Abstract
Purpose:
Nuclear weapons testing in the northern Marshall Islands between 1946 and 1958 resulted in ionizing radiation (IR) exposure of the thousands of Marshallese. Furthermore, numerous islands were contaminated by radioactive fallout. Significant increases in cancer and metabolic syndrome incidences have been reported among Marshallese, and potential for further increases looms due to the latency of radiation-induced health effects. The purpose of this study was to investigate the genetic and epigenetic effects of exposure to IR that could be associated with radiation-induced disease among the Northwest Arkansas (NWA) Marshallese.
Materials and Methods:
We performed analysis of chromosomal aberrations and DNA methylation based on residential and exposure history of NWA Marshallese.
Results:
Analysis of chromosomal aberrations demonstrated higher incidence of genetic rearrangements in women with self-reported history of radiation exposure (95% CI: 0.10, 1.22; p=.022). Further clustering of study participants based on their residential history demonstrated that participants who spent substantial amounts of time (≥ 6 months) in the northern atolls (thus, in the proximity of nuclear tests) before 1980 had more chromosomal aberrations than their peers who lived only in the southern atolls (95% CI: 0.08, −0.95; p=.021), and that this difference was driven by women. A relationship between the time spent in the northern atolls and increase in chromosomal aberrations was observed: 0.31 increase in chromosomal aberrations for every 10 years spent at northern atolls (95% CI: 0.06, 0.57; p=.020). Finally, significant inverse correlations between the chromosomal aberrations and the extent of DNA methylation of four LINE-1 elements L1PA2, L1PA16, L1PREC1, and L1P4B were identified.
Conclusions:
The results of this study provide first evidence of the presence of stable genetic and epigenetic rearrangements in peripheral lymphocytes of NWA Marshallese and warrant further studies to analyze the role of radiation exposure in health disparities experienced by this Pacific Island nation.
Keywords: chromosomal aberrations, DNA methylation, health disparities, ionizing radiation, LINE-1, Marshallese
Introduction
Nuclear weapons testing in the northern Marshall Islands between 1946 and 1958 resulted in ionizing radiation (IR) exposure of the thousands of Marshallese (Land et al. 2010). Furthermore, numerous islands were contaminated by radioactive fallout. Significant increases in cancer and metabolic syndrome incidences have been reported among Marshallese, and potential for further increases looms due to the latency of radiation-induced health effects (Land et al. 2010; Simon and Bouville 2010; Simon et al. 2010). Increased cancer rates and the development of metabolic syndrome in this population of Marshallese are related to acute exposures from the radioactive fallout, premature resettlement to some of the still contaminated atolls, and long-term consumption of contaminated locally grown food (Robison and Stone 1992; Robison et al. 2006; Robison and Hamilton 2010; Simon et al. 2010; Bordner et al. 2016). Indeed, residual radioactivity in the environment led to chronic intake of fallout radionuclides for many years after their deposition occurred. Of particular importance, soil and ground waters were contaminated with 137Cs, which has a half-life of 30.1 years (the period during which half of the radioactivity is lost) and which comprises 95–99% of total radioactivity associated with nuclear tests performed in the Marshall Islands (Simon et al. 2010; Bordner et al. 2016). Soil of the atolls in the Marshall Islands is characterized by poor levels of potassium, a natural competitor for 137Cs, resulting in much higher than expected root uptake of the latter (Robison and Hamilton 2010). Radioecological remediation programs to eliminate 137Cs from the soil were not introduced until the 1980s, lengthening the period of exposure. Furthermore, the most recent studies report that a number of locations in northern Marshall Islands still experience levels of radioactive plutonium in the soil above the permissible levels set by a memorandum of understanding between the United States and the Government of the RMI (Abella et al. 2019). Therefore, many RMI residents were continuously exposed to IR via not only residual external gamma radiation but also internal exposure by way of ingestion of contaminated ground water and locally sourced foods (Robison and Stone 1992; Robison et al. 2006; Robison and Hamilton 2010).
A large group of Marshallese relocated to Northwest Arkansas (NWA) in the late 1970s through early 1990s; as a result, 12,000 to 15,000 Marshallese currently reside in NWA. Multiple studies indicate health disparities in the NWA Marshallese population, such as disproportionally high levels of metabolic syndrome and diabetes, with rates of over 20% among the NWA Marshallese compared to 9.4% in Americans (Felix et al. 2017a; Felix et al. 2017b). These disparities are attributed largely to variations in genetic architecture and socioeconomic factors, such as the lack of medical insurance. (McElfish et al. 2017). To date, however, no data exist demonstrating the role of previous radiation exposures in the etiology of disease in this group of people.
Both genetic and epigenetic alterations act in concert to contribute to the development of disease, including cancer (Norppa et al. 2006; Hanahan and Weinberg 2011). Cytogenetic alterations (i.e., changes in chromosome structure/number) are a type of genetic alteration that contributes to development and progression of cancer (Hagmar et al. 1994; Norppa et al. 2006). Global DNA hypomethylation is an epigenetic alteration most frequently observed in human cancers (Hanahan and Weinberg 2011). The loss of DNA methylation primarily stems from repetitive elements (RE), such as long interspersed nuclear element 1 (LINE-1), which comprises ~20% of the human genome. LINE-1 is a retrotransposon whose activity is suppressed by DNA methylation (Miousse and Koturbash 2015; Jones 2012). Both chromosomal aberrations and loss of DNA methylation are generally accepted as “hallmarks” of cancer (Hanahan and Weinberg 2011).
Cytogenetic damage and epigenetic alterations are potently induced by IR (Aypar et al. 2014; Miousse et al. 2014; Prior et al. 2016; Tanaka et al. 2016). High levels of chromosomal aberrations were reported in the Republic of Marshall Islands (RMI) residents 10 years post-exposure to IR (Lisco and Conard 1967). Another study of Japanese fisherman who were as far as 1,200 km from the hypocenter clearly demonstrated that these cytogenetic aberrations persisted for at least six decades (Tanaka et al. 2016). Studies in experimental systems indicate that exposure to IR leads to stable alterations in LINE-1 DNA methylation in hematopoietic cells (Miousse et al. 2014), including peripheral lymphocytes (Koturbash et al. 2007). Thus, accumulating data indicate that IR-induced cytogenetic aberrations and altered LINE-1 DNA methylation are persistent by nature.
The goal of this study was to investigate the presence of persistent genetic and epigenetic alterations associated with previous IR exposure in former residents of the RMI. For this purpose, we consented 38 NWA Marshallese who were in the Marshall Islands between 1946 and 1958 – the period of nuclear weapons testing at this location. We further analyzed the levels of chromosomal aberrations and DNA methylation of REs in the plasma lymphocytes of the study participants.
Materials and Methods
Study Participants Recruitment and Enrollment
Thirty-eight NWA Marshallese study participants who were present at RMI between 1946 and 1958 (nuclear testing period) and who resettled in NWA after 1980 were recruited and consented for the study. Consent from study participants was received prior to initiation of the study. Gender and underlying health conditions were not exclusion criteria for this study as long as participants could be interviewed and provide blood specimens. A structured questionnaire was used to administer the interview by trained medical staff (the questionnaire can be found in Supplementary Document 1). The questionnaire was also translated and available in Marshallese for all participants. The interview included a residential history which included specific islands within RMI. Calendar year and important life events were used to help participants recall the location of residence. Study participant were also asked about past occupations and medical exposures to IR. A structured questionnaire was used to assess dietary patterns, lifestyle factors, and health history. Finally, all study participants were asked to provide blood samples at the North Street Clinic, UAMS NW campus, Fayetteville, AR. Peripheral blood was drawn (30 ml, into sodium heparinized vacutainers) by a professional healthcare worker. The samples were then either transferred same day to the University of Arkansas for Medical Sciences Little Rock Campus for cytogenetic analysis or stored at −80 °C conditions for further epigenetic analysis upon processing with Lymphoprep™ (Stemcell Technologies, Cambridge, MA, USA) following the manufacturer’s recommendations. All studies were reviewed and approved by the University of Arkansas for Medical Sciences institutional review board (IRB, Protocol # 207491).
Cell culture
Peripheral blood was collected in a sodium heparin vacutainer and transported to the laboratory at room temperature. Samples were cultured for 48 h with RPMI 1640 Medium (Gibco, USA) supplemented with 20% fetal bovine serum (Gibco, USA) and 0.1% phytohemaglutinin (PHA, Gibco, USA) at 37 °C in a 5% CO2 humidified incubator.
