ABSTRACT
Veillonella species are abundant members of the human oral microbiome with multiple interspecies commensal relationships. Examining the distribution patterns of Veillonella species across the oral cavity is fundamental to understanding their oral ecology. In this study, we used a combination of pangenomic analysis and oral metagenomic information to clarify Veillonella taxonomy and to test the site specialist hypothesis for the Veillonella genus, which contends that most oral bacterial species are adapted to live at specific oral sites. Using isolate genome sequences combined with shotgun metagenomic sequence data, we showed that Veillonella species have clear, differential site specificity: Veillonella parvula showed strong preference for supra- and subgingival plaque, while closely related V. dispar, as well as more distantly related V. atypica, preferred the tongue dorsum, tonsils, throat, and hard palate. In addition, the provisionally named Veillonella sp. Human Microbial Taxon 780 showed strong site specificity for keratinized gingiva. Using comparative genomic analysis, we identified genes associated with thiamine biosynthesis and the reductive pentose phosphate cycle that may enable Veillonella species to occupy their respective habitats.
IMPORTANCE Understanding the microbial ecology of the mouth is fundamental for understanding human physiology. In this study, metapangenomics demonstrated that different Veillonella species have clear ecological preferences in the oral cavity of healthy humans, validating the site specialist hypothesis. Furthermore, the gene pool of different Veillonella species was found to be reflective of their ecology, illuminating the potential role of vitamins and carbohydrates in determining Veillonella distribution patterns and interspecies interactions.
KEYWORDS: Veillonella, pangenomics, metagenomics, metapangenomics, tropism, site specialists, niche partitioning
INTRODUCTION
The genus Veillonella is comprised of anaerobic, Gram-negative cocci (1, 2) that are abundant in the human oral cavity. Their physiology is characterized by active fermentation of organic acids, primarily lactic acid, and inability to use carbohydrates or amino acids as an energy source (3, 4). Because of their ability to consume lactic acid, Veillonella organisms are thought to establish commensal relationships with Streptococcus and other organisms that generate lactic acid from the metabolism of carbohydrates (5, 6). Despite the potential importance of the Veillonella genus for taxon-taxon interaction, little is known about the genetic variation between species or strains that may be critical for adaptation of Veillonella to different niches within the mouth. An understanding of oral Veillonella ecology requires delving deeper into the genomic differences that underlie physiological differences between individual species in the sites they inhabit.
In a recent review of the literature (7), we advanced the site specialist hypothesis of the oral microbiome to summarize the evidence that separate sites in the mouth, such as the teeth, tongue dorsum, and buccal mucosa, while bathed in the same saliva, are profoundly different in the microbiotas they support. These differences consist not merely of shifts in the proportions of the same taxa but of colonization by substantially different taxa. Although most genera of oral bacteria are represented throughout the human oral cavity, many species have strong tropisms for specific sites in the mouth (8). To date, support for the site specialist hypothesis is based primarily on 16S rRNA gene sequence data (8–10) as well as older studies with cultivated organisms (11, 12). These approaches have limitations, as 16S rRNA gene phylogenies assign species designations based on a small region of a single gene, and cultivation-based studies examine only a few species. A broader basis is necessary for evaluating the site specialist hypothesis.
Currently, taxonomic assignment of species within the genus Veillonella rests on multiple marker genes. Seven species of human oral Veillonella, 5 named and 2 unnamed, are currently recognized in the expanded Human Oral Microbiome Database (eHOMD; http://www.homd.org) (13, 14), and 11 more species are recognized in the List of Prokaryotic Names with Standing in Nomenclature (15), 6 of which are human and 5 of which are of animal origin. Although membership in the genus Veillonella can be determined with high certainty from 16S rRNA gene sequences, discrimination between some of the members at the species level has been problematic because of low dissimilarity between 16S rRNA genes of different species (16, 17) and because of heterogeneity of the four 16S rRNA gene copies within a single cell (18). These difficulties have led investigators to introduce the use of additional marker gene sequences (19) to discriminate species.
Circumvention of these limitations, resolution of taxonomic uncertainties, and insight into the gene drivers of niche adaptation can be attained by a “metapangenomics” approach (20), which brings together pangenome analysis and environmental metagenomic information. The pangenome, the sum of all genes found across members of a given group, reveals the functional essence and diversity held within that group (21, 22). Although the pangenome contains the theoretical palette of traits available, by itself it does not say which are actually realized in a particular environment. Complementary to the pangenome is whole-genome shotgun metagenomic information obtained from environmental samples. Mapping short-read metagenomic data to the pangenome makes it possible to estimate the relative abundance of environmental populations without the limitations of cultivation or assembly (23–25). This combination of metagenomes and pangenomes can demonstrate the distribution of taxa across environmental habitats, thus indicating environment-specific selection pressures. Functional annotation can suggest genes putatively required for site adaptation.
In this study, we used a metapangenomics approach to assess the distribution of natural Veillonella populations in the healthy human oral cavity and identify genes that characterize human and oral Veillonella species. We showed that Veillonella species have clear, differential site specificities, validating the site specialist hypothesis. We then identified specific genes associated with key functions that may facilitate habitat specialization for different Veillonella species. Our findings provide a first step for identifying the molecular underpinnings that drive Veillonella distribution in the oral microbiome.
RESULTS
Veillonella pangenome, phylogeny, and ANI.
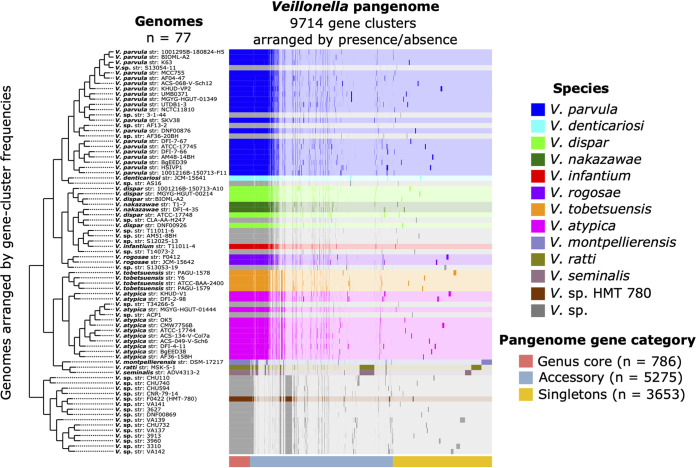
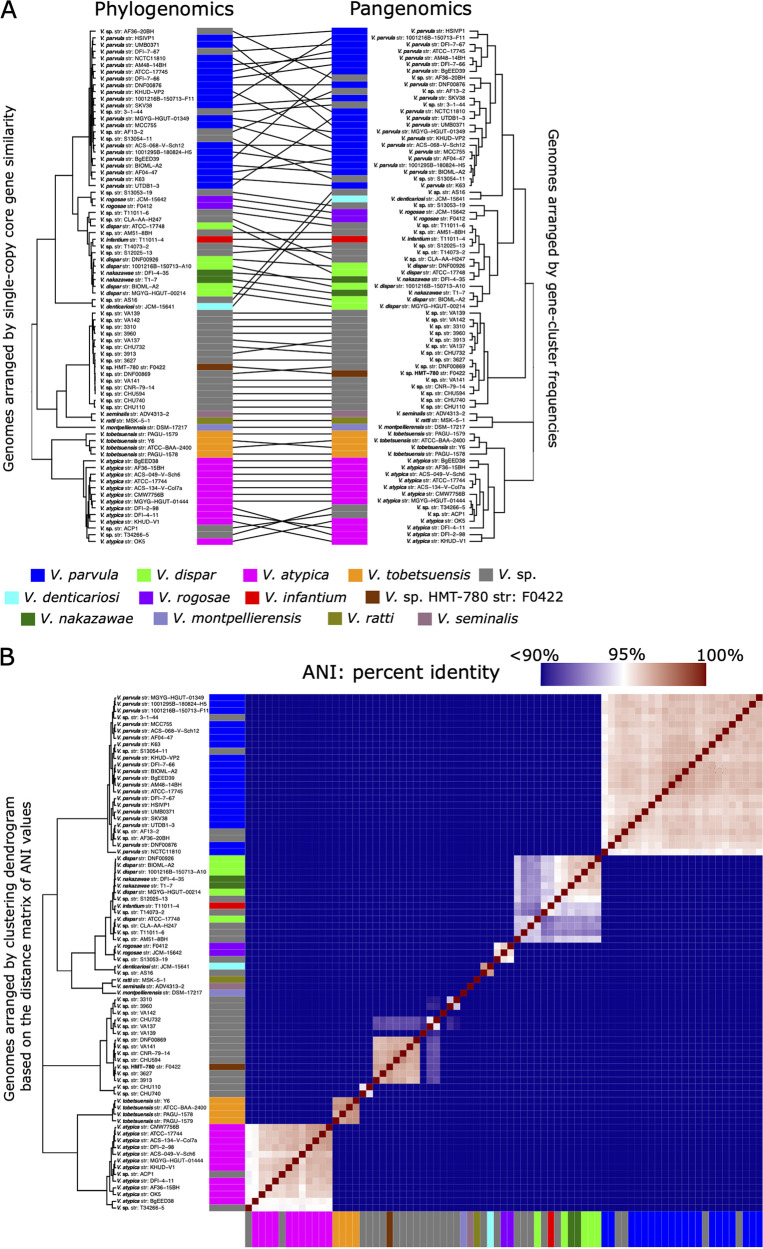
