In the January 2015 issue of PNAS, He et al. reported the isolation and characterization of the first Saccharibacteria (formerly known as TM7) strain from the human oral cavity (1). Named Nanosynbacter lyticus TM7x—this first cultured representative of Saccharibacteria displays an intriguing biology: It has a small cell size (200 to 300 nm in diameter) with a reduced genome and enjoys an obligate epiparasitic lifestyle by living on the surface of its bacterial host. Later work from the Banfield group showed that Saccharibacteria belongs to the candidate phyla radiation (CPR), a large bacterial linage with over 70 phyla accounting for more than a quarter of microbial diversity with predicted symbiotic lifestyle similar to that of TM7x (2–4). The discovery of CPR organisms has greatly expanded the bacterial tree of life (5); however, we still have only a limited number of cultured representatives due to their recalcitrance to conventional cultivation. This severely limits exploration of the biology and ecological roles of these mysterious bacteria. Expanding cultured representatives of CPR organisms for in-depth study is essential for the illumination of this so-called "microbial dark matter."
In the current issue of PNAS, Xie et al. (6) report the isolation of the first insect-associated Saccharibacteria, named Leucosynbacter cicadicola TM7i (TM7i), achieved by capturing in situ Saccharibacteria–host association guided by epicPCR (Emulsion, Paired Isolation, and Concatenation PCR). The study offers a high-throughput method for identifying candidate host bacteria, elevating our ability to cultivate CPR organisms. Furthermore, using high-resolution live cell imaging, the study also provides strong evidence demonstrating the crucial role of the CPR-encoded type IV pili (T4P) in its symbiotic lifestyle.
EpicPCR-Assisted Identification of TM7 Host Bacteria
One of the critical challenges in studying Saccharibacteria and other CPR bacteria is in obtaining stable cultured representatives to allow in-depth physiological and pathological characterization in the laboratory. Culture-independent metagenomic data and imaging analysis of natural samples indicate that Saccharibacteria and other CPR are submicron bacteria with reduced genomes and a likely symbiotic lifestyle (7–9). The symbiotic nature of these nanosized bacteria makes it challenging, if not impossible to isolate them as a pure culture for laboratory study, since so far, the cultivated representatives of Saccharibacteria can only persist and propagate when cocultured with their host bacteria (10). Furthermore, except for Mycolasynbacter amalyticus JR1, which is a cultured representative of environmental Saccharibacteria with lytic ability against its bacterial host (11), all the cultivated Saccharibacteria strains are derived from the human oral cavity.
The most commonly used approach in isolating Saccharibacteria is the “baiting” method (10), which is based on the initial observation that free-floating TM7x cells can attach to and infect naive host bacteria (12). In this method, a panel of potential host bacteria is chosen based on their co-occurrence with Saccharibacteria within the same microbial sample, and infected with Saccharibacteria cells which are collected by filtering the sample through a submicron filter paper. In the presence of suitable hosts, Saccharibacteria will initiate a symbiotic interaction and eventually form stable binary coculture with their host bacteria, or, for virulent TM7 strains with a lytic ability against their host, host bacteria can be periodically added to maintain the coculture (10). Attention has been given to Actinobacteria when choosing putative host bacteria in the baiting method based on the observation that all the isolated Saccharibacteria strains prefer Actinobacteria as hosts (13) and microbial association data from metagenomic analyses (7). However, limiting host bacterial candidates to Actinobacteria is likely biased and could miss the true host bacteria for some of the Saccharibacteria species. Furthermore, without spatial proximity information, the selection of putative host bacteria for baiting is neither targeted nor efficient.
Cross et al. developed a reverse genomics approach that uses genomic information of Saccharibacteria to engineer antibodies to immunolabel and isolate Saccharibacteria together with its physically associated host bacteria via fluorescence-activated cell sorting (14). This approach allows more effective and targeted isolation of Saccharibacteria strains. However, it demands bioinformatics capability and involves multiple steps including antibody synthesis and flow cytometry.
In the article published in this PNAS issue, taking advantage of the close physical interaction between Saccharibacteria and their host bacteria as a result of symbiosis, Xie et al. present a high-throughput approach to identify Saccharibacteria–host bacterial candidates by detecting the presence of Saccharibacteria–host bacterial associations within a community. This was achieved by repurposing epicPCR, which was originally developed for linking functional genes with phylogenetic markers, to capture in situ Saccharibacteria–host physical associations (15). Identifying potential host bacteria with intimate physical interaction with Saccharibacteria revealed by epicPCR further directs the effort toward isolating these candidate hosts, which is less challenging compared with the direct isolation of Saccharibacteria–host symbionts. Using this method, they identify and isolate the candidate host bacterium, Leucobacter aridicollis J1 from Cicadae Periostracum–associated microbiome. By further employing the baiting method using L. aridicollis J1 as “bait,” they successfully isolate the first insect-associated TM7, named L. cicadicola TM7i.
