Abstract
Natural killer (NK) cells are unique innate immune cells that mediate anti‐viral and anti‐tumor responses. Thus, they might hold great potential for cancer immunotherapy. NK cell adoptive immunotherapy in humans has shown modest efficacy. In particular, it has failed to demonstrate therapeutic efficiency in the treatment of solid tumors, possibly due in part to the immunosuppressive tumor microenvironment (TME), which reduces NK cell immunotherapy's efficiencies. It is known that immune checkpoints play a prominent role in creating an immunosuppressive TME, leading to NK cell exhaustion and tumor immune escape. Therefore, NK cells must be reversed from their dysfunctional status and increased in their effector roles in order to improve the efficiency of cancer immunotherapy. Blockade of immune checkpoints can not only rescue NK cells from exhaustion but also augment their robust anti‐tumor activity. In this review, we discussed immune checkpoint blockade strategies with a focus on chimeric antigen receptor (CAR)‐NK cells to redirect NK cells to cancer cells in the treatment of solid tumors.
Keywords: natural killer cell, immune checkpoint, chimeric antigen receptor‐natural killer cell, immunotherapy, tumor
List of abbreviations
- mAb
monoclonal antibody
- CTLA‐4
cytotoxic T‐lymphocyte associated protein
- PD‐1
programmed cell death protein 1
- ICIs
immune checkpoint inhibitors
- LAK
lymphokine‐activated killer
- TILs
tumor‐infiltrating lymphocytes
- TCRs
T cell receptors
- CARs
chimeric antigen receptors
- FDA
Food and Drug Administration
- GVHD
graft‐versus‐host disease
- TME
tumor microenvironment
- NK
natural killer
- BM
bone marrow
- LNs
lymph nodes
- NCR
natural cytotoxicity receptor
- PB
peripheral blood
- IFN‐γ
interferon gamma
- MHC‐I
major histocompatibility complex class I
- KIRs
killer cell immunoglobulin‐like receptors
- NKG2A
natural killer group 2 A
- LIRs
leukocyte immunoglobin‐like receptors
- Ig
immunoglobulin
- HLA
human leukocyte antigen
- ILTs
immunoglobin‐like transcripts
- TIGIT
T cell immunoreceptor with Ig and ITIM domains
- LAG‐3
lymphocyte activation gene‐3
- TIM‐3
T cell immunoglobulin‐ and mucin‐domain‐containing molecule‐3
- DNAM‐1
DNAX accessory molecule‐1
- FasL
Fas ligand
- TRAIL
tumor necrosis factor‐related apoptosis‐inducing ligand
- ADCC
antigen‐dependent cell‐mediated cytotoxicity
- TNF‐α
tumor necrosis factor‐α
- GM‐CSF
granulocyte macrophage colony‐stimulating factor
- MIP
macrophage inflammatory protein
- PRRs
pattern recognition receptors
- aKIRs
activating KIRs
- iKIRs
inhibitory KIRs
- ITIM
immunoreceptor tyrosine‐based inhibitory motif
- MM
multiple myeloma
- EGFR
epidermal growth factor receptor
- HMGB1
high mobility group box 1
- Gal‐9
galactin‐9
- CEACAM‐1
carcinoembryonic antigen‐related cell adhesion molecule 1
- ITT
immunoreceptor tyrosine tail
- FGL1
fibrinogen‐like protein 1
- PTLD
post‐transplant lymphoproliferative disease
- TM
transmembrane
- scFvs
single‐chain variable fragments
- DARPins
designed ankyrin repeat proteins
- ITAMs
immunoreceptor tyrosine‐based activation motifs
- TRUCKs
T cells redirected for universal cytokine‐mediated killing
- ACT
adoptive cell therapy
- GvT
graft‐versus‐tumor
- HSCT
hematopoietic stem cell transplantation
- AML
acute myeloid leukemia
- HPCs
hematopoietic progenitor cells
- iPSCs
induced pluripotent stem cells
- hESCs
human embryonic stem cells
- UCB
umbilical cord blood
- CSCs
cancer stem cells
- TGF‐β
transforming growth factor‐β
- PBMCs
peripheral blood mononuclear cells
- EBV
Epstein‐Barr Virus
- CMV
cytomegalovirus
- aAPCs
artificial antigen‐presenting cells
- Eomes
eomesodermin
- iC9
inducible caspase 9
- G‐CSF
granulocyte colony stimulating factor
- FLT‐3L
FMS‐like tyrosine kinase 3 ligand
- EBs
embryonic bodies
- GBM
glioblastoma
- CRISPR
clustered regularly interspaced short palindromic repeats
- BMP4
bone morphogenetic protein 4
- VEGF
vascular endothelial growth factor
- ZFN
zinc finger nuclease
- GPC3
glypican‐3
- PSCA
prostate stem cell antigen
- HCC
hepatocellular carcinoma
- CCCR
chimeric costimulatory converting receptor
- TNBC
triple negative breast cancer
- ADAM17
A Disintegrin And Metalloproteinase‐17
- rAAV6
recombinant adeno‐associated virus serotype 6
- ABE
adenine base editor
- DTCR
dual targeting chimeric receptor
- PDAC
pancreatic ductal adenocarcinoma
- CIS
cytokine‐inducible SH2‐containing protein
- DNMT1
DNA methyltransferase 1
- TAP‐1
transporter associated with antigen processing 1
- TALENs
transcriptional activator‐like effector nucleases
- NPs
nanoparticles
- CTC
circulating tumor cell
- BiKEs
bi‐specific natural killer cell engagers
- TriKEs
tri‐specific natural killer cell engagers
- NSCLC
non‐small cell lung cancer
- SCLC
small cell lung cancer
- IAP
integrin‐associated protein
- TSP‐1
thrombospondin‐1
- SIRPα
signal regulatory protein alpha
- HNSCC
head and neck squamous cell carcinoma
- LDH
lactate dehydrogenase
- Bi‐apt
bi‐specific aptamer
- KI
knock in
- HI
hepatic intrasinusoidal
- EMA
European Medicines Agency
- MMPs
matrix metalloproteinases
- CRS
cytokine release syndrome
- SCF
stem cell factor
- NB
neuroblastoma
- GSDME
gasdermin E
- IND
investigational new drug
- GMP
Good Manufacturing Practice
- PD‐L1
programmed death‐ligand 1
1. BACKGROUND
Following cardiovascular disease, cancer is the second cause of mortality worldwide [1]. As a result, several treatment options have been developed to restrict disease development, including surgery, chemotherapy, radiation, targeted therapy, and immunotherapy. The relevance of new cancer treatment approaches, such as immunotherapy, has been recognized due to the negative effects of traditional cancer treatment methods. Although cancers use a variety of tactics to delay, alter or stop immune responses, various strategies have been devised to improve the immune response. Generally, cancer immunotherapy is classified into passive and active approaches. In passive immunotherapy, the host's immune response is not stimulated directly. This approach involves the administration of ex vivo‐generated cells or molecules (tumor‐specific monoclonal antibodies (mAbs), recombinant cytokines, and adoptive cell transfer) to patients. On the other hand, active immunotherapy promotes the patient's immune response and leads to effector immunological responses [2]. Active cancer immunotherapy includes Abs targeting immune checkpoints, oncolytic viruses, and vaccines (such as peptide vaccines, DC vaccines, and allogeneic whole cell vaccines) [2, 3].
A breakthrough in immunotherapy was achieved when James P. Allison and Tasuku Honjo received the Nobel Prize in Physiology or Medicine for their discovery of new targets for cancer therapy. They discovered two critical immune checkpoints, including the cytotoxic T‐lymphocyte‐associated protein (CTLA‐4) and programmed cell death protein 1 (PD‐1) as T cells brake [4]. By inhibiting or blocking these checkpoints, they hoped that T cells would be able to find cancer cells and kill them.
Natural killer (NK) cells are unique innate immune cells that play crucial roles in anti‐viral and anti‐tumor responses. This suggests that they might hold great promise for cancer immunotherapy. NK cell adoptive immunotherapy in humans has shown modest efficacy [5]. It has failed to demonstrate therapeutic efficacy in the treatment of solid tumors in particular [6, 7]. This is due in part to the immunosuppressive tumor microenvironment (TME), which reduces NK cell immunotherapy's efficiencies. It is known that immune checkpoints play a prominent role in creating an immunosuppressive TME, leading to NK cell exhaustion and tumor immune escape. Therefore, NK cells must be reversed from their dysfunctional status and increased in their effector roles in order to improve the efficiency of cancer immunotherapy. Blockade of immune checkpoints can not only rescue NK cells from exhaustion but also augment their robust anti‐tumor activity [8].
Immune checkpoint inhibitors (ICIs), which are mAbs, have been shown to be effective in clinical trials, defeating tumor immune evasion mechanisms [9]. While some patients significantly respond to ICIs, most cancers are either resistant at first or develop resistance after the initial response. Therefore, treatment strategies must be developed to overcome ICI resistance and other approaches.
Activation of NK cells by Abs has been a hot research topic in recent years. Because cancer is a multifactorial disease, targeting two or more of the involved molecules will aid in effective treatment. Bi/tri‐specific Abs, which encompass two or three target‐binding units, have been explored as a therapeutic point of view in cancer immunotherapy [10]. Bi and tri‐specific natural killer cell engagers (BiKEs and TriKEs) are bi/tri‐specific Abs that specifically target NK cell CD16 and tumor antigens. In this setting, CD16 interaction causes NK cells to become more cytotoxic and improves their anti‐cancer actions [11, 12]. Therefore, there is no competition for binding to CD16 with the physiologic serum IgG compared to therapeutic mAbs [13, 14, 15]. Due to their smaller size, BiKEs and TriKEs may have better biodistribution than mAbs, particularly in the treatment of solid tumors [16, 17, 18]. In addition, they are non‐immunogenic, have rapid clearance properties, and can be engineered quickly to target known tumor antigens.
Aptamers, which are short single‐strand oligonucleotides (single‐stranded DNA or RNA), are emerging as a promising molecular tool in targeted immunotherapy. They encompass Ab‐mimicking functions and can attach to specific targets and regulate biological processes. They are more stable than Abs, smaller, less immunogenic, and can be produced without the use of biological systems [19, 20]. Aptamers are currently divided into three groups in cancer immunotherapy: aptamers against immunoregulatory factors, aptamers against cancer‐specific proteins, and aptamers used as a drug delivery system for anti‐cancer agents [21].
In immune cell therapy, which is an adoptive immunotherapy strategy, large numbers of autologous or allogeneic immune cells are injected into a patient to target cancer cells. In clinical trials, immune cell therapies using non‐genetically modified lymphokine‐activated killer cells (LAKs) and tumor‐infiltrating lymphocytes (TILs) showed relatively minor benefits [22]. Therefore, engineered immune cells have been created to express antigen‐specific T cell receptors (TCRs) or chimeric antigen receptors (CARs). Adoptive cell therapy (ACT) with T cells expressing CARs has been widely used to redirect autologous T cells’ specificity against lymphoid malignancies. While CAR‐T cell treatment has been demonstrated to be highly beneficial in individuals with hematologic malignancies, it has not been proven to be effective in patients with solid tumors. Furthermore, large‐scale usage of CAR‐T cells confronts various hurdles, including the generation of autologous products from patients, which takes several weeks to manufacture, and the inability to produce clinical doses of CAR‐T cells from T cell lymphopenia patients who have been highly pretreated [23]. In addition, graft‐versus‐host disease (GVHD) can occur even in human leukocyte antigen (HLA)‐matched patients when an allogeneic source is used [24]. Antigen escape, trafficking and tumor infiltration, immunosuppressive TME, toxicities [25], and sub‐optimal persistence and potency are some of the other constraints [26]. Although many innovative strategies to improve the function and to overcome the toxicity of CAR‐T cells are being explored [27, 28], an alternative approach is to use other immune cell types that are more efficient and safe, such as NK cells. Figure 1 shows blockade of immune checkpoints to improves anti‐tumor efficacy of NK cells. In this review, we discuss strategies for immune checkpoint blockade with a focus on CAR‐NK cells to redirect NK cells to cancer cells in the treatment of solid tumors.
FIGURE 1.
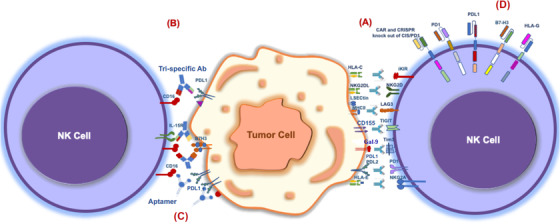
Blockade of immune checkpoints improves anti‐tumor efficacy of NK cells. NK cell dysfunction is caused by multiple factors in the tumor microenvironment (TME), as demonstrated by the upregulation of inhibitory checkpoint receptors and the downregulation of activating receptors on tumor‐infiltrating NK cells, resulting in the exhaustion of these cells. (A) Immune checkpoint inhibitors (ICIs), (B) Bi and tri‐specific natural killer cell engagers (BiKEs and TriKEs), (C) Aptamers, and (D) Chimeric antigen receptor (CAR)‐NK cells targeting immune checkpoints would restore NK cell antitumor activity. (The purple triangle and red circle on the tumor cell indicate tumor antigens [in part B and C]). Abbreviations: Ab, antibody; iKIRs, inhibitory killer cell immunoglobulin‐like receptors; NKG2A, natural killer group 2 A; TIM‐3, T cell immunoglobulin‐ and mucin‐domain‐containing molecule‐3; TIGIT, T cell immunoreceptor with Ig and ITIM domains; LAG‐3, lymphocyte activation gene‐3; PD‐1, programmed cell death protein 1; PD‐L1, programmed death‐ligand 1; HLA, human leukocyte antigen; CAR, chimeric antigen receptor; CRISPR, clustered regularly interspaced short palindromic repeats; CIS, cytokine‐inducible SH2‐containing protein; MHC‐II, major histocompatibility complex class II; IL‐15R, IL‐15 receptor; NKG2D, natural killer group 2 D.
2. WHAT ARE NK CELLS?
In 1976, NK cells were defined as a defense against pathogen invasion and malignant transformation [29]. These cells were initially thought to develop in the bone marrow (BM), but recent evidence suggests that they can also develop and mature in secondary lymphoid tissues, including tonsils, lymph nodes (LNs), and spleen [30]. These lymphocytes are located in the blood, BM, LNs, skin, gut, tonsils, liver, uterus (during pregnancy), and lungs [31]. NK cells are licensed and avoid attacking normal self‐cells through a process known as education, which helps them acquire functional maturation and self‐tolerance during development [32]. NK cells are CD3‒CD56+ lymphocytes in humans, whereas they are recognized in C57BL/6 and SJL mice by the presence of NK1.1 and natural cytotoxicity receptor 1 (NCR1 or NKp46), as well as CD49b but not CD3 [33] and in mice strains without NK1.1 expression (e.g., BALB/c mice), CD49b is used to identify NK cells [33, 34]. Human NK cells are further divided depending on the density of CD56 on the cell surface, which differ in phenotype, tissue placement, and immunological effects: CD56bright CD16dim NK cells (usually known as immature NK cells), constitute about 5%‐15% of total peripheral blood (PB) NK cells, preferably reside in secondary lymphoid organs, such as LNs, produce cytokines (especially interferon gamma (IFN‐γ)) and play a key role in immunomodulation; CD56dimCD16bright NK cells (also known as mature and cytotoxic NK cells), make up about 90% of NK cells in PB and primarily mediate NK cells immune function [35]. CD56 is not only a marker for NK cells, but it also plays a significant role in their final differentiation because its blockade by mAbs impedes the transition from CD56bright to CD56dim, decreasing their cytotoxic capacity [36]. Furthermore, almost all human NK cells express the activating receptor NKp46 [37, 38].
It is the balance of germline‐encoded activating and inhibitory receptors (without undergoing somatic recombination) that controls whether NK cells are activated or inhibited. The activation of NK cells is usually initiated in one of two ways: “missing‐self” recognition or “induced‐self” recognition [39, 40, 41, 42]. “Missing‐self” recognition occurs when the target cells downregulate or lose surface expression of major histocompatibility complex class I (MHC‐I) molecules, which is usually associated with virally infected cells or cellular transformation to evade T cell anti‐tumor immunity. This induces NK cell activation by reducing inhibitory signals from the MHC‐I‐binding killer cell immunoglobulin‐like receptors (KIRs) or CD94/natural killer group 2 A (NKG2A) in human and Ly49 receptors in mice. Subsequent studies have revealed that the absence of MHC expression is neither sufficient nor necessary for NK activation, and signaling from activating receptors is also required. “Induced‐self” recognition occurs when stress or virus‐related ligands on target cells engage with germ‐line activating receptors, resulting in activating signals.
Leukocyte immunoglobin‐like receptors (LIRs), NKG2A, and some KIRs are inhibitory receptors on the surface of NK cells that detect MHC‐I [43, 44, 45, 46, 47, 48]. KIRs, which belong to the immunoglobulin (Ig) superfamily, include inhibitory and activating receptors and detect classical HLA‐A, ‐B, and ‐C (HLA class Ia) in different ways [43, 44, 46]. LIRs, also known as immunoglobin‐like transcripts (ILTs), are Ig superfamily members that include both activating and inhibitory receptors [49, 50, 51]. From a total of 11 LIR members, 5 inhibitory receptors, including LIRB1‐5, have been identified [52]. Among them, LIRB1 (ILT2) and LIRB2 (ILT4), in addition to other ligands, recognize HAL‐G as their main ligand, resulting in immunological tolerance [50, 53]. NKG2A, another inhibitory receptor, belongs to the C‐type lectin family of receptors and is a member of the CD94/NKG2 family of receptors, which includes A, B, C, D, E, F, and H with activating or inhibitory potential. NKG2A pairs with CD94 to form the NKG2A/CD94 receptor and recognizes non‐classical MHC‐I molecule, HLA‐E, as the ligand [54]. In addition, emerging immune checkpoints such as CTLA‐4 and PD‐1 that are from the B7‐CD28 family of receptors, T cell immunoreceptor with Ig and ITIM domains (TIGIT), CD96, lymphocyte activation gene‐3 (LAG‐3), and T cell immunoglobulin‐and mucin‐domain‐containing molecule‐3 (TIM‐3) have been identified to mediate NK cell dysfunction in the TME [55]. On the other hand, dominant activating receptors of NK cells, including NCRs (NKp46, NKp30, and NKp44), NKG2D, and DNAX accessory molecule‐1 (DNAM‐1, also known as CD226), recognize ligands on the surface of virus‐infected or malignant cells [56, 57, 58]. Coreceptors such as 2B4 (CD244), NKp80, NTB‐A, and CD59 are also expressed and begin their function when co‐engaged with other activating receptors [57, 59].
The effector function of NK cells is similar to that of CD8+ cytotoxic T cells, but NK cells do not require prior antigen exposure or MHC class I limitation to carry out their functions [60]. When exposed to their targets, NK cells can cause cytotoxicity through a variety of mechanisms, including expression of Fas ligand (FasL) and tumor necrosis factor‐related apoptosis‐inducing ligand (TRAIL) [61], the release of preformed cytotoxic granules containing perforin and granzyme B [62, 63, 64], secretion of extracellular vesicles, such as exosomes, with cytotoxic potential [65], and participation in antigen‐dependent cell‐mediated cytotoxicity (ADCC) by detecting IgG Abs attached to the tumor cell surface by low‐affinity Fc activating receptor CD16 (or FcγRIII) [66]. NK cells are also polyfunctional and can secrete multiple cytokines and chemokines, such as IFN‐γ, tumor necrosis factor‐α (TNF‐α), granulocyte‐macrophage colony‐stimulating factor (GM‐CSF), IL‐5, IL‐10, IL‐13, macrophage inflammatory protein (MIP)‐1α, MIP‐1β, IL‐8, and CCL5 (RANTES) that help modulate other innate and adaptive immune cells function [67, 68, 69]. NK cells express cytokine receptors (including IL‐2R, IL‐12R, IL‐15R, IL‐18R and IL‐21R) that allow them to respond to cytokines secreted by other cells [70]. Also, other receptors, such as chemokine receptors (including CXCR1‐4 and CCR5) [71], and a diverse repertoire of pattern recognition receptors (PRRs), such as TLRs, are expressed by NK cells and play an important role in their migration and pathogen recognition, respectively [72]. NK cells have recently been revealed to have adaptive immune cell properties, especially memory‐like responses [73, 74, 75, 76, 77, 78, 79]. Figure 2 shows a schematic diagram of the types and functions of NK cells for killing tumors.
FIGURE 2.
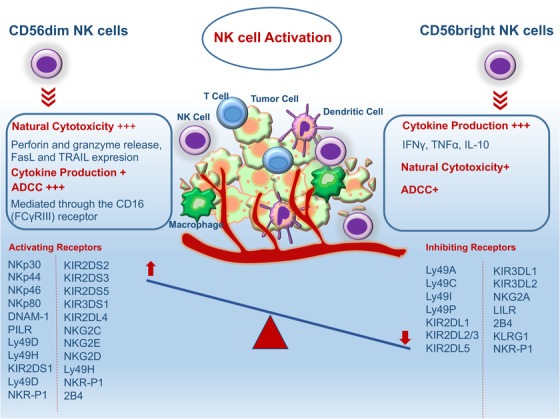
Schematic diagram of types and functions of NK cells for killing tumors. It is the balance of germline‐encoded activating and inhibitory receptors that control whether NK cells are activated or inhibited. Abbreviations: NK, natural killer; ADCC, antigen‐dependent cell‐mediated cytotoxicity; FasL, Fas ligand; TRAIL, tumor necrosis factor‐related apoptosis‐inducing ligand; KIR, killer cell immunoglobulin‐like receptor; NKG2, natural killer group 2; DNAM‐1, DNAX accessory molecule‐1; LILR, leukocyte immunoglobulin‐like receptors; KLRG1, killer cell lectin like receptor G1; IFN‐γ, interferon gamma; TNF‐α, tumor necrosis factor‐α.
In naive NK cells, ligation of individual activating receptors (except for CD16) is insufficient to activate them; cytokine exposure is necessary to preactivate them [80]. Cytokines such as IL‐2, IL‐15, IL‐12, IL‐18, IL‐21, and type I IFNs all play an important role in regulating NK cell activation, maturation, and survival. Improved NK cell performance has been achieved by administering cytokines in vivo or pre‐treating NK cells before adoptive transfer [81]. In NK cells, IL‐12 enhances signaling from activating receptors [82]. IL‐2 increases NKG2D expression [83], whereas IL‐15 is a powerful driver of NK cell differentiation and proliferation, and is shown to improve NK cell viability ex vivo when combined with IL‐2 [84]. The production of IFN‐γ by NK cells was demonstrated to be stimulated by IL‐18, which also provides co‐stimulatory activation [85], while the maturation of NK cells without promoting proliferation was enhanced by IL‐21 [86]. The proinflammatory cytokine type I IFN potentially leads to the pre‐activation of NK cells, readying them for activation by activating receptors [87]. Combination of IL‐15 with IL‐12 and IL‐18 results in significant biological alterations and a status reminiscent of memory NK cells, including improved function upon re‐stimulation, hyper‐responsiveness to IL‐2, increased IFN‐γ production and increased cytotoxicity [87, 88, 89].
3. TARGETING OF NK CELL‐BASED IMMUNE CHECKPOINTS BY mAbs
3.1. KIRs
Based on the number of extracellular Ig‐like domains and a cytoplasmic tail that dictate the function of the molecule, KIRs are divided into two categories: activating KIRs (aKIRs) and inhibitory KIRs (iKIRs). aKIRs have a shorter cytoplasmic tail than iKIRs and lack an immunoreceptor tyrosine‐based inhibitory motif (ITIMs). Binding of a positively charged lysine residue to the KARAP/DAP12 adapter molecule results in activated NK cell‐mediated lysis [90, 91]. iKIRs contain two (KIR2DL) or three (KIR3DL) extracellular Ig domains that confer specificity to HLAs. As a result, they can detect changes in HLA molecules caused by a viral infection or cancer [92]. iKIRs are upregulated in tumor malignancy, while aKIRs are downregulated in multiple tumors, such as breast cancer, lymphoma, and non‐small cell lung cancer (NSCLC) [93, 94]. These alterations reduce NK cell activation and anti‐cancer activity, allowing the tumor to evade immunosurveillance. The anti‐tumor activity of NK cells may be induced by mAbs that inhibit the interactions of iKIRs by blocking the signaling pathway. Thus, several anti‐KIR mAbs, including those targeting KIR2DL1‐3, have been evaluated in several clinical trials for treating patients with leukemia, lymphoma, multiple myeloma (MM) and some solid tumors [95]. Lirilumab (IPH2101, formerly called 1‐7F9) is a humanized IgG4 mAb that blocks KIR2DL1, KIR2DL2, KIR2DL3, KIR2DS1 and KIR2DS2 interacting with HLA‐C [96]. However, lirilumab's results in many clinical trials were disappointing. Increasingly, clinical trials of combination blocking strategies are being conducted.
