Abstract
Background:
Following detection, rupture risk assessment for intracranial aneurysms (IAs) is critical. Towards molecular prognostics, we hypothesized that circulating blood RNA expression profiles are associated with IA risk.
Methods:
We performed RNA sequencing on 68 blood samples from IA patients. Here, patients were categorized as either high- or low-risk by assessment of aneurysm size (≥5mm=high-risk) and Population Hypertension Age Size Earlier subarachnoid hemorrhage Site (PHASES) score (≥1=high-risk). Modified F-statistics and Benjamini-Hochberg false discovery rate correction was performed on transcripts per million-normalized gene counts. Protein-coding genes expressed in ≥50% of samples with q-value<0.05 and absolute fold-change ≥2 were considered significantly differentially expressed. Bioinformatics in Ingenuity Pathway Analysis was performed to understand the biology of risk-associated expression profiles. Association was assessed between gene expression and risk via Pearson correlation analysis. Linear discriminant analysis models using significant genes were created and validated for classification of high-risk cases.
Results:
We analyzed transcriptomes of 68 IA patients. In these cases, 31 IAs were large (≥5 mm) while 26 IAs had a high PHASES score. Based on size, 36 genes associated with high-risk IAs, and two were correlated with the size measurement. Alternatively, based on PHASES score, 76 genes associated with high-risk cases, and 9 of them showed significant correlation to the score. Similar ontological terms were associated with both gene profiles, which reflected inflammatory signaling and vascular remodeling. Prediction models based on size and PHASES stratification were able to correctly predict IA risk status, with >80% testing accuracy for both.
Conclusions:
Here, we identified genes associated with IA risk, as quantified by common clinical metrics. Preliminary classification models demonstrated feasibility of Assessing IA risk using whole blood expression.
1.0. BACKGROUND
Intracranial aneurysms (IAs) are pathological outpouchings within cerebral vasculature that are present in about 3–6% of the general population [1]. Aneurysmal rupture is the predominant cause of non-traumatic subarachnoid hemorrhage and has high mortality and morbidity rates (disability rate >50% among survivors) [2–4]. Upon discovery of an unruptured IA, neurointerventionalists must decide whether it should be treated. This decision is critical, since rupture rates are low (annual risk of rupture ∼ 0.5–1% [1]) and complication risks can be significant. The ability to stratify rupture risk, such that dangerous IAs can be treated immediately while less dangerous ones can be monitored, is crucial.
Currently, clinicians use IA size [5–8] (≥ 5 mm[9], or 7 mm[10]) and the patient’s age/health to determine if treatment is warranted. These cutoffs were based on longitudinal prospective studies showing that larger IAs were more likely to rupture [6]. Recently, other clinical metrics, such as the PHASES (Population, Hypertension, Age, Size of IA, Earlier subarachnoid hemorrhage, and Site of IA) score, which use aneurysm characteristics and patient demographics, have been developed in attempts to better stratify risk [7]. However, these metrics, and others like them, require information derived from digital subtraction angiography (DSA), an invasive, expensive, and potentially risky imaging procedure and do not consider the biology of the patient. A non-invasive molecular diagnostic that can assess IA risk could help with patient triage and disease management, as well as patient monitoring during watchful waiting or following treatment.
In previous studies, we investigated molecular changes in the circulating blood that are associated with the presence of IAs in the Circle of Willis [11–17]. Transcriptome profiling revealed distinct expression signatures of the disease, which have broadly been shown to reflect inflammatory cell activation, chemotaxis, and dysregulated inflammatory responses. Using whole blood RNA expression profiles from n=34 patients with IA and n=33 IA-free controls (confirmed on DSA), we identified 18 genes that distinguished patients harboring IAs with an accuracy of 85% and an area under the receiver operating characteristic curve (AUC) of 0.91 in an independent, n=20 cohort [16]. During these studies, we also found that certain molecular signatures delineated IAs sub-populations by size [17]. This led us to hypothesize that patients harboring more dangerous, rupture-prone IAs have distinct circulating gene expression profiles.
Here, we tested our hypothesis by performing whole blood gene expression profiling on blood from unruptured IA patients. To train and test machine learning classifiers of high- and low-risk, we stratified the IA cases based on both IA size (5 mm threshold) and the PHASES score (1%, 5-year risk threshold). Model genes were subjected to bioinformatics analyses via Ingenuity Pathway Analysis (IPA), in order to understand the biologic underpinnings of risk expression profiles. Further, expression of key differentially expressed genes was verified by quantitative polymerase change reaction (qPCR).
2.0. METHODS
2.1. Patient Enrollment
This study was approved by the University at Buffalo Human Research Institutional Review Board (study number 030474433). Written informed consent was obtained from all subjects and the study was carried out in accordance with the approved protocols. Patients at Gates Vascular Institute (Buffalo, NY) receiving cerebral DSA for IA were prospectively enrolled in this study between December 2013 and September 2018. Indications for imaging included confirmation of IAs detected on noninvasive imaging or follow-up imaging of previously-identified IAs.
2.2. Whole Blood Collection and RNA Processing
A volume of 2.5 mL of blood was taken from the femoral access sheath during DSA and transferred into a PAXgene blood RNA tube (PreAnalytiX, Hombrechtikon, Switzerland). Total RNA was extracted using the PAXgene Blood RNA kit (Qiagen, Venlo, Limburg, Netherlands) according to manufacturer’s instructions. Globin mRNA was removed by magnetic-bead capture with the GLOBINclear kit (Ambion, Austin, TX, USA) following manufacturer’s instructions. RNA purity and concentration were assessed by absorbance at 260 nm, then precisely measured by the Quant-iT RiboGreen Assay (Invitrogen, Carlsbad, CA) and the Agilent 2100 BioAnalyzer RNA 6000 Pico Chip (Agilent, Las Vegas, NV) before sequencing. Samples of acceptable purity (260/280 ratio of ∼2) and integrity (RIN ≥ 6.0) were used for RNA sequencing (RNAseq).
2.3. RNA Sequencing
For sequencing, RNA libraries were prepared using the Illumina TruSeq stranded total RNA gold kit (Illumina, San Diego, CA). All samples underwent RNAseq on the Illumina NovaSeq6000 or the HiSeq2500 System (Illumina) in a series of two batches. Samples were demultiplexed with Bcl2Fastq. Per-cycle basecall files generated by the NovaSeq6000 were converted to pre-read FASTQ files using bclfastq version 2.20.0.422 using default parameters. The quality of the sequencing was reviewed using FastQC v.0.11.5. Potential contamination detected using FastQ Screen v.0.11.1. Genomic alignments were performed using HISAT2 v.2.1.0 using default parameters. NCBI reference GRCh38 was used for the reference genome and gene annotation set. Sequence alignments were compressed and sorted into binary alignment map files using samtools v.1.3. Mapped reads for genomic features were counted using Subread featureCounts v.1.6.2 using the parameters -s 2 –g gene_id–t exon–Q 60; the annotation file specified with—a was the NCBI GRCh38 reference from Illumina iGenomes. ComBat-seq (in R) was used to correct raw counts of protein-coding genes with a sum >0 across all samples on a comparison-wise basis, for any bias introduced by sequencing in different batches. Corrected counts were then normalized as transcripts per million (TPM).
2.4. Differential Expression Analysis
To identify gene expression patterns that are different between high- and low-risk IA, we dichotomized samples using 2 risk metrics. We first dichotomized our data based on IA size, using ≥5 mm as a threshold [9, 10] (for cases with multiple IAs, the size of the largest IA was considered). As a second risk metric, we calculated the PHASES score for each case, using ≥1.0 as the cutoff between high-and low-risk (for cases with multiple IAs, the IA with the highest score was considered). Differential gene expression analysis was performed on TPM values for each high- and low-risk comparison, using modified F statistics to assess differential variation in the mean on a gene-by-gene basis. Adjustment for multiple hypothesis testing was performed using Benjamini-Hochberg false discovery rate (FDR) correction. Genes expressed in at least 50% of samples (TPM>0) with a false discovery rate (FDR)-corrected p-value (q-value) <0.05, and with an absolute fold-change in mean expression ≥2 were considered significantly differentially expressed for each comparison. To visualize how differential expression separated high- and low-risk cases, we performed principal component analysis and one minus Pearson correlation hierarchical clustering, using the prcomp packages in R and the Broad Institute’s Morpheus application, respectively.
