PURPOSE
Total cell-free DNA (cfDNA) and tumor-derived cfDNA (ctDNA) can be used to study tumor-derived genetic aberrations. We analyzed the diagnostic and prognostic potential of cfDNA and ctDNA, obtained from pediatric patients with rhabdomyosarcoma.
METHODS
cfDNA was isolated from diagnostic plasma samples from 57 patients enrolled in the EpSSG RMS2005 study. To study the diagnostic potential, shallow whole genome sequencing (shWGS) and cell-free reduced representation bisulphite sequencing (cfRRBS) were performed in a subset of samples and all samples were tested using droplet digital polymerase chain reaction to detect methylated RASSF1A (RASSF1A-M). Correlation with outcome was studied by combining cfDNA RASSF1A-M detection with analysis of our rhabdomyosarcoma-specific RNA panel in paired cellular blood and bone marrow fractions and survival analysis in 56 patients.
RESULTS
At diagnosis, ctDNA was detected in 16 of 30 and 24 of 26 patients using shallow whole genome sequencing and cfRRBS, respectively. Furthermore, 21 of 25 samples were correctly classified as embryonal by cfRRBS. RASSF1A-M was detected in 21 of 57 patients. The presence of RASSF1A-M was significantly correlated with poor outcome (the 5-year event-free survival [EFS] rate was 46.2% for 21 RASSF1A-M‒positive patients, compared with 84.9% for 36 RASSF1A-M‒negative patients [P < .001]). RASSF1A-M positivity had the highest prognostic effect among patients with metastatic disease. Patients both negative for RASSF1A-M and the rhabdomyosarcoma-specific RNA panel (28 of 56 patients) had excellent outcome (5-year EFS 92.9%), while double-positive patients (11/56) had poor outcome (5-year EFS 13.6%, P < .001).
CONCLUSION
Analyzing ctDNA at diagnosis using various techniques is feasible in pediatric rhabdomyosarcoma and has potential for clinical use. Measuring RASSF1A-M in plasma at initial diagnosis correlated significantly with outcome, particularly when combined with paired analysis of blood and bone marrow using a rhabdomyosarcoma-specific RNA panel.
INTRODUCTION
Rhabdomyosarcoma, the most common sarcoma among children and adolescents, accounts for approximately 3% of pediatric tumors.1 Despite considerable research regarding treatment and risk stratification, 1 of 3 patients will experience relapse.2-6 The use of liquid biopsies in pediatric patients is drawing growing interest.7,8 Our group reported that the presence of rhabdomyosarcoma-derived mRNA in the cellular fraction of peripheral blood (PB) and bone marrow (BM) at initial diagnosis is correlated with poor outcome and could potentially improve current risk stratification.9 Studies on other pediatric solid tumors demonstrated cell-free DNA (cfDNA) analysis from plasma to provide added value for diagnostics, prognostics, and response monitoring.10-16 In rhabdomyosarcoma, the presence of tumor-derived cfDNA (ctDNA) has been shown to correlate to tumor burden throughout treatment in a few small case series.17,18 ctDNA can be studied using various techniques, using genetic aberrations present in rhabdomyosarcoma. The alveolar subtype has a tumor-driving fusion between the PAX3 or PAX7 gene and the FOXO1 gene. Epigenetic analyses revealed distinct methylation profiles in alveolar and embryonal rhabdomyosarcoma, allowing for the classification of cases into fusion-positive versus fusion-negative tumors.19,20 Van Paemel et al21 showed that these distinct methylation patterns can be detected in ctDNA from diagnostic plasma, using cell-free reduced representation bisulphite sequencing (cfRRBS) to correctly classify rhabdomyosarcoma as either the embryonal or alveolar subtype. Copy number aberrations (CNAs) have been found to occur in several chromosomes.3,22 These can be analyzed in cfDNA by shallow whole genome sequencing (shWGS).23 Recently, Van Paemel et al16 showed that shWGS data from cfDNA can be complementary to CNA analysis on the primary tumor.
CONTEXT
Key Objective
In pediatric rhabdomyosarcoma, the use of liquid biopsies can assist in generating a more comprehensive view of the molecular landscape of the tumor. We explore different methods for analysis of cell-free DNA (cfDNA) from plasma by cell-free reduced representation bisulphite sequencing, shallow whole genome sequencing, and droplet digital polymerase chain reaction for RASSF1A methylation (RASSF1-M). Furthermore, we study whether combining cfDNA analyses with detection of rhabdomyosarcoma-specific RNA in the cellular fraction of blood and bone marrow (BM) has a complementary value.
Knowledge Generated
Both cell-free reduced representation bisulphite sequencing and shallow whole genome sequencing have diagnostic potential, whereas the presence of RASSF1A-M at diagnosis correlates to poor survival, especially in patients testing positive for rhabdomyosarcoma-specific RNA in cells from blood and BM.
Relevance
Analysis of cfDNA through different molecular approaches can be of additional value to current clinical risk stratification, especially the detection of RASSF1A-M in cfDNA and rhabdomyosarcoma-specific RNA in paired blood and BM.
However, cfDNA typically contains a relatively small amount of ctDNA; the remaining cfDNA is derived from healthy cells, which can cause high background noise and limit the ability to detect a tumor-derived signal.24 To overcome this, a tumor-specific assay can be used, such as droplet digital polymerase chain reaction (ddPCR) which is highly sensitive and less expensive.25 A target suited for analysis by ddPCR is methylation of the tumor-suppressor gene RASSF1A; this gene has been shown to be silenced by methylation in several adult26 and pediatric27-30 tumors. Moreover, methylated RASSF1A (RASSF1A-M) has been detected in cfDNA in patients with neuroblastoma.10,31 Recently, we developed a methylation-specific enzyme-based approach involving ddPCR to detect RASSF1A-M in several pediatric solid tumors, including rhabdomyosarcoma.14
Here, we report the detection of ctDNA in plasma of patients with rhabdomyosarcoma for diagnostic purposes, such as cfRBBS and shWGS. Furthermore, we study the prognostic potential of RASSF1A-M detection in cfDNA and measure the added value of combining RASSF1A-M ctDNA detection with our rhabdomyosarcoma-specific mRNA panel in paired BM and PB samples.
METHODS
Patients and Sample Collection
Plasma samples were collected prospectively from the same cohort described in our previous article,9 consisting of all patients included in the Dutch Minimal Residual Disease add-on study within the EpSSG RMS2005 trial (EudraCT number: 2005-000217-35) from 2013 through July 2019. Informed consent was given via the EpSSG RMS2005 trial until 2017. From 2017, consent was provided if the patients/caretakers consented to the collection of samples for biobanking. PB was collected in EDTA tubes (Becton-Dickinson, Franklin Lakes, NJ) and processed within 24 hours. Plasma was obtained by centrifuging the blood samples at 1,375×g for 10 minutes and stored at −20°C until further processing. Matched tumor material was not available.
