Abstract
The plant-associated microbiome is a key component of plant systems, contributing to their health, growth, and productivity. The application of machine learning (ML) in this field promises to help untangle the relationships involved. However, measurements of microbial communities by high-throughput sequencing pose challenges for ML. Noise from low sample sizes, soil heterogeneity, or technical factors can impact the performance of ML. Additionally, the compositional and sparse nature of these datasets can impact the predictive accuracy of ML. We review recent literature from plant studies to illustrate that these properties often go unmentioned. We expand our analysis to other fields to quantify the degree to which mitigation approaches improve the performance of ML and describe the mathematical basis for this. With the advent of accessible analytical packages for microbiome data including learning models, researchers must be familiar with the nature of their datasets.
Keywords: Machine learning, Deep Learning, Plant-associated microbiome, compositional data analysis
Organisms as a system: the plant holobiont
The last two decades have marked a paradigm shift in our understanding of biological systems, from thinking of organisms as entities with clearly delimited boundaries to the concept of the holobiont, which defines individual phenotypes as a product of interactions between host and associated microbial species [1]. In this context, the role of the microbiome is as crucial as the biological processes underway within the host. A recent interdisciplinary effort from experts in the field expanded the definition of microbiome to include all microbial communities occupying a defined habitat, their properties, and their interactions [2,3].
The interpretation of biological communities as holobionts naturally extends to plants as well [4], and the combined insight within all plant microbial domains has outlined their paramount role: unique communities are associated with increased plant growth through nutrient fixation, protection against biotic and abiotic stressors, changes in composition of secondary metabolites, and well-defined growth stages [5]. The recently-introduced concept of synthetic microbial communities (SynComs), formulated to mimic a simplified version of a known beneficial microbial cohort and which was shown to display desirable impacts when inoculated on a plant, further substantiates the importance of the plant microbiome [6]. Several influential reviews have been published on plant-microbiome interactions [7–9]. We encourage the interested reader to consult these sources to fully grasp the depth of the topic.
A friend and a foe: microbiome data is complex and high-dimensional
Large-scale microbiome studies require the collection of several types of high-dimensional data, often high throughput sequencing of DNA (metagenomics) or RNA (metatranscriptomics), or analysis of secreted bioactive compounds (metabolomics) [10]. As is common with all -omics disciplines, this produces large datasets which, if interrogated correctly, can yield meaningful insight. However, their analysis requires the utilization of sophisticated statistical methodologies and computational techniques. It is also necessary to select analytical models that are suited to the underlying structure and peculiarities of the dataset while retaining sufficient biological interpretability.
The advent of machine learning (ML) and deep learning (DL) approaches has proven vital to microbiome research [10–12]. ML approaches can aid in dimensionality reduction [13], clustering of amplicon sequences or microbial community types, and classification of multi-omics data [14]. Further, DL approaches typically aim to harness the complexity of multidimensional datasets to gather insight on host-microbiome interactions [14]. DL algorithms, such as convolutional neural networks (CNN) use either microbial phylogeny (as is the case PopPhy-CNN [15] or Ph-CNN [16]), or OTU tables (as in MetaNN [17]) as a starting point to predict host phenotype. ML algorithms can be used to associate specific microbial communities with unique phenotypes, or for predictive purposes (e.g. to infer a particular phenotype from an associated microbiome), focusing either on a single trait or multiple outcomes (as is the case with multi-task prediction [18], which is gaining relevance in microbiome studies [19]). In the field of plant science, most endeavors can be considered association studies, and recent efforts have employed ML or DL approaches to investigate pathogen-microbiome association [20,21], explore nitrogen fixation ability [22,23], and assess the impact of the microbiome on crop productivity [24,25]. The use of microbial profiles to predict phenotypical characteristics, rather popular in human microbiome studies [26,27], is gaining momentum in plant science: recent studies have focused on soil health, using microbiome profiling to predict physicochemical properties of the soil such as pH, concentration of individual nutrients, organic matter and surface hardness [28,29]. Other studies have focused on the plant host, using the concentration and type of microbial communities to predict yield in foxtail millet [30], soybean [31] and potato [32]. In Table 1, we provide the reader with an outline of some relevant publications on ML in plant microbiome studies from the last two years.
Table 1:
manuscripts that use ML/DL in plant (or soil) microbiome datasets in the period 2020-2022. Abbreviations: SVM = support vector machine, RF = random forest, ITS = internal transcribed spacer, CLR = centered log-ratio, TSS = total-sum scaling,
| Broad research topic | Specific research question | Quantification method | ML Method | Findings | Compositionality Normalized | Compositionality Discussed |
|---|---|---|---|---|---|---|
| Plant-pathogen interaction | Identification of biomarkers associated with ginseng rusty root [98] | 16S rRNA seq | SVM | 30 biomarkers associated with the disease | No | No |
| Phenotype/Functional prediction | Identification of generalist and specialized core taxa across plant holobiont in rice cultivars [99] | 16S rRNA seq | RF | 15 species identified as present across microhabitats | No | No |
| Predicting potato yield from sample microbiome [32] | 16S rRNA/ ITS seq | RF | 70 principal components (from OTU table) predict yield level above or below threshold. >80% accuracy | Yes (CLR) | No | |
| SynCom | Identification of general patterns of bacterial community composition in disease suppressive soils. [100] | SVM, RF, Logistic Regression | Distinguish between disease-conducive and disease-suppressive bacterial communities. Identified 28 genera enriched in disease- suppressive soils and 21 biomarkers in disease conducive soils. | No | No | |
| Soil microbiome relevance to determine functionality of root nodule symbiosis and healthy plant growth [101] | 16S rRNA seq | SVM | Pseudomonas most prevalent in non-rhizobiales ASVs, were characteristic of healthy plants, but absent in starved Lotus Burtii | Yes (SparCC) | No | |
| Multiscale modeling of microbiome | Explore the dependence of predictive models on spatial scale and taxonomic scale across soil microbiome [102] | 16S rRNA seq | Markov Models | Predictability in soil microbial abundances increases with spatial scales within different microbial communities in Neon dataset | Yes (rescaling, TSS) | Yes |
| Soil physics and chemistry | Predict physical and chemical variable from microbiome within soil samples [28] | 16S rRNA seq | RF | Microbial composition can predict land use (indigenous, exotic, horticulture, grassland) and physicochemical variables with ~50–90% accuracy | No | No |
| Predict soil health metrics (CASH framework) based on microbial communities present within sample [29] | 16S rRNA seq | RF SVM | Microbial family/genus/ASVs can predict soil health score metrics (CASH framework) with 60–80% accuracy | Yes (regularization) | Yes |
In agreement with this expansion, a recent article by members of the “Machine Learning for Microbiome” action of the European Cooperation in Science and Technology program highlights that a crucial priority of the field is the development of tools accessible to clinical and research personnel, who may not possess the skillset required to develop analytical models [14]. Indeed, currently available software suites like QIIME2 [33], Calypso [34], MicrobiomeAnalyst [35], Mothur [36], and Mixomics [37] provide deployment-ready and user-friendly ML implementation. Within this shift in user base, it is important that researchers in the field be aware of the structure of their data. Like other -omics disciplines, microbiome sequencing datasets are noisy, compositional, and sparse and, if not treated accordingly, the accuracy of the resulting analyses can be impacted.
