Significance
Circulating tumor environment-derived extracellular vesicles (cTME-EVs) are a small portion of EVs in blood co-existing with those derived from other tissues; the accurate identification of cTME-EVs could facilitate both fundamental research and clinical liquid biopsy of cancer. In this work, we report the double-switch pH-low insertion peptide (D-S pHLIP) system, which allows selective enrichment of cTME-EVs from blood serum for comprehensive transcriptomic analysis. Plentiful differentially expressed genes and hub genes were identified, demonstrating the high performance of this technology. This work contributes to the design of pHLIP variants for specific functions and the discovery of tumor diagnostic biomarkers and therapeutic targets.
Keywords: pHLIP, conformational switch, computational design, extracellular vesicle, tumor microenvironment
Abstract
Circulating tumor microenvironment-derived extracellular vesicles (cTME-EVs) are gaining considerable traction in cancer research and liquid biopsy. However, the study of cTME-EVs is largely limited by the dearth of a general isolation technique to selectively enrich cTME-EVs from biological fluids for downstream analysis. In this work, we broke through this dilemma by presenting a double-switch pH-low insertion peptide (D-S pHLIP) system to exclusively harvest cTME-EVs from the blood serum of tumor mouse models. This D-S pHLIP system consists of a highly sensitive pH-driven conformational switch (pKa ≈ 6.8) that allows specific installation of D-S pHLIP on the EV membranes in TME (pH 6.5 to 6.8) and a unique hook-like switch to “lock” the peptide securely on the cTME-EVs during the systemic circulation. The D-S pHLIP-anchored cTME-EVs were magnetically enriched and then analyzed with high-resolution messenger RNA sequencing, by which more than 18 times the number of TME-related differentially expressed genes and 10 times the number of hub genes were identified, compared with those achieved by the gold-standard ultracentrifugation. This work could revolutionize basic TME research as well as clinical liquid biopsy for cancer.
Extracellular vesicles (EVs), which carry rich molecular cargos (e.g., proteins, lipids, metabolites, and nucleic acids) of their parental cells, are regarded as the mediators of intercellular communication (1). In tumor microenvironment (TME), bidirectional communication between tumor cells and diverse tumor-associated cells (TACs) via EV trafficking plays a crucial role in the sustained growth, invasion, and metastasis of cancer (2). Therefore, circulating EVs derived from TME (termed cTME-EVs, including both tumor cell- and TAC-derived EVs) have emerged as ideal non-invasive biomarkers, thus prompting the development of basic TME-associated research and clinical diagnosis. However, cTME-EVs have not been fully exploited due to the lack of efficient means to selectively enrich them from bodily fluids for downstream analysis.
Currently available approaches for EV enrichment, such as ultracentrifugation (UC) (3), polymer-based precipitation (4), filtration (5), and size exclusion chromatography (6), are unable to discriminate cTME-EVs from the multitudinous co-existing EV counterparts derived from other tissues, resulting in the cover-up of cancer signatures. At present, cancer-specific molecules such as aptamers (7) and antibodies (8, 9) have been introduced for the immune-enrichment of tumor cell-derived EVs, such as the recently reported lipid patch microarrays (10) and lipid-protein capture systems on herringbone microfluidic chips (11). However, these biomarker-dependent strategies are largely limited by the huge heterogeneity of biomarker expression on the EV surfaces (12, 13). Epithelial cell adhesion molecule (EpCAM), for example, is the most common biomarker used for magnetic enrichment of tumor cell-derived EVs in the commercial EpCAM Exo-Flow Capture Kit; this biomarker is only expressed in less than 50% of these EVs (14). In addition, there are a dozen kinds of TACs (typically immune cells, endothelial cells, and fibroblasts) alongside tumor cells in TME, which also constitute cancer signatures (15, 16). However, TAC-derived EVs cannot be enriched by the cancer biomarker-based kits, resulting in the loss of vast indispensable molecular information reflecting TME. It is therefore highly desirable to develop a general platform for the selective and efficient enrichment of cTME-EVs from biological fluids irrespective of cancer types.
TME is characteristic of weak acidity, which is commonly recognized as one of the significant cancer signatures (17). This property has shed light on the development of a variety of diagnostic and therapeutic strategies for cancer (18). The pH-low insertion peptide (pHLIP) (19) is wildly studied as a TME-targeting agent that can fold by a pH-driven switch upon acidification to insert into membrane-like structures (20). However, wild-type (WT) pHLIP cannot be used as a handle for selective harvesting of cTME-EVs for the following two reasons. First, WT pHLIP (pKa ≈ 6.2) (21) cannot respond optimally to TME (pH 6.5 to 6.8) (22), which means low TME-targeting ability. Second, and more importantly, WT pHLIP will disassociate from the cTME-EV membranes during systemic circulation (pH 7.2 to 7.4), resulting from the pH-driven conformational conversion of the peptide back to its initial unstructured state. This reversible conformational change would be the essential cause, which makes WT pHLIP impossible to capture cTME-EVs in blood samples.
In this work, we present a general platform capable of selectively enriching cTME-EVs from the blood plasma of diverse tumor mouse models. We performed a series of computational designs on pHLIP to optimize its inherent pH-driven switch to efficiently target TME and create a pH-driven hook-like switch to prevent the dissociation of pHLIP from cTME-EVs in systemic circulation. We investigated the performance of this double-switch pHLIP (D-S pHLIP) system by molecular dynamic (MD) simulations, free-energy calculations, in vitro and in vivo experiments, and performed high-resolution transcriptomic sequencing for the selectively enriched cTME-EVs. This D-S pHLIP-based platform shows excellent potential in excavating comprehensive and precise cancer-related information.
Results
Design Strategy for D-S pHLIP.
