Abstract
RNA modification is manifested as chemically altered nucleotides, widely exists in diverse natural RNAs, and is closely related to RNA structure and function. Currently, mRNA-based vaccines have received great attention and rapid development as novel and mighty fighters against various diseases including cancer. The achievement of RNA vaccines in clinical application is largely attributed to some methodological innovations including the incorporation of modified nucleotides into the synthetic RNA. The selection of optimal RNA modifications aimed at reducing the instability and immunogenicity of RNA molecules is a very critical task to improve the efficacy and safety of mRNA vaccines. This review summarizes the functions of RNA modifications and their application in mRNA vaccines, highlights recent advances of mRNA vaccines in cancer immunotherapy, and provides perspectives for future development of mRNA vaccines in the context of personalized tumor therapy.
Keywords: RNA modification, RNA methylation, mRNA cancer vaccine, Immunogenicity, Immunotherapy, Combination therapy
Introduction
Vaccines serve as highly specialized biological agents that can provide active adaptive immunity to specific infectious diseases [1]. Vaccination has a long history for humans. In the 1790s, a rural doctor named Edward Jenner made a great discovery by the utilization of the cowpox virus preparation from a milkmaid as a vaccine against smallpox [2]. Subsequent study suggested that efficacious vaccines may work in two ways: One is to induce an immune mechanism for prophylactically preventing potential infection, and the other is to provide a treatment when infection has already occurred [3]. A milestone technology for vaccine development is the mRNA vaccine, which takes full advantage of the advancement in the fields of molecular biology and immunology and has been regarded as a form of gene therapy to some extent [4].
Back in the early 1990s, many studies attempted to apply mRNA to new therapeutics [5]. It was not until 2005 that a team of researchers at the University of Pennsylvania pioneered mRNA technology when they demonstrated that the activation of certain immune cells could be ablated through modifying RNA with 5-methylcytosine (m5C), N6-methyladenosine (m6A), 5-methyluridine (m5U), 2-thiouridine (s2U), or pseudouridine (Ψ) [6], laying a solid foundation for future mRNA-based therapies.
Currently, RNA modifications have become a critical and indispensable factor for designing and developing new and highly efficient mRNA vaccines. With the global pandemic of coronavirus disease 2019 (COVID-19), many pharmaceutical companies, such as Moderna, Pfizer/BioNTech, CureVac and Arctrus, developed mRNA-based COVID-19 vaccine candidates [5], with or without RNA modification technology. Notably, the vaccines produced by Moderna and Pfizer/BioNTech both contained the modified base (N1-methylpseudouridine, m1Ψ) and achieved a protective efficacy greater than 90% [7–9]. By contrast, the unmodified CureVac vaccine showed only 47% protection against coronavirus infection [9, 10]. These clinical observations reflected in part the contribution of RNA modifications to the protective efficacy of mRNA vaccines.
Immunotherapy is one of the most promising strategies for cancer treatment. As a representative approach to cancer immunotherapy, tumor vaccines have attracted increasing attention in recent years [11]. Functionally they can be divided into two categories: preventive and therapeutic vaccines [11]. The former is used in healthy individuals to induce immune memory and thereby prevent morbidity from certain cancers [11]. The latter is used for disease management by strengthening or reactivating the tumor patient's own immune system [11]. It is worth noting that mRNA vaccines have become a popular form of cancer vaccine because they provide antigen delivery and innate immune activation-mediated co-stimulation in a spatiotemporally aligned manner [12]. At present, mRNA vaccines targeting a variety of tumors are in clinical trials, including renal cell carcinoma, brain tumor, melanoma, prostate cancer, lung cancer, gastrointestinal tumor and AML. Compelling evidence shows that RNA modifications, such as m5C and Ψ, play an important role in the development of tumor vaccines [13–15]. In theory, numerous modifications that affect RNA structure and function could be used in cancer vaccines (Fig. 1), but few have actually been used. Therefore, exploring more RNA modifications aimed at improving the vaccine efficacy and safety is a meaningful task in future development of tumor vaccines. In this review, we describe the functions of RNA modifications and their application in mRNA vaccines, emphasize recent advances of mRNA vaccines in cancer immunotherapy, and look ahead to the future prospects of RNA modification in the development of novel mRNA cancer vaccines.
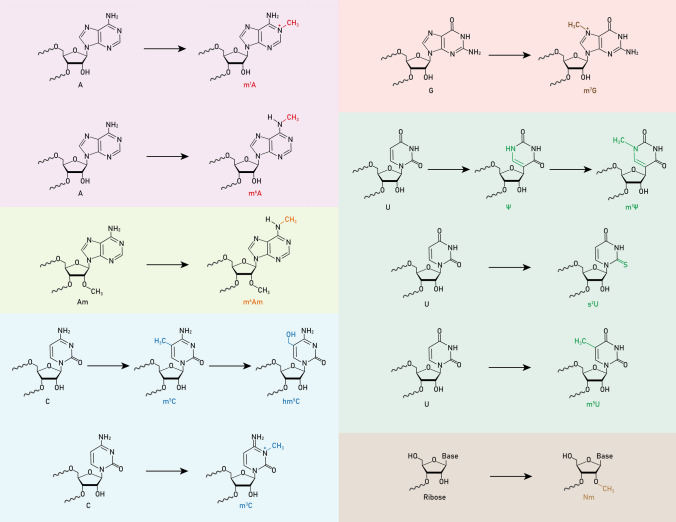
Fig. 1.
Chemical structures of common RNA modifications
Principles of developing mRNA vaccine
The interdisciplinary combination of techniques derived from molecular biology and immunology has greatly facilitated the researchers’ ability to design and produce mRNA vaccines. The basic principle is that mRNA molecules encoding antigens are delivered into the subject and subsequently translated in vivo into intracellular, membrane-bound, or secreted antigens that will elicit potent immune responses [16, 17] (Fig. 2). To this end, a series of in vitro experiments should be well accomplished. Specifically, the target antigenic sequence is selected in silico and synthesized as a DNA template, whereby the mRNA can be transcribed on a large scale [16, 17]. Considering that mRNA molecules are manufactured in a cell-free fashion, any available gene sequences could be regarded as potential mRNA vaccine candidates, which thus can be efficiently, rapidly and cost-effectively tested and assessed in the laboratory using animal models [16–18].
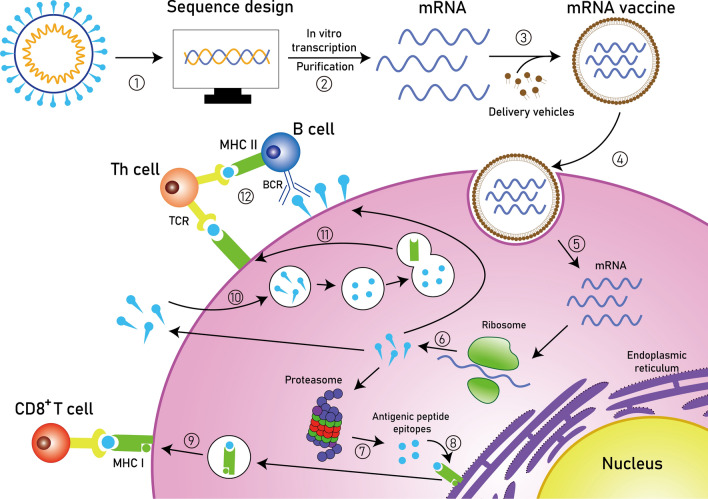
Fig. 2.