Despite the necessity of accounting for these properties, explicit discussion of how best to deal with noise, compositionality, and sparsity in plant microbiome studies is missing. Many recent endeavors fail to adopt measures that account for these properties, and those that do rarely explain their choices in terms of the impacts of noise, sparsity, and compositionality. Even a recent review on the application of deep learning to microbiome data [38] fails to mention these properties of microbiome datasets. In Table 1, we highlight whether compositionality was addressed within the studies, and if that was done so explicitly (i.e., if compositionality was mentioned). Most of the reported studies did not explicitly mention compositionality, and only 4 out of 8 employed some form of compositionally-aware normalization strategy. Consequently, the purpose of this review is to explicitly highlight structural characteristics of microbiome datasets so their influence can be more easily identified, and to recognize which fundamental assumptions of ML methods are likely to be violated by them. In doing this, we hope to facilitate bias mitigation through ML method selection and the application of adequate normalization approaches.
The nature of the problem: microbiome data are noisy, compositional, and sparse
A major challenge for microbiome studies is noise, which can arise from low sample sizes and soil heterogeneity. Both endophytic and rhizospheric root microbiomes are highly influenced by their surrounding soil, due to intrinsically heterogeneous physical and chemical micro-environments present within the latter [39–43]. Spurious heterogeneity in microbiome data can also arise from technical factors such as sample collection, storage, processing, DNA extraction, PCR amplification and sequencing or simply from stochastic events during plant colonization [44–48]. Heterogeneity related to the aforementioned reasons can severely impact the performance of machine learning algorithms.
A starting point to deal with the unwanted variation is exploratory data analysis via ordination plots. Unconstrained ordination (MDS, NMDS etc.) will detect anomalies in the data set, such as outlier samples, confounding factors and noise that arises from the technical issues. Variation that is visualized with the ordination plot can be estimated and accounted for using methods developed for dealing with batch effects, such as SVA, ComBat, and removeBatchEffect [49–51], although there are some reservations regarding the applicability of these methods to microbiome data [44]. Further, because the variation of interest can be masked by high variability and high correlation structure among variables unrelated to group differences, unconstrained ordination should be followed by a constrained ordination (CAP, CCA etc.). Though this may not provide information about the within-group variability, it can reveal location differences among the groups, which enables the researcher to cluster different groups by their microbiome profiles.
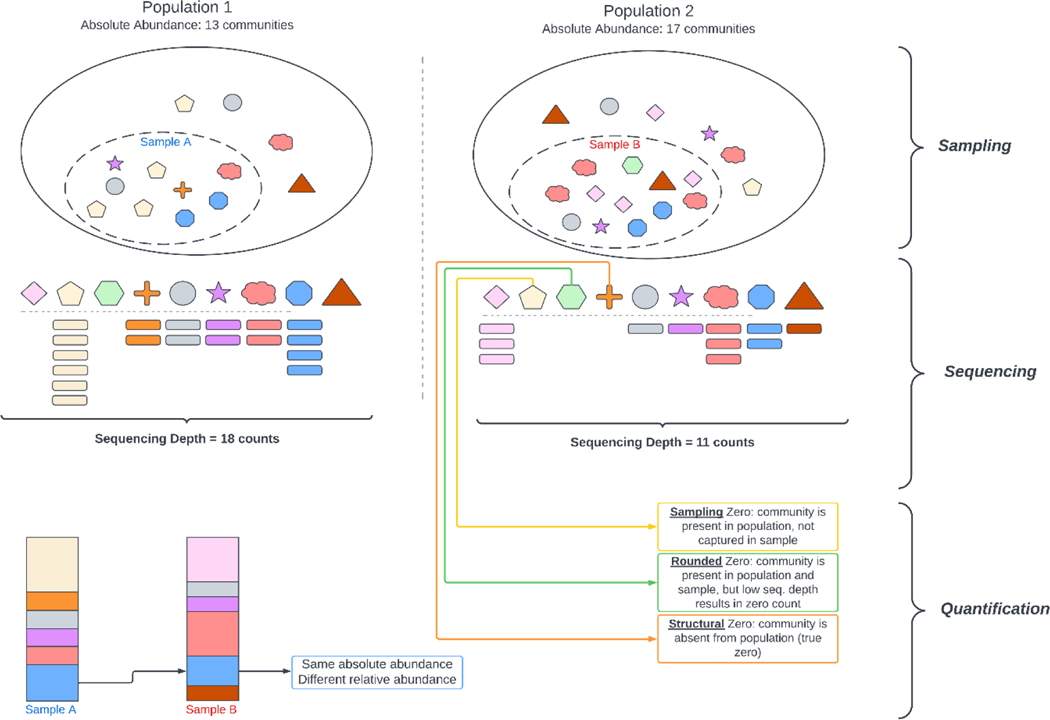
A second source of potential bias is associated with the sequencing technique employed. Most microbiome studies are carried out using high-throughput sequencing (HTS) platforms, , using either bulk transcripts from a group of cells or DNA/RNA from a single cell [52,53] as an experimental sample. As such, the information gathered for each biological sample is limited by the capacity of the sequencing instrument to deliver reads (known as sequencing depth) [54]. As the total number of reads per sample is a finite quantity, changes in abundance of one sequence will affect abundance of others within that sequencing run. Consequently, HTS-based assays are only representative of relative microbial abundance within each sample, and uninformative of their absolute abundance (Figure 1). When individual components of a set can be represented as ratios of a total, they are termed compositional [55]. Microbiome datasets are inherently compositional [56], whether obtained via bulk sequencing or through novel single-cell prokaryotic sequencing methods such as microSPLIT [52] and PETRIseq [53]. The compositional nature of microbiome data brings about several features that must be acknowledged throughout their analysis. Compositional datasets display a bias towards negative correlation [55]. This is because the sum of the feature values is an arbitrary constant and as a result, the correlation coefficients between one element and the rest of the set must add up to −1 [55]. Further, it was observed that correlation between elements of a partial subset of the composition is fundamentally different than the correlation between those same elements when computed with the complete composition. This is a property known as subcompositional incoherence [57]. Analytical methods based on correlation are inevitably affected by these properties.
