Abstract
Caffeine is an important functional component in tea, which has the effect of excitement and nerve stimulation, but excessive intake can cause insomnia and dysphoria. Therefore, the production of tea with low-caffeine content can meet the consumption needs of certain people. Here, in addition to the previous alleles of the tea caffeine synthase (TCS1) gene, a new allele (TCS1h) from tea germplasms was identified. Results of in vitro activity analysis showed that TCS1h had both theobromine synthase (TS) and caffeine synthase (CS) activities. Site-directed mutagenesis experiments of TCS1a, TCS1c, and TCS1h demonstrated that apart from the 225th amino acid residue, the 269th amino acid also determined the CS activity. GUS histochemical analysis and dual-luciferase assay indicated the low promoter activity of TCS1e and TCS1f. In parallel, insertion and deletion mutations in large fragments of alleles and experiments of site-directed mutagenesis identified a key cis-acting element (G-box). Furthermore, it was found that the contents of purine alkaloids were related to the expression of corresponding functional genes and alleles, and the absence or presence and level of gene expression determined the content of purine alkaloids in tea plants to a certain extent. In summary, we concluded TCS1 alleles into three types with different functions and proposed a strategy to effectively enhance low-caffeine tea germplasms in breeding practices. This research provided an applicable technical avenue for accelerating the cultivation of specific low-caffeine tea plants.
Introduction
Tea plant (Camellia sinensis (L.) O. Kuntze) is an economical beverage plant, which has been cultivated in more than 60 countries [1]. Tea, which is processed from young shoots, has become the second most popular beverage after water. Its popularity is closely correlated with its diverse biochemical constituents, including theanine, catechins, and purine alkaloids. Among them, purine alkaloids (3.0%–6.0% dry weight), as the characteristic secondary metabolites of tea, primarily contain caffeine (1,3,7-trimethylxanthine), theobromine (3,7-dimethylxanthine), and occasionally theacrine (1,3,7,9-tetramethyluric acid) with a bitter taste and resistance to some biotic and abiotic stresses [2, 3]. However, the types and concentrations of the above-mentioned purine alkaloids vary greatly among tea plants [2, 4]. Caffeine (2.5%–5.0%) is the main purine alkaloid in the cultivated C. sinensis (L.) O. Kuntze and some C. taliensis (W. W. Smith) Melchior. Only a few special tea plants in ‘cocoa tea’ (CCT) (C. ptilophyll Chang, a caffeine-free tea plant from Guangdong, China) and ‘Kucha’ (i.e. bitter tea) contain high theobromine or theacrine, respectively [5, 6].
Caffeine is an important biochemical component affecting the quality and function of tea. It exhibits not only antioxidant [7] and anti-inflammatory [8] activities, but also protection against various diseases, such as cardiovascular diseases [9], obesity [10], and cancers [11]. However, chronic or excessive caffeine consumption could lead to addiction, insomnia [12], migraines [13], and other side effects [14]. In addition, children, adolescents, pregnant women, and those sensitive to caffeine must limit or reduce their intake to avoid adverse effects [15]. For pregnant women, in particular, prenatal caffeine exposure could affect offspring development, induce cognitive impairment, and increase susceptibility to disease in adulthood [14]. Therefore, the creation of low-caffeine tea (caffeine <1.0%) has attracted extensive attention. Multiple industrial processes such as solvent extraction, supercritical fluid extraction with carbon dioxide (CO2), microbial degradation, and enzymatic degradation were adopted in de-caffeinate [16]. However, breeding low-caffeine tea cultivars remains an efficient and safe method compared with industrial processes; some progress has been made in the breeding of low-caffeine tea plants [17]. A functional marker for caffeine-free tea plants had been developed for low-caffeine breeding, while it needed to be detected by gene sequencing [18]. Moreover, breeding has been greatly hindered because of the difficulties in breeding and poor adaptability of special tea plants, such as CCT. Therefore, investigating the differential enrichment mechanisms of purine alkaloids among various tea plants will facilitate the cultivation of low-caffeine tea cultivars.
The core synthetic pathway of caffeine involves three methylation steps and one de-nucleoside reaction. N-methyltransferases (NMTs) play a key role in regulating the biosynthesis of caffeine in plants [19]. Notably, CsNMT17 (tea caffeine synthase gene, TCS1) is expressed with higher expression levels than other NMT genes, suggesting its predominant roles in caffeine biosynthesis [20]. TCS1 is the most critical NMT enzyme in the caffeine biosynthesis pathway, which catalyzes the methylation of N-3 and N-1 [6, 21]. The first NMT gene to be cloned was TCS1 [22], after which five homologous genes of TCSs (TCS2–6) and seven allelic variations of TCS1 (a, b, c, d, e, f, g) were isolated from tea plants [21, 23]. In addition, TCS1i has been found in a unique tea plant, namely, ‘Hongyacha’ (HYC) [24]. Although tea plants had multiple NMT genes related to caffeine synthesis, many of them had no function or low expression [1, 25, 26]. Several allelic variations with markedly different enzyme activities or expression levels are found in a few specific tea germplasms [21]. NMTs involving purine alkaloid biosynthesis from different genera evolve independently at least two times [27–29]. It made NMTs of different genera less similar and also generated a diversity of variations in TCS1 sequences resulting in diverse function of TCS1. Previous studies already showed that the 225th amino acid residue of TCS1a plays an important role in substrate specificity, and its mutation directly determined whether it had CS activity or not [21, 30]. However, TCS1 alleles are not sufficiently investigated at present, and the expression regulation and genetic mechanism of different TCS1 alleles remain unclear, thereby limiting the effective utilization of specific tea germplasms. For example, in thepractice of crossbreeding, practical difficulties such as parents not meeting the flowering period, incompatibility, and poor adaptability are often encountered. Therefore, finding more tea plants carrying specific TCS1 alleles could enhance the breeding success rate.
