PURPOSE
Gastroenteropancreatic neuroendocrine carcinomas (GEP-NEC) are rare and have a poor prognosis. Most GEP-NEC are diagnosed with metastatic disease, with only minor biopsies available for molecular diagnostics. We assessed the applicability of liquid biopsies for molecular profiling of GEP-NEC.
MATERIALS AND METHODS
We performed massive parallel sequencing of 76 cancer-related genes in circulating tumor DNA from 50 patients with advanced GEP-NEC and compared findings to previous analyses of solid tumor biopsies from the same patients. Plasma samples were collected before therapy, and the median time span between blood and tissue sampling was 25 days.
RESULTS
We detected 178 somatic mutations in the liquid biopsies, 127 (71%) were also detected in the solid biopsies, whereas 51 (29%) were unique to the liquid biopsies. In the same 76 genes, we previously detected 199 somatic mutations (single nucleotide variants) in solid biopsies, of which 127 (64%) were also now detected in liquid biopsies. In exploratory subgroup assessments, concordance was higher in patients with liver metastases (P = 1.5 × 10–5) and increasing with level of liver involvement (P = 1.2 × 10–4). The concordance was similar between GEP-NEC with different primary sites, except being lower in esophageal cases (P = .001). Concordance was not associated with tumor mutation burden. Tumor tissue mutations also detected in liquid biopsies was lower for MSI (40%) versus MSS tumors (70%; P = 7.8 × 10–4). We identified potentially targetable mutations in plasma of 26 (52%) of patients with GEP-NEC; nine patients (18%) had potentially targetable mutation detected only in liquid biopsies.
CONCLUSION
Liquid biopsy analyses may be an applicable alternative to solid biopsies in GEP-NEC. Liquid biopsies may add additional mutations compared with tumor biopsies alone and could be useful for biomarker assessment in clinical trials for these patients.
INTRODUCTION
Gastroenteropancreatic-neuroendocrine carcinomas (GEP-NEC) have a particularly poor prognosis.1-4 Molecular information that may aid in classification, estimation of prognosis and, importantly, treatment selection is limited and usually not considered in clinical practice. We recently provided an extensive molecular characterization of a large series of GEP-NEC tumor samples5 and found distinct molecular differences to small-cell lung cancer, questioning the rationale for extrapolating treatment strategies from small-cell lung cancer to GEP-NEC.6,7 At the same time, we found that a high fraction (66%) of GEP-NEC harbor potentially targetable mutations, opening new possibilities for use of these alterations as biomarkers and implementation of more personalized treatments.
CONTEXT
Key Objective
We aimed to analyze the mutational spectrum in liquid biopsies from patients with gastroenteropancreatic neuroendocrine carcinoma (GEP-NEC). Furthermore, we wanted to compare the findings with corresponding data from matched solid tumor biopsies.
Knowledge Generated
We found a generally good concordance between mutations detected in liquid biopsies and matched solid tumor biopsies from patients with GEP-NEC. The concordance varied with primary tumor site and was associated with disease burden.
Relevance
Patients with GEP-NEC have a very poor prognosis, and molecular data are limited. Our findings indicate that liquid biopsies can be used for mutation analyses in much needed biomarker-driven clinical trials for these patients.
With only one exception,8 mutational studies on GEP-NEC, so far, have been performed on tumor tissue gained from resection specimens and biopsies. This approach has several theoretical shortcomings: resected specimens underlie a selection bias, including mainly patients with locoregional disease, excluding the cases with aggressive metastatic disease, where only a small biopsy is performed. This is especially relevant for GEP-NEC since around two third of cases present with metastatic disease at the time of diagnosis.9 An additional bias of biomarker assessment in biopsies is the region of sampling, which may not capture all subclones of a heterogenous tumor.
Application of liquid biopsies as a noninvasive strategy for assessing biomarkers has been described in detail for many cancer forms (for overview, see Ref. 10). In theory, all subclones of a heterogenous tumor may contribute to the circulating tumor DNA (ctDNA) pool, and analyses of liquid biopsies should therefore represent all subclones in the tumor. In addition to the potential to capture a more complete set of subclones, liquid biopsies may be advantageous in settings where the tumor is located at a site preventing tissue biopsy.
The representativity of a liquid biopsy for the mutational spectrum in a given tumor varies widely between different cancer types and according to tumor load within cancer types.10 For GEP-NEC, this topic has so far only been addressed in a small study including 24 patients, of which six cases had matched solid tumor tissue for analysis.11 Thus, there is an urgent need to assess the applicability of liquid biopsies for future biomarker-driven clinical trials and potential targeted treatments for GEP-NEC.
In this study, we performed massive parallel sequencing of ctDNA of plasma samples from a selected set of 50 patients from our previous study on tissue biopsies.5 We provide an overview of the mutational landscape of ctDNA in GEP-NEC and a detailed assessment of the concordance between data from solid and liquid biopsies.
MATERIALS AND METHODS
Patients and Samples
Patients diagnosed with high-grade gastroenteropancreatic neuroendocrine neoplasms (GEP-NEN) from 2013 to 2017 in Nordic centers were prospectively included in the Nordic neuroendocrine carcinoma registry, according to inclusion criteria previously described5 and outlined in Appendix 1. Centralized pathologic re-evaluation was performed.
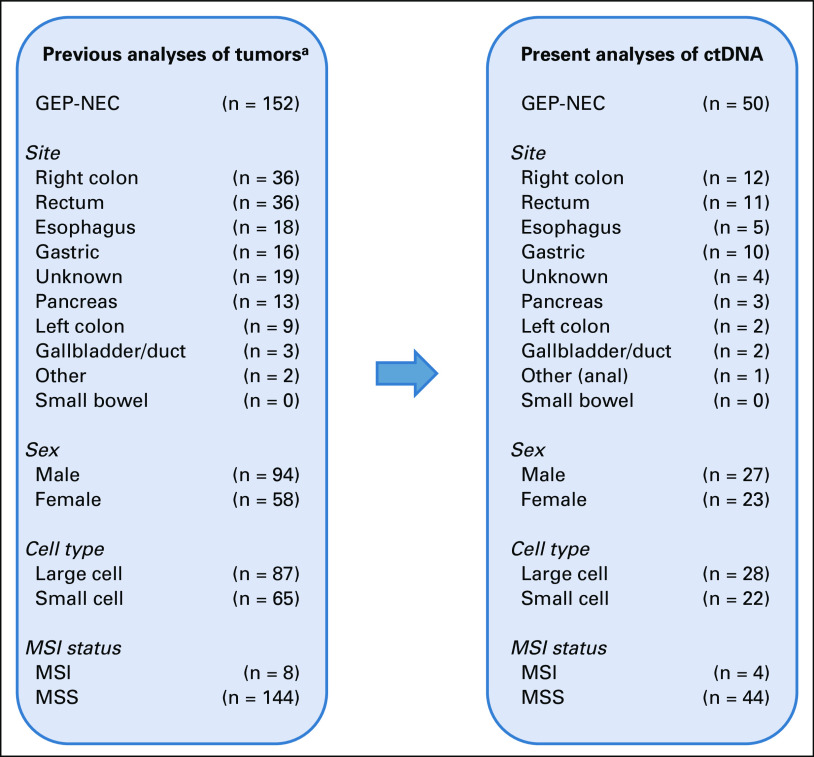
For the present analyses, 50 patients with GEP-NEC were included, all among the 152 GEP-NEC previously characterized for molecular alterations in tumor tissue and normal cells (WBC).5 Patients were selected for the present analyses to provide representative groups of tumors with respect to primary tumor sites, sex, cell type, and microsatellite instability (MSI) status and/or on the basis of high or low tumor mutation burden (TMB; Appendix Fig A1) as well as tumor cell fraction > 20% in the solid biopsies. All blood samples were collected before any treatment, and the median time between tumor biopsy and blood sample was 25 days (Appendix 1). General demographics for the selected patients are given in Table 1 and Appendix Figure A1, with details on individual patients and selection criteria in the Data Supplement. No preliminary data on potential effect sizes in comparisons between subgroups were available. Thus, this study was exploratory. Survival was followed for a minimum of 32 months (median, 43 months) or until death.
TABLE 1.
Patient Characteristics
The research protocol was institutional review board–approved by the responsible ethics committees in Norway (REK vest 2012/940) and Sweden (REC Uppsala Dnr 2012/285). All sample donors have signed informed written consent.