So far, Saccharibacteria is the only phylum among the CPR group with cultivated representatives that can be stably maintained in laboratory conditions. Increasing evidence from culture-independent studies indicates that symbiotic, particularly episymbiotic, lifestyle is likely widespread among CPR bacteria (7). Thus, combined with the established baiting method, the epicPCR-assisted approach for identifying candidate host bacteria for Saccharibacteria in situ will enable the isolation of more Saccharibacteria from diverse environments. It will also help to culture other CPR organisms, particularly those that share a similar episymbiotic lifestyle to TM7x for in-depth study of their physiology, lifestyle, and ecological role, as well as their impact on host health and diseases for those mammalian-associated CPR bacteria.
High-Resolution Live Cell Imaging Reveals T4P-Mediated Symbiotic Association between TM7 and Host Bacteria
One of the fundamental questions related to the episymbiotic nature of Saccharibacteria is how they achieve initial contact with the host bacteria, eventually leading to the establishment of the symbiotic relationship. Direct visualization of the physical associations between CPR organisms and their host bacteria in natural environments using high-resolution microscopy has frequently revealed the presence of pili-like structures on CPR bacterial surface (7, 8). Meanwhile, comparative genomic analysis revealed well-conserved T4P-encoding gene clusters associated with Saccharibacteria and other CPR organisms (9, 16). T4P are bacterial surface-associated dynamic filamentous appendages with diverse functions, including DNA uptake, twitching motility, and cellular adherence among others (17). T4P encoded by CPR have long been suspected of mediating their physical interaction with host bacteria (7, 8). However, due to the symbiotic lifestyle, no effective genetic tools have been developed thus far for CPR, and the role of T4P in symbiosis cannot be confirmed using a conventional mutagenesis-based approach. Meanwhile, snapshots of high-resolution imaging cannot provide convincing evidence of the involvement of T4P in the dynamic physical association process. Thus, the role of T4P in mediating symbiotic interaction remains to be determined.
Xie et al. present a high throughput approach to identify Saccharibacteria host bacterial candidates by detecting the presence of Saccharibacteria-host bacterial associations within a community.
In this article, Xie et al. first reveal the presence of pili-like appendages on the surface of TM7i by transmission electron microscopy. Using high-resolution real-time live cell imaging, they further demonstrate that the presence of host bacteria significantly triggers the movement of TM7i, while the addition of quercetin, a PilB inhibitor that prevents T4P extrusion, greatly inhibits TM7i twitching motility. And the inhibited motility of TM7i was restored upon quercetin removal.
The more convincing data in supporting the T4P-enabled twitching motility in TM7i come from their nanometer-resolution fluorescence imaging of TM7 symbionts using structured illumination microscopy. The imaging directly captures TM7i’s pili in motion, showing the active extrusion and retraction of pili accompanied by the active translocation of TM7i cells. Furthermore, they interrogate the effect of T4P-dependent twitching motility on the episymbiotic lifestyle of TM7i. Using long-term microscopy monitoring, they record the proliferation of TM7i when physically associated with its bacterial host. The addition of quercetin suppresses the motility and inhibits the propagation of TM7i cells as a result of drastically reduced host-associated TM7i. This high-resolution real-time imaging provides compelling data emphasizing the crucial role of TM7i-encoded T4P in mediating its initial physical association with host bacteria and impacting its episymbiotic lifestyle.
While the most definitive evidence needs to come from genetic studies when effective genetic tools are available for Saccharibacteria, the high-resolution live cell imaging data presented by Xie et al. provide thus far the most convincing data on the critical role of T4P in Saccharibacteria’s symbiotic lifestyle. Meanwhile, the finding on the role of Saccharibacteria T4P may well apply to other CPR organisms considering their well-conserved T4P and symbiotic lifestyle similar to that of Saccharibacteria (9, 16).
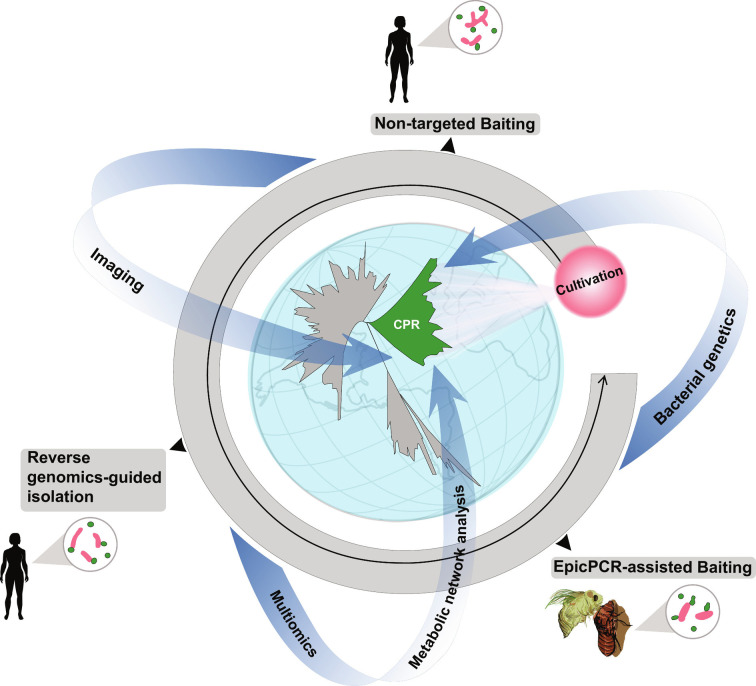
Dubbed "microbial dark matter", CPR organisms are omnipresent yet remain poorly understood. Effective isolation methods as described by Xie et al. will greatly facilitate the cultivation of CPR organisms from diverse environments for comprehensive study under well-controlled laboratory conditions. Meanwhile, high-resolution imaging techniques, such as electron microscopy and real-time live cell imaging (6), combined with bioinformatics-based metabolic network analysis (18) and multiomics studies (19, 20) will help shed light on the intricate symbiotic relationship between CPR and their hosts. More effort should be made to develop genetic tools that can be applied to CPR for a detailed mechanistic understanding of their unique biology at the gene level (Fig. 1).