3.2. CD94/NKG2A
Monalizumab is a humanized IgG4 Ab that blocks NKG2A binding to its HLA‐E ligand, which is overexpressed in tumor cells, as well as initiates an NK and CTL‐mediated immune response against cancer cells [97]. The findings of published studies from phase I clinical trial NCT02459301 show that monalizumab prescribed in patients with advanced gynecological cancers with minimal therapeutic toxicity is well tolerated [98]. In phase II of the NCT02643550 clinical trial, monalizumab and cetuximab, an epidermal growth factor receptor (EGFR) inhibitor, were tested in patients with squamous cell carcinoma of the head and neck. According to preliminary findings, the most common adverse effects were fatigue, fever, and headache, while the objective response rate was 31% [99]. In clinical trials, NCT05061550, NCT03822351, NCT03794544 and NCT03833440, the efficacy of monalizumab plus anti‐PD‐L1 blocking mAb (durvalumab) was studied. In several trials, monalizumab and anti‐PD‐L1 have demonstrated to have a synergistic effect on cancers that express HLA‐E and PD‐L1 [55].
3.3. TIM‐3
TIM‐3 is an inhibitory receptor of the TIM protein family that can detect multiple ligands. The extracellular regions of TIMs include a variable IgV domain that binds to the high mobility group box 1 (HMGB1) and galactin‐9 (Gal‐9). It also detects phosphatidylserine and carcinoembryonic antigen‐related cell adhesion molecule 1 (CEACAM‐1) [100, 101]. The intracellular domain of TIMs consists of 5 conserved tyrosine residues that interact with several components of the TCR complex [102]. TIM‐3 is considered a marker for the maturation or activation of NK cells because it is expressed on almost all mature CD56dimCD16+ NK cells. NK cell‐mediated cytotoxicity can also be inhibited by TIM‐3 cross‐linking. Upregulation of circulating and/or tumor‐infiltrating TIM‐3+ NK cells has been elevated in various types of malignant tumors such as gastric cancer, lung cancer, follicular B‐cell non‐Hodgkin lymphoma, and colorectal cancer [103, 104]. Anti‐TIM‐3 Abs such as Sym023, LY3321367, MGB453, TSR‐022, and BGB‐A425 alone or in combination with anti‐PD‐1 or anti‐LAG‐3 Abs in patients with advanced solid tumors (NCT02608268) are being evaluated. Previous studies have shown that increased TIM‐3 expression on NK cells improves cytotoxicity and IFN‐γ production [105, 106]. Therefore, due to the use of TIM‐3 blockade, there is a risk of decreased NK cell‐mediated cytolysis in pancreatic cancer cell lines, so caution is required [107]. Anti‐Gal‐9 Abs, on the other hand, reduce IFN‐γ production in NK cells in response to acute myeloid leukemia (AML) blast cells, complicating the effects of TIM‐3 inhibition [108]. Therefore, more studies need to be done to comprehend the role of TIM‐3 and its potential application in the management of NK cell‐mediated malignancy.
3.4. TIGIT and CD96
TIGIT and CD96 are inhibitory receptors that compete for PVR (CD155) and Nectin‐2 (CD112) binding with the activating receptor DNAM‐1 [100]. TIGIT and CD96 share the same ligand and are composed of an immunoreceptor tyrosine tail (ITT) and an ITIM. ITT‐like motifs play an important role in inhibiting signals. TIGIT interaction with CD155 induces phosphorylation via Fyn and Lck, which recruits SHIP1 in T cells and reduces signaling pathways in immune cells. Human NK cytotoxicity and cytokine production are also prevented by this interaction [100]. TIGIT and its ligands are highly expressed in a variety of cancers, and TIGIT expression has been demonstrated to decrease the activity of NK and CD8+ T cells in colorectal cancer patients as well as being linked to the depletion of tumor‐infiltrated NK cells in a mouse model of colon cancer [109]. CD155 is highly expressed in many types of tumor cells. TIGIT inhibition could help to reverse NK cell exhaustion and restore efficient anti‐tumor immunity. Moreover, NK cells in TME express lower levels of PD1 and CTLA‐4 in comparison to T cells. Therefore, it has been suggested that PD‐1 Ab treatment is less effective on NK cells than anti‐TIGIT blockade as a supplemental treatment because it improves tumor clearance and provides substantial immunity in tumor rechallenge models [109]. PVR expression, on the other hand, has been linked to a poor prognosis in many solid tumors, including colon, breast, lung, and pancreatic cancers. Accordingly, the PVR/TIGIT immune checkpoint axis has been proposed as a novel target for cancer immunotherapy, and several TIGIT‐targeting clinical trials are currently undergoing regulatory approval [110]. Tiragolumab, an anti‐TIGIT Ab, is currently being evaluated in combination with atezolizumab (anti‐PD‐L1) for use in small cell lung cancer (SCLC) (NCT04256421) and NSCLC (NCT04513925 and NCT03563716). In addition, two other anti‐TIGIT Abs, ociperlimab and domvanalimab, are being investigated in combination with tislelizumab (anti‐PD‐1) in phase III and zimberelimab (anti‐PD‐1) in phase II in solid malignancies, respectively (NCT04746924 and NCT04791839). The role of CD96 in NK cells is relatively less clear. One study found that using CD96 Ab‐mediated blockade increases IFN‐γ production by NK cells and controls cancer in a mouse model of metastatic lung tumor [111, 112, 113]. However, the efficiency of CD96 Ab‐mediated blockade on the performance of NK cells and their effects on human cancer patients remains unknown. Therefore, further studies are required to grasp its potential as a target molecule for immunotherapy.
3.5. LAG‐3
LAG‐3 is a member of the Ig family of receptors with inhibitory properties that are structurally similar to CD4; however, it binds to MHC class II molecules with a higher affinity than CD4. Fibrinogen‐like protein 1 (FGL1) is a recently identified ligand for LAG‐3 [114]. LSECtin, a member of the DC‐SIGN family, has also been described as a potential ligand for LAG‐3‐expressing immune cells [115]. LAG‐3 is expressed as an upregulated molecule on T and NK cells but is also expressed on other immune cells, including TILs, Treg cells, iNKT cells, B cells, and DCs [116, 117]. High expression of LAG‐3 has been indicated in patients with breast cancer, gastric cancer, B‐cell lymphoma, and lung cancer [118]. Because of LAG‐3's role in T cell exhaustion and Treg inhibitory activity, targeting it in combination with anti‐PD‐1 has a synergistic effect on T cell function restoration [119, 120]. However, the precise role of LAG‐3 in regulating NK cell function has yet to be determined, demanding further investigation. Notably, Abs that block the LAG‐3 pathway are not able to induce human NK cell cytotoxicity, or even soluble LAG‐3, able to bind to MHC‐II molecules, has no effect on the NK cells’ ability to kill cancer cells [121]. However, targeting LAG‐3 in immunotherapy may be beneficial because of its effect on the effective function of T and NK cells. Eftilagimod alpha (IMP321), a soluble version of the recombinant LAG‐3‐Ig fusion protein, activates human NK cells to produce IFN‐γ and TNF‐α in vitro and has been utilized as a safety aid for immunization against several diseases and malignancies. IMP321 has been investigated as a monotherapy for advanced renal cell carcinoma and in combination with chemotherapy for metastatic breast cancer in clinical trials [122, 123]. Relatlimab (BMS‐986016) is a new anti‐LAG‐3 blocking Ab that is currently being evaluated in phase I clinical trials, such as solid tumors and lymphomas (NCT03489369) or in combination with other ICIs such as nivolumab and ipilimumab in advanced cancer (NCT02488759) or either alone or combined with PD‐1 blockade, in various cancers, such as in patients with advanced solid tumors (NCT05134948 and NCT01968109).
3.6. PD‐1/PD‐L1
PD‐1 is expressed on various immune cells, including NK cells. PD‐1's cognate ligands are PD‐L1 (B7‐H1) and PD‐L2 (B7‐DC), which suppress immune cells such as T cells and NK cells, resulting in immunological escape [124]. PD‐1 expression in mature NK cells, CD56dim NKG2A−KIR+ CD57+, is found in one‐fourth of healthy people's PB but not in CD56bright NK cells [125]. PD‐1 overexpression on NK cells is related to NK cell dysfunction in cancer patients with ovarian cancer, Kaposi sarcoma and MM, presumably due to the presence of MHC‐deficient tumor cells [126, 127, 128]. In comparison to PD‐1− NK cells, PD‐1+ NK cells are functionally exhausted [129]. The blockade of PD‐L1 on hepatocellular carcinoma (HCC) cells did not significantly increase the cytotoxicity of hepatic intrasinusoidal (HI) NK cells. [130]. Also, in organoid culture, colorectal cancer cells were effectively killed by primary human‐activated NK cells, regardless of PD‐L1 expression [131]. On the other hand, anti‐PD‐1 inhibition may induce the production of cytokines like IFN‐γ, which may indirectly boost NK cells [59]. NK cell function is restored using anti‐PD‐1 mAbs. In mice, tumor‐infiltrating NK cells express PD‐1, and PD‐1 blocking in NK cells promoted an anti‐tumor immune response [132]. In post‐transplant lymphoproliferative disease (PTLD) patients, disruption of the PD‐1 pathway enhances IFN‐γ release but has no impact on cytotoxicity, demonstrating a relative dependence on the PD‐1 pathway and calling for additional research to determine the function of PD‐1 inhibition in NK cells [133]. PD‐1 blockade, on the other hand, has been demonstrated to improve NK cell‐based anti‐tumor responses. The safety and efficacy of NK cells in combination with anti‐PD‐1 or PD‐L1 are being evaluated in clinical trials NCT03841110, NCT04847466 and NCT03439501. Anti‐PD‐L1 Abs such as atezolizumab, avelumab, and durvalumab have been approved for use as monotherapy or in combination with other ICIs in the treatment of certain types of tumors. By overcoming T cell exhaustion, the use of an anti‐PD‐L1 mAb promises to increase the effectiveness of cancer treatment. However, the function of PD‐L1 on NK cells and the anti‐PD‐L1 mAb effects on PD‐L1+ NK cells remain unknown. One study found that NK cell function in some tumors was enhanced by induction of PD‐L1 by AKT signaling on NK cells and prevented cell exhaustion [134]. Also, another study found that NK cells induce ADCC against PD‐L1‐positive tumors in the presence of anti‐PD‐L1 mAbs, indicating the efficacy of NK cell immunotherapy in targeting cancers with high PD‐L1 expression in combination with anti‐PD‐L1 mAbs [135]. Avelumab is a fully human IgG1 anti‐PD‐L1 Ab, with the ability to induce ADCC, was evaluated in patients with non‐resectable advanced mesothelioma with or without PD‐L1 expression in phase I of the NCT01772004 clinical trial. The findings revealed that avelumab was both effective and safe in clinical trials. The results of the NCT01693562 clinical trial show that durvalumab has clinical benefits in enhancing NK cell‐specific genes expression, which leads to NK cell priming of the adaptive immunity response.
3.7. CD73
During hypoxia, apoptosis, and inflammatory response in TME, the levels of ATP and adenosine as a derivative of ATP are increased dramatically [136, 137]. CD39 and 5’‐nucleotidase CD73, which are highly expressed on a variety of cells, including T cells, tumor cells, stromal cells, Tregs, and MDSCs, mediate ATP catabolism in the tumor environment [138, 139]. Interestingly, NK cells in the TME achieve the CD73 molecule and contribute to immune suppression through adenosine production. In this state, adenosine accumulation inhibits NK cell activation by binding to A2ARs on the surface of NK cells [140]. Therefore, CD73 inhibition and A2A receptor blocking have been considered effective treatments to decrease tumor metastasis and improve survival. Recently, anti‐CD73 mAbs (oleculumab, NZV930) alone or in combination with other immunosuppressive drugs like anti‐PD‐1 and A2AR antagonists have been assessed in the treatment of different solid tumors in several phase I/II studies (NCT03381274, NCT03454451 and NCT03549000). However, the impact of anti‐CD73 treatment on NK cell function requires more detailed studies.
3.8. CD47
CD47 was first identified on the leukocytes’ surface as a part of the β3 integrin pathway. This molecule, which is also known as integrin‐associated protein (IAP), is a glycoprotein that belongs to the Ig superfamily [141]. In addition to integrins, CD47 interaction with thrombospondin‐1 (TSP‐1) and signal regulatory protein alpha (SIRPα) has been indicated in previous studies [142, 143]. The inhibitory function of CD47 is performed through engagement with SIRPα and TSP‐1 molecules, which is accompanied with suppression of phagocytic cells, T cells, and NK cells [144, 145]. Kim et al. [144] have shown that high CD47‐expressing head and neck squamous cell carcinoma (HNSCC) cell lines induce lower NK cell cytotoxicity compared to low‐expressing cells. Findings of another study have indicated the inhibitory effects of CD47 in NK cells are independent of SIRPα [146]. In several phase I and II clinical trials magrolimab (Hu5F9‐G4), an anti‐CD47 Ab, is under investigation in various combinations with other therapeutic agents such as cetuximab and atezolizumab [55]. Nevertheless, there are still unknown aspects worth investigating to deepen our understanding of the CD47 targeting effects on NK cells.
3.9. B7‐H3 (CD276)
The B7 homolog 3 protein (B7‐H3) is an immune checkpoint that regulates the function of T and NK cells [147]. Overexpression of B7‐H3 in several cancers has been indicated and is related to tumor metastasis and evasion [148]. Upregulation of B7‐H3 suppresses NK mediated cell lysis [149]. The expression of this molecule by the tumor vasculature has also been indicated in previous studies [150]. B7‐H3 has been proposed as a suitable target for cancer immunotherapy [148]. Several phase I/II clinical trials have been conducted to assess the impact of anti‐B7‐H3 in combination with other mAbs in different cancers. To improve the ADCC function of NK cells, B7‐H3 binding Fc‐optimized humanized IgG1 mAb (enoblituzumab) has currently been designed. It has been shown to suppress tumor development in B7‐H3 positive renal and bladder carcinoma [151]. In an ongoing dose‐escalating phase I study, the safety and efficacy of a Fc‐optimized humanized anti‐B7‐H3 mAb (MGA271) have been indicated [152]. The pharmacological characteristics of enoblituzumab and also its safety, efficacy, and anti‐tumor capacity are being assessed in a phase I study (NCT02982941). The safety, anti‐tumor effect, and immunogenicity of enoblituzumab given before radical prostatectomy are being evaluated in a phase II study (NCT02923180). Table 1 shows clinical studies of ICIs for the treatment of solid tumors.
TABLE 1.
Clinical studies of ICIs for the treatment of solid tumors
| Immune checkpoint | Clinical trials identifier | Phase | Status | Malignancy | Combinations | Ab/drug | Number enrolled |
|---|---|---|---|---|---|---|---|
| KIRs | NCT03203876 | I | Completed | Advanced and/or metastatic solid tumors |
Nivolumab Ipilimumab |
Lirilumab (BMS‐986015) | 10 |
| NCT01750580 | I | Completed | Advanced tumor | Ipilimumab | Lirilumab (BMS‐986015) | 22 | |
| NCT01714739 | I/II | Completed | Advanced solid tumors |
Nivolumab Ipilimumab |
Lirilumab (BMS‐986015) | 337 | |
| NCT03532451 | I | Active, not recruiting | Bladder cancer | Nivolumab | Lirilumab | 43 | |
| NCT03347123 | I/II | Terminated | Advanced or metastatic malignancies |
Epacadostat Nivolumab Ipilimumab |
Lirilumab | 11 | |
| NCT03341936 | II | Active, not recruiting | Squamous cell carcinoma of the head and neck | Nivolumab | Lirilumab | 29 | |
| CD94/NKG2A | NCT02459301 | I | Completed | Gynecologic cancer | Single | Monalizumab (IPH2201) | 59 |
| NCT02671435 | I/II | Completed | Advanced solid tumors | Durvalumab | Monalizumab (IPH2201) | 383 | |
| NCT02643550 | I/II | Active, not recruiting | Head and neck neoplasms |
Cetuximab Anti‐PD(L)1 |
Monalizumab (IPH2201) | 143 | |
| NCT05061550 | II | Recruiting | Non‐small cell lung cancer |
Durvalumab Oleclumab Chemotherapy |
Monalizumab (IPH2201) | 140 | |
| NCT05221840 | III | Recruiting | Non‐small cell lung cancer |
Durvalumab Oleclumab Placebo |
Monalizumab (IPH2201) | 999 | |
| NCT02331875 | I/II | Terminated | Squamous cell carcinoma of the oral cavity |
Standard Surgery Postsurgical Adjuvant Therapy |
Monalizumab (IPH2201) | 3 | |
| NCT03822351 | II | Active, not recruiting | Non‐small cell lung cancer |
Durvalumab Oleclumab |
Monalizumab (IPH2201) | 189 | |
| NCT03833440 | II | Recruiting | Non‐small cell lung cancer |
Durvalumab Oleclumab Ceralasertib Docetaxel |
Monalizumab (IPH2201) | 120 | |
| NCT03088059 | II | Recruiting | Squamous cell carcinoma of the head and neck |
Afatinib Palbociclib Durvalumab Niraparib |
Monalizumab (IPH2201) | 340 | |
| TIM‐3 | NCT03744468 | I/II | Recruiting | Advanced or metastatic solid tumors | Tislelizumab | BGB‐A425 | 162 |
| NCT03680508 | II | Recruiting | Liver cancer |
Dostarlimab (TSR‐042) |
Cobolimab (TSR‐022) | 42 | |
| NCT02608268 | I/II | Active, not recruiting | Advanced malignancies |
Decitabine Spartalizumab (PDR001) |
Sabatolimab (MBG453) | 252 | |
| NCT03961971 | I | Recruiting | Glioblastoma multiforme |
Anti‐PD‐1 Stereotactic radiosurgery |
Sabatolimab (MBG453) | 15 | |
| NCT03099109 | I | Active, not recruiting | Solid tumor | Anti‐PD‐L1 (LY3300054) | LY3321367 | 275 | |
| NCT03489343 | I | Completed | Solid tumor malignancies or lymphomas | Single | Sym023 | 24 | |
| NCT02817633 | I | Recruiting | Neoplasms |
Nivolumab Docetaxel Cisplatin Carboplatin Pemetrexed TSR‐033 TSR‐042 |
TSR‐022 | 369 | |
| TIGIT | NCT02794571 | I | Recruiting | Advanced/metastatic tumors |
Atezolizumab Carboplatin Cisplatin Pemetrexed Paclitaxel Etoposide Capecitabine Bevacizumab Pembrolizumab |
Tiragolumab | 660 |
| NCT04256421 | III | Active, not recruiting | Small cell lung cancer |
Atezolizumab Carboplatin Etoposide Placebo |
Tiragolumab | 490 | |
| NCT03119428 | I | Terminated |
Locally advanced cancer metastatic cancer |
Nivolumab | OMP‐313M32 | 33 | |
| NCT03563716 | II | Active, not recruiting | Non‐small cell lung cancer |
Atezolizumab Placebo |
Tiragolumab | 135 | |
| NCT03628677 | I | Active, not recruiting | Solid Tumor | Zimberelimab (AB122) | Domvanalimab (AB154) | 74 | |
| NCT05211895 | III | Recruiting | Non‐small cell lung cancer |
Durvalumab Placebo |
Domvanalimab (AB154) | 860 | |
| LAG‐3 | NCT00349934 | I | Completed | Metastatic breast cancer | Paclitaxel | Eftilagimod alpha (IMP321) | 33 |
| NCT03252938 | I | Recruiting | Solid Tumors | Avelumab | Eftilagimod alpha (IMP321) | 45 | |
| NCT04811027 | II | Recruiting | Squamous cell carcinoma of the head and neck | Pembrolizumab | Eftilagimod alpha (IMP321) | 154 | |
| NCT03625323 | II | Active, not recruiting |
Non‐small cell lung cancer Squamous head and neck cancer |
Pembrolizumab | Eftilagimod alpha | 189 | |
| NCT02614833 | II | Completed | Metastatic breast carcinoma |
Paclitaxel Placebo |
Eftilagimod alpha | 242 | |
| NCT02996110 | II | Active, not recruiting | Advanced renal cell carcinoma |
Nivolumab Ipilimumab BMS‐986205 BMS‐813160 |
Relatlimab | 155 | |
| NCT02750514 | II | Terminated | Advanced non‐small cell lung cancer |
Nivolumab Ipilimumab Dasatinib BMS‐986205 |
Relatlimab | 295 | |
| NCT02935634 | II | Active, not recruiting | Advanced gastric cancer |
Nivolumab Ipilimumab BMS‐986205 Rucaparib |
Relatlimab | 190 | |
| NCT04095208 | II | Recruiting |
Soft tissue sarcoma adult Advanced cancer |
Nivolumab | Relatlimab | 67 | |
| NCT03489369 | I | Completed | Solid tumor Lymphoma | Single | Sym022 | 15 | |
| NCT03044613 | I | Active, not recruiting | Gastro/esophageal cancer |
Nivolumab Carboplatin Paclitaxel Radiation |
Relatlimab | 32 | |
| NCT03623854 | II | Recruiting | Advanced chordoma | Nivolumab | Relatlimab | 20 | |
| NCT03459222 | I/II | Recruiting | Solid tumor |
Nivolumab Ipilimumab BMS‐986205 |
Relatlimab | 255 | |
| NCT03493932 | I | Active, not recruiting | Glioblastoma | Nivolumab | Relatlimab | 20 | |
| NCT02658981 | I | Active, not recruiting | Glioblastoma, recurrent brain neoplasm | Nivolumab or urelumab | Relatlimab | 63 | |
| NCT02966548 | I | Active, not recruiting | Solid tumor | Nivolumab | Relatlimab | 35 | |
| PD‐1 | NCT03590054 | I | Active, not recruiting | Advanced solid tumor | Abexinostat | Pembrolizumab | 42 |
| NCT03815084 | I | Unknown | Solid tumor | DC‐NK Cells | Pembrolizumab | 100 | |
| NCT03841110 | I | Recruiting | Advanced solid tumor |
iPSC‐derived NK Cells Nivolumab Atezolizumab Cyclophosphamide Fludarabine IL‐2 |
Pembrolizumab Nivolumab |
37 | |
| NCT02660034 | I | Completed | Solid tumor |
BGB‐290 (PARP inhibitor) |
Tislelizumab | 229 | |
| NCT03707808 | I | Completed | Solid tumor |
CD1c (BDCA‐1)+myeloid DC Ipilimumab Avelumab |
Nivolumab | 9 | |
| NCT03958097 | II | Unknown | Non‐small cell lung cancer | NK Cells | PD‐1 Ab | 20 | |
| NCT04080804 | II | Recruiting | Head and neck squamous cell carcinoma |
Relatlimab (anti‐LAG‐3 Ab) Ipilimumab (anti‐CTLA‐4 Ab) |
Nivolumab | 60 | |
| NCT02913313 | I/II | Recruiting | Advanced solid tumor |
Ipilimumab (anti‐CTLA‐4 Ab) BMS‐986207 (anti‐TIGIT Ab) |
Nivolumab | 241 | |
| NCT04047862 | I | Recruiting | Advanced solid tumor |
BGB‐A1217 (anti‐TIGIT Ab) Pemetrexed Paclitaxel Nab paclitaxel Carboplatin Cisplatin Etoposide 5fluorouracil Oxaliplatin Capecitabine |
Tislelizumab | 444 | |
| NCT02857166 | I | Completed | Advanced solid tumors | Single | Toripalimab | 24 | |
| NCT03374007 | I | Recruiting | Solid tumor lymphoma | Single | Geptanolimab(GB226) | 72 | |
| NCT03468751 | I | Unknown | Solid tumors | Single | HLX10 | 30 | |
| NCT03474640 | I | Active, not recruiting | Advanced malignancies | Single | Toripalimab | 198 | |
| NCT02715284 | I | Recruiting | Advanced solid tumors | Single | Dostarlimab (TSR‐042) | 740 | |
| PD‐L1 | NCT03518606 | I/II | Active, not recruiting | Advanced solid tumors |
Tremelimumab Metronomic vinorelbine |
Durvalumab | 150 |
| NCT03430466 | II | Terminated | Breast cancer |
Tremelimumab Fulvestrant |
Durvalumab | 1 | |
| NCT03084471 | III | Active, not recruiting | Advanced solid tumors | Tremelimumab | Durvalumab | 867 | |
| NCT03217747 | I/II | Active, not recruiting | Advanced malignancies |
Utomilumab PF‐04518600 (Ivuxolimab) Radiation Therapy |
Avelumab | 173 | |
| NCT02862275 | I | Recruiting | Active hematologic or solid tumor malignancies | Single | Atezolizumab | 40 | |
| NCT03212404 | I | Recruiting | Advanced cancers | Single | CK‐301 | 500 | |
| NCT03588650 | I | Completed | Advanced solid tumors | Single | HLX20 | 30 | |
| NCT03275597 | I | Active, not recruiting | Non‐small cell lung cancer |
Tremelimumab Stereotactic body radiotherapy |
Durvalumab | 17 | |
| CD73 | NCT03381274 | Ib/II | Active, not recruiting | Non‐small cell lung cancer |
Osimertinib AZD4635 |
Oleclumab | 43 |
| NCT03454451 | I/Ib | Active, not recruiting | Advanced cancers |
Ciforadenant Pembrolizumab |
CPI‐006 | 378 | |
| NCT03549000 | I/Ib | Active, not recruiting | Advanced malignancies |
PDR001 NIR178 |
NZV930 | 127 | |
| CD47 | NCT02953782 | Ib/II | Completed | Solid tumors and advanced colorectal cancer | Cetuximab | Magrolimab (Hu5F9‐G4) | 78 |
| NCT03869190 | Ib/II | Recruiting | Urothelial carcinoma |
Atezolizumab Enfortumab Vedotin Niraparib Hu5F9‐G4 Tiragolumab Sacituzumab Govitecan Tocilizumab Cisplatin Gemcitabine |
Magrolimab (Hu5F9‐G4) | 645 | |
| B7‐H3 | NCT02923180 | II | Active, not recruiting | Localized intermediate and high‐risk prostate cancer | Single | Enoblituzumab (MGA271) | 33 |
| NCT02982941 | I | Completed | Malignant solid tumors | Single | Enoblituzumab | 25 |
Abbreviations: KIRs, killer cell immunoglobulin‐like receptors; NKG2A, natural killer group 2 A; TIM‐3, T cell immunoglobulin‐ and mucin‐domain‐containing molecule‐3; TIGIT, T cell immunoreceptor with Ig and ITIM domains; LAG‐3, lymphocyte activation gene‐3; PD‐1, programmed cell death protein 1; PD‐L1, programmed death‐ligand 1; NK, natural killer; iPSCs, induced pluripotent stem cells; PARP, poly (ADP‐ribose) polymerase; Ab, antibody.
