PURPOSE
PTEN-associated clinical syndromes such as Cowden syndrome (CS) increase cancer risk and have historically been diagnosed based upon phenotypic criteria. Because not all patients clinically diagnosed with CS have PTEN pathogenic variants (PVs), and not all patients with PTEN PVs have been clinically diagnosed with CS, the cancer risk conferred by PTEN PVs calculated from cohorts of patients with clinical diagnoses of CS/CS-like phenotypes may be inaccurate.
METHODS
We assessed a consecutive cohort of 727,091 individuals tested clinically for hereditary cancer risk, with a multigene panel between September 2013 and February 2022. Multivariable logistic regression models accounting for personal and family cancer history, age, sex, and ancestry were used to quantify disease risks associated with PTEN PVs.
RESULTS
PTEN PVs were detected in 0.027% (193/727,091) of the study population, and were associated with a high risk of female breast cancer (odds ratio [OR], 7.88; 95% CI, 5.57 to 11.16; P = 2.3 × 10−31), endometrial cancer (OR, 13.51; 95% CI, 8.77 to 20.83; P = 4.2 × 10−32), thyroid cancer (OR, 4.88; 95% CI, 2.64 to 9.01; P = 4.0 × 10−7), and colon polyposis (OR, 31.60; CI, 15.60 to 64.02; P = 9.0 × 10−22). We observed modest evidence suggesting that PTEN PVs may be associated with ovarian cancer risk (OR, 3.77; 95% CI, 1.71 to 8.32; P = 9.9 × 10−4). Among patients with similar personal/family history and ancestry, every 5-year increase in age of diagnosis decreased the likelihood of detecting a PTEN PV by roughly 60%.
CONCLUSION
We demonstrate that PTEN PVs are associated with significantly increased risk for a range of cancers. Together with the observation that PTEN PV carriers had earlier disease onset relative to otherwise comparable noncarriers, our results may guide screening protocols, inform risk-management strategies, and warrant enhanced surveillance approaches that improve clinical outcomes for PTEN PV carriers, regardless of their clinical presentation.
BACKGROUND
The tumor-suppressor phosphatase PTEN antagonizes the phosphatidylinositol 3-kinase/AKT signaling pathway that regulates apoptosis, cell-cycle arrest, and other cellular pathways.1-3 Heterozygous germline pathogenic variants (PVs) in PTEN cause PTEN hamartoma tumor syndrome (PHTS). Multiple clinically diagnosed disorders, including Cowden syndrome (CS), Bannayan-Riley-Ruvalcaba syndrome (BRRS), and Proteus-like syndrome, have been associated with germline PV in PTEN.4-9 Individuals clinically diagnosed with CS may or may not have an underlying germline PTEN PV.10-13 Indeed, PTEN PVs are only found in approximately 30%-35% of individuals with a clinical diagnosis of CS/CS-like and approximately 60% of individuals considered to have BRRS.14 By definition, all PTEN PV carriers have a molecular diagnosis of PHTS, whether or not they meet diagnostic criteria for CS, BRRS, Proteus, or Proteus-like syndrome.
CONTEXT
Key Objective
Cancer risks associated with germline pathogenic variants (PVs) in PTEN have been established from patients ascertained for Cowden or related syndromes, potentially resulting in inaccurate estimates of risk. This work describes cancer risks and cancer types associated with PTEN PVs in a large hereditary cancer screening cohort using a previously established methodology.
Knowledge Generated
PTEN PVs are associated with a high risk of female breast, endometrial, and thyroid cancers, as well as colon polyposis, and may be associated with ovarian cancer risk. PTEN PV carriers have an earlier age of onset than otherwise comparable individuals without PTEN PVs.
Relevance
These findings may help guide screening and risk-management strategies for individuals with PTEN PVs, regardless of clinical presentation.
Patients molecularly diagnosed with PHTS have an elevated risk of benign and malignant tumors,11 yet quantifying the cancer risk directly attributable to PTEN PVs has been challenging. Early analyses of PTEN cancer risk focused exclusively on patients with a clinical diagnosis of CS, but such evaluation does not fully reflect the cancer risk of a PTEN PV because many patients clinically diagnosed with CS do not have PTEN PVs.4,9 Later analyses that assessed cancer risk in the subset of patients with CS known to harbor PTEN PVs also may not be accurate because not all patients with PTEN PVs are diagnosed with CS.10,14
With the widespread utilization of gene-panel sequencing in patients who have known cancer status, it is now possible to analyze the cancer risk associated with PTEN PVs irrespective of a patient's clinical presentation. Here, we assessed the cancer risk conferred by PTEN PVs in > 700,000 patients, who met broad criteria for clinical panel testing, by using a multivariable logistic regression framework that isolates the impact of PTEN PVs after accounting for clinical factors.
