Abstract
The narrow-headed vole, collared lemming and common vole were the most abundant small mammal species across the Eurasian Late Pleistocene steppe-tundra environment. Previous ancient DNA studies of the collared lemming and common vole have revealed dynamic population histories shaped by climatic fluctuations. To investigate the extent to which species with similar adaptations share common evolutionary histories, we generated a dataset comprised the mitochondrial genomes of 139 ancient and 6 modern narrow-headed voles from several sites across Europe and northwestern Asia covering approximately the last 100 thousand years (kyr). We inferred Bayesian time-aware phylogenies using 11 radiocarbon-dated samples to calibrate the molecular clock. Divergence of the main mtDNA lineages across the three species occurred during marine isotope stages (MIS) 7 and MIS 5, suggesting a common response of species adapted to open habitat during interglacials. We identified several time-structured mtDNA lineages in European narrow-headed vole, suggesting lineage turnover. The timing of some of these turnovers was synchronous across the three species, allowing us to identify the main drivers of the Late Pleistocene dynamics of steppe- and cold-adapted species.
Keywords: paleoclimate, climate change, interstadials, habitat, mitochondrial DNA, Pleistocene
1. Introduction
There is growing evidence that the Late Pleistocene climatic and environmental oscillations have been a major factor shaping the evolutionary histories of many mammalian species [1–3]. Climate and environmental change in the Northern Hemisphere have led to dramatic demographic and genetic changes, either causing the extinction of whole genetic lineages and species or engendering a significant change in their ranges over a short period of time and large areas [4–6]. Most of the genetic studies have focused on megafauna species, such as mammoths, bears, horses and others [2,4,7]. However, less is known about the Late Pleistocene evolutionary histories of small mammals. The notable exceptions are the ancient DNA investigations of the collared lemming (Dicrostonyx torquatus) [8–11] and the common vole (Microtus arvalis) [12,13]. These studies revealed that both species experienced several mtDNA lineage replacements across most of the European continent during marine isotope stage (MIS) 3 and MIS 2 [8,9,13].
The narrow-headed vole (genus Stenocranius Kashchenko, 1901) is distinguished from all other voles by a high and narrow brain capsule. Stenocranius was originally proposed as a subgenus of Microtus Schrank, 1798 but in recent years phylogenetic reconstructions supported the sister relationship of Stenocranius and Lasiopodomys Lataste, 1887 [14] and Stenocranius has been accepted as a subgenus of the latter. However, the fossil record suggests an early split (ca 1.2 Ma) of Stenocranius from the Allophaiomys stock [15] and recent mitogenomic analyses showed divergence of Stenocranius and Lasiopodomys similar to the divergence between many genera within Arvicolini [16]. We, therefore, follow Kryštufek & Shenbrot [17] and recognize a genus status for Stenocranius. Narrow-headed vole are presently only found in Asia, in the belt of arctic tundra and forest-tundra in the north and the steppes and forest-steppes at the centre of the continent. These animals inhabit open areas, such as tundra, steppes and meadows [17]. Their current distribution in Asia is considered the result of habitat fragmentation during the Holocene [18]. The range of Stenocranius was much larger when treeless steppe-tundra was the dominant biome during the Pleistocene and included most of Northern Eurasia. The westernmost part of their former range reached the British Isles [19] and southern France [20]. Recent genetic investigations have shown that the Pleistocene narrow-headed voles were a complex of three cryptic species, including Stenocranius anglicus (Hinton, 1910) in Europe [21] and Stenocranius gregalis (Pallas, 1779) and Stenocranius raddei (Poljakov, 1881) in Asia [22]. The divergence of these three lineages was estimated to have occurred between 250 and 200 kyr ago by adopting the substitution rate of mtDNA cytochrome b from other vole species [21]. These substitution rates were estimated using recent calibration points, either directly radiocarbon-dated specimens [23] or biogeographic events [24]. Despite this, the estimated divergence time of the European and Asiatic populations has challenged the widely accepted biogeographic hypothesis, assuming that all narrow-headed-voles retreated from Europe during the interglacial periods and suggested that they survived the Eemian interglacial (MIS 5e, ca 128–115 ka) in European refugia [21].
The narrow-headed vole, and other rodents adapted to open habitats and/or cold climates, such as the collared lemming and common vole, frequently made up to 90% of the Late Pleistocene small mammal assemblages across Western and Central Europe [20,25–27] and constitute a key element of steppe-tundra environments. Investigations into their evolutionary and phylogeographic history would enhance our understanding of species' responses to climate change and demonstrate to what extent species with similar environmental requirements share the same evolutionary pathways.
Here, we generated more than 100 mitogenomes from Late Pleistocene skeletal remains and reconstructed time-calibrated phylogenies to explore the putative effects of climatic and environmental changes on the dynamics of narrow-headed vole populations.
2. Material and methods
(a) . Samples
Specimens identified as Stenocranius sp. based on the morphology of the occlusal surface of the first lower molar were obtained from several sites across Europe and Asia. Before the genetic analyses the occlusal surface of the m1 tooth was photographed at the Institute of Systematic and Evolution of Animals, Polish Academy of Science.
(b) . DNA extraction, library preparation, enrichment and sequencing
Genetic analyses were performed in the Laboratory of Paleogenetics and Conservation Genetics, Centre of New Technologies, University of Warsaw. To minimize the probability of contamination, all pre-polymerase chain reaction (PCR) library preparation steps were carried out in a dedicated ancient DNA laboratory separated from the post-PCR area. This laboratory has positive air pressure, and is UV irradiated after each use; researchers wore full protective suits.
Vole teeth were flushed twice with ultra-pure water in a sterile tube and crushed with a pipette tip. The DNA was extracted using a silica bead-based protocol optimized for retrieving ultrashort DNA molecules [28,29]. A negative control was processed with each batch of 15 samples to monitor possible contamination. One-third of each DNA extract was converted into double-indexed and either double- or single-stranded DNA sequencing libraries following the protocols proposed by [30] or [31], respectively. In each case, the double indexing scheme was introduced during 19 cycles of indexing PCR, using either AmpliTaq Gold or AccuPrime Pfx DNA Polymerase.
The libraries were enriched for vole mtDNA using a custom DNA bait produced from a range of vole species and following a protocol proposed by [32]. Up to five libraries were pooled in a single enrichment reaction which was carried out twice at 65°C for 22–24 h. After each round, the library pools were amplified in triplicate for 10–15 cycles using Herculase II Fusion Polymerase. The enriched library pools were combined, quantified using Qubit 4 and sequenced on an Illumina NextSeq550 platform (MID output, 2 × 75 bp kit). Additional details are provided in the electronic supplementary material, text S1.1–1.4 and table S1.