4. BiKEs AND TriKEs
The GTB‐4550 TriKE molecule (CD16a/IL‐15/PD‐L1) is being investigated in preclinical studies for application in the treatment of solid cancers. This TriKE specifically targets PD‐L1+ tumors, and the IL‐15 part of it enhances NK cell activation and proliferation [153]. The preclinical studies of CD16/B7‐H3 BiKE [154] and CD16a/IL‐15/B7‐H3 TriKE [153] are also ongoing. Additionally, a humanized TriKE (EGFR, CD16a, and PD‐L1) with a shared light chain has recently been developed [155]. Therefore, in addition to activating NK cells and inhibiting inhibitory molecules, this TriKE will likely impede the growth and survival of tumor cells.
5. APTAMER
Passariello et al. [156] have investigated the impact of anti‐CTLA4 Ab (ipilimumab) and anti‐EGFR CL4 nuclease‐resistant RNA‐aptamer combinatorial treatment on the anti‐cancer function of immune cells, including NK cells. They constructed the Fc region of ipilimumab mAb with the amino‐terminated CL4 aptamer and produced an immunoconjugate (CL4‐ipilimumab). CL4‐ipilimumab activated NK cells and induced IL‐2 and IFN‐γ production by these cells more efficiently than the parental mAb used alone or in combination with the CL4 aptamer, according to their findings. Also, NK cells’ cytotoxic function against SK‐BR‐3 tumor cells increased in the presence of CL4‐ipilimumab with a higher lactate dehydrogenase (LDH) release by tumor cells. They have suggested that expression of CTLA‐4 affects the anti‐tumor activity of NK cells and CL4‐ipilimumab restores it. In another study, Zhang et al. [157] produced dual aptamer‐equipped NK cells (T‐P NK cells) by using TLS11a aptamer targeting HepG2 cells (hepatocellular carcinoma cell line) and PD‐L1‐specific aptamer without genetic modification. They observed significant cytotoxicity against HepG2 cells following treatment with T‐P NK cells and hypothesized that the ease of NK cell‐target interaction likely boosts the NK cells’ capacity to identify the target cells. Simultaneously, PD1‐PD‐L1 interaction blockade by PD‐L1 aptamer improves NK cell cytotoxicity. They tracked T‐P NK cells in tumor‐bearing mice and found significant tumor penetration and accumulation of T‐P NK cells. Moreover, increased IFN‐γ production by T‐P NK cells enhanced the expression of PD‐L1 on cancer cells. To improve NK cell migration into the tumor environment, CP‐bi‐apt, a stable CD16/PD‐L1 bi‐specific aptamer, has been designed. CP‐bi‐apt has increased NK cell cytotoxicity and promoted NK cell accumulation in the tumor site [158]. Bi‐apts seem to be effective in cancer immunotherapy for activating NK cells, but more preclinical and trial research is needed before aptamers can be used as immunotherapy drugs.
6. CAR‐NK CELL THERAPY
While NK cell therapy has a lot of benefits, it also has a number of drawbacks that restrict its effectiveness. NK cells have a short lifespan without cytokine assistance, lasting only one to two weeks. Ex vivo expansion and activation of a limited number of cells is also required, and NK cells may be vulnerable to the immunosuppressive TME, which could limit their effector function and trafficking. Exosome shedding, proteolytic cleavage, internalization and degradation, decreased transcript stability, decreased translation, differential glycosylation and lipidation, increased intracellular retention, impaired protein maturation and refolding, and affecting splicing and alternative adenylation are all pathways by which tumors reduce activating receptor ligand expression [159]. Tumors can potentially evade the function of NK cells by upregulating MHC, which engages their inhibitory receptors [160]. Engineering advancements have helped to overcome some of these constraints. One of these, genetic alterations of NK cells with CAR constructs, has received a lot of attention recently.
CAR is an artificially modified fusion protein. It can recognize antigens in the absence of MHC presentation, unlike innate TCRs [161]. CAR‐NK cells have extracellular antigen recognition domain, hinge, transmembrane (TM) and intracellular domains just like CAR‐T cells. T cells that have been genetically engineered to produce CAR can directly detect the CAR‐targeted antigen, enabling them to activate, proliferate, release cytokines, and exhibit cytotoxicity against tumor cells that express the CAR‐specific antigen.
Single‐chain variable fragments (scFvs), which include variable sections of both the light and heavy chains of a target‐reactive Ig, are separated by a flexible linker in the extracellular binding domain of a CAR [162]. Alternative binding domains, such as ligands (ICAM‐1‐specific CAR‐T cells built using the I domain derived from LFA‐1) [163], physiological receptors (chimeric PD‐1 receptor) [164], peptides (using NK1 as a targeting moiety for generating MET‐specific CAR) [165], nanobodies (single domain antibodies (VHH)), [166] and DARPins (designed ankyrin repeat proteins) are still being investigated in preclinical studies [167]. These recognition sequences establish CAR specificity and affinity for target antigens. Evidence suggests that the antigen binding affinity and stability of the construct are affected by the order of the variable fragments and the length of the linker. Both epitope location and abundance should be considered while constructing scFv [168]. The CAR's hinge domain connects the extracellular domain to the TM domain, providing it enough orientation and flexibility to bind to tumor antigens and influence CAR‐NK cell activity. The size of the hinge area has been shown to affect the CAR‐T cell function. Longer hinges allow for more flexibility in membrane‐proximal antigens, whereas shorter hinges suffice for membrane‐distal antigens [169, 170]. According to several studies, CAR‐T cells with a shorter hinge region had more anti‐tumor efficacy [171, 172]. A TM domain is located between the hinge and the intracellular signaling domain, which is required for surface expression and stability of the receptor. The structure of the intracellular signaling domain, which consists of immunoreceptor tyrosine‐based activation motifs (ITAMs), determines the intensity of the CAR‐NK activation signal. The backbone of all CAR generations is the same.
Based on the structure and composition of the endodomain, the evolution of CARs during the last three decades can be approximately divided into five generations [173]. In the first generation of CARs, a single CD3ζ intracellular domain was presented. Early experiments of first‐generation CAR‐T cells demonstrated low cytotoxicity and proliferation due to the lack of co‐stimulatory (e.g., CD27, CD28, OX40 [CD134], 4‐1BB [CD137]) and cytokine (e.g. IL‐2) signaling [173, 174]. The second generation of CARs was generated by adding a costimulatory domain to the intracellular signaling domain, such as parts of CD28 or 4‐1BB, to increase T cell proliferation and cytotoxicity [175, 176, 177]. A third intracellular signaling sequence with an additional co‐stimulatory domain, such as OX40 or 4‐1BB, was added to the third generation of CARs to improve on the second generation [173]. In contrast to the second‐generation CAR, there is no consistent evidence that performance has increased in the third‐generation CAR. The CAR‐mediated effector functions can be dramatically influenced by the type of co‐stimulatory domain used. Co‐activation of 4‐1BB leads to the formation of T memory cells, whereas co‐activation of CD28 leads to enhanced T cell activation and expansion [178]. T cells redirected for universal cytokine‐mediated killing (TRUCKs), the fourth generation of CARs, are based on the second generation, but they release transgenic cell products that are expressed either constitutively or inducible when the CAR is activated to induce tumor death [179, 180]. A fifth generation of CARs is currently being investigated; these are based on the second generation of CARs but have a truncated cytoplasmic IL‐2 receptor β (IL‐2Rβ)‐chain domain with a STAT3 binding site, which effectively provides all three synergistic signals needed for complete T cell activation and proliferation [181]. SUPRA CAR, Tandem CAR, Dual CAR, split CAR, and SynNotch CAR are among the newer generations of CARs [182, 183].
6.1. CAR‐NK cell advantages in tumor immunotherapy
The use of allogeneic haploidentical NK cells in ACT has been clinically proven to be safe, without the risk of causing GVHD. Using NK cells, an increased graft‐versus‐tumor (GvT) response has been seen after hematopoietic stem cell transplantation (HSCT) for AML [184, 185]. T cells have been proven to be less effective at driving CARs than NK cells. CAR‐expressing NK cells are deemed safer in clinical application than CAR‐T cells, as evidenced by the results of many clinical trials, and NK cell immunotherapy is a safe and practical therapeutic method [186]. In some phase I/II trials, allogeneic NK cell administration has been well tolerated and has not caused GVHD or other severe unwanted events [187, 188, 189] if the infused product is sufficiently depleted of T cells, indicating that NK cells are general CAR drivers that are not restricted to autologous cells. Unlike CAR‐T cells, CAR‐NK cells have a short lifespan in circulation; therefore, there is a low chance of on‐target/off‐tumor damage to normal organs [190]. NK cells also generate different cytokines from T cells. When stimulated, they produce IFN‐γ and GM‐CSF, whereas CAR‐T cells produce proinflammatory cytokines such as TNF‐α, IL‐1, and IL‐6 [191, 192]. While their anti‐cancer activity is based on the CAR‐specific mechanism, which involves the identification of tumor‐related antigens via scFv, NK cells also eliminate malignant cells by detecting a variety of ligands through their receptors like NCRs (NKp46, NKp44, and NKp30), NKG2D, and DNAM‐1 [193]. In addition, CD16 facilitates ADCC by NK cells [194]. CAR‐modified NK cells are therefore capable of effectively eliminating heterogeneous tumors in which some tumor cells do not express CAR‐targeted antigens through both CAR‐dependent and NK cell receptor‐dependent mechanisms. The CAR‐NK cells do not need to be matched to the patient and can be derived from autologous or allogeneic sources, including PB [195], BM‐derived hematopoietic progenitor cells (HPCs) [196], induced pluripotent stem cells (iPSCs) [197], human embryonic stem cells (hESCs) [198], umbilical cord blood (UCB) [199], post‐partum placenta [200], NK cell lines (such as NK‐92) [201] or memory‐like NK cells [87], each with its own advantages and disadvantages (Figure 3). Compared to the manufacturing of autologous CAR‐T cells, the development of “off‐the‐shelf” allogenic NK cells takes less time. Additionally, CAR‐NK therapy can be used to treat solid tumors more effectively because of its lower expression of PD‐1 [202]. The resistance of cancer stem cells (CSCs) to chemotherapy, radiotherapy, and immunotherapy can lead to relapses and metastasis. So, since CSCs are quiescent and express low levels of MHC‐I, NK cells can target and kill them by default [203, 204].
FIGURE 3.
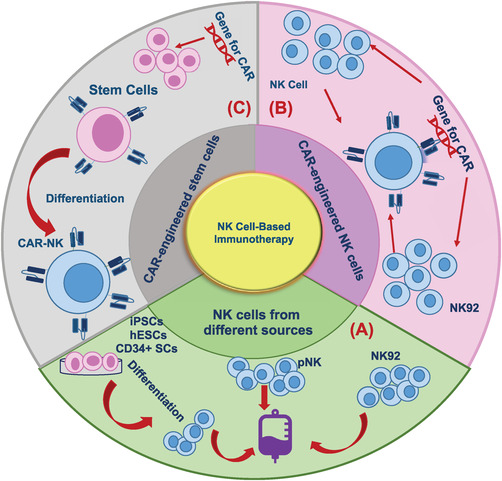
The schematic diagram of the various sources for CAR‐NK cell production. (A) NK cells from peripheral blood mononuclear cells (PBMCs), umbilical cord blood (UCB), post‐partum placenta, and memory‐like NK cells are used in NK therapy. It is also possible to differentiate NK cells from CD34+ hematopoietic progenitor cells (HPCs). It has recently become more attractive to use induced pluripotent stem cells (iPSCs) for NK cell products. (B) Production of CAR‐engineered NK cells by delivery of CAR construct gene into NK cells and NK‐92 cell lines. (C) A variety of CAR‐expressing vectors can be used to genetically engineer stem cells, and these cells can be differentiated into CAR‐NK cells. Abbreviations: NK, natural killer; CAR, chimeric antigen receptor; iPSCs, induced pluripotent stem cells; hESCs, human embryonic stem cells; SCs, stem cells; pNK, peripheral blood NK.
6.2. NK cell sources of CAR generation
Autologous NK cells activated by IL‐2 were employed in early trials of NK cell immunotherapy. Autologous NK cell infusions in combination with IL‐2 were examined in other trials after high‐dose chemotherapy conditioning [205, 206]. However, this approach led to poor clinical outcomes due to the inhibition of autologous NK cells because ligands presented by self HLA molecules on tumor cells bound to KIR on NK cells [205, 206]. Additionally, IL‐2 induces the expansion of Tregs, which can inhibit NK cell expansion indirectly by depriving them of IL‐2 or directly by inhibiting NK cell function in a transforming growth factor (TGF)‐β‐dependent manner [207, 208]. Endogenous NK cells may not retain sufficient cytotoxicity against cancer cells because of the overall immunosuppressive tumor milieu with impaired functions [209]. Another aspect contributing to this limitation could be the difficulty of obtaining pure and enhanced expansion of autologous NK cells from cancer patients. Furthermore, autologous NK cells are vulnerable to the presence of resistant tumor cells, allowing tumor cells that are resistant to autologous NK cells to survive and develop [210]. Patients who received NK cell infusions were heavily pretreated before their cells were collected and used, which might have adversely affected their function and expansion [211]. While it is also possible that anti‐KIR Abs may block KIR‐HLA matching in order to increase NK function [96], the transfer of allogeneic or haploidentical NK cells with KIR ligand‐mismatch prevents this suppression due to the “missing self” recognition of tumor cells [184]. Allogeneic NK cells may also be less likely to be rejected by the recipient's alloreactive T cells.
6.2.1. PB‐NK cells
The PB‐NK cells are phenotypically mature and highly functional. However, they need blood or leukapheresis from a healthy person who is willing to donate. Even though PB‐NK cells are readily available, their low transduction efficiency, coupled with poor expansion, limits their application [212]. However, this source has certain limitations. The process of selecting an appropriate allogeneic NK cell donor is time‐consuming, which may cause the patient's treatment to be delayed. The number of peripheral blood mononuclear cells (PBMCs) that can be extracted and the proportion of NK cells contained in the PBMCs vary considerably among donors, all of which can have an impact on the amount of purified NK cells available for transplant. Furthermore, NK cell phenotypes and functions differ between donors (depending on age, sex, weight, and other factors), reducing consistency in the expansion rate and final product of NK cells.
6.2.2. UCB‐NK cells
Another interesting source of NK cells, UCB, has a number of advantages, such as its ready availability “off‐the‐shelf”. UCB collection is easy and harmless to the mother or baby, and they are almost never infected with Epstein‐Barr Virus (EBV) or cytomegalovirus (CMV). UCB is easily frozen [213]. The benefit of UCB banks is the ability to select donors with certain HLA types and specific NK receptor profiles. UCB also has fewer T cells than PB, and these cells have a naive phenotype, lowering the risk of GVHD from contaminated T cells infused into patients via an NK cell product infusion [214]. The NK cells derived from the UCB are younger and have a greater potential for proliferating than those derived from the PB [215]. By using feeder cells and cytokines, UCB‐NK cells can be expanded and genetically modified to express CARs. Because of their higher proliferative capacity, NK cells from this source are easier to engineer, as demonstrated in the first reported clinical trial of CAR‐NK cells [216]. In UCB, NK cells comprise up to 30% of lymphocytes [217], with a higher proportion of CD56bright cells and hence inferior cytotoxic capabilities [215, 217, 218]. Unexpanded UCB‐derived NK cells have some limitations, including availability in low numbers due to the UCB unit's small volume and immature function [217]. UCB‐NK cells express lower levels of certain adhesion molecules, along with KIRs, CD16, perforin, and granzyme B, compared to PB‐NK cells, as well as higher levels of inhibitory molecules, including NKG2A [219]. One study used Good Manufacturing Practice (GMP)‐grade K562‐based artificial antigen‐presenting cells (aAPCs) that express membrane‐bound IL‐21 and 4‐1BB ligand to develop a GMP‐compliant procedure that reliably produces therapeutically relevant amounts of GMP‐grade NK cells from a CB unit for the purposes of adoptive immunotherapy [199]. When activated and expanded ex vivo, CB‐derived NK cells express the full array of activating and inhibitory receptors, strongly express eomesodermin (Eomes) and T‐bet, two factors needed to mature NK cells, and have cytotoxic properties similar to PB‐NK cells [220, 221]. However, unlike PBMCs, CAR‐NK cells produced from UCBs are not homogeneous, making standardisation challenging [222].
6.2.3. NK cell lines
In most clinical trials with CAR‐NK cells, the NK‐92 cell line is used because it shows clinical benefits and has minimal side effects, and it is the only one that has been approved for clinical application [223, 224]. It demonstrates unlimited proliferation in vitro and is less sensitive to repeated freeze/thaw cycles [225]. These properties will make it easier and cheaper to manufacture “off‐the‐shelf” CAR‐NK products for clinical use. Furthermore, they are a genetically modifiable NK cell population. In addition, NK‐92 cells have higher transduction efficiency than primary NK cells [226, 227]. However, this cell line has some limitations: neither CD16 (are hence unable to trigger ADCC) nor NKp44 are expressed due to their tumor cell line origin, they must be irradiated before infusion and they can't be expanded in vivo, owing to the lethal irradiation required before their infusion [228], rendering them unsuitable for CAR‐NK cell therapy. Genetically modifying NK‐92 cells to express the high‐affinity V158 variant of the FcγRIIIa/CD16a (termed haNKTM) and endogenous, intracellularly retained IL‐2 helped overcome these cytotoxic limitations [224, 229]. NK‐92 also lacks some KIRs, with the exception of KIR2DL4 (CD158d), which may promote GVHD [230, 231, 232, 233]. A recent study has suggested that the host PBMCs might show cytotoxicity against CAR‐transduced NK‐92 cells [234]. Moreover, they are EBV‐positive and exhibit multiple cytogenetic abnormalities similar to those of NK lymphoma [235]. While repeated infusions of irradiated NK‐92 cells are possible and may be utilized to overcome their short‐term persistence, such an approach is likely to result in the production of Abs and cellular immunity against the allogeneic cell line, as well as faster rejection with each infusion. Since IL‐2 is essential to the growth and survival of these cells in culture, it must be accompanied by any additional stimulation factors [225, 236]. NK‐92MI cells were created by genetically engineering NK‐92 cells to produce soluble IL‐2, thus eliminating the need for exogenous IL‐2. In addition to NK‐92 cells, other human cell lines have also been evaluated as alternatives, such as NKL, KHYG‐1, YTS or NKG [237]. Irradiation reduces the anti‐tumor potency of this product when compared to NK cells from other sources, and it may not be clinically relevant.
6.2.4. NK cells derived from stem cells
NK cell products derived from UCB stem cells, hESCs and iPSCs are currently being tested in clinical trials. Because ESCs are more difficult to obtain and their applications raise ethical concerns, they are used less frequently than CB stem cells and iPSCs. It is possible to obtain large numbers of NK cells by differentiating them from CD34+ HPCs. This approach provides virtually unlimited numbers of homogeneous NK cells, as well as the advantage of allowing their genetic manipulation over primary NK cells. CD34+ HPCs can be isolated from BM, mobilized PB, or UCB, expanded and then differentiated into mature NK cells in a culture system using a cocktail of cytokines [238]. As a result, a more homogeneous and well‐defined NK cell product can be produced. Although PB apheresis after granulocyte colony‐stimulating factor (G‐CSF) stimulation is a well‐established alternative to extracting CD34+ progenitors from BM, it may influence the NK cell phenotype [239]. It was previously common to use BM CD34+ cells for the generation of NK cells, but UCB CD34+ cells have become more frequently used as UCB is easier to obtain and contains higher levels of HSCs [240, 241, 242, 243, 244, 245]. In this way, various combinations of growth factor and cytokine mixtures, BM stroma cells, and culture media with or without animal or human serum were used in the different studies. oNKord, an allogeneic partial HLA‐matched NK cell product derived from UCB CD34+ progenitors, has been designated as an orphan medication by the European Medicines Agency (EMA) and the Food and Drug Administration (FDA) for treating AML patients who are not candidates for allogeneic stem cell transplantation. CD56+CD3− NK cells produced from this process are largely similar to PB‐NK cells, expressing activating receptors, and exhibiting potent cytotoxicity against leukemia cells in vitro and in vivo [246]. On the other hand, it has been shown that while CD34+ cells from CB can be employed to produce a homogeneous population of CD56+ NK cells on a large scale, they do not appear to be as mature as PB‐NK cells [238] and have little ADCC activity [247] even after expansion, necessitating additional measures to improve potency for clinical application. In addition, the differentiation process is labor‐intensive, requiring frequently changing media. In order to develop clinically applicable NK cell expansion protocols, shorter expansion times and less operator involvement are likely to be preferred.