METHODS
Study Population Participants
We examined clinical and genetic records from a consecutive cohort of patients who underwent hereditary cancer testing between September 2013 and February 2022. Patients were eligible for inclusion if they were age 18 years or older at the time of testing and negative for PVs on cancer-associated genes other than PTEN. Patients were excluded from analysis if they were submitted from states that disallow use of genetic data after completion of genetic testing, if they submitted an incomplete test request form (TRF), or if they had multigene panel testing after receiving negative test results from a limited gene panel. Analyses were restricted to patients with full panel sequencing to ensure a homogenous study population. This study was conducted according to a study protocol that was approved by the Advarra Institutional Review Board (Pro00036775) with a waiver of informed consent. Further details are provided in Appendix 1.
Hereditary Cancer Testing
Genetic testing was performed in a Clinical Laboratory Improvement Amendments– and College of American Pathology–approved laboratory (Myriad Genetic Laboratories Inc, Salt Lake City, UT). The hereditary cancer panel was composed of 25-35 cancer-associated genes; the initial multigene panel test included 25 genes (APC, ATM, BARD1, BMPR1A, BRCA1, BRCA2, BRIP1, CDH1, CDK4, CHEK2, MLH2, MSH2, MSH6, MUTYH, NBN, P14ARF, P16, PALB2, PMS2, PTEN, RAD51C, RAD51D, SMAD4, STK11, and TP53). Subsequent additions to the panel test in 2016 and 2019 included GREM1, HOXB13, POLD1, POLE, AXIN2, GALNT12, MSH3, NTHL1, RNF43, and RPS20. This next-generation sequencing assay has been detailed previously.15,16 Sequencing and large rearrangement analysis was performed for all genes evaluated except HOXB13, POLD1, and POLE, for which only sequencing is performed, and EPCAM and GREM1, in which only large rearrangement analysis is performed.
Variant classification was performed using the American College of Molecular Genetics and Genomics and Association for Molecular Pathology guidelines, as well as previously described statistical variant classification methods.17-20 Variants with a laboratory classification of deleterious or suspected deleterious were considered pathogenic.
Statistical Methods
All analyses were conducted using R version 4.1 or higher.21 CIs were calculated from Wald statistics. P values were derived from Wald statistics and reported as two-sided.
Association With Cancer Risk
We quantified disease risks associated with PTEN using a previously described multivariable logistic regression methodology.22 Disease risks were estimated as adjusted odds ratios (ORs), with 95% CIs. These adjusted ORs represent the relative risks associated with PTEN PVs after accounting for other risk factors and may be interpreted as the fold-increase in risk for a PTEN PV carrier compared with a noncarrier who is identical with respect to age, personal/family cancer history, and ancestry. The adjusted ORs presented here should be consistent with the ORs that would be obtained from a population-based study using either multivariable regression or matched case-control analysis to adjust for age, personal/family cancer history, and ancestry.22-24 As PTEN PVs are associated with childhood mortality,25 the ORs in this study—where only patients age ≥ 18 years were included—represent risks for PTEN carriers who survive to adulthood.
A threshold of five or more disease-affected PTEN PV carriers was prespecified as the minimum data required to investigate association with a specific disease using multivariable regression. We constructed a separate multivariable logistic regression model for each disease that met the minimum data threshold. Models testing association with female-specific cancers were restricted to female patients. For each disease, we coded disease status (affected or unaffected) as the dependent variable. Independent variables included PTEN PV status, age, sex (where applicable), ancestry, and personal and family histories associated with hereditary breast and ovarian cancer (HBOC), Lynch syndrome, and adenomatous polyposis colon cancer syndrome. Family history variables were coded as numeric counts of diagnoses, weighted according to degree of relatedness: we used a weight of 0.5 for each first-degree relative (FDR) and 0.25 for each second-degree relative (SDR). Further details regarding coding of variables are provided in Appendix 1.
