Abstract
Extracellular vesicles (EV) are membranous particles secreted by all cells and found in body fluids. Established EV contents include a variety of RNA species, proteins, lipids and metabolites that are considered to reflect the physiological status of their parental cells. However, to date, little is known about cell‐type enriched EV cargo in complex EV mixtures, especially in urine. To test whether EV secretion from distinct human kidney cells in culture differ and can recapitulate findings in normal urine, we comprehensively analysed EV components, (particularly miRNAs, long RNAs and protein) from conditionally immortalised human kidney cell lines (podocyte, glomerular endothelial, mesangial and proximal tubular cells) and compared to EV secreted in human urine. EV from cell culture media derived from immortalised kidney cells were isolated by hydrostatic filtration dialysis (HFD) and characterised by electron microscopy (EM), nanoparticle tracking analysis (NTA) and Western blotting (WB). RNA was isolated from EV and subjected to miRNA and RNA sequencing and proteins were profiled by tandem mass tag proteomics. Representative sets of EV miRNAs, RNAs and proteins were detected in each cell type and compared to human urinary EV isolates (uEV), EV cargo database, kidney biopsy bulk RNA sequencing and proteomics, and single‐cell transcriptomics. This revealed that a high proportion of the in vitro EV signatures were also found in in vivo datasets. Thus, highlighting the robustness of our in vitro model and showing that this approach enables the dissection of cell type specific EV cargo in biofluids and the potential identification of cell‐type specific EV biomarkers of kidney disease.
Keywords: exosomes, extracellular vesicles, glomerular endothelial cells, kidney, mesangial cells, miRNA, podocytes, proteomics, proximal tubule cells, RNA
1. INTRODUCTION
Extracellular Vesicles (EV) are cell‐derived small membranous particles involved in cell‐to‐cell communication (Yáñez‐Mó et al., 2015). As EV often retain protein, lipids and nucleic acids from their cell types of origin, they can reflect the underlying pathophysiological state of those cells (Barreiro & Holthofer, 2017; Iraci et al., 2016; Yuana et al., 2013) and since they are present in body fluids, they are considered a promising source of non‐invasive biomarkers for numerous diseases (Dickhout & Koenen, 2018; Liang et al., 2021; Pang et al., 2020; Xu et al., 2020).
EV in the urine has gained particular interest as non‐invasive biomarkers of kidney disease. They are secreted by (and contain molecules from) all cell types constituting the nephron (Erdbrügger & Le, 2016). Numerous studies have begun to characterise uEV in kidney health and in disease (Braun et al., 2020; Ghai et al., 2018; Sonoda et al., 2019), although the methodologies to isolate and measure EV have been limited (Barreiro & Holthofer, 2017; Merchant et al., 2017).
Notably, due to the complexity and heterogeneity of uEV, it can be difficult to define the origin of the parental cells. As such, the profiles of uEV secreted by individual kidney cell types, and how their content directly reflects cellular protein/nucleic acid content, are not yet understood. Such knowledge would not only help to understand injury sites and target the cell for biological intervention but also design useful in vitro model systems. Some attempts have been made to identify cell (Connor et al., 2020; Müller‐Deile et al., 2019) and tissue (Baker et al., 2017; Bazzell et al., 2018; Habuka et al., 2014) enriched miRNAs, mRNA or proteins and their contributions to biofluids composition (Cui & Cui, 2020; Hulstaert et al., 2020). Only recently, the contribution of cell and tissue derived EV to biofluids (plasma) was explored by gene expression signatures and deconvolution (Li et al., 2020). However, to date little is known about cell‐type enriched EV cargo in urine.
Conditionally‐immortalised kidney cell culture models have provided useful tools in studying molecular responses of individual kidney cell types (Audzeyenka et al., 2021; Grigorieva et al., 2019; Lau et al., 2012; Lay et al., 2017; Mutsaers et al., 2013; Ramnath & Satchell, 2020; Ramnath et al., 2014; Saleem et al., 2002; Wright et al., 2019) . Furthermore, the ease in which cell culture models can be ‘up‐scaled’ to provide sufficient starting material when isolating EV and studying their cargo has obvious benefits.
Using our recently characterised EV isolation workflows (Barreiro et al., 2020), we here characterised the EV cargo from conditionally‐immortalised human kidney cell lines to provide a clearer understanding of the composition and biology of EV secreted by distinct kidney cell types. We further cross‐compare our findings from human cell lines with those from human uEV and kidney biopsies to further establish the utility of these models in studying human uEV signatures.
2. MATERIALS AND METHODS
The experimental workflow is shown in Figure 1.
FIGURE 1.
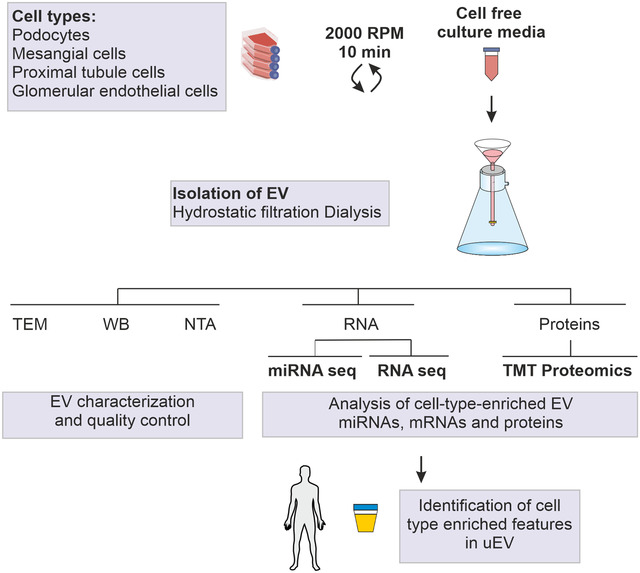
Experimental workflow: EV were isolated from 50 ml of cell free culture media by hydrostatic filtration dialysis. Vesicles were characterised by TEM, WB and NTA. Isolated total RNA was subjected to miRNA‐ and RNA‐sequencing (n = 5 per cell type). Isolated proteins were subjected to quantitative proteomics (TMT approach). Omics dataset were used to define cell‐type‐enriched EV features. Lists of enriched features were tested in human uEV datasets. EV, extracellular vesicles; miRNA‐seq, miRNA sequencing; NTA, nanoparticle tracking analysis; RNA‐seq, RNA sequencing; TMT, tandem mass tag; TEM, transmission electron microscopy; uEV, urinary extracellular vesicles; WB, Western blotting
2.1. Cell culture
Conditionally‐immortalised human cell lines were grown to 70% confluence at 33°C with 5% CO2 before thermo‐switching to 37°C in 5% CO2, allowing them to differentiate for 9–12 days. Conditionally immortalised human podocytes (POD) (Saleem et al., 2002) and mesangial cells (MC) (Sarrab et al., 2011) were maintained in RPMI‐1640 containing L‐glutamine and NaHCO3, supplemented with 10% or 20% FBS, respectively (Gibco). Conditionally immortalised human glomerular endothelial cells (GEC), (Satchell et al., 2006) were maintained in Endothelial Cell Growth Medium‐2, containing microvascular SingleQuots Supplement Pack in 5% FBS (Lonza). Conditionally immortalised human proximal tubular cells (PTC) (Wilmer et al., 2010) were maintained in DMEM‐HAM F‐12 (Lonza) containing 36 ng/ml hydrocortisone (Sigma), 10 ng/ml EGF (Sigma) and 40 pg/ml Tri‐iodothyronine (Sigma) and 10% FBS. Prior to study all cells were washed and incubated with 50 ml exosome‐depleted FBS (Gibco) for 24 h. Conditioned media was centrifuged for 10 min at 2000 g to remove cell debris and stored at –80 before EV harvest.
2.2. Extracellular vesicles isolation
Cell free culture media (CCM) samples were incubated at 37°C in a water bath and vortexed for 30 s. EV isolation by hydrostatic filtration dialysis was performed as described previously with some modifications (Barreiro et al., 2020; Musante et al., 2014). Briefly, 50 ml of CCM was poured into a cellulose ester (CE) dialysis membrane made with molecular weight cut‐off (MWCO) 1000 kDa (Repligen Corp., Waltham, MA). The sample was first concentrated to 5–6 ml. Thereafter, the membrane was refilled with 100 ml of deionised water to wash remaining analytes below the MWCO. When the sample inside the membrane reached about 1 ml, it was collected in protein or DNA LowBind tubes (Eppendorf, Hamburg, Germany) and stored at –80°C. The HFD device was connected to a vacuum pump at a constant pressure of –20 kPa.
2.3. Nanoparticle tracking analysis (NTA)
NTA was used to assess the concentration and size distribution of the isolated EV (Gardiner et al., 2013). EV samples were analysed using Nanosight model LM14 (Malvern Instruments Ltd, Malvern, UK) equipped with blue (404 nm, 70 mW) laser and SCMOS camera (Hamamatsu photonics K.K., Hamamatsu, Japan). To obtain 40–100 particles/view samples were diluted in 0.1 μm filtered (Millex VV, Millipore) DPBS. Five 30‐s videos were recorded using camera level 14 and analysed with NTA software 3.0 (Malvern Instruments Ltd) with the detection threshold 5 and screen gain at 10.
2.4. Electron microscopy
Negative staining was performed as previously described (Barreiro et al., 2020; Puhka et al., 2017). Briefly, 5 μl of sample (equivalent to 0.5–1 ml of CCM) was loaded on Formvar/Carbon 200 mesh TH, Copper grids (Ted Pella Inc., Redding, CA), fixed with 2% PFA (Electron Microscopy Sciences, Hatfield, PA), negatively stained with 2% neutral uranyl acetate and embedded in methyl cellulose uranyl acetate mixture (1.8/0.4%). Images were viewed using Jeol JEM‐1400 (Jeol Ltd, Tokyo, Japan) operating at 80 kV.
2.5. Sample concentration and protein quantification
EV samples to be used for negative staining and Western blotting were 4X concentrated using Amicon Ultra‐0.5 ml Centrifugal Filters 10k (Merck Millipore) following manufacturer's instructions.
Protein concentration was measured as previously described (Barreiro et al., 2020) using Micro BCA™ Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA).
2.6. Western blotting
Western blotting was performed as described previously (Barreiro et al., 2020). Briefly, 20 ug of EV samples were mixed with 5X Laemmli buffer, denatured, and loaded to 4%–20% Mini‐PROTEAN® TGX Stain‐Free Protein Gels (Bio‐Rad laboratories Inc., Hercules, CA). After transference, membranes were blocked and incubated with primary antibodies (TSG‐101, Santa Cruz Biotechnology SC‐7964, dilution 1:200; CD63 MEM, ExBio 11‐343‐C100, dilution 1:500; CD81, Santa Cruz Biotechnology SC‐166029, dilution 1:200) overnight at +4°C. Afterwards, membranes were incubated with secondary antibody. Secondary antibody (Rabbit anti‐mouse, HRP, Jackson Immuno Research 315‐035‐045, dilution 1:6000) was detected using Pierce™ ECL Plus Western Blotting Substrate (Thermo Fisher Scientific) and ChemiDoc Imager system (Bio‐Rad laboratories Inc.).
2.7. RNA isolation
Total RNA from 400 ul of EV samples (equivalent to 20 ml of CCM) was isolated using TRIzol LS (Life Technologies Corp., Carlsbad, CA) following manufacturer's instructions. After phase separation, we used miRNeasy mini kit (Qiagen, Hilden, Germany) following manufacturer's protocol for Purification of Total RNA, Including Small RNAs, from Animal Cells. RNA was eluted in 30 μl of RNase free water. RNA concentration and profile were analyzed using Agilent RNA 6000 Pico kit (Agilent Technologies, Santa Clara, CA).
2.8. RNA sequencing
The RNA cDNA libraries were prepared from 2 ng of total RNA using SMART‐Seq® stranded kit (Takara BIo Inc., Mountain View, CA).
miRNA libraries were prepared using the QIAseq miRNA Library Kit (Qiagen) following the manufacturer's protocol, starting with ∼1 ng of input RNA for each sample. Quality control and concentrations of individual libraries were assessed with Bioanalyzer 2100 Instrument and High Sensitivity DNA Kit (Agilent Technologies).
miRNA and RNA sequencing was performed on the Illumina HiSeq 4000 platform (Illumina Inc.) as 85 bp cycle single end read.
2.9. RNAseq data analysis
Illumina reads from mRNA were converted to the industry standard FASTQ format and aligned to the human reference genome from Ensembl 84 using the STAR v2.5.2a aligner on default settings (Dobin et al., 2013). For increased alignment accuracy, the STAR genome index was generated to include splice junction annotations with the options ‘–sjdbGTFfile’ and ‘–sjdbOverhang 49′. The quality of the RNA‐Seq samples was verified with FASTQC version 0.11.2 and RNASeQC version 1.18 (DeLuca et al., 2012). The featureCounts tool of the subRead package version 1.5.0 (Liao et al, 2014) was used to quantify the per‐gene read counts.
For the miRNA, adapter sequences were trimmed via CutAdapt v.1.16 and UMI reads were collapsed with a customised script. The trimmed reads were mapped against the human reference genome hg19 using STAR version 2.5.2a. Reads 16–26 nucleotides in length were filtered (samtools v0.1.18) to quantify mature miRNAs and reads mapping to the miRNAs annotated in miRBase release 20 were counted applying featureCounts v.1.6.2.
Filtering and normalisation: Protein‐coding genes from the RNA‐seq data were studied. For noise filtering, we kept only genes with log2 cpm (counts per million) ≥5 from at least one library in the analysis. The RNA‐seq read count data were normalised with the TMM (trimmed mean of M‐values) method from the edgeR R package (v3.22.5) (Robinson & Oshlack, 2010) and transformed to log2 cpm with the voom method from the limma R package (v3.38.3) (Law et al., 2014). For miRNA, we considered those with non‐empty mapping in at least one library. The miRNA‐seq log2 count data were normalised with the cyclic loess method also from limma.
2.10. Tandem mass tag proteomics
EV secreted by a matched number of cells (95,000) was evaporated to dryness for each sample. As cell type and culture conditions can affect, for example, cell growth and number of EV secreted and currently no standardised guidelines on EV proteomics normalisation are in place, we considered that the best approach to answer our research question was to isolate proteins from EV secreted by same number of cells (instead of doing the isolation from same amount of proteins or same amount of EV). Each pellet was resuspended in 50 μl RIPA buffer (25 mM Tris‐HCl pH 7.6, 150 mM NaCl, 1% NP‐40, 1% sodium deoxycholate, 0.1% SDS) plus proteinase inhibitors (cOmplete™ Protease Inhibitor Cocktail, Sigma‐Aldrich). The samples were centrifuged at 14,000 g for 15 min and the supernatant removed. The protein concentration was measured with Bradfords reagent (Bio‐Rad, UK) using 10 μl of sample as per the manufacturer's instructions.
Isolated proteins were denatured, reduced, alkylated and tryptic digested following manufacturer's instructions.
For peptide labelling, 41 μl anhydrous acetonitrile was added to room temp TMT reagents, they were left to dissolve for 5 min with occasional vortexing, the tube was centrifuged to collect the label then 41 μl added to each sample. Samples were incubated for 1 h at room temperature. A total of 8 μl of 5% hydroxylamine was added to quench the labelling reaction and incubated for 15 min at room temperature. For each experiment all samples were combined and evaporated to dryness, resuspended in 5% formic acid and then desalted using a SepPak cartridge according to the manufacturer's instructions (Waters, Milford, MA). Eluate from the SepPak cartridge was again evaporated to dryness and resuspended in buffer A (20 mM ammonium hydroxide, pH 10) prior to fractionation by high pH reversed‐phase chromatography using an Ultimate 3000 liquid chromatography system (Thermo Scientific). In brief, the sample was loaded onto an XBridge BEH C18 Column (130Å, 3.5 μm, 2.1 mm × 150 mm, Waters, UK) in buffer A and peptides eluted with an increasing gradient of buffer B (20 mM Ammonium Hydroxide in acetonitrile, pH 10) from 0%–95% over 60 min. The resulting fractions (15) were evaporated to dryness and resuspended in 1% formic acid prior to analysis by nano‐LC MSMS using an Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific).
2.11. Proteomics data analysis
The raw data files were processed and quantified using Proteome Discoverer software v2.1 (Thermo Scientific) and searched against the UniProt Human database (downloaded September 2018: 152,927 entries) using the SEQUEST HT algorithm. Peptide precursor mass tolerance was set at 10 ppm, and MS/MS tolerance was set at 0.6 Da. Search criteria included oxidation of methionine (+15.9949) as a variable modification and carbamidomethylation of cysteine (+57.0214) and the addition of the TMT mass tag (+229.163) to peptide N‐termini and lysine as fixed modifications. Searches were performed with full tryptic digestion and a maximum of 2 missed cleavages were allowed. The reverse database search option was enabled and all protein data was filtered to satisfy false discovery rate (FDR) of 5%.
The proteins with missing measurement in each EV preparation were ignored. The obtained proteomics scaled abundance data in log2 were normalised with the quantile method from the limma R package.
2.12. Characterisation of cell‐type‐enriched EV features
A variant of the one‐way analysis of variance (ANOVA) method was used to characterise miRNAs, mRNAs and proteins enriched in the four EV preparations. We exploited the oneway.test function in the stats R package (v3.5.1) (Welch, 1951), which tested whether multiple samples from normal distributions had the same mean value without assumption of equal variances between samples. Each sample contains measurements (normalised counts or abundances) of each mRNA, miRNA or protein in the corresponding quintuplicates for RNA‐seq and miRNA‐seq, or triplicates for proteomics from each EV preparation. It is noteworthy that only less than 5% of the features violated the assumption of normality with the Shapiro–Wilk test (Royston, 1982) despite the limited sample sizes. Pairwise t‐tests were also performed to test for significant difference between all the pairs of samples, yielding p‐values that are subsequently corrected for multiple testing (FDR) (Benjamini & Hochberg, 1995). Each sample is associated to the maximum FDR against the remaining samples. A log2 fold‐change of each feature in one EV is also computed as the log2 ratio of average measurement in the corresponding EV against the average over all the others. A feature is considered as enriched in one EV preparation if it satisfies (i) log2 fold‐change is positive; (ii) oneway.test p‐value is less than 0.05 and (iii) only the corresponding cell type has FDR <0.05.
2.13. Comparison of cell‐type‐enriched EV features to previous studies and uEV datasets
Datasets used to compare the cell‐type‐enriched EV features to human kidney expression data and to assess their presence in human uEV are detailed in Table S1. Gene/protein functions in Tables 2 and 3 were retrieved from Uniprot (UniProtConsortium, 2021) (https://www.uniprot.org/) and Humanmine (Kalderimis et al., 2014; Smith et al., 2012) (https://www.humanmine.org/humanmine) (both accessed on 20.05.21). Opentargets (Koscielny et al., 2017; Ochoa et al., 2021) (https://www.opentargets.org/) (accession date: 25.05.21), and Ingenuity pathway analysis (IPA) knowledge base (Qiagen) (accession date: 17.05.21) were used to retrieve genes associated with kidney disease. Vesiclepedia (Kalra et al., 2012) (http://microvesicles.org/) was used to retrieve miRNAs, mRNAs and proteins that had been reported as part of EV preparations (accession date 23.04.21). Bulk miRNA sequencing data from kidney was obtained from miRNATissueAtlas2 (Keller et al., 2022) (https://ccb‐web.cs.uni‐saarland.de/tissueatlas2) (accession date: 06.05.22).
TABLE 2.
