Abstract
Nirmatrelvir is the main component of Paxlovid, an oral antiviral drug approved for the treatment of COVID-19 caused by SARS-COV-2 infection. Nirmatrelvir targets the main protease (Mpro), which is substantially conserved among different coronaviruses. Here, our molecular docking analysis indicates comparable affinity of nirmatrelvir binding to the Mpro enzymes of SARS-CoV-2 and three seasonal coronaviruses (OC43, 229E and NL63). However, in cell culture models, we found that nirmatrelvir potently inhibited SARS-CoV-2, OC43 and 229E, but not NL63. The insensitivity of NL63 to nirmatrelvir treatment was demonstrated at both viral replication and infectious titer levels. The antiviral activity of nirmatrelvir against OC43 and 229E was further confirmed in human airway organoids. The combination of nirmatrelvir and molnupiravir exerted differential patterns of antiviral response against OC43 and 229E. These results revealed disparities in the ability of nirmatrelvir to inhibit different coronaviruses, and caution against repurposing of nirmatrelvir as a pan-coronavirus treatment.
Keywords: Nirmatrelvir, Pan-coronavirus, Antiviral activity, Disparity
Coronaviruses are enveloped, single-stranded, positive-sense RNA viruses that widely circulate among mammals and birds. There are currently seven types of coronaviruses known to infect humans and cause diseases (Ma et al., 2020). The three highly pathogenic members are SARS-CoV, SARS-CoV-2 and MERS-CoV. The four seasonal members—OC43, NL63, 229E and HKU1— generally cause only the common cold, but severe complications may occur in vulnerable populations (Li et al., 2021). Given the clinical need for treating severely infected patients irrespective of the type of coronavirus, an ideal scenario would be the identification of pan-coronavirus inhibitors (Wang et al., 2021). These agents are potent inhibitors of currently circulating human coronaviruses and are likely to be effective against new species that emerge in the future, thus constituting a key approach in pandemic preparedness.
Coronaviruses encode a relatively conserved protease known as the main protease (Mpro) or 3C-like protease (3CLpro). Mpro processes the viral polyproteins into functional proteins to support the life cycle in host cells (Antonopoulou et al., 2022). Given its vital role and the conservation among different coronaviruses, Mpro represents an excellent target for the development of pan-coronavirus inhibitors (Dai et al., 2020; Mengist et al., 2020; Yoshino et al., 2020). Nirmatrelvir (PF-07321332) is an orally bioavailable SARS-CoV-2 Mpro inhibitor (Owen et al., 2021; Singh et al., 2022), and is the main ingredient of Paxlovid, an oral antiviral drug approved for treating COVID-19. The potent antiviral activity of nirmatrevir against ancestral and emerging variants of SARS-CoV-2 has been extensively demonstrated in experimental models (Li et al., 2022b). The efficacy of Paxlovid has been demonstrated in clinical trials to reduce the risk of severe COVID-19 or death (Najjar-Debbiny et al., 2022; Vassilopoulos and Mylonakis, 2022). Here, we aim to comprehensively assess the potential pan-coronavirus antiviral activity of nirmatrelvir.
