Abstract
We have attempted to explore further the involvement of complement components in the host COVID-19 (Coronavirus disease-19) immune responses by targeted genotyping of COVID-19 adult patients and analysis for missense coding Single Nucleotide Polymorphisms (coding SNPs) of genes encoding Alternative pathway (AP) components. We have identified a small group of common coding SNPs in Survivors and Deceased individuals, present in either relatively similar frequencies (CFH and CFI SNPs) or with stark differences in their relative abundance (C3 and CFB SNPs). In addition, we have identified several sporadic, potentially protective, coding SNPs of C3, CFB, CFD, CFH, CFHR1 and CFI in Survivors. No coding SNPs were detected for CD46 and CD55. Our demographic analysis indicated that the C3 rs1047286 or rs2230199 coding SNPs were present in 60 % of all the Deceased patients (n = 25) (the rs2230199 in 67 % of all Deceased Males) and in 31 % of all the Survivors (n = 105, p = 0.012) (the rs2230199 in 25 % of all Survivor Males). When we analysed these two major study groups using the presence of the C3 rs1047286 or rs2230199 SNPs as potential biomarkers, we noticed the complete absence of the protective CFB rs12614 and rs641153 coding SNPs from Deceased Males compared to Females (p = 0.0023). We propose that in these individuals, C3 carrying the R102G and CFB lacking the R32W or the R32Q amino acid substitutions, may contribute to enhanced association dynamics of the C3bBb AP pre-convertase complex assembly, thus enabling the exploitation of the activation of the Complement Alternative pathway (AP) by SARS-CoV-2.
Keywords: SARS-CoV-2, COVID-19, Complement, Alternative pathway, C3, CFB, Coding SNPs, Host genomics
1. Introduction
Since the beginning of the COVID-19 (Coronavirus disease 2019) outbreak a large number of studies have attempted to dissect the complex molecular and cellular aspects of the SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) induced pathophysiology (Yuki et al., 2020, Varghese et al., 2020). Advanced efforts were carried out to understand the dynamic plasticity of the virus (Plante et al., 2021, Jungreis et al., 2021) and explore the multidimensional host immune responses during infection (Ramlall et al., 2020, Kaneko et al., 2020, Yao et al., 2021, Sette and Crotty, 2021). As aspects of the COVID-19 pathology resemble clinical features traditionally associated with complementopathies (Holter et al., 2020, Noris et al., 2020, Marcos-Jiménez et al., 2021, Zinellu and Mangoni, 2021, Siggins et al., 2022, Gavriilaki and Brodsky, 2020), it became apparent early on that the Complement system (Carroll and Sim, 2011) may influence viral sensing and inflammation dynamics during infection (Java et al., 2020). Therefore, there has been a rationale under discussion for targeting Complement in COVID-19 (Polycarpou et al., 2020, Risitano et al., 2020, Mastellos et al., 2020).
Targeting complement activation in COVID-19 has been recently discussed more extensively, as several elegant studies have showed that SARS-CoV-2 can directly activate Complement by engaging multiple of its pattern recognition components (Ramlall et al., 2020, Kulkarni and Atkinson, 2020, Yu et al., 2020, Wilk, 2020, Ali et al., 2021, Götz et al., 2022, Bosmann, 2021, Ma et al., 2021, Yan et al., 2021, Georg et al., 2022, Satyam et al., 2021, Defendi et al., 2021). The activation of the Alternative pathway by the spike glycoprotein S has been of particular interest as it promotes entry of the virus into cells and is a major antigenic target for B cell responses (Yu et al., 2020, Walls et al., 2020, Renhong et al., 2020, Wu et al., 2021, Watanabe et al., 2020). Furthermore, the potential exploitation of the activation of the Alternative pathway via the amplification loop by SARS-CoV-2 is of even greater particular interest, because it can explain to a considerable extent the gradual diminishment of Complement responses during the systemic establishment of the virus, especially in the severe cases of infection (Marcos-Jiménez et al., 2021, Zinellu and Mangoni, 2021, Sinkovits et al., 2021, Siggins et al., 2022).
The evidence-based involvement of Complement as a mediator of the overall immune responses highlights it as a considerable potential regulator of the clinical outcomes of infection among individuals. Although predicting and/or assessing the tuning of the human systemic immune responses genetically and/or epigenetically is a daunting task, advanced efforts during the pandemic attempted to identify host genetic sources of susceptibility to severe infection. One major group of efforts attempted to identify intracellular host factors of susceptibility by whole-genome CRISPR (clustered regularly interspaced short palindromic repeats) knock out screening approaches using in vitro cell models (Daniloski et al., 2021, Schneider et al., 2021, Wang et al., 2021).
The other major group of efforts aimed to identify, by targeted or holistic approaches, Single Nucleotide Polymorphisms (SNPs) that could further promote prognostic and/or diagnostic applications as well as in-depth reverse genetics mechanistic studies (“Genomewide Association Study of Severe Covid-19 with Respiratory Failure,” 2020; “Mapping the human genetic architecture of COVID-19,” 2021; Gavriilaki et al., 2021, Ramlall et al., 2020, Genomewide Association Study of Severe Covid-19 with Respiratory Failure, 2020, Mapping the human genetic architecture of COVID-19, 2021, Zhang et al., 2020). These cited efforts revealed complex patterns of genetic mosaicity that could be linked with various layers and processes of the host responses such as the Type I IFN (interferon) responses (Bastard et al., 2020, Martin-Sancho et al., 2021, King and Sprent, 2021), as well as the Complement (Ramlall et al., 2020, Gavriilaki et al., 2021) and coagulation cascades (Ramlall et al., 2020).
The Complement SNPs findings published by Ramlall et al., 2020 study were of particular interest, because a subsequent refining metadata analysis identified a few coding SNPs (coding SNPs rs2230199 and rs1047286 for C3 and rs45574833 for C4BP) that could structurally explain the deregulation of the Alternative pathway in severe cases of the disease (Tsiftsoglou, 2021). These concepts have been made possible as the Biochemistry of Complement has been intensely investigated for decades and the majority of the structures of Complement components have now been solved in high resolution (Sim et al., 2016, Geisbrecht et al., 2022). In addition, precision technological advances in modern molecular biology and genetics have begun to bridge genetic mosaicity with molecular structures and phenotypes in Complement (Liszewski and Atkinson, 2015, Łukawska et al., 2018, Rodríguez de Córdoba, 2022).
The findings discussed above have prompted us to explore further the involvement of Complement components in host COVID-19 immune responses. In this study, we present the relative distribution patterns and frequencies of missense coding SNPs as revealed by the targeted genotyping of 130 COVID-19 adult patients for genes encoding the Alternative pathway (AP) components C3, C5, CFB, CFD, CFH, CFHR1, CFI, CD46 and CD55. We have identified 8 common coding SNPs in Survivors and Deceased individuals, present in either relatively similar frequencies (CFH and CFI SNPs) or with considerable differences in their relative frequencies (C3 and CFB SNPs). Moreover, we identified 24 sporadic and potentially protective coding SNPs of C3, CFB, CFD, CFH, CFHR1 and CFI in Survivors. Our demographic analysis of the common coding SNPs revealed patterns of their distribution that can be potentially associated with deregulated Complement Alternative pathway (AP) responses during infection by SARS-CoV-2.
2. Materials and methods
2.1. Study population
We prospectively studied consecutive patients that met the following inclusion criteria: 1) adult, 2) of Caucasian origin, 3) hospitalized with COVID-19 in our referral centres during the first, second and third waves of the pandemic (April-May 2020, September-October 2020 and November-January 2021) Diagnosis was confirmed by reverse-transcriptase polymerase chain reaction (RT-PCR). COVID-19 severity was assessed based on World Health Organization’s (WHO) criteria. Additional data on patients’ history and course were recorded by treating physicians that followed patients up to discharge or death. The final analysis included only patients with available data on clinical course and outcome. The study was approved by Institutional Review Boards (IRBs) of the recruiting centres (G. Papanicolaou Hospital at Thessaloniki, Greece and Attikon Hospital at Athens, Greece) and conducted according to the Declaration of Helsinki.
2.2. Genomic analysis
Genomic DNA was isolated from peripheral blood samples using the QIAamp DNA Mini Blood (Qiagen, Hilden, Germany). It was analyzed using Next Generation Sequencing (NGS) with a customized complement-related gene panel that included CFB, CFD, CFH, CFHR1, CFI, C3, C5, CD46 and CD55, as previously published in a study of adult patients by the team of the Hematology Clinic of Papanikolaou Hospital (Gavriilaki et al., 2021). The probes and primers were designed in DesignStudio (Illumina, San Diego, CA, USA) to cover all exonic regions spanning 15 bases into introns (98 % coverage). 10 ng of genomic DNA was used to build the libraries (MiniSeq, Illumina). Libraries were quantified using Qubit (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced on a MiniSeq System in a 2 × 150 bp run (Illumina). Sequence quality was initially assessed using Illumina tools. As the original sequencing was of high quality, each sample was processed independently, in order to properly map the sequences against the human reference genome. Variations with an allelic frequency higher than 20 % were analyzed. Both Ensembl and Refseq resources were used for annotation of the output files. The clinical significance of the variants was preliminary evaluated with the NCBI ClinVar database (Landrum et al., 2018) and then interpreted with the NCBI dbSNP and UniProt databases as well as the international published literature during further refined analysis. The interpretation aimed at identifying missense coding variants of interest that can contribute to complement dysregulation through protein–protein interactions in characterized complex formations.
2.3. Data mining and clustering
We first identified all the detected missense coding SNPs of the genes encoding the Alternative pathway (AP) components C3, C5, CFB, CFD, CFH, CFHR1, CFI, CD46 and CD55 from each annotated output patient genotyping profile file and validated them with publicly available databases (Sherry et al., 2001, Landrum et al., 2018). The sum of identified coding SNPs was then clustered in two major groups based on the clinical outcome of the infection. One major group included the coding SNPs from patients who were hospitalized and eventually deceased (Deceased group), and the other included the coding SNPs from patients who eventually survived (Survivors group) and developed a range of mild to severe symptoms that required hospitalization in an Intensive Care Unit (ICU). Initially, we maintained the data from the Survivors group in two separate subgroups based on the severity of the disease (whether hospitalization in ICU was necessary or not), but for unbiased comparisons and simplicity reasons, we merged both sets into a unified Survivors group. Within this frame, the sum of the coding SNPs identified in our study, was segregated using as a sole criterium the terminal clinical outcome of infection, that of survival or not. For every coding SNP identified, its relative frequency (%) was calculated by dividing the number of times the coding SNP is represented within each group, with the total number of patients included in each group. For the sum of the coding SNPs identified for both major groups, analysis effort was put towards identifying common groups of SNPs, as well as unique SNPs for each group. For direct table comparison purposes, all the coding SNPs identified the Deceased and the Survivors groups were ranked from the most to the least abundant SNP.
2.4. Coding SNPs demographics
In order to identify any patterns of biologically interesting coding SNPs distributions that could be associated with disease severity, we analysed the demographics of all the common coding SNPs found in the Deceased and the Survivors groups and clustered them by gender (M/F) and age (<60 or ≥ 60 yrs). The relative SNP frequency (%) in each subgroup was calculated by dividing the number of patients carrying each coding SNP, with the total number of patients included in each subgroup. As we realized that the CFB coding SNPs rs12614 (R32W) and rs641153 (R32Q) encode different amino acid substitutions for the same position R32 (Sherry et al., 2001, Landrum et al., 2018) (Table 2), we decided to include both in our demographics analysis separately, although the rs641153 SNP was detected only in the Survivors group and at a lower frequency compared to the rs12614 (Table 1 ).
Table 2.
All the Complement missense coding SNPs identified in the current study.
 |
Table 1.
Human Complement missense coding SNPs detected in SARS-CoV-2 patients who deceased in ICU or survived after infection.
 |
To explore whether the presence of the C3 rs1047286 and rs2230199 coding SNPs is functionally associated with disease severity in particular, we further stratified our two major patient groups, the Deceased and the Survivors groups, by the presence of the C3 rs1047286 and/or the rs2230199 as biomarkers and analysed the demographics of the common coding SNPs distributions in those individuals using the same criteria as above.
2.5. Statistical analysis
Differences between groups were compared using chi-square test (with Bonferroni’s correction when needed) for categorical variables and Student t-test or Mann-Whitney U test for quantitative variables, as appropriate. Statistical tests were 2-sided with p-values ≤ 0.05 denoting statistical significance. The Hardy-Weinberg Equilibrium (HWE) was tested via the χ2-test for goodness of fit using a web program (https://ihg.gsf.de/cgi-bin/hw/hwa1.pl). For simplicity reasons, only statistically significant p-values are included in the result sections. Analysis was performed on SPSS 22.0 (IBM SPSS Statistics for Windows, Version 22.0. Armonk, NY: IBM Corp) software.