PSCs, such as ESCs and iPSCs, have the ability to self‐renew while maintaining pluripotency and could provide limitless supplies for target cells [248]. It has been established that clinical‐scale NK cell generation can be achieved using hESC or iPSC [198, 249, 250]. The use of commonly available hESC/iPSC cell lines is often preferred to the use of stem cells from BM biopsy, G‐CSF mobilization, or human embryos [251, 252]. To differentiate these cell sources into NK cells, xenogeneic stromal feeder cell lines [253] or human spin embryoid bodies (EBs) [197, 254, 255] are required, which eliminate xenogeneic cells for more defined NK cell development conditions and can therefore be scaled up clinically. hESC/iPSC were centrifuged to form spin EBs [256], which gave rise to HPCs expressing CD34 and CD45, which were then differentiated into mature NK cells by using a specific cytokine cocktail. The choice of target donor somatic cell type and the reprograming protocol, which includes the nature and combination of genes as well as the method used to deliver transcription factors into somatic cells, have an impact on the pluripotency and differentiation abilities of reprogramed cells in iPS cells [257]. NK cells derived from iPSC/hESC expressed common NK cell markers, such as KIRs, CD16, NKp44, NKp46, NKG2D, and TRAIL, and were cytotoxic against several hematological and solid tumor cells in vitro [198, 249]. NK cells produced from iPSCs or hESCs combine the advantages of PB‐NK and NK‐92 cells, as they have a similar phenotype to PB‐NK cells and are a homogeneous population. NK cells derived from the H9 hESC cell line had a lower allogeneic immunological response, but they had a more mature cytotoxic profile than CB‐NK cells [249]. In contrast to PB‐NK cells, iPSC‐derived NK cells, similar to UCB‐NK cells, have an immature phenotype, with lower KIR and CD16 expression and higher NKG2A expression [197, 254, 258]. Because of their unlimited proliferation capacity, iPSCs have emerged as an attractive source of CAR‐NK cells [259]. Using iPSCs to produce NK cells allows for more gene modification, repeat dosing, and the generation of standardized products, allowing for more successful treatment of refractory solid tumors [260]. Another advantage of using iPSC‐NK products over iPSC‐T cell therapies is that iPSC‐NK products can be truly “off‐the‐shelf” because they don't require HLA matching between donors and patients.
There are still obstacles to overcome before iPSC‐NK cells can be safely employed to generate CAR‐NK for clinical usage. iPSC‐derived cells are always capable of malignant transformation and have the potential to be immunogenic, causing effector cell death or even adverse immunological reactions like cytokine release storms. The 5‐week biomanufacturing time for iPSC‐NK cells, on the other hand, could be a barrier to establishing and maintaining iPSC‐NK cell banks, especially if genetic modifications such as CARs are required. It would be useful to shorten and streamline the production process.
6.2.5. Memory‐like NK cells
Memory‐like NK cells were first discovered as a result of CMV infection. CMV was observed to cause the expansion of an NK cell subpopulation with CD94/NKG2C overexpression as well as decreased expression of PD‐1, TIGIT, and NKG2A [261, 262]. When activated, CD56dim/CD16bright/CD94/NKG2C cells showed enhanced proliferation, degranulation, and elevated IFN‐γ and TNF‐α production [263]. In addition to living longer in vivo, CMV‐induced adaptive memory NK cells can withstand the suppressive effects of Tregs and MDSCs [264, 265]. Incubation of NK cells with IL‐12, IL‐15, and IL‐18 for 16 hours stimulates the generation of memory‐like NK cells with abilities and features comparable to CMV‐induced cells [87]. On the other hand, IL‐12 has been shown to promote NKG2A expression and inhibit NK cell activation [266]. Cytokine‐induced memory‐like NK cells demonstrated an excellent safety profile, expanded in vivo, and resulted in remission in 44 percent of evaluable patients with AML in a phase I trial [89]. According to preclinical research, inserting CAR into a memory‐like NK cell improves its survival and cytotoxicity [267].
6.2.6. Post‐partum placenta
Placenta‐derived cells contain a proportion of NK cell progenitors with the ability to develop into NK cells during the maturation and expansion stages. After a 21‐day NK culture of placenta‐isolated NKs, an average of 1.2 × 109 NK cells with an 80% viability rate was obtained [200]. Derived placental‐NK cells are largely similar to UCB‐NK cells phenotypically and functionally. The FDA has approved CYNK‐001, an allogeneic “off‐the‐shelf” cell therapy enriched for CD56+/CD3− NK cells generated from human placental CD34+ cells, as an investigational new drug (IND) for the treatment of glioblastoma (GBM).
6.2.7. CAR‐engineered stem cells
A CAR can be expressed by transfecting stem cells with the genetic sequence. It is a newly developed method that allows stem cells to differentiate into CAR‐NK cells after being engineered. Stem and progenitor cells can be cryopreserved and have better transduction effectiveness than mature NK cells. Engineered stem cells could potentially produce a cellular product that is “off‐the‐shelf,” eliminating the necessity for a personalized, patient‐specific product. Performing gene engineering or gene editing on a small number of stem cells could reduce the amount of gene‐engineering/editing materials, which could be cost‐limiting, and allow for the most efficient gene engineering/editing, which could be passed down to subsequent divisions and final cell products. Therefore, there is no need to use viral vectors to transfect NK cells, avoiding high cell mortality from viral infection and uncontrollable variation of gene insertion and expression. Furthermore, the advancement of stem cell‐engineered cellular therapy will create new opportunities for product upscaling and banking. In addition, gene‐engineering technologies like clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 can be employed to boost the anti‐cancer properties of CAR‐engineered stem cells. The genetic changes will be evident in differentiated immune cells since these cells are non‐differentiated [258, 268]. One study successfully introduced CD19‐targeted CARs into CD34+ progenitors followed by NK differentiation. Gene transfer had no effect on hematopoietic differentiation and cell proliferation when transduced at 1‐2 copies/cell [269]. In media supplemented with stem cell factor (SCF), bone morphogenetic protein 4 (BMP4), and vascular endothelial growth factor (VEGF), genetically engineered iPSCs can stably differentiate into HPCs. A substantial number of highly homogeneous CAR‐NK cells may be produced for therapeutic usage after activation using aAPCs and stimulation with IL‐15, IL‐7, SCF, and FMS‐like tyrosine kinase 3 ligand (FLT‐3L) [197, 254, 258]. Only one CAR‐engineered iPSC cell is required for this approach to produce a large number of highly homogeneous CAR‐NK cell products for therapeutic usage. Li et al. [255] used a CAR genetic sequence to transduce iPSCs. In the xenograft model of ovarian cancer, iPSC‐derived CAR‐NK cells showed prolonged survival and better anti‐tumor efficacy than iPSC‐derived CAR‐modified T cells and non‐CAR‐bearing immune cells. In another study, researchers used zinc finger nuclease (ZFN) technology to genetically modify human iPSCs to insert a cDNA encoding an anti‐EpCAM‐CAR into the adeno‐associated virus integration site 1, a “safe harbour” for transgene insertion into the human genome, and then differentiated the modified iPSCs into CAR‐expressing iPSC‐derived NK (iNK) cells. The CAR‐iNK cells' lytic activity against NK cell‐resistant, EpCAM‐positive cancer cells was validated in an in vitro cytotoxicity experiment but not against EpCAM‐positive normal cells, confirming the CAR‐iNK cells' retained tolerability towards normal cells [270]. A recent study found that anti‐glypican 3 (GPC3) CAR‐modified NK/ILC cells made from iPSC had a strong effect against tumors and helped people with ovarian cancer live longer without causing acute systemic toxicity [271]. In a recent phase I trial, iPSC was employed for an “off‐the‐shelf” CAR‐NK product (called FT596) to treat CD19‐expressing B cell malignancies (NCT04245722). iPS cell‐derived NK cell products that have been genetically modified are being tested in people with different types of cancers (NCT03841110 and NCT04023071). In conclusion, it is not clear currently, which source is optimal for creating CAR‐NK cells with the most potent activity.
6.3. CAR constructs in NK cells
Most CAR‐NK cell research has so far relied on CAR constructs developed for CAR‐T cells. Novel CAR constructs for NK cells specifically have been developed. However, diverse CAR constructs have various effects on cytotoxicity and cytokine production in NK cells [272, 273]. T and NK cells share signal domains such as CD3ζ and 4‐1BB, but NK cells lack some costimulatory domains (e.g., CD28) and the CD8α TM domain. Both T and NK cells rely on CD3 for signaling and activation [274]. Despite the fact that CD28 is one of the most commonly used costimulatory domains in CAR‐T cells [275], its role in NK cell function is less clear [276]. Despite this, when it was combined with CD3ζ in a second generation ErbB2‐specific CAR‐NK‐92, it performed better than a CD3ζ construct alone and was similar to the 4‐1BB‐CD3ζ CAR against ErbB2‐expressing tumor cells [228]. Another study found that NK‐92 cells transduced with a CD19‐CAR that expressed CD28‐CD3ζ had better cytotoxicity against CD19‐positive targets than cells that expressed a 4‐1BB‐CD3ζ‐containing CAR [277]. Both primary NK cells and CAR‐NK cell lines use a CD8α hinge region. CAR‐NK cells also used other spacers, such as CD28, IgG Fc domains, and DAP12 [278]. Some of the most commonly used CAR‐NK TM parts have been adapted from CD3ζ, CD8 and CD28, but others have been explored as well (e.g., NKG2D, 2B4, DNAM‐1). Currently, CD8α and CD28‐derived TM are prevalent in primary CAR‐NK cells, while CD28 is the preferred TM region in CAR‐NK cell lines [278].
It is critical to build optimized CAR constructs based on NK cell‐specific signal activation properties in order to improve the activation and cytolytic capacities of NK cells. Typically, CAR‐NK cell studies use first‐generation CARs, which have a single signaling domain comprising CD3ζ, FcεR1γ, or DAP‐12 [279, 280, 281]. A second‐ and third‐generation CAR uses one or two costimulatory domains in conjunction with CD3ζ to improve cytotoxicity. NK cell‐specific adaptor molecules such as DAP12 have ITAMs. DAP10, as the NKG2D receptor's adaptor protein, has not ITAM and can enhance and stabilize NKG2D membrane expression. According to one study, anti‐CD19‐CAR construct incorporating the intracellular CD3ζ component or DAP10 showed that the anti‐CD19‐ζ‐CAR caused more NK cell‐mediated cytotoxicity than the anti‐CD19‐DAP10‐CAR [282]. DAP12 was found to be preferable to CD3 as an intracellular domain for NKG2D‐engineered primary NK cells. In this study, anti‐prostate stem cell antigen (PSCA)‐CAR‐NK cells containing DAP12 showed improved cytotoxicity, cytokine production, and tumor eradication in a mouse model [280, 283]. Furthermore, as compared to anti‐CD5‐CAR‐NK cells created with a 4‐1BB‐containing construct, anti‐CD5‐CAR‐NK cells engineered with a construct including the 2B4 intracellular domain were observed to have greater anti‐tumor activity [284]. In comparison to NK‐92 cells transduced with the CD28‐containing CAR construct, anti‐GPC3‐CAR‐NK‐92 cells engineered with a construct containing NK cell‐associated DNAM‐1 and/or 2B4 costimulatory domains displayed enhanced proliferative capacity, persistence, and cytotoxicity against HCC cells. The maximum cytolytic activity was seen in CAR‐NK‐92 cells that contained both the DNAM‐1 and 2B4 costimulatory domains [285]. To find the best CAR design for NK cells, Li and colleagues used NK‐92 cells to produce ten distinct anti‐mesothelin CARs and tested their impact on NK cell‐mediated killing. According to the reports, only three CARs (CAR4, CAR7, and CAR9) with an NKG2D TM domain and a 2B4 co‐stimulatory domain showed the greatest improvement in anti‐tumor activity. The capacity of these NKG2D TM and 2B4 carrying CARs to activate signaling by recruiting endogenous DAP10 under the circumstances of mesothelinhigh A1847 activation could be one reason for their better performance [255]. In an osteosarcoma xenograft mouse model, a receptor called NKG2D‐DAP10‐CD3ζ, containing the NKG2D ectodomain and intracellular DAP10 and CD3ζ costimulatory domains, has been shown to greatly augment the activation and anti‐tumor capacity of primary NK cells. CAR‐NK cells expressing the activating receptor NKG2D were designed by Parihar and colleagues to target human MDSCs. They demonstrated that NKG2D‐CAR‐NK cells could efficiently reduce human MDSCs in vitro and in a mouse xenograft model grafted with human neuroblastoma and MDSCs. Additionally, CAR‐NK cells release proinflammatory cytokines and chemokines that might enhance the infiltration and function of subsequently infused CAR‐T cells in the mouse model. For solid tumors, CAR‐NK cells have been suggested to be combined with T cell‐based therapies [286]. A CAR construct against PD‐1 using the NKG2D TM domain fused to 4‐1BB and DAP10 found DAP10 to be superfluous, and the same construct lacking DAP10 showed improved NK cell function [287]. Based on these results, it is evident that more studies are needed to optimize NK cell‐specific CAR constructs. In addition, similar strategies can be used to design other NK activating receptor‐based CAR, with the advantage that a single CAR could be used to attack several different types of cancer.
6.4. Preclinical studies of CAR‐NK cells targeting immune checkpoints for the treatment of solid tumors
6.4.1. PD‐1
In the study of Guo et al. [287], different chimeric receptors that mediate robust NK cell signaling have been designed using NKG2D. The extracellular domain of the inhibitory receptor PD‐1 was used in these NKG2D signaling‐based chimeric receptors to counteract the immunological escape mediated by PD‐1 ligands in solid tumors. They used the PD‐1 ectodomain and NKG2D TM domain as a backbone to rationally design four chimeric receptors by TM protein structure modeling. PNBB, a chimeric receptor that was constructed by fusing the human PD‐1 extracellular domain to the hinge region, TM and cytoplasmic domains of human NKG2D and human 4‐1BB cytoplasmic domain, showed stable surface expression on NK‐92 cells and induced significant in vitro cytotoxicity against various tumor cells. As a result, this strategy now offers a promising method for the rational design of NK cell‐tailored chimeric receptors with NKG2D signaling. One study created a novel chimeric costimulatory converting receptor (CCCR) by rationally combining the extracellular domain of PD‐1, the TM and cytoplasmic domains of NKG2D, and the cytoplasmic domain of 4‐1BB created based on the study of Guo et al. [287]. This NK‐tailored CCCR was able to switch the negative PD‐1 signal into an activating signal, reversing PD‐1's immune suppressive effects. When compared to untransduced NK‐92 cells, the CCCR‐modified NK‐92 (CCCR‐NK‐92) cells retained their typical NK cell features and displayed enhanced anti‐tumor activity against human lung cancer H1299 cells in vitro. The rapid clearance of H1299 cells was due to the significant gasdermin E (GSDME)‐dependent pyroptosis caused by CCCR‐NK‐92 cells. In a lung cancer xenograft model, CCCR‐NK‐92 cells dramatically inhibited tumor growth. Their results highlighted the promising immunotherapeutic potential of using NK‐tailored CCCR‐engineered NK‐92 cells for the treatment of human lung cancer [288]. Li et al. [289] developed a structure‐based rational design approach and created a novel Dual Targeting Chimeric Receptor (DTCR) PD1‐DAP10/NKG2D, which consists of the ectodomain of PD‐1 fused to a key co‐stimulatory receptor DAP10, and subsequently harnessed the activating receptor NKG2D as the second recognition receptor, to assess solid tumor cell killing capacity. Increased PD‐1 and NKG2D cell surface expression in NK‐92 cells after retroviral DTCR transduction promoted strong cytotoxicity against human gastric cell SGC‐7901. TNF‐α and TRAIL levels increased significantly after stimulation of the chimeric receptor DTCR, which can cause apoptosis in SGC‐7901 cells. DTCR‐NK‐92 cells had significant anti‐tumor activity in the solid tumor cell SGC‐7901‐ bearing mouse model. In another study, a CAR‐NK‐92 cell line, with an IL‐15Rα‐sushi/IL‐15 complex and a PD‐1 signal inverter, was created and named SP (Sushi‐IL15‐PD1). cDNA fragments of a CAR encoded IL‐15Rα‐sushi, TM domain of CD8, signal transduction zone of 4‐1BB, and CD3ζ, 2A signal peptide, PD1 extracellular region, and IL‐15. They showed that CAR expression allowed SP cells to proliferate independently of IL‐2 and become more resistant to nutrition‐starvation‐induced apoptosis. Meanwhile, SP cells were more effective in pancreatic ductal adenocarcinoma (PDAC) cell killing assays than NK‐92 in vitro and in vivo, and there was a positive correlation between the killing capability of SP cells and PD‐L1 expression in pancreatic cancer cells. The adhesion, responsiveness, degranulation efficiency, targeted delivery of cytotoxic granule content, and cytotoxicity of SP cells were considerably stronger compared to NK‐92. Finally, the SP cell line is a promising adoptive immunotherapy cell line that could be useful as an adjuvant treatment for pancreatic cancer, particularly in patients with high PD‐L1 expression [290]. In another study, CRISPR/Cas9 was used to develop a highly efficient method for editing the genome of human NK cells to knock out inhibitory signaling molecules. Up to 90% of primary PB‐NK cells were efficiently edited by their method. They showed highly efficient knockout of A disintegrin and metalloproteinase‐17 (ADAM17) and PD1 (PDCD1) genes that have a functional impact on NK cells and demonstrated that these gene‐edited NK cells have considerably improved activity, cytokine production, and cancer cell cytotoxicity. In addition, they were able to expand cells to clinically relevant numbers without losing their activity. They also demonstrated that their CRISPR/Cas9 method can be used to efficiently knock in (KI) genes by delivering homologous recombination template DNA using recombinant adeno‐associated virus serotype 6 (rAAV6). Their platform represents a feasible method for producing engineered primary NK cells as a universal cancer immunotherapy [291]. They suggested that this simple method for stable gene KI provides a platform for the delivery of other relevant cargo, including CARs and self‐stimulating cytokine receptors.
6.4.2. PD‐L1
Other researchers discovered that PD‐L1 t‐haNK cells expressed PD‐L1‐targeting CAR (composed of an extracellular scFv that binds to PD‐L1, a TM domain, and a FcεRIγ intracellular domain for downstream signaling) and CD16, retained the expression of native NK receptors, and had a high content of granzyme and perforin granules. They showed that irradiated PD‐L1 t‐haNK cells could lyse 20 of the 20 human cancer cell lines studied in vitro, including triple‐negative breast cancer (TNBC), lung, urogenital, and stomach cancer cells. The cytotoxicity of PD‐L1 t‐haNK cells was correlated to the PD‐L1 expression of the tumor targets and could be improved by pretreating the targets with IFN‐γ. It was found that irradiated PD‐L1 t‐haNK cells inhibited the growth of engrafted TNBC, lung, and bladder tumors in NSG mice. The combination of PD‐L1 t‐haNK cells with N‐803 (formerly ALT‐803), an IL‐15 super agonist complexed with IL‐15Rα−Sushi‐Fc fusion protein and anti‐PD‐1 Ab resulted in superior tumor growth control of engrafted oral cavity squamous carcinoma tumors in C57BL/6 mice. Also, when cocultured with human PBMCs, PD‐L1 t‐haNK cells lysed MDSCs preferentially but not other immune cell types. This study provides a rationale for using these cells in clinical trials [292]. haNKs were engineered by Robbins et al. [293] to express a CAR targeting PD‐L1. This second‐generation CAR included a scFv derived from NANT‐601, and an IgG1 mAb targeting PD‐L1, along with a CD8 hinge, a CD28 TM domain, and an intracellular FcεR1γ signaling domain. They demonstrated that haNKs engineered to express a PD‐L1‐CAR killed a panel of human and murine head and neck cancer cells at low effector‐to‐target ratios in a PD‐L1‐dependent fashion. Treatment of syngeneic tumors led to CD8 and PD‐L1‐dependent tumor rejection or growth inhibition and a reduction in myeloid cells endogenously expressing high levels of PD‐L1. Also, treatment of xenograft tumors resulted in PD‐L1‐dependent inhibition of tumor growth. In the PB of patients with head and neck cancer, PD‐L1‐CAR‐haNKs reduced levels of macrophages and other myeloid cells endogenously expressing high PD‐L1. Therefore, clinical study of PD‐L1‐CAR‐haNKs is recommended. Another study developed experimental models to investigate mechanisms of T cell escape and demonstrated that resistance to T cell killing can be overcome by adding NK cells engineered to express a CAR targeting PD‐L1 developed in two previous studies [292, 293]. In engineered models of tumor heterogeneity, PD‐L1‐CAR‐engineered NK cells (PD‐L1 t‐haNKs) inhibited the clonal selection of T cell‐resistant tumor cells seen with T cell treatment alone in multiple models. T cell treatment of heterogeneous cancer cell populations in vitro and in vivo resulted in IFN‐γ release and subsequent PD‐L1 upregulation on tumor cells that escaped T cell killing due to defects in antigen processing and presentation, priming escape cell populations for PD‐L1 dependent killing by PD‐L1 t‐haNKs. These findings shed light on the processes underpinning the synergistic anti‐cancer effectiveness of T cell‐based immunotherapy that result in IFN‐γ production, PD‐L1 overexpression on T cell escape cells, and the utilization of PD‐L1‐CAR‐engineered NK cells to target and eradicate resistant tumor cell populations [294]. In another study, PD‐L1‐targeted CAR was generated and introduced into T, NK, or NK‐92 cells. They generated a new atezolizumab‐based scFv and combined it with a standard second‐generation CAR backbone comprised of the IgG4 hinge region, CD28 TM and signaling portions, and the CD3ζ signaling domain. PD‐L1‐CAR effector cells degranulated and produced cytokines in response to PD‐L1high MDA‐MB‐231 cells but not to PD‐L1low MCF‐7 cells. However, in long‐term killing assays, both MDA‐MB‐231 and MCF‐7 cells have been killed by the PD‐L1‐CAR cells, but with a delay in the case of PD‐L1low MCF‐7 cells. Notably, the coculture of MCF‐7 cells with activated PD‐L1‐CAR cells led to bystander induction of PD‐L1 expression on MCF‐7 cells and to the unique self‐amplifying effect of the PD‐L1‐CAR cells. In tumor xenograft models, PD‐L1‐CAR‐T cells were active not only against MDA‐MD‐231 and MCF‐7‐PD‐L1, but also against MCF‐7‐pLVX (PD‐L1low/−) cells. Significantly, PD‐L1‐CAR cells exhibited potent cytotoxic effects against non‐malignant MCF‐10A, HMEC, and BM‐MSC cells, but not against HEK293T cells that initially did not express PD‐L1 and did not respond to the stimulation. Finally, they discovered that HER‐2‐CAR‐T cells stimulate expression of PD‐L1 on MCF‐7 cells and hence accelerate the functionality of PD‐L1‐CAR‐T cells when used in combination [295].
Combining genetically engineered immune cells with ICIs is an intriguing strategy. A number of studies have shown that co‐administration of PD‐1/PD‐L1 blockers with CAR‐T cell therapy enhances anti‐tumor activity and prevents T cell exhaustion in preclinical models [296] and patients receiving mesothelin‐based CAR‐T cell therapy [297]. CAR‐T cells were further engineered to produce and secrete an anti‐PD‐1 scFv for targeting and boosting CAR‐T cell activity in order to reduce the systemic effects of ICIs therapy [298]. Cherkassky et al. [299] successfully co‐transduced anti‐MSLN‐CAR‐T cells with vectors expressing shRNAs targeting PD‐1 genes. As a result of cotransduction with PD‐1 shRNAs and subsequent downregulation of PD‐1 intrinsic levels, CAR‐T cells were able to further expand and kill MSLN/PD‐L1‐positive cancer cells. Reduced N‐linked glycosylation of PD‐1 may reduce PD‐1 expression and relieve its inhibitory effects on CAR‐T cells. Therefore, a preclinical investigation using an adenine base editor (ABE) to decrease PD‐1 suppression by changing the glycosylated residue in CAR‐T cells downregulated the expression of PD‐1 in CAR‐T cells and improved cytotoxic functions in vitro and in vivo [300]. These strategies can also be used for CAR‐NK cells.