Sequencing and Mutation Calling
For liquid biopsy analyses, 4 mL of EDTA-plasma was subject to DNA isolation and library preparation using the Avenio ctDNA Expanded Kit (Roche; Basel, Switzerland), covering 77 cancer genes (Data Supplement; Appendix 1). In brief, library quality control was performed with Qubit dsDNA HS Assay and Agilent Fragment Analyzer before sequencing on an Illumina NextSeq 500, using the 300-cycle NextSeq High Output Kit (Illumina; San Diego, CA). Sequencing yielded an average of 60,442,281 mapped reads per sample, corresponding to a depth of 11,539×.
Data were analyzed with the AVENIO ctDNA Analysis Software (version 2.0.0) with default settings. Point mutations (single nucleotide variants [SNVs]) in coding regions of the 77 genes were considered. As postprocessing filters, we applied variant allele frequency (VAF) > 0.0025 and population minor allele frequency (MAF) < 0.02 (EXAC or 1000 Genome Project databases). Furthermore, variants were filtered against previously published data from matched WBC.5
Annotation of mutations as targetable was performed as described in our previous analysis of solid biopsies5 (a list of predefined targetable mutations is provided in the Data Supplement) and with a separate assessment applying the OncoKB database.13 In the latter assessment, a conservative approach was taken, including only mutations satisfying level 1 (US Food and Drug Administration–approved drugs) or level 2 (standard care).
Sequencing and mutation calling of the solid tumor biopsies from the patients included in this study has been reported previously5 and summary metrics for the 50 patients included in this study are described in Appendix 1.
Concordance Analysis
Among the 77 genes covered by the Avenio ctDNA Expanded Kit, 76 were also included in the 360 gene panel applied for solid biopsies and WBC. Thus, all concordance assessments between liquid and solid biopsies were performed on the basis of 76 genes.
Statistics
Statistical analyses were performed using SPSS v.28.0.1.0 (Appendix 1). All P values are given as two-sided, and P values from Fisher exact tests are given as two-sided and cumulative. P < .05 was considered significant.
RESULTS
Mutation Spectrum in Liquid Biopsies From Patients With GEP-NEC
To assess the mutational status in liquid biopsies from patients with GEP-NEC, we performed targeted massive parallel sequencing of 77 cancer genes, covered by the Avenio ctDNA Expanded Kit, in plasma from 50 selected patients. Overall, we detected 202 mutations, yielding an average of 4.0 mutations/patient (median, 4 mutations/patient; range, 0-13 mutations/patient; Appendix Fig A2).
We had germline information available for all patients, from a previous study on solid biopsies5 and therefore went on to filter all data from liquid biopsy against variants called in WBC. Doing so, 23 of the 202 variants were found in WBC, leaving 179 variants to be deemed true somatic variants. This yielded an average of 3.6 mutations per patient (median, 3 mutations per patient; range, 0-13 mutations per patient, with at least one mutation detected in the ctDNA of 44 of the 50 patients).
In concordance with our previous analysis of solid tumors,5 the most frequently altered genes were TP53 (60%), KRAS (28%), APC (26%), RB1 (24%), and BRAF (20%; Fig 1).
FIG 1.
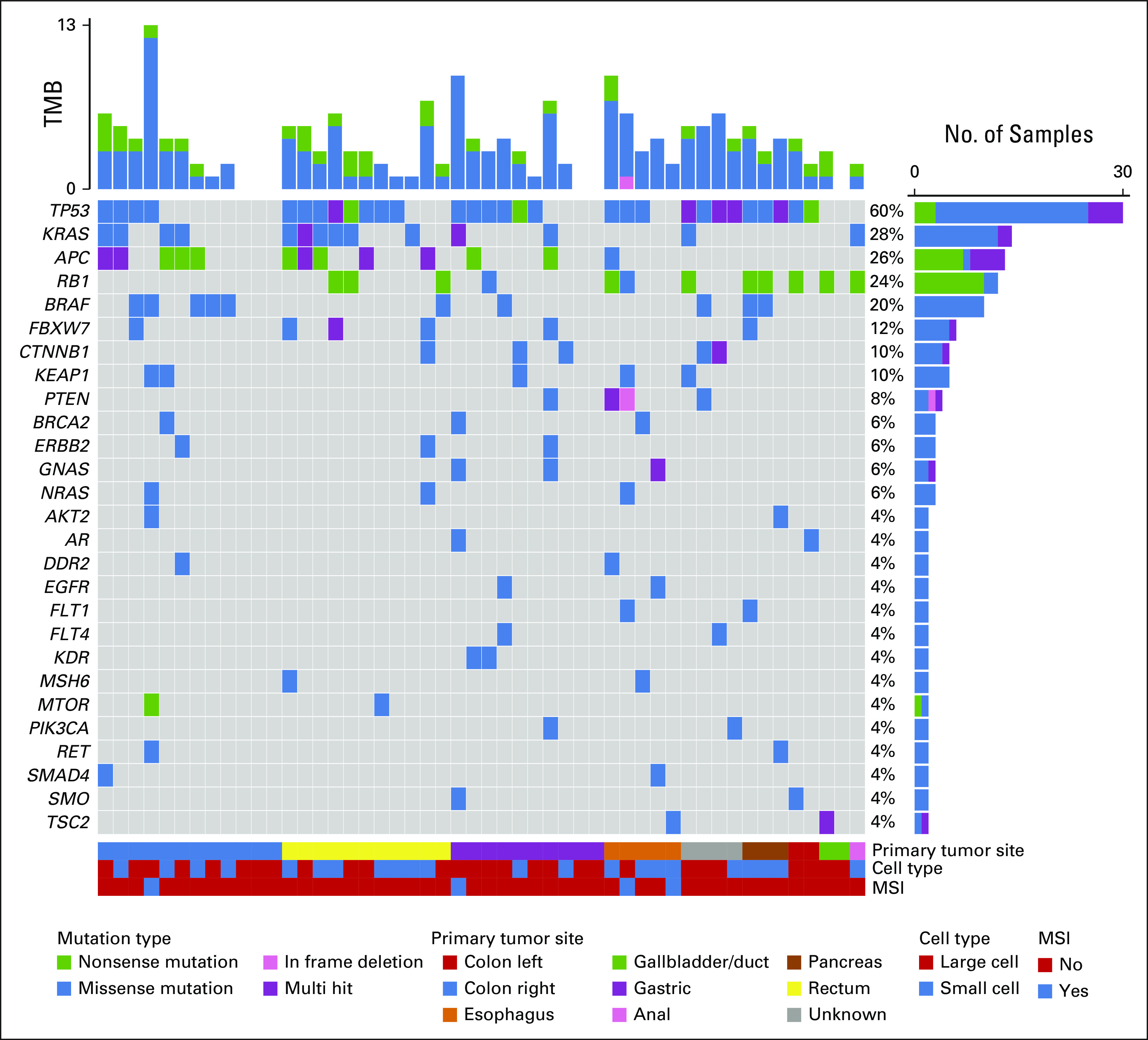
Mutation spectrum in liquid biopsies from patients with GEP-NEC. The oncoplot shows the most frequently altered genes (rows) in liquid biopsies from 50 patients with GEP-NEC (columns). Upper panel shows the mutational burden per sample, with colors indicating mutation type for individual mutations. Percentages on the right represent mutation frequency per gene, with colors indicating mutation types (and a separate, purple color for multiple mutations in the same gene in the same patient). The panel under the oncoplot area is composed of three single-row heatmaps showing in order, from top to bottom, primary tumor site, cell type, and MSI status. Multihit indicates that more than one mutation occurs in the same gene, in the same patient. Mutations presented represent the findings in liquid biopsies after additional filtering against WBC-data (see Methods). GEP-NEC, gastroenteropancreatic neuroendocrine carcinoma, MSI, microsatellite instable; TMB, tumor mutation burden.
Concordance Between Liquid and Solid Biopsies
We performed a direct assessment of concordance between mutations detected in liquid biopsies and mutations previously detected in matching solid biopsies.5 This was possible in 76/77 genes covered in our analyses of liquid biopsies. The only nonoverlapping gene was PMS2. One patient had a PMS2 mutation. Thus, 178 of the 179 variants remained for comparison with solid biopsy data.