CfRRBS and shWGS
We performed cfRRBS21 and shWGS16,32 on cfDNA as described and validated previously. In brief, cfDNA was isolated from 200 μL of plasma as described previously.16,21,32 For shWGS, the modified copy number profile abnormality score was calculated in order to quantify the copy number tumor burden present in the cfDNA.16 On the basis of 80 healthy volunteers, the level corresponding to a 1% false discovery rate was set at 0.355 for shWGS.
ddPCR Assay for Measuring RASSF1A-M
For ddPCR, cfDNA was isolated from plasma samples using the Quick-cfDNA Serum & Plasma kit (Zymo Research, Irvine, CA). The RASSF1A-M ddPCR assay was performed using double digestion with the methylation-sensitive restriction enzymes Hhal and Bsh1236I BstUI; Thermo Fisher Scientific, Waltham, MA) using a thermocycler T100 and QX200 reader (Bio-Rad, Hercules, CA) as described previously.14 The sequences and concentrations of the primers and probes, cycling conditions, and analyses were performed as described previously, with the threshold for RASSF1A-M positivity per sample set at ≥ 14 copies/ml and ≥ 4 RASSF1A-M–positive droplets, as determined in 18 healthy pediatric and 22 adult control plasmas.14 The percentage of RASSF1A-M was calculated relative to total RASSF1A. On the basis of the plasma volume available (ranging from 150 µL to 1 mL), different amounts of plasma were used to isolate cfDNA. To correct for variations in the amount of input plasma, cfDNA is reported in ng/mL plasma. In all ddPCR assays, total cfDNA was determined using the reference gene ACTB. Since there was no matched tumor material available, we used data on RASSF1A-M in rhabdomyosarcoma tumors from published data sets from Clay et al,33 Koelsche et al,34 and specifically requested data from Seki et al.20 Data from Clay and Koelsche were analyzed in R2.35 We focused on hypermethylation of the promotor region of RASSF1A as this is typically hypermethylated in cancer.36 We calculated the mean beta value and report the range of the beta values.37,38
Detection of Rhabdomyosarcoma-Specific mRNA Using an RNA Panel
Rhabdomyosarcoma-specific mRNA was detected in the cellular fractions of matched diagnostic patient PB and BM samples using our previously reported 11-marker RNA panel.9 The RNA panel was considered positive if either PB or BM was positive.
Statistical Analysis
Statistical analyses were performed using SPSS version 23. Figures were generated using GraphPad Prism version 8. The correlation between continuous variables was determined using Pearson's test. Continuous variables were analyzed using the nonparametric Mann-Whitney U test, and two or more groups were analyzed using the Kruskal-Wallis test. Independence between two categorical variables was determined using the nonparametric Pearson chi-square test. Event-free survival (EFS) and overall survival (OS) were estimated using the Kaplan-Meier approach, and differences in survival were analyzed using the log-rank test. Differences were considered significant at P < .05.
RESULTS
Patient and Sample Characteristics
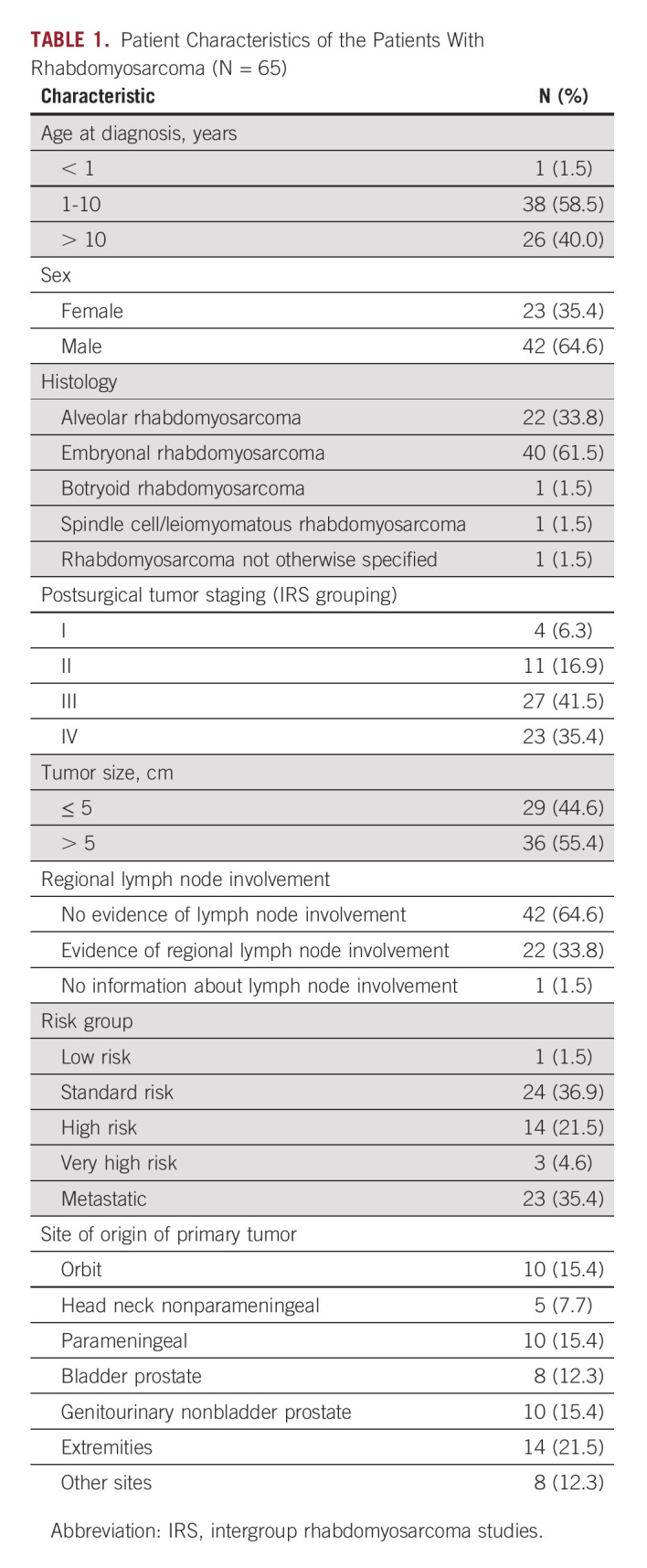
We collected a total of 152 plasma samples from 65 patients, treated according to the EpSSG RMS2005 protocol; diagnostic plasma samples were available for 57 patients. The patient characteristics, assigned risk group and tumor histology, are summarized in Table 1. The median follow-up was 4.21 years (range, 0.34-10.60 years).
TABLE 1.