Fig. 1.
Expanding cultured representatives for studying CPR organisms. Effective isolation methods for targeted cultivation are critical in CPR research. The application of high-resolution microscopies, such as electron microscopy and real-time live cell imaging, combined with bioinformatics-based metabolic network analysis and multiomics, as well as bacterial genetics, will help shed light on the unique biology and the ecological impact of these omnipresent "microbial dark matter".
Acknowledgments
I would like to express my gratitude to Dr. Susan R. Rittling for her thoughtful editing of the manuscript. This work was supported by the National Institute of Dental and Craniofacial Research of the NIH under awards 1R01DE023810 and 1R01DE030943.
Author contributions
X.H. wrote the paper.
Competing interest
The author declares no competing interest.
Footnotes
See companion article, “Type IV pili trigger episymbiotic association of Saccharibacteria with its bacterial host,” 10.1073/pnas.2215990119.
References
- 1.He X., et al. , Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc. Natl. Acad. Sci. U.S.A. 112, 244–249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hug L. A., et al. , A new view of the tree of life. Nat. Microbiol. 1, 16048 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Brown C. T., et al. , Unusual biology across a group comprising more than 15% of domain bacteria. Nature. 523, 208–211 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Castelle C. J., Banfield J. F., Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell. 172, 1181–1197 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Hug L. A., et al. , A new view of the tree of life. Nat. Microbiol. 1, 16048 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Xie B., et al. , Type IV pili trigger episymbiotic association of Saccharibacteria with its bacterial host. Proc. Natl. Acad. Sci. U.S.A. 119, e2215990119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He C., et al. , Genome-resolved metagenomics reveals site-specific diversity of episymbiotic CPR bacteria and DPANN archaea in groundwater ecosystems. Nat. Microbiol. 6, 354–365 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luef B., et al. , Diverse uncultivated ultra-small bacterial cells in groundwater. Nat. Commun. 6, 6372 (2015). [DOI] [PubMed] [Google Scholar]
- 9.McLean J. S., et al. , Acquisition and adaptation of ultra-small parasitic reduced genome bacteria to mammalian hosts. Cell Rep. 32, 107939 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bor B., et al. , Insights obtained by culturing Saccharibacteria with their bacterial hosts. J. Dent. Res. 99, 685–694 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batinovic S., et al. , Cocultivation of an ultrasmall environmental parasitic bacterium with lytic ability against bacteria associated with wastewater foams. Nat. Microbiol. 6, 703–711 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Bor B., et al. , Rapid evolution of decreased host susceptibility drives a stable relationship between ultrasmall parasite TM7x and its bacterial host. Proc. Natl. Acad. Sci. U.S.A. 115, 12277–12282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nie J., et al. , Strain-level variation and diverse host bacterial responses in episymbiotic Saccharibacteria. mSystems 7, e0148821 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cross K. L., et al. , Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat. Biotechnol. 37, 1314–1321 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spencer S. J., et al. , Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. ISME J. 10, 427–436 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meheust R., et al. , The distinction of CPR bacteria from other bacteria based on protein family content. Nat. Commun. 10, 4173 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craig L., Forest K. T., Maier B., Type IV pili: Dynamics, biophysics and functional consequences. Nat. Rev. Microbiol. 17, 429–440 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Bernstein D. B., Dewhirst F. E., Segre D., Metabolic network percolation quantifies biosynthetic capabilities across the human oral microbiome. Elife 8, e39733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendrickson E. L., et al. , Transcriptome of epibiont Saccharibacteria nanosynbacter lyticus strain TM7x during the establishment of symbiosis. J. Bacteriol. 204, e0011222 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giannone R. J., et al. , Proteomic characterization of cellular and molecular processes that enable the Nanoarchaeum equitans–Ignicoccus hospitalis relationship. PLoS One 6, e22942 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]