Abstract
Tuberculosis (TB) pathologic lesions in rhesus macaques resemble those in humans. The expression levels of several host TB candidate genes in the peripheral blood mononuclear cells (PBMCs) of six rhesus macaques experimentally infected with Mycobacterium tuberculosis were quantified pre-infection and at several dates post-infection. Quantitative measures of TB histopathology in the lungs including: granuloma count, granuloma size, volume of granulomatous and non-granulomatous lesions, and direct bacterial load, were used as the outcomes of a multi-level Bayesian regression model in which expression levels of host genes at various dates were used as predictors. The results indicate that the expression levels of TR4, CD40, CD40L, FAS (CD95) and TNF in PBMC were associated with quantitative measures of the severity of TB histopathologic lesions in the lungs of the study animals. Moreover, no reliable association between the expression levels of IFNE in PBMCs and the severity of TB lesions in the lungs of the study animals was found. In conclusion, PBMC expression profiles derived from the above-listed host genes might be appropriate biomarkers for probabilistic diagnosis and/or prognosis of TB severity in rhesus macaques.
Keywords: Tuberculosis (TB), rhesus macaques, gene expression, host genes, histopathologic lesions, TR4, IFNE
1. Introduction
Tuberculosis (TB) is a zoonotic disease in primates caused by Mycobacterium tuberculosis (MTB). TB can exist in vivo in either active or latent form. Approximately 30% of the human population worldwide is latently infected with Mycobacterium tuberculosis (MTB), 5-10% of whom progress to active TB [1]. Nonhuman primates (NHPs), particularly rhesus and cynomolgus macaques infected by TB, exhibit clinical symptoms and histopathologic lesions similar to those of humans, making them more appropriate animal models for the study of human TB pathogenesis than other laboratory animals, such as mice [2, 3, 4]. However, since NHPs are not genetically homogenous, unlike the mice used in biomedical research, the results of case-control studies of TB in NHPs might be confounded by the genetic heterogeneity of the study animals [5, 6].
This genetic heterogeniety in NHP study populations, which may lead to variation in clinical and histopathologic outcomes for TB, could, however, facilitate the identification of host genes (e.g. inducible nitric oxide synthase [iNOS] and nuclear factor kappa-light-chain-enhancer of activated B cells [NFKB]) that play key roles in various immune response against MTB in NHPs [7, 8]. Genes with altered expression levels as a result of TB exposure may be useful as host markers for probabilistic TB diagnosis in both humans and NHPs. Previous studies in human populations, in vivo mouse model experiments, and in vitro experiments, have identified several host candidate genes associated with susceptibility to TB [9]. Most of these genes encode key components in the innate immune response against MTB. These genes are represented in the following four categories: 1) genes encoding receptors that detect MTB (e.g. Toll-like receptor 2 [TLR2] and peroxisome proliferator-activated receptor gamma [PPARG]), 2) genes encoding proinflammatory cytokines and chemokines (e.g. interferon gamma [IFNG] and chemokine C-C motif ligand 2 [CCL2]), 3) genes encoding anti-inflammatory cytokines (e.g. interleukin-10 [IL10]), and 4) genes encoding enzymes that play key roles in intracellular immunity and the elimination of MTB inside macrophages (e.g. iNOS) [8, 10]. Current TB tests such as the tuberculin skin test (TST) and IFNG based assays lack sensitivity and/or specificity, and are logistically difficult for patients and/or laboratories involved. For example, rhesus macaques with several consecutive negative TST results are sometimes found to be TB positive following pathologic investigations at necropsy [11, 12].
The goal of this study is to investigate the associations between the expression levels of a selected group of host innate immune response genes in the peripheral blood mononuclear cells (PBMC) of rhesus macaques (pre-and post-MTB infection) and the severity of TB pathogenesis. The selection of the host candidate genes in the present study was based on a comprehensive literature review and includes: PPARG [13], nuclear receptor subfamily 2 group C member 2 (NR2C2) also known as testicular receptor 4 (TR4) [13], IFNE [14, 15, 16], CD40 [17, 18], chemokine C-X-C motif ligand 10 (CXCL10) also known as IFNG-inducible protein 10 (IP10) [19], CSF2 [20, 21], CD40L [22, 23, 24], IL7 [25, 26, 27], IL15 [28, 29], FAS (CD95) [30], IL23A [31, 32], IL12A [33, 34, 35, 36, 24], IL13 [37], IL4 [38, 39, 40, 36], IL5, solute carrier family 11 member 1 (SLC11A1) also known as natural resistance-associated macrophage protein (NRAMP1) [41, 42, 43, 44, 45, 46], TLR2 [47, 48], FASL [49], IL17A [50, 51], CCL2 [52, 53, 54, 55], tumor necrosis factor-α (TNF) [34, 56], nitric oxide synthase 2A (NOS2A) also known as iNOS [57, 58, 10, 59, 60], and dendritic cell-specific intercellular adhesion molecule-3-Grabbing non-integrin (DCSIGN) also known as CD209 [61, 62].
Previous studies involving human subjects, in vivo mouse model experiments, and in vitro cell cultures have shown that the expression of these genes may be associated with TB pathogenesis. For instance, it has been recently shown through in vitro experiments that PPARG and TR4 play key roles in the pathogenesis and survival of MTB within macrophages [13]. However, few in vivo studies have demonstrated the role of these new candidate genes in MTB pathogenesis. The role of some other genes in the above list have been described in in vitro and in vivo model experiments.