Patient Characteristics of the Patients With Rhabdomyosarcoma (N = 65)
Diagnostic Potential of Various Molecular Techniques for Detecting ctDNA
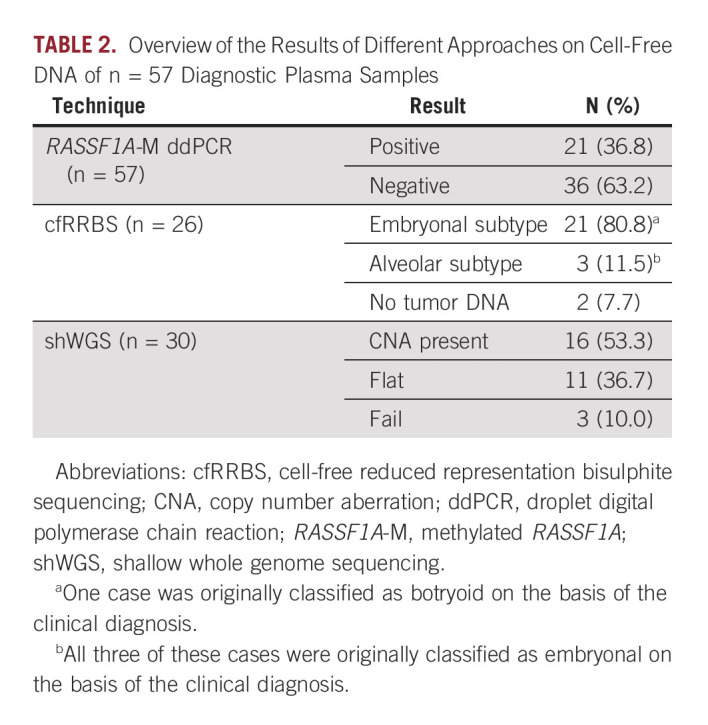
First, total cfDNA levels at diagnosis were determined by measuring ACTB using ddPCR for all samples. No significant differences in total cfDNA levels were observed between patients with respect to tumor histology, risk group, localized versus metastatic disease, tumor size, or event-free survival (Data Supplement). Next, we examined the feasibility to detect ctDNA using cfRRBS, shWGS, and ddPCR (Table 2, Data Supplement). Overall, in 39 of 57 patients (68.4%), at least one of these techniques detected ctDNA in diagnostic plasma samples. Please note that cfRRBS and shWGS were tested on a subset of samples.
TABLE 2.
Overview of the Results of Different Approaches on Cell-Free DNA of n = 57 Diagnostic Plasma Samples
Methylation profiling for diagnostic classification.
As negative control, cfRRBS was performed on 31 samples from healthy controls, all classified correctly as normal (Data Supplement). We applied cfRRBS to diagnostic samples from 24 patients with the embryonal subtype, 1 with botryoid subtype, and 1 with alveolar subtype, successfully detecting rhabdomyosarcoma DNA in 24 of these 26 samples (92.3% of cases). Twenty of these samples were correctly identified as embryonal tumors. Three cases with embryonal histology were classified as alveolar, one case of botryoid rhabdomyosarcoma was classified as embryonal, and no tumor DNA was detected in two samples (one alveolar and one embryonal).
CNAs.
We performed shWGS on 30 plasma samples and obtained a median copy number profile abnormality score of 0.35 (range, 0.27-3.94; Data Supplement). In three cases (two embryonal and one alveolar), the analysis failed (Table 2, Data Supplement). Twelve embryonal cases (7 of 12 metastatic) and four alveolar (all metastatic) cases had CNAs, while 10 embryonal cases and one botryoid case had no CNA. Most CNAs were detected in patients with metastatic disease, and 7 of 16 (43.8%) patients with detectable CNAs suffered from an event.
Methylated RASSF1A.
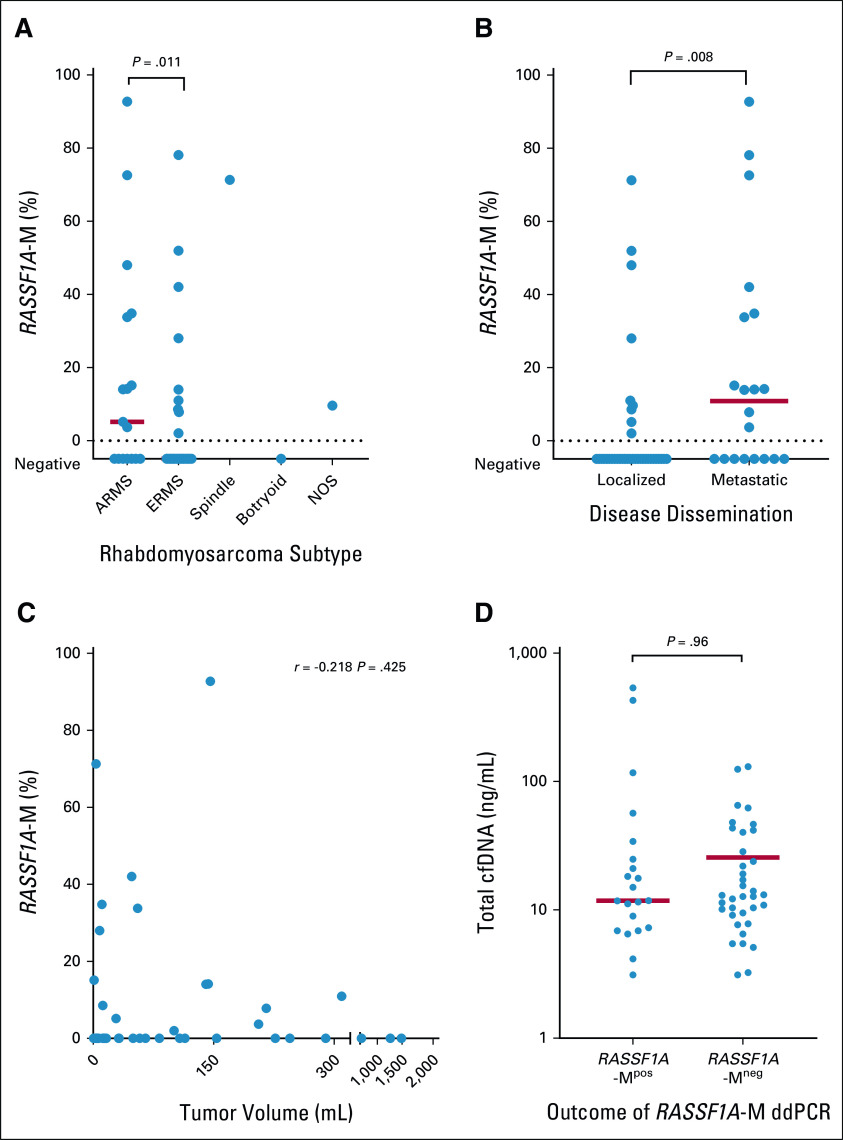
Using data from Clay et al,33 Koelsche et al,34 and Seki et al,20 the mean beta value of RASSF1A-M was 0.550 (range, 0.032-0.933; Data Supplement). We next examined the presence of RASSF1A-M in plasma using ddPCR. Methylated RASSF1A was detectable in 21 of 57 diagnostic plasma samples; 9 of 37 embryonal cases, 10 of 17 alveolar cases, 1 of 1 spindle case, 0 botryoid case, and one not-otherwise-specified case, with a median RASSF1A-M concentration of 2.46 ng/mL (range, 0.22-273.11 ng/mL). In patients with alveolar tumors and metastatic disease, methylated RASSF1A was more frequently detected, compared with embryonal histology (P = .014) and localized disease (RASSF1A-M–positive in 9 of 37 patients with localized, 12 of 20 patients with metastatic disease P = .008). The total level of RASSF1A-M varied widely within the RASSF1A-M–positive samples and was correlated with tumor histology (Data Supplement). To correct for variations in total cfDNA, we calculated the percentage of RASSF1A-M relative to total RASSF1A for each patient, yielding a median percentage of 15.1% (range, 2.0%-92.7%) for the RASSF1A-M‒positive samples. Although metastatic and alveolar tumors more often show the presence of RASSF1A-M in cfDNA, the RASSF1A-M percentage in positive samples was similar in alveolar and embryonal tumors (P = .55) and in localized and metastatic cases (P = .35; Figs 1A and 1B). We found no correlation between tumor size at diagnosis and either total RASSF1A-M (r = 0.132 and P = .64; Data Supplement) or the percentage of RASSF1A-M (r = –.229 and P = .41; Fig 1C). Finally, we found no difference in total cfDNA levels (ACTB) between RASSF1A-M‒positive and RASSF1A-M‒negative cases (P = .96; Fig 1D).
