Abstract
Fusarium oxysporum (Fo) is ubiquitous in soil and forms a species complex of pathogenic and putatively non-pathogenic strains. Pathogenic strains cause disease in over 150 plant species. Fusarium oxysporum f. sp. ciceris (Foc) is a major fungal pathogen causing Fusarium wilt in chickpeas (Cicer arietinum). In some countries such as Australia, Foc is a high-priority pest of biosecurity concern. Specific, sensitive, robust and rapid diagnostic assays are essential for effective disease management on the farm and serve as an effective biosecurity control measure. We developed and validated a novel and highly specific PCR and a LAMP assay for detecting the Indian Foc race 1 based on a putative effector gene uniquely present in its genome. These assays were assessed against 39 Fo formae speciales and found to be specific, only amplifying the target species, in a portable real-time fluorometer (Genie III) and qPCR machine in under 13 min with an anneal derivative temperature ranging from 87.7 to 88.3 °C. The LAMP assay is sensitive to low levels of target DNA (> 0.009 ng/µl). The expected PCR product size is 143 bp. The LAMP assay developed in this study was simple, fast, sensitive and specific and could be explored for other Foc races due to the uniqueness of this marker to the Foc genome.
Subject terms: Genetics, Molecular biology
Introduction
Chickpea (Cicer arietinum) is an economically important crop worldwide and plays an important nutritional role in the diets of millions of people, especially in developing countries, providing an essential source of protein, calcium, iron, phosphorus, and other minerals1. It is the second-largest cultivated legume crop after dry beans (Phaseolus vulgaris) globally2. It is grown throughout tropical, subtropical and temperate regions in South and West Asia, East and North Africa, southern Europe, North and South America, and Australia2. Asian countries contribute to 83% of global chickpea production. Approximately 12 million tonnes of chickpea are produced annually, with India contributing approximately 64%, followed by Australia holding 7% of the global share1. In 2021, approximately 500,000 Ha was planted in Australia, producing 732,000 tonnes. Most grains were exported, fetching AUS$543 million in foreign earnings3.
Global chickpea production is highly dependent on various biotic and abiotic stresses. One of the critical biotic stresses, Fusarium wilt, causes significant economic losses ranging from 10 to 40% in many countries but has the potential to cause complete crop loss under disease-favourable conditions4–7. It is caused by the soil and seed-borne fungal pathogen Fusarium oxysporum (Fo) forma specialis (f. sp.) ciceris (Foc). Fusarium oxysporum f. sp. ciceris is a member of the Fo species complex (FOSC), a soil-borne fungus comprising pathogenic and putatively non-pathogenic strains. Plant pathogenic Fo strains cause vascular wilt and cortical rot in many agricultural crop species. They are classified into host-specific forms (formae speciales, ff. spp.) and are often further subdivided into races based on their capacity to infect different cultivars of a plant species.
Fusarium wilt in chickpea was initially reported in India by Butler in 1918, but its aetiology was not correctly determined until 1940 by Padwick8. It is widespread in most chickpea-growing areas of Asia, Africa, southern Europe and the Americas but is considered to be absent from Australia9. Eight physiological races (0, 1A, 1B/C, 2, 3, 4, 5 and 6) based on disease reactions on 10 chickpea differential cultivars have been reported globally in Foc10. There are two pathotypes based on aboveground symptoms: one causing yellowing and the other causing wilting11,12. Races 1A, 2, 3, 4, 5 and 6 induce wilting symptoms, including severe chlorosis and flaccidity combined with vascular discolouration followed by plant death. Races 0 and 1B/C are less virulent than the other races and induce yellowing symptoms11,13. The eight races have distinct geographic distributions, with race 1A more widely distributed across India, Mediterranean regions and California14. Races 2, 3 and 4 are found in India and Ethiopia15–17 while races 0, 1B/C, 5 and 6 are found mainly in the Mediterranean regions and the USA16,18,19.
Due to high genetic variation in the Foc population in India, Dubey et al.20 in 2012 developed a new set of chickpea differential cultivars to identify the Indian Foc population and characterised them into eight new races (races 1–8). How these newly Indian characterised races compare to the globally characterised races is not yet known. Isolates belonging to the eight Indian races were found to belong to the same vegetative compatibility group (VCG)21. Vegetative compatibility grouping is a system for genetic diversity analysis and classification, as proposed by Puhalla22. Isolates collected globally, irrespective of geographical distribution, race and symptom type, were also found to be from a single VCG23. Isolates belonging to the same VCG can exchange genetic information via heterokaryosis and a parasexual cycle24, hence are clonally related and genetically similar.
Disease symptoms can develop at any stage of plant growth and affected plants may be grouped in patches or appear spread across the field8,25. Disease symptoms in highly susceptible cultivars can develop 25 days after sowing, including flaccidity of individual leaves followed by a dull-green discolouration, desiccation and collapse of the whole plant (designated ‘early wilt’)8. The symptoms are usually more conspicuous at the onset of flowering, 6–8 weeks after sowing and can also appear up to the podding stage (‘late wilt’)26. Late wilted plants exhibit drooping of the petioles, rachis and leaflets, followed by yellowing and foliage necrosis26. Initially, drooping is observed in the upper part of the plant, but within a few days, the entire plant collapses26. Roots and stems of affected plants develop a dark-brown discoloration of xylem tissues visible upon vertical split or when cross-sectioned26.
