ABSTRACT
Oropharyngeal candidiasis (OPC), which has a high incidence in immunocompromised and denture stomatitis patients, is commonly caused by Candida albicans infection and in some cases develops into disseminated candidiasis throughout the throat and esophagus, resulting in high mortality. New drugs are needed to combat OPC because of the limited treatment options currently available and increasing resistance to existing drugs. Here, we confirmed that riboflavin (RF), a cofactor of flavin adenine mononucleotide and flavin adenine dinucleotide, has broad-spectrum anti-Candida activity. The formation of C. albicans hyphae and biofilm was inhibited by RF. Mechanistically, RF disrupted membrane and cell wall integrity, as well as promoting reactive oxygen species and pyruvate accumulation. Furthermore, RF targeted multiple essential pathways via functional disruption of thiamine and RF metabolic pathways, central carbon metabolism, and ribosome metabolism. Similar to the results in vitro, the inhibitory effect of RF on C. albicans hyphae was confirmed in a mouse model of OPC. Moreover, after 5 consecutive days of intraperitoneal injection, RF exhibited therapeutic efficacy, as demonstrated by phenotype investigation, the fungal burden, and histopathological analysis. These findings revealed that RF exerts a multifaceted anti-Candida effect and has potential benefits in the treatment of OPC.
IMPORTANCE Candida species are common pathogens in fungal infections, causing mucosal infection and invasive infection in immunodeficient patients. Given the limited classes of drugs and resistance to these drugs, new antifungal agents need to be developed. Drug repurposing is a potential method for antifungal drug development. This study demonstrated that riboflavin (RF) exhibited broad-spectrum anti-Candida activity. RF affected multiple targets involving the membrane and cell wall integrity, the accumulation of reactive oxygen species and pyruvate, and the altered metabolic pathways in C. albicans. Moreover, RF exhibited efficacy in the treatment of C. albicans in an oropharyngeal candidiasis mouse model. Taken together, the antifungal activity and the promising clinical application of RF were highlighted.
KEYWORDS: antifungal agent, Candida species, fungal metabolism, oropharyngeal candidiasis, riboflavin
INTRODUCTION
Candida albicans is a common opportunistic fungal pathogen that causes superficial cutaneous and mucosal infections, as well as invasive infections, known as candidiasis and candidemia, respectively (1, 2). C. albicans symbiotically inhabits the oral cavity of healthy individuals. However, it can become pathogenic, causing oropharyngeal candidiasis (OPC) in immunocompromised individuals (especially those with AIDS, diabetes, cancer, or organ transplantation) (3). The exacerbation and recurrence of OPC provide favorable conditions for the occurrence of candidemia and increase the difficulty of treatment of the primary disease (4). In addition, the emergence of coronavirus has further increased the morbidity associated with Candida infection, including in OPC patients (5, 6).
At present, only four classes of drugs (azoles, polyenes, echinocandins, and flucytosine) are available for the treatment of pathogenic fungal infections. These agents target the fungal cell membrane, cell wall, or DNA/RNA (7). However, the widespread use of antifungal agents, as well as the limitation of drug classes and targets, has caused various levels of fungal resistance. The emergence of drug-resistant species, including Candida auris, has also further threatened the treatment of pathogenic fungi (8). Moreover, both fungi and human hosts are eukaryotes (9), which further complicates the development of uniquely targeted antifungal agents. The time and costs involved in the development of new classes of drugs have meant that no new antifungal agents have been approved in the 20th century (10, 11). Therefore, drug repurposing is gaining increased attention as an alternative method for antifungal drug development.
Riboflavin (RF), also known as vitamin B2, is an intracellular precursor of flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) and is involved in fatty acid oxidation, methylation, mitochondrial electron transport, nucleotide synthesis, and redox reaction processes (12). Most mammals, including humans, are unable to synthesize RF. RF derived from the diet is absorbed and metabolized in the human gut (13). RF deficiency results in cleft lip, glossitis, and oral mucosal edema; it can also affect gastrointestinal and brain function, as well as the metabolism of other vitamins (such as folate, niacin, pyridoxine, and vitamin B12) (13). Besides its classic role, RF also has application prospects in the field of anti-infection (14). Since RF is a photosensitizer, RF-mediated photodynamic therapy has an inhibitory effect on microbial keratitis (14), as well as infections caused by Escherichia coli, Listeria monocytogenes, Plasmodium falciparum, Staphylococcus aureus, and dengue virus (15–19). Furthermore, the level of RF was reported to be reduced in patients infected with human papillomavirus 16/18 or Helicobacter pylori (20, 21). Moreover, RF has a synergistic effect with fluconazole (FCZ) in treating vaginal candidiasis (22). However, the direct mechanism of this anti-Candida effect and the application of RF in OPC have not been reported. Considering these previous promising studies, the in vitro and in vivo anti-Candida effects of RF were examined in this study. Our results suggested that RF has a broad-spectrum anti-Candida effect in vitro by generating reactive oxygen species (ROS) and targeting cellular metabolic and ribosomal pathways. Moreover, in vivo, RF reduced the fungal burden in an OPC murine model, and the adhesion to and invasion of oral epithelial cells by C. albicans were decreased after RF treatment.
RESULTS
Broad-spectrum activity of RF inhibits Candida growth in vitro.
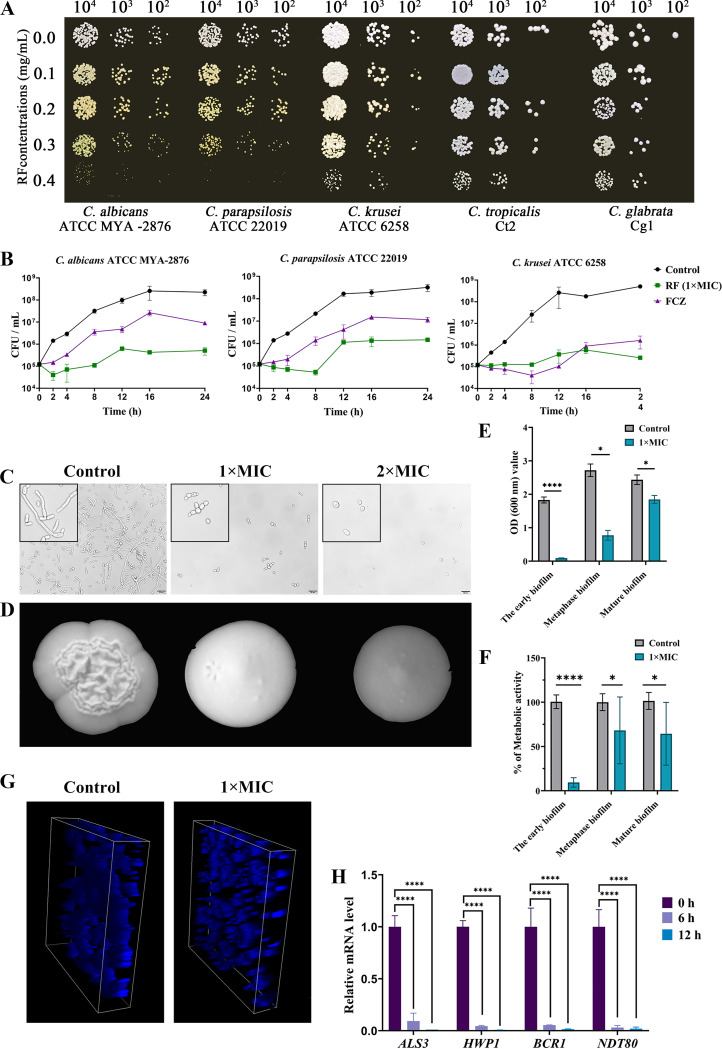
The MICs of RF for four standard strains and five clinical isolates were determined. RF exhibited broad-spectrum activity against Candida species, with an MIC of 0.4 mg/mL. With the dose of RF increased, the anti-Candida effect became more significant in a spot assay (Fig. 1A). A growth curve assay further characterized the fungicidal effects of RF. FCZ was used as a positive-control drug. Compared with the control group, 1× MIC of RF inhibited more than 90% of C. albicans, Candida krusei, and Candida parapsilosis grwoth between 2 and 24 h (Fig. 1B), and the antifungal activity of 1× MIC of RF was more effective than that of the FCZ group (64 μg/mL).
FIG 1.
The inhibitory effect on cell growth and hyphal and biofilm formation of Candida species. (A) The growth of Candida species on YPD agar plates with different concentrations of RF. (B) Growth curves of standard strains C. albicans ATCC MYA-2876, C. krusei ATCC 6258, and C. parapsilosis ATCC 22019 treated with 0.4 mg/mL of RF. (C) Hyphal formation was evaluated in RPMI 1640 plus 10% (vol/vol) FBS liquid medium, and uniformly enlarged images are presented in the black boxes on the left side. Bar, 20 μm. (D) Hyphal formation was evaluated on YPD plus 10% (vol/vol) FBS agar plates. (E) The biomass of C. albicans biofilm was observed by a crystal violet assay. (F) Metabolic activity of C. albicans biofilm was determined by a 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide assay. The results are presented as relative percentages. (G) The three-dimensional structure of C. albicans biofilm was stained with calcofluor white and observed by confocal laser scanning microscopy. (H) The expression of biofilm-related genes was determined. C. albicans treated with 1× MIC of RF was incubated for 0, 6, and 12 h before RNA extraction. 1× MIC, 0.4 mg/mL of RF; 2× MIC, 0.8 mg/mL of RF. Data were analyzed by a t test or one-way analysis of variance (one-way ANOVA) (ns [not significant], P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
RF inhibits Candida hypha and biofilm formation in vitro.
