Abstract
Cerebral white matter hyperintensities on MRI are markers of cerebral small vessel disease, a major risk factor for dementia and stroke. Despite the successful identification of multiple genetic variants associated with this highly heritable condition, its genetic architecture remains incompletely understood. More specifically, the role of DNA methylation has received little attention.
We investigated the association between white matter hyperintensity burden and DNA methylation in blood at ∼450 000 cytosine-phosphate-guanine (CpG) sites in 9732 middle-aged to older adults from 14 community-based studies. Single CpG and region-based association analyses were carried out. Functional annotation and integrative cross-omics analyses were performed to identify novel genes underlying the relationship between DNA methylation and white matter hyperintensities.
We identified 12 single CpG and 46 region-based DNA methylation associations with white matter hyperintensity burden. Our top discovery single CpG, cg24202936 (P = 7.6 × 10−8), was associated with F2 expression in blood (P = 6.4 × 10−5) and co-localized with FOLH1 expression in brain (posterior probability = 0.75). Our top differentially methylated regions were in PRMT1 and in CCDC144NL-AS1, which were also represented in single CpG associations (cg17417856 and cg06809326, respectively). Through Mendelian randomization analyses cg06809326 was putatively associated with white matter hyperintensity burden (P = 0.03) and expression of CCDC144NL-AS1 possibly mediated this association. Differentially methylated region analysis, joint epigenetic association analysis and multi-omics co-localization analysis consistently identified a role of DNA methylation near SH3PXD2A, a locus previously identified in genome-wide association studies of white matter hyperintensities. Gene set enrichment analyses revealed functions of the identified DNA methylation loci in the blood–brain barrier and in the immune response. Integrative cross-omics analysis identified 19 key regulatory genes in two networks related to extracellular matrix organization, and lipid and lipoprotein metabolism. A drug-repositioning analysis indicated antihyperlipidaemic agents, more specifically peroxisome proliferator-activated receptor-alpha, as possible target drugs for white matter hyperintensities.
Our epigenome-wide association study and integrative cross-omics analyses implicate novel genes influencing white matter hyperintensity burden, which converged on pathways related to the immune response and to a compromised blood–brain barrier possibly due to disrupted cell–cell and cell–extracellular matrix interactions. The results also suggest that antihyperlipidaemic therapy may contribute to lowering risk for white matter hyperintensities possibly through protection against blood–brain barrier disruption.
Keywords: epigenome-wide association study, white matter hyperintensities, cerebral small vessel disease, integrative cross-omics analysis, blood–brain barrier dysfunction
Yang et al. investigate the association between epigenetic changes and white matter hyperintensities—markers of cerebral small vessel disease—in 9732 adults. Integrative cross-omics analyses implicate blood–brain barrier dysfunction and the immune response in WMH burden, and suggest antihyperlipidemic agents as a potential treatment.
Introduction
Cerebral white matter hyperintensities (WMH) on MRI are indicative of cerebral small vessel disease (cSVD) and are part of the spectrum of brain vascular injury that affects cognitive function, also known as vascular contributions to cognitive impairment and dementia.1,2 While the pathophysiology of WMH is little understood and probably heterogeneous, it probably has ischaemic and neurodegenerative origins.1 Historical pathology data suggested chronic ischaemia resulting in demyelination and axonal loss as an underlying mechanism; however, neuroimaging data point to blood–brain barrier (BBB) dysfunction, dysfunctional blood flow linked with impaired cerebrovascular autoregulation, vascular stiffness, periarteriolar inflammation and more recently protein deposition (i.e. amyloid angiopathy).2 Genetics play a significant role in WMH with a heritability estimated from 54% to 80%3–7; however, the genetic variants identified in association studies explain only ∼29% of WMH variance.8,9 Epigenetic changes such as DNA methylation (DNAm), which regulate gene expression, have emerged as another key component of the genetic architecture of complex traits.10 Unlike DNA sequence variation, which remains unchanged throughout life, DNAm is plastic and highly sensitive to changes in the environment and ageing.10,11 To date, its role in cSVD has received little attention. We hypothesized that there may be patterns of DNAm associated with WMH that are common across all populations. We also hypothesized that the interplay between genotype, epigenotype and risk factor exposure underlies cSVD aetiology and used an integrated analytic framework to identify such relationships.
Materials and methods
Overview
This study comprised five analytic parts to implicate novel genes and gene networks in WMH aetiology (Fig. 1). First, we performed an epigenome-wide association analysis to identify DNAm loci, both cytosine-phosphate-guanine (CpG) sites and differentially methylated regions (DMRs), associated with WMH burden. The identified DNAm loci were then annotated for regulatory features, pathways and association with other traits. Second, we investigated the contribution of genetic variation to variation in DNAm at the identified CpGs and used Mendelian randomization (MR) techniques to test for causal association with WMH burden and for the mediating role of expression of nearby genes. Third, we examined the role of DNAm at established WMH genome-wide association study (GWAS) loci. Fourth, we integrated gene expression and expression quantitative trait loci (eQTL) data to prioritize candidate genes associated with the identified CpGs. Last, we performed integrative cross-omics analyses to derive WMH-associated genes networks and their key drivers and to reposition drug targets.
Figure 1.
Overview of the study analytic scheme.
Study subjects
The sample included 9732 middle-aged to older adults of European and African ancestry from 14 community-based studies. Our discovery sample includes 5715 subjects of European ancestry (n = 4610) and of African ancestry (n = 1105) from Atherosclerosis Risk in Communities (ARIC),12 Biobanking and BioMolecular resources Research Infrastructure,13 Cardiovascular Health Study (CHS),14 Coronary Artery Risk Development in Young Adults (CARDIA),15 Framingham Heart Study (FHS) offspring study,16,17 Genetic Epidemiology Network of Arteriopathy (GENOA) study,18 Lothian Birth Cohort 1936,19,20 Rotterdam Study21,22 and Study of Health in Pomerania (SHIP).23 To replicate our findings, we accessed data on 3398 subjects from the Alzheimer’s Disease Neuroimaging Initiative,24,25 FHS third generation study,26 the Older Australian Twin Study27,28 and the Rhineland Study.29 Additionally, we included a secondary replication sample (n = 619) from the BRIDGET Consortium.30 Subjects with history of stroke or dementia were excluded. Details about participating studies and study-specific ethics statements are provided in the Supplementary material. Each study obtained written informed consent from all participants and approval from the appropriate institutional review boards.
WMH burden measurements
Brain MRI was taken in the same or the closest subsequent visit to the visit in which DNAm was measured. In each study, MRI scans were performed and interpreted using standardized procedures without reference to demographic or clinical information. The field strength of the scanners used ranged mostly from 1.5 to 3.0 T. T1-, T2- and/or proton-density-weighted scans were obtained for all participants. Most studies used a fully automated segmentation method to quantify WMH burden. MRI procedures and WMH quantification in each study are detailed in the Supplementary material.
DNAm profiling
DNAm levels were measured at ∼450 K CpGs from whole blood samples with the Illumina Infinium Human 450 Methylation BeadChip in most participating cohorts. The GENOA study measured methylation levels at ∼27 K CpGs with the Illumina Infinium HumanMethylation 27 BeadChip, entirely covered by the Human 450 BeadChip. CARDIA, SHIP-TREND, Alzheimer’s Disease Neuroimaging Initiative and Rhineland Study used the Illumina MethylationEPIC BeadChip with a denser coverage of CpGs (∼850 K). Each study independently performed quality control for DNAm data, complying with the agreed minimum quality control guidelines; CpGs with >95% of samples with a detection P < 0.01 and samples with >95% of CpGs with a detection P < 0.01 were selected. DNAm values were then standardized using an intra-array normalization method. The BRIDGET Consortium measured DNAm levels using Hi-seq bisulphate sequencing, and DNAm sites with sample coverage <95% were excluded. Details of DNAm data collection and processing in participating studies are presented in the Supplementary material.
Cohort-level epigenome-wide association analyses
We tested association between DNAm level (untransformed beta values) and WMH burden [ln(WMH+1)] using a linear mixed regression model by ancestry group adjusted for age, sex, study site if applicable, total (intra)cranial volume (cm3), white blood cell proportion (%)31 and within-ancestry principal components as fixed effects and technical covariates (i.e. plate, chip-position, row and column) as random effects. In FHS, family structure is also adjusted as a random effect. Multi-ancestry studies with a small number of subjects in each ancestry, namely CHS and CARDIA, performed a pooled-ancestry analysis that also adjusted for ancestry group as fixed effects. Additionally, subgroup analyses by hypertension status were conducted. Hypertension was defined if either systolic or diastolic blood pressure (SBP or DBP) is >140 or 90 mmHg, respectively, or if a subject was taking any antihypertensive medication at the time of MRI measurement. In the BRIDGET study, we tested the association of DNAm with an extreme-SVD phenotype defined as excessive WMH volume with or without brain infarcts accounting for age, sex, country, the sequencing read counts and sample relatedness.32 DNAm measurements and statistical models used in participating studies are described in Supplementary material.
Epigenome-wide meta-analysis and replication analysis
We combined EWAS results based on sample-size-weighted z-score-based fixed-effect method in METAL33 because WMH was measured on different scales in the various cohorts and because our primary aim was to identify novel DNAm loci for WMH burden rather than estimate effect sizes of methylation probes.34 Hypertensive and normotensive subgroup meta-analyses and ancestry-specific meta-analyses (excluding CHS and CARDIA) were also performed. Study-specific results were corrected for inflation during meta-analysis if inflation was detected [genomic inflation factor (λ) > 1.0]. An association was considered as significant if P was smaller than Bonferroni threshold (∼1.2 × 10−7). A less stringent threshold was also set as 1.0 × 10−5 to detect suggestive associations. CpGs on sex chromosomes were not considered because our analytic plan did not account for hemi-methylation on the X chromosome due to chromosome X inactivation in females. Cross-reactive CpGs reported by Chen et al.35 and those showing evidence of heterogeneity (Cochran Q P-value < 0.05) were removed from the results post hoc. In the replication samples, associations for the identified CpGs were tested. CpGs were considered replicated if they were significant at the Bonferroni threshold (0.05 / the number of the CpGs). We plotted epigenetic associations in cis (±50 kb) using R ‘coMET’ package.36
Annotation of regulatory features and traits
We scored genomic positions of the identified CpGs according to RegulomeDB’s37 ranking criteria ranging from one (likely to affect binding and linked to expression of a gene target) to five (minimal binding evidence) and also computed a probability score within a range from zero to one (the most likely to be a regulatory variant). CpGs at the locations with significant regulatory features (rank category one or two, and probability score ≥0.9) are discussed. We also identified enhancers or promoters mapped to CpGs using the database of genome-wide enhancer-to-gene or promoter-to-gene associations computed based on five elements: eQTLs, eRNA co-expression, transcription factor co-expression, capture Hi-C and gene target distance (GeneHancer DB).38 Identified CpGs were also searched in EWAS catalogue39 and EWAS atlas40 to identify associated traits reported in previous EWAS. Last, to examine possible correlations among the CpGs, Spearman correlations were calculated in 906 European ancestry and 639 African ancestry subjects from the ARIC study.
