Significance
Genetic and phenotypic changes that occur during infection may alter a strain’s propensity for virulence; however, our current understanding of how passage through a mammalian host alters Cryptococcus neoformans remains limited. In this study, we leverage a clinical strain and the suspected avian-source strain to better understand the changes that manifest during human passage. In addition, we characterize the genetic and phenotypic changes arising from passage of the source strain through mice. Notably, we find similar genetic changes in the human and mouse-passaged isolates and identify organ-specific differences arising from murine passage. This unique strain set allows us to explore the genetic and phenotypic plasticity of Cryptococcus neoformans in response to selective pressures across host environments.
Keywords: Cryptococcus, fungus, genomics
Abstract
A pet cockatoo was the suspected source of Cryptococcus neoformans recovered from an immunocompromised patient with cryptococcosis based on molecular analyses available in 2000. Here, we report whole genome sequence analysis of the clinical and cockatoo strains. Both are closely related MATα strains belonging to the VNII lineage, confirming that the human infection likely originated from pet bird exposure. The two strains differ by 61 single nucleotide polymorphisms, including eight nonsynonymous changes involving seven genes. To ascertain whether changes in these genes are selected for during mammalian infection, we passaged the cockatoo strain in mice. Remarkably, isolates obtained from mouse tissue possess a frameshift mutation in one of the seven genes altered in the human sample (LQVO5_000317), a gene predicted to encode an SWI–SNF chromatin-remodeling complex protein. In addition, both cockatoo and patient strains as well as mouse-passaged isolates obtained from brain tissue had a premature stop codon in a homologue of ZFC3 (LQVO5_004463), a predicted single-zinc finger containing protein, which is associated with larger capsules when deleted and reverted to a full-length protein in the mouse-passaged isolates obtained from lung tissue. The patient strain and mouse-passaged isolates show variability in virulence factors, with differences in capsule size, melanization, rates of nonlytic expulsion from macrophages, and amoeba predation resistance. Our results establish that environmental strains undergo genomic and phenotypic changes during mammalian passage, suggesting that animal virulence can be a mechanism for genetic change and that the genomes of clinical isolates may provide a readout of mutations acquired during infection.
Cryptococcus neoformans is a human pathogenic fungus that is a major cause of life-threatening meningoencephalitis (1). Cryptococcosis is more common in patients with impaired immune systems, although occasional disease occurs in individuals with no apparent immune deficits. Cryptococcus neoformans infection is first thought to be acquired in childhood (2) and is either cleared or can become latent to reactivate if impaired immunity occurs later in life (3). However, disease primarily follows exposure to contaminated environmental sources in adults, but the ubiquity and high genetic diversity of this fungus complicate the identification of point sources. Restriction enzyme polymorphism analysis of patient and environmental samples in New York City revealed that some clinical isolates shared the same restriction patterns and were thus indistinguishable from local infection sources (4); however, such analysis lacked the precision to reveal point sources, particularly given that the disease often develops slowly and can be the result of latent, distantly acquired infection (3). A recent investigation of a cryptococcosis outbreak in a Scottish hospital revealed how difficult it is to make associations between clinical and geographically and temporally matched environmental samples (5).
In 2000, we reported on the case of an immunosuppressed patient with cryptococcosis who had a pet cockatoo (6). Cryptococcus was also recovered from the cockatoo guano, which was not unexpected as bird guano is a common environmental reservoir for Cryptococcus neoformans (7, 8). Both the patient and bird guano strains were indistinguishable using molecular tools available at the time, resulting in the first example of cryptococcal infection traced from a point source (6). In subsequent years, additional cases of Cryptococcus neoformans infections potentially linked to pet birds were reported (9, 10). Since the original report, genome sequencing has become routine, and there is now a wealth of information available on Cryptococcus neoformans genomes from varied source isolates (11). In this study, we compared the genome sequences of the patient and cockatoo strains and passaged the cockatoo strain in mice to identify genetic and phenotypic changes resulting from mammalian infection. The results validate the earlier conclusion that the clinical and cockatoo strain sequences were closely related and that the cockatoo was the likely source of infection. We also find evidence of similar genome evolution in mouse-passaged and patient strains. These results implicate genomic loci in virulence, identify functionally similar genomic changes arising from mammalian infection, and characterize phenotypic differences between mouse-passaged isolates that appear to be organ specific.
Results
Genomic Analysis of Patient and Cockatoo Strains.
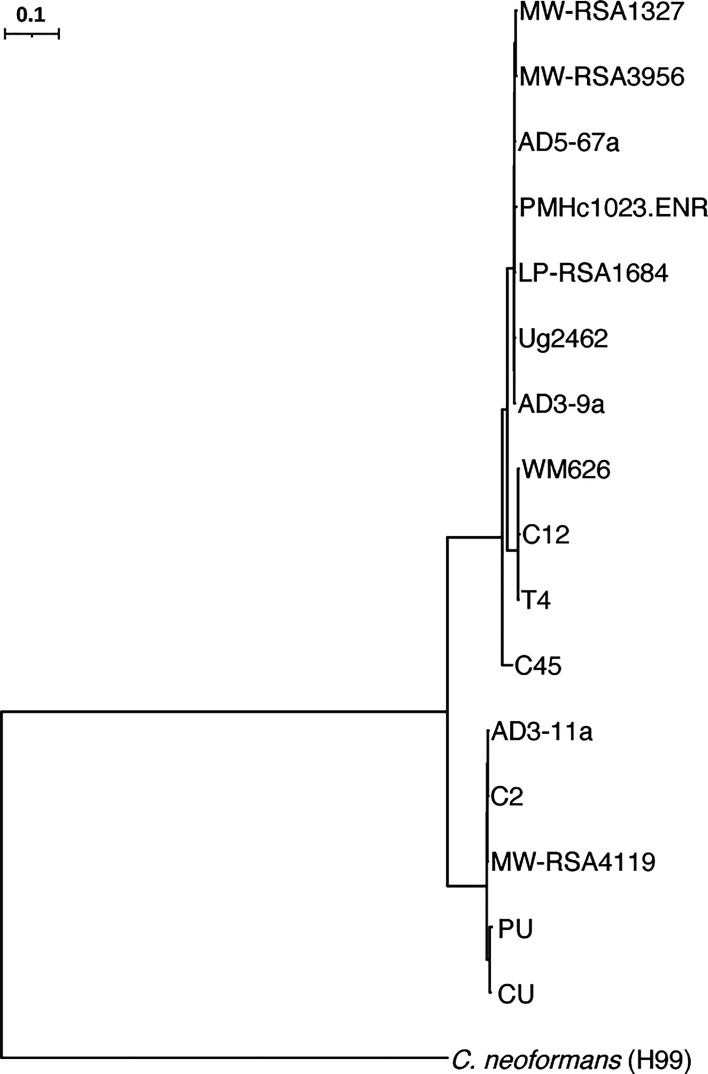
To determine the relatedness of the previously reported strains that were isolated from patient cerebrospinal fluid (PU, patient unpassaged) and guano of the patient’s pet cockatoo (CU, cockatoo unpassaged) (6), we aligned Illumina reads for both strains to the Cryptococcus neoformans H99 reference genome (12), and variants were called to identify single nucleotide polymorphisms (SNPs) and insertion/deletion events (indels). Based on a phylogenetic analysis of these samples and a subset of 238 additional whole genome sequences chosen to represent the lineages VNIa, VNIb, VNIc, VNBI, VNBII, and VNII, previously described by Desjardins et al. (13), we assigned the PU and CU strains to the globally detected but least common VNII lineage confirming their close relationship and supporting a role of the CU strain in the patient’s cryptococcosis (Figs. 1 and 2 and SI Appendix, Fig. S1). With these data, we also determined that both strains possess the MATα mating type. Utilizing Oxford Nanopore Technologies (ONT) read data, we then generated genome assemblies for both the PU and CU strains, consisting of complete telomere-to-telomere sequences for each chromosome, polished with Illumina reads. To identify variants, we compared the two assemblies with nucmer (MUMmer). We identified seven genes with nonsynonymous variants between the two assemblies, all impacting genes with homologues in the Cryptococcus neoformans var. grubii (H99) genome (Table 1). Of these genes, 2 (LQVO5_002184 and LQVO5_000317) had Cryptococcus neoformans H99 homologues (CNAG_06273 and CNAG_00342) that are repressed during titan cell formation and murine cryptococcal infection, respectively (14, 15). For both strains, we were able to generate full-length chromosomal assemblies consisting of 14 chromosomes, with rRNA content residing on chromosome 4 and chromosome 15 representing the mitochondrion, with equivalent gene sets and high levels of identity between the two assemblies (99.99%), indicating high levels of relatedness (SI Appendix, Fig. S2A). We did not detect any structural differences between the CU and PU assemblies or any chromosomal duplications or copy number variations (CNVs) between the two. When these assemblies were compared with the Cryptococcus neoformans H99 reference assembly, they shared sequence identities of 97.7% to the H99 reference (SI Appendix, Fig. S2B).
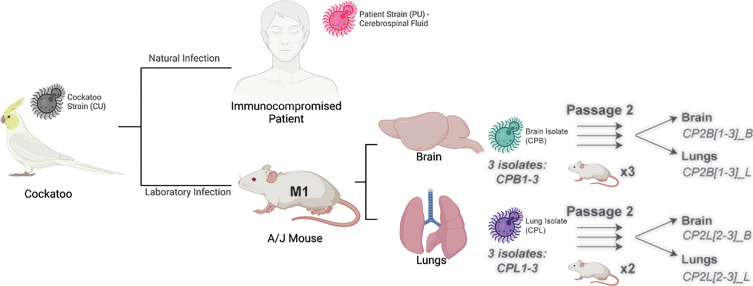
Fig. 1.
