The lysate was centrifuged, supernatant was collected and protein concentration was estimated using BCA protein assay kit (Pierce) according to manufacturer’s protocol. Cell lysate containing equal amount of total protein was incubated with primary antibodies at at 40C overnight. Protein A/G agarose beads were added later for another 2 hr. The immunoprecipitates were collected, washed with wash buffer containing 0.5% Triton X-100. Finally, the samples were diluted with 6X SDS PAGE loading buffer, boiled for 10 min and subjected to Western blotting.
Subcellular fractionation
Cells were washed with ice cold PBS; collected by centrifugation at 1000 rpm for 5 min and resuspended in cytoplasm extraction buffer (10 mM HEPES pH 7.5, 60 mM KCl, 1 mM EDTA, 0.1% NP-40, 1 mM DTT and protease inhibitor cocktail) for 5 min on ice. The cell lysate was centrifuges at 1000 rpm for 5 min at 4ᴏ C and cytoplasmic fraction was separated in a new tube. The nuclei were washed in cytoplasm extraction buffer without NP-40 and resuspended in nucleus extraction buffer (20 mM Tris-HCl pH 8, 400 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol and protease inhibitor cocktail). The extracts were incubated on ice for 10 min. Both cytoplasmic and nuclear extracts were centrifuged at 14000 rpm for 15 min at 4ᴏ C and transferred to new tubes.
Flow cytometry
Trypsinized cells were pelleted, washed two times with ice cold FACS buffer (0.1% glucose in PBS) and resuspended in ice cold 70% ethanol while vortexing. Cells were fixed by incubating overnight at 40C, washed with PBS and incubated with 1 ml propidium iodide (PI) staining solution (50 μg/ml PI, 0.1 mg/ml RNase A in FACS buffer) for 30 min. Cells were analyzed in a FACSCalibur (BD Biosciences) flow cytometer and the data were analyzed by ModFit 3.0 software (BD Biosciences). Ten thousand events were analyzed for each sample.
Apoptosis assay
Trypsinized cells were washed twice with ice cold PBS and stained with Annexin V-Alexa Fluor 488 antibody and propidium iodide (Molecular Probes, Thermo Fisher) according to the manufacturer’s instruction. The samples were divided in two parts. One half was subjected to FACSCalibur (BD Biosciences) flow cytometer. The other part was subjected to confocal fluorescence microscopy using Alexa Fluor 488 and PI filters.
Immunofluorescence (IF) staining and microscopy
HeLa and PC3 cells were grown on poly D-lysine coated coverslips, washed with PBS, fixed in 4% paraformaldehyde at room temperature for 30 min, washed three times with 1X PBS, blocked in 5% normal goat serum containing 0.3% Triton X-100 for 1 hr at room temperature. The cells were incubated with the primary antibodies diluted in antibody dilution buffer (1% BSA, 0.3% Triton X-100 in 1X PBS) overnight at 40C, washed three times in 1X PBS, incubated with secondary antibodies diluted in antibody dilution buffer for 1 hr at room temperature. The cells were counterstained with DAPI (1 μg/ml) and the coverslips were mounted in ProLong Gold antifade reagent (Invitrogen). The cells were analyzed using an Olympus Fluoview (FV1000) confocal microscope using 60 X/1.45 objectives.
Statistical analysis
Data analysis was done using the Students’ t-test for comparison between two groups (two-tailed). All statistical analyses were done using GraphPad Prism IV software (GraphPad Software, La Jolla, CA, USA). All values are represented as mean ± standard estimated mean (SEM) of at least three independent experiments. A p value < 0.05 was considered statistically significant (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, ns: not significant).
Results
Elevated PKD2 activity during mitotic entry of the cell cycle
First, we examined the expression and activity of PKD (refers to PKD2 and PKD3 throughout the text) in different cell cycle stages. PKD2 and PKD3 are major isoforms expressed in HeLa cells (Fig. S1), similar to the prostate cancer cell line PC3, but differed from LNCaP cells (19). HeLa and PC3 cells were treated with nocodazole, a microtubule destabilizing agent that activates the spindle assembly checkpoint and causes cell arrest in start of M phase (prometaphase), and the activity of PKD2 was assessed after releasing from nocodazole using the phospho-PKD2Ser876 (p-PKD2) antibody. As shown in Fig. 1A (top panel) and Fig. 1B, p-PKD2 was detected in G2/M-arrested HeLa cells following nocodazole synchronization and persisted till the end of mitosis. This was accompanied by increased cyclin B1, a marker for G2 and M phases of the cell cycle, and phospho-Histone H3Ser10 (pHH3) which was detectable in G2, peaks during mitosis and disappears when cells enters in G1 (Fig. 1A and S2). Similar results were obtained with PC3 cells where activation of PKD2 was marked by increased levels of p-PKD2 after release from nocodazole arrest (Fig. 1A, bottom panel). To determine if the activation is specific to G2/M, PKD2 activation was measured in G1-S synchronized HeLa cells which progressed to M phase. As shown in Fig. 1C–D, when cells were released from a double-thymidine (DT) block that synchronizes cells at G1-S border, PKD2 phosphorylation gradually elevated, starting at 4 h when cells entered in late S phase peaked around 10 h in late G2, and persisted throughout mitosis till 14 h after thymidine release. The highest activity was detected at G2/M border and M phase. This pattern correlated well with increased cyclin B1 and pHH3. As controls, PKD2 remained the same under both conditions.
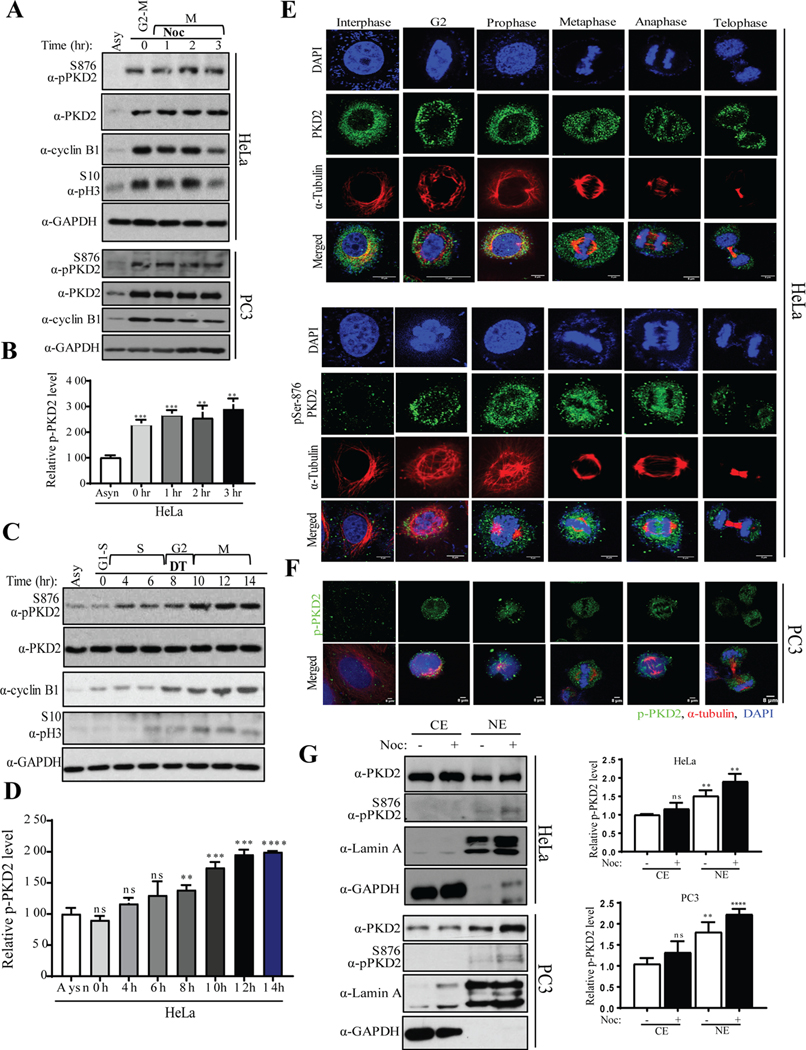
Figure. 1.