2.5. RT-qPCR Verification
For verification of RNAseq, we performed quantitative reverse transcription polymerase chain reaction (RT-qPCR) on select genes that were differentially expressed between high-and low-risk cases, as defined by both size and PHASES. Primers for these genes and HPRT1 and GPI (housekeeping genes) were designed in Primer3 software and Primer BLAST (NCBI, Bethesda, MD) to have an ∼60–65°C melting temperature, have length of 15–25 nucleotides, and produce products of 50–250 bps (see Supplemental Table 1). We performed RT-qPCR described in detail elsewhere [18, 19], using the SuperScript III Reverse Transcriptase kit (ThermoFisher, Waltham, MA) and the qScript One-Step SYBR Green Master Mix kit (Quantabio, Beverly, MA, USA). Triplicate reactions of 25 μL (with 10 ng of cDNA and 0.02 μM primers) were run on a StepOne Real-Time PCR System (ThermoFisher, Waltham, MA). Gene-specific amplification was demonstrated by a single peak on the dissociation melt curve. Ct values for the housekeeping genes were recorded for all samples, and average expression differences were calculated via the 2-ΔΔCt method.
2.6. Bioinformatics Analyses
To understand the biology of identified molecular risk profiles, we performed bioinformatics analyses on differentially expressed genes via IPA [20]. We studied terms enriched in significantly differentially expressed genes by disease and biological function term analysis. We considered terms with a Benjamini-Hochberg p-value<0.05 and at least 5 input genes. IPA was also used to generate networks of potential gene interactions by mapping each gene’s identifier to its corresponding gene object in the Ingenuity Knowledge Base and overlaying them onto identified molecular networks in the database. Gene networks were algorithmically generated based on their “connectivity” derived from known interactions between the products of these genes. Networks with p-scores >21 were considered significant.
2.7. Classification Model Generation
To explore the diagnostic potential of each set of differentially expressed genes to distinguish high-risk cases from low-risk cases, we developed a linear discriminant analysis (LDA) model using the scikit-learn machine learning library in Python v3.8.5. TPM data for the differentially expressed genes was used as input, and samples were randomly split into training (n=48) and testing (n=20) groups. The model was evaluated in training by calculating the area under the receiver operating characteristic (ROC) curve (AUC). We calculated accuracy and used a confusion matrix to determine model sensitivity and specificity in testing group. To reduce selection bias, the classification model creation process was repeated for 100 random iterations of the testing/training split, and all performance metrics were reported as averages.
To assess the most informative genes in the model, we tested their linear association with the risk metrics by calculating Pearson’s correlation coefficient (PCC) (Wald’s test used for significance testing). We stratified the correlation strength as follows: 1 ≥ |PCC| ≥ 0.8 represented ‘very strong’ correlation, 0.79 ≥ |PCC| ≥ 0.6 represented ‘strong’ correlation, 0.59 ≥ |PCC| ≥ 0.4 represented ‘moderate’ correlation, 0.39 ≥ |PCC| ≥ 0.2 represented ‘weak’ correlation, and |PCC| < 0.19 represented a ‘very weak’ or no correlation [21]. Genes with a p-value<0.05 and a PCC>0.3 were reported.
3.0. RESULTS
3.1. Study Population
We analyzed 93 whole blood samples from patients with IAs, and excluded 25 samples due to additional potentially confounding cerebrovascular or inflammatory conditions. The demographics and comorbidities of the remaining 68 patient samples we analyzed is presented in Table 1. For risk assessment, 31 had large (≥5 mm) high-risk IAs (37 had small), and 26 had a risky PHASES score >1(42 were <1) (See Supplemental Table 2 for patient data of the high- and low-risk cohorts when stratified by each metric). In the entire cohort, IA size (maximum diameter on DSA) ranged from 1 to 19 mm. The 68 RNA samples had an average 260/280 of 1.98 and average RNA integrity number of 8.46. In RNAseq, an average of 46.39 million sequences per sample and an average alignment rate of 96.63% were obtained. For all samples, RNA quality and sequencing metrics are reported in Supplemental Table 3.
Table 1:
Patient and Aneurysm Characteristics.*
| Patient Characteristics | |
|---|---|
|
| |
| Age (average years ± s.d.) | 57.3 ± 12.0 |
| Smoking (n/ntotal) | 19/68 (27.9%) |
| Hypertension (n/ntotal) | 30/68 (44.1%) |
| Family history of IA (n/ntotal) | 9/68 (13.2%) |
| Patients with multiple IAs (n/ntotal) | 15/68 (22.1%) |
| Aneurysm Characteristics | |
|
| |
| IA size (average mm ± s.d.) | 4.6 ± 3.2 |
| IA location (n/ntotal) | |
| ACA/ACom | 9/89 (10.1%) |
| BA/BT | 7/89 (7.9%) |
| ICA | 44/89 (49.4%) |
| MCA | 13/89 (14.6%) |
| PCA/PCom | 16/41 (18.0%) |
These clinical factors were retrieved from patients’ medical records. With the exception of age, these datapoints were quantified as binary data points. Abbreviations: IA=intracranial aneurysm, n=number, s.d.=standard deviation, ACA=anterior cerebral artery, ACom=anterior communicating artery, BA=basilar artery, BT=basilar terminus, ICA=internal carotid artery, MCA=middle cerebral artery, PCA=posterior cerebral artery, PCom=posterior communicating artery
3.2. Differentially Expressed Genes between High- and Low-Risk Cases
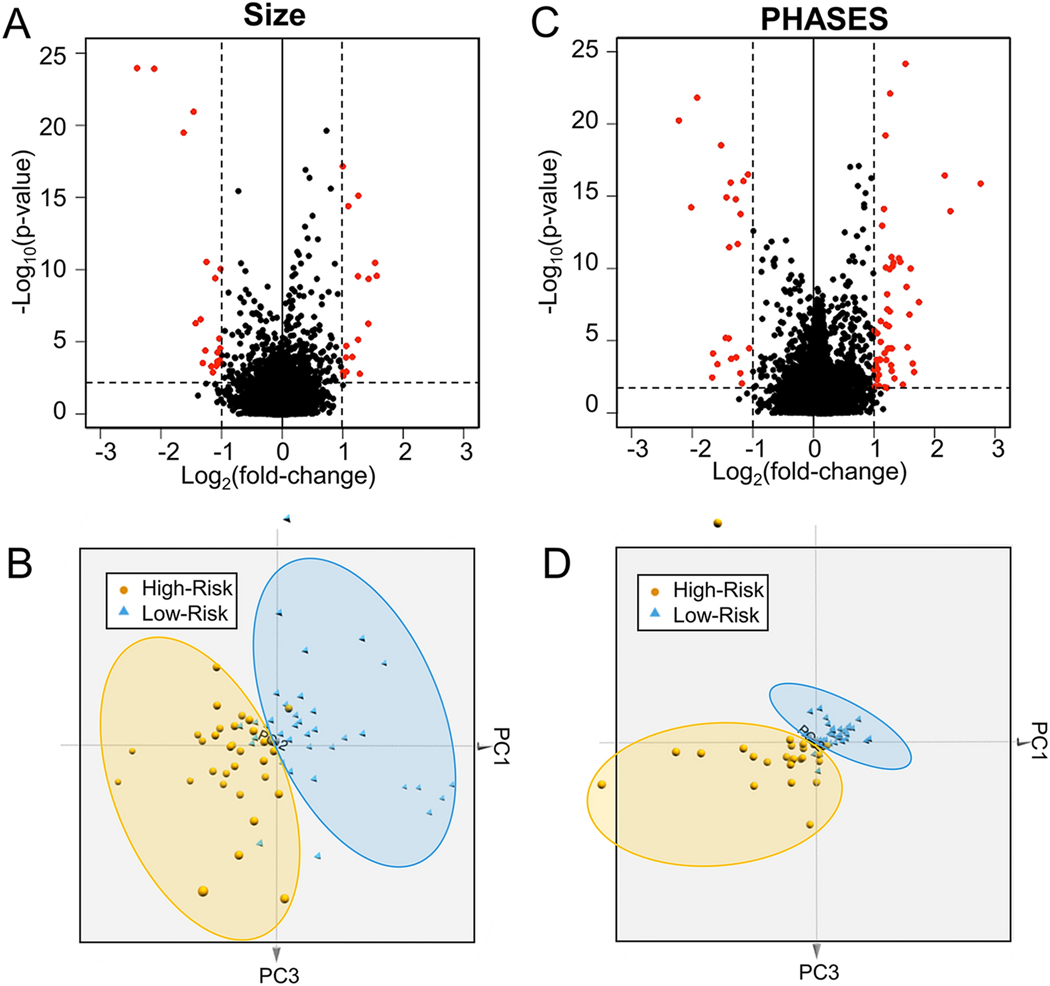
From our RNA sequencing data, we queried 66,023 transcripts, 47,084 of which had detectable expression. Of these, 18,738 (39.80%) were protein-coding genes and 14,902 of which had expression in ≥50% of the samples. The volcano plots in Figure 1 shows expression differences between the high-and low-risk IA patients in terms of average fold-change in expression and significance level. We found 36 significantly differentially expressed genes (q-value<0.05, an absolute fold-change≥2) between cases with large and small IAs (16 increased and 20 decreased) and 76 significantly differentially expressed genes between those with high and low PHASES score (54 increased and 22 decreased). The principal component analysis in Figure 1 shows how these sets of differentially expressed genes were able to cluster the high- and low-risk samples in each analysis. For size, one large IA (≥5 mm) case was grouped with the low-risk IAs, and 5 low-risk cases were grouped with the high-risk ones. For PHASES, one high-risk case was grouped with the low-risk IAs, and 2 low-risk cases were grouped with the high-risk ones.