Association With Age of Diagnosis
We investigated whether PTEN PV carriers tended to be diagnosed at earlier ages than noncarriers who were similar with respect to personal/family history and ancestry. For each phenotype that showed significant association with PTEN, we constructed a multivariable logistic regression model restricted to the subcohort of patients affected by that specific disease. PTEN PV status was the dependent variable in each disease model. Independent variables for age at diagnosis, ancestry, sex (where applicable), and personal/family history were coded as above for tests of association with disease risk.
Association With Familial Cancer
We tested increased rates of family cancer history among PTEN PV carriers compared with noncarriers with similar clinical features. For this analysis, we constructed a single multivariable logistic regression model on the basis of the entire study cohort. The dependent variable was PTEN PV status. Independent variables for age, ancestry, sex, and personal/family cancer history were coded as above for tests of association with cancer risk. ORs for association of PTEN PV status with familial disease were reported per one-half unit of the weighted relative count described above and represent the fold-increase in likelihood of detecting a PTEN PV because of one affected FDR or two affected SDRs. We examined all diseases for which five or more PTEN PV carriers reported at least one affected FDR or SDR.
RESULTS
We identified 727,091 patients who met study eligibility criteria. Clinical characteristics are detailed in Table 1. The study cohort was predominantly composed of female patients (699,209 [96.2%]) who were referred for genetic testing because of suspected HBOC syndrome. Nearly one third (226,120 [31.1%]) of patients reported a personal history of cancer. Most patients (689,692 [94.6%]) reported a FDR or SDR affected by a cancer associated with HBOC, Lynch syndrome, or adenomatous polyposis colon cancer syndrome.
TABLE 1.
Patient Characteristics
Pathogenic PTEN variants were detected in 193 (0.027%) study subjects. Eight additional patients carried PTEN PVs but were excluded from the study cohort because they had a concurrent PV in a second hereditary cancer gene (Appendix Table A1). PTEN PV carriers were more frequently affected by benign or malignant neoplasms than noncarriers (69.4% v 31.1%) and tended to be younger (median age, 41 v 46 years) at the time of multigene panel testing (Table 1). Details regarding prevalence and types of cancers affecting study subjects and their families are tabulated by PTEN status in Appendix Table A2. Details regarding the distribution of age at diagnosis are provided in Appendix Table A3.
Association With Disease Risk
We had sufficient data to evaluate seven neoplastic phenotypes for association with PTEN: ductal invasive breast cancer, ductal carcinoma in situ (DCIS), endometrial cancer, thyroid cancer, colon polyposis (defined as ≥ 20 colon polyps), and nonpolyposis colorectal cancer (defined as colon or rectal cancer with < 20 colon polyps). We defined colon polyposis and nonpolyposis colorectal cancer as in previous work22 on the basis of different genetic syndromes associated with colon polyposis versus hereditary nonpolyposis colon cancer. We further evaluated risk associated with overall female breast cancer, defined as any occurrence of ductal invasive breast cancer, lobular invasive breast cancer, or DCIS.
PVs in PTEN were associated with a high risk of overall female breast cancer (OR, 7.88; 95% CI, 5.57 to 11.16; P = 2.3 × 10−31), ductal invasive breast cancer (OR, 7.55; 95% CI, 5.24 to 10.88; P = 1.7 × 10−27), DCIS (OR, 11.56; 95% CI, 6.52 to 20.50; P = 5.6 × 10−17), endometrial cancer (OR, 13.51; 95% CI, 8.77 to 20.83; P = 4.2 × 10−32), thyroid cancer (OR, 4.88; 95% CI:, 2.64 to 9.01; P = 4.0 × 10−7), and colon polyposis (OR, 31.60; CI, 15.60 to 64.02; P = 9.0 × 10−22). We observed modest evidence suggesting association with risk of ovarian cancer (OR, 3.77; 95% CI, 1.71 to 8.32; P = 9.9 × 10−4). PTEN PVs were not associated with risk of nonpolyposis colorectal cancer (OR, 1.31; 95% CI, 0.48- to 3.59; P = .60). The results from multivariable regression models testing association of PTEN with disease risk are detailed in Table 2 and Appendix Table A4.
TABLE 2.