PKD is activated during mitotic entry. A) HeLa and PC3 cells were synchronized with nocodazole (100 ng/ml) for 16 hr and mitotic cells were released into fresh medium for indicated times. Cell extracts from each sample were subjected to Western blot analysis to analyze activation of PKD2. B) Densitometric quantification of phospho-PKD2S876 from the immunoblot of HeLa was performed. C) Western blot analysis of indicated proteins after the HeLa cells were released from a double thymidine (DT) block was performed. D) Densitometric quantification of phospho-PKD2S876 from the immunoblot of C was performed. Different phases of cell cycle are marked (G1, S, G2 and M). E) HeLa cells were subjected to immunofluorescence (IF) staining to visualize co-localization PKD2 or phospho-PKD2S876 with α-tubulin during different phases of cell cycle. F) PC3 cells were subjected to IF staining to visualize co-localization of phospho-PKD2S876 with α-tubulin during different phases of cell cycle. DAPI was used to stain the nucleus. Scale bar indicates 8μm. GAPDH was used as loading control in A and C. G) HeLa and PC3 cells were synchronized with nocodazole (100 ng/ml) for 16 hr and mitotic cells were collected by shake-off. Cytoplasmic (Cyto) and nuclear (Nuc) fractions were prepared and the samples were analyzed by Western blotting with indicated antibodies (left panel). Relative amount of phospho-PKD2S876 was quantified (right panel). The graphs show average of three independent experiments with error bars representing standard estimated mean (SEM) (** p < 0.01, *** p < 0.001, **** p < 0.0001, ns: not significant).
The intracellular distribution of PKD2 was then analyzed at different stages of the cell cycle by immunofluorescence (IF) staining. In HeLa cells, PKD2 was primarily localized in the cytoplasm and perinuclear zone in interphase. When cells progress through cell cycle, it exhibited scattered punctate staining that are minimally overlapped with mitotic bodies (spindle fibers and midbody) and excluded from condensed chromatin (Fig. 1E, top panel). PKD2 phosphorylation was low or undetectable during interphase and significantly increased during G2, sustained throughout prophase, metaphase and anaphase, before returning to baseline when the cells entered telophase. To corroborate these results in cancer, PC3 cells were stained in similar fashion. Similar results were found when different stages of the cell cycle in PC3 cells were imaged using antibodies against phospho-PKD2Ser876 (green) and α-tubulin (red) (Fig. 1F). It is well documented that PKD, upon activation, accumulates in the nucleus (20). Indeed, we observed elevated active PKD in the nucleus during G2 and early prophase when the nucleus was still intact, further attesting that PKD2 was activated during G2 and mitosis. We further validated that phospho-PKD2 is increased in the nucleus during early mitosis by cell fractionation (Fig. 1G, left panel). HeLa and PC3 cells were arrested at the start of M phase by nocodazole and cytoplasmic (Cyto) or nuclear (Nuc) fractions were prepared. The samples were analyzed by Western blotting using phospho-PKD2 antibody and the results showed that, indeed, active PKD2 levels were elevated in early mitosis (Fig. 1G, right panel). Lamin A was blotted as a marker for the nuclear fraction. Taken together, these results suggested that PKD is active during G2 and mitosis and may play an important role in cell cycle regulation.
Inactivation or knockdown of PKD2 causes delay in G2/M transition
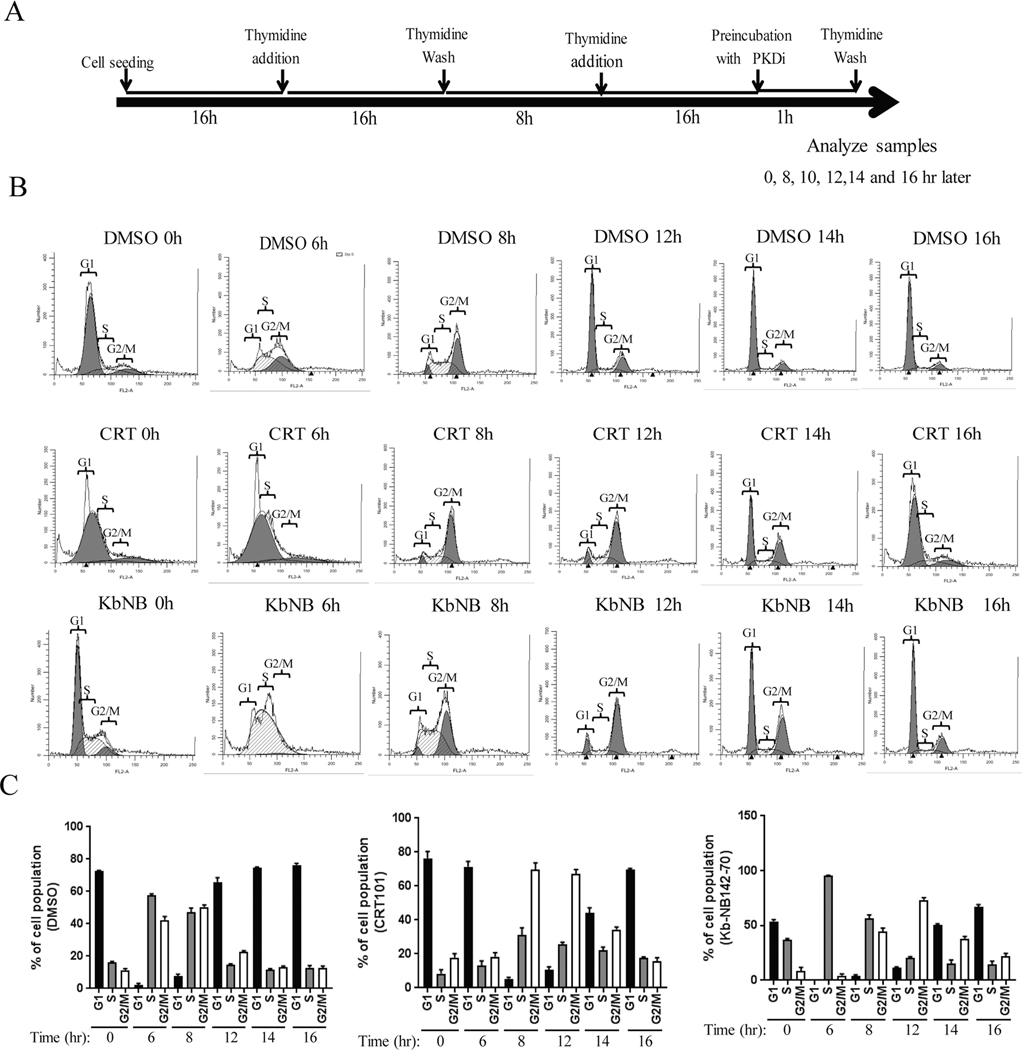
The activation profile of PKD2 suggests that PKD2 may regulate G2 and M phase cell cycle progression. To test this hypothesis, cells with altered PKD2 expression and activity were synchronized by DT at G1-S border and released (Fig. 2A, 3A); cell cycle progression was analyzed by flow cytometry. As shown in Fig. 2B–C, after thymidine release, cells reached G2/M at 8 h, and by 12 h, cells resumed normal distribution. In contrast, cells treated with the PKD inhibitor CRT0066101 (CRT101) reached G2/M at 8 h, stayed after 12 h, and by 14 h there remained a large G2/M population as compared to the control. Inactivation of PKD by another PKD inhibitorkb-NB142–70 (kb-NB)) gave rise to similar results (Fig. 2B, kb-NB). Quantification analysis confirmed prolonged G2/M phase of cell cycle in cells treated with PKC inhibitors (Fig. 2C). Thus, inactivation PKD2 prolonged G2/M phase and resulted in delayed mitotic entry in cells.
