Abstract
Australia's cervical screening program transitioned from cytology to HPV-testing with genotyping for HPV16/18 in Dec’2017. We investigated whether program data could be used to monitor HPV vaccination program impact (commenced in 2007) on HPV16/18 prevalence and compared estimates with pre-vaccination benchmark prevalence. Pre-vaccination samples (2005–2008) (n = 1933; WHINURS), from 25 to 64-year-old women had been previously analysed with Linear Array (LA). Post-vaccination samples (2013-2014) (n = 2989; Compass pilot), from 25 to 64-year-old women, were analysed by cobas 4800 (cobas), and by LA for historical comparability. Age standardised pre-vaccination HPV16/18 prevalence was 4.85% (95%CI:3.81–5.89) by LA; post-vaccination estimates were 1.67% (95%CI:1.21–2.13%) by LA, 1.49% (95%CI:1.05–1.93%) by cobas, and 1.63% (95%CI:1.17–2.08%) for cobas and LA testing of non-16/18 cobas positives (cobas/LA). Age-standardised pre-vaccination oncogenic HPV prevalence was 15.70% (95%CI:13.79–17.60%) by LA; post-vaccination estimates were 9.06% (95%CI:8.02–10.09%) by LA, 8.47% (95%CI:7.47–9.47%) by cobas and cobas/LA. Standardised rate ratios between post-vs. pre-vaccination rates were significantly different for HPV16/18, non-16/18 HPV and oncogenic HPV: 0.34 (95%CI:0.23–0.50), 0.68 (95%CI:0.55–0.84) and 0.58 (95%CI:0.48–0.69), respectively. Additional strategies (LA for all cobas positives; combined cobas and LA results on all samples) had similar results. If a single method is applied consistently, it will provide important data on relative changes in HPV prevalence following vaccination.
Keywords: HPV-Based screening, Cobas 4800, Linear array, Surveillance, HPV vaccination, Prevalence
Highlights
-
•
HPV vaccine effects can be monitored by population surveillance of HPV infections.
-
•
We assessed whether data from HPV screening can be used for population surveillance.
-
•
LA HPV prevalence estimates were compared to cobas 4800 and cobas/LA estimates.
-
•
HPV prevalence varied slightly between testing strategies.
-
•
Additional HPV typing data from screening could be used for long-term surveillance.
1. Introduction
Australia implemented a fully government-funded National HPV Vaccination Program, routinely offering the quadrivalent HPV vaccine through schools, to girls aged 12–13 years since 2007 (plus a 2-year catch-up 2007–2009 for women up to 26 years) and to boys aged 12–13 years since 2013. In 2018, 2 doses of the nonavalent HPV vaccine replaced the 3-dose quadrivalent vaccine schedule. The program has achieved high coverage in young girls and modest coverage among young women in the catch-up (80% nationally for 3-doses in 15-year-olds in 2017 [1], 59–68% in 18–19 year-olds and 30–39% in 20–26 year-olds in 2009 [2]). Substantial reductions in anogenital warts and incidence of high-grade cervical abnormalities [3,4] have been reported in women age-eligible for vaccination [5] as well as a drop of 92% in the prevalence of vaccine-targeted HPV types among 18–35-year-old women (in 2015) compared to pre-vaccination [6]. Due to the change in vaccine type and dose schedule in 2018, and as Australia monitors its progress towards cervical cancer elimination [7], there is a need for continuous and systematic monitoring of the cervical cancer control program, including surveillance of type-specific HPV prevalence within the population.
In December 2017, Australia's National Cervical Screening Program (NCSP) transitioned from 2-yearly cytology-based screening to 5-yearly primary HPV nucleic acid screening with partial genotyping for women aged 25–74 years [8]. To support partial genotyping for screening, clinical assays calibrated to detect oncogenic HPV genomes (types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68 with or without HPV66 as per Australian NCSP) at a clinical threshold that correlate with cervical disease have been approved for use. Most regulatory-approved assays for clinical use distinguish between a small number of individual oncogenic HPV types, while generating a pooled result for other oncotypes. In contrast, assays used in epidemiological studies detect individual oncogenic and non-oncogenic (‘low risk types’) HPV types with higher analytic sensitivity [9]. HPV-based cervical screening now offers the potential for routinely collected high-volume data for monitoring the impact of HPV vaccination, with the major caveat being that HPV detection for screening purposes doesn't aim to detect all HPV present, only oncogenic HPV at levels correlated with the presence of disease. Thus, a major question is whether these data can provide sufficient sensitivity to detect vaccine-relevant epidemiological changes over time.
We used post-vaccination cervical samples from the Compass pilot trial to compare all-age and age-specific oncogenic HPV prevalence estimates for various HPV groupings, based on the cobas 4800 HPV assay (cobas), a clinical assay used in the current NCSP in Australia. We also compared post-vaccination estimates to pre-vaccination benchmarking prevalence based on data from the WHINURS study, using the Linear Array® (LA) HPV genotyping test (Roche Molecular Diagnostics, Pleasnaton, CA, USA) for historical comparability, to confirm the impact of the HPV vaccination program. Furthermore, we assessed prevalence estimates based on combined testing of LA and cobas (in 3 different algorithms) to exemplify the potential use of additional typing data from routinely collected samples from the NCSP for long-term surveillance of infection prevalence in Australia. Finally, we determined all-age and age-specific prevalence of individual oncogenic HPV types in post-vaccination samples.
2. Materials and methods
2.1. Study populations
For the pre-vaccination era, we used data from WHINURS (Women's HPV Indigenous Non-Indigenous Urban Rural Study), a cross-sectional study with adequate representation of women from more remote locations and from Indigenous women which has been described previously [10,11]. 2620 women presenting for routine Pap smear cytology, aged 18–66 years, were recruited from 34 sites across all states and the Northern Territory of Australia between April 2005 and February 2008. Cervical specimens were collected by clinicians into PreservCyt for ThinPrep cytology. Prevalence data used for the current study are based on the analysis of 1933 unvaccinated women aged 25–64 years by LA. WHINURS was approved by 34 site ethics committees and all participants provided informed consent.
For the post-vaccination era, cervical samples were obtained from Compass pilot, the first phase of a large randomised controlled trial of 5-yearly HPV screening versus 2.5-yearly liquid-based cytology (LBC) screening, acting as a sentinel experience for the primary HPV screening program [12]. Briefly, consenting women aged 25–64 years presenting for routine screening at 47 primary practices in Victoria, Australia, provided a cervical sample (collected by a healthcare professional) and were randomised at VCS Pathology at a 1:2:2 allocation to (i) image-read LBC screening with HPV triage of low-grade cytology, (ii) HPV screening, with those HPV16/18 positive referred to colposcopy and with LBC triage for non-16/18 oncogenic types, or (iii) HPV screening with those HPV16/18 positive referred to colposcopy and with dual-stained cytology triage for non-16/18 HPV types. A total of 5006 eligible women were recruited from October 2013 to November 2014. LBC samples were collected in ThinPrep media (Hologic, Marlborough, MA, US). The current study includes 3101 samples from the HPV arms of the pilot. Of note, samples from vaccinated participants would have received the quadrivalent HPV vaccine. Approval for the conduct of this study was obtained from Bellberry human research ethics committee (HREC) (ethics reference ID: 2015-08-579), a national, unaffiliated, non-institution-based HREC, and the Royal Australian College of General Practitioners National Research and Evaluation Ethics Committee (ethics reference: NREEC 13–005). All participants provided written informed consent.