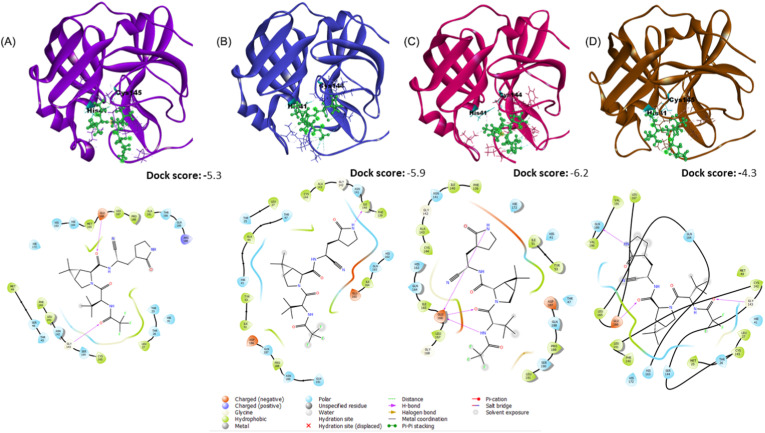
Mechanistically, nirmatrelvir firmly binds to SARS-CoV-2 Mpro and disrupts the His41–Cys145 catalytic dyad (Ahmad et al., 2021). We first comparatively evaluated the binding activity of nirmatrelvir to the Mpro of SARS-CoV-2, 229E, NL63 and OC43 by molecular docking. These protein structures of Mpro were retrieved from public database or modelled in silico (Supplement methods; Fig. S1). These structures were submitted to the GLIDE module of Schrodinger suite (Friesner et al., 2006) for docking with nirmatrelvir. The GLIDE results indicate that nirmatrelvir has the highest binding affinity towards Mpro of NL63 (Dock Score: −6.2), followed by Mpro of 229E, SARS-CoV-2 and OC43 (Fig. 1 ; Table S1). The 2D interaction analyses in maestro showed that His41 and Cys144/145 of all the Mpro structures are involved in the interaction with the ligand molecule (Fig. 1, Table S1). Apart from these two identical residues, each coronavirus has unique residues to interact with nirmatrelvir. Some of these residues are in the same position but appeared as different amino acids among different coronaviruses (Table S2). Docking analysis in AutoDock Vina (Trott and Olson, 2010) revealed similar results (Fig. S2). These in silico analyses support the pan-coronavirus antiviral activity of nirmatrelvir.
Fig. 1.
Molecular docking of the main protease of SARS-CoV-2, 229E, NL63 and OC43 with nirmatrelvir.
The interacting interface of the receptor with the ligand and its corresponding 2D interaction with the Mpro of (A) SARS-CoV-2, (B) 229E, (C) NL63 and (D) OC43 along with the dock scores. Interactions of the catalytic dyad residues present on the receptor (His41 and Cys144/145) with the ligand are highlighted. Corresponding bonding patterns are shown in different color codes.
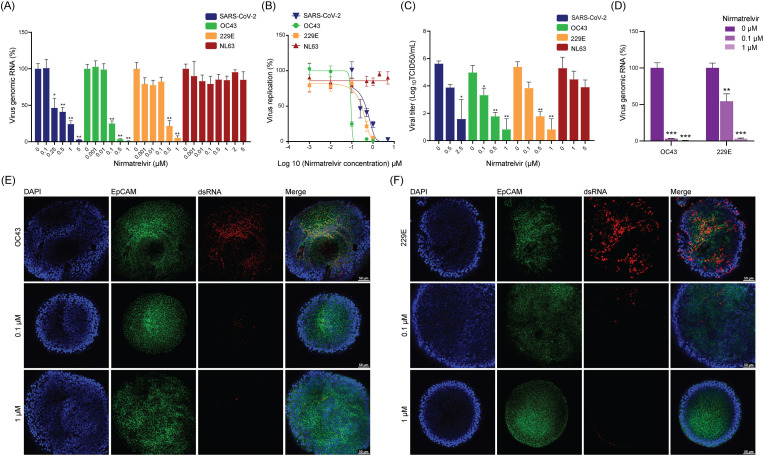
Next, we attempted to validate the pan-coronavirus antiviral activity of nirmatrelvir in widely used cell culture models. Intracellular viral RNA levels and infectious titer of produced viruses in the medium were quantified by qRT-PCR and TCID50 assay, respectively. A potent and dose-dependent inhibition of SARS-CoV-2 in Calu-3 cells, and OC43 and 229E in Huh7 cells were observed. For example, nearly complete inhibition of SARS-CoV-2 replication was observed by treatment with 5 μM nirmatrelvir for 48 h, whereas complete inhibition of OC43 and 22E was achieved already at 1 μM concentration (Fig. 2 A). The half maximum effective concentrations (EC50) of nirmatrelvir against SARS-CoV-2, OC43 and 229E were 0.45 μM, 0.09 μM and 0.29 μM, respectively (Fig. 2B), without major effect on cell viability (Fig. S3B). This potent inhibition was further confirmed by a reduction of up to 2–3 log10 infectious virus titers on SARS-CoV-2, OC43 and 229E (Fig. 2C).