DMR analysis
We performed a DMR analysis to identify a group of CpGs that collectively influence WMH burden using two specific methods, Comb-p41 and DMRCate,42 accounting for their spatial correlations. Briefly, Comb-P detects regional enrichment of low Ps at varying distance using the Stouffer–Liptak–Kechris correction for adjacent Ps.41 DMRcate models Gaussian kernel smoothing within predefined distance (1 kbp in this study) and collapses contiguous significant CpGs (P < 0.05) after multiple testing correction. DMR identified by both Comb-P (Šidák P < 0.05) and DMRCate (FDR < 0.05) was considered significant. To replicate, individual association Ps were pooled at each identified DMRs using DMRCate in the replication samples.
Gene set enrichment analysis of WMH-associated epigenetic loci
Identified CpGs and DMRs were tested for enrichment in gene sets from MSigDB c5 gene ontology database43,44 and KEGG pathway database,45 using ‘gsameth’ and ‘gsaregion’ functions built in R ‘missMethyl’ package.46
Shared epigenetics with blood pressure
BP is an influential risk factor for WMH.47–49 To investigate the shared epigenetics between WMH burden and BP, we performed a pairwise multivariate association test using summary statistics from a previous EWAS of SBP and DBP.50 CpGs associated with both traits were tested against the null hypothesis H0: βWMH = βSBP or DBP = 0. The test uses Z-scores for each trait and estimates multivariate test statistics accounting for the trait correlation calculated on the basis of the null associations (trait-specific P > 1 × 10−5). This method is implemented in the ‘metaUSAT’ software.51 To avoid false positive associations driven entirely by one trait, we included CpGs showing significance (P < 0.001) for both traits. Bonferroni threshold was set at 8.33 × 10−3 (=0.05/6) on the basis of the number of associations tested.
Heritability analysis and GWAS of WMH-associated CpGs
Inter-individual variation in DNAm may result from differences in environmental exposures, stochastic variation or genetic influences. To examine the contribution of genetic variation to variation in DNAm at the identified CpGs, we estimated the narrow-sense heritability (h2meth) in the FHS Offspring Cohort subjects (n = 2377) adjusting for age, sex, blood cell counts, principal components and technical covariates. Body mass index (BMI) and smoking status were additionally corrected in sensitivity analyses.
To further identify genetic variants associated with DNAm levels at the WMH-associated CpGs, we performed GWAS in ARIC European ancestry subjects (n = 984). Genotypes were measured with Affymetrix 6.0 array and imputed from 1000 Genome phase one version three reference using MaCH v.1.0.16. Variants were excluded if minor allele frequency <0.01, sample call rate <95% or imputation quality <0.3. The untransformed methylation beta value was tested for genetic association adjusting for age, sex, top 42 methylation principal components and blood cell components.
Bi-directional MR analysis of the identified CpGs and WMH burden
To determine whether the WMH-associated DNAm level is a causal factor for WMH burden (Forward-MR) or a secondary outcome of WMH burden (Reverse-MR), we performed a bi-directional two-sample MR analysis52 for the identified CpGs with at least three instrumental variables (IVs). We identified methylation quantitative trait loci (mQTL) associations in cis (±1 Mb) from the FHS study (n = 4170) that had been validated using ARIC data (n = 963).53 Those mQTLs were clumped at linkage disequilibrium r2 < 0.05 for independence. For WMH, the UK Biobank GWAS summary statistics (n = 11 226)54 was downloaded from the Cerebrovascular Disease Knowledge Portal (http://www.cerebrovascularportal.org/) on 1 September 2019. Reverse-MR analysis was performed using eight clumped genome-wide significant associations (linkage disequilibrium r2 < 0.05). Since the FHS mQTL study shares only significant associations in cis,53 we used the mQTL association statistics from ARIC European ancestry subjects for reverse-MR analysis.
For each CpG in both directions, causal association was tested on the basis of the IVW method in the R package ‘TwoSampleMR’.55 To validate the MR result, sensitivity analyses based on weighted median and MR Egger methods, and built-in tests for pleiotropy and heterogeneity were also performed. For existence of pleiotropy (MR Egger intercept test P < 0.05), the Egger regression estimate was assessed instead of the IVW estimate.
Mediating effect of in cis genes between CpGs and WMH burden
To investigate whether expression of nearby genes mediates the relationship between the identified CpG and WMH burden, a two-step MR analysis was performed. We tested the directional relationships: (i) from ‘the exposure (CpGs)’ to ‘the mediator (gene expression)’ (step one); and (ii) from ‘the mediator (gene expression)’ to ‘the outcome (WMH burden)’ (step two) using the identified mQTL IVs, the WMH GWAS associations54 and eQTL associations from the GTEx version eight brain eQTL data accessed via eQTL Catalogue (https://www.ebi.ac.uk/eqtl/) on 12 November 2020. Among available GTEx brain tissues, cortex (n = 205), frontal cortex (n = 175), cerebellum (n = 209), cerebellar hemisphere (n = 175) and caudate basal ganglia (n = 194) were selected. MR association based on the IVW method was again tested and sensitivity analysis was also performed. Gene expression with IVW P < 0.05 at both steps was considered as a potential mediator in the association between the identified CpG and WMH burden.
Cis-acting genes associated with the identified CpGs in blood
To functionally annotate the identified CpGs, we tested associations with gene expression in blood in long-range53cis-regions (±5 Mb) in 1966 and 728 European ancestry subjects from FHS and Rotterdam Study, respectively. Expression of the nearest gene/mRNA was regressed on DNAm β score at the CpG adjusting for age, sex, population structure and family structure (FHS only), blood cell counts and technical covariates. Technical covariates and family structure were modelled as random effects. In sensitivity analyses, smoking status and BMI were added to the model. Estimates from two studies were then combined for each gene using the sample-sized based meta-analysis method in METAL.33
Genes co-localizing with the identified CpGs in brain
To investigate cis-acting genes co-localized with the identified CpGs in the brain, we performed a multiple-trait co-localization (moloc) analysis using brain QTL data. Before this analysis, we examined the inter-individual correlations between DNAm levels in whole blood and in prefrontal cortex at the identified CpGs, using publicly available data.56 For CpGs with significant correlation (P < 0.05) between blood and prefrontal cortex, we tested the posterior probabilities for full co-localization (PPFC) that multiple traits (DNAm, gene expression and WMH burden) share causal variants at each locus, given the data. We used coloc priors of 1 × 10−5. We identified EA-specific GWAS associations8 and brain mQTL (n = 543) and eQTL (n = 534) associations accessed via http://mostafavilab.stat.ubc.ca/xqtl/.57 If PPFC is >0.7, we considered the gene is significantly co-localized with CpG and WMH burden. Moloc analysis was performed using the R package ‘moloc’.58
Epigenetic regulation of known GWAS loci
We next investigated the role of DNAm at established WMH GWAS loci, which may not have been detectable at the genome-wide significance threshold. Among 26 loci reported in the latest WMH GWAS,8 we mapped 450 K-array CpGs to 21 loci. EWAS associations at each of these 21 loci were pooled using the Brown’s method (implemented in the package ‘poolr’) adjusting for dependence among CpGs.59 For dependency information, we calculated correlation among CpGs in the GWAS loci using ARIC methylation data (906 European ancestry and 639 African ancestry subjects). A GWAS locus with combined P was considered significant if P was smaller than Bonferroni-adjusted threshold (0.05/number of loci tested).
Alternatively, we performed a moloc analysis at the 21 GWAS loci, again using the GWAS and brain QTL data.8,57 With the priors of 1 × 10−5, we considered genes with a PPFC >70% as convincingly co-localized with DNAm and WMH burden.
Identification of biological pathways using multi-dimensional data integration
Integrating multi-omics associations for WMH may boost power to identify novel genes influencing WMH burden. We integrated genetic,8 transcriptomic60 and epigenetic GWAS of WMH using the R package ‘mergeomics’ (version 1.2).61 To reduce noise in the GWAS data, the top 50% of genetic associations8 were included and pruned at r2 < 0.5 based on HapMap3 linkage disequilibrium information as recommended.61 For transcriptomic associations, we used the recent WMH transcriptome-wide association study (TWAS) results.60 For epigenetic associations, we used our discovery EWAS. Markers were primarily mapped to the nearest genes. For CpGs, cis-acting genes reported in the MesaEpiGenomics study62 were additionally annotated. For each GWAS, EWAS and TWAS, we tested marker-level enrichment with hierarchical permutation size of 20 000 on the basis of biological pathways from pre-defined public databases: KEGG,45 REACTOME,63 Biocarta64 and the gene ontology knowledgebase.65,66 Then, we meta-analysed the enriched gene sets from association studies and identified the WMH-associated gene sets (FDR-adjusted P < 0.05).
To describe the regulatory network of the identified gene sets and identify its local hub genes, we performed a weighted key driver analysis (wKDA) using the web-based software Mergeomics version 2.0.67 Gene regulatory network was constructed using in-house brain-specific Bayesian network (minimum hub overlap 0.33 and directed edge type)68 and visualized via Cytoscape version 3.8.2.69
We also conducted an overlap-based drug-repositioning analysis ‘PharmOmics’ based on the identified key driver genes (FDR < 0.05) to predict potential drugs or small molecules targeting WMH.70 PharmOmics comprises a curated drug signature database covering 941 drugs, constructed from transcriptomic data across >20 tissues from rat, human and mouse. For our analysis, we selected drug signatures from relevant tissues (in vivo human transcriptome data in cardiovascular and nervous system, and in vitro transcriptome data from murine oligodendroglial precursor cells), and examined the overlap between these drug signature genes and key driver genes from our identified WMH-associated gene sets.
Data availability
The data that support the findings of this study are included in this paper. Full EWAS summary statistics are available in dbGaP at phs000930.v9.p1.
Results
Identification of epigenetic changes associated with WMH burden
Study sample characteristics
In the discovery sample, the mean age ranges from 49.7 years in SHIP to 74.6 in CHS. Sex ratios are balanced in all studies except for GENOA study, which has 72.8% female. ARIC, CARDIA and CHS have both European ancestry and African ancestry subjects, other studies consist of single ancestry subjects (African or European ancestry). In the primary replication sample, subjects from FHS third generation and Rhineland Study (mean age 47.1 and 54.1 years, respectively), which compose 86.2% of the replication study, are younger than most discovery studies and show relatively smaller median WMH burden (0.34 in the FHS third generation study and 0.40 in the Rhineland Study). All subjects in the replication studies are of European ancestry. Demographic characteristics of participating cohorts are shown in Supplementary Table 1.
