Genetic, epigenetic, and cellular adaptations contribute to posttherapy relapse in patients with normal karyotype acute myeloid leukemia.
Abstract
To better understand clonal and transcriptional adaptations after relapse in patients with acute myeloid leukemia (AML), we collected presentation and relapse samples from six normal karyotype AML cases. We performed enhanced whole-genome sequencing to characterize clonal evolution, and deep-coverage single-cell RNA sequencing on the same samples, which yielded 142,642 high-quality cells for analysis. Identifying expressed mutations in individual cells enabled us to discriminate between normal and AML cells, to identify coordinated changes in the genome and transcriptome, and to identify subclone-specific cell states. We quantified the coevolution of genetic and transcriptional heterogeneity during AML progression, and found that transcriptional changes were significantly correlated with genetic changes. However, transcriptional adaptation sometimes occurred independently, suggesting that clonal evolution does not represent all relevant biological changes. In three cases, we identified cells at diagnosis that likely seeded the relapse. Finally, these data revealed a conserved relapse-enriched leukemic cell state bearing markers of stemness, quiescence, and adhesion.
Significance:
These data enabled us to identify a relapse-enriched leukemic cell state with distinct transcriptional properties. Detailed case-by-case analyses elucidated the complex ways in which the AML genome, transcriptome, and immune microenvironment interact to evade chemotherapy. These analyses provide a blueprint for evaluating these factors in larger cohorts.
This article is highlighted in the In This Issue feature, p. 1
Introduction
Intratumoral genetic heterogeneity is a general characteristic of cancer that has been recognized for over four decades (1, 2). Genetic heterogeneity arises through repeated cycles of mutation and selection (or drift) that yield mutationally distinct subclones of cells within the same tumor. In acute myeloid leukemia (AML), early studies used karyotyping to characterize genetic heterogeneity and its temporal dynamics (2), while next-generation sequencing technologies, including single-cell DNA sequencing, have enabled detailed genetic analyses of subclonal architecture and its evolution (3–7). One theme arising from these studies is that relapsed AMLs often derive from rare, mutationally distinct subclones present at diagnosis. Furthermore, functional studies have revealed that individual subclones can have distinct phenotypes (8, 9), and it is now well appreciated that intratumoral genetic heterogeneity provides the raw material for natural selection and is associated with the development of drug resistance and poor survival (1, 2, 10, 11).
A growing body of work suggests that transcriptional heterogeneity also contributes to tumor adaptability and potential for evolution, and may be particularly relevant in AML due to its low mutation burden (3–5, 12). Our study of paired presentation–relapse samples using bulk RNA sequencing (RNA-seq) did not reveal canonical transcriptional changes associated with relapse after chemotherapy (6), demonstrating the need for techniques such as single-cell RNA-seq (scRNA-seq) to characterize the transcriptional features of relapse. scRNA-seq has revealed extensive transcriptional heterogeneity in numerous tumor types, including AML (13–17). Moreover, several studies have combined scRNA-seq with single-cell mutation detection to identify subclone-specific transcriptional cell states (16–20), strengthening the evidence that transcriptional and genetic heterogeneity interact to drive the tumor's response to chemotherapy.
In this study, we performed enhanced whole-genome sequencing (eWGS) and ultradeep scRNA-seq on paired presentation–relapse samples from six normal karyotype patients with AML (the most common cytogenetic presentation of AML), four normal bone marrow samples, and three additional time points from one patient. These data enabled us to address several questions that are not yet tractable using other methods. First, we asked whether the extent of genetic evolution is quantitatively correlated with the extent of transcriptional evolution. Second, we sought to identify presentation-enriched or relapse-enriched cell states. Third, we asked whether we could retrospectively identify subpopulations of cells in the presentation leukemias that likely seeded the relapses and whether these populations were distinct with respect to genetics, cell state, or both; we also asked how often genetically defined subclones have unique expression signatures. Fourth, we used pseudotemporal ordering to infer the timing and order of expression changes in each case. Finally, in samples with appreciable numbers of T cells at presentation and relapse, we assessed changes in the T-cell compartment.
We deliberately focused on a relatively small number of cases representing typical AML relapses and a range of clonal architectures because of the complexity and expense of the requisite data set, which involved eWGS, bulk RNA-seq, ultra-deep scRNA-seq, and the use of dual cDNA libraries for many samples. This set of six case reports of AML relapse illustrates the diverse ways in which the cancer genome, transcriptome, and microenvironment interact to contribute to therapy evasion. This is a rich data resource for the community that should serve as a foundation for many additional studies.
Results
Selection of AML Cases and Data Generation
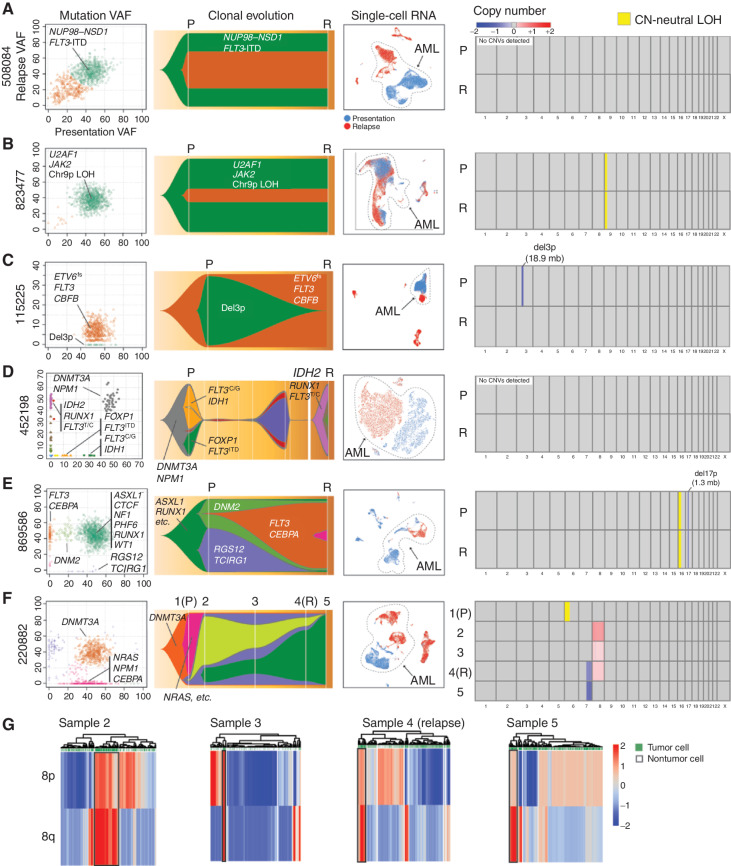
We selected a small set of normal karyotype AML samples for deep study, representing a broad spectrum of disease, with different initiating mutations, clonal architectures, initial therapies, and relapse presentations (i.e., refractory disease, postchemotherapy relapse, and postallogeneic transplant relapse; Table 1). Matched AML and normal skin samples were required at presentation, so that all mutations could be definitively identified as somatically acquired. Cases having cryovials with excellent viability at presentation and relapse were required for the scRNA-seq studies. Five cases relapsed after documented remissions, whereas one (508084) was refractory to initial therapy. These cases exhibited a range of genetic adaptations, from minimal to extensive. Detailed clinical summaries are provided in the Supplementary Materials and Methods. eWGS (16) was performed for normal skin, presentation AML samples pretreatment, and relapsed AML samples. Somatic mutations were identified and used to infer the clonal evolution of each case (Fig. 1; Supplementary Table S1). Figure 1 summarizes the genomic data generated for these cases and groups them according to their major mode of adaptation: Two cases, 508084 (A) and 823477 (B), exhibited little genetic or transcriptional adaptation. Cases 115225 (C), 452198 (D), 869586 (E), and 220882 (F) showed appreciable genetic and transcriptional adaptation.
Table 1.
Summary of patients, clinical features, and putative driver mutations
| UPN, FAB, age, gender | 115225, indeterminate, 22 y, M | 220882, M2, 71 y, F | 452198, M5, 55 y, M | 508084, M4, 38 y, M | 823477, M0, 51 y, M | 869586, M4, 23 y, M |
| Mean founding clone VAF (P, R) | 47.8, 7.2 | 49.4, 40.1 | 46.9, 47.2 | 46.2, 45.2 | 47.9, 37.9 | 48.0, 44.8 |
| Bone marrow blast % (P, R) | >90, 27 | 82, 11 | 97, 95 | 53, 49 | 72, 63 | 51, 54 |
| Induction (I), consolidation (C), time to relapse (TR) | I: 7+3 + midostaurin | I: 7+3 | I: 7+3 | I: 7+3 | I: 7+3 (Ida+AraC) | I: 7+3+3 (AraC, daunorubicin, etoposide) |
| C: HiDAC × 3 + midostaurin | C: 5-Aza × 2 cycles | C: HiDAC × 4 | TR: Day 49 (refractory disease) | C: HiDAC with Ida × 4 | C: HiDAC + etoposide, autologous transplant | |
| TR: 28 mo.a | TR: 245 days (4th banking) | TR1: 16 mo., then matched sib allo | TR: Day 242 | TR: 7 mo. | ||
| TR2: 108 mo. (sample used) | ||||||
| Founding clone mutations | ETV6 R103fs, FLT3 (ITD, 6 bp), CBFBD87ins, del3p (18.9 Mb, includes FOXP1) | IDH2 R140Q | DNMT3A R882H | NUP98–NSD1, FLT3-ITD | JAK2 V617F with 9p UPD, U2AF1S34F | ASXL1 G645fs, CTCFR457a, NF1R1306a, PHF6G288V, RUNX1G164fs, WT1A382fs, chr16 CN-neutral LOH, del17p (1.3 Mb, includes NF1) |
| DNMT3A R882H | NPMc | IGF2R G971S | ||||
| NPMc | ELANE Q194H | POL1 D16 | ||||
| Subclonal mutations | WT1 S383fs | NRAS G12 CEBPA Q209 a , ASXL1G645fs, CN-neutral LOH of 6p | FLT3-ITD | No recognized drivers | No recognized drivers | No recognized drivers |
| IDH1 R132H | ||||||
| FOXP1 e11+1 | ||||||
| FLT3 R835H | ||||||
| Relapse-specific mutations | DNMT3A R882H (probably underlying clonal hematopoiesis selected for by chemo) | del7q, trisomy 8 | IDH2 R140Q | Nothing unique | Nothing unique | CEBPA F6fs, FLT3Q577E |
| RUNX1 P339fs | ||||||
| FLT3 R835G | ||||||
| Cells (P; R) | 17,288; 11,965 | 8,703; 14,787 | 5,667; 7,382 | 14,281; 9,531 | 18,099; 13,153 | 6,808; 2,438 |
| Cells with SNV/indel (P; R) | 646; 329 | 11,787; 2,398 | 1,067; 2,790 | 417; 1,226 | 8,827; 7,500 | 7,162; 8,335 |
| Unique SNVs/indels in single cells (P; R) | 51; 50 | 269; 193 | 19; 24 | 12; 23 | 74; 76 | 162; 150 |
AraC, cytarabine; CN-neutral LOH, copy number–neutral loss of heterozygosity; F, female; FAB, French–American–British subtype; 5-Aza, 5-azacytidine; indel, insertion/deletion; HiDAC, high-dose cytarabine; Ida, idarubicin; M, male; P, presentation; R, relapse; sib allo, allogeneic bone marrow transplant using a sibling donor; SNV, single-nucleotide variant; UPN, unique patient number; VAF, variant allele frequency.