Since the course of TB infection in rhesus macaques is similar to that in humans, and it is possible to perform controlled MTB experimental infection in rhesus macaques, the change in expression levels of various genes in the PBMCs of the laboratory animals can be assayed at different dates pre-and post-infection. The genes with highly divergent expression levels (pre- and post-infection) as a function of the presence and severity of TB lesions in lungs may be useful as host biomarkers in probabilistic TB diagnostic methods, especially for the diagnosis of latent TB cases in which direct isolation of MTB can not be easily conducted.
In this study, six rhesus macaques were experimentally infected with MTB [63]. Blood samples from the infected animals were obtained pre-infection and at different dates (every 1-2 months) after MTB infection. The study animals were euthanized approximately six months post-infection and gross necropsy and histopathological investigations were conducted. A quantitative assessment of the severity of lesions in the lungs and other organs was determined using several measures of TB pathogenesis: 1) number of granulomas, 2) size of the largest granuloma, 3) volume of the lung tissue exhibiting granulomatous lesions, 4) volume of the lung tissue exhibiting non-granulomatous lesions, and 5) a direct count of bacterial load in the lungs, in colony forming units (CFUs). A comprehensive morphometric/quantitative description of the histopathologic lesions of MTB in the lungs of all study animals was previously reported in detail [63].
We identify several host TB candidate genes whose upregulation or downregulation in the PBMCs of the study animals is associated with the severity of lesions in the lung tissues of the study animals, as discussed below. Similar approaches for identification of the expression profiles of candidate genes have been previously used for prognosis of cancer and autoimmune diseases [64].
2. Results
2.1. Model Results
We find consistent secular trends across animals in the expression levels of CCL2, FASL, IL10, and TNF as TB infection develops over time (Figure 1). This provides evidence that experimental infection with TB can lead to changes in the expression levels of these genes. As such, a multi-gene panel which assays expression levels of these genes might be useful for distinguishing non-infected macaques from infected ones. However, these results are not sufficient to comment on the relationship between gene expression levels and TB severity; to do that, we must explicitly model the relationship between gene expression and TB severity at time of euthanasia, as in Figure 2.
Figure 1:
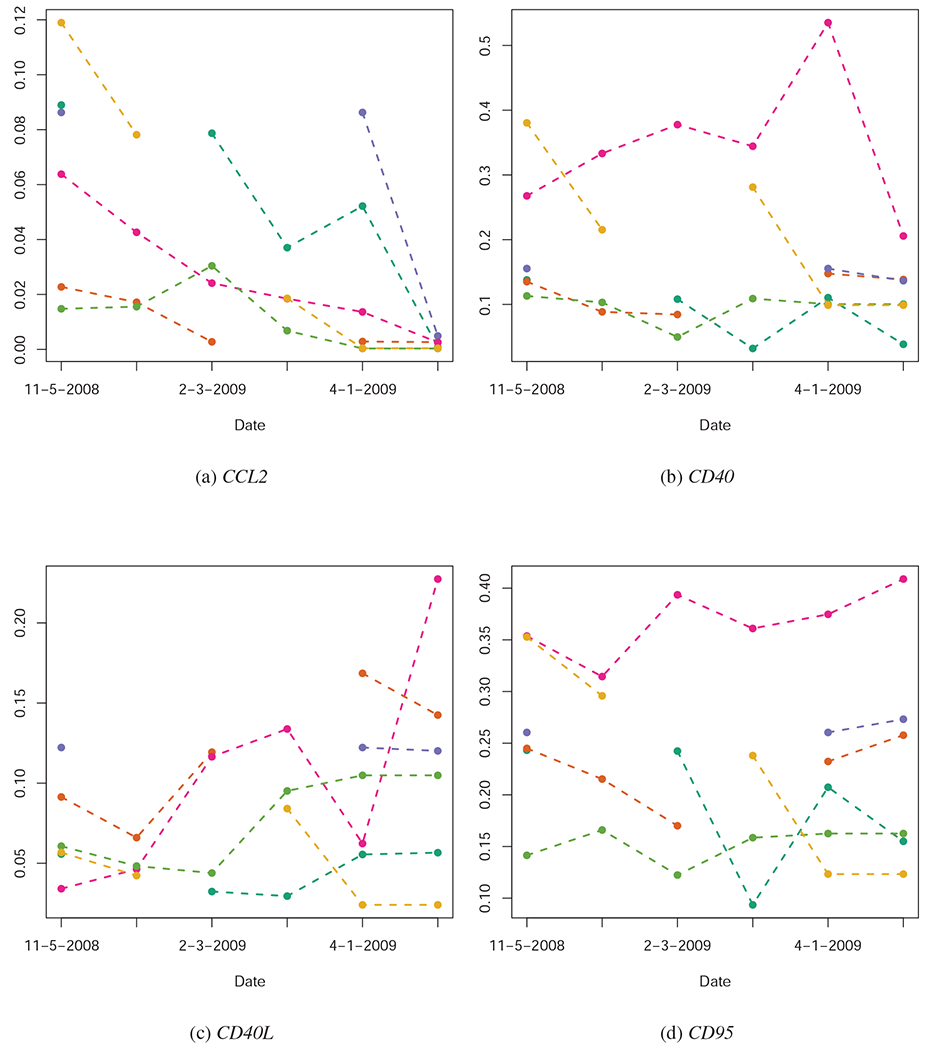
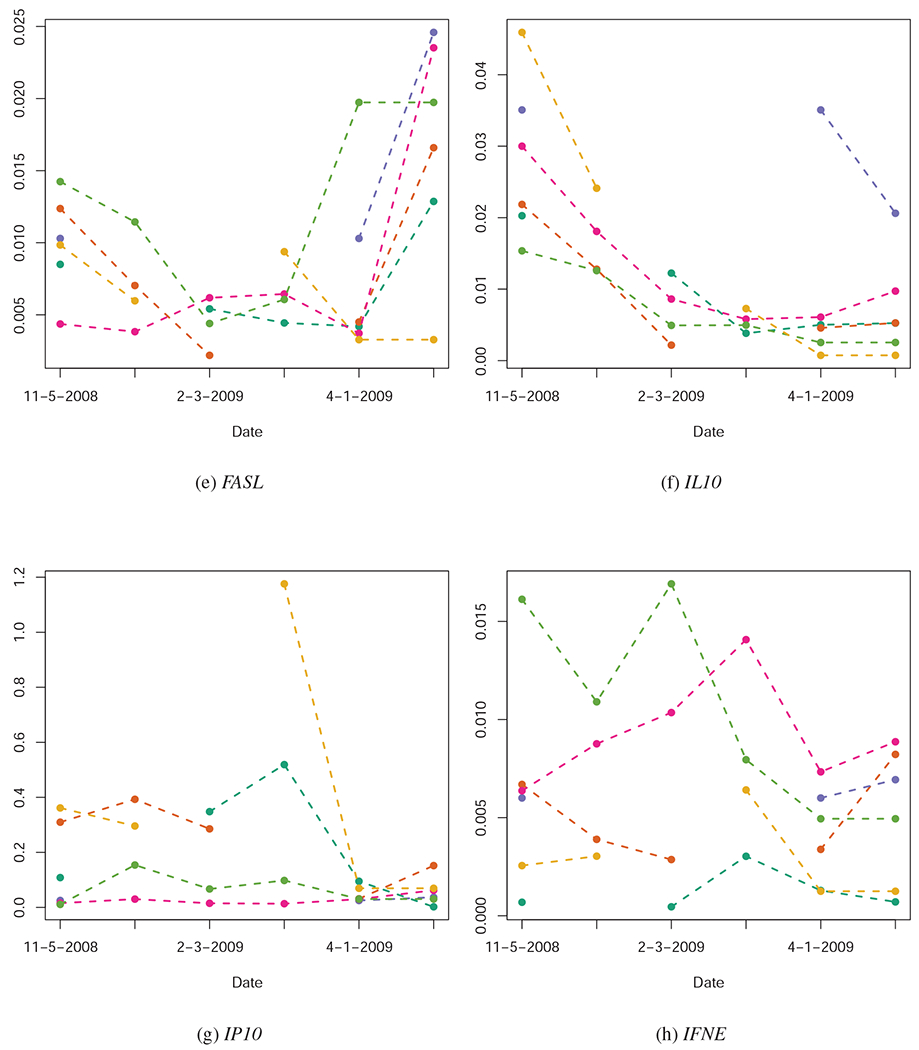
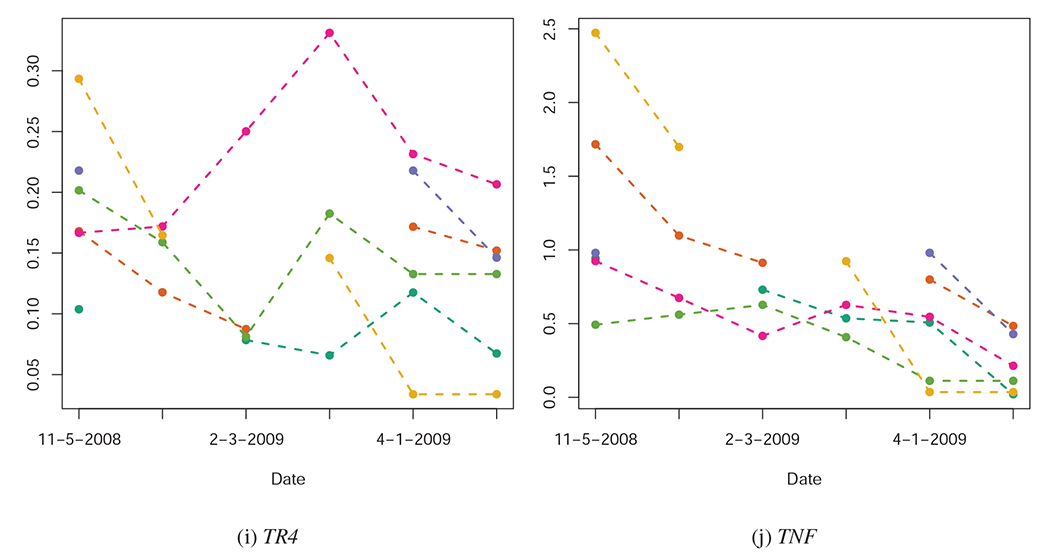
Quantified expression levels (Y-axis) of several of the genes investigated in this study, pre-infection (11-05-2008), at several dates post-infection (01-05-2009, 02-03-2009, 03-02-2009, 04-01-2009), and at time of euthanasia. Each animal’s gene expression over time is represented by a unique color. Quantification could not be completed for all animals at all dates, leading to some cases of missing data. Across these sub-figures, we note that some genes (e.g. TNF) show consistent temporal trends across animals; other genes, however, show no overall temporal pattern in expression, even though their expression levels may be indicative of the severity TB infection (See Figure 2).
Figure 2:
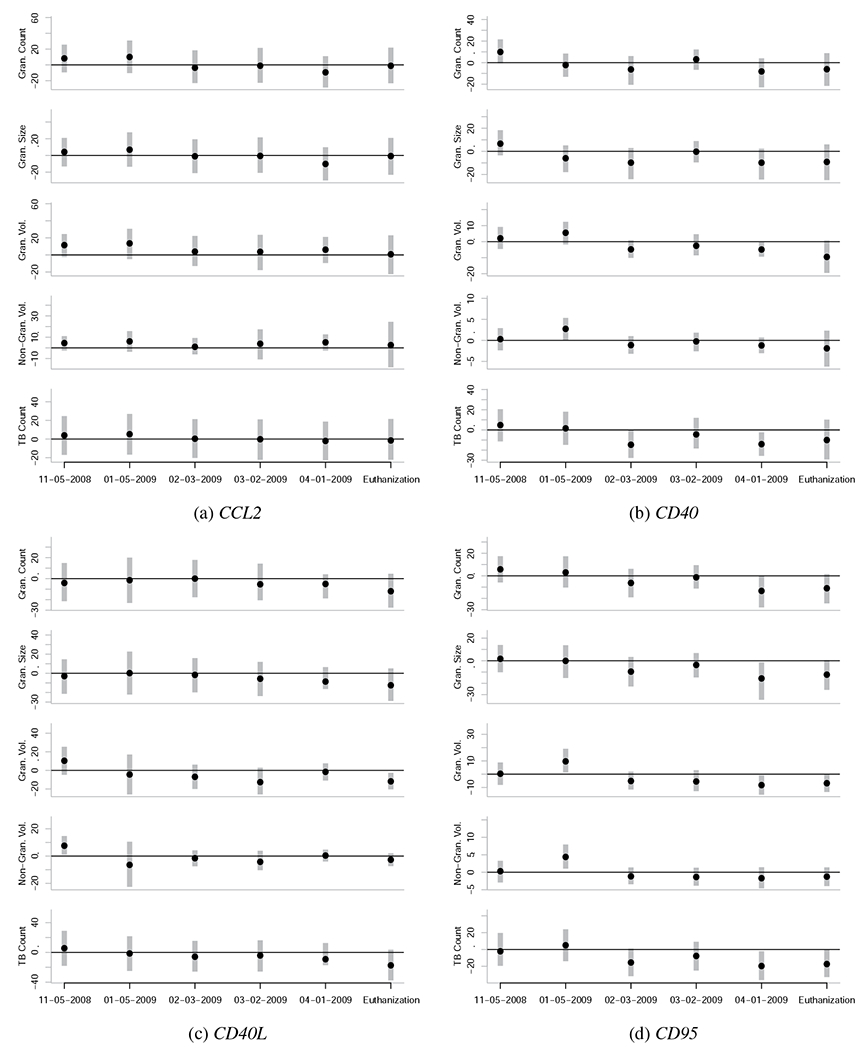
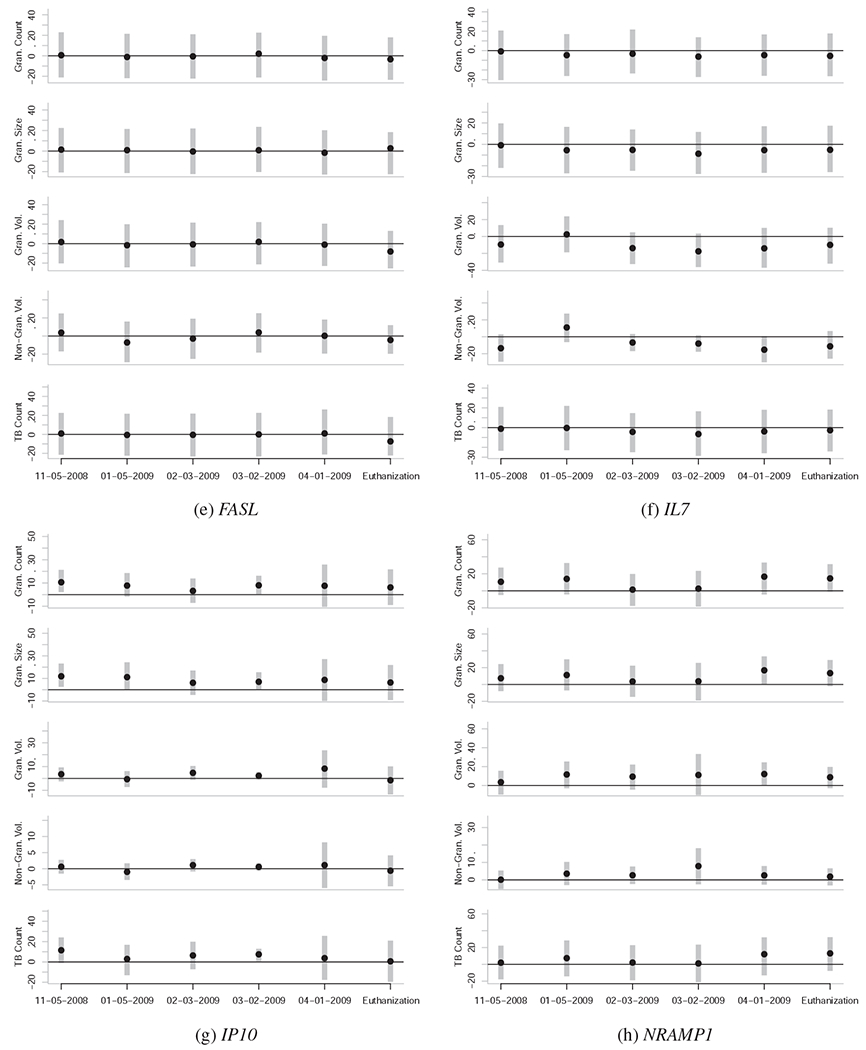
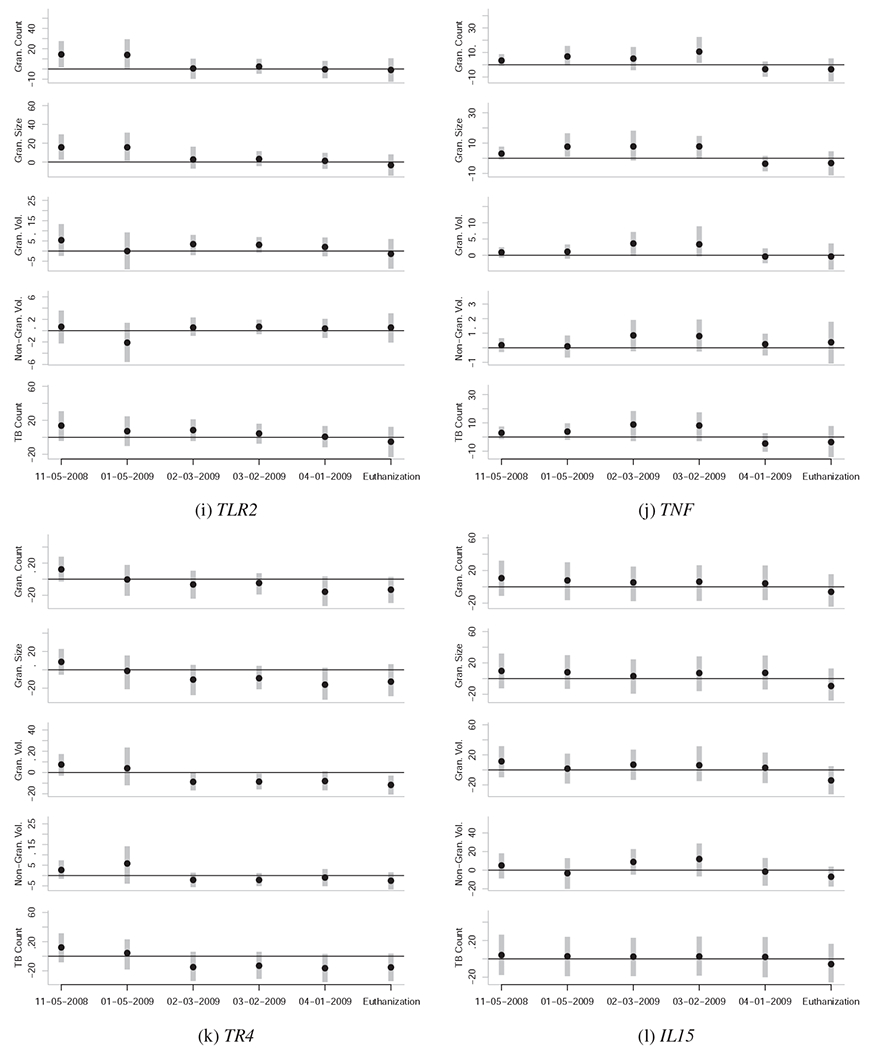
Caterpillar plots representing the associations between gene expression levels at various dates and quantitative measures of TB severity at euthanasia. Each subfigure is unique to a given gene, and each subplot within each subfigure is unique to a given quantitative measure of TB severity at euthanasia (vertical axis) for a given date of gene expression quantification (horizontal axis). The black points represent the posterior mean and the vertical grey bars indicate the central 95% posterior credibility intervals. The horizontal black lines represent the value of zero, or no effect. Null effects are typically close to zero, with confidence intervals that overlap zero. Reliable effects have confidence intervals that don’t overlap zero, or do so only slightly. To be confident that an effect is actually non-zero, it is also useful to see if the effect is observed across the majority of the five outcome variables, and if the effect is part of a temporal trend.
Elevated expression levels of IP10 (CXCL10), TLR2, TR4, and TNF in the PBMCs of the study animals before MTB infection are reliably associated with the severity of TB lesions and the bacterial count of MTB in the lungs of the study animals months later (Figure 2). These genes can be categorized as the group of genes whose expression levels pre-infection in this sample are associated with susceptibility to TB.
Further, we find that the relationship between expression levels of CD40, CD40L, CD95, TNF, and especially TR4 changes from the pre-infection state to the post-infection state (Figure 2). These genes can be categorized as the group of genes whose expression levels may be indicative of TB severity.
PPARG, CSF2, IL23A, IL12A, IL13, IL4, IL5, IL17A, and iNOS, could not be quantified in all animals (we could not distinguish between a true lack of expression and problems in quantification), so these genes were excluded from the study.