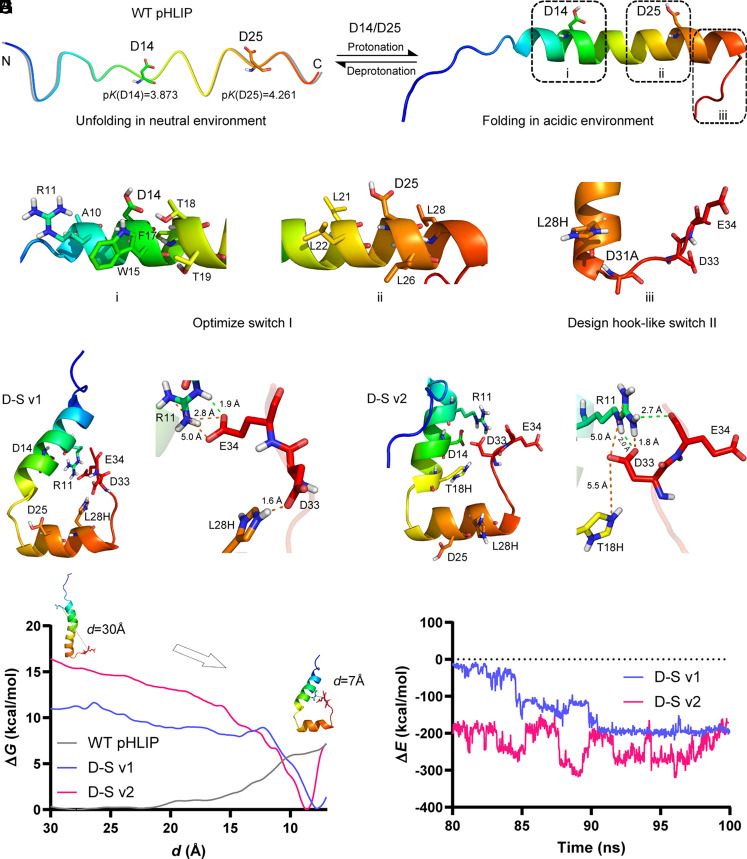
As depicted in Fig. 1 A and B, D14 and D25 are the two key residues that control the folding of pHLIP (23). The pK (24) values of the two residues were, respectively, calculated to be 3.873 and 4.261 by H++ software. Compared with D14, D25 possesses a higher pK value, indicating that D25 obtains H proton preferably, thereby governing the folding response of the peptide (i.e., pKa). Additionally, the relevant pH sensitivity can be mediated by D14. H++ software is regarded as an accurate and advisable tool to predict the effect of a point mutation on the pK of a nearby site. Therefore, to improve the pKa of pHLIP from the wide-type 6.2 to ideally 6.8 for better targeting TME, a feasible strategy is to increase the pK value of D25 from approximately 4.2 to 4.8. Based on this assumption, we screened the residues that are closely associated with the pK values of D14 (i region, Fig. 1C) and D25 (ii region, Fig. 1D).
Fig. 1.
D-S pHLIP design. (A) The unfolding state of WT pHLIP in neutral environment. Two critical residues, D14 and D25 with their calculated pK values. (B) The folding state of WT pHLIP in acidic environment. Design regions for switch I (i and ii) and switch II (iii). (C and D) Residues that influence the pK values of D14 and D25. (E) A double mutation, L28H/D31A for the hook-like switch II. (F and G) Structural snapshots of the two D-S variants, i.e., L28H/D31A/L21F/L22G (D-S v1) and L28H/D31A/L21F/L22G/T18H (D-S v2) generated from the trajectories of their 100 ns MD simulations. Electrostatic and hydrogen bonding interactions are colored by orange and green dotted lines, respectively. (H and I) Calculations of free-energy and interaction-energy for WT pHLIP and two D-S variants. d denotes the center-of-mass distance between the two residues of D33 and E34 in the C-terminal and R11 nearby the N-terminal. In (A–G), nonpolar hydrogens are hidden. N and C in (A) denote the N- and C-terminal of the peptide, respectively, wherein the C-terminal will insert into the phospholipid bilayer structure after folding.
With the aid of H++ software, we designed more than 200 variants by virtual saturation mutation on these sites and further built the relevant three-dimensional structures to calculate the mutation effect on the pK increment of D25 and D14 (SI Appendix, Tables S1 and S2). The optimal pK value of D14 achieves 4.752 (about 1 pK unit improvement) by T18Q mutation, while that of D25 is still below 4.7. Multiple mutations were, therefore, employed to further increase the pK value of D25 (SI Appendix, Table S3). As summarized in SI Appendix, Table S4, the six variants (L21W/L22W, L21W/L26N, L21F/L22W/L26N, L21Y/L22W/L26N, L21F/L22G, and L21W/L22T/L26N) were selected as the promising residues for D25, in which the pK values range from 4.8 to 5.2.
To avoid the dissociation of pHLIP from the captured EVs, we proposed a hook-like switch II, where L28, at the bottom of the alpha-helix nearby the C-terminal, was mutated to His (Fig. 1E). Given the calculated pK value of L28H (6.563, see SI Appendix, Table S5), the nitrogen (Nε) in L28H is anticipated to obtain a proton in or below neutral environments. L28H will, therefore, become a positively-charged residue to attract the negatively-charged residues D31, D33, and E34 in the C-terminal. To form an ideal hook-like structure, D31 was mutated to Ala to facilitate the interactions between L28H and D33/E34. As provided in SI Appendix, Table S5, we integrated the mutation strategy of the hook-like switch II with the promising variants of switch I and calculated the pK values of ionizable residues in the peptides. We finally selected four D-S variants, i.e., L28H/D31A/L21F/L22G (named D-S v1), L28H/D31A/L21F/L22G/T18H (named D-S v2), L28H/D31A/L21F/L22G/T18F (named D-S v3), and L28H/D31A/L21F/L22G/T18Q (named D-S v4), and further investigated the performance of the hook-like switch II in each assembly by MD simulations. In D-S v1 and D-S v2 assemblies, we found that the two negatively-charged residues, D33 and E34, were attracted by L28H via electrostatic interactions at the outset of the simulations, causing the bending of the loop in the C-terminal into a small “hook” (see Movies S1 for D-S v1 and S2 for D-S v2). Interestingly, D33 and E34 further interacted with the positively-charged residue, R11, which was unexpected but more desirable. As depicted in Fig. 1 F and G, the distinctive hook-like structures were formed in these two variants. By contrast, no hook-like structures were formed in the D-S v3 and D-S v4 assemblies.
We next performed free-energy calculations to quantify the barriers against forming hook-like structures in these peptides. There are approximately 6 kcal/mol for WT pHLIP, no more than 1 kcal/mol for the D-S v1, and no free-energy barrier for the D-S v2 to change their “upright” conformations into the hook-like ones (Fig. 1H). These results indicate that it is difficult for WT pHLIP to form a hook-like structure spontaneously, while it is easy for the two variants. Additionally, we calculated the interaction energies between the interactional charged residues to quantify the intensity of the “hooks” in the two variants (Fig. 1I). Given the intense interaction energies, we reasoned that the hook-like structures are very steady. Furthermore, the interaction energy of D-S v2 is much more potent than that of D-S v1, owing to the additional T18H mutation.
Fluorescence and Spectral Studies.
