Abstract
Tumor-associated macrophages (TAMs) are closely related to tumorigenesis and metastasis of multiple cancer types. The infiltration of TAMs is used for predicting the prognosis of cancers, including colorectal cancer (CRC). However, the density and prognostic significance of M1 and M2 TAM phenotypes in the intratumor versus the invasive front (IF) are largely unknown in CRC. In this study, CD68 was selected as a general marker of TAMs, CD11c, NOS2 and CXCL10 as markers for M1 phenotype and CD163, CD206, CD115 as markers for M2 phenotype. Firstly, immunohistochemistry staining and double-labeling immunofluorescence staining showed that M1 molecular markers (NOS2, CXCL10, CD11c) were lowly expressed at both IF and intratumor, while M2 molecular markers (CD163, CD206, CD115) were highly expressed mainly at IF. Moreover, we also demonstrated that three M1 molecular markers including NOS2, CXCL10 and CD11c were correlated to each other. Meanwhile, three M2 molecular markers including CD163, CD206, and CD115 were also correlated to each other. Patients with low expression of three M1 molecular markers (NOS2/CXCL10/CD11c) exhibited low overall survival (OS) rate, whereas patients with high expression of three M2 molecular markers (CD163/CD206/CD115) exhibited low OS rate. We also observed that the prognostic value of treble markers combination (NOS2/CXCL10/CD11c or CD163/CD206/CD115) was superior to that of single marker. Together, our results reveal the combination of treble TAMs markers (NOS2/CXCL10/CD11c or CD163/CD206/CD115) could better evaluate the prognosis of CRC patients, which might be used as a more comprehensive method for predicting the prognosis of CRC patients.
Keywords: Colorectal cancer, Tumor-associated macrophages, Heterogeneity, Tumor invasive front, Tumor microenvironment
Abbreviations: CRC, colorectal cancer; TME, the tumor microenvironment; TAMs, tumor-associated macrophages; IF, invasive front; LPS, Lipopolysaccharide; TGF-β, transforming growth factor-β; IFN-γ, interferon-γ
1. Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in the world and the second leading cause of cancer death worldwide [1]. Traditional methods do not allow precise prediction of prognosis for the patients after surgical removal of the primary tumor, there is an acute need for biomarkers capable of distinguishing patients with poor or good prognosis [2]. Recent studies suggest that the tumor microenvironment (TME) plays a pivotal role in CRC progression and is considered as a prognostic biomarker [3]. Tumor progression is driven not only by cancer cells themselves, but also by the mesenchymal and inflammatory cells in tumor environment (TME) [4]. Tumor-associated macrophages (TAMs) represent the main immune cells population in TME in most types of cancer. A lot of clinical and experimental evidence shows that TAMs are involved in the process of tumor occurrence, growth, invasion and metastasis [5]. Previous studies have pointed out that TAMs possess significant diagnostic and prognostic values in cancers including head-neck squamous cell carcinoma [6], breast cancer [7], pancreatic cancer [8], hepatocellular carcinoma [9], and CRC [10]. However, the distribution patterns of TAMs and TAM phenotypes between intratumor and tumor invasive front (IF) in CRC remain unexplored. It has been proposed that TAMs harbor either classically activated macrophages (M1 type) or alternatively activated macrophages (M2 type) in tumor tissues. In most tumors, TAMs are primarily M2 macrophages. Many studies showed a correlation between macrophage density and poor patient prognosis [11]. However, there is a controversy on correlation between macrophage density and CRC patient survival. The role of TAMs in CRC progression remains elusive [12]. These controversies may be the consequence of the insufficient markers and techniques used to identify TAMs. Therefore, we attempted to establish a more effective method for evaluating the prognostic values of M1 subtype and M2 subtype TAMs in CRC.
2. Materials and method
2.1. Clinical specimens
Formalin-fixed human CRC specimens were collected from the department of pathology, Nanfang hospital, southern medical university, China. No patients received any preoperative chemotherapy or radiation therapy before surgical resection. All these cases were clinically and histologically diagnosed as CRC. The study was approved by the Ethics Committee of Nanfang hospital, southern medical university, China. Before collecting these clinical materials for research purposes, we obtained informed consent from all patients.
2.2. Immunohistochemistry (IHC) staining
The sections were immersed in EDTA antigen recovery buffer and heated with microwaves. The slides were blocked with peroxidase blocker. According to the standard IHC protocol, the mouse anti-human CD68 (ZSGB Bio, China), rabbit anti-human NOS2 (LifeSpan, USA), rabbit anti-human CXCL10 (Proteintech, USA), rabbit anti-human CD11c (Proteintech, USA), rabbit anti-human CD163 (ZSGB Bio., China), mouse anti-human CD206 (Abcam, USA) and rabbit anti-human CD115 (Abcam, USA) and secondary antibodies (Abcam, USA) were used to perform IHC staining using the paraffin sections. IHC images of CD68, NOS2, CXCL10, CD11c, CD163, CD206 and CD115 staining were captured by light microscopy.
The numbers of CD68-, NOS2-, CXCL10-, CD11c-, CD163-, CD206-, and CD115-positive cells were counted from five independent high-power microscopic fields (400 × ) along the tumor IF [13,14]. The specimens were evaluated twice by two different researchers who were blind to the prognosis or clinicopathological variables.
2.3. Double-labeling immunofluorescence staining
The sections were dewaxed and hydrated to exposure in 3% H2O2 for 15 min to block endogenous peroxidase. After repairing by citric acid antigen, the sections were blocked with goat serum for 30 min and then incubated with the primary antibodies of CD68&CD163, CD68&CD206, CD68&CD115, CD68&NOS2, CD68&CXCL10, CD68&CD11c overnight at 4 °C. Then the fluorescent secondary antibody (Abcam, USA) was added to the slides and incubated at room temperature in the dark for 1 h. DAPI was applied to counter-stain the nuclei for 10 min in the dark. Finally, the slides were observed, and the images were taken under a confocal microscope.