aAll samples had eWGS and scRNA-seq performed from bone marrow samples pretreatment (day 0) and then at one or more additional time points. For 115225, the second sample was from 28 months (relapse after remission); for 220882, downstream samples were analyzed at days 34 (postinduction), 149 (“remission”), 245 (relapse), and 287 (refractory postsalvage); for 452198, the second sample was from month 108 (relapse post–allogeneic transplant); for 508084, the second sample was from day 49 (refractory disease postinduction); for 823477, the second sample was from day 242 (relapse postremission); and for 869586, the second sample was obtained at 7 months (relapse postremission).
Figure 1.
Paired presentation–relapse AML cases grouped by primary mode of postchemotherapy adaptation. For each case in A–F, the following data types are shown from left to right: (1) subclonal composition inferred from eWGS data, and plotted with SciClone, where the x- and y-axes show the variant allele frequencies (VAF) of all somatic mutations (each represented by a dot) in the presentation and relapse samples, respectively. Selected driver mutations are labeled for each case, but they are not comprehensive due to size constraints. The colors of the dots correspond to subclones, which are represented by the same colors in the Fish plots to the right of each panel. (2) Subclonal evolution visualized as Fish plots, where each color represents the founding clone or a subclone, and the height of the (sub)clone represents the percentage of the tumor comprised by that (sub)clone. Selected driver mutations are shown. (3) Uniform Manifold Approximation and Projection (UMAP) and t-distributed Stochastic Neighbor Embedding (t-SNE) representations of scRNA-seq data, with AML cells circled and labeled (blue: presentation cells, red: relapse cells). (4) Copy-number variant (CNV) regions and copy number–neutral loss-of-heterozygosity (CN-neutral LOH) regions identified from comparisons of tumor and matched normal WGS data. Chromosome numbers are shown under the boxes. Colors indicate deletions (blue) and amplifications (red), expressed as change relative to the matched normal control, per the color scale in the legend. All five male patients had one copy of X, and 220882, who was female, had two copies of X. Regions of CN-neutral LOH are designated in yellow. P denotes presentation, and R denotes relapse at times of sample collection. Disease status at each sample time is indicated in the Table 1 footnote. G, CONICSmat results for the chromosome 8 trisomy for 220882 are shown for samples 2 to 5, with putative AML cells enclosed in black boxes. Each column represents a cell, and each row a region of chromosome 8 (8p or 8q).
cDNA libraries for scRNA-seq were prepared using the 5′ 10X Chromium platform (in duplicate for seven samples), and sequenced to an average depth of 150,000 reads per cell (more than three times the standard depth). The 5′ chemistry and high-depth sequencing were used to increase the sensitivity of detection of expressed mutations. A total of 142,642 AML cells and 9,601 normal bone marrow cells were evaluated after filtering for high-quality cells, yielding 9,509 cells per AML sample, on average. These datasets have been deposited in dbGaP. Sequencing metrics are available in Supplementary Table S2; single-nucleotide variants (SNV) and insertions/deletions (indel) in each cell are available in Supplementary Table S3.
We first distinguished AML and nonmalignant cells by identifying cells that expressed somatic, AML-specific mutations. For this, we first used the eWGS data for de novo mutation discovery and subclone inference (Supplementary Table S1), and then used cb_sniffer and CONICSmat to identify mutations in the scRNA-seq reads of individual cells. cb_sniffer is a Python-based tool we developed (16) for the identification of SNVs and small indels in 10X generated single-cell data; CONICSmat (21) is an R package for identification of copy-number variants (CNV) in single-cell data. SNVs and indels are identifiable in a subset of cells that have abundant unique molecular identifiers (UMI), uniform cDNA coverage, proximity to the 5′ end, and/or high expression of the mutant gene(s) in question, with a false-positive rate under 1% (16). In this sample set, the number of AML cells with expressed somatic mutations ranged from several hundred to several thousand cells per sample (Table 1), and we used these to infer the AML status of coclustered cells. Each cell containing identifiable mutations was assigned to the subclone containing those mutations. CNVs were not used for inference of clonal architecture or subclonal membership (and they were detected in only three of the cases; see Fig. 1). Finally, cell type and differentiation state were inferred by matching each cell to its nearest normal cell type using SingleR (22) trained on the DMAP database (23) and the Human Cell Atlas (24). On the basis of mutation status, coclustering, and inferred differentiation state, the majority of the cells in these samples were AML cells, with very few normal myeloid cells.
Relationship between Genetic and Transcriptional Adaptation
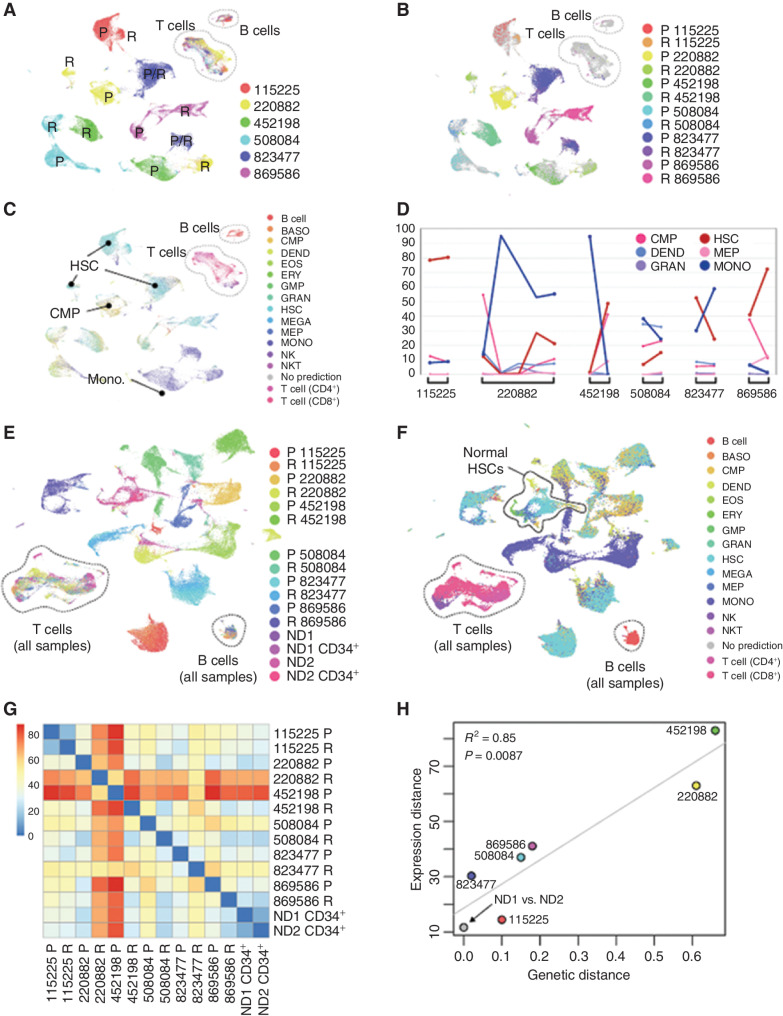
We first generated an overview of intersample relationships using scRNA-seq data (Fig. 2A and B). As observed in previous studies, AML cells clustered by sample, but normal cells [B, T, and natural killer (NK) cells] clustered by cell type. The majority of AML cells were hematopoietic stem cell (HSC)–like (38%), monocytic (30%), or common myeloid progenitor (CMP)–like (16%), but there was substantial lineage diversity within and among leukemias (Fig. 2C). All patients displayed lineage shifts between presentation and relapse. Although the proportion of HSCs and monocytic cells was anticorrelated across samples (Pearson correlation = −0.74; cell-type proportions shown in Supplementary Table S4), the malignant populations did not consistently shift in the same direction (Fig. 2D). When AML samples were analyzed together with normal donor–derived bone marrow, nonmalignant lymphocytes from patients with AML coclustered with those from normal donors (Fig. 2E). In contrast, malignant cells did not cocluster with their cognate normal donor cell types [e.g., malignant hematopoietic stem/progenitor cells (HSPC) were different from normal HSPCs; Fig. 2F].
Figure 2.
Overview of intersample relationships among diverse presentation and relapse AMLs. A, All-sample Uniform Manifold Approximation and Projection (UMAP) colored by unique patient number (UPN; patient) with presentation (P) and relapse (R) samples indicated. B, Mutation-containing cells colored by sample of origin. C, Cells colored by nearest normal inferred lineage using singleR and the DMAP database. D, Lineage shifts between time points, showing those lineages that comprise more than 5% of the cells in at least one sample. Differentiated monocyte-like cells (dark blue) were consistently anticorrelated with HSC-like cells (dark red). E, UMAP representation of cells from 12 AML samples and 4 normal bone marrow samples, colored by sample. F, UMAP from E, with cells colored by nearest normal hematopoietic lineage. G, Pairwise expression distances among all sample pairs (Wasserstein metric computed on the first 10 principal components). H, Relationship between genetic and transcriptional evolution: Expression distance between P and R samples for each patient (vertical axis) are plotted against the genetic distance between P and R samples for each patient (horizontal axis). Normal donors were excluded from the regression calculation. BASO, basophil; DEND, dendritic cell; EOS, eosinophil; ERY, erythrocyte; GMP, granulocyte–monocyte progenitor; GRAN, granulocyte; MEGA, megakaryocyte; MEP, megakaryocyte–erythroid progenitor; MONO, monocyte; ND, normal donor; NKT, natural killer T cell.