To visually represent the strength and direction of the relationship between gene expression levels and TB pathogenesis, we plot the values of the regression coefficient estimates in Figure 2. The numerical values and posterior confidence intervals of these parameter estimates are included in the Supplementary Materials.
2.2. Information Theoretic Model Comparison
The sample size of animals in this study is small (N = 6); however, independent assessments of TB severity were conducted in each of (L = 6) lung lobes (left caudal, left cranial, right accessory, right cranial, right caudal, and right middle), allowing for the relationship between gene expression and TB pathogenesis to be assessed in six semi-independent tissues per animal, meaning the actual sample size of outcome data is 36. Prior to the development of mutli-level models and Bayesian analysis, issues of pseudoreplication would rendered nuanced analysis of such data unfeasible [65]. However, by developing a custom multi-level statistical model, we can analyze the data using methods designed specifically for cases where observations (eg. granuloma counts, max granuloma size, and granuloma volume per lung lobe) are clustered into higher-level units (animals), which may introduce correlations in outcomes at the lower level.
To test if use of gene expression data actually improve predictive inference relative to a null intercept-only model, we use information theoretic model comparison [66]. These methods allow us to demonstrate that models using gene expression information have significantly better predictive ability than the null model (Figure 3). This analysis provides evidence that although our sample size is small in terms of animals, there is strong enough signal in our data to make model-based predictive inference possible.
Figure 3:
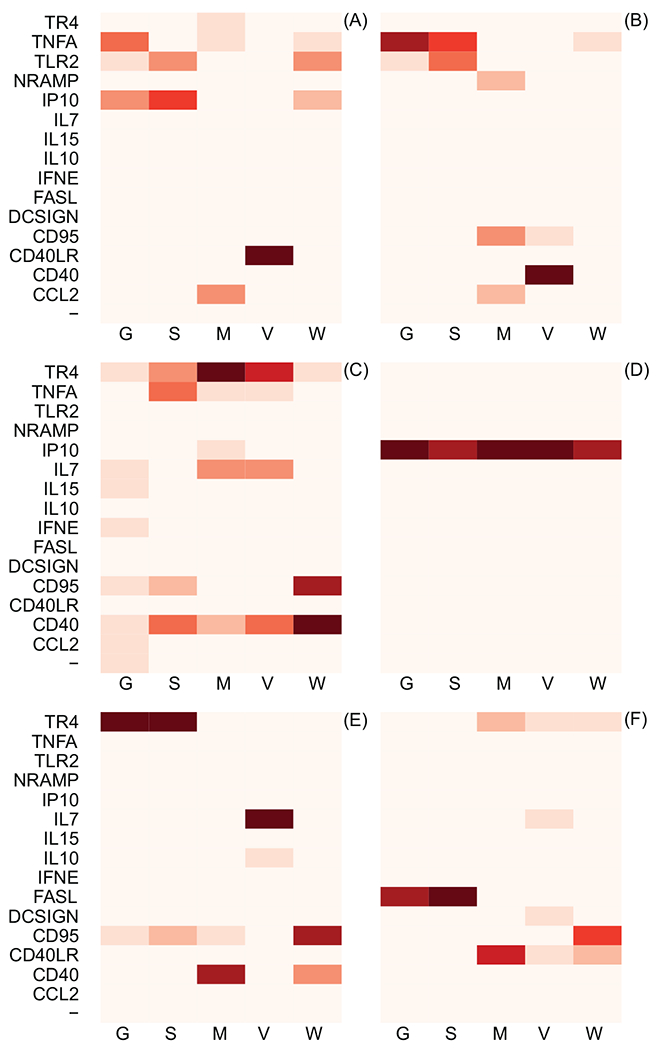
Results of formal model comparison using WAIC. Each block represents a date when gene expression levels were quantified: A) 11/05/2008, B) 01/05/2009, C) 02/03/2009, D) 03/02/2009, E) 04/01/2009, F) Time of euthanasia. Each vertical column illustrates the information theoretic support that the specified gene (on the vertical axis) is the gene whose expression levels best predict a proxy of TB severity (on the horizontal axis). The column labels: G, S, M, V, and W, correspond to granuloma count in sections of lung, max granuloma size, granulomatous lesion volume, non-granulomatous lesion volume, and TB cell count, respectively. Increasing intensity of the red color indicates increasing information theoretic support. The quantitative results of the information theoretic model comparison used to generate this figure are included as Supplementary Materials. The “-” symbol indicates the performance of the null model, which does not include gene expression levels as a predictor; it should be noted that the null model has almost no information theoretic support in any of the model comparisons, meaning that use of gene expression levels to predict TB severity almost universally improves predictive inference. Consistent support for a given gene across proxies for TB severity and time periods provides good evidence that there exists a relationship between TB pathogensis and gene expression levels.
We use information theoretic model comparison to assess the relative utility of gene expression levels at each date, in each measure of TB pathogenesis. The results of model comparison are represented visually in Figure 3, and numerically in the Supplementary Materials. Across all measures and time periods, the null model has almost no information theoretic support, which provides strong evidence that use of gene expression levels improves predictive inference concerning TB severity. There is some variation in which genes have the most predictive ability at each date (for example, TNF expression levels shortly after infection are among the best predictors of TB severity, but TR4 and CD40 expression levels at later dates are better predictors of TB severity). There are also genes which are consistently of little use in predicting TB severity (e.g. IFNE, IL10, and IL15).
2.3. Gene by Gene Profiles
Elevated expression levels of TNF in the PBMCs of the study animals pre-infection is associated with increased severity of TB infection at time of the euthanasia of the study animals 5-6 month post-infection (Figure 2j). However at the date of 4-01-2009 and at necropsy, TNF expression levels were inversely associated with TB severity (Figure 2j).
A similar pattern was observed for CD40 and CD40L, whose expression levels become inversely associated with TB severity at euthanasia around 02-03-2009 or 03-02-2009 (Figures 2b and 2c). CD95 expression levels become inversely associated with TB severity at 02-03-2009 (Figure 2d). As such, the expression levels of these four genes may contain information on the underlying severity of TB infection, and may be useful in probabilistically estimating TB severity from PBMC-based expression assays.
Elevated expression levels of CCL2 pre-infection (11-05-2008) and (to some extent) one month post-infection (01-05-2009) show a weak but positive association with TB severity at time of the euthanasia; however, no reliable association between CCL2 expression and TB severity after 01-05-2009 was observed (Figure 2a).
The expression levels of IP10 both pre-infection (11-05-2008) and post-infection (01-05-2009 to 03-02-2009) show a strong positive association with TB severity at euthanasia (Figure 2g).
The expression levels of IL7 and IL15 show no reliable signs of association with TB severity at euthanasia (Figures 2f and 2l).