Pangenome construction identified distinct genomic groups within the Veillonella genus (Fig. 1). One clade included all named Veillonella species along with genomes identified in the National Center for Biotechnology Information (NCBI) database as Veillonella spp. that clustered within it, while a second clade consisted of the provisionally named Veillonella sp. Human Microbial Taxon 780 (HMT-780) along with Veillonella species genomes that clustered with it. The clade including named species clustered into three principal groups: group I, the Veillonella parvula group, consisted of V. parvula, V. denticariosi, and their cognate Veillonella species genomes; group II, the V. dispar group, consisted of V. dispar, V. infantium, V. rogosae, and their cognate Veillonella species genomes; and group III, the V. atypica group, contained the genomes of V. atypica, V. tobetsuensis, and the genomes identified in the NCBI database as Veillonella species that clustered with them. Genomes of V. montpellierensis, V. ratti, and V. seminalis, which were isolated from human nonoral sites (vagina, gut, and semen), did not cluster with any of the human oral genomes. We tested the congruence of these pangenomic groupings with phylogeny by generating a phylogenomic tree based on single-copy core gene clusters (SCG phylogeny). The SCG phylogeny matched well to the pangenome, although the organizations of genomes within certain clades were slightly different (Fig. 2A). One difference was the placement of V. denticariosi, and its single cognate Veillonella species genome, relative to the V. parvula and V. dispar groups. In the SCG phylogeny, V. denticariosi was an outgroup of the V. parvula clade, whereas in the pangenome, V. denticariosi was an outgroup of the V. parvula and V. dispar clades. We further assessed the taxonomic affiliations of Veillonella genomes by evaluating the average nucleotide identity (ANI) between all pairs of genomes. ANI is a similarity index between a given pair of genomes that can be used to identify genomes from the same species (26). A threshold of 95% ANI is often used to define bacterial species (27). The genomes of most species formed separate monophyletic clades within which all genomes shared >95% ANI, apart from a group containing V. dispar, V. nakazawae, V. infantium, and several genomes with no species designation in the NCBI database (Fig. 2B; see also Table S2 in the supplemental material). Within this group, application of a 95% ANI criterion would separate the type strain, V. dispar ATCC 17748, from the other 4 genomes named “V. dispar” in the NCBI database, while these 4 genomes share >95% identity with V. nakazawae. The taxonomy of this group may merit revision or reassignment. Overall, all three measures of genome relatedness (pangenomic gene content, phylogenomics, and ANI) agreed on the basic grouping of oral Veillonella genomes.
FIG 1.
Human oral Veillonella genus pangenome constructed from available NCBI RefSeq genomes (n = 77). For all genomes, open reading frames (ORFs) were predicted, NCBI blastp was used to calculate amino acid sequence similarity between all possible gene pairs, and a Markov cluster algorithm was used to cluster similar sequences to identify homologous genes (i.e., gene clusters). The 9,714 distinct gene clusters of the pangenome include 786 core genes that occur in every genome, 3,653 singleton genes that occur in only a single genome, and 5,275 accessory genes that occur in more than one but not all genomes. Gene clusters are colored by species and arranged based on their presence or absence across the genomes. Genomes are hierarchically clustered based on gene cluster frequency (i.e., the number of representatives of each gene cluster present in each genome), which is shown by the dendrogram on the left. This pangenomic analysis results in distinct groups by species and predicts the species identities of the unnamed Veillonella genomes (V. sp.).
FIG 2.
(A) Phylogenomic and pangenomic tree comparisons of Veillonella reference genomes cluster genomes into the same species-level groups. Rectangle color indicates species. The phylogenomic tree was constructed using maximum likelihood with 496 concatenated single-copy core genes. The pangenomic tree was constructed using the gene (n = 146,660) frequencies present in each genome. Lines connect rectangles that represent the same genome. (B) Average nucleotide identity (ANI) comparison of Veillonella reference genomes included in the pangenome. ANI represents the genome-level similarity at the nucleotide level between any two genomes and reveals distinct species, corroborating the results of the pangenome clustering. Rectangle color indicates species as in panel A. Color scale indicates genome percent similarity; 100% is red, 95% is white, and 90% and below is blue.
Distribution of Veillonella genomes across human oral sites.
Mapping of metagenomic reads to the pangenome can reveal the relative distribution of species and individual genes across environments. We analyzed the distribution of Veillonella species across the oral cavity of healthy subjects by mapping metagenomic sequence data from the Human Microbiome Project (HMP) for individual sites onto our curated Veillonella reference genome set. In total, we mapped over 2,500,000,000 quality-filtered metagenomic short reads from 686 samples across nine oral sites (buccal mucosa [n = 183], supragingival plaque [n = 210], subgingival plaque [n = 19], dorsum of tongue [n = 220], hard palate [n = 1], palatine tonsil [n = 19], throat [n = 13], and saliva [n = 7]). Overall, based on the proportion of metagenomic reads that successfully mapped to Veillonella reference genomes, Veillonella organisms were abundant in the oral cavity, varying from 3% to 10% depending on site and accounting, on average, for 6.50% ± 0.72% (mean ± standard error [SE]) of the total metagenomic reads across all oral sites.
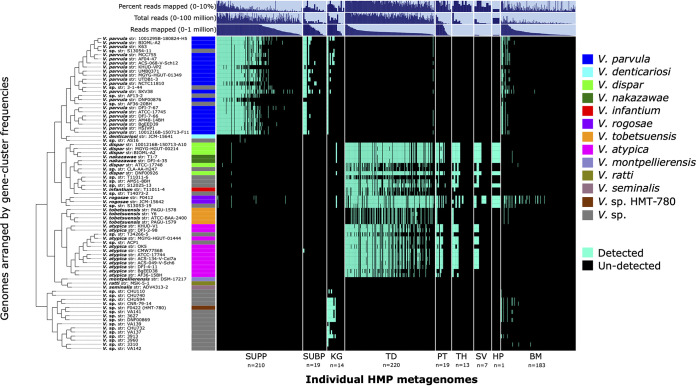
We classified genomes as detected if there was at least 1× depth of coverage for at least 50% of nucleotides for a given genome. Based on this detection metric, we found that human and oral Veillonella genomic groups are site specialists (Fig. 3). Reads from supragingival and subgingival plaque samples mapped almost exclusively to genomes of the V. parvula group, whereas reads from tongue dorsum, palatine tonsil, throat, and saliva samples mapped to the V. atypica and V. dispar groups. Genomes in the Veillonella sp. HMT-780 group were not detected in reads from supra- and subgingival plaque or tongue dorsum, but did recruit reads from keratinized gingiva and some buccal mucosa samples. Reads in some keratinized gingiva and buccal mucosa samples also mapped to V. parvula. The V. denticariosi genome, along with its cognate Veillonella species genome, was detected in only two supragingival plaque samples. The nonoral V. montpellierensis, V. ratti, and V. seminalis negative-control genomes were not detected in any metagenomic samples, thus supporting the integrity of our mapping results. Mapping results were independent of donor gender.
FIG 3.
Detection plot of Veillonella species and strains in 686 Human Microbiome Project (HMP) metagenomic samples from 9 major oral sites. Each row displays the detection of a genome across all samples. A genome is detected (cyan) if at least 50% of its nucleotides have at least 1× coverage. If the genome is not detected in a sample, it is represented by a black bar. Samples are ordered by oral site and then by decreasing number of reads mapped to the set of genomes. From left to right, oral sites are supragingival plaque (SUPP), subgingival plaque (SUBP), keratinized gingiva (KG), (TD), palatine tonsil (PT), throat (TH), saliva (SV), hard palate (HP), and buccal mucosa (BM). Additional data are shown for total mapped reads, total sample reads, and percentage of reads mapped, which may be taken as a measure of genus abundance per metagenomic sample.