Figure 1: Differential Gene Expression Analysis.
Volcano plots demonstrate the differential expression between low- and high-risk groups based on risk stratification by either (A) aneurysm size or (B) PHASES score. Differentially expressed genes in red have a q-value<0.05 and an absolute fold-change≥2. Three-dimensional principal component analysis demonstrates separation between low- and high-risk samples based on both (C) aneurysm size and (D) PHASES score. Abbreviations: PC=principal component, PHASES=Population, Hypertension, Age, Size, Earlier subarachnoid hemorrhage, Site
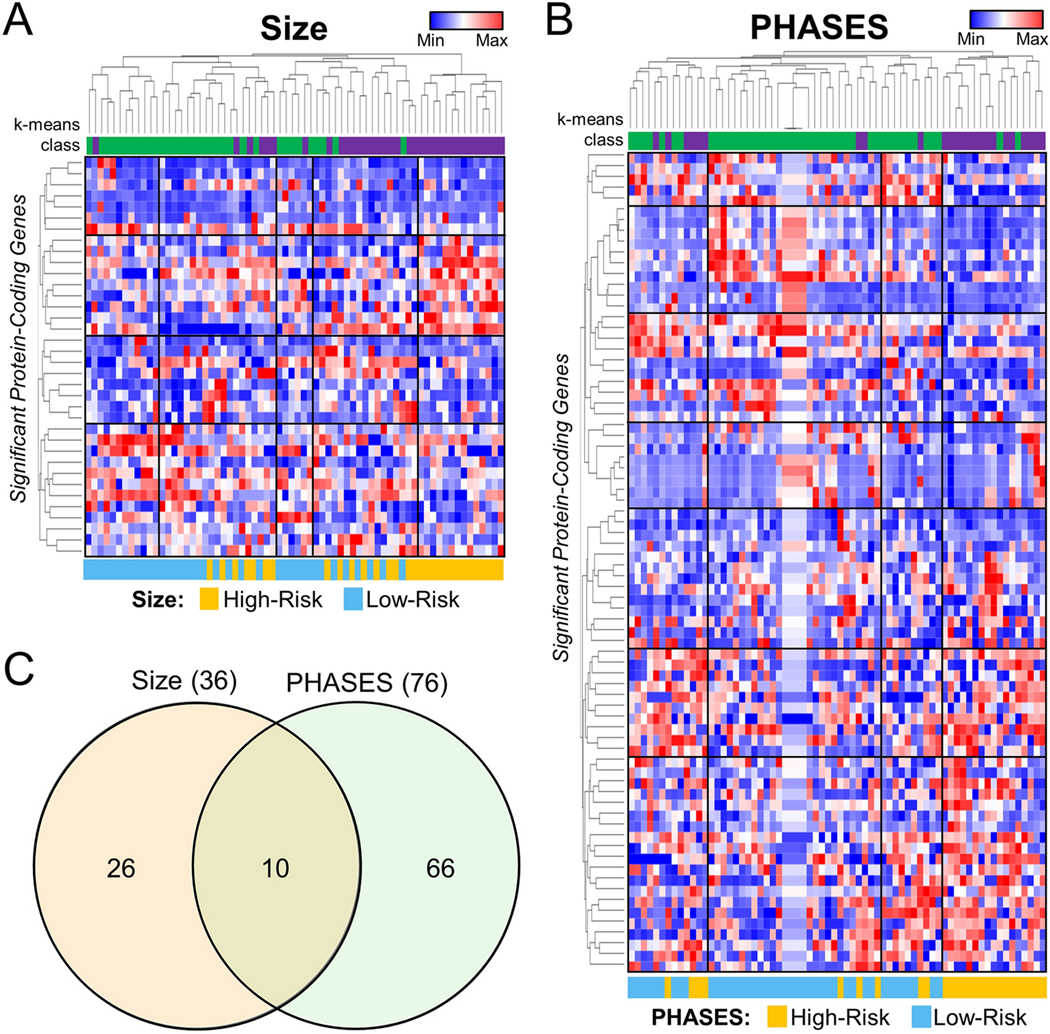
Additional unsupervised analysis was performed by hierarchal and k-means clustering, as shown adjacent to the heatmaps in Figure 2. The k-means clustering using the differentially expressed genes for each analysis showed correct grouping of high- and low-risk samples. 84% (57/68) of the samples were clustered to their proper group with respect to size and 88% (60/68) of the samples were clustered to their proper group with respect to PHASES. Such groupings on PCA and hierarchical clustering were not observed when all detected protein-coding genes were used (Supplemental Figure 1). Overall, 10 genes were shared between the two analyses, as demonstrated in the Venn diagram (detailed plots of average TPMs for each of these 10 genes are shown in Supplemental Figure 2). See Supplemental Table 4 for all significant genes and their associated fold-changes and q-values). Based on an analysis by ProteINSIDE, 34 of the 102 unique differentially expressed genes (33.33%) were potentially secreted by cells (genes that were called signal peptide with a TargetP score of 1 or 2). Thus, their protein products may enter the blood stream and be detectable by immunoassay.
Figure 2: Visualization of Differential Expression.
Heatmaps generated using TPM expression data of significant genes show broad differences between low- and high-risk groups whether risk was classified by (A) aneurysm size or (B) PHASES score. Hierarchical clustering separated the two groups with the majority of low-risk samples on the left of the heatmap (log-transformed, normalized TPMs) and the majority of high-risk cases at the right. Furthermore, K means clustering grouped 84% of samples in the correct cohort by size, and 88% of samples in the correct cohort by PHASES. C). Venn Diagram shows 10 genes were identified as differentially expressed upon stratification of cases by either aneurysm size or PHASES score. Abbreviations: Min=minimum, Max=maximum, PHASES= Population, Hypertension, Age, Size, Earlier subarachnoid hemorrhage, Site
To verify expression differences we found, RT-qPCR analysis was performed on 6 of the 10 significant genes shared between the differential expression analyses (DEFA1, FN1, HBA1, MSLN, RPL3L, and USP9Y) in a subset of 30 of the patients. Supplemental Figure 3 shows that the expression differences between high-and low-risk cases (defined by both size and PHASES) for RNAseq and qPCR in that dataset. Largely, expression differences were detected to be of the same direction and of similar magnitudes when comparing RNAseq to qPCR (differences highlighted by an “*”). The only prominent exception was the expression of MSLN. These differences may be due to the fact that the qPCR testing was only done in a subset of patients, that RNA sequencing has a higher dynamic range, or that the primer pairs may not have had efficiency=1.0.
3.3. Bioinformatics Analyses
Genes differentially expressed between small and large IAs (≥5 mm threshold) were significantly enriched for disease and biological function terms related to the cardiovascular system, like angiogenesis, atherosclerosis, and occlusion of blood vessel. There were also multiple terms associated with cellular movement, including chemotaxis and movement of myeloid cells. We observed enrichment of similar terms in the differentially expressed genes based on PHASES stratification. These included angiogenesis, atherosclerosis, cell movement of myeloid cells, and cell movement of phagocytes. Unique terms associated with PHASES-derived differentially expressed genes were enrichment of cellular movement and/or infiltration of many immune cells (leukocytes, neutrophils, phagocytes) terms. Interestingly, systemic lupus erythematosus was also enriched in this gene set, which may reflect the burgeoning relationship between IA growth/rupture and SLE that has been reported in the literature [22, 23]. Supplemental Table 5 shows all significant disease and biological function terms for both comparisons.