Metaphase arrest and chromosome preparation
For arresting cells at the metaphase stage of the cell cycle, colcemid (Gibco, USA) at a final concentration of 0.25 μg/ml was added for 20 minutes before harvest. Cells were then centrifuged at 400× g at room temperature. Supernatant was removed, and cell pellet was slowly resuspended in 37 °C pre-warmed hypotonic solution (75 mM KCl) and incubated for 10 min. After hypotonic treatment, cells were fixed by gently adding 500 μL of acetomethanol fixative (5:2 Methanol:Acetic acid) with gentle and thorough agitation. Cells were then centrifuged for at 1000 rpm (10 min) and the formed cell pellet was resuspended carefully in 2 mL of supernatant. This was followed by addition of fixative in a dropwise manner (total volume 4 mL) with constant agitation. After that, additional 4 mL of fixative were added and the cells were kept at room temperature (20 min). Two additional washes were performed, after which the cells were dropped onto pre-cleaned glass slides.
G-banding
In order to prepare G-banded chromosomes, the trypsin–Giemsa staining was used. Briefly, the slides were baked at 66 °C (12 h) followed by treatment with 0.025% trypsin (1 min), and then rinsed with Tyrode’s buffer (Sigma, St. Louis, MO, USA), and stained with Giemsa (Sigma) (5 min). Using the standard G-banding technique, karyotype integrity was determined according to the International System for Human Cytogenetic Nomenclature (ISCN). Images were acquired using Zeiss Imager.Z2 microscope, and karyotyped using the GenASIs Case Data Manager system, version 7.2.2.40970 © ASI 2013 application. Up to 20 cells from each group were photographed and analyzed.
Locus-specific FISH
Slides were aged at room temperature overnight and soaked in 2×SSC at room temperature (5 min), then dehydrated in room temperature with graded ethanol series—70%, 80%, and 100% respectively. Slides were then denatured with pre-warmed (71 °C) denaturation solution for 1 min. Slides were placed directly into a cold graded ethanol series (70%, 85%, and 100% respectively) for approximately 2 minutes each. Slides remained in the 100% ethanol solution for 1 minute prior to probe application, and then removed and blown dry to prevent uneven or inadequate drying. FISH probe aliquots were denatured in micro centrifuge tubes, secured into a floatation device, and placed into a water bath heated to 75 °C (5 min). Following slide denaturation, 10 μL of appropriate denatured probe mixture was applied to the slide, covered with a 22 × 22 mm coverslip and sealed with rubber cement. Slides were placed in a humidified chamber box, and allowed to hybridize at 37 °C (12 h) protected from light. Slides were then washed at 45 °C with 50% formamide/2X SSC followed by 2×SSC. After, the slides were dipped in distilled water, immediately blown and counterstained with DAPI.
Image acquisition
A Zeiss Imager.Z2 microscope (Gottingen, Germany) at 100× magnification along with GenASIs software (Applied Spectral Imaging, Carlsbad, CA, USA) was used to acquire and analyze images of hybridized metaphase spreads.
Repetitive elements DNA methylation analysis
DNA was isolated from frozen lymphocytes using the AllPrep micro kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. The REs families’ consensus sequences were obtained from the Genetic Information Research Institute (GIRI) Database: http://www.girinst.org/. Thirty two REs were identified for this study: 17 LINE-1 elements, 8 Long Terminal Repeats (LTR)/Human Endogenous Retroviruses (HERV), 3 Transposons, 3 REs associated with centromeric satellite DNA, and 1 Short Interspersed Nucleotide Element (SINE/Alu). The list of REs as well as forward and reverse primers are provided in Supplementary Table 1. The 5’-UTRs of the REs were analyzed using NEBcutter® (http://nc2.neb.com/NEBcutter2/) and the most frequent CpG sites that can are cleaved by the methylation-sensitive restriction enzymes (AciI, BstUI, HhaI, HpaII, and SmaI) were identified. Analysis of DNA methylation was performed as previously described (Prior et al. 2016). The change in methylation was calculated using the following equation:
The “control” group consisted of individuals from the Southern Atolls, who self-identified as non-exposed, and, arbitrarily, men.
Statistical analyses
We used means and standard deviations (SDs) to summarize quantitative characteristics of the participants and percentages for categorical characteristics. When evaluating the extent to which radiation exposure-related variables related to chromosomal aberrations and DNA methylation, we started with models that included gender, the exposure-related variable, and the interaction of the two; all models accounted for whether the participant was an ever-smoker or never-smoker. In the absence of any gender × exposure interaction effect (at the p < .30 level due to the small sample size), we dropped the interaction term leaving only gender, the exposure-related variable, and ever-smoker indicator. We left gender in the model regardless of its contribution so as to provide estimated gender effects. We considered three exposure-related variables: history of exposure (yes/no); substantial amounts of time spent in the northern atolls (yes/no); and time spent in the northern atolls. For the two dichotomous variables, we used analysis of variance (ANOVA) models; for the continuous variable, we used regression models. Beyond these analyses, we also estimated correlations of DNA methylation with chromosomal aberrations.
We report key results with 95% confidence intervals. Because we investigated many outcomes (e.g., there were 32 elements for which we measured DNA methylation), we estimated the positive False Discovery Rate (pFDR) for the significant results we found (Storey, 2003). Linear models and correlation analyses were conducted in SAS/STAT software, version 9.4 (SAS Institute). The pFDRs were estimated with custom code in R version 4.1.0. Figure were produced in R version 4.1.0 and GraphPad Prism.
Results
Characteristics of the study population
Out of 38 consented study participants, 37 study participants answered a structured questionnaire, and 36 study participants provided blood samples. The characteristics of the study population are provided in Table 1. The cohort was composed of 13 men and 24 women , with mean age at the time of consent of 69.1±5.0 years in men and 67.3±4.7 years in women. 13 of 37 study participants (35%) were categorized into the “self-reported exposure” cohort, comprising those who either spent more than 4 weeks at one of the 4 most contaminated atolls (Bikini, Rongelap, Enewetak, and Utrik), and/or with a history of radiation exposure, (i.e., being part of the DOE, 177, or 4 Atoll programs, and/or were said to be present or living in the zone of radiation contamination or high background radiation, and/or to be exposed to, and/or who received treatment against radiation poisoning). The rest of the study participants were categorized into the “without self-reported exposure” cohort.
Table 1.
Characteristics of the study population.
| Characteristic | Men | Women | Overall |
|---|---|---|---|
| Number of participants | 13 | 24 | 37 |
| Age | 69.1 ± 5.0 | 67.3 ± 4.7 | |
| Self-reported Exposure Status | |||
| Self-reported exposure | 4 (30.8%) | 9 (37.5%) | 13 |
| Study participant was present on Bikini, Enewetak, Utrik (Utrok, Utirik), or Rongelap atolls after 1946 for 4 weeks or more |
2 (15.4%) |
4 (16.7%) |
6 |
| Study participant was part of the DOE, 177, or 4 Atolls Programs | 1 (7.6%) | 4 (16.7%) | 5 |
| Study participant was indicated by a physician to have been present or living in the zone of radiation contamination or high background radiation |
2 (15.4%) |
5 (20.8%) |
7 |
| Study participant was indicated by a physician to have been exposed to radiation | 0 (0%) | 3 (12.5%) | 3 |
| Study participant was indicated by a physician to have been experiencing the symptoms of radiation sickness | 1a (7.6%) | 3 (12.5%) | 4 |
| Study participant received any treatment for radiation poisoning | 1 (7.6%) | 3 (12.5%) | 4 |
| Without self-reported exposure (answered “no” to all above questions) | 9 (69.2%) | 15 (62.5%) | 24 |
| Local produce consumption | 13 (100%) | 24 (100%) | 37 |
| Breadfruit | 11 (84.6%) | 12 (50%) | 23 |
| Pandanus | 9 (69.2%) | 8 (33.3%) | 17 |
| Coconut | 7 (53.8%) | 6 (25%) | 13 |
| Other (taro, bananas, papaya) | 8 (61.5%) | 8 (33.3%) | 16 |
| Items not specified | 2 (15.4%) | 12 (50%) | 14 |
| Fish/seafood | 13 (100%) | 22 (91.7%) | 35 |
| Imported canned/processed foods | 3 (23.1%) | 15 (62.5%) | 18 |
| Water | |||
| Rain/fresh water | 13 (100%) | 24 (100%) | 37 |
| Well water | 6 (46.2%) | 21 (87.5%) | 27 |
| Coconut water/milk | 11 (84.6%) | 18 (75%) | 29 |
| Tobacco usage | |||
| Never smoked | 9 (69.2%) | 22 (91.7%) | 31 |
| Quit | 4 (30.8%) | 1 (4.15%) | 5 |
| Life-time smoker | 0 (0%) | 1 (4.15%) | 1 |
| Among “Self-reported exposure” | |||
| Never smoked | 2 (15.4%) | 7 (29.2%) | 9 |
| Quit | 2 (15.4%) | 0 (0%) | 2 |
| Life-time smoker | 0 (0%) | 1 (4.2%) | 1 |
| Among “Without self-reported exposure” | |||
| Never smoked | 7 (53.8%) | 15 (62.5%) | 22 |
| Quit | 2 (15.4%) | 1 (4.15%) | 3 |
| Life-time smoker | 0 (0%) | 0 (0%) | 0 |
| Cancer history (therapy, yes/no) | 1 (7.6%) / no | 0 (0%) | 1 |
| “Self-reported exposure” | 1 (7.6%) | 0 (0%) | 1 |
| “Without self-reported exposure” | 0 (0%) | 0 (0%0 | 0 |
Notes:
Study participant could not provide a definite answer on the question (response was “maybe”).