2.4. Bioinformatics analysis
GSE17538 dataset from GEO database (https://www.ncbi.nlm.nih.gov/gds) was used for analyzing the correlation between the interested genes expression and overall survival (OS) of CRC patients [[15], [16], [17]]. The dataset was derived by using the Affymetrix Human Genome U133 Plus 2.0 Array. Expression values of the genes of interest were extracted from the expression profiling data, sorted from low to high, and divided into low and high expression groups. The threshold defining low or high expression is determined by consulting with the protein expressions obtained from IHC. Patient OS curves in these two groups were analyzed by using Kaplan-Meier survival analysis.
2.5. Statistical analysis
SPSS software for windows version 13.0 was used for statistical analyses. An unpaired two-tailed Student's t-test was used to compare two groups with normal distribution. Statistical comparison among multi-groups were analyzed with Kruskal-Wallis test. Spearman's correlation test was applied to analyze correlated expression levels of TAMs markers in CRC tissue. Kaplan-Meier survival curves were plotted and log-rank test was performed. Cox regression was used for univariate and multivariate analyses with HRs and 95% confidence intervals (CI). Only factors with P < 0.05 in the univariate analysis were included in the multivariate analysis. To obtain the best prognostic efficacy, The cut-off values of TAMs as prognostic biomarkers were determined bys using X-Tile Software (Yale University, version 3.6.1). All the data were presented as mean ± standard deviation (SD) of the mean unless specified otherwise. P < 0.05 in all cases were considered to be significant. *P < 0.05, **P < 0.01, ***P < 0.001.
3. Results
3.1. CD68+ TAMs or CD163+ TAMs accumulated at the IF
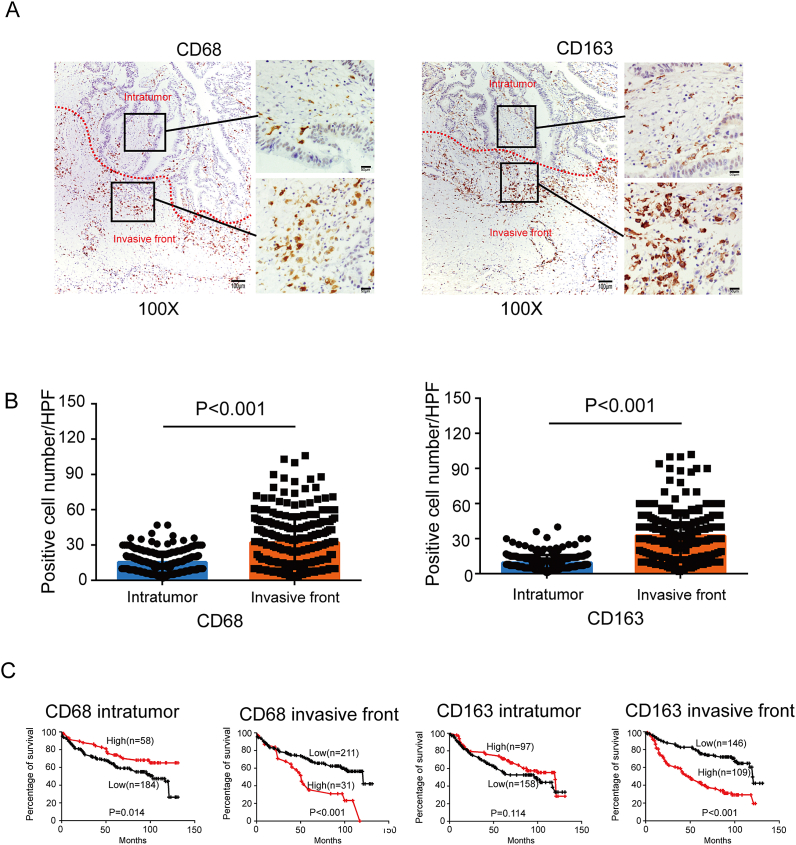
To study the distribution of TAMs at tumor site, we performed immunohistochemical staining of CD68 and CD163 in paraffin-embedded tissues. CD68 is a pan marker for macrophages and CD163 is usually considered as a M2 macrophage marker. In serial sections, CD68+ TAMs or CD163+ TAMs were found in the intratumor and IF, but often more prominent at the IF (Fig. 1A). 242 cases of CRC exhibited that there were averagely 32 CD68+ TAMs located at the IF. However, only 15 CD68+ TAMs were found in the intratumoral regions (Fig. 1B left). Among 255 samples, the number of CD163+ cells at the IF (32.71 ± 1.205) was about 3 times higher than that in intratumor (9.525 ± 0.3981) (Fig. 1B right). So CD68+ and CD163+ cells are predominantly found within the tumor IF.
Fig. 1.
The spatial density of CD68+ and CD163+ macrophages in CRC tissue. A, Paraffin-embedded CRC were stained with an anti-CD68 and anti-CD163 antibody. The micrographs at higher magnification show the stained IF and intratumor. B, Graphical illustration of statistical CD68+ and CD163+ macrophages distribution in intratumor and IF in CRC patients. C, OS curves for the density of CD68+ and CD163+ macrophages in intratumor and IF. For CD68+ TAM density in the intratumor, ≥22/HPF was defined as high and <22/HPF was defined as low. For CD68+ TAM density at the IF, ≥58/HPF was defined as high and <58/HPF was defined as low. For CD163+ TAM density in the intratumor, ≥10/HPF was defined as high and <10/HPF was defined as low. For CD163+ TAM density at the IF, ≥35/HPF was defined as high and <35/HPF was defined as low. Kaplan-Meier plots and curves were compared through Log-rank test.
3.2. Higher CD68+ TAM density or CD163+ TAM density at the IF is associated with decreased OS
For CD68+ TAM density in intratumor, ≥22/HPF was defined as high and <22/HPF was defined as low. For CD68+ TAM density at the IF, ≥58/HPF was defined as high and <58/HPF was defined as low. For CD163+ TAM density in intratumor, ≥10/HPF was defined as high and <10/HPF was defined as low. For CD163+ TAM density at the IF, ≥35/HPF was defined as high and <35/HPF was defined as low. Details are shown in Supplementary Figure 1-4. Patients with higher CD68+ TAM density in the intratumor showed better OS (P = 0.014), whereas patients with more CD68+ cells at the IF had poorer OS (P < 0.001) (Fig. 1C left). CD163+ TAM density in the intratumor was not a significant prognostic biomarker for OS (P = 0.114). However, patients with high CD163+ TAM density at the IF had significantly worse OS (P < 0.001) than those with low density (Fig. 1C right).
