Significance
Skeletal muscle fibers are highly compartmentalized syncytial cells with many myonuclei. Here, we show that sarcolemma-associated dystrophin protein is organized in focal and nonmobile nuclear domains of ~80 µm size, which we called “basal sarcolemmal dystrophin units (BSDUs).” Organization in BSDUs persists even following therapeutic genomic correction of the murine mdx-23 mutation. Focal restoration of dystrophin in BSDUs precludes more widespread distribution when only some nuclear dystrophin-encoding loci are edited. In contrast, modest correction of the mutation at transcript level results in fully compartmentalized dystrophin restoration including high expression at myotendinous junctions that efficiently attenuates myofiber injury. Our results highlight the importance of understanding the local pharmacodynamics of therapeutics for multinucleated muscle fibers.
Keywords: dystrophin-EGFP, nuclear domain, myotendinous junction, mdx mouse
Abstract
Dystrophin is essential for muscle health: its sarcolemmal absence causes the fatal, X-linked condition, Duchenne muscular dystrophy (DMD). However, its normal, spatial organization remains poorly understood, which hinders the interpretation of efficacy of its therapeutic restoration. Using female reporter mice heterozygous for fluorescently tagged dystrophin (DmdEGFP), we here reveal that dystrophin distribution is unexpectedly compartmentalized, being restricted to myonuclear-defined sarcolemmal territories extending ~80 µm, which we called “basal sarcolemmal dystrophin units (BSDUs).” These territories were further specialized at myotendinous junctions, where both Dmd transcripts and dystrophin protein were enriched. Genome-level correction in X-linked muscular dystrophy mice via CRISPR/Cas9 gene editing restored a mosaic of separated dystrophin domains, whereas transcript-level Dmd correction, following treatment with tricyclo-DNA antisense oligonucleotides, restored dystrophin initially at junctions before extending along the entire fiber—with levels ~2% sufficient to moderate the dystrophic process. We conclude that widespread restoration of fiber dystrophin is likely critical for therapeutic success in DMD, perhaps most importantly, at muscle–tendon junctions.
The loss of sarcolemmal dystrophin in the X-linked condition Duchenne muscular dystrophy (DMD) causes necrosis of muscle fibers, which over time leads to muscle wasting, paresis, and premature death (1–3). To date, there is no cure. Therapeutic gene editing of the disease-associated DMD gene or antisense-induced alternative splicing of its transcript can restore dystrophin by targeted excision/skipping of exons that harbor premature termination codons (PTCs), or that disrupt the reading frame (following deletion or duplication) (4). We previously developed two strategies for antisense-mediated exon skipping: tricyclo-DNA (tcDNA) oligomers and adeno associated virus (AAV)-U7 gene therapy (5, 6). Although both strategies restored high levels of skeletal muscle dystrophin, we found that only tcDNA completely normalized serum creatine kinase (CK) activity, a biomarker of muscle fiber necrosis and dystrophic processes. CK activity decreased but remained elevated after AAV-U7 therapy, suggesting that ongoing muscle fiber damage might be responsible for the observed progressive loss of the AAV genome and further implying that dystrophin restoration might only be temporary (7, 8). The myofiber is a multinuclear syncytium of about 100 myonuclei per mm length in humans (9, 10), and this differential response to therapy (despite similar initial magnitude of restored dystrophin) led us to question whether the multinucleate nature of myofibers might result in distinct subcellular spatial domains, potentially influencing the pharmacodynamics of directed therapies.
Dystrophin is an essential component of the sarcolemmal dystrophin-associated protein complex (DAPC), which links the myofiber cytoskeleton with the basal lamina and extracellular matrix (11). Sarcolemmal dystrophin confers mechanical stability to skeletal muscle fibers during contraction (1), particularly at the myotendinous junction (MTJ) (12), where junctional myonuclei express a specific set of genes encoding junctional enriched proteins (13–16).
The organization of dystrophin distribution at subcellular levels along the entire fiber sarcolemma remains underexplored, although some insight was derived following a study of female DMD carriers. In female mammals (carrying two X chromosomes), random inactivation of one X chromosome occurs early in embryonic development, with subsequent gene expression being near exclusively from the other (17–21). This epigenetic mark is retained during proliferation (22) and within myogenic lineages during differentiation into myofibers. X-chromosomal inactivation (XCI) occurs usually at random (23). Carriers of a mutated DMD allele are thus generally expected to express functional dystrophin from only half of their myonuclei leading to mosaic dystrophin expression patterns (24–28), although skewed XCI is more frequent in carriers than those in the general population (29–31).
The presence of dystrophin protein within revertant fibers (RFs) is another interesting phenomenon that provides insights into subcellular expression. RFs have been examined in detail in the mdx-23 DMD mouse model because of their high prevalence (32–34). RFs arise due to restoration of an open reading frame via spontaneous skipping of the mutated Dmd exon 23 with neighboring exons (to varying extents), with consequent production of internally truncated but functional protein (35).
The organization of the myofiber syncytium can be studied using murine extensor digitorum longus (EDL) muscles, from which intact myofibers can be readily isolated for ex vivo studies (36). We recently generated a DmdEGFP reporter mouse, which harbors an in-frame insertion of the EGFP coding sequence beyond the last Dmd codon, in exon 79 (37). The resulting dystrophin-enhanced green fluorescent protein (EGFP) fusion protein allows live imaging of dystrophin in vivo and ex vivo, where the protein is found in the expected distribution within skeletal, heart, and smooth muscles, with shorter dystrophin-EGFP isoforms also observed as expected in the retina and brain (37). Importantly, DmdEGFP mice had no muscle phenotype and DAPC members were normally present, indicating that the dystrophin-EGFP is fully functional. We also generated a DmdEGFP-mdx mouse line carrying the mdx-23 nonsense mutation in cis (38), with concomitant absence of the dystrophin-EGFP full-length isoform, to facilitate live imaging of therapeutic dystrophin restoration.
Here, we explored the subcellular spatial organization of dystrophin in these novel reporter mouse models and examined the influence of different dystrophin-restoring therapeutic strategies on local pharmacodynamics in single muscle fibers.
Results
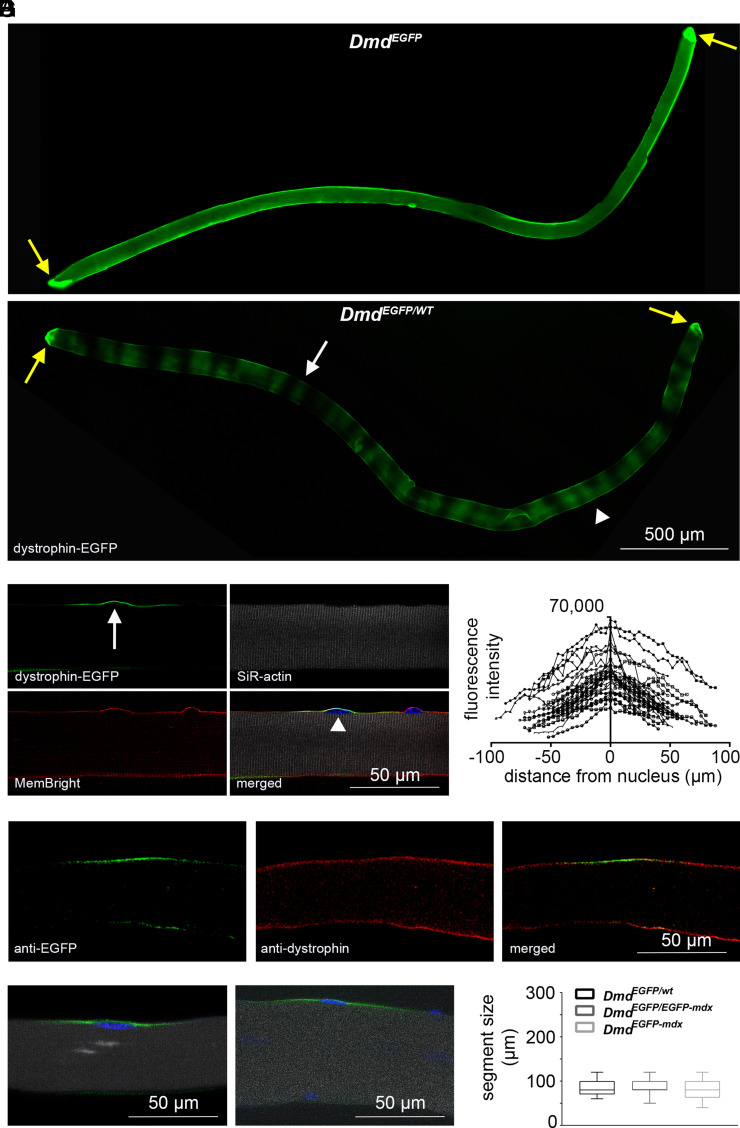
Live imaging of freshly isolated EDL muscle fibers from DmdEGFP mice, both male and female, revealed a continuous distribution of the dystrophin-EGFP fusion protein along the sarcolemma (Fig. 1A). To examine how dystrophin-EGFP is spatially arranged within skeletal muscle myofibers, we exploited random X chromosome inactivation in heterozygous female mice, which harbor one DmdEGFP allele and one wild-type Dmd allele (DmdEGFP/WT mice) (37). Live imaging of these heterozygous EDL muscle fibers revealed a mosaic of positive and negative dystrophin-EGFP segments (Fig. 1A), whereas live staining of the membrane with MemBright or immunostaining using an antibody against the C-terminal domain of dystrophin revealed a continuous signal, confirming the integrity of the sarcolemma and uniformity of dystrophin distribution (Fig. 1 B and D). The smallest dystrophin-EGFP segments consistently extended to similar lengths (~80 µm) along the longitudinal fiber axis, corresponding to about 40 sarcomeres of 2 µm each (Fig. 1 B, D, and G): we termed such minimal segments the “basal sarcolemmal dystrophin unit” (BSDU). Each BSDU had a myonucleus at its center (Fig. 1B), and 3D reconstruction showed that these territories extended a similar distance circumferentially around the myofiber (~50% of the fiber perimeter) (SI Appendix, Fig. S1A). Quantification of sarcolemmal fluorescence intensities along the longitudinal fiber axis showed the highest dystrophin-EGFP values at the center of each BSDU (in the vicinity of the nucleus), tapering toward the periphery, confirming the visual observations (Fig. 1C). Maximum fluorescence intensities varied up to fivefold (Fig. 1C); however, the distribution pattern of fluorescence remained consistent within individual units. Taken together, these results suggest that the BSDU represents a nuclear domain for dystrophin.