Passaging scheme and relationship between the Cryptococcus neoformans strains and isolates. The cockatoo and patient strains had been kept frozen since the prior study was completed in 2000 (6). These are distinguished in that they were not passaged and are labeled cockatoo strain (CU) and patient strain (PU), where the U stands for unpassaged. The mouse-passaged isolates come from a laboratory infection and are labeled CPB and CPL for cockatoo-passaged brain and cockatoo-passaged lungs, respectively, to denote the mouse organ from where they were recovered. Cryptococcus neoformans recovered from patient and cockatoo are referred to as strains to denote different origins, while those recovered from mouse tissues are referred to as isolates. Three isolates from passage 1 were then passaged a second time into six separate mice, and a single second passage isolate was harvested from the brain and lungs of each second passage mouse. The mouse reinfected with CPL1 did not survive the anesthesia during infection.
Fig. 2.
VNII phylogeny. SNP-based maximum likelihood phylogeny for PU, CU, and VNII strains rooted to the Cryptococcus neoformans reference strain H99. Illumina reads for the VNII strains shown were aligned to the Cryptococcus neoformans H99 reference genome to identify SNPs across the genome, which were used to infer a phylogeny with RAxML.
Table 1.
Genes with nonsynonymous variants between patient (PU) and cockatoo (CU) strains
| CU (VNII) gene | H99 (VNI) homologue | Variant (CU > PU) | Function |
|---|---|---|---|
| LQVO5_000317 | CNAG_00342 | 270C > G | KOG: SWI–SNF chromatin-remodeling complex protein |
| LQV05_001277 | CNAG_05000 | 1835T > C | EC: exo-alpha-sialidase |
| LQVO5_004692 | CNAG_05713 | 194C > G | RNA polymerase II elongator complex protein 1 |
| LQVO5_005253 | CNAG_02201 | 448C > G | KOG: serine/threonine protein kinase |
| LQVO5_005443 | CNAG_03102 | 122C > T | Hypothetical protein |
| LQVO5_006755 | CNAG_01999 | 34C > T | KOG: alpha-SNAP protein, membrane fusion |
| LQVO5_002184 | CNAG_06273 | 217C > A 223C > T | Pfam: ribosome biogenesis regulatory protein (RRS1) |
Mouse Passage of Cockatoo (CU) Strain.
The identification of genomic changes when comparing the CU and PU strains suggested that the PU differences may have occurred during human infection. Consequently, we wondered whether passage of the CU strain in mice would result in similar genomic changes. A female A/J mouse was infected intravenously with the CU strain and observed for signs of illness, but none were apparent. Hence, at day 43, the mouse was euthanized, the brain and lungs were harvested and homogenized, and the suspensions plated, which yielded 816 and 78 colony-forming unit (CFU)/mg in the brain and lungs, respectively. Three individual colonies were isolated from the brain (CPB1-3, cockatoo-passaged brain) and lungs (CPL1-3, cockatoo-passaged lungs) for sequencing and phenotypic analysis (Fig. 1). These isolates were then each submitted to a second round of passage, and isolates were again recovered from the brain and lungs. We noted increased fungal burdens for the CPB1-3 isolates but decreased burdens for the CPL1-3 isolates. The average fungal burdens of the twice-passaged isolates were 6,468, 1,616, 6, and 1 CFU/mg for CP2B_L, CP2B_B, CP2L_L, and CP2L_B, respectively (SI Appendix, Table S1). These isolates were sequenced and studied for phenotypic characteristics.
Genomic Analysis of Mouse-Passaged Isolates.
To identify variants arising from passage of the CU strain and the CPB or CPL isolates in mice, we aligned Illumina data generated for all mouse-evolved isolates (first and second passages), to our CU genome assembly. Samples aligned with an average coverage of 775× across the CU reference. After the initial round of passage, we found that a frameshift variant in one of the seven genes altered in the patient strain, LQVO5_000317 (Table 1). The frameshift in LQVO5_000317 was present in all mouse-evolved isolates from both brain and lung tissues (CPB1-3 and CPL1-3) (Table 2). The commonality of this variant across brain and lung isolates suggests that the mutation was acquired at a common site prior to dissemination. This gene is a homologue of CNAG_00342 in the Cryptococcus neoformans H99 (VNI) genome, which is predicted via eukaryotic orthologous groupings to function as an SWI–SNF chromatin-remodeling complex protein (KOG2510). This gene is down-regulated in a murine lung infection model (15), consistent with loss-of-function mutations appearing tolerated during mammalian infection.
Table 2.
Variants in patient strain (PU) and mouse-evolved isolates (CPB and CPL)
| VNII gene | CNAG | Variant type | CNAG function | Desjardins collection (13) | |||
|---|---|---|---|---|---|---|---|
| Sample | Lineage* | Variant type | Source | ||||
| LQVO5_000317 | 00342 | Frameshift (270delC) | KOG: SWI–SNF chromatin-remodeling complex protein | 3 | VNI (1), VNII (1), and VNB (1) | Frameshift | Clinical |
| LQVO5_004463 | 05940 | Premature stop codon lost (639A > T) | Pfam: ZFC3, zinc finger, C2H2 type | 24 | VNI (5) and VNB (19) | Frameshift | Clinical (50%) and environmental (50%) |
*Strain frequency for each lineage is listed in parentheses.
A second variant unique to the mouse-passaged lung isolates (CPL1-3) resulted in the loss of a premature stop codon in LQVO5_004463. This premature stop codon is present in the CU, PU, and mouse-passaged brain isolates (CPB1-3) but appears to have reverted to wild type in the CPL isolates. When LQVO5_004463 is extended through the loss of this premature stop codon, the resulting gene encodes a full-length protein comparable in sequence and structure with its Cryptococcus neoformans H99 homologue, CNAG_05940 (SI Appendix, Fig. S3). CNAG_05940 is predicted to function as a zinc finger domain protein (ZFC3). Zfc3 has a single predicted zinc finger and a striking number of serine and threonine residues, totaling 21.9% of all residues present in CNAG_05940. The predicted structure of this protein includes long intrinsically disordered regions that may transition to structured regions upon binding to a substrate (SI Appendix, Fig. S3C). Deletion of CNAG_05940 in Cryptococcus neoformans results in significantly increased capsule content (16), and this gene is thought to be a target of the virulence-implicated transcription factors Gat201 and Liv3 and the capsule and titan cell regulating transcription factor Pdr802 (17, 18). The presence of variants impacting common genes in both patient and cockatoo-derived mouse isolates is consistent with the hypothesis that the patient’s Cryptococcus neoformans infection was also derived from the pet cockatoo.
To assess the frequency of loss-of-function mutations in the SWI–SNF and ZFC3 homologues among clinical and environmental Cryptococcus isolates, we looked for loss-of-function variants in 387 published isolates from both patient and environmental sources (13). We found three of the 286 clinical samples (from the lineages VNI, VNII, and VNB) with frameshift variants in CNAG_00342 (SWI–SNF) and 24 samples with frameshift variants present in CNAG_05940 (ZFC3) (Table 2). VNI and VNB isolates from both clinical and environmental sources are impacted by frameshift variants in CNAG_05940. We did not find any missense or loss/gain-of-function variants within the isolates that underwent a second round of passage when compared with their CPB and CPL progenitor isolates. Therefore, to identify large-scale genomic variation across all isolates, we looked for evidence of aneuploidy and CNV based on sequence coverage. In the CPL isolates that underwent a second round of passage, we identified a single region that was uniquely duplicated in isolates recovered from the brain and lungs (CP2L_L and CP2L_B). This region encodes two genes, LQV05_002052 and LQV05_002053, homologues of CNAG_00128 and CNAG_00127. Both are hypothetical proteins, and CNAG_00128 is predicted to be an ER–Golgi vesicle-tethering protein (KOG0946). Of note, CNAG_00128 is repressed during titan cell formation (14). We found no evidence of aneuploidy arising in response to human, bird, or murine passage.
CNAG_00342 and CNAG_05940 Expression in Published Datasets.
To further probe the role of these genes altered in the patient and mouse isolates, we analyzed eight publicly deposited RNA-seq datasets of Cryptococcus neoformans strains H99 and KN99α (Table 3). These databases were selected because they represented transcriptional studies done in conditions that resemble the expected conditions during infection. We found that both CNAG_00342 and CNAG_05940 were significantly differentially regulated (P < 0.05) under conditions related to those expected during animal passage and infection, namely 37 °C, increased CO2, in vitro infection of macrophages, or in vivo infection of rabbit cerebrospinal fluid (CSF). Both genes were most frequently up-regulated in H99 strains and down-regulated in KN99α under stress conditions, with the caveat that many of the H99 strains were exposed to ambient CO2 levels, while both KN99α strains were exposed to 5% CO2, indicating strain and condition-specific changes that highlight variability in expression.
Table 3.
Metadata from RNA-seq datasets, only including log2FC values with P < 0.05
| Reference PMID* | Strain | Conditions | Duration | CNAG_00342 log2FC | CNAG_05940 log2FC |
|---|---|---|---|---|---|
| 31666517 | H99 | 37 °C, YPD | 1 h | – | – |
| 28376087 | H99 | 37 °C, YPD | 2 h | 0.91 | – |
| 24743168 | H99 | 37 °C, YPD | Grown at 37 °C until stationary | – | – |
| 31860441_CSF | H99 | Rabbit CSF | 24 h | 2.76 | 2.50 |
| 31860441 | H99 | 37 °C, 5%, CO2, macrophage | 24 h | 6.30 | 4.40 |
| 22174677 | H99 | 37 °C, 5% CO2, DMEM | 1.5 h | – | – |
| 27094327 | KN99α | 37 °C, 5% CO2, DMEM | 1.5 h | – | −1.14 |
| 27094327_TM | KN99α | 37 °C, 5% CO2, DMEM | 1.5, 3, 8, and 24 h | 0.37 | −0.57 |
| 33688010 | KN99α | 37 °C, 5% CO2, DMEM | 24 h | −4.75 | −3.23 |
*Dataset 31860441_CSF indicates analysis of Cryptococcus neoformans samples from cerebrospinal fluid in vivo infections, opposed to cryptococcal samples incubated in the described conditions in vitro. Dataset 27094327_TM indicates analysis of an array of time points as a time course model, opposed to analysis of a single time point closest to the other datasets for consistency.
