PURPOSE
The PAOLA-1/ENGOT-ov25 trial of maintenance olaparib plus bevacizumab for newly diagnosed advanced high-grade ovarian cancer demonstrated a significant progression-free survival (PFS) benefit over placebo plus bevacizumab, particularly in patients with homologous recombination deficiency (HRD)–positive tumors. We explored whether mutations in non-BRCA1 or BRCA2 homologous recombination repair (non–BRCA HRRm) genes predicted benefit from olaparib plus bevacizumab in PAOLA-1.
METHODS
Eight hundred and six patients were randomly assigned (2:1). Tumors were analyzed using the Myriad MyChoice HRD Plus assay to assess non–BRCA HRRm and HRD status; HRD was based on a genomic instability score (GIS) of ≥ 42. In this exploratory analysis, PFS was assessed in patients harboring deleterious mutations using six non–BRCA HRR gene panels, three devised for this analysis and three previously published.
RESULTS
The non–BRCA HRRm prevalence ranged from 30 of 806 (3.7%) to 79 of 806 (9.8%) depending on the gene panel used, whereas 152 of 806 (18.9%) had non‐BRCA1 or BRCA2 mutation HRD-positive tumors. The majority of tumors harboring non–BRCA HRRm had a low median GIS; however, a GIS of > 42 was observed for tumors with mutations in five HRR genes (BLM, BRIP1, RAD51C, PALB2, and RAD51D). Rates of gene-specific biallelic loss were variable (0% to 100%) in non–BRCA HRRm tumors relative to BRCA1-mutated (99%) or BRCA2-mutated (86%) tumors. Across all gene panels tested, hazard ratios for PFS (95% CI) ranged from 0.92 (0.51 to 1.73) to 1.83 (0.76 to 5.43).
CONCLUSION
Acknowledging limitations of small subgroup sizes, non–BRCA HRRm gene panels were not predictive of PFS benefit with maintenance olaparib plus bevacizumab versus placebo plus bevacizumab in PAOLA-1, irrespective of the gene panel tested. Current gene panels exploring HRRm should not be considered a substitute for HRD determined by BRCA mutation status and genomic instability testing in first-line high-grade ovarian cancer.
INTRODUCTION
Approximately 70% of patients with newly diagnosed ovarian cancer (OC) will experience relapse in the first three years,1 and OC is the eighth most common cause of cancer-related mortality among women worldwide.2 Therapeutic advances, including poly (ADP-ribose) polymerase (PARP) inhibitors, alone or in combination with bevacizumab, have improved patient outcomes significantly.
CONTEXT
Key Objective
The phase III randomized PAOLA-1 trial established maintenance olaparib plus bevacizumab as a standard of care for newly diagnosed advanced ovarian cancer (OC), particularly in patients with homologous recombination deficiency (HRD)–positive tumors. However, current understanding of how mutations in non-BRCA genes involved in homologous recombination repair (non–BRCA HRRm) affect sensitivity to poly (ADP-ribose) polymerase inhibition is incomplete. To our knowledge, we report the first data exploring the clinical relevance of non–BRCA HRRm to predict benefit from poly (ADP-ribose) polymerase inhibition plus bevacizumab in newly diagnosed advanced OC.
Knowledge Generated
In PAOLA-1, non–BRCA HRRm captured a small proportion of HRD-positive tumors. Non–BRCA HRRm gene panels were not predictive of progression-free survival benefit with maintenance olaparib plus bevacizumab versus placebo plus bevacizumab, irrespective of the gene panel tested.
Relevance
Current gene panels exploring non–BRCA HRRm should not be considered a substitute for HRD determined by BRCA mutation status and genomic instability testing in newly diagnosed advanced OC.
PARP inhibitors prevent repair of single-stranded breaks in DNA, generating double-stranded breaks that cannot be repaired accurately in tumors with homologous recombination deficiency (HRD),3 which is an important biomarker associated with maintenance PARP inhibitor efficacy in newly diagnosed advanced OC. Three first-line phase III studies have demonstrated the benefit of PARP inhibition beyond patients with a BRCA1 or BRCA2 mutation (BRCAm).4-6 In patients with HRD-positive tumors (including BRCAm or with a genomic instability score [GIS] of ≥ 42) in the PAOLA-1 study of maintenance olaparib plus bevacizumab versus placebo plus bevacizumab, the hazard ratio (HR; 95% CI) for progression-free survival (PFS) was 0.33 (0.25 to 0.45), with a median PFS of 37.2 versus 17.7 months. By contrast, the HR among patients with HRD-negative or HRD-unknown tumors was 0.92 (0.72 to 1.17).4 A PFS benefit was also seen in patients with HRD-positive tumors in the other two phase III trials that evaluated the PARP inhibitors niraparib and veliparib.5,6
HRD testing may include testing for a BRCAm and for genomic instability to evaluate the consequence of HRD beyond BRCAm. Different HRD tests use different methodologies and different genomic markers such as genome-wide loss of heterozygosity (LOH), telomeric allelic imbalance, and large-scale state transitions. It has been hypothesized that tumor mutations in genes beyond BRCA1/BRCA2 that are implicated in homologous recombination repair (HRR) may also be predictive of PARP inhibitor benefit in OC7 and could be detected using gene panel–based assays. However, clinical data clarifying the role of non-BRCA homologous recombination repair gene mutations (HRRm) in newly diagnosed OC are lacking; furthermore, multiple genes are implicated in HRR, with no consensus regarding the optimal HRR gene panel to identify PARP inhibitor sensitivity in OC.
We analyzed data from the PAOLA-1 study to explore mutations in non-BRCA genes involved in homologous recombination repair (non–BRCA HRRm) as a predictive biomarker of benefit from maintenance olaparib plus bevacizumab for patients with newly diagnosed advanced high-grade OC.
METHODS
Study Design
PAOLA-1 (ClinicalTrials.gov identifier: NCT02477644) was a phase III, randomized, double-blind, international trial (primary data cutoff: March 22, 2019); the study design, patient inclusion criteria, and HRD testing methodology have been previously described in detail.4 Briefly, adults with newly diagnosed advanced (stage III-IV) high-grade serous or endometrioid OC, primary peritoneal cancer, or fallopian tube cancer who had no evidence of disease or clinical complete or partial response after first-line treatment with platinum-based chemotherapy plus bevacizumab were randomly assigned (2:1) to receive either maintenance olaparib (300 mg twice a day) plus bevacizumab (15 mg/kg every 3 weeks) or maintenance placebo plus bevacizumab.