Figure 1. The emergence of compositionality and sparsity in high-throughput sequencing-based microbiome studies.
We present a hypothetical comparison of two microbial populations with distinct absolute abundances (all values are arbitrary units). Investigators collect and sequence one sample per population, which results in some species with low abundance (Yellow Pentagon in Population 2) to be excluded from the collected sample, leading to a value of zero for said community. Unequal sequencing depths lead to non-quantification of other species (Green Hexagon in Population 2) uniquely due to the lack of corresponding sequencing counts. Finally, the compositional nature of the experimental setup and the resulting dataset leads to observed changes caused by a difference in relative abundance only, leading to bias in differential abundance compared to absolute changes.
The field of compositional data analysis focuses on developing normalization strategies that are appropriate to this data type [55,58–61]. Individual data points can be scaled to the total sum of all elements (a procedure called closure or Total Sum Scaling) and presented as proportion of the total [55,62]. More commonly, compositional data are linearly transformed by taking the log of either a ratio between each element and the geometric mean of the composition (called centered log-ratio, clr, [55,58,60]), the log of the ratio between each element and a set element in the composition (additive log-ratio, alr, [55,58,60]), or through the more complex isometric log-ratio (ilr) [61]. Log-ratio transformations are widely used in microbiome studies [56], though the choice of which transformation to employ has been subject of discussion [63,64]. Most importantly however, many readily available analytical platforms for microbiome data (such as the aforementioned QIIME2, Mothur, and MicrobiomeAnalyst) provide users with several options for log-ratio transformations.
In addition to being noisy and compositional, taxonomic abundance tables (OTU/ASV tables) have a high proportion of “zero-count” values, a property known as sparsity [10]. The roots of this phenomenon are twofold: first, microbial communities are heterogeneous, with many organisms detected in only few samples across the datasets, and few organisms responsible for most of the sequencing reads. Second, sequencing depth is uneven between samples, which causes low-count microbes to not be detected in samples with low sequencing depth [65]. This is exacerbated by the compositional nature of the data, as a highly-abundant taxon may artificially reduce the counts of other taxa below the detection threshold of the sequencer, by virtue of sequencing depth being a finite quantity.
Sparsity causes taxonomic tables to adopt distributions that are right-skewed, with considerable point mass at zero. This results in data that must be analyzed using methods that are robust to non-normality [10]. The analysis is further complicated when considering log-ratio transformations, because log(0) is undefined. Zeros can be removed by adding a small arbitrary pseudo-count to all values. The result of the transformation, however, can be impacted by the value of the pseudo-count [62]. Replacement of zero values through imputation has also been proposed, though the appropriate strategy and validity of this procedure are not entirely clear [66,67]. Finally, a series of models which are robust against excess zeros have been employed for microbiome datasets, such as zero-inflated negative binomial [68], zero-inflated beta-binomial [69], zero-inflated Poisson [70], hurdle models [71] and zero-aware mixture models [72], although these models do not inherently account for compositionality.
Noise, compositionality and sparsity can affect the accuracy of ML/DL models
Spurious sources of noise, sparsity and compositionality in microbiome datasets have tangible impacts on the use of ML and DL models. Reducing unwanted variability prior to the implementation of ML algorithms, e.g. by using SVA and ComBat, may improve inference accuracy in datasets with confounders [73]. Further, when multi-omics data is available, this can be integrated with tools such as MOFA [74] and DIABLO from MixOmics [37]. Using this framework for unsupervised integration, microbiome signatures of continuous gradients and/or discrete clusters in relationship with other biological layers can be revealed, which can mitigate the noise of microbiome data sets. This can consequently improve the performance of downstream applications. While comparative studies on the use of said integration tools in microbiomics have not been published, a recent effort focusing on multi-omics data (gene expression, whole-exome sequencing mutations, copy number variation and protein abundance) for several cancer lines indicated favorable results from both DIABLO and MOFA [75].
The constrained correlation structure typical of compositional data, combined with a right-skewed distribution caused by sparsity, may violate assumptions of regression analysis such as absence of multicollinearity and normality [76]. Further, the use of rarefaction (a form of subsampling without replacement [77]) to normalize sequencing depth may lead to unequal variances between elements across samples, violating the assumption of homoscedasticity [10]. Consequently, the accuracy of regression-based ML models such as linear and logistic regression may suffer because they rely on assumptions of data distribution that do not hold true for compositional sequencing datasets. Some authors have mitigated these issues by employing a regularization method. Dong and collaborators [78] used L1-regularized multinomial logistic regression on transformed counts in a study on the human microbiome and its relation to Parkinson’s disease. Another study by Lin and collaborators [79] used L1 regularization for the linear log-contrast model (originally proposed by Aitchison and Bacon-Shone [80]), which was chosen to deal with compositionality. Application of the regularized model to a dataset linking gut microbiome composition and dietary patterns in humans indicated an improvement in performance. Other recent efforts seem to suggest that L2-regularized logistic regression can considerably improve prediction accuracy compared to non-regularized regression as shown in a recent study on human microbiome data to predict colonic neoplasias [81].
Certain nonlinear predictors, such as Decision Trees and Random Forests (RF), are broadly regarded as being unaffected by multicollinearity and distribution shape [82] and are shown to perform well in microbiome studies [81,83]. Within plant studies, RF classifiers have garnered interest and are being adopted in endeavors focused on plant-microbiome interaction. RF classifiers were recently applied to microbiome sequencing data to predict soil health [29]. This resulted in a prediction accuracy around 83% after hyperparameter optimization, indicating agreement between classification output and true soil health. The model was also able to predict the importance of individual microbes to soil health based on their impact on classification performance, showing that groups of microbes with known importance to soil health were important predictors for the model. However, the discarding of microbe abundances containing zero values to mitigate sparsity posed challenges for prediction accuracy, which the authors indicate could be solved with greater sequencing depth. A further study by Chang et al. [31] applied RF classifiers to predict crop yield in combination with metagenome-wide association studies (MWAS). The RF classifier was able to predict crop productivity categories of high yield and low yield from soil composition with 79% accuracy, and key microbes (as identified by the classifier) matched those identified by MWAS, including known nitrogen-fixing bacteria.
