Abstract
Brain metastasis (BM) represents the single most severe neurological complication of systemic cancer. The prognosis of patients with BM is poor, irrespective of the implemented treatment. The present study performed a systematic review of the literature using three online databases (PubMed, Scopus and Web of Science). Recently, a number of small RNA molecules, the microRNAs (miRNAs/miRs), have attracted increasing scientific attention. Members of the miR-200 family, which includes five miRNAs (miR-141, miR-200a, miR-200b, miR-200c and miR-429) appear to play pivotal roles in cancer initiation and metastasis. Indeed, a systematic review of the pertinent literature revealed that miR-200 family members regulate the brain metastatic cascade, particularly by modulating epithelial-to-mesenchymal transition. That holds true for the major representatives of BM, including lung and breast cancer, as well as for other less frequent secondary lesions originating from melanoma and the gastrointestinal tract. Therefore, the miRNAs may serve as potential diagnostic and/or prognostic markers, and under specific circumstances, as invaluable therapeutic targets. However, the available clinical evidence is relatively limited. A number of studies have suggested that the miR-200 family members are accurate prognostic markers of survival and resistance to chemotherapy in patients with breast cancer. Similarly, they may prove helpful in differentiating a metastatic lesion from a malignant glioma, or a hemangioblastoma from a renal cell carcinoma in patients with von Hippel Lindau syndrome, based on a cerebrospinal fluid sample. However, currently, there is no known therapeutic role for miR-200 family members in the setting of BM.
Keywords: brain metastasis, miR-200 family, endothelial-to-mesenchymal transition, zinc finger E-box-binding homeobox-1, pathogenesis, prognosis, therapy
Introduction
Brain metastasis (BM) represents the single most severe neurological complication of systemic cancer. In addition, BM constitutes a major source of morbidity and mortality, accounting for as much as 60% of intraparenchymal brain tumors (1,2). The brain parenchyma may harbor secondary neoplasms from each malignant tumor (2,3). However, metastases from lung and breast cancers, and melanoma predominate in adults (4-6). Almost 50% of lung tumors, 25% of breast neoplasms and 20% of melanomas have seeded the brain by the time of diagnosis (4-6). Regardless of treatment, the prognosis of patients with BM is poor (4-7). Therefore, scientific interest has focused on the underlying molecular pathways and the genetic signatures in order to identify novel drugs which may lead to improved outcomes (4-6).
Recently, a number of small RNA molecules, known as microRNAs (miRNAs/miRs) have attracted increasing scientific attention. miRNAs are natural, small, non-coding RNA gene products, ~22 nucleotides in length, with a characteristic hairpin structure (8). They apppear to regulate a number of target genes involved in the control of development, proliferation, apoptosis and stress response by repressing the translation or regulating the degradation of messenger transcripts through complementary binding (9-12).
Additional evidence indicates that genes coding for miRNAs play a crucial role in almost every step of carcinogenesis, including cell proliferation, angiogenesis, migration, colonization, metastasis and immunosuppression, functioning either as oncogenes or tumor suppressors (13-18). The miR-200 family, which includes five miRNAs (miR-141, miR-200a, miR-200b, miR-200c and miR-429), is among the most extensively studied miRNAs (18-22). Its members appear to play pivotal roles in cancer initiation and metastasis (19-23).
To the best of our knowledge, the role of the miR-200 family in BM has not been adequately described and summarized. The present systematic review focuses on the role of miR-200 family members in the pathogenetic cascade of BM. In addition, their implications in cancer subtype classification, as diagnostic or prognostic markers, drug-response predictors, and as potential therapeutic targets in BM are discussed.
Data and methods
Two authors (AGB and GF) performed a systematic review of the literature, using three online databases (PubMed, Scopus and Web of Science). The search included the terms ‘brain metastasis’ OR ‘cerebral metastasis’ AND ‘miR-200 family’ OR ‘miR-200a’ OR ‘miR-200b’ OR ‘miR-200c’ OR ‘miR-141’ OR ‘miR-249’ AND ‘colorectal cancer’ OR ‘lung cancer’ OR ‘breast cancer’ OR ‘melanoma’ OR ‘gastric cancer’ OR ‘renal cancer’ OR ‘ovarian cancer’. Additional studies were discovered in the reference lists of the collected studies.