Definitions and Assays
This analysis used targeted next-generation sequencing on tumor samples to assess six panels of genes with direct or indirect roles in HRR (three designed for this analysis and three previously published). Tumor mutations may be germline or somatic in origin. Details on the methodology are provided in the Data Supplement.
Tumor BRCAm (tBRCAm) was defined as a deleterious or suspected deleterious tumor mutation in the BRCA1 and/or BRCA2 genes, irrespective of non–BRCA HRRm. Non–BRCA HRRm was defined as the presence of a deleterious or suspected deleterious tumor mutation in one or more other genes involved in HRR and the absence of a deleterious or suspected deleterious mutation in BRCA1 and/or BRCA2. The criteria for definition of deleterious or suspected deleterious alterations are provided in the Data Supplement. Before primary data cutoff, a subgroup of patients with a non–BRCA HRRm was determined using a predefined panel of 13 genes involved in HRR. To investigate whether expanding the gene panel to a larger number of genes involved in HRR improved the predictive value of non–BRCA HRRm, an expanded panel, including five additional genes involved in HRR, was formed. Further post hoc analyses using a restricted panel of five selected HRR genes with the highest median GIS and additional published gene panels (those used in Study 19,8 NOVA,9 and ARIEL310 trials) were also conducted. Where published gene panels were assessed, analysis was limited to genes also included in the 108-gene panel defined by Myriad: this gene panel, and the HRR genes included in each panel are shown in the Data Supplement. Mutation prevalence was evaluated for individual genes; patients with mutations in more than one HRR gene were classified as having co-occurring HRR gene mutations.
For the analyses in non–BRCA HRRm subgroups reported herein, tumors were analyzed using the MyChoice HRD Plus assay (Myriad Genetic Laboratories Inc, Salt Lake City, UT). HRD-positive status was defined as a GIS of ≥ 42 on the basis of this assay. HRD-negative status was defined as a GIS of < 42 and the absence of a tumor mutation in BRCA1/2 on the basis of this assay. HRD-unknown were cases where a GIS could not be determined. This GIS encompasses LOH, telomeric allelic imbalance, and large-scale state transitions. Detailed methods specific to the MyChoice HRD Plus assay, including Myriad genetic variant classification and gene-specific zygosity assessment, are shown in the Data Supplement.
PFS was defined as the time from random assignment until investigator-assessed disease progression (modified RECIST version 1.1) or death.
Statistical Considerations
This was an exploratory post hoc analysis of the PAOLA-1 trial. PFS was evaluated using the Kaplan-Meier method. HRs and 95% CIs for PFS subgroup analyses were calculated using a Cox proportional hazards model that included treatment, subgroup, and the subgroup by treatment interaction. HRs for PFS were only calculated if there were at least 20 events per subgroup.
RESULTS
HRR Mutation Prevalence and HRD Status
In total, 806 patients were randomly assigned in PAOLA-1. As previously published,4 387 (48.0%) had HRD-positive tumors by GIS ≥ 42 and/or tBRCAm; 235 (29.2%) had a tBRCAm according to Myriad testing, and 152 (18.9%) had non-BRCAm HRD-positive tumors. Two hundred and seventy-seven patients (34.4%) had HRD-negative tumors (GIS < 42), and 142 (17.6%) had tumors with unknown GIS. The overall patient disposition is shown in the Data Supplement.
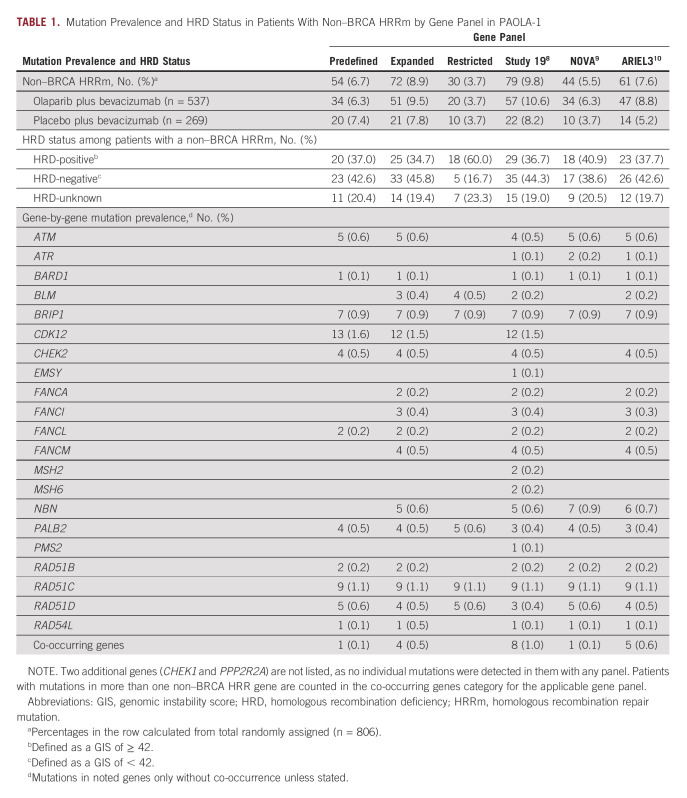
Across the six gene panels used in this analysis, the number of patients harboring a non–BRCA HRRm was 54 (6.7%) in the predefined gene panel (comprised of 13 non–BRCA HRR genes, 11 of which were found to be mutated), 72 (8.9%) in the expanded gene panel that included five additional genes reported to be involved in HRR, and 30 (3.7%) in the restricted gene panel (comprised of 5 of 16 mutated genes with the highest GIS in the expanded gene panel). Among the previously published panels, the number of patients harboring a non–BRCA HRRm was 44 (5.5%) with the gene panel used in NOVA, 61 (7.6%) with the gene panel used in ARIEL3, and 79 (9.8%) with the gene panel used in Study 19 (Table 1). The gene prevalence and mutually exclusive nature of BRCA1 and BRCA2 mutations and the detected non–BRCA HRRm across the six gene panels are shown in Figure 1A.
TABLE 1.
Mutation Prevalence and HRD Status in Patients With Non–BRCA HRRm by Gene Panel in PAOLA-1
FIG 1.

