Abstract
Background
Glioblastoma (GBM) is a highly lethal malignancy for which neoangiogenesis serves as a defining hallmark. The anti-VEGF antibody, bevacizumab, has been approved for the treatment of recurrent GBM, but resistance is universal.
Methods
We analyzed expression data of GBM patients treated with bevacizumab to discover potential resistance mechanisms. Patient-derived xenografts (PDXs) and cultures were interrogated for effects of phosphofructokinase-1, muscle isoform (PFKM) loss on tumor cell motility, migration, and invasion through genetic and pharmacologic targeting.
Results
We identified PFKM as a driver of bevacizumab resistance. PFKM functions dichotomize based on subcellular location: cytosolic PFKM interacted with KIF11, a tubular motor protein, to promote tumor invasion, whereas nuclear PFKM safeguarded genomic stability of tumor cells through interaction with NBS1. Leveraging differential transcriptional profiling, bupivacaine phenocopied genetic targeting of PFKM, and enhanced efficacy of bevacizumab in preclinical GBM models in vivo.
Conclusion
PFKM drives novel molecular pathways in GBM, offering a translational path to a novel therapeutic paradigm.
Keywords: bevacizumab, DNA damage and repair, invasion, PFKM
Graphical Abstract
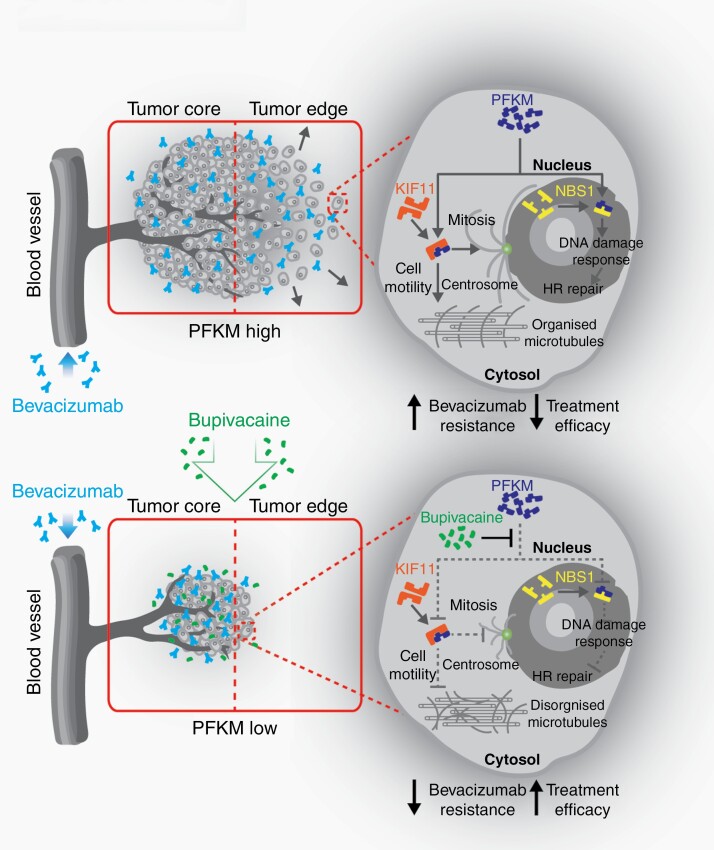
Highly invasive and DNA repair-proficient GBM cells with high PFKM expression in the tumor periphery evade bevacizumab therapy by invading normal brain parenchyma. Loss of PFKM impairs survival and invasion and exacerbates genomic instability of GBM cells, thereby potentiating the effect of bevacizumab therapy.
Key Points.
Cytosolic PFKM binds to KIF11 to maintain GBM cytoskeleton assembly and invasion.
Nuclear PFKM regulates DNA damage responses in GBM cells.
The anesthetic, bupivacaine, phenocopies the effects of targeting PFKM in reducing tumor growth and augments the efficacy of bevacizumab.
Importance of the Study.
PFK family members regulate glycolysis to promote tumor growth. Here, we report novel, non-metabolic functions of PFKM that promote tumor resistance to bevacizumab. PFKM functions dichotomize based on molecular location to augment cellular invasion and DNA repair, supporting PFKM as a therapeutic target. We identified bupivacaine as a repurposed pharmacologic agent that counteracts bevacizumab resistance in preclinical GBM models. Our observation supports a prospective future where therapies involving angiogenesis inhibition are pivotal to integrate drugs targeting the resistance mechanism to offer long-term therapeutic efficacy.
Glioblastoma (GBM) ranks among the most lethal of all human malignancies, with conventional therapy offering only palliation. While GBM has undergone extensive molecular characterization, genomic-informed precision medicine has provided little benefit for patients.1 Vascular proliferation represents a canonical histologic feature of GBM, and vascular endothelial growth factor A (VEGF-A) overexpression in endothelial cells are detected in early studies of GBM.2 Thus, anti-angiogenic therapies have been tested in numerous clinical trials with mixed success. Bevacizumab, which blocks circulating VEGF-A ligand function, is FDA approved for recurrent or progressive GBM,3 demonstrating frequent radiographic response but failing to increase overall survival in clinical trials.
Resistance to bevacizumab and anti-angiogenics has been linked to a diverse spectrum of molecular and cellular mechanisms that may inform novel therapeutic strategies. Our previous work implied the limited impact of bevacizumab-mediated VEGF-A blockage may reflect ongoing autocrine VEGF-VEGFR2 signaling on the surface of GBM cells to promote tumor growth.4,5 In a subset of patients failing bevacizumab, recurrent tumor growth patterns reflect increased invasion.6 GBM evades the growth inhibitory effects of bevacizumab through induction of MET signaling and progresses to a mesenchymal phenotype reminiscent of an epithelial-to-mesenchymal transition.7 Additional mechanisms include metabolic reprogramming and changes to the immune landscapes of treated tumors. Novel therapeutic paradigms have been developed based on these observations, but have yet to improve patient outcome, suggesting additional efforts may augment tumor control.
Based on this background, we hypothesized that transcriptional profiles of GBM surgical biopsy specimens derived from patients treated with bevacizumab would uncover additional evasion mechanisms whereby GBM biology may be revealed. In contrast to prior studies that have sought resistance mechanisms as an entry into combined therapies, we were interested in using bevacizumab resistance as a discovery platform to reveal novel tumor biology and therapeutic discovery.
Materials and Methods
Antibodies, expression constructs, buffers, and bioinformatic analysis are found in the Supplementary Information.
Cell Culture
T4121 and T10 GBM cells were obtained in accordance with approval from the Duke University Institutional Review Board and the Danish National Ethical Committee (H-3-2009-136_63114), respectively. IN1123 cells were a gift from Dr. Nakano (University of Alabama). GBM models were cultured as neurospheres in serum-free medium supplemented with EGF (epidermal growth factor)/FGF (fibroblast growth factor). Cells were authenticated by STR profiling and tested negative for mycoplasma. Normal human astrocytes (NHA; purchased from 3H Biomedical).
Inhibitors, Supplements, and Therapeutics
Sigma-Aldrich: CDK1 inhibitor (CDKi) RO-3306 and leptomycin. Roche: bevacizumab. AstraZeneca: bupivacaine. Ionizing radiation (IR) was administered via an x-ray source that delivers a rate of 0.04 Gy/s (YXLON).
