Significance
Filoviruses EBOV and MARV are zoonotic, emerging pathogens that cause sporadic and global outbreaks of severe hemorrhagic fever. As filoviruses can establish persistent infections in immune-privileged sites inaccessible to antibody therapy, and their re-emergence can result in long-term sequelae and death, identifying virus–host interactions that contribute to their transmission and pathogenesis will reveal new opportunities and targets for antiviral development and intervention. Here, we describe how the mTORC1/CASA axis functions to regulate and restrict the egress of EBOV particles by targeting the main driver of filovirus budding—the VP40 matrix protein, which may represent a new host defense strategy in the ongoing arms race between virus and host.
Keywords: filovirus, ebola, bag3, mTORC1, autophagy
Abstract
The filovirus VP40 protein directs virion egress, which is regulated either positively or negatively by select VP40–host interactions. We demonstrate that host BAG3 and HSP70 recognize VP40 as a client and inhibit the egress of VP40 virus-like particles (VLPs) by promoting degradation of VP40 via Chaperone-assisted selective autophagy (CASA). Pharmacological inhibition of either the early stage formation of the VP40/BAG3/HSP70 tripartite complex, or late stage formation of autolysosomes, rescued VP40 VLP egress back to WT levels. The mechanistic target of rapamycin complex 1 (mTORC1) is a master regulator of autophagy, and we found that surface expression of EBOV GP on either VLPs or an infectious VSV recombinant virus, activated mTORC1. Notably, pharmacological suppression of mTORC1 signaling by rapamycin activated CASA in a BAG3-dependent manner to restrict the egress of both VLPs and infectious EBOV in Huh7 cells. In sum, our findings highlight the involvement of the mTORC1/CASA axis in regulating filovirus egress.
Ebola (EBOV) and Marburg (MARV) viruses are virulent pathogens that cause severe hemorrhagic disease in humans and non-human primates (1), and these emerging pathogens remain a global public health threat. An in-depth understanding of the filovirus–host interplay will reveal mechanistic and fundamental knowledge of the virus lifecycle, as well as identify new targets and strategies for antiviral development (2). Toward this end, we have focused on the late stage of filovirus budding mediated by the VP40 matrix protein and the key roles of select host interactors that either positively or negatively regulate the egress of virus particles (3, 4). For example, we and others have shown that several members of the HECT (Homologous to the E6-AP carboxyl terminus) family of E3 ubiquitin ligases (e.g., Nedd4, Itch, and WWP1) and members of the ESCRT (Endosomal Sorting Complexes Required for Transport) pathway (e.g., Tsg101) are hijacked by VP40 to enhance the egress (pinching-off) and spread of both VP40 virus-like particles (VLPs) and infectious virions (5–8). On the other hand, a growing list of host interactors is emerging that appear to negatively regulate budding, including co-chaperone BAG3 (Bcl2-associated athanogene 3) (9), transcription coactivators YAP/TAZ (Yes-associated protein/transcriptional coactivator with PDZ-binding motif) (10, 11) and WWOX (WW domain containing oxidoreductase) (12, 13). Some of the best-characterized positive or negative host regulators of egress interact with the highly conserved PPxY Late (L) domain motif of VP40 via one or more of their modular WW-domains (14). Indeed, the filoviral PPxY/host WW-domain interface mimics and competes with similar host–host PPxY/WW-domain interactions and this modular mimicry has the potential to significantly affect both the virus lifecycle and the host cell environment.
We first identified BAG3 as a specific VP40 interactor using a WW-domain array screen (9). BAG3 has multiple modular domains including a single WW-domain, and BAG3 is the central regulator of Chaperone-assisted selective autophagy (CASA); a noncanonical macroautophagy pathway that specifically recognizes and targets client proteins to aggresomes for autophagic lysosomal degradation as a means of regulating cellular and protein homeostasis (15–21). Autophagy in general is an evolutionarily conserved cellular process that maintains protein homoeostasis, and it often serves as a cellular defense strategy to capture and clear invading pathogens (22–25). Indeed, several reports have linked filovirus infection to autophagy and specific autophagy-associated proteins, such as Beclin1, ATG7 (autophagy related 7) and LC3B (microtubule-associated protein light chain 3) (26–29).
While we demonstrated previously that BAG3 interacted with VP40 and inhibited VP40 VLP egress, the underlying mechanism of this inhibition remained to be elucidated. Here, we reveal that the mechanism by which BAG3 and the CASA pathway restrict filovirus egress involves BAG3-mediated targeting of VP40 for autophagic sequestration and degradation. We demonstrate that pharmacological intervention at early and late stages of the CASA pathway can reverse the inhibitory effect on budding and restore VP40 VLP egress back to WT levels. These functional assays correlated well with visualization of VP40 via confocal microscopy at the plasma membrane (PM) or within aggresomes/autophagosomes/autolysosomes under various treatment conditions and in the absence or presence of BAG3. We also find that the mTORC1/CASA axis regulates EBOV infection, in that EBOV glycoprotein (GP) can activate mTORC1, while pharmacological suppression of mTORC1 by rapamycin treatment facilitates CASA-mediated inhibition of egress of both VP40 VLPs and live authentic EBOV in Huh7 cells. In sum, our findings provide insights into the mechanism by which mTORC1 signaling and the CASA pathway may be entwined with late stages of the filovirus lifecycle, which may reveal new targets for antiviral development.
Results
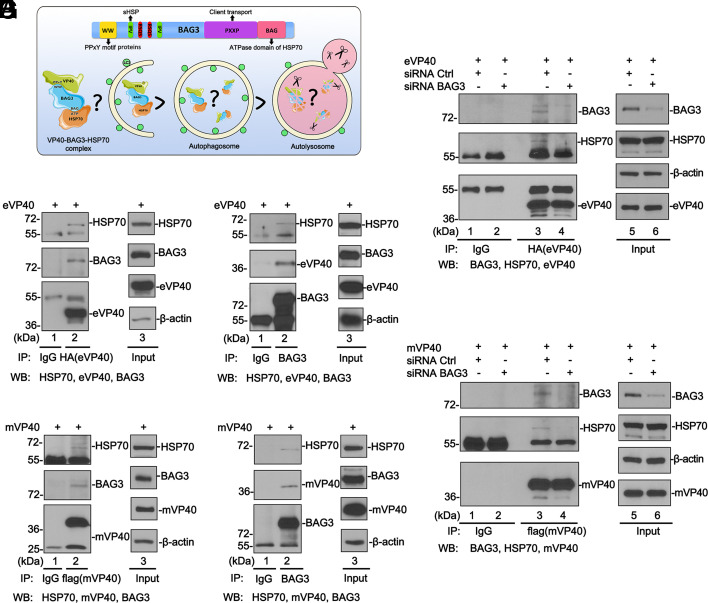
Filovirus VP40 Is a Client of the BAG3–HSP70 Complex.
