Abstract
Oral inflammatory diseases such as apical periodontitis are common bacterial infectious diseases that may affect the periapical alveolar bone tissues. A protective process occurs simultaneously with the inflammatory tissue destruction, in which mesenchymal stem cells (MSCs) play a primary role. However, a systematic and precise description of the cellular and molecular composition of the microenvironment of bone affected by inflammation is lacking. In this study, we created a single-cell atlas of cell populations that compose alveolar bone in healthy and inflammatory disease states. We investigated changes in expression frequency and patterns related to apical periodontitis, as well as the interactions between MSCs and immunocytes. Our results highlight an enhanced self-supporting network and osteogenic potential within MSCs during apical periodontitis-associated inflammation. MSCs not only differentiated toward osteoblast lineage cells but also expressed higher levels of osteogenic-related markers, including Sparc and Col1a1. This was confirmed by lineage tracing in transgenic mouse models and human samples from oral inflammatory-related alveolar bone lesions. In summary, the current study provides an in-depth description of the microenvironment of MSCs and immunocytes in both healthy and disease states. We also identified key apical periodontitis-associated MSC subclusters and their biomarkers, which could further our understanding of the protective process and the underlying mechanisms of oral inflammatory-related bone disease. Taken together, these results enhance our understanding of heterogeneity and cellular interactions of alveolar bone cells under pathogenic and inflammatory conditions. We provide these data as a tool for investigators not only to better appreciate the repertoire of progenitors that are stress responsive but importantly to help design new therapeutic targets to restore bone lesions caused by apical periodontitis and other inflammatory-related bone diseases.
Research organism: Human, Mouse
Introduction
Oral diseases, particularly dental caries and periodontal diseases, affect 3.5 billion people worldwide (Disease et al., 2018). Untreated dental caries can directly lead to pulp necrosis and periapical lesions, resulting in apical periodontitis (AP). Individuals with at least one tooth affected by AP comprise up to 52% of cases, indicating that AP is a highly prevalent disease (Tibúrcio-Machado et al., 2021). Inflammation in the oral cavity can lead to destruction of surrounding periapical tissues and resorption of hard tissues, a consequence of the unbalanced interaction between infection and the immune response (Gazivoda et al., 2009; Márton and Kiss, 2014). Restoring and regenerating the destroyed periapical alveolar bone structures have always been a challenging task in clinical practice. Active inflammation, the tissue injury and the protective process all occur simultaneously in the setting of chronic AP (Márton and Kiss, 2014). Importantly, there is a complex assemblage of immune cell types involved in the pathogenesis, highlighting the importance of polymorphonuclear leukocytes, lymphocytes, and monocyte/macrophages in periapical defense (Braz-Silva et al., 2019; Nair, 2004). Notably, an increasing number of studies report the involvement of mesenchymal stem cells (MSCs) in the protective action that occurs during oral inflammatory diseases, whereby MSCs exert immunomodulatory effects and have regenerative potential (Li et al., 2014; Márton and Kiss, 2000; Nair, 2004). MSC markers such as CD44, CD73, CD90, CD106, and STRO-1 have been observed in human periapical inflammatory tissues (Estrela et al., 2019; Liao et al., 2011). Cells isolated from the inflamed periapical region were able to produce colony-forming unit-fibroblasts (CFU-Fs) with high-osteogenic capacity. It is also reported that interference with MSC mobilization toward the periapex region in an AP mouse model led to enlargement of lesions, accompanied by decreased wound healing markers and increased inflammatory cytokines (Araujo-Pires et al., 2014). These findings indicate the involvement of MSCs in the repair and regeneration of oral inflammatory-related bone lesions. They also suggest that MSCs present promising targets for treating bone lesions, with great potential for modulating inflammation and promoting tissue regeneration. However, most studies have surveyed whole tissues to understand the transcriptomic and cellular profile of these diseases. Specific cell populations and their regulatory molecules, as well as the interaction among different cell populations, remain far from clear. The advances in single-cell technologies offer an unbiased approach for identifying heterogeneous cell subsets, tracking the trajectories of distinct cell clusters and uncovering regulatory relationships between genes (Hwang et al., 2018; Tang et al., 2009). In this study, we collected mandibular alveolar bone samples from control and AP in mice and subjected them to single-cell RNA sequencing (scRNA-seq). The atlas of the mandibular alveolar bone explored the distinct cell subsets and their expression profiles relevant to AP. We also investigated the relationship between MSCs and immune cell subsets. The results reveal the role of a subset of MSCs in inflammation, which showed increased frequency and which formed a self-supporting network. Moreover, MSCs exhibited upregulated osteogenic potential, which was confirmed in transgenic mouse models and human patients with chronic AP. These results advance our understanding of heterogeneity and interactions of alveolar bone cells in the pathogenesis of inflammatory-related bone diseases. Defining key cellular subsets such as MSCs and their biomarkers in inflamed tissue will be important for identifying new therapeutic targets for oral inflammatory-related bone diseases.
Results
Single-cell transcriptional profiling identified 15 discrete populations in homeostasis and chronic AP samples
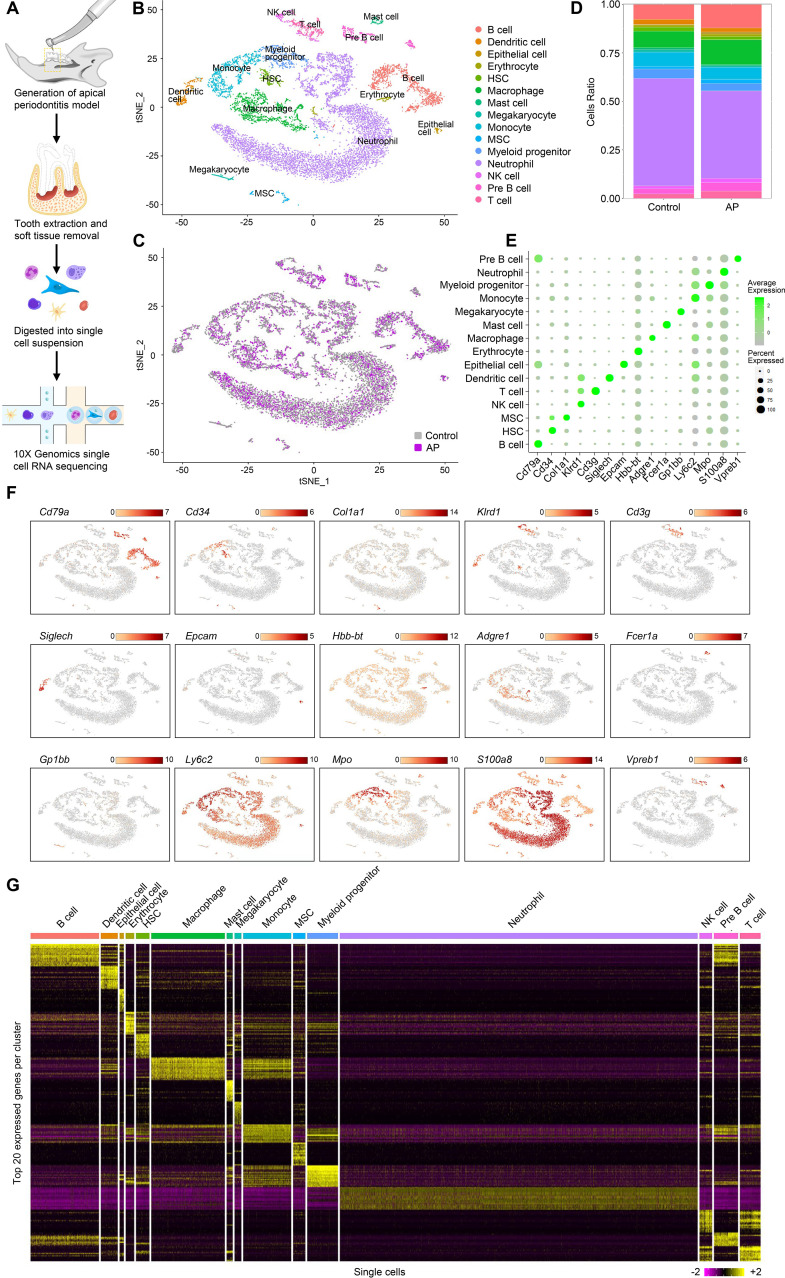
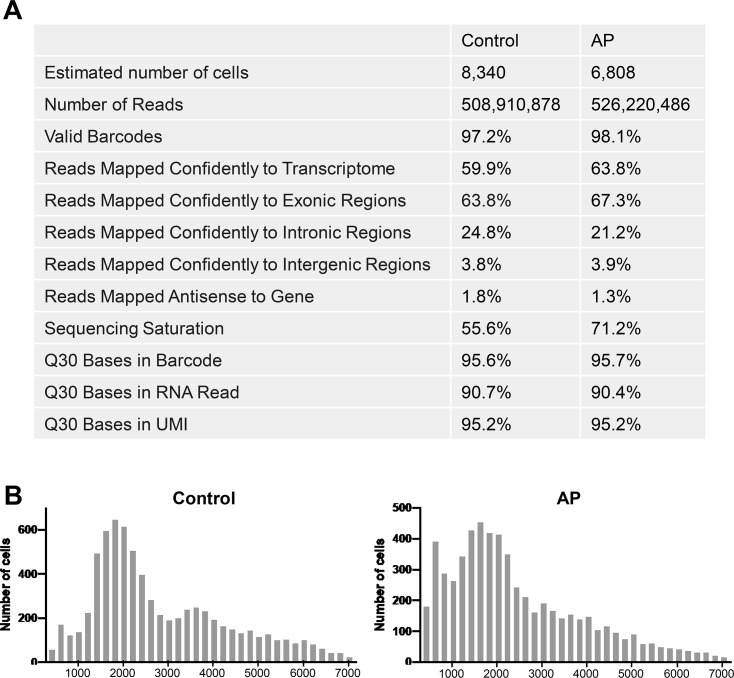
Individual cells were isolated from alveolar bone of healthy mice and mice with AP. We modeled AP using a well-established AP mouse model in which the mandibular first molar pulp was exposed and subsequently developed chronic AP over a 3 wk period (Taira et al., 2019). Bar-coded cDNA libraries from individual cells were obtained using the 10× Genomics Chromium Controller platform (Zheng et al., 2017; Figure 1A). The combined libraries from healthy and AP alveolar bone contained 15,148 individual cells. The median value of feature_RNA was between 1000 and 2000 (Figure 1—figure supplement 1B). After quality control filtering and removal of the batch effect between batches, the t-stochastic neighbor embedding (t-SNE) method was applied to reduce the dimensionality. Seurat’s unbiased cluster detection algorithm identified 15 distinct cell populations (Figure 1B and C). Cluster-specific transcripts were utilized to annotate cell types with classic markers as described in a previous study (Lin et al., 2021). These included B cell (Cd79a), hematopoietic stem cell (HSC) (Cd34), MSC (Col1a1), natural killer (NK) cell (Klrd1), T cell (Cd3g), dendritic cell (Siglech), epithelial cell (Epcam), erythrocyte (Hbb-bt), macrophage (Adgre1), mast cell (Fcer1a), megakaryocyte (Gp1bb), monocyte (Ly6c2), myeloid progenitor (Mpo), neutrophil (S100a8), and pre-B cell (Vpreb1; Figure 1E and F). The top 20 enriched genes in each defined cluster were identified and compared (Figure 1G).
Figure 1. Identification of the single-cell atlas of alveolar bone using scRNA-sequencing (scRNA-seq) and unbiased clustering.
(A) Schematic diagram of the experimental design. (B–C) t-Stochastic neighbor embedding (t-SNE) representation of aligned gene expression data in single cells extracted from mandibles of control mice (n=8340) and apical periodontitis (AP) mice (n=6808) showing 15 distinct clusters and cellular origin. (D) Relative abundance of 15 cell populations composing alveolar bone under healthy and AP conditions. (E) Expression of gene markers in distinct cell types. (F) Gene expression patterns projected onto t-SNE plots of marker genes. Scale: log-transformed gene expression. (G) Heatmap showing the 20 most upregulated genes (ordered by decreasing Padj value) in each cluster defined in B. Scale: log2 fold change.