FIG 1.
Methylated RASSF1A (RASSF1A-M) in diagnostic plasma samples of patients with rhabdomyosarcoma. The percentage of cell-free methylated RASSF1A (RASSF1A-M) is calculated according to total RASSF1A copies at diagnosis in patients: (A) with different subtypes, (B) with localized and metastatic disease, and (C) plotted against tumor volume at diagnosis. (D) Level of cfDNA (quantified by beta-actin [ACTB]) at diagnosis in plasma samples with detectable RASSF1A-M and with no detectable RASSF1A-M; note that the y-axis is plotted on a log scale. In this figure, each symbol represents an individual patient, and the red horizontal lines represent the median values. Tumor size was determined by MRI, CT scan, or ultrasonography. ARMS, alveolar rhabdomyosarcoma; cfDNA, cell-free DNA; CT, computed tomography; ddPCR, droplet digital polymerase chain reaction; ERMS, embryonal rhabdomyosarcoma; MRI, magnetic resonance imaging; NOS, not otherwise specified.
Cell-Free RASSF1A-M Correlates With Poor Outcome
We examined whether the detection of ctDNA in 57 diagnostic plasma samples was associated with patient outcome. Eleven of 21 (52.3%) RASSF1A-M–positive patients suffered from an event.
The 5-year EFS rate was 46.2% for the RASSF1A-M‒positive patients, compared with 84.9% for the RASSF1A-M‒negative patients (P = .001; Fig 2A), and the 5-year OS rate was 55.7% for the RASSF1A-M‒positive patients, compared with 100% for the RASSF1A-M‒negative patients (P < .001; Fig 2B). The prognostic value of detecting RASSF1A-M at diagnosis was attributed almost exclusively to patients with metastasized disease (Figs 2C and 2D, Data Supplement).
FIG 2.
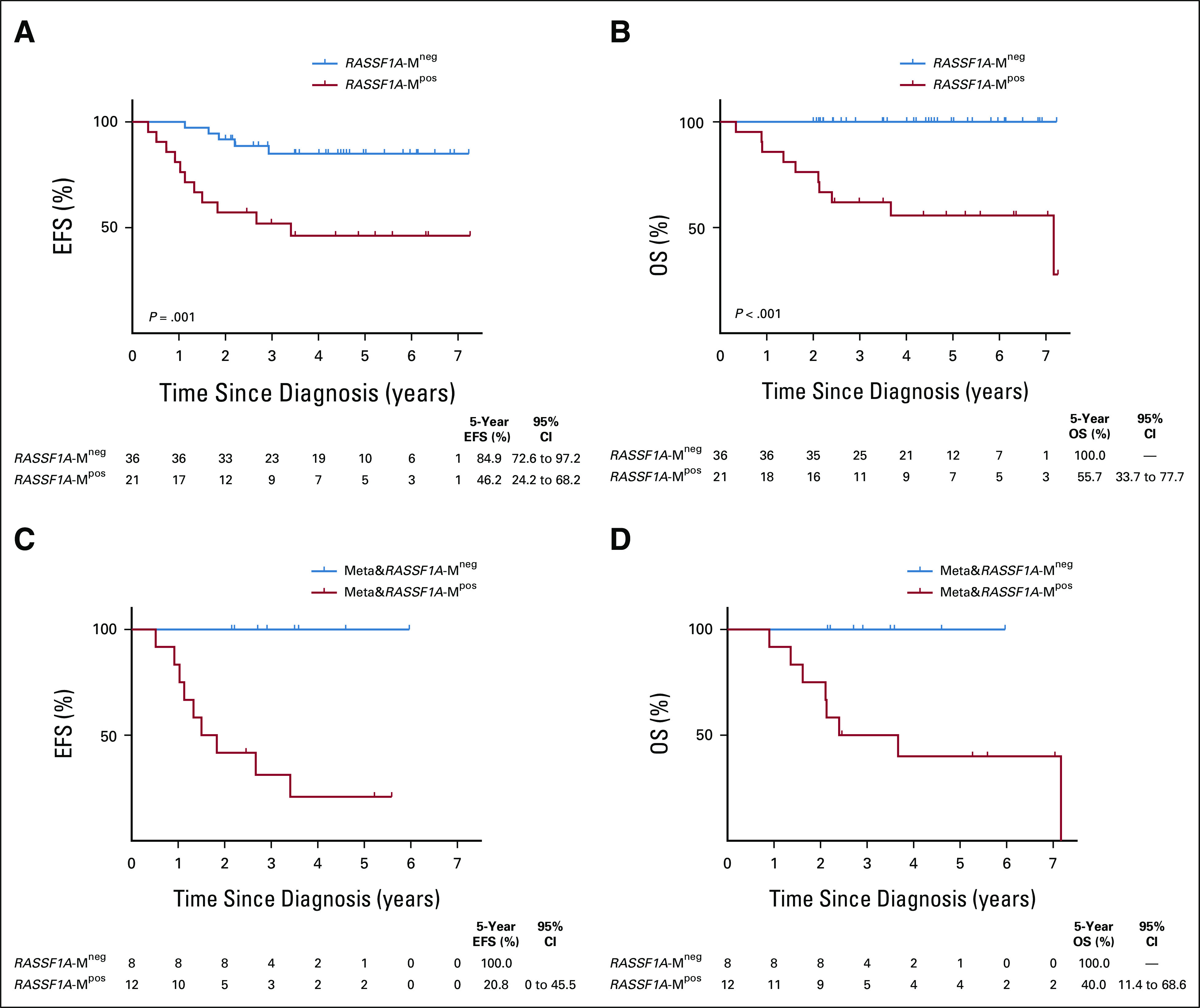
Survival outcome defined by detection of cell-free methylated RASSF1A (RASSF1A-M) at diagnosis. (A and B) EFS and OS, respectively, of patients with no detectable methylated RASSF1A in the diagnostic plasma (RASSF1A-Mneg; n = 36) and patients with detectable methylated RASSF1A in the diagnostic plasma (RASSF1A-Mpos; n = 21). (C and D) EFS and OS of RASSF1A-M‒negative patients (n = 8) and RASSF1A-M‒positive patients (n = 12) with metastatic disease. Shown below each plot is the number of patients at each time point and 5-year survival with the 95% CI. cfDNA, cell-free DNA; EFS, event-free survival; OS, overall survival.