In this study, we systematically revealed TCS1 allele variations. Using the TCS1-InDel marker, a novel allele was identified from 673 tea accessions. The CDS sequence of different alleles was isolated to construct the recombinant protein and study the new key active sites of caffeine synthase (CS); the promoter activities of the genes were determined by β-glucuronidase (GUS) staining of stable transgenic Arabidopsis thaliana and the dual-luciferase reporter assay system. By performing site-directed mutagenesis experiments, G-Box was found to be a key cis-acting element affecting the promoter activities of different TCS1 alleles. Based on the breeding practice of low caffeine in the past 10 years, efficiently innovating new germplasms with low caffeine was proposed. These results will promote the understanding of caffeine biosynthesis in tea plants, revise previous conclusions and innovate specific tea germplasms through rare allelic variations.
Results
Screening and analysis of novel alleles of TCS1 in tea resources
Different genetic background of low-caffeine resources were newly discovered in Yunnan and its adjacent region, which indicates that there may be new TCS1 allelic variations in tea resources [31, 32]. To deeply excavate more novel TCS1 alleles, genomic DNA of 673 accessions of tea genetic resources was collected from Yunnan and the National Germplasm Hangzhou Tea Repository in Zhejiang. The previously acquired TCS1-InDel marker [21] was used to find tea plants carrying different rare TCS1 alleles (Fig. S1, see online supplementary material). Consequently, a novel allele of 338 bp, namely, TCS1h, was isolated from a few special tea plants (Tables S1 and S2, see online supplementary material). Until now, combined with previous studies, a total of nine TCS1 alleles with evident sequence differences have been found in natural tea plants.
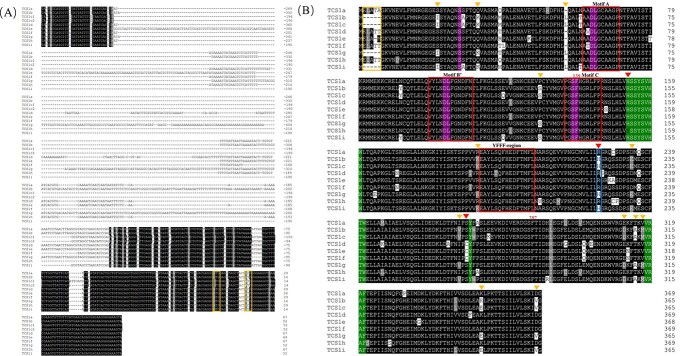
Further analysing the sequence differences, the sequence alignment of the 5′ upstream regulatory region showed that the initiation codon ‘ATG’ of TCS1g, TCS1i, TCS1b, and TCS1c was 15 bp later than that of TCS1h, TCS1a, TCS1d, TCS1e, and TCS1f(Fig. 1A; Table S3; see online supplementary material). The amino acid sequence similarity of TCS1h with TCS1a, TCS1d, TCS1e, and TCS1f was more than 92.12% (Table S4, see online supplementary material). TCS1a, TCS1d, TCS1e, TCS1f, and TCS1h compared to TCS1b, TCS1c, TCS1g, and TCS1i had 13 amino acid residue substitutions. We also found that the amino acid at the 225th position of five alleles, including TCS1a, TCS1d, TCS1e, TCS1f, and TCS1h, was arginine (Arg), whereas the corresponding amino acid of the other four alleles was histidine (His) (Fig. 1B).
Figure 1.
Comparison of TCS1 allele promoter sequences and amino acid sequences. A Comparison of promoter sequences between TCS1 alleles. The yellow box marks the initiation codon (ATG). B Comparison of amino acid sequences of TCS1h and other TCS1s. The proposed SAM-binding motifs (A, B′, and C) and conserved region that are nominated as the ‘YFFF-region’ are shown in red empty boxes. The amino acid residues are marked in a blue empty box, and they played a critical role in substrate recognition. The nominated amino acids in substrate binding are indicated by purple (SAM) and red (methyl acceptor) boxes. The TCS1b, TCS1c, TCS1g, and TCS1i variant sites compared with TCS1a, TCS1d, TCS1e, TCS1f, and TCS1h are indicated by a brownish yellow triangle. The important mutation sites are marked with red triangles. The TCS1b, TCS1c, TCS1g, and TCS1i deletion motif ‘ELATA’ is marked with a yellow empty box.
Functional identification of proteins encoded by different TCS1 alleles
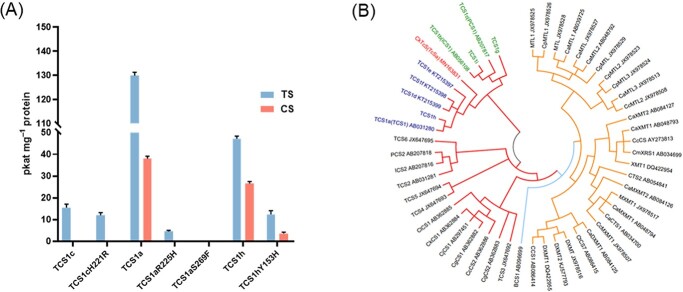
For the particular differences in the TCS1h amino acid sequence, further analysis of enzyme activity was required. In the presence of methyl donor S-adenosyl-L-methionine (SAM), the recombinant enzyme preparation was incubated with the substrate 7-methylxanthine or theobromine. HPLC detection revealed that TCS1h had similar functions to TCS1a, TCS1d, TCS1e, and TCS1f, and it had both theobromine synthase (TS) and CS activities. The CS/TS activity ratio (56.3%) was higher than that of TCS1a (29.3%, Fig. 2A; Fig. S2, see online supplementary material).