AP led to significant changes in frequency and transcriptional expression of cell populations
All the identified cell clusters were present in both AP and control samples, but there were significant differences in the cellular compositions of particular clusters. T cell, B cell, NK cell, macrophage, epithelial cell, and MSC had significantly increased frequency in AP samples. Neutrophil, myeloid progenitor, monocyte, megakaryocyte, mast cell, HSC, and dendritic cell were markedly decreased (Figures 2A and 1D).
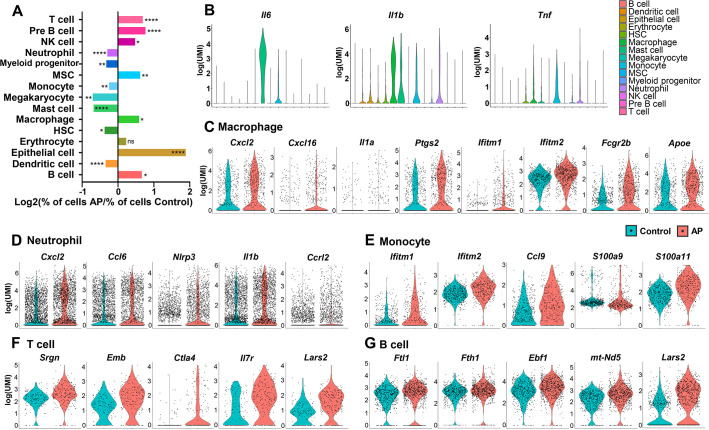
Figure 2. Changes in frequency and transcriptional expression pattern in each cell population from control and apical periodontitis (AP) groups.
(A) Bar plot of cells per cluster (AP versus control). Normalization to overall number of inputs per condition. Fisher’s exact test with Bonferroni correction was used. *p<0.05, **p<0.01, and ****p<0.0001. All data were exhibited as mean ± SEM. (B) Violin plots of cluster-specific expression of representative genes. (C–G) Violin plots showing genes that significantly changed in each cluster from control and AP.
AP is a complex inflammatory process involving innate and adaptive immune responses (Cotti et al., 2014). A variety of inflammatory cells such as neutrophils, mast cells, monocytes, macrophages, and lymphocytes are involved in periapical lesions, highlighting the direct involvement of the immune response in the pathogenesis of AP (Nair, 2004). Neutrophils are important components in the acute phase of AP as a first line of defense. But they are also important in the progression of AP by interacting with microorganisms, leading to tissue damage and chemotaxis (Braz-Silva et al., 2019). Single-cell differential expression analysis revealed that the most significantly enriched genes in neutrophils were various proinflammatory chemokines and cytokines. These included C-X-C motif chemokine ligand 2 (Cxcl2), C-C motif chemokine ligand 6 (Ccl6), NLR family pyrin domain containing 3 (Nlrp3), and Interleukin-1β (Il1b). Notably, we found that C-C motif chemokine receptor like 2 (Ccrl2) was upregulated in neutrophils during AP (Figure 2D). It is responsible for the innate defense against pathogens and is also involved in the regulation of neutrophil migration (Del Prete et al., 2017; Kolaczkowska and Kubes, 2013; Mantovani et al., 2011).
Mast cells, monocytes, and macrophages have critical roles in the inflammatory infiltrate during chronic AP (Braz-Silva et al., 2019). The production of Interleukin-6 (Il6) was present in these cell populations with the highest expression level in mast cells. The pro-inflammatory cytokine IL-1β is a key mediator of host response to microbial infection and is associated with the persistence of AP (Morsani et al., 2011; Ng et al., 2008). We found Il1b transcripts in a series of cell types, such as monocyte, macrophage, mast cell, and neutrophil. Of these, macrophages had the highest Il1b expression. Another major cytokine, tumor necrosis factor (Tnf; Cotti et al., 2014), was detected in immunoresponsive cell clusters, such as monocyte, macrophage, mast cell, myeloid progenitor, neutrophil, and HSC, with the highest expression observed in the monocyte population (Figure 2B).
Furthermore, gene signatures from monocytes showed that the interferon-induced transmembranes (IFITMs) protein 1 and 2 (Ifitm1 and Ifitm2; Figure 2E) were upregulated the most during AP. These factors have been associated with signal transduction of anti-inflammation activity in the immune system (Yánez et al., 2020). We also detected upregulated expression levels of Ccl9 in the monocyte population from AP. Ccl9 is an important cytokine and is involved in the survival of osteoclasts during the destruction of the periapical bone (Silva et al., 2007). Also, genes coding for pro-inflammatory calcium-binding S100 family proteins such as S100a9 and S100a11 had increased expression in the AP monocyte cluster (Figure 2E). Previous reports demonstrated that macrophages are capable of secreting pro- and anti-inflammatory substances which act on the development and repair of the AP lesions (Italiani and Boraschi, 2014; Shapouri-Moghaddam et al., 2018). Indeed, several genes encoding pro-inflammatory mediators, including Cxcl2, Cxcl16, Il1a, and Ptgs2, were upregulated in macrophages from AP samples (Figure 2C). Expression of anti-inflammatory-associated genes such as Ifitm1 and Ifitm2 was significantly increased in the AP macrophage cluster compared to control cells. Furthermore, Fcγ receptor IIB (Fcgr2b) was markedly upregulated in macrophages from AP samples. Fcgr2b is expressed in most tissue-resident macrophages (Gautier et al., 2012) and functions to inhibit Fcγ-dependent phagocytosis. It also inhibits release of cytokines such as IL-6, TNF-α, IL-1α, as well as neutrophil chemotactants (Clatworthy and Smith, 2004; Espéli et al., 2016). In addition, expression of Apolipoprotein E (Apoe), which can suppress the pro-inflammatory response (Jofre-Monseny et al., 2007), was significantly increased in the macrophage population (Figure 2C). These data indicated the activation of anti-inflammatory factors by macrophages during local inflammation by AP.
The major classes of lymphocytes are T lymphocytes, B lymphocytes, and the NK cells. T and B lymphocytes comprise the majority of the inflammatory infiltrate in AP (Graunaite et al., 2012). A significant increase in the expression of inflammatory-associated genes, such as Srgn, Emb, Ctla4, and Il7r could be observed in the AP T cell population (Figure 2F). In the AP B lymphocytes cluster, inflammation-responsive genes (Fth1, Ftl1, Ebf1, and mt-Nd5) were upregulated (Figure 2G). Interestingly, Lars2, the gene encoding a mitochondrial leucyl tRNA synthase (Carminho-Rodrigues et al., 2020; ’t Hart et al., 2005), was significantly upregulated in T and B lymphocytes, indicating changes in mitochondrial metabolism in both clusters.
Inflammation induces osteoclasts differentiation leading to periapical alveolar bone destruction
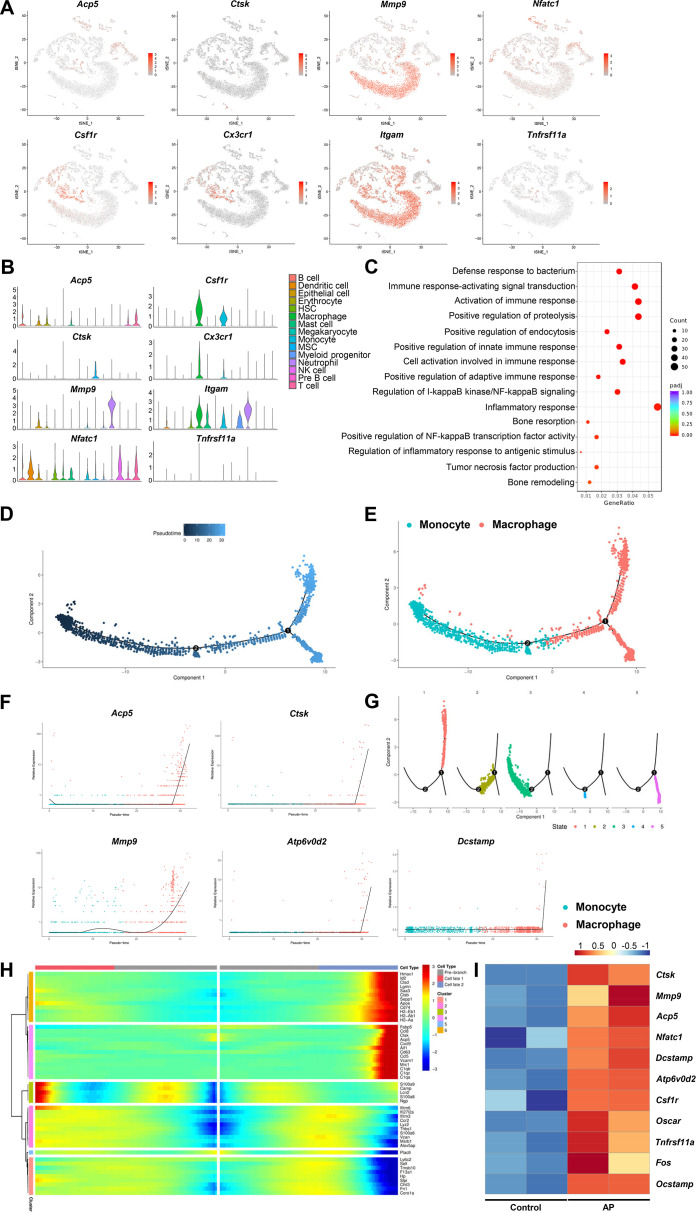
Bone destruction is a major pathological factor in chronic inflammatory diseases such as AP. Various cytokines including TNF-α, IL-1α, and IL-6 were released by immunocytes to recruit the osteoclast precursors and induce the maturation of osteoclasts (Lyu et al., 2022). We have detected osteoclast markers including Ctsk, Acp5, Mmp9, and Nfatc1 by scRNA-seq. Moreover, Csfr1, Cx3cr1, Itgam, and Tnfrs11a were used to identify osteoclast precursors. Markers of osteoclast and osteoclast precursors were highly expressed in the clusters of monocyte and macrophage (Figure 3A and B). Gene Ontology (GO) analysis showed that inflammation related immune reactions and bone resorption activity were significantly enriched in macrophage cluster (Figure 3C). To further study the differential trajectory of osteoclasts, pseudotime analysis was performed for the clusters of macrophage and monocyte. Two independent branch points were determined, and five monocyte/macrophage subclusters were scattered at different branches in the developmental tree (Figure 3D and G). The results showed that the monocyte cluster differentiated into the macrophage cluster (Figure 3E). During this trajectory, the gene expression pattern across pseudotime showed that osteoclastic genes, such as Ctsk, Acp5, Mmp9, Atp6v0d2, and Dcstamp, were progressively elevated (Figure 3F). Of note, we have observed a branch which was highly positive for Ctsk and Acp5 (Figure 3H), indicating the mature osteoclasts were differentiated from monocyte/macrophage lineage and contributed to inflammatory bone resorption during AP. We have also analyzed the expression of osteoclast related genes using the bulk RNA-seq library built on mandibular samples extracted from mice with AP. Markers of osteoclast and osteoclast precursors were significantly upregulated, confirming the osteoclasts activity in the inflammatory-related bone lesion (Figure 3I).
Figure 3. Inflammatory-related bone resorption under apical periodontitis (AP) situation.
(A) The expression levels of markers of osteoclasts and osteoclast precursors. (B) Violin plots of the expression of osteoclastogenesis genes. (C) Gene Ontology (GO) enrichment analysis of the biological functions of macrophage cluster. (D) Trajectory order of the monocyte/macrophage populations by pseudotime value. (E) The differentiation trajectory of monocyte and macrophage clusters presented on a t-stochastic neighbor embedding (t-SNE) visualization. (F) The expression patterns of osteoclast markers during the trajectory of monocyte/macrophage populations. (G) Distribution of monocytes/macrophages on the developmental tree by clusters. (H) Heatmap of differential genes of three states. (I) Heatmap of genes associated with osteoclastogenesis in bulk RNA-seq analysis.