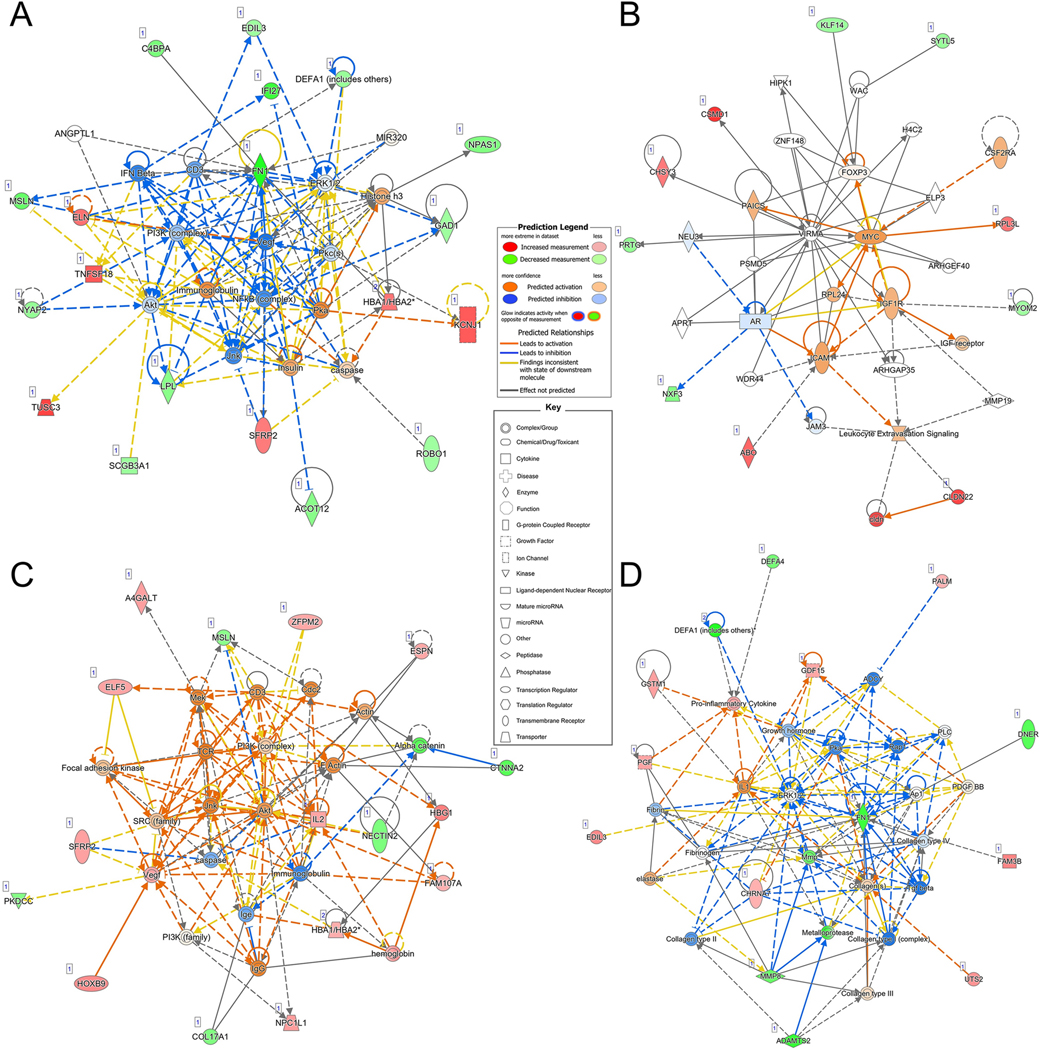
Network analysis was performed to examine potential interactions between the gene sets and terms identified in the Ingenuity Knowledge Base. As shown in Figure 3A-B, there were 2 significant networks of genes (score>21) associated with size-based risk. The first network (Figure 3A, P-score=50) was associated with “embryonic development”, “organismal development”, and “tissue development”, with inflammatory signaling molecule nodes, including NFKB, ERK1/2, and PI3K. The second network (Figure 3B, P-score of 22) was associated with “organ development”, “tissue development”, and “tissue morphology” processes, and had nodes of interaction surrounding MYC and VIRMA. As shown in Figure 3C-D, there were also 2 significant networks of genes (score>21) associated with PHASES-based risk. The first network (Figure 3C, P-score=34) was enriched for “connective tissue disorders”, “organismal development”, and “organismal injury and abnormalities” ontologies. PI3K was a central node of this network along with Akt. The second network (Figure 3D, P-score=29) was associated with “cellular movement”, “hematological system development and function”, and “immune cell trafficking” and had central genes of ERK1/2, FN1, MMP, and collagens. Supplemental Table 6 presents the details of all Networks in Figure 3.
Figure 3: Ingenuity Pathway Analysis Results.
Networks derived from Ingenuity Pathway Analysis based on differentially expressed genes associated with risk as stratified by aneurysm size or PHASES score. For input differentially expressed genes, red indicates increased expression in high-risk aneurysm cases, while green denotes decreased expression; the color intensity reflects fold-change. Non-differentially expressed genes with known interactions are not colored. Direct and indirect relationships are shown by solid and dashed lines, respectively. There were 2 significant networks associated with the differentially expressed genes identified using aneurysm size as a risk metric. A). The first network, p-score of 50, reflected embryonic development, organismal development, tissue development and had a main node at NFKB. B). The second network had a score of 22 and its top diseases and functions were organ development, tissue development, and tissue morphology. C). The most significant network created using DEGs identified when assessing risk by PHASES score had a pscore of 34 and reflected connective tissue disorders, organismal development, and organismal injury and abnormalities. D). The second network with a score of 29 was associated with cellular movement, hematological system development and function, and immune cell trafficking. Abbreviations: PHASES=Population, Hypertension, Age, Size, Earlier subarachnoid hemorrhage, Site
3.4. Classification Models of IA Risk
To investigate if the sets of differentially expressed genes could delineate high-risk cases from low-risk cases, we estimated the predictive performance of an LDA model over 100 iterations. The resultant risk models (based on size and based on PHASES score) had high training performance, with average AUCs across 100 randomizations of 0.99 (95% CI: 0.985–0.988) and 1.0 (95% CI: 1.0–1.0), respectively. The estimated testing performance was also high, with an average accuracy of 82% for the size model and 88% for the PHASES model. The size model had an average sensitivity of 0.83 and an average specificity of 0.81, while the PHASES model had an average sensitivity and specificity of 0.78 and 0.95 respectively. See Table 2 for a summary of model performances. For LDA model codes, see Supplemental Table 7.
Table 2.