Novel DNAm loci are associated with WMH burden
In the discovery sample, we identified a novel epigenome-wide significant association between WMH burden and level of DNAm at cg24202936 (Z = 5.38, P = 7.58 × 10−8) in SEPTIN7P11. Associations at cg24202936 in each study are presented in a forest plot (Supplementary Fig. 1) and regional associations within 50 kb are presented with annotations (Supplementary Fig. 2). At the suggestive significance threshold of 1 × 10−5, we identified 11 additional loci (Table 1). The associations remained significant (P < 0.05/12 = 4.17 × 10−3) after adjusting for BMI, smoking status and SBP and DBP. Quantile–quantile and Miami plots are presented in Supplementary Figs 3 and 4. All subsequent analyses focus on these 12 CpGs, which are referred to as ‘target CpGs’. None of the target CpGs associations were replicated in independent samples and a meta-analysis of the discovery and replication samples showed significant heterogeneity in many of the resulting associations, which was not present in the discovery cohorts (Supplementary Table 2). Target CpGs showed consistent associations with WMH in subgroup analyses by ancestry and hypertension status (Supplementary Tables 3 and 4). Cg06450373 in CDH18 (P = 6.48 × 10−8) was identified in normotensive subjects (Supplementary Table 5); but not replicated. In a gene set enrichment analysis on discovered CpGs (P < 1.0 × 10−5), ‘cell–cell junction organization’ was identified as the top pathway [P = 1.32 × 10−3, false discovery rate (FDR) = 0.32].
Table 1.
Single-CpG associations with WMH burden in the discovery sample (P < 1 × 10–5)
| CpG | Chr:Position (hg19) | Nearest gene | Reduced model | Full model | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | Z | P | Q | FDR | n | Z | P | |||
| cg24202936 | 11:50257256 | SEPTIN7P11 | 5359 | 5.38 | 7.58 × 10–8 | 0.03 | 0.04 | 4930 | 5.28 | 1.30 × 10–7 |
| cg17417856 | 19:50191637 | PRMT1;ADM5 | 4917 | –4.95 | 7.42 × 10–7 | 0.15 | 0.28 | 4526 | –4.40 | 1.11 × 10–5 |
| cg01506471 | 7:3990479 | SDK1 | 5359 | –4.81 | 1.52 × 10–6 | 0.21 | 0.3 | 4930 | –4.00 | 6.41 × 10–5 |
| cg14547240 | 4:15428750 | C1QTNF7 | 5359 | –4.71 | 2.48 × 10–6 | 0.25 | 0.3 | 4930 | –4.17 | 3.10 × 10–5 |
| cg21547371 | 3:52869521 | MUSTN1 | 5359 | –4.65 | 3.30 × 10–6 | 0.25 | 0.3 | 4930 | –4.06 | 4.95 × 10–5 |
| cg03116124 | 1:231293208 | TRIM67 | 5129 | –4.64 | 3.54 × 10–6 | 0.25 | 0.31 | 4700 | –4.58 | 4.63 × 10–6 |
| cg06809326 | 17:20799526 | CCDC144NL-AS1 | 5359 | 4.57 | 4.80 × 10–6 | 0.28 | 0.34 | 4930 | 3.44 | 5.88 × 10–4 |
| cg13476133 | 7:44185646 | GCK | 5359 | 4.55 | 5.46 × 10–6 | 0.28 | 0.36 | 4930 | 4.03 | 5.65 × 10–5 |
| cg14133539 | 9:104568 | FOXD4 | 4917 | –4.53 | 5.98 × 10–6 | 0.28 | 0.38 | 4526 | –4.45 | 8.41 × 10–6 |
| cg17577122 | 22:19511967 | CLDN5 | 5359 | 4.50 | 6.88 × 10–6 | 0.29 | 0.4 | 4930 | 4.79 | 1.68 × 10–6 |
| cg23586595 | 4:84034390 | PLAC8 | 5359 | 4.45 | 8.45 × 10–6 | 0.32 | 0.43 | 4930 | 3.93 | 8.36 × 10–5 |
| cg23054394 | 3:140784675 | SPSB4 | 5359 | –4.42 | 9.88 × 10–6 | 0.34 | 0.45 | 4930 | –4.01 | 6.07 × 10–5 |
The reduced model is adjusted for age, sex, study site (if applicable), total (intra)cranial volume (cm3), white blood cell proportion (%), technical covariates and genetic principal components. The full model is additionally adjusted for BMI, smoking status and systolic and diastolic blood pressure measures. Chr = chromosome; EA = European ancestry; FDR = local false discovery rate value; n = number of subjects tested for the CpG; P = P-value; Q = Q-value; SE = standard error; Z = Z-score.
Annotated regulatory functions of target CpGs
We found significant regulatory features from RegulomeDB at the genomic positions of cg24202936 (rank 2b and score 0.93), and cg06809326 (rank 2b and score 0.91) (Supplementary Table 6). Cg24202936 resides near a transcriptional starting site (0.2 kb upstream), and identified as a transcriptional factor binding site computationally annotated with 20 genes (Supplementary Table 6). Previously reported EWAS traits associated with target CpGs are presented in Supplementary Table 7. In particular, cg24202936 was previously reported associated with HIV infection.71 Cg06450373, cg031161214, cg01506471 and cg14547240 were correlated each other in both ancestries with weak to moderate r (0.23 to 0.55) (Supplementary Fig. 5). In African ancestry, cg23586595 showed weak but significant correlations with cg13476133 (r = 0.32), cg03116124 (r = −0.42) and cg14547240 (r = −0.36). No correlated CpG (|r| > 0.3) was identified for our top CpG, cg24202936, in both ancestries.
WMH-associated DMRs are enriched in immune response-related pathways
We identified 46 DMRs in associations with WMH burden (Supplementary Table 8). Notably, one DMR was in SH3PXD2A, previously identified in GWAS.8,72–74 Identified DMRs were enriched in several gene ontologies, including signal transducer and activator of transcription family protein binding (FDR = 4.91 × 10−3) and defence response to virus (FDR = 5.68 × 10−3), which are related to the immune response (Supplementary Table 9). Of the 46 identified DMRs, PRMT1, ABAT, BHMT2, C11orf21, IZUMO1, C5orf66, ENPEP, SLC35F3, FBXO47, SLC45A4, KCTD16, KITLG and UCN3 were replicated (Supplementary Table 8). Of note, ENPEP, SLC35F3 and SLC45A4 were previously reported in BP GWAS.75–81
Shared epigenetic loci between WMH and BP
At the Bonferroni-corrected threshold (P < 8.33 × 10−3), we identified six CpGs associated with both WMH burden and BP (Supplementary Table 10). For WMH-DBP, cg23291754 in MOBKL1A (P = 2.38 × 10−7) and cg24372586 in GNL1 (P = 7.84 × 10−7) were identified. For WMH-SBP, cg00711496 in CDC42BPB (P = 1.99 × 10−7), cg04987734 in C19orf76; PRMT1 (P = 3.09 × 10−7), cg00934987 in SEPT4 (P = 1.07 × 10−6) and cg18770635 in KLHDC7B (P = 1.68 × 10−6) were identified.
Heritability of the WMH-associated CpGs
Significant h2meth was estimated for cg17417856 (40.4%, P = 1.37 × 10−8), cg06809326 (26.5%, P = 1.03 × 10−4), cg23586595 (24.2%, P = 1.47 × 10−3), cg17577122 (14.3%, P = 2.80 × 10−2) and cg24202936 (15.5%, P = 1.34 × 10−2) (Table 2). Additional adjustment for BMI and smoking status did not significantly modify these estimates. In GWAS of the target CpGs in the ARIC European ancestry sample, we observed significant cis-genetic influence on cg06809326, cg13476133 and cg24202936 (Supplementary Fig. 6). This result agrees with a previous publication that included the same dataset.53
Table 2.
Heritability estimates of WMH-associated CpGs
| CpG | Nearest gene | Reduced model | Full model | ||||
|---|---|---|---|---|---|---|---|
| h 2 met h | SE | P | h 2 met h | SE | P | ||
| cg0150647 | SDK1 | 0.02 | 0.07 | 0.38 | 0.01 | 0.07 | 0.42 |
| cg03116124 | TRIM67 | 0.01 | 0.07 | 0.45 | 0.01 | 0.07 | 0.47 |
| cg06809326 | CCDC144NL | 0.26 | 0.07 | 1.03 × 10–4a | 0.27 | 0.07 | 9.51 × 10–5a |
| cg13476133 | GCK | 0.09 | 0.07 | 0.11 | 0.09 | 0.07 | 0.12 |
| cg14133539 | FO × D4 | 0.08 | 0.07 | 0.14 | 0.07 | 0.07 | 0.17 |
| cg14547240 | C1QTNF7 | 0.06 | 0.07 | 0.20 | 0.06 | 0.07 | 0.18 |
| cg17417856 | PRMT1;ADM5 | 0.40 | 0.08 | 1.37 × 10–8a | 0.40 | 0.08 | 3.06 × 10–8a |
| cg17577122 | CLDN5 | 0.14 | 0.08 | 2.80 × 10–2 | 0.15 | 0.08 | 2.27 × 10–2 |
| cg21547371 | MUSTN1 | 0.00 | – | 0.50 | 0.00 | 0.50 | |
| cg23054394 | SPSB4 | 0.00 | – | 0.50 | 0.00 | 0.50 | |
| cg23586595 | PLAC8 | 0.24 | 0.08 | 1.47 × 10–3a | 0.23 | 0.08 | 2.51 × 10–3a |
| cg24202936 | LOC441601 | 0.15 | 0.07 | 1.34 × 10–2 | 0.16 | 0.07 | 1.17 × 10–2 |
h 2 meth = the narrow-sense heritability; SE = standard error. Reduced model is adjusted for age, sex, blood cell counts, principal components of the ancestry and technical covariates. Full model is additionally adjusted for BMI and smoking.
Significant after adjustment for multiple testing burden.
Mendelian randomization analyses between target CpGs and WMH burden
Forward two-sample multiple IV MR analysis was performed for two target CpGs, cg06809326 and cg24202936, which have at least three independent cis-mQTL IVs in Huan et al.53 (Supplementary Table 11). We found a marginally significant causal relationship from cg06809326 to WMH burden (P = 2.91 × 10−2). Higher methylation level at the locus is associated with greater WMH burden {odds ratio (OR) [95% confidence interval (CI)] = 1.39 (1.03, 1.87)}. Evidence was lacking for horizontal pleiotropy (P = 0.41) or heterogeneity (P = 0.42) (Supplementary Table 12). In reverse-MR analysis, evidence that WMH causally influence methylation levels at any of the target CpGs was lacking (Supplementary Tables 12 and 13).