Top 10 cell‐type‐enriched EV mRNA also detected in human urinary extracellular vesicles
| Gene name | description | Function | Association with diseases | References | Open targets | IPA |
|---|---|---|---|---|---|---|
| Podocytes | ||||||
| PLEKHB2 | pleckstrin homology domain containing B2/evectin‐2 | Involved in retrograde transport of recycling endosomes. | Role in proliferation of metastatic cells by activating Yes‐associated protein in cardiomyocytes. | (Matsudaira et al., 2017) | yes | |
| COBLL1 | cordon‐bleu WH2 repeat protein like 1 | Associated with prostate cancer progression. It stimulates androgen receptor signalling pathway and modulates the cell morphology. Also, its upregulation is associated with chronic lymphocytic leukaemia reduced survival. Associated with apoptosis and fibrosis in cardiomyoblasts. | (Plešingerová et al., 2018; Takayama et al., 2018; Xiong et al., 2021) | no | ||
| MRPL57 | mitochondrial ribosomal protein L57 | Component of the mitochondrial large ribosomal subunit. | no | |||
| LCP1 | lymphocyte cytosolic protein 1 | Actin‐binding protein. | Downregulation reduces motility and proliferation of breast cancer cells in vitro and in a mouse model. Involved in Hypereosinophilia by activating mTORC2/Akt. Candidate circulating biomarker for kidney cancer. | (Ma et al., 2020; Pillar et al., 2019; Su Kim et al., 2013), | yes | Renal cancer |
| GNG4 | G protein subunit gamma 4 | Subunit required for the guanine nucleotide‐binding proteins GTPase activity, for replacement of GDP by GTP, and for G protein‐effector interaction. | Upregulated in liver cancer, promotes cell proliferation and adhesion. Downregulation in colon cancer cells reduced proliferation and migration and invasion. Association with cognitive decline as a consequence of aging. | (Bonham et al., 2018; Liang et al., 2021; Tanaka et al., 2021) | yes | |
| WSB1 | WD repeat and SOCS box containing 1 | Probable substrate‐recognition component of a SCF‐like ECS (Elongin‐Cullin‐SOCS‐box protein) E3 ubiquitin ligase complex which mediates the ubiquitination and subsequent proteasomal degradation of target proteins. | In cancer, WSB1 upregulates hypoxia inducible factors by pVHL ubiquitination and proteosomal degradation. It is also associated with thyroid hormone homeostasis and immune regulation. | (Haque et al., 2016; Kim et al., 2015) | yes | |
| ALDH6A1 | aldehyde dehydrogenase 6 family member A1 | Plays a role in the valine and pyrimidine catabolic pathways. | Downregulation in renal cell carcinoma which was associated with poor survival. Also survival predictive value for metastatic prostate cancer. | (Cho et al., 2018; Lu et al., 2020) | yes | |
| ORC4 | origin recognition complex subunit 4 | Subunit of the ORC, which is essential for the initiation of the DNA replication in eukaryotic cells. | Postulated as a gene which could be involved in eGFR for kidney disease of diverse origin. | (Morris et al., 2019) | yes | |
| STYXL1 | serine/threonine/tyrosine interacting like 1 | STYXL1 is an inactive phosphatase. It binds to G3BP1 and inhibits the formation of G3BP1‐induced stress granules. | Upregulated in glioma and hepatocellular carcinoma. Upregulation is associated with poor prognosis | (Tomar et al., 2019; J. Z. Wu, Jiang, Lin & Liu, 2020) | yes | Renal clear cell adenocarcinoma |
| MESANGIAL CELLS | ||||||
| KRT19 | keratin 19 | Intermediate filament Involved in the organisation of myofibers. | Associated with increased risk of developing tumours in kidney failure. Upregulated in diverse models of tubular injury. High expression in breast and thyroid cancer. | (Djudjaj et al., 2016; Martínez‐Camberos et al., 2022; Sarlos et al., 2018) | yes | Autosomal dominant polycystic kidney disease |
| TXNIP | thioredoxin interacting protein | Protects cells from oxidative stress by regulating of cellular redux status. | Role in kidney fibrosis related to aging. Upregulated by hyperglycaemia in kidney. Knock‐out attenuates DKD histopathological changes in a streptozotocin model of diabetes. | (De Marinis et al., 2016; Shah et al., 2015) | yes | Oxidative stress response, damage of renal glomerulus, failure of kidney, injury of podocytes |
| MT‐ND2 | mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 2 | Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I). | Associates with higher risk to develop nephropathy in T2D. Mutations associated with maternal transmitted T2D. | (Jiang et al., 2021; X. Yang, Zhang, Ma, Zhao & Lyu, 2015) | yes | |
| MT‐ND1 | mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 1 | Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) | Mutations associated with maternal transmitted T2D. In colorectal cancer, circulating levels are associated with progression and response to chemotherapy. | (Jiang et al., 2021) | yes | Chromophobe renal cell carcinoma |
| PABPC1 | poly(A) binding protein cytoplasmic 1 | PABPC1 binding to poly(A) promotes ribosome recruitment and translation initiation. It is also involved in poly(A) shortening. | Upregulated in oesophageal squamous cell carcinoma, which promotes angiogenesis and malignant progression. Upregulated in gastric cancer and hepatocellular carcinoma where can act as a prognostic biomarker. Mutations of these genes were found in renal cancers. | (An et al., 2021; Yamamoto et al., 2021; YuFeng & Ming, 2020; Y. Zhang et al., 2022) | yes | Renal cancer |
| S100A11 | S100 calcium binding protein A11 | It may play a role in cell motility, invasion and tubulin polymerisation | level of expression in primary clear cell renal cell carcinoma can predict disease free survival. Regulates cell proliferation, invasion and migration in renal cell carcinoma through EGFR/Akt. | (Gabril et al., 2016; Liu et al., 2017) | yes | Renal cell carcinoma |
| RPS27A | ribosomal protein S27a | Part od the 40s ribosomal subunit | Associated with cervical cancer prognosis. In in lung adenocarcinoma cells regulates apoptosis, proliferation and cell cycle. It may control microglia activation in neurodegenerative diseases. | (Khayer et al., 2020; Li et al., 2022; Wang et al., 2021) | yes | |
| ATP5F1A | ATP synthase F1 subunit alpha, mitochondrial | Subunit of mitochondrial ATP synthase | In HeLa cells, overexpression of ATP5F1A regulated the expression of genes related to innate immune response, angiogenesis and collagen catabolic processed. It also regulated the alternative splicing of genes involved in glucose homeostasis. Associated with clear cell renal cell carcinoma prognosis. | (Song et al., 2021; Yuan et al., 2018) | yes | |
| CNDP2 | carnosine dipeptidase 2 | It is a nonspecific dipeptidase, hydrolyses a variety of dipeptides including L‐carnosine | Variants of these genes are associated with risk of kidney disease in T2D. Upregulated in substantia nigra pars compacta during Parkinson disease. Role in cancer growth and metastasis of ovarian cancer. | (Ahluwalia et al., 2011; Licker et al., 2012; Zhang et al., 2019) | yes | |
| SKP1 | S‐phase kinase associated protein 1 | Component of the SCF complexes, participates in ubiquitination of specific protein substrates for their degradation at the proteosome. | Role in colon cancer cell proliferation. Role in the progression of hepatocellular carcinoma. | (Zhai et al., 2017; Zhu et al., 2021) | yes | |
| Proximal tubule cells | ||||||
| GPX3 | glutathione peroxidase 3 | It protects cells against oxidative damage by catalysing the reduction of organic hydroperoxides and hydrogen peroxide by glutathione. | Downregulated in a mouse model of DN. Associated with cardiac disease. Acts as a tumour suppressor in kidney cell lines | (An et al., 2018; Morito et al., 2014; Pang et al., 2018) | yes | Chromophobe renal cell carcinoma, papillary renal cell carcinoma |
| ASCC2 | activating signal cointegrator 1 complex subunit 2 | As part of the ASCC complex, it is involved in DNA damage repair. As part of the ASC‐1 complex plays a role enhancing NF‐kappa‐B, SRF and AP1 transactivation. | Associated with coronary artery disease. In cancer, somatic mutations interferes with ASCC2‐ASCC3 interaction, which disturbs DNA repair. | (Jia et al., 2020; Y. Yang & Xu, 2021) | yes | |
| B2M | beta‐2‐microglobulin | Component of the class I major histocompatibility complex. | Associated with acute kidney injury severity. Indicator of tubular injury and glomerular filtration. | (Barton et al., 2018; Grubb et al., 1985) | yes | Acute kidney injury, acute renal allograft rejection, DN, ostertag type, injury of renal glomerulus |
| DAP | death associated protein | Dap is a positive regulator of apoptosis induced by interferon‐gamma and a negative regulator of autophagy. | In breast cancer, role in cell migration, proliferation and adhesion. | (Wazir et al., 2015) | yes | |
| MARCKSL1 | Myristoylated alanine‐rich C kinase substrate‐like 1 | It plays a role in the formation of adherens function. It controls cell movement through the regulation of cytoskeleton homeostasis. | Downregulated in DN. Anti‐angiogenic function preventing the VEGF phosphorylation mediated by Akt/PDK‐1/mTOR pathway. | (Kim et al., 2016; Tang et al., 2020) | yes | |
| BASP1 | brain abundant membrane attached signal protein 1 | Membrane bound protein with several transient phosphorylation sites and PEST motifs. PEST motif is associated with proteins that have a short intracellular half‐life. | Mediates albumin induced apoptosis in tubular cells. In podocytes, pro‐apoptotic in DN. | (Sanchez‐Niño et al., 2010; Zhang et al., 2021) | yes | Renal cell carcinoma |
| OSBP | oxysterol binding protein | It is a lipid transporter which delivers sterol to the Golgi in exchange for PI4P to the endoplasmic reticulum. It is also believed to transport sterols from lysosomes to the nucleus. | Plays a role in insulin granule formation in rat insulinoma cell line. Role in regulating the proatherogenic protein profilin‐1 under diabetic like conditions. | (Hussain et al., 2018; Romeo & Kazlauskas, 2008) | yes | |
| LMNA | lamin A/C | Component of the nuclear lamina. | Some variants are associated to cardiac disease. Mutation of these gene is associated with proteinuric renal disease and membranous glomerulonephritis. | (Fountas et al., 2017; Fujita & Hatta, 2018; Lazarte et al., 2022) | yes | Renal clear cell adenocarcinoma |
| PDLIM5 | PDZ and LIM domain 5 | It binds protein kinases to the Z‐disk in striated muscles. It may have a role in cardiomyocyte expansion and in preventing postsynaptic growth of excitatory synapses. | Recruited to integrin adhesion complexes in Podocyte. Polymorphisms of this gene are associated with T2D and hypertension. | (Maier et al., 2021; Owusu et al., 2017) | yes | |
| STARD10 | StAR related lipid transfer domain containing 10 | The protein encoded by this gene transfer phospholipids. It can transfer phosphatidylcholine and phosphatidylethanolamine between membranes. | Deletion of STARD10 impairs insulin secretion. Upregulated in breast cancer. | (Carrat et al., 2017; Olayioye et al., 2004) | yes | |
| Glomerular endothelial cells | ||||||
| TMSB4X | thymosin beta 4 X‐linked | Actin sequestering protein that inhibits actin polymerisation. It is also involved in cell proliferation, migration and differentiation. | Reduces lung inflammation and fibrosis induced by LPS. Administration of TMSB4X reduced the kidney damage in a model od DN. | (Tian et al., 2022; Zhu et al., 2015) | yes | |
| FDFT1 | farnesyl‐diphosphate farnesyltransferase 1 | Role in cholesterol biosynthesis. Enzyme that catalyses the dimerisation of 2 farnesyl pyrophosphate groups to form squalene. | Antitumoral effect by inhibiting AKT/mTOR/HIF1α pathway during fasting in colorectal cancer. Associated with fibrosis progression in chronic hepatitis C. | (Stättermayer et al., 2014; Weng et al., 2020) | yes | Cell death of kidney cell lines, metastatic renal cancer, renal cell carcinoma, renal osteodystrophy |
| SELENOW | selenoprotein W | It plays a role as a glutathione‐dependent antioxidant and may have a redox function | During nervous system development, it Protects neurons against oxidative stress. | (Chung et al., 2009) | yes | Adhesion of multiple myeloma endothelial cells |
| SNCG | synuclein gamma | It plays a role in neurofilament network integrity and may modulate the keratin network in skin | Protection against ER stress by regulating autophagy in colon cancer cells. Promotes cancer metastasis. Downregulation in endometrial cancer cells prevented proliferation, migration and invasion, and promoted apoptosis. | (Liu et al., 2022; Ni et al., 2021; Ye et al., 2021) | yes | |
| GLUL | glutamate‐ammonia ligase | It is involved in ammonia and glutamate detoxification, acid‐base homeostasis, cell signalling and cell proliferation. | In early DN, GLUL is downregulated in podocytes. Associated with cardiovascular disease in T2D. | (Look AHEAD Research Group, 2016; Wei et al., 2021) | yes | Early stage DN, migration of endothelial cells, migration of vascular endothelial cells, sprouting of vascular endothelial cells |
| ARHGDIB | Rho GDP dissociation inhibitor beta | Regulates the GDP/GTP exchange reaction of the Rho proteins by inhibiting the dissociation of GDP from them, and the subsequent binding of GTP to them. | Development of antibodies against ARHGDIB protein are associated with kidney graft loss. Upregulated in breast cancer and associated with prognosis. | (Kamburova et al., 2019; Wang et al., 2020) | yes | |
| GTF3A | general transcription factor IIIA | It is involved in ribosomal large subunit biogenesis. | Dysregulated in secondary hyperparathyroidism. | (Santamarea et al., 2005) | yes | |
| CALCOCO2 | calcium binding and coiled‐coil domain 2 | It acts as a receptor for ubiquitin‐coated bacteria and plays and a role in innate immunity by mediating macroautophagy. | Dysregulated in dilated cardiomyopathy with increased autophagy. Role in mitophagy. | (Gil‐Cayuela et al., 2019; Yamano & Youle, 2020) | no | |
| RBM3 | RNA binding motif protein 3 | It enhances global protein synthesis. It is Induced by cold shock and low oxygen tension. | Role in inducing the proliferation of hepatocellular carcinoma. In hypoxic conditions, promotes neural stem cells proliferation. | (Miao & Zhang, 2020; Yan et al., 2019) | yes | Renal cancer |
| TCEAL4 | transcription elongation factor A like 4 | Involved in transcription modulation in a promoter context‐dependent manner. | Downregulated in gestational diabetes. Down regulated in anaplastic thyroid cancer. | (Akaishi et al., 2006; Alur et al., 2021) | yes | Renal clear cell adenocarcinoma |
Note: Top 10 mRNAs were selected based on their level of expression in urinary extracellular vesicles dataset. Functions (as reported in uniprot, humanmine and bibliography) and examples of association to kidney disease and/or other diseases (evaluated using open targets kidney disease entries, ingenuity pathway analysis (IPA) and bibliography).
Abbreviations: AP1, activator protein 1; ASC1, activating signal cointegrator 1 complex; BAX, Bcl‐2‐associated X; BCL‐2, B‐cell lymphoma 2; DN, diabetic nephropathy; EGFR, epidermal growth facrtor receptor; eGFR, estimated glomerular filtration rate; G3BP1, Ras GTPase‐activating protein‐binding protein 1; GDP, guanosine diphosphate; GTP, guanosine triphosphate; JAK, Janus kinases; mTOR, mammalian target of rapamycin; mTORC2, mammalian target of rapamycin complex 2; NADH, reduced nicotinamide adenine dinucleotide; NF‐kappa‐B, nuclear factor kappa beta; ORC, origin recognition complex; PDK‐1, 3‐phosphoinositide dependent protein kinase 1; PEST motif, Proline, glutamic acid, serine and threonine; PI3K, phosphoinositide 3‐kinase; PI4P, phosphatidylinositol‐4‐phosphate; PTEN, phosphatase and tensin homolog; SCF complex, Skp, Cullin, F‐box containing complex; SRF, serum response factor; STAT3, signal transducer and activator of transcription protein 3; T2D, type 2 diabetes; VEGF, vascular endothelial growth factor.
TABLE 3.
Cell‐type‐enriched EV proteins also detected in human urinary extracellular vesicles
| Gene name | Description | Function (Uniprot and/or Humanmine) | Disease association | Association with kidney disease | References |
|---|---|---|---|---|---|
| Podocytes | |||||
| PSMB2 | Proteasome subunit beta type‐2 | Component of the 20S core proteasome complex which is involved in the proteolytic degradation of most intracellular proteins. | knock‐down suppresses hepatocellular carcinoma cell proliferation and invasion. Up‐regulated in muscle tissue from burnt patients where correlated with cellular respiration rates. | no | (Ogunbileje et al., 2016; Tan et al., 2018) |
| CUL2 | Cullin‐2 | Component of Cul2‐RING ubiquitin ligase complex, which mediate the ubiquitination of target proteins. | Required for the activity of hypoxia‐inducible factor and vasculogenesis. | no | (Maeda et al., 2008) |
| C3 | Complement C3 | Plays a central role in the activation of the complement system. | Highly expressed in the kidneys of T2D rats with DN; deposits in glomeruli from individuals with C3 Glomerulonephritis. Protects pancreatic β‐cells against pro‐inflammatory cytokines induced apoptosis. | yes | (Dos Santos et al., 2017; Huang et al., 2019; Kelly et al., 2015; Sethi et al., 2012) |
| CSTA | Cystatin‐A | Intracellular thiol proteinase inhibitor. | Up‐regulated in hypoxic cells, anti‐apoptotic effect.Role in maintaining cell‐cell adhesion. Upregulated in several epithelial‐derived malignancies, including squamous cell carcinoma. | no | (Gupta et al., 2015; Shou et al., 2020) |
| KRT14 | Keratin, type I cytoskeletal 14 | Involved in promoting KRT5‐KRT14 filaments to self‐organise into large bundles and enhances the mechanical properties involved in resilience of keratin intermediate filaments in vitro. | Expressed in progenitor cells of salivary glands, trachea, prostate, bladder and lung. Regulates epidermal homeostasis and barrier function. | no | (Abashev et al., 2017; Guo et al., 2020) |
| PLBD2 | Putative phospholipase B‐like 2 | Presence in autolysosomes in human aged brains (accumulate undegraded proteins and lipids) may be associated with Parkinson disease. | no | (Zucca et al., 2018) | |
| ERP44 | Endoplasmic reticulum resident protein 44 | Acts as a chaperone in the secretory pathway. It is activated by pH. | ER resident chaperone protein, has been implicated in the modulation of ER stress; its depletion worsen ER stress and aggravates DN in db/db mice. Elevated in urine from individuals with T1D | yes | (Pang et al., 2018; Van et al., 2017) |
| MESANGIAL CELLS | |||||
| CKM | Creatine kinase | Cytoplasmic enzymewith a role in energy homeostasis. | Hyperoxia‐responsive, Nrf2‐dependent in lung. | no | (Zervou et al., 2017) |
| CLSTN2 | Calsyntenin‐2 | May modulate calcium‐mediated postsynaptic signals. | Involved in homophilic cell adhesion via plasma membrane adhesion molecules. Positively correlated with Immune Infiltration level and immune markers in colon adenocarcinomas. Hyperoxia‐responsive, Nrf2‐dependent in lung. | no | (Cho et al., 2012; Wu et al., 2020) |
| CRIM1 | Cysteine‐rich motor neuron 1 protein | May have a functionin tissue development by interacting with members of the transforming growth factor beta family. | In a gene‐trap mouse line, abnormal glomerular development (enlarged capillary loops, podocyte effacement and mesangiolysis) was observed. Involved in endothelial maintenance and integrity. CRIM1 loss contributes also to defects in the extraglomerular vasculature | yes | (Wilkinson et al., 2007; Wilkinson et al., 2009) |
| Proximal tubule cells | |||||
| HEXB | Beta‐hexosaminidase subunit beta | Hydrolyses glycoconjugates. | Mutations cause Sandhoff disease (lysosomal storage disorder) and accumulation of ganglioside GM2 in brain kidney and liver. Knockout associated with apoptosis in the central nervous system | yes | (J. Q. Huang et al., 1997; Merscher & Fornoni, 2014) |
| EEF1G | Elongation factor 1‐gamma | Member of the elongation factor‐1 complex (responsible for the enzymatic delivery of aminoacyl tRNAs to the ribosome). | Upregulated in kidney clear cell carcinoma. Positive regulator of autophagy. Required for autophagy signalling upstream of mTORC1 | yes | (Dengjel et al., 2012; Hassan et al., 2018) |
| PPP1CA | Serine/threonine‐protein phosphatase PP1‐alpha catalytic subunit | Subunit from protein phosphatase 1, which dephosphorylate several biological targets. | Associated with renal control of ion homeostasis in distal convoluted tubule. May have a role in the regulation of ciliary structure and function. | no | (Luo et al., 2019; Penton et al., 2019) |
| PPP1CB | Serine/threonine‐protein phosphatase PP1‐beta catalytic subunit | Subunit from protein phosphatase 1, which dephosphorylate several biological targets. | Catalytic subunit of protein phosphatase 1. Its deletion causes remodelling of the heart, interstitial fibrosis and contractile dysregulation. | no | (Liu et al., 2015; Penton et al., 2019) |
| BASP1 | Brain acid soluble protein 1 | Membrane protein that has several transient phosphorylation sites and PEST sequences. PEST are associated with proteins that have a short intracellular half‐life. | Mediates glucose‐induced and albumin‐induced apoptosis in tubular cells. proapoptotic factor in DN and possibly also in hypertensive nephropathy | yes | (Sanchez‐Niño et al., 2015; Sanchez‐Niño et al., 2010) |
| HNRNPUL2 | Heterogeneous nuclear ribonucleoprotein U‐like protein 2 | no | |||
| Glomerular endothelial cells | |||||
| EFEMP1 | EGF containing fibulin‐like extracellular matrix protein 1 isoform 1 | Extracellular matrix glycoprotein member (member of the fibulin family). | Pro‐angiogenic. Overexpression in HUVECs increased Tube formation and proliferation. Overexpressed in IgA nephropathy. Reduces phosphate‐induced vascular smooth muscle cell calcification by Inhibition of oxidative stress. Activates MAPK and Akt pathways in pancreatic carcinoma cells by binding to EGFR. | no | (Camaj et al., 2009; Cheng et al., 2020; Luong et al., 2018; Paunas et al., 2019) |
| PSAP | Prosaposin | involved in the catabolism of glycosphingolipids with short oligosaccharide groups | Increased in urine from children with urinary tract infection. dysregulated in urine from individuals with Fabry nephropathy. Elevated in the urine of T1D children. | yes | (Forster et al., 2019; Manwaring et al., 2013; Riccio et al., 2019) |
| PTX3 | Pentraxin‐related protein PTX3 | Plays a role in the regulating inflammatory reactions, angiogenesis and tissue remodeling. | PTX3 increased the respiratory capacity in human endothelial cells. Induced vascular dysfunction and changes in endothelial layer in vitro. PTX3 inhibits acute renal injury‐induced interstitial fibrosis by suppressing IL‐6/Stat3 pathway in mice. Increased circulating levels associated with lower GFR in the elderly. | yes | (Carrizzo et al., 2015; Carrizzo et al., 2019; Sjöberg et al., 2016; Xiao et al., 2014) |
| LTBP2 | Latent‐transforming growth factor beta‐binding protein 2 | Extracellular matrix protein with multiple domains involved in diverse functions such as member of the TGF‐beta latent complex, as a structural component of microfibrils, and a role in cell adhesion. | Predictor for acute kidney injury and major adverse kidney events after open heart surgery. LTBP2 can upregulate TGF‐β1 expression and secretion in fibroblasts which may be significant in connective tissue disorders. | yes | (Haase et al., 2014; Sideek et al., 2017) |
| TXNRD1 | Thioredoxin reductase 1, cytoplasmic | Member of the thioredoxin (Trx) system. TrxRs reduce thioredoxins and other substrates. Plays a role in redox homoeostasis. | TXNRD1 levels were high in a model of gestational diabetes in rats. Upregulated in high glucose treated HMVEC. Alleviates tubular damage and interstitial fibrosis by inducing the expression of genes that protects against oxidative stress. | yes | (Fu et al., 2021; Nezu & Suzuki, 2020; Patel et al., 2013) |
| CSPG4 | Chondroitin sulphate proteoglycan 4 | Proteoglycan involved in cell proliferation and migration. It is a cell surface receptor for collagen alpha 2(VI), which may facilitate the mobility in this substrate. | In human gliomas induces cell proliferation and migration. In gliomas it is highly expressed in pericytes where has a role in neoangiogenesis. Involved in fibrogenic/adipogenic differentiation in skeletal muscle tissues | no | (Schiffer et al., 2018; Takeuchi et al., 2016) |
Note: Functions (as reported in uniprot, humanmine and bibliography) and examples of association to kidney disease and/or other diseases.