The formation of Candida hyphae plays an important role in the invasion and damage of epithelial and endothelial cells, as well as in causing bloodstream infections (23, 24). The effect of RF on C. albicans hyphal formation under a hypha-induced medium was elucidated. Hypha cells and both tubular and multicellular forms of C. albicans were inhibited by RF in liquid medium (Fig. 1C). The antihypha capacity of RF was further confirmed by the transition from crenulated to smooth colonies on solid medium (Fig. 1D).
Filamentous development is critical for biofilm formation, and biofilm formation on medical devices is the main cause of nosocomial fungal infections (25, 26). The effect of RF on biofilm formation was further evaluated. A crystal violet (CV) assay and a 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT; Macklin, Shanghai, China) assay were used to analyze the effect of RF on the formation, development, and maturity of biofilms. As shown in Fig. 1E and F, 1× MIC of RF seriously affects the early biofilm formation and has a minor effect on destroying the mature biofilm. To better define the effect of RF on biofilm formation, confocal laser scanning microscopy (CLSM; Leica, Beijing, China) investigation was performed and revealed that C. albicans cells were compact and connected into sheets in the absence of RF treatment. Conversely, cells treated with RF showed low density and could not connect into sheets (Fig. 1G). Subsequently, the transcription levels of genes related to hyphal and biofilm formation were evaluated by reverse transcription-quantitative PCR (RT-qPCR). Hyphal-formation-specific genes (including ALS3, HGT2, and HWP1) and a biofilm development-specific gene (BCR1) were analyzed. As expected, all genes were downregulated after RF treatment (Fig. 1H). Moreover, to further confirm the anti-hyphae and anti-biofilm activity against other Candida species, a similar effect was observed on C. parapsilosis ATCC 22019 hyphal and biofilm formation after RF treatment (see Fig. S1 in the supplemental material).
RF treatment damages Candida cell wall integrity.
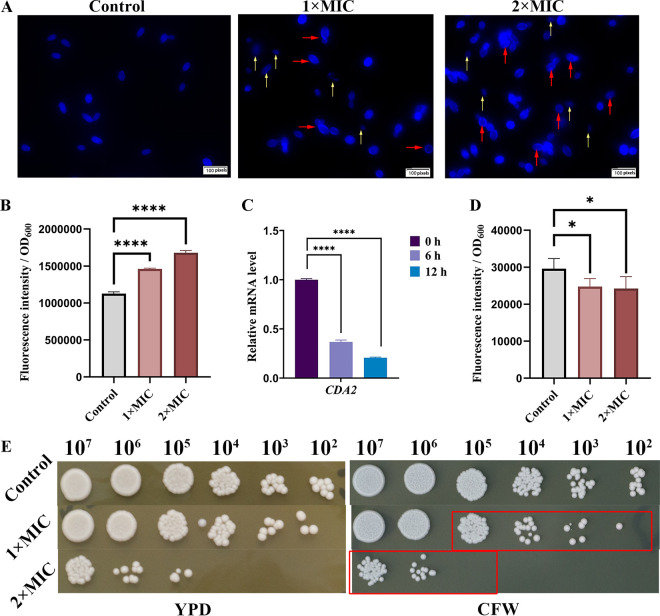
The fungal cell wall is involved in cell morphological maintenance and polarized growth (27). Candida can survive by altering its cell wall composition and structure in changed environments (28). As shown in Fig. 2A, calcofluor white (CFW; Sigma, Shanghai, China) was homogeneously distributed in yeast cells in the absence of RF treatment. However, more CFW was aggregated and distributed in the outer layer of C. albicans cells, especially at the budding end following RF treatment (red arrows). Even the elliptical structure of some yeast cells was disrupted (yellow arrows). Furthermore, investigation of CFW fluorescence indicated that the chitin content was increased by RF (Fig. 2B), and the expression of CDA2 (encoding chitin deacetylase, which catalyzes chitin to chitosan [29, 30]) was obviously downregulated, implying that the increase in chitin occurred as a result of the blocked consumption pathway (Fig. 2C). In addition, the aniline blue staining assay showed that total glucan levels were decreased following RF treatment (Fig. 2D). Investigation of the susceptibility of C. albicans cells to RF under CFW stress conditions also showed that pretreatment with RF inhibited cell growth (Fig. 2E, red rectangle). These data suggest that RF remodels the C. albicans cell wall structure.
FIG 2.
The effect of RF on the C. albicans cell wall. (A) C. albicans with or without RF treatment was stained with calcofluor white (CFW) and observed under a fluorescence microscope. CFW binds fungal chitin and produces blue fluorescence. Yeast cells with a disrupted elliptical structure are indicated by yellow arrows. Chitin deposition is indicated by red arrows. Images were captured at ×60 magnification using a fluorescence microscope. (B) Chitin levels were quantified through the fluorescence intensity of cells stained with CFW. (C) The expression of the gene encoding chitin deacetylase (CDA2) was determined. (D) Total glucan levels were quantified through the fluorescence intensity of cells stained with aniline blue. (E) C. albicans with or without RF treatment was spotted onto YPD agar containing 50 μg/mL of CFW and incubated for 24 h. Data were analyzed by one-way ANOVA (ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
RF treatment alters Candida cell membrane permeability.
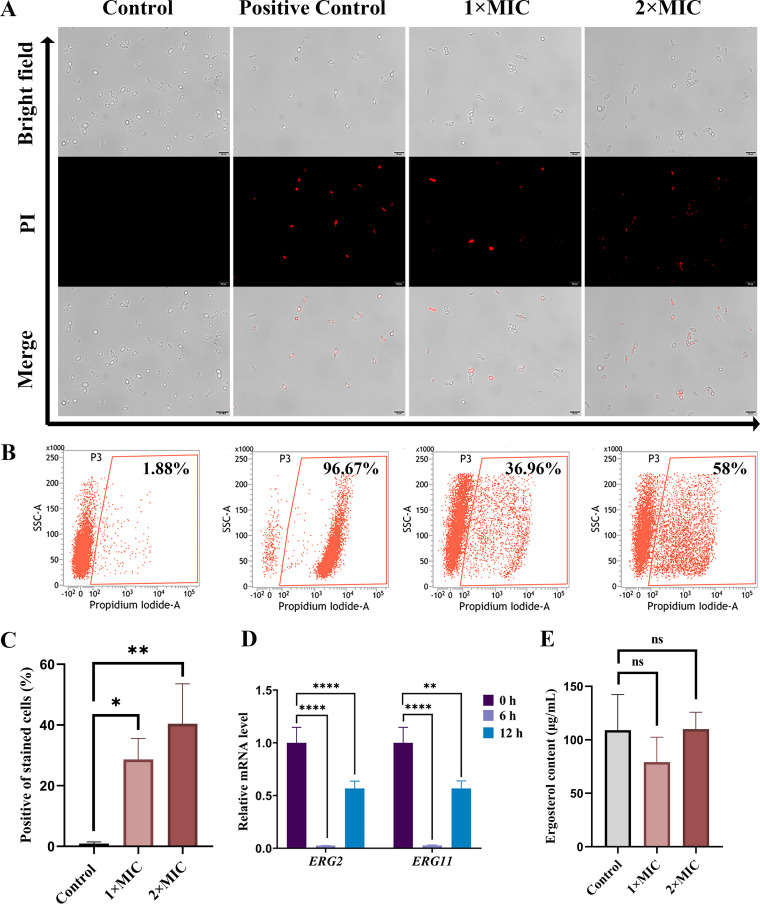
The transport of RF has been reported to be associated with the cell membrane in Bacillus subtilis (25). To further explore the effect of RF on the structure of Candida, its cell membrane integrity was evaluated. Propidium iodide (PI) enters cells through a damaged cell membrane and binds to nucleic acids, producing red fluorescence. As shown in Fig. 3A, the number of red cells increased after RF treatment, while there were no red cells in the control group. Similarly, a flow cytometry assay further confirmed that 36.96% and 58.00% of C. albicans cells were stained fluorescently following RF treatment with 1× MIC and 2× MIC, respectively (Fig. 3B and C). It is worth noting that cells with damaged cell membranes cannot survive after being spread onto yeast extract-peptone-dextrose (YPD) agar (Fig. 1B). Additionally, there is a similar effect on the C. parapsilosis ATCC 22019 cell membrane (Fig. S2). Ergosterol, the main component of the Candida cell membrane, has become a focus of the development of antifungal drugs (31). In this study, the genes involved in the ergosterol synthesis pathway (including ERG2 and ERG11) were analyzed by RT-qPCR. As shown in Fig. 3D, the expression of ERG2 and ERG11 was downregulated after RF treatment. However, compared with the control group, the content of ergosterol in C. albicans showed no statistically significant difference after treatment with 1× MIC and 2× MIC of RF (Fig. 3E). In short, RF enhanced cell membrane permeability without affecting the ergosterol content.
FIG 3.
The effect of RF on the C. albicans cell membrane. (A and B) C. albicans cells with or without RF treatment were stained with propidium iodide (PI) and analyzed by fluorescence microscopy (A) and flow cytometry (B). Bar, 20 μm. DIC, differential inference contrast; SSC, side scatter. (C) Histogram analysis shows the percentage of PI-positive cells. (D) The expression of genes involved in ergosterol synthesis was determined. (E) Histogram analysis shows the ergosterol concentration of C. albicans cells. Data were analyzed by one-way ANOVA (ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
RF treatment induces ROS accumulation.