Figure. 2.
Inactivation of PKD by CRT101 and kb-NBinduces a delay in G2-M progression. A) Schematic representation of experimental design is shown. B) Flow cytometry analysis of cells treated with DMSO, CRT101 and kb-NB were performed according to A. C) Summarizing graph of the distribution of different phases of cell cycle are shown (right panels). The graphs show average of three independent experiments with error bars representing SEM.
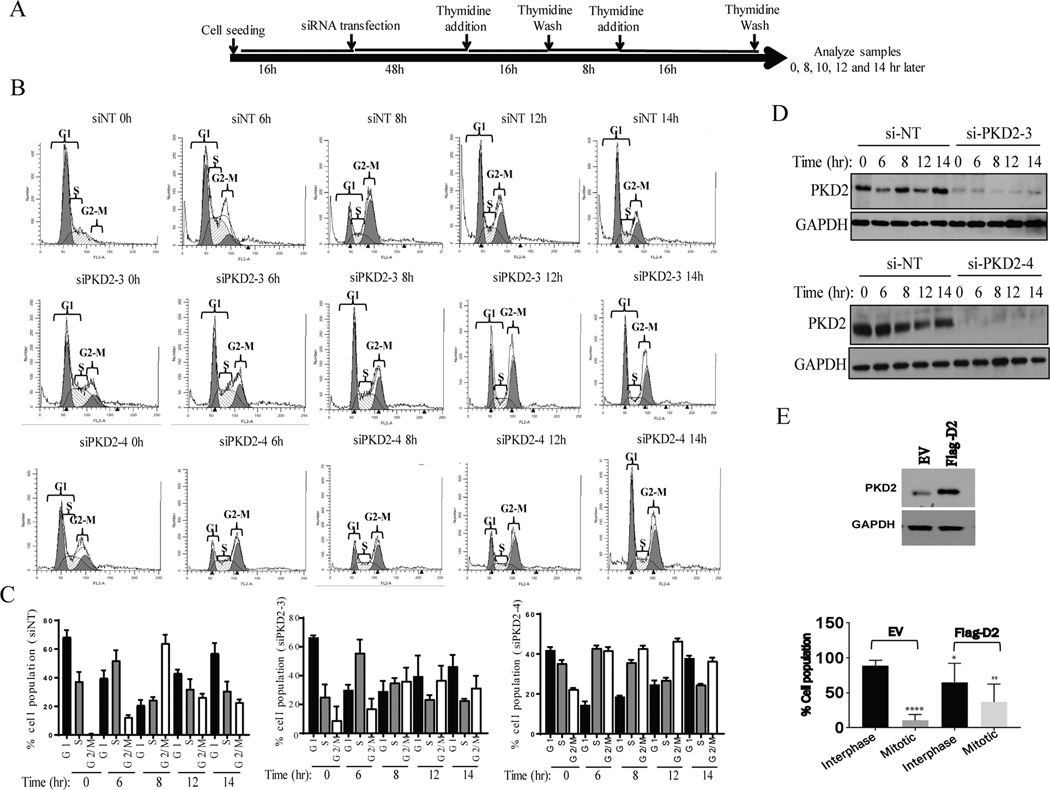
Figure. 3.
Abrogation of PKD2 expression delays G2-M progression. A) Schematic representation of experimental design is shown. B) Flow cytometry analysis of cells transfected with control non-targeting siRNA (siNT) and two siRNAs against PKD2 was performed according to A. C) Summarizing graph of the distribution of different phases of cell cycle are shown. The graphs show average of three independent experiments with error bars representing SEM. D) Western blot analysis of the samples from A was performed to confirm knockdown of PKD2. GAPDH was used as loading control. E) PKD2 was ectopically overexpressed using a plasmid expressing Flag-PKD2 (top panel) and percentage of interphase and mitotic cell population was counted (bottom panel). The graphs show average of three independent experiments with error bars representing SEM (* p<0.5, ** p < 0.01, **** p < 0.0001).
To determine that PKD2 activity was indeed required or G2/M transition, cells transfected with PKD2 siRNAs were synchronized to G1-S border by DT and released; cell cycle progression was monitored by flow cytometry (Fig. 3A). Our data showed that cells transfected with non-targeting siRNA reached G2/M around 8 h and returned to normal distribution at about 12–14 h. In contrast, cells transfected with two PKD2 siRNAs (Fig. 3B) progressed slower through the cell cycle and peaked at G2/M phase around 12 h and remained till after 14 h (Fig. 3C), indicating that PKD2-depleted cells progressed through G2/M boundary at much slower rate than control cells. Knockdown of PKD2 using two siRNAs in these samples were confirmed by Western blotting (Fig. 3D). To corroborate these findings, we examined HeLa cells with overexpressed PKD2. As shown in Fig. 3E, overexpression of PKD2 significantly increased the number of cells entering mitosis. Taken together, our data indicate that PKD2 was activated in G2/M and its activity was required for mitotic entry.
Abrogation of PKD2 expression sensitizes cells to mitotic catastrophe and cell death
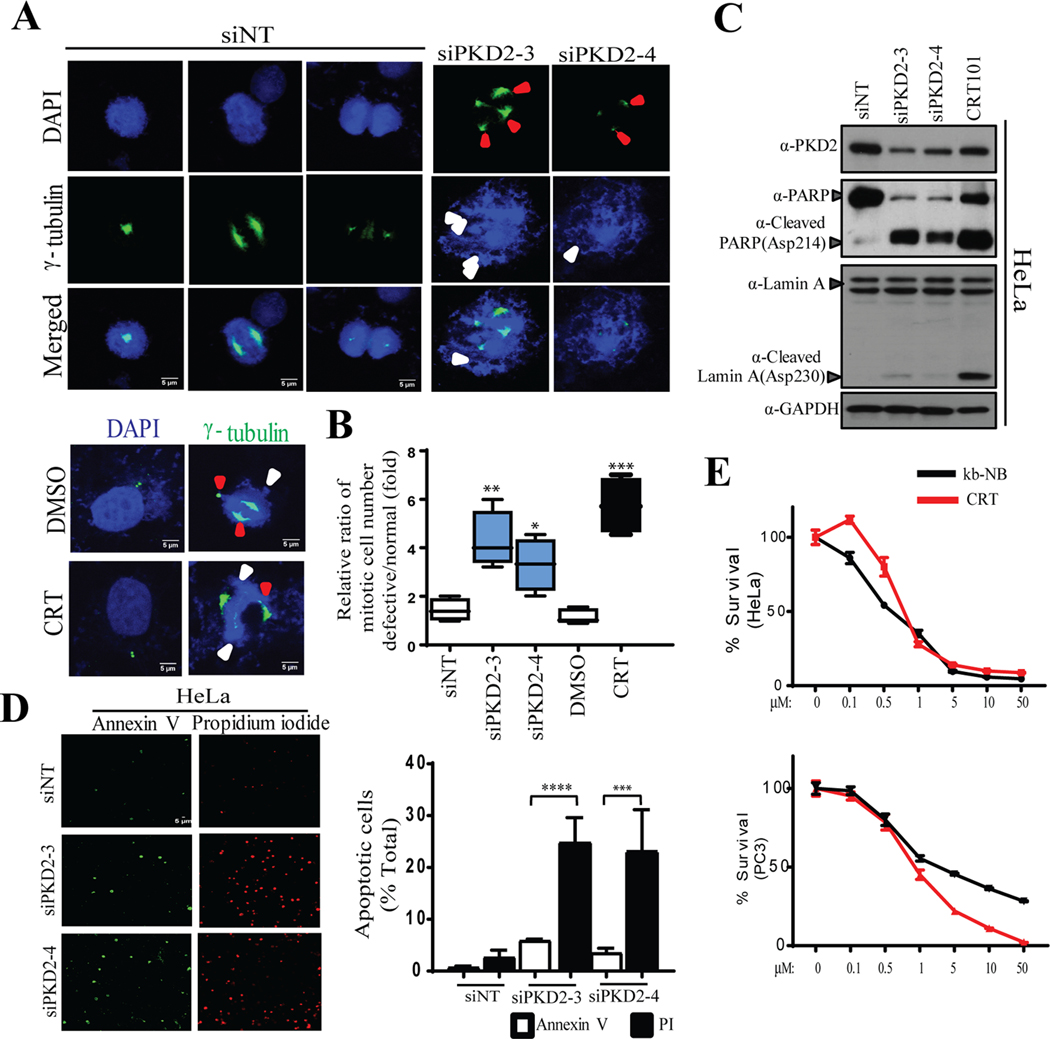
Cells experience prolonged G2 arrest and delayed mitotic entry may eventually enter into mitosis, however, they often suffer from aberrant mitosis known as mitotic catastrophe, and die subsequently by apoptosis (21). Here, we examined if mitotic catastrophe was associated with loss/inactivation of PKD2 and delayed G2/M transition. Knockdown or inactivation of PKD2 using two siRNAs or PKD inhibitor CRT101, respectively, induced a series of mitotic defects including chromosomal miss-alignment at metaphase plane, abnormal chromosomal arrangement (Fig. 4A, white arrows) and formation of multiple centrosomes (Fig. 4A, red arrows). Quantification analysis indicated that depletion or inactivation of PKD2 significantly increased cell population with defective mitosis (Fig. 4B). It is well known that mitotic catastrophe leads to apoptosis followed by PARP and Lamin A cleavage (22, 23). Analysis of PKD2-depleted or PKD-inactivated HeLa cells showed expression of cleaved and PARPAsp214 Lamin AAsp230, indicative of apoptotic response (Fig. 4C). To further this analysis, cells transfected with siNT and siPKD2s were stained with Annexin V-Alexa Fluor 488/propidium iodide (PI) and analyzed by flow cytometry (Fig. 4D, left panel). Knockdown of PKD2 caused significant cell death (PI-positive) with elevated apoptotic population (Annexin V position) as compared to the control samples (siNT) (Fig. 4D, right panel). Treatment of both HeLa and PC3 cells using increasing concentrations of CRT101 and kb-NB reduced cell viability in a concentration-dependent manner (Fig. 4E). Taken together, our data indicated that G2/M deficiencies caused by PKD2 depletion or inhibition could result in mitotic catastrophe and cell death followed by apoptosis.