Figure 2.
Functional studies of proteins encoded by different TCS1 alleles and comparison of phylogenetic trees of N-methyltransferases. A Activity assay results of TCS1 recombinant proteins (n = 3). B Phylogenetic tree of N-methyltransferase coding region sequences related to purine alkaloids in Camellia (red), Coffea (yellow), and Theobroma (blue) is performed by the MEGA Program. Green, blue, and red groups indicate TS, CS, and TcS (theacrine synthase gene) classes.
Phylogenetic tree analysis was performed using cDNA sequences of NMTs related to caffeine synthesis in Camellia, Coffea, and Theobroma(Fig. 2B). Combining the above differences in enzyme activities, the result showed that NMTs of different species clustered independently, and the different allelic variations of TCS1 and TcS (theacrine synthase gene) [33] in tea plants could be clustered into three discriminative branches: TS, CS, and TcS classes. TCS1b, TCS1c, TCS1g, and TCS1i were clustered in the TS class, and their expressed proteins only had TS activity. TCS1h was clustered with TCS1a, TCS1d, TCS1e, and TCS1f into the CS class, and the expressed proteins of this class had both TS and CS activities.
To explore key sites affecting enzyme activity of TCS1, we performed site-directed mutagenesis on amino acids at the 153th [34], 221th or 225th, and 269th positions. These were the residues affecting the TCS1 activity [21] of the proteins encoded by three TCS1 alleles (TCS1a, TCS1c, and TCS1h) (Fig. 2A). The result showed that the activity of TS decreased significantly, and the activity of CS disappeared after the mutation of Arg225His in TCS1a. The activities of TS and CS decreased to less than 0.5 pkat mg−1 protein through the mutation of the 269th residue from serine (Ser) to proline (Pro) (this mutation did not exist in tea plants). When the 221th residue of TCS1c was mutated from His to Arg, it still did not restore CS activity, and it only had TS activity, indicating that other sites also affected the CS activity of TCS1c. When the 153th residue of TCS1h was mutated from tyrosine (Tyr) to His, the activities of TS and CS decreased significantly, and the ratio of the CS/TS activity was decreased (29.0%). These results indicated that the 225th and 269th amino acids are the key residues that determined whether or not TCS1 has CS activity, and the 153th amino acid significantly affected the TCS1 activity.
Analysis of purine alkaloids content and TCS1 expression in tea germplasms
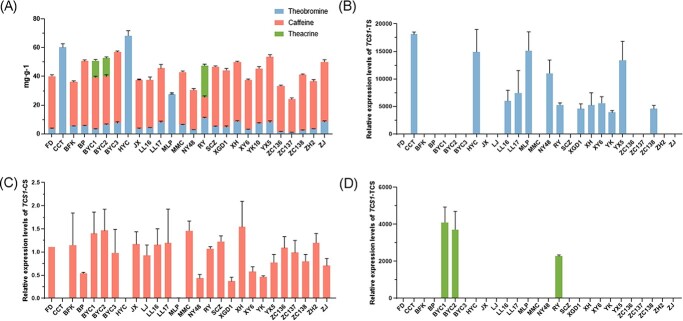
To verify the correlation between the above three different discriminative branches and the content of purine alkaloids in tea plants, we determined purine alkaloids of 27 accessions of tea germplasms with diverse genetic backgrounds by ultra-high performance liquid chromatography (UPLC, Fig. 3A) before clarifying the respective expression level of alleles with distinct functions, and the gene expression level was detected by quantitative real-time PCR (qRT-PCR, Figs 3B, C, and D). These results show that in ‘Malipo’ (MLP) (a wild tea plant from Yunnan, China), CCT, and HYC containing high or only theobromine, the expression level of TCS1 alleles of the CS class was extremely low or not detected, which also proved the specificity of the primers. The tea plants contained theobromine and caffeine, when TCS1 alleles of the CS class were expressed with high levels. In BYC1, BYC2, and RY containing theacrine, the expression level of TcS was high, whereas those that do not contain theacrine were extremely low. Therefore, we conceived that the contents of purine alkaloids in tea plants were related to the expression of corresponding functional genes and alleles, and the absence or presence and level of gene expression determined the content of purine alkaloids in tea plants to a certain extent.
Figure 3.
Expression level analysis of TCS1 and TcS in 27 accession tea plants with diverse genetic background. A Purine alkaloid contents in 27 accessions. B–D Determination of TCS1 allele expression levels in 27 accessions of tea plants by qRT-PCR. Primer pairs TCS1-TS, TCS1-CS, and TcS were used to detect gene whose encoded protein had only theobromine synthase (TS), both TS and caffeine synthase (CS), and TcS (theacrine synthase) activities, respectively.