As the Marshallese Islands represent a remote location, residents were mostly dependent on locally grown produce and locally caught seafood, especially in the middle of the 20th century. The answers provided for the offered questionnaire have further confirmed the following: 100% of recruited participants reported consumption of locally grown produce, 100% of men and 92% of women reported consumption of fish and/or seafood. At the same time, only 23% of men and 65% of women reported consumption of imported canned/processed foods. As for water resources, 46% of men and 88% of women reported consumption of well water while at the Marshall Islands. Additionally, 85% of men and 75% of women reported consumption of coconut milk or coconut water as another source of drinking water (Table 1). All these findings suggest high potential of chronic low-dose internal radiation exposure associated with consumption of contaminated local foods and drinking water.
Analysis of chromosomal aberrations in “self-reported exposure” vs “without self-reported exposure” participants
Increasing evidence indicates that genetic and epigenetic alterations in concert contribute to disease development. For instance, both genetic and epigenetic alterations are considered hallmarks of cancer (Norppa et al. 2006; Hanahan and Weinberg 2011). Cytogenetic alterations (i.e., changes in chromosome structure) are structural permutations understood to be the result of improper rejoining of DNA double-strand breaks induced by oxidative stress factors--including IR. Importantly, IR-induced chromosomal aberrations can persist decades after the initial radiation exposure. Therefore, we then sought to analyze the presence of chromosomal aberrations in peripheral lymphocytes of NWA Marshallese study participants.
There were two exceptional participants (1 male, 1 female) with a high frequency of aberrations (>3). One of them, a 67-year-old man, did not report exposure to radiation, but both of his parents were suffering from “thyroid disease” (as reported by the participant himself). Given a spike in thyroid cancers in the Marshallese population in the 1970s (Takahashi et al. 2003), it is plausible to hypothesize that exposure to IR was a major etiological factor. The second exceptional participant was a woman with life-long smoking history. Smoking is a known genotoxic stressor and is a strong inducer of chromosomal aberrations. Therefore, those two study participants were excluded from the analysis presented below; however, the results that include all study participants are presented in the Supplementary Materials to allow the reader access to all data.
Chromosomal aberrations were investigated using the standard G-banding technique with further visualization of specific loci using the FISH technology (Figure 1). From the initial analysis, comparisons of study participants with “self-reported exposure” vs “without self-reported exposure” depended on the participant’s gender (interaction p=.015). The mean aberration rate in women without self-reported exposure was 1.88, whereas the mean for those with self-reported exposure was 0.66 higher (95% CI: 0.10, 1.22; p=.022). For men, those without self-reported exposure had an aberration rate of 2.10 while the rate in the self-reported exposure group was 0.53 lower (95% CI: −0.24, 1.30; p=.168) (Figure 2). The aberration rates in ever-smokers was estimated to be 0.12 higher than in never-smokers (95% CI: −0.51, 0.75). The results after excluding the two exceptional participants were similar, except for the comparison of exposed vs unexposed women which was no longer significant at the 0.05 level: 0.53 higher (95% CI: −0.03, 1.08; p=.061).
Figure 1.
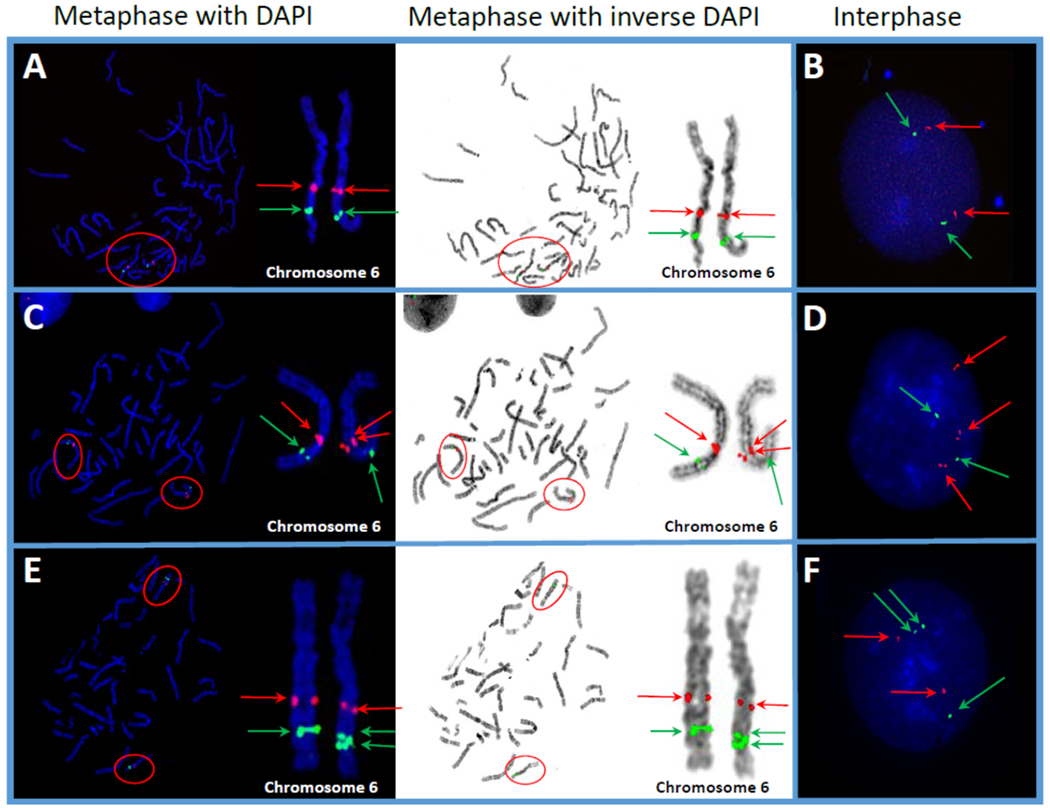
Photomicrographs representing normal and various anomalies in the long arm (q) of chromosome 6 in Marshallese study participants. A cocktail of FISH probes for two different loci,6q21 (encompassing SEC63 gene, red signals) and 6q23 (encompassing MYB gene, green signals) was used to identify cytogenetic abnormalities involving these two loci on chromosome 6. Red arrows indicate the SEC63 gene and green arrows indicate the MYB gene. Red ovals indicate human chromosome 6, featured in magnified view. (A) Metaphase spread with both DAPI and inverse DAPI, showing 2 positive hybridizations of both loci, illustrating an apparently normal signal pattern. (B) Interphase nucleus showing 2 positive hybridizations for both loci, indicating an apparently normal signal pattern. (C) Metaphase spread with both DAPI and inverse DAPI, showing positive hybridization for 3 red signals and 2 green signals, illustrating a duplication of the SEC63 gene locus. (D) Interphase nucleus also showing positive hybridization for 3 red signals and only 2 green signals, indicating a duplication of the SEC63 gene locus. (E) Metaphase spread with both DAPI and inverse DAPI showing positive hybridization for 2 red signals and 3 green signals, indicating a duplication of the MYB gene locus. (F) Interphase nucleus also showing positive hybridization for 2 red signals and 3 green signals, indicating a duplication of the MYB gene locus.
Figure 2.