We next asked whether genetic and transcriptional responses to chemotherapy were correlated. To this end, we quantified the “genetic distance” between each presentation–relapse pair as the fraction of all mutations in both samples that were in the founding clone. To compare the “expression distance” between sample pairs, we computed the Wasserstein metric (25) on the first 10 principal components using the “transport” R package (Fig. 2G). The results indicated that genetic and transcriptional distance are highly correlated [R2 = 0.85, P = 0.0087; Fig. 2H; notably, expression distance corresponds closely to physical distances in the Uniform Manifold Approximation and Projection (UMAP)]. We also calculated the Wasserstein expression distance between the two normal CD34-enriched samples, which were taken from different individuals. These were transcriptionally more similar than any AML pair from the same patient (Fig. 2G and H), demonstrating that AML samples from the same patient were always more different than normal cells from different individuals, highlighting the extensive transcriptional remodeling that occurs in response to chemotherapy. Finally, when cells containing observable mutations were analyzed separately, the same pairwise intersample relationships were observed: The correlation in Wasserstein metrics was 0.96 (Pearson correlation, P = 0.0024; Supplementary Fig. S1A–S1D).
Each AML Sample Expresses a Unique Combination of Transcriptional Programs
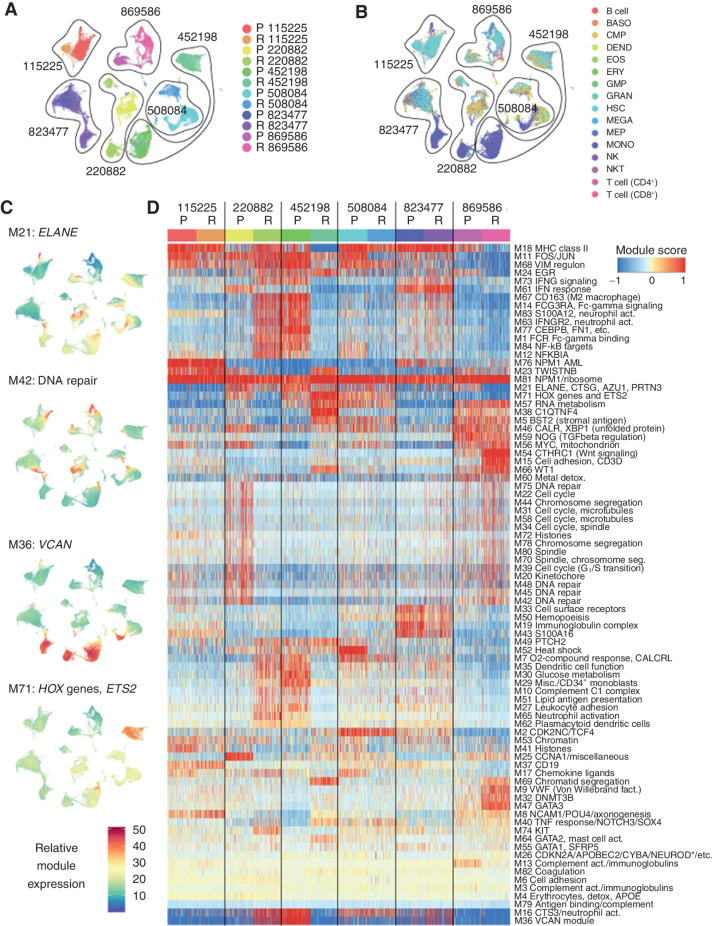
Initial analysis of the scRNA-seq data indicated that certain groups of genes tend to be coexpressed, and potentially coregulated, across conditions and samples. After excluding T cells and B cells (Fig. 3A and B), we used weighted gene correlation network analysis (WGCNA; ref. 26) to identify these gene modules and derive a more concise representation of AML transcriptomes and their dynamics. WGCNA identified 83 distinct expression modules, containing an average of 21.7 genes (min = 6, max = 379, median = 13; Fig. 3C and D; Supplementary Table S5). We annotated each module according to its functional enrichment, or if the module contained too few genes for enrichment analysis, by selecting key genes that have well-documented functions. Most modules corresponded to groups of genes that are known or generally assumed to be coexpressed [e.g., the MHC class II genes (module 18), FOS and JUN (module 11), interferon response (module 61), HOX genes (module 71), histone genes (multiple modules), genes involved in chromosome segregation (multiple modules), and ribosomal genes (module 81)]. In addition, WGCNA identified several less well-understood regulons, such as MYC and mitochondrial genes (module 56), lipid antigen presentation (module 51), oxygen response (module 7), and glucose metabolism (module 30).
Figure 3.
Individual AML samples express unique combinations of gene modules. A, UMAP representation of scRNA-seq data from all AML samples generated after excluding lymphocyte clusters (T cells, B cells, and NK cells), with cells colored by sample. These cells were used to identify gene modules using the WGCNA algorithm. B, Cells colored by inferred lineage using DMAP data. C, UMAPs colored according to the single-cell expression of selected representative modules. Top to bottom: module 21 (ELANE, etc.), module 42 (DNA repair), module 36 (VCAN), and module 71 (HOX genes and ETS2). D, Clustered heatmap of WGCNA gene module expression (labeled “Module score” in the key) in individual cells from each sample. *NEUROD indicates that multiple NEUROD genes were present. act., activation; BASO, basophil; DEND, dendritic cell; detox., detoxification; EOS, eosinophil; ERY, erythrocyte; fact. factor; GMP, granulocyte–monocyte progenitor; GRAN, granulocyte; MEGA, megakaryocyte; MEP, megakaryocyte–erythroid progenitor; Misc., miscellaneous; MONO, monocyte; NKT, natural killer T cell; P, presentation; R, relapse; seg, segregation.
These gene modules are a useful tool for downstream analysis of single-cell AML data. For each cell and each module, we calculated a “module expression” score, which showed that some modules vary within samples, while others vary between samples (Fig. 3C). We then clustered the modules, which provided a concise representation of the biological processes active in each cancer (Fig. 3D). As expected, biologically related modules coclustered (e.g., cell-cycle, chromosome segregation, histone, and DNA metabolism genes). However, each AML expressed a unique combination of modules, and no module was entirely presentation or relapse specific. Moreover, some relapse samples were actually more transcriptionally similar to presentation samples from other patients—for instance, 220882-R resembled 452198-P, and 220882-P resembled 869586-R.
Identification of a Relapse-Enriched Leukemic Cell State
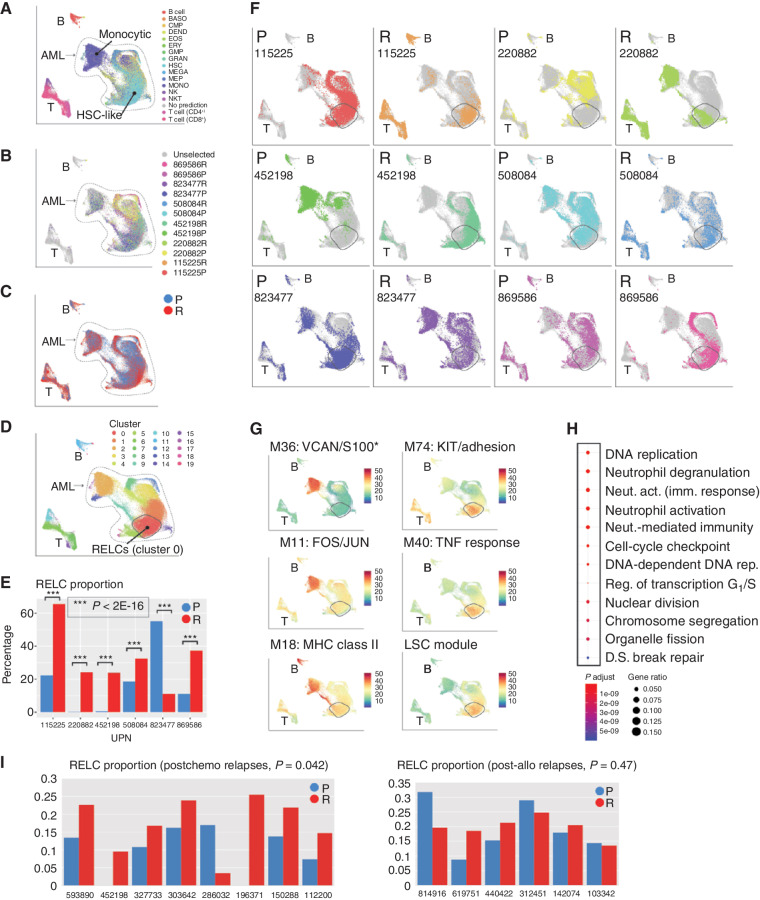
We hypothesized that AMLs may evolve toward a shared cell state in response to the selective pressures imposed by chemotherapy. These may be obscured by the more obvious patient-specific signatures observed above, so we used a data integration workflow (27) to computationally minimize differences among patients while preserving differences between time points. This indicated that malignant cells from all samples lie along a spectrum of differentiation, with each sample consisting of two main subpopulations: one of relatively differentiated cells (“monocytic,” Fig. 4A) and one of less-differentiated cells (“HSC-like”). Somatic AML-associated mutations were present throughout both (Fig. 4B). Next, we asked whether any region of expression space contains predominantly presentation or relapse cells (Fig. 4C), and found that one region, corresponding to graph-based “cluster 0” (Fig. 4D), is significantly enriched for relapse cells in every patient but one (P < 2 × 10−16, Fisher exact test; Fig. 4E and F). The exception, 823477, was already in a relapse-like state at presentation, with a large proportion of cells in cluster 0, and changed in the opposite direction after treatment (Fig. 4F). This case was unique in the cohort, in that it displayed very few genetic or transcriptional changes at relapse.
