Summary
Microglia are the brain’s resident immune cells, performing surveillance to promote homeostasis and healthy functioning. While microglial chemical signaling is well-studied, mechanical cues regulating their function are less well-understood. Here, we investigate the role of the mechanosensitive ion channel Piezo1 in microglia migration, pro-inflammatory cytokine production, and stiffness sensing. In Piezo1 knockout transgenic mice, we demonstrated the functional expression of Piezo1 in microglia and identified genes whose expression was consequently affected. Functional assays revealed that Piezo1 deficiency in microglia enhanced migration toward amyloid β-protein, and decreased levels of pro-inflammatory cytokines produced upon stimulation by lipopolysaccharide, both in vitro and in vivo. The phenomenon could be mimicked or reversed chemically using a Piezo1-specific agonist or antagonist. Finally, we also showed that Piezo1 mediated the effect of substrate stiffness-induced migration and cytokine expression. Altogether, we show that Piezo1 is an important molecular mediator for microglia, its activation modulating microglial migration and immune responses.
Subject areas: Molecular neuroscience, Immunology, Cell biology
Graphical abstract
Highlights
-
•
Piezo1 is a highly expressed mechanosensitive ion channel in microglia
-
•
Piezo1 regulates microglia migration both in vivo and in vitro
-
•
Piezo1 regulates microglia immune response challenged by LPS both in vivo and in vitro
-
•
Microglial Piezo1 modulates stiffness-induced microglia migration and immune response
Molecular neuroscience; Immunology; Cell biology
Introduction
Microglia, as the resident innate immune cells of the CNS, play pivotal roles in maintaining brain homeostasis, pathogen defense, and repairing injuries.1 Microglia can recognize pathogens via pathogen-associated molecular patterns (PAMPs) on their surface2 and migrate to the sites of damage along a chemo-attractant gradient.3 They are able to secrete chemicals, e.g., inflammatory cytokines4 and neurotrophic factors5 in order to protect the brain. In addition to responding to biochemical stimuli, physical cues including tissue stiffness and mechanical stimulation are also thought to contribute to microglial activity.6,7,8 Previous studies have shown that stiff surgical implants in the CNS led to a localized foreign body reaction (FBR) by enhancing inflammatory activation of glial cells.7 Setting aside the artificial manipulation of CNS microenvironmental stiffness, brain tissue is itself mechanically heterogeneous,9 and these mechanical properties may alter with age10 or pathological conditions.11,12,13,14 Thus, microglia are exposed to varying mechanical conditions through their life cycle, but little is known about molecular mechanism how these stimuli may modulate microglial activity.
A growing body of evidence implicates the roles of the cytoskeleton, cell adhesion and Rho signaling in the process of mechanosensation and mechanotransduction.15 However, located in lipid bilayers of cell membranes, mechanically gated ion channels serve as primary mediators of cellular mechanosensation, and merit deeper examination as well.16 Upon an increase in membrane tension, mechanically activated ion channels undergo conformational changes and allow the flow of ions across the membrane, sensing and transducing external physical stimuli into electrochemical activity that further influences cell signaling and behavior.17,18 Of particular note is Piezo1, a mechanosensitive cation ion channel that detects mechanical forces with high sensitivity and broad dynamic range.19 Piezo1 has been reported to be expressed at high levels in endothelial cells and microglia of the brain, according to previous RNA sequencing results.20 Substantial recent research reveals that Piezo1 transduces mechanical cues to cells21 and is involved in many physiological and pathological processes.22,23,24,25,26,27 Therefore, Piezo1 is a potential mechanosensor and may be involved in coordinating the functions of microglia. In light of this evidence, we hypothesized that Piezo1 may play important role in the functioning of microglia.
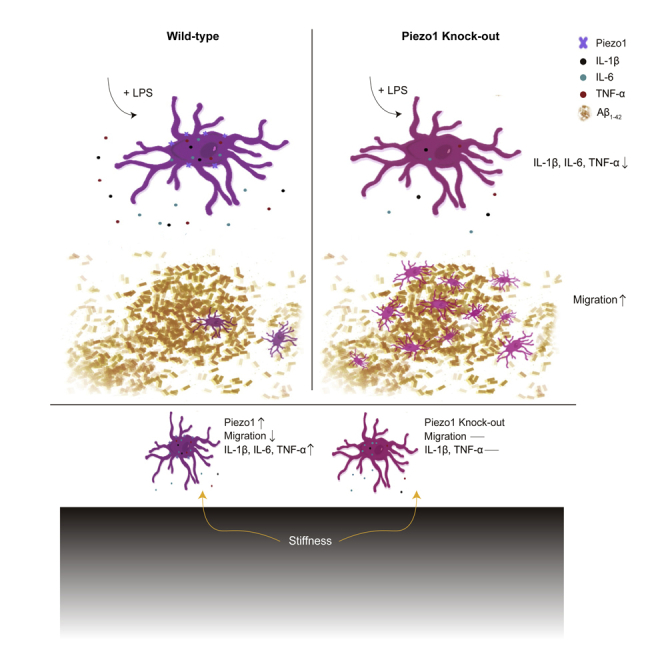
In the present study, microglia from mouse brains, in vivo and ex vivo, and a mouse microglial cell line were used as models for examining Piezo1 expression and its role in microglial functions. RT-PCR analysis showed that Piezo1 was expressed at high levels in primary microglia compared to other mechanosensitive ion channels. Gene clusters affected by the conditional knockout (KO) of Piezo1 in microglia were identified by RNA sequencing (RNAseq) and were found to include genes associated with cell motility and extracellular matrix composition. Functional analyses confirmed that Piezo1 deficiency in microglia resulted in enhanced migration ability toward Aβ1-42, a major component of Alzheimer’s Disease (AD) plaques. Moreover, activation of Piezo1 could suppress the migration of microglia, while blockade of Piezo1 enhanced it. We also found that microglia lacking Piezo1 showed decreased pro-inflammatory cytokine production (IL-1β, IL-6, and TNF-α) when challenged by lipopolysaccharide (LPS). Activation of Piezo1 by Yoda1, a Piezo1-specific agonist, increased the production of these pro-inflammatory cytokines by microglia when stimulated by LPS. Variation in microglial substrate stiffness was mimicked in vitro through modifiable gels, and we found that both, the expected stiffness-dependent reduction in migration and increase in pro-inflammatory cytokine production by microglia, were Piezo1-dependent. Interestingly, we also found the expression of Piezo1 was itself enhanced in microglia cultured on stiffer substrates. In summary, our study reveals that Piezo1 is a mechanosensor of stiffness in microglia, and that its activation modulates the migratory tendencies and immune responses of microglia.
Results
Piezo1 is functionally expressed in microglial cells
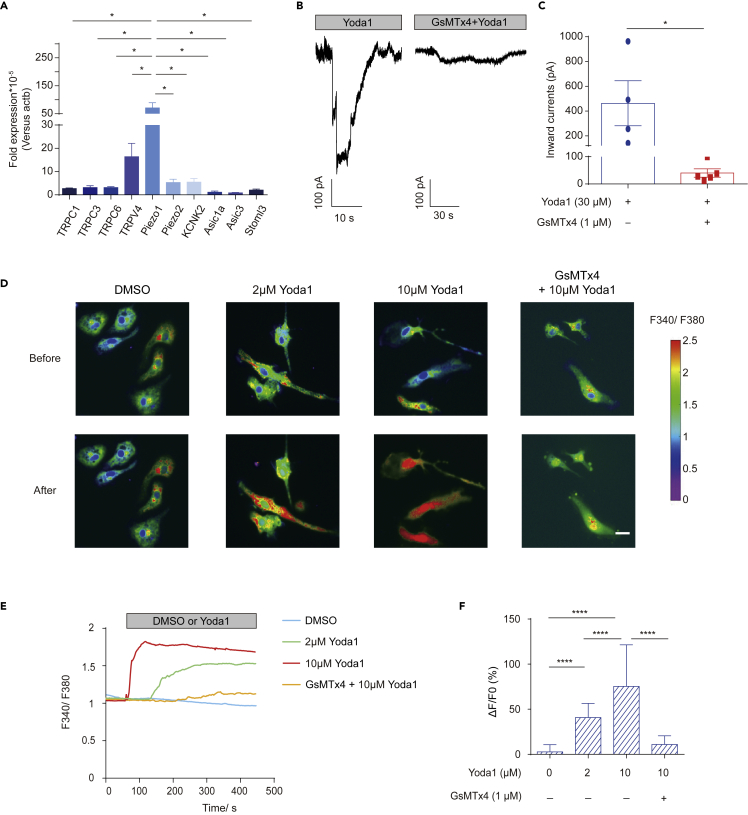
We first attempted to confirm the expression of Piezo1 in primary microglia. Microglia from C57BL/6J mouse pups at postnatal day 3 (P3) were isolated by magnetic-activated cell sorting (MACS) and the purity was checked using flow cytometry (Figure S1). The expression of various well-established mechanosensitive ion channels was then compared through reverse transcription followed by qPCR (RT-qPCR).28,29,30,31,32 mRNA for Piezo1 was the most abundantly expressed of the genes tested, and its expression was significantly higher than its homolog Piezo221 (Figure 1A). Functional analysis of Piezo1 in primary microglia cultured for 5 days was performed by patch clamping cells treated with a Piezo1-specific agonist Yoda133 and the Piezo1 blocker GsMTx4.34 30 μM Yoda1 could induce a strong inward current in microglia (462.7 ± 182.1 pA), but pre-treatment with 1 μM GsMTx-4 significantly reduced the currents induced (40.4 ± 15.36 pA) (Figures 1B and 1C). Piezo1 being a cation channel with high permeability to Ca2+, calcium imaging was performed on cells loaded with a fluorescent indicator (Fura-2). We found that Yoda1 at different concentrations (2 and 10 μM) could induce robust intracellular calcium increase in primary microglia, and adding GsMTx4 significantly decreased the induced Ca2+ influx induced (Figures 1D-1F).
Figure 1.
Piezo1 is expressed in primary microglia
(A) The relative expression of mechanically activated ion channel mRNA in primary microglia was analyzed by real-time RT-PCR using the ΔΔCT method. Bars represent mean ± SEM N = 3 independent experiments. Two-tailed unpaired t-test. ∗p < 0.05.
(B) Representative traces of Yoda1-induced inward currents in primary microglia recorded at −60 mV under the indicated conditions.
(C) Scatterplots of Yoda1-induced inward currents in the indicated conditions. Bars represent mean ± SEM N = 4 in the Yoda1 group, n = 5 in the GsMTx4+Yoda1 group. Two-tailed unpaired t-test. ∗p < 0.05.
(D) Representative live-cell imaging of primary microglia loaded with Fura-2 at rest state (upper panel) and at the time of maximum fluorescence (lower panel). Scale bar = 20 μm
(E) Representative calcium imaging traces of Fura-2 (F340/F380 ratio) in primary microglia treated with different Yoda1 and/or GsMTx4 concentrations.