Using the same three IVs, we also investigated whether cg6809326 is causally associated with expression of nearby genes (step one). Two cis transcripts were annotated to this CpG in GTEx version eight data.82 They both encode a long non-coding RNA designated as CCDC144NL and CCDC144NL-AS1, and we identified one IV for both transcripts. In all five brain tissues, we found evidence of causal association between cg06809326 and both CCDC144NL and CCDC144NL-AS1 (Supplementary Table 14). In step two, a marginal association between CCDC144NL and WMH burden was observed in caudate basal ganglia and cortex (step one P = 1.11 × 10−3 and step two P = 3.94 × 10−2 in caudate basal ganglia; step one P = 1.21 × 10−3 and step two P = 4.28 × 10−2 in cortex).
DNAm at established GWAS loci and WMH burden
We estimated the combined effect of DNAm at each locus from our EWAS results at the 21 established GWAS loci.8 Consistent with our DMR results, CpGs at the GWAS locus SH3PXD2A were jointly associated with WMH (P = 8.48 × 10−3), but evidence of DNAm effects on WMH at other loci was lacking (Supplementary Table 15). We also conducted a multiple-trait co-localization analysis (moloc)58 of brain mQTL and expression QTL (eQTL),57 and WMH-associated single nucleotide polymorphisms (SNPs). At 17 out of the 21 GWAS loci, we identified significant co-localization evidence (PPFC > 0.7) (Supplementary Table 16 and Supplementary Fig. 7). At eight loci, the SNPs with the highest PPFC were the sentinel SNPs in the GWAS.
Candidate genes implicated by gene-expression associations with the target CpGs
At the Bonferroni threshold (6.93 × 10−5 = 0.05/722 cis genes in ±5 Mb of the target CpGs), we identified significant associations between cg23586595 and PLAC8 (P = 2.98 × 10−7) and between cg24202936 and F2 (P = 6.39 × 10−5) (Table 3). Adjusting for additional covariates (smoking status and BMI) did not change these associations.
Table 3.
Cis genes (±5 Mb) whose expression is significantly associated with identified CpGs
| CpG | Gene | Region (hg19) | n | Z reduced | P reduced | Z full | P full |
|---|---|---|---|---|---|---|---|
| cg23586595 | PLAC8 | 4:84011211–84138405 | 2687 | −5.13 | 2.98 × 10–7 | −5.11 | 3.27 × 10–7 |
| cg24202936 | F2 | 11:46740749–46761054 | 1963 | −4.00 | 6.39 × 10–5 | −4.01 | 6.04 × 10–5 |
Z-scores and P-values from the reduced and full model are presented. Reduced model is adjusted for age, sex, blood cell counts, principal components of the ancestry and technical covariates. Full model is additionally adjusted for BMI and smoking. PLAC8 = placenta-associated 8; F2 = coagulation factor II.
Cg24202936, cg01506471 and cg06809326 showed significant correlation estimates (|r| > 0.3) between blood and brain (r = 0.33, 0.87 and 0.57, respectively) (Supplementary Table 17) and, thus, were tested for co-localization. We found that mQTLs for cg24202936 and WMH GWAS SNPs colocalize with FOLH1 expression in dorsolateral prefrontal cortex (DLPFC) (PPFC = 0.75) (Supplementary Table 18). Also, suggestive evidence existed for co-localization of cg06809326 mQTLs, CCDC144NL-AS1 eQTLs and WMH SNPs (PPFC = 0.69).
Integrative cross-omics analysis
Integrative cross-omics analysis identifies novel gene regulatory networks
At FDR <0.05, we identified 576 WMH-associated gene sets enriched from the integrated data of GWAS, EWAS and TWAS out of 12 303 gene sets from curated databases.45,63–66 Top associated gene sets includes ‘regulation of actin cytoskeleton’ (P = 1.14 × 10−45, 211 genes), ‘telomeres, telomerase, cellular ageing and immortality’ (P = 1.10 × 10−35, 18 genes), ‘integrin-mediated cell surface interactions’ (P = 3.17 × 10−34, 84 genes), ‘thrombin signalling through proteinase activated receptors’ (P = 1.41 × 10−33, 32 genes) and ‘Nef protein mediated CD4 down-regulation’ (P = 4.70 × 10−32, nine genes). All enriched pathways with FDR P < 0.05 are listed in Supplementary Table 19.
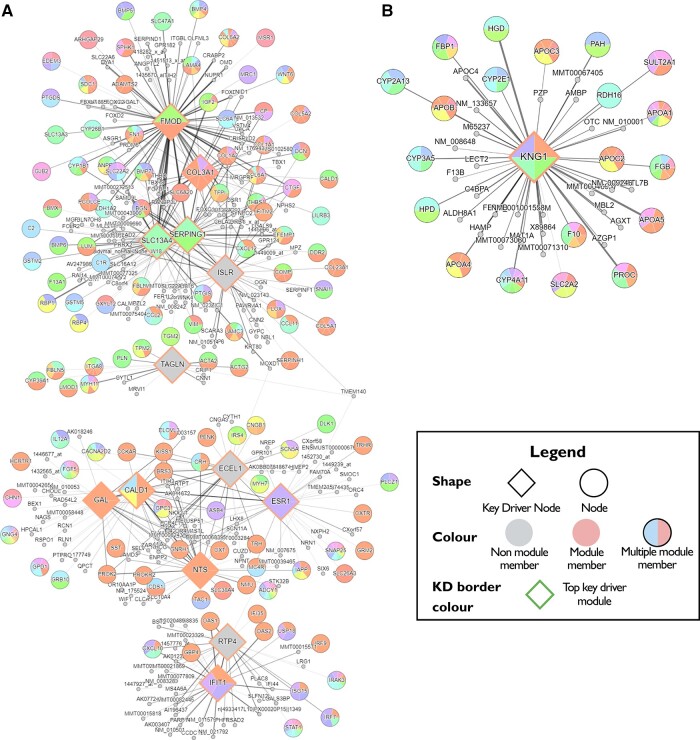
We derived two WMH burden-associated gene networks in brain. The first network is comprised of four subnetworks. Five key driver genes (FMOD, COL3A1, SERPING1, SLC13A4 and ISLR) represent a subnetwork of ‘extracellular matrix (ECM) organization, ECM–receptor interaction, focal adhesion and collagen formation’. Additionally, three related subnetworks, ‘smooth muscle contraction’ with key driver TAGLN; ‘G-protein-coupled receptor ligand binding’ with key drivers GAL, ECEL1, ESR1 and NTS; and ‘cytokine signalling in immune system’ with key drivers IFIT1 and RTP4, make up the network (Fig. 2 and Supplementary Table 20). We also identified an independent second network associated with ‘lipid and lipoprotein metabolism’, with key driver gene KNG1. Genes included in each subnetwork are presented in Supplementary Table 21.
Figure 2.
WMH-associated gene networks. WMH-associated genes based on multi-molecular evidence are organized around the 19 key driver genes. (A) WMH-associated network consisting of four subnetworks—extracellular matrix (ECM) organization (FMOD, COL3A1, SEPING1, SLC13A4 and ISLR); smooth muscle contraction (TAGLN); G-protein-coupled receptor ligand binding (GAL, ECEL1, ESR1 and NTS) and cytokine signalling in immune system (IFIT1 and RTP4). (B) WMH-associated network of lipid and lipoprotein metabolism (KNG1). Key drivers and associated gene networks identified in the Mergeomics analysis are coloured in orange. Neighbouring genes are grouped into networks and labelled in random colours.
Overlap-based drug-repositioning analysis of WMH-associated genes
Using drug signatures derived from in vivo cardiovascular and nervous system data, we predicted antihyperlipidaemic drugs, including PPAR-α (peroxisome proliferator-activated receptor-alpha) agonist ‘fenofibrate’, as the top therapeutic target. Using drug signatures derived from murine oligodendroglial precursor cells data, we predicted several small molecules, including a glycogen synthase kinase inhibitor and a phenylalanyl tRNA synthetase inhibitor that may have therapeutic potential for Alzheimer’s disease83 and autoimmune diseases,84 respectively (Supplementary Table 22).
Discussion
This first EWAS of WMH burden in 9732 middle-aged to older adults from 14 community-based cohorts identified several novel epigenetic loci. Although we could not independently replicate the association of single CpGs with WMH, probably due to a limited sample size and differences between the discovery and replication sample, functional annotation and bioinformatic analyses provided strong supportive evidence. Moreover, powerful DMR analyses identified 46 DMRs of which 13 were replicated. Integrative analyses of multi-omics information also suggested novel gene networks with key drivers and potential drug targets for WMH.