Linear Discriminant Analysis Prediction Model Performance.*
| IA risk stratified by size | IA risk stratified by PHASES | |
|---|---|---|
|
Avg. Training AUC
(95% CI) |
0.986 (0.985–0.988) |
1 (1.00–1.00) |
|
Avg. Testing Accuracy
(95% CI) |
0.820 (0.804–0.835) |
0.883 (0.872–0.894) |
|
Avg. Sensitivity
(95% CI) |
0.834 (0.809–0.860) |
0.778 (0.753–0.802) |
|
Avg. Specificity
(95% CI) |
0.807 (0.784–0.830) |
0.953 (0.939–0.967) |
Abbreviations: Avg.=average, IA=intracranial aneurysm, CI=confidence interval
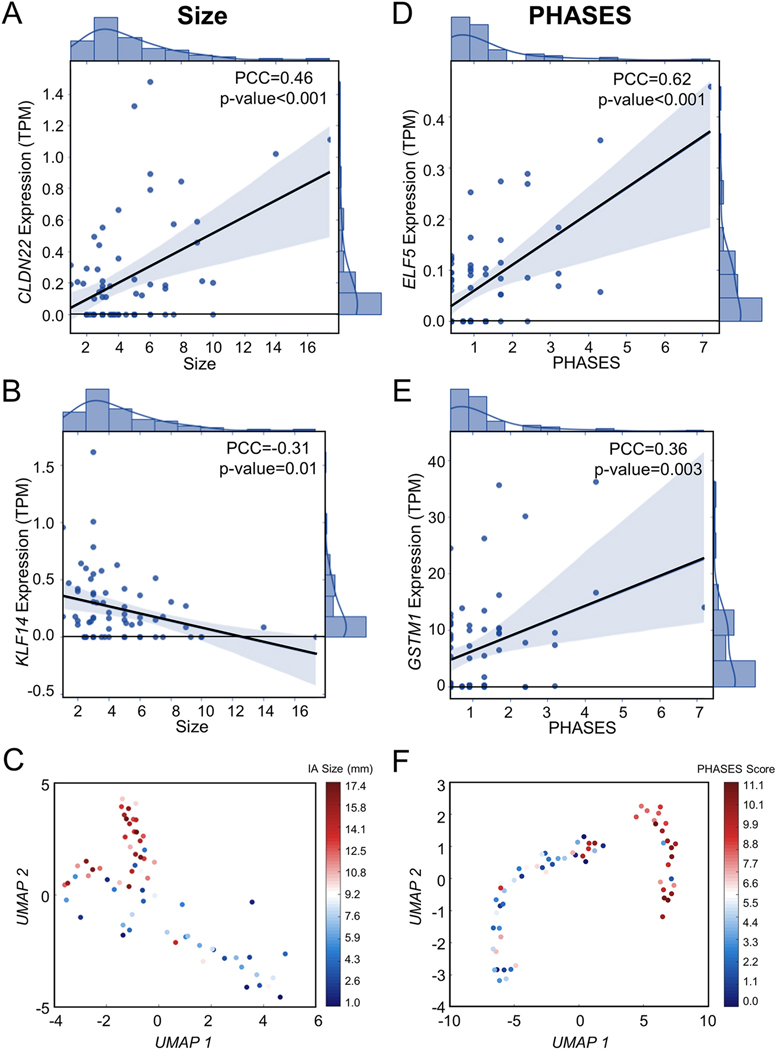
Pearson correlation analysis was performed to determine the most informative, linearly risk-related genes in the model. This analysis found that 2 genes were significantly (p-value<0.05 and PCC>0.30) linearly related to IA size (CLDN22, KLF14). Additionally, 9 genes were significantly linearly related to PHASES score (CLGN, DDX3Y, ELF5, GSTM1, LOXL4, PALM, RIMBP2, USP9Y, UTS2). Correlation plots for the top 2 most strongly correlated genes with size and PHASES are shown in Figure 4 (Supplemental Figure 4 shows the remaining correlation plots for PHASES).
Figure 4: Correlation of Differentially Expressed Genes with IA Risk.
Two genes were significantly correlated with IA size (PCC>0.3, p-value<0.05). A). CLDN22 had a moderate, positive correlation (PCC=0.46) while (B) KLF14 had a weak, negative correlation (PCC=−0.31). Per these criteria, expression levels of 9 genes were correlated with PHASES score. The top 2 most significant are shown here (see Supplemental Figure 2 for correlation plots of the remaining genes). C). ELF5 had a strong, positive correlation with a PCC of 0.62. D). GSTM1 had a weak, positive correlation (PCC=0.36). Abbreviations: PCC=Pearson correlation coefficient, TPM=transcripts per million, PHASES=Population, Hypertension, Age, Size, Earlier subarachnoid hemorrhage, Site
Furthermore, we used Seurat [24] and UMAP [25] to perform dimensionality reduction and visualize how the samples (based on all DEGs) distribute in terms of size and PHASES, as an unbiased way of measuring dose-response to our outcome variable. Here, we completed parametrized, density-based clustering of the 68 samples based on collective expression of the DEGs (36 for IA size and 76 for PHASES). IA size and PHASES score were withheld from this unsupervised analysis. A 2D projection of the data, based on the top 2 UMAP components (UMAP 1 and UMAP 2) was visualized, and the holdout feature (IA size or PHASES) was superimposed as a scaled colormap, with red indicating higher risk and blue indicating lower risk. As shown in Figure 4, the collective expression of the DEGs was able to separate patients into high (larger IA, higher PHASES) and low (smaller IA, lower PHASES) IA rupture risk groups, with largest IA (and highest PHASES) and smallest IA (and lowest PHASES) cases at either end of the groupings.
4.0. DISCUSSION
When an IA is detected, clinicians must weigh the risk of rupture and the risk of treatment complications. However, clinical risk assessment metrics all rely, in one way or another, on medical imaging, typically by invasive DSA, and do not consider the biology of the patient. A rupture risk blood test could work in concert with current clinical diagnosis paradigms by providing clinicians with a non-invasive, biologic method to assess aneurysm risk or a means to monitor patients more frequently during watchful waiting or after intervention. Over the past few decades, researchers have investigated the potential of blood-based biomarkers for intracranial aneurysm rupture risk. A review by Hussain et al. [26] highlighted seven blood-based biomarker studies for ruptured IAs, the majority of which focused on adhesion molecules such as E-selectin [27, 28] and ICAM [29, 30]. Others examined hormone levels, including T3 and T4 [31, 32], and S-100 protein levels [33]. Clearly, multiple proteins were correlated with outcomes and had increased levels in ruptured cases, but since many of these proposed biomarkers are also dysregulated in other pathophysiological states, they may not be sensitive enough to aneurysm [34–36]. Recent studies have taken advantage of the power of transcriptome profiling to screen for multitudes of potential RNA expression markers that may be associated with many different facets of the disease. Indeed, reports have shown differences in mRNA expression, as well as differences in noncoding elements, like miRNAs [37] [38] and circular RNAs [39], between individuals with unruptured and ruptured IAs. However, these efforts have identified classifiers of rupture status (i.e., discriminants of IAs that are already ruptured), rather than which unruptured IAs are at higher risk of rupturing.
In this study, we performed transcriptome analysis on whole blood from patients with IA to identify genes related to IA rupture risk in unruptured cases. To do this, we assessed rupture propensity, by using two common clinical metrics, IA size and PHASES score. Based on differential expression analysis, there were 36 significant genes associated with risk assessed by IA size, and 76 significant genes associated with risk assessed by PHASES score (10 genes overlapped). Using these size- and PHASES-associated genes, we trained and tested LDA prediction models, which both achieved >80% average testing accuracy. While the risk model that classified cases by PHASES had higher testing accuracy (88.3% vs 82.0% for size-based risk classification), it had lower sensitivity (77.8% vs. 83.4%), which is less ideal for identifying all truly rupture-prone IAs. We suspect the genes in these models were able to robustly classify higher-risk IAs because they reflected important, ongoing biological processes related to IA natural history and rupture.
Our bioinformatics analyses show that differentially expressed, IA risk genes were related to two, broad biologic phenomena, namely inflammatory cell movement/trafficking and vascular remodeling. It is widely known that inflammation plays a crucial role throughout IA natural history [40, 41]. Immune cells infiltrate the aneurysm wall and coordinate inflammatory processes, cytotoxic responses, and destructive remodeling [42, 43], and are increasingly prevalent as the lesion grows and progresses to rupture [44–48]. Numerous model genes that were increased in high-risk IA patients, such as GDF15, IL2, and TNFSF18, play roles in cytokine activity and immune signaling. Specifically, GDF15 encodes a ligand of the TGF- β family and is upregulated in response to inflammation and oxidative stress, important processes in IA [49]. Expression of GDF15 is rapidly induced in macrophages by TGF-β, TNFα, and IL-1 (other key molecules in IA that are present as nodes in our network analyses), and has been shown to be vasoprotective in stroke models[50]. Other model genes reflect increased production of immune cell populations in patients with IA. For example, decreased PVRL2 (aka NECTIN2), which can act as a co-inhibitor of T cell function, could indicate increased T cell proliferation in high-risk IAs. Increased IL2, which is produced by activated T lymphocytes, also indicates increased proliferation of immune cells. Indeed, such expression patterns were related to cell movement of myeloid cells and phagocytes and cell movement of granulocytes, leukocytes, and neutrophils in our IPA analysis.
In addition to inflammatory cell infiltration and responses, many other model genes were related to vascular remodeling. During aneurysm growth and rupture, inflammatory infiltrates, along with resident, pro-inflammatory smooth muscle cells, coordinate destructive remodeling events. These processes are accompanied by destruction of cell adhesions, matrix degradation and turnover, disruption of the internal elastic lamina, and lipid accumulation, in response to changing blood flow and wall stress forces. The genes selected for our models were demonstrated by IPA analysis to reflect disease and biological function terms such as, angiogenesis, atherosclerosis, vasculogenesis, and development of vasculature, and to interact with networks that included nodes at FN1, ICAM, and actin. Our data generally reflected dysregulation of such remodeling processes. For example, several genes involved in eutrophic tissue growth, like LOXL4 and ELN (encoding elastin) that initiate collagen and elastin linking, were increased, while genes encoding degradative proteins, like MMP8 and ADAMTS2 (a disintegrin) that degrades extracellular matrix, were decreased. On the other hand, some genes involved in decreased matrix integrity, like MMRN2 and TMPRSS9, that can act to negatively affect angiogenesis, were increased, while other structural components, like FN1 (encoding fibronectin) and CTNNA2 were decreased. These complex expression patterns in circulating blood cells might suggest immune priming for regulating vascular structure turnover following IA wall infiltration, which appears to be greater in cases with higher rupture risk.