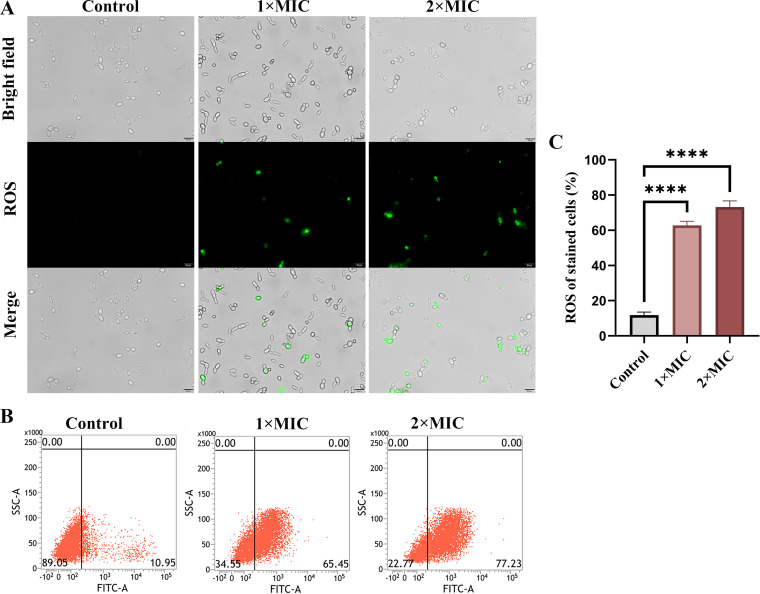
ROS are superoxides produced in mitochondria, and excessive levels of intracellular ROS affect the integrity of cell walls and cell membranes (32). Thus, ROS production was measured. As shown in Fig. 4A, significantly increased fluorescence was observed after treatment with RF. Flow cytometry further confirmed that 65.45% and 77.23% of cells were ROS positive after RF treatment with 1× MIC and 2× MIC, respectively, while only 10.95% of cells were ROS positive in the control group (Fig. 4B). These results suggested that RF markedly promotes the excessive production of ROS (Fig. 4C).
FIG 4.
The effect of RF on reactive oxygen species (ROS) production. (A) The accumulation of ROS green fluorescence was observed. ROS oxidizes DCFH-DA to DCF, emitting green fluorescence. Bar, 20 μm. (B) The relative percentage of ROS-positive cells was shown by flow cytometry. FITC, fluorescein isothiocyanate. (C) Histogram analysis showed the percentage of ROS-positive cells. Data were analyzed by ANOVA (ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
RF induces coenzyme metabolic disorder.
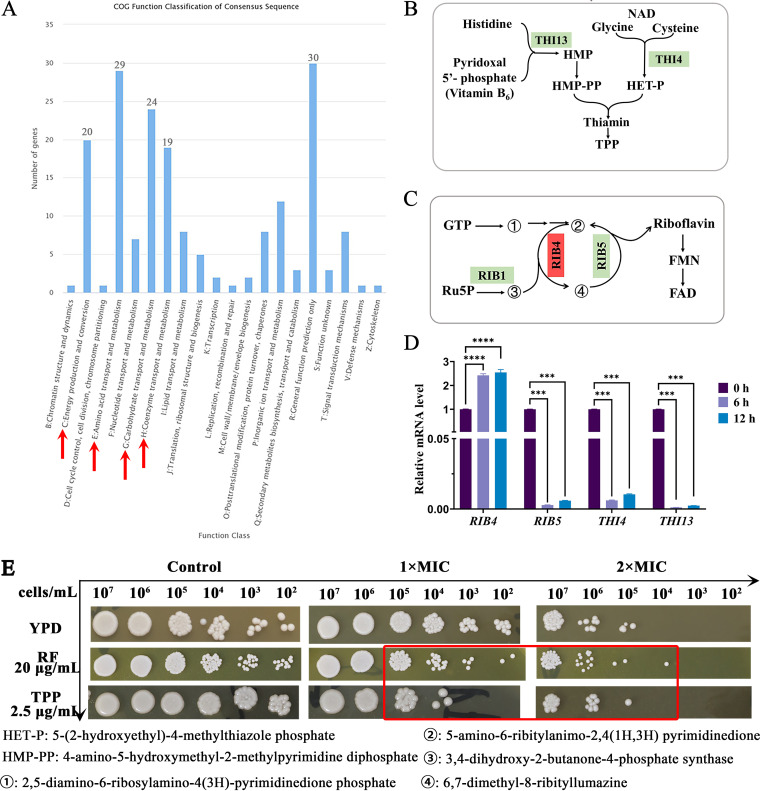
The accumulation of ROS induces protein, lipid, and nucleic acid damage, resulting in C. albicans metabolic disorder and cell death (33). To analyze the mechanism of action of RF in greater depth, RNA-sequencing (RNA-Seq) was performed. The results showed that 563 genes were upregulated and 469 genes were downregulated. The downregulated differentially expressed genes were further analyzed and were found to be enriched in energy production and conversion, amino acid transport and metabolism, and carbohydrate transport and metabolism (Fig. 5A). Unexpectedly, genes involved in coenzyme transport and metabolism were most significantly altered, especially those in the coenzyme thiamine and RF metabolism pathways. Therefore, the expression of key genes in the thiamine (THI4 and THI13, Fig. 5B) and RF (RIB4, and RIB5, Fig. 5C) metabolic pathways was subsequently analyzed. The results showed that exogenous RF significantly decreased the expression of RIB5, THI4, and THI13, while the expression of RIB4 was increased by RF treatment (Fig. 5D). RF-treated C. albicans was spotted onto YPD medium containing thiamine pyrophosphate (TPP) and RF, which are the metabolites of the coenzyme metabolic pathway. As shown in Fig. 5E, the growth of RF-treated C. albicans was inhibited following the addition of TPP or RF stress (red rectangle). These data suggested that RF induces thiamine and RF metabolic disorder in C. albicans.
FIG 5.
The effect of RF on C. albicans coenzyme metabolism. (A) Clusters of orthologous groups analysis of downregulated differentially expressed genes was performed, and the main enrichment pathways are indicated by red arrows. (B and C) Schematic model of the endogenous thiamine metabolism pathway (B) and the endogenous riboflavin metabolism pathway (C). Genes encoding proteins in red were upregulated, while genes encoding proteins in green were downregulated. (D) The expression of genes involved in the thiamine and RF metabolic pathways was analyzed. (E) C. albicans cells with or without RF treatment were spotted onto YPD agar plates containing exogenous thiamine or RF. TPP, thiamine pyrophosphate. Data were analyzed by ANOVA (ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
RF affects central carbohydrate metabolism.
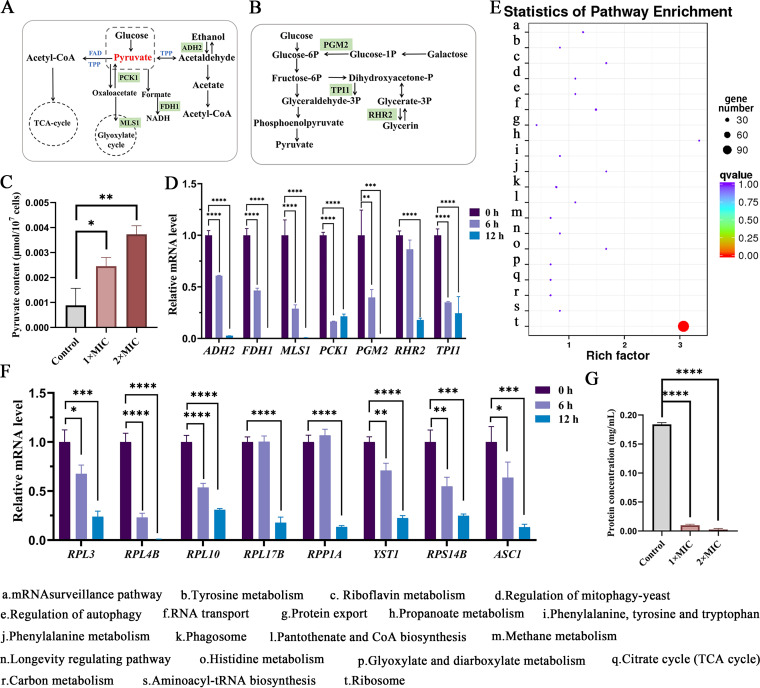
Coenzymes FAD and TPP, as members of the pyruvate oxidative decarboxylase complex, are involved in the enzymatic conversion of pyruvate to acetyl coenzyme A (34, 35) (Fig. 6A and B). It has been confirmed that pyruvate accumulation is the first line of fungal cell defense against ROS caused by heat stress (36). Thus, the content of pyruvate was measured. As shown in Fig. 6C, the concentration of pyruvate was increased in C. albicans after treatment with RF. In addition, pyruvate is an important regulator of central carbohydrate metabolism in the cell, linking the glycolytic pathway to the tricarboxylic acid cycle (34). Suppression in central carbohydrate metabolism was confirmed by RNA-Seq (Fig. 5A) and the downregulation of related genes (including ADH2, FDH1, MLS1, PCK1, PGM2, RHR2, and TPI1) (Fig. 6D). These results implied that exogenous RF targets internal RF and the thiamine metabolic pathway, increasing the concentration of pyruvate, which in turn induces central carbohydrate metabolic disorders in C. albicans.