Fig. 2.
Antiviral activity of nirmatrelvir against four coronaviruses in cell culture models.
(A) The effects of nirmatrelvir on intracellular viral RNA levels of SARS-CoV-2 in Calu-3 cells, OC43 and 229E in Huh7 cells and NL63 in LLC-MK2 cells (n = 4–9). (B) Virus replication curves of these coronaviruses in Calu-3, Huh7 and LLC-MK2 cells treated with nirmatrelvir (n = 4–9). (C) TCID50 assay quantifying titers of secreted infectious virus particles at 48 h post-treatment of nirmatrelvir (n = 4–5). (D) The effects of nirmatrelvir on intracellular viral RNA levels of OC43 and 229E in human airway organoids (hAOs) (n = 5). (E & F) Immunofluorescence staining of OC43 and 229E viral dsRNA, cytomembrane marker EpCAM and DAPI in hAOs.
Surprisingly, by qRT-PCR quantification of viral genomic RNA, nirmatrelvir showed limited inhibition on NL63 replication in LLC-MK2 cells (Fig. 2A and B). Consistently, we observed only mild inhibitory effect of nirmatrelvir on NL63 production in the culture medium as quantified by TCID50 assay (Fig. 2C). In human intestinal Caco2 cells infected with NL63, nirmatrelvir did not show any significant antiviral activity at concentrations ranging from 0 to 5 μM (Fig. S3A).
Primary human airway organoids (hAOs) represent an advanced model for coronavirus infection and antiviral drug testing (Han et al., 2022; Li et al., 2022a, 2022b). To validate the distinct potency of nirmatrelvir against different coronaviruses in cell culture models, we inoculated hAOs with the three seasonal coronaviruses. A potent inhibition of viral RNA replication was observed in 229E and OC43 infected organoids treated with nirmatrelvir (Fig. 2D), whereas no inhibition was observed in NL63 infected organoids by the treatment (Fig. S3C). Immunofluorescence staining of the viral double-stranded RNA (dsRNA) confirmed the potent antiviral effect of nirmatrelvir against 229E and OC43 (Fig. 2E and F).
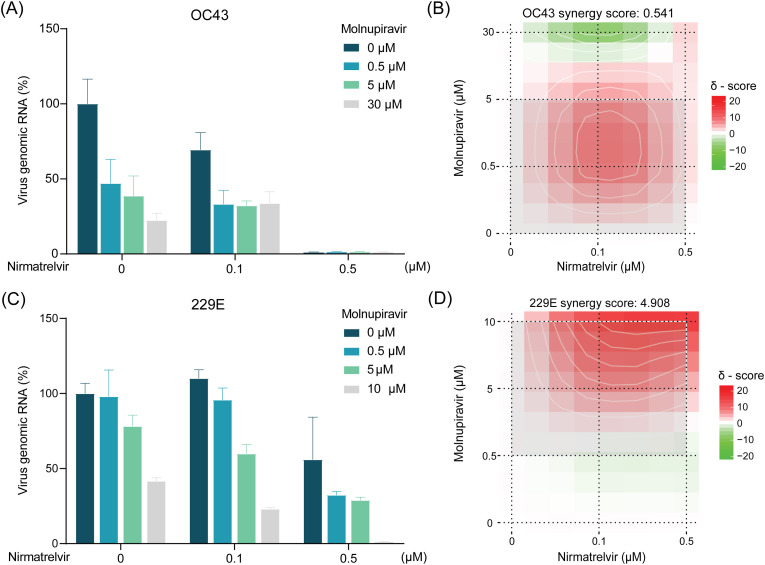
Antiviral monotherapies are commonly found to be suboptimal in clinical settings, whereas combination therapies are increasingly being explored for treating viral infections in the clinic. Combining antiviral therapies with complementary antiviral mechanisms often exhibit synergism. Furthermore, these antiviral combinations could prevent the development of drug-resistant strains by entirely halting viral replication, an advantage seldom achieved with monotherapies (Ianevski et al., 2022b). Molnupiravir, a small-molecule prodrug of the nucleoside derivative N-hydroxycytidine (NHC) targeting the viral RNA-dependent RNA polymerase (RdRp), is the first approved oral antiviral drug for treating COVID-19 (Jayk Bernal et al., 2022). The combinational treatment