Principles of synthetic mRNA pharmacology. Step 1: After obtaining the pathogen genome, the target antigen sequence is designed and then inserted into the plasmid DNA vector. Step 2: The linearized plasmid DNA template is used for in vitro transcription and the resulting synthetic mRNA is purified. Step 3: The mRNA vaccine is prepared by encapsulating the purified mRNA with delivery vehicles. Step 4: The mRNA vaccine is taken up by endocytosis. Step 5: The target mRNA is released into the cytoplasm. Step 6: The mRNA is translated into protein by the ribosome. Step 7: The protein product is degraded into antigenic peptide epitopes by the proteasome complex. Step 8: The antigenic epitopes are loaded onto MHC class I molecules in the endoplasmic reticulum. Step 9: MHC class I molecules present antigenic peptides to CD8+ T lymphocytes. Step 10: Alternatively, the protein product is secreted and then taken up by the cell, followed by a degradation process in the endosome. Step 11: The antigenic fragments are presented on the cell surface to Th cells by MHC class II molecules. Step 12: Th cells stimulate B cells to produce neutralizing antibodies against circulating pathogens. MHC, major histocompatibility complex; BCR, B cell receptor; TCR, T cell receptor; Th cell, T helper cell
Selection of the optimal RNA modification is an important consideration for in vitro synthesis of mRNA vaccines. RNA modifications can abrogate the immunogenicity of synthetic mRNA molecules by bypassing immune activation pathways (Fig. 3). During the body's natural immunity to foreign pathogens (e.g., virus), the innate immune system senses multiple pathogen-associated molecular patterns (PAMPs) through specific pattern recognition receptors (PRRs), two types of which are Toll-like receptors (TLRs) and RIG-I-like receptors (RLRs) [19]. TLRs are type I transmembrane proteins [20, 21], in which TLR3, TLR7, and TLR8 are expressed in dendritic cells (DCs) and can recognize viral-derived double-stranded RNA (dsRNA) or single-strand RNA (ssRNA) in the endosome, activating multiple signaling cascades that promote type I interferon (IFN) production [22, 23]. However, in a variety of cells other than DCs, the key viral sensors appear to be the RLRs [24]. Two representative RLRs (RIG-I and MDA5) can recognize viral RNA present in the cytoplasm [25] and then interact with MAVS to induce the expression of type I IFN through the activation of IRF3 or NF-κB [25–27]. In addition, dsRNA produced during viral infection can also stimulate PKR activity, leading to eIF2α phosphorylation and subsequent antiviral responses [28].
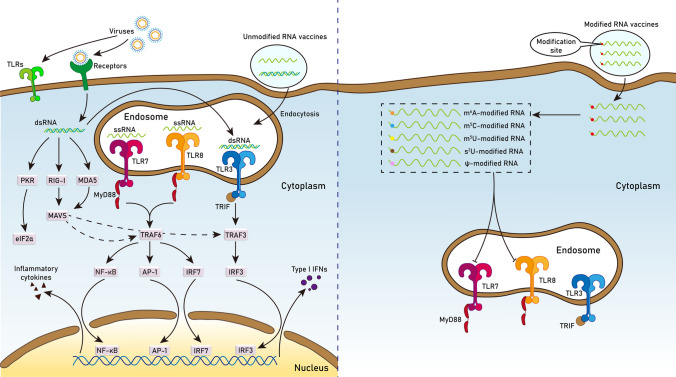
Fig. 3.
Schematic depicting the mechanism by which RNA modifications abrogate the immunogenicity of synthetic mRNA molecules in the cytoplasm
For unmodified RNA vaccines, the in vitro transcription process produces not only the desired full-length ssRNA but also dsRNA byproduct [29], and both ssRNA and dsRNA can induce immune responses via the above-mentioned pathways [22]. However, modified nucleotides can reduce the immunogenicity of in vitro transcribed RNAs in two ways. First, they inhibit the formation of dsRNA byproduct, thereby improving the purity of ssRNA. Specifically, Ψ, m1Ψ, and m5C are able to reduce the production of dsRNA byproduct, but not the MDA5-stimulatory activity of the dsRNA byproduct [30]. Second, they inhibit the activation of PRRs. It has been found that RNAs with m6A, m5C, m5U, s2U or Ψ modifications do not activate TLR7/8, and those with m6A and s2U do not activate TLR3 [6]. Notably, s2U and Ψ can also decrease the activity of RIG-I [31]. In addition, modified nucleotides affect RNA stability, structure, and intramolecular interactions [32]. For instance, Ψ modification can improve the translational capacity and stability of mRNA [33]. Taken together, enhanced properties conferred by modified nucleotides make synthetic RNA molecules effective and safe vehicles for vaccination.
RNA modification in mRNA vaccines
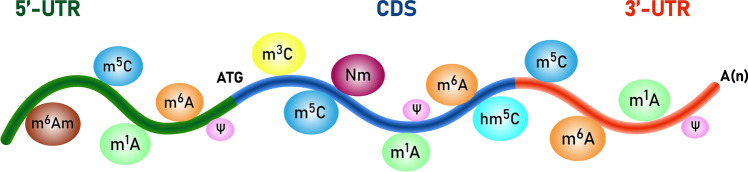
To date, more than 150 modifications have been discovered in all kinds of RNA molecules, including mRNA [34]. The functional domains of mRNA usually contain coding sequence (CDS), 5'-untranslated region (UTR), and 3'-UTR [35], and the properties of these structures are closely related to the effectiveness of mRNA vaccines. Therefore, we focused on characterizing the various RNA modifications that occur in these regions, so that the knowledge built on them will benefit the design of highly stable and efficacious mRNA vaccines (Fig. 4).
Fig. 4.
Schematic representation of the types of modified nucleotides on native mRNA
Ψ and m1Ψ
Ψ (pseudouridine) is the most abundant RNA modification [36] that is introduced in RNA by pseudouridine synthases or PUS enzymes [37]. It acts as a multifunctional player involved in RNA stability, translation and immunogenicity. Specifically, based on thermodynamic data, a report showed that Ψ can stabilize RNA duplexes when replacing U and generating Ψ-A, Ψ-U and Ψ-C pairs, which relies on type of base pair, the location of the Ψ in the duplex, and type and direction of adjacent Watson–Crick pairs [38]. Another study revealed that pseudouridylation (conversion of uridine into Ψ) of stop codons can inhibit translation termination both in vitro and in vivo and the resulting Ψ-containing codons encode for amino acids with similar properties [39]. In addition, incorporation of Ψ into in vitro-transcribed mRNA can improve the stability and translation of mRNA while evading the attack of the body's immune system by inhibiting the immunogenicity of mRNA [33]. These properties make Ψ a very popular modified nucleotide that has been used in various mRNA vaccines, for example, against melanoma [13, 14]. As for the method to detect Ψ, a high-throughput sequencing technology called Ψ-seq was developed in 2014, which can map Ψ sites at a transcriptome-wide scale with high resolution, confirming that Ψ is ubiquitous in different types of RNAs including mRNA, snoRNA and rRNA [36]. In the same year, the pseudouridine site identification sequencing (PSI-seq) technology was developed, using 1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate to modify Ψ and forcing reverse transcription to specifically stop at Ψ sites, thereby detecting Ψ modification with single-base resolution [40]. Notably, high-throughput sequencing methods for identifying Ψ have advanced significantly in the last few years. In 2022, bisulfite-induced deletion sequencing (BID-seq) was developed to capture quantitative information about Ψ at a single-base resolution [41]. Relying on this approach, the researchers revealed that the TRUB1 protein is the main mRNA Ψ ‘writer’ and confirmed the role of Ψ in promoting stop codon readthrough in vivo [41]. Due to the long read length, third-generation sequencing (TGS) technologies, mainly represented by Oxford Nanopore Technologies (ONT) and Pacific Biosciences (PacBio), are being used to interrogate various RNA biological processes at the single-molecule level. Based on ONT direct RNA sequencing data, two research groups demonstrated the utility of TGS in detecting Ψ modification and built their respective computational tools to predict Ψ modifications and evaluate the modification stoichiometry [42, 43]. Applying different stimuli to cell models, one group identified heat-sensitive Ψ-modified sites in small nuclear RNAs [42], and the other group found IFN inducible Ψ-modified sites in IFN-stimulated mRNAs, consistent with a role for Ψ in enabling efficacy of mRNA vaccines [43].
Because m1Ψ can reduce the immunogenicity of mRNA and exert context-dependent effects on mRNA translation [44], it is also favored by mRNA vaccine developers. It has been reported that (m5C/)m1Ψ-modified mRNA can lead to reduced innate immunogenicity in part by increasing its ability to evade activation of TLR3 and downstream immune signaling [45]. Notably, m1Ψ has a brief history of use in the preparation of mRNA vaccines. A potent m1Ψ-modified mRNA vaccine was developed against Zika virus (ZIKV) in 2017, which induced strong and long-lasting neutralizing antibody responses in mice and nonhuman primates [46]. As an example of mRNA vaccines for autoimmune diseases, delivery of m1Ψ-modified mRNA encoding disease-associated self-antigens can suppress multiple sclerosis in different mouse models by reducing effector T cells and developing regulatory T cell populations [47]. These studies carried out in animal models confirmed the application potential of m1Ψ modification in mRNA vaccines, providing confidence in translating the work from animal to human clinical studies. It must be emphasized that the m1Ψ modification is currently used in human COVID-19 mRNA vaccines to make the vaccines more effective [44].
