Abstract
In critically colonised wounds, many of the signs of infection are often absent, and delayed healing may be the only clinical sign. The prevention of critical colonisation is important, but its pathophysiology has not yet been elucidated. We have previously reported that dysbiotic microbiota dissimilar to the peri‐wound skin microbiota may develop in critically colonised wounds. To investigate the role of dysbiotic microbiota, this study aimed to develop a critically colonised wound model by transplantation of dysbiotic microbiota. To transplant microbiota, a bacterial solution (dysbiosis group) or with Luria‐Bertani medium (commensal group) was inoculated to full‐thickness wounds of rats. The bacterial solution was prepared by anaerobically culturing bacteria from donor rats on an artificial dermis in Luria‐Bertani medium for 72 hours. As a result, the degree of the change in the microbial similarity between pre‐ and post‐transplantation of microbiota was significantly higher in the dysbiosis group (P < .001). No signs of infection were observed in any rat in either group. The wound area in the dysbiosis group was significantly larger (P < .001), and there was a significant infiltration of neutrophils (P < .001). All rats of the dysbiosis group represented the clinical features of critically colonised wounds. Furthermore, there were significantly fewer regulatory T cells in the wounds of the dysbiosis group. This is the first study to develop a novel animal model that represents the clinical features of critically colonised wounds and will be useful in investigating the pathogenesis of critical colonisation via regulatory T cells.
Keywords: 16S ribosomal RNA, chronic wounds, hard‐to‐heal wounds, microbiome, wound infection
1. INTRODUCTION
Critical colonisation, which is a transition state between bacterial colonisation and invasive wound infection, lacks the macroscopic sign of infection but causes delayed wound healing. 1 Biofilm on the wound surface is likely to cause this wound status, 2 and care to disrupt and remove the wound biofilm and prevent its re‐formation is recommended. 3 However, wounds are constantly exposed to environmental bacteria and commensal skin microbiota, 4 such care alone is not enough to prevent critical colonisation because keeping the wound sterile is difficult. On the other hand, the composition of the wound microbiota, which is formed by the dissemination of external bacteria, has been pointed out to be associated with the healing status. 5 In addition, in our previous study of the microbiota of patients with pressure injuries, we found that the microbiota of the critically colonised wounds was dissimilar to that of the peri‐wound skin. 6 This indicates that critical colonisation may be related to dysbiosis, the formation of microbiota in the wound that deviates from the patient's unique skin commensal microbiota. Thus, Interventions targeting the wound microbiota are necessary to prevent critical colonisation. However, the mechanism by which the dysbiosis wound microbiota induces excessive inflammation and consequently delays healing (ie, critical colonisation) has not been clarified.
To investigate the role of dysbiotic wound microbiota on the establishment of critical colonisation, in vivo animal models are required as microbiota cannot be artificially modulated in human wounds. Several animal models that mimic critically colonised wounds and bacteria‐induced delayed healing have been reported previously. 7 , 8 , 9 , 10 However, these models are associated with several limitations, including the use of a single pathogenic bacterial solution or lipopolysaccharide. Thus, these models cannot accurately simulate colonisation by dysbiotic microbiota, which consists of multiple bacteria. Additionally, interventions other than bacterial administration have also been used to delay healing, including inducing diabetes or placing a ring around the wound. However, these interventions make it difficult to investigate the sole effects of the microbiota. Therefore, it was necessary to develop a model using skin commensal microbiota to represent a critically colonised wound with dysbiotic microbiota.
Regulatory T cells (Tregs) may be involved in the delayed healing caused by dysbiotic wound microbiota. Tregs play a major role in maintaining immunological unresponsiveness to self‐antigens and commensal organisms and in suppressing excessive immune responses deleterious to the host. 11 Immediately after injury, the wound surface is sterile, but it is subsequently exposed to large amounts of bacteria through bacterial dissemination from the peri‐wound skin. In the intestines and skin of germ‐free mice, there was a greater accumulation of Tregs in the tissues than in those of specific pathogen‐free mice. 12 , 13 In addition, activated Tregs can accumulate in skin tissues, allowing the colonisation of commensal bacteria. 14 Contrarily, in previous studies, excessive inflammation was not suppressed and wound healing was delayed in Treg‐depleted mice compared with the control group. 15 Inflammation may be caused not only by the injury but also by colonisation of the wound surface by skin commensal bacteria. These results suggest that Tregs may be involved in the formation of commensal microbiota in wounds and prevent delayed healing. However, when the wound is exposed to dysbiotic microbiota, the role of Tregs is not clear. To investigate the role of the dysbiotic microbiota, this study first aimed to develop an in vivo animal critically colonised wound model with dysbiotic microbiota originating from skin commensal microbiota, and then investigate Treg in the wound.
2. MATERIALS AND METHODS
2.1. Animals
A total of 26 healthy 6‐month‐old male Sprague Dawley rats, weighing 500 to 600 g, were used in this study, with 6 of them serving as donor rats for dysbiotic microbiota transplantation. The remaining 20 rats were used as recipient rats and acclimatised for 7 days before removing hair under specific‐pathogen‐free conditions in the local animal facility. Rats were maintained under standard conditions with a temperature of 23 ± 2°C and humidity of 45 ± 10%, 12 hours/12 hours light/dark cycle, and ad libitum feeding and drinking. All animals were obtained from Japan SLC (Shizuoka, Japan). The study protocols were approved by the Animal Experimental Committee of the author's university. All animals were treated according to the Guide for the Care and Use of Laboratory Animals of the National Institute of Health (NIH).