We identified a novel epigenetic locus, cg24204936, mapping to a pseudogene SEPTIN7P11. Functional integration revealed two candidate genes whose expression may be influenced by variation in DNAm at this locus: F2 in blood and FOLH1 in DLPFC. Prothrombin encoded by F2 plays an essential role in blood clot formation, angiogenesis, tissue repair and vascular integrity. A prothrombotic state or circulating prothrombin has been reported for symptomatic cSVD,85,86 WMH and stroke.87,88 However, it remains unclear whether coagulation plays a major role in the aetiology of WMH or is secondary to injury to the cerebral small vessels and white matter.89FOLH1 encodes glutamate carboxypeptidase II that catalyses the hydrolysis of N-acetylaspartylglutamate. An elevated level of N-acetylaspartylglutamate in the CSF has been reported in two patients with almost complete absence of myelin in the CNS90 and has been proposed as a diagnostic biomarker for rare diseases of the white matter.91
An epigenetic locus mapping to PRMT1, which encodes a protein arginine N-methylase, was identified in single-CpG and DMR analyses and also as a shared epigenetics locus with BP. The biological link between DNAm at PRMT1 and WMH burden may involve pathways related to endothelial dysfunction, which have previously been implicated in WMH aetiology.92PRMT1, a predominant member of the PRMT family, methylates histone and non-histone proteins to regulate various cellular functions.93PRMT1 is essential for the development of neurons, astrocytes and oligodendrocytes and is critical for myelin formation.94 PRMTs also catalyse the formation of ADMA (asymmetric dimethylarginine), which reduces nitric oxide production, promotes endothelial dysfunction in the BBB and triggers the immune response in atherosclerosis.92,95,96 Higher ADMA levels have been repeatedly associated with cSVD and its monogenic form, cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy.97–103
Single-CpG association combined with functional genomic analyses and DMR analyses identified a novel epigenetic locus near CCDC144NL;CCDC144NL-AS1 (coiled-coil domain containing 144 family and its antisense RNA1). Cg06809326 is under strong cis-genetic control and brain expression of its nearest gene, CCDC144NL;CCDC144NL-AS1, may mediate the association between DNAm and WMH burden (Supplementary Fig. 8). A TWAS of WMH using blood gene-expression data60 did not report a significant association for CCDC144NL;CCDC144NL-AS1 expression, possibly due to its low expression in blood. CCDC144NL-AS1 encodes a long non-coding mRNA transcript that controls expression of target genes by acting as a molecular sponge for various regulatory miRNAs.104–109In vitro studies have uncovered several of its target genes with potentially relevant function to cSVD. These include matrix metalloproteinases MMP2 and MMP9,108 F-actin and vimentin,110 and transforming growth factor beta (TGF-β)-activated kinase 1 (TAK1).106 MMP2 and MMP9 can damage the BBB by triggering recruitment of immune cells111 and have been implicated in white matter injury and cSVD.112 F-actin plays an important role in maintaining the shape of endothelial cells and the integrity of the BBB.113 Disturbed TGF-β signalling has been implicated in the pathogenesis of several monogenic forms of cSVD.114–117 Deficiency of TAK1 in mouse brain endothelial cells resulted in endothelial cell death, small vessel rarefaction and disruption of the BBB.118
A central role of endothelial dysfunction, possibly resulting in a compromised BBB, in WMH burden119 is further suggested by identified DNAm associations in genes involved in cell junctions. Claudins are integral membrane proteins that comprise tight junctions specifically in brain microvascular endothelial120 cells and that regulate BBB permeability.121 Claudin-5 mapped to cg17577122 is the most enriched tight junction protein in the BBB, and its dysfunction has been implicated in neurodegenerative and neuroinflammatory diseases, and cSVD.122–126 A recent DMR analysis using DLPFC DNAm levels also identified CLDN5 to be associated with cognitive decline.127 In normotensive subjects, we identified a CpG in cadherin 18 (CDH18) that encodes an adherens junction protein, which mediates calcium-dependent cell–cell adhesion. CDH18 is also involved in cell junction organization process and in cell signalling pathways including G-proteins signalling together with F2.128
Our DMR analysis, aggregated epigenetic associations using Brown’s method59 and moloc analysis using brain QTL data consistently identified an epigenetic association at a known WMH GWAS locus, SH3PXD2A (SH3 and PX-domain-containing protein 2A).8,72–74 Several genome-wide associations with WMH-related traits have also been reported at SH3PXD2A, including white matter microstructure129 and stroke.130,131SH3PXD2A encodes an adaptor protein (TKS5) involved in the formation of podosomes that act as sites of close contact to as well as degradation of ECM.132 Gene set enrichment of identified DNAm loci and integrative cross-omics analyses collectively point to a central role of the ECM in WMH burden. One of the two WMH burden networks identified through the Mergeomics approach centred around key driver genes involved in ECM organization and function and the top associated module was ‘regulation of actin cytoskeleton’. Notably, actin polymerization and disassembly of junctional proteins within microvascular endothelial cells were shown to play a key role in early BBB disruption in a murine model.133
Another network includes genes that function in lipid and lipoprotein metabolism and our overlap-based drug-repositioning analyses suggested antihyperlipidaemic drugs as potential drug targets. A recent MR analysis showed that genetically increased high-density lipoprotein cholesterol level was associated with lower WMH volume and lower risk of small vessel stroke.134 Statin therapy for cSVD has also been regarded as promising since individuals with high WMH burden typically carry higher vascular risk factors. Few randomized clinical trials assessing the effect of lipid lowering on WMH progression have been conducted and they have generally provided mixed results.135–137 While they suggest a possible role of statins, in particular rosuvastatin, in preventing WMH progression, the lack of high-quality data prevents strong evidence-based recommendation at this time.138 It has been postulated that statins improve endothelial function and stabilize the BBB in cSVD.139,140 Studies that investigated membrane proteins including phospholipid flippase (ATP11B) and aquaporin-4 showed that the loss of these proteins cause pathological features of cSVD including endothelial cell dysfunction with reduced tight junctions, nitric oxide, oligodendrocyte progenitor cell maturation block and microglial activation.126,141
Finally, this study provides further emphasis concerning the long-observed perivascular inflammation as an additional crucial player in cSVD pathology and provides a possible explanation. Interestingly, gene set enrichment analyses identified a possible role of the defence response to viral infection with several DMR-associated genes related to interferon gamma signalling and the innate immune response (DTX3L-PARP9, BNIP3 and IFITM1). Our top associated CpG has been previously reported in an EWAS of chronic HIV infection71 and our drug-repositioning analysis also identified a HIV antiviral as a possible drug target. Several studies have reported that people with HIV are at higher risk of an increased burden of WMH compared to uninfected controls.142,143
Several limitations of our study must be acknowledged. First, many of our EWAS discoveries were not independently confirmed. Since a series of functional analyses showed biological relevance, we suspect that the lack of replication may stem from the limited size of the replication sample and from differences between the discovery and replication samples as hinted by the increased heterogeneity in the DNAm association observed in the meta-analysis (Supplementary Table 2). Indeed, variation in WMH burden was smaller in the replication studies than in the discovery studies perhaps due to the younger age of the participants. The younger cohorts, CARDIA (n = 277) with a mean age 53.9 years and SHIP (n = 214) with a mean age 49.7 years, make up only 8.59% of the discovery sample; whereas the Rhineland Study with a mean age 54.1 years and FHS third generation cohort with a mean age of 47.1 years, make up over 86% of the replication sample. Replication of several WMH-associated loci identified through more powerful DMR analyses further underscore an underpowered replication study for single-CpG associations. Additional studies are needed to confirm the findings presented here. Second, we conducted a subgroup analysis stratified by hypertension status, but statistical power in each stratum was limited. A more ideal design to study this and other modifiable risk factors of cSVD will be a longitudinal study or a stratified association study of a larger sample size. Similarly, our study was not sufficiently powered to examine ancestry-specific associations of DNAm with WMH and possible ancestry difference in epigenetic patterns could not be investigated. Third, we did not adjust for additional lifestyle factors or comorbidities to maximize our sample size by minimizing the number of covariates in the models. Our primary goal was to identify novel DNAm loci associated with WMH burden and we cannot exclude the possibility that the identified loci may reflect, in part, variation in those risk factors. Fourth, the currently publicly available brain QTL data are limited to cis-regions of omics markers and, thus, our in silico bioinformatics analyses were restricted only to the CpGs with substantial cis-acting genetic influence. For example, cg17417856 in PRMT1 had a strong heritability estimate (h2 = 0.40, P = 1.37 × 10−8) but was not followed-up because it was under polygenic control. Last, the study was conducted in blood and cell type-specific associations, most notably in brain, may have been missed. To extrapolate the findings in blood to brain, we assessed the correlation with DNAm in brain, and used available brain QTL data. Due to the difficulties of getting both brain DNAm and MRI data from a large population-based sample, an EWAS of WMH burden using brain DNAm may not be easy to achieve. However, findings from this large blood-based study may provide a basis for an epigenetic candidate gene study in the brain.
Supplementary Material
Acknowledgements
The authors thank the staff and participants of ARIC study, Biobanking and BioMolecular resources Research Infrastructure, CARDIA, CHS, FHS, GENOA, LBC1936 (Lothian Birth Cohort), Rotterdam Study, SHIP, Alzheimer’s Disease Neuroimaging Initiative, Rhineland Study, Older Australian Twin Study and BRIDGET for their pivotal contributions. We acknowledge collaborative contributions from the neuroCHARGE working group. We also acknowledge Dr Dan Levy’s team (Drs Roby Joehanes and Tianxiao Huan) for the mQTL and eQTL data used in this study. A full set of study-specific acknowledgements is provided in the Supplementary material.
Contributor Information
Yunju Yang, Brown Foundation Institute of Molecular Medicine, McGovern Medical School, University of Texas Health Science at Houston, Houston, TX 77030, USA.
Maria J Knol, Department of Epidemiology, Erasmus MC University Medical Center, 3015 GD, Rotterdam, The Netherlands.
Ruiqi Wang, Department of Biostatistics, Boston University School of Public Health, Boston, MA 02118, USA.
Aniket Mishra, University of Bordeaux, Inserm, Bordeaux Population Health Research Center, Team VINTAGE, UMR 1219, F-33000 Bordeaux, France.
Dan Liu, Population Health Sciences, German Centre for Neurodegenerative Diseases (DZNE), 53127 Bonn, Germany.
Michelle Luciano, Department of Psychology, University of Edinburgh, Edinburgh, EH8 9JZ, UK.
Alexander Teumer, Institute for Community Medicine, University Medicine Greifswald, Greifswald 17475, Germany; German Centre for Cardiovascular Research (DZHK), Partner Site Greifswald, Greifswald 17475, Germany; Department of Population Medicine and Lifestyle Diseases Prevention, Medical University of Bialystok, Bialystok, 15-269, Poland.
Nicola Armstrong, Mathematics and Statistics, Curtin University, 6845 Perth, Australia.
Joshua C Bis, Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA 02115, USA.
Min A Jhun, Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, MI 48104, USA.
Shuo Li, Department of Biostatistics, Boston University School of Public Health, Boston, MA 02118, USA.
Hieab H H Adams, Department of Epidemiology, Erasmus MC University Medical Center, 3015 GD, Rotterdam, The Netherlands; Department of Radiology and Nuclear Medicine, Erasmus MC University Medical Center, 3015 GD, Rotterdam, The Netherlands.
Nasir Ahmad Aziz, Population Health Sciences, German Centre for Neurodegenerative Diseases (DZNE), 53127 Bonn, Germany; Department of Neurology, Faculty of Medicine, University of Bonn, 53127 Bonn, Germany.
Mark E Bastin, Centre for Clinical Brain Sciences, Department of Neuroimaging Sciences, University of Edinburgh, Edinburgh, EH8 9AB, UK.
Mathieu Bourgey, Canadian Centre for Computational Genomics, McGill University, Montréal, Quebec, Canada H3A 0G1; Department for Human Genetics, McGill University Genome Centre, McGill University, Montréal, Quebec, Canada H3A 0G1.
Jennifer A Brody, Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA 02115, USA.
Stefan Frenzel, Department of Psychiatry and Psychotherapy, University Medicine Greifswald, Greifswald 17475, Germany.
Rebecca F Gottesman, Stroke Branch, National Institutes of Neurological Disorders and Stroke, Bethesda, MD 20814, USA.
Norbert Hosten, Department of Radiology and Neuroradiology, University Medicine Greifswald, 17475 Greifswald, Germany.
Lifang Hou, Department of Preventive Medicine, Northwestern University, Chicago, IL 60611, USA.
Sharon L R Kardia, Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, MI 48104, USA.
Valerie Lohner, Population Health Sciences, German Centre for Neurodegenerative Diseases (DZNE), 53127 Bonn, Germany.
Pascale Marquis, Canadian Centre for Computational Genomics, McGill University, Montréal, Quebec, Canada H3A 0G1; Department for Human Genetics, McGill University Genome Centre, McGill University, Montréal, Quebec, Canada H3A 0G1.
Susana Muñoz Maniega, Centre for Clinical Brain Sciences, Department of Neuroimaging Sciences, University of Edinburgh, Edinburgh, EH8 9AB, UK.
Claudia L Satizabal, Glenn Biggs Institute for Alzheimer's and Neurodegenerative Diseases and Department of Population Health Sciences, UT Health San Antonio, San Antonio, TX 78229, USA; The Framingham Heart Study, Framingham, MA 01701, USA; Department of Neurology, Boston University School of Medicine, Boston, MA 02115, USA.