FIG 6.
The effect of RF on C. albicans central carbon metabolism and ribosome metabolism. (A and B) A schematic model of central carbon metabolism (A) and glycometabolism (B). Genes encoding proteins in red were upregulated, while genes encoding proteins in green were downregulated. TCA, tricarboxylic acid. (C) The concentration of pyruvate was determined. (D) The expression of genes involved in central carbon metabolism was determined. (E) Kyoto Encyclopedia of Genes and Genomes analysis of upregulated differentially expressed genes. (F) The expression of genes involved in ribosome metabolism was determined. (G) Histogram analysis shows the concentration of total protein with or without RF treatment. 1× MIC, 0.4 mg/mL of RF; 2× MIC, 0.8 mg/mL of RF. Data were analyzed by ANOVA (ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
RF affects ribosome metabolism.
The upregulated differentially expressed genes were also analyzed, and 134 out of 563 upregulated genes were enriched in ribosome metabolism (Fig. 6E). To confirm this finding, eight genes were selected for additional RT-qPCR analysis. Ribosomes consist of a large 60S subunit (encoded by RPL3, RPL4B, RPL10, RPL17B, and RPP1A) and a small 40S subunit (encoded by YST1, RPS14B, and ASC1). The expression of all of these genes was obviously downregulated following RF treatment for 6 or 12 h, with the exception of RPP1A (Fig. 6F). Ribosomes are responsible for reading mRNA to synthesize protein (37). Therefore, we measured the total protein content of C. albicans following RF treatment and found it to be significantly decreased (Fig. 6G), indicating that RF induced the dysfunction of ribosome metabolism.
RF improved the symptoms of OPC in vivo.
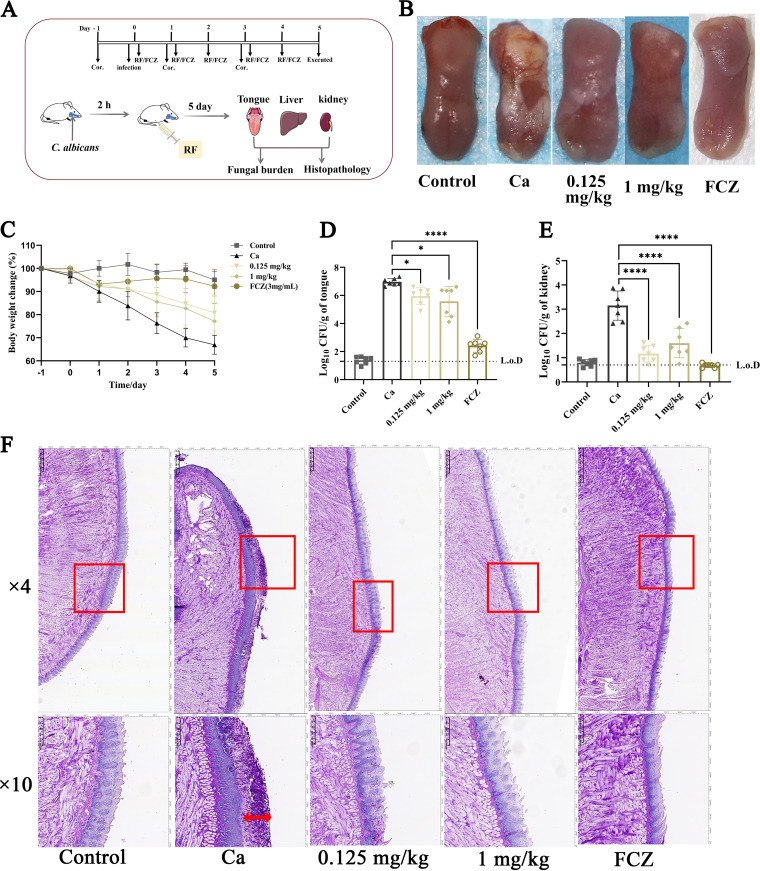
The efficacy of RF in vivo was investigated in an OPC murine model. First, immunosuppressed mice were infected with C. albicans. After 2 h of incubation, RF or normal saline was injected intraperitoneally (Fig. 7A). As shown in Fig. 7B, the back of the tongue was ruddy and smooth in the control group, whereas a thick white plaque was evident on the dorsum of the tongue in the Ca group (OPC mice treated with normal saline). After administering 0.125 or 1 mg of RF per kg of body weight or 13.6 mg/kg of FCZ for 5 days, the white plaques were thinner and showed less coverage. In addition, the loss in body weight in the RF-treated group was less than that in the Ca group (Fig. 7C). Importantly, compared with the Ca group, there was a statistically significant reduction in the fungal burden on the tongue and kidneys in the RF-treated group (Fig. 7D and E); however, there was no statistical difference in the liver (data not shown). To assess the curative efficacy of RF in vivo, histopathological changes on the mouse tongue were evaluated by periodic acid-Schiff (PAS) staining. In the Ca group, the tongue epithelium covered a large number of hypha cells destroying the papillae (Fig. 7F). In contrast, fewer hyphal cells and thin and loose biofilm were found in the tongue epithelium of the RF-treated group. In addition, for the RF-treated group, damage of the tongue papillae was relieved (Fig. 7F). These results indicated that RF has an antifungal effect in an OPC murine model.
FIG 7.
The effect of RF on an oropharyngeal candidiasis (OPC) mouse model. (A) A schematic model of the experiment. Mice were injected with immunosuppressant cortisone acetate on days −1, 1, and 3. Mice were infected with C. albicans and continuously treated with normal saline, RF, or fluconazole (FCZ) for 5 days. All mice were euthanized on day 5, and the tongue, kidney, and liver were collected. (B) Visual analysis of the phenotype of mouse tongues. Control group, immunodeficient and uninfected mice that were continuously treated with sterile saline; Ca group, immunodeficient and infected mice that were continuously treated with sterile saline; 0.125- and 1-mg/kg groups, immunodeficient and infected mice that were continuously treated with 0.125 and 1 mg/kg of RF, respectively; FCZ, immunodeficient and infected mice that were continuously treated with 13.6 mg/kg of FCZ. (C) Changes in the body weight of mice over 5 consecutive days. (D and E) Fungal burden on the tongue (D) and kidneys (E). L.o.D., limit of detection. (F) Histopathological analysis by periodic acid-Schiff staining. In the image showing an ×4 magnification, the area in the red rectangle is shown at ×10 magnification below. The damage to tongue epithelial cells caused by C. albicans is indicated by red arrows. Data were analyzed by ANOVA (ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
DISCUSSION
The prevalence of novel pathogens has been on the rise over the past few decades, as evidenced by the emergence of novel coronavirus COVID-19/SARS-CoV-2, Ebola viruses, Middle East respiratory viruses, C. auris, and novel mutant bacteria (38–40). However, drug development has not kept pace with the rise of novel pathogens and the resistance to antimicrobial drugs, which provides an opportunity for drug repurposing (11). Drug repurposing is the search for new uses or characteristics from approved drugs, such as molecular targets, mode of action, or pharmacological effects (11). Such a strategy reduces the risk, time taken, and cost expended on new drug development. Recent studies have emphasized the important role of B vitamins in the human nervous system, immune response, and tumor development (41–43). In addition, B vitamins also have value in patients with kidney disease, sepsis, and COVID-19 (44–46). Our previous review summarized the anti-infection effect of RF and suggested that RF may have direct antimicrobial effects (14). Here, in vitro and in vivo phenotypic investigations confirmed that RF has anti-Candida ability. In vitro, RF is able to inhibit biofilm formation. Adhesion, which is the first step in biofilm formation, was also found to be reduced after RF treatment (see Fig. S3A and B in the supplemental material).
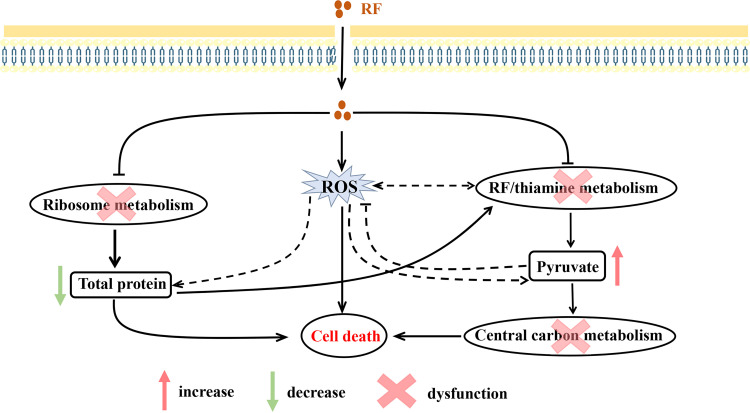
Low doses of ROS are signaling molecules involved in the normal physiological functions and development of fungi, while excessive ROS doses can lead to pathological processes or cell death through their toxic effects (47). In this study, RF disrupted the membrane and cell wall integrity, resulting in the ROS accumulation. Although mitochondria are one of the main sources of ROS production, our investigation found that RF did not affect the mitochondrial respiratory chain (Fig. S4). The production of ROS may connect total metabolism via thiamine metabolism, RF metabolism, and ribosome metabolism caused by exogenous RF (Fig. 8) (48–51). More specifically, exogenous RF targets the thiamine and RF metabolic pathways, thus disrupting central carbon metabolism. The production of ROS was accompanied by an accumulation of pyruvate, which is consistent with previous work (36, 52). Ribosome metabolism was also affected by exogenous RF via the production of ROS. These data were confirmed by the downregulation of ribosome-related genes and a reduction in the total protein concentration (Fig. 6). Moreover, our findings were consistent with those of a previous investigation which showed the damaging effect of ROS on the structure and activity of proteins (50, 53). Thus, the accumulation of ROS induced by RF may contribute to the dysfunction of total metabolism. Taken together, our biochemical and transcriptome results revealed the multiple mechanisms of action of RF against C. albicans, leading to damage to the membrane and cell wall integrity, the accumulation of ROS, disordered central carbon metabolism, and protein damage (Fig. 8).