3. Results
3.1. Study population and identification of common and unique coding SNPs
We prospectively studied 130 hospitalized COVID-19 patients in total, from whom 25 eventually deceased (nD = 25) (Deceased group), while the other 105 survived (nS = 105) (Survivors group). Median age was 62 years (range 18–93). Reported comorbidities included: essential hypertension (72 %), obesity (69 %), dyslipidemia (63 %), diabetes mellitus (32 %) and other disorders (11 %). No history of genetic disorder was recorded.
Among the extended diversity of the SNPs that we managed to detect, we identified several coding SNPs of the genes encoding the Complement components C3, CFB, CFD, CFH, CFHR1 and CFI, but interestingly none for the genes encoding the regulators CD46 and CD55 as shown in Table 1, Table 2. In the two major groups of our study, the Survivors and the Deceased groups, we identified 8 common as well as 26 unique coding SNPs. Among them, 24 potentially protective were found only in the Survivors group and just 2 potentially hazardous only in the Deceased group (p = 0.042, Table 1). The patients Survivors group contained several unique SNPs for CFB, CFH, CFI, C3, CFD and CFHR1 that were absent from those in the Deceased group. The vast majority of all the unique coding SNPs identified were sporadic in terms of abundance and their considerable larger number in the Survivors group might be associated with increased natural protection in some individuals.
3.2. Common coding SNPs with similar and different frequencies
Our analysis of the common group of 8 coding SNPs, revealed initially the presence of 4 common SNPs present in both groups in very similar abundances (Table 1). These include the rs11098044 of CFI as well as the rs1061170, rs800292 and rs1065489 of CFH. The SNPs rs11098044 of CFI (T300A) and rs1061170 of CFH (Y402H in CCP7) were detected in fairly high abundances in both groups of patients, 100 % and > 80 %, respectively, while the SNPs rs800292 (V62I in CCP1) and rs1065489 (E936D in CCP16) of CFH in lower abundances (∼30 %) (Table 1, Table 2).
Our further analysis of the common coding SNPs between the Deceased and Survivors groups, revealed the presence of the 3 common SNPs, the C3 rs2230199 and rs1047286 and the CFB rs12614 in considerable abundance in both groups, but with markedly different frequencies (Table 1). Our data indicate that the frequencies of each of these two C3 SNPs of interest are both nearly double (∼2X) in the Deceased group as compared to the Survivors group (p < 0.001, Table 1). Although the relative abundance of the CFB rs12614 coding SNP is mildly greater in the Survivors group (29 % compared to 20 %), we noticed that when we added the relative abundances of rs12614 and rs641153 together in the Survivors group (as they both encode for different amino acid substitutions of R32 in CFB), then the merged frequency of rs12614 (29 %) and rs641153 (12 %) is also almost double (∼2X) in the Survivors group (41 %) compared to the Deceased group (20 %) (p < 0.001, Table 1).
3.3. Common coding SNP demographics
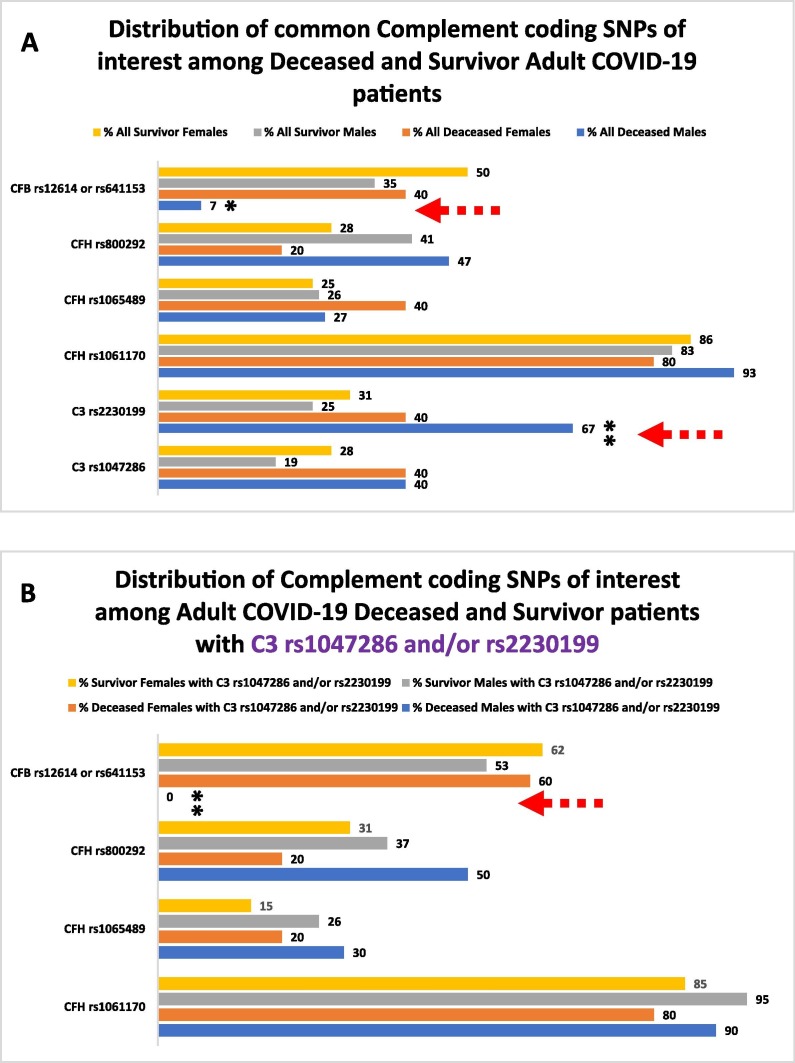
When we analysed all the demographic distributions of our common coding SNPs, we observed a significantly higher frequency of the C3 rs1047286 or rs2230199 SNPs in the Male (M) patients of the Deceased compared to the Survivors subgroup (67 % vs 28 %, p = 0.008, Table 3 ). Within this subgroup, these SNPs appeared predominantly present in older patients (≥60 yrs), while their age-related distribution frequencies appeared more balanced in the other three subgroups (Table 3). We also noticed the very low presence of the CFB rs12614 SNP in the Deceased M subgroup (7 %) compared to the Deceased F subgroup (40 %). In addition, the corresponding joint frequencies of the CFB rs12614 or rs641153 SNP entries were significantly higher in the Survivors M (35 %, p = 0.041).
Table 3.
Demographics of the common coding SNPs distributions in all the patients examined in this study.
 |
To identify any potential associations between the presence of the C3 rs1047286 and rs2230199 coding SNPs and disease severity in patients, we took a step forward and analysed our two major study groups further, the Deceased and Survivors groups, using the two C3 SNPs as potential biomarkers (Table 4 ). We discovered that the patients with the C3 rs1047286 and/or rs2230199 SNPs corresponded to 60 % of all the Deceased and 31 % of all the Survivors patients, respectively (p = 0.012). In the C3 selected subgroup of Deceased M, the rs2230199 SNP is strongly predominant over the rs1047286 SNP (100 % vs 60 %) and highly abundant in older patients (≥60 yrs) (Table 4). In the same cohort, the CFH rs1061170 SNP was found similarly highly predominant in older patients (≥60 yrs), while the overall frequencies of the three common CFH coding SNPs of our study appeared similar in the 3 remaining C3 rs1047286 and/or rs2230199 stratified subgroups (Table 4). Interestingly, we could not detect the CFB rs12614 coding SNP in this selected patient population of Deceased M compared to F (60 %, p = 0.023), while there is considerable enrichment for the CFB rs12614 or rs641153 entries in the Survivors M (53 %) and F (62 %) subgroups (Table 4).
Table 4.
Demographics of common coding SNPs distributions in patients with the C3 rs1047286 and/or rs2230199.
 |
4. Discussion
The current study stemmed from our urgent initiative during the COVID-19 pandemic to explore further the potential contributions of the Complement system to the immune responses against SARS-CoV-2. Despite the surge in volume of published studies and the significant advances carried out towards the exploration of the molecular basis of pathophysiology, it has been a challenging task to identify potential cornerstones that can rationally explain the population wide impact of the infection on an individual basis. In this study, we stepped on our recent findings (Gavriilaki et al., 2021) and observations (Tsiftsoglou, 2021) and attempted to decipher the patterns and distributions of SNPs encoding amino acid substitutions in Complement components historically associated with the mechanics of the Alternative pathway (AP) (Sim et al., 2016, Liszewski and Atkinson, 2022, Harrison et al., 2022).
Our focus on the coding SNPs did not aim to reduce the potential importance of other non-coding gene regulatory elements that may be involved in important regulatory processes, but was chosen after careful consideration towards explaining on biochemical grounds, potential differential capacities of the individual immune responses to defend and tackle the viral infection. In order to apply this targeted analytic approach, we have not included additional variables, other than the clinical outcome of the infection. Among the two major groups of study, the Survivors and the Deceased groups, we identified several unique and sporadic, in terms of frequencies, coding SNPs that some maybe protective or hazardous for the host, respectively (Table 1, Table 2). Some of the encoded amino acid substitutions (Table 2) may alter the functional properties of the encoded Complement components directly or in surface complex formations. Such changes may influence the activation threshold dynamics of the Alternative pathway (AP) and to a further extent the clinical outcome of the infection (Harrison et al., 2022).
The involvement of the C3 rs2230199 and rs1047286 coding SNPs in the molecular pathophysiology of the COVID-19 was first reported in the Ramlall et al., 2020 study (Ramlall et al., 2020) and their potential contributions to the disease on a biochemical basis was then discussed in detail by Tsiftsoglou (Tsiftsoglou, 2021). The rs2230199 introduced R102G (R80G) polymorphism was first reported in 1967 and has been described in the literature to distinguish the C3S (Slow, R80) and the C3F (Fast, G80) protein isoforms (Wieme and Demeulenaere, 1967, Alper and Propp, 1968, Botto et al., 1990). The three positively charged amino acids of C3, R72, R80 and K82 of the MG1 domain, along with the four negatively charged amino acids D1007, E1008, E1010, E1013 of the TED domain, form the internal thioester bond that is critical for the reactivity of C3 with nucleophilic groups (Law and Dodds, 1997, Sim et al., 2022). The C3F isoform, containing neutral glycine instead of the positively charged arginine has potentially reduced properties with regards to the stability of its thioester and its potency to react with potential complement activator surfaces (Rodriguez et al., 2015). It also exhibits a weaker ability to bind to CFH which is the major circulating cofactor for the CFI mediated downregulation of the C3bBb AP convertase (Rodriguez et al., 2015, Soames and Sim, 1997, Sim and Tsiftsoglou, 2004, Roversi et al., 2011, Nilsson et al., 2011, Janssen et al., 2006, Torreira et al., 2009). The common C3S/F comprises approximately 98 % of all C3 phenotypes (Łukawska et al., 2018). Carriers of the C3F allele have been reported to be at higher risk for developing Acute Macular Degeneration (AMD) (Yates et al., 2007) and various types of glomerulonephritis namely membranoproliferative glomerulonephritis type II (MPGN II) that is presently known as dense deposit disease (DDD) (Abrera-Abeleda et al., 2011), IgA nephropathy (IgAN) (Rambausek et al., 1987) and systemic vasculitis (SV) (Persson et al., 2013).
The second C3 point mutation corresponding to rs1047286 in the exon 9 of the beta chain, the P314L (P292L) has also been historically characterized as HAV4-1+/- (Table 1) (Koch and Behrendt, 1986, Botto et al., 1990). The P292 (HAV 4-1-) residue is located in the MG3 domain of C3 that contains the major binding site for the activated complement factor B (Bb). Published structural studies have showed that the polymorphic protein isoform HAV4-1+ (L292) exhibits stronger ability to bind to Bb (Torreira et al., 2009, Forneris et al., 2010). This practically suggests that the HAV4-1+ (L292) gain-of-function isoform may form a hyperactive C3bBb AP convertase that may be also more resilient to inactivation by complement factor I (FI) in the presence of a cofactor such as complement factor H (FH) (Rodríguez De Córdoba et al., 2004, Ferreira et al., 2010) or C4BP (Blom et al., 2003). It has been estimated that 98 % of C3S allele carriers is HAV4-1- and only 2 % is HAV4-1+; in contrast, 90 % carriers of the C3F allele is HAV4-1 + and 10 % is HAV4-1- (Botto et al., 1990).
The frequency of the C3F minor allele of the rs2230199 SNP varies among different populations, and is the highest in Caucasian (18 %) and the lowest in Asian (1 %) (Bazyar et al., 2012). Our independent database investigation indicated that compared to Europeans, the minor alleles of the rs2230199 and rs1047286 C3 coding SNPs of interest are rare in some world populations, in particular the African and East Asian populations (1000 Genomes database). This remarkable genetic variability could be the result of evolutionary population adaptation to local microbial challenges and/or viral outbreaks over extended periods of time (Ermini et al., 2012, Souilmi et al., 2021). Such variability could be important for the regulation of the Alternative pathway (AP) in the molecular pathophysiology of COVID-19 and the resilience to severe infection among different populations (Holers, 2008).