Figure 3—figure supplement 1. The expression pattern of markers of M1-like macrophages and M2-like macrophages in macrophage cluster.
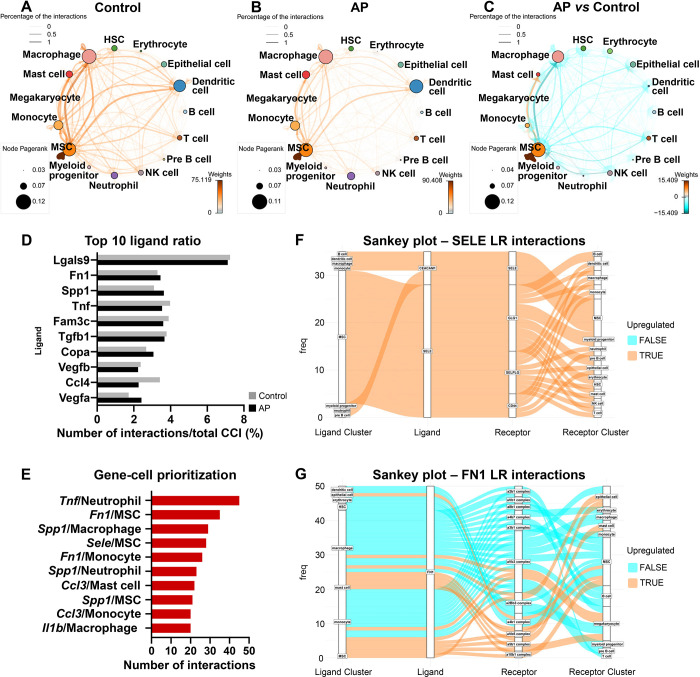
AP leads to reduced transcriptionally inferred cellular interactions with an increased self-supporting network in MSCs
We next sought to characterize the cell-cell communication related to the perturbation of signaling pathways detected in the AP samples by employing CellphoneDB (Efremova et al., 2020; Nagai et al., 2021). We identified a close interaction among MSC, macrophage, and dendritic cells under homeostasis conditions (Figure 4A). A similarly close communication was found among MSC, macrophage, and dendritic cells under inflammatory conditions (Figure 4B). Next, we compared the differential cell-cell interaction (CCI) network between AP and control samples using CrossTalkeR (Nagai et al., 2021). The results suggested that AP is associated with an overall decrease in cellular interactions. However, MSC intercellular communication with mast cells and monocytes is upregulated and accompanied by the highest number of interactions within MSCs themselves (Figure 4C). These results indicate that, although cell populations lose their normal physiological interactions, MSCs were able to establish a self-interacting network and coordinate with certain types of cells during chronic AP. Next, we ranked the individual ligands by the number of their interactions. Inflammatory-related proteins (Lgals9, Tnf, and Ccl4), extracellular matrix protein (Fn1), and protein involved in biomineralization (Spp1) were among the highest interactions. Also, Tgfb1, Vegfb, and Vegfa were in the top 10 most abundant ligands (Figure 4D). Bar plots were also generated to display the top 10 upregulated gene/cell pairs, showing that inflammation associated genes (Tnf/neutrophil, Ccl3/mast cell, Ccl3/monocyte, and Il1b/macrophage) and matrix related genes (Sele/MSC, Fn1/MSC, and Fn1/monocyte) were the most influential ligands during AP when compared to control state (Figure 4E). We used a Sankey plot to further focus on MSC-related interactions (Figure 4F). The results indicated that Sele was primarily directed by MSC toward the MSC cluster via multiple receptors including Glg1, Selplg, and Cd44. Moreover, Fn1 was secreted by MSC, monocyte, macrophage, and mast cells toward MSCs (Figure 4G). Of note, Sele and Fn1 are important cell adhesion molecules that mediate cell homing and migration (Frenette et al., 1998; To and Midwood, 2011). This was in accordance with the increased frequency of the MSC cluster (Figure 2A) as well as the upregulated cellular interaction among MSCs and other cell populations.
Figure 4. Apical periodontitis (AP) suppressed transcriptionally inferred cellular interactions and increased a self-supporting network within the mesenchymal stem cell (MSC) cluster.
(A–C) Network plot of ligand-receptor activity in control (A), AP (B), and AP versus control (C). (D) Bar plot of top 10 most abundant ligands in all inferred ligand-receptor interactions. (E) Ranking of ligand/source regarding communication gains in AP state. (F and G) Sankey plot listing all predicted source, receptor, and receiver interactions associated with Fn1 and Sele.
scRNA-seq based identification of AP-associated MSC population
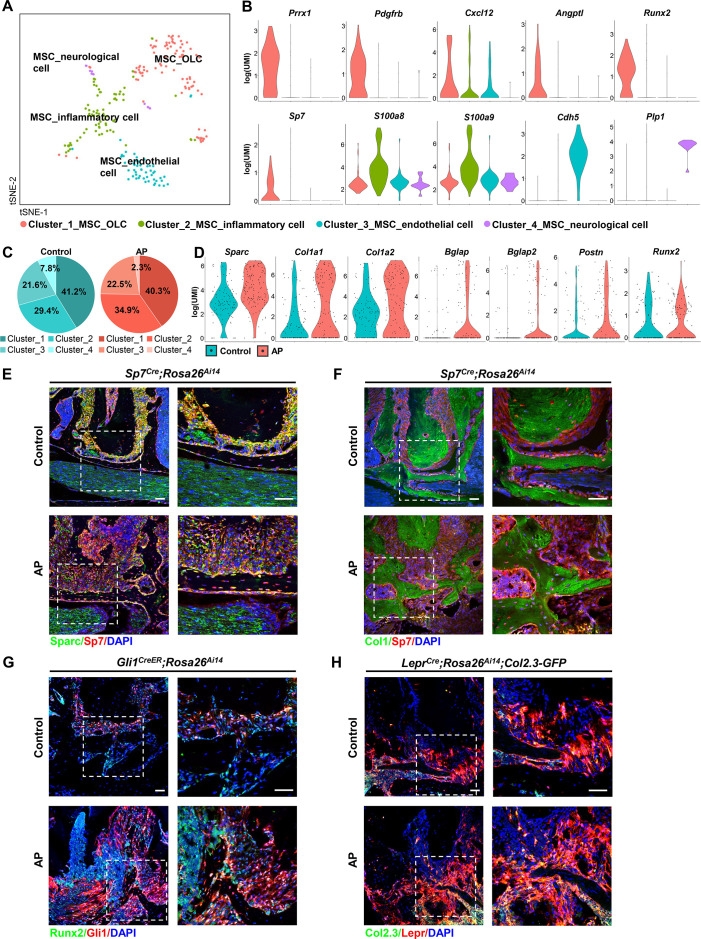
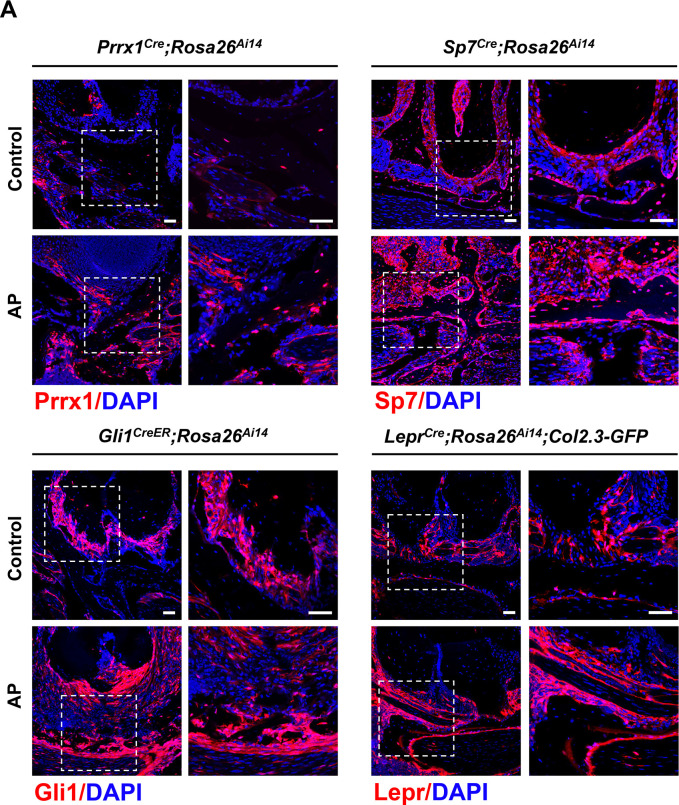
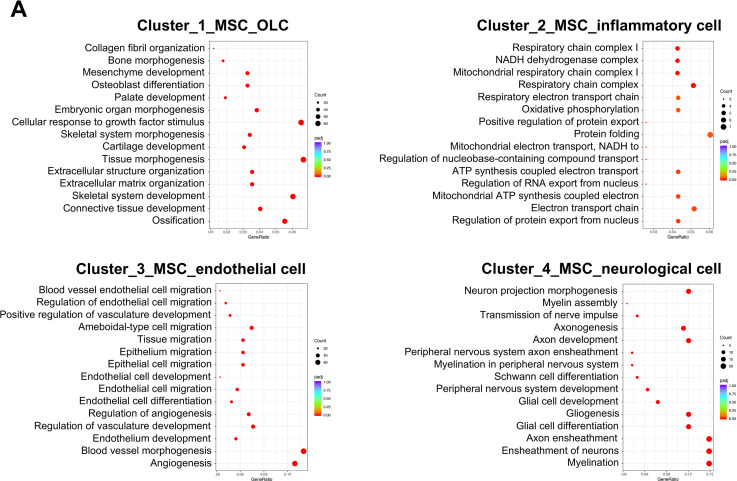
MSCs represented the non-immune cell population in alveolar bone, constituting 1.76% of total identified cells. This cell population could be decomposed into four subclusters (Figure 5A). The most dominant subcluster was characterized by high expression of Prrx1, platelet-derived growth factor receptor β (Pdgfrb), and hematopoiesis supporting factors such as C-X-C motif chemokine 12 (Cxcl12) and angiopoietin (Angpt1). It was also characterized by osteogenic-related markers, such as Runt-related transcription factor 2 (Runx2), Sp7, and was thus classified as the MSC_osteolineage cells (OLCs). The other subclusters were identified as MSC_endothelial (Cdh5+), MSC_neurological (Plp1+), and MSC_inflammatory (S100a8/S100a9+; Figure 5A and B). The cell composition of the MSC cluster indicated an expansion of MSC_inflammatory subcluster during AP, whereas the MSC_neurological subcluster was reduced (Figure 5C). Of note, we performed lineage tracing experiments using Prrx1Cre, Sp7Cre, LeprCre, and Gli1CreER transgenic mouse models to identify stem cells in alveolar bone and the surrounding periodontal tissues (Cui et al., 2020; Fan et al., 2022; Men et al., 2020; Zhang et al., 2020). The results suggested increased numbers of Prrx1+ and Sp7+ MSC_OLCs as well as upregulated Gli1+ and Lepr+ periodontal ligament stem cells (PDLSCs) in AP lesions (Figure 5—figure supplement 1A). We compared the MSC marker genes, identified above, among the four subpopulations. Their respective GO enrichment analysis confirmed that there are four specific MSC populations. MSC_OLCs were enriched for ossification, connective tissue development, skeletal system development, and osteoblast differentiation. MSC_endothelial cells displayed enrichment for angiogenesis, blood vessel morphogenesis, and endothelium development. MSC_neurological cells were enriched for myelination, ensheathment of neurons, and axon ensheathment. GO term analyses suggested highly specialized functional features of MSC_inflammatory cells, including regulation of protein export from nucleus, electron transport chain, mitochondrial ATP synthesis coupled electron transport, and oxidative phosphorylation (Figure 5—figure supplement 2A).
Figure 5. Identification and characterization of apical periodontitis (AP)-associated mesenchymal stem cell (MSC) population and its subclusters.