FIG 8.
Schematic diagram of the mechanism of action of RF against C. albicans in vitro. Exogenous RF destroys the membrane and cell wall integrity, increasing the accumulation of ROS in C. albicans. Meanwhile, exogenous RF targets the coenzyme (including endogenous RF and thiamine metabolism) and ribosome metabolism, resulting in total metabolic disorders. These effects together lead to the death of C. albicans. Specifically, repression of endogenous RF and thiamine metabolism caused by exogenous RF induces the accumulation of pyruvate (red arrow), which therefore affects central carbon metabolism. Exogenous RF induces ribosome metabolism disorders, inducing a block in protein synthesis (green arrow). This further aggravates the disorder in thiamine metabolism, since histidine is the substrate for thiamine synthesis. Moreover, ROS are capable of inducing protein damage and interacting with endogenous RF and thiamine metabolism (35, 48–51). The accumulating pyruvate may defend against ROS (36). However, in the presence of exogenous RF, the relationship between ROS and endogenous RF or thiamine metabolism, proteins, and pyruvate (dashed line) requires further study.
Central carbon metabolism plays an important role in fungal cells in obtaining energy and sustaining their survival. Our results suggested that exogenous RF may target endogenous thiamine and RF metabolism, resulting in a central carbohydrate metabolic disorder. Moreover, the growth of RF and thiamine metabolism-repressed C. albicans was not restored by the addition of extra RF or TPP, and extra RF or TPP had only a minor inhibitory effect on C. albicans (Fig. 5E). This was contrary to previous results showing that strains mutated in the RF or thiamine metabolic pathways would recover growth after supplementation with RF or thiamine (54, 55). Based on our findings, we speculated that exogenous RF may influence a key factor in the vitamin regulatory system (including endogenous synthesis and exogenous uptake) of C. albicans. However, further investigations are needed to confirm this key factor. Importantly, these results indicated that the endogenous RF and thiamine metabolic pathways could provide favorable targets for the development of antimicrobial drugs. Reasons for this include that thiamine and RF metabolism specifically exists in pathogens and does not exist in humans (14), and these metabolic pathways are essential for pathogen virulence (56). In fact, other B-group vitamins are important cofactors for fungal cells, including niacin (VB3), pantothenic acid (VB5), pyridoxine (VB6), biotin (VB7), folate (B9), and cobalamin (VB12), which are also potential candidates for the development of antifungal drugs (56).
In vivo, the inhibitory effect of RF on C. albicans virulence factors, including hyphae and biofilms, was also observed in the phenotype of tongues and histopathological analysis in an OPC mouse model. Additionally, the results of histopathological analysis and fungal burden qualitatively confirmed that the infection of mice was reduced by RF (Fig. 7). Interestingly, the effective concentration of RF (0.125 or 1 mg/kg) in vivo is much lower than that of FCZ (13.6 mg/kg). RF treatment showed a 2-order-of-magnitude reduction in kidney fungal burden, which was superior to the 1-order-of-magnitude reduction in the tongue. The metabolism of RF is inextricably linked to the kidney (12), suggesting that understanding the absorption and transport of RF is vital to studying the mechanism of anti-infection activity in vivo. In a mouse ulcer model, RF alleviated oral ulcers by exerting antioxidant and antibacterial activity and reducing the local secretion of inflammatory factors (57). Previously, 20 to 100 mg/kg of RF combined with azithromycin was shown to reduce the expression of tumor necrosis factor alpha (TNF-α), gamma interferon (IFN-γ), interleukin-6 (IL-6), and monocyte chemoattractant protein 1 (MCP-1) in S. aureus infection (58). In addition, mucosa-associated invariant T cells are reportedly involved in the pathogenesis of oral diseases and can be activated by RF in an MR1-dependent manner (59). These data imply that RF may play a strong role in immune regulation in vivo. However, the mechanism of action of the direct anti-Candida effect and the immune reaction to RF in the OPC mouse model require further investigation.
In summary, the present study reveals the anti-Candida effect of RF in vitro and in vivo. Explorations of the anti-Candida mechanisms in vitro confirmed a multitarget effect. Among the repressed metabolic pathways, the thiamine and RF metabolic pathways were uniquely affected by the action of RF, which provides insight into the metabolic targets of RF.
MATERIALS AND METHODS
Strains, media, and chemicals.
Nine Candida species were used in this study. Standard strains C. albicans ATCC MYA-2876, C. albicans ATCC 90028, C. parapsilosis ATCC 22019, and C. krusei ATCC 6258 were purchased from the American Type Culture Collection (ATCC). Clinical isolates C. parapsilosis Cp1 and Cp2, Candida tropicalis Ct2 and Ct3, and Candida glabrata Cg1 were from the affiliated hospital of Southwest Medical University. RF (Sigma, Shanghai, China), RF injection (commercially available), and RPMI 1640 medium (HyClone, Chengdu, China) were purchased as reagents. Fungal cells were cultured and maintained in YPD medium (1% yeast extract, 2% peptone, and 2% glucose).
Antifungal susceptibility assay.
According to CLSI standard M27-A3, a microdilution broth susceptibility assay was conducted to determine the MIC (60). Briefly, the activated yeast cells were adjusted to 0.5 × 104 to 2.5 × 104 cells/mL and 100 μL of yeast suspension was added into 96-well plates. To each well, 100 μL of working solution ranging from 0.0 to 1.6 mg/mL of RF (diluted in RPMI 1640 medium) was added. The plate was incubated for 24 h at 37°C, and then the optical density at 600 nm (OD600) was measured. The MIC that suppressed fungal growth by 90% was defined as the lowest concentration.
Next, a spot assay determined the inhibitory activity on the solid medium. The activated cells were adjusted to 1 × 102 to 3 × 102, 103, and 104 cells/mL, and then 3 μL was spotted onto YPD agar, which contained 0.1, 0.2, 0.3, or 0.4 mg/mL RF. Phosphate-buffered saline (PBS) was added to the YPD plate as a control. After incubation for 48 h at 37°C, images of the colonies were recorded using a digital camera.
Growth curve assay.
The strains grown overnight were diluted to 5 × 105 cells/mL in YPD medium, containing 0.4 mg/mL of RF or 64 μg/mL of FCZ. The control was free of RF and FCZ. The cells were incubated at 37°C with constant shaking (200 rpm). At 0, 2, 4, 8, 12, 16, and 24 h, cells were harvested and washed with PBS before being spread onto YPD agar medium. Samples supplemented with water were considered controls. The fungal CFU were determined after being incubated for 24 h at 37°C.
Hyphal formation.
The activated yeast cells were adjusted to a final concentration of 1 × 106 to 3 × 106 cells/mL in RPMI 1640 plus 10% (vol/vol) fetal bovine serum (FBS) liquid medium or YPD plus 10% (vol/vol) FBS solid medium containing 1× MIC or 2× MIC of RF. Sterile water was added as a control group. Liquid and solid media were incubated for 6 h and 5 days at 37°C, respectively.
Biofilm assay.
Biofilm susceptibility was measured by CV and XTT assays (61). In brief, the activated yeast cells were suspended in RPMI 1640 (1 × 106 to 3 × 106 cells/mL) and 200 μL was added to 96-well plates. For the initial phase, cultures were preincubated for 1.5 h and RF was added for 6 h at 37°C. Then, the supernatant was removed and incubation was continued for 45.5 h. For the developmental phase, cultures were preincubated for 12 h and RF was added for 6 h at 37°C. Then, the supernatant was removed and incubation was continued for 36 h. For the maturation phase, cultures were preincubated for 48 h and RF was added for 6 h at 37°C. The biofilm biomass and activity were measured by the CV and XTT methods, as follows. (i) For the CV method, 100 μL of 15% CV was added to each well and incubated at room temperature for 10 min. After washing with PBS, 200 μL of 15% acetic acid was added, and after 30 min at room temperature, the OD600 was measured. (ii) For the XTT method, 1 mg/mL of XTT (in PBS) and 0.4 mM menadione (in ethanol) were prepared in a 5:1 ratio. To each well, 200 μL PBS and 12 μL XTT-menadione solution were added, and after 2 h of incubation at 37°C in the dark, the OD490 was measured.
Confocal laser scanning microscopy.