(F) Statistical analysis of ΔF/F0 (%) in primary microglia under different Yoda1 and/or GsMTx4 concentrations. Bars represent the mean ± SD N = 3 independent experiments; 20-30 cells analyzed per experiment. Two-tailed Mann-Whitney U test. ∗∗∗∗p < 0.0001.
We also tested the calcium response patterns in a commonly used mouse microglial cell line, BV2. Yoda1 significantly increased intracellular calcium levels in BV2, and the responses were dose-dependent, but GsMTx4 (1 μM) inhibited the calcium influx induced by 10 μM Yoda1 (Figures S2A-S2C). Thus, we concluded that Piezo1 was expressed and that the channels were functional, in the cellular models of microglia chosen for our experiments.
Piezo1 deficiency affects the expression profiles of genes
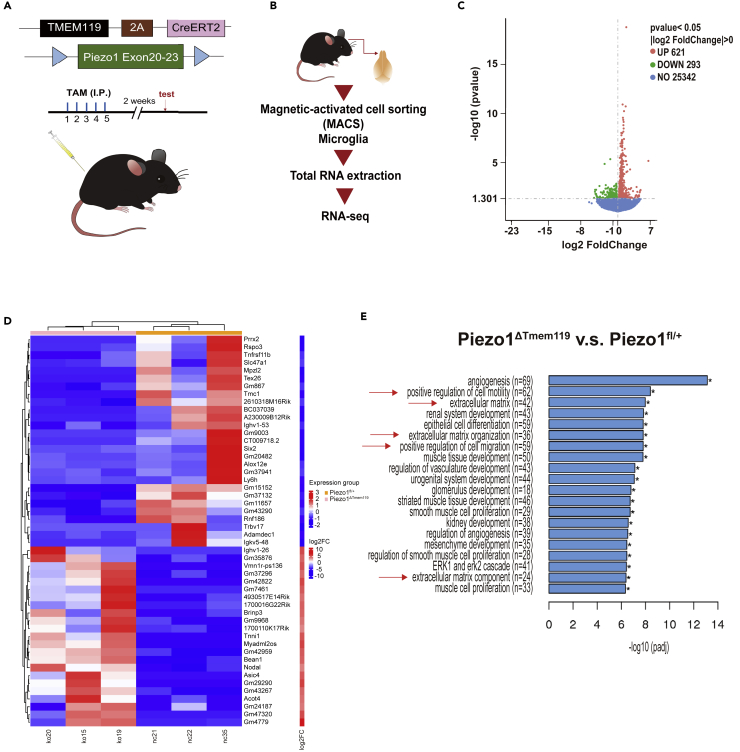
Piezo1 has been documented to play key roles in some important physiological functions, e.g., lymphatic valve formation,24 neuronal sensing of blood pressure,35 and red blood cell volume homeostasis.26 Having confirmed the robust expression of Piezo1 in primary microglia, we were interested in exploring the role of Piezo1 in regulating microglial functions. Knockout or knockdown models of genes are a crucial way to study their roles in various cells. Global deletion of the Piezo1 gene is known to be lethal to embryos36; thus, we instead generated transgenic mice using a conditional knockout model. A mouse line having Piezo1 conditionally depleted from Tmem119-expressing CNS microglia was generated, of the genotype Piezo1flox/flox Tmem119CreERT2/+ (Piezo1ΔTmem119) (Figure 2A). Mice of the genotype Piezo1flox/+Tmem119CreERT2/+ (Piezo1fl/+) were used as a control group for comparison. Genotyping and Piezo1 knockout validation was done using PCR to confirm the successful deletion (Figure S3). To evaluate the consequences of Piezo1 loss, primary microglia were isolated from the brains of adult mice by MACS (Figure 2B). Total RNA was extracted from microglia immediately after isolation and was commercially analyzed for transcriptional profile change using RNAseq. The analysis revealed that 621 genes were upregulated and 293 were downregulated in Piezo1ΔTmem119 compared to the control (Figures 2C and 2D). A gene ontology (GO) pathway analysis was performed, and the top 20 significantly enriched terms were compiled. The analysis revealed several clusters of dysregulated genes, including genes responsible for extracellular matrix organization and regulating cellular motility and migration (Figure 2E). We thus found evidence that Piezo1 is involved in regulating cell motility, a key aspect of microglial behavior.
Figure 2.
Piezo1 deficiency in microglia affects the expression profiles of genes
(A) Schematic illustration of the Piezo1 conditional knock out mice and treatment scheme with tamoxifen.
(B) Schematic illustration of RNA-seq preparation procedure.
(C) Volcano plot showing Piezo1ΔTmem119 vs. Piezo1fl/+ differentially expressed genes by RNA-seq based on primary microglia isolated from Piezo1 conditional knock out mice.
(D) Heatmap showing the top 50 differentially expressed genes, Piezo1ΔTmem119 vs. Piezo1fl/+.
(E) The top 20 significantly enriched terms in the gene ontology (GO) enrichment analysis.
Piezo1 regulates the migration ability of microglia
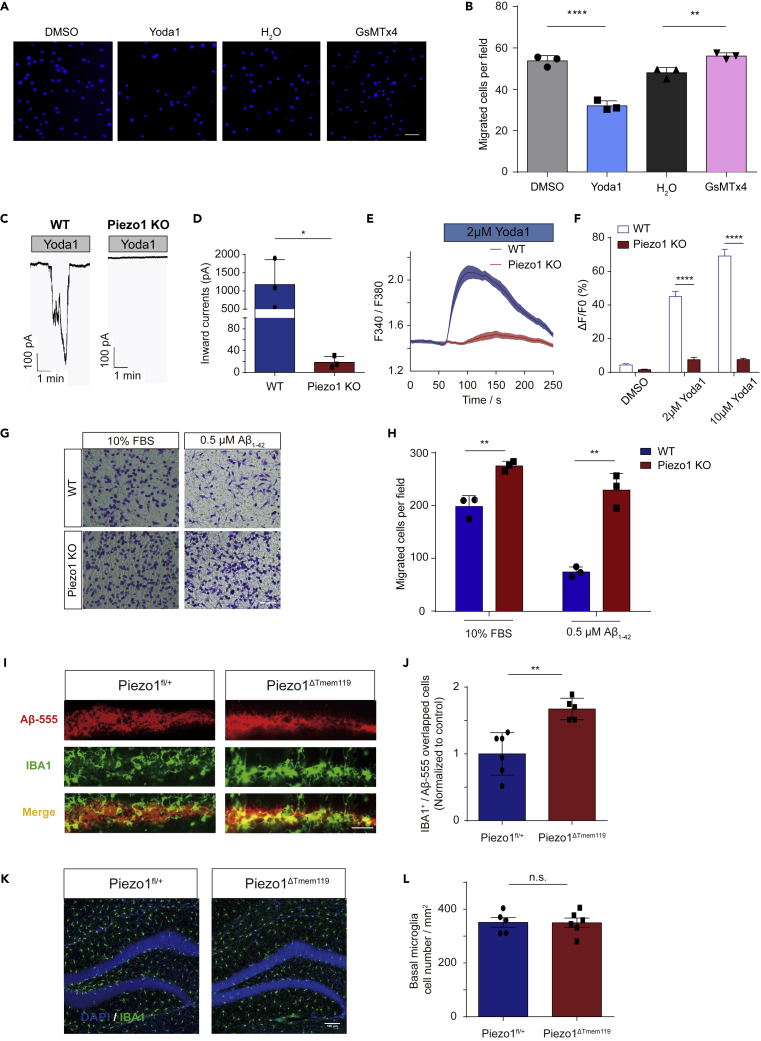
Based on the RNAseq data, we were interested in directly investigating the impact of Piezo1 on microglial migration. We first tested whether the migration of primary microglia could be affected by Piezo1 channel activity using a transwell assay. 10% FBS was used as a chemo-attractant for primary microglia seeded in the upper chamber of a membrane insert. 1‰ DMSO, Yoda1 (10 μM) 1‰ H2O, or GsMTx4 (10 μM) was added to the primary microglial medium to modify the functioning of Piezo1. We found that the addition of Yoda1 (32.07 ± 6.135 cells) significantly suppressed the migratory tendency of primary microglia compared to its control (DMSO) (53.73 ± 9.610 cells toward 10% FBS), whereas adding GsMTx4 (56.13 ± 8.132 cells) enhanced the cells’ movement compared to its control (H2O) (48.00 ± 6.719 cells) (Figures 3A and 3B). Similar migration patterns were observed with the BV2 cell line (Figures S4A and S4B). Thus, we found that chemical activation or blockade of the Piezo1 channel could affect the ability of microglia to migrate.
Figure 3.
Piezo1 regulates the migration ability of microglia both in vivo and in vitro
(A) Representative images of migrated primary microglia stained with DAPI, treated with 1% DMSO, Yoda1 (10 μM) 1% H2O, and GsMTx4 (10 μM) respectively, attracted by 10% FBS. Scale bar = 50um.
(B) Quantification of migrated cell numbers treated with 1% DMSO, Yoda1 (10 μM) 1% H2O, and GsMTx4 (10 μM) attracted by 10% FBS. Bars represent mean ± SD Repeated 3 times. Two-tailed unpaired t test. ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
(C) Representative traces of 30 μM Yoda1 induced inward currents in BV2-WT and BV2-Piezo1 KO cells recorded at −60 mV in the indicated conditions.
(D) Quantification of 30 μM Yoda1-induced inward currents in BV2-WT or BV2-Piezo1 KO cells. Bars represent mean ± SD N = 3 cells for each group. Two-tailed unpaired t test. ∗p < 0.05.
(E) Representative time course of calcium fluorescence in BV2-WT and BV2 - Piezo1 KO cells treated with 2 μM Yoda1.
(F) Statistical analysis of ΔF/F0 (%) in BV2-WT and BV2-Piezo1 KO cells under different Yoda1 concentrations. Bars represent mean ± SEM N = 3 experiments; 30-40 cells were analyzed for each experiment. Two-tailed Mann-Whitney U test. ∗∗∗∗p < 0.0001.
(G) Representative images of migrated BV2-WT or BV2-Piezo1 KO cells stained by crystal violet, attracted by FBS or Aβ1-42 respectively. Scale bar = 100μm.
(H) Quantification of migrated BV2-WT or BV2-Piezo1 KO cell numbers attracted by FBS or Aβ1-42 respectively. Bars represent mean ± SD Repeated 3 times. Two-tailed unpaired t test. ∗∗p < 0.01.
(I) Representative images of Aβ1-42 injected regions in the dentate gyrus (DG) region of Piezo1fl/+ and Piezo1ΔTmem119 mouse groups respectively. Scale bar = 50 μm
(J) Quantified average values for total Aβ-555/IBA1 positive (IBA1+) cells, normalized to control group (n = 6 in Piezo1fl/+ group, n = 5 in Piezo1ΔTmem119 group). Bars represent mean ± SD Two-tailed unpaired t test. ∗∗p < 0.01.