RNA methylation
RNA methylation is one of the most common RNA modifications, which harbors multiple modification sites, including m5C, N1-methyladenosine (m1A), m6A, and 7-methylguanosine (m7G), 1-methylguanosine (m1G), and 2-methylguanosine (m2G) [48]. Previous studies have confirmed that m5C can promote mRNA export by affecting ALYREF, a kind of mRNA export adaptor [49]. In addition, m6A can affect mRNA translation and decay through controlling m6A-related proteins like YTHDF2 and YTHDF3 [50]. As in m1Ψ, other RNA methylations are gaining attention as new candidates for mRNA vaccine development. For example, Starostina et al. have tested different mRNA vaccine variants modified with m5C or m6A against influenza virus [51]. As technology iterates and evolves, a variety of methods have been developed to detect RNA methylation, such as liquid chromatography tandem-mass spectrometry (LC–MS/MS) [52, 53], methylated RNA immunoprecipitation sequencing (MeRIP-seq) [54], N6-methyladenosine sequencing (m6A-seq) [55, 56], and m6A individual-nucleotide-resolution cross-linking and immunoprecipitation sequencing (miCLIP-seq) [57]. To get rid of the dependence on antibodies for the detection of m6A, several novel techniques have recently been developed by three independent research teams [58–60]. Meyer reported DART-Seq (deamination adjacent to RNA modification targets), which can map thousands of m6A sites in cellular RNAs with as little as 10 nanograms of total RNA as input [58]. This method employs a strategy where m6A-adjacent cytidines can be edited by fusing the cytidine deaminase APOBEC1 to the m6A-binding YTH domain and the resulting C-to-U edits are detected using RNA-seq [58]. In 2022, Hu et al. developed m6A-selective allyl chemical labeling and sequencing (m6A-SAC-seq) to map global m6A sites in whole transcriptome at single-base resolution and revealed the dynamics of cell-state-specific m6A sites during human hematopoietic stem and progenitor cell differentiation into monocytes [59]. In the same year, Liu et al. presented an unbiased and convenient method for the absolute quantification of m6A at single-base resolution using glyoxal and nitrite-mediated deamination of unmethylated adenosines (GLORI) [60]. With the GLORI, they successfully identified 176,642 m6A sites in the HEK293T transcriptome and also provided a quantitative landscape of the m6A methylome in response to stress [60].
5'-cap
The 5'-cap (m7GpppN) is a characteristic structure for eukaryotic mRNAs, which is formed by linking m7G to the 5'-end of RNA-polymerase II transcripts through a 5ʹ-5ʹ-triphosphate linkage (ppp) [61, 62]. Specific sites in the 5'-cap can be methylated by methyltransferase (MTase)-transferred methyl groups from S-adenosyl-L-methionine or functional moieties from non-natural analogs [63, 64]. It has been reported that cap-specific adenosine methyltransferase (CAPAM, a methyltransferase responsible for methylation of the N6 position of the adenosine initiation nucleotide)-dependent methylation and propargylation have exactly the opposite effects on translation—namely the methylation greatly reduces translation but the propargylation maintains translation [64]. Moreover, propargylated mRNA can cause a stronger immune response [64]. There is study that developed a set of biotin-labeled cap analogs, which have been shown to not only promote translation in vitro but also resist degradation by the RNA decapping enzyme human decapping enzyme 2 (hDcp2) [65]. Except for the normal 5'-cap structure, some RNAs have a "reverse" cap where the m7G is adjacent to the body of the RNA [66]. Such reverse caps in conventional in vitro-synthesized mRNAs can result in a reduction of translational efficiency [67]. Stepinski et al. designed two novel cap analogs that cannot be incorporated in the reverse orientation and are thus called anti-reverse cap analogs (ARCAs) [67]. ARCA-capped transcripts have the same size and homogeneity as those produced by m7GpppG or GpppG and show higher translation efficiency than m7GpppG-capped transcripts [67]. As an important detection tool, CapQuant is a system-level mass spectrometry-based technology that can accurately quantify various types of 5'-cap [68]. Given that chemical modification of the 5'-cap structure significantly affects RNA stability and translation efficiency, it is also a critical factor in optimizing the design of RNA vaccines.
Poly(A) tail
The poly(A) tail, almost found on every mRNA in eukaryotes, is associated with the translation and stability of mRNA [69, 70]. Removal of the poly(A) tail is the first and rate-limiting step in the mRNA decay pathway [71, 72]. There has evidence that an evolutionarily conserved protein, poly(A)-binding protein (PAB), can bind to the poly(A) tail, not only affecting the activity of deadenylating nuclease but also stimulating translation [72, 73]. Interestingly, one report suggested that PAB also has an intrinsic property of stabilizing RNA, and the main or only role of poly(A) tail in mRNA stability is to bring PAB to mRNA [74]. In addition, it has been found that the poly(A) tail of appropriate length can serve as an identity element for mRNA nuclear export, because that the poly(A) tail can either increase RNA length or provide a platform for recruitment of mRNA export factors [75]. Many studies have shown that a number of modification sites exist in the poly(A) tail region. The uridylation of the poly(A) tail was found to promote mRNA decay [76], while the guanylation could protect mRNA from rapid deadenylation [77]. A novel RNA-seq technology, called poly(A) inclusive RNA isoform sequencing (PAIso-seq), showed that 17% of mRNAs contained non-A residues in the poly(A) tails in mouse GV oocytes [78]. In addition to PAIso-seq, there have been several methods developed for deciphering sequence features of the poly(A) tail, such as TAIL-seq [79, 80] and poly(A)-tail length profiling by sequencing (PAL-seq) [80]. To sum up, modification of the poly(A) tail should be an important consideration in controlling the stability and translation of mRNA vaccine.
5'- and 3'-UTRs
There are two UTRs at the 5' and 3' ends of the mature mRNA. They do not encode proteins, but have regulatory biological functions on mRNA. The 5'-UTR acts as a controller of mRNA translation initiation in eukaryotes [81]. Different forms of RNA modifications can be found in 5'-UTR sequences, including m1A, m6A, N6,2'-O-dimethyladenosine (m6Am), m5C and Ψ [48, 82, 83]. The 3'-UTR is an essential regulatory region for diverse mRNA processes, such as nuclear export, RNA stability, polyadenylation, subcellular localization, and mRNA translation and decay [54, 84]. As such, any modifications that occur in this region may affect gene expression by altering RNA fate [84]. Much evidence shows that several modifications including m6A, m5C and Ψ are prevalent within the 3'-UTR [54, 83, 85]. Due to the pivotal role of 5'- and 3'-UTRs in mRNA functionalization, their RNA modification types provide an important reference model for the in vitro synthesis of mRNA vaccines similar to natural mRNA.
Application of mRNA vaccines in cancer immunotherapy
As an emerging therapeutic strategy for different types of tumors, cancer vaccines mainly induce or enhance tumor-specific immunity by acting on key target proteins such as tumor-associated antigens (TAAs) and cancer neoantigens [86]. The mRNA-based techniques provide a new and promising avenue for the design and development of novel cancer vaccines. There are already numerous clinical trials of mRNA vaccines targeting the following five tumors (Table 1).
Table 1.