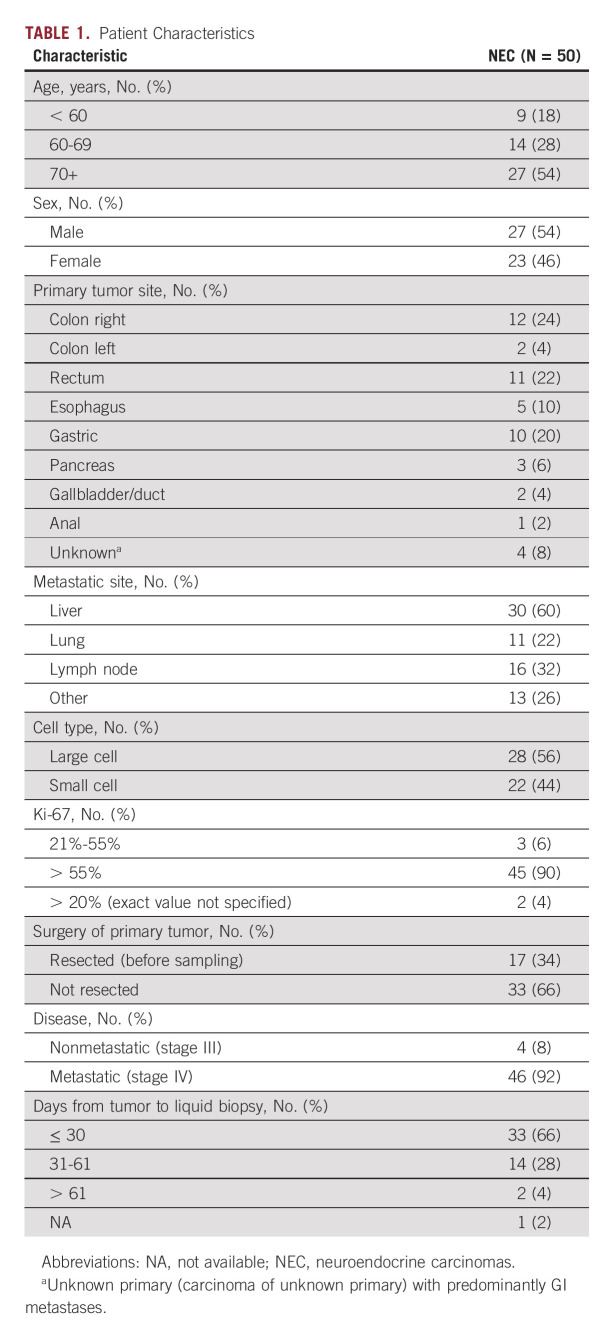
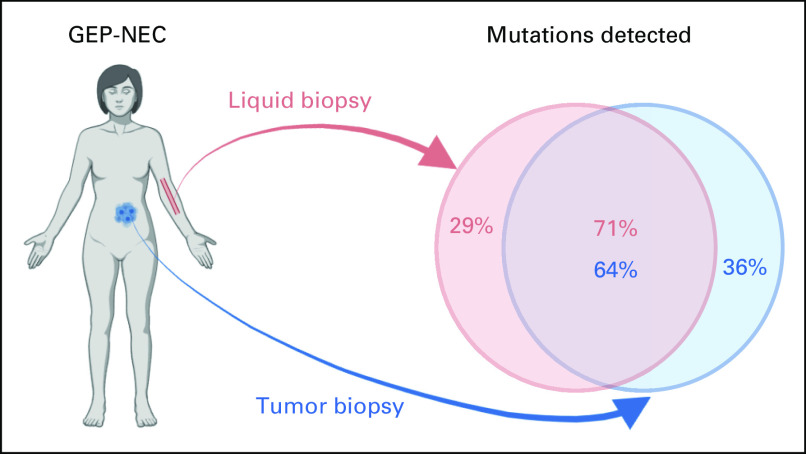
Of the 178 mutations detected in liquid biopsies, 127 (71%) were also detected in the solid biopsies, whereas 51 (29%) were only found in liquid biopsies. Conversely, in the same 76 genes, we previously detected 199 mutations (SNVs) in solid biopsies and, among these, 127 (64%) were also found in liquid biopsies (Fig 2A).
FIG 2.
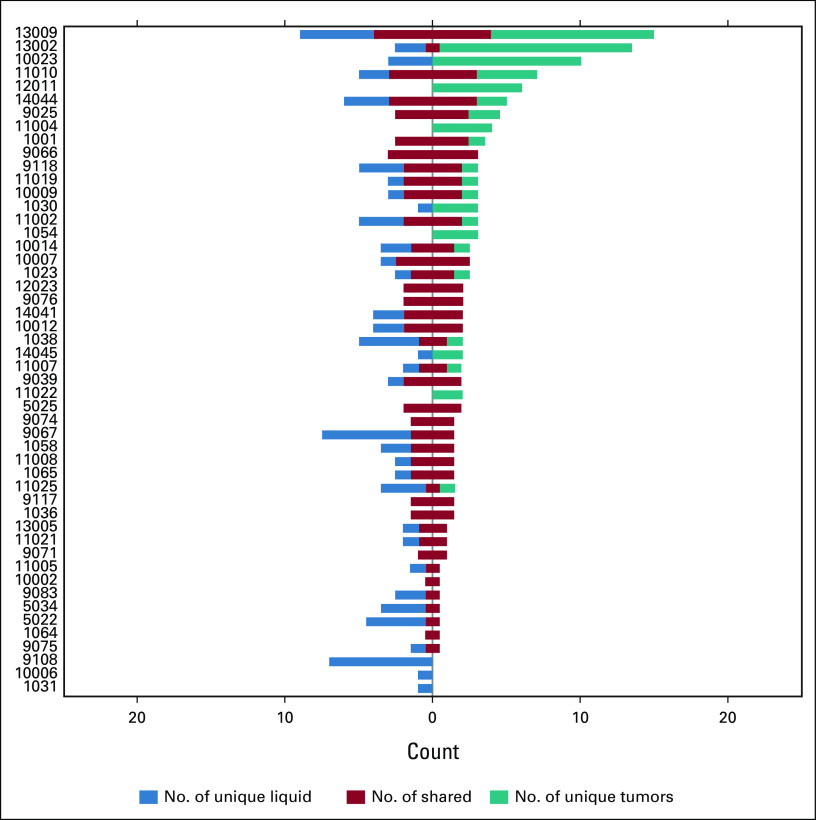
Concordance between liquid and solid biopsies. (A) Venn diagram illustrating the total numbers of mutations detected in liquid and solid biopsies across all 50 analyzed patients with GEP-NEC. Blue circles indicate numbers of mutations detected in liquid biopsies (solid line, with filtering against WBC data; dotted line, without filtering against WBC data) and the teal circle indicate the numbers of mutations detected in solid biopsies. Numbers indicate data from 76 genes where mutation status was assessed in both liquid and solid biopsies (ie, excluding data for PMS2 from the 77-gene Avenio panel). aWithout filtering of variants seen in WBC. (B) Co-bar plot illustrating the concordance and differences in mutations detected in liquid and solid biopsies from each of the 50 analyzed patients. Each horizontal bar represents one patient (as indicated on the y-axis). The blue part of each bar illustrates the number of mutations (x-axis) uniquely detected in liquid biopsies. The red part indicates shared mutations (detected both in liquid and solid biopsies), while the teal part illustrates mutation uniquely detected in solid biopsies. Data presented represent the findings in liquid biopsies after additional filtering against WBC data (see Methods). Orange arrows indicate MSI-positive cases, while purple arrows indicate cases with locally advanced stage III disease without metastases. GEP-NEC, gastroenteropancreatic neuroendocrine carcinoma; MSI, microsatellite instable.
On average, each patient thus had one mutation uniquely detected in liquid biopsies and 1.4 mutations uniquely detected in solid biopsies, while the average number of mutations detected in both samples was 2.5. However, assessing these data in individual patients, we found a large variability between patients (Fig 2B). Of the 50 patients, 15 (30%) had identical data from liquid and solid biopsies, while one patient had unique mutations in both liquid and solid biopsies but no overlapping mutations (germline single nucleotide polymorphisms (SNP) status confirmed samples were from the same individual). Furthermore, in three patients, we detected unique mutations in liquid samples but zero mutations in solid biopsy. Conversely, in six patients, we found unique mutations in solid biopsy but zero mutations in the liquid sample. On average, across the 50 patients, the concordance between mutations in liquid and solid biopsies was 57%.
In a real-life setting, data from WBC may not be available when analyzing solid tumor biopsies or plasma samples. In an exploratory analysis, we set the WBC-filtered data from solid biopsies as baseline and simulated a scenario where data from WBC were not available for plasma analyses. In such a scenario, the fraction of variants called in liquid biopsies that could be confirmed as somatic in solid biopsies was 63% (127/201 mutations; Fig 2A and Appendix Fig A3).