2.2. Laboratory procedures
For WHINURS, sample analysis was conducted as part of other studies with laboratory methods described elsewhere [10,11]; only the resulting HPV prevalence data were used for the current study. Briefly in terms of lab methods, HPV positive samples identified either by the AMPLICOR DNA test (Roche Molecular Systems) or by an “in-house” PGMY09/11-based HPV consensus PCR/ELISA, were genotyped using LA with previously reported modifications [13,14]. Samples were analysed at the World Health Organization Regional HPV Reference Laboratory, Melbourne, Australia.
For Compass pilot, samples were tested with cobas 4800 HPV assay (cobas) (Roche Molecular Diagnostics, Pleasanton, CA, USA) on the same or subsequent day after laboratory receipt in 2013/2014 according to manufacturer's instructions. The cobas 4800 HPV assay is a real-time PCR assay that detects 14 oncogenic HPV types: HPV 16 and HPV 18 individually and a pool of 12 other HPV types (HPV 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68). The remaining sample was concentrated and stored at – 80 °C until use.
In 2019, frozen samples were thawed and resuspended in ThinPrep medium to its original sample volume. This sample was then re-tested by cobas 4800 HPV assay to confirm reproducibility of the original results obtained in 2013/2014.
The leftover extracted material was then analysed by LA to enable comparison between WHINURS pre-vaccination prevalence data (analysed by LA) and Compass data. Samples were treated with the LA buffer with the addition of 10 μL of a 1 M Tris-HCL pH 7.4 solution (ThermoFisher, MA, US) containing 0.09% sodium azide (Sigma-Aldrich, MI, US) [14]. Extracted DNA from all samples was then genotyped according to manufacturer's instructions using LA. Briefly, PCR was performed in a 100 μL volume, using 50 μL of the LA-HPV master mix and 50 μL of the extracted sample from the cobas 4800 system (x 480 module) as the DNA template. The BeeBlot (Bee Robotics Ltd., Gwynedd, United Kingdom), an automated platform for the washing and hybridization steps required for strip-based assays, was used for the hybridization component of the LA assay [13]. Laboratory analyses were conducted at VCS Pathology (Australian Centre for the Prevention of Cervical Cancer, Melbourne, Victoria). It should be noted that LA was assessed against the WHO LabNet Proficiency Panel 2017 and was found to perform above expectations based on the manufacturers limits of detection, with no false positive results.
2.3. Statistical analysis
Pre-vaccination HPV prevalence in 10-year age groups (25–34, 35–44, 45–54, and 55–64 years) and overall were determined based on LA. Post-vaccination HPV prevalence for the same age groups and overall were determined based on: (i) LA testing, (ii) cobas testing; (iii) cobas testing of all samples followed by retesting by LA of samples returning a cobas non-16/18 HPV positive result (Table 1).
Table 1.
Testing methods used to determine HPV prevalence in the Compass pilot study samples.a
| Monitoring Strategy | Technology | Samples included in HPV prevalence estimation | Comments |
|---|---|---|---|
| LA | LA | All LA oncogenic positive samples | |
| cobas | cobas | All cobas positive samples | |
| cobas/LA | cobas | All cobas positive samples counted as HPV positive | Only samples testing non-16/18 HPV positive with cobas are re-tested by LA. If LA returns a HPV16 or/and 18 result then the cobas non-16/18 HPV positive sample contributes to HPV16 or 16/18 prevalence. |
| AND | Samples testing cobas non-16/18 HPV positive and LA negative contribute towards non-16/18 HPV prevalence | ||
| LA | Samples with cobas result of non-16/18 HPV positive are counted as HPV16/18 if LA returns a HPV16/18 result |
Additional combined testing strategies were also explored (Algorithm A and B) as presented in appendix Table S1.
Type-specific prevalence was determined in a hierarchical manner for the following oncogenic HPV groupings, where possible, as follows: (a) HPV 16 (with or without the inclusion of other oncogenic HPV types); (b) HPV 16/18 (samples testing positive for HPV16 and/or HPV18, with or without the inclusion of other oncogenic HPV types); (c) non-16/18 HPV types (samples testing positive for one or more of the oncogenic HPV genotypes HPV 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68, excluding HPV 16 and 18); (d) seven oncogenic HPV types in the nonavalent vaccine (samples testing positive for one or more HPV genotypes 16,18, 31, 33, 45, 52, and 58); (e) five oncogenic non-16/18 HPV types in the nonavalent vaccine (samples testing positive for HPV 31, 35, 45, 52 and 58); and (f) all oncogenic HPV types (samples testing positive for oncogenic HPV types including 16 or 18 as detected by cobas). Prevalence for HPV types included in the nonavalent vaccine could not be determined for WHINURS due to pooled data for non-16/18 HPV types; similarly, for cobas, positivity for non-16/18 HPV positive samples is pooled and cannot distinguish between individual HPV types.
As mentioned earlier, LA was used to enable comparison between WHINURS pre-vaccination prevalence data (analysed by LA) and Compass data. As LA is no longer manufactured (commercially unavailable since January 01, 2020), for the purpose of this study the use of LA should be viewed as an example of an assay that provides additional genotyping information. From the combined testing, an LA positive result of HPV16/18 for a cobas positive non-16/18 sample would replace the cobas result due to LA's lower limit of detection for HPV16 [[16], [17], [18]] and the hierarchical manner by which type-specific prevalence was determined (see above). Two additional monitoring strategies combining cobas/LA testing were also considered in order to explore comparability of results across the various testing approaches. Results for these strategies are presented in supplementary materials as they are unlikely to be used in common practice due to the additional costs involved. Algorithm A involves testing of all samples by cobas and LA, and oncogenic HPV prevalence determined by results from either technology, while algorithm B assumes cobas testing of all samples followed by retesting with LA of all samples returning any cobas HPV positive result (see Appendix, Table S1).
Prevalence estimates presented in the results are based on analyses of samples conducted in 2019 only. Cobas results obtained in 2014 on the same samples analysed in 2019, are presented in the appendix with a comparison to the 2019 results and relevant commentary.
To enable comparison between pre-vaccination prevalence estimates derived from WHINURS and post-vaccination estimates based on Compass data, we age standardised them using the Australian Standard Population of June 30, 2001 according to the recommendation by the Australian Bureau of Statistics [15] and for consistency with methodology used in the Australian Cervical Screening Monitoring Reports by the Australian Institute of Health and Welfare [16]. The age-standardised prevalence rates (aPRs) were then used to calculate the standardised rate ratio (SRR) within HPV groupings. SRRs were also used for comparing post-vaccination estimates between LA and the other strategies. Cohen's kappa statistic was used to estimate agreement between cobas results and LA results for all samples analysed. We also investigated the agreement between cobas results from 2013/14 vs. results from 2019 (see supplementary materials). All analyses were conducted using Stata software, version 17.0 (Stata Corp, College Station, Tx, USA).
3. Results
Pre-vaccination HPV prevalence in 10-year age groups determined by LA, based on 1933 samples (WHINURS), is presented in Table 2. Highest HPV16/18 prevalence is observed in the youngest age group (25–34 years), decreasing in older age groups; the most noticeable decrease in prevalence is observed between the 25–34 and the 35–44 year age groups. Similar observations can be made for all oncogenic HPV and other HPV groupings. The aPR for all-age HPV16/18 was 4.85% (95%CI 3.81–5.89), for non-16/18 HPV types was 10.85% (95%CI 9.17–12.52), and for oncogenic HPV was 15.70% (95%CI 13.79–17.60).
Table 2.