(A) t-Stochastic neighbor embedding (t-SNE) representation an unsupervised clustering of single cells within the MSC cluster. (B) Violin plots of MSC subcluster-specific expression of representative genes. (C) The percentages of four subclusters of MSC population were quantified in control and AP groups. (D) Violin plots depict the changes in the expression of top upregulated genes in the MSC cluster. (E and F) Immunofluorescence double staining of Sparc (E) and Col1 (F) in Sp7-expressing osteoprogenitors. Scale bar, 50 μm. (G) Immunofluorescence double staining of Runx2 in Gli1+ periodontal stem cells (PDLSCs). Scale bar, 50 μm. (H) Lineage tracing analysis of Lepr+ PDLSCs and Col2.3+ osteoblasts. Scale bar, 50 μm.
Figure 5—figure supplement 1. Red fluorescent protein expression in Prrx1Cre;Rosa26Ai14, Sp7Cre;Rosa26Ai14, Gli1CreER;Rosa26Ai14, and LeprCre;Rosa26Ai14;Col2.3-GFP mouse models.
Figure 5—figure supplement 2. Gene Ontology (GO) enrichment analysis of the biological functions of mesenchymal stem cell (MSC) subclusters.
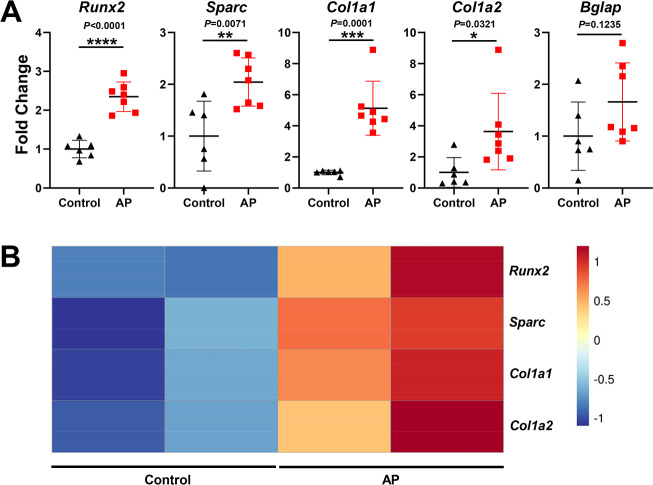
Figure 5—figure supplement 3. Real-time quantitative PCR (qPCR) and bulk RNA-seq analysis of osteogenic markers during apical periodontitis (AP) situation.
AP stimulates MSC differentiation toward osteoblast lineage cells
We next compared the transcriptome data from the MSC cluster between AP and control groups. Among the top upregulated genes, we observed a significant difference in the single cell expression levels of OLC-associated transcripts, such as secreted protein that is acidic and rich in cysteine (Sparc), Col1a1, Col1a2, Bglap, Bglap2, and Postn, accompanied by a trend toward increased Runx2 expression (Figure 5D). We verified their expressions by real-time quantitative PCR and bulk RNA sequence analysis. The results revealed that Sparc, Col1a1, Col1a2, and Runx2 were significantly upregulated, and Bglap tended to increase during AP (Figure 5—figure supplement 3A, B). In vivo experiments confirmed the upregulated expression of osteogenic-related markers, such as Sparc and Col1a1 in the AP group. There were increased numbers of Sparc+/Sp7+ or Col1+/Sp7+ cells embedded in the bone matrix, implying the protective function of MSC to differentiate into osteoprogenitors and osteoblasts under AP conditions (Figure 5E and F). Lineage tracing analysis further revealed that Gli1+ PDLSCs migrated to AP lesions and differentiated into Runx2+-osteoblasts (Figure 5G). Moreover, using LeprCre;Rosa26Ai14;Col2.3-GFP mouse model, we identified more Lepr+/Col2.3+ cells in the alveolar bone, confirming that inflammation could stimulate the progenitor cells in the adjacent bone and periodontium differentiating toward osteoblasts, thus contributing to the protective actions during AP (Figure 5H).
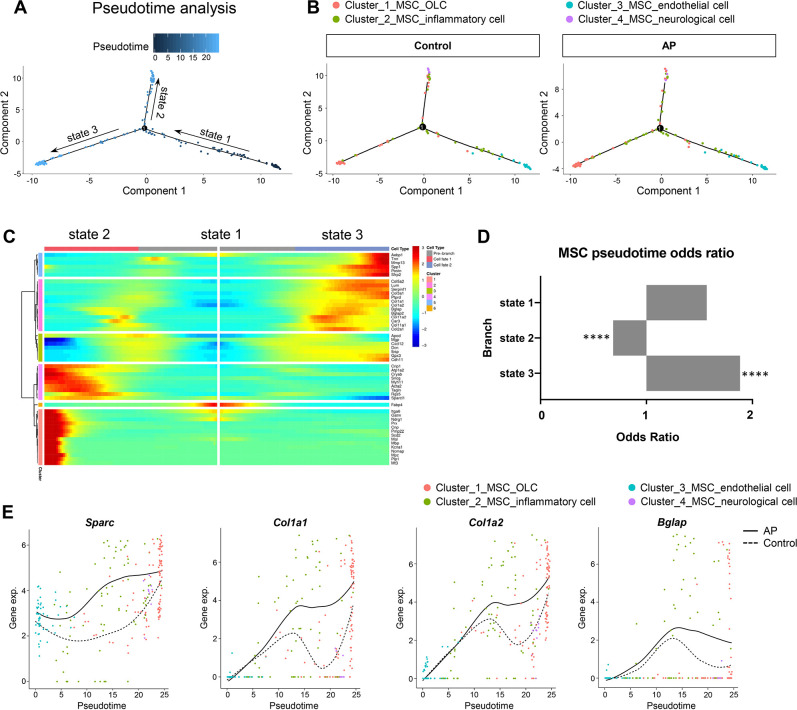
We investigated the heterogeneity of MSCs by applying branch expression analysis modeling (BEAM) and corresponding pseudotime analysis implemented in Monocle 2 (Qiu et al., 2017a). The results revealed three distinct states. Of these, state 2 and state 3 represented two diverse differentiated cell populations (terminal branch; Figure 6A). We then examined the characteristics of the MSCs with respect to their specific states. We plotted the estimated pseudotime of each cluster cell in the state space, and the results indicate that starting with MSC_endothelial cells, MSCs were able to differentiate into MSC_OLCs and MSC_neurological cells. MSC_inflammatory cells may correspond to an intermediate state (Figure 6B). Interestingly, MSC_OLCs in state 3, as osteogenic primed MSCs, exhibited the highest tendency toward osteoblastic differentiation with upregulated expressions of Col1a1, Col1a2, Bglap, Spp1, and Postn (Figure 6C). Notably, this MSC population increased significantly in the AP group (Figure 6D). A comparison of the dynamics of the gene expression pattern across pseudotime revealed upregulation of osteogenic genes such as Sparc, Col1a1, Col1a2, and Bglap during inflammation when compared to the homeostatic state (Figure 6E). Thus, the MSC subcluster exhibited gene expression levels of osteogenic markers that increased during AP. These results were in accordance with our previous single-cell analysis of gene expression in the AP group (Figure 5D–H).
Figure 6. Apical periodontitis (AP) stimulates mesenchymal stem cell (MSC) differentiation toward osteoblast lineage cells.
(A) Pseudotime analysis of the MSC population. (B) Reconstructed cell differentiation trajectory of MSC populations in control and AP groups. (C) Heatmap of differential genes of three states. (D) Bar plot of numerical changes between AP and control in respective states as identified in pseudotime analysis. Fisher’s exact test with Bonferroni correction was used. ****p<0.0001. All data were exhibited as mean ± SEM. (E) Comparison between the pseudotime gene trajectories of MSC subpopulations showed upregulation of Sparc, Col1a1, Col1a2, and Bglap. Black line indicates AP, and dotted lines indicate control.
Higher OLC-associated marker levels were identified in alveolar bone from patients with AP
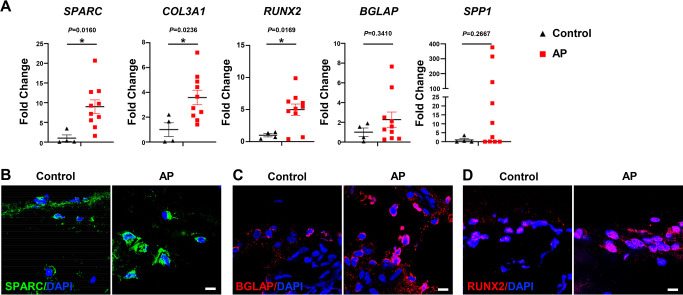
We next determined whether osteogenesis within the MSC population was altered in human patients with AP. The expressions of SPARC, COL3A1, and RUNX2 were significantly enhanced in alveolar bone from patients with AP, accompanied by increases in SPP1 and BGLAP (Figure 7A). Moreover, immunostaining detected a more extensive distribution of SPARC, BGLAP, and RUNX2 in AP alveolar bone marrow, confirming higher osteogenic potential under AP conditions (Figure 7B–D). Upregulation of OLC-associated markers in AP lesions from patients is consistent with our previous results, verifying an augmentation of osteogenic characteristics within the MSC subcluster in alveolar bone marrow under AP conditions.
Figure 7. Human alveolar bone in apical periodontitis (AP) is associated with higher osteolineage subcluster (OLC)-associated transcripts.
(A) Gene expression of SPARC, COL3A1, RUNX2, SPP1, and BGLAP in samples of healthy individuals and AP patients showed an upregulated expression in AP group. n=4 in healthy individuals and n=10 in AP patients. *p<0.05. All data are shown as the mean ± SEM. (B–C) Immunofluorescence staining of SPARC, BGLAP, and RUNX2 in bone marrow of human alveolar bone exhibited high osteogenic protein levels in AP. n=4. Scale bar, 10 μm.
In summary, the current study elucidated the characterization of MSCs and immunoresponsive cells under healthy and chronic AP conditions, including unforeseen heterogeneity in their states of activation. We believe that this analysis presents a comprehensive transcriptomic view of the alveolar bone microenvironment during AP. These results could pave the way for designing new therapeutic approaches by targeting MSCs to restore the alveolar bone lesions caused by AP and other inflammatory-related bone diseases.
Discussion
In this study, we employed scRNA-seq technology for an unbiased assessment of cell populations in alveolar bone marrow from mice with induced chronic AP. We identified 15 discrete cell clusters, each with unique transcriptional signatures and putative functions during AP progression. Immune cells accounted for the majority of the cell components in the mandibular microenvironment. Cell-to-cell communication analysis revealed that among the multiple cell types, the MSC population had the most interaction with itself and other immune cells under inflammatory conditions. Furthermore, we identified four main subclusters within the MSC population. The transcriptional profiles of each subcluster from both the homeostatic and disease states were significantly different. We used the expression profile and the pseudotime analysis to demonstrate that MSCs were stimulated by inflammation and differentiated toward osteogenic lineage cells in order to protect the alveolar bone. We confirmed the enriched genes and proteins associated with osteogenesis in AP-associated alveolar bone from both the murine models and human patients. Our results define the bone marrow microenvironment in a homeostatic state and under chronic AP, highlighting the protective action of MSCs in regeneration of alveolar bone lesions.
The oral cavity is one of the most complex microenvironments where host and microbes interacts closely (Hasturk et al., 2012). Oral inflammatory diseases such as AP is a complex disease characterized by the simultaneous presence of inflammation, destructive immunoreactions, and healing processes (Márton and Kiss, 2000; Márton and Kiss, 2014). A dense infiltration of immunocompetent cells is present in periradicular lesions. Extensive studies have examined these cells by immunostaining and gene expression techniques to determine their specific properties in terms of numbers, morphology, and function (Graunaite et al., 2012). These results provide a deeper understanding of immune cells and their inflammatory mediators. Yet the extent of their heterogeneity and the distinct markers of cell subsets have remained unexplored.