Impact of Disease and Mutational Burden
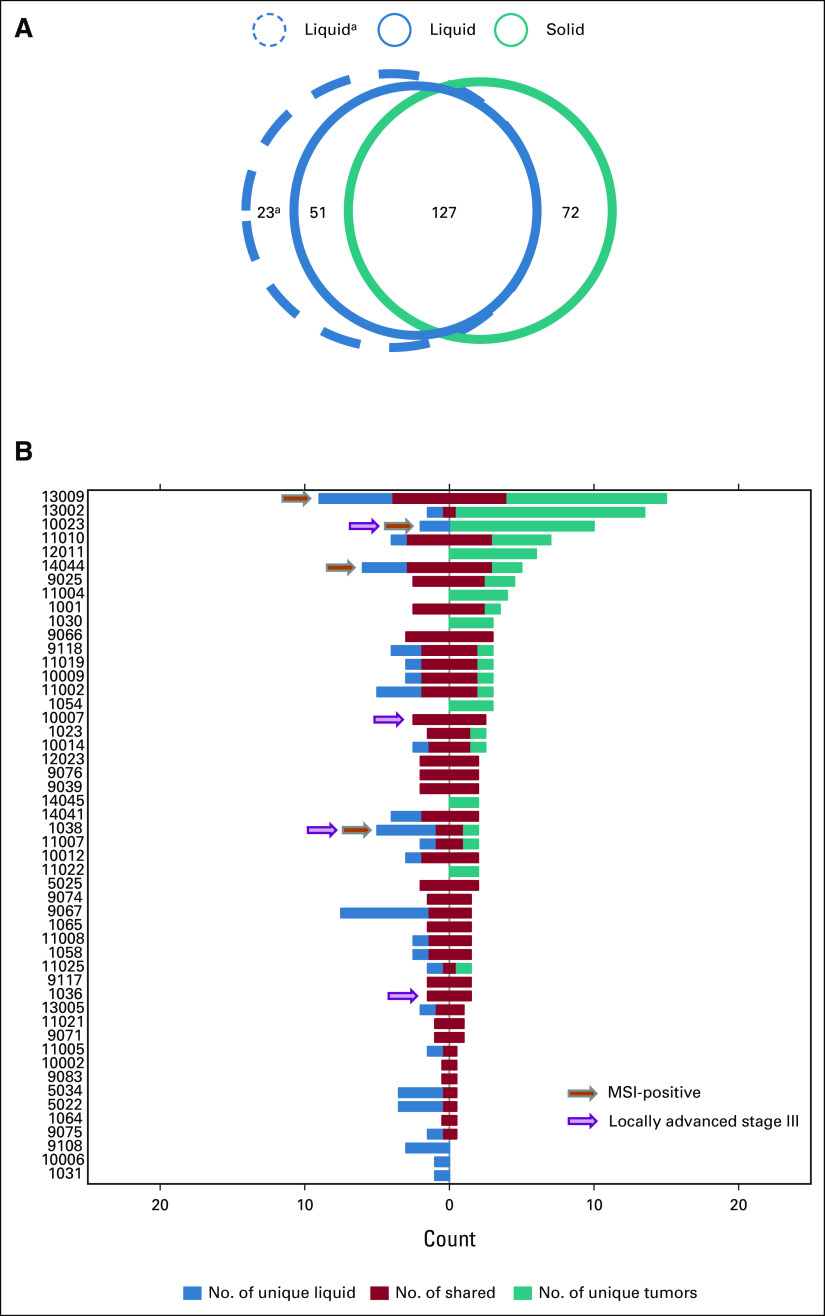
We used presence or absence of liver metastases as a measure for disease burden in patients. In patients with established liver metastases (n = 30), we found a significantly higher proportion of mutations in tumors to also be detected in the liquid biopsies (78%) compared with in patients without liver metastases (48%; P = 1.5 × 10–5; Fig 3A). Stratifying patients into subgroups according to degree of liver involvement, we found a significant trend of increasing proportion of tumor mutations detected in liquid biopsies, with increasing disease burden (Ptrend = 1.2 × 10–4; Fig 3A). However, there was a large variability between individual patients, with a range from 0% to 100% concordance, within both patient groups (with or without liver metastases; Data Supplement).
FIG 3.
Concordance between liquid and solid biopsies in subgroups of patients with GEP-NEC. (A) Co-bar plot illustrating the difference in concordance of mutations detected in liquid and solid biopsies between patients with and without liver metastases as well as subgroups with different levels of liver affected by disease. The blue part of each horizontal bar illustrates the fraction of mutations (x-axis) uniquely detected in liquid biopsies. The red part indicates the fraction of shared mutations (detected both in liquid and solid biopsies), while the teal part illustrates the fraction of mutations uniquely detected in solid biopsies. Note that the percentages (x-axis) is given as percentage of all three categories of mutations, while the percentages mentioned in the main text (78% and 48%) relate to fractions of mutations in solid tumor biopsies that was also detected in liquid biopsies, ie, for that particular comparison, the sum of the red and teal categories were set to 100%. Data presented in the co-bars represent fraction of mutations, while n for each category refers to the number of patients. Among 30 patients with liver metastases, data on level of liver involvement were available for 21. (B) Co-bar plot illustrating the difference in concordance of mutations detected in liquid and solid biopsies between patients with GEP-NEC of different primary sites. Color keys are as given for panel A. GEP-NEC, gastroenteropancreatic neuroendocrine carcinoma.
One may expect the landscape of mutations in ctDNA to be dominated by major subclones in the tumor. Supporting this, we found that mutations detected both in tumor and in ctDNA had an average VAF of 0.42 in the tumors, while those mutations that were unique to tumor had a slightly lower average VAF of 0.38 (P = .033; Fig 4A). In liquid biopsies, the average VAF of those mutations detected both in liquid and tumor biopsies was 0.28, while the average was 0.03 for those unique to liquid biopsies (P < .001; Fig 4B).
FIG 4.
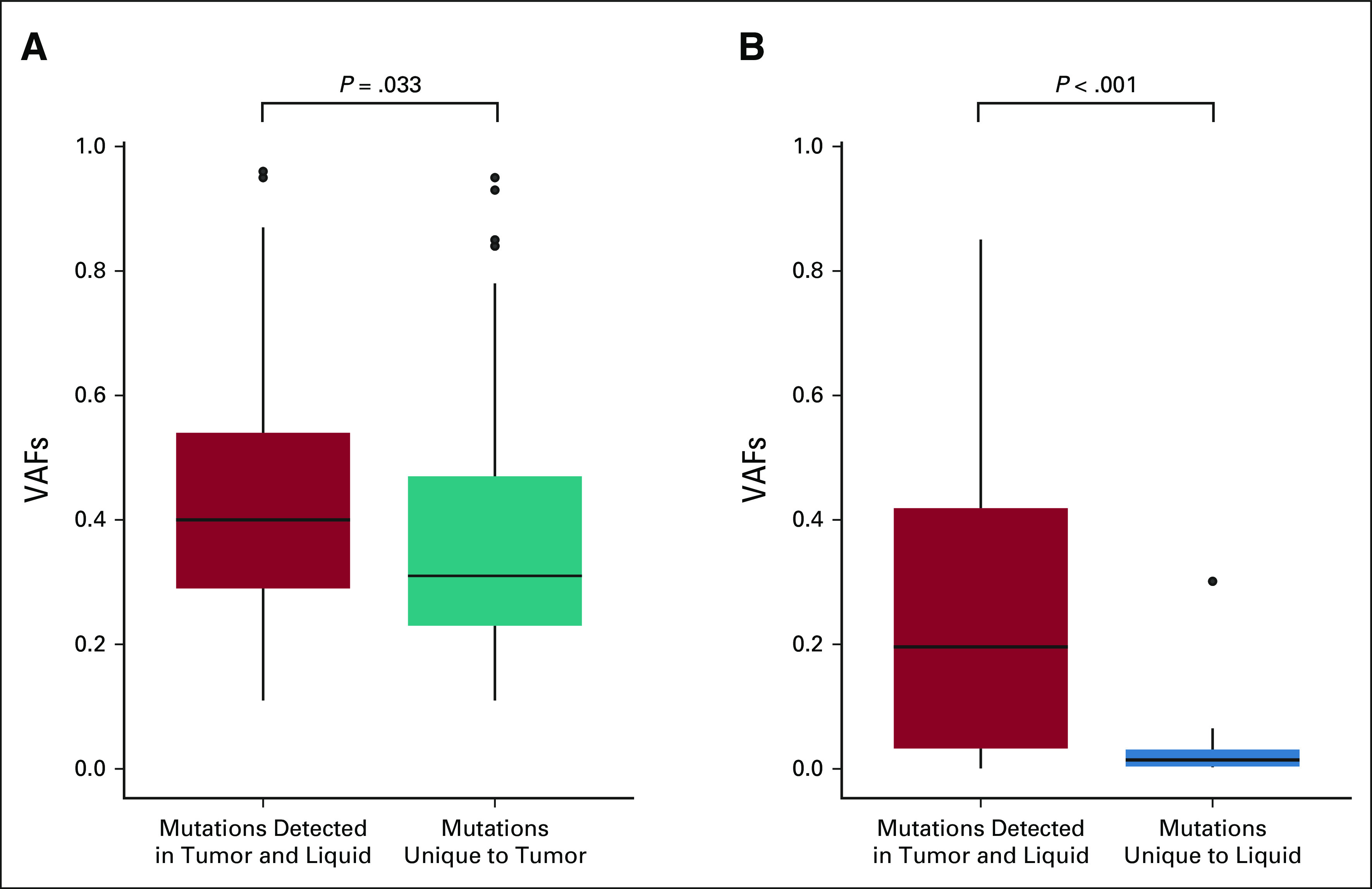
Subclone size and contribution to circulating tumor DNA in patients with GEP-NEC. (A) Box plot illustrates the VAFs for mutations detected in solid tumor tissue. The higher tumor VAFs among those mutations detected in both solid and liquid biopsies (red) compared with those uniquely detected in solid biopsies (teal) indicates that larger tumor subclones are better represented in liquid biopsies than in smaller subclones, in GEP-NEC. (B) Box plot illustrates the VAFs for mutations detected in liquid biopsies. The higher tumor VAFs among those mutations detected in both liquid and tumor biopsies (red) compared with those uniquely detected in liquid biopsies (blue) indicates that liquid biopsies may detect mutations from smaller subclones, not captured by solid tumor biopsies, in GEP-NEC. GEP-NEC, gastroenteropancreatic neuroendocrine carcinoma; VAF, variant allele frequency.