List of clinical trials of mRNA vaccines against five types of cancer
| Cancer Types | Status | Phases | Study Type | References |
|---|---|---|---|---|
| Brain cancer; Neoplasm metastases | Completed | Phase 1 | Interventional | NCT02808416 [87] |
| GBM; Brain tumor | Completed | Phase 1 & 2 | Interventional | NCT00846456 [88] |
| Malignant neoplasms of brain | Active, not recruiting | Phase 1 | Interventional | NCT00639639 [89] |
| Malignant glioma; Astrocytoma; GBM | Completed | Phase 1 | Interventional | NCT02529072 |
| Adult GBM | Recruiting | Phase 1 | Interventional | NCT04573140 |
| Recurrent central nervous system neoplasm | Completed | Phase 1 | Interventional | NCT00890032 |
| High grade glioma; Diffuse intrinsic pontine glioma | Recruiting | Phase 1 & 2 | Interventional | NCT04911621 |
| Metastatic NSCLC; NSCLC | Completed | Phase 1 & 2 | Interventional | NCT03164772 |
| Esophageal cancer; NSCLC | Unknown status | NA | Interventional | NCT03908671 |
| NSCLC | Completed | Phase 1 & 2 | Interventional | NCT00923312 [90] |
| Ewings sarcoma; NSCLC; Liver cancer | Completed | Phase 1 | Interventional | NCT01061840 |
| Malignant melanoma | Completed | Phase 1 & 2 | Interventional | NCT00204516 |
| Melanoma | Terminated | Phase 1 & 2 | Interventional | NCT01944709 |
| Melanoma; Colon cancer; Gastrointestinal cancer; Genitourinary cancer; Hepatocellular cancer | Terminated | Phase 1 & 2 | Interventional | NCT03480152 [91] |
| Melanoma | Not yet recruiting | Phase 1 | Interventional | NCT05264974 |
| Malignant melanoma | Completed | Phase 1 & 2 | Interventional | NCT01278940 |
| Metastatic malignant melanoma | Terminated | Phase 1 & 2 | Interventional | NCT00961844 |
| Melanoma | Active, not recruiting | Phase 1 | Interventional | NCT02410733 [92] |
| Melanoma | Active, not recruiting | Phase 2 | Interventional | NCT03897881 |
| Melanoma stage III; Melanoma stage IV; Unresectable melanoma | Recruiting | Phase 2 | Interventional | NCT04526899 |
| Melanoma stage III or IV | Completed | Phase 1 & 2 | Interventional | NCT00243529 |
| Melanoma | Completed | Phase 1 | Interventional | NCT01456104 |
| Melanoma | Completed | Phase 1 & 2 | Interventional | NCT01530698 |
| Breast cancer; Malignant melanoma | Completed | Phase 1 | Interventional | NCT00978913 |
| Melanoma | Completed | Phase 1 | Interventional | NCT01066390 [93] |
| Melanoma | Completed | Phase 2 | Interventional | NCT02285413 [94] |
| Recurrent melanoma; Stage IV melanoma | Terminated | Phase 2 | Interventional | NCT00087373 |
| Hormonal refractory prostate cancer | Completed | Phase 1 & 2 | Interventional | NCT00831467 [90] |
| Prostate cancer | Active, not recruiting | Phase 1 & 2 | Interventional | NCT01197625 |
| Prostate cancer | Completed | Phase 1 & 2 | Interventional | NCT01278914 |
| Metastatic prostate cancer | Withdrawn | Phase 1 & 2 | Interventional | NCT01153113 |
| Prostatic neoplasms | Completed | Phase 2 | Interventional | NCT01446731 |
| Prostate carcinoma | Terminated | Phase 2 | Interventional | NCT02140138 |
| RCC | Unknown status | Phase 1 & 2 | Interventional | NCT02787915 |
Data from https://clinicaltrials.gov/ (Accessed January 19, 2023); NA: Not available
Renal cell carcinoma (RCC)
RCC is one of the most common cancers in the world, accounting for 5% of all new diagnoses in men and 3% in women, according to Cancer Statistics 2022 [95]. At present, the treatment of RCC is mainly surgery, supplemented by regular follow-up, and new treatment options such as vascular endothelial growth factor inhibitors and tyrosine kinase inhibitors are also emerging [96]. Since RCC is classified as an immunogenic tumor, exploring its immunotherapeutic approach has become one of the research hotspots in the field of RCC [97]. It has been confirmed that therapy using RCC RNA-transfected DCs is not only feasible and safe, but also stimulates the expansion of tumor-specific polyclonal T cells in vivo [98]. Another research established a generic DC vaccine strategy in which RNA prepared from a well-characterized highly immunogenic RCC cell line (RCC-26) was used as a source of TAAs for loading of DCs [99]. In addition, when the number of CD4+/CD25+ Tregs in metastatic RCC patients is decreased with the recombinant IL-2 diphtheria toxin conjugate DAB389IL-2, RNA-transfected DC vaccine can drastically boost the stimulation of tumor-specific T cell responses [100]. An mRNA vaccine consisting of RNAs encoding six different TAAs has been reported to induce T-cell responses against multiple TAA epitopes, thereby contributing to prolonging overall survival in patients with metastatic RCC [97]. Of note, high-throughput data mining provides a powerful approach to discover more potentially effective neoantigens for RCC mRNA vaccine development. For example, analysis of The Cancer Genome Atlas (TCGA)-kidney renal clear cell carcinoma (KIRC) dataset identified four genes (TOP2A, NCF4, FMNL1, and DOK3) that were upregulated, mutated, and positively correlated with survival and antigen-presenting cells [101].
Brain tumor
Brain tumors are defined as tumors that grow intracranially. Brain and other nervous system tumors are the leading cause of cancer death in the United States for men under 40 and women under 20, according to Cancer Statistics 2022 [95]. The current mainstay of treatment for tumors is surgical resection, supplemented by radiation and/or chemotherapy [102, 103]. However, due to the complexity of brain structure and the possibility of tumor metastasis, surgical resection is sometimes difficult to achieve the desired effect, and thus, some researchers have tried to use immunotherapy to treat brain tumors. A study has demonstrated the feasibility and safety of the monocyte-derived dendritic cells pulsed with tumor RNA (DCRNA vaccine) in pediatric patients with recurrent brain tumors [104]. Glioma, the most common primary intracranial tumor, accounts for 81% of malignant brain tumors [105]. There are many studies on glioma immunotherapy through analyzing the relevant data. Four tumor antigens (TCF12, TP53, C3, and IDH1) were found to be associated with poor prognosis and infiltration of antigen-presenting cells [106], while those (ANXA5, FKBP10, MSN, and PYGL) with good prognosis [107]. For glioblastoma (GBM), the most common and aggressive malignant brain tumor in adults, Rose et al. identified 11 specific potential targets for immunotherapy strategies by comparing surfaceomes between GBM cells and astrocytes [108]. In another study, ARPC1B and HK3 were revealed as potential antigens for the development of GBM mRNA vaccines [109]. For diffuse glioma, Zhou et al. suggested that COL1A2, SAMD9 and KDR can be used as potential antigens for developing mRNA vaccines [110]. Overall, these findings provide numerous candidate target proteins awaiting experimental and clinical validation for the design and development of effective brain tumor RNA vaccines.
Melanoma
Melanoma is an aggressive tumor that originates in melanocytes [111]. In the event of malignant transformation, the primary tumor is prone to metastasize, posing a huge threat to the life of the patient [111]. Melanoma ranks fifth in estimated new cases in both men and women, according to Cancer Statistics 2022 [95]. There are already several treatments for melanoma, such as surgical resection, chemotherapy, targeted therapy, and immunotherapy. Three classes of proteins displayed on major histocompatibility complex (MHC) class I proteins on the surface of melanoma cells, including TAAs, tumor-specific antigens and melanoma differentiation antigens, are able to warn the immune system that the cells are diseased [112]. Two research groups demonstrated the efficacy of TAA-based mRNA vaccines using the same RNA modifications (m5C and Ψ), showing that these vaccines were able to induce robust immune responses in melanoma mouse models [13, 14]. Specifically, one group aimed at reducing the mRNA immunogenicity and increasing the expression levels of antigens using the modified nucleotides [13], while the other group found no significant difference in the CD8 T cell levels in mice immunized with modified mRNA relative to controls, but significantly higher CD8 T cell levels in mice treated with unmodified mRNA relative to controls [14]. In addition, there are some studies using combination therapy strategies to increase the antitumor effect [113, 114]. For example, Li et al. revealed that the combination of cytosine-phosphate-guanosine-oligodeoxynucleotides (CpG-ODNs) and mRNA vaccines modified by N1-methylpseudo-UTP and cap 1 analogs could be a promising candidate approach for immunostimulatory sequence-based therapeutic strategies [113]. The good news is that a commercial vaccine FixVac (BNT111) has been investigated in an ongoing, first-in-human, dose-escalation clinical trial in advanced melanoma patients, which can be intravenously administered against four non-mutated TAAs and shows a favorable safety profile and preliminary antitumor responses, either alone or in combination with immune checkpoint inhibitor therapy [92, 115]. More importantly, this vaccine was modified by uridine, which increased the immunostimulatory effect and was optimized for enhanced pharmacological activity [115].