Heat Tolerance.
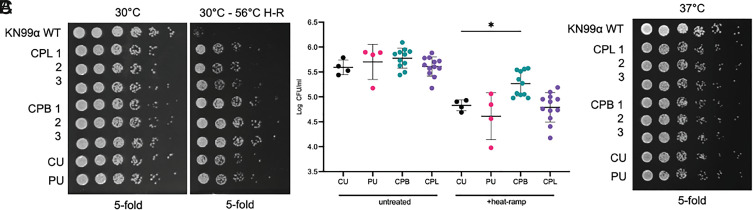
To determine whether resistance to cell death may be related to virulence in these strains, we compared the mouse-passaged isolates with the patient strain in a cell death assay that has been previously demonstrated to induce gene-dependent cell death in Saccharomyces cerevisiae and more recently in Cryptococcus neoformans (19, 20). To assess cell death susceptibility, a transient sublethal heat-ramp (not heat shock) was applied to all isolates, and survival was determined by CFUs when plated at 30 °C. Interestingly, all mouse, cockatoo, and patient strains were death resistant when compared with the laboratory strain, KN99α (Fig. 3A). Among the isolates tested, the three mouse-passaged brain isolates (CPB1-3) were significantly more death resistant than the cockatoo (CU) strain (P = 0.0239, ANOVA with Dunnett’s multiple comparisons test) (Fig. 3B). However, cell death resistance does not appear to reflect a gain of heat tolerance at body temperature as all isolates grew indistinguishably when untreated samples were plated on SAB agar and incubated at 37 °C (Fig. 3C). Although the laboratory strain KN99α may be slightly more robust than the CU, PU, CPB, and CPL isolates at both 30 °C and 37 °C, no differences in CFU number or size among the isolates were observed, suggesting that heat-ramp cell death resistance is not directly correlated with the ability to grow at high temperature.
Fig. 3.
Survival of the various Cryptococcus neoformans strains and isolates determined by application of heat-ramp cell death stimulus (H–R). The cockatoo passage brain isolates (CPB) recovered from mice infected with the cockatoo strain (CU) were more resistant to cell death when compared with the original cockatoo strain (CU). (A) Survival after a cell death stimulus [10 min linear 30 °C to 56 °C H-R] was determined by CFUs on SAB agar plates incubated for 2 d at 30 °C. (B) Quantification of survival for (A) calculated as log CFU/mL of untreated and heat-ramp–treated strains from four independent experiments. Statistical significance was determined by ordinary one-way ANOVA with Dunnett’s multiple comparisons test, *P = 0.0239. (C) Growth of the same isolates (no heat-ramp) spotted on SAB and incubated for 2 d at 37 °C (C) in two independent experiments; no differences between isolates were detected.
Growth In Vitro and Virulence Factor Characterization.
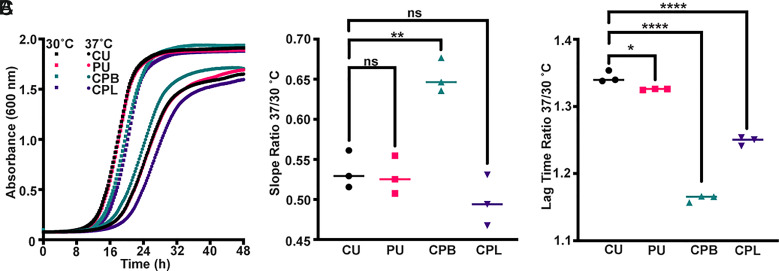
We analyzed both the CU and PU strains and the CPB and CPL isolates for growth in vitro (Fig. 4) and measured four phenotypes known to be associated with virulence factors: capsule, melanin, urease, and phospholipase (Figs. 5 and 6). All four isolates grew well in culture, with the CPB isolate recovered from brain tissue growing faster than the others (Fig. 4). All isolates exhibited activities associated with each of these virulence factors, but there were subtle differences observed.
Fig. 4.
Comparison of Cryptococcus neoformans cockatoo, human, and mouse-passaged derivatives' growth at 30 °C and 37 °C. (A) Growth curves of Cryptococcus neoformans strains (CU, PU, CPB1, and CPL1) cultured in Sabouraud dextrose broth at the indicated temperatures plotted as the mean of three biological replicates. (B) Comparison of growth rates at the two temperatures expressed as a ratio of linear phase slopes indicates a significant growth advantage at 37 °C for the mouse-passaged brain isolate compared with the other strains and isolates. (C) Both the patient strain and mouse-derived isolates show a significant decrease in the length of lag time at 37 °C compared with 30 °C. Statistical significance was determined using an ordinary one-way ANOVA (ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
Fig. 5.
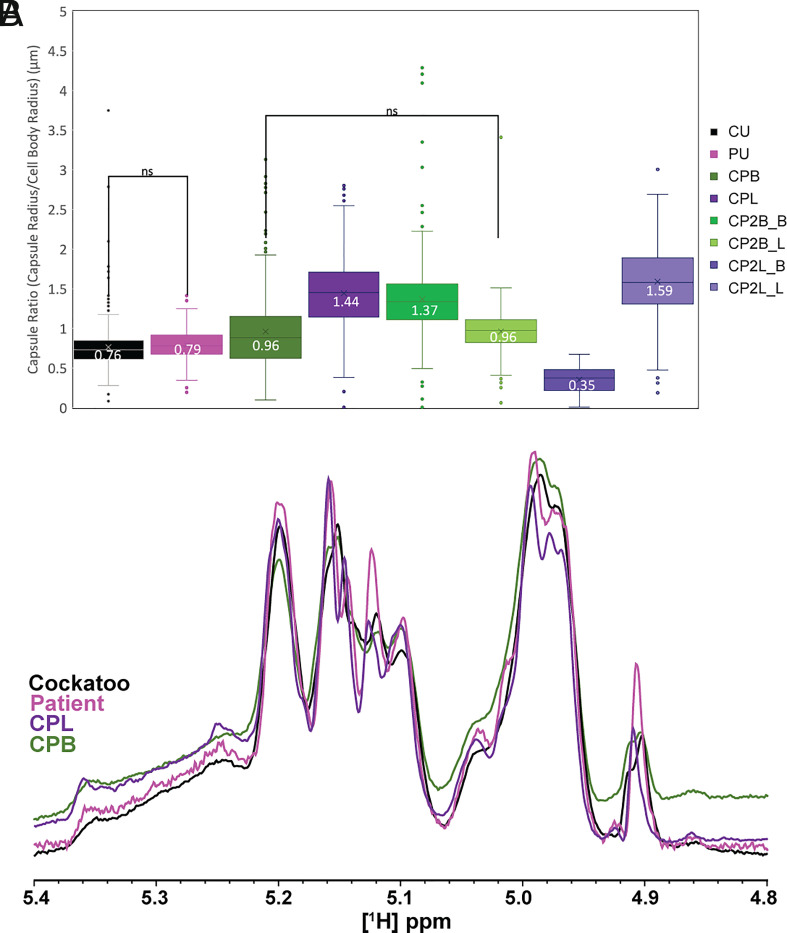
Capsular characterization of unpassaged cockatoo and patient strains and mouse-passaged isolates. (A) Capsule radius analysis of India ink–stained cryptococcal cells performed by automated quantitative capsule analysis (QCA) program of Cryptococcus neoformans cockatoo, patient, and first and second passage lung and brain isolates. Difference between groups was assessed by two-way ANOVA with unequal variance, and statistical difference between groups was determined by Tukey’s multiple comparisons test. For each sample set, >300 cells were examined. Box-and-whisker plots include the minimum value, first quartile, median, third quartile, and maximum value of each dataset with the mean value enumerated. (B) 1D [1H] NMR analysis of EPS isolated from cockatoo, patient, and first mouse-passaged lung and brain isolates of Cryptococcus neoformans. The overlaid anomeric region of each proton spectrum, which contains the structural reporter groups of the dominant polysaccharide glucuronoxylomannan (GXM), is displayed.
Fig. 6.
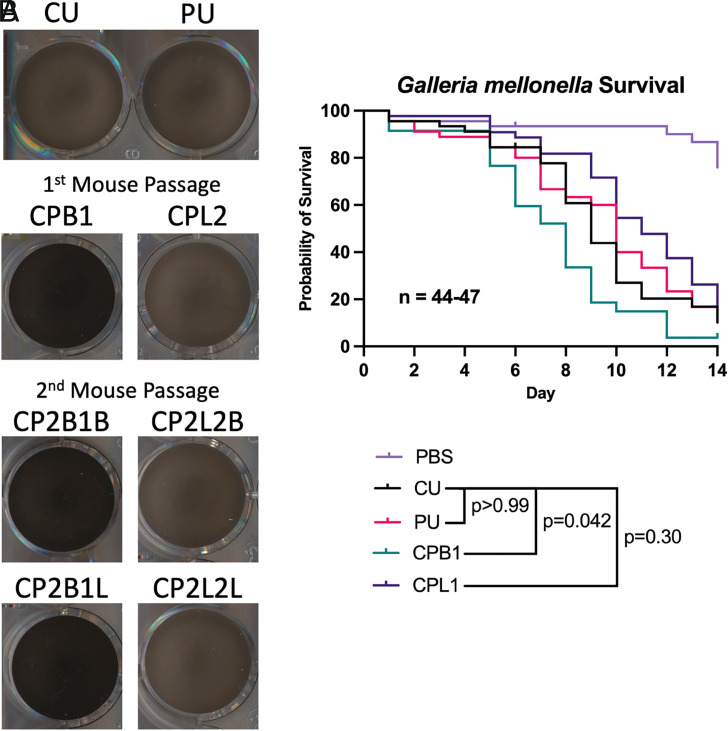
Changes in virulence factor activity associated with passaging the cockatoo strain in mice. (A) Melanization in liquid media is greatly enhanced in the CPB isolates after growth at 30 °C for 5 d. There are no major differences in melanization between the PU strain or CPL isolates, which generally show melanization consistent with the parental strain (CU). (B) There is no statistical difference in virulence between the CU and PU strains in the G. mellonella model system. However, the CPB1 isolate from the mouse had significantly enhanced virulence when compared with the parental CU strain. The log-rank Mantel–Cox test was performed using GraphPad Prism and corrected for multiple comparisons using the Bonferroni method.