We previously identified co-chaperone protein BAG3 as a negative regulator of VP40-mediated egress, and the physical interaction between the single WW-domain of BAG3 and the VP40 PPxY motif was required for the negative regulatory activity (9). BAG3 mediates a noncanonical macroautophagy pathway termed CASA in which BAG3 interacts and cooperates with HSP70 (heat shock protein 70) and small HSP to deliver targeted client proteins for autophagic–lysosomal degradation (15, 16, 19, 30). Here, we sought to determine whether VP40 is a client of the HSP70/BAG3 (HB) complex (Fig. 1A). To test this, we first used co-immunoprecipitation (co-IP) assays to determine whether VP40 associates with the endogenous HB complex in HEK293T cells. We found that endogenous BAG3 and HSP70 were both detected in either eVP40 (Fig. 1B) or mVP40 precipitates (Fig. 1D). Similarly, HSP70 and eVP40 (Fig. 1C) or mVP40 (Fig. 1E) were detected in BAG3 precipitates. These results show that BAG3/HSP70/VP40 form a complex in HEK293T cells.
Fig. 1.
Filovirus VP40 interacts with CASA components in a BAG3-dependent manner. (A) Diagram showing the modular domains of BAG3 and the CASA pathway. (B and D) Co-IP assay detecting both endogenous BAG3 and HSP70 in eVP40 or mVP40 precipitates. (C and E) Co-IP assay detecting eVP40, mVP40, or HSP70 in BAG3 precipitates. (F and G) siRNA knockdown of endogenous BAG3 abolishes VP40/BAG3/HSP70 complex formation. The 55 kDa band is IgG heavy chain, the 25 kDa band is IgG light chain.
We reasoned that BAG3 was likely the central mediator of this complex formation since it interacts directly with VP40 PPxY motif via its WW-domain and directly with HSP70 via its BAG domain. To test this, we used an siRNA approach to knock down endogenous BAG3 in HEK293T cells, followed by exogenous expression of eVP40 (Fig. 1F) or mVP40 (Fig. 1G) and co-IP analysis. Following knockdown of endogenous BAG3, HSP70 was no longer detected in VP40 precipitates (Fig. 1 E and F, lanes 4), suggesting that BAG3 expression is required for the formation of the tri-protein complex. Together, these results demonstrate that BAG3 directs the formation of the tripartite complex of VP40/BAG3/HSP70, and suggest that VP40 may be a client of the CASA pathway (Fig. 1A).
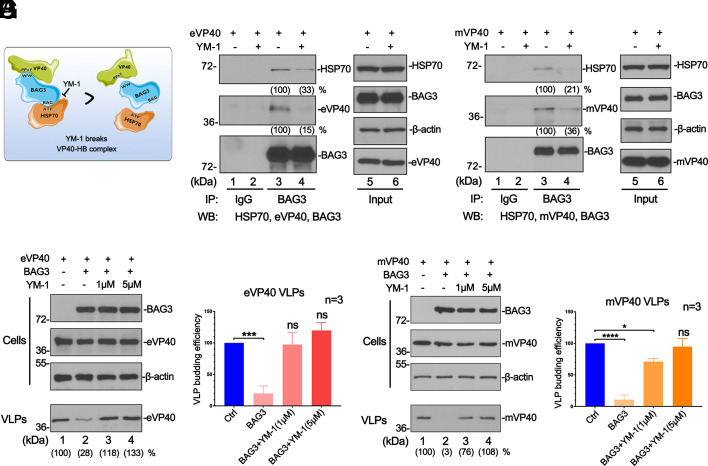
Breaking the BAG3-HSP70 Complex Releases VP40 and Rescues VLP Egress.
If the HB complex plays a role in directing the VP40 client to the aggresome for degradation via the autophagic–lysosomal pathway, then we postulated that disruption of the HB complex should release VP40 and rescue budding of VP40 VLPs (Fig. 2A). To test this, we used YM-1, a compound that targets the ATPase domain of HSP70 and breaks its interaction with BAG3 (31). First, we asked whether YM-1 could inhibit the physical formation of the VP40/BAG3/HSP70 tripartite complex in HEK293T. Using co-IP analysis, we found that YM-1 significantly inhibited the interaction between BAG3 and HSP70 as expected (Fig. 2 B and C, lanes 3 and 4), and in doing so, also impaired the interaction between BAG3 and eVP40 (Fig. 2B, lanes 3 and 4) or mVP40 (Fig. 2C, lanes 3 and 4). These findings indicated that YM-1 was capable of disrupting the physical interactions required to form the VP40/BAG3/HSP70 complex. We then performed a VLP budding assay in the absence or presence of YM-1 to determine whether physical disruption of the VP40/BAG3/HSP70 complex would lead to functional recovery of VP40 VLP egress. Notably, YM-1 completely reversed the inhibitory effect of the HB complex on VLP budding leading to the rescue of VP40 VLP budding back to control levels (Fig. 2 D–G). Moreover, the treatment with YM-1 boosted the egress of eVP40 and mVP40 VLPs in both HEK 293T and Huh7 cells in a dose-dependent manner (SI Appendix, Fig. S1). Thus, the inhibitory effect of the HB complex on VP40 VLP budding is reversible, and disruption of this early stage of the CASA pathway can restore VP40 VLP egress back to control levels.
Fig. 2.
YM-1 disrupts the VP40/BAG3/HSP70 complex and rescues VP40 VLP egress. (A) Diagram of YM-1 disrupting the VP40/BAG3/HSP70 complex. (B and C) Co-IP assay showing reduced levels of HSP70 and VP40 detected in BAG3 precipitates in the presence of YM-1 compared to DMSO control. (D–G) eVP40 and mVP40 VLP budding assays in HEK293T cells and VLP quantification (bar graphs from three independent experiments) in the absence (DMSO) or presence of YM-1. Statistical significance was analyzed by one-way ANOVA. ns: not significant, *P < 0.05, ***P < 0.001, ****P < 0.0001.
Pharmacological Blockage of the Autophagy Flux Rescues VP40 VLP Egress.