RNA Interference
shRNA control, shPFKM #1 and #2 (Sigma-Aldrich) targeting phosphofructokinase-1 muscle isoform (PFKM) were produced using PAX2/VSV-G packaging system and calcium phosphate transfection kit (Clontech). Lentiviral particles were concentrated using PEG-it Virus Precipitation Solution (SBI Biotech). Stealth siRNA negative control and siRNA PFKM were also used (Invitrogen) (Supplementary Table 2).
Public Datasets
RNA-seq data were obtained for the BELOB8 and AVAglio trial.9,10
Immunohistochemistry
Paraffin-embedded animal brains and human tumor tissues were sectioned at 3 µm. Following antigen retrieval, sections were treated with 1.5% H2O2 and probed with respective antibodies (Supplementary Table 1). Sections were incubated with HRP-conjugated secondary antibody (EnVision+) and visualized using DAB/chromogen. Nuclei were counterstained with Mayer’s hematoxylin. For Supplementary Figure 1D, WHO grade II (n = 24), III (n = 18), and IV (n = 72) archival tumor specimens were analyzed. An independent cohort of 23 WHO grade IV (GBM) specimens was used to compare PFKM expression between tumor core and periphery (Figure 1D).
Fig. 1.
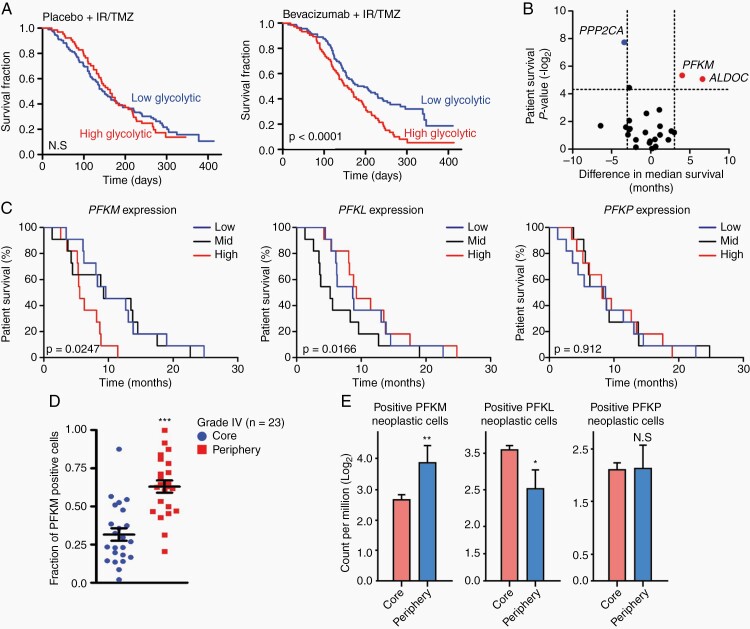
PFKM expression informs poor survival in bevacizumab-treated glioblastoma. (A) High glycolytic rate profile informs poor outcome of patients receiving bevacizumab in the AVAglio dataset. (B) PFKM and ALDOC expression levels informed survival of GBM patients under bevacizumab therapy. Data presented as median survival difference. (C) High PFKM but not PFKL and PFKP expression levels inform the survival of GBM patients in BELOB trial. (D) Immunohistochemistry shows elevated PFKM expression in WHO grade IV glioma (GBM) tumor periphery (n = 23). (E) Darmanis’s single-cell RNA-seq dataset shows PFKM but not PFKP or PFKL expression is elevated in invading neoplastic cells.
Significance is determined by log-rank test (A, C) or t test (D, E) and where applicable, data are shown as mean ± SEM (*P < .05, **P < .01, ***P < .001).
Wound Closure Assay
4-7 × 104 cells were seeded on Geltrex (Gibco) coated plate. Cells were nutrient-deprived 24 hours prior to treatment. Samples were scratched with a Woundmaker tool (Sartorius). Migration was analyzed at 24 hours by normalizing to 0 hours and calculated as a percentage of wound closure.
PFKM Activity Assay
PFKM kinase activity was measured according to the manufacturer’s protocol (Abcam) using 1 × 106 cells. Reaction-mix lacking substrate component determined background fluorescence. Plates were incubated at 37°C and measured at OD = 450 nm. PFKM kinase activity was calculated as the amount of NADH (nmol) generated per 1 × 106 cells.
Invasion Assay
Cells (1 × 103) were seeded in Geltrex-coated 96-well plate (Sigma-Aldrich) and spun down to form spheres.11 The following day, additional Geltrex was added, and plates were incubated at 37°C before adding serum-free medium (time, 0 hours). Cell invasion was analyzed by ImageJ at 24 hours.
Immunofluorescence
GBM cells grown on coverslips were fixed with 4% paraformaldehyde (PFA) (PFKM-GFP cells were fixed with methanol) and following permeabilization stained using indicated primary antibody (Supplementary Table 1). Nuclei were counterstained with 1 µg/mL DAPI (4′,6-diamidino-2-phenylindole)/PBS (phosphate-buffered saline) (Sigma-Aldrich). Images were acquired by LSM800 confocal (Zeiss) or ScanR (Olympus) high-content screening station. Proximity ligation assay (PLA) was performed by using the Duolink fluorescence protocol (Sigma-Aldrich).
RNA Sequencing
GBM (T4121 and T10) cells with silenced PFKM (shPFKM-pool, shPFKM #1 and #2 in 1:1 ratio) or treated with (24 hours) bupivacaine (GI50) were subjected to total RNA isolation (RNeasy plus kit, QIAGEN), library preparation and paired-end sequencing (BGI) to identify overlapping transcriptional changes associated with PFKM knockdown and bupivacaine treatment.
Cell-Cycle Assay
Cells were fixed in 70% ethanol, blocked in 5% BSA (bovine serum albumin)/PBS followed by staining with a conjugated H3Ser10-Alexa 488 antibody in 0.1% BSA of 0.1% Triton X-100/PBS and counterstained with DAPI prior to acquisition on FACSVerse (BD).
DNA Repair Assay
To determine homologous recombination (HR) repair, 1 × 106 cells were co-transfected with 1.5 µg of non-functioning HR plasmid (pDRGFP) and 3 µg of I-Sce1 meganuclease expression plasmid (pCMV-I-SceI).12 DNA repair was determined by detecting fluorescent positive cells with FACSVerse.
Cell Fractionation, Immunoprecipitation, Immunoblotting
Cells (1 × 107) received hypotonic treatment followed by centrifugation.13 Supernatant was collected as cytosol lysate. Pellet was then resuspended, layered onto sucrose gradient solution, and centrifuged. Supernatant was discarded and nuclear pellet lysed. For immunoprecipitation (IP), 1-5 mg of pre-cleared lysate was incubated with 1 µg of antibody or IgG control overnight together with 10 µL of protein G (Dynabeads). Next, lysates and protein antibody-bounded beads were separated with DynaMag (Invitrogen). IP-pulldowns or protein extracts (25-50 µg) were loaded onto SDS-polyacrylamide gels, electrophoresed, transferred onto nitrocellulose membrane, and incubated with primary and species-specific secondary antibodies. Chemiluminescence reagent (Cytica) was used for protein detection via ChemiDoc imaging system (BioRad).