with molnupiravir and nirmatrelvir has been shown synergistic antiviral activity against both ancestral and Omicron variant of SARS-CoV-2 (Li et al., 2022b). Here, we explored the combination of molnupiravir and nirmatrelvir with a series of doses, and visualized the potential synergistic activity by SynergyFinder 3.0 tool (Ianevski et al., 2022a). We found this combination resulted in moderate synergism in Huh7 cells with 229E (synergy score: 4.908) (Fig. 3 C and D), whereas no synergistic effect was observed in Huh7 cells infected with OC43 (Fig. 3A and B).
Fig. 3.
Combinational effects of nirmatrelvir and molnupiravir against OC43 and 229E coronaviruses.
(A) The antiviral effects of combining nirmatrelvir and molnupiravir in OC43-infected Huh7 cells based on intracellular viral RNA levels (n = 4–7). (B) Synergy distribution of pairwise combination of nirmatrelvir and molnupiravir in OC43-infected Huh7 cells (n = 4–7). (C) The antiviral effects of combining nirmatrelvir and molnupiravir in 229E-infected Huh7 cells based on intracellular viral RNA levels (n = 4–9). (D) Synergy distribution of pairwise combination of nirmatrelvir and molnupiravir in 229E-infected Huh7 cells (n = 4–9).
A recent study reported that nirmatrelvir can potently inhibit seasonal coronaviruses OC43, NL63 and 229E, advocating it as a broad-spectrum coronavirus inhibitor (Weil et al., 2022). However, their conclusion was solely based on immunodetection of the viral nucleocapsid protein by in-cell ELISA, a new method that they developed for quantification of seasonal coronavirus infection. In our in silico analysis, we also observed high affinities of nirmatrelvir binding to the Mpro of the four coronaviruses including NL63 (Fig. 1). However, in cell culture and hAOs models infected with these viruses, by quantifying both viral RNA levels as an indicator of replication and infectious virus production, we demonstrated distinct potency of nirmatrelvir with minimal inhibitory activity on NL63. Although in silico analysis identified comparable affinities of nirmatrelvir to Mpro of these coronaviruses, different residues in each coronavirus contacting with nirmatrelvir may contribute to the distinct inhibitory activity. The NL63 strain used in this study was originally isolated from a patient in the Netherland in 2006 (Amsterdam, GenBank: DQ445912.1). We performed sanger sequencing and identified an R221C (Arginine to Cysteine) mutation in Mpro sequence, compared to the original sequence and other strains (Fig. S4). However, this R221C is not involved in the interaction with nirmatrelvir according to molecular docking (Table S2), which does not explain the insensitivity of NL63 to nirmatrelvir treatment.