Association of PTEN Pathogenic Variant Status With Disease Risk
Association With Age of Diagnosis
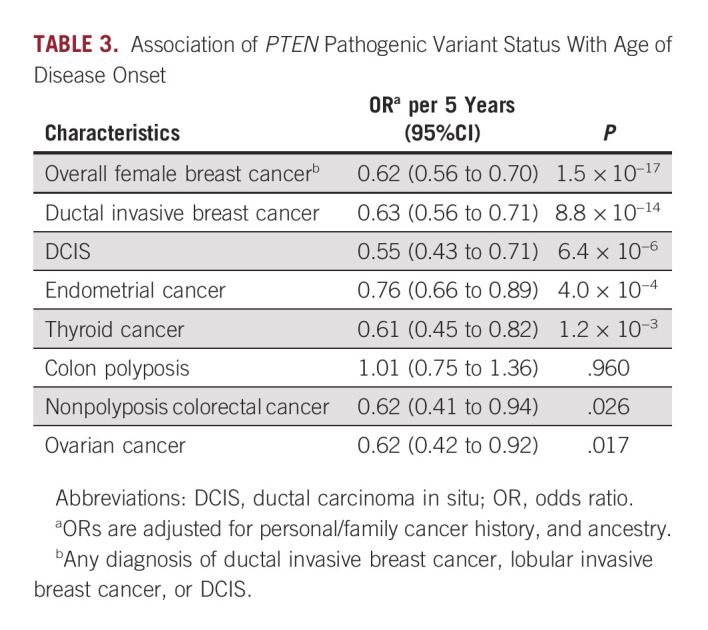
For most PTEN-associated diseases, we found that PV carriers were diagnosed at earlier ages than noncarriers with similar clinical characteristics (Table 3 and Appendix Table A5). Among patients with breast cancer who were equivalent with respect to personal/family history and ancestry, every 5-year increase in age of diagnosis decreased the likelihood of detecting a PTEN PV by a factor of roughly 62% (OR per 5 years, 0.62; 95% CI, 0.56 to 0.70; P = 1.5 × 10−17). Similar associations between PTEN carrier status and age of diagnosis were observed for ductal invasive breast cancer, DCIS, endometrial cancer, thyroid cancer, and ovarian cancer (Table 3). Although PTEN PV carriers had substantially increased risk of colon polyposis, they did not have earlier onset than noncarriers (OR per 5 years, 1.01; 95% CI, 0.75 to 1.36; P = .96). Further details are provided in Table 3.
TABLE 3.
Association of PTEN Pathogenic Variant Status With Age of Disease Onset
Association With Familial Cancer
We had sufficient data to evaluate whether PTEN carriers had stronger family histories than noncarriers with similar clinical characteristics for 11 different hereditary diseases (Table 4). For most hereditary diseases, we found that patients with PVs in PTEN did not have stronger family histories than noncarriers who were equivalent with respect to sex, personal history, age, and ancestry (Table 4 and Appendix Table A6). We observed marginal significance for association between PTEN PV status and familial kidney cancer: the likelihood of detecting a PTEN PV increased by roughly 2.28-fold because of each FDR or 2 SDRs affected by kidney cancer (OR, 2.28; 95% CI, 1.16 to 4.47; P = .017). No other familial diseases were more common among PTEN PV carriers than among noncarriers with similar clinical features.
TABLE 4.
Association of PTEN Pathogenic Variant Status With Familial Cancer

Sex-Specific Sensitivity Analyses
For diseases that affect both males and females, and for which we had adequate data, we conducted sensitivity analyses by retesting associations within subcohorts defined by sex. We found no evidence of sex-specific differences in disease risk (Appendix Table A4), age of diagnosis (Appendix Table A5), or association with familial cancer (Appendix Table A6).
DISCUSSION
Here, we quantified the risk conferred by PTEN PVs on a range of cancers in a cohort containing > 700,000 patients screened with a clinical-grade multigene sequencing panel. To focus specifically on PTEN PV carriers, our analysis differed from preceding studies in two key aspects: first, patient inclusion in our cohort was independent of a clinical diagnosis for CS or CS-like phenotype or the PTEN risk calculator11; second, we used multivariable logistic regression analysis to isolate the impact of PTEN PVs after accounting for other clinical parameters. Our results suggest that, irrespective of clinical diagnosis, PTEN PVs significantly increase the risk for female breast cancer, endometrial cancer, thyroid cancer, and colon polyposis. After adjusting for other factors, we found that PTEN PVs were associated with earlier disease onset. Additionally, our observation of little to no association between PTEN PVs and familial cancer, despite being sufficiently powered to see such associations, suggests that many PTEN PVs arise de novo, consistent with previous reports.26 Together, we expect these findings to better inform patients, their providers, and clinical-management guidelines about the impact of PVs in PTEN identified via sequencing.