Cell Viability
Cells were seeded in 96-well plate, treated as indicated, and assessed for viability using CellTiter-Glo (Promega) or Incucyte 72 hours later. Viability was calculated by normalizing to untreated control. GI50 was calculated using non-linear regression.
Alkaline Comet Assay
Cells were resuspended in 0.5% low melting agarose (Sigma-Aldrich) and spread onto 1% ultra-pure agarose-coated glass slides. Slides were lysed, neutralized, electrophoresed, and dehydrated in 96% ethanol prior to mounting in TE (Tris-EDTA) buffer with SYBR Gold (Invitrogen). DNA tails were imaged by an Axiovert fluorescence microscope (Zeiss) and scored using Comet Assay IV software.
In vivo Studies
Orthotopic and subcutaneous animal survival studies were performed as previously described.14 Detailed methodology is provided in the Supplementary Information.
Statistical Analysis
Unless stated otherwise, GraphPad determined statistical significance from three independent experiments. Where appropriate, comparisons were conducted using Mantel-Cox test, two-tailed Student’s t test, or two-way ANOVA followed by post hoc analysis. For correlation analysis, Spearman’s rank correlation test was performed. Statistical significance is represented with P-values (*P < .05; **P < .01; ***P < .001).
Results
PFKM Expression Informs Poor Survival in Bevacizumab-treated GBM Patients
Based on the hypothesis that bevacizumab-resistant tumors may reveal novel molecular mechanisms underlying tumor biology, we interrogated gene expression profiles from GBM patients enrolled in the AVAglio phase III trial (bevacizumab plus temozolomide and radiotherapy in newly diagnosed GBM patients).10 We reasoned that using data from newly diagnosed patients would minimize the complicating effects of prior therapies (eg, radiation and chemotherapy) that would be present in recurrent patients.9 Poor response to bevacizumab correlated with high glycolytic rate and poor survival outcome (Figure 1A). Next, we interrogated the BELOB trial dataset8 and analyzed gene expression profiles from patients treated with bevacizumab monotherapy arm to identify causative genes in the glycolysis pathway. Based on survival differences between low and high glycolytic patient cohorts, the muscle isoform of PFKM and aldolase C (ALDOC) were identified as key regulators of the glycolysis pathway (Figure 1B; Supplementary Figure 1A and B). Confirming this discovery, ALDOC has been previously shown to mediate glioma invasion.15 PFKM is a muscle-specific isoenzyme of PFK-1 that regulates the first step of metabolic process.16 Among patients treated with bevacizumab on the BELOB trial, high PFKM tumor expression portended a worsened survival compared to tumors with PFKM low or medium expression. In contrast, high tumor expression of the two other PFK-1 isoenzymes, PFKP (platelet) and PFKL (liver), did not inform prognosis (Figure 1C). Furthermore, independent analysis using the TCGA GBM dataset showed that high PFKM correlated with worse survival of GBM patients treated with bevacizumab (Supplementary Figure 1C). Immunohistochemistry (IHC) analysis of PFKM protein expression found elevated levels in GBM (WHO grade IV) compared to WHO grade II and III gliomas (Supplementary Figure 1D). Further examination showed that PFKM expression was markedly increased in the tumor periphery, suggesting a possible role in tumor invasion (Figure 1D; Supplementary Figure 1E). Comparative single-cell RNA sequencing profiling using the Darmanis dataset17 confirmed that PFKM, but not PFKL or PFKP, was preferentially expressed in the invasive region (Figure 1E).
PFKM Promotes GBM Motility and Invasion
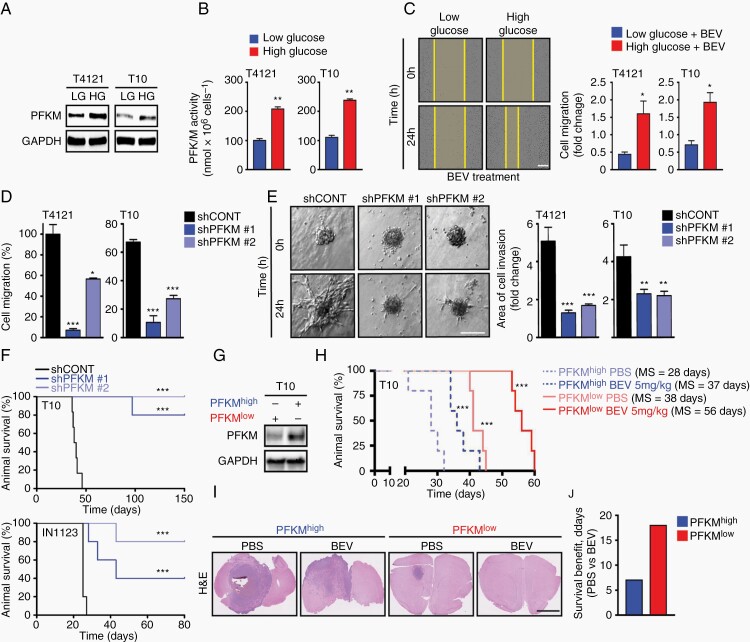
As PFKM expression was enriched in the tumor periphery, we hypothesized that GBMs co-opt PFKM activity to invade into normal tissues and acquire nutrients. As angiogenesis is crucial for tumors to accelerate metabolic activity during tumor expansion, we mimicked the response of patient-derived GBM cells to different levels of glucose, revealing induction of PFKM protein and activity with high glucose levels (Figure 2A and B). VEGF canonically induces endothelial cell proliferation to promote tumor growth,18 but we and others have demonstrated that GBM cells express VEGF receptors and respond directly to VEGF ligands.4,19 Bevacizumab-mediated VEGF sequestration decreased cellular proliferation of GBM cells with diminished efficacy in the presence of high glucose concentration (Supplementary Figure 2A). Invasion into the normal brain is a universal feature of gliomas. Bevacizumab reduced the motility of GBM cells, as measured by a wound assay, with the effects attenuated by high glucose (Figure 2C). Loss of function for PFKM was achieved through short-hairpin RNAs (shPFKM #1 and #2) and associated with diminished PFKM kinase activity (Supplementary Figure 2B and C). PFKM-silencing induced stress fiber formation, destabilization of tubulin structures (Supplementary Figure 2D), and lowered migratory as well as invasive capacity of GBM cells (Figure 2D and E). Mice intracranially implanted with GBM cells with silenced PFKM displayed longer survival (Figure 2F). To validate the clinical relevance of PFKM expression in response to bevacizumab, we intracranially implanted mice with subclones of T10 GBM model exhibiting either high (PFKMhigh, n = 5) or low PFKM (PFKMlow, n = 5) expression (Figure 2G). While bevacizumab extended the survival of both PFKMhigh and PFKMlow mice, a greater survival benefit (fold increase in median survival) and reduced tumor burden (H&E) was observed in the cohort of PFKMlow-injected mice (Figure 2H–J).
Fig. 2.