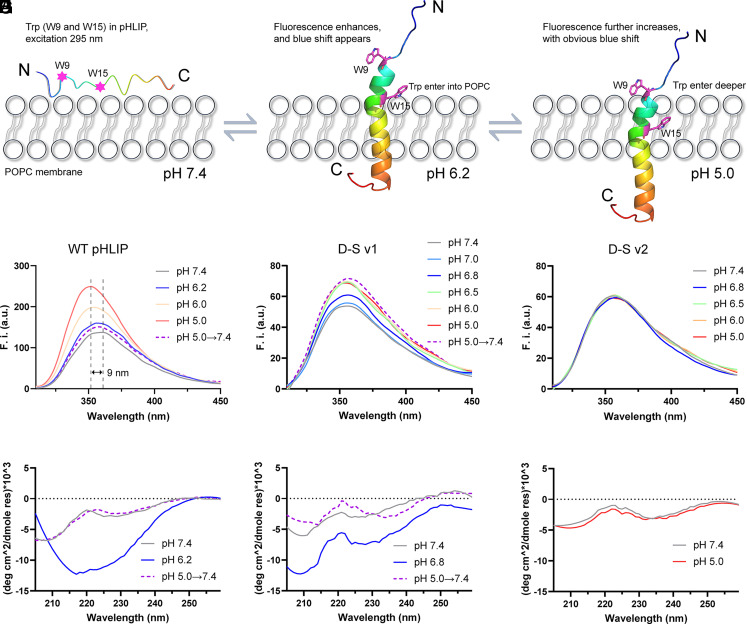
To investigate the actual performance of the D-S variants, we examined the insertion behaviors of these peptides using 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) model membrane by recording the Trp fluorescence (W9 and W15 in pHLIP, excitation at 295 nm). In a neutral environment, the unstructured pHLIP could suspend in solutions or interact with the membrane surface (Fig. 2A). When the pH condition decreases from neutral to acidic, WT pHLIP will fold at its folding-response value and start to insert into the POPC membrane (Fig. 2B), leading to the enhancement of fluorescence intensity. Furthermore, as the acidity rises, the Trp residues will move into the bilayer’s hydrophobic core (Fig. 2C). As a result, vigorous fluorescence intensity and clear blue shift occur owing to the minimal water quenching of Trp emission in the hydrophobic region (23).
Fig. 2.
The Trp fluorescence and corresponding CD spectra of WT pHLIP, D-S v1, and D-S v2 during membrane insertion. (A–C) Schematic diagram of the insertion behaviors. W9 and W15 in WT pHLIP are shown in sticks. The C atoms are colored in purple, and all the hydrogen atoms are hidden. Trp fluorescence and corresponding CD results for (D and G) WT pHLIP, (E and H) D-S v1, and (F and I) D-S v2, respectively. WT pHLIP and D-S v1 start to fold and insert into POPC at pH 6.2 and 6.8, respectively. The fluorescence intensity and CD signals in the D-S v2 group did not change under different pH conditions.
As illustrated in Fig. 2D, with the decrease of pH values, the Trp fluorescence intensity of WT pHLIP enhances gradually and reaches the maximum at pH 5.0, along with a clear blue-shift of emission (9 nm). The pKa value of WT pHLIP is calculated to be approximately 6.2 by using the Henderson–Hasselbalch equation (25) via plotting the shift of the maximum emission wavelength versus pH values (SI Appendix, Fig. S1A), which is in accordance with the value reported in previous works (21, 25). When the pH values are raised from 5.0 to 7.4, the fluorescence intensity decreases significantly (purple dotted line), implying that the WT pHLIP moves from the hydrophobic layer back to the membrane surface. This reversibility has also been announced by previous works (21, 23). Additionally, the relevant circular dichroism (CD) spectra verify that WT pHLIP folds at pH 6.2 and unfolds at pH 7.4 and pH 5.0→7.4 (Fig. 2G).
As depicted in Fig. 2E, the fluorescence intensity of D-S v1 starts to elevate visibly by pH 6.8 treatment and reaches the maximum at pH 6.5 to 5.0. However, compared with WT pHLIP, the overall fluorescence intensity of D-S v1 is low, and the degree of blue shift is small (2 nm). As discussed above, both fluorescence intensity and blue shift are positively associated with the depth of peptide insertion into the membrane. Therefore, we analyzed the hydrophobic property of the peptides by ProtScale. The results indicate that the hydrophobicity of D-S v1 is a little lower than that of WT pHLIP (SI Appendix, Fig. S2), which may weaken the interaction degree of the peptide with the membrane and impair the depth of insertion. Furthermore, the hook-like switch II will cause the C-terminal of D-S v1 to fold back toward the N-terminal instead of inserting into the membrane deeper (Fig. 1F), which could be the fundamental reason why the overall fluorescence intensity of D-S v1 is lower than that of WT pHLIP. The slight blue shift makes it impossible to calculate the pKa value for D-S v1 using the current Henderson–Hasselbalch equation. Therefore, we plotted the fluorescence changes of the maximal intensity versus pH, by which the pKa value of D-S v1 was determined to be approximately 6.8 (SI Appendix, Fig. S1B), which agrees well with the pK calculation result. This result was further confirmed by the CD experiment (Fig. 2H).
It should be noted that the fluorescence intensity of D-S v1 at pH 5.0→7.4 is as high as that at pH 5.0 (Fig. 2E, purple dotted line), and this peptide remained folding at the condition of pH 5.0→7.4 (Fig. 2H). These results imply that once D-S v1 has inserted in the membranes at acidic conditions, it can be locked firmly, even when the pH is switched back to neutral. This irreversible insertion, contrary to WT pHLIP, is ascribed to the expected performance of the hook-like switch II. In view of the results of Fig. 1 F and I, we conjecture that the strong electrostatic interactions between the positively-charged R11/L28H and the negatively-charged D33/E34 enable the peptide to be “locked” securely on the membrane. As shown in Fig. 2 F and I, D-S v2 has lost the ability of pH-driven conformational change. According to the results of Fig. 1I, we infer that the electrostatic attraction between the charged residues in D-S v2 may be too strong to separate, thereby hindering the folding process. The fluorescence and CD experiments demonstrated that only D-S v1 meets the requirements of high-sensitivity TME targeting and irreversible installation on cTME-EVs, i.e., i) folding at pH 6.8; and ii) “locking” firmly in the membrane after pH returning to neutral.
D-S pHLIP Insertion in the Membranes of Cells and EVs.