Abbreviations: DN, diabetic nephropathy; EGFR, epidermal growth factor receptor; ER, endoplasmic reticulum; GFR, glomerular filtration rate; HMVEC, human microvascular endothelial cells; HUVECs, human umbilical vein endothelial cells; MAPK, mitogen‐activated protein kinase; mTORC1, mammalian target of rapamycin complex 1; T1D, Type 1 diabetes; T2D, Type 2 diabetes; TGF‐β1, transforming growth factor beta 1.
2.14. Data availability
RNA and miRNA sequencing raw data as well as TMT proteomics data are deposited on the NCBI BioProject PRJNA905899.
3. RESULTS
3.1. Characterisation and quality control of kidney cell line EV preparations
EV isolated by hydrostatic filtration dialysis were characterised by EM, NTA and WB.
EM negative staining revealed typical round and collapsed membrane bounded vesicles of various sizes and with diverse electron density content in all kidney cell types (Figure 2a). NTA profiles demonstrated enrichment of EV between 100 and 300 nm (Figure 2b). Particle mean size and concentration did not differ significantly between cell types (Figure 2c,d). Notably, EV protein markers (TSG101, CD63 and CD81) were present in all isolated EV preparations (Figure 2e). Podocyte (POD), proximal tubule cell (PTC) and MC isolates had an RNA peak between 25 and 200 nt while GEC isolates presented a peak between 25 and 1000 nt (Figure 2f). EV RNA biotype distribution showed variability between cell types, that is, POD isolates had high content of miscellaneous RNA, PTC isolates contained more miRNAs compared to the other cell types, MC isolates had more mitochondrial and protein coding RNA while GEC isolates had higher content of sense intronic, processed transcript and long intergenic non‐coding RNA (Figure S1).
FIGURE 2.

EV preparations characterisation and quality control. (a) Electron microscopy micrographs of EV samples isolated by HFD. (b) Representative NTA profiles of uEV total RNA. (c) Diameter mean per cell type EV evaluated by NTA. (d) Particle yield per ml of cell free culture media as measured by NTA. (e) Western blotting. Immunodetection of EV markers (TSG101, CD63 and CD81). (f) Representative RNA electropherograms obtained by Agilent RNA 6000 Pico Kit. Black arrows: extracellular vesicles. GEC, glomerular endothelial cells; EV, extracellular vesicles; MC, mesangial cells; NTA, nanoparticle tracking analysis; POD, podocytes; PTC, proximal tubule cells
3.2. Kidney cell enriched EV miRNAs, RNAs and proteins
We next investigated the EV features which were enriched in each kidney cell type. Our Principal component analysis (PCA) from each of the three omics data sets in the different cell types revealed striking variation between EV cargo derived from GEC vs the other three cell types (Figure 3). This variation caused a bias towards GEC derived EV when studying enriched features in each EV isolate. Therefore, parallel comparative analysis, without GEC derived EV, to identify enriched features in each of the three other cell type derived EV, were performed and cell‐type enriched EV feature lists were generated for each analysis (i.e., including and excluding GEC‐EV (Figure S1).
FIGURE 3.
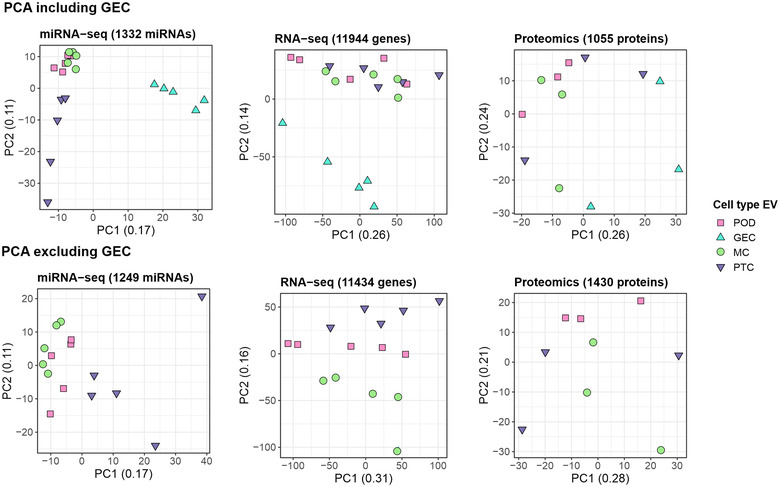
Principal component analysis (PCA) on miRNA‐seq, RNA‐seq and proteomics data from the four extracellular vesicles (EV) isolates. (a) PCA including GEC, (b) PCA excluding GEC in order to avoid GEC bias. GEC, glomerular endothelial cells; MC, mesangial cells; POD, podocytes; PTC, proximal tubule cells
A total of 16, 26, 21 and 70 enriched EV miRNAs (Tables S2 and S3; Figure S1), 80, 391, 218 and 132 enriched EV mRNAs (Tables S4 and S5; Figure S1) and 13, 8, 4 and 15 enriched EV proteins (Figure 6, Tables S6 and S7; Figure S2) were identified in POD, PTC, MC and GEC isolates, respectively. For the next sections, we refer to podocytes, proximal tubule cells, mesangial cell and glomerular endothelial cell EV‐enriched features as POD‐EV, PTC‐EV and MC‐EV and GEC‐EV plus the feature we analysed, that is, podocyte EV enriched miRNAs are referred as POD‐EV miRNAs. When we refer to cell‐type‐enriched features but without specifying the cell type, we use EV plus the feature we analyse, that is, cell‐type‐enriched EV miRNAs are referred as EV‐miRNAs.
FIGURE 6.
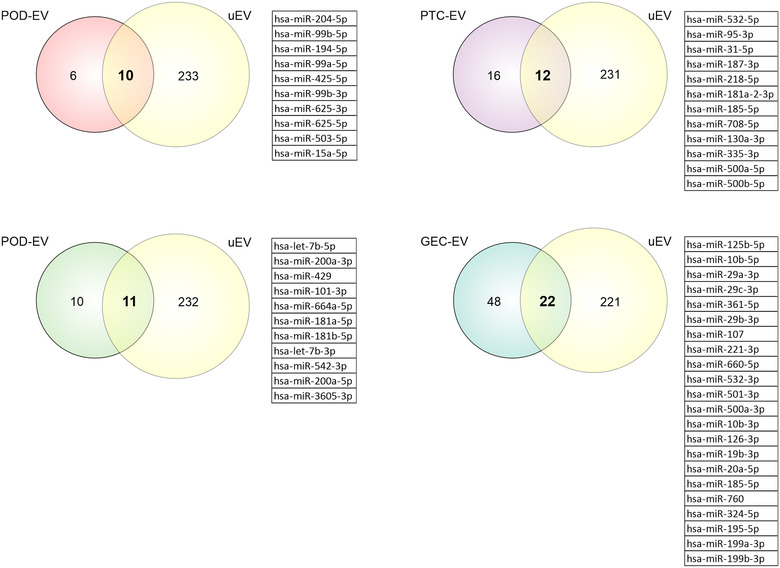
Identification of cell‐type‐enriched extracellular vesicles (EV) miRNAs in urinary extracellular vesicles (uEV). Venn diagrams depict overlapping between cell‐type‐enriched EV miRNAs (POD‐EV, PTC‐EV, MC‐EV and GEC‐EV miRNAs) and human uEV miRNAs. GEC, glomerular endothelial cells; MC, mesangial cells; POD, podocytes; PTC, proximal tubule cells
3.3. Pathway enrichment analysis
To identify important biological pathways in the different cell type enriched EV‐features, gene‐set enrichment analysis (GSEA) was performed. Pathways were considered enriched if p‐value <0.05 and q‐value <0.1 in at least two omics analysis in any cell type.
Enrichment analysis of Gene Ontology (GO) biological processes showed that POD‐EV miRNAs were enriched in cell growth and maintenance and a set of miRNAs and mRNAs were associated with cell adhesion (Figure 4). PTC‐EV miRNAs and mRNAs were enriched in epithelial to mesenchymal transition, response to stress, signal transduction, cell growth and maintenance. A set of PTC‐EV proteins were associated with cell growth and maintenance and cell adhesion (Figure 4). MC‐EV miRNAs were enriched in biological processes related to epithelial to mesenchymal transition and responses to stress and signal transduction (also mRNAs) (Figure 4). GEC‐EV miRNAs and proteins were enriched in response to stress while GEC‐EV mRNAs were enriched in processes related to cell growth and maintenance and cell adhesion (Figure 4). A full list of enriched biological processes is shown in Figure S3. Analysis of GO molecular function enrichment (Figure 5) revealed that GEC‐EV miRNAs, mRNAs and proteins were enriched in growth factor binding, collagen binding, structural extracellular matrix constituents and miRNAs and proteins in heparin binding. PTC‐EV miRNAs and proteins were enriched in cadherin binding, cell adhesion mediator activity while only miRNAs were enriched in collagen binding, heparin binding and structural extracellular matrix constituent. No significant molecular function terms were found for POD‐EV and MC‐EV miRNAs, mRNAs or proteins.
FIGURE 4.

Gene ontology biological process enrichment analysis with multi‐omics data. Heatmap depicts selected enriched biological processes per cell and molecule type. Analysis was done using cell‐type‐enriched EV features (POD‐EV, PTC‐EV, MC‐EV and GEC‐EV). GGEC, glomerular endothelial cells; MC, mesangial cells; POD, podocytes; PTC, proximal tubule cells
FIGURE 5.

Gene ontology molecular function enrichment analysis with multi‐omics data. Heatmap depicts selected enriched molecular functions per cell and molecule type. Analysis was done using cell‐type‐enriched EV features (POD‐EV, PTC‐EV, MC‐EV and GEC‐EV). GEC, glomerular endothelial cells; MC, mesangial cells; POD, podocytes; PTC, proximal tubule cells
3.4. Assessment of identified cell‐type‐enriched EV features in human datasets
Our identified EV features enriched in the four kidney cell types were benchmarked against different human datasets reported in the literature. The comparison was done separately for each type of feature (EV‐miRNAs, EV‐mRNAs and EV‐proteins) for each cell type (POD, MC, GEC and PTC). The number of cell‐type‐enriched EV features detected in a dataset is presented as percentage range, that is, the values correspond to cell types with the lowest and highest percentage of EV‐features overlapping with a particular dataset. To see the results for each cell type separately please refer to the respective figures and/or tables.
We initially cross‐referenced the ‘vesiclepedia’ (Kalra et al., 2012) database to assess evidence that EV‐features had previously been reported as EV cargo. We found that 69%–81% of EV‐miRNAs, 71%–85% of EV‐mRNAs and 100% of EV‐proteins had been reported previously as present in EV from diverse biofluids (Tables S8–S10).
We next compared our cell‐type‐enriched EV transcriptomic profiles with those derived from human urine samples. For miRNA and mRNA, we used our previously published data (Barreiro et al., 2020) comprising uEV mi‐ and mRNA sequencing datasets of healthy controls and individuals with type‐1 diabetes and macroalbuminuria (n = 10). Of note, from Barreiro et al. (2020) we retrieved only the datasets from uEV isolated by hydrostatic filtration dialysis. To work with RNAs consistently found in uEV, we selected miRNAs and mRNAs with raw counts ≥5 in at least 80% of the samples. We found that 31%–63% of the EV‐miRNAs and 61%–85% of the EV‐mRNAs were also detected in human uEV (Figures 6 and 7, Tables S8 and S9).
FIGURE 7.
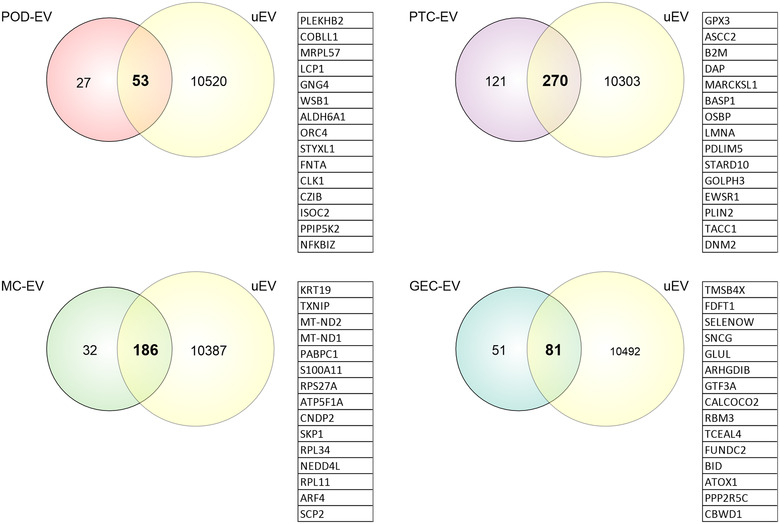
Identification of cell‐type‐enriched extracellular vesicles (EV) genes in urinary extracellular vesicles (uEV). Venn diagrams depict overlapping between cell‐type‐enriched EV mRNAs (POD‐EV, PTC‐EV, MC‐EV and GEC‐EV mRNAs) and human uEV mRNAs. Top 15 mRNA (by expression in uEV) are listed for each cell type. GEC, glomerular endothelial cells; MC, mesangial cells; POD, podocytes; PTC, proximal tubule cells
To compare EV‐proteins to those from uEV, several published proteomics datasets were analysed (Braun et al., 2020; Gonzales et al., 2009; Oeyen et al., 2018; Xu et al., 2019) (see Table S1 for details). As the majority of the published datasets reported proteins by gene name without including unique identifiers, for example, UNIPROT ID, we could only compare EV‐proteins with associated gene name from our lists (see Table S10 for UNIPROT IDs without associated gene name in our cell‐type‐enriched EV protein lists). We found that 58%–67% of the EV‐proteins have previously been detected in human uEV (Figure 8).
FIGURE 8.
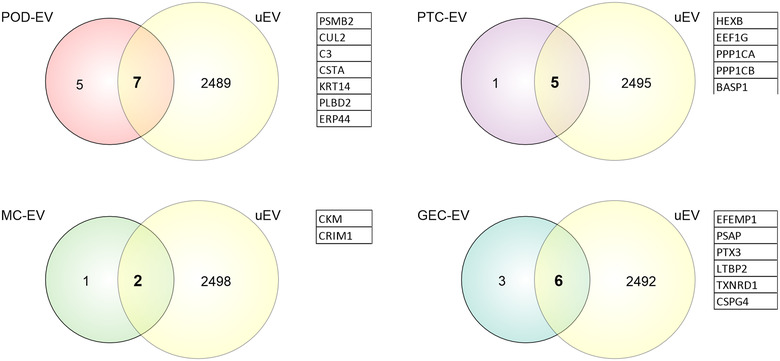
Identification of cell‐type‐enriched extracellular vesicles (EV) proteins in urinary extracellular vesicles (uEV). Venn diagrams depict overlapping between cell‐type‐enriched EV proteins (POD‐EV, PTC‐EV, MC‐EV and GEC‐EV proteins) and human uEV proteins. GEC, glomerular endothelial cells; MC, mesangial cells; POD, podocytes; PTC, proximal tubule cells
With the aim of verifying kidney expression of the cell‐type‐enriched EV features, we compared them to miRNA, mRNA and proteins found in human kidney biopsies. Studies selected included bulk small RNA sequencing (miRNATissueAtlas2), bulk RNA sequencing (Levin et al., 2020), proteomics data (Miyamoto et al., 2007; Yoshida et al., 2012) and single cell sequencing (Wu et al., 2018). We found that 57%–69% of the EV‐miRNAs were expressed in human kidneys (Table S8). Regarding mRNAs we found that 91%–94% of the EV‐mRNA were found in human kidneys (Table S9). For proteins, between 0% and 84% of the EV‐proteins have been described in human glomeruli. Protein results showed high variability between datasets and between cell types (Table S10). To ascertain whether EV‐enriched genes were expressed by the same cell types in human kidneys, we explored scRNA‐seq data (see Table S1). We found that 9%–29% of the EV‐mRNAs expression pattern was concordant with human cell type expression pattern.
Finally, we verified if any of our identified EV‐uEV (EV features that were also found in human uEV) molecules had previously been described in pathways associated to kidney disease and/or other diseases. Interestingly, we found several of them were, for example, miR‐29a‐3p (Wang et al., 2012), thioredoxin interacting protein (Huang et al., 2016), and complement component 3 (Kelly et al., 2015) as detailed in Tables 1, 2, 3, which reveals a potential to qualify our findings functionally from cells in vitro to in vivo.
TABLE 1.
Cell‐type‐enriched EV miRNA also detected in human urinary extracellular vesicles
| miRNA | Functions associated to disease | Association with kidney disease | References |
|---|---|---|---|
| Podocytes | |||
| miR‐204‐5p | highly expressed in the kidney; miR‐204‐5p plays a prominent role in safeguarding the kidneys against common causes of chronic renal injury; urinary exosomal level as biomarker for Xp11.2 translocation renal cell carcinoma | yes | (Cheng et al., 2020; Kurahashi et al., 2019) |
| miR‐99b‐5p | miR‐99b‐5p is associated with response to tyrosine kinase inhibitor treatment in clear cell renal cell carcinoma patients | yes | (Lukamowicz‐Rajska et al., 2016) |
| miR‐194‐5p | miR‐194‐5p was significantly downregulated in patient urine exosomes, in murine Pkd1 cystic kidneys and in human PKD1 cystic kidney tissue | yes | (Magayr et al., 2020) |
| miR‐99a‐5p | miR‐99a‐5p was upregulated in macro‐albuminuric patients compared with normo‐albuminuric and micro‐albuminuric patients. Transfection of miR‐99a‐5p in cultured human podocytes downregulated mTOR protein expression and downregulated the podocyte injury marker vimentin | yes | (Uil et al., 2021) |
| miR‐425‐5p | miR‐425‐5p is a potential predictor of extreme response to tyrosine kinase inhibitors in renal cell cancer | yes | (Garrigós et al., 2020) |
| miR‐99b‐3p | miR‐99b‐3p promotes angiotensin II‐induced cardiac fibrosis in mice by targeting GSK‐3β | no | (Yu et al., 2021) |
| miR‐625‐3p | miR‐625‐3p promotes migration and invasion and reduces apoptosis of clear cell renal cell carcinoma | yes | (Zhao et al., 2019) |
| miR‐625‐5p | involved in regulation of Wnt/β‐catenin and NF‐kappa‐B activation | no | (Tang et al., 2019) |
| miR‐503‐5p | miR‐503‐5p functions as an oncogene in various cancer diseases, such as oral squamous cell carcinoma | no | (Fei et al., 2020) |
| miR‐15a‐5p | decreased urinary exosomal level in incipient T2D kidney disease | yes | (Xie et al., 2017) |
| Mesangial cells | |||
| let‐7b‐5p | TGF‐β1‐regulated miRNA which is associated with an increased risk of rapid progression to end‐stage renal disease | yes | (Pezzolesi et al., 2015) |
| miR‐200a‐3p | MicroRNA‐200a‐3p suppresses tumor proliferation and induces apoptosis by targeting SPAG9 in renal cell carcinoma | yes | (Wang et al., 2016) |
| miR‐429 | increased urinary exosomal level upon acute kidney injury | yes | (Sonoda et al., 2019) |
| miR‐101‐3p | potential earyl biomarker for acute kidney injury | yes | (Aguado‐Fraile et al., 2015) |
| miR‐664a‐5p | decreased serum exosomal level in patients with pancreatic ductal adenocarcinoma | no | (Wang et al., 2021) |
| miR‐181a‐5p | miR‐181a‐5p regulates the proliferation and apoptosis of glomerular mesangial cells by targeting KLF6 | yes | (X. Liang & Xu, 2020) |
| let‐7b‐3p | involved in the pathogenesis of chronic thromboembolic pulmonary hypertension | no | (Gong et al., 2021) |
| miR‐542‐3p | MiR‐542‐3p drives renal fibrosis by targeting AGO1 in vivo and in vitro | yes | (Li et al., 2020) |
| miR‐200a‐5p | increased urinary exosomal level upon acute kidney injury; miR‐200a Prevents renal fibrogenesis through repression of TGF‐β2 expression | yes | (Sonoda et al., 2019; Wang et al., 2011) |
| miR‐3605‐3p | miscellaneous | no | |
| Proximal tubule cells | |||
| miR‐532‐5p | miR‐532‐5p suppresses renal cancer cell proliferation by disrupting the ETS1‐mediated positive feedback loop with the KRAS‐NAP1L1/P‐ERK axis; increased urinary exosomal miR‐532‐5p level in intermediate‐risk prostate cancer | yes | (Kim et al., 2021; Zhai et al., 2018) |
| miR‐95‐3p | invoved in angiogenesis; increased in ectosomes obtained from patients with T2D | no | (Stępień et al., 2018) |
| miR‐31‐5p | miR‐31‐5p acts as a tumor suppressor in renal cell carcinoma by targeting cyclin‐dependent kinase 1 | yes | (Li et al., 2019) |
| miR‐187‐3p | Mesenchymal Stem/Stromal Cells Increase Cardiac MIR‐187‐3P Expression in Polymicrobial Animal Model of Sepsis | no | (Ektesabi et al., 2021) |
| miR‐218‐5p | miR‐218‐5p is expressed in endothelial progenitor cells and contributes to the development and repair of the kidney microvasculature | yes | (Wang et al., 2020) |
| miR‐181a‐2‐3p | miR‐181a‐2‐3p alleviates the apoptosis of renal tubular epithelial cell via targeting GJB2 in sepsis‐induced acute kidney injury | yes | (Yi et al., 2021) |
| miR‐185‐5p | miR‐185‐5p ameliorates endoplasmic reticulum stress and renal fibrosis by downregulation of ATF6 | yes | (Yuan et al., 2020) |
| miR‐708‐5p | considered as a therapeutic agent against metastatic lung cancer | no | (Wu et al., 2016) |
| miR‐130a‐3p | miR‐130a‐3p inhibition protects against renal fibrosis in vitro via the TGF‐β1/Smad pathway by targeting SnoN | yes | (Ai et al., 2020) |
| miR‐335‐3p | aldosterone regulates miR‐335‐3p in the cortical collecting duct to alter sodium transport | yes | (Edinger et al., 2014) |
| miR‐500a‐5p | involved in epithelial‐mesenchymal transition | no | (Tang et al., 2020) |
| miR‐500b‐5p | involved in phenotypic switching in vascular smooth muscle cells and is involved in the pathogenesis of aortic dissection. | no | (Wang et al., 2021) |
| Glomerular endothelial cells | |||
| miR‐125b‐5p | decreased urinary exosomal miRNA level in individuals with diabetic kidney disease | Yes | (Zang et al., 2019) |
| miR‐10b‐5p | miR‐10a‐5p has been associated with disease progression, proliferation and invasion of renal cell carcinoma | yes | (Kowalik et al., 2017) |
| miR‐29a‐3p | acts anti‐fibrotic, mainly by targeting different types of collagen via a Smad3‐dependent mechanism, thus leading to decreased ECM accumulation; Linagliptin, a DPP‐4 inhibitor used as an anti‐diabetic drug, can also confer renal protection and decrease fibrosis in a mouse model of DN by inducing miR‐29a, which targets DPP‐4 | yes | (Assmann et al., 2018; Kanasaki et al., 2014) |
| miR‐29c‐3p | acts anti‐fibrotic, mainly by targeting different types of collagen via a Smad3‐dependent mechanism, thus leading to decreased ECM accumulation; linagliptin and telmisartan‐induced restorative effects on miR‐29c expression were reflected in urinary exosomes | yes | (Assmann et al., 2018; Delić et al., 2020) |
| miR‐29b‐3p | acts anti‐fibrotic, mainly by targeting different types of collagen via a Smad3‐dependent mechanism, thus leading to decreased ECM accumulation | yes | (Assmann et al., 2018) |
| miR‐199a‐3p | acts as a tumor suppressor in clear cell renal cell carcinoma and acts in a pro‐fibrotic manner in chronic diabetic kidney disease; telmisartan and linagliptin suppressed the induction of miR‐199a‐3p | yes | (Delić et al., 2020; Liu et al., 2018) |
| miR‐199b‐3p | miscellaneous | no | |
| miR‐361‐5p | involved in regulation of epithelial‐mesenchymal transition | no | (Yin et al., 2020) |
| miR‐107 | increased urinary exosomal miRNA level in clinical responders (lupus nephritis patients) | yes | (Garcia‐Vives et al., 2020) |
| miR‐221‐3p | increased urinary level after drug‐induced kidney injury | yes | (Chorley et al., 2021) |
| miR‐660‐5p | miR‐660‐5p is associated with cell migration, invasion, proliferation and apoptosis in renal cell carcinoma | yes | (He et al., 2018) |
| miR‐532‐3p | decreased expression in kidneys obtained from patients with progressive chronic kidney disease | yes | (Rudnicki et al., 2016) |
| miR‐501‐3p | high expression levels of exosome miR‐501‐3p contribute to arteriosclerotic changes | no | (Toyama et al., 2021) |
| miR‐500a‐3p | miR‐500a‐3P alleviates kidney injury by targeting MLKL‐mediated necroptosis in renal epithelial cells | yes | (L. Jiang et al., 2019) |
| miR‐10b‐3p | miscellaneous | no | |
| miR‐126‐3p | relevant for DKD pathogenesis and endothelial to mesenchymal transition | yes | (Wang et al., 2019) |
| miR‐19b‐3p | exosomal miRNA‐19b‐3p of tubular epithelial cells promotes M1 macrophage activation in kidney injury | yes | (Lv et al. 2020) |
| miR‐20a‐5p | miR‐20a‐5p is enriched in hypoxia‐derived tubular exosomes and protects against acute tubular injury | yes | (Yu et al., 2020) |
| miR‐185‐5p | reduces endoplasmic reticulum stress and fibrosis in vitro and in a mouse model of kidney fibrosis. | yes | (Yuan et al., 2020) |
| miR‐760 | increased urinary exosomal microRNA signatures in nephrotic, biopsy‐proven DN | yes | (Lee et al., 2020) |
| miR‐324‐5p | miR‐324‐5p inhibits lipopolysaccharide‐induced proliferation of rat glomerular mesangial cells by regulating the Syk/Ras/c‐fos pathway | yes | (Wang et al., 2020) |
| miR‐195‐5p | miR‐195‐5p alleviates acute kidney injury through repression of inflammation and oxidative stress by targeting vascular endothelial growth factor A | yes | (Chen et al., 2017) |
Note: Functions associated with disease (reported in bibliography).