The identified CFB rs12614 and rs641153 coding SNPs introduce two different amino acid substitutions of same residue R32. The rs12614 SNP corresponds to the R32W mutation, while the rs641153 SNP refers to the R32Q mutation. Both have been studied in the recent years as sources of protection in Complement mediated pathologies such as AMD (Montes et al., 2009, Hughes et al., 2011, Heurich et al., 2011, Pilotti et al., 2020) and the atypical Hemolytic Uremic syndrome (aHUS) (de Córdoba et al., 2011, Marinozzi et al., 2014). The R32 residue is located in the Ba fragment of the pre-activated CFB that is later released upon activation of the C3bBb pre-convertase by CFD (Arlaud et al., 1998, Sim and Tsiftsoglou, 2004). Although the Ba fragment is released upon activation of the pre-convertase, it harbors a weak, but important low affinity interaction site for C3b, that contributes to the overall avidity of CFB for C3b and the assembly of the pre-convertase (Arlaud et al., 1998, Torreira et al., 2009, Forneris et al., 2010, Marinozzi et al., 2014). The more recent high resolution structural studies confirmed an early model based on mutagenesis studies, proposing that the initial binding of CFB to activator-attached C3b is mediated by two-low affinity sites, one on Ba and the other on the vWFA domain (Arlaud et al., 1998). Both the CFB rs12614 and rs641153 introduced substitutions weaken the low affinity interaction and contribute to the disruption of the dynamics that contribute to the C3bBb pre-convertase assembly (Montes et al., 2009, de Córdoba et al., 2011, Marinozzi et al., 2014).
If the AP pre-convertase cannot be efficiently assembled on an activator surface, the activator-attached C3b is sequentially cleaved by complement factor I (FI) in the presence of a cofactor. In this context, the CFB rs12614 and rs641153 coding SNPs act protectively for the host, as they weaken or structurally ‘counterbalance’ the effects of the C3 rs2230199 and rs1047286 coding SNPs that promote the formation of a potential hyperactive C3bBb AP convertase. This convertase may be more resilient to inactivation by complement factor I (FI) in the presence of a suitable cofactor such as CFH. A hyperactive C3bBb AP convertase can contribute to increased kinetics of local or systemic Complement consumption during the viral infection. This ‘molecular loophole’ may enable SARS-CoV-2 to exploit the Alternative activation pathway amplification loop introducing C3 consumption. This can practically result in lower generation of iC3b, C3dg and C3d which are major opsonins of the complement system (Carroll and Isenman, 2012, Morgan et al., 2011, Xue et al., 2017). Impaired opsonization during infection can contribute to poor immune synergies of complement with the various professional immune surveillance cell populations that include phagocytes, neutrophils and B cells (Carroll and Isenman, 2012, Zarantonello et al., 2022). We propose that in individuals, with C3 carrying the R102G and CFB lacking the R32W or the R32Q amino acid substitutions, the association dynamics of the C3bBb AP pre-convertase complex assembly are enhanced and therefore enable the exploitation of the activation of the Complement Alternative pathway (AP) by SARS-CoV-2. Interestingly, several pathogens have evolved several elegant mechanisms to evade complement via the AP by expressing enzymes and competitors that degrade AP components and/or block the assembly of C3 convertases (Shaughnessy et al., 2023). Therefore, the combinatorial use of the C3 rs2230199 and the CFB rs12614, rs641153 coding SNPs as molecular biomarkers can potentially distinguish a relatively large cohort (∼70 %) of individual males with a strong predisposition to severe infection (Table 3, Table 4 and Fig. 1 A, B). In the context of the variant distributions discussed earlier and in terms of population genetics, individuals carrying the C3 R102G amino acid substitution would be considerably fewer in the African and East Asian populations that exhibit low penetrance of the minor allele C3F of rs2230199.
Fig. 1.
Graph visualised data derived from Tables 3 (A) and 4 (B), respectively. The red dashed arrows indicate the most striking differences observed for the frequencies of C3 rs2230199 and CFB rs12614, rs641153 among the Deceased and Survivor Adult COVID-19 patients examined. *For comparisons between deceased and survivors with p-values < 0.05, **For comparisons between male and female patients with p-values < 0.05.
It should be noted that the landscape of COVID-19 infection is constantly evolving with new virus variants and added rounds of vaccinations that influence the clinical outcomes of the disease. Nevertheless, we present robust findings in a homogeneous Caucasian population that are in accordance and expand previous knowledge. Some of the comorbidities reported for this study group, may influence the severity of the disease and aspects of the responses to medication through epigenetic gene regulatory mechanisms, but herein we decided to keep our study focused on the findings of germline genetic variation. Moreover, our findings further support the notion behind complement therapeutics in COVID-19, especially the ones targeting the components of the AP, primarily C3 (Sim et al., 2022, Skendros et al., 2022, Pedersen et al. 2020), CFB as well as CFD (Schubart et al., 2022, Dreismann et al., 2023). In particular, safety and efficacy has been shown for terminal complement inhibition with the anti-C5 eculizumab (Diurno et al., 2020), C3 inhibition with the AMY-101 (Mastaglio et al., 2020), C1 inhibition with conestat alpha (Urwyler et al., 2020), and lectin pathway inhibition with the anti-MASP-2 narsoplimab (Rambaldi et al., 2020). Importantly, when C3 inhibition was compared to terminal inhibition, a broader involvement of C3 in COVID-19 thrombo-inflammation was suggested (Mastellos et al., 2020).
Overall, we consider these findings important, because at the protein level they support our argument about the exploitation of the activation of the AP by SARS-CoV-2, particularly at the level of C3. In the context of the COVID-19 molecular pathology, this can gradually lead to complement deregulation, poor opsonization, weakening of the immune clearance mechanisms and sustained inflammation resulting in various host tissue damage responses in severe cases of infection. Further studies are needed to better understand the potential molecular, cellular and clinical implications of our findings.
Sources of funding
The genetic analysis was supported by an independent investigator driven grant (Pfizer Pharmaceuticals) as well as by COVID-19 research funding by the Prefecture of Central Macedonia in Thessaloniki, Greece.
Author contributions statement
EG conceptualized and designed the genetic analysis study, SAT designed and carried out the analysis of the coding SNPs and drafted the manuscript. SAT and EG have contributed equally in this study. VK, CV, AV, IK, EK, MB recruited patients and recorded data. TT, EEK, MK, PGP, AP, FC, MC, DC, ER, AP performed laboratory procedures for samples preparation and genetic testing. DTB, AT, DS, IS, SK, and AA contributed in research design, edited and approved the paper.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imbio.2023.152351.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data Availability: The authors declare that data supporting the findings of this study are available within the paper and its supplementary information files.
References
- Abrera-Abeleda M.A., Nishimura C., Frees K., Jones M., Maga T., Katz L.M., Zhang Y., Smith R.J.H. Allelic variants of complement genes associated with dense deposit disease. J. Am. Soc. Nephrol. 2011;22:1551–1559. doi: 10.1681/ASN.2010080795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Y.M., Ferrari M., Lynch N.J., Yaseen S., Dudler T., Gragerov S., Demopulos G., Heeney J.L., Schwaeble W.J. Lectin pathway mediates complement activation by SARS-CoV-2 proteins. Front. Immunol. 2021;12:2645. doi: 10.3389/fimmu.2021.714511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper C.A., Propp R.P. Genetic polymorphism of the third component of human complement (C’3) J. Clin. Invest. 1968;47:2181–2191. doi: 10.1172/JCI105904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlaud, G.J., Volanakis, J.E., Thielens, N.M., Narayana, S.V.L., Rossi, V., Xu, Y., 1998. The Atypical Serine Proteases of the Complement System. pp. 249–307. https://doi.org/10.1016/s0065-2776(08)60609-4. [PubMed]