(c) . Processing the sequencing data
Sequencing reads were demultiplexed using bcl2fastq. Overlapping reads were collapsed and adaptor and quality trimmed using AdapterRemoval v. 2 [33]. The reads were mapped to the S. gregalis mtDNA genome (MN199170.1) using the bwa mem algorithm with the default settings. As the mapping efficiency was low due to high sequence divergence between extant Asiatic and ancient European specimens, we constructed two additional mtDNA references using the most complete sequences of European S. anglicus. In some cases, we performed competitive mapping using concatenated vole and human mtDNA genomes as a reference to remove human DNA contamination [34]. Duplicates and low mapping quality (mapq < 30) reads were removed using samtools. Variants and consensus sequences were called using the bcftools mpileup and ivar call commands. Positions with coverage lower than 3 were masked, and bases supported by less than 75% of the reads were coded with the appropriate IUPAC symbol. MapDamage v. 2.08 [35] was used to assess the damage patterns and length distribution of the DNA molecules. Additional details are provided in electronic supplementary material, text S1.5.
(d) . Radiocarbon dating
We attempted to radiocarbon date 12 specimens ranging in weight from 21.5 to 102 mg. Collagen was extracted and quality was assessed in the Department of Human Evolution at the Max Planck Institute for Evolutionary Anthropology (Leipzig, Germany) following the protocol for less than 100 mg bone samples described in [36]. Samples were combusted and converted to graphite using the AGE system [37] and dated using the MICADAS system [38] Radiocarbon dates were calibrated in OxCal v. 4.4 [39] using the IntCal20 [40] calibration curve. See electronic supplementary material, text S1.6 for further details.
(e) . Data analysis
The mtDNA sequences of the ancient and modern narrow-headed vole were aligned using MAFFT v. 7.407 [41]. The best-fitting partitioning scheme was found using PartitionFinder v. 2 [42]. Phylogenetic analyses were performed in BEAST v. 1.10.4 [43]. We used the Bayesian evaluation of temporal signal (BETS) [44] and the date-randomisation test [45] on the dataset consisting of all dated and modern sequences (n = 17) to determine whether there was enough temporal resolution to calibrate the molecular clock. Next, we performed a leave-one-out analysis on the directly dated specimens to check the accuracy of the molecular age estimate. In this analysis, we estimated the age of each directly dated specimen using all of the other dated specimens to calibrate the molecular clock. Then, we estimated the age of each undated specimen separately using directly dated specimens to calibrate the molecular clock. In each of the 128 separate BEAST analyses, we set the gamma prior (shape = 2; scale = 50 000) on the age of the undated specimen and increased the operator weight of the age estimate to 5. Finally, we ran a joint analysis using all of the sequences and setting the lognormal priors on the ages of undated specimens. The priors were set to match the posteriors of the age estimates from the individual analyses. See electronic supplementary material, text S1.7–1.8 and tables S2–S6 for more details.
To include the full previously recognized mtDNA diversity of Asiatic narrow-headed voles, we also reconstructed the phylogeny using an 882 bp fragment of mtDNA cytochrome b. This dataset was comprised 311 sequences (139 from ancient and 172 from extant specimens). See electronic supplementary material, text S1.9 and tables S7–S9, S11, for more details.
3. Results
We recovered near complete (greater than 70% sites with a minimum coverage of 3 reads) mtDNA genomes from 139 specimens from 35 sites across Europe and western Asia. Together with the sequences of extant voles available from repositories, our dataset consisted of 145 mitogenomic sequences (electronic supplementary material, tables S8 and S9; figure 1). All ancient specimens yielded short inserts and an elevated level of deamination at the ends of the DNA molecules typical of ancient DNA (electronic supplementary material, table S8). Of the 12 specimens large enough for radiocarbon dating, 11 yielded collagen of sufficient quantity and quality for AMS dating (electronic supplementary material, table S10). The mtDNA sequences of these 11 specimens and the six extant specimens constituted a directly dated dataset used to estimate the ages of the remaining specimens. BETS analysis and the date-randomization test indicated that, the ‘dated’ dataset had a sufficient temporal signal to calibrate the molecular clock (electronic supplementary material, text S1.7, figure S1 and table S3). In the leave-one-out analysis, the 95% highest posterior density (HPD) intervals of the ages estimated for 10 of 11 specimens overlapped with their radiocarbon age. In most cases, the estimates were accurate, with the difference between the medians of the estimated ages and the radiocarbon ages ranged between 0.4% and 35% (median difference, 4.6%) of the latter (electronic supplementary material, figure S2). As a result, the age of the undated specimens estimated using the molecular approach was usually highly consistent with the stratigraphic position of the specimens and with the age estimates obtained for other species from the same sites and layers. The exception was some of the European specimens from the Early Holocene layers whose ages were slightly but systematically overestimated compared to their stratigraphic position (electronic supplementary material, table S8). However, the youngest estimated ages (ca 14 ka; electronic supplementary material, table S1), corresponded well with the beginning of the Bølling–Allerød interstadial (14.7–12.9 ka), suggesting earlier extinction of the narrow-headed vole from central Europe. We also note that in the final joint phylogenetic analysis of the 145 mitogenomes the MCMC chains for age estimates of six specimens did not converge (effective sample size [ESS] < 200) in both replicates. However, the age parameters were reliable (ESS > 200) in individual analyses of all of these samples, and yielded values very similar to those obtained in the final joint analysis (electronic supplementary material, text S1.8 and table S6); thus, we consider these estimates to be reliable.
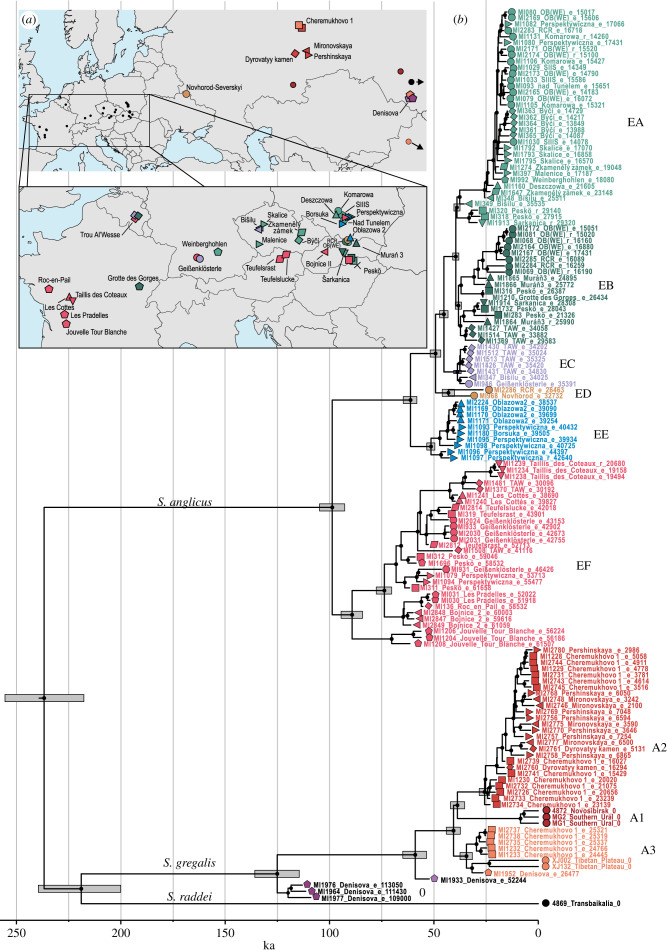
Figure 1.