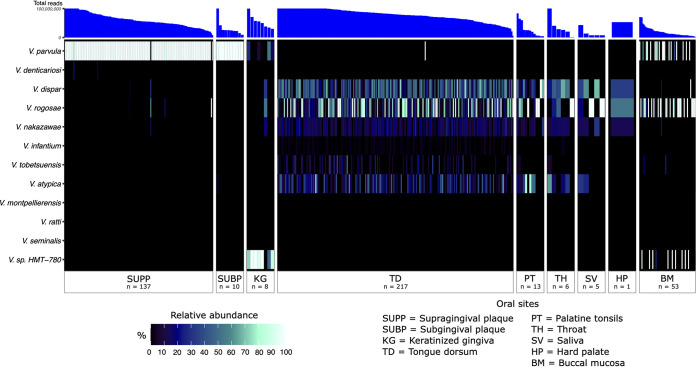
Complementary to detection is abundance, which can reveal potentially large differences in the number of reads recruited (i.e., depth of coverage). For each metagenome, we analyzed the relative abundance of each Veillonella genome by evaluating the mean depth of coverage of each detected Veillonella genome relative to the total depth of coverage of all detected Veillonella genomes. Summing this abundance metric across all genomes in a species group revealed the species that are dominant in each oral habitat. Generally, the distribution patterns of Veillonella species in the healthy human oral cavity as measured by this abundance metric are similar to those of the detection metric (Fig. 4). Taken together, our results reveal that Veillonella species are specialized to ecologically related sites: V. parvula to both supra- and subgingival plaque; V. dispar, V. atypica, V. rogosae, V. tobetsuensis, and V. nakazawae to tongue dorsum, throat, and tonsils; and Veillonella sp. HMT-780 to keratinized gingiva.
FIG 4.
Heat map showing the relative abundances of Veillonella species with respect to the set of reference genomes across 9 major oral sites: supragingival plaque, subgingival plaque, keratinized gingiva, tongue dorsum, palatine tonsil, throat, saliva, hard palate, and buccal mucosa. The relative abundance for each species represents the sum of relative abundances of each reference genome from that species, calculated from the mean depth of coverage across nucleotide positions in the 2nd and 3rd quartiles (the interquartile range) after nucleotides were ranked by their depth of coverage and then divided by the sum of mean coverages of all genomes within a metagenomic sample. The rows and columns correspond to individual species and Human Microbiome Project metagenomic samples, respectively. Only metagenomes in which at least one reference genome was detected (i.e., at least 50% of its nucleotides have at least 1× coverage) were included. The number of metagenomes for each oral site in which a species was detected is listed. For clearer visualization, we inflated the width of oral sites with small sample sizes. Metagenomes are in descending order for each oral site by the total number of quality filtered reads.
Competitive read mapping provides limited information about the correspondence between the reference genomes and the strain-level composition of the metagenome. Reads whose best matches are equidistant to multiple reference genomes will be distributed at random among these genomes; therefore, the fact that multiple reference genomes from a single species pass the detection threshold does not necessarily signify that multiple strains of that species are present in the metagenome. However, differences in genome-level mapping can indicate which sequenced genomes are most representative of the Veillonella populations in the samples. The mapping results for each reference genome indicated that not all strains of a given species were equally detected in the oral cavity (Fig. 3). Some V. parvula, V. dispar, and V. atypica reference genomes recruited fewer reads than others, reflecting their poor resemblance to the populations of these three species present in the samples. Overall, genomes within each clade showed similar distribution patterns across oral sites, indicating a lack of subspecialization to different oral sites by strains within a species.
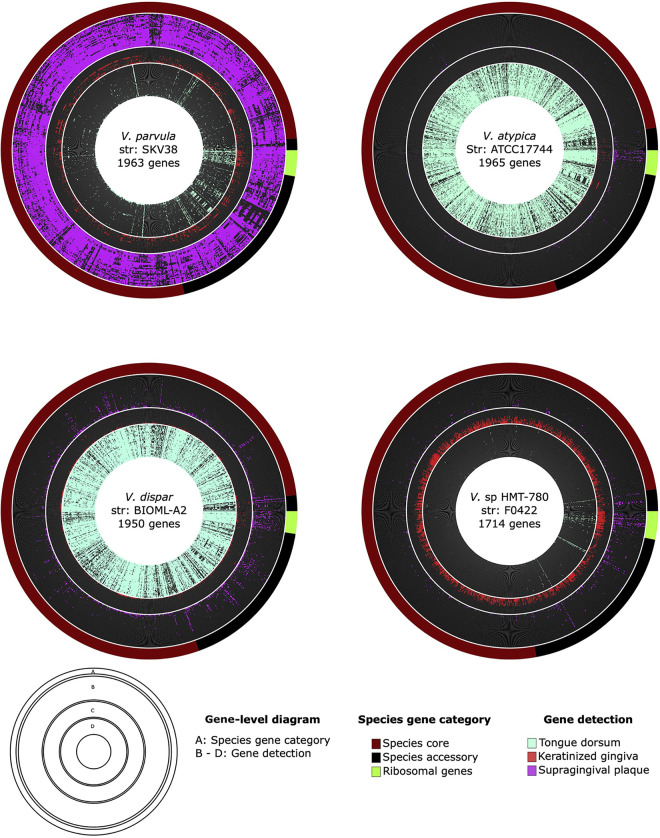
Inspection of gene-level coverage can be used to assess whether coverage is distributed evenly across a genome, as would be expected if the presence and absence of genes in the reference genome is an accurate representative of genomes in the mouth. Having found clear patterns of species site specialization using our genome detection and relative-abundance metrics, we then further examined coverage at a gene-level scale. We classified a gene as detected if at least 90% of its nucleotides had at least 1× coverage; this 90% threshold is stringent yet allows for hypervariable regions that may differ between reference genomes and environmental genomes. Fig. 5 shows radial gene-level detection maps of four genomes (V. parvula SKV38, V. dispar BIOML-A2, V. atypica ATCC 17744, and Veillonella sp. HMT-780) that had high relative abundance within their preferred sites (supragingival plaque, tongue dorsum, tongue dorsum, and keratinized gingiva, respectively). These gene detection maps show that for each genome, a large proportion of genes were detected in only one of the three oral sites. Three genomes, those of V. parvula SKV38, V. dispar BIOML-A2, and V. atypica ATCC 17744, showed preference for oral sites for which many metagenomic samples with high numbers of reads were available. For the 30 metagenomic samples with greatest median coverage as shown in Fig. 5, gene detection was clearly distributed evenly across the genome, with 78% ± 7% (mean ± 1 standard deviation) of genes detected on average within the preferred oral site. One genome, that of Veillonella sp. HMT-780, attracted reads specifically from keratinized gingival samples. For this site, only 14 metagenomic samples were available, and most of them had few total reads compared to samples from other oral sites, limiting the detection sensitivity (Fig. 3). Consequently, the proportion of genes detected for Veillonella sp. HMT-780 was lower and their evenness across the genome was less evident. However, like for the other genomes, gene-level detection was site specific. Also apparent in the gene detection maps is that a small proportion of genes attracted reads from multiple habitats. Many of these genes were annotated with highly conserved functions (e.g., ribosomal genes, cell division, and DNA replication) or transposable elements and thus may represent cross-mapping to different taxa that share high similarity in these genes or share elements that have been horizontally acquired. In sum, gene-level coverage analysis confirmed the site tropisms suggested by the whole-genome-level coverage analysis and extended that analysis by identifying individual genes present in specific oral sites.
FIG 5.
Gene-level detection diagrams for select Veillonella genomes that showed clear patterns of site specialization. Here, we display radial gene-level detection maps for four genomes showing the detection of genes across a subset of the top 30 metagenomic samples ranked by median coverage from 2 oral sites (supragingival plaque and tongue dorsum) and all available 14 metagenomic samples from keratinized gingiva (red). We included V. parvula strain SKV38, V. dispar strain BIOML-A2, V. atypica strain ATCC 17744, and Veillonella sp. HMT-780 strain F0422 to represent species that were the most abundant based on mean coverage within their respective preferred oral site. A gene was classified as detected within an oral site when at least 90% of the nucleotides of the gene had at least 1× coverage. Genes are ordered according to the gene category for the respective species as indicated in the outermost layer for each diagram. A group of ribosomal genes translated using hidden Markov models via the Anvi’o program anvi-run-hmms are indicated in bright green.
Functional analysis of detected Veillonella genomes across human oral sites.