Indeed, several of the differentially expressed model genes have also been identified to be differentially expressed in aneurysm tissue. Kleinloog et al. [51] identified 1,489 differentially expressed genes between ruptured and unruptured IA tissues, which were associated with enriched immune response and lysosome pathways. Interestingly, CTNNA2 (encoding a catenin involved in cell adhesion), which was lower in high-risk cases in our study, was found to be downregulated in ruptured IAs. Similarly, Nakaoka et al. [52] compared gene expression in early ruptured IAs to unruptured IAs and identified 1,046 differentially expressed genes. These genes were associated with inflammatory responses, immune responses, and phagocytosis. Both LPL (encoding Phospholipase A1) and NPAS1 (encoding a PAS-family transcription factor) were found to be downregulated in ruptured IA tissue, and were also lower in high-risk IAs in our study. Proteomics studies on IA tissues have also shown gene products of model gene to be differentially present in ruptured IA tissue. Sharma et al. [53] showed decreased DEFA1 (defensin alpha 1) and FN1 in ruptured IAs. Both were also decreased in the blood of high-risk cases in the current study. We also previously demonstrated that FN1 was further decreased in the whole blood of IA patients compared to IA-free controls [16], while DEFA1 was lower in peripheral blood mononuclear cells of patients with IA, compared to IA-free controls [15]. These findings may indicate that some peripheral changes in blood cells of patients with higher-risk IAs permeate into the IA tissue. They also may show that some peripheral expression aberrations may progressively change as IAs develop towards rupture (e.g., FN1 and DEFA1 expression). Future studies investigating expression in paired IA tissue and blood samples would enable a better understanding of this phenomenon. Additional work in animal models will also likely be needed to unravel the mechanistic roles of such genes and the interplay between blood expression and tissue pathology.
This study has several limitations. First, our samples were collected at a single center. Multi-centered studies are underway, which will help eliminate potential selection bias. Moreover, patients were enrolled for a one-time blood sample collection, which precludes longitudinal studies investigating the biomarker’s relationship to rupture. Second, cohorts in this study were not matched, and thus some DEGs may also be related to covariates. A factorial MANCOVA[54] with evaluation of group-covariate interactions (factors included sex, smoking status, hypertension, IA family history) for the PHASES and size DEGs revealed that most differential expression was not related to covariates, as only 26% of PHASES DEGs and 22% of size DEGs had covariate interaction (see Supplemental Table 8). Also, since patients with other vascular or inflammatory conditions were excluded, the resultant models may not work in populations with such confounders. Third, the sample size of our study was relatively small. This may be one reason that the identified significantly DEGs do not largely overlap with other IA blood profiling studies. Future studies in larger, independent, multicenter datasets are needed to assess true biomarker performance and identify robust trends. Fourth, our population contained IAs located mostly in the anterior Circle of Willis, which could have confounded our results. Fifth, additional validation studies in large independent cohorts using qPCR are needed to determine which differentially expressed genes would be the most reliable markers as the agreement in the small cohort examined here was not overwhelming. Lastly, IA size and PHASES score are only surrogates of IA rupture risk (publications have even suggested that PHASES and other clinical metrics may underestimate rupture risk [55–58]). More accurate prediction models will be needed to better delineate high- from low-risk IAs. Other IA features (e.g., irregularity or presence of blebs) and more other clinical metrics could be assessed. However, longitudinal studies identifying circulating expression changes in patients with unstable, growing IAs (which are truly at higher risk for rupturing) are needed, and are currently underway.
5.0. CONCLUSIONS
In conclusion, transcriptome profiling on whole blood from patients with IA enabled us to successfully identify gene expression signatures associated with greater aneurysmal rupture risk. Machine learning models based on the expression of these signatures was able to correctly classify high-risk IAs from low-risk ones with >80% accuracy in our data. The genes used in these models generally reflect biological processes that have been shown to be critical in IA growth and rupture, namely inflammatory signaling and trafficking, and vascular remodeling. In the future, these models could be used to assess rupture risk prior to imaging and assist in more frequent monitoring during watchful waiting. A blood-based diagnostic to assess IA risk could facilitate better, more proactive IA management and treatment planning, ultimately reducing the number of ruptures. In the future, we are planning to conduct large, multi-center studies with heterogeneous populations to validate our biomarker and determine the patient population that can be best served by this biomarker. Furthermore, we intend to conduct animal studies to understand how the genes we have identified mechanistically influence aneurysm development and progression towards rupture, which may create opportunities for nonsurgical treatments.
Supplementary Material
Acknowledgements
The authors thank the patients who participated in this study. Special thanks also to Brandon Marzullo, MS and Jonathan Bard, MS for RNAseq data analysis assistance, and Jennifer Gay, CCRP and Elizabeth Bommaraju for study protocol management. This work was performed in part at the New York State Center of Excellence in Bioinformatics and Life Sciences’ Genomics and Bioinformatics Core.
Funding
Research reported in this publication was supported, in part, by the Brain Aneurysm Foundation (VMT), and by the National Institute of Neurological Disorders and Stroke of the NIH under award number 1 R43 NS115314-01 to Neurovascular Diagnostics, Inc (VMT). Additional support from the National Heart, Lung, and Blood Institute of the NIH under award number 1R01HL152270-01 (AC) and a UCLA Exploratory Research Grant (AC).
Availability of data and materials
The datasets used in the current study are available from the corresponding author upon reasonable request.
Footnotes
DECLARATIONS
Ethics approval and consent to participate
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Human Research Institutional Review Board of the University at Buffalo (study number 030474433). Informed consent was obtained from all subjects involved in the study and the study was carried out in accordance with the approved protocols.
Consent for publication
No specific patient information or images are given in the manuscript.
Competing Interests
KEP—None.; AC— Awardee of NIH grant, 1R01HL152270-01 and a UCLA Exploratory Research Grant.; BAS—None.; LC—None.; SSV—None.; MW—None.; AM–None.; AAD—None.; J-KB—None.; MM—None.; KVS—Consulting/teaching: Canon Medical Systems Corporation, Penumbra Inc., Medtronic, Jacobs Institute. Co-Founder: Neurovascular Diagnostics, Inc..; AHS—Financial Interest/Investor/Stock Options/Ownership: Adona Medical, Inc., Amnis Therapeutics, BlinkTBI, Inc, Boston Scientific Corp (for purchase of Claret Medical), Buffalo Technology Partners, Inc., Cardinal Consultants, LLC, Cerebrotech Medical Systems, Inc, Cognition Medical, Endostream Medical, Ltd, Imperative Care, Inc., International Medical Distribution Partners, Neurovascular Diagnostics, Inc., Q’Apel Medical, Inc., Radical Catheter Technologies, Inc., Rebound Therapeutics Corp. (Purchased 2019 by Integra Lifesciences, Corp), Rist Neurovascular, Inc., Sense Diagnostics, Inc., Serenity Medical, Inc., Silk Road Medical, Spinnaker Medical, Inc., StimMed, Synchron, Three Rivers Medical, Inc., Vastrax, LLC, VICIS, Inc., Viseon, Inc. Consultant/Advisory Board: Amnis Therapeutics, Boston Scientific, Canon Medical Systems USA, Inc., Cerebrotech Medical Systems, Inc., Cerenovus, Corindus, Inc., Endostream Medical, Ltd, Imperative Care, Inc., Integra LifeSciences Corp., Medtronic, MicroVention, Minnetronix Neuro, Inc., Northwest University – DSMB Chair for HEAT Trial, Penumbra, Q’Apel Medical, Inc., Rapid Medical, Rebound Therapeutics Corp., Serenity Medical, Inc., Silk Road Medical, StimMed, Stryker, Three Rivers Medical, Inc., VasSol, W.L. Gore & Associates. National PI/Steering Committees: Cerenovus LARGE Trial and ARISE II Trial, Medtronic SWIFT PRIME and SWIFT DIRECT Trials, MicroVention FRED Trial & CONFIDENCE Study, MUSC POSITIVE Trial, Penumbra 3D Separator Trial, COMPASS Trial, INVEST Trial. Research Grants: Co-investigator: NIH/NINDS 1R01NS091075 Virtual Intervention of Intracranial Aneurysms; Role: Co-Principal Investigator NIH-NINDS R21 NS109575-01 Optimizing Approaches to Endovascular Therapy of Acute Ischemic Stroke.; VMT—Principal investigator: National Science Foundation Award No. 1746694 and NIH NINDS award R43 NS115314-0. Awardee of a Brain Aneurysm Foundation grant, a Center for Advanced Technology grant, and a Cummings Foundation grant. Co-founder: Neurovascular Diagnostics, Inc., QAS.AI, Inc.