The cockatoo strain (CU) and patient strain (PU) did not vary significantly with regard to capsule ratio (capsule radius/cell body radius) (P = 0.15); however, the mouse-passaged CPL and CPB isolates displayed significantly larger capsule (Tukey’s multiple comparisons test, P < 0.0001) (Fig. 5A). Interestingly, when these isolates were passaged for a second time through mice, isolates obtained from their primary organ (brain-to-brain and lung-to-lung) displayed significantly increased capsule size (CP2B1_B vs. CPB, P < 0.0001) (CP2L2_L vs. CPL, P < 0.0001), whereas isolates obtained from the alternate organ displayed unchanged (CP2B1_L vs. CPB, P = ns) or reduced capsule size (CP2L2_B vs CPL, P < 0.0001). When grown in titan cell–inducing media, brain-derived isolates displayed smaller cell body diameters than other strains and isolates (SI Appendix, Fig. S4), in line with previous findings which demonstrate that Cryptococcus neoformans adopts a smaller cell morphology in brain tissue (21, 22). We did not observe any correlation between cell body enlargement and copy number of LQV05_002052, homologue of CNAG_00128, despite the repression of CNAG_00128 during titan cell formation.
Since a prior analysis of sequential isolates recovered from individual patients showed changes in polysaccharide structure (23), we analyzed their exopolysaccharide (EPS) by NMR, which revealed variation in O-acetylation content such that CU (37.46) > PU (26.15) > CPL (23.01) > CPB (19.50) (SI Appendix, Fig. S5). Further analysis of the structural reporter group (SRG) 1H NMR resonances corresponding to backbone mannose anomeric protons, which provide a spectral signature for the repeating triad of glucuronoxylomannan (4.8 to 5.4 ppm) (24), showed the same peak set for the CU and PU strains and CPB and CPL isolates (4.91, 4.97, 4.99, 5.10, 5.12, 5.15. 5.16, and 5.20) (Fig. 5B), implying conservation of EPS structure in these strains and isolates. Together, these data suggest that the size of the capsule and acetylation patterns, but not the primary sequence of the polysaccharides, are altered by these mouse passages.
Isolates from the brain of the CU-infected mouse, CPB1-3, melanized faster when compared with the other isolates, including the parental CU strain, the PU strain, and the CPL1-3 isolates from the lungs of the mouse (Fig. 6A and SI Appendix, Fig. S6B). The first and second passage brain isolates (CPB and CP2B_L/B) were more melanized than all other unpassaged strains and lung-derived isolates when grown at 30 °C in liquid media. For phospholipase, the size of the precipitation zone varied across isolates and increased with the time of incubation. After 72 h of incubation at 30 °C, the isolate recovered from the double passage through the murine lungs (CP2L2_L) presented significantly higher (P = 0.045, t test) phospholipase activity than the original CU strain, as well as significant differences between all samples (P = 0.028, ANOVA). (SI Appendix, Fig. S7A). In contrast, urease activity was comparable between unpassaged strains and first passage isolates. However, significant decreases in urease activity were observed for repassaged brain isolates recovered from the brain and lungs (P = 0.0042 (CP2B1_B) and P = 0.0481 (CP2B1_L)) and the repassaged lung isolates recovered from the lungs (P = 0.0217) (SI Appendix, Fig. S7B).
Virulence in Galleria mellonella.
To identify strain differences in virulence, we tested the CU, PU, CPB, and CPL isolates in the invertebrate model G. mellonella. This model has been used previously to compare virulence of Cryptococcus isolates, which roughly correlates to virulence in mammalian models (25, 26). We found no statistical differences between the virulence of the CU and PU strains. There was enhanced virulence of the CPB1 brain isolate when compared with its parental CU strain (P = 0.042, log-rank Mantel–Cox test) and a slight reduction in virulence of the CPL1 lung isolate (Fig. 6B). These results are consistent with increased virulence displayed by the brain and inositol-conditioned isolates (27). While all strains and isolates tested were virulent, these results show subtle changes in virulence for an insect host.
Interactions with Macrophages.
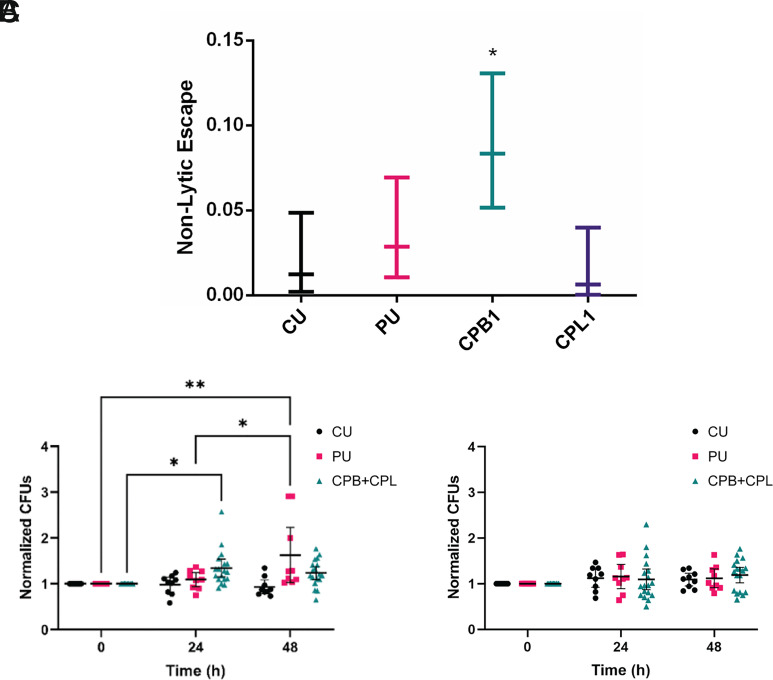
The interaction of Cryptococcus neoformans with macrophages is unusual in that ingestion results in transient intracellular residence, which may be followed by nonlytic exocytosis whereby the fungal cell exits the phagocytic cell without lysing the latter (28, 29). Given that this process involves a complex choreography of cellular events that must occur in synchrony, we considered it a sensitive indicator of Cryptococcus neoformans–macrophage interaction and examined its frequency for the CU, PU, CPL, and CPB isolates. The results show that the CPB1 isolate displayed a significantly higher frequency of nonlytic exocytosis relative to the other strains and isolates (P = 0.013, test of equal proportions with Bonferroni correction) (Fig. 7A).
Fig. 7.
Interactions with phagocytic cells. (A) Nonlytic escape frequency of each strain during BMDM infection. CPB was the only isolate with a significantly increased frequency of nonlytic escape compared with the original CU strain. *P < 0.05 via a test of equal proportions with Bonferroni correction. Normalized CFU counts of passaged or unpassaged Cryptococcus neoformans isolates or strains following coculture with (B) A. castellanii in A.c. buffer or (C) culture in A.c. buffer alone for 0, 24, and 48 h. Data were normalized to the average CFU count at 0 h for each of the three biological replicates for each strain. Bars represent the mean normalized CFU count with 95% CI. n = 9 for PU and CU experiments and n = 18 for CPB + CPL experiments; mouse-passaged strains (CPB and CPL) were combined to increase statistical power and compare the overall effect of mouse passage with that of PU and CU. *P < 0.05 and **P = 0.0019.
Interactions with Amoeba.
Since amoebas are predators of Cryptococcus neoformans and amoeba–fungal interactions have been proposed to select for traits that function during mammalian virulence, we evaluated whether passage through human and mice affected the interaction with Acanthamoeba castellanii (30). Fungal resistance to amoeba predation was investigated with a Cryptococcus neoformans survival assay. Our analysis revealed differences in the growth dynamics between the strains during culture with amoebas (SI Appendix, Fig. S8A). No significant differences were observed between the unpassaged strains and murine-passaged isolates from individual organs (SI Appendix, Fig. S8B), but as a whole, isolates and strains that underwent passage through human or murine hosts (PU and CPB + CPL) displayed increased survival in the presence of amoebas (P = 0.039, n.s. (CPB + CPL; 24 and 48 h), P = n.s., 0.002 (PU; 24 and 48 h), two-way ANOVA) (Fig. 7B). Unlike the PU and CPB + CPL strains, CFU counts for the CU strain slightly decreased over the course of 48 h. Control experiments in A. castellanii (A.c.) buffer did not demonstrate any obvious growth differences between strains (Fig. 7C).
Discussion
A comparison of the patient and cockatoo strains revealed that they are genetically and phenotypically very similar and likely derived from each other. Given that the patient strain came from an immunocompromised patient with cryptococcosis and the cockatoo strain came from bird excreta in the home of the patient, the inference was made that the human infection was a result of exposure to the bird as the point source of the infection (6). Although psittacine birds can develop cryptococcosis, their association with Cryptococcus neoformans is usually in the form of saprophytic growth in their excrement (31). However, the prior case report could not rule out the possibility that the patient had acquired the infection first and then infected the bird in some form, possibly through cough during a pulmonary phase of disease. Although patient-to-bird transmission was, and is, considered unlikely, mice with experimental cryptococcosis contaminate their cage bedding, indicating that mammalian hosts with systemic infection can shed Cryptococcus neoformans (32). However, our finding that some of the genes impacted by mutations observed when the cockatoo strain was passaged in mice are also altered in the patient strain suggests that these were selected in mammalian hosts, which in turn supports the original conclusion that the transmission was from the bird to the patient.