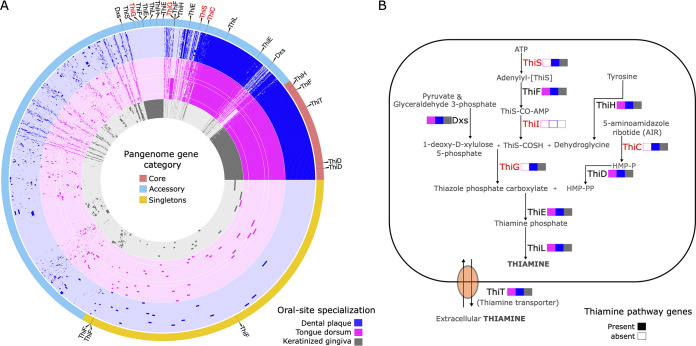
Metapangenomics can be used to reveal genetic variation that drives community structure of the oral microbiome by identifying gene functions that are core to a group of genomes at a specific oral site and absent from the genomes of other sites. Based on their detection and abundance profiles (Fig. 3 and Fig. S1), we grouped Veillonella genomes according to their site preferences. Three metabolic modules were significantly enriched in supragingival plaque and keratinized gingiva genomes compared to tongue dorsum genomes based on an adjusted q value threshold of 0.01 (Table S3). This included two modules associated with the thiamine biosynthesis pathway (vitamin B1; KEGG module identifiers [ID] M00896 and M00127) and the reductive pentose phosphate cycle (KEGG module ID M00167), which were consistently incomplete in tongue dorsum genomes (Table S3). Closer inspection of the presence and absence of individual genes that make up the thiamine biosynthesis pathway revealed that the genomes of V. dispar, V. atypica, V. tobetsuensis, and V. rogosae, all of which appear to be tongue dorsum specialists, are missing genes that play an important role in the thiazole and hydroxymethylpyrimidine phosphate moiety complexes required to produce thiamine de novo (Fig. 6). Additionally, both dental plaque and tongue dorsum specialists had a complete thiamine salvage pathway (KEGG module ID M00899), which was missing entirely from keratinized gingiva specialists. In a similar light, closer inspection of module completeness for the reductive pentose phosphate cycle revealed that V. atypica and V. tobetsuensis genomes are missing fructose-1,6-bisphosphatase II (GlpX), which is present in the V. parvula and V. dispar genomes (Table S3).
FIG 6.
(A) A pangenome of human oral Veillonella reference genomes (n = 72) was constructed for supragingival plaque, keratinized gingiva, and tongue dorsum genomes. Species and strains not detected in these oral sites were excluded. Gene cluster arrangement, genome color, and genome arrangement are like the previously constructed pangenome (Fig. 1). Thiamine pathway-associated genes are indicated with a black line in the pangenome gene category outer layer and pointed out by a black triangle. (B) The diagram illustrates the enzymes, metabolites, and transporters involved in the thiamine pathway in human oral Veillonella species. The presence of a gene is indicated with a filled color box; empty boxes indicate that no gene was found in any genome assigned to the respective oral site. The colors designate oral sites, and genomes were grouped into an oral site based on detection and relative abundance patterns (Fig. 3 and 4).
Keratinized gingiva specialists (i.e., Veillonella sp. HMT-780) were distinguished from both dental plaque and tongue dorsum genomes by the enrichment of three metabolic pathways associated with carbohydrate metabolism, including the oxidative-phase pentose phosphate pathway (M00006), which converts glucose-6-phosphate to ribulose-5-phosphate, and two pathways associated with galactose metabolism: the galactose degradation Leloir pathway (M00632) and a nucleotide sugar biosynthesis (NSB) pathway that converts galactose to UDP-galactose (M00554). The Leloir pathway enzymes include aldose 1-epimerase (GalM), galactokinase (GalK), galactose-1-phosphate uridylyltransferase (GalT), and UDP-galactose 4-epimerase (GalE), which are responsible for the conversion of galactose to glucose-1-phosphate. The NSB pathway enzymes include a subset of the Leloir pathway: GalT and GalE. Closer inspection of all functional annotations in our data set (i.e., NCBI clusters of orthologous genes [COGs], Pfam, and KEGG) revealed that Veillonella dental plaque and tongue dorsum specialists lack GalM and GalK but have GalT and GalE. All Veillonella genomes in our data set have the genes required to metabolize fructose via the Embden-Meyerhof-Parnas (EMP) pathway, as well as a complete tagatose-6-phosphate pathway, in which d-galactose or lactose is converted to d-galactose-6-phosphate and further metabolized through tagatose derivatives (28).
In addition to assessing the completeness of metabolic pathways, we analyzed functional associations derived from the NCBI COG20, Pfam, and KEGG databases to identify gene functions that were significantly enriched in certain Veillonella groups (Table S4). Noteworthy enriched gene functions were as follows: eight functions were significantly enriched in both supragingival plaque and keratinized gingiva compared to tongue dorsum genomes, the top three of which included thiamine biosynthesis genes: the sulfur carrier protein (ThiS), thiazole synthase (ThiG), and 4-amino-2-methyl-5-hydroxymethylpyrimidine synthase (ThiC). Only a single function, magnetosome formation protein MamQ, was enriched in both tongue dorsum and keratinized gingiva compared to supragingival plaque genomes. Magnetosomes are organelles comprising magnetic mineral crystals surrounded by a phospholipid bilayer that are thought to play an important role in cell movement to optimize growth and survival (29). Thirty-two functions were significantly enriched in both dental plaque and tongue compared to keratinized gingiva genomes, including beta-lactamase (LemA), which is a protein that confers resistance to antibiotics, namely, penicillins (30). Adhesins play an important role in biofilm formation and may be a factor contributing to Veillonella species-level niche partitioning in the oral microbiome (31). However, we did not find evidence of differential enrichment of any genes directly associated with adherence, such as adhesins, between the genomes of Veillonella site specialists.
The pangenome was constructed using a set of isolate reference genomes that were dereplicated based on a 98% ANI threshold, which may result in the loss of information on variability in the accessory gene content among representatives from the same species. To evaluate the potential cost of removing genomes, we constructed additional pangenomes using genome sets dereplicated at 99% and 99.5% ANI. Raising the ANI threshold increased the size of the pangenome by 2 and 5 genomes, respectively (Fig. S1 and S2). Increasing the ANI threshold also modestly decreased the number of core and singleton gene clusters and increased the number of gene clusters categorized as accessory (Fig. S3). Inspection of the singleton gene clusters shows that many of the additional protein sequences are functionally associated with mobile elements or annotated as hypothetical proteins (Table S5). One genome (V. dispar strain BIOML-A3) that was added to the 99.5% ANI threshold pangenome contained a relatively large number of singleton gene clusters, for many of which the top blast hit was a protein sequence from the bacterial genus Eggerthella and likely reflects bacterial contamination in the genome. We did not find any evidence of additional genes associated with vitamin biosynthesis or carbohydrate metabolism that would alter our previous results. We thus conclude that the 98% ANI threshold for genome dereplication did not adversely affect the results of our analysis.
DISCUSSION
Metapangenomics can provide information on ecological distribution and genetic variation among oral bacterial species and strains. Using this approach, we found that human oral Veillonella species demonstrated strong patterns of site specialization, such that individual species were most abundant in either dental plaque, keratinized gingiva, or the tongue dorsum, throat, and tonsils. Analysis of unique or overrepresented genes and functions among site-specific genome groups identified genes associated with carbohydrate metabolism and two key metabolic pathways, including thiamine biosynthesis and the pentose phosphate cycle. These genes associated with adaptation to an oral site may enable different Veillonella species to occupy different habitats and thus help explain their oral site tropisms.
Our approach takes advantage of a cornucopia of publicly available data to systematically analyze the genomic content of Veillonella species in the oral microbiome within an ecological framework of niche partitioning across oral habitats. The combination of pangenomic analysis using genome sequences from cultivars and oral metagenomic information provided unique species-, strain-, and gene-level insights. First, because marker genes, such as the 16S rRNA gene, fail to distinguish some closely related Veillonella species (16, 17), metapangenomics improved identification of species based on genomic coherence and enabled identification of strains for in vitro model systems that more accurately represent the oral community. Second, our approach goes beyond characterization of a species’ site tropism and analyzes gene distribution and abundance across genomes and in samples from the mouth. The relevance of cultivars to most of the organisms living in the mouth is generally not known, and successful mapping depends on the set of reference genomes being sufficiently similar at the nucleotide level to environmental genomes. Nonetheless, we obtained extensive mapping, resulting in relative abundance estimates similar to those obtained by 16S rRNA gene sequencing (7). This validates the overall relevance of cultivars, but follow-up mechanistic studies using experimental manipulations are necessary to validate our findings.