Fluorescence resonance energy transfer (FRET) assay is a robust technique for identifying molecular interactions (Piston and Kremers, 2007). The interactions between nirmatrelvir and the Mpro of various coronaviruses including SARS-CoV-2, MERS-CoV, 229E, OC43 and NL63 have been well characterized by FRET-based biochemical assays in a previous study. However, in their study, the antiviral activity of nirmatrelvir was only validated in experimental models of SARS-CoV-2, MERS-CoV and 229E, but not OC43 and NL63 infection (Owen et al., 2021). Considering the limited inhibitory effects of nirmatrelvir against NL63 infection in our in vivo models, we highlight the need of validating the antiviral activity in bona fide experimental models, in addition to assays such as FRET or molecular docking.
In summary, molecular docking predicted high affinity of nirmatrelvir towards the Mpro enzymes of SARS-CoV-2, OC43, 229E and NL63. However, the bona fide antiviral activity in cell culture and organoid models varies dramatically with minimal activity against NL63. Although we are unable to elucidate the underlying mechanisms explaining the disparities between the molecular docking and experimental results, we urge caution in defining nirmatrelvir as a pan-coronavirus inhibitor and repurposing it for treating all coronavirus patients.
Author contributions
Conceptualization, J.L. and Y.W., P.L., and Q.P.; Methodology, J.L., Y.W., K.S., R.A. and M.L.; Formal analysis, J.L., Y.W., K.S and R.A.; Writing-Original Draft, J.L., K.S, R.A.; Funding acquisition, P.L. and Q.P.; Writing-Review & Editing, P.L., M.B. and Q.P.; J.L. and P.L. have accessed and verified the data. All authors read and approved the final version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This research is supported by funding of a VIDI grant (No. 91719300) from the Netherlands Organization for Scientific Research and the Dutch Cancer Society Young Investigator Grant (10140) to Q.P.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2023.105555.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
No data was used for the research described in the article.
References
- Ahmad B., Batool M., Ain Q.U., Kim M.S., Choi S. Exploring the binding mechanism of PF-07321332 SARS-CoV-2 protease inhibitor through molecular dynamics and binding free energy simulations. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22179124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonopoulou I., Sapountzaki E., Rova U., Christakopoulos P. Inhibition of the main protease of SARS-CoV-2 (M(pro)) by repurposing/designing drug-like substances and utilizing nature's toolbox of bioactive compounds. Comput. Struct. Biotechnol. J. 2022;20:1306–1344. doi: 10.1016/j.csbj.2022.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Han Y., Yang L., Lacko L.A., Chen S. Human organoid models to study SARS-CoV-2 infection. Nat. Methods. 2022;19:418–428. doi: 10.1038/s41592-022-01453-y. [DOI] [PubMed] [Google Scholar]
- Ianevski A., Giri A.K., Aittokallio T. SynergyFinder 3.0: an interactive analysis and consensus interpretation of multi-drug synergies across multiple samples. Nucleic Acids Res. 2022;50:W739–W743. doi: 10.1093/nar/gkac382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianevski A., Yao R., Simonsen R.M., Myhre V., Ravlo E., Kaynova G.D., Zusinaite E., White J.M., Polyak S.J., Oksenych V., Windisch M.P., Pan Q., Lastauskiene E., Vitkauskiene A., Matukevicius A., Tenson T., Bjoras M., Kainov D.E. Mono- and combinational drug therapies for global viral pandemic preparedness. iScience. 2022;25 doi: 10.1016/j.isci.2022.104112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayk Bernal A., Gomes da Silva M.M., Musungaie D.B., Kovalchuk E., Gonzalez A., Delos Reyes V., Martin-Quiros A., Caraco Y., Williams-Diaz A., Brown M.L., Du J., Pedley A., Assaid C., Strizki J., Grobler J.A., Shamsuddin H.H., Tipping R., Wan H., Paschke A., Butterton J.R., Johnson M.G., De Anda C., Group M.O.