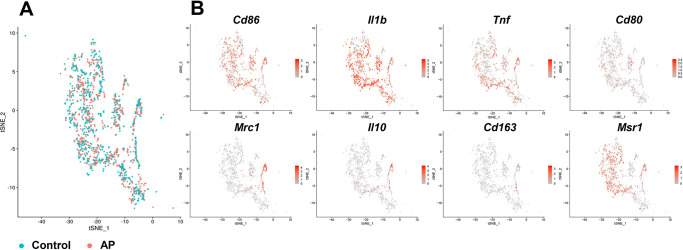
Our scRNA-seq analysis supplements knowledge of the landscape of bone marrow immunoreactions during oral inflammatory diseases. For example, the pro-inflammatory cytokines (IL-1β, IL-6, and TNF-α) have been recognized as key mediators associated with the persistence of the disease (Morsani et al., 2011). We found high expression of Il1b and Il6 in the mast cell cluster and Tnf in the monocyte population. The release of these substances is directly related to both inflammatory events and bone resorption (Bracks et al., 2014). Moreover, other inflammatory cells, including neutrophils, macrophages, and lymphocytes, present specific expression patterns of various cytokines and chemokines. In particular, a significant upregulation of Cxcl2 expression was identified in neutrophil and macrophage populations. Cxcl2 was detected in endodontic infections (de Gomes and Herrera, 2018) and is one of the most critical chemokines for neutrophil recruitment at a site of inflammation (Kobayashi, 2008; Silva et al., 2007). Increased levels of Cxcl2 have been reported to be associated with bone-resorptive activity in periapical lesions (AlShwaimi et al., 2013), highlighting the pro-inflammatory role of Cxcl2 in the progression of AP. Furthermore, we detected several new anti-inflammatory factors such as Ifitm1 and Ifitm2 in monocyte/macrophage subsets. Ifitm1 and Ifitm2 encode for a family of small transmembrane proteins localized in the plasma and endolysosomal membranes. Although the function of Ifitm has not been assessed in the context of AP, there is considerable evidence suggesting that Ifitm1 and Ifitm2 can be stimulated by interferons and exert an anti-inflammatory function in innate and adaptive immunity (Liao et al., 2019; Yánez et al., 2020). It is notable that the frequencies of macrophage, T cell, B cell, NK cell, epithelial cell, and MSCs were significantly increased in AP samples. Being one of the most significant components of the innate immune system, macrophages function to remove the pathogenic substances and dead cells by phagocytosis (Lu et al., 2021). They can be divided into two subsets, M1-like macrophages and M2-like macrophages. The M1 type is termed as the proinflammatory one, which is Cd86, Cd80, Il1b, and Tnf positive. M1-like macrophages also work as antigen presenting cells to interact with T helper 1 (Th1) cells (Song et al., 2022). Moreover, they release TNF-α, IL-1α, IL-6, and prostaglandins to increase the resorptive intensity in AP (Paula-Silva et al., 2020). By contrast, M2-like macrophages display immunoregulatory functions. They are positive for Cd163, Cd206, Msr1, and Il10. The analysis of macrophage subsets showed the M1-like macrophage accounted for the vast majority in AP lesions. The expression pattern of M2 markers were also presented in macrophage cluster (Figure 3—figure supplement 1A, B). Lymphocytes including T cells and B cells are crucial in the adaptive immune response of AP (Wang et al., 2022). Th1 cells are believed to be pro-inflammatory, while Th2 cells usually function as the immunosuppressive subcluster. Tregs are another important type of T cells related to the regenerative process of AP. They can release IL-10 and tissue growth factor (TGF)-β to ameliorate the local inflammation. Tregs also function as the suppressor of T cell response and restrict the progression of AP. Different types of B cells act conversely during AP process. It was reported that antigen-activated B cells and switched memory B cells increased RANKL expression then stimulated osteoclast generation and alveolar bone resorption (Han et al., 2018; Settem et al., 2021). In contrast, the plasma cells play a fundamental role in tissue repair by producing antibodies (Han et al., 2018; Polanco et al., 2021). NK cells exert a crucial function in host defense by rapidly producing cytokines and upregulating the expression of cell death-inducing molecules to eliminate infected cells (Kratzmeier et al., 2023). CD57+ NK cells were presented in chronic AP lesions, and upregulated NK cells were observed in periapical granulomas compared with radicular cysts (Silva et al., 2017). Proliferating epithelium is frequently present in inflamed apical lesions, which is usually developed from epithelial rests of Malassez (Langeland et al., 1977; Lin et al., 2007). It is an important protective bioactivity since these epithelial tissues aim to build a barrier to prevent the diffusion of microorganisms. Increased number of epithelial cells is highly connected with the progression toward periapical cyst, the connective tissue capsule of which is lined by a non-keratinized stratified squamous epithelium (Schulz et al., 2009). Our scRNA-seq analysis has revealed novel transcriptional signatures of specific cell populations which were not visible with bulk analysis. Further studies will be required to investigate the contribution of these factors to the immune response and inflammation.
Cell-cell communications mediated by ligand-receptor complexes are crucial to control diverse biological processes, including development, differentiation, and inflammation (Efremova et al., 2020). We employed CellPhoneDB to demonstrate previously unrecognized intercellular communication among all cell subsets, including close crosstalk among MSCs, macrophages, and dendritic cells. More importantly, CrossTalkeR enabled us to identify cell communication differences between two biological states: disease versus homeostasis (Nagai et al., 2021). Our results suggest that most ligand-receptor interactions were markedly reduced during AP. Alterations in cell-cell communication have also been described for multiple tissues and organs under other disease, injury, and infection conditions (Armingol et al., 2021). The shifts in the pattern of CCIs in the alveolar bone microenvironment may imply reorganization of the network in response to inflammation. It is worth noting that MSCs possessed the highest number of interactions with themselves and other cell types under both homeostasis and AP conditions, indicating MSCs act as a hub in CCIs. Thus, alterations of MSCs may substantially affect the biological behaviors of other cell types in both healthy and diseased states, further underscoring the importance of MSCs in alveolar bone marrow during inflammation.
In most cases of chronic AP, periapical bone destruction is self-limiting in which a new equilibrium has been established between root canal pathogens and anti-infective defense mechanisms (Wang et al., 2011). The mechanisms that underly the protective actions of immune cells in apical inflammatory tissues have been extensively studied (Márton and Kiss, 2000; Márton and Kiss, 2014; Nair, 2004; Stashenko, 1990). Yet, the kinetics for the healing of these lesions by regenerating alveolar bone is less well understood.
MSCs are recognized as a promising resource for periapical bone healing in the treatment of oral inflammatory diseases. Several lines of evidence support this tenet. The evoked-bleeding step in revascularization procedures for regenerative endodontic treatment leads to a substantial influx of MSCs, confirming the existence and regenerative properties of MSCs in the periapical region. Indeed, several dental MSCs are present in the periapical tissues, including MSCs derived from alveolar bone, periodontal ligament, and apical papilla (Zhao and Chai, 2015). More importantly, MSCs were found to be present in inflamed periapical tissues in adult patients with AP. Evidence showed that human apical papilla and its resident cells from teeth with pulp necrosis and AP could survive and retain their stemness in an inflammatory environment (Chrepa et al., 2017). MSCs found in the periapical region were positive for typical stem cell markers, such as STRO-1, CD90, CD73, CD105, CD146, and SOX2 (Chrepa et al., 2015; Estrela et al., 2017; Liao et al., 2011). These progenitors were able to form CFU-Fs, illustrating a typical MSC feature. However, our understanding of the characteristics of MSCs in inflamed periapical tissues remains at an early stage. In this study, we explored the heterogeneity and expression signature of MSCs in the presence of inflammation. Several key observations were only possible by study at the single-cell level. First, we identified four main subclusters in the MSC population: MSC_OLCs, MSC_endothelial cells, MSC_neurological cells, and MSC_inflammatory cells. The first three clusters were consistent with previously described MSC populations in alveolar bone marrow (Lin et al., 2021). These MSC subclusters have distinct differentiation potentials and may be associated with the pathogenesis of AP. Importantly, we noted that the proportion of MSC_inflammatory cells in AP was higher and accompanied by significantly increased expression of S100a8/a9. These factors are involved with Ca2+ binding which is significantly increased during inflammatory processes (Wang et al., 2018). Recent studies have found that although MSCs do not express S100a8/a9 under healthy conditions, they do upregulate its expression in disease states (Leimkühler et al., 2021). The higher S100a8/a9 expression in the MSC_inflammatory cell subcluster suggests that S100a8/a9 might be a candidate for detecting an inflammatory-associated MSC state in AP. Activated by inflammatory bone destruction, these MSCs with increased osteogenic potentials may rescue the bone resorption process, which reach the equilibrium between bone formation and resorption then drive the progression of AP into stable states (Márton and Kiss, 2014). Since the pathological stimuli exists constantly, the protective actions can alleviate the bone loss to some extent. In clinical practice, root canal therapy (RCT) aims to disinfect and remove the pathogenic factors, which make the protective activities overweigh the destructive ones (Lin et al., 2009). The bone lesions of AP patients receiving RCT usually fully recovered with resolution of radiolucency after the inflammation is controlled in apical area (Soares et al., 2006). The healing of AP lesion is highly correlated with the osteogenic potential of inflamed MSCs (Lin et al., 2009). The differentiative traits of these cells and their role in inflamed periapical tissue remain to be explored. Deeper understanding of the features of MSCs in inflammatory microenvironment and the regulatory network underlying this situation will shed light on the MSC-based regenerative medicine for AP and other inflammatory bone diseases.
An important finding in the present study is the increased osteogenic potential in the MSC population during inflammatory-associated bone lesions. Several lines of evidence support this conclusion. The overall size of the MSC population was significantly increased and was accompanied by upregulated expression of key osteogenic-related genes. Moreover, pseudotime analysis detected a specific state progression toward osteogenesis in the MSC cluster. The numerical changes in this state were statistically higher in AP samples when compared to control. MSCs in this state harbored the highest osteoblastic differentiation potential, along with upregulated expressions of Sparc, Col1a1, Col1a2, Runx2, and Bglap. More importantly, we confirmed this observation in animal models and human patients with AP. By using a lineage tracing strategy, we identified increased percentages of Prrx1+, Gli1+, Lepr+ MSC, and Sp7+ progenitor cells with higher expression of Sparc, Col1a1, and Runx2 in the alveolar bone region surrounding periapical lesions. We show, for the first time, upregulated SPARC, RUNX2, COL3A1, and BGLAP in alveolar bone from human patients with AP. It is notable that Sparc was the most upregulated marker in the MSC population from AP samples. Sparc is one of the most abundant non-collagenous proteins expressed in bone. It has a critical role in osteoblast differentiation, procollagen processing, and assembly as well as mineralization (Rosset and Bradshaw, 2016). SPARC mutations have been identified in patients with osteogenesis imperfecta and idiopathic osteoporosis (Delany et al., 2008; Mendoza-Londono et al., 2015). Sparc null mice exhibit decreased osteoblast number and activity along with reduced bone formation rate (Delany et al., 2003). Recent research using scRNA-seq revealed high levels of SPARC expression in human periodontal MSCs, highlighting the potential role of SPARC in oral MSC populations (Pagella et al., 2021). Notably, Sparc is also a molecule closely related to inflammation. Overexpressed Sparc is observed in inflammatory pulp cells, experimental glomerulonephritis, and ovarian cancer associated inflammation (Dimitrova-Nakov et al., 2014; Pichler et al., 1996; Said et al., 2008). Both pro-inflammatory and anti-inflammatory properties of Sparc have been reported (Said et al., 2008; Sangaletti et al., 2011; Tanaka et al., 2018; Toba et al., 2015). Sparc may act in two ways by activating different pathways depending on context. It therefore creates a microenvironment suitable for tissue remodeling and repair at different stages (Ghanemi et al., 2020). Given the upregulation of Sparc observed in AP mouse models and human patients, we believe that the MSC population was stimulated by inflammation and underwent osteogenesis with a significant function in bone matrix assembly and mineralization, thus serving as an important protective component in healing the bone lesions in inflammation.