Figure 4.
Use of interpatient data integration to identify a relapse-enriched leukemia cell (RELC) expression signature in multiple patients. A, UMAP representation of scRNA-seq data from all AML samples colored by inferred cell lineage, with the differentiated (“monocytic”) and undifferentiated (“HSC-like”) subpopulations indicated. B, Mutation-containing cells colored according to sample of origin. C, Cells colored according to time point (blue, presentation; red, relapse). D, Cells colored according to graph-based cluster assignment, with the relapse enriched cluster, “cluster 0,” indicated. E, Proportion of each sample in cluster 0 (blue, presentation; red, relapse). F, Each sample plotted individually. G, Selected modules that exhibit heterogeneous expression. Top row, representative modules that are more highly expressed in differentiated cells. Bottom row, modules that are more highly expressed in relapse-enriched cluster 0. *S100 indicates that multiple S100 genes were present. H, Functional enrichment, calculated and plotted using the clusterProfiler R package, of genes downregulated (log2 fold change ≤ –1.5) in cluster 0 compared with other HSC-rich clusters. I, RELC proportions in presentation and relapse samples from an independent cohort analyzed using bulk RNA-seq. Left, postchemotherapy cases; right, postallogeneic transplant cases. RELC proportions were inferred using CIBERSORTx (see Methods). act., activation; BASO, basophil; DEND, dendritic cell; D.S., double-strand; EOS, eosinophil; ERY, erythrocyte; fact. factor; GMP, granulocyte–monocyte progenitor; GRAN, granulocyte; imm., immune; LSC, leukemic stem cell; MEGA, megakaryocyte; MEP, megakaryocyte–erythroid progenitor; MONO, monocyte; Neut., neutrophil; NKT, natural killer T cell; P, presentation; R, relapse; Reg., regulation; rep., replication; UPN, unique patient number.
Cluster 0 is primarily composed of HSC-like cells (Fig. 4B), and we characterized it further using several approaches. First, we scored each cell for expression of each WGCNA module and plotted that score on the UMAP. Most modules displayed clear differences in expression between the differentiated and undifferentiated populations, and were more highly expressed in the former. This includes module 18, composed almost entirely of MHC class II genes (Fig. 4G). However, several modules (e.g., modules 74 and 40) were most highly expressed in cluster 0 (Fig. 4G). Module 74 is enriched for genes involved in cell migration (9/24 genes) and contains KIT, a receptor tyrosine kinase important for hematopoiesis and stem cell maintenance. Module 40 is enriched for genes associated with TNF binding (2/13), JAK–STAT signaling (3/13), and cell adhesion (4/13) and contains SOX4, an oncogenic transcription factor that interacts with TP53 and is implicated in CEBPA-mutated AML (28). In other cancer types, overexpression of SOX4 is thought to induce stem-like phenotypes and associated resistance to chemotherapy (29). We combined these related modules into a relapse-specific “metamodule,” which is enriched for genes associated with cell migration [q = 0.0030 (Benjamini–Hochberg correction), 12/37: CEACAM6, ADGRG1/GPR56, PRSS3, TNFRSF18, APOB, IGF1, KITLG, TNFSF18, NET1, KIT, LOX, CXCL9], cell adhesion [q = 0.0030 (Benjamini–Hochberg correction), 11/37: CEACAM6, ADGRG1, PRSS2, TNFRSF18, IGF1, KITLG, TNFSF18, NRXN2, NET1, KIT, CDH9], as well as numerous categories related to TNF response [q = 0.0083 (Benjamini–Hochberg correction), 5/37: TNFRSF18, LTB, APOB, TNFSF18, TNFRSF4]. Complete metamodule functional enrichment results are available in Supplementary Table S6.
Second, we performed a Wilcoxon rank-sum differential gene expression test on the patient-corrected data to identify genes that are overexpressed in cluster 0 (all differentially expressed gene results available in Supplementary Table S6). The top genes (fold change ≥ 1.5) are enriched for leukocyte migration and cell adhesion [q = 0.008 (Benjamini–Hochberg correction), 6/23: TNFRSF18, ANGPT1, CD9, KIT, CD34, IGHM]. More generally, many relapse-enriched genes are involved in adhesion to, or generation of, the extracellular matrix (ADGRG1, PXDN, LOX, CD34) or adhesion to endothelial cells (CEACAM6, CD9). Several have dual roles in adhesion and stem cell maintenance (KIT, ADGRG1/GPR56). CD34, a marker of HSPCs, may be involved in the attachment of stem cells to the bone marrow stroma (30). We also performed this analysis using the uncorrected data, and the results were nearly identical in terms of functional enrichment, but different in the range and distribution of log2 fold changes and the gene ranking (Supplementary Table S6). The top 200 most upregulated genes by fold change were most highly enriched for the biological processes “leukocyte migration,” “cell migration,” and “cell adhesion.”
Third, we examined a 17-gene leukemic stem cell signature that has been associated with poor prognosis and a higher risk of relapse, which includes several genes that we identified as relapse-specific with WGCNA or DEG analysis (CD34, NYNRIN, and ADGRG1/GPR56; ref. 31). This signature also displays higher expression in cluster 0 than elsewhere (Fig. 4G).
Fourth, we compared cluster 0 to the other HSC-rich clusters (1, 3, 4, and 5) using the patient-corrected data and the Wilcoxon rank-sum test. Notably, cluster 0 displayed more downregulated genes than upregulated genes, and these were dramatically downregulated (up to 90-fold; Supplementary Table S6). Using the Toppfun tool, genes downregulated in cluster 0 (log2 fold change ≤ 1.5) were strikingly enriched for cell cycle–related processes, such as nuclear division (66 genes, q = 2.1 × 10−20); cell-cycle process (75 genes, q = 2.1 × 10−20); DNA biosynthetic process (41 genes, q = 6.5 × 10−20); and mitotic cell cycle (59 genes, q = 4 × 10−19). In addition, they were enriched for genes that encode proteins that interact with numerous regulators of cell-cycle progression and DNA replication, including CDK1 (59 genes, q = 5.99 × 10−20), CCNA2 (30 genes, q = 6.27 × 10−14), H2BC8 (37 genes, q = 3.35 × 10−13), CDK (66 genes, q = 3.07 × 10−12), and many others. Another notable interactor was the hypoxia-inducible factor EGLN3, which interacts with the largest number of genes in this cluster (89 genes, q = 5.17 × 10−10). Enrichment analysis by the clusterProfiler R package showed a mix of functions related to cell cycle and neutrophil activation (Fig. 4H), the latter reflecting the relative lack of differentiation markers in these cells. We also performed the Wilcoxon rank-sum test using the uncorrected data. The ordering of genes and the range and distribution of log2 fold changes differed, but functional enrichments were similar. The top 200 most downregulated genes were enriched for biological processes associated with immune cell maturation (e.g., “neutrophil activation”) and DNA replication.
Taken together, these results suggest that relapsed AML may be enriched for quiescent stem-like cells (32) that may adhere to endothelial cells, or the extracellular matrix within the bone marrow stroma. We refer to these cells as relapse-enriched leukemia cells (RELC).
Confirmation of RELCs in an Independent Cohort
We next tested whether the RELC signature can be generalized to an independent cohort of 16 presentation–relapse pairs analyzed with bulk RNA-seq (6), 14 of which were not included in this study. Of these, eight relapses were obtained after treatment with chemotherapy and six after post–allogeneic transplant. We used the CIBERSORTx algorithm to estimate the proportion of each sample corresponding to the RELC population and observed the same pattern in the postchemotherapy samples: Seven contained a larger RELC fraction than at presentation; the outlier contained very few malignant cells at relapse (Fig. 4I). The pattern was not observed in the posttransplant samples, which are dominated by different transcriptional features (6).
Distinguishing RELCs from Normal CD34+ Cells
To compare RELCs to a broader data set including normal CD34+ cells, we rederived the RELC population using the approach above, this time including four normal bone marrow samples (two of whole bone marrow and two of sorted CD34+ cells from the same donors; Supplementary Fig. S2A). This again revealed a cluster (“cluster 0”) that was more prevalent in the relapses and overlapped the original RELCs (Supplementary Fig. S2B–S2E). We refer to this cluster as RELCprime (“RELCp”). Most normal CD34+ cells were not part of the RELC/RELCp populations, indicating that RELCs and normal CD34+ cells are not equivalent. To compare the RELCp cells with other clusters containing normal CD34+ cells (as well as other AML cells), we performed a differential expression analysis (analogous to that performed above for RELC cells vs. HSC-like clusters). Seventeen times as many genes were downregulated (and dramatically so) than upregulated in RELCp, and these were markedly enriched for cell cycle–related processes (Supplementary Fig. S2F; Supplementary Table S6). Twenty-two genes were more highly expressed in RELCp than in the other clusters containing normal CD34+ cells (Padj = 0, log fold change ≥ 1), 16 of which were also RELC specific. The top gene is Adhesion G Protein-Coupled Receptor G1 (ADGRG1), and the cell adhesion genes CD34 and CD9 are ranked fourth and sixth, respectively, corroborating the previous result that adhesion genes are upregulated in the RELC/RELCp population (Supplementary Table S6).
One finding from the integrated analyses presented above is that genetic adaptation and transcriptional adaptation are correlated. We observed strikingly different degrees of adaptation among these six cases (Figs. 1A–F and 2H): four cases (452198, 220882, 115225, and 869586) displayed appreciable genetic and transcriptional adaptation, one case (508084) displayed primarily transcriptional adaptation, and one case (823477) underwent little of either. To better understand the relationships between the underlying genetic and transcriptional changes, we examined each case in detail as described below.