(K) Representative images of IBA1 positive cells in DG area of Piezo1fl/+ and Piezo1ΔTmem119 brain slice respectively (blue: DAPI, green: IBA1). Scale bar = 100 μm
(L) Quantification of microglia numbers per mm2 around DG area in Piezo1fl/+ and Piezo1ΔTmem119 mice respectively (n = 5 in Piezo1fl/+ group, n = 6 in Piezo1ΔTmem119 group). Bars represent mean ± SEM Two-tailed unpaired t test. n.s., no significance.
We next created a BV2-Piezo1 knockout (KO) stable cell line using CRISPR-Cas9 technology. We used a patch clamp to confirm that BV2-Piezo1 KO cells were insensitive to Yoda1 stimulation compared to BV2-WT control cells (Figure 3C). Inward current induced by 30 μM Yoda1 in BV2-Control cells was 1176 ± 394.8 pA while it was much lower (18.67 ± 6.333 pA) in BV2-Piezo1 KO cells (Figure 3D). The resting membrane potential of BV2 cells with Piezo1 KO was not obviously different from BV2-WT control cells (Figure S2D) indicating that Piezo1 knockout did not itself alter the cells’ general electrical activity and health. Calcium imaging also showed that BV2-Piezo1 KO cells were insensitive to Yoda1 stimulation (2 μM and 10 μM) (Figures 3E and 3F). Piezo1 depletion was also found to significantly enhance the cells’ migration toward both FBS and Aβ1-42 compared to the control (Figures 3G and 3H).
The cell migration process requires close control and coordination of actin dynamics, and dysfunctional actin signaling has been known to lead to aberrant cell migration.37 Hence, we also examined the effect of Piezo1 KO on the polymerization of actin. Yoda1 was seen to significantly enhance actin polymerization in BV2-WT control cells, but this effect was abolished in KO cells (Figures S5A and S5B). Interestingly, Piezo1 KO was found to reduce basal actin polymerization levels. Thus, knocking out Piezo1 in BV2 cells was found to significantly affect the cells’ electrical responses, calcium dynamics, and tendency to migrate without altering their background electrical activity and health.
The role of Piezo1 in mediating the migration ability of microglia was further examined in vivo with a simple chemoattraction assay. The oligomer Aβ1-42 HiLyte Fluor 555 was injected into the dentate gyrus (DG) of the hippocampus as an attractant. Counting IBA1-positive cells inside the Aβ1-42 -555 signal regions, many more microglia were found to have migrated to the DG in Piezo1ΔTMEM119 mice than control Piezo1fl/+ mice (Figures 3I and 3J). In mice without the injection of the chemoattractant, the conditional knockout of Piezo1 did not affect the microglia numbers in their basal state (Figures 3K and 3L). Thus, we found that Piezo1 expression and activation regulated the ability of microglia to migrate toward attractant stimuli in vitro and in vivo.
Piezo1 regulates the immune response of microglia
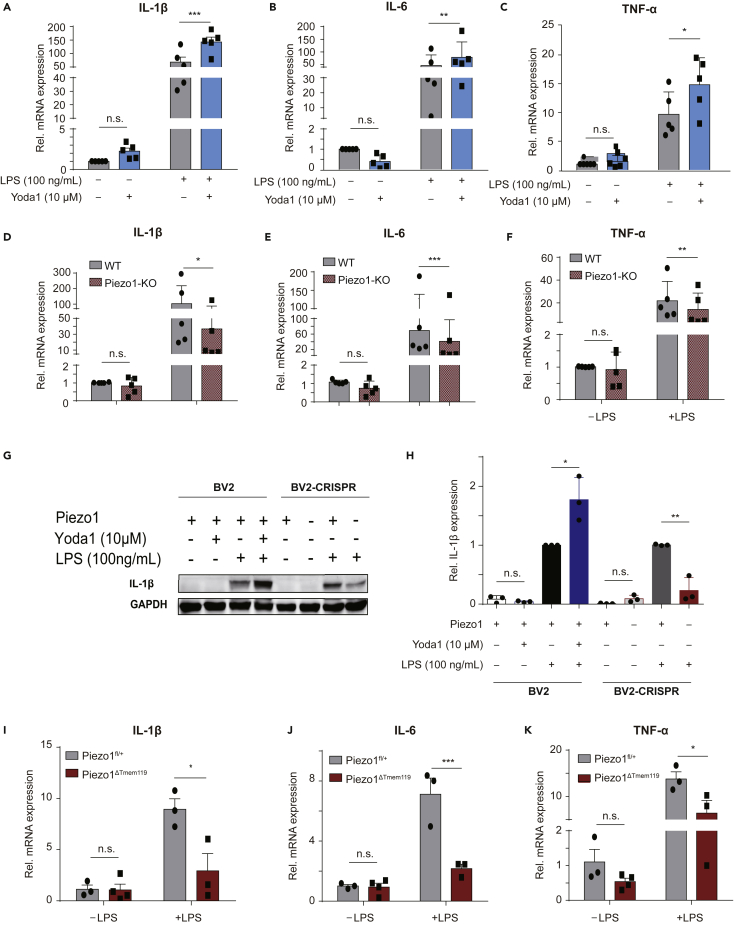
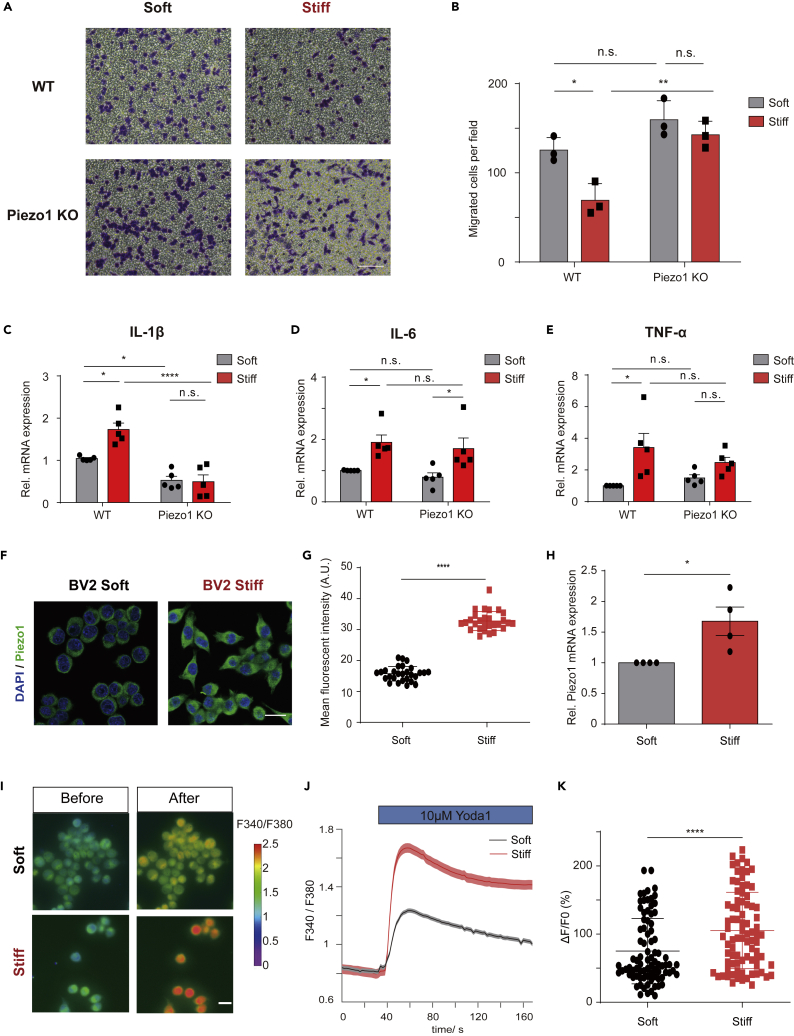
Since microglia are key players in the primary immune response of the CNS, we also wanted to investigate whether Piezo1 was involved in the microglial immune response. BV2-Piezo1 KO and WT control cell lines were stimulated with Yoda1 and/or LPS for 24 h, LPS being a toxin used to trigger the inflammatory response. The expression levels of some important pro-inflammatory cytokines were then monitored by RT-qPCR. In BV2 cells stimulated with LPS, adding Yoda1 increased IL-1β, IL-6, and TNF-α mRNA expression levels (Figures 4A-4C). However, BV2-Piezo1 KO cells expressed significantly lower levels of these cytokines even when stimulated with LPS (Figures 4D-4F). Interestingly, LPS treatment was found to increase Piezo1 mRNA expression (Figure S6). Western blotting for IL-1β further backed up the finding that loss of Piezo1 expression significantly reduced the ability of microglia to response to LPS stimulation by producing cytokines (Figures 4G and 4H). We further tested the immune response mediation by Piezo1 in vivo using our transgenic mouse model. The cortex tissues of mice were evaluated for cytokine expression levels after intraperitoneal LPS injection (0.5 mg/kg) for 4 h. Mice with the conditional Piezo1 KO in microglia showed lower expression of these cytokines when challenged by LPS, but not in the basal state (Figures 4I-4K). Thus we found that Piezo1 plays an important role in mediating the LPS-induced neuroinflammatory response of microglia.
Figure 4.
Piezo1 regulates the production of pro-inflammatory cytokines by microglia when challenged by LPS both in vivo and in vitro
(A-C) Relative mRNA expression of IL-1β, IL-6, and TNF-α in BV2-WT cells, exposed to Yoda1 (10 μM) or not, in basal state or stimulated by LPS (100 ng/mL), normalized to BV2 basal control state. Bars represent mean ± SD N = 5 independent experiments. two-way ANOVA, n.s., no significance, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D-F) Relative mRNA expression of IL-1β, IL-6, and TNF-α of BV2-WT and BV2-Piezo1 KO cells in basal state or stimulated by LPS (100 ;ng/mL), normalized to BV2-WT basal state. Bars represent mean ± SD N = 5 independent experiments. two-way ANOVA, n.s., no significance, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(G) Representative western blot images for IL-1 β and GAPDH in indicated conditions.
(H) Quantification of IL-1 β protein levels normalized to GAPDH compared with BV2-WT treated with LPS group. Bars represent mean ± SD N = 3 independent experiments. Two-tailed unpaired T test. n.s., no significance, ∗p < 0.05, ∗∗p < 0.01.
(I-K) Relative mRNA expression of IL-1β, IL-6, and TNF-α in the cortex of Piezo1fl/+ and Piezo1ΔTMEM119 mice in basal state or after the administration of LPS (n = 3 in Piezo1fl/+ -LPS group, n = 4 in Piezo1ΔTmem119 -LPS group, n = 3 in Piezo1fl/+ +LPS group, n = 3 in Piezo1ΔTmem119 + LPS group). Bars represent mean ± SEM For panel A-F and I-K. two-way ANOVA, n.s., no significance, ∗p < 0.05, ∗∗∗p < 0.001.