Our observation of significantly increased risk of endometrial cancer associated with PTEN PVs (OR, 13.51; 95% CI, 8.77 to 20.83; P = 4.2 × 10−32) aligns with previous studies and underscores the importance of consideration of endometrial screening in PV carriers.27-29 In an analysis of individuals who met one of several eligibility criteria for PHTS, the lifetime risk of developing endometrial cancer was 28%, and relative risk was observed to dramatically increase around age 25 years.28 In our study, the median age of diagnosis was 37 years (range, 33-48.5 years). Currently, the National Comprehensive Cancer Network (NCCN) recommends individualized screening on the basis of personal and family history, as well as consideration of beginning endometrial screenings by age 35 years.12 The significance of our regression results and the early age of onset observed herein suggest endometrial screening for all women with a PV in PTEN should be considered.
Our results in female breast cancer support current guidelines. We identified a significant female breast cancer OR of 7.88 (95% CI, 5.57 to 11.16; P = 2.3 × 10−31), which is consistent with previous reports in HBOC cohorts22,30 and consistent in direction with PTEN-associated disease cohorts albeit smaller in size likely because of overestimation of risk on the basis of methodology in these individuals.27,28 Additionally, the median age of onset of 40 years (range, 35-45 years) is consistent with previous reports.22,30 Multiple studies have confirmed that CS-affected individuals are more likely to have malignant breast neoplasm if they also harbor a PTEN PV compared with those without PTEN PVs.4 Furthermore, the previously reported average age of breast cancer diagnosis in individuals with a PTEN PV is 36-46 years.30 Collectively, the results from our group and others support the current NCCN recommendations to start a screening regimen with clinical examinations at or before age 25 years, to conduct breast imaging at or before age 35 years, and to discuss prophylactic mastectomy with individuals harboring a germline PV in PTEN.
In our study, males were more highly represented in the PTEN PV cohort (9.3%; 18/193) than in the full testing cohort (3.8%; 27,931/727,962), yet too few had cancer diagnoses to fully assess PV-associated cancer risk. Notably, it was previously shown that males comprised 32.4%-44.6% of patients who met CS relaxed criteria (pathognomonic criteria, or at least two criteria, either major or minor) and had an underlying pathogenic PTEN variant.11 Although male breast cancer in an individual with a germline PTEN PV has previously been reported,31 there was no evidence for increased risk in a study of more than 3,000 probands with CS, and male breast cancer was not significantly associated with PTEN in that cohort.11 Despite a significantly elevated lifetime risk of thyroid cancer, men with germline PTEN PVs continue to be difficult to ascertain because of a lower lifetime incidence of sentinel cancers typically associated with PHTS that would lead to molecular assessment (eg, breast cancer). When compared with females with germline PTEN PVs, males have propensity for developing cancers in sites not traditionally related to CS.32
We observed a significant thyroid cancer risk associated with PTEN PVs, consistent with previous findings in patients with CS, among whom 11% had epithelial cell thyroid carcinoma as their sentinel cancer.33 In our study, the median age of diagnosis was 35 years (range, 18.5-39 years). However, studies have revealed earlier onset of thyroid cancer,32,34 with one specifically finding increased risk in pediatric patients.34 A prospective study of patients with CS (or Cowden-like syndrome) and a PTEN, SDH, or KLLN PV found that 16.7% presented with thyroid cancer before age 18 years.34 Although the NCCN currently recommends screening starting at age 7 years, it is important that individuals continue to follow these screening recommendations, as later ages of onset were observed in the cohort presented here than have been previously reported. It should be noted that the gene panel used in the current study does not include several well-characterized genes associated with a hereditary predisposition to medullary thyroid cancer, such as RET or CDKN1B. This might have led some patients with a personal or family history of undefined thyroid cancer to use a different diagnostic laboratory offering and could potentially result in an under-representation of patients with a personal and/or family history of thyroid cancer in our cohort.
Although our patient cohort was large and diverse in many respects (eg, ethnicity, cancer type, etc), it was also not a random cross-section of the population. For instance, most patients were tested because they met criteria for HBOC syndrome or Lynch syndrome. To isolate the impact of PTEN PV status, we used a multivariable logistic regression analysis that accounts for clinical factors. To ensure the statistical validity of these analyses, we only performed calculations for a given cancer if at least five patients were diagnosed with that cancer and carried PTEN PVs. Inaccuracies on the TRF could also influence our results: we typically cannot access health records that permit confirmation of the content reported on the TRF. A further limitation of the TRF is that it does not ascertain polyp histology, limiting the ability to characterize polyposis disease.