2.2. Preparation of dysbiotic microbiota transplantation
Because necrotic tissue is associated with the relative abundance of anaerobes and alteration of wound microbiota, 16 , 17 , 18 we hypothesised that dysbiotic wound microbiota is formed by necrotic tissue. Necrotic tissue in pressure injuries is basically granulation tissue (ie, collagen) lacking viable cells and blood flow, and this inspired me to use an artificial dermis that is composed of collagen sponges. Prior to acclimatisation, 6 × 8 cm2 full‐thickness tissues were obtained from the dorsal skin of donor rats after hair removal using a shaver. The collected tissues were homogenised and the remaining skin fibres were removed using the Steriflip filter device (Millipore, Bedford, Massachusetts). The filtrate was centrifuged for 10 min at 1500g to pellet the bacteria. After removing the supernatant, the pellet was resuspended in phosphate‐buffered saline (PBS). The bacterial suspension was then added to the artificial dermis, cut into 1.6 cm2, and cultured anaerobically with Luria‐Bertani (LB) medium in a 3.5 cm diameter dish for 72 hours. All culture solutions and cultures were collected using a cell scraper and homogenised with PBS. After centrifugation, the bacterial pellet was washed with PBS and resuspended with LB medium to adjust the optical density (600 nm) to 1.0 (about 1.0 × 108 cells/mL).
2.3. Reproducibility of dysbiotic microbiota in the bacterial solution
To evaluate the reproducibility of the dysbiotic microbiota, the bacterial solution was prepared from each of the six donor rats and assigned to an ID from #1 to #6. Reproducibility was evaluated from four perspectives: bacterial count, predominant bacterial in the microbiota, alpha diversity (species richness within a single microbiota), and beta diversity (a similarity in the microbiota between different environments). The criteria were that the coefficient of variation of the logarithmically converted bacterial count was less than 0.05 and that the predominant bacteria were consistent in all bacterial solutions. The criterion for alpha diversity was that the coefficient of variation of the value of the index was less than 0.05. The criterion for beta diversity was that the mean value of the index was less than 0.3 because the beta diversity of skin microbiota among individual rats was approximately 0.3 in the pre‐tests.
2.4. Treatment
The requirements of the critical colonisation model in this study are as follows: (a) no signs of infection, (b) delayed healing, and (c) infiltration of neutrophils in response to bacterial invasion. 19 In addition, to eliminate the possibility that healing is delayed as a result of high bioburden, a model with a similar bacterial count in a wound that was colonised by non‐dysbiotic skin commensal bacteria was also developed for comparison (Figure 1).
FIGURE 1.
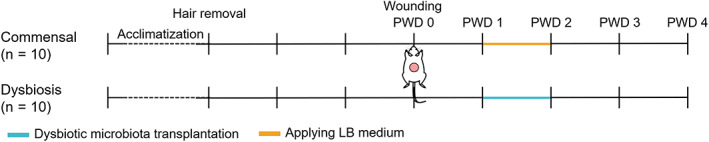
Experimental protocols and animal groups. LB medium, Luria‐Bertani medium; PWD, post‐wounding day
After 3 days of hair removal using a shaver and depilatory cream under anaesthesia (isoflurane inhalation solution, 2%, 0.2 L/min, Pfizer, Tokyo, Japan), a full‐thickness excisional 2.0 cm diameter wound was created on the dorsal skin of recipient rats. The center of the wounds was located on the posterior median line and between the middle of the greater trochanter and the axilla. The wounds were covered with Duoactive CGF (Conva Tec Japan Co. Ltd., Tokyo, Japan). The wounds were washed with sterile normal saline and the dressing changed daily until tissue sampling.
A piece of gauze (1.5 cm2) was incubated with a 2 mL bacterial solution of dysbiotic microbiota in a dish with a diameter of 3.5 cm at 37°C for 1.5 hours for the dysbiosis group (n = 10). In the commensal group (n = 10), an equal amount of LB medium was used instead of the bacterial solution, representing the dissemination of skin commensal microbiota to the wound and its colonisation. The incubated gauze was applied onto the wound bed on the post‐wounding day (PWD) 1, after wound washing, to transplant the microbiota, and removed on PWD 2.
2.5. Sample collection
Images of each wound were recorded using a digital camera (RX100II, Sony Corporation, Tokyo, Japan) and obtained next to a colour chart (CASMATCH, Bear Medic Co., Tokyo, Japan). Wound samples were collected daily before wound washing, PWD 1 to 4, from the wound bed using the Levine technique. 20 Skin samples were collected from the surface of a 3 × 3 cm square area, by swabbing twice with the Z stroke technique on PWD 1. 21 All samples were obtained using a flock swab (Puritan) soaked in saline with 0.1% Tween‐20 and stored at −80°C until DNA extraction. Extracted DNA was used for bacterial count of the wound surface and for detecting microbiota composition.
The rats were sacrificed using CO2 gas on PWD 4. Tree specimens were taken from each wound site using surgical instruments: a specimen for histological examination, gene expression analysis, and bacterial count. One of the specimens was immersed in 10% formalin (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) for 2 days. Another was stored in RNAlater (Sigma‐Aldrich Co.) until RNA extraction. The other was immediately taken to the nitrogen tank, frozen, and stored at −80°C until bacterial DNA extraction. Extracted DNA was used for the bacterial count in the tissue.