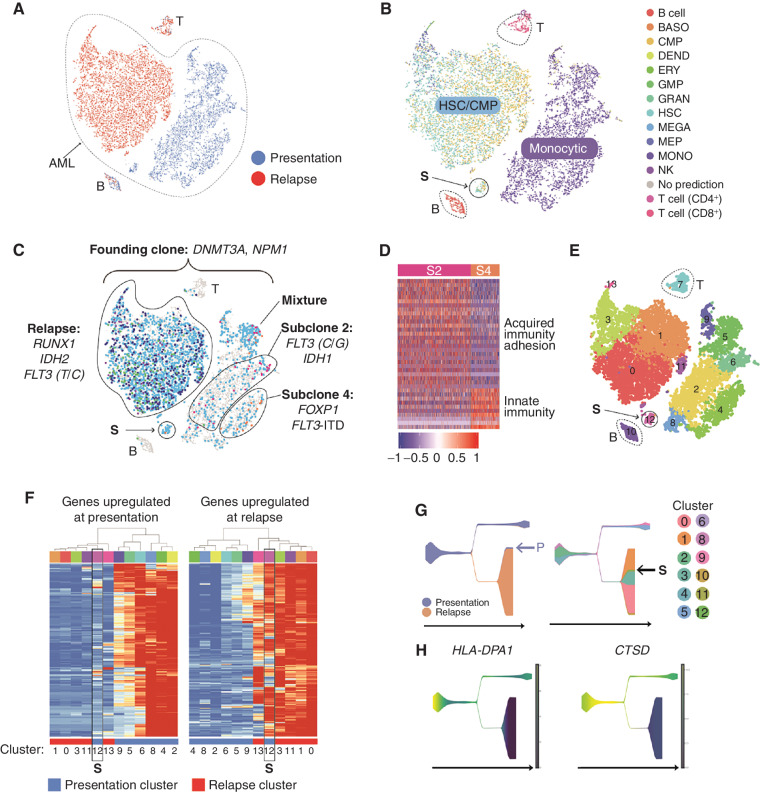
UPN 452198: Coordinated Changes in Mutations and Cell State, Distinct Subclonal Cell States, and a Transcriptionally Defined Link between the Presentation and Relapse AMLs
UPN (unique patient number) 452198 is an extensively studied patient who relapsed twice, once after chemotherapy (not studied here) and once after allogeneic stem cell transplant; we have previously reported many findings from this patient (3, 6, 8, 33); the scRNA-seq data presented here was generated specifically for this study. This AML exhibited complex adaptations in terms of clonal evolution (Fig. 1D) and cell state (Fig. 5A and B). The presentation and relapse samples [which were both highly enriched for AML cells, based on founding clone variant allele frequencies (VAF)] are transcriptionally distinct, and lineage inference revealed a shift from monocyte-like cells at presentation to a mixture of stem-like cells resembling HSCs and CMPs at relapse—observations that were confirmed by flow cytometric and morphologic analysis (8). Because the relapse sample was almost completely comprised of AML cells (>95%), contaminating cells from the allogeneic donor did not affect the analysis. The presentation sample contained two dominant, mutationally defined subclones (subclones 2 and 4) marked by distinct gene expression profiles. Subclone 2 was enriched for genes associated with acquired immunity and cell adhesion, and subclone 4 was enriched for innate immunity markers (Fig. 5C and D). Because chemotherapy eradicated both subclones, these expression differences were probably not key to the evolution of this cancer. Instead, the data are consistent with the possibility that the relapse was seeded by a small, transcriptionally distinct population of cells present at diagnosis (labeled “S” in Fig. 5B, C, and E–G). Six methods were used to assess the relationship of population S to the presentation and relapse samples, including t-distributed Stochastic Neighbor Embedding (t-SNE) and UMAP dimensionality reductions; pseudotemporal ordering using Monocle V2, Monocle V3, and STREAM; and hierarchical clustering of the gene expression profiles in each graph-based cluster. All of these data suggest that population S was intermediate between the presentation and relapse samples, which differed in several respects. Specifically, genes downregulated at relapse were enriched for IFN response and MHC class II genes, whereas genes upregulated at relapse were enriched for proliferation and protein translation, and included many canonical AML-associated genes. We used pseudotemporal ordering to infer the order and timing of the expression changes that occurred between presentation and relapse, and found that the MHC class II gene HLA-DPA1 and the CTSD gene were probably among the first genes to change during the evolution of this cancer, with similar expression in cluster 12 and the relapse cells (Fig. 5G and H).
Figure 5.
A, t-distributed Stochastic Neighbor Embedding (tSNE) plot with cells colored according to time point (blue, presentation; red, relapse). B, Cells colored by inferred lineage. C, Cells with detectable mutations colored according to the subclone assignment of the mutation(s) (turquoise, founding clone; pink, subclone 2; purple, subclone 3; orange, subclone 4; blue, subclone 8; and green, subclone 9). The population believed to have seeded the relapse is labeled “S.” Key driver mutations are indicated. D, Genes that are differentially expressed in subclone 2 compared with subclone 4 (fold change ≥ 1.5, Padj ≤ 0.05). E, Cells colored according to graph-based cluster assignment. S is cluster 12. F, Heatmap representing the expression of genes (rows) that are differentially expressed in presentation and relapse samples (Padj ≤ 0.05, top and bottom 2.5% based on fold change). Each column represents a graph-based cluster defined in E, with the cluster number indicated below, and the time point indicated with a colored bar (blue, presentation; red, relapse). Expression data were averaged across cells in each cluster. Left, genes that are more highly expressed at presentation. These were enriched for gene ontology (GO) terms such as myeloid immune response, MHC class II, IFNG response, and respiratory burst, as well as binding sites for PU.1 and IRF1. Right, genes that are more highly expressed at relapse. These are enriched for GO terms including proliferation, nucleosomes, and translation and include key AML genes (e.g., FLT3, RUNX1, NPM1, KDM5B, MYC, MYB, GATA2, XBP1, EGR1, SOX4, HOXA9). Cluster 12, that is, population S, is highlighted with a black box in each. G, Pseudotemporal ordering obtained using STREAM. Left, states colored according to time point (blue, presentation; orange, relapse). Right, states colored according to the graph-based cluster assignment of the constituent cells (from F), with the seed population S indicated. H, Two of the genes that change early in pseudotime and display a relapse-specific expression pattern, HLA-DPA1 and CTSD. BASO, basophil; DEND, dendritic cell; ERY, erythrocyte; fact. factor; GMP, granulocyte–monocyte progenitor; GRAN, granulocyte; MEGA, megakaryocyte; MEP, megakaryocyte–erythroid progenitor; MONO, monocyte.
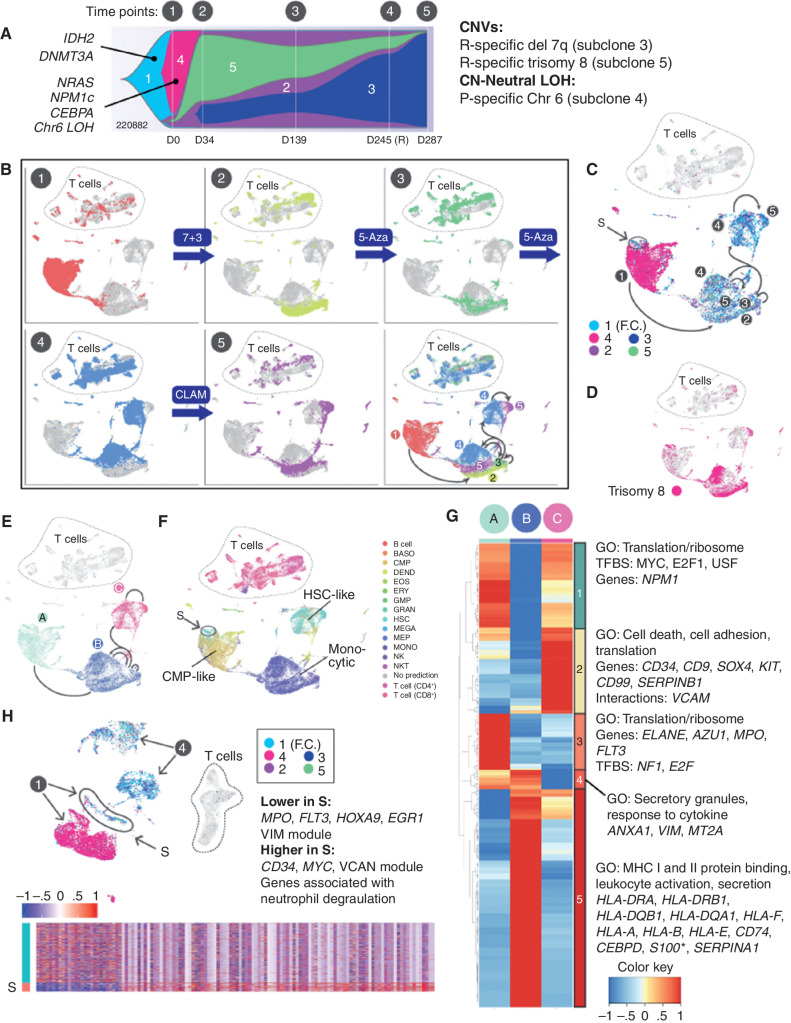
UPN 220882: Coordinated Genetic, Transcriptional, Morphologic, and Microenvironmental Adaptation
For UPN 220882, data from five time points delineated a series of coordinated genetic, transcriptional, morphologic, and microenvironmental responses to four different courses of chemotherapy. It also revealed a transitional cell population present at diagnosis, defined by distinct mutations and cell state, which persisted through treatment and may have evolved into the relapse. Each time point displayed unique transcriptional and genetic features (Fig. 6A–D). Dimensionality reduction using the UMAP algorithm indicated that the five time points spanned three main cell states (Fig. 6E), each characterized by a unique combination of genomic alterations and gene expression signatures. As expected, all three states contained founding clone mutations, but they differed with respect to additional mutations and the expression level of the initiating mutation, DNMT3AR882H (discussed further below). These cell states evolved over time; in particular, the fourth time point (obtained at relapse) represented a transitional state composed of two distinct subpopulations, one similar to the earlier samples, and the other more similar to the final, chemoresistant sample.
Figure 6.