Glioblastoma invasion depends on PFKM. (A, B) Exposure (24 hours) of high glucose (HG: 5 µg/mL) but not low glucose (LG: 1 µg/mL) induces PFKM protein and activity in GBM cells. Loading control: GAPDH. (C) HG treatment (HG: 5 µg/mL) increases migratory capacity of GBM cells exposed to bevacizumab (BEV: 50 µg/mL) in a wound healing assay. LG (1 µg/mL). Data shown are normalized to 0 hours. Scale bar = 150 µm. (D) PFKM-silencing inhibits GBM cell migration in wound healing assay. (E) PFKM-silencing impairs GBM cell invasion in a 3D spheroid assay. Data shown are normalized to 0 hours. Scale bar = 100 µm. (F) PFKM-silencing (shPFKM#1 & shPFKM#2 vs shCONT) extends the survival for tumor-bearing mice (n = 5 per arm). (G) Immunoblot analysis of PFKM protein level in PFKMhigh and PFKMlow T10 sub-cell lines used in (H). (H) Kaplan-Meier survival analysis of animals intracranially injected with PFKMhigh or PFKMlow T10 cells and treated intraperitoneally with PBS or BEV (5 mg/kg). (I) Representative H&E staining of coronal section of brains from study shown in (H). Scale bar = 2.5 µm. (J) Bevacizumab (BEV: 5 mg/kg) treatment drives superior survival benefits in mice intracranially implanted with PFKMlow T10 cells.
Significance is determined by log-rank test (F, H) or t test (B–E), where data are shown as mean ± SEM (*P < .05, **P < .01, ***P < .001).
Cytoplasmic PFKM-KIF11 Interaction Promotes Invasion and Mitosis in GBM Cells
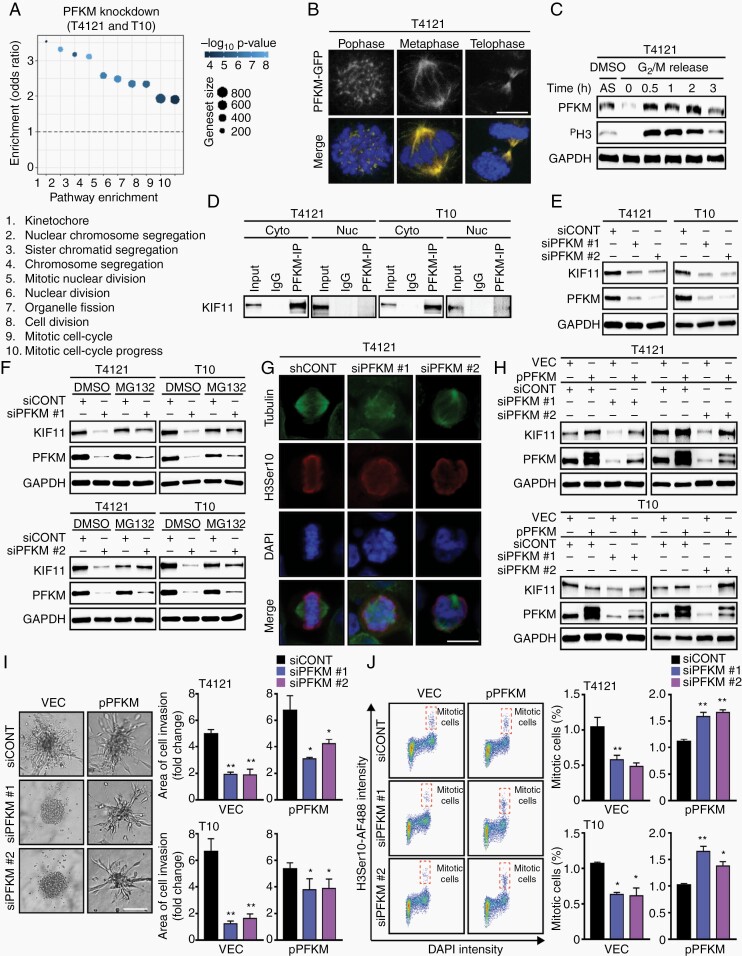
Next, we employed differential RNA sequencing (RNA-seq) in which downregulated genes were subjected to an unsupervised pathway analysis, which revealed the top 10 scoring pathways were associated with aberrant mitosis (Figure 3A). We GFP-tagged PFKM in T4121 GBM cells (PFKM-GFP) to determine cellular localization. Confocal microscopy identified strong association of PFKM with the mitotic spindle (Figure 3B). Synchronization of T4121 GBM cells confirmed higher expression of PFKM during G2/M phase (Figure 3C). Co-immunoprecipitation followed by mass spectrometry analysis identified KIF11 as an interacting partner (Supplementary Figure 3A and B). Protein-protein interaction between PFKM and KIF11 occurred exclusively in the cytosol as demonstrated by PLA and co-immunoprecipitation experiments in cytosolic and nuclear protein fractions of GBM cells (Figure 3D; Supplementary Figure 3C). PFKM knockdown in GBM cells induced proteasome-mediated degradation of KIF11 protein (Figure 3E and F), resulting in impaired cytoskeleton assembly and mitotic catastrophe (Figure 3G). Ectopic expression of exogenous PFKM rescued KIF11 stability (Figure 3H; Supplementary Figure 3D), as well as invasion and mitotic progression (Figure 3I and J). Collectively, these results demonstrate that PFKM binds and stabilizes the KIF11 molecular motor protein to drive GBM proliferation and invasion.
Fig. 3.
PFKM-KIF11 interaction is critical for invasion and mitosis in glioblastoma. (A) Pathway enrichment analysis using genes downregulated (log2fc ≥ 2 and adjusted P ≤ .05) after PFKM-silencing in T10 and T4121 GBM cells. (B) Confocal microscopy confirms ectopically expressed PFKM-GFP to localize to mitotic spindle. Nuclei counterstained by DAPI. Scale bar = 3 µm. (C) Immunoblot analysis of T4121 cells shows increased PFKM level in mitosis. Asynchronous cells (AS); cells synchronized at G2 phase, released and collected at indicated time points. PH3 as mitotic marker. Loading control: GAPDH. (D) Immunoprecipitation of PFKM followed by immunoblot analysis confirms its interaction with KIF11 in the cytosolic fraction (Cyto) and not in the nuclear fraction (Nuc). (E) GBM cells with silenced PFKM show decreased KIF11 protein expression. Loading control: GAPDH. (F) MG132 (10 µM; proteasomal inhibitor) treatment (6 hours) restores KIF11 protein levels in GBM cells with silenced PFKM. Loading control: GAPDH. (G) PFKM loss in GBM cells leads to a mitotic catastrophe. Tubulin, H3Ser10 (red). Nuclei were counterstained with DAPI. Scale bar = 5 µm. (H) Ectopic PFKM expression rescues KIF11 degradation in PFKM-silenced GBM cells. Loading control: GAPDH. (I) Ectopic PFKM expression restores invasion capacity of PFKM-silenced GBM cells. Scale bar = 100 µm. (J) Ectopic PFKM expression rescues mitotic phenotype in PFKM-silenced GBM cells as shown by FACS analysis.
Significance is determined by t test (I, J), and data are shown as mean ± SEM (*P < .05, **P < .01).