Fig. 1.
Organization of dystrophin in basal sarcolemmal units. Live imaging of single fibers freshly isolated from EDL muscles except for the fiber in D. (A) Epifluorescence microscopy of DmdEGFPand DmdEGFP/WT fibers. Example of a small dystrophin-EGFP-positive segment (white arrow). Example of a larger segment with varying dystrophin levels (white arrowhead). Yellow arrows point to MTJs. (B) Confocal microscopy of a DmdEGFP/WTmyofiber section. Native dystrophin-EGFP (green), SiR-actin (gray), MemBright (red), Hoechst (blue). BSDU (arrow). Central nucleus (arrowhead). (C) Quantification of fluorescence intensities of BSDUs from DmdEGFP/WT mice (n = 30 segments of fibers from 3 mice). (D) Confocal microscopy of a DmdEGFP/WTmyofiber section after immunocytochemistry using anti-EGFP (green) and anti-dystrophin (DYS2, red) antibodies. (E and F) Confocal microscopy of a DmdEGFP/EGFP-mdx myofiber section (E) and DmdEGFP-mdx myofiber section (F). Native dystrophin-EGFP (green), SiR-actin (gray), Hoechst (blue). (G) The length of BSDUs was measured in myofibers from DmdEGFP/WT mice (n = 35 myofibers), DmdEGFP/EGFP-mdx mice (n = 114 myofibers), and DmdEGFP-mdxmice (n = 86 myofibers). Data are presented as Whisker–Tukey boxplots. Boxes indicate interquartile ranges (IQR), whiskers indicate (1.5 × IQR).
BSDUs were never observed in strictly alternating blocks of positive and negative segments along the entire fiber, though dystrophin-EGFP-positive (and negative) segments of DmdEGFP/WT fibers were often larger than 80 µm in size (Fig. 1A). Such different segment sizes were likely caused by a stochastic distribution of maternal or paternal X chromosome inactivated myonuclei during fiber formation, giving occasional clusters of adjacent nuclei with the same inactivated X chromosome. Moreover, these larger segments often had varying intensities, suggesting a degree of overlap between BSDUs (Fig. 1A). The sarcolemmal extension of the nuclear domains is usually considered as adjacent units, expressed as total fiber surface divided by the number of myonuclei. By modeling EDL myofibers as cylinders, using previously published parameters (39) (~50 µm diameter, 3.8 mm length, 260 myonuclei) gives a minimal territory of ~2,300 µm2 per myonucleus. The ~80 µm diameter measured here for BSDUs however gives an area closer to 5,000 µm2, approximately twice the theoretical core domain size: this strongly supports an overlapping organization of BSDUs, an arrangement that would ensure a uniform sarcolemmal dystrophin distribution, as found in homozygous DmdEGFP (Fig. 1A), guaranteeing sarcolemmal stability.
The X chromosome-linked muscular dystrophy (mdx) mouse is a commonly used murine model of DMD (2, 40). We recently introduced the mdx-23 nonsense mutation into the DmdEGFP allele to generate the DmdEGFP-mdxmouse line, allowing live imaging of spontaneously or therapeutically restored dystrophin-EGFP (38). We here crossed DmdEGFP-mdxwith DmdEGFP mice and obtained female carriers of the mdx-23 mutation in the F1 generation (DmdEGFP/EGFP-mdx). We examined the distribution of sarcolemmal dystrophin-EGFP in these female carriers, where ~50% of myonuclei would be expected to generate no dystrophin protein. We postulated that such local deficiencies in dystrophin might affect the size and properties of BSDUs generated by adjacent DmdEGFP-active (dystrophin-EGFP positive) myonuclei. Single fibers from DmdEGFP/EGFP-mdx EDL muscles consisted of positive and negative dystrophin-EGFP-positive segments as expected (SI Appendix, Fig. S2A), however measured BSDU parameters were essentially unchanged: minimal dystrophin-EGFP territories were approximately 80 µm in length, with a gradient of fluorescence intensity focused around a subsarcolemmal nucleus, which we assumed being DmdEGFP active (Fig. 1 E and G and SI Appendix, Fig. S1B). This suggests that BSDUs do not exhibit significant compensatory changes in response to dystrophin deficiency in adjacent nuclear domains and that excursion of dystrophin protein from each expressing nucleus is finite [concomitantly, the previously observed increase in dystrophin expression with increasing age in female mdx carrier mice must be dependent on a different, yet unexplored, mechanism (41)].
We also analyzed the distribution of dystrophin-EGFP in the so-called RFs, which spontaneously arise in otherwise dystrophin-deficient hemizygous male and homozygous female DmdEGFP-mdx mice (38). Examined at the single fiber level, most RFs exhibited only one small revertant (EGFP-positive) segment (Fig. 1F). Less frequently, we found RFs with repeated revertant segments that were usually short and interspersed with large negative segments (SI Appendix, Fig. S2B). The size and shape of BSDUs in RF segments were largely consistent with those of heterozygous females (Fig. 1G and SI Appendix, Fig. S1C), suggesting that most revertant segments corresponded to a single nuclear domain. Further, immunolabeling of revertant segments of EDL fibers from classical mdx-23 mice revealed BSDUs that were comparable to those from DmdEGFP-mdx mice, suggesting that the dystrophin-EGFP fusion protein does not alter the size or behavior of BSDUs (SI Appendix, Fig. S1D).
The MTJ is a specialized myofiber compartment adapted for longitudinal force transmission: absence of dystrophin at the MTJ destabilizes the fiber and reduces resistance to tear stress (12). The cellular and molecular mechanisms that result in the previously observed accumulation of dystrophin at the junctional site are still unknown (42). In vivo, in situ, and ex vivo imaging of DmdEGFP muscle fibers revealed high dystrophin-EGFP levels at fiber tips compared with low interjunctional dystrophin-EGFP levels ( Figs. 1A and 2 A–C). High levels of dystrophin were located at the insertion sites of muscle fibers into tendons (Fig. 2 A and B and SI Appendix, Fig. S3 A–C). Coimmunostaining against collagen XXII, a MTJ marker (43), confirmed that dystrophin indeed accumulated in MTJs from DmdEGFP mice as well as C57BL/6 mice (SI Appendix, Fig. S3 A and B). 3D reconstruction suggested that these high levels of dystrophin-EGFP were associated with digital protrusions (Movie S1), typical of the membrane folding at MTJs (44). Higher levels of dystrophin-EGFP (when compared to the mid-fiber region) were always present at the fiber tips (n = 90 fibers from 9 DmdEGFP mice). Similar increased dystrophin at MTJ was found in isolated myofibers from adult rat EDL and flexor digitorum brevis (FDB) muscles (SI Appendix, Fig. S3E). Increased junctional dystrophin-EGFP was associated with increased subjunctional actin accumulation, as shown in situ, ex vivo, and on cryosections (Fig. 2B and SI Appendix, Fig. S3 C and D), consistent with the attributed importance of actin–dystrophin interaction for junctional mechanotransduction (45). Curiously, MTJs in different mosaic states were often only partially dystrophin-EGFP positive, suggesting that junctional dystrophin is also organized into units corresponding to nuclear domain areas (Fig. 2D and SI Appendix, Fig. S3F). Dystrophin-EGFP accumulation in MTJs can partly be attributed to an optical effect of junctional membrane folding, as shown by the parallel increased junctional accumulation of the membrane marker MemBright in fibers from DmdEGFP and DmdEGFP-mdx mice (Fig. 2 C and D); however, the increase in MTJ membrane area is not accompanied by concomitant increase in myonuclear number [as shown by others (46)], implying higher local dystrophin synthesis by these junctional nuclei. Indeed, RNA sequencing of single myonuclei revealed higher Dmd transcription in MTJ nuclei (SI Appendix, Fig. S4), and RNA-fluorescence in situ hybridization (FISH) similarly confirmed the local enrichment of Dmd transcripts at the MTJ in single fibers of wild-type EDL muscles (Fig. 2E).