Pre-vaccination HPV prevalence (WHINURS) by LA and post-vaccination prevalence (Compass pilot) by 10-year age groups as determined by LA, cobas and cobas/LA.ⱡ
| HPV type | Age | Pre-vaccination |
Post-vaccination |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LA |
LA |
cobas (2019) |
cobas/LAa |
|||||||
| +ve/N | Pr | N | +ve | Pr (95%CI) | +ve | Pr (95%CI) | +ve | Pr (95%CI) | ||
|
HPV16 |
25–34 | 77/1010 | 7.62% (6.06–9.44) | 727 | 8 | 1.10% (0.48–2.16) | 8 | 1.10% (0.48–2.16) | 9 | 1.24% (0.57–2.34) |
| 35–44 | 11/578 | 1.90% (0.95–3.38) | 935 | 13 | 1.39% (0.74–2.37) | 11 | 1.18% (0.59–2.10) | 13 | 1.39% (0.74–2.37) | |
| 45–54 | 5/270 | 1.85% (0.60–4.27) | 794 | 8 | 1.01% (0.44–1.98) | 6 | 0.76% (0.28–1.64) | 6 | 0.76% (0.28–1.64) | |
| 55–64 | 1/75 | 1.33% (0.03–7.21) | 533 | 5 | 0.94% (0.31–2.18) | 3 | 0.56% (0.12–1.64) | 4 | 0.75% (0.20–1.91) | |
| Total | 94/1933 | 4.86% (3.95–5.92) | 2989 | 34 | 1.14% (0.79–1.59) | 28 | 0.94% (0.62–1.35) | 32 | 1.07% (0.73–1.51) | |
| aPR 3.38% (95%CI 2.55–4.32), SE (aPR): 0.42% |
aPR 1.13% (95%CI 0.75–1.51), SE (aPR): 0.19% |
aPR 0.94% (95%CI 0.59–1.29), SE (aPR): 0.18% |
aPR 1.07% (95%CI -.70-1.51) SE (aPR): 0.19% |
|||||||
|
HPV16/18 |
25–34 | 104/1010 | 10.30% (8.49–12.34) | 727 | 13 | 1.79% (0.96–3.04) | 16 | 2.20% (1.26–3.55) | 17 | 2.34% (1.37–3.72) |
| 35–44 | 17/578 | 2.94% (1.72–4.67) | 935 | 21 | 2.25% (1.40–3.41) | 17 | 1.82% (1.06–2.90) | 19 | 2.03% (1.23–3.16) | |
| 45–54 | 7/270 | 2.59% (1.05–5.27) | 794 | 10 | 1.26% (0.61–2.30) | 8 | 1.01% (0.44–1.98) | 8 | 1.01% (0.44–1.98) | |
| 55–64 | 2/75 | 2.67% (0.32–9.30 | 533 | 6 | 1.13% (0.41–2.43) | 3 | 0.56% (0.12–1.64) | 4 | 0.75% (0.20–1.91) | |
| Total | 130/1933 | 6.73% (5.65–7.93) | 2989 | 50 | 1.67% (1.24–2.20) | 44 | 1.47% (1.07–1.97) | 48 | 1.61% (1.19–2.12) | |
| aPR 4.85% (95%CI 3.81–5.89), SE (aPR): 0.53% |
aPR 1.67% (95%CI 1.21–2.13), SE (aPR): 1.21% |
aPR 1.49% (95%CI 1.05–1.93), SE (aPR): 0.22% |
aPR 1.63% (95%CI 1.17–2.08), SE (aPR): 0.23% |
|||||||
|
Non-16/18 HPV types |
25–34 | 172/1010 | 17.03% (14.76–19.49) | 727 | 105 | 14.44% (11.97–17.21) | 105 | 14.44% (11.97–17.21) | 104 | 14.31% (11.84–17.06) |
| 35–44 | 59/578 | 10.21% (7.86–12.97) | 935 | 62 | 6.63% (5.12–8.42) | 59 | 6.31% (4.84–8.06) | 57 | 6.10% (4.65–7.83) | |
| 45–54 | 16/270 | 5.93% (3.42–9.45) | 794 | 32 | 4.03% (2.77–5.64) | 23 | 2.90% (1.84–4.31) | 23 | 2.90% (1.84–4.31) | |
| 55–64 | 7/75 | 9.33% (3.84–18.29) | 533 | 13 | 2.44% (1.30–4.13) | 12 | 2.25% (1.17–3.90) | 11 | 2.06% (1.03–3.66) | |
| Total | 254/1933 | 13.14% (11.67–14.73) | 2989 | 212 | 7.09% (6.20–8.07) | 199 | 6.66% (5.79–7.61) | 195 | 6.52% (5.66–7.47) | |
| aPR 10.85% (95%CI 9.17–12.52), SE (aPR): 0.85% |
aPR 7.39% (95%CI 6.45–8.34), SE (aPR): 0.48% |
aPR 6.97% (95%CI 6.05–7.90), SE (aPR): 0.47% |
aPR 6.84% (95%CI 5.93–7.75), SE (aPR): 0.47% |
|||||||
|
HPV types in 9V vaccine (16,18, 31, 33, 45, 52, 58) |
25–34 | 727 | 59 | 8.12% (6.24–10.34) | 57 | 7.84% (5.99–10.04) | ||||
| 35–44 | 935 | 55 | 5.88% (4.46–7.59) | 47 | 5.03% (3.72–6.63) | |||||
| 45–54 | Not applicableb | 794 | 22 | 2.77% (1.74–4.17) | Not applicableb | 16 | 2.02% (1.16–3.25) | |||
| 55–64 | 533 | 13 | 2.44% (1.30–4.13) | 10 | 1.88% (0.90–3.42) | |||||
| Total | 2989 | 149 | 4.98% (4.23–5.83) | 130 | 4.35% (3.65–5.14) | |||||
| aPR 5.09% (95%CI 4.30–5.89), SE (aPR): 0.41% |
aPR 4.48% (95%CI 3.73–5.23), SE (aPR): 0.38% |
|||||||||
|
Non-16/18 HPV in 9V vac(31,33, 45,52,58) |
25–34 | 727 | 46 | 6.33% (4.67–8.35) | 40 | 5.50 (3.96–7.42) | ||||
| 35–44 | Not applicableb | 935 | 34 | 3.64% (2.53–5.04) | Not applicableb | 28 | 2.99 (2.00–4.30) | |||
| 45–54 | 794 | 12 | 1.51% (0.78–2.63) | 8 | 1.01 (0.44–1.98) | |||||
| 55–64 | 533 | 7 | 1.31% (0.53–2.69) | 6 | 1.13 (0.41–2.43) | |||||
| Total | 2989 | 99 | 3.31% (2.70–4.02) | 82 | 2.74 (2.13–3.39) | |||||
| aPR 3.43% (95%CI 2.76–4.09), SE (aPR): 0.34% |
aPR 2.85% (95%CI 2.24–3.46), SE (aPR): 0.31% |
|||||||||
| All onco-genic HPV | 25–34 | 276/1010 | 27.33% (24.60–30.19) | 727 | 118 | 16.23% (13.62–19.12) | 121 | 16.64% (14.01–19.55) | 121 | 16.64% (14.01–19.55) |
| 35–44 | 76/578 | 13.15% (10.50–16.18) | 935 | 83 | 8.88% (7.13–10.89) | 76 | 8.13% (6.46–10.07) | 76 | 8.13% (6.46–10.07) | |
| 45–54 | 23/270 | 8.52% (5.48–12.51) | 794 | 42 | 5.29% (3.84–7.08) | 31 | 3.90% (2.67–5.50) | 31 | 3.90% (2.67–5.50) | |
| 55–64 | 9/75 | 12.00% (5.64–21.56) | 533 | 19 | 3.56% (2.16–5.51) | 15 | 2.81% (1.58–4.60) | 15 | 2.81% (1.58–4.60) | |
| Total | 384/1933 | 19.87% (18.11–21.72) | 2989 | 262 | 8.77% (7.78–9.84) | 243 | 8.13% (7.17–9.17) | 243 | 8.13% (7.17–9.17) | |
| aPR 15.70% (95%CI 13.79–17.60), SE (aPR): 0.97% | aPR 9.06% (95%CI 8.02–10.09), SE (aPR): 0.53% | aPR 8.47% (95%CI 7.47–9.47), SE (aPR): 0.51% | aPR 8.47% (95%CI 7.47–9.47), SE (aPR): 0.51% | |||||||
aPR: age-standardised prevalence rate; LA: linear array; N: number; Pr: prevalence; SE: standard error; +ve: positive; 9V: nonavalent.
