Abstract
Background
Dendritic cells (DC), the most potent professional antigen presenting cells capable of effective cross-presentation, have been demonstrated to license T helper cells to induce antitumor immunity in solid tumors. Specific DC subtypes are recruited to the injured brain by microglial chemokines, locally adapting to distinct transcriptional profiles. In isocitrate dehydrogenase (IDH) type 1 mutant gliomas, monocyte-derived macrophages have recently been shown to display an attenuated intratumoral antigen presentation capacity as consequence of the local accumulation of the oncometabolite R-2-hydroxyglutarate. The functionality and the contribution of DC to the IDH-mutant tumor microenvironment (TME) remains unclear.
Methods
Frequencies and intratumoral phenotypes of human DC in IDH-wildtype (IDHwt) and -mutant high-grade gliomas are comparatively assessed by transcriptomic and proteomic profiling. DC functionality is investigated in experimental murine glioblastomas expressing the model antigen ovalbumin. Single-cell sequencing-based pseudotime analyses and spectral flow cytometric analyses are used to profile DC states longitudinally.
Results
DC are present in primary and recurrent high-grade gliomas and interact with other immune cell types within the TME. In murine glioblastomas, we find an IDH-status-associated major histocompatibility class I-restricted cross-presentation of tumor antigens by DC specifically in the tumor but not in meninges or secondary lymphoid organs of tumor-bearing animals. In single-cell sequencing-based pseudotime and longitudinal spectral flow cytometric analyses, we demonstrate an IDH-status-dependent differential, exclusively microenvironmental education of DC.
Conclusions
Glioma-associated DCs are relevantly abundant in human IDHwt and mutant tumors. Glioma IDH mutations result in specifically educated, dysfunctional DCs via paracrine reprogramming of infiltrating monocytes, providing the basis for combinatorial immunotherapy concepts against IDH mutant gliomas.
Keywords: cDC1, cDC2, dendritic cell, glioblastoma, glioma microenvironment, IDH mutation, R-2-HG
Key Points.
DC are present in primary and recurrent high-grade gliomas and interact with other immune cell types within the TME.
Glioma-associated DC show limited antigen presentation capacity to T cells.
Glioma-educated DC demonstrate an immature cellular state in IDH-mutated gliomas.
Importance of the Study.
Tumor antigen-presenting dendritic cells (DC) are considered as key cell type for the initiation of antitumor immune responses by priming peripheral T cells. In early clinical trials, DC vaccination approaches showed some signs of efficacy in gliomas. Using recent advances in single-cell sequencing technologies and reference-based mapping, we show that in glioma the biological function of DC is not restricted to priming peripheral T cells. Importantly, blood-borne monocytes themselves accumulate intratumorally during glioma progression and experience a glioma genotype-dependent DC education resulting in diverging immunological capacities. Differential intratumoral genotype-specific DC states provide opportunities to rationally design novel combinatorial immunotherapies for malignant gliomas.
High-grade gliomas (HGG), including isocitrate dehydrogenase (IDH)-wildtype (IDHwt) glioblastomas (GBMs) and WHO grade 3 and 4 IDH-mutant (IDHmut) oligodendrogliomas and astrocytomas, are aggressive tumors originating from the brain with generally poor prognosis.1 There is substantial evidence that different immune cell infiltrates within the tumor microenvironment (TME) are associated with therapy response and prognosis in HGG. Overall, the TME in HGG is considered to be highly immunosuppressive, which highlights the need for TME modulation to improve tumor immunotherapy.2,3 However, there is a substantial need for insights into the complex cellular and molecular interplay between the different cell types in the TME,4,5 including tumor-associated macrophages (TAM) and tumor-infiltrating dendritic cells (DC). While TAM are widely recognized to contribute to T cell suppression and to promote epithelial-to-mesenchymal transition of glioma cells,3,6 there is no evidence for a significant functional role of intratumoral DC in solid tumors to date.
Recent work has shown the transcriptional dynamics of the glioma-associated myeloid cell compartment during disease progression, specifically revealing distinct TAM subsets, which have been shown to be conserved across species and may be relevant targets in the context of immunotherapy.7 We have previously shown that the metabolically perturbed TME inherent to IDH-mutant HGG can be targeted to modulate and reverse its immunosuppressive effects on infiltrating immune cells.8,9
However, the relevance and therapeutic potential of tumor-associated DC within the complex TME of HGG remain poorly understood, with some limited evidence that DC are present in human HGG.4 Here, we aim at deciphering monocyte-to-DC differentiation and the function of glioma-infiltrating DC subsets by defining longitudinal signatures of glioma DC education. By assessment of DC cellular states and putative tumor-genotype dependence, we demonstrate a predictive and prognostic impact of HGG infiltration by DC.
Methods
Ethical Approval for Human Samples
Ethical approval for the analysis of human gliomas and GBM was obtained from the Heidelberg Medical Faculty Ethics Committee (Reference number S-064/2008, March 31, 2008) and the Freiburg Medical Faculty Ethics Committee (Reference number 472/15). Research-only buffy coat formulations from healthy donors were purchased from the Institute of Clinical Transfusion Medicine and Cell Therapy (IKTZ) Heidelberg.
Animal Research Approval
All animal experiments were performed under research approvals issued by the Regierungspräsidium (Administrative Council) Karlsruhe according to the German Animals Welfare Act.
Experimental Animals
C57BL/6J wt mice were purchased from Charles River. Mice were bred according to local regulatory authorities (breeding approval reference Z124I02). All mice were 7–10 weeks of age at use. Mice were kept under specific pathogen-free conditions at the animal facility of the DKFZ Heidelberg.
Cell Lines
Murine glioma cell line GL261 was obtained from the Division of Cancer Treatment and Diagnosis, National Cancer Institute, MD, USA. They were cultured in Dulbecco's Modified Eagle Medium (DMEM, Sigma-Aldrich), supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich), 100 U/ml Penicillin, and 100 μg/ml Streptomycin (Invitrogen) (full DMEM). The GL261-SIINFEKL cell line was provided by M. Kilian and cultured in full DMEM supplemented with 9µg/ml blasticidin (Gibco). GL261-IDHwt and -IDHR132H cell lines were described previously.8
Isolation of Murine Tumor-Infiltrating or Spleen-Derived Immune Cells
Murine GL261-containing brain hemispheres were excised, washed in HBSS (Sigma-Aldrich), and cut into small pieces before tissue disruption in HBSS supplemented with 50 µg/ml Liberase D for 0.5 h under slow rotation at 37 °C. Dispersed tissue was mashed through a 100 µm and 70 μm cell strainer and lymphocytes. For GL261 samples, myelin removal was performed using myelin removal beads II (Miltenyi Biotec; 130-096) or by percoll density gradient as described in Supplementary Methods. Murine splenocytes were isolated by homogenization using a cell strainer and ACK lysis. CD3+ and CD11c+ cells were purified by flow cytometry. In some cases, cells were treated with Brefeldin to prevent secretion of cytokines, chemokines, and other secretory proteins before analysis. If required, single-cell suspensions were labeled with CellTrace Far Red according to the manufacturer’s instructions.
Generation of Human Monocyte-Derived DC
Human monocyte-derived DC were generated by magnetic cell separation of CD14+ monocytes from human peripheral blood mononuclear cells using CD14+ magnetic beads and large selection (LS) positive selection columns (Miltenyi Biotech) according to the manufacturer’s instructions. Cells were counted and resuspended in Roswell Park Memorial Institute medium-1640 (RPMI-1640, PAN-Biotech) with 10% human serum AB (Sigma-Aldrich), 100 U/ml Penicillin, and 100 μg/ml Streptomycin (Invitrogen) and 2 mM L-Glutamine (Invitrogen). 100 ng/ml recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF, Peprotech) and 50 ng/ml recombinant human interleukin-4 (IL-4; Peprotech) were added to the medium, and cells were incubated for 3 days. If not mentioned otherwise, naïve DC were matured on day 3 with 1000 U/ml recombinant human tumor necrosis factor alpha (TNF-α), 10 ng/ml recombinant human IL-1β, 10 ng/ml recombinant human IL-6, and 10 μg/ml recombinant human Prostaglandin E2 (all Peprotech) for 24 h and stimulated with 100 ng/ml LPS (Sigma-Aldrich) and 100 ng/ml recombinant human IFN-γ (Peprotech) overnight. If treated with R-2-Hydroxyglutarate in vitro, cells were treated on day 3 overnight and matured or stimulated subsequently.