Prostate cancer
Prostate cancer is the most common malignancy among men in western countries with high mortality. According to Cancer Statistics 2022, prostate cancer ranks first in estimated new cases and second in estimated deaths from the male population [95]. In addition to traditional radical prostatectomy, many non-surgical treatments are widely practiced, including chemotherapy, radiation therapy, ablative therapy, androgen-deprivation therapy and immunotherapy [116]. Although mRNA vaccines against prostate cancer have been studied, the vaccines have disadvantages such as high cost of gold particles, unstable mRNA, and limited large-scale production of mRNA-transfected DCs in vitro [117]. To address these issues, the recombinant bacteriophage MS2 virus-like particles were designed to encapsulate target mRNA, resulting in an easy-to-prepare, non-toxic, ribonuclease-resistant vaccine that elicited potent humoral and cellular immune responses and delayed tumor growth [117]. Another study developed an adjuvant-pulsed mRNA vaccine modified by m5C and Ψ and revealed that co-delivery of the modified mRNA and palmitic acid-conjugated Resiquimod (C16-R848) could promote the recruitment of CD8+ T cells to tumors and enhance the overall antitumor response, improving the therapeutic and preventive efficacy of the vaccine [15]. Notably, two vaccines, CV9103 and CV9104, based on the novel RNActive® technology, are currently available for the treatment of prostate cancer patients and have been shown to be highly specific, safe and effective [118]. In addition, some studies have revealed new targets for mRNA vaccines. For instance, eight mutated antigens (KLHL17, CPT1B, IQGAP3, LIME1, YJEFN3, KIAA1529, MSH5, and CELSR3) were found to be overexpressed in prostate adenocarcinoma and associated with poor prognosis [119]. Furthermore, based on distinct clinical, molecular, and cellular characteristics, this study classified prostate adenocarcinoma into three immune subtypes (PIS1, PIS2, and PIS3) and suggested that patients with PIS2 and PIS3 were more suitable for vaccination [119].
Lung cancer
Lung cancer remains the leading cause of cancer-related death worldwide. The disease can be divided into two major histological subtypes: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), which account for approximately 15% and 85% of cases, respectively [120]. RNA vaccines for NSCLC are already in clinical trials. For example, CV9201, a cancer immunotherapy based on RNActive® technology encoding five NSCLC antigens, was well tolerated and elicited a detectable immune response [121]. Furthermore, in phase IIa, median progression-free and overall survival for patients were 5.0 months (95% CI 1.8–6.3) and 10.8 months (8.1–16.7) from first administration, respectively [121]. Another clinical trial evaluated the safety and tolerability of BI1361849 (CV9202), a self-adjuvanted protamine formulated mRNA-based active cancer immunotherapy encoding six NSCLC-associated antigens, showing that it was well tolerated in combination with local radiation [122]. In addition, several studies have used bioinformatic approaches to screen and identify potential tumor antigens for the development of RNA vaccines for lung adenocarcinoma (LUAD), the most common histological subtype of lung cancer [123]. Two genes, KLRG1 and CBFA2T3, were identified to be associated with prognosis in LUAD patients and positively correlated with the infiltration of antigen-presenting cells (APCs) [124]. Another seven genes including GPRIN1, MYRF, PLXNB2, SLC9A4, TRIM29, UBA6, and XDH can serve as potential immune biomarker candidates to activate the immune response [125].
Future prospects and challenges of mRNA vaccines
Increasing basic research and clinical applications show that mRNA vaccines have many advantages over other types of vaccines. Firstly, mRNA vaccines have the excellent safety. Unlike DNA and viral vaccines, mRNA is a non-infectious nucleic acid substance and once delivered to the cytoplasm, the mRNA is translated immediately [61, 126]. Since mRNA vaccines do not enter the nucleus and cannot integrate into the genome, there is no risk of causing genetic mutations in recipients [61, 126]. Like native mRNAs, the activity of in vitro transcribed mRNAs is transient, and they are fully degraded by physiological metabolic pathways [61]. Secondly, mRNA vaccines are relatively simple to design and inexpensive to produce [61, 127]. Once the sequence information of the mRNA of a new target gene is obtained, its corresponding mRNA vaccine can theoretically be designed and synthesized very quickly [127]. In the field of tumor research, there are a large number of high-throughput sequencing data and clinical case resources that can be used to screen and identify valuable target gene candidates for mRNA vaccines. Thirdly, there are already some measures to enhance the thermostability of mRNA vaccines with or without major changes in the formulation [128]. For instance, the lyophilized vaccines are stable for 36 months at 5–25 °C, free from the dependence on low temperature conditions during long-distance transportation and long-term storage [126].
Despite the above-mentioned advantages and rapid advance of mRNA vaccines, there are still some gaps between their application status and our expectations. Specifically, much of the mRNA vaccine research currently underway is primarily tested and evaluated in animal models rather than humans [129], which means that whether the vaccine can elicit an immune response in humans is unclear, so further clinical trials are urgently needed to verify the safety and efficacy of the mRNA vaccines.
In order to fully realize the potential of mRNA vaccines to treat diseases, especially tumors, another issue is how to choose a biocompatible and safe delivery system so that the synthetic RNA molecules can avoid being cleared by off-target organs, can reach the correct tissue, can interact with the desired cell type in the complex tissue microenvironment, can be taken up by endocytosis, and can eventually get out of the endosome [130]. Lipid nanoparticles (LNPs) are currently the most trending delivery vehicles for in vivo application, which can increase mRNA entrapment efficiency, prevent mRNA degradation before release, promote endosomal escape, and control the site and timing of mRNA vaccine delivery in the body [131, 132]. In a clinical trial (NCT03480152), LNPs have been utilized to encapsulate personalized mRNA vaccine against melanoma [91]. An emerging advantage of LNPs is that organ-specific targeting can be readily achieved by altering their lipid structures [133]. In one example, changes in the alkyl chain length of a lipid led to selective delivery of mRNA to the liver or spleen [133, 134]. Intriguingly, LNP charge can also affect mRNA delivery [130]. For example, researchers redirected hepatotropic LNP to the lung via adding a cationic lipid to the LNP [130, 135]. Although these advances offer valuable information for the future design of tissue-targeted mRNA therapeutics, the urgency for mRNA cancer vaccines is to expedite testing of the delivery efficiency of different LNP formulations in animal models and to increase the number of vaccines entering clinical trials in LNP formulations.
It is believed that the property of the selected antigen also affects the quality of the elicited immune response [129], and therefore, numerous bioinformatic analyses have been actively focused on mining potential new tumor-associated antigens for mRNA vaccines, but relevant experimental validation is lacking. In fact, high-throughput sequencing technologies coupled with bioinformatics tools may offer great help in improving the specificity of mRNA vaccines in the context of personalized tumor therapy. For example, most (up to 70%) human genes generate many forms of transcript variants [136], which may be translated into different isoforms. Moreover, given the possibility of population-specific and individual-specific isoforms, there is a need to obtain reliable full-length transcript sequence information from the target individuals for precise design of mRNA vaccines, which can be achieved via TGS technologies, such as ONT RNA-seq and PacBio Iso-Seq.
Last but not least, based on the lessons and experiences of mRNA-modified vaccines for infectious diseases, modified nucleosides and sequence engineering should be applied to the development of more cancer mRNA vaccines as soon as possible. Furthermore, we have entered an era where various combinations of RNA modifications will be detectable at the transcript level in an individual using TGS techniques, especially ONT direct RNA sequencing [42]. If the functional effects of these combinatorial RNA modifications on mRNA stability, structure and translation can be well interpreted, this will open up a huge possibility for customizing unique mRNA vaccines that best fit an individual's immune system and thus greatly facilitate the application of mRNA vaccines in cancer immunotherapy.
Conclusions
In summary, we reviewed and discussed the functions of RNA modifications and their applications in mRNA vaccines and emphasized recent advances of mRNA vaccines in cancer immunotherapy. More importantly, we provided future research directions to explore the functions of combinatorial RNA modifications at the transcript level as a basis for the development of novel mRNA vaccines in the context of individualized tumor therapy.
Acknowledgements
Not applicable.
Authors’ contribution
XW and YM designed, wrote and edited the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by National Natural Science Foundation of China (No. 81870130).
Declarations
Competing interest
The authors declare that they have no competing interests.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Availability of data and materials
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ndwandwe D, Wiysonge CS. COVID-19 vaccines. Curr Opin Immunol. 2021;71:111–6. doi: 10.1016/j.coi.2021.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maruyama X, Tucker JB. The once and future threat of smallpox. In: Naval War College Review. U.S. Naval War College. 2002. https://digital-commons.usnwc.edu/nwc-review/vol55/iss2/11. Accessed 11 Aug 2021.