Abbreviations: AGO1, argonaute RISC component 1; ATF6, activating transcription factor 6; COL I/III, collagen I/III; DKD, diabetic kidney disease; DPP‐4, dipeptidyl peptidase‐4; ECM, extracellular matrix; ETS1, ETS proto‐oncogene 1; GJB2, gap junction protein beta 2; GSK‐3β, glycogen synthase kinase 3 beta; KLF6, Kruppel Like factor 6; KRAS, KRAS proto‐oncogene; MLKL, mixed lineage kinase domain like pseudokinase; mTOR, mammalian target of rapamycin; NAP1L1, nucleosome assembly protein 1 like 1; NF‐kappa‐B, nuclear factor kappa beta; P‐ERK, phospho‐ERK; Pkd1, polycystic kidney disease 1; SnoN, nuclear Smad‐interacting protein; SPAG9, sperm associated antigen 9; Syk, spleen associated tyrosine kinase; T2D, Type 2 diabetes; TGF‐β1, transforming growth factor beta 1; TGF‐β2, transforming growth factor beta 2.
4. DISCUSSION
The potential of EV as carriers of biomarkers for diverse diseases is high, which reflects in the large number of studies that are currently searching for EV biomarkers, especially in biofluids. Urinary EV are an attractive approach to search for non‐invasive biomarkers of urinary tract diseases, as urine is easy to collect. Most of the approaches are based on bulk isolation and processing of vesicles and hence, it is not possible to identify which cell types are affected. Understanding which cell types contribute to disturbances in EV profiles in complex biofluids is highly relevant to identify pharmacological targets but also to decide and follow up pharmacological interventions. Moreover, as proposed for miRNAs (Chorley et al., 2021; Koturbash et al., 2015), cell type EV signatures could be used in, for example, clinical trials to define which region of the kidney are drug effects most likely derived from and safety and efficacy. Some attempts have been made to shed light to this topic. One approach which has shown promise involves single cell EV isolation (Nikoloff et al., 2021), but this is still in its infancy. Moreover, it is not clear if the RNA and protein yield would be sufficient for profiling applications. Another approach used earlier for plasma derived EV is deconvolution (Li et al., 2020), which consists of using cell sequencing datasets (or tissue bulk RNA sequencing datasets) to derive cell/tissue signature matrices and using deconvolution algorithms to predict, for example, the cell/tissue origin of EV mixtures. However, at present it is not yet clear how expression of EV contents correlates with the expression in their cells of origin, that is, correlation or anti‐correlation. A recent study that compared the transcriptome of prostate cancer tissue to paired uEV samples, showed that while the complexity for mRNA between sample types was comparable, a set of mRNAs were significantly enriched in uEV (Almeida et al., 2022). Moreover, the same study showed that uEV were enriched in mature cytoplasmic mRNA, which was confirmed also in vitro (Almeida et al., 2022). Hence, the applicability of tissue/single cell signatures for deconvolution are not straightforward.
Here, we present an in vitro approach to identify cell‐type enriched EV signatures with applicability to research on human uEV. In our approach, we isolated EV from cell culture media from four kidney cell types and profiled miRNA, mRNA and proteins. Thereafter, we defined cell‐type‐enriched EV RNAs and proteins and compared them to human datasets.
Vesicles of typical morphology and variable sizes were identified in all EV preparations. We did not find differences in EV size and concentration between cell types. In contrast, we did find that GEC Bioanalyzer profile differ from the other cell types. Thus, we explored if differences in the RNA biotype distribution could explain the wider RNA peak in the GEC Bioanalyzer profiles. GEC‐EV had a higher percentage of long intergenic non‐coding RNA and sense intronic RNAs compared to other cell types. For both RNA types, size exceeds 200 nt, which could explain at least partly the differences in Bioanalyzer RNA profiles. In addition, previous reports have mentioned differences in Bioanalyzer RNA profiles between cell types and EV types (Crescitelli et al., 2013).
We successfully derived lists of miRNA, mRNA and proteins enriched in EV isolated from different cell types. Firstly, we characterised the list of features by enrichment analyses and we found that most of the cell type EV shared differentially represented gene ontology terms related to cell adhesion, cell growth and maintenance as well as response to stress. As expected, not all the pathways were shared, for example, PTC‐EV and MC‐EV were enriched in epithelial to mesenchymal transition and signal transduction and PTC‐EV and GEC‐EV shared structural extracellular matrix constituent and heparin and collagen binding. We then compared our lists to in‐vivo datasets, that is, human uEV transcriptomics and kidney biopsy data (bulk transcriptomics and proteomics and single cell sequencing). Between 31%–85% and 44%–94% of the EV RNAs (miRNA and mRNA) and proteins were found in human uEV datasets and bulk (bulk RNA sequencing or proteomics) biopsy data, respectively. On the contrary, none of the three MC‐enriched EV proteins were found in proteomics datasets from kidney biopsies. In addition, the comparison of our lists to single cell sequencing data showed that less than 30% of EV enriched mRNAs had previously been detected using this technology. The variability on the number of features occurring in human datasets could arise from factors such as low translatability between in vitro models and in vivo data or specific cell sorting of some of the features to EV (Carnino et al., 2020; Garcia‐Martin et al., 2022; O'grady et al., 2022). In addition, more abundant features from other cell types (including in EV derived from non‐kidney cell types) could mask the expression of some of the cell‐type‐enriched features. This possibility could be assessed by targeted detection methods such as qPCR. Moreover, the missing cell‐type‐enriched mRNAs in the single cell sequencing data could be partly due to technical limitations, for example, high level of drop‐outs (lack of detection of transcripts due to low efficiency in mRNA capture or amplification) (Haque et al., 2017). Taken together, since many of the features were found in human datasets and importantly in human uEV, our in vitro model showed to be robust. Moreover, our approach appears to be more specific than deconvolution because our datasets consist of cell type specific EV instead of single cell sequencing. Furthermore, our approach can be easily scaled up to yield enough EV isolates and perform different omics from the same sample.
The limitations of our study include a low samples number, especially for proteomics profiling, which could be one of the reasons for the low number of enriched EV proteins per cell type. In addition, we included only a subset of kidney cells that contribute to uEV heterogeneity (POD, PTC, MC, and GEC) and we did not include urothelial cells. However, our line of research is focused on diabetic nephropathy and POD, PTC, MC and GEC are key targets of this disease (reviewed in (Jefferson et al., 2008)). Thus, major disturbances in uEV feature profiles will most likely occur in these cells. Finally, EV isolates can contain contaminants, that is, non‐vesicular RNAs or proteins (Théry et al., 2018). We found that most of the features in our EV signatures had been described previously as part of EV isolates in urine or other biofluids, but we cannot discard that not all the features are enclosed in vesicles.
In summary, we propose here an in vitro approach to study cell type enriched EV cargo that could be translated to in vivo findings. Our profiles of EV enriched miRNAs, mRNAs and proteins by cell type, specifically the ones that were also found in uEV, could be applied to understand cell type contribution to complex EV mixtures such as urine and could translate to improve diagnostics. Moreover, as our in vitro model showed to be robust, the approach presented here could be further developed to find cell type specific biomarker for kidney disease such as diabetic kidney disease.
AUTHOR CONTRIBUTIONS
Karina Barreiro: Conceptualization; Formal analysis; Investigation; Methodology; Validation; Visualization; Writing – original draft. Abigail C. Lay: Conceptualization; Investigation; Methodology; Validation; Writing – original draft. German Leparc: Formal analysis; Software; Writing – review & editing. Van Du T. Tran: Formal analysis; Software; Visualization; Writing – original draft. Marcel Rosler: Formal analysis; Software; Visualization; Investigation; Writing – review & editing. Lusyan Dayalan: Investigation; Writing – review & editing. Frederic Burdet: Supervision; Writing – review & editing. Mark Ibberson: Resources; Supervision; Writing – review & editing. Richard J. M. Coward: Conceptualization; Funding acquisition; Resources; Supervision; Writing – original draft. Tobias B. Huber: Funding acquisition; Writing – review & editing. Bernhard K. Krämer: Supervision; Writing – review & editing. Denis Delic: Conceptualization; Resources; Supervision; Writing – original draft. Harry Holthofer: Conceptualization; Funding acquisition; Project administration; Resources; Supervision; Writing – original draft.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
Supporting information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
ACKNOWLEDGEMENTS
We thank Dr Maija Puhka, Research coordinator and Head of EV and HiPrep cores at the Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki for training and guidance in EV work. We thank the Electron Microscopy Unit of the Institute of Biotechnology, University of Helsinki for providing laboratory facilities and the EV Core of University of Helsinki for performing NTA. We thank Dr Kate Heesom and Dr Marieangela Wilson for guidance and for performing the TMT proteomics at The University of Bristol Proteomics Facility. This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No 115974. The JU receives support from the European Union's Horizon 2020 research and innovation programme and EFPIA and JDRF. Any dissemination of results reflects only the author's view; the JU is not responsible for any use that may be made of the information it contains.
Open access funding enabled and organized by Projekt DEAL.
Barreiro, K. , Lay, A. C. , Leparc, G. , Tran, V. D. T. , Rosler, M. , Dayalan, L. , Burdet, F. , Ibberson, M. , Coward, R. J. M. , Huber, T. B. , Krämer, B. K. , Delic, D. , & Holthofer, H. (2023). An in vitro approach to understand contribution of kidney cells to human urinary extracellular vesicles. Journal of Extracellular Vesicles, 12, e12304. 10.1002/jev2.12304
REFERENCES
- Abashev, T. M. , Metzler, M. A. , Wright, D. M. , & Sandell, L. L. (2017). Retinoic acid signaling regulates Krt5 and Krt14 independently of stem cell markers in submandibular salivary gland epithelium. Developmental Dynamics, 246(2), 135–147. 10.1002/dvdy.24476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguado‐Fraile, E. , Ramos, E. , Conde, E. , Rodríguez, M. , Martín‐Gómez, L. , Lietor, A. , Candela, Á. , Ponte, B. , Liaño, F. , & García‐Bermejo, M. L. (2015). A pilot study identifying a set of microRNAs as precise diagnostic biomarkers of acute kidney injury. PLoS ONE, 10(6), e0127175. 10.1371/journal.pone.0127175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahluwalia, T. S. , Lindholm, E. , & Groop, L. C. (2011). Common variants in CNDP1 and CNDP2, and risk of nephropathy in type 2 diabetes. Diabetologia, 54(9), 2295–2302. 10.1007/s00125-011-2178-5 [DOI] [PubMed] [Google Scholar]
- Ai, K. , Zhu, X. , Kang, Y. , Li, H. , & Zhang, L. (2020). miR‐130a‐3p inhibition protects against renal fibrosis in vitro via the TGF‐β1/Smad pathway by targeting SnoN. Experimental and Molecular Pathology, 112, 104358. 10.1016/j.yexmp.2019.104358 [DOI] [PubMed] [Google Scholar]
- Akaishi, J. , Onda, M. , Okamoto, J. , Miyamoto, S. , Nagahama, M. , Ito, K. , Yoshida, A. , & Shimizu, K. (2006). Down‐regulation of transcription elogation factor A (SII) like 4 (TCEAL4) in anaplastic thyroid cancer. BMC cancer, 6, 260. 10.1186/1471-2407-6-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida, A. , Gabriel, M. , Firlej, V. , Martin‐Jaular, L. , Lejars, M. , Cipolla, R. , Petit, F. , Vogt, N. , San‐Roman, M. , Dingli, F. , Loew, D. , Destouches, D. , Vacherot, F. , De La Taille, A. , Théry, C. , & Morillon, A. (2022). Urinary extracellular vesicles contain mature transcriptome enriched in circular and long noncoding RNAs with functional significance in prostate cancer. Journal of Extracellular Vesicles, 11(5), e12210. 10.1002/jev2.12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alur, V. , Raju, V. , Vastrad, B. , Tengli, A. , Vastrad, C. , & Kotturshetti, S. (2021). Integrated bioinformatics analysis reveals novel key biomarkers and potential candidate small molecule drugs in gestational diabetes mellitus. Bioscience Reports, 41(5), BSR20210617. 10.1042/bsr20210617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, B. C. , Choi, Y.‐D. , Oh, I.‐J. , Kim, J. H. , Park, J.‐I. , & Lee, S.‐W. (2018). GPx3‐mediated redox signaling arrests the cell cycle and acts as a tumor suppressor in lung cancer cell lines. PLoS ONE, 13(9), e0204170. 10.1371/journal.pone.0204170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, T. , Deng, L. , Wang, Y. , Yang, Z. , Chai, C. , Ouyang, J. , Lu, X. , & Zhang, C. (2021). The prognostic impacts of PABPC1 expression on gastric cancer patients. Future Oncology, 17(33), 4471–4479. 10.2217/fon-2021-0101 [DOI] [PubMed] [Google Scholar]
- Assmann, T. S. , Recamonde‐Mendoza, M. , De Souza, B. M. , Bauer, A. C. , & Crispim, D. (2018). MicroRNAs and diabetic kidney disease: Systematic review and bioinformatic analysis. Molecular and Cellular Endocrinology, 477, 90–102. 10.1016/j.mce.2018.06.005 [DOI] [PubMed] [Google Scholar]
- Audzeyenka, I. , Rachubik, P. , Typiak, M. , Kulesza, T. , Topolewska, A. , Rogacka, D. , Angielski, S. , Saleem, M. A. , & Piwkowska, A. (2021). Hyperglycemia alters mitochondrial respiration efficiency and mitophagy in human podocytes. Experimental Cell Research, 407(1), 112758. 10.1016/j.yexcr.2021.112758 [DOI] [PubMed] [Google Scholar]
- Baker, M. A. , Davis, S. J. , Liu, P. , Pan, X. , Williams, A. M. , Iczkowski, K. A. , Gallagher, S. T. , Bishop, K. , Regner, K. R. , Liu, Y. , & Liang, M. (2017). Tissue‐specific microRNA expression patterns in four types of kidney disease. Journal of the American Society of Nephrology, 28(10), 2985–2992. 10.1681/asn.2016121280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro, K. , Dwivedi, O. P. , Leparc, G. , Rolser, M. , Delic, D. , Forsblom, C. , Groop, P.‐H. , Groop, L. , Huber, T. B. , Puhka, M. , & Holthofer, H. (2020). Comparison of urinary extracellular vesicle isolation methods for transcriptomic biomarker research in diabetic kidney disease. Journal of Extracellular Vesicles, 10(2), e12038. 10.1002/jev2.12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro, K. , & Holthofer, H. (2017). Urinary extracellular vesicles. A promising shortcut to novel biomarker discoveries. Cell and Tissue Research, 369(1), 217–227. 10.1007/s00441-017-2621-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, K. T. , Kakajiwala, A. , Dietzen, D. J. , Goss, C. W. , Gu, H. , & Dharnidharka, V. R. (2018). Using the newer kidney disease: Improving global outcomes criteria, beta‐2‐microglobulin levels associate with severity of acute kidney injury. Clinical Kidney Journal, 11(6), 797–802. 10.1093/ckj/sfy056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzell, B. G. , Rainey, W. E. , Auchus, R. J. , Zocco, D. , Bruttini, M. , Hummel, S. L. , & Byrd, J. B. (2018). Human urinary mRNA as a biomarker of cardiovascular disease. Circulation: Genomic and Precision Medicine, 11(9), e002213. 10.1161/circgen.118.002213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. , & Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological), 57(1), 289–300. Retrieved from http://www.jstor.org/stable/2346101 [Google Scholar]
- Bonham, L. W. , Evans, D. S. , Liu, Y. , Cummings, S. R. , Yaffe, K. , & Yokoyama, J. S. (2018). Neurotransmitter pathway genes in cognitive decline during aging: Evidence for GNG4 and KCNQ2 genes. American Journal of Alzheimers Disease and Other Dementias, 33(3), 153–165. 10.1177/1533317517739384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, F. , Rinschen, M. , Buchner, D. , Bohl, K. , Plagmann, I. , Bachurski, D. , Richard Späth, M. , Antczak, P. , Göbel, H. , Klein, C. , Lackmann, J. ‐. W. , Kretz, O. , Puelles, V. G. , Wahba, R. , Hallek, M. , Schermer, B. , Benzing, T. , Huber, T. B. , Beyer, A. , … Müller, R.‐U. (2020). The proteomic landscape of small urinary extracellular vesicles during kidney transplantation. Journal of Extracellular Vesicles, 10(1), e12026. 10.1002/jev2.12026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camaj, P. , Seeliger, H. , Ischenko, I. , Krebs, S. , Blum, H. , De Toni, E. N. , Faktorova, D. , Jauch, K.‐W. , & Bruns, C. J. (2009). EFEMP1 binds the EGF receptor and activates MAPK and Akt pathways in pancreatic carcinoma cells. Biological Chemistry, 390(12), 1293–1302. 10.1515/bc.2009.140 [DOI] [PubMed] [Google Scholar]
- Carnino, J. M. , Ni, K. , & Jin, Y. (2020). Post‐translational modification regulates formation and cargo‐loading of extracellular vesicles. Frontiers in Immunology, 11, 948. 10.3389/fimmu.2020.00948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrat, G. R. , Hu, M. , Nguyen‐Tu, M.‐S. , Chabosseau, P. , Gaulton, K. J. , Van De Bunt, M. , Siddiq, A. , Falchi, M. , Thurner, M. , Canouil, M. , Pattou, F. , Leclerc, I. , Pullen, T. J. , Cane, M. C. , Prabhala, P. , Greenwald, W. , Schulte, A. , Marchetti, P. , Ibberson, M. , … Rutter, G. A. (2017). Decreased STARD10 expression is associated with defective insulin secretion in humans and mice. American Journal of Human Genetics, 100(2), 238–256. 10.1016/j.ajhg.2017.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrizzo, A. , Lenzi, P. , Procaccini, C. , Damato, A. , Biagioni, F. , Ambrosio, M. , Amodio, G. , Remondelli, P. , Del Giudice, C. , Izzo, R. , Malovini, A. , Formisano, L. , Gigantino, V. , Madonna, M. , Puca, A. A. , Trimarco, B. , Matarese, G. , Fornai, F. , & Vecchione, C. (2015). Pentraxin 3 induces vascular endothelial dysfunction through a P‐selectin/matrix metalloproteinase‐1 pathway. Circulation, 131(17), 1495–1505. discussion 1505. 10.1161/circulationaha.114.014822 [DOI] [PubMed] [Google Scholar]