Molecular diagnostic assays for Foc are highly desirable as the determination of the organism to the taxonomic level of f. sp. using nonmolecular methods is costly in terms of time and resources. However, horizontal gene transfer between strains in the FOSC has resulted in a polyphyletic origin of host specificity in most ff. spp., which prevents molecular identification of strains based on conserved, vertically inherited genes27. Pathogenic strains may share higher sequence similarity of conserved genes with putatively non-pathogenic strains or other ff. spp.
Currently, two studies have developed molecular markers for diagnostics of Foc. One is a loop-mediated isothermal amplification (LAMP) assay based on a conserved gene, translation elongation factor 1-alpha (EF1α)28. The aim was to discriminate Fo from other fungal pathogens that infect chickpeas and therefore, the assay is not specific to Foc but distinguishes Fo from other fungi. The other is a conventional PCR assay based on random amplified polymorphic DNA (RAPD) fragments, resulting in sequence characterized amplified region (SCAR) markers29. SCAR markers are suboptimal for f. sp. discrimination because they are based on genomic regions that are not necessarily required for virulence. Furthermore, as they can be localized anywhere on the genome, there is often little to no sequence data available in the public databases for comparison with other sequences. With the amount of genomic data currently available in the public domain, it has been determined that the markers used by Jiménez-Gasco and Jiménez-Díaz29 to identify Foc races 0, 1A, 5 and 6 contain a fragment that is identical to the Impala transposon30. The impala elements in strains of different ff. spp. are highly similar31. Molecular markers based on such regions could produce false-positive results.
Effectors are functional elements in the pathogen-host interaction and have shown very limited sequence diversity between strains of the same f. sp., making them potential markers for host-specific pathogenicity32. Effector proteins in Fo and in other fungal pathogens may have pathogenicity roles and act as enzymes, toxins, transcription factors, elicitors or virulence factors33. One of the main groups of effector genes in Fo encodes for proteins that are secreted in xylem (SIX1-SIX14)34–37, but the pathogenicity role has been experimentally validated for only a few members of that group38,39. Recent studies have shown that host specificity in Fo is governed by the suite of effector genes33,40. Both presence-absence polymorphisms and the individual effector gene sequence predict a strain’s host range32,33. These genes, therefore, should be used as molecular markers for the discrimination of ff. spp. within the FOSC. Effectors have successfully been used as molecular markers within the FOSC to discriminate Fo f. sp. lycopersici41 and Cucurbitaceae-affecting strains32 from other ff. spp. New Fo genomes are being constantly assembled and made publicly available, making whole-genome comparative studies the new gold standard for identifying molecular markers for diagnostics.
The spread of Foc race 1 in India is not known; however, it has been confirmed to be present in Andhra Pradesh and Karnataka states20,21. Indian Foc race 1 has been more intensively studied than any other races. These studies have identified the molecular mechanisms behind the chickpea-Foc1 interactions42 and have used Foc race 1 for comparative genomics studies to identify conditionally dispensable sequences in legume-infecting Fo ff. spp.43. These studies all used Foc race 1 strain 38-1 obtained from ICRISAT, India. Therefore, a well-curated genomic sequence for strain 38-1 exists on the NCBI GenBank and single-spored cultures of Foc race 1 strain 38-1 are maintained at ICRISAT, India. Genomic DNA of Foc race 1 strain 38-1 was imported from ICRISAT, India, specifically for use in this study.
The aim of this study was to utilise a comparative genomics approach through the use of OrthoFinder44 to identify an effector gene unique to Indian Foc race 1 and to develop a specific, sensitive and rapid LAMP and a PCR assay that can selectively discriminate isolates of Indian Foc race 1 from other FOSC ff. spp.
Materials and methods
Fungal isolates and genomes
We created a database of 356 Fo genomes of 39 ff. spp. of exotic and endemic isolates. One hundred and fifty genomes of 33 ff. spp. were retrieved from the NCBI GenBank. These ff. spp., with the number of isolates in parentheses, are: albedinis (1), apii (3), capsici (1), cepae (3), ciceris (1), conglutinans (6), coriandrii (2), cubense (6), cucumerinum (9), fragariae (19), gladioli (3), koae (1), lagenariae (4), lilii (1), lini (5), luffae (2), lycopersici (8), matthiolae (1), medicaginis (1), melongenae (2), melonis (10), momordicae (2), mori (1), narcissi (2), nicotianae (4), niveum (12), pisi (1), radicis-cucumerinum (3), radicis-lycopersici (1), raphani (1), sesami (1), spinaciae (10), tulipae (1), vasinfectum (10) (Supplementary Table 1). An additional 12 genomes only identified as Fo were included; four of these 12 genomes were of non-pathogenic Fo isolated from chickpeas from Ethiopia (Supplementary Table 1).
Two hundred and six cultures of Fo already present in Australia (henceforth called endemic) were sourced from culture collections across the country (Victorian Plant Pathogen Herbarium (VPRI), Royal Botanic Gardens (RBG) Sydney, NSW Plant Pathology and Mycology Herbarium (DAR), NSW Department of Primary Industries (DPI), Melbourne University and South Australia Research and Development Institute (SARDI)), and their genomes generated to add to the database. One hundred and four isolates were sequenced and assembled as part of this study, while genomes of other isolates were generated as part of a previous study27. Seventy of these isolates are of Fo isolated from diseased horticultural plants, 14 isolates are from soil from the natural ecosystems and 122 isolates are of 17 ff. spp.. These ff. spp., with the number of isolates in parentheses, are basilici (5), canariensis (13), conglutinans (1), cubense (5), dianthi (1), fragariae (3), lycopersici (9), medicaginis (2), melonis (9), niveum (7), passiflorae (5), pisi (35), spinaciae (1), tracheiphilum (1), tulipae (4), vasinfectum (17) and zingiberi (4) (Supplementary Table 1). Purified Foc race 1 genomic DNA of strain 38–1 was sourced from ICRISAT, India, to be used as a positive control and the genome was sequenced and assembled as part of this study.