In the cohort with 242 patients, univariate analysis showed that OS was associated with CD68+ TAM density at the IF, age, pathologic T stage and distant metastasis (Table 1). Logistic multivariate regression revealed that the density of CD68+ TAMs at the IF was independently associated with CRC OS (Table 2). In the cohort with 255 patients, univariate analysis showed that OS was associated with CD163+ TAM density at the IF, age, tumor size, tumor differentiation, pathologic T stage, lymph node metastasis and distant metastasis (Table 3). Logistic multivariate regression revealed that the density of CD163+ TAMs at the IF was independently associated with CRC OS (Table 4). Taken together, we identified CD68+ and CD163+ TAM density at the IF as independent prognostic factors of disease outcome in patients with CRC. However, CD68+ and CD163+ TAM density in the intratumor were not significant prognostic biomarkers.
Table 1.
Univariate Cox regression analysis for overall survival in 242 CRC patients.
| Variable | B | SE | Wald | P value | HR | 95.0% CI |
|
|---|---|---|---|---|---|---|---|
| Lower | Upper | ||||||
| CD68 intratumor | −0.022 | 0.011 | 3.769 | 0.052 | 0.978 | 0.957 | 1.000 |
| CD68 IF | −0.011 | 0.004 | 7.486 | 0.006 | 1.011 | 1.003 | 1.019 |
| Age | −0.861 | 0.279 | 9.535 | 0.002 | 0.423 | 0.245 | 0.730 |
| Gender | −0.256 | 0.201 | 1.621 | 0.203 | 0.774 | 0.522 | 1.148 |
| Tumor size | −0.108 | 0.194 | 0.308 | 0.579 | 0.898 | 0.613 | 1.314 |
| Differentiation | −0.154 | 0.143 | 1.152 | 0.283 | 0.857 | 0.647 | 1.136 |
| T stage | 1.604 | 0.213 | 56.828 | <0.001 | 4.971 | 3.276 | 7.543 |
| N stage | 0.701 | 0.192 | 13.357 | 0.364 | 2.017 | 1.384 | 2.938 |
| Distant metastasis | 1.341 | 0.205 | 42.781 | <0.001 | 3.823 | 2.558 | 5.713 |
Table 2.
Multivariate Cox regression analysis for overall survival in 242 CRC patients.
| Variable | B | SE | Wald | P value | HR | 95.0% CI |
|
|---|---|---|---|---|---|---|---|
| Lower | Upper | ||||||
| CD68 IF | 0.014 | 0.004 | 11.684 | 0.001 | 1.014 | 1.006 | 1.023 |
| Age | −1.031 | 0.285 | 13.051 | <0.001 | 0.357 | 0.204 | 0.624 |
| T stage | 1.300 | 0.231 | 31.805 | <0.001 | 3.670 | 2.336 | 5.767 |
| N stage | 0.383 | 0.201 | 3.631 | 0.057 | 1.467 | 0.989 | 2.177 |
| Distant metastasis | 0.675 | 0.235 | 8.268 | 0.004 | 1.963 | 1.240 | 3.109 |
Only factors with P < 0.05 in univariate analysis were included into multivariate analysis.
HR: hazard ratio; CI: confidence interval.
Table 3.
Univariate Cox regression analysis for overall survival in 255 CRC patients.
| Variable | B | SE | Wald | P value | HR | 95.0% CI |
|
|---|---|---|---|---|---|---|---|
| Lower | Upper | ||||||
| CD163 intratumor | −0.020 | 0.016 | 1.701 | 0.192 | 0.980 | 0.950 | 1.010 |
| CD163 IF | 0.025 | 0.004 | 34.141 | <0.001 | 1.025 | 1.016 | 1.033 |
| age | −0.727 | 0.250 | 8.441 | 0.004 | 0.483 | 0.296 | 0.789 |
| gender | −0.338 | 0.191 | 3.121 | 0.077 | 0.713 | 0.491 | 1.038 |
| Tumor size | 0.463 | 0.184 | 6.315 | 0.012 | 1.590 | 1.107 | 2.282 |
| Differentiation | 0.290 | 0.136 | 4.589 | 0.032 | 1.337 | 1.025 | 1.744 |
| T stage | 1.149 | 0.184 | 39.098 | <0.001 | 3.154 | 2.200 | 4.521 |
| N stage | 0.847 | 0.183 | 21.373 | <0.001 | 2.333 | 1.629 | 3.341 |
| Distant metastasis | 2.105 | 0.196 | 115.712 | <0.001 | 8.209 | 5.593 | 12.046 |
Table 4.
Multivariate Cox regression analysis for overall survival in 255 CRC patients.
| Variable | B | SE | Wald | P value | HR | 95.0% CI |
|
|---|---|---|---|---|---|---|---|
| Lower | Upper | ||||||
| CD163 IF | 0.018 | 0.004 | 19.120 | <0.001 | 1.018 | 1.010 | 1.026 |
| age | −0.845 | 0.258 | 10.730 | 0.001 | 0.430 | 0.259 | 0.712 |
| Tumor size | 0.018 | 0.198 | 0.009 | 0.926 | 1.019 | 0.691 | 1.501 |
| Differentiation | 0.084 | 0.134 | 0.387 | 0.534 | 1.087 | 0.835 | 1.415 |
| T stage | 0.379 | 0.182 | 4.344 | 0.037 | 1.461 | 1.023 | 2.087 |
| N stage | −0.022 | 0.233 | 0.009 | 0.924 | 0.978 | 0.620 | 1.544 |
| Distant metastasis | 1.979 | 0.270 | 53.672 | <0.001 | 7.234 | 4.261 | 12.283 |
Only factors with P < 0.05 in univariate analysis were included into multivariate analysis.
HR: hazard ratio; CI: confidence interval.