- 3.Sell S. How vaccines work: immune effector mechanisms and designer vaccines. Expert Rev Vaccines. 2019;18(10):993–1015. doi: 10.1080/14760584.2019.1674144. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Zhang Z, Luo J, Han X, Wei Y, Wei X. mRNA vaccine: a potential therapeutic strategy. Mol Cancer. 2021;20(1):33. doi: 10.1186/s12943-021-01311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaviria M, Kilic B. A network analysis of COVID-19 mRNA vaccine patents. Nat Biotechnol. 2021;39(5):546–8. doi: 10.1038/s41587-021-00912-9. [DOI] [PubMed] [Google Scholar]
- 6.Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23(2):165–75. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Baden LR, El Sahly HM, Essink B, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384(5):403–16. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N Engl J Med. 2020;383(27):2603–15. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbier AJ, Jiang AY, Zhang P, Wooster R, Anderson DG. The clinical progress of mRNA vaccines and immunotherapies. Nat Biotechnol. 2022;40(6):840–54. doi: 10.1038/s41587-022-01294-2. [DOI] [PubMed] [Google Scholar]
- 10.CureVac. CureVac provides update on phase 2b/3 trial of first-generation COVID-19 vaccine candidate, CVnCoV. 2021. https://www.curevac.com/en/curevac-provides-update-on-phase-2b-3-trial-of-first-generation-covid-19-vaccine-candidate-cvncov/. Accessed 28 Dec 2022.
- 11.Igarashi Y, Sasada T. Cancer vaccines: toward the next breakthrough in cancer immunotherapy. J Immunol Res. 2020;2020:5825401. doi: 10.1155/2020/5825401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beck JD, Reidenbach D, Salomon N, et al. mRNA therapeutics in cancer immunotherapy. Mol Cancer. 2021;20(1):69. doi: 10.1186/s12943-021-01348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Zhang L, Xu Z, Miao L, Huang L. mRNA vaccine with antigen-specific checkpoint blockade induces an enhanced immune response against established melanoma. Mol Ther. 2018;26(2):420–34. doi: 10.1016/j.ymthe.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oberli MA, Reichmuth AM, Dorkin JR, et al. Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 2017;17(3):1326–35. doi: 10.1021/acs.nanolett.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam MA, Rice J, Reesor E, et al. Adjuvant-pulsed mRNA vaccine nanoparticle for immunoprophylactic and therapeutic tumor suppression in mice. Biomaterials. 2021;266:120431. doi: 10.1016/j.biomaterials.2020.120431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kowalzik F, Schreiner D, Jensen C, Teschner D, Gehring S, Zepp F. mRNA-based vaccines. Vaccines (Basel) 2021 doi: 10.3390/vaccines9040390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maruggi G, Zhang C, Li J, Ulmer JB, Yu D. mRNA as a transformative technology for vaccine development to control infectious diseases. Mol Ther. 2019;27(4):757–72. doi: 10.1016/j.ymthe.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaudhary N, Weissman D, Whitehead KA. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat Rev Drug Discov. 2021;20(11):817–38. doi: 10.1038/s41573-021-00283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu G, Xia Z, Deng F, et al. Inducible LGALS3BP/90K activates antiviral innate immune responses by targeting TRAF6 and TRAF3 complex. PLoS Pathog. 2019;15(8):e1008002. doi: 10.1371/journal.ppat.1008002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rock FL, Hardiman G, Timans JC, Kastelein RA, Bazan JF. A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci U S A. 1998;95(2):588–93. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurst J, Von Landenberg P. Toll-like receptors and autoimmunity. Autoimmun Rev. 2008;7(3):204–8. doi: 10.1016/j.autrev.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 22.Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 23.Creagh EM, O’neill LA. TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol. 2006;27(8):352–7. doi: 10.1016/j.it.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Kato H, Sato S, Yoneyama M, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23(1):19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 25.Nakhaei P, Genin P, Civas A, Hiscott J. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin Immunol. 2009;21(4):215–22. doi: 10.1016/j.smim.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Xu C, Peng Y, Zhang Q, Xu XP, Kong XM, Shi WF. USP4 positively regulates RLR-induced NF-kappaB activation by targeting TRAF6 for K48-linked deubiquitination and inhibits enterovirus 71 replication. Sci Rep. 2018;8(1):13418. doi: 10.1038/s41598-018-31734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122(5):669–82. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 28.Zhang S, Sun Y, Chen H, et al. Activation of the PKR/eIF2alpha signaling cascade inhibits replication of Newcastle disease virus. Virol J. 2014;11:62. doi: 10.1186/1743-422X-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mu X, Hur S. Immunogenicity of in vitro-transcribed RNA. Acc Chem Res. 2021;54(21):4012–23. doi: 10.1021/acs.accounts.1c00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mu X, Greenwald E, Ahmad S, Hur S. An origin of the immunogenicity of in vitro transcribed RNA. Nucleic Acids Res. 2018;46(10):5239–49. doi: 10.1093/nar/gky177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hornung V, Ellegast J, Kim S, et al. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–7. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 32.D'ascenzo L, Popova AM, Abernathy S, Sheng K, Limbach PA, Williamson JR. Pytheas: a software package for the automated analysis of RNA sequences and modifications via tandem mass spectrometry. Nat Commun. 2022;13(1):2424. doi: 10.1038/s41467-022-30057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kariko K, Muramatsu H, Welsh FA, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16(11):1833–40. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boccaletto P, Stefaniak F, Ray A, et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022;50(D1):D231–D5. doi: 10.1093/nar/gkab1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shabalina SA, Ogurtsov AY, Spiridonov NA. A periodic pattern of mRNA secondary structure created by the genetic code. Nucleic Acids Res. 2006;34(8):2428–37. doi: 10.1093/nar/gkl287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz S, Bernstein DA, Mumbach MR, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159(1):148–62. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uddin MB, Wang Z, Yang C. Dysregulations of functional RNA modifications in cancer, cancer stemness and cancer therapeutics. Theranostics. 2020;10(7):3164–89. doi: 10.7150/thno.41687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kierzek E, Malgowska M, Lisowiec J, Turner DH, Gdaniec Z, Kierzek R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 2014;42(5):3492–501. doi: 10.1093/nar/gkt1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karijolich J, Yu YT. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011;474(7351):395–8. doi: 10.1038/nature10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lovejoy AF, Riordan DP, Brown PO. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One. 2014;9(10):e110799. doi: 10.1371/journal.pone.0110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai Q, Zhang LS, Sun HL, et al. Quantitative sequencing using BID-seq uncovers abundant pseudouridines in mammalian mRNA at base resolution. Nat Biotechnol. 2022 doi: 10.1038/s41587-022-01505-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Begik O, Lucas MC, Pryszcz LP, et al. Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat Biotechnol. 2021;39(10):1278–91. doi: 10.1038/s41587-021-00915-6. [DOI] [PubMed] [Google Scholar]
- 43.Huang S, Zhang W, Katanski CD, et al. Interferon inducible pseudouridine modification in human mRNA by quantitative nanopore profiling. Genome Biol. 2021;22(1):330. doi: 10.1186/s13059-021-02557-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nance KD, Meier JL. Modifications in an emergency: the role of N1-methylpseudouridine in COVID-19 vaccines. ACS Cent Sci. 2021;7(5):748–56. doi: 10.1021/acscentsci.1c00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andries O, Mc Cafferty S, De Smedt SC, Weiss R, Sanders NN, Kitada T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J Control Release. 2015;217:337–44. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 46.Pardi N, Hogan MJ, Pelc RS, et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543(7644):248–51. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krienke C, Kolb L, Diken E, et al. A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis. Science. 2021;371(6525):145–53. doi: 10.1126/science.aay3638. [DOI] [PubMed] [Google Scholar]
- 48.Lan Q, Liu PY, Haase J, Bell JL, Huttelmaier S, Liu T. The critical role of RNA m(6)A methylation in cancer. Cancer Res. 2019;79(7):1285–92. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 49.Yang X, Yang Y, Sun BF, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27(5):606–25. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi H, Wang X, Lu Z, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27(3):315–28. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Starostina EV, Sharabrin SV, Antropov DN, et al. Construction and immunogenicity of modified mRNA-vaccine variants encoding influenza virus antigens. Vaccines (Basel) 2021 doi: 10.3390/vaccines9050452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–7. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149(7):1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–6. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 56.Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8(1):176–89. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- 57.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12(8):767–72. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meyer KD. DART-seq: an antibody-free method for global m(6)A detection. Nat Methods. 2019;16(12):1275–80. doi: 10.1038/s41592-019-0570-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu L, Liu S, Peng Y, et al. m(6)A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat Biotechnol. 2022;40(8):1210–9. doi: 10.1038/s41587-022-01243-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu C, Sun H, Yi Y, et al. Absolute quantification of single-base m(6)A methylation in the mammalian transcriptome using GLORI. Nat Biotechnol. 2022 doi: 10.1038/s41587-022-01487-9. [DOI] [PubMed] [Google Scholar]