HPV types for cobas; similarly, results provided by WHINURS for non-16/18 types were pooled.
ⱡ Results for algorithms A and B are presented in Supplementary Table S3.
Cobas/LA: cobas all samples/LA for cobas positive non-16/18HPV samples only.
Prevalence for HPV types included in the nonavalent vaccine could not be determined as positivity for non-16/18 HPV positive samples is pooled and cannot distinguish between individual.
There were 3101 post-vaccination samples (Compass pilot) available for HPV testing. 112 samples were subsequently excluded (see Appendix for details) leaving 2898 samples with results for cobas and LA. Oncogenic HPV was detected in 243 (8.39%) samples by cobas, and in 262 (9.04%) samples by LA. There was substantial agreement (97%, κ = 0.79) between cobas and LA for oncogenic HPV detection (see appendix, Table S2).
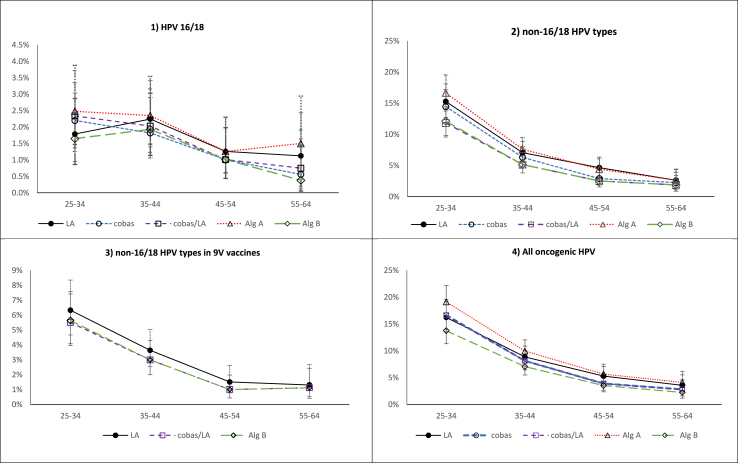
Table 2 presents age-specific and all-age post-vaccination HPV prevalence based on LA, cobas and cobas/LA, for various HPV groupings. Post-vaccination LA-determined HPV16/18 crude prevalence was low (1.79%) in 25–34-year-olds and comparable to prevalence in 35–44-year-olds (2.25%). Decreasing prevalence was observed among the older age groups (Fig. 1). For non-16/18 HPV types, and all oncogenic HPV types, age-specific HPV prevalence estimates indicate a decreasing number of infections with increasing age (Fig. 1). Similar trends in prevalence by age group were also observed for cobas, cobas/LA and algorithms A and B.
Fig. 1.
Prevalence estimates by 10-year age groups based on Linear Array, cobas 4800, cobas/LA and (algorithms A and B) for 1) HPV 16/18, 2) non-16/18 HPV types, 3) non-16/18 HPV types in 9 V vaccine and 4) any oncogenic HPV type.
Post-vaccination LA-determined aPR for HPV16/18 was 1.67% (95%CI 1.21–2.13), 7.39% (95%CI 6.45–8.34) for non-16/18 HPV types and 9.06% (95%CI 8.02–10.09) for oncogenic HPV. SRRs of post-vs. pre-vaccination aPRs for HPV16/18, non-16/18 HPV and for oncogenic HPV were 0.34 (95%CI 0.23–0.50), 0.68 (95%CI 0.55–0.84) and 0.58 (95%CI 0.48–0.69), respectively, indicating statistically significant differences between estimates from these two time points (Table 3).
Table 3.
Standardised rate ratios based on age-standardised prevalence rates comparing pre- and post-vaccination LA estimates and post-vaccination estimates between LA, cobas and other monitoring strategies.
| HPV type | Standardised Rate Ratios |
||||
|---|---|---|---|---|---|
| LA (post-vaccination) vs. WHINURS LA (pre-vaccination) (95%CI) | cobas vs. LA (95%CI) | cobas/LA vs. LA (95%CI) | Alg. A vs LA (95%CI) | Alg. B vs. LA (95%CI) | |
| HPV16 | 0.33 (0.21–0.52) | 0.83 (0.50–1.34) | 0.95 (0.59–1.53) | 1.15 (0.73–1.83) | 0.79 (0.48–1.31) |
| HPV16/18 | 0.34 (0.23–0.59) | 0.90 (0.60–1.34) | 0.98 (0.66–1.45) | 1.17 (0.81–1.71) | 0.80 (0.53–1.21) |
| Non-16/18 HPV types | 0.68 (0.55–0.84) | 0.94 (0.79–1.13) | 0.93 (0.77–1.11) | 1.14 (0.96–1.35) | 0.79 (0.65–0.96) |
| HPV types in 9V vaccine (16,18, 31, 33, 45, 52, 58) | Not applicable* | Not applicable * | 0.88 (0.70–1.10) | Not applicable * | 0.60 (0.47–0.77) |
| Non-16/18 HPV in 9V vaccine (31,33, 45,52,58) | Not applicable* | Not applicable * | 0.83 (0.62–1.11) | Not applicable * | 0.84 (0.63–1.12) |
| All oncogenic HPV | 0.58 (0.48–0.69) | 0.93 (0.79–1.10) | 0.93 (0.79–1.10) | 1.14 (0.98–1.34) | 0.79 (0.67–0.94) |
*Prevalence for HPV types included in the nonavalent vaccine could not be determined as positivity for non-16/18 HPV positive samples is pooled and cannot distinguish between individual HPV types for cobas; similarly, results provided by WHINURS for non-16/18 types were pooled.
The post-vaccination aPR for HPV16/18 by LA [1.67 (95% CI1.21–2.13)] was similar to estimates by cobas [aPR: 1.49 (95%CI 1.05–1.93)], and cobas/LA [aPR: 1.63 (1.17–2.08)]. The aPR for oncogenic HPV by LA [aPR: 9.06 (95%CI 8.02–10.09)] was also comparable to the aPR by cobas and cobas/LA [aPR: 8.47 (95%CI 7.47–9.47)]]. Comparable results were also obtained for oncogenic types in the nonavalent vaccine [cobas/LA vs. LA: SRR = 0.88 (0.70–1.10)] and non-16/18 HPV types in the nonavalent vaccine [cobas/LA vs LA: SRR = 0.83 (0.62–1.11)]. Estimates obtained for algorithms A and B were also in line with the results above (Appendix, Table S3).