Among the 50 tumors, four were MSI-high (Fig 2B; Data Supplement). The fraction of mutations in tumor tissue that was also detected in liquid biopsies was lower for these four patients (40%) compared with microsatellite stable (MSS) tumors (70%; P = 7.8 × 10–4), while the fraction of mutation uniquely detected in liquid biopsies (47%) was higher than in MSS tumors (25%; P = .026).
Four cases had locally advanced disease stage III without metastases. Two of these cases had 100% concordance between liquid and solid biopsies, while one had 0% concordance. Thus, these cases revealed a concordance pattern similar to the metastatic cases (Fig 2B).
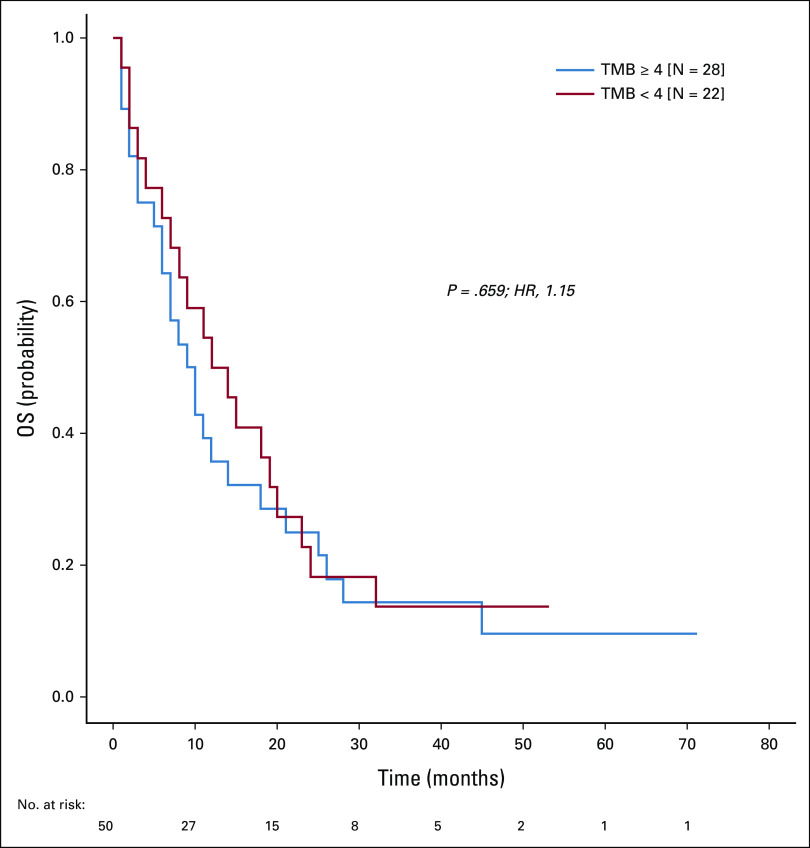
Assessing the TMB, we found strong correlations between the total number of mutations per patient and each of the parameters number of shared mutations, mutations unique to liquid biopsies, and mutations unique to solid biopsies (P = 7.0 × 10–6, 7.3 × 10–4, and 2.0 × 10–4, respectively; Appendix Fig A4). Notably, TMB did not affect overall survival (Appendix Fig A5).
Liquid Versus Solid Biopsies in Subgroups of GEP-NEC
We performed stratified assessments according to different primary tumor sites. Assessing the subgroups, with five or more cases in each primary site (Table 1), some numerical differences were observed (Fig 3B), but these did not reach statistical significance. The exception was the finding that esophageal cases had a significantly higher proportion of mutations uniquely detected in liquid biopsies compared with the other subgroups (43%; P = .001). Although this must be interpreted with caution, because of the low number of esophageal cases (n = 5), it was not a covariate of high proportion of cases with liver metastases, since this was lower (40%) among the esophageal cases compared with the other subgroups (62%).
Detection of Targetable Mutations in Liquid Biopsies
We previously found 66% of solid biopsies from GEP-NEC tumors to harbor potentially targetable mutations.5 Applying the same classification of mutations as potentially targetable or not, as previously done,5 we here found 40 mutations, in 26 patients, to be potentially targetable. Thus, 26 of 50 (52%) tumors harbored potentially targetable mutations within the 77 gene panel (Data Supplement). Notably, 25 of the 40 potentially targetable mutations were also detected in solid biopsies, while 15 were detected only in the liquid biopsies. Thus, a total of nine (18%) patients had potentially targetable mutation detected only in liquid biopsies.
We also performed a stringent re-evaluation of all mutations, identifying those listed as currently established biomarkers (level 1 or 2; Data Supplement) in the OncoKB database. In this analysis, 15 mutations, in 15 patients, were identified in OnkoKB. Thus, 15 of 50 patients (30%) had OncoKB-validated targetable mutations in the ctDNA. Notably, three of these 15 mutations were uniquely detected in the liquid biopsies, while 12 were also found in the tumors.
Parallel Evolution Detected in Liquid Biopsies
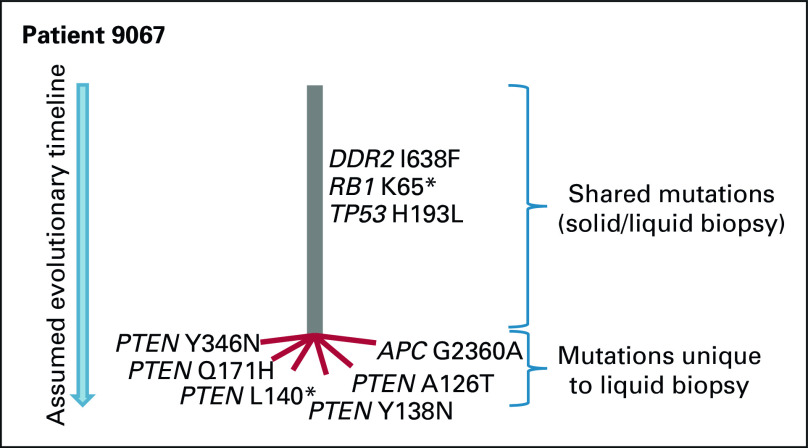
In the liquid biopsies, we found 15/50 patients (30%) to harbor more than one mutation in one or more genes, indicating parallel evolution (Fig 1). The most frequent cases were observed among frequently mutated genes (TP53, KRAS, and APC). However, the case indicating the strongest patient-specific selection pressure (case 9067) had five different PTEN mutations (Fig 5). Assessing three of these mutations that were located in close proximity to each other in PTEN exon 5 (codons 126, 138, and 140), they were observed on different sequencing reads, supporting that they occurred as independent, parallel events.
FIG 5.
Case (patient 9067) exemplifying parallel evolution in GEP-NEC. Mutations are drawn in an assumed phylogenetic tree, based in molecular data from patient 9067. Gray line (trunk) indicates the (presumably early) mutations shared between solid and liquid biopsy, while the red lines (branches) indicate mutations uniquely detected in the liquid biopsy. As one of two patients with PTEN mutations uniquely detected in liquid biopsy, among the 50 analyzed cases, patient 9067 had five different parallel PTEN mutations detected in liquid biopsy. GEP-NEC, gastroenteropancreatic neuroendocrine carcinoma.