To demonstrate further that the BAG3-mediated macroautophagy pathway restricts VP40 VLP egress, we sought to determine whether disruption of stages of the CASA pathway downstream of the initial formation of the BAG3–client complex would also lead to the rescue of VP40 VLP egress (Fig. 3A). Here, we utilized autophagy inhibitor chloroquine (CQ) to block the late stage of fusion between the autophagosome and lysosome (32) and then evaluated whether CQ treatment reversed the inhibitory function of BAG3 and the HB complex. Briefly, HEK 293T cells were co-transfected with BAG3 and eVP40 (Fig. 3 B and D) or mVP40 (Fig. 3 C and E) in the absence or presence of increasing concentrations of CQ, and VP40 levels were quantified in VLPs. We observed that VP40 VLP budding was inhibited in the presence of BAG3 alone as expected; however, upon addition of CQ, egress of both eVP40 and mVP40 VLPs was rescued back to control levels (Fig. 3 B and C, compare lanes 2 to 4; Fig. 3 D and E). To further understand the mechanism underlying this phenomenon, we used a co-IP assay to evaluate the formation of the VP40-HB complex with/without CQ treatment, and our results showed that CQ did not affect the physical interactions among VP40, BAG3, and HSP70 (SI Appendix, Fig. S2). Given that VP40 was sequestered away from the PM in the presence of BAG3 alone (9), we postulated that VP40 may be re-localized back to the PM in the presence of CQ resulting in enhanced VLP egress. To test this, we isolated cytosol and PM fractions from HEK293T cells co-expressing BAG3 and eVP40 (Fig. 3F) or mVP40 (Fig. 3G) in the absence or presence of CQ. Notably, while ectopic expression of BAG3 markedly decreased the amount of VP40 in the PM fraction, the amount of VP40 in the PM fraction was enhanced in CQ-treated cells (Fig. 3 F and G, lanes 4 to 6). Taken together, our findings suggest that a functional CASA pathway is necessary for BAG3 induced-inhibition of VLP egress, and that pharmacological inhibition of either early (BAG3-client complex formation) or late (autophagy flux) stages of this pathway can overcome the inhibitory effect on VLP egress.
Fig. 3.
CQ blocks the last stage of the CASA pathway and rescues VP40 VLP egress. (A) Schematic diagram of CQ blocking the autophagy flux to rescue VP40 VLP budding. (B–E) eVP40 and mVP40 VLP budding assays and VLP quantification (bar graphs) from HEK293T cells exogenously expressing BAG3 and in the absence or presence of CQ. Statistical significance was analyzed by one-way ANOVA. ns: not significant, *P < 0.05, ***P < 0.001, ****P < 0.0001. (F and G) eVP40 and mVP40 levels were quantified by NIH Image-J from the cytosol or PM fractions of HEK293T cells under the indicated conditions. β-actin and Na/K ATPase served as loading controls for the cytosol and PM fractions, respectively.
BAG3 Sequesters VP40 Aggregates in Autophagosomes.
Next, we used confocal microscopy to visualize the association of VP40, BAG3, and the CASA pathway by detecting the intracellular localization of VP40, BAG3, and three autophagic marker proteins (LC3, p62, and ATG5) in Huh7 cells (Fig. 4). We exogenously expressed human-LC3-CFP and YFP-eVP40 with either vector alone (Fig. 4 A–C), or BAG3-mCherry (Fig. 4 D–K), and samples were examined using live cell confocal microscopy. We observed the typical pattern of eVP40 in eVP40 + LC3 expressing cells of enriched localization of eVP40 in PM protrusions and abundant filamentous VLP formation at the cell periphery (Fig. 4B). We observed LC3 evenly distributed in the cytoplasm and nuclei (Fig. 4A) of these cells (suggestive of a base level of cellular autophagy) with no detectable co-localization with eVP40 (Fig. 4C). Upon expression of BAG3, we observed colocalization of LC3, eVP40, and BAG3 in perinuclear puncta (Fig. 4 G–K), suggesting that BAG3 mediated re-localization and sequestration of eVP40 into aggresomes (Ags) and autophagosomes (APs).
Fig. 4.
Confocal Microscopy showing that BAG3 sequesters and delivers VP40 into LC3-containing autophagosomes. (A–K) Confocal images of Huh7 cells expressing the indicated combinations of proteins including LC3-CFP (cyan), YFP-eVP40 (green), BAG3-mCherry (red). Panel J and zoomed-in panel K highlight the colocalization of LC3, BAG3, and eVP40 in aggregates and autophagosomes. (L–Q) Representative confocal images of Huh7 cells co-expressing LC3-mCherry (red) and GFP-eVP40 (green) in the absence or presence of rapamycin (200 nM). Graphs R and S, Quantification of LC3-containing autophagosomes (APs) measured by the numbers of LC3 puncta per cell (R), and the percentage of cells showing colocalization of VP40 aggregates (Ags) and LC3 APs (S). n ≥ 75 cells were scored for each condition from three independent experiments. Statistical significance was determined by Welch’s t test, ****P < 0.0001, **P < 0.01. (T–W) Confocal images of rapamycin (200 nM)-treated Huh7 cells co-expressing LC3-CFP (cyan), YFP-eVP40 (green) and p62-mCherry (red). (Scale bar, 10 µm in main panels and 2 µm in Inset panels.)
Given that CASA is a specific pathway that belongs to macroautophagy, we next sought to determine whether endogenous BAG3 and activation of the autophagy flux facilitates sequestration of eVP40 into autophagosomes. Toward this end, we first confirmed the association of eVP40 and the HB complex in Huh7 cells by co-IP (SI Appendix, Fig. S3). Next, Huh7 cells co-expressing GFP-eVP40 and LC3-mCherry were mock-treated or treated with autophagy inducer rapamycin, and then observed using live cell confocal microscopy. Again, the typical pattern of eVP40 was observed in control cells with basal levels of autophagy (Fig. 4 L–N). Cells treated with rapamycin displayed enhanced formation of LC3-containing puncta, indicative of the activation of autophagy flux in these cells (Fig. 4 P and R). In addition, we observed altered localization of eVP40 from the PM to perinuclear LC3-containing autophagosomes (Fig. 4 O–Q and S), as well as LC3-marked autophagosomes gathered at the cell periphery (SI Appendix, Fig. S4A). To further confirm that VP40 is sequestered in autophagic vesicles, we detected selective autophagy substrate p62, and phagophore membrane protein ATG5. Indeed, we observed colocalization of VP40 with LC3 and P62 (Fig. 4 T–W), and with ATG5 (SI Appendix, Fig. S4 B and C) in puncta upon the treatment with rapamycin. In sum, rapamycin-induced autophagy and concomitant sequestration of eVP40 away from the PM and into autophagosomes.