- Bastard, P., Rosen, L.B., Zhang, Q., Michailidis, E., Hoffmann, H.H., Zhang, Y., Dorgham, K., Philippot, Q., Rosain, J., Béziat, V., Manry, J., Shaw, E., Haljasmägi, L., Peterson, P., Lorenzo, L., Bizien, L., Trouillet-Assant, S., Dobbs, K., de Jesus, A.A., Belot, A., Kallaste, A., Catherinot, E., Tandjaoui-Lambiotte, Y., Le Pen, J., Kerner, G., Bigio, B., Seeleuthner, Y., Yang, R., Bolze, A., Spaan, A.N., Delmonte, O.M., Abers, M.S., Aiuti, A., Casari, G., Lampasona, V., Piemonti, L., Ciceri, F., Bilguvar, K., Lifton, R.P., Vasse, M., Smadja, D.M., Migaud, M., Hadjadj, Jérome, Terrier, B., Duffy, D., Quintana-Murci, L., van de Beek, D., Roussel, L., Vinh, D.C., Tangye, S., Haerynck, F., Dalmau, D., Martinez-Picado, J., Brodin, P., Nussenzweig, M.C., Boisson-Dupuis, S., Rodríguez-Gallego, C., Vogt, G., Mogensen, T.H., Oler, A.J., Gu, J., Burbelo, P.D., Cohen, J.I., Biondi, A., Bettini, L.R., DÁngio, M., Bonfanti, P., Rossignol, P., Mayaux, J., Rieux-Laucat, F., Husebye, E.S., Fusco, F., Ursini, M.V., Imberti, L., Sottini, A., Paghera, S., Quiros-Roldan, E., Rossi, C., Castagnoli, R., Montagna, D., Licari, A., Marseglia, G.L., Duval, X., Ghosn, J., Tsang, J.S., Goldbach-Mansky, R., Kisand, K., Lionakis, M.S., Puel, A., Zhang, S.Y., Holland, S.M., Gorochov, G., Jouanguy, E., Rice, C.M., Cobat, A., Notarangelo, L.D., Abel, L., Su, H.C., Casanova, J.L., Arias, A.A., Boisson, B., Boucherit, S., Bustamante, J., Chbihi, M., Chen, J., Chrabieh, M., Kochetkov, T., Le Voyer, T., Liu, D., Nemirovskaya, Y., Ogishi, M., Papandrea, D., Patissier, C., Rapaport, F., Roynard, M., Vladikine, N., Woollett, M., Zhang, P., Kashyap, A., Ding, L., Bosticardo, M., Wang, Q., Ochoa, S., Liu, H., Chauvin, S.D., Stack, M., Koroleva, G., Bansal, N., Dalgard, C.L., Snow, A.L., Abad, J., Aguilera-Albesa, S., Akcan, O.M., Darazam, I.A., Aldave, J.C., Ramos, M.A., Nadji, S.A., Alkan, G., Allardet-Servent, J., Allende, L.M., Alsina, L., Alyanakian, M.A., Amador-Borrero, B., Amoura, Z., Antolí, A., Arslan, S., Assant, S., Auguet, T., Azot, A., Bajolle, F., Baldolli, A., Ballester, M., Feldman, H.B., Barrou, B., Beurton, A., Bilbao, A., Blanchard-Rohner, G., Blanco, I., Blandinières, A., Blazquez-Gamero, D., Bloomfield, M., Bolivar-Prados, M., Borie, R., Bousfiha, A.A., Bouvattier, C., Boyarchuk, O., Bueno, M.R.P., Bustamante, J., Agra, J.J.C., Calimli, S., Capra, R., Carrabba, M., Casasnovas, C., Caseris, M., Castelle, M., Castelli, F., de Vera, M.C., Castro, M. V., Catherinot, E., Chalumeau, M., Charbit, B., Cheng, M.P., Clavé, P., Clotet, B., Codina, A., Colkesen, Fatih, Colkesen, Fatma, Colobran, R., Comarmond, C., Corsico, A.G., Dalmau, D., Darley, D.R., Dauby, N., Dauger, S., de Pontual, L., Dehban, A., Delplancq, G., Demoule, A., Di Sabatino, A., Diehl, J.L., Dobbelaere, S., Durand, S., Eldars, W., Elgamal, M., Elnagdy, M.H., Emiroglu, M., Erdeniz, E.H., Aytekin, S.E., Euvrard, R., Evcen, R., Fabio, G., Faivre, L., Falck, A., Fartoukh, M., Faure, M., Arquero, M.F., Flores, C., Francois, B., Fumadó, V., Fusco, F., Solis, B.G., Gaussem, P., Gil-Herrera, J., Gilardin, L., Alarcon, M.G., Girona-Alarcón, M., Goffard, J.C., Gok, F., González-Montelongo, R., Guerder, A., Gul, Y., Guner, S.N., Gut, M., Hadjadj, Jérôme, Haerynck, F., Halwani, R., Hammarström, L., Hatipoglu, N., Hernandez-Brito, E., Holanda-Peña, M.S., Horcajada, J.P., Hraiech, S., Humbert, L., Iglesias, A.D., Íñigo-Campos, A., Jamme, M., Arranz, M.J., Jordan, I., Kanat, F., Kapakli, H., Kara, I., Karbuz, A., Yasar, K.K., Keles, S., Demirkol, Y.K., Klocperk, A., Król, Z.J., Kuentz, P., Kwan, Y.W.M., Lagier, J.C., Lau, Y.L., Le Bourgeois, F., Leo, Y.S., Lopez, R.L., Leung, D., Levin, M., Levy, M., Lévy, R., Li, Z., Linglart, A., Lorenzo-Salazar, J.M., Louapre, C., Lubetzki, C., Luyt, C.E., Lye, D.C., Mansouri, Davood, Marjani, M., Pereira, J.M., Martin, A., Pueyo, D.M., Martinez-Picado, J., Marzana, I., Mathian, A., Matos, L.R.B., Matthews, G. V., Mayaux, J., Mège, J.L., Melki, I., Meritet, J.F., Metin, O., Meyts, I., Mezidi, M., Migeotte, I., Millereux, M., Mirault, T., Mircher, C., Mirsaeidi, M., Melián, A.M., Martinez, A.M., Morange, P., Mordacq, C., Morelle, G., Mouly, S., Muñoz-Barrera, A., Nafati, C., Neves, J.F., Ng, L.F.P., Medina, Y.N., Cuadros, E.N., Gonzalo Ocejo-Vinyals, J., Orbak, Z., Oualha, M., Özçelik, T., Hammarström, Q.P., Parizot, C., Pascreau, T., Paz-Artal, E., de Diego, Rebeca Pérez, Philippe, A., Philippota, Q., Planas-Serra, L., Ploin, D., Poissy, J., Poncelet, G., Pouletty, M., Quentric, P., Raoult, D., Rebillat, A.S., Reisli, I., Ricart, P., Richard, J.C., Rivet, N., Rivière, J.G., Blanch, G.R., Rodrigo, C., Rodriguez-Gallego, C., Rodríguez-Palmero, A., Romero, C.S., Rothenbuhler, A., Rozenberg, F., del Prado, M.Y.R., Riera, J.S., Sanchez, O., Sánchez-Ramón, S., Schluter, A., Schmidt, M., Schweitzer, C.E., Scolari, F., Sediva, A., Seijo, L.M., Sene, D., Senoglu, S., Seppänen, M.R.J., Ilovich, A.S., Shahrooei, Mohammad, Smadja, D., Sobh, A., Moreno, X.S., Solé-Violán, J., Soler, C., Soler-Palacín, P., Stepanovskiy, Y., Stoclin, A., Taccone, F., Tandjaoui-Lambiottea, Y., Taupin, J.L., Tavernier, S.J., Terrier, B., Thumerelle, C., Tomasoni, G., Toubiana, J., Alvarez, J.T., Trouillet-Assanta, S., Troya, J., Tucci, A., Ursini, M.V., Uzunhan, Y., Vabres, P., Valencia-Ramos, J., van Den Rym, A.M., Vandernoot, I., Vatansev, H., Vélez-Santamaria, V., Viel, S., Vilain, C., Vilaire, M.E., Vincent, A., Voiriot, G., Vuotto, F., Yosunkaya, A., Young, B.E., Yucel, F., Zannad, F., Zatz, M., Belota, A., Foti, G., Bellani, G., Citerio, G., Contro, E., Pesci, A., Valsecchi, M.G., Cazzaniga, M., Bole-Feysot, C., Lyonnet, S., Masson, C., Nitschke, P., Pouliet, A., Schmitt, Y., Tores, F., Zarhrate, M., Abela, L., Andrejak, C., Angoulvant, F., Bachelet, D., Basmaci, R., Behillil, S., Beluze, M., Benkerrou, D., Bhavsar, K., Bompart, F., Bouadma, L., Bouscambert, M., Caralp, M., Cervantes-Gonzalez, M., Chair, A., Coelho, A., Couffignal, C., Couffin-Cadiergues, S., D’ortenzio, E., da Silveira, C., Debray, M.P., Deplanque, D., Descamps, D., Desvallées, M., Diallo, A., Diouf, A., Dorival, C., Dubos, F., Duval, X., Eloy, P., Enouf, V.V.E., Esperou, H., Esposito-Farese, M., Etienne, M., Ettalhaoui, N., Gault, N., Gaymard, A., Ghosn, J., Gigante, T., Gorenne, I., Guedj, J., Hoctin, A., Hoffmann, I., Jaafoura, S., Kafif, O., Kaguelidou, F., Kali, S., Khalil, A., Khan, C., Laouénan, C., Laribi, S., Le, M., Le Hingrat, Q., Le Mestre, S., Le Nagard, H., Lescure, F.X., Lévy, Y., Levy-Marchal, C., Lina, B., Lingas, G., Lucet, J.C., Malvy, D., Mambert, M., Mentré, F., Mercier, N., Meziane, A., Mouquet, H., Mullaert, J., Neant, N., Noret, M., Pages, J., Papadopoulos, A., Paul, C., Peiffer-Smadja, N., Petrov-Sanchez, V., Peytavin, G., Picone, O., Puéchal, O., Rosa-Calatrava, M., Rossignol, B., Rossignol, P., Roy, C., Schneider, M., Semaille, C., Mohammed, N.S., Tagherset, L., Tardivon, C., Tellier, M.C., Téoulé, F., Terrier, O., Timsit, J.F., Treoux, T., Tual, C., Tubiana, S., van der Werf, S., Vanel, N., Veislinger, A., Visseaux, B., Wiedemann, A., Yazdanpanah, Y., Abelc, L., Alcover, A., Aschard, H., Astrom, K., Bousso, P., Bruhns, P., Cumano, A., Demangel, C., Deriano, L., Santo, J. Di, Dromer, F., Eberl, G., Enninga, J., Fellay, J., Gomperts-Boneca, I., Hasan, M., Hercberg, S., Lantz, O., Mouquet, H., Patin, E., Pellegrini, S., Pol, S., Rausell, A., Rogge, L., Sakuntabhai, A., Schwartz, O., Schwikowski, B., Shorte, S., Tangy, F., Toubert, A., Touvier, M., Ungeheuer, M.N., Albert, M.L., Duffy, D., Quintana-Murci, L., Alavoine, L., Amat, K.K.A., Behillil, S., Bielicki, J., Bruijning, P., Burdet, C., Caumes, E., Charpentier, C., Coignard, B., Costa, Y., Couffin-Cadiergues, S., Damond, F., Dechanet, A., Delmas, C., Descamps, D., Duval, X., Ecobichon, J.L., Enouf, V., Espérou, H., Frezouls, W., Houhou, N., Ilic-Habensus, E., Kafif, O., Kikoine, J., Le Hingrat, Q., Lebeaux, D., Leclercq, A., Lehacaut, J., Letrou, S., Lina, B., Lucet, J.C., Malvy, D., Manchon, P., Mandic, M., Meghadecha, M., Motiejunaite, J., Nouroudine, M., Piquard, V., Postolache, A., Quintin, C., Rexach, J., Roufai, L., Terzian, Z., Thy, M., Tubiana, S., van der Werf, S., Vignali, V., Visseaux, B., Yazdanpanah, Y., van Agtmael, M., Algera, A.G., van Baarle, F., Bax, D., Beudel, M., Bogaard, H.J., Bomers, M., Bos, L., Botta, M., de Brabander, J., Bree, G., Brouwer, M.C., de Bruin, S., Bugiani, M., Bulle, E., Chouchane, O., Cloherty, A., Elbers, P., Fleuren, L., Geerlings, S., Geerts, B., Geijtenbeek, T., Girbes, A., Goorhuis, B., Grobusch, M.P., Hafkamp, F., Hagens, L., Hamann, J., Harris, V., Hemke, R., Hermans, S.M., Heunks, L., Hollmann, M.W., Horn, J., Hovius, J.W., de Jong, M.D., Koning, R., van Mourik, N., Nellen, J., Paulus, F., Peters, E., van der Poll, T., Preckel, B., Prins, J.M., Raasveld, J., Reijnders, T., Schinkel, M., Schultz, M.J., Schuurman, A., Sigaloff, K., Smit, M., Stijnis, C.S., Stilma, W., Teunissen, C., Thoral, P., Tsonas, A., van der Valk, M., Veelo, D., Vlaar, A.P.J., de Vries, H., van Vugt, M., Joost Wiersinga, W., Wouters, D., Zwinderman, A.H., van de Beek, D., Abelb, L., Iuti, F., Muhsen, S. Al, Al-Mulla, F., Anderson, M.S., Arias, A.A., Feldman, H.B., Bogunovic, D., Bolze, A., Bondarenko, A., Bousfiha, A.A., Brodin, P., Bryceson, Y., Bustamante, C.D., Butte, M., Chakravorty, S., Christodoulou, J., Cirulli, E., Condino-Neto, A., Cooper, M.A., Dalgard, C.L., DeRisi, J.L., Desai, M., Drolet, B.A., Espinosa, S., Fellay, J., Flores, C., Franco, J.L., Gregersen, P.K., Haerynck, F., Hagin, D., Halwani, R., Heath, J., Henrickson, S.E., Hsieh, E., Imai, K., Itan, Y., Karamitros, T., Kisanda, K., Ku, C.L., Lau, Y.L., Ling, Y., Lucas, C.L., Maniatis, T., Mansouri, Davoud, Marodi, L., Milner, J.D., Mironska, K., Morio, T., Notarangeloa, L.D., Novelli, G., Novelli, A., O’Farrelly, C., Okada, S., Ozcelik, T., de Diego, Rebeca Perez, Planas, A.M., Prando, C., Pujol, A., Quintana-Murci, L., Renia, L., Renieri, A., Rodríguez-Gallego, C., Sancho-Shimizu, V., Sankaran, V., Barrett, K.S., Shahrooei, Mohammed, Snow, A., Soler-Palacín, P., Spaan, A.N., Turvey, S., Uddin, F., Uddin, M.J., van de Beek, D., Vazquez, S.E., Vinh, D.C., von Bernuth, H., Washington, N., Zawadzki, P., Sua, H.C., Casanovaa, J.L., 2020. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science (80-.). 370. https://doi.org/10.1126/SCIENCE.ABD4585.