DISCUSSION
The existing molecular data for GEP-NEC are very limited, and there is a large need for better data to improve classification and for identification of potentially new therapeutic strategies. We recently performed a genetic characterization of tissue biopsies from 181 GEP-NEN including 152 GEP-NEC.5 In this study, in an effort to extend the molecular understanding of these tumors, we built on our previous data from solid biopsies and performed assessment of genetic alterations in the circulation in a subset of the same patients.
For GEP-NEC, the topic of liquid biopsies has previously only been addressed in a single study. Gerard et al,11 including 24 patients, for whom matched solid tumor tissue was available for 6 cases, found a 44% concordance between ctDNA and tumor tissue. In our present study, among all mutations previously called in solid biopsies, we detected 64% in the analyses of liquid biopsies. Among mutations in the liquid biopsies, 71% were also detected in solid biopsies, whereas 51 (29%) were unique to liquid biopsies (Appendix Fig A6). Although this concordance was good, we also observed differences that could have multiple explanations. For mutations found in solid but not liquid biopsies, this could be due to particular subclones in the tumor that are either smaller than other subclones, or for some unknown reason shed less genetic material to circulation. For mutations found in liquid biopsies but not in solid biopsies, these may reflect subclones present, but not captured, in solid biopsies. Notably, although spatial distribution of subclones has not been studied in GEP-NEC, in other carcinomas, subclones may grow infiltrating in each other in some patients, while subclones in other patients grow in more geographically restricted areas,12 affecting representativeness of biopsies. An additional possibility is that some patients harbor additional lesions and/or distant metastases where alternative subclones will not contribute to the mutational spectrum in a solid biopsy but may do so in liquid biopsies.
Disease burden has been found to affect the representativeness of liquid biopsies in most cancer types.10 We used presence and extent of liver metastases as a marker for disease burden. We found a better concordance between liquid and solid biopsies in patients with liver metastases, and less unique mutations in tumor with increasing percentage of liver involvement. Notably, we also found a generally lower VAF for mutations detected uniquely in tumors compared with those detected in both tumors and liquid biopsies, indicating that intraindividually, the size of subclones may influence detection of mutations in liquid biopsies. We also found that liquid biopsies seem to detect mutations in small subclones that are difficult to capture in solid tumor biopsies.
We previously reported a high fraction (66%) of neuroendocrine carcinoma tumors to harbor potentially targetable mutations, opening new possibilities for treatment strategies for these tumors.5 An important question is whether such mutations are also detectable in liquid biopsies. In our present analysis, we applied a smaller gene panel, covering 77 genes, contrasting our previous assessment of 360 genes in the solid biopsies. In addition, the present panel precluded assessment of copy-number alterations and MSI status. Even with these limitations, we found potentially targetable mutations in 52% of the samples. Importantly, nine (18%) patients had potentially targetable mutations detected only in liquid biopsies, indicating that at least in some GEP-NEC cases, liquid biopsies may provide additional information, beyond solid biopsies, to guide future treatment choices. Overall, our present findings indicate that liquid biopsy may be used for biomarker assessments to guide therapy choice for many patients with GEP-NEC.
A limitation of this study is that the mutation calling in liquid biopsies was performed for SNVs only. Currently, an indel calling option for the present analysis is not yet available and we have therefore not compared the detection of indels between liquid and solid biopsies. However, we have no reason to believe that the concordance between liquid and solid biopsies would be different for indels than for SNVs. Another limitation is that we analyzed a single liquid biopsy per patient. In a future real-life setting, we imagine the application of liquid biopsies in GEP-NEC management to include longitudinal sampling for monitoring disease progression and subclonal dynamics. However, our present study provides a basis, indicating that such assessments of ctDNA are highly feasible.
We find liquid biopsies from patients with GEP-NEC to represent a good alternative material for biomarker analyses. We found some mutations uniquely detected in solid biopsies and others uniquely detected in liquid biopsies; however, in a real-life setting, it may be too resource-demanding to perform both analyses for all patients. Liquid biopsies can be used clinically to characterize tumors in cases where the possibility of solid biopsy is precluded. Since liquid biopsy analyses may add additional mutations compared with tumor biopsy analyses alone, liquid biopsy may also be an option in cases where no targetable mutations are detected in the solid tumor biopsy.
ACKNOWLEDGMENT
A part of this work was performed in the Mohn Cancer Research Laboratory. The authors would like to thank Liv Sylvi Meyer for technical assistance.
APPENDIX 1. Supplementary Materials and Methods
Patients and Samples
Patients diagnosed with high grade (HG) gastroenteropancreatic (GEP)-NEN during 2013-2017 in Nordic centers were prospectively included in the Nordic NEC registry. Inclusion criteria were as follows: histopathologically confirmed metastatic or unresectable locally advanced high-grade neuroendocrine neoplasm (Ki-67>20%) with gastroenteropancreatic primary or unknown primary (carcinoma of unknown primary) with predominantly GI metastases. Clinical information, tumor tissue, plasma, and blood cells for matched non-neoplastic DNA were collected for each case before start of chemotherapy. Histologic sections (hematoxylin/eoisin, chromogranin A, synaptophysin, and Ki-67) were collected and subject to centralized pathologic re-evaluation by an expert NEN pathologist. Subsequently, an additional blinded pathology review was done by three NEN pathologists for all cases meeting the following criteria: neuroendocrine tumor (NET) G3 or non–small-cell NEC with a Ki-67 ≤ 55% or uncertain morphology since these are the cases where pathology assessment separating NET G3 from NEC is important. Difficult cases were finally discussed during a virtual consensus meeting. On the basis of the criteria above, 181 cases were previously included in a study that aimed to assess the molecular landscape in solid biopsies of HG GEP NEN, of which 152 were GEP neuroendocrine carcinomas (GEP NECs).5
For the present analyses, 50 patients with GEP-NEC were included. All of these were among the 152 GEP-NECs previously characterized for molecular alterations in the tumor tissue and normal cells (WBC). Thus, for all 50 in this study, we had data both from liquid and solid biopsies as well as from WBC. Patients were selected for the present analyses to provide representative groups of tumors with respect to different primary tumor sites, sex, cell type, and MSI status (Appendix Fig A1). At the same time, the samples were selected on the basis of particularly high mutation burden or particularly low mutation burden as well as tumor cell fraction in the solid tumor biopsies of at least 20%, to ensure a proper basis for comparison.
For one patient (case 11004), exact dates for sampling were not available. For the remaining 49 patients, all blood samples were collected before any treatment. For one patient (case 10007), the blood sample was collected before start of neoadjuvant chemotherapy, whereas the tumor tissue specimen was collected after neoadjuvant therapy at surgery. For all remaining patients, all biopsies were taken before any treatment, and tumor biopsies were taken first, with subsequent collection of blood samples. The median time from tumor biopsy to blood sampling was 25 days, with a range from six to 524 days. Notably, only two patients had long time spans (437 and 524 days, because of initial surgical resection for loco-regional disease and plasma sampling at recurrence). For the rest, the range was six to 61 days (Table 1; Data Supplement).
General demographic information on the selected patients is given in Table 1 and Appendix Fig A1, with further details on individual patients and selection criteria in the Data Supplement. Notably, no preliminary data on potential effect sizes in comparisons between subgroups were available. Thus, this study must be regarded as exploratory, not on the basis of formal power estimates.
Among the 50 patients, survival was followed for a minimum of 32 months or until death (median, 43 months; range, 32-71 months or until death).
The study was institutional review board–approved by the responsible ethics committees in Norway (REK vest 2012/940) and Sweden (REC Uppsala Dnr 2012/285). All sample donors signed informed written consent.
Sequencing and Mutation Calling
For liquid biopsy analyses, 4 mL of EDTA-plasma was subject to DNA isolation and subsequent library preparation using the Avenio ctDNA Expanded Kit (Roche) according to the manufacturer's instructions, covering 77 cancer-related genes (Data Supplement). Individual enriched libraries were quantified with Qubit dsDNA HS Assay Kit, and their profile was assessed with the Agilent Fragment Analyzer System using the Agilent HS NGS Fragment Kit (1-6000bp). Twelve purified libraries per run were pooled and sequenced on an Illumina NextSeq 500 (Illumina, San Diego, CA), using the 300-cycle NextSeq High Output Kit, in paired-end mode (2 × 151 cycles). This resulted in an average of 60,442,281 mapped reads per sample, corresponding to an average sequencing depth of 11,539×.