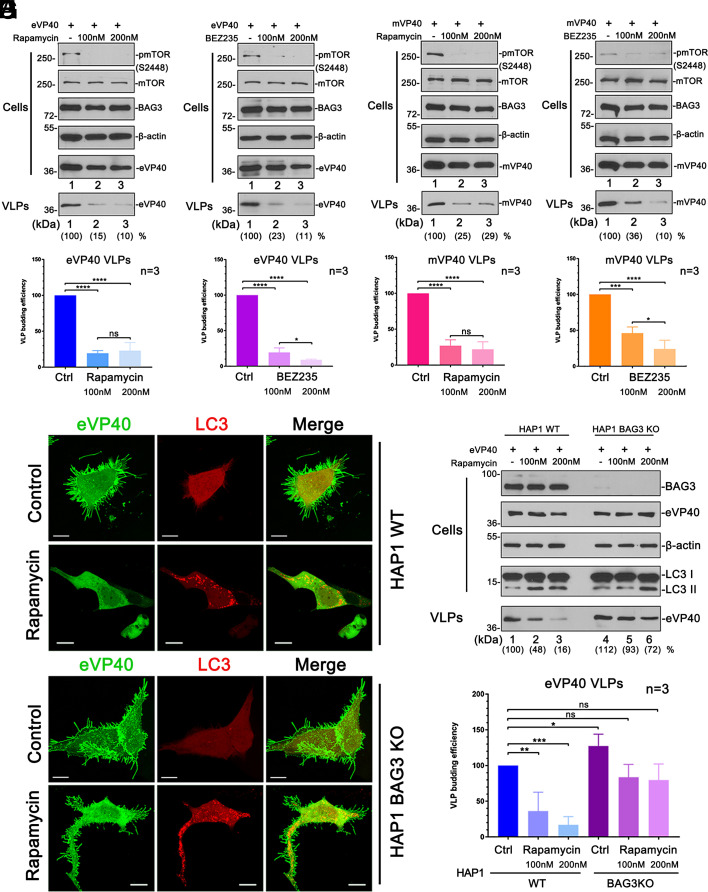
Pharmacological Suppression of mTORC1 Inhibits VP40 VLP Budding via the CASA Pathway.
Since activation of autophagy flux led to the sequestration of VP40 into APs, we reasoned that VLP budding would be inhibited under these conditions as well. The mechanistic target of rapamycin complex 1 (mTORC1) is the upstream gateway which modulates autophagy, and thus inhibition of mTOCRC1 activates autophagy (23, 33). Hence, we utilized rapamycin or compound BEZ235 to suppress mTOR signaling and then evaluated their impact on the egress of VP40 VLPs. Huh7 cells were transfected with eVP40 (Fig. 5 A and B) or mVP40 (Fig. 5 C and D) in the absence or presence of increasing concentrations of rapamycin or BEZ235. VP40 levels were quantified in VLPs, and suppression of mTORC1 was verified by quantifying the levels of phosphorylated (Ser2448) mTOR (34, 35) (Fig. 5 A–D). We observed that rapamycin and BEZ235 blocked the activation of mTORC1 (Fig. 5 A–D, Cells, pmTOR and mTOR), and notably, inhibited the egress of VP40 VLPs (Fig. 5 A–D and SI Appendix, Fig. S5).
Fig. 5.
mTOR inhibitors rapamycin and BEZ-235 restrict egress of VP40 VLPs via CASA. (A–D) eVP40 and mVP40 VLP budding assays in Huh7 cells and VP40 VLP quantification (bar graphs) in the absence or presence of either rapamycin (A and C) or BEZ-235 (B and D). Statistical significance was analyzed by one-way ANOVA. ns: not significant, *P < 0.05, ***P < 0.001, ****P < 0.0001. (E and F) Confocal images of HAP1 WT (E) and BAG3 KO (F) cells co-expressing LC3-mCherry (red) and GFP-eVP40 (green) with or without rapamycin treatment (200 nM) (Scale bar, 10 µm.). (G and H) eVP40 VLP budding assays in HAP1 WT and BAG3 KO cells with the treatment of rapamycin (G) and VLP quantification (H). Statistical significance was analyzed by one-way ANOVA. ns: not significant, *P < 0.05, **P < 0.01, ***P < 0.001.
To further illustrate the central role of BAG3 in regulating the intersection of mTORC1 signaling, the CASA pathway, and VP40-mediated budding, we used a commercially available BAG3 KO HAP1 cell line. We used confocal microscopy to demonstrate that rapamycin treatment did induce the formation of LC3-containing APs in both HAP1 WT and BAG3 KO cells; however, egress of eVP40 VLPs from BAG3 WT cells was inhibited more significantly than that from BAG3 KO cells (Fig. 5 E and F). Notably, eVP40 VLP projections at the PM were prominent in the BAG3 KO cells, but were largely absent in the HAP1 WT cells. The results obtained via imaging correlated well with those observed from eVP40 VLP budding assays in HAP1 WT and BAG3 KO cells under the same rapamycin treatment conditions (Fig. 5 G and H).
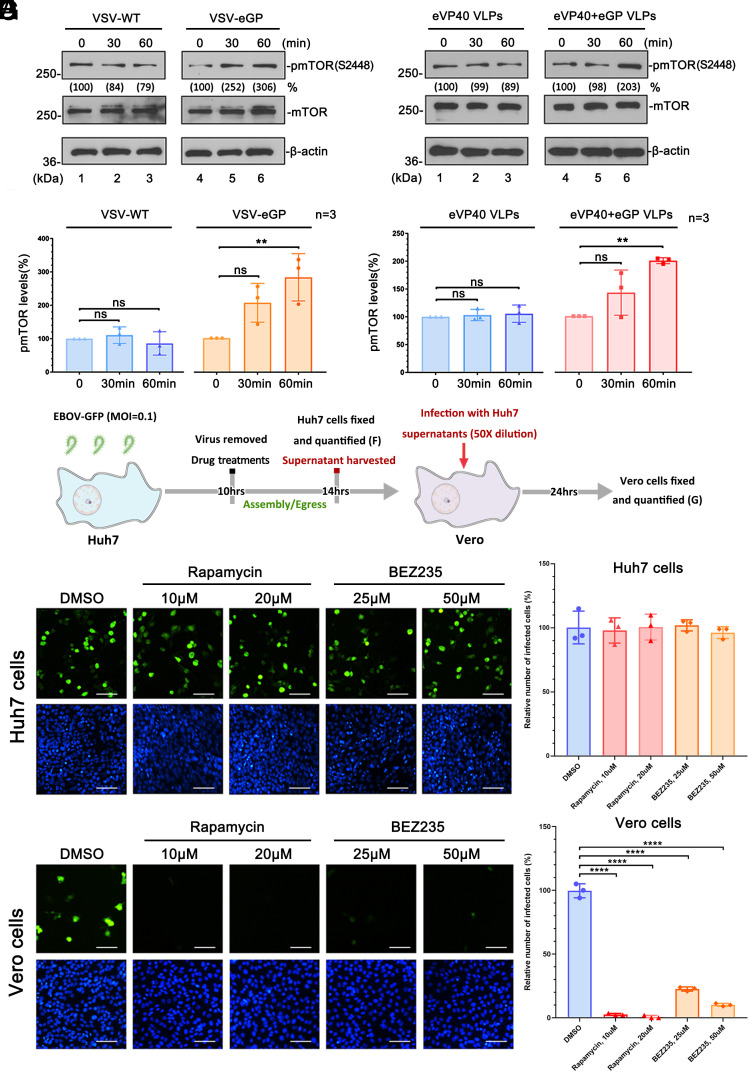
Pharmacological Suppression of mTORC1 Restricts Egress of Infectious EBOV.