As indicated earlier in this review, discussion of the impact of compositionality and sparsity in microbiome studies is relatively limited, especially outside of specialized reviews on the topic. Consequently, our understanding of the impact of compositionality (and of the approaches chosen to mitigate it) on ML and DL performance is rather limited. However, a series of authors have recently performed and published comparative analyses on the topic using datasets which, though not identical to microbiome data, possess many of the same qualities (large, compositional, and sparse) and can provide insight on our question of interest. We provide a summary of these findings and indicate the degree of similarity to microbiome datasets.
When compositionality is accounted for by log-ratio transformation, the choice of transformation can impact the performance of the model. A recent effort by Tolosana-Delgado and collaborators to characterize the effects of log-ratio transformation on simulated data illustrated that the performance of a RF model was impacted by the type of log-ratio transformation chosen, with pairwise log-ratios (pwlr) performing significantly better than clr or ilr, and non-transformed data having the largest out-ofbag error estimate overall [84]. This may be due, in part, to the fact that pwlr transformations generate more features than clr and ilr. The same article indicates that the accuracy of regression-based models would not be affected by the choice of log-ratio transformation, which follows from the fact that any log-ratio transformation is a linear transformation of log-transformed feature values.
Limited information is available on the sensitivity of ML/DL models of experimental data to compositional normalization strategies and sparsity, but initial reports seem to indicate an impact. A study on chemical profiling of honey and saffron (both resulting in compositional datasets) found lower misclassification rates whenever log-ratio transformations (clr, ilr, and a modified ilr) were applied to the data compared to raw counts, leading to improved classification using multiple ML models (artificial neural networks, ANN), linear discriminant analysis (LDA), or K-nearest neighbors (KNN) [85]. However, the honey dataset, aside from being compositional, had a minor degree of sparsity. To account for this, the authors used several zero replacement strategies operating under the assumption that all zeros within the datasets were due to components being present below the detection limit of the instrument, rather than truly absent, which is a valid assumption in the context of chemical analysis. However, the subject of zero-replacement is debated in microbiome research [66,67,86], and the likely scenario is that some taxa are actually absent from the sample (structural zero), while others are not detected due to low concentration and shallow sequencing (rounded zero - Figure 1). It is important to note that current state-of-the-art imputation methods in microbiome studies exist, and hinge on numerically determining the likelihood of a zero-count being biologically relevant (structural, rather than rounded). This has been attempted using a combination of taxon information and between-sample variability, modeled either through a penalized linear regression (mbImpute [87]) or zero-inflated probabilistic PCA (mbDenoise [88]). Crucially, this is a fundamentally different approach from zero replacement in single-cell RNA sequencing, where methods like SAVER treat all zeros equally, with the creators of SAVER recommending removing extremely low-abundance genes at the beginning [89]. Recent comparisons of these methods seem to indicate better performance of mbDenoise over mbImpute and SAVER [88], though this seems to vary depending on the dataset. Further, no studies have explored the impact of either of these methods on ML algorithms.
Taxonomic abundance tables often contain rare OTUs, which contribute to sparsity and are likely not informative. Further, the combination of a pseudo-count and log-ratio transformation could potentially introduce spurious signals in OTUs with a large proportion of zeros. Removing such OTUs (prevalence-based filtering) is a common practice (see e.g. [90]). Note that the alr- and pwlr-transformed abundances of the remaining features are preserved after filtering, while this is not the case for clr. The impact of such filtering on ML procedures is not well-studied. In one review [91], the use of PERFect [92], which carries out statistical tests to identify noninformative OTUs, and the contaminant-removal procedure decontam [93] on the predictive accuracy of RF was examined. Applying both procedures before fitting a random forest was found to have little effect on the area under the receiver operating characteristic curve (AUC), but fewer features were used. The models were most likely fitted on the raw counts since no transformation was mentioned. Notably, filtering and contaminant removal reduced lab-to-lab variability in the data. This could improve the ability of ML models fitted on data from one lab to predict outcomes on data from another.
Deep learning models are a class of artificial neural network approaches that use the network architecture to perform representation learning, and are known as “deep” models due to the large number of network layers involved [94]. As the choice of compositionally aware transformation can have significant impacts on the performance of DL models, recent efforts in related disciplines have focused on integrating log-ratio normalization within the DL workflow. DeepCoDA, an analytical framework for high-dimensional compositional health data, uses a log-bottleneck module to allow the model to automatically select the best log-ratio transformation [95]. Other strategies use learning-based approaches to determine the most meaningful pairwise log-ratio relationships in a computationally-efficient manner [96,97]. Although these approaches are not specific to plant studies, they indicate that the quality and rigor of ML/DL approaches in plants would benefit from integrating processes to select and compare compositionally aware normalization strategies.
Conclusion
Spurious noise, compositionality and sparsity of sequencing-based microbiome datasets can represent pitfalls for the implementation of machine learning algorithms. Our analysis of the most recent endeavors in the field indicates that most plant microbiome studies employing machine learning approaches do not explicitly mention compositionality and sparsity, even when methods are used to account for their impact. In this review, we have provided an overview of strategies to account for how noise, compositionality and sparsity and the consequences that those strategies can have on ML algorithms, whether the associated microbiome is used as a predictor or as an outcome. Additionally, we have outlined a series of steps throughout the pipeline at which investigators can explore approaches to these characteristics and assess the resulting impact on the predictions of their learning algorithms. Ultimately, appropriate strategies will depend on the biological relationships being modeled and will likely be unique to each study. Similarly, model selection will have to consider the tradeoff between the increased performance and adaptability offered by nonlinear predictors and the simplicity and interpretability of linear predictors. We situate our work within a larger context in the conversation about the structure of microbiome data, one in which we are confident that studies focusing on plant-microbe interactions will benefit from participating.