Activity analysis of TCS1 promoters and identification of the key cis-acting element
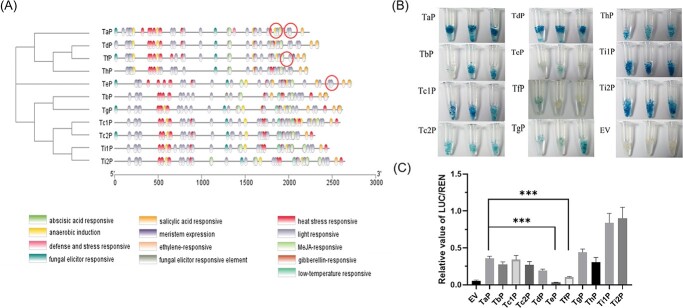
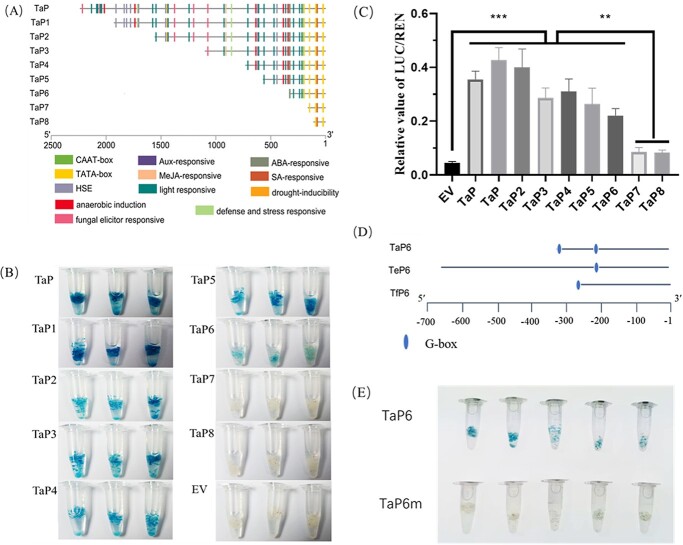
The sequences of different TCS1 promoters are fairly variable (Fig. 1A), which may lead to significant differences in transcription levels. To investigate the mechanism of gene differential expressions, we cloned the promoters of different alleles including TCS1h from specific tea germplasms (Tables S1 and S5, see online supplementary material). Among them, two different TCS1 promoter sequences were amplified from tea plants containing TCS1c and TCS1i alleles. The 5′ end sequences of the 11 different TCS1 alleles (sequences before the start codon ATG) were defined as the responding promoters, namely, TaP, TbP, Tc1P, Tc2P, TdP, TeP, TfP, TgP, ThP, Ti1P, and Ti2P. Although amplified with the same primers, they varied widely in sequences from 2193 to 2715 bp. The variation of their similarity ranged from 78.6% to 98.8% (Table S6, see online supplementary material). It showed the number of different functional cis-acting elements (except for the TATA box and CAAT box) and the distribution on the promoter sequence of different TCS1 alleles (Fig. 4A; Fig. S3, see online supplementary material). Each promoter had elements related to light response. TbP, Tc1P, Tc2P, TgP, Ti1P, and Ti2P had cis-acting elements with low-temperature response, whereas TaP, TdP, TeP, TfP, and ThP did not have it. In addition, a segment of the Tc1P sequence was found at the 3′ end with a large number of elements, whereas Tc2P with high similarity did not have this feature.
Figure 4.
Cis-acting elements and promoter activity analysis of different TCS1 alleles. A Distribution of cis-acting elements across the promoter sequences of different TCS1 alleles. Predictive analysis with online software PlantCARE. B GUS histochemical staining of T3 generation Arabidopsis thaliana transduced with different promoters of TCS1 alleles. Red circles indicate important variations in the light-response element. C Determination of the promoter activity of different TCS1 alleles by dual-luciferase assay (n = 6); the difference in promoter activity was determined by detecting the ratio of firefly luciferase and renilla luciferase activities with an empty vector as the control; ***P < 0.001. EV, empty vector.
To verify promoter activities, we recombined promoters of different TCS1 alleles (greater than 2100 bp) into the PBI1101.3-GUS plus vector and transformed into A. thaliana (Fig. 4B). Based on the histochemical staining results of T3 transgenic A. thaliana, the promoter activities of TCS1e and TCS1f were significantly lower than the remaining promoters. Dual-luciferase assays (Fig. 4C) also demonstrated that TCS1e and TCS1f promoters had low activities. The results also suggest that molecular mechanism of low caffeine in MLP may be for the low transcription level of TCS1f, though TCS1f encoding protein with CS activity.
To further figure out the internal mechanism of different caffeine content in tea plants, based on the ubiquitous TCS1 allele in tea plants, we deleted the TCS1a promoter (TaP) at the 5′ end. As shown in Fig. 5A, the deletion of the TaP 5′ end fragment results in a gradual decrease in the number and types of elements. GUS histochemical analysis and dual-luciferase assay conferred that a segment between −331 and −178 was the basic region for starting TCS1a transcription (Fig. 5B and C). Due to the low activities of promoters, we further compared the promoter sequences of TCS1e and TCS1f with other TCS1 alleles and found that large fragments of insertion or deletion mutations occurred in them, and a G-box (CACGTG) cis-acting element was missing (Fig. 5D). Through the experiments of site-directed mutagenesis, we found that the activity almost disappeared, when a G-box cis-acting element (near the 3′ end) in the 331 bp TCS1a promoter sequence was mutated (Fig. 5E). In summary, we believed that the key cis-element in the promoter had a certain influence on the activity of different promoters, thereby affecting the caffeine content in different tea plants.
Figure 5.
Promoter activity analysis of the key regions and cis-elements of truncate promoters of TCS1a. A Distribution of cis-acting elements on the TCS1a promoter sequence with deletions of different lengths. B GUS histochemical staining of T3 generation of Arabidopsis thaliana transfected with 5′ end fragment deletion of TCS1a promoter. C Determination of the promoter activity of different deleted fragments of the TCS1a promoter (n ≥ 6); ***P < 0.001, **P < 0.01. D Schematic representation of large indel mutations in TCS1e and TCS1f and the absence of a G-Box cis-element. E Comparison of GUS histochemical staining between TaP6 and mutant promoter. TaP6 represents the 331 bp of the TCS1a promoter, TeP6 and TfP6 represent the corresponding promoter sequences of TCS1e and TCS1f, and TaP6m represents a G-Box element near the 3′ end mutated with one base (G-214A). EV indicates empty vector.