Nuclear PFKM Engages DNA Damage Response to Ensure Genome Stability
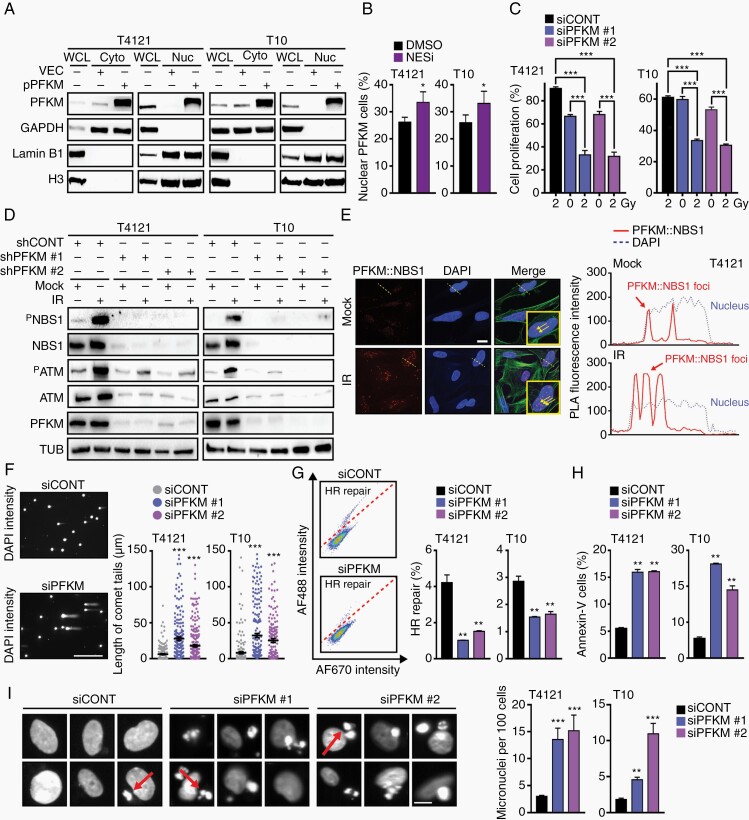
Subcellular protein fractionation experiments confirmed the presence of endogenous as well as ectopically expressed PFKM in the nucleus of GBM cells (nucPFKM; Figure 4A; Supplementary Figure 4A). This observation was also supported by an in silico analysis of the PFKM sequence, which confirmed the presence of nuclear localization signals (NLS, Supplementary Figure 4B). We validated the functionality of NLS in PFKM through the exposure of GBM cells to a nuclear export signal inhibitor, leptomycin (NESi) or the NLS inhibitor, ivermectin (NLSi) which are both commonly used to study nucleo-cytoplasmic protein translocation. Leptomycin caused nuclear PFKM accumulation (Figure 4B) while ivermectin led to PFKM sequestration in the cytosol of GBM cells (Supplementary Figure 4C). Collectively, these data support the nuclear localization of PFKM.
Fig. 4.
Nuclear PFKM regulates DNA damage response to ensure genome stability. (A) Subcellular fractionation followed by immunoprecipitation confirms nuclear localization of ectopically expressed PFKM. Cytosolic marker: GAPDH; nuclear marker: Lamin B1 and H3. (B) PFKM accumulates in the nucleus after leptomycin (NESi; 2 nM) treatment of GBM cells. (C) GBM cells with silenced PFKM show higher sensitivity to IR (2 Gy) as assessed by cell proliferation measurement at 72-hour post-IR. (D) PFKM-silencing impairs IR-induced DDR activation (PATM and PNBS1) in GBM cells. Loading control: tubulin (TUB). (E) PLA assay shows PFKM::NBS1 interaction in the nucleus of GBM cells. Fluorescence intensity peaks (right graph) marked the co-localization (proximity) of PFKM and NBS1 along the dotted line across the selected cell. Scale bar = 10 µm. (F) Representative images of comet tails in GBM cells transfected with siCONT or siPFKM. DSBs quantification was determined 48-hour post-IR by measuring comet tail length (~200 tails per condition was measured). Scale bar = 100 µm. (G) Homologous recombination (HR) repair (GFP-positive cells above diagonal line) rates are lower in GBM cells with silenced PFKM. (H) FACS-based quantification of Annexin V-positive cells (%) shows increased apoptosis in GBM cells with silenced PFKM. (I) Microscopy analysis of GBM cells with silenced PFKM shows higher micronuclei (MN) formation using DAPI staining. Quantification was based on ~1000 cells and presented the number of micronuclei per 100 cells. Images of GBM cells with MN (arrows). Scale bar = 5 µm.
Significance is determined by t test (B, C, and F–I), and data are shown as mean ± SEM (*P < .05, **P < .01, ***P < .001).
Given the disruption of the mitotic spindle and cell-cycle arrest that was driven by PFKM loss, we interrogated the activation of the DNA damage response (DDR) pathway. Supporting the role of nuclear PFKM (nucPFKM) in the DDR, PFKM knockdown impaired activation of the DDR and enhanced sensitivity of GBM cells to IR (Figure 4C and D). NBS1 is a primary sensor for radiation-induced DNA double-strand breaks (DSBs) and two independent assays (IP and PLA) confirmed an interaction of nucPFKM with NBS1 in the nucleus of GBM cells (Figure 4E; Supplementary Figure 4D). Next, we assessed the impact of PFKM loss on DSBs accumulation and found that PFKM loss induced DNA damage (DSB count, comet tail moment) and impaired DSB repair via HR repair (Figure 4F and G). This enhanced genomic instability translated into apoptosis measured by increased micronuclei (MN) count and frequency of Annexin V-positive cells (Figure 4H and I). Collectively, PFKM displays a distinct nuclear localization and regulation of the DDRs to protect GBM from genomic instability and cell death by apoptosis.
Bupivacaine Phenocopies PFKM Loss
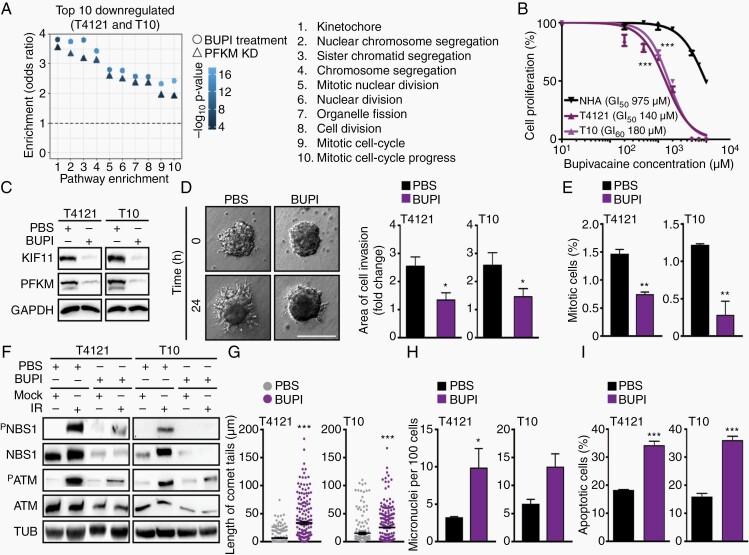
To determine whether the above-described phenotypes associated with PFKM loss are due to its role in glycolysis, we attempted to rescue the loss of PFKM by treating with fructose 1,6-bisphosphate (F1,6P), a downstream metabolite (Supplementary Figure 5A),20 which failed to fully rescue the proliferation defects of PFKM-depleted GBM cells (Supplementary Figure 5B). The exposure of GBM cells to 2-deoxy-d-glucose (2-DG; a glucose analog that cannot undergo further glycolysis)21 effectively inhibited PFKM kinase activity but did not impact KIF11 or NBS1 protein levels (Supplementary Figure 5C and D) altogether suggesting that this role of PFKM is independent from its metabolic activity. In a search for agents that would mimic the PFKM loss phenotype, such as invasion and DDR activation, we interrogated RNA-seq data of GBM cells with silenced PFKM and sought drug treatments that overlapped in differential gene expression, revealing the local anesthetic, bupivacaine (Figure 5A). The top 10 downregulated pathways upon PFKM depletion or bupivacaine treatment were identical and strongly correlated (Supplementary Figure 5E). We examined the inhibitory effects of bupivacaine on GBM cells and NHAs, as measured by concentrations that induced 50% growth inhibition (GI50), revealing preferential sensitivity of GBM cells (T4121 GI50 = 140 µM; T10 GI50 = 180 µM) relative to NHAs (GI50 = 975 µM) (Figure 5B). Furthermore, bupivacaine treatment reduced both PFKM and KIF11 protein expression, induced stress fiber formation, and impaired the migratory potential of GBM cells (Figure 5C and D; Supplementary Figure 5F and G). Bupivacaine monotherapy decreased the mitotic index (Figure 5E) and sensitized GBM cells to radiation as demonstrated by impaired DDR activation (Figure 5F), DNA damage accumulation, increased MN formation, and cell death (Figure 5G–I; Supplementary Figure 5H–J).