Phylogeny of last glacial period narrow-headed voles. (a) Sampling locations for the vole specimens. Colours and symbols correspond to mtDNA lineages and sampling sites. (b) Tip-calibrated phylogeny of narrow-headed voles based on the mtDNA genomes. Dots at nodes indicate posterior probability greater than 0.95. Grey bars represent 95% HPD intervals of the estimated divergence times. Tips are annotated with sample ID, sampling site and median of estimated (‘e’ prefix) or radiocarbon age (‘r’ prefix). Sites abbreviations: TAW, Trou Al'Wesse; RCR, Rock overhang in Cisowa Rock; SIIIS, Shelter in Smoleń III; OB(WE), Obłazowa (Western Entrance).
(a) . Diversification of the narrow-headed vole mtDNA lineages
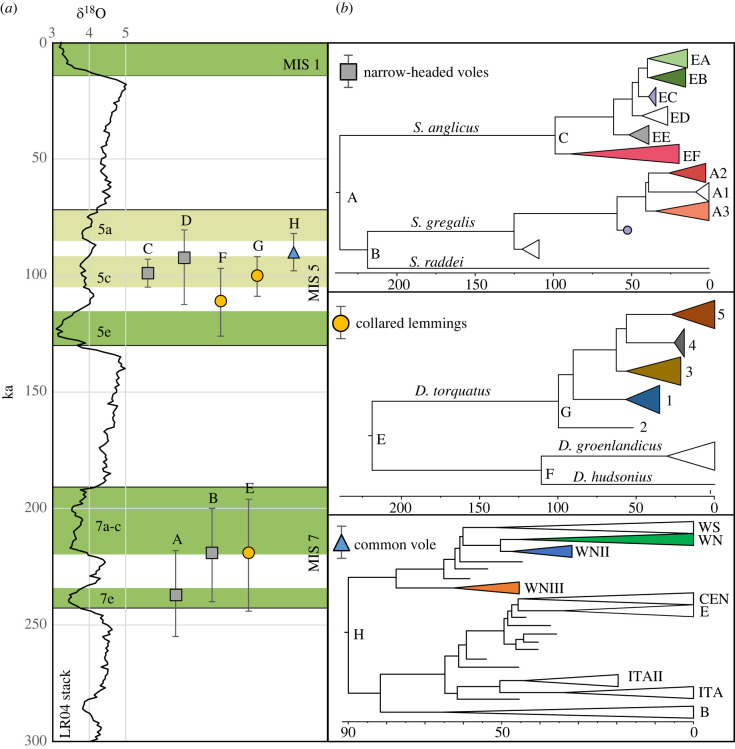
The Bayesian phylogeny based on mitochondrial genomes recovered three divergent lineages corresponding to cryptic species identified previously, including S. anglicus in Western and Central Europe, and S. gregalis and S. raddei in Asia [21,22,46]. The three lineages diverged at similar times during the MIS 7 interstadial (243–191 ka). The divergence of the S. anglicus and S. gregalis/S. raddei lineages was estimated to be 233 ka (95% HPD: 253–213 ka) and that of the S. gregalis and S. raddei lineages was 216 ka (95% HPD: 236–193 ka; figure 1).
The S. anglicus lineage further diversified about 98 ka (95% HPD: 104–92 ka) during MIS 5c (∼GS-23; ca 104–88 ka). Our mitogenomic tree does not reflect the complete diversity of Asiatic S. gregalis, but the tree based on the mtDNA cytochrome b and including all extant lineages of this species had a similar tMRCA of 95 ka (95% HPD: 112–80 ka; electronic supplementary material, text S1.9, tables S11 and S12, figure S3). The three specimens from layers 14 and 12.3 of the Denisova cave with ages estimated using the molecular approach to be between 116 and 107 ka fell outside the mtDNA diversity of all of the last glacial period (ca 113–11.7 ka) S. gregalis specimens. The estimated age of the specimen from layer 12.3 was in very good agreement with the dating of this layer (146–121 ka), while the estimated ages of the two specimens from layer 14 were slightly younger than the estimated time of deposition of this layer (202–163 ka) [47].
(b) . Temporal population structure and dynamics of the Asiatic narrow-headed vole
The geographical scope of our S. gregalis mitogenomic dataset was limited and included four sites in the Northern and Middle Urals and one in Altai. However, the phylogeny based on mtDNA cytochrome b, which included 172 modern specimens, was consistent with previous studies and consisted of three main lineages (A–C). Lineage A was further subdivided into six subclades (A1–A6) (electronic supplementary material, figure S3). The diversification of these sublineages was estimated to occur between 51 and 28 ka. A single specimen from layer 11.3 in Denisova cave was placed in a sister position to the A lineage. Its age was estimated to be about 52 (95% HPD: 58–46) ka only slightly younger than the estimated deposition period of this layer (100–61 ka). Among the ancient specimens, those older than ca 23 ka belonged to lineage A3, which is currently limited to Altai. Lineage A3 was replaced by A2 ca 23 ka in the Northern and Middle Urals and the latter persisted there until the Middle Holocene. Currently, A2 occurs mostly to the north, on the Yamal Peninsula (electronic supplementary material, figure S4). This finding supports previous reports suggesting that the range of Asiatic narrow-headed voles became fragmented during the Holocene [18].
(c) . Temporal population structure and dynamics of the European narrow-headed vole
We observed six S. anglicus mtDNA lineages in Europe, which we labelled EA–EF (figure 1). The EF lineage was distributed across most of Europe since at least 60 ka and until ca 44–42 ka (figures 1 and 3). Then, its range decreased and eventually, it was limited to the territory of what is now southwestern France where it persisted until at least 17 ka. Lineage EE had a limited geographical range, as it was only found at three sites from what is modern-day Poland (Obłazowa 2, Perspektywiczna layer 7c and Borsuka layer VI) among specimens with ages estimated to be between 44.4 and 38.5 ka. Lineage ED was attributed to only two specimens from two sites in what is modern-day Poland and Ukraine (Novgorod Severskyi and a rock overhang at Cisowa Rock). In both cases, the estimated ages were significantly older than suggested by the site's stratigraphy. Lineage EC was found in western and central Europe (Trou Al'Wesse, Geißenklösterle and Bišilu) in specimens with ages estimated to be between 35.4 and 34.2 ka. Lineages EB and EA diverged about 40 ka. The oldest specimens from these two lineages had similar ages and came from western (Trou Al'Wesse, Belgium, ca 34 ka) and central Europe (Bišilu, Czechia, ca 35 ka), respectively. Lineage EB appeared in central Europe 28 ka when its range started to overlap with the EA lineage, which remained restricted to central Europe.