The inclusion criteria were the following: i) The study examined the role of the miR-200 family or any of its members; ii) one of the study outcomes was related to cerebral metastasis; iii) the study involved adult human subjects, animal models or cells in culture, iv) the study provided sufficient data relevant to the context of the present systematic review; and v) the study was written in the English language. On the contrary, studies were excluded if they were not written in the English language, were irrelevant to the miR-200 family or to BM, involved subjects that did not provide data relevant to the present study. Additionally, review studies, editorials and underpowered studies were excluded (less than five cases).
Two authors (AGB and GF) extracted variable data from each eligible study, including the name of the first author and year of publication, the hosting country, the miRNA(s) under study, the primary site of BM, the type of tissue sample, and other information pertinent to the present systematic review (Table I).
Table I.
Characteristics of the studies included in the data synthesis.
| Author, year | Country | Malignancy | Sample type | Assay | Sample size | Survival | miRNA | Comments | (Refs.) |
|---|---|---|---|---|---|---|---|---|---|
| Choi et al, 2011 | Korea | Lung cancer cervical adenocarcinoma | Tissue | Luciferase reporter | 40 | OS | miR-200b | miR200b downregulates VEGF signaling through VEGF and its receptors and may be used as an angiogenesis inhibitor | (19) |
| Roybal et al, 2011 | USA | Lung cancer | Tissue | Dual-luciferase reporter, RT- qPCR | 27 | RFS | miR-200b/c/429 | miR-200 blocks lung cancer invasion and metastasis through Flt1/VEGFR1 | (24) |
| Byun et al, 2019 | Korea | Lung cancer | Tissue | RT-qPCR | 19 | OS | miR-200c | miR-200c inhibits lung cancer cell migration by HIF-1α downregulation | (25) |
| Gregory et al, 2008 | Australia | Breast cancer | Tissue | RT-qPCR | 15 | NR | miR-200a, miR- 200b, miR-200c, miR-141, miR- 429, miR-205 | The miR-200 family control epithelial to mesenchymal transition via ZEB1 and SIP1 | (26) |
| Chen et al, 2014 | USA | Lung cancer | Tissue | RT-qPCR | 230 | NR | miR-200 | The miRNA-200/ZEB1 axis control of PD-L1 expression regulates metastasis | (27) |
| Gibbons et al, 2009 | USA | Lung cancer | Tissue | RT-qPCR | 76 | NR | miR-200 | miR-200 changes in response to contextual extracellular cues regulate metastasis | (28) |
| Park et al, 2008 | USA | Ovarian cancer | Tissue | RT-qPCR | 59 | NR | miR-200 | The miR-200 family regulates the epithelial phenotype of cancer cells through ZEB1 and ZEB2 | (29) |
| Pecot et al, 2013 | USA | Ovarian cancer, renal cancer, breast cancer, lung cancer | Tissue | RT-qPCR | 579 474 325 | OS | miR-200 | The miR-200 family regulates tumor angiogenesis | (30) |
| Gravgaard et al, 2012 | Denmark | Breast cancer | Tissue | RT-qPCR | 47 | NR | miR-200 miR-9 | The miR-200 family is differentially expressed in primary and metastatic breast cancer | (31) |
| Debeb et al, 2016 | USA | Breast cancer | Tissue | RT-qPCR | 105 | OS and RFS | miR-141 | miR-141 controls of brain metastasis after breast cancer | (32) |
| Duhachek- Muggy et al, 2015 | USA | Breast cancer | Tissue | RT-qPCR | 100 | RFS | miR-29 and miR- 200 | miR-200 family regulates ADAM12-L 3'UTR directly | (33) |
| Shao et al, 2019 | China | Breast cancer | Tissue | RT-qPCR | 103 | OS and RFS | miR-200a, miR- 210 7 miR-451 | miR-200a and miR-210 could predict chemotherapy resistance in metastatic breast cancer | (34) |