Prevalence of HRRm within HRD-positive, HRD-negative, and HRD-unknown tumors. (A) OncoPrint of mutated HRR genes with annotated positive, negative, and unknown status for HRD and tBRCAm. Percentages on the basis of the total number of enrolled patients in PAOLA-1 (n = 806). Genes are arranged in descending order of median GIS. HRD-positive was defined as a GIS of ≥ 42, and HRD-negative as a GIS of < 42. (B) Expanded panel. The expanded panel included mutated genes in the predefined panel plus five additional selected genes involved in homologous recombination repair. HRD-positive was defined as a GIS of ≥ 42, and HRD-negative as a GIS of < 42. BRCAm, BRCA1, and/or BRCA2 mutation; GIS, genomic instability score; HRD, homologous recombination deficiency; HRRm, homologous recombination repair mutation; tBRCAm, tumor BRCAm.
Of the non-BRCAm HRD-positive (GIS ≥ 42) population in PAOLA-1, using the expanded gene panel, only 25 of 152 (16.4%) had a non–BRCA HRRm. Patients with a non–BRCA HRRm were not particularly enriched in HRD-positive tumors (n = 25 of 806; 3.1%) versus HRD-negative tumors (n = 33 of 806; 4.1%) or unknown HRD status (n = 14 of 806; 1.7%; Fig 1B). These patterns were generally consistent regardless of the HRR gene panel analyzed (Data Supplement).
Biallelic inactivation of HRR genes is more likely to ensure loss of function of HRR proteins, as shown by pan-tumor studies examining BRCA1/BRCA2.11 The distributions of GIS and proportion of biallelic loss relative to heterozygous alterations (ie, loss of both alleles relative to one allele) for the expanded panel are shown in Figure 2. The five highest median GISs of the 16 genes were for BLM (median [interquartile range (IQR)] GIS, 75.5 [74.8-76.3]), BRIP1 (median [IQR] GIS, 56.0 [46.5-62.8]), RAD51C (median [IQR] GIS, 55.0 [45.0-59.0]), PALB2 (median [IQR] GIS, 53.0 [39.0-65.0]), and RAD51D (median [IQR] GIS, 51.5 [46.0-56.5]; Fig 2A).
FIG 2.

Distribution of GISs and proportion of biallelic loss and heterozygous alterations. (A) Distribution of GISs and gene-specific zygosity per HRR gene in BRCA1, BRCA2, and the expanded panel. Of non-BRCA genes analyzed in the expanded gene panel, the genes with the highest median GISs were BLM, BRIP1, RAD51C, PALB2, and RAD51D, forming the basis of the restricted gene panel. Patients with mutations in more than one gene (ie, co-occurring genes) are excluded from this analysis. (B) Proportion of biallelic loss and heterozygous alterations per gene where gene-specific zygosity could be assessed for BRCA1, BRCA2, and the expanded panel. Patients with mutations in more than one gene (ie, co-occurring genes) are excluded from this analysis. BRCAm, BRCA1, and/or BRCA2 mutation; GIS, genomic instability score; HRRm, homologous recombination repair mutation; tBRCAm, tumor BRCAm.
Where gene-/locus-specific zygosity could be assessed, biallelic loss was observed in the majority of patients with a tBRCAm: in 128 of 129 (99.2%) with a tumor BRCA1 mutation and 55 of 64 (85.9%) with a tumor BRCA2 mutation. However, the prevalence of biallelic loss in other genes among patients with a non–BRCA HRRm ranged from 0% to 100% (Fig 2B).
Progression-Free Survival
Investigator-assessed PFS using the six different gene panels is shown among patients with a non–BRCA HRRm in Figure 3. The HR (95% CI) for PFS (olaparib plus bevacizumab v placebo plus bevacizumab) using the predefined 13-gene panel was 0.95 (0.49 to 1.94; Fig 3A). In the expanded panel, the HR for PFS was 1.01 (0.55 to 1.95; Fig 3B). In the restricted panel, the HR for PFS was not calculated, as there were fewer than 20 events per subgroup (11 and six events in the olaparib plus bevacizumab and placebo plus bevacizumab arms, respectively); 2-year PFS rates (on the basis of Kaplan-Meier estimates) were 47.7% and 45.0% in each arm, respectively (Fig 3C). When interrogating the gene panel used in Study 19,8 the HR for PFS was 0.92 (0.51 to 1.73; Fig 3D); when interrogating the panel used in the NOVA study,9 the HR was 1.83 (0.76 to 5.43; Fig 3E); when interrogating the gene panel used in the ARIEL3 study,10 the HR was 1.35 (0.65 to 3.14; Fig 3F). Per-patient PFS using the predefined panel varied by individual gene mutation, as shown in the Data Supplement.
FIG 3.

PFS for patients with non-BRCA homologous recombination repair gene mutation detected using six different gene panels: (A) predefined panel, (B) expanded panel, (C) restricted panel, (D) panel used in Study 19, (E) panel used in the NOVA study, and (F) panel used in the ARIEL3 study. The expanded panel included five additional genes involved in homologous recombination repair; the restricted panel used the five genes with the highest genomic instability scores. bev, bevacizumab; HR, hazard ratio; NC, not calculated (< 20 events); PFS, progression-free survival.
PFS by HRD status among patients with a non–BRCA HRRm as defined by the expanded panel, which includes the largest number of patients, is shown in Figure 4. Among patients with HRD-positive tumors (GIS ≥ 42), the median PFS was 28.1 months with olaparib plus bevacizumab versus 17.7 months with placebo plus bevacizumab; among patients with HRD-negative or unknown tumors, the median PFS was 16.1 months with olaparib plus bevacizumab versus 16.6 months with placebo plus bevacizumab.
FIG 4.

PFS for patients with mutations in non-BRCA genes involved in homologous recombination repair using the expanded panel by HRD status: (A) HRD-positive (GIS of ≥ 42) tumors and (B) HRD-negative (GIS of < 42) or unknown tumors. bev, bevacizumab; GIS, genomic instability score; HRD, homologous recombination deficiency; PFS, progression-free survival.
DISCUSSION
In this exploratory subgroup analysis of PAOLA-1, our data suggest that non–BRCA HRRm status does not appear to be a useful biomarker of PFS benefit from maintenance olaparib plus bevacizumab compared with placebo plus bevacizumab in patients with newly diagnosed advanced OC.