3.3. TAMs accumulated at the IF are primarily M2-like macrophages
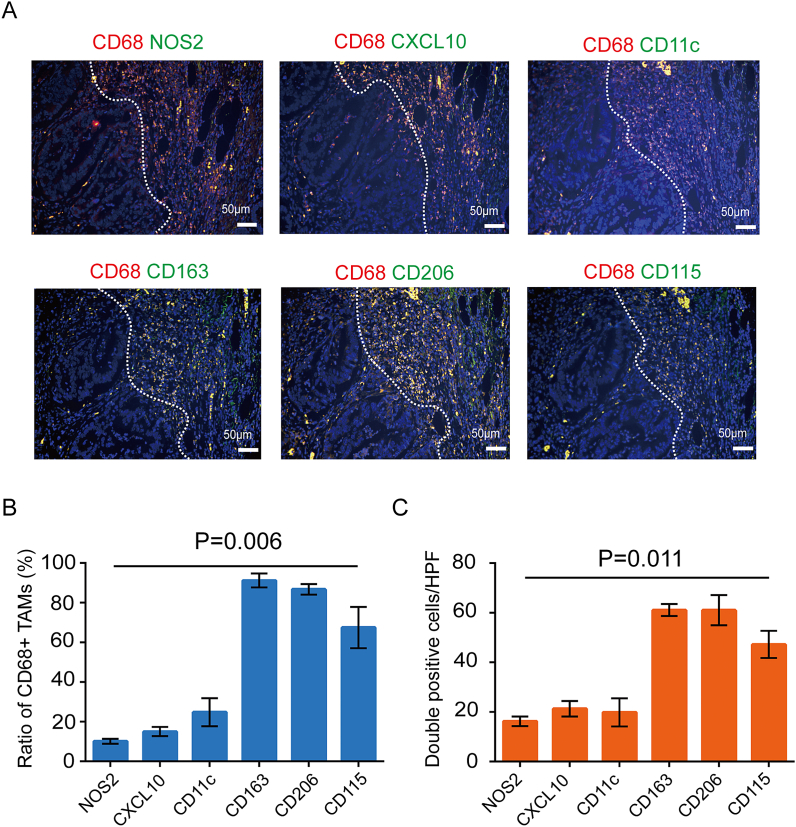
Due to the heterogeneity of TAMs, single marker is not enough to define the subtype for M1 or M2 TAMs. To separate the distinct populations of macrophages in CRC, CD11c, NOS2 and CXCL10 were selected as markers for M1 phenotype and CD163, CD206, CD115 as markers for M2 phenotype. Because most TAMs located at the IF, so we took double immunofluorescent staining to detect the infiltration of different macrophage subtypes (M1 and M2) at the IF region. Intriguingly, we observed that most TAMs, particularly those CD68+/CD163+, CD68+/CD206+ or CD68+/CD115+ double positive cells, were present at the IF whereas CD68+/NOS2+, CD68+/CXCL10+ or CD68+/CD11c+ double positive cells were barely visualized. (Fig. 2). These results indicated that CD163+, CD206+ and CD115+ M2 TAMs showed a dominant density compared with NOS2+, CXCL10+ and CD11c+ M1 TAMs at IF.
Fig. 2.
Double immunofluorescent staining for CD68 and M1 macrophage marker (NOS2, CXCL10 or CD11c) or M2 marker (CD163, CD206 or CD115) at the IF of serial sections from the same CRC clinical sample. A, The representative immunofluorescent staining images of IF samples (scale bar 50 μm). B, The ration with positive CD163, CD206, CD115, NOS2, CXCL10 and CD11c in CD68+ macrophages at IF in CRC tissue. C, The number of CD68+ CD163+, CD68+ CD206+, CD68+ CD115+, CD68+ NOS2+, CD68+ CXCL10+, CD68+ CD11c+ cells at IF in CRC tissue.
3.4. Association among M1 molecular markers (NOS2, CXCL10, CD11c) and M2 molecular markers (CD163, CD206, CD115)
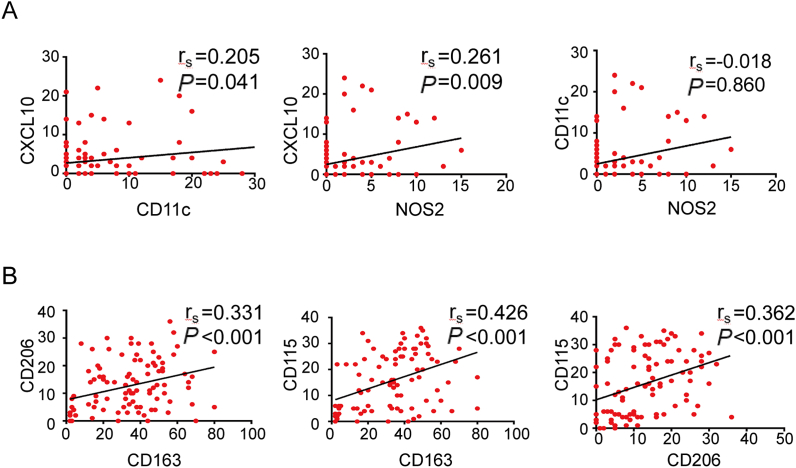
We conducted spearman's correlation analyses among M1 molecular markers (NOS2, CXCL10, CD11c), as well as M2 molecular markers (CD163, CD206, CD115) in a cohort with 100 CRC patients. The result revealed positive correlations among the density of NOS2+, CXCL10+ and CD11c+ cells and among the CD163+, CD206+ and CD115+ cells. The correlation coefficients were 0.205, 0.261 and −0.018, respectively (Fig. 3A). Similarly, the correlation coefficients between CD163 and CD206, CD163 and CD115, CD206 and CD115 were 0.331, 0.426 and 0.362, respectively (P < 0.001) (Fig. 3B). Similar results were obtained when analyzing in 224 CRC patients from a public clinical microarray database of Data GSE17538. As shown in Supplementary Figure 5A, three M1 molecular markers including NOS2, CXCL10 and CD11c were correlated to each other (Rs = 0.307, 0.122, 0.186). Meanwhile, three M2 molecular markers including CD163, CD206, and CD115 were also correlated to each other (Rs = 0.869, 0.858, 0.795; Supplementary Figure 5B).
Fig. 3.
Correlation analyses between two M1 macrophage markers (NOS2, CXCL10 or CD11c) or M2 markers (CD163, CD206 or CD115). A, Correlation between the number of NOS2+ or CXCL10+ and CD11c+ cells in 100 CRC patients. B, Correlation between the number of CD163+ or CD204+ and CD115+ cells in 100 CRC patients. Statistically significant differences between groups were determined by Spearman's rank correlation.