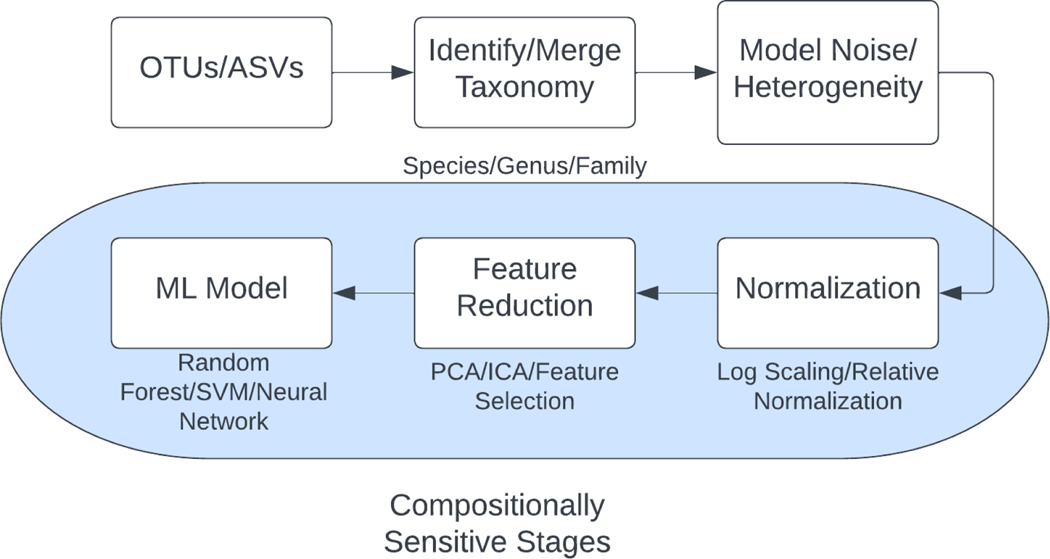
Figure 2. Typical Steps in Preparing Microbiome Sequencing Data for ML.
We show a common sequence of processes done to prepare microbiome sequencing data for use in machine learning models, with the stages of this preparatory process where the impacts of compositionally need to be accounted for and may be mitigated through selection of appropriate techniques.
Acknowledgements
SB was funded by the National Institute of Health (NIH 1R01HD093041-01). MG was funded by the National Science Foundation (DGE-1746939) and the National Institute of Health (NIH T32 GM008776). MC, IJ, and TA were funded by the Novo Nordisk Foundation (NNF19SA0059362).
Cranos Williams reports financial support was provided by Novo Nordisk Inc. Max Gordon reports financial support was provided by National Science Foundation. Sebastiano Busato reports financial support was provided by National Institute of Health. Stig Andersen reports financial support was provided by Novo Nordisk Inc. Meenal Chaudhari reports financial support was provided by Novo Nordisk Inc. Ib Jensen reports financial support was provided by Novo Nordisk Inc. Turgut Akyol reports financial support was provided by Novo Nordisk Inc.
Footnotes
Declaration of Competing Interest
The authors have no conflicts of interest to declare.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* special interest ** outstanding interest
- 1.Simon J-C, Marchesi JR, Mougel C, Selosse M-A: Host-microbiota interactions: from holobiont theory to analysis. Microbiome 2019, 7:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whipps JM, Lewis K, Cooke R: Mycoparasitism and plant disease control. Fungi in biological control systems 1988,
- 3.Berg G, Rybakova D, Fischer D, Cernava T, Vergès M- CC, Charles T, Chen X, Cocolin L, Eversole K, Corral GH, et al. : Microbiome definition re-visited: old concepts and new challenges. Microbiome 2020, 8:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A: The importance of the microbiome of the plant holobiont. New Phytologist 2015, 206:1196–1206. [DOI] [PubMed] [Google Scholar]
- 5.Berg G, Rybakova D, Grube M, Köberl M: The plant microbiome explored: implications for experimental botany. J Exp Bot 2016, 67:995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Souza RSC, Armanhi JSL, Arruda P: From Microbiome to Traits: Designing Synthetic Microbial Communities for Improved Crop Resiliency. Frontiers in Plant Science 2020, 11. * Review of the use of synthetic microbial communities (SynCom) as inoculants for agriculture. In depth discussion of biological aspects of the technique, as well as computational methods for screening and identification of beneficial colonies.
- 7.Lebeis SL: Greater than the sum of their parts: characterizing plant microbiomes at the community-level. Current Opinion in Plant Biology 2015, 24:82–86. [DOI] [PubMed] [Google Scholar]
- 8.Song C, Jin K, Raaijmakers JM: Designing a home for beneficial plant microbiomes. Current Opinion in Plant Biology 2021, 62:102025. [DOI] [PubMed] [Google Scholar]
- 9.Song S, Liu Y, Wang NR, Haney CH: Mechanisms in plant–microbiome interactions: lessons from model systems. Current Opinion in Plant Biology 2021, 62:102003. [DOI] [PubMed] [Google Scholar]
- 10. Jiang D, Armour CR, Hu C, Mei M, Tian C, Sharpton TJ, Jiang Y: Microbiome Multi-Omics Network Analysis: Statistical Considerations, Limitations, and Opportunities. Frontiers in Genetics 2019, 10. * Review on the integration of multiple -omics datasets pertaining to microbiome data. Discussion of crucial aspects of microbiome datasets (compositionality, sparsity, heterogeneity, normalization) as highlighted in the present manuscript. Further discussion on the role of network analysis to model microbiome data.