Fig. 2.
Compartmentalization of dystrophin at the MTJ. (A) In vivo multiphoton intravital microscopy of the TA muscle of DmdEGFP mouse. Left image shows native dystrophin-EGFP. MTJ (white arrows). The Right image shows dystrophin-EGFP together with second harmonic generation (SHG) to highlight the collagen fibrils of the tendon (yellow asterisks). (B) In situ confocal microscopy shows TA muscles from DmdEGFP mice. Dystrophin-EGFP (green), SiR-actin (red), SHG (gray). SiR-actin at MTJ level (white arrows). Tendon (yellow asterisk). (C and D) Ex vivo epifluorescence microscopy shows live single myofibers freshly isolated from EDL muscles of DmdEGFP mice (C) and DmdEGFP-mdxmice (D). MemBright accumulated at the MTJ in the presence (white arrow) or absence (white arrowheads) of dystrophin-EGFP. A small MTJ segment from the DmdEGFP-mdxmice expressed revertant dystrophin-EGFP (yellow arrow) (E) Confocal microscopy of an isolated EDL fiber from wild-type mice after RNA-FISH to detect Dmd transcripts. The tip of the fiber appears on the left side. 5′-Dmd probe (green) and DAPI-stained nuclei (blue). Nascent transcripts are detected with the 5′-Dmd probe and appear as large intranuclear foci (white arrowheads), while smaller sarcoplasmic foci concentrate mostly at the fiber tip (yellow arrows).
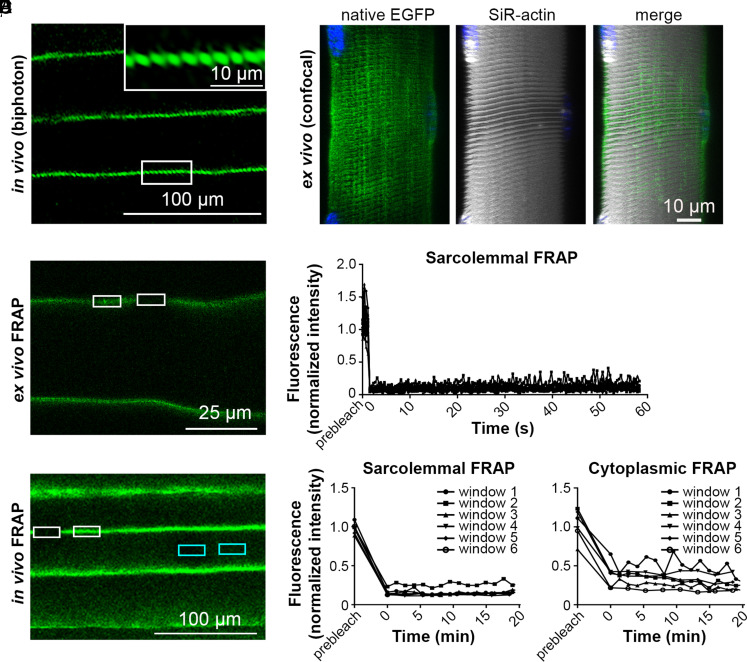
Sarcolemmal dystrophin-EGFP displayed a “woven” pattern after in vivo and ex vivo imaging, and regular repeats were visible along the longitudinal axis of the myofibers, corresponding in size and position to the sarcoplasmic striation stained with SiR-actin (Fig. 3 A and B and Movie S1). Live imaging thus confirmed the previously described costameric organization of dystrophin (47), suggesting specific sarcolemmal anchoring at these sites and consequent lateral immobility. We tested this hypothesis by measuring fluorescence recovery after photobleaching (FRAP) in isolated EDL myofibers from DmdEGFP mice (Fig. 3 C and D). Measurements of basal sarcoplasmic fluorescence were low and comparable with extracellular noise, arguing against the existence of a mobile, soluble dystrophin pool that might equilibrate with sarcolemmal dystrophin: this suggested that the behavior of dystrophin within adult mammalian myofibers differs from that within developing myofibers in zebrafish embryos where rapid turnover of cytoplasmic mobile dystrophin is reported (48). Sarcolemmal fluorescence in defined longitudinal regions conversely decreased sharply following intentional bleaching and no gradient in fluorescence recovery was observed after repeated intensity measurements when normalized to unbleached sarcolemmal stretches (Fig. 3 C and D). The tendency of isolated fibers to contract following bleaching precluded measurements longer than 60 s; thus, we repeated the FRAP assay in vivo. No recovery of sarcolemmal fluorescence was observed even 20 min after bleaching (Fig. 3 E and F). This suggests that dystrophin from the unbleached sarcolemma did not move into the bleached zone, as observed previously with immobile dystrophin in zebrafish MTJs (48). In contrast to the isolated ex vivo fibers, we detected weak sarcoplasmic fluorescence in vivo; however, this also did not recover after bleaching (Fig. 3 E and F), suggesting that sarcolemmal dystrophin in neighboring fibers is the most likely source for this background signal (a phenomenon that is largely unavoidable when FRAP is performed in solid tissue). Taken together, our results suggest that dystrophin is not freely mobile along the sarcolemma, explaining (at least in part) its apparent territorial restriction to defined BSDUs.
Fig. 3.
Sarcolemmal dystrophin is organized in a costameric pattern and lacks lateral mobility. (A) In vivo multiphoton microscopy of hindlimb muscle of DmdEGFP mice revealed a costameric pattern of dystrophin-EGFP. The Inset shows the sarcolemma in higher magnification. (B) Three dimensional reconstruction of ex vivo confocal microscopy of live EDL fibers from heterozygous DmdEGFP-mdx/EGFP mice. Dystrophin-EGFP (in green) and SiR-actin (gray). (C) Ex vivo confocal microscopy of live single fibers and (E) in vivo confocal microscopy of hindlimb muscle after photobleaching of selected regions with a high-intensity laser pulse. Rectangles show ROIs where EGFP fluorescence intensity was measured for sarcolemmal (white) and cytoplasmic (cyan) EGFP in bleached and nonbleached regions. (D and F) Normalized fluorescence intensity curves of bleached/unbleached ROIs for FRAP recovery up to 1 min (D) and 20 min (F) after bleaching of dystrophin-EGFP in sarcolemma (D and F) and cytoplasm (F) [four isolated fibers, n = 12 ROIs (D); two mice, n = 6 ROIs for cytoplasmic or sarcolemmal EGFP (F)].
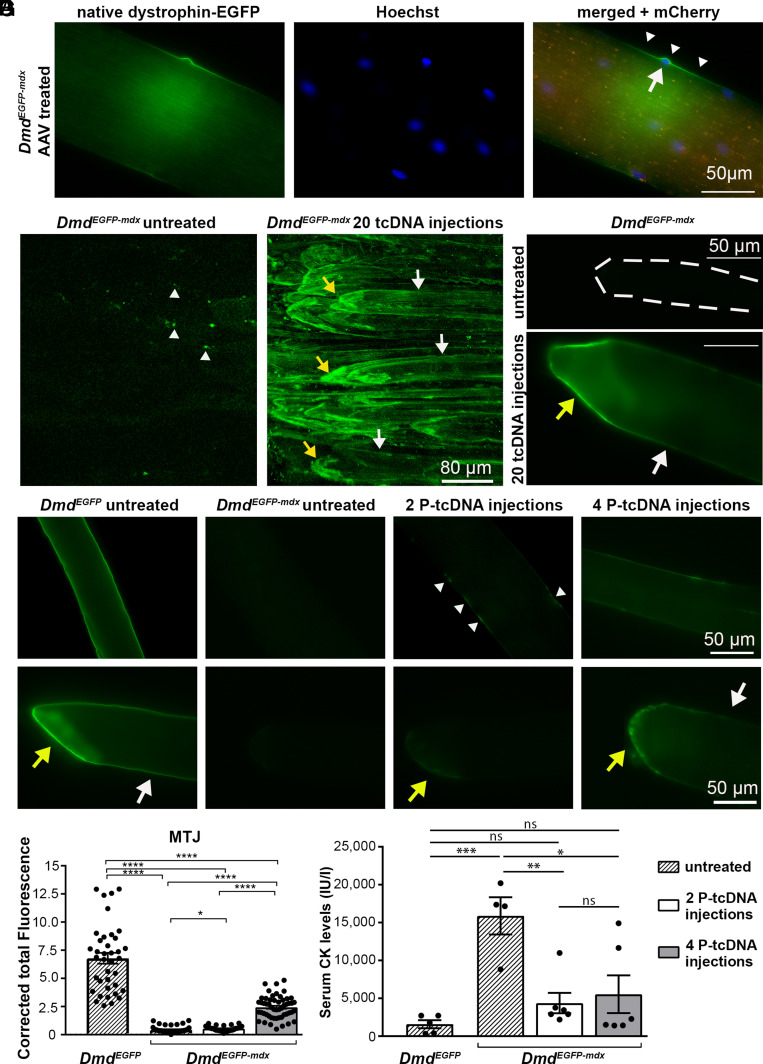
We next investigated the pharmacodynamics of dystrophin following therapeutic restoration at the genomic- (via gene editing) or transcript- (via antisense tcDNA) level. First, we targeted Dmd DNA by injecting 4.6e10 vg AAV-SaCas9-sgRNA22 in combination with AAV-mCherry-sgRNA23 vectors into the tibialis anterior (TA) muscle group of adult DmdEGFP-mdx mice, to induce excision of the mutant Dmd exon 23 containing the PTC. Seven weeks after AAV injection, large numbers of myofibers expressed cytoplasmic diffusible mCherry, demonstrating successful transduction. Accordingly, dystrophin-EGFP was also restored in these mCherry-positive myofibers but crucially, restoration appeared in a segmental fashion (Fig. 4A). Dystrophin-positive segments were randomly distributed, approximately 80 µm in length, and centered around individual myonuclei with EGFP fluorescence extending outward in a gradient (Fig. 4A and SI Appendix, Fig. S5 A and B). We also frequently found partial recovery of dystrophin-EGFP at the MTJs (SI Appendix, Fig. S5C). These results indicate that AAV-mediated gene editing restored dystrophin within independent and separate BSDUs.