Generation of Murine Monocyte-Derived DC
Murine monocyte-derived DC were generated from bone marrow (BM)-derived monocytes by culturing BMDMs in Roswell Park Memorial Institute medium-1640 (RPMI-1640, PAN-Biotech) with 10% FBS (Sigma-Aldrich), 100 U/ml Penicillin, and 100 μg/ml Streptomycin (Invitrogen), 2 mM L-Glutamine (Invitrogen), supplemented with 20 ng/ml recombinant murine GM-CSF and 20 ng/ml recombinant murine IL-4 (both Peprotech) for 6 days. If not stated otherwise, cells were stimulated on day 7 with 100 ng/ml LPS (Sigma-Aldrich) and 100 ng/ml murine IFN-γ (Peprotech) overnight.
Cellular Co-culture Assays
For co-culture assays, antigen-presenting cells (APCs) from C57/BL6J mice were generated as described above and seeded into 48-well plates before treatment with indicated substances. DCs were pulsed with respective OVA peptides (p257-264: SIINFEKL for OT-I T cells and MHC-I/CD8+-restricted immune response and p323-339: ISQAVHAAHAEINEAGR for OT-II T cells and MHC-II/CD4+-restricted immune response as well as the full OVA peptides for both MHC-I/CD8+ and MHC-II/CD4+-restricted immune responses) for 24 h and thoroughly washed with phosphate-buffered saline (PBS, Sigma-Aldrich). Cells were then resuspended in T cell medium containing Roswell Park Memorial Institute medium-1640 (RPMI-1640, PAN-Biotech) with 10% FBS (Sigma-Aldrich), 100 U/ml Penicillin and 100 μg/ml Streptomycin (Invitrogen) and 2 mM l-Glutamine (Invitrogen). Antigen-specific T cells from sex- and age-matched OT-I or OT-II donor mice were added and co-culture was incubated for 48 h (OT-I T cells) or 72 h (OT-II T cells), respectively. Co-cultures were recalled with the respective peptides 5 h before further analysis.
Tumor Cell Inoculation
1 × 105 GL261 or GL261-OVA or GL261-IDHwt/mut tumor cells in 2 µl sterile PBS (Sigma-Aldrich) were stereotactically implanted into the right hemisphere of 7-10 weeks old female C57BL/6J mice (coordinates: 2 mm right lateral of the bregma and 1 mm anterior to the coronal suture with an injection depth of 3mm below the dural surface) using a 10 µl Hamilton micro-syringe driven by a fine step stereotactic device (Stoelting).
Murine Sample Preparation
Mice were sacrificed by overdosing anesthesia. For BM isolation, hind limbs were segregated, and femurs and tibias were exposed. After removal of the spleen, mice were cardially perfused with PBS and tumor-bearing hemisphere was excised. In some cases, meninges were prepared by removing meninges from the dura with sharp tweezers under a sectioning microscope. BM cells were harvested from femurs and tibias by flushing the bones with PBS. Fibrous material from BM and spleen was removed by filtration or meshing through a 40 µm cell strainer, respectively. Erythrocytes were lysed with lysis buffer (150 mM NH4Cl, 10 mM KHCO3, 2 mM NaEDTA) for 3 min at room temperature followed by addition of PBS (Life Technologies) containing 2 % FBS (Sigma-Aldrich). Total cell numbers were determined by trypan blue exclusion.
Spectral Flow Cytometry
First, cells were stained with a live/dead marker by incubating 3 × 106 cells with 1:1000 v/v Live/dead blue (Life Technologies) in 100 µl PBS for 30 min at room temperature. The following washing and incubation steps were performed with fluorescence activated cell sorting (FACS) buffer (PBS [Life technologies] containing 1% FBS [Sigma-Aldrich], 0.09 % NaN3 [Sigma-Aldrich], and 2 mM EDTA [Merck]). Blocking of Fc receptors was done before staining by incubating cells at 4 °C for 20 min with CD16/CD32 mAb either purified or labeled with PerCp/Cy5.5 (progenitor panel). After centrifugation cells were stained with all extracellular antibodies simultaneously at 4 °C for 20 min. The antibodies used are listed in Supplemental Table S1. We used spectral flow cytometry, which allows the use of fluorochromes with similar excitation and emission maxima in a single panel by measuring the entire emission spectrum of a fluorochrome and then unmixing the spectrum to identify individual fluorochromes, to simultaneously detect 28 cellular proteins After washing, cells were fixed with FoxP3 Fix/Perm buffer set (BioLegend) and stored over night at 4 °C in the dark. Intracellular staining was performed in FoxP3 Fix/Perm buffer at room temperature for 30 min. Samples were acquired on an AURORA spectral flow cytometer (Cytek Biosciences) and analyzed using OMIQ software (OMIQ).
Mass Cytometry Data Processing and Analysis
Mass cytometry data was processed and analyzed as described before. Briefly, immune phenotypes of human tumor-infiltrating lymphocytes were visualized using reduced-dimensional (2D) t-distributed stochastic neighbor embedding (SNE) maps generated according to the expression level of all markers used in the panel. For meta-clustering analysis, FlowSOM/ConsensusClusterPlus (doi: 10.12688/f1000research.11622.3) was used with 100 initial SOM-grid points and maxim of 25 meta-clusters. Based on visual inspection of t-SNE plots and heat maps generated at each merging step, for each parent subset a final number of meta-clusters was chosen that merged clusters into populations with consistent phenotypes.
Comparisons of protein expression between conditions was conducted on clusters that contained at least 0.05% of all cells per condition to ensure robust comparisons (>31 cells for antibody panel A and >22 cells for antibody panel B). Clusters that were below this threshold were not considered for visualization and analysis. Pairwise testing was performed using the Kruskal-Wallis test followed by a Dunn post-hoc test with multiple testing adjustment according to the Benjamini-Hochberg method.
Fluorescence-Activated Cell Sorting of Murine Samples
Brain tumor-bearing hemispheres were mechanically dissected, digested with liberase D (50 µg/ml) and meshed through a cell strainer to obtain a single-cell suspension. Myelin was removed using a continuous 30% Percoll gradient. The cell suspension was stained with the following antibodies after blocking of Fc receptors (anti-mouse CD16/32 Antibody, BioLegend, San Diego, USA): anti-CD45.2 (clone 30-F11, BioLegend, San Diego, USA) and fixable viability dye eFluor 780 (Invitrogen), at dilutions according to the manufacturer’s instructions. Cells were sorted under sterile conditions on a BD FACS Aria Fusion equipped with the following lasers: 405 nm; 488 nm; 561 nm; 640 nm, using an 85 µm nozzle and 4-way purity mode.
Multiplex Cytokine Array of Tumor Interstitial Fluid
Murine GL261-containing brain hemispheres were excised, washed in HBSS (Sigma-Aldrich), and cut into small pieces before tissue disruption in HBSS supplemented with 50 µg/ml Liberase D for 0.5 h under slow rotation at 37 °C. Tumor interstitial fluid was isolated from tissue and cells by filtration. For cytokine detection, interstitial fluid was diluted 1:5 in PBS and measured using the Bio-Plex Pro Mouse Cytokine 23-plex Assay (#M60009RDPD, Bio-Rad) in a 96-well plate assay format that includes premixed coupled magnetic beads, detection antibodies, standards, reagents, and diluents for detecting IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-17A, Eotaxin, G-CSF, GM-CSF, IFN-γ, KC, MCP-1 (MCAF), MIP-1α, MIP-1β, RANTES, TNF-α. Assay was performed according to the manufacturer’s instructions. Single cytokine ELISAs of murine IL-6, IL-13, and IL-4 (Invitrogen) were performed according to the manufacturer’s protocol.
RNA Isolation and cDNA Synthesis
For isolation of RNA, cells were lysed directly after sorting without prior washing using TRIzol reagent (Thermo Fisher Scientific) and by passing the cell lysate through a pipette tip repeatedly. After 5 min incubation time to allow complete dissociation of nucleoprotein complexes, supernatant was transferred to 1.5 ml tubes and 200 μl chloroform (Carl Roth) were added. Tubes were shaken vigorously and gradient centrifugation was performed at 12.000g for 15 min at 4 °C to allow for precipitation of protein and other contaminants. The colorless RNA-containing upper phase was transferred to a clean 1.5 ml tube and mixed with an equal volume of 70% ethanol to obtain a final concentration of 35% ethanol. Solution was then vortexed and transferred to a spin cartridge and processed according to the manufacturer’s instructions (TRIzol Plus RNA Purification Kit, Thermo Fisher Scientific). RNA concentration was measured on a NanoDrop 2000 device (Thermo Fisher Scientific) and further purified for further analysis using the RNeasy MinElute Cleanup Kit (QIAGEN). 1 μg of total mRNA was used for cDNA-synthesis using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to manufacturer’s instructions.