Fig. 5.
Bupivacaine phenocopies PFKM loss to potentiate the effect of bevacizumab. (A) Pathway enrichment analysis using overlapping up- and downregulated genes in GBM cells with silenced PFKM and treated with bupivacaine (BUPI) (log2fc ≥ 2 and adjusted P ≤ .05) de-regulated mitosis and cell division among the top 10 scoring hits. (B) GBM cells show higher sensitivity to bupivacaine compared to normal human astrocytes (NHA). GI50 was assessed at 72-hour post-treatment. (C) Bupivacaine treatment (24 hours) of GBM cells decreases PFKM and KIF11 protein expression. Loading control: GAPDH. (D) Bupivacaine treatment (24 hours) impairs GBM cell invasion. Quantification was performed by normalizing to 0 hours. Scale bar = 100 µm. (E) Bupivacaine treatment of GBM cells lowers mitotic index (% of pH3-positive cells). Nuclei were counterstained with DAPI. (F) Bupivacaine treatment (24 hours) of GBM cells impairs IR-induced DDR activation (PATM and PNBS1) Loading control: TUB. (G) Comet assay shows increased DSB formation in GBM cells treated with bupivacaine for 24 hours (~200 tails per condition was measured). (H) Bupivacaine treatment (24 hours) of GBM cells increases micronuclei (MN) formation. MN quantification was based on ~1000 cell count and presented as a number of identified MN per 100 cells. (I) FACS-based quantification of Annexin V-positive cells (%) shows higher apoptotic rate in GBM cells treated with bupivacaine for 24 hours.
Significance is determined by t test (B, D, E, G–I), and data are shown as mean ± SEM (*P < .05, **P < .01, ***P < .001).
Bupivacaine Impairs Glioblastoma Growth and Augments Bevacizumab Efficacy In Vivo
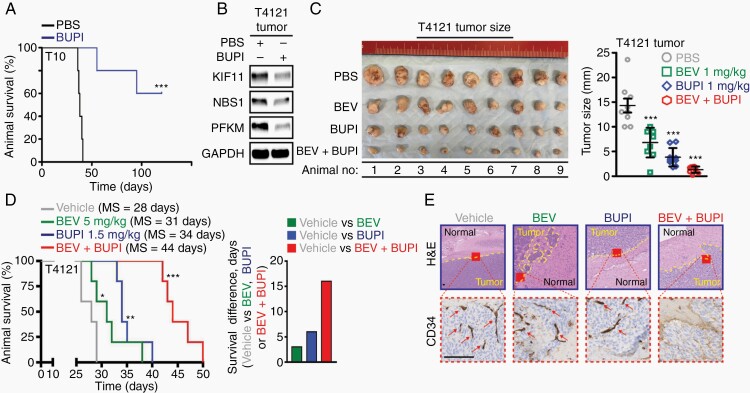
Based on the in vitro anti-tumor properties of bupivacaine, we interrogated the efficacy of bupivacaine against tumor growth in vivo. Pre-treatment of GBM cells with bupivacaine prior to their implantation displayed extended survival compared to those implanted with GBM cells exposed to vehicle control (Figure 6A). As we initially identified PFKM as a driver of bevacizumab resistance and found bupivacaine to phenocopy PFKM loss, we hypothesized that bupivacaine augments anti-tumor efficacy of bevacizumab. First, we confirmed that bupivacaine reduces PFKM expression in vivo (Figure 6B) and then assessed its effect on tumor growth alone or in combination with bevacizumab using a subcutaneous T4121 xenograft model. As shown in Figure 6C, intratumoral (i.t.) injection of bupivacaine in combination with bevacizumab (i.p.) led to a superior tumor growth inhibition compared to either of the monotherapies alone. To validate this observation in a clinically relevant orthotopic PDX model (T4121), we mounted a cranial screw guide on the head of immunocompromised animals to allow for the intratumoral administration of bupivacaine. Here, the co-administration of bupivacaine (i.t.) with bevacizumab (i.p.) showed a marked survival benefit compared to either monotherapies or vehicle control (Figure 6D; Supplementary Figure 6A). In addition to a significant survival benefit and decreased tumor burden, IHC analysis confirmed reduced invasion and vascularization (CD34) in remnant tumors of mice receiving the combination therapy (Figure 6E; Supplementary Figure 6B). Collectively, these results suggest bupivacaine as a potential combination partner to improve the efficacy of bevacizumab in GBM therapy.
Fig. 6.
Bupivacaine impairs glioblastoma growth and augments bevacizumab efficacy in vivo. (A) Bupivacaine pre-treatment (GI50 for 24 hours) in vitro reduces tumorigenicity of T10 GBM cells intracranially implanted in the frontal lobe of immunocompromised mice (n = 5 per arm). (B) Bupivacaine treatment (intratumoral injection at 1 mg/kg on alternate days for 2 weeks) reduces PFKM, KIF11, and NBS1 expression in GBM tumors grown in immunocompromised mice. Loading control: GAPDH. Combinational bupivacaine and bevacizumab therapy inhibited tumor growth with a greater efficacy than either of the monotherapies in subcutaneous (C) as well as (D) in orthotopic GBM xenograft model. Treatment was given on alternate days for 3 weeks: (i) vehicle PBS intraperitoneally and vehicle artificial cerebral spinal fluid intratumorally via a guide screw; (ii) 5 mg/kg bevacizumab/BEV intraperitoneally; (iii) 1.5 mg/kg bupivacaine/BUPI intratumorally via a guide screw; and (iv) the combination of bevacizumab/BEV and bupivacaine/BUPI. (E) H&E analysis and immunohistochemistry assessment of CD34 biomarker in representative coronal brain sections confirmed reduced tumor burden, parenchymal invasion, and vascularization (arrows) in animals co-treated with bevacizumab and bupivacaine. Scale bar = 100 µm.
Significance is determined by log-rank test (A, D) or t test (C), where data are shown as mean ± SEM (*P < .05, **P < .01, ***P < .001).