Because of the ongoing battle between the virus and host, we reasoned that if suppression of mTORC1 by rapamycin led to inhibition of VP40 VLP egress, then mTORC1 signaling may be beneficial for virus production and possibly activated upon virus infection. To test this, Huh7 cells were incubated with VSV-WT or a recombinant infectious vesicular stomatitis virus (VSV) expressing EBOV GP (VSV-eGP) in place of VSV-G for 30 or 60 min, and phosphorylation (activation) of mTOR was assessed by Western blotting (Fig. 6 A and B). We observed a significant enhancement of phosphorylation of mTOR in the presence of VSV-eGP, but not VSV-WT (Fig. 6 A and B and SI Appendix, Fig. S6 A and B). To confirm this finding, we performed a similar experiment using purified VLPs containing either eVP40 alone, or eVP40 + eGP in place of live virus. Indeed, we observed a significant enhancement of phosphorylation of mTOR in the presence of eVP40 + eGP VLPs, but not eVP40 VLPs alone (Fig. 6 C and D and SI Appendix, Fig. S6 C and D). Together, these results suggest that phosphorylation of mTOR and likely activation of mTORC1 signaling (34, 35) is triggered specifically by eGP (present on the surface of either VLPs or live virus) binding to Huh7 cells.
Fig. 6.
mTOR inhibitors rapamycin and BEZ-235 restrict egress of infectious EBOV. (A–D) Detection and quantification of phosphorylation levels of mTOR in Huh7 cells incubated with VSV-WT or VSV-eGP (A and B) and eVP40 VLPs or eVP40+eGP VLPs (C and D). Statistical significance was analyzed by one-way ANOVA. ns: not significant, ***P < 0.001. (E) Schematic timeline of EBOV-GFP infection protocol and virus harvest. (F) Confocal images and quantification (bar graph) of EBOV-GFP infected Huh7 cells at 14 h.p.i. in the absence or presence of rapamycin or BEZ-235. (G) Confocal images and quantification (bar graph) of Vero cells infected with EBOV-GFP-containing supernatants that were harvested from Huh7 cells under the indicated conditions (Scale bar, 50 µm.)
Next, we sought to determine whether suppression of mTORC1 would inhibit the egress of infectious EBOV as it did eVP40 VLPs. Huh7 cells were incubated with EBOV encoding green fluorescent protein (EBOV-GFP) at an MOI of 0.1, and the inoculum was removed at 10 h.p.i. (Fig. 6E). Cells were then treated with dimethyl sulfoxide (DMSO) alone, or with rapamycin or BEZ235 for 14 h (Fig. 6 E and F), and virions in the Huh7 supernatants were quantified by titrating on Vero cells (Fig. 6G). Since eVP40 localizes to the PM as early as 12 h.p.i. in infected cells (36), the treatments would target EBOV egress. We observed that EBOV infected equally well in Huh7 cells treated with DMSO alone, or those treated with either rapamycin or BEZ235 (Fig. 6F), showing that the treatments did not affect the early stages of EBOV lifecycle. In contrast, the generation of EBOV virions from Huh7 cells treated with either mTOR inhibitor was reduced by >95% for rapamycin and >75% for BEZ235 treatment (P < 0.0001), as compared to DMSO control (Fig. 6G and SI Appendix, Fig. S7A). These findings indicate that specific inhibition of mTORC1 with either blocker potently inhibited the egress of live EBOV without any detectable cytotoxicity at the concentrations tested (SI Appendix, Fig. S7 B–D).
mTORC1/CASA Axis Targets VP40 to Autophagic–Lysosomal Sequestration and Degradation.
We next sought to investigate whether the mTORC1/CASA axis can direct VP40 into autolysosomes for degradation. Toward this end, we first utilized a double fluorescent LC3 reporter construct to verify the formation of autolysosomes (ALs)/autophagosomes (APs) (37) under different treatment conditions (Fig. 7 A and B). Briefly, Huh7 cells expressing LC3-eGFP-mCherry were observed via live cell confocal microscopy, and as expected, we found that treatment with rapamycin increased the formation of ALs, while CQ blocked the autophagy cascade by preventing fusion of APs with lysosomes (Fig. 7A).
Fig. 7.
mTORC1/CASA targets VP40 into autolysosomes. (A and B) LC3-eGFP-mCherry is a dual-fluorescing autophagy monitoring reporter that will be predominantly yellow (autophagosomes; APs) under conditions of neutral pH due to low autophagic flux (control) or an autophagy blockade (CQ treatment), whereas LC3-eGFP-mCherry will be predominantly red (autolysosomes; ALs) under conditions of acidic pH due to high autophagic flux (rapamycin treatment). (C and D) eVP40 VLP budding assay and VLP quantification (bar graph) in Huh7 cells treated as indicated. Statistical significance was analyzed by one-way ANOVA. ns: not significant, ****P < 0.0001. (E–G) Confocal images of Huh7 cells co-expressing GFP-eVP40 (green) and LC3-mCherry (red) and either mock-treated (control), treated with rapamycin (200 nM) alone, or treated with rapamycin (200 nM) plus CQ (10 µM). Cells were labeled with lysotracker-deep red (magenta). The Inset panels highlight the colocalization of eVP40 aggregates with LC3-marked lysotracker-labeled puncta under treatment with autophagy inducer, rapamycin. (Scale bar, 10 µm in main panels and 2 µm in Inset panels.) (H and I) Representative CHX (10 µg/mL) chase assay and cellular eVP40 quantification. Statistical significance was analyzed by one-way ANOVA. ns: not significant, ***P < 0.001, ****P < 0.0001.
We next asked whether the formation of ALs was necessary for rapamycin-mediated inhibition of VLP budding. To test this, Huh7 cells transfected with eVP40 were either mock-treated, or treated with CQ alone, rapamycin alone, or CQ + rapamycin, and the levels of eVP40 were quantified in cell extracts and VLPs. We found that while rapamycin alone consistently blocked VLP egress, the addition of CQ restored VLP budding to levels comparable to controls (Fig. 7 C and D). To further illustrate the mechanistic role of ALs in rapamycin-mediated inhibition of budding, Huh7 cells co-expressing GFP-eVP40 and LC3-mCherry were either mock-treated, treated with rapamycin alone, or with rapamycin + CQ. Cells were then labeled with lysotracker and observed using live cell confocal microscopy. As before, we observed a typical pattern of eVP40 localization, and the lack of ALs in control cells (Fig. 7E). Treatment with rapamycin sequestered eVP40 into aggregates colocalizing with LC3 puncta, and interestingly, a portion of these puncta was labeled with lysotracker (Fig. 7F), implying that rapamycin caused sequestration of eVP40 in ALs (Movies S1 and S2). In contrast, we observed a significant decrease in the lysotracker signal in the presence of CQ, and notably, eVP40 re-localized to the PM, and exhibited robust VLP formation and egress at the cell periphery (Fig. 7G).