- Bazyar N., Azarpira N., Khatami S.R., Galehdari H. The investigation of allele and genotype frequencies of human C3 (rs2230199) in south Iranian population. Mol. Biol. Rep. 2012;39:8919–8924. doi: 10.1007/s11033-012-1759-9. [DOI] [PubMed] [Google Scholar]
- Blom A.M., Kask L., Dahlbäck B. CCP1-4 of the C4b-binding protein α-chain are required for factor I mediated cleavage of complement factor C3b. Mol. Immunol. 2003;39:547–556. doi: 10.1016/S0161-5890(02)00213-4. [DOI] [PubMed] [Google Scholar]
- Bosmann M. Complement control for COVID-19. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abj1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto M., Fong K.Y., So A.K., Koch C., Walport M.J. Molecular basis of polymorphisms of human complement component C3. J. Exp. Med. 1990;172:1011–1017. doi: 10.1084/jem.172.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll M.C., Isenman D.E. Regulation of Humoral Immunity by Complement. Immunity. 2012 doi: 10.1016/j.immuni.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll M.V., Sim R.B. Complement in health and disease. Adv. Drug Deliv. Rev. 2011 doi: 10.1016/j.addr.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Daniloski Z., Jordan T.X., Wessels H.H., Hoagland D.A., Kasela S., Legut M., Maniatis S., Mimitou E.P., Lu L., Geller E., Danziger O., Rosenberg B.R., Phatnani H., Smibert P., Lappalainen T., tenOever B.R., Sanjana N.E. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell. 2021;184:92–105.e16. doi: 10.1016/j.cell.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Córdoba S.R., Harris C.L., Morgan B.P., Llorca O. Lessons from functional and structural analyses of disease-associated genetic variants in the complement alternative pathway. Biochim. Biophys. Acta - Mol. Basis Dis. 2011;1812:12–22. doi: 10.1016/j.bbadis.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Defendi F., Leroy C., Epaulard O., Clavarino G., Vilotitch A., Le Marechal M., Jacob M.-C., Raskovalova T., Pernollet M., Le Gouellec A., Bosson J., Poignard P., Roustit M., Thielens N., Dumestre-Pérard C., Cesbron J. Complement alternative and mannose-binding lectin pathway activation is associated with COVID-19 mortality. Front. Immunol. 2021;12:3675. doi: 10.3389/fimmu.2021.742446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diurno, F., Numis, F.G., Porta, G., Cirillo, F., Maddaluno, S., Ragozzino, A., Negri, P.D.E., Gennaro, C.D.I., Pagano, A., Allegorico, E., Bressy, L., Bosso, G., Ferrara, A., Serra, C., Montisci, A., D’Amico, M., Lo Morello, S.S., Costanzo, G.D.I., Tucci, A.G., Marchetti, P., Vincenzo, U.D.I., Sorrentino, I., Casciotta, A., Fusco, M., Buonerba, C., Berretta, M., Ceccarelli, M., Nunnari, G., Diessa, Y., Cicala, S., Facchini, G., 2020. Eculizumab treatment in patients with COVID-19: Preliminary results from real life ASL Napoli 2 Nord experience. Eur. Rev. Med. Pharmacol. Sci. https://doi.org/10.26355/EURREV_202004_20875. [DOI] [PubMed]
- Dreismann A.K., Hallam T.M., Tam L.C., Nguyen C.V., Hughes J.P., Ellis S., Harris C.L. Gene targeting as a therapeutic avenue in diseases mediated by the complement alternative pathway. Immunol. Rev. 2023;313:402–419. doi: 10.1111/imr.13149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermini L., Wilson I.J., Goodship T.H.J., Sheerin N.S. Complement polymorphisms: geographical distribution and relevance to disease. Immunobiology. 2012;217:265–271. doi: 10.1016/j.imbio.2011.07.020. [DOI] [PubMed] [Google Scholar]
- Ferreira V.P., Pangburn M.K., Cortés C. Complement control protein factor H: the good, the bad, and the inadequate. Mol. Immunol. 2010 doi: 10.1016/j.molimm.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneris F., Ricklin D., Wu J., Tzekou A., Wallace R.S., Lambris J.D., Gros P. Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science 80. 2010;330:1816–1820. doi: 10.1126/science.1195821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavriilaki E., Asteris P.G., Touloumenidou T., Koravou E.E., Koutra M., Papayanni P.G., Karali V., Papalexandri A., Varelas C., Chatzopoulou F., Chatzidimitriou M., Chatzidimitriou D., Veleni A., Grigoriadis S., Rapti E., Chloros D., Kioumis I., Kaimakamis E., Bitzani M., Boumpas D., Tsantes A., Sotiropoulos D., Sakellari I., Kalantzis I.G., Parastatidis S.T., Koopialipoor M., Cavaleri L., Armaghani D.J., Papadopoulou A., Brodsky R.A., Kokoris S., Anagnostopoulos A. Genetic justification of severe COVID-19 using a rigorous algorithm. Clin. Immunol. 2021;226 doi: 10.1016/j.clim.2021.108726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavriilaki E., Brodsky R.A. Complementopathies and precision medicine. J. Clin. Invest. 2020;130:2152–2163. doi: 10.1172/JCI136094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbrecht B.V., Lambris J.D., Gros P. Complement component C3: A structural perspective and potential therapeutic implications. Semin. Immunol. 2022 doi: 10.1016/j.smim.2022.101627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomewide Association Study of Severe Covid-19 with Respiratory Failure, 2020. . N. Engl. J. Med. 383, 1522–1534. https://doi.org/10.1056/nejmoa2020283. [DOI] [PMC free article] [PubMed]
- Georg P., Astaburuaga-García R., Bonaguro L., Brumhard S., Michalick L., Lippert L.J., Kostevc T., Gäbel C., Schneider M., Streitz M., Demichev V., Gemünd I., Barone M., Tober-Lau P., Helbig E.T., Hillus D., Petrov L., Stein J., Dey H.P., Paclik D., Iwert C., Mülleder M., Aulakh S.K., Djudjaj S., Bülow R.D., Mei H.E., Schulz A.R., Thiel A., Hippenstiel S., Saliba A.E., Eils R., Lehmann I., Mall M.A., Stricker S., Röhmel J., Corman V.M., Beule D., Wyler E., Landthaler M., Obermayer B., von Stillfried S., Boor P., Demir M., Wesselmann H., Suttorp N., Uhrig A., Müller-Redetzky H., Nattermann J., Kuebler W.M., Meisel C., Ralser M., Schultze J.L., Aschenbrenner A.C., Thibeault C., Kurth F., Sander L.E., Blüthgen N., Sawitzki B. Complement activation induces excessive T cell cytotoxicity in severe COVID-19. Cell. 2022;185:493–512.e25. doi: 10.1016/j.cell.2021.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz M.P., Skjoedt M.O., Bayarri-Olmos R., Hansen C.B., Pérez-Alós L., Jarlhelt I., Benfield T., Rosbjerg A., Garred P. Lectin pathway enzyme MASP-2 and downstream complement activation in COVID-19. J. Innate Immun. 2022 doi: 10.1159/000525508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R.A., Harris C.L., Thurman J.M. The complement alternative pathway in health and disease—activation or amplification? Immunol. Rev. 2022 doi: 10.1111/imr.13172. [DOI] [PubMed] [Google Scholar]
- Heurich M., Martínez-Barricarte R., Francis N.J., Roberts D.L., Rodríguez De Córdoba S., Morgan B.P., Harris C.L. Common polymorphisms in C3, factor B, and factor H collaborate to determine systemic complement activity and disease risk. Proc. Natl. Acad. Sci. U. S. A. 2011;108:8761–8766. doi: 10.1073/pnas.1019338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holers V.M. The spectrum of complement alternative pathway-mediated diseases. Immunol. Rev. 2008 doi: 10.1111/j.1600-065X.2008.00641.x. [DOI] [PubMed] [Google Scholar]
- Holter J.C., Pischke S.E., de Boer E., Lind A., Jenum S., Holten A.R., Tonby K., Barratt-Due A., Sokolova M., Schjalm C., Chaban V., Kolderup A., Tran T., Gjølberg T.T., Skeie L.G., Hesstvedt L., Ormåsen V., Fevang B., Austad C., Müller K.E., Fladeby C., Holberg-Petersen M., Halvorsen B., Müller F., Aukrust P., Dudman S., Ueland T., Andersen J.T., Lund-Johansen F., Heggelund L., Dyrhol-Riise A.M., Mollnes T.E. Systemic complement activation is associated with respiratory failure in COVID-19 hospitalized patients. Proc. Natl. Acad. Sci. U. S. A. 2020;117:25018–25025. doi: 10.1073/pnas.2010540117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A.E., Mullan G.M., Bradley D.T. Complement factor B polymorphism 32W protects against agerelated macular degeneration. Mol. Vis. 2011;17:983–988. [PMC free article] [PubMed] [Google Scholar]
- Janssen B.J.C., Christodoulidou A., McCarthy A., Lambris J.D., Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444:213–216. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- Java A., Apicelli A.J., Kathryn Liszewski M., Coler-Reilly A., Atkinson J.P., Kim A.H.J., Kulkarni H.S. The complement system in COVID-19: Friend and foe? JCI Insight. 2020;5 doi: 10.1172/jci.insight.140711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungreis I., Sealfon R., Kellis M. SARS-CoV-2 gene content and COVID-19 mutation impact by comparing 44 Sarbecovirus genomes. Nat. Commun. 2021;12:1–20. doi: 10.1038/s41467-021-22905-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko N., Kuo H.H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S., Piechocka-Trocha A., Lefteri K., Osborn M., Bals J., Bartsch Y.C., Bonheur N., Caradonna T.M., Chevalier J., Chowdhury F., Diefenbach T.J., Einkauf K., Fallon J., Feldman J., Finn K.K., Garcia-Broncano P., Hartana C.A., Hauser B.M., Jiang C., Kaplonek P., Karpell M., Koscher E.C., Lian X., Liu H., Liu J., Ly N.L., Michell A.R., Rassadkina Y., Seiger K., Sessa L., Shin S., Singh N., Sun W., Sun X., Ticheli H.J., Waring M.T., Zhu A.L., Alter G., Li J.Z., Lingwood D., Schmidt A.G., Lichterfeld M., Walker B.D., Yu X.G., Padera R.F., Pillai S. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell. 2020 doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C., Sprent J. Dual Nature of Type I Interferons in SARS-CoV-2-Induced Inflammation. Trends Immunol. 2021;42:312–322. doi: 10.1016/j.it.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C., Behrendt N. A novel polymorphism of human complement component C3 detected by means of a monoclonal antibody. Immunogenetics. 1986;23:322–325. doi: 10.1007/BF00398796. [DOI] [PubMed] [Google Scholar]
- Kulkarni H.S., Atkinson J.P. Targeting complement activation in COVID-19. Blood. 2020;136:2000–2001. doi: 10.1182/BLOOD.2020008925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W., Karapetyan K., Katz K., Liu C., Maddipatla Z., Malheiro A., McDaniel K., Ovetsky M., Riley G., Zhou G., Holmes J.B., Kattman B.L., Maglott D.R. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law S.K.A., Dodds A.W. The internal thioester and the covalent binding properties of the complement proteins C3 and C4. Protein Sci. 1997 doi: 10.1002/pro.5560060201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszewski K., Atkinson J.P. Complement regulators in human disease: Lessons from modern genetics. J. Intern. Med. 2015 doi: 10.1111/joim.12338. [DOI] [PubMed] [Google Scholar]
- Liszewski M.K., Atkinson J.P. Alternative pathway activation: Ever ancient and ever new. Immunol. Rev. 2022 doi: 10.1111/imr.13132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łukawska E., Polcyn-Adamczak M., Niemir Z.I. The role of the alternative pathway of complement activation in glomerular diseases. Clin. Exp. Med. 2018;18:297–318. doi: 10.1007/s10238-018-0491-8. [DOI] [PubMed] [Google Scholar]
- Ma L., Sahu S.K., Cano M., Kuppuswamy V., Bajwa J., McPhatter J., Pine A., Meizlish M.L., Goshua G., Chang C.H., Zhang H., Price C., Bahel P., Rinder H., Lei T., Day A., Reynolds D., Wu X., Schriefer R., Rauseo A.M., Goss C.W., O’Halloran J.A., Presti R.M., Kim A.H., Gelman A.E., Dela Cruz C.S., Lee A.I., Mudd P.A., Chun H.J., Atkinson J.P., Kulkarni H.S. Increased complement activation is a distinctive feature of severe SARS-CoV-2 infection. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abh2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapping the human genetic architecture of COVID-19, 2021. . Nature 1–8. https://doi.org/10.1038/s41586-021-03767-x. [DOI] [PMC free article] [PubMed]