| Schickel et al, 2010 | USA | Lung cancer | Tissue | RT-qPCR | 60 | NR | miR-200 | miR-200c induces apoptosis through CD95 by regulating FAP-1 | (35) |
| Mueller et al, 2009 | Germany | Melanoma | Tissue | RT-qPCR | 64 | NR | miR-17-5p, miR- 222, miR-181a, miR-194, miR-22 & miR-373 | The miR-200 family members are up- or downregulated in primary and metastatic melanoma | (36) |
| Rosenfeld et al, 2008 | Israel, USA, Korea, Germany | Bladder cancer, breast cancer, colon cancer, endometrial cancer, renal cancer, lung cancer, ovarian cancer, gastric cancer, sarcoma | Tissue | RT-qPCR | 253 | NR | miR-27b; miR- 181b; miR-29a; miR-345; miR- 29c; miR-182; miR-205; miR- 152; miR-187; miR-29b; miR- 214; miR-19b | The miR-200 family members could be used in an algorithm to identify the origin of cancers of unknown origin | (37) |
| Elson-Schwab et al, 2010 | UK, USA | Melanoma | Tissue | RT-qPCR | 254 | OS | miR-200 | The miR-200 family members are involved in melanoma invasion | (38) |
| Zhong et al, 2016 | China | Lung cancer Colon cancer Pancreatic cancers | Tissue microarray | RT-qPCR | 381 | OS | miR-200 | KRAS activation downregulates endogenous mir-200 expression in tumor initiation and progression | (39) |
| Zhong et al, 2020 | China | Gastric cancer | Tissue | RT-qPCR | 68 | OS | miR-141 | LINC00242/miR-141/FOXC1 axis promotes oncogenesis in gastric cancer | (40) |
| Youn et al, 2019 | Korea | Gastric cancer | Tissue | RT-qPCR | 38 | OS | Long non-coding RNA N-BLR | Migration and invasion of a gastric adenocarcinoma is promoted by N-BLR | (41) |
OS, overall survival; DFS, disease-free survival; PFS, progression-free survival; RFS, relapse-free survival; NR, not reported; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; VEGF, vascular endothelial growth factor; Flt1, FMS-like tyrosine kinase; HIF-1α, hypoxia-inducible factor 1-α; ZEB1, zinc finger E-box binding homeobox 1; SIP1, Smad interacting protein 1; ADAM12, ADAM12, a disintegrin and metalloproteinase domain-containing protein-12; 3'UTR, 3' untranslated region; FAP-1, fibroblast activation protein-1; KRAS, Kirsten renin-angiotensin system; LINC, long intergenic non-protein coding; FOXC1, Forkhead box carcinogen 1.
In anticipation of the limited amount of quantitative data, the evidence obtained was summarized into a narrative review. The evidence was stratified according to the site of the primary cancer and was supervised by the senior author (NT). The screening strategy for the selection of the articles is presented in Fig. 1.
Figure 1.
Screening strategy for the selection of the articles included in the present systematic review.
Results
Lung cancer
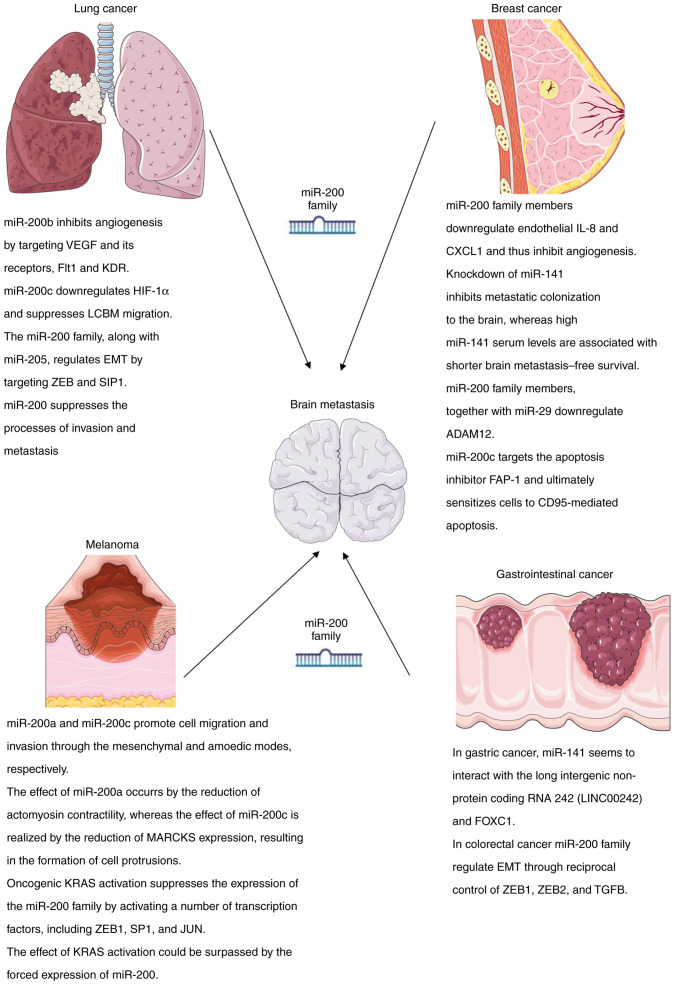
miR-200 family members are involved in at least four steps of lung cancer-associated BM (LCBM) development. Under normal conditions, miR-200b functions as an angiogenesis inhibitor by targeting VEGF and its receptors, FMS-like tyrosine kinase and kinase insert domain receptor (18,24). Similarly, miR-200c downregulates hypoxia-inducible factor-1α, a key regulator of hypoxia-related angiogenesis and LCBM migration (25). In addition, the miR-200 family, along with miR-205, regulates epithelial-to-mesenchymal transition (EMT) by targeting zinc finger E-box-binding homeobox (ZEB)1 and Smad interacting protein-1(26). In conjunction with ZEB1, this largely accounts for the observed intratumoral immunosuppression, required for BM survival and immune escape (27). Finally, the induced expression of miR-200 appears to suppress the processes of invasion and metastasis (28).