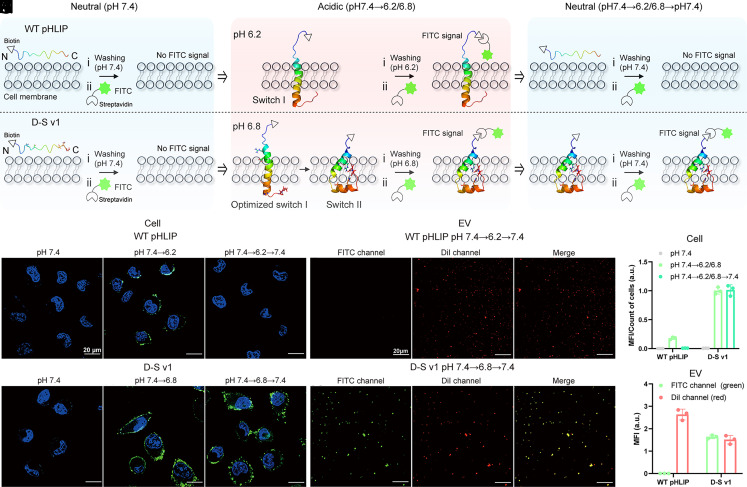
We then investigated the insertion behaviors of WT pHLIP and D-S v1 on human lung tumor cells (A549) and related EVs (the schematic diagram is shown at the top of Fig. 3). The peptides were conjugated with biotin tags (peptides@biotin), which can be recognized by the fluorescein isothiocyanate (FITC)-modified streptavidin (FITC@SA) for membrane labeling. As depicted in Fig. 3 A–D, and 3G, there are no FITC signals (green fluorescence) on the cell surfaces, indicating that WT pHLIP and D-S v1 were unable to insert into tumor cells in a neutral environment (pH 7.4). When the pH conditions changed to acidic, the FITC signals were detected in both groups (Fig. 3 B, E–H). These results suggest that the peptides have folded at their own folding-response values and inserted into the cell membranes. When the pH condition switched back to neutral again (7.4→6.2→7.4), no FITC signals were detected on the cell membranes in the WT pHLIP group (Fig. 3 C and F). However, strong green fluorescence was detected in the D-S v1 group with pH treatment of 7.4→6.8→7.4 (Fig. 3I), indicating that D-S v1 was retained on the cell membranes with the assistance of the hook-like switch II.
Fig. 3.
Insertion states of WT pHLIP and D-S v1 in the membranes of tumor cells and corresponding EVs under different pH treatments. (A–C) Schematic diagram of the interactions between the pHLIP peptides and the biological membranes under different pH treatments. The molecular structure of WT pHLIP unfolds at 7.4, folds at pH 7.4→6.2 to insert into membranes, and move out under the treatment of pH 7.4→6.2→7.4; D-S v1 unfolds at 7.4, folds at pH 7.4→6.8 to insert into membranes, and maintains folding after pH 7.4→6.8→7.4 treatment. The biotin-tagged pHLIP is anchored on the membranes and then labeled by the FITC-modified streptavidin (FITC@SA) for fluorescence imaging. (D–I), Cell imaging experiments. Cell nuclei are labelled with DAPI (blue fluorescence). The green fluorescence in cell membranes denotes that the peptides have inserted into cells in the acid environment (E and H), or “locked” on cells after switching the pH back to neutral (I). (J and K) EV imaging experiments. The EV membranes are stained by DiI (red fluorescence). The merged yellow fluorescence denotes that D-S v1 still “locked” on EV membranes after pH 7.4→6.8→7.4 treatment. (L and M) Fluorescence quantification data for the cell and EV results. MFI denotes mean fluorescence intensity. The results are expressed as mean ± SEM (n = 3 biological replicates).
We further investigated whether these peptides can be retained on the A549-derived EV membranes under the pH 7.4→6.2/6.8→7.4 transition. As shown in Fig. 3J, the FITC signals in the EVs of the WT pHLIP group were undetectable, confirming the release of the FITC-tagged WT pHLIP from the EV surfaces at neutral conditions. In contrast, in the D-S v1 group, strong FITC signals were found to merge with the red fluorescence of DiI, which is commonly used for staining EV membranes (Fig. 3K). The quantification data for the cells and EVs are shown in Fig. 3 L and M, respectively. The particle size, concentration, and morphological features of EVs detected by nanoparticle tracking analysis (NTA) and transmission electron microscope are, respectively, shown in SI Appendix, Fig. S3. These results verify the insertion capability of D-S v1 at the cellular and EV levels.
After incubating the cells with D-S v1@biotin under the pH 7.4→6.8→7.4 treatment, the cells kept growing in the fresh culture medium. To investigate the dynamic distribution of D-S v1 inserted into cell membranes, we prolonged the incubation time to 8 h. The FITC signals on the cell surfaces were found to decrease gradually; simultaneously, the FITC signals of the cell-secreted EVs (as validated by the merged fluorescence with DiI) increased with the time course (SI Appendix, Fig. S4). These results imply that the peptides inserted into the cell membranes were taken away by the continuously secreted EVs. Therefore, we reason that there may be two modes of D-S v1 installation on cTME-EVs: i) D-S v1 directly inserts into the cTME-EVs that already exist in TME; and ii) D-S v1 inserts into tumor cells and TACs first and then transfers to their secreted EVs.
To assess the cell viability and cytotoxic effects, various concentrations of D-S v1 (0, 5, 10, 20, 30, and 40 μM) were incubated with RAW264.7 cells at different pH values and then tested by Cell Counting Kit-8 and MTT assay kits, respectively. When the concentrations of D-S v1 were below 10 μM under both pH 6.8 and 7.4 conditions, the cells retained over 95% viability and D-S v1 showed negligible cytotoxicity (<4%) (SI Appendix, Fig. S5). When the D-S v1 concentrations were above 20 μM, the cell viability declined gradually, along with the manifestation of cytotoxicity. There is no significant difference between the two pH conditions. In light of these results, the D-S pHLIP shows trivial cytotoxic effects at concentrations lower than 10 μM.
Selective Enrichment of cTME-EVs from the Tumor Model Mice.
We demonstrated the tumor-targeting capability of the D-S pHLIP system through intraperitoneally (i.p.) injecting Cy5.5-conjugated D-S v1 (D-S v1@Cy5.5) into the opposite side of the lung adenocarcinoma in model mice for fluorescence imaging in vivo. At 24 h post-injection, intense D-S v1@Cy5.5 signals were found at the tumor site while nearly no signals were detected on the muscle of the non-tumor site, and the tumor-to-muscle fluorescence ratios were determined to be 2.2 ± 0.3 (n = 3, SI Appendix, Fig. S6). These in vivo imaging results support the positive tumor-targeting ability of this D-S pHLIP variant.