Our present work and previous research from ours and other groups elucidate a clear role for PTEN PVs in increasing cancer risk. Less clear is the precise way particular PTEN genotypes affect a patient's phenotype, and this is an important topic for future work. Thorough examination of risk modifiers (eg, family history of specific cancers), tumor histologic subtypes, and particular DNA variants (eg, single-nucleotide polymorphisms, indels, and copy-number variants)35 will likely provide key insights into the highly variable clinical manifestation of PTEN-associated disorders.
ACKNOWLEDGMENT
The authors would like to thank Dale Muzzey, PhD, for assistance with manuscript editing, as well as Elizabeth Cogan, PhD, and Sarah Ratzel, PhD, for assistance with manuscript editing and preparation, and Nydia Santiago Neal for assistance with concept and design.
APPENDIX 1. Supplemental Material
Study Population
Patients were eligible for inclusion if they were age 18 years or older at the time of hereditary cancer testing and negative for pathogenic variants (PVs) in cancer-associated genes other than PTEN. Patients were excluded from analysis if they had variants of uncertain significance or low-penetrance variants in any cancer-associated genes, if test results indicated mosaicism in PTEN, if they submitted an incomplete test request form, if they had multigene panel testing after receiving negative test results from a limited gene panel, or if they submitted from states that disallow use of genetic data after completion of genetic testing (Alaska, Colorado, Florida, New York, New Hampshire, Oregon, Oklahoma, South Dakota, and Minnesota).
Coding of Variables in Logistic Regression Analysis
PTEN PV carrier status was coded as a binary variable. Patients with a PTEN variant classified as deleterious or suspected deleterious were coded as PV-positive. Patients were coded as PV-negative if only benign polymorphisms or no variants were detected.
Age was coded in years as a continuous variable. For each model, age at the time of genetic testing was used for patients unaffected by the cancer used as the outcome variable. Age at diagnosis of cancer was used for patients affected by the cancer used as the outcome variable.
Ancestries were coded as quantitative variables representing fractions of reported ancestries. For example, a patient who listed only Ashkenazi ancestry was coded with an Ashkenazi value of 1.0, and zero for all other ancestries. A patient who reported Asian and African ancestries was coded with Asian and African values of 0.5, and zero for all other ancestries. The ancestry White was used in place of ancestries listed as Central/Eastern Europe, Western/Northern Europe, and White/non-Hispanic. Ancestry variables included Ashkenazi Jewish, Asian, Black/African, Hispanic/Latino, Middle Eastern, Native American, White, and other.
Personal cancer variables were coded as binary (ever or never affected). Separate variables were coded for male breast cancer, ductal carcinoma in situ (DCIS), ductal invasive breast cancer, lobular invasive breast cancer, endometrial cancer, pancreatic cancer, gastric cancer, nonpolyposis colorectal cancer (defined as colon cancer or rectal cancer with < 20 colon polyps), colon polyposis (defined as ≥ 20 colon polyps), kidney cancer, melanoma, ovarian cancer, prostate cancer, and thyroid cancer. Patients with DCIS in addition to ductal invasive breast cancer were recoded to only be considered as having ductal invasive breast cancer. A personal history of overall female breast cancer was defined as an occurrence of ductal invasive breast cancer, DCIS, and/or lobular invasive breast cancer.
Familial cancers were coded as numeric counts of diagnoses, weighted according to degree of relatedness. We used a weight of 0.5 for each first-degree relative and 0.25 for each second-degree relative. All models included family history variables for each cancer type listed above.
TABLE A1.
Genes With Concurrent Mutations in Patients With Germline PTEN Pathogenic Variants

TABLE A2.
Prevalence of Personal and Familial Disease by PTEN PV Status
TABLE A3.
Distribution of Age at Diagnosis by PTEN PV Status
TABLE A4.
Association of PTEN Pathogenic Variant Status With Disease Risk Within Each Sexa

TABLE A5.
Association of PTEN Pathogenic Variant Status With Age of Disease Onset Within Each Sexa

TABLE A6.