As there is no consensus definition of non–BRCA HRRm among trials in OC and gene panels vary, non–BRCA HRRm was defined using six different gene panels in this analysis of PAOLA-1. Each of these non–BRCA HRR gene panels captured a smaller proportion of patients with newly diagnosed advanced OC (3.7% to 9.8%) than HRD testing by genomic instability excluding BRCAm (18.9%). Importantly, non–BRCA HRRm was not predictive of PFS benefit using any of the six gene panels, with HRs (95% CI) for PFS ranging from 0.92 (0.51 to 1.73) to 1.83 (0.76 to 5.43) depending on the gene panel used. The lack of predictive power of non–BRCA HRRm observed in this analysis might not have been an expected result, given clinical data in platinum-sensitive relapsed (PSR) OC that suggest a benefit can still be observed with PARP inhibition in patients with a non–BRCA HRRm. The HR (95% CI) for PFS among patients with a non–BRCA HRRm in Study 19 was 0.21 (0.04 to 0.86; n = 21),8 in NOVA was 0.31 (0.13 to 0.77; n = 41),9 and in ARIEL3 was 0.21 (0.09 to 0.50; n = 43).13 This difference between the predictive value in recurrent disease and first-line therapy might be caused by the selection of patients with response to at least two prior lines of platinum-based chemotherapy before maintenance PARP inhibition in the recurrent setting.8,10,12
It is possible that HRR mechanisms beyond the established BRCA-mediated HRR, or beyond HRRm detected by the gene panels tested, are present in first-line OC. For example, BRCA1 hypermethylation and RAD51C hypermethylation are present in approximately 11% and 3% of OC, respectively,14,15 BRCA1 hypermethylation is also associated with high levels of genomic instability,8,16 and BRCA1 hypermethylation and RAD51C hypermethylation have been reported to be associated with PARP inhibitor response.17,18 However, epigenetic changes may be more easily reversed than genetic mutations; a recent analysis found that 24% of ovarian tumors lost BRCA1 hypermethylation during treatment.19 Therefore, it is possible that hypermethylation may account for a meaningful proportion of tumors with high genomic instability and that these tumors are sensitive to PARP inhibition in the first-line setting; however, reversion of hypermethylation with treatment may lead to PARP inhibitor resistance in subsequent lines of therapy.
The functional significance of somatic gene mutations in HRR may also differ in patients with newly diagnosed OC who have received only one prior line of systemic therapy and whose true platinum sensitivity is unknown, compared with patients who have known late PSR disease and who might have received multiple lines of therapy. Although some available data suggest that the levels of genomic instability in a tumor do not appear to undergo notable changes between the archival and baseline samples,20 using an LOH cutoff of 14% (since refined to 16%),10 17 of 50 patients (34%) in ARIEL2 who had low levels (< 14%) of LOH in their archival biopsy had high levels of LOH in their pretreatment (within 28 days before first dose of rucaparib) biopsy, and of those 17 patients, five had a response to PARP inhibition.21
The challenge of interpreting somatic alterations in genes related to homologous recombination is compounded by the wide range of genes reported to be involved in HRR. For example, loss-of-function mutations in PTEN have been reported to be associated with PARP inhibitor sensitivity in endometrial carcinomas and glioblastomas,22-24 but the precise role of PTEN in OC is controversial25-27 and data regarding its role in newly diagnosed OC are lacking. The heterogeneity underlying genome maintenance mechanisms in OC necessitates context-specific clinical evaluation and validation of specific biomarkers. Furthermore, the low frequency and indeterminate functional relevance of specific mutations in non–BRCA HRR genes add to the challenges of a single-gene approach. Interestingly, of the non–BRCA HRR genes analyzed here, loss of function of RAD51C and RAD51D has been suggested to confer sensitivity to PARP inhibition in PSR OC; preclinical work has suggested that loss of RAD51C methylation or secondary RAD51C/RAD51D reversion mutations may induce PARP inhibitor resistance.28,29 Among the 12 patients in our analysis with RAD51C/RAD51D mutations in the olaparib arm, there was a wide range of observed PFS, and no clear pattern was seen between RAD51C/RAD51D mutations and PFS in PAOLA-1. Further investigation of the role of specific biomarkers in the context of PARP inhibition in newly diagnosed OC is required; efforts are ongoing to identify further potential markers of underlying tumor HRD in OC.
HRD as determined by genomic instability testing is an important predictive biomarker of PARP inhibitor treatment benefit, including in non-BRCAm patients. Among patients with HRD-positive tumors (with a GIS ≥ 42 and/or BRCAm) in PAOLA-1, the HR for PFS with maintenance olaparib plus bevacizumab versus placebo plus bevacizumab was 0.33 (95% CI, 0.25 to 0.45), and for patients with HRD-positive tumors (with a GIS ≥ 42 excluding BRCAm), the HR was 0.43 (95% CI, 0.28 to 0.66).4 Across all phase III studies in the first-line OC setting, the greatest benefit from maintenance PARP inhibition was seen in patients with HRD-positive tumors (with or without a BRCAm).4-6,30 In this study, even among a small population of patients with a non–BRCA HRRm, median PFS was longer among patients with HRD-positive tumors (GIS ≥ 42) after the addition of maintenance olaparib to bevacizumab (median PFS 28.1 v 17.7 months) although small subgroup sizes limit interpretation.
It is notable that in this study, there was a wide range of GIS observed in patients with non–BRCA HRRm, and only 6 of 16 genes involved in HRR analyzed had a median GIS ≥ 42 (BLM, BRIP1, RAD51C, PALB2, RAD51D, and RAD51B in descending order of GIS). This is consistent with recent pan-tumor analyses that suggest that biallelic loss of BRCA1/BRCA2 greatly distinguishes tumors in terms of HRD, with biallelic loss of other HRR genes having less of an impact.16 The European experts consensus statement on HRD testing in first-line OC noted a high level of agreement that if non-BRCA tumor mutation status is examined, RAD51C, RAD51D, BRIP1, PALB2, and BARD1 could be considered.31 The utility of testing for ATM, CHEK1, CHEK2, ATR, BLM, and CDK12 mutations in OC is yet to be established.31,32 It should also be noted that although biallelic loss was observed in the majority of patients with a tBRCAm (99.2% of those with a BRCA1 mutation and 85.9% of those with a BRCA2 mutation), this trend was not consistent between genes among patients with a non–BRCA HRRm.
Limitations of this post hoc analysis include small subgroup sizes and imbalances between treatment arms on an individual gene level, which limit precision on point estimates of HRs for evaluation of HRRm as a predictive biomarker for PFS in the subgroups analyzed. In analyses of PFS among patients with a non–BRCA HRRm irrespective of GIS reported here, subgroup sizes ranged from 30 to 79, depending on the gene panel used. Although small, this is comparable with prior reports in the PSR setting: subgroups of patients with a non–BRCA HRRm in Study 19,8 NOVA,9 and ARIEL313 were 21, 41, and 43, respectively. Furthermore, owing to small subgroup sizes, these data do not exclude a predictive effect with the addition of olaparib to bevacizumab, which may be associated with mutations in particular individual genes, for example, BRIP1, RAD51C, or RAD51D.33 Furthermore, prospective research is needed to evaluate individual non–BRCA HRR gene mutations as predictors of PARP inhibitor response, in particular, those genes that are strongly correlated with high GIS and biallelic loss.