Breast cancer
There is strong evidence to indicate that there is an inverse association between the transforming growth factor (TGF)-1-induced EMT and all five members of the miR-200 family, by targeting the E-cadherin repressors, ZEB1 and ZEB2 (26,29). Additional evidence indicates that the miR-200 family members downregulate endothelial interleukin-8 and chemokine (C-X-C motif) ligand-1 (CXCL1) and thus inhibit angiogenesis (30). The differential expression of the majority of members of the miR-200 family in primary and metastatic tissue indicates that they are actively involved in the metastatic process (31). In particular, the knockdown of miR-141 has been shown to inhibit metastatic colonization to the brain, whereas high miR-141 serum levels have been shown to be associated with a shorter BM-free survival (32). In addition, miR-200 family members, together with miR-29, are involved in the progression of breast cancer-associated BM (BCBM) by the direct downregulation of a disintegrin and metalloproteinase domain-containing protein-12, which has prognostic and chemopredictive values in the management of BCBM (33). Indeed, Shao et al (34) demonstrated that plasma miR-200a could accurately predict the resistance to chemotherapy in patients with metastatic breast cancer. Furthermore, it has been found that miR-200c targets the apoptosis inhibitor, Fas-associated phosphatase 1, and ultimately sensitizes cells to CD95-mediated apoptosis (35).
Melanoma
The differential expression of the miR-200 family members was noted in melanoma-associated BM (MBM). To begin with, miR-200 levels was found to be higher in melanoma cells than in normal melanocytes (36). At the same time, miR-200 was found to be expressed at higher levels in lung metastases and at lower levels in BM when compared to the primary lesion (37). Additional differences were noted between the actions of different miR-200 family members. miR-200a and miR-200c promoted cell migration and invasion through the mesenchymal and amoedic modes, respectively (38). The effect of miR-200a occurred by the reduction of actomyosin contractility, whereas the effect of miR-200c was realized by the reduction of myristoylated alanine-rich C-kinase substrate expression, resulting in the formation of cell protrusions (39). In addition, MBM is largely driven by oncogenic KRAS activation, which actively promotes cell survival and EMT (39). As a matter of fact, KRAS activation suppresses the expression of the miR-200 family by activating a number of transcription factors, including ZEB1, SP1 and JUN (39). In turn, the effect of KRAS activation could be surpassed by the forced expression of miR-200(39).
Gastrointestinal tract cancer
In gastric cancer, miR-141 appears to interact with the long intergenic non-protein coding RNA 242 (LINC00242) and forkhead box C-1 to enhance cancer progression (40), whereas in colorectal cancer, the members of the miR-200 family regulate EMT through reciprocal control of ZEB1, ZEB2 and TGFB (41).
The roles of miR-200 family members in BM from lung cancer, breast cancer, melanoma and gastrointestinal cancer are illustrated in Fig. 2.
Figure 2.