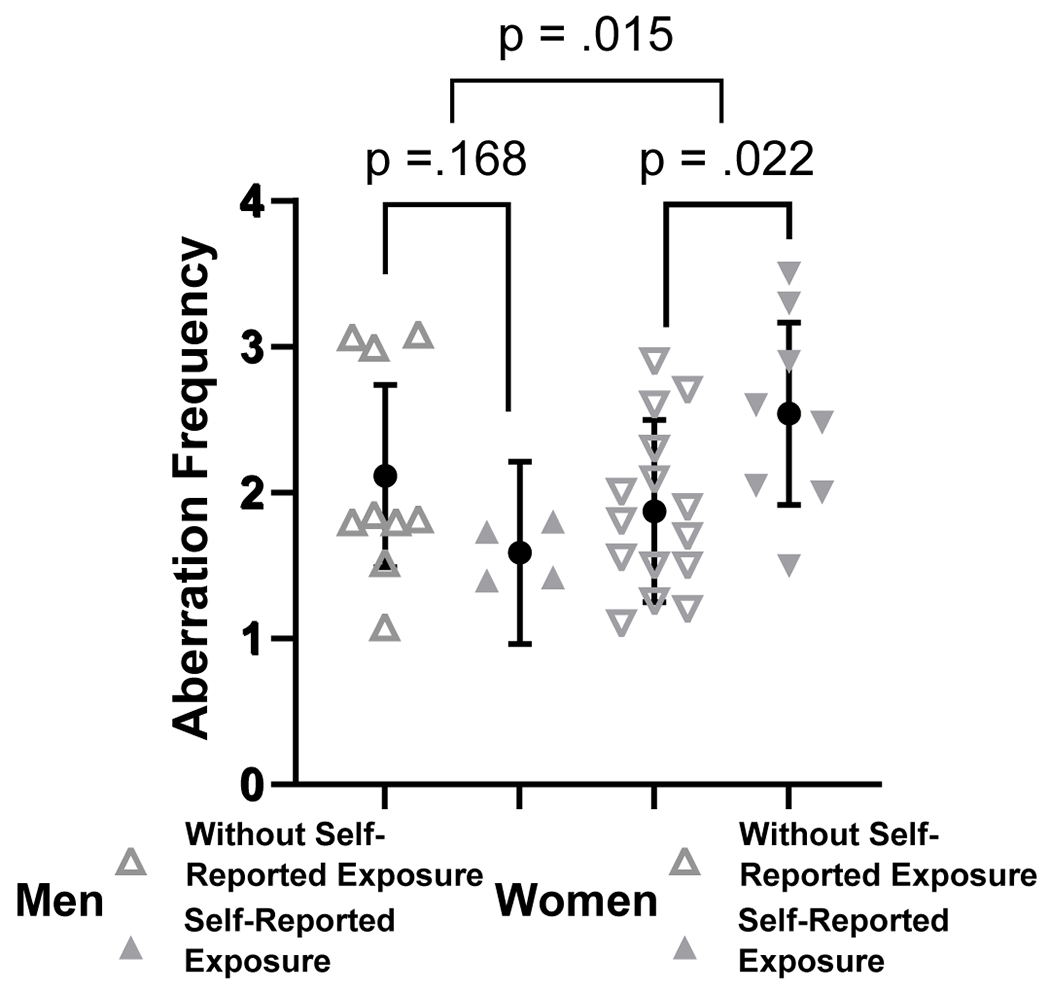
Analysis of chromosomal aberrations in peripheral lymphocytes of NWA Marshallese (“self-reported radiation exposure” vs “without self-reported radiation exposure”). Chromosomal aberration frequency was analyzed with a two-way ANOVA. Brackets above the data indicate which groups are being compared. Data are represented as mean ± pooled SD.
Analysis of chromosomal aberrations based on residential history
Because categorizing study participants into the self-reported or without self-reported groups was not substantiated by documentation, some errors could occur [i.e., one study participant could not re-collect with certainty whether the symptoms he was experiencing were considered by a physician as symptoms of radiation sickness or not (please, see Table 1)]. On the other hand, given the underestimated energy yields of some nuclear tests (i.e., Castle Bravo) and mistakes in calculation of the directions of the radioactive plume migration, we hypothesized that not everyone who was exposed was diagnosed with radiation sickness, were identified as having been exposed to IR, or received treatment against radiation poisoning. Therefore, using a detailed residential history that was available to us for every study participant, we attempted the analysis of chromosomal aberrations based on the proximity to the nuclear tests. First, we divided the Marshall Islands into northern atolls (Ujae, Lae Kwajalein, Erikub, Wotje, Mejit, and all others located north of these) and southern atolls (Lib, Namu, Maloelap, and all those located south of these) (Figure 3). Subsequently, based on the residential histories, we dichotomized participants into those who had spent at least 6 months in the northern atolls before 1980, and those who had not. Of the 25 participants without self-reported exposure, only 8 (32%) had spent at least 6 months in the northern atolls before 1980; and among the 12 participants self-reported as exposed, 9 (75%) were in the northern atolls group.
Figure 3.
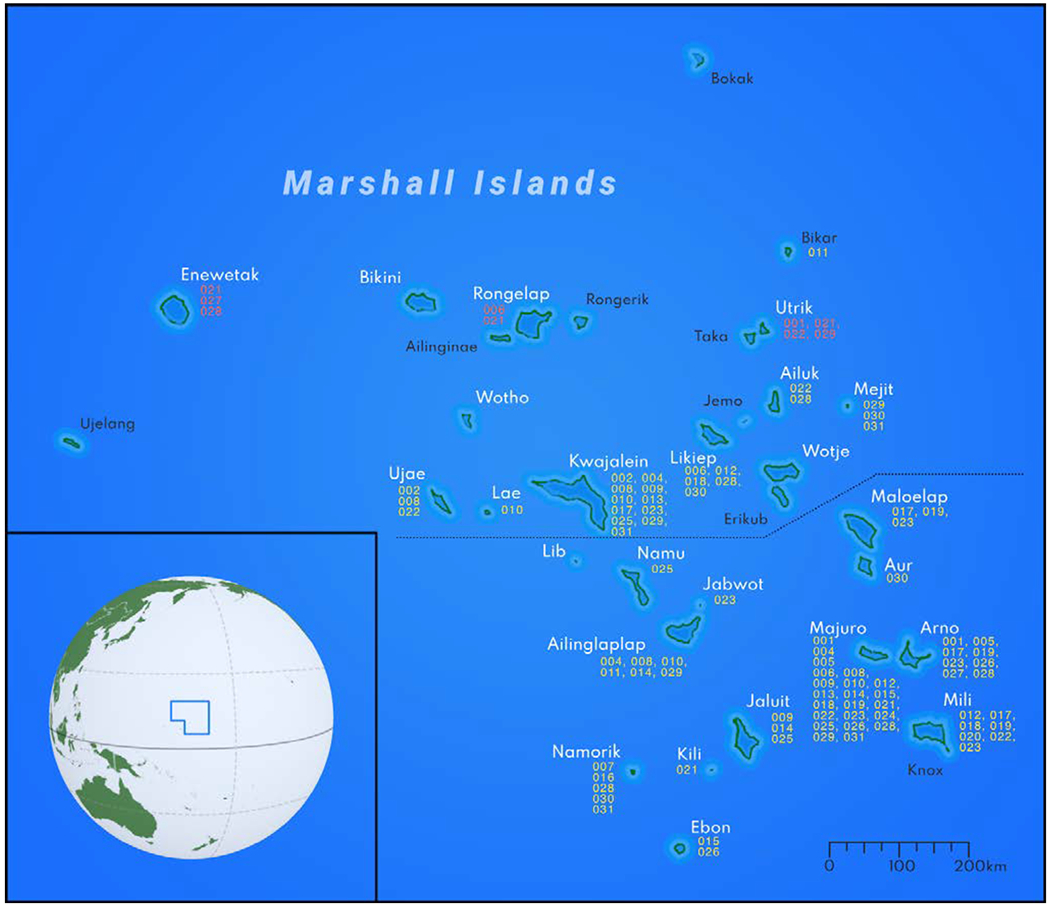
Map of Marshall Islands and residential history of the study participants.
The reasons for selecting 1980 as the cut-off year were manifold. First, major clean-up efforts to remove the topsoil (top 15 cm) on a number of atolls happened between 1977 and 1979 (Robison and Hamilton 2010). Second, programs for introduction of potassium – a natural 137Cs competitor – were initiated in the 1980s, thus significantly reducing the root uptake of the radioactive element up to 95% (Robison and Hamilton 2010). Third, the late 1970s to the early 1980s were characterized by the first large wave of emigration of Marshallese to the United States, which subsequently prevented further exposures to radioactive elements of many of the study participants.