Figure. 4.
Knockdown of PKD2 causes mitotic catastrophe followed by cell death. A) IF staining of cells transfected with control non-targeting siRNA (siNT) and two siRNAs against PKD2 (upper panel) or DMSO/CRT101 (bottom panel) was performed to visualize γ-tubulin (green). DAPI was used to stain the nucleus. Red and white arrow heads indicate defects in spindle pole formation and misaligned chromosomes, respectively. B) Quantification of cell population harboring mitotic defects after the cells were treated with either si-PKD2 (top panel) or CRT101 (bottom panel) was performed. C) Western blot was done to show cleavage of PARP/Lamin A upon depletion or inactivation of PKD2 in HeLa. GAPDH served as loading control. D) Microscopic analysis of cells transfected with siNT or two siPKD2s after co-staining with Annexin V-Alexa Fluor 488 and propidium iodide (PI) was performed (left panel). Quantification of cell population positive for Annexin V-Alexa Fluor 488 and PI (right panel) was performed. E) Hela and PC3 cells were treated with different concentrations of PKD inhibitors (CRT101, kb-NB) and cultured for 72 hr. MTT assay was performed to assess cell viability. The graphs show average of three independent experiments with error bars representing SEM (*** p < 0.001, **** p < 0.0001).
Inactivation of PKD2 causes degradation of Aurora A, an effect that can be suppressed by overexpression of Aurora A
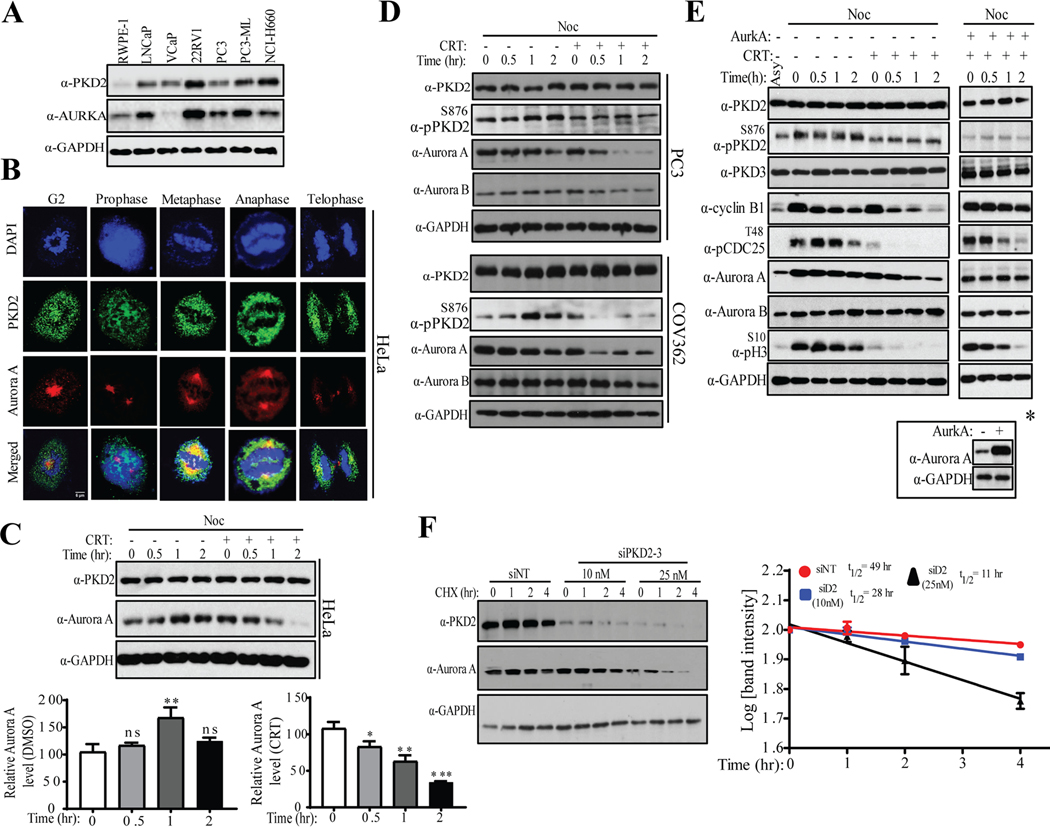
We showed that knockdown or inactivation of PKD2 caused prolonged G2/M transition, delay in mitotic entry, and mitotic catastrophe (Figs. 2–4). In the order to gain insights into the role of PKD in cell cycle, we sought to identify its downstream targets. Through a whole genome RNA-seq analysis conducted on cancer cells treated with CRT101 and kb-NB, we identified AURKA/Aurora A kinase as one of the top candidate genes altered by inhibition of PKD. Aurora A is a master regulator of G2/M and mitosis (12, 24–26). First, using normal prostate epithelial cell line RWPE-1 and prostate adenocarcinoma (PAC) cell lines, we sought to identify if expression level of PKD2 correlates with that of Aurora A kinase. As shown in Fig. 5A, we observed positive correlation between PKD2 and Aurora A in these cell lines. Next, we subjected HeLa cells to IF staining and checked if PKD2 and Aurora A co-localize at different stages of mitosis. Our confocal microscopy results showed that a significant portion of PKD2 (green) co-localizes with Aurora A (red) during mitosis (Fig. 5B), raising a strong possibility that PKD2 might contribute to cell cycle by regulating Aurora A. We examined the expression levels of Aurora A upon inactivation or depletion of PKD. HeLa cells were arrested at the start of M phase by nocodazole, released from the block by mitotic shake off and turnover of Aurora A protein levels was analyzed in the absence or presence of CRT101 by Western blotting. The results showed that CRT101 caused rapid downregulation of Aurora A with < 50% remaining 2 h after CRT101 treatment (Fig. 5C). To corroborate this finding in context of cancer, we treated PC3 prostate adenocarcinoma and COV362 ovarian epithelial-endometroid carcinoma cell lines as described above and checked Aurora A turnover by Western blotting (Fig. 5D). As expected, Aurora A was rapidly downregulated by CRT101 in these two cell lines.