Farzaneh A Sorond, Department of Neurology, Feinberg School of Medicine, Northwestern University, Chicago, IL 60611, USA.
Maria C Valdés Hernández, Centre for Clinical Brain Sciences, Department of Neuroimaging Sciences, University of Edinburgh, Edinburgh, EH8 9AB, UK.
Cornelia M van Duijn, Department of Epidemiology, Erasmus MC University Medical Center, 3015 GD, Rotterdam, The Netherlands; Nuffield Department of Population Health, Oxford University, Oxford, OX3 7LF, UK.
Meike W Vernooij, Department of Epidemiology, Erasmus MC University Medical Center, 3015 GD, Rotterdam, The Netherlands; Department of Radiology and Nuclear Medicine, Erasmus MC University Medical Center, 3015 GD, Rotterdam, The Netherlands.
Katharina Wittfeld, Department of Psychiatry and Psychotherapy, University Medicine Greifswald, Greifswald 17475, Germany; German Center for Neurodegenerative Diseases (DZNE), Site Rostock/Greifswald, 17475 Rostock, Germany.
Qiong Yang, Department of Biostatistics, Boston University School of Public Health, Boston, MA 02118, USA; The Framingham Heart Study, Framingham, MA 01701, USA.
Wei Zhao, Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, MI 48104, USA.
Eric Boerwinkle, Human Genetics Center, School of Public Health, University of Texas Health Science at Houston, Houston, TX 77030, USA; Human Genome Sequencing Center, Baylor College of Medicine, Houston, TX 77030, USA.
Daniel Levy, The Framingham Heart Study, Framingham, MA 01701, USA; Population Sciences Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD 20814, USA.
Ian J Deary, Department of Psychology, University of Edinburgh, Edinburgh, EH8 9JZ, UK.
Jiyang Jiang, Centre for Healthy Brain Ageing, School of Psychiatry, University of New South Wales, Sydney, NSW 2052, Australia.
Karen A Mather, Centre for Healthy Brain Ageing, School of Psychiatry, University of New South Wales, Sydney, NSW 2052, Australia; Neuroscience Research Australia, Sydney, NSW 2031, Australia.
Thomas H Mosley, The Memory Impairment Neurodegenerative Dementia (MIND) Research Center, University of Mississippi Medical Center, Jackson, MS 39216, USA.
Bruce M Psaty, Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA 02115, USA; Department of Epidemiology, University of Washington, Seattle, WA 98104, USA.
Perminder S Sachdev, Centre for Healthy Brain Ageing, School of Psychiatry, University of New South Wales, Sydney, NSW 2052, Australia; Neuropsychiatric Institute, The Prince of Wales Hospital, University of New South Wales, Randwick, NSW 2031, Australia.
Jennifer A Smith, Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, MI 48104, USA.
Nona Sotoodehnia, Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA 02115, USA.
Charles S DeCarli, Department of Neurology and Center for Neuroscience, University of California at Davis, Sacramento, CA 95816, USA.
Monique M B Breteler, Population Health Sciences, German Centre for Neurodegenerative Diseases (DZNE), 53127 Bonn, Germany; Institute for Medical Biometry, Informatics and Epidemiology (IMBIE), Faculty of Medicine, University of Bonn, 53127 Bonn, Germany.
M Arfan Ikram, Department of Epidemiology, Erasmus MC University Medical Center, 3015 GD, Rotterdam, The Netherlands.
Hans J Grabe, Department of Psychiatry and Psychotherapy, University Medicine Greifswald, Greifswald 17475, Germany; German Center for Neurodegenerative Diseases (DZNE), Site Rostock/Greifswald, 17475 Rostock, Germany.
Joanna Wardlaw, Centre for Clinical Brain Sciences, Department of Neuroimaging Sciences, University of Edinburgh, Edinburgh, EH8 9AB, UK.
W T Longstreth, Department of Epidemiology, University of Washington, Seattle, WA 98104, USA; Department of Neurology, University of Washington, Seattle, WA 98104, USA.
Lenore J Launer, Intramural Research Program, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA.
Sudha Seshadri, Glenn Biggs Institute for Alzheimer's and Neurodegenerative Diseases and Department of Population Health Sciences, UT Health San Antonio, San Antonio, TX 78229, USA; The Framingham Heart Study, Framingham, MA 01701, USA; Department of Neurology, Boston University School of Medicine, Boston, MA 02115, USA.
Stephanie Debette, University of Bordeaux, Inserm, Bordeaux Population Health Research Center, Team VINTAGE, UMR 1219, F-33000 Bordeaux, France; Department of Neurology, Boston University School of Medicine, Boston, MA 02115, USA; CHU de Bordeaux, Department of Neurology, F-33000 Bordeaux, France.
Myriam Fornage, Brown Foundation Institute of Molecular Medicine, McGovern Medical School, University of Texas Health Science at Houston, Houston, TX 77030, USA; Human Genetics Center, School of Public Health, University of Texas Health Science at Houston, Houston, TX 77030, USA.
Funding
We thank all study participants for contributing to this work. This project was largely supported by US National Institutes of Health grant R01NS087541 with additional support from grants U01AG052409 and R01HL105756. A full set of study-specific funding sources is provided in the Supplementary material. R.F.G. is supported by the NINDS Intramural Research Program.
Competing interests
C.S. is a consultant of Novartis pharmaceuticals on a safety study for heart failure. J.W. is supported by the UK Dementia Research Institute, which is funded by the UK Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK. B.M.P. serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. H.J.G. has received travel grants and speaker’s honoraria from Fresenius Medical Care, Neuraxpharm, Servier and Janssen Cilag as well as research funding from Fresenius Medical Care.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Pantoni L. Cerebral small vessel disease: From pathogenesis and clinical characteristics to therapeutic challenges. Lancet Neurol. 2010;9:689–701. [DOI] [PubMed] [Google Scholar]
- 2. Wardlaw JM, Benveniste H, Williams A. Cerebral vascular dysfunctions detected in human small vessel disease and implications for preclinical studies. Annu Rev Physiol. 2022;84:409–434. [DOI] [PubMed] [Google Scholar]
- 3. Carmelli D, DeCarli C, Swan GE, et al. Evidence for genetic variance in white matter hyperintensity volume in normal elderly male twins. Stroke. 1998;29:1177–1181. [DOI] [PubMed] [Google Scholar]
- 4. Atwood LD, Wolf PA, Heard-Costa NL, et al. Genetic variation in white matter hyperintensity volume in the Framingham Study. Stroke. 2004;35:1609–1613. [DOI] [PubMed] [Google Scholar]
- 5. Kochunov P, Glahn D, Winkler A, et al. Analysis of genetic variability and whole genome linkage of whole-brain, subcortical, and ependymal hyperintense white matter volume. Stroke. 2009;40:3685–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Turner ST, Jack CR, Fornage M, Mosley TH, Boerwinkle E, de Andrade M. Heritability of leukoaraiosis in hypertensive sibships. Hypertension. 2004;43:483–487. [DOI] [PubMed] [Google Scholar]
- 7. Duperron MG, Tzourio C, Sargurupremraj M, et al. Burden of dilated perivascular spaces, an emerging marker of cerebral small vessel disease, is highly heritable. Stroke. 2018;49:282–287. [DOI] [PubMed] [Google Scholar]
- 8. Sargurupremraj M, Suzuki H, Jian X, et al. Cerebral small vessel disease genomics and its implications across the lifespan. Nat Commun. 2020;11:6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jian X, Satizabal CL, Smith AV, et al. Exome chip analysis identifies low-frequency and rare variants in MRPL38 for white matter hyperintensities on brain magnetic resonance imaging. Stroke. 2018;49:1812–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Handel AE, Ebers GC, Ramagopalan SV. Epigenetics: Molecular mechanisms and implications for disease. Trends Mol Med. 2010;16:7–16. [DOI] [PubMed] [Google Scholar]
- 11. Jaenisch R, Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. [DOI] [PubMed] [Google Scholar]
- 12. Wright JD, Folsom AR, Coresh J, et al. The ARIC (Atherosclerosis Risk in Communities) Study: JACC focus seminar 3/8. J Am Coll Cardio. 2021;77:2939–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. BBMRI Stakeholder’s Forum . BBMRI: A Step Closer—Stakeholder’s Forum Report. Accessed 14 February 2022. https://www.bbmri-eric.eu/wp-content/uploads/2016/07/stakeholders-forum-report-a-step-closer-a4.pdf
- 14. Fried LP, Borhani NO, Enright P, et al. The cardiovascular health study: Design and rationale. Ann Epidemiol. 1991;1:263–276. [DOI] [PubMed] [Google Scholar]
- 15. Friedman GD, Cutter GR, Donahue RP, et al. CARDIA: Study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol. 1988;41:1105–1116. [DOI] [PubMed] [Google Scholar]
- 16. Dawber TR, Kannel WB. The Framingham Study. An epidemiological approach to coronary heart disease. Circulation. 1966;34:553–555. [DOI] [PubMed] [Google Scholar]
- 17. Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study. Design and preliminary data. Prev Med. 1975;4:518–525. [DOI] [PubMed] [Google Scholar]
- 18. Daniels PR, Kardia SLR, Hanis CL, et al. Familial aggregation of hypertension treatment and control in the Genetic Epidemiology Network of Arteriopathy (GENOA) study. Am J Med. 2004;116:676–681. [DOI] [PubMed] [Google Scholar]
- 19. Wardlaw JM, Bastin ME, Valdés Hernández MC, et al. Brain aging, cognition in youth and old age and vascular disease in the Lothian Birth Cohort 1936: Rationale, design and methodology of the imaging protocol. Int J Stroke. 2011;6:547–559. [DOI] [PubMed] [Google Scholar]
- 20. Deary IJ, Gow AJ, Taylor MD, et al. The Lothian Birth Cohort 1936: A study to examine influences on cognitive ageing from age 11 to age 70 and beyond. BMC Geriatr. 2007;7:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ikram MA, Brusselle G, Ghanbari M, et al. Objectives, design and main findings until 2020 from the Rotterdam Study. Eur J Epidemiol. 2020;35:483–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ikram MA, van der Lugt A, Niessen WJ, et al. The Rotterdam Scan Study: Design update 2016 and main findings. Eur J Epidemiol. 2015;30:1299–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Volzke H, Alte D, Schmidt CO, et al. Cohort profile: The study of health in Pomerania. Int J Epidemiol. 2011;40:294–307. [DOI] [PubMed] [Google Scholar]
- 24. Khachaturian ZS. Perspective on the Alzheimer’s Disease Neuroimaging Initiative: Progress report and future plans. Alzheimers Dement. 2010;6:199–201. [DOI] [PubMed] [Google Scholar]
- 25. Petersen RC, Aisen PS, Beckett LA, et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI): Clinical characterization. Neurology. 2010;74:201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Splansky GL, Corey D, Yang Q, et al. The third generation cohort of the National Heart, Lung, and Blood Institute’s Framingham Heart Study: Design, recruitment, and initial examination. Am J Epidemiol. 2007;165:1328–1335. [DOI] [PubMed] [Google Scholar]
- 27. Sachdev PS, Lee T, Lammel A, et al. Cognitive functioning in older twins: The Older Australian Twins Study. Australas J Ageing. 2011;30:17–23. [DOI] [PubMed] [Google Scholar]
- 28. Sachdev PS, Lammel A, Trollor JN, et al. A comprehensive neuropsychiatric study of elderly twins: The Older Australian Twins Study. Twin Res Hum Genet. 2009;12:573–582. [DOI] [PubMed] [Google Scholar]
- 29. Breteler MMB, Wolf H. P2-135: The Rhineland Study: A novel platform for epidemiologic research into Alzheimer disease and related disorders. Alzheimers Dement. 2014;10:P520. [Google Scholar]
- 30. University of Bordeaux . BRIDGET: BRain Imaging, cognition, Dementia and next generation GEnomics. Accessed February 14, 2022. https://bridget.u-bordeaux.fr/Project.