REFERENCES
- 1.Rinkel GJE, Djibuti M, Algra A, van Gijn J. Prevalence and Risk of Rupture of Intracranial Aneurysms. Stroke. 1998. 1998/01/01;29(1):251–6. [DOI] [PubMed] [Google Scholar]
- 2.de Rooij NK, Linn FH, van der Plas JA, Algra A, Rinkel GJ. Incidence of subarachnoid haemorrhage: a systematic review with emphasis on region, age, gender and time trends. J Neurol Neurosurg Psychiatry. 2007. Dec;78(12):1365–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olafsson E, Hauser WA, Gudmundsson G. A population-based study of prognosis of ruptured cerebral aneurysm: mortality and recurrence of subarachnoid hemorrhage. Neurology. 1997. May;48(5):1191–5. [DOI] [PubMed] [Google Scholar]
- 4.Bederson JB, Connolly ES Jr., Batjer HH, Dacey RG, Dion JE, Diringer MN, et al. Guidelines for the management of aneurysmal subarachnoid hemorrhage: a statement for healthcare professionals from a special writing group of the Stroke Council, American Heart Association. Stroke. 2009. Mar;40(3):994–1025. [DOI] [PubMed] [Google Scholar]
- 5.Varble N, Tutino Vincent M, Yu J, Sonig A, Siddiqui Adnan H, Davies Jason M, et al. Shared and Distinct Rupture Discriminants of Small and Large Intracranial Aneurysms. Stroke. 2018. 2018/04/01;49(4):856–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiebers DO. Unruptured intracranial aneurysms: natural history, clinical outcome, and risks of surgical and endovascular treatment. The Lancet. 2003. 2003/07/12/;362(9378):103–10. [DOI] [PubMed] [Google Scholar]
- 7.Greving JP, Wermer MJ, Brown RD Jr., Morita A, Juvela S, Yonekura M, et al. Development of the PHASES score for prediction of risk of rupture of intracranial aneurysms: a pooled analysis of six prospective cohort studies. (1474–4465 (Electronic)). [DOI] [PubMed] [Google Scholar]
- 8.Investigators UJ. The Natural Course of Unruptured Cerebral Aneurysms in a Japanese Cohort. New England Journal of Medicine. 2012. 2012/06/28;366(26):2474–82. [DOI] [PubMed] [Google Scholar]
- 9.Murayama Y, Takao H, Ishibashi T, Saguchi T, Ebara M, Yuki I, et al. Risk Analysis of Unruptured Intracranial Aneurysms. Stroke. 2016. 2016/02/01;47(2):365–71. [DOI] [PubMed] [Google Scholar]
- 10.Wermer MJH, van der Schaaf IC, Algra A, Rinkel GJE. Risk of Rupture of Unruptured Intracranial Aneurysms in Relation to Patient and Aneurysm Characteristics. Stroke. 2007. 2007/04/01;38(4):1404–10. [DOI] [PubMed] [Google Scholar]
- 11.Tutino VM, Poppenberg KE, Li L, Shallwani H, Jiang K, Jarvis JN, et al. Biomarkers from circulating neutrophil transcriptomes have potential to detect unruptured intracranial aneurysms. Journal of translational medicine. 2018;16(1):373-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tutino VM, Poppenberg KE, Damiano RJ, Patel TR, Waqas M, Dmytriw AA, et al. Characterization of Long Non-coding RNA Signatures of Intracranial Aneurysm in Circulating Whole Blood. Molecular Diagnosis & Therapy. 2020. 2020/12/01;24(6):723–36. [DOI] [PubMed] [Google Scholar]
- 13.Tutino VM, Poppenberg KE, Jiang K, Jarvis JN, Sun Y, Sonig A, et al. Circulating neutrophil transcriptome may reveal intracranial aneurysm signature. PLOS ONE. 2018;13(1):e0191407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poppenberg KE, Tutino VM, Li L, Waqas M, June A, Chaves L, et al. Classification models using circulating neutrophil transcripts can detect unruptured intracranial aneurysm. Journal of translational medicine. 2020. 2020/10/15;18(1):392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tutino VM, Zebraski HR, Rajabzadeh-Oghaz H, Waqas M, Jarvis JN, Bach K, et al. Identification of Circulating Gene Expression Signatures of Intracranial Aneurysm in Peripheral Blood Mononuclear Cells. Diagnostics. 2021;11(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poppenberg KE, Li L, Waqas M, Paliwal N, Jiang K, Jarvis JN, et al. Whole blood transcriptome biomarkers of unruptured intracranial aneurysm. PLOS ONE. 2020;15(11):e0241838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tutino VM, Lu Y, Ishii D, Poppenberg KE, Rajabzadeh-Oghaz H, Siddiqui AH, et al. Aberrant Whole Blood Gene Expression in the Lumen of Human Intracranial Aneurysms. Diagnostics. 2021;11(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tutino VM FS, Chien A, Patel TR, Monteiro A, Rai HH, Dmytriw AA, Chaves LD, Waqas M, Levy EI, Poppenberg KE, Siddiqui AH. Gene expression profiles of ischemic stroke clots retrieved by mechanical thrombectomy are associated with disease etiology. J NeuroIntervent Surg. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tutino VM, Fricano S, Frauens K, Patel TR, Monteiro A, Rai HH, et al. Isolation of RNA from Acute Ischemic Stroke Clots Retrieved by Mechanical Thrombectomy. Genes. 2021;12(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krämer A, Green J, Pollard J Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics (Oxford, England). 2014;30(4):523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans JD. Straightforward statistics for the behavioral sciences. Pacific Grove: Brooks/Cole Pub. Co.; 1996. [Google Scholar]
- 22.Graffeo CS, Tanweer O, Nieves CF, Belmont HM, Izmirly PM, Becske T, et al. Rapid aneurysm growth and rupture in systemic lupus erythematosus. Surg Neurol Int. 2015;6:9-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JH, Lee SW, Choi CH, Ko JK. Does Systemic Lupus Erythematosus Increase the Risk of Procedure-Related Complication in Endovascular Treatment of Intracranial Aneurysm? Yonsei Med J. 2020;61(5):441–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nature biotechnology. 2015;33(5):495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McInnes L, Healy J, Melville J. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv preprint arXiv:180203426. 2018. [Google Scholar]
- 26.Hussain S, Barbarite E, Chaudhry NS, Gupta K, Dellarole A, Peterson EC, et al. Search for Biomarkers of Intracranial Aneurysms: A Systematic Review. World neurosurgery. 2015. Nov;84(5):1473–83. [DOI] [PubMed] [Google Scholar]
- 27.Tanriverdi T, Sanus GZ, Ulu MO, Tureci E, Uzun H, Aydin S, et al. Serum and cerebrospinal fluid concentrations of E-selectin in patients with aneurysmal subarachnoid hemorrhage. Brazilian journal of medical and biological research. 2005;38(11):1703–10. [DOI] [PubMed] [Google Scholar]
- 28.Jia W, Wang R, Zhao J, Liu IY, Zhang D, Wang X, et al. E-Selectin Expression Increased in Human Ruptured Cerebral Aneurysm Tissues. Canadian journal of neurological sciences. 2011;38(6):858–62. [DOI] [PubMed] [Google Scholar]
- 29.Mack WJ, Mocco J, Hoh DJ, Huang J, Choudhri TF, Kreiter KT, et al. Outcome prediction with serum intercellular adhesion molecule-1 levels after aneurysmal subarachnoid hemorrhage. Journal of Neurosurgery. 2002. 01 Jan. 2002;96(1):71–5. [DOI] [PubMed] [Google Scholar]