In 27 samples, both shWGS and RASSF1A-M were performed (Data Supplement). In six patients, shWGS was positive while RASSF1A-M was negative, and only one patient suffered from an event, while 6 of 10 double-positive patients suffered from an event, suggesting that the presence of both RASSF1A-M and ctDNA by shWGS may be more prognostic than detection of ctDNA by shWGS alone.
We next examined whether combining RASSF1A-M detection with detection of rhabdomyosarcoma-specific mRNA (on the basis of our previously published mRNA panel9) tested in 56 matched diagnostic PB and BM samples could improve the predictive value. Rhabdomyosarcoma-specific mRNA was detected in 18 of 56 PB and/or BM samples (8 of 18 tested positive on conventional BM histology, Data Supplement). Five-year EFS ranged from 92.9% to 13.6% for RASSF1A-Mneg/mRNA panelneg and RASSF1A-Mpos/mRNA panelpos (P = .006) and 5 years OS from 100% to 36.4% for RASSF1A-Mneg/mRNA panelneg and RASSF1A-Mpos/mRNA panelpos, respectively (P < .001; Figs 3A and 3B).
FIG 3.
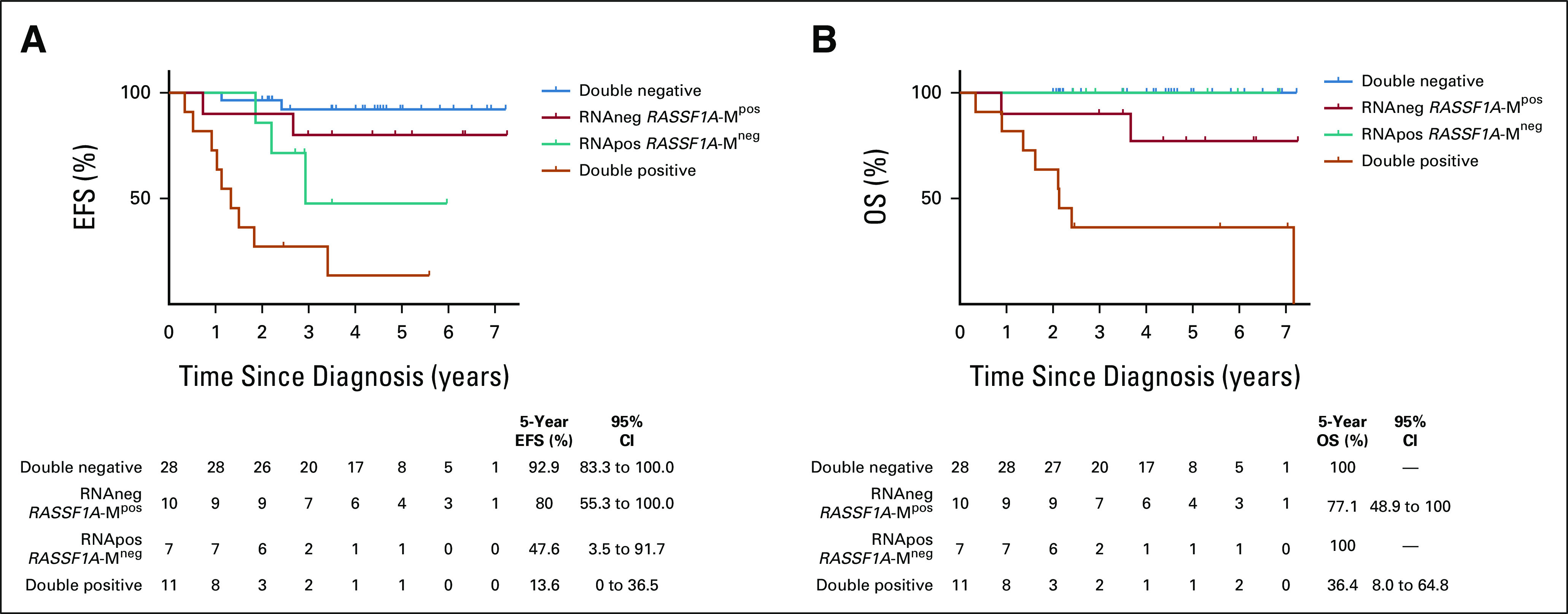
Survival outcome defined by detection of cell-free methylated RASSF1A (RASSF1A-M) from plasma and rhabdomyosarcoma-specific RNA in blood and bone marrow at diagnosis. (A and B) EFS and OS of 56 patients on the basis of the absence or presence of rhabdomyosarcoma-specific RNA (RNA-negative and RNA-positive, respectively) combined with RASSF1A-M status. Shown below each plot is the number of patients at each time point and 5-year survival with the 95% CI. OS, overall survival; EFS, event-free survival.