-O.S. Molnupiravir for oral treatment of covid-19 in nonhospitalized patients. N. Engl. J. Med. 2022;386:509–520. doi: 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Ikram A., Peppelenbosch M.P., Ma Z., Pan Q. Systematically mapping clinical features of infections with classical endemic human coronaviruses. Clin. Infect. Dis. 2021;73:554–555. doi: 10.1093/cid/ciaa1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Wang Y., Lamers M.M., Lavrijsen M., Iriondo C., de Vries A.C., Rottier R.J., Peppelenbosch M.P., Haagmans B.L., Pan Q. Recapitulating infection, thermal sensitivity and antiviral treatment of seasonal coronaviruses in human airway organoids. EBioMedicine. 2022;81 doi: 10.1016/j.ebiom.2022.104132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Wang Y., Lavrijsen M., Lamers M.M., de Vries A.C., Rottier R.J., Bruno M.J., Peppelenbosch M.P., Haagmans B.L., Pan Q. SARS-CoV-2 Omicron variant is highly sensitive to molnupiravir, nirmatrelvir, and the combination. Cell Res. 2022;32:322–324. doi: 10.1038/s41422-022-00618-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Li P., Ji Y., Ikram A., Pan Q. Cross-reactivity towards SARS-CoV-2: the potential role of low-pathogenic human coronaviruses. Lancet Microbe. 2020;1:e151. doi: 10.1016/S2666-5247(20)30098-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengist H.M., Fan X., Jin T. Designing of improved drugs for COVID-19: crystal structure of SARS-CoV-2 main protease M(pro) Signal Transduct. Targeted Ther. 2020;5:67. doi: 10.1038/s41392-020-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najjar-Debbiny R., Gronich N., Weber G., Khoury J., Amar M., Stein N., Goldstein L.H., Saliba W. Effectiveness of Paxlovid in reducing severe COVID-19 and mortality in high risk patients. Clin. Infect. Dis. 2022 doi: 10.1093/cid/ciac443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., Gibson S.A., Greasley S.E., Hurst B.L., Kadar E.P., Kalgutkar A.S., Lee J.C., Lee J., Liu W., Mason S.W., Noell S., Novak J.J., Obach R.S., Ogilvie K., Patel N.C., Pettersson M., Rai D.K., Reese M.R., Sammons M.F., Sathish J.G., Singh R.S.P., Steppan C.M., Stewart A.E., Tuttle J.B., Updyke L., Verhoest P.R., Wei L., Yang Q., Zhu Y. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- Piston D.W., Kremers G.J. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem. Sci. 2007;32:407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Singh R.S.P., Toussi S.S., Hackman F., Chan P.L., Rao R., Allen R., Van Eyck L., Pawlak S., Kadar E.P., Clark F., Shi H., Anderson A.S., Binks M., Menon S., Nucci G., Bergman A. Innovative randomized phase I study and dosing regimen selection to accelerate and inform pivotal COVID-19 trial of nirmatrelvir. Clin. Pharmacol. Ther. 2022;112:101–111. doi: 10.1002/cpt.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilopoulos A., Mylonakis E. In patients with COVID-19 at risk for severe disease, nirmatrelvir + ritonavir reduced hospitalization or death. Ann. Intern. Med. 2022;175:JC63. doi: 10.7326/J22-0038. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li P., Solanki K., Li Y., Ma Z., Peppelenbosch M.P., Baig M.S., Pan Q. Viral polymerase binding and broad-spectrum antiviral activity of molnupiravir against human seasonal coronaviruses. Virology. 2021;564:33–38. doi: 10.1016/j.virol.2021.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil T., Lawrenz J., Seidel A., Munch J., Muller J.A. Immunodetection assays for the quantification of seasonal common cold coronaviruses OC43, NL63, or 229E infection confirm nirmatrelvir as broad coronavirus inhibitor. Antivir. Res. 2022;203 doi: 10.1016/j.antiviral.2022.105343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino R., Yasuo N., Sekijima M. Identification of key interactions between SARS-CoV-2 main protease and inhibitor drug candidates. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-69337-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.