- Carrizzo, A. , Procaccini, C. , Lenzi, P. , Fusco, C. , Villa, F. , Migliarino, S. , De Lucia, M. , Fornai, F. , Matarese, G. , Puca, A. A. , & Vecchione, C. (2019). PTX3: An inflammatory protein modulating ultrastructure and bioenergetics of human endothelial cells. Immunity & Ageing, 16, 4. 10.1186/s12979-019-0144-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Wang, L. , Yao, X. , Chen, H. , Xu, C. , Tong, L. , Shah, A. , Huang, T. , Chen, G. , Chen, J. , Liu, T.‐L. , Li, X.‐T. , Zheng, J.‐H. , & Li, L. (2017). miR‐195‐5p is critical in REGγ‐mediated regulation of wnt/β‐catenin pathway in renal cell carcinoma. Oncotarget, 8(38), 63986–64000. 10.18632/oncotarget.19256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, L. , Chen, C. , Guo, W. , Liu, K. , Zhao, Q. , Lu, P. , Yu, F. , & Xu, X. (2020). EFEMP1 overexpression contributes to neovascularization in age‐related macular degeneration. Frontiers in pharmacology, 11, 547436. 10.3389/fphar.2020.547436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. , Wang, D. , Wang, F. , Liu, J. , Huang, B. , Baker, M. A. , Yin, J. , Wu, R. , Liu, X. , Regner, K. R. , Usa, K. , Liu, Y. , Zhang, C. , Dong, L. , Geurts, A. M. , Wang, N. , Miller, S. S. , He, Y. , & Liang, M. (2020). Endogenous miR‐204 protects the kidney against chronic injury in hypertension and diabetes. Journal of the American Society of Nephrology, 31(7), 1539–1554. 10.1681/asn.2019101100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H.‐Y. , Van Houten, B. , Wang, X. , Miller‐Degraff, L. , Fostel, J. , Gladwell, W. , Perrow, L. , Panduri, V. , Kobzik, L. , Yamamoto, M. , Bell, D. A. , & Kleeberger, S. R. (2012). Targeted deletion of nrf2 impairs lung development and oxidant injury in neonatal mice. Antioxid Redox Signaling, 17(8), 1066–1082. 10.1089/ars.2011.4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, S. Y. , Kang, S. , Kim, D. S. , Na, H. J. , Kim, Y. J. , Choi, Y. D. , & Cho, N. H. (2018). HSP27, ALDH6A1 and prohibitin act as a trio‐biomarker to predict survival in late metastatic prostate cancer. Anticancer Research, 38(11), 6551–6560. 10.21873/anticanres.13021 [DOI] [PubMed] [Google Scholar]
- Chorley, B. N. , Ellinger‐Ziegelbauer, H. , Tackett, M. , Simutis, F. J. , Harrill, A. H. , Mcduffie, J. , Atabakhsh, E. , Nassirpour, R. , Whiteley, L. O. , Léonard, J.‐F. , Carswell, G. K. , Harpur, E. , Chen, C. L. , & Gautier, J.‐C. (2021). Urinary miRNA biomarkers of drug‐induced kidney injury and their site specificity within the nephron. Toxicological Sciences, 180(1), 1–16. 10.1093/toxsci/kfaa181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, Y. W. , Jeong, D. , Noh, O. J. , Park, Y. H. , Kang, S. I. , Lee, M. G. , Lee, T.‐H. , Yim, M. B. , & Kim, I. Y. (2009). Antioxidative role of selenoprotein W in oxidant‐induced mouse embryonic neuronal cell death. Molecules and Cells, 27(5), 609–613. 10.1007/s10059-009-0074-3 [DOI] [PubMed] [Google Scholar]
- Connor, K. L. , Teenan, O. , Cairns, C. , Banwell, V. , Thomas, R. A. B. , Rodor, J. , Finnie, S. , Pius, R. , Tannahill, G. M. , Sahni, V. , Savage, C. O. S. , Hughes, J. , Harrison, E. M. , Henderson, R. B. , Marson, L. P. , Conway, B. R. , Wigmore, S. J. , & Denby, L. (2020). Identifying cell‐enriched miRNAs in kidney injury and repair. JCI Insight, 5(24), e140399. 10.1172/jci.insight.140399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescitelli, R. , Lässer, C. , Szabó, T. G. , Kittel, A. , Eldh, M. , Dianzani, I. , Buzás, E. I. , & Lötvall, J. (2013). Distinct RNA profiles in subpopulations of extracellular vesicles: Apoptotic bodies, microvesicles and exosomes. Journal of Extracellular Vesicles, 2, 20677. 10.3402/jev.v2i0.20677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, C. , & Cui, Q. (2020). The relationship of human tissue microRNAs with those from body fluids. Scientific Reports, 10(1), 5644. 10.1038/s41598-020-62534-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marinis, Y. , Cai, M. , Bompada, P. , Atac, D. , Kotova, O. , Johansson, M. E. , Garcia‐Vaz, E. , Gomez, M. F. , Laakso, M. , & Groop, L. (2016). Epigenetic regulation of the thioredoxin‐interacting protein (TXNIP) gene by hyperglycemia in kidney. Kidney International, 89(2), 342–353. 10.1016/j.kint.2015.12.018 [DOI] [PubMed] [Google Scholar]
- Delić, D. , Wiech, F. , Urquhart, R. , Gabrielyan, O. , Rieber, K. , Rolser, M. , Tsuprykov, O. , Hasan, A. A. , Krämer, B. K. , Baum, P. , Köhler, A. , Gantner, F. , Mark, M. , Hocher, B. , & Klein, T. (2020). Linagliptin and telmisartan induced effects on renal and urinary exosomal miRNA expression in rats with 5/6 nephrectomy. Scientific Reports, 10(1), 3373. 10.1038/s41598-020-60336-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca, D. S. , Levin, J. Z. , Sivachenko, A. , Fennell, T. , Nazaire, M.‐D. , Williams, C. , Reich, M. , Winckler, W. , & Getz, G. (2012). RNA‐SeQC: RNA‐seq metrics for quality control and process optimization. Bioinformatics, 28(11), 1530–1532. 10.1093/bioinformatics/bts196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengjel, J. , Høyer‐Hansen, M. , Nielsen, M. O. , Eisenberg, T. , Harder, L. M. , Schandorff, S. , Farkas, T. , Kirkegaard, T. , Becker, A. C. , Schroeder, S. , Vanselow, K. , Lundberg, E. , Nielsen, M. M. , Kristensen, A. R. , Akimov, V. , Bunkenborg, J. , Madeo, F. , Jäättelä, M. , & Andersen, J. S. (2012). Identification of autophagosome‐associated proteins and regulators by quantitative proteomic analysis and genetic screens. Molecular & Cellular Proteomics, 11(3), M111014035. 10.1074/mcp.M111.014035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickhout, A. , & Koenen, R. R. (2018). Extracellular vesicles as biomarkers in cardiovascular disease; Chances and risks. Frontiers in Cardiovascular Medicine, 5, 113. 10.3389/fcvm.2018.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djudjaj, S. , Papasotiriou, M. , Bülow, R. D. , Wagnerova, A. , Lindenmeyer, M. T. , Cohen, C. D. , Strnad, P. , Goumenos, D. S. , Floege, J. , & Boor, P. (2016). Keratins are novel markers of renal epithelial cell injury. Kidney International, 89(4), 792–808. 10.1016/j.kint.2015.10.015 [DOI] [PubMed] [Google Scholar]
- Dos Santos, R. S. , Marroqui, L. , Grieco, F. A. , Marselli, L. , Suleiman, M. , Henz, S. R. , Marchetti, P. , Wernersson, R. , & Eizirik, D. L. (2017). Protective role of complement C3 against cytokine‐mediated β‐cell apoptosis. Endocrinology, 158(8), 2503–2521. 10.1210/en.2017-00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A. , Davis, C. A. , Schlesinger, F. , Drenkow, J. , Zaleski, C. , Jha, S. , Batut, P. , Chaisson, M. , & Gingeras, T. R. (2013). STAR: ultrafast universal RNA‐seq aligner. Bioinformatics. Oxford, England, 29(1), 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger, R. S. , Coronnello, C. , Bodnar, A. J. , Laframboise, W. A. , Benos, P. V. , Ho, J. , Johnson, J. P. , & Butterworth, M. B. (2014). Aldosterone regulates microRNAs in the cortical collecting duct to alter sodium transport. Journal of the American Society of Nephrology, 25(11), 2445–2457. 10.1681/asn.2013090931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ektesabi, A. M. , Mori, K. , Tsoporis, J. N. , Vaswani, C. M. , Gupta, S. , Walsh, C. , Varkouhi, A. K. , Mei, S. H. J. , Stewart, D. J. , Liles, W. C , Marshall, J. C. , Hu, P. , Parker, T. G. , & Dos Santos, C. C. (2021). Mesenchymal stem/stromal cells increase cardiac miR‐187‐3p expression in a polymicrobial animal model of sepsis. Shock (Augusta, Ga.), 56(1), 133–141. 10.1097/shk.0000000000001701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdbrügger, U. , & Le, T. H. (2016). Extracellular vesicles in renal diseases: More than novel biomarkers? Journal of the American Society of Nephrology, 27(1), 12–26. 10.1681/asn.2015010074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, Y. , Shan, W. , & Chen, X. (2020). MiR‐503‐5p functions as an oncogene in oral squamous cell carcinoma by targeting Smad7. Histology and Histopathology, 35(8), 893–901. 10.14670/hh-18-220 [DOI] [PubMed] [Google Scholar]
- Forster, C. S. , Haffey, W. D. , Bennett, M. , Greis, K. D. , & Devarajan, P. (2019). Identification of urinary CD44 and prosaposin as specific biomarkers of urinary tract infections in children with neurogenic bladders. Biomark Insights, 14, 117727191983557. 10.1177/1177271919835570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountas, A. , Giotaki, Z. , Dounousi, E. , Liapis, G. , Bargiota, A. , Tsatsoulis, A. , & Tigas, S. (2017). Familial partial lipodystrophy and proteinuric renal disease due to a missense c.1045C > T LMNA mutation. Endocrinology, Diabetes & Metabolism Case Reports, 2017, 10.1530/edm-17-0049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, S. , Fu, S. , Ma, X. , Yang, X. , & Ling, J. (2021). miR‑875‑5p regulates IR and inflammation via targeting TXNRD1 in gestational diabetes rats. Molecular Medicine Reports, 23(5), 303. 10.3892/mmr.2021.11942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, K. , & Hatta, K. (2018). Membranous glomerulonephritis with an LMNA mutation. CEN Case Reports, 7(1), 98–100. 10.1007/s13730-018-0303-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabril, M. , Girgis, H. , Scorilas, A. , Rotondo, F. , Wala, S. , Bjarnason, G. A. , Ding, Q. , Evans, A. , Tawedrous, E. , Pasic, M. , Finelli, A. , Al‐Haddad, S. , & Yousef, G. M. (2016). S100A11 is a potential prognostic marker for clear cell renal cell carcinoma. Clinical & Experimental Metastasis, 33(1), 63–71. 10.1007/s10585-015-9758-6 [DOI] [PubMed] [Google Scholar]
- Garcia‐Martin, R. , Wang, G. , Brandão, B. B. , Zanotto, T. M. , Shah, S. , Kumar Patel, S. , Schilling, B. , & Kahn, C. R. (2022). MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature, 601(7893), 446–451. 10.1038/s41586-021-04234-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Vives, E. , Solé, C. , Moliné, T. , Vidal, M. , Agraz, I. , Ordi‐Ros, J. , & Cortés‐Hernández, J. (2020). The Urinary Exosomal miRNA Expression Profile is Predictive of Clinical Response in Lupus Nephritis. International Journal of Molecular Sciences, 21(4), 1372. 10.3390/ijms21041372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, C. , Ferreira, Y. J. , Dragovic, R. A. , Redman, C. W. G. , & Sargent, I. L. (2013). Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. Journal of Extracellular Vesicles, 2, 19671. 10.3402/jev.v2i0.19671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigós, C. , Molina‐Pinelo, S. , Meléndez, R. , Espinosa, M. , Lerma, A. , Taron, M. , García‐Donas, J. , Rodriguez‐Antona, C. , & Duran, I. (2020). MicroRNAs as potential predictors of extreme response to tyrosine kinase inhibitors in renal cell cancer. Urologic Oncology, 38(7), 640.e23–640.e29. 10.1016/j.urolonc.2020.01.012 [DOI] [PubMed] [Google Scholar]
- Ghai, V. , Wu, X. , Bheda‐Malge, A. , Argyropoulos, C. P. , Bernardo, J. F. , Orchard, T. , Galas, D. , & Wang, K. (2018). Genome‐wide profiling of urinary extracellular vesicle microRNAs associated with diabetic nephropathy in type 1 diabetes. Kidney International Reports, 3(3), 555–572. 10.1016/j.ekir.2017.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil‐Cayuela, C. , López, A. , Martínez‐Dolz, L. , González‐Juanatey, J. R. , Lago, F. , Roselló‐Lletí, E. , Rivera, M. , & Portolés, M. (2019). The altered expression of autophagy‐related genes participates in heart failure: NRBP2 and CALCOCO2 are associated with left ventricular dysfunction parameters in human dilated cardiomyopathy. PLoS ONE, 14(4), e0215818. 10.1371/journal.pone.0215818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, J. , Yang, Y. , Wang, J. , Li, Y. , Guo, X. , Huang, Q. , Kuang, T. , Yang, S. , Li, J. , & Miao, R. (2021). Expression of miR‐93‐5p as a potential predictor of the severity of chronic thromboembolic pulmonary hypertension. BioMed Research International, 2021, 1. 10.1155/2021/6634417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales, P. A. , Pisitkun, T. , Hoffert, J. D. , Tchapyjnikov, D. , Star, R. A. , Kleta, R. , Wang, N. S. , & Knepper, M. A. (2009). Large‐scale proteomics and phosphoproteomics of urinary exosomes. Journal of the American Society of Nephrology, 20(2), 363–379. 10.1681/asn.2008040406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieva, I. V. , Oszwald, A. , Grigorieva, E. F. , Schachner, H. , Neudert, B. , Ostendorf, T. , Floege, J. , Lindenmeyer, M. T. , Cohen, C. D. , Panzer, U. , Aigner, C. , Schmidt, A. , Grosveld, F. , Thakker, R. V. , Rees, A. J. , & Kain, R. (2019). A novel role for GATA3 in mesangial cells in glomerular development and injury. Journal of the American Society of Nephrology, 30(9), 1641–1658. 10.1681/asn.2018111143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb, A. , Simonsen, O. , Sturfelt, G. , Truedsson, L. , & Thysell, H. (1985). Serum concentration of cystatin C, factor D and beta 2‐microglobulin as a measure of glomerular filtration rate. Acta Medica Scandinavica, 218(5), 499–503. 10.1111/j.0954-6820.1985.tb08880.x [DOI] [PubMed] [Google Scholar]
- Guo, Y. , Redmond, C. J. , Leacock, K. A. , Brovkina, M. V. , Ji, S. , Jaskula‐Ranga, V. , & Coulombe, P. A. (2020). Keratin 14‐dependent disulfides regulate epidermal homeostasis and barrier function via 14‐3‐3σ and YAP1. Elife, 9, 10.7554/eLife.53165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, A. , Nitoiu, D. , Brennan‐Crispi, D. , Addya, S. , Riobo, N. A. , Kelsell, D. P. , & Mahoney, M. G. (2015). Cell cycle‐ and cancer‐associated gene networks activated by Dsg2: Evidence of cystatin A deregulation and a potential role in cell‐cell adhesion. PLoS ONE, 10(3), e0120091. 10.1371/journal.pone.0120091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase, M. , Bellomo, R. , Albert, C. , Vanpoucke, G. , Thomas, G. , Laroy, W. , Verleysen, K. , Kropf, S. , Kuppe, H. , Hetzer, R. , & Haase‐Fielitz, A. (2014). The identification of three novel biomarkers of major adverse kidney events. Biomarkers in Medicine, 8(10), 1207–1217. 10.2217/bmm.14.90 [DOI] [PubMed] [Google Scholar]
- Habuka, M. , Fagerberg, L. , Hallström, B. M. , Kampf, C. , Edlund, K. , Sivertsson, Å. , Yamamoto, T. , Pontén, F. , Uhlén, M. , & Odeberg, J. (2014). The kidney transcriptome and proteome defined by transcriptomics and antibody‐based profiling. PLoS ONE, 9(12), e116125. 10.1371/journal.pone.0116125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque, A. , Engel, J. , Teichmann, S. A. , & Lönnberg, T. (2017). A practical guide to single‐cell RNA‐sequencing for biomedical research and clinical applications. Genome Medicine, 9(1), 75. 10.1186/s13073-017-0467-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque, M. , Kendal, J. K. , Macisaac, R. M. , & Demetrick, D. J. (2016). WSB1: From homeostasis to hypoxia. Journal of Biomedical Science, 23(1), 61. 10.1186/s12929-016-0270-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan, M. K. , Kumar, D. , Naik, M. , & Dixit, M. (2018). The expression profile and prognostic significance of eukaryotic translation elongation factors in different cancers. PLoS ONE, 13(1), e0191377. 10.1371/journal.pone.0191377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, T. , Chen, P. , Jin, L. , Hu, J. , Li, Y. , Zhou, L. , Yang, S. , Mao, X. , Gui, Y. , Chen, Y. , & Lai, Y. (2018). miR‑660‑5p is associated with cell migration, invasion, proliferation and apoptosis in renal cell carcinoma. Molecular Medicine Reports, 17(1), 2051–2060. 10.3892/mmr.2017.8052 [DOI] [PubMed] [Google Scholar]
- Huang, C. , Zhang, Y. , Kelly, D. J. , Tan, C. Y. R. , Gill, A. , Cheng, D. , Braet, F. , Park, J.‐S. , Sue, C. M. , Pollock, C. A. , & Chen, X.‐M. (2016). Thioredoxin interacting protein (TXNIP) regulates tubular autophagy and mitophagy in diabetic nephropathy through the mTOR signaling pathway. Scientific Reports, 6, 29196. 10.1038/srep29196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J.‐Q. , Trasler, J. M. , Igdoura, S. , Michaud, J. , Hanai, N. , & Gravel, R. A. (1997). Apoptotic cell death in mouse models of GM2 gangliosidosis and observations on human Tay‐Sachs and Sandhoff diseases. Human Molecular Genetics, 6(11), 1879–1885. 10.1093/hmg/6.11.1879 [DOI] [PubMed] [Google Scholar]
- Huang, Y. , Xu, J. , Wu, X. , Chen, X. , Bai, X. , Zhuang, Y. , Fang, J. , & Lin, X. (2019). High expression of complement components in the kidneys of type 2 diabetic rats with diabetic nephropathy. Frontiers in Endocrinology (Lausanne), 10, 459. 10.3389/fendo.2019.00459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulstaert, E. , Morlion, A. , Avila Cobos, F. , Verniers, K. , Nuytens, J. , Vanden Eynde, E. , Yigit, N. , Anckaert, J. , Geerts, A. , Hindryckx, P. , Jacques, P. , Brusselle, G. , Bracke, K. R. , Maes, T. , Malfait, T. , Derveaux, T. , Ninclaus, V. , Van Cauwenbergh, C. , Roelens, K. , … Mestdagh, P. (2020). Charting extracellular transcriptomes in the human biofluid RNA atlas. Cell Reports, 33(13), 108552. 10.1016/j.celrep.2020.108552 [DOI] [PubMed] [Google Scholar]
- Hussain, S. S. , Harris, M. T. , Kreutzberger, A. J. B. , Inouye, C. M. , Doyle, C. A. , Castle, A. M. , Arvan, P. , & Castle, J. D. (2018). Control of insulin granule formation and function by the ABC transporters ABCG1 and ABCA1 and by oxysterol binding protein OSBP. Molecular Biology of the Cell, 29(10), 1238–1257. 10.1091/mbc.E17-08-0519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraci, N. , Leonardi, T. , Gessler, F. , Vega, B. , & Pluchino, S. (2016). Focus on extracellular vesicles: Physiological role and signalling properties of extracellular membrane vesicles. International Journal of Molecular Sciences, 17(2), 171. 10.3390/ijms17020171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, J. A. , Shankland, S. J. , & Pichler, R. H. (2008). Proteinuria in diabetic kidney disease: A mechanistic viewpoint. Kidney International, 74(1), 22–36. 10.1038/ki.2008.128 [DOI] [PubMed] [Google Scholar]
- Jia, J. , Absmeier, E. , Holton, N. , Pietrzyk‐Brzezinska, A. J. , Hackert, P. , Bohnsack, K. E. , Bohnsack, M. T. , & Wahl, M. C. (2020). The interaction of DNA repair factors ASCC2 and ASCC3 is affected by somatic cancer mutations. Nature Communications, 11(1), 5535. 10.1038/s41467-020-19221-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L. , Liu, X.‐Q. , Ma, Q. , Yang, Q. , Gao, L. , Li, H.‐D. , Wang, J.‐N. , Wei, B. , Wen, J. , Li, J. , Wu, Y.‐G. , & Meng, X.‐M. (2019). hsa‐miR‐500a‐3P alleviates kidney injury by targeting MLKL‐mediated necroptosis in renal epithelial cells. Faseb Journal, 33(3), 3523–3535. 10.1096/fj.201801711R [DOI] [PubMed] [Google Scholar]
- Jiang, Z. , Teng, L. , Zhang, S. , & Ding, Y. (2021). Mitochondrial ND1 T4216C and ND2 C5178A mutations are associated with maternally transmitted diabetes mellitus. Mitochondrial DNA Part A: DNA Mapping, Sequencing, and Analysis, 32(2), 59–65. 10.1080/24701394.2020.1856101 [DOI] [PubMed] [Google Scholar]