Figure. 5.
Inactivation or knockdown of PKD2 causes downregulation of Aurora A kinase. A) Western blot analysis was performed to determine expression patterns of PKD2 and Aurora A in designated cell lines. B) HeLa cells were stained with antibodies against PKD2 (green) and Aurora A (red). IF staining was performed to visualize co-localization of PKD2 and Aurora A at different stages of cell cycle. Scale bar indicates 8μm. DAPI was used to stain the nucleus. C) HeLa cells were synchronized at the start of M phase by nocodazole, released from the block and mitotic cells were cultured for indicated times with or without CRT101. Downregulation of Aurora A was analyzed by Western blotting (top panel). Quantification of Aurora A protein levels by densitometry scanning from immunoblots of was performed (bottom panel). D) PC3 and COV362 cells were treated as described in C and turnover of Aurora A protein levels were analyzed by Western blotting. The expression levels of Aurora B were analyzed as control. E) HeLa Cells were arrested at the start of M phase by nocodazole (noc), released in fresh medium containing DMSO or CRT101 for indicated times and cell extracts from each samples were analyzed by Western blotting using indicated antibodies (left panel). Aurora A was ectopically overexpressed and the cells were released from nocodazole block in fresh medium containing CRT101 for indicated times. Cell extracts from each sample were analyzed by Western blotting using indicated antibodies. * Overexpression of Aurora A was confirmed by Western blotting. F) Cells were transfected by si-NT (control) and two different concentrations of si-PKD2. 48 hr post-transfection, cells were treated with cycloheximide for indicated times and cell extracts from each sample were analyzed by Western blotting using indicated antibodies (left panel). GAPDH served as loading control. Half-life of Aurora A protein was measured from the samples of F (right panel). The graphs show average of three independent experiments with error bars representing SEM (* p<0.5, ** p < 0.01, *** p < 0.001, ns: not significant).
Meanwhile, the downregulation of Aurora A coupled with time-dependent decline of cyclin B1, p-CDC25T48 and p-HH3, which are in line with a critical role of PKD activity in G2/M transition and mitotic progression (Fig. 5E, left panel). In comparison, the expression levels of Aurora B did not significantly change (Figs 5D–E, Aurora B), indicating a selective effect of CRT101 on Aurora A degradation. Importantly, when ectopically overexpressed, Aurora A completely reversed the effect of CRT101 on the cell cycle markers (Fig. 5E, right panel; Fig. S3). Thus, Aurora A mediated the effect of PKD inhibition on G2/M transition. To corroborate these findings, Aurora A protein stability was assessed in cells transfected with siPKD2–3 (siD2) and treated with cycloheximide (CHX) that blocks protein synthesis. In the control cells (siNT), the half-life (T1/2) of Aurora A was 49 h. Partial knockdown of PKD2 reduced T1/2 of Aurora A about 28 hr, while nearly complete knockdown further reduced it to 11 hr (Fig. 5F). Similarly, knockdown of PKD3 with two siRNAs also resulted in downregulation of Aurora A (Fig. S5). These results identify Aurora A as a key mediator of PKD2-regulated cell cycle progression, and the activation of PKD2 at G2/M stabilizes Aurora A kinase and promotes mitotic entry.
PKD2 and Aurora A localize at the centrosome and knockdown of PKD2 inhibits centrosome separation during G2
Aurora A has been shown to localize at mitotic apparatus and contribute to G2/M transition and mitotic progression (13, 22, 27, 28). Aurora A resides in the centrosome and regulates centrosome duplication and separation, inhibition of Aurora A arrests cells in G2 and blocks mitotic entry. Here, we sought to determine if the regulation of Aurora A by PKD also took place at the centrosome and whether it affects centrosome function.
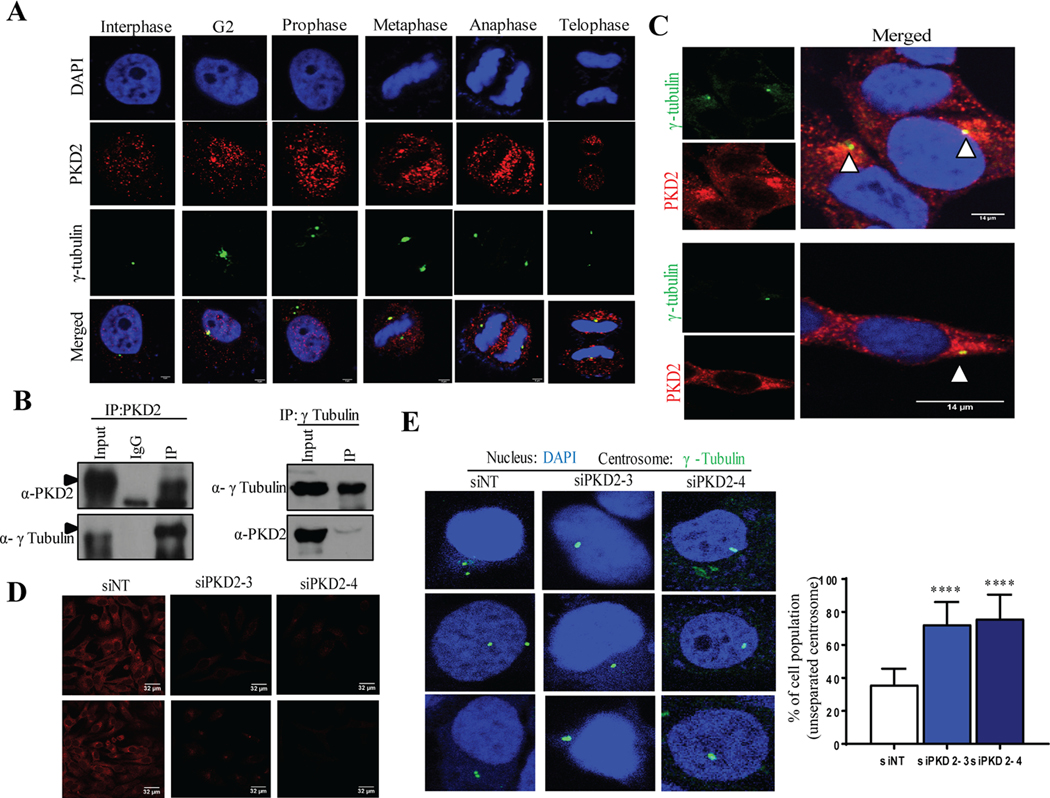
HeLa cells were co-stained with PKD2 and γ-tubulin, a marker of the centrosome. As shown in Fig. 6A, PKD2 (red) was found to co-localize with γ-tubulin (green) at different stages of cell cycle, and the co-localization was most prominent in the G2 stage. Next, cells were synchronized to the stat of M phase by nocodazole and the interaction of PKD2 and γ-tubulin were examined by reciprocal immunoprecipitation. As shown in Fig. 6B, γ-tubulin was detected in immunoprecipitated PKD2 complex, and vice versa. Thus, a portion of PKD2 was present on the centrosome and PKD2 directly interacts with γ-tubulin at the centrosome during G2/M, which was further confirmed by IF staining (Fig. 6C). Next, we examined whether PKD2 might directly interact with Aurora A. Aurora A is known to localize at the centrosome (13, 26, 29, 30). Our data confirmed that Aurora A (red) co-localized with γ-tubulin (green) during G2 and mitosis (Fig. S6). By co-staining for PKD2 and Aurora A, we also found significant co-localization of PKD2 (green) and Aurora A (red) during G2 and mitosis (Fig. 5A). Co-localization of PKD2 and Aurora A on the centrosome raised a possibility that they might physically interact. However our follow-up study indicated that PKD2 failed to co-immunoprecipitate with Aurora A in G2/M-synchronized cells (Fig. S7). Despite the lack of direct interaction, the co-localization of PKD2 and Aurora A support the potential functional link between the two proteins in centrosome. Similar co-localization pattern was observed when the intracellular distributions of PKD3 and Aurora A were analyzed during G2 and mitosis (Fig. S7). Aurora A is crucial for centrosome maturation and separation during G2 (30). Hence, we questioned whether loss of Aurora A by depletion of PKD2 caused any defect in centrosome separation. To do so, cells were transfected with two siPKD2s followed by IF staining of γ-tubulin to visualize the centrioles. Knockdown of PKD2 was confirmed by IF staining (red, Fig. 6D). Our results showed that depletion of PKD2 resulted in defects in centriole separation (Fig. 6E, left panel) and significantly increased cell population harboring unseparated centrosome (Fig. 6E, right panel). Taken together, these results suggest that PKD positively regulates stabilization of Aurora A and centrosome separation to promote G2/M progression.