In summary, we profiled the transcriptome of alveolar bone marrow single cells from control and AP samples, providing novel insights into the inflammatory biomarkers associated with the pathogenesis of oral inflammatory diseases. The interplay among various cell populations highlights the role of MSC subsets and the therapeutic potential of MSCs in healing bone lesions. These results demonstrate marked heterogeneity in the alveolar bone marrow microenvironment and provide a basis for considering MSC-based treatment for inflammatory-related bone diseases.
Materials and methods
Animal experiments
Gt(ROSA)26Sortm14(CAG-tdTomato)Hze mice (Rosa26Ai14; Cat# JAX:007914), Sp7Cre (Cat# JAX:006361), Prrx1Cre (Cat# JAX:005584), and Gli1CreER (Cat# JAX:007913) mice were purchased from Jackson Laboratory. Col2.3-GFP and LeprCre mice were kindly provided by Dr. Bo O. Zhou (Institute of Biochemistry and Cell Biology). Hybrid mice were generated to mark Sp7-, Prrx1-, Lepr-, and Gli1-expressing cells by crossing Sp7Cre, Prrx1Cre, LeprCre, and Gli1CreER with Rosa26Ai14 respectively. LeprCre;Rosa26Ai14;Col2.3-GFP mice were generated by crossing LeprCre;Rosa26Ai14 mice with Col2.3-GFP mice. Gli1CreER;Rosa26Ai14 mice (at postnatal day 50) were injected intraperitoneally with tamoxifen (Sigma-Aldrich) at dosage of 2.5 mg/10 g body weight every 2 d for three times. Wild-type mice were purchased from Chengdu Dossy Biological Technology Co., Ltd. All animal experiments were carried out in accordance with the guidelines of the Institutional Animal Care and Use Committee at the State Key Laboratory of Oral Diseases, Sichuan University (WCHSIRB-D-2021–339).
AP mouse model
The AP model was generated using 8-wk-old mice as described previously (Taira et al., 2019). In brief, the pulp chambers of the mandibular first molars were opened by a high-speed handpiece with #1/4 carbide round bur under direct visualization by a stereoscopic microscope (Leica). A #10 endodontic K file was then used to probe the root canals. The teeth were then exposed to the oral environment for 3 wk.
Single-cell sample preparation and sequencing
20 C57/B6 male mice with AP or control mice were used to acquire single-cell suspensions. Mandibles were dissected under a stereo microscope (Leica). Specifically, soft tissues, molars, incisors, and bone from behind the condyle were removed. Then, mandibular alveolar bone tissue was cut into small pieces (<1 mm3) and digested with 3 mg/mL collagenase type I (Gibco) and 4 mg/mL dispase II (Sigma) for 60 min at 37°C as previously described (Cui et al., 2020; Yamaza et al., 2011). Red blood cells were lysed with 1 mL Red Cell Lysis Solution (Biosharp). Cells were centrifuged and resuspended in 1% BSA solution. The final concentration of cells was 1 × 106 cells/mL. Cellular suspensions were loaded into Chromium microfluidic chips and barcoded with a 10× Chromium Controller (10× Genomics). RNA from the barcoded cells was subsequently reverse-transcribed and sequencing libraries constructed with reagents from a Chromium Single Cell 3’ v3 reagent kit (10× Genomics) and sequenced with the NovaSeq system (Illumina).
Preprocessing of scRNA-seq data
We used Cellranger (v3.1, 10× Genomics) to compare reads to the mouse genome mm10 and for detection of cells using default parameters. Next, we used the Seurat package (v3.1) for further analysis of the scRNA-seq. A gene with less than 3 cells, mitochondrial genes higher than 20%, cells with less than 200 expressed genes, and cells with erythrocyte gene expression higher than 5% were filtered out. The Seurat package was used for data normalization and dimensional reduction. Principal component analysis was based on the highly variable genes, and subclusters of cells were identified using specific gene expression in each group.
Pseudotime analysis
The cluster identified as MSC was loaded into R environment and then the R package Monocle 2 was used to infer the trajectory and reduce the dimensional space to discover the state transitions of MSCs (Qiu et al., 2017b). Each cell’s pseudotime was automatically calculated, and the branch was assigned on the principal graph. BEAM was run to analyze the hub genes in branches, and top 50 genes were visualized with the plot_genes_branched_heatmap function.
CCI analysis
CellPhoneDB v.2.0 was performed to predict enriched cellular interaction between different cell populations according to the expression of a receptor by one state and a ligand by another one (Efremova et al., 2020). Receptors and ligands were analyzed when they were expressed in at least 10% cells of the corresponding cluster in mouse data. Subsequently, CrossTalkeR was used to visualize the ligand-receptor networks, which identified relevant ligands, receptors, and cell types contributing to changes in cellular crosstalk when contrasting AP and homeostasis states (Nagai et al., 2021). We then ranked the individual ligand by their number of interactions and normalized to the number of all possible CCIs to identify the most influential ligand. Interactions associated with Fn1 and Sele were visualized via Sankey plot.
Bulk RNA-seq analysis
Healthy and AP mandibles were used to extract total RNA and analyzed with bulk RNA-seq. PowerLyzer 24 Homogenizer (Qiagen) was used for lysis and homogenization of alveolar bone samples, and Trizol (Invitrogen) was used for mRNA extraction according to the manufacturer’s protocol. NanoDrop ND-1000 (Thermo fisher Scientific) was used to quantify RNA concentration. Sequencing libraries were generated using the NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, USA), and index codes were added to correlate sequences to each sample. The library preparations were sequenced on an Illumina Novaseq 6000 (LC-Bio Technology Co., Ltd.).
Quantitative real-time PCR
PrimeScript RT reagent kit (Takara) was used to reverse transcribed RNA, and qRT-PCR was performed by SybrGreen Supermix (Bio-Rad Laboratories). Relative expression was calculated using a 2ΔΔCT method with Actb for normalization. All primers are listed in the Supplementary file 1.
Collection of human alveolar bone
This study was permitted by the Ethical Committees of the West China Hospital of Stomatology, Sichuan University (WCHSIRB-D-2021–292). Written informed consent was obtained from all patients. Patients who were characterized with periapical rarefaction in radiography, suffered with a failed root canal treatment, and needed endodontic surgery were recruited as subjects. Patients with periodontitis, vertical root fracture, suspected odontogenic tumor, systematic diseases, or a history of antibiotic, antiviral, or immunosuppressive therapy for 3 mo were excluded. 10 human alveolar bone samples were collected from discards during the apicoectomy surgery. Normal apical bone tissue was harvested from patients who required a bone repair procedure. There was no significant difference in age or gender between the control group and the patients with AP. The samples were rinsed by sterile PBS. Subsequently, part of the samples was quick-frozen with liquid nitrogen and stored at –80°C for mRNA extraction. Others were fixed by 4% paraformaldehyde (PFA) and decalcified by 20% EDTA for immunostaining.
Immunostaining
Immunofluorescent staining was performed to assess the expression of proteins in the region of the AP lesion. Mandibles from Sp7Cre;Rosa26Ai14, Prrx1Cre;Rosa26Ai14, LeprCre;Rosa26Ai14;Col2.3-GFP, and Gli1CreER;Rosa26Ai14 mice were dissected and fixed in 4% paraformaldehyde for 3 d. Subsequently, samples were decalcified in 20% EDTA (pH 7.5) and embedded in Tissue-Tek O.C.T Compound (Sakura) and cut into 8 μm sections using CryoStar NX50 (Thermo fisher Scientific). Slides were permeabilized with 0.3% Triton X-100 (Beyotime) for 15 min, blocked with 5% BSA for 1 hr, incubated with anti-Sparc (1:100, R&D, AF942) or anti-Collagen I (1:200, Abcam, ab21286) or anti-Bglap (1:200, Abcam, ab93876) or anti-Runx2 (1:200, Abcam, ab23981) overnight at 4°C, and then incubated with Alexa fluor 488 (1:1000, Invitrogen, A11070) for 1 hr at room temperature. 4',6-diamidino-2-phenylindole dihydrochloride (DAPI) (Vector) was used as a nuclei counterstain. An Olympus confocal microscope FV3000 (Olympus) was used to capture images.
Statistical analysis
GraphPad Prism 9.0 (GraphPad Software Inc) was used for statistical analysis. Unpaired two-tailed Student’s t test was used in two-group comparisons. All data were exhibited as mean ± SEM. p-Values<0.05 were considered statistically significant for all analyses.
Acknowledgements
This work was supported by NSFC grants 81800928, 81901040, 82222015 and 82171001, the Young Elite Scientist Sponsorship Program by CAST (No. 2020 QNRC001 and 2018QNR001), the Sichuan Science and Technology Program (No. 2019YJ0054), Research Funding from West China School/Hospital of Stomatology Sichuan University (No. RCDWJS2021-1), State Key Laboratory of Oral Diseases Open Funding Grant SKLOD-R013.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Quan Yuan, Email: yuanquan@scu.edu.cn.
Chenchen Zhou, Email: chenchenzhou5510@scu.edu.cn.
Di Chen, Chinese Academy of Sciences, China.
Carlos Isales, Augusta University, United States.
Funding Information
This paper was supported by the following grants:
National Natural Science Foundation of China 81800928 to Yi Fan.
National Natural Science Foundation of China 81901040 to Chenchen Zhou.
National Natural Science Foundation of China 82171001 to Chenchen Zhou.
Young Elite Scientist Sponsorship Program by CAST 2020QNRC001 to Chenchen Zhou.
Young Elite Scientist Sponsorship Program by CAST 2018QNR001 to Yi Fan.
Sichuan Science and Technology Program 2019YJ0054 to Yi Fan.
Research Funding from West China School/Hospital of Stomatology Sichuan University RCDWJS2021-1 to Yi Fan.
State Key Laboratory of Oral Diseases Open Funding Grant Sichuan University SKLOD-R013 to Yi Fan.
National Natural Science Foundation of China 82222015 to Chenchen Zhou.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Data curation, Formal analysis, Supervision, Funding acquisition, Validation, Investigation, Methodology, Writing - original draft, Writing - review and editing.
Formal analysis, Validation, Investigation, Methodology, Writing - original draft.
Data curation, Formal analysis, Validation, Investigation, Methodology.
Data curation, Investigation, Methodology, Writing - original draft.
Resources, Validation.
Supervision, Validation, Writing - original draft, Writing - review and editing.
Conceptualization, Resources, Data curation, Supervision, Funding acquisition, Writing - original draft, Project administration, Writing - review and editing.
Conceptualization, Data curation, Formal analysis, Supervision, Funding acquisition, Validation, Investigation, Methodology, Writing - original draft, Project administration, Writing - review and editing.
Ethics
Human subjects: This study was permitted by the Ethical Committees of the West China Hospital of Stomatology, Sichuan University (Permit Number: WCHSIRB-D-2021-292). Written informed consent was obtained from all patients.
All animal experiments were carried out in accordance with the guidelines of the Institutional Animal Care and Use Committee at the State Key Laboratory of Oral Diseases, Sichuan University (Permit Number: WCHSIRB-D-2021-339).
Additional files
Data availability
All data generated or analysed during this study are included in the manuscript and supporting file; Source Data files have been provided for Figures 1, 2, 3, 4, 5, 6, 7, Figure1-figure supplement 1 and Figure5-figure supplement 3. Sequence data are deposited in the NCBI Gene Expression Omnibus under accession numbers GSE212975 and GSE221990.