- Kalderimis, A. , Lyne, R. , Butano, D. , Contrino, S. , Lyne, M. , Heimbach, J. , Hu, F. , Smith, R. , Štěpán, R. , Sullivan, J. , & Micklem, G. (2014). InterMine: Extensive web services for modern biology. Nucleic Acid Research, 42, W468–W472. 10.1093/nar/gku301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra, H. , Simpson, R. J. , Ji, H. , Aikawa, E. , Altevogt, P. , Askenase, P. , Bond, V. C. , Borràs, F. E. , Breakefield, X. , Budnik, V. , Buzas, E. , Camussi, G. , Clayton, A. , Cocucci, E. , Falcon‐Perez, J. M. , Gabrielsson, S. , Gho, Y. S. , Gupta, D. , Harsha, H. C. , … Mathivanan, S. (2012). Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. Plos Biology, 10(12), e1001450. 10.1371/journal.pbio.1001450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamburova, E. G. , Gruijters, M. L. , Kardol‐Hoefnagel, T. , Wisse, B. W. , Joosten, I. , Allebes, W. A. , Van Der Meer, A. , Hilbrands, L. B. , Baas, M. C. , Spierings, E. , Hack, C. E. , Van Reekum, F. E. , Van Zuilen, A. D. , Verhaar, M. C. , Bots, M. L. , Drop, A. C. A. D. , Plaisier, L. , Melchers, R. C. A. , Seelen, M. A. J. , … Otten, H. G. (2019). Antibodies against ARHGDIB are associated with long‐term kidney graft loss. American Journal of Transplantation, 19(12), 3335–3344. 10.1111/ajt.15493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanasaki, K. , Shi, S. , Kanasaki, M. , He, J. , Nagai, T. , Nakamura, Y. , Ishigaki, Y. , Kitada, M. , Srivastava, S. P. , & Koya, D. (2014). Linagliptin‐mediated DPP‐4 inhibition ameliorates kidney fibrosis in streptozotocin‐induced diabetic mice by inhibiting endothelial‐to‐mesenchymal transition in a therapeutic regimen. Diabetes, 63(6), 2120–2131. 10.2337/db13-1029 [DOI] [PubMed] [Google Scholar]
- Keller, A. , Gröger, L. , Tschernig, T. , Solomon, J. , Laham, O. , Schaum, N. , Wagner, V. , Kern, F. , Schmartz, G. P. , Li, Y. , Borcherding, A. , Meier, C. , Wyss‐Coray, T. , Meese, E. , Fehlmann, T. , & Ludwig, N. (2022). miRNATissueAtlas2: An update to the human miRNA tissue atlas. Nucleic Acid Research, 50(D1), D211–D221. 10.1093/nar/gkab808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, K. J. , Liu, Y. , Zhang, J. , & Dominguez, J. H. (2015). Renal C3 complement component: Feed forward to diabetic kidney disease. American Journal of Nephrology, 41(1), 48–56. 10.1159/000371426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayer, N. , Mirzaie, M. , Marashi, S.‐A. , & Jalessi, M. (2020). Rps27a might act as a controller of microglia activation in triggering neurodegenerative diseases. PLoS ONE, 15(9), e0239219. 10.1371/journal.pone.0239219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, B.‐R. , Lee, S.‐H. , Park, M.‐S. , Seo, S.‐H. , Park, Y.‐M. , Kwon, Y.‐J. , & Rho, S.‐B. (2016). MARCKSL1 exhibits anti‐angiogenic effects through suppression of VEGFR‐2‐dependent Akt/PDK‐1/mTOR phosphorylation. Oncology Reports, 35(2), 1041–1048. 10.3892/or.2015.4408 [DOI] [PubMed] [Google Scholar]
- Kim, J. J. , Lee, S. B. , Jang, J. , Yi, S.‐Y. , Kim, S.‐H. , Han, S.‐A. , Lee, J.‐M. , Tong, S.‐Y. , Vincelette, N. D. , Gao, B. , Yin, P. , Evans, D. , Choi, D. W. , Qin, B. , Liu, T. , Zhang, H. , Deng, M. , Jen, J. , Zhang, J. , … Lou, Z. (2015). WSB1 promotes tumor metastasis by inducing pVHL degradation. Genes & Development, 29(21), 2244–2257. 10.1101/gad.268128.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. Y. , Shin, H. , Moon, H. W. , Park, Y. H. , Park, J. , & Lee, J. Y. (2021). Urinary exosomal microRNA profiling in intermediate‐risk prostate cancer. Scientific Reports, 11(1), 7355. 10.1038/s41598-021-86785-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koscielny, G. , An, P. , Carvalho‐Silva, D. , Cham, J. A. , Fumis, L. , Gasparyan, R. , Hasan, S. , Karamanis, N. , Maguire, M. , Papa, E. , Pierleoni, A. , Pignatelli, M. , Platt, T. , Rowland, F. , Wankar, P. , Bento, A. P. , Burdett, T. , Fabregat, A. , Forbes, S. , … Dunham, I. (2017). Open targets: A platform for therapeutic target identification and validation. Nucleic Acid Research, 45(D1), D985–D994. 10.1093/nar/gkw1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash, I. , Tolleson, W. H. , Guo, L. , Yu, D. , Chen, S. , Hong, H. , Mattes, W. , & Ning, B. (2015). microRNAs as pharmacogenomic biomarkers for drug efficacy and drug safety assessment. Biomarkers in medicine, 9(11), 1153–1176. 10.2217/bmm.15.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalik, C. G. , Palmer, D. A. , Sullivan, T. B. , Teebagy, P. A. , Dugan, J. M. , Libertino, J. A. , Burks, E. J. , Canes, D. , & Rieger‐Christ, K. M. (2017). Profiling microRNA from nephrectomy and biopsy specimens: Predictors of progression and survival in clear cell renal cell carcinoma. Bju International, 120(3), 428–440. 10.1111/bju.13886 [DOI] [PubMed] [Google Scholar]
- Kurahashi, R. , Kadomatsu, T. , Baba, M. , Hara, C. , Itoh, H. , Miyata, K. , Endo, M. , Morinaga, J. , Terada, K. , Araki, K. , Eto, M. , Schmidt, L. S. , Kamba, T. , Linehan, W. M , & Oike, Y. (2019). MicroRNA‐204‐5p: A novel candidate urinary biomarker of Xp11.2 translocation renal cell carcinoma. Cancer Science, 110(6), 1897–1908. 10.1111/cas.14026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, G. J. , Godin, N. , Maachi, H. , Lo, C.‐S. , Wu, S.‐J. , Zhu, J.‐X. , Brezniceanu, M.‐L. , Chénier, I. , Fragasso‐Marquis, J. , Lattouf, J.‐B. , Ethier, J. , Filep, J. G. , Ingelfinger, J. R. , Nair, V. , Kretzler, M. , Cohen, C. D. , Zhang, S.‐L. , & Chan, J. S. D. (2012). Bcl‐2‐modifying factor induces renal proximal tubular cell apoptosis in diabetic mice. Diabetes, 61(2), 474–484. 10.2337/db11-0141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law, C. W. , Chen, Y. , Shi, W. , & Smyth, G. K. (2014). voom: Precision weights unlock linear model analysis tools for RNA‐seq read counts. Genome Biology, 15(2), R29. 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay, A. C. , Hurcombe, J. A. , Betin, V. M. S. , Barrington, F. , Rollason, R. , Ni, L. , Gillam, L. , Pearson, G. M. E. , Østergaard, M. V. , Hamidi, H. , Lennon, R. , Welsh, G. I. , & Coward, R. J. M. (2017). Prolonged exposure of mouse and human podocytes to insulin induces insulin resistance through lysosomal and proteasomal degradation of the insulin receptor. Diabetologia, 60(11), 2299–2311. 10.1007/s00125-017-4394-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarte, J. , Jurgens, S. J. , Choi, S. H. , Khurshid, S. , Morrill, V. N. , Weng, L.‐C. , Nauffal, V. , Pirruccello, J. P. , Halford, J. L. , Hegele, R. A. , Ellinor, P. T. , Lunetta, K. L. , & Lubitz, S. A. (2022). LMNA Variants and Risk of Adult‐Onset Cardiac Disease. Journal of the American College of Cardiology, 80(1), 50–59. 10.1016/j.jacc.2022.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W.‐C. , Li, L.‐C. , Ng, H.‐Y. , Lin, P.‐T. , Chiou, T. T.‐Y. , Kuo, W.‐H. , & Lee, C.‐T. (2020). Urinary exosomal microRNA signatures in nephrotic, biopsy‐proven diabetic nephropathy. Journal of Clinical Medicine, 9(4), 1220. 10.3390/jcm9041220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, A. , Reznichenko, A. , Witasp, A. , Liu, P. , Greasley, P. J. , Sorrentino, A. , Blondal, T. , Zambrano, S. , Nordström, J. , Bruchfeld, A. , Barany, P. , Ebefors, K. , Erlandsson, F. , Patrakka, J. , Stenvinkel, P. , Nyström, J. , & Wernerson, A. (2020). Novel insights into the disease transcriptome of human diabetic glomeruli and tubulointerstitium. Nephrology, Dialysis, Transplantation, 35(12), 2059–2072. 10.1093/ndt/gfaa121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Zhang, H. , Huang, G. , Bing, Z. , Xu, D. , Liu, J. , Luo, H. , & An, X. (2022). Loss of RPS27a expression regulates the cell cycle, apoptosis, and proliferation via the RPL11‐MDM2‐p53 pathway in lung adenocarcinoma cells. Journal of Experimental & Clinical Cancer Research, 41(1), 33. 10.1186/s13046-021-02230-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Bao, H. , Zhang, K. , Yang, X. , Liu, X. , Li, P. , Li, Q. , & Chen, W. (2020). MiR‐542‐3p drives renal fibrosis by targeting AGO1 in vivo and in vitro. Life Sciences, 255, 117845. 10.1016/j.lfs.2020.117845 [DOI] [PubMed] [Google Scholar]
- Li, Y. , He, X. , Li, Q. , Lai, H. , Zhang, H. , Hu, Z. , Li, Y. , & Huang, S. (2020). EV‐origin: Enumerating the tissue‐cellular origin of circulating extracellular vesicles using exLR profile. Computational and Structural Biotechnology Journal, 18, 2851–2859. 10.1016/j.csbj.2020.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Quan, J. , Chen, F. , Pan, X. , Zhuang, C. , Xiong, T. , Zhuang, C. , Li, J. , Huang, X. , Ye, J. , Zhang, F. , Zhang, Z. , & Gui, Y. (2019). MiR‐31‐5p acts as a tumor suppressor in renal cell carcinoma by targeting cyclin‐dependent kinase 1 (CDK1). Biomedicine & Pharmacotherapy, 111, 517–526. 10.1016/j.biopha.2018.12.102 [DOI] [PubMed] [Google Scholar]
- Liang, L. , Huang, J. , Yao, M. , Li, L. , Jin, X.‐J. , & Cai, X.‐Y. (2021). GNG4 promotes tumor progression in colorectal cancer. Journal of oncology, 2021, 1. 10.1155/2021/9931984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X. , & Xu, W. (2020). miR‐181a‐5p regulates the proliferation and apoptosis of glomerular mesangial cells by targeting KLF6. Experimental and Therapeutic Medicine, 20(2), 1121–1128. 10.3892/etm.2020.8780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y. , Lehrich, B. M. , Zheng, S. , & Lu, M. (2021). Emerging methods in biomarker identification for extracellular vesicle‐based liquid biopsy. Journal of Extracellular Vesicles, 10(7), e12090. 10.1002/jev2.12090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y. , Smyth, G. K. , & Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. Oxford, England, 30(7), 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Licker, V. , Côte, M. , Lobrinus, J. A. , Rodrigo, N. , Kövari, E. , Hochstrasser, D. F. , Turck, N. , Sanchez, J.‐C. , & Burkhard, P. R. (2012). Proteomic profiling of the substantia nigra demonstrates CNDP2 overexpression in Parkinson's disease. Journal of Proteomics, 75(15), 4656–4667. 10.1016/j.jprot.2012.02.032 [DOI] [PubMed] [Google Scholar]
- Liu, J. , Liu, B. , Guo, Y. , Chen, Z. , Sun, W. , Gao, W. , Wu, H. , & Wang, Y. (2018). MiR‐199a‐3p acts as a tumor suppressor in clear cell renal cell carcinoma. Pathology, Research and Practice, 214(6), 806–813. 10.1016/j.prp.2018.05.005 [DOI] [PubMed] [Google Scholar]
- Liu, J. , Shao, T. , Zhang, J. , Liu, Q. , Hua, H. , Zhang, H. , Wang, J. , Luo, T. , Shi, Y. E. , & Jiang, Y. (2022). Gamma synuclein promotes cancer metastasis through the MKK3/6‐p38MAPK cascade. International Journal of Biological Sciences, 18(8), 3167–3177. 10.7150/ijbs.69155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. , Miao, L. , Liu, Y. , Qi, A. , Xie, P. , Chen, J. , & Zhu, H. (2017). S100A11 regulates renal carcinoma cell proliferation, invasion, and migration via the EGFR/Akt signaling pathway and E‐cadherin. Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine, 39(5), 101042831770533. 10.1177/1010428317705337 [DOI] [PubMed] [Google Scholar]
- Liu, R. , Correll, R. N. , Davis, J. , Vagnozzi, R. J. , York, A. J. , Sargent, M. A. , Nairn, A. C. , & Molkentin, J. D. (2015). Cardiac‐specific deletion of protein phosphatase 1β promotes increased myofilament protein phosphorylation and contractile alterations. Journal of Molecular and Cellular Cardiology, 87, 204–213. 10.1016/j.yjmcc.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Look AHEAD Research Group . (2016). Prospective association of GLUL rs10911021 with cardiovascular morbidity and mortality among individuals with type 2 diabetes: The look AHEAD study. Diabetes, 65(1), 297–302. 10.2337/db15-0890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J. , Chen, Z. , Zhao, H. , Dong, H. , Zhu, L. , Zhang, Y. , Wang, J. , Zhu, H. , Cui, Q. , Qi, C. , Wang, S. , Chen, S. , & Shao, J. (2020). ABAT and ALDH6A1, regulated by transcription factor HNF4A, suppress tumorigenic capability in clear cell renal cell carcinoma. Journal of Translational Medicine, 18(1), 101. 10.1186/s12967-020-02268-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukamowicz‐Rajska, M. , Mittmann, C. , Prummer, M. , Zhong, Q. , Bedke, J. , Hennenlotter, J. , Stenzl, A. , Mischo, A. , Bihr, S. , Schmidinger, M. , Vogl, U. , Blume, I. , Karlo, C. , Schraml, P. , & Moch, H. (2016). MiR‐99b‐5p expression and response to tyrosine kinase inhibitor treatment in clear cell renal cell carcinoma patients. Oncotarget, 7(48), 78433–78447. 10.18632/oncotarget.12618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, C. , Wu, M. , Su, X. , Yu, F. , Brautigan, D. L. , Chen, J. , & Zhou, A. J. (2019). Protein phosphatase 1α interacts with a novel ciliary targeting sequence of polycystin‐1 and regulates polycystin‐1 trafficking. Faseb Journal, 33(9), 9945–9958. 10.1096/fj.201900338R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong, T T. D. , Schelski, N. , Boehme, B. , Makridakis, M. , Vlahou, A. , Lang, F. , Pieske, B. , Alesutan, I. , & Voelkl, J. (2018). Fibulin‐3 attenuates phosphate‐induced vascular smooth muscle cell calcification by inhibition of oxidative stress. Cellular Physiology and Biochemistry, 46(4), 1305–1316. 10.1159/000489144 [DOI] [PubMed] [Google Scholar]
- Ma, G. , Gezer, D. , Herrmann, O. , Feldberg, K. , Schemionek, M. , Jawhar, M. , Reiter, A. , Brümmendorf, T. H. , Koschmieder, S. , & Chatain, N. (2020). LCP1 triggers mTORC2/AKT activity and is pharmacologically targeted by enzastaurin in hypereosinophilia. Molecular Carcinogenesis, 59(1), 87–103. 10.1002/mc.23131 [DOI] [PubMed] [Google Scholar]
- Maeda, Y. , Suzuki, T. , Pan, X. , Chen, G. , Pan, S. , Bartman, T. , & Whitsett, J. A. (2008). CUL2 is required for the activity of hypoxia‐inducible factor and vasculogenesis. Journal of Biological Chemistry, 283(23), 16084–16092. 10.1074/jbc.M710223200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magayr, T. A. , Song, X. , Streets, A. J. , Vergoz, L. , Chang, L. , Valluru, M. K. , Yap, H. L. , Lannoy, M. , Haghighi, A. , Simms, R. J. , Tam, F. W. K. , Pei, Y. , & Ong, A. C. M. (2020). Global microRNA profiling in human urinary exosomes reveals novel disease biomarkers and cellular pathways for autosomal dominant polycystic kidney disease. Kidney International, 98(2), 420–435. 10.1016/j.kint.2020.02.008 [DOI] [PubMed] [Google Scholar]
- Maier, J. I. , Rogg, M. , Helmstädter, M. , Sammarco, A. , Schilling, O. , Sabass, B. , Miner, J. H. , Dengjel, J. , Walz, G. , Werner, M. , Huber, T. B. , & Schell, C. (2021). EPB41L5 controls podocyte extracellular matrix assembly by adhesome‐dependent force transmission. Cell Reports, 34(12), 108883. 10.1016/j.celrep.2021.108883 [DOI] [PubMed] [Google Scholar]
- Manwaring, V. , Heywood, W. E. , Clayton, R. , Lachmann, R. H. , Keutzer, J. , Hindmarsh, P. , Winchester, B. , Heales, S. , & Mills, K. (2013). The identification of new biomarkers for identifying and monitoring kidney disease and their translation into a rapid mass spectrometry‐based test: Evidence of presymptomatic kidney disease in pediatric Fabry and type‐I diabetic patients. Journal of Proteome Research, 12(5), 2013–2021. 10.1021/pr301200e [DOI] [PubMed] [Google Scholar]
- Martínez‐Camberos, A. , Alvarez‐Arrazola, M. , Arámbula‐Meraz, E. , Romero‐Quintana, J. , Luque‐Ortega, F. , Romo‐Martinez, E. , Sánchez‐Urbina, R. , Cedano‐Prieto, D. , González‐Castillo, A. , & García‐Magallanes, N. (2022). Dysregulation of KRT19, TIMP1, and CLDN1 gene expression is associated with thyroid cancer. Biochemical and Biophysical Research Communications, 617(Pt 1), 55–59. 10.1016/j.bbrc.2022.05.093 [DOI] [PubMed] [Google Scholar]
- Matsudaira, T. , Mukai, K. , Noguchi, T. , Hasegawa, J. , Hatta, T. , Iemura, S.‐I. , Natsume, T. , Miyamura, N. , Nishina, H. , Nakayama, J. , Semba, K. , Tomita, T. , Murata, S. , Arai, H. , & Taguchi, T. (2017). Endosomal phosphatidylserine is critical for the YAP signalling pathway in proliferating cells. Nature Communications, 8(1), 1246. 10.1038/s41467-017-01255-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant, M. L. , Rood, I. M. , Deegens, J. K. J. , & Klein, J. B. (2017). Isolation and characterization of urinary extracellular vesicles: Implications for biomarker discovery. Nature Reviews Nephrology, 13(12), 731–749. 10.1038/nrneph.2017.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merscher, S. , & Fornoni, A. (2014). Podocyte pathology and nephropathy ‐ sphingolipids in glomerular diseases. Frontiers in Endocrinology (Lausanne), 5, 127. 10.3389/fendo.2014.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, X. , & Zhang, N. (2020). Role of RBM3 in the regulation of cell proliferation in hepatocellular carcinoma. Experimental and Molecular Pathology, 117, 104546. 10.1016/j.yexmp.2020.104546 [DOI] [PubMed] [Google Scholar]
- Miyamoto, M. , Yoshida, Y. , Taguchi, I. , Nagasaka, Y. , Tasaki, M. , Zhang, Y. , Xu, B. , Nameta, M. , Sezaki, H. , Cuellar, L. M. , Osawa, T. , Morishita, H. , Sekiyama, S. , Yaoita, E. , Kimura, K. , & Yamamoto, T. (2007). In‐depth proteomic profiling of the normal human kidney glomerulus using two‐dimensional protein prefractionation in combination with liquid chromatography‐tandem mass spectrometry. Journal of Proteome Research, 6(9), 3680–3690. 10.1021/pr070203n [DOI] [PubMed] [Google Scholar]
- Morito, N. , Yoh, K. , Ojima, M. , Okamura, M. , Nakamura, M. , Hamada, M. , Shimohata, H. , Moriguchi, T. , Yamagata, K. , & Takahashi, S. (2014). Overexpression of Mafb in podocytes protects against diabetic nephropathy. Journal of the American Society of Nephrology, 25(11), 2546–2557. 10.1681/asn.2013090993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, A. P. , Le, T. H. , Wu, H. , Akbarov, A. , Van Der Most, P. J. , Hemani, G. , Smith, G. D. , Mahajan, A. , Gaulton, K. J. , Nadkarni, G. N. , Valladares‐Salgado, A. , Wacher‐Rodarte, N. , Mychaleckyj, J. C. , Dueker, N. D. , Guo, X. , Hai, Y. , Haessler, J. , Kamatani, Y. , Stilp, A. M. , … Franceschini, N. (2019). Trans‐ethnic kidney function association study reveals putative causal genes and effects on kidney‐specific disease aetiologies. Nature Communications, 10(1), 29. 10.1038/s41467-018-07867-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musante, L. , Tataruch, D. , Gu, D. , Benito‐Martin, A. , Calzaferri, G. , Aherne, S. , & Holthofer, H. (2014). A simplified method to recover urinary vesicles for clinical applications, and sample banking. Scientific Reports, 4, 7532. 10.1038/srep07532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsaers, H. A. M. , Wilmer, M. J. G. , Reijnders, D. , Jansen, J. , Van Den Broek, P. H. H. , Forkink, M. , Schepers, E. , Glorieux, G. , Vanholder, R. , Van Den Heuvel, L. P. , Hoenderop, J. G. , & Masereeuw, R. (2013). Uremic toxins inhibit renal metabolic capacity through interference with glucuronidation and mitochondrial respiration. Biochimica Et Biophysica Acta, 1832(1), 142–150. 10.1016/j.bbadis.2012.09.006 [DOI] [PubMed] [Google Scholar]