Roles of miR-200 family members in brain metastases from lung cancer, breast cancer, melanoma and gastrointestinal cancer. Parts of the figure were drawn using images from Servier Medical Art. Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License (https://creativecommons.org/licenses/by/3.0/). VEGF, endothelial growth factor; KDR, kinase insert domain receptor; Flt1, FMS-like tyrosine kinase; HIF-1α, hypoxia-inducible factor-1α; EMT, epithelial-to-mesenchymal transition; ZEB1, zinc finger E-box-binding homeobox-1; ZEB2, zinc finger E-box-binding homeobox-2; SIP1, smad interacting protein-1; IL-8, interleukin-8; CXCL1, chemokine (C-X-C motif) ligand-1; ADAM12, a disintegrin and metalloproteinase domain-containing protein-12; FAP-1, Fas-associated phosphatase 1; MARCKS, myristoylated alanine-rich C-kinase substrate; SP-1, specificity protein 1; KRAS, Kirsten rat sarcoma virus; FOXC1, forkhead box C-1; TGFB, transforming growth factor-B.
Discussion
The invasion-metastasis cascade is a sequential and selective process involving multiple steps. Among these are EMT, the invasion of the basement membrane, the intravasation of escapers, the protection of tumor cell emboli from physical stress and the host's immune response, the extravasation of survivors at distant loci, penetration through the blood-brain barrier, and metastasis ‘seeding-and-soiling’ (22,42). The present systematic review demonstrated that the miR-200 family members hold crucial regulatory positions in almost every step of BM as regards the most common primary cancers.
The findings of the present systematic review are in agreement with the pertinent literature. Huang et al (20) conducted a meta-analysis on the role of miR-200 family members in cancer patient survival. They revealed that there was a strong association between the miR-200 family member blood and tissue levels and the overall prognosis (20). In another review, Balachandran et al (43) examined the role of the most critical miRs in primary and metastatic brain malignancies, and analyzed the roles of the 10 most crucial miRs. Even though the miR-200 family members were not included in the list of top 10 miRs, they admitted that they could play a significant role, both in primary and metastatic brain lesions (43). Similarly, the miR-200 family members were included among potential diagnostic and therapeutic targets in the management of lung and breast cancer BM (42,44). It is worth mentioning that miR-141 and miR-429 have been found to be associated with malignant glioma development and progression (45).
Given the pivotal role of miR-200 family members in the development of cancer metastasis, it is evident that they may play a critical role in the management of patients with BM. They could act as potential cancer subtype classifiers, diagnostic and drug-response indicators, prognostic markers and therapeutic targets. However, despite the abundance of research in the experimental field, the miR-200 family has not been thoroughly studied in the clinical setting. Currently, there are two ongoing registered trials focusing on the utilization of miR-200 as a potential marker, at least to the best of our knowledge. The clinical trial with registration no. NCT02581098 is investigating the role of miR-200b as a potential barrier to wound healing in diabetic patients, whereas the trial with registration no. NCT03457935 investigates the role of miR in the early detection of pulmonary fibrosis.
To date, the miR-200 family members have been considered mostly for diagnostic or prognostic purposes in relation to metastatic brain cancer. Teplyuk et al (46) recognized that they could differentiate a BM from a malignant glioblastoma based on the cerebrospinal levels of miR-200 family members. Debeb et al (32) examined the association of serum miR-141 levels in 105 patients with breast cancer and reported that high levels were an independent predictor of metastasis-free, progression-free and overall survival. It has been proposed that miR-141 may be used as a biomarker and therapeutic target in the treatment of BCBM (32). In another study, Shao et al (34) reported that the sensitivity and specificity of miR-200a in predicting chemotherapeutic resistance in the brain cancer metastases of patients were as high as 94 and 77%, respectively, and therefore could be used as a treatment response predictor. Venneti et al (47) suggested that miR-200a could distinguish hemangioblastomas from metastatic clear cell renal carcinomas in the central nervous system, particularly in patients with von Hippel-Lindau syndrome. Minn et al (48) demonstrated that the expression of miR-200 family members in patients with gastric adenocarcinoma was an independent predictor of BM (48). There is additional evidence to suggest that members of the miR-200 family may be used in a battery of tests to assess outcome prognosis, including metastatic potential (49,50).
The role of miR-200 family members as therapeutic targets has not been extensively studied. Fu et al (51) reported that NPV-LDE-225 (Erismodegib) inhibited the EMT and self-renewal of glioblastoma-initiating cells, offering a potential therapeutic alternative. The effect was mediated by upregulating E-cadherin and inhibiting N-cadherin, Snail, Slug and Zeb1 levels through the modulation of the levels of miR-200 family members (51). However, neither the use of Erismodegib in the setting of BM nor its safety in clinical practice have been elucidated or documented to date, at least to the best of our knowledge.