All-age and age-specific HPV prevalence estimates among HPV groupings determined by LA tended to be higher compared to estimates by cobas and cobas/LA, indicated by the ratio of estimates being over 1 (Appendix, Table S4). Overall, ratios tended to increase with age, being highest for 45–54 or 55-64-year-olds, depending on the monitoring strategy, driven by the lower detection of HPV16/18 by cobas compared to LA.
Post-vaccination, the highest type-specific HPV prevalence for non-16/18 HPV types detected by LA was HPV39 in 25–34 year-olds (26/727), HPV66 in 35–44 (15/936) and 45-54 year-olds (11/795), and HPV33/52 (3/533) in 55–64 year-olds (Table 4). The highest type-specific HPV prevalence by cobas/LA, was HPV39 (25/727), HPV31 (13/935), HPV 66 (8/794) and HPV 52 (3/533) for 25–34, 35–44, 45–54 and 55-64 year-olds, respectively (Table 4; full list of type-specific prevalence by LA see Table S5 in Appendix). Comparing LA to cobas/LA, and considering the within-age strata, the three most prevalent non-16/18 HPV types were similar among participants.
Table 4.
Oncogenic HPV types in order of rank and HPV type number in the Compass pilot by 10-year age group as detected by Linear Array alone and cobas/LA.
| HPV Rank order | Age group 25-34 y (n = 727) |
Age group 35-44 y (n = 936) |
Age group 45-54 y (n = 795) |
Age group 55-64 y (n = 533) |
Age group 25-64 y (n = 2989) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LA | Pr (%) | Cobas/LA | Pr (%) | LA | Pr (%) | Cobas/LA | Pr (%) | LA | Pr (%) | Cobas/LA | Pr (%) | LA | Pr (%) | Cobas/LA | Pr (%) | LA | Pr (%) | Cobas/LA | Pr (%) | |
| 1 | 39 | 3.58 | 39 | 3.44 | 66 | 1.69 | 31 | 1.39 | 66 | 1.38 | 66 | 1.01 | 16 | 0.94 | 52 | 0.56 | 66 | 1.54 | 39 | 1.34 |
| 2 | 51 | 3.30 | 51 | 2.61 | 16 | 1.39 | 16 | 1.28 | 16 | 1.01 | 16 | 0.76 | 33 | 0.56 | 16 | 0.38 | 39 | 1.40 | 66 | 1.07 |
| 3 | 66 | 2.48 | 52 | 1.93 | 31 | 1.39 | 66 | 1.07 | 51 | 1.01 | 39 | 0.63 | 52 | 0.56 | 33 | 0.38 | 51 | 1.27 | 31 | 0.94 |
| 4 | 52 | 2.20 | 53 | 1.79 | 52 | 1.07 | 39 | 0.96 | 39 | 0.75 | 45 | 0.63 | 51 | 0.38 | 58 | 0.38 | 16 | 1.14 | 16 | 0.90 |
| 5 | 31 | 1.65 | 66 | 1.79 | 39 | 0.96 | 52 | 0.86 | 45 | 0.63 | 56 | 0.50 | 58 | 0.38 | 31 | 0.19 | 52 | 1.14 | 51 | 0.90 |
| 6 | 59 | 1.65 | 31 | 1.65 | 18 | 0.85 | 18 | 0.64 | 52 | 0.63 | 51 | 0.38 | 66 | 0.38 | 35 | 0.19 | 31 | 0.94 | 52 | 0.87 |
| 7 | 45 | 1.38 | 59 | 1.51 | 33 | 0.75 | 45 | 0.64 | 56 | 0.50 | 18 | 0.25 | 18 | 0.19 | 39 | 0.19 | 45 | 0.74 | 45 | 0.70 |
| 8 | 16 | 1.10 | 45 | 1.38 | 45 | 0.75 | 56 | 0.64 | 18 | 0.38 | 31 | 0.25 | 31 | 0.19 | 51 | 0.19 | 59 | 0.70 | 59 | 0.64 |
| 9 | 58 | 1.10 | 68 | 1.10 | 56 | 0.75 | 59 | 0.64 | 59 | 0.38 | 33 | 0.25 | 35 | 0.19 | 56 | 0.19 | 33 | 0.60 | 56 | 0.57 |
| 10 | 68 | 1.10 | 16 | 0.96 | 59 | 0.64 | 58 | 0.53 | 31 | 0.25 | 35 | 0.25 | 39 | 0.19 | 66 | 0.19 | 56 | 0.60 | 18 | 0.43 |
| 11 | 35 | 0.96 | 35 | 0.96 | 35 | 0.53 | 33 | 0.43 | 33 | 0.25 | 59 | 0.25 | 56 | 0.19 | 18 | 0.00 | 18 | 0.57 | 33 | 0.43 |
| 12 | 33 | 0.83 | 58 | 0.83 | 58 | 0.53 | 51 | 0.43 | 35 | 0.25 | 52 | 0.13 | 45 | 0.00 | 45 | 0.00 | 58 | 0.53 | 58 | 0.43 |
| 13 | 56 | 0.83 | 56 | 0.83 | 51 | 0.43 | 68 | 0.21 | 58 | 0.13 | 58 | 0.00 | 59 | 0.00 | 59 | 0.00 | 35 | 0.50 | 35 | 0.37 |
| 14 | 18 | 0.69 | 33 | 0.69 | 68 | 0.21 | 35 | 0.11 | 68 | 0.13 | 68 | 0.00 | 68 | 0.00 | 68 | 0.00 | 68 | 0.37 | 68 | 0.33 |
Cobas/LA: cobas testing of all samples and LA re-testing of cobas non-16/18 positive samples only; LA: linear array; Pr: prevalence; y; years.
4. Discussion
In this study we found consistency of HPV positivity results between LA, cobas and combined cobas/LA monitoring strategies when analysing post-vaccination cervical samples. Age-specific HPV prevalence patterns were consistent between strategies; total oncogenic HPV prevalence and non-16/18 oncogenic HPV prevalence decreased with increasing age whereas HPV16/18 prevalence was comparable between 25-34 and 35-44 year-olds (accounting for the wide confidence intervals) in Compass pilot, decreasing in older age groups. The fall in detection of HPV16/18 in the younger age group was not observed in pre-vaccination data which showed higher rates of HPV 16/18 detection in 25–34-year olds. Individual prevalence estimates for oncogenic HPV prevalence, HPV16/18 and non-16/18 HPV types were overall higher by LA compared to cobas, and cobas/LA as confirmed by their ratio. This ratio increased with increasing age, with the highest ratio observed in women aged 45–64 years, which was driven by lower detection of HPV by cobas.
Comparable age-specific HPV prevalence patterns between LA, cobas and combined testing monitoring strategies support the potential usefulness of routinely collected cobas test results from primary HPV screening for additional examination of extended typing information to monitor vaccine impact. Previous studies have reported good agreement between LA and cobas technologies for the detection of HPV16/18 and other oncogenic HPV genotypes (≥90% agreement) [17,18]. However, a comparison of prevalence estimates for population surveillance using cervical screening samples has not been previously assessed. In a national US study, type-specific HPV prevalence by LA was compared to type-specific prevalence by LA on samples that first tested positive with the Digene Hybrid Capture 2 (HC-2) clinical test [9]. The relative prevalence of individual HPV types was reported to be similar for both approaches, but absolute prevalence estimates varied widely [HPV16/18 prevalence: 6.2% (LA) vs 2.4% (HC-2/LA); oncogenic HPV: 23.7% (LA) vs 7.3% (HC-2/LA)]. This could be due to the use of HC-2 which is a non-PCR HPV test and the analysis of self-collected cervico-vaginal specimens instead of clinician-collected samples. A meta-analysis found that HPV testing of self-collected samples using signal-based assays is less sensitive than testing clinician-collected samples whereas PCR-based HPV assays show comparable sensitivity on self-collected and clinician-collected samples [19].