2.6. Macroscopic observations
The wound area was measured with image analysis software (ImageJ version 1.8.0, NIH, Bethesda, Maryland) and calculated relative to the wound area of PWD 1. To quantify the degree of redness around the wound site (within 2 mm of the wound edge), the erythema index was quantified, based on digital image analysis for colour brightness. 22
2.7. Next‐generation DNA sequencing
In this experiment, full‐length 16S rRNA gene amplicon sequencing was used to identify bacteria at the species level and to represent the composition of the microbiota more precisely. Bacterial DNA was extracted with the QIAamp DNA Mini Kit (Qiagen N.V., Venlo, Netherlands) as previously described. 4 10 ng of DNA from each sample were used in polymerase chain reaction (PCR) with 16S primers 27 F and 1492 R included in the 16S Barcoding Kit 1‐24 (SQK‐16S024; Oxford Nanopore Technologies, Oxford) for amplification of the near full‐length bacterial 16S rRNA gene. Sequencing was performed using the flow cell (FLO‐MIN106D; Oxford Nanopore Technologies) on the MinION portable sequencer (Oxford Nanopore Technologies). Basecalling was conducted on the online EPI2ME platform (Oxford Nanopore Technologies) and diversity analysis was performed using Qiime. 23 For the analysis of alpha diversity and the evaluation of reproducibility, the samples were rarefied at 21 000 depths (minimum read number among all samples) to compare the diversity under the same conditions followed by a calculation of the phylogenetic diversity index. The alpha diversity index was measured 10 times. Beta diversity was calculated using the weighted UniFrac dissimilarity index. To evaluate the reproducibility of dysbiotic microbiota, the values of all pairs from #1 to #6 were calculated. To evaluate the in vivo model, the value of beta diversity index between the skin microbiota collected on PWD 1 and the wound microbiota at each time point was investigated. The difference in the values between PWD 1 and 2, which is before and after microbiota transplantation, was calculated.
2.8. Histological analysis
The wound samples preserved in formalin were embedded in paraffin blocks. Subsequently, serial sections (3 μm thick) were cut for Haematoxylin and eosin (HE) staining (Muto Pure Chemicals, Tokyo, Japan) to evaluate the granulation tissue and necrotic tissue. The tissue samples were also stained by immunohistochemistry for myeloperoxidase (MPO), a marker of neutrophils, and Forkhead box protein P3 (FOXP3), a marker of Tregs. For evaluation of neutrophil infiltration, the tissue sections were incubated with primary antibody (anti‐MPO heavy chain antibody, 1:50 dilution, Santa Cruz Biotechnology, Dallas, Texas) overnight at 4°C after inactivation of endogenous peroxidase and blocking non‐specific staining. Next, the sections were incubated with a secondary antibody labelled with horseradish peroxidase (Histofine Simple Stain Rat MAX PO [G], Nichirei Biosciences, Tokyo, Japan) for 30 min at room temperature. Tissue examination was performed based on the presence of neutrophils in the center of the granulation tissue of each section (rating scale: 0 = few; 1 = occasional; 2 = mild; 3 = moderate; 4 = abundant). To detect Tregs in the tissue, an anti‐FOXP3 antibody (1:500, Abcam plc, Cambridge) was used. The number of positive cells identified within a single field of view was counted using a ×20 image of the granulation tissue of the stained tissue, and the mean of five times was calculated. 3,3′‐Diaminobenzidine (FUJIFILM Wako Pure Chemical Corporation) was used for visualisation of both, MPO and FOXP3. Haematoxylin was used for counterstaining.
2.9. Bacterial count
Bacterial count of the bacterial solution, swab, and tissue was measured using the culture‐independent targeted gene real‐time PCR with Bacteria (tuf gene) Quantitative PCR Kit (Takara Bio Inc., Shiga, Japan). 24 The reaction was performed with 5 μL of template DNA in a total volume of 25 μL, according to the manufacturer's protocol (Takara Bio). All amplifications were run on the Mx3000P QPCR System (Agilent Technologies, Santa Clara, California) and performed in triplicate.
2.10. Real‐time reverse transcription (RT)‐PCR
Total RNA was extracted from wound tissue stored in RNAlater. RNA extraction was performed using the TANBead® Nucleic Acis Extraction Kit according to the manufacturer's protocol (Taiwan Advanced Nanotech Inc., Taoyuan, Taiwan). cDNA synthesis was performed using the MJ Mini™ Thermal Cycler (Bio‐Rad, Hercules, California) and the High‐Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific, Waltham, Massachusetts). The real‐time PCR analysis was performed in the Mx3000P QPCR System (Agilent Technologies), using TaqMan Master Mix and the following TaqMan primers and probes (Applied Biosystems™): IL10 (Rn00563409_m1), Ctla4 (Rn01437152_m1), Garp (Rn01510975_m1), and Foxp3 (Rn01525092_m1). 18S ribosomal RNA gene was used as an internal control to normalise the samples. Measurements were performed in triplicate, and samples for which the threshold cycle (Ct) values were not obtained in more than two out of three reactions were considered to have no detectable gene expression. If the gene was detected in all rats of all groups, and the expression level of the target gene was calculated according to the −ΔCt formula. 25
2.11. Statistical analysis
All data were analysed with STATA/SE 15.0 (StataCorp LP, College Station, Texas) and presented as median and interquartile range (IQR). Significance was determined at P < .05. The Wilcoxon rank‐sum test was used to determine whether there are differences in the medians of the two groups.