Culture growth and DNA extraction
Endemic Fo cultures were single-spored using the method described by Burgess et al.45 and cultures were grown and mycelia were harvested as described by Achari et al.27. Genomic DNA was extracted using Wizard® Genomic DNA Purification Kit (www.promega.com). The quality of the genomic DNA was assessed using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). DNA samples with 260/280 ratios of ~ 1.8 were used for downstream library preparation.
Sequencing and genome assembly
Paired-end libraries were prepared for endemic isolates and Foc genomic DNA using the Illumina Nextera XT DNA library prep kit according to the manufacturer’s protocols. These libraries were sequenced using Illumina NovaSeq 6000. Fastq sequence files generated from the sequencing run were filtered using fastp, filtering sequences based on a minimum length of 50 bp and removal of adaptors46. Low-quality reads (< Q20) from the fastq sequence files were also filtered using fastp46. The reads were de novo assembled using SPAdes version 3.7.147. Contigs < 5X coverage and < 200 bp were removed. The quality and completeness of the assembled genomes were measured using BUSCO as described by Achari, et al.27 and assemblies having greater than 97% of 3725 core Sordariomycete genes as estimated by BUSCO were kept in the database for comparative genomic analysis.
Identification of the molecular marker
All the genomes in the database were annotated using Augustus48. The amino acid sequences of each genome were used for comparative genomic analysis using OrthoFinder44. The OrthoFinder divided the 356 genomes into orthogroups. An orthogroup is a gene family or clade of genes defined at a specific taxonomic level44. These orthogroups were assessed across the 356 genomes and a single orthogroup unique to Foc was identified as the Foc molecular marker. The gene number of the Foc-specific orthogroup was used to retrieve the amino acid and nucleotide sequence from the Foc genome gff file.
LAMP and PCR primer design
Novel LAMP and end-point conventional PCR primers were designed targeting the Foc unique molecular marker. PCR and LAMP primers were designed and analysed using Primer 349 in Geneious and Primer Explorer V5 software (http://primerexplorer.jp), respectively. The LAMP primers included: two outer primers (forward primer, F3; backward primer, B3), two inner primers (forward inner primer, FIP (F1c and F2); backward inner primer, BIP (B1c and B2) and two loop primers (forward loop primer, FL; backward loop primer, BL). The specificity of the primers was further tested using the NCBI Primer-BLAST on Refseq mRNA against Fusarium as an organism. Primers for both LAMP and PCR were synthesized by Merck (https://www.sigmaaldrich.com). Primer sequences and their relative positions on the Foc molecular marker gene are shown in Table 1 and Fig. 1, respectively.
Table 1.
Details of the primers designed based on the putative effector gene found to be unique to Fusarium oxysporum f. sp. ciceris.
| Primer type | Primer | Length of primer (bp) | Sequence (5′-3′) |
|---|---|---|---|
| LAMP-Foc | F3 | 20 | CCCAGCGAAGTACTTGAACC |
| B3 | 20 | TGAACTAGGAGGGGGTTGAT | |
| FIP (F1c and F2) | 42 | CTGAGGCCATCGAACCAGTCTTGTCCGCAAACTACCCGAC | |
| BIP (B1c and B2) | 41 | AGGACTACTGCAAGGAGCCGGCGACAAGCGGTCAAGAGAAG | |
| LF | 20 | CTGTGCTTCAGATGGGTCAAG | |
| LB | 18 | AAGTGAGCGAGGCCGACT | |
| Conventional end-point PCR | |||
| Foc | Forward (FP) | 20 | CGATGGCCTCAGCGATTCAT |
| Reverse (RP) | 20 | CTCTGCGAGCCAGTGAACTA | |
| ITS | ITS1 | 19 | TCCGTAGGTGAACCTGCGG |
| ITS4 | 20 | TCCTCCGCTTATTGATATGC | |
Figure 1.
Schematic representation of position and sequence of the designed primer sets within the nucleotide sequence of the putative effector gene of Fusarium oxysporum f. sp. ciceris used for (a) LAMP and (b) PCR assays.
Development of the LAMP assay
The primer ratio (F3/B3: FIP/BIP: LF/LB) used for the assay was 1:8:2 with the final concentrations of 0.2 µM, 1.6 µM and 0.4 µM for F3/B3, FIP/BIP and LF/LB. LAMP reactions were performed and optimised in the Genie III triplicates and repeated three times. The assay was run using WarmStart® LAMP Kit (DNA & RNA) (New England BioLabs Inc.) in a 25 μL reaction volume. The components of the reaction were 12.5 μL of LAMP 2X Master Mix, 0.5 μl Fluorescent dye (50X), 2.5 μl LAMP Primer Mix (10X), 8.5 μl water and 2 μl (5 ng/ul) DNA. 2 μL of ultrapure water was used as no template control (NTC). LAMP amplification reactions were run at 65 °C for 30 min, followed by an annealing analysis from 95 to 60 °C with ramping at 0.05 °C per second that allowed the generation of derivative melting curves. The approximate running time was 40 min.
Upon completion of the run, the amplification and anneal derivative curves were visualised on the Genie III screen to ensure amplification occurred as expected. No amplification supported by flat amplification lines are expected from non-target species and NTCs. The time to amplification (minutes) and anneal derivative temperature (°C) were recorded from the results table. The run details such as the date and the run number of each assay completed on the Genie III were recorded for ease of run file transfer and analysis using a PC version of the software Genie Explorer version V2.0.6.3. The blue channel of the Genie III was used for visualising LAMP assays in this study.