Stiffness-mediated microglial migration and activation are mediated by Piezo1
Given that Piezo1 is involved in mechanosensation in many cell types and physiological functions,22,23,26,27,36,38 we reasoned that Piezo1 would play such a role in microglia as well. To investigate the role of Piezo1 in microglia sensing of stiffness, to regulate migration ability and cytokine production of microglia, BV2-WT control and BV2-Piezo1 KO cells were plated onto soft (0.6 kPa) or stiff (35 kPa) polyacrylamide gel substrates (as previously described39) for 48 h. The cells’ migration toward 10% FBS and expression of inflammatory cytokines were then compared between groups detaching cells from the PA gel using 2 mM EDTA. We found that increased substrate stiffness reduced the migration of BV2-WT cells but not in BV2-Piezo1 KO cells (Figures 5A and 5B). We also found that greater substrate stiffness could augment the expression of pro-inflammatory factors (IL-1β, IL-6, and TNF-α) in BV2-WT cells, but IL-1β and TNF-α expression did not increase in BV2-Piezo1 KO cells (Figures 5C-5E). These data indicate that Piezo1 is involved in stiffness-induced microglial migration and activation process.
Figure 5.
Stiffness-dependent Piezo1 expression/activity modulates microglia migration and pro-inflammatory cytokines production
(A) Representative images of migrated BV2-WT or Piezo1-KO cells pre-cultured for 48h in PA gel of varying stiffness (Soft: 0.6 KPa, Stiff: 35 KPa) stained by crystal violet. Scale bar = 100um.
(B) Quantification of migrated cell numbers of BV2-WT or Piezo1-KO cells pre-cultured for 48h in PA gel of different stiffness (Soft: 0.6 KPa, Stiff: 35 KPa). Bars represent mean ± SD N = 3 independent experiments. two-way ANOVA, n.s., no significance, ∗p < 0.05, ∗∗p < 0.01.
(C-E) Relative IL-1β, IL-6, and TNF-α mRNA expression of BV2-WT and BV2-Piezo1 KO seeded on surfaces of indicated stiffness (Soft: 0.6 KPa, Stiff: 35 KPa). Bars represent mean ± SEM N = 5 independent experiments. two-way ANOVA, n.s., no significance; ∗p < 0.05, ∗∗∗∗p < 0.0001.
(F) Immunofluorescence confocal images of Piezo1 (green) in BV2 cell lines seeding on different stiffness of PA gel (Soft: 0.6 KPa, Stiff: 35 KPa). Scale bar = 20 μm
(G) Quantification of Piezo1 mean fluorescent intensity (MFI) of BV2 cells seeding on different stiffness of PA gel. Bars represent mean ± SD Repeated 3 times, around 30 cells were analyzed for each experiment. Two-tailed unpaired t test. ∗∗∗∗p < 0.0001.
(H) Relative Piezo1 mRNA levels of BV2 cell lines seeding on different stiffness of PA gel. Bars represent mean ± SEM N = 4 independent experiments. Two-tailed unpaired t test. ∗p < 0.05.
(I) Representative live-cell imaging of BV2 cells loaded with Fura2 calcium indicator treated with Yoda1 (10 μM) at time 0 (left panel) and at the time of maximum fluorescence (right panel) seeding on soft and stiff substrate respectively. Scale bar = 20 μm
(J) Representative calcium imaging traces of Fura-2 (F340/F380 ratio) in BV2 cells seeding on soft (black curve) and stiff substrates (red curve) treated with Yoda1 (10 μM).
(K) Statistical analysis of ΔF/F0 (%) of BV2 cells seeding on soft (black) and stiff substrates (red) induced by Yoda1 (10 μM). Bars represent mean ± SD N = 3 independent experiments, 20-30 cells were analyzed for each experiment. Two-tailed Mann-Whitney U test. ∗∗∗∗p < 0.0001.
An unexpected aspect of the soft/stiff substrate culture was that an increase in stiffness was also correlated with increased Piezo1 expression. BV2 cells on the stiffer substrate showed significantly increased Piezo1 expression after 48 h of culture in both protein (Figures 5F and 5G) and mRNA level (Figure 5H). The specificity of the Piezo1 antibody used here was verified using constructed BV2 Piezo1 KO cells first (Figure S7). Stiff-cultured cells also showed a much greater calcium response when stimulated with 10 μM Yoda1 than did soft-cultured cells (Figures 5I-5K), which also points toward the increased expression or activation of Piezo1. Enhanced expression of Piezo1 in the stiffer substrate was also observed in primary microglia (Figures S8A and S8B). Thus, we found that Piezo1 was a mediator for microglial response to environmental stiffness cues, while also itself being upregulated in microglia in response to varying mechanical conditions.
Discussion
In the CNS, microglia constantly encounter mechanically heterogeneous regions of varying cellular density and matrix composition. This implies the presence of a suite of cellular machinery to sense and transduce these cues into cellular signaling. In the present study, we identify a role for the mechanically activated ion channel Piezo1 in regulating microglial functions, i.e., migration, pro-inflammatory cytokines production, and stiffness sensing. We found that Piezo1 is highly expressed in primary microglia. Microglia from mice with conditional Piezo1 knockout exhibited enhanced migration ability toward injected Aβ1-42 and reduced pro-inflammatory cytokines production after stimulation by LPS. These phenomena were also observed in vitro using constructed BV2-WT and BV2-Piezo1 KO stable cell lines. In vitro, we found that activation and blockade of the Piezo1 channel reversibly regulated the migration tendencies of microglia. Furthermore, Yoda1 could enhance pro-inflammatory cytokines production of microglia stimulated by LPS, but similarly activating Piezo1 in unstimulated microglia had little effect. We also found that substrate stiffness modulates Piezo1 expression, and regulates microglia migration and pro-inflammatory cytokines production (IL-1β, TNF-α) enhancement was affected by Piezo1 depletion. Together, our study reveals the critical roles of Piezo1 in modulating microglial functions and indicates its significance in maintaining brain health.
Ca2+-mediated signals are a common signal transduction pathway in all living cells, including microglia.40 Our study shows Piezo1 is an important mediator of calcium influx in microglia, through which extracellular signals are transduced. Microglia have been shown to be very sensitive to brain damage, and rapidly generate localized calcium signals after brain damage.41 The Ca2+ activities of microglia are also known to be altered in some pathological conditions, such as in patients with Alzheimers,42 and researchers have shown amyloid plaques to activate microglia through Ca2+ influx.43 A previous study using a TgF344-AD transgenic model also found that Piezo1 expression level in astrocytes increases around stiff amyloid plaques.44 Thus, microglial Ca2+ channels are a logical candidate for research into treatments for brain pathologies. Indeed, the blockade of calcium ion channels as a therapeutic strategy for AD has been explored by applying isradipine, a blocker of the Ca(v)1.2 channel.45 Given that mechanical cues are altered in the brain during the progression of several pathological conditions in which microglia are involved, e.g., Alzheimer’s disease,11,46 aberrant Piezo1 expression, and function may be a contributor to microglial calcium signaling dysfunction in such conditions as well. Thus, the role of Piezo1 in microglia, regulated by changing microenvironment stiffness of the brain, merits further study using appropriate animal disease models.
Resting microglia are highly dynamic and can be rapidly activated to participate in pathological responses, including migrating to affected sites, releasing various inflammatory molecules, and clearing cellular debris47,48 to maintain brain health. Therefore, migration is a critical first step for microglia to function after injury or inflammatory stimuli. We found that the inhibition of Piezo1 promotes the migration of microglia toward a chemoattractant. It is known that microglia in their resting state have a small cell body and radiating thin branches, motile and sensitive to environmental changes. Once a pathological event is detected, microglia are activated and specific signaling, through receptors and kinases, is initiated to induce the enlargement of cell bodies and retraction of processes.3 Previous studies report that Piezo1 inhibition promotes cell migration in several biological contexts. Holt et al. found that epidermal-specific Piezo1 knock-out mice exhibited faster wound closure than WT mice through the regulation of keratinocytes.49 Yu et al. reported that Piezo1 KO significantly enhanced cell migration via decreased cell adhesion, cell stiffness, and contractility.50 Moreover, Piezo1 has also been found to form protein complexes with the extracellular matrix and cytoskeleton which could regulate the cell motility and transformation.51 Interestingly, in our study we observed that F-actin, which plays a crucial role maintaining cell stiffness,52 was down-regulated in the Piezo1 KO group. Taken together, a plausible explanation may be that the lack of microglial Piezo1 may disrupt the extracellular matrix-cytoskeleton-Piezo1 complex, making the cell membrane softer and making migration toward the chemoattractant easier.
Numerous studies have already shown that microglia tend to migrate upon encountering chemoattractive stimuli.3 Our data add the important detail that microglial Piezo1 can sense substrate stiffness (Figures 5A and 5B) and plays a part in regulating microglial migration. Thus mechanical cues exerted on microglia, thus, also merit exploration, such as in aging10 and some pathological conditions.11,12,13,14 One issue requiring resolution is that Jantti et al. report Yoda1 to promote microglial migration,53 in apparent contradiction to our findings. To somewhat mimic the physiological environment of microglial migration, we adopted a transwell assay, with cells in the upper chamber and chemoattractant in the lower chamber, giving the assay a 3D aspect.54 This is very different from the 2D would healing assay, without chemoattractant, in Jantti’s work. Microglia in their resting state are constantly mobile, surveilling their surrounding environment. Jantti et al. showed that the activation of Piezo1 increases migration in a wound healing assay, which may tell us what happens when microglial Piezo1 is activated in the absence of a chemoattractant. Further, the choice of cellular model (primary mouse microglia and BV2 cells, vs pluripotent stem cell-derived microglia-like cells) could be another possible factor explaining the discrepancy. Thus, we are of the opinion that the conclusions from these two studies are not, in fact, contradictory.
Piezo1 also played an important role in the microglial production of pro-inflammatory cytokines (IL-1β, IL-6, TNF-α), which are involved in the pathogenesis of many neurodegenerative diseases.55 Enhanced stiffness, mediated by Piezo1, was also associated with increased pro-inflammatory cytokine production in microglia. Calcium homeostasis is known to be closely associated with microglial activation, capable of activating the inflammasome,56 inhibiting LPS-induced activation57 and contributing to neuroinflammation and AD pathology.58 It is hence possible that greater brain tissue stiffness could activate Piezo1 and the subsequent calcium influx could trigger microglial cytokine production. Previous research shows that both LPS and amyloid plaques upregulate Piezo1 in astrocytes.44,59 In our study, we also find LPS increased Piezo1 levels in BV2 cells, perhaps indicating a role for Piezo1 in the microglial response to pathological conditions. It is plausible Piezo1 in microglia around amyloid plaques could be upregulated as well, similar to astrocytic Piezo1,44 and this question should be addressed in future studies.