UPN 220882, a time course of concerted genetic, transcriptional, and microenvironmental evolution. A, Fish plot with additional details redrawn from Fig. 1F, showing evolution of clonal architecture across the time points sampled (labeled 1–5 above the diagram). Subclones are labeled 1 to 5 within the Fish plot, with known AML driver mutations indicated. Copy number–altered regions and copy number–neutral loss-of-heterozygosity (CN-neutral LOH) regions, and their subclonal locations, are shown on the right. B, UMAP representations of scRNA-seq data for all time points, with cells colored according to the indicated time point. Treatments given between time points are indicated. The final panel shows all time points combined, with arrows indicating directions of transcriptional shifts. The same arrows are shown in C and E for reference. C, Cells from all time points with detectable mutations are colored according to the clonal assignment of the mutation(s), matching the colors in A: turquoise, founding clone 1 (F.C.); pink, subclone 4; purple, subclone 2; green, subclone 5; and blue, subclone 3. Presentation population S, which is genetically and transcriptionally distinct, and which likely seeded the relapse, is circled. D, Cells from all time points containing the subclonal trisomy 8 are shown in pink. E, Cells from all time points, demonstrating the three main cell states identified using the UMAP algorithm. Green, state A; blue, state B; pink, state C. F, Cells from all time points colored and labeled by SingleR lineage inference using DMAP database. Presentation population S is circled. G, Genes that are differentially expressed among the cell states (Padj ≤ 0.05, fold change ≥ 1.5). Each column represents one of the cell states A, B, or C, and expression values are averaged across cells within each state. *S100 indicates that multiple S100 genes were present. H, Analysis restricted to the presentation (time point 1) versus relapse (time point 4) samples. Cells from each time point are labeled with arrows. Cells with detectable mutations are colored according to the subclone assignment of the mutation(s), as shown within the inset, using the same color scheme as shown in C. Presentation cells that contain relapse mutations and putatively seeded the relapse are labeled S. Bottom, heatmap of genes that are differentially expressed between S and the remainder of the presentation cells; enrichments and key genes are indicated (Padj ≤ 0.05, fold change ≥ 1.5). BASO, basophil; CLAM, clofarabine, cytarabine, and mitoxantrone; DEND, dendritic cell; EOS, eosinophil; ERY, erythrocyte; fact. factor; 5-Aza, 5-azacytidine; GMP, granulocyte–monocyte progenitor; GO, gene ontology; GRAN, granulocyte; MEGA, megakaryocyte; MEP, megakaryocyte–erythroid progenitor; MONO, monocyte; NKT, natural killer T cell; P, presentation; R, relapse; TFBS, transcription factor binding site.
We next characterized these three cell states (A, B, and C; Fig. 6E), which showed lineage shifts that occurred in concert with the genetic changes, and corresponded to the morphology and behavior of the cells (Fig. 6F). State A (CMP-like), characterized by high DNMT3AR882H expression and subclone 4 mutations, was represented by the presentation sample. State B (monocytic), was marked by trisomy 8, mutations from subclones 2, 3, and 5, and low expression of DNMT3AR882H and encompassed time points 2 and 3 (both samples had morphologic features of remission). State C (HSC-like), containing mutations from subclones 2, 3, and 5, highly expressed DNMT3AR882H; was comprised of cells from the 4th and 5th time points (relapse and refractory disease); and expressed the RELC expression signature. Genes that were differentially expressed (Wilcoxon rank-sum test) among these states fell into five main expression patterns (Fig. 6G). Notably, state B exhibited high expression of many MHC class I and II genes, and state C exhibited high expression of several RELC genes noted earlier (e.g., CD9, CD34, SOX4, KIT, and CD99), consistent with the relapse/refractory nature of the disease at these time points.
Of note, we also identified a distinct cell population from the presentation sample that appears to have given rise to the relapse (labeled S in Fig. 6C, F, and H). In this case, that population was genetically and transcriptionally distinct, containing relapse-specific mutations (from subclones 2 and 5), and it exhibited a stem-like expression signature. We examined this population with greater resolution by directly comparing the 1st (presentation) and 4th (relapse) time points (Fig. 6H). Compared with the rest of the presentation sample, population S exhibited increased expression of CD34, MYC, and additional genes that were enriched for “neutrophil degranulation” (using a Wilcoxon rank-sum test). Population S was more stem-like than the rest of the presentation sample, and evolved genetically and transcriptionally at relapse, when it expressed a RELC transcriptional signature.
We also observed coordinated evolution of the AML and its immune microenvironment (Supplementary Fig. S3), and we speculate here about the potential meaning of these findings: The presentation sample contained few cytotoxic T cells, suggesting that the AML may have been invisible to the immune system (Supplementary Fig. S3A–S3C). As the disease progressed (samples 3 and 4), both the AML and T-cell compartments had undergone extensive changes in transcriptional signatures: Cytotoxic T cells were abundant, and exhibited markers of activation and exhaustion, suggesting that they had recognized the malignancy but failed to kill it (Supplementary Fig. S3C–S3E). The AML cells had shifted to a more differentiated phenotype (state B, monocytic), and highly expressed MHC class I and class II genes (Supplementary Fig. S3F). In addition, the AML cells expressed high levels of SERPINA1 and SERPINB9, suggesting possible resistance to granzyme-mediated killing (Supplementary Fig. S3G). By the final time point, T cells were diminished in number, and AML cells lost expression of MHC genes and serpins.
UPN 115225: Transcriptional Impact of a Dominant Subclone with Coupled Genetic and Transcriptional Evolution
In this case, the dominant subclone at presentation (subclone 2; Supplementary Fig. S4A) was eliminated by treatment, resulting in a relapse AML that contained only founding clone mutations. This subclone did not contain known driver SNVs but did contain a single copy, 18.9 Mb chromosome 3 deletion that included the FOXP1 gene. The relapse exhibited a unique cell state, revealing the transcriptional impact of the dominant subclone (Supplementary Fig. S4B–S4D). Specifically, the relapse sample was comprised of a single, relapse-specific cluster (cluster 1; Supplementary Fig. S4E), which genetically represented the leukemia as it existed prior to the acquisition of the mutations in the dominant subclone. The relapse was unique in its high expression of POU4F1, SOX4, CD34, and CD99, among others. Similarly, a pseudotemporal trajectory derived using STREAM (Supplementary Fig. S4F) showed that the relapse comprised one branch characterized by high POU4F1 and NCAM1 (CD56) expression (Supplementary Fig. S4G). POU4F1 encodes a transcription factor that is thought to regulate SOX4 in the context of AML (34). Differential expression analysis comparing presentation and relapse showed that cell adhesion genes were significantly upregulated in the relapse sample (Supplementary Fig. S4H and S4I).
UPN 869586: Genetic and Transcriptional Adaptation, and a Mutationally and Transcriptionally Defined Link between Presentation and Relapse
This case (Supplementary Fig. S5A) contains a genetically and transcriptionally distinct cell population at diagnosis that may have evolved into the relapse. Single-cell mutation mapping showed that there were two main cell states at presentation with different frequencies of relapse-specific and relapse-enriched mutations (Supplementary Fig. S5B–S5D). Specifically, subclone 5 was present at low VAF (15%–40% of cells) at presentation and became the dominant subclone at relapse (Supplementary Fig. S5A). In addition, several mutations in subclone 2 (relapse-specific according to bulk eWGS data) were revealed by scRNA-seq to be present at diagnosis (FLT3 p.Q577E and a mutation in the TUBB 3′ untranslated region). The two cell states also corresponded to distinct graph-based cell clusters and distinct cell lineages (Supplementary Fig. S5D and S5E), suggesting that these mutations shifted the presentation cells from a CD34+ myeloid state to a CD34+ multilineage progenitor state. Pseudotemporal ordering suggested that within the subclone 5–high cell state, cluster 14 contained the transitional cells that evolved into clusters 7 and 8, characterized by a relapse cell state (Supplementary Fig. S5F). Cluster 14 was also enriched for actively cycling cells (Supplementary Fig. S5G). Pairwise differential expression analysis revealed that the presentation sample more highly expressed MHC class II and genes associated with neutrophil/granulocyte activation, suggesting again that “immune escape” may have been relevant at relapse.
UPN 823477: T-cell Response and Subtle Transcriptional Adaptation
This AML did not change genetically at relapse (Supplementary Fig. S6A), and transcriptional changes were subtle compared with the other patients (Supplementary Fig. S6B–S6D). However, changes in the T-cell compartment were readily apparent: CD8+ T-cell abundance increased more than 3-fold from presentation (1.7% of the sample) to relapse (6% of the sample; shown in relation to normal donor T cells in Supplementary Fig. S6E and S6F). This was accompanied by an increase in the expression of key cytotoxicity and exhaustion markers (GZMB, PRF1, PDCD1, TIGIT, EOMES, and TOX), with terminally differentiated, exhausted CD8 T cells present in the relapse sample only (Supplementary Fig. S6F and S6G). Concomitantly, the AML exhibited a marked increase in cells expressing the serine protease inhibitors SERPINA1 (Supplementary Fig. S6H) and, to a lesser extent, SERPING1. Twelve percent of presentation cells versus 34% of relapse cells had this serpin expression signature (Supplementary Fig. S6B, S6D, and S6H); we speculate that this finding may have been relevant for protecting AML cells from the granzymes of cytotoxic T cells (35, 36). Notably, this case was the outlier with respect to RELC abundance, underscoring its largely tumor-extrinsic response to treatment.
UPN 508084: Transcriptional Adaptation to Chemotherapy
This AML was refractory to initial chemotherapy; the posttreatment sample was acquired 49 days after diagnosis. As expected, given the short time frame, the presentation and posttreatment malignancies exhibited no discernable genetic differences (Supplementary Fig. S7A). However, the posttreatment tumor displayed striking transcriptional changes (Supplementary Fig. S6B and S6C) more resembling a stem cell–like state: lineage inference indicated that the percentage of HSC-like cells doubled from presentation (7%) to relapse (15%; Supplementary Fig. S6D), and CytoTRACE (37) analysis revealed that the posttreatment cells had greater differentiation potential than the presentation cells (Supplementary Fig. S6E).