Discussion
Some progress had been made in the alleles of TCS1, but it had abundant allelic variation due to the independent and recent rapid evolution mechanism of NMTs in tea plants [29]. Through the deeply functional identification of TCS1 alleles, it showed that the diversity of TCS1 sequence leads to the variations of TCS1 enzyme activity or transcription level, affecting the different distribution patterns of purine alkaloids in tea plants. Previous studies indicated that the mutation of His at the 225th position to Arg in TCS1 would alter the steric hindrance of enzyme binding to substrates [21]. Therefore, the 225th amino acid residue was Arg or His, determining whether or not TCS1 had CS activity. In addition, this study found that the 221th amino acid residue (corresponding to the 225th amino acid residue of TCS1a) of TCS1c with only TS activity was mutated from His to Arg through site-directed mutagenesis, and it indicated that more than one sites affected the CS activity of TCS1c. The important role of the 269th amino acid residue in TCS1 in activity and substrate recognition was demonstrated by the mutation of Ser269Cys [21], but no mutation with a ring structure (such as Pro) was observed at 269th amino acid in tea plants. Through an interesting artificial mutation (Ser269Pro), the nearly disappeared enzymatic activity of TCS1 indicated that the 269th amino acid of TCS1 was the key active site determined by the CS activity.
Tea plants containing TCS1e or TCS1h also contain at least one other allele with the same function. Therefore, it is not feasible to analyse the expression levels of TCS1e or TCS1h by qRT-PCR. Thus, we investigated the difference in promoter activity of different TCS1 alleles to analyse the internal mechanism of the difference in expression levels of different alleles. The low activities of the promoters TCS1e and TCS1f were confirmed by transient and stable transformation. Deletion of the 5′ end fragment essentially identified a basic activity region (between −331 and −178). Sequence alignment revealed that TCS1e and TCS1f had a large InDel mutation, and one G-Box cis-acting element was missing. The activity of the 331 bp TCS1a promoter was nearly abolished during a single-base mutation in the G-box cis-acting element (near the 3′ end). In conclusion, G-Box was a cis-acting element that critically affects the TCS1 promoter activity, which may also lead to significantly lower activities of TCS1e and TCS1f than other alleles. This finding provided an important basis for the follow-up transcriptional regulation studies of TCS1.
Based on cluster analysis and the above-mentioned results, nine TCS1 alleles are classified into three types: type I, the coding proteins only have TS activity and high promoter activities (TCS1b, TCS1c, TCS1g, and TCS1i); type II, the coding proteins have both TS and CS activities, but low promoter activities (TCS1e and TCS1f); and type III, the coding proteins have TS and CS activity, and the activities of promoter are high (TCS1a, TCS1d, and TCS1h). When tea plants contained only TCS1 alleles of types I and II, they are low in caffeine, such as CCT (TCS1c), HYC (TCS1i), and MLP (TCS1b and TCS1f). Although these alleles with the ability to innovate low-caffeine tea germplasms were proposed in 2016 [21], previous cross-breeding practices had shown that low-caffeine breeding could not be achieved by aggregating TS alleles (TCS1b, TCS1c, TCS1g, and TCS1i). For example, no low-caffeine individual appeared in the offspring of ‘Hualing 7’ × CCT-F1(Fig. S4, see online supplementary material) and another three similar hybrid combination populations. Therefore, low-caffeine breeding aims to ensure that the offspring of the hybrid tea plants do not contain the CS allele with a high transcript level. In wild tea plants without caffeine, such as HYC and CCT, only TS alleles were contained. Theobromine and caffeine contents of the offspring HYC × CCT-F1 (the F1 individuals of CCT opening pollination) showed that this hybrid method could obtain low-caffeine offspring (Fig. S5, see online supplementary material). However, F1 offsprings of CCT, HYC, and MLP have poor adaptability, poor growth potential, and low survival rate. Using the specific genetic resource RY (containing one low transcription level allele TCS1e with CS activity) as the parent of low-caffeine breeding, it shows strong adaptability and similar growth potential to cultivated tea plants, and is more conducive to the rapid breeding of low-caffeine cultivars. Therefore, the development and utilization of TCS1e were more important.
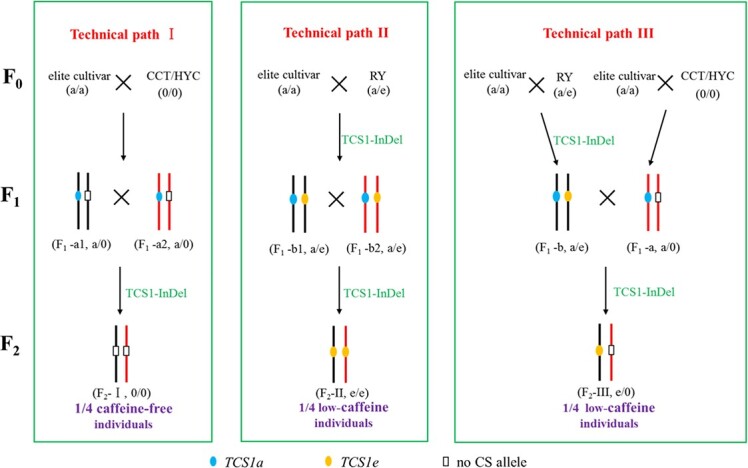
According to the above-mentioned results and breeding practice of the past 10 years, combined with the growth potential and adaptability of the F1 generation, we proposed three high-efficiency breeding strategies with high theobromine and low caffeine (Fig. 6). Path I is the use of wild-specific resources, such as HYC and CCT, which contain only the TS alleles to cross with elite cultivars and obtain F1 offspring with better growth potential. Then, caffeine-free tea plants will be obtained by crossing between F1 offspring. Path II is aggregating TCS1e in RY with lower transcript levels. F1 offspring with better growth potential is obtained by crossing RY with elite cultivar. Path III is the mutual crossing of the two above-mentioned types of F1 offspring, innovating different high-theobromine and low-caffeine tea plants. The TCS1-Indel molecular marker can be used to rapidly screen different TCS1 genotypes of hybrid offspring and assist in the selection of target individuals. Wild tea plants rich in theacrine can also be used to innovate new germplasms with high theacrine and low caffeine [33].