- Müller‐Deile, J. , Dannenberg, J. , Liu, P. , Lorenzen, J. , Nyström, J. , Thum, T. , & Schiffer, M. (2019). Identification of cell and disease specific microRNAs in glomerular pathologies. Journal of Cellular and Molecular Medicine, 23(6), 3927–3939. 10.1111/jcmm.14270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezu, M. , & Suzuki, N. (2020). Roles of Nrf2 in protecting the kidney from oxidative damage. International Journal of Molecular Sciences, 21(8), 2951. 10.3390/ijms21082951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, M. , Zhao, Y. , & Wang, X. (2021). Suppression of synuclein gamma inhibits the movability of endometrial carcinoma cells by PI3K/AKT/ERK signaling pathway. Genes Genomics, 43(6), 633–641. 10.1007/s13258-021-01080-5 [DOI] [PubMed] [Google Scholar]
- Nikoloff, J. M. , Saucedo‐Espinosa, M. A. , Kling, A. , & Dittrich, P. S. (2021). Identifying extracellular vesicle populations from single cells. Proceedings of the National Academy of Sciences of the United States of America, 118(38), 10.1073/pnas.2106630118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'grady, T. , Njock, M.‐S. , Lion, M. , Bruyr, J. , Mariavelle, E. , Galvan, B. , Boeckx, A. , Struman, I. , & Dequiedt, F. (2022). Sorting and packaging of RNA into extracellular vesicles shape intracellular transcript levels. BMC Biology, 20(1), 72. 10.1186/s12915-022-01277-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa, D. , Hercules, A. , Carmona, M. , Suveges, D. , Gonzalez‐Uriarte, A. , Malangone, C. , Miranda, A. , Fumis, L. , Carvalho‐Silva, D. , Spitzer, M. , Baker, J. , Ferrer, J. , Raies, A. , Razuvayevskaya, O. , Faulconbridge, A. , Petsalaki, E. , Mutowo, P. , Machlitt‐Northen, S. , Peat, G. , … Mcdonagh, E. . M. (2021). Open targets platform: Supporting systematic drug‐target identification and prioritisation. Nucleic Acid Research, 49(D1), D1302–D1310. 10.1093/nar/gkaa1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeyen, E. , Van Mol, K. , Baggerman, G. , Willems, H. , Boonen, K. , Rolfo, C. , Pauwels, P. , Jacobs, A. , Schildermans, K. , Cho, W. C. , & Mertens, I. (2018). Ultrafiltration and size exclusion chromatography combined with asymmetrical‐flow field‐flow fractionation for the isolation and characterisation of extracellular vesicles from urine. Journal of Extracellular Vesicles, 7(1), 1490143. 10.1080/20013078.2018.1490143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunbileje, J. O. , Porter, C. , Herndon, D. N. , Chao, T. , Abdelrahman, D. R. , Papadimitriou, A. , Chondronikola, M. , Zimmers, T. A. , Reidy, P. T. , Rasmussen, B. B. , & Sidossis, L. S. (2016). Hypermetabolism and hypercatabolism of skeletal muscle accompany mitochondrial stress following severe burn trauma. American Journal of Physiology. Endocrinology and Metabolism, 311(2), E436–E448. 10.1152/ajpendo.00535.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olayioye, M. A. , Hoffmann, P. , Pomorski, T. , Armes, J. , Simpson, R. J. , Kemp, B. E. , Lindeman, G. J. , & Visvader, J. E. (2004). The phosphoprotein StarD10 is overexpressed in breast cancer and cooperates with ErbB receptors in cellular transformation. Cancer Research, 64(10), 3538–3544. 10.1158/0008-5472.Can-03-3731 [DOI] [PubMed] [Google Scholar]
- Owusu, D. , Pan, Y. , Xie, C. , Harirforoosh, S. , & Wang, K.‐S. (2017). Polymorphisms in PDLIM5 gene are associated with alcohol dependence, type 2 diabetes, and hypertension. Journal of Psychiatric Research, 84, 27–34. 10.1016/j.jpsychires.2016.09.015 [DOI] [PubMed] [Google Scholar]
- Pang, B. , Zhu, Y. , Ni, J. , Thompson, J. , Malouf, D. , Bucci, J. , Graham, P. , & Li, Y. (2020). Extracellular vesicles: The next generation of biomarkers for liquid biopsy‐based prostate cancer diagnosis. Theranostics, 10(5), 2309–2326. 10.7150/thno.39486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, P. , Abbott, M. , Abdi, M. , Fucci, Q.‐A. , Chauhan, N. , Mistri, M. , Proctor, B. , Chin, M. , Wang, B. , Yin, W. , Lu, T.‐S. , Halim, A. , Lim, K. , Handy, D. E. , Loscalzo, J. , & Siedlecki, A. M. (2018). Pre‐clinical model of severe glutathione peroxidase‐3 deficiency and chronic kidney disease results in coronary artery thrombosis and depressed left ventricular function. Nephrology, Dialysis, Transplantation, 33(6), 923–934. 10.1093/ndt/gfx304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, X. , Zhang, Y. , Shi, X. , Li, D. , & Han, J. (2018). ERp44 depletion exacerbates ER stress and aggravates diabetic nephropathy in db/db mice. Biochemical and Biophysical Research Communications, 504(4), 921–926. 10.1016/j.bbrc.2018.09.037 [DOI] [PubMed] [Google Scholar]
- Patel, H. , Chen, J. , Das, K. C. , & Kavdia, M. (2013). Hyperglycemia induces differential change in oxidative stress at gene expression and functional levels in HUVEC and HMVEC. Cardiovasc Diabetol, 12, 142. 10.1186/1475-2840-12-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunas, F. T. I. , Finne, K. , Leh, S. , Osman, T. A.‐H. , Marti, H.‐P. , Berven, F. , & Vikse, B. E. (2019). Characterization of glomerular extracellular matrix in IgA nephropathy by proteomic analysis of laser‐captured microdissected glomeruli. BMC Nephrology [Electronic Resource], 20(1), 410. 10.1186/s12882-019-1598-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penton, D. , Moser, S. , Wengi, A. , Czogalla, J. , Rosenbaek, L. L. , Rigendinger, F. , Faresse, N. , Martins, J. R. , Fenton, R. A. , Loffing‐Cueni, D. , & Loffing, J. (2019). Protein phosphatase 1 inhibitor‐1 mediates the cAMP‐dependent stimulation of the renal NaCl cotransporter. Journal of the American Society of Nephrology, 30(5), 737–750. 10.1681/asn.2018050540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzolesi, M. G. , Satake, E. , Mcdonnell, K. P. , Major, M. , Smiles, A. M. , & Krolewski, A. S. (2015). Circulating TGF‐β1‐regulated miRNAs and the risk of rapid progression to ESRD in type 1 diabetes. Diabetes, 64(9), 3285–3293. 10.2337/db15-0116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillar, N. , Polsky, A. L. , & Shomron, N. (2019). Dual inhibition of ABCE1 and LCP1 by microRNA‐96 results in an additive effect in breast cancer mouse model. Oncotarget, 10(21), 2086–2094. 10.18632/oncotarget.26747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plešingerová, H. , Janovská, P. , Mishra, A. , Smyčková, L. , Poppová, L. , Libra, A. , Plevová, K. , Ovesná, P. , Radová, L. , Doubek, M. , Pavlová, Š. , Pospíšilová, Š. , & Bryja, V. (2018). Expression of COBLL1 encoding novel ROR1 binding partner is robust predictor of survival in chronic lymphocytic leukemia. Haematologica, 103(2), 313–324. 10.3324/haematol.2017.178699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhka, M. , Nordberg, M ‐ E. , Valkonen, S. , Rannikko, A. , Kallioniemi, O. , Siljander, P. , & Af Hällström, T. M. (2017). KeepEX, a simple dilution protocol for improving extracellular vesicle yields from urine. European Journal of Pharmaceutical Sciences, 98, 30–39. 10.1016/j.ejps.2016.10.021 [DOI] [PubMed] [Google Scholar]
- Ramnath, R. , Foster, R. R. , Qiu, Y. , Cope, G. , Butler, M. J. , Salmon, A. H. , Mathieson, P. W. , Coward, R. J. , Welsh, G. I. , & Satchell, S. C. (2014). Matrix metalloproteinase 9‐mediated shedding of syndecan 4 in response to tumor necrosis factor α: A contributor to endothelial cell glycocalyx dysfunction. Faseb Journal, 28(11), 4686–4699. 10.1096/fj.14-252221 [DOI] [PubMed] [Google Scholar]
- Ramnath, R. D. , & Satchell, S. C. (2020). Glomerular endothelial cells: Assessment of barrier properties in vitro. Methods in Molecular Biology, 2067, 145–151. 10.1007/978-1-4939-9841-8_11 [DOI] [PubMed] [Google Scholar]
- Riccio, E. , Sabbatini, M. , Capuano, I. , & Pisani, A. (2019). Early biomarkers of fabry nephropathy: A review of the literature. Nephron, 143(4), 274–281. 10.1159/000502907 [DOI] [PubMed] [Google Scholar]
- Robinson, M. D. , & Oshlack, A. (2010). A scaling normalization method for differential expression analysis of RNA‐seq data. Genome biology, 11(3), R25. 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo, G. R. , & Kazlauskas, A. (2008). Oxysterol and diabetes activate STAT3 and control endothelial expression of profilin‐1 via OSBP1. Journal of Biological Chemistry, 283(15), 9595–9605. 10.1074/jbc.M710092200 [DOI] [PubMed] [Google Scholar]
- Royston, J. P. (1982). An extension of Shapiro and Wilk's W test for normality to large samples. Journal of the Royal Statistical Society. Series C (Applied Statistics), 31(2), 115–124. 10.2307/2347973 [DOI] [Google Scholar]
- Rudnicki, M. , Perco, P. , D′Haene, B. , Leierer, J. , Heinzel, A. , Mühlberger, I. , Schweibert, N. , Sunzenauer, J. , Regele, H. , Kronbichler, A. , Mestdagh, P. , Vandesompele, J. , Mayer, B. , & Mayer, G. (2016). Renal microRNA‐ and RNA‐profiles in progressive chronic kidney disease. European Journal of Clinical Investigation, 46(3), 213–226. 10.1111/eci.12585 [DOI] [PubMed] [Google Scholar]
- Saleem, M. A. , O'hare, M. J. , Reiser, J. , Coward, R. J. , Inward, C. D. , Farren, T. , Xing, C. Y. , Ni, L. , Mathieson, P. W. , & Mundel, P. (2002). A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. Journal of the American Society of Nephrology, 13(3), 630–638. 10.1681/asn.V133630 [DOI] [PubMed] [Google Scholar]
- Sanchez‐Niño, M. D. , Fernandez‐Fernandez, B. , Perez‐Gomez, M. V. , Poveda, J. , Sanz, A. B. , Cannata‐Ortiz, P. , Ruiz‐Ortega, M. , Egido, J. , Selgas, R. , & Ortiz, A. (2015). Albumin‐induced apoptosis of tubular cells is modulated by BASP1. Cell Death & Disease, 6(2), e1644. 10.1038/cddis.2015.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez‐Niño, M. D. , Sanz, A. B. , Lorz, C. , Gnirke, A. , Rastaldi, M. P. , Nair, V. , Egido, J. , Ruiz‐Ortega, M. , Kretzler, M. , & Ortiz, A. (2010). BASP1 promotes apoptosis in diabetic nephropathy. Journal of the American Society of Nephrology, 21(4), 610–621. 10.1681/asn.2009020227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamarea, Í. , Elvarez‐Hernendez, D. , Jofre, R. , Polo, J. R. , Menerguez, J. , & Cannata‐Andea, J. B. (2005). Progression of secondary hyperparathyroidism involves deregulation of genes related to DNA and RNA stability. Kidney International, 67(6), 2267–2279. 10.1111/j.1523-1755.2005.00330.x [DOI] [PubMed] [Google Scholar]
- Sarlos, D. P. , Peterfi, L. , Szanto, A. , & Kovacs, G. (2018). Shift of keratin expression profile in end‐stage kidney increases the risk of tumor development. Anticancer Research, 38(9), 5217–5222. 10.21873/anticanres.12845 [DOI] [PubMed] [Google Scholar]
- Sarrab, R. M. , Lennon, R. , Ni, L. , Wherlock, M. D. , Welsh, G. I. , & Saleem, M. A. (2011). Establishment of conditionally immortalized human glomerular mesangial cells in culture, with unique migratory properties. American Journal of Physiology. Renal Physiology, 301(5), F1131–F1138. 10.1152/ajprenal.00589.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchell, S. C. , Tasman, C. H. , Singh, A. , Ni, L. , Geelen, J. , Von Ruhland, C. J. , O'hare, M. J. , Saleem, M. A. , Van Den Heuvel, L. P. , & Mathieson, P. W. (2006). Conditionally immortalized human glomerular endothelial cells expressing fenestrations in response to VEGF. Kidney International, 69(9), 1633–1640. 10.1038/sj.ki.5000277 [DOI] [PubMed] [Google Scholar]
- Schiffer, D. , Mellai, M. , Boldorini, R. , Bisogno, I. , Grifoni, S. , Corona, C. , Bertero, L. , Cassoni, P. , Casalone, C. , & Annovazzi, L. (2018). The significance of chondroitin sulfate proteoglycan 4 (CSPG4) in human gliomas. International Journal of Molecular Sciences, 19(9), 2724. 10.3390/ijms19092724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi, S. , Fervenza, F. C. , Zhang, Y. , Zand, L. , Vrana, J. A. , Nasr, S. H. , Theis, J. D. , Dogan, A. , & Smith, R. J. H. (2012). C3 glomerulonephritis: Clinicopathological findings, complement abnormalities, glomerular proteomic profile, treatment, and follow‐up. Kidney International, 82(4), 465–473. 10.1038/ki.2012.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, A. , Xia, L. , Masson, E. A. Y. , Gui, C. , Momen, A. , Shikatani, E. A. , Husain, M. , Quaggin, S. , John, R. , & Fantus, I. G. (2015). Thioredoxin‐interacting protein deficiency protects against diabetic nephropathy. Journal of the American Society of Nephrology, 26(12), 2963–2977. 10.1681/asn.2014050528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou, Y. , Yang, L. , Yang, Y. , Zhu, X. , Li, F. , & Xu, J. (2020). Identification of signatures of prognosis prediction for melanoma using a hypoxia score. Frontiers in Genetics, 11, 570530. 10.3389/fgene.2020.570530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideek, M. , Smith, J. , Menz, C. , Adams, J. , Cowin, A. , & Gibson, M. (2017). A central bioactive region of LTBP‐2 stimulates the expression of TGF‐β1 in fibroblasts via Akt and p38 signalling pathways. International Journal of Molecular Sciences, 18(10), 2114. 10.3390/ijms18102114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöberg, B. , Qureshi, A. R. , Heimbürger, O. , Stenvinkel, P. , Lind, L. , Larsson, A. , Bárány, P. , & Ärnlöv, J. (2016). Association between levels of pentraxin 3 and incidence of chronic kidney disease in the elderly. Journal of Internal Medicine, 279(2), 173–179. 10.1111/joim.12411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R. N. , Aleksic, J. , Butano, D. , Carr, A. , Contrino, S. , Hu, F. , Lyne, M. , Lyne, R. , Kalderimis, A. , Rutherford, K. , Stepan, R. , Sullivan, J. , Wakeling, M. , Watkins, X. , & Micklem, G. (2012). InterMine: A flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics, 28(23), 3163–3165. 10.1093/bioinformatics/bts577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Ba, Y. , Wang Ma, F. , Wei Ma, Y. , Chen Ba, D. , & Deng Ba, G. (2021). ATP5A1 participates in transcriptional and posttranscriptional regulation of cancer‐associated genes by modulating their expression and alternative splicing profiles in HeLa cells. Technology in Cancer Research & Treatment, 20, 153303382110391. 10.1177/15330338211039126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda, H. , Lee, B. R. , Park, K.‐H. , Nihalani, D. , Yoon, J.‐H. , Ikeda, M. , & Kwon, S.‐H. (2019). miRNA profiling of urinary exosomes to assess the progression of acute kidney injury. Scientific Reports, 9(1), 4692. 10.1038/s41598-019-40747-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stępień, E. Ł. , Durak‐Kozica, M. , Kamińska, A. , Targosz‐Korecka, M. , Libera, M. , Tylko, G. , Opalińska, A. , Kapusta, M. , Solnica, B. , Georgescu, A. , Costa, M. C. , Czyżewska‐Buczyńska, A. , Witkiewicz, W. , Małecki, M. T. , & Enguita, F. J. (2018). Circulating ectosomes: Determination of angiogenic microRNAs in type 2 diabetes. Theranostics, 8(14), 3874–3890. 10.7150/thno.23334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stättermayer, A. F. , Rutter, K. , Beinhardt, S. , Wrba, F. , Scherzer, T.‐M. , Strasser, M. , Hofer, H. , Steindl‐Munda, P. , Trauner, M. , & Ferenci, P. (2014). Role of FDFT1 polymorphism for fibrosis progression in patients with chronic hepatitis C. Liver International, 34(3), 388–395. 10.1111/liv.12269 [DOI] [PubMed] [Google Scholar]
- Su Kim, D. , Choi, Y. D. , Moon, M. , Kang, S. , Lim, J.‐B. , Kim, K. M. , Park, K. M. , & Cho, N. H. (2013). Composite three‐marker assay for early detection of kidney cancer. Cancer Epidemiology and Prevention Biomarkers, 22(3), 390–398. 10.1158/1055-9965.Epi-12-1156 [DOI] [PubMed] [Google Scholar]
- Takayama, K.‐I. , Suzuki, T. , Fujimura, T. , Takahashi, S. , & Inoue, S. (2018). COBLL1 modulates cell morphology and facilitates androgen receptor genomic binding in advanced prostate cancer. Proceedings of the National Academy of Sciences of the United States of America, 115(19), 4975–4980. 10.1073/pnas.1721957115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, S. , Nakano, S.‐I. , Nakamura, K. , Ozoe, A. , Chien, P. , Yoshihara, H. , Hakuno, F. , Matsuwaki, T. , Saeki, Y. , Takahashi, S.‐I. , Yamanouchi, K. , & Nishihara, M. (2016). Roles of chondroitin sulfate proteoglycan 4 in fibrogenic/adipogenic differentiation in skeletal muscle tissues. Experimental Cell Research, 347(2), 367–377. 10.1016/j.yexcr.2016.08.023 [DOI] [PubMed] [Google Scholar]
- Tan, S. , Li, H. , Zhang, W. , Shao, Y. , Liu, Y. , Guan, H. , Wu, J. , Kang, Y. , Zhao, J. , Yu, Q. , Gu, Y. , Ding, K. , Zhang, M. , Qian, W. , Zhu, Y. , Cai, H. , Chen, C. , Lobie, P. E. , Zhao, X. , … Zhu, T. (2018). NUDT21 negatively regulates PSMB2 and CXXC5 by alternative polyadenylation and contributes to hepatocellular carcinoma suppression. Oncogene, 37(35), 4887–4900. 10.1038/s41388-018-0280-6 [DOI] [PubMed] [Google Scholar]
- Tanaka, H. , Kanda, M. , Miwa, T. , Umeda, S. , Sawaki, K. , Tanaka, C. , Kobayashi, D. , Hayashi, M. , Yamada, S. , Nakayama, G. , Koike, M. , & Kodera, Y. (2021). G‐protein subunit gamma‐4 expression has potential for detection, prediction and therapeutic targeting in liver metastasis of gastric cancer. British Journal of Cancer, 125(2), 220–228. 10.1038/s41416-021-01366-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, B. , Xu, Q. , Xuan, L. , Wang, H. , Zhang, H. , Wang, X. , & Kang, P. (2019). Circ 0001434 RNA reduces inflammation in acute lung injury model through Wnt/β‐catenin and NF‐κB by miR‐625‐5p. International Journal of Clinical and Experimental Pathology, 12(9), 3290–3300. [PMC free article] [PubMed] [Google Scholar]
- Tang, S. , Wang, X. , Deng, T. , Ge, H. , & Xiao, X. (2020). Identification of C3 as a therapeutic target for diabetic nephropathy by bioinformatics analysis. Scientific Reports, 10(1), 13468. 10.1038/s41598-020-70540-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, W. , Hong, L. , Dai, W. , Li, J. , Zhu, H. , Lin, J. , Yang, Q. , Wang, Y. , Lin, Z. , Liu, M. , Xiao, Y. , Zhang, Y. , Wu, X. , Wang, J. , Chen, Y. , Hu, H. , Liu, S. , Wang, J. , & Xiang, L. (2020). MicroRNA‑500a‑5p inhibits colorectal cancer cell invasion and epithelial‑mesenchymal transition. International Journal of Oncology, 56(6), 1499–1508. 10.3892/ijo.2020.5015 [DOI] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J.‐M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7(1), 1535750. 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Z. , Yao, N. , Wang, F. , & Ruan, L. (2022). Thymosin β4 suppresses LPS‐induced murine lung fibrosis by attenuating oxidative injury and alleviating inflammation. Inflammation, 45(1), 59–73. 10.1007/s10753-021-01528-6 [DOI] [PubMed] [Google Scholar]
- Tomar, V. S. , Baral, T. K. , Nagavelu, K. , & Somasundaram, K. (2019). Serine/threonine/tyrosine‐interacting‐like protein 1 (STYXL1), a pseudo phosphatase, promotes oncogenesis in glioma. Biochemical and Biophysical Research Communications, 515(1), 241–247. 10.1016/j.bbrc.2019.05.093 [DOI] [PubMed] [Google Scholar]