Although pangenomic analysis is typically performed at the species level, analysis at the genus level provides several benefits. First, the clustering of genomes identified in databases only to the genus level (i.e., Veillonella) with genomes identified to the species level greatly augments the number of genomes in the pangenome analysis, thus enabling a greater representation of species and strain genetic variation. This point is most dramatically exemplified for the Veillonella sp. HMT-780, for which the single genome available in the NCBI database was augmented by 14 additional genomes that clustered with it. Other oral species were also augmented: V. atypica by 2 genomes, V. parvula by 4 genomes, V. rogosae by 2 genomes, and V. dispar by 5 genomes. Analysis of other oral genera by pangenomics has resulted in similar findings (32). Second, relationships among the species comprising the genus can be readily visualized. For example, the difference in gene content between the Veillonella sp. HMT-780 group and the other Veillonella oral genomes is much greater than the differences between any of the other oral genomes, indicating that human oral Veillonella can be divided into two major clades, one defined by Veillonella sp. HMT-780 and the other containing the V. parvula, V. dispar, and V. atypica groups. The last three groups share a large set of core genes and are distinguished from each other by small sets of unique species core genes. Third, possible species misclassifications become apparent. For example, our results revealed that the V. dispar and V. nakazawae genomes were similar in gene content and had nearly identical distribution patterns in the human oral cavity. V. nakazawae was originally classified as a novel species by comparing two strains of previously unknown Gram-negative cocci, T1-7T and S6-16, to a single V. dispar type strain, ATCC 17748, in terms of catalase production, partial dnaK, rpoB, and gltA sequences, average ANI, and digital DNA-DNA hybridization (dDDH) values (33). However, our analysis suggests that V. nakazawae should be considered a later synonym of V. dispar. In addition to V. nakazawae, our analysis included genomes for two species not listed in eHOMD (http://www.homd.org): V. tobetsuensis and V. infantium. Our results from both phylogenomics and ANI indicate that V. tobetsuensis is a sister group to V. atypica, whereas V. infantium clusters within V. dispar and, like V. nakazawae, should be considered a later heterotypic synonym of V. dispar unless the V. dispar cluster is subdivided into distinct species.
Comparative genomic analysis can be used to reveal the potential molecular underpinnings that drive Veillonella site specialization in the oral microbiome. One important factor in determining species colonization is the availability of macronutrients, such as carbohydrates, fats, and proteins. Veillonella organisms are known to rely on by-products of other heterotrophic bacteria, primarily pyruvate and lactate, as a source of energy (34–36) and are thought to be unable to use carbohydrates or amino acids as an energy source (3, 4). Our findings suggest that Veillonella species that inhabit different oral sites metabolize lactate and carbohydrates in slightly different ways. First, the gene for lactate racemase (LarA), which enables the consumption of both l- and d-enantiomers of lactate in highly anoxic environments (37, 38), was enriched in tongue dorsum specialists compared to supragingival plaque and keratinized gingiva specialists. Lactate is mostly found in its natural l-form, whereas d-lactate is a by-product of some fermentation processes (39) and lactic acid-producing bacteria (LAB) such as streptococci and lactobacilli (40). Thus, LarA may enhance the ability of Veillonella tongue dorsum specialists to utilize localized pockets of high concentrations of d-lactate produced by LAB on the tongue. However, the genes for both l-lactate dehydrogenase (LdhL) and d-lactate dehydrogenase (LdhD), which convert l-lactate and d-lactate to pyruvate and back, respectively, were found in all oral Veillonella species in our data set regardless of site preference. Consequently, LarA may act as a rescue enzyme to ensure d-lactate metabolism under physiological conditions where LdhD production is not sufficient. Second, in anaerobic environments C5 sugars are released as by-products of aerobic bacteria and subsequently taken up by anaerobic bacteria (41). In this study, we found that Veillonella tongue specialists are further differentiated from dental plaque specialists by having an incomplete pathway for the reductive pentose phosphate cycle, although the pathway is lacking only in V. atypica and V. tobetsuensis, a subset of tongue specialists.
We found that the tagatose-6-phosphate pathway (i.e., Lac genes) is conserved in all oral Veillonella species. In this pathway, galactose metabolism occurs by transforming galactose-6-phosphate to glycolysis intermediates via a series of enzymes: galactose-6-phosphate isomerase, tagatose-6-phosphate kinase, and tagatose diphosphate aldolase. A complete tagatose-6-phosphate pathway suggests that oral Veillonella could utilize galactose as a source of energy in addition to lactate. Additionally, Veillonella keratinized gingiva specialists (HMT-780 group) also have a complete Leloir pathway (i.e., Gal genes) not found in Veillonella tongue dorsum and dental plaque specialists. This suggests that Veillonella HMT-780 has an enhanced capability to metabolize galactose compared to other oral Veillonella species. Our results support a previously published comparative pangenomic analysis of eight human oral Veillonella species that used one genome for each species and found that oral Veillonella organisms have conserved pathways that utilize carbohydrates other than lactate as an energy source, namely, fructose and tagatose (13). Expanding on this finding, here we propose that Veillonella HMT-780 and other members of the Veillonella genus may have evolved to utilize galactose as an additional carbon source.
Another important factor determining species colonization is the availability of micronutrients, such as inorganic cofactors (e.g., zinc ions) and coenzymes that maintain normal metabolism. Our findings indicate that Veillonella tongue specialists lack the ability to synthesize the vitamin thiamine de novo. Thiamine plays a major role as a coenzyme in many essential metabolic pathways, including glycolysis, the pentose phosphate cycle, and the citric acid cycle (42). Bacteria can either synthesize thiamine de novo or acquire precursors or mature thiamine via transport from their environment. Thiamine synthesis requires the independent production of two major precursors, including the thiazole and pyrimidine moieties (42). To produce thiazole, a sulfur carrier protein (ThiS) provides an initial source of a sulfur group, which is combined with dehydroglycine and deoxy-d-xylulose-phosphate (DXP) with the help of thiazole synthase (ThiG). Tongue dorsum Veillonella organisms lack ThiS and ThiG and are thus unable to synthesize the thiazole moiety de novo, whereas dental plaque and keratinized gingiva specialist Veillonella spp. possess the full pathway.
The evolutionary loss of costly genes required to produce thiamine is expected where an organism could obtain thiamine or its precursors through other less costly means (43). Concentrations of free thiamine in the human body are thought to be exceptionally low due to tight sequestration of the vitamin, efficient recycling (44), and the rapid degradation brought on by physical and chemical factors such as UV radiation, temperature, and pH (45, 46). Thus, for thiamine synthesis to be lost, a reliable source of thiamine must be present in the immediate environment of the microbe. The distance over which a nutrient can be provided depends on its diffusion constant and the rate at which it is taken up by intervening cells, but it is generally less than 20 μmol when uptake rates are high (47). Therefore, we hypothesize that the tongue dorsum environment allows Veillonella to acquire thiamine or the thiazole precursor from other bacteria in its immediate neighborhood, whereas the dental plaque and keratinized gingiva environments do not.
The complexity of the microbial community of the oral cavity suggests that interactions with other bacterial taxa may be an important determinant of Veillonella colonization patterns. Veillonella species may compete with other bacteria for nutrients or may form commensal or mutualistic interactions that stimulate growth and survival of one or more interactors. With respect to thiamine, a recent systematic genome assessment of B-vitamin biosynthesis found that approximately one-third of oral bacterial species are capable of de novo thiamine synthesis (48), indicating that different taxa may encode different portions of the thiamine pathway. Thus, Veillonella tongue specialists may acquire thiamine or precursor chemicals from neighboring bacteria. Indeed, the thiamine transporter ThiT (49) is present in all oral Veillonella genomes analyzed in this study. A recent spatial analysis using fluorescence in situ hybridization has shown that the taxon Rothia mucilaginosa, which is capable of de novo thiamine synthesis, is present in tongue dorsum bacterial consortia in high abundance (50). This finding suggests that Veillonella tongue specialists may acquire thiamine from nearby R. mucilaginosa. Conversely, Veillonella dental plaque specialists may have developed the ability to synthesize thiamine de novo to cope with a lack of freely available thiamine or its precursors in the environment. Indeed, it appears that dominant dental plaque taxa, including Streptococcus, Actinomyces, and Rothia dentocariosa (7), lack the ability to produce thiamine de novo (48), although in Corynebacterium, which has been described as a base of microbial community structure and interactions in dental plaque (7), thiamine synthesis appears to be conserved (48). These studies highlight the potential for vitamins to play a critical role in driving microbiome dynamics and ultimately the distribution of Veillonella in the human oral cavity.
In summary, our systematic metapangenomics approach has established the oral site-specific tropisms of Veillonella species, has provided insight into the genetic determinants of the ecology of the genus, and has illuminated the potential role of vitamins and carbohydrates in determining Veillonella distribution patterns and interspecies interactions. Spatial niche partitioning has been discovered for multiple taxa within the oral microbiome (8, 32, 51), as well as different microbial species across a large diversity of habitats, such as riverine floodplains, soil, and plastic debris in the ocean (52–54). Our results indicate a need for further research into whether the presence or absence of pathways for synthesis of certain nutrients is a determining factor in defining the niche space of a bacterium or represents evolutionary change resulting from life in the niche. The oral microbiome, with its diverse range of bacterial taxa living on a variety of distinct habitats, such as teeth, tongue, cheeks, and gums, which differ in chemistry, topography, and stability, provides a useful system for testing ecological theories about the causes and consequences of niche partitioning.