3.5. Correlation between TAMs density at IF and patients’ survival
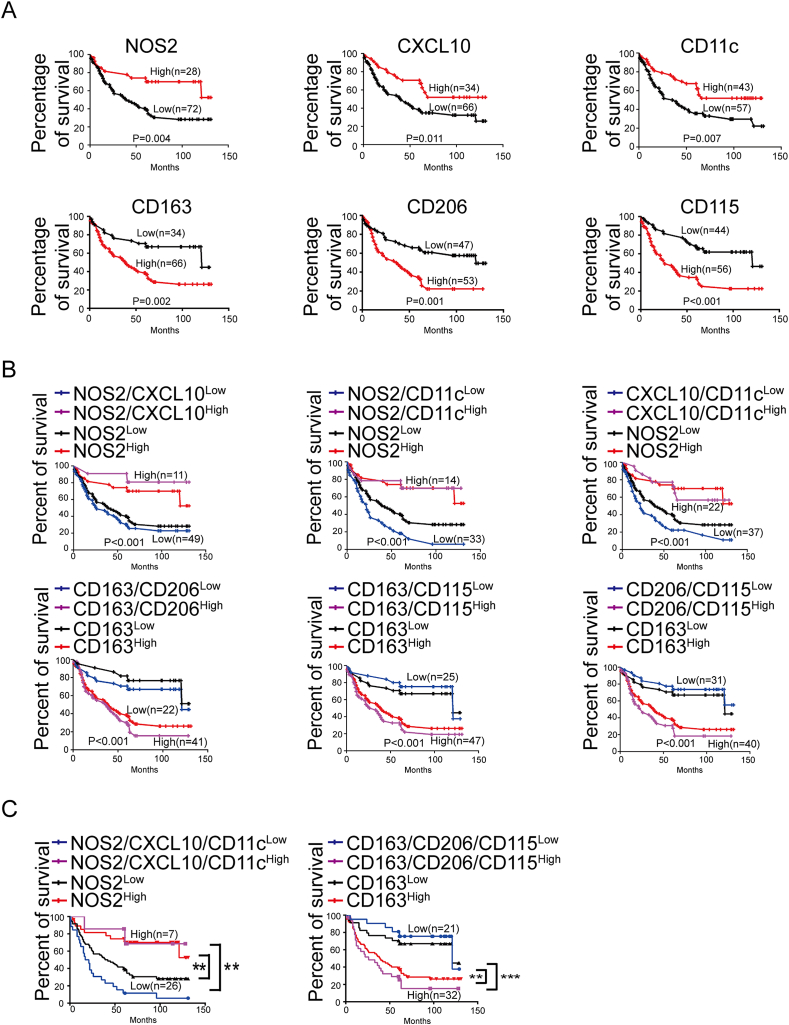
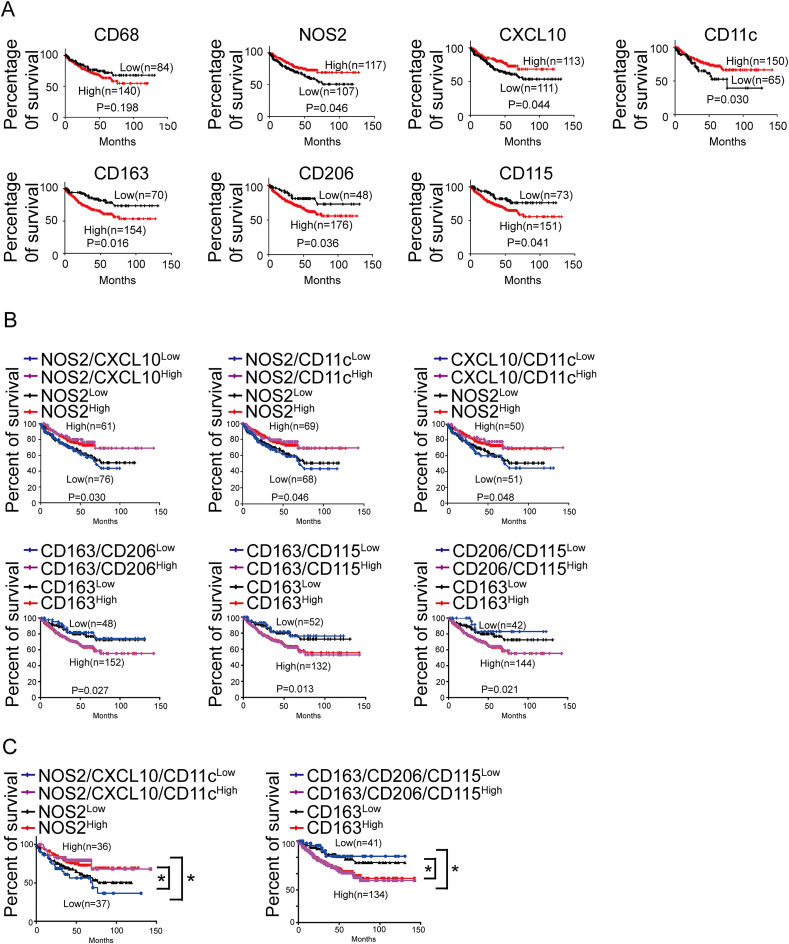
For NOS2+ TAM density at the IF, ≥2/HPF was defined as high and <2/HPF was defined as low. For CXCL10+ TAM density at the IF, ≥4/HPF was defined as high and <4/HPF was defined as low. For CD11c+ TAM density at the IF, ≥3/HPF was defined as high and <3/HPF was defined as low. For CD163+ TAM density at the IF, ≥23/HPF was defined as high and <23/HPF was defined as low. For CD206+ TAM density at the IF, ≥13/HPF was defined as high and <13/HPF was defined as low. For CD115+ TAM density at the IF, ≥14/HPF was defined as high and <14/HPF was defined as low. Details are shown in Supplementary Figure 6-11. The prognostic effect of every marker of TAMs on CRC patients’ OS was compared between patients with high and low TAMs density at IF in a cohort of 100 CRC specimens. Kaplan-Meier survival analysis showed patients with a higher density of NOS2+, CXCL10+ or CD11c+ cells showed better survival (P = 0.004; P = 0.011; P = 0.007), whereas patients with more CD163+, CD206+ or CD115+ cells had poor survival (P = 0.002; P = 0.001; P < 0.001) (Fig. 4A). Such associations were much clearer when analyses were performed by combining two markers. Patients with positive coexpression of CD163/CD206, CD163/CD115, CD206/CD115 at IF had the shortest OS times, whereas patients with positive coexpression of NOS2/CXCL10, NOS2/CD11c, CXCL10/CD11c expression pattern had the highest OS rate. The combination of two markers for prognosis was better than single marker (Fig. 4B).