The biomass and three-dimensional structure of the biofilm were analyzed by CLSM (62). The activated fungal cells were resuspended in fresh RPMI 1640 at a final concentration of 1 × 106 to 3 × 106 cells/mL. Then, 3 mL of fungal cells was added to 6-well plates with circular microscope cover glasses in the bottom of the wells. After incubation for 90 min at 37°C, RF was added to a final concentration of 1× MIC and continuously incubated for 6 h. For the control groups, sterile water was added. Subsequently, plates were washed with PBS and samples were incubated with fresh RPMI 1640 medium for 24 h at 37°C. The biofilm was washed with PBS and stained with 1 mL of CFW for 3 min in the dark at room temperature. The circular microscope cover glasses were then transferred into antifade mounting medium, before being placed upside down on a glass slide, and the biofilm was observed through a TCS SP8 CLSM.
Cell wall assay to determine cell wall integrity and glucan and chitin content.
The fungal cells were treated with 1× MIC or 2× MIC of RF for 12 h at 37°C with shaking at 200 rpm. Sterile water was added as a control. Cells were harvested and washed with PBS, and then the fungal suspension was treated in three ways. (i) To the fungal suspension, 3 μL of 1% KOH and 7 μL of CFW were added, and after 2 to 3 min in the dark at room temperature, the cell morphology was observed through a DP80 fluorescence microscope (Olympus, Tokyo, Japan). (ii) The fungal suspension was adjusted to 1 × 102 to 3 × 102, 1 × 103, and 1 × 104 cells/mL and then spotted onto YPD agar medium containing 50 μg/mL of CFW. After being incubated for 5 days at 37°C, the colonies were recorded using a digital camera. (iii) The fungal suspension was adjusted to 5 × 107 cells/mL, and then the OD600 was measured. To determine the chitin content, CFW (at a final concentration of 3.5 μg/mL) was added and staining was carried out at 37°C for 10 min. The fluorescence intensity was measured at 365-nm excitation and 435-nm emission wavelengths on a BioTek Varioskan Synergy H1 plate reader (Thermo Fisher Scientific, Shanghai, China). To determine the total glucan content, aniline blue (1%) was added and incubated at 80°C for 15 min in the dark. Fluorescence intensity was measured at 398-nm excitation and 508-nm emission wavelengths.
PI staining.
The cell membrane integrity was determined by PI (Solarbio, Beijing, China) staining (60), an ergosterol content assay, and gene expression analysis. Sample collection was the same as for the cell wall integrity assay. The cells were resuspended to 0.5 × 107 to 1.0 × 107 cells/mL. Then, PI (10 μg/mL) was added and incubated for 30 min at 37°C in the dark, followed by PBS washing. The cell membrane permeability was determined using a FACSAria flow cytometer (BD Biosciences, NJ, USA) and an inverted fluorescence microscope. The blank group was not treated with PI. The positive-control group was pretreated with hydrogen peroxide for 20 min at 37°C.
HPLC assay.
The ergosterol content of Candida was evaluated by high-performance liquid chromatography (HPLC) (1260 Infinity II; Agilent). In brief, after centrifuging and washing, the wet samples were adjusted to 0.5 g. Then, 25 mL of methanol was added to samples and the total weight was recorded, followed by an ultrasound for 1 h. Following cooling to room temperature, methanol was added to the original weight and the solution was filtered through a 0.45-μm Millipore filter and stored at −20°C. The ergosterol was determined at 283 nm by HPLC.
ROS level measurement.
The fresh fungal cells (1 × 106 cells/mL) were treated with 1× MIC or 2× MIC RF for 6 h at 37°C with constant shaking (200 rpm). Samples were resuspended to 0.5 × 107 to 1.0 × 107 cells/mL in PBS and stained with 10 μM dichlorodihydrofluorescein diacetate (DCFH-DA) (Sigma, Shanghai, China) for 30 min at 37°C. After centrifuging and removing excess DCFH-DA, cells were resuspended. A FACSAria flow cytometer and inverted fluorescence microscope were used to analyze the ROS levels.
Spot assay.
The fresh fungal cells (1 × 106 cells/mL) were treated with 1× MIC or 2× MIC RF for 12 h at 37°C with constant shaking (200 rpm). Cells were washed with PBS and diluted over a gradient. To investigate the effect of RF on C. albicans RF and thiamine metabolism, 3 μL of fungal suspension was spotted onto the YPD agar (containing RF or thiamine pyrophosphate) and cultured for 2 to 4 days at 37°C. To assess the effect of RF on the ability for carbon source utilization, a gradient dilution of the fungal suspension was spotted onto YPD agar (glucose source) or glycerol agar (1% yeast extract, 2% peptone, 2% glycerol, and 2% agar), and cultured for 2 to 4 days at 37°C.
RNA sequencing.
C. albicans ATCC MYA-2876 (1 × 107 cells/mL) was treated with 1× MIC RF or without RF for 12 h at 37°C with constant shaking (200 rpm). Samples were collected by centrifugation for 2 min at 12,000 × g and 4°C. Then, total RNA was isolated using yeast processing reagent (TaKaRa, Dalian, China). The transcriptome data were processed by Biomarker (Beijing and Qingdao, China) using Oxford Nanopore Technologies Long Read Processing (63). The standard for the screening of differentially expressed genes was a |fold change| of ≥2 and a false-discovery rate of ≤0.01.
RT-qPCR analysis.
The activated C. albicans ATCC MYA-2876 was adjusted to 1 × 107 cells/mL and grown in YPD liquid medium at 37°C with shaking at 200 rpm. Samples were collected at 0, 6, and 12 h. Total RNA was isolated as previously described. The PrimeScript RT reagent kit with genomic DNA (gDNA) eraser and TB Green Premix Ex Taq II (TaKaRa, Beijing, China) were used to generate reverse cDNA and for RT-qPCR. The transcript of the β-actin (ACT1) gene was used as an internal standard. All primers are shown in Table S1 in the supplemental material. The threshold cycle (2−ΔΔCT) method was used to determine the relative change in gene expression (64).
Total protein assay.
C. albicans ATCC MYA-2876 (1 × 107 cells/mL) was treated with 1× MIC or 2× MIC RF or without RF for 12 h at 37°C with constant shaking (200 rpm). Samples were collected by centrifugation, washed three times, and adjusted to 1 × 108 cells/mL. Total protein was collected by a yeast total protein extraction kit (Sangon Biotech, Shanghai, China). Subsequently, the protein level was tested by an enhanced bicinchoninic acid (BCA) protein assay kit (Beyotime, Shanghai, China).
Quantification of the pyruvate concentration.
C. albicans ATCC MYA-2876 was cultured as described for the total protein assay. Samples were adjusted to 2 × 107 cells/mL and 5 × 107 cells/mL for the determination of pyruvate and acetyl coenzyme A (acetyl-CoA), respectively. Samples were collected by centrifugation and ground with liquid nitrogen, and then the concentration of pyruvate was quantified by a pyruvic acid content assay kit (Boxbio, Beijing, China).
Antifungal effect in vivo.
All experimental protocols were approved by the Southwest Medical University Institutional Animal Care and Use Committee (2020540). C57BL/6J male mice (6 to 8 weeks old; SiPeiFu, Beijing, China) were fed at 25 ± 1°C for 1 week before the experiment. The mice were assigned to five groups of eight mice each. One day before infection, and on the 1st and 3rd days after infection, mice were subcutaneously injected with 200 μL of the immunosuppressant cortisone acetate (225 mg/kg), which was dissolved in normal saline containing 0.5% Tween 80 (65, 66). On the day of infection, all groups were narcotized by intraperitoneal injection with pentobarbital (50 mg/kg) and the dorsum of the tongue, free of redness and hemorrhage, was scratched with a scalpel. Subsequently, a cotton ball saturated with 100 μL of C. albicans ATCC (2 × 108 cells/mL) was placed in the oral cavity for 90 min. Two hours after infection, RF (0.125 or 1 mg/kg) or FCZ (13.6 mg/kg) was intraperitoneally injected into the treatment group, and normal saline was intraperitoneally injected into the control group and the Ca (infection) group. When the intraperitoneal injection volume is 100 μL, 0.125 mg/kg and 1 mg/kg of RF correspond to 1/16× MIC and 1/2× MIC of RF. As previously described (65), 13.6 mg/kg of FCZ was used to treat oropharyngeal candidiasis in mice. The mice were injected once every 24 h for 5 consecutive days. After the last treatment, the mice were euthanized and the tongue, kidney, and liver tissues were collected for determination of the fungal load and pathological analysis.
Statistical analysis.
The in vitro experiments and RNA-Seq were performed three independent experiments, except the animal experiments. The differences between the groups were compared by a t test or one-way analysis of variance (ANOVA) followed by a least significant difference (LSD) test or Tamhane T2 test using IBM SPSS Statistics 26 (IBM SPSS Inc., Chicago, IL, USA). P values of <0.05 were considered statistically significant. GraphPad Prism 9.0 software was used to generate all of the figures.
Data availability.
Sequence data were deposited in the Beijing Institute of Genomics Genome Sequence Archive (accession no. PRJCA007860).
ACKNOWLEDGMENTS
This research was supported financially by the Science and Technology Project of Sichuan (23NSFSC1320, 23NSFSC4982, and 2022NSFSC1539) and Luzhou (2021-JYJ-73), the Technology Strategic Cooperation Project of Luzhou Municipal People’s Government-Southwest Medical University (2020LZXNYDJ38 and 2020LZXNYDJ23), and the Foundation of Southwest Medical University (2021ZKMS008, 2022QN042, 2022QN085, 2022QN102, and 2022QN118).
Footnotes
Supplemental material is available online only.
Contributor Information
Wenbi Chen, Email: cwb298@163.com.
Zhangyong Song, Email: szy83529@163.com.