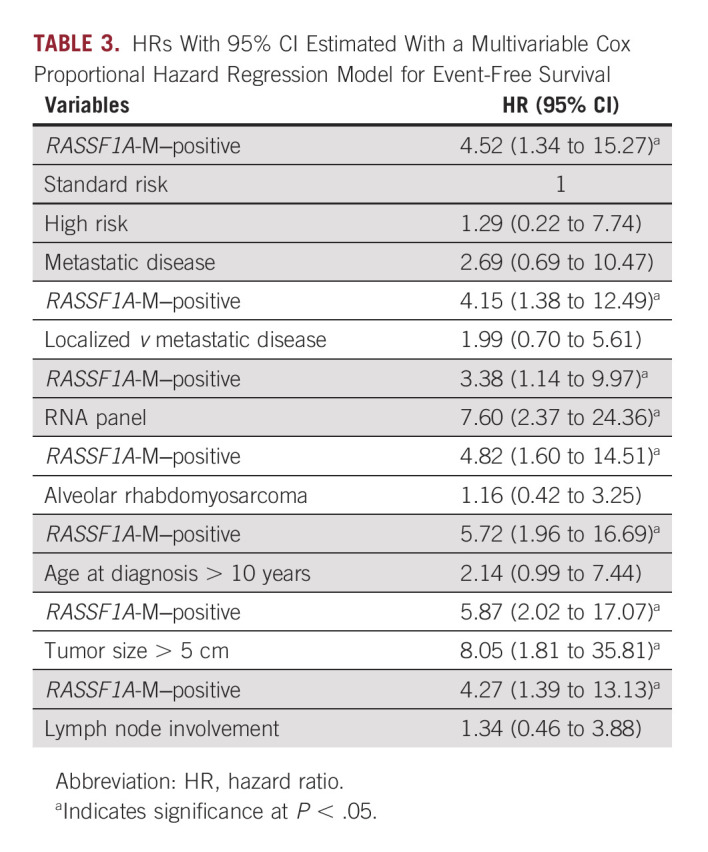
To validate the association of RASSF1A-M to clinical outcome, we performed univariate and multivariable Cox regression analyses for EFS (Data Supplement and Table 3, respectively). In the multivariable model, only RASSF1A-M, RNA panel, and tumor size larger than 5 cm had a significant effect on outcome. The known EpSSG RMS2005 risk group classification, metastatic disease, alveolar subtype, over 10 years of age, and lymph node involvement were not significantly associated with outcome in our multivariable model. Finally, OS could not be analyzed due to the low number of events in this cohort.
TABLE 3.
HRs With 95% CI Estimated With a Multivariable Cox Proportional Hazard Regression Model for Event-Free Survival
RASSF1A-M During Treatment and Clinical Follow-Up
For 33 patients, a total of 95 samples drawn during primary treatment and/or subsequent clinical follow-up were available. RASSF1A-M was measured in the follow-up samples only if the patient was RASSF1A-M‒positive at diagnosis or—if a diagnostic sample was not available—at relapse. Among the 23 patients for whom samples were collected during primary treatment, only two patients (Data Supplement) were RASSF1A-M‒positive after two cycles of chemotherapy, but RASSF1A-M‒negative in all subsequent samples. In eight patients, RASSF1A-M was measured in a sample taken during a clinical event (five at first relapse, two at second relapse, and one at progressive disease during primary treatment). Five of these eight samples were RASSF1A-M‒positive (three at first relapse and two at second relapse); no samples at initial diagnosis were available for these five patients. After initiating relapse therapy, all subsequent samples from these patients were RASSF1A-M‒negative. The sample taken from the patient at progressive disease (patient RMS133) was RASSF1A-M‒negative, and no previous plasma samples were available for this patient.
DISCUSSION
On the basis of our findings, we propose that each cfDNA-based technique can address a specific clinical need, ranging from assisting at initial tumor diagnosis to fine-tuning of risk stratification. In our cohort, cfRRBS proved its potential as a highly sensitive method for identifying rhabdomyosarcoma-derived cfDNA at initial diagnosis, and the majority was classified correctly as embryonal. Van Paemel et al21 found that cfRRBS was also able to correctly identify alveolar ctDNA. Thus, cfRRBS can provide added value at initial diagnosis, particularly if the ability to perform a tumor biopsy is restricted by clinical features such as tumor location or the patient's condition and when the ability to distinguish between other types of pediatric solid tumors is important.21
We detected CNAs in 53.3% of samples analyzed by shWGS, mostly metastatic cases. On the basis of the literature, CNAs are present in nearly all fusion-negative rhabdomyosarcomas20,39 and in approximately one third of all fusion-positive rhabdomyosarcomas.39,40 We detected CNAs in the cfDNA of only half of the patients with fusion-negative tumors. This relatively low rate may have been due in part to contamination of the cfDNA with genomic DNA, as the protocol for drawing and storing blood was not standardized, which can lower the sensitivity to detect CNA.16 Van Paemel et al noted that performing shWGS on cfDNA can provide additional value with respect to analyzing CNAs in the primary tumor, resulting in a more complete overview of the patient's genetic landscape and bypassing any potential heterogeneity within the tumor and/or metastatic lesions. This is important to consider when designing further studies.
On the basis of the previous reports, demonstrating feasibility to use RASSF1A-M ddPCR as a tumor-specific marker with a high specificity due to extremely low background in plasma from healthy controls,14,15 we studied RASSF1A-M ddPCR in cfDNA of patients with rhabdomyosarcoma. One of the limitations of this study was the absence of paired primary tumor samples. However, the presence of RASSF1A-M, as extracted from data published by several groups,20,33,34 indicated the potential to detect RASSF1A-M in primary tumors, with admittedly a large variation in the level of RASSF1A-M. Still, for the patients in our cohort who were RASSF1A-M‒negative, on the basis of cfDNA obtained at diagnosis, we were unable to determine whether this was due to absence of RASSF1A methylation or no detectable ctDNA. This is underlined by the 18 samples testing negative for RASSF1A-M, in which ctDNA was detected by cfRRBS and/or shWGS. Future studies should include matching tumor material to establish the contribution of different approaches for cfDNA analysis. Nonetheless, we were able to detect RASSF1A-M in cfDNA in 36% of diagnostic samples and found a strong correlation between RASSF1A-M positivity and event-free survival and OS. Importantly, this predictive value was obtained almost exclusively in the group of patients with metastatic disease. This finding might suggest that more aggressive tumors contain methylated RASSF1A and deserves further investigations in a follow-up study, including matching primary tumor material. Interestingly, in the samples that were tested by both shWGS and RASSF1A-M, the results suggest that detection of ctDNA by both methods may be more prognostic than detection of ctDNA by shWGS alone. This should be studied further in a larger cohort.
As we previously showed rhabdomyosarcoma-specific RNA detection in PB and/or BM at diagnosis to detect additional disseminated disease and to correlate with the outcome,9 we now showed that combining mRNA and ctDNA (RASSF1A-M) in paired diagnostic samples identifies patients with very good and very poor outcome. Our multivariable analysis revealed that combining the cfDNA RASSF1A-M assay with rhabdomyosarcoma-specific RNA detection in PB and BM samples provides an even better tool for discriminating between low-risk patients and patients with a poor prognosis. Given the relatively small number of patients in our cohort, however, we were unable to investigate the effect of adding both RASSF1A-M and the RNA panel to established prognostic factors, particularly in the EpSSG RMS2005 risk group; nevertheless, our results can form a starting point for future studies involving a prospective cohort.