- 61.Sahin U, Kariko K, Tureci O. mRNA-based therapeutics–developing a new class of drugs. Nat Rev Drug Discov. 2014;13(10):759–80. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 62.Curry S, Kotik-Kogan O, Conte MR, Brick P. Getting to the end of RNA: structural analysis of protein recognition of 5' and 3' termini. Biochim Biophys Acta. 2009;1789(9–10):653–66. doi: 10.1016/j.bbagrm.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 63.Muthmann N, Spacek P, Reichert D, Van Dulmen M, Rentmeister A. Quantification of mRNA cap-modifications by means of LC-QqQ-MS. Methods. 2022;203:196–206. doi: 10.1016/j.ymeth.2021.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Dulmen M, Muthmann N, Rentmeister A. Chemo-enzymatic modification of the 5' cap maintains translation and increases immunogenic properties of mRNA. Angew Chem Int Ed Engl. 2021;60(24):13280–6. doi: 10.1002/anie.202100352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bednarek S, Madan V, Sikorski PJ, Bartenschlager R, Kowalska J, Jemielity J. mRNAs biotinylated within the 5' cap and protected against decapping: new tools to capture RNA-protein complexes. Philos Trans R Soc Lond B Biol Sci. 2018 doi: 10.1098/rstb.2018.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pasquinelli AE, Dahlberg JE, Lund E. Reverse 5' caps in RNAs made in vitro by phage RNA polymerases. RNA. 1995;1(9):957–67. [PMC free article] [PubMed] [Google Scholar]
- 67.Stepinski J, Waddell C, Stolarski R, Darzynkiewicz E, Rhoads RE. Synthesis and properties of mRNAs containing the novel "anti-reverse" cap analogs 7-methyl(3'-O-methyl)GpppG and 7-methyl (3'-deoxy)GpppG. RNA. 2001;7(10):1486–95. [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J, Alvin Chew BL, Lai Y, et al. Quantifying the RNA cap epitranscriptome reveals novel caps in cellular and viral RNA. Nucleic Acids Res. 2019;47(20):e130. doi: 10.1093/nar/gkz751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Passmore LA, Coller J. Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat Rev Mol Cell Biol. 2022;23(2):93–106. doi: 10.1038/s41580-021-00417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim H, Lee JH, Lee Y. Regulation of poly(A) polymerase by 14–3-3epsilon. EMBO J. 2003;22(19):5208–19. doi: 10.1093/emboj/cdg486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen CY, Shyu AB. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol Cell Biol. 2003;23(14):4805–13. doi: 10.1128/MCB.23.14.4805-4813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Korner CG, Wahle E. Poly(A) tail shortening by a mammalian poly(A)-specific 3'-exoribonuclease. J Biol Chem. 1997;272(16):10448–56. doi: 10.1074/jbc.272.16.10448. [DOI] [PubMed] [Google Scholar]
- 73.Gray NK, Coller JM, Dickson KS, Wickens M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 2000;19(17):4723–33. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Coller JM, Gray NK, Wickens MP. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 1998;12(20):3226–35. doi: 10.1101/gad.12.20.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fuke H, Ohno M. Role of poly (A) tail as an identity element for mRNA nuclear export. Nucleic Acids Res. 2008;36(3):1037–49. doi: 10.1093/nar/gkm1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lim J, Ha M, Chang H, et al. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell. 2014;159(6):1365–76. doi: 10.1016/j.cell.2014.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lim J, Kim D, Lee YS, et al. Mixed tailing by TENT4A and TENT4B shields mRNA from rapid deadenylation. Science. 2018;361(6403):701–4. doi: 10.1126/science.aam5794. [DOI] [PubMed] [Google Scholar]
- 78.Liu Y, Nie H, Liu H, Lu F. Poly(A) inclusive RNA isoform sequencing (PAIso-seq) reveals wide-spread non-adenosine residues within RNA poly(A) tails. Nat Commun. 2019;10(1):5292. doi: 10.1038/s41467-019-13228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chang H, Lim J, Ha M, Kim VN. TAIL-seq: genome-wide determination of poly(A) tail length and 3' end modifications. Mol Cell. 2014;53(6):1044–52. doi: 10.1016/j.molcel.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 80.Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508(7494):66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leppek K, Das R, Barna M. Functional 5' UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol. 2018;19(3):158–74. doi: 10.1038/nrm.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Davalos V, Blanco S, Esteller M. SnapShot: messenger RNA modifications. Cell. 2018;174(2):498–e1. doi: 10.1016/j.cell.2018.06.046. [DOI] [PubMed] [Google Scholar]
- 83.Squires JE, Patel HR, Nousch M, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40(11):5023–33. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Conne B, Stutz A, Vassalli JD. The 3' untranslated region of messenger RNA: a molecular 'hotspot' for pathology? Nat Med. 2000;6(6):637–41. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 85.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515(7525):143–6. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khan P, Siddiqui JA, Lakshmanan I, et al. RNA-based therapies: a cog in the wheel of lung cancer defense. Mol Cancer. 2021;20(1):54. doi: 10.1186/s12943-021-01338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang QT, Nie Y, Sun SN, et al. Tumor-associated antigen-based personalized dendritic cell vaccine in solid tumor patients. Cancer Immunol Immunother. 2020;69(7):1375–87. doi: 10.1007/s00262-020-02496-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vik-Mo EO, Nyakas M, Mikkelsen BV, et al. Therapeutic vaccination against autologous cancer stem cells with mRNA-transfected dendritic cells in patients with glioblastoma. Cancer Immunol Immunother. 2013;62(9):1499–509. doi: 10.1007/s00262-013-1453-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Batich KA, Mitchell DA, Healy P, Herndon JE, 2nd, Sampson JH. Once, twice, three times a finding: reproducibility of dendritic cell vaccine trials targeting cytomegalovirus in glioblastoma. Clin Cancer Res. 2020;26(20):5297–303. doi: 10.1158/1078-0432.CCR-20-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fotin-Mleczek M, Duchardt KM, Lorenz C, et al. Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity. J Immunother. 2011;34(1):1–15. doi: 10.1097/CJI.0b013e3181f7dbe8. [DOI] [PubMed] [Google Scholar]
- 91.Cafri G, Gartner JJ, Zaks T, et al. mRNA vaccine-induced neoantigen-specific T cell immunity in patients with gastrointestinal cancer. J Clin Invest. 2020;130(11):5976–88. doi: 10.1172/JCI134915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sahin U, Oehm P, Derhovanessian E, et al. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature. 2020;585(7823):107–12. doi: 10.1038/s41586-020-2537-9. [DOI] [PubMed] [Google Scholar]
- 93.Wilgenhof S, Van Nuffel AMT, Benteyn D, et al. A phase IB study on intravenous synthetic mRNA electroporated dendritic cell immunotherapy in pretreated advanced melanoma patients. Ann Oncol. 2013;24(10):2686–93. doi: 10.1093/annonc/mdt245. [DOI] [PubMed] [Google Scholar]
- 94.Boudewijns S, Bloemendal M, De Haas N, et al. Autologous monocyte-derived DC vaccination combined with cisplatin in stage III and IV melanoma patients: a prospective, randomized phase 2 trial. Cancer Immunol Immunother. 2020;69(3):477–88. doi: 10.1007/s00262-019-02466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 96.Li YZ, Zhu HC, Du Y, Zhao HC, Wang L. Silencing lncRNA SLC16A1-AS1 induced ferroptosis in renal cell carcinoma through miR-143-3p/SLC7A11 signaling. Technol Cancer Res Treat. 2022;21:15330338221077803. doi: 10.1177/15330338221077803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rittig SM, Haentschel M, Weimer KJ, et al. Long-term survival correlates with immunological responses in renal cell carcinoma patients treated with mRNA-based immunotherapy. Oncoimmunology. 2016;5(5):e1108511. doi: 10.1080/2162402X.2015.1108511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Su Z, Dannull J, Heiser A, et al. Immunological and clinical responses in metastatic renal cancer patients vaccinated with tumor RNA-transfected dendritic cells. Cancer Res. 2003;63(9):2127–33. [PubMed] [Google Scholar]
- 99.Geiger C, Regn S, Weinzierl A, Noessner E, Schendel DJ. A generic RNA-pulsed dendritic cell vaccine strategy for renal cell carcinoma. J Transl Med. 2005;3:29. doi: 10.1186/1479-5876-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dannull J, Su Z, Rizzieri D, et al. Enhancement of vaccine-mediated antitumor immunity in cancer patients after depletion of regulatory T cells. J Clin Invest. 2005;115(12):3623–33. doi: 10.1172/JCI25947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu H, Zheng X, Zhang S, et al. Tumor antigens and immune subtypes guided mRNA vaccine development for kidney renal clear cell carcinoma. Mol Cancer. 2021;20(1):159. doi: 10.1186/s12943-021-01465-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.World Health Organization. Cancer. 2022. https://www.who.int/health-topics/cancer#tab=tab_3. Accessed 01 Aug 2022.