6.4.3. B7‐H3 (CD276)
The B7‐H3‐CAR was generated by Grote et al. [301] by conjugating a B7‐H3 clone m851‐derived scFv on a second‐generation CAR backbone incorporating a CD8 hinge domain, a CD28 TM domain and the cytoplasmic CD28 co‐stimulatory as well as CD3ζ signaling domains. NK‐92 cells engineered with B7‐H3‐CAR could eliminate neuroblastoma (NB) cells in vitro specifically and long‐term while sparing B7‐H3‐negative cancer cells. In addition, B7‐H3‐CAR‐NK‐92 cells demonstrated enhanced cytotoxicity in a 3D NB spheroid model recapitulating in vivo morphology as well as cell connectivity, polarity, gene expression, and tissue architecture, thus bridging the gap between in vitro and in vivo studies. A multitude of NK effector molecules were produced by B7‐H3‐CAR‐NK‐92 cells as well as pro‐inflammatory cytokines that stimulated the immune system. A stable surface expression of B7‐H3‐CAR and cytotoxic effector function was observed for more than six months. Grote et al. [302] also used these CAR‐NK‐92 cells that target B7‐H3, which is abundantly expressed in melanoma, and tested their effectivity in vitro in the presence of low pH, hypoxia, and other features of the TME that affect anti‐tumor responses. Additionally, CRISPR/Cas9‐induced disruption of NKG2A was tested for its potential to enhance the anti‐tumor effect of NK‐92. These B7‐H3‐CAR‐NK‐92 cells had the ability to cytolyze melanoma cell lines while being able to overcome the immunosuppressive effects typically exerted by TME. Cytotoxicity of CAR‐NK‐92 cells was not further improved by knocking out NKG2A. This study concluded that B7‐H3‐CAR‐NK‐92 cells are a promising cellular product for treating melanoma and beyond due to the cytotoxic properties of CARs targeting B7‐H3 and the capabilities of an “off‐the‐shelf” NK‐92 cell line unaffected by many negative factors associated with the TME. To improve NK cell functions, one study generated NK‐92MI cells carrying anti‐B7‐H3‐CAR by lentiviral transduction. The anti‐B7‐H3‐CAR was comprised of an anti‐B7‐H3 scFv derived from 8H9 Ab, followed by the CD8 TM region, the intracellular domains of 4‐1BB and CD3ζ. The expression of anti‐B7‐H3‐CAR drastically enhanced the cytotoxicity of NK‐92MI cells against B7‐H3‐positive tumor cells. Perforin and granzyme B secretions, as well as CD107a expression, were significantly increased in anti‐B7‐H3‐CAR‐NK‐92MI cells. Furthermore, anti‐B7‐H3‐CAR‐NK‐92MI cells efficiently inhibited tumor growth in mouse xenografts of NSCLC and significantly increased mouse survival days when compared to unmodified NK‐92MI cells [303].
6.4.4. HLA‐G
Another study established a novel CAR strategy comprised of the HLA‐G scFv, TM and cytosolic domains of KIR2DS4, the P2A (porcine teschovirus‐1 2A) sequence, the full‐length DAP12 sequence, and inducible caspase 9 (iC9) (FK506‐binding protein 12 fused to caspase‐9). HLA‐G‐CAR‐transduced NK cells showed effective cytolysis of breast, brain, pancreatic, and ovarian cancer cells in vitro, as well as decreased xenograft tumor growth with extended median survival in orthotopic mouse models. In tumor coculture assays, the anti‐HLA‐G scFv moiety increases Syk/Zap70 activation of NK cells, implying a reversal of the HLA‐G‐mediated immunosuppression and therefore, restoration of native NK cytolytic functions. Tumor expression of HLA‐G can be further promoted using low‐dose chemotherapy, which when combined with anti‐HLA‐G‐CAR‐NK, leads to extensive tumor ablation both in vitro and in vivo. This upregulation of tumor HLA‐G involves the inhibition of DNA methyltransferase 1 (DNMT1) and demethylation of transporters associated with antigen processing 1 promoter (TAP‐1) to trigger the translocation of HLA‐G [304].
6.4.5. Cytokine‐induced SH2‐containing protein (CIS)
Another study developed a strategy that couples targeting of the CIS protein, a major negative regulator of interleukin IL‐15 signaling, by CRISPR/Cas9 gene editing with fourth generation ‘armored’ CAR encoded iC9.CAR19.CD28‐zeta‐2A‐IL‐15 in CB‐derived NK cells. This combined strategy boosted NK cell effector function via enhancing the Akt/mTORC1 axis and c‐MYC signaling and led to increased aerobic glycolysis. When tested in a lymphoma mouse model, this combined approach enhanced NK cell anti‐tumor activity more than either alteration alone, eliminating lymphoma xenografts with no signs of any measurable toxicity. They concluded that targeting a cytokine checkpoint further improves the anti‐tumor activity of IL‐15 secreting armored CAR‐NK cells by enhancing their metabolic fitness and anti‐tumor activity. This combined approach could pave the way for the next generation of cancer immunotherapy using NK cells [305]. This strategy could also be used against solid tumors.
6.5. Clinical studies of CAR‐NK cells targeting immune checkpoints for the treatment of solid tumors
Because pre‐clinical data support the potential therapeutic benefits of CAR‐NK‐92 and PD‐L1 t‐haNK cell therapy, these novel cell therapies have a strong rationale for clinical application. A phase I clinical trial (NCT03656705) is being conducted to evaluate the safety and effects of CCCR‐modified NK‐92 (CCCR‐NK‐92) infusion with a starting dose of 1 × 107‐1 × 108, twice a week, that will be administered intravenously over 1 h in previously treated advanced NSCLC. NCT04050709 is a phase I study to assess the safety and preliminary efficacy of PD‐L1 t‐haNK and to determine the maximal tolerated dose and designate the recommended phase II dose in subjects with locally advanced or metastatic solid cancers. In a phase II, three‐cohort (2 randomized and 1 single‐arm), open‐label study (NCT04390399) the comparative efficacy and overall safety of standard‐of‐care chemotherapy versus standard‐of‐care chemotherapy in combination with aldoxorubicin HCl, N‐803, and PD‐L1 t‐haNK in subjects with locally advanced or metastatic pancreatic cancer are evaluated. Another phase II study (NCT04847466) is testing the effectiveness of irradiated PD‐L1 t‐haNK cells combined with anti‐PD‐1 (pembrolizumab) and N‐803 in people with advanced forms of gastric or head and neck cancer. The efficacy of PD‐L1 t‐haNK with a PD‐1/PD‐L1 checkpoint inhibitor and N‐803 in patients with solid tumors that have progressed and/or relapsed after PD‐1/PD‐L1 ICIs is also being determined in a phase IIb, multicohort, open‐label multicenter study (NCT03228667). In another phase Ib/II open‐label study (NCT04927884), the safety and efficacy of sacituzumab govitecan‐hziy (an Ab‐drug conjugate) in combination with chemoimmunotherapy (cyclophosphamide, N‐803, and PD‐L1 t‐haNK) in subjects with TNBC after at least 2 prior treatments for metastatic disease is being evaluated. Table 2 shows clinical studies of CAR‐NK cells targeting immune checkpoints for the treatment of solid tumors.
TABLE 2.
Clinical studies of CAR‐NK cells targeting immune checkpoints for the treatment of solid tumors
| No. NCT | Status | Phase | Target | Cancer | NK source | Combination agent |
|---|---|---|---|---|---|---|
| NCT03656705 | Enrolling by invitation | I | PD‐L1 | Non‐small cell lung cancer | CCCR‐NK‐92 cells | Not applicable |
| NCT04050709 | Active, not recruiting | I | PD‐L1 | Locally advanced or metastatic solid cancers | haNK | Not applicable |
| NCT04390399 | Recruiting | II | PD‐L1 | Pancreatic cancer | haNK |
N‐803 Aldoxorubicin HCl Nab‐paclitaxel Gemcitabine Cyclophosphamide 5‐fluorouracil Leucovorin Stereotactic Body Radiation Therapy Irinotecan liposome |
| NCT04847466 | Recruiting | II | PD‐L1 | Recurrent/metastatic gastric or head and neck cancer | haNK |
N‐803 Pembrolizumab |
| NCT03228667 |
Active, not recruiting |
II | PD‐L1 | Non‐small cell lung cancer, small cell lung cancer, urothelial carcinoma, head and neck squamous cell carcinoma, merkel cell carcinoma, melanoma, renal cell carcinoma, gastric cancer, cervical cancer, hepatocellular carcinoma, microsatellite instability, mismatch repair deficiency, and colorectal cancer | haNK |
N‐803 Pembrolizumab Nivolumab Atezolizumab Avelumab Durvalumab |
| NCT04927884 | Recruiting | I/II | PD‐L1 | Advanced triple‐negative breast cancer | haNK |
N‐803 Sacituzumab govitecan‐hziy Cyclophosphamide |
Abbreviations: PD‐L1, programmed death‐ligand 1; CCCR, chimeric costimulatory converting receptor; hank, high‐affinity natural killer.
7. Conclusion and future perspectives
Although checkpoint inhibitor therapy has been demonstrated to restore anti‐tumor immune responses, a single checkpoint disruption does not necessarily result in NK or T cell function, and many cancers develop initial resistance or adapt to become resistant to subsequent treatments [306]. To combat this resistance, researchers are currently investigating combinations of ICIs. A growing body of evidence suggests that this approach has the potential to improve anti‐tumor activity and response rates dramatically [307]. In clinical trials and/or animal models, checkpoint inhibitors may also be used with other therapies such as radiation therapy, targeted therapy, chemotherapy, epigenetic modulators, TLR agonists, or other immunotherapies. It's also worth noting that systemic administration of ICIs medications can exacerbate immune‐related side effects [308]. As a result, alternate treatments with fewer side effects must be developed.
Currently, the BiKE and TriKE constructs exhibit great translational potential, but efforts are being made to further improve their efficacy. The efficacy of BiKEs and TriKEs may be limited by CD16 expression because by activating NK cells through CD16, matrix metalloproteinases (MMPs), such as ADAM‐17, rapidly clip CD16 from their surface [309, 310, 311, 312]. Therefore, BiKE/TriKE function may be enhanced by co‐treatment with the ADAM‐17 inhibitor [313]. Another approach is targeting other receptors on NK cells to circumvent the CD16 problem [314]. BiKE‐mediated NK cell function may also be enhanced by the co‐administration of cytokines such as IL‐15, IL‐2, IL‐21, and IL‐12 [314].
Compared to Abs, aptamers are still in a nascent stage, meaning further development is required to ensure widespread adoption. Clinical trials testing aptamers’ therapeutic effects are needed to accurately determine their clinical potential for cancer treatment. Although aptamer‐based cancer therapies have huge potential, their disadvantages, such as serum instability and rapid renal clearance, may limit their application. Therefore, modifications are needed to enhance their in vivo half‐life and their superior therapeutic benefits [21].
Based on a growing amount of evidence, CAR‐NK cell‐based cancer therapy has emerged as a promising and advanced research area. However, the field of CAR‐NK therapy is still in its infancy, and many questions remain unresolved. To mention a few, these include determining the best NK cell source for immunotherapy, enhancing persistence capacity, determining the best protocol for introducing foreign genetic material into NK cells due to NK cells’ resistance to genetic engineering and optimizing them using viral vectors, nonviral alternatives such as electroporation, nucleofection, lipofection, transposon systems, trogocytosis or nanoparticle treatment; and genome editing using CRISPR/Cas9, ZFNs or transcriptional activator‐like effector nucleases (TALENs). Also, determining CARs with the optimum structure for improving NK cell proliferation, activation, cytokine production, and cytolytic activity, the best ex vivo expansion method for acquiring a homogeneous suspension of memory‐like, unexhausted NK cells from harvested NK cells to obtain clinically relevant numbers, incorporating a suicide gene into CAR‐NK cells as an important point for safety concerns and best freezing media to improve the preservation of NK cells are other challenges in the development of NK cell‐based immunotherapy.
Targeting lentiviral to identify distinct surface markers as entrance receptors have enabled precise in vivo gene delivery into certain cell types [315, 316]. Engineering human NK‐targeted vectors capable of efficient and highly selective gene delivery into human NK cells in vivo would have significant implications for adoptive immunotherapy development in the future. The targeted vectors will be employed as a therapeutic tool in this situation, and the cells will not need to be prepared or expanded ex vivo (Figure 4A). Exosomes, which are released by immune cells, can affect tumors or the anti‐tumor immune response. Exosomes from NK cells contain FasL, granulysin, perforin, and granzyme A and B, which have cytotoxic properties against tumor cells [317, 318, 319]. Researchers have previously demonstrated the safety and efficacy of CAR exosomes in treating tumors. A recent study showed that purified CAR‐T cell exosomes expressed CARs, and these CAR+ exosomes expressed a high level of cytotoxic molecules and were cytotoxic to tumor cells. As opposed to CAR‐T cells, CAR exosomes did not express PD‐1, and their anti‐tumor effect could not be weakened by recombinant PD‐L1 treatment. CAR exosomes were relatively safe compared to CAR‐T therapy in a preclinical in vivo model of cytokine release syndrome (CRS) [320]. Exosomes have been shown in multiple studies to have several other benefits, including accessibility and storage, the ability to pass through the blood–brain and blood–tumor barriers, the ability to load various effector molecules, and the ability to combine other therapeutic modalities [321, 322] making them potential alternatives to cell therapies. As a result, exosomes expressing CAR derived from CAR‐NK cells may offer superior benefits to CAR‐NK cells, opening a novel cell‐free immunotherapy pathway that has yet to be explored (Figure 4B). The nanometer size of NK extracellular vesicles as well as their ability to survive in acidic TME makes CAR‐NK cell‐derived extracellular vesicles a promising treatment for solid tumors. In addition, a novel biomimetic platform, nanoparticles (NPs) camouflaged in the cell membranes, mimics some membrane functions of the cells from which these membranes are derived in biological systems. It is also suggested to design biomimetic nanostructures by coating NPs with NK‐secreted exosomes containing identifying markers (e.g., CD226) and cell‐death markers (e.g., TRAIL, FAS, and perforin/granzyme B) for targeting and killing circulating tumor cells (CTCs) [323]. It is desired to develop exosome‐biomimetic nanoparticles with good biocompatibility and high drug loading for the treatment of cancer.
FIGURE 4.
Targeting lentiviral for identifying NK cell surface receptor and CAR‐NK cell‐derived exosome as future directions in CAR‐NK cell therapy. (A) Targeting lentiviral to identify distinct surface markers on NK cells in vivo as entrance receptors would have significant implications for adoptive immunotherapy development in the future. (B) Exosomes expressing CAR derived from CAR‐NK cells may offer superior benefits to CAR‐NK cells, opening a novel cell‐free immunotherapy pathway and suggesting promising treatment for solid tumors. Abbreviations: NK, natural killer; CAR, chimeric antigen receptor; IFN‐γ, interferon gamma.
It is expected that a variety of combinatorial approaches will be developed in the near future to improve NK cell active tumor targeting, persistence and trafficking, higher cytotoxicity while maintaining safety, and improved resistance to immunosuppressive TME. Also, more research needs to be conducted on the safety, tolerability, and effectiveness of checkpoint inhibitors and their combinations. To identify appropriate patient populations for specific therapies, it is important to understand the immune phenotypes of tumors, lymphocyte infiltration, and TME composition. This could assist with deciding which therapies or combinations will be the most effective. Furthermore, identification of biomarkers for the prediction and monitoring of the response to specific therapies or the appropriate combination of therapies is essential.
DECLARATIONS
AUTHOR CONTRIBUTIONS
All authors contributed to manuscript conception and drafting of the manuscript. All authors provided critical review and revisions, and approved the final version of the manuscript.
COMPETING INTERESTS
The authors declare that they have no conflicts of interest.
FUNDING
This work was supported by Isfahan University of Medical Sciences, Isfahan, Iran (grant No. 140170).
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
ACKNOWLEDGEMENTS
Not applicable.
Ghaedrahmati F, Esmaeil N, Abbaspour M. Targeting immune checkpoints: how to use natural killer cells for fighting against solid tumors. Cancer Communications. 2023;43:177–213. 10.1002/cac2.12394
Contributor Information
Farhoodeh Ghaedrahmati, Email: farhoodeh68@yahoo.com.
Nafiseh Esmaeil, Email: nafesm5@gmail.com, Email: n_esmaeil@med.mui.ac.ir.
Maryam Abbaspour, Email: mary.abbaspour2013@gmail.com.
REFERENCES
- 1. Wang H, Naghavi M, Allen C, Barber RM, Bhutta ZA, Carter A, et al. Global, regional, and national life expectancy, all‐cause mortality, and cause‐specific mortality for 249 causes of death, 1980‐2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet North Am Ed. 2016;388(10053):1459‐544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Papaioannou NE, Beniata OV, Vitsos P, Tsitsilonis O, Samara P. Harnessing the immune system to improve cancer therapy. Ann Transl Med. 2016;4(14):261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang Y, Wang M, Wu HX, Xu RH. Advancing to the era of cancer immunotherapy. Cancer Commun. 2021;41(9):803‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Altmann DM. A Nobel Prize‐worthy pursuit: cancer immunology and harnessing immunity to tumour neoantigens. Immunology. 2018;155(3):283‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Daher M, Rezvani K. Next generation natural killer cells for cancer immunotherapy: the promise of genetic engineering. Curr Opin Immunol. 2018;51:146‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farag SS, Caligiuri MA. Human natural killer cell development and biology. Blood Rev. 2006;20(3):123‐37. [DOI] [PubMed] [Google Scholar]
- 7. Pross HF, Lotzova E. Role of natural killer cells in cancer. Nat Immun. 1993;12(4‐5):279‐92. [PubMed] [Google Scholar]
- 8. Zhang C, Liu Y. Targeting NK cell checkpoint receptors or molecules for cancer immunotherapy. Front Immunol. 2020;11:1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ma L, Diao B, Huang Z, Wang B, Yu J, Meng X. The efficacy and possible mechanisms of immune checkpoint inhibitors in treating non‐small cell lung cancer patients with epidermal growth factor receptor mutation. Cancer Commun. 2021;41(12):1314‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moon D, Tae N, Park Y, Lee S‐W, Kim DH. Development of bispecific antibody for cancer immunotherapy: focus on T cell engaging antibody. Immune Network. 2022;22(1):e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kellner C, Bruenke J, Horner H, Schubert J, Schwenkert M, Mentz K, et al. Heterodimeric bispecific antibody‐derivatives against CD19 and CD16 induce effective antibody‐dependent cellular cytotoxicity against B‐lymphoid tumor cells. Cancer Lett. 2011;303(2):128‐39. [DOI] [PubMed] [Google Scholar]
- 12. Shin HG, HR Yang, Yoon A, Lee S. Bispecific Antibody‐Based Immune‐Cell Engagers and Their Emerging Therapeutic Targets in Cancer Immunotherapy. Int J Mol Sci. 2022;23(10):5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Preithner S, Elm S, Lippold S, Locher M, Wolf A, da Silva AJ, et al. High concentrations of therapeutic IgG1 antibodies are needed to compensate for inhibition of antibody‐dependent cellular cytotoxicity by excess endogenous immunoglobulin G. Mol Immunol. 2006;43(8):1183‐93. [DOI] [PubMed] [Google Scholar]
- 14. Baselga J, Albanell J. Mechanism of action of anti‐HER2 monoclonal antibodies. Ann Oncol. 2001;12:S35‐S41. [DOI] [PubMed] [Google Scholar]
- 15. Berinstein N, Grillo‐Lopez A, White C, Bence‐Bruckler I, Maloney D, Czuczman M, et al. Association of serum Rituximab (IDEC–C2B8) concentration and anti‐tumor response in the treatment of recurrent low‐grade or follicular non‐Hodgkin's lymphoma. Ann Oncol. 1998;9(9):995‐1001. [DOI] [PubMed] [Google Scholar]
- 16. Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23(9):1126‐36. [DOI] [PubMed] [Google Scholar]
- 17. Chames P, Van Regenmortel M, Weiss E, Baty D. Therapeutic antibodies: successes, limitations and hopes for the future. Br J Pharmacol. 2009;157(2):220‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat Rev Cancer. 2012;12(4):278‐87. [DOI] [PubMed] [Google Scholar]
- 19. Ray P, Viles KD, Soule EE, Woodruff RS. Application of aptamers for targeted therapeutics. Arch Immunol Ther Exp (Warsz). 2013;61(4):255‐71. [DOI] [PubMed] [Google Scholar]
- 20. Pei X, Zhang J, Liu J. Clinical applications of nucleic acid aptamers in cancer. Mol Clin Oncol. 2014;2(3):341‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou G, Wilson G, Hebbard L, Duan W, Liddle C, George J, et al. Aptamers: A promising chemical antibody for cancer therapy. Oncotarget. 2016;7(12):13446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weber JS. At the Bedside: Adoptive cell therapy for melanoma—clinical development. J Leukocyte Biol. 2014;95(6):875‐82. [DOI] [PubMed] [Google Scholar]
- 23. Allen ES, Stroncek DF, Ren J, Eder AF, West KA, Fry TJ, et al. Autologous lymphapheresis for the production of chimeric antigen receptor T cells. Transfusion (Paris). 2017;57(5):1133‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goulmy E. Human minor histocompatibility antigens: new concepts for marrow transplantation and adoptive immunotherapy. Immunol Rev. 1997;157(1):125‐40. [DOI] [PubMed] [Google Scholar]
- 25. Sterner RC, Sterner RM. CAR‐T cell therapy: current limitations and potential strategies. Blood Cancer J. 2021;11(4):1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Petty AJ, Heyman B, Yang Y. Chimeric antigen receptor cell therapy: overcoming obstacles to battle cancer. Cancers. 2020;12(4):842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tian Y, Li Y, Shao Y, Zhang Y. Gene modification strategies for next‐generation CAR T cells against solid cancers. J Hematol Oncol. 2020;13:1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu S, Yi M, Qin S, Wu K. Next generation chimeric antigen receptor T cells: safety strategies to overcome toxicity. Mol Cancer. 2019;18(1):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Herberman RB, Holden HT, Ting C‐C, Lavrin DL, Kirchner H. Cell‐mediated immunity to leukemia virus‐and tumor‐associated antigens in mice. Cancer Res. 1976;36(2 Part 2):615‐21. [PubMed] [Google Scholar]
- 30. Scoville SD, Freud AG, Caligiuri MA. Modeling human natural killer cell development in the era of innate lymphoid cells. Front Immunol. 2017;8:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ferlazzo G, Carrega P. Natural killer cell distribution and trafficking in human tissues. Front Immunol. 2012;3:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. He Y, Tian Z. NK cell education via nonclassical MHC and non‐MHC ligands. Cell Mol Immunol. 2017;14(4):321‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walzer T, Bléry M, Chaix J, Fuseri N, Chasson L, Robbins SH, et al. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc Natl Acad Sci. 2007;104(9):3384‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Walzer T, Jaeger S, Chaix J, Vivier E. Natural killer cells: from CD3− NKp46+ to post‐genomics meta‐analyses. Curr Opin Immunol. 2007;19(3):365‐72. [DOI] [PubMed] [Google Scholar]