The initial ANOVA having northern islands (yes/no), gender, their interaction, and ever-smoking as factors provided no evidence of a northern islands × gender interaction (p=.463); we thus dropped the interaction and refit the aberration data with northern islands, gender, and ever-smoking as factors (Figure 4B). Those participants who spent 6 or more months in the northern islands had an aberration rate of 2.81, while the aberration rate of participants in the southern group was 0.28 lower (95% CI: −0.76, −0.20; p=.248). Regarding gender, the aberration rates differed only by 0.10 (95% CI: −0.41, 0.60) between men and women. Ever-smokers were estimated to have 0.10 more aberrations (95% CI: −0.59, 0.78) than never-smokers. When we excluded the two exceptional participants (both of them where from the southern atolls group and the only study participants from the southern atolls with a frequency of aberrations >3), the inferences on the interaction, gender main effect, and smoking effect remained similar. Importantly, those spending time in the northern islands were estimated to have aberration frequencies that were 0.52 higher than in their peers in the southern atolls (95% CI: 0.08, 0.95; p=.021) (Figure 4A).We then regressed aberration rates on time spent in the northern atolls (Figure 5). There was no evidence that the slopes on time differed between the genders in the initial model (interaction p=.416); thus, we dropped the interaction term. In the model that accounted for time in the northern atolls while controlling for gender and smoking history, every 10 years spent in the northern atolls before 1980 corresponded to a 0.23 increase in aberration rate (95% CI: 0.06, 0.52; p=.118). Regarding gender, men and women differed very little: 0.07 (95% CI: −0.43, 0.57). Ever-smokers were estimated to have 0.15 more aberrations than never-smokers (95% CI: −0.51, 0.82). When excluding the two exceptional participants who spent no time in the northern atolls, every 10 years spent in the northern atolls corresponded to a significant 0.31 increase in aberration rate (95% CI: 0.06, 0.57; p=.020); other inferences on gender and smoking were similar.
Figure 4.
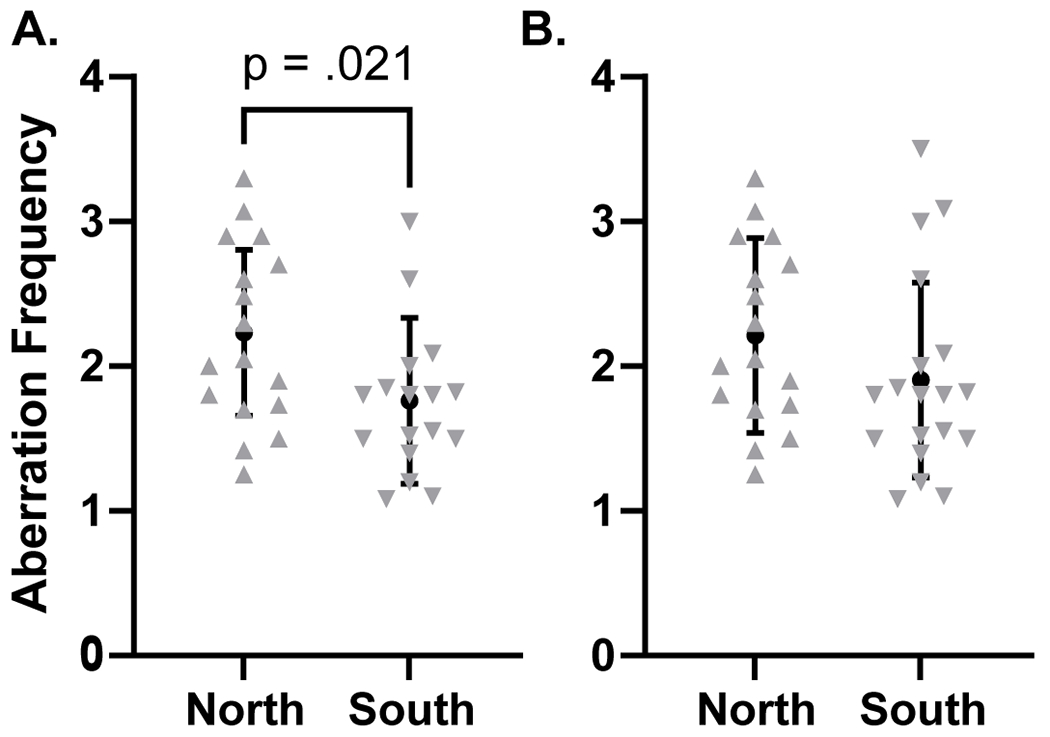
Analysis of chromosomal aberrations in peripheral lymphocytes of NWA Marshallese (“north” vs “south” scenario). Chromosomal aberration frequency was analyzed with a t-test. A compares aberration frequency between those living on the North Atoll and those living on the South Atoll, while excluding two exceptional individuals (see text for explanation). B shows the same comparison without removing the two individuals from the analysis. Data are represented as mean ± pooled SD.
Figure 5.
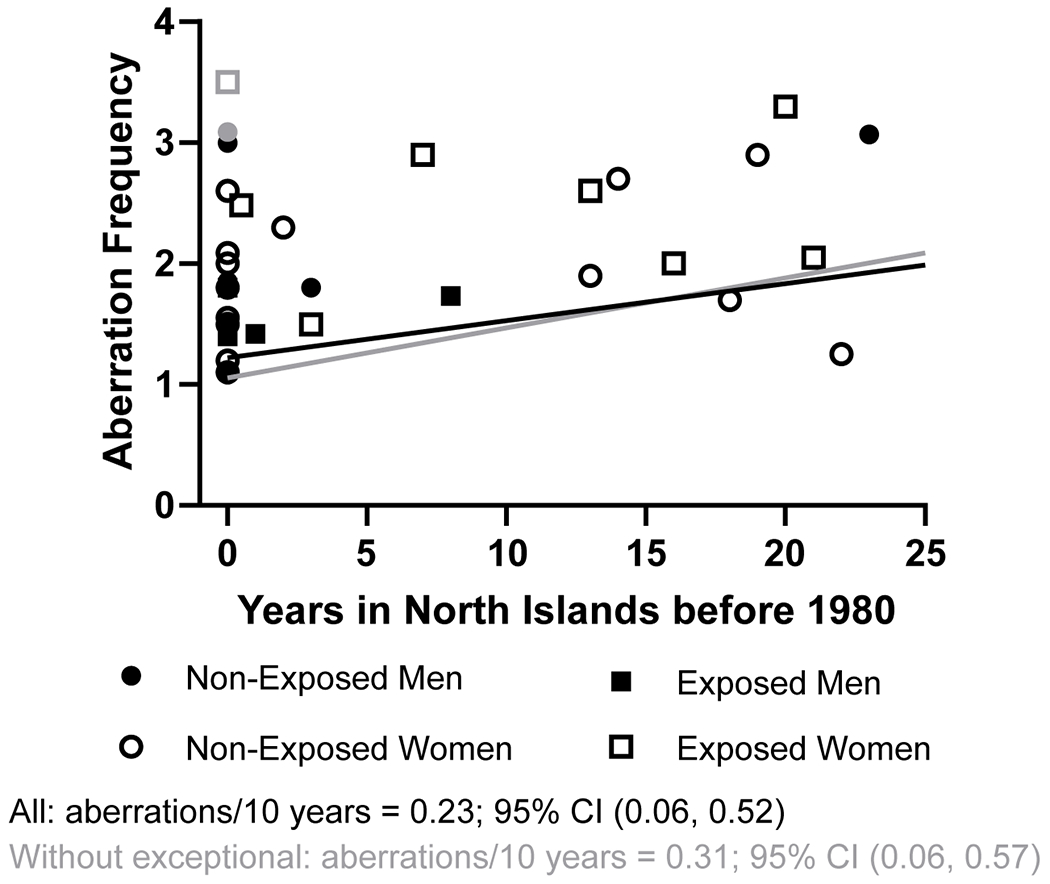
Estimation of aberration frequency per 10 years spent on the northern atolls. Symbols in gray indicate two exceptional individuals (see text for more information), and the regression line in gray is the result of excluding those from the analysis. The regression line in black includes all individuals.
DNA methylation analysis
A number of studies have previously attempted to investigate the effect of environmental exposures on DNA methylation using the REs, and LINE-1 in particular, as a substrate. Given the abundance of REs, this would provide knowledge on changes in DNA methylation on the global scale as well as could result in development of novel robust biomarkers of exposure. These studies, however, often were limited by the analysis of DNA methylation within the ORF-1 of LINE-1 elements, which, in turn are poor in CpG sites. It is known, however, that the vast majority of potential sites for LINE-1 DNA methylation are sited within the 5’-UTR.
Due to advances in computational biology, it became possible to classify LINE-1 elements based on their evolutionary age and sequences (Khan et al. 2006). This allowed us to utilize a recently developed by us assay for simultaneous analysis of DNA methylation of various LINE-1 families (Prior et al. 2016; Miousse et al. 2017).