We evaluated re-testing of cobas non-16/18 positive samples by LA and found that post-vaccination prevalence estimates were comparable to estimates by cobas (and LA) for all HPV groupings, with no evidence of substantial difference as indicated by overlapping confidence intervals. We present this combined cobas/LA monitoring strategy as an example of how additional genotyping information from samples used in routine screening could be sensitive enough to show trends in HPV prevalence over time. LA was used for historical comparability with pre-vaccination prevalence data which is important for benchmarking. In terms of cobas, although the cobas 4800 system was used in our study, results would be generalisable to cobas 6800 and 8800 systems. We previously undertook a clinical validation of the Roche cobas HPV test used on the above mentioned high-throughput systems compared to the reference Roche cobas 4800 HPV assay [20]. Results indicated that the cobas HPV test is of equivalent performance (statistically non-inferior) for the purpose of detection of clinically relevant HPV infection as the cobas 4800 HPV assay, although there may be a possibility for the cobas HPV test to be more sensitive for the detection of HPV16 based on data in that study and a comparison of the limits of detection of the two assays. In Australia, assays other than cobas have been approved for nucleic acid amplification testing of samples within the screening program as long as they meet the National Pathology Accreditation Advisory Council requirements [21]. Examples of approved assays which can provide more detailed identification of individual oncogenic genotypes include the Abbott Alinity m HPV test, the BD Onclarity HPV assay and the Seegene Anyplex II HR HPV assay.
We observed that the ratio of LA over cobas HPV 16/18 prevalence increased with age, driven by an apparent lower detection of HPV types by cobas compared to LA. The highest ratio was observed in women aged 45–64 years. Intermittently detected persistent infections with low viral loads have previously been reported in older women [22] without cytological presentations of precancer or cancer [23]. As LA has a higher sensitivity, detecting copies of viral DNA at a lower threshold than cobas, higher prevalence maybe due to LA detecting non-clinically significant infections. Indeed, the assays have different limits of detection, defined as the level of HPV DNA in the sample that yields positive test results in ≥95% of the replicates; for example LA has a limit of detection of 200 copies/mL and 1200 copies/mL for HPV16 and 18, respectively whereas cobas 4800 corresponding limits of detection are 600 copies/mL for HPV16 and 18 [[24], [25], [26]]. LA was selected as a genotyping test for historical comparability because it was used in WHINURS, from which our pre-vaccination data was obtained, as well as other vaccine effectiveness studies for HPV genotype prevalence surveillance [[27], [28], [29]].
Consistent with previous evidence from Australia [6,30], compared to pre-vaccination estimates, we observed a decrease in HPV 16/18 prevalence post-vaccination among vaccine-eligible 25–34-year-olds demonstrating the significant impact of the HPV vaccine on targeted HPV infections. Prevalence in this age group was low (crude PR: 1.65–2.48%) and comparable to prevalence in the unvaccinated 35–44 age group (crude PR: 1.82–2.35%). These findings are in agreement with previous results from a post-vaccination Australian study [31] confirming the effects of vaccination in driving HPV16/18 infections down. In contrast, non-16/18 HPV type prevalence in our study was higher in 25–34 than the 35-44-year-olds, as shown previously [31]. In addition, we also observed a drop in SRR between pre- and post-vaccination aPRs in non-HPV16/18 types, with confidence intervals less than unity. A cross-protective effect of the quadrivalent vaccine against other HPV types could be a possible explanation. A systematic review of randomised controlled trials and observational studies reported consistent effectiveness of the quadrivalent vaccine against HPV 31 and 45 [32].
As expected, post-vaccination prevalence of non-16/18 HPV types is high, especially among women aged 25–34 yeas [crude PR:12.10%–16.64% (including algorithm A and B data)], including prevalence of non-16/18 HPV types in the nonavalent vaccine (crude PR: 5.50%–6.33% in 25–34 year olds). With vaccination of successive cohorts with the nonavalent vaccine, prevalence of targeted non-16/18 HPV types is expected to decrease gradually, however, there will still be a risk of cervical abnormalities developing from infections with (less) oncogenic HPV types not included in the vaccine. We found that non-vaccine targeted HPV types 39, 51 and 66 ranked among the most prevalent oncogenic types in women of all ages. Caution is needed in interpreting the ranking results, due to the low numbers of positive cases, small sample sizes within specific-type groups and consequent uncertainty.
Our study is subject to potential limitations. We compared pre-vaccination prevalence estimates from WHINURS to post-vaccination estimates from Compass pilot. Although both studies were conducted in Australia, WHINURS recruited women attending health clinics across Australia, whereas Compass pilot participants were recruited only from the state of Victoria. Differences in population sampling between studies may have contributed to differences between pre- and post-vaccination prevalence estimates. It should also be noted that prevalence estimates for the WHINURS cohort in this study cannot be directly compared with estimates from previous WHINURS publications due to different age groups and age ranges used between analyses. Secondly, post-vaccination prevalence estimates from Compass pilot samples may not be generalisable nationally. However, HPV vaccine coverage in Victorian females aged 18–26 years during the catch-up (2007–2009) was comparable to the national average [33]. Also, we didn't know the HPV vaccination status of Compass participants which would have enabled stratification of results by vaccination status. Linkage between Compass and the Australian Immunisation Register has been initiated and will be completed in due course. Thirdly, in this study we used clinician-collected samples though universal access to self-collection is now an option for all screening participants (since July 2022) [34]. Differences in sampling methods may impact prevalence surveys and should be considered in any future studies for population surveillance. In a previous study, we found high observed agreement in HPV16/18 results and good agreement for non-16/18 HPV oncogenic results between self-collected and practitioner-collected specimens using six PCR-based HPV assays [35].
In conclusion, population surveillance of type-specific HPV prevalence could be used to monitor long-term effects of HPV vaccination including declines in targeted types, herd effects, impact of program features (e.g. catch-ups programs), changes in vaccine schedule/or type, addition of male vaccination, and monitoring for possible type-replacement effects. We evaluated cobas, LA and 3 monitoring strategies combining the two technologies, demonstrating similar age-specific prevalence patterns for all oncogenic HPV types, HPV 16/18, and non-16/18 HPV types and prevalence ratios of LA over cobas and combined testing strategies. Findings from this study support the use of routinely collected data from primary HPV-based screening programs for long-term monitoring of HPV infection prevalence in screened populations.
Author statement
Karen Canfell, Marion Saville, David Hawkes: conceptualization, methodology. Khurran Karrim: sample analysis; Suzanne Garland, Julia Brotherton: provision on WHINURS data; Lara Roeke: Compass Pilot project management; Michael Caruana: data curation and formal statistical analysis; David Hawkes, Marion Saville, Karen Canfell: resources; Karen Canfell, Phillip Castle, Julia Brotherton: supervision; Louiza Velentzis: original draft and visualization; all authors: writing, review and editing; Marion Saville, Karen Canfell: funding acquisition.