Fig. 4.
Experimental restoration of dystrophin-EGFP. (A) Epifluorescence microscopy shows single live myofibers isolated from EDL muscles of DmdEGFP-mdx mouse after injection of AAV-SaCas9-sgRNA22 and AAV-mCherry-sgRNA23 vectors. Dystrophin-EGFP in green (Left image), Hoechst in blue (Middle image), mCherry in red (Right merged image). Restored BSDU (arrowheads) and central myonucleus (arrow). (B) In vivo multiphoton microscopy of hindlimb muscle from untreated DmdEGFP-mdx control mice (Left image) with autofluorescent macrophages (white arrowheads) and muscle from tcDNA-treated mice (Right image) with restored dystrophin-EGFP at the MTJ (yellow arrows) and interjunctional sarcolemma (white arrows). (C) Epifluorescence microscopy of a single live myofiber from untreated DmdEGFP-mdx mice (Top image; dotted line outlines fiber) and after tcDNA treatment (Bottom image). MTJ (yellow arrow) and interjunctional sarcolemma (white arrow). (D and E) Epifluorescence microscopy with identical microscopy settings of live single myofibers from EDL muscles of untreated DmdEGFP mice, untreated DmdEGFP-mdx mice, and after two P-tcDNA and after four P-tcDNA injections of DmdEGFP-mdx mice (from Left to Right) of interjunctional sarcolemma (D) and fiber tips (E). Interspersed dystrophin-EGFP patches (white arrowheads), MTJ (yellow arrows), and interjunctional sarcolemma (white arrows). (F) The corrected total intensity of native dystrophin-EGFP fluorescence was measured and compared between the different genotypes and conditions at the MTJ level. The dot plots show the values of the analyzed areas with a histogram of the average and SEM (between 8 and 38 fibers were analyzed). P values were calculated using the Mann–Whitney test. (G) Serum CK levels were quantified in DmdEGFP mice in comparison with untreated and treated DmdEGFP-mdx mice. All values are expressed as mean ± SEM. Points represent individual mice from each group (n = 4 to 6 mice). Statistical analyses were performed using one-way ANOVA followed by Sidak multiple comparison test (ns = not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Next, we treated DmdEGFP-mdx mice for 20 wk with tcDNA at a dose of 200 mg/kg, a dosing regimen we have used previously (5). This approach gave robust exon skipping (as determined via qPCR) and ~50% dystrophin restoration levels via western blot (SI Appendix, Fig. S6 A–D), which was associated with normalization of histopathological signs (SI Appendix, Fig. S7A). Remarkably, in vivo and ex vivo imaging revealed low levels of homogeneously distributed dystrophin-EGFP at interjunctional sarcolemma and high levels in the MTJs, suggesting that tcDNA treatment produces modest restoration in essentially all BSDUs (Fig. 4 B and C and SI Appendix, Fig. S7B). Furthermore, serum CK activities completely normalized (SI Appendix, Fig. S6E).
We recently prepared palmitoyl-conjugated tricyclo-DNA (P-tcDNA) oligomers in order to improve pharmacokinetics (49). We observed the onset of dystrophin restoration after two or four once-weekly systemic injections of 50 mg/kg P-tcDNA (mice were analyzed 14 d after the last injection). qPCR showed that Dmd-exon 23 skipping was ~3% after two injections in the TA muscle, increasing to ~8% after four injections, with similarly low levels found in other muscles (SI Appendix, Fig. S8 A and B). Western blot showed recovery of ~2% and ~6% dystrophin-EGFP after two and four injections, respectively (SI Appendix, Fig. S8 C–E). In isolated EDL myofibers following only two injections, sarcolemmal dystrophin-EGFP was detected (faintly) in rare, sporadic patches, a few micrometers in size, whereas higher levels of dystrophin-EGFP were already present at the MTJ (Fig. 4 D–F), suggesting modest but widespread initiation of dystrophin restoration. After four injections, recovery at the MTJ increased, and a continuous low intensity of sarcolemmal dystrophin-EGFP became visible along the entire fiber (Fig. 4 D–F), again suggesting low but ubiquitous restoration of dystrophin across all BSDUs. Furthermore, while young adult untreated DmdEGFP-mdx had markedly elevated serum CK activities (15.886 ± 2.455 IU/L, compared with a mean of 1.902 ± 532 IU/L in DmdEGFP controls), as few as two or four P-tcDNA injections were sufficient to significantly reduce CK activities (to 4.885 ± 1.343 IU/L or 5.534 ± 2.490, respectively) (Fig. 4G).
Absence of dystrophin causes deficiency of DAPC members in DMD (50). We thus investigated whether dystrophin restoration via tcDNA normalized levels of the DAPC member β-sarcoglycan in a similar manner. β-sarcoglycan was distributed at high levels at MTJs from DmdEGFP control muscles, where it colocalized with collagen XXII and with dystrophin (SI Appendix, Fig. S9A). Lower levels were found at nonjunctional sarcolemma (SI Appendix, Fig. S9A). β-sarcoglycan levels were markedly lower (though not zero) in untreated DmdEGFP-mdx mice: protein remained faintly visible at MTJs, but was almost absent at sarcolemmal level (SI Appendix, Fig. S9B). Two tcDNA injections did not result in an observable restoration of β-sarcoglycan, while four tcDNA injections modestly increased protein levels at the MTJs when compared with untreated DmdEGFP-mdx mice. Twenty injections of tcDNA restored β-sarcoglycan to levels comparable to DmdEGFP control muscles, both at MTJs and nonjunctional sarcolemma (SI Appendix, Fig. S9B).
The domain-restricted nature of dystrophin in mature myofibers prompted us to examine the behavior during myofiber development in vitro. We, therefore, generated myofibers using primary mouse myoblasts isolated from DmdEGFP and DmdEGFP-mdx mice, either cultured individually, or cocultured at 1:7, 1:3, and 1:1 ratios (12.5%, 25%, and 50% DmdEGFP myoblasts, 87.5%, 75%, and 50% DmdEGFP-mdx myoblasts, respectively) (n = 3 for each condition) (SI Appendix, Fig. S10). Following 7 d of differentiation, thin (~5 µm in diameter) but striated myofibers were observed: these cells were multinucleated, with individual nuclei typically separated from one another by ~100 µm (SI Appendix, Fig. S10). Sarcolemma-enriched dystrophin-EGFP expression was widespread in myofibers derived from DmdEGFP myoblasts (SI Appendix, Fig. S10A), while (as expected) no signal was found in myofibers derived from DmdEGFP-mdx myoblasts (SI Appendix, Fig. S10B). In DmdEGFP/DmdEGFP-mdx myoblast cocultures, irrespective of whether 1:7, 1:3, or 1:1 mix, dystrophin-EGFP-positive and dystrophin-EGFP-negative segments were often present within the same myofiber, suggesting that heterokaryons had formed (SI Appendix, Fig. S10C). Dystrophin-EGFP-positive regions varied in length, with their shortest segments measuring a mean of ~60 µm and the dystrophin-EGFP-positive segments typically positioning close to a myonuclei (n = 33 observations) (SI Appendix, Fig. S10 C and D). These results suggest that sarcolemmal dystrophin-EGFP is also regional in in vitro differentiated myofibers, resembling BSDUs of adult myofibers.