- 35. Campbell KS, Hasegawa J. Natural killer cell biology: an update and future directions. J Allergy Clin Immunol. 2013;132(3):536‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wagner JA, Rosario M, Romee R, Berrien‐Elliott MM, Schneider SE, Leong JW, et al. CD56 bright NK cells exhibit potent antitumor responses following IL‐15 priming. J Clin Invest. 2017;127(11):4042‐58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Caligiuri MA. Human natural killer cells. Blood, The Journal of the American Society of Hematology. 2008;112(3):461‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells—a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13(2):145‐9. [DOI] [PubMed] [Google Scholar]
- 39. Ljunggren H‐G, Kärre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237‐44. [DOI] [PubMed] [Google Scholar]
- 40. Joncker NT, Raulet DH. Regulation of NK cell responsiveness to achieve self‐tolerance and maximal responses to diseased target cells. Immunol Rev. 2008;224(1):85‐97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kärre K. Natural killer cell recognition of missing self. Nat Immunol. 2008;9(5):477‐80. [DOI] [PubMed] [Google Scholar]
- 42. Shifrin N, Raulet DH, Ardolino M, editors. NK cell self tolerance, responsiveness and missing self recognition. Seminars in Immunology; 2014: Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wagtmann N, Rajagopalan S, Winter CC, Peruui M, Long EO. Killer cell inhibitory receptors specific for HLA‐C and HLA‐B identified by direct binding and by functional transfer. Immunity. 1995;3(6):801‐9. [DOI] [PubMed] [Google Scholar]
- 44. Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari MC, et al. Receptors for HLA class‐I molecules in human natural killer cells. Annu Rev Immunol. 1996;14(1):619‐48. [DOI] [PubMed] [Google Scholar]
- 45. Moretta L, Mingari MC, Pende D, Bottino C, Biassoni R, Moretta A. The molecular basis of natural killer (NK) cell recognition and function. J Clin Immunol. 1996;16(5):243‐53. [DOI] [PubMed] [Google Scholar]
- 46. Moretta A, Biassoni R, Bottino C, Pende D, Vitale M, Poggi A, et al. Major histocompatibility complex class I‐specific receptors on human natural killer and T lymphocytes. Immunol Rev. 1997;155(1):105‐17. [DOI] [PubMed] [Google Scholar]
- 47. Bottino C, Biassoni R, Millo R, Moretta L, Moretta A. The human natural cytotoxicity receptors (NCR) that induce HLA class I‐independent NK cell triggering. Hum Immunol. 2000;61(1):1‐6. [DOI] [PubMed] [Google Scholar]
- 48. Godal R, Bachanova V, Gleason M, McCullar V, Yun GH, Cooley S, et al. Natural killer cell killing of acute myelogenous leukemia and acute lymphoblastic leukemia blasts by killer cell immunoglobulin‐like receptor–negative natural killer cells after NKG2A and LIR‐1 blockade. Biol Blood Marrow Transplant. 2010;16(5):612‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kang X, Kim J, Deng M, John S, Chen H, Wu G, et al. Inhibitory leukocyte immunoglobulin‐like receptors: immune checkpoint proteins and tumor sustaining factors. Cell Cycle. 2016;15(1):25‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang F, Zheng J, Kang X, Deng M, Lu Z, Kim J, et al. Inhibitory leukocyte immunoglobulin‐like receptors in cancer development. Sci China Life Sci. 2015;58(12):1216‐25. [DOI] [PubMed] [Google Scholar]
- 51. Rouas‐Freiss N, Moreau P, LeMaoult J, Carosella ED. The dual role of HLA‐G in cancer. J Immunol Res. 2014;2014:359748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hogan L, Bhuju S, Jones DC, Laing K, Trowsdale J, Butcher P, et al. Characterisation of bovine leukocyte Ig‐like receptors. PLoS One. 2012;7(4):e34291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Navarro F, Llano M, Bellón T, Colonna M, Geraghty DE, López‐Botet M. The ILT2 (LIR1) and CD94/NKG2A NK cell receptors respectively recognize HLA‐G1 and HLA‐E molecules co‐expressed on target cells. Eur J Immunol. 1999;29(1):277‐83. [DOI] [PubMed] [Google Scholar]
- 54. Borrego F, Kabat J, Kim D‐K, Lieto L, Maasho K, Peña J, et al. Structure and function of major histocompatibility complex (MHC) class I specific receptors expressed on human natural killer (NK) cells. Mol Immunol. 2002;38(9):637‐60. [DOI] [PubMed] [Google Scholar]
- 55. Khan M, Arooj S, Wang H. NK cell‐based immune checkpoint inhibition. Front Immunol. 2020;11:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, et al. Activating receptors and coreceptors involved in human natural killer cell‐mediated cytolysis. Annu Rev Immunol. 2001;19:197‐223. [DOI] [PubMed] [Google Scholar]
- 57. Moretta L, Montaldo E, Vacca P, Del Zotto G, Moretta F, Merli P, et al. Human natural killer cells: origin, receptors, function, and clinical applications. Int Arch Allergy Immunol. 2014;164(4):253‐64. [DOI] [PubMed] [Google Scholar]
- 58. Vitale M, Cantoni C, Della Chiesa M, Ferlazzo G, Carlomagno S, Pende D, et al. An Historical Overview: The Discovery of How NK Cells Can Kill Enemies, Recruit Defense Troops, and More. Front Immunol. 2019;10:1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chiossone L, Vienne M, Kerdiles YM, Vivier E. Natural killer cell immunotherapies against cancer: checkpoint inhibitors and more. Semin Immunol. 2017;31:55‐63. [DOI] [PubMed] [Google Scholar]
- 60. Abel AM, Yang C, Thakar MS, Malarkannan S. Natural killer cells: development, maturation, and clinical utilization. Front Immunol. 2018;9:1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sonar S, Lal G. Role of tumor necrosis factor superfamily in neuroinflammation and autoimmunity. Front Immunol. 2015;6:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Martínez‐Lostao L, Anel A, Pardo J. How do cytotoxic lymphocytes kill cancer cells? Clin Cancer Res. 2015;21(22):5047‐56. [DOI] [PubMed] [Google Scholar]
- 63. Topham NJ, Hewitt EW. Natural killer cell cytotoxicity: how do they pull the trigger? Immunology. 2009;128(1):7‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pardo J, Balkow S, Anel A, Simon MM. Granzymes are essential for natural killer cell‐mediated and perf‐facilitated tumor control. Eur J Immunol. 2002;32(10):2881‐6. [DOI] [PubMed] [Google Scholar]
- 65. Jong AY, Wu C‐H, Li J, Sun J, Fabbri M, Wayne AS, et al. Large‐scale isolation and cytotoxicity of extracellular vesicles derived from activated human natural killer cells. J Extracell Vesicles. 2017;6(1):1294368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mellor JD, Brown MP, Irving HR, Zalcberg JR, Dobrovic A. A critical review of the role of Fc gamma receptor polymorphisms in the response to monoclonal antibodies in cancer. J Hematol Oncol. 2013;6(1):1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bluman EM, Bartynski KJ, Avalos BR, Caligiuri MA. Human natural killer cells produce abundant macrophage inflammatory protein‐1 alpha in response to monocyte‐derived cytokines. J Clin Invest. 1996;97(12):2722‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Roda JM, Parihar R, Magro C, Nuovo GJ, Tridandapani S, Carson WE. Natural killer cells produce T cell–recruiting chemokines in response to antibody‐coated tumor cells. Cancer Res. 2006;66(1):517‐26. [DOI] [PubMed] [Google Scholar]
- 69. Fauriat C, Long EO, Ljunggren H‐G, Bryceson YT. Regulation of human NK‐cell cytokine and chemokine production by target cell recognition. Blood, The Journal of the American Society of Hematology. 2010;115(11):2167‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Romee R, Leong JW, Fehniger TA. Utilizing cytokines to function‐enable human NK cells for the immunotherapy of cancer. Scientifica. 2014;2014:205796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lima M, Leander M, Santos M, Santos AH, Lau C, Queirós ML, et al. Chemokine receptor expression on normal blood CD56+ NK‐cells elucidates cell partners that comigrate during the innate and adaptive immune responses and identifies a transitional NK‐cell population. J Immunol Res. 2015;2015:839684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Noh J‐Y, Yoon SR, Kim T‐D, Choi I, Jung H. Toll‐like receptors in natural killer cells and their application for immunotherapy. J Immunol Res. 2020;2020:2045860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. O'Sullivan TE, Sun JC, Lanier LL. Natural Killer Cell Memory. Immunity. 2015;43(4):634‐45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rapp M, Wiedemann GM, Sun JC. Memory responses of innate lymphocytes and parallels with T cells. Semin Immunopathol. 2018;40(4):343‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pal M, Schwab L, Yermakova A, Mace EM, Claus R, Krahl AC, et al. Tumor‐priming converts NK cells to memory‐like NK cells. Oncoimmunology. 2017;6(6):e1317411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wu LS, Wang JY. Warm up, cool down, and tearing apart in NK cell memory. Cell Mol Immunol. 2018;15(12):1095‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. O'Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell‐ and B cell‐independent adaptive immunity mediated by natural killer cells. Nat Immunol. 2006;7(5):507‐16. [DOI] [PubMed] [Google Scholar]
- 78. Paust S, Gill HS, Wang BZ, Flynn MP, Moseman EA, Senman B, et al. Critical role for the chemokine receptor CXCR6 in NK cell‐mediated antigen‐specific memory of haptens and viruses. Nat Immunol. 2010;11(12):1127‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nikzad R, Angelo LS, Aviles‐Padilla K, Le DT. Human natural killer cells mediate adaptive immunity to viral antigens. Sci Immunol. 2019;4(35):eaat8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bryceson YT, March ME, Ljunggren H‐G, Long EO. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood. 2006;107(1):159‐66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nayyar G, Chu Y, Cairo MS. Overcoming resistance to natural killer cell based immunotherapies for solid tumors. Front Oncol. 2019;9:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Oka N, Markova T, Tsuzuki K, Li W, El‐Darawish Y, Pencheva‐Demireva M, et al. IL‐12 regulates the expansion, phenotype, and function of murine NK cells activated by IL‐15 and IL‐18. Cancer Immunol Immunother. 2020;69(9):1699‐712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sarkar S, Germeraad WT, Rouschop KM, Steeghs EM, Mv Gelder, Bos GM, et al. Hypoxia induced impairment of NK cell cytotoxicity against multiple myeloma can be overcome by IL‐2 activation of the NK cells. PLoS One. 2013;8(5):e64835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Siegler U, Meyer‐Monard S, Jörger S, Stern M, Tichelli A, Gratwohl A, et al. Good manufacturing practice‐compliant cell sorting and large‐scale expansion of single KIR‐positive alloreactive human natural killer cells for multiple infusions to leukemia patients. Cytotherapy. 2010;12(6):750‐63. [DOI] [PubMed] [Google Scholar]
- 85. Chaix J, Tessmer MS, Hoebe K, Fuséri N, Ryffel B, Dalod M, et al. Cutting edge: Priming of NK cells by IL‐18. J Immunol. 2008;181(3):1627‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ozaki K, Kikly K, Michalovich D, Young PR, Leonard WJ. Cloning of a type I cytokine receptor most related to the IL‐2 receptor β chain. Proc Natl Acad Sci. 2000;97(21):11439‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Romee R, Schneider SE, Leong JW, Chase JM, Keppel CR, Sullivan RP, et al. Cytokine activation induces human memory‐like NK cells. Blood, The Journal of the American Society of Hematology. 2012;120(24):4751‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Leong JW, Chase JM, Romee R, Schneider SE, Sullivan RP, Cooper MA, et al. Preactivation with IL‐12, IL‐15, and IL‐18 induces CD25 and a functional high‐affinity IL‐2 receptor on human cytokine‐induced memory‐like natural killer cells. Biol Blood Marrow Transplant. 2014;20(4):463‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Romee R, Rosario M, Berrien‐Elliott MM, Wagner JA, Jewell BA, Schappe T, et al. Cytokine‐induced memory‐like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci Transl Med. 2016;8(357):357ra123‐357ra123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Moretta A, Bottino C, Pende D, Tripodi G, Tambussi G, Viale O, et al. Identification of four subsets of human CD3‐CD16+ natural killer (NK) cells by the expression of clonally distributed functional surface molecules: correlation between subset assignment of NK clones and ability to mediate specific alloantigen recognition. J Exp Med. 1990;172(6):1589‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Donatelli SS, Djeu JY. Immunological Sculpting: Natural Killer‐Cell Receptors and Ligands. Cancer Immunotherapy: Elsevier; 2013. p. 115‐27. [Google Scholar]
- 92. Pende D, Falco M, Vitale M, Cantoni C, Vitale C, Munari E, et al. Killer Ig‐like receptors (KIRs): their role in NK cell modulation and developments leading to their clinical exploitation. Front Immunol. 2019:1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sun H, Sun C. The rise of NK cell checkpoints as promising therapeutic targets in cancer immunotherapy. Front Immunol. 2019:2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cornillet M, Jansson H, Schaffer M, Hertwig L, Berglin L, Zimmer CL, et al. Imbalance of genes encoding natural killer immunoglobulin‐like receptors and human leukocyte antigen in patients with biliary cancer. Gastroenterology. 2019;157(4):1067‐80. e9. [DOI] [PubMed] [Google Scholar]
- 95. Kim N, Kim HS. Targeting checkpoint receptors and molecules for therapeutic modulation of natural killer cells. Front Immunol. 2018:2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kohrt HE, Thielens A, Marabelle A, Sagiv‐Barfi I, Sola C, Chanuc F, et al. Anti‐KIR antibody enhancement of anti‐lymphoma activity of natural killer cells as monotherapy and in combination with anti‐CD20 antibodies. Blood, The Journal of the American Society of Hematology. 2014;123(5):678‐86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. van Hall T, André P, Horowitz A, Ruan DF, Borst L, Zerbib R, et al. Monalizumab: inhibiting the novel immune checkpoint NKG2A. J Immunother Cancer. 2019;7(1):1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Tinker AV, Hirte HW, Provencher D, Butler M, Ritter H, Tu D, et al. Dose‐ranging and cohort‐expansion study of Monalizumab (IPH2201) in patients with advanced gynecologic malignancies: a trial of the Canadian Cancer Trials Group (CCTG): IND221. Clin Cancer Res. 2019;25(20):6052‐60. [DOI] [PubMed] [Google Scholar]
- 99. André P, Denis C, Soulas C, Bourbon‐Caillet C, Lopez J, Arnoux T, et al. Anti‐NKG2A mAb is a checkpoint inhibitor that promotes anti‐tumor immunity by unleashing both T and NK cells. Cell. 2018;175(7):1731‐43. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Anderson AC, Joller N, Kuchroo VK. Lag‐3, Tim‐3, and TIGIT: co‐inhibitory receptors with specialized functions in immune regulation. Immunity. 2016;44(5):989‐1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. He Y, Cao J, Zhao C, Li X, Zhou C, Hirsch FR. TIM‐3, a promising target for cancer immunotherapy. Onco Targets Ther. 2018;11:7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Monney L, Sabatos CA, Gaglia JL, Ryu A, Waldner H, Chernova T, et al. Th1‐specific cell surface protein Tim‐3 regulates macrophage activation and severity of an autoimmune disease. Nature. 2002;415(6871):536‐41. [DOI] [PubMed] [Google Scholar]
- 103. Wang Z, Zhu J, Gu H, Yuan Y, Zhang B, Zhu D, et al. The clinical significance of abnormal Tim‐3 expression on NK cells from patients with gastric cancer. Immunol Invest. 2015;44(6):578‐89. [DOI] [PubMed] [Google Scholar]
- 104. Komita H, Koido S, Hayashi K, Kan S, Ito M, Kamata Y, et al. Expression of immune checkpoint molecules of T cell immunoglobulin and mucin protein 3/galectin‐9 for NK cell suppression in human gastrointestinal stromal tumors. Oncol Rep. 2015;34(4):2099‐105. [DOI] [PubMed] [Google Scholar]
- 105. So EC, Khaladj‐Ghom A, Ji Y, Amin J, Song Y, Burch E, et al. NK cell expression of Tim‐3: First impressions matter. Immunobiology. 2019;224(3):362‐70. [DOI] [PubMed] [Google Scholar]
- 106. Ndhlovu LC, Lopez‐Vergès S, Barbour JD, Jones RB, Jha AR, Long BR, et al. Tim‐3 marks human natural killer cell maturation and suppresses cell‐mediated cytotoxicity. Blood, The Journal of the American Society of Hematology. 2012;119(16):3734‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Van Audenaerde JR, De Waele J, Marcq E, Van Loenhout J, Lion E, Van den Bergh JM, et al. Interleukin‐15 stimulates natural killer cell‐mediated killing of both human pancreatic cancer and stellate cells. Oncotarget. 2017;8(34):56968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Folgiero V, Cifaldi L, Pira GL, Goffredo BM, Vinti L, Locatelli F. TIM‐3/Gal‐9 interaction induces IFNγ‐dependent IDO1 expression in acute myeloid leukemia blast cells. J Hematol Oncol. 2015;8(1):1‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zhang Q, Bi J, Zheng X, Chen Y, Wang H, Wu W, et al. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti‐tumor immunity. Nat Immunol. 2018;19(7):723‐32. [DOI] [PubMed] [Google Scholar]
- 110. Gorvel L, Olive D. Targeting the “PVR–TIGIT axis” with immune checkpoint therapies. F1000Res. 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Hodgins JJ, Khan ST, Park MM, Auer RC, Ardolino M. Killers 2.0: NK cell therapies at the forefront of cancer control. J Clin Invest. 2019;129(9):3499‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Roman Aguilera A, Lutzky VP, Mittal D, Li X‐Y, Stannard K, Takeda K, et al. CD96 targeted antibodies need not block CD96‐CD155 interactions to promote NK cell anti‐metastatic activity. Oncoimmunology. 2018;7(5):e1424677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Chan CJ, Martinet L, Gilfillan S, Souza‐Fonseca‐Guimaraes F, Chow MT, Town L, et al. The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat Immunol. 2014;15(5):431‐8. [DOI] [PubMed] [Google Scholar]
- 114. Wang J, Sanmamed MF, Datar I, Su TT, Ji L, Sun J, et al. Fibrinogen‐like protein 1 is a major immune inhibitory ligand of LAG‐3. Cell. 2019;176(1‐2):334‐47. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Xu F, Liu J, Liu D, Liu B, Wang M, Hu Z, et al. LSECtin expressed on melanoma cells promotes tumor progression by inhibiting antitumor T‐cell responses. Cancer Res. 2014;74(13):3418‐28. [DOI] [PubMed] [Google Scholar]
- 116. Kisielow M, Kisielow J, Capoferri‐Sollami G, Karjalainen K. Expression of lymphocyte activation gene 3 (LAG‐3) on B cells is induced by T cells. Eur J Immunol. 2005;35(7):2081‐8. [DOI] [PubMed] [Google Scholar]
- 117. Juno JA, Stalker AT, Waruk JL, Oyugi J, Kimani M, Plummer FA, et al. Elevated expression of LAG‐3, but not PD‐1, is associated with impaired iNKT cytokine production during chronic HIV‐1 infection and treatment. Retrovirology. 2015;12(1):1‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Shi A‐P, Tang X‐Y, Xiong Y‐L, Zheng K‐F, Liu Y‐J, Shi X‐G, et al. Immune Checkpoint LAG3 and Its Ligand FGL1 in Cancer. Front Immunol. 2022;12:785091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Burton BR, Britton GJ, Fang H, Verhagen J, Smithers B, Sabatos‐Peyton CA, et al. Sequential transcriptional changes dictate safe and effective antigen‐specific immunotherapy. Nat Commun. 2014;5(1):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Catakovic K, Klieser E, Neureiter D, Geisberger R. T cell exhaustion: from pathophysiological basics to tumor immunotherapy. Cell Commun Signal. 2017;15(1):1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Marhelava K, Pilch Z, Bajor M, Graczyk‐Jarzynka A, Zagozdzon R. Targeting negative and positive immune checkpoints with monoclonal antibodies in therapy of cancer. Cancers. 2019;11(11):1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Brignone C, Escudier B, Grygar C, Marcu M, Triebel F. A phase I pharmacokinetic and biological correlative study of IMP321, a novel MHC class II agonist, in patients with advanced renal cell carcinoma. Clin Cancer Res. 2009;15(19):6225‐31. [DOI] [PubMed] [Google Scholar]
- 123. Brignone C, Gutierrez M, Mefti F, Brain E, Jarcau R, Cvitkovic F, et al. First‐line chemoimmunotherapy in metastatic breast carcinoma: combination of paclitaxel and IMP321 (LAG‐3Ig) enhances immune responses and antitumor activity. J Transl Med. 2010;8(1):1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Chang W‐S, Kim J‐Y, Kim Y‐J, Kim Y‐S, Lee J‐M, Azuma M, et al. Cutting edge: Programmed death‐1/programmed death ligand 1 interaction regulates the induction and maintenance of invariant NKT cell anergy. J Immunol. 2008;181(10):6707‐10. [DOI] [PubMed] [Google Scholar]
- 125. Pesce S, Greppi M, Tabellini G, Rampinelli F, Parolini S, Olive D, et al. Identification of a subset of human natural killer cells expressing high levels of programmed death 1: A phenotypic and functional characterization. J Allergy Clin Immunol. 2017;139(1):335‐46. e3. [DOI] [PubMed] [Google Scholar]
- 126. Tabellini G, Benassi M, Marcenaro E, Coltrini D, Patrizi O, Ricotta D, et al. Primitive neuroectodermal tumor in an ovarian cystic teratoma: natural killer and neuroblastoma cell analysis. Case Rep Oncol. 2014;7(1):70‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Beldi‐Ferchiou A, Lambert M, Dogniaux S, Vély F, Vivier E, Olive D, et al. PD‐1 mediates functional exhaustion of activated NK cells in patients with Kaposi sarcoma. Oncotarget. 2016;7(45):72961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Benson DM Jr, Bakan CE, Mishra A, Hofmeister CC, Efebera Y, Becknell B, et al. The PD‐1/PD‐L1 axis modulates the natural killer cell versus multiple myeloma effect: a therapeutic target for CT‐011, a novel monoclonal anti–PD‐1 antibody. Blood, The Journal of the American Society of Hematology. 2010;116(13):2286‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Della Chiesa M, Pesce S, Muccio L, Carlomagno S, Sivori S, Moretta A, et al. Features of memory‐like and PD‐1+ human NK cell subsets. Front Immunol. 2016;7:351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Hwang S, Han J, Baek J‐S, Tak E, Song G‐W, Lee S‐G, et al. Cytotoxicity of human hepatic intrasinusoidal CD56bright natural killer cells against hepatocellular carcinoma cells. Int J Mol Sci. 2019;20(7):1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Lanuza PM, Vigueras A, Olivan S, Prats AC, Costas S, Llamazares G, et al. Activated human primary NK cells efficiently kill colorectal cancer cells in 3D spheroid cultures irrespectively of the level of PD‐L1 expression. Oncoimmunology. 2018;7(4):e1395123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Hsu J, Hodgins JJ, Marathe M, Nicolai CJ, Bourgeois‐Daigneault M‐C, Trevino TN, et al. Contribution of NK cells to immunotherapy mediated by PD‐1/PD‐L1 blockade. J Clin Invest. 2018;128(10):4654‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Wiesmayr S, Webber SA, Macedo C, Popescu I, Smith L, Luce J, et al. Decreased NKp46 and NKG2D and elevated PD‐1 are associated with altered NK‐cell function in pediatric transplant patients with PTLD. Eur J Immunol. 2012;42(2):541‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Dong W, Wu X, Ma S, Wang Y, Nalin AP, Zhu Z, et al. The mechanism of Anti–PD‐L1 antibody efficacy against PD‐L1–Negative tumors identifies NK cells expressing PD‐L1 as a cytolytic effector. Cancer Discov. 2019;9(10):1422‐37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Park J‐E, Kim S‐E, Keam B, Park H‐R, Kim S, Kim M, et al. Anti‐tumor effects of NK cells and anti‐PD‐L1 antibody with antibody‐dependent cellular cytotoxicity in PD‐L1‐positive cancer cell lines. J Immunother Cancer. 2020;8(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Di Virgilio F, Sarti AC, Falzoni S, De Marchi E, Adinolfi E. Extracellular ATP and P2 purinergic signalling in the tumour microenvironment. Nat Rev Cancer. 2018;18(10):601‐18. [DOI] [PubMed] [Google Scholar]