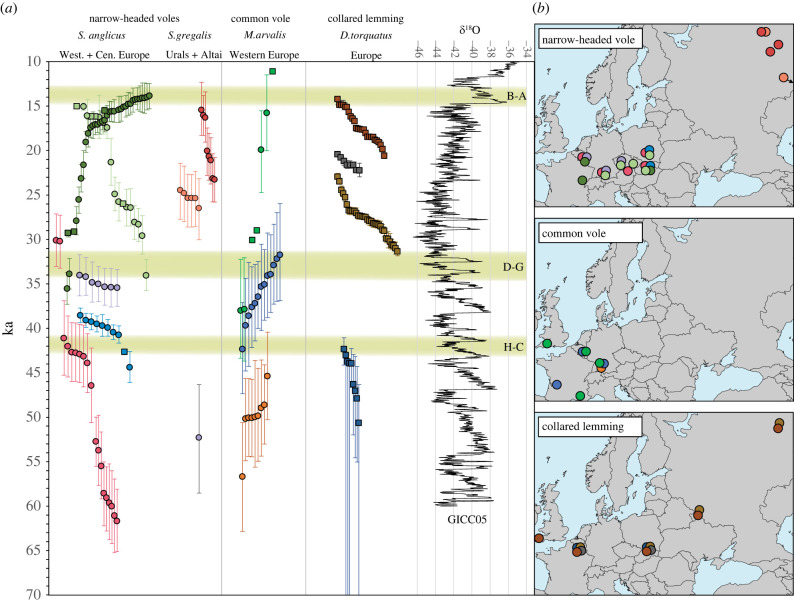
Figure 3.
Temporal distribution of the mtDNA lineages across four steppe- and cold-adapted rodent species (a). Circles represent medians and whiskers are the 95% HPDs of the specimen ages estimated using the tip-dating approach; squares represent medians of the calibrated radiocarbon ages with 95.4% probability ranges. In the case of narrow-headed vole data from the present-day western France, where lineage EF was present continuously, was omitted. The phylogenetic relationships between the presented mtDNA lineages are shown in figure 2b. Data for the collared lemming are from [8,9] and for the common vole from [12]. The oxygen isotope record (δ18O V-SMOW) from the Greenland Ice cores (GICC05) [62] is given on the right side. Green stripes denote the main interstadials identified from the palynological records within the last 50 kyr (H-C: Hengelo-Charbon; D-G: Denekamp-Grand Bois; B-A: Bølling–Allerød). (b) Geographical ranges of the mtDNA lineages presented in (a). Nearby localities were merged for better visibility.
4. Discussion
(a) . Impact of the interglacial periods on cold-adapted small mammals
Our results based on ancient mitogenomic data indicate that the divergence of the three main narrow-headed vole lineages occurred during the MIS 7 interstadial (243–191 ka). These divergence estimates are much younger than those of S. gregalis and S. raddei based on the fossil calibration [16,46]. This is likely to be due to time dependence of molecular rate estimates [48]. There is compelling evidence that the estimates of evolutionary rates are negatively correlated with the age of calibration points and the use of fossil calibration may lead to overestimation of recent divergence times [49].
Our estimates are very similar to the estimated divergence of the two main lineages of collared lemming, Eurasian (D. torquatus) and North American (D. groenlandicus/D. hudsonius), based on ancient mitogenomes (figure 2). The available palynological records from MIS 7 suggest the expansion of mixed coniferous-broadleaved forest across Europe, from southern France [51] to the Eastern European Plain [52] possibly forcing narrow-headed voles to retreat to refugia in Europe and Asia, although the location of these areas is unknown. The sea level during the warm phases of MIS 7 (MIS7a–c, e) was only marginally lower than it is today [53], resulting in flooding of the Bering Strait and the separation of the Eurasian and American collared lemming populations.
Figure 2.
Divergence times of the main lineages within the three species groups. (a) Divergence times (medians and 95% HPDs) plotted against the benthic foraminifera δ18O record (LR04 stack; [50]). Interglacial and interstadial periods mentioned in the text are marked with green strips. (b) Schematic mitochondrial phylogenies of the narrow-headed voles, collared lemmings and common vole. Letters at the nodes correspond to the divergence times in (a). The age of diversification of modern S. gregalis lineages (D) was taken from the mtDNA cytochrome b phylogeny (see electronic supplementary material, figure S3 and text S1.9). Symbols at tips indicate the main mtDNA lineages within each species. Colours indicate mtDNA lineages presented in figure 3. Data for collared lemming are from [11] and for common vole from [12].
The estimated age of diversification of the main mtDNA lineages found in the European and Asiatic narrow-headed voles from the last glacial period was similar to the estimated diversification of the main lineages within common voles [12] and Eurasian collared lemmings [11] that occurred during MIS 5c (Brørup interstadial; ∼GI 23) (figure 2).
Temperate deciduous vegetation developed across Europe during Brørup (MIS 5c) [54,55]. The presence of Quercus followed by Carpinus woodlands was evidenced in France. A high percentage of Betula pollen was detected in Northern Germany, followed by Pinus with admixtures of Picea and Larix. In central Europe, the development of Betula forests followed by Betula–Pinus and Pinus–Picea–Larix phases has been observed in the palynological records. The development of dense forests over vast areas of Europe may have led to fragmentation of the populations adapted to open, steppe habitats, leading to the divergence and formation of new lineages.
Taken together, these observations suggest a common response of species adapted to open habitats in interglacial environments. Similar contraction during the Eemian interglacial period followed by expansion during the last glacial period was recently identified in a range of steppe insect and plant species [56].
As mentioned in the Introduction, the long-held notion that narrow-headed voles withdrew completely from Europe during the Eemian (and penultimate interglacial periods) cannot be maintained due to the high divergence of the European narrow-headed voles from Asian populations shown previously [21]. Narrow-headed voles can theoretically survive unfavourable interglacial conditions in mountain/upland areas or in the far north, where open and relatively humid environments persisted. However, the fossil record does not provide evidence for the species in Europe at this time, despite the examination of over 90 sites of Eemian age in Europe [57]. S. cf anglicus was only found in two sites in Hungary and one in Poland although Eemian dates could not be confirmed. By contrast, S. anglicus appeared in great numbers at the very beginning of the last glaciation during MIS 5d–a in various parts of Europe, such as the Rhone Valley and the Massif Central in France [58], the Ach Valley in Germany [59] and the Kraków-Częstochowa Upland of Poland [60]. This may suggest that the narrow-headed vole survived the Eemian interglacial in several refugia, and therefore their rapid re-colonization of Europe was possible as soon as the climatic cooling offered a suitable environment for its expansion. This would confirm the proposed idea of cryptic refugia for continental adapted species in more oceanic areas in the past [61].