Declaration of interests
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: KC and MS are co-PIs, and MC is an investigator, of an investigator-initiated trial of cervical screening, Compass, run by the Australian Centre for Prevention of Cervical Cancer (ACPCC), a government-funded not-for-profit charity. Compass receives infrastructure support from the Australian government and the ACPCC has received equipment and a funding contribution from Roche Molecular Diagnostics, USA. KC is also co-PI on a major implementation program Elimination of Cervical Cancer in the Western Pacific which has received support from the Minderoo Foundation and the Frazer Family Foundation and equipment donations from Cepheid Inc. KC receives contract funding from Commonwealth Department of Health, Australia to her institution for work to monitor the safety of the National Cervical Screening Program. KC also receives support for a range of other Australian and international government projects including support from philanthropic organizations, WHO, and government agencies related to cervical cancer control.
MS, DH, JMLB, and KAK state receiving free testing kit donations for research purposes from some of the following comparies: Roche, Seegene, Abbott, Becton Dickinson, Cepheid, AusDiganostics, Copan, Qiagen and Atila Biosystems. MAS receives salary support via fellowship grants from the National Health and Medical Research Council (NHMRC) of Australia and Cancer Institute NSW and contracts paid to her institution (the Daffodil Centre) with the Commonwealth Department of Health (Australia) and National Screening Unit (New Zealand). MS holds NHMRC grants for 5 projects, is the Director for Cancer Council Australia and Co-chair of HPV test characteristics expert panel and consultant for Cancer Care Ontario. DH reports payment to his institution by Roche for presentations, lectures, etc; conference registration fees paid by Roche, Abbott and Qiagen and membership of the Quality and Safety monitoring committee for the Australian National Cervical Screening Program (unpaid). PEC reports receipt of HPV tests and assays at a reduced or no cost for research only from Roche, Atila, Cepheid, Becton Dickinson, and Arbor Vita. SMG reports an an NHMRC Leadership Investigator grant, an investigator-initiated grant to her institution on HPV in young women; lecture fees from Merck for work performed in personal time; participation in the Global Advisory Board on HPV by Merck; being past and inaugural president of the Asia Oceania research 396 organization on Genital Infections and Neoplasia (AOGIN) and Vice President of the International Papillomavirus Society (IPVS) (unpaid). CMW reports grants to her institutions from the US National Institute of Allergy and Infectious Diseases, the US National Cancer Institute, Hologic and Becton Dickinson; receiving reagents and equipment from Roche Molecular Systems and Roche/Ventana Medical Systems through her institution outside the submitted work and personal fees from Becton Dickinson outside the submitted work. CDW is Deputy Chair of The Australian Centre for the Prevention of Cervical Cancer (formerly VCS Foundation Pty Ltd); owns shares in CSL Pty Ltd; has received Honoraria from BioGen, Merck and Seqirus and sponsorship to attend EOGIN 2019 from Seqirus; is a member of the Quality and Safety Monitoring Committee of the National Cervical Screening Program. LSV and JT have no conflicts of interests to declare.
Acknowledgements
We would like to thank all participants of the WHINURS and Compass pilot study for contributing to this research.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tvr.2023.200255.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
The data that has been used is confidential.
References
- 1.Australian Government, Department of Health Historical human papillomavirus (HPV) immunisation coverage rates. Historical reports showing vaccination coverage with 3 doses of the HPV vaccine for adolescents turning 15 years of age by year for each state and territory. https://www.health.gov.au/sites/default/files/documents/2019/12/historical-human-papillomavirus-hpv-immunisation-coverage-rates-females--2017.pdf
- 2.Australian Government, Department of Health National HPV vaccination coverage for the female catch up cohort by year of age. Historical reports showing national human papillomavirus (HPV) vaccination coverage for the female catch up cohort by dose number (1, 2 or 3) and single age cohort (12–26 years) for the specified year. https://www.health.gov.au/sites/default/files/documents/2019/12/national-hpv-vaccination-coverage-for-the-female-catch-up-cohort-by-year-of-age-2009--final-data_0.pdf
- 3.Drolet M., Bénard É., Pérez N., Brisson M. HPV Vaccination Impact Study Group, Population-level impact and herd effects following the introduction of human papillomavirus vaccination programmes: updated systematic review and meta-analysis. Lancet. 2019;394:497–509. doi: 10.1016/S0140-6736(19)30298-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith M.A., Liu B., McIntyre P., Menzies R., Dey A., Canfell K. Fall in genital warts diagnoses in the general and indigenous Australian population following implementation of a national human papillomavirus vaccination program: analysis of routinely collected national hospital data. J. Infect. Dis. 2015;211:91–99. doi: 10.1093/infdis/jiu370. [DOI] [PubMed] [Google Scholar]
- 5.Australian Institute of Health and Welfare National cervical screening program monitoring report 2019. 2019. https://www.aihw.gov.au/getmedia/fcacac12-cd05-4325-88bc-5529a61b53f3/aihw-can-132.pdf.aspx?inline=true Cat. no. CAN 132. Canberra: AIHW.
- 6.Machalek D.A., Garland S.M., Brotherton J.M.L., Bateson D., McNamee K., Stewart M., Skinner S.R., Liu B., Cornall A.M., Kaldor J.M., Tabrizi S.N. Very low prevalence of vaccine human papillomavirus types among 18- to 35-year old Australian women 9 Years following implementation of vaccination. J. Infect. Dis. 2018;217:1590–1600. doi: 10.1093/infdis/jiy075. [DOI] [PubMed] [Google Scholar]
- 7.NHMRC Centre of Research Excellence in Cervical Cancer Control Cervical Cancer Elimination Progress Report: Australia's progress towards the elimination of cervical cancer as a public health problem. 2021. https://www.cervicalcancercontrol.org.au/wp-content/uploads/2021/03/2021-C4-CRE-Elim-Report.pdf
- 8.Cancer Council Australia Cervical Cancer Screening Guidelines Working Party National Cervical Screening Program: guidelines for the management of screen-detected abnormalities, screening in specific populations and investigation of abnormal vaginal bleeding. https://www.cancer.org.au/clinical-guidelines/cervical-cancer-screening/?title=Guidelines:Cervical_cancer/Screening Sydney: Cancer Council Australia.
- 9.Meites E., Lin C., Unger E.R., Steinau M., Patel S., Markowitz L.E., Hariri S. Can clinical tests help monitor human papillomavirus vaccine impact? Int. J. Cancer. 2013;133:1101–1106. doi: 10.1002/ijc.28115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garland S.M., Brotherton J.M.L., Condon J.R., McIntyre P.B., Stevens M.P., Smith D.W., Tabrizi S.N., WHINURS study group Human papillomavirus prevalence among indigenous and non-indigenous Australian women prior to a national HPV vaccination program. BMC Med. 2011;9:104. doi: 10.1186/1741-7015-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brotherton J.M.L., Condon J., McIntyre P.B., Tabrizi S.N., Malloy M., Garland S.M. Human papillomavirus prevalence to age 60 years amongst Australian women pre-vaccination. Sex. Health. 2015;12:353–359. doi: 10.1071/SH15035. [DOI] [PubMed] [Google Scholar]
- 12.Canfell K., Caruana M., Gebski V., Darlington-Brown J., Heley S., Brotherton J.M.L., Gertig D., Jennett C.J., Farnsworth A., Tan J., Wrede C.D., Castle P.E., Saville M M. Cervical screening with primary HPV testing or cytology in a population of women in which those aged 33 years or younger had previously been offered HPV vaccination: results of the COMPASS pilot randomised trial. PLoS Med. 2017;14 doi: 10.1371/journal.pmed.1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevens M.P., Garland S.M., Tabrizi S.N. Human papillomavirus genotyping using a modified linear array detection protocol. J. Virol. Methods. 2016;135:124–126. doi: 10.1016/j.jviromet.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Stevens M.P., Garland S.M., Tabrizi S.N. Validation of an automated detection platform for use with the Roche linear array human papillomavirus genotyping test. J. Clin. Microbiol. 2008;46:3813–3816. doi: 10.1128/JCM.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Australian Bureau of Statistics 3101.0-Australian demographic statistics. Mar 2013. https://www.abs.gov.au/ausstats/abs@.nsf/Lookup/3101.0Feature+Article1Mar%202013 30th January 2023.