Specificity of detection of the LAMP assay
When designing an assay, it is imperative to test close relatives of the target pathogen to ensure cross-reactions do not cause a false positive result. The LAMP specificity assay was performed with template fungal DNA from 17 other Fo ff. spp. (basilici, canariensis, conglutinans, cubense, dianthi, fragariae, lycopersici, medicaginis, melonis, niveum, passiflorae, pisi, spinaciae, tracheiphilum, tulipae, vasinfectum and zingiberi). Only a single isolate per f. sp. was used for specificity testing. The reaction was run in triplicate and the experiment was repeated twice. Since there were more than eight samples, this assay was run on a qPCR machine using the same assay conditions described previously for the Genie III run.
Once the run was completed, the amplification and anneal derivative curves were visualised on the qPCR screen to ensure that amplification had occurred as expected. As described above, non-target species and NTCs are expected to have flat amplification lines. The time of amplification (minutes) and anneal derivative temperature (°C) were recorded from the results table. The eds file of the run was transferred and analysed using a PC version of QuantStudio Design and Analysis software.
Sensitivity of the LAMP assay
A five-fold serial dilution of genomic Foc DNA was prepared using ultrapure water. Starting DNA concentration was quantified using a Quantus Fluorometer (www.promega.com). The DNA was serially diluted using ultrapure water from 9 to 0.00009 ng/µl (1:1 to 1:100,000). The sensitivity of the LAMP assay was tested using the serially diluted DNA on a qPCR machine. The experiment was run twice in triplicates.
Conventional polymerase chain reaction (PCR) assay
PCR was executed using Veriti (Applied Biosystems, ThermoFisher Scientific) in a 25 µl reaction volume which included the following components: 12.5 µl of Hot Start Taq 2X Master Mix (New England Biolabs), 1.25 µl of forward and reverse primers, 2 µl (10 ng) of template DNA and 8 µl of ultrapure water. The Foc-specific PCR was screened against 17 other ff. spp. (basilici, canariensis, conglutinans, cubense, dianthi, fragariae, lycopersici, medicaginis, melonis, niveum, passiflorae, pisi, spinaciae, tracheiphilum, tulipae, vasinfectum and zingiberi). Only one isolate per f. sp. was used for PCR. Ultrapure water was used as an NTC. Different cycling PCR parameters were trialled to identify the optimum PCR cycling conditions. After amplification, 3 μl of gel loading dye was added to each sample and resolved on 2% agarose gel in 0.5X TBE buffer. The gel electrophoresis was run at 100 V for an hour. The size of amplified DNA fragments was estimated with a hyperladder™ 100 bp (Bioline). The expected amplicon size was 143 bp. Internal transcribed spacer (ITS) universal primers ITS1/ITS4, designed to amplify a ribosomal DNA fragment of approximately 544 bp was used as an internal control for Fo.
Results
Identification of Foc-specific molecular marker
The molecular marker identified for detection of Foc race 1 is a 378 bp gene which encodes for a protein of 125 amino acids in length. Specificity of the gene to the Foc genome was confirmed through in silico and in vitro experimentations. In silico analysis was carried out using BLASTn and BLASTx with default settings against the nucleotide and protein databases on the NCBI GenBank and using BLASTn (evalue: 1E-10) against the genomes in the database created in this study. Specificity testing on LAMP and PCR assay has shown that this gene was found to be uniquely present in the Foc genome. This gene was confirmed as an effector gene using EffectorP versions 1.0, 2.0 and 3.0 (https://effectorp.csiro.au/)50–52 with default settings.
Specificity and sensitivity of Foc race 1 LAMP assay
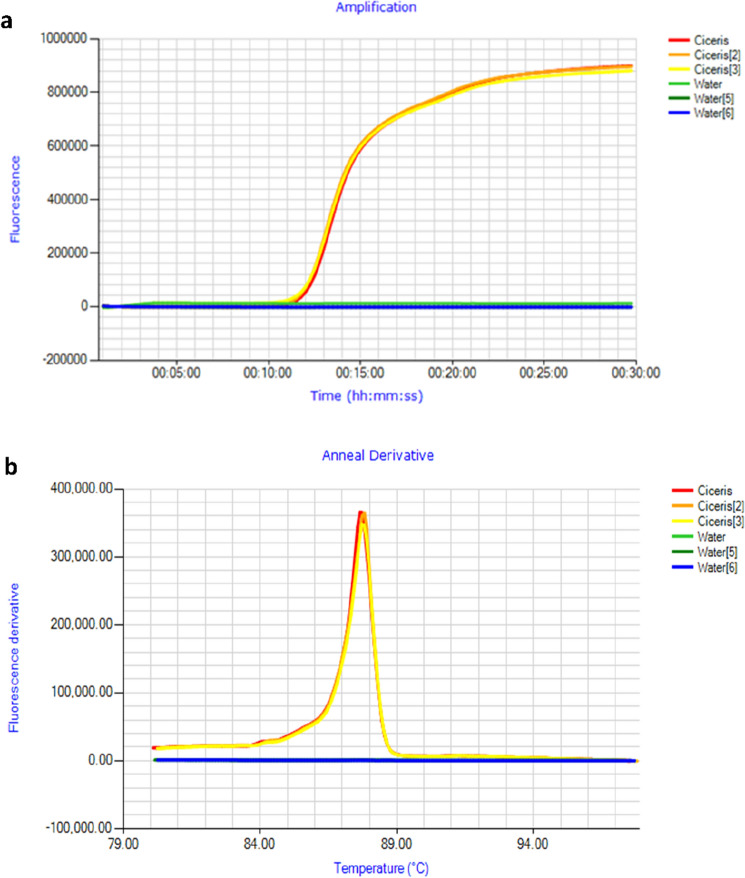
The Foc race 1 LAMP assay on Genie III produced amplification of the target f. sp. DNA in less than 13 min with an average anneal derivative temperature of 87.7 °C (Fig. 2). There was no amplification of the NTC. The LAMP specificity assay was performed with template fungal DNA from 17 other Fo ff. spp. (basilici, canariensis, conglutinans, cubense, dianthi, fragariae, lycopersici, medicaginis, melonis, niveum, passiflorae, pisi, spinaciae, tracheiphilum, tulipae, vasinfectum and zingiberi). At optimum conditions, no positive amplification was observed in the case of other ff. spp. The average time to amplify three replicates of Foc genomic DNA on a qPCR machine was 10 min and 51 s, with an average anneal derivative temperature of 88.3 °C.