To assess autophagic–lysosomal degradation of VP40, we used a cycloheximide (CHX) chase assay (38) to quantify cellular VP40 levels under different treatment conditions (Fig. 7 H and I). Huh7 cells were transfected with eVP40 for 20 h and then mock-treated (DMSO), or treated with CHX alone, CHX + rapamycin, or CHX + rapamycin + CQ for 3 or 6 h. We observed that CHX treatment for 3 and 6 h blocked de novo expression of exogenous eVP40 (Fig. 7H, compare lanes 1 and 2, and 5 and 6). Notably, degradation of eVP40 was significantly enhanced in cells co-treated with CHX and rapamycin, but not in cells co-treated with CHX + rapamycin + CQ (Fig. 7H, lanes 2 to 4 and lanes 6 to 8; Fig. 7I). Importantly, endogenous p62, an indicator of autophagic flux, was also degraded similarly with eVP40 under the same conditions, and thus served as a key internal control (Fig. 7H, lanes 3 and 7). In sum, these results support our conclusion that eVP40 is subjected to autophagic–lysosomal degradation (SI Appendix, Fig. S8 and Movie S3) as a client of CASA in cells treated with rapamycin, and that the formation of autolysosomes contributes to both the sequestration and degradation of eVP40.
Discussion
In the ongoing arms race between virus and host, each side is continuously adapting and developing new strategies to ensure survival. Autophagy is an evolutionarily conserved cell survival mechanism that is crucial for maintaining cell and tissue homeostasis under conditions of stress or tension, as well as being a strategy to resist invasion by pathogens (22, 24, 25). Autophagy itself is regulated by mTORC1; a protein complex that regulates/activates cellular protein synthesis required for cell growth and proliferation (23, 33). Here, we reveal how the mTORC1/autophagy axis functions to regulate and restrict the egress of EBOV particles by targeting the main driver of filovirus budding—the VP40 matrix protein.
We previously identified BAG3 as novel host interactor of filovirus VP40 that negatively regulated the egress of VP40 VLPs in a WW-domain/PPxY-dependent manner (9). Notably, co-chaperone protein BAG3 is an essential and central regulator for the initiation of the CASA pathway; a stress-/tension-induced pathway that regulates protein turnover and tissue homeostasis that are critical for cell adhesion, migration, and proliferation (15–17, 19–21, 39). Indeed, we provide evidence here that BAG3 recognizes VP40 as a client and directs VP40 into the CASA pathway to inhibit egress, likely as part of an innate host defense strategy triggered by virus infection. It remains to be determined whether post-translational modifications of VP40 (e.g., ubiquitination) play a role in mediating its recruitment by BAG3/HSP70. We demonstrated that BAG3 directs the formation of a tripartite complex composed of VP40/BAG3/HSP70. Importantly, we show that the formation of the HB complex is essential for BAG3’s inhibitory effect on the egress, since disruption of the HB complex pharmacologically using compound YM-1 released VP40 from the HB complex and rescued VP40 VLP budding. Thus, we concluded that the early stage of CASA (i.e., the physical formation of the client complex) is a prerequisite for sequestration of VP40 away from the site of budding at the PM and inhibition of VLP egress. We further demonstrated that pharmacological disruption of the more downstream stage of CASA (i.e., client aggregation and autophagic–lysosomal degradation) (15, 21) by CQ resulted in the rescue of VP40 VLP budding. Interestingly, CQ completely eliminated the budding inhibition induced by ectopic expression of BAG3, leading to relocalization of VP40 back to the PM. Additionally, CQ did not affect the formation of the VP40/BAG3/HSP70 tripartite complex, but rather we observed BAG3 co-localizing with VP40 at the PM following CQ treatment (Fig. 3 F and G), implying that the initial sequestration of VP40 away from PM by BAG3 was dependent on a functional CASA pathway.
To further illustrate how the CASA pathway restricts VP40 VLP budding, we visualized VP40, BAG3, and LC3-marked autophagosomes by confocal microscopy and found that ectopic expression of BAG3-induced VP40 to colocalize and aggregate with BAG3 in the perinuclear region. Notably, most of these aggregates were sequestered within vesicles determined to be autophagosomes as judged by the presence of three markers of autophagic vesicles (LC3, p62, and ATG5). If selective macroautophagy does indeed contribute to sequestration of VP40, then we reasoned that induction of cellular autophagy should facilitate targeting of VP40 by the CASA pathway. While mTORC1 regulates cell growth, proliferation, and protein synthesis, it is also the gateway modulator of autophagy, since suppression of mTORC1 is required for autophagy induction (23, 33). Indeed, activation of autophagy by mTORC1 inhibitor rapamycin-induced sequestration of VP40 in aggresomes and autophagosomes, implying that pharmacological induction of autophagy flux facilitated targeting of VP40 as a client of endogenous BAG3. Given that rapamycin-induced sequestration of VP40 into autophagosomes, we reasoned that rapamycin would also block VP40 VLP budding as well. Indeed, we showed that mTOR inhibitors rapamycin and BEZ235 not only potently blocked VP40 VLP egress, but also blocked the egress of infectious EBOV. Importantly, the mechanism by which the mTOR inhibitors blocked VP40-mediated egress requires expression of endogenous BAG3, since the inhibitory effect was not evident in BAG3 KO cells (Fig. 5 E–H).
Since mTORC1 activates protein synthesis for cell growth and proliferation, and its suppression induces autophagy, we postulated that EBOV may activate mTORC1 signaling to stimulate a cellular environment favorable for productive infection. Notably, the process of macropinocytosis by which EBOV enters cells, is known to activate mTORC1 (40–42). Interestingly, we found that EBOV GP expressed on the surface of VLPs and an infectious VSV recombinant-activated mTORC1 signaling. EBOV has been reported to induce the PI3K/AKT pathway for efficient entry (43, 44). As a downstream effector of the PI3K/AKT pathway (45, 46), the activation of mTORC1 by eGP will promote protein synthesis and repress cellular autophagy, both of which would be beneficial for post-entry stages of the filovirus lifecycle. Indeed, treatment with rapamycin or BEZ235 potently blocked release of VLPs and infectious EBOV, implying that mTORC1 signaling is required for productive and efficient EBOV infection. It should be noted that BEZ235 is a dual inhibitor targeting PI3K and mTORC1/C2 (46), while rapamycin is an acute inhibitor of mTORC1 (33, 35), and therefore, our results imply that mTORC1 is likely a central modulator of the EBOV life cycle. Similarly, HIV-1 envelope activates mTORC1 to shutdown autophagy in DCs, and thus impairs the innate and adaptive immune responses (47, 48). Notably, EBOV and MARV also interfere with DCs by inhibiting their ability to mature and stimulate T cells (49, 50), which may occur in part via mTORC1 activation by eGP.