Figure 6.
The strategy for enhancing high-theobromine and low-caffeine tea germplasms.
Materials and methods
Plant materials
A total of 673 tea accessions (Table S7, see online supplementary material) were used for screening rare TCS1 allelic variations, including 200 accessions (primarily C. sinensis (L.) O. Kuntze and C. sinensis var. assamica (Masters) Kitamura) collected from the China National Germplasm Hangzhou Tea Repository (Hangzhou, China) in Zhejiang province and 473 accessions (primarily C. taliensis (W.W. Smith) Melchior, C. sinensis var. assamica (Masters) Kitamura, and the intermediate types of them) collected from Lincang city, Yunnan province, China, were used as experimental materials. A total of 27 typical tea accessions with clear genetic background information, as described in our previous study [35], were used for purine alkaloid determination and specific allele expression analysis. They were collected from their native habitat and are currently maintained in the China National Germplasm Hangzhou Tea Repository (Hangzhou, China).
Detection of purine alkaloids and UPLC conditions
The biochemical samples were ground by using a micropipette grinder (Verder Shanghai Instruments and Equipment Co., Ltd, Shanghai, China) and assayed using UPLC. The content of caffeine, theobromine, and theacrine in the samples was determined, and each sample was independently repeated three times. The pretreatment of the sample and the UPLC measurement conditions were referred to the determination method in our previous study [36].
Screening for novel alleles by using the TCS1-InDel marker
Based on the previous study [21], we designed specific primer pairs for the identification of TCS1 allelic variation: TCS1PInDel-F and TCS1PInDel-R (Table S2, see online supplementary material). PCR amplification was performed using KOD-Plus-Neo (Toyobo, Co., Ltd, Osaka, Japan) with a 25 μL reaction system under the following conditions: 94°C for 2 min; 30 cycles of 94°C denaturation for 10 s, 51°C annealing for 25 s, 72°C extension for 45 s, extension at 72°C for 5 min.
Sequence analysis
Sequence assembly and alignment used DNAMAN (v9.0) and ClustalW with BoxShade in EXPASY to output homologous alignment results. A phylogenetic tree was constructed in MEGA (v7.0) using the Neighbor-joining method and 1000 repeat bootstrap. The gene structure was visualized using Gene Structure Display Server (v2.0). Gene sequence information was visualized using TBtools.
Gene expression analysis by qRT-PCR
Total RNA was extracted using the plant RNA kit (Aidlab Biotechnologies Co., Ltd, Beijing, China) following the manufacturer’s recommendations. The purity and concentration of RNA was certified using agarose gel electrophoresis and a spectrophotometer (Implen, CA, USA). First-strand cDNA was synthesized using the FastKing cDNA First-Strand synthesis kit (Tiangen Biotech Co., Ltd, Beijing, China) according to the manufacturer’s protocols (Table S3, see online supplementary material). Cs18S was used as the reference gene. Table S2 (see online supplementary material) lists the specific primer pairs for qRT-PCR of TCS1-CS, TCS1-TS, and TcS designed according to 3′ end difference. Real-time PCR was similar to that presented in our recent study [33].
Protein purification and activity assay of recombinant TCS1 alleles
The full-length coding region of TCS1h was inserted into the pMAL-c5x vector (NEB, Beijing, China). The recombinant expression vector was introduced into BL21 (DE3) pLys cells (TransGen Biotechnologies Co., Beijing, China). The target protein was purified by using the fusion protein label (MBP) on the expression vector, and an empty load was used as the negative control. The fusion expression of the target gene in Escherichia coli, the detection of the target protein, and the quantification of the purified protein were investigated as described by Jin et al. [21]. Each protein was determined in triplicate, and the activity was determined by HPLC.
Promoter activity assay of TCS1
The promoter activity assay consists of the stable A. thaliana transformation and tobacco dual-luciferase reporter assay. The TCS1 promoter sequence was recombined into PBI1101.3-GUS plus vector to construct a GUS expression vector. The successfully ligated binary expression plasmids were transferred into EHA105 competent cells (BioMed, BC303–01) using the freeze–thaw method. T3 generation of transgenic positive plants was obtained after A. thaliana inflorescence infection, and then GUS histochemical staining was performed. These GUS staining experiments were conducted on the basis of the instructions of the GUS staining kit (Gcloning Co., Ltd, Beijing, China). The TCS1 promoter sequence was recombined into the pGreenII0800-Luc vector, and then the plasmids verified by sequencing were transformed into GV3101 (pSoup-19). The dual-luciferase activity assay was in accordance with our recent study [33].
Statistical analyses
Data are presented as mean ± standard deviation (SD) for the relative expression of TCS1 and relative value of the ratio of firefly luciferase and Renilla luciferase activities. Statistical significance of differences among groups was determined by Student’s t-test using SPSS (Chicago, IL, USA). Graphing was performed using GraphPad Prism 8.
Supplementary Material
Contributor Information
Yi Wang, Key Laboratory of Biology, Genetics and Breeding of Special Economic Animals and Plants, Ministry of Agriculture and Rural Affairs; Tea Research Institute of the Chinese Academy of Agricultural Sciences, Hangzhou 310008, China.