Figure 2.
Optimised LAMP assay performed on genomic Fusarium oxysporum f. sp. ciceris DNA (a) Amplification profile, with positive samples amplifying in < 13 min. (b) Anneal derivative of LAMP amplicons, with an average anneal derivative of 87.7 °C.
A five-fold serial dilution of genomic Foc DNA was prepared using ultrapure water. We successfully detected amplification of all the replicates until the three-fold dilution, up to 0.009 ng/µl of DNA. Only one replicate was amplified for samples with 0.0009 ng/µl of DNA, while there was no amplification from any replicates with 0.00009 ng/µl of DNA. The amplification time became slower in a predictable manner as DNA template concentrations reduced, showing a strong relationship between increased amplification times and decreasing DNA concentrations (Table 2).
Table 2.
The average amplification time and anneal derivative for serial dilutions of Fusarium oxysporum f. sp. ciceris genomic DNA.
| DNA concentration | Average time to amplification (m:s) | Average anneal derivative (°C) |
|---|---|---|
| 9 × 10−1 | 11:36 | 88.2 |
| 9 × 10−2 | 12:19 | 88.1 |
| 9 × 10−3 | 16:06 | 88.0 |
Detection of Foc race 1 using PCR
The optimum cycling parameters were initial denaturing for 30 s at 95 °C; 28 cycles of [denaturing at 95 °C for 30 s, annealing at 60 °C for 30 s, extension at 72 °C for 30 s] and final extension at 72 °C for 1 min. The primer pair Foc-FP/Foc-RP amplified a unique DNA fragment of approximately 143 bp in Foc race 1 but did not yield amplification products for any of the 17 other ff. spp. (Fig. 3). Internal transcribed spacer (ITS) universal primers ITS1/ITS4, designed to amplify a ribosomal DNA fragment of approximately 544 bp, yielded positive PCR reactions for all the isolates (data not shown).
Figure 3.
Agarose gel showing amplification products from PCR using genomic Fusarium oxysporum f. sp. ciceris (Foc) DNA. Lane 2 with Foc DNA amplified showing a product size of 143 bp. There is no amplification from any other ff. spp.
Discussion
Loop-mediated isothermal amplification (LAMP) is becoming a reliable and robust method for detecting and identifying a variety of phytopathogens. Isothermal amplification with easy detection of amplifications makes LAMP a simple-to-operate and easy-to-read molecular diagnostic tool for laboratory and in-field settings. Several LAMP-based diagnostic kits and assays have been developed targeting a range of pathogens53. Many technical advances have been made over the years to satisfy the demands of the molecular diagnostic industry for more specificity, sensitivity, efficiency, and rapidity in diagnostics54.
Fusarium oxysporum f. sp. ciceris is a high-priority exotic grain pest of biosecurity importance to Australia. Specific, sensitive, robust and rapid identification of Foc is necessary for effective biosecurity control. Effector genes are known to be involved in pathogenicity and determine host-specificity. We used a comparative genomics approach to comprehensively screen 356 Fo genomes of 39 ff. spp. to identify a putative effector gene uniquely present in Foc race 1. PCR and LAMP assays were developed that were very specific and sensitive to Indian Foc race 1.
Two previous Foc diagnostic assays have been published. A LAMP assay was developed based on the translation elongation factor (TEF) gene28. This assay is not specific to Foc but distinguishes Fo amongst other pathogens causing diseases on chickpea plants. If a soil sample is screened for the presence of Foc before planting chickpeas, this assay will produce a positive amplification irrespective of Foc being absent because Fo is ubiquitously present in the soil as non-pathogenic strains, and the assay is generic to Fo and not specific to Foc.
A Foc PCR diagnostic assay was developed in 2003 based on an impala transposon29,30 and is the test endorsed for use in Australia’s NDP36, the National Diagnostic Protocol for Fusarium oxysporum f. sp. ciceris9. However, this assay was not comprehensively screened against other ff. spp.29. There are five subfamilies of impala transposon identified in Fo, and most of these are present in multiple copies in several strains with different host specificities55. Using an impala transposon as a molecular marker may lead to unspecific amplifications and consequently a false positive diagnosis. Genome sequences of all ff. spp. obtained from NCBI GenBank had an incomplete transposon sequence except for the two strains of f. sp. lini (F282- JABJUB010000001.1 and F329- JABJUE010000001.1). The sequence region where the forward and reverse primers bind was present in two pieces in the genomic sequence, with the middle sequence region missing (Supplementary Fig. 2). The forward and reverse primers and the f. sp. lini sequences showed no nucleotide sequence difference at the binding sites (Supplementary Fig. 2). Since the transposon sequence was incomplete for most of the ff. spp., we ran the PCR assay against some ff. spp. and found the assay not to be specific to Foc. Their assay will amplify isolates belonging to ff. spp.: lycopersici, tulipae, canariensis, medicaginis, passiflorae, tracheiphilum, pisi-race 5, dianthi and vasinfectum (Australian endemic strains) (Supplementary Fig. 3) and lini (Supplementary Fig. 2).