The expression levels of IL10 and IFNE show no relationship to TB severity at euthanasia (Figure 3).
The pre-infection (11-05-2008) expression levels of TR4 are positively associated with the severity of TB infection 5-6 months later. Furthermore, the expression levels of TR4 become strongly inversely associated with TB severity by 02-03-2009, and remain this way until the animals were euthanized (Figure 2k).
Likewise, pre-infection (11-05-2008) expression levels of TLR2 are positively associated with the severity of TB infection 5-6 months later (Figure 2i). However, the relationship remains positive until 03-02-2009, and thereafter shows no relationship with TB severity.
NRAMP1 expression levels pre-infection show no reliable association with TB severity (Figure 2h); however, expression levels of NRAMP1 post-infection show a marginally reliable positive association with TB severity, especially in the months immediately prior to euthanization (04-01-2009 to euthanization).
Finally, expression levels of DC-SIGN failed to show signs of association with TB severity (Figure 3).
2.4. Histopathology
Figures 4 and 5 present the histopathology of granulomatous lesions in right caudal lobe of lungs in the study animals using hematoxylin and eosin and then trichrome blue staining, respectively. In general, granulomas with central area of necrosis and/or mineralized cavities are observed. Additionally, the margins of macrophages, lymphocytes, and rare multinuclear giant cells are surrounded by fibrous connective tissue. Smaller granulomas are noticed within and outside of the capsules. These results indicate that the encapsulation seems to be breached. The areas of necrosis were extensive in two animals that exhibited severe TB, necessitating euthanasia before scheduled termination of the study.
Figure 4:
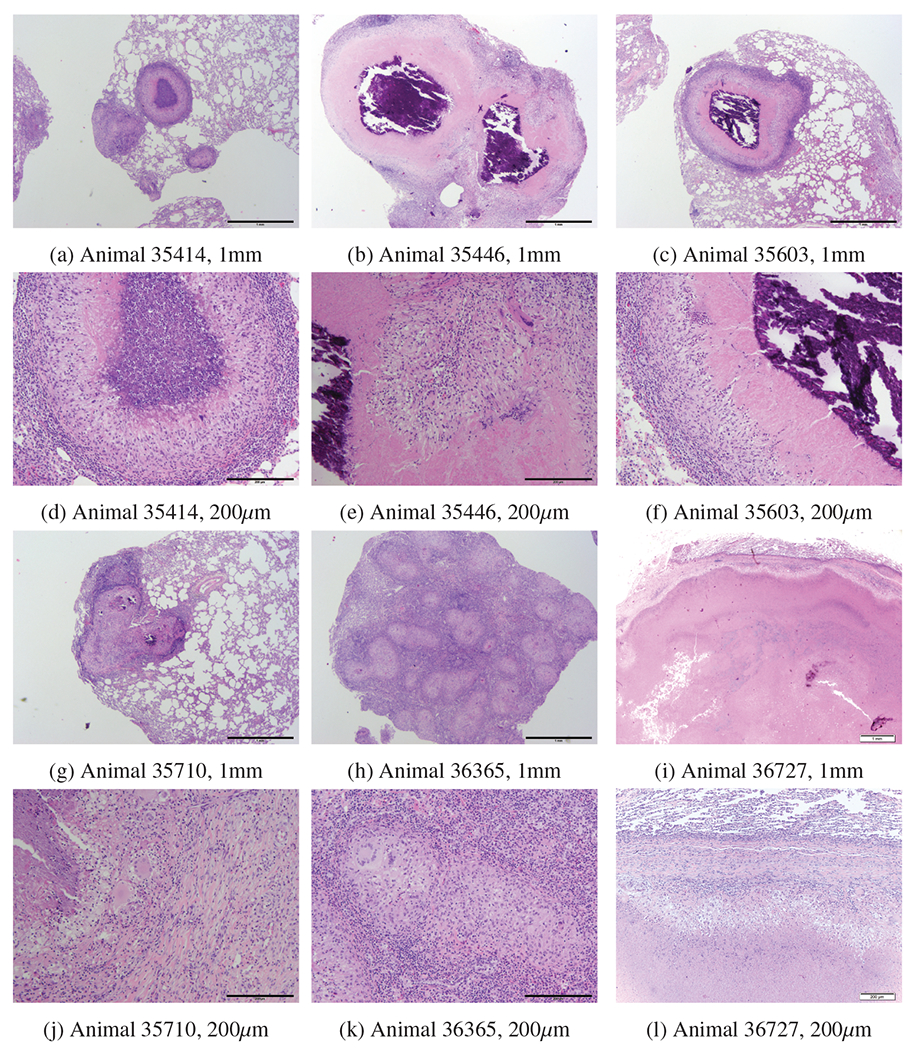
Multifocally in the lung, there were individual and coalescing granulomas which ranged from approximately 200 to greater than 1000 um in diameter. Granulomas were characterized by central areas which varied from having closely apposed epithelioid macrophages to necrosis consisting of intense eosinophilia, karyorrhexis, and karyolysis with basophilic hyalinized foci of mineralization (35446, b) or without basophilic hyalinized foci of mineralization (35414, a). The central region of granulomas were surrounded by areas of necrosis in animals that exhibited severe and active TB (c) or fibrosis in animals that exhibitted less severe disease and more successfully controlled the infection (d). Mainly at the periphery of granulomas there was a primarily mononuclear inflammatory cell infiltrate composed of varying numbers of lymphocytes, plasma cells, histiocytes and rare multinucleated giant cells which were more sparsely distributed. An outer rim of fibrosis (encapsulation) delineated the granulomas from pulmonary parenchyma. Degree of severity was based on the percent of pulmonary parenchyma affected by this chronic inflammatory process, size and number of granuloma. 35710 was least affected; granulomas for this case were also smaller, see [63]. 36727 was most extensively affected with granulomas also in eyes and brain.
Figure 5:
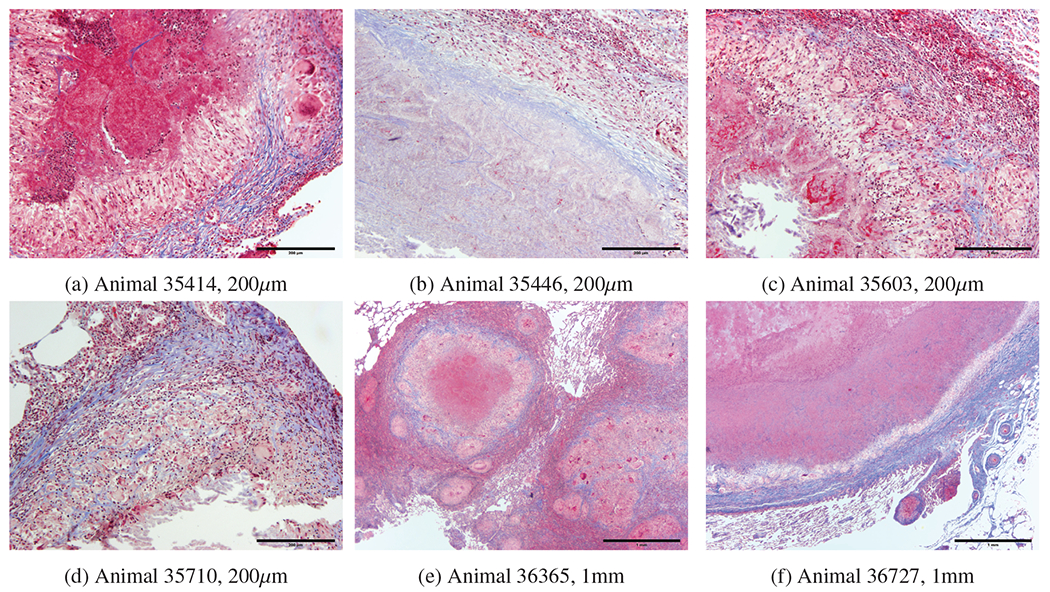
a) 35414 central area of caseous necrosis with margin of macrophages, multinucleated giant cells, and lymphocytes surrounded by fibrous connective tissue. b) 35446 central area of necrosis surrounded by fibrous connective tissue. c) 35603 central cavity containing mineralized material with margin of necrotic cells, surrounded by macrophages, distinct multinucleated giant cells and lymphocytes surrounded by patchy multifocal areas of fibrous connective tissue. d) 35710 central cavity containing mineralized material surrounded by macrophages, distinct multinucleated giant cells and lymphocytes surrounded by distinct area of fibrous connective tissue. e) 36365 central area of extensive necrosis surrounded by macrophages, lymphocyte, and distinct multinucleated giant cells surrounded by distinct fibrous connective tissue. f) 36727 central area of extensive necrosis with margine of macrophages, lymphocytes multinucleated giant cells surrounded by distinct fibrous connective tissue.
3. Discussion
The results of this study indicate that the expression levels of some host candidate genes both pre- and post-infection are associated with the severity of TB outcomes. Moreover, histopatholgical investigations found presence of TB lesions in organs other than the lungs (in brain and eye tissue). Quantitative morphometric measures of the histopathologic changes in the lungs of the study animals and other quantitative measures of severity of TB infection (e.g. bacterial load in lungs) were used as the outcomes of a statistical model in which the expression levels of the candidate genes in PBMCs were predictors. We have identified several genes (including the previously unverified TR4) whose expression levels in PBMCs post-infection are indicative of the severity of TB pathogenesis. These genes may thus be of use in a multi-gene expression panel for the probabilistic estimation of the likelihood of TB infection. No reliable association between the expression level of IFNE in PBMCs and the severity of TB lesions in the lungs of the study animals was found.
3.1. Gene Expression Levels as Biomarkers for TB Infection
Our results suggest that the genes TR4, CD40, CD40L, CD95, IL10, and TNF may be of use in the creation of a gene-expression panel for probabilistic estimation of TB infection. The information on some of the genes such as lipid sensing nuclear receptors (e.g. TR4) and IFN-ε are novel in an in vivo setting.
The study of TB pathogenesis in cynomolgus macaques may be better suited to study of human TB than the rhesus macaques used herein. Uniquely, cynomolgus macaques can progress to latent tuberculosis if infected with a low dose of MTB. This animal model has been utilized to investigate and categorize latent and active MTB infections at the molecular and histopathologic levels [67, 5, 2]. We did not have access to samples from TB infected cynomolgus macaques, however. It would be beneficial to compare and contrast our results to studies in which cynomolgus macaques are experimentally infected with TB, possibly using varying doses MTB at inoculation.
Studies similar to ours can not be conducted in human subjects due to ethical limitations; studies of gene expression in NHPs experimentally infected with TB are thus one of the best models for human TB [68]. However, findings in studies of gene expression and TB in NHPs can be compared to similar studies of human TB cases using other available (yet lower resolution) measures of TB severity (e.g. chest X-ray etc).
In this paper, we have not provided any evidence that the pattern of change in gene expression levels observed in this study is unique to TB infection as opposed to infection with a different pathogen. We recognize the necessity of future studies to investigate the uniqueness of gene expression changes in the above listed genes. However, we suspect that although the expression levels of any one gene are unlikely to be diagnostic of TB infection to the exclusion of other pathogens, an expression panel that includes many genes may be able to make such distinctions. Below we place our findings in the context of gene-specific research programs.