- 30.Witkowska AM, Borawska MH, Socha K, Kochanowicz J, Mariak Z, Konopka M. TNF-α and sICAM-1 in intracranial aneurismal rupture. Archivum Immunologiae et Therapiae Experimentalis. 2009. 2009/04/02;57(2):137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mangieri P, Suzuki K, Ferreira M, Domingues L, Casulari LA. Evaluation of pituitary and thyroid hormones in patients with subarachnoid hemorrhage due to ruptured intracranial aneurysm. Arquivos de neuro-psiquiatria. 2003;61(1):14–9. [DOI] [PubMed] [Google Scholar]
- 32.Casulari LA, Mangieri P, Naves LA, Suzuki K, Ferreira M, Domingues L. Nonthyroidal illness syndrome in patients with subarachnoid hemorrhage due to intracranial aneurysm. Arquivos de neuropsiquiatria. 2004;62(1):26–32. [DOI] [PubMed] [Google Scholar]
- 33.Wiesmann M, Missler U, Hagenström H, Gottmann D. S-100 protein plasma levels after aneurysmal subarachnoid haemorrhage. Acta Neurochirurgica. 1997. 1997/12/01;139(12):1155–60. [DOI] [PubMed] [Google Scholar]
- 34.Ray L, Khemka VK, Behera P, Bandyopadhyay K, Pal S, Pal K, et al. Serum Homocysteine, Dehydroepiandrosterone Sulphate and Lipoprotein (a) in Alzheimer’s Disease and Vascular Dementia. Aging Dis. 2013;4(2):57–64. [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J, Han L, Xu X, Tang H, Wang H, Wei B. Serum biomarkers VEGF-C and IL-6 are associated with severe human Peripheral Artery Stenosis. Journal of inflammation (London, England). 2015;12:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimura H, Okada O, Tanabe N, Tanaka Y, Terai M, Takiguchi Y, et al. Plasma Monocyte Chemoattractant Protein-1 and Pulmonary Vascular Resistance in Chronic Thromboembolic Pulmonary Hypertension. American Journal of Respiratory and Critical Care Medicine. 2001. 2001/07/15;164(2):319–24. [DOI] [PubMed] [Google Scholar]
- 37.Korostynski M, Morga R, Piechota M, Hoinkis D, Golda S, Dziedzic T, et al. Inflammatory Responses Induced by the Rupture of Intracranial Aneurysms Are Modulated by miRNAs. Molecular Neurobiology. 2020. 2020/02/01;57(2):988–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Supriya M, Christopher R, Indira Devi B, Bhat DI, Shukla D. Circulating MicroRNAs as Potential Molecular Biomarkers for Intracranial Aneurysmal Rupture. Molecular Diagnosis & Therapy. 2020. 2020/06/01;24(3):351–64. [DOI] [PubMed] [Google Scholar]
- 39.Huang Q, Sun Y, Huang Q, Zeng Y, Lin S, Huang S, et al. Association Between Circular RNAs and Intracranial Aneurysm Rupture Under the Synergistic Effect of Individual Environmental Factors. Front Neurol. 2021;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chalouhi N, Ali MS, Jabbour PM, Tjoumakaris SI, Gonzalez LF, Rosenwasser RH, et al. Biology of intracranial aneurysms: role of inflammation. Journal of cerebral blood flow and metabolism. 2012;32(9):1659–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tulamo R, Frösen J, Hernesniemi J, Niemelä M. Inflammatory changes in the aneurysm wall: a review. Journal of NeuroInterventional Surgery. 2010;2(2):120. [DOI] [PubMed] [Google Scholar]
- 42.Yokoi T, Saito M, Yoshimura Y, Tsuji K, Nozaki K. Cerebral aneurysms and inflammation. Neuroimmunology and Neuroinflammation. 2015;2:55–8. [Google Scholar]
- 43.Meng H, Tutino VM, Xiang J, Siddiqui A. High WSS or Low WSS? Complex Interactions of Hemodynamics with Intracranial Aneurysm Initiation, Growth, and Rupture: Toward a Unifying Hypothesis. American Journal of Neuroradiology. 2014;35(7):1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chyatte D, Bruno G, Desai S, Todor DR. Inflammation and Intracranial Aneurysms. Neurosurgery. 1999;45(5):1137–47. [DOI] [PubMed] [Google Scholar]
- 45.Frösen J, Tulamo R, Paetau A, Laaksamo E, Korja M, Laakso A, et al. Saccular intracranial aneurysm: pathology and mechanisms. Acta Neuropathologica. 2012. 2012/06/01;123(6):773–86. [DOI] [PubMed] [Google Scholar]
- 46.Frösen J, Piippo A, Paetau A, Kangasniemi M, Niemelä M, Hernesniemi J, et al. Remodeling of Saccular Cerebral Artery Aneurysm Wall Is Associated With Rupture. Stroke. 2004;35(10):2287. [DOI] [PubMed] [Google Scholar]
- 47.Kataoka K, Taneda M, Asai T, Kinoshita A, Ito M, Kuroda R. Structural Fragility and Inflammatory Response of Ruptured Cerebral Aneurysms. Stroke. 1999. 1999/07/01;30(7):1396–401. [DOI] [PubMed] [Google Scholar]
- 48.Tutino VM, Zebraski HR, Rajabzadeh-Oghaz H, Chaves L, Dmytriw AA, Siddiqui AH, et al. RNA Sequencing Data from Human Intracranial Aneurysm Tissue Reveals a Complex Inflammatory Environment Associated with Rupture. Molecular Diagnosis & Therapy. 2021. 2021/11/01;25(6):775–90. [DOI] [PubMed] [Google Scholar]
- 49.Starke RM, Chalouhi N, Jabbour PM, Tjoumakaris SI, Gonzalez LF, Rosenwasser RH, et al. Critical role of TNF-α in cerebral aneurysm formation and progression to rupture. Journal of neuroinflammation. 2014;11:77-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ago T, Sadoshima J. GDF15, a Cardioprotective TGF-β Superfamily Protein. Circulation research. 2006. 2006/02/17;98(3):294–7. [DOI] [PubMed] [Google Scholar]
- 51.Kleinloog R, Verweij Bon H, van der Vlies P, Deelen P, Swertz Morris A, de Muynck L, et al. RNA Sequencing Analysis of Intracranial Aneurysm Walls Reveals Involvement of Lysosomes and Immunoglobulins in Rupture. Stroke. 2016. 2016/05/01;47(5):1286–93. [DOI] [PubMed] [Google Scholar]
- 52.Nakaoka H, Tajima A, Yoneyama T, Hosomichi K, Kasuya H, Mizutani T, et al. Gene Expression Profiling Reveals Distinct Molecular Signatures Associated With the Rupture of Intracranial Aneurysm. Stroke. 2014;45(8):2239. [DOI] [PubMed] [Google Scholar]
- 53.Sharma T, Datta KK, Kumar M, Dey G, Khan AA, Mangalaparthi KK, et al. Intracranial Aneurysm Biomarker Candidates Identified by a Proteome-Wide Study. OMICS: A Journal of Integrative Biology. 2020. 2020/08/01;24(8):483–92. [DOI] [PubMed] [Google Scholar]
- 54.Field A, Miles J, Field Z. Discovering statistics using R: Sage publications; 2012. [Google Scholar]
- 55.Feghali J, Gami A, Xu R, Jackson CM, Tamargo RJ, McDougall CG, et al. Application of unruptured aneurysm scoring systems to a cohort of ruptured aneurysms: are we underestimating rupture risk? Neurosurgical review. 2021. 2021/12/01;44(6):3487–98. [DOI] [PubMed] [Google Scholar]
- 56.Foreman PM, Hendrix P, Harrigan MR, Fisher WS, Vyas NA, Lipsky RH, et al. PHASES score applied to a prospective cohort of aneurysmal subarachnoid hemorrhage patients. Journal of Clinical Neuroscience. 2018. 2018/07/01/;53:69–73. [DOI] [PubMed] [Google Scholar]
- 57.Pagiola I, Mihalea C, Caroff J, Ikka L, Chalumeau V, Iacobucci M, et al. The PHASES score: To treat or not to treat? Retrospective evaluation of the risk of rupture of intracranial aneurysms in patients with aneurysmal subarachnoid hemorrhage. Journal of Neuroradiology. 2020. 2020/09/01/;47(5):349–52. [DOI] [PubMed] [Google Scholar]
- 58.Neyazi B, Sandalcioglu IE, Maslehaty H. Evaluation of the risk of rupture of intracranial aneurysms in patients with aneurysmal subarachnoid hemorrhage according to the PHASES score. Neurosurgical review. 2019. 2019/06/01;42(2):489–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in the current study are available from the corresponding author upon reasonable request.