The comparison of patient and cockatoo genomes confirmed their close relationship and revealed amino acid changes in seven proteins. There were two nonsynonymous changes in the predicted ribosome regulatory protein (LQVO5_002184, homologue of the H99 gene CNAG_06273), and a gene repressed during titan cell formation (14). One variant is found in the predicted SWI–SNF chromatin-remodeling complex (LQVO5_000317, homologue of the H99 gene CNAG_00342), a protein that is down-regulated during murine cryptococcal infection (15), is involved in cryptococcal morphogenesis (33), and is implicated in nitrosative stress response in the plant pathogenic fungus Fusarium graminearum (34).
To ascertain whether we could reproduce some of the changes observed in the patient strain relative to the cockatoo strain, we passaged the latter in mice. In all the initial mouse-passaged isolates, we observed one frameshift variant that impacts the same gene (LQVO5_000317, homologue of the H99 gene CNAG_00342) observed in the patient strain. A second variant impacting only the initial mouse-passaged isolates collected from lung tissue results in the reversion of a premature stop codon for LQVO5_004463, a homologue of CNAG_05940 (ZFC3). ZFC3 is a transcription factor that is repressed during titan cell induction and is involved in proliferation within the brain in addition to being a target of the transcription factors Gat201, Liv3, and Pdr802, a regulator of titan cell formation (17, 18, 35). Truncation of the protein encoded by LQVO5_000317 is likely to impact the function of this gene and reduce expression levels of the target genes. During the second round of passage, we observed CNV in the lung-derived isolates, specifically duplication of a region encoding two hypothetical proteins (LQV05_002052 and LQV05_002053). LQV05_002052 is predicted to function as an ER–Golgi vesicle-tethering protein, the homologue of which (CNAG_00128) has been shown to be repressed during titan cell formation in gene expression experiments (14). However, we did not see a causal relationship between LQV05_002052 or LQVO5_000317 and cell body enlargement, as lung-passaged isolates displayed cell body sizes similar to the CU and PU strains, after growth in titan cell–inducing media.
The comparison of clinical and environmental isolates for genomic differences such as the ones found in this study revealed multiple instances of frameshift variants in both CNAG_00342 and CNAG_05940 in isolates spanning the lineages VNI, VNII, and VNB (13). How these genetic changes affect virulence and pathogenesis is a question for future studies. In this regard, a comparison of the virulence of 10 clinical and 11 environmental Cryptococcus neoformans isolates in mice revealed that seven clinical isolates and only 1 environmental isolate caused lethal infection (36). Considering these results in the light of our findings suggest that the clinical strains in that study were perhaps more virulent because of genetic changes that were selected for by human passage.
The occurrence of shared Cryptococcus neoformans genetic changes across experimental and patient isolates suggests that fungal replication in mammalian hosts may select for specific changes. Multiple studies have reported genetic changes during infection; analysis of serial isolates from patients shows the emergence of chromosome rearrangements, ploidy alterations, SNPs, insertions, and deletions (37) and even the reversion of an SNP responsible for attenuating virulence in vivo but not in vitro (38). One mechanism of mutagenesis is transposon mobilization during infection (39); however, we saw no evidence of transposon mobilization upon mammalian passage here. Cryptococcus neoformans replicates within macrophages in vivo (40), and internalized fungal cells are exposed to oxygen- and nitrogen-derived radicals that can be mutagenic (41). A recent study of C. albicans during in vitro and in vivo passage suggested a higher mutation rate in vivo (42), and this suggests that microbes capable of prolonged residence in the human host can acquire genetic changes, such that the capacity for virulence could provide a shortcut to greater genetic variation. Indeed, both genetic and phenotypic variation of the infecting pathogen have been observed for individual hosts infected with Cryptococcus (21, 43–46). Due to the initial sampling strategy of the CU and PU strains, we have been unable to explore the full diversity of cryptococcal genotypes and phenotypes that might have been present in the patient and cockatoo. Despite this, we have seen notable variability in phenotype following the passage of a single strain across mammalian systems.
The comparison of Cryptococcus neoformans characteristics associated with virulence among cockatoo, patient, and mouse-passaged isolates in this study revealed subtle phenotypic changes. There were differences in average capsule size between isolates recovered from the mouse lungs and brain, but NMR analysis of the major polysaccharide component revealed no major alterations in the primary sequence. The finding of organ-related differences in capsule size is consistent with prior reports (21). However, the finding that the GXM structure was unchanged contrasted with the prior observation that serial clinical isolates from patients with persistent infection manifested changes in the polysaccharide structure (25), suggesting that for the CU strain studied here, the polysaccharide structure was more stable. The major difference in the polysaccharide structures involved the extent of mannose-6-O-acetylation (Man-6-OAc). We observe the greatest Man-6-OAc in the CU isolate and relatively less in PU and both the primary mouse-passaged isolates (CPB and CPL). Man-6-OAc has been shown to modulate serological activity and antibody binding (47, 48); however, Man-6-OAc levels have not been directly correlated with virulence. It is possible that the differences in acetylation we see here may be contributing to the differences in virulence noted in the G. mellonella model.
While no major differences were observed in urease activity between the CU and PU strains and the primary mouse-passaged isolates, we saw moderate decreases in urease activity across isolates following the second round of passage. We also noted modest changes in phospholipase activity for lung isolates following a second round of passage. These subtle changes may reflect the co-opting of these virulence factors for pathogenesis rather than for their usual environmental function.
Mouse-passaged isolates and patient strains grew better at higher temperatures than the cockatoo strain, possibly reflecting a period of adaptation to thermal mammalian conditions. Since the mouse-passaged isolates recovered from lung tissue (CPL) did not show this phenotype, we cannot attribute this to simple thermal adaptation to mammalian temperatures. Furthermore, the comparison of genetic variants between CPB and CPL isolates did not reveal genetic changes known to confer increased thermal stability. Consequently, the most likely explanation for increased thermal tolerance is epigenetic change, possibly associated with organ-specific alteration of metabolic states, which may allow for greater survival during rapid heating (49–52). Although a mechanistic investigation of this phenomenon is beyond the scope of this paper, we note that if this phenomenon occurs in nature, it could provide environmental fungi capable of mammalian infection with a mechanism for rapid thermal adaptation that could increase their fitness during climate change.
In addition, the mouse-passaged isolates recovered from brain tissue exhibited faster melanization and increased rates of nonlytic macrophage escape and killed moth larvae faster, consistent with a relative gain of virulence during animal passage. We also noted higher fungal burdens for the CP2B isolates compared with the CPB isolates. This is also consistent with a gain of virulence during passage; however, we caution against comparing fungal burdens across different time points as these differences may reflect the alternate stages of cryptococcal clearance for different time points. Consistent with our findings, studies that focused on the virulence of isolates conditioned in inositol, brain, and lung environments have shown that inositol-rich environments enhance virulence in a murine model (27). When pooled amoeba interaction data from all murine-passaged isolates are compared with those of unpassaged strains, increased resistance to amoeba predation is observed, in agreement with previous findings that the passage of environmental isolates in amoebas results in the emergence of pleiotropic phenotypes (53). However, the effect is small and disappears when comparing isolates from individual organs with the unpassaged CU strain, possibly due to loss of statistical power. Since Entamoeba spp. and slime molds can occur in bird feces (54, 55), the CU strain may already be close to maximally resistant to amoeba predation. These interstrain and interisolate phenotypic differences highlight the tremendous variation apparent in closely related Cryptococcus neoformans strains, a phenomenon that contributes to virulence (56, 57) and is also apparent in pleiotropic variants generated by amoeba predation (53) and phenotypic switching (58). Overall, these results indicate that the cryptococcal genome is highly malleable such that genetic changes can accumulate rapidly.
In summary, genomic analysis of cockatoo, human, and mouse-passaged isolates strongly supports our earlier inference that human infection resulted from exposure to a pet cockatoo. In this study, the comparison of cryptococcal genomes of the incident bird and patient strains with mouse-passaged isolates revealed the occurrence of common genetic changes during mammalian passage. Previously, we showed that passage of Cryptococcus neoformans in mice promotes the appearance of electrophoretic karyotypes (59). Similarly, in the ascomycete C. albicans, infection was associated with larger changes in heterozygosity during murine passage (42) and human infection (60) that occur in vitro. The capacity for virulence in pathogenic microbial species is not without cost as evident by genome reduction and host specialization (61). Findings that mammalian infection can promote genomic changes in both Cryptococcus neoformans and C. albicans suggest that the capacity for virulence can provide a mechanism for more rapid evolutionary change through selection and adaptation in the mammalian host.
Materials and Methods
Resource Availability.
Requests for strains used in this study should be directed to Arturo Casadevall.
Experimental Model and Subject Details.
Cryptococcus neoformans strains.
The original patient and cockatoo strains were described by Nosanchuk et al. (6). For both strains, a single colony was isolated and stored frozen at −80 °C; they have been maintained this way since last studied. This study involved the analysis of the original clinical and cockatoo strains and mouse-passaged cockatoo isolates.
Mouse passage studies.
An inoculum of 2 × 105 freshly grown Cryptococcus neoformans (CU) cells from the cryopreserved samples obtained from cockatoo guano was administered by retro-orbital intravenous injection to a female A/J mouse. Infections were administered to the mouse under xylazine and ketamine anesthesia, 10 mg/kg and 100 mg/kg, respectively. The mouse was observed daily for signs of cryptococcosis symptoms and eventually euthanized 43 d postinfection. The brain and lungs were aseptically removed and homogenized in a total volume of 2 mL sterile PBS, and dilutions of 10−4, 10−3, 10−2, 10−1, and 100 were plated onto Yeast Extract-Peptone-Dextrose (YPD) agar plates supplemented with 1% Pen/Strep (Gibco 15140122) to determine CFUs in each organ. The brain and lung homogenates contained 8.16 × 102 and 7.88 × 101 CFU/mg, respectively. Cryptococcus neoformans isolates each from the brain and lungs in the first mouse passage (CPB/CPL) were again cultured in YPD liquid media and used to reinfect five new female A/J mice. Isolates from the brain and lungs of each mouse (CP2B1-3_B/L and CP2L2-3_B/L) were collected 15 d after the second infection by the same protocol as above. Fungal burden in each organ after passage is tabulated in SI Appendix, Table S1.