- 137. Khair DO, Bax HJ, Mele S, Crescioli S, Pellizzari G, Khiabany A, et al. Combining immune checkpoint inhibitors: established and emerging targets and strategies to improve outcomes in melanoma. Front Immunol. 2019;10:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Stagg J, Smyth M. Extracellular adenosine triphosphate and adenosine in cancer. Oncogene. 2010;29(39):5346‐58. [DOI] [PubMed] [Google Scholar]
- 139. Harmon C, Robinson MW, Hand F, Almuaili D, Mentor K, Houlihan DD, et al. Lactate‐mediated acidification of tumor microenvironment induces apoptosis of liver‐resident NK cells in colorectal liver metastasis. Cancer Immunol Res. 2019;7(2):335‐46. [DOI] [PubMed] [Google Scholar]
- 140. Chambers AM, Wang J, Dao TN, Lupo KB, Veenhuis P, Ayers MG, et al. Functional expression of CD73 on human natural killer cells. Cancer Immunol Immunother. 2022;1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Oldenborg P‐A. CD47: a cell surface glycoprotein which regulates multiple functions of hematopoietic cells in health and disease. Int Sch Res Notices. 2013;2013:614619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Brown E, Hooper L, Ho T, Gresham H. Integrin‐associated protein: a 50‐kD plasma membrane antigen physically and functionally associated with integrins. J Cell Biol. 1990;111(6):2785‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Gao Q, Chen K, Gao L, Zheng Y, Yang Y‐G. Thrombospondin‐1 signaling through CD47 inhibits cell cycle progression and induces senescence in endothelial cells. Cell Death Dis. 2016;7(9):e2368‐e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Kim MJ, Lee J‐C, Lee J‐J, Kim S, Lee SG, Park S‐W, et al. Association of CD47 with natural killer cell‐mediated cytotoxicity of head‐and‐neck squamous cell carcinoma lines. Tumor Biology. 2008;29(1):28‐34. [DOI] [PubMed] [Google Scholar]
- 145. Liu X, Pu Y, Cron K, Deng L, Kline J, Frazier WA, et al. CD47 blockade triggers T cell–mediated destruction of immunogenic tumors. Nat Med. 2015;21(10):1209‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Pierson BA, Gupta K, Hu WS, Miller JS. Human natural killer cell expansion is regulated by thrombospondin‐mediated activation of transforming growth factor‐beta 1 and independent accessory cell‐derived contact and soluble factors. Blood. 1996;87(1):180‐9. [PubMed] [Google Scholar]
- 147. Hofmeyer KA, Ray A, Zang X. The contrasting role of B7‐H3. Proc Natl Acad Sci. 2008;105(30):10277‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Yang S, Wei W, Zhao Q. B7‐H3, a checkpoint molecule, as a target for cancer immunotherapy. Int J Biol Sci. 2020;16(11):1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Janakiram M, Shah UA, Liu W, Zhao A, Schoenberg MP, Zang X. The third group of the B7‐CD 28 immune checkpoint family: HHLA 2, TMIGD 2, B7x, and B7‐H3. Immunol Rev. 2017;276(1):26‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Kraan J, van den Broek P, Verhoef C, Grunhagen DJ, Taal W, Gratama J‐W, et al. Endothelial CD276 (B7‐H3) expression is increased in human malignancies and distinguishes between normal and tumour‐derived circulating endothelial cells. Br J Cancer. 2014;111(1):149‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Loo D, Alderson RF, Chen FZ, Huang L, Zhang W, Gorlatov S, et al. Development of an Fc‐Enhanced Anti–B7‐H3 Monoclonal Antibody with Potent Antitumor ActivityDevelopment of Fc‐Enhanced Anti–B7‐H3 Monoclonal Antibody. Clin Cancer Res. 2012;18(14):3834‐45. [DOI] [PubMed] [Google Scholar]
- 152. Powderly J, Cote G, Flaherty K, Szmulewitz RZ, Ribas A, Weber J, et al. Interim results of an ongoing Phase I, dose escalation study of MGA271 (Fc‐optimized humanized anti‐B7‐H3 monoclonal antibody) in patients with refractory B7‐H3‐expressing neoplasms or neoplasms whose vasculature expresses B7‐H3. J Immunother Cancer. 2015;3(2):1‐2.25648675 [Google Scholar]
- 153. Demaria O, Gauthier L, Debroas G, Vivier E. Natural killer cell engagers in cancer immunotherapy: Next generation of immuno‐oncology treatments. Eur J Immunol. 2021;51(8):1934‐42. [DOI] [PubMed] [Google Scholar]
- 154. Liu J, Yang S, Cao B, Zhou G, Zhang F, Wang Y, et al. Targeting B7‐H3 via chimeric antigen receptor T cells and bispecific killer cell engagers augments antitumor response of cytotoxic lymphocytes. J Hematol Oncol. 2021;14(1):1‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Bogen JP, Carrara SC, Fiebig D, Grzeschik J, Hock B, Kolmar H. Design of a trispecific checkpoint inhibitor and natural killer cell engager based on a 2+ 1 common light chain antibody architecture. Front Immunol. 2021;12:669496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Passariello M, Camorani S, Vetrei C, Ricci S, Cerchia L, De Lorenzo C. Ipilimumab and its derived EGFR aptamer‐based conjugate induce efficient NK cell activation against cancer cells. Cancers. 2020;12(2):331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Zhang D, Zheng Y, Lin Z, Liu X, Li J, Yang H, et al. Equipping natural killer cells with specific targeting and checkpoint blocking aptamers for enhanced adoptive immunotherapy in solid tumors. Angew Chem. 2020;132(29):12120‐6. [DOI] [PubMed] [Google Scholar]
- 158. Zheng A, Du Y, Wang Y, Zheng Y, Ning Z, Wu M, et al. CD16/PD‐L1 bi‐specific aptamer for cancer immunotherapy through recruiting NK cells and acting as immunocheckpoint blockade. Mol Ther‐Nucleic Acids. 2022;27:998‐1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Schmiedel D, Mandelboim O. NKG2D ligands–critical targets for cancer immune escape and therapy. Front Immunol. 2018:2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Classen CF, Falk CS, Friesen C, Fulda S, Herr I, Debatin K‐M. Natural killer resistance of a drug‐resistant leukemia cell line, mediated by up‐regulation of HLA class I expression. Haematologica. 2003;88(5):509‐21. [PubMed] [Google Scholar]
- 161. Lim WA, June CH. The principles of engineering immune cells to treat cancer. Cell. 2017;168(4):724‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Maher J, Brentjens RJ, Gunset G, Rivière I, Sadelain M. Human T‐lymphocyte cytotoxicity and proliferation directed by a single chimeric TCRζ/CD28 receptor. Nat Biotechnol. 2002;20(1):70‐5. [DOI] [PubMed] [Google Scholar]
- 163. Park S, Shevlin E, Vedvyas Y, Zaman M, Park S, Hsu Y‐MS, et al. Micromolar affinity CAR T cells to ICAM‐1 achieves rapid tumor elimination while avoiding systemic toxicity. Sci Rep. 2017;7(1):1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Lynch A, Hawk W, Nylen E, Ober S, Autin P, Barber A. Adoptive transfer of murine T cells expressing a chimeric‐PD 1‐Dap10 receptor as an immunotherapy for lymphoma. Immunology. 2017;152(3):472‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Thayaparan T, Petrovic RM, Achkova DY, Zabinski T, Davies DM, Klampatsa A, et al. CAR T‐cell immunotherapy of MET‐expressing malignant mesothelioma. Oncoimmunology. 2017;6(12):e1363137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Jamnani FR, Rahbarizadeh F, Shokrgozar MA, Mahboudi F, Ahmadvand D, Sharifzadeh Z, et al. T cells expressing VHH‐directed oligoclonal chimeric HER2 antigen receptors: towards tumor‐directed oligoclonal T cell therapy. Biochimica et Biophysica Acta (BBA)‐General Subjects. 2014;1840(1):378‐86. [DOI] [PubMed] [Google Scholar]
- 167. Hammill JA, VanSeggelen H, Helsen CW, Denisova GF, Evelegh C, Tantalo DG, et al. Designed ankyrin repeat proteins are effective targeting elements for chimeric antigen receptors. J Immunother Cancer. 2015;3(1):1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Guedan S, Calderon H, Posey AD Jr, Maus MV. Engineering and design of chimeric antigen receptors. Mol Ther‐Methods Clin Dev. 2019;12:145‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Guest RD, Hawkins RE, Kirillova N, Cheadle EJ, Arnold J, O'Neill A, et al. The role of extracellular spacer regions in the optimal design of chimeric immune receptors: evaluation of four different scFvs and antigens. J Immunother. 2005;28(3):203‐11. [DOI] [PubMed] [Google Scholar]
- 170. James SE, Greenberg PD, Jensen MC, Lin Y, Wang J, Till BG, et al. Antigen sensitivity of CD22‐specific chimeric TCR is modulated by target epitope distance from the cell membrane. J Immunol. 2008;180(10):7028‐38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Hudecek M, Lupo‐Stanghellini M‐T, Kosasih PL, Sommermeyer D, Jensen MC, Rader C, et al. Receptor affinity and extracellular domain modifications affect tumor recognition by ROR1‐specific chimeric antigen receptor T cells. Clin Cancer Res. 2013;19(12):3153‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Hombach A, Heuser C, Gerken M, Fischer B, Lewalter K, Diehl V, et al. T cell activation by recombinant FcεRI γ‐chain immune receptors: an extracellular spacer domain impairs antigen‐dependent T cell activation but not antigen recognition. Gene Ther. 2000;7(12):1067‐75. [DOI] [PubMed] [Google Scholar]
- 173. Zhang C, Liu J, Zhong JF, Zhang X. Engineering car‐t cells. Biomark Res. 2017;5(1):1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Smith AJ, Oertle J, Warren D, Prato D. Chimeric antigen receptor (CAR) T cell therapy for malignant cancers: summary and perspective. J Cell Immunother. 2016;2(2):59‐68. [Google Scholar]
- 175. Acuto O, Michel F. CD28‐mediated co‐stimulation: a quantitative support for TCR signalling. Nat Rev Immunol. 2003;3(12):939‐51. [DOI] [PubMed] [Google Scholar]
- 176. Hombach A, Wieczarkowiecz A, Marquardt T, Heuser C, Usai L, Pohl C, et al. Tumor‐specific T cell activation by recombinant immunoreceptors: CD3ζ signaling and CD28 costimulation are simultaneously required for efficient IL‐2 secretion and can be integrated into one combined CD28/CD3ζ signaling receptor molecule. J Immunol. 2001;167(11):6123‐31. [DOI] [PubMed] [Google Scholar]
- 177. Finney HM, Lawson AD, Bebbington CR, Weir ANC. Chimeric receptors providing both primary and costimulatory signaling in T cells from a single gene product. J Immunol. 1998;161(6):2791‐7. [PubMed] [Google Scholar]
- 178. Jain MD, Davila ML. Concise review: emerging principles from the clinical application of chimeric antigen receptor T cell therapies for B cell malignancies. Stem Cells. 2018;36(1):36‐44. [DOI] [PubMed] [Google Scholar]
- 179. Chmielewski M, Kopecky C, Hombach AA, Abken H. IL‐12 release by engineered T cells expressing chimeric antigen receptors can effectively Muster an antigen‐independent macrophage response on tumor cells that have shut down tumor antigen expression. Cancer Res. 2011;71(17):5697‐706. [DOI] [PubMed] [Google Scholar]
- 180. Chmielewski M, Abken H. TRUCKs: the fourth generation of CARs. Expert Opin Biol Ther. 2015;15(8):1145‐54. [DOI] [PubMed] [Google Scholar]
- 181. Kagoya Y, Tanaka S, Guo T, Anczurowski M, Wang C‐H, Saso K, et al. A novel chimeric antigen receptor containing a JAK–STAT signaling domain mediates superior antitumor effects. Nat Med. 2018;24(3):352‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Tahmasebi S, Elahi R, Khosh E, Esmaeilzadeh A. Programmable and multi‐targeted CARs: a new breakthrough in cancer CAR‐T cell therapy. Clin Transl Oncol. 2021;23(6):1003‐19. [DOI] [PubMed] [Google Scholar]
- 183. Klingemann H. Are natural killer cells superior CAR drivers? Oncoimmunology. 2014;3(4):e28147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295(5562):2097‐100. [DOI] [PubMed] [Google Scholar]
- 185. Porrata LF, Inwards DJ, Ansell SM, Micallef IN, Johnston PB, Gastineau DA, et al. Early lymphocyte recovery predicts superior survival after autologous stem cell transplantation in non‐Hodgkin lymphoma: a prospective study. Biol Blood Marrow Transplant. 2008;14(7):807‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Becker PS, Suck G, Nowakowska P, Ullrich E, Seifried E, Bader P, et al. Selection and expansion of natural killer cells for NK cell‐based immunotherapy. Cancer Immunol Immunother. 2016;65(4):477‐84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Ciurea SO, Schafer JR, Bassett R, Denman CJ, Cao K, Willis D, et al. Phase 1 clinical trial using mbIL21 ex vivo–expanded donor‐derived NK cells after haploidentical transplantation. Blood, The Journal of the American Society of Hematology. 2017;130(16):1857‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Concha‐Benavente F, Kansy B, Moskovitz J, Moy J, Chandran U, Ferris RL. PD‐L1 mediates dysfunction in activated PD‐1+ NK cells in head and neck cancer patients. Cancer Immunol Res. 2018;6(12):1548‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Sakamoto N, Ishikawa T, Kokura S, Okayama T, Oka K, Ideno M, et al. Phase I clinical trial of autologous NK cell therapy using novel expansion method in patients with advanced digestive cancer. J Transl Med. 2015;13(1):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Zhang Y, Wallace DL, De Lara CM, Ghattas H, Asquith B, Worth A, et al. In vivo kinetics of human natural killer cells: the effects of ageing and acute and chronic viral infection. Immunology. 2007;121(2):258‐65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Dranoff G. GM‐CSF‐based cancer vaccines. Immunol Rev. 2002;188(1):147‐54. [DOI] [PubMed] [Google Scholar]
- 192. Harrison C. Calming the cytokine storm. Nat Rev Drug Discovery. 2010;9(5):360‐1. [DOI] [PubMed] [Google Scholar]
- 193. Han B, Mao F‐y, Zhao Y‐l, Lv Y‐p, Teng Y‐s, Duan M, et al. Altered NKp30, NKp46, NKG2D, and DNAM‐1 expression on circulating NK cells is associated with tumor progression in human gastric cancer. J Immunol Res. 2018;2018:6248590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Hatjiharissi E, Xu L, Santos DD, Hunter ZR, Ciccarelli BT, Verselis S, et al. Increased natural killer cell expression of CD16, augmented binding and ADCC activity to rituximab among individuals expressing the FcγRIIIa‐158 V/V and V/F polymorphism. Blood, The Journal of the American Society of Hematology. 2007;110(7):2561‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Miller JS, Soignier Y, Panoskaltsis‐Mortari A, McNearney SA, Yun GH, Fautsch SK, et al. Successful adoptive transfer and in vivo expansion of human haploidentical NK cells in patients with cancer. Blood. 2005;105(8):3051‐7. [DOI] [PubMed] [Google Scholar]
- 196. Mrózek E, Anderson P, Caligiuri MA. Role of interleukin‐15 in the development of human CD56+ natural killer cells from CD34+ hematopoietic progenitor cells. Blood. 1996;87(7):2632‐40. [PubMed] [Google Scholar]
- 197. Knorr DA, Ni Z, Hermanson D, Hexum MK, Bendzick L, Cooper LJ, et al. Clinical‐scale derivation of natural killer cells from human pluripotent stem cells for cancer therapy. Stem Cells Transl Med. 2013;2(4):274‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Woll PS, Martin CH, Miller JS, Kaufman DS. Human embryonic stem cell‐derived NK cells acquire functional receptors and cytolytic activity. J Immunol. 2005;175(8):5095‐103. [DOI] [PubMed] [Google Scholar]
- 199. Shah N, Martin‐Antonio B, Yang H, Ku S, Lee DA, Cooper LJ, et al. Antigen presenting cell‐mediated expansion of human umbilical cord blood yields log‐scale expansion of natural killer cells with anti‐myeloma activity. PLoS One. 2013;8(10):e76781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Kang L, Voskinarian‐Berse V, Law E, Reddin T, Bhatia M, Hariri A, et al. Characterization and ex vivo expansion of human placenta‐derived natural killer cells for cancer immunotherapy. Front Immunol. 2013;4:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Cheng M, Zhang J, Jiang W, Chen Y, Tian Z. Natural killer cell lines in tumor immunotherapy. Front Med. 2012;6(1):56‐66. [DOI] [PubMed] [Google Scholar]
- 202. Alvarez M, Simonetta F, Baker J, Morrison AR, Wenokur AS, Pierini A, et al. Indirect impact of PD‐1/PD‐L1 blockade on a murine model of NK cell exhaustion. Front Immunol. 2020;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Grossenbacher SK, Canter RJ, Murphy WJ. Natural killer cell immunotherapy to target stem‐like tumor cells. J Immunother Cancer. 2016;4(1):1‐3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Ames E, Canter RJ, Grossenbacher SK, Mac S, Chen M, Smith RC, et al. NK cells preferentially target tumor cells with a cancer stem cell phenotype. J Immunol. 2015;195(8):4010‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Miller JS, Tessmer‐Tuck J, Pierson BA, Weisdorf D, McGlave P, Blazar BR, et al. Low dose subcutaneous interleukin‐2 after autologous transplantation generates sustained in vivo natural killer cell activity. Biol Blood Marrow Transplant. 1997;3(1):34‐44. [PubMed] [Google Scholar]
- 206. Burns L, Weisdorf D, DeFor T, Vesole D, Repka T, Blazar B, et al. IL‐2‐based immunotherapy after autologous transplantation for lymphoma and breast cancer induces immune activation and cytokine release: a phase I/II trial. Bone Marrow Transplant. 2003;32(2):177‐86. [DOI] [PubMed] [Google Scholar]
- 207. Gasteiger G, Hemmers S, Firth MA, Le Floc'h A, Huse M, Sun JC, et al. IL‐2–dependent tuning of NK cell sensitivity for target cells is controlled by regulatory T cells. J Exp Med. 2013;210(6):1167‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Ghiringhelli F, Ménard C, Terme M, Flament C, Taieb J, Chaput N, et al. CD4+ CD25+ regulatory T cells inhibit natural killer cell functions in a transforming growth factor–β–dependent manner. J Exp Med. 2005;202(8):1075‐85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Smyth MJ, Hayakawa Y, Takeda K, Yagita H. New aspects of natural‐killer‐cell surveillance and therapy of cancer. Nat Rev Cancer. 2002;2(11):850‐61. [DOI] [PubMed] [Google Scholar]
- 210. Cantoni C, Huergo‐Zapico L, Parodi M, Pedrazzi M, Mingari MC, Moretta A, et al. NK cells, tumor cell transition, and tumor progression in solid malignancies: new hints for NK‐based immunotherapy? J Immunol Res. 2016;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Veluchamy JP, Kok N, van der Vliet HJ, Verheul HM, de Gruijl TD, Spanholtz J. The rise of allogeneic natural killer cells as a platform for cancer immunotherapy: recent innovations and future developments. Front Immunol. 2017;8:631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Szmania S, Lapteva N, Garg T, Greenway A, Lingo J, Nair B, et al. Ex vivo expanded natural killer cells demonstrate robust proliferation in vivo in high‐risk relapsed multiple myeloma patients. J Immunother. 2015;38(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Munoz J, Shah N, Rezvani K, Hosing C, Bollard CM, Oran B, et al. Concise review: umbilical cord blood transplantation: past, present, and future. Stem Cells Transl Med. 2014;3(12):1435‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Mehta RS, Shpall EJ, Rezvani K. Cord blood as a source of natural killer cells. Front Med. 2016;2:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Dalle J‐H, Menezes J, Wagner É, Blagdon M, Champagne J, Champagne MA, et al. Characterization of cord blood natural killer cells: implications for transplantation and neonatal infections. Pediatr Res. 2005;57(5):649‐55. [DOI] [PubMed] [Google Scholar]
- 216. Liu E, Marin D, Banerjee P, Macapinlac HA, Thompson P, Basar R, et al. Use of CAR‐transduced natural killer cells in CD19‐positive lymphoid tumors. N Engl J Med. 2020;382(6):545‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Luevano M, Daryouzeh M, Alnabhan R, Querol S, Khakoo S, Madrigal A, et al. The unique profile of cord blood natural killer cells balances incomplete maturation and effective killing function upon activation. Hum Immunol. 2012;73(3):248‐57. [DOI] [PubMed] [Google Scholar]
- 218. Tanaka H, Kai S, Yamaguchi M, Misawa M, Fujimori Y, Yamamoto M, et al. Analysis of natural killer (NK) cell activity and adhesion molecules on NK cells from umbilical cord blood. Eur J Haematol. 2003;71(1):29‐38. [DOI] [PubMed] [Google Scholar]
- 219. Sarvaria A, Jawdat D, Madrigal JA, Saudemont A. Umbilical cord blood natural killer cells, their characteristics, and potential clinical applications. Front Immunol. 2017;8:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Gill S, Vasey AE, De Souza A, Baker J, Smith AT, Kohrt HE, et al. Rapid development of exhaustion and down‐regulation of eomesodermin limit the antitumor activity of adoptively transferred murine natural killer cells. Blood, The Journal of the American Society of Hematology. 2012;119(24):5758‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, et al. Effector and memory CD8+ T cell fate coupled by T‐bet and eomesodermin. Nat Immunol. 2005;6(12):1236‐44. [DOI] [PubMed] [Google Scholar]
- 222. Fang F, Xiao W, Tian Z. NK cell‐based immunotherapy for cancer. Semin Immunol. 2017;31:37‐54. [DOI] [PubMed] [Google Scholar]
- 223. Tonn T, Schwabe D, Klingemann HG, Becker S, Esser R, Koehl U, et al. Treatment of patients with advanced cancer with the natural killer cell line NK‐92. Cytotherapy. 2013;15(12):1563‐70. [DOI] [PubMed] [Google Scholar]
- 224. Klingemann H, Boissel L, Toneguzzo F. Natural killer cells for immunotherapy–advantages of the NK‐92 cell line over blood NK cells. Front Immunol. 2016;91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Gong J‐H, Maki G, Klingemann HG. Characterization of a human cell line (NK‐92) with phenotypical and functional characteristics of activated natural killer cells. Leukemia. 1994;8(4):652‐8. [PubMed] [Google Scholar]
- 226. Nowakowska P, Romanski A, Miller N, Odendahl M, Bonig H, Zhang C, et al. Clinical grade manufacturing of genetically modified, CAR‐expressing NK‐92 cells for the treatment of ErbB2‐positive malignancies. Cancer Immunol Immunother. 2018;67(1):25‐38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Tang X, Yang L, Li Z, Nalin AP, Dai H, Xu T, et al. First‐in‐man clinical trial of CAR NK‐92 cells: safety test of CD33‐CAR NK‐92 cells in patients with relapsed and refractory acute myeloid leukemia. Am J Cancer Res. 2018;8(6):1083. [PMC free article] [PubMed] [Google Scholar]
- 228. Schönfeld K, Sahm C, Zhang C, Naundorf S, Brendel C, Odendahl M, et al. Selective inhibition of tumor growth by clonal NK cells expressing an ErbB2/HER2‐specific chimeric antigen receptor. Mol Ther. 2015;23(2):330‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Jochems C, Hodge JW, Fantini M, Fujii R, Maurice YM. An NK cell line (haNK) expressing high levels of granzyme and engineered to express the high affinity CD16 allele. Oncotarget. 2016;7(52):86359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Maki G, Klingemann H‐G, Martinson JA, Tam YK. Factors regulating the cytotoxic activity of the human natural killer cell line, NK‐92. J Hematother Stem Cell Res. 2001;10(3):369‐83. [DOI] [PubMed] [Google Scholar]