- Toyama, K. , Igase, M. , Spin, J. M. , Abe, Y. , Javkhlant, A. , Okada, Y. , Wagenhäuser, M. U. , Schelzig, H. , Tsao, P. S. , & Mogi, M. (2021). Exosome miR‐501‐3p elevation contributes to progression of vascular stiffness. Circulation Reports, 3(3), 170–177. 10.1253/circrep.CR-20-0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uil, M. , Hau, C. M. , Ahdi, M. , Mills, J. D. , Kers, J. , Saleem, M. A. , Florquin, S. , Gerdes, V. E. A. , Nieuwland, R. , & Roelofs, J. J. T. H. (2021). Cellular origin and microRNA profiles of circulating extracellular vesicles in different stages of diabetic nephropathy. Clinical Kidney Journal, 14(1), 358–365. 10.1093/ckj/sfz145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProtConsortium . (2021). UniProt: The universal protein knowledgebase in 2021. Nucleic Acid Research, 49(D1), D480–D489. 10.1093/nar/gkaa1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van, J. A. D. , Scholey, J. W. , & Konvalinka, A. (2017). Insights into diabetic kidney disease using urinary proteomics and bioinformatics. Journal of the American Society of Nephrology, 28(4), 1050–1061. 10.1681/asn.2016091018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B. , Koh, P. , Winbanks, C. , Coughlan, M. T. , Mcclelland, A. , Watson, A. , Jandeleit‐Dahm, K. , Burns, W. C. , Thomas, M. C. , Cooper, M. E. , & Kantharidis, P. (2011). miR‐200a prevents renal fibrogenesis through repression of TGF‐β2 expression. Diabetes, 60(1), 280–287. 10.2337/db10-0892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B. , Komers, R. , Carew, R. , Winbanks, C. E. , Xu, B. , Herman‐Edelstein, M. , Koh, P. , Thomas, M. , Jandeleit‐Dahm, K. , Gregorevic, P. , Cooper, M. E. , & Kantharidis, P. (2012). Suppression of microRNA‐29 expression by TGF‐β1 promotes collagen expression and renal fibrosis. Journal of the American Society of Nephrology, 23(2), 252–265. 10.1681/asn.2011010055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Wang, J. , Cui, W. , Liu, Y. , Zhou, H. , Wang, Y. , Chen, X. , Chen, X. , & Wang, Z. (2021). Serum exosomal miRNA‐1226 as potential biomarker of pancreatic ductal adenocarcinoma. OncoTargets and Therapy, 14, 1441–1451. 10.2147/ott.S296816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Zhu, X. , Qin, X. , Jiang, H. , Gao, Y. , & Gao, J. (2020). [miR‐324‐5p inhibits lipopolysaccharide‐induced proliferation of rat glomerular mesangial cells by regulating the Syk/Ras/c‐fos pathway]. Nan Fang Yi Ke Da Xue Xue Bao, 40(11), 1571–1578. 10.12122/j.issn.1673-4254.2020.11.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L.‐P. , Gao, Y.‐Z. , Song, B. , Yu, G. , Chen, H. , Zhang, Z.‐W. , Yan, C.‐F. , Pan, Y.‐L. , & Yu, X.‐Y. (2019). MicroRNAs in the progress of diabetic nephropathy: A systematic review and meta‐analysis. Evid Based Complement Alternat Med, 2019, 1. 10.1155/2019/3513179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Cai, Y. , Fu, X. , & Chen, L. (2021). High RPS27A expression predicts poor prognosis in patients with HPV type 16 cervical cancer. Frontiers in Oncology, 11, 752974. 10.3389/fonc.2021.752974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. , Liu, Q. , Wang, Y. , Piao, H. , Zhu, Z. , Li, D. , Wang, T. , & Liu, K. (2021). LINC01278 sponges miR‐500b‐5p to regulate the expression of ACTG2 to control phenotypic switching in human vascular smooth muscle cells during aortic dissection. Journal of the American Heart Association, 10(9), e018062. 10.1161/jaha.120.018062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Bi, X. , Huang, X. , Wang, B. , Guo, Q. , & Wu, Z. (2020). Systematic investigation of biomarker‐like role of ARHGDIB in breast cancer. Cancer Biomark, 28(1), 101–110. 10.3233/cbm-190562 [DOI] [PubMed] [Google Scholar]
- Wang, X. , Jiang, F. , Song, H. , Li, X. , Xian, J. , & Gu, X. (2016). MicroRNA‐200a‐3p suppresses tumor proliferation and induces apoptosis by targeting SPAG9 in renal cell carcinoma. Biochemical and Biophysical Research Communications, 470(3), 620–626. 10.1016/j.bbrc.2016.01.095 [DOI] [PubMed] [Google Scholar]
- Wang, X. , Liu, J. , Yin, W. , Abdi, F. , Pang, P. D. , Fucci, Q.‐A. , Abbott, M. , Chang, S. L. , Steele, G. , Patel, A. , Mori, Y. , Zhang, A. , Zhu, S. , Lu, T.‐S. , Kibel, A. S. , Wang, B. , Lim, K. , & Siedlecki, A. M. (2020). miR‐218 expressed in endothelial progenitor cells contributes to the development and repair of the kidney microvasculature. American Journal of Pathology, 190(3), 642–659. 10.1016/j.ajpath.2019.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wazir, U. , Sanders, A. J. , Wazir, A. , Baig, R. M. , Jiang, W. G. , Ster, I. C. , Sharma, A. K. , & Mokbel, K. (2015). Effect of the knockdown of death‐associated protein 1 expression on cell adhesion, growth and migration in breast cancer cells. Oncology Reports, 33(3), 1450–1458. 10.3892/or.2014.3686 [DOI] [PubMed] [Google Scholar]
- Wei, Y. , Gao, X. , Li, A. , Liang, M. , & Jiang, Z. (2021). Single‐nucleus transcriptomic analysis reveals important cell cross‐talk in diabetic kidney disease. Frontiers of Medicine (Lausanne), 8, 657956. 10.3389/fmed.2021.657956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, B. L. (1951). On the comparison of several mean values: An alternative approach. Biometrika, 38(3‐4), 330–336. 10.1093/biomet/38.3-4.330 [DOI] [Google Scholar]
- Weng, M.‐L. , Chen, W.‐K. , Chen, X.‐Y. , Lu, H. , Sun, Z.‐R. , Yu, Q. , Sun, P.‐F. , Xu, Y.‐J. , Zhu, M.‐M. , Jiang, N. , Zhang, J. , Zhang, J.‐P. , Song, Y.‐L. , Ma, D. , Zhang, X.‐P. , & Miao, C.‐H. (2020). Fasting inhibits aerobic glycolysis and proliferation in colorectal cancer via the Fdft1‐mediated AKT/mTOR/HIF1α pathway suppression. Nature Communications, 11(1), 1869. 10.1038/s41467-020-15795-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, L. , Gilbert, T. , Kinna, G. , Ruta, L.‐A. , Pennisi, D. , Kett, M. , & Little, M. H. (2007). Crim1KST264/KST264 mice implicate Crim1 in the regulation of vascular endothelial growth factor‐A activity during glomerular vascular development. Journal of the American Society of Nephrology, 18(6), 1697–1708. 10.1681/asn.2006091012 [DOI] [PubMed] [Google Scholar]
- Wilkinson, L. , Gilbert, T. , Sipos, A. , Toma, I. , Pennisi, D. J. , Peti‐Peterdi, J. , & Little, M. H. (2009). Loss of renal microvascular integrity in postnatal Crim1 hypomorphic transgenic mice. Kidney International, 76(11), 1161–1171. 10.1038/ki.2009.345 [DOI] [PubMed] [Google Scholar]
- Wilmer, M. J. , Saleem, M. A. , Masereeuw, R. , Ni, L. , Van Der Velden, T. J. , Russel, F. G. , Mathieson, P. W. , Monnens, L. A. , Van Den Heuvel, L. P. , & Levtchenko, E. N. (2010). Novel conditionally immortalized human proximal tubule cell line expressing functional influx and efflux transporters. Cell and Tissue Research, 339(2), 449–457. 10.1007/s00441-009-0882-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, R. D. , Dimou, P. , Northey, S. J. , & Beresford, M. W. (2019). Mesangial cells are key contributors to the fibrotic damage seen in the lupus nephritis glomerulus. Journal of Inflammation (London), 16, 22. 10.1186/s12950-019-0227-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H. , Malone, A. F. , Donnelly, E. L. , Kirita, Y. , Uchimura, K. , Ramakrishnan, S. M. , Gaut, J. P. , & Humphreys, B. D. (2018). Single‐cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. Journal of the American Society of Nephrology, 29(8), 2069–2080. 10.1681/asn.2018020125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. Z. , Jiang, N. , Lin, J. M. , & Liu, X. (2020). STYXL1 promotes malignant progression of hepatocellular carcinoma via downregulating CELF2 through the PI3K/Akt pathway. European Review for Medical and Pharmacological Sciences, 24(6), 2977–2985. 10.26355/eurrev_202003_20662 [DOI] [PubMed] [Google Scholar]
- Wu, M. , Lou, W. , Lou, M. , Fu, P. , & Yu, X.‐F. (2020). Integrated analysis of distant metastasis‐associated genes and potential drugs in colon adenocarcinoma. Frontiers in Oncology, 10, 576615. 10.3389/fonc.2020.576615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X. , Liu, T. , Fang, O. , Dong, W. , Zhang, F. , Leach, L. , Hu, X. , & Luo, Z. (2016). MicroRNA‐708‐5p acts as a therapeutic agent against metastatic lung cancer. Oncotarget, 7(3), 2417–2432. 10.18632/oncotarget.6594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y. , Yang, N. , Zhang, Q. , Wang, Y. , Yang, S. , & Liu, Z. (2014). Pentraxin 3 inhibits acute renal injury‐induced interstitial fibrosis through suppression of IL‐6/Stat3 pathway. Inflammation, 37(5), 1895–1901. 10.1007/s10753-014-9921-2 [DOI] [PubMed] [Google Scholar]
- Xie, Y. , Jia, Y. , Cuihua, X. , Hu, F. , Xue, M. , & Xue, Y. (2017). Urinary exosomal microRNA profiling in incipient type 2 diabetic kidney disease. Journal of Diabetes Research, 2017, 1. 10.1155/2017/6978984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, X. , Liu, J. , He, Q. , Dai, R. , Zhang, H. , Cao, Z. , Liao, Y. , Liu, B. , Zhou, Y. , Chen, J. , Liu, J. , & Chen, M. (2021). Long non‐coding RNA NORAD aggravates acute myocardial infarction by promoting fibrosis and apoptosis via miR‐577/COBLL1 axis. Environmental Toxicology, 36(11), 2256–2265. 10.1002/tox.23339 [DOI] [PubMed] [Google Scholar]
- Xu, K. , Liu, Q. , Wu, K. , Liu, L. , Zhao, M. , Yang, H. , Wang, X. , & Wang, W. (2020). Extracellular vesicles as potential biomarkers and therapeutic approaches in autoimmune diseases. Journal of Translational Medicine, 18(1), 432. 10.1186/s12967-020-02609-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. , Barreiro, K. , Musante, L. , Kretz, O. , Lin, H. , Zou, H. , Huber, T. B. , & Holthofer, H. (2019). Management of Tamm‐Horsfall protein for reliable urinary analytics. Proteomics ‐ Clinical Applications, 13(6), 1900018. 10.1002/prca.201900018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, T. , Kato, T. , Hatano, K. , Kawashima, A. , Ujike, T. , Fukuhara, S. , Kiuchi, H. , Imamura, R. , Ibuki, N. , Kiyotani, K. , Kurashige, M. , Morii, E. , Fujita, K. , Nonomura, N. , & Uemura, M. (2021). Genomic and pathological characterization of multiple renal cell carcinoma regions in patient with tuberous sclerosis complex: A case report. Frontiers in Oncology, 11, 691996. 10.3389/fonc.2021.691996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano, K. , & Youle, R. J. (2020). Two different axes CALCOCO2‐RB1CC1 and OPTN‐ATG9A initiate PRKN‐mediated mitophagy. Autophagy, 16(11), 2105–2107. 10.1080/15548627.2020.1815457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, J. , Goerne, T. , Zelmer, A. , Guzman, R. , Kapfhammer, J. P. , Wellmann, S. , & Zhu, X. (2019). The RNA‐binding protein RBM3 promotes Neural Stem Cell (NSC) proliferation under hypoxia. Frontiers in Cell and Developmental Biology, 7, 288. 10.3389/fcell.2019.00288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez‐Mó, M. , Siljander, P. R.‐M. , Andreu, Z. , Bedina Zavec, A. , Borràs, F. E. , Buzas, E. I. , Buzas, K. , Casal, E. , Cappello, F. , Carvalho, J. , Colás, E. , Cordeiro‐Da Silva, A. , Fais, S. , Falcon‐Perez, J. M. , Ghobrial, I. M. , Giebel, B. , Gimona, M. , Graner, M. , Gursel, I. , … De Wever, O. (2015). Biological properties of extracellular vesicles and their physiological functions. Journal of Extracellular Vesicles, 4, 27066. 10.3402/jev.v4.27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Zhang, Y. , Ma, Y. , Zhao, Q. , & Lyu, J. (2015). Effect of mitochondrial DNA 5178 C/A polymorphism on risks for type 2 diabetes mellitus and its complications. Zhonghua Yi Xue Yi Chuan Xue Za Zhi = Zhonghua Yixue Yichuanxue Zazhi = Chinese Journal of Medical Genetics, 32(6), 855–860. 10.3760/cma.j.issn.1003-9406.2015.06.023 [DOI] [PubMed] [Google Scholar]
- Yang, Y. , & Xu, X. (2021). Identification of key genes in coronary artery disease: An integrative approach based on weighted gene co‐expression network analysis and their correlation with immune infiltration. Aging (Albany NY), 13(6), 8306–8319. 10.18632/aging.202638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Q. , Peng, Y. , Huang, F. , Chen, J. , Xu, Y. , Li, Y. , Liu, S. , & Huang, L. (2021). γ‐synuclein is closely involved in autophagy that protects colon cancer cell from endoplasmic reticulum stress. Anti‐cancer agents in medicinal chemistry, 21(17), 2385–2396. 10.2174/1871520621666210119093327 [DOI] [PubMed] [Google Scholar]
- Yi, H.‐X. , Jiang, S.‐Y. , Yu, L.‐H. , Chen, K. , Yang, Z.‐X. , & Wu, Q. (2021). MicroRNA 181a‐2‐3p alleviates the apoptosis of renal tubular epithelial cells via targeting GJB2 in sepsis‐induced acute kidney injury. Molecular and Cellular Biology, 41(7), e0001621. 10.1128/mcb.00016-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, L.‐C. , Xiao, G. , Zhou, R. , Huang, X.‐P. , Li, N.‐L. , Tan, C.‐L. , Xie, F.‐J. , Weng, J. , & Liu, L.‐X. (2020). MicroRNA‐361‐5p inhibits tumorigenesis and the EMT of HCC by targeting twist1. BioMed Research International, 2020, 1. 10.1155/2020/8891876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, Y. , Nameta, M. , Kuwano, M. , Zhang, Y. , Bo, X. , Magdeldin, S. , Cui, Z. , Fujinaka, H. , Yaoita, E. , Tomonaga, T. , & Yamamoto, T. (2012). Proteomic approach to human kidney glomerulus prepared by laser microdissection from frozen biopsy specimens: Exploration of proteome after removal of blood‐derived proteins. Proteomics ‐ Clinical Applications, 6(7‐8), 412–417. 10.1002/prca.201200016 [DOI] [PubMed] [Google Scholar]
- Yu, W. , Zeng, H. , Chen, J. , Fu, S. , Huang, Q. , Xu, Y. , Xu, A. , Lan, H.‐Y. , & Tang, Y. (2020). miR‐20a‐5p is enriched in hypoxia‐derived tubular exosomes and protects against acute tubular injury. Clinical Science (London, England: 1979), 134(16), 2223–2234. 10.1042/cs20200288 [DOI] [PubMed] [Google Scholar]
- Yu, Y.‐H. , Zhang, Y.‐H. , Ding, Y.‐Q. , Bi, X.‐Y. , Yuan, J. , Zhou, H. , Wang, P.‐X. , Zhang, L.‐L. , & Ye, J.‐T. (2021). MicroRNA‐99b‐3p promotes angiotensin II‐induced cardiac fibrosis in mice by targeting GSK‐3β. Acta Pharmacologica Sinica, 42(5), 715–725. 10.1038/s41401-020-0498-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, L. , Chen, L. , Qian, K. , Wang, G. , Lu, M. , Qian, G. , Cao, X. , Jiang, W. , Xiao, Y. , & Wang, X. (2018). A novel correlation between ATP5A1 gene expression and progression of human clear cell renal cell carcinoma identified by co‑expression analysis. Oncology Reports, 39(2), 525–536. 10.3892/or.2017.6132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Q. , Xu, T. , Chen, Y. , Qu, W. , Sun, D. , Liu, X. , & Sun, L. (2020). MiR‐185‐5p ameliorates endoplasmic reticulum stress and renal fibrosis by downregulation of ATF6. Laboratory Investigation, 100(11), 1436–1446. 10.1038/s41374-020-0447-y [DOI] [PubMed] [Google Scholar]
- Yuana, Y. , Sturk, A. , & Nieuwland, R. (2013). Extracellular vesicles in physiological and pathological conditions. Blood Reviews, 27(1), 31–39. 10.1016/j.blre.2012.12.002 [DOI] [PubMed] [Google Scholar]
- Yufeng, Z. , & Ming, Q. (2020). Expression and prognostic roles of PABPC1 in hepatocellular carcinoma. International Journal of Surgery, 84, 3–12. 10.1016/j.ijsu.2020.10.004 [DOI] [PubMed] [Google Scholar]
- Zang, J. , Maxwell, A. P. , Simpson, D. A. , & Mckay, G. J. (2019). Differential expression of urinary exosomal microRNAs miR‐21‐5p and miR‐30b‐5p in individuals with diabetic kidney disease. Scientific Reports, 9(1), 10900. 10.1038/s41598-019-47504-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervou, S. , Whittington, H. J. , Ostrowski, P. J. , Cao, F. , Tyler, J. , Lake, H. A. , Neubauer, S. , & Lygate, C. A. (2017). Increasing creatine kinase activity protects against hypoxia/reoxygenation injury but not against anthracycline toxicity in vitro. PLoS ONE, 12(8), e0182994. 10.1371/journal.pone.0182994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, H. , Shi, Y. , Chen, X. , Wang, J. , Lu, Y. , Zhang, F. , Liu, Z. , Lei, T. , & Fan, D. (2017). CacyBP/SIP promotes the proliferation of colon cancer cells. PLoS ONE, 12(2), e0169959. 10.1371/journal.pone.0169959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, W. , Ma, J. , Zhu, R. , Xu, C. , Zhang, J. , Chen, Y. , Chen, Z. , Gong, D. , Zheng, J. , Chen, C. , Li, S. , Li, B. , Huang, Y. , Xue, W. , & Zheng, J. (2018). MiR‐532‐5p suppresses renal cancer cell proliferation by disrupting the ETS1‐mediated positive feedback loop with the KRAS‐NAP1L1/P‐ERK axis. British Journal of Cancer, 119(5), 591–604. 10.1038/s41416-018-0196-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. Q. , Yang, H. Q. , Yang, S. Q. , Wang, Y. , Chen, X. J. , Lu, H. S. , & Zhao, L. P. (2019). CNDP2 acts as an activator for human ovarian cancer growth and metastasis via the PI3K/AKT pathway. Technology in Cancer Research & Treatment, 18, 153303381987477. 10.1177/1533033819874773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Chen, C. , Liu, Z. , Guo, H. , Lu, W. , Hu, W. , & Lin, Z. (2022). PABPC1‐induced stabilization of IFI27 mRNA promotes angiogenesis and malignant progression in esophageal squamous cell carcinoma through exosomal miRNA‐21‐5p. Journal of Experimental & Clinical Cancer Research, 41(1), 111. 10.1186/s13046-022-02339-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Xu, C. , Ye, Q. , Tong, L. , Jiang, H. , Zhu, X. , Huang, L. , Lin, W. , Fu, H. , Wang, J. , Persson, P. B. , Lai, E. Y. , & Mao, J. (2021). Podocyte apoptosis in diabetic nephropathy by BASP1 activation of the p53 pathway via WT1. Acta Physiologica (Oxford), 232(1), e13634. 10.1111/apha.13634 [DOI] [PubMed] [Google Scholar]
- Zhao, L. , Liu, K. , Pan, X. , Quan, J. , Zhou, L. , Li, Z. , Lin, C. , Xu, J. , Xu, W. , Guan, X. , Li, H. , Ni, L. , Gui, Y. , & Lai, Y. (2019). miR‐625‐3p promotes migration and invasion and reduces apoptosis of clear cell renal cell carcinoma. American journal of translational research, 11(10), 6475–6486. [PMC free article] [PubMed] [Google Scholar]
- Zhu, H. , Zhang, H. , Pei, Y. , Liao, Z. , Liu, F. , Su, C. , Liu, Y. , Dong, R. , Song, J. , Zhang, X. , Fan, Y. , Liang, H. , Zhang, B. , & Chen, X. (2021). Long non‐coding RNA CCDC183‐AS1 acts AS a miR‐589‐5p sponge to promote the progression of hepatocellular carcinoma through regulating SKP1 expression. Journal of Experimental & Clinical Cancer Research, 40(1), 57. 10.1186/s13046-021-01861-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J. , Su, L.‐P. , Zhou, Y. , Ye, L. , Lee, K.‐O. , & Ma, J.‐H. (2015). Thymosin β4 attenuates early diabetic nephropathy in a mouse model of type 2 diabetes mellitus. American Journal of Therapeutics, 22(2), 141–146. 10.1097/MJT.0b013e3182785ecc [DOI] [PubMed] [Google Scholar]
- Zucca, F. A. , Vanna, R. , Cupaioli, F. A. , Bellei, C. , De Palma, A. , Di Silvestre, D. , Mauri, P. , Grassi, S. , Prinetti, A. , Casella, L. , Sulzer, D. , & Zecca, L. (2018). Neuromelanin organelles are specialized autolysosomes that accumulate undegraded proteins and lipids in aging human brain and are likely involved in Parkinson's disease. NPJ Parkinson's Disease, 4, 17. 10.1038/s41531-018-0050-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Data Availability Statement
RNA and miRNA sequencing raw data as well as TMT proteomics data are deposited on the NCBI BioProject PRJNA905899.