Figure. 6.
PKD2 positively regulates centriole separation during late G2 by co-localizing with centrosome. A-C) HeLa cell was subjected to IF staining to visualize co-localization of PKD2 (red) and γ-tubulin (green). B) Co-immunoprecipitation assay was carried out to assess physical interaction between PKD2 and γ-tubulin. D) Knockdown of PKD2 by two siRNAs was confirmed by IF staining. E) Centrioles were visualized by IF staining of γ-tubulin in cells transfected with siNT or two siPKD2s (left panel). Cell population harboring unseparated centrioles was quantified (right panel). At least 100 cells were counted and graph shows average number of cells with error bars representing SEM (**** p < 0.0001). DAPI was used to stain the nucleus.
PKD2 interferes with Fbxw7-mediated ubiquitination and downregulation of Aurora A
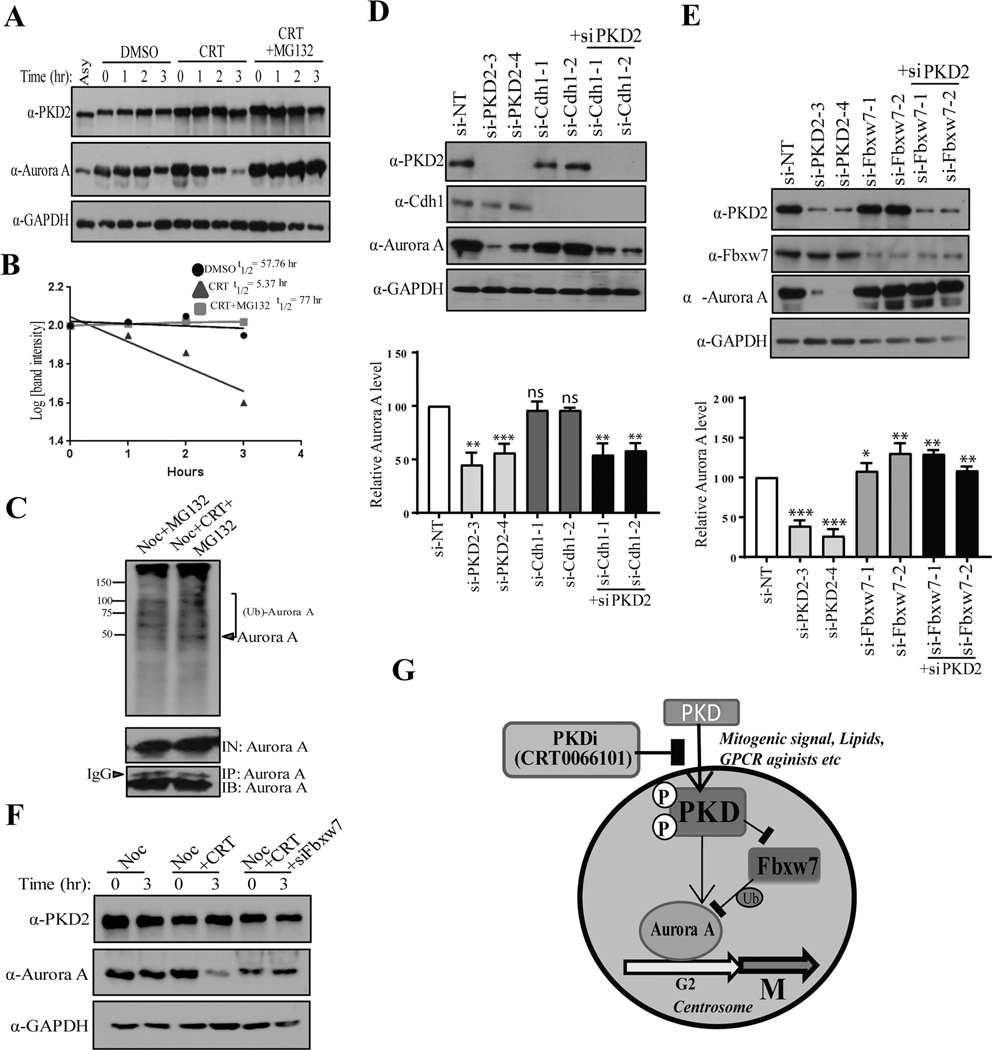
To assess if PKD regulates Aurora A degradation through the proteasome degradation pathway, cells were synchronized with nocodazole, released in fresh medium containing CRT101 in the presence or absence of a proteasome inhibitor MG132, and the levels of Aurora A were examined by immunoblotting. Our data showed that Aurora A levels were dampened within 3 hr of CRT101 treatment and MG132 completely inhibited CRT101-induced Aurora A degradation (Fig. 7A). The half-life of Aurora A was measured in the presence of CHX. As shown in Fig. 7B, treatment of CRT101 reduced the half-life of Aurora A protein to about 5 hr, whereas, MG132 restored its half-life like control sample (nocodazole only) (Fig. 7B). These results demonstrated that CRT101 directed Aurora A to the proteasomal pathway for its degradation. To further corroborate this finding, G2/M arrested cells were released in medium containing MG132 with or without CRT101, and cell extracts from the samples were subjected to IP using anti-Aurora A antibody, followed by immunoblotting with anti-ubiquitin antibody (Fig. 7C). We found that CRT101 increased ubiquitination of Aurora A (Fig. 7C, top panel) confirming that Aurora A is downregulated via ubiquitination-dependent proteasomal degradation pathway upon treatment with CRT101. Aurora A is targeted by Cdh1 and F-box protein Fbxw7 (15, 31) for degradation. Therefore, to gain insight into PKD-mediated downregulation of Aurora A, we knocked down PKD2, Cdh1 and Fbxw7 using site-specific siRNAs and analyzed the turnover of Aurora A protein levels, as shown in Fig, 7D. Knockdown of PKD2 downregulated Aurora A, whereas, depletion of Cdh1 did not significantly alter Aurora A protein level (Fig. 7D). Interestingly, when Fbxw7 was depleted in PKD2-knocked down cells, Aurora A level was significantly restored as compared to siPKD2 (Fig. 7E). To further corroborate our findings, cells were transfected with non-targeting siRNA (siNT) or siFbxw7, synchronized at the start of M phase by nocodazole, treated with CRT101 and cell extracts were analyzed by Western blotting to monitor Aurora A degradation (Fig. 7F). The immunoblot showed that Aurora A was downregulated within 3 hr of CRT101 treatment and this turnover was inhibited by Fbxw7 knockdown. These results suggested that PKD protects Aurora A from Fbxw7-mediated ubiquitination and proteasomal degradation in order to regulate G2/M transition.