MATERIALS AND METHODS
The following analyses were conducted primarily using the Anvi’o v7 platform (55) with Python v3.7.9.
Veillonella reference genomes.
To assess whether Veillonella species are site specialists or generalists in the human oral cavity, we first developed a reference genome set representative of natural Veillonella populations in the human oral microbiome. We obtained publicly available sequenced genomes of Veillonella species from the National Center for Biotechnology Information (NCBI) database (all genomes downloaded 15 December 2021) and selected genomes that were of sufficient quality to be designated reference sequences (RefSeq). Of 113 genomes meeting this criterion, 11 were excluded because they were isolated from a nonhuman host species and 2 were excluded because they were duplicate strains. Of the resulting 100 genomes, 49 had species names that matched species contained in eHOMD (http://www.homd.org), including Veillonella sp. HMT-780; 20 genomes belonged to species not listed in eHOMD and 31 genomes were identified in NCBI only to the genus level, i.e., Veillonella. Information about genome genus, species, strain, BioSample, BioProject, isolation host, isolation site, RefSeq status, type strain, disease association, and submitter can be found in Table S1.
We then further filtered the set of reference genomes so that each genome included in the set shared no more than 98% average nucleotide identity (ANI) with any other genome, as well as had a completeness of ≥90% and a contamination level of <5% estimated by CheckM (56). ANI between genomes was estimated using the Anvi’o program anvi-compute-genome-similarity with the program pyANI (v0.2.10) and ANIblastall, which used the blastn method (57). The result was 46 high-quality reference genomes from 8 species that represent the diversity of Veillonella within the human oral microbiome, as well as 28 genomes identified only to the genus level. In addition, we included 3 high -quality genomes that represent Veillonella species isolated from human hosts but not known to be present in the human oral cavity, including V. montpellierensis, V. ratti, and V. seminalis. At the time of this study, the taxonomy check for the V. ratti genome was identified in the NCBI database as inconclusive and the genome may actually be a V. seminalis genome (58). These nonoral genomes function as a negative control for downstream analyses. The total number of genomes used to construct a genus-level Veillonella pangenome was 77.
Constructing an oral Veillonella pangenome.
Pangenome construction was adapted from previously developed methods (20, 32) and carried out using the Anvi’o platform. First, all contigs within each reference genome that were <300 nucleotides (nt) were removed and noncanonical nucleotide letters were replaced with the letter “N.” Each genome was then converted into an Anvi’o-compatible contig database using anvi-gen-contigs-db. Open reading frames (ORFs; here also referred to as genes) were identified for each genome using Prodigal (v2.6.3) (59). Functional annotation of ORFs was done using the scripts anvi-run-hmms to find bacterial single-copy genes (Bacteria71 SCG set) (60, 61) with hidden Markov model (HMM) profiles, anvi-run-ncbi-cogs using blastp (v2.10.1+) to annotate with the cluster of orthologous genes (COGs) database (version COG20) (62), and anvi-run-pfams and anvi-run-kegg-kofams with hmmscan from HMMER (v3.3.1) to functionally annotate with Pfams (v34.0) (63) and KOfams/KEGG modules (v97.0) (64), respectively. To construct the pangenome, blastp was used to compute amino acid identity between all possible ORF pairs, and weak matches were removed by employing the –minbit criterion of 0.5. A Markov cluster algorithm (MCL) was then used to group ORFs into putatively homologous groups termed gene clusters (mcl-inflation = 10), and amino acid sequences within gene clusters were aligned with MUSCLE (v3.8.1551) (65). Hierarchical clustering was performed across gene clusters and genomes using Euclidean distance and Ward linkage.
Veillonella phylogeny.
To assess how well the pangenome organized the Veillonella reference genomes, we constructed a phylogenetic tree based on the concatenated amino acid sequences of single-copy core genes present in all Veillonella reference genomes. To construct the phylogenetic tree, we selected informative single-copy core gene clusters from the pangenome using anvi-get-sequences-for-gene-clusters (parameters: --min-num-genomes-gene-cluster-occurs 77 --max-num-genes-from-each-genome 1 --concatenate-gene-clusters), which resulted in 496 single-copy gene clusters that contained a total of 38,192 genes. Genes were concatenated into a single fasta file, trimAl (v1.2) (66) was used to remove all columns with gaps in more than 50% of the sequences, and IQ-tree (v1.6.12) (67) was used to build a phylogenetic tree using maximum likelihood and the Whelan and Goldman (WAG) method with 1,000 bootstrap replicates.
Distribution of Veillonella genomes across human oral sites.
The metagenomes used in this study were obtained from the Human Microbiome Project portal (https://portal.hmpdacc.org/) using the following search parameters: oral sites (buccal mucosa [n = 183], supragingival plaque [n = 210], subgingival plaque [n = 19], dorsum of tongue [n = 220], hard palate [n = 1], palatine tonsil [n = 19], throat [n = 13], and saliva [n = 7]), Healthy Human Study (HHS), fastq files (FASTQ), and whole-genome sequencing (wgs_raw_seq_set). Metagenomes consisted of ~100-bp paired-end reads sequenced from samples collected from nine oral sites in phases I and II of the Human Microbiome Project. Quality filtering was performed using iu-filter-quality-minoche (68), which keeps high-quality reads from Illumina sequencing data based on recommendations from Minoche et al. (69).
Individual metagenomic samples were competitively mapped against a concatenated file of the reference genome set using bowtie2 v2.4.1 (70) with the “--very-sensitive,” “--end-to-end,” and “--no-unal” flags. Each successfully mapped read was assigned only to a single genome that provided the closest match. We used Samtools v1.9 (71) to generate BAM files from the read alignment data generated by bowtie2. We then used anvi-single-profile to create an Anvi’o single-profile database for each metagenome’s BAM file. Single profiles for each sample were merged for each oral site using anvi-merge, thus generating a profile database for each oral site. For each metagenome, we used anvi-summarize to extract the mapping metrics of mean depth of coverage and breadth of coverage for reads aligned to each genome. Breadth of genome coverage is defined as the proportion of nucleotides covered at least 1×, and a genome was detected within a metagenome if the breadth of coverage was at least 50%, following recently developed procedures (20, 72). For each metagenome, the relative abundance of each genome was calculated by first averaging the depth of coverage (the number of reads mapped to a nucleotide position) across nucleotide positions in the 2nd and 3rd quartiles (the interquartile range) for the reference genome and then dividing by the total mean depth of coverage in the interquartile range for all reference genomes.
Functional analysis of detected Veillonella genomes across human oral sites.
Based on the detection and abundance profiles from metagenomic mapping, reference genomes that showed clear signs of site specialization were assigned to an oral site. We then used the Anvi’o function anvi-estimate-metabolism (parameters: --kegg-output-modes modules) to predict the metabolic capabilities of each of the selected genomes. The function determines which enzymes are present in a genome based on KEGG orthologs (KOs) and calculates the percentage of steps in each metabolic pathway (module) that are present in each genome. We used the default module completion threshold of 0.75, which scores a module as “complete” within a genome when at least 75% of the KOs in a module are present. We then used the Anvi’o script anvi-compute-metabolic-enrichment to identify significantly enriched complete modules associated with one set of genomes compared to another. Briefly, the script uses a generalized linear model with a logit link function to obtain enrichment scores and adjusted q values for each combination of pairwise group comparisons. We also used the Anvi’o script anvi-compute-functional-enrichment to identify significantly enriched functional annotations from the NCBI COG, KEGG, and Pfam databases, independent of metabolic pathway completeness, and associated with genomes from an oral site.
Data availability.
The raw data used in this study are publicly available at NIH GenBank and RefSeq (https://www.ncbi.nlm.nih.gov/genome/) for genomes and Human Microbiome Project for metagenomes (https://portal.hmpdacc.org/). Code used for analyses is available on GitHub (https://github.com/FatherofEverest/Site-specialization-of-human-oral-Veillonella-species.git).
ACKNOWLEDGMENTS
We thank Jennifer Gundrum for preliminary Veillonella pangenomic analysis. We thank Rich Fox for expert systems administration and assistance with using the Josephine Bay Paul Center servers at the Marine Biological Laboratory. We thank A. Murat Eren and the Anvi’o team for their assistance, consultations, and support in the use of Anvi’o.
This work was supported by NIH grants R01 DE030136, R01 DE022586, and 2R01 DE016937.