Discussion
Microenvironmental cues, such as limited access to nutrients, hypoxia, and necrosis, dictate the metabolic pathway choice in GBM.22,23 Bevacizumab treatment induces a metabolic switch to anaerobic glycolysis in GBM and enhances the invasion of GBM cells.24 PFK-1 is the pacemaker of glycolysis and executes its functions through multiple tetrameric isozymic forms consisting of three isoforms: muscle (PFKM), liver (PFKL), and platelet (PFKP), with variable composition dictated by tissue and cell type.25,26 Here, we identify PFKM as a driver of bevacizumab resistance in GBM and mechanistically interrogate its non-canonical roles dictated by subcellular localization. While our data implicate the cytosolic PFKM to play a role in invasion and cytoskeleton assembly, nuclear PFKM is found to safeguard the genomic stability of GBM cells. PFKM is not the first PFK-1 isoform found to play a non-glycolytic role as Lee et al showed previously that PFKP binds and phosphorylates AKT on Ser386 thereby promoting tumorigenesis.27 Pyruvate kinase M2 (PKM2) was among the first glycolytic enzymes shown to regulate cell-cycle progression and tumorigenesis and its loss has been associated with similar phenotypes as those observed herein for PFKM. While cytosolic PKM2 regulates tumorigenesis by stabilizing mutant EGFR protein via direct interaction, nuclear PKM2 was shown to regulate the expression of numerous proglycolytic enzymes and G1-S phase transition of the cell cycle.28–31
Aberrant DNA repair is associated with metabolic reprogramming, in general, and enrichment of glycolytic-gluconeogenesis proteins, in particular.32,33 Consistent with this concept, glycolytic enzymes, such as PKM2 and PFKFB3, are implicated in DNA repair.34,35 Here, we found nuclear PFKM is critical for DDR activation and the error-free repair of DSBs via HR. In the absence of PFKM, GBM cells fail to respond to radiation-induced DNA damage and in consequence die by apoptosis.
Preferential expression of PFKM in the tumor periphery is concordant with previous reports on glycolytic enzymes, such as ALDOC,15 which was among the candidates identified through our in silico screen. Previously, ALDOC expression was found elevated in invading GBM cells under hypoxia. While ALDOC knockdown impaired the invasion of GBM cells in vitro, in vivo experiments showed accelerated tumorigenesis and shorter survival of tumor-bearing mice.36 In contrast to ALDOC, we show that PFKM-silencing negatively impacts the viability of GBM cells both in vitro and in vivo. To further dissect the mechanistic underpinnings of PFKM-regulated invasion, cell-cycle regulation, and mitotic spindle assembly, we find PFKM to stabilize the molecular motor protein KIF11 by interacting with it in the cytosol of GBM cells. These findings build on our previous reports on KIF11 regulating GBM cell motility and invasion.5
Retrospective analysis and data from experimental studies point toward a potential beneficial effect of the local anesthetics regarding clinical outcome—that is, overall and/or recurrence-free survival in patients undergoing cancer surgery.37 Lidocaine sensitizes breast cancer and GBM to cisplatin.38,39 Here, we show that bupivacaine, another local anesthetic, phenocopies PFKM loss and inhibits the survival, motility, and invasion of GBM cells and extends the survival of tumor-bearing mice. In vitro effects of bupivacaine on GBM cells are consistent with previous reports in breast, ovarian, and prostate cancers, where clinically relevant concentrations inhibited their survival.40,41 The in vivo data presented in this study underline the clinical relevance of our findings and show that bupivacaine monotherapy reduces the tumorigenicity of GBM cells in vivo. As high PFKM levels predicted poor prognosis of GBM patients undergoing bevacizumab therapy, the superior efficacy of bupivacaine and bevacizumab combination compared to either of the drugs alone, provides the proof of concept and validates our initial hypothesis. Optimization of drug efficacy in the brain requires understanding of the local exposure to unbound drug at the site of action, which is impacted by the blood-brain barrier (BBB). The work by Mateus et al suggests that bupivacaine penetrates the BBB.42 To avoid systemic toxicity, localized drug delivery could be considered. A pharmaceutical grade liposomal bupivacaine formulation has been clinically approved for pain management (Exparel)43 and could be potentially explored for local delivery at the surgical resected site of GBM.
Encouraged by our findings, we believe that PFKM may serve as an attractive therapeutic target for GBM. The early evidence provided here suggests that PFKM inhibition therapy can potentiate the efficacy of bevacizumab and warrants further investigation to fulfill the therapeutic promise of this early preclinical study.
Supplementary Material
Acknowledgments
We thank Dr. Jiri Bartek, Jr. for providing bupivacaine.
Contributor Information
Yi Chieh Lim, Danish Cancer Society, Denmark.
Kamilla E Jensen, Danish Cancer Society, Denmark.
Diana Aguilar-Morante, Danish Cancer Society, Denmark.
Lina Vardouli, Danish Cancer Society, Denmark.
Kristoffer Vitting-Seerup, Danish Cancer Society, Denmark; Department of Health Technology, Danish Technical University, Denmark.
Ryan C Gimple, Department of Medicine, Division of Regenerative Medicine, University of California San Diego, La Jolla, CA, USA.
Qiulian Wu, Department of Medicine, Division of Regenerative Medicine, University of California San Diego, La Jolla, CA, USA.
Henriette Pedersen, Danish Cancer Society, Denmark.
Kirstine J Elbaek, Danish Cancer Society, Denmark.
Irina Gromova, Danish Cancer Society, Denmark.
Robert Ihnatko, Institute of Pathology, University Medical Center, Goettingen University, Germany.
Bjarne W Kristensen, Department of Pathology, Odense University Hospital, Denmark.
Jeanette K Petersen, Department of Pathology, Odense University Hospital, Denmark; Department of Clinical Research, University of Southern Denmark, Denmark.
Jane Skjoth-Rasmussen, Department of Neurosurgery, Copenhagen University Hospital, Denmark.
William Flavahan, Department of Pathology and Center for Cancer Research, Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA.
Jeremy N Rich, Department of Medicine, Division of Regenerative Medicine, University of California San Diego, La Jolla, CA, USA; UPMC Hillman Cancer Center, University of Pittsburgh, Pittsburgh, PA, USA.
Petra Hamerlik, Danish Cancer Society, Denmark.
Funding
This work was supported by the Novo Nordisk (NNF17OC0026056, NNF19OC0058676 to P.H), Danish Cancer Society (R146-A9511, R148-A10151 to P.H), and National Institutes of Health (CA217065 to R.C.G; CA197718, CA238662, NS103434 to J.N.R.).
Conflict of interest statement. Petra Hamerlik is currently a full-time employee of AstraZeneca UK. All other authors declare no conflicts.
Authorship statement. Concept and design: Y.C.L., J.N.R., and P.H. Data acquisition: Y.C.L., K.E.J., D.A.M., Q.W., L.V., I.G., R.I., B.W.K., J.K.P., H.P., K.J.E., and P.H. Data analysis and interpretation: Y.C.L., W.F., D.A.M., K.E.J., K.V.S., R.C.G., J.K.P., J.S.R., and P.H. Manuscript writing: Y.C.L., D.A.M., P.H., and J.N.R. Study supervision: P.H.