3.1.1. TR4 and PPARG
Previous in vitro studies have shown that LSNRs (e.g. PPARG and TR4) play a key role in the survival of MTB in macrophages [13, 69]. In this study, we evaluated whether the expression levels of two LSNR genes (TR4 and PPARG) in PBMCs of MTB infected rhesus macaques are associated with the severity of TB infection in the lungs. To the best of our knowledge, there have been no previous in vivo studies of these genes as predictive host markers of TB severity in NHPs or humans. Although we could not successfully quantify PPARG expression in the PBMCs of all the study animals, we did successfully quantify the expression levels of TR4 in the PBMCs of all study animals.
We observe that elevated pre-infection expression levels of TR4 are reliably associated with increased severity of TB at time of euthanasia; however, as infection develops over time, we observe an inverse relationship develop between TR4 expression and TB severity. These results suggest that upregulation of TR4 post-infection may play a role in attenuating TB pathogenesis. Furthermore, our results suggest that TR4 may have a significant role as a new host biomarker/gene for inclusion in a diagnostic gene expression panel for TB infection.
3.1.2. CD40, CD40L, and CD95
It has been previously reported that CD40 plays a key role in immune response against MTB and host genetic mutations can increase TB susceptibility by imparing CD40 production [24]. Recent findings show that MTB infection induces expression of CD40 in macrophages [70] and that the expression level of CD40 in PBMCs increases with age in human infants, as the innate response against MTB develops [18].
Some in vitro and in vivo experiments indicate that CD40L induces an antimicrobial response to MTB infection [22, 23]; the results of the present study also suggest that the expression levels of CD40L in the PBMCs of the TB infected animals are inversely associated with the severity of tuberculosis, agreeing with the results from some previous studies in the mouse model [71, 17].
Additionally, MTB-reactive γδ-T cells have been shown to express high levels of FAS (CD95) [72]. Our findings are thus in concordance with those from other studies of TB pathogenesis in animal models, which reported the importance of FAS (CD95) in a robust immune response against MTB [73, 74, 75, 76].
The inverse associations we observe develop between the expression levels of CD40, CD40L, and FAS (CD95) in PBMCs and the severity of TB in the study animals suggests that they might be appropriate host biomarkers for inclusion in an expression profile for the prediction of MTB infection in rhesus macaques.
3.1.3. iNOS
Previous studies have indicated that the expression level of iNOS at the site of granulomatous lesions in lung tissue, alveolar epithelial cells, and macrophages is relatively high in TB-infected individuals; specifically, the expression level of iNOS in the lungs of mice infected with MTB is high in the early phases of TB infection, during the formation of granulomas, and low in the late phases [10]. Since previous studies have shown that the expression levels of iNOS in the lung tissue of TB-infected/Bcg vaccinated macaques is significantly elevated, and since iNOS is thought to play an important role in intra-celluar immunity and the elimination of MTB in macrophages through the production of peroxynitrite, its expression levels in the PBMCs of study animals was also evaluated through the use of two (human and rhesus macaque) NOS2A probes [77, 78, 79, 80]. Unfortunately, the expression levels of iNOS could not be successfully quantitfied in the PBMCs of all rhesus macaques included in the present study. As such, future research is needed in order to evaluate the relationship between the expression level of iNOS in PBMCs of TB infected rhesus macaques and the severity of TB. Alternatively, research could focus on the development of methods that detect iNOS and/or nitric oxide (NO) metabolites [81].
3.1.4. IP10
It has been previously shown that polymorphisms in the promoter of IP10 are associated with susceptibility to TB [82]. Moreover, IP10 has been previously considered as a host candidate gene for TB susceptibility and a key marker for TB diagnosis [83, 84]. The results of this study indicate that the expression level of IP10 (CXCL10) in PBMCs of TB infected rhesus macaques both pre- and shortly post-infection may be associated with increased TB severity.
3.1.5. NRAMP1
The occurrence of mutations in the NRAMP1 gene and resultant susceptibility to TB has been extensively studied over the last 20 years. It was originally shown that mutations in the coding region of NRAMP1 caused susceptibility to some Mycobacterial infections but not to Mycobacterium tuberculosis [42, 43, 44, 46, 45]. Recent studies, however, indicate an important role of NRAMP1 in susceptibility to MTB [85, 86, 87], but there are not many studies that validate the predictive value of NRAMP1 expression for estimating TB severity. The results of our study indicate that expression levels of NRAMP1 post-infection show some positive association with TB severity, especially in the months immediately prior to euthanasia. The expression profile of NRAMP1, together with other genes that show an association with severity of TB lesions in the lung, might be valuable for prognosis of TB in NHPs and humans.
3.1.6. IFNE
Although IFNG has been extensively used as one of the most important host immune genes for TB pathogenesis, to the best of our knowledge there is not much information on the predictive role of IFNE for TB diagnosis and/or prognosis purposes. While the expression of IFNG has been previously shown to have predictive value in the diagnosis of TB in both humans and macaques, this study failed to find a relationship between the expression levels of the recently identified IFNE in the PBMCs of the study animals and severity of TB infection, suggesting that IFNE may not be an appropriate host gene/biomarker for the diagnosis of TB. Our study animals all exhibited the active form of TB and it would be useful to investigate the predictive value of IFNE in cynomolgus macaque that can develop latent TB when infected with low dose of MTB [5]. This will help us better understand the predictive value of this gene in latent TB cases.
3.1.7. Other Genes
Although several other host genes investigated in this study have been found to be functionally important genes or biomarkers in predicting severity of disease progression [see 88, 54] and expression levels of some of them in human samples (e.g. in Bronchioalveolar lavage and sputum) have been shown to correlate with the status of TB infection [36], we did not find associations between the expression levels of these genes in PBMCs and severity of TB in rhesus macaques.
There are several other immune-related candidate genes that could be included in a study with a larger sample size to assess the predictive ability of gene expression for TB diagnosis and prognosis. Killer immunoglobulin-like receptor (KIR) genes are one of many genes that could be included in a more comprehensive study [89]. Although some of the genes investigated in our study do not show reliable bivariate associations with the severity of TB in NHPs, they might have significant roles in predicting TB outcomes when used in combination with other genes or when considering gene-gene interactions in the model. A larger sample size is required to investigate whether considering gene-gene interactions improves the predictive ability of gene expression for diagnosis and/or prognosis of TB in NHPs.
In conclusion, while the expression levels of the recently identified member of the interferon family, IFNE, was not reliably associated with severity of TB infection, the results of this study indicate that expression levels of TR4, CD40, CD40L, FASC (D95), and TNF, in the PBMCs of MTB-infected rhesus macaques are associated with severity of disease progression, and, as such, exhibit substantial potential as host biomarkers for inclusion in a diagnostic and/or prognostic expression profile for TB infection.
4. Methods
4.1. Study Subjects
Tissue samples remaining from a study of NHPs experimentally infected with TB at the California National Primate Research Center (CNPRC) for other purposes were used for this study [63]. The study subjects were six male rhesus macaques that were simian retrovirus D-negative and SIV-negative (four of pure Indian ancestry, two of mixed Indian-Chinese ancestry) and were colony-bred from the California National Primate Research Center (CNPRC) at the University of California, Davis [63, 90]. All study animals were housed and cared for according to the standards detailed in the Guide for the Care and Use of Laboratory Animals (National Research Council 1996). Protocols for the maintenance of animals were approved by the UC Davis Institutional Animal Care and Use Committee (IACUC). Animals were handled in accordance with Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) Standards [63].
4.2. MTB inoculation, blood sampling, and necropsy
As was explained previously [63], the study animals were inoculated with 500 CFU of MTB (Erdman strain, lot K01) in 3 ml of sterile saline through bronchospcopic inoculation using a BF-XP60 fiberoptic bronchoscope (Olympus, Tokyo, Japan) [63]. Blood samples from the study animals were drawn before MTB inoculation, as well as after inoculation at approximately one month intervals. PBMCs were isolated from whole blood immediately after blood samples were taken, and were stored in a −80°C freezer. PBMCs were isolated from blood samples one month before MTB inoculation (11-05-2008), at several dates post-inoculation (01-05-2009, 02-03-2009, 03-02-2009, 04-01-2009, and at the time of each animal’s euthanasia). These PBMCs were used for the gene expression analysis in this study. However, since two of the six study animals progressed to severe TB and exhibited extreme clinical symptoms, they were euthanized for ethical reasons 9 weeks prior to the euthanasia of the other four study animals.
4.3. PBMC isolation and total RNA extraction
The PBMCs were isolated using Ficoll following standard methods. Frozen PBMCs isolated from the blood samples of the study animals were processed at the Biological Safety Level 3 laboratory at the Center for Comparative Medicine (CCM) at UC Davis. Each vial of frozen PBMC contained approximately 107 cells. These cells were defrosted and rapidly subjected to RNA extraction. The freezing buffer was washed off the PBMCs using 15 ml of Classical Gibco media (RPMI 1640, Life Technologies, NY), for each sample. Lysis buffer was added to the tubes containing PBMCs and the RNA was was extracted following the RNeasy Mini Kit protocol (Qiagen Inc., Valencia, CA). The extracted RNA was stored overnight in a −20°C freezer after adding 10 μL of NaOH and 250 μL of 100% ethanol for precipitation.
4.4. cDNA synthesis and real-time polymerase chain reaction (q-PCR)
The extracted RNA samples were quantified prior to cDNA synthesis following the manufacturers’ guidelines. Normalized concentrations of RNA samples were used for cDNA synthesis and q-PCR reactions. The primers for IFNE (Hs00703565-s1), IL10 (Hs00174086 m1), CD40 (Rh02621776-m1), CXCL10 (Rh02788357-m1), IL17A (Hs00174383-m1), CD40L (Rh02787995-m1), IL12A (Hs00168405-m1), TLR2 (Rh02787279-s1), CSF2 (Rh02621728-m1), IL7 (Rh02621732-m1), IL15 (Rh02621777-m1), PPARG (Rh02787676-m1), TR4 (Hs00991824-m1), IL23A (Rh02872166-m1), FAS (CD95) (Rh02787979-m1), IFNG (Hs00989291-m1), NRAMP1 (Rh02829150-mH), IL13 (Rh03043053-m1), IL4 (Rh02789319-m1), IL5 (Rh03456659-u1), FASL (Rh02621724-m1), CCL2 ( Rh02787889-mH), CD209 (Rh02788045-m1), TNF (Rh02789783-m1), and two different primers (Hs00167257-m1) and (Rh02829281-m1) for NOS2A were obtained from Applied Biosystems (Foster City, CA). The primers for GAPDH (forward, GCACCACCAACTGCTTAGCACC; reverse, TCTTCTGGGTGGCAGTGATG; probe, TCGTGGGAAGGACTCATGACCACAGTCC) were designed, optimized, and validated by the Lucy Whittier Molecular Core Lab at the University of California Davis. The reaction was performed on the ABI ViiA 7 sequence detector (Applied Biosystems, CA) in tubes containing RT-qPCR master mix. Expression levels of all samples were normalized to that of a housekeeping gene, GAPDH (glycealdehyde-3-phosphate dehydrogenase), and expressed as the relative difference.