Gustavo H. Goldman, Universidade de Sao Paulo
REFERENCES
- 1.McCarty TP, White CM, Pappas PG. 2021. Candidemia and invasive candidiasis. Infect Dis Clin North Am 35:389–413. doi: 10.1016/j.idc.2021.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Strickland AB, Shi M. 2021. Mechanisms of fungal dissemination. Cell Mol Life Sci 78:3219–3238. doi: 10.1007/s00018-020-03736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.d’Enfert C, Kaune AK, Alaban LR, Chakraborty S, Cole N, Delavy M, Kosmala D, Marsaux B, Frois-Martins R, Morelli M, Rosati D, Valentine M, Xie Z, Emritloll Y, Warn PA, Bequet F, Bougnoux ME, Bornes S, Gresnigt MS, Hube B, Jacobsen ID, Legrand M, Leibundgut-Landmann S, Manichanh C, Munro CA, Netea MG, Queiroz K, Roget K, Thomas V, Thoral C, Van den Abbeele P, Walker AW, Brown AJP. 2021. The impact of the fungus-host-microbiota interplay upon Candida albicans infections: current knowledge and new perspectives. FEMS Microbiol Rev 45:fuaa060. doi: 10.1093/femsre/fuaa060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Liu Y, Yang X, Li C, Song Z. 2022. The oral microbiota: community composition, influencing factors, pathogenesis, and interventions. Front Microbiol 13:895537. doi: 10.3389/fmicb.2022.895537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salehi M, Ahmadikia K, Mahmoudi S, Kalantari S, Jamalimoghadamsiahkali S, Izadi A, Kord M, Dehghan Manshadi SA, Seifi A, Ghiasvand F, Khajavirad N, Ebrahimi S, Koohfar A, Boekhout T, Khodavaisy S. 2020. Oropharyngeal candidiasis in hospitalised COVID-19 patients from Iran: species identification and antifungal susceptibility pattern. Mycoses 63:771–778. doi: 10.1111/myc.13137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moser D, Biere K, Han B, Hoerl M, Schelling G, Chouker A, Woehrle T. 2021. COVID-19 impairs immune response to Candida albicans. Front Immunol 12:640644. doi: 10.3389/fimmu.2021.640644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y, Puumala E, Robbins N, Cowen LE. 2021. Antifungal drug resistance: molecular mechanisms in Candida albicans and beyond. Chem Rev 121:3390–3411. doi: 10.1021/acs.chemrev.0c00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pristov KE, Ghannoum MA. 2019. Resistance of Candida to azoles and echinocandins worldwide. Clin Microbiol Infect 25:792–798. doi: 10.1016/j.cmi.2019.03.028. [DOI] [PubMed] [Google Scholar]
- 9.Sun S, Hoy MJ, Heitman J. 2020. Fungal pathogens. Curr Biol 30:R1163–R1169. doi: 10.1016/j.cub.2020.07.032. [DOI] [PubMed] [Google Scholar]
- 10.Perfect JR. 2017. The antifungal pipeline: a reality check. Nat Rev Drug Discov 16:603–616. doi: 10.1038/nrd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, Doig A, Guilliams T, Latimer J, McNamee C, Norris A, Sanseau P, Cavalla D, Pirmohamed M. 2019. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov 18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 12.Suwannasom N, Kao I, Pruß A, Georgieva R, Bäumler H. 2020. Riboflavin: the health benefits of a forgotten natural vitamin. Int J Mol Sci 21:950. doi: 10.3390/ijms21030950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thakur K, Tomar SK, Singh AK, Mandal S, Arora S. 2017. Riboflavin and health: a review of recent human research. Crit Rev Food Sci Nutr 57:3650–3660. doi: 10.1080/10408398.2016.1145104. [DOI] [PubMed] [Google Scholar]
- 14.Lei J, Xin C, Xiao W, Chen W, Song Z. 2021. The promise of endogenous and exogenous riboflavin in anti-infection. Virulence 12:2314–2326. doi: 10.1080/21505594.2021.1963909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halili F, Arboleda A, Durkee H, Taneja M, Miller D, Alawa KA, Aguilar MC, Amescua G, Flynn HW, Jr, Parel JM. 2016. Rose bengal- and riboflavin-mediated photodynamic therapy to inhibit methicillin-resistant Staphylococcus aureus keratitis isolates. Am J Ophthalmol 166:194–202. doi: 10.1016/j.ajo.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Banerjee S, Ghosh D, Vishakha K, Das S, Mondal S, Ganguli A. 2020. Photodynamic antimicrobial chemotherapy (PACT) using riboflavin inhibits the mono and dual species biofilm produced by antibiotic resistant Staphylococcus aureus and Escherichia coli. Photodiagnosis Photodyn Ther 32:102002. doi: 10.1016/j.pdpdt.2020.102002. [DOI] [PubMed] [Google Scholar]
- 17.Faddy HM, Fryk JJ, Watterson D, Young PR, Modhiran N, Muller DA, Keil SD, Goodrich RP, Marks DC. 2016. Riboflavin and ultraviolet light: impact on dengue virus infectivity. Vox Sang 111:235–241. doi: 10.1111/vox.12414. [DOI] [PubMed] [Google Scholar]
- 18.Mansjo M, Johansson J. 2011. The riboflavin analog roseoflavin targets an FMN-riboswitch and blocks Listeria monocytogenes growth, but also stimulates virulence gene-expression and infection. RNA Biol 8:674–680. doi: 10.4161/rna.8.4.15586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akompong T, Ghori N, Haldar K. 2000. In vitro activity of riboflavin against the human malaria parasite Plasmodium falciparum. Antimicrob Agents Chemother 44:88–96. doi: 10.1128/AAC.44.1.88-96.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aili A, Hasim A, Kelimu A, Guo X, Mamtimin B, Abudula A, Upur H. 2013. Association of the plasma and tissue riboflavin levels with C20orf54 expression in cervical lesions and its relationship to HPV16 infection. PLoS One 8:e79937. doi: 10.1371/journal.pone.0079937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matnuri M, Zheng C, Sidik D, Bai G, Abdukerim M, Abdukadier A, Ahmat K, Ma Y, Eli M. 2015. Correlation analysis of riboflavin, RFT2 and Helicobacter pylori in gastric carcinoma. Int J Clin Exp Pathol 8:13339–13345. [PMC free article] [PubMed] [Google Scholar]
- 22.Sun MG, Huang Y, Xu YH, Cao YX. 2017. Efficacy of vitamin B complex as an adjuvant therapy for the treatment of complicated vulvovaginal candidiasis: an in vivo and in vitro study. Biomed Pharmacother 88:770–777. doi: 10.1016/j.biopha.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Sudbery PE. 2011. Growth of Candida albicans hyphae. Nat Rev Microbiol 9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- 24.Yang W, Yan L, Wu C, Zhao X, Tang J. 2014. Fungal invasion of epithelial cells. Microbiol Res 169:803–810. doi: 10.1016/j.micres.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Nobile CJ, Johnson AD. 2015. Candida albicans biofilms and human disease. Annu Rev Microbiol 69:71–92. doi: 10.1146/annurev-micro-091014-104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandra J, Mukherjee PK. 2015. Candida biofilms: development, architecture, and resistance. Microbiol Spectr 3. doi: 10.1128/microbiolspec.MB-0020-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knafler HC, Smaczynska-de Rooij II, Walker LA, Lee KK, Gow NAR, Ayscough KR. 2019. AP-2-dependent endocytic recycling of the chitin synthase Chs3 regulates polarized growth in Candida albicans. mBio 10:e02421-18. doi: 10.1128/mBio.02421-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ibe C, Munro CA. 2021. Fungal cell wall proteins and signaling pathways form a cytoprotective network to combat stresses. J Fungi 7:739. doi: 10.3390/jof7090739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Degani G, Ragni E, Botias P, Ravasio D, Calderon J, Pianezzola E, Rodriguez-Pena JM, Vanoni MA, Arroyo J, Fonzi WA, Popolo L. 2016. Genomic and functional analyses unveil the response to hyphal wall stress in Candida albicans cells lacking beta(1,3)-glucan remodeling. BMC Genomics 17:482. doi: 10.1186/s12864-016-2853-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bemena LD, Min K, Konopka JB, Neiman AM. 2021. A conserved machinery underlies the synthesis of a chitosan layer in the Candida chlamydospore cell wall. mSphere 6:e00080-21. doi: 10.1128/mSphere.00080-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharya S, Esquivel BD, White TC. 2018. Overexpression or deletion of ergosterol biosynthesis genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae. mBio 9:e01291-18. doi: 10.1128/mBio.01291-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu Q, Zhang B, Li J, Zhang B, Wang H, Li M. 2016. Endoplasmic reticulum-derived reactive oxygen species (ROS) is involved in toxicity of cell wall stress to Candida albicans. Free Radic Biol Med 99:572–583. doi: 10.1016/j.freeradbiomed.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 33.Zorov DB, Juhaszova M, Sollott SJ. 2014. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev 94:909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandai D, Yin Z, Selway L, Stead D, Walker J, Leach MD, Bohovych I, Ene IV, Kastora S, Budge S, Munro CA, Odds FC, Gow NA, Brown AJ. 2012. The evolutionary rewiring of ubiquitination targets has reprogrammed the regulation of carbon assimilation in the pathogenic yeast Candida albicans. mBio 3:e00495-12. doi: 10.1128/mBio.00495-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lian J, Zhao H. 2016. Functional reconstitution of a pyruvate dehydrogenase in the cytosol of Saccharomyces cerevisiae through lipoylation machinery engineering. ACS Synth Biol 5:689–697. doi: 10.1021/acssynbio.6b00019. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, St Leger RJ, Fang W. 2017. Pyruvate accumulation is the first line of cell defense against heat stress in a fungus. mBio 8:e01284-17. doi: 10.1128/mBio.01284-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brar GA, Weissman JS. 2015. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol 16:651–664. doi: 10.1038/nrm4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng YL, Salim CK, Chu JJH. 2021. Drug repurposing for COVID-19: approaches, challenges and promising candidates. Pharmacol Ther 228:107930. doi: 10.1016/j.pharmthera.2021.107930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonne C, Alstrup AKO. 2019. Thousands of Danish children find ten new bacteria species. Nature 567:31. doi: 10.1038/d41586-019-00765-y. [DOI] [PubMed] [Google Scholar]