(b) . Narrow-headed vole population dynamics during the last glacial period
Our phylogenetic reconstruction suggests a highly dynamic history of the European narrow-headed vole through the last glacial period. We identified six mtDNA lineages in western and central Europe. Most of them appeared consecutively within the limits of our resolution, suggesting genetic turnover across the region. Some of the lineages (e.g. EE) had limited geographical distributions. This is likely to be an effect of sampling bias and the short temporal range of these lineages. Nevertheless, the full geographical scope of the recorded turnovers remains to be described. Most of the turnover in the mtDNA lineages of the European narrow-headed vole, the collared lemming and the common vole occurred at the end of MIS 3 between ca 45 and 30 ka (figure 3). The limited data available for the Asiatic S. gregalis, with divergence times for the A1–A6 lineages estimated to be between 51 and 28 ka, also suggest a major reorganization of genetic diversity during this period. This is broadly consistent with the cluster of megafaunal extinctions and mtDNA lineage turnovers reported previously at the end of MIS 3, between 40 and 26 ka [5,63–65], including that of the Neanderthals [66]. The main cause of the megafauna turnovers has been argued to be the abrupt warming at the onset of the Greenland Interstadials (GI), particularly around GI7–GI5. The role of humans, exacerbating the impact of climate change, has also been emphasized [5,65], as no turnovers were detected before modern humans appeared in Europe ca 45 ka. The direct impact of Palaeolithic humans on the vole and lemming populations across Eurasia has been considered negligible. The concurrence of mtDNA lineage turnovers in small and large mammals at the end of MIS 3 suggest that the climate and environmental conditions may have been the sole drivers of the observed population dynamics.
The resolution of the molecular estimates of the ages of the specimens is limited, although a comparison of the temporal distribution of the mtDNA lineages of the three rodent species suggests that populations of all three rodent species were affected at similar times about 45–40 ka and about 32 ka (figure 3). These findings generally coincide with the timing of the interstadials identified in the European pollen records; Hengelo–Charbon dated to about 43–41 ka cal BP [54,67] and Denekamp–Grand Bois dated to about 36–33 ka cal BP [54,55]. The correlation of these interstadials with the Greenland ice core records is challenging due to the relatively wide date error ranges and that Hengelo–Charbon is usually associated with Greenland Interstadial 11 (GI-11; ca 43.3–42.2 ka) or GI-10 (ca 41.5–40.8 ka) and Denekamp–Grand Bois is associated with GI-8 (38.2–36.6 ka) [54,68]. Both interstadials were characterized by the emergence of forests, although especially during the Hengelo–Charbon period the landscape remained relatively open albeit the steppe-tundra biome significantly altered [54,55]. Another factor that may have contributed to the observed dynamics was a period of weakend magnetic field before the Laschamps geomagnetic inversion. It has been suggested as a cause of major environmental rearrangements ca 42 ka. A similar paleomagnetic anomaly, the Mono Lake event, that occurred about 34 ka was hypothesized to cause similar effects [69]. Nevertheless, the main cause of these events, the synchronous faunal turnovers and local extinctions observed across species of different sizes and occupying different trophic levels, support a major ecosystem rearrangement at that time.
Following the theory of a major impact of rapid environmental change on animal populations, few megafaunal turnovers were recorded around the Last Glacial Maximum (LGM; ca 23–19 ka), when the climate was harsh but stable compared to the end of MIS 3 [5]. By contrast, studies on collared lemmings have previously shown two mtDNA lineage turnovers during the LGM [8,9]. Our data suggest another turnover in the S. gregalis populations from the Northern and Middle Urals, estimated to be about 24–23 ka. No such turnover was visible in European populations, but we noted that all specimens younger than 21 ka coalesced around 24 ka, suggesting a reduction in the population followed by expansion around this time. The causes of these turnovers remain unclear. However, there are reports, confirmed by direct radiocarbon dating, of species described by some as temperate such as brown bears (Ursus arctos) at medium and high latitudes in Europe during the LGM [2,70]. It has been hypothesized that a steppe-tundra environment may have been replaced by more temperate habitat for short periods [2]. Some climate variability between 27 and 21 ka cal BP, which was not apparent in the Greenland δ18O records, was detected in the Nussloch loess sequence, indicating several short phases of milder climate [68]. The accumulation of radiocarbon dates from organic material reflecting a warmer period about 23 ka was also detected in metanalysis of radiocarbon dates from Polish sites [71]. This may explain the published results of brown bear and collared lemming as well as those for the narrow-headed vole reported here.
Interestingly, the highly dynamic history of the narrow-headed vole populations observed in central and part of western Europe did not extend to what is now western France, where lineage EF was continuously present from at least 61 to 19 ka and plausibly until the extirpation of narrow-headed voles from Europe at the beginning of Late Glacial, and/or the Pleistocene–Holocene transition. This observation suggests that during MIS 3 and MIS 2 the environmental conditions in this region were more stable and/or favourable for narrow-headed voles than in other parts of Europe. Unfortunately, no genetic data of other small mammalian species are available from the region to determine whether the observed population continuity was a common phenomenon across open habitat and/or cold-adapted species. A recent reconstruction, based on the small mammal fossil record, revealed the general stability of the rodent communities between 50 and 19 ka in the region [20], although such reconstructions have limited power to detect rapid population turnover.
Acknowledgements
We thank Prof. Aleksandr V. Borodin (IPAE UB RAS) for his help in obtaining samples. We also acknowledge the late Rebbeca Miller, director of Trou Al'Wesse excavations and the Agence Wallonne du Patrimoine (AWaP), as the main funding institution of the work at the site. All samples were studied with the permission of the relevant authorities.
Contributor Information
Mateusz Baca, Email: m.baca@cent.uw.edu.pl.
Adam Nadachowski, Email: nadachowski@isez.pan.krakow.pl.
Data accessibility
The consensus mtDNA sequences generated in this study have been deposited in GenBank under Accession numbers OM809882–OM810020. The mtDNA sequence alignments used to reconstruct the mitogenome and cytochrome phylogenies have been deposited in Dryad Digital Repository and can be accessed at https://doi.org/10.5061/dryad.kh1893295 [72]. The mitochondrial alignments generated in this study have been deposited in the European Nucleotide Archive under project number PRJEB56307.
The data are provided in electronic supplementary material [73].
Authors' contributions
M.B.: conceptualization, data curation, formal analysis, funding acquisition, investigation, supervision, visualization, writing—original draft; D.P.: data curation, investigation, writing—review and editing; A.K.A.: resources; K.B.: investigation; N.J.C.: resources; H.F.: data curation, investigation, writing—review and editing; T.F.: resources; M.G.: investigation; I.H.: resources, writing—review and editing; M.V.K.: resources; Mag.K.: resources, writing—review and editing; Mar.K.: investigation, resources, writing—review and editing; L.L.: resources; A.L.: investigation, resources, writing—review and editing; L.C.M.: resources, writing—review and editing; D.N.: resources; P.N.: resources; J.P.: resources; L.R.: resources; S.E.R.: resources; A.R.: resources, writing—review and editing; N.V.S.: resources; M.S.: resources; J.R.S.: resources, writing—review and editing; T.S.: resources; S.T.: methodology, resources; J.W.: resources; A.N.: conceptualization, funding acquisition, project administration, resources, supervision, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We have no competing interests.