Australian biosecurity measures restrict the importation of live cultures of exotic plant pathogens of biosecurity significance to Australia. We were only able to source genomic DNA of Foc race 1 strain 38-1 from ICRISAT, India, and this served as a model for all of the Foc races. Our Foc molecular marker has been comprehensively screened against 39 ff. spp. as well as endemic Fo present in natural ecosystems in Australia and non-pathogenic Fo isolated from chickpea plants. We were unable to in silico test these assays on other Foc races as genomic data for other Foc races are currently not present in the public domain. However, since this effector gene was uniquely present in the Foc race 1 genome, and all the Foc races belong to the same VCG, we anticipate that it may detect other Foc races. Furthermore, in silico analysis has shown the absence of this effector gene in the genomes of non-pathogenic Fo (Foxy_EtdFoc: 22, 36, 191 and 136) isolated from chickpea, suggesting that this marker will differentiate pathogenic and non-pathogenic Fo isolated from chickpea plants. We have developed a novel LAMP and PCR assay which is specific and sensitive to Indian Foc race 1 and can be field deployable.
Supplementary Information
Acknowledgements
We are grateful to so many people who played a significant role in ensuring the success of this project. Firstly, we would like to thank Professor Brendan Rodoni for seeking funding for this project and Dr. Fiona Contable for being the Agriculture Victoria project leader. Secondly, a heartfelt appreciation to Drs Linda Zheng, Yuhong Liu, Arati Agarwal and Peter Mee on sharing their knowledge and experiences on designing the LAMP assays. Finally, without the isolates, the project could not have been carried out this comprehensively, hence a heartful appreciation to Drs Yu Pei Tan, Jordan Bailey, Edward Liew, Paul Taylor, Sophia Callaghan and Duy Le for providing Fusarium oxysporum cultures from their collections.
Author contributions
J.E. and S.A. developed the project concept. S.A., R.M. and J.E. planned the experimental design. S.A., R.M. and J.E. planned the experimental design. S.A. performed the experiments, analysed the data, created the figures and drafted and edited the manuscript. J.E. edited the manuscript. M.S. supplied the Foc DNA. All the authors approved the final version.
Funding
The Boosting Diagnostics project is supported by Grains Research and Development Corporation, through funding from the Australian Government Department of Agriculture, Fisheries and Forestry as part of its Rural R&D for Profit program and along with Cotton Research and Development Corporation, Hort Innovation Australia, Wine Australia, Sugar Research Australia and Forest and Wood Products Australia. The Commonwealth of Australia and the State Government of Victoria co-funded the research.
Data availability
Genomes of the endemic Australian isolates that were sequenced for this project are available on the NCBI GenBank with Accession numbers JAMSCL000000000-JAMWEW000000000 under bioproject PRJNA846078. GenBank accession numbers are given of other genomes retrieved from the NCBI GenBank.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-29730-6.
References
- 1.Merga B, Haji J. Economic importance of chickpea: Production, value, and world trade. Cogent Food Agric. 2019;5:1615718. doi: 10.1080/23311932.2019.1615718. [DOI] [Google Scholar]
- 2.FAO. in World Food and Agriculture—Statistical Yearbook 2019., (Rome, 2019).
- 3.ABARES. In Agricultural commodity statistics- Rural commodities-pulses (2021).
- 4.Sharma M, et al. Genome wide transcriptome profiling of Fusarium oxysporum f. sp. Ciceris conidial germination reveals new insights into infection-related genes. Sci. Rep. 2016;6:37353. doi: 10.1038/srep37353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halila, M. & Strange, R. Identification of the causal agent of wilt of chickpea in Tunisia as Fusarium oxysporum f. sp. re-pr race 0. Phytopathologia Mediterranea, 67–74 (1996).
- 6.Navas-Cortés JA, Hau B, Jiménez-Díaz RM. Yield loss in chickpeas in relation to development of Fusarium wilt epidemics. Phytopathology. 2000;90:1269–1278. doi: 10.1094/PHYTO.2000.90.11.1269. [DOI] [PubMed] [Google Scholar]
- 7.Arvayo-Ortiz RM, Esqueda M, Acedo-Felix E, Sanchez A, Gutierrez A. Morphological variability and races of Fusarium oxysporum f. sp. ciceris associated with chickpea (Cicer arietinum) crops. Am. J. Agric. Biol. Sci. 2011;6:114–121. doi: 10.3844/ajabssp.2011.114.121. [DOI] [Google Scholar]
- 8.Haware M. Fusarium wilt and other important diseases of chickpea in the Mediterranean area. Sémin. 1990;9:61–64. [Google Scholar]
- 9.Cunnington, J., Lindbeck, K. & Jones, R. H. in National diagnostic protocol for the detection of Fusarium wilt of Chickpea (Fusarium oxysporum f. sp. ciceris). (Plant Health Australia, Canberra, Australia, 2007).