Animal studies.
An A/J female mouse 5 to 8 wk of age was obtained from The Jackson Laboratory (Bar Harbor, ME). Animal experiments were conducted in accordance with the policies and with the approval of the Johns Hopkins University Institutional Animal Care and Use Committee (protocol MO21H124). Mice were euthanized using CO2 asphyxiation.
Macrophage nonlytic event quantification.
Bone marrow–derived murine macrophages (BMDMs) were harvested from the hind leg bones of a 6-wk-old C57BL/6 female mouse from The Jackson Laboratory and were differentiated by seeding in 10-cm tissue culture–treated dishes in Dulbecco's Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS), 1% nonessential amino acids, 1% penicillin–streptomycin, 2 mM Glutamax, 1% HEPES buffer, 20% L-929 cell-conditioned supernatant, and 0.1% beta-mercaptoethanol for 6 d at 37 °C and 9.5% CO2. BMDMs were used for experiments within 5 d of differentiation. BMDMs were activated with LPS (0.5 μg/mL) and IFN-γ (10 ng/mL) for 16 h prior to experiments. The medium was then refreshed, and the BMDMs were infected with opsonized Cryptococcus neoformans at an MOI of one and then imaged every 2 min for 24 h on a Zeiss Axiovert 200M inverted scope at 37 °C with 9.5% CO2. Nonlytic events were quantified by determining the outcome of each infected macrophage throughout the 24-h period, and 95% confidence intervals were estimated with a test of proportions. Statistical significance was determined with a test of equal proportions and Bonferroni correction.
Amoeba fungal killing assays.
A modified version of the fungal killing assay described previously (62) was performed to determine the ability of different fungal strains to resist killing by amoebas. Acanthamoeba castellanii (ATCC 30234) were washed twice with Dulbecco’s phosphate-buffered solution (DPBS), counted, and diluted in A.c. buffer [0.8 mM CaCl2, 4 mM MgSO4, 2.5 mM NaHPO4, 2.5 mM KH2PO4, 0.1% sodium citrate, and 0.05 mM Fe(NH4)2(SO4)2]. Amoebas were seeded in 96-well plates at 104 cells/well and incubated at 25 °C for 2 h. The same volume of A.c. buffer alone was added to wells as a control. Cryptococcus neoformans strains were inoculated the day prior in liquid Sabouraud media (SAB) and grown at 30 °C with rotation. Cryptococcus neoformans strains were washed twice with DPBS and similarly counted and diluted in A.c. buffer. Wells containing amoebas and control wells were inoculated with Cryptococcus neoformans at an MOI of one and incubated at 25 °C for 0, 24, or 48 h. At each time interval, amoebas were lysed by pulling the suspension seven times through a 27-gauge needle, and lysates were serially diluted. Three 10 μL samples of diluted lysates were plated on SAB agar with 1% penicillin/streptomycin and incubated at 30 °C for 48 h, and colonies were enumerated to CFUs. CFU counts were normalized to the average CFU count of each biological replicate at 0 h, and a two-way ANOVA with Tukey’s multiple comparisons test were performed using GraphPad Prism to determine significance.
Virulence in G. mellonella.
G. mellonella were infected with CU, PU, CPB, and CBL isolates as previously described (63). Briefly, final instar larvae ranging between approximately 175 and 225 mg were injected with 10 µL of Cryptococcus neoformans culture grown to the stationary phase in YPD media, washed twice in PBS, and diluted to a concentration of 107 cells/mL. Survival of larvae and pupae was monitored daily for 14 d, with death being defined by lack of movement in response to the stimulus of a pipette tip. Statistical significance was determined with the log-rank Mantel–Cox test, corrected for multiple comparisons using the Bonferroni method, via GraphPad Prism.
Titan cell induction.
Cryptococcus neoformans cultures were inoculated from frozen stocks in 5 mL Sabouraud dextrose broth (BD Difco) at 30 °C and grown for 24 h. The cultures were then washed three times with 50 mM MOPS buffer, and cell densities were counted. Each culture was then seeded into 200 µL of titan cell medium (TCM; 90% 50 mM MOPS buffer, 5% Sabouraud broth, 5% FBS, and 0.1% 15 mM sodium azide) in sterile 96 plates and incubated for 16 h at 37 °C and 5% CO2. The cells were collected from each well, washed three times in MOPS buffer, and mixed with India ink for cell morphology measurements.
India ink staining and capsule analysis.
For capsule size measurements, Cryptococcus neoformans cells were grown in capsule-inducing minimal media (29.4 mM KH2PO4, 10 mM MgSO4*7H2O, 13 mM glycine, 15 mM glucose, and 3 µM thiamine, pH 5.5), washed and mixed with India ink, and imaged on an Olympus AX70 microscope using QImaging Retiga 1300 digital camera and QCapture Suite V2.46 software (QImaging). Capsule measurements were calculated using the exclusion zone produced with India ink and Quantitative Capture Analysis program as previously described (64). A minimum of 100 yeast cells were measured for each strain and condition. Statistical differences between groups were assessed by two-way ANOVA with unequal variance, and statistical difference between groups was determined by Tukey’s multiple comparisons test.
Growth curves.
Cryptococcus neoformans strains were recovered from cryopreserved stocks by growth in YPD for 48 h at 30 °C and then subcultured in triplicate into Sabouraud dextrose broth (BD Difco) at a density of 1 × 105 cells/mL in wells of a 96-well plate. The plate was incubated at 30 or 37 °C with orbital shaking in a SpectraMax iD5 plate reader (Molecular Devices), and absorbance at 600 nm was read at 15-min intervals. Absorbance was plotted against time using GraphPad Prism software, and the linear region of each curve was analyzed by simple linear regression to derive the slope and lag time (x-intercept).
EPS preparation.
Cryptococcus neoformans CU, PU, CPB, and CPL isolates were grown in YPD for 48 h and subcultured to modified minimal media (MMM) for NMR analysis, which also induces capsule growth. Cells were grown in MMM for 3 d, and the EPS was harvested by centrifuging cells (10 min at 3494 × g) and sterile filtration of the culture supernatant (0.45-nm filtration). Any remaining media components were removed, and EPS was concentrated by passage through a 3-kDa molecular weight cutoff (MWCO) Centricon filtration unit. The >3-kDa fraction of EPS was then characterized by NMR.
NMR analysis.
1D 1H NMR data were collected on a Bruker Avance II (600 MHz) equipped with a triple resonance, TCI cryogenic probe, and z-axis pulsed field gradients. Spectra were collected at 60 °C, with 128 scans and a free induction decay size of 84,336 points. Standard Bruker pulse sequences were used to collect the 1D data (p3919gp and zggpw5). Data were processed in TopSpin (Bruker version 4.1.3) by truncating the FID to 8,192 points, apodizing with a squared cosine bell window function and zero filling to 65,536 points. Relative peak integration and 1H frequency referencing were performed in TopSpin using d6-DSS internal standard (10 µL/510 µL sample).
DNA Preparation and genomic sequencing.
ONT sequencing libraries were prepared with genomic DNA from the CU and PU strains using the Ligation Sequencing Kit (SQK-LSK109) with the Native Barcoding Kit (EXP-NBD103) according to the manufacturer instructions (ONT, Oxford, UK). Illumina sequencing libraries were prepared using the Nextera Flex DNA Library Prep Kit (Illumina), and sequencing was performed with a MiSeq using v2 2 × 150 chemistry. For genomic sequencing of CPL and CPB isolates, three individual colonies were selected from brain and lung CFU plates and used to seed 100 mL YPD cultures, which were allowed to grow for 48 h at 30 °C with rotation. Genomic DNA was isolated from each culture following the protocol described by Velegraki et al. (65). Briefly, after a 48-h growth period, Cryptococcus neoformans cells were collected by centrifugation, frozen at −80 °C overnight, and then subsequently lyophilized overnight. Glass beads (0.5 mm diameter, BioSpec Products, catalog no. 11079105z) were added to the dry cell pellets and vortexed into a powder. DNA was then extracted with a CTAB buffer (100 mM Tris, pH 7.5, 700 mM NaCl, 10 mM EDTA, 1% CTAB, and 1% beta-mercaptoethanol) at 65 °C for 30 min. The solutions were cooled and then extracted with chloroform, then isopropanol, and then 70% ethanol. The DNA pellet was then resuspended in 1 mL sterile water and treated with 20 μg RNase for 30 min at 37 °C. Finally, the genomic DNA was further purified using the DNeasy PowerClean CleanUp Kit (Qiagen 12877-50) for genomic sequencing. 100 microliter aliquots of 19 to 30 ng/μL DNA were used in the sequencing reactions. DNA was sheared to 250 bp using a Covaris LE instrument and adapted for Illumina sequencing as described by Fisher et al. (66). Libraries were sequenced on a HiSeq X10 generating 150-bp paired reads.
Assembly and genomic analysis.