3. RESULTS
3.1. Reproducibility of dysbiotic microbiota in the bacterial solution
The median bacterial count in the bacterial solution was 2.37 × 109 copy/g (IQR: 1.45‐3.62 × 109 Figure 2A). The coefficient of variation of the logarithmically converted bacterial count was 0.04. In all samples, Enterococcus faecalis was the predominant bacterium in the microbiota (Figure 2B). The median value of the phylogenetic diversity index was 1168.8 (IQR: 1132.5‐1197.2, Figure 2C), and the coefficient of variation was 0.047. The median value of the weighted UniFrac dissimilarity index for all pairs was 0.200 (IQR: 0.192‐0.201, Figure 2D).
FIGURE 2.
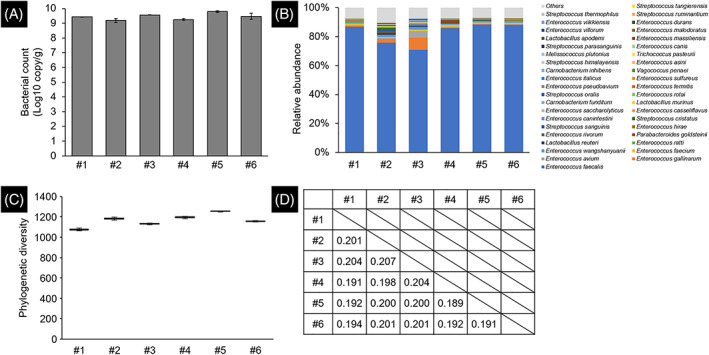
Reproducibility of dysbiotic microbiota in the bacterial solution. (A) Bacterial count was estimated from the copy number of tuf gene. The error bar represents the SE. (B) Microbial composition was shown using the relative abundance of bacteria classified at the species level (top 40). (C) Alpha diversity was evaluated using the Phylogenetic Diversity index. The error bar represents the SE. (D) Beta diversity was evaluated using the weighted UniFrac dissimilarity index
3.2. Requirements of in vivo models
Microbiotas of the skin on PWD 1 and wounds on PWD 1 to 4 were identified (Figure 3A). In both the commensal and dysbiosis groups, the predominant bacteria in the skin and wounds were different among rats. The difference in the weighted UniFrac index value between PWD 1 and PWD 2 in the dysbiosis group was significantly higher compared with the commensal group (P < .001, Figure 3B).
FIGURE 3.
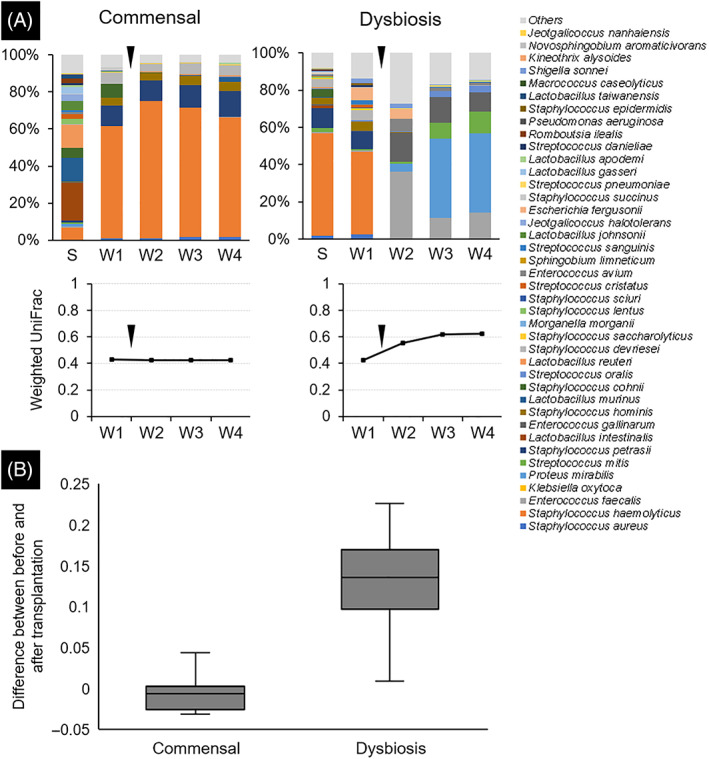
Composition and diversity of the microbiota. Swab samples collected from skin and wounds were used for the 16S rRNA gene analysis to identify the microbiota. (A) One case from each of the commensal and dysbiosis groups was presented. The bar graph shows the composition of the microbiota at each site. The line graph shows the temporal change of microbial similarity between the skin sample collected on post‐wounding day 1 and the wound sample at each time point. Black triangles indicated the time point of microbiota transplantation. (B) The difference in the values of the weighted UniFrac dissimilarity index between the post‐wounding day 1 and 2 values (before and after microbiota transplantation) was calculated. S, skin samples collected on PWD1; W, wound samples
On PWD 0, the median body weights of rats in the commensal and dysbiosis groups were 551 g (IQR: 520‐574) and 543.5 g (IQR: 533‐573), respectively, with no significant differences (P = .71). No animals died during the experiment.
In the commensal group, the wound bed was bright red and the formation of granulation tissue was observed on PWD 4. In the dysbiosis group, slough formation in the wound beds began on PWD 2 and gradually spread to cover the wound beds (Figure 4A). No signs of infection were observed in any rat in either group. No significant differences in the erythema index value on PWD 4 were detected between the groups (P = .60, Figure 4B). The mean wound area in the dysbiosis group was significantly larger than that in the commensal group (PWD 2, P = .01; PWD 3, P < .001; PWD 4, P < .001) (Figure 4C).
FIGURE 4.