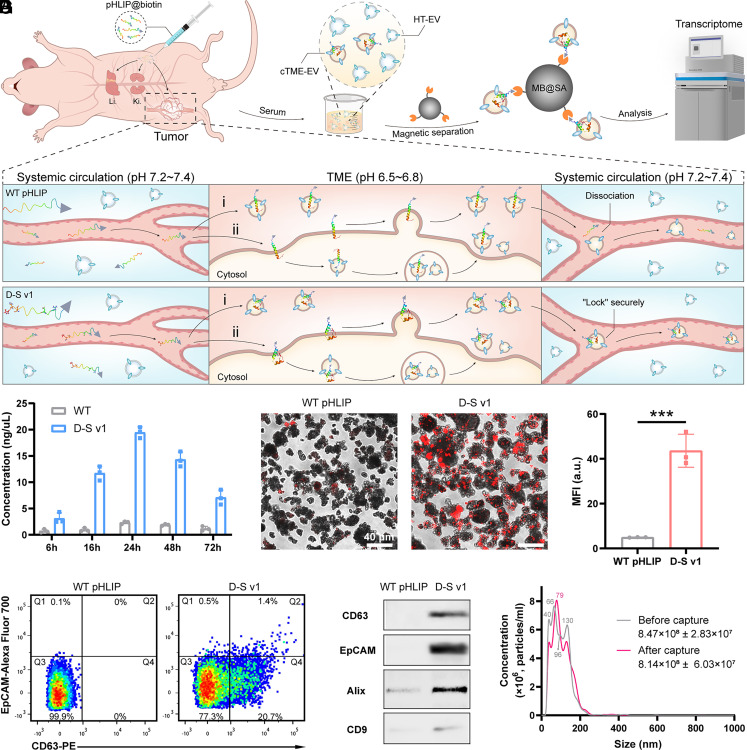
To verify the availability of this D-S pHLIP system for selective enrichment of cTME-EVs from the serum of animal models, the peptides@biotin were i.p. injected into the tumor model mice. The blood sera of the mice were collected at 6, 16, 24, 48, and 72 h post-injection and the cTME-EVs installed by the peptides@biotin were harvested using streptavidin-modified magnetic beads (namely MB@SA). The schematic diagram of pHLIP installation on cTME-EVs, total RNA analysis of the captured cTME-EVs, confocal fluorescent imaging, flow cytometry (FCM), Western blot (WB), and NTA characterizations are depicted in Fig. 4. From the results of total RNA detection (Fig. 4B), the D-S pHLIP group harvested many more cTME-EVs than the WT pHLIP group, especially at 24 h. At that optimal time point, the fluorescence intensities between the D-S and WT pHLIP groups have no significant difference (P > 0.05, n = 3) in the liver and kidney, suggesting that they show the similar metabolism pathway (SI Appendix, Fig. S7). The results also confirm that the D-S pHLIP group shows higher fluorescence intensity on the tumor than the WT pHLIP group (P < 0.001, n = 3). The higher targeting capability of D-S pHLIP is ascribed to its optimized folding-response value (pKa ≈ 6.8).
Fig. 4.
Selective enrichment of cTME-EVs from tumor-bearing mice via the D-S pHLIP system. (A) Schematic diagram of installing biotin-tagged pHLIP on cTME-EVs for magnetic enrichment and transcriptomic analysis. (B–G) Identification of the captured cTME-EVs by (B) RNA detection, (C and D) confocal fluorescent imaging, (E) FCM, (F) WB, and (G) NTA. In (A), Li. and Ki. denote liver and kidney, respectively. In TME, i and ii denote the two modes of D-S v1 installation on cTME-EVs. In (B), WT pHLIP@biotin and D-S v1@biotin were injected into the lung cancer model mice, and the blood was withdrawn after 6, 16, 24, 48, and 72 h. In (C), the EV membranes were dyed by DiI (red fluorescence). In (D), MFI denotes the mean fluorescence intensity. ***P < 0.001. In (E), intense CD63 and EpCAM signals were detected in the D-S v1 group. In (F), clear biomarker bands were detected in the D-S v1 group, while those in the WT pHLIP group were too weak to be detected. In (G), after capture, the sizes of the cTME-EVs at 40 nm and 130 nm are reduced, and those at 66 nm and 96 nm appear to merge into the size at 79 nm before capture. The results are expressed as mean ± SEM (n = 3 biological replicates).
We next investigated the dynamic distribution of the biotin-tagged D-S v1 in the major organs of tumor model mice. The organs were collected at three representative time points (6, 24, and 72 h) and stained with FITC@SA, followed by recording the fluorescent images ex vivo (SI Appendix, Fig. S8). Significant signals were enriched in the liver and kidney at 6 h, suggesting that the peptides were rapidly metabolized by the body after i.p. injection. Comparing the results at 24 h to those at 6 h, the signals on the tumor increased obviously (P = 0.008, n = 3) while those on the other organs continued to decline. At 72 h, the signals on most organs—aside from the kidney—decreased obviously, demonstrating that the peptides were being eliminated from the body via the urinary system. The metabolism of D-S v1 is similar to that of WT pHLIP in terms of tumor-targeting time and clearance rate (26).
Confocal fluorescent imaging was employed to further identify the captured cTME-EVs. To do this, DiI was used to stain the EV membranes. As shown in Fig. 4C, high-density strong fluorescent dots (red) were detected on the MB surfaces in the D-S v1 group, suggesting that the D-S v1@biotin has been installed on the cTME-EVs and collected by MB@SA efficiently. By contrast, a few weak fluorescent dots were observed in the WT pHLIP group. The quantification data show that the fluorescence intensity in the D-S v1 group is approximately ninefold higher than that of the WT pHLIP group (Fig. 4D). We then confirmed the cTME-EVs by measuring the protein biomarkers with FCM and WB experiments. The results in Fig. 4E indicate no fluorescence signals of CD63 (a specific biomarker of EV, Q4 area) in the WT pHLIP group. In contrast, intense CD63 signals (20.7% proportion, Q4 area) were detected in the D-S v1 group. Furthermore, the fluorescence signals of EpCAM (a specific biomarker of cancer) merged with CD63 were also detected in the D-S v1 group (1.4% proportion, Q2 area), suggesting that the D-S pHLIP system has captured the tumor cell-derived EVs, which account for approximately 6.8% of all the captured ones. In addition to CD63 and EpCAM, the bands for the other two EV biomarkers (Alix and CD9) were detected in the D-S v1 group by WB experiments (Fig. 4F). We infer that the weak signals in the WT pHLIP group may result from the minimal retaining of WT pHLIP@biotin on the EV surfaces or slight nonspecific adsorption of EVs on the MBs. Additionally, we demonstrated that i) the MB@SA shows very weak nonspecific adsorption toward the EVs, and ii) D-S v1 can hardly capture EVs from a healthy mouse model, suggesting its tumor specificity (SI Appendix, Fig. S9).