4.5. Histopathology
Paraffin embedded blocks containing tissue samples from the right caudal lobe of the lungs of all the study animals were used to prepare histopathology slides using hemtoxyline and eosin or trichrome blue staining at the Anatomic Pathology Service of the William R. Pritchard Veterinary Medical Teaching Hospital, University of California, Davis.
The Supplementary Material contains the raw data on TB outcomes for each animal, in each lung lobe, for each measure of TB severity; see GranulomaCount.csv, GranulomaSize.csv, GranulomaVolume.csv, NonGranulomaVolume.csv, and CellCount.csv. Definitions for the values in these data sets are provided below.
4.6. Statistical Analysis
4.6.1. Model Definition
We have five outcome variables per animal, n ∈ {1, …, N}, per lung lobe, l ∈ {1 … L}, which are: 1) a semiquantitative granuloma count, G[n,l] ∈ {0, 1}, where a value of 1 indicates more than 10 granulomas per sample or a miliary pattern, and 0 indicates 10 or fewer granulomas; 2) a semiquantitative characterization of the size of the largest granuloma, S [n,l] ∈ {0, 1}, where a value of 1 indicates a granuloma size of 3mm or greater, and a value of 0 indicates a size of less than 3mm; 3) the log of the volume of granuloma lesions ; 4) the log of the volume of non-granuloma lesions ; and 5) the log of Mycobacterium tuberculosis load (CFU), . A small constant was added to volume data to ensure that the log of 0 was not taken. Each of these outcome variables is taken directly from previously published work [63].
As is the case with the non-independence of TB pathogenesis across lung lobes within animals, our outcome variables within lung lobes are non-independent. Each of these outcome variables, however, contain slightly different information, in that differing genetic factors might have implications for the count versus size of granulomas, or implications for the nature of lesions as granulomatous versus non-granulomatous. We model these outcomes jointly using a multi-level hybrid Gaussian-Bernoulli regression model, so that the correlations, if any, between the regression parameters for each outcome variable can be estimated and used to improve inference within each sub-model. We use Hamiltonian Marcov Chain Monte Carlo (MCMC) simulation, implemented using the Stan v2.2 C++ library, to fit our model [91].
At each MCMC iteration we model:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
where λ[n,l], θ[n,l], ψ[n,l], ϕ[n,l], and κ[n,l] are defined as functions of the expression levels, X[n,t], of a given gene, in a given animal, n, at a given point in time, t:
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
It should be noted that a single predictor in animal n, the gene expression level, X[n,t] in PBMCs, is used to predict each outcome variable in each lung lobe. It should also be noted that the intercept (β[1,l], β[3,l], …, β[9,l]) and slope (β[2,l], β[4,l], …, β[10,l]) coefficients have unique values for each lobe, l. This allows for the relationship between gene expression and presence of granuloma to vary by lung lobe (an effect which is clearly visible in the raw data distributions). Finally, we note that we refit the model iteratively for each combination of gene and time, t, to yield an estimation of the usefulness of the expression levels of a given gene at a given time to predict the severity and form of M. tuberculosis infection at post-mortem analysis.
At each model iteration, we use a hierarchical model structure to estimate the value of the coefficient vector, β[l], while simultaneously pooling information across lung lobes, to yield coefficient estimates that describe the average relationship between gene expression levels and pathogenesis across lung lobe types, μβ. We model:
| (11) |
where Σ is a variance-covariance matrix which allows for partial pooling of information across the parameters within and between each of the sub-models included in our analysis.
To complete the fully Bayesian model definition, we define weakly informative priors on each cell of the mean parameter vector, μβ:
| (12) |
We define a prior on the variance-covariance matrix Σ, using a correlation matrix, ρ, multiplied on both sides by diagonal matrices of value ξ to scale the size of the variance terms:
| (13) |
We use weakly regularizing (half) Cauchy priors on the elements of ξ:
| (14) |
and the correlation matrix is given a prior of:
| (15) |
which places an approximately uniform density across the off diagonal of the matrix. The LKJcorr distribution is describe by [92], and is used in the Stan programming language as an alternative to Wishart-based variance-covariance matrix parameterizations, which Gelman and colleagues argue are often too restrictive for general use [93].
The standard deviation parameters for the Gaussian models are given weakly regularizing (half) Cauchy distributions as priors:
| (16) |
| (17) |
| (18) |
4.6.2. Model Comparison
We compare models using the Watanabe-Akaike information criterion (WAIC) [66], which is a more fully-Bayesian generalization of the standard Akaike information theoretic criteria, AIC. Computed WAIC is defined as:
| (19) |
The computed log pointwise posterior predictive density, lppd, is defined as:
| (20) |
where q ∈ {1, …, Q} references the index of simulations from the posterior distribution, with Q being the total number of saved simulations. The effective number of parameters, pE is computed as:
| (21) |
where the symbol represents the function to calculate the sample variance over the posterior simulations. The symbol Ψ[n,l,q] represents the model predictions (eg. λ[n,l], θ[n,l], etc …), and the symbol Y[n,l] represents the model data (eg. G[n,l], S[n,l], etc …).
Supplementary Material
Highlights.
The expression levels of NR2C2(TR4), CD40, CD40L, Fas (CD95) and TNF-α in PBMC were associated with quantitative measures of the severity of TB histopathologic lesions in the lungs of the study animals.
No reliable association between the expression levels of IFN-ε in PBMCs and the severity of TB lesions in the lungs of the study animals was found.
In conclusion, PBMC expression profiles derived from the above-listed host genes might be appropriate biomarkers for probabilistic diagnosis and/or prognosis of TB severity in rhesus macaques.
5. Acknowledgments
This research was funded by NIH grant #RR025871 and the Clinical and Translational Science Center’s T32 program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].W. H. Organization, Global Tuberculosis Report 2013, World Health Organization, 2013. [Google Scholar]
- [2].Walsh GP, Tan EV, Cruz ECD, Abalos RM, Villahermosa LG, Young LJ, Cellona RV, Narareno JB, Horwitz MA, The Philippine cynomolgus monkey (Macaca fasicularis) provides a new nonhuman primate model of tuberculosis that resembles human disease, Nature medicine 2 (4) (1996) 430–436. [DOI] [PubMed] [Google Scholar]
- [3].Capuano SV, Croix DA, Pawar S, Zinovik A, Myers A, Lin PL, Bissel S, Fuhrman C, Klein E, Flynn JL, Experimental Mycobacterium tuberculosis infection of cynomolgus macaques closely resembles the various manifestations of human M. tuberculosis infection, Infection and immunity 71 (10) (2003) 5831–5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Barclay W, Anacker R, Brehmer W, Leif W, Ribi E, Aerosol-induced tuberculosis in subhuman primates and the course of the disease after intravenous BCG vaccination, Infection and immunity 2 (5) (1970) 574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Flynn J, Capuano S, Croix D, Pawar S, Myers A, Zinovik A, Klein E, Non-human primates: a model for tuberculosis research, Tuberculosis 83 (1) (2003) 116–118. [DOI] [PubMed] [Google Scholar]
- [6].Ross CT, Weise JA, Bonnar S, Nolin D, Satkoski Trask J, Smith DG, Ferguson B, Ha J, Kubisch HM, Vinson A, et al. , An empirical comparison of short tandem repeats (STRs) and single nucleotide polymorphisms (SNPs) for relatedness estimation in Chinese rhesus macaques (Macaca mulatta), American journal of primatology 76 (4) (2014) 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Marino S, Cilfone NA, Mattila JT, Linderman JJ, Flynn JL, Kirschner DE, Macrophage polarization drives granuloma outcome during Mycobacterium tuberculosis infection, Infection and immunity 83 (1) (2015) 324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Roodgar M, Ross CT, Kenyon NJ, Marcelino G, Smith DG, Inducible nitric oxide synthase (iNOS) regulatory region variation in non-human primates, Infection, Genetics and Evolution 31 (2015) 236–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Möller M, Hoal EG, Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis, Tuberculosis 90 (2) (2010) 71–83. [DOI] [PubMed] [Google Scholar]
- [10].Hernandez-Pando R, Schön T, Orozco E, Serafin J, Estrada-Garcia I, Expression of inducible nitric oxide synthase and nitrotyrosine during the evolution of experimental pulmonary tuberculosis, Experimental and Toxicologic Pathology 53 (4) (2001) 257–265. [DOI] [PubMed] [Google Scholar]
- [11].Kessler M, Brown R, Intraocular granuloma associated with disseminated tuberculosis in a rhesus monkey (Macaca mulatta), The Journal of Zoo Animal Medicine (1979) 122–124. [Google Scholar]
- [12].Zumpe D, Silberman M, Michael R, Unusual outbreak of tuberculosis due to mycobacterium bovis in a closed colony of rhesus monkeys (macaca mulatta)., Laboratory animal science 30 (2 Pt 1) (1980) 237–240. [PubMed] [Google Scholar]
- [13].Mahajan S, Dkhar HK, Chandra V, Dave S, Nanduri R, Janmeja AK, Agrewala JN, Gupta P, Mycobacterium tuberculosis modulates macrophage lipid-sensing nuclear receptors PPARγ and TR4 for survival, The Journal of Immunology 188 (11) (2012) 5593–5603. [DOI] [PubMed] [Google Scholar]
- [14].Cooper AM, Dalton DK, Stewart TA, Griffin J, Russell D, Orme I, Disseminated tuberculosis in interferon gamma gene-disrupted mice, The Journal of experimental medicine 178 (6) (1993) 2243–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR, An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection., The Journal of experimental medicine 178 (6) (1993) 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Takeda K, Kawai T, Nakazawa Y, Komuro H, Shoji K, Morita K, Katsuta T, Yamamoto M, Miyairi I, Ohya Y, et al. , Augmentation of antitubercular therapy with interferon-γ in a patient with dominant partial interferon-γ receptor 1 deficiency, Clinical Immunology. [DOI] [PubMed] [Google Scholar]