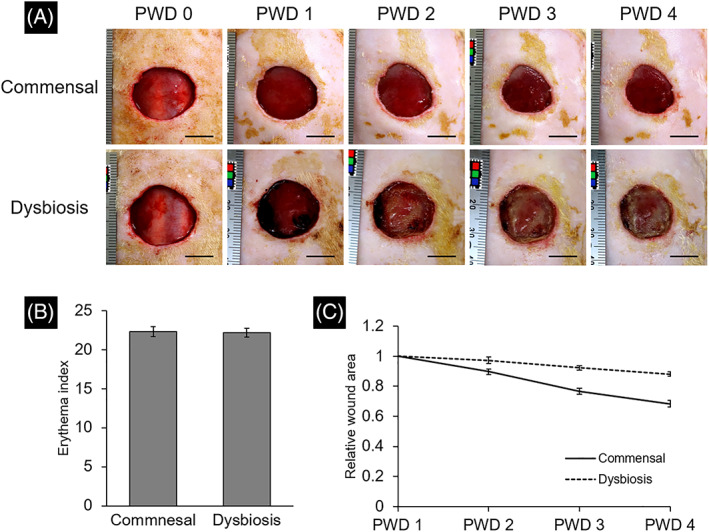
Alpha diversity of wound and peri‐wound skin microbiota. (A) The wound appearance was recorded daily. The scale bar indicates 1 cm in the photograph. (B) The erythema index was quantified, based on the digital images taken on post‐wounding day 4. Results are expressed as the mean ± SE. (C) Temporal change in relative wound area adjusted by the value of post‐wounding day 1 was shown. Results are expressed as mean ± SE. PWD, post‐wounding day
Histological analysis of tissues collected on PWD 4 showed the proliferation of granulation tissue, and a small amount of necrotic tissue was formed on the wound surface in all samples of the commensal group (Figure 5A,B). Neutrophil infiltration was also observed in the granulation tissue (Figure 6A). In contrast, the necrotic tissue was thicker, and the collagen fibres were thinner and looser in the dysbiosis group. Thrombosis was observed in the granulation tissue (Figure 5C,D). Neutrophils infiltrated the entire granulation tissue (Figure 6B). There was a significant difference in the level of neutrophil infiltration between the commensal and dysbiosis groups (P < .001, Figure 6C). Temporal change of bacterial count on the wound surface was measured using bacterial DNA extracted from swabs (Figure 6D). There was no significant difference in the bacterial counts at PWD 1, 3, and 4 (P = .34, .29, and .94, respectively), but there was a significant difference at PWD 2 (P < .01). The median bacterial count in the tissue on PWD 4 in the commensal and dysbiosis groups was 1.19 × 107 copy/g (IQR: 0.42‐1.99 × 107) and 3.82 × 107 copy/g (IQR: 1.13‐5.96 × 107), respectively. No significant differences were observed (P = .07, Figure 6E).
FIGURE 5.
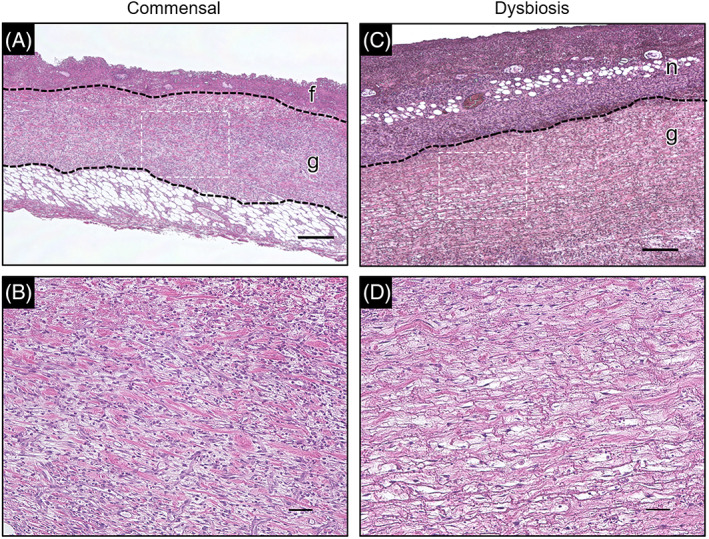
Haematoxylin and eosin staining of the wound tissue. Pathophysiological analysis was performed using haematoxylin and eosin staining of paraffin sections prepared from post‐wounding day 4 wound tissue. (A,C) Lower magnification images of the commensal and dysbiosis groups. Regions enclosed in the dotted line are at higher magnification (B,D). (f,g,n) The fibrin membrane, granulation tissue, and necrotic tissue, respectively. Magnification: ×4.0. Scale bar = 300 μm. (B,D) Higher magnification images of the granulation tissue. Magnification: ×20. Scale bar = 50 μm
FIGURE 6.