- 31. Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mishra A, Chauhan G, Violleau MH, et al. Association of variants in HTRA1 and NOTCH3 with MRI-defined extremes of cerebral small vessel disease in older subjects. Brain. 2019;142:1009–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Willer CJ, Li Y, Abecasis GR. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pereira TV, Patsopoulos NA, Salanti G, Ioannidis JPA. Discovery properties of genome-wide association signals from cumulatively combined data sets. Am J Epidemiol. 2009;170:1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen YA, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the illumina infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martin TC, Yet I, Tsai PC, Bell JT. coMET: Visualisation of regional epigenome-wide association scan results and DNA co-methylation patterns. BMC Bioinformatics. 2015;16:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fishilevich S, Nudel R, Rappaport N, et al. Genehancer: Genome-wide integration of enhancers and target genes in GeneCards. Database. 2017;2017:bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Battram T, Yousefi P, Crawford G, et al. The EWAS Catalog: A database of epigenome-wide association studies. Wellcome Open Res. 2022;7:41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li M, Zou D, Li Z, et al. EWAS Atlas: A curated knowledgebase of epigenome-wide association studies. Nucleic Acids Res. 2019;47:D983–D988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pedersen BS, Schwartz DA, Yang IV, Kechris KJ. Comb-p: Software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics. 2012;28:2986–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peters TJ, Buckley MJ, Statham AL, et al. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin. 2015;8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Phipson B, Maksimovic J, Oshlack A. Missmethyl: An R package for analyzing data from illumina’s HumanMethylation450 platform. Bioinformatics. 2016;32:286–288. [DOI] [PubMed] [Google Scholar]
- 47. Debette S, Schilling S, Duperron MG, Larsson SC, Markus HS. Clinical significance of magnetic resonance imaging markers of vascular brain injury: A systematic review and meta-analysis. JAMA Neurol. 2019;76:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Prins ND, Scheltens P. White matter hyperintensities, cognitive impairment and dementia: An update. Nat Rev Neurol. 2015;11:157. [DOI] [PubMed] [Google Scholar]
- 49. Prins ND, van Dijk EJ, den Heijer T, et al. Cerebral small-vessel disease and decline in information processing speed, executive function and memory. Brain. 2005;128:2034–2041. [DOI] [PubMed] [Google Scholar]
- 50. Richard MA, Huan T, Ligthart S, et al. DNA methylation analysis identifies loci for blood pressure regulation. Am J Hum Genet. 2017;101:888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ray D, Boehnke M. Methods for meta-analysis of multiple traits using GWAS summary statistics. Genet Epidemiol. 2018;42:134–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pierce BL, Burgess S. Efficient design for Mendelian randomization studies: Subsample and 2-sample instrumental variable estimators. Am J Epidemiol. 2013;178:1177–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huan T, Joehanes R, Song C, et al. Genome-wide identification of DNA methylation QTLs in whole blood highlights pathways for cardiovascular disease. Nat Commun. 2019;10:4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Traylor M, Tozer DJ, Croall ID, et al. Genetic variation in PLEKHG1 is associated with white matter hyperintensities (n = 11,226). Neurology. 2019;92:e749–e757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hemani G, Zheng J, Elsworth B, et al. The MR-base platform supports systematic causal inference across the human phenome. eLife. 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hannon E, Lunnon K, Schalkwyk L, Mill J. Interindividual methylomic variation across blood, cortex, and cerebellum: Implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 2015;10:1024–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ng B, White CC, Klein HU, et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat Neurosci. 2017;20:1418–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Giambartolomei C, Zhenli Liu J, Zhang W, et al. A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics. 2018;34:2538–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brown MB. 400: A method for combining non-independent, one-sided tests of significance. Biometrics. 1975;31:987–992. [Google Scholar]
- 60. Lin H, Satizabal C, Xie Z, et al. Whole blood gene expression and white matter hyperintensities. Mol Neurodegener. 2017;12:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shu L, Zhao Y, Kurt Z, et al. Mergeomics: Multidimensional data integration to identify pathogenic perturbations to biological systems. BMC Genomics. 2016;17:874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu Y, Ding J, Reynolds LM, et al. Methylomics of gene expression in human monocytes. Hum Mol Genet. 2013;22:5065–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jassal B, Matthews L, Viteri G, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–D503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nishimura D. Biocarta. Biotech Softw Internet Rep. 2001;2:117–120. [Google Scholar]
- 65. Carbon S, Douglass E, Good BM, et al. The gene ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Arneson D, Bhattacharya A, Shu L, Makinen VP, Yang X. Mergeomics: A web server for identifying pathological pathways, networks, and key regulators via multidimensional data integration. BMC Genomics. 2016;17:722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhu J, Lum PY, Lamb J, et al. An integrative genomics approach to the reconstruction of gene networks in segregating populations. Cytogenet Genome Res. 2004;105:363–374. [DOI] [PubMed] [Google Scholar]
- 69. Shannon P, Markiel A, Ozier O, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen YW, Diamante G, Ding J, et al. Pharmomics: A species- and tissue-specific drug signature database and online tool for drug repurposing. iScience. 2022;25(4):104052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gross AM, Jaeger PA, Kreisberg JF, et al. Methylome-wide analysis of chronic HIV infection reveals five-year increase in biological age and epigenetic targeting of HLA. Mol Cell. 2016;62:157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Persyn E, Hanscombe KB, Howson JMM, Lewis CM, Traylor M, Markus HS. Genome-wide association study of MRI markers of cerebral small vessel disease in 42,310 participants. Nat Commun. 2020;11:2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Verhaaren BFJ, Debette S, Bis JC, et al. Multiethnic genome-wide association study of cerebral white matter hyperintensities on MRI. Circ Cardiovasc Genet. 2015;8:398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Armstrong NJ, Mather KA, Sargurupremraj M, et al. Common genetic variation indicates separate causes for periventricular and deep white matter hyperintensities. Stroke. 2020;51:2111–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Giri A, Hellwege JN, Keaton JM, et al. Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nat Genet. 2019;51:51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kichaev G, Bhatia G, Loh PR, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet. 2019;104:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sakaue S, Kanai M, Tanigawa Y, et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat Genet. 2021;53:1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Warren HR, Evangelou E, Cabrera CP, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017;49:403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Takeuchi F, Akiyama M, Matoba N, et al. Interethnic analyses of blood pressure loci in populations of east Asian and European descent. Nat Commun. 2018;9:5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kato N, Takeuchi F, Tabara Y, et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011;43:531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhu Z, Wang X, Li X, et al. Genetic overlap of chronic obstructive pulmonary disease and cardiovascular disease-related traits: A large-scale genome-wide cross-trait analysis. Respir Res. 2019;20:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. The GTEX Consortium . The GTEx consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lauretti E, Dincer O, Praticò D. Glycogen synthase kinase-3 signaling in Alzheimer’s disease. Biochim Biophys Acta Mol Cell Res. 2020;1867:118664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kwon NH, Fox PL, Kim S. Aminoacyl-tRNA synthetases as therapeutic targets. Nat Rev Drug Discov. 2019;18:629–650. [DOI] [PubMed] [Google Scholar]
- 85. Tomimoto H, Akiguchi I, Ohtani R, et al. The coagulation-fibrinolysis system in patients with leukoaraiosis and Binswanger disease. Arch Neurol. 2001;58:1620–1625. [DOI] [PubMed] [Google Scholar]
- 86. Wiseman S, Marlborough F, Doubal F, Webb DJ, Wardlaw J. Blood markers of coagulation, fibrinolysis, endothelial dysfunction and inflammation in lacunar stroke versus non-lacunar stroke and non-stroke: Systematic review and meta-analysis. Cerebrovasc Dis. 2014;37:64–75. [DOI] [PubMed] [Google Scholar]
- 87. Nagai M, Hoshide S, Kario K. Association of prothrombotic status with markers of cerebral small vessel disease in elderly hypertensive patients. Am J Hypertens. 2012;25:1088–1094. [DOI] [PubMed] [Google Scholar]
- 88. Kario K, Yano Y, Matsuo T, Hoshide S, Eguchi K, Shimada K. Additional impact of morning haemostatic risk factors and morning blood pressure surge on stroke risk in older Japanese hypertensive patients. Eur Heart J. 2011;32:574–580. [DOI] [PubMed] [Google Scholar]
- 89. Markus HS, Hunt B, Palmer K, Enzinger C, Schmidt H, Schmidt R. Markers of endothelial and hemostatic activation and progression of cerebral white matter hyperintensities. Stroke. 2005;36:1410–1414. [DOI] [PubMed] [Google Scholar]
- 90. Wolf NI, Willemsen MAAP, Engelke UF, et al. Severe hypomyelination associated with increased levels of N-acetylaspartylglutamate in CSF. Neurology. 2004;62:1503. [DOI] [PubMed] [Google Scholar]
- 91. Mochel F, Boildieu N, Barritault J, et al. Elevated CSF N-acetylaspartylglutamate suggests specific molecular diagnostic abnormalities in patients with white matter diseases. Biochim Biophys Acta. 2010;1802:1112–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wardlaw JM, Smith C, Dichgans M. Small vessel disease: Mechanisms and clinical implications. Lancet Neurol. 2019;18:684–696. [DOI] [PubMed] [Google Scholar]