Interestingly, substrate stiffness could also regulate Piezo1 expression in microglia and induce subsequent changes in microglial function (Figure 5). This phenomenon suggests Piezo1 could be a key molecule mediating between microenvironment stiffness and microglial function. In the present article, we performed a limited exploration of how Piezo1 expression is altered in pathological conditions, focusing on its role as a mechanosensor. However, this is a topic that merits deeper study, as the regulation of Piezo1 will naturally redound to the cells’ mechanosensation abilities and all the ensuing signaling implications. Chronic inflammation could be a consequence of AD pathology that further exacerbates the deleterious effects exerted by Aβ and tau.55 Mechanically stiff β-amyloid plaques may activate Piezo1 activity due to its mechanosensing role. Thus, this raises the possibility of Piezo1 as a druggable target for the treatment of AD. Several studies have already described the roles of mechanosensitive ion channels in regulating brain diseases, e.g., TRPC1,60 TRPC3, TRPC6,61,62 TRPV4.63 Given Piezo1’s early role in initiating cell signaling and its possible participation in mediating crucial microglial processes, it could be involved in neurodegenerative diseases besides AD as well. Thus, the study of microglial Piezo1 could improve our understanding of brain diseases in general.
Limitations of the study
This study describes the role of mechanosensitive ion channel Piezo1 in microglia response to disturbed homeostasis in the brain and in vitro. We used primary microglia and BV2 microglial cell line as our in vitro model. Specifically, we constructed a BV2 Piezo1 knock-out cell line using CRISPR technology and the in vitro functional assays (migration and cytokines production) was based on this cell model. We did not evaluate the functions of primary microglia lack of Piezo1, which lays the limitations here. Besides, the role of microglial Piezo1 in brain diseases deserves further study. We did not include any disease model in this study. Nevertheless, the findings in our study could provide some clues for the study in the context of brain diseases.
STAR★Methods
Key resources table
| REAGENTS or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Iba1 rabbit monoclonal primary antibody | Cell Signaling Technology | Cat. # 17198S; RRID: AB_2820254 |
| Iba1 goat polyclonal primary antibody | Novus Biologicals | Cat. # NB100-1028 |
| Piezo1 rabbit polyclonal primary antibody | Proteintech | Cat. # 15939-1-AP; RRID: AB_2231460 |
| Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 488 | Thermo-Fisher Scientific | Cat. # A-11008; RRID: AB_143165 |
| Anti-IL-1 beta primary antibody | Abcam | Cat. # ab254360 |
| Goat anti-Rabbit IgG (H + L) Secondary Antibody, HRP | Thermo-Fisher Scientific | Cat. # 31460; RRID: AB_228341 |
| Donkey anti-goat Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo-Fisher Scientific | Cat. # A-11055; RRID: AB_2534102 |
| Donkey anti-rabbit Adsorbed Secondary Antibody, Alexa Fluor 555 | Thermo-Fisher Scientific | Cat. # A-32572 |
| CD11b Antibody, anti-mouse, Vio® Bright FITC, REAfinity™ | Miltenyi Biotec | Cat. # 130-113-243; RRID: AB_2726049 |
| CD45 Antibody, anti-mouse, APC, REAfinity™ | Miltenyi Biotec | Cat. # 130-110-660; RRID:AB_2658221 |
| Bacterial and virus strains | ||
| DH5-α | Thermo-Fisher Scientific | Cat. # 18265017 |
| Chemicals, peptides, and recombinant proteins | ||
| Yoda1 | Sigma-Aldrich | Cat. # SML1558 |
| GsMTx-4 | Abcam | Cat. # ab141871 |
| Fura-2 AM | Invitrogen | Cat. #F1221 |
| Pluronic F-127 | Invitrogen | Cat. #P3000MP |
| Poly-L-Lysine | Sigma-Aldrich | Cat. #P2636 |
| DMEM/F-12 | Gibco | Cat. # 10565018 |
| DMEM | Gibco | Cat. # 10566016 |
| Fetal Bovine Serum | Gibco | Cat. # A3840402 |
| Penicillin-Streptomycin | Gibco | Cat. # 15140122 |
| Puromycin | Life Technologies | Cat. # A1113803 |
| Beta-Amyloid (1–42), HiLyte™ Fluor 555-labeled | ANASPEC | Cat. # AS-60480-01 |
| Goat serum | Gibco | Cat. # 16210072 |
| RIPA lysis buffer | Thermo-Fisher Scientific | Cat. # 89900 |
| Halt™ Protease and Phosphatase Inhibitor Cocktail | Thermo-Fisher Scientific | Cat. # 78446 |
| Mounting Medium With DAPI-Aqueous, Fluoroshield | Abcam | Cat. # ab104139 |
| Ammonium persulfate | Thermo Fisher Scientific | Cat. # 17874 |
| TEMED | Thermo Fisher Scientific | Cat. #17919 |
| Sulfo-SANPAH | Thermo Fisher Scientific | Cat. #A35395 |
| Critical commercial assays | ||
| Neural Tissue Dissociation Kit (P) | Miltenyi Biotec | Cat. 130-092-628 |
| CD11b Micro-Beads Ultra-Pure, mouse | Miltenyi Biotec | Cat. 130-126-725 |
| Lipofectamine™ 3000 transfection reagent | Thermo-Fisher Scientific | Cat. L3000015 |
| MiniBEST Universal RNA Extraction Kit | Takara | Cat. # 9767 |
| PrimeScript RT Master Mix | Takara | Cat. # RR036A |
| SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) | Takara | Cat. # RR820A |
| Pierce™ Rapid Gold BCA Protein Assay Kit | Thermo-Fisher Scientific | Cat. # A53225 |
| Green fluorescent phalloidin conjugate | Abcam | Cat. # ab112125 |
| 6.5 mm Transwell with 8.0 μm pore polycarbonate membrane insert, TC-treated | Corning | Cat. # 3422 |
| Deposited data | ||
| RNA-seq raw data | This paper | SRA database from NCBI: PRJNA919004 |
| Experimental models: Cell lines | ||
| Mouse BV2 Microglia Cell Line | Gift from Bo Peng lab from Fudan University | RRID: CVCL_0182 |
| Experimental models: Organisms/strains | ||
| Transgenic Mouse: Piezo1flox/flox | The Jackson Laboratories | JAX: 029,213 |
| Transgenic Mouse: Tmem119-2A-CreERT2 | The Jackson Laboratories | JAX: 031,820 |
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000,664 |
| Recombinant DNA | ||
| MLM3636 vector | Addgene | Plasmid #43860 |
| pX459 v2.0 | Addgene | Plasmid #62988 |
| Software and algorithms | ||
| ImageJ | N/A | https://imagej.nih.gov/ij |
| MATLAB | The MathWorks, Inc., MA, USA | https://www.mathworks.com/products/matlab.html |
| GraphPad Prism 8.0 | GraphPad Software, CA, USA | https://www.graphpad.com |
Resource availability
Lead contact
Further information and request for reagents and resources should be addressed to, and will be fulfilled by, the lead contact, Lei Sun (lei.sun@polyu.edu.hk).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
All animal procedures were approved by the Animal Subjects Ethics Sub-Committee (ASESC) of the Hong Kong Polytechnic University, and were performed in compliance with the guidelines of the Department of Health - Animals (Control of Experiments) of the Hong Kong S.A.R. government. Piezo1flox/flox (Jackson Laboratories Stock No. 029213) and Tmem119-2A-CreERT2 (Jackson Laboratories Stock No: 031820) mice were used to generate Piezo1 microglia conditional knock-out mice. Offspring transgenic female mice started tamoxifen administration at 7 weeks old and rested for two weeks after last day of injection.
Method details
Transgenic mouse line generation and genotyping
Generation of Piezo1ΔTMEM119 mice was accomplished through breeding Piezo1flox/flox (Jackson Laboratories Stock No. 029213) and Tmem119-2A-CreERT2 (Jackson Laboratories Stock No: 031820) mice together to generate progeny that were heterozygous for both genes. The generated heterozygotes were then bred with Piezo1flox/flox mice to generate Piezo1flox/floxTmem119Cre/+ (Piezo1ΔTmem119) and Piezo1flox/+Tmem119Cre/+ (Piezo1fl/+) mice. Mice were housed in a 12-h light/12-h dark cycle, with temperatures between 20 °C and 24 °C and 40–70% humidity with food and water available ad libitum. Tamoxifen (Sigma-Aldrich, T5648) was dissolved in corn oil containing 5% ethanol at 20 mg/mL under vortex at times for several hours in the dark. At time of administration, the mice weighed 19–26 g and intraperitoneal (i.p.) injection with 100 μL of the 20 mg/mL tamoxifen solution for 5 consecutive days. After administration of tamoxifen for at least 2 weeks, mice were used for different in vivo assays. Mice tail samples were genotyped using the following primers suggested by Jackson Laboratories: Piezo1- 28247- forward: GCC TAG ATT CAC CTG GCT TC; Piezo1-28248- reverse: GCT CTT AAC CAT TGA GCC ATC T; TMEM119-16504: ATC GCA TTC CTT GCA AAA GT; TMEM119-42648: CAG TAT GTG GGG TCA CTG AAG A; TMEM119-42649: ACT TGG GGA GAT GTT TCC TG using Phire Tissue Direct PCR Master Mix (F170S, Thermo Scientific) with the following cycling conditions: Initial denaturation 98°C for 5 min, followed by 10 cycles of 98°C for 5 s, 65°C for 5 s, −0.5 C per cycle decrease, 68°C for 20 s, followed by 30 cycles of 98°C for 5 s, 60°C for 5 s, 72°C for 20 s, followed by a final hold of 72°C for 1 min. Reactions were separated on 2% agarose gels yielding the following band sizes: Cre +/−: 280bp and 378bp, Cre −/−: 378bp, Piezo1 fl/fl: 380bp, Piezo1 fl/+: 380bp and 188bp. Successful knockout Piezo1 were validated by examining DNA from mice brains using the following primers: Piezo1- 28,247- forward: GCC TAG ATT CAC CTG GCT TC; Piezo1 KO- reverse: AGG TTG CAG GGT GGC ATG GCT CTT TTT. An obvious Piezo1 KO band was found in 230bp but none in control mice group. Brain samples were also collected for testing Piezo1 mRNA levels by qPCR using the following primers: Piezo1-192-forward: CTCTGGCCTAACTACTGTCT; Piezo1-192-reverse: GATAAGGTTGGTGGAGTTGG.
Cell culture
The BV2 cell line was a gift from Dr. Bo Peng’s group. Cells were cultured in DMEM (10566016, Gibco), 10% fetal bovine serum (A3840402, Gibco), 1% Penicillin-Streptomycin (15140122, Gibco) at 37°C inside a standard cell culture incubator containing 5% CO2.
Primary microglia isolation
For primary microglia isolation, P3 pups of C57BL/6J mice were sacrificed, followed by dissociation and isolation using the CD11b isolation kit according to the manufacturer’s protocol (130-092-628 and 130-126-725, Miltenyi Biotec). Primary microglia purity was examined by flow cytometry after staining by Cd11b-FITC (130-113-243, Miltenyi Biotec) and Cd45-APC (130-110-660, Miltenyi Biotec). Dishes were pre-coated with 50 μg/μl Poly-L-Lysine and cells were then plated on the dishes at a density of 105 cells/Ml and cultured in DMEM/F12 (10565018, Gibco), 10% fetal bovine serum (A3840402, Gibco), 1% Penicillin-Streptomycin (15140122, Gibco) at 37°C inside a standard cell culture incubator containing 5% CO2.