Association of PTEN Pathogenic Variant Status With Familial Cancer Within Each Sex
Shelly Cummings
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Travel, Accommodations, Expenses: Myriad Genetics
Andrew Alfonso
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Elisha Hughes
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Brent Mabey
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Research Funding: Myriad Genetics
Nanda Singh
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Patents, Royalties, Other Intellectual Property: Royalties for gene patents through the University of Utah, my former employer
Charis Eng
Stock and Other Ownership Interests: Family Care Path Inc
Patents, Royalties, Other Intellectual Property: I am the inventor of MyLegacy/MyFHH (a web-based, patient-entered family health history tool with risk algorithms and clinical decision support pushed to the point of care). This invention is utlized in Cleveland Clinic spin off company, Family Care Path Inc (see above)
Uncompensated Relationships: Family Care Path Inc
No other potential conflicts of interest were reported.
Footnotes
S.C., A.A., and E.H. contributed equally to this work as first authors.
AUTHOR CONTRIBUTIONS
Conception and design: Shelly Cummings, Elisha Hughes, Matt Kucera, Charis Eng
Collection and assembly of data: Shelly Cummings, Elisha Hughes, Matt Kucera, Brent Mabey, Nanda Singh
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Shelly Cummings
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Travel, Accommodations, Expenses: Myriad Genetics
Andrew Alfonso
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Elisha Hughes
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Brent Mabey
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Research Funding: Myriad Genetics
Nanda Singh
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Patents, Royalties, Other Intellectual Property: Royalties for gene patents through the University of Utah, my former employer
Charis Eng
Stock and Other Ownership Interests: Family Care Path Inc
Patents, Royalties, Other Intellectual Property: I am the inventor of MyLegacy/MyFHH (a web-based, patient-entered family health history tool with risk algorithms and clinical decision support pushed to the point of care). This invention is utlized in Cleveland Clinic spin off company, Family Care Path Inc (see above)
Uncompensated Relationships: Family Care Path Inc
No other potential conflicts of interest were reported.
REFERENCES
- 1.Steck PA, Pershouse MA, Jasser SA, et al. : Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet 15:356-362, 1997 [DOI] [PubMed] [Google Scholar]
- 2.Maehama T, Dixon JE: The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3, 4, 5-trisphosphate. J Biol Chem 273:13375-13378, 1998 [DOI] [PubMed] [Google Scholar]
- 3.Stambolic V, Suzuki A, de la Pompa JL, et al. : Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 95:29-39, 1998 [DOI] [PubMed] [Google Scholar]
- 4.Marsh DJ, Coulon V, Lunetta KL, et al. : Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum Mol Genet 7:507-515, 1998 [DOI] [PubMed] [Google Scholar]
- 5.Marsh DJ, Kum JB, Lunetta KL, et al. : PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum Mol Genet 8:1461-1472, 1999 [DOI] [PubMed] [Google Scholar]
- 6.Orloff MS, Eng C: Genetic and phenotypic heterogeneity in the PTEN hamartoma tumour syndrome. Oncogene 27:5387-5397, 2008 [DOI] [PubMed] [Google Scholar]
- 7.Yehia L, Eng C: PTEN hamartoma tumor syndrome, in Adam M, Ardinger H, Pagon R (eds): GeneReviews. Seattle, WA, University of Washington, 2021 [PubMed] [Google Scholar]
- 8.Nelen MR, Padberg GW, Peeters EAJ, et al. : Localization of the gene for Cowden disease to chromosome 10q22–23. Nat Genet 13:114-116, 1996 [DOI] [PubMed] [Google Scholar]
- 9.Liaw D, Marsh DJ, Li J, et al. : Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet 16:64-67, 1997 [DOI] [PubMed] [Google Scholar]
- 10.Pilarski R, Stephens JA, Noss R, et al. : Predicting PTEN mutations: An evaluation of Cowden syndrome and Bannayan-Riley-Ruvalcaba syndrome clinical features. J Med Genet 48:505-512, 2011 [DOI] [PubMed] [Google Scholar]
- 11.Tan MH, Mester J, Peterson C, et al. : A clinical scoring system for selection of patients for PTEN mutation testing is proposed on the basis of a prospective study of 3042 probands. Am J Hum Genet 88:42-56, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daly MB, Pilarski R, Berry M, et al. : NCCN clinical practice guidelines in oncology, genetic/familial high-risk assessment: Breast, ovarian, and pancreatic (version 2.2022). NCCN clinical practice guidelines in oncology web site. 2022. https://www.nccn.org/professionals/physician_gls/pdf/genetics_screening.pdf [DOI] [PubMed]