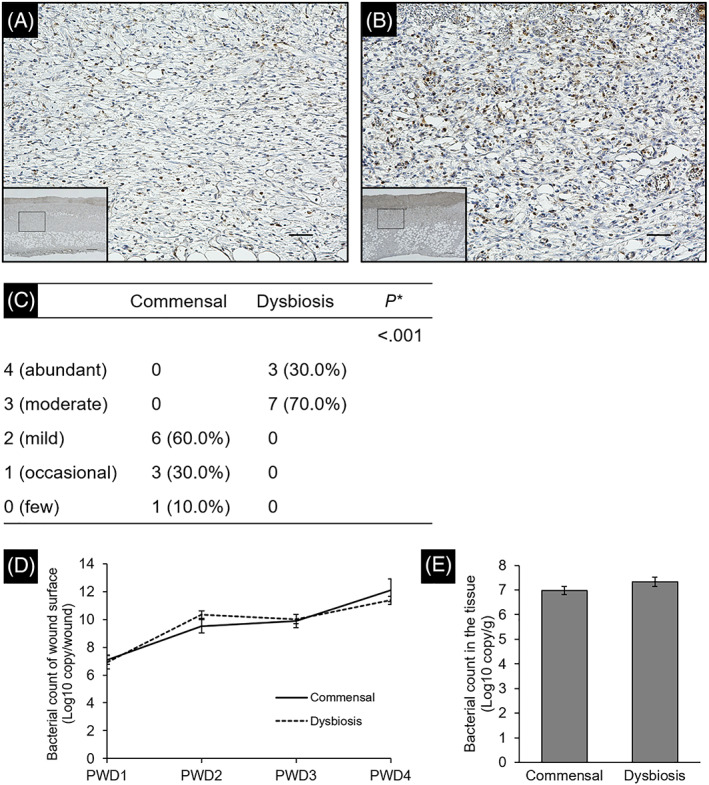
Evaluation of wound inflammation based on the infiltration of neutrophils. Wound tissue was stained with anti‐myeloperoxidase heavy chain antibody to confirm infiltration of neutrophils. The images of granulation tissue in the commensal group (A) and dysbiosis group (B) were shown. Region enclosed in solid box in lower magnification (×4.0, Scale bar = 300 μm) is at higher magnification images (×20, Scale bar = 50 μm). (C) Data show the number of rats assigned to each class. *Wilcoxon rank sum test. (D,E) The bacterial count of the wound surface and wound tissue was determined based on the copy number of tuf gene. Each error bar represents the SE
3.3. Characteristics of the in vivo models
Localization of Tregs was evaluated by immunohistochemical staining using an anti‐FOXP3 antibody. FOXP3‐positive cells were identified in the granulation tissues of both groups (Figure 7A,B). The median number of Tregs in the commensal and dysbiosis groups was 5.5 (IQR: 5.0‐6.8) and 1.0 (IQR: 0.6‐1.4), respectively, with significant differences (P < .001). Gene expression analysis using RNA extracted from wound tissues showed that the expression of Foxp3 and Ctla4 were significantly lower in the dysbiosis group than in the commensal group (both P = .01, Figure 7C). There were no significant differences in the expression of IL10 and Garp between the two groups (P = .82 and P = .17, respectively).
FIGURE 7.
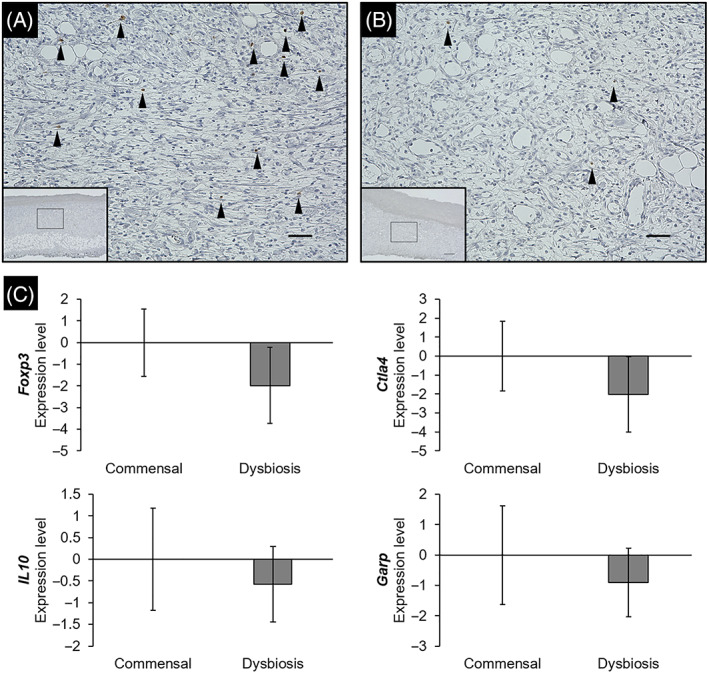
Localization of FOXP3 in tissues and expression level for Treg‐related genes. Wound tissue was stained with an anti‐FOXP3 antibody to confirm the localization of regulatory T cells. The images of granulation tissue in the commensal group (A) and dysbiosis group (B) were shown. Region enclosed in solid box in lower magnification (×4.0, Scale bar = 300 μm) is at higher magnification images (×20, Scale bar = 50 μm). Black arrows: regulatory T cells. (C) The expression level was quantified by a real‐time reverse transcription‐polymerase chain reaction. Each error bar represents the SE. FOXP3, Forkhead box P3.
4. DISCUSSION
This was the first study to develop an in vivo animal critically colonised wound model by transplantation of dysbiotic microbiota originating from skin commensal microbiota. In this model, wounds transplanted with dysbiotic microbiota showed delayed healing and neutrophil infiltration in response to bacterial invasion without signs of infection, similar to critically colonised wounds in clinical settings. In addition, there were significantly fewer Tregs in the granulation tissue of the dysbiotic microbiota‐transplanted wounds.