Data were analyzed with the AVENIO ctDNA Analysis Software program (version 2.0.0) with default parameter settings for the expanded panel. Only point mutations (SNVs) in the coding regions of the 77 genes in the panel were considered. As further postprocessing filters, we applied a VAF filter of > 0.0025 to exclude technical sequencing noise. To exclude SNPs, only variants with MAF < 0.02 in the EXAC or the 1000 Genomes Project databases were considered. Furthermore, variants were filtered against sequencing information available from matched WBCs (previously published in our study on solid biopsies5).
Annotation of mutations as targetable was performed as described in our previous analysis of solid biopsies of NEC5 (a list of predefined targetable mutations is provided in the Data Supplement and with a separate assessment applying the OncoKB database).
Sequencing and Mutation Calling of Matched Solid Biopsies and WBCs
Sequencing and mutation calling of the solid tumor biopsies from the patients included in this study has been reported previously.5 In brief, targeted massive parallel sequencing was performed on DNA from formalin-fixed paraffin-embedded tumor tissue and matched WBCs as normal tissue. Targeted enrichment was performed using baits against an in-house panel of 360 cancer-related genes.12 Libraries were sequenced on a MiSeq instrument (Illumina) to an average depth of 136× for the tumors and 165× for normal blood (in the original report5): for the subset of patients in this study, the average depth was 120× (range, 57× to 184×) for the tumors and 175× (range, 82× to 256×) for the normal blood.
Concordance Analysis
Among the 77 genes covered by the Avenio ctDNA Expanded Kit, 76 were also included in the 360 gene panel applied for solid biopsies and WBCs. The only gene not included in the 360 gene panel was PMS2. Thus, all concordance assessments between data from liquid and solid biopsies were performed on the basis of 76 genes.
Statistics
Potential differences in distribution of variables between groups were assessed by Fisher exact test. Potential correlations between continuous variables were assessed by calculation of Spearman's rho. Comparison of continuous variables between groups was performed by Mann-Whitney rank test. P < .05 was considered significant. All P values are given as two-sided, and P values from Fisher exact tests are given as two-sided and cumulative. Statistical analyses were performed using SPSS v.28.0.1.0.
FIG A1.
Summary of samples included in our previous analysis of solid tumor biopsies (n = 152; left panel) and the distribution across the same parameters within the subset selected for liquid biopsy analyses (n = 50; right panel). aVenizelos et al.5 ctDNA, circulating tumor DNA; GEP-NEC, gastroenteropancreatic neuroendocrine carcinoma; MSI, microsatellite instable; MSS, microsatellite stable.
FIG A2.
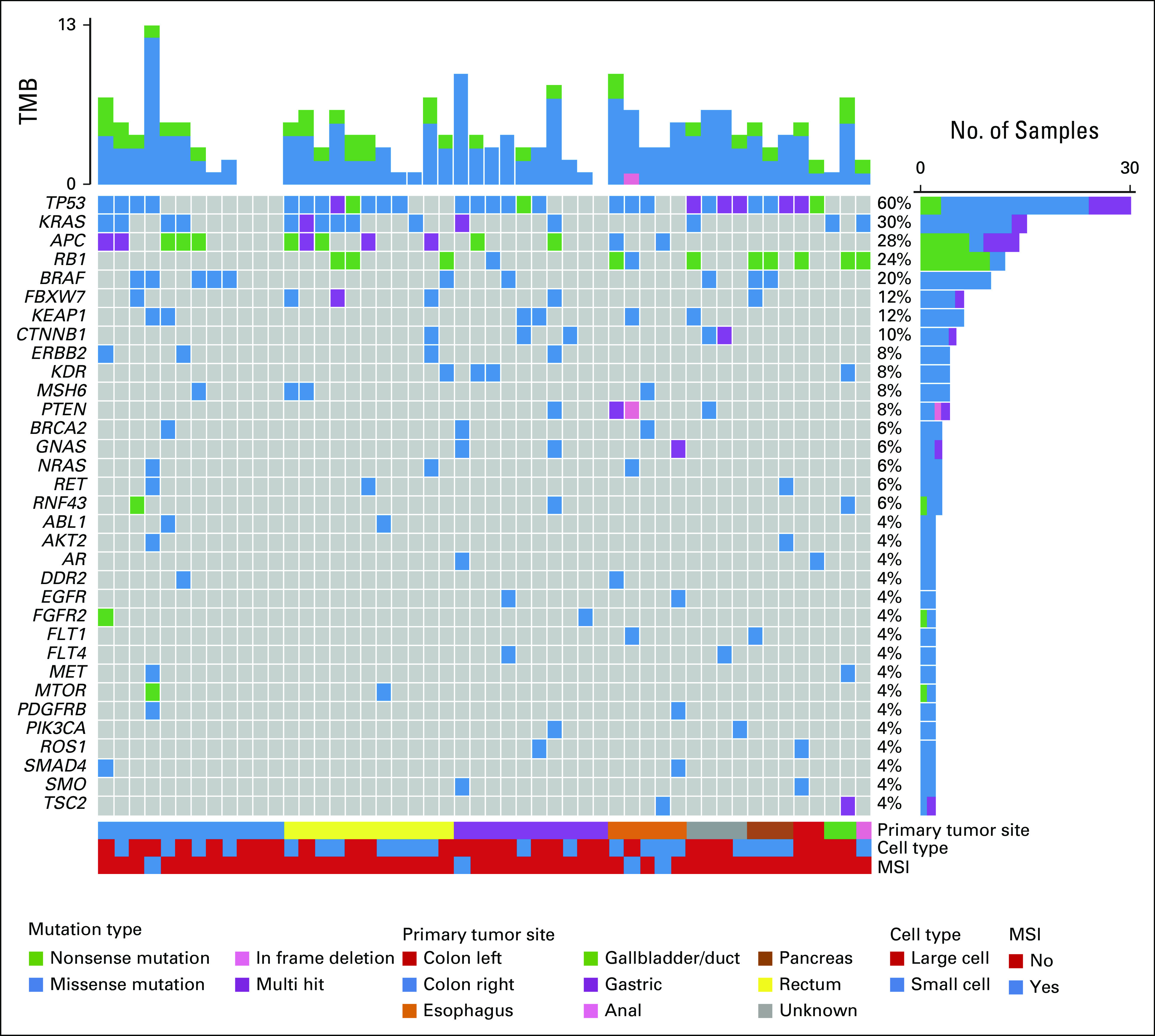
Mutation spectrum in liquid biopsies from patients with GEP-NEC. The oncoplot shows the most frequently altered genes (rows) in liquid biopsies from 50 patients with GEP-NEC (columns). Upper panel shows the mutational burden per sample. Percentages on the right represent mutations' frequency per gene. The panel under the oncoplot area is composed of three single-row heatmaps showing in order, from top to bottom, primary tumor site, cell type, and MSI status. Multihit indicates that more than one mutation occurs in the same gene, in the same patient. Mutations presented represent the findings in liquid biopsies without filtering against WBC data (see Methods). For data with WBC-based filtering, see main Figure 1. GEP-NEC, gastroenteropancreatic neuroendocrine carcinoma; MSI, microsatellite instable; TMB, tumor mutation burden.
FIG A3.
Concordance between liquid and solid biopsies in individual patients. Co-bar plots illustrating the concordance and differences in mutations detected in liquid and solid biopsies from each of the 50 analyzed patients. Each horizontal bar represents one patient (as indicated on the y-axis). The blue part of each bar illustrates the number of mutations (x-axis) uniquely detected in liquid biopsies. The red part indicates shared mutations (detected both in liquid and solid biopsies), while the teal part illustrates mutation uniquely detected in solid biopsies. Data presented represent the findings in liquid biopsies without filtering against WBC data (see Methods). For data with WBC-based filtering, see main Figure 2.
FIG A4.