Analysis of 32 REs revealed a significant difference between the study participants with self-reported exposure and without self-reported exposure only for L1P4e LINE-1 element, when including all individuals (2.083 vs −1.984, p = .0445) and after excluding the two exceptional individuals (2.163 vs −2.268, p = .031) (Supplementary Figure 1). When comparing exposures in men, excluding the exceptional individual, HERVK11I methylation was increased in the group of self-reported exposure men (7.420 vs 0.144, p = .014) (Supplementary Figure 1). Conversely, in women, when including all individuals, HERVK11I methylation was decreased in women with self-reported exposure (0.703 vs 7.158, p = .031) (Supplementary Figure 2). However, this difference was lost when the exceptional individual was excluded (Supplementary Figure 2).
We next compared DNA methylation status between individuals from the northern vs southern atolls. When including all participants, there were no significant differences in LINE1 methylation by location or when analyzing by location and gender (Supplementary Figure 3). However, when the two exceptional individuals were excluded, there was a significant increase in HERV11I methylation in men only (7.168 vs 0.270, p = .039) (Supplementary Figure 3).
Some studies indicate that an altered epigenetic profile may be associated with or even predispose subjects to a higher degree of DNA damage. Taking this into consideration, we next performed a correlative analysis between the levels of chromosomal aberrations and the extent of DNA methylation for every RE analyzed in this study. Interestingly, we identified 3 elements, all of which were retrotransposons LINE-1 [L1PA13; L1PA2; L1PREC1], and whose extent of DNA methylation was significantly inversely correlated with the level of chromosomal aberrations among the study participants, with Pearson r-values between −0.35 and −0.51 (three p < .032) (Figure 6A). When excluding the two exceptional participants, 4 LINE-1 retrotransposons were identified: [L1PA2; L1PA16; L1PREC1; L1P4b] (four p < .028) (Figure 6B).
Figure 6.
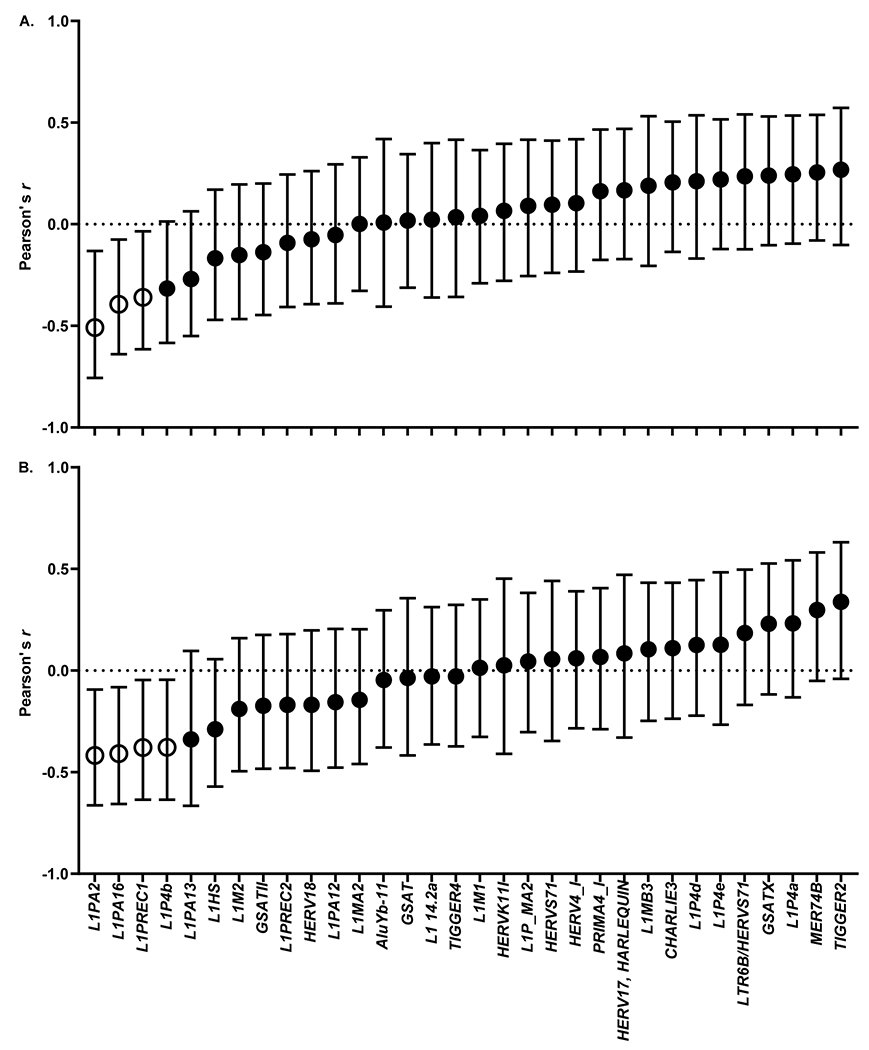
Correlation of LINE-1 DNA methylation status with chromosomal aberration frequency. (A) All individuals. (B) Excluding two exceptional individuals. Data are presented as Pearson’s r-value ± 95% CI. Open symbols indicate a confidence interval that does not include zero.
In the analyses of chromosomal aberrations and DNA methylation, 12 of the 334 tests of exposure effects were significant at the 0.05 level. The pFDR was 0.93 meaning that 11 of the 12 may be false discoveries. We also performed the same 334 tests excluding the two exceptional participants, finding 17 significant results, of which 11 may be false discoveries (pFDR = 0.68). For the correlations of DNA methylation with chromosomal aberrations, we found 3 of 32 correlations to be significant. Of these 3, 1 may be a false discovery (pFDR = 0.48). When we estimated the same correlations excluding the 2 exceptional participants, there were 4 significant correlations with possibly 1 false discovery (pFDR = 0.17).
Discussion
In our study, all participants reported consumption of locally grown food, including pandanus (69.2% of men and 33.3% of women) and coconut (53.8% of men and 25% of women) – the fruits in which high levels of 137Cs were detected on a number of atolls as late as in 2017 (Topping et al., 2019).
The evaluation of persistent damage in chromosome structure, particularly in lymphocytes of human peripheral blood, provides critical information about disease risks related to IR exposure. In these regards, two studies are of particular interest. The first is the study reporting chromosomal aberrations in blood lymphocytes of Marshallese 10 years after exposure (Lisco and Conard 1967). The second study, a cytogenetic analysis conducted in Japanese fishermen who were 420–1,200 km from the nuclear test hypocenter, demonstrated elevated frequency of stable and unstable chromosomal aberrations, relative to age-matched controls, a staggering 60 years after the exposure (Tanaka et al., 2016). Here, for the first time to our knowledge, we report the presence of stable chromosomal aberrations in NWA Marshallese decades after potential exposure(s) to IR. While our study lacked control study population, clustering study participants into self-reported exposure and without self-reported exposure groups (based on medical history) allowed us to determine significantly increased levels of chromosomal aberrations in NWA Marshallese women who reported history of exposure to IR. Importantly, analysis by clustering the study participants into “northern atolls” and “southern atolls” groups (based on residential history) confirmed these findings, demonstrating higher levels of chromosomal aberrations in former residents of northern atolls – geographically proximal to nuclear tests – and further confirmed that this difference was driven by women. It is unclear at this point as to why women demonstrated higher levels of cytogenetic damage than men, and future studies are needed using larger cohorts and genetically related control study populations (i.e., Hawaii Marshallese – a group of Marshallese that relocated to Hawaii during the WW II and, thus, did not experience exposure to IR due to nuclear tests at the Marshall Islands).
IR is a known risk factor for cancer, and damage to DNA caused by IR, exhibited in the form of stable chromosomal aberrations, is considered one of the central mechanisms in radiation-induced carcinogenesis. Therefore, it is plausible to hypothesize that stable chromosomal aberrations observed in NWA Marshallese may predispose them to higher cancer incidence. While there is no data on cancer incidence among the NWA Marshallese, studies have demonstrated that RMI residents have excess risks for cancer, particularly thyroid and hematological malignancies (Takahashi et al. 2003; Land et al. 2010; Simon et al. 2010). Importantly, a recent study reported that total cancer incidence-based mortality among women in the US-affiliated Pacific Islands were the highest at RMI with 120.3 per 100,000 inhabitants compared to Palau (107.7), Guam (72.2), American Samoa (42.8), Commonwealth of Northern Mariana Islands (19.0), and Federated States of Micronesia (23.2) (Van Dyne et al. 2020). The latter data may be the first evidence of connection between the presence of stable chromosomal aberrations among the NWA Marshallese women observed in our study and high cancer incidence and mortality observed among Marshallese women.