Discussion
Clonal evolution and intratumoral transcriptional heterogeneity are associated with relapse and treatment failure in numerous cancers (1, 2, 10, 11). Our recent study of AML relapse using bulk RNA-seq highlighted the importance of approaches that capture this heterogeneity (6). In this study, we used WGS and scRNA-seq to analyze paired presentation and relapse samples from six patients representing a variety of AML subtypes, driver mutations, subclonal architectures, and treatments. This revealed heterogeneity in cell states within and between tumors, dramatic transcriptional shifts between time points, and putative coordinated changes in the tumor and immune microenvironment. Using the WGCNA algorithm to derive a concise representation of gene expression variation in AML, we found that every tumor expresses a unique combination of transcriptional programs. The diverse treatments received by these patients may contribute to their varied molecular trajectories.
Despite the genetic, clinical, and transcriptional diversity of these patients, we identified a relapse-enriched leukemic cell (RELC) population bearing transcriptional markers of quiescence, stemness, and cell adhesion, and corroborated it in bulk RNA-seq data from an independent patient cohort. These results suggest that many relapsed AML cases may be enriched for noncycling, stem-like cells that adhere to epithelial cells or the extracellular matrix within the bone marrow stroma. This may promote a stem-like phenotype and/or provide a protective environment that renders RELCs inaccessible to chemotherapeutic agents, many of which target cycling cells, or antitumor immune responses (32, 38, 39). These results support the leukemia stem cell hypothesis (40), but additional studies will be required to validate these findings.
In addition, these data strengthen the idea that immune escape plays a role in AML relapse for some patients. We previously identified MHC class II gene downregulation as a contributing factor in posttransplant relapse immune evasion (6, 41), but these data suggest that it might be a more general phenomenon that was not discernable using bulk RNA-seq. Notably, the RELCs exhibit low MHC class II expression compared with the more highly differentiated cell populations within these leukemias. In addition, UPN 220882, which we sampled five times over the course of treatment, exhibited a transient increase in MHC class II expression that coincided with an increase in the differentiation state of the malignant cells (resembling morphologic remission), as well as an increase in cytotoxic T-cell abundance, suggesting that a functional immune response may have been induced with chemotherapy. This suggests that dynamic MHC class II expression in leukemias may be relevant for immune recognition and the killing of AML cells by cytotoxic lymphocytes.
We introduced a distance metric from the field of transport theory (the Wasserstein metric) that demonstrated that clonal and transcriptional adaptation are significantly correlated. However, there was substantial case-to-case variation in the underlying molecular details. We integrated eWGS and scRNA-seq data to examine these findings further, first asking whether genetically defined subclones can have unique expression signatures. We found multiple subclones with clear expression signatures—stronger evidence than we presented in our initial investigation of this phenomenon (16). Further work using genetically defined cohorts will be required to determine whether particular mutation combinations drive particular cell states.
The identification of individual AML cells with expressed mutations also enabled us to trace relapse to small populations of cells that were present at time of diagnosis. Out of four cases that displayed posttreatment genetic changes (115225, 220882, 452198, and 869586), we identified “transitional” populations with a unique cell state in three (220882, 452198, and 869586). In two cases (220882 and 869586), this population was also genetically distinct from other presentation cells. In all three patients, these populations were less differentiated than the rest of the presentation cells, consistent with the stem-like features of the RELC population; further work using additional samples will be required to assess the generality of this phenomenon.
This data often revealed interactions between clonal and transcriptional heterogeneity, manifested as heterogeneous expression of mutated genes (Supplementary Fig. S8). This may represent an additional means to generate heterogeneity for an expressed mutation. For instance, heterogeneous expression of a gene that is mutated in the founding clone (i.e., present in every cell) may lead to heterogeneous expression of the mutant protein, thereby introducing an additional layer of heterogeneity within a cancer (Supplementary Fig. S8).
Taken together, the relationships among genetics, cell state, and microenvironment were unique to each patient, revealing the remarkably diverse mechanisms that AML cells can employ to adapt to and evade therapy. Functional validation of many of the hypotheses generated here will require new approaches, new tools, and many additional studies. Regardless, the information from this cohort of patients with relapsed AML should provide a rich resource for the community that will serve as a foundation for future functional and computational studies.
Methods
Institutional Review Board Approval
Samples were acquired after informed consent under an Institutional Review Board–approved protocol 201011766, “Tissue acquisition for analysis of genetic progression factors in hematologic diseases,” which explicitly allows for WGS from banked AML samples.
Data Production
eWGS, germline SNP detection, somatic variant detection, and bulk RNA-seq and analysis were performed as previously described (16). eWGS is WGS with enriched coverage: median exome coverage of 322×, median genome coverage of 42×. Subclone identification was performed as previously described using the SciClone algorithm. Mutations and their subclone assignments are provided in Supplementary Table S1. scRNA-seq sample preparation and sequencing were performed as described (Supplementary Materials and Methods).
scRNA-seq Data Analysis
The feature-barcode matrix for each sample was generated from raw sequence data using Cellranger V3.0 (42) and the GRCh38 human transcriptome reference. For analyses involving two samples (presentation and relapse), samples were aggregated using the Cellranger “aggr” command. Using Seurat V3 (27), features expressed in fewer than 10 cells were excluded, as were cells containing fewer than 100 or more than 4,000 features, or more than 10% mitochondrial transcripts. Counts were then log-normalized and scaled to 1 × 104 counts/cell. The most variable features were identified using a variance stabilizing transformation (“vst” function with 0.1 ≤ mean.cutoff ≤ 8 and 1 ≤ dispersion.cutoff ≤ Inf). A cell-cycle score was assigned to each cell using the CellCycleScoring function, and the resulting “S.Score” and “G2M.Score” values were “regressed out” using the ScaleData function. Principal component analysis (PCA) was performed on variable features, and elbow plots and JackStraw analysis was performed to assess the contribution of each principal component to the data. Principal components were chosen separately for each patient using the elbow plot and JackStraw analysis, ensuring that any statistically significant component contributing 2% or more of the standard deviation was included. The number of components ranged from 15 to 30. t-SNE and UMAP layouts and nearest-neighbor graphs were generated using the chosen number of components. Graph-based cell clustering was performed with cluster resolution 0.7. Cluster-specific genes were identified using the Wilcoxon rank-sum test with default parameters.
For analyses involving all patients, samples were aggregated using Seurat V3. Although multiple libraries were generated for some samples, only one library per sample was used in this particular analysis. Features expressed in fewer than 10 cells were excluded, as were cells containing fewer than 700 features or more than 10% mitochondrial transcripts. Due to the heterogeneity of AML samples, a sample-specific UMI filtering threshold was imposed: Cells with more UMIs than the cell in the 93rd percentile were removed (based on the estimated doublet rate in Chromium data). Counts were then log-normalized and scaled to 1 × 106 counts/cell. The most variable features were identified using a variance stabilizing transformation (“vst” function with 0.1 ≤ mean.cutoff ≤ 8 and 1 ≤ dispersion.cutoff ≤ Inf). A cell-cycle score was assigned to each cell using the CellCycleScoring function, and the resulting “S.Score” and “G2M.Score” values were “regressed out” using the ScaleData function. PCA was performed on variable features, and elbow plots and JackStraw analysis was performed to assess the contribution of each principal component to the data. t-SNE and UMAP layouts and nearest-neighbor graphs were generated using the top 50 components. Graph-based cell clustering was performed with cluster resolution 0.7, and cluster-specific genes were identified using the Wilcoxon rank-sum test with default parameters. To minimize interpatient differences, a “data set” was defined as all samples from the same patient, and the Seurat FindAnchors and IntegrateData functions were used to integrate the data sets.
For the five time point analysis of UPN 220882, all libraries were aggregated using Seurat V3 and analyzed as above with several modifications: Cells containing fewer than 200 or more than 4,000 features, or more than 10% mitochondrial transcripts, were removed. Variable selection, normalization, scaling, and removal of cell-cycle signal were performed using the recently implemented “SCTransform” function. (In our hands, SCTransform is comparable to the older VST, normalization, and scaling steps.)
Cell-Type Inference
Cell-type inference was performed by SingleR (22) using gene expression data from “The Human Cell Atlas bone marrow single-cell interactive web portal” (24) and the built-in Novershtern DMAP database (23). To use the Human Cell Atlas data with SingleR, raw gene expression counts were first log-normalized and clustered. All lineage predictions were confirmed using established sets of marker genes. When we selected (or excluded) B cells, T cells, and NK cells, we excluded entire clusters that were annotated as such and were underenriched for somatic mutations.
Genetic and Transcriptional Distance Calculations
Genetic distance between two samples was calculated as the number of mutations shared in all cells (i.e., the founding clone mutations) divided by the total number of mutations in both samples. The transcriptional distance between two samples was calculated as the Wasserstein distance (25) between the probability distributions represented by those two samples. Projections of the cells' expression profiles onto the first 10 principal components were used to generate the probability distributions. These calculations were performed using the “transport” R package (43).
Differential Gene Expression and Functional Enrichment
Differential gene expression analysis between presentation and relapse samples was done after removing nonmalignant cells using Wilcoxon rank-sum test in Seurat. Heatmaps and functional enrichment for significantly differentially expressed genes (q-value ≤ 0.05) was done using only the top and bottom 2.5% of genes based on log fold change. Gene ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment for these genes was done using clusterProfiler (44). Overrepresented transcription factor binding motifs were identified using RcisTarget (45); “hg19–500bp-upstream-7species.mc8nr” was used as the reference for motif ranking, and “motifAnnotations_hgnc” was used as the reference for motif annotation. In all other analyses, functional enrichment was performed using ToppFun, with all data sources selected and the default background set (46).
Pseudotemporal ordering was performed using a variety of methods (Supplementary Materials and Methods).
WGCNA and Module Enrichment
WGCNA (26) was performed using UMAP coordinates calculated in PCA space in Seurat. The adjacency matrix was calculated using the PCA rotation scores in WGCNA with a soft power of 6. The adjacency matrix was further transformed to the Topological Overlap matrix. The Topological Overlap matrix was then clustered using hierarchical clustering. The clustering dendrogram was cut using the “dynamictreecut” function (deepSplit = 4, mergeCutHeight = 0.25). The minimum module size was 5. Module eigengene values were then calculated for each gene in order to assign genes to modules. Module expression for each Seurat graph-based cluster was represented using a normalized, averaged expression z-score across all cells in the cluster, and visualized with ComplexHeatmap (47). Enrichment for gene ontology terms, molecular signatures, genomic positions, cell-type markers and immune system pathways was calculated using anRichment (48) for all modules.