NTA was finally employed to investigate the particle size and concentration of EVs before and after magnetic enrichment from the mouse serum samples (Fig. 4G). The mean concentration of total EVs in the serum samples is 8.47 × 108 particles/mL; after magnetic capturing, the mean concentration of the rest EVs decreases to be 8.14 × 108 particles/mL. The mean concentration of the captured EVs was calculated to be 3.3 × 107 particles/mL, accounting for 3.9% of the total EVs. The sizes of the EVs are distributed from 30 to 200 nm, meaning that majority of the vesicles are exosomes (1). Additionally, given the TME-responsive feature of D-S pHLIP, this technology can be amenable to other cancer types. For instance, the similar capture results were achieved from the mice with breast cancer. The corresponding RNA detection, FCM, and NTA results confirmed the broad applicability of this D-S pHLIP-based platform (SI Appendix, Fig. S10).
Transcriptomic Profiling of the Yielded cTME-EVs.
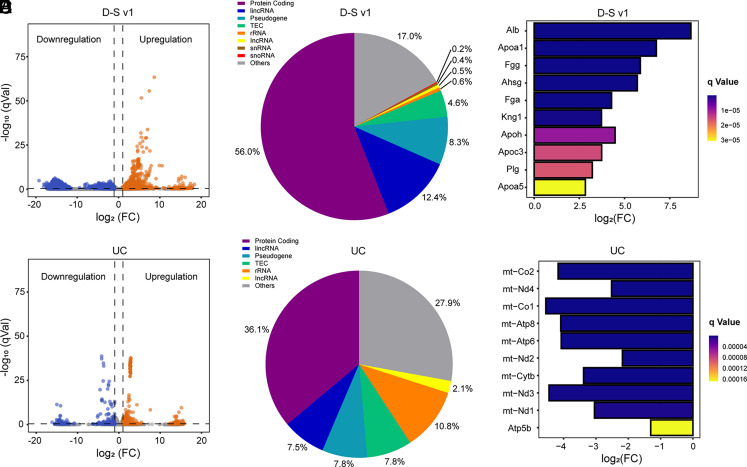
We then took advantage of transcriptome to further verify the availability of this isolation platform at the genetic level and reveal the potential cellular origin of these captured cTME-EVs. UC, the gold-standard approach for EV isolation in various settings, was employed as a control to isolate EVs from the mice with lung cancer and healthy ones (SI Appendix, Fig. S11). SI Appendix, Fig. S12 indicates that all the samples have good biological repeatability. We then calculated the differentially expressed genes (DEGs) for the two groups of tumor samples. 6,256 DEGs, including 448 upregulated genes and 5,808 downregulated genes, were identified in the D-S v1 group, while only 332 DEGs (243 upregulated and 89 downregulated) were found in the UC group (SI Appendix, Fig. S13). Apart from number, the volcano plots (Fig. 5 A and D) show that the DEGs in the D-S v1 group possess more significant differences than that of the UC group. Among the detected RNAs, the protein-coding messenger RNAs (mRNAs) and long intergenic non-coding RNAs (lincRNAs) were identified as the most abundant biotypes in the D-S v1 group (Fig. 5B), while mRNAs and ribosomal RNAs (rRNAs) are abundant in the UC group (Fig. 5E). To date, the source tracking of EVs is significant but remains challenging (27). Therefore, to trace the potential cell sources of the captured EVs, we matched the top 50 DEGs (SI Appendix, Fig. S14) against the exosomal biomarker database–ExoCarta (http://exocarta.org/index.html). The traceable top DEGs in the D-S v1 group originate from both tumor cells and TACs (SI Appendix, Table S6), wherein the latter mainly includes mast cells, fibroblasts, and macrophages that play important roles in many cancers (16, 28, 29). Additionally, the traceable top DEGs in the UC group are mainly from mast cells and fibroblasts (SI Appendix, Table S7). Obviously, both the quantity and cancer relationship of the traceable genes in the UC group are much lower than that in the D-S v1 group.
Fig. 5.
Transcriptomic profiling of the yielded cTME-EVs enriched by D-S v1 and UC. Volcano plots showing DEGs in (A) D-S v1 and (D) UC groups. q < 0.05 and a fold change (FC) >2 are used to define upregulation (red) and downregulation (blue), while the gray ones are not significant. Reads of the distribution of RNA biotypes from DEGs in (B) D-S v1 and (E) UC groups. The differential expression degrees of hub genes in (C) D-S v1 and (F) UC groups.
Gene ontology analysis shows that the DEGs in the D-S v1 group mainly participate in the biological processes of signal transduction and G protein-coupled receptor (GPCR) signaling pathway to perform molecular functions of protein binding, metal ion binding, and GPCR activity (SI Appendix, Fig. S15). Many diseases including cancers are associated with GPCRs, which are targeted by a variety of modern drugs (30, 31). Therefore, this D-S pHLIP system may have the potential to search for TME-associated therapeutic targets of cancer. In the UC group, the DEGs mainly participate in biological processes of oxidation-reduction process and immune system to perform molecular functions of protein binding and metal ion binding. We then analyzed the protein–protein interaction (PPI) network for these DEGs (SI Appendix, Fig. S16), by which the top 10 hub genes were identified (SI Appendix, Fig. S17). In the D-S v1 group, the hub genes are Apoa5, Plg, Apoc3, Apoh, Kng1, Fga, Ahsg, Fgg, Apoa1, and Alb, wherein four of them (Apoa5, Apoc3, Apoh, and Apoa1) belong to the apolipoprotein (Apo) family. Furthermore, Fig. 5C indicates that all the hub genes are significantly upregulated, especially Apos. Previous works have demonstrated that the Apo levels are associated with many cancerous risks and can be used to predict tumor progression (32–34). Furthermore, the level of Plg was found to be upregulated in bladder carcinoma patients (35). Fga, Fgg (36), and Ahsg (37) were employed as biomarkers for prostate cancer and gastric cancer, respectively. However, the association of Kng1 and Alb with cancers has rarely been reported, which may serve as tumor biomarkers or therapeutic targets. Collectively, the significantly upregulated hub genes in the lung cancer models have great potential to be used as the combined diagnostic biomarkers and may further be employed to develop relevant therapeutic targets. In the UC group, all the hub genes are downregulation (Fig. 5F). Furthermore, the differential expression level is much lower than that of the D-S v1 group. The most significant one, Atp5b, was reported to relate to cancers (38), while the other hub gens are all ribosomal RNA.