- 10.Caballo C, et al. Candidate genes expression profiling during wilting in chickpea caused by Fusarium oxysporum f. sp. ciceris race 5. PLoS ONE. 2019;14:e0224212–e0224212. doi: 10.1371/journal.pone.0224212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trapero-Casas AJ-DR. Fungal wilt and root rot diseases of chickpea in southern Spain. Phytopathology. 1985;75:1146–1151. doi: 10.1094/Phyto-75-1146. [DOI] [Google Scholar]
- 12.Jiménez-Fernández D, Landa B, Kang S, Jiménez-Díaz R, Navas-Cortés J. Quantitative and microscopic assessment of compatible and incompatible interactions between chickpea cultivars and Fusarium oxysporum f. sp. ciceris races. PLoS ONE. 2013;8:e61360. doi: 10.1371/journal.pone.0061360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiménez-Díaz R, Alcala-Jimenez A, Hervás A, Trapero-Casas J. Pathogenic variability and host resistance in the Fusarium oxysporum f. sp. Ciceris-Cicer arietinum pathosystem. Hod. Rosl. Aklim. Nasienn. 1993;37:87–94. [Google Scholar]
- 14.Jiménez-Gasco MDM, Milgroom MG, Jiménez-Díaz RM. Stepwise evolution of races in Fusarium oxysporum f. sp. ciceris inferred from fingerprinting with repetitive DNA sequences. Phytopathology®. 2004;94:228–235. doi: 10.1094/phyto.2004.94.3.228. [DOI] [PubMed] [Google Scholar]
- 15.Haware MP, Nene YL. Symptomless carriers of the chickpea wilt Fusarium. Plant Dis. 1982;66:250–251. doi: 10.1094/PD-66-250. [DOI] [Google Scholar]
- 16.Jiménez-Gasco DMM, Pérez-Artés E, Jiménez-Diaz RM. Identification of pathogenic races 0, 1B/C, 5, and 6 of Fusarium oxysporum f. sp. ciceris with random amplified polymorphic DNA (RAPD) Eur. J. Plant Pathol. 2001;107:237–248. doi: 10.1023/A:1011294204630. [DOI] [Google Scholar]
- 17.Shehabu M, Ahmed S, Sakhuja PK. Pathogenic variability in Ethiopian isolates of Fusarium oxysporum f. sp. ciceris and reaction of chickpea improved varieties to the isolates. Int. J. Pest Manag. 2008;54:143–149. doi: 10.1080/09670870701780153. [DOI] [Google Scholar]
- 18.Bayraktar H, Dolar FS. Pathogenic variability of Fusarium oxysporum f. sp. ciceris isolates from chickpea in Turkey. Pak. J. Bot. 2012;44:821–823. [Google Scholar]
- 19.Mohamed OE, Hamwieh A, Ahmed S, Ahmed NE. Genetic variability of Fusarium oxysporum f. sp. ciceris population affecting chickpea in the Sudan. J. Phytopathol. 2015;163:941–946. doi: 10.1111/jph.12396. [DOI] [Google Scholar]
- 20.Dubey SC, Priyanka K, Singh V, Singh B. Race profiling and molecular diversity analysis of Fusarium oxysporum f. sp. ciceris causing wilt in chickpea. J. Phytopathol. 2012;160:576–587. doi: 10.1111/j.1439-0434.2012.01954.x. [DOI] [Google Scholar]
- 21.Saabale P, Dubey S. Pathogenicity and vegetative compatibility grouping among Indian populations of Fusarium oxysporum f. sp. ciceris causing chickpea wilt. Phytoparasitica. 2014;42:465–473. doi: 10.1007/s12600-014-0383-8. [DOI] [Google Scholar]
- 22.Puhalla JE. Classification of strains of Fusarium oxysporum on the basis of vegetative compatibility. Can. J. Bot. 1985;63:179–183. doi: 10.1139/b85-020. [DOI] [Google Scholar]
- 23.Moncada, A., Jiménez-Díaz, R. & Artés, E. Vegetative compatibility groups in Fusarium oxysporum f. sp. ciceris and F. oxysporum non‐pathogenic to chickpea. J. Phytopathol.157, 729–735. doi:10.1111/j.1439-0434.2009.01562.x (2009).
- 24.Rodrigues de Carvalho C, Cristina Mendes-Costa M. Vegetative compatibility and heterokaryon formation between different isolates of Colletotrichum lindemuthianum by using the nit mutant system. Braz. J. Microbiol. 2011;42:346–353. doi: 10.1590/s1517-83822011000100044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nene, Y. L. & Reddy, M. V. in The Chickpea (eds M.C. Saxena & K.B Singh) 233e270. (CAB Int, 1987).
- 26.Jimenez-Díaz, R. M., Basallote-Ureba, M. J. & Rapoport, H. in Vascular wilt diseases of plants Vol. H28 (eds E.C. Tjamos & C. Beckman) 113e121 (Springer-Verlag, 1989).