- 40.Jeffery-Smith A, Taori SK, Schelenz S, Jeffery K, Johnson EM, Borman A, Candida auris Incident Management Team, Manuel R, Brown CS. 2018. Candida auris: a review of the literature. Clin Microbiol Rev 31:e00029-17. doi: 10.1128/CMR.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sechi G, Sechi E, Fois C, Kumar N. 2016. Advances in clinical determinants and neurological manifestations of B vitamin deficiency in adults. Nutr Rev 74:281–300. doi: 10.1093/nutrit/nuv107. [DOI] [PubMed] [Google Scholar]
- 42.Elmadfa I, Meyer AL. 2019. The role of the status of selected micronutrients in shaping the immune function. Endocr Metab Immune Disord Drug Targets 19:1100–1115. doi: 10.2174/1871530319666190529101816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scaranti M, Cojocaru E, Banerjee S, Banerji U. 2020. Exploiting the folate receptor alpha in oncology. Nat Rev Clin Oncol 17:349–359. doi: 10.1038/s41571-020-0339-5. [DOI] [PubMed] [Google Scholar]
- 44.Capelli I, Cianciolo G, Gasperoni L, Zappulo F, Tondolo F, Cappuccilli M, La Manna G. 2019. Folic acid and vitamin B12 administration in CKD, why not? Nutrients 11:383. doi: 10.3390/nu11020383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Obi J, Pastores SM, Ramanathan LV, Yang J, Halpern NA. 2020. Treating sepsis with vitamin C, thiamine, and hydrocortisone: exploring the quest for the magic elixir. J Crit Care 57:231–239. doi: 10.1016/j.jcrc.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Batista KS, Cintra VM, Lucena PAF, Manhaes-de-Castro R, Toscano AE, Costa LP, Queiroz M, de Andrade SM, Guzman-Quevedo O, Aquino JS. 2022. The role of vitamin B12 in viral infections: a comprehensive review of its relationship with the muscle-gut-brain axis and implications for SARS-CoV-2 infection. Nutr Rev 80:561–578. doi: 10.1093/nutrit/nuab092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gessler NN, Aver’yanov AA, Belozerskaya TA. 2007. Reactive oxygen species in regulation of fungal development. Biochemistry (Mosc) 72:1091–1109. doi: 10.1134/s0006297907100070. [DOI] [PubMed] [Google Scholar]
- 48.Chen YT, Yang KX, Dai ZY, Yi H, Peng XX, Li H, Chen ZG. 2022. Repressed central carbon metabolism and its effect on related metabolic pathways in cefoperazone/sulbactam-resistant Pseudomonas aeruginosa. Front Microbiol 13:847634. doi: 10.3389/fmicb.2022.847634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolak N, Tomasi M, Kozik A, Rapala-Kozik M. 2015. Characterization of thiamine uptake and utilization in Candida spp. subjected to oxidative stress. Acta Biochim Pol 62:445–455. doi: 10.18388/abp.2015_1044. [DOI] [PubMed] [Google Scholar]
- 50.Han J, Gao P, Zhao S, Bie X, Lu Z, Zhang C, Lv F. 2017. iTRAQ-based proteomic analysis of LI-F type peptides produced by Paenibacillus polymyxa JSa-9 mode of action against Bacillus cereus. J Proteomics 150:130–140. doi: 10.1016/j.jprot.2016.08.019. [DOI] [PubMed] [Google Scholar]
- 51.Ye JZ, Su YB, Lin XM, Lai SS, Li WX, Ali F, Zheng J, Peng B. 2018. Alanine enhances aminoglycosides-induced ROS production as revealed by proteomic analysis. Front Microbiol 9:29. doi: 10.3389/fmicb.2018.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu C, Zhang X, Fang W. 2019. Increasing pyruvate concentration enhances conidial thermotolerance in the entomopathogenic fungus Metarhizium robertsii. Front Microbiol 10:519. doi: 10.3389/fmicb.2019.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ezraty B, Gennaris A, Barras F, Collet JF. 2017. Oxidative stress, protein damage and repair in bacteria. Nat Rev Microbiol 15:385–396. doi: 10.1038/nrmicro.2017.26. [DOI] [PubMed] [Google Scholar]
- 54.Demuyser L, Palmans I, Vandecruys P, Van Dijck P. 2020. Molecular elucidation of riboflavin production and regulation in Candida albicans, toward a novel antifungal drug target. mSphere 5:e00714-20. doi: 10.1128/mSphere.00714-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garfoot AL, Zemska O, Rappleye CA. 2014. Histoplasma capsulatum depends on de novo vitamin biosynthesis for intraphagosomal proliferation. Infect Immun 82:393–404. doi: 10.1128/IAI.00824-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meir Z, Osherov N. 2018. Vitamin biosynthesis as an antifungal target. J Fungi 4:72. doi: 10.3390/jof4020072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gu Y, Huang Y, Qiu Z, Xu Z, Li D, Chen L, Jiang J, Gao L. 2020. Vitamin B2 functionalized iron oxide nanozymes for mouth ulcer healing. Sci China Life Sci 63:68–79. doi: 10.1007/s11427-019-9590-6. [DOI] [PubMed] [Google Scholar]
- 58.Mal P, Dutta K, Bandyopadhyay D, Basu A, Khan R, Bishayi B. 2013. Azithromycin in combination with riboflavin decreases the severity of Staphylococcus aureus infection induced septic arthritis by modulating the production of free radicals and endogenous cytokines. Inflamm Res 62:259–273. doi: 10.1007/s00011-012-0574-z. [DOI] [PubMed] [Google Scholar]
- 59.Jiang Q, Wang F, Yang JY, Zhou G. 2022. MAIT cells and their implication in human oral diseases. Inflamm Res 71:1041–1054. doi: 10.1007/s00011-022-01600-3. [DOI] [PubMed] [Google Scholar]
- 60.Seyedjavadi SS, Khani S, Eslamifar A, Ajdary S, Goudarzi M, Halabian R, Akbari R, Zare-Zardini H, Imani FA, Amani J, Razzaghi-Abyaneh M. 2019. The antifungal peptide MCh-AMP1 derived from Matricaria chamomilla inhibits Candida albicans growth via inducing ROS generation and altering fungal cell membrane permeability. Front Microbiol 10:3150. doi: 10.3389/fmicb.2019.03150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shin DS, Eom YB. 2019. Zerumbone inhibits Candida albicans biofilm formation and hyphal growth. Can J Microbiol 65:713–721. doi: 10.1139/cjm-2019-0155. [DOI] [PubMed] [Google Scholar]
- 62.Del Rio M, de la Canal L, Pinedo M, Mora-Montes HM, Regente M. 2019. Effects of the binding of a Helianthus annuus lectin to Candida albicans cell wall on biofilm development and adhesion to host cells. Phytomedicine 58:152875. doi: 10.1016/j.phymed.2019.152875. [DOI] [PubMed] [Google Scholar]
- 63.Marchet C, Lecompte L, Da Silva C, Cruaud C, Aury J-M, Nicolas J, Peterlongo P. 2019. De novo clustering of long reads by gene from transcriptomics data. Nucleic Acids Research 47:e2. doi: 10.1093/nar/gky834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roselletti E, Sabbatini S, Ballet N, Perito S, Pericolini E, Blasi E, Mosci P, Cayzeele DA, Monari C, Vecchiarelli A. 2019. Saccharomyces cerevisiae CNCM I-3856 as a new therapeutic agent against oropharyngeal candidiasis. Front Microbiol 10:1469. doi: 10.3389/fmicb.2019.01469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen L, Qu S, Yang K, Liu M, Li YX, Keller NP, Zeng X, Tian J. 2020. Perillaldehyde: a promising antifungal agent to treat oropharyngeal candidiasis. Biochem Pharmacol 180:114201. doi: 10.1016/j.bcp.2020.114201. [DOI] [PubMed] [Google Scholar]
- 66.Kamai Y, Kubota M, Kamai Y, Hosokawa T, Fukuoka T, Filler SG. 2001. New model of oropharyngeal candidiasis in mice. Antimicrob Agents Chemother 45:3195–3197. doi: 10.1128/AAC.45.11.3195-3197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1-S4. Download spectrum.03801-22-s0001.pdf, PDF file, 0.5 MB (482.1KB, pdf)
Data Availability Statement
Sequence data were deposited in the Beijing Institute of Genomics Genome Sequence Archive (accession no. PRJCA007860).