Discussion
The phenomenon of muscle syncytial proteins being organized in nuclear domains was first observed by Pavlath et al. in human–mouse heterokaryons (51). The nuclear domain of dystrophin, however, remained underexplored, although segmentally restricted distribution was found in RFs (35) following transplantation of dystrophin-competent myoblasts into mdx mice (52) and in female mdx carrier mice (53). Here, we provide conclusive evidence that sarcolemmal dystrophin is organized into highly focal and nonmobile nuclear domains of ~80 µm in size, which we called BSDUs.
Although the localized distribution of dystrophin has until now remained largely ignored, various groups have examined organization of other proteins in syncytial muscle cells. Pioneering work from Blau et al. suggested the existence of factors that diffused between nuclei of heterokaryons (54). Ralston and Hall then provided evidence for the widespread diffusion of cytoplasmically soluble beta-galactosidase (β-gal), and translocation of nuclear localizing β-gal in myotubes (55). Yang et al. and others demonstrated β-gal translocation over distances up to 150 µm in vivo (56–58). However, it remained unclear whether such translocation involved protein or RNA diffusion. Bajanca et al. overexpressed human dystrophin-EGFP fusion protein in embryonic zebrafish muscle and demonstrated, using FRAP, the existence of three distinct dystrophin pools: one immobile and sarcolemmal, one diffusible and cytosolic, and a third, “mobile-bound” pool, capable of rapid exchange between sarcolemma and cytosol (48). In our FRAP experiments, however, we were only able to reveal the presence of immobile sarcolemma-bound dystrophin-EGFP (although we cannot exclude the existence of a diffusible cytoplasmic fraction that was below the level of detection).
We here showed that Dmd transcripts were present in myonuclei and cytoplasm, confirming previous work (59–61), whereas García-Rodriguez et al. found a primarily nuclear localization (62). Denes et al. and Pinheiro et al. demonstrated that cytosolic transport of notably giant RNA species such as Dmd required microtubules (60, 61). Denes et al. studied EDL myofibers and described the highest density of Dmd transcripts close to the myonuclei, which tapered toward zero over a ~40 to 50 µm distance (60). These RNA distributions correspond well with the BSDUs identified in our work, where the highest dystrophin protein levels are found in the immediate vicinity of the myonucleus but then decline over a similar distance. In contrast, Pinheiro et al. found a more uniform distribution of Dmd transcripts longitudinally in myofibers that were generated in vitro (61). We here demonstrated that sarcolemmal dystrophin-EGFP was regional in in vitro myofiber cultures generated from mixed DmdEGF and DmdEGFP-mdx myoblasts: these resembled the BSDUs of adult myofibers. However, in the absence of a suitable nuclear marker to ascertain the position of respective nuclei in a heterokaryon, we cannot unequivocally confirm the spatial relationship between dystrophin-competent nuclei and dystrophin-positive segments. Moreover, it is challenging to fully differentiate mature myofibers in culture: it will thus be interesting to explore how BSDUs develop in newly formed or regenerating myofibers in vivo. Finally, we cannot exclude the possibility that the EGFP-tagged Dmd alters RNA properties (such as stability or transport), which might influence BSDU size. This seems unlikely however, given that we show that the BSDU size of RFs from DmdEGFP-mdx mice is similar to those in mdx mice, excluding a major effect of EGFP.
In addition to translocation of RNA and protein, myonuclei themselves can be mobile: Gache et al. showed that while only a fraction of myonuclei move in myotubes, these can nevertheless achieve velocities of ~0.3 µm/min (63). In cultured isolated myofibres, Iyer et al. measured markedly lower velocities (~0.03 µm/min) (64), though given the ~24 d half-life of dystrophin protein (65), a unidirectional velocity of even 0.03 µm/min would be sufficient to impact the BSDU size. Iyer et al. further showed that myonuclei from undamaged wild-type fibers appeared to move in an essentially random manner, however remaining within a small (~10 μm diameter) region over the 10-h measurement period (though mdx myonuclei were observed to move faster and further and in a more directional fashion) (64). Roman et al. recently demonstrated directed myonuclear movement to sites of fiber damage (66); however, it is at present unclear how these findings would translate to a dystrophic context. Our data suggest that the BSDU architecture of RFs or after AAV-mediated gene editing is not significantly different from that within healthy muscle, implying that BSDUs might be robust to damage-associated myonuclear motility.
We found that peak EGFP fluorescence intensity within individual BSDUs varied by as much as fivefold, though intensity did not alter BSDU size. Sardone et al. also observed variation in sarcolemmal dystrophin intensities between fibers (67), though differences were more modest than those found here, as might be expected (analysis of transverse sections will sample contributions from multiple BSDUs). Nuclear movement might also contribute to variation in peak BSDU intensities: static myonuclei could locally increase dystrophin and thus cause sharper peaks than mobile ones. Alternatively, transcriptional and translational activities might vary between individual nuclei, a hypothesis that remains to be explored. Kinoshita et al. demonstrated that higher dystrophin levels were associated with increased nuclear domain size: mdx mice transplanted with myoblasts isolated from Tg-MDA mice (Tg-MDA, for mouse creatine kinase [MCK] promoter Dystrophin complementary DNA [cDNA] plus polyAdenylation. Transgenic mice which overexpress dystrophin 50-fold) exhibited fibers with dystrophin-positive segments three times longer (~1,300 μm) than those found following transplant of myoblasts expressing wild-type levels of dystrophin (~440 μm) (52). Of note, Kinoshita et al. assumed that only one transplanted myoblast fused per fiber and thus defined emerging dystrophin segments as nuclear domains, but multiple fusions could not be excluded. Elevated dystrophin expression might also enhance fiber survival, facilitating subsequent fusion events.
We here observed a mosaic distribution of dystrophin-EGFP in female mice heterozygous for the DmdEGFP allele, enabling measurement of BSDU size. Our data are consistent with expression from only a single Dmd locus per myonucleus, and indeed van Putten et al. showed that nonrandom XCI in mdx-XistΔhs mice correlated with the extent of dystrophin expression (53), but we cannot empirically assert that the mosaic distribution of dystrophin shown here corresponds to the XCI pattern. Future studies, combining in situ labeling of nuclear Dmd-EGFP transcripts with dystrophin-EGFP immunocytochemistry, might address this issue.
Samitt and Bonilla first reported on the high levels of dystrophin at MTJ in adult human muscle (42). As we show here, fibers isolated from DmdEGFP EDL muscles invariably exhibited high levels of dystrophin-EGFP at both tips, and three dimensional reconstructions of these tips suggested that this dystrophin was associated with digital membrane protrusions [in agreement with the ultrastructural description of MTJs at the tips of isolated fibers by Trotter et al. using scanning electron microscopy (44)]. Further, dystrophin enrichment was readily detected in vivo, in situ, and after histology at positions where fibers connect to the intramuscular tendon region, which we confirmed using collagen XXII as a separate MTJ marker (43). Accordingly, we conclude that dystrophin does indeed exhibit marked MTJ-associated enrichment. Recent single-nuclei transcriptomics identified a specific subtype of MTJ myonuclei (13–16), and we confirmed higher numbers of Dmd transcripts in these regions. Bassett et al. demonstrated a junctional distribution of Dys mRNA and dystrophin protein in zebrafish embryos (68). Further experiments are now required to ascertain whether these regional-specific increases in transcript levels are indeed associated with higher transcription by MTJ myonuclei (or increased stability of mature transcripts). Alternatively, long-range, longitudinal transport of dystrophin transcripts might result in accumulation at fiber tips due to the physical disruption imposed by the fiber terminus (a “traffic jam” phenomenon) or due to targeted shuttling. We have shown however that dystrophin within fiber tips also exhibits mosaic, domain-restricted behavior, suggesting that BSDU organization persists in this compartment, arguing in favor of locally transcribed Dmd.
Law and Tidball and Hakim et al. showed that loss of dystrophin caused ultrastructural defects and age-associated weakening of MTJs in mdx mice (12, 45). Here, we showed that short-term treatment with tcDNA initiated dystrophin restoration preferentially at MTJs, which was associated with a reduction in CK (suggesting that MTJs regained mechanical properties, a hypothesis that remains to be ascertained by future work). Longer treatment with tcDNA restored dystrophin and DAPC member β-sarcoglycan both at the MTJ and interjunctional sarcolemma, suggesting restoration of DAPC across all BSDUs in a manner consistent with canonical expression. Also, 50% dystrophin restoration via tcDNA was sufficient to normalize CK activity, suggesting protection from myofiber necrosis. This is consistent with previous results showing that >10% restoration levels in mdx mouse are sufficient for normalization of serum biomarkers following AON-induced exon skipping (5, 49, 69, 70), whereas such normalization was not found when comparable levels of dystrophin were present in mosaic fashion, within mdx-XistΔhs mice (53). García-Rodriguez et al. demonstrated that the mdx mutation caused a fourfold reduction in Dmd transcripts due to transcriptional repression by H3K9 methylation (62). We have not explored whether epigenetic DNA modifications limit the efficacy of therapeutic exon skipping and dystrophin restoration in BSDUs and this remains an area for future study.