Fig. 4.
TAMs are associated with OS of 100 CRC patients. A, Kaplan-Meier survival analyses for the association of the expressions of pan-monocyte/macrophage marker (CD68), M1 macrophage markers (NOS2, CXCL10 and CD11c) or M2 macrophage markers (CD163, CD206 and CD115) with patient OS. B, Kaplan-Meier survival analyses for the association of concurrent double expressions of M1 markers (NOS2 and CXCL10, NOS2 and CD11c, CXCL10 and CD11c) or M2 markers (CD163 and CD206, CD163 and CD115, CD206 and CD115) with patient OS. C, Kaplan-Meier survival analyses for the concurrent triple expressions of M1 macrophage markers (NOS2, CD11c and CXCL10) or M2 macrophage markers (CD163, CD206 and CD115) with patient OS. **P < 0.01, ***P < 0.001.
To examine whether three markers for M1 or M2 macrophage is more effective than using single marker, we analyzed the correlation between treble molecular markers and the OS rate. Patients with low coexpression of three M1 molecular markers (NOS2/CXCL10/CD11c) exhibited lowest OS rate. Whereas patients with low coexpression of three M2 molecular markers (CD163/CD206/CD115) exhibited highest OS rate (Fig. 4C). Moreover, we compared the prognostic values of single marker (NOS2 or CD163) and treble markers. It could be observed that the prognostic value of treble markers was superior to that of single marker. To further verify our findings, bioinformatics analysis was used to extract more data. The GSE17538 dataset from GEO database was downloaded and 224 CRC patients were enrolled. Based on the Kaplan-Meier survival analysis, we confirmed that usage of treble markers was superior to single marker for the prediction of prognosis (Fig. 5A–C).
Fig. 5.
Bioinformatic analyses for the expressions of macrophage marker genes and OS of 224 CRC patients using the expression profiling dataset of GSE17538. A, Kaplan-Meier survival analyses for the association between expression of pan-monocyte/macrophage marker (CD68), M1 macrophage markers (NOS2, CXCL10 and CD11c) or M2 macrophage markers (CD163, CD206 and CD115) and patient OS. B, Kaplan-Meier survival analyses for the association between concurrent double expression of M1 markers (NOS2 and CXCL10, NOS2 and CD11c, CXCL10 and CD11c) or M2 markers (CD163 and CD206, CD163 and CD115, CD206 and CD115) and patient OS. C, Kaplan-Meier survival analyses for the association between concurrent triple expression of M1 macrophage markers (NOS2, CXCL10 and CD11c) or M2 macrophage markers (CD163, CD206 and CD115) and patient OS. *P < 0.05.
In the cohort with 100 patients, univariate analysis showed that OS was associated with NOS2+, CXCL10+, CD11c+, CD163+, CD206+, CD115+ TAM density at the IF, pathologic T stage, lymph node metastasis and distant metastasis (Supplementary Table 1). Logistic multivariate regression revealed that the density of CD11c+ TAMs, CD206+, CD115+ TAM density at the IF were independently associated with CRC OS (Supplementary Table 2-7). Together, these findings indicated that the combination of treble TAMs markers at IF is rather indicative for OS of CRC patients.
4. Discussion
The tumor IF is a very important area for prognostic determination of CRC. Some scientists attempted to add CD163+ TAMs density at the IF of primary tumor to primary tumor immune score [18]. But up to now no conclusions have been drawn. Furthermore, how would the TAMs that located at tumor IF contribute to prognosis in CRC need further investigation. In this study, we confirmed that TAMs were mainly distributed at IF of CRC and most of them were M2 subtype. M1 subtype and M2 subtype are two major polarization states of TAMs. It has been documented that M1 subtype and M2 subtype TAMs possess almost opposite anti- and pro-tumoral activity respectively [18]. M1 type TAMs are mainly involved in Th1 type immune response. They are induced by bacteria, lipopolysaccharide (LPS) or interferon-γ (IFN-γ), with high expression of MHC class II molecules, IL-12, TNF-α and B7 molecules [19]. M1 type TAMs commonly produce a variety of toxic intermediate products, such as nitric oxide (NO) and reactive oxygen intermediates (ROIs) [20], and therefore have the ability to kill pathogens and tumor cells. M2 type TAMs are induced by factors such as IL-4, IL-13, IL-10 or glucocorticoids and produce a range of anti-inflammatory cytokines, specifically transforming growth factor (TGF)-β and IL-10 [21]. They are characterized by scavenger, angiogenic, and pro-invasive properties, thus contributing to tumor growth, invasion, and metastasis [22]. In most cancers, TAMs are polarized to the M2 phenotype, which are known to promote tumor progression and to be associated with a poor prognosis in numerous cancers. Therefore, the density of total TAMs can't be used as the only indicator for tumor progression. In recent years, the phenotypes of TAMs have been proven to be critical TAMs-associated parameters for diagnosis and prognosis of patients with cancers [23].
Previous study has found that TAMs played protective role for the CRC patients and interfered with primary tumor growth and metastasis [24]. In addition, the TAMs’ heterogeneity in CRC patients has also been evaluated. In immunogenic types of CRC, patients are observed with high levels of infiltrating CD68+ TAMs [25]. Due to the great heterogeneity of TAMs in CRC patients, the prognostic role of different types of TAMs infiltration has attracted the attention of researchers. CD68 is the major biomarker for the quantification of total TAMs amounts. Li et al. [26] reported that high density of TAMs in CRC tissues is significantly associated with favorable 5-year OS. Low levels of CD68+ TAMs at IF and tumor stroma are found to be associated with more advanced status of CRC, while high amount of TAMs is found in patients with good prognosis [27]. Zhou et al. [28] reported that high density of CD68+ macrophages at IF of tumor is associated with lower metastasis and higher 5-year survival rate.