The following datasets were generated:
Fan Y, Lyu P, Yuan Q, Zhou C. 2022. Single-cell gene expression of control mandibular bone marrow cells and mandibular bone marrow cells under apical periodontitis. NCBI Gene Expression Omnibus. GSE212975
Fan Y, Lyu P, Yuan Q, Zhou C. 2023. RNA-seq analysis of RNA extracted from control alveolar bone samples and alveolar bone under apical periodontitis. NCBI Gene Expression Omnibus. GSE221990
References
- AlShwaimi E, Berggreen E, Furusho H, Rossall JC, Dobeck J, Yoganathan S, Stashenko P, Sasaki H. Il-17 receptor a signaling is protective in infection-stimulated periapical bone destruction. Journal of Immunology. 2013;191:1785–1791. doi: 10.4049/jimmunol.1202194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo-Pires AC, Biguetti CC, Repeke CE, de Rodini CO, Campanelli AP, Trombone APF, Letra A, Silva RM, Garlet GP. Mesenchymal stem cells as active prohealing and immunosuppressive agents in periapical environment: evidence from human and experimental periapical lesions. Journal of Endodontics. 2014;40:1560–1565. doi: 10.1016/j.joen.2014.02.012. [DOI] [PubMed] [Google Scholar]
- Armingol E, Officer A, Harismendy O, Lewis NE. Deciphering cell-cell interactions and communication from gene expression. Nature Reviews. Genetics. 2021;22:71–88. doi: 10.1038/s41576-020-00292-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracks IV, Armada L, Gonçalves LS, Pires FR. Distribution of mast cells and macrophages and expression of interleukin-6 in periapical cysts. Journal of Endodontics. 2014;40:63–68. doi: 10.1016/j.joen.2013.09.037. [DOI] [PubMed] [Google Scholar]
- Braz-Silva PH, Bergamini ML, Mardegan AP, De Rosa CS, Hasseus B, Jonasson P. Inflammatory profile of chronic apical periodontitis: a literature review. Acta Odontologica Scandinavica. 2019;77:173–180. doi: 10.1080/00016357.2018.1521005. [DOI] [PubMed] [Google Scholar]
- Carminho-Rodrigues MT, Klee P, Laurent S, Guipponi M, Abramowicz M, Cao-van H, Guinand N, Paoloni-Giacobino A. LARS2-perrault syndrome: a new case report and literature review. BMC Medical Genetics. 2020;21:109. doi: 10.1186/s12881-020-01028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrepa V, Henry MA, Daniel BJ, Diogenes A. Delivery of apical mesenchymal stem cells into root canals of mature teeth. Journal of Dental Research. 2015;94:1653–1659. doi: 10.1177/0022034515596527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrepa V, Pitcher B, Henry MA, Diogenes A. Survival of the apical papilla and its resident stem cells in a case of advanced pulpal necrosis and apical periodontitis. Journal of Endodontics. 2017;43:561–567. doi: 10.1016/j.joen.2016.09.024. [DOI] [PubMed] [Google Scholar]
- Clatworthy MR, Smith KGC. Fcgammariib balances efficient pathogen clearance and the cytokine-mediated consequences of sepsis. The Journal of Experimental Medicine. 2004;199:717–723. doi: 10.1084/jem.20032197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotti E, Schirru E, Acquas E, Usai P. An overview on biologic medications and their possible role in apical periodontitis. Journal of Endodontics. 2014;40:1902–1911. doi: 10.1016/j.joen.2014.08.013. [DOI] [PubMed] [Google Scholar]
- Cui C, Bi R, Liu W, Guan S, Li P, Song D, Xu R, Zheng L, Yuan Q, Zhou X, Fan Y. Role of PTH1R signaling in prx1+ mesenchymal progenitors during eruption. Journal of Dental Research. 2020;99:1296–1305. doi: 10.1177/0022034520934732. [DOI] [PubMed] [Google Scholar]
- de Gomes B, Herrera DR. Etiologic role of root canal infection in apical periodontitis and its relationship with clinical symptomatology. Brazilian Oral Research. 2018;32:e69. doi: 10.1590/1807-3107bor-2018.vol32.0069. [DOI] [PubMed] [Google Scholar]
- Delany AM, Kalajzic I, Bradshaw AD, Sage EH, Canalis E. Osteonectin-null mutation compromises osteoblast formation, maturation, and survival. Endocrinology. 2003;144:2588–2596. doi: 10.1210/en.2002-221044. [DOI] [PubMed] [Google Scholar]
- Delany AM, McMahon DJ, Powell JS, Greenberg DA, Kurland ES. Osteonectin/SPARC polymorphisms in caucasian men with idiopathic osteoporosis. Osteoporosis International. 2008;19:969–978. doi: 10.1007/s00198-007-0523-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Prete A, Martínez-Muñoz L, Mazzon C, Toffali L, Sozio F, Za L, Bosisio D, Gazzurelli L, Salvi V, Tiberio L, Liberati C, Scanziani E, Vecchi A, Laudanna C, Mellado M, Mantovani A, Sozzani S. The atypical receptor CCRL2 is required for CXCR2-dependent neutrophil recruitment and tissue damage. Blood. 2017;130:1223–1234. doi: 10.1182/blood-2017-04-777680. [DOI] [PubMed] [Google Scholar]
- Dimitrova-Nakov S, Baudry A, Harichane Y, Kellermann O, Goldberg M, Dr ès Sciences Naturelles Pulp stem cells: implication in reparative dentin formation. Journal of Endodontics. 2014;40:S13–S18. doi: 10.1016/j.joen.2014.01.011. [DOI] [PubMed] [Google Scholar]
- Disease GBD, Injury I, Prevalence C. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the global burden of disease study 2017. Lancet. 2018;392:1789–1858. doi: 10.1016/S0140-6736(18)32279-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nature Protocols. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- Espéli M, Smith KGC, Clatworthy MR. FcγRIIB and autoimmunity. Immunological Reviews. 2016;269:194–211. doi: 10.1111/imr.12368. [DOI] [PubMed] [Google Scholar]
- Estrela C, Freitas Silva BS, Silva JA, Yamamoto-Silva FP, Pinto-Júnior DDS, Gomez RS. Stem cell marker expression in persistent apical periodontitis. Journal of Endodontics. 2017;43:63–68. doi: 10.1016/j.joen.2016.09.002. [DOI] [PubMed] [Google Scholar]
- Estrela C, Carmo Souza PO, Barbosa MG, Aburad de Carvalhosa A, Batista AC, Pinto Júnior DDS, Yamamoto-Silva FP, de Freitas Silva BS. Mesenchymal stem cell marker expression in periapical abscess. Journal of Endodontics. 2019;45:716–723. doi: 10.1016/j.joen.2019.03.009. [DOI] [PubMed] [Google Scholar]
- Fan Y, Cui C, Rosen CJ, Sato T, Xu R, Li P, Wei X, Bi R, Yuan Q, Zhou C. Klotho in osx+-mesenchymal progenitors exerts pro-osteogenic and anti-inflammatory effects during mandibular alveolar bone formation and repair. Signal Transduction and Targeted Therapy. 2022;7:155. doi: 10.1038/s41392-022-00957-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenette PS, Subbarao S, Mazo IB, von Andrian UH, Wagner DD. Endothelial selectins and vascular cell adhesion molecule-1 promote hematopoietic progenitor homing to bone marrow. PNAS. 1998;95:14423–14428. doi: 10.1073/pnas.95.24.14423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, Mazloom AR, Ma’ayan A, Chua W-J, Hansen TH, Turley SJ, Merad M, Randolph GJ, Immunological Genome Consortium Gene-Expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nature Immunology. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazivoda D, Dzopalic T, Bozic B, Tatomirovic Z, Brkic Z, Colic M. Production of proinflammatory and immunoregulatory cytokines by inflammatory cells from periapical lesions in culture. Journal of Oral Pathology & Medicine. 2009;38:605–611. doi: 10.1111/j.1600-0714.2009.00788.x. [DOI] [PubMed] [Google Scholar]
- Ghanemi A, Yoshioka M, St-Amand J. Secreted protein acidic and rich in cysteine and inflammation: another homeostatic property? Cytokine. 2020;133:155179. doi: 10.1016/j.cyto.2020.155179. [DOI] [PubMed] [Google Scholar]
- Graunaite I, Lodiene G, Maciulskiene V. Pathogenesis of apical periodontitis: a literature review. Journal of Oral & Maxillofacial Research. 2012;2:e1. doi: 10.5037/jomr.2011.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Jin Y, Miao Y, Shi T, Lin X. Switched memory B cells promote alveolar bone damage during periodontitis: an adoptive transfer experiment. International Immunopharmacology. 2018;62:147–154. doi: 10.1016/j.intimp.2018.07.003. [DOI] [PubMed] [Google Scholar]
- Hasturk H, Kantarci A, Van Dyke TE. Oral inflammatory diseases and systemic inflammation: role of the macrophage. Frontiers in Immunology. 2012;3:118. doi: 10.3389/fimmu.2012.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B, Lee JH, Bang D. Single-Cell RNA sequencing technologies and bioinformatics pipelines. Experimental & Molecular Medicine. 2018;50:1–14. doi: 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiani P, Boraschi D. From monocytes to M1/M2 macrophages: phenotypical vs. Functional Differentiation. Front Immunol. 2014;5:514. doi: 10.3389/fimmu.2014.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofre-Monseny L, Loboda A, Wagner AE, Huebbe P, Boesch-Saadatmandi C, Jozkowicz A, Minihane A-M, Dulak J, Rimbach G. Effects of APOE genotype on macrophage inflammation and heme oxygenase-1 expression. Biochemical and Biophysical Research Communications. 2007;357:319–324. doi: 10.1016/j.bbrc.2007.03.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y. The role of chemokines in neutrophil biology. Frontiers in Bioscience. 2008;13:2400–2407. doi: 10.2741/2853. [DOI] [PubMed] [Google Scholar]
- Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nature Reviews. Immunology. 2013;13:159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- Kratzmeier C, Singh S, Asiedu EB, Webb TJ. Current developments in the preclinical and clinical use of natural killer T cells. BioDrugs. 2023;37:57–71. doi: 10.1007/s40259-022-00572-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeland K, Block RM, Grossman LI. A histopathologic and histobacteriologic study of 35 periapical endodontic surgical specimens. Journal of Endodontics. 1977;3:8–23. doi: 10.1016/s0099-2399(77)80217-3. [DOI] [PubMed] [Google Scholar]
- Leimkühler NB, Gleitz HFE, Ronghui L, Snoeren IAM, Fuchs SNR, Nagai JS, Banjanin B, Lam KH, Vogl T, Kuppe C, Stalmann USA, Büsche G, Kreipe H, Gütgemann I, Krebs P, Banz Y, Boor P, Tai EWY, Brümmendorf TH, Koschmieder S, Crysandt M, Bindels E, Kramann R, Costa IG, Schneider RK. Heterogeneous bone-marrow stromal progenitors drive myelofibrosis via a druggable alarmin axis. Cell Stem Cell. 2021;28:637–652. doi: 10.1016/j.stem.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Jiang C-M, An S, Cheng Q, Huang Y-F, Wang Y-T, Gou Y-C, Xiao L, Yu W-J, Wang J. Immunomodulatory properties of dental tissue-derived mesenchymal stem cells. Oral Diseases. 2014;20:25–34. doi: 10.1111/odi.12086. [DOI] [PubMed] [Google Scholar]