We conclude that dystrophin is localized to discrete and well-defined regions (BSDUs) governed by individual myonuclei. In normal muscle, these BSDUs overlap to ensure continuous dystrophin distribution and concomitant fiber protection, and BSDUs further specialize at muscle–tendon boundaries, likely to meet greater physical demands. We further suggest that modest but uniformly distributed restoration of dystrophin is likely to be of more therapeutic benefit to DMD patients than high-level correction in only a subset of myonuclei.
Materials and Methods
Mouse Models.
All animal experiments were conducted according to National and European legislation and institutional guidelines for the care and use of laboratory animals approved by the French government (Ministère de l’Enseignement Supérieur et de la recherche, authorization APAFiS). Mice were bred in the Plateforme 2Care animal facility, UFR Simone Veil—Santé, Université de Versailles Saint-Quentin-en-Yvelines, and were maintained in a standard 12-h light/12-h dark cycle with free access to food and water. The mice were weaned after 4 wk postnatally, and two to five individuals were housed per cage. To generate heterozygous DmdEGFP/+ female mice, hemizygous male DmdEGFP/y mice (37) were crossed with wild-type female C57BL/6 mice purchased from Charles River Laboratories (USA). To generate heterozygous DmdEGFP-mdx/EGFP females, male DmdEGFP-mdx/y mice (38) were crossed with homozygous DmdEGFP/EGFP female mice. For clarity, homozygous females and hemizygous males are designated as DmdEGFPand DmdEGFP-mdx, respectively, as previously described (37, 38). Sprague Dawley rats were purchased (Janvier Labs). All the animal studies were performed on adult animals.
Gene Editing.
pAAV-SaCas9-sgRNA22 and pAAV-mCherry-sgRNA23 were cloned as previously described (38). Adenovirus-free rAAV2/9 viral preparations were generated by packaging AAV2-ITR recombinant genomes in AAV9 capsids, using a three plasmid transfection protocol (38). Viral suspensions (50 µL) containing the combination of rAAV2/9-SaCas9-sgRNA22 and rAAV2/9-mCherry-sgRNA23 at a titer of 4.6e10 vg each were injected intramuscularly into the TA muscle of 6/7-wk-old DmdEGFP-mdx mice. The mice were killed 7 wk postinjection and muscles were harvested as described below.
Antisense Oligonucleotide (AON) Treatment.
tcDNA and P-tcDNA AON (SQY Therapeutics, France) were used to target the donor splice site of murine Dmd exon 23 as previously described (5, 49). Adult 6-wk-old DmdEGFP-mdxmice, under general anesthesia using 1.5 to 2% isoflurane, were given tcDNA via systemic injection into the retro-orbital sinus in three experimental protocols: i) two injections of P-tcDNA at 50 mg/kg once weekly; ii) four injections of P-tcDNA at 50 mg/kg once weekly; and iii) 20 injections of tcDNA at 200 mg/kg once weekly. The mice were killed 2 wk following the last injection and muscles were harvested as described below.
Tissue Preparation, Histology, and Immunocytochemistry.
The muscles were mounted on 6% Tragacanth gum 292 (Sigma-Aldrich), snap frozen in liquid nitrogen-cooled isopentane, and stored at −80 °C.
Hematoxylin-eosin staining was performed using a multistainer LEICA ST5020 following standard procedures. Images were recorded using a digital slide scanner (Leica Biosystems) and analyzed using ImageScope software (Leica Biosystems).
Isolated muscle fibers were prepared from EDL muscle as described by us (38) and others (36). The isolated live myofibers were either immediately mounted on slides for direct EGFP-fluorescence imaging or mounted following a 10-min incubation with SiR-actin (1:1,000 dilution, Spirochrome), MemBright (1:1,000 dilution, 550 Lipilight by MemBright, Idylle), and Hoechst (1:1,000 dilution, Sigma-Aldrich) in isolation medium [FluoroBrite Dulbecco's Modified Eagle's Medium (DMEM, ThermoFisher) supplemented with 1% penicillin/streptomycin, 4 mM L-glutamine (ThermoFisher), and 1% sodium pyruvate (ThermoFisher)]. Isolated fibers from adult rat EDL and FDB muscles were prepared using the protocol for enzymatic digestion as described (38).
For immunocytochemistry, muscle fibers were fixed with 4% PFA directly after enzymatic isolation. For mechanical fiber isolation, EDL muscles of C57BL/6 mice were fixed in 4% paraformaldehyde (PFA) for 30 min, then washed in phosphate buffered saline (PBS), and fibers were separated using blunt glass needles. For immunohistochemistry, cryosections were fixed with 4% PFA for 10 min. Immunostaining was performed as previously described (36) using primary antibodies against C-terminal part of dystrophin (1:10 dilution, NCL-DYS2, mouse IgG1, Leica Biosystems; or 1:500 dilution, RB-9024-P, rabbit pAb IgG, ThermoFisher Scientific), GFP (1:500 dilution, ab13970, chicken pAb IgG, Abcam), Col XXII (1:100 and 1:250 dilution for isolated fibers and cryosections, respectively, ab121846, rabbit pAb IgG, Abcam), and β-sarcoglycan (1:50 dilution, NCL-L-b-SARC, mouse IgG1, Leica Biosystems), incubated overnight at 4 °C, followed by incubation for 1 h at room temperature with fluorochrome-labeled secondary antibodies (1:400, Alexa Fluor goat anti-mouse IgG1-555/633, goat anti-chicken IgG-555/647, and goat anti-rabbit IgG (H+L)-488/555/633). Fibers were then mounted on slides using Fluoromount-G (SouthernBiotech).
Fluorescence Imaging.
Fluorescent images of freshly isolated or stained myofibers, as well as freshly cut or stained muscle sections, were acquired with either an epifluorescence microscope or a confocal microscope as specified in the figure legends. Epifluorescence imaging was conducted with a Zeiss Axio Imager microscope equipped with Orca camera (Hamamatsu) and AxioVision software (Carl Zeiss) for isolated fibers and Olympus IX83 microscope equipped with Orca R2 camera (Hamamatsu) and CellSens software (Evident/Olympus) for in vitro generated fibers. Confocal imaging was conducted with a Leica SP8 white light laser scanning confocal microscope with LAS X software (Leica Biosystems).
Intravital and in situ muscle fluorescence images were acquired using a combined confocal/multiphoton point-scanning microscope (SP8 MP-Leica microsystem, Wetzlar) as previously described (38). Briefly, for intravital imaging, a longitudinal skin and subcutaneous incision was made laterally in the pelvic limb, distal to the femorotibial joint to expose the underlying TA muscle. The mice were kept at 37 °C in the microscope’s incubation chamber and the exposed muscle was stably positioned on a coverslip fixed to the sample holder to lower motion artifacts. For in situ-ex vivo confocal imaging, TA muscles were dissected and incubated with SiR-actin (as described above) and weighed down using metallic coins in µ-Dish35mm, high, Glass Bottom (Ibidi) filled with PBS to prevent dehydration. The 3D video was obtained using Imaris software (Bitplane AG).
Serum Analysis.
Analyses of serum CK activities were performed in the pathology laboratory at the Mary Lyon Centre, Medical Research Council, Harwell, Oxfordshire, UK.
FRAP.
FRAP was performed to bleach dystrophin-EGFP in regions of interest (ROIs) at sarcolemmal and cytoplasmic positions either intravitally (as described above) or on live isolated fibers from DmdEGFP mice. FRAP experiments were conducted by using a combined confocal/multiphoton point-scanning microscope (SP8 MP-Leica Microsystems). Images were obtained before and after photobleaching in the 488-channel at 4% and 14% laser power, for isolated fibers and intravital muscle, respectively. ROIs were bleached by 100% laser power in the 488-nm channel for 6 to 7 iterations during a total of 1.6 to 2 s. FRAP image acquisition time is described for each experiment in the figure legends. Pixel intensities of the ROIs were acquired using Icy software. Control ROIs of an unbleached dystrophin-EGFP area (as illustrated in the figures) were also acquired as an internal control of photobleaching during imaging and for normalization of the pixel intensity of FRAP ROIs at the same time points.
Quantification of Sarcolemmal Dystrophin-EGFP.
The size of small individual dystrophin-positive areas (BSDUs) was measured on single fibers of EDL muscles from experimental mice as detailed in the text using a stage micrometer (S12, Graticules Optics Ltd.). The length of BSDUs was shown on live fibers for representative images, and quantification was performed on fixed single fibers that were immunostained against EGFP or dystrophin as described above. The extremities of BSDU were defined as the position at which intensities dropped to background levels.