- 231. Cooley S, Xiao F, Pitt M, Gleason M, McCullar V, Bergemann TL, et al. A subpopulation of human peripheral blood NK cells that lacks inhibitory receptors for self‐MHC is developmentally immature. Blood, The Journal of the American Society of Hematology. 2007;110(2):578‐86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Faure M, Long EO. KIR2DL4 (CD158d), an NK cell‐activating receptor with inhibitory potential. J Immunol. 2002;168(12):6208‐14. [DOI] [PubMed] [Google Scholar]
- 233. Suck G, Odendahl M, Nowakowska P, Seidl C, Wels WS, Klingemann HG, et al. NK‐92: an ‘off‐the‐shelf therapeutic'for adoptive natural killer cell‐based cancer immunotherapy. Cancer Immunol Immunother. 2016;65(4):485‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Bergman H, Sissala N, HÄgerstrand H, Lindqvist C. Human NK‐92 Cells Function as Target Cells for Human NK Cells–Implications for CAR NK‐92 Therapies. Anticancer Res. 2020;40(10):5355‐9. [DOI] [PubMed] [Google Scholar]
- 235. MacLeod RA, Nagel S, Kaufmann M, Greulich‐Bode K, Drexler HG. Multicolor‐FISH analysis of a natural killer cell line (NK‐92). Leuk Res. 2002;26(11):1027‐33. [DOI] [PubMed] [Google Scholar]
- 236. Chrobok M, Dahlberg CI, Sayitoglu EC, Beljanski V, Nahi H, Gilljam M, et al. Functional assessment for clinical use of serum‐free adapted NK‐92 cells. Cancers. 2019;11(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Rafei H, Daher M, Rezvani K. Chimeric antigen receptor (CAR) natural killer (NK)‐cell therapy: leveraging the power of innate immunity. Br J Haematol. 2021;193(2):216‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Spanholtz J, Preijers F, Tordoir M, Trilsbeek C, Paardekooper J, De Witte T, et al. Clinical‐grade generation of active NK cells from cord blood hematopoietic progenitor cells for immunotherapy using a closed‐system culture process. PLoS One. 2011;6(6):e20740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Giuliani M, Giron‐Michel J, Negrini S, Vacca P, Durali D, Caignard A, et al. Generation of a novel regulatory NK cell subset from peripheral blood CD34+ progenitors promoted by membrane‐bound IL‐15. PLoS One. 2008;3(5):e2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Kiessling R, Klein E, Wigzell H. “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol. 1975;5(2):112‐7. [DOI] [PubMed] [Google Scholar]
- 241. Lotzová E, Savary CA, Champlin RE. Genesis of human oncolytic natural killer cells from primitive CD34+CD33‐ bone marrow progenitors. J Immunol. 1993;150(12):5263‐9. [PubMed] [Google Scholar]
- 242. Giarratana MC, Vergé V, Schmitt C, Bertho JM, Kobari L, Barret C, et al. Presence of primitive lymphoid progenitors with NK or B potential in ex vivo expanded bone marrow cell cultures. Exp Hematol. 2000;28(1):46‐54. [DOI] [PubMed] [Google Scholar]
- 243. Yu Y, Hagihara M, Ando K, Gansuvd B, Matsuzawa H, Tsuchiya T, et al. Enhancement of human cord blood CD34+ cell‐derived NK cell cytotoxicity by dendritic cells. J Immunol. 2001;166(3):1590‐600. [DOI] [PubMed] [Google Scholar]
- 244. Kalberer CP, Siegler U, Wodnar‐Filipowicz A. Human NK cell development in NOD/SCID mice receiving grafts of cord blood CD34+ cells. Blood. 2003;102(1):127‐35. [DOI] [PubMed] [Google Scholar]
- 245. Perez SA, Sotiropoulou PA, Gkika DG, Mahaira LG, Niarchos DK, Gritzapis AD, et al. A novel myeloid‐like NK cell progenitor in human umbilical cord blood. Blood. 2003;101(9):3444‐50. [DOI] [PubMed] [Google Scholar]
- 246. Cany J, van der Waart AB, Spanholtz J, Tordoir M, Jansen JH, van der Voort R, et al. Combined IL‐15 and IL‐12 drives the generation of CD34+‐derived natural killer cells with superior maturation and alloreactivity potential following adoptive transfer. Oncoimmunology. 2015;4(7):e1017701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Lehmann D, Spanholtz J, Osl M, Tordoir M, Lipnik K, Bilban M, et al. Ex vivo generated natural killer cells acquire typical natural killer receptors and display a cytotoxic gene expression profile similar to peripheral blood natural killer cells. Stem Cells Dev. 2012;21(16):2926‐38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861‐72. [DOI] [PubMed] [Google Scholar]
- 249. Woll PS, Grzywacz B, Tian X, Marcus RK, Knorr DA, Verneris MR, et al. Human embryonic stem cells differentiate into a homogeneous population of natural killer cells with potent in vivo antitumor activity. Blood, The Journal of the American Society of Hematology. 2009;113(24):6094‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Ni Z, Knorr DA, Clouser CL, Hexum MK, Southern P, Mansky LM, et al. Human pluripotent stem cells produce natural killer cells that mediate anti‐HIV‐1 activity by utilizing diverse cellular mechanisms. J Virol. 2011;85(1):43‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Fujisaki H, Kakuda H, Imai C, Mullighan CG, Campana D. Replicative potential of human natural killer cells. Br J Haematol. 2009;145(5):606‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Lapteva N, Durett AG, Sun J, Rollins LA, Huye LL, Fang J, et al. Large‐scale ex vivo expansion and characterization of natural killer cells for clinical applications. Cytotherapy. 2012;14(9):1131‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Zeng J, Tang SY, Toh LL, Wang S. Generation of “off‐the‐shelf” natural killer cells from peripheral blood cell‐derived induced pluripotent stem cells. Stem Cell Reports. 2017;9(6):1796‐812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Hermanson DL, Bendzick L, Pribyl L, McCullar V, Vogel RI, Miller JS, et al. Induced Pluripotent Stem Cell‐Derived Natural Killer Cells for Treatment of Ovarian Cancer. Stem Cells. 2016;34(1):93‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Li Y, Hermanson DL, Moriarity BS, Kaufman DS. Human iPSC‐Derived Natural Killer Cells Engineered with Chimeric Antigen Receptors Enhance Anti‐tumor Activity. Cell Stem Cell. 2018;23(2):181‐92.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Ng ES, Davis RP, Azzola L, Stanley EG, Elefanty AG. Forced aggregation of defined numbers of human embryonic stem cells into embryoid bodies fosters robust, reproducible hematopoietic differentiation. Blood. 2005;106(5):1601‐3. [DOI] [PubMed] [Google Scholar]
- 257. Eguizabal C, Zenarruzabeitia O, Monge J, Santos S, Vesga MA, Maruri N, et al. Natural killer cells for cancer immunotherapy: pluripotent stem cells‐derived NK cells as an immunotherapeutic perspective. Front Immunol. 2014;5:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Saetersmoen ML, Hammer Q, Valamehr B, Kaufman DS, Malmberg KJ. Off‐the‐shelf cell therapy with induced pluripotent stem cell‐derived natural killer cells. Semin Immunopathol. 2019;41(1):59‐68. [DOI] [PubMed] [Google Scholar]
- 259. Valamehr B, Robinson M, Abujarour R, Rezner B, Vranceanu F, Le T, et al. Platform for induction and maintenance of transgene‐free hiPSCs resembling ground state pluripotent stem cells. Stem Cell Reports. 2014;2(3):366‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Hermanson DL, Kaufman DS. Utilizing chimeric antigen receptors to direct natural killer cell activity. Front Immunol. 2015;6:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD, et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity. 2015;42(3):443‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, et al. Epigenetic modification and antibody‐dependent expansion of memory‐like NK cells in human cytomegalovirus‐infected individuals. Immunity. 2015;42(3):431‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Foley B, Cooley S, Verneris MR, Curtsinger J, Luo X, Waller EK, et al. Human cytomegalovirus (CMV)‐induced memory‐like NKG2C+ NK cells are transplantable and expand in vivo in response to recipient CMV antigen. J Immunol. 2012;189(10):5082‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Sarhan D, Cichocki F, Zhang B, Yingst A, Spellman SR, Cooley S, et al. Adaptive NK cells with low TIGIT expression are inherently resistant to myeloid‐derived suppressor cells. Cancer Res. 2016;76(19):5696‐706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Sarhan D, Hippen KL, Lemire A, Hying S, Luo X, Lenvik T, et al. Adaptive NK cells resist regulatory T‐cell suppression driven by IL37. Cancer Immunol Res. 2018;6(7):766‐75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Saez‐Borderias A, Romo N, Magri G, Gumá M, Angulo A, López‐Botet M. IL‐12‐dependent inducible expression of the CD94/NKG2A inhibitory receptor regulates CD94/NKG2C+ NK cell function. J Immunol. 2009;182(2):829‐36. [DOI] [PubMed] [Google Scholar]
- 267. Gang M, Marin ND, Wong P, Neal CC, Marsala L, Foster M, et al. CAR‐modified memory‐like NK cells exhibit potent responses to NK‐resistant lymphomas. Blood. 2020;136(20):2308‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Lee JM. When CAR Meets Stem Cells. Int J Mol Sci. 2019;20(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. De Oliveira SN, Ryan C, Giannoni F, Hardee CL, Tremcinska I, Katebian B, et al. Modification of hematopoietic stem/progenitor cells with CD19‐specific chimeric antigen receptors as a novel approach for cancer immunotherapy. Hum Gene Ther. 2013;24(10):824‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Tang SY, Zha S, Du Z, Zeng J, Zhu D, Luo Y, et al. Targeted integration of EpCAM‐specific CAR in human induced pluripotent stem cells and their differentiation into NK cells. Curr Stem Cell Res Ther. 2021;12(1):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Ueda T, Kumagai A, Iriguchi S, Yasui Y, Miyasaka T, Nakagoshi K, et al. Non–clinical efficacy, safety and stable clinical cell processing of induced pluripotent stem cell‐derived anti–glypican‐3 chimeric antigen receptor‐expressing natural killer/innate lymphoid cells. Cancer Sci. 2020;111(5):1478‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Zheng L, Ren L, Kouhi A, Khawli LA, Hu P, Kaslow HR, et al. A humanized Lym‐1 CAR with novel DAP10/DAP12 signaling domains demonstrates reduced tonic signaling and increased antitumor activity in B‐cell lymphoma models. Clin Cancer Res. 2020;26(14):3694‐706. [DOI] [PubMed] [Google Scholar]
- 273. Kotanides H, Sattler RM, Lebron MB, Carpenito C, Shen J, Li J, et al. Characterization of 7A5: a human CD137 (4‐1BB) receptor binding monoclonal antibody with differential agonist properties that promotes antitumor immunity. Mol Cancer Ther. 2020;19(4):988‐98. [DOI] [PubMed] [Google Scholar]
- 274. Moingeon P, Lucich JL, McConkey DJ, Letourneur F, Malissen B, Kochan J, et al. CD3 zeta dependence of the CD2 pathway of activation in T lymphocytes and natural killer cells. Proc Natl Acad Sci. 1992;89(4):1492‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Chen X, Allan DS, Krzewski K, Ge B, Kopcow H, Strominger JL. CD28‐stimulated ERK2 phosphorylation is required for polarization of the microtubule organizing center and granules in YTS NK cells. Proc Natl Acad Sci. 2006;103(27):10346‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Lang S, Vujanovic NL, Wollenberg B, Whiteside TL. Absence of B7. 1‐CD28/CTLA‐4‐mediated co‐stimulation in human NK cells. Eur J Immunol. 1998;28(3):780‐6. [DOI] [PubMed] [Google Scholar]
- 277. Oelsner S, Friede ME, Zhang C, Wagner J, Badura S, Bader P, et al. Continuously expanding CAR NK‐92 cells display selective cytotoxicity against B‐cell leukemia and lymphoma. Cytotherapy. 2017;19(2):235‐49. [DOI] [PubMed] [Google Scholar]
- 278. Gong Y, Klein Wolterink RG, Wang J, Bos GM, Germeraad WT. Chimeric antigen receptor natural killer (CAR‐NK) cell design and engineering for cancer therapy. J Hematol Oncol. 2021;14(1):1‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Zhang C, Oberoi P, Oelsner S, Waldmann A, Lindner A, Tonn T, et al. Chimeric antigen receptor‐engineered NK‐92 cells: an off‐the‐shelf cellular therapeutic for targeted elimination of cancer cells and induction of protective antitumor immunity. Front Immunol. 2017;8:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Töpfer K, Cartellieri M, Michen S, Wiedemuth R, Müller N, Lindemann D, et al. DAP12‐based activating chimeric antigen receptor for NK cell tumor immunotherapy. J Immunol. 2015;194(7):3201‐12. [DOI] [PubMed] [Google Scholar]
- 281. Müller N, Michen S, Tietze S, Töpfer K, Schulte A, Lamszus K, et al. Engineering NK cells modified with an EGFRvIII‐specific chimeric antigen receptor to overexpress CXCR4 improves immunotherapy of CXCL12/SDF‐1α‐secreting glioblastoma. J Immunother. 2015;38(5):197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Imai C, Iwamoto S, Campana D. Genetic modification of primary natural killer cells overcomes inhibitory signals and induces specific killing of leukemic cells. Blood. 2005;106(1):376‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Xiao L, Cen D, Gan H, Sun Y, Huang N, Xiong H, et al. Adoptive transfer of NKG2D CAR mRNA‐engineered natural killer cells in colorectal cancer patients. Mol Ther. 2019;27(6):1114‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Xu Y, Liu Q, Zhong M, Wang Z, Chen Z, Zhang Y, et al. 2B4 costimulatory domain enhancing cytotoxic ability of anti‐CD5 chimeric antigen receptor engineered natural killer cells against T cell malignancies. J Hematol Oncol. 2019;12(1):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Huang Y, Zeng J, Liu T, Xu Q, Song X, Zeng J. DNAM1 and 2B4 costimulatory domains enhance the cytotoxicity of anti‐GPC3 chimeric antigen receptor‐modified natural killer cells against hepatocellular cancer cells in vitro. Cancer Manag Res. 2020;12:3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Parihar R, Rivas C, Huynh M, Omer B, Lapteva N, Metelitsa LS, et al. NK cells expressing a chimeric activating receptor eliminate MDSCs and rescue impaired CAR‐T cell activity against solid tumors. Cancer Immunol Res. 2019;7(3):363‐75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Guo C, Wang X, Zhang H, Zhi L, Lv T, Li M, et al. Structure‐based rational design of a novel chimeric PD1‐NKG2D receptor for natural killer cells. Mol Immunol. 2019;114:108‐13. [DOI] [PubMed] [Google Scholar]
- 288. Lu C, Guo C, Chen H, Zhang H, Zhi L, Lv T, et al. A novel chimeric PD1‐NKG2D‐41BB receptor enhances antitumor activity of NK92 cells against human lung cancer H1299 cells by triggering pyroptosis. Mol Immunol. 2020;122:200‐6. [DOI] [PubMed] [Google Scholar]
- 289. Li M, Zhi L, Yin M, Guo C, Zhang H, Lu C, et al. A novel bispecific chimeric PD1‐DAP10/NKG2D receptor augments NK92‐cell therapy efficacy for human gastric cancer SGC‐7901 cell. Biochem Biophys Res Commun. 2020;523(3):745‐52. [DOI] [PubMed] [Google Scholar]
- 290. Xu DL, He YQ, Xiao B, Si Y, Shi J, Liu XA, et al. A Novel Sushi‐IL15‐PD1 CAR‐NK92 Cell Line With Enhanced and PD‐L1 Targeted Cytotoxicity Against Pancreatic Cancer Cells. Front Oncol. 2022;12:726985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Pomeroy EJ, Hunzeker JT, Kluesner MG, Lahr WS, Smeester BA, Crosby MR, et al. A genetically engineered primary human natural killer cell platform for cancer immunotherapy. Mol Ther. 2020;28(1):52‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Fabian KP, Padget MR, Donahue RN, Solocinski K, Robbins Y, Allen CT, et al. PD‐L1 targeting high‐affinity NK (t‐haNK) cells induce direct antitumor effects and target suppressive MDSC populations. J Immunother Cancer. 2020;8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Robbins Y, Greene S, Friedman J, Clavijo PE, Van Waes C, Fabian KP, et al. Tumor control via targeting PD‐L1 with chimeric antigen receptor modified NK cells. Elife. 2020;9:e54854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Lee MY, Robbins Y, Sievers C, Friedman J, Sater HA, Clavijo PE, et al. Chimeric antigen receptor engineered NK cellular immunotherapy overcomes the selection of T‐cell escape variant cancer cells. J Immunother Cancer. 2021;9(3):e002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Bajor M, Graczyk‐Jarzynka A, Marhelava K, Burdzinska A, Muchowicz A, Goral A, et al. PD‐L1 CAR effector cells induce self‐amplifying cytotoxic effects against target cells. J Immunother Cancer. 2022;10(1):e002500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Yoon DH, Osborn MJ, Tolar J, Kim CJ. Incorporation of immune checkpoint blockade into chimeric antigen receptor T cells (CAR‐Ts): combination or built‐in CAR‐T. Int J Mol Sci. 2018;19(2):340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Adusumilli PS, Zauderer MG. A Phase I Trial of Regional Mesothelin‐Targeted CAR T‐cell Therapy in Patients with Malignant Pleural Disease, in Combination with the Anti‐PD‐1 Agent Pembrolizumab. Cancer Discov. 2021;11(11):2748‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Li S, Siriwon N, Zhang X, Yang S, Jin T, He F, et al. Enhanced cancer immunotherapy by chimeric antigen receptor–modified T cells engineered to secrete checkpoint inhibitors. Clin Cancer Res. 2017;23(22):6982‐92. [DOI] [PubMed] [Google Scholar]
- 299. Cherkassky L, Morello A, Villena‐Vargas J, Feng Y, Dimitrov DS, Jones DR, et al. Human CAR T cells with cell‐intrinsic PD‐1 checkpoint blockade resist tumor‐mediated inhibition. J Clin Invest. 2016;126(8):3130‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Shi X, Zhang D, Li F, Zhang Z, Wang S, Xuan Y, et al. Targeting glycosylation of PD‐1 to enhance CAR‐T cell cytotoxicity. J Hematol Oncol. 2019;12(1):1‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Grote S, Chan KCH, Baden C, Bösmüller H, Sulyok M, Frauenfeld L, et al. CD276 as a novel CAR NK‐92 therapeutic target for neuroblastoma. Advances in Cell and Gene Therapy. 2021;4(1):e105. [Google Scholar]
- 302. Grote S, Ureña‐Bailén G, Chan KC‐H, Baden C, Mezger M, Handgretinger R, et al. In Vitro Evaluation of CD276‐CAR NK‐92 Functionality, Migration and Invasion Potential in the Presence of Immune Inhibitory Factors of the Tumor Microenvironment. Cells. 2021;10(5):1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Yang S, Cao B, Zhou G, Zhu L, Wang L, Zhang L, et al. Targeting B7‐H3 immune checkpoint with chimeric antigen receptor‐engineered natural killer cells exhibits potent cytotoxicity against non‐small cell lung cancer. Front Pharmacol. 2020;11:1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Jan C‐I, Huang S‐W, Canoll P, Bruce JN, Lin Y‐C, Pan C‐M, et al. Targeting human leukocyte antigen G with chimeric antigen receptors of natural killer cells convert immunosuppression to ablate solid tumors. J Immunother Cancer. 2021;9(10):e003050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Daher M, Basar R, Gokdemir E, Baran N, Uprety N, Nunez Cortes AK, et al. Targeting a cytokine checkpoint enhances the fitness of armored cord blood CAR‐NK cells. Blood. 2021;137(5):624‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Sharma P, Hu‐lieskovan S, Wargo J, Ribas A. Cancer JC, Angeles L. Primary, Adaptive and Acquired Resistance to Cancer Immunotherapy. Cell. 2017;168(4):707‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Seliger B. Combinatorial approaches with checkpoint inhibitors to enhance anti‐tumor immunity. Front Immunol. 2019;10:999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Ramos‐Casals M, Brahmer JR, Callahan MK, Flores‐Chávez A, Keegan N, Khamashta MA, et al. Immune‐related adverse events of checkpoint inhibitors. Nat Rev Dis Primers. 2020;6(1):1‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Harrison D, Phillips JH, Lanier L. Involvement of a metalloprotease in spontaneous and phorbol ester‐induced release of natural killer cell‐associated Fc gamma RIII (CD16‐II). J Immunol. 1991;147(10):3459‐65. [PubMed] [Google Scholar]
- 310. Borrego F, Lopez‐Beltran A, Pena J, Solana R. Downregulation of Fcγ receptor IIIAα (CD16‐II) on natural killer cells induced by anti‐CD16 mAb is independent of protein tyrosine kinases and protein kinase C. Cell Immunol. 1994;158(1):208‐17. [DOI] [PubMed] [Google Scholar]
- 311. Grzywacz B, Kataria N, Verneris MR. CD56dimCD16+ NK cells downregulate CD16 following target cell induced activation of matrix metalloproteinases. Leukemia. 2007;21(2):356‐9. [DOI] [PubMed] [Google Scholar]
- 312. Liu Q, Sun Y, Rihn S, Nolting A, Tsoukas PN, Jost S, et al. Matrix metalloprotease inhibitors restore impaired NK cell‐mediated antibody‐dependent cellular cytotoxicity in human immunodeficiency virus type 1 infection. J Virol. 2009;83(17):8705‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Wiernik A, Foley B, Zhang B, Verneris MR, Warlick E, Gleason MK, et al. Targeting Natural Killer Cells to Acute Myeloid Leukemia In Vitro with a CD16× 33 Bispecific Killer Cell Engager and ADAM17 InhibitionTargeting NK Cells to AML with a BiKE and ADAM17 Inhibitor. Clin Cancer Res. 2013;19(14):3844‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Felices M, Lenvik TR, Davis ZB, Miller JS, Vallera DA. Generation of BiKEs and TriKEs to improve NK cell‐mediated targeting of tumor cells. Natural Killer Cells: Springer; 2016. p. 333‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Anliker B, Abel T, Kneissl S, Hlavaty J, Caputi A, Brynza J, et al. Specific gene transfer to neurons, endothelial cells and hematopoietic progenitors with lentiviral vectors. Nat Methods. 2010;7(11):929‐35. [DOI] [PubMed] [Google Scholar]
- 316. Buchholz CJ, Friedel T, Büning H. Surface‐engineered viral vectors for selective and cell type‐specific gene delivery. Trends Biotechnol. 2015;33(12):777‐90. [DOI] [PubMed] [Google Scholar]
- 317. Lugini L, Cecchetti S, Huber V, Luciani F, Macchia G, Spadaro F, et al. Immune surveillance properties of human NK cell‐derived exosomes. J Immunol. 2012;189(6):2833‐42. [DOI] [PubMed] [Google Scholar]
- 318. Wu C‐H, Li J, Li L, Sun J, Fabbri M, Wayne AS, et al. Extracellular vesicles derived from natural killer cells use multiple cytotoxic proteins and killing mechanisms to target cancer cells. J Extracell Vesicles. 2019;8(1):1588538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Zhu L, Kalimuthu S, Gangadaran P, Oh JM, Lee HW, Baek SH, et al. Exosomes derived from natural killer cells exert therapeutic effect in melanoma. Theranostics. 2017;7(10):2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Fu W, Lei C, Liu S, Cui Y, Wang C, Qian K, et al. CAR exosomes derived from effector CAR‐T cells have potent antitumour effects and low toxicity. Nat Commun. 2019;10(1):1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Lu J, Wu J, Tian J, Wang S. Role of T cell‐derived exosomes in immunoregulation. Immunol Res. 2018;66(3):313‐22. [DOI] [PubMed] [Google Scholar]
- 322. Torralba D, Baixauli F, Villarroya‐Beltri C, Fernández‐Delgado I, Latorre‐Pellicer A, Acín‐Pérez R, et al. Priming of dendritic cells by DNA‐containing extracellular vesicles from activated T cells through antigen‐driven contacts. Nat Commun. 2018;9(1):1‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Dianat‐Moghadam H, Mahari A, Heidarifard M, Parnianfard N, Pourmousavi‐Kh L, Rahbarghazi R, et al. NK cells‐directed therapies target circulating tumor cells and metastasis. Cancer Lett. 2021;497:41‐53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