- Liao J, Al Shahrani M, Al-Habib M, Tanaka T, Huang GT-J. Cells isolated from inflamed periapical tissue express mesenchymal stem cell markers and are highly osteogenic. Journal of Endodontics. 2011;37:1217–1224. doi: 10.1016/j.joen.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Goraya MU, Yuan X, Zhang B, Chiu SH, Chen JL. Functional involvement of interferon-inducible transmembrane proteins in antiviral immunity. Frontiers in Microbiology. 2019;10:1097. doi: 10.3389/fmicb.2019.01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LM, Huang GT-J, Rosenberg PA. Proliferation of epithelial cell rests, formation of apical cysts, and regression of apical cysts after periapical wound healing. Journal of Endodontics. 2007;33:908–916. doi: 10.1016/j.joen.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Lin LM, Ricucci D, Lin J, Rosenberg PA. Nonsurgical root canal therapy of large cyst-like inflammatory periapical lesions and inflammatory apical cysts. Journal of Endodontics. 2009;35:607–615. doi: 10.1016/j.joen.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Lin W, Li Q, Zhang D, Zhang X, Qi X, Wang Q, Chen Y, Liu C, Li H, Zhang S, Wang Y, Shao B, Zhang L, Yuan Q. Mapping the immune microenvironment for mandibular alveolar bone homeostasis at single-cell resolution. Bone Research. 2021;9:17. doi: 10.1038/s41413-021-00141-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Xu Y, Liu Q, Zhang Q. Mesenchymal stem cell-macrophage crosstalk and maintenance of inflammatory microenvironment homeostasis. Frontiers in Cell and Developmental Biology. 2021;9:681171. doi: 10.3389/fcell.2021.681171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu P, Song Y, Bi R, Li Z, Wei Y, Huang Q, Cui C, Song D, Zhou X, Fan Y. Protective actions in apical periodontitis: the regenerative bioactivities led by mesenchymal stem cells. Biomolecules. 2022;12:1737. doi: 10.3390/biom12121737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nature Reviews. Immunology. 2011;11:519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- Márton IJ, Kiss C. Protective and destructive immune reactions in apical periodontitis. Oral Microbiology and Immunology. 2000;15:139–150. doi: 10.1034/j.1399-302x.2000.150301.x. [DOI] [PubMed] [Google Scholar]
- Márton IJ, Kiss C. Overlapping protective and destructive regulatory pathways in apical periodontitis. Journal of Endodontics. 2014;40:155–163. doi: 10.1016/j.joen.2013.10.036. [DOI] [PubMed] [Google Scholar]
- Men Y, Wang Y, Yi Y, Jing D, Luo W, Shen B, Stenberg W, Chai Y, Ge W-P, Feng JQ, Zhao H. Gli1+ periodontium stem cells are regulated by osteocytes and occlusal force. Developmental Cell. 2020;54:639–654. doi: 10.1016/j.devcel.2020.06.006. [DOI] [PubMed] [Google Scholar]
- Mendoza-Londono R, Fahiminiya S, Majewski J, Tétreault M, Nadaf J, Kannu P, Sochett E, Howard A, Stimec J, Dupuis L, Roschger P, Klaushofer K, Palomo T, Ouellet J, Al-Jallad H, Mort JS, Moffatt P, Boudko S, Bächinger H-P, Rauch F, Care4Rare Canada Consortium Recessive osteogenesis imperfecta caused by missense mutations in SPARC. American Journal of Human Genetics. 2015;96:979–985. doi: 10.1016/j.ajhg.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsani JM, Aminoshariae A, Han YW, Montagnese TA, Mickel A. Genetic predisposition to persistent apical periodontitis. Journal of Endodontics. 2011;37:455–459. doi: 10.1016/j.joen.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai JS, Leimkühler NB, Schaub MT, Schneider RK, Costa IG. CrossTalkeR: analysis and visualization of ligand-receptorne tworks. Bioinformatics. 2021;37:4263–4265. doi: 10.1093/bioinformatics/btab370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair PNR. Pathogenesis of apical periodontitis and the causes of endodontic failures. Critical Reviews in Oral Biology and Medicine. 2004;15:348–381. doi: 10.1177/154411130401500604. [DOI] [PubMed] [Google Scholar]
- Ng YL, Mann V, Rahbaran S, Lewsey J, Gulabivala K. Outcome of primary root canal treatment: systematic review of the literature -- part 2. influence of clinical factors. International Endodontic Journal. 2008;41:6–31. doi: 10.1111/j.1365-2591.2007.01323.x. [DOI] [PubMed] [Google Scholar]
- Pagella P, de Vargas Roditi L, Stadlinger B, Moor AE, Mitsiadis TA. A single-cell atlas of human teeth. IScience. 2021;24:102405. doi: 10.1016/j.isci.2021.102405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paula-Silva FWG, Ribeiro-Santos FR, Petean IBF, Manfrin Arnez MF, de Almeida-Junior LA, de Carvalho FK, da Silva LAB, Faccioli LH. Root canal contamination or exposure to lipopolysaccharide differentially modulate prostaglandin E 2 and leukotriene B 4 signaling in apical periodontitis. Journal of Applied Oral Science. 2020;28:e20190699. doi: 10.1590/1678-7757-2019-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler RH, Bassuk JA, Hugo C, Reed MJ, Eng E, Gordon KL, Pippin J, Alpers CE, Couser WG, Sage EH, Johnson RJ. Sparc is expressed by mesangial cells in experimental mesangial proliferative nephritis and inhibits platelet-derived-growth-factor-medicated mesangial cell proliferation in vitro. The American Journal of Pathology. 1996;148:1153–1167. [PMC free article] [PubMed] [Google Scholar]
- Polanco XBJ, Bertasso AS, Silveira HA, Yamamoto de Almeida L, Almeida LKY, da Silva RAB, da Silva LAB, de Rossi A, Nelson-Filho P, León JE. IgG4-positive plasma cells are more often detected in chronic periapical lesions arising from permanent rather than primary teeth. International Endodontic Journal. 2021;54:682–692. doi: 10.1111/iej.13464. [DOI] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. Single-cell mrna quantification and differential analysis with census. Nature Methods. 2017a;14:309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nature Methods. 2017b;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosset EM, Bradshaw AD. Sparc/Osteonectin in mineralized tissue. Matrix Biology. 2016;52–54:78–87. doi: 10.1016/j.matbio.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said NA, Elmarakby AA, Imig JD, Fulton DJ, Motamed K. Sparc ameliorates ovarian cancer-associated inflammation. Neoplasia. 2008;10:1092–1104. doi: 10.1593/neo.08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangaletti S, Tripodo C, Cappetti B, Casalini P, Chiodoni C, Piconese S, Santangelo A, Parenza M, Arioli I, Miotti S, Colombo MP. Sparc oppositely regulates inflammation and fibrosis in bleomycin-induced lung damage. The American Journal of Pathology. 2011;179:3000–3010. doi: 10.1016/j.ajpath.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz M, von Arx T, Altermatt HJ, Bosshardt D. Histology of periapical lesions obtained during apical surgery. Journal of Endodontics. 2009;35:634–642. doi: 10.1016/j.joen.2009.01.024. [DOI] [PubMed] [Google Scholar]
- Settem RP, Honma K, Chinthamani S, Kawai T, Sharma A. B-cell RANKL contributes to pathogen-induced alveolar bone loss in an experimental periodontitis mouse model. Frontiers in Physiology. 2021;12:722859. doi: 10.3389/fphys.2021.722859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapouri-Moghaddam A, Mohammadian S, Vazini H, Taghadosi M, Esmaeili S-A, Mardani F, Seifi B, Mohammadi A, Afshari JT, Sahebkar A. Macrophage plasticity, polarization, and function in health and disease. Journal of Cellular Physiology. 2018;233:6425–6440. doi: 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- Silva TA, Garlet GP, Fukada SY, Silva JS, Cunha FQ. Chemokines in oral inflammatory diseases: apical periodontitis and periodontal disease. Journal of Dental Research. 2007;86:306–319. doi: 10.1177/154405910708600403. [DOI] [PubMed] [Google Scholar]
- Silva LAB, Sá MAR, Melo RA, Pereira JDS, da Silveira ÉJD, da Miguel MCC. Analysis of CD57+ natural killer cells and CD8+ T lymphocytes in periapical granulomas and radicular cysts. Brazilian Oral Research. 2017;31:e106. doi: 10.1590/1807-3107BOR-2017.vol31.0106. [DOI] [PubMed] [Google Scholar]
- Soares J, Santos S, Silveira F, Nunes E. Nonsurgical treatment of extensive cyst-like periapical lesion of endodontic origin. International Endodontic Journal. 2006;39:566–575. doi: 10.1111/j.1365-2591.2006.01109.x. [DOI] [PubMed] [Google Scholar]
- Song Y, Li X, Huang D, Song H. Macrophages in periapical lesions: potential roles and future directions. Frontiers in Immunology. 2022;13:949102. doi: 10.3389/fimmu.2022.949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stashenko P. Role of immune cytokines in the pathogenesis of periapical lesions. Endodontics & Dental Traumatology. 1990;6:89–96. doi: 10.1111/j.1600-9657.1990.tb00400.x. [DOI] [PubMed] [Google Scholar]
- Taira TM, Lima V, Prado DS, Silva TA, Issa JPM, da Silva LAB, Zamboni DS, Cunha FQ, Fukada SY. NLRP12 attenuates inflammatory bone loss in experimental apical periodontitis. Journal of Dental Research. 2019;98:476–484. doi: 10.1177/0022034518820289. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Takagi T, Naito Y, Uchiyama K, Hotta Y, Toyokawa Y, Ushiroda C, Hirai Y, Aoi W, Higashimura Y, Mizushima K, Okayama T, Katada K, Kamada K, Ishikawa T, Handa O, Itoh Y. Secreted protein acidic and rich in cysteine functions in colitis via IL17A regulation in mucosal CD4+ T cells. Journal of Gastroenterology and Hepatology. 2018;33:671–680. doi: 10.1111/jgh.13842. [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. Mrna-Seq whole-transcriptome analysis of a single cell. Nature Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- ’t Hart LM, Hansen T, Rietveld I, Dekker JM, Nijpels G, Janssen GMC, Arp PA, Uitterlinden AG, Jørgensen T, Borch-Johnsen K, Pols HAP, Pedersen O, van Duijn CM, Heine RJ, Maassen JA. Evidence that the mitochondrial leucyl trna synthetase (LARS2) gene represents a novel type 2 diabetes susceptibility gene. Diabetes. 2005;54:1892–1895. doi: 10.2337/diabetes.54.6.1892. [DOI] [PubMed] [Google Scholar]
- Tibúrcio-Machado CS, Michelon C, Zanatta FB, Gomes MS, Marin JA, Bier CA. The global prevalence of apical periodontitis: a systematic review and meta-analysis. International Endodontic Journal. 2021;54:712–735. doi: 10.1111/iej.13467. [DOI] [PubMed] [Google Scholar]
- To WS, Midwood KS. Plasma and cellular fibronectin: distinct and independent functions during tissue repair. Fibrogenesis & Tissue Repair. 2011;4:21. doi: 10.1186/1755-1536-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba H, de Castro Brás LE, Baicu CF, Zile MR, Lindsey ML, Bradshaw AD. Secreted protein acidic and rich in cysteine facilitates age-related cardiac inflammation and macrophage M1 polarization. American Journal of Physiology. Cell Physiology. 2015;308:C972–C982. doi: 10.1152/ajpcell.00402.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang R, Xiong H, Peng B. The involvement of platelet-derived growth factor-A in the course of apical periodontitis. International Endodontic Journal. 2011;44:65–71. doi: 10.1111/j.1365-2591.2010.01798.x. [DOI] [PubMed] [Google Scholar]
- Wang S, Song R, Wang Z, Jing Z, Wang S, Ma J. S100A8/A9 in inflammation. Front Immunol. 2018;9:1298. doi: 10.3389/fimmu.2018.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yang F, Qiu Y, Ye L, Song D, Huang D. The potential roles of T cells in periapical lesions. Journal of Endodontics. 2022;48:70–79. doi: 10.1016/j.joen.2021.09.016. [DOI] [PubMed] [Google Scholar]
- Yamaza T, Ren G, Akiyama K, Chen C, Shi Y, Shi S. Mouse mandible contains distinctive mesenchymal stem cells. Journal of Dental Research. 2011;90:317–324. doi: 10.1177/0022034510387796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yánez DC, Ross S, Crompton T. The IFITM protein family in adaptive immunity. Immunology. 2020;159:365–372. doi: 10.1111/imm.13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zhang S, Wang J, Li Q, Xue H, Sheng R, Xiong Q, Qi X, Wen J, Fan Y, Zhou BO, Yuan Q. LepR-expressing stem cells are essential for alveolar bone regeneration. Journal of Dental Research. 2020;99:1279–1286. doi: 10.1177/0022034520932834. [DOI] [PubMed] [Google Scholar]
- Zhao H, Chai Y. Stem cells in teeth and craniofacial bones. Journal of Dental Research. 2015;94:1495–1501. doi: 10.1177/0022034515603972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH. Massively parallel digital transcriptional profiling of single cells. Nature Communications. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]