- 103.Nabors LB, Portnow J, Ahluwalia M, et al. Central nervous system cancers, version 3.2020, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Netw. 2020;18(11):1537–70. doi: 10.6004/jnccn.2020.0052. [DOI] [PubMed] [Google Scholar]
- 104.Caruso DA, Orme LM, Neale AM, et al. Results of a phase 1 study utilizing monocyte-derived dendritic cells pulsed with tumor RNA in children and young adults with brain cancer. Neuro Oncol. 2004;6(3):236–46. doi: 10.1215/S1152851703000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a "state of the science" review. Neuro Oncol. 2014;16(7):896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ma S, Ba Y, Ji H, Wang F, Du J, Hu S. Recognition of tumor-associated antigens and immune subtypes in glioma for mRNA vaccine development. Front Immunol. 2021;12:738435. doi: 10.3389/fimmu.2021.738435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhong H, Liu S, Cao F, et al. Dissecting tumor antigens and immune subtypes of glioma to develop mRNA vaccine. Front Immunol. 2021;12:709986. doi: 10.3389/fimmu.2021.709986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rose M, Cardon T, Aboulouard S, et al. Surfaceome proteomic of glioblastoma revealed potential targets for immunotherapy. Front Immunol. 2021;12:746168. doi: 10.3389/fimmu.2021.746168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ye L, Wang L, Yang J, et al. Identification of tumor antigens and immune landscape in glioblastoma for mRNA vaccine development. Front Genet. 2021;12:701065. doi: 10.3389/fgene.2021.701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhou Q, Yan X, Zhu H, et al. Identification of three tumor antigens and immune subtypes for mRNA vaccine development in diffuse glioma. Theranostics. 2021;11(20):9775–90. doi: 10.7150/thno.61677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bidram M, Zhao Y, Shebardina NG, et al. mRNA-based cancer vaccines: a therapeutic strategy for the treatment of melanoma patients. Vaccines (Basel). 2021 doi: 10.3390/vaccines9101060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Davis LE, Shalin SC, Tackett AJ. Current state of melanoma diagnosis and treatment. Cancer Biol Ther. 2019;20(11):1366–79. doi: 10.1080/15384047.2019.1640032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li Q, Ren J, Liu W, Jiang G, Hu R. CpG oligodeoxynucleotide developed to activate primate immune responses promotes antitumoral effects in combination with a neoantigen-based mRNA cancer vaccine. Drug Des Devel Ther. 2021;15:3953–63. doi: 10.2147/DDDT.S325790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shadbad MA, Hajiasgharzadeh K, Derakhshani A, et al. From melanoma development to RNA-modified dendritic cell vaccines: highlighting the lessons from the past. Front Immunol. 2021;12:623639. doi: 10.3389/fimmu.2021.623639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.BioNTech. Pipeline & Products. 2022. https://www.biontech.com/int/en/home/pipeline-and-products/pipeline.html. Accessed 15 Nov 2022.
- 116.Evans AJ. Treatment effects in prostate cancer. Mod Pathol. 2018;31(S1):S110–21. doi: 10.1038/modpathol.2017.158. [DOI] [PubMed] [Google Scholar]
- 117.Li J, Sun Y, Jia T, Zhang R, Zhang K, Wang L. Messenger RNA vaccine based on recombinant MS2 virus-like particles against prostate cancer. Int J Cancer. 2014;134(7):1683–94. doi: 10.1002/ijc.28482. [DOI] [PubMed] [Google Scholar]
- 118.Rausch S, Schwentner C, Stenzl A, Bedke J. mRNA vaccine CV9103 and CV9104 for the treatment of prostate cancer. Hum Vaccin Immunother. 2014;10(11):3146–52. doi: 10.4161/hv.29553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zheng X, Xu H, Yi X, et al. Tumor-antigens and immune landscapes identification for prostate adenocarcinoma mRNA vaccine. Mol Cancer. 2021;20(1):160. doi: 10.1186/s12943-021-01452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. 2015;16(4):e165–72. doi: 10.1016/S1470-2045(14)71180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sebastian M, Schroder A, Scheel B, et al. A phase I/IIa study of the mRNA-based cancer immunotherapy CV9201 in patients with stage IIIB/IV non-small cell lung cancer. Cancer Immunol Immunother. 2019;68(5):799–812. doi: 10.1007/s00262-019-02315-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Papachristofilou A, Hipp MM, Klinkhardt U, et al. Phase Ib evaluation of a self-adjuvanted protamine formulated mRNA-based active cancer immunotherapy, BI1361849 (CV9202), combined with local radiation treatment in patients with stage IV non-small cell lung cancer. J Immunother Cancer. 2019;7(1):38. doi: 10.1186/s40425-019-0520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu F, Chen JX, Yang XB, et al. Analysis of lung adenocarcinoma subtypes based on immune signatures identifies clinical implications for cancer therapy. Mol Ther Oncolytics. 2020;17:241–9. doi: 10.1016/j.omto.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xu R, Lu T, Zhao J, Wang J, Peng B, Zhang L. Identification of tumor antigens and immune subtypes in lung adenocarcinoma for mRNA vaccine development. Front Cell Dev Biol. 2022;10:815596. doi: 10.3389/fcell.2022.815596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang Y, Tan H, Yu T, Chen X, Jing F, Shi H. Potential immune biomarker candidates and immune subtypes of lung adenocarcinoma for developing mRNA vaccines. Front Immunol. 2021;12:755401. doi: 10.3389/fimmu.2021.755401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ilyichev AA, Orlova LA, Sharabrin SV, Karpenko LI. mRNA technology as one of the promising platforms for the SARS-CoV-2 vaccine development. Vavilovskii Zhurnal Genet Sel. 2020;24(7):802–7. doi: 10.18699/VJ20.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Weng Y, Li C, Yang T, et al. The challenge and prospect of mRNA therapeutics landscape. Biotechnol Adv. 2020;40:107534. doi: 10.1016/j.biotechadv.2020.107534. [DOI] [PubMed] [Google Scholar]
- 128.Uddin MN, Roni MA. Challenges of storage and stability of mRNA-based COVID-19 vaccines. Vaccines (Basel). 2021 doi: 10.3390/vaccines9091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sandbrink JB, Shattock RJ. RNA vaccines: a suitable platform for tackling emerging pandemics? Front Immunol. 2020;11:608460. doi: 10.3389/fimmu.2020.608460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Paunovska K, Loughrey D, Dahlman JE. Drug delivery systems for RNA therapeutics. Nat Rev Genet. 2022;23(5):265–80. doi: 10.1038/s41576-021-00439-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tenchov R, Bird R, Curtze AE, Zhou Q. Lipid nanoparticles-from liposomes to mRNA vaccine delivery, a landscape of research diversity and advancement. ACS Nano. 2021 doi: 10.1021/acsnano.1c04996. [DOI] [PubMed] [Google Scholar]
- 132.Wu Z, Li T. Nanoparticle-mediated cytoplasmic delivery of messenger RNA vaccines: challenges and future perspectives. Pharm Res. 2021;38(3):473–8. doi: 10.1007/s11095-021-03015-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hou X, Zaks T, Langer R, Dong Y. Lipid nanoparticles for mRNA delivery. Nat Rev Mater. 2021;6(12):1078–94. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu S, Cheng Q, Wei T, et al. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR-Cas gene editing. Nat Mater. 2021;20(5):701–10. doi: 10.1038/s41563-020-00886-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kauffman KJ, Oberli MA, Dorkin JR, et al. Rapid, single-cell analysis and discovery of vectored mRNA transfection in vivo with a loxP-flanked tdTomato reporter mouse. Mol Ther Nucleic Acids. 2018;10:55–63. doi: 10.1016/j.omtn.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.De La Grange P, Dutertre M, Correa M, Auboeuf D. A new advance in alternative splicing databases: from catalogue to detailed analysis of regulation of expression and function of human alternative splicing variants. BMC Bioinform. 2007;8:180. doi: 10.1186/1471-2105-8-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.