References
- 1. Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu KV, Chang JP, Parachoniak CA, et al. VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell. 2012;22(1):21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gilbert MR, Dignam JJ, Armstrong TS, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Michaelsen SR, Staberg M, Pedersen H, et al. VEGF-C sustains VEGFR2 activation under bevacizumab therapy and promotes glioblastoma maintenance. Neuro Oncol. 2018;20(11):1462–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Venere M, Horbinski C, Crish JF, et al. The mitotic kinesin KIF11 is a driver of invasion, proliferation, and self-renewal in glioblastoma. Sci Transl Med. 2015;9(304):304ra143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Keunen O, Johansson M, Oudin A, et al. Anti-VEGF treatment reduces blood supply and increases tumor cell invasion in glioblastoma. Proc Natl Acad Sci USA. 2011;108(9):3749–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. [DOI] [PubMed] [Google Scholar]
- 8. Taal W, Oosterkamp HM, Walenkamp AME, et al. Single-agent bevacizumab or lomustine versus a combination of bevacizumab plus lomustine in patients with recurrent glioblastoma (BELOB trial): a randomised controlled phase 2 trial. Lancet Oncol. 2014;15(9):943–953. [DOI] [PubMed] [Google Scholar]
- 9. Nowosielski M, Ellingson BM, Chinot OL, et al. Radiologic progression of glioblastoma under therapy—an exploratory analysis of AVAglio. Neuro Oncol. 2018;20(4):557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chinot OL, Wick W, Mason W, et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):709–722. [DOI] [PubMed] [Google Scholar]
- 11. Vinci M, Box C, Eccles SA. Three-dimensional (3D) tumor spheroid invasion assay. J Vis Exp. 2015;( 99):e52686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lim YC, Ensbey KS, Offenhauser C, et al. Simultaneous targeting of DNA replication and homologous recombination in glioblastoma with a polyether ionophore. Neuro Oncol. 2020;22(2):216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shaiken TE, Opekun AR. Dissecting the cell to nucleus, perinucleus and cytosol. Sci Rep. 2014;4:4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Michaelsen SR, Staberg M, Pedersen H, et al. VEGF-C sustains VEGFR2 activation under bevacizumab therapy and promotes glioblastoma maintenance. Neuro Oncol. 2018;20(11):1462–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang CS, Hawley SA, Zong Y, et al. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature. 2017;548(7665):112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Webb BA, Forouhar F, Szu FE, et al. Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations. Nature. 2015;523(7558):111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Darmanis S, Sloan SA, Croote D, et al. Single-cell RNA-seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 2017;21(5):1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bernatchez PN, Soker S, Sirois MG. Vascular endothelial growth factor effect on endothelial cell proliferation, migration, and platelet-activating factor synthesis is Flk-1-dependent. J Biol Chem. 1999;274(43):31047–31054. [DOI] [PubMed] [Google Scholar]
- 19. Lu-Emerson C, Duda DG, Emblem KE, et al. Lessons from anti-vascular endothelial growth factor and anti-vascular endothelial growth factor receptor trials in patients with glioblastoma. J Clin Oncol. 2015;33(10):1197–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seki Y, Sato K, Kono T, Akiba Y. Two types of phosphofructokinase-1 differentially regulate the glycolytic pathway in insulin-stimulated chicken skeletal muscle. Comp Biochem Phys B. 2006;143(3):344–350. [DOI] [PubMed] [Google Scholar]
- 21. Zhao J, Ma Y, Zhang Y, et al. Low-dose 2-deoxyglucose and metformin synergically inhibit proliferation of human polycystic kidney cells by modulating glucose metabolism. Cell Death Discov. 2019;5:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Flavahan WA, Wu Q, Hitomi M, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16(10):1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han T, Kang D, Ji D, et al. How does cancer cell metabolism affect tumor migration and invasion? Cell Adh Migr. 2013;7(5):395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fack F, Espedal H, Keunen O, et al. Bevacizumab treatment induces metabolic adaptation toward anaerobic metabolism in glioblastomas. Acta Neuropathol. 2015;129(1):115–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mor I, Cheung EC, Vousden KH. Control of glycolysis through regulation of PFK1: old friends and recent additions. Cold Spring Harb Symp Quant Biol. 2011;76:211–216. [DOI] [PubMed] [Google Scholar]
- 26. Moreno-Sanchez R, Rodriguez-Enriquez S, Marin-Hernandez A, Saavedra E. Energy metabolism in tumor cells. FEBS J. 2007;274(6): 1393–1418. [DOI] [PubMed] [Google Scholar]
- 27. Lee JH, Liu R, Li J, et al. Stabilization of phosphofructokinase 1 platelet isoform by AKT promotes tumorigenesis. Nat Commun. 2017;8(1):949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang W, Xia Y, Hawke D, et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150(4):685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Palsson-McDermott EM, Curtis AM, Goel G, et al. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the Warburg effect in LPS-activated macrophages. Cell Metab. 2015;21(1):65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang W, Xia Y, Ji H, et al. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature. 2011;480(7375):118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alves-Filho JC, Palsson-McDermott EM. Pyruvate kinase M2: a potential target for regulating inflammation. Front Immunol. 2016;7:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marzano V, Santini S, Rossi C, et al. Proteomic profiling of ATM kinase proficient and deficient cell lines upon blockage of proteasome activity. J Proteomics. 2012;75(15):4632–4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schneider JG, Finck BN, Ren J, et al. ATM-dependent suppression of stress signaling reduces vascular disease in metabolic syndrome. Cell Metab. 2006;4(5):377–389. [DOI] [PubMed] [Google Scholar]
- 34. Gustafsson NMS, Farnegardh K, Bonagas N, et al. Targeting PFKFB3 radiosensitizes cancer cells and suppresses homologous recombination. Nat Commun. 2018;9(1):3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sizemore ST, Zhang M, Cho JH, et al. Pyruvate kinase M2 regulates homologous recombination-mediated DNA double-strand break repair. Cell Res. 2018;28(11):1090–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kathagen-Buhmann A, Schulte A, Weller J, et al. Glycolysis and the pentose phosphate pathway are differentially associated with the dichotomous regulation of glioblastoma cell migration versus proliferation. Neuro Oncol. 2016;18(9):1219–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Muller SD, Ziegler JSH, Piegeler T. Local anesthetics and recurrence after cancer surgery—what’s new? A narrative review. J Clin Med. 2021;10(4):719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fan X, Yang H, Zhao C, et al. Local anesthetics impair the growth and self-renewal of glioblastoma stem cells by inhibiting ZDHHC15-mediated GP130 palmitoylation. Stem Cell Res Ther. 2021;12(1):107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li K, Yang J, Han X. Lidocaine sensitizes the cytotoxicity of cisplatin in breast cancer cells via up-regulation of RARβ2 and RASSF1A demethylation. Int J Mol Sci. 2014;15(12):23519–23536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li R, Xiao C, Liu H, et al. Effects of local anesthetics on breast cancer cell viability and migration. BMC Cancer. 2018;18(1):666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xuan W, Zhao H, Hankin J, et al. Local anesthetic bupivacaine induced ovarian and prostate cancer apoptotic cell death and underlying mechanisms in vitro. Sci Rep. 2016;6:26277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mateus A, Matsson P, Artursson P. A high-throughput cell-based method to predict the unbound drug fraction in the brain. J Med Chem. 2014;57(7):3005–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Torchilin VP. Recent advances with liposomes as pharmaceutical carriers. Nat Rev Drug Discov. 2005;4(2):145–160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.