CRISPR knockout (KO) Piezo1 cell cloning
To generate Piezo1 knockout (KO) cell lines, single guide RNAs (sgRNAs) ACGCTTCAATGCTCTCTCGC targeting the second exon of mouse piezo1 as reference64 was inserted into MLM3636 vector (Addgene Plasmid #43860). SgRNA inserted MLM3636 and Cas9 expression plasmid pX459 v2.0 (Addgene Plasmid #62988) were transfected into BV2 cells with Lipofectamine™ 3000 transfection reagent (L3000015, Thermo-Fisher Scientific). MLM3636 and pX459 v2.0 (Addgene Plasmid #62988) vectors were transfected into BV2 cells to create BV2-WT control cells. These two transfected BV2 cell lines were selected with 1ug/mL puromycin (A1113803, Life Technologies). After selection, live cells were trypsinized, diluted as 5 cell/mL and then seeded on 96-well plate at an average density of 0.5cell/well for growing to obtain single clones. The successful knockout clone was verified using Sanger sequencing, patch clamp and calcium imaging.
Intracellular calcium imaging
Cells were plated on confocal dishes (200,350, SPL Life Sciences). Prior to recording, cells were incubated with 2 μM Fura-2 (F1221, Invitrogen) at 37°C for 30 min dispersed in Margo solution with 0.2% w/v Pluronic F-127. Cells were then washed twice and incubated for another 15 min. Inhibitors were added in this step as needed. Experiments were performed at room temperature using Nikon Eclipse Ti2-E microscope. Intracellular calcium concentration was indicated as the ratio of Fura-2 emission (510 nm) intensities for 340 and 380 nm excitation.
Patch clamp
Cells were seeded in culture dishes before patch clamp recording. Borosilicate glass-made patch pipettes (Vitrex, Modulohm A/S, Herlev, Denmark), were pulled with micropipette puller (P-97, Sutter Instrument Co., USA) to a resistance of 2–5 MΩ before being filled with KCl pipette solution (in mM): KCl 138, NaCl 10, MgCl2 1 and HEPES 10 with D-manitol compensated for osm290. Inward current was recorded with a data acquisition system (DigiData 1322A, Axon Instruments) and an amplifier (Axopatch-200B, Axon Instruments, Foster City, CA, USA). The command voltages were controlled by a computer equipped with pClamp Version 9 software. For cell resting membrane potential (RMP) test, cells were placed on bath solution (in mM): NaCl 130, MgCl2 2, KCl 4.5, Glucose 10, HEPES 20, and CaCl2 2, pH 7.4. When the whole-cell Giga seal was formed and the capacitance of cell was measured, the RMP of cells was recorded when the injection current was set at zero. Afterward, the inward currents induced by Yoda1 was recorded by voltage clamp gap free recording mode.
RNA isolation and RT-qPCR
RNA was extracted using the MiniBEST Universal RNA Extraction Kit (Cat No. 9767, TaKaRa). cDNA syntheses were generated using PrimeScript RT Master Mix (RR036A, TaKaRa). qPCR was performed using SYBR Premix Ex Taq II (Tli RNaseH Plus) (RR820A, TaKaRa) and cycled in the CFX96 Touch Deep Well Real-Time PCR system under standard conditions. The ΔΔCt method was used to calculate gene expression. Primer sequences are provided in the table (Table S1).
RNA sequencing and data analysis
RNA sequencing and informatics analysis were performed by Novogene Company (Beijing, China). RNA integrity was assessed with the Agilent 2100 Bioanalyzer (Agilent Technologies). Libraries were barcoded, quantified using the NEBNext Ultra RNA Library Prep Kit for Illumina (Cat No. 7530, New England Biolabs (NEB)). Quantified libraries were pooled and sequenced on Illumina platforms, according to effective library concentration and data amount. The clustering of the index-coded samples was performed according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina platform and paired-end reads were generated. Raw data (raw reads) of fastq format were first processed through in-house perl scripts. Clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N and low quality reads from raw data. At the same time, Q20, Q30 and GC content the clean data were calculated. All the downstream analyses were based on the clean data with high quality. Reference genome and gene model annotation files were downloaded from genome website directly. Index of the reference genome was built using Hisat2 v2.0.5 and paired-end clean reads were aligned to the reference genome using Hisat2 v2.0.5. Feature Counts v1.5.0-p3 was used to count the reads numbers mapped to each gene. Differential expression analysis of two groups (3 biological replicates per condition) was performed using the DESeq2 R package. Genes with an adjusted p value ≤0.05 found by DESeq2 were assigned as differentially expressed. Gene Ontology (GO) enrichment analysis of differentially expressed genes was implemented by the cluster Profiler R package, in which gene length bias was corrected. GO terms with corrected p value less than 0.05 were considered significantly enriched by differential expressed genes.
Cell migration transwell assay
Primary microglia and BV2 cells migration assay were tested in a transwell assay (COSTAR 24-well plate with inserts, 8-μm pore, Corning, NY). 600 μL medium together with the attractant- 10% FBS or 0.5 μM Aβ1-42 were added below the membrane inserts. Drugs were added to top chamber before 5 × 104 cells in 100 μL media were added. Migration was assayed for 20 h inside the incubator. Subsequently, inserts were removed and washed three times with PBS. Primary microglia were then fixed with 4% PFA for 10min and stained with DAPI for 30min, while BV2 cells were fixed and stained with 0.5% crystal violet for 20min. Stationary cells were removed from the top with cotton tips. Five random fields were selected for imaging, and the migrated cells were quantified using ImageJ software.
Stereotactic injection of Aβ1-42
Beta-Amyloid (1–42), HiLyte Fluor 555-labeled (Cat. AS-60480-01, ANASPEC) were dissolved in DMSO to obtain a 1 mM stock, resuspended in PBS at 100 μM, and incubated at 37 °C for 1 day to promote Aβ1-42 aggregation. Mice were anesthetized by i.p. injection of a mixture of ketamine (100 mg/kg), xylazine (10 mg/kg), 2 μL of Aβ1-42-555 was distributed into DG area of the hippocampus (AP -2 mm; ML -1.5 mm; DV 2 mm) using stereotaxic equipment (RWD Life Science Co., Ltd). After recovery from surgery, animals were returned to their home cages. Post surgery (16 h), mice were anesthetized and perfused for subsequent experiments.
For histological analyses of microglia migration toward Aβ1-42-555, brains were perfused with ice-cold PBS, fixed in 4% PFA and immersed in 4% PFA for another 4 h. Brains were then sectioned in 50 μM thickness by Vibrating Microtome (Leica VT1200S). Sections were washed, then permeabilized with 0.3% Triton X-100 (Sigma) in PBS with 5% Goat serum (16210072, Gibco) and blocked for 1 h. The microglia-specific antibody IBA1 (Rabbit mAb 17198S, CST) was incubated in blocking buffer overnight at 4°C followed by intensive washing and incubation with a Goat anti-rabbit Adsorbed Secondary Antibody, Alexa Fluor 488 (A-11008, Thermo-Fisher Scientific) for 2 h. Sections were washed again and mounted with Mounting Medium With DAPI-Aqueous, Fluoroshield (ab104139, Abcam). Data acquisition was performed using a Nikon Eclipse Ti2-E Live-cell Fluorescence Imaging System in z stack model.
Western blotting
Cells were rinsed with PBS before being exposed to lysis buffer, a combination of RIPA lysis buffer (89,900, Thermo-Fisher Scientific) and 1% Halt Protease and Phosphatase Inhibitor Cocktail (78,446, Thermo-Fisher Scientific). Cells were scraped to obtain the adhered cells and the lysate was collected. The lysate was put on ice for another 30 min to achieve thorough lysis. Lysate was spun at 13,000 rpm for 20 min at 4°C and the supernatant was collected. Samples were quantified using Pierce Rapid Gold BCA Protein Assay Kit (A53225, Thermo-Fisher Scientific) and aliquots of 50 μg of protein were prepared.The proteins were denatured through the use of a loading buffer supplemented with 5% 2-mercaptoethanol at 95°C for 10 min before each sample was loaded. Gel electrophoresis resulted in the separation of proteins before being transferred onto nitrocellulose membranes using the Thermo Scientific Pierce G2 Fast Blotter. Following electroblotting, the membranes were blocked using 5% nonfat milk in TBST for 1 h at room temperature. Membranes were then gently washed with TBST and incubated with Anti-IL-1 beta primary antibody (ab254360, Abcam) at 4°C overnight with continuous rotation. Additional washes, 10 min 3 times in TBST followed. Before the membranes were probed with Goat anti-Rabbit IgG (H + L) Secondary Antibody, HRP (31,460, Thermo-Fisher Scientific) diluted at 1:5000 at room temperature for 1 h. The membrane was then washed in TBST and immersed into a chemiluminescent HRP substrate solution (Bio-Rad) and imaged using a ChemiDoc MP System (Bio-Rad).
Immunofluorescent staining of cells
Primary microglia were washed with PBS 3 times, then fixed with 4% PFA for 15 min. Cells were then washed with PBS and permeabilized with 0.1% Triton X-100 in PBS for 20 min. After that, cells were washed with PBS and blocked with 5% BSA for 1 h at room temperature. Cells were then incubated with Piezo1 primary antibody (15939-1-AP, Proteintech) diluted 1:200 and Iba1 primary antibody (NB100-1028, Novus Biologicals) diluted 1:200 at 4°C overnight and washed with PBS for 3 times. After that, cells were incubated with 1:1000 donkey anti-goat Adsorbed Secondary Antibody, Alexa Fluor 488 (A-11055, Thermo-Fisher Scientific) and donkey anti-rabbit Adsorbed Secondary Antibody, Alexa Fluor 555 (A-32572, Thermo-Fisher Scientific) at room temperature for 1 h. Cells were finally washed with PBS for 3 times and sealed with mounting medium with DAPI.
Polyacrylamide gel preparation
Polyacrylamide gels were fabricated in a three-layer assembly. A thin film of 0.1M sodium hydroxide was allowed to form on the coverslips at approximately 65°C. The entire surface of each coverslip was then immersed with a sufficient volume of (3-aminopropyl) triethoxysilane (Sigma-Aldrich, Saint Louis, MO) for 10 min at room temperature and dark condition. The (3-aminopropyl) triethoxysilane was then completely rinsed off to prevent precipitation before 0.5% (v/v) glutaraldehyde (Sigma-Aldrich, Saint Louis, MO) in PBS was added onto the silanized coverslips for 20 min under dark condition. Fluids were removed by suction before the amino-silanized coverslips were air-dried and sterilized with 70% ethanol for 16 h prior to gel preparation.