We observed that rapamycin treatment resulted in an increase in autolysosome formation, sequestration of a portion of VP40 into aggregates and autolysosomes, and degradation of VP40 as determined by chycloheximide chase assays. Importantly, the addition of CQ resulted in an autophagy blockage, reversed sequestration of VP40, and prevented degradation of VP40 by autophagic flux leading to rescue of VLP egress. These findings confirm that eVP40 is a client of CASA and the host mTORC1/CASA axis plays a key role in regulating eVP40 localization, trafficking, and egress. It is tempting to speculate that the role for BAG3 and CASA in restricting egress of EBOV may be of biological significance in cells that are either not productively infected by EBOV, or in cells that become persistently infected. For example, EBOV infection of CD4+ T cells, which express high levels of BAG3, often results in an abortive infection and leads to ER stress-induced autophagy (28). Lastly, pharmacological intervention targeting the mTORC1/CASA axis may represent a promising antiviral strategy. Indeed, rapamycin and BEZ235 have been reported to function as potential antivirals against HIV-1 (47, 51, 52), middle east respiratory syndrome coronavirus (MERS-CoV) (53), influenza A virus (IV/IAV) (54, 55), LCMV (56), and Rotavirus (57). It would be interesting to determine whether the mTOR inhibitors could be combined with host-oriented PPxY budding inhibitors (58) as a potential combination therapy.
Materials and Methods
Plasmids, Antibodies, and Reagents.
The pcDNA6 myc-His-BAG3 plasmid was kindly provided by K. Khalili (Temple University). Plasmid pDEST-mCherry-BAG3 was kindly provided by E. Sjøttem (University of Tromsø), pcDNA3.1-mCherry-hLC3B was a gift from David Rubinsztein (Addgene plasmid # 40827), pEX-CFP-hLC3B was a gift from Isei Tanida (Addgene plasmid # 24985), pDEST-CMV mCherry-GFP-LC3B was a gift from Robin Ketteler (Addgene plasmid # 123230).
Cells.
HEK293T, Huh7, and Vero cells were maintained in Dulbecco's Modified Eagle Medium (DMEM) (Corning), HAP1 WT cells (kindly provided by K. Chandran, Albert Einstein College of Medicine) and HAP1-BAG3 KO cells (Horizon Discovery) were maintained in Iscove's Modified Dulbecco's Medium (IMDM) (Corning), supplemented with 10% FBS (Gibco), penicillin (100 U/mL)/streptomycin (100 μg/mL) (Invitrogen), and the cells were maintained at 37 °C in a humidified 5% CO2 incubator. The HAP1-BAG3 KO cells contain an 11-base pair deletion in the coding exon of BAG3.
Viruses.
All experiments with infectious EBOV were performed in the biosafety level 4 (BSL-4) laboratory at the Texas Biomedical Research Institute, San Antonio, TX. The recombinant EBOV variant Mayinga expressing GFP (EBOV-GFP) (NCBI accession number KF_990213) was used.
Immunoprecipitation, siRNA, VLP Budding, Cell Fractionation, Pharmacological, Live Cell Imaging, and Virus Infection assays are described in more detail in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Rapamycin induces autophagic sequestration and degradation of eVP40. eVP40 VLP egress in untreated Huh7 cells (control).
eVP40 forms aggregates and becomes sequestered in LC3 marked autophagic vesicles in rapamycin treated Huh7 cells.
eVP40 aggregates are degraded within LC3 marked autophagic vesicles in rapamycin treated Huh7 cells. Huh7 cells exogenous expressing GFP-eVP40 and hLC3-Cherry were mock treated or treated with rapamycin (200nM) for 14 hours and imaged via confocal microscopy. A total of 100 frames were collected with a time interval of 6s for 10min. Still images are shown in Fig. S8C. The video is displayed as 10 frames per second. Scale bars, 5 μm.
Acknowledgments
We thank S. Becker, E. Sjøttem, J. Lavoie, and K. Khalili for kindly providing reagents. We thank members of the Harty lab for fruitful discussions and suggestions on this work, and the Penn Vet Imaging Core for confocal microscopy. Funding was provided in part by NIH grants AI138052, AI139392, AI153815, and EY031465 to R.N.H.; and AI154336 and AI151717 and to O.S. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Author contributions
J.L., O.S., and R.N.H. designed research; J.L., M.A.D., and O.S. performed research; J.L., O.S., and R.N.H. analyzed data; and J.L. and R.N.H. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. C.F.B. is a guest editor invited by the Editorial Board.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Mahanty S., Bray M., Pathogenesis of filoviral haemorrhagic fevers. Lancet. Infect. Dis. 4, 487–498 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Hoenen T., Groseth A., Feldmann H., Therapeutic strategies to target the Ebola virus life cycle. Nat. Rev. Microbiol. 17, 593–606 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Noda T., et al. , Assembly and budding of Ebolavirus. PLoS Pathog. 2, e99 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartlieb B., Weissenhorn W., Filovirus assembly and budding. Virology 344, 64–70 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Harty R. N., Brown M. E., Wang G., Huibregtse J., Hayes F. P., A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: Implications for filovirus budding. Proc. Natl. Acad. Sci. U.S.A. 97, 13871–13876 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Licata J. M., et al. , Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: Involvement of host proteins TSG101 and VPS-4. J. Virol. 77, 1812–1819 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han Z., et al. , ITCH E3 ubiquitin ligase interacts with Ebola virus VP40 to regulate budding. J. Virol. 90, 9163–9171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han Z., et al. , Ubiquitin ligase WWP1 interacts with Ebola virus VP40 to regulate egress. J. Virol. 91, e00812-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang J., et al. , Chaperone-mediated autophagy protein BAG3 negatively regulates Ebola and Marburg VP40-mediated egress. PLoS Pathog. 13, e1006132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han Z., et al. , Angiomotin regulates budding and spread of Ebola virus. J. Biol. Chem. 295, 8596–8601 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han Z., et al. , Modular mimicry and engagement of the Hippo pathway by Marburg virus VP40: Implications for filovirus biology and budding. PLoS Pathog. 16, e1008231 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang J., et al. , Angiomotin counteracts the negative regulatory effect of host WWOX on Viral PPxY-mediated egress. J. Virol. 95, e00121–21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang J., Ruthel G., Freedman B. D., Harty R. N., WWOX-mediated degradation of AMOTp130 negatively affects egress of filovirus VP40 virus-like particles. J. Virol. 96, e0202621 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shepley-McTaggart A., Fan H., Sudol M., Harty R. N., Viruses go modular. J. Biol. Chem. 295, 4604–4616 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamerdinger M., Kaya A. M., Wolfrum U., Clement A. M., Behl C., BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 12, 149–156 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behl C., BAG3 and friends: Co-chaperones in selective autophagy during aging and disease. Autophagy 7, 795–798 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Rosati A., Graziano V., De Laurenzi V., Pascale M., Turco M. C., BAG3: A multifaceted protein that regulates major cell pathways. Cell Death Dis. 2, e141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minoia M., et al. , BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta: Implications for a proteasome-to-autophagy switch. Autophagy 10, 1603–1621 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Behl C., Breaking BAG: The co-chaperone BAG3 in health and disease. Trends Pharmacol. Sci. 37, 672–688 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Sturner E., Behl C., The role of the multifunctional BAG3 protein in cellular protein quality control and in disease. Front. Mol. Neurosci. 