We declare no conflicts of interest.
All authors participated in the design of the study. J.J.G. performed the analysis. J.J.G., G.G.B., and J.L.M.W. drafted the manuscript. All authors critically revised the manuscript and approved the final version.
Footnotes
Supplemental material is available online only.
Contributor Information
Jonathan J. Giacomini, Email: jgiacomini@forysth.org.
Justin R. Kaspar, Ohio State University
REFERENCES
- 1.Prévot AR. 1933. “Étude de systématique bactérienne. I. Lois générales. II. Cocci anaérobies.” Annales des Sciences Naturelles Zoologie et Biologie Animale 15:23–260. [Google Scholar]
- 2.Veillon A, Zuber A. 1898. Recherches sur quelques microbes strictement anaérobies et leur rôle en pathologie. Arch Med Exp 10:517–545. [Google Scholar]
- 3.Delwiche EA, Pestka JJ, Tortorello ML. 1985. The veillonellae: gram-negative cocci with a unique physiology. Annu Rev Microbiol 39:175–193. doi: 10.1146/annurev.mi.39.100185.001135. [DOI] [PubMed] [Google Scholar]
- 4.Rogosa M. 1964. The genus Veillonella I: general cultural, ecological, and biochemical considerations. J Bacteriol 87:162–170. doi: 10.1128/jb.87.1.162-170.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mashima I, Nakazawa F. 2014. The influence of oral Veillonella species on biofilms formed by Streptococcus species. Anaerobe 28:54–61. doi: 10.1016/j.anaerobe.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Palmer RJ, Jr, Diaz PI, Kolenbrander PE. 2006. Rapid succession within the Veillonella population of a developing human oral biofilm in situ. J Bacteriol 188:4117–4124. doi: 10.1128/JB.01958-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mark Welch JL, Dewhirst FE, Borisy GG. 2019. Biogeography of the oral microbiome: the site-specialist hypothesis. Annu Rev Microbiol 73:335–358. doi: 10.1146/annurev-micro-090817-062503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eren AM, Borisy GG, Huse SM, Mark Welch JL. 2014. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci USA 111:E2875–E2884. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huse SM, Ye Y, Zhou Y, Fodor AA. 2012. A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS One 7:e34242. doi: 10.1371/journal.pone.0034242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mark Welch JL, Utter DR, Rossetti BJ, Mark Welch DB, Eren AM, Borisy GG. 2014. Dynamics of tongue microbial communities with single-nucleotide resolution using oligotyping. Front Microbiol 5:568. doi: 10.3389/fmicb.2014.00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbons RJ, Socransky SS, De Araujo WC, Van Houte J. 1964. Studies of the predominant cultivable microbiota of dental plaque. Arch Oral Biol 9:365–370. doi: 10.1016/0003-9969(64)90069-X. [DOI] [PubMed] [Google Scholar]
- 12.Gordon DF, Jr, Gibbons RJ. 1966. Studies of the predominant cultivable micro-organisms from the human tongue. Arch Oral Biol 11:627–632. doi: 10.1016/0003-9969(66)90229-9. [DOI] [PubMed] [Google Scholar]
- 13.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu W-H, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol 192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, Lemon KP. 2018. New insights into human nostril microbiome from the expanded Human Oral Microbiome Database (eHOMD): a resource for the microbiome of the human aerodigestive tract. mSystems 3:e00187-18. doi: 10.1128/mSystems.00187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parte AC, Carbasse JS, Meier-Kolthoff JP, Reimer LC, Göker M. 2020. List of Prokaryotic Names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol 70:5607–5612. doi: 10.1099/ijsem.0.004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byun R, Carlier J-P, Jacques NA, Marchandin H, Hunter N. 2007. Veillonella denticariosi sp. nov., isolated from human carious dentine. Int J Syst Evol Microbiol 57:2844–2848. doi: 10.1099/ijs.0.65096-0. [DOI] [PubMed] [Google Scholar]
- 17.Takeshita T, Nakano Y, Kumagai T, Yasui M, Kamio N, Shibata Y, Shiota S, Yamashita Y. 2009. The ecological proportion of indigenous bacterial populations in saliva is correlated with oral health status. ISME J 3:65–78. doi: 10.1038/ismej.2008.91. [DOI] [PubMed] [Google Scholar]
- 18.Marchandin H, Teyssier C, de Buochberg MS, Jean-Pierre H, Carriere C, Jumas-Bilak E. 2003. Intra-chromosomal heterogeneity between the four 16S rRNA gene copies in the genus Veillonella: implications for phylogeny and taxonomy. Microbiology (Reading) 149:1493–1501. doi: 10.1099/mic.0.26132-0. [DOI] [PubMed] [Google Scholar]
- 19.Arif N, Do T, Byun R, Sheehy E, Clark D, Gilbert SC, Beighton D. 2008. Veillonella rogosae sp. nov., an anaerobic, Gram-negative coccus isolated from dental plaque. Int J Syst Evol Microbiol 58:581–584. doi: 10.1099/ijs.0.65093-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delmont TO, Eren AM. 2018. Linking pangenomes and metagenomes: the Prochlorococcus metapangenome. PeerJ 6:e4320. doi: 10.7717/peerj.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O’Connor KJB, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc Natl Acad Sci USA 102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vernikos G, Medini D, Riley DR, Tettelin H. 2015. Ten years of pan-genome analyses. Curr Opin Microbiol 23:148–154. doi: 10.1016/j.mib.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 23.Donati C, Zolfo M, Albanese D, Tin Truong D, Asnicar F, Iebba V, Cavalieri D, Jousson O, De Filippo C, Huttenhower C, Segata N. 2016. Uncovering oral Neisseria tropism and persistence using metagenomic sequencing. Nat Microbiol 1:16070. doi: 10.1038/nmicrobiol.2016.70. [DOI] [PubMed] [Google Scholar]
- 24.Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. 2017. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res 27:626–638. doi: 10.1101/gr.216242.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yassour M, Jason E, Hogstrom LJ, Arthur TD, Tripathi S, Siljander H, Selvenius J, Oikarinen S, Hyöty H, Virtanen SM, Ilonen J, Ferretti P, Pasolli E, Tett A, Asnicar F, Segata N, Vlamakis H, Lander ES, Huttenhower C, Knip M, Xavier RJ. 2018. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe 24:146–154.e4. doi: 10.1016/j.chom.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Figueras MJ, Beaz-Hidalgo R, Hossain MJ, Liles MR. 2014. Taxonomic affiliation of new genomes should be verified using average nucleotide identity and multilocus phylogenetic analysis. Genome Announc 2:e00927-14. doi: 10.1128/genomeA.00927-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olm MR, Crits-Christoph A, Diamond S, Lavy A, Matheus Carnevali PB, Banfield JF. 2020. Consistent metagenome-derived metrics verify and delineate bacterial species boundaries. mSystems 5:e00731-19. doi: 10.1128/mSystems.00731-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Rooijen RJ, van Schalkwijk S, de Vos WM. 1991. Molecular cloning, characterization, and nucleotide sequence of the tagatose 6-phosphate pathway gene cluster of the lactose operon of Lactococcus lactis. J Biol Chem 266:7176–7181. doi: 10.1016/S0021-9258(20)89626-4. [DOI] [PubMed] [Google Scholar]
- 29.Lower BH, Bazylinski DA. 2013. The bacterial magnetosome: a unique prokaryotic organelle. J Mol Microbiol Biotechnol 23:63–80. doi: 10.1159/000346543. [DOI] [PubMed] [Google Scholar]
- 30.Zeng X, Lin J. 2013. Beta-lactamase induction and cell wall metabolism in Gram-negative bacteria. Front Microbiol 4:128. doi: 10.3389/fmicb.2013.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cross BW, Ruhl S. 2018. Glycan recognition at the saliva-oral microbiome interface. Cell Immunol 333:19–33. doi: 10.1016/j.cellimm.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Utter DR, Borisy GG, Eren AM, Cavanaugh CM, Mark Welch JL. 2020. Metapangenomics of the oral microbiome provides insights into habitat adaptation and cultivar diversity. Genome Biol 21:293. doi: 10.1186/s13059-020-02200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mashima I, Theodorea CF, Djais AA, Kunihiro T, Kawamura Y, Otomo M, Saitoh M, Tamai R, Kiyoura Y. 2021. Veillonella nakazawae sp. nov., an anaerobic Gram-negative coccus isolated from the oral cavity of Japanese children. Int J Syst Evol Microbiol 71:e004583. doi: 10.1099/ijsem.0.004583. [DOI] [PubMed] [Google Scholar]