Except for evaluating the prognostic value of total amount of TAMs in CRC, there are some studies focused on the prognostic value of M2 type TAMs, which are rather indicative for the negative prognosis of patients with CRC [24]. For instance, high expression of stromal CD163 at tumor IF in CRC patients is significantly associated with various clinical characteristics and correlated with poor recurrence-free survival (RFS) [29]. In these studies, the infiltration of M2 type TAMs is wildly used, while the accurate definition of M2 type TAMs has not been investigated. Thus, the TAMs polarization is hard to evaluate. Moreover, single M2-specific marker, especially CD163, is usually used to reflect the infiltration of M2 type TAMs. However, the biomarkers for different subtypes are multifarious, such as M1-specific CXCL10, CD11c, CD80, HLA-DR, NOS2, and M2-specific CD115, CD163, CD169, CD204, CD206, CCL8, COX-2, IGF1, MARCO, MMP-9, stabilin-1 [24,30]. With the addition of their variable expression profiles in different individuals, usage of single marker is not sufficient.
Here we combined several markers to enhance the specificity to identify specific subsets and we utilized CD68 as a general marker of TAMs, CD11c, NOS2, CXCL10 as markers for M1 phenotype and CD163, CD206, CD115 as markers for M2 phenotype [[31], [32], [33]]. The transmembrane scavenger receptor CD163 is expressed exclusively in monocytes (low expression) and macrophages (high expression). It is often used as an M2 TAM marker [34]. CD206 is a C-type lectin, also known as the macrophage mannose receptor, which is expressed by tissue macrophages [13]. The macrophage colony-stimulating factor (M-CSF) receptor (also known as CSF1R, CD115), could support metastasis formation by promotion of tumor angiogenesis, M2 polarization of macrophages [35]. NOS2 (nitric oxide synthase 2), as a marker of M1 macrophages in tumor islets, was associated with an improved prognosis for patients with CRC [36]. NOS2 and CXCL10 were also used as M1 macrophage markers in breast cancer [37]. The M1 macrophages with CD11c positive expression, may be able to perform complement-mediated cytotoxicity and phagocytosis [38].
Our results indicated that TAMs accumulated at the tumor IF of CRC and the main subpopulation was M2 type TAMs. We could conclude that low expression of M1 markers and high expression of M2 markers were associated with tumor aggressiveness. Regarding to the considerable roles of these markers, we speculated that combination of these three M1-specific markers or three M2-specific markers might be better than single marker to be indicative for CRC progression. The speculation was further confirmed when we found that the three M1-specific markers or three M2-specific markers were significantly associated with each other in CRC patients. Further Kaplan-Meier survival analysis showed that low expression of three M1 molecular markers (NOS2/CXCL10/CD11c) was correlated with low OS rate, while low expression of three M2 molecular markers (CD163/CD206/CD115) was correlated with high OS rate. It could be observed that the prognostic value of treble markers was superior to that of single marker. These findings suggested that combination of these three M1-specific markers or three M2-specific markers is a more precise method for predicting prognosis of CRC patients.
In summary, here we established a more comprehensive method for predicting the prognosis of CRC patients using a combination of three M1-specific markers (NOS2, CXCL10, and CD11c) and three M2-specific markers (CD163, CD206, and CD115). Our results proved that the method is more precise than conventional method with single marker.
Author contribution statement
Xia Wang, Xiao-yan Wang, Zu-guo Li: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper. Tian-jiao Yuwen: Contributed reagents, materials, analysis tools or data; Wrote the paper. Yan Zhong: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by National Natural Science Foundation of China [No. 81672437, 82273355], Natural Science Foundation of Guangdong Province [2022A1515012638].
Data availability statement
Data will be made available on request.
Declaration of competing interest
The authors declare no conflict of interest.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e13211.
Contributor Information
Zu-Guo Li, Email: lizg@smu.edu.cn.
Xiao-Yan Wang, Email: wangxiaoyan639@163.com.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Sung Hyuna, Ferlay Jacques, Rebecca L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Brenner H., Kloor M., Pox C.P. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 3.Mazzoccoli G. Colorectal cancer prognosis and PPARδ/β expression in the tumormicroenvironment. J. Gastroenterol. 2014;49(3):564–565. doi: 10.1007/s00535-013-0913-z. [DOI] [PubMed] [Google Scholar]
- 4.Coffelt S.B., de Visser K.E. Cancer: inflammation lights the way to metastasis. Nature. 2014;507(7490):48–49. doi: 10.1038/nature13062. [DOI] [PubMed] [Google Scholar]
- 5.Pathria P., Louis T.L., Varner J.A. Targeting tumor-associated macrophages in cancer. Trends Immunol. 2019;40(4):310–327. doi: 10.1016/j.it.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Troiano G., Caponio V.C.A., Adipietro I., et al. Prognostic significance of CD68(+) and CD163(+) tumor associated macrophages in head and neck squamous cell carcinoma: a systematic review and meta-analysis. Oral Oncol. 2019;93:66–75. doi: 10.1016/j.oraloncology.2019.04.019. [DOI] [PubMed] [Google Scholar]
- 7.Zhao X., Qu J., Sun Y., et al. Prognostic significance of tumor-associated macrophages in breast cancer: a meta-analysis of the literature. Oncotarget. 2017;8:30576–30586. doi: 10.18632/oncotarget.15736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu M., Guan R., Hong W., et al. Prognostic value of tumor-associated macrophages in pancreatic cancer: a meta-analysis. Cancer Manag. Res. 2019;11:4041–4058. doi: 10.2147/CMAR.S196951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J., Chang L., Zhang X., Zhou Z., Gao Y. Meta-analysis of the prognosticand clinical value of tumor-associated macrophages in hepatocellular carcinoma. J. Invest. Surg. 2021;34:297–306. doi: 10.1080/08941939.2019.1631411. [DOI] [PubMed] [Google Scholar]
- 10.Yang Z., Zhang M., Peng R., et al. The prognostic and clinicopathological value of tumor-associated macrophages in patients with colorectal cancer: a systematic review and meta-analysis. Int. J. Colorectal Dis. 2020;35(9):1651–1661. doi: 10.1007/s00384-020-03686-9. [DOI] [PubMed] [Google Scholar]