- 16.Australian Institute of Health and Welfare Cancer screening - reports. https://www.aihw.gov.au/reports-data/health-welfare-services/cancer-screening/reports
- 17.Gage J.C., Sadorra M., Lamere B.J., Kail R., Aldrich C., Kinney W., Fetterman B., Lorey T., Schiffman M., Castle P.E. PaP Cohort Study Group, Comparison of the cobas Human Papillomavirus (HPV) test with the hybrid capture 2 and linear array HPV DNA tests. J. Clin. Microbiol. 2012;50:61–65. doi: 10.1128/JCM.05989-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castel P.E., Sadorra M., Lau T., Aldrich C., Garcia F.A.R., Kornegay J. Evaluation of a prototype real-time PCR assay for carcinogenic human papillomavirus (HPV) detection and simultaneous HPV genotype 16 (HPV16) and HPV18 genotyping. J. Clin. Microbiol. 2009;47:3344–3347. doi: 10.1128/JCM.00725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arbyn M., Verdoodt F., Snijders P.J., Verhoef V.M., Suonio E., Dillner L., S> Minozzi C., Bellisario, Banzi R., Zhao F.H., Hillemanns P., Anttila A A. Accuracy of human papillomavirus testing on self-collected versus clinician-collected samples: a meta-analysis. Lancet Oncol. 2014;15:172–183. doi: 10.1016/S1470-2045(13)70570-9. [DOI] [PubMed] [Google Scholar]
- 20.Saville M., Sultana F., Malloy M.J., Velentzis L.S., Caruana M., Ip E.K.O., Keung M.H.T., Canfell K., Brotherton J.M.L., Hawkes D. Clinical validation of the cobas HPV test on the cobas 6800 system for the purpose of cervical screening. J. Clin. Microbiol. 2019;57 doi: 10.1128/JCM.01239-18. e01239–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Australian Government, Department of Health. 2019. The Requirements for Laboratories Reporting Tests for the National Cervical Screening Program. second ed.. https://www1.health.gov.au/internet/main/publishing.nsf/Content/npaac-cervical-screening (accessed 27 September 2022).
- 22.Winer R.L., Xi L.F., Shen Z., Stern J.E., Newman L., Feng Q., Hughes J.P., Koutsky L.A. Viral load and short-term natural history of type-specific oncogenic human papillomavirus infections in a high-risk cohort of midadult women. Int. J. Cancer. 2014;134:1889–1898. doi: 10.1002/ijc.28509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castle P.E., Rodríguez A.C., Burk R.D., Herrero R., Wacholder S., Hildesheim A., Morales J., Rydzak G., Schiffman M. Proyecto Epidemiológico Guanacaste Group, Long-term persistence of prevalently detected human papillomavirus infections in the absence of detectable cervical precancer and cancer. J. Infect. Dis. 2011;203:814–822. doi: 10.1093/infdis/jiq116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linear Array HPV Genotyping Test Kit Insert, Doc Rec. 10.0.
- 25.Rao A., Young S., Erlich H., Boyle S., Krevolin M., Sun R., Apple R., Behrens C. Development and characterization of the cobas human papillomavirus test. J. Clin. Microbiol. 2014;51:1478–1484. doi: 10.1128/JCM.03386-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cornall A.M., Poljak M., Garland S.M., Phillips S., Machalek D.A., Tan J.H., Quinn M.A., Tabrizi Sn S.N. HPV genotype-specific concordance between EuroArray HPV, Anyplex II HPV28 and Linear Array HPV Genotyping test in Australian cervical samples. Papillomavirus. Res. 2017;4:79–84. doi: 10.1016/j.pvr.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poljak M., Cuzick J., Kocjan B.J., Iftner T., Dillner J., Arbyn M M. Nucleic acid tests for the detection of alpha human papillomaviruses. Vaccine. 2012;30:F100–F106. doi: 10.1016/j.vaccine.2012.04.105. [DOI] [PubMed] [Google Scholar]
- 28.Garland S.M., Cornall A.M., Brotherton J.M.L., Wark J.D., Malloy M.J., Tabrizi S.N., on behalf of the VACCINE study group Final analysis of a study assessing genital HPV genoprevalance in young Australian women, following eight years of a national vaccination program. Vaccine. 2018;36:3221–3230. doi: 10.1016/j.vaccine.2018.04.080. [DOI] [PubMed] [Google Scholar]
- 29.Phillips S., Cornall A.M., Machalek D.A., Garland S.M., Bateson D., Garefalakis S.N., Tabrizi S.N. Comparison of the Roche Cobas® 4800 HPV assay to Roche amplicor for detection of high-risk human papillomavirus. J.Clin. Microbiol. Infect. 2016;35:1305–1307. doi: 10.1007/s10096-016-2665-1. [DOI] [PubMed] [Google Scholar]
- 30.Tabrizi S.N., Brotherton J.M., Kaldor J.M., Skinner S.R., Cummins E., Liu B., Bateson D., McNamee K., Garefalakis M., Garland S.M. Fall in human papillomavirus prevalence following a national vaccination program. J. Infect. Dis. 2012;206:1645–1651. doi: 10.1093/infdis/jis590. [DOI] [PubMed] [Google Scholar]
- 31.Brotherton J.M., Hawkes D., Sultana F., Malloy M.J., Machalek D.A., Smith M.A., Garland S.M., Saville M M. Age-specific HPV prevalence among 116,052 women in Australia's renewed cervical screening program: a new tool for monitoring vaccine impact, Vaccine. 2019;37:412–416. doi: 10.1016/j.vaccine.2018.11.075. [DOI] [PubMed] [Google Scholar]
- 32.Brown D.R., Joura E.A., Yen G.P., Kothari S., Luxembourg A., Saah A., Walia A., Perez G., Khoury H., Badgley D., Stanley M. Systematic literature review of cross-protective effect of HPV vaccines based on data from randomized clinical trials and real-world evidence. Vaccine. 2021;39:2224–2236. doi: 10.1016/j.vaccine.2020.11.076. [DOI] [PubMed] [Google Scholar]
- 33.Brotherton J.M., Budd A., Rompotis C., Bartlett N., Malloy M.J., Anderson R.L., Coulter K.A., Couvee P.W., Steel N., Ward G.H., Saville M. Is one dose of human papillomavirus vaccine as effective as three? A national cohort analysis. Papillomavirus Res. 2019;8 doi: 10.1016/j.pvr.2019.100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Australian Government Department of health. Self collection to increase choice within the national cervical screening program. https://www.health.gov.au/news/self-collection-to-increase-choice-within-the-national-cervical-screening-program
- 35.Saville M., Hawkes D., Keung M., Ip E., Silvers J., Sultana F., Malloy M.J., Velentzis L.S., Canfel K., Wrede C.D., Brotherton J. Analytical performance of HPV assays on vaginal self-collected vs practitioner-collected cervical samples: the SCoPE study. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that has been used is confidential.