- 11.Jasner Y, Belogolovski A, Ben-Itzhak M, Koren O, Louzoun Y: Microbiome Preprocessing Machine Learning Pipeline. Frontiers in Immunology 2021, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SJ, Rho M: Multimodal deep learning applied to classify healthy and disease states of human microbiome. Sci Rep 2022, 12:824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghannam RB, Techtmann SM: Machine learning applications in microbial ecology, human microbiome studies, and environmental monitoring. Computational and Structural Biotechnology Journal 2021, 19:1092–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreno-Indias I, Lahti L, Nedyalkova M, Elbere I, Roshchupkin G, Adilovic M, Aydemir O, Bakir-Gungor B, Santa Pau EC, D’Elia D, et al. : Statistical and Machine Learning Techniques in Human Microbiome Studies: Contemporary Challenges and Solutions. Frontiers in Microbiology 2021, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reiman D, Metwally AA, Sun J, Dai Y: PopPhy-CNN: A Phylogenetic Tree Embedded Architecture for Convolutional Neural Networks to Predict Host Phenotype From Metagenomic Data. IEEE Journal of Biomedical and Health Informatics 2020, 24:2993–3001. [DOI] [PubMed] [Google Scholar]
- 16.Fioravanti D, Giarratano Y, Maggio V, Agostinelli C, Chierici M, Jurman G, Furlanello C: Phylogenetic convolutional neural networks in metagenomics. BMC Bioinformatics 2018, 19:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo C, Marculescu R: MetaNN: accurate classification of host phenotypes from metagenomic data using neural networks. BMC Bioinformatics 2019, 20:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhan X, Tong X, Zhao N, Maity A, Wu MC, Chen J: A small-sample multivariate kernel machine test for microbiome association studies. Genetic Epidemiology 2017, 41:210–220. [DOI] [PubMed] [Google Scholar]
- 19.Magnúsdóttir S, Thiele I: Modeling metabolism of the human gut microbiome. Current Opinion in Biotechnology 2018, 51:90–96. [DOI] [PubMed] [Google Scholar]
- 20. Jiang G, Zhang Y, Gan G, Li W, Wan W, Jiang Y, Yang T, Zhang Y, Xu Y, Wang Y, et al.: Exploring rhizo-microbiome transplants as a tool for protective plant-microbiome manipulation. ISME COMMUN 2022, 2:1–10. ** The paper studies the rhizosphere microbiome transplant with model pathogen strain R. solanacearum (bacterial wilt disease) on donor species of eggplant and recipient species of tomato.
- 21.Poncheewin W, van Diepeningen AD, van der Lee TAJ, Suarez-Diez M, Schaap PJ: Classification of the plant-associated lifestyle of Pseudomonas strains using genome properties and machine learning. Sci Rep 2022, 12:10857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quides KW, Atamian HS: A microbiome engineering framework to evaluate rhizobial symbionts of legumes. Plant Soil 2021, 463:631–642. [Google Scholar]
- 23. Higdon SM, Huang BC, Bennett AB, Weimer BC: Identification of Nitrogen Fixation Genes in Lactococcus Isolated from Maize Using Population Genomics and Machine Learning. Microorganisms 2020, 8. * Provides hypothetical analysis of pathway for bio-nitrogen fixation in Sierra Mixe maize through lactococci strains using random forest and GWAS.
- 24.Yue H, Banerjee S, Liu C, Ren Q, Zhang W, Zhang B, Tian X, Wei G, Shu D: Fertilizing-induced changes in the nitrifying microbiota associated with soil nitrification and crop yield. Science of The Total Environment 2022, 841:156752. [DOI] [PubMed] [Google Scholar]
- 25.Xiong C, Zhu Y-G, Wang J-T, Singh B, Han L-L, Shen J-P, Li P-P, Wang G-B, Wu C-F, Ge A-H, et al. : Host selection shapes crop microbiome assembly and network complexity. New Phytologist 2021, 229:1091–1104. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Y-H, Gallins P: A Review and Tutorial of Machine Learning Methods for Microbiome Host Trait Prediction. Front Genet 2019, 10:579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saulnier DM, Riehle K, Mistretta T-A, Diaz M-A, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, et al. : Gastrointestinal Microbiome Signatures of Pediatric Patients With Irritable Bowel Syndrome. Gastroenterology 2011, 141:1782–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hermans SM, Buckley HL, Case BS, Curran-Cournane F, Taylor M, Lear G: Using soil bacterial communities to predict physico-chemical variables and soil quality. Microbiome 2020, 8:79. ** Bacterial community composition is used to predict land use (indigenous forests, exotic forest plantations, horticulture, or pastoral grasslands) in soil samples using Random Forest, with ~85% prediction accuracy. Within site type, the model can also predict physicochemical properties (pH, carbon, nitrogen, microporosity and others).
- 29. Wilhelm RC, van Es HM, Buckley DH: Predicting measures of soil health using the microbiome and supervised machine learning. Soil Biology and Biochemistry 2022, 164:108472. ** Authors use microbiome composition of ~1000 soil samples to predict 12 metrics of soil health (part of the Comprehensive Assessment of Soil Health (CASH) framework), using L2-regularized Support Vector Machine and Random Forest.
- 30.Jin T, Wang Y, Huang Y, Xu J, Zhang P, Wang N, Liu X, Chu H, Liu G, Jiang H, et al. : Taxonomic structure and functional association of foxtail millet root microbiome. Gigascience 2017, 6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang H-X, Haudenshield JS, Bowen CR, Hartman GL: Metagenome-Wide Association Study and Machine Learning Prediction of Bulk Soil Microbiome and Crop Productivity. Frontiers in Microbiology 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Imam N, Belda I, García-Jiménez B, Duehl AJ, Doroghazi JR, Almonacid DE, Thomas VP, Acedo A: Local Network Properties of Soil and Rhizosphere Microbial Communities in Potato Plantations Treated with a Biological Product Are Important Predictors of Crop Yield. mSphere 2021, 6:e00130–21. ** Authors train a machine learning algorithm (Random Forest) to predict potato yield from microbial abundance. Their model is able to predict whether the yield will be above or below 30 t/ha using a set of 70 principal components, based on microbial abundance and other factors.