Figure. 7.
PKD interferes with Fbxw7-mediated ubiquitination of Aurora kinase A. A) Western blot analysis was performed to show that CRT101-mediated downregulation of Aurora A was proteasome-dependent. B) Half-life of Aurora A protein was measured from the immunoblot of A. C) HeLa cells were synchronized at the start of M phase by nocodazole and preincubated with MG132 for 30 mins. DMSO (control) or CRT101 was added to the culture and the cells were further incubated for 3 hr. Aurora A was pulled down by IP and the protein was analyzed by Western blotting to assess its ubiquitination by CRT101. Downregulation of Aurora A by knockdown of PKD2 and Cdh1 (D) or PKD2 and Fbxw7 (E) was analyzed by Western blotting (top panel). Quantification of Aurora A protein levels was performed by densitometry analysis (bottom panel). F) Western blot analysis depicting that CRT101-mediated downregulation of Aurora A is inhibited by knockdown of Fbxw7. GAPDH was used as loading control. G) A model of regulation of Aurora A protein stability by PKD. The graph represents average of at least three independent experiments with error bars representing SEM (* p<0.5, ** p < 0.01, *** p < 0.001, ns: not significant).
Discussion
PKD as a key target of the second messenger DAG regulates many important cellular functions including cell proliferation. Aberrant expression and activation of PKD have been demonstrated in many cancers and are implicated in oncogenic transformation and tumor progression. PKD2 in particular has been shown to promote tumor cell proliferation in human cancer including carcinoma of the prostate (8). However, the mechanisms through which it regulates tumor cell proliferation have not been well defined. In this study, we identified PKD2 as a cell cycle regulator that promotes G2/M transition and mitotic entry. PKD2 is activated during G2 and M phases of the cell cycle. It is localized at the centrosome and upon activation regulates centrosome duplication and separation, a key event for the cells to exit G2 and enter mitosis. Mechanistically, we further demonstrated that PKD activity was required for Aurora A stability during G2/M, and it acts through Fbxw7 to protect Aurora A from ubiquitination and proteasomal degradation. Our study has identified PKD2 as a potential G2/M checkpoint kinase. Its aberrant activation of PKD2 may contribute to premature mitotic entry and genome instability that drive tumorigenesis.
A key finding from our study is that PKD is active during G2 and its activity is sustained throughout mitosis (Fig. 1F). Increased PKD2 activity in G2/M was supported by: 1) immunoblotting results showing elevated active form of PKD2 in cells entering G2 and progressing through mitosis (Fig. 1A–D), 2) IF staining results showing increased active PKD2 staining in cell nuclei during G2 (Fig. 1) and prophase of mitosis, indicative of PKD activation (20). Our finding that activation of PKD is crucial for G2/M cell cycle progression agrees with the report by Papazyan et al. that demonstrated a possible connection of PKD activation during mitosis and its role in cell cycle (32). The effects of PKD inhibitors are in line with previous reports of anti-cancer effects of PKDis. CRT0066101 (CRT101) in particular has been shown to block cell proliferation by inducing G2/M arrest, downregulation of mitotic regulatory proteins and induction of apoptosis in colorectal (33), bladder (34), pancreatic (35) and prostate (36) cancer cell lines. Our lab has also shown that kb-NB142–70, another novel PKD inhibitor causes dramatic G2/M cell cycle arrest in PC3 prostate cancer cells (37). Both of these two PKD inhibitors were also shown to inhibit HeLa cell proliferation (Fig. S3). Collectively, our study has identified PKD as a crucial cell cycle regulator that is activated during G2/M and promotes mitotic entry. Moreover, in this study, we have provided possible mechanistic insights to PKD inhibitor-mediated cell cycle arrest, downregulation of cell cycle regulators and inhibition of cell proliferation, which reflected a positive role of PKD in promoting G2/M transition by modulating Aurora A protein stability.
Aurora A modulates the function of several cell cycle regulators critical for G2/M transition and mitosis, such as PLK1, CDK1/cyclin B and CDC25C (18) and is essential for key mitotic events, such as centrosome maturation and separation (30), mitotic entry and formation of mitotic spindles (26). Aurora A must be degraded to ensure proper mitotic exit. During late anaphase, it loses its interaction with TPX2 and targeted by anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase for proteasomal degradation (16). Aurora A is also downregulated by Fbxw7, a p53-dependent tumor suppressor (15). Mechanistically, Fbxw7 physically interacts with Aurora A and facilitates the ubiquitination-mediated proteasomal degradation in a Gsk3β-dependent manner. Aberrant expression and proteostasis of Aurora A have been linked to many cancers, thus enabling it as a promising target for therapeutic interventions in cancer (18). Our study has revealed for the first time, a functional link between PKD and Aurora A kinase in G2/M progression and mitotic entry of the cell cycle. We have shown evidence that PKD is critical for late G2 cells to enter mitosis, and that abrogation of its activity delays G2/M transition (Figs 2–3). We have shown that loss of PKD has profound effects on cell cycle, such as, 1) rapid downregulation of mitotic regulators including cyclin B1, phospho-CDC25T48 (Fig. S5), 2) induction of mitotic catastrophe characterized by defects in spindle formation, apoptosis and cell death, 3) defects in centriole separation in late G2 and most importantly, 4) degradation of Aurora A by Fbxw7-mediated proteasomal degradation. Cdh1 is a component of Anaphase-Promoting Complex that is responsible for substrate degradation during anaphase of mitosis (38). Whereas, Fbxw7 (α, β and γ) expression spans all cell cycle stages (39), which may be a potential cause for siFbxw7, not siCdh1 to be effective against Aurora A turnover in an asynchronous culture where mitotic cell population is significantly low. Therefore, it is conceivable that a cell harboring inactive or no PKD2 eventually lose Aurora A at the protein level and displays somewhat an “Aurora A null” phenotype (40).
Another significant finding of our study is that PKD2 resides on the centrosome during G2/M and controls centriole separation. We showed that Aurora A and PKD2 co-localized in centrosome in early G2 and throughout G2/M. It is known that the maturation and separation of centrosomes are dependent on the recruitment and activation of Aurora A kinase on centrosomes(41). It is unclear if PKD2 plays a direct role in recruiting Aurora A to the centrosomes. We did not detect direct interaction between PKD2 and Aurora A although both proteins colocalize in the centrosomes and bind to γ-tubulin. Several possibilities may explain this result: (1) the interaction was limited to the centrosomes, which is difficult to detect using whole cell lysates, (2) the interaction may be low affinity and transient like many protein kinases and substrates; (3) the regulation may be indirect through other regulators. Although Fbxw7 was found to mediate the regulation of Aurora A by PKD, it remains to be determined if PKD directly regulates Fbxw7 activity. Functionally, it is evident that PKD activity modulates the availability/stability of Aurora A at the centrosomes. Thus, inhibition of PKD results in degradation of Aurora A, which may impede the maturation and separation of centrosomes, and thereby block the entry of cells into mitosis. Kienzle et al. have shown that PKD positively controls Golgi complex fragmentation at G2 phase of the cell cycle via Raf-MEK1 pathway, thus, enabling cells to enter mitosis (42). This study is in line with our findings since centrosome duplication and separation is an early step that required for Golgi fragmentation in G2 (43). Thus, the alteration of Aurora A stability and the resulting defects in centrosome separation may be linked to Golgi fragmentation that occurs at G2 phase. Beyond Aurora A, the localization of PKD2 in centrosome, the major microtubule-organizing center of the cell, raises the possibility that it might modulate other centrosomal proteins to ensure tight regulation of cell cycle.