The present systematic review is characterized by some limitations which should be mentioned. One of the limitations of the present systematic review was that it was based on experimental animal models using tumor cells growing in cultures. These models may not have the same metastatic potential and properties as in real-life circumstances. Therefore, their results cannot be safely extrapolated into clinical practice. Another limitation of the present systematic review is that it is based on quantitative data. Unfortunately, the gathered evidence is characterized by significant heterogeneity in the adopted methodology and reported outcomes, which prohibits further quantitative analysis. Furthermore, the gathered evidence was characterized by a limited number of clinical studies on the potential role of miR-200 family members in BM diagnosis and management.
In conclusion, members of the miR-200 family play a crucial role in the development of BM. This evidence is derived from heterogeneous experimental research studies. On the other hand, miR-200 family members may play a significant role in the diagnosis, treatment and prognosis assessment of BM. However, there is a significant paucity of clinical studies in the pertinent literature. Therefore, further high-quality clinical studies are required before establishing the clinical role of these regulatory molecules.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
GF, AGB and VEG conceptualized the study. VEG, DAS, IT, PP, EA, AAF and NT analyzed the literature data, and wrote and prepared the draft of the manuscript. DAS and AGB provided critical revisions. All authors contributed to manuscript revision and have read and approved the final version of the manuscript. GF and AGB confirm the authenticity of all the raw data.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had no personal involvement in the reviewing process, or any influence in terms of adjudicating on the final decision, for this article. The other authors declare that they have no competing interests.
References
- 1.Clouston PD, DeAngelis LM, Posner JB. The spectrum of neurological disease in patients with systemic cancer. Ann Neurol. 1992;31:268–273. doi: 10.1002/ana.410310307. [DOI] [PubMed] [Google Scholar]
- 2.Soffietti R, Cornu P, Delattre JY, Grant R, Graus F, Grisold W, Heimans J, Hildebrand J, Hoskin P, Kalljo M, et al. EFNS Guidelines on diagnosis and treatment of brain metastases: Report of an EFNS Task Force. Eur J Neurol. 2006;13:674–681. doi: 10.1111/j.1468-1331.2006.01506.x. [DOI] [PubMed] [Google Scholar]
- 3.Nussbaum ES, Djalilian HR, Cho KH, Hall WA. Brain metastases. Histology, multiplicity, surgery, and survival. Cancer. 1996;78:1781–1788. [PubMed] [Google Scholar]
- 4.Achrol AS, Rennert RC, Anders C, Soffietti R, Ahluwalia MS, Nayak L, Peters S, Arvold ND, Harsh GR, Steeg PS, Chang SD. Brain metastases. Nat Rev Dis Primers. 2019;5(5) doi: 10.1038/s41572-018-0055-y. [DOI] [PubMed] [Google Scholar]
- 5.Boire A, Brastianos PK, Garzia L, Valiente M. Brain metastasis. Nat Rev Cancer. 2020;20:4–11. doi: 10.1038/s41568-019-0220-y. [DOI] [PubMed] [Google Scholar]
- 6.Gavrilovic IT, Posner JB. Brain metastases: Epidemiology and pathophysiology. J Neurooncol. 2005;75:5–14. doi: 10.1007/s11060-004-8093-6. [DOI] [PubMed] [Google Scholar]
- 7.Fidler IJ. The biology of brain metastasis: Challenges for therapy. Cancer J. 2015;21:284–293. doi: 10.1097/PPO.0000000000000126. [DOI] [PubMed] [Google Scholar]
- 8.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 11.Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Sontheimer EJ, Carthew RW. Silence from within: Endogenous siRNAs and miRNAs. Cell. 2005;122:9–12. doi: 10.1016/j.cell.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 13.Barbato S, Solaini G, Fabbri M. MicroRNAs in oncogenesis and tumor suppression. Int Rev Cell Mol Biol. 2017;333:229–268. doi: 10.1016/bs.ircmb.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Frixa T, Donzelli S, Blandino G. Oncogenic MicroRNAs: Key players in malignant transformation. Cancers (Basel) 2015;7:2466–2485. doi: 10.3390/cancers7040904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Zhang Z, Chen F, Hu T, Peng W, Gu Q, Sun Y. The diverse oncogenic and tumor suppressor roles of microRNA-105 in cancer. Front Oncol. 