- [17].Lazarevic V, Myers AJ, Scanga CA, Flynn JL, CD40, but not CD40L, is required for the optimal priming of t cells and control of aerosol M. tuberculosis infection, Immunity 19 (6) (2003) 823–835. [DOI] [PubMed] [Google Scholar]
- [18].Shey MS, Nemes E, Whatney W, de Kock M, Africa H, Barnard C, van Rooyen M, Stone L, Riou C, Kollmann T, et al. , Maturation of innate responses to mycobacteria over the first nine months of life, The Journal of Immunology (2014) 1400062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Juffermans NP, Verbon A, van Deventer SJ, van Deutekom H, Belisle JT, Ellis ME, Speelman P, van der Poll T, Elevated chemokine concentrations in sera of human immunodeficiency virus HIV-seropositive and HIV-seronegative patients with tuberculosis: a possible role for Mycobacterial lipoarabinomannan, Infection and immunity 67 (8) (1999) 4295–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gonzalez-Juarrero M, Hattle JM, Izzo A, Junqueira-Kipnis AP, Shim TS, Trapnell BC, Cooper AM, Orme IM, Disruption of granulocyte macrophage-colony stimulating factor production in the lungs severely affects the ability of mice to control Mycobacterium tuberculosis infection, Journal of leukocyte biology 77 (6) (2005) 914–922. [DOI] [PubMed] [Google Scholar]
- [21].Szeliga J, Daniel DS, Yang C-H, Sever-Chroneos Z, Jagannath C, Chroneos ZC, Granulocyte-macrophage colony stimulating factor-mediated innate responses in tuberculosis, Tuberculosis 88 (1) (2008) 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Klug-Micu GM, Stenger S, Sommer A, Liu PT, Krutzik SR, Modlin RL, Fabri M, CD40 ligand and Interferon-γ induce an antimicrobial response against Mycobacterium tuberculosis in human monocytes, Immunology 139 (1) (2013) 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tripathi MK, Yasir M, Gurjar VS, Bose P, Dubey A, Shrivastava R, Insights from the molecular docking of hydrolytic products of methyl-iso-cyanate (MIC) to inhibition of human immune proteins, Interdisciplinary Sciences: Computational Life Sciences (2015) 1–8. [DOI] [PubMed] [Google Scholar]
- [24].Filipe-Santos O, Bustamante J, Haverkamp MH, Vinolo E, Ku C-L, Puel A, Frucht DM, Christel K, Von Bernuth H, Jouanguy E, et al. , X-linked susceptibility to mycobacteria is caused by mutations in nemo impairing cd40-dependent il-12 production, The Journal of experimental medicine 203 (7) (2006) 1745–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Maeurer MJ, Trinder P, Hommel G, Walter W, Freitag K, Atkins D, Störkel S, Interleukin-7 or Interleukin-15 enhances survival of Mycobacterium tuberculosis-infected mice, Infection and immunity 68 (5) (2000) 2962–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Feske M, Nudelman RJ, Medina M, Lew J, Singh M, Couturier J, Graviss EA, Lewis DE, Enhancement of human antigen-specific memory t-cell responses by Interleukin-7 may improve accuracy in diagnosing tuberculosis, Clinical and Vaccine Immunology 15 (10) (2008) 1616–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rane L, Rahman S, Magalhaes I, Ahmed R, Spångberg M, Kondova I, Verreck F, Andersson J, Brighenti S, Maeurer M, Increased (6 exon) Interleukin-7 production after M. tuberculosis infection and soluble Interleukin-7 receptor expression in lung tissue, Genes and immunity 12 (7) (2011) 513–522. [DOI] [PubMed] [Google Scholar]
- [28].Zissel G, Bäumer I, Schlaak M, Müller-Quernheim J, In vitro release of Interleukin-15 by broncho-alveolar lavage cells and peripheral blood mononuclear cells from patients with different lung diseases., European cytokine network 11 (1) (2000) 105–12. [PubMed] [Google Scholar]
- [29].Abebe F, Mustafa T, Nerland A, Bjune G, Cytokine profile during latent and slowly progressive primary tuberculosis: a possible role for Interleukin-15 in mediating clinical disease, Clinical & Experimental Immunology 143 (1) (2006) 180–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Turner J, D’souza C, Pearl J, Marietta P, Noel M, Frank A, Appelberg R, Orme I, Cooper A, CD8-and CD95/95L-dependent mechanisms of resistance in mice with chronic pulmonary tuberculosis, American journal of respiratory cell and molecular biology 24 (2) (2001) 203–209. [DOI] [PubMed] [Google Scholar]
- [31].Khader SA, Bell GK, Pearl JE, Fountain JJ, Rangel-Moreno J, Cilley GE, Shen F, Eaton SM, Gaffen SL, Swain SL, et al. , IL-23 and IL-17 in the establishment of protective pulmonary CD4+T cell responses after vaccination and during Mycobacterium tuberculosis challenge, Nature immunology 8 (4) (2007) 369–377. [DOI] [PubMed] [Google Scholar]
- [32].Khader SA, Pearl JE, Sakamoto K, Gilmartin L, Bell GK, Jelley-Gibbs DM, Ghilardi N, Cooper AM, et al. , IL-23 compensates for the absence of IL-12p70 and is essential for the IL-17 response during tuberculosis but is dispensable for protection and antigen-specific IFN-γ responses if IL-12p70 is available, The Journal of Immunology 175 (2) (2005) 788–795. [DOI] [PubMed] [Google Scholar]
- [33].Zhang M, Gately M, Wang E, Gong J, Wolf S, Lu S, Modlin R, Barnes P, Interleukin-12 at the site of disease in tuberculosis, Journal of Clinical Investigation 93 (4) (1994) 1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Flynn J, Goldstein MM, Triebold KJ, Sypek J, Wolf S, Bloom BR, Il-12 increases resistance of BALB/c mice to Mycobacterium tuberculosis infection., The Journal of Immunology 155 (5) (1995) 2515–2524. [PubMed] [Google Scholar]
- [35].Hölscher C, Atkinson RA, Arendse B, Brown N, Myburgh E, Alber G, Brombacher F, A protective and agonistic function of il-12p40 in mycobacterial infection, The Journal of Immunology 167 (12) (2001) 6957–6966. [DOI] [PubMed] [Google Scholar]
- [36].Nolan A, Fajardo E, Huie ML, Condos R, Pooran A, Dawson R, Dheda K, Bateman E, Rom WN, Weiden MD, Increased production of IL-4 and IL-12p40 from bronchoalveolar lavage cells are biomarkers of Mycobacterium tuberculosis in the sputum, PloS one 8 (3) (2013) e59461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Seah G, Scott G, Rook G, Type 2 cytokine gene activation and its relationship to extent of disease in patients with tuberculosis, Journal of Infectious Diseases 181 (1) (2000) 385–389. [DOI] [PubMed] [Google Scholar]
- [38].Orme IM, Roberts AD, Griffin JP, Abrams J, Cytokine secretion by CD4 T lymphocytes acquired in response to Mycobacterium tuberculosis infection, The Journal of Immunology 151 (1) (1993) 518–525. [PubMed] [Google Scholar]
- [39].Collins HL, Schaible UE, Kaufmann SH, Early IL-4 induction in bone marrow lymphoid precursor cells by Mycobacterial lipoarabinomannan, The Journal of Immunology 161 (10) (1998) 5546–5554. [PubMed] [Google Scholar]
- [40].Benjamin R, Banerjee A, Sunder SR, Gaddam S, Valluri VL, Banerjee S, Discordance in CD4+ T-cell levels and viral loads with co-occurrence of elevated peripheral TNF-α and IL-4 in newly diagnosed HIV-TB co-infected cases, PloS one 8 (8) (2013) e70250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Li X, Yang Y, Zhou F, Zhang Y, Lu H, Jin Q, Gao L, SLC11A1 (NRAMP1) polymorphisms and tuberculosis susceptibility: updated systematic review and meta-analysis, PLoS One 6 (1) (2011) e15831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Malo D, Vogan K, Vidal S, Hu J, Cellier M, Schurr E, Fuks A, Bumstead N, Morgan K, Gros P, Haplotype mapping and sequence analysis of the mouse Nramp gene predict susceptibility to infection with intracellular parasites, Genomics 23 (1) (1994) 51–61. [DOI] [PubMed] [Google Scholar]
- [43].Vidal SM, Malo D, Vogan K, Skamene E, Gros P, Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg, Cell 73 (3) (1993) 469–485. [DOI] [PubMed] [Google Scholar]
- [44].North RJ, LaCourse R, Ryan L, Gros P, Consequence of Nramp1 deletion to Mycobacterium tuberculosis infection in mice, Infection and immunity 67 (11) (1999) 5811–5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bellamy R, Ruwende C, Corrah T, McAdam KP, Whittle HC, Hill AV, Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africans, New England Journal of Medicine 338 (10) (1998) 640–644. [DOI] [PubMed] [Google Scholar]
- [46].Gruenheid S, Pinner E, Desjardins M, Gros P, Natural resistance to infection with intracellular pathogens: the Nramp1 protein is recruited to the membrane of the phagosome, The Journal of experimental medicine 185 (4) (1997) 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Underhill DM, Ozinsky A, Smith KD, Aderem A, Toll-like receptor-2 mediates mycobacteria-induced proinflammatory signaling in macrophages, Proceedings of the National Academy of Sciences 96 (25) (1999) 14459–14463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Means TK, Wang S, Lien E, Yoshimura A, Golenbock DT, Fenton MJ, Human toll-like receptors mediate cellular activation by Mycobacterium tuberculosis, The Journal of Immunology 163 (7) (1999) 3920–3927. [PubMed] [Google Scholar]
- [49].Mustafa T, Phyu S, Nilsen R, Bjune G, Jonsson R, Increased expression of Fas ligand on Mycobacterium tuberculosis infected macrophages: a potential novel mechanism of immune evasion by Mycobacterium tuberculosis?, Inflammation 23 (6) (1999) 507–521. [DOI] [PubMed] [Google Scholar]
- [50].Ling WL, Wang LJ, Pong JC, Lau AS, Li JC, A role for Interleukin-17A in modulating intracellular survival of Mycobacterium bovis bacillus Calmette-Guérin in murine macrophages, Immunology 140 (3) (2013) 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wareham AS, Tree JA, Marsh PD, Butcher PD, Dennis M, Sharpe SA, Evidence for a role for Interleukin-17, Th17 cells and iron homeostasis in protective immunity against tuberculosis in cynomolgus macaques, PloS one 9 (2) (2014) e88149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Friedland JS, Shattock RJ, Griffin GE, Phagocytosis of Mycobacterium tuberculosis or particulate stimuli by human monocytic cells induces equivalent monocyte chemotactic protein-1 gene expression, Cytokine 5 (2) (1993) 150–156. [DOI] [PubMed] [Google Scholar]
- [53].Kipnis A, Basaraba RJ, Orme IM, Cooper AM, Role of chemokine ligand 2 in the protective response to early murine pulmonary tuberculosis, Immunology 109 (4) (2003) 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hasan Z, Cliff JM, Dockrell HM, Jamil B, Irfan M, Ashraf M, Hussain R, CCL2 responses to Mycobacterium tuberculosis are associated with disease severity in tuberculosis, PLoS One 4 (12) (2009) e8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ganachari M, Guio H, Zhao N, Flores-Villanueva PO, Host gene-encoded severe lung TB: from genes to the potential pathways, Genes and immunity 13 (8) (2012) 605–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gautam US, Mehra S, Ahsan MH, Alvarez X, Niu T, Kaushal D, Role of TNF in the altered interaction of dormant Mycobacterium tuberculosis with host macrophages, PloS one 9 (4) (2014) e95220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kwon OJ, Kim JH, Kim HC, Suh GY, Park JW, Chung MP, Kim H, Rhee CH, Nitric oxide expression in airway epithelial cells in response to tubercle bacilli stimulation, Respirology 3 (2) (1998) 119–124. [DOI] [PubMed] [Google Scholar]