Yu-Fei Liu, Key Laboratory of Biology, Genetics and Breeding of Special Economic Animals and Plants, Ministry of Agriculture and Rural Affairs; Tea Research Institute of the Chinese Academy of Agricultural Sciences, Hangzhou 310008, China; Yunnan Provincial Key Laboratory of Tea Science, Tea Research Institute, Yunnan Academy of Agricultural Sciences, 2 Jingnan Road, Menghai, Yunnan 666201, China.
Meng-Yuan Wei, Key Laboratory of Biology, Genetics and Breeding of Special Economic Animals and Plants, Ministry of Agriculture and Rural Affairs; Tea Research Institute of the Chinese Academy of Agricultural Sciences, Hangzhou 310008, China.
Chen-Yu Zhang, Key Laboratory of Biology, Genetics and Breeding of Special Economic Animals and Plants, Ministry of Agriculture and Rural Affairs; Tea Research Institute of the Chinese Academy of Agricultural Sciences, Hangzhou 310008, China.
Jie-Dan Chen, Key Laboratory of Biology, Genetics and Breeding of Special Economic Animals and Plants, Ministry of Agriculture and Rural Affairs; Tea Research Institute of the Chinese Academy of Agricultural Sciences, Hangzhou 310008, China.
Ming-Zhe Yao, Key Laboratory of Biology, Genetics and Breeding of Special Economic Animals and Plants, Ministry of Agriculture and Rural Affairs; Tea Research Institute of the Chinese Academy of Agricultural Sciences, Hangzhou 310008, China.
Liang Chen, Key Laboratory of Biology, Genetics and Breeding of Special Economic Animals and Plants, Ministry of Agriculture and Rural Affairs; Tea Research Institute of the Chinese Academy of Agricultural Sciences, Hangzhou 310008, China.
Ji-Qiang Jin, Key Laboratory of Biology, Genetics and Breeding of Special Economic Animals and Plants, Ministry of Agriculture and Rural Affairs; Tea Research Institute of the Chinese Academy of Agricultural Sciences, Hangzhou 310008, China.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31670685), Zhejiang Provincial Natural Science Foundation of China (LZ22C160008), Key Scientific and Technological Grant of Zhejiang for Breeding New Agricultural Varieties (2021C02067) to J.-Q.J., the Chinese Academy of Agricultural Sciences through the Agricultural Science and Technology Innovation Program (CAASASTIP-2021-TRICAAS), the China Agricultural Research System of MOF and MARA (CARS-19) to L.C., the Zhejiang Provincial Natural Science Foundation of China (LQ20C160010) to J.-D.C. The authors thank Dr Penghui Li of Anhui Agricultural University, Professor Kang Wei of TRICAAS, and three anonymous reviewers for their valuable suggestions and critical peer review.
Author contributions
J.-Q.J. and L.C. conceived and designed the research. J.-Q.J., Y.-F.L., M.-Y.W., Y.W., and C.-Y.Z. performed the experiments. Y.W. and J.-D.C. analysed the data. Y.W., J.-Q.J. and C.-Y.Z. wrote the manuscript, M.-Z.Y. and L.C. edited and contributed to the content.
Data availability
All data supporting the findings of this study are available within the paper and within its supplementary materials published online.
Conflict of interest
All authors declare that they have no conflict of interest.
Supplementary data
Supplementary data is available at Horticulture Research online.
References
- 1. Xia EH, Zhang HB, Sheng Jet al. . The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol Plant. 2017;10:866–77. [DOI] [PubMed] [Google Scholar]
- 2. Ashihara H, Sano H, Crozier A. Caffeine and related purine alkaloids: biosynthesis, catabolism, function and genetic engineering. Phytochemistry. 2008;69:841–56. [DOI] [PubMed] [Google Scholar]
- 3. Zhang SR, Jin JQ, Chen JDet al. . Purine alkaloids in tea plants: component, biosynthetic mechanism and genetic variation. Bev Plant Res. 2022;2:13. [Google Scholar]
- 4. Ashihara H, Kato M, Crozier A. Distribution, biosynthesis and catabolism of methylxanthines in plants. Handb Exp Pharmacol. 2011;200:11–31. [DOI] [PubMed] [Google Scholar]
- 5. Jin JQ, Zhou CY, Ma CLet al. . Identification on purine alkaloids of representative tea germplasms in China. J Plant G Res 2014;15:279–85. [Google Scholar]
- 6. Ashihara H, Kato M, Ye CX. Biosynthesis and metabolism of purine alkaloids in leaves of cocoa tea (camellia ptilophylla). J Plant Res. 1998;111:599–604. [Google Scholar]
- 7. Machado ML, Arantes LP, da Silveira TLet al. . Ilex paraguariensis extract provides increased resistance against oxidative stress and protection against amyloid beta-induced toxicity compared to caffeine in Caenorhabditis elegans. Nutr Neurosci. 2021;24:697–709. [DOI] [PubMed] [Google Scholar]
- 8. Madeira MH, Boia R, Ambrósio AFet al. . Having a coffee break: the impact of caffeine consumption on microglia-mediated inflammation in neurodegenerative diseases. Mediat Inflamm. 2017;2017:4761081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Said MA, Vegte YJ, Verweij Net al. . Associations of observational and genetically determined caffeine intake with coronary artery disease and diabetes mellitus. J Am Heart Assoc. 2020;9:e016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tabrizi R, Saneei P, Lankarani KBet al. . The effects of caffeine intake on weight loss: a systematic review and dos-response meta-analysis of randomized controlled trials. Crit Rev Food Sci Nutr. 2019;59:2688–96. [DOI] [PubMed] [Google Scholar]