Funding
This research was supported by the Polish National Science Centre grants nos: 2017/25/B/NZ8/02005 to A.N. and 2020/39/B/NZ8/02853 to M.B. Fieldwork at Roc-en-Pail (France) was granted by the French Ministry of Culture (MCC) through the Pays-de-la-Loire Regional Archaeology Service (DRAC/SRA) and in 2016 by the Mécène & Loire Foundation (http://www.mecene-et-loire.fr/). Fieldwork at Teufelslucken (Austria) was granted by the Geological Survey of Lower Austria (grant no. BD1-G-5306/001-2019). Marie Soressi was supported by the NWO VICI award (VI.C.191.O70). Mar.K. was supported by Russian Science Foundation grant 22-14-00332. Loïc Lebreton has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 101034349 and the State Research Agency of the Spanish Ministry of Science and Innovation through the Program Maria de Maeztu Unit of Excellence (grant no. CEX2019-000945-M).
References
- 1.Campos PF, et al. 2010. Ancient DNA analyses exclude humans as the driving force behind late Pleistocene musk ox (Ovibos moschatus) population dynamics. Proc. Natl Acad. Sci. USA 107, 5675-5680. ( 10.1073/pnas.0907189107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ersmark E, et al. 2019. Genetic turnovers and northern survival during the last glacial maximum in European brown bears. Ecol. Evol. 9, 5891-5905. ( 10.1002/ece3.5172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palkopoulou E, et al. 2013. Holarctic genetic structure and range dynamics in the woolly mammoth. Proc. R. Soc. B 280, 20131910. ( 10.1098/rspb.2013.1910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lorenzen ED, et al. 2011. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature 479, 359-364. ( 10.1038/nature10574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper A, Turney C, Hughen KA, Barry W, McDonald HG, Bradshaw CJA. 2015. Abrupt warming events drove Late Pleistocene Holarctic megafaunal turnover. Science 349, 1-8. ( 10.1126/science.aac4315) [DOI] [PubMed] [Google Scholar]
- 6.Baca M, Nadachowski A, Lipecki G, Mackiewicz P, Marciszak A, Popović D, Socha P, Stefaniak K, Wojtal P. 2017. Impact of climatic changes in the Late Pleistocene on migrations and extinction of mammals in Europe: four case studies. Geol. Q. 61, 291-304. ( 10.1101/090878) [DOI] [Google Scholar]
- 7.Fellows YJA, et al. 2017. Central European Woolly Mammoth population dynamics: insights from Late Pleistocene mitochondrial genomes. Sci. Rep. 7, 17714. ( 10.1038/s41598-017-17723-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brace S, et al. 2012. Serial population extinctions in a small mammal indicate Late Pleistocene ecosystem instability. Proc. Natl Acad. Sci. USA 109, 20 532-20 536. ( 10.1073/pnas.1213322109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palkopoulou E, et al. 2016. Synchronous genetic turnovers across Western Eurasia in Late Pleistocene collared lemmings. Glob. Change Biol. 22, 1710-1721. ( 10.1111/gcb.13214) [DOI] [PubMed] [Google Scholar]
- 10.Prost S, Smirnov N, Fedorov VB, Sommer RS, Stiller M, Nagel D, Knapp M, Hofreiter M. 2010. Influence of climate warming on arctic mammals? New insights from ancient DNA studies of the collared lemming Dicrostonyx torquatus. PLoS ONE 5, e10447. ( 10.1371/journal.pone.0010447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lord E, et al. 2022. Population dynamics and demographic history of Eurasian collared lemmings. BMC Ecol. Evol. 22, 126. ( 10.1186/s12862-022-02081-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baca M, et al. 2023. Ancient DNA reveals interstadials as a driver of the common vole population dynamics during the last glacial period. J. Biogeogr. 50, 183-196. ( 10.1111/jbi.14521) [DOI] [Google Scholar]
- 13.Baca M, et al. 2020. Diverse responses of common vole (Microtus arvalis) populations to Late Glacial and Early Holocene climate changes – evidence from ancient DNA. Quat. Sci. Rev. 233, 106239. ( 10.1016/j.quascirev.2020.106239) [DOI] [Google Scholar]
- 14.Abramson NI, Lebedev VS, Tesakov AS, Bannikova AA. 2009. Supraspecies relationships in the subfamily Arvicolinae (Rodentia, Cricetidae): an unexpected result of nuclear gene analysis. Mol. Biol. 43, 834-846. ( 10.1134/s0026893309050148) [DOI] [PubMed] [Google Scholar]
- 15.Maul LC, Markova AK. 2007. Similarity and regional differences in Quaternary arvicolid evolution in Central and Eastern Europe. Quat. Int. 160, 81-99. ( 10.1016/j.quaint.2006.09.010) [DOI] [Google Scholar]
- 16.Abramson NI, Bodrov SY, Bondareva OV, Genelt-Yanovskiy EA, Petrova TV. 2021. A mitochondrial genome phylogeny of voles and lemmings (Rodentia: Arvicolinae): evolutionary and taxonomic implications. PLoS ONE 16, 1-28. ( 10.1371/journal.pone.0248198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kryštufek B, Shenbrot G. 2022. Voles and lemmings (Arvicolinae) of the palaearctic region. Maribor, Slovenia: University of Maribor, University Press. [Google Scholar]
- 18.Smirnov NG, Kuz'mina E, Golovachev IB, Fadeeva TV. 2007. The narrow-skulled vole (Microtus gregalis Pall.) in the dynamics of zonal rodent communities of northern Eurasia. Russ. J. Ecol. 38, 106-111. ( 10.1134/S1067413607020075) [DOI] [Google Scholar]
- 19.Suttcliffe AJ, Kowalski K. 1976. Pleistocene rodents of the British Isles. Bull. Br. Mus. (Nat. Hist.) Geol. 27, 31-147. [Google Scholar]
- 20.Royer A, Montuire S, Legendre S, Discamps E, Jeannet M, Lécuyer C. 2016. Investigating the influence of climate changes on rodent communities at a regional-scale (MIS 1–3, Southwestern France). PLoS ONE 11, e0145600. ( 10.1371/journal.pone.0145600) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baca M, Popović D, Lemanik A, Baca K, Horáček I, Nadachowski A. 2019. Highly divergent lineage of narrow-headed vole from the Late Pleistocene Europe. Sci. Rep. 9, 17799. ( 10.1038/s41598-019-53937-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrova T V., Tesakov AS, Kowalskaya YM, Abramson NI. 2016. Cryptic speciation in the narrow-headed vole Lasiopodomys (Stenocranius) gregalis (Rodentia: Cricetidae). Zool. Scr. 45, 618-629. ( 10.1111/zsc.12176) [DOI] [Google Scholar]