Mouse 10× Single-Cell RNA Sequencing
10x libraries were prepared from FACS-purified CD45+ cells in a single run. Read alignment and transcript quantification were conducted using cellRanger v3.1.0. The resulting counts files were analyzed using Seurat v3 [10.1016/j.cell.2019.05.031] dataset integration workflow. The counts table was filtered for features expressed by at least five cells and cells with at least 500 detected features, corresponding to the arguments min.cells = 5 and min.features = 500 in the CreateSeuratObject function call. The data was scaled and normalized using the SCTranform function [10.1186/s13059-019-1874-1] with the function set to return 10 000 variable features and regress out the percentage of mitochondrial genes, corresponding to the arguments, variable.features n = 10 000, return.only.var.genes = F, and vars.to.regress = ”percent.mt”. For dataset integration, 10 000 variable features were used with batch-effect associated features containing the following patterns in their name filtered out Jun, Fos, Gm, Rpl, Rps, Atf3, Zfp36, AY, Egr, Hsp, Malat1, Xist, mt-, Hist, Socs3. These genes were subsequently removed from the RunPCA function. Subsequently, Uniform Manifold Approximation and Projection and shared nearest neighbors graph construction were performed on the top 15 principal components. Clusters were identified with the resolution set to 2.5.
Pseudotime Analysis of Single-Cell Transcriptomes
Pseudotime trajectory analysis of the single-cell RNA-Seq data was conducted using the StemID2 functionality of RaceID3 and the FateID R package with default settings [10.1016/j.stem.2016.05.010, 10.1038/nmeth.4662]. First, a lineage tree was computed using the nearest neighbor mode (nmode = TRUE) with default parameters of StemID2. Then, a list of significant links determined in the previous step was chosen based on the underlying question. A filtered gene expression matrix was obtained through the getfdata function of the FateID R package and used as input for the pseudotime gene expression analysis of cells along the given list of links. Genes expressed at <2 normalized transcripts in at least 10 cells in mice were filtered out using the filterset FateID function. Genes with similar gene expression profiles were grouped into modules on a self-organizing map using the getsom FateID function with the minimal size of the modules set to 3 and the correlation threshold to 0.85. With the help of the procsom function, modules on the self-organizing maps were grouped into larger modules that were used for visualization and downstream gene ontology term and other analyses.
Analysis of Human GBM-Associated Immune Cells
CellPhoneDB v2.1.7 was used with the default parameters. Human 10× counts and metadata from newly diagnosed and recurrent GBM were downloaded from https://www.brainimmuneatlas.org/. Gene symbols were converted to ENSEMBL IDs using the clusterProfiler R package v4.0.5.10 CellPhoneDB analysis and visualization was run from the command line using default settings. The extraction of relevant celltype interactions and genes was achieved using the tidyverse v1.3.1 R package collection in R v4.1.0.
Data Visualization
Tabular data from the analyses above were processed using the tidyverse suite of packages [https://CRAN.R-project.org/package=tidyverse] and visualized in the R programming environment (see below) using the ggplot2 package [see below].
Statistics
Unless otherwise mentioned, data are represented as individual values or as mean ± SEM. Group sizes (n) and applied statistical tests are indicated in figure legends. Significance was assessed by either unpaired t-test analysis, paired t-test analysis, or one-way ANOVA analysis with Tukey post hoc testing as indicated in figure legends. Survival was analyzed by log-rank Mantel-Cox test. Spearman correlation was applied for all correlation analysis and the Kaplan-Meier method was used to examine survival differences. Statistics were calculated using GraphPad Prism 9.0.
Results
DC Infiltration and Interaction in HGG
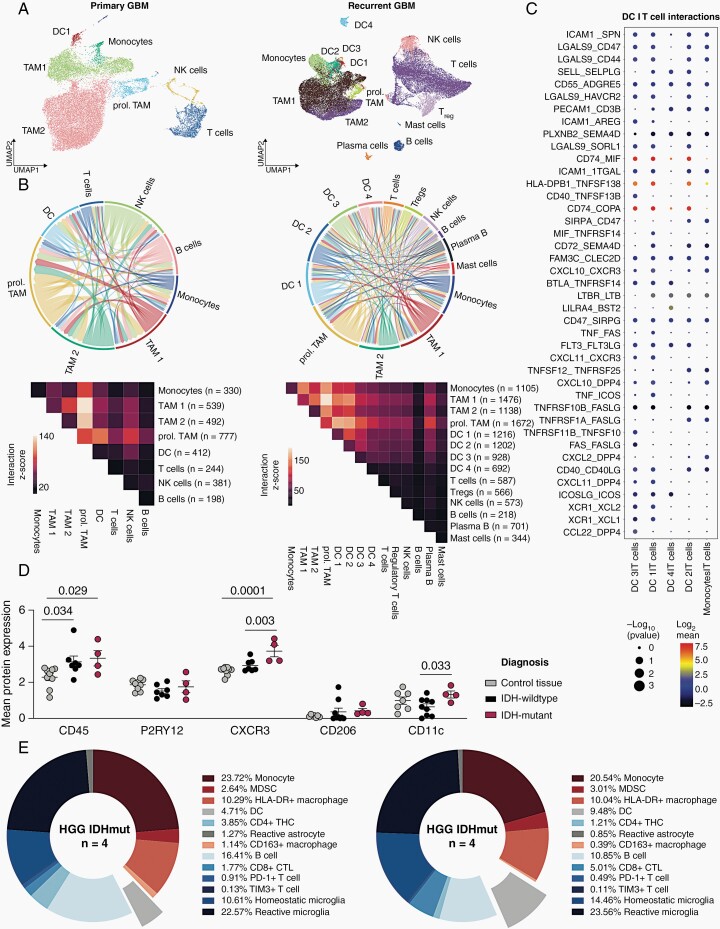
To investigate the relevance and interaction dynamics of infiltrating DC in GBM, we performed cell-cell interaction analyses on publicly available single-cell RNA-sequencing (scRNA-seq) datasets, which include newly diagnosed and recurrent late-stage GBM (Figure 1A and B; Supplementary Figure 1)7,11. While in newly diagnosed GBM, DC seemed to be predominantly interacting within the myeloid compartment and showed very limited interaction with T and B cells (Figure 1B), there was increasing predicted interaction between T cells and DC in the TME of late-stage, recurrent disease. In these tissues, we found the strongest predicted cell–cell interactions to be a direct myeloid cell crosstalk between TAM subsets and DC as well as moderate predicted cell–cell interactions between T cells with TAM and DC, while B cells did not show potent intercellular interactions. Specifically, DC cluster 1 (DC1), corresponding to conventional DC type 1 (cDC1), cluster 2 (DC2), corresponding to migratory DCs, and cluster 3 (DC3), corresponding to plasmacytoid DCs (pDC), show strong cell–cell interaction with conventional T cells, Tregs, and NK cells (Figure 1B). Analysis of co-expressed interaction partner molecules revealed that—in addition to canonical mediators of APCs, such as CD40—CD40LG and SELL (encoding selectin L)—SELPLG (encoding selectin P ligand)—the most highly co-expressed molecules were CD74 on DC1-3 clusters and its ligand macrophage migration inhibitory factor (MIF) on T cells, which has recently been described as chemokine that might be responsible for the migration of pathogenic B cells and immunosuppressive myeloid cells as well as their aberrant proliferation.12,13 DC1 and DC3 clusters exhibited interaction to T cells via XCR1 which suggests the ability to perform cross-presentation of antigens on MHC class I (Figure 1C; Supplementary Figure 2).14 Next, we assessed protein levels of candidates attributed to immune cell infiltration using our previously generated CyTOF dataset of IDHmut and IDHwt HGG.8 The antibody panel used targeted 36 proteins enriched in immune cells.15 Unsupervised clustering was performed of n = 62 239 cells (n = 25 928 control brain tissue; n = 16 764 IDHmut HGG; n = 19 547 IDHwt HGG).8 While tumor samples were enriched for immune cells (CD45) compared to control brain tissue, there was significantly higher expression of CD11c and CXCR3 in IDHmut HGG compared to IDHwt HGG (Figure 1D). This is reflected by a higher relative abundance of DC in IDHmut HGG (9.48%) compared to IDHwt HGG (4,71%) as predicted by reference-based cellular mapping (Figure 1E). Notably, homeostatic microglia were also enriched in the IDHmut HGG cohort (Figure 1E). In summary, we demonstrate that there is a relevant infiltration of DC in human HGG and hypothesize that there is interaction between T cells and tumoral DC subsets in the glioma microenvironment that intensifies in late-stage tumors.