- Marcos-Jiménez A., Sánchez-Alonso S., Alcaraz-Serna A., Esparcia L., López-Sanz C., Sampedro-Núñez M., Mateu-Albero T., Sánchez-Cerrillo I., Martínez-Fleta P., Gabrie L., del Campo Guerola L., Rodríguez-Frade J.M., Casasnovas J.M., Reyburn H.T., Valés-Gómez M., López-Trascasa M., Martín-Gayo E., Calzada M.J., Castañeda S., de la Fuente H., González-Álvaro I., Sánchez-Madrid F., Muñoz-Calleja C., Alfranca A. Deregulated cellular circuits driving immunoglobulins and complement consumption associate with the severity of COVID-19 patients. Eur. J. Immunol. 2021;51:634–647. doi: 10.1002/eji.202048858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinozzi M.C., Vergoz L., Rybkine T., Ngo S., Bettoni S., Pashov A., Cayla M., Tabarin F., Jablonski M., Hue C., Smith R.J., Noris M., Halbwachs-Mecarelli L., Donadelli R., Fremeaux-Bacchi V., Roumenina L.T. Complement factor b mutations in atypical hemolytic uremic syndrome-disease-relevant or benign? J. Am. Soc. Nephrol. 2014;25:2053–2065. doi: 10.1681/ASN.2013070796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Sancho L., Lewinski M.K., Pache L., Stoneham C.A., Yin X., Becker M.E., Pratt D., Churas C., Rosenthal S.B., Liu S., Weston S., De Jesus P.D., O’Neill A.M., Gounder A.P., Nguyen C., Pu Y., Curry H.M., Oom A.L., Miorin L., Rodriguez-Frandsen A., Zheng F., Wu C., Xiong Y., Urbanowski M., Shaw M.L., Chang M.W., Benner C., Hope T.J., Frieman M.B., García-Sastre A., Ideker T., Hultquist J.F., Guatelli J., Chanda S.K. Functional landscape of SARS-CoV-2 cellular restriction. Mol. Cell. 2021;81:2656–2668.e8. doi: 10.1016/j.molcel.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastaglio S., Ruggeri A., Risitano A.M., Angelillo P., Yancopoulou D., Mastellos D.C., Huber-Lang M., Piemontese S., Assanelli A., Garlanda C., Lambris J.D., Ciceri F. The first case of COVID-19 treated with the complement C3 inhibitor AMY-101. Clin. Immunol. 2020;215 doi: 10.1016/j.clim.2020.108450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastellos D.C., Pires da Silva B.G.P., Fonseca B.A.L., Fonseca N.P., Auxiliadora-Martins M., Mastaglio S., Ruggeri A., Sironi M., Radermacher P., Chrysanthopoulou A., Skendros P., Ritis K., Manfra I., Iacobelli S., Huber-Lang M., Nilsson B., Yancopoulou D., Connolly E.S., Garlanda C., Ciceri F., Risitano A.M., Calado R.T., Lambris J.D. Complement C3 vs C5 inhibition in severe COVID-19: Early clinical findings reveal differential biological efficacy. Clin. Immunol. 2020;220 doi: 10.1016/j.clim.2020.108598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes T., Tortajada A., Morgan B.P., De Córdoba S.R., Harris C.L. Functional basis of protection against age-related macular degeneration conferred by a common polymorphism in complement factor B. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4366–4371. doi: 10.1073/pnas.0812584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan H.P., Schmidt C.Q., Guariento M., Blaum B.S., Gillespie D., Herbert A.P., Kavanagh D., Mertens H.D.T., Svergun D.I., Johansson C.M., Uhrín D., Barlow P.N., Hannan J.P. Structural basis for engagement by complement factor H of C3b on a self surface. Nat. Struct. Mol. Biol. 2011;18:463–471. doi: 10.1038/nsmb.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson S.C., Sim R.B., Lea S.M., Fremeaux-Bacchi V., Blom A.M. Complement factor I in health and disease. Mol. Immunol. 2011 doi: 10.1016/j.molimm.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Noris M., Benigni A., Remuzzi G. The case of complement activation in COVID-19 multiorgan impact. Kidney Int. 2020 doi: 10.1016/j.kint.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson U., Gullstrand B., Pettersson Å., Sturfelt G., Truedsson L., Segelmark M. A candidate gene approach to ANCA-associated vasculitis reveals links to the C3 and CTLA-4 genes but not to the IL1-Ra and Fcγ-RIIa genes. Kidney Blood Press. Res. 2013;37:641–648. doi: 10.1159/000355744. [DOI] [PubMed] [Google Scholar]
- Pilotti C., Greenwood J., Moss S.E. Functional evaluation of AMD-associated risk variants of complement Factor B. Investig. Ophthalmol. Vis. Sci. 2020;61 doi: 10.1167/IOVS.61.5.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Mitchell B.M., Plante K.S., Debbink K., Weaver S.C., Menachery V.D. The variant gambit: COVID-19’s next move. Cell Host Microbe. 2021 doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polycarpou A., Howard M., Farrar C.A., Greenlaw R., Fanelli G., Wallis R., Klavinskis L.S., Sacks S. Rationale for targeting complement in COVID-19. EMBO Mol. Med. 2020;12 doi: 10.15252/emmm.202012642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaldi A., Gritti G., Micò M.C., Frigeni M., Borleri G., Salvi A., Landi F., Pavoni C., Sonzogni A., Gianatti A., Binda F., Fagiuoli S., Di Marco F., Lorini L., Remuzzi G., Whitaker S., Demopulos G. Endothelial injury and thrombotic microangiopathy in COVID-19: treatment with the lectin-pathway inhibitor narsoplimab. Immunobiology. 2020;225 doi: 10.1016/j.imbio.2020.152001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambausek M., Van den Wall Bake A.W.L., Schumacher-Ach R., Spitzenberg R., Rother U., Van Es L.A., Ritz E. Genetic polymorphism of C3 and Bf in IgA nephropathy. Nephrol. Dial. Transplant. 1987;2:208–211. doi: 10.1093/oxfordjournals.ndt.a091543. [DOI] [PubMed] [Google Scholar]
- Ramlall V., Thangaraj P.M., Meydan C., Foox J., Butler D., Kim J., May B., De Freitas J.K., Glicksberg B.S., Mason C.E., Tatonetti N.P., Shapira S.D. Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection. Nat. Med. 2020;26:1609–1615. doi: 10.1038/s41591-020-1021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renhong Y., Yuanyuan Z., Yaning L., Lu X., Yingying G., Qiang Z. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 80- 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risitano A.M., Mastellos D.C., Huber-Lang M., Yancopoulou D., Garlanda C., Ciceri F., Lambris J.D. Complement as a target in COVID-19? Nat. Rev. Immunol. 2020;20:343–344. doi: 10.1038/s41577-020-0320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez de Córdoba S. Genetic variability shapes the alternative pathway complement activity and predisposition to complement-related diseases. Immunol. Rev. 2022 doi: 10.1111/imr.13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez De Córdoba S., Esparza-Gordillo J., Goicoechea De Jorge E., Lopez-Trascasa M., Sánchez-Corral P. The human complement factor H: Functional roles, genetic variations and disease associations. Mol. Immunol. 2004;41:355–367. doi: 10.1016/j.molimm.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Rodriguez E., Nan R., Li K., Gor J., Perkins S.J. A revised mechanism for the activation of complement C3 to C3b: A molecular explanation of a disease-associated polymorphism. J. Biol. Chem. 2015;290:2334–2350. doi: 10.1074/jbc.M114.605691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roversi P., Johnson S., Caesar J.J.E., McLean F., Leath K.J., Tsiftsoglou S.A., Morgan B.P., Harris C.L., Sim R.B., Lea S.M. Structural basis for complement factor I control and its disease-associated sequence polymorphisms. Proc. Natl. Acad. Sci. U. S. A. 2011 doi: 10.1073/pnas.1102167108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyam A., Tsokos M.G., Brook O.R., Hecht J.L., Moulton V.R., Tsokos G.C. Activation of classical and alternative complement pathways in the pathogenesis of lung injury in COVID-19. Clin. Immunol. 2021;226 doi: 10.1016/j.clim.2021.108716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Luna J.M., Hoffmann H.H., Sánchez-Rivera F.J., Leal A.A., Ashbrook A.W., Le Pen J., Ricardo-Lax I., Michailidis E., Peace A., Stenzel A.F., Lowe S.W., MacDonald M.R., Rice C.M., Poirier J.T. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell. 2021;184:120–132.e14. doi: 10.1016/j.cell.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubart A., Flohr S., Junt T., Eder J. Low-molecular weight inhibitors of the alternative complement pathway. Immunol. Rev. 2022;313 doi: 10.1111/imr.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaughnessy J., Chabeda A., Lewis L.A., Ram S. Alternative pathway amplification and infections. Immunol. Rev. 2023;313:162–180. doi: 10.1111/imr.13160. [DOI] [PubMed] [Google Scholar]
- Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. DbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggins, M.K., Davies, K., Fellows, R., Thwaites, R.S., Baillie, J.K., Semple, M.G., Openshaw, P.J.M., Zelek, W.M., Harris, C.L., Morgan, B.P., Baillie, J.K., Semple, M.G., Openshaw, P.J.M., Carson, G., Alex, B., Bach, B., Barclay, W.S., Bogaert, D., Chand, M., Cooke, G.S., Docherty, A.B., Dunning, J., da Silva Filipe, A., Fletcher, T., Green, C.A., Harrison, E.M., Hiscox, J.A., Ho, A.Y.W., Horby, P.W., Ijaz, S., Khoo, S., Klenerman, P., Law, A., Lim, W.S., Mentzer, A.J., Merson, L., Meynert, A.M., Noursadeghi, M., Moore, S.C., Palmarini, M., Paxton, W.A., Pollakis, G., Price, N., Rambaut, A., Robertson, D.L., Russell, C.D., Sancho‐Shimizu, V., Scott, J.T., de Silva, T., Sigfrid, L., Solomon, T., Sriskandan, S., Stuart, D., Summers, C., Tedder, R.S., Thomson, E.C., Thompson, A.A.R., Thwaites, R.S., Turtle, L.C.W., Gupta, R.K., Zambon, M., Hardwick, H., Donohue, C., Lyons, R., Griffiths, F., Oosthuyzen, W., Norman, L., Pius, R., Drake, T.M., Fairfield, C.J., Knight, S.R., Mclean, K.A., Murphy, D., Shaw, C.A., Dalton, J., Girvan, M., Saviciute, E., Roberts, S., Harrison, J., Marsh, L., Connor, M., Halpin, S., Jackson, C., Gamble, C., Leeming, G., Law, A., Wham, M., Clohisey, S., Hendry, R., Scott‐Brown, J., Greenhalf, W., Shaw, V., McDonald, Sara, Keating, S., Ahmed, K.A., Armstrong, J.A., Ashworth, M., Asiimwe, I.G., Bakshi, S., Barlow, S.L., Booth, L., Brennan, B., Bullock, K., Catterall, B.W.A., Clark, J.J., Clarke, E.A., Cole, S., Cooper, L., Cox, H., Davis, C., Dincarslan, O., Dunn, C., Dyer, P., Elliott, A., Evans, A., Finch, L., Fisher, L.W.S., Foster, T., Garcia‐Dorival, I., Greenhalf, W., Gunning, P., Hartley, C., Jensen, R.L., Jones, C.B., Jones, T.R., Khandaker, S., King, K., Kiy, R.T., Koukorava, C., Lake, A., Lant, S., Latawiec, D., Lavelle‐Langham, L., Lefteri, D., Lett, L., Livoti, L.A., Mancini, M., McDonald, Sarah, McEvoy, L., McLauchlan, J., Metelmann, S., Miah, N.S., Middleton, J., Mitchell, J., Moore, S.C., Murphy, E.G., PenriceRandal, R., Pilgrim, J., Prince, T., Reynolds, W., Ridley, P.M., Sales, D., Shaw, V.E., Shears, R.K., Small, B., Subramaniam, K.S., Szemiel, A., Taggart, A., Tanianis‐Hughes, J., Thomas, Jordan, Trochu, E., van Tonder, L., Wilcock, E., Zhang, J.E., Flaherty, L., Maziere, N., Cass, E., Carracedo, A.D., Carlucci, N., Holmes, A., Massey, H., Murphy, L., Wrobel, N., McCafferty, S., Morrice, K., MacLean, A., Adeniji, K., Agranoff, D., Agwuh, K., Ail, D., Aldera, E.L., Alegria, A., Angus, B., Ashish, A., Atkinson, D., Bari, S., Barlow, G., Barnass, S., Barrett, N., Bassford, C., Basude, S., Baxter, D., Beadsworth, M., Bernatoniene, J., Berridge, J., Best, N., Bothma, P., Chadwick, D., Brittain‐Long, R., Bulteel, N., Burden, T., Burtenshaw, A., Caruth, V., Chadwick, D., Chambler, D., Chee, N., Child, J., Chukkambotla, S., Clark, T., Collini, P., Cosgrove, C., Cupitt, J., Cutino‐Moguel, M., Dark, P., Dawson, C., Dervisevic, S., Donnison, P., Douthwaite, S., DuRand, I., Dushianthan, A., Dyer, T., Evans, C., Eziefula, C., Fegan, C., Finn, A., Fullerton, D., Garg, S., Garg, S., Garg, A., GkraniaKlotsas, E., Godden, J., Goldsmith, A., Graham, C., Hardy, E., Hartshorn, S., Harvey, D., Havalda, P., Hawcutt, D.B., Hobrok, M., Hodgson, L., Hormis, A., Jacobs, M., Jain, S., Jennings, P., Kaliappan, A., Kasipandian, V., Kegg, S., Kelsey, M., Kendall, J., Kerrison, C., Kerslake, I., Koch, O., Koduri, G., Koshy, G., Laha, S., Laird, S., Larkin, S., Leiner, T., Lillie, P., Limb, J., Linnett, V., Little, J., Lyttle, M., MacMahon, M., MacNaughton, E., Mankregod, R., Masson, H., Matovu, E., McCullough, K., McEwen, R., Meda, M., Mills, G., Minton, J., Mirfenderesky, M., Mohandas, K., Mok, Q., Moon, J., Moore, E., Morgan, P., Morris, C., Mortimore, K., Moses, S., Mpenge, M., Mulla, R., Murphy, M., Nagel, M., Nagarajan, T., Nelson, M., O’Shea, M.K., Otahal, I., Ostermann, M., Pais, M., Palmieri, C., Panchatsharam, S., Papakonstantinou, D., Paraiso, H., Patel, B., Pattison, N., Pepperell, J., Peters, M., Phull, M., Pintus, S., Pooni, J.S., Post, F., Price, D., Prout, R., Rae, N., Reschreiter, H., Reynolds, T., Richardson, N., Roberts, M., Roberts, D., Rose, A., Rousseau, G., Ryan, B., Saluja, T., Shah, A., Shanmuga, P., Sharma, A., Shawcross, A., Sizer, J., Shankar‐Hari, M., Smith, R., Snelson, C., Spittle, N., Staines, N., Stambach, T., Stewart, R., Subudhi, P., Szakmany, T., Tatham, K., Thomas, Jo, Thompson, C., Thompson, R., Tridente, A., TupperCarey, D., Twagira, M., Ustianowski, A., Vallotton, N., Vincent‐Smith, L., Visuvanathan, S., Vuylsteke, A., Waddy, S., Wake, R., Walden, A., Welters, I., Whitehouse, T., Whittaker, P., Whittington, A., Papineni, P., Wijesinghe, M., Williams, M., Wilson, L., Cole, S., Winchester, S., Wiselka, M., Wolverson, A., Wootton, D.G., Workman, A., Yates, B., Young, P., 2022. Alternative pathway dysregulation in tissues drives sustained complement activation and predicts outcome across the disease course in COVID‐19. Immunology. https://doi.org/10.1111/imm.13585.