- 93. Wei H, Mundade R, Lange KC, Lu T. Protein arginine methylation of non-histone proteins and its role in diseases. Cell Cycle. 2014;13:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hashimoto M, Murata K, Ishida J, Kanou A, Kasuya Y, Fukamizu A. Severe hypomyelination and developmental defects are caused in mice lacking protein arginine methyltransferase 1 (PRMT1) in the central nervous system. J Biol Chem. 2016;291:2237–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Dowsett L, Higgins E, Alanazi S, Alshuwayer NA, Leiper FC, Leiper J. ADMA: A key player in the relationship between vascular dysfunction and inflammation in atherosclerosis. J Clin Med. 2020;9:3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Watson CP, Pazarentzos E, Fidanboylu M, Padilla B, Brown R, Thomas SA. The transporter and permeability interactions of asymmetric dimethylarginine (ADMA) and L-arginine with the human blood-brain barrier in vitro. Brain Res. 2016;1648:232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Janes F, Cifù A, Pessa ME, et al. ADMA As a possible marker of endothelial damage. A study in young asymptomatic patients with cerebral small vessel disease. Sci Rep. 2019;9:14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Pikula A, Böger RH, Beiser AS, et al. Association of plasma ADMA levels with MRI markers of vascular brain injury: Framingham Offspring Study. Stroke. 2009;40:2959–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Guan J, Yan C, Gao Q, et al. Analysis of risk factors in patients with leukoaraiosis. Medicine. 2017;96:e6153–e6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Khan U, Hassan A, Vallance P, Markus HS. Asymmetric dimethylarginine in cerebral small vessel disease. Stroke. 2007;38:411–413. [DOI] [PubMed] [Google Scholar]
- 101. Notsu Y, Nabika T, Bokura H, et al. Evaluation of asymmetric dimethylarginine and homocysteine in microangiopathy-related cerebral damage. Am J Hypertens. 2009;22:257–262. [DOI] [PubMed] [Google Scholar]
- 102. Gao Q, Fan Y, Mu LY, Ma L, Song ZQ, Zhang YN. S100b and ADMA in cerebral small vessel disease and cognitive dysfunction. J Neurol Sci. 2015;354:27–32. [DOI] [PubMed] [Google Scholar]
- 103. Rufa A, Blardi P, de Lalla A, et al. Plasma levels of asymmetric dimethylarginine in cerebral autosomal dominant arteriopathy with subcortical infarct and leukoencephalopathy. Cerebrovasc Dis. 2008;26:636–640. [DOI] [PubMed] [Google Scholar]
- 104. Guo L, Peng Y, Meng Y, et al. Expression profiles analysis reveals an integrated miRNA-lncRNA signature to predict survival in ovarian cancer patients with wild-type BRCA1/2. Oncotarget. 2017;8:68483–68492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. He J, Guan J, Liao S, et al. Long noncoding RNA CCDC144NL-AS1 promotes the oncogenicity of osteosarcoma by acting as a molecular sponge for microRNA-490-3p and thereby increasing HMGA2 expression. Onco Targets Ther. 2021;14:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 106. Fan H, Ge Y, Ma X, et al. Long non-coding RNA CCDC144NL-AS1 sponges miR-143-3p and regulates MAP3K7 by acting as a competing endogenous RNA in gastric cancer. Cell Death Dis. 2020;11:521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhang L, Chi B, Chai J, et al. LncRNA CCDC144NL-AS1 serves as a prognosis biomarker for non-small cell lung cancer and promotes cellular function by targeting miR-490-3p. Mol Biotechnol. 2021;63:933–940. [DOI] [PubMed] [Google Scholar]
- 108. Zhang Y, Zhang H, Wu S. LncRNA-CCDC144NL-AS1 promotes the development of hepatocellular carcinoma by inducing WDR5 expression via sponging miR-940. J Hepatocell Carcinoma. 2021;8:333–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Niu P, Yao B, Wei L, Zhu H, Fang C, Zhao Y. Construction of prognostic risk prediction model based on high-throughput sequencing expression profile data in childhood acute myeloid leukemia. Blood Cells Mol Dis. 2019;77:43–50. [DOI] [PubMed] [Google Scholar]
- 110. Zhang C, Wu W, Zhu H, et al. Knockdown of long noncoding RNA CCDC144NL-AS1 attenuates migration and invasion phenotypes in endometrial stromal cells from endometriosis. Biol Reprod. 2019;100:939–949. [DOI] [PubMed] [Google Scholar]
- 111. Anwar MM, Özkan E, Gürsoy-Özdemir Y. The role of extracellular matrix alterations in mediating astrocyte damage and pericyte dysfunction in Alzheimer’s disease: A comprehensive review. Eur J Neurosci. 2022;56(9):5453–5475. [DOI] [PubMed] [Google Scholar]
- 112. Montaner J, Ramiro L, Simats A, et al. Matrix metalloproteinases and ADAMs in stroke. Cell Mol Life Sci. 2019;76:3117–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Prasain N, Stevens T. The actin cytoskeleton in endothelial cell phenotypes. Microvasc Res. 2009;77:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Hara K, Shiga A, Fukutake T, et al. Association of HTRA1 mutations and familial ischemic cerebral small-vessel disease. New Engl J Med. 2009;360:1729–1739. [DOI] [PubMed] [Google Scholar]
- 115. Kast J, Hanecker P, Beaufort N, et al. Sequestration of latent TGF-β binding protein 1 into CADASIL-related Notch3-ECD deposits. Acta Neuropathol Commun. 2014;2:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Hamel E. Cerebral circulation: Function and dysfunction in Alzheimer’s disease. J Cardiovasc Pharm. 2015;65:317–324. [DOI] [PubMed] [Google Scholar]
- 117. Smahi A, Courtois G, Vabres P, et al. Genomic rearrangement in NEMO impairs NF-κB activation and is a cause of incontinentia pigmenti. Nature. 2000;405:466–472. [DOI] [PubMed] [Google Scholar]
- 118. Ridder DA, Wenzel J, Müller K, et al. Brain endothelial TAK1 and NEMO safeguard the neurovascular unit. J Exp Med. 2015;212:1529–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wardlaw JM, Makin SJ, Valdés Hernández MC, et al. Blood-brain barrier failure as a core mechanism in cerebral small vessel disease and dementia: Evidence from a cohort study. Alzheimers Dement. 2017;13:634–643. [Google Scholar]
- 120. Sukriti S, Tauseef M, Yazbeck P, Mehta D. Mechanisms regulating endothelial permeability. Pulm Circ. 2014;4:535–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Lippmann ES, Azarin SM, Kay JE, et al. Derivation of blood-brain barrier endothelial cells from human pluripotent stem cells. Nat Biotechnol. 2012;30:783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Greene C, Hanley N, Campbell M. Claudin-5: Gatekeeper of neurological function. Fluids Barriers CNS. 2019;16:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Yang Y, Kimura-Ohba S, Thompson JF, et al. Vascular tight junction disruption and angiogenesis in spontaneously hypertensive rat with neuroinflammatory white matter injury. Neurobiol Dis. 2018;114:95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Berndt P, Winkler L, Cording J, et al. Tight junction proteins at the blood-brain barrier: Far more than claudin-5. Cell Mol Life Sci. 2019;76:1987–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Bailey EL, Wardlaw JM, Graham D, Dominiczak AF, Sudlow CLM, Smith C. Cerebral small vessel endothelial structural changes predate hypertension in stroke-prone spontaneously hypertensive rats: A blinded, controlled immunohistochemical study of 5- to 21-week-old rats. Neuropathol Appl Neurobiol. 2011;37:711–726. [DOI] [PubMed] [Google Scholar]
- 126. Rikesh RM, Sophie Q, Silvie RR, et al. Reversal of endothelial dysfunction reduces white matter vulnerability in cerebral small vessel disease in rats. Sci Transl Med. 2018;10:eaam9507. [DOI] [PubMed] [Google Scholar]
- 127. Hüls A, Robins C, Conneely KN, et al. Brain DNA methylation patterns in CLDN5 associated with cognitive decline. Biol Psychiat. 2021;91:389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Belinky F, Nativ N, Stelzer G, et al. Pathcards: Multi-source consolidation of human biological pathways. Database. 2015;2015:bav006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Zhao B, Zhang J, Ibrahim JG, et al. Large-scale GWAS reveals genetic architecture of brain white matter microstructure and genetic overlap with cognitive and mental health traits (n = 17,706). Mol Psychiatr. 2021;26:3943–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Malik R, Chauhan G, Traylor M, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50:524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Ishigaki K, Akiyama M, Kanai M, et al. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat Genet. 2020;52:669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Murphy DA, Courtneidge SA. The ‘ins’ and ‘outs’ of podosomes and invadopodia: Characteristics, formation and function. Nat Rev Mol Cell Biol. 2011;12:413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Shi Y, Zhang L, Pu H, et al. Rapid endothelial cytoskeletal reorganization enables early blood-brain barrier disruption and long-term ischaemic reperfusion brain injury. Nat Commun. 2016;7:10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Georgakis MK, Malik R, Anderson CD, Parhofer KG, Hopewell JC, Dichgans M. Genetic determinants of blood lipids and cerebral small vessel disease: Role of high-density lipoprotein cholesterol. Brain. 2020;143:597–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Zhang H, Cui Y, Zhao Y, et al. Effects of sartans and low-dose statins on cerebral white matter hyperintensities and cognitive function in older patients with hypertension: A randomized, double-blind and placebo-controlled clinical trial. Hypertens Res. 2019;42:717–729. [DOI] [PubMed] [Google Scholar]
- 136. Guo Y, Li Y, Liu X, et al. Assessing the effectiveness of statin therapy for alleviating cerebral small vessel disease progression in people ≥75 years of age. BMC Geriatr. 2020;20:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. ten Dam VH, van den Heuvel DMJ, van Buchem MA, et al. Effect of pravastatin on cerebral infarcts and white matter lesions. Neurology. 2005;64:1807–1809. [DOI] [PubMed] [Google Scholar]
- 138. Wardlaw JM, Debette S, Jokinen H, et al. ESO Guideline on covert cerebral small vessel disease. Eur Stroke J. 2021;6:CXI–CLXII. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Zhang CE, Wong SM, van de Haar HJ, et al. Blood–brain barrier leakage is more widespread in patients with cerebral small vessel disease. Neurology. 2017;88:426. [DOI] [PubMed] [Google Scholar]
- 140. Giannopoulos S, Katsanos AH, Tsivgoulis G, Marshall RS. Statins and cerebral hemodynamics. J Cereb Blood Flow Metab. 2012;32:1973–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Holland PR, Searcy JL, Salvadores N, et al. Gliovascular disruption and cognitive deficits in a mouse model with features of small vessel disease. J Cereb Blood Flow Metab. 2015;35:1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Moulignier A, Savatovsky J, Assoumou L, et al. Silent cerebral small-vessel disease is twice as prevalent in middle-aged individuals with well-controlled, combination antiretroviral therapy–treated human immunodeficiency virus (HIV) than in HIV-uninfected individuals. Clin Infect Dis. 2018;66:1762–1769. [DOI] [PubMed] [Google Scholar]
- 143. Mina Y, Wu T, Hsieh HC, et al. Association of white matter hyperintensities with HIV Status and vascular risk factors. Neurology. 2021;96:e1823–e1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are included in this paper. Full EWAS summary statistics are available in dbGaP at phs000930.v9.p1.