- 33.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. : Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019, 37:852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zakrzewski M, Proietti C, Ellis JJ, Hasan S, Brion M-J, Berger B, Krause L: Calypso: a user-friendly web-server for mining and visualizing microbiome–environment interactions. Bioinformatics 2017, 33:782–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chong J, Liu P, Zhou G, Xia J: Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc 2020, 15:799–821. [DOI] [PubMed] [Google Scholar]
- 36.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. : Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Applied and Environmental Microbiology 2009, 75:7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohart F, Gautier B, Singh A, Cao K-AL: mixOmics: An R package for ‘omics feature selection and multiple data integration. PLOS Computational Biology 2017, 13:e1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng Z, Zhang J, Li J, Zhang X: Application of Deep Learning in Plant–Microbiota Association Analysis. Frontiers in Genetics 2021, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bickel S, Or D: Soil bacterial diversity mediated by microscale aqueous-phase processes across biomes. Nat Commun 2020, 11:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou J, Xia B, Treves DS, Wu L-Y, Marsh TL, O’Neill RV, Palumbo AV, Tiedje JM: Spatial and Resource Factors Influencing High Microbial Diversity in Soil. Applied and Environmental Microbiology 2002, 68:326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang G, Or D: Hydration dynamics promote bacterial coexistence on rough surfaces. ISME J 2013, 7:395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bach EM, Williams RJ, Hargreaves SK, Yang F, Hofmockel KS: Greatest soil microbial diversity found in micro-habitats. Soil Biology and Biochemistry 2018, 118:217–226. [Google Scholar]
- 43.Vos M, Wolf AB, Jennings SJ, Kowalchuk GA: Micro-scale determinants of bacterial diversity in soil. FEMS Microbiology Reviews 2013, 37:936–954. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, LêCao K-A: Managing batch effects in microbiome data. Briefings in Bioinformatics 2020, 21:1954–1970. [DOI] [PubMed] [Google Scholar]
- 45.Brooks JP, Edwards DJ, Harwich MD, Rivera MC, Fettweis JM, Serrano MG, Reris RA, Sheth NU, Huang B, Girerd P, et al. : The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiology 2015, 15:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lauder AP, Roche AM, Sherrill-Mix S, Bailey A, Laughlin AL, Bittinger K, Leite R, Elovitz MA, Parry S, Bushman FD: Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 2016, 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW: Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 2014, 12:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sinha R, Abnet CC, White O, Knight R, Huttenhower C: The microbiome quality control project: baseline study design and future directions. Genome Biol 2015, 16:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leek JT, Storey JD: Capturing Heterogeneity in Gene Expression Studies by Surrogate Variable Analysis. PLOS Genetics 2007, 3:e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK: limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 2015, 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson WE, Li C, Rabinovic A: Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8:118–127. [DOI] [PubMed] [Google Scholar]
- 52.Kuchina A, Brettner LM, Paleologu L, Roco CM, Rosenberg AB, Carignano A, Kibler R, Hirano M, DePaolo RW, Seelig G: Microbial single-cell RNA sequencing by split-pool barcoding. Science 2021, 371:eaba5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blattman SB, Jiang W, Oikonomou P, Tavazoie S: Prokaryotic single-cell RNA sequencing by in situ combinatorial indexing. Nat Microbiol 2020, 5:1192–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zaheer R, Noyes N, Ortega Polo R, Cook SR, Marinier E, Van Domselaar G, Belk KE, Morley PS, McAllister TA: Impact of sequencing depth on the characterization of the microbiome and resistome. Sci Rep 2018, 8:5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aitchison J: The Statistical Analysis of Compositional Data. Journal of the Royal Statistical Society: Series B (Methodological) 1982, 44:139–160. [Google Scholar]
- 56.Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ: Microbiome Datasets Are Compositional: And This Is Not Optional. Front Microbiol 2017, 8:2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Filzmoser P, Hron K: Correlation Analysis for Compositional Data. Math Geosci 2008, 41:905. [Google Scholar]
- 58.Aitchison J: Principles of Compositional Data Analysis. Lecture Notes-Monograph Series 1994, 24:73–81. [Google Scholar]
- 59.Aitchison J: Principal component analysis of compositional data. Biometrika 1983, 70:57–65. [Google Scholar]
- 60.Pawlowsky-Glahn V, Egozcue JJ: Compositional data and their analysis: an introduction. Geological Society, London, Special Publications; 2006, 264:1–10. [Google Scholar]
- 61.Egozcue JJ, Pawlowsky-Glahn V, Mateu-Figueras G, Barceló-Vidal C: Isometric Logratio Transformations for Compositional Data Analysis. Mathematical Geology 2003, 35:279–300. [Google Scholar]
- 62.Costea PI, Zeller G, Sunagawa S, Bork P: A fair comparison. Nat Methods 2014, 11:359–359. [DOI] [PubMed] [Google Scholar]
- 63.Greenacre M, Grunsky E: The isometric logratio transformation in compositional data analysis: a practical evaluation. 2019,
- 64.Greenacre M, Martínez-Álvaro M, Blasco A: Compositional Data Analysis of Microbiome and Any-Omics Datasets: A Validation of the Additive Logratio Transformation. Frontiers in Microbiology 2021, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paulson JN, Stine OC, Bravo HC, Pop M: Differential abundance analysis for microbial marker-gene surveys. Nat Methods 2013, 10:1200–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martín-Fernández J-A, Hron K, Templ M, Filzmoser P, Palarea-Albaladejo J: Bayesian-multiplicative treatment of count zeros in compositional data sets. Statistical Modelling 2015, 15:134–158. [Google Scholar]
- 67.Gloor GB, Macklaim JM, Vu M, Fernandes AD: Compositional uncertainty should not be ignored in high-throughput sequencing data analysis. Austrian Journal of Statistics 2016, 45:73–87. [Google Scholar]
- 68.Zhang X, Yi N: NBZIMM: negative binomial and zero-inflated mixed models, with application to microbiome/metagenomics data analysis. BMC Bioinformatics 2020, 21:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu T, Gallins P, Zhou Y-H: A zero-inflated beta-binomial model for microbiome data analysis. Stat 2018, 7:e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu L, Paterson AD, Turpin W, Xu W: Assessment and Selection of Competing Models for Zero-Inflated Microbiome Data. PLOS ONE 2015, 10:e0129606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu M-C, Pavlicova M, Nunes EV: Zero-Inflated and Hurdle Models of Count Data with Extra Zeros: Examples from an HIV-Risk Reduction Intervention Trial. The American Journal of Drug and Alcohol Abuse 2011, 37:367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ji X, Tsao D, Bai K, Tsao M, Xing L, Zhang X: scAnnotate: an automated cell type annotation tool for single-cell RNA-sequencing data. 2022, doi: 10.1101/2022.02.19.481159. [DOI] [PMC free article] [PubMed]