Mutations in non–BRCA HRR genes account for a small proportion of HRD-positive tumors overall (GIS ≥ 42; 6.5% [n = 25 of 387] and 4.7% [n = 18 of 387] using the expanded and restricted panels, respectively), and for a large proportion of tumors, the mechanism of HRD has not been determined. Further research is needed, including full evaluation of the range of methodologies used to delineate the mechanisms of HRD in these cases and to reduce the number of HRD-unknown tumors.
Finally, patients in PAOLA-1 received bevacizumab along with initial platinum-based chemotherapy. It is unclear if use of bevacizumab as part of initial therapy affects response to maintenance PARP inhibition, and caution must be taken when extrapolating these results to PARP inhibitor monotherapy settings.
In conclusion, to our knowledge, these are the first data exploring the clinical relevance of non–BRCA HRRm as a predictive biomarker of benefit from PARP inhibition in combination with bevacizumab in newly diagnosed advanced OC. This exploratory subgroup analysis of the PAOLA-1 study suggests that non–BRCA HRRm detected using the gene panels in this analysis is not predictive of PFS benefit with maintenance olaparib plus bevacizumab versus placebo plus bevacizumab in patients with newly diagnosed advanced OC. Mutation analysis using current HRRm gene panels did not have utility beyond tBRCAm for selecting patients who may benefit from maintenance olaparib plus bevacizumab in PAOLA-1 and should not be considered a substitute for genomic instability testing to determine HRD beyond tBRCAm.
ACKNOWLEDGMENT
We thank the patients and families for making this trial possible and the investigators and clinical study teams who participated in this trial. Support with statistical analysis was provided by Phil Rowe, MSc, at AstraZeneca. Medical writing support was provided by Adam Gill, MRes, at Cence, funded by AstraZeneca and Merck Sharp & Dohme LLC (a subsidiary of Merck & Co. Inc., Rahway, NJ, USA).
Eric Pujade-Lauraine
Employment: Arcagy-Gineco
Honoraria: AstraZeneca, GlaxoSmithKline
Consulting or Advisory Role: AstraZeneca, Roche, Merck, Incyte
Research Funding: AstraZeneca (Inst)
Other Relationship: Arcagy-Gineco
Jessica Brown
Employment: AstraZeneca
Stock and Other Ownership Interests: AstraZeneca
Alan Barnicle
Employment: AstraZeneca
Stock and Other Ownership Interests: AstraZeneca
Patents, Royalties, Other Intellectual Property: Patent pending for the early stage project with the current employer
Travel, Accommodations, Expenses: AstraZeneca
Jonathan Wessen
Employment: AstraZeneca
Stock and Other Ownership Interests: AstraZeneca
Travel, Accommodations, Expenses: AstraZeneca
Pierre Lao-Sirieix
Employment: AstraZeneca (I), Autolus
Stock and Other Ownership Interests: AstraZeneca (I), Autolus
Patents, Royalties, Other Intellectual Property: Named author on the patent licensed to Medtronic. Not received any payments yet
Steven W. Criscione
Employment: AstraZeneca/MedImmune
Stock and Other Ownership Interests: AstraZeneca/MedImmune
Andreas du Bois
Consulting or Advisory Role: AstraZeneca, Roche/Genentech, Clovis Oncology, GlaxoSmithKline/Tesaro, Mersana, Zodiac Pharma, Amgen, Novartis
Domenica Lorusso
Consulting or Advisory Role: PharmaMar, AstraZeneca, Clovis Oncology, GlaxoSmithKline, MSD, Genmab, Amgen, Seattle Genetics, Immunogen, Merck Serono, Oncoinvent, Corcept Therapeutics, Sutro Biopharma
Speakers' Bureau: AstraZeneca, Clovis Oncology, GlaxoSmithKline, MSD, PharmaMar
Research Funding: PharmaMar (Inst), Clovis Oncology (Inst), GlaxoSmithKline (Inst), MSD (Inst), AstraZeneca (Inst), Genmab (Inst), Seattle Genetics (Inst), Immunogen (Inst), Incyte (Inst), Novartis (Inst), Roche (Inst)
Travel, Accommodations, Expenses: Roche, PharmaMar, AstraZeneca, Clovis Oncology, GlaxoSmithKline
Uncompensated Relationships: Gynecological Cancer InterGroup
Ignacio Romero
Consulting or Advisory Role: Clovis Oncology, GlaxoSmithKline, AstraZeneca Spain, Roche
Speakers' Bureau: Roche, AstraZeneca, PharmaMar, GlaxoSmithKline
Research Funding: Roche (Inst), GlaxoSmithKline (Inst)
Travel, Accommodations, Expenses: PharmaMar, AstraZeneca Spain, GlaxoSmithKline, Clovis Oncology, Roche
Edgar Petru
Honoraria: Roche, AstraZeneca, Daiichi Sankyo/Astra Zeneca
Consulting or Advisory Role: Roche, Daiichi Sankyo/Astra Zeneca
Travel, Accommodations, Expenses: Roche, AstraZeneca/Daiichi Sankyo
Ignace Vergote
Consulting or Advisory Role: AstraZeneca, Elevar Therapeutics, Genmab, Immunogen, Jazz Pharmaceuticals, Mersana, MSD, Novocure, Sotio, Verastem, Zentalis, Roche, Agenus, Eisai, Novartis, Seattle Genetics, Akeso Biopharma, Bristol Myers Squibb, Deciphera, Exelixis, GlaxoSmithKline, Karyopharm Therapeutics, Oncoinvent, OncXerna, Regeneron, Sanofi
Research Funding: Roche (Inst), Amgen (Inst), Oncoinvent (Inst)
Travel, Accommodations, Expenses: Karyopharm Therapeutics, Genmab, Novocure
Nicoletta Colombo