- 11.Fakih Marwan, Ouyang Ching, Wang Chongkai, et al. Immune overdrive signature in colorectal tumor subset predicts poor clinical outcome. J. Clin. Invest. 2019;129(10):4464–4476. doi: 10.1172/JCI127046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H., Tian T., Zhang J. Tumor-associated macrophages (TAMs) in colorectal cancer (CRC): from mechanism to therapy and prognosis. Int. J. Mol. Sci. 2021;22(16):8470. doi: 10.3390/ijms22168470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rafiul Haque A.S.M., Moriyama Masafumi, Kubota Keigo, et al. CD206+ tumor-associated macrophages promote proliferation and invasion in oral squamous cell carcinoma via EGF production. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-51149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng Q.Y., Chang W.J., Mao Y.H., et al. Tumor-associated macrophages as prognostic and predictive biomarkers for postoperative adjuvant chemotherapy in patients with stage II colon cancer. Clin. Cancer Res. 2019;25(13):3896–3907. doi: 10.1158/1078-0432.CCR-18-2076. [DOI] [PubMed] [Google Scholar]
- 15.Barrett T., Troup D.B., Wilhite S.E., et al. NCBI GEO: mining millions of expression profiles--database and tools. Nucleic Acids Res. 2005;33(Database issue):D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeman T.J., Smith J.J., Chen X., et al. Smad4-mediated signaling inhibits intestinal neoplasia by inhibiting expression of beta-catenin. Gastroenterology. 2012;142(3):562–571. doi: 10.1053/j.gastro.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith J.J., Deane N.G., Wu F., et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138(3):958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinto M.L., Rios E., Duraes C., et al. The two faces of tumor-associated macrophages and their clinical significance in colorectal cancer. Front. Immunol. 2019;10:1875. doi: 10.3389/fimmu.2019.01875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glass C.K., Natoli G. Molecular control of activation and priming in macrophages. Nat. Immunol. 2016;17(1):26–33. doi: 10.1038/ni.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Kaiyue, Lin Kangjia, Li Xiaoyan, et al. Redefining tumor-associated macrophage subpopulations and functions in the tumor microenvironment. Front. Immunol. 2020;11:1731. doi: 10.3389/fimmu.2020.01731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mantovani A., Sica A., Sozzani S., et al. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25(12):677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 22.Condeelis J., Pollard J.W. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124(2):263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 23.Jayasingam S.D., Citartan M., Thang T.H., et al. Evaluating the polarization of tumor-associated macrophages into M1 and M2 phenotypes in human cancer tissue: technicalities and challenges in routine clinical practice. Front. Oncol. 2019;9:1512. doi: 10.3389/fonc.2019.01512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larionova I., Tuguzbaeva G., Ponomaryova A., et al. Tumor-associated macrophages in human breast, colorectal, lung, ovarian and prostate cancers. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.566511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roelands J., Kuppen P.J.K., Vermeulen L., et al. Immunogenomic classification of colorectal cancer and therapeutic implications. Int. J. Mol. Sci. 2017;18(10) doi: 10.3390/ijms18102229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J., Li L., Li Y., et al. Tumor-associated macrophage infiltration and prognosis in colorectal cancer: systematic review and meta-analysis. Int. J. Colorectal Dis. 2020;35(7):1203–1210. doi: 10.1007/s00384-020-03593-z. [DOI] [PubMed] [Google Scholar]
- 27.Roelands J., Kuppen P.J.K., Vermeulen C L., et al. Relationships between tumor-associated macrophages and clinicopathological factors in patients with colorectal cancer. Anticancer Res. 2002;22(6C):4291–4296. PMID: 12553072. [PubMed] [Google Scholar]
- 28.Zhou Q., Peng R.Q., Wu X.J., et al. The density of macrophages in the invasive front is inversely correlated to liver metastasis in colon cancer. J. Transl. Med. 2010;8:13. doi: 10.1186/1479-5876-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei C., Yang C., Wang S., et al. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol. Cancer. 2019;18(1):64. doi: 10.1186/s12943-019-0976-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martínez V.G., Rubio C., Martínez-Fernández M., et al. BMP4 induces M2 macrophage polarization and favors tumor progression in bladder cancer. Clin. Cancer Res. 2017;23(23):7388–7399. doi: 10.1158/1078-0432.CCR-17-1004. [DOI] [PubMed] [Google Scholar]
- 31.Wan Shanshan, Zhao Ende, Kryczek Ilona, et al. Tumor-associated macrophages produce interleukin 6 and signal via STAT3 to promote expansion of human hepatocellular carcinoma stem cells. Gastroenterology. 2014;147:1393–1404. doi: 10.1053/j.gastro.2014.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez F.O., Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joyce J.A., Pollard J.W. Microenvironmental regulation of metastasis. Nat. Rev. Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skytthe Maria K., Graverse Jonas Heilskov, Moestrup Søren K. Targeting of CD163+ macrophages in inflammatory and malignant diseases. Int. J. Mol. Sci. 2020;21(15):5497. doi: 10.3390/ijms21155497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Francesca Maria Consonni. Bleve Augusto, Grazia Totaro Maria, et al. Heme catabolism by tumor-associated macrophages controls metastasis formation. Nat. Immunol. 2021;22(5):595–606. doi: 10.1038/s41590-021-00921-5. [DOI] [PubMed] [Google Scholar]
- 36.Edin Sofia, Wikberg Maria L., Dahlin Anna M., et al. The distribution of macrophages with a M1 or M2 phenotype in relation to prognosis and the molecular characteristics of colorectal cancer. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0047045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheuk I.W., Chen J., Siu M., et al. Resveratrol enhanced chemosensitivity by reversing macrophage polarization in breast cancer. Clin. Transl. Oncol. 2022;24(5):854–863. doi: 10.1007/s12094-021-02731-5. [DOI] [PubMed] [Google Scholar]
- 38.Wang Hui, Zhou Junjie, Li Jun, et al. A study of multinucleated giant cells in esophageal cancer. Clin. Immunol. 2021;222 doi: 10.1016/j.clim.2020.108600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.