Wound healing process in the dysbiosis group was significantly delayed compared with that of the commensal group, even although the bacterial count of the tissue was similar. Regarding bacterial counts on the wound surface, there was a significant difference in PWD 2. However, swabs were collected before wound washing in this study; thus, the data at PWD 2 included not only the colonising bacteria but also the floating bacteria in the bacterial solution adhering to the wound surface. In addition, there was no significant difference in bacterial counts after PWD 3, suggesting that there was no clinically relevant difference in the bacterial counts that colonised after microbiota transplantation. Thus, delayed healing was likely caused by dysbiosis rather than by high bacterial counts. Besides, there were no signs of infection in any of the wounds, but the extent of neutrophil infiltration was significantly higher in the dysbiosis group than in the commensal group. These results indicate that the wounds in the dysbiosis group were consistent with the features of critically colonised clinical wounds. Furthermore, the temporal change in the wound area was consistent with the critically colonised wound model reported in a previous study. 7 Therefore, this study successfully developed a highly repeatable model of critically colonised wounds. In addition, there were no adverse events or deaths in any of the rats during experiments, and the clinical features were reproducible.
The microbiota of each bacterial solution also showed high reproducibility, although the skin commensal bacteria were collected from different donor rats. To prepare the bacterial solution, bacteria were collected from the donor rats without an acclimatisation period. This was to prevent contamination with pathogenic bacteria during the acclimatisation period, as bacterial contamination has been reported from the housing environment. 26 This was necessary consideration in order to maintain high reproducibility. In addition, the microbiota in the bacterial solution in this study was dominated by E. faecalis. In a previous study, a single inoculation of 2.8 × 106 CFU/wound of E. faecalis OG1RF caused severe wound infections. 27 Therefore, to mimic critical colonisation, a single inoculation with pathogenic E. faecalis is inappropriate, and it may be important to use skin commensal microbiota.
In the dysbiosis group, the number of Tregs in the wound tissue was significantly lower than in the commensal group. These results may indicate that Tregs were induced in the wounds of commensal microbiota, but not when the wounds were exposed to a dysbiotic microbiota. Therefore, to clarify the relationship between Tregs and wound microbiota, it should be confirmed whether excessive inflammation occurs and healing is delayed in wounds of commensal and dysbiotic microbiota when Tregs are suppressed. Further studies are called to confirm whether the induction of Treg suppression leads to delayed healing because inflammation is not inhibited in response to the colonisation of skin commensal bacteria in the wound.
In this study, temporal changes in wound microbiota were not uniform. In recent years, faecal microbiota transplantation has been widely used to correct the intestinal microbiota, and many reports recommend administering antibiotics before transplantation to facilitate the colonisation of microbiota from the donor. 28 , 29 In contrast, this study performed dysbiotic microbiota transplantation without antibiotic treatment because complete absence of wound bacteria is not possible. As a result, the colonisation of the transplanted bacteria may have been inhibited by any bacteria that colonised before transplantation. However, in the dysbiosis group, the microbial similarity between the wound and skin was significantly decreased after transplantation, and wound healing was delayed. It is thought that sufficient changes in microbiota can be induced without antibiotic treatment. In addition, even when the microbiota changed immediately after transplantation, healing was delayed. This result suggests that maintaining dysbiotic microbiota is not important for delayed healing.
In this model, microbiota transplantation was performed by applying gauze incubated with a bacterial solution to the wound. Inoculation with the bacterial solution was used to create a critically colonised wound model as carried out in a previous study. 7 However, this method inhibits the natural process of microbiota formation, and inoculation with a needle can cause tissue damage. In contrast, no tissue damage was reported in another model in which wound infection occurred using gauze application. 30 Therefore, the transplantation method used in this model is suitable for representing clinical cases. In addition, this model does not require artificial pretreatments, such as induction of diabetes or immunosuppression; therefore, it is suitable for pathological analysis.
The limitation in this study was that the number of cells was not counted using fluorescence‐activated cell sorting on wound tissue specimens as a result of the small number of Tregs in the tissues. Instead, immunohistochemistry was used to detect Tregs in the tissues and to examine the differences in Treg localization between the groups.
In conclusion, we developed an in vivo animal model that represents a critically colonised wound by transplanting dysbiotic microbiota into the wound. This model showed increased inflammation and delayed healing than the model in which the wound microbiota was formed through only the dissemination of skin commensal microbiota. Furthermore, there were significantly fewer Tregs in the wounds of the dysbiosis group. Therefore, experiments on Treg suppression using this model should be conducted to confirm whether excessive inflammation occurs and healing is delayed in wounds of commensal and dysbiotic microbiota.
CONFLICT OF INTEREST
Takeo Minematsu and Sofoklis Koudounas belong to the Department of Skincare Science, which receives financial support from Saraya Co., Ltd.
ACKNOWLEDGEMENTS
We would like to thank Editage (http://www.editage.com) for editing and reviewing this manuscript in the English language. We are grateful to Sanai Tomida for excellent technical assistance. All persons designated as authors qualify for authorship. Each author has participated sufficiently in the work and takes public responsibility for the content.