Employment: Sarepta Therapeutics (I)
Honoraria: Roche/Genentech, AstraZeneca, GlaxoSmithKline, MSD Oncology, Clovis Oncology, Pfizer, Amgen, Immunogen, Novartis, Pfizer, Mersana, Eisai, Advaxis, Nuvation Bio
Consulting or Advisory Role: Roche/Genentech, AstraZeneca, Clovis Oncology, Pfizer, MSD Oncology, GlaxoSmithKline, Immunogen, Pfizer, Mersana, Eisai, Advaxis, Nuvation Bio
Sakari Hietanen
Consulting or Advisory Role: GSK, AstraZeneca, MSD
Speakers' Bureau: AstraZeneca, GSK
Barbara Schmalfeldt
Honoraria: Roche, Clovis Oncology, GlaxoSmithKline, AstraZeneca, MSD Oncology
Consulting or Advisory Role: AstraZeneca, GlaxoSmithKline, Clovis Oncology, MSD Oncology, Roche Pharma AG
Research Funding: AstraZeneca, Roche Pharma, GSK, MSD, Clovis Oncology (Inst)
Travel, Accommodations, Expenses: Roche, AstraZeneca, GSK, MSD
Sandro Pignata
Honoraria: AstraZeneca, Roche, PharmaMar, Tesaro, Pfizer, MSD
Consulting or Advisory Role: AstraZeneca, Roche, PharmaMar, Pfizer, Tesaro, Clovis Oncology
Research Funding: Roche (Inst), AstraZeneca (Inst), MSD (Inst), Pfizer (Inst)
Cristina Martín Lorente
Consulting or Advisory Role: AstraZeneca, GlaxoSmithKline, MSD Oncology, Clovis Oncology
Speakers' Bureau: GlaxoSmithKline, AstraZeneca, MSD Oncology
Travel, Accommodations, Expenses: GlaxoSmithKline, AstraZeneca, MSD Oncology, Clovis Oncology
Ingo B. Runnebaum
Consulting or Advisory Role: GlaxoSmithKline
Isabelle Ray-Coquard
Honoraria: Roche, PharmaMar, AstraZeneca, Clovis Oncology, Tesaro, MSD Oncology, Genmab, AbbVie, Pfizer, Bristol Myers Squibb, GlaxoSmithKline, Deciphera, Mersana, Amgen, Advaxis, OxOnc, Seattle Genetics, MacroGenics, Agenus, Sutro Biopharma, Novartis, Daiichi Sankyo
Consulting or Advisory Role: Pfizer, AbbVie, Genmab, Roche, AstraZeneca, Tesaro, Clovis Oncology, PharmaMar, MSD Oncology, Bristol Myers Squibb, Deciphera, Mersana, GlaxoSmithKline, Agenus, MacroGenics, Seattle Genetics, BMS, Novartis, Novocure, Ose pharma, Daichi
Research Funding: MSD Oncology, BMS, Roche/Genentech (Inst)
Travel, Accommodations, Expenses: Roche, AstraZeneca, Tesaro, PharmaMar, GlaxoSmithKline, Clovis Oncology, BMS, Advaxis
Uncompensated Relationships: Arcagy-Gineco, French National Cancer Institute (INCA), Italian Health Authorities, German Health Authorities, Belgium Health Authorities
No other potential conflicts of interest were reported.
DISCLAIMER
The sponsor was involved in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, and approval of the manuscript; and decision to submit the manuscript for publication.
PRIOR PRESENTATION
Topline findings from these analyses were presented at the Society for Gynecologic Oncology (SGO) 2021 virtual meeting, March 19–26, 2021.
SUPPORT
Supported by ARCAGY Research, AstraZeneca, Merck Sharp & Dohme LLC (a subsidiary of Merck & Co Inc, Rahway, NJ), and F. Hoffmann-La Roche.
CLINICAL TRIAL INFORMATION
DATA SHARING STATEMENT
Data underlying the findings described in this manuscript may be obtained in accordance with AstraZeneca's data-sharing policy described at https://astrazenecagrouptrials.pharmacm.com/ST/Submission/Disclosure. Data for studies directly listed on Vivli can be requested through Vivli at www.vivli.org. Data for studies not listed on Vivli could be requested through Vivli at https://vivli.org/members/enquiries-about-studies-not-listed-on-the-vivli-platform/. AstraZeneca Vivli member page is also available outlining further details: https://vivli.org/ourmember/astrazeneca/.
AUTHOR CONTRIBUTIONS
Conception and design: Eric Pujade-Lauraine, Jessica Brown, Alan Barnicle, Andreas du Bois, Isabelle Ray-Coquard
Administrative support: Cristina Martín Lorente
Provision of study materials or patients: Andreas du Bois, Ignacio Romero, Edgar Petru, Ignace Vergote, Sakari Hietanen, Magali Provansal, Barbara Schmalfeldt, Sandro Pignata, Cristina Martín Lorente, Dominique Berton, Ingo B. Runnebaum, Isabelle Ray-Coquard
Collection and assembly of data: Jessica Brown, Alan Barnicle, Pierre Lao-Sirieix, Steven W. Criscione, Andreas du Bois, Domenica Lorusso, Ignacio Romero, Edgar Petru, Hiroyuki Yoshida, Ignace Vergote, Sakari Hietanen, Magali Provansal, Barbara Schmalfeldt, Ingo B. Runnebaum, Isabelle Ray-Coquard
Data analysis and interpretation: Eric Pujade-Lauraine, Jessica Brown, Alan Barnicle, Jonathan Wessen, Pierre Lao-Sirieix, Steven W. Criscione, Ignace Vergote, Nicoletta Colombo, Barbara Schmalfeldt, Sandro Pignata, Cristina Martín Lorente, Dominique Berton, Isabelle Ray-Coquard
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Eric Pujade-Lauraine
Employment: Arcagy-Gineco
Honoraria: AstraZeneca, GlaxoSmithKline
Consulting or Advisory Role: AstraZeneca, Roche, Merck, Incyte
Research Funding: AstraZeneca (Inst)
Other Relationship: Arcagy-Gineco
Jessica Brown
Employment: AstraZeneca
Stock and Other Ownership Interests: AstraZeneca
Alan Barnicle
Employment: AstraZeneca
Stock and Other Ownership Interests: AstraZeneca