2019;9(518) doi: 10.3389/fonc.2019.00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 17.Zhou K, Liu M, Cao Y. New Insight into microRNA functions in cancer: Oncogene-microRNA-Tumor suppressor gene network. Front Mol Biosci. 2017;4(46) doi: 10.3389/fmolb.2017.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koutsaki M, Libra M, Spandidos DA, Zaravinos A. The miR-200 family in ovarian cancer. Oncotarget. 2017;8:66629–66640. doi: 10.18632/oncotarget.18343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi YC, Yoon S, Jeong Y, Yoon J, Baek K. Regulation of vascular endothelial growth factor signaling by miR-200b. Mol Cells. 2011;32:77–82. doi: 10.1007/s10059-011-1042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang GL, Sun J, Lu Y, Liu Y, Cao H, Zhang H, Calin GA. MiR-200 family and cancer: From a meta-analysis view. Mol Aspects Med. 2019;70:57–71. doi: 10.1016/j.mam.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Humphries B, Yang C. The microRNA-200 family: Small molecules with novel roles in cancer development, progression and therapy. Oncotarget. 2015;6:6472–6498. doi: 10.18632/oncotarget.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langley RR, Fidler IJ. The biology of brain metastasis. Clin Chem. 2013;59:180–189. doi: 10.1373/clinchem.2012.193342. [DOI] [PubMed] [Google Scholar]
- 23.Uhlmann S, Zhang JD, Schwäger A, Mannsperger H, Riazalhosseini Y, Burmester S, Ward A, Korf U, Wiemann S, Sahin O. miR-200bc/429 cluster targets PLCgamma1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene. 2010;29:4297–4306. doi: 10.1038/onc.2010.201. [DOI] [PubMed] [Google Scholar]
- 24.Roybal JD, Zang Y, Ahn YH, Yang Y, Gibbons DL, Baird BN, Alvarez C, Thilaganathan N, Liu DD, Saintigny P, et al. miR-200 Inhibits lung adenocarcinoma cell invasion and metastasis by targeting Flt1/VEGFR1. Mol Cancer Res. 2011;9:25–35. doi: 10.1158/1541-7786.MCR-10-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byun Y, Choi YC, Jeong Y, Lee G, Yoon S, Jeong Y, Yoon J, Baek K. MiR-200c downregulates HIF-1α and inhibits migration of lung cancer cells. Cell Mol Biol Lett. 2019;24(28) doi: 10.1186/s11658-019-0152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 27.Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, Zhang X, Yi X, Dwyer D, Lin W, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5(5241) doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, Thilaganathan N, Du L, Zhang Y, Pertsemlidis A, Kurie JM. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pecot CV, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C, Wu S, Han HD, Shah MY, Rodriguez-Aguayo C, et al. Tumour angiogenesis regulation by the miR-200 family. Nat Commun. 2013;4(2427) doi: 10.1038/ncomms3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gravgaard KH, Lyng MB, Laenkholm AV, Søkilde R, Nielsen BS, Litman T, Ditzel HJ. The miRNA-200 family and miRNA-9 exhibit differential expression in primary versus corresponding metastatic tissue in breast cancer. Breast Cancer Res Treat. 2012;134:207–217. doi: 10.1007/s10549-012-1969-9. [DOI] [PubMed] [Google Scholar]
- 32.Debeb BG, Lacerda L, Anfossi S, Diagaradjane P, Chu K, Bambhroliya A, Huo L, Wei C, Larson RA, Wolfe AR, et al. miR-141-Mediated regulation of brain metastasis from breast cancer. J Natl Cancer Inst. 2016;108(djw026) doi: 10.1093/jnci/djw026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duhachek-Muggy S, Zolkiewska A. ADAM12-L is a direct target of the miR-29 and miR-200 families in breast cancer. BMC Cancer. 2015;15(93) doi: 10.1186/s12885-015-1108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao B, Wang X, Zhang L, Li D, Liu X, Song G, Cao H, Zhu J, Li H. Plasma microRNAs predict chemoresistance in patients with metastatic breast cancer. Technol Cancer Res Treat. 2019;18(1533033819828709) doi: 10.1177/1533033819828709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schickel R, Park SM, Murmann AE, Peter ME. miR-200c regulates induction of apoptosis through CD95 by targeting FAP-1. Mol Cell. 2010;38:908–915. doi: 10.1016/j.molcel.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueller DW, Rehli M, Bosserhoff AK. miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J Invest Dermatol. 2009;129:1740–1751. doi: 10.1038/jid.2008.452. [DOI] [PubMed] [Google Scholar]