For quantification of dystrophin-EGFP fluorescence intensities, images were acquired using the same acquisition parameters between samples. The fluorescence intensity of basal dystrophin units was quantified using Adobe Photoshop 2020 software. Four-micrometer-square ROIs were laid along the sarcolemma of the entire basal segments and fluorescence intensity was determined for each ROI. Background fluorescence was determined from three ROIs outside the fibers.
Fluorescence intensity of dystrophin-EGFP in MTJs of single fibers from DmdEGFP mice, from untreated DmdEGFP-mdx mice, and from P-tcDNA-treated DmdEGFP-mdx mice, was measured using Image J software. From each image, 5 to 7 different ROIs of the same size were selected from MTJ. Background fluorescence was determined from three ROIs in the cytoplasmic region of each fiber.
The corrected total cell fluorescence (CTCF) was calculated from extracted values as:
CTCF = (integrated density) − ([area of selected cell] × [mean fluorescence of background readings]). Background value was subtracted from the average of CTCF value of MTJ for each individual fiber.
RNAscope In Situ Hybridization (ISH).
RNAscope (ACDbio) was performed essentially as previously published (59). EDL single fiber ISH used the method of Kann and Krauss (71) with Mm-Dmd (452801), targeting the 5′ (exons 2 to 10) region of the dp427 transcript. TSA-Cy3 (Akoya Biosciences) was used for labeling (1:500 dilution).
Statistical Analysis.
Data were analyzed with GraphPad Prism 7 software (GraphPad). Values for replicates (“n”) are indicated in the legends. Experimental groups were compared using nonparametric Mann–Whitney U tests for two-group comparisons and ANOVA tests for comparison of three or more groups followed by multiple comparison tests as specified in the figure legends. Significance values were set at *P < 0.05, **P < 0.001, ***P < 0.0001, and ****P < 0.00001.
Additional Materials and Methods.
Additional methods regarding supplementary figures are available in the SI Appendix. The latter include protocols for RNA and western blot analysis as well as primary myofiber cultures.
Supplementary Material
Appendix 01 (PDF)
Dystrophin-EGFP distribution at the myotendinous junction. Video depicts 3D reconstruction of dystrophin-EGFP at the level of a myotendinous junction following ex vivo confocal microscopy of a freshly isolated single myofiber from EDL muscle of a 6-month-old DmdEGFP mouse.
Acknowledgments
We thank Thomas Bestetti, Chloé Dambrune, Karima Relizani, and Cécile Gastaldi for technical assistance. We thank Dr. Feng Zhang for providing the plasmid pX601-AAV-CMV::NLS-SaCas9-NLS-3xHA-bGHpA;U6::BsaI-sgRNA. We also thank Frederic De Leeuw for assistance with multiphoton imaging and Matthieu Dos Santos for preliminary RNA scope experiments. This work was supported by the Association Monegasque contre les Myopathies (AMM, Monaco), the Université Franco-Allemande (CDFA-06-11, UFA), the Association Française contre les Myopathies (AFM, France), and the Agence National de la Recherche (ANR, France).
Author contributions
I.R., L.G., and H.A. designed research; L.G. designed tcDNA antisense oligonucleotides; A.M., A.S., O.N.P., J.H., M.M., M.V.P., I.R., T.M., A.G., S.F., M.S., C.L.-B., and H.A. performed research; T.T. and I.R. contributed new reagents/analytic tools; A.M., A.S., O.N.P., J.H., S.F., and H.A. analyzed data; M.V.P. and M.S. created the DmdEGFP and DmdEGFP-mdx reporter mice; and A.M., A.S., J.H., R.J.P., and H.A. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. E.G. is a guest editor invited by the Editorial Board.
Data, Materials, and Software Availability
Data were extracted from ref. 15 on ArrayExpress and are available under accession number E-MTAB-862 (https://shiny.mdc-berlin.de/MyoExplorer/).
Supporting Information
References
- 1.Davies K. E., Nowak K. J., Molecular mechanisms of muscular dystrophies: Old and new players. Nat. Rev. Mol. Cell Biol. 7, 762–773 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Hoffman E. P., Brown R. H., Kunkel L. M., Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell 51, 919–928 (1987). [DOI] [PubMed] [Google Scholar]
- 3.Koenig M., Monaco A. P., Kunkel L. M., The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell 53, 219–228 (1988). [DOI] [PubMed] [Google Scholar]
- 4.Schneider A.-F. E., Aartsma-Rus A., Developments in reading frame restoring therapy approaches for Duchenne muscular dystrophy. Expert Opin. Biol. Ther. 21, 343–359 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Goyenvalle A., et al. , Functional correction in mouse models of muscular dystrophy using exon-skipping tricyclo-DNA oligomers. Nat. Med. 21, 270–275 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Goyenvalle A., et al. , Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping. Science 306, 1796–1799 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Goyenvalle A., et al. , Rescue of severely affected dystrophin/utrophin-deficient mice through scAAV-U7snRNA-mediated exon skipping. Hum. Mol. Genet. 21, 2559–2571 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Hir M., et al. , AAV genome loss from dystrophic mouse muscles during AAV-U7 snRNA-mediated exon-skipping therapy. Mol. Ther. J. Am. Soc. Gene Ther. 21, 1551–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cristea A., et al. , Effects of aging and gender on the spatial organization of nuclei in single human skeletal muscle cells. Aging Cell 9, 685–697 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Psilander N., et al. , Effects of training, detraining, and retraining on strength, hypertrophy, and myonuclear number in human skeletal muscle. J. Appl. Physiol. (1985) 126, 1636–1645 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Ervasti J. M., Ohlendieck K., Kahl S. D., Gaver M. G., Campbell K. P., Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature 345, 315–319 (1990). [DOI] [PubMed] [Google Scholar]
- 12.Hakim C. H., Grange R. W., Duan D., The passive mechanical properties of the extensor digitorum longus muscle are compromised in 2- to 20-mo-old mdx mice. J. Appl. Physiol. (1985) 110, 1656–1663 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chemello F., et al. , Degenerative and regenerative pathways underlying Duchenne muscular dystrophy revealed by single-nucleus RNA sequencing. Proc. Natl. Acad. Sci. U. S. A. 117, 29691–29701 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dos Santos M., et al. , Single-nucleus RNA-seq and FISH identify coordinated transcriptional activity in mammalian myofibers. Nat. Commun. 11, 5102 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim M., et al. , Single-nucleus transcriptomics reveals functional compartmentalization in syncytial skeletal muscle cells. Nat. Commun. 11, 6375 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrany M. J., et al. , Single-nucleus RNA-seq identifies transcriptional heterogeneity in multinucleated skeletal myofibers. Nat. Commun. 11, 6374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gartler S. M., Riggs A. D., Mammalian X-chromosome inactivation. Annu. Rev. Genet. 17, 155–190 (1983). [DOI] [PubMed] [Google Scholar]
- 18.Lyon M. F., Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190, 372–373 (1961). [DOI] [PubMed] [Google Scholar]
- 19.Monk M., Harper M. I., Sequential X chromosome inactivation coupled with cellular differentiation in early mouse embryos. Nature 281, 311–313 (1979). [DOI] [PubMed] [Google Scholar]
- 20.Lyon M. F., X-chromosome inactivation and developmental patterns in mammals. Biol. Rev. Camb. Philos. Soc. 47, 1–35 (1972). [DOI] [PubMed] [Google Scholar]
- 21.Mak W., et al. , Reactivation of the paternal X chromosome in early mouse embryos. Science 303, 666–669 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Barakat T. S., Gribnau J., X chromosome inactivation in the cycle of life. Development 139, 2085–2089 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Amos-Landgraf J. M., et al. , X chromosome-inactivation patterns of 1,005 phenotypically unaffected females. Am. J. Hum. Genet. 79, 493–499 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arahata K., et al. , Mosaic expression of dystrophin in symptomatic carriers of Duchenne’s muscular dystrophy. N. Engl. J. Med. 320, 138–142 (1989). [DOI] [PubMed] [Google Scholar]
- 25.Arahata K., et al. , Immunostaining of skeletal and cardiac muscle surface membrane with antibody against Duchenne muscular dystrophy peptide. Nature 333, 861–863 (1988). [DOI] [PubMed] [Google Scholar]
- 26.Clerk A., et al. , Characterisation of dystrophin in carriers of Duchenne muscular dystrophy. J. Neurol. Sci. 102, 197–205 (1991). [DOI] [PubMed] [Google Scholar]
- 27.Schmidt-Achert M., Fischer P., Müller-Felber W., Mudra H., Pongratz D., Heterozygotic gene expression in endomyocardial biopsies: A new diagnostic tool confirms the Duchenne carrier status. Clin. Investig. 71, 247–253 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Schmidt-Achert M., Fischer P., Pongratz D., Myocardial evidence of dystrophin mosaic in a Duchenne muscular dystrophy carrier. Lancet 340, 1235–1236 (1992). [PubMed] [Google Scholar]