Mutant Cell Identification
SNVs and indels were discovered using eWGS and then identified in single cells using cb_sniffer, a Python script that identifies mutant scRNA-seq reads (https://github.com/sridnona/cb_sniffer), with default parameters (16). To identify the FLT3-ITD in single cells, scRNA-seq fastq files were first aligned to a custom reference containing the exact sample-specific FLT3-ITD using Cellranger V3. Cells containing one or more detected mutations were assigned to the founding clone or subclone associated with those mutations. For nested subclones, the cell was assigned to the smallest (most nested) subclone. In rare cases where mutations in single cells corresponded to multiple non-nested (i.e., inconsistent) subclones, cells were not assigned to any subclone. Copy-number alterations were identified using CONICSmat (21).
Differentiation potential was calculated for each patient using Cytotrace with default parameters (37).
Data Availability
eWGS, bulk RNA-seq, and scRNA-seq data generated during this study are available in dbGaP (https://www.ncbi.nlm.nih.gov/gap/) with the primary accession code phs000159.v11.p5 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000159.v11.p5); specific accession codes are in Supplementary Table S2.
Authors' Disclosures
P. Westervelt reports personal fees from Pfizer outside the submitted work. J. DiPersio reports other support from Magenta Therapeutics, WuGen Therapeutics, and Rivervest outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
A.A. Petti: Conceptualization, data curation, formal analysis, investigation, writing–original draft. S.M. Khan: Formal analysis, investigation. Z. Xu: Formal analysis. N. Helton: Resources. C.C. Fronick: Resources. R. Fulton: Resources, data curation. S.M. Ramakrishnan: Formal analysis. S. Nonavinkere Srivatsan: Formal analysis. S.E. Heath: Resources. P. Westervelt: Resources. J.E. Payton: Resources. M.J. Walter: Conceptualization, resources. D.C. Link: Conceptualization, resources. J. DiPersio: Conceptualization, resources. C. Miller: Conceptualization, data curation, formal analysis, writing–review and editing. T.J. Ley: Conceptualization, supervision, investigation, project administration, writing–review and editing.
Supplementary Material
Acknowledgments
This work was supported by an NIH-NCI Program Project Grant (PPG) “Genomics of Acute Myeloid Leukemia” PO1CA101937 [principal investigator (PI): T.J. Ley]: A.A. Petti, N. Helton, R. Fulton, S.E. Heath, P. Westervelt, J.E. Payton, M.J. Walter, D.C. Link, J. DiPersio, C.A. Miller, T.J. Ley; NIH-NCI R35CA197561 (PI: T.J. Ley); NIH-NCI Washington University Paul Calabresi Career Development Award for Clinical Oncology K12 CA167540 (PI: R. Govindan, J. DiPersio): A.A. Petti; NCI R50 CA211782 (PI: C. Miller): C. Miller; NIH-NCI Specialized Program of Research Excellence (SPORE) in Leukemia P50CA171963 (PI: D.C. Link): A.A. Petti, M.J. Walter, D.C. Link, J. DiPersio, T.J. Ley); and Barnes Jewish Hospital Foundation (T.J. Ley).
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
References
- 1. Nowell PC. The clonal evolution of tumor cell populations. Science 1976;194:23–8. [DOI] [PubMed] [Google Scholar]
- 2. Testa JR, Mintz U, Rowley JD, Vardiman JW, Golomb HM. Evolution of karyotypes in acute nonlymphocytic leukemia. Cancer Res 1979;39:3619–27. [PubMed] [Google Scholar]
- 3. Ley TJ. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SLet al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts NDet al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016;374:2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Christopher MJ, Petti AA, Rettig MP, Miller CA, Chendamarai E, Duncavage EJet al. Immune escape of relapsed AML cells after allogeneic transplantation. N Engl J Med 2018;379:2330–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miles LA, Bowman RL, Merlinsky TR, Csete IS, Ooi AT, Durruthy-Durruthy Ret al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 2020;587:477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klco JM, Spencer DH, Miller CA, Griffith M, Lamprecht TL, O'Laughlin Met al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell 2014;25:379–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meyer M, Reimand J, Lan X, Head R, Zhu X, Kushida Met al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc Natl Acad Sci U S A 2015;112:851–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Landau DA, Carter SL, Getz G, Wu CJ. Clonal evolution in hematological malignancies and therapeutic implications. Leukemia 2014;28:34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mroz EA, Tward AD, Pickering CR, Myers JN, Ferris RL, Rocco JW. High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer 2013;119:3034–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Campbell PJ, Getz G, Korbel JO. Pan-cancer analysis of whole genomes. Nature 2020;578:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak Ket al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 2016;539:309–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald Cet al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell 2017;169:750–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Galen P, Hovestadt V, Wadsworth Ii MH, Hughes TK, Griffin GK, Battaglia Set al. Single-cell RNA-Seq reveals AML hierarchies relevant to disease progression and immunity. Cell 2019;176:1265–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Petti AA, Williams SR, Miller CA, Fiddes IT, Srivatsan SN, Chen DYet al. A general approach for detecting expressed mutations in AML cells using single cell RNA-sequencing. Nat Commun 2019;10:3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nam AS, Kim KT, Chaligne R, Izzo F, Ang C, Taylor Jet al. Somatic mutations and cell identity linked by Genotyping of Transcriptomes. Nature 2019;571:355–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Giustacchini A, Thongjuea S, Barkas N, Woll PS, Povinelli BJ, Booth CAGet al. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat Med 2017;23:692–702. [DOI] [PubMed] [Google Scholar]
- 19. Rodriguez-Meira A, Buck G, Clark SA, Povinelli BJ, Alcolea V, Louka Eet al. Unravelling intratumoral heterogeneity through high-sensitivity single-cell mutational analysis and parallel RNA sequencing. Mol Cell 2019;73:1292–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zachariadis V, Cheng H, Andrews N, Enge M. A highly scalable method for joint whole-genome sequencing and gene-expression profiling of single cells. Mol Cell 2020;80:541–53. [DOI] [PubMed] [Google Scholar]
- 21. Müller S, Cho A, Liu SJ, Lim DA, Diaz A. CONICS integrates scRNA-seq with DNA sequencing to map gene expression to tumor sub-clones. Bioinformatics 2018;34:3217–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu Aet al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 2019;20:163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey MEet al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 2011;144:296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hay SB, Ferchen K, Chetal K, Grimes HL, Salomonis N. The Human Cell Atlas bone marrow single-cell interactive web portal. Exp Hematol 2018;68:51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wasserstein LN. Markov processes over denumerable products of spaces describing large systems of automata. Probl Inform Transmission 1969;5:47–52. [Google Scholar]
- 26. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rdet al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang H, Alberich-Jorda M, Amabile G, Yang H, Staber PB, Di Ruscio Aet al. Sox4 is a key oncogenic target in C/EBPalpha mutant acute myeloid leukemia. Cancer Cell 2013;24:575–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lourenço AR, Coffer PJ. SOX4: joining the master regulators of epithelial-to-mesenchymal transition? Trends Cancer 2017;3:571–82. [DOI] [PubMed] [Google Scholar]
- 30. Healy L, May G, Gale K, Grosveld F, Greaves M, Enver T. The stem cell antigen CD34 functions as a regulator of hemopoietic cell adhesion. Proc Natl Acad Sci U S A 1995;92:12240–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova Net al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 2016;540:433–7. [DOI] [PubMed] [Google Scholar]
- 32. Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes Met al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 2000;287:1804–8. [DOI] [PubMed] [Google Scholar]
- 33. Griffith M, Miller CA, Griffith OL, Krysiak K, Skidmore ZL, Ramu Aet al. Optimizing cancer genome sequencing and analysis. Cell Syst 2015;1:210–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fortier JM, Payton JE, Cahan P, Ley TJ, Walter MJ, Graubert TA. POU4F1 is associated with t(8;21) acute myeloid leukemia and contributes directly to its unique transcriptional signature. Leukemia 2010;24:950–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaiserman D, Bird PI. Control of granzymes by serpins. Cell Death Differ 2010;17:586–95. [DOI] [PubMed] [Google Scholar]
- 36. Jiang L, Wang YJ, Zhao J, Uehara M, Hou Q, Kasinath Vet al. Direct tumor killing and immunotherapy through anti-SerpinB9 therapy. Cell 2020;183:1219–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gulati GS, Sikandar SS, Wesche DJ, Manjunath A, Bharadwaj A, Berger MJet al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science 2020;367:405–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nervi B, Ramirez P, Rettig MP, Uy GL, Holt MS, Ritchey JKet al. Chemosensitization of acute myeloid leukemia (AML) following mobilization by the CXCR4 antagonist AMD3100. Blood 2009;113:6206–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rashidi A, DiPersio JF. Targeting the leukemia-stroma interaction in acute myeloid leukemia: rationale and latest evidence. Ther Adv Hematol 2016;7:40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997;3:730–7. [DOI] [PubMed] [Google Scholar]
- 41. Toffalori C, Zito L, Gambacorta V, Riba M, Oliveira G, Bucci Get al. Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat Med 2019;25:603–11. [DOI] [PubMed] [Google Scholar]
- 42. Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson Ret al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schuhmacher DBB, Gottschlich C, Hartmann V, Heinemann F, Schmitzer B. transport: Computation of optimal transport plans and Wasserstein distances. R package version 0.12–2; 2020.
- 44. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans Get al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 2017;14:1083–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 2009;37( Web Server):W305–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847–9. [DOI] [PubMed] [Google Scholar]
- 48. Langfelder P, Miller JA, Horvath S. anRichment: annotation and enrichment functions.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
eWGS, bulk RNA-seq, and scRNA-seq data generated during this study are available in dbGaP (https://www.ncbi.nlm.nih.gov/gap/) with the primary accession code phs000159.v11.p5 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000159.v11.p5); specific accession codes are in Supplementary Table S2.