For the middle gel layers of differing stiffness, a prepolymer mixture was prepared as shown in the table below. Polymerization of the prepolymer mixture was carried out by adding 10 μL of 10% ammonium persulfate (17,874, Thermo Fisher Scientific), 2 μL of TEMED (17,919, Thermo Fisher Scientific) and sufficient distilled water to yield a final volume of 1000 μL. The resulting prepolymer-catalyst mixture was dropped onto hydrophobic amino-silanized coverslips. Coverslips were then transferred to the surfaces of the polyacrylamide gels during polymerization. After 30 min, the top layer was gently peeled off and washed three times with HEPES solution.
| 40% acrylamide (μL) | 2% N′-N′-methylenebisacrylamide (μL) | Distilled water(μl) | |
|---|---|---|---|
| 0.6 kPa | 75 | 30 | 895 |
| 35 kPa | 250 | 150 | 600 |
For the top layer, the mixture- 5 μL 50 μg/μL Sulfo-SANPAH (A35395, Thermo Fisher Scientific) in 1 mL PBS were prepared first. 100 μL mixture were dropped on the gel for each well and put the six-well plate under the UV light for 15min. The gels were then washed with PBS for 3 times and repeat UV light for 15min. The gels were then washed again and 250 μL 50ug/mL poly-L-lysine in 50 mM HEPES (pH = 8.5) were dropped onto the gel for each well and store at 4°C overnight.
Actin polymerization staining
Green fluorescent phalloidin conjugate (ab112125, Abcam) was used to stain F-actin according to the manufacturer’s recommendation. Cells were then washed with PBS 3 times and sealed with mounting medium containing DAPI. Data acquisition was performed using a Leica TCS SPE Confocal Microscope.
Quantification and statistical analysis
Statistical tests were performed using GraphPad Prism 8, which was also used to prepare the graphs. Bars charts represent the mean of the indicated number of experiments or subjects, unless otherwise indicated. Details of the statistical analyses performed for each figure are provided in the figure legends. A p value of <0.05 or below was considered statistically significant for all experiments.
Acknowledgments
This work was supported by the Hong Kong Research Grants Council General Research Fund (15104520, 15102417, and 15326416), Hong Kong Innovation Technology Fund (MRP/018/18X and MHP/014/19), Key-Area Research and Development Program of Guangdong Province (2018B030331001), internal funding from the Hong Kong Polytechnic University (1-ZE1K and 1-ZVW8), and Research Institute of Smart Ageing (1-CD76). The authors would like to thank the facility and technical support from the University Research Facility in Life Sciences (ULS) and the University Research Facility in Behavioral and Systems Neuroscience (UBSN) of The Hong Kong Polytechnic University. We thank to Dr. Michael Yuen from ULS for instructing us in primary microglia isolation. We thank to Mr. Kai Tang for instructing us in PA gel fabrication.
Author contributions
T.Z. and L.S. designed the research. T.Z., S.K., Y.W., H.C., JJ.Z., C.P.C., XY.Z., T.L. performed research, T.Z., JH.G., K.F.W., XH.H., MY.Y. analyzed data. T.Z., S.K., and L.S. wrote the article.
Declaration of interests
The authors declare no competing interests.
Published: January 16, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.105993.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request, unless it is protected by law.
This paper does not report original code.
The RNA-sequencing data associated with this project are deposited in the NCBI Sequence Read Archive (SRA) database: PRJNA919004.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Matcovitch-Natan O., Winter D.R., Giladi A., Vargas Aguilar S., Spinrad A., Sarrazin S., Ben-Yehuda H., David E., Zelada González F., Perrin P., et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 2016;353:aad8670. doi: 10.1126/science.aad8670. [DOI] [PubMed] [Google Scholar]
- 2.Kigerl K.A., de Rivero Vaccari J.P., Dietrich W.D., Popovich P.G., Keane R.W. Pattern recognition receptors and central nervous system repair. Exp. Neurol. 2014;258:5–16. doi: 10.1016/j.expneurol.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan Y., Xie L., Chung C.Y. Signaling pathways controlling microglia chemotaxis. Mol. Cells. 2017;40:163–168. doi: 10.14348/molcells.2017.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aloisi F. Immune function of microglia. Glia. 2001;36:165–179. doi: 10.1002/glia.1106. [DOI] [PubMed] [Google Scholar]
- 5.Nakajima K., Tohyama Y., Maeda S., Kohsaka S., Kurihara T. Neuronal regulation by which microglia enhance the production of neurotrophic factors for GABAergic, catecholaminergic, and cholinergic neurons. Neurochem. Int. 2007;50:807–820. doi: 10.1016/j.neuint.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Bollmann L., Koser D.E., Shahapure R., Gautier H.O.B., Holzapfel G.A., Scarcelli G., Gather M.C., Ulbricht E., Franze K. Microglia mechanics: immune activation alters traction forces and durotaxis. Front. Cell. Neurosci. 2015;9:363. doi: 10.3389/fncel.2015.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moshayedi P., Ng G., Kwok J.C.F., Yeo G.S.H., Bryant C.E., Fawcett J.W., Franze K., Guck J. The relationship between glial cell mechanosensitivity and foreign body reactions in the central nervous system. Biomaterials. 2014;35:3919–3925. doi: 10.1016/j.biomaterials.2014.01.038. [DOI] [PubMed] [Google Scholar]
- 8.Ayata P., Schaefer A. Innate sensing of mechanical properties of brain tissue by microglia. Curr. Opin. Immunol. 2020;62:123–130. doi: 10.1016/j.coi.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elkin B.S., Azeloglu E.U., Costa K.D., Morrison B., 3rd Mechanical heterogeneity of the rat hippocampus measured by atomic force microscope indentation. J. Neurotrauma. 2007;24:812–822. doi: 10.1089/neu.2006.0169. [DOI] [PubMed] [Google Scholar]
- 10.Segel M., Neumann B., Hill M.F.E., Weber I.P., Viscomi C., Zhao C., Young A., Agley C.C., Thompson A.J., Gonzalez G.A., et al. Niche stiffness underlies the ageing of central nervous system progenitor cells. Nature. 2019;573:130–134. doi: 10.1038/s41586-019-1484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy M.C., Huston J., 3rd, Jack C.R., Jr., Glaser K.J., Manduca A., Felmlee J.P., Ehman R.L. Decreased brain stiffness in Alzheimer's disease determined by magnetic resonance elastography. J. Magn. Reson. Imaging. 2011;34:494–498. doi: 10.1002/jmri.22707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riek K., Millward J.M., Hamann I., Mueller S., Pfueller C.F., Paul F., Braun J., Infante-Duarte C., Sack I. Magnetic resonance elastography reveals altered brain viscoelasticity in experimental autoimmune encephalomyelitis. Neuroimage. Clin. 2012;1:81–90. doi: 10.1016/j.nicl.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schregel K., Wuerfel E., Garteiser P., Gemeinhardt I., Prozorovski T., Aktas O., Merz H., Petersen D., Wuerfel J., Sinkus R. Demyelination reduces brain parenchymal stiffness quantified in vivo by magnetic resonance elastography. Proc. Natl. Acad. Sci. USA. 2012;109:6650–6655. doi: 10.1073/pnas.1200151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Streitberger K.J., Sack I., Krefting D., Pfüller C., Braun J., Paul F., Wuerfel J. Brain viscoelasticity alteration in chronic-progressive multiple sclerosis. PLoS One. 2012;7:e29888. doi: 10.1371/journal.pone.0029888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohashi K., Fujiwara S., Mizuno K. Roles of the cytoskeleton, cell adhesion and rho signalling in mechanosensing and mechanotransduction. J. Biochem. 2017;161:245–254. doi: 10.1093/jb/mvw082. [DOI] [PubMed] [Google Scholar]
- 16.Lim C.G., Jang J., Kim C. Cellular machinery for sensing mechanical force. BMB Rep. 2018;51:623–629. doi: 10.5483/BMBRep.2018.51.12.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mobasheri A., Carter S.D., Martín-Vasallo P., Shakibaei M. Integrins and stretch activated ion channels; putative components of functional cell surface mechanoreceptors in articular chondrocytes. Cell Biol. Int. 2002;26:1–18. doi: 10.1006/cbir.2001.0826. [DOI] [PubMed] [Google Scholar]
- 18.Wang J.H.C., Thampatty B.P. An introductory review of cell mechanobiology. Biomech. Model. Mechanobiol. 2006;5:1–16. doi: 10.1007/s10237-005-0012-z. [DOI] [PubMed] [Google Scholar]
- 19.Syeda R., Florendo M.N., Cox C.D., Kefauver J.M., Santos J.S., Martinac B., Patapoutian A. Piezo1 channels are Inherently mechanosensitive. Cell Rep. 2016;17:1739–1746. doi: 10.1016/j.celrep.2016.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O'Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N., et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coste B., Mathur J., Schmidt M., Earley T.J., Ranade S., Petrus M.J., Dubin A.E., Patapoutian A. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330:55–60. doi: 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pathak M.M., Nourse J.L., Tran T., Hwe J., Arulmoli J., Le D.T.T., Bernardis E., Flanagan L.A., Tombola F. Stretch-activated ion channel Piezo1 directs lineage choice in human neural stem cells. Proc. Natl. Acad. Sci. USA. 2014;111:16148–16153. doi: 10.1073/pnas.1409802111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romac J.M.J., Shahid R.A., Swain S.M., Vigna S.R., Liddle R.A. Piezo1 is a mechanically activated ion channel and mediates pressure induced pancreatitis. Nat. Commun. 2018;9:1715. doi: 10.1038/s41467-018-04194-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nonomura K., Lukacs V., Sweet D.T., Goddard L.M., Kanie A., Whitwam T., Ranade S.S., Fujimori T., Kahn M.L., Patapoutian A. Mechanically activated ion channel PIEZO1 is required for lymphatic valve formation. Proc. Natl. Acad. Sci. USA. 2018;115:12817–12822. doi: 10.1073/pnas.1817070115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X., Wanggou S., Bodalia A., Zhu M., Dong W., Fan J.J., Yin W.C., Min H.K., Hu M., Draghici D., et al. A feedforward mechanism mediated by mechanosensitive ion channel Piezo1 and tissue mechanics promotes glioma aggression. Neuron. 2018;100:799–815.e7. doi: 10.1016/j.neuron.2018.09.046. [DOI] [PubMed] [Google Scholar]
- 26.Cahalan S.M., Lukacs V., Ranade S.S., Chien S., Bandell M., Patapoutian A. Piezo1 links mechanical forces to red blood cell. Elife. 2015;4:e07370. doi: 10.7554/eLife.07370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solis A.G., Bielecki P., Steach H.R., Sharma L., Harman C.C.D., Yun S., de Zoete M.R., Warnock J.N., To S.D.F., York A.G., et al. Mechanosensation of cyclical force by PIEZO1 is essential for innate immunity. Nature. 2019;573:69–74. doi: 10.1038/s41586-019-1485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi M., Du F., Liu Y., Li L., Cai J., Zhang G.F., Xu X.F., Lin T., Cheng H.R., Liu X.D., et al. Glial cell-expressed mechanosensitive channel TRPV4 mediates infrasound-induced neuronal impairment. Acta Neuropathol. 2013;126:725–739. doi: 10.1007/s00401-013-1166-x. [DOI] [PubMed] [Google Scholar]