Fig. 1.
Complexity and connectivity of the immune microenvironment in human GBM. (A) Uniform Manifold Approximation and Projection (UMAP) map of single immune cells in human high-grade gliomas at primary diagnosis (left) and recurrence (right). Cell subsets are indicated by color code and annotation. (B) Circos plots (top) and heatmaps (bottom) representing receptor-ligand interaction analyses by CellPhoneDB in newly diagnosed (n = 7 patients) and recurrent (n = 4 patients) GBM. Interaction z-score shown. Immune cell subsets defined as in Ref. 7. (C) Dot plot representation of top 40 receptor–ligand interactions based on molecule co-expression and reference-based cell subsets. Mean relative expression of both interaction partners (dot color) and interaction P-value (dot size) shown. P-values are derived from one-sided permutation tests and refer to the enrichment of the interacting ligand–receptor pair in each of the interacting pairs of cell types. GBM cohort from A to B. (D and E). CyTOF surface protein analysis on sorted glioma-infiltrating myeloid cells. n = 62 239 cells (n = 25 928 control brain tissue; n = 16 764 IDHmut HGG; n = 19 547 IDHwt HGG). (C) Dot plots representing individual patient specimens shown. One-way ANOVA + Fisher’s LSD test used for statistical testing. (E) Immune microenvironment composition based on reference-based prediction of immune cell subsets in IDHwt HGG (n = 9, left) and IDHmut HGG (n = 4, right) CyTOF cohort.
Antigen Presentation by Glioma-Associated DC
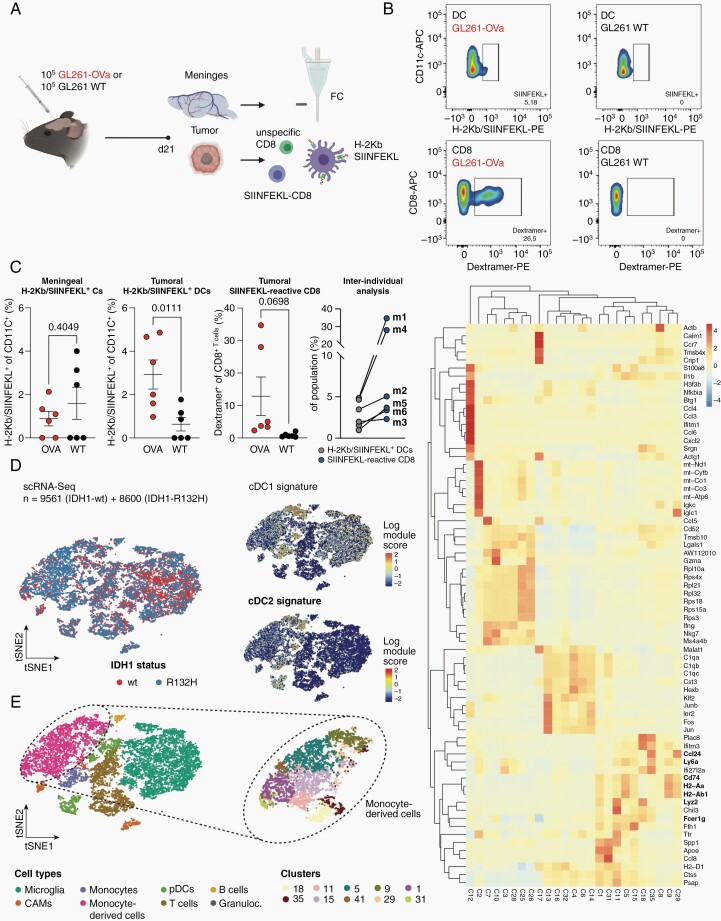
We next investigated if there is relevant antigen presentation by DC in experimental gliomas and where glioma-associated antigen-presenting DC are located. We thus used a syngeneic immunocompetent murine glioma model overexpressing full-length ovalbumin (Ova) as a model antigen comprising both major histocompatibility class I (MHC-I) and major histocompatibility class II (MHC-II) epitopes (GL261-Ova), and isolated DC and T cells from intracranial tumor fractions and meninges (Figure 2A–C). Ova-expressing tumor-bearing C57BL/6 mice spontaneously generate SIINFEKL-reactive CD8+ T cells. Accordingly, we identified tumor-specific CD8+ T cells by SIINFEKL-dextramer staining (Figure 2B). Tumor-associated DC (CD11c+/MHC-II+) were tested for cross-presentation of the MHC-I-restricted Ova epitope SIINFEKL by flow cytometry. At day 21 after tumor inoculation, we did not find specific DC presenting the tumor-specific antigen in the meninges.16 However, SIINFEKL-presenting DC were found in Ova-expressing brain tumors, and tumor-infiltrating SIINFEKL-reactive CD8+ T cells were detectable at very high frequencies in some mice, suggestive of functional antigen presentation by tumoral DC via MHC-I (Figure 2C). Of note, the mice with the highest abundance of SIINFEKL-presenting DC (m1, m4) demonstrated the corresponding highest abundance of tumor-reactive T cells, underlining the relevance of antigen presentation by glioma-infiltrating DC in this model (Figure 2C).
Fig. 2.
HGG-infiltrating dendritic cells exhibit limited antigen presentation via MHC class I. (A) Experimental overview. GL261-OVA or wildtype control cells were implanted in n = 12 C57BL6/J mice. 28 days post implantation, tumors and meninges were isolated and subjected to flow cytometry of SIINFEKL-reactive T cells and SIINFEKL-presenting DCs (CD11c+/MHC-II +). (B) Representative flow cytometry gating of H-2Kb-SIINFEKL-presenting DCs and SIINFEKL-dextramer-positive T cells isolated from syngeneic gliomas. Left column: GL261-Ova; Right column: GL261-WT. (C) left: Quantification of H-2Kb-SIINFEKL-presenting DCs and SIINFEKL-dextramer-positive T cells in meninges and glioma samples. n = 6 biological replicates per group. P-values are derived from paired student’s t-tests. right: inter-individual analysis of matched SIINFEKL-presenting DCs and SIINFEKL-dextramer-positive T cells from (left). Mouse IDs indicated. (D–F) Single-cell RNA-Seq of CD45+ cells from IDHmut and IDHwt GL261 mouse experimental gliomas. t-SNE maps are color-coded for (D) the genotype of the glioma and (E) the identified cell types based on marker gene expression. Median expression of cDC1 marker gene signature (Itgae, Xcr1, Sept3, Gcsam, Clec9a, Pianp, Ffar4, A503099J19Rik, Plpp1, Cxx1a, Ifi205, Cyp8b1, Tlr3, Snx2) and cDC2 signature (CD209a, Tnfsf9, Tnip3, Kcne3) shown in (D). Analysis was conducted on n = 7910 cells from IDH-mutated experimental glioma and n = 8835 cells from IDH-wildtype experimental glioma. (F) Heatmap representation of differentially regulated genes across all clusters. Clusters 5, 9, 11, 15, 18 represent cells on the monocyte-to-DC trajectory. Relative gene expression shown.