An interesting finding from our study is the dynamics of ctDNA. Before our study, we hypothesized that ctDNA would still be present during primary treatment and decrease slowly, tracking the decrease in tumor burden. However, in our rhabdomyosarcoma cohort, we found that most of the samples were negative for ctDNA after the first course of chemotherapy. This rapid transition to a ctDNA-negative state is consistent with the results reported by Klega et al18 who found that most samples were negative for ctDNA before the second course of chemotherapy. Thus, an interesting question is whether performing earlier sampling and obtaining multiple samples during the first 2 weeks after the start of treatment would reveal the presence of ctDNA and—if so—would lead to the development of a prognostic marker, similar to the marker for minimal residual disease developed for use in leukemia.41,42
In conclusion, we demonstrate the feasibility to study ctDNA in pediatric rhabdomyosarcoma by different approaches. The choice of a given technique will depend on whether the underlying question is diagnostic or prognostic. We show that the presence of methylated RASSF1A in cfDNA is associated with poor outcome and can be used to improve risk stratification at diagnosis. Furthermore, we show that combining detection of methylated RASSF1A in plasma with analysis of tumor-specific RNA in blood and bone marrow identified patients with good versus poor outcome.
ACKNOWLEDGMENT
We thank Masafumi Seki for sharing detailed data regarding RASSF1A methylation from their published cohort. We thank Maisa Renata Ferro dos Santos from Ghent University for the analysis of cfRRBS data from healthy controls.
Johannes H.M. Merks
Consulting or Advisory Role: Bayer, GlaxoSmithKline, Merck KGaA
Gudrun Schleiermacher
Honoraria: BMS
Research Funding: Bristol Myers Squibb (Inst), Pfizer (Inst), MSDavenir (Inst), Roche (Inst)
Travel, Accommodations, Expenses: Roche, Roche
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented as a poster at the International Society of Pediatric Oncology (SIOP) 2021 Congress, virtual, October 21-24, 2021.
SUPPORT
N.L., L.Z.-K., and J.S. were supported by KiKa (Children Cancer Free; grant number 312). O.R. was supported by the children's cancer charity Alice's Arc.
G.A.M.T. and J.S. contributed equally to this work.
AUTHOR CONTRIBUTIONS
Conception and design: Nathalie S.M. Lak, Lieke M.J. van Zogchel, Gudrun Schleiermacher, Janet Shipley, Johannes H.M. Merks, Max M. van Noesel, C. Ellen van der Schoot, Godelieve A.M. Tytgat, Janine Stutterheim
Financial support: Godelieve A.M. Tytgat, Janine Stutterheim
Administrative support: Nathalie S.M. Lak, Max M. van Noesel, Godelieve A.M. Tytgat
Provision of study materials or patients: Charlotte Vandeputte, Katleen De Preter, Gudrun Schleiermacher, Johannes H.M. Merks, Godelieve A.M. Tytgat
Collection and assembly of data: Nathalie S.M. Lak, Lieke M.J. van Zogchel, Lily Zappeij-Kannegieter, Ahmad Javadi, Ruben van Paemel, Charlotte Vandeputte, Katleen De Preter, Bram De Wilde, Mathieu Chicard, Gudrun Schleiermacher, Olivia Ruhen, Johannes H.M. Merks, Max M. van Noesel, C. Ellen van der Schoot, Godelieve A.M. Tytgat, Janine Stutterheim
Data analysis and interpretation: Nathalie S.M. Lak, Lieke M.J. van Zogchel, Ruben van Paemel, Katleen De Preter, Yasmine Iddir, Gudrun Schleiermacher, Janet Shipley, Marta Fiocco, Johannes H.M. Merks, Max M. van Noesel, C. Ellen van der Schoot, Godelieve A.M. Tytgat, Janine Stutterheim
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Johannes H.M. Merks
Consulting or Advisory Role: Bayer, GlaxoSmithKline, Merck KGaA
Gudrun Schleiermacher
Honoraria: BMS
Research Funding: Bristol Myers Squibb (Inst), Pfizer (Inst), MSDavenir (Inst), Roche (Inst)
Travel, Accommodations, Expenses: Roche, Roche
No other potential conflicts of interest were reported.
REFERENCES
- 1.Princess Máxima Center for Pediatric Oncology Annual Report 2020 : https://www.prinsesmaximacentrum.nl/storage/configurations/prinsesmaximacentrumnl/files/hetprinsesmaximacentrum-jaarverslag_cenr2020_06-21_s_interactive.pdf
- 2.Gartrell J, Pappo A: Recent advances in understanding and managing pediatric rhabdomyosarcoma. F1000Res 9:F1000 Faculty Rev-685, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skapek SX, Ferrari A, Gupta AA, et al. : Rhabdomyosarcoma. Nat Rev Dis Primers 5:1, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oberlin O, Rey A, Lyden E, et al. : Prognostic factors in metastatic rhabdomyosarcomas: Results of a pooled analysis from United States and European cooperative groups. J Clin Oncol 26:2384-2389, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oberlin O, Rey A, Sanchez de Toledo J, et al. : Randomized comparison of intensified six-drug versus standard three-drug chemotherapy for high-risk nonmetastatic rhabdomyosarcoma and other chemotherapy-sensitive childhood soft tissue sarcomas: Long-term results from the International Society of Pediatric Oncology MMT95 study. J Clin Oncol 30:2457-2465, 2012 [DOI] [PubMed] [Google Scholar]
- 6.Chisholm JC, Marandet J, Rey A, et al. : Prognostic factors after relapse in nonmetastatic rhabdomyosarcoma: A nomogram to better define patients who can be salvaged with further therapy. J Clin Oncol 29:1319-1325, 2011 [DOI] [PubMed] [Google Scholar]
- 7.Van Paemel R, Vlug R, De Preter K, et al. : The pitfalls and promise of liquid biopsies for diagnosing and treating solid tumors in children: A review. Eur J Pediatr 179:191-202, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersson D, Fagman H, Dalin MG, et al. : Circulating cell-free tumor DNA analysis in pediatric cancers. Mol Aspects Med 72:100819, 2020 [DOI] [PubMed] [Google Scholar]
- 9.Lak NSM, Voormanns TL, Zappeij-Kannegieter L, et al. : Improving risk stratification for pediatric patients with rhabdomyosarcoma by molecular detection of disseminated disease. Clin Cancer Res 27:5576-5585, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Zogchel LMJ, van Wezel EM, van Wijk J, et al. : Hypermethylated RASSF1A as circulating tumor DNA marker for disease monitoring in neuroblastoma. JCO Precision Oncol 4:291-306, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jimenez I, Chicard M, Colmet-Daage L, et al. : Circulating tumor DNA analysis enables molecular characterization of pediatric renal tumors at diagnosis. Int J Cancer 144:68-79, 2019 [DOI] [PubMed] [Google Scholar]