Discussion
Nowadays, several long-standing issues are difficult to solve in EV-based liquid biopsy and cancer research (15). For instance, how to discriminate the weak cancer-related signatures from the intense background noise of normal human physiology especially at the early stages of cancer? How to decipher the contributions of TME to tumor development and to further develop TME-associated diagnostic and therapeutic strategies? The comprehensive identification of cTME-EVs by developing selective enrichment technology provides unprecedented opportunities to attack the aforesaid issues.
pHLIP is a widely exploited TME-targeting agent for tumor-targeted imaging and drug delivery (39). However, its targeting ability to TME is limited by its relatively low folding-responsive value (pKa ≈ 6.2). The development of engineered pHLIP with higher folding-responsive values (ideally pKa ≈ 6.8) that can be sensitively activated in TME (pH 6.5 to 6.8) is crucial for achieving higher tumor targeting efficiency. To improve its responsiveness to TME, the structure of pHLIP has been engineered using traditional mutation screening (40) and non-canonical amino acids (21), which unfortunately suffer from complexity and time-consuming. Recently, computational means, such as the pharmacokinetic model (20) and kinetics (41), were introduced as practical strategies to advance the development of pHLIP variants.
We herein present a computational tactic to design hundreds of virtual variants via pK calculations on the key regions of pHLIP to efficiently respond TME. This general strategy enables us to evaluate the potential mutation effects in silico in a straightforward, high-efficiency, and time-saving manner. Additionally, to selectively enrich cTME-EVs from bodily fluids, a pH-driven conformational switch should be designed to conjugate with pHLIP to prevent their dissociation in the neutral systemic circulation. Although computational designs have made significant progress in engineering proteins, creating conformational switch with specific functions is generally considered full of challenges (42). Here, the hook-like switch was tied in the C-terminal of D-S pHLIP and its tight binding with cTME-EVs was simulated through computational tests using MD simulations, free energy calculations, and interactional energy calculations.
In our study, cTME-EVs include the EVs derived from tumor cells and TACs in TME, which were confirmed by transcriptome analysis. To date, a variety of tumor biomarkers (e.g., EpCAM) have been identified on the tumor cell-derived EVs. Based on this fact, numerous antibodies or aptamers targeting the tumor biomarkers have been developed to enrich the tumor cell-derived EVs from biofluids for discovering potential cancer biomarkers. For instance, Choi et al. have employed antibodies against CD9, CD63, CD81, and EpCAM to enrich lung tumor cell-derived EVs from serum (43). However, the EVs enriched by these approaches are tumor cell-derived EVs other than TAC-derived ones. The TAC-derived EVs are highly heterogeneous and have rarely been studied due to the lack of effective tools to isolate them from biofluids. The proposed D-S pHLIP system, to our knowledge, represents the approach that is able to selectively enrich both tumor cell-derived EVs and TAC-derived EVs from blood serum. The analysis of cTME-EVs could not only achieve the comprehensive information of the important relationships between tumor and TME, but also uncover the roles of TAC-derived EVs in the tumor development and metastasis. Moreover, the secretion of TAC-derived EVs could be influenced by tumor cell-derived EVs to produce a complex TME. In combination with multiple omics analysis, this D-S pHLIP system would prompt the development of EV-based cancer diagnosis and therapy.
Except for the acidic foci in tumors, pHLIP can target the acidic inflammatory sites as well (26). In many established cancers, inflammation has been demonstrated as a hallmark of cancer and contributes to cancer progression substantially (44–46). Therefore, the EVs derived from cancer inflammation sites can be captured by our system to offer rich cancer signatures, aiding more comprehensive cancer diagnosis and personalized treatment. It is worth noting that the EVs derived from the non-cancer-related inflammatory sites in patients with multiple complications may influence the specificity of cTME-EVs. By coupling EVs with other classical biomarkers, such as free proteins and circulating DNA, these complex diseases could be discriminated accurately.
The proposed D-S pHLIP system breaks through the dilemma of selective enrichment of cTME-EVs from complex bodily fluids in broad cancer types. Given the excellent conformational switches, D-S pHLIP can be further engineered by simply replacing the biotin in the N-terminal with dyes or drugs, which will show great potential in tumor imaging and TME-targeted drug delivery.
Materials and Methods
The D-S pHLIP was computationally designed via H++ software of 3.2 version and SWISS-MODEL and assessed by MD simulations, free-energy calculations, and interaction energy calculations. pHLIP variants were synthesized and purified by GL Biochem Ltd. Liposome preparation. The WT pHLIP and variants were characterized by high-performance liquid chromatography and mass spectrometry. Detailed descriptions of fluorescence and CD measurements, in vitro and in vivo experiments, and transcriptome analysis are presented in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Conformational changes of D-S v1 based on its trajectory in 100 ns MD simulation.
Conformational changes of D-S v2 based on its trajectory in 100 ns MD simulation.
Acknowledgments
This study was supported by the National Key R&D Program of China (2019YFA0210100), the National Natural Science Foundation of China (21775075, 21977053 to D.L., and 22005157 to Z.Z.), China Postdoctoral Science Foundation (2021M701788), and Frontiers Science Center for Cell Responses, Nankai University.