Besides carcinogenic potential, it is increasingly recognized that exposure to IR can also cause gross metabolic rearrangements exhibited as alterations in the intestinal microbiome, host metabolism, and gene expression that, in turn, lead to the development of metabolic syndrome. For instance, exposure to IR caused long-term metabolic rearrangements, elevated glucose levels, persistently increased leptin and IGF1 in serum, an activated leptin-IGF1 signaling axis, and increased body weight in experimental systems (Suman et al. 2015; Suman et al. 2016; Casero et al. 2017; Ewing et al. 2021). Studies also demonstrate the development of metabolic syndrome, exhibited as impaired glucose homeostasis/insulin resistance, dyslipidemia, and overweight/obesity among long-term childhood cancer survivors who received radiotherapy as a treatment modality (Rutter and Rose 2007; van Waas et al. 2010; Casco and Soto-Vega 2016; Chueh et al. 2017). Furthermore, evacuation associated with the disaster may itself have had a tremendous influence on the development of metabolic syndrome. For instance, evacuees in the case of the Fukushima Daiichi Nuclear Power Plant disaster had higher body mass index, waist circumference, triglycerides, and fasting plasma glucose after the disaster than non-evacuees (Hashimoto et al. 2017). Additionally, the incidence of diabetes was 1.61 times higher in evacuees than in non-evacuees (Satoh et al. 2017). These findings resonate well with accumulating evidence on a number of health disparities in the NWA Marshallese population, such as disproportionally high levels of metabolic syndrome and diabetes with rates of over 20% among the NWA Marshallese compared to 9.4% in Americans (Felix et al. 2017a; Felix et al. 2017b). While our data are not sufficient to establish a direct link with previous exposures to IR and metabolic syndrome and diabetes, it will certainly set the stage for future large-scale studies that will allow analysis of the role of IR in health disparities experienced by NWA Marshallese.
Epigenetic alteration is another recently recognized signature of exposure to IR, as effects of IR on DNA methylation have received much interest in the last two decades (Miousse et al. 2017). Loss of global DNA methylation, the most frequent epigenetic alteration caused by exposure to IR is also a generally-accepted epigenetic hallmark of cancer (Hanahan and Weinberg 2011). This global loss of DNA primarily stems from REs, the most abundant of which is retrotransposon LINE-1 that comprises nearly 20% of mammalian genomes (Miousse et al. 2015). Methylation of LINE-1 DNA is a primary mechanism for silencing of this RE and preventing its unwanted retrotransposition that further results in insertional mutagenesis and genome amplification (Jones et al. 2012). Accumulating evidence suggests that previous exposures to IR may be indicated by the DNA methylation status of LINE-1 elements in peripheral blood (Koturbash et al. 2007; Koturbash 2018). Here, we investigated the DNA methylation profiles of the 32 most abundant human REs, mostly comprised of LINE-1 elements and human endogenous retroviruses (HERVs). Despite a large number of evaluated RE’s, only a couple of them were found to be significantly differentially methylated between the two groups - L1P4e (with self-reported exposure vs without self-reported exposure) and HERVK11I (for both with and without self-reported exposures and “northern atoll” vs “southern atoll” scenarios). This could be explained by an overall small cohort of study participants as well as a high degree of inter-individual variability in methylation of LINE-1 elements. Nevertheless, the differential methylation status of HERVK11I observed in both scenarios and a significant difference in its methylation between women and men may suggest that it might be a promising marker of previous radiation exposure. Future studies built on larger cohorts will be needed to further investigate its potential.
Of particular interest was identification of four LINE-1 elements whose DNA methylation status was inversely correlated with levels of chromosomal aberrations observed in a study participant (i.e., the lower extent of LINE-1 methylation was observed, the higher levels of chromosomal aberrations were observed). It has been postulated previously that radiation-induced alterations in DNA methylation of transcriptionally active REs may lead to chromosomal aberrations and genomic instability and eventual development of cancer (Morgan 2003; Aypar et al. 2011; Morgan 2012; Baulch et al. 2014; Prior et al. 2016). To our knowledge, this is the first time when the locus-specific DNA methylation status and levels of chromosomal aberrations from the same specimen were analyzed and such correlation was demonstrated.
In conclusion, analysis of chromosomal aberrations in NWA Marshallese demonstrated higher incidence of genetic rearrangements in women with self-reported radiation exposure. Further clustering of study participants based on residential history demonstrated that those participants who spent substantial amounts of time (≥ 6 months) in the northern atolls before 1980 had more chromosomal aberrations than their peers who lived only in the southern atolls, and this relationship increased with time spent in the northern atolls. Finally, while high inter-individual variability masked altered DNA methylation as a result of exposure, significant correlations between the chromosomal aberrations and DNA methylation of four LINE-1 elements were identified. Future studies, including those that will involve participants of Hawaii Marshallese (e.g., those Marshallese who had no radiation exposure in their history) in larger cohorts are warranted to further dissect the role of radiation exposure in health disparities experienced by this Pacific Islander nation.
Supplementary Material
Acknowledgements
Research reported in this publication was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM109005, and Arkansas Center for Health Disparities via National Institute on Minority Health and Health Disparities under grant number 5U54MD002329-12. The authors would like to extend their gratitude to all study participants and the members of the Marshallese Community of Northwest Arkansas. The authors would like to thank Mr. Eldon Alik, the Council General of the Republic of Marshall Islands in Springdale for his enthusiastic support of this study. We are thankful to Ralph Wilmoth and the entire team of the Northwest Clinic. We are thankful to Dr. Kristy R. Kutanzi for critical reading of the manuscript and to Christopher Fettes for proofreading and editing this manuscript.
Funding
Research reported in this publication was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM109005, and Arkansas Center for Health Disparities via National Institute on Minority Health and Health Disparities under grant number 5U54MD002329-12.
Footnotes
Disclosure statement
The authors declare that they have no known competing financial interests or personal relationship that could have appeared to influence the work reported in this paper.
Data availability statement
The data that support the findings of this study are available on request from the corresponding author (IK).
References
- Abella MKIL, Molina MR, Nikolić-Hughes I, Hughes EW, Ruderman MA. 2019. Background gamma radiation and soil activity measurements in the northern Marshall Islands. Proc Natl Acad Sci USA. 116(31):15425–15434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aypar U, Morgan WF, Baulch JE. 2011. Radiation-induced epigenetic alterations after low and high LET irradiations. Mutat Res. 707(1-2):24–33. [DOI] [PubMed] [Google Scholar]
- Baulch JE, Aypar U, Waters KM, Yang AJ, Morgan WF. 2014. Genetic and epigenetic changes in chromosomally stable and unstable progeny of irradiated cells. PLoS One. 9(9):e107722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz RL, Tian E, Sadhukhan R, Zhou D, Hauer-Jensen M, Pathak R. 2019. Identification of novel breakpoints for locus- and region-specific translocations in 293 cells by molecular cytogenetics before and after irradiation. Sci Rep. 9(1):10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordner AS, Crosswell DA, Katz AO, Shah JT, Zhang CR, Nikolic-Hughes I, Hughes EW, Ruderman MA. 2016. Measurement of background gamma radiation in the northern Marshall Islands. Proc Natl Acad Sci USA. 113(25):6833–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casco S, Soto-Vega E. 2016. Development of Metabolic Syndrome Associated to Cancer Therapy: Review. Horm Cancer. 7(5-6):289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero D, Gill K, Sridharan V, Koturbash I, Nelson G, Hauer-Jensen M, Boerma M, Braun J, Cheema AK. 2017. Space-type radiation induces multimodal responses in the mouse gut microbiome and metabolome. Microbiome. 5(1):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chueh HW, Yoo JH. 2017. Metabolic syndrome induced by anticancer treatment in childhood cancer survivors. Ann Pediatr Endocrinol Metab. 22(2):82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing LE, Skinner CM, Pathak R, Yee EU, Krager K, Gurley PC, Melnyk S, Boerma M, Hauer-Jensen M, Koturbash I. 2021. Dietary Methionine Supplementation Exacerbates Gastrointestinal Toxicity in a Mouse Model of Abdominal Irradiation. Int J Radiat Oncol Biol Phys. 109(2):581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix H, Rowland B, Long CR, Narcisse MR, Piel M, Goulden PA, McElfish PA. 2018. Diabetes Self-Care Behaviors Among Marshallese Adults Living in the United States. J Immigr Minor Health 20(6):1500–1507. [DOI] [PubMed] [Google Scholar]