- 27.Achari SR, et al. Phylogenetic relationship between Australian Fusarium oxysporum isolates and resolving the species complex using the multispecies coalescent model. BMC Genom. 2020;21:248. doi: 10.1186/s12864-020-6640-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosh R, Tarafdar A, Sharma M. Rapid detection of Fusarium oxysporum f. sp. ciceris from disease infested chickpea fields by loop-mediated isothermal amplification. Indian Phytopathol. 2016;69:47–50. [Google Scholar]
- 29.Jiménez-Gasco MDM, Jiménez-Díaz R. Development of a specific polymerase chain reaction-based assay for the identification of Fusarium oxysporum f. sp. ciceris and its pathogenic races 0, 1A, 5, and 6. Phytopathology. 2003;93:200–209. doi: 10.1094/phyto.2003.93.2.200. [DOI] [PubMed] [Google Scholar]
- 30.Langin T, Capy P, Daboussi M-J. The transposable element impala, a fungal member of the Tc1-mariner superfamily. Mol. Gen. Genet. MGG. 1995;246:19–28. doi: 10.1007/BF00290129. [DOI] [PubMed] [Google Scholar]
- 31.Aurélie H-V, Thierry L, Marie-Josée D. Evolutionary history of the impala transposon in Fusarium oxysporum. Mol. Biol. Evol. 2001;18:1959–1969. doi: 10.1093/oxfordjournals.molbev.a003736. [DOI] [PubMed] [Google Scholar]
- 32.van Dam P, de Sain M, Ter Horst A, van der Gragt M, Rep M. Comparative genomics-based markers: Discrimination of host-specificity in Fusarium oxysporum. Appl. Environ. Microbiol. 2018;84:e01868–e01817. doi: 10.1128/AEM.01868-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Dam P, et al. Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ. Microbiol. 2016;18:4087–4102. doi: 10.1111/1462-2920.13445. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt SM, et al. MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum. BMC Genom. 2013;14:119. doi: 10.1186/1471-2164-14-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma L, et al. The AVR2–SIX5 gene pair is required to activate I-2-mediated immunity in tomato. New Phytol. 2015;208:507–518. doi: 10.1111/nph.13455. [DOI] [PubMed] [Google Scholar]
- 36.Gawehns F, et al. The effector repertoire of Fusarium oxysporum determines the tomato xylem proteome composition following infection. Front. Plant Sci. 2015;6:967. doi: 10.3389/fpls.2015.00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rep MS, S. et al. in Fungal Genetics Reports (ed M. S. Sachs) (Fungal Genetics Stock Center, 2015).
- 38.Rep M, et al. A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 2004;53:1373–1383. doi: 10.1111/j.1365-2958.2004.04177.x. [DOI] [PubMed] [Google Scholar]
- 39.de Sain M, Rep M. The role of pathogen-secreted proteins in fungal vascular wilt diseases. Mol. Plant Microbe Interact. 2015;16:23970. doi: 10.3390/ijms161023970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Achari SR, et al. Investigating the effector suite profile of Australian Fusarium oxysporum isolates from agricultural and natural ecosystems. Plant Pathol. 2020;00:1–10. doi: 10.1111/ppa.13303. [DOI] [Google Scholar]
- 41.Lievens B, Houterman PM, Rep M. Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. Fems Microbiol. Lett. 2009;300:201–215. doi: 10.1111/j.1574-6968.2009.01783.x. [DOI] [PubMed] [Google Scholar]
- 42.Gupta, S., Bhar, A., Chatterjee, M. & Das, S. Fusarium oxysporum f. sp. re-pr race 1 induced redox state alterations are coupled to downstream defense signaling in root tissues of chickpea (Cicer arietinum L.). PloS one8, e73163–e73163. 10.1371/journal.pone.0073163 (2013). [DOI] [PMC free article] [PubMed]
- 43.Williams AH, et al. Comparative genomics and prediction of conditionally dispensable sequences in legume infecting Fusarium oxysporum formae speciales facilitates identification of candidate effectors. BMC Genom. 2016;17:191. doi: 10.1186/s12864-016-2486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emms DM, Kelly S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:1–14. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burgess LW, Summerell BA, Bullock S, Gott KP, Backhouse D. Laboratory Manual for Fusarium Research. 3. University of Sydney; 1994. [Google Scholar]
- 46.Chen S, Zhou Y, Chen Y, Gu J. fastp: An ultra-fast all-in-one FASTQ re-processor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bankevich A, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stanke M, Schöffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinform. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Untergasser A, et al. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sperschneider J, et al. Advances and challenges in computational prediction of effectors from plant pathogenic fungi. PluS Pathog. 2015;11:e1004806. doi: 10.1371/journal.ppat.1004806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sperschneider J, Dodds PN, Gardiner DM, Singh KB, Taylor JM. Improved prediction of fungal effector proteins from secretomes with EffectorP 20. Mol. Plant Pathol. 2018;19:2094–2110. doi: 10.1111/mpp.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sperschneider J, Dodds PN. EffectorP 3.0: Prediction of apoplastic and cytoplasmic effectors in fungi and oomycetes. Mole. Plant Microbe Interact®. 2022;35:146–156. doi: 10.1094/mpmi-08-21-0201-r. [DOI] [PubMed] [Google Scholar]
- 53.Garg N, Ahmad FJ, Kar S. Recent advances in loop-mediated isothermal amplification (LAMP) for rapid and efficient detection of pathogens. Curr. Res. Microb. Scie. 2022;3:100120. doi: 10.1016/j.crmicr.2022.100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong Y-P, Othman S, Lau Y-L, Radu S, Chee H-Y. Loop-mediated isothermal amplification (LAMP): A versatile technique for detection of micro-organisms. J. Appl. Microbiol. 2018;124:626–643. doi: 10.1111/jam.13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hua-Van A, Langin T, Daboussi M-J. Evolutionary history of the impala transposon in Fusarium oxysporum. Mol. Biol. Evol. 2001;18:1959–1969. doi: 10.1093/oxfordjournals.molbev.a003736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomes of the endemic Australian isolates that were sequenced for this project are available on the NCBI GenBank with Accession numbers JAMSCL000000000-JAMWEW000000000 under bioproject PRJNA846078. GenBank accession numbers are given of other genomes retrieved from the NCBI GenBank.