- Sim R.B., Tsiftsoglou S.A. Proteases of the complement system. Biochem. Soc. Trans. 2004 doi: 10.1042/BST0320021. [DOI] [PubMed] [Google Scholar]
- Sim R.B., Schwaeble W., Fujita T. Complement research in the 18th–21st centuries: Progress comes with new technology. Immunobiology. 2016 doi: 10.1016/j.imbio.2016.06.011. [DOI] [PubMed] [Google Scholar]
- Sim R., Twose T.M., Sim E., Reid K.B.M. Intrinsic chemical reactivity of activated human complement component C3: A historical glimpse into research during 1979–1980 on the covalent binding properties of C3, C4 and alpha-2 macroglobulin. Immunobiology. 2022;227 doi: 10.1016/j.imbio.2022.152209. [DOI] [PubMed] [Google Scholar]
- Sinkovits G., Mező B., Réti M., Müller V., Iványi Z., Gál J., Gopcsa L., Reményi P., Szathmáry B., Lakatos B., Szlávik J., Bobek I., Prohászka Z.Z., Förhécz Z., Csuka D., Hurler L., Kajdácsi E., Cervenak L., Kiszel P., Masszi T., Vályi-Nagy I., Prohászka Z. Complement overactivation and consumption predicts in-hospital mortality in SARS-CoV-2 infection. Front. Immunol. 2021;12:866. doi: 10.3389/fimmu.2021.663187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skendros P., Germanidis G., Mastellos D.C., Antoniadou C., Gavriilidis E., Kalopitas G., Samakidou A., Liontos A., Chrysanthopoulou A., Ntinopoulou M., Kogias D., Karanika I., Smyrlis A., Cepaityte D., Fotiadou I., Zioga N., Mitroulis I., Gatselis N.K., Papagoras C., Metallidis S., Milionis H., Dalekos G.N., Willems L., Persson B., Manivel V.A., Nilsson B., Connolly E.S., Iacobelli S., Papadopoulos V., Calado R.T., Huber-Lang M., Risitano A.M., Yancopoulou D., Ritis K., Lambris J.D. Complement C3 inhibition in severe COVID-19 using compstatin AMY-101. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abo2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soames C.J., Sim R.B. Interactions between human complement components factor H, factor I and C3b. Biochem. J. 1997;326:553–561. doi: 10.1042/bj3260553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souilmi Y., Lauterbur M.E., Tobler R., Huber C.D., Johar A.S., Moradi S.V., Johnston W.A., Krogan N.J., Alexandrov K., Enard D. An ancient viral epidemic involving host coronavirus interacting genes more than 20,000 years ago in East Asia. Curr. Biol. 2021;31:3504–3514.e9. doi: 10.1016/j.cub.2021.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torreira E., Tortajada A., Montes T., De Córdoba S.R., Llorca O. 3D structure of the C3bB complex provides insights into the activation and regulation of the complement alternative pathway convertase. Proc. Natl. Acad. Sci. U. S. A. 2009;106:882–887. doi: 10.1073/pnas.0810860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiftsoglou S.A. SARS-CoV-2 associated Complement genetic variants possibly deregulate the activation of the Alternative pathway affecting the severity of infection. Mol. Immunol. 2021;135:421–425. doi: 10.1016/j.molimm.2021.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwyler P., Moser S., Charitos P., Heijnen I.A.F.M., Rudin M., Sommer G., Giannetti B.M., Bassetti S., Sendi P., Trendelenburg M., Osthoff M. Treatment of COVID-19 with conestat alfa, a regulator of the complement, contact activation and kallikrein-kinin system. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.02072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese P.M., Tsolaki A.G., Yasmin H., Shastri A., Ferluga J., Vatish M., Madan T., Kishore U. Host-pathogen interaction in COVID-19: Pathogenesis, potential therapeutics and vaccination strategies. Immunobiology. 2020;225 doi: 10.1016/j.imbio.2020.152008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Simoneau C.R., Kulsuptrakul J., Bouhaddou M., Travisano K.A., Hayashi J.M., Carlson-Stevermer J., Zengel J.R., Richards C.M., Fozouni P., Oki J., Rodriguez L., Joehnk B., Walcott K., Holden K., Sil A., Carette J.E., Krogan N.J., Ott M., Puschnik A.S. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell. 2021;184:106–119.e14. doi: 10.1016/j.cell.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;80-.). 369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieme R.J., Demeulenaere L. Genetically determined electrophoretic variant of the human complement component C′;3. Nature. 1967;214:1042–1043. doi: 10.1038/2141042a0. [DOI] [PubMed] [Google Scholar]
- Wilk C.M. Coronaviruses hijack the complement system. Nat. Rev. Immunol. 2020;20:350. doi: 10.1038/s41577-020-0314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.C., Arthur C.M., Wang J., Verkerke H., Josephson C.D., Kalman D., Roback J.D., Cummings R.D., Stowell S.R. The SARS-CoV-2 receptor-binding domain preferentially recognizes blood group A. Blood Adv. 2021;5:1305–1309. doi: 10.1182/bloodadvances.2020003259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X., Wu J., Ricklin D., Forneris F., Di Crescenzio P., Schmidt C.Q., Granneman J., Sharp T.H., Lambris J.D., Gros P. Regulator-dependent mechanisms of C3b processing by factor i allow differentiation of immune responses. Nat. Struct. Mol. Biol. 2017;24:643–651. doi: 10.1038/nsmb.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B., Freiwald T., Chauss D., Wang L., West E., Mirabelli C., Zhang C.J., Nichols E.M., Malik N., Gregory R., Bantscheff M., Ghidelli-Disse S., Kolev M., Frum T., Spence J.R., Sexton J.Z., Alysandratos K.D., Kotton D.N., Pittaluga S., Bibby J., Niyonzima N., Olson M.R., Kordasti S., Portilla D., Wobus C.E., Laurence A., Lionakis M.S., Kemper C., Afzali B., Kazemian M. SARS-CoV-2 drives JAK1/2-dependent local complement hyperactivation. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abg0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Bora S.A., Parimon T., Zaman T., Friedman O.A., Palatinus J.A., Surapaneni N.S., Matusov Y.P., Cerro Chiang G., Kassar A.G., Patel N., Green C.E.R., Aziz A.W., Suri H., Suda J., Lopez A.A., Martins G.A., Stripp B.R., Gharib S.A., Goodridge H.S., Chen P. Cell-type-specific immune dysregulation in severely ill COVID-19 patients. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2020.108590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J.R.W., Sepp T., Matharu B.K., Khan J.C., Thurlby D.A., Shahid H., Clayton D.G., Hayward C., Morgan J., Wright A.F., Armbrecht A.M., Dhillon B., Deary I.J., Redmond E., Bird A.C., Moore A.T. Complement C3 variant and the risk of age-related macular degeneration. N. Engl. J. Med. 2007;357:553–561. doi: 10.1056/nejmoa072618. [DOI] [PubMed] [Google Scholar]
- Yu J., Yuan X., Chen H., Chaturvedi S., Braunstein E.M., Brodsky R.A. Direct activation of the alternative complement pathway by SARS-CoV-2 spike proteins is blocked by factor D Inhibition. Blood. 2020;136:2080–2089. doi: 10.1182/BLOOD.2020008248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: A review. Clin. Immunol. 2020;215 doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarantonello A., Revel M., Grunenwald A., Roumenina L.T. C3-dependent effector functions of complement. Immunol. Rev. 2022 doi: 10.1111/imr.13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Liu, Z., Moncada-Velez, M., Chen, J., Ogishi, M., Bigio, B., Yang, R., Arias, A.A., Zhou, Q., Han, J.E., Ugurbil, A.C., Zhang, P., Rapaport, F., Li, J., Spaan, A.N., Boisson, B., Boisson-Dupuis, S., Bustamante, J., Puel, A., Ciancanelli, M.J., Zhang, S.Y., Béziat, V., Jouanguy, E., Abel, L., Cobat, A., Casanova, J.L., Bastard, P., Korol, C., Rosain, J., Philippot, Q., Chbihi, M., Lorenzo, L., Bizien, L., Neehus, A.L., Kerner, G., Seeleuthner, Y., Manry, J., Le Voyer, T., Boisson, B., Boisson-Dupuis, S., Bustamante, J., Puel, A., Zhang, S.Y., Béziat, V., Jouanguy, E., Abel, L., Cobat, A., Bastard, P., Rosain, J., Philippot, Q., Chbihi, M., Lorenzo, L., Bizien, L., Neehus, A.L., Kerner, G., Seeleuthner, Y., Manry, J., Le Voyer, T., Boisson, B., Boisson-Dupuis, S., Bustamante, J., Puel, A., Zhang, S.Y., Béziat, V., Jouanguy, E., Abel, L., Cobat, A., Le Pen, J., Schneider, W.M., Razooky, B.S., Hoffmann, H.H., Michailidis, E., Rice, C.M., Sabli, I.K.D., Hodeib, S., Sancho-Shimizu, V., Bilguvar, K., Ye, J., Maniatis, T., Bolze, A., Arias, A.A., Arias, A.A., Zhang, Y., Notarangelo, L.D., Su, H.C., Zhang, Y., Notarangelo, L.D., Su, H.C., Onodi, F., Korniotis, S., Karpf, L., Soumelis, V., Bonnet-Madin, L., Amara, A., Dorgham, K., Gorochov, G., Smith, N., Duffy, D., Moens, L., Meyts, I., Meade, P., García-Sastre, A., Krammer, F., Corneau, A., Masson, Cecile, Schmitt, Yohann, Schlüter, A., Pujol, A., Khan, T., Marr, N., Fellay, J., Fellay, J., Fellay, J., Roussel, L., Vinh, D.C., Shahrooei, Mohammad, Shahrooei, Mohammad, Alosaimi, M.F., Alsohime, F., Hasanato, R., Mansouri, Davood, Mansouri, Davood, Mansouri, Davood, Al-Saud, H., Almourfi, F., Al-Mulla, F., Al-Muhsen, S.Z., Al Turki, S., Al Turki, S., van de Beek, D., Biondi, A., Bettini, L.R., D’Angio, M., Bonfanti, P., Imberti, L., Sottini, A., Paghera, S., Quiros-Roldan, E., Rossi, C., Oler, A.J., Tompkins, M.F., Alba, C., Dalgard, C.L., Vandernoot, I., Smits, G., Goffard, J.C., Migeotte, I., Haerynck, F., Soler-Palacin, P., Martin-Nalda, A., Colobran, R., Morange, P.E., Keles, S., Çölkesen, F., Ozcelik, T., Yasar, K.K., Senoglu, S., Karabela, Ş.N., Rodríguez-Gallego, C., Rodríguez-Gallego, C., Novelli, G., Hraiech, S., Tandjaoui-Lambiotte, Y., Tandjaoui-Lambiotte, Y., Duval, X., Laouénan, C., Duval, X., Laouénan, C., Laouénan, C., Snow, A.L., Dalgard, C.L., Milner, J.D., Mogensen, T.H., Spaan, A.N., Bustamante, J., Ciancanelli, M.J., Maniatis, T., Soumelis, V., Nussenzweig, M., Nussenzweig, M., García-Sastre, A., García-Sastre, A., García-Sastre, A., Lifton, R.P., Lifton, R.P., Lifton, R.P., Foti, G., Bellani, G., Citerio, G., Contro, E., Pesci, A., Valsecchi, M.G., Cazzaniga, M., Abad, J., Blanco, I., Rodrigo, C., Aguilera-Albesa, S., Akcan, O.M., Darazam, I.A., Aldave, J.C., Ramos, M.A., Nadji, S.A., Alkan, G., Allardet-Servent, J., Allende, L.M., Alsina, L., Alyanakian, M.A., Amador-Borrero, B., Mouly, S., Sene, D., Amoura, Z., Mathian, A., Antolí, A., Blanch, G.R., Riera, J.S., Moreno, X.S., Arslan, S., Assant, S., Auguet, T., Azot, A., Bajolle, F., Bustamante, J., Lévy, R., Oualha, M., Baldolli, A., Ballester, M., Feldman, H.B., Barrou, B., Beurton, A., Bilbao, A., Blanchard-Rohner, G., Blandinières, A., Rivet, N., Blazquez-Gamero, D., Bloomfield, M., Bolivar-Prados, M., Clavé, P., Borie, R., Bosteels, C., Lambrecht, B.N., van