Patents, Royalties, Other Intellectual Property: Patent pending for the early stage project with the current employer
Travel, Accommodations, Expenses: AstraZeneca
Jonathan Wessen
Employment: AstraZeneca
Stock and Other Ownership Interests: AstraZeneca
Travel, Accommodations, Expenses: AstraZeneca
Pierre Lao-Sirieix
Employment: AstraZeneca (I), Autolus
Stock and Other Ownership Interests: AstraZeneca (I), Autolus
Patents, Royalties, Other Intellectual Property: Named author on the patent licensed to Medtronic. Not received any payments yet
Steven W. Criscione
Employment: AstraZeneca/MedImmune
Stock and Other Ownership Interests: AstraZeneca/MedImmune
Andreas du Bois
Consulting or Advisory Role: AstraZeneca, Roche/Genentech, Clovis Oncology, GlaxoSmithKline/Tesaro, Mersana, Zodiac Pharma, Amgen, Novartis
Domenica Lorusso
Consulting or Advisory Role: PharmaMar, AstraZeneca, Clovis Oncology, GlaxoSmithKline, MSD, Genmab, Amgen, Seattle Genetics, Immunogen, Merck Serono, Oncoinvent, Corcept Therapeutics, Sutro Biopharma
Speakers' Bureau: AstraZeneca, Clovis Oncology, GlaxoSmithKline, MSD, PharmaMar
Research Funding: PharmaMar (Inst), Clovis Oncology (Inst), GlaxoSmithKline (Inst), MSD (Inst), AstraZeneca (Inst), Genmab (Inst), Seattle Genetics (Inst), Immunogen (Inst), Incyte (Inst), Novartis (Inst), Roche (Inst)
Travel, Accommodations, Expenses: Roche, PharmaMar, AstraZeneca, Clovis Oncology, GlaxoSmithKline
Uncompensated Relationships: Gynecological Cancer InterGroup
Ignacio Romero
Consulting or Advisory Role: Clovis Oncology, GlaxoSmithKline, AstraZeneca Spain, Roche
Speakers' Bureau: Roche, AstraZeneca, PharmaMar, GlaxoSmithKline
Research Funding: Roche (Inst), GlaxoSmithKline (Inst)
Travel, Accommodations, Expenses: PharmaMar, AstraZeneca Spain, GlaxoSmithKline, Clovis Oncology, Roche
Edgar Petru
Honoraria: Roche, AstraZeneca, Daiichi Sankyo/Astra Zeneca
Consulting or Advisory Role: Roche, Daiichi Sankyo/Astra Zeneca
Travel, Accommodations, Expenses: Roche, AstraZeneca/Daiichi Sankyo
Ignace Vergote
Consulting or Advisory Role: AstraZeneca, Elevar Therapeutics, Genmab, Immunogen, Jazz Pharmaceuticals, Mersana, MSD, Novocure, Sotio, Verastem, Zentalis, Roche, Agenus, Eisai, Novartis, Seattle Genetics, Akeso Biopharma, Bristol Myers Squibb, Deciphera, Exelixis, GlaxoSmithKline, Karyopharm Therapeutics, Oncoinvent, OncXerna, Regeneron, Sanofi
Research Funding: Roche (Inst), Amgen (Inst), Oncoinvent (Inst)
Travel, Accommodations, Expenses: Karyopharm Therapeutics, Genmab, Novocure
Nicoletta Colombo
Employment: Sarepta Therapeutics (I)
Honoraria: Roche/Genentech, AstraZeneca, GlaxoSmithKline, MSD Oncology, Clovis Oncology, Pfizer, Amgen, Immunogen, Novartis, Pfizer, Mersana, Eisai, Advaxis, Nuvation Bio
Consulting or Advisory Role: Roche/Genentech, AstraZeneca, Clovis Oncology, Pfizer, MSD Oncology, GlaxoSmithKline, Immunogen, Pfizer, Mersana, Eisai, Advaxis, Nuvation Bio
Sakari Hietanen
Consulting or Advisory Role: GSK, AstraZeneca, MSD
Speakers' Bureau: AstraZeneca, GSK
Barbara Schmalfeldt
Honoraria: Roche, Clovis Oncology, GlaxoSmithKline, AstraZeneca, MSD Oncology
Consulting or Advisory Role: AstraZeneca, GlaxoSmithKline, Clovis Oncology, MSD Oncology, Roche Pharma AG
Research Funding: AstraZeneca, Roche Pharma, GSK, MSD, Clovis Oncology (Inst)
Travel, Accommodations, Expenses: Roche, AstraZeneca, GSK, MSD
Sandro Pignata
Honoraria: AstraZeneca, Roche, PharmaMar, Tesaro, Pfizer, MSD
Consulting or Advisory Role: AstraZeneca, Roche, PharmaMar, Pfizer, Tesaro, Clovis Oncology
Research Funding: Roche (Inst), AstraZeneca (Inst), MSD (Inst), Pfizer (Inst)
Cristina Martín Lorente
Consulting or Advisory Role: AstraZeneca, GlaxoSmithKline, MSD Oncology, Clovis Oncology
Speakers' Bureau: GlaxoSmithKline, AstraZeneca, MSD Oncology
Travel, Accommodations, Expenses: GlaxoSmithKline, AstraZeneca, MSD Oncology, Clovis Oncology
Ingo B. Runnebaum
Consulting or Advisory Role: GlaxoSmithKline
Isabelle Ray-Coquard
Honoraria: Roche, PharmaMar, AstraZeneca, Clovis Oncology, Tesaro, MSD Oncology, Genmab, AbbVie, Pfizer, Bristol Myers Squibb, GlaxoSmithKline, Deciphera, Mersana, Amgen, Advaxis, OxOnc, Seattle Genetics, MacroGenics, Agenus, Sutro Biopharma, Novartis, Daiichi Sankyo
Consulting or Advisory Role: Pfizer, AbbVie, Genmab, Roche, AstraZeneca, Tesaro, Clovis Oncology, PharmaMar, MSD Oncology, Bristol Myers Squibb, Deciphera, Mersana, GlaxoSmithKline, Agenus, MacroGenics, Seattle Genetics, BMS, Novartis, Novocure, Ose pharma, Daichi
Research Funding: MSD Oncology, BMS, Roche/Genentech (Inst)
Travel, Accommodations, Expenses: Roche, AstraZeneca, Tesaro, PharmaMar, GlaxoSmithKline, Clovis Oncology, BMS, Advaxis
Uncompensated Relationships: Arcagy-Gineco, French National Cancer Institute (INCA), Italian Health Authorities, German Health Authorities, Belgium Health Authorities
No other potential conflicts of interest were reported.