Whole genome assemblies were generated for the CU and PU strains with ONT long reads via Canu v2.1.1 (genome size 20 Mb) (67), followed by short-read polishing via medaka v0.8.1 (1X) (https://github.com/nanoporetech/medaka) and pilon v1.23 (3X) (68). SNPs, insertions, and deletions between the CU and PU assemblies were then identified using nucmer v3.1 (69), with variants in centromeric and telomeric regions removed prior to downstream analysis. To identify variants in the mouse-passaged isolates, Illumina reads for CPL and CPB samples were aligned to the CU assembly with BWA-MEM v0.7.17 (70), and variants were called with our publicly available GATK v4 pipeline (https://github.com/broadinstitute/fungal-wdl/tree/master/gatk4). After calling, variants were filtered on the following parameters: QD < 2.0, QUAL < 30.0, SOR > 3.0, FS > 60.0 (indels > 200), MQ < 40.0, GQ < 50, alternate allele percentage = 0.8, and DP < 10. All variants were annotated with SNPeff, v4.3t (71). To identify strain lineage, reads for the CU and PU samples were aligned to the Cryptococcus neoformans var. grubii H99 reference genome (GCA_000149245.3) with BWA-MEM v0.7.17 (70), and variants were called and filtered as described above. A maximum likelihood phylogeny was estimated using segregating SNP sites present in one or more isolates, allowing ambiguity in a maximum of 10% of samples, with RAxML v8.2.12 (72) rapid bootstrapping (GTRCAT substitution model), and visualized with ggtree (R 3.6.0) (73). Aneuploidies were visualized using funpipe (coverage analysis) v0.1.0 (https://github.com/broadinstitute/funpipe); transposon mobilization was assessed through whole genome alignment of the CU and PU assemblies with nucmer v3.1 to identify alignment gaps, and CNV was assessed using CNVnator v0.3 (74).
Urease activity assay.
Cryptococcus neoformans strains and isolates were first grown in YPD for 48 h at 30 °C. Urea broth comprising 10 mM KH2PO4, 0.1% Bacto Peptone (Difco), 0.1% D-glucose, 0.5% NaCl, 2% urea, and 0.03 mM phenol red, as described by Roberts et al. (75), was inoculated with PBS-washed cells at a density of 1 × 106 cells/mL for each strain in triplicate. After incubation for 16 h at 30 °C, increased pH of culture media that is indicative of ammonium production due to urease activity was detected by measuring absorbance at 560 nm over a 6-h time course. Absorbance readings that had been corrected by subtraction of media-only background absorbance were plotted against time. Urease activity rates were derived by a simple linear regression model, and data were analyzed for statistical significance using an ordinary one-way ANOVA with GraphPad Prism 9 software.
Cryptococcus neoformans melanization assay.
Cryptococcus neoformans strains and isolates were first grown in YPD liquid media at 30 °C until the cultures were in the stationary phase. Cells were washed twice in PBS, and 40 µL of washed culture was added to 2 mL minimal media with 1 mM L-3,4-dihydroxyphenylalanine (L-DOPA) and grown for 5 d at 30 °C in a polystyrene 12-well plate. Cultures were imaged directly using a Cannon9000F scanner. Alternatively, 1 × 106 PBS-washed cells were spotted onto L-DOPA agar in triplicate. Plates were incubated at 30 °C or 37 °C and then photographed after the indicated number of days using a 12-megapixel camera. Color images were converted to gray scale using Adobe Photoshop; pigmentation intensities were quantified using Image Studio Lite software and graphed using GraphPad Prism software. Statistical significance was determined with the ordinary one-way ANOVA test via GraphPad Prism.
Phospholipase activity.
Extracellular phospholipase activity in Cryptococcus neoformans strains and isolates was tested by the modified method reported by Chen et al. (76). Egg yolk agar medium was created based on Difco Sabouraud dextrose agar media with 8% egg yolk, 1 M sodium chloride, and 0.05 M calcium chloride. After overnight growth in YPD media, 3 μL of each strain, with a total of 10,000 cells, was spotted onto egg yolk agar medium. Each strain and isolate were tested on five separate plates and incubated at 30 °C. After 72 h, colonies were photographed and measured with ImageJ software. Activity of phospholipase was analyzed by the ratio of colony diameter to total precipitation diameter, where a ratio equal to 1.0 indicates a lack of phospholipase activity. Statistical significance was determined by an unpaired t test via GraphPad Prism.
Heat-ramp and thermal stability analysis.
Strains and isolates were maintained at −80 °C in glycerol, streaked onto Sabouraud dextrose (SAB) (BD Difco) agar, and incubated at 30 °C for 48 h prior to heat-ramp cell death assays. SAB broth was inoculated from growth on plates to equal densities (OD 0.1) and incubated at 30 °C for 18 h in stationary 96-well plates. Isolates were resuspended and diluted 1:5 in fresh SAB broth, and 100 μL was treated with a linear 30 to 56 °C heat-ramp stress over 10 min in a water bath with agitation (Lauda). Untreated and heat-ramp–treated strains were immediately spotted (5 μL in SAB) in fivefold serial dilutions on SAB agar and incubated at 30 °C for 48 h to assess viability. CFUs before and after heat-ramp assays were enumerated to calculate relative survival. To determine growth differences at high temperature, untreated strains were spotted on SAB agar and incubated at 37 °C for 48 h. Statistical significance was determined with the ANOVA test via GraphPad Prism.
RNA-seq analysis.
RNA-seq datasets were downloaded from the GEO (Gene Expression Omnibus) database from the following deposited datasets: GSE162851, GSE136879, GSE93005, PRJEB4169, GSE32049, GSE32228, GSE121183, GSE60398, and GSE66510. Where necessary, raw data were reanalyzed by bowtie2 (2.3.5) (77) alignment to the most recent Cryptococcus neoformans H99 or KN99α genome (fungibd.org), count matrices generated with HTSeq (1.99.2) (78) and RNA-seq analysis with Bioconductor DESeq2 (1.22.2) (79).
Protein structure prediction and glycosylation analysis.
Protein structure predictions were generated with AlphaFold2 v2.1.0 (reduced BFD database) (80). Intrinsically disordered regions were identified with IUPred (81), and glycosylation predictions were made with GPP (82).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This project has been funded in part with Federal funds from the National Institute of Allergy and Infectious Diseases, NIH, Department of Health and Human Services under award U19AI110818 to the Broad Institute. C.A.C. is a CIFAR fellow in the Fungal Kingdom program. A.C. was supported in part by NIH grants AI052733, AI15207, and HL059842. S.A.M. was supported in part by NIH grants T32AI007417 and R01AI162381.
Author contributions
S.A.M., N.G., and A.C. designed research; P.S.-C., S.A.M., N.G., R.P.B., Q.D., Y.F., M.S.F., G.G., S.G., M.K., D.F.Q.S., P.S., M.P.W., and A.C. performed research; S.M.L., J.D.N., and S.S. contributed new reagents/analytic tools; P.S.-C., S.A.M., R.P.B., Q.D., Y.F., J.M.H., W.T., and C.A.C. analyzed data; and P.S.-C., S.A.M., and A.C. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
Reviewers: J.C.P., University at Buffalo - Downtown Campus; and F.H.S., University of Notre Dame.
Contributor Information
Christina A. Cuomo, Email: cuomo@broadinstitute.org.
Arturo Casadevall, Email: acasade1@jhu.edu.
Data, Materials, and Software Availability
Genome data can be accessed via accession PRJNA783275 (83).
Supporting Information
References
- 1.Casadevall A., Perfect J. R., Cryptococcus neoformans (American Society for Microbiology, 1998). [Google Scholar]
- 2.Goldman D. L., et al. , Serologic evidence for Cryptococcus neoformans infection in early childhood. Pediatrics 107, E66 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Alanio A., Dormancy in Cryptococcus neoformans: 60 years of accumulating evidence. J. Clin. Invest. 130, 3353–3360 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Currie B. P., Freundlich L. F., Casadevali A., Restriction fragment length polymorphism analysis of Cryptococcus neoformans isolates from environmental (pigeon excreta) and clinical sources in New York City. J. Clin. Microbiol. 32, 1188–1192 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farrer R. A., et al. , Genomic epidemiology of a Cryptococcus neoformans case cluster in Glasgow, Scotland, 2018. Microb. Genom. 7, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nosanchuk J. D., et al. , Evidence of zoonotic transmission of Cryptococcus neoformans from a pet cockatoo to an immunocompromised patient. Ann. Intern. Med. 132, 205–208 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Littman M. L., Schneierson S. S., Cryptococcus neoformans in pigeon excreta in New York City. Am. J. Hyg. 69, 49–59 (1959). [DOI] [PubMed] [Google Scholar]
- 8.Kielstein P., Hotzel H., Schmalreck A., Khaschabi D., Glawischnig W., Occurrence of Cryptococcus spp. in excreta of pigeons and pet birds. Mycoses 43, 7–15 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Shrestha R., Stoller J., Honari G., Procop G., Gordon S., Pneumonia due to Cryptococcus neoformans in a patient receiving infliximab: Possible zoonotic transmission from a pet cockatiel. Respir. Care 43, 606–608 (2004). [PubMed] [Google Scholar]
- 10.Lagrou K., et al. , Zoonotic transmission of Cryptococcus neoformans from a magpie to an immunocompetent patient. J. Intern. Med. 257, 385–388 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Cuomo C. A., Rhodes J., Desjardins C. A., Advances in Cryptococcus genomics: Insights into the evolution of pathogenesis. Mem. Inst. Oswaldo Cruz 113, e170473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janbon G., et al. , Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 10, e1004261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desjardins C. A., et al. , Population genomics and the evolution of virulence in the fungal pathogen Cryptococcus neoformans. Genome Res. 27, 1207–1219 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trevijano-Contador N., et al. , Cryptococcus neoformans can form titan-like cells in vitro in response to multiple signals. PLoS Pathog. 14, e1007007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H., et al. , Unveil the transcriptional landscape at the Cryptococcus-host axis in mice and nonhuman primates. PLoS Negl. Trop. Dis. 13, e0007566 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung K. W., et al. , Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nat. Commun. 6, 6757 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Homer C. M., et al. , Intracellular action of a secreted peptide required for fungal virulence. Cell Host Microbe 19, 849–864 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reuwsaat J. C. V., et al. , The transcription factor pdr802 regulates titan cell formation and pathogenicity of Cryptococcus neoformans. mBio 12, 1–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teng X., et al. , Gene-dependent cell death in yeast. Cell Death Dis. 2, e188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stolp Z. D., et al. , Yeast cell death pathway requiring AP-3 vesicle trafficking leads to vacuole/lysosome membrane permeabilization. Cell Rep. 39, 110647 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denham S. T., et al. , A dissemination-prone morphotype enhances extrapulmonary organ entry by Cryptococcus neoformans. Cell Host Microbe 30, 1382-1400.e8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rivera J., Feldmesser M., Cammer M., Casadevall A., Organ-dependent variation of capsule thickness in Cryptococcus neoformans during experimental murine infection. Infect. Immun. 