Braeckel, E., Bousfiha, A.A., Bouvattier, C., Vincent, A., Boyarchuk, O., Bueno, M.R.P., Castro, M. V., Matos, L.R.B., Zatz, M., Agra, J.J.C., Calimli, S., Capra, R., Carrabba, M., Fabio, G., Casasnovas, C., Vélez-Santamaria, V., Caseris, M., Falck, A., Poncelet, G., Castelle, M., Castelli, F., de Vera, M.C., Catherinot, E., Chalumeau, M., Toubiana, J., Charbit, B., Li, Z., Pellegrini, S., Cheng, M.P., Clotet, B., Codina, A., Colkesen, F., Çölkesen, F., Colobran, R., Comarmond, C., Dalmau, D., Dalmau, D., Darley, D.R., Dauby, N., Dauger, S., Le Bourgeois, F., Levy, M., de Pontual, L., Dehban, A., Delplancq, G., Demoule, A., Diehl, J.L., Dobbelaere, S., Durand, S., Mircher, C., Rebillat, A.S., Vilaire, M.E., Eldars, W., Elgamal, M., Elnagdy, M.H., Emiroglu, M., Erdeniz, E.H., Aytekin, S.E., Euvrard, R., Evcen, R., Faivre, L., Fartoukh, M., Philippot, Q., Faure, M., Arquero, M.F., Flores, C., Flores, C., Flores, C., Flores, C., Francois, B., Fumadó, V., Fumadó, V., Fumadó, V., Fusco, F., Ursini, M.V., Solis, B.G., de Diego, Rebeca Pérez, van Den Rym, A.M., Gaussem, P., Gil-Herrera, J., Gilardin, L., Alarcon, M.G., Girona-Alarcón, M., Goffard, J.C., Gok, F., Yosunkaya, A., González-Montelongo, R., Íñigo-Campos, A., Lorenzo-Salazar, J.M., Muñoz-Barrera, A., Guerder, A., Gul, Y., Guner, S.N., Gut, M., Hadjadj, J., Haerynck, F., Halwani, R., Hammarström, L., Hatipoglu, N., Hernandez-Brito, E., Heijmans, C., Holanda-Peña, M.S., Horcajada, J.P., Hoste, L., Hoste, E., Hraiech, S., Humbert, L., Mordacq, C., Thumerelle, C., Vuotto, F., Iglesias, A.D., Jamme, M., Arranz, M.J., Jordan, I., Jorens, P., Kanat, F., Kapakli, H., Kara, I., Karbuz, A., Yasar, K.K., Senoglu, S., Keles, S., Demirkol, Y.K., Klocperk, A., Król, Z.J., Kuentz, P., Kwan, Y.W.M., Lagier, J.C., Lau, Y.L., Leung, D., Leo, Y.S., Young, B.E., Lopez, R.L., Levin, M., Linglart, A., Loeys, B., Louapre, C., Lubetzki, C., Luyt, C.E., Lye, D.C., Mansouri, Davood, Marjani, M., Pereira, J.M., Martin, A., Soler-Palacín, P., Pueyo, D.M., Martinez-Picado, J., Marzana, I., Matthews, G. V., Mayaux, J., Parizot, C., Quentric, P., Mège, J.L., Raoult, D., Melki, I., Meritet, J.F., Metin, O., Mezidi, M., Migeotte, I., Taccone, F., Millereux, M., Mirault, T., Mirsaeidi, M., Melián, A.M., Martinez, A.M., Morange, P., Morelle, G., Naesens, L., Nafati, C., Neves, J.F., Ng, L.F.P., Medina, Y.N., Cuadros, E.N., Gonzalo Ocejo-Vinyals, J., Orbak, Z., Özçelik, T., Pan-Hammarström, Q., Pascreau, T., Paz-Artal, E., Philippe, A., Planas-Serra, L., Schluter, A., Ploin, D., Viel, S., Poissy, J., Pouletty, M., Reisli, I., Ricart, P., Richard, J.C., Rivière, J.G., Rodriguez-Gallego, C., Rodriguez-Gallego, C., Rodríguez-Palmero, A., Romero, C.S., Rothenbuhler, A., Rozenberg, F., del Prado, M.Y.R., Sanchez, O., Sánchez-Ramón, S., Schmidt, M., Schweitzer, C.E., Scolari, F., Sediva, A., Seijo, L.M., Seppänen, M.R.J., Ilovich, A.S., Slabbynck, H., Smadja, D.M., Sobh, A., Solé-Violán, J., Soler, C., Stepanovskiy, Y., Stoclin, A., Tandjaoui-Lambiotte, Y., Taupin, J.L., Tavernier, S.J., Terrier, B., Tomasoni, G., Alvarez, J.T., Trouillet-Assant, S., Troya, J., Tucci, A., Uzunhan, Y., Vabres, P., Valencia-Ramos, J., van de Velde, S., van Praet, J., Vandernoot, I., Vatansev, H., Vilain, C., Voiriot, G., Yucel, F., Zannad, F., Belot, A., Bole-Feysot, C., Lyonnet, S., Masson, Cécile, Nitschke, P., Pouliet, A., Schmitt, Yoann, Tores, F., Zarhrate, M., Shahrooei, Mohammad, Abel, L., Andrejak, C., Angoulvant, F., Bachelet, D., Bhavsar, K., Bouadma, L., Chair, A., Couffignal, C., Silveira, C. Da, Debray, M.P., Duval, X., Eloy, P., Esposito-Farese, M., Ettalhaoui, N., Gault, N., Ghosn, J., Gorenne, I., Hoffmann, I., Kafif, O., Kali, S., Khalil, A., Laouénan, C., Laribi, S., Le, M., Le Hingrat, Q., Lescure, F.X., Lucet, J.C., Mentré, F., Mullaert, J., Peiffer-Smadja, N., Peytavin, G., Roy, C., Schneider, M., Mohammed, N.S., Tagherset, L., Tardivon, C., Tellier, M.C., Timsit, J.F., Trioux, T., Tubiana, S., Basmaci, R., Behillil, S., Beluze, M., Benkerrou, D., Dorival, C., Meziane, A., Téoulé, F., Bompart, F., Bouscambert, M., Gaymard, A., Lina, B., Rosa-Calatrava, M., Terrier, O., Caralp, M., Cervantes-Gonzalez, M., D’Ortenzio, E., Puéchal, O., Semaille, C., Coelho, A., Diouf, A., Hoctin, A., Mambert, M., Couffin-Cadiergues, S., Deplanque, D., Descamps, D., Visseaux, B., Desvallées, M., Khan, C., Diallo, A., Mercier, N., Paul, C., Petrov-Sanchez, V., Dubos, F., Enouf, V.V.E., Mouquet, H., Esperou, H., Jaafoura, S., Papadopoulos, A., Etienne, M., Gigante, T., Rossignol, B., Guedj, J., Le Nagard, H., Lingas, G., Neant, N., Kaguelidou, F., Lévy, Y., Wiedemann, A., Lévy, Y., Wiedemann, A., Levy-Marchal, C., Malvy, D., Noret, M., Pages, J., Picone, O., Rossignol, P., Tual, C., Veislinger, A., van der Werf, S., Vanel, N., Yazdanpanah, Y., Alavoine, L., Costa, Y., Duval, X., Ecobichon, J.L., Frezouls, W., Ilic-Habensus, E., Leclercq, A., Lehacaut, J., Letrou, S., Mandic, M., Nouroudine, M., Quintin, C., Rexach, J., Tubiana, S., Vignali, V., Amat, K.K.A., Behillil, S., Enouf, V., van der Werf, S., Bielicki, J., Bruijning, P., Burdet, C., Burdet, C., Caumes, E., Charpentier, C., Damond, F., Descamps, D., Le Hingrat, Q., Visseaux, B., Coignard, B., Couffin-Cadiergues, S., Delmas, C., Espérou, H., Roufai, L., Dechanet, A., Houhou, N., Kafif, O., Kikoine, J., Manchon, P., Piquard, V., Postolache, A., Terzian, Z., Lebeaux, D., Lina, B., Lucet, J.C., Malvy, D., Meghadecha, M., Motiejunaite, J., Thy, M., van Agtmael, M., Bomers, M., Chouchane, O., Geerlings, S., Goorhuis, B., Grobusch, M.P., Harris, V., Hermans, S.M., Hovius, J.W., Nellen, J., Peters, E., van der Poll, T., Prins, J.M., Reijnders, T., Schinkel, M., Sigaloff, K., Stijnis, C.S., van der Valk, M., van Vugt, M., Joost Wiersinga, W., Algera, A.G., van Baarle, F., Bos, L., Botta, M., de Bruin, S., Bulle, E., Elbers, P., Fleuren, L., Girbes, A., Hagens, L., Heunks, L., Horn, J., van Mourik, N., Paulus, F., Raasveld, J., Schultz, M.J., Smit, M., Stilma, W., Thoral, P., Tsonas, A., de Vries, H., Bax, D., Cloherty, A., Beudel, M., Brouwer, M.C., Koning, R., van de Beek, D., Bogaard, H.J., de Brabander, J., de Bree, G., Bugiani, M., Geerts, B., Hollmann, M.W., Preckel, B., Veelo, D., Geijtenbeek, T., Hafkamp, F., Hamann, J., Hemke, R., de Jong, M.D., Schuurman, A., Teunissen, C., Vlaar, A.P.J., Wouters, D., Zwinderman, A.H., Abel, L., Aiuti, A., Muhsen, S. Al, Al-Mulla, F., Anderson, M.S., Arias, A.A., Feldman, H.B., Bogunovic, D., Itan, Y., Bolze, A., Cirulli, E., Barrett, K.S., Washington, N., Bondarenko, A., Bousfiha, A.A., Brodin, P., Bryceson, Y., Bustamante, C.D., Butte, M., Casari, G., Chakravorty, S., Christodoulou, J., Le Mestre, S., Condino-Neto, A., Cooper, M.A., Dalgard, C.L., David, A., DeRisi, J.L., DeRisi, J.L., Desai, M., Drolet, B.A., Espinosa, S., Fellay, J., Flores, C., Franco, J.L., Gregersen, P.K., Haerynck, F., Hagin, D., Halwani, R., Heath, J., Henrickson, S.E., Hsieh, E., Imai, K., Karamitros, T., Kisand, K., Ku, C.L., Lau, Y.L., Ling, Y., Lucas, C.L., Maniatis, T., Mansouri, Davoud, Marodi, L., Milner, J., Mironska, K., Morio, T., Notarangelo, L.D., Su, H.C., Novelli, A., Novelli, G., O’Farrelly, C., Okada, S., Ozcelik, T., de Diego, Rebeca Perez, Planas, A.M., Prando, C., Pujol, A., Quintana-Murci, L., Renia, L., Renieri, A., Rodríguez-Gallego, C., Sancho-Shimizu, V., Sankaran, V., Shahrooei, Mohammed, Snow, A., Soler-Palacín, P., Spaan, A.N., Tangye, S., Turvey, S., Uddin, F., Uddin, M.J., Uddin, M.J., van de Beek, D., Vazquez, S.E., Vinh, D.C., von Bernuth, H., Zawadzki, P., Jing, H., Tung, W., Meguro, K., Shaw, E., Jing, H., Tung, W., Shafer, S., Zheng, L., Zhang, Z., Kubo, S., Chauvin, S.D., Meguro, K., Shaw, E., Lenardo, M., Luthers, C.R., Bauman, B.M., Shafer, S., Zheng, L., Zhang, Z., Kubo, S., Chauvin, S.D., Lenardo, M., Lack, J., Karlins, E., Hupalo, D.M., Rosenberger, J., Sukumar, G., Wilkerson, M.D., Zhang, X., 2020. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science (80-). 370. https://doi.org/10.1126/SCIENCE.ABD4570.
- Zinellu A., Mangoni A.A. Serum complement C3 and C4 and COVID-19 severity and mortality: a systematic review and meta-analysis with meta-regression. Front. Immunol. 2021 doi: 10.3389/fimmu.2021.696085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, H.; Jensen, R. K.; Jensen, J. M. B.; Fox, R.; Pedersen, D. V.; Olesen, H. G.; Hansen, A. G.; Christiansen, D.; Mazarakis, S. M. M.; Lojek, N.; Hansen, P.; Gadeberg, T. A. F.; Zarantonello, A.; Laursen, N. S.; Mollnes, T. E.; Johnson, M. B.; Stevens, B.; Thiel, S.; Andersen, G. R. A Complement C3–Specific Nanobody for Modulation of the Alternative Cascade Identifies the C-Terminal Domain of C3b as Functional in C5 Convertase Activity. J. Immunol. 2020, 205 (8), 2287–2300. 10.4049/jimmunol.2000752. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data Availability: The authors declare that data supporting the findings of this study are available within the paper and its supplementary information files.