- 23.Martínková N, et al. 2013. Divergent evolutionary processes associated with colonization of offshore islands. Mol. Ecol. 22, 5205-5220. ( 10.1111/mec.12462) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herman JS, Searle JB. 2011. Post-glacial partitioning of mitochondrial genetic variation in the field vole. Proc. R. Soc. B 278, 3601-3607. ( 10.1098/rspb.2011.0321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhodes SE, Conard NJ. 2021. A quantitative paleoclimatic reconstruction of the non-analogue environment of oxygen isotope stage 3: new data from small mammal records of southwestern Germany. Archaeol. Anthropol. Sci. 13, 216-234. ( 10.1007/s12520-021-01363-8) [DOI] [Google Scholar]
- 26.Nadachowski A. 1989. Origin and history of the present rodent fauna in Poland based on fossil evidence. Acta Theriol. 34, 37-53. ( 10.4098/at.arch.89-2). [DOI] [Google Scholar]
- 27.Horáček I, Ložek V. 1988. Palaeozoology and the Mid-European Quaternary past: scope of the approach and selected results. Rozpravy ČSAV, r. MPV (Praha) 98(4), 1-106. [Google Scholar]
- 28.Dabney J, et al. 2013. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl Acad. Sci. USA 110, 15 758-15 763. ( 10.1073/pnas.1314445110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohland N, Glocke I, Aximu-Petri A, Meyer M. 2018. Extraction of highly degraded DNA from ancient bones, teeth and sediments for high-throughput sequencing. Nat. Protoc. 13, 2447-2461. ( 10.1038/s41596-018-0050-5) [DOI] [PubMed] [Google Scholar]
- 30.Meyer M, Kircher M. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 5, t5448. ( 10.1101/pdb.prot5448) [DOI] [PubMed] [Google Scholar]
- 31.Gansauge MT, Aximu-Petri A, Nagel S, Meyer M. 2020. Manual and automated preparation of single-stranded DNA libraries for the sequencing of DNA from ancient biological remains and other sources of highly degraded DNA. Nat. Protoc. 15, 2279-2300. ( 10.1038/s41596-020-0338-0) [DOI] [PubMed] [Google Scholar]
- 32.Horn S. 2012. Target enrichment via DNA hybridization capture. In Ancient DNA. Methods and protocols. (eds Shapiro B, Hofreiter M), pp. 189-195. Totowa, NJ: Humana Press. [Google Scholar]
- 33.Schubert M, Lindgreen S, Orlando L. 2016. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes 9, 1-7. ( 10.1186/s13104-016-1900-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feuerborn TR, et al. 2020. Competitive mapping allows for the identification and exclusion of human DNA contamination in ancient faunal genomic datasets. BMC Genom. 21, 1-10. ( 10.1186/s12864-020-07229-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. 2013. MapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682-1684. ( 10.1093/bioinformatics/btt193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fewlass H, Tuna T, Fagault Y, Hublin JJ, Kromer B, Bard E, Talamo S. 2019. Pretreatment and gaseous radiocarbon dating of 40–100 mg archaeological bone. Sci. Rep. 9, 1-11. ( 10.1038/s41598-019-41557-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wacker L, Němec M, Bourquin J. 2010. A revolutionary graphitisation system: fully automated, compact and simple. Nucl. Instrum. Methods Phys. Res. B 268, 931-934. ( 10.1016/j.nimb.2009.10.067) [DOI] [Google Scholar]
- 38.Synal HA, Stocker M, Suter M. 2007. MICADAS: a new compact radiocarbon AMS system. Nucl. Instrum. Methods Phys. Res. B 259, 7-13. ( 10.1016/j.nimb.2007.01.138) [DOI] [Google Scholar]
- 39.Bronk Ramsey C. 2009. Bayesian analysis of radiocarbon dates. Radiocarbon 51, 337-360. ( 10.2458/azu_js_rc.v51i1.3494) [DOI] [Google Scholar]
- 40.Reimer PJ, et al. 2020. The IntCal20 northern hemisphere radiocarbon age calibration curve (0–55 cal kBP). Radiocarbon 62, 725-757. ( 10.1017/rdc.2020.41) [DOI] [Google Scholar]
- 41.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772-780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, msw260. ( 10.1093/molbev/msw260) [DOI] [PubMed] [Google Scholar]
- 43.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, 1-5. ( 10.1093/ve/vey016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duchene S, Lemey P, Stadler T, Ho SYW, Duchene DA, Dhanasekaran V, Baele G. 2020. Bayesian evaluation of temporal signal in measurably evolving populations. Mol. Biol. Evol. 37, 3363-3379. ( 10.1093/molbev/msaa163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duchêne S, Duchêne D, Holmes EC, Ho SYW. 2015. The performance of the date-randomization test in phylogenetic analyses of time-structured virus data. Mol. Biol. Evol. 32, 1895-1906. ( 10.1093/molbev/msv056) [DOI] [PubMed] [Google Scholar]
- 46.Petrova TV, Zakharov ES, Samiya R, Abramson NI. 2015. Phylogeography of the narrow-headed vole Lasiopodomys (Stenocranius) gregalis (Cricetidae, Rodentia) inferred from mitochondrial cytochrome b sequences: an echo of Pleistocene prosperity. J. Zool. Syst. Evol. Res. 53, 97-108. ( 10.1111/jzs.12082) [DOI] [Google Scholar]
- 47.Jacobs Z, et al. 2019. Timing of archaic hominin occupation of Denisova Cave in southern Siberia. Nature 565, 594-599. ( 10.1038/s41586-018-0843-2) [DOI] [PubMed] [Google Scholar]
- 48.Ho SYW, Lanfear R, Bromham L, Phillips MJ, Soubrier J, Rodrigo AG, Cooper A. 2011. Time-dependent rates of molecular evolution. Mol. Ecol. 20, 3087-3101. ( 10.1111/j.1365-294X.2011.05178.x) [DOI] [PubMed] [Google Scholar]
- 49.Ho SYW, Saarma U, Barnett R, Haile J, Shapiro B. 2008. The effect of inappropriate calibration: three case studies in molecular ecology. PLoS ONE 3, e1615. ( 10.1371/journal.pone.0001615) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lisiecki LE, Raymo ME. 2005. A Pliocene-Pleistocene stack of 57 globally distributed benthic δ18O records. Paleoceanography 20, PA1003. ( 10.1029/2004PA001071) [DOI] [Google Scholar]
- 51.Reille M, De Beaulieu JL, Svobodova H, Andrieu-Ponel V, Goeury C. 2000. Pollen analytical biostratigraphy of the last five climatic cycles from a long continental sequence from the Velay region (Massif Central, France). J. Quat. Sci. 15, 665-685. () [DOI] [Google Scholar]
- 52.Velichko AA, Faustova MA, Pisareva VV, Gribchenko YUN, Sudakova NG, Lavrentiev N V. 2011. Glaciations of the East European Plain. Distribution and chronology. Dev. Quat. Sci. 15, 337-359. ( 10.1016/B978-0-444-53447-7.00026-X) [DOI] [Google Scholar]