- [58].Garcia I, Guler R, Vesin D, Olleros ML, Vassalli P, Chvatchko Y, Jacobs M, Ryffel B, Lethal Mycobacterium bovis Bacillus Calmette Guerin infection in nitric oxide synthase 2-deficient mice: cell-mediated immunity requires nitric oxide synthase 2, Laboratory investigation 80 (9) (2000) 1385–1397. [DOI] [PubMed] [Google Scholar]
- [59].Kapoor N, Narayana Y, Patil SA, Balaji KN, Nitric oxide is involved in Mycobacterium bovis Bacillus Calmette-Guérin-activated Jagged1 and Notch1 signaling, The Journal of Immunology 184 (6) (2010) 3117–3126. [DOI] [PubMed] [Google Scholar]
- [60].Gupta D, Sharma S, Singhal J, Satsangi AT, Antony C, Natarajan K, Suppression of TLR2-induced IL-12, reactive oxygen species, and inducible nitric oxide synthase expression by Mycobacterium tuberculosis antigens expressed inside macrophages during the course of infection, The Journal of Immunology 184 (10) (2010) 5444–5455. [DOI] [PubMed] [Google Scholar]
- [61].Tailleux L, Schwartz O, Herrmann J-L, Pivert E, Jackson M, Amara A, Legres L, Dreher D, Nicod LP, Gluckman JC, et al. , DC-SIGN is the major Mycobacterium tuberculosis receptor on human dendritic cells, The Journal of experimental medicine 197 (1) (2003) 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Geijtenbeek TB, van Vliet SJ, Koppel EA, Sanchez-Hernandez M, Vandenbroucke-Grauls CM, Appelmelk B, van Kooyk Y, Mycobacteria target DC-SIGN to suppress dendritic cell function, The Journal of experimental medicine 197 (1) (2003) 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Luciw PA, Oslund KL, Yang X.-w., Adamson L, Ravindran R, Canfield DR, Tarara R, Hirst L, Christensen M, Lerche NW, et al. , Stereological analysis of bacterial load and lung lesions in nonhuman primates (rhesus macaques) experimentally infected with Mycobacterium tuberculosis, American Journal of Physiology-Lung Cellular and Molecular Physiology 301 (5) (2011) L731–L738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Alizadeh AA, Ross DT, Perou CM, van de Rijn M, Towards a novel classification of human malignancies based on gene expression patterns, The Journal of pathology 195 (1) (2001) 41–52. [DOI] [PubMed] [Google Scholar]
- [65].Millar RB, Anderson MJ, Remedies for pseudoreplication, Fisheries Research 70 (2) (2004) 397–407. [Google Scholar]
- [66].Gelman A, Hwang J, Vehtari A, Understanding predictive information criteria for Bayesian models, Statistics and Computing (2013) 1–20. [Google Scholar]
- [67].Lin PL, Pawar S, Myers A, Pegu A, Fuhrman C, Reinhart TA, Capuano SV, Klein E, Flynn JL, Early events in Mycobacterium tuberculosis infection in cynomolgus macaques, Infection and immunity 74 (7) (2006) 3790–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kanthaswamy S, Ng J, Ross CT, Trask JS, Smith DG, Buffalo VS, Fass JN, Lin D, Identifying human-rhesus macaque gene orthologs using heterospecific SNP probes, Genomics 101 (1) (2013) 30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Dkhar HK, Nanduri R, Mahajan S, Dave S, Saini A, Somavarapu AK, Arora A, Parkesh R, Thakur KG, Mayilraj S, et al. , Mycobacterium tuberculosis keto-mycolic acid and macrophage nuclear receptor TR4 modulate foamy biogenesis in granulomas: a case of a heterologous and noncanonical ligand-receptor pair, The Journal of Immunology 193 (1) (2014) 295–305. [DOI] [PubMed] [Google Scholar]
- [70].Xu Y, Yang E, Huang Q, Ni W, Kong C, Liu G, Li G, Su H, Wang H, PPE57 induces activation of macrophages and drives Th1-type immune responses through TLR2, Journal of Molecular Medicine (2015) 1–18. [DOI] [PubMed] [Google Scholar]
- [71].Campos-Neto A, Ovendale P, Bement T, Koppi TA, Fanslow WC, Rossi MA, Alderson MR, Cutting edge: CD40 ligand is not essential for the development of cell-mediated immunity and resistance to Mycobacterium tuberculosis, The Journal of Immunology 160 (5) (1998) 2037–2041. [PubMed] [Google Scholar]
- [72].Stenger S, Modlin RL, T cell mediated immunity to Mycobacterium tuberculosis, Current opinion in microbiology 2 (1) (1999) 89–93. [DOI] [PubMed] [Google Scholar]
- [73].Roberts A, Orme IM, CD95 expression in aged mice infected with tuberculosis, Infection and immunity 66 (10) (1998) 5036–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Mustafa T, Wiker HG, Mørkve O, Sviland L, Differential expression of mycobacterial antigen MPT64, apoptosis and inflammatory markers in multinucleated giant cells and epithelioid cells in granulomas caused by Mycobacterium tuberculosis, Virchows Archiv 452 (4) (2008) 449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].El-Masry S, Lotfy M, Nasif W, El-Kady I, Al-Badrawy M, Elevated serum level of interleukin IL-18, interferon IFN-γ and soluble Fas in patients with pulmonary complications in tuberculosis, Acta microbiologica et immunologica Hungarica 54 (1) (2007) 65–77. [DOI] [PubMed] [Google Scholar]
- [76].Mustafa T, Mogga SJ, Mfinanga SG, Mørkve O, Sviland L, Significance of Fas and Fas ligand in tuberculous lymphadenitis, Immunology 114 (2) (2005) 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Roodgar M, Lackner A, Kaushal D, Sankaran S, Dandekar S, Satkoski Trask J, Drake C, Smith DG, Expression levels of 10 candidate genes in lung tissue of vaccinated and TB-infected cynomolgus macaques, Journal of medical primatology 42 (3) (2013) 161–164. [DOI] [PubMed] [Google Scholar]
- [78].Mattila JT, Ojo OO, Kepka-Lenhart D, Marino S, Kim JH, Eum SY, Via LE, Barry CE, Klein E, Kirschner DE, et al. , Microenvironments in tuberculous granulomas are delineated by distinct populations of macrophage subsets and expression of nitric oxide synthase and arginase isoforms, The Journal of Immunology 191 (2) (2013) 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Choi H-S, Rai PR, Chu HW, Cool C, Chan ED, Analysis of nitric oxide synthase and nitrotyrosine expression in human pulmonary tuberculosis, American journal of respiratory and critical care medicine 166 (2) (2002) 178–186. [DOI] [PubMed] [Google Scholar]
- [80].Roy S, Sharma S, Sharma M, Aggarwal R, Bose M, Induction of nitric oxide release from the human alveolar epithelial cell line A549: an in vitro correlate of innate immune response to Mycobacterium tuberculosis, Immunology 112 (3) (2004) 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Alvarez-Puebla M, Olaguibel RJ, Almudevar E, Echegoyen A, de Esteban CB, Cambra K , Cutoff point for exhaled nitric oxide corresponding to 3% sputum eosinophils., Journal of investigational allergology & clinical immunology 25 (2) (2014) 107–111. [PubMed] [Google Scholar]
- [82].Tang NL-S, Fan HPY, Chang KC, Ching JKL, Kong KPS, Yew WW, Kam KM, Leung CC, Tam CM, Blackwell J, et al. , Genetic association between a chemokine gene CXCL-10 (IP-10, interferon gamma inducible protein 10) and susceptibility to tuberculosis, Clinica Chimica Acta 406 (1) (2009) 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kasprowicz VO, Mitchell JE, Chetty S, Govender P, Huang K, Fletcher HA, Webster DP, Brown S, Kasmar A, Millington K, et al. , A molecular assay for sensitive detection of pathogen-specific T-cells, PloS one 6 (8) (2011) e20606–e20606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Ahmed Yassin M, Petrucci R, Garie KT, Harper G, Arbide I, Aschalew M, Merid Y, Kebede Z, Bawazir AA, Abuamer NM, et al. , Can interferon-gamma or interferon-gamma-induced-protein-10 differentiate tuberculosis infection and disease in children of high endemic areas?, PLoS ONE 6 (9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Simeone R, Sayes F, Song O, Gröschel MI, Brodin P, Brosch R, Majlessi L, Salgame P, Cytosolic access of Mycobacterium tuberculosis: Critical impact of phagosomal acidification control and demonstration of occurrence in vivo., PLoS pathogens 11 (2) (2015) e1004650–e1004650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Spigelman M, Donoghue HD, Abdeen Z, Ereqat S, Sarie I, Greenblatt CL, Pap I, Szikossy I, Hershkovitz I, Bar-Gal GK, et al. , Evolutionary changes in the genome of Mycobacterium tuberculosis and the human genome from 9000 years bp until modern times, Tuberculosis. [DOI] [PubMed] [Google Scholar]
- [87].Wu L, Deng H, Zheng Y, Mansjö M, Zheng X, Hu Y, Xu B, An association study of NRAMP1, VDR, MBL and their interaction with the susceptibility to tuberculosis in a Chinese population, International Journal of Infectious Diseases 38 (2015) 129–135. [DOI] [PubMed] [Google Scholar]
- [88].Higgins DM, Sanchez-Campillo J, Rosas-Taraco AG, Higgins JR, Lee EJ, Orme IM, Gonzalez-Juarrero M, Relative levels of M-CSF and GM-CSF influence the specific generation of macrophage populations during infection with Mycobacterium tuberculosis, The Journal of Immunology 180 (7) (2008) 4892–4900. [DOI] [PubMed] [Google Scholar]
- [89].Braun K, Wolfe J, Kiazyk S, Sharma MK, Evaluation of host genetics on outcome of tuberculosis infection due to differences in killer immunoglobulin-like receptor gene frequencies and haplotypes, BMC genetics 16 (1) (2015) 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Kanthaswamy S, Trask JS, Ross CT, Kou A, Houghton P, Smith DG, Lerche N, A large-scale SNP-based genomic admixture analysis of the captive rhesus macaque colony at the California National Primate Research Center, American journal of primatology 74 (8) (2012) 747–757. [DOI] [PubMed] [Google Scholar]
- [91].Stan Development Team, Stan: A C++ library for probability and sampling, version 2.2 (2013). URL http://mc-stan.org/
- [92].Lewandowski D, Kurowicka D, Joe H, Generating random correlation matrices based on vines and extended onion method, Journal of multivariate analysis 100 (9) (2009) 1989–2001. [Google Scholar]
- [93].Tokuda T, Goodrich B, Van Mechelen I, Gelman A, Tuerlinckx F, Visualizing distributions of covariance matrices, Tech. rep., Technical report, University of Leuwen, Belgium and Columbia University, USA. URL http://www.stat.columbia.edu/~gelman/research/unpublished/Visualization.pdf.(Cited on pages 114, 116, 117 and 119.) (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