- 29.Matthews P. M., et al. , Muscle X-inactivation patterns and dystrophin expression in Duchenne muscular dystrophy carriers. Neuromuscul. Disord. 5, 209–220 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Juan-Mateu J., et al. , Prognostic value of X-chromosome inactivation in symptomatic female carriers of dystrophinopathy. Orphanet J. Rare Dis. 7, 82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brioschi S., et al. , Genetic characterization in symptomatic female DMD carriers: Lack of relationship between X-inactivation, transcriptional DMD allele balancing and phenotype. BMC Med. Genet. 13, 73 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danko I., Chapman V., Wolff J. A., The frequency of revertants in mdx mouse genetic models for Duchenne muscular dystrophy. Pediatr. Res. 32, 128–131 (1992). [DOI] [PubMed] [Google Scholar]
- 33.Echigoya Y., et al. , Mutation types and aging differently affect revertant fiber expansion in dystrophic mdx and mdx52 mice. PloS One 8, e69194 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffman E. P., Morgan J. E., Watkins S. C., Partridge T. A., Somatic reversion/suppression of the mouse mdx phenotype in vivo. J. Neurol. Sci. 99, 9–25 (1990). [DOI] [PubMed] [Google Scholar]
- 35.Lu Q. L., et al. , Massive idiosyncratic exon skipping corrects the nonsense mutation in dystrophic mouse muscle and produces functional revertant fibers by clonal expansion. J. Cell Biol. 148, 985–996 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim C. L., Ling K.-H., Cheah P.-S., Isolation, cultivation and immunostaining of single myofibers: An improved approach to study the behavior of satellite cells. J. Biol. Methods 5, e87 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petkova M. V., et al. , Characterization of a Dmd (EGFP) reporter mouse as a tool to investigate dystrophin expression. Skelet. Muscle 6, 25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petkova M. V., et al. , Live-imaging of revertant and therapeutically restored dystrophin in the DmdEGFP-mdx mouse model for Duchenne muscular dystrophy. Neuropathol. Appl. Neurobiol. 46, 602–614 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Amthor H., et al. , Muscle hypertrophy driven by myostatin blockade does not require stem/precursor-cell activity. Proc. Natl. Acad. Sci. U.S.A. 106, 7479–7484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sicinski P., et al. , The molecular basis of muscular dystrophy in the mdx mouse: A point mutation. Science 244, 1578–1580 (1989). [DOI] [PubMed] [Google Scholar]
- 41.Watkins S. C., Hoffman E. P., Slayter H. S., Kunkel L. M., Dystrophin distribution in heterozygote MDX mice. Muscle Nerve 12, 861–868 (1989). [DOI] [PubMed] [Google Scholar]
- 42.Samitt C. E., Bonilla E., Immunocytochemical study of dystrophin at the myotendinous junction. Muscle Nerve 13, 493–500 (1990). [DOI] [PubMed] [Google Scholar]
- 43.Koch M., et al. , A novel marker of tissue junctions, collagen XXII. J. Biol. Chem. 279, 22514–22521 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trotter J. A., Samora A., Baca J., Three-dimensional structure of the murine muscle-tendon junction. Anat. Rec. 213, 16–25 (1985). [DOI] [PubMed] [Google Scholar]
- 45.Law D. J., Tidball J. G., Dystrophin deficiency is associated with myotendinous junction defects in prenecrotic and fully regenerated skeletal muscle. Am. J. Pathol. 142, 1513–1523 (1993). [PMC free article] [PubMed] [Google Scholar]
- 46.Bruusgaard J. C., Liestøl K., Ekmark M., Kollstad K., Gundersen K., Number and spatial distribution of nuclei in the muscle fibres of normal mice studied in vivo. J. Physiol. 551, 467–478 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minetti C., Beltrame F., Marcenaro G., Bonilla E., Dystrophin at the plasma membrane of human muscle fibers shows a costameric localization. Neuromuscul. Disord. 2, 99–109 (1992). [DOI] [PubMed] [Google Scholar]
- 48.Bajanca F., et al. , In vivo dynamics of skeletal muscle Dystrophin in zebrafish embryos revealed by improved FRAP analysis. eLife 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Relizani K., et al. , Palmitic acid conjugation enhances potency of tricyclo-DNA splice switching oligonucleotides. Nucleic Acids Res. 50, 17–34 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohlendieck K., et al. , Duchenne muscular dystrophy: Deficiency of dystrophin-associated proteins in the sarcolemma. Neurology 43, 795–800 (1993). [DOI] [PubMed] [Google Scholar]
- 51.Pavlath G. K., Rich K., Webster S. G., Blau H. M., Localization of muscle gene products in nuclear domains. Nature 337, 570–573 (1989). [DOI] [PubMed] [Google Scholar]
- 52.Kinoshita I., Vilquin J. T., Asselin I., Chamberlain J., Tremblay J. P., Transplantation of myoblasts from a transgenic mouse overexpressing dystrophin prduced only a relatively small increase of dystrophin-positive membrane. Muscle Nerve 21, 91–103 (1998). [DOI] [PubMed] [Google Scholar]
- 53.van Putten M., et al. , The effects of low levels of dystrophin on mouse muscle function and pathology. PloS One 7, e31937 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blau H. M., Chiu C. P., Webster C., Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell 32, 1171–1180 (1983). [DOI] [PubMed] [Google Scholar]
- 55.Ralston E., Hall Z. W., Transfer of a protein encoded by a single nucleus to nearby nuclei in multinucleated myotubes. Science 244, 1066–1069 (1989). [DOI] [PubMed] [Google Scholar]
- 56.Yang J., Ontell M. P., Kelly R., Watkins S. C., Ontell M., Limitations of nls beta-galactosidase as a marker for studying myogenic lineage or the efficacy of myoblast transfer. Anat. Rec. 248, 40–50 (1997). [DOI] [PubMed] [Google Scholar]
- 57.Blaveri K., et al. , Patterns of repair of dystrophic mouse muscle: Studies on isolated fibers. Dev. Dyn. 216, 244–256 (1999). [DOI] [PubMed] [Google Scholar]
- 58.Heslop L., et al. , Transplanted primary neonatal myoblasts can give rise to functional satellite cells as identified using the Myf5nlacZl+ mouse. Gene Ther. 8, 778–783 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Hildyard J. C. W., Rawson F., Wells D. J., Piercy R. J., Multiplex in situ hybridization within a single transcript: RNAscope reveals dystrophin mRNA dynamics. PloS One 15, e0239467 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Denes L. T., Kelley C. P., Wang E. T., Microtubule-based transport is essential to distribute RNA and nascent protein in skeletal muscle. Nat. Commun. 12, 6079 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pinheiro H., et al. , mRNA distribution in skeletal muscle is associated with mRNA size. J. Cell Sci. 134, jcs256388 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.García-Rodríguez R., et al. , Premature termination codons in the DMD gene cause reduced local mRNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 117, 16456–16464 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gache V., Gomes E. R., Cadot B., Microtubule motors involved in nuclear movement during skeletal muscle differentiation. Mol. Biol. Cell 28, 865–874 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iyer S. R., et al. , Altered nuclear dynamics in MDX myofibers. J. Appl. Physiol. (1985) 122, 470–481 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Novak J. S., et al. , Interrogation of dystrophin and dystroglycan complex protein turnover after exon skipping therapy. J. Neuromuscul. Dis. 8, S383–S402 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roman W., et al. , Muscle repair after physiological damage relies on nuclear migration for cellular reconstruction. Science 374, 355–359 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Sardone V., et al. , A novel high-throughput immunofluorescence analysis method for quantifying dystrophin intensity in entire transverse sections of Duchenne muscular dystrophy muscle biopsy samples. PloS One 13, e0194540 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bassett D. I., et al. , Dystrophin is required for the formation of stable muscle attachments in the zebrafish embryo. Development 130, 5851–5860 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Relizani K., et al. , Efficacy and safety profile of tricyclo-DNA antisense oligonucleotides in Duchenne muscular dystrophy mouse model. Mol. Ther. Nucleic Acids 8, 144–157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Westering T. L. E., et al. , Uniform sarcolemmal dystrophin expression is required to prevent extracellular microRNA release and improve dystrophic pathology. J. Cachexia Sarcopenia Muscle 11, 578–593 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kann A. P., Krauss R. S., Multiplexed RNAscope and immunofluorescence on whole-mount skeletal myofibers and their associated stem cells. Development 146, dev179259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dystrophin-EGFP distribution at the myotendinous junction. Video depicts 3D reconstruction of dystrophin-EGFP at the level of a myotendinous junction following ex vivo confocal microscopy of a freshly isolated single myofiber from EDL muscle of a 6-month-old DmdEGFP mouse.
Data Availability Statement
Data were extracted from ref. 15 on ArrayExpress and are available under accession number E-MTAB-862 (https://shiny.mdc-berlin.de/MyoExplorer/).