- 12.Chicard M, Boyault S, Colmet Daage L, et al. : Genomic copy number profiling using circulating free tumor DNA highlights heterogeneity in neuroblastoma. Clin Cancer Res 22:5564-5573, 2016 [DOI] [PubMed] [Google Scholar]
- 13.Chicard M, Colmet-Daage L, Clement N, et al. : Whole-exome sequencing of cell-free DNA reveals temporo-spatial heterogeneity and identifies treatment-resistant clones in neuroblastoma. Clin Cancer Res 24:939-949, 2018 [DOI] [PubMed] [Google Scholar]
- 14.van Zogchel LMJ, Lak NSM, Verhagen OJHM, et al. : Novel Circulating Hypermethylated RASSF1A ddPCR for liquid biopsies in patients with pediatric solid tumors. JCO Precis Oncol 5:1738-1748, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lobo J, van Zogchel LMJ, Nuru MG, et al. : Combining hypermethylated RASSF1A detection using ddPCR with miR-371a-3p testing: An improved panel of liquid biopsy biomarkers for testicular germ cell tumor patients. Cancers (Basel) 13:5228, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Paemel R, Vandeputte C, Raman L, et al. : The feasibility of using liquid biopsies as a complementary assay for copy number aberration profiling in routinely collected paediatric cancer patient samples. Eur J Cancer 160:12-23, 2022 [DOI] [PubMed] [Google Scholar]
- 17.Eguchi-Ishimae M, Tezuka M, Kokeguchi T, et al. : Early detection of the PAX3-FOXO1 fusion gene in circulating tumor-derived DNA in a case of alveolar rhabdomyosarcoma. Genes Chromosomes Cancer 58:521-529, 2019 [DOI] [PubMed] [Google Scholar]
- 18.Klega K, Imamovic-Tuco A, Ha G, et al. : Detection of somatic structural variants enables quantification and characterization of circulating tumor DNA in children with solid tumors. JCO Precis Oncol 10.1200/PO.17.0028, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun W, Chatterjee B, Wang Y, et al. : Distinct methylation profiles characterize fusion-positive and fusion-negative rhabdomyosarcoma. Mod Pathol 28:1214-1224, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seki M, Nishimura R, Yoshida K, et al. : Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nat Commun 6:7557, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Paemel R, De Koker A, Vandeputte C, et al. : Minimally invasive classification of paediatric solid tumours using reduced representation bisulphite sequencing of cell-free DNA: A proof-of-principle study. Epigenetics 16:196-208, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parham DM, Barr FG: Classification of rhabdomyosarcoma and its molecular basis. Adv Anat Pathol 20:387-397, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Koker R, Van Paemel R, De Wilde B, et al. : A versatile method for circulating cell-free DNA methylome profiling by reduced representation bisulfite sequencing. https://www.biorxiv.org/content/10.1101/663195v2, 2019 [Google Scholar]
- 24.Wan JCM, Massie C, Garcia-Corbacho J, et al. : Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat Rev Cancer 17:223-238, 2017 [DOI] [PubMed] [Google Scholar]
- 25.Vessies DCL, Greuter MJE, van Rooijen KL, et al. : Performance of four platforms for KRAS mutation detection in plasma cell-free DNA: ddPCR, Idylla, COBAS z480 and BEAMing. Sci Rep 10:8122, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grawenda AM, O'Neill E: Clinical utility of RASSF1A methylation in human malignancies. Br J Cancer 113:372-381, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim S, Yang MH, Park JH, et al. : Inactivation of the RASSF1A in osteosarcoma. Oncol Rep 10:897-901, 2003 [PubMed] [Google Scholar]
- 28.Wagner KJ, Cooper WN, Grundy RG, et al. : Frequent RASSF1A tumour suppressor gene promoter methylation in Wilms' tumour and colorectal cancer. Oncogene 21:7277-7282, 2002 [DOI] [PubMed] [Google Scholar]
- 29.Honda S, Miyagi H, Suzuki H, et al. : RASSF1A methylation indicates a poor prognosis in hepatoblastoma patients. Pediatr Surg Int 29:1147-1152, 2013 [DOI] [PubMed] [Google Scholar]
- 30.Astuti D, Agathanggelou A, Honorio S, et al. : RASSF1A promoter region CpG island hypermethylation in phaeochromocytomas and neuroblastoma tumours. Oncogene 20:7573-7577, 2001 [DOI] [PubMed] [Google Scholar]
- 31.Stutterheim J, Ichou FA, den Ouden E, et al. : Methylated RASSF1a is the first specific DNA marker for minimal residual disease testing in neuroblastoma. Clin Cancer Res 18:808-814, 2012 [DOI] [PubMed] [Google Scholar]
- 32.Van Roy N, Van Der Linden M, Menten B, et al. : Shallow whole genome sequencing on circulating cell-free DNA allows reliable noninvasive copy-number profiling in neuroblastoma patients. Clin Cancer Res 23:6305-6314, 2017 [DOI] [PubMed] [Google Scholar]
- 33.Clay MR, Patel A, Tran Q, et al. : Methylation profiling reveals novel molecular classes of rhabdomyosarcoma. Sci Rep 11:22213, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koelsche C, Schrimpf D, Stichel D, et al. : Sarcoma classification by DNA methylation profiling. Nat Commun 12:498, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R2 : Genomics Analysis and Visualization Platform. http://r2.amc.nl
- 36.Raos D, Ulamec M, Katusic Bojanac A, et al. : Epigenetically inactivated RASSF1A as a tumor biomarker. Bosn J Basic Med Sci 21:386-397, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niu H, Yang J, Yang K, et al. : The relationship between RASSF1A promoter methylation and thyroid carcinoma: A meta-analysis of 14 articles and a bioinformatics of 2 databases (PRISMA). Medicine (Baltimore) 96:e8630, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu G, Zhou X, Xing J, et al. : Identification of RASSF1A promoter hypermethylation as a biomarker for hepatocellular carcinoma. Cancer Cell Int 20:547, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L, Shern JF, Wei JS, et al. : Clonality and evolutionary history of rhabdomyosarcoma. PLoS Genet 11:e1005075, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lynn M, Shah N, Conroy J, et al. : A study of alveolar rhabdomyosarcoma copy number alterations by single nucleotide polymorphism analysis. Appl Immunohistochem Mol Morphol 22:213-221, 2014 [DOI] [PubMed] [Google Scholar]
- 41.Laughton SJ, Ashton LJ, Kwan E, et al. : Early responses to chemotherapy of normal and malignant hematologic cells are prognostic in children with acute lymphoblastic leukemia. J Clin Oncol 23:2264-2271, 2005 [DOI] [PubMed] [Google Scholar]
- 42.van Dongen JJ, Seriu T, Panzer-Grumayer ER, et al. : Prognostic value of minimal residual disease in acute lymphoblastic leukaemia in childhood. Lancet 352:1731-1738, 1998 [DOI] [PubMed] [Google Scholar]