REFERENCES
- 1.Ledermann JA, Raja FA, Fotopoulou C, et al. : Newly diagnosed and relapsed epithelial ovarian carcinoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 24:vi24-vi32, 2013. (suppl 6) [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, et al. : Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394-424, 2018 [DOI] [PubMed] [Google Scholar]
- 3.O'Connor MJ: Targeting the DNA damage response in cancer. Mol Cell 60:547-560, 2015 [DOI] [PubMed] [Google Scholar]
- 4.Ray-Coquard I, Pautier P, Pignata S, et al. : Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med 381:2416-2428, 2019 [DOI] [PubMed] [Google Scholar]
- 5.González-Martín A, Pothuri B, Vergote I, et al. : Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med 381:2391-2402, 2019 [DOI] [PubMed] [Google Scholar]
- 6.Coleman RL, Fleming GF, Brady MF, et al. : Veliparib with first-line chemotherapy and as maintenance therapy in ovarian cancer. N Engl J Med 381:2403-2415, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCabe N, Turner NC, Lord CJ, et al. : Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res 66:8109-8115, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Hodgson DR, Dougherty BA, Lai Z, et al. : Candidate biomarkers of PARP inhibitor sensitivity in ovarian cancer beyond the BRCA genes. Br J Cancer 119:1401-1409, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mirza MR, Feng B, Shan M, et al. : Elucidation of PARP inhibitor activity in BRCAwt recurrent ovarian cancer by HRR mutational gene profile analysis. Poster presented at ASCO 2019, Chicago, IL, May 31-June 4, 2019
- 10.Coleman RL, Oza AM, Lorusso D, et al. : Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 390:1949-1961, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maxwell KN, Wubbenhorst B, Wenz BM, et al. : BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nat Commun 8:319, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mirza MR, Monk BJ, Herrstedt J, et al. : Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med 375:2154-2164, 2016 [DOI] [PubMed] [Google Scholar]
- 13.O'Malley DM, Coleman RL, Oza AM, et al. : Results from the phase 3 study ARIEL3: Mutations in non-BRCA homologous recombination repair genes confer sensitivity to maintenance treatment with the PARP inhibitor rucaparib in patients with recurrent platinum-sensitive high-grade ovarian carcinoma. Mol Cancer Ther 17, 2018. (1 suppl; abstr LB-A12) [Google Scholar]
- 14.The Cancer Genome Atlas Research Network : Integrated genomic analyses of ovarian carcinoma. Nature 474:609-615, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunningham JM, Cicek MS, Larson NB, et al. : Clinical characteristics of ovarian cancer classified by BRCA1, BRCA2, and RAD51C status. Sci Rep 4:4026, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rempel E, Kluck K, Beck S, et al. : Pan-cancer analysis of genomic scar patterns caused by homologous repair deficiency (HRD). NPJ Precis Oncol 6:36, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondrashova O, Topp M, Nesic K, et al. : Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat Commun 9:3970, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swisher EM, Kwan TT, Oza AM, et al. : Molecular and clinical determinants of response and resistance to rucaparib for recurrent ovarian cancer treatment in ARIEL2 (Parts 1 and 2). Nat Commun 12:2487, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elazezy M, Prieske K, Kluwe L, et al. : BRCA1 promoter hypermethylation on circulating tumor DNA correlates with improved survival of patients with ovarian cancer. Mol Oncol 15:3615-3625, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel JN, Braicu I, Timms KM, et al. : Characterisation of homologous recombination deficiency in paired primary and recurrent high-grade serous ovarian cancer. Br J Cancer 119:1060-1066, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swisher EM, Lin KK, Oza AM, et al. : Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): An international, multicentre, open-label, phase 2 trial. Lancet Oncol 18:75-87, 2017 [DOI] [PubMed] [Google Scholar]
- 22.Dedes KJ, Wetterskog D, Mendes-Pereira AM, et al. : PTEN deficiency in endometrioid endometrial adenocarcinomas predicts sensitivity to PARP inhibitors. Sci Transl Med 2:53ra75, 2010 [DOI] [PubMed] [Google Scholar]
- 23.Philip CA, Laskov I, Beauchamp MC, et al. : Inhibition of PI3K-AKT-mTOR pathway sensitizes endometrial cancer cell lines to PARP inhibitors. BMC Cancer 17:638, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McEllin B, Camacho CV, Mukherjee B, et al. : PTEN loss compromises homologous recombination repair in astrocytes: Implications for glioblastoma therapy with temozolomide or poly(ADP-ribose) polymerase inhibitors. Cancer Res 70:5457-5464, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pennington KP, Walsh T, Harrell MI, et al. : Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res 20:764-775, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martins FC, de Santiago I, Trinh A, et al. : Combined image and genomic analysis of high-grade serous ovarian cancer reveals PTEN loss as a common driver event and prognostic classifier. Genome Biol 15:526, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martins FC, Couturier DL, Paterson A, et al. : Clinical and pathological associations of PTEN expression in ovarian cancer: A multicentre study from the Ovarian Tumour Tissue Analysis Consortium. Br J Cancer 123:793-802, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondrashova O, Nguyen M, Shield-Artin K, et al. : Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov 7:984-998, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurley RM, McGehee CD, Nesic K, et al. : Characterization of a RAD51C-silenced high-grade serous ovarian cancer model during development of PARP inhibitor resistance. NAR Cancer 3:zcab028, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore K, Colombo N, Scambia G, et al. : Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med 379:2495-2505, 2018 [DOI] [PubMed] [Google Scholar]
- 31.Vergote I, Gonzalez-Martin A, Ray-Coquard I, et al. : European experts consensus: BRCA/homologous recombination deficiency testing in first-line ovarian cancer. Ann Oncol 33:276-287, 2022 [DOI] [PubMed] [Google Scholar]
- 32.Poti A, Gyergyak H, Nemeth E, et al. : Correlation of homologous recombination deficiency induced mutational signatures with sensitivity to PARP inhibitors and cytotoxic agents. Genome Biol 20:240, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suszynska M, Ratajska M, Kozlowski P: BRIP1, RAD51C, and RAD51D mutations are associated with high susceptibility to ovarian cancer: Mutation prevalence and precise risk estimates based on a pooled analysis of ∼30, 000 cases. J Ovarian Res 13:50, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data underlying the findings described in this manuscript may be obtained in accordance with AstraZeneca's data-sharing policy described at https://astrazenecagrouptrials.pharmacm.com/ST/Submission/Disclosure. Data for studies directly listed on Vivli can be requested through Vivli at www.vivli.org. Data for studies not listed on Vivli could be requested through Vivli at https://vivli.org/members/enquiries-about-studies-not-listed-on-the-vivli-platform/. AstraZeneca Vivli member page is also available outlining further details: https://vivli.org/ourmember/astrazeneca/.