- 34.Distler W, Kröncke A. 1980. Acid formation by mixed cultures of cariogenic strains of Streptococcus mutans and Veillonella alcalescens. Arch Oral Biol 25:655–658. doi: 10.1016/0003-9969(80)90096-5. [DOI] [PubMed] [Google Scholar]
- 35.Distler W, Kröncke A. 1981. Acid formation by mixed cultures of dental plaque bacteria Actinomyces and Veillonella. Arch Oral Biol 26:123–126. doi: 10.1016/0003-9969(81)90081-9. [DOI] [PubMed] [Google Scholar]
- 36.Mikx FHM, Van der Hoeven JS. 1975. Symbiosis of Streptococcus mutans and Veillonella alcalescens in mixed continuous cultures. Arch Oral Biol 20:407–410. doi: 10.1016/0003-9969(75)90224-1. [DOI] [PubMed] [Google Scholar]
- 37.Desguin B, Goffin P, Viaene E, Kleerebezem M, Martin-Diaconescu V, Maroney MJ, Declercq J-P, Soumillion P, Hols P. 2014. Lactate racemase is a nickel-dependent enzyme activated by a widespread maturation system. Nat Commun 5:3615. doi: 10.1038/ncomms4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alfano M, Cavazza C. 2020. Structure, function, and biosynthesis of nickel-dependent enzymes. Protein Sci 29:1071–1089. doi: 10.1002/pro.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright MR, Jamali F. 1993. Methods for the analysis of enantiomers of racemic drugs application to pharmacological and pharmacokinetic studies. J Pharmacol Toxicol Methods 29:1–9. doi: 10.1016/1056-8719(93)90044-f. [DOI] [PubMed] [Google Scholar]
- 40.Ewaschuk JB, Naylor JM, Zello GA. 2005. d-Lactate in human and ruminant metabolism. J Nutr 135:1619–1625. doi: 10.1093/jn/135.7.1619. [DOI] [PubMed] [Google Scholar]
- 41.Schink B, Stams A, Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. 2006. The prokaryotes: a handbook on the biology of bacteria. Springer, New York, NY. [Google Scholar]
- 42.Jurgenson CT, Begley TP, Ealick SE. 2009. The structural and biochemical foundations of thiamin biosynthesis. Annu Rev Biochem 78:569–603. doi: 10.1146/annurev.biochem.78.072407.102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morris JJ, Lenski RE, Zinser ER. 2012. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. mBio 3:e00036-12. doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kraft C, Angert E. 2017. Competition for vitamin b1 (thiamin) structures numerous ecological interactions. Q Rev Biol 92:151–168. doi: 10.1086/692168. [DOI] [PubMed] [Google Scholar]
- 45.Voelker AL, Taylor LS, Mauer LJ. 2021. Effect of pH and concentration on the chemical stability and reaction kinetics of thiamine mononitrate and thiamine chloride hydrochloride in solution. BMC Chem 15:47. doi: 10.1186/s13065-021-00773-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voelker AL, Miller J, Running CA, Taylor LS, Mauer LJ. 2018. Chemical stability and reaction kinetics of two thiamine salts (thiamine mononitrate and thiamine chloride hydrochloride) in solution. Food Res Int 112:443–456. doi: 10.1016/j.foodres.2018.06.056. [DOI] [PubMed] [Google Scholar]
- 47.Dal Co A, van Vliet S, Kiviet DJ, Schlegel S, Ackermann M. 2020. Short-range interactions govern the dynamics and functions of microbial communities. Nat Ecol Evol 4:366–375. doi: 10.1038/s41559-019-1080-2. [DOI] [PubMed] [Google Scholar]
- 48.Magnúsdóttir S, Ravcheev D, de Crécy-Lagard V, Thiele I. 2015. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front Genet 6:148. doi: 10.3389/fgene.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodionov DA, Hebbeln P, Eudes A, ter Beek J, Rodionova IA, Erkens GB, Slotboom DJ, Gelfand MS, Osterman AL, Hanson AD, Eitinger T. 2009. A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol 191:42–51. doi: 10.1128/JB.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilbert SA, Welch JLM, Borisy GG. 2020. Spatial ecology of the human tongue dorsum microbiome. Cell Rep 30:4003–4015.e3. doi: 10.1016/j.celrep.2020.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McLean AR, Torres-Morales J, Dewhirst FE, Borisy GG, Mark Welch JL. 2022. Site-tropism of streptococci in the oral microbiome. Mol Oral Microbiol 37:229–243. doi: 10.1111/omi.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peipoch M, Miller SR, Antao TR, Valett HM. 2019. Niche partitioning of microbial communities in riverine floodplains. Sci Rep 9:16384. doi: 10.1038/s41598-019-52865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH. 2010. Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J 4:337–345. doi: 10.1038/ismej.2009.122. [DOI] [PubMed] [Google Scholar]
- 54.Dussud C, Meistertzheim AL, Conan P, Pujo-Pay M, George M, Fabre P, Coudane J, Higgs P, Elineau A, Pedrotti ML, Gorsky G, Ghiglione JF. 2018. Evidence of niche partitioning among bacteria living on plastics, organic particles and surrounding seawaters. Environ Pollut 236:807–816. doi: 10.1016/j.envpol.2017.12.027. [DOI] [PubMed] [Google Scholar]
- 55.Eren AM, Kiefl E, Shaiber A, Veseli I, Miller SE, Schechter MS, Fink I, Pan JN, Yousef M, Fogarty EC, Trigodet F, Watson AR, Esen ÖC, Moore RM, Clayssen Q, Lee MD, Kivenson V, Graham ED, Merrill BD, Karkman A, Blankenberg D, Eppley JM, Sjödin A, Scott JJ, Vázquez-Campos X, McKay LJ, McDaniel EA, Stevens SLR, Anderson RE, Fuessel J, Fernandez-Guerra A, Maignien L, Delmont TO, Willis AD. 2021. Community-led, integrated, reproducible multi-omics with anvi’o. Nat Microbiol 6:3–6. doi: 10.1038/s41564-020-00834-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pritchard L, Cock P, Esen Ö. 2019. pyani v0. 2.8: average nucleotide identity (ANI) and related measures for whole genome comparisons.
- 58.Aujoulat F, Bouvet P, Jumas-Bilak E, Jean-Pierre H, Marchandin H. 2014. Veillonella seminalis sp. nov., a novel anaerobic Gram-stain-negative coccus from human clinical samples, and emended description of the genus Veillonella. Int J Syst Evol Microbiol 64:3526–3531. doi: 10.1099/ijs.0.064451-0. [DOI] [PubMed] [Google Scholar]
- 59.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K. 2016. A new view of the tree of life. Nat Microbiol 1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 61.Lee MD. 2019. GToTree: a user-friendly workflow for phylogenomics. Bioinformatics 35:4162–4164. doi: 10.1093/bioinformatics/btz188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Galperin MY, Wolf YI, Makarova KS, Vera Alvarez R, Landsman D, Koonin EV. 2021. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res 49:D274–D281. doi: 10.1093/nar/gkaa1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, Finn RD, Bateman A. 2021. Pfam: the protein families database in 2021. Nucleic Acids Res 49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eren AM, Vineis JH, Morrison HG, Sogin ML. 2013. A filtering method to generate high quality short reads using Illumina paired-end technology. PLoS One 8:e66643. doi: 10.1371/journal.pone.0066643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Minoche AE, Dohm JC, Himmelbauer H. 2011. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol 12:R112. doi: 10.1186/gb-2011-12-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Olm MR, Crits-Christoph A, Bouma-Gregson K, Firek BA, Morowitz MJ, Banfield JF. 2021. inStrain profiles population microdiversity from metagenomic data and sensitively detects shared microbial strains. Nat Biotechnol 39:727–736. doi: 10.1038/s41587-020-00797-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S5. Download spectrum.04042-22-s0001.xlsx, XLSX file, 0.5 MB (565.5KB, xlsx)
Supplemental material. Download spectrum.04042-22-s0002.pdf, PDF file, 7.7 MB (7.8MB, pdf)
Supplemental material. Download spectrum.04042-22-s0003.xlsx, XLSX file, 0.03 MB (31.1KB, xlsx)
Supplemental material. Download spectrum.04042-22-s0004.xlsx, XLSX file, 0.02 MB (18KB, xlsx)
Supplemental material. Download spectrum.04042-22-s0005.xlsx, XLSX file, 0.1 MB (130.3KB, xlsx)
Data Availability Statement
The raw data used in this study are publicly available at NIH GenBank and RefSeq (https://www.ncbi.nlm.nih.gov/genome/) for genomes and Human Microbiome Project for metagenomes (https://portal.hmpdacc.org/). Code used for analyses is available on GitHub (https://github.com/FatherofEverest/Site-specialization-of-human-oral-Veillonella-species.git).