66, 5027–5030 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cherniak R., Morris L. C., Belay T., Spitzer E. D., Casadevall A., Variation in the structure of glucuronoxylomannan in isolates from patients with recurrent Cryptococcal meningitis. Infect. Immun. 63, 1899–1905 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cherniak R., Valafar H., Morris L. C., Valafar F., Cryptococcus neoformans chemotyping by quantitative analysis of 1H nuclear magnetic resonance spectra of glucuronoxylomannans with a computer-simulated artificial neural network. Clin. Diagn. Lab. Immunol. 5, 146–159 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Firacative C., Duan S., Meyer W., Galleria mellonella model identifies highly virulent strains among all major molecular types of Cryptococcus gattii. PLoS One 9, e105076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mylonakis E., et al. , Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect. Immun. 73, 3842–3850 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y., et al. , Inositol metabolism regulates capsule structure and virulence in the human pathogen Cryptococcus neoformans. mBio 12, e0279021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma H., Croudace J. E., Lammas D. A., May R. C., Expulsion of live pathogenic yeast by macrophages. Curr. Biol. 16, 2156–2160 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Alvarez M., Casadevall A., Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr. Biol. 16, 2161–2165 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Fu M. S., Casadevall A., Divalent metal cations potentiate the predatory capacity of amoeba for Cryptococcus neoformans. Appl. Environ. Microbiol. 84, e01717-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guilherme Augusto M. G., Grandi F., Are all psittacine birds carriers of Cryptococcus neoformans? Mem. Inst. Oswaldo Cruz 106, 781 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Nosanchuk J. D., Mednick A., Shi L., Casadevall A., Experimental murine cryptococcal infection results in contamination of bedding with Cryptococcus neoformans. Contemp. Top. Lab. Anim. Sci. 42, 9–12 (2003). [PubMed] [Google Scholar]
- 33.Lin J., et al. , Transcription factor Znf2 coordinates with the chromatin remodeling SWI/SNF complex to regulate cryptococcal cellular differentiation. Commun. Biol. 2, 412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jian Y., et al. , Interplay of two transcription factors for recruitment of the chromatin remodeling complex modulates fungal nitrosative stress response. Nat. Commun. 12, 2576 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee K. T., et al. , Fungal kinases and transcription factors regulating brain infection in Cryptococcus neoformans. Nat. Commun. 11, 1521 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Litvintseva A. P., Mitchell T. G., Most environmental isolates of Cryptococcus neoformans var. grubii (serotype A) are not lethal for mice. Infect. Immun. 77, 3188–3195 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y., et al. , Microevolution of Serial Clinical Isolates of Cryptococcus neoformans var . grubii and C. gattii. mBio 8, e00166-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magditch D. A., Liu T. B., Xue C., Idnurm A., DNA mutations mediate microevolution between host-adapted forms of the pathogenic fungus Cryptococcus neoformans. PLoS Pathog. 8, e1002936 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gusa A., et al. , Transposon mobilization in the human fungal pathogen Cryptococcus is mutagenic during infection and promotes drug resistance in vitro. Proc. Natl. Acad. Sci. U.S.A. 117, 9973–9980 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feldmesser M., Kress Y., Novikoff P., Casadevall A., Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect. Immun. 68, 4225–4237 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiseman H., Halliwell B., Damage to DNA by reactive oxygen and nitrogen species: Role in inflammatory disease and progression to cancer. Biochem. J. 313, 17–29 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ene I. V., et al. , Global analysis of mutations driving microevolution of a heterozygous diploid fungal pathogen. Proc. Natl. Acad. Sci. U.S.A. 115, E8688–E8697 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandes K. E., et al. , Phenotypic variability correlates with clinical outcome in Cryptococcus isolates obtained from botswanan hiv/aids patients. mBio 9, e02016-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fries B. C., Taborda C. P., Serfass E., Casadevall A., Phenotypic switching of Cryptococcus neoformans occurs in vivo and influences the outcome of infection. J. Clin. Invest. 108, 1639 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Charlier C., et al. , Capsule structure changes associated with Cryptococcus neoformans crossing of the blood-brain barrier. Am. J. Pathol. 166, 421–432 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desnos-Ollivier M., et al. , Mixed infections and in vivo evolution in the human fungal pathogen Cryptococcus neoformans. mBio 1, 91–101 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belay T., Cherniak R., Kozel T. R., Casadevall A., Reactivity patterns and epitope specificities of anti-Cryptococcus neoformans monoclonal antibodies by enzyme-linked immunosorbent assay and dot enzyme assay. Infect. Immun. 65, 718 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kozel T. R., et al. , Antigenic and biological characteristics of mutant strains of Cryptococcus neoformans lacking capsular O acetylation or Xylosyl Side Chains. Infect. Immun. 71, 2868 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polacheck I., Platt Y., Aronovitch J., Catecholamines and virulence of Cryptococcus neoformans. Infect. Immun. 58, 2919–2922 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldman D., Lee S. C., Casadevall A., Pathogenesis of pulmonary Cryptococcus neoformans infection in the rat. Infect. Immun. 62, 4755–4761 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldman D. L., Casadevall A., Cho Y., Lee S. C., Cryptococcus neoformans meningitis in the rat. Lab. Invest. 75, 759–770 (1996). [PubMed] [Google Scholar]
- 52.Huffnagle G. B., Yates J. L., Lipscomb M. F., Immunity to a pulmonary Cryptococcus neoformans infection requires both CD4+ and CD8+ T cells. J. Exp. Med. 173, 793–800 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu M. S., et al. , Amoeba predation of Cryptococcus neoformans results in pleiotropic changes to traits associated with virulence. mBio 12, e00567-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martínez-Díaz R. A., Herrera S., Castro A., Ponce F., Entamoeba sp. (Sarcomastigophora: Endamoebidae) from ostriches (Struthio camelus) (Aves: Struthionidae). Vet. Parasitol. 92, 173–179 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Suthers H. B., Ground-feeding migratory songbirds as cellular slime mold distribution vectors. Oecologia 65, 526–530 (1985). [DOI] [PubMed] [Google Scholar]
- 56.Gerstein A. C., et al. , Identification of pathogen genomic differences that impact human immune response and disease during Cryptococcus neoformans infection. mBio 10, e01440-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fernandes K. E., et al. , Phenotypic variability correlates with clinical outcome in Cryptococcus isolates obtained from Botswanan HIV/AIDS patients. mBio 9, e02016-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guerrero A., Jain N., Wang X., Fries B. C., Cryptococcus neoformans variants generated by phenotypic switching differ in virulence through effects on macrophage activation. Infect. Immun. 78, 1049–1057 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fries B. C., Chen F., Currie B. P., Casadevall A., Karyotype instability in Cryptococcus neoformans infection. J. Clin. Microbiol. 34, 1531–1534 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forche A., et al. , Rapid phenotypic and genotypic diversification after exposure to the oral host Niche in Candida albicans. Genetics 209, 725–741 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Casadevall A., Pirofski L. A., Benefits and costs of animal virulence for microbes. mBio 10, e00863-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Malliaris S. D., Steenbergen J. N., Casadevall A., Cryptococcus neoformans var. gattii can exploit Acanthamoeba castellanii for growth. Med. Mycol. 42, 149–158 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Stempinski P., Smith D., Casadevall A., Cryptococcus neoformans virulence assay using a galleria mellonella larvae model system. Bio. Protoc. 12 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dragotakes Q., Casadevall A., Automated measurement of cryptococcal species polysaccharide capsule and cell body. J. Vis. Exp. 11, 56957 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Velegraki A., Kambouris M., Kostourou A., Chalevelakis G., Legakis N. J., Rapid extraction of fungal DNA from clinical samples for PCR amplification. Med. Mycol. 37, 69–73 (1999). [PubMed] [Google Scholar]
- 66.Fisher S., et al. , A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 12, R1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koren S., et al. , Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walker B. J., et al. , Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Delcher A. L., Phillippy A., Carlton J., Salzberg S. L., Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30, 2478–2483 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H., Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint] (2013). 10.48550/arxiv.1303.3997. Accessed 25 August 2022. [DOI]
- 71.Cingolani P., et al. , A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu G., Smith D. K., Zhu H., Guan Y., Lam T. T. Y., ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36 (2017). [Google Scholar]
- 74.Abyzov A., Urban A. E., Snyder M., Gerstein M., CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome. Res. 21, 974–984 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roberts G. D., Horstmeier C. D., Land G. A., Foxworth J. H., Rapid urea broth test for yeasts. J. Clin. Microbiol. 7, 584–588 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen S. C. A., Muller M., Zhou Jin Zhong, Wright L. C., Sorrell T. C., Phospholipase activity in Cryptococcus neoformans: A new virulence factor? J. Infect. Dis. 175, 414–420 (1997). [DOI] [PubMed] [Google Scholar]
- 77.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anders S., Pyl P. T., Huber W., HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mészáros B., Erdös G., Dosztányi Z., IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 46, W329–W337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hamby S. E., Hirst J. D., Prediction of glycosylation sites using random forests. BMC Bioinform. 9, 500 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Septhon-Clark P., Cryptococcus neoformans VNII assembly. BioProject. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA783275/. Deposited 7 April 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Genome data can be accessed via accession PRJNA783275 (83).