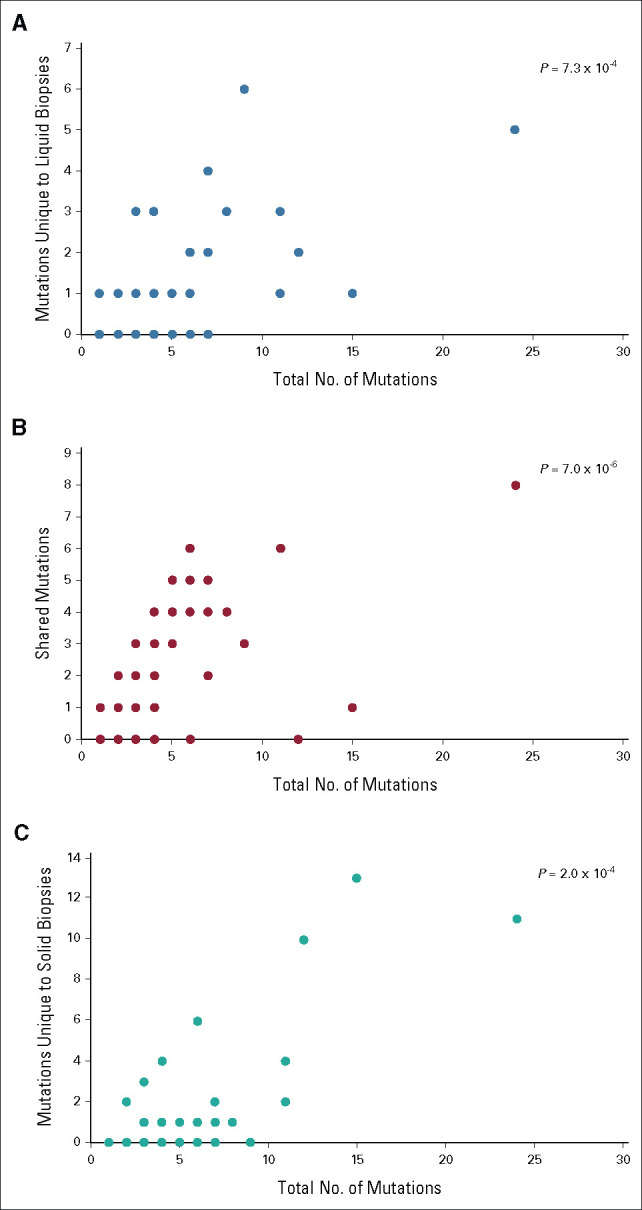
Tumor mutation burden. Scatter plots illustrating that (A) mutations unique to liquid biopsies, (B) mutations shared between liquid and solid biopsies, and (C) mutations unique to solid biopsies were all strongly correlated with the total number of mutations detected in each patient, in gastroenteropancreatic neuroendocrine carcinoma.
FIG A5.
Overall survival in patients with low versus high mutation burden in liquid biopsies. Kaplan-Meier curves illustrate survival for patients with mutation count in the liquid biopsy below median (four mutations; red line) and above or equal to median (blue line). HR, hazard ratio; TMB, tumor mutation burden.
FIG A6.
Overall concordance in detected mutations between liquid- and tumor biopsies in patients with gastroenteropancreatic neuroendocrine carcinoma.
Stian Knappskog
Honoraria: AstraZeneca, Pierre Fabre, Pfizer
Research Funding: AstraZeneca (Inst), Pfizer (Inst)
Patents, Royalties, Other Intellectual Property: Patent EP2389450 A1, Patent WO 2012/010661
Tobias Grob
Research Funding: Roche Sequencing Solutions (Inst)
Ursula Amstutz
Honoraria: Roche Sequencing Solutions
Geir Olav Hjortland
Honoraria: MSD (Inst), Bristol Myers Squibb Norway (Inst)
Consulting or Advisory Role: Bristol Myers Squibb Norway (Inst)
Research Funding: Bristol Myers Squibb Norway (Inst)
Hege Elvebakken
Stock and Other Ownership Interests: CRISPR Therapeutics
Halfdan Sorbye
Honoraria: Ipsen, Pierre Fabre, SAM NORDIC
Consulting or Advisory Role: Hutchnson, AAA/Endocyte/Novartis
Aurel Perren
Stock and Other Ownership Interests: Illumina, Roche
Honoraria: Roche
Travel, Accommodations, Expenses: Roche
No other potential conflicts of interest were reported.
SUPPORT
Supported by The Norwegian Cancer Society, The Western Norwegian Health Region, Novartis, and Ipsen.
H. S. and A.P. contributed equally to this work as last authors.
DATA SHARING STATEMENT
The data supporting the findings are available from the corresponding author upon reasonable request and upon appropriate ethics approvals.
AUTHOR CONTRIBUTIONS
Conception and design: Stian Knappskog, Halfdan Sorbye, Aurel Perren
Financial support: Stian Knappskog
Administrative support: Stian Knappskog, Christian Kersten, Aurel Perren
Provision of study materials or patients: Stian Knappskog, Geir Olav Hjortland, Christian Kersten, Anna Sundlöv, Halfdan Sorbye
Collection and assembly of data: Stian Knappskog, Tobias Grob, Ursula Amstutz, Geir Olav Hjortland, Inger Marie B. Lothe, Christian Kersten, Eva Hofsli, Anna Sundlöv, Hege Elvebakken, Herish Garresori, Johanna Svensson, Halfdan Sorbye, Aurel Perren
Data analysis and interpretation: Stian Knappskog, Tobias Grob, Andreas Venizelos, Christian Kersten, Anne Couvelard, Halfdan Sorbye, Aurel Perren
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Stian Knappskog
Honoraria: AstraZeneca, Pierre Fabre, Pfizer
Research Funding: AstraZeneca (Inst), Pfizer (Inst)
Patents, Royalties, Other Intellectual Property: Patent EP2389450 A1, Patent WO 2012/010661
Tobias Grob
Research Funding: Roche Sequencing Solutions (Inst)
Ursula Amstutz
Honoraria: Roche Sequencing Solutions
Geir Olav Hjortland
Honoraria: MSD (Inst), Bristol Myers Squibb Norway (Inst)
Consulting or Advisory Role: Bristol Myers Squibb Norway (Inst)
Research Funding: Bristol Myers Squibb Norway (Inst)
Hege Elvebakken
Stock and Other Ownership Interests: CRISPR Therapeutics
Halfdan Sorbye
Honoraria: Ipsen, Pierre Fabre, SAM NORDIC
Consulting or Advisory Role: Hutchnson, AAA/Endocyte/Novartis
Aurel Perren
Stock and Other Ownership Interests: Illumina, Roche
Honoraria: Roche
Travel, Accommodations, Expenses: Roche
No other potential conflicts of interest were reported.
REFERENCES
- 1.Heetfeld M, Chougnet CN, Olsen IH, et al. : Characteristics and treatment of patients with G3 gastroenteropancreatic neuroendocrine neoplasms. Endocr Relat Cancer 22:657-664, 2015 [DOI] [PubMed] [Google Scholar]
- 2.Sorbye H, Welin S, Langer SW, et al. : Predictive and prognostic factors for treatment and survival in 305 patients with advanced gastrointestinal neuroendocrine carcinoma (WHO G3): The NORDIC NEC Study. Ann Oncol 24:152-160, 2013 [DOI] [PubMed] [Google Scholar]
- 3.Walter T, Tougeron D, Baudin E, et al. : Poorly differentiated gastro-entero-pancreatic neuroendocrine carcinomas: Are they really heterogeneous? Insights from the FFCD-GTE national cohort. Eur J Cancer 79:158-165, 2017 [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi T, Machida N, Morizane C, et al. : Multicenter retrospective analysis of systemic chemotherapy for advanced neuroendocrine carcinoma of the digestive system. Cancer Sci 105:1176-1181, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venizelos A, Elvebakken H, Perren A, et al. : The molecular characteristics of high-grade gastroenteropancreatic neuroendocrine neoplasms. Endocr Relat Cancer 29:1-14, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Carbonero R, Sorbye H, Baudin E, et al. : ENETS consensus guidelines for high-grade gastroenteropancreatic neuroendocrine tumors and neuroendocrine carcinomas. Neuroendocrinology 103:186-194, 2016 [DOI] [PubMed] [Google Scholar]
- 7.Strosberg JR, Coppola D, Klimstra DS, et al. : The NANETS consensus guidelines for the diagnosis and management of poorly differentiated (high-grade) extrapulmonary neuroendocrine carcinomas. Pancreas 39:799-800, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zakka K, Nagy R, Drusbosky L, et al. : Blood-based next-generation sequencing analysis of neuroendocrine neoplasms. Oncotarget 11:1749-1757, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dasari A, Mehta K, Byers LA, et al. : Comparative study of lung and extrapulmonary poorly differentiated neuroendocrine carcinomas: A SEER database analysis of 162, 983 cases. Cancer 124:807-815, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan JCM, Massie C, Garcia-Corbacho J, et al. : Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat Rev Cancer 17:223-238, 2017 [DOI] [PubMed] [Google Scholar]
- 11.Gerard L, Garcia J, Gauthier A, et al. : ctDNA in neuroendocrine carcinoma of gastroenteropancreatic origin or of unknown primary: The CIRCAN-NEC Pilot Study. Neuroendocrinology 111:951-964, 2021 [DOI] [PubMed] [Google Scholar]
- 12.Yates LR, Gerstung M, Knappskog S, et al. : Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 21:751-759, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. doi: 10.1200/PO.17.00011. 10.1200/PO.17.00011, 2017 Chakravarty D, Gao J, Phillips SM, et al: OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the findings are available from the corresponding author upon reasonable request and upon appropriate ethics approvals.