- 53.Spratt RM, Lisiecki LE. 2016. A Late Pleistocene sea level stack. Clim. Past 12, 1079-1092. ( 10.5194/cp-12-1079-2016) [DOI] [Google Scholar]
- 54.Helmens KF. 2014. The Last Interglacial-Glacial cycle (MIS 5–2) re-examined based on long proxy records from central and northern Europe. Quat. Sci. Rev. 86, 115-123. ( 10.1016/j.quascirev.2013.12.012) [DOI] [Google Scholar]
- 55.Gutier F, Andrieu-Ponel V, de Beaulieu J-L, Cheddadi R, Calvez M, Ponel P, Reille M, Keller T, Goeury C. 2003. The last climatic cycles in Western Europe: a comparison between long continuous lacustrine sequences from France and other terrestrial records. Quat. Int. 111, 59-74. ( 10.1016/S1040-6182(03)00015-6) [DOI] [Google Scholar]
- 56.Kirschner P, et al. 2022. Congruent evolutionary responses of European steppe biota to late Quaternary climate change. Nat. Commun. 13, 1-11. ( 10.1038/s41467-022-29267-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Markova A, Puzachenko A. 2018. Preliminary analysis of European small mammal faunas of the Eemian interglacial: species composition and species diversity at a regional scale. Quaternary 1, 1-21. ( 10.3390/quat1020009) [DOI] [Google Scholar]
- 58.Foury Y, Desclaux E, Daujeard C, Defleur A, Moncel M-H, Raynal J-P. 2016. Evolution des faunes de rongeurs en moyenne vallée du Rhône (rive droite, Ardèche, France) au cours du pléistocène moyen final et du pléistocène supérieur ancien, du mis 6 au mis 4. Quaternaire 27, 55-79. ( 10.4000/quaternaire.7527) [DOI] [Google Scholar]
- 59.Luzi E, Blanco-Lapaz Á, Rhodes SE, Conard NJ. 2022. Paleoclimatic and paleoenvironmental reconstructions based on the small vertebrates from the Middle Paleolithic of Hohle Fels Cave, SW Germany. Archaeol. Anthropol. Sci. 5. ( 10.1007/s12520-022-01568-5) [DOI] [Google Scholar]
- 60.Socha P. 2014. Rodent palaeofaunas from Biśnik Cave (Kraków-Częstochowa Upland, Poland): Palaeoecological, palaeoclimatic and biostratigraphic reconstruction. Quat. Int. 326–327, 64-81. ( 10.1016/j.quaint.2013.12.027) [DOI] [Google Scholar]
- 61.Stewart JR, Lister AM, Barnes I, Dalén L. 2010. Refugia revisited: individualistic responses of species in space and time. Proc. R. Soc. B 277, 661-671. ( 10.1098/rspb.2009.1272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Svensson A, et al. 2008. A 60 000 year Greenland stratigraphic ice core chronology. Clim. Past 4, 47-57. ( 10.5194/cp-4-47-2008) [DOI] [Google Scholar]
- 63.Kosintsev P, et al. 2019. Evolution and extinction of the giant rhinoceros Elasmotherium sibiricum sheds light on late Quaternary megafaunal extinctions. Nat. Ecol. Evol. 3, 31-38. ( 10.1038/s41559-018-0722-0) [DOI] [PubMed] [Google Scholar]
- 64.Soubrier J, et al. 2016. Early cave art and ancient DNA record the origin of European bison. Nat. Commun. 7, 13158. ( 10.1038/ncomms13158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rey-Iglesia A, et al. 2021. Exploring the phylogeography and population dynamics of the giant deer (Megaloceros giganteus) using Late Quaternary mitogenomes. Proc. R. Soc. B 288, 20201864. ( 10.1098/rspb.2020.1864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Higham T, et al. 2014. The timing and spatiotemporal patterning of Neanderthal disappearance. Nature 512, 306-309. ( 10.1038/nature13621) [DOI] [PubMed] [Google Scholar]
- 67.Vandenberghe J, van der Plicht J. 2016. The age of the Hengelo interstadial revisited. Quat. Geochronol. 32, 21-28. ( 10.1016/j.quageo.2015.12.004) [DOI] [Google Scholar]
- 68.Moine O, Antoine P, Hatté C, Landais A, Mathieu J, Prud'homme C, Rousseau DD. 2017. The impact of Last Glacial climate variability in west-European loess revealed by radiocarbon dating of fossil earthworm granules. Proc. Natl Acad. Sci. USA 114, 6209-6214. ( 10.1073/pnas.1614751114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cooper A, et al. 2021. A global environmental crisis 42,000 years ago. Science 374, 811-818. ( 10.1126/science.abi8330) [DOI] [PubMed] [Google Scholar]
- 70.Marciszak A, Schouwenburg C, Lipecki G, Talamo S, Shpansky A, Malikov D, Gornig W. 2019. Steppe brown bear Ursus arctos ‘priscus’ from the Late Pleistocene of Europe. Quat. Int. 534, 158-170. ( 10.1016/j.quaint.2019.02.042) [DOI] [Google Scholar]
- 71.Michczyńska DJ, Dzieduszyńska DA, Petera-Zganiacz J, Wachecka-Kotkowska L, Krzyszkowski D, Wieczorek D, Ludwikowska-Kędzia M, Gębica P, Starkel L. 2022. Climatic oscillations during MIS 3–2 recorded in the sets of 14C and OSL dates—a study based on data from Poland. Radiocarbon 64(6), 1373-1386. ( 10.1017/RDC.2022.69) [DOI] [Google Scholar]
- 72.Baca M, et al. 2023. Aligned and curated mtDNA sequences from Baca et al. 2023; Ancient DNA of narrow-headed vole reveal common features of the Late Pleistocene population dynamics in cold-adapted small mammals. Dryad Digital Repository. ( 10.5061/dryad.kh1893295) [DOI] [PMC free article] [PubMed]
- 73.Baca M, et al. 2023. Ancient DNA of narrow-headed vole reveal common features of the Late Pleistocene population dynamics in cold-adapted small mammals. Figshare. ( 10.6084/m9.figshare.c.6423382) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Baca M, et al. 2023. Aligned and curated mtDNA sequences from Baca et al. 2023; Ancient DNA of narrow-headed vole reveal common features of the Late Pleistocene population dynamics in cold-adapted small mammals. Dryad Digital Repository. ( 10.5061/dryad.kh1893295) [DOI] [PMC free article] [PubMed]
- Baca M, et al. 2023. Ancient DNA of narrow-headed vole reveal common features of the Late Pleistocene population dynamics in cold-adapted small mammals. Figshare. ( 10.6084/m9.figshare.c.6423382) [DOI] [PMC free article] [PubMed]
Data Availability Statement
The consensus mtDNA sequences generated in this study have been deposited in GenBank under Accession numbers OM809882–OM810020. The mtDNA sequence alignments used to reconstruct the mitogenome and cytochrome phylogenies have been deposited in Dryad Digital Repository and can be accessed at https://doi.org/10.5061/dryad.kh1893295 [72]. The mitochondrial alignments generated in this study have been deposited in the European Nucleotide Archive under project number PRJEB56307.
The data are provided in electronic supplementary material [73].