- 13.Eng C: Will the real Cowden syndrome please stand up: Revised diagnostic criteria. J Med Genet 37:828-830, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pilarski R: PTEN hamartoma tumor syndrome: A clinical overview. Cancers (Basel) 11:844, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yurgelun MB, Allen B, Kaldate RR, et al. : Identification of a variety of mutations in cancer predisposition genes in patients with suspected Lynch syndrome. Gastroenterology 149:604-613.e20, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Judkins T, Leclair B, Bowles K, et al. : Development and analytical validation of a 25-gene next generation sequencing panel that includes the BRCA1 and BRCA2 genes to assess hereditary cancer risk. BMC Cancer 15:215, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, et al. : Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405-424, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowles KR, Mancini-DiNardo D, Coffee B, et al. : Hereditary cancer testing challenges: Assembling the analytical pieces to solve the patient clinical puzzle. Future Oncol 15:65-79, 2019 [DOI] [PubMed] [Google Scholar]
- 19.Eggington JM, Bowles KR, Moyes K, et al. : A comprehensive laboratory-based program for classification of variants of uncertain significance in hereditary cancer genes. Clin Genet 86:229-237, 2014 [DOI] [PubMed] [Google Scholar]
- 20.Pruss D, Morris B, Hughes E, et al. : Development and validation of a new algorithm for the reclassification of genetic variants identified in the BRCA1 and BRCA2 genes. Breast Cancer Res Treat 147:119-132, 2014 [DOI] [PubMed] [Google Scholar]
- 21.R Development Core Team : R: A Language and Environment for Statistical Computing. Vienna, Austria, R Foundation for Statistical Computing, 2022 [Google Scholar]
- 22.Kurian AW, Hughes E, Handorf EA, et al. : Breast and ovarian cancer penetrance estimates derived from germline multiple-gene sequencing results in women. JCO Precis Oncol 10.1200/PO.16.00066, 2017 [DOI] [PubMed] [Google Scholar]
- 23.Rothman KJ, Greenland S, Lash TL: Modern Epidemiology, Volume 3. Philadelphia, PA, Wolters Kluwer Health/Lippincott Williams & Wilkins, 2008 [Google Scholar]
- 24.Rajamani S: Eliminating Bias in Cancer Risk Estimates a Simulation Study [Dissertation]. University of Utah, Salt Lake City, UT, 2016 [Google Scholar]
- 25.Plamper M, Gohlke B, Woelfle J: PTEN hamartoma tumor syndrome in childhood and adolescence—A comprehensive review and presentation of the German pediatric guideline. Mol Cell Pediatr 9:1-12, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mester J, Eng C: Estimate of de novo mutation frequency in probands with PTEN hamartoma tumor syndrome. Genet Med 14:819-822, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bubien V, Bonnet F, Brouste V, et al. : High cumulative risks of cancer in patients with PTEN hamartoma tumour syndrome. J Med Genet 50:255-263, 2013 [DOI] [PubMed] [Google Scholar]
- 28.Tan MH, Mester JL, Ngeow J, et al. : Lifetime cancer risks in individuals with germline PTEN mutations. Clin Cancer Res 18:400-407, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gammon A, Jasperson K, Champine M: Genetic basis of Cowden syndrome and its implications for clinical practice and risk management. Appl Clin Genet 9:83-92, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nusbaum R, Vogel KJ, Ready K: Susceptibility to breast cancer: Hereditary syndromes and low penetrance genes. Breast Dis 27:21-50, 2006 [DOI] [PubMed] [Google Scholar]
- 31.Fackenthal JD, Marsh DJ, Richardson AL, et al. : Male breast cancer in Cowden syndrome patients with germline PTEN mutations. J Med Genet 38:159-164, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riegert-Johnson DL, Gleeson FC, Roberts M, et al. : Cancer and Lhermitte-Duclos disease are common in Cowden syndrome patients. Hered Cancer Clin Pract 8:6, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ngeow J, Stanuch K, Mester JL, et al. : Second malignant neoplasms in patients with Cowden syndrome with underlying germline PTEN mutations. J Clin Oncol 32:1818-1824, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ngeow J, Mester J, Rybicki LA, et al. : Incidence and clinical characteristics of thyroid cancer in prospective series of individuals with Cowden and Cowden-like syndrome characterized by germline PTEN, SDH, or KLLN alterations. J Clin Endocrinol Metab 96:E2063-E2071, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yehia L, Seyfi M, Niestroj L-M, et al. : Copy number variation and clinical outcomes in patients with germline PTEN mutations. JAMA Netw Open 3:e1920415, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]