- Felix H, Li X, Rowland B, Long CR, Yeary KHK, McElfish PA. 2017. Physical Activity and Diabetes-related Health Beliefs of Marshallese Adults. Am J Health Behav 41(5):553–560. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell. 144(5):646–674. [DOI] [PubMed] [Google Scholar]
- Hashimoto S, Nagai M, Fukuma S, Ohira T, Hosoya M, Yasumura S, Satoh H, Suzuki H, Sakai A, Ohtsuru A, Kawasaki Y, Takahashi A, Ozasa K, Kobashi G, Kamiya K, Yamashita S, Fukuhara SI, Ohto H, Abe M; Fukushima Health Management Survey Group. 2017. Influence of Post-disaster Evacuation on Incidence of Metabolic Syndrome. J Atheroscler Thromb. 24(3):327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond [review]. Nat Rev Genet. 13(7):484–492. [DOI] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. 2006. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 16(1):78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash I, Boyko A, Rodriguez-Juarez R, McDonald RJ, Tryndyak VP, Kovalchuk I, Pogribny IP, Kovalchuk O. 2007. Role of epigenetic effectors in maintenance of the long-term persistent bystander effect in spleen in vivo. Carcinogenesis. 28(8):1831–1838. [DOI] [PubMed] [Google Scholar]
- Koturbash I 2017. LINE-1 in response to exposure to ionizing radiation. Mob Genet Elements. 7(6):e1393491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land CE, Bouville A, Apostoaei I, Simon SL. 2010. Projected lifetime cancer risks from exposure to regional radioactive fallout in the Marshall Islands. Health Phys. 99(2):201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisco H, Conard RA. 1967. Chromosome studies on Marshall Islanders exposed to fallout radiation. Science. 157(3787):445–447. [DOI] [PubMed] [Google Scholar]
- McElfish PA, Narcisse MR, Long CR, Ayers BL, Hawley NL, Aitaoto N, Riklon S, Su LJ, Ima SZ, Wilmoth RO, Schulz TK, Kadlubar S. 2017. Leveraging community-based participatory research capacity to recruit Pacific Islanders into a genetics study. J Community Genet. 8(4): 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, Shao L, Chang J, Feng W, Wang Y, Allen AR, Turner J, Stewart B, Raber J, Zhou D, Koturbash I. 2014. Exposure to low-dose (56)Fe-ion radiation induces long-term epigenetic alterations in mouse bone marrow hematopoietic progenitor and stem cells. Radiat Res. 182(1):92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, Chalbot MC, Lumen A, Ferguson A, Kavouras IG, Koturbash I. 2015. Response of transposable elements to environmental stressors. Mutat Res Rev Mutat Res. 765:19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, Koturbash I. 2015. The Fine LINE: Methylation Drawing the Cancer Landscape. Biomed Res Int. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, Kutanzi KR, Koturbash I. 2017. Effects of ionizing radiation on DNA methylation: from experimental biology to clinical applications. Int J Radiat Biol. 93(5):457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan WF. 2003. Non-targeted and delayed effects of exposure to ionizing radiation: II. Radiation-induced genomic instability and bystander effects in vivo, clastogenic factors and transgenerational effects. Radiat Res. 159(5):581–596. [DOI] [PubMed] [Google Scholar]
- Morgan WF. 2012. Non-targeted and Delayed Effects of Exposure to Ionizing Radiation: I. Radiation-Induced Genomic Instability and Bystander Effects In Vitro. Radiat Res. 178(2):Av223–Av36. [DOI] [PubMed] [Google Scholar]
- Norppa H, Bonassi S, Hansteen IL, Hagmar L, Strömberg U, Rössner P, Boffetta P, Lindholm C, Gundy S, Lazutka J, Cebulska-Wasilewska A, Fabiánová E, Srám RJ, Knudsen LE, Barale R, Fucic A. 2006. Chromosomal aberrations and SCEs as biomarkers of cancer risk [review]. Mutat Res. 600(1-2):37–45. [DOI] [PubMed] [Google Scholar]
- Prior S, Miousse IR, Nzabarushimana E, Pathak R, Skinner C, Kutanzi KR, Allen AR, Raber J, Tackett AJ, Hauer-Jensen M, Nelson GA, Koturbash I. 2016. Densely ionizing radiation affects DNA methylation of selective LINE-1 elements. Environ Res. 150:470–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison WL, Stone EL. 1992. The effect of potassium on the uptake of 137Cs in food crops grown on coral soils: coconut at Bikini Atoll. Health Phys. 62(6):496–511. Erratum in: Health Phys. 63(5):604. [DOI] [PubMed] [Google Scholar]
- Robison WL, Stone EL, Hamilton TF, Conrado CL. 2006. Long-term reduction in (137)Cs concentration in food crops on coral atolls resulting from potassium treatment. J Environ Radioact. 88(3):251–266. [DOI] [PubMed] [Google Scholar]
- Robison WL, Hamilton TF. 2010. Radiation doses for Marshall Islands Atolls affected by U.S. nuclear testing: all exposure pathways, remedial measures, and environmental loss of (137)Cs. Health Phys. 98(1):1–11. [DOI] [PubMed] [Google Scholar]
- Rutter MM, Rose SR. 2007. Long-term endocrine sequelae of childhood cancer. Curr Opin Pediatr. 19(4):480–487. Review. [DOI] [PubMed] [Google Scholar]
- Satoh H, Ohira T, Nagai M, Hosoya M, Sakai A, Yasumura S, Ohtsuru A, Kawasaki Y, Suzuki H, Takahashi A, Sugiura Y, Shishido H, Hayashi Y, Takahashi H, Kobashi G, Ozasa K, Hashimoto S, Ohto H, Abe M, Kamiya K, Fukushima Health Management Survey Group. 2017. Evacuation is a risk factor for diabetes development among evacuees of the Great East Japan earthquake: A 4-year follow-up of the Fukushima Health Management Survey. Diabetes Metab. 45(3):312–315. [DOI] [PubMed] [Google Scholar]
- Simon SL, Robison WL. 1997. A compilation of nuclear weapons test detonation data for U.S. Pacific ocean tests. Health Phys. 73(1):258–264. [DOI] [PubMed] [Google Scholar]
- Simon SL, Bouville A, Land CE, Beck HL. 2010. Radiation doses and cancer risks in the Marshall Islands associated with exposure to radioactive fallout from Bikini and Enewetak nuclear weapons tests: summary. Health Phys. 99(2):105–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon SL, Bouville A, Melo D, Beck HL, Weinstock RM. 2010. Acute and chronic intakes of fallout radionuclides by Marshallese from nuclear weapons testing at Bikini and Enewetak and related internal radiation doses. Health Phys. 99(2):157–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon SL, Bouville A. 2015. Health effects of nuclear weapons testing. Lancet. 386(9992):407–409. [DOI] [PubMed] [Google Scholar]
- Storey JD. 2003. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Statist. 31(6): 2013–2035 (December 2003). DOI: 10.1214/aos/1074290335. [DOI] [Google Scholar]
- Suman S, Kallakury BV, Fornace AJ Jr, Datta K. 2015. Protracted upregulation of leptin and IGF1 is associated with activation of PI3K/Akt and JAK2 pathway in mouse intestine after ionizing radiation exposure. Int J Biol Sci. 11(3):274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suman S, Kumar S, Fornace AJ, Datta K. 2016. Space radiation exposure persistently increased leptin and IGF1 in serum and activated leptin-IGF1 signaling axis in mouse intestine. Sci Rep. 6:31853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Schoemaker MJ, Trott KR, Simon SL, Fujimori K, Nakashima N, Fukao A, Saito H. 2003. The relationship of thyroid cancer with radiation exposure from nuclear weapon testing in the Marshall Islands. J Epidemiol. 13(2):99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Ohtaki M, Hoshi M. 2016. Chromosome aberrations in Japanese fishermen exposed to fallout radiation 420-1200 km distant from the nuclear explosion test site at Bikini Atoll: report 60 years after the incident. Radiat Environ Biophys. 55(3):329–337. [DOI] [PubMed] [Google Scholar]
- Topping CEW, Abella MKIL, Berkowitz ME, Molina MR, Nikolić-Hughes I, Hughes EW, Ruderman MA. 2019. In situ measurement of cesium-137 contamination in fruits from the northern Marshall Islands. Proc Natl Acad Sci USA. 116(31):15414–15419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyne EA, Saraiya M, White A, Novinson D, Senkomago V, Buenconsejo-Lum L. 2020. Cancer Mortality in the US-Affiliated Pacific Islands, 2008-2013. Hawaii J Health Soc Welf. 79(6 Suppl 2):99–107. [PMC free article] [PubMed] [Google Scholar]
- van Waas M, Neggers SJ, Pieters R, van den Heuvel-Eibrink MM. 2010. Components of the metabolic syndrome in 500 adult long-term survivors of childhood cancer. Ann Oncol. 21(5):1121–1126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author (IK).