10, 177 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klimek C., Kathage B., Wördehoff J., Höhfeld J., BAG3-mediated proteostasis at a glance. J. Cell Sci. 130, 2781–2788 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Lennemann N. J., Coyne C. B., Catch me if you can: The link between autophagy and viruses. PLoS Pathog. 11, e1004685 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galluzzi L., et al. , Molecular definitions of autophagy and related processes. EMBO J. 36, 1811–1836 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi Y., Bowman J. W., Jung J. U., Autophagy during viral infection–A double-edged sword. Nat. Rev. Microbiol. 16, 341–354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmad L., Mostowy S., Sancho-Shimizu V., Autophagy-virus interplay: From cell biology to human disease. Front. Cell Dev. Biol. 6, 155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiramel A. I., Dougherty J. D., Nair V., Robertson S. J., Best S. M., FAM134B, the selective autophagy receptor for endoplasmic reticulum turnover, inhibits replication of Ebola virus strains Makona and Mayinga. J. Infect. Dis. 214, S319–S325 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shtanko O., Reyes A. N., Jackson W. T., Davey R. A., Autophagy-associated proteins control Ebola virus internalization into host cells. J. Infect. Dis. 218, S346–S354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Younan P., et al. , Ebola virus-mediated T-lymphocyte depletion is the result of an abortive infection. PLoS Pathog. 15, e1008068 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang B., et al. , Protein disulfide isomerases (PDIs) negatively regulate Ebolavirus structural glycoprotein expression in the endoplasmic reticulum (ER) via the autophagy-lysosomal pathway. Autophagy 18, 2350–2367 (2022), 10.1080/15548627.2022.2031381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meriin A. B., et al. , Hsp70-Bag3 complex is a hub for proteotoxicity-induced signaling that controls protein aggregation. Proc. Natl. Acad. Sci. U.S.A. 115, E7043–E7052 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colvin T. A., et al. , Hsp70-Bag3 interactions regulate cancer-related signaling networks. Cancer Res. 74, 4731–4740 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mauthe M., et al. , Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14, 1435–1455 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y. C., Guan K. L., mTOR: A pharmacologic target for autophagy regulation. J. Clin. Invest. 125, 25–32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiang G. G., Abraham R. T., Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J. Biol. Chem. 280, 25485–25490 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Hoeffer C. A., Klann E., mTOR signaling: At the crossroads of plasticity, memory and disease. Trends Neurosci. 33, 67–75 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nanbo A., Watanabe S., Halfmann P., Kawaoka Y., The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells. Sci. Rep. 3, 1206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizushima N., Yoshimori T., Levine B., Methods in mammalian autophagy research. Cell 140, 313–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sha Z., Zhao J., Goldberg A. L., Measuring the overall rate of protein breakdown in cells and the contributions of the ubiquitin-proteasome and autophagy-lysosomal pathways. Methods Mol. Biol. 1844, 261–276 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulbricht A., et al. , Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr. Biol. 23, 430–435 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Yoshida S., Pacitto R., Yao Y., Inoki K., Swanson J. A., Growth factor signaling to mTORC1 by amino acid-laden macropinosomes. J. Cell Biol. 211, 159–172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pacitto R., Gaeta I., Swanson J. A., Yoshida S., CXCL12-induced macropinocytosis modulates two distinct pathways to activate mTORC1 in macrophages. J. Leukocyte Biol. 101, 683–692 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida S., Pacitto R., Inoki K., Swanson J., Macropinocytosis, mTORC1 and cellular growth control. Cell Mol. Life Sci. 75, 1227–1239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saeed M. F., Kolokoltsov A. A., Freiberg A. N., Holbrook M. R., Davey R. A., Phosphoinositide-3 kinase-Akt pathway controls cellular entry of Ebola virus. PLoS Pathog. 4, e1000141 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kindrachuk J., et al. , Ebola virus modulates transforming growth factor beta signaling and cellular markers of mesenchyme-like transition in hepatocytes. J. Virol. 88, 9877–9892 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karar J., Maity A., PI3K/AKT/mTOR pathway in angiogenesis. Front. Mol. Neurosci. 4, 51 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Porta C., Paglino C., Mosca A., Targeting PI3K/Akt/mTOR signaling in cancer. Front. Oncol. 4, 64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanchet F. P., et al. , Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity 32, 654–669 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghislat G., Lawrence T., Autophagy in dendritic cells. Cell Mol. Immunol. 15, 944–952 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bosio C. M., et al. , Ebola and Marburg viruses replicate in monocyte-derived dendritic cells without inducing the production of cytokines and full maturation. J. Infect. Dis. 188, 1630–1638 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Mohamadzadeh M., Chen L., Schmaljohn A. L., How Ebola and Marburg viruses battle the immune system. Nat. Rev. Immunol. 7, 556–567 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Besnard E., et al. , The mTOR complex controls HIV latency. Cell Host Microbe 20, 785–797 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campbell G. R., et al. , Induction of autophagy by PI3K/MTOR and PI3K/MTOR/BRD4 inhibitors suppresses HIV-1 replication. J. Biol. Chem. 293, 5808–5820 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kindrachuk J., et al. , Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob. Agents Chemother. 59, 1088–1099 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smallwood H. S., et al. , Targeting metabolic reprogramming by influenza infection for therapeutic intervention. Cell Rep. 19, 1640–1653 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuss-Duerkop S. K., et al. , Influenza virus differentially activates mTORC1 and mTORC2 signaling to maximize late stage replication. PLoS Pathog. 13, e1006635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Urata S., Ngo N., de la Torre J. C., The PI3K/Akt pathway contributes to arenavirus budding. J. Virol. 86, 4578–4585 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yin Y., et al. , PI3K-Akt-mTOR axis sustains rotavirus infection via the 4E-BP1 mediated autophagy pathway and represents an antiviral target. Virulence 9, 83–98 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han Z., et al. , Compound FC-10696 inhibits egress of Marburg Virus. Antimicrob. Agents Chemother. 65, e0008621 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Rapamycin induces autophagic sequestration and degradation of eVP40. eVP40 VLP egress in untreated Huh7 cells (control).
eVP40 forms aggregates and becomes sequestered in LC3 marked autophagic vesicles in rapamycin treated Huh7 cells.
eVP40 aggregates are degraded within LC3 marked autophagic vesicles in rapamycin treated Huh7 cells. Huh7 cells exogenous expressing GFP-eVP40 and hLC3-Cherry were mock treated or treated with rapamycin (200nM) for 14 hours and imaged via confocal microscopy. A total of 100 frames were collected with a time interval of 6s for 10min. Still images are shown in Fig. S8C. The video is displayed as 10 frames per second. Scale bars, 5 μm.
Data Availability Statement
All study data are included in the article and/or SI Appendix.