- 11. Tonkaboni A, Lotfibakhshaiesh N, Danesh Pet al. . Evaluation of inhibitory effects of caffeine on human carcinoma cells. Nutr Cancer. 2021;73:1998–2002. [DOI] [PubMed] [Google Scholar]
- 12. Panagiotou M, Meijer M, Meijer JHet al. . Effects of chronic caffeine consumption on sleep and the sleep electroencephalogram in mice. J Psychopharmacol. 2019;33:122–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alstadhaug KB, Ofte HK, Müller KIet al. . Sudden caffeine withdrawal triggers migraine-a randomized controlled trial. Front Neurol. 2020;11:1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saimaiti A, Zhou DD, Li Jet al. . Dietary sources, health benefits, and risks of caffeine. Crit Rev Food Sci Nutr. 2022;May 16. [DOI] [PubMed] [Google Scholar]
- 15. Dam RM, Hu FB, Willett WC. Coffee, caffeine, and health. N Engl J Med. 2020;383:369–78. [DOI] [PubMed] [Google Scholar]
- 16. Xu YQ, Yin JF, Yuan HBet al. . A review on the technique of tea decaffeination. J Tea Sci. 2008;28. [Google Scholar]
- 17. Ogino A, Tanaka J, Taniguchi Fet al. . Detection and characterization of caffeine-less tea plants originated from interspecific hybridization. Breed Sci. 2009;59:277–83. [Google Scholar]
- 18. Ogino A, Taniguchi F, Yoshida Ket al. . A new DNA marker CafLess-TCS1 for selection of caffeine-less tea plants. Breed Sci. 2019;69:393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kato M, Kitao N, Ishida Met al. . Expression for caffeine biosynthesis and related enzymes in Camellia sinensis. Z Naturforsch C J Biosci. 2010;65:245–56. [DOI] [PubMed] [Google Scholar]
- 20. Zhang Y, Fu J, Zhou Qet al. . Metabolite profiling and transcriptome analysis revealed the conserved transcriptional regulation mechanism of caffeine biosynthesis in tea and coffee plants. J Agric Food Chem. 2022;70:3239–51. [DOI] [PubMed] [Google Scholar]
- 21. Jin JQ, Yao MZ, Ma CLet al. . Natural allelic variations of TCS1 play a crucial role in caffeine biosynthesis of tea plant and its related species. Plant Physiol Biochem. 2016;100:18–26. [DOI] [PubMed] [Google Scholar]
- 22. Kato M, Mizuno K, Crozier Aet al. . Caffeine synthase gene from tea leaves. Nature. 2000;406:956–7. [DOI] [PubMed] [Google Scholar]
- 23. Liu YF, JIin JQ, Yao MZet al. . Screening, cloning and functional research of the rare allelic variation of caffeine synthase gene (TCS1g) in tea plants. Sci Agr Sin. 2019;52:1772–83. [Google Scholar]
- 24. Jin JQ, Chai YF, Liu YFet al. . Hongyacha, a naturally caffeine-free tea plant from Fujian, China. J Agric Food Chem. 2018;66:11311–9. [DOI] [PubMed] [Google Scholar]
- 25. Yoneyama N, Morimoto H, Ye CXet al. . Substrate specificity of N-methyltransferase involved in purine alkaloids synthesis is dependent upon one amino acid residue of the enzyme. Mol Gen Genomics. 2006;275:125–35. [DOI] [PubMed] [Google Scholar]
- 26. Jin JQ, Yao MZ, Ma CLet al. . Cloning and sequence analysis of the N-methyltransferase gene family involving in caffeine biosynthesis of tea plant. J Tea Sci. 2014;34:188–94. [Google Scholar]
- 27. Ishida M, Kitao N, Mizuno Ket al. . Occurrence of theobromine synthase genes in purine alkaloid-free species of camellia plants. Planta. 2009;229:559–68. [DOI] [PubMed] [Google Scholar]
- 28. Denoeud F, Carretero-Paulet L, Dereeper Aet al. . The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science. 2014;345:1181–4. [DOI] [PubMed] [Google Scholar]
- 29. Pichersky E, Lewinsohn E. Convergent evolution in plant specialized metabolism. Annu Rev Plant Biol 2011;62:549–66. [DOI] [PubMed] [Google Scholar]
- 30. Jin L, Bhuiya MW, Li Met al. . Metabolic engineering of Saccharomyces cerevisiae for caffeine and theobromine production. PLoS One. 2014;9:e105368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhu B, Chen LB, Lu Met al. . Caffeine content and related gene expression: novel insight into caffeine metabolism in camellia plants containing low, normal, and high caffeine concentrations. J Agric Food Chem. 2019;67:3400–11. [DOI] [PubMed] [Google Scholar]
- 32. Teng J, Yan C, Zeng Wet al. . Purification and characterization of theobromine synthase in a theobromine-enriched wild tea plant (camellia gymnogyna Chang) from Dayao mountain, China. Food Chem. 2020;311:125875. [DOI] [PubMed] [Google Scholar]
- 33. Zhong H, Wang Y, Qu FRet al. . A novel TcS allele conferring the high-theacrine and low-caffeine traits and having potential use in tea plant breeding. Hortic Res. 2022;9:uhac191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jin JQ, Yao MZ, Ma CLet al. . Association mapping of caffeine content with TCS1 in tea plant and its related species. Plant Physiol Biochem. 2016;105:251–9. [DOI] [PubMed] [Google Scholar]
- 35. Jin JQ, Dai WD, Zhang CYet al. . Genetic, morphological, and chemical discrepancies between Camellia sinensis (L.) O. Kuntze and its close relatives. J Food Compost Anal. 2022;108:104417. [Google Scholar]
- 36. Liu QS, QU F, WEI Met al. . The genetic variation of the chemical components of the ′Jinxuan′ × ′Zijuan′ F1 segregating population based on UPLC. J Tea Sci. 2022;42:29–40. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and within its supplementary materials published online.