- 29.Echeverry S., Rodriguez M.J., Torres Y.P. Transient receptor potential channels in microglia: roles in physiology and disease. Neurotox. Res. 2016;30:467–478. doi: 10.1007/s12640-016-9632-6. [DOI] [PubMed] [Google Scholar]
- 30.Cojocaru A., Burada E., Bălșeanu A.T., Deftu A.F., Cătălin B., Popa-Wagner A., Osiac E. Roles of microglial ion channel in neurodegenerative diseases. J. Clin. Med. 2021;10:1239. doi: 10.3390/jcm10061239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konno M., Shirakawa H., Iida S., Sakimoto S., Matsutani I., Miyake T., Kageyama K., Nakagawa T., Shibasaki K., Kaneko S. Stimulation of transient receptor potential vanilloid 4 channel suppresses abnormal activation of microglia induced by lipopolysaccharide. Glia. 2012;60:761–770. doi: 10.1002/glia.22306. [DOI] [PubMed] [Google Scholar]
- 32.Ohana L., Newell E.W., Stanley E.F., Schlichter L.C. The Ca2+ release-activated Ca2+ current (I(CRAC)) mediates store-operated Ca2+ entry in rat microglia. Channels. 2009;3:129–139. doi: 10.4161/chan.3.2.8609. [DOI] [PubMed] [Google Scholar]
- 33.Syeda R., Xu J., Dubin A.E., Coste B., Mathur J., Huynh T., Matzen J., Lao J., Tully D.C., Engels I.H., et al. Chemical activation of the mechanotransduction channel Piezo1. Elife. 2015;4:e07369. doi: 10.7554/eLife.07369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bae C., Sachs F., Gottlieb P.A. The mechanosensitive ion channel Piezo1 is inhibited by the peptide GsMTx4. Biochemistry. 2011;50:6295–6300. doi: 10.1021/bi200770q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng W.Z., Marshall K.L., Min S., Daou I., Chapleau M.W., Abboud F.M., Liberles S.D., Patapoutian A. PIEZOs mediate neuronal sensing of blood pressure and the baroreceptor reflex. Science. 2018;362:464–467. doi: 10.1126/science.aau6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J., Hou B., Tumova S., Muraki K., Bruns A., Ludlow M.J., Sedo A., Hyman A.J., McKeown L., Young R.S., et al. Piezo1 integration of vascular architecture with physiological force. Nature. 2014;515:279–282. doi: 10.1038/nature13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaks M., Giannone G., Rottner K. Actin dynamics in cell migration. Essays Biochem. 2019;63:483–495. doi: 10.1042/EBC20190015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gudipaty S.A., Lindblom J., Loftus P.D., Redd M.J., Edes K., Davey C.F., Krishnegowda V., Rosenblatt J. Mechanical stretch triggers rapid epithelial cell division through Piezo1. Nature. 2017;543:118–121. doi: 10.1038/nature21407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang T.Y., Chen C., Lee M., Chang Y.C., Lu C.H., Lu S.T., Wang D.Y., Wang A., Guo C.L., Cheng P.L. Paxillin facilitates timely neurite initiation on soft-substrate environments by interacting with the endocytic machinery. Elife. 2017;6:e31101. doi: 10.7554/eLife.31101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Möller T. Calcium signaling in microglial cells. Glia. 2002;40:184–194. doi: 10.1002/glia.10152. [DOI] [PubMed] [Google Scholar]
- 41.Eichhoff G., Brawek B., Garaschuk O. Microglial calcium signal acts as a rapid sensor of single neuron damage in vivo. Biochim. Biophys. Acta. 2011;1813:1014–1024. doi: 10.1016/j.bbamcr.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 42.McLarnon J.G., Choi H.B., Lue L.F., Walker D.G., Kim S.U. Perturbations in calcium-mediated signal transduction in microglia from Alzheimer's disease patients. J. Neurosci. Res. 2005;81:426–435. doi: 10.1002/jnr.20487. [DOI] [PubMed] [Google Scholar]
- 43.Combs C.K., Johnson D.E., Cannady S.B., Lehman T.M., Landreth G.E. Identification of microglial signal transduction pathways mediating a neurotoxic response to amyloidogenic fragments of beta-amyloid and prion proteins. J. Neurosci. 1999;19:928–939. doi: 10.1523/JNEUROSCI.19-03-00928.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Velasco-Estevez M., Mampay M., Boutin H., Chaney A., Warn P., Sharp A., Burgess E., Moeendarbary E., Dev K.K., Sheridan G.K. Infection augments expression of mechanosensing Piezo1 channels in amyloid Plaque-reactive astrocytes. Front. Aging Neurosci. 2018;10:332. doi: 10.3389/fnagi.2018.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Copenhaver P.F., Anekonda T.S., Musashe D., Robinson K.M., Ramaker J.M., Swanson T.L., Wadsworth T.L., Kretzschmar D., Woltjer R.L., Quinn J.F. A translational continuum of model systems for evaluating treatment strategies in Alzheimer's disease: isradipine as a candidate drug. Dis. Model. Mech. 2011;4:634–648. doi: 10.1242/dmm.006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy M.C., Jones D.T., Jack C.R., Jr., Glaser K.J., Senjem M.L., Manduca A., Felmlee J.P., Carter R.E., Ehman R.L., Huston J., 3rd Regional brain stiffness changes across the Alzheimer's disease spectrum. Neuroimage. Clin. 2016;10:283–290. doi: 10.1016/j.nicl.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kreutzberg G.W. Microglia: a sensor for pathological events in the CNS. Trends Neurosci. 1996;19:312–318. doi: 10.1016/0166-2236(96)10049-7. [DOI] [PubMed] [Google Scholar]
- 48.Koizumi S., Shigemoto-Mogami Y., Nasu-Tada K., Shinozaki Y., Ohsawa K., Tsuda M., Joshi B.V., Jacobson K.A., Kohsaka S., Inoue K. UDP acting at P2Y6 receptors is a mediator of microglial phagocytosis. Nature. 2007;446:1091–1095. doi: 10.1038/nature05704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holt J.R., Zeng W.Z., Evans E.L., Woo S.H., Ma S., Abuwarda H., Loud M., Patapoutian A., Pathak M.M. Spatiotemporal dynamics of PIEZO1 localization controls keratinocyte migration during wound healing. Elife. 2021;10:e65415. doi: 10.7554/eLife.65415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Y., Wu X., Liu S., Zhao H., Li B., Zhao H., Feng X. Piezo1 regulates migration and invasion of breast cancer cells via modulating cell mechanobiological properties. Acta Biochim. Biophys. Sin. 2021;53:10–18. doi: 10.1093/abbs/gmaa112. [DOI] [PubMed] [Google Scholar]
- 51.Wang J., Jiang J., Yang X., Zhou G., Wang L., Xiao B. Tethering Piezo channels to the actin cytoskeleton for mechanogating via the cadherin-beta-catenin mechanotransduction complex. Cell Rep. 2022;38:110342. doi: 10.1016/j.celrep.2022.110342. [DOI] [PubMed] [Google Scholar]
- 52.Tavares S., Vieira A.F., Taubenberger A.V., Araújo M., Martins N.P., Brás-Pereira C., Polónia A., Herbig M., Barreto C., Otto O., et al. Actin stress fiber organization promotes cell stiffening and proliferation of pre-invasive breast cancer cells. Nat. Commun. 2017;8:15237. doi: 10.1038/ncomms15237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jäntti H., Sitnikova V., Ishchenko Y., Shakirzyanova A., Giudice L., Ugidos I.F., Gómez-Budia M., Korvenlaita N., Ohtonen S., Belaya I., et al. Microglial amyloid beta clearance is driven by PIEZO1 channels. J. Neuroinflammation. 2022;19:147. doi: 10.1186/s12974-022-02486-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Decaestecker C., Debeir O., Van Ham P., Kiss R. Can anti-migratory drugs be screened in vitro? A review of 2D and 3D assays for the quantitative analysis of cell migration. Med. Res. Rev. 2007;27:149–176. doi: 10.1002/med.20078. [DOI] [PubMed] [Google Scholar]
- 55.Smith J.A., Das A., Ray S.K., Banik N.L. Role of pro-inflammatory cytokines released from microglia in neurodegenerative diseases. Brain Res. Bull. 2012;87:10–20. doi: 10.1016/j.brainresbull.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee G.S., Subramanian N., Kim A.I., Aksentijevich I., Goldbach-Mansky R., Sacks D.B., Germain R.N., Kastner D.L., Chae J.J. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature. 2012;492:123–127. doi: 10.1038/nature11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang B.R., Chang P.C., Yeh W.L., Lee C.H., Tsai C.F., Lin C., Lin H.Y., Liu Y.S., Wu C.Y.J., Ko P.Y., et al. Anti-neuroinflammatory effects of the calcium channel blocker nicardipine on microglial cells: implications for neuroprotection. PLoS One. 2014;9:e91167. doi: 10.1371/journal.pone.0091167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng J., Dong Y., Ma J., Pan R., Liao Y., Kong X., Li X., Li S., Chen P., Wang L., et al. Microglial Calhm2 regulates neuroinflammation and contributes to Alzheimer's disease pathology. Sci. Adv. 2021;7:eabe3600. doi: 10.1126/sciadv.abe3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Velasco-Estevez M., Rolle S.O., Mampay M., Dev K.K., Sheridan G.K. Piezo1 regulates calcium oscillations and cytokine release from astrocytes. Glia. 2020;68:145–160. doi: 10.1002/glia.23709. [DOI] [PubMed] [Google Scholar]
- 60.Wu J., Ryskamp D., Birnbaumer L., Bezprozvanny I. Inhibition of TRPC1-dependent store-operated calcium entry improves synaptic stability and motor performance in a mouse model of Huntington's disease. J. Huntingtons Dis. 2018;7:35–50. doi: 10.3233/JHD-170266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J., Lu R., Yang J., Li H., He Z., Jing N., Wang X., Wang Y. TRPC6 specifically interacts with APP to inhibit its cleavage by gamma-secretase and reduce Abeta production. Nat. Commun. 2015;6:8876. doi: 10.1038/ncomms9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen X., Lu M., He X., Ma L., Birnbaumer L., Liao Y. TRPC3/6/7 knockdown protects the brain from cerebral ischemia injury via astrocyte apoptosis inhibition and effects on NF-DB translocation. Mol. Neurobiol. 2017;54:7555–7566. doi: 10.1007/s12035-016-0227-2. [DOI] [PubMed] [Google Scholar]
- 63.Wang Z., Zhou L., An D., Xu W., Wu C., Sha S., Li Y., Zhu Y., Chen A., Du Y., et al. TRPV4-induced inflammatory response is involved in neuronal death in pilocarpine model of temporal lobe epilepsy in mice. Cell Death Dis. 2019;10:386. doi: 10.1038/s41419-019-1612-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Del Mármol J.I., Touhara K.K., Croft G., MacKinnon R. Piezo1 forms a slowly-inactivating mechanosensory channel in mouse embryonic stem cells. Elife. 2018;7:e33149. doi: 10.7554/eLife.33149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request, unless it is protected by law.
This paper does not report original code.
The RNA-sequencing data associated with this project are deposited in the NCBI Sequence Read Archive (SRA) database: PRJNA919004.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.