Author contributions
Z.Z. and D.L. designed research; Z.Z., X.L., and Z.Y. performed research; Z.Z., X.L., Z.Y., and D.L. analyzed data; and Z.Z. and D.L. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. X.O.B. is a guest editor invited by the Editorial Board.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Marar C., Starich B., Wirtz D., Extracellular vesicles in immunomodulation and tumor progression. Nat. Immunol. 22, 560–570 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu R., et al. , Extracellular vesicles in cancer-implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 15, 617–638 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Tauro B. J., et al. , Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon tumor cell line LIM1863-derived exosomes. Methods (San Diego, Calif.) 56, 293–304 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Liu X., et al. , pH-mediated clustering of exosomes: Breaking through the size limit of exosome analysis in conventional flow cytometry. Nano Lett. 21, 8817–8823 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Davies R. T., et al. , Microfluidic filtration system to isolate extracellular vesicles from blood. Lab Chip 12, 5202–5210 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Mol E. A., Goumans M.-J., Doevendans P. A., Sluijter J. P. G., Vader P., Higher functionality of extracellular vesicles isolated using size-exclusion chromatography compared to ultracentrifugation. Nanomed. Nanotechnol. 13, 2061–2065 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Wu L., et al. , Aptamer-based detection of circulating targets for precision medicine. Chem. Rev. 121, 12035–12105 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Park J., et al. , An integrated magneto-electrochemical device for the rapid profiling of tumour extracellular vesicles from blood plasma. Nat. Biomed. Eng. 5, 678–689 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melo S. A., et al. , Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 523, 177–182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H. Y., et al. , Rapid capture of cancer extracellular vesicles by lipid patch microarrays. Adv. Mater. 33, e2008493 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L., et al. , Tim4-functionalized HBEV-chip by isolating plasma-derived phosphatidylserine-positive small extracellular vesicles for pan-cancer screening. Adv. Mater. Technol. 7, 2101115–2101127 (2022). [Google Scholar]
- 12.Keller L., Pantel K., Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat. Rev. Cancer 19, 553–567 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Dagogo-Jack I., Shaw A. T., Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 15, 81–94 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Takao M., Nagai Y., Ito M., Ohba T., Flow cytometric quantitation of EpCAM-positive extracellular vesicles by immunomagnetic separation and phospholipid staining method. Genes Cells 23, 963–973 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Crosby D., et al. , Early detection of cancer. Science 375, eaay9040 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Chen Y., McAndrews K. M., Kalluri R., Clinical and therapeutic relevance of cancer-associated fibroblasts. Nat. Rev. Clin. Oncol. 18, 792–804 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Wang S., et al. , A tumor microenvironment–induced absorption red-shifted polymer nanoparticle for simultaneously activated photoacoustic imaging and photothermal therapy. Sci. Adv. 7, eabe3588 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt J. F., Rath P., Rothschild K. J., Engelman D. M., Spontaneous, pH-dependent membrane insertion of a transbilayer alpha-helix. Biochemistry 36, 15177–15192 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Svoronos A. A., Engelman D. M., Pharmacokinetic modeling reveals parameters that govern tumor targeting and delivery by a pH-Low Insertion Peptide (pHLIP). Proc. Natl. Acad. Sci. U.S.A. 118, e2016605118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onyango J. O., et al. , Noncanonical amino acids to improve the pH response of pHLIP insertion at tumor acidity. Angew. Chem. Int. Ed. 54, 3658–3663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., et al. , A nanoparticle-based strategy for the imaging of a broad range of tumours by nonlinear amplification of microenvironment signals. Nat. Mater. 13, 204–212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Musial-Siwek M., Karabadzhak A., Andreev O. A., Reshetnyak Y. K., Engelman D. M., Tuning the insertion properties of pHLIP. Biochim. Biophys. Acta 1798, 1041–1046 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onufriev A., Case D. A., Ullmann G. M., A novel view of pH titration in biomolecules. Biochemistry 40, 3413–3419 (2001). [DOI] [PubMed] [Google Scholar]
- 25.An M., Wijesinghe D., Andreev O. A., Reshetnyak Y. K., Engelman D. M., pH-(low)-insertion-peptide (pHLIP) translocation of membrane impermeable phalloidin toxin inhibits tumor cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 107, 20246–20250 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andreev O. A., et al. , Mechanism and uses of a membrane peptide that targets tumors and other acidic tissues in vivo. Proc. Natl. Acad. Sci. U.S.A. 104, 7893–7898 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Q., et al. , The genetic source tracking of human urinary exosomes. Proc. Natl. Acad. Sci. U.S.A. 118, e2108876118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ju M. J., et al. , Combination of peritumoral mast cells and T-regulatory cells predicts prognosis of hepatocellular carcinoma. Cancer Sci. 100, 1267–1274 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eissmann M. F., et al. , IL-33-mediated mast cell activation promotes gastric cancer through macrophage mobilization. Nat. Commun. 10, 2735 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cherezov V., et al. , High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palczewski K., et al. , Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289, 739–745 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Borgquist S., et al. , Apolipoproteins, lipids and risk of cancer. Int. J. Cancer 138, 2648–2656 (2016). [DOI] [PubMed] [Google Scholar]
- 33.His M., et al. , Prospective associations between serum biomarkers of lipid metabolism and overall, breast and prostate cancer risk. Eur. J. Epidemiol. 29, 119–132 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Hsu M. C., et al. , The dyslipidemia-associated SNP on the APOA1/C3/A5 gene cluster predicts post-surgery poor outcome in Taiwanese breast cancer patients: A 10-year follow-up study. BMC Cancer 13, 330 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tateno T., Ichinose A., Expression of plasminogen-related gene B varies among normal tissues and increases in cancer tissues. FEBS Lett. 445, 31–35 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Davalieva K., et al. , Proteomics analysis of urine reveals acute phase response proteins as candidate diagnostic biomarkers for prostate cancer. Proteome Sci. 13, 2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi F., et al. , Identification of serum proteins AHSG, FGA and APOA-I as diagnostic biomarkers for gastric cancer. Clin. Proteom. 15, 18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu M., et al. , Integrated analyses reveal the multi-omics and prognostic characteristics of ATP5B in breast cancer. Front. Genet. 12, 652474 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Z., et al. , An acidic-microenvironment-driven DNA nanomachine enables specific ATP imaging in the extracellular milieu of tumor. Adv. Mater. 31, 1901885 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Weerakkody D., et al. , Family of pH (low) insertion peptides for tumor targeting. Proc. Natl. Acad. Sci. U.S.A. 110, 5834–5839 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slaybaugh G., Weerakkody D., Engelman D. M., Andreev O. A., Reshetnyak Y. K., Kinetics of pHLIP peptide insertion into and exit from a membrane. Proc. Natl. Acad. Sci. U.S.A. 117, 12095–12100 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyken S. E., et al. , De novo design of tunable, pH-driven conformational changes. Science 364, 658–664 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi E. S., et al. , CD5L as an extracellular vesicle-derived biomarker for liquid biopsy of lung cancer. Diagnostics 11, 620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diakos C. I., Charles K. A., McMillan D. C., Clarke S. J., Cancer-related inflammation and treatment effectiveness. Lancet Oncol. 15, e493–e503 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Crusz S. M., Balkwill F. R., Inflammation and cancer: Advances and new agents. Nat. Rev. Clin. Oncol. 12, 584–596 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Coussens L. M., Werb Z., Inflammation and cancer. Nature 420, 860–867 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Conformational changes of D-S v1 based on its trajectory in 100 ns MD simulation.
Conformational changes of D-S v2 based on its trajectory in 100 ns MD simulation.
Data Availability Statement
All study data are included in the article and/or SI Appendix.