- 37.Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 38.Elson-Schwab I, Lorentzen A, Marshall CJ. MicroRNA-200 family members differentially regulate morphological plasticity and mode of melanoma cell invasion. PLoS One. 2010;5(e13176) doi: 10.1371/journal.pone.0013176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong X, Zheng L, Shen J, Zhang D, Xiong M, Zhang Y, He X, Tanyi JL, Yang F, Montone KT, et al. Suppression of MicroRNA 200 family expression by oncogenic KRAS Activation promotes cell survival and epithelial-mesenchymal transition in KRAS-Driven cancer. Mol Cell Biol. 2016;36:2742–2754. doi: 10.1128/MCB.00079-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong X, Yu X, Wen X, Chen L, Gu N. Activation of the LINC00242/miR-141/FOXC1 axis underpins the development of gastric cancer. Cancer Cell Int. 2020;20(272) doi: 10.1186/s12935-020-01369-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Youn YH, Byun HJ, Yoon JH, Park CH, Lee SK. Long Noncoding RNA N-BLR upregulates the migration and invasion of gastric adenocarcinoma. Gut Liver. 2019;13:421–429. doi: 10.5009/gnl18408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanchan RK, Siddiqui JA, Mahapatra S, Batra SK, Nasser MW. microRNAs orchestrate pathophysiology of breast cancer brain metastasis: Advances in therapy. Mol Cancer. 2020;19(29) doi: 10.1186/s12943-020-1140-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balachandran AA, Larcher LM, Chen S, Veedu RN. Therapeutically significant MicroRNAs in primary and metastatic brain malignancies. Cancers (Basel) 2020;12(2534) doi: 10.3390/cancers12092534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahn YH, Ko YH. Diagnostic and therapeutic implications of microRNAs in non-small cell lung cancer. Int J Mol Sci. 2020;21(8782) doi: 10.3390/ijms21228782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng L, Fu J, Ming Y. The miR-200 family: Multiple effects on gliomas. Cancer Manag Res. 2018;10:1987–1992. doi: 10.2147/CMAR.S160945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teplyuk NM, Mollenhauer B, Gabriely G, Giese A, Kim E, Smolsky M, Kim RY, Saria MG, Pastorino S, Kesari S, Krichevsky AM. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol. 2012;14:689–700. doi: 10.1093/neuonc/nos074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venneti S, Boateng LA, Friedman JR, Baldwin DA, Tobias JW, Judkins AR, Mourelatos Z, Lal P. MiRNA-9 and MiRNA-200a distinguish hemangioblastomas from metastatic clear cell renal cell carcinomas in the CNS. Brain Pathol. 2012;22:522–529. doi: 10.1111/j.1750-3639.2011.00551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minn YK, Lee DH, Hyung WJ, Kim JE, Choi J, Yang SH, Song H, Lim BJ, Kim SH. MicroRNA-200 family members and ZEB2 are associated with brain metastasis in gastric adenocarcinoma. Int J Oncol. 2014;45:2403–2410. doi: 10.3892/ijo.2014.2680. [DOI] [PubMed] [Google Scholar]
- 49.Maierthaler M, Benner A, Hoffmeister M, Surowy H, Jansen L, Knebel P, Chang-Claude J, Brenner H, Burwinkel B. Plasma miR-122 and miR-200 family are prognostic markers in colorectal cancer. Int J Cancer. 2017;140:176–187. doi: 10.1002/ijc.30433. [DOI] [PubMed] [Google Scholar]
- 50.Yu C, Wan H, Shan R, Wen W, Li J, Luo D, Wan R. The prognostic value of the MiR-200 family in colorectal cancer: A meta-analysis with 1882 patients. J Cancer. 2019;10:4009–4016. doi: 10.7150/jca.27529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu J, Rodova M, Nanta R, Meeker D, Van Veldhuizen PJ, Srivastava RK, Shankar S. NPV-LDE-225 (Erismodegib) inhibits epithelial mesenchymal transition and self-renewal of glioblastoma initiating cells by regulating miR-21, miR-128, and miR-200. Neuro Oncol. 2013;15:691–706. doi: 10.1093/neuonc/not011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.