Kunimitsu M, Nakagami G, Minematsu T, Koudounas S, Sanada H. An in vivo critically colonised wound model with dysbiotic wound microbiota. Int Wound J. 2023;20(3):648‐658. doi: 10.1111/iwj.13906
Funding information Japan Society for the Promotion of Science, Grant/Award Numbers: JP20H04010, JP20J11635; Japanese Society of Pressure Ulcers; Leave a Nest Co., Ltd.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Edwards R, Harding KG. Bacteria and wound healing. Curr Opin Infect Dis. 2004;17(2):91‐96. [DOI] [PubMed] [Google Scholar]
- 2. James GA, Swogger E, Wolcott R, et al. Biofilms in chronic wounds. Wound Repair Regen. 2008;16(1):37‐44. [DOI] [PubMed] [Google Scholar]
- 3. Murphy C, Atkin L, Dissemond J, et al. Defying hard‐to‐heal wounds with an early antibiofilm intervention strategy: ‘wound hygiene’. J Wound Care. 2019;28(12):818‐822. [DOI] [PubMed] [Google Scholar]
- 4. Kunimitsu M, Nakagami G, Kitamura A, et al. Dissemination of microbiota between wounds and the beds of patients with pressure injuries: a cross‐sectional study. Wound Pract Res. 2021;29(2):70‐76. [Google Scholar]
- 5. Xu Z, Hsia HC. The impact of microbial communities on wound healing: a review. Ann Plast Surg. 2018;81(1):113‐123. [DOI] [PubMed] [Google Scholar]
- 6. Kunimitsu M, Nakagami G, Kitamura A, et al. Relationship between healing status and microbial dissimilarity in wound and peri‐wound skin in pressure injuries: a prospective cohort study. under review. 2022. [DOI] [PubMed]
- 7. Ueda K, Akase T, Nakagami G, et al. A possible animal model for critical colonisation. J Wound Care. 2010;19(7):295‐300. [PubMed] [Google Scholar]
- 8. Zhao G, Hochwalt PC, Usui ML, et al. Delayed wound healing in diabetic (db/db) mice with Pseudomonas aeruginosa biofilm challenge: a model for the study of chronic wounds. Wound Repair Regen. 2010;18(5):467‐477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crompton R, Williams H, Ansell D, et al. Oestrogen promotes healing in a bacterial LPS model of delayed cutaneous wound repair. Lab Invest. 2016;96(4):439‐449. [DOI] [PubMed] [Google Scholar]
- 10. Brandenburg KS, Calderon DF, Kierski PR, Czuprynski CJ, Mcanulty JF. Microbial pathogenesis novel murine model for delayed wound healing using a biological wound dressing with Pseudomonas aeruginosa bio fi lms. Microb Pathogenes. 2018;122(March):30‐38. [DOI] [PubMed] [Google Scholar]
- 11. Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133(5):775‐787. [DOI] [PubMed] [Google Scholar]
- 12. Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous clostridium species. Science. 2011;331(6015):337‐341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Naik S, Bouladoux N, Wilhelm C, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337(6098):1115‐1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Scharschmidt TC, Vasquez KS, Truong HA, et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity. 2015;43(5):1011‐1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nosbaum A, Prevel N, Truong HA, et al. Cutting edge: regulatory T cells facilitate cutaneous wound healing. J Immunol. 2016;196(5):2010‐2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Malone M, Johani K, Jensen SO, et al. Next generation DNA sequencing of tissues from infected diabetic foot ulcers. EBioMedicine. 2017;21:142‐149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Verbanic S, Shen Y, Lee J, Deacon JM, Chen IA. Microbial predictors of healing and short‐term effect of debridement on the microbiome of chronic wounds. NPJ Biofilms Microbiomes. 2020;6(21):1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kalan L, Meisel J, Loesche MA, et al. Strain‐ and species‐level variation in the microbiome of diabetic wounds is associated with clinical outcomes and therapeutic efficacy. Cell Host Microbe. 2019;25(5):641‐655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. White RJ, Cutting KF. Critical colonization—the concept under scrutiny. Ostomy Wound Manag. 2006;52(11):50‐56. [PubMed] [Google Scholar]
- 20. Levine NS, Lindberg RB, Mason AD, Pruitt BA. The quantitative swab culture and smear: a quick, simple method for determining the number of viable aerobic bacteria on open wounds. J Trauma. 1976;16(2):89‐94. [PubMed] [Google Scholar]
- 21. Rushing J. Obtaining a wound culture specimen. Nursing. 2007;37(11):18. [DOI] [PubMed] [Google Scholar]
- 22. Asada M, Nakagami G, Minematsu T, et al. Novel models for bacterial colonization and infection of full‐thickness wounds in rats. Wound Repair Regen. 2012;20(4):601‐610. [DOI] [PubMed] [Google Scholar]
- 23. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high‐throughput community sequencing data. Nat Methods. 2010;7(5):335‐336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tanaka Y, Takahashi H, Simidu U, Kimura B. Design of a new universal real‐time PCR system targeting the tuf gene for the enumeration of bacterial counts in food. J Food Prot. 2010;73(4):670‐679. [DOI] [PubMed] [Google Scholar]
- 25. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402‐408. [DOI] [PubMed] [Google Scholar]
- 26. Pritzkow S, Morales R, Lyon A, Concha‐Marambio L, Urayama A, Soto C. Efficient prion disease transmission through common environmental materials. J Biol Chem. 2008;293(9):3363‐3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chong KKL, Tay WH, Janela B, et al. Enterococcus faecalis modulates immune activation and slows healing during wound infection. J Infect Dis. 2017;216(12):1644‐1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fang H, Fu L, Wang J. Protocol for fecal microbiota transplantation in inflammatory bowel disease: a systematic review and meta‐analysis. Biomed Res Int. 2018;2018:8941340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kassam Z, Lee CH, Yuan Y, Hunt RH. Fecal microbiota transplantation for clostridium difficile infection: systematic review and meta‐analysis. Am J Gastroenterol. 2013;108(4):500‐508. [DOI] [PubMed] [Google Scholar]
- 30. Kitamura A, Nakagami G, Minematsu T, Kunimitsu M, Sanada H. Establishment of an infected wound model in rats with a natural route of bacterial application. J Nurs Sci Eng. 2021;9:1‐8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