- 73.Wang S, McCormick TH, Leek JT: Methods for correcting inference based on outcomes predicted by machine learning. Proceedings of the National Academy of Sciences 2020, 117:30266–30275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Argelaguet R, Velten B, Arnol D, Dietrich S, Zenz T, Marioni JC, Buettner F, Huber W, Stegle O: Multi‐Omics Factor Analysis—a framework for unsupervised integration of multi‐omics data sets. Mol Syst Biol 2018, 14:e8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cai Z, Poulos RC, Liu J, Zhong Q: Machine learning for multi-omics data integration in cancer. iScience 2022, 25:103798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Osborne J, Waters E: Four assumptions of multiple regression that researchers should always test. Practical Assessment, Research, and Evaluation 2019, 8. [Google Scholar]
- 77.Hughes JB, Hellmann JJ: The Application of Rarefaction Techniques to Molecular Inventories of Microbial Diversity. In Methods in Enzymology. . Academic Press; 2005:292–308. [DOI] [PubMed] [Google Scholar]
- 78.Dong M, Li L, Chen M, Kusalik A, Xu W: Predictive analysis methods for human microbiome data with application to Parkinson’s disease. PLOS ONE 2020, 15:e0237779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin W, Shi P, Feng R, Li H: Variable selection in regression with compositional covariates. Biometrika 2014, 101:785–797. [Google Scholar]
- 80.Aitchison J, Bacon-Shone J: Log contrast models for experiments with mixtures. Biometrika 1984, 71:323–330. [Google Scholar]
- 81.Topçuoğlu BD, Lesniak NA, Ruffin MT, Wiens J, Schloss PD: A Framework for Effective Application of Machine Learning to Microbiome-Based Classification Problems. mBio 2020, 11:e00434–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matsuki K, Kuperman V, Van Dyke JA: The Random Forests statistical technique: An examination of its value for the study of reading. Sci Stud Read 2016, 20:20–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang X-W, Liu Y-Y: Comparative study of classifiers for human microbiome data. Medicine in Microecology 2020, 4:100013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tolosana-Delgado R, Talebi H, Khodadadzadeh M, van den Boogaart KG: On machine learning algorithms and compositional data. In Proceedings of the 8th International Workshop on Compositional Data Analysis (CoDaWork2019): Terrassa, 3–8 June, 2019, 2019, ISBN 978–84-947240–2-2, págs. 172–175. . Universitat Politècnica de Catalunya; 2019:172–175. [Google Scholar]
- 85.Templ M, Templ B: Statistical Analysis of Chemical Element Compositions in Food Science: Problems and Possibilities. Molecules 2021, 26:5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rivera-Pinto J, Egozcue JJ, Pawlowsky-Glahn V, Paredes R, Noguera-Julian M, Calle ML: Balances: a New Perspective for Microbiome Analysis. mSystems 2018, 3:e00053–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiang R, Li WV, Li JJ: mbImpute: an accurate and robust imputation method for microbiome data. Genome Biology 2021, 22:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zeng Y, Li J, Wei C, Zhao H, Tao W: mbDenoise: microbiome data denoising using zero-inflated probabilistic principal components analysis. Genome Biology 2022, 23:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang M, Wang J, Torre E, Dueck H, Shaffer S, Bonasio R, Murray JI, Raj A, Li M, Zhang NR: SAVER: gene expression recovery for single-cell RNA sequencing. Nat Methods 2018, 15:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Callahan BJ, Sankaran K, Fukuyama JA, McMurdie PJ, Holmes SP: Bioconductor Workflow for Microbiome Data Analysis: from raw reads to community analyses. 2016, doi: 10.12688/f1000research.8986.2. [DOI] [PMC free article] [PubMed]
- 91.Cao Q, Sun X, Rajesh K, Chalasani N, Gelow K, Katz B, Shah VH, Sanyal AJ, Smirnova E: Effects of Rare Microbiome Taxa Filtering on Statistical Analysis. Frontiers in Microbiology 2021, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smirnova E, Huzurbazar S, Jafari F: PERFect: PERmutation Filtering test for microbiome data. Biostatistics 2019, 20:615–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ: Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018, 6:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.LeCun Y, Bengio Y, Hinton G: Deep learning. Nature 2015, 521:436–444. [DOI] [PubMed] [Google Scholar]
- 95.Quinn T, Nguyen D, Rana S, Gupta S, Venkatesh S: DeepCoDA: personalized interpretability for compositional health data. In Proceedings of the 37th International Conference on Machine Learning. PMLR; 2020:7877–7886. [Google Scholar]
- 96.Coenders G, Greenacre M: Three approaches to supervised learning for compositional data with pairwise logratios. arXiv:211108953 [cs, stat] 2021, [DOI] [PMC free article] [PubMed]
- 97.Gordon-Rodriguez E, Quinn TP, Cunningham JP: Learning sparse log-ratios for high-throughput sequencing data. Bioinformatics 2022, 38:157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kang G-U, Ibal JC, Lee S, Jang MH, Park Y-J, Kim M-C, Park T-H, Kim M-S, Kim R-H, Shin J-H: Alteration of the Soil Microbiota in Ginseng Rusty Roots: Application of Machine Learning Algorithm to Explore Potential Biomarkers for Diagnostic and Predictive Analytics. J Agric Food Chem 2021, 69:8298–8306. ** This manuscript models the plant pathogen interaction in ginseng root using machine learning algorithm, SVM and correlation metrics, SPIEC-EASI that identifies 30 biomarkers associated with Ginseng rusty root.
- 99. Guo J, Ling N, Li Y, Li K, Ning H, Shen Q, Guo S, Vandenkoornhuyse P: Seed-borne, endospheric and rhizospheric core microbiota as predictors of plant functional traits across rice cultivars are dominated by deterministic processes. New Phytologist 2021, 230:2047–2060. ** Identifies the core and generalist species across plant holobiont in rice cultivars and uses random forest regressor to predict plant functional phenotype.
- 100. Zhang Z, Zhang Q, Cui H, Li Y, Xu N, Lu T, Chen J, Penuelas J, Hu B, Qian H: Composition identification and functional verification of bacterial community in disease-suppressive soils by machine learning. Environ Microbiol 2022, doi: 10.1111/1462-2920.15902. ** Identifies general patterns of bacterial community composition using disease suppressive and disease conducive soils; validated strains through experimental follow up.
- 101. Crosbie DB, Mahmoudi M, Radl V, Brachmann A, Schloter M, Kemen E, Marín M: Microbiome profiling reveals that Pseudomonas antagonises parasitic nodule colonisation of cheater rhizobia in Lotus. New Phytologist 2022, 234:242–255. ** The manuscript aids in design of synthetic community to understand the root nodule symbiosis in healthy and starved Lotus Burtii
- 102. Averill C, Werbin ZR, Atherton KF, Bhatnagar JM, Dietze MC: Soil microbiome predictability increases with spatial and taxonomic scale. Nat Ecol Evol 2021, 5:747–756. ** Develops large fungi and bacteria prediction models at spatial and taxonomic level. This paper also addresses to the issue of compositionality in the Neon, microbiome dataset.