In summary, our results provide strong evidence for a novel function of PKD2 in regulating G2/M transition, mitotic entry and cell cycle progression. PKD2 does so by protecting critical cell cycle regulators including Aurora A kinase, an essential mitotic regulator from Fbxw7-mediated proteasomal degradation (Fig. 7G). The regulation of Aurora A by PKD2 provides mechanistic insights to the oncogenic effects of these kinases and may have important therapeutic implications in cancer.
Supplementary Material
Implications: PKD2 is a novel cell cycle regulator that promotes G2/M transition by modulating Aurora A kinase stability in cancer cells and suggests the PKD2/Aurora A kinase regulatory axis as new therapeutic targets for cancer treatment.
Acknowledgments
Financial support:
This work was supported in part by the National Institute of Health grant Department of Defense award PC150190 (Wang).
Footnotes
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declaration of interests:
The authors declare no competing interests.
References
- 1.Otto T, Sicinski P, Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer 17, 93–115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellwanger K, Hausser A, Physiological functions of protein kinase D in vivo. IUBMB Life 65, 98–107 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Rozengurt E, Rey O, Waldron RT, Protein kinase D signaling. J Biol Chem 280, 13205–13208 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Rykx A. et al. , Protein kinase D: a family affair. FEBS Letters 546, 81–86 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Guo J, Gertsberg Z, Ozgen N, Sabri A, Steinberg SF, Protein kinase D isoforms are activated in an agonist-specific manner in cardiomyocytes. J Biol Chem 286, 6500–6509 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durand N, Borges S, Storz P, Protein Kinase D Enzymes as Regulators of EMT and Cancer Cell Invasion. J Clin Med 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong C, Jin ZG, Protein kinase C-dependent protein kinase D activation modulates ERK signal pathway and endothelial cell proliferation by vascular endothelial growth factor. J Biol Chem 280, 33262–33269 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy A, Ye J, Deng F, Wang QJ, Protein kinase D signaling in cancer: A friend or foe? Biochim Biophys Acta 1868, 283–294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang QJ, PKD at the crossroads of DAG and PKC signaling. Trends Pharmacol Sci 27, 317–323 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Bischoff JR, Plowman GD, The Aurora/Ipl1p kinase family: regulators of chromosome segregation and cytokinesis. Trends Cell Biol 9, 454–459 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Chan F. et al. , Mechanism of action of the Aurora kinase inhibitor CCT129202 and in vivo quantification of biological activity. Mol Cancer Ther 6, 3147–3157 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Vader G, Lens SM, The Aurora kinase family in cell division and cancer. Biochim Biophys Acta 1786, 60–72 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Barr AR, Gergely F, Aurora-A: the maker and breaker of spindle poles. J Cell Sci 120, 2987–2996 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Nigg EA, Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol 2, 21–32 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Kwon YW et al. , Pten regulates Aurora-A and cooperates with Fbxw7 in modulating radiation-induced tumor development. Mol Cancer Res 10, 834–844 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindon C, Grant R, Min M, Ubiquitin-Mediated Degradation of Aurora Kinases. Front Oncol 5, 307 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kollareddy M, Dzubak P, Zheleva D, Hajduch M, Aurora kinases: structure, functions and their association with cancer. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 152, 27–33 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Yan M. et al. , Aurora-A Kinase: A Potent Oncogene and Target for Cancer Therapy. Med Res Rev 36, 1036–1079 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Deng F, Singh SV, Wang QJ, Protein kinase D3 (PKD3) contributes to prostate cancer cell growth and survival through a PKCepsilon/PKD3 pathway downstream of Akt and ERK 1/2. Cancer Res 68, 3844–3853 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Rey O, Sinnett-Smith J, Zhukova E, Rozengurt E, Regulated nucleocytoplasmic transport of protein kinase D in response to G protein-coupled receptor activation. J Biol Chem 276, 49228–49235 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Bucher N, Britten CD, G2 checkpoint abrogation and checkpoint kinase-1 targeting in the treatment of cancer. Br J Cancer 98, 523–528 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura M. et al. , Mitotic catastrophe and cell death induced by depletion of centrosomal proteins. Cell Death Dis 4, e603 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castedo M. et al. , Cell death by mitotic catastrophe: a molecular definition. Oncogene 23, 2825–2837 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Hirota T. et al. , Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell 114, 585–598 (2003). [DOI] [PubMed] [Google Scholar]
- 25.He L. et al. , Identification of Aurora-A as a direct target of E2F3 during G2/M cell cycle progression. J Biol Chem 283, 31012–31020 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Fu J, Bian M, Jiang Q, Zhang C, Roles of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res 5, 1–10 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Dutertre S. et al. , Phosphorylation of CDC25B by Aurora-A at the centrosome contributes to the G2-M transition. J Cell Sci 117, 2523–2531 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Jang CY, Coppinger JA, Seki A, Yates JR 3rd, Fang G, Plk1 and Aurora A regulate the depolymerase activity and the cellular localization of Kif2a. J Cell Sci 122, 1334–1341 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukasiewicz KB, Lingle WL, Aurora A, centrosome structure, and the centrosome cycle. Environ Mol Mutagen 50, 602–619 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Dutertre S, Descamps S, Prigent C, On the role of aurora-A in centrosome function. Oncogene 21, 6175–6183 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Taguchi S. et al. , Degradation of human Aurora-A protein kinase is mediated by hCdh1. FEBS Lett 519, 59–65 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Papazyan R. et al. , Protein kinase D isozymes activation and localization during mitosis. Exp Cell Res 314, 3057–3068 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei N, Chu E, Wipf P, Schmitz JC, Protein kinase d as a potential chemotherapeutic target for colorectal cancer. Mol Cancer Ther 13, 1130–1141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li QQ et al. , Protein kinase D inhibitor CRT0066101 suppresses bladder cancer growth in vitro and xenografts via blockade of the cell cycle at G2/M. Cell Mol Life Sci, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harikumar KB et al. , A novel small-molecule inhibitor of protein kinase D blocks pancreatic cancer growth in vitro and in vivo. Mol Cancer Ther 9, 1136–1146 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tandon M. et al. , New pyrazolopyrimidine inhibitors of protein kinase d as potent anticancer agents for prostate cancer cells. PLoS One 8, e75601 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lavalle CR et al. , Novel protein kinase D inhibitors cause potent arrest in prostate cancer cell growth and motility. BMC Chem Biol 10, 5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pesin JA, Orr-Weaver TL, Regulation of APC/C activators in mitosis and meiosis. Annu Rev Cell Dev Biol 24, 475–499 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sionov RV, Netzer E, Shaulian E, Differential regulation of FBXW7 isoforms by various stress stimuli. Cell Cycle 12, 3547–3554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasai K. et al. , Targeted disruption of Aurora A causes abnormal mitotic spindle assembly, chromosome misalignment and embryonic lethality. Oncogene 27, 4122–4127 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Cervigni RI, Barretta ML, Persico A, Corda D, Colanzi A, The role of Aurora-A kinase in the Golgi-dependent control of mitotic entry. Bioarchitecture 1, 61–65 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kienzle C. et al. , PKD controls mitotic Golgi complex fragmentation through a Raf-MEK1 pathway. Mol Biol Cell 24, 222–233 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corda D, Barretta ML, Cervigni RI, Colanzi A, Golgi complex fragmentation in G2/M transition: An organelle-based cell-cycle checkpoint. IUBMB Life 64, 661–670 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Lukasiewicz KB, Lingle WL, Aurora A, centrosome structure, and the centrosome cycle. Environ Mol Mutagen 50, 602–619 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.