To investigate the DC landscape in gliomas in vivo, we performed scRNA-seq of FACS-purified CD45+ cells isolated from late-stage IDHmut and IDHwt GL261 mouse experimental gliomas comprising microglia, TAM, monocyte-derived cells, plasmacytoid DC, and T and B cells (Figure 2D and E; Supplementary Figure 3A).8 Furthermore, we identified cells expressing cDC1 and cDC2 marker genes, mainly in the monocyte-derived cell compartment (Figure 2D). Here, we identified 10 distinct subclusters of monocytes and monocyte-derived cells with variable expression of monocyte markers, such as Ly6a, Ccl24, Lyz2 as well as markers delineating APC function including CD74, H2-Aa, H2-Ab1 (Figure 2E and F). Subcluster 18 co-expressed markers of mature DC with Fcer1. Activated Fcer1-expressing dendric cells have been previously reported to induce T helper 1 cell proliferation and cytokine production (Figure 2F).17
Differential Education of Glioma-Associated DC
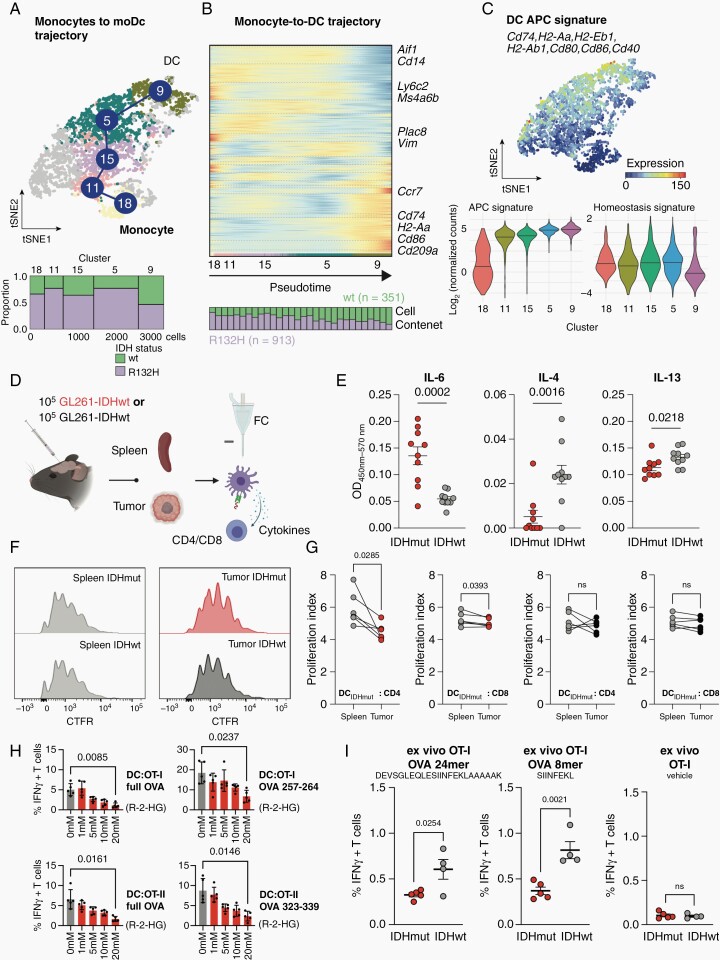
To further elucidate differentiation fates of glioma-infiltrating monocytes, we performed pseudotime trajectory analyses on the APC compartment, setting blood-borne monocytes (cluster 18) as root node (Figure 3A and B). Indeed, we observed a trajectory spanning from hematogenic monocytes to mature, antigen-presenting DC located in cluster 9 (Figure 3A). Trajectory analyses suggested a monocyte-to-DC differentiation block that was specifically inherent to IDHmut tumors. Conversely, predominantly IDHwt tumor-derived clusters were dominated by DC upregulating molecules of the MHC and co-stimulatory molecules, including CD74, H2-Aa, and CD86. IDHwt-derived DC also expressed CD209a, the murine homologue of human DC-specific ITAM-3-grabbing non-integrin (DC-SIGN) that is relevant for DC rolling along the blood endothelium and T cell activation (Figure 3B). Increased expression level of an antigen presentation capacity (APC) gene signature including CD74, H2-Aa, and CD86 was observed along the trajectory (Figure 3C). In summary, glioma-infiltrating monocyte-derived DC are aligned to a differentiation trajectory that can be used to derive signatures reflective of differential DC education according to tumoral IDH-mutation status. IDHmut-educated DC exhibit an immature cellular state, as evidenced by low frequencies of APChigh DC. Multiplexed cytokine profiling of experimental glioma interstitial fluid revealed largely balanced concentrations of pro-immunogenic cytokines IL-6, TNF-α, and interferon-γ (IFN-γ) between IDHmut and IDHwt tumors (Figure 3D; Supplementary Figure 3B). Although here, the cellular origin of cytokines remains unknown, similarly, tumor-derived increased levels of IL-6 have been reported upon alpha thalassemia/mental retardation syndrome X-linked (ATRX) loss in experimental IDHmut tumors.18 In contrast, IL-4 and IL-13 were significantly enriched in the microenvironment of IDHwt gliomas (Supplementary Figure 3B), which we were able to validated by single cytokine ELISA of tumor microfluid (Figure 3E).19 It was previously shown that tumor cells release IL-6, which inhibits the differentiation of CD34+ cells into DC and promotes their commitment toward monocytic lineage with a poor APC function. It was further demonstrated that IL-4 and IL-13 reverse the inhibitory effect of tumor cells on DC differentiation.20 We, therefore, hypothesize that the IL-4 and IL-13-abundant microenvironment of IDHwt gliomas allows for the differentiation of infiltrating monocytes into antigen-presenting DC (Figure 3E). Considering this, we reasoned that IDHmut-educated DC are affected in their APC. To assess whether this dysfunctional phenotype of DC in IDHmut gliomas impacts T cell function, we isolated tumoral and splenic DC from mice bearing IDHmut and IDHwt experimental GL261 gliomas and performed ex vivo co-cultures with splenocyte-derived autologous murine T cells (Figure 3F and G). CD4+ T helper cells and CD8+ T cells showed reduced proliferation when co-cultured with tumoral versus splenic DC from IDHmut-bearing mice (Figure 3F and G). As IDHmut HGG were shown to modulate their immune microenvironment via paracrine R-2-Hydroxyglutarate (R-2-HG),8,9,21 we next assessed the mediators of restrained T-cell activation by DC after R-2-HG-exposure. In line with recent reports, following exposure to R-2-HG for 48h, human monocyte-derived DC demonstrated a tolerogenic phenotype with significantly reduced expression of IL-6 as well as MHC-II (human leukocyte antigen (HLA)-DR). In contrast, the two immunosuppressive mediators mannose receptor (CD206) and programmed cell death 1 ligand 1 were moderately induced in DC when exposed to exogenous R-2-HG (Supplementary Figure 3C and D). Consequently, activation and IFN-γ secretion of T cells decreased with increasing R-2-HG exposure of murine DC in a dose-dependent manner after co-culture of antigen-specific T cells with peptide-loaded and R-2-HG-treated DC (Figure 3H). As the reduced APC of ex vivo isolated IDHmut-associated DC was evident and similar in 8-mer MHC-I-restricted peptides as well as 24-mers that require prior uptake and antigen processing, we hypothesize that unlike the protein expression of class I and class II MHC, the antigen presentation machinery of DC is not significantly affected in IDHmut gliomas (Figure 3I). We thus confirm the occurrence of DC with a poor APC that limit T cell effector functions as a consequence of R-2-HG-mediated reprogramming in IDHmut experimental gliomas.
Fig. 3.
IDH status-dependent differential education of monocyte-derived dendritic cells. (A) Top: t-SNE representation of monocyte-derived macrophages and moDCs color-coded for RaceID cluster. Analysis was conducted on n = 913 cells from IDH-mutated (IDHmut) and n = 351 cells from IDH-wildtype (IDHwt) experimental HGG. Bottom: Marimekko plot of cluster-wise cellular abundance between IDHmut and IDHwt gliomas (B) Illustration of distinct trajectories from monocytes to monocyte-derived DCs. Heatmaps represent gene expression along the respective trajectory with the cell composition along each trajectory shown below (25 cells are binned for each stacked column). The StemID2 algorithm was utilized. (C) Top: Median expression of DC antigen presentation capacity (APC) signature. Marker genes of the signature indicated. Bottom: Violin plots showing cluster-wise cumulative gene expression of the APC and Homeostasis signature in cells extracted from HGG. Median and probability density smoothed by a kernel density estimator shown. (D–G) Functional ex vivo DC: T cell activation assay. (D) Experimental overview. DCs were isolated from intracranial experimental HGG and spleens by FACS and co-cultured with antigen-specific CD4 and CD8 T cells. (E) Cytokine ELISA of indicated cytokines measured in tumor interstitial fluid isolated from IDHmut and IDHwt experimental HGG at late stage (d21). OD450nm-570nm ± SEM shown for each cytokine. (F) Representative flow cytometry-based histograms of T cell proliferation via CellTrace Far Red (CFTR) staining of CD8+ T cells. (G) Quantification of differential T cell proliferation in ex vivo DC: T cell co-culture. Proliferation index calculated from CFTR-staining. P-values are derived from paired student’s t-tests. (H) Flow cytometry-based quantifications of intracellular Interferon-gamma (IFN-γ) of OT-I and OT-II T cells co-cultured with syngeneic DCs after exposure to varying concentrations of paracrine R-2-HG or vehicle. DCs were pulsed with full Ovalbumin (OVA) protein or indicated OVA peptides to elicit OT-I or OT-II restricted T cell responses. P-values are derived from paired student’s t-tests. (I) Flow cytometry-based quantifications of intracellular Interferon-gamma (IFN-γ) of OT-I T cells co-cultured with glioma-derived immune cells from IDHmut or IDHwt tumors. Co-culture was pulsed with indicated OVA 24mer or 8mer peptides to elicit OT-I T cell responses. P-values are derived from unpaired student’s t-tests.
Prognostic Impact of Glioma DC Education Signatures
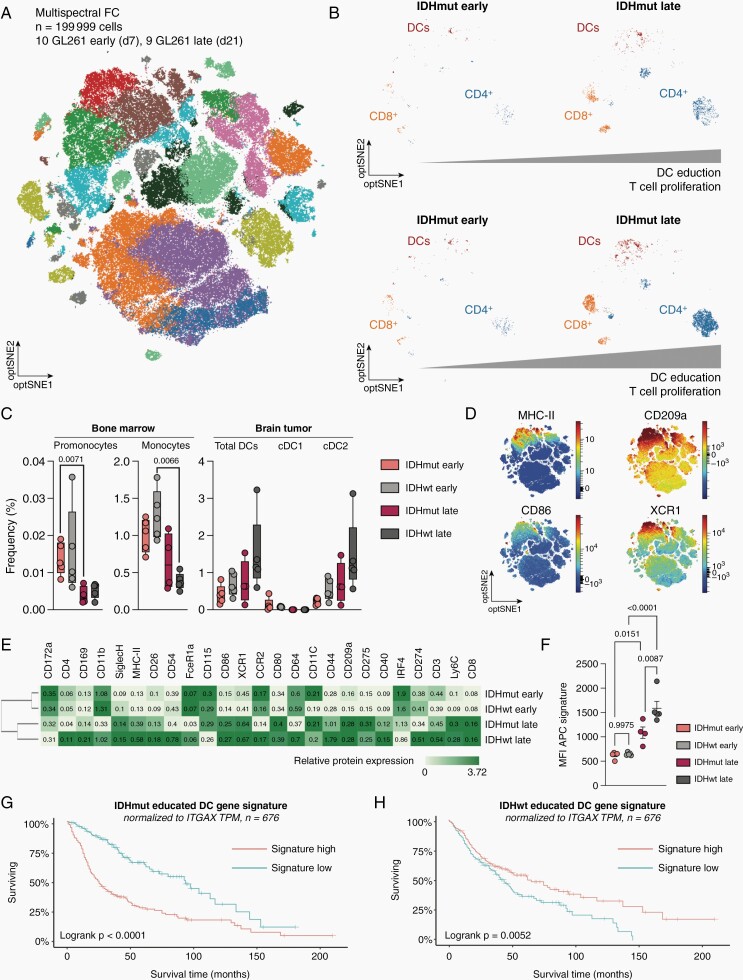
We next aimed to longitudinally reconstruct our pseudotime glioma DC education signatures and validate them on a protein level. This was done by generating a high dimensional protein atlas of 199,999 immune cells isolated from 19 murine experimental gliomas at two-time points and corresponding bone marrows and analyzed by spectral flow cytometry using 40 lineage- and DC-subtype-specific antigens (Figure 4A; Supplementary Figures 3 and 4). To allow for temporal resolution, immune infiltrates were isolated from early (d7 postinjection) and late-stage (d21 postinjection) tumors and integrated into one dataset. Here, we observed a higher infiltration of DC in IDHwt compared to IDHmut gliomas over time, which was accompanied by a similarly increased abundance of CD4+ and CD8+ T cells (Figure 4B). Furthermore, abundance of monocytic precursors decreased in the corresponding bone marrow throughout the course of disease, irrespective of tumor genotype. This was accompanied by an increase in overall DC and specifically cDC2 cells (CD11c+/MHC-II+/CD172a+/XCR1−) abundance in the tumor, which was more pronounced in IDHwt gliomas. In contrast, there were limited to none tumoral cDC1 cells (CD11c+/MHC-II+/CD172a−/XCR1+) found by spectral flow cytometry (Figure 4C). Importantly, cDC1 perform cross-presentation of antigens to MHC-I and initiate type I cytotoxic immune responses, which correlates with the limited CD8+ T cell infiltration in murine and human gliomas (Figures 1A–E and 4B). We found pronounced surface protein expression of APC markers CD86 and MHC-II (H-2 IAb) as well as education signature genes CD209a and XCR1 in the dataset, allowing for the reconstruction of our defined glioma DC education signatures on a protein level (Figure 4D, Supplementary Figure 5). In fact, we observed considerable dynamics in the expression of DC identity-defining proteins from early to late-stage glioma-infiltrating DC that aligned with the transcriptomic monocyte-to-DC trajectory (Figures 3B and C and 4E). In early-stage tumors, infiltrating monocyte-derived, lineage-negative cells were immature and co-expressed markers of infiltrating monocytes, such as CD172a, IRF4, CD115, and CCR2. As these cells did not express CD40, CD86, or CD275, they remained functionally immature (Figure 4E).22 We could further validate differential protein expression of DC education signature genes CD86, MHC-II, CD209a in IDHwt-derived DC. Strikingly, monocyte-derived cells isolated from late-stage IDHmut tumors showed significantly reduced APC marker protein expression compared to IDHwt educated cells. However, they were defined by monocytic and IDHmut glioma DC signature markers such as CD172a, CD115, IRF4, and Ly6C. Taken together, the phenotype of IDHmut-educated monocyte-derived cells is characterized as an intermediate state between immature monocyte-derived cells in early-stage gliomas and fully functional DC found in late-stage IDHwt tumors (Figure 4E). We have confirmed the significantly reduced expression of the DC antigen presentation protein signature in late-stage IDHmut tumors compared to IDHwt tumors (Figure 4F). The discrepancy in monocyte-to-DC differentiation between IDHwt and IDHmut tumors thus results in distinct glioma education signatures according to tumoral IDH-mutation status. These signatures can be reconstructed on both transcriptome and proteome level. Interestingly, high expression of the IDHwt glioma DC education signature normalized to ITGAX (CD11c) expression was associated with higher OS in GBM, whereas high expression of the IDHmut glioma DC education signature was associated with lower OS in IDHmut HGG, demonstrating the importance of cellular state analysis and not only cellular numbers in such complex immunosuppressive diseases (Figure 4G and H).
Fig. 4.
Longitudinal characterization of hematogenic DC infiltration and education protein signature in experimental gliomas. (A) Multispectral flow cytometry of n = 19 C57BL6/J mice bearing experimental HGG at early (d7 postinjection) or late-stage (d21 postinjection). Optical SNE (optSNE) map shown. opt-SNE maps are color-coded by the identified cell types based on marker gene expression. (B) opt-SNE representation of DCs, CD4+ T cells and CD8+ T cells in GL261 IDHmut and IDHwt tumors over time. Bar represents increase in normalized T cell abundance over time. (C) Quantification of cell subtype abundance in bone marrow and brain tumor samples according to IDH mutation status. Individual values and boxplot with median and 5-95 percentile shown. P-values are derived from paired student’s t-tests. (D) optSNE density plot representations of indicated cell surface markers. Relative expression shown. (E) Heatmap representing median cell surface marker expression in indicated early-stage and late-stage conditions. Median relative expression indicated. (F) Mean fluorescence intensity of cumulative protein expression of the APC signature in DCs extracted from IDHwt and IDHmut gliomas. One-way ANOVA + Fisher’s LSD test used for statistical testing. (G–H). Kaplan-Meier survival estimates analysis of (B) averaged IDHmut glioma DC education signature and (G) averaged IDHwt glioma DC education signature. TCGA GBM cohort (n = 676 patients).
Discussion
HGG are a complex group of primary central nervous system (CNS) malignancies in adults with a generally poor prognosis. The currently limited therapy repertoire in gliomas has made the development of novel therapies targeting their suppressive immune microenvironment of paramount importance. Due to their unique interaction with both the innate and adaptive immune system,14 there is particular interest in developing glioma therapies that target DC. As professional antigen processing and presenting cells, DC play a key role in the initiation of antitumor immune responses but at the same time represent important integrators of microenvironmental signals. Typically, phagocytosis of tumor cells following immunogenic cell death activates and matures DC.14 However, (1) IL-10 production by macrophages, leading to reduced IL-12 production by cDC1,23 (2) LXRa-driven trapping of DC within the hostile TME,24 (3) intratumoral immunosuppressive IDO1-upregulation and tryptophan starvation,25 and (4) an interaction of FLT3L and vascular endothelial growth factor (VEGF), negatively impacting cDC differentiation,26 are few examples of multiple DC modulating mechanisms that lead to inefficient acquisition of effector states and DC-driven immunosuppressive conditioning of the TME. With the emerging field of single-cell sequencing, a more granular view on DC in the peripheral blood or secondary lymphoid organs and tissues in human and murine systems, respectively, has been gained. Historically, DC have been separated into four subgroups, that is, cDC1, cDC2, mo-DCs, and pDCs. scRNA-seq from blood-derived DC using the Smart-Seq2 protocol identified six distinct DC cell clusters with some overlap with canonical subtypes.27 More recently, four additional cDC2-subgroups that are conserved between human and mouse, have been described in late-stage human and murine GBM, when combining single-cell sequencing and CITE-Seq.7 However, the specific role of DC in the setting of CNS tumors is still being elucidated, while current studies suggest a complex interplay between DC, microglia, T cells, and tumor cells in the TME.2,4,8 Furthermore, DC are typically not found in normal brain parenchyma, but are instead present in vascular-rich compartments such as the choroid plexus and meninges.
Here, we demonstrate longitudinal infiltration of human and experimental murine gliomas by distinct populations of monocyte-derived DC. In contrast to peripheral DC, these glioma-infiltrating DC exhibit limited APC. We show that monocytes are differentially educated in IDHmut and IDHwt tumors resulting in distinct phenotypical states. The phenotype of IDHmut-educated DC is characterized as an intermediate state between immature monocyte-derived cells in early-stage gliomas and the fully functional DC found in late-stage IDHwt tumors. Monocyte-to-DC differentiation is further impaired in IDH-mutant glioma by paracrine R-2-hydroxyglutarate. We demonstrate that paracrine R-2-hydroxyglutarate delays DC maturation and specifically suppresses MHC class I/II-mediated antigen (cross-)presentation and co-stimulation by IL-6, which translates to reduced T cell activating capacities. The discrepancy in monocyte-to-DC differentiation between IDHwt and IDHmut tumors results in distinct glioma DC education signatures according to tumoral IDH mutation status. We derived IDH-status-dependent DC education signatures from transcriptomic trajectories and validated these signatures on a protein level. We provide evidence that this glioma subtype-specific education of transmigrated DC is prognostically relevant in human gliomas and identify dysfunctional DC as a target for immunotherapy in IDH-mutant tumors. An obvious limitation of our study represents the unmet need of IDH-mutant mouse glioma models that truly recapitulate IDH-dependent gliomagenesis. Herewith, at least in part, some observational discrepancies between human and murine systems can be explained. Further research is needed to elucidate the mechanisms of recruitment and fate commitment of monocytes infiltrating CNS tumors and to identify molecularly driven therapy targets in the glioma immune microenvironment to enhance treatment response in this generally immunotherapy-refractory disease.
Supplementary Material
Acknowledgments
We acknowledge the support of the DKFZ Genomics and Proteomics Core Facility. We acknowledge the support by the Flow Cytometry Core Facility at the German Cancer Research Center and the Flow Cytometry Core Facility at the Medical Faculty Mannheim of the Heidelberg University.
Contributor Information
Mirco Friedrich, DKTK Clinical Cooperation Unit Neuroimmunology and Brain Tumor Immunology, German Cancer Research Center (DKFZ), Heidelberg, Germany; Department of Hematology, Oncology and Rheumatology, Heidelberg University Hospital , Heidelberg, Germany; Faculty of Biosciences, Heidelberg University, Heidelberg, Germany.
Markus Hahn, DKTK Clinical Cooperation Unit Neuroimmunology and Brain Tumor Immunology, German Cancer Research Center (DKFZ), Heidelberg, Germany; Department of Hematology, Oncology and Rheumatology, Heidelberg University Hospital , Heidelberg, Germany; Faculty of Biosciences, Heidelberg University, Heidelberg, Germany; Department of Neurology, MCTN, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany.
Julius Michel, DKTK Clinical Cooperation Unit Neuroimmunology and Brain Tumor Immunology, German Cancer Research Center (DKFZ), Heidelberg, Germany; Department of Neurology, MCTN, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany.
Roman Sankowski, Department of Neuropathology, Freiburg University Hospital, Freiburg, Germany.
Michael Kilian, DKTK Clinical Cooperation Unit Neuroimmunology and Brain Tumor Immunology, German Cancer Research Center (DKFZ), Heidelberg, Germany; Department of Neurology, MCTN, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany.
Niklas Kehl, DKTK Clinical Cooperation Unit Neuroimmunology and Brain Tumor Immunology, German Cancer Research Center (DKFZ), Heidelberg, Germany; Department of Neurology, MCTN, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany.
Manina Günter, Dendritic Cells in Infection and Cancer, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Theresa Bunse, DKTK Clinical Cooperation Unit Neuroimmunology and Brain Tumor Immunology, German Cancer Research Center (DKFZ), Heidelberg, Germany; Department of Neurology, MCTN, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany.
Stefan Pusch, Department of Neuropathology, Heidelberg University Hospital, Heidelberg, Germany; DKTK Clinical Cooperation Unit Neuropathology, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Andreas von Deimling, Department of Neuropathology, Heidelberg University Hospital, Heidelberg, Germany; DKTK Clinical Cooperation Unit Neuropathology, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Wolfgang Wick, Department of Neurology, Heidelberg University Hospital, Heidelberg, Germany; DKTK Clinical Cooperation Unit Neurooncology, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Stella E Autenrieth, Dendritic Cells in Infection and Cancer, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Marco Prinz, Department of Neuropathology, Freiburg University Hospital, Freiburg, Germany.
Michael Platten, DKTK Clinical Cooperation Unit Neuroimmunology and Brain Tumor Immunology, German Cancer Research Center (DKFZ), Heidelberg, Germany; Department of Neurology, MCTN, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany; Helmholtz Institute of Translational Oncology (HI-TRON), Mainz, Germany; Immune Monitoring Unit, National Center for Tumor Diseases (NCT), Heidelberg, Germany ; DKFZ Hector Cancer Institute at the University Medical Center Mannheim, Mannheim, Germany.
Lukas Bunse, DKTK Clinical Cooperation Unit Neuroimmunology and Brain Tumor Immunology, German Cancer Research Center (DKFZ), Heidelberg, Germany; Department of Neurology, MCTN, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany.
Funding
M.F. was supported by BMBF and the Ministry of Science Baden-Württemberg within the framework of the Excellence Strategy of the Federal and State Governments of Germany and the José Carreras Leukemia Foundation (DJCLS 01ZI/2022). M.K. was supported by the Helmholtz International Graduate School (HIGS). R.S. was funded by the Berta-Ottenstein-Programme for Clinician Scientists, Faculty of Medicine, University of Freiburg and Else Kröner Fresenius Foundation. A fellowship from the Mildred-Scheel doctoral program of the German Cancer Aid was given to J.M.; N.K. was supported by a doctoral stipend as part of the DFG Research Training Group (RTG) 2099 “Hallmarks of Skin Cancer”. M.Pr. was supported by the Sobek Foundation, the Ernst-Jung Foundation, the DFG - SFB 992, SFB1160, SFB 1479, SFB/TRR167, Reinhart-Koselleck-Grant, Gottfried Wilhelm Leibniz-Prize and the Ministry of Science, Research and Arts, Baden-Wuerttemberg (Sonderförderlinie “Neuroinflammation”), DFG - Germany’s Excellence Strategy (CIBSS – EXC-2189 – Project 668 ID390939984).
This work was supported by grants from the Helmholtz Gemeinschaft, Zukunftsthema “Immunology and Infection” (ZT0027, WP3); the Ministry of Science, Research and Arts, Baden-Wuerttemberg (Sonderförderlinie “Neuroinflammation”); the German Ministry of Education and Science (BMBF) NCT 3.0 program “Precision immunotherapy of brain tumors” and the DKTK program; the German Research Foundation (DFG) - Project ID 404521405, SFB1389-B01 - UNITE Glioblastoma, Project ID 394046768, SFB1366-C01—Vascular Control of Organ Function, and Project ID 406052676, PL-315/5-1 - Impact of dietary Tryptophan on the gut microbiome and autoimmune neuroinflammation; and the German Cancer Aid (projects 70112399 and 70113515) to M.Pl.
Grants from the DFG - SFB1389-B03 to L.B. and S.P. and from the Dr. Rolf M. Schwiete Foundation to M.Pl. and L.B. Grants from the Medical Faculty Mannheim of Heidelberg University; the Medical Faculty Heidelberg of Heidelberg University; the Else Kröner Fresenius Foundation and the Swiss Cancer Foundation to L.B.
The authors gratefully acknowledge the data storage service SDS@hd supported by the Ministry of Science, Research and the Arts Baden-Württemberg (and the German Research Foundation (DFG) through (INST 35/1314-1 FUGG and INST 35/1503-1 FUGG).
Conflict of interest statement. T.B., W.W., and M.Pl. are inventors and patent-holders on “Peptides for use in treating or diagnosing IDHR132H positive cancers” (EP2800580B1). S.P. and A.v.D. are eligible to royalties as co-inventors of BAY 1436032 and are patent holders of “Means and methods for the determination of (D)-2-hydroxyglutarate (D2HG)” (WO2013127997A1). The other authors declare no conflict of interest.
Authorship statement. M.F. designed and performed experiments, analyzed and interpreted data, and wrote the paper. M.F. and M.H. performed in vitro experiments. M.F., M.H., M.K. and N.K. performed in vivo experiments. R.S. performed comparative analyses of single-cell transcriptomics and CyTOF datasets. J.M. analyzed data. T.B. interpreted data and wrote the paper. M.G. and S.E.A. established and performed spectral flow cytometry. S.P. performed R-2-HG measurements. W.W., A.v.D., M.Pr., and M.Pl. were involved in study design and data interpretation. L.B. conceptualized the study, interpreted data, and wrote the paper.
Data Availability
Mouse bulk and single-cell RNA-seq data that support the findings of this study was downloaded from the Gene Expression Omnibus (GEO) under the accession code GSE166218 (mouse 10× data). Human GBM-associated immune cell data was downloaded from https://www.brainimmuneatlas.org/. Mass cytometry was downloaded from the FlowRepository: https://flowrepository.org/id/FR-FCM-Z3G7. TCGA datasets were downloaded from gliovis.bioinfo.cnio.es. The DC education signature is distilled from the trajectory analysis shown in Figure 3B and reflects the top regulated genes among the monocyte-to-DC trajectory Aif1, Cd14, Ly6c2, Plac8, Vim, Ccr7, Cd74, Cd86, CD209a, respectively, their human isoforms. All other data supporting the findings of this study are available from the corresponding author on reasonable request: l.bunse@dkfz.de.
Code Availability
Custom code for the transcriptomic and proteomic analyses can be found under: https://github.com/rsankowski/friedrich-et-al-IDHwt-mut-micr.git and https://github.com/rsankowski/antunes_GBMm_cellphonedb.git.
References
- 1. Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. J Am Med Assoc. 2013; 310(17):1842–1850. [DOI] [PubMed] [Google Scholar]
- 2. Friebel E, Kapolou K, Unger S, et al. Single-cell mapping of human brain cancer reveals tumor-specific instruction of tissue-invading leukocytes. Cell. 2020; 181(17):1626–1642.e20. [DOI] [PubMed] [Google Scholar]
- 3. Locarno C V., Simonelli M, Carenza C, et al. Role of myeloid cells in the immunosuppressive microenvironment in gliomas. Immunobiology. 2020; 225(1):151853. [DOI] [PubMed] [Google Scholar]
- 4. Klemm F, Maas RR, Bowman RL, et al. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell. 2020; 181(7): 1643–1660.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Venteicher AS, Tirosh I, Hebert C, et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science (1979). 2017; 355(6332):eaai8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hara T, Chanoch-Myers R, Mathewson ND, et al. Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma. Cancer Cell. 2021; 39(6):779–792.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pombo Antunes AR, Scheyltjens I, Lodi F, et al. Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat Neurosci. 2021; 24(4):595–610. [DOI] [PubMed] [Google Scholar]
- 8. Friedrich M, Sankowski R, Bunse L, et al. Tryptophan metabolism drives dynamic immunosuppressive myeloid states in IDH-mutant gliomas. Nature Cancer. 2021; 2(7):723–740. [DOI] [PubMed] [Google Scholar]
- 9. Bunse L, Pusch S, Bunse T, et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat Med. 2018; 24:1192–1203 [DOI] [PubMed] [Google Scholar]
- 10. Yu G, Wang LG, Han Y, He QY. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS J Integr Biol. 2012; 16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat Protocols. 2020; 15:1484–1506 [DOI] [PubMed] [Google Scholar]
- 12. Klasen C, Ohl K, Sternkopf M, et al. MIF promotes B cell chemotaxis through the receptors CXCR4 and CD74 and ZAP-70 signaling. J Immunol. 2014; 192(11):5273–5284. [DOI] [PubMed] [Google Scholar]
- 13. Klasen C, Ziehm T, Huber M, et al. LPS-mediated cell surface expression of CD74 promotes the proliferation of B cells in response to MIF. Cell Signal. 2018; 46:32–42. doi: 10.1016/j.cellsig.2018.02.010 [DOI] [PubMed] [Google Scholar]
- 14. Wculek SK, Cueto FJ, Mujal AM, et al. Dendritic cells in cancer immunology and immunotherapy. Nat Rev Immunol. 2020; 20(1):7–24. [DOI] [PubMed] [Google Scholar]
- 15. Bottcher C, Schlickeiser S, Sneeboer MAM, et al. Human microglia regional heterogeneity and phenotypes determined by multiplexed single-cell mass cytometry. Nat Neurosci. 2019; 22(1):78–90. [DOI] [PubMed] [Google Scholar]
- 16. Hu X, Deng Q, Ma L, et al. Meningeal lymphatic vessels regulate brain tumor drainage and immunity. Cell Res. 2020; 30(3):229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boltjes A, van Wijk F. Human dendritic cell functional specialization in steady-state and inflammation. Front Immunol. 2014; 5:31. doi: 10.3389/fimmu.2014.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu C, Wang K, Damon C, et al. ATRX loss promotes immunosuppressive mechanisms in IDH1 mutant glioma. Neuro-Oncology. 2022;24(6):888-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luo XL, Dalod M. The quest for faithful in vitro models of human dendritic cells types. Mol Immunol. 2020; 12:3. [DOI] [PubMed] [Google Scholar]
- 20. Menetrier-Caux C, Thomachot MC, Alberti L, Montmain G, Blay JY. Il-4 prevents the blockade of dendritic cell differentiation induced by tumor cells. Cancer Res. 2001; 61(7):3096–3104. [PubMed] [Google Scholar]
- 21. Friedrich M, Bunse L, Wick W, Platten M. Perspectives of immunotherapy in isocitrate dehydrogenase-mutant gliomas. Curr Opin Oncol. 2018; 30(6):368–374. [DOI] [PubMed] [Google Scholar]
- 22. Segura E, Amigorena S. Inflammatory dendritic cells in mice and humans. Trends Immunol. 2013; 34(9):440–445. [DOI] [PubMed] [Google Scholar]
- 23. Ruffell B, Chang-Strachan D, Chan V, et al. Macrophage IL-10 Blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell. 2014; 26(5):623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Villablanca EJ, Raccosta L, Zhou D, et al. Tumor-mediated liver X receptor-α activation inhibits CC chemokine receptor-7 expression on dendritic cells and dampens antitumor responses. Nat Med. 2010; 16(1):98–105. [DOI] [PubMed] [Google Scholar]
- 25. Munn DH, Mellor AL. IDO in the tumor microenvironment: inflammation, counter-regulation, and tolerance. Trends Immunol. 2016; 37(3):193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barry KC, Hsu J, Broz ML, et al. A natural killer–dendritic cell axis defines checkpoint therapy–responsive tumor microenvironments. Nat Med. 2018; 24(8):1178–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Villani AC, Satija R, Reynolds G, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science (1979) 2017;356(6335):eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mouse bulk and single-cell RNA-seq data that support the findings of this study was downloaded from the Gene Expression Omnibus (GEO) under the accession code GSE166218 (mouse 10× data). Human GBM-associated immune cell data was downloaded from https://www.brainimmuneatlas.org/. Mass cytometry was downloaded from the FlowRepository: https://flowrepository.org/id/FR-FCM-Z3G7. TCGA datasets were downloaded from gliovis.bioinfo.cnio.es. The DC education signature is distilled from the trajectory analysis shown in Figure 3B and reflects the top regulated genes among the monocyte-to-DC trajectory Aif1, Cd14, Ly6c2, Plac8, Vim, Ccr7, Cd74, Cd86, CD209a, respectively, their human isoforms. All other data supporting the findings